Abstract
When we transduced pancreatic cancers with sgRNAs that targeted 2–16 target sites in the human genome, we found that increasing the number of CRISPR-Cas9 target sites produced greater cytotoxicity, with >99% growth inhibition observed by targeting only 12 sites. However, cell death was delayed by 2–3 weeks after sgRNA transduction, in contrast to the repair of double strand DNA breaks (DSBs) that happened within 3 days after transduction. To explain this discrepancy, we used both cytogenetics and whole genome sequencing to interrogate the genome. We first detected chromatid and chromosome breaks, followed by radial formations, dicentric, ring chromosomes, and other chromosomal aberrations that peaked at 14 days after transduction. Structural variants (SVs) were detected at sites that were directly targeted by CRISPR-Cas9, including SVs generated from two sites that were targeted, but the vast majority of SVs (89.4%) were detected elsewhere in the genome that arose later than those directly targeted. Cells also underwent polyploidization that peaked at day 10 as detected by XY FISH assay, and ultimately died via apoptosis. Overall, we found that the simultaneous DSBs induced by CRISPR-Cas9 in pancreatic cancers caused chromosomal instability and polyploidization that ultimately led to delayed cell death.
Keywords: CRISPR-Cas9, double strand break (DSB), genomic instability, chromosomal instability, cell death
Introduction
Double strand DNA breaks (DSBs) are known to be the most lethal among all DNA lesions (1,2). This cytotoxicity originates from the DNA damage response mechanism that either senses and repairs DSBs, or triggers a permanent exit from the cell cycle via senescence or cell death. The toxicity of DSBs is exploited by the majority of conventional anti-cancer strategies such as radiation therapy and chemotherapy, which rely on DNA damage-induced cell death (1). Although human cells have multiple DSB repair mechanisms, DSBs remain dangerous to cell survival as they directly disrupt the integrity of the DNA and erroneous processing of these breaks can lead to genomic rearrangements characteristic of chromosomal instability (CIN) (1–3). CIN has been linked to tumor evolution, therapeutic resistance, and cell death (4). Novel strategies to exploit the fitness cost associated with CIN while modulating its role in phenotypic adaptation remain elusive and an active area of investigation.
Studies using enzymes, such as I-SceI and CRISPR-Cas9, which are capable of inducing DSBs at specific locations, have shown a positive correlation between increasing number of DSBs and growth inhibition (5–7). CRISPR-Cas9 induced DSBs have been associated with chromothripsis (5), chromosomal rearrangements (8), and cytotoxicity (9–11). Recent studies have reported upregulation of p53 pathway in Cas9-expressing cells and that p53 expression inhibited genetic perturbations in these cells, suggesting that TP53 mutation status has to be considered while studying the effect of CRISPR-Cas9 induced DSBs on cytotoxicity (12–14).
Initially, we investigated the relationship between the number of DSBs and cytotoxicity using CRISPR-Cas9 by targeting 2–16 sites in the non-coding regions of the human genome. The goal was to determine the minimum number of DSBs required to kill pancreatic cancer (PC) cells that harbor TP53 missense mutations, as PC is the third leading cause of cancer death in the United States with a dismal 5-year survival rate of 11.5% (15). More than 90% of PCs are pancreatic ductal adenocarcinomas (PDACs) (16), and approximately 70% of PDACs contain inactivating mutations in the TP53 gene (17). We predicted that higher number of CRISPR-Cas9 target sites would lead to more cytotoxicity and this could be observed within days after the induction of DSBs, since apoptosis, the main cell death mechanism caused by DSBs, occurs within hours (18,19). However, we were surprised to find that cell death was delayed, manifesting 2–3 weeks after the induction of a small number of DSBs. To study the mechanism behind this temporal delay, we implemented both whole genome sequencing and conventional cytogenetics, and found that the accumulation of chromosomal instability events and polyploidization preceded eventual cell death.
Results
Increased number of CRISPR-Cas9 induced DSBs and inhibited cell growth
To determine the minimum number of DSBs required to eliminate cancer cells, we designed multi-target sgRNAs that contain 2–16 target sites in the non-coding regions of the human genome to minimize risk of confounding our interpretations with gene essentiality-linked cytotoxicity (see Methods, table S1). Negative controls included two non-targeting sgRNAs that have no target sites in the human genome, while positive controls included three sgRNAs that target repetitive elements in the human genome (table S1). Two sgRNAs targeting HPRT1 gene were also designed for functional testing of Cas9 activity, as inactivation of the HPRT1 gene leads to resistance to 6-thioguanine. We selected two PDAC cell lines, Panc10.05 and TS0111, which both harbor inactivating TP53 mutations (homozygous I255N and homozygous C275Y, respectively) to make Cas9-expressing cell lines. After documenting functional Cas9 activity (fig. S1A), we transduced negative and positive control sgRNAs into parental, dead Cas9 (dCas9)-expressing, and Cas9-expressing cell lines, and found that growth inhibition was only observed in Cas9-expressing cell lines transduced with positive control sgRNAs (with >600 target sites in the human genome), indicating that DSBs played a role in the growth inhibition observed.
To test the hypothesis that growth inhibition would increase with the number of simultaneously induced DSBs, we transduced the multi-target sgRNAs into Cas9-expressing cells and performed clonogenicity assays. We established that growth inhibition increased as a function of the number of target sites (from 0 to 16 target sites) using two assays, alamarBlue cell viability assay (Fig. 1A; least squares non-linear regressions, R2 Panc10.05 = 0.80, TS0111 = 0.83) and colony counting (Fig. 1B; R2 Panc10.05 = 0.72, TS0111 = 0.73). Bland-Altman tests revealed that the two assays were concordant (Bias 5.84, 95% CI −7.70–19.37). We noted variability in this dose-response relationship which may be due to sgRNA targeting efficiency, single nucleotide variant present on target sites, or other factors.
