Abstract
Recent studies have described a DNA damage tolerance pathway choice that involves a competition between PrimPol-mediated repriming and fork reversal. Screening different translesion DNA synthesis (TLS) polymerases by the use of tools for their depletion, we identified a unique role of Pol ι in regulating such a pathway choice. Pol ι deficiency unleashes PrimPol-dependent repriming, which accelerates DNA replication in a pathway that is epistatic with ZRANB3 knockdown. In Pol ι–depleted cells, the excess participation of PrimPol in nascent DNA elongation reduces replication stress signals, but thereby also checkpoint activation in S phase, triggering chromosome instability in M phase. This TLS-independent function of Pol ι requires its PCNA-interacting but not its polymerase domain. Our findings unravel an unanticipated role of Pol ι in protecting the genome stability of cells from detrimental changes in DNA replication dynamics caused by PrimPol.
Downregulation of DNA Pol ι augments repriming at nascent DNA, diminishes checkpoint and triggers chromosome instability.
INTRODUCTION
DNA damage is frequent in cells, and DNA repair mechanisms are not efficient enough to prevent the encounter of replication forks with damaged DNA (1). The highly processive replicative DNA polymerases stall at these locations as they cannot accommodate damaged DNA in their active sites (2). Then, specialized DNA polymerases, capable of using damaged DNA as replication templates, are recruited to the replisome to perform translesion DNA synthesis (TLS) across damaged DNA, safeguarding DNA replication continuity (2). Different specialized DNA polymerases are critical for TLS across specific DNA lesions. For example, polymerase (Pol) eta (η) from the Y family of DNA polymerases is essential for TLS across cyclobutane pyrimidine dimers (CPDs) (2), while Pol kappa (κ) is required for TLS across benzo[a]pyrenediol-epoxide (BPDE)–induced DNA lesions (3), and it promotes DNA synthesis and replication stress recovery at stalled forks under conditions of nucleotide deprivation (4). Pol zeta (ζ), from the B family of DNA polymerases, can bypass and extend tracks initiated by other specialized polymerases (3). REV1 is central for switching from replicative DNA polymerases to TLS polymerases at forks that encounter DNA lesions (3) and may have a role as a platform during TLS (3). On the other hand, PrimPol promotes DNA replication-restart downstream of DNA lesions (5–7), including replication traverse of DNA interstrand cross-links (8). While a crucial role for each specialized DNA polymerase during DNA replication events across at least one type of DNA lesion has been identified, Pol iota (ι) is considered a backup enzyme with no essential role in TLS (9).
During the past years, the role of specialized polymerases in TLS-independent DNA replication events has emerged. For example, we have recently proposed a role of Pol ι in fork reversal (10). Pol κ was reported to increase the abundance of single-stranded DNA/double-stranded DNA (ssDNA/dsDNA) hybrids at replication forks to promote checkpoint activation (11). Specialized polymerases also participate in DNA repair, as is the case for Pol η and Pol κ in nucleotide excision repair (12–15), Pol ζ in DNA interstrand cross-link repair (16), or Pol ι in base excision repair, yet the latter to a very limited extent (17). They also contribute to DNA replication under unperturbed conditions, for example, Pol η is required to duplicate DNA at hard-to-replicate regions (18). These results indicate that the role of specialized DNA polymerases in the DDR (DNA damage response) exceeds their involvement in TLS. Given their positive contribution to different aspects of DDR, it is expected that the loss of any specialized DNA polymerase should cause a similar increment of the levels of DNA damage–induced replication stress (4, 19).
