Abstract
Sirtuin 6 (SIRT6) is a multifaceted protein deacetylase/deacylase and a major target for small-molecule modulators of longevity and cancer. In the context of chromatin, SIRT6 removes acetyl groups from histone H3 in nucleosomes, but the molecular basis for its nucleosomal substrate preference is unknown. Our cryo–electron microscopy structure of human SIRT6 in complex with the nucleosome shows that the catalytic domain of SIRT6 pries DNA from the nucleosomal entry-exit site and exposes the histone H3 N-terminal helix, while the SIRT6 zinc-binding domain binds to the histone acidic patch using an arginine anchor. In addition, SIRT6 forms an inhibitory interaction with the C-terminal tail of histone H2A. The structure provides insights into how SIRT6 can deacetylate both H3 K9 and H3 K56.
The structure of the SIRT6 deacetylase/nucleosome complex suggests how the enzyme acts on both histone H3 K9 and K56 residues.
INTRODUCTION
Sirtuins are evolutionarily conserved metabolic sensor enzymes that use reduced form of nicotinamide adenine dinucleotide (NAD+) as a coenzyme (1, 2). A critical role of sirtuins in aging was first suggested by studies of yeast Sir2 (3). In mammals, depletion of the SIRT6 (sirtuin 6) sirtuin results in shortened life spans, while SIRT6 overexpression extended life spans (4–7). In addition, SIRT6 is associated both with tumor suppression and tumorigenesis in different cancers (8). These aging and cancer-related phenotypes are linked to the ability of SIRT6 to deacylate substrates such as tumor necrosis factor–α (9) and to deacetylate histone H3K9ac and H3K56ac on nucleosomes (10, 11). Histone H3K9ac is a mark of transcriptionally active promoters (12), and histone H3 K9 acetylation plays an important role in DNA repair and telomere maintenance. Crystal structures and biochemical studies have explored how SIRT6 binds a peptide substrate, its NAD+ cofactor, and allosteric effectors (9, 13–20). However, the molecular basis for the sequence preference of SIRT6 to deacetylate histone H3 at positions K9 and K56 (10, 21, 22) is not known. Nor do we understand the structural basis for the preference of SIRT6 to deacetylate nucleosomes at H3 K9 and K56 over free histones (23). To address these deficiencies, we have determined the structure of SIRT6 in complex with its nucleosome substrate. The multivalent interactions between SIRT6 and both histone and DNA components of the nucleosome provide insights into how SIRT6 can deacetylate exposed H3 K9 and occluded H3 K56 residues.
RESULTS
SIRT6-nucleosome structure determination
We obtained 2.7- to 3.1-Å-resolution cryo–electron microscopy (cryo-EM) maps of SIRT6 in complex with a 172–base pair (bp) nucleosome containing a 26-bp DNA extension on one end (Fig. 1 and figs. S1 and S2). Although our sample was prepared in the presence of the SNF2h adenosine triphosphatase (ATPase) subunit, which SIRT6 recruits to sites of DNA damage (24), we do not see density for SNF2h. This suggests that the ATPase may engage with either the nucleosome and/or SIRT6 in a transient or flexible manner. We also do not see density for the intrinsically disordered C-terminal domain of SIRT6, which was previously reported to bind nucleosomal DNA (25). This does not exclude the possibility that dynamic interactions between the SIRT6 C-terminal domain and nucleosomal DNA were not resolved by single-particle cryo-EM. Our data also yielded a subset of particles containing two copies of SIRT6 simultaneously bound to opposite faces of the nucleosome (fig. S2). The density for the SIRT6 bound to the face associated with the DNA extension appears much stronger relative to the density for the SIRT6 on the opposite face without extended DNA, suggesting that the presence of extended DNA stabilizes the positioning of SIRT6 on the nucleosome.
Fig. 1. Overview of SIRT6 nucleosome structure.
(A) The 3.07-Å cryo-EM Coulomb potential density map of structure. (B) Cartoon representation of structure.
Overview of SIRT6-nucleosome complex
Our study shows that the SIRT6 deacetylase domain forms multivalent interactions with the nucleosome via the nucleosome acidic patch, the H3 N-terminal histone tail, the C-terminal H2A tail, and nucleosomal DNA (Fig. 1). The structure of SIRT6 is very similar in the absence or presence of the nucleosome except for residues in and around the SIRT6 NAD+ binding loop and separately, the 10 residues that form the SIRT6-specific extended loop in its zinc-binding domain (13). The orientation of the SIRT6 zinc-binding motif with respect to its catalytic Rossman-fold domain is unchanged upon nucleosome binding. In contrast, the nucleosome is partially unwrapped, with DNA displaced from the extended end of the nucleosome to accommodate SIRT6-DNA interactions.
SIRT6-nucleosome acidic patch interaction
Previous biochemical experiments indicate that SIRT6 engages with a nucleosome through the acidic patch formed by histones H2A and H2B (25), and hydrogen-deuterium exchange mass spectrometry experiments suggested that an N-terminal helix of SIRT6 (residues 28 to 43) may participate in nucleosome interactions (25). However, our structure shows that SIRT6 binds to the acidic patch using the extended loop (167TVAKARGLRA176) (Fig. 2) in its zinc-binding domain that is absent in other sirtuins (13). In the previous hydrogen-deuterium exchange experiments (25), peptides for zinc-binding domain were not observed, which likely prevented assessing whether this region interacts with the nucleosome. The zinc-binding domain can be thought of as a pipe that fills the shallow ditch formed by the H2A/H2B acidic patch. These contacts are mediated by a combination of hydrophilic and van der Waals interactions. Prominent among these interactions is SIRT6 R175, which makes an arginine anchor interaction with H2A acidic patch residues E61, D90, and E92. In addition, other SIRT6 basic residues interact with the acidic patch. SIRT6 R172 constitutes a type 1 variant arginine and interacts with H2A E56 and H2B Q44 and E110, while SIRT6 R178 acts as an atypical arginine to interact with H2A D90 and E92 following the arginine acidic patch nomenclature proposed by McGinty and Tan (26). In addition, SIRT6 K170 makes ionic interactions with H2A E64. These contacts are consistent with biochemical data where mutation of H2A residues E61, E64, D90, and E92 to alanines resulted in weaker SIRT6 binding (25). Mutating each of these SIRT6 basic side chains (K170, R172, R175, and R178) to alanine reduced binding to nucleosomes in an electrophoretic mobility shift assay and, for a subset of mutants analyzed, also in time-resolved FRET (fluorescence resonance energy transfer) assays (Fig. 3). The SIRT6(R175) mutant shows the largest effect with six- to ninefold weaker binding consistent with a critical role of this residue as an arginine anchor. Furthermore, the SIRT6(R175A) has little or no H3K9ac nucleosomal deacetylation activity, corroborating the importance of the R175 arginine anchor for SIRT6 enzymatic activity (Fig. 4, A and C).