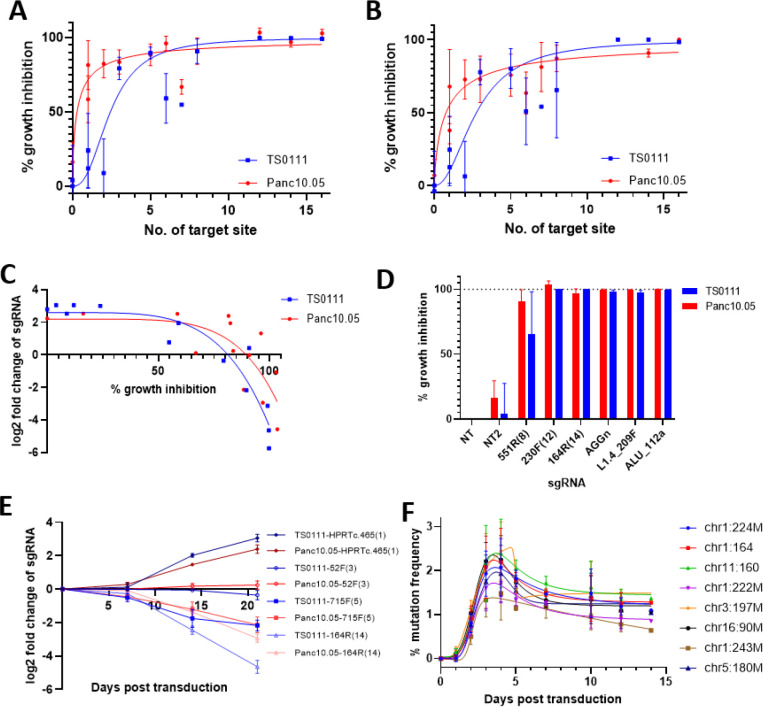
Fig. 1. Increased number of CRISPR-Cas9 targets sites led to an increase in growth inhibition and delayed cell death.
(A-B) Growth inhibition as a function of the number of CRISPR-Cas9 target sites in the human genome for two Cas9-expressing pancreatic cancer (PC) cell lines as detected by (A) alamarBlue cell viability reagent (least squares non-linear regressions; R2 Panc10.05=0.80, TS0111=0.83) and (B) colony counting via phase microscopy (R2 Panc10.05=0.72, TS0111=0.73). N=3; mean ± SEM are shown. (C) Correlation of growth inhibition results collected from sgRNA tag survival and alamarBlue cell viability assay. Spearman r for Panc10.05 = −0.83, P=0.0008; for TS0111 = −0.92, P<0.0001. (D) Comparison of growth inhibitions (via alamarBlue assay) in the two PC cell lines for negative control sgRNAs (NT and NT2), 12- and 14-target sgRNAs (230F(12) and 164R(14), respectively), and positive control sgRNAs (AGGn, L1.4_209F, ALU_112a). N=3; mean ± SEM are shown. (E) sgRNA tag survival of various sgRNAs as a function of time. N=3; mean ± SEM are shown. (F) Mutation frequency at 8 different target loci of Panc10.05 Cas9-expressing cells transduced with 164R(14) at various time points. N=3; mean ± SEM are shown; bell-shaped least squares regression; R2 = 0.60–0.74. Relatively low percentages were due to the lack of selection of transduced cells.
As an independent measure of growth inhibition, we assessed sgRNA tag survival in the same two cell lines on the premise that sgRNAs that were lethal to cells would be eliminated from the pool of tags, while sgRNAs with little to no toxicity should be enriched in the pool. We transduced all the multi-target sgRNAs as a pool into Cas9-expressing cell lines and measured fold change of the sgRNA tags every 7 days for 21 days after transduction. We found that in general, sgRNAs with a higher number of target sites demonstrated a greater degree of sgRNA tag loss (fig. S1C). We noted some variability in this relationship, suggesting that other factors also played a role in this relationship. Notably, the parental cell lines (without Cas9 expression) transduced with sgRNAs didn’t show any significant growth inhibition (fig. S1C), indicating that Cas9 was required to produce the cytotoxicity observed with increased target sites. We then compared the sgRNA tag survival results to the results obtained from the growth inhibition assay and found that the data were highly correlated (Spearman r for Panc10.05 = −0.83, P < 0.001; TS0111 = −0.92, P < 0.0001). This shows that the sgRNA tag survival was a surrogate of growth inhibition especially when the growth inhibition exceeded 70% (Fig. 1C). Interestingly, sgRNAs that targeted 12 or more sites consistently showed >99% elimination in all assays, and the magnitude was comparable to positive control sgRNAs (Fig. 1D, fig. S1E–F).
To determine whether the growth inhibition observed could be generalized to other PDAC cell lines, we cloned Cas9-sgRNA expression vectors, verified cutting activity in cell lines tested (fig. S2A), and performed sgRNA tag survival on four other PDAC cell lines. We observed a general inverse correlation between the number of sgRNA target sites and sgRNA fold change, showing that the dose-response effect could be observed in a number of PDACs (fig. S2B). There was some variation in the magnitude of inhibition among different cell lines, indicating some level of cell line-specific effect from the multi-targeting sgRNAs. Overall, we found that an increased number of CRISPR-Cas9 target sites increased growth inhibition, and this relationship was generalizable across different PDAC cell lines.
Off-target activity did not significantly contribute to growth inhibition
To investigate whether the growth inhibition observed was influenced by off-target activities of the sgRNAs, we performed whole genome sequencing (WGS) analyses on the surviving colonies, if we were able to obtain them, from the clonogenicity assay and used two different approaches to check for off-target activity. Surviving colonies from clonogenicity assays were subcultured for another month before DNA was extracted for WGS. Notably, we couldn’t obtain colonies from the 12- and 16-cutter treated Panc10.05 cells, and 8–14 cutters treated TS0111 cells from any of the replicates, indicating significant cytotoxicity conferred by these multi-target sgRNAs (Table 1). Through manual inspection on the Integrative Genomics Viewer (IGV (20)) of on- and potential off-target sites suggested by CRISPOR (21), we found that >95% of mutations came from on-target sites, and only 28% of 1-mismatch sites were mutated among all the potential off-target sites (1–4 mismatches, Table 1, table S2–4). To validate our findings from WGS analyses, we performed deep sequencing on all the 1- and 2-mismatch sites and found that only 1-mismatch sites were mutated but not at 2-mismatch sites (fig. S3A–D). Our data showed that the predicted number of target sites is a reliable surrogate of the actual number of mutated sites in surviving colonies (fig. S3E). We also found that >30% of the on-target sites of 176R(7) have single nucleotide variants present in both cell lines, so we excluded 176R(7) from our subsequent analyses to prevent confounding interpretations (data not shown).
Table 1.