We reasoned that a side-by-side comparison, based on the levels of a DNA damage–induced global DDR marker, such as is the case for γH2AX, would indicate the extent of augmentation of replication stress that results from the loss of each specialized DNA polymerase. Hence, γH2AX accumulation was assessed in cancer cells depleted from each of the abovementioned DNA polymerases and treated with the widely used chemotherapeutic agent cisplatin (CDDP) (16). γH2AX increased in cells depleted of most DNA polymerases tested, with the notable exception of Pol ι, whose depletion caused an unexpected reduction in γH2AX and other stress markers. Such alterations depended on the excess participation of PrimPol in DNA replication. In the absence of Pol ι, PrimPol promoted repriming during nascent DNA elongation and consequent accumulation of postreplicative ssDNA gaps. Such a change in the DNA damage tolerance pathway favored a premature and checkpoint-blind S phase finalization, followed by augmented levels of under-replicated DNA in the M phase, aberrant anaphases, and micronuclei. Together, these results unraveled the role of Pol ι in coupling DNA elongation rates and checkpoint activation, which prevents rapid Sphase progression and untimely M phase progression to prevent the accumulation of genomically unstable cells.
RESULTS
γH2AX up-regulation after DNA damage is impaired in Pol ι–depleted cells
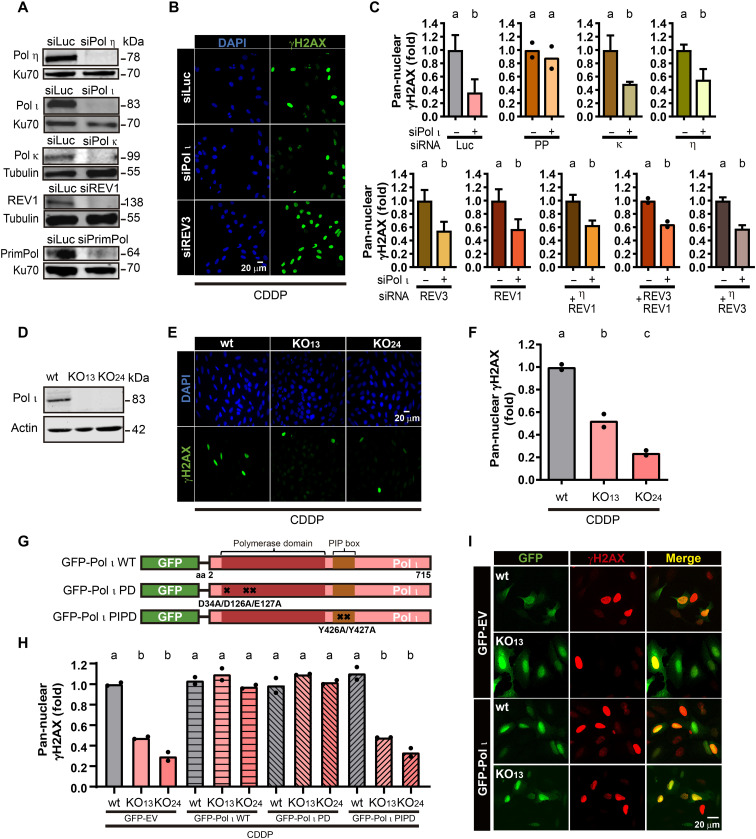
To explore the level of redundancy between different specialized DNA polymerases, we transiently down-regulated the ones with a reported role in DNA damage tolerance: η (2), ι (10), REV1 (3), κ (3), REV3 (3), and PrimPol (8) in U2OS cells using small interfering RNA (siRNA) technology. Down-regulations were validated using antibodies against the indicated endogenously expressed DNA polymerases (Fig. 1A) or quantitative reverse transcription polymerase chain reaction (qRT-PCR) and also verified after transfecting green fluorescent protein (GFP)–tagged versions of the indicated DNA polymerases (fig. S1A). We focused our analysis on CDDP as it is broadly used for cancer treatment and is a well-known trigger for TLS activation. In agreement with a previous report (20), CDDP causes a steep up-regulation of γH2AX in the vast majority of the cells, and the accumulation of massive replication stress revealed as pan-nuclear γH2AX in a portion of the cells (fig. S1B). We focused our initial analysis on the levels of pan-nuclear γH2AX, and samples were evaluated 24 hours after CDDP treatment. As previously reported (20), in control samples (Luc-siRNA transfection), CDDP treatment caused a notable accumulation of pan-nuclear and focal γH2AX (fig. S1B). In agreement with the known contribution of these polymerases to the DDR, single or combined depletions of most polymerases increased the percentage of cells with pan-nuclear accumulation of γH2AX, albeit to different extents (fig. S1C and representative images in Fig. 1B). These results imply that replication stress increases when these specialized DNA polymerases are missing, suggesting a contribution to the DDR post-CDDP. Notably, however, Pol ι depletion caused the opposite effect on the levels of pan-nuclear γH2AX (fig. S1C and representative images in Fig. 1B), which was more clearly evidenced when the effect of Pol ι knockdown was expressed relative to the induction of pan-nuclear γH2AX in control samples (siLuc) or double knockdown relative to single DNA polymerase knockdown (Fig. 1C). This means that Pol ι, opposite to other specialized DNA polymerases, augments (rather than prevents) replication stress post-CDDP. Notably, such