Abstract
Hundreds of pathogenic variants of mitochondrial DNA (mtDNA) have been reported to cause mitochondrial diseases, which still lack effective treatments. It is a huge challenge to install these mutations one by one. We repurposed the DddA-derived cytosine base editor to incorporate a premature stop codon in the mtProtein-coding genes to ablate mitochondrial proteins encoded in the mtDNA (mtProteins) instead of installing pathogenic variants and generated a library of both cell and rat resources with mtProtein depletion. In vitro, we depleted 12 of 13 mtProtein-coding genes with high efficiency and specificity, resulting in decreased mtProtein levels and impaired oxidative phosphorylation. Moreover, we generated six conditional knockout rat strains to ablate mtProteins using Cre/loxP systemhttp://www.ratresource.com. Mitochondrially encoded ATP synthase membrane subunit 8 and NADH:ubiquinone oxidoreductase core subunit 1 were specifically depleted in heart cells or neurons, resulting in heart failure or abnormal brain development. Our work provides cell and rat resources for studying the function of mtProtein-coding genes and therapeutic strategies.
The DdCBE was repurposed to conditionally knock out mitochondrial protein-coding genes in rats.
INTRODUCTION
In mammals, the mitochondrion contains multiple copies of mitochondrial DNA (mtDNA), which is essential to mitochondrial biogenesis and function (1–3). Mutations in mtDNA result in various mitochondrial diseases, usually involved in the heart, nervous system, and skeletal muscles (4). These diseases are mostly heteroplasmic with heterogeneous phenotypes and variable onset ages (4). Hundreds of pathogenic mtDNA mutations have been reported. So far, no curative treatments are available because of the absence of suitable cellular and animal models (5–8). Mitochondrial genome modification is a challenge because (i) one mammal cell contains hundred to thousand copies of mtDNA, whereby cell may often tolerate relatively high levels of mtDNA mutations without affecting mitochondrial functions (1, 2, 4); (ii) mitochondria are resistant to nucleic acid (DNA or RNA) entry, resulting in that CRISPR system–based gene editing tools cannot be applied for mtDNA manipulation (9); and (iii) DNA nucleases, such as zinc finger nucleases (ZFNs) and transcription activator–like effector nucleases (TALENs), could been engineered to target and degrade mutant mtDNA rather than to generate models with mutant mtDNA (5).
Recently, a bacterial-derived toxin named DddA, which can catalyze the conversion of cytosine to uracil within double-strand DNA, has been applied to genetically modify mtDNA in human cells (10), zebrafish (11), mice (12, 13), and rats (14) by fusion with transcription activator-like effector (TALE). However, it is cumbersome to install hundreds of mutations one by one in cellular and animal models because (i) it is expensive, laborious, and time-consuming and (ii) only a few of sites could be precisely installed by DddA-derived cytosine base editors (DdCBE) and transcription-activator-like effector linked deaminases (TALEDs) (15); the remaining variants, including indels, are hard to be modeled. During evolution, most mitochondrial genes were transferred to the nucleus, leaving 13 protein-coding genes and 24 noncoding genes in mammalian mitochondria. Mutations in these noncoding genes (transfer RNA and ribosomal RNA) can disturb the translation of the 13 protein-coding genes (16, 17). We reasoned that knockout (KO) of the 13 mitochondrial protein-coding genes is more feasible than installing all or partial pathogenic variants of mtDNA.
Rat is an optimal model for cardiac and nerve physiology studies compared with mouse (18, 19); meanwhile, the two organs are more susceptible to mtDNA mutations due to high energy consumptions. In previous studies, we succeeded in installation of pathogenic variants in rats (14) by DdCBE-mediated mtDNA editing and demonstrated that rats with installed mutations could resemble human mitochondrial diseases. In this study, we expanded the application of DdCBE to ablate mitochondrial proteins encoded in the mtDNA (mtProteins) by incorporating a premature stop codon into the mtDNA-encoding genes in cells and rats and investigated the roles of mtProteins in the heart and brain. First, Atp8 and Nd1 were selected for targeting in rat C6 cells. We observed nearly abolished mitochondrially encoded ATP synthase membrane subunit 8 (ATP8) and NADH:ubiquinone oxidoreductase core subunit 1 (ND1) expression and notably impaired oxidative phosphorylation (OXPHOS) in corresponding targeted cells. Second, to avoid the impairment of embryonic development due to the depletion of mtProteins throughout the body, the Cre/loxP system was used to achieve mtProtein ablation in a spatial-temporal manner.
Conditional KO (cKO) of Atp8 and Nd1 in rat heart impaired cardiac function and mitochondrial structure, resulting in pups’ death within 2 weeks after birth. Conditional depletion of ATP8 and ND1 in rat brain also led to death within 2 weeks, featured with abnormal brain development. Last, we applied this strategy to ablate all rat mtProteins. Except ND4L, which has no swappable codons, all of 12 mtProteins could be depleted or nearly depleted in C6 cells. Furthermore, six cKO rat lines—including Nd1, Nd2, and Nd5 for complex I; Cytb for complex III; Cox3 for complex IV; and Atp8 for complex V—were generated and preserved in the Rat Resource Center of China (www.ratresource.com), allowing researchers to study the biological function of mtProteins and develop potential treatments for mitochondrial disorders.
RESULTS
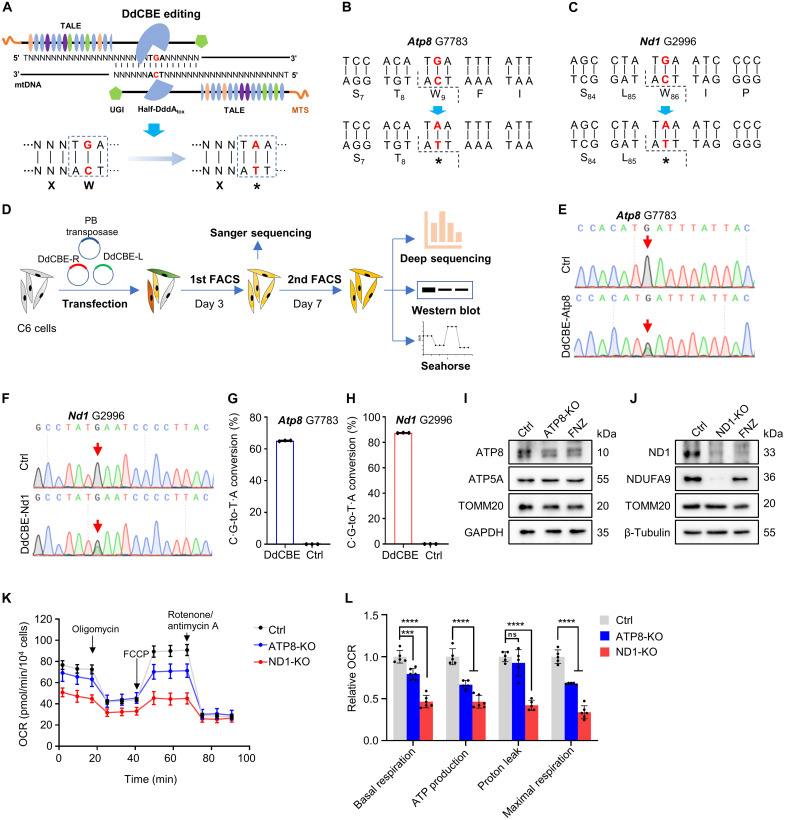
DdCBE-mediated ATP8 and ND1 ablation in rat C6 cells
Previously, we have applied DdCBE-mediated C·G-to-T·A conversion to install human pathogenic mtDNA mutation in rats (14). Here, we intended to use DdCBE-mediated mtDNA editing to ablate mtProtein by swapping Trp codon TGA with stop codon TAA through deaminating C to T on the opposite strand (Fig. 1A). We first selected the G7783 site in rat Atp8 and G2996 site in rat Nd1 for targeting (Fig. 1, B and C, and fig. S1, A and B). Four DdCBE pairs [left G1333C (L1333C) + right G1333N (R1333N), L1397C + R1397N, L1333N + R1333C, and L1397N + R1397C] were designed for each targeting site to test their editing efficiency in C6 cells. The DdCBE plasmids were assembled as described using the Golden Gate strategy (11). The L1333C + R1333N pair yielded the highest editing activity at both target sites and was selected for the following experiments (fig. S1C).
Fig. 1. DdCBE-mediated ATP8/ND1 KO in rat C6 cells.
(A) Strategy of KO mtProteins by introducing the stop codon using DdCBE. (B and C) The DdCBE-mediated C·G-to-T·A conversion at Atp8 G7783 (B) and Nd1 G2996 (C). (D) The work flow for DdCBE-mediated mtProteins KO in C6 cells. (E and F) Sanger sequencing chromatogram of spacer for Atp8 G7783 site (E) and Nd1 G2996 site (F) at day 3 after the first-round enrichment [first fluorescence-activated cell sorting (FACS)] of double-positive C6 cells. (G and H) The frequency of the DdCBE-mediated C·G-to-T·A conversion at Atp8 G7783 (G) and Nd1 G2996 (H) sites was analyzed using deep sequencing at day 7 after the second-round enrichment (second FACS). n = 3 technical replicates. Data were presented as means ± SD. Untreated C6 cells were used as control (Ctrl). (I and J) The protein levels of ATP8 (I) and ND1 (J) were detected by Western blot in rat C6 cells treated with DdCBE or flunarizine (FNZ). (K) Mitochondrial respiratory capacity was indicated by the oxygen consumption rate (OCR) in Ctrl and ATP8-/ND1-KO C6 cells. (L) Relative values of OXPHOS parameters from (K). n = 5 for technical replicates. Data were presented as means ± SD. ns, not significant; ***P ≤ 0.001 and ****P ≤ 0.0001, by one-way analysis of variance (ANOVA) test paired with a Tukey’s post hoc test.
