Abstract
Parkinson’s disease (PD) is a progressive neurodegenerative disorder. However, cell type–dependent transcriptional regulatory programs responsible for PD pathogenesis remain elusive. Here, we establish transcriptomic and epigenomic landscapes of the substantia nigra by profiling 113,207 nuclei obtained from healthy controls and patients with PD. Our multiomics data integration provides cell type annotation of 128,724 cis-regulatory elements (cREs) and uncovers cell type–specific dysregulations in cREs with a strong transcriptional influence on genes implicated in PD. The establishment of high-resolution three-dimensional chromatin contact maps identifies 656 target genes of dysregulated cREs and genetic risk loci, uncovering both potential and known PD risk genes. Notably, these candidate genes exhibit modular gene expression patterns with unique molecular signatures in distinct cell types, highlighting altered molecular mechanisms in dopaminergic neurons and glial cells including oligodendrocytes and microglia. Together, our single-cell transcriptome and epigenome reveal cell type–specific disruption in transcriptional regulations related to PD.
Single-nucleus transcriptome and epigenome uncover cell type–specific gene dysregulation in Parkinson’s disease.
INTRODUCTION
Parkinson’s disease (PD) is a chronic, progressive neurodegenerative disorder accompanying both motor and nonmotor symptoms including resting tremor, bradykinesia, rigidity, and other nonmotor symptoms. Pathologically, PD is characterized by dopaminergic neuronal loss, abnormal protein deposition of Lewy bodies and Lewy neurites, and neuroinflammation in the substantia nigra (SN) (1). With regard to its etiology, genetic studies for familial forms of PD, which accounts for ~10% of patients with PD, have identified critical causal genetic factors (2, 3). Large-scale genome-wide association studies (GWAS) have further identified up to 90 genomic loci associated with PD (4–8). However, the identified genetic risk factors explain only 30% of familial and 3 to 5% of sporadic PD cases, ascribed to the complex genetic predisposition associated with the disease susceptibility (8, 9). The interpretation of these genetic variants is hindered because specific cell types in which these genetic variants exert their function are unknown. Further, the functional mechanism by which these variants contribute to disease susceptibility is still elusive, as most of them are located in noncoding sequences. The disease risk variants located in noncoding regions may disrupt transcription factor (TF) binding and cause a perturbation in cis-regulatory activity (10).
The growing recognition that perturbations in cis-regulatory elements (cREs) involve in disease-specific gene expression and colocalize with many noncoding genetic variants provides a rationale for in-depth investigation of epigenome associated with PD (10, 11). Although a systematic examination of cREs in PD is scarce, a global dysregulation of acetylated histone H3 lysine 27 (H3K27ac) landscape and a localization of PD GWAS genes proximal to the dysregulated cREs have been reported in the prefrontal cortex (12). Further, the advent of single-nucleus sequencing approach has allowed the investigation of epigenome landscape across individual brain cell types. A recent study reported the association of genetic variants of Alzheimer’s disease (AD) and PD at region- and cell type–specific cREs in healthy brains (13). Another study has characterized AD-associated dysregulation in chromatin accessibility at sub–cell type level, identifying cell type–specific cRE candidates (14). In line with this, there is a tremendous demand for cell type–resolved investigation of aberrant cis-regulatory regions in PD with respect to PD-specific gene expression.
As the function of cREs is dependent on the long-range chromatin interaction with a target promoter located over large genomic distances, the identification of cRE-to-promoter relationships is challenging. To this end, high-throughput chromosome conformation capture methods, including chromatin interaction anaysis with paired-end tag (ChIA-PET), Hi-C, promoter-capture Hi-C, HiChIP, and proximity ligation-assisted ChIP-seq (PLAC-seq), have allowed the investigation of genome-wide chromatin interactions (11, 15–18) and substantially advanced our view on the regulatory role of cREs regulating distal target gene expression. Recent integrative analyses of long-range chromatin interactome in healthy brain tissues have identified putative target genes of AD and PD genetic variants (13, 19). Nevertheless, the connection between cREs and altered gene expression in PD is largely unknown because of the lack of high-resolution three-dimensional (3D) chromatin contact maps available in the SN region of PD and control individuals.
To determine how cell type–dependent dysregulation in cis-regulatory regions affects molecular mechanisms related to PD pathogenesis in the context of 3D chromatin interactions, we conducted integrative analyses using multiomics data generated from the SN, the brain region most affected by PD. Single-nucleus sequencing of RNA (snRNA-seq) and chromatin accessibility (snATAC-seq) established cell type–resolved transcriptome and epigenome for both PD and control SN. Analysis of global H3K27ac signals from chromatin immunoprecipitation followed by sequencing (ChIP-seq) identified a set of dysregulated cREs, which were annotated on the basis of active cell type. By integrating PD GWAS variants, we confirmed a strong association between the cell type–resolved epigenome and the PD-associated genetic components and identified specific TF binding motifs that are disrupted by the genetic variants. Furthermore, we generated high-resolution 3D chromatin contact maps to effectively expand potential PD candidate genes by inferring putative targets of dysregulated cREs and PD GWAS–single-nucleotide polymorphisms (SNPs). Notably, modular expression patterns of the putative target genes resolved heterogeneous molecular pathways involved in PD, in which we annotated each cluster with a unique biological property and responsible cell type. Our findings shed light on the complex molecular characteristics of PD pathogenesis and expand candidate genes through mapping the target genes of PD-specific noncoding sequences in a cell type–resolved manner.
RESULTS
Single-nucleus profiling of transcriptome and chromatin accessibility in the human SN of control and PD cases
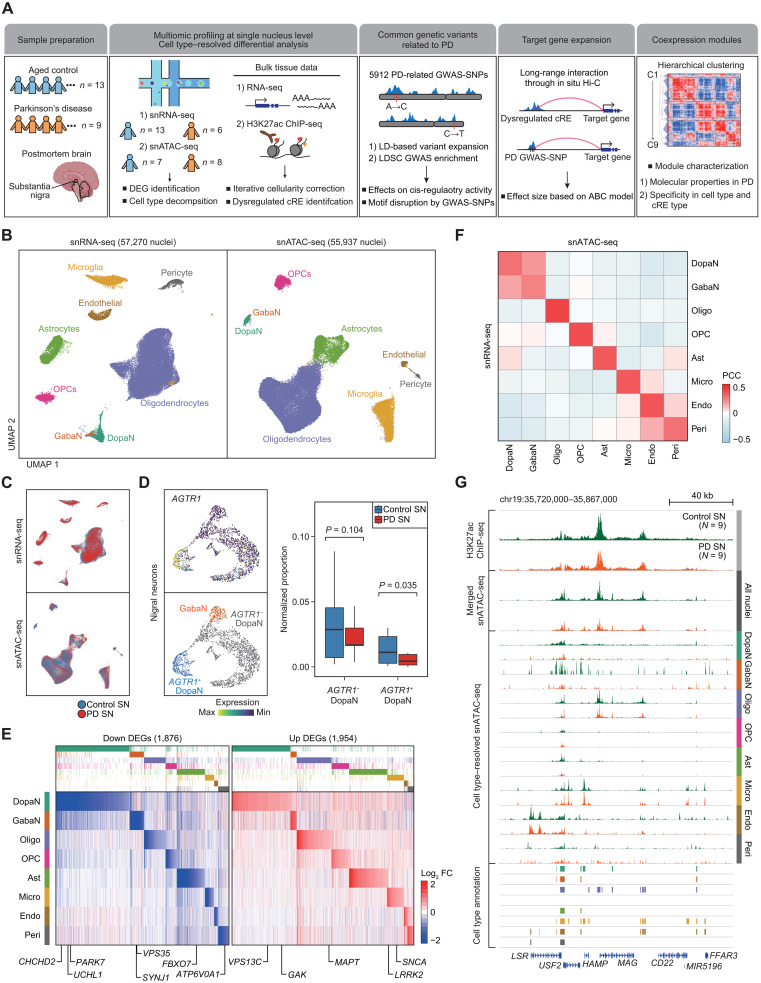
To dissect the disease-specific gene regulations in PD, we conducted integrative multiomics analysis on the transcriptome and chromatin accessibility for individual cell types present in the human SN (Fig. 1A). We performed snRNA-seq (10x Genomics v3) to characterize PD-associated changes in transcriptome at single-nucleus resolution in 19 flash-frozen postmortem SN specimens (late-stage PD = 6 and control = 6) and incorporated 7 raw snRNA-seq data for control SN (table S1) (20). In parallel, we conducted snATAC-seq (10x Genomics v1) on 13 postmortem SN specimens (late-stage PD = 8 and control = 5) and integrated 2 additional SN samples (13) to characterize cell type–resolved accessible chromatin (table S1). The SN specimens used for the experiments were obtained from Alzheimer’s Disease Research Center (ADRC) at the University of California, San Diego (fig. S1A). A rigorous quality control yielded a final set of 57,270 nuclei (34,638 controls and 22,632 PD nuclei) for snRNA-seq and 55,937 nuclei (26,074 controls and 29,863 PD nuclei) for snATAC-seq (Fig. 1, B and C, and table S2).
Fig. 1. Single-nucleus profiling of transcriptomic and epigenomic landscape in the human SN.
(A) A schematic of the study design, illustrating the preparation of sequencing-based omics data and downstream computational analysis. (B) Uniform Manifold Approximation and Projection (UMAP) embeddings of quality control–passed nuclei for snRNA-seq (left) and snATAC-seq (right). The nuclei were annotated into dopaminergic neurons (DopaNs), GABAergic neurons (GabaN), oligodendrocytes (Oligo), oligodendrocyte precursor cells (OPCs), astrocytes (Ast), microglia (Micro), endothelial cells (Endo), and pericytes (Peri), based on cell type markers. (C) UMAP embeddings of nuclei colored by pathological status, where red and blue indicate nuclei from PD and control SN, respectively. (D) UMAP embeddings of nigral neurons illustrating sub-DopaN populations with or without the expression of AGTR (left), along with box plots showing the normalized nucleus proportion of AGTR+ and AGTR− DopaNs between control and PD cases based on snRNA-seq data. (E) A heatmap by log2 fold change (FC) of cell type–resolved snRNA-seq reads (PD/control), illustrating 1876 down-regulated DEGs (left) and 1954 up-regulated DEGs (right), with the annotation of known PD risk genes. (F) A heatmap showing Pearson’s correlation coefficients (PCC) between snATAC-seq gene activity scores and snRNA-seq gene expression across the cell types present in the human SN. (G) Genome browser tracks of H3K27ac ChIP-seq signals and pseudo-bulk snATAC-seq signals resolved according to each cell type for PD (red) and control (green) groups, along with tracks indicating the positions of cell type–resolved cREs. The signals for ChIP-seq and cell type–resolved snATAC-seq were normalized by the total reads mapped in cREs.
We followed a standard Seurat (21) and Signac (22) framework to process snRNA-seq and snATAC-seq data, respectively. Unbiased clustering of the nuclei and projecting them on the Uniform Manifold Approximation and Projection (UMAP) dimensions did not show particular segregation by potential confounders (sex, age of death, postmortem interval, and doublet score) in both snRNA-seq and snATAC-seq clusters (figs. S1, B and C, and S2). On the basis of known marker genes, we profiled all major cell types in the SN, including neurons (SYT1), oligodendrocytes (Oligo; MAG and MOBP), oligodendrocyte precursor cells (OPCs; PDGFRA), astrocytes (Ast; AQP4 and GFAP), microglia (Micro; CD74 and RUNX1), endothelial cells (Endo; CLDN5), and pericytes (Peri; PDGFRB) (figs. S3A and S4A). Subclustering of neuronal populations allowed the annotation of dopaminergic neurons (DopaNs; TH and SLC6A3) as a distinct cluster from GABAergic neurons (GabaNs; GAD1 and GAD2) (figs. S3B and S4B). Cellular composition analysis between PD and control groups indicated that PD cases present a more prominent reduction in AGTR+ DopaNs (one-sided Welch t test, P = 0.035) compared to the AGTR− population (one-sided Welch t test, P = 0.104), in line with a recent report (23) that PD-associated neuronal degeneration is more actively associated in specific sup-DopaNs expressing AGTR (Fig. 1D and fig. S4, C and D).
Further, we identified 3830 PD-associated differentially expressed genes (DEGs; down-regulated = 1876 and up-regulated = 1954) (table S3) by iteratively performing a differential analysis in individual cell types based on donor-based pseudo-bulk (Fig. 1E). The correlation analysis of PD and control SN at the individual level showed that our samples are generally well clustered together on the basis of PD diagnosis (fig. S5A) and DEGs are consistently represented by all PD donors without a sample bias (figs. S5B and S6A). The majority (65.1%) of the identified DEGs was found to be SN specific, when matched with differential genes obtained from comparing with other brain parts (hippocampus and frontal cortex), by incorporating 545 bulk brain RNA-seq results from GTEx portal (v8).
Down-regulated DEGs were overrepresented by cell type–specific Gene Ontology (GO) pathways relevant to PD, as exemplified by mitochondrial function in DopaNs, neurogenesis regulation in OPCs and astrocytes, and immune responses in microglia (fig. S6B). Enriched biological processes in up-regulated DEGs exhibited recurrent cellular processes in multiple cell types, including macroautophagy (DopaN and Micro), protein folding and stabilization (Oligo and Endo), and cellular differentiation (OPC and Ast) (fig. S6B). We found that 12 of 20 well-known PD risk genes (24) were included in DEGs, which were concentrated in DopaNs (UCHL1, PARK7, CHCHD2, VPS13C, and GAK), microglia (SNCA, LRRK2, VPS13C, and GAK), and oligodendrocytes (MAPT and FBXO7) (Fig. 1E and fig. S6C). The results indicate that our transcriptomic profiling of PD and control SN at the single-nucleus level effectively recapitulates PD biology and highlight the pathogenic association of these cell types in PD.
We established cell type–resolved cis-regulatory landscape in PD and control SN by decomposing the snATAC-seq reads according to cell type annotation. Seurat’s label transfer algorithm based on gene expression and gene activity score using cell type marker genes indicated a close linkage between identical clusters of snRNA-seq and snATAC-seq nuclei (fig. S7A). We also confirmed that the snRNA-seq and snATAC-seq showed a high correlation across the cell types (Fig. 1F) and across individual samples (fig. S7B). In total, 128,724 cREs were identified from pseudo-bulk snATAC-seq chromatin accessibility, which were assigned to the corresponding cell type (Fig. 1G and fig. S7C). A small fraction of cREs (1.12%) was commonly annotated by all cell types, which suggests that a high degree of cell type specificity was captured by the dynamic cRE repertoire (fig. S7D).