a stimulatory effect of Pol ι on pan-nuclear γH2AX accumulation is also revealed under conditions of concomitant depletion of other specialized polymerases (Fig. 1C and fig. S1C), implying that the effect of Pol ι on pan-nuclear γH2AX accumulation is independent of other specialized polymerases. These observations were verified using a second siRNA (fig. S1D), Pol ι knockout (KO) clones generated in U2OS cells using CRISPR-Cas9 technology (Fig. 1, D to F, and fig. S1E), and Pol ι depletion in HCT116 and RPE-1 cells [fig. S1, F and G; note that in RPE-1 cells (fig. S1G), pan-nuclear γH2AX was barely detectable, which is why γH2AX intensity was analyzed]. Impaired accumulation of γH2AX in two independent Pol ι KO clones was rescued when transiently introducing exogenous Pol ι and a polymerase-dead (PD) mutant but not a mutant that cannot interact with proliferating cell nuclear antigen (PCNA) (Fig. 1, G to I). These mutants were previously described in (21) and (22), respectively, were similarly expressed after transfection (percentages of transfection of 30 to 40%), and were validated in their capacity to form nuclear foci after ultraviolet (UV) treatment (fig. S2, A and B). These data confirmed the requirement of Pol ι for optimal γH2AX accumulation and indicated that such a role of Pol ι is associated with its loading to PCNA but not its ability to synthesize DNA. The focal organization of 53BP1, another replication stress marker, was also impaired in Pol ι knockdown cells (fig. S2, C and D). In conclusion, Pol ι depletion mitigated the accumulation of pan-nuclear γH2AX and 53BP1 foci after CDDP treatment.
Fig. 1. Depletion of Pol ι mitigates pan-nuclear γH2AX accumulation after CDDP treatment.
(A) Western blot showing the down-regulation of the indicated DNA polymerases in U2OS samples. Ku70 and tubulin were used as loading controls. (B) Representative fields showing pan-nuclear γH2AX in U2OS cells treated with CDDP (6 μg/ml). (C) Fold change in the number of U2OS nuclei with pan-nuclear γH2AX; at least 450 nuclei per condition were quantified in two or three independent experiments. Statistical analysis for this and all experiments in this article: values labeled with the same letter are not significantly different. Significant differences have a P < 0.05 or lower; see Materials and Methods for a detailed explanation (statistics: Student’s t test was performed for each pair of samples). (D) Western blot of samples from Pol ι wild-type control (wt) and Pol ι KO U2OS clones developed with a Pol ι antibody and actin was used as a loading control. (E) Representative images showing pan-nuclear γH2AX staining in control (wt) and Pol ι KO U2OS clones KO13 and KO24 treated with CDDP (12 μg/ml). (F) Fold changes in pan-nuclear γH2AX–positive cells in control (wt) and Pol ι KO U2OS cells treated with CDDP (12 μg/ml). More than 300 nuclei per sample were analyzed. Dots represent the individual values of two independent experiments [statistics: one-way analysis of variance (ANOVA) and Tukey post-test]. (G) Schematic representations of GFP–Pol ι wild-type (WT) and point mutations that disrupt the polymerase and the PCNA-binding domains. PD, polymerase dead; PIPD, PCNA-interacting domain dead. (H) Fold changes in pan-nuclear γH2AX in control (wt) and Pol ι KO U2OS cells transfected with GFP-EV (empty vector), GFP–Pol ι WT, PD, and PIPD, treated with CDDP (12 μg/ml). More than 200 GFP-positive cells per sample were analyzed. Dots represent the individual values of two independent experiments (statistics: one-way ANOVA and Tukey post-test). (I) Representative images from the experiment shown in (H).