Fig. 2. Interactions of SIRT6 zinc-binding domain with nucleosome acidic patch shown in cartoon representation.
Side chains of key residues are shown in stick representation.
Fig. 3. Nucleosome binding of wild-type SIRT6 and SIRT6 containing point mutations in zinc-binding domain.
(A) Quantification of electrophoresis mobility shift nucleosome binding assay (EMSA) for wild-type (WT) and mutant SIRT6. (B) Results of time-resolved FRET nucleosome binding assay for wild type and mutant SIRT6. (C) Dissociation constants for wild-type and mutant SIRT6 determined by EMSA and time-resolved FRET. NA, not analyzed.
Fig. 4. H3 K9Ac nucleosome deacetylation activity of wild-type SIRT6 and arginine anchor mutant SIRT6(R175A).
(A to C) Representative Western blots for the H3K9ac, H3K56ac, and H3K27ac deacetylation reactions for wild-type SIRT6 shown on top, and H3K9ac and H3K56ac deacetylation reactions for SIRT6(R175) shown at the bottom. (D to F) Corresponding plots for quantification of SIRT6-nucleosome deacetylation Western blots with SD error bars (n = 3).
SIRT6-H2A tail interaction
In most nucleosome complex structures, the H2A C-terminal tail is disordered beyond K118 or K119. In our SIRT6-nucleosome structure, weak density for approximately 10 more residues of the histone H2A C-terminal tail is visible and tracks upward to interact with the SIRT6 catalytic domain (Fig. 5A). This interaction was not predicted in previous SIRT6 studies, and to the best of our knowledge, this is the first example of a chromatin enzyme interacting with the H2A C-terminal tail. To test the importance of the H2A C-terminal tail interaction, we reconstituted nucleosomes with histone H2A lacking the C-terminal tail and tested the ability of SIRT6 to deacetylate H3K9ac on these nucleosomes. Intriguingly, we observe increased SIRT6 H3K9ac deacetylation activity on nucleosomes lacking the H2A C-terminal tail (Fig. 5B). This suggests that the H2A tail may have an inhibitory role in regulating SIRT6 deacetylation activity on the nucleosome.
Fig. 5. Inhibitory interactions of histone H2A C-terminal tail near SIRT6 allosteric binding pocket.
(A) The histone H2A C-terminal binds to SIRT6 proximal to allosteric activators {MDL-801, modeled from two different structures [Protein Data Bank (PDB) 5Y2F and 6XV1]} and an allosteric inhibitor (catechin gallate, PDB 6QCJ). The modeled product analog, 2′-O-acyl-ADP-ribose, adopts a well-defined conformation in these three SIRT6-allosteric effector structures. The Cα positions of the H2A C-terminal residues 119 to 128 are shown as yellow spheres. Same color codes as for Fig. 2. (B) Deletion of the H2A C-terminal tail (residues 120 to 130) enhances SIRT6 nucleosomal H3K9ac deacetylase activity (normalized against histone H2B). Representative Western blot data (top) and plot for histone deacetylase (HDAC) assays with SD error bars shown (n = 3).
Our SIRT6-nucleosome sample used for structural determination lacks NAD+, which is a necessary cofactor for SIRT6 deacetylation activity (10). In comparison to crystal structures of SIRT6 bound to adenosine diphosphate (ADP)–ribose (9, 13–20), we notice that the density for SIRT6 residues 64 to 80 is very weak in our map, although they are resolved in the crystal structures without nucleosomes (Fig. 5A and fig. S3). This region of SIRT6 contains residue R65, which makes extensive contacts with the adenosine ribose and pyrophosphate of ADP-ribose in solved crystal structures and is important for the activation of SIRT6 for catalysis (27).The lack of defined density in our map suggests that this region is flexible in the absence of a bound cofactor and only becomes stabilized after cofactor binding. Previous biochemical data showing that R65 mediates a necessary conformational change for activation support this interpretation (27).
SIRT6-nucleosome DNA interactions
A previous study suggested that the disordered SIRT6 C-terminal repeat domain (CTD) can bind nucleosomal DNA (25). While we do not observe the SIRT6 CTD, we find instead that the SIRT6 catalytic domain interacts with nucleosomal DNA. We observe multiple arginine residues within the SIRT6 catalytic domain (R205, R220, R231, R232, and R248) that contact nucleosomal DNA near the entry-exit site of the nucleosome. R205, R231, and R232 contact the DNA phosphate backbone at superhelical location 6 (SHL+6), while R248 binds across the major groove to the opposite DNA strand phosphate backbone at SHL+7 just beyond the nucleosome core (Fig. 6A). The polar residue, N224, in the vicinity may also have a role in the interaction. These interactions are made only to the DNA phosphate backbone with no apparent contacts to the nucleotide bases. To determine the contribution of these residues to SIRT6-nucleosome binding, we generated three sets of SIRT6 mutants: an R205E/R231E/R232E triple mutant, an R220E/N224D double mutant, and an R248E point mutant. SIRT6(R248E) binding to nucleosomes was severely impaired (~12-fold weaker versus wild type), and the SIRT6 triple and double mutants were no longer able to bind nucleosomes (Fig. 6B). These results suggest that the SIRT6 globular domain interactions with nucleosomal DNA play a critical role in stabilizing the SIRT6-nucleosome complex. We also generated and visualized two new SIRT6-nucleosome complexes, containing shorter DNA constructs (147 and 145 bp; figs. S4 and S5). These reconstructions revealed substantially the reduced quality of SIRT6, thereby further suggesting that DNA-SIRT6 interactions play an important role in the stabilization of the complex.
Fig. 6. The SIRT6 catalytic domain interacts with nucleosomal DNA.
(A) Interactions of SIRT6 catalytic domain with nucleosomal DNA showing SIRT6 residues proximal to DNA. Same color code as for Fig. 2. (B) Quantification of electrophoresis mobility shift nucleosomal binding assay for wild-type (WT) and SIRT6 mutated in DNA binding residues.