We cloned L1333C and R1333N into the PiggyBac (PB) transposon vectors with enhanced green fluorescent protein (EGFP) or mCherry tag, respectively (fig. S1D). To ablate mtProteins, the PB-DdCBE pair was cotransfected with PB transposase into rat C6 cells. We evaluated the edit efficiency with Sanger sequencing at day 3 posttransfection and at day 7 posttransfection with deep sequencing (Fig. 1D). Sanger sequencing results showed that DdCBE mediated C·G-to-T·A conversion efficiently at the expected target sites (Fig. 1, E and F). Although bystander editing events were observed around target loci (Atp8 and Nd1) in deep sequencing results, they do not affect the formation of the DdCBE-induced premature stop codon (fig. S1, E and F). Deep sequencing results showed that the editing efficiency at the Atp8 G7783 site and Nd1 G2996 site was up to 65.13 ± 0.32% (Fig. 1G) and 87.47 ± 0.2% (Fig. 1H), respectively. To assess whether the protein levels of targeted genes are disturbed after incorporating a premature stop codon via DdCBE, we detected the protein expression of ATP8 and ND1 in ATP8-KO and ND1-KO C6 cells after the second round of enrichment on day 7 (Fig. 1D). Cells treated with flunarizine (FNZ), which can decrease the mitochondrial content, were used as the positive control. We observed that the protein levels of targeted genes markedly decreased. Compared to the control cells, ATP8 protein decreased by ~75% in ATP8-KO C6 cells (Fig. 1I and fig. S1G) and ND1 decreased by ~79% in ND1-KO C6 cells (Fig. 1J and fig. S1H). These results were further confirmed using rho zero (ρ0) C6 cells as control, which were generated by treatment of ethidium bromide (EB) and 2′,3′-dideoxycytidine (ddC) (fig. S1, I and J) (20). It has been reported that mutations in mtProteins generally impair the assembly or stability of OXPHOS complexes (21, 22). In our edited C6 cells, NDUFA9, a member subunit of complex I, decreased upon ND1 depletion, while ATP5, a member of complex V, was not obviously affected upon ATP8 depletion. Consistently, the basal and uncoupled oxygen consumption rates (OCRs) were reduced significantly in Atp8 and Nd1 targeted cells (Fig. 1, K and L), indicating that the function of mitochondria was impaired. Our data suggest that DdCBE can be repurposed to KO mitochondrial protein-coding genes in rat C6 cells.
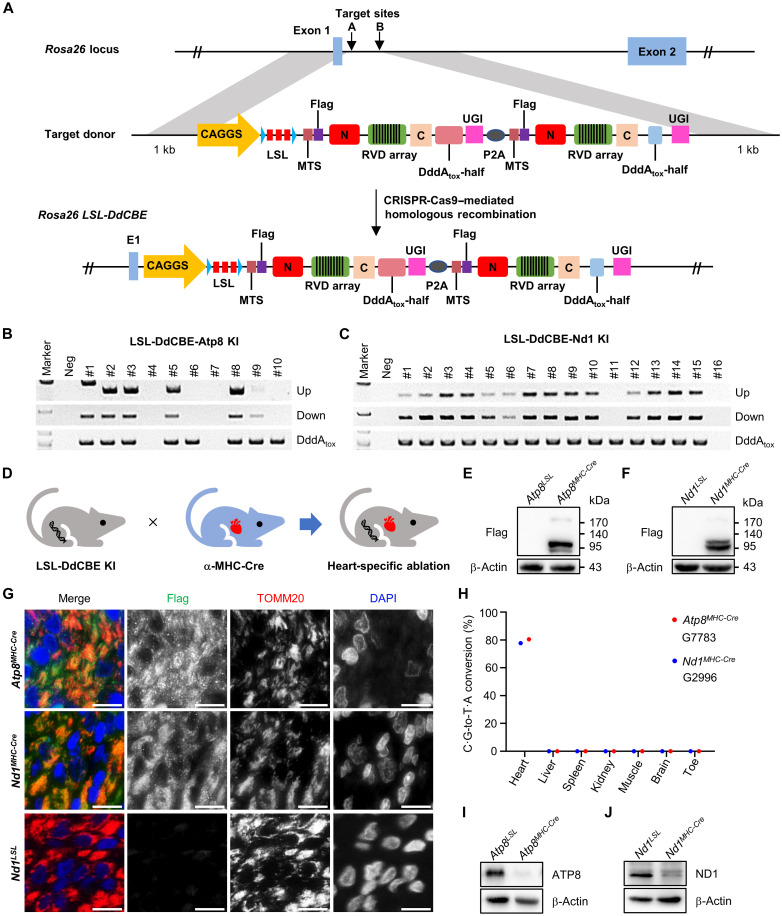
Generation of LSL-DdCBE-Atp8 and LSL-DdCBE-Nd1 knockin rats
Encouraged by the results in C6 cells, we tried to apply this DdCBE-mediated KO strategy to rats. mtProteins are the core subunit of the respiratory chain complex and are vital for mitochondria function maintenance (23–25). Depletion of the mtProteins throughout the body may lead to embryonic lethality. To avoid this, we used the Cre/loxP system to ablate mtProteins in a spatial-temporal manner. We first constructed a conditional expression vector by placing a loxP-stop-loxP cassette (LSL) between the CAG promoter and 2A peptide-linked DdCBE pair (LSL-DdCBE) (fig. S2A) and then cotransfected it with or without Cre expression vector to the C6 cells. The DdCBE pair’s protein could be detected in a Cre-dependent manner (fig. S2, B and C). Consistently, the DdCBE-mediated C·G-to-T·A conversions at Atp8 G7783 and Nd1 G2996 were detected with frequencies of 20 and 30%, respectively, in the presence of Cre recombinase (fig. S2, D and E).
To generate the cKO rats, we cloned the LSL-DdCBE into a vector with rat Rosa26 homology arms to construct the LSL-DdCBE-Atp8 and LSL-DdCBE-Nd1 donors and incorporated them into rat Rosa26 locus (LSL-DdCBE KI) via CRISPR-Cas9–assisted homologous recombination as previously described (Fig. 2A) (26). For LSL-DdCBE-Atp8 targeting, 44 injected eggs were transferred, and 10 pups were born (table S1). Five of 10 (50%) rats harbored the correct recombination at rat Rosa26 locus identified by primer pairs flanking the 3′ and 5′ homology arms (Fig. 2B and table S2). For LSL-DdCBE-Nd1 targeting, a total of 44 injected eggs were transferred to two pseudopregnant female rats, and 16 pups were born (table S1). Fourteen of 16 (88%) rats were detected with the correct recombination (Fig. 2C and table S2). We further confirmed the correct homologous recombination events using Sanger sequencing (fig. S3, A and B). The F0 rats #2 from LSL-DdCBE-Atp8 and #3 from LSL-DdCBE-Nd1 were crossed with wild-type rats to test the germline transmission of LSL-DdCBE knockin (KI). The results showed that these KI elements could be transmitted to the next generation (fig. S3, C and D). In F1 rats, the LSL cassette is intact, and no editing event could be detected at Atp8 G7783 and Nd1 G2996 sites among various tissues, indicating that the LSL cassette entirely blocks the expression of the DdCBE pair without Cre recombinase (fig. S3, E to H).
Fig. 2. DdCBE-mediated ATP8/ND1 cKO in rats.
(A) The incorporation of LSL-DdCBE into rat Rosa26 locus using CRISPR-Cas9–mediated homologous recombination. (B and C) Genotyping of LSL-DdCBE-Atp8 KI (B) and LSL-DdCBE-Nd1 KI (C) F0 rats by PCR with upstream primer pair (Up band), downstream primer pair (Down band), and internal primer pair (DddAtox band). (D) The mtProteins KO in the heart tissue induced by crossing LSL-DdCBE KI rat with α-MHC-Cre rat. (E and F) The DdCBE expression in the heart tissue of Atp8MHC-Cre (E) and Nd1MHC-Cre (F) rat was detected by Western blot using the anti-Flag antibody. (G) The localization of DdCBE in mitochondrial of Atp8MHC-Cre and Nd1MHC-Cre heart tissues. The anti-Flag (green) for DdCBE, anti-TOMM20 for mitochondria (red), and 4′,6-diamidino-2-phenylindole (DAPI) for nucleus (blue). Scale bars, 10 μm. (H) The frequency of DdCBE-mediated C·G-to-T·A conversion at Atp8 G7783 and Nd1 G2996 sites in the heart, liver, spleen, kidney, muscle, brain, and toe of Atp8MHC-Cre and Nd1MHC-Cre rats was analyzed by deep sequencing. (I and J) Protein level of ATP8 (I) and ND1 (J) in tissues of Atp8MHC-Cre and Nd1MHC-Cre was detected by Western blot, respectively.
To test whether this cKO strategy works in vivo, we crossed LSL-DdCBE KI rats with α–major histocompatibility complex (MHC)–Cre rats, a rat strain expressing Cre recombinase in myocardial cells specifically (Fig. 2D and fig. S4, A and B). In double-positive (Atp8MHC-Cre and Nd1MHC-Cre) rat, we detected the Cre-induced LSL deletion specifically occurred in the heart tissue by polymerase chain reaction (PCR) (fig. S4, C and D, and table S2) and Sanger sequencing (fig. S4, E and F). We also detected the expression of the DdCBE cassette in the heart tissues of Atp8MHC-Cre and Nd1MHC-Cre rats by anti-Flag antibody (Fig. 2, E and F). The DdCBE ectopic expression in mitochondria was further confirmed by colocalization with the mitochondrial member protein TOMM20 (Fig. 2G). Sanger sequencing and deep sequencing results showed DdCBE-mediated high mtDNA editing efficiency at Atp8 G7783 and Nd1 G2996 sites specifically in the heart tissue (Fig. 2H and fig. S4, G and H). The editing efficiency of Atp8MHC-Cre and Nd1MHC-Cre rats was up to 80.43 and 77.67%, respectively (Fig. 2H). Consistently, the protein level of ATP8 or ND1 was observed as nearly abolished in the heart tissues of Nd1MHC-Cre or Atp8MHC-Cre rats, respectively (Fig. 2, I and J). Our data suggest that DdCBE-mediated mtDNA C·G-to-T·A conversion can be repurposed to deplete mtProteins in a tissue-specific manner in vivo.