Different checkpoint-associated markers are down-regulated in Pol ι–depleted cells
To test whether the positive contribution of Pol ι to the accumulation of pan-nuclear γH2AX and 53BP1 nuclear foci was restricted to CDDP treatment, we transiently silenced Pol ι or used the KO clones and applied treatments that activate DDR via distinct lesions such as after exposure to hydroxyurea or UV irradiation. Again, Pol ι depletion caused a reduction in pan-nuclear γH2AX and 53BP1 nuclear foci accumulation after those treatments (fig. S3).
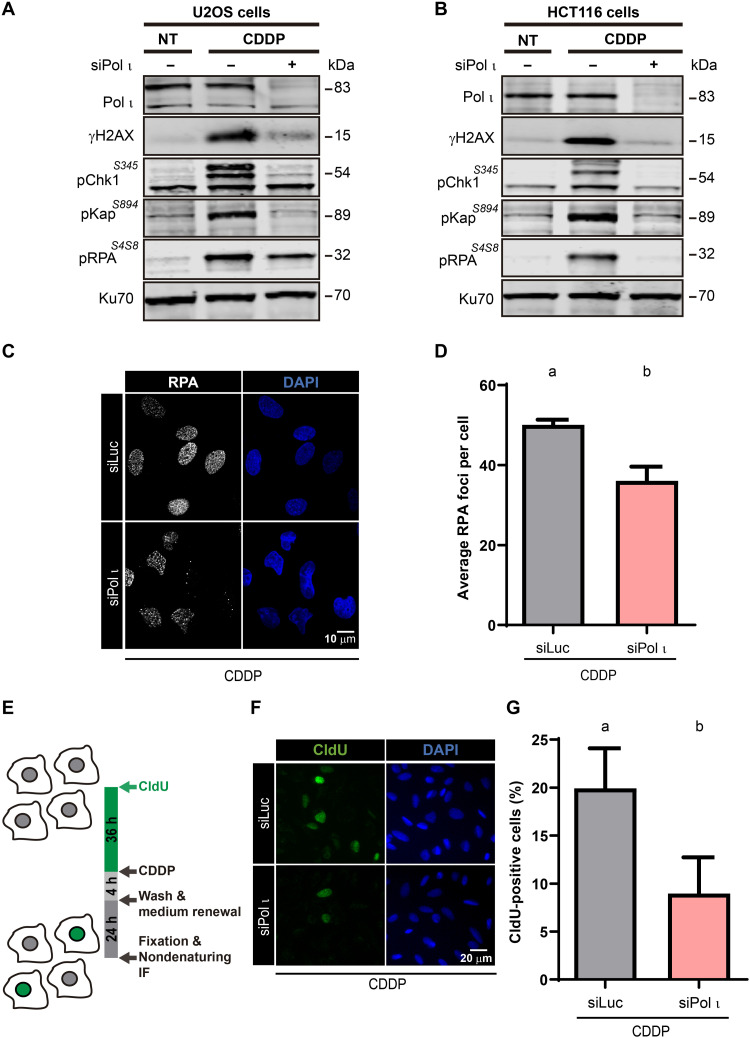
The DDR markers assessed above accumulate due to the activation of checkpoint sensors and effectors (23). Therefore, we wondered if Pol ι depletion affects other DDR proteins phosphorylated by checkpoint sensors and effector kinases. The phosphorylation of different marker proteins after CDDP treatment was reduced in Pol ι–depleted cells when compared to control samples, including the phosphorylation of Chk1 at Ser345, which revealed ATR activation (24), and the phosphorylation of KAP1 at Ser824 and RPA at Ser4/8, which are markers of double-strand break (DSB)–triggered signaling (Fig. 2A) (25, 26). Results obtained with the KO clones were in agreement with those generated using siPol ι (fig. S4A). We also detected similar defects in other cell lines subjected to Pol ι depletion (Fig. 2B and fig. S4, B to E). Together, these results demonstrate that Pol ι depletion or elimination causes a global down-regulation of DDR markers and suggest a common cause for the down-regulation of such DDR markers. We, therefore, evaluated whether Pol ι knockdown or KO ruled over the levels of CDDP-induced RPA foci in U2OS cells (Fig. 2, C and D, and fig. S4, F and G). This result suggests that Pol ι promotes the accumulation of ssDNA. To more directly test this possibility, we performed a 36-hour 5-chloro-2′-deoxyuridine (CldU) or 5-bromo-2′-deoxyuridine (BrdU) incorporation followed by a nondenaturing detection of the thymidine analog (Fig. 2E) (20), which revealed the accumulation of stretches of ssDNA in Pol ι–depleted (Fig. 2, F and G) or KO cells (fig. S4H). Together, these results suggest that the focal accumulation of RPA and its downstream signals are reduced at an early stage in Pol ι–deficient cells, implying a requirement of Pol ι for the accumulation of ssDNA during DNA replication.