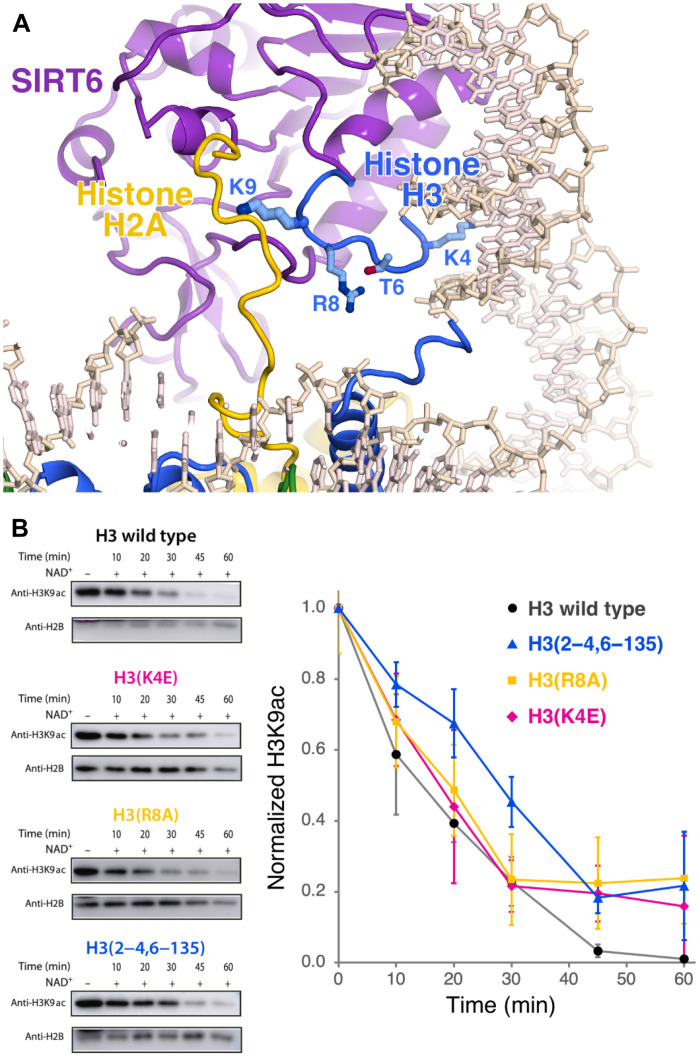
Positioning of H3 substrate residues
We have built H3 residues 3 to 12 into relatively weak density occupying the same peptide substrate binding site in previous SIRT6/myristoylated H3 peptide crystal structures (9, 15). In our structural model, the conformation of the H3 tail substrate is similar but not identical to the SIRT6/myristoylated H3 peptide structures with the H3 K9 side chain positioned essentially equivalent to the myristoylated H3 K9. The H3 tail substrate is sandwiched between the SIRT6 active site and nucleosomal DNA, allowing the H3 K4 side chain to fill the DNA minor groove at SHL 6.5 (Fig. 7A). We wondered whether this H3 K4-DNA interaction and potential interactions made by H3 residues between K4 and K9 might account for the H3 K9 histone deacetylase (HDAC) specificity of SIRT6. We therefore assayed the HDAC activity of SIRT6 on H3 K9 acetylated nucleosomes also containing H3 mutated at specific tail residues. Mutating the H3 K4 to glutamic acid slightly decreased the ability of SIRT6 to deacetylate H3 K9 (Fig. 7B) in nucleosomes. Similar modest adverse effects on SIRT6 H3 K9 deacetylation were observed when the H3 R8 side chain, which might interact with SIRT6, was removed and when one residue between H3 K4 and K9, Q6, was deleted. These results suggest a role of H3 residues 4 to 8 in SIRT6 deacetylation of H3 K9, but further investigation will be needed to determine what defines the specificity of SIRT6 for H3 K9.
Fig. 7. SIRT6 nucleosomal substrate specificity.
(A) Cartoon and stick representation of SIRT6 binding of H3 tail around substrate residue K9. (B) Point mutations and deletions of H3 tail residues that interact with SIRT6 adversely affect SIRT6 nucleosomal histone H3K9Ac deacetylase activity. Representative Western blots are shown to the left and the plot for the HDAC assays is shown to the right with SD error bars (n = 3).
Although we observe H3 K9 in the SIRT6 active site, H3 K56 is far from SIRT6 active site (Fig. 8). In agreement with previous biochemical results (23), we observe that SIRT6 deacetylates H3K56ac albeit less efficiently compared to H3K9ac (Fig. 4, B and E versus A and D). Our structural observation of H3 K56 being distant from the SIRT6 active site raises the question of how SIRT6 accesses H3 K56 for deacetylation (discussed further below). We find that the H3 K56 deacetylation activity of SIRT6 is dependent on the SIRT6 R175 arginine anchor since the SIRT6(R175A) point mutant appears to have little or no H3 K56 deacetylase activity (Fig. 4, B and E). We further note that while a previous study proposed that nucleosomal H3K27ac is also a substrate for SIRT6 (28), we find that SIRT6 does not deacetylate nucleosomal H3K27ac (Fig. 4, C and F).
Fig. 8. Cartoon and stick representation of SIRT6-nucleosome complex shows that histone H3 K56 is exposed but is at least 25 Å from H3 K9 and the SIRT6 catalytic site.
Same color code as for Fig. 2.
DISCUSSION
We have determined the cryo-EM structure of the SIRT6 HDAC in complex with the nucleosome. Our structure explains how SIRT6 deacetylates histone H3 K9 in its physiological nucleosome substrate more efficiently than in peptide substrates. We observe that SIRT6 binds at the nucleosome entry-exit site, using its globular domain to pry DNA from the histone octamer. This action is facilitated by SIRT6’s multivalent interactions with both histone and DNA components of the nucleosome using its catalytic and zinc-binding domains, which are rigidly attached to each other. On one end, SIRT6 uses its zinc-binding domain to dock onto the histone acidic patch. On the other end, SIRT6’s catalytic domain binds to nucleosomal DNA at the entry-exit site replacing contacts otherwise made by the histone H3 N-terminal helix and thus partially exposing this helix. This release also allows the histone H3 N-terminal tail region to position the target H3 K9 side chain into the SIRT6 catalytic site while simultaneously inserting the H3 K4 side chain into the nucleosome DNA minor groove at SHL 6.5. Thus, SIRT6’s multiple interactions with the nucleosome likely facilitate productive binding of the H3 tail for catalysis.
The histone dimer acidic patch is a frequent target of chromatin enzymes and factors with an arginine anchor often used to bind this nucleosomal patch. SIRT6 uses R175 in the zinc-binding domain as an arginine anchor to bind to the histone dimer acidic patch, with the arginine side chain matching the tightly clustered conformation of arginine anchors in other chromatin complexes (26). The critical importance of the R175 arginine anchor for SIRT6 function is confirmed by the reduction in binding affinity to nucleosomes and apparent complete loss of H3 K9 deacetylase activity. We also find that mutations of SIRT6 zinc-binding domain basic residues K170, R172, and R178 adversely affect nucleosome binding, consistent with their interactions with the histone dimer acidic patch observed in our cryo-EM structure.