Identification of dysregulated cREs in distinct cell types
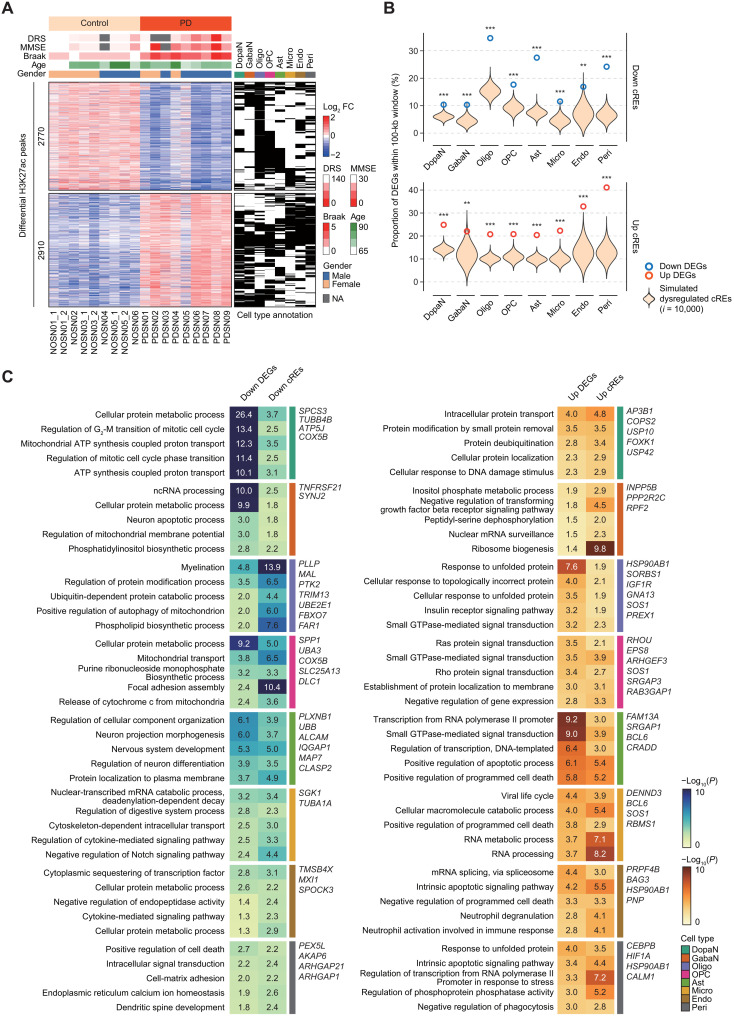
Next, we questioned the presence of global dysregulation in the noncoding regulatory landscape associated with PD in individual cell types. While the single-nucleus sequencing approach provides valuable insights into cell type specificity in the brain, it is challenging to effectively identify disease-specific dysregulated cREs due to its inherent sparsity. To compensate for these limitations, we incorporated high-quality bulk H3K27ac ChIP-seq results (late-stage PD = 9 and control = 9) using the SN specimens (fig. S7E). Given a high consistency between H3K27ac ChIP-seq and pseudo-bulk snATAC-seq signals across the cREs (Fig. 1G and fig. S7F), we implemented an iterative cellularity correction approach to resolving cellular heterogeneity in our bulk H3K27ac ChIP-seq results (fig. S8). Then, we performed a quasi-likelihood test in EdgeR (25, 26) to identify 5680 dysregulated cREs [down-regulated = 2770 and up-regulated = 2910; Benjamini-Hochberg (BH)–adjusted Q < 0.05] (Fig. 2A and table S4). Down-regulated cREs (0.93%) were found to be cell type–common, retaining a similar level of cell type specificity as the overall cREs, with the cell types dominantly annotated in oligodendrocytes, OPCs, and astrocytes. A higher rate of cell type–common cREs (5.01%) was found in up-regulated cREs, which may be ascribed to the recurrent cellular processes associated with up-regulated DEGs.
Fig. 2. Dysregulation of cREs shaping PD-specific gene expression.
(A) A heatmap of 2770 down-regulated and 2910 up-regulated cREs by log2 fold change of normalized H3K27ac ChIP-seq reads divided by the mean of all samples, along with binarized annotation of cell types. DRS, dementia rating scale; MMSE, mini mental state of examination; NA, not available. (B) Enrichment analysis for colocalization between DEGs and dysregulated cREs in a distance genomic window (100 kb). Violin plots represent the expected proportion of DEGs harbored by simulated cREs with 10,000 permutations. The observed proportion of DEGs is shown in bold donuts (blue for down-regulated DEGs and dark orange for up-regulated DEGs). The statistical significance was calculated based on empirical testing (**P < 0.01 and ***P < 0.001). (C) Top 5 enriched GO biological pathways for down-regulated (left) and up-regulated (right) DEGs commonly represented by genes annotated by dysregulated cREs through Genomic Regions Enrichment of Annotations Tool (GREAT) for individual cell types. DEGs within PD-related pathways that are also supported by dysregulated cREs are labeled. ATP, adenosine 5′-triphosphate.
To validate the regulatory role of the identified dysregulated cREs, we evaluated the genomic localization of DEGs with respect to cis-regulatory dysregulation in a cell type–specific manner. Our results showed that the PD-specific DEGs compared to controls were preferentially colocalized with dysregulated cREs in PD cases in a 100-kb genomic window compared to random expectations (Fig. 2B). In addition, GO analysis for DEGs and dysregulated cREs presented shared molecular pathways implicated in PD pathogenesis in a cell type–resolved manner (Fig. 2C). Together, the findings suggest that genome-wide dysregulation in the cis-regulatory landscape is highly associated with the aberrant gene expression specific to PD.
Identification of potential PD genes associated with dysregulated cREs and PD genetic risk variants
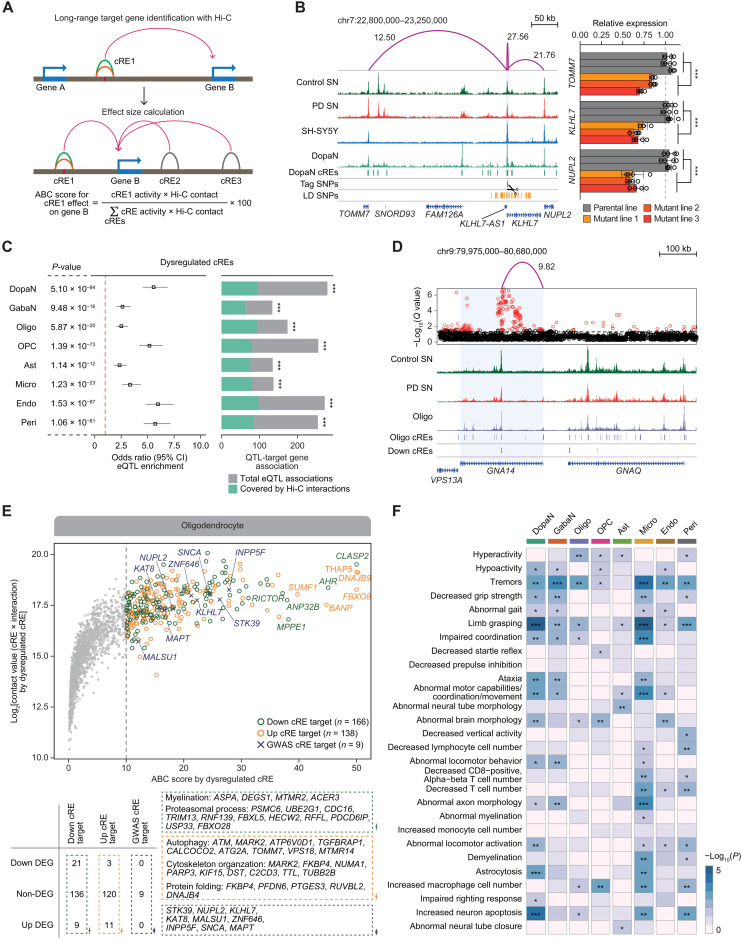
Given the strong link of dysregulated cis-regulatory regions with PD-specific gene expression, we aimed to systematically decipher the gene regulatory circuitry by implementing the “activity-by-contact” (ABC) model (27). It is an experimentally proven model, in which the quantitative effect of a cRE over its target gene expression is determined by the activity of the cRE weighted by the interaction frequencies to its promoter (Fig. 3A). To this end, we performed in situ Hi-C experiment using 11 SN specimens (late-stage PD = 5 and control = 6) and sequenced a total of 5.16 billion mapped reads to obtain 1.61 billion valid cis read pairs (744 million for PD SN and 870 million for control SN), generating unbiased, all-to-all 3D chromatin contact maps for PD and control SN (fig. S9A). Using Fit-Hi-C (28), we identified 1.42 million and 1.03 million long-range chromatin interactions in 5-kb resolution within a 1–megabase pair (Mbp) window for PD and control SN, respectively (Q < 0.01; union = 1.87 million) (fig. S9B).
Fig. 3. Target gene inference for noncoding regulatory sequences through 3D chromatin contacts.
(A) The illustrative approaches to identifying long-range interaction target genes using high-resolution 3D chromatin contact map and to characterizing the cis-gene regulation circuitry by calculating the effect size of all cRE-to-gene promoter associations within 1 Mb based on ABC model. (B) Left: H3K27ac ChIP-seq tracks for control SN, PD SN, and SH-SY5Y neuroblastoma, along with pseudo-bulk chromatin accessibility for DopaNs. Additional tracks indicate the positions of DopaN cREs and PD GWAS-SNPs. Significant long-range chromatin interactions are shown in purple arcs with the corresponding ABC score. Right: Bar plots with dots indicating TOMM7, KLHL7, and NUPL2 RNA levels in the parental SH-SY5Y cells and three independent mutant clones. Each clone has three biological replicates (Welch two-sample t test, ***P < 0.001). (C) Forest plots showing the enrichment of eQTLs in dysregulated cREs (left) and the proportion of eQTL-target gene associations matched with Hi-C interactions (right). The P value and odd ratio were calculated using two-sided Fisher’s exact test for eQTL enrichment, and the significance of target gene overlap was calculated on the basis of hypergeometric test (***P < 0.001). CI, confidence interval. (D) An example of a down-regulated cRE in oligodendrocytes whose significant chromatin interactions to a target gene promoter is supported by eQTL associations, with H3K27ac ChIP-seq tracks for PD and control SN and pseudo-bulk chromatin accessibility for oligodendrocytes (Oligo). Significant long-range chromatin interactions are shown in purple arcs with the corresponding ABC score. (E) Top: A scatter plot illustrating the putative target genes for oligodendrocyte with an ABC score threshold of 10. Bottom: Putative target genes were categorized on the basis of DEG status and the representative GO biological pathways. (F) A heatmap illustrating the enrichment of 656 putative PD genes for 28 neurological, movement, and immune symptoms potentially associated with PD symptoms (*P < 0.05, **P < 0.01, and ***P < 0.001).
To assess the quality and validity of the identified interactions, we first confirmed the marked enrichment of these interactions at promoters (11.05%) and cREs (53.64%) (fig. S9, C and D). In addition, the total chromatin contacts for a gene accounting all chromatin interactions anchored to its gene promoter strongly correlated with gene expression in all cell types (fig. S9E). We experimentally validated promoter-to-cRE relationships identified on the basis of significant interactions by disrupting a cRE harboring PD GWAS-SNPs through CRISPR-Cas9–mediated genome editing using SH-SY5Y neuroblastoma (Fig. 3B and fig. S10). The quantitative mRNA expression analysis on the genes (TOMM7, KLHL7, and NUPL2) linked to the PD GWAS-SNP–harboring cRE, showed a significant reduction in gene expression after the induction of the genetic mutation (Fig. 3B). Last, the integration of expression quantitative trait loci (eQTL) associations for the human SN from GTEx portal (v7) showed a marked enrichment of eQTLs in dysregulated cREs and PD GWAS-SNPs, and a significant portion of target genes identified by the ABC model were validated (Fig. 3, C and D, and fig. S11). In sum, our data strongly support the regulatory effects of dysregulated cREs and PD GWAS-SNPs on target gene expression by means of chromatin interactions.
With the ABC model applied to our high-resolution chromatin contact map, we quantified all cRE-target gene relationships with respect to their contribution to target gene expression within 1-Mb window. A total of 656 target genes of dysregulated cREs and PD GWAS-SNPs were identified in a cell type–specific manner (DopaN = 165, GabaN = 191, Oligo = 300, OPC = 231, Ast = 233, Micro = 223, Endo = 235, and Peri = 201; table S5) based on an ABC score threshold greater than 10 (equivalent to 10% contribution in overall chromatin contacts for a gene). These putative target genes were highly cell type–specific, with a considerable portion of the target genes (52.13%) assigned to only one or two cell types (fig. S12A). The enriched biological pathways of up-regulated cREs indicated a recurrent representation of autophagy (DopaN with SBF2 and KLHL22; Oligo with MARK2, ATG2A, and TOMM7; and Micro with PTGES3 and RUVBL2) and protein folding (DopaN with ABCA7 and Oligo with PFDN6 and DNAJB4) in multiple cell types (Fig. 3E and fig. S12, B and C). Conversely, cell type–specific biological pathways implicated in PD pathogenesis were enriched by the target genes of down-regulated cREs, including learning or memory (DopaN; ATP8A1 and ARL6IP5), myelination (Oligo; DEGS1, MTMR2, and ACER3), and protein lipidation (Micro; MPPE1, ATG10, and ZDHHC20) (Fig. 3E and fig. S12, B and C). Last, a gene set enrichment analysis of putative PD genes based on MGI mammalian phenotype database showed that, among the 75 enriched phenotype ontologies (P < 0.05), 28 of them were neurological, movement, and immune phenotypes with a potential link to PD-related symptoms (fig. S12D and table S6). When the gene set analysis was iterated in each cell type, we found varying cell types causing each of these phenotypes (Fig. 3F). Together, our findings strongly suggest that the identified putative target genes of dysregulated cREs and PD GWAS-SNPs have notable implications in PD pathogenesis and reaffirm that PD is a highly heterogeneous disorder with the involvement of diverse cell types and risk genes.
Association of PD susceptibility with cell type–resolved cis-regulatory landscape
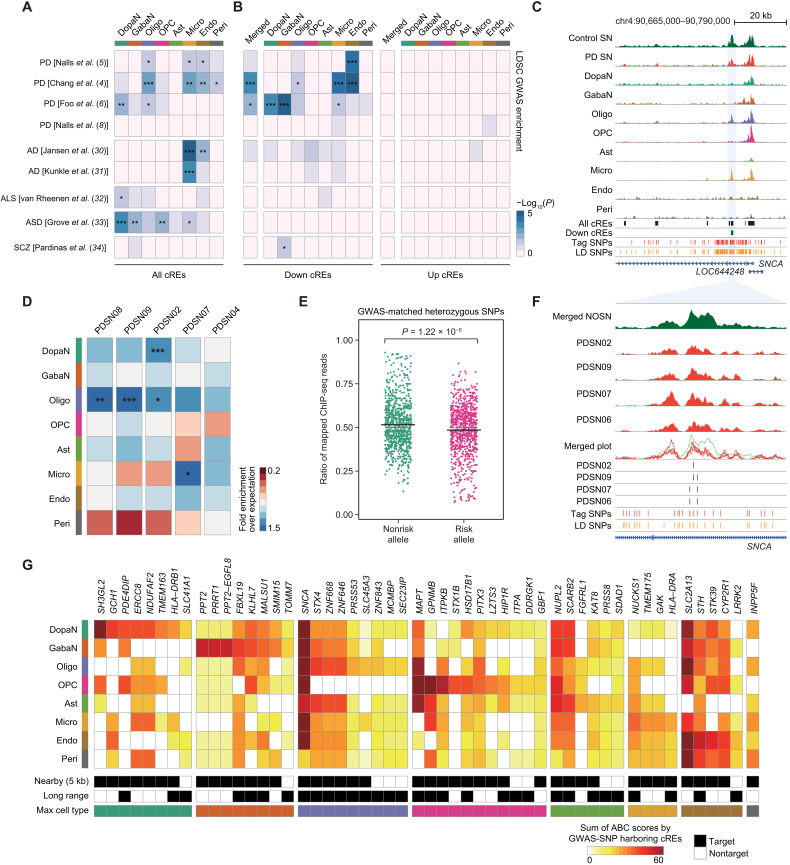
On the basis of the gene regulatory circuitry identified by 3D chromatin contact maps, a putative function of PD risk variants in the gene regulation program was annotated in individual cell types. To examine the association of common genetic variants related to PD in cis-regulatory regions (10), we collected and analyzed 5912 genetic variants from four PD GWAS summary statistics (P < 5 × 10−8) (4–6, 8). We found that 61.69% of these genetic variants were associated with cREs by linkage disequilibrium (LD; r2 > 0.8) (fig. S13A). The enrichment of PD-related SNPs in cell type–resolved cREs was assessed by implementing LD score (LDSC) regression analysis of heritability (29). We incorporated five additional GWAS summary statistics of other neurological and psychiatric disorders (table S7) (30–34). Oligodendrocyte cREs showed a strong association in three PD GWAS studies (4–6) (Fig. 4A). Microglia and endothelial cREs indicated enrichment in two PD GWAS studies (4, 5), and DopaN cREs were exclusively enriched in the PD GWAS conducted in East Asian cases (6). Our finding shows that PD etiology involves more diverse cellular properties than AD, whose GWAS-SNPs are most heavily linked to microglia cREs (13, 14, 19). Further, the GWAS heritability analysis on dysregulated regulatory regions suggests that the PD risk variants were specifically enriched in down-regulated cREs (Fig. 4B), which is well illustrated by a down-regulated cRE located in the vicinity of SNCA (Fig. 4C). This highlights the notion that genetic predisposition related to PD is associated with down-regulated cis-regulatory landscape.