Fig. 2. Pol ι depletion impairs checkpoint activation.
(A) Western blot analysis of U2OS cells transfected with the indicated siRNAs treated with CDDP (6 μg/ml) or mock treatment (NT). Checkpoint effectors (γH2AX, pChk1S345, pKapS894, and pRPAS4S8) were detected with specific antibodies. Ku70 was used as a loading control. (B) Western blot analysis of HCT116 cells treated and processed as in (A). (C) Representative fields of U2OS cells treated with CDDP (6 μg/ml) and subjected to RPA immunodetection. (D) Average number of RPA foci (mean ± SD) from the experiments shown in (C). More than 250 cells per sample were analyzed in three independent experiments (statistics: one-way ANOVA and Tukey post-test). (E) Schematic representation of the protocol used to quantify ssDNA under nondenaturing conditions. (F) Representative fields of cells treated as described in (E). (G) Percentages (mean ± SD) of CldU-positive U2OS cells transfected with control siRNA (siLuc) or siPol ι and treated with CDDP (6 μg/ml). More than 400 nuclei per sample were analyzed in three independent experiments (statistics: Student’s t test).
Pol ι depletion promotes nascent DNA synthesis in a manner that is independent of the up-regulation of TLS or NER events
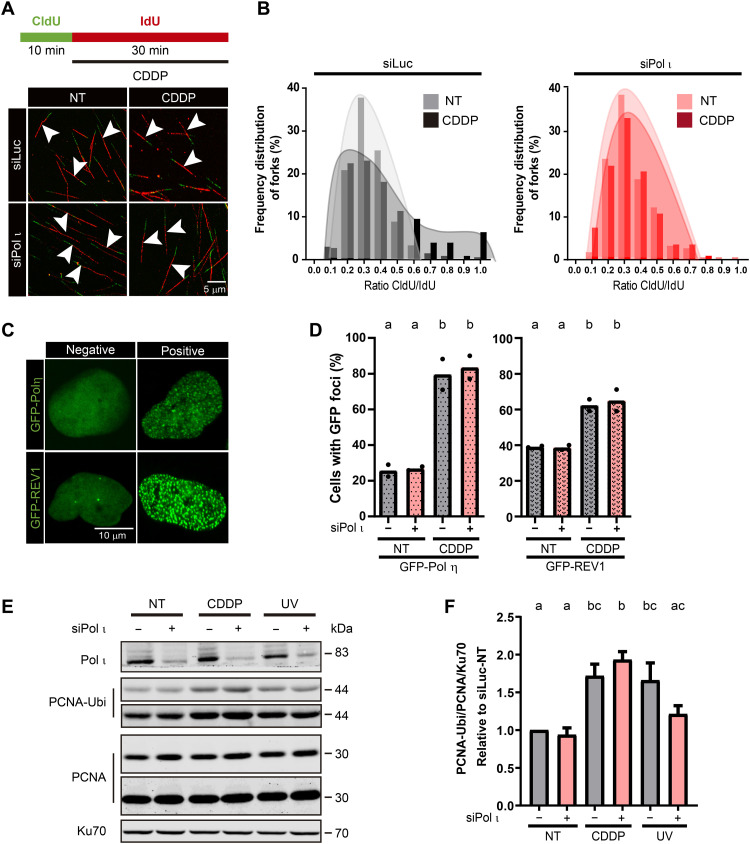
To understand the mechanism by which Pol ι contributes to checkpoint activation, we first analyzed the DNA replication dynamics in control and Pol ι–depleted samples. As a consequence of the stalling of replication forks, the accumulation of DNA damage is expected to cause the shortening of nascent replication tracks and the spreading of undamaged/damaged track ratios obtained from dual-colored DNA molecules (27). Accumulated evidence indicates that the ratios between a first track reflecting unperturbed replication and a second track reflecting replication of damaged DNA should increase and spread when TLS polymerases are lost (27, 28). Accordingly, we performed the fiber assay using 10-min CldU followed by 30-min 5-iodo-2′-deoxyuridine (IdU) (Fig. 3A). As anticipated, in control samples, in which the replication speed for CldU and IdU is unaltered, the CldU/IdU ratio was ≅0.33, revealing a constant speed during incorporation times of 10 and 30 min (Fig. 3B and fig. S5A). As expected, in siLuc control samples, CDDP treatment reduced the length of the second track, increasing the ratios with the additional spreading of the values as