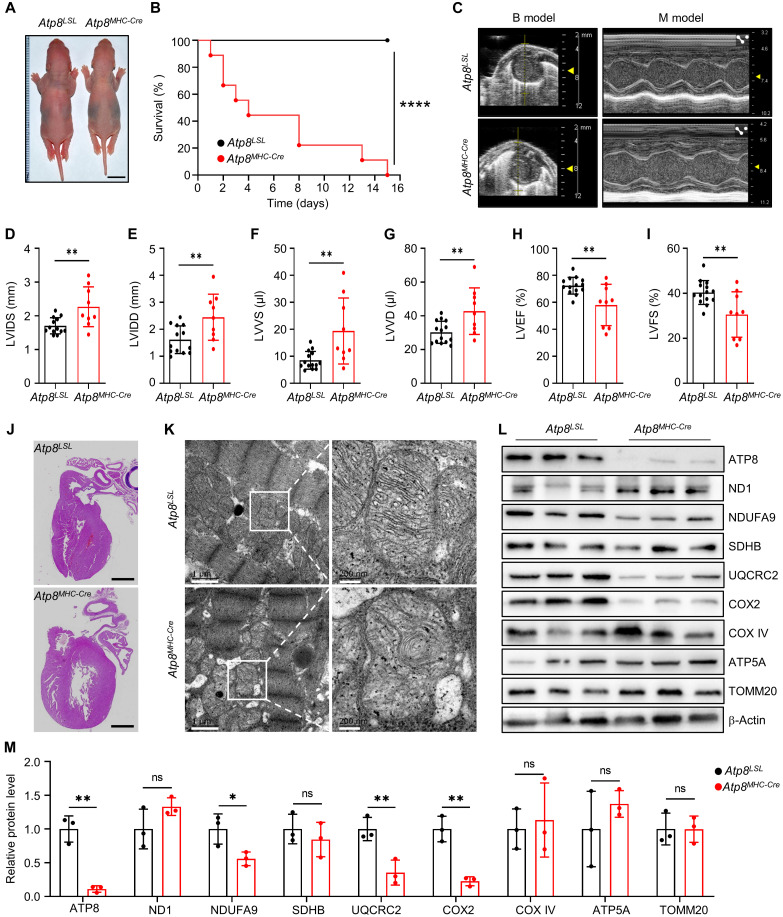
Heart-specific ablation of ATP8/ND1 in rats
Clinically, the patient with a nonsense mutation (p. Trp55X) in mitochondrial ATP8 gene has been reported with a slender figure, apical hypertrophic cardiomyopathy, and neuropathy (27). Hence, we first characterized the phenotype of Atp8MHC-Cre rats. The retardation of growth was observed in Atp8MHC-Cre rats (Fig. 3A). All of them died within 15 days after birth (Fig. 3B). Using echocardiography, we observed that the cardiac structure and function of Atp8MHC-Cre rats exhibited dilated cardiomyopathy phenotypes at 3 days of age, with enlarged chambers and weakened contraction (Fig. 3C). These changes were further verified by increased left ventricular (LV) diameter at the end of systole and diastole (LVIDS and LVIDD) (Fig. 3, D and E), increased LV volume at the end of systole and diastole (LVVS and LVVD) (Fig. 3, F and G), and decreased in LV ejection fraction (LVEF) and fractional shortening (LVFS) (Fig. 3, H and I). These phenotypes reproduce the clinical symptom of the patient. We further performed pathological analyses for the heart of Atp8MHC-Cre rats. Consistently, Atp8MHC-Cre rats histologically showed enlarged LV (Fig. 3J). We next used transmission electron microscopy (TEM) to examine the mitochondrial morphology and ultrastructure. Dilated mitochondria and evident cristae degeneration in the mitochondrial matrix were observed in the heart of Atp8MHC-Cre rats (Fig. 3K). Upon ATP8 depletion, the protein levels of NDUFA9 (complex I subunit), UQCRC2 (complex III subunit), and COX2 (complex IV core subunit) also evidently decreased (Fig. 3, L and M), suggesting that ATP8 ablation impaired the assembly or stability of these respiratory chain complexes.
Fig. 3. Heart-specific depletion of ATP8 causes heart failure in rats.
(A) The whole-body image of 3-day-old Atp8LSL and Atp8MHC-Cre pups. Scale bars, 1 cm. (B) The survival curve of Atp8LSL and Atp8MHC-Cre rats. n = 10 biological replicates for each group. ****P ≤ 0.0001 by log-rank test. (C) The snapshot of echocardiography for Atp8LSL and Atp8MHC-Cre rat. (D to I) The cardiac structure and function parameters of Atp8LSL and Atp8MHC-Cre rats calculated from echocardiography in (C). LVIDS (D), LVIDD (E), LVVS (F), (LVVD) (G), LVEF (H), and LVFS (I). n = 14 for the Atp8LSL group and n = 9 for the Atp8MHC-Cre group. Data were presented as means ± SD. **P ≤ 0.01 by Student’s unpaired two-tailed t test. (J) The H&E image of heart tissue from Atp8LSL and Atp8MHC-Cre rats. Scale bars, 1 mm. (K) The TEM image of mitochondria in heart tissues of Atp8LSL and Atp8MHC-Cre rats. (L) The mitochondrial protein level was detected by Western blot in heart tissues of Atp8LSL and Atp8MHC-Cre rats. (M) Relative protein level of mitochondrial proteins from (L). n = 3 biological replicates for each group. Data were presented as means ± SD. *P ≤ 0.05 and **P ≤ 0.01 by Student’s unpaired two-tailed t test. ns, not significant.
For Nd1MHC-Cre rats, we also observed retardation of growth (fig. S5A). The survival curve data showed that neonatal deaths of Nd1MHC-Cre pups occurred within 2 weeks (fig. S5B). Then, we analyzed the cardiac structure and function of Nd1MHC-Cre rats at 7 days of age. The data showed that Nd1MHC-Cre rats exhibited dilated cardiomyopathy phenotypes with larger chambers and decreased contraction (fig. S5C). These changes were verified by increased LVIDS, LVIDD, LVVS, and LVVD (fig. S5, D to G), as well as decreased LVEF and LVFS (fig. S5, H and I). The structure changes were further verified by hematoxylin and eosin (H&E) staining (fig. S5J). The dilated mitochondria and evident cristae degeneration were also observed in the heart of Nd1MHC-Cre rats (fig. S5K). Along with the nearly abolished ND1 expression, the protein levels of NDUFA9, SDHB, COX2, ATP8, and ATP5A were decreased significantly (fig. S5, L and M). Together, these results suggest that ATP8 or ND1 ablation in the heart results in mitochondria dysfunction and heart failure in rats.
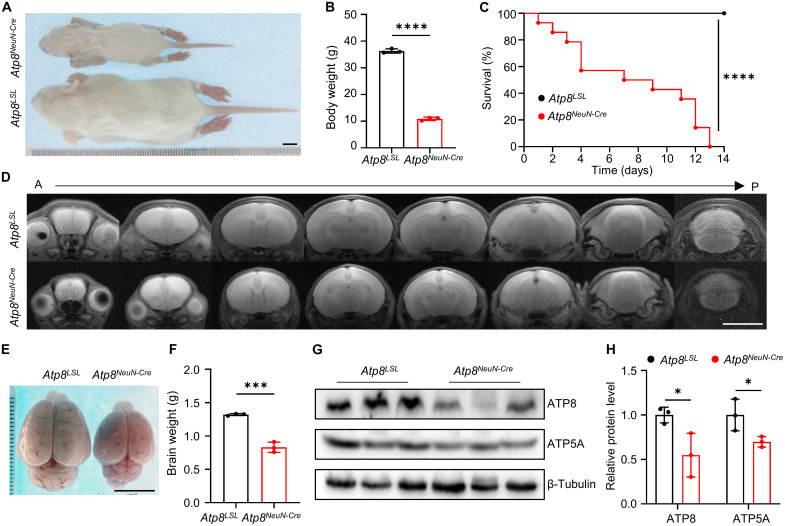
Neuron-specific deletion of ATP8/ND1 in rats
Encouraged by the successful depletion of ATP8/ND1 in rat heart tissues, we next crossed LSL-DdCBE-Atp8 and LSL-DdCBE-Nd1 rats with a neuron-specific Cre strain (NeuN-Cre) to KO Atp8 and Nd1 in a neuron-specific manner, respectively. Ablation of ATP8 in neurons led to a smaller body size (Fig. 4A), decreased body weight (Fig. 4B), and death within 2 weeks (Fig. 4C). Compared with the control group, magnetic resonance imaging (MRI) results of ATP8NeuN-Cre brains showed no notable difference on T2-weighted images (Fig. 4D). Through dissection, we found that the brains of ATP8NeuN-Cre rats were smaller and lighter than that of control rats (Fig. 4, E and F). As expected, the decreased protein level of ATP8 was detected in the brain of ATP8NeuN-Cre rats (Fig. 4, G and H). Similar phenotypes were also displayed in Nd1NeuN-Cre rats, including small body size (fig. S6A), decreased body weight (fig. S6B), and death within 2 weeks (fig. S6C). However, an abnormal signal on T2-weighted images was observed including high signal infiltrates in lateral ventricle and cerebellar atrophy in the brain of Nd1NeuN-Cre rats (fig. S6, D to F). The protein levels of ND1 and NDUFA9 decreased by 50% in the brain of Nd1NeuN-Cre rats (fig. S6, G and H). Together, our data suggest that ND1 or ATP8 ablation in neuron impairs the development of brain.
Fig. 4. Neuron-specific KO ATP8 results in postnatal death in rats.
(A) The whole-body image of Atp8LSL and Atp8NeuN-Cre rat. Scale bar, 1 cm. (B) The body weight of Atp8LSL and Atp8NeuN-Cre rats. n = 3 biological replicates for each group. Data were presented as means ± SD. ****P ≤ 0.0001 by Student’s unpaired two-tailed t test. (C) The survival curve of Atp8LSL and Atp8NeuN-Cre rats. n = 10 biological replicates for each group. ****P ≤ 0.0001 by log-rank test. (D) The brain MRI image of Atp8LSL and Atp8NeuN-Cre rats from anterior (A) to posterior (P). Scale bar, 1 cm. (E) The brain images of Atp8LSL and Atp8NeuN-Cre rats. Scale bar, 1 cm. (F) The brain weight of Atp8LSL and Atp8NeuN-Cre rats. n = 3 biological replicates for each group. Data were presented as means ± SD. ***P ≤ 0.001 by Student’s unpaired two-tailed t test. (G) The protein level of ATP8 and ATP5A in brain tissues of Atp8LSL and Atp8NeuN-Cre rats were detected by Western blot. (H) The relative protein level of ATP8 and ATP5A from (G). n = 3 biological replicates for each group. Data were presented as means ± SD. *P ≤ 0.05 by Student’s unpaired two-tailed t test.