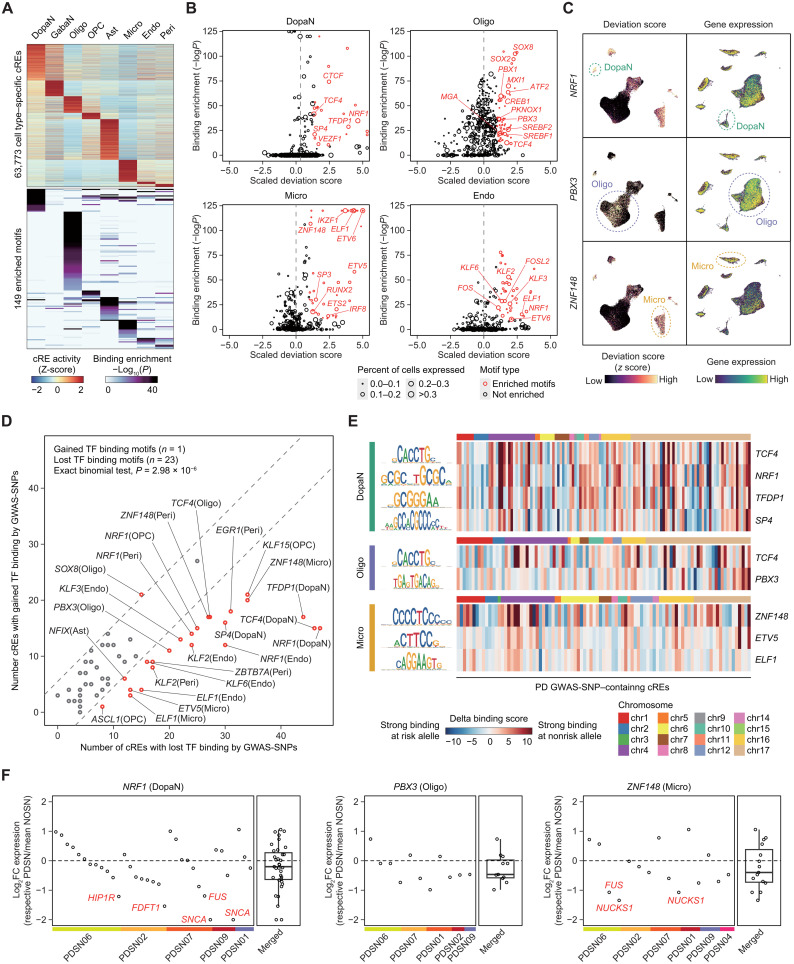
Fig. 4. Characterization of PD GWAS-SNPs based on cell type–resolved epigenomic landscape.
(A and B) Heatmaps illustrating the LDSC GWAS-SNP enrichment for neurological and psychiatric disorders in cell type–resolved cREs (A) and dysregulated cREs (B). ALS, amyotrophic lateral sclerosis; ASD, autism spectrum disorder; SCZ, schizophrenia. P values were derived from the LDSC enrichment testing (*P < 0.05, **P < 0.01, and ***P < 0.001). (C) H3K27ac ChIP-seq tracks for PD and control SN and cell type–resolved pseudo-bulk snATAC-seq signals in the SNCA locus. Additional tracks indicate the positions of all and down-regulated cREs and PD GWAS-SNPs. (D) Heatmap showing the fold enrichment of PD GWAS-matched genetic variants identified from each PD donor on cell type–resolved cis-regulatory landscape. The statistical enrichment of variants on each cell type cREs was calculated on the basis of exact binomial test (*P < 0.05, **P < 0.01, and ***P < 0.001). (E) Jitter plots describing the ratio of ChIP-seq reads mapped to the risk and nonrisk alleles of PD GWAS-matched heterozygous SNPs identified in PD donors. The black horizontal bars indicate the mean of each group. The statistical significance was calculated on the basis of two-sided Welch two-sample t test. (F) Epigenome browser tracks for H3K27ac ChIP-seq signals of control and four PD cases, along with GWAS-matched variants identified in each PD case and the PD-related tag and LD-expanded GWAS-SNPs in the SNCA locus. The ChIP-seq signals are normalized by the total reads mapped in cREs. (G) A heatmap describing the putative target genes of GWAS-SNP–harboring cREs identified on the basis of significant chromatin interactions. The ABC scores are calculated iteratively on the basis of the cell type–resolved epigenome to describe cell type–specific activation of key pathogenic genes by the PD GWAS-SNPs. Fifty-two genes with the sum of ABC scores by the GWAS-SNP–harboring cREs greater than 20 are shown.
We further examined the potential mechanistic role of PD GWAS-SNPs at individual level. For each donor, we identified variants in the cis-regulatory region using ChIP-seq data. When the genetic variants identified from each PD and control individual were matched with PD GWAS-SNPs, a higher number of variants (one-sided Wilcoxon rank sum test, P = 0.0385) overlapped with PD GWAS-SNP in PD cases compared to controls (fig. S13B). Further, the GWAS-matched variants in each PD donor were enriched in DopaNs, oligodendrocytes, and microglia, in line with the GWAS enrichments obtained from stratified LDSC regression (Fig. 4D). Then, we tested the effect of altered genetic variations in cis-regulatory activity by calculating allelic biases in GWAS variants found in PD cases. The number of ChIP-seq reads aligned to the risk alleles of PD GWAS variants showed a significant reduction compared to nonrisk alleles (two-sided Welch two-sample t test, P < 1.22 × 10−5) (Fig. 4E). The cis-regulatory effects of PD GWAS–matched variants are well represented in the SNCA locus, where PD cases contained GWAS variants in the down-regulated cRE proximal to the SNCA promoter (Fig. 4F). These results imply that the PD GWAS-SNPs are associated with altered cRE activities and that our dysregulated cREs represent aberrant regulatory features pertinent to PD susceptibility.
In addition, our target gene analysis based on 3D chromatin interactions and PD GWAS-SNPs unraveled important regulatory associations involving known PD genes and annotated specific cell types in which these associations are active. For example, the association of PD GWAS-SNPs to SH3GL2 and GCH1 was highly specific to DopaNs, while SNCA was prominent in most cell types (Fig. 4G). MAPT was dominantly associated with oligodendrocytes, OPCs, and astrocytes. GPNMB was active in OPCs, astrocytes, and microglia, while SCARB2 was substantially associated with neurons and astrocytes (Fig. 4G). Our data substantially facilitate the interpretation regarding the regulatory effects of PD GWAS-SNPs and link known risk factors of PD with the responsible cell types.
TF binding alterations induced by PD GWAS variants
To investigate the effect of PD GWAS-SNPs on TF binding motifs, we first identified 149 TFs whose binding motifs are highly enriched (P < 1.0 × 10−10) in cell type–specific cREs (Fig. 5A). By applying chromVAR (35), we computed deviation scores in per-cell basis and further selected 60 enriched TFs (scaled deviation score > 1 and percent of nuclei expressed > 0.1) that are highly active and expressed in their respective cell type (DopaN = 6, GabaN = 2, Oligo = 12, OPC = 4, Ast = 12, Micro = 9, Endo = 8, and Peri = 7) (Fig. 5B, fig. S13C, and table S8). UMAP representation of snRNA-seq and snATAC-seq nuclei based on motif activity and expression showed that the enriched TFs exhibit a substantial level of cell type specificity (Fig. 5C).
Fig. 5. Altered motif binding affinity for active TFs in the PD GWAS-SNP–containing cREs.
(A) Heatmaps describing the z-transformed activity of cell type–specific cREs (top) and the enrichment of binding motifs in the cell type–specific cREs (bottom). Cell type–specific cREs were identified on the basis of Wilcoxon BH-adjusted P < 0.05, log2FC > 1, and percent of nucleus detected > 0.05, comparing each cell type to the background of all other cell types. (B) Scatter plots illustrating the −log10 hypergeometric enrichment of TFs and the scaled chromVAR deviation score, depicting TF motif activity. The size of each data point represents the percent of nuclei expressed, and TFs with minimum expression detected greater than 10% were selected as enriched TFs. (C) UMAP embeddings of snATAC-seq (left) and snRNA-seq (right) nuclei illustrating z-transformed deviation score and gene expression of enriched TFs including NRF1, PBX3, and ZNF148. (D) Scatter plots of enriched motifs illustrating GWAS-SNP–containing cREs with gain or loss of TF binding as a result of PD-associated genetic variations. For each GWAS-SNP–containing cRE, a difference in binding score greater than 3 between risk and nonrisk alleles of GWAS-SNPs was used to define gained and lost TF binding. The dashed gray lines represent the difference in the number of cREs with gained and lost TF binding of 5. (E) Heatmaps showing the delta binding scores of individual PD GWAS-SNP–containing cREs across the genome, for enriched TFs in DopaNs, oligodendrocytes, and microglia. (F) Scatter plots describing the log2 fold change expression (a respective PD donor over the mean of control samples) for target genes of cREs with disruption in binding motifs for respective TFs by GWAS-matched donor variants based on cumulative ABC score greater than 1. The name of target genes with log2 fold change less than −1 is labeled in red. PDSN, Parkinson's disease SN; NOSN, control SN.
To study individual TF binding motifs that are altered by PD GWAS-SNPs, we calculated TF binding scores for each of the GWAS-SNP–containing cRE in both risk and nonrisk alleles of GWAS-SNPs. The evaluation of delta binding scores showed that GWAS variants on cis-regulatory regions most often act toward TF binding disruption, as 23 of the 60 enriched motifs exhibited an overall reduction in TF binding affinity by the GWAS-SNPs, as opposed to only one motif with a gained TF binding affinity (exact binomial test, P = 2.98 × 10−6) (Fig. 5D and table S9). Specifically, NRF1 (DopaN, and OPC), TFDP1 (DopaN), TCF4 (DopaN and Oligo), PBX3 (Oligo), ZNF148 (Micro), and KLF2 (Endo) were associated with GWAS-SNP–induced binding disruption (Fig. 5E and fig. S13D). We also found that the down-regulated cRE at the SNCA locus can potentially be explained by the binding disruption of these TFs, where the delta binding score was mostly positive (i.e., TF motif disruption) for NRF1 (DopaN) = 6.30, PBX3 (Oligo) = 3.84, TCF4 (Oligo) = 2.69, and ZNF148 (Micro) = 7.92. Next, to examine the transcriptional effects of GWAS variants on the disrupted motifs, we collected genetic variants in cREs that match with PD GWAS-SNPs from each donor and identified the putative target genes of the disrupted motifs using the chromatin interaction map (cumulative ABC score > 1). The expression of the target genes showed a remarkable down-regulation in each patient with PD for most of the TFs that were associated with SNP-mediated motif disruption (Fig. 5F and fig. S13E). Together, our analysis provides a global view on how individual PD GWAS-SNPs influence the TF binding behavior and a biological rationale for the enrichment of PD GWAS-SNPs in the down-regulated cREs and their reduced regulatory activities.
Modular expression patterns of potential PD genes resolve complex molecular characteristics of PD
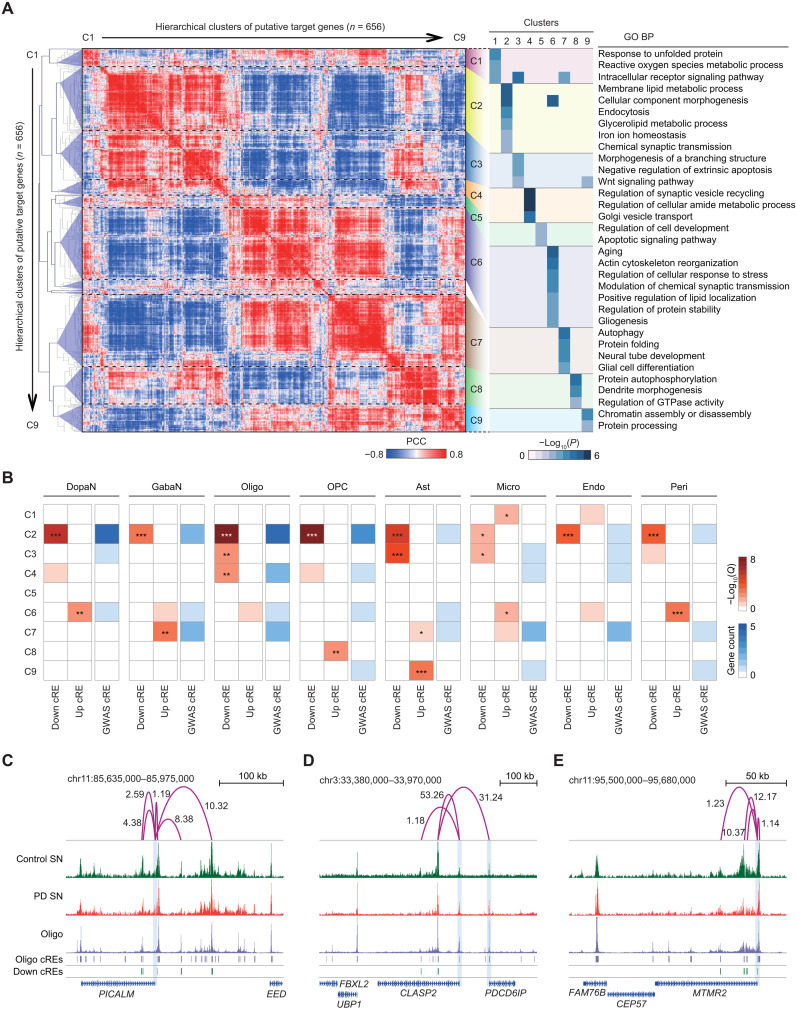
Since a number of potential PD genes have been identified by inferring the target genes of dysregulated cREs and PD GWAS-SNPs, we attempted to characterize the complex molecular pathways underlying the neuropathology based on the potential PD genes and identify the responsible cell type associated with each pathway. To this end, we examined the dynamic modular gene expression patterns of the 656 potential PD genes using the cellularity-corrected bulk RNA-seq dataset from 16 SN specimens (late-stage PD = 8 and control = 8). The hierarchical clustering of gene expression correlation across these samples displayed a modular pattern with nine distinct clusters (from C1 to C9) with notable biological processes involved in PD pathogenesis (Fig. 6A and table S10). In addition, these biological pathways were represented by cell type–specific gene regulatory relationships. For example, genes involved in C1 (response to unfolded proteins and reactive oxygen species) were specifically targeted by microglia cREs (Fig. 6B). Alternatively, the C3 genes represented by negative regulation of apoptosis were selectively enriched in oligodendrocytes and astrocytes, implicating their neuroprotective function toward the loss of nigral neurons. Unexpectedly, C2 contained multiple cellular processes associated with PD pathogenesis, including endocytosis, lipid metabolism, iron homeostasis, and synaptic function (36–38), and harbored many target genes of PD GWAS-SNPs, highlighting the possibility that these cellular pathways are highly associated with PD pathogenesis (Fig. 6B). The genes in C2 were enriched with down-regulated cRE target genes from all cell types, which is consistent with our finding in the heritability analysis that PD-related genetic variations are associated with down-regulated cREs from diverse cell types. Furthermore, through our target gene analysis, we pin-pointed potential links to PD candidates while confirming the connections of known PD risk genes. For example, the endocytosis-related genes in C2 identified CLASP2, PDCD6IP, MTMR2, and PICALM, in addition to SCARB2 and INPP5F, whose genetic variations are highly related to PD pathogenesis (Fig. 6, C to E) (5, 39, 40).
Fig. 6. Analysis of modular gene expression patterns across putative PD genes.
(A) Left: A hierarchical clustering of 656 putative PD genes based on similarities in gene expression between 16 bulk RNA-seq samples from control and PD SN. The color intensity indicates PCC of putative target gene expression between pair-wise samples. Nine distinct clusters were identified by a linkage distance (0.66) threshold in the dendrogram. Right: A heatmap describing the enrichment of GO biological pathways (BP). Each entry indicates −log10(P) of GO biological processes in the corresponding cluster. (B) Heatmaps illustrating the enrichment of cell type–resolved target genes with respect to the nine coexpression clusters. For each cell type, the putative target genes were categorized into three groups based on the type of cRE connected (down- and up-regulated cREs or GWAS-SNP–harboring cRE). The enrichment level was calculated on the basis of one-sided exact binomial test with BH multiple testing correction (*Q < 0.05, **Q < 0.01, and ***Q < 0.001). (C to E) H3K27ac ChIP-seq tracks for PD and control SN and pseudo-bulk chromatin accessibility for oligodendrocytes (Oligo) for a genomic locus containing PICALM (C), CLASP2 and PDCD6IP (D), and MTMR2 (E). Additional tracks indicate positions of Oligo and down-regulated cREs. The signals for ChIP-seq and cell type–resolved snATAC-seq are normalized by the total reads mapped in cREs. Significant long-range chromatin interactions are shown in purple arcs with the corresponding ABC score.