Number of CRISPR-Cas9 induced scissions from WGS of surviving TS0111 and Panc10.05 colonies.
| sgRNA | Number of predicted perfect target sites1 | Number of potential off-target sites2 | Number of mutated sites in TS01113 | Number of mutated sites in Panc10.053 | Total number of Cas9-induced cuts in Panc10.055 |
|---|---|---|---|---|---|
| NT | 0 | 0-1 | 0-0-0 | 0-0-0 | 0-0-0 |
| NT2 | 0 | 0-0 | 0-0-0 | 0-0-0 | 0-0-0 |
| HPRTc.80 | 1 | 0-2 | 1-0-0 | 1-0-0 | 1-0-0 |
| HPRTc.465 | 1 | 0-2 | 1-0-0 | 1-0-0 | 1-0-0 |
| 531F(2) | 2 | 4-1 | 2-0-0 | 2-0-0 | 3-0-0 |
| 52F(3) | 3 | 0-0 | 3-0-0 | 3-0-0 | 4-0-0 |
| 715F(5) | 5 | 2-1 | 5-1-04 | 5-1-0 | 9-2-0 |
| 451F(6) | 6 | 0-1 | 6-0-0 | 6-0-0 | 12-0-0 |
| 176R(7) | 7 | 2-1 | 6-1-0 | 6-0-0 | 10-0-0 |
| 551R(8) | 8 | 2-1 | NA | 7-0-0 | 12-0-0 |
| 230F(12) | 12 | 8-1 | NA | NA | NA |
| 164R(14) | 14 | 5-2 | NA | 13-3-04 | 21-5-0 |
| 676F(16) | 16 | 2-6 | 16-1-0 | NA | NA |
Number of perfect matches in CRISPOR using the GRCh38 human reference genome, including both canonical (NGG) and non-canonical (NGA/NAG) PAMs.
From CRISPOR, number of 1 and 2 mismatches (1mm-2mm).
Matched or mismatched sites that are mutated from analysis of two resistant colonies for each sgRNA, using an average variant allele frequency cutoff of 10%. Numbers are shown as 0mm-1mm-2mm.
Only one colony could be obtained.
The number of mutated sites that incorporates copy number of the target for Panc10.05 cell line based on hg19.
NA: not available since no resistant colonies could be obtained.
As an alternative approach to look for potential off-target sites comprehensively, we identified all novel InDels and structural variants (SVs) from WGS data of Panc10.05 surviving colonies and checked for homology of sequences surrounding the mutations or breakpoints to the corresponding sequence of the sgRNA transduced into these colonies. None of the sequences surrounding the mutations have fewer than 5mm compared to the original sgRNA sequence, suggesting that none of the novel mutations were a result of off-targeting by CRISPR-Cas9 (table S5).
We also used B-allele frequencies to quantify the copy number of each mutated site, both on- and off-targets, and found that the number of predicted target sites correlated significantly with the number of mutated sites (fig. S3E; Pearson r = 0.98, P < 0.0001). Overall, our results indicate that the growth inhibition observed was due to the small number of DSBs that were directly targeted instead of widespread off-targeting events.
Simultaneous CRISPR-Cas9 targeting led to delayed cell death
Interestingly, most of the reduction in sgRNA tag counts of high-cutters (e.g. 12-cutter and 14-cutter) did not occur in the first 7 days post-transduction of sgRNAs, but between days 7 and 21 (Fig. 1E, fig. S4A–C). This pattern was specific to Cas9-expressing cell lines, as parental cell lines transduced with the same sgRNA pool exhibited no difference in sgRNA fold changes across different time points (fig. S4A–C). Clonogenicity assays performed with different dilutions of cell seeding on 96-well plates (see Methods) also showed that growth inhibition was more apparent in 1:1000 dilution cultures, in which they were cultured for 1–2 months compared to 1–2 weeks in 1:10 dilution cultures (fig. S4D–E).
To investigate whether the temporal delay in growth inhibition was a result of delayed production of DSBs by CRISPR-Cas9, we transduced the 14-cutter sgRNA into Panc10.05 Cas9-expressing cells and quantified the mutation frequency at 8 on-target sites over the course of two weeks. We found that scission occurred over the course of days and peaked around days 3–5 (Fig. 1F, R2 = 0.60–0.74), consistent with other recent observations (22,23). This suggests that most DSBs were produced within the first few days post transduction of sgRNAs. Overall, our results showed that the multiple scission events did not immediately and directly trigger cell death, pointing to a different mechanism that was contributing to the temporal delay in growth inhibition.
Cytogenetics analyses revealed ongoing chromosomal instability
We hypothesized that the mechanism of cell death was likely not due to DNA damage responses that were immediately and directly triggered by multiple CRISPR-Cas9 scissions, but was rather caused by a slower process which ultimately led to cell death. To examine whether there were any structural changes to the chromosomes after multiple CRISPR-Cas9 scissions, we performed cytogenetic analyses on cells harvested from 0–21 days post transduction of sgRNA at 3–4 day intervals using a detailed chromosome breakage assay (Fig. 2A–D, fig. S5B–D). TS0111 Cas9-expressing cell line was chosen due to its simpler karyotype than Panc10.05 at baseline (fig. S5A), and the sgRNA transduced was a 14-cutter, 164R(14), which had manifested significant growth inhibition in the previous assays for dose-response studies.
Fig. 2. Cytogenetics and whole genome sequencing revealed ongoing chromosomal instability.
(A-D) Chromosome breakage analyses of TS0111-Cas9-EGFP cells transduced with 164R(14) and harvested on (A) day 1, (B) day 3, (C) day 10, and (D) day 16 post transduction of 164R(14). Metaphase images of representative cells are shown and karyotypic alterations indicated by arrows. (E) Cytogenetic changes (events per 100 metaphase cells) of TS0111-Cas9-EGFP cells as a function of time. (F) Breakpoints on dicentric, tricentric, and ring chromosomes categorized by their locations, whether at targeted or non-targeted sites. (G) Break-apart FISH probe results for one of the 164R(14) target sites on 1q41 analyzed on day 14 post transduction of TS0111-Cas9-EGFP cells. (H) WGS of Panc10.05-Cas9-EGFP surviving clones after treatment with multi-target sgRNAs analyzed to identify structural variants (SVs). SVs were categorized by whether they resulted from 2 sites that were targeted (green), 1 site that was targeted (red) or whether they were completely novel (no sites targeted, blue). Number in parenthesis indicates the number of perfect target site in human genome. N=2 except for 164R(14) (N=1); mean ± SEM are shown.
We found that on the first day after transduction of 164R(14), multiple chromosome and chromatid breaks were detected along with radial formations, which increased over time (Fig. 2A, E). Other chromosomal aberrations also accumulated over time, including formation of ring, dicentric and tricentric chromosomes, telomere-telomere associations, chromosome pulverizations, and endomitosis (Fig. 2B–E, fig. S5C–D). Most of these aberrations peaked at day 14, but they were still observed at high numbers from day 14 to 21, suggesting ongoing occurrence of breakage events and chromosomal instability (Fig. 2E). We also analyzed breakpoints on dicentric and tricentric chromosomes to determine whether they occurred at CRISPR-Cas9 targeted or non-targeted regions based on chromosomal band locations of the sgRNA target sequences. Although SVs at targeted regions predominated at early time points and decreased over a time, the majority of SVs occurred at non-targeted regions and peaked at day 14 (Fig. 2F), consistent with ongoing chromosomal rearrangements. While most targeted regions were located at telomeric regions, 61.5% of novel SVs identified at non-targeted regions were also located at telomeric regions (fig. S5E). These results indicated that targeting multiple regions at telomeric ends led to ongoing chromosomal rearrangements, which led to more SVs found near telomeric regions.