Unlike H3 K9 which resides in the unstructured H3 N-terminal tail, H3 K56 is located on the H3 N-terminal helix which interacts with nucleosomal DNA in the absence of SIRT6. It was previously unclear how SIRT6 would then be able to access the H3 K56 side chain for catalysis. Our structure shows that SIRT6 prying apart nucleosomal DNA from the histone octamer also exposes the H3 N-terminal helix (Fig. 8). Conversely, acetylation of H3 K56 increases unwrapping of entry-exit nucleosomal DNA (29, 30). In our structure, the alpha carbons of H3 K56 and H3 K9 are 25 Å apart and it is clear that, in this state, H3 K56 cannot reach the SIRT6 active site. We propose that the conformational flexibility in the 10-residue insertion of the SIRT6 zinc-finger domain might allow SIRT6 to pivot as a rigid body about the histone acidic patch to approach the histone core. This hypothesis is supported by our finding that H3 K56 deacetylation of SIRT6 depends on the R175 arginine anchor which interacts with the histone acidic patch. This positioning and the possible unwinding of the H3 N-terminal helix might allow H3 K56 to enter the SIRT6 catalytic site. The additional distortions necessary for this to occur could explain the lower deacetylase activity on H3 K56 substrates.
The H2A C-terminal tail interacts with a SIRT6 short helix adjacent to both the SIRT6 NAD+ binding loop (residues 55 to 65) and the allosteric binding pocket targeted by both activators MDL-801 and quercetin activators and the catechin gallate inhibitor (Fig. 5A). For this reason and our observation of the inhibitory effect of the SIRT6-H2A tail interaction, we suspect that this structure represents SIRT6 bound to a nucleosome in an inhibited, nonactivated state. Since the unbound structure of SIRT6 is largely unchanged when activated or inhibited by the allosteric effectors, it is possible or even likely that activation of SIRT6 will involve local conformational changes near the catalytic site without major changes in how SIRT6 binds to the nucleosome via interactions with the histone acidic patch or nucleosomal DNA. This underscores the significance of the SIRT6 allosteric binding pocket as a target for drug discovery and the importance of using physiological nucleosome substrates for characterizing the effect of drug candidates on the enzymatic activity of SIRT6.
Our structural and biochemical studies explain how multivalent interactions of SIRT6 with both histone and DNA components of the nucleosome enable SIRT6 to deacetylate H3K9ac. Note that the SIRT6 nucleosome used for our structural studies did not include NAD+ or a cofactor analog. We suspect this is not a serious limitation since the SIRT6 catalytic domain adopts the same structure in the presence or absence of cofactor (13, 31). In terms of substrate specificity, our structure suggested possible mechanistic roles for specific H3 residue side chains N-terminal to the H3 K9 target residue. However, the modest effect of mutating these residues on the H3K9ac deacetylase activity of SIRT6 suggests either a concerted role for these N-terminal residues or a more complicated explanation for why SIRT6 targets H3 K9 versus other lysines present in the unstructured H3 tail. The relatively weak density for the H3 tail peptide also limits our confidence in the quality of the structure in this particular region. Our SIRT6-nucleosome atomic model provides the structural framework for further studies to understand H3 K9 specificity and remaining issues such how SIRT6 engages H3 K56 for deacetylation and the role of the H2A tail in SIRT6 function.
MATERIALS AND METHODS
Nucleosome preparation
Three DNA constructs centered on the Widom 601 sequence (32) were used for nucleosome assembly; these constructs consisted of 145 (0-145-0), 147 (1-145-1), and 172 (1-145-26) bp of DNA. Recombinant H3 and H4 Xenopus laevis histones and H2A and H2B human histones were expressed, purified, and reconstituted with each DNA construct as described previously (33), including anion exchange purification of the nucleosomes. Acetylated H3 X. laevis histones expressed in Escherichia coli by amber suppression acetyl lysine incorporation (29) were prepared in-house or purchased from the Histone Source (https://histonesource-colostate.nbsstore.net).
Protein purification
The gene coding for the full-length human SIRT6 (UniProtKB: Q8N6T7) was cloned into pST50Tr (34) vector with an N-terminal Gly-Ser-Ser-hexahistidine (His6). Gly-Ser-Ser-(His6)-SIRT6 was expressed in BL21(DE3) pLysS E. coli cells at 23°C. Bacterial cells were lysed by sonication, and the crude lysate was centrifuged at 36,000g for 40 min at 4°C. The protein was purified by metal affinity chromatography (TALON resin, Clontech), the affinity tag was removed using tobacco etch virus (TEV) protease, and the protein was further purified by Source S cation-exchange chromatography (Cytiva).
His6-tagged human SNF2h was expressed and purified as previously described with minor modifications (35). Briefly, His6-SNF2h was expressed in BL21(DE3) Rosetta E. coli cells at 18°C. Cells were lysed via sonication, and Ni–nitrilotriacetic acid affinity chromatography was used to isolate His6-SNF2h from the clarified lysate. TEV protease was used to remove the His6-tag, and the untagged SNF2h was passed through a HiTrapQ column (Cytiva) to remove contaminating DNA. The protein was then run over a HiLoad Superdex200 column (Cytiva), and pure fractions were pooled, aliquoted, and stored at −80°C.
Cryo-EM sample preparation
SIRT6 was reconstituted in reconstitution buffer [20 mM HHepes (pH 7.5), 75 mM NaCl, and 1 mM dithiothreitol (DTT)] at 0.8:1 enzyme:nucleosome ratio with the 145- and 147-bp nucleosomes and at 2.2:1 ratio with the 172-bp nucleosomes. SNF2h was also included in the SIRT6–172-bp nucleosome sample at 1.1-fold excess over nucleosome.
The SIRT6–147-bp nucleosome sample was cross-linked in reconstitution buffer with 0.05% glutaraldehyde. The sample was incubated on ice for 10 min and then quenched with 100 mM tris-HCl (pH 7.5). The SIRT6–145-bp/SIRT6–172-bp nucleosome samples were first purified using a Superdex 200 column (GE HealthCare) and then stabilized using the GraFix method (36). Light buffer [20 mM Hepes (pH 7.5), 75 mM NaCl, 1 mM DTT, and 10% glycerol] and heavy buffer [20 mM Hepes (pH 7.5), 75 mM NaCl, 1 mM DTT, 40% glycerol, and 0.15% glutaraldehyde] were used to generate a 10 to 40% glycerol gradient with a 0 to 0.15% glutaraldehyde gradient. Fractions from the gradient were checked via native polyacrylamide gel electrophoresis (PAGE). Fractions containing the SIRT6-nucleosome complex were concentrated and buffer-exchanged into EM buffer [12.5 mM Hepes (pH 7.5), 60 mM KCl, 1.5% glycerol, and 1 mM DTT for SIRT6–172-bp nucleosome and 20 mM Hepes (pH 7.5), 75 mM NaCl, and 1 mM DTT for SIRT6–145-bp nucleosome]. The final concentration of the SIRT6-nucleosome samples was ~3 μM.