From C6 to C9, we found PD-associated biological features targeted by up-regulated cREs pertaining to a specific cellular identity (Fig. 6B). We found that C6 represented fundamental pathogenic pathways (aging, cytoskeleton organization, stress response, modulation of synaptic activity, protein stability, and gliogenesis) related to DopaNs and microglia, while containing PD risk genes such as MAPT, BAX, and TOMM7 (41). In C7, cellular processes such as glial cell differentiation and neural tube development were activated in astrocytes, in all likelihood, as a response mechanism to PD progression. Last, target genes in C8 indicate guanosine triphosphatase (GTPase)–mediated signaling and protein autophosphorylation are active in OPCs as a result of autophagy accumulation in PD SN (42, 43).
Together, our analysis on the modular expression pattern demonstrates that the heterogeneous molecular features in the advanced PD cases are well explained by the target genes of dysregulated cREs and PD GWAS-SNPs. Although common genetic variations found in GWASs explain only a fraction of PD pathogenesis in individual PD cases, most of the PD-related cellular processes were represented by our target genes based on the integration of epigenetic features. The strong association between PD genetic variants and down-regulated cREs supports the notion that cellular processes linked to PD are coordinated by the combined effects of genetic and epigenetic aberrations. On the basis of these findings, we propose that a genetic predisposition and epigenetic dysregulation are two indistinguishable modes of gene regulation that contribute to PD pathogenesis (fig. S14).
DISCUSSION
Major progresses have been made in the past decades furthering the understanding of molecular mechanisms and risk genes involved in PD pathogenesis. While evidence has revealed diverse PD-related cellular properties including protein quality control, autophagy-lysosome pathway, mitochondria homeostasis, lipid metabolism, synaptic toxicity, and neuroinflammation (36–38, 44), our knowledge is still limited to fully explain the molecular causes of sporadic PD cases. A thorough characterization of the dynamic and interactive role of individual cell types during PD development is essential to broaden the scope of PD heritability. In this regard, the current study provides important insights into PD pathogenesis by identifying the cell type–resolved dysregulation of cis-regulatory landscape and by characterizing the perturbed molecular pathways based on the relevant cell types. Through this effort, we identified 656 known and potential PD candidate genes, which demonstrate the heterogeneous molecular characteristics implicated in PD.
Our cell type–specific cross-investigation of PD and control SN repeatedly shows that oligodendrocytes and microglia are the key cell types associated with PD pathogenesis, based on three lines of evidence. First, a large proportion of known PD genes were included in PD-associated DEGs from oligodendrocytes and microglia. Among the 20 PD risk genes (24), oligodendrocytes and microglia each contained two and four PD risk genes as DEGs (MAPT and FBXO7 for oligodendrocytes and SNCA, LRRK2, VPS13C, and GAK for microglia), respectively, while other glial cell types (OPCs, endothelial cells, and pericytes) had none or one PD risk genes as DEGs. Second, our GWAS heritability analysis showed a specific enrichment of PD GWAS-SNPs in oligodendrocyte and microglia cREs. While a significant enrichment was found in at least two sets of PD GWAS statistics for oligodendrocytes and microglia, other glial cell types (astrocytes and OPCs) presented a low correlation with PD GWAS-SNPs. This finding was further supported by the individual SNPs identified from our PD donors, where PD GWAS–matched variants from each PD case were concentrated mostly on oligodendrocyte and microglia cREs. Third, coexpression modules highly associated with PD pathogenesis were specifically represented by oligodendrocyte and microglia. For instance, genes in C1 (response to unfolded proteins, and reactive oxygen species) were almost exclusively associated with microglia’s up-regulated cREs in PD SN, and genes in C2 (endocytosis, lipid metabolism, iron homeostasis, and synaptic function) were most strongly associated with down-regulated cRE target genes of oligodendrocyte, along with its GWAS-SNP targets. Overall, our analysis suggests that oligodendrocytes and microglia are closely linked to PD pathogenesis.
Our findings suggest that PD is a highly heterogeneous disorder. The GWAS-SNP enrichment test of PD heritability showed that PD involves far more diverse cellular properties than AD, whose enrichment is limited predominantly in microglia. We also found that heterogeneity exists among the four PD GWAS statistics used in this study, where SNPs from three PD GWAS showed a varying LDSC enrichment pattern across DopaNs, oligodendrocytes, microglia, and endothelial cells. The low correlation of PD GWAS-SNPs by Nalls et al. (8) with overall cis-regulatory regions may be ascribed to their heavy localization in nonintergenic regions, and other types of regulatory signals (e.g., different histone marks) are required to precisely characterize the PD GWAS-SNPs. by Nalls et al. (8). Nevertheless, it is clear that the common variants identified by GWAS exhibit a regulatory function in a cell type–specific manner. However, most of the previous genomics-based studies for complex traits did not sufficiently address this critical issue. We conducted cell type annotation of key genetic variants by overlaying them onto the cell type–resolved epigenome and demonstrated that the PD variants are highly associated with genome regulatory elements and likely conduct a regulatory role in cell type–specific manner, together with dysregulated cREs. In this regard, our results provide a unique perspective on the PD heritability that the previous genomics investigations were not able to address.
Our analysis provides biological insights into PD GWAS-SNPs regarding their mode of action on disease-specific cREs. Although GWAS variants do not always cause a loss in regulatory activity, we found a consistent association of PD GWAS-SNPs with reduced cis-regulatory activities, as evidenced by the enrichment of GWAS-SNPs in down-regulated cREs and the reduced ChIP-seq reads on the GWAS-matched variants from PD donors. This finding was further supported by our motif analysis, which showed that the risk variants in cREs have a higher chance of causing cell type–specific disruption in TF binding. Thus, the overall loss of function in disease-specific cREs may be linked to putative risk genes involved in PD pathogenesis.
We present a unique multiomics data integration centering on high-resolution 3D chromatin interactome. The implementation of ABC model effectively combined cell type–resolved epigenome and transcriptome while accounting for chromatin contacts and quantitatively defined the regulatory effects of all cREs to the surrounding genes. The integrative analysis based on cell type–resolved 3D epigenome bridged the gap between key risk genetic variants and candidate genes, greatly advancing our view on the regulatory mechanisms involving PD. Our 3D epigenome analysis revealed specific cis-gene regulations that modulate PD risk genes (SNCA, MAPT, SCARB2, GCH1, BAG3, and INPP5F), while identifying candidate genes that are associated with PD-implicated biological processes. The present work emphasizes the role of noncoding regulatory elements in understanding PD and provides additional insights into molecular mechanisms related to the perturbed epigenomic landscape. This computational framework incorporating omics data integration is highly applicable in other complex human disorders.
The single-nucleus sequencing strategy is a highly advanced technique allowing the investigation of individual cell populations. However, because of the insufficient coverage obtained for each nucleus, the single-nucleus assays allow a differential analysis in only a small fraction of genes. The detection rate of DNA fragments is far less for snATAC-seq because of the limited copies of DNA to capture, in comparison to snRNA-seq. This inherent sparsity hinders robust identification of epigenomic dysregulation for a complex disease to its entirety. In this aspect, our work shows that the integration of bulk assays with a corresponding single-nucleus dataset may provide a solution. Our single-nucleus datasets offered a suitable reference for cellularity correction method for the bulk sequencing data, and the ratio of sequenced reads mapped to cell type markers effectively addressed the fraction of each cell type across the samples. This shows that the sequencing data generated from bulk tissues may still hold substantial value as a resource when integrated with appropriate single-nucleus data.
The difficulty in interpreting postmortem tissue data lies in the unresolved cause and effect signature by the pathology. Studies pin-pointing the epigenomic changes before the motor phase of PD (Braak stages 1 and 2) or studies leveraging Braak stage–specific samples may further elucidate the molecular mechanism underlying the pathogenic progression of PD. In addition, the chromatin conformation capture method conducted at a single-nucleus level will allow the cell type–specific investigation of chromatin interactome. The application of single-cell Hi-C technology to clinical brain samples of neurodegenerative disorders may better portray the disease-related gene regulatory circuitry in a cell type–specific manner. Establishing the complete 3D epigenome may play an important part, especially in effective personalized therapeutics, in light of the recent success in restoring clinical symptoms of PD by the implantation of patient-derived dopaminergic progenitor cells (45). Further understanding of pathogenic mechanisms in glial cells may be required to prevent neurodegeneration in patient-specific manner. Nevertheless, the present delineation of PD-specific aberration in cis-genome regulation, coupled with high-resolution chromatin interaction maps, substantially broadened the scope for disease-specific gene regulation mechanisms and expanded potential therapeutic candidate genes for PD. The present work conveying cell type–resolved noncoding regulatory elements lays the ground for further understanding of the gene regulatory network involved in complex genetic disorders.
MATERIALS AND METHODS
Collection of human brain samples
Flash-frozen postmortem tissues from the human SN were acquired for PD and control subjects from the ADRC at the University of California, San Diego (table S1). The main cause of death for most of the donors was bronchopneumonia and cardiovascular failure, although obtaining the information at individual level was restricted. The difference in age of death between PD and control cases was calculated to be insignificant (Welch two-sample t test, P = 0.803). Samples from the left mid-frontal cortex were fixed in 4% paraformaldehyde, and those from the right mid-frontal cortex were stored in liquid nitrogen for experiments. Formalin-fixed brains were sectioned at 40 μm and processed for pathohistological examination by hematoxylin and eosin, and pathological scoring was assessed according to Braak stages (46). Institutional Review Board approval was obtained from Korea Advanced Institute of Science and Technology for the use of these brain tissues.
snRNA-seq and snATAC-seq
About 15 mg of tissue was placed in an extra-thick tissue processing tube (Covaris, 520140) while being kept in liquid nitrogen and repeatedly hammered to produce frozen tissue powder. The frozen tissue was transferred to a lysis buffer of 0.2% Triton X-100, a protease inhibitor (Roche, 04-693-159-001), 1 mM dithiothreitol (Sigma-Aldrich, D9779), RNasin (0.2 U/μl; Promega, N211B), and 2% bovine serum albumin (BSA) (Sigma-Aldrich, SRE0036) in phosphate-buffered saline (PBS). The sample was pipetted 10 times, rotated for 5 min at 4°C, and centrifuged at 500g for 5 min. The pellet was resuspended in a sorting buffer of 1 mM EDTA, RNasin (0.2 U/μl), and 2% BSA in PBS and passed through a 30-μm strainer (Sysmex, 04-0042-2316) to remove excessive debris. The nuclei were stained with DRAQ7 (1:100; Cell Signaling Technology, 7406) for snRNA-seq and with 4′,6-diamidino-2-phenylindole (10 μg/ml; Sigma-Aldrich, 32670) for snATAC-seq. Between 100,000 and 150,000 nuclei were sorted using a MoFlo Astrios EQ sorter (Beckman Coulter, laser/filter) into a collection tube with RNasin (1 U/μl) and 5% BSA in PBS. Sorted nuclei were centrifuged at 1000g for 15 min at 4°C, and the supernatant was removed. Nuclei were resuspended in resuspension buffer [RNasin (0.2 U/μl) and 1% BSA in PBS] and counted using a cell counter (Cellometer Auto 2000, Nexcelom Biosicence) with acridine orange/propidium iodide (AO/PI) staining.
A subset of snRNA-seq and snATAC-seq libraries was generated by pooling the nuclei from two or three different donors with a matched pathological state and demultiplexed on the basis of individual genetic backgrounds using souporcell (47). We generated snRNA-seq libraries using the Chromium Single Cell 3′ Library & Gel Bead Kit v3 (10x Genomics). For snATAC-seq libraries, the isolated nuclei were subject to permeabilization in a lysis buffer with tris-HCl (pH 8.0), 10 mM NaCl, 3 mM MgCl2, 0.1% Tween 20, 0.1% NP-40 substitute (Sigma-Aldrich, 74385), 0.01% digitonin (Thermo Fisher Scientific, BN2006), and 1% BSA. After washing, the nuclei were resuspended in nuclei buffer, and snATAC-seq libraries were generated using the Chromium Single Cell ATAC Library & Gel Bead Kit v1 (10x Genomics). Quality control for DNA libraries was performed using Agilent Tape Station 4200 with a high-sensitivity D5000 kit. The libraries were sequenced in a paired-end mode using Illumina HiSeq 4000 and MGI DNBSEQ-G400 platform.
snRNA-seq data processing
In addition to the single-nucleus data that we generated from the SN specimens from ADRC, raw snRNA-seq data generated from healthy SN specimens (GSE140231) (20) was downloaded and processed in parallel (table S1). To demultiplex the data for pooled libraries, the nuclei were clustered on the basis of individual genetic variants using souporcell (47) and assigned to the corresponding donor by matching the genetic variants obtained from bulk RNA-seq data. The genetic variants from bulk RNA-seq data were obtained using freebayes (-iXu -C 2 -q 20 -n 3 -E 1 -m 30 --min-coverage 20 --pooled-continuous --skip-coverage 100000). The number of demultiplexed nuclei from each pooled library is described in table S2. The feature-barcode matrix was generated using cellranger count (10x Genomics, v3.0.2), aligning the sequenced reads to the human reference genome (hg19; 10x Cell Ranger reference GRCh37 v3.0.0). Then, the count matrices were aggregated by cellranger aggr function across the samples with default parameters. The following analysis was performed using Seurat R package v4.0.5 (21). Nuclei with fewer than 200 or greater than 10,000 genes detected were filtered from the snRNA-seq dataset. Low-quality nuclei with mapped reads in the mitochondrial genes greater than 10% were removed. Doublets were identified using Scrublet (48), and nuclei with doublet score greater than 0.4 were excluded from analysis. The snRNA-seq data were integrated to correct for technical differences across individual samples. For this, the feature-barcode matrix was individually normalized by the total read count and log-transformed, and top 5000 variable genes were selected for each sample using Seurat’s FindVariableFeatures function. Data integration was conducted on the basis of anchors identified using FindIntegrationAnchors function. The aligned nuclei were scaled, and principal components analysis (PCA) was conducted on the scaled expression matrix. The top 45 principal components (PCs) were used to compute Shared Nearest Neighbors (SNNs), which were then used to cluster nuclei based on the Louvain algorithm (resolution = 1.5) in FindClusters function in Seurat R package. The top 45 PCs were used for the UMAP embedding.
We selected 3000 highly variable genes by the vst method in Seurat R package v4.0.5 (21). Harmony (v1.0) (49) was used to correct the technical variations across the samples in the PCA dimensions. The first five dimensions were used to build SNN graph, which was clustered using the Louvain algorithm (resolution = 1.0). These dimensions were used to visualize neuronal nuclei in UMAP dimensions. Cell type markers for DopaNs (TH and SLC6A3) and GabaNs (GAD1 and GAD2) were used to assign subneuronal identity for individual subclusters. The nucleus-level expression signals were imputed using MAGIC (v2.0.3) (50) for UMAP visualization of cell type markers in snRNA-seq clusters. To build a reference for cell type–dependent transcriptome in the SN, we generated a count matrix from the feature-barcode matrix using the cellular identity annotated for all nuclei. The count matrix was then quantile-normalized, and the ratio of normalized reads based on individual cell types was computed to represent the gene expression ratio (ER) among different cell types. The cell types whose gene ER is greater than 10% were annotated as the active cell type for the corresponding gene.
snATAC-seq data processing
In addition to the single-nucleus data we generated from the SN specimens from ADRC, raw snATAC-seq data generated from postmortem SN specimens (GSE147672) (13) was downloaded and processed in parallel (table S1). To demultiplex the data for pooled libraries, the nuclei were clustered on the basis of individual genetic variants using souporcell (47), using the same parameters (--min_alt 8 --min_ref 8 --no_umi True --skip_remap True --ignore True) used in the study of Fiskin et al. (51). The genetic variants from bulk ChIP-seq data were obtained using freebayes (-iXu -C 2 -q 20 -n 3 -E 1 -m 30 --min-coverage 20 --pooled-continuous --skip-coverage 100000). The number of demultiplexed nuclei from each pooled library is described in table S2. The feature-barcode matrix was generated using the cellranger-atac count (10x Genomics, v1.1.0), and aligning the sequenced reads was aligned to the human reference genome (hg19). Then, the count matrices were aggregated by cellranger-atac aggr function with default parameters. The following analysis was performed using Signac R package v1.4.0 (22). Nucleosome signal was computed with Signac’s NucleosomeSignal function. Low-quality nuclei were removed from snATAC-seq dataset based on the following criteria: fewer than 2000 or greater than 30,000 fragments mapped to peak regions, less than 15% of reads in peak regions, nucleosome signal greater than 10, and transcription start site (TSS) enrichment less than 2. Doublets were identified using Scrublet (48) and excluded from analysis. We selected top 50% most common features as the variable features and performed latent semantic indexing (LSI) dimensionality reduction by implementing term frequency–inverse document frequency transformation, followed by singular value decomposition. Then, reciprocal LSI projection was conducted to identify integration anchors for each sample, and the snATAC-seq data were integrated across the samples using low-dimensional cell embeddings with Signac’s IntegrateEmbeddings function. The identical method used in snRNA-seq for graph-based clustering and nonlinear dimension reduction by UMAP was applied to the snATAC-seq dataset. Gene activity scores were computed for protein coding genes by summing snATAC-seq reads mapped in the gene body and the promoter (5-kb upstream to TSS) using GeneActivity function in Signac R package. We used Signac’s label-transfer algorithm with default parameters using cell type marker genes identified on the basis of snRNA-seq data.