compared to the mock treatment (Fig. 3B and fig. S5B). While it could be speculated that shortening of the second track is the consequence of augmented template DNA breakage at DNA damage sites, previous work from our laboratory indicates that DNA shortening after DNA damage is not due to fork breakage, as DNA counterstaining revealed that only a small fraction (less than 10%) of forks localized on discontinuous DNA after DNA damage (28). Also, the DNA track length was insignificantly modified when excluding the broken fibers from the quantification (28). Notably, in Pol ι–depleted samples, the ratio was maintained at 0.33 even after CDDP treatment (fig. S5C) without spreading the track ratios in treated versus untreated cells (Fig. 3B). Notably, these results suggest that replication barriers do not cause fork stalling or DNA breaks in Pol ι–depleted samples.
Fig. 3. Pol ι depletion facilitates nascent DNA synthesis across replication barriers.
(A) Top: Labeling protocol used for the fiber assay in this and all figures that follow, if not indicated otherwise. Bottom: Representative DNA fiber spread fields of U2OS cells transfected with siLuc or siPol ι with or without CDDP (1 μM) exposure. White arrows indicate single DNA fibers that were quantified. (B) Distribution of CldU/IdU ratios (frequency) of U2OS cells transfected with siLuc or siPol ι with or without CDDP (1 μM) treatment. At least 180 fibers per condition were quantified in total in two independent experiments. (C) Representative images of U2OS nuclei transfected with GFP–Pol η or GFP-REV1 with or without focal organization of the indicated DNA polymerases. (D) Percentage (mean) of U2OS cells with nuclear foci of GFP–Pol η and GFP-REV1. Samples were transfected with siLuc and siPol ι, treated with CDDP (6 μg/ml). Two hundred GFP-positive nuclei were quantified. Dots represent individual values of two independent experiments (statistics: one-way ANOVA and Tukey post-test). (E) Western blot analysis of samples from control and Pol ι–depleted U2OS either mock-treated (NT) or treated with CDDP or UV irradiation. PCNA ubiquitination (Ubi) and PCNA-specific antibodies were used. Ku70 was used as a loading control. Pol ι levels from the same samples were developed on a separate membrane. (F) Quantification of PCNA ubiquitination from four independent Western blot gels (statistics: one-way ANOVA and Tukey post-test).