Cryo-EM grids of the complexes were prepared using an established procedure (37). Specifically, 3.5 μl of the concentrated sample was applied to holey carbon Quantifoil 2/2 Cu200 (172-bp sample) or 1.2/1.3 Cu300 (145/147-bp samples) mesh grids in a FEI Vitrobot Mark IV maintained at 4°C with 100% humidity. The sample was blotted for 3.5 s with a blot force of −1 and then plunge-frozen into liquid ethane.
Cryo-EM data collection
The SIRT6–147-bp nucleosome dataset was collected at the Penn State Cryo-Electron Microscopy Facility on a Titan Krios operated at 300 keV and equipped with an FEI Falcon 3 direct electron detector operated in counting mode. Five hundred twenty-four movies were collected at ×59,000 magnification, which corresponds to 1.14 Å/pixel, at a defocus range of −0.5 to −2.5 μm with accumulated exposure (~58 e/Å2) divided between 44 frames (table S1).
The SIRT6–145-bp nucleosome dataset was collected at the Pacific Northwest Cryo-EM Center on a Titan Krios operated at 300 keV and equipped with a Gatan K3 direct electron detector and an energy filter (20-eV slit). A total of 7730 movies were collected in counting mode at ×81,000 magnification, which corresponds to 1.059 Å/pixel, at a defocus range of −0.8 to −2.2 μm with accumulated exposure (~50 e/Å2) divided between 44 frames (table S1).
The SIRT6–172-bp nucleosome dataset was collected at the National Cancer Institute using a Titan Krios operated at 300 keV and equipped with a Gatan K3 direct electron detector and an energy filter (20-eV slit). The data were collected over the course of two separate sessions at ×81,000 magnification, which corresponds to 1.08 Å/pixel (0.54 Å/pixel in super-resolution mode), at a defocus range of −1.0 to −2.2 μm with accumulated exposure (~50 e/Å2) fractionated into 40 super-resolution frames. In total, 11,872 movies were collected over the two sessions (5122 movies in the first session and 6750 movies in the second session; table S1).
Cryo-EM data processing
The SIRT6–172-bp nucleosome dataset was imported into RELION v3.1 (38, 39). Using UCSF MotionCor2 v1.4.1 (40) within RELION, raw movies were motion-corrected, binned twice to 1.08 Å/pixel, and the resulting dose-weighted micrographs (41) were imported into cryoSPARC (42). Patch CTF estimation (multi) was used to estimate defocus values. A nucleosome map was used to generate templates for template picking of particles; these particles were then extracted using a 300-pixel box Fourier-binned to 100 pixels (resulting in 3.24 Å/pixel). Two-dimensional (2D) classification was performed and classes containing obvious junk were removed. Ab initio reconstruction was performed with the remaining particles to generate input classes for heterogeneous refinement. Subsequently, a cleaned dataset of nucleosomal particles was subjected to numerous rounds of 3D classification, yielding a single class with a strong density for SIRT6. To further the quality of the complex, 3D classification without alignment in RELION was performed with a mask focused on the globular domain of SIRT6 and the adjacent DNA. A class with 71,603 particles that represented the most stable positioning of SIRT6 on the nucleosome visually was selected. These particles were refined using nonuniform refinement (43) in cryoSPARC, as well as with RELION and with cisTEM (44). All reconstructions looked reasonable, but the cisTEM reconstruction was used for further interpretation.
Subsequently, Bayesian polishing was performed in RELION to further improve the quality of the map (45). Two rounds of Bayesian polishing and CTF refinement were performed using 1,007,638 particles from an earlier 2.83-Å consensus reconstruction. This improved the resolution of the consensus reconstruction to 2.63 Å. Using these optimized particles, the 71,603-particle subset was refined to 3.07 Å with the improved quality of SIRT6 (figs. S1 and S2 and table S1).
The 147-bp dataset containing 524 movies was first motion-corrected using cryoSPARC’s Patch motion correction (multi). Defocus values were calculated using Patch CTF estimation (multi), and 398,186 particles were blob-picked using a sphere with a radius of 120 to 140 Å and extracted in a 256-pixel box. Five models were generated using ab initio reconstruction, and the particles were subjected to heterogeneous refinement. One class was selected on the basis of the presence of SIRT6 and refined to 4.84 Å using nonuniform refinement. These particles were then subjected to classification with the best reconstruction from the previous step seeded three times. One of the resulting maps exhibited a more stable SIRT6-nucleosome complex and was subjected to nonuniform refinement that yielded a reconstruction at 5.0 Å. Using these particles, we re-extracted the subset in a larger 300-pixel box; this yielded 40,834 particles that we refined the 4.9-Å final resolution using non-uniform refinement (new) (fig. S4 and table S1).
The SIRT6–145-bp nucleosome dataset was processed as described for SIRT6–147-bp nucleosome datasets, except for motion correction—here, we used UCSF MotionCor2 instead. We obtained two final reconstructions, differing in their SIRT6-interacting DNA: (i) 3.28-Å reconstruction from 31,802 particles (fig. S5, C to E and I, and table S1) and (ii) 3.27-Å reconstruction from 34,737 particles (fig. S5, F to H and J, and table S1).
The final resolutions reported were calculated using Fourier shell correlation and assessed at the 0.143 cutoff following gold standard refinement (46). All file format conversions between cryoSPARC and RELION, as well as particle stack preparation for cisTEM refinement, were performed using UCSF pyem v0.5 (47).
Model building and refinement
Protein Data Bank (PDB) 3LZ0 (48) was used as a starting model for the nucleosome, and 3PKI (13) and 7CL0 (19) were used as starting models for SIRT6. The models were rigid-body fitted into the 3.07-Å reconstruction using the “fit in map” function in UCSF ChimeraX (49) and then further optimized in Coot (50).
A combination of 3D Coulomb potential maps from the 172-bp dataset was used to construct the final model. Using the highest resolved 2.63-Å reconstruction in Coot, histones and core DNA were optimized and the SIRT6 acidic patch–interacting loop was constructed. The 3.07-Å reconstruction was then used to position and adjust the SIRT6 globular domain, zinc-binding domain, histone H2A C-terminal tail, histone H3 N-terminal tail, and the DNA overhang. The final model was refined using phenix.real_space_refine (51) with secondary structure, Ramachandran, and rotamer restraints. Last, the model was validated manually in Coot and with Molprobity (52) using comprehensive validation (cryo-EM) in Phenix. Model statistics are reported in table S2. The model contains H2A(11-120), H2B(27-122), H3(36-134), H4(19-102), 149-bp DNA, SIRT6(14-293), and one copy of the H3(3-12) N-terminal tail in the SIRT6 active site.