Major cell types in the SN were assigned to each snATAC-seq cluster based on the identical set of known marker genes used in snRNA-seq processing. The neuronal population was subclustered to identify DopaNs and GabaNs. First, we selected features with fragments detected in more than 10 nuclei as the variable features. Then, harmony (v1.0) (49) was used to correct the technical variations across the samples in the LSI dimensions. The first five dimensions were used to build SNN graph, which was clustered using the Louvain algorithm (resolution = 1.0). These dimensions were used to visualize neuronal nuclei in UMAP dimensions. Cell type markers for DopaNs (TH and SLC6A3) and GabaNs (GAD1 and GAD2) were used to assign subneuronal identity for individual subclusters. The nucleus-level gene activity scores were imputed using MAGIC (v2.0.3) (50) for UMAP visualization of cell type markers in snATAC-seq clusters.
We generated cell type–resolved BAM files for each sample and merged the BAM files according to the cell type. Peak calling by MACS2 identified 240,354 peaks (P < 0.05). Through a manual inspection of pseudo-bulk signals in the epigenome browser, we collected top 128,724 peaks based on the abundance of snATAC-seq reads and defined them as cREs. To build a reference for cell type–dependent epigenome in the SN, we generated a count matrix by conducting bedtools (v2.29.1) coverage function for the cell type–resolved BAM files. The count matrix was then quantile-normalized, and the ratio of normalized reads based on individual cell types was computed to represent the cRE activity among different cell types. The cell types whose cRE activity ratio is greater than 10% were annotated as the active cell type for the corresponding cRE.
Identification of cell type–dependent differential features
To identify cell type markers for snRNA-seq and snATAC-seq data, we used FindAllMarkers function in Seurat R package to identify cell type markers with the model-based analysis of single-cell transcriptomics algorithm (52) based on the two data modalities, leveraging the gene expression levels (snRNA-seq) and gene activity scores (snATAC-seq). It computes differential genes by iteratively contrasting one cell type to all, and the genes that satisfy BH-adjusted P < 0.05, a log2 fold change of >0.25, and an expression detected in at least 10% of nuclei were defined as cell type markers. These cell type markers were used for cellularity correction and label transfer between snRNA-seq and snATAC-seq clusters.
To compute DEGs between PD and control SN, we obtained raw RNA counts from each snRNA-seq library and merged the counts from a same donor to construct a donor-based pseudo-bulk for individual cell types. The raw counts were normalized by the total count, and the batch effect from different snRNA-seq data source was removed by using ComBat function in sva R package (53). We identified cell type–specific DEGs by applying EdgeR likelihood ratio test (25, 26) on the pseudo-bulk using a threshold of BH-adjusted P < 0.05, a log2 fold change of >0.5, and a percentage of nuclei expressed >0.1 in either control or PD SN nuclei.
Bulk RNA-seq and data processing
About 40 mg of tissue was placed in an extra-thick tissue processing tube (Covaris, 520140) while being kept in liquid nitrogen and repeatedly hammered to produce frozen tissue powder. Total RNA was extracted using NucleoSpin RNA XS (Macherey-Nagel, 740902). RNA-seq libraries were prepared using TruSeq stranded mRNA library prep kit (Illumina, 20020594). External RNA controls consortium (ERCC) RNA spike-in mixes (Thermo Fisher Scientific, 4456740) were included for quality assurance. RNA-seq libraries were sequenced in a paired-end mode using Illumina HiSeq 4000 and MGI DNBSEQ-G400 platform.
Paired-end reads were aligned to the reference genome (hg19 with ERCC) using STAR software v2.7.5 with default parameters. The raw read counts were quantified with RSEM based on a gene list obtained from GENCODE v38 by selecting protein coding genes and long noncoding RNAs (lncRNAs) with confidence levels 1 and 2 (n = 21,151). The count values were merged into a count matrix and quantile-normalized using preprocessCore R package across the samples. The cellular heterogeneity in individual samples was assessed with unique marker genes, and the overall gene expression pattern was adjusted iteratively on the basis of relative gene ERs across the cell types. Technical variations from experimental and sequencing batches were corrected using ComBat function in sva R package (53). To identify APOE isoform for each patient with PD, we conducted genotype profiling by implementing freebayes on the bulk RNA-seq samples, and the APOE isoform (ε2, ε3, and ε4) was determined on the basis of rs429358 and rs7412 genotype.
To identify the degree of intersection between PD-associated DEGs and SN tissue-specific expression, we incorporated the expression count matrix for bulk RNA-seq data encompassing 197 hippocampus, 209 frontal cortex, and 139 SN. The expression of SN tissues was compared iteratively with hippocampus and frontal cortex based on edgeR threshold of BH-adjusted P < 0.05, and the intersection of DEGs obtained from hippocampus and frontal cortex was compared to PD-associated DEGs identified by our snRNA-seq data.
Chromatin immunoprecipitation sequencing
To conduct H3K27ac ChIP-seq experiments from flash-frozen SN tissues, about 40 mg of tissue was placed in an extra-thick tissue processing tube (Covaris, 520140) while being kept in liquid nitrogen and repeatedly hammered into frozen tissue powder. The tissue sample was cross-linked in a cross-linking buffer of 100 mM NaCl, 0.1 mM EDTA, 5 mM Hepes (pH 8.0), and 1% formaldehyde for 10 min at room temperature. The cross-linking was quenched with 125 mM glycine for 5 min on a rotation and washed twice with ice-cold PBS. The samples were passed through a 30-μm strainer (Sysmex, 04-0042-2316) to remove excessive debris and suspended in SDS lysis buffer of 1% SDS, 50 mM tris-HCl (pH8.0), 10 mM EDTA, and protease inhibitor (Roche, 04-693-159-001). Chromatin fragmentation was performed by sonication (Covaris, S220) in the volume of 100 μl to obtain mono-, di-, and trinucleosome size chromatin. After centrifugation at 12,000g for 15 min at 4°C, the sonicated chromatin in supernatant was diluted 10 times with dilution buffer to achieve final concentration of 0.1% Triton X-100, 0.1% SDS, 150 mM NaCl, 15 mM tris-HCl (pH 8.0), 1 mM EDTA, and protease inhibitor for ChIP. The sonicated chromatin in supernatant was incubated with protein Dynabead (Thermo Fisher Scientific, 10001D) coated with anti-H3K27ac antibody (Active Motif, 39133) for 4 hours at 4°C with rotation, while a fraction of the input chromatin was stored to be used as an input control. The chromatin-antibody-bead complex was subjected to serial washing with varying salt concentrations optimized for the antibody used. The immunoprecipitated complex was treated with ribonuclease A (QIAGEN, 19101) and reverse–cross-linked overnight at 68°C. The immunoprecipitated DNA was recovered using AMPure XP beads (Beckman Coulter, A63881), and ChIP-seq libraries were prepared using NEBNext Ultra II DNA library Prep Kit [New England Biolabs (NEB), E7645] following the manufacturer’s instructions. The ChIP-seq libraries were sequenced in paired-end mode Illumina HiSeq 4000 and MGI DNBSEQ-G400 platform.
Quantification of cRE activity
The sequenced DNA reads from ChIP-seq libraries were mapped to the human reference genome (hg19) using Burrows-Wheeler aligner (BWA)-mem (ver. 0.7.17, “-M” option). Reads with a low alignment quality (MAPQ < 10) were removed, and polymerase chain reaction (PCR) duplicates were discarded using Picard (v2.6.0). We computed the number of ChIP-seq reads aligned in the 128,724 cREs identified on the basis of snATAC-seq data using bedtools (v2.29.1) coverage function. The read counts were merged into a count matrix and quantile-normalized using preprocessCore R package across the samples. The cellular heterogeneity in individual samples was assessed with unique marker cREs, and the global cRE activities were adjusted iteratively on the basis of relative cRE activity ratios across the cell types. Technical variations from experimental and sequencing batches were corrected using ComBat function in sva R package (53). We used the quasi-likelihood F test in EdgeR (25, 26) to identify dysregulated cREs with a multitest corrected (BH method) significance threshold (adjusted P < 0.05). We annotated active cell types for each dysregulated cRE based on the cell type reference obtained from snATAC-seq data.
Assessment and adjustment of cellular heterogeneity
To investigate the cellular heterogeneity in the bulk RNA-seq data generated from the SN tissues, we used 100 unique cell type marker genes for each cell type based on the cell type–resolved transcriptome identified by snRNA-seq data. For individual samples, we computed the sum of reads mapped to the marker genes from each cell type and the ratio of these summed reads across cell types to evaluate the relative composition of cell populations within the bulk data. The identified composition of reads in the unique marker genes considerably matched the cellular compositions obtained from single-nucleus sequencing datasets. For each cell type, we calculated the mean of relative compositions computed from the bulk samples, and then a relative cellular fraction (RCF) was obtained for a given cell type in a sample by dividing its relative composition to the mean. Then, cellularity-adjusted value (CAV) for gene i was computed on the basis of the RCF of a sample and the ER among eight cell types present in the SN as the following:
where {Cj} = {DopaN, GabaN, Oligo, OPC, Ast, Micro, Endo, and Peri}. This procedure was repeated three times until the variation of cellular compositions was minimal across the samples.
To examine the cellular heterogeneity in the bulk H3K27ac ChIP-seq data, we used the top 200 unique cell type marker cREs for each cell type based on the cell type–resolved chromatin accessibility identified by snATAC-seq data. Identical to the method applied to the bulk RNA-seq dataset, we computed the ratio of summed reads mapped to cell type marker cREs for individual samples, and the mean of relative compositions was calculated for each cell type. Then, the RCF was calculated for each cell type in a sample, and CAV for cRE i was computed on the basis of the RCF of the sample and the cRE activity ratio (CR) among eight cell types present in the SN as the following:
where {Cj} = {DopaN, GabaN, Oligo, OPC, Ast, Micro, Endo, and Peri}. This procedure was repeated three times until the variation of cellular compositions was minimal across the samples.
Colocalization of DEGs in proximity of dysregulated cREs
To examine the regulatory effect of dysregulated cREs on the surrounding genes, we conducted an enrichment test to measure the number of DEGs harbored by the dysregulated cREs in a given genomic window (100 kb), compared to random expectations. We created two sets of random groups, simulating both dysregulated cREs and DEGs. First, simulated cREs were generated by creating a set of genome coordinates that match the dysregulated cREs in number, size, and chromosome. Simulated DEGs were created by random gene sampling from the total gene set. The enrichment was measured on the basis of iterative trials (n = 10,000) considering the degree of DEGs colocalized in 100-kb window. The statistical significance was calculated in the form of empirical testing. The test was performed independently with respect to cell types and the type of cRE dysregulation.
PD GWAS-SNP imputation based on LD structure
PD-related GWAS-SNPs were collected from four GWAS summary statistics (4–6, 8), and tag GWAS-SNPs with the significance threshold (P < 5 × 10−8) were selected for downstream analysis. We expanded the GWAS-SNPs using LD structure. LDSCs were calculated using PLINK for five different populations, including African, American, East Asian, European, and South Asian, from 1000 genome phase 3 data. For each tag SNP, we identified associated SNPs that share a tight LDSC (r2 > 0.8) in at least three ethnic groups.
LDSC regression of disease heritability
To determine whether cell type–resolved cREs and dysregulated cREs are enriched with heritability of specific neurological and psychiatric disorders, we applied LDSC regression analysis (29, 54). We used four GWAS summary statistics for PD (4–6, 8). We obtained five additional GWAS summary statistics for AD (30, 31), amyotrophic lateral sclerosis (32), autism spectrum disorder (33), and schizophrenia (34). The cell type–resolved cREs and dysregulated cREs were tested for enrichment of heritability while controlling for the full baseline model.
Assessment of allelic bias on cRE activities for PD GWAS–matched variants
To determine genetic variations on cis-regulatory regions, we conducted genotype profiling by implementing freebayes on the bulk ChIP-seq samples. Then, the variants from each individual were matched with LD-expanded PD GWAS-SNPs. To assess the effects of PD GWAS-SNPs in cis-regulatory activity, we calculated the allelic bias on the heterozygous GWAS variants found in PD cases. For this, we first collected mapped ChIP-seq reads containing the variant position and compared the number of sequenced reads mapped to risk and nonrisk (reference) alleles of GWAS-SNPs. To rule out the alignment bias between reference and alternative alleles, we generated synthetic 100-bp reads covering the variant position. By performing alignment with reads in both risk and nonrisk alleles, we estimated variant-dependent alignment bias. We discarded variants showing BH-adjusted P < 0.05 from the binomial test in downstream analysis. The PD donors containing PD GWAS-SNPs greater than 100 were used to test the enrichment with cis-regulatory landscape across the cell types.
In situ Hi-C library preparation and data processing
In situ Hi-C experiments were performed on the SN tissues from PD and control cases (table S1). About 50 mg of tissue was placed in an extra-thick tissue processing tube (Covaris, 520140) and repeatedly hammered while being frozen in liquid nitrogen to produce tissue powder. The pulverized tissue was cross-linked with 1% formaldehyde. In situ Hi-C was conducted on the basis of the previously reported protocol with minor modifications (55). Cross-linked cells were lysed with 10 nM tris-HCl (pH 8.0), 10 mM NaCl, and 0.2% IGEPAL CA-630 (Sigma-Aldrich, 18896) and digested with 100 U of MboI (NEB, R0147). Digested fragments were labeled with biotin-14-dCTP (Invitrogen, 19518018) and proximally ligated with T4 DNA Ligase (NEB, M0202), followed by reverse–cross-linking with proteinase K (2 μg/μL; NEB, P8102), 1% SDS, and 500 mM NaCl overnight at 68°C. The DNA fragments were purified with AMPure XP beads (Beckman Coulter, A63880) and subjected to sonication (Covaris, S220). The ligated DNA fragments were pulled down with Dynabeads MyOne streptavidin T1 beads (Invitrogen, 65602) with thorough washing. Hi-C libraries were prepared manually by performing DNA end repair, removal of un-ligated ends, adenosine addition at 3′ end (NEB, M0212), ligation of Illumina indexed adapters (NEB, M2200), and PCR amplification (Thermo Fisher Scientific, F549). The number of cycles for PCR amplification was determined on the basis of KAPA library quantification kit (KAPA, KK4854). The Hi-C libraries were then subjected to deep sequencing in paired-end mode using Illumina HiSeq 4000 and X platforms.
The sequencing output from Hi-C libraries was mapped to the reference genome (hg19) using BWA-mem (“–M” option). In-house scripts were used to remove low-quality reads (MAPQ <10), the reads that span ligation sites, chimeric reads, and self-interacting reads (two fragments located within 5 kb). The chimeric reads were removed since they are biproducts of ligation chemistry in Hi-C library construction and cannot be properly processed by paired-end BWA-mem command. The read pairs were merged together as paired-end aligned BAM files, and PCR duplicates were removed with Picard (v2.6.0).
Significant chromatin interaction calling
Statistically significant, long-range chromatin interactions were identified at 5-kb resolution using Fit-Hi-C, as previously described (28). We created merged Hi-C BAM files with respect to control and PD status, converted them into an input format for Fit-Hi-C in each chromosome, and used the default Fit-Hi-C code to calculate the interaction significance between two genomic coordinates in 1-Mbp distance. A significance threshold (Q < 0.01) was used to define significant chromatin interactions. We defined the union of chromatin interactions obtained from PD and control SN as a general interaction set and classified promoter- and cRE-associated chromatin interactions by determining whether either bin of a chromatin interaction contained a TSS or a cRE. We labeled them as “none” if the bin in the chromatin interaction contained no regulatory element.