To visually confirm that the SVs found at targeted regions were direct results of CRISPR-Cas9 targeting, we designed and performed a break-apart fluorescence in situ hybridization (FISH) assay on one of the target sites to improve our ability to observe genomic rearrangements (fig. S6A). While simple rearrangements were observed at early time points (fig. S6A), more complex rearrangements were observed at later time points (Fig. 2G, fig. S6B). The number of cells with abnormal FISH patterns increased over time and peaked at day 14 (fig. S6C), demonstrating ongoing chromosomal rearrangements that originated from a CRISPR-Cas9 scission event. These results confirmed that the chromosomal rearrangements detected were not mainly due to inherent CIN of cancer cells and tumor heterogeneity, but were directly contributed by CRISPR-Cas9 scission events. Overall, our results showed that multiple CRISPR-Cas9 induced DSBs resulted in an increase in karyotypic abnormalities and SVs that peaked around 14 days after transduction, instead of the time close to the initial induction of the DSBs.
Bioinformatics analyses found increased DSBs led to more SVs
As a complementary method to study the effects of multiple CRISPR-Cas9 targeting on chromosome integrity, we analyzed the surviving colonies from our clonogenicity assays using WGS to identify, categorize, and quantify novel SVs. This approach would allow us to not only study the effect of DSB repair at both targeted and non-targeted regions at high resolution, but also inspect for potential off-target activities through sequence homology analysis. We used an SV detection software, manta (24), to identify SVs in surviving colonies previously transduced with multi-target sgRNAs, followed by visual inspection of all identified SVs on IGV for validation and categorization. Our data showed that novel SVs increased as a function of the number of sgRNA target sites (Fig. 2H), and this finding has been corroborated by using a different SV caller, Trellis (25) (fig. S7A). Most of the novel SVs were not directly contributed by CRISPR-Cas9 targeting, as CRISPR-Cas9 target sites or potential off-target sites were not found at or near breakpoints located at non-targeted regions (Fig. 2H, table S5). E.g. for the 14-cutter sgRNA, only 7.7% of SVs were produced from two sites that were directly targeted, and 2.9% were produced where one site was targeted, while the majority (89.4%) were found at non-targeted sites. This finding supports the concept of ongoing chromosomal instability as most novel SVs were found at non-targeted regions. Moreover, the increase in SVs was mostly contributed by increases in deletions and translocations (fig. S7B), demonstrating that deletions and translocations were produced from simultaneous targeting of multiple sites in the genome.
As an additional measure to check for CRISPR-Cas9 off-target activity, we compared SVs found at non-targeted regions between individual colonies transduced with the same sgRNA. We found that SVs in non-targeted regions were unique to each colony, indicating that these were not results of off-target activities. We found one instance of a shared novel SV but the breakpoint differed from the sgRNA sequence by 13 mismatches, therefore the shared novel SV was likely present in the bulk cell line at a low level prior to selection by cloning. In summary, WGS analyses showed that increasing CRISPR-Cas9 targeting produced more SVs, and the majority of SVs arose at non-targeted regions, both supporting the concept of ongoing chromosomal instability resulting from multiple CRISPR-Cas9 targeting.
Polyploidization and delayed cell death
During the course of our experiments, to our surprise, we found that cells responded to the 14-cutter sgRNA, 164R(14), by becoming polyploid, manifesting as extremely large nuclei or multinucleated giant cells (Fig. 3A, fig. S8A–B). Metaphase images of transduced cells also showed that chromosome number increased after transduction and that the cells were clearly polyploid by day 10 (Fig. 3B), with cells commonly containing >100 chromosomes. As TS0111 is a female cell line, we confirmed polyploidization using XY FISH by counting cells with >6 copies of X chromosomes (Fig. 3C). Our results showed that polyploidy peaked at day 10 and decreased by day 21.
Fig. 3. Polyploidization and apoptosis after treatment with 164R(14).
(A) Panc10.05-Cas9-EGFP cells transduced with NT2 or 164R(14), and stained with wheat germ agglutinin (WGA; green) and Hoechst 33342 (blue) 14 days after transduction. White arrow indicates a large nucleus and yellow arrows indicate multiple nuclei in a single cell. (B) Metaphase images of cells pre-transduction (day 0), day 7 and day 10 after transduction of TS0111-Cas9-EGFP cells with 164R(14). Each box contains an image of one representative cell. (C) Number of TS0111-Cas9-EGFP cells with >6 X chromosomes over time using XY FISH. (D) Apoptosis analysis of Panc10.05-Cas9-EGFP after treatment with 164R(14) or control (NT2) using Annexin V flow cytometry assay, showing an increase in apoptotic cells by 164R(14) on days 7 (Paired t test, two-tailed, P=0.009) and 14 (P=0.020) after treatment with 164R(14) compared to NT2, and decreased by day 21 (P=0.14). N=3; mean ± SEM are shown.
Additionally, we assayed for apoptosis markers in cells transduced with 164R(14) to determine if apoptosis was involved in the cell death mechanism. We found that the proportion of apoptotic cells stained with annexin V increased on days 7 and 14 compared to cells transduced with a non-targeting sgRNA, and decreased by day 21 (Fig. 3D, fig. S8C). TUNEL (terminal deoxynucleotidyl transferase dUTP nick end labeling) staining, on the other hand, showed decreased TUNEL signal on day 14 and increased signal on day 21 (fig. S8D). As annexin V stained for intermediate stages of apoptosis while TUNEL stained for late stages of apoptosis, the mixed results from both assays could suggest that most of the cells on day 21 were dead or nearly dead. This was consistent with our experience as we were consistently not able to harvest enough day 21 cells for flow cytometry analysis due to the extremely low number of living cells remaining.