Electrophoresis mobility shift nucleosome binding assay
Ten-microliter reactions of SIRT6 variants at indicated concentrations were incubated with 100 nM wild-type nucleosome core particles for 30 min at 30°C in 5× binding buffer [25 mM Hepes (pH 7.3), 150 mM NaCl, 4.5 mM MgCl2, 1 mM DTT, 0.01% Tween 20, and bovine serum albumin (BSA) (0.1 mg/ml)]. Reactions were quenched with 2 μl of 50% glycerol and electrophoresed on 4.5% native PAGE gels at 100 V, 4°C.
Time-resolved FRET nucleosome binding assay
Time-resolved FRET nucleosome binding assays were performed as described by Wesley et al. (53). Acceptor mixtures were prepared by mixing ULight α-6xHIS acceptor antibody (PerkinElmer) with 6xHIS-tagged SIRT6 variants at a ratio of 1:20 and serially diluting across 13 concentrations in H75 buffer [20 mM Hepes (pH 7.5), 75 mM NaCl, 5 mM DTT, 5% glycerol, 0.01% NP-40, 0.01% CHAPS, and BSA (100 μg/ml)]. Two-time donor mixtures were prepared by mixing 4 nM streptavidin-Eu (PerkinElmer) with or without 2 nM nucleosomes containing 177 bp of Widom 601 DNA (31 + 145 + 1) with a 5′ biotin group on the 31-bp extension. Samples were prepared in 384-well plates by mixing 5 μl of 2× donor mixtures with 5 μl of acceptor mixtures at each dilution. Fluorescence signals were acquired at room temperature in a Victor Nivo multimode fluorescent plate reader (PerkinElmer) using an excitation filter at 320 nm and emission filters at 615 and 665 nm. Emission signals at 615 and 665 nm were measured simultaneously following a 100-μs delay. Kd (dissociation constant) values were determined from triplicate titrations of each SIRT6 variant and are reported as means ± SEM.
Histone deacetylation assays
Wild-type SIRT6 (500 or 125 nM) and SIRT6(R175A) (200 nM) were incubated with nucleosomes (125 nM) containing either acetylated H3 K9, H3 K56, or H3 K27, and/or with C-terminally truncated H2A [H2A(2-119)] at 37°C for varying time points in HDAC reaction buffer [25 mM Hepes (pH 7.3), 49 mM NaCl, 4.5 mM MgCl2, and 1 mM DTT]. Reactions were quenched with the addition of 2× SDS sample buffer, boiled, and then electrophoresed on a 12% SDS-PAGE gel. Proteins on the gel were transferred to a polyvinylidene difluoride membrane (Millipore, IPFL 00010), which was first blocked with 10% milk, and then probed with either 1:10,000 anti-H3K9ac antibody (Active Motif, #39038), 1:1000 anti-H3K56ac antibody (Active Motif, #39133), or 1:3000 anti-H3K27ac antibody (Active Motif, #39082), and also probed with 1:3000 anti-H2B antibody (Abcam, #64039). Western blots were quantified using ImageJ (54) or Bio-Rad Image Lab. Control experiments show similar SIRT6 deacetylase activity using nucleosomes reconstituted from H3K9ac protein prepared in-house or purchased from the Histone Source.
Acknowledgments
We thank S.-Y. Kuan and E. Baron at Penn State for nucleosome preparations, A. Wier at NCI and T. Humphreys at PNCC for assistance with EM data collection, J. Cho and C. Bator of the Penn State Cryo-Electron Microscopy Facility for assistance with EM screening and data collection, and J. Gray at Penn State for assistance with training on the T20 for negative stain EM.
Funding: This work was supported by National Institutes of Health (NIH) grant F32GM137463 (U.S.C.), NIH grant T32BM107000 (A.R.B.), NIH grant R35GM122519 (C.L.P.), and NIH grant R35GM127034 (S.T.). Research reported in this publication was supported by the Office of the Director, NIH, under award number S10OD026822-01. This project is funded, in part, under a grant from the Pennsylvania Department of Health using Tobacco CURE Funds. The Department specifically disclaims responsibility for any analyses, interpretations or conclusion. This research was, in part, supported by the National Cancer Institute’s National Cryo-EM Facility at the Frederick National Laboratory for Cancer Research under contract HSSN261200800001E. A portion of this research was supported by NIH grant U24GM129547 and performed at the PNCC at OHSU and accessed through EMSL, a DOE Office of Science User Facility sponsored by the Office of Biological and Environmental Research. The figures for this manuscript were generated using PyMOL and UCSF ChimeraX. UCSF ChimeraX is developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, with support from NIH R01-GM129325 and the Office of Cyber Infrastructure and Computational Biology, National Institute of Allergy and Infectious Diseases.
Author contributions: Conceptualization: C.L.P., S.T., and J.-P.A. Methodology: U.S.C., O.R., A.R.B., J.Z., E.M.L., J.L.F., C.L.P., S.T., and J.-P.A. Investigation: U.S.C., O.R., A.R.B., J.Z., E.M.L., J.L.F., and J.-P.A. Visualization: S.T. and J.-P.A. Funding acquisition: U.S.C., C.L.P., S.T., and J.-P.A. Project administration: C.L.P., S.T., and J.-P.A. Supervision: C.L.P., S.T., and J.-P.A. Writing—original draft: U.S.C., S.T., and J.-P.A. Writing—review and editing: O.R., A.R.B., C.L.P., S.T., and J.-P.A.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The atomic coordinates of the SIRT6-nucleosome complex have been deposited to the RCSB PDB with PDB ID 8G57. The cryo-EM Coulomb potential map for the best-resolved SIRT6-nucleosome complex was deposited in the Electron Microscopy Data Bank as EMD-29735. The raw cryo-EM data were deposited in EMPIAR (EMPIAR-11427).