Another way to evaluate TLS efficiency is to study the focal organization of TLS polymerases. After DNA damage formation, specialized polymerases accumulate into nuclear foci that colocalize with replication factories (29); such relocation of specialized polymerases to sites of DNA synthesis indicates TLS activation (19). When a specialized TLS polymerase is lost, other TLS polymerases attempt to compensate for their functional loss (30, 31). However, Pol ι depletion did not change the efficiency of other specialized polymerases’ recruitment to such nuclear locations after exposure to CDDP (Fig. 3, C and D), UV irradiation (fig. S5D), or hydroxyurea (fig. S5E), suggesting that the contribution of Pol ι to the replication dynamics is not because of a role of Pol ι in TLS. Another marker of TLS activation is PCNA monoubiquitination (32), but Pol ι depletion did not cause an up-regulation of this TLS parameter (Fig. 3, E and F). Hence, Pol ι depletion was not compensated by the accumulation of TLS markers.
Another possibility that could explain faster replication speed is the more efficient removal of DNA lesions by nucleotide excision repair (NER) (12), which is the DNA replication mechanism that removes most CDDP- and UV-induced DNA lesions (33). We, therefore, analyzed the repair kinetics of the UV-induced DNA damage CPDs in control versus Pol ι–depleted samples. CPD removal was not affected by the levels of Pol ι (fig. S5, F and G). Hence, Pol ι knockdown disfavors the stalling of forks at DNA lesions in a manner independent of its participation in TLS or NER events.
Pol ι depletion leads to faster replication and decreased replication stress markers under untreated conditions
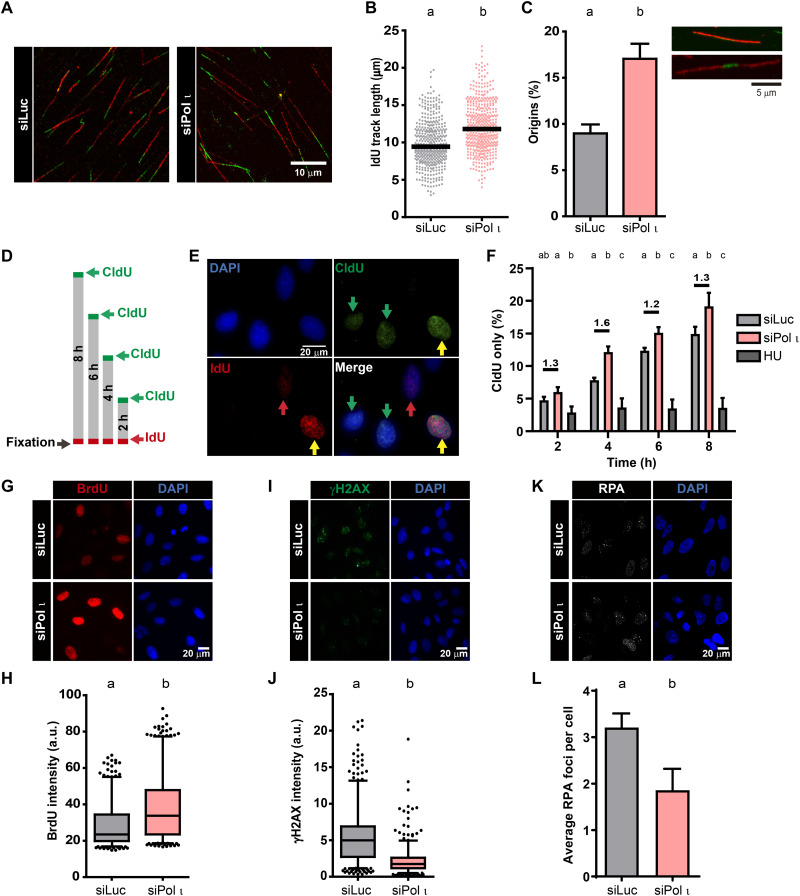
We then moved forward to analyze the replication program of Pol ι–depleted cells under unperturbed conditions and noticed longer DNA tracks when Pol ι was depleted or knocked out (approximately 25% increase in track length; see Fig. 4, A and B, and fig. S6, A and B). Accelerated nascent DNA elongation in Pol ι KO cells was reverted by transfection of exogenous Pol ι, demonstrating the specificity for Pol ι (fig. S6C). These data suggest that Pol ι depletion unleashes nascent DNA elongation in a manner that is independent of exogenous DNA damage. Furthermore, we found an epistatic effect of simultaneous depletion of Pol ι and p53 on restricting excessive DNA elongation (fig. S7, A and B). This result suggests that Pol ι activity at replication forks is linked to functions of the p53 tumor suppressor, in agreement with our previous reports (10, 34).