Calculation of ABC score
We applied a conceptually identical framework described in the ABC model (27), in which the quantitative effect of a cRE to a target gene depends on the frequency with which it contacts its promoter multiplied by the activity of the cRE. Briefly, ABC score for the effect of cRE i on gene j was measured as the following:
The ABC scores for all cRE to gene relationships within 1-Mb window were computed for individual cell types. The cRE activity was defined by the normalized snATAC-seq reads in the given cell type, and the contact frequency was represented by Hi-C contact frequency between the two bins containing the cRE and TSS of the target gene in 5-kb resolution. The position of a cRE was determined by the 5-kb genomic bin, in which the center of the cRE was located. When two cREs were present in one 5-kb bin, the sum of these cREs was used. Contact value was defined by cRE activity multiplied by contact frequency.
Integration of eQTL dataset from the human SN
The significant and all-paired eQTL associations from the human SN were downloaded from GTEx portal (v7). The significant eQTLs defined by GTEx (Q < 0.05) were first overlapped with dysregulated cREs, and the proportion of significant eQTLs in dysregulated cREs was compared to expectation to calculate the eQTL enrichment via two-sided Fisher’s exact test. The LD-expanded PD GWAS-SNPs in cis-regulatory region were matched with significant eQTLs, and the significance in enrichment compared to expectation was calculated identically. Then, the dysregulated cREs and PD GWAS-SNPs that matched with eQTLs were used to test the degree of overlap between target genes of eQTL association and significant chromatin interactions, by considering the number of eQTLs matched with the chromatin interactions. The significance in the overlap between eQTL and Hi-C target genes was calculated by performing hypergeometric test.
Putative target gene identification for dysregulated cREs and PD GWAS-SNPs
Putative target genes of dysregulated cREs were identified iteratively for each cell type based on the following criteria: (i) Dysregulated cRE and its target gene are connected by a significant interaction, (ii) dysregulated cRE and its target gene are annotated as an active cell type in the cell type–resolved transcriptomic and epigenomic reference, (iii) ABC score of dysregulated cRE to target gene relationship is greater than 10, and (iv) the Pearson’s correlation coefficient (PCC) between cRE activity and target gene expression across the samples based on bulk sequencing data is greater than 0.3. Putative target genes of PD GWAS-SNPs were identified for individual cell types based on the following data: (i) SNP-harboring cRE and its target gene are connected by a significant interaction, (ii) SNP-harboring cRE and its target gene are annotated as an active cell type in the cell type–resolved transcriptomic and epigenomic reference, and (iii) ABC score of SNP-harboring cRE to target gene relationship is greater than 10.
GO and mammalian phenotype analyses
EnrichR was used to identify biological processes for cell type–specific DEGs based on GO Biological Processes 2018 database. Similarly, Genomic Regions Enrichment of Annotations Tool (GREAT) was used to determine biological processes that are associated with dysregulated cREs. We used enrichR to identify mammalian phenotype overrepresented in putative target genes of dysregulated cREs and PD GWAS-SNPs based on MGI mammalian phenotype database (56). Among 4601 mammalian phenotype level 4 (2021) ontologies, we found 75 terms were enriched with the significance threshold of P < 0.05 and a gene count of >7. Then, 28 phenotype ontologies representing neurological, movement, and immune symptoms were manually selected by neuropathology experts. The full list of enriched phenotype ontologies and its associated gene set is provided in table S6. Phenotypes with no biological relevance to human diseases, including “lethality,” “death,” and “no abnormal phenotype,” were excluded.
CRISPR-Cas9 genome editing
We validated gene-to-regulatory sequence relationships identified by our Hi-C chromatin interactions. CRISPR-Cas9–mediated genome editing was conducted using ribonucleoprotein (RNP) delivery method in the SH-SY5Y cell line. Three CRISPR RNAs (crRNAs), along with trans-activating RNA (tracrRNA), were synthesized in vitro (Integrated DNA Technologies). To create a RNP complex, a guide RNA and tracrRNA were annealed, mixed with Cas9 nuclease (Enzynomics, M058HL), and incubated for 15 min at room temperature. The RNP complex was transfected into cells by electroporation with Neon transfection 10-μl kit (Thermo Fisher Scientific, MPK1096). To measure the efficiency of genome editing in each guide RNA, we performed targeted deep sequencing. For this, the genomic DNA was extracted from the transfected cells, and the target sites were amplified by PCR subsequently. Indices and sequencing adaptors were attached by additional PCR. High-throughput sequencing was performed using Illumina MiniSeq (San Diego, CA, USA). The mutation frequencies and patterns were analyzed using the Cas-Analyzer program implemented in CRISPR RGEN Tools (www.rgenome.net/). The cells were separated into single clones by serial dilutions on 96-well plates. After sufficient growth of each clone, the genotype was confirmed by conducting a Sanger sequencing of the target region from both directions. We selected mutant clones with the largest mutation size from each of the three guide RNAs from Sanger sequencing results and purified total RNA for reverse transcription quantitative PCR (RT-qPCR) to measure the relative mRNA expression levels of putative target genes. crisprRNA #1, TCTTGTGTGAAGAAACCCGTTGG; crisprRNA #2, GCCCAAACCGAAGCCCCCAAAGG; crisprRNA #3, AGCAACTCTCCTCCCTTTGGGGG; genotyping, TCGTCTGCCGAGGATGTA (forward) and AATTTCACGAATGCACCACAC (reverse); RT-qPCR primers: GAPDH, CCACTCCTCCACCTTTGACG (forward) and TTCGTTGTCATACCAGGAAATGAG (reverse); TOMM7, CGGAATGCCTGAACCAACT (forward) and GCCTTGTGCCATCCAACTA (reverse); KLHL7, CAGCAAGAAGAAGACCGAGAAG (forward) and GCAAGAACAACACGATGAGCAG (reverse); NUPL2, AAGTTTGGGAGTCGTCGGGA (forward) and CTTTTACGTCAGAGAGCAGAGC (reverse).
Analysis of motif disruption by PD GWAS-SNPs
To determine enriched motifs and TFs, we first obtained cell type–specific cREs using Signac’s FindAllMarkers function based on a threshold of Wilcoxon BH-adjusted P < 0.05, a log2 fold change of >1, and a minimum percent of nucleus detected >0.05, evaluating cRE activity in one cell type to the background of all other cell types. Next, Signac’s AddMotifs function was used to add motif information to each cRE using JASPAR2020 CORE vertebrate nonredundant database. The enrichment of binding motifs in cell type–specific cREs was calculated by performing the hypergeometric test in Signac’s FindMotifs function, and the motifs with a significant binding enrichment (P < 1 × 10−10) were selected. We also computed the deviation scores to evaluate the motif activity in per-cell basis by running chromVAR (35). Then, the TFs with a scaled deviation score greater than 1 and RNA detected at least 10% of nuclei were selected to determine TFs that are highly active and expressed in each cell type.
To examine the association of PD GWAS-SNPs with TF binding disruption, we constructed a synthetic genome containing the risk alleles of PD GWAS-SNPs. Motif binding scores of enriched TFs were computed on each GWAS-SNP–containing cRE using FIMO (57) for both risk and nonrisk (reference) alleles. The default P value threshold of FIMO was changed to 0.99 to account for all binding possibilities, and the binding score for each GWAS-SNP–containing cRE was defined by the sum of −log10(P) of all binding combinations within the cRE. The delta binding score was calculated by the difference in binding score between risk and nonrisk alleles for each GWAS-SNP–containing cRE, and a delta binding score greater than 3 was used to define cREs with gained and lost TF binding. Next, to evaluate the transcriptional effect of the disrupted motifs, we collected GWAS-matched genetic variants obtained from each PD donor and identified cREs with disrupted binding (delta binding score > 3). Then, the putative target genes of these disrupted motifs were identified on the basis of the chromatin interaction map using a threshold of cumulative ABC score > 1.
Analysis of modular gene expression patterns
For the 656 putative target genes obtained, a correlation matrix where each entry indicates a similarity score between two putative target genes was prepared by computing the PCC based on 16 bulk RNA-seq samples. The correlation matrix was subjected to a hierarchical clustering (Pearson correlation metric with average linkage), which presented nine distinct gene clusters at a dendrogram height threshold of 0.65. Enriched biological processes of protein-coding genes in clusters from C1 to C9 were determined using Metascape (v3.5). The enrichment of target genes based on the cell type and the type of dysregulated cREs in each cluster was evaluated using the one-sided exact binomial test. The corresponding significance values were multiple testing corrected for the number of cell type annotations.
Acknowledgments
We thank the members of Jung laboratory, B. Ren, and A. Singleton for their support and critical suggestions throughout the course of this work.
Funding: This work was funded by the Korean Ministry of Health and Welfare (HI19C0256 to I.J.), the Ministry of Science and ICT through the National Research Foundation in the Republic of Korea (2020R1A2C4001464 and 2022R1A5A1026413 to I.J.), Suh Kyungbae Foundation (to I.J.), and the Ministry of Education through the National Research Foundation in the Republic of Korea (2022R1I1A1A0106292912 to A.J.L.).
Author contributions: A.J.L., C.K., E.M., and I.J. conceived the study. A.J.L., S.P., J.J., and J.E. performed the sequencing library preparation. A.J.L. and K.J. performed molecular experiments. A.J.L. conducted the bioinformatics analysis with assistance from S.P., J.J., B.C., and D.Y. C.K. and E.M. provided the human brain samples. C.K. and R.A.R. performed histopathologic quality control for clinical specimens. C.K., E.M., S.-J.L., S.J.C., and J.C. contributed to the result interpretation. A.J.L. prepared the manuscript with assistance from C.K., E.M., and I.J. All authors read and commented on the manuscript.
Competing interests: S.-J.L. is a founder and CEO of Neuramedy Co. Ltd. The authors declare that they have no other competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. All raw sequencing data are deposited in the GEO database under accession number GSE148434. The additional data for seven snRNA-seq libraries from the human SN used in the analysis can be downloaded from GEO (GSE140231). The snATAC-seq data derived from two human SN specimens are available under GEO accession number GSE147672. All other data and materials can be provided by the corresponding authors pending scientific review and a completed material transfer agreement. Requests for all other data and materials should be submitted to ijung@kaist.ac.kr. Custom code supporting this work has been uploaded to Zenodo (https://zenodo.org/record/6979139).
Supplementary Materials
This PDF file includes:
Figs. S1 to 14
Legends for tables S1 to S10
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S10
REFERENCES AND NOTES
- 1.J. Jankovic, Parkinson’s disease: Clinical features and diagnosis. J. Neurol. Neurosurg. Psychiatry 79, 368–376 (2008). [DOI] [PubMed] [Google Scholar]
- 2.A. B. Singleton, M. Farrer, J. Johnson, A. Singleton, S. Hague, J. Kachergus, M. Hulihan, T. Peuralinna, A. Dutra, R. Nussbaum, S. Lincoln, A. Crawley, M. Hanson, D. Maraganore, C. Adler, M. R. Cookson, M. Muenter, M. Baptista, D. Miller, J. Blancato, J. Hardy, K. Gwinn-Hardy, α-Synuclein locus triplication causes Parkinson’s disease. Science 302, 841 (2003). [DOI] [PubMed] [Google Scholar]
- 3.V. Bonifati, P. Rizzu, M. J. van Baren, O. Schaap, G. J. Breedveld, E. Krieger, M. C. Dekker, F. Squitieri, P. Ibanez, M. Joosse, J. W. van Dongen, N. Vanacore, J. C. van Swieten, A. Brice, G. Meco, C. M. van Duijn, B. A. Oostra, P. Heutink, Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science 299, 256–259 (2003). [DOI] [PubMed] [Google Scholar]
- 4.D. Chang, M. A. Nalls, I. B. Hallgrimsdottir, J. Hunkapiller, M. van der Brug, F. Cai; International Parkinson’s Disease Genomics Consortium; 23andMe Research Team, G. A. Kerchner, G. Ayalon, B. Bingol, M. Sheng, D. Hinds, T. W. Behrens, A. B. Singleton, T. R. Bhangale, R. R. Graham, A meta-analysis of genome-wide association studies identifies 17 new Parkinson’s disease risk loci. Nat. Genet. 49, 1511–1516 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.M. A. Nalls, N. Pankratz, C. M. Lill, C. B. Do, D. G. Hernandez, M. Saad, A. L. DeStefano, E. Kara, J. Bras, M. Sharma, C. Schulte, M. F. Keller, S. Arepalli, C. Letson, C. Edsall, H. Stefansson, X. Liu, H. Pliner, J. H. Lee, R. Cheng; International Parkinson’s Disease Genomics Consortium (IPDGC); Parkinson’s Study Group (PSG) Parkinson’s Research: The Organized GENetics Initiative (PROGENI); 23andMe; GenePD; NeuroGenetics Research Consortium (NGRC); Hussman Institute of Human Genomics (HIHG); Ashkenazi Jewish Dataset Investigator; Cohorts for Health and Aging Research in Genetic Epidemiology (CHARGE); North American Brain Expression Consortium (NABEC); United Kingdom Brain Expression Consortium (UKBEC); Greek Parkinson’s Disease Consortium; Alzheimer Genetic Analysis Group, M. A. Ikram, J. P. Ioannidis, G. M. Hadjigeorgiou, J. C. Bis, M. Martinez, J. S. Perlmutter, A. Goate, K. Marder, B. Fiske, M. Sutherland, G. Xiromerisiou, R. H. Myers, L. N. Clark, K. Stefansson, J. A. Hardy, P. Heutink, H. Chen, N. W. Wood, H. Houlden, H. Payami, A. Brice, W. K. Scott, T. Gasser, L. Bertram, N. Eriksson, T. Foroud, A. B. Singleton, Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat. Genet. 46, 989–993 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.J. N. Foo, L. C. Tan, I. D. Irwan, W. L. Au, H. Q. Low, K. M. Prakash, A. Ahmad-Annuar, J. Bei, A. Y. Chan, C. M. Chen, Y. C. Chen, S. J. Chung, H. Deng, S. Y. Lim, V. Mok, H. Pang, Z. Pei, R. Peng, H. F. Shang, K. Song, A. H. Tan, Y. R. Wu, T. Aung, C. Y. Cheng, F. T. Chew, S. H. Chew, S. A. Chong, R. P. Ebstein, J. Lee, S. M. Saw, A. Seow, M. Subramaniam, E. S. Tai, E. N. Vithana, T. Y. Wong, K. K. Heng, W. Y. Meah, C. C. Khor, H. Liu, F. Zhang, J. Liu, E. K. Tan, Genome-wide association study of Parkinson’s disease in East Asians. Hum. Mol. Genet. 26, 226–232 (2017). [DOI] [PubMed] [Google Scholar]