Discussion
Genomic instability is a hallmark of cancer (26). However, its implications on therapeutic responses have been contradictory, as some studies have associated higher genomic instability with better therapeutic response while others have linked it to therapeutic resistance (4,27,28). It has been proposed that there is a tolerance limit of genomic instability in cancer cells, such that at an extreme level, it becomes lethal to cells (29). In our study, we showed that few CRISPR-Cas9 induced DSBs caused chromosomal instability that accumulated over time, which led to polyploidization and eventual cell death. This showed that the simultaneous production of a small number of DSBs overwhelmed the integrity of the DSB repair system and generated CIN that resulted in cell death. However, it is important to note that based on our sgRNA design strategy, most of our sgRNA target sites were located near telomeres, which could have contributed to the cell death observed. Multi-target sgRNA treated cells exhibited similar CIN features to those observed in cells undergoing telomere crisis, such as massive chromosomal rearrangements and endoreduplication, which resulted in a high rate of cell death (30,31). We also demonstrated through WGS that cells treated with sgRNAs with 2 to 16 target sites in the human genome exhibited chromosomal rearrangements, indicating that cutting at telomeric or near-telomeric regions could lead to gradual cell death. A potential avenue could be to explore the efficacy of using CRISPR-Cas9 to engineer a telomere crisis for cell killing in a TP53 mutant background.
Additionally, polyploidization of cancer cells has been associated with therapeutic resistance and tumor repopulation, as it enables more phenotypic variations compared to non-polyploid cells, thus allowing the formation of different therapeutic resistance phenotypes (32–34). However, this does not explain why we could not obtain surviving colonies from cells treated by some of our multi-target sgRNAs that had been grown for about three months post transduction. We also did not observe significant polyploidization by Cas9-expressing cells alone or cells treated with non-targeting sgRNAs, suggesting that Cas9 expression itself does not cause polyploidization of cancer cells. As both damaged telomeres and polyploidy have been associated with increased risk of chromothripsis (35,36), we suspect that the polyploidization of multi-target sgRNA treated cells allowed more genomic instability that led to lethal consequences instead of providing a permissive background for tumorigenicity. We also found very few SVs at the CRISPR-Cas9 target sites in our surviving colonies though we observed translocations as a result of CRISPR-Cas9 scission in our break-apart FISH assay, suggesting that the surviving colonies might have lower levels of CIN compared to the rest of the non-surviving cells. Potential future studies should look into whether the surviving colonies obtained were progenies of polyploid cells, and whether they were outliers in terms of CIN rate compared to the rest of the cell population.
We also found significant advantages in integrating cytogenetics analyses and whole genome sequencing while studying CIN for a few reasons: (1) cytogenetics analyses are more likely to reflect the “truth” as the chromosomes are visually inspected for presence of breakpoints and karyotypic aberrations, while SV-calling software is more prone to false positives and negatives, especially at repetitive regions (data not shown), due to various artifacts such as mapping errors (37); (2) WGS analyses can pinpoint the nucleotide sequence of breakpoints and allow us to examine for potential off-target activity at higher resolution, while cytogenetics analyses can only provide the approximate location of breakpoints; (3) cytogenetic analyses allow us to witness the formation of chromosomal aberrations that are known to directly contribute to CIN due to mitotic errors, while WGS analyses heavily rely on the parameter settings to accurately call out structural abnormalities; (4) WGS analyses can capture smaller variations such as small indels and single nucleotide variant, features that will be overlooked by cytogenetics approaches due to resolution issues. Although significant progress has been made to improve SV callers, cytogenetics analyses remain valuable due to the accuracy of their findings.
More importantly, we demonstrated that while we could eliminate cancer cells with relatively few CRISPR-Cas9 cuts, a critical design feature was to allow cells to grow for a few weeks, since cell elimination did not occur in the first 7 days. This could have serious implications on experimental designs, as an experiment with a shorter time frame could unintentionally lead to false negative results if cell death is the only measured outcome. As studies have shown that cells with wild-type TP53 have limited genome editing efficiency and that TP53-mediated DNA damage response and cell cycle arrest were activated during CRISPR-Cas9 genome editing (12–14), it would be interesting to examine whether the delayed cell death observed in this study was specific to cells with TP53 mutations and absent in TP53 wild-type cells.
In summary, we demonstrated that a small number of DSBs at non-coding regions could lead to delayed PDAC cell death, and this cytotoxicity was driven by the accumulation of chromosomal instability events and polyploidization (Fig. 4). As novel therapies are urgently needed for PDAC patients due to their low survival rate, this study could potentially be exploited as a new cell killing strategy to target PDAC.
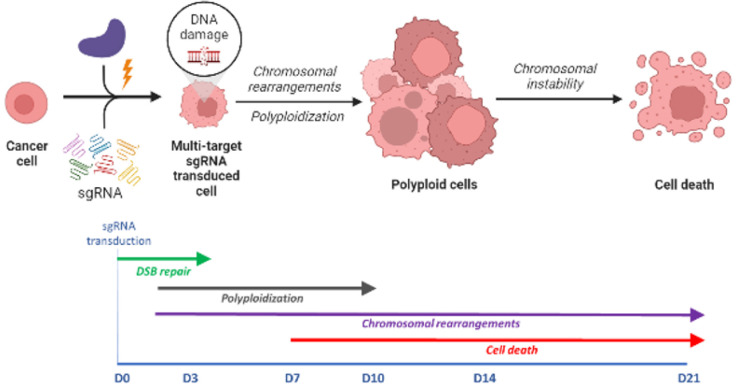
Fig. 4. Mechanistic timeline of delayed cell death after multiple CRISPR-Cas9 induced double strand breaks.
Cytotoxicity in multi-target sgRNA-transduced PC cells occurred following the induction of multiple DSBs, and their repair resulted in chromosomal rearrangements, polyploidization, and ultimately cell death.
Methods
Multitarget sgRNA design
Chromosome range was entered into CRISPOR (21) 2kb at a time starting at chr1:0–2000 and ending at chr1:100,248,000–100,250,000 based on hg19 and hg38, respectively. sgRNAs that have 2–16 perfect target sites were selected from the pool of sgRNA options generated by CRISPOR based on the following criteria: (1) none of the perfect target sites and potential off-target sites target exons; (2) Doench’16 efficiency score (38) is >50%, and (3) the number of off-targets that have no mismatches in the 12bp adjacent to the PAM (SEED region) is <10. Sequences of non-targeting control sgRNAs were obtained from Doench et al. (38) (NT) and Chiou et al. (39) (NT2). HPRT1 sgRNAs (1-cutters) were designed using CRISPOR. Positive control sgRNAs were designed by either putting together a trinucleotide sequence (AGGn) or by inserting LINE-1 and Alu element sequences to CRISPOR.