Fig. 4. Pol ι depletion speeds up S phase and reduces replication stress.
(A) Representative DNA fibers from U2OS cells. (B) IdU track length quantification. At least 280 fibers per condition were analyzed in three independent experiments. Median is shown in black (statistics: Mann-Whitney U test). (C) Percentage of origin firing (mean ± SD) analyzed in (B) [(red-green-red + red only fibers)/total fibers]. At least 500 fibers per condition were analyzed in three independent experiments (statistics: Student’s t test). (D) Schematic representation of experiments in (F). Cells were labeled with CldU (15′, green boxes), incubated with fresh medium (gray boxes), and labeled with IdU (15′, red boxes). (E) Representative field analyzed in (F). Green arrows: Cells transiting S phase during the CldU but not the IdU pulse. Red arrows: Cells entering S phase during the incorporation of the IdU pulse. Yellow arrows: Cells incorporating both analogs. (F) Quantification of cells incorporating CldU only (green arrows) in two (for 0.75 mM HU) or three (for siLuc and siPol ι) independent experiments. At least 300 cells per sample were analyzed per condition. Number above each bar indicates their fold difference (statistics: each time point was compared using a one-way ANOVA and Tukey post-test). (G) Representative fields of U2OS cells pulse-labeled with BrdU for 15 min before fixation. (H) Quantification of BrdU intensity (box and whiskers plot with 5 to 95% confidence interval). At least 350 positive BrdU nuclei per condition were analyzed in three independent experiments (statistics: Mann-Whitney U test). (I) Representative fields of γH2AX intensity in U2OS cells. (J) Quantification of γH2AX intensity (box and whiskers plot with 5 to 95% confidence interval). At least 500 nuclei per condition were analyzed in two independent experiments (statistics: Mann-Whitney U test). (K) Representative fields revealing RPA foci in U2OS cells. (L) Quantification (mean ± SD) of RPA foci per cell. At least 80 nuclei per sample were analyzed in three independent experiments (statistics: Student’s t test).
In addition to altering fork speed, Pol ι depletion also dysregulated origin firing (Fig. 4C). Given that we observed longer tracks and increased origin firing, we evaluated if the concomitant augmentation of both parameters modulated the length of the S phase (35). To do so, we applied two pulses of thymidine analogs with increasing interval lengths between pulses. The faster the transition through the S phase, the higher the proportion of cells marked with the first analog only (CldU only %). Using this approach, we observed that Pol ι–depleted cells were at least 1.2-fold faster at transiting S phase than control samples (Fig. 4, D to F). In agreement with these results, the global rate of DNA synthesis, as measured by total BrdU incorporation (Fig. 4, G and H), was higher in Pol ι–depleted samples. While the up-regulation of fork elongation speed was reported to correlate with increased replication stress in other settings (36), replication stress markers were lower in Pol ι–depleted samples than in control samples. This was the case when analyzing γH2AX intensity (Fig. 4, I and J), RPA foci (Fig. 4, K and L), and checkpoint activation markers (fig. S4, C to E), implying that the modulation of the replication dynamics and RPA-derived signaling observed after Pol ι depletion is triggered under unperturbed conditions and maintained after DNA damage. Hence, Pol ι depletion increases global DNA replication rates impairing basal replication stress signaling.
Unleashed DNA replication speed after Pol ι depletion causes chromosome instability