- 7.S. J. Chung, S. M. Armasu, J. M. Biernacka, K. J. Anderson, T. G. Lesnick, D. N. Rider, J. M. Cunningham, J. Eric Ahlskog, R. Frigerio, D. M. Maraganore, Genomic determinants of motor and cognitive outcomes in Parkinson’s disease. Parkinsonism Relat. Disord. 18, 881–886 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.M. A. Nalls, C. Blauwendraat, C. L. Vallerga, K. Heilbron, S. Bandres-Ciga, D. Chang, M. Tan, D. A. Kia, A. J. Noyce, A. Xue, J. Bras, E. Young, R. von Coelln, J. Simón-Sánchez, C. Schulte, M. Sharma, L. Krohn, L. Pihlstrøm, A. Siitonen, H. Iwaki, H. Leonard, F. Faghri, J. R. Gibbs, D. G. Hernandez, S. W. Scholz, J. A. Botia, M. Martinez, J.-C. Corvol, S. Lesage, J. Jankovic, L. M. Shulman, M. Sutherland, P. Tienari, K. Majamaa, M. Toft, O. A. Andreassen, T. Bangale, A. Brice, J. Yang, Z. Gan-Or, T. Gasser, P. Heutink, J. M. Shulman, N. W. Wood, D. A. Hinds, J. A. Hardy, H. R. Morris, J. Gratten, P. M. Visscher, R. R. Graham; 23andMe Research Team; System Genomics of Parkinson’s Disease Consortium; International Parkinson’s Disease Genomics Consortium , Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: A meta-analysis of genome-wide association studies. Lancet Neurol. 18, 1091–1102 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.C. Blauwendraat, M. A. Nalls, A. B. Singleton, The genetic architecture of Parkinson’s disease. Lancet Neurol. 19, 170–178 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.M. T. Maurano, R. Humbert, E. Rynes, R. E. Thurman, E. Haugen, H. Wang, A. P. Reynolds, R. Sandstrom, H. Qu, J. Brody, A. Shafer, F. Neri, K. Lee, T. Kutyavin, S. Stehling-Sun, A. K. Johnson, T. K. Canfield, E. Giste, M. Diegel, D. Bates, R. S. Hansen, S. Neph, P. J. Sabo, S. Heimfeld, A. Raubitschek, S. Ziegler, C. Cotsapas, N. Sotoodehnia, I. Glass, S. R. Sunyaev, R. Kaul, J. A. Stamatoyannopoulos, Systematic localization of common disease-associated variation in regulatory DNA. Science 337, 1190–1195 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.I. Jung, A. Schmitt, Y. Diao, A. J. Lee, T. Liu, D. Yang, C. Tan, J. Eom, M. Chan, S. Chee, Z. Chiang, C. Kim, E. Masliah, C. L. Barr, B. Li, S. Kuan, D. Kim, B. Ren, A compendium of promoter-centered long-range chromatin interactions in the human genome. Nat. Genet. 51, 1442–1449 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.L. Toker, G. T. Tran, J. Sundaresan, O. B. Tysnes, G. Alves, K. Haugarvoll, G. S. Nido, C. Dolle, C. Tzoulis, Genome-wide histone acetylation analysis reveals altered transcriptional regulation in the Parkinson’s disease brain. Mol. Neurodegener. 16, 31 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.M. R. Corces, A. Shcherbina, S. Kundu, M. J. Gloudemans, L. Fresard, J. M. Granja, B. H. Louie, T. Eulalio, S. Shams, S. T. Bagdatli, M. R. Mumbach, B. Liu, K. S. Montine, W. J. Greenleaf, A. Kundaje, S. B. Montgomery, H. Y. Chang, T. J. Montine, Single-cell epigenomic analyses implicate candidate causal variants at inherited risk loci for Alzheimer’s and Parkinson’s diseases. Nat. Genet. 52, 1158–1168 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.S. Morabito, E. Miyoshi, N. Michael, S. Shahin, A. C. Martini, E. Head, J. Silva, K. Leavy, M. Perez-Rosendahl, V. Swarup, Single-nucleus chromatin accessibility and transcriptomic characterization of Alzheimer’s disease. Nat. Genet. 53, 1143–1155 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.M. J. Fullwood, M. H. Liu, Y. F. Pan, J. Liu, H. Xu, Y. B. Mohamed, Y. L. Orlov, S. Velkov, A. Ho, P. H. Mei, E. G. Chew, P. Y. Huang, W. J. Welboren, Y. Han, H. S. Ooi, P. N. Ariyaratne, V. B. Vega, Y. Luo, P. Y. Tan, P. Y. Choy, K. D. Wansa, B. Zhao, K. S. Lim, S. C. Leow, J. S. Yow, R. Joseph, H. Li, K. V. Desai, J. S. Thomsen, Y. K. Lee, R. K. Karuturi, T. Herve, G. Bourque, H. G. Stunnenberg, X. Ruan, V. Cacheux-Rataboul, W. K. Sung, E. T. Liu, C. L. Wei, E. Cheung, Y. Ruan, An oestrogen-receptor-alpha-bound human chromatin interactome. Nature 462, 58–64 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.E. Lieberman-Aiden, N. L. van Berkum, L. Williams, M. Imakaev, T. Ragoczy, A. Telling, I. Amit, B. R. Lajoie, P. J. Sabo, M. O. Dorschner, R. Sandstrom, B. Bernstein, M. A. Bender, M. Groudine, A. Gnirke, J. Stamatoyannopoulos, L. A. Mirny, E. S. Lander, J. Dekker, Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.M. R. Mumbach, A. J. Rubin, R. A. Flynn, C. Dai, P. A. Khavari, W. J. Greenleaf, H. Y. Chang, HiChIP: Efficient and sensitive analysis of protein-directed genome architecture. Nat. Methods 13, 919–922 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.R. Fang, M. Yu, G. Li, S. Chee, T. Liu, A. D. Schmitt, B. Ren, Mapping of long-range chromatin interactions by proximity ligation-assisted ChIP-seq. Cell Res. 26, 1345–1348 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.A. Nott, I. R. Holtman, N. G. Coufal, J. C. M. Schlachetzki, M. Yu, R. Hu, C. Z. Han, M. Pena, J. Xiao, Y. Wu, Z. Keulen, M. P. Pasillas, C. O’Connor, C. K. Nickl, S. T. Schafer, Z. Shen, R. A. Rissman, J. B. Brewer, D. Gosselin, D. D. Gonda, M. L. Levy, M. G. Rosenfeld, G. McVicker, F. H. Gage, B. Ren, C. K. Glass, Brain cell type–specific enhancer-promoter interactome maps and disease-risk association. Science 366, 1134–1139 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.D. Agarwal, C. Sandor, V. Volpato, T. M. Caffrey, J. Monzon-Sandoval, R. Bowden, J. Alegre-Abarrategui, R. Wade-Martins, C. Webber, A single-cell atlas of the human substantia nigra reveals cell-specific pathways associated with neurological disorders. Nat. Commun. 11, 4183 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Y. Hao, S. Hao, E. Andersen-Nissen, W. M. Mauck III, S. Zheng, A. Butler, M. J. Lee, A. J. Wilk, C. Darby, M. Zager, P. Hoffman, M. Stoeckius, E. Papalexi, E. P. Mimitou, J. Jain, A. Srivastava, T. Stuart, L. M. Fleming, B. Yeung, A. J. Rogers, J. M. McElrath, C. A. Blish, R. Gottardo, P. Smibert, R. Satija, Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.T. Stuart, A. Srivastava, S. Madad, C. A. Lareau, R. Satija, Single-cell chromatin state analysis with Signac. Nat. Methods 18, 1333–1341 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.T. Kamath, A. Abdulraouf, S. J. Burris, J. Langlieb, V. Gazestani, N. M. Nadaf, K. Balderrama, C. Vanderburg, E. Z. Macosko, Single-cell genomic profiling of human dopamine neurons identifies a population that selectively degenerates in Parkinson’s disease. Nat. Neurosci. 25, 588–595 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.K. J. Billingsley, S. Bandres-Ciga, S. Saez-Atienzar, A. B. Singleton, Genetic risk factors in Parkinson’s disease. Cell Tissue Res. 373, 9–20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.M. D. Robinson, D. J. McCarthy, G. K. Smyth, edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.D. J. McCarthy, Y. Chen, G. K. Smyth, Differential expression analysis of multifactor RNA-seq experiments with respect to biological variation. Nucleic Acids Res. 40, 4288–4297 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.C. P. Fulco, J. Nasser, T. R. Jones, G. Munson, D. T. Bergman, V. Subramanian, S. R. Grossman, R. Anyoha, B. R. Doughty, T. A. Patwardhan, T. H. Nguyen, M. Kane, E. M. Perez, N. C. Durand, C. A. Lareau, E. K. Stamenova, E. L. Aiden, E. S. Lander, J. M. Engreitz, Activity-by-contact model of enhancer-promoter regulation from thousands of CRISPR perturbations. Nat. Genet. 51, 1664–1669 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.F. Ay, T. L. Bailey, W. S. Noble, Statistical confidence estimation for Hi-C data reveals regulatory chromatin contacts. Genome Res. 24, 999–1011 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.B. K. Bulik-Sullivan, P. R. Loh, H. K. Finucane, S. Ripke, J. Yang; Schizophrenia Working Group of the Psychiatric Genomics Consortium, N. Patterson, M. J. Daly, A. L. Price, B. M. Neale, LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.I. E. Jansen, J. E. Savage, K. Watanabe, J. Bryois, D. M. Williams, S. Steinberg, J. Sealock, I. K. Karlsson, S. Hagg, L. Athanasiu, N. Voyle, P. Proitsi, A. Witoelar, S. Stringer, D. Aarsland, I. S. Almdahl, F. Andersen, S. Bergh, F. Bettella, S. Bjornsson, A. Braekhus, G. Brathen, C. de Leeuw, R. S. Desikan, S. Djurovic, L. Dumitrescu, T. Fladby, T. J. Hohman, P. V. Jonsson, S. J. Kiddle, A. Rongve, I. Saltvedt, S. B. Sando, G. Selbaek, M. Shoai, N. G. Skene, J. Snaedal, E. Stordal, I. D. Ulstein, Y. Wang, L. R. White, J. Hardy, J. Hjerling-Leffler, P. F. Sullivan, W. M. van der Flier, R. Dobson, L. K. Davis, H. Stefansson, K. Stefansson, N. L. Pedersen, S. Ripke, O. A. Andreassen, D. Posthuma, Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat. Genet. 51, 404–413 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.B. W. Kunkle, B. Grenier-Boley, R. Sims, J. C. Bis, V. Damotte, A. C. Naj, A. Boland, M. Vronskaya, S. J. van der Lee, A. Amlie-Wolf, C. Bellenguez, A. Frizatti, V. Chouraki, E. R. Martin, K. Sleegers, N. Badarinarayan, J. Jakobsdottir, K. L. Hamilton-Nelson, S. Moreno-Grau, R. Olaso, R. Raybould, Y. Chen, A. B. Kuzma, M. Hiltunen, T. Morgan, S. Ahmad, B. N. Vardarajan, J. Epelbaum, P. Hoffmann, M. Boada, G. W. Beecham, J. G. Garnier, D. Harold, A. L. Fitzpatrick, O. Valladares, M. L. Moutet, A. Gerrish, A. V. Smith, L. Qu, D. Bacq, N. Denning, X. Jian, Y. Zhao, M. Del Zompo, N. C. Fox, S. H. Choi, I. Mateo, J. T. Hughes, H. H. Adams, J. Malamon, F. Sanchez-Garcia, Y. Patel, J. A. Brody, B. A. Dombroski, M. C. D. Naranjo, M. Daniilidou, G. Eiriksdottir, S. Mukherjee, D. Wallon, J. Uphill, T. Aspelund, L. B. Cantwell, F. Garzia, D. Galimberti, E. Hofer, M. Butkiewicz, B. Fin, E. Scarpini, C. Sarnowski, W. S. Bush, S. Meslage, J. Kornhuber, C. C. White, Y. Song, R. C. Barber, S. Engelborghs, S. Sordon, D. Voijnovic, P. M. Adams, R. Vandenberghe, M. Mayhaus, L. A. Cupples, M. S. Albert, P. P. De Deyn, W. Gu, J. J. Himali, D. Beekly, A. Squassina, A. M. Hartmann, A. Orellana, D. Blacker, E. Rodriguez-Rodriguez, S. Lovestone, M. E. Garcia, R. S. Doody, C. Munoz-Fernadez, R. Sussams, H. Lin, T. J. Fairchild, Y. A. Benito, C. Holmes, H. Karamujic-Comic, M. P. Frosch, H. Thonberg, W. Maier, G. Roshchupkin, B. Ghetti, V. Giedraitis, A. Kawalia, S. Li, R. M. Huebinger, L. Kilander, S. Moebus, I. Hernández, M. I. Kamboh, R. Brundin, J. Turton, Q. Yang, M. J. Katz, L. Concari, J. Lord, A. S. Beiser, C. D. Keene, S. Helisalmi, I. Kloszewska, W. A. Kukull, A. M. Koivisto, A. Lynch, L. Tarraga, E. B. Larson, A. Haapasalo, B. Lawlor, T. H. Mosley, R. B. Lipton, V. Solfrizzi, M. Gill, W. T. Longstreth Jr., T. J. Montine, V. Frisardi, M. Diez-Fairen, F. Rivadeneira, R. C. Petersen, V. Deramecourt, I. Alvarez, F. Salani, A. Ciaramella, E. Boerwinkle, E. M. Reiman, N. Fievet, J. I. Rotter, J. S. Reisch, O. Hanon, C. Cupidi, A. G. A. Uitterlinden, D. R. Royall, C. Dufouil, R. G. Maletta, I. de Rojas, M. Sano, A. Brice, R. Cecchetti, P. S. George-Hyslop, K. Ritchie, M. Tsolaki, D. W. Tsuang, B. Dubois, D. Craig, C. K. Wu, H. Soininen, D. Avramidou, R. L. Albin, L. Fratiglioni, A. Germanou, L. G. Apostolova, L. Keller, M. Koutroumani, S. E. Arnold, F. Panza, O. Gkatzima, S. Asthana, D. Hannequin, P. Whitehead, C. S. Atwood, P. Caffarra, H. Hampel, I. Quintela, A. Carracedo, L. Lannfelt, D. C. Rubinsztein, L. L. Barnes, F. Pasquier, L. Frolich, S. Barral, B. McGuinness, T. G. Beach, J. A. Johnston, J. T. Becker, P. Passmore, E. H. Bigio, J. M. Schott, T. D. Bird, J. D. Warren, B. F. Boeve, M. K. Lupton, J. D. Bowen, P. Proitsi, A. Boxer, J. F. Powell, J. R. Burke, J. S. K. Kauwe, J. M. Burns, M. Mancuso, J. D. Buxbaum, U. Bonuccelli, N. J. Cairns, A. McQuillin, C. Cao, G. Livingston, C. S. Carlson, N. J. Bass, C. M. Carlsson, J. Hardy, R. M. Carney, J. Bras, M. M. Carrasquillo, R. Guerreiro, M. Allen, H. C. Chui, E. Fisher, C. Masullo, E. A. Crocco, C. DeCarli, G. Bisceglio, M. Dick, L. Ma, R. Duara, N. R. Graff-Radford, D. A. Evans, A. Hodges, K. M. Faber, M. Scherer, K. B. Fallon, M. Riemenschneider, D. W. Fardo, R. Heun, M. R. Farlow, H. Kolsch, S. Ferris, M. Leber, T. M. Foroud, I. Heuser, D. R. Galasko, I. Giegling, M. Gearing, M. Hüll, D. H. Geschwind, J. R. Gilbert, J. Morris, R. C. Green, K. Mayo, J. H. Growdon, T. Feulner, R. L. Hamilton, L. E. Harrell, D. Drichel, L. S. Honig, T. D. Cushion, M. J. Huentelman, P. Hollingworth, C. M. Hulette, B. T. Hyman, R. Marshall, G. P. Jarvik, A. Meggy, E. Abner, G. E. Menzies, L. W. Jin, G. Leonenko, L. M. Real, G. R. Jun, C. T. Baldwin, D. Grozeva, A. Karydas, G. Russo, J. A. Kaye, R. Kim, F. Jessen, N. W. Kowall, B. Vellas, J. H. Kramer, E. Vardy, F. M. LaFerla, K. H. Jockel, J. J. Lah, M. Dichgans, J. B. Leverenz, D. Mann, A. I. Levey, S. Pickering-Brown, A. P. Lieberman, N. Klopp, K. L. Lunetta, H. E. Wichmann, C. G. Lyketsos, K. Morgan, D. C. Marson, K. Brown, F. Martiniuk, C. Medway, D. C. Mash, M. M. Nothen, E. Masliah, N. M. Hooper, W. C. McCormick, A. Daniele, S. M. McCurry, A. Bayer, A. N. McDavid, J. Gallacher, A. C. McKee, H. van den