Supplementary Materials
This PDF file includes:
Figs. S1 to S5
Tables S1 and S2
REFERENCES AND NOTES
- 1.R. H. Houtkooper, E. Pirinen, J. Auwerx, Sirtuins as regulators of metabolism and healthspan. Nat. Rev. Mol. Cell Biol. 13, 225–238 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Z. Mei, X. Zhang, J. Yi, J. Huang, J. He, Y. Tao, Sirtuins in metabolism, DNA repair and cancer. J. Exp. Clin. Cancer Res. 35, 182 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.M. Kaeberlein, M. McVey, L. Guarente, The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 13, 2570–2580 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.R. Mostoslavsky, K. F. Chua, D. B. Lombard, W. W. Pang, M. R. Fischer, L. Gellon, P. Liu, G. Mostoslavsky, S. Franco, M. M. Murphy, K. D. Mills, P. Patel, J. T. Hsu, A. L. Hong, E. Ford, H. L. Cheng, C. Kennedy, N. Nunez, R. Bronson, D. Frendewey, W. Auerbach, D. Valenzuela, M. Karow, M. O. Hottiger, S. Hursting, J. C. Barrett, L. Guarente, R. Mulligan, B. Demple, G. D. Yancopoulos, F. W. Alt, Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell 124, 315–329 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Y. Kanfi, S. Naiman, G. Amir, V. Peshti, G. Zinman, L. Nahum, Z. Bar-Joseph, H. Y. Cohen, The sirtuin SIRT6 regulates lifespan in male mice. Nature 483, 218–221 (2012). [DOI] [PubMed] [Google Scholar]
- 6.A. Roichman, S. Elhanati, M. A. Aon, I. Abramovich, A. Di Francesco, Y. Shahar, M. Y. Avivi, M. Shurgi, A. Rubinstein, Y. Wiesner, A. Shuchami, Z. Petrover, I. Lebenthal-Loinger, O. Yaron, A. Lyashkov, C. Ubaida-Mohien, Y. Kanfi, B. Lerrer, P. J. Fernández-Marcos, M. Serrano, E. Gottlieb, R. de Cabo, H. Y. Cohen, Restoration of energy homeostasis by SIRT6 extends healthy lifespan. Nat. Commun. 12, 3208 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.J. R. Taylor, J. G. Wood, E. Mizerak, S. Hinthorn, J. Liu, M. Finn, S. Gordon, L. Zingas, C. Chang, M. A. Klein, J. M. Denu, V. Gorbunova, A. Seluanov, J. D. Boeke, J. M. Sedivy, S. L. Helfand, Sirt6 regulates lifespan in Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 119, e2111176119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.V. Desantis, A. Lamanuzzi, A. Vacca, The role of SIRT6 in tumors. Haematologica 103, 1–4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.H. Jiang, S. Khan, Y. Wang, G. Charron, B. He, C. Sebastian, J. Du, R. Kim, E. Ge, R. Mostoslavsky, H. C. Hang, Q. Hao, H. Lin, SIRT6 regulates TNF-α secretion through hydrolysis of long-chain fatty acyl lysine. Nature 496, 110–113 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.E. Michishita, R. A. McCord, E. Berber, M. Kioi, H. Padilla-Nash, M. Damian, P. Cheung, R. Kusumoto, T. L. A. Kawahara, J. C. Barrett, H. Y. Chang, V. A. Bohr, T. Ried, O. Gozani, K. F. Chua, SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature 452, 492–496 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.S. Kugel, J. L. Feldman, M. A. Klein, D. M. Silberman, C. Sebastián, C. Mermel, S. Dobersch, A. R. Clark, G. Getz, J. M. Denu, R. Mostoslavsky, Identification of and Molecular Basis for SIRT6 loss-of-function point mutations in cancer. Cell Rep. 13, 479–488 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.K. Karmodiya, A. R. Krebs, M. Oulad-Abdelghani, H. Kimura, L. Tora, H3K9 and H3K14 acetylation co-occur at many gene regulatory elements, while H3K14ac marks a subset of inactive inducible promoters in mouse embryonic stem cells. BMC Genomics 13, 424 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.P. W. Pan, J. L. Feldman, M. K. Devries, A. Dong, A. M. Edwards, J. M. Denu, Structure and biochemical functions of SIRT6. J. Biol. Chem. 286, 14575–14587 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.W. You, D. Rotili, T. M. Li, C. Kambach, M. Meleshin, M. Schutkowski, K. F. Chua, A. Mai, C. Steegborn, Structural basis of Sirtuin 6 activation by synthetic small molecules. Angew. Chem. Int. Ed. 56, 1007–1011 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Z. Huang, J. Zhao, W. Deng, Y. Chen, J. Shang, K. Song, L. Zhang, C. Wang, S. Lu, X. Yang, B. He, J. Min, H. Hu, M. Tan, J. Xu, Q. Zhang, J. Zhong, X. Sun, Z. Mao, H. Lin, M. Xiao, Y. E. Chin, H. Jiang, Y. Xu, G. Chen, J. Zhang, Identification of a cellularly active SIRT6 allosteric activator. Nat. Chem. Biol. 14, 1118–1126 (2018). [DOI] [PubMed] [Google Scholar]
- 16.W. You, C. Steegborn, Structural basis of Sirtuin 6 inhibition by the hydroxamate trichostatin A: Implications for protein deacylase drug development. J. Med. Chem. 61, 10922–10928 (2018). [DOI] [PubMed] [Google Scholar]
- 17.W. You, C. Steegborn, Structural basis for activation of human sirtuin 6 by fluvastatin. ACS Med. Chem. Lett. 11, 2285–2289 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.W. You, C. Steegborn, Binding site for activator MDL-801 on SIRT6. Nat. Chem. Biol. 17, 519–521 (2021). [DOI] [PubMed] [Google Scholar]
- 19.Z. Huang, J. Zhao, W. Deng, Y. Chen, J. Shang, K. Song, L. Zhang, C. Wang, S. Lu, X. Yang, B. He, J. Min, H. Hu, M. Tan, J. Xu, Q. Zhang, J. Zhong, X. Sun, Z. Mao, H. Lin, M. Xiao, Y. E. Chin, H. Jiang, H. Shen, Y. Xu, G. Chen, J. Zhang, Reply to: Binding site for MDL-801 on SIRT6. Nat. Chem. Biol. 17, 522–523 (2021). [DOI] [PubMed] [Google Scholar]