Depletion of all mtProteins in rat C6 cells
Encouraged by the success of DdCBE-mediated ATP8 and ND1 depletion in both cells and rats, we set out to KO all the remaining mtProteins in C6 cells. Besides editing Trp (W) codon TGA (W stop), we also intended to edit Gln (Q) codon CAA to generate TAA stop codon (Q stop) by deaminating the C base (Fig. 5A). On the basis of the above two strategies (W stop and Q stop), we designed the DdCBEs for all mtDNA-coding genes, except Nd4l because no swappable codons suit these strategies (Fig. 5B and fig. S7). The W stop strategy was used in Nd2, Nd4, Nd5, Nd6, Cytb, Cox1, Cox3, and Atp6 (Fig. 5B and fig. S7). The Q stop strategy was used in Nd3 and Cox2 (Fig. 5B and fig. S7). As the same for Nd1 and Atp8 targeting, we tested the editing efficiency of four DdCBE pairs for each targeting site in C6 cells (fig. S8) and cloned the DdCBE pair with best performance into the PB vector with fluorescent tags (fig. S1D). After cotransfection with PB transposase and enrichment following the procedures used for Nd1 and Atp8 targeting, we collected the edited C6 cells for Sanger sequencing (fig. S9A), deep sequencing (fig. S9B), Western blot, and Seahorse assay. The deep sequencing result showed that the editing efficiency was up to 99.29 ± 0.006% for Nd2 (Fig. 5C), 97.65 ± 0.032% for Nd3 (Fig. 5D), 99.24 ± 0.42% for Nd4 (Fig. 5E), 94.32 ± 0.017% for Nd5 (Fig. 5F), 48.07 ± 1.162% for Nd6 (Fig. 5G), 50.48 ± 1.081% for Cytb (Fig. 5H), 97.33 ± 0.084% for Cox1 (Fig. 5I), 100 ± 0% for Cox2 (Fig. 5J), 95.81 ± 0.162% for Cox3 (Fig. 5K), and 63.24 ± 0.394% for Atp6 (Fig. 5L). Furthermore, we detected significantly decreased targeted protein expression in 8 of the 10 mtProteins described above, except ND6 and ATP6, which have no commercial antibodies available (Fig. 5, C to L, and fig. S10, A to J). To characterize the ND6-KO and ATP6-KO cell lines, we detected the protein level of ND5 and ATP8 instead of ND6 and ATP6, respectively. We observed a significantly decreased ND5 expression in ND6-KO cell lines (Fig. 5G and fig. S10D) and significantly decreased ATP8 in ATP6-KO cell lines (Fig. 5L and fig. S10J). The Seahorse results also showed reduced basal and uncoupled respiration rates in the targeted cells (Fig. 5, C to L, and fig. S10, K to N), except in ND5-, ND6-, and ATP6-KO cells, where only maximal respiration decreased significantly. These strategies were further validated by the second target sites for Cox1, Cox2, and Cox3 (fig. S11, A and B). After C6 cell transfection and enrichment, Sanger sequencing (fig. S11, C to E) and Western blot (fig. S11, F to K) were performed for the collected cells. The significantly decreased protein expression of COX1 (fig. S11, F and G), COX2 (fig. S11, H and I), and COX3 (fig. S11, J and K) was observed in the corresponding targeted cells, respectively. Our data indicate that DdCBE-mediated mtDNA editing can be repurposed to KO all rat mtDNA protein-coding genes except Nd4l.
Fig. 5. DdCBE-mediated KO of rat mtProteins in C6 cells.
(A) Strategy of mtDNA-encoded protein depletion by DdCBE-introduced stop codon. The W codon TGA was changed to TAA stop codon (W stop). The Q codon CAA was changed to TAA stop codon (Q stop). (B) Location of introduced stop codon at mtProteins. (C to L) The identification of DdCBE-mediated mtProtein depletion in ND2-KO (C), ND3-KO (D), ND4-KO (E), ND5-KO (F), ND6-KO (G), CYTB-KO (H), COX1-KO (I), COX2-KO (J), COX3-KO (K), and ATP6-KO (L) C6 cells by deep sequencing (left), Western blot (middle), and Seahorse assay (right). For deep sequencing, n = 3 technical replicates. For Seahorse assay, n = 5 technical replicates for Ctrl and n = 6 technical replicates for each KO cell line. Data were presented as means ± SD.
A cKO rat resource for mitochondrial protein-coding genes
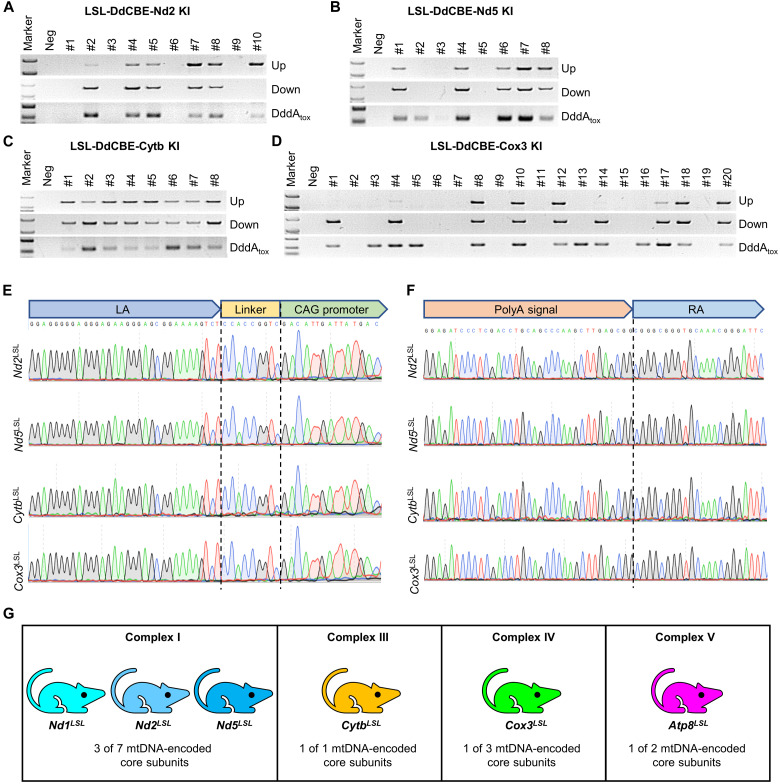
To generate mtProteins KO rat resource, we selected the validated target sites for Nd2, Nd5, Cytb, and Cox3 to produce LSL-DdCBE KI rat models. The CRISPR-Cas9–mediated LSL-DdCBE recombination at the Rosa26 locus was verified by PCR (Fig. 6, A to D) and Sanger sequencing (Fig. 6, E to F). In total, we established six rat lines—including LSL-DdCBE-Nd1, LSL-DdCBE-Nd2, and LSL-DdCBE-Nd5 for complex I; LSL-DdCBE-Cytb for complex III; LSL-DdCBE-Cox3 for complex IV; and LSL-DdCBE-Atp8 for complex V—covering four respiratory chain complexes (Fig. 6G). These rat lines were preserved in the Rat Resource Center of China (www.ratresource.com) (table S3). Our work provides a valuable resource for studying the biological function and disease mechanism of mtProteins in a spatial-temporal manner in vivo.
Fig. 6. A cKO rat resource for mtProteins.
(A to D) Genotyping of LSL-DdCBE-Nd2 KI (A), LSL-DdCBE-Nd5 KI (B), LSL-DdCBE-Cytb KI (C), and LSL-DdCBE-Cox3 KI (D) F0 rats by PCR with upstream primer pair (Up band), downstream primer pair (Down band), and internal primer pair (DddAtox band). (E and F) The recombination sites were confirmed by Sanger sequencing for upstream primer pair (E) and downstream primer pair (F) in LSL-DdCBE-Nd2 KI, LSL-DdCBE-Nd5 KI, LSL-DdCBE-Cytb KI, and LSL-DdCBE-Cox3 KI rats. (G) Six cKO rat strains covering four respiratory chain complexes were generated in this study.
Off-target mutagenesis in mitochondrial genome and nuclear genome
Our previous work observed a relatively low off-target effect in DdCBE-mediated editing in rat mitochondrial genome (14). Recently, it has been reported that DdCBE could result in off-target editing in the nuclear genome in human cell lines (10) and mouse embryos (28). Here, we selected two rats with high on-target editing efficiency (Atp8MHC-Cre #7 and #28) to analyze the off-target effect of DdCBE in mitochondrial and nuclear genomes.
To profile the off-target activity of DdCBEs in the entire mitochondrial genome, we performed whole mtDNA sequencing of the ear and heart tissues from the same rat in parallel. Off-target edits could be detected with editing efficiency of less than 8% in the heart, but not in the ear (fig. S12A). Twenty-nine and 42 off-target sites (OTS) with conversion rates of more than 1% were detected in the two rats, and 28 OTS are the common sites (fig. S12B). We did not note a strong tC motif editing preference for DdCBE-Atp8 (fig. S12C). Compared with the transient expression of DdCBE in early embryos reported by our group previously (14), the sustained expression of DdCBE mediated by Cre recombinase seemed to induce a slightly higher off-target editing, although the mutation loads of these OTS were not high enough to display phenotypes.