Cell viability and clonogenicity assay
Cells were seeded for 24 hours before the media was replaced to contain 10ug/mL of polybrene. Lentivirus of MOI 10 was added into the media and transduction took place for 18–20 hours. The media was then removed, washed once with PBS, and replaced with media that contained 5ug/mL blasticidin. After 48 hours, the cells were split into two 96-well plates (one with 1:10 dilution and one with 1:1000 dilution of the original cultures) with media that contained both 5ug/mL blasticidin and 1ug/mL puromycin for selection. When cells in non-targeting controls reached full confluence, colonies were counted based on phase microscopy observation in 1:1000 dilution cultures. Then, 10uL of alamarBlue Cell Viability Reagent (ThermoFisher) was added to 90uL cell culture medium per well on 96-well plates. The plates were incubated at 37°C for 3 or 24 hours, depending on cell lines, and transferred to BMG POLARstar Optima microplate reader for fluorescence reading. Excitation was set at 544nm and emission at 590nm, with a gain of 1000 and required value of 90%.
sgRNA tag survival assay
Cells were seeded for 24 hours before the media was replaced to contain 10ug/mL of polybrene. Lentivirus of MOI 0.1 was added into the media and transduction took place for 18–20 hours. The media was then removed, washed once with PBS, and replaced with media that contained 5ug/mL blasticidin. After 24 hours, approximately 1 million cells were collected for day 1 timepoint, and the remaining cells were subjected to both 5ug/mL blasticidin and 1ug/mL puromycin selections simultaneously. Cells were collected on day 7, 14, and 21 post-transduction, and along with day 1 cells, genomic extractions were performed using QIAamp UCP DNA Micro Kit (QIAGEN) by following manufacturer’s protocol. sgRNA library was prepared by amplifying the sgRNA target region from gDNAs using next generation sequencing (NGS) primers provided by Joung et al. (40), based on the protocol outlined in the paper, and sent for NGS (Primers Table 1). Read counts of each sgRNA were extracted from FASTQ files and were put through the MAGeCK (41) pipeline to obtain sgRNA fold change.
WGS of surviving colonies
Genomic DNA was extracted from surviving colonies of clonogenicity assay using QIAamp UCP DNA Micro Kit (QIAGEN) by following manufacturer’s protocol. SKCCC Experimental and Computational Genomics Core sent the samples to New York Genome Center (NYGC) for WGS with an Illumina HiSeq 2000 using the TruSeq DNA prep kit. Sequencing was carried out so as to obtain 30X coverage from 2×100bp paired-end reads. FASTQ files were aligned to both hg19 and hg38 using bwa v0.7.7 (42) to create BAM files. The default parameters were used. Picard-tools1.119 (http://broadinstitute.github.io/picard/) was used to add read groups as well as remove duplicate reads. GATK v3.6.0 (43) base call recalibration steps were used to create a final alignment file.
Time-course PCR
Panc10.05-Cas9-EGFP cells were transduced with 164R(14) sgRNA and cultured without antibiotic selection. Cell pellets were collected at various time points for gDNA extraction using QIAamp UCP DNA Micro Kit (QIAGEN) by following manufacturer’s protocol. Primers were designed for 8 perfect target regions of the 164R(14) sgRNA for PCR and NGS (Primers Table 2). Quantification of mutation frequency of all target sites was done using the CRISPResso2 (44) pipeline.
Chromosome breakage assay
The TS0111-Cas9-EGFP cells plated at 5 × 105 /ml were treated with 164R(14), a 14-cutter sgRNA, and harvested at 0, 1, 3, 7, 10, 14, 16 and 21 days. Colcemid (0.01 μg/ml) was added 20 hours before harvesting. Cells were then exposed to 0.075 M KCl hypotonic solution for 30 minutes, fixed in 3:1 methanol:acetic acid and stained with Leishman’s for 3 minutes. For each treatment, one hundred consecutive analyzable metaphases were analyzed for induction of chromosome abnormalities including chromosome/chromatid breaks and exchanges.
1q41 Break-apart FISH assay
FISH was performed on the TS0111-Cas9-EGFP cells before and after 164R(14) sgRNA treatment (from 0, 1, 3, 7, 10, 14, 16 and 21 days) using RP11-14B15 and RP11-120E23 probes flanking a 1q41 sgRNA cut according to the manufacturer’s protocol (Empiregenomics Inc., Williamsville, NY). The RP11-14B15 probe is for the 5’ (centromeric) side of the 1q41 sgRNA cut and in Spectrum Orange. The RP11-120E23 probe is for the 3’ (telomeric) side of the 1q41 sgRNA cut and in Spectrum Green. For these probes, an overlapping red/green or fused yellow signal represents the normal pattern, and separate red and green signals indicate the presence of a rearrangement. The normal cutoff was calculated based on the scoring of the TS0111-Cas9-EGFP cells before sgRNA treatment (day 0). The normal cutoff for an analysis of 500 cells with the 1q41 break-apart probe set is calculated using the Microsoft Excel β inverse function, = BETAINV (confidence level, false-positive cells plus 1, number of cells analyzed). This formula calculates a one-sided upper confidence limit for a specified percentage proportion based on an exact computation for a binomial distribution assessment. The normal cutoff for the 1q41 break-apart probe set is 0.6% (for a 95% confidence level). For each time point, a total of 500 nuclei were visually evaluated with fluorescence microscopy using a Zeiss Axioplan 2, with MetaSystems imaging software (MetaSystems, Medford, MA), to determine percentages of abnormal cells.
SV identification and quantification
From the WGS BAM files of surviving colonies, Manta v0.29.6 was used to call somatic SVs between the sample and the control, in which the control is the Panc10.05-Cas9-EGFP non-transduced cell line. The default parameters were used. Variants were annotated according to UCSC refseq annotations using an in-house script. The list of SVs generated were then individually, visually inspected on IGV to validate its presence in sample and absence in control. Novel SVs were quantified using SVs that have passed the manual screening.
For SV identification using Trellis (45), we performed analysis on the Joint High Performance Computing Exchange, a 64 bit Linux Red Hat cluster, hosted at the Johns Hopkins Bloomberg School of Public Health. We used Bowtie2 (45), with default settings, to align the paired end, 2 × 151 bp, Fastq files to Hg19. We indexed the aligned files with samtools version 1.14 (46) and used the resulting bam files as input to the R program Trellis for rearrangement detection (25). The Trellis code was customized to prevent removal of aligned read-pairs containing at least one read with a map quality below 30. This modification enabled rearrangements to be detected within low complexity reference sequence, a change necessary to detect rearrangements overlapping our target loci, all of which comprised sequences that were repeated multiple times within the reference genome. Trellis input settings included five minimum tags per cluster, 100 bp gap width between reads within a cluster, 10k bp maximum cluster size, and 10k bp minimum read-pair separation, and no automatic removal of genomic loci with previous annotation of publicly available samples indicating germline rearrangements. A secondary set of filters was applied to the primary Trellis results to remove likely artifacts. The secondary filters removed candidate rearrangements with mean map quality scores < 1, read-pair count 40, at least one junction in the Y chromosome, Trellis annotation indicating a copy number change (either an amplification or deletion) and rearrangements junctions appearing in at least one of the two negative controls.