- 20.W. You, W. Zheng, S. Weiss, K. F. Chua, C. Steegborn, Structural basis for the activation and inhibition of Sirtuin 6 by quercetin and its derivatives. Sci. Rep. 9, 19176 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.E. Michishita, R. A. McCord, L. D. Boxer, M. F. Barber, T. Hong, O. Gozani, K. F. Chua, Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6. Cell Cycle 8, 2664–2666 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.L. Tasselli, Y. Xi, W. Zheng, R. I. Tennen, Z. Odrowaz, F. Simeoni, W. Li, K. F. Chua, SIRT6 deacetylates H3K18ac at pericentric chromatin to prevent mitotic errors and cellular senescence. Nat. Struct. Mol. Biol. 23, 434–440 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.R. Gil, S. Barth, Y. Kanfi, H. Y. Cohen, SIRT6 exhibits nucleosome-dependent deacetylase activity. Nucleic Acids Res. 41, 8537–8545 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.D. Toiber, F. Erdel, K. Bouazoune, D. M. Silberman, L. Zhong, P. Mulligan, C. Sebastian, C. Cosentino, B. Martinez-Pastor, S. Giacosa, A. D’Urso, A. M. Näär, R. Kingston, K. Rippe, R. Mostoslavsky, SIRT6 recruits SNF2H to DNA break sites, preventing genomic instability through chromatin remodeling. Mol. Cell 51, 454–468 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.W. H. Liu, J. Zheng, J. L. Feldman, M. A. Klein, V. I. Kuznetsov, C. L. Peterson, P. R. Griffin, J. M. Denu, Multivalent interactions drive nucleosome binding and efficient chromatin deacetylation by SIRT6. Nat. Commun. 11, 5244 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.R. K. McGinty, S. Tan, Principles of nucleosome recognition by chromatin factors and enzymes. Curr. Opin. Struct. Biol. 71, 16–26 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.M. A. Klein, J. M. Denu, Biological and catalytic functions of sirtuin 6 as targets for small-molecule modulators. J. Biol. Chem. 295, 11021–11041 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.W. W. Wang, Y. Zeng, B. Wu, A. Deiters, W. R. Liu, A chemical biology approach to reveal Sirt6-targeted histone H3 sites in nucleosomes. ACS Chem. Biol. 11, 1973–1981 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.H. Neumann, S. M. Hancock, R. Buning, A. Routh, L. Chapman, J. Somers, T. Owen-Hughes, J. van Noort, D. Rhodes, J. W. Chin, A method for genetically installing site-specific acetylation in recombinant histones defines the effects of H3 K56 acetylation. Mol. Cell 36, 153–163 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.J. C. Shimko, J. A. North, A. N. Bruns, M. G. Poirier, J. J. Ottesen, Preparation of fully synthetic histone H3 reveals that acetyl-lysine 56 facilitates protein binding within nucleosomes. J. Mol. Biol. 408, 187–204 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.J. Zhang, Z. Huang, K. Song, Sirt6 apo structure (2017); 10.2210/pdb5X16/pdb. [DOI]
- 32.P. T. Lowary, J. Widom, New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 276, 19–42 (1998). [DOI] [PubMed] [Google Scholar]
- 33.K. Luger, T. J. Rechsteiner, T. J. Richmond, Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 304, 3–19 (1999). [DOI] [PubMed] [Google Scholar]
- 34.S. Tan, R. C. Kern, W. Selleck, The pST44 polycistronic expression system for producing protein complexes in Escherichia coli. Protein Expr. Purif. 40, 385–395 (2005). [DOI] [PubMed] [Google Scholar]
- 35.J. D. Leonard, G. J. Narlikar, A nucleotide-driven switch regulates flanking DNA length sensing by a dimeric chromatin remodeler. Mol. Cell 57, 850–859 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.B. Kastner, N. Fischer, M. M. Golas, B. Sander, P. Dube, D. Boehringer, K. Hartmuth, J. Deckert, F. Hauer, E. Wolf, H. Uchtenhagen, H. Urlaub, F. Herzog, J. M. Peters, D. Poerschke, R. Lührmann, H. Stark, GraFix: Sample preparation for single-particle electron cryomicroscopy. Nat. Methods 5, 53–55 (2008). [DOI] [PubMed] [Google Scholar]
- 37.X. Li, P. Mooney, S. Zheng, C. R. Booth, M. B. Braunfeld, S. Gubbens, D. A. Agard, Y. Cheng, Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat. Methods 10, 584–590 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.S. H. W. Scheres, RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.J. Zivanov, T. Nakane, B. O. Forsberg, D. Kimanius, W. J. H. Hagen, E. Lindahl, S. H. W. Scheres, New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.S. Q. Zheng, E. Palovcak, J. P. Armache, K. A. Verba, Y. Cheng, D. A. Agard, MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.T. Grant, N. Grigorieff, Measuring the optimal exposure for single particle cryo-EM using a 2.6 Å reconstruction of rotavirus VP6. eLife 4, e06980 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.A. Punjani, J. L. Rubinstein, D. J. Fleet, M. A. Brubaker, CryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 43.A. Punjani, H. Zhang, D. J. Fleet, Non-uniform refinement: Adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods 17, 1214–1221 (2020). [DOI] [PubMed] [Google Scholar]
- 44.T. Grant, A. Rohou, N. Grigorieff, cisTEM, user-friendly software for single-particle image processing. eLife 7, e35383 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.J. Zivanov, T. Nakane, S. H. W. Scheres, A Bayesian approach to beam-induced motion correction in cryo-EM single-particle analysis. IUCrJ. 6, 5–17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.P. B. Rosenthal, R. Henderson, Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333, 721–745 (2003). [DOI] [PubMed] [Google Scholar]
- 47.D. Asarnow, E. J. Palovcak, Y. Cheng, asarnow/pyem: UCSF pyem v0.5. Zenodo (2019).
- 48.D. Vasudevan, E. Y. D. Chua, C. A. Davey, Crystal structures of nucleosome core particles containing the “601” strong positioning sequence. J. Mol. Biol. 403, 1–10 (2010). [DOI] [PubMed] [Google Scholar]
- 49.E. F. Pettersen, T. D. Goddard, C. C. Huang, E. C. Meng, G. S. Couch, T. I. Croll, J. H. Morris, T. E. Ferrin, UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.P. Emsley, B. Lohkamp, W. G. Scott, K. Cowtan, Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.P. V. Afonine, B. K. Poon, R. J. Read, O. V. Sobolev, T. C. Terwilliger, A. Urzhumtsev, P. D. Adams, Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Struct. Biol. 74, 531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.C. J. Williams, J. J. Headd, N. W. Moriarty, M. G. Prisant, L. L. Videau, L. N. Deis, V. Verma, D. A. Keedy, B. J. Hintze, V. B. Chen, S. Jain, S. M. Lewis, W. B. Arendall, J. Snoeyink, P. D. Adams, S. C. Lovell, J. S. Richardson, D. C. Richardson, MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.N. A. Wesley, A. Skrajna, H. C. Simmons, G. R. Budziszewski, D. N. Azzam, A. P. Cesmat, R. K. McGinty, Time resolved-fluorescence resonance energy transfer platform for quantitative nucleosome binding and footprinting. Protein Sci. 31, e4339 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.C. A. Schneider, W. S. Rasband, K. W. Eliceiri, NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S5
Tables S1 and S2