To assess the nuclear off-target effects of DdCBE, we performed whole-genome sequencing of the ear and heart from Atp8MHC-Cre #7 rat, which harbored a higher on-target editing than Atp8MHC-Cre #28. To reduce false positives, we first used LoFreq, Mutect2, and Strelka to call single-nucleotide variants (SNVs) and considered the overlap SNVs of the three algorithms as the actual variants. Eleven and 0 SNVs were detected in the hearts and ears of Atp8MHC-Cre rats, respectively (fig. S12D). To verify the SNVs identified in heart samples, we performed Sanger sequencing for SNVs with C·G-to-A·T conversions and found that the two putative OTS could be false positives because of the repeat sequences around them (fig. S12E). Among the three algorithms, LoFreq yielded the minimum number of SNVs, resulting in only a dozen SNVs being detected after overlap. To obtain more SNVs for further analysis of the nuclear off-target effects of DdCBE, we only used Mutect2 and Strelka to call SNVs. A total of 198 and 86 SNVs were identified in the heart and ear, respectively, without enrichment for C·G-to-A·T conversions (fig. S12F), suggesting that no obvious nuclear off-target event was detected in Atp8MHC-Cre rat.
DISCUSSION
Gene KO is an essential tool for understanding complex disease progress, which allows us to dissect pathological processes and gene functions involved (29–31). Gene KO in animals enables us to study gene functions in an entirety. Our understanding of mitochondrial gene functions is limited by the shortage of related gene KO animals (5). A substantial obstacle to develop gene KO animal lies in the fact that mitochondrial genome is genetically not accessible (32). CRISPR-Cas has been proven as a powerful tool for nuclear gene editing but not for the mitochondrial genome, mainly because single guide RNAs cannot be efficiently delivered to mitochondria (33, 34). Protein-guided DNA target endonuclease, such as ZFNs and TALENs, could be used to target mutant mtDNA and result in degeneration of them rather than build disease animal models (35, 36). A chimera, composed of a mitochondrial precursor protein chemically linked to a morpholino, was used to target specific mitochondrial mRNA and result in mtProtein silence. This approach only induces mtProtein silence in purified mitochondria but is unsuitable for mammal cells and animals (37). In another report, the small interfering RNAs were used to silence targeted mitochondrial transcripts but only had a recordable effect on relatively unstable proteins such as mtProteins of the complex IV subunit (38). Recently, an engineered cytidine deaminase named DdCBE has been proven to induce mtDNA C·G-to-T·A conversion in human cells and various animals (10–14).
Rat has many advantages over mouse, which thereby serves an optimal disease model especially in neurological and cardiovascular studies (18, 19). Meanwhile, the brain and heart are energy-intensive organs that are sensitive to mutations in mtDNA. Therefore, rat models with mitochondrial mutations are well suited to study human mitochondrial diseases. In our study, (i) DdCBE was repurposed to knock out mitochondrial protein-coding genes via incorporating a premature stop codon in rats. We obtained nearly homoplasmic KO most of the mitochondrial protein-coding genes in rat C6 cells. This strategy also works in the mouse cell line in a latest report (39), (ii) using the Cre/loxP system to generate six cKO rat strains to ablate mtProteins in a precisely controlled spatial-temporal manner. Our functional studies with conditional depletion of ATP8 and ND1 in rat heart or neuron caused heart failure or abnormal brain development. This strategy allowed us to obtain a batch of mtProtein KO rats in expected tissue or cell types; (iii) a cKO rat resource containing six rat strains was generated and deposited in the Rat Resource Center of China (www.ratresource.com) for researchers interested in mitochondrial diseases. Our established strategy and rat models allow temporal, spatial, and cell type–specific KO mtProteins to study their functional role in an organ-specific or cell type–specific manner under physiological and pathological conditions.
Mitochondria dysfunction has been repeatedly reported in heart diseases and neurological disorders (40–45), but the underlying mechanisms are still not fully understood (43, 46–50). In this study, our established heart-specific KO Atp8 (Atp8MHC-Cre) and Nd1 (Nd1MHC-Cre) rats displayed growth retardation, short life span, dilated cardiomyopathy, and impaired mitochondria structure. Our work suggests that ATP8 or ND1 is essential in cardiac function maintenance. Although a similar phenotype was observed, the protein levels of mitochondrial proteins were different. For example, we detected decreased NDUFA9, UQCRC2, and COX2 expression in Atp8MHC-Cre rats (Fig. 3, L and M), while the significantly declined NDUFA9, COX2, and ATP8 along with nearly abolished ND1 expression were observed in the heart of Nd1MHC-Cre rats (fig. S5, L and M). These results indicated the distinct mechanisms involved, which need further investigation. A similar phenotype was also observed in ATP8 and ND1 neuron-specific KO rats.
DdCBE causes off-target effects in cells (10) and animals (28). Here, we studied the off-target effects in Cre-mediated DdCBE targeting rats. Our results indicated that the Cre-mediated DdCBE-sustained expression induced more off-target edits in the whole mitochondrial genome compared with the transient expression of DdCBE in early embryos. Using whole nuclear genome sequencing, we found that the identified SNVs exhibited no enrichment for any base conversion either in the heart or in ear tissues, indicating that no obvious nuclear off-target edits were detected. Although we performed whole-genome sequencing with more than 30× coverage per sample, we could not exclude that the low frequency of nuclear off-target editing occurred, which may be omitted because of population averaging.
Moreover, the Cre/loxP system permits DdCBE expression in tissue- or cell type–specific manner. However, the limitations of this system also need to be considered, such as the unexpected transient expression of Cre recombinase in animals (51–53). The expression of DdCBE will not be closed once activated; the continuous expression of DdCBE would result in more severe disease phenotype and off-target events. In addition, the shortage of diverse rat strains of Cre also limits its wide application. More precise and sensitive methods should be developed to analyze the DdCBE-mediated off-target in mitochondrial and nuclear genomes. Furthermore, new mtDNA editing tools with broadened targeting scope, improved editing efficiency, and high specificity need to be developed.
In summary, we manipulated the mtProteins’ expression by introducing a premature stop codon into the open reading frame of rat mtDNA-coding genes using DdCBE-mediated C·G-to-T·A conversion. In vitro, except ND4L, we knocked out all rat mtProteins in C6 cells and detected depleted mtProtein expression and impaired mitochondrial OXPHOS. In vivo, six cKO rat lines covering four complexes of the OXPHOS were generated, which allows KO of these mtProteins in a tissue- or cell type–specific manner. Furthermore, we detected impaired cardiac function and brain development by KO of Nd1 and Atp8 in rat heart and neuron. Thus, our work provides strategies and a valuable resource for studying the biological function and disease mechanisms of mtProteins and for developing potential treatments for mitochondrial disorders.
MATERIALS AND METHODS
Plasmids construction
DdCBE vectors were assembled by the Golden Gate strategy using a repeat variable di-residues (RVD) library as described before (11, 14). The complete set of plasmids for assembling DdCBE can be obtained from Addgene (Shen Lab DdCBE Kit #1000000212). The assembled plasmids were identified by PCR using RVD seq Fwd/RVD seq Rev primers in table S2 and further confirmed by Sanger sequencing. All sequence information of DdCBE vectors is listed in sequence S1. Left DdCBE and Right DdCBE of each DdCBE pair were digested with Not I-HF [R3189L, New England Biolabs (NEB)] and Pme I (R0560L, NEB) and cloned into PB-CAG-EGFP-P2A-MTS and PB-CAG-mCherry-P2A-MTS backbone, respectively, to construct PB expression plasmids. DdCBE pair linked by P2A was cloned into the Rosa26-HR-CAG-LSL plasmid backbone (26) by Afl II and Nhe I sites to construct Rosa26-HR-CAG-LSL-DdCBE donor plasmids. All constructed plasmids were verified by Sanger sequencing. The DdCBE pairs for KO of mitochondrial protein-coding genes have been deposited to Addgene (ID, 198838-198861).
Cell culture, nucleofection, and sorting
C6 cells (American Type Culture Collection, CCL-107) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (A1896701, Gibco) supplemented with 10% fetal bovine serum (FBS) (10099141C, Gibco) at 37°C with 5% CO2 and were detected without mycoplasma contamination by PCR test. C6 ρ0 cells were generated using EB and ddC treatment as previously described (20). Briefly, wild-type C6 cells were exposed to 10 μM EB (E8751, Sigma-Aldrich) and 16 μM ddC (D5782, Sigma-Aldrich) in DMEM supplemented with uridine (50 μg/ml; U3003, Sigma) and pyruvate (0.1 mg/ml; 11360070, Gibco), with a daily change of the culture medium. Cells were cultured in the above medium at 37°C, 5% CO2, and 90% humidity for 21 consecutive days. The mitochondria clearance efficiency was detected by PCR of mtDNA.
For DdCBE pair screening, C6 cells were cotransfected with 400 ng of left DdCBE and 400 ng of right DdCBE using the SF Cell Line 4D-Nucleofector X Kit (V4XC-2024, Lonza) according to the manufacturer’s protocol. The nucleofected cells were seeded on the 12-well plate and supplemented with puromycin (0.6 μg/ml) after 24 hours after nucleofection. Cells were collected and genome-extracted for Sanger sequencing and deep sequencing.
For fluorescence-activated cell sorting (FACS), 1 × 106 C6 cells were cotransfected with 1 μg of PB-EGFP-DdCBE, 1 μg of PB-mCherry-DdCBE, and 0.5 μg of helper PB transposase (PBase) plasmids using the SF Cell Line 4D-Nucleofector X Kit (V4XC-2024, Lonza). The nucleofected cells were seeded on the 6-cm plate and supplemented with uridine (5 μg/ml; U3003, Sigma-Aldrich) and 3.3 mM sodium pyruvate (11360070, Gibco). EGFP and mCherry double-positive cells were sorted using a FACSAria Fusion sorter (BD Bioscience) with sterile operation at day 3 and day 7.
For FNZ treatment, the C6 cells were cultured in DMEM supplied with 10% FBS, uridine (15 mg/ml), 3.3 mM sodium pyruvate, and 15 mM FNZ (S2030, Selleck). After 7 days, the treated cells were collected and used for Western blotting.
OCR analysis
The OCR of control and DdCBE-targeted C6 cells was measured using the Seahorse XFe96 Extracellular Flux Analyzer (Agilent). Briefly, 2 × 104 cells were seeded on Seahorse XF96 cell culture plates (101085-004, Agilent) for 24 hours. The Mito stress protocol was applied with the use of 1 μM oligomycin, 1.5 μM FCCP (carbonyl cyanide p-trifluoromethoxyphenylhydrazone), and 0.5 μM rotenone + 0.5 μM antimycin A, which were optimized for C6 cells. Data normalization was achieved by cell counting using a CyQUANT cell proliferation assay kit (Invitrogen, C7027) according to the manufacturer’s protocol.