Cell membrane and genomic staining
Alexa Fluor 488 conjugate of wheat germ agglutinin (WGA; ThermoFisher) was used to stain cell membrane on fixed cells according to manufacturer’s protocol. Hoechst 33342 stain was used to stain genomic content by incubating the cells in Hoechst 33342 for 10 minutes in room temperature before covering the cell with mounting media.
XY FISH assay
FISH was performed on the TS0111-Cas9-EGFP cells before and after a 14-cutter sgRNA treatment (from 0, 1, 3, 7, 10, 14, 16 and 21 days) using X/Y centromere FISH probes according to the manufacturer’s protocol (Abbott Molecular Inc., Des Plaines, IL). For each time point, a total of 200 nuclei were visually evaluated with fluorescence microscopy using a Zeiss Axioplan 2, with MetaSystems imaging software (MetaSystems, Medford, MA), to determine copy number of the X chromosome.
Apoptosis assays
Cells were detached using Accutase and stained with Annexin V binding antibodies and propidium iodide using BioLegend’s APC Annexin V Apoptosis Detection Kit, according to the manufacturer’s protocol. Fluorescence was quantified using Attune NxT Flow Cytometer. Cells were also plated on black with clear flat bottom 96-well plates and stained with both TUNEL and Hoechst using Cell Meter Live Cell TUNEL Apoptosis Assay Kit (Red Fluorescence), according to the manufacturer’s protocol (AAT Bioquest). BMG POLARstar Optima microplate reader was used for fluorescence reading. For TUNEL measurement, excitation was set at 544nm and emission at 590nm, with a gain of 1000 and required value of 90%. For Hoechst 33342, excitation was set at 490nm and emission at 520nm, with a gain of 1700 and required value of 90%. Final calculation was done based on a formula used by Daniel and DeCoster (47).
Statistical analysis
The appropriate statistical tests were performed in GraphPad Prism (Version 9.2.0). The statistical models used were stated in results and legends of figures. For all statistically significant results, * indicates p<0.05, ** indicates p<0.01, *** indicates p<0.001, and **** indicates p<0.0001.
Supplementary Material
Statement of significance.
Using whole genome sequencing and conventional cytogenetics, we discovered that CRISPR-Cas9 cuts led to genomic instability, including chromosomal rearrangements and polyploidization, and ultimately to delayed cell death, 2–3 weeks after the induction of CRISPR-Cas9 DSBs.
Acknowledgements
We thank the generosity of the patients who consented to providing their samples. We also acknowledge Drs. Leslie Cope, Chien-Fu Hung, Sarah Wheelan, Ming-Tseh Lin, Elizabeth Jaffee, Lei Zheng, Feyruz V. Rassool, Richard Burkhart, Christine Iacobuzio-Donahue, Jessica Gucwa, Qingfeng Zhu, Shiwen Peng, and Toni Seppulla, in addition to Aparna Pallavajjala for helpful discussions. We thank Jacqueline Tang, Ada Tam, Raluca Yonescu, Emily Adams, Lisa Haley, Anuj Gupta, Lee Blosser, Nicholas Ionta, Suping Chen, Kai Calder, Jormoh Bellepu, Jordyn Winters, and Nori Leybengrub for outstanding technical assistance.
Financial support:
The Stringer foundation (JRE), Susan Wojcicki and Dennis Troper (JRE), The Sol Goldman Pancreatic Cancer Research Center (JRE), National Institutes of Health grant P50CA62924 (Dr. Alison P. Klein), National Institutes of Health grant P30CA006973 (Dr. William G. Nelson).
Footnotes
Conflict of interest disclosures: Kirsten Bowland, Drs. Teh, Roberts, and Eshleman, and Johns Hopkins University have filed a provisional patent with the USPTO. Johns Hopkins University owns equity in Delfi Diagnostics. Dr. Scharpf is a founder of and holds equity in Delfi Diagnostics. He also serves as the Head of Data Science. This arrangement has been reviewed and approved by Johns Hopkins University in accordance with its conflict of interest policies.
Supplementary Data
Data availability
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials. Plasmids constructed had been deposited at Addgene. Plasmids expressing various specific sgRNAs are available upon request from the corresponding author. Sequencing data will be uploaded to the recommended archive.
References
- 1.Nickoloff JA, Sharma N, Taylor L. Clustered DNA Double-Strand Breaks: Biological Effects and Relevance to Cancer Radiotherapy. Genes [Internet]. 2020;11. Available from: 10.3390/genes11010099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Trenner A, Sartori AA. Harnessing DNA Double-Strand Break Repair for Cancer Treatment. Front Oncol. 2019;9:1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iliakis G, Mladenov E, Mladenova V. Necessities in the Processing of DNA Double Strand Breaks and Their Effects on Genomic Instability and Cancer. Cancers [Internet]. 2019;11. Available from: 10.3390/cancers11111671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sansregret L, Vanhaesebroeck B, Swanton C. Determinants and clinical implications of chromosomal instability in cancer. Nat Rev Clin Oncol. 2018;15:139–50. [DOI] [PubMed] [Google Scholar]
- 5.Schipler A, Mladenova V, Soni A, Nikolov V, Saha J, Mladenov E, et al. Chromosome thripsis by DNA double strand break clusters causes enhanced cell lethality, chromosomal translocations and 53BP1-recruitment. Nucleic Acids Res. 2016;44:7673–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aguirre AJ, Meyers RM, Weir BA, Vazquez F, Zhang C-Z, Ben-David U, et al. Genomic Copy Number Dictates a Gene-Independent Cell Response to CRISPR/Cas9 Targeting. Cancer Discov. 2016;6:914–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Munoz DM, Cassiani PJ, Li L, Billy E, Korn JM, Jones MD, et al. CRISPR Screens Provide a Comprehensive Assessment of Cancer Vulnerabilities but Generate False-Positive Hits for Highly Amplified Genomic Regions. Cancer Discov. 2016;6:900–13. [DOI] [PubMed] [Google Scholar]
- 8.Kosicki M, Tomberg K, Bradley A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat Biotechnol. 2018;36:765–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kwon T, Ra JS, Lee S, Baek I-J, Khim KW, Lee EA, et al. Precision targeting tumor cells using cancer-specific InDel mutations with CRISPR-Cas9. Proc Natl Acad Sci U S A [Internet]. 2022;119. Available from: 10.1073/pnas.2103532119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang L, Güell M, Niu D, George H, Lesha E, Grishin D, et al. Genome-wide inactivation of porcine endogenous retroviruses (PERVs). Science. 2015;350:1101–4. [DOI] [PubMed] [Google Scholar]
- 11.Kuscu C, Parlak M, Tufan T, Yang J, Szlachta K, Wei X, et al. CRISPR-STOP: gene silencing through base-editing-induced nonsense mutations. Nat Methods. 2017;14:710–2. [DOI] [PubMed] [Google Scholar]