Bussche, M. Mesulam, C. Brayne, B. L. Miller, S. Riedel-Heller, C. A. Miller, J. W. Miller, A. Al-Chalabi, J. C. Morris, C. E. Shaw, A. J. Myers, J. Wiltfang, S. O’Bryant, J. M. Olichney, V. Alvarez, J. E. Parisi, A. B. Singleton, H. L. Paulson, J. Collinge, W. R. Perry, S. Mead, E. Peskind, D. H. Cribbs, M. Rossor, A. Pierce, N. S. Ryan, W. W. Poon, B. Nacmias, H. Potter, S. Sorbi, J. F. Quinn, E. Sacchinelli, A. Raj, G. Spalletta, M. Raskind, C. Caltagirone, P. Bossù, M. D. Orfei, B. Reisberg, R. Clarke, C. Reitz, A. D. Smith, J. M. Ringman, D. Warden, E. D. Roberson, G. Wilcock, E. Rogaeva, A. C. Bruni, H. J. Rosen, M. Gallo, R. N. Rosenberg, Y. Ben-Shlomo, M. A. Sager, P. Mecocci, A. J. Saykin, P. Pastor, M. L. Cuccaro, J. M. Vance, J. A. Schneider, L. S. Schneider, S. Slifer, W. W. Seeley, A. G. Smith, J. A. Sonnen, S. Spina, R. A. Stern, R. H. Swerdlow, M. Tang, R. E. Tanzi, J. Q. Trojanowski, J. C. Troncoso, V. M. Van Deerlin, L. J. Van Eldik, H. V. Vinters, J. P. Vonsattel, S. Weintraub, K. A. Welsh-Bohmer, K. C. Wilhelmsen, J. Williamson, T. S. Wingo, R. L. Woltjer, C. B. Wright, C. E. Yu, L. Yu, Y. Saba, A. Pilotto, M. J. Bullido, O. Peters, P. K. Crane, D. Bennett, P. Bosco, E. Coto, V. Boccardi, P. L. DeJager, A. Lleo, N. Warner, O. L. Lopez, M. Ingelsson, P. Deloukas, C. Cruchaga, C. Graff, R. Gwilliam, M. Fornage, A. M. Goate, P. Sanchez-Juan, P. G. Kehoe, N. Amin, N. Ertekin-Taner, C. Berr, S. Debette, S. Love, L. J. Launer, S. G. Younkin, J. F. Dartigues, C. Corcoran, M. A. Ikram, D. W. Dickson, G. Nicolas, D. Campion, J. Tschanz, H. Schmidt, H. Hakonarson, J. Clarimon, R. Munger, R. Schmidt, L. A. Farrer, C. Van Broeckhoven, M. C. O’Donovan, A. L. DeStefano, L. Jones, J. L. Haines, J. F. Deleuze, M. J. Owen, V. Gudnason, R. Mayeux, V. Escott-Price, B. M. Psaty, A. Ramirez, L. S. Wang, A. Ruiz, C. M. van Duijn, P. A. Holmans, S. Seshadri, J. Williams, P. Amouyel, G. D. Schellenberg, J. C. Lambert, M. A. Pericak-Vance; Alzheimer Disease Genetics Consortium (ADGC); European Alzheimer’s Disease Initiative (EADI); Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium (CHARGE); Genetic and Environmental Risk in AD/Defining Genetic, Polygenic and Environmental Risk for Alzheimer’s Disease Consortium (GERAD/PERADES) , Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Abeta, tau, immunity and lipid processing. Nat. Genet. 51, 414–430 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.W. van Rheenen, A. Shatunov, A. M. Dekker, R. L. McLaughlin, F. P. Diekstra, S. L. Pulit, R. A. van der Spek, U. Vosa, S. de Jong, M. R. Robinson, J. Yang, I. Fogh, P. T. van Doormaal, G. H. Tazelaar, M. Koppers, A. M. Blokhuis, W. Sproviero, A. R. Jones, K. P. Kenna, K. R. van Eijk, O. Harschnitz, R. D. Schellevis, W. J. Brands, J. Medic, A. Menelaou, A. Vajda, N. Ticozzi, K. Lin, B. Rogelj, K. Vrabec, M. Ravnik-Glavac, B. Koritnik, J. Zidar, L. Leonardis, L. D. Groselj, S. Millecamps, F. Salachas, V. Meininger, M. de Carvalho, S. Pinto, J. S. Mora, R. Rojas-Garcia, M. Polak, S. Chandran, S. Colville, R. Swingler, K. E. Morrison, P. J. Shaw, J. Hardy, R. W. Orrell, A. Pittman, K. Sidle, P. Fratta, A. Malaspina, S. Topp, S. Petri, S. Abdulla, C. Drepper, M. Sendtner, T. Meyer, R. A. Ophoff, K. A. Staats, M. Wiedau-Pazos, C. Lomen-Hoerth, V. M. Van Deerlin, J. Q. Trojanowski, L. Elman, L. McCluskey, A. N. Basak, C. Tunca, H. Hamzeiy, Y. Parman, T. Meitinger, P. Lichtner, M. Radivojkov-Blagojevic, C. R. Andres, C. Maurel, G. Bensimon, B. Landwehrmeyer, A. Brice, C. A. Payan, S. Saker-Delye, A. Dürr, N. W. Wood, L. Tittmann, W. Lieb, A. Franke, M. Rietschel, S. Cichon, M. M. Nöthen, P. Amouyel, C. Tzourio, J. F. Dartigues, A. G. Uitterlinden, F. Rivadeneira, K. Estrada, A. Hofman, C. Curtis, H. M. Blauw, A. J. van der Kooi, M. de Visser, A. Goris, M. Weber, C. E. Shaw, B. N. Smith, O. Pansarasa, C. Cereda, R. Del Bo, G. P. Comi, S. D’Alfonso, C. Bertolin, G. Soraru, L. Mazzini, V. Pensato, C. Gellera, C. Tiloca, A. Ratti, A. Calvo, C. Moglia, M. Brunetti, S. Arcuti, R. Capozzo, C. Zecca, C. Lunetta, S. Penco, N. Riva, A. Padovani, M. Filosto, B. Muller, R. J. Stuit; PARALS Registry; SLALOM Group; SLAP Registry; FALS Sequencing Consortium; SLAGEN Consortium; NNIPPS Study Group, I. Blair, K. Zhang, E. P. McCann, J. A. Fifita, G. A. Nicholson, D. B. Rowe, R. Pamphlett, M. C. Kiernan, J. Grosskreutz, O. W. Witte, T. Ringer, T. Prell, B. Stubendorff, I. Kurth, C. A. Hubner, P. N. Leigh, F. Casale, A. Chio, E. Beghi, E. Pupillo, R. Tortelli, G. Logroscino, J. Powell, A. C. Ludolph, J. H. Weishaupt, W. Robberecht, P. Van Damme, L. Franke, T. H. Pers, R. H. Brown, J. D. Glass, J. E. Landers, O. Hardiman, P. M. Andersen, P. Corcia, P. Vourc’h, V. Silani, N. R. Wray, P. M. Visscher, P. I. de Bakker, M. A. van Es, R. J. Pasterkamp, C. M. Lewis, G. Breen, A. Al-Chalabi, L. H. van den Berg, J. H. Veldink, Genome-wide association analyses identify new risk variants and the genetic architecture of amyotrophic lateral sclerosis. Nat. Genet. 48, 1043–1048 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.J. Grove, S. Ripke, T. D. Als, M. Mattheisen, R. K. Walters, H. Won, J. Pallesen, E. Agerbo, O. A. Andreassen, R. Anney, S. Awashti, R. Belliveau, F. Bettella, J. D. Buxbaum, J. Bybjerg-Grauholm, M. Bækvad-Hansen, F. Cerrato, K. Chambert, J. H. Christensen, C. Churchhouse, K. Dellenvall, D. Demontis, S. De Rubeis, B. Devlin, S. Djurovic, A. L. Dumont, J. I. Goldstein, C. S. Hansen, M. E. Hauberg, M. V. Hollegaard, S. Hope, D. P. Howrigan, H. Huang, C. M. Hultman, L. Klei, J. Maller, J. Martin, A. R. Martin, J. L. Moran, M. Nyegaard, T. Nærland, D. S. Palmer, A. Palotie, C. B. Pedersen, M. G. Pedersen, T. dPoterba, J. B. Poulsen, B. S. Pourcain, P. Qvist, K. Rehnström, A. Reichenberg, J. Reichert, E. B. Robinson, K. Roeder, P. Roussos, E. Saemundsen, S. Sandin, F. K. Satterstrom, G. D. Smith, H. Stefansson, S. Steinberg, C. R. Stevens, P. F. Sullivan, P. Turley, G. B. Walters, X. Xu; Autism Spectrum Disorder Working Group of the Psychiatric Genomics Consortium; BUPGEN; Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium; 23andMe Research Team, K. Stefansson, D. H. Geschwind, M. Nordentoft, D. M. Hougaard, T. Werge, O. Mors, P. B. Mortensen, B. M. Neale, M. J. Daly, A. D. Børglum, Identification of common genetic risk variants for autism spectrum disorder. Nat. Genet. 51, 431–444 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.A. F. Pardinas, P. Holmans, A. J. Pocklington, V. Escott-Price, S. Ripke, N. Carrera, S. E. Legge, S. Bishop, D. Cameron, M. L. Hamshere, J. Han, L. Hubbard, A. Lynham, K. Mantripragada, E. Rees, J. H. MacCabe, S. A. McCarroll, B. T. Baune, G. Breen, E. M. Byrne, U. Dannlowski, T. C. Eley, C. Hayward, N. G. Martin, A. M. McIntosh, R. Plomin, D. J. Porteous, N. R. Wray, A. Caballero, D. H. Geschwind, L. M. Huckins, D. M. Ruderfer, E. Santiago, P. Sklar, E. A. Stahl, H. Won, E. Agerbo, T. D. Als, O. A. Andreassen, M. Baekvad-Hansen, P. B. Mortensen, C. B. Pedersen, A. D. Borglum, J. Bybjerg-Grauholm, S. Djurovic, N. Durmishi, M. G. Pedersen, V. Golimbet, J. Grove, D. M. Hougaard, M. Mattheisen, E. Molden, O. Mors, M. Nordentoft, M. Pejovic-Milovancevic, E. Sigurdsson, T. Silagadze, C. S. Hansen, K. Stefansson, H. Stefansson, S. Steinberg, S. Tosato, T. Werge, G. Consortium, C. Consortium, D. A. Collier, D. Rujescu, G. Kirov, M. J. Owen, M. C. O’Donovan, J. T. R. Walters, Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat. Genet. 50, 381–389 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.A. N. Schep, B. Wu, J. D. Buenrostro, W. J. Greenleaf, chromVAR: Inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat. Methods 14, 975–978 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.L. Gan, M. R. Cookson, L. Petrucelli, A. R. La Spada, Converging pathways in neurodegeneration, from genetics to mechanisms. Nat. Neurosci. 21, 1300–1309 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.A. Elkouzi, V. Vedam-Mai, R. S. Eisinger, M. S. Okun, Emerging therapies in Parkinson disease - repurposed drugs and new approaches. Nat. Rev. Neurol. 15, 204–223 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.I. Alecu, S. A. L. Bennett, Dysregulated lipid metabolism and its role in α-synucleinopathy in Parkinson’s disease. Front. Neurosci. 13, 328 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.H. Michelakakis, G. Xiromerisiou, E. Dardiotis, M. Bozi, D. Vassilatis, P. M. Kountra, G. Patramani, M. Moraitou, D. Papadimitriou, E. Stamboulis, L. Stefanis, E. Zintzaras, G. M. Hadjigeorgiou, Evidence of an association between the scavenger receptor class B member 2 gene and Parkinson’s disease. Mov. Disord. 27, 400–405 (2012). [DOI] [PubMed] [Google Scholar]
- 40.C. Blauwendraat, K. Heilbron, C. L. Vallerga, S. Bandres-Ciga, R. von Coelln, L. Pihlstrom, J. Simon-Sanchez, C. Schulte, M. Sharma, L. Krohn, A. Siitonen, H. Iwaki, H. Leonard, A. J. Noyce, M. Tan, J. R. Gibbs, D. G. Hernandez, S. W. Scholz, J. Jankovic, L. M. Shulman, S. Lesage, J. C. Corvol, A. Brice, J. J. van Hilten, J. Marinus; 23andMe Research Team, J. Eerola-Rautio, P. Tienari, K. Majamaa, M. Toft, D. G. Grosset, T. Gasser, P. Heutink, J. M. Shulman, N. Wood, J. Hardy, H. R. Morris, D. A. Hinds, J. Gratten, P. M. Visscher, Z. Gan-Or, M. A. Nalls, A. B. Singleton; International Parkinson’s Disease Genomics Consortium (IPDGC) , Parkinson’s disease age at onset genome-wide association study: Defining heritability, genetic loci, and α-synuclein mechanisms. Mov. Disord. 34, 866–875 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.J. Bové, M. Martínez-Vicente, B. Dehay, C. Perier, A. Recasens, A. Bombrun, B. Antonsson, M. Vila, BAX channel activity mediates lysosomal disruption linked to Parkinson disease. Autophagy 10, 889–900 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.D. M. Arduino, A. R. Esteves, S. M. Cardoso, Mitochondria drive autophagy pathology via microtubule disassembly: A new hypothesis for Parkinson disease. Autophagy 9, 112–114 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.J. Obergasteiger, G. Frapporti, P. P. Pramstaller, A. A. Hicks, M. Volta, A new hypothesis for Parkinson’s disease pathogenesis: GTPase-p38 MAPK signaling and autophagy as convergence points of etiology and genomics. Mol. Neurodegener. 13, 40 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.E. C. Hirsch, S. Hunot, Neuroinflammation in Parkinson’s disease: A target for neuroprotection? Lancet Neurol. 8, 382–397 (2009). [DOI] [PubMed] [Google Scholar]
- 45.J. S. Schweitzer, B. Song, T. M. Herrington, T. Y. Park, N. Lee, S. Ko, J. Jeon, Y. Cha, K. Kim, Q. Li, C. Henchcliffe, M. Kaplitt, C. Neff, O. Rapalino, H. Seo, I. H. Lee, J. Kim, T. Kim, G. A. Petsko, J. Ritz, B. M. Cohen, S. W. Kong, P. Leblanc, B. S. Carter, K. S. Kim, Personalized iPSC-derived dopamine progenitor cells for Parkinson’s disease. N. Engl. J. Med. 382, 1926–1932 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.H. Braak, E. Braak, Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 82, 239–259 (1991). [DOI] [PubMed] [Google Scholar]
- 47.H. Heaton, A. M. Talman, A. Knights, M. Imaz, D. J. Gaffney, R. Durbin, M. Hemberg, M. K. N. Lawniczak, Souporcell: Robust clustering of single-cell RNA-seq data by genotype without reference genotypes. Nat. Methods 17, 615–620 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.S. L. Wolock, R. Lopez, A. M. Klein, Scrublet: Computational identification of cell doublets in single-cell transcriptomic data. Cell Syst. 8, 281–291.e9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.I. Korsunsky, N. Millard, J. Fan, K. Slowikowski, F. Zhang, K. Wei, Y. Baglaenko, M. Brenner, P. R. Loh, S. Raychaudhuri, Fast, sensitive and accurate integration of single-cell data with harmony. Nat. Methods 16, 1289–1296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.D. van Dijk, R. Sharma, J. Nainys, K. Yim, P. Kathail, A. J. Carr, C. Burdziak, K. R. Moon, C. L. Chaffer, D. Pattabiraman, B. Bierie, L. Mazutis, G. Wolf, S. Krishnaswamy, D. Pe’er, Recovering gene interactions from single-cell data using data diffusion. Cell 174, 716–729.e27 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.E. Fiskin, C. A. Lareau, L. S. Ludwig, G. Eraslan, F. Liu, A. M. Ring, R. J. Xavier, A. Regev, Single-cell profiling of proteins and chromatin accessibility using PHAGE-ATAC. Nat. Biotechnol. 40, 374–381 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.G. Finak, A. McDavid, M. Yajima, J. Deng, V. Gersuk, A. K. Shalek, C. K. Slichter, H. W. Miller, M. J. McElrath, M. Prlic, P. S. Linsley, R. Gottardo, MAST: A flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 16, 278 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.J. T. Leek, W. E. Johnson, H. S. Parker, A. E. Jaffe, J. D. Storey, The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28, 882–883 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.H. K. Finucane, Y. A. Reshef, V. Anttila, K. Slowikowski, A. Gusev, A. Byrnes, S. Gazal, P. R. Loh, C. Lareau, N. Shoresh, G. Genovese, A. Saunders, E. Macosko, S. Pollack, C. Brainstorm, J. R. B. Perry, J. D. Buenrostro, B. E. Bernstein, S. Raychaudhuri, S. McCarroll, B. M. Neale, A. L. Price, Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat. Genet. 50, 621–629 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.S. S. Rao, M. H. Huntley, N. C. Durand, E. K. Stamenova, I. D. Bochkov, J. T. Robinson, A. L. Sanborn, I. Machol, A. D. Omer, E. S. Lander, E. L. Aiden, A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.C. L. Smith, J. T. Eppig, The mammalian phenotype ontology: Enabling robust annotation and comparative analysis. Wiley Interdiscip. Rev. Syst. Biol. Med. 1, 390–399 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.C. E. Grant, T. L. Bailey, W. S. Noble, FIMO: Scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to 14
Legends for tables S1 to S10
Tables S1 to S10