Animals
All Sprague-Dawley rats used in this project were purchased from Beijing Vital River Laboratories Animal Technology Co. Ltd. and bred in a specific pathogen-free facility. All animal procedure and experiment were approved by the Institutional Animal Care and Use Committee (IACUC) of the Institute of Laboratory Animal Science, Chinese Academy of Medical Sciences and Peking Union Medical College (IACUC-MYW21006). All established rats in this study were preserved to the Rat Resource Center of China (www.ratresource.com).
Rat zygotes collection and manipulation
The rat zygotes were collected and manipulated for injection as described previously (54). Briefly, the 4- to 6-week-old female rats were given 40 IU of pregnant mare serum gonadotropin (G4877, Sigma-Aldrich) and 40 IU of human chorionic gonadotropin (9002-61-3, Sigma-Aldrich) by intraperitoneal injection with a time interval of 72 hours. Then, the injected female rats were mated with male rats immediately. Then, we collected eggs from superovulated female rats and cultured them in KSOM (MR-121-D, Millipore) at 37°C and 5% CO2 for microinjection. The editing mixture was injected into the cytoplasm and nucleus of rat zygotes using a microinjection system (Nikon) following the instruction. The injected zygotes were transferred to pseudopregnant Sprague-Dawley rats for live birth.
DNA extraction and genotyping
The nuclear genomic DNA and mitochondrial genomic DNA of C6 cells were extracted using QuickExtract DNA Extraction Solution (Lucigen). The rat nuclear genomic DNA and mitochondrial genomic DNA from collected tissues were extracted using the EasyPure Genomic DNA Kit (TransGen). The target regions were amplified using PCR and further confirm by Sanger sequencing and deep sequencing. Primers used in this study are listed in table S2.
Echocardiography analysis
The rat heart structure and function were evaluated by echocardiography analysis as described previously (14). Briefly, the Nd1MHC-CRE and Atp8MHC-CRE rats were anesthetized with 2% isoflurane (RWD) at 0.6 liter/min of O2 flow. The echocardiographic observation was performed using a Vevo3100 ultrasound machine with an MX400 probe (30 MHz) (VisualSonics, Fujifilm). Measurements of at least three continuous cardiac cycles were recorded.
Magnetic resonance imaging
In vivo animal MRI was performed using a 7.0-T small-animal MRI system (Varian, Palo Alto, CA, USA). During imaging, the rats were anesthetized with medical oxygen (0.6 liter/min) containing 2% isoflurane. T2-weighted MRI images in coronal plane were acquired with a fast spin echo (fsems) sequence using the following parameters: FOV (field of view) = 35 by 35 mm; no slices = 20; slice thickness = 1 mm (zero slice gap); TR (repetition time) = 3500 ms; TE (echo time) = 18.0 ms; image matrix = 256 by 256; number of averages = 10.
Transmission electron microscopy
The mitochondrial structure of the cardiac muscle was observed by TEM as previously described (55). The fresh heart tissues were collected and fixed in 2.5% glutaraldehyde (P1126, Solarbio) after perfusion with ice-cold PBS. Then, the fixed tissues were steeped in 1% osmium tetroxide (05500, Sigma-Aldrich) for 2 hours. Following dehydration with a serial alcohol gradient, samples were embedded in Epon 812 resin (TAAB), sectioned into approximately 90-nm slices, and then stained with uranyl acetate and lead citrate. The slices were observed and imaged using TEM (JEM-1400, JEOL, Japan).
Immunofluorescent staining
The immunofluorescent staining was performed using heart paraffin sections. The primary antibodies anti-Flag (1:100; 66008-4-lg, Proteintech) and anti-TOMM20 (1:200; 11802-1-AP, Proteintech) were incubated overnight at 4°C following deparaffinizing, rehydrating, permeabilizing, and blocking. The fluorescent secondary antibodies anti-mouse immunoglobulin G (IgG) 488 (1:200; A-11001, Invitrogen) and anti-rabbit IgG 555 (1:200; A-21428, Invitrogen) were used to detect the primary antibodies. The slides were sealed with 4′,6-diamidino-2-phenylindole sealing tablets and photographed using PANNORAMIC 250 Flash III DX (3DHISTECH Ltd., Hungary).
H&E staining
Heart tissues collected from the euthanized rats were fixed in 10% formalin and embedded in paraffin followed by 3- to 4-μm section preparation. After dewaxing and rehydrating, the tissue sections were stained with H&E according to methods described previously (56). H&E-stained slides were scanned with ANNORAMIC 250 Flash III DX and imaged using CaseViewer 3.0 software (3DHISTECH Ltd., Hungary).
Western blot
Total proteins of cells or tissues were extracted using radioimmunoprecipitation assay lysis buffer supplemented with protease inhibitor cocktail (P1006, Beyotime). Proteins were separated by 10 or 12% SDS–polyacrylamide gel electrophoresis following standard procedures after quantification with a bicinchoninic acid (BCA) protein assay kit (P0012S, Beyotime, China). The following primary antibodies were used: anti-ND1 (1:1000; 19703-1-AP, Proteintech), anti-ND2 (1:1000; 19704-1-AP, Proteintech), anti-ND3 (1:500; NBP2-93832, Novus), anti-ND4 (1:1000; NBP2-93530, Novus), anti-ND5 (1:2000; ab233100, Abcam), anti-NDUFA9 (1:1000; ab14713, Abcam), anti-SDHB (1:200; ab14714, Abcam), anti-CYTB (1:2000; 55090-1-AP, Proteintech), anti-UQCRC2 (1:1000; ab14745, Abcam), anti-COX1 (1:1000; A17889, ABclonal), anti-COX2 (1:1000; 55070-1-AP, Proteintech), anti-COX3 (1:1000; A9939, ABclonal), anti-COX IV (1:1000; 11242-1-AP, Proteintech), anti-ATP8 (1:1000; A17890, ABclonal), anti-ATP5A (1:1000; ab176569, Abcam), anti-Flag (1:1000; 66008-4-lg, Proteintech), anti-TOMM20 (1:2000; 11802-1-AP, Proteintech), anti–glyceraldehyde phosphate dehydrogenase (1:5000; HRP-60004, Proteintech), anti–β-tubulin (1:5000; 10094-1-AP, Proteintech), and anti–β-actin (1:5000; HRP-60008, Proteintech). Horseradish peroxidase–conjugated secondary antibodies (ZB-2301, ZB-2305, ZSGB-BIO) were diluted in 1:5000. Protein signals were detected using an enhanced ECL Western blot substrate (P0018S, Beyotime) and visualized by a GelDoc XR+ system (Bio-Rad).
Deep sequencing and analysis
The target region was first amplified with barcoded primers (first-round PCR, PCR1) using the Phanta Max Super-Fidelity DNA Polymerase (P505, Vazyme). Then, the PCR1 products were pooled with equal amount and purified by gel extraction or exonuclease I digestion to remove traces of barcoded primers, followed by the second-round PCR (PCR2) using index primers (N321/N322, Vazyme). Libraries were purified using DNA Clean Beads (N411, Vazyme) for sequencing using Illumina NovaSeq platform. Barcoded primers used for PCR1 are listed in table S2. For data analysis, the trimmed reads were aligned to the mitochondrial genome reference sequence of Rattus norvegicus (NC_001665) using bowtie2 with default parameters. The alignment results were converted to bam format by Samtools. Samtools mpileup was used to detect C-to-T or G-to-A conversion. Sites with conversion rate ≥ 1% in any untreated sample were identified as SNP and excluded for further analysis.
Whole mtDNA sequencing and data analysis
Two overlapping fragments around 8 kb each were amplified by long-range PCR and purified by gel extraction. The two fragments were pooled with equal amount and subjected to library preparation using the TruePrep DNA Library Prep Kit V2 for Illumina (TD501, Vazyme) or KAPA HyperPlus Kit (KR1145) according to the manufacturer’s instruction. The libraries were purified using DNA Clean Beads by 0.5×/0.15× double size selection. Libraries were pooled and sequenced using the Illumina NovaSeq platform. Primers used long-range PCR are listed table S2. For data analysis, the trimmed reads were aligned to the mitochondrial genome reference sequence of R. norvegicus (NC_001665) using bowtie2 with default parameters. The alignment results were converted to bam format by Samtools. Samtools mpileup was used to detect C-to-T or G-to-A conversion.
The following sites were excluded before analysis: (i) SNP sites of R. norvegicus obtained from variation VCF in Ensemble database, (ii) the sites of which C·G-to-T·A variation over 1% in any untreated sample, (iii) the evident SNP sites of which C·G-to-T·A variation over 90% in any sample, and (iv) sites within the DdCBE spacing region. The average off-target editing frequency was then calculated independently for each biological replicate of each treatment condition as the following: The sum events of C·G-to-T·A conversion were divided by the total coverage of these sites.
Whole-genome sequencing and data analysis
Genomic DNA extracted from the heart and ear was subjected to library preparation using the KAPA HyperPlus Kit (KR1145) according to the manufacturer’s instruction. Whole-genome sequencing was performed at a mean coverage of 30× using the Illumina NovaSeq platform. The data analysis was performed as described in the previous study (28) with minor modifications. Briefly, the trimmed reads were mapped to the rat reference genome (mRatBN7.2) by BWA (v0.7.12). Picard Tools (v2.3.0) was used to reorder, sort, and add read groups and mark duplicates of the aligned BAM files. Then, Strelka (v2.9.10), LoFreq (v2.1.2), and Mutect2 (v3.8.1) were used to identify the genome-wide de novo variants with high confidence. The variants were identified in the mapped BAM file of heart with ear from the same rat as control. In addition, we called variants in ear with heart from the same rat as control. LoFreq is an ultrasensitive variant caller to call somatic variants (57). Mutect2 uses a Bayesian classifier to detect somatic mutations with even low allele fractions (58). Strelka uses a mixture model–based estimation with high speed to detect SNVs (59). Different algorithms apply different models and have their own preferences to variants. Hence, to reduce false positives, only SNVs identified by all the three algorithms or two (Strelka and Mutect2) were used for the further analysis. Variants were removed if they overlapped with (i) repeat regions and microsatellite sequences downloaded from the UCSC Genome Browser (http://genome.ucsc.edu/) and (ii) genetic variant sites reported in the dbSNP151 database (www.ncbi.nlm.nih.gov/snp/).