- 12.Ihry RJ, Worringer KA, Salick MR, Frias E, Ho D, Theriault K, et al. p53 inhibits CRISPR-Cas9 engineering in human pluripotent stem cells. Nat Med. 2018;24:939–46. [DOI] [PubMed] [Google Scholar]
- 13.Haapaniemi E, Botla S, Persson J, Schmierer B, Taipale J. CRISPR–Cas9 genome editing induces a p53-mediated DNA damage response. Nat Med. Nature Publishing Group; 2018;24:927–30. [DOI] [PubMed] [Google Scholar]
- 14.Enache OM, Rendo V, Abdusamad M, Lam D, Davison D, Pal S, et al. Cas9 activates the p53 pathway and selects for p53-inactivating mutations. Nat Genet. 2020;52:662–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cancer of the Pancreas - Cancer Stat Facts [Internet]. SEER. [cited 2023 Feb 7]. Available from: https://seer.cancer.gov/statfacts/html/pancreas.html [Google Scholar]
- 16.Kleeff J, Korc M, Apte M, La Vecchia C, Johnson CD, Biankin AV, et al. Pancreatic cancer. Nat Rev Dis Primers. 2016;2:16022. [DOI] [PubMed] [Google Scholar]
- 17.AACR Project GENIE Consortium. AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discov. 2017;7:818–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elmore S. Apoptosis: a review of programmed cell death. Toxicol Pathol. 2007;35:495–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saraste A, Pulkki K. Morphologic and biochemical hallmarks of apoptosis. Cardiovasc Res. 2000;45:528–37. [DOI] [PubMed] [Google Scholar]
- 20.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Concordet J-P, Haeussler M. CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 2018;46:W242–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brinkman EK, Chen T, de Haas M, Holland HA, Akhtar W, van Steensel B. Kinetics and Fidelity of the Repair of Cas9-Induced Double-Strand DNA Breaks. Mol Cell. 2018;70:801–13.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zou RS, Marin-Gonzalez A, Liu Y, Liu HB, Shen L, Dveirin RK, et al. Massively parallel genomic perturbations with multi-target CRISPR interrogates Cas9 activity and DNA repair at endogenous sites. Nat Cell Biol. 2022;24:1433–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen X, Schulz-Trieglaff O, Shaw R, Barnes B, Schlesinger F, Källberg M, et al. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics. 2016;32:1220–2. [DOI] [PubMed] [Google Scholar]
- 25.Papp E, Hallberg D, Konecny GE, Bruhm DC, Adleff V, Noë M, et al. Integrated Genomic, Epigenomic, and Expression Analyses of Ovarian Cancer Cell Lines. Cell Rep. 2018;25:2617–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanahan D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022;12:31–46. [DOI] [PubMed] [Google Scholar]
- 27.Ye CJ, Sharpe Z, Heng HH. Origins and Consequences of Chromosomal Instability: From Cellular Adaptation to Genome Chaos-Mediated System Survival. Genes [Internet]. 2020;11. Available from: 10.3390/genes11101162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simonetti G, Bruno S, Padella A, Tenti E, Martinelli G. Aneuploidy: Cancer strength or vulnerability? Int J Cancer. 2019;144:8–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andor N, Maley CC, Ji HP. Genomic Instability in Cancer: Teetering on the Limit of Tolerance. Cancer Res. 2017;77:2179–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dewhurst SM. Chromothripsis and telomere crisis: engines of genome instability. Curr Opin Genet Dev. 2020;60:41–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davoli T, de Lange T. Telomere-driven tetraploidization occurs in human cells undergoing crisis and promotes transformation of mouse cells. Cancer Cell. 2012;21:765–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu J, Erenpreisa J, Sikora E. Polyploid giant cancer cells: An emerging new field of cancer biology. Semin Cancer Biol. 2022;81:1–4. [DOI] [PubMed] [Google Scholar]
- 33.Moein S, Adibi R, da Silva Meirelles L, Nardi NB, Gheisari Y. Cancer regeneration: Polyploid cells are the key drivers of tumor progression. Biochim Biophys Acta Rev Cancer. 2020;1874:188408. [DOI] [PubMed] [Google Scholar]
- 34.Coward J, Harding A. Size Does Matter: Why Polyploid Tumor Cells are Critical Drug Targets in the War on Cancer. Front Oncol. 2014;4:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cortés-Ciriano I, Lee JJ-K, Xi R, Jain D, Jung YL, Yang L, et al. Comprehensive analysis of chromothripsis in 2,658 human cancers using whole-genome sequencing. Nat Genet. 2020;52:331–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mardin BR, Drainas AP, Waszak SM, Weischenfeldt J, Isokane M, Stütz AM, et al. A cell-based model system links chromothripsis with hyperploidy. Mol Syst Biol. 2015;11:828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mahmoud M, Gobet N, Cruz-Dávalos DI, Mounier N, Dessimoz C, Sedlazeck FJ. Structural variant calling: the long and the short of it. Genome Biol. 2019;20:246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34:184–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chiou S-H, Winters IP, Wang J, Naranjo S, Dudgeon C, Tamburini FB, et al. Pancreatic cancer modeling using retrograde viral vector delivery and in vivo CRISPR/Cas9-mediated somatic genome editing. Genes Dev. 2015;29:1576–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Joung J, Konermann S, Gootenberg JS, Abudayyeh OO, Platt RJ, Brigham MD, et al. Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat Protoc. 2017;12:828–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li W, Xu H, Xiao T, Cong L, Love MI, Zhang F, et al. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 2014;15:554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM [Internet]. arXiv [q-bio.GN]. 2013. Available from: http://arxiv.org/abs/1303.3997 [Google Scholar]
- 43.Van der Auwera GA, O’Connor BD. Genomics in the Cloud. O’Reilly Media, Inc.; [Google Scholar]
- 44.Clement K, Rees H, Canver MC, Gehrke JM, Farouni R, Hsu JY, et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat Biotechnol. 2019;37:224–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Daniel B, DeCoster MA. Quantification of sPLA2-induced early and late apoptosis changes in neuronal cell cultures using combined TUNEL and DAPI staining. Brain Res Brain Res Protoc. 2004;13:144–50. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials. Plasmids constructed had been deposited at Addgene. Plasmids expressing various specific sgRNAs are available upon request from the corresponding author. Sequencing data will be uploaded to the recommended archive.