Statistical analyses
All experimental data were analyzed using Microsoft Excel (2019) and GraphPad Prism (8.3.0) software (GraphPad Software LLC). The statistical details for each experiment were provided in the figure legends.
Acknowledgments
We would like to thank Y. Wang of the Institute of Cardiovascular Sciences and Key Laboratory of Molecular Cardiovascular Sciences, Peking University and J. Gong of the Institute of Laboratory Animal Science, Chinese of Academy of Medical Science for critical comments and suggestions.
Funding: This work was supported by National Key R&D Program of China 2021YFC2700600 (to B.S.) and 2022YFF0710702 (to L.Z.), Funds for Creative Research Groups of China 82221005 (to B.S.), National Natural Science Foundation of China 31970796 (to B.S.), CAMS Innovation Fund for Medical Sciences of China CIFMS 2021-I2M-1-024 (to Y.M.) and 2021-I2M-1-034 (to F.G.), Beijing Municipal Natural Science Foundation Grant M21004 (to Y.M.), and 111 Project of Ministry of Education B20095 (to Y.M.).
Author contributions: Conceptualization: Y.M. and B.S. Methodology: Y.M. and B.S. Investigation: L.T., X.Q., W.K., J.J., D.L., X.Z., Y.W., S.W., W.D., X.S., W.C., J.W., K.L., Y.X., L.G., F.G., K.G., C.L., C.W., Z.H., L.Z., and X.G. Supervision: Y.M. and B.S. Writing—original draft: Y.M., B.S., X.Q., and L.T. Writing—review and editing: Y.M., B.S., X.Q., and L.T. Funding acquisition: Y.M., B.S., L.Z., and F.G.
Competing interests: Y.M. is an inventor on patent application (202211313920.X, China) submitted by the Institute of Laboratory Animal Science, Chinese Academy of Medical Sciences that covers generation of cKO mitochondrial protein-coding genes in rats. The other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The high-throughput sequencing data generated in this study have been deposited to the National Center of Biotechnology Information (NCBI) Sequence Read Archive database (accession code, PRJNA889450). Materials generated in this study can be requested from the corresponding authors [mayuanwu@cnilas.org (Y.M.) or binshen@njmu.edu.cn (B.S.)]. The plasmids of DdCBE pairs for KO of mitochondrial protein-coding genes have been deposited in Addgene (ID, 198838-198861). All established rat models were preserved in the Rat Resource Center of China (www.ratresource.com).
Supplementary Materials
This PDF file includes:
Figs. S1 to S12
Tables S1 to S3
Sequences S1 and S2
REFERENCES AND NOTES
- 1.C. Bruser, J. Keller-Findeisen, S. Jakobs, The TFAM-to-mtDNA ratio defines inner-cellular nucleoid populations with distinct activity levels. Cell Rep. 37, 110000 (2021). [DOI] [PubMed] [Google Scholar]
- 2.C. Kukat, C. A. Wurm, H. Spåhr, M. Falkenberg, N. G. Larsson, S. Jakobs, Super-resolution microscopy reveals that mammalian mitochondrial nucleoids have a uniform size and frequently contain a single copy of mtDNA. Proc. Natl. Acad. Sci. U.S.A. 108, 13534–13539 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.D. J. Pagliarini, S. E. Calvo, B. Chang, S. A. Sheth, S. B. Vafai, S. E. Ong, G. A. Walford, C. Sugiana, A. Boneh, W. K. Chen, D. E. Hill, M. Vidal, J. G. Evans, D. R. Thorburn, S. A. Carr, V. K. Mootha, A mitochondrial protein compendium elucidates complex I disease biology. Cell 134, 112–123 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.N. J. Lake, A. G. Compton, S. Rahman, D. R. Thorburn, Leigh syndrome: One disorder, more than 75 monogenic causes. Ann. Neurol. 79, 190–203 (2016). [DOI] [PubMed] [Google Scholar]
- 5.J. D. Barrera-Paez, C. T. Moraes, Mitochondrial genome engineering coming-of-age. Trends Genet. 38, 869–880 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.N. A. Khan, M. Auranen, I. Paetau, E. Pirinen, L. Euro, S. Forsström, L. Pasila, V. Velagapudi, C. J. Carroll, J. Auwerx, A. Suomalainen, Effective treatment of mitochondrial myopathy by nicotinamide riboside, a vitamin B3. EMBO Mol. Med. 6, 721–731 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.S. C. Johnson, M. E. Yanos, E. B. Kayser, A. Quintana, M. Sangesland, A. Castanza, L. Uhde, J. Hui, V. Z. Wall, A. Gagnidze, K. Oh, B. M. Wasko, F. J. Ramos, R. D. Palmiter, P. S. Rabinovitch, P. G. Morgan, M. M. Sedensky, M. Kaeberlein, mTOR inhibition alleviates mitochondrial disease in a mouse model of Leigh syndrome. Science 342, 1524–1528 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Y. Yamada, Satrialdi, M. Hibino, D. Sasaki, J. Abe, H. Harashima, Power of mitochondrial drug delivery systems to produce innovative nanomedicines. Adv. Drug Deliv. Rev. 154-155, 187–209 (2020). [DOI] [PubMed] [Google Scholar]
- 9.P. A. Gammage, C. T. Moraes, M. Minczuk, Mitochondrial genome engineering: The revolution may not be CRISPR-Ized. Trends Genet. 34, 101–110 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Z. Lei, H. Meng, L. Liu, H. Zhao, X. Rao, Y. Yan, H. Wu, M. Liu, A. He, C. Yi, Mitochondrial base editor induces substantial nuclear off-target mutations. Nature 606, 804–811 (2022). [DOI] [PubMed] [Google Scholar]
- 11.J. Guo, X. Zhang, X. Chen, H. Sun, Y. Dai, J. Wang, X. Qian, L. Tan, X. Lou, B. Shen, Precision modeling of mitochondrial diseases in zebrafish via DdCBE-mediated mtDNA base editing. Cell Discov. 7, 78 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.J. Guo, X. Chen, Z. Liu, H. Sun, Y. Zhou, Y. Dai, Y.'. Ma, L. He, X. Qian, J. Wang, J. Zhang, Y. Zhu, J. Zhang, B. Shen, F. Zhou, DdCBE mediates efficient and inheritable modifications in mouse mitochondrial genome. Mol. Ther. Nucleic Acids 27, 73–80 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.P. Silva-Pinheiro, P. A. Nash, L. van Haute, C. D. Mutti, K. Turner, M. Minczuk, In vivo mitochondrial base editing via adeno-associated viral delivery to mouse post-mitotic tissue. Nat. Commun. 13, 750 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.X. Qi, X. Chen, J. Guo, X. Zhang, H. Sun, J. Wang, X. Qian, B. Li, L. Tan, L. Yu, W. Chen, L. Zhang, Y. Ma, B. Shen, Precision modeling of mitochondrial disease in rats via DdCBE-mediated mtDNA editing. Cell Discov. 7, 95 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.S. I. Cho, S. Lee, Y. G. Mok, K. Lim, J. Lee, J. M. Lee, E. Chung, J. S. Kim, Targeted A-to-G base editing in human mitochondrial DNA with programmable deaminases. Cell 185, 1764–1776.e12 (2022). [DOI] [PubMed] [Google Scholar]
- 16.B. Bornstein, J. A. Mas, C. Patrono, M. A. Fernández-Moreno, E. González-Vioque, Y. Campos, R. Carrozzo, M. A. Martín, P. del Hoyo, F. M. Santorelli, J. Arenas, R. Garesse, Comparative analysis of the pathogenic mechanisms associated with the G8363A and A8296G mutations in the mitochondrial tRNALys gene. Biochem. J. 387, 773–778 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.J. V. Leonard, A. H. Schapira, Mitochondrial respiratory chain disorders I: Mitochondrial DNA defects. Lancet 355, 299–304 (2000). [DOI] [PubMed] [Google Scholar]
- 18.J. R. Homberg, M. Wöhr, N. Alenina, Comeback of the rat in biomedical research. ACS Chem. Nerosci. 8, 900–903 (2017). [DOI] [PubMed] [Google Scholar]
- 19.P. M. Iannaccone, H. J. Jacob, Rats! Dis. Model. Mech. 2, 206–210 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Y. Jin, G. Luan, J. Li, H. Wang, Z. Wang, B. Bai, Effect of mtDNA depletion from C6 glioma cells and characteristics of the generated C6ρ0 cells. Mol. Med. Rep. 23, 265 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.D. A. Stroud, E. E. Surgenor, L. E. Formosa, B. Reljic, A. E. Frazier, M. G. Dibley, L. D. Osellame, T. Stait, T. H. Beilharz, D. R. Thorburn, A. Salim, M. T. Ryan, Accessory subunits are integral for assembly and function of human mitochondrial complex I. Nature 538, 123–126 (2016). [DOI] [PubMed] [Google Scholar]
- 22.G. Yang, T. Zhao, S. Lu, J. Weng, X. Zeng, T1121G point mutation in the mitochondrial gene COX1 suppresses a null mutation in ATP23 required for the assembly of yeast mitochondrial ATP synthase. Int. J. Mol. Sci. 23, 2327 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.J. L. Pohjoismaki, S. Goffart, The role of mitochondria in cardiac development and protection. Free Radic. Biol. Med. 106, 345–354 (2017). [DOI] [PubMed] [Google Scholar]
- 24.M. R. Duchen, Mitochondria in health and disease: Perspectives on a new mitochondrial biology. Mol. Aspects Med. 25, 365–451 (2004). [DOI] [PubMed] [Google Scholar]
- 25.M. R. Chiaratti, B. M. Garcia, K. F. Carvalho, T. S. Machado, F. K. da Silva Ribeiro, C. H. Macabelli, The role of mitochondria in the female germline: Implications to fertility and inheritance of mitochondrial diseases. Cell Biol. Int. 42, 711–724 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Y. Ma, L. Yu, S. Pan, S. Gao, W. Chen, X. Zhang, W. Dong, J. Li, R. Zhou, L. Huang, Y. Han, L. Bai, L. Zhang, L. Zhang, CRISPR/Cas9-mediated targeting of the Rosa26 locus produces Cre reporter rat strains for monitoring Cre-loxP-mediated lineage tracing. FEBS J. 284, 3262–3277 (2017). [DOI] [PubMed] [Google Scholar]
- 27.A. I. Jonckheere, M. Hogeveen, L. G. Nijtmans, M. van den Brand, A. J. Janssen, J. H. Diepstra, F. van den Brandt, L. van den Heuvel, F. A. Hol, T. G. Hofste, L. Kapusta, U. Dillmann, M. G. Shamdeen, J. A. Smeitink, R. J. Rodenburg, A novel mitochondrial ATP8 gene mutation in a patient with apical hypertrophic cardiomyopathy and neuropathy. J. Med. Genet. 45, 129–133 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Y. Wei, Z. Li, K. Xu, H. Feng, L. Xie, D. Li, Z. Zuo, M. Zhang, C. Xu, H. Yang, E. Zuo, Mitochondrial base editor DdCBE causes substantial DNA off-target editing in nuclear genome of embryos. Cell Discov. 8, 27 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.P. Silva-Pinheiro, M. Minczuk, The potential of mitochondrial genome engineering. Nat. Rev. Genet. 23, 199–214 (2022). [DOI] [PubMed] [Google Scholar]
- 30.S. Rath, R. Sharma, R. Gupta, T. Ast, C. Chan, T. J. Durham, R. P. Goodman, Z. Grabarek, M. E. Haas, W. H. W. Hung, P. R. Joshi, A. A. Jourdain, S. H. Kim, A. V. Kotrys, S. S. Lam, J. G. McCoy, J. D. Meisel, M. Miranda, A. Panda, A. Patgiri, R. Rogers, S. Sadre, H. Shah, O. S. Skinner, T. L. To, M. A. Walker, H. Wang, P. S. Ward, J. Wengrod, C. C. Yuan, S. E. Calvo, V. K. Mootha, MitoCarta3.0: An updated mitochondrial proteome now with sub-organelle localization and pathway annotations. Nucleic Acids Res. 49, D1541–D1547 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.N. Pfanner, B. Warscheid, N. Wiedemann, Mitochondrial proteins: From biogenesis to functional networks. Nat. Rev. Mol. Cell Biol. 20, 267–284 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.A. N. Patananan, T. H. Wu, P. Y. Chiou, M. A. Teitell, Modifying the mitochondrial genome. Cell Metab. 23, 785–796 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.R. Prakash, A. Kannan, Mitochondrial DNA modification by CRISPR/Cas system: Challenges and future direction. Prog. Mol. Biol. Transl. Sci. 178, 193–211 (2021). [DOI] [PubMed] [Google Scholar]
- 34.T. Amai, T. Tsuji, M. Ueda, K. Kuroda, Development of a mito-CRISPR system for generating mitochondrial DNA-deleted strain in Saccharomyces cerevisiae. Biosci. Biotechnol. Biochem. 85, 895–901 (2021). [DOI] [PubMed] [Google Scholar]
- 35.C. V. Pereira, S. R. Bacman, T. Arguello, U. Zekonyte, S. L. Williams, D. R. Edgell, C. T. Moraes, mitoTev-TALE: A monomeric DNA editing enzyme to reduce mutant mitochondrial DNA levels. EMBO Mol. Med. 10, e8084 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.P. A. Gammage, J. Rorbach, A. I. Vincent, E. J. Rebar, M. Minczuk, Mitochondrially targeted ZFNs for selective degradation of pathogenic mitochondrial genomes bearing large-scale deletions or point mutations. EMBO Mol. Med. 6, 458–466 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.L. D. Cruz-Zaragoza, S. Dennerlein, A. Linden, R. Yousefi, E. Lavdovskaia, A. Aich, R. R. Falk, R. Gomkale, T. Schöndorf, M. T. Bohnsack, R. Richter-Dennerlein, H. Urlaub, P. Rehling, An in vitro system to silence mitochondrial gene expression. Cell 184, 5824–5837.e15 (2021). [DOI] [PubMed] [Google Scholar]
- 38.K. Gao, M. Cheng, X. Zuo, J. Lin, K. Hoogewijs, M. P. Murphy, X. D. Fu, X. Zhang, Active RNA interference in mitochondria. Cell Res. 31, 219–228 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.P. Silva-Pinheiro, C. D. Mutti, L. Van Haute, C. A. Powell, P. A. Nash, K. Turner, M. Minczuk, A library of base editors for the precise ablation of all protein-coding genes in the mouse mitochondrial genome. Nat. Biomed. Eng., (2022); 10.1038/s41551-022-00968-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.M. G. Rosca, C. L. Hoppel, Mitochondrial dysfunction in heart failure. Heart Fail. Rev. 18, 607–622 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.S. W. Ballinger, Mitochondrial dysfunction in cardiovascular disease. Free Radic. Biol. Med. 38, 1278–1295 (2005). [DOI] [PubMed] [Google Scholar]
- 42.A. Kumar, Editorial (Thematic selection: Mitochondrial dysfunction & neurological disorders). Curr. Neuropharmacol. 14, 565–566 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.L. Wang, Z. Yang, X. He, S. Pu, C. Yang, Q. Wu, Z. Zhou, X. Cen, H. Zhao, Mitochondrial protein dysfunction in pathogenesis of neurological diseases. Front. Mol. Neurosci. 15, 974480 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.C. E. Toomey, W. E. Heywood, J. R. Evans, J. Lachica, S. N. Pressey, S. C. Foti, M. al Shahrani, K. D’Sa, I. P. Hargreaves, S. Heales, M. Orford, C. Troakes, J. Attems, E. Gelpi, M. Palkovits, T. Lashley, S. M. Gentleman, T. Revesz, K. Mills, S. Gandhi, Mitochondrial dysfunction is a key pathological driver of early stage Parkinson's. Acta Neuropathol. Commun. 10, 134 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.R. H. Swerdlow, J. K. Parks, S. W. Miller, R. E. Davis, J. B. Tuttle, P. A. Trimmer, J. P. Sheehan, J. P. Bennett, W. D. Parker, Origin and functional consequences of the complex I defect in Parkinson's disease. Ann. Neurol. 40, 663–671 (1996). [DOI] [PubMed] [Google Scholar]
- 46.A. Takeuchi, S. Matsuoka, Integration of mitochondrial energetics in heart with mathematical modelling. J. Physiol. 598, 1443–1457 (2020). [DOI] [PubMed] [Google Scholar]
- 47.E. Y. Adashi, D. S. Rubenstein, J. A. Mossman, E. A. Schon, I. G. Cohen, Mitochondrial disease: Replace or edit? Science 373, 1200–1201 (2021). [DOI] [PubMed] [Google Scholar]
- 48.K. J. Dunham-Snary, S. W. Ballinger, GENETICS. mitochondrial-nuclear DNA mismatch matters. Science 349, 1449–1450 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.C. S. Palmer, A. J. Anderson, D. Stojanovski, Mitochondrial protein import dysfunction: Mitochondrial disease, neurodegenerative disease and cancer. FEBS Lett. 595, 1107–1131 (2021). [DOI] [PubMed] [Google Scholar]
- 50.A. Jishi, X. Qi, Altered mitochondrial protein homeostasis and proteinopathies. Front. Mol. Neurosci. 15, 867935 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.M. A. McLellan, N. A. Rosenthal, A. R. Pinto, Cre-loxP-mediated recombination: General principles and experimental considerations. Curr. Protoc. Mouse Biol. 7, 1–12 (2017). [DOI] [PubMed] [Google Scholar]
- 52.H. Kim, M. Kim, S. K. Im, S. Fang, Mouse Cre-LoxP system: General principles to determine tissue-specific roles of target genes. Lab. Anim. Res. 34, 147–159 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.X. Wang, Cre transgenic mouse lines. Methods Mol. Biol. 561, 265–273 (2009). [DOI] [PubMed] [Google Scholar]
- 54.Y. Ma, X. Zhang, B. Shen, Y. Lu, W. Chen, J. Ma, L. Bai, X. Huang, L. Zhang, Generating rats with conditional alleles using CRISPR/Cas9. Cell Res. 24, 122–125 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Y. Ling, X. Yang, X. Zhang, F. Guan, X. Qi, W. Dong, M. Liu, J. Ma, X. Jiang, K. Gao, J. Li, W. Chen, S. Gao, X. Gao, S. Pan, J. Wang, Y. Ma, D. Lu, L. Zhang, Myocardium-specific Isca1 knockout causes iron metabolism disorder and myocardial oncosis in rat. Life Sci. 297, 120485 (2022). [DOI] [PubMed] [Google Scholar]
- 56.X. Y. Jiang, F. F. Guan, J. X. Ma, W. Dong, X. L. Qi, X. Zhang, W. Chen, S. Gao, X. Gao, S. Pan, J. Z. Wang, Y. W. Ma, L. F. Zhang, D. Lu, Cardiac-specific Trim44 knockout in rat attenuates isoproterenol-induced cardiac remodeling via inhibition of AKT/mTOR pathway. Dis. Model. Mech. 16, dmm.049444 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.M. A. Field, G. Burgio, A. Chuah, J. al Shekaili, B. Hassan, N. al Sukaiti, S. J. Foote, M. C. Cook, T. D. Andrews, Recurrent miscalling of missense variation from short-read genome sequence data. BMC Genomics 20, 546 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.R. Bao, L. Huang, J. Andrade, W. Tan, W. A. Kibbe, H. Jiang, G. Feng, Review of current methods, applications, and data management for the bioinformatics analysis of whole exome sequencing. Cancer Inform. 13, 67–82 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.D. L. Cameron, L. Di Stefano, A. T. Papenfuss, Comprehensive evaluation and characterisation of short read general-purpose structural variant calling software. Nat. Commun. 10, 3240 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S12
Tables S1 to S3
Sequences S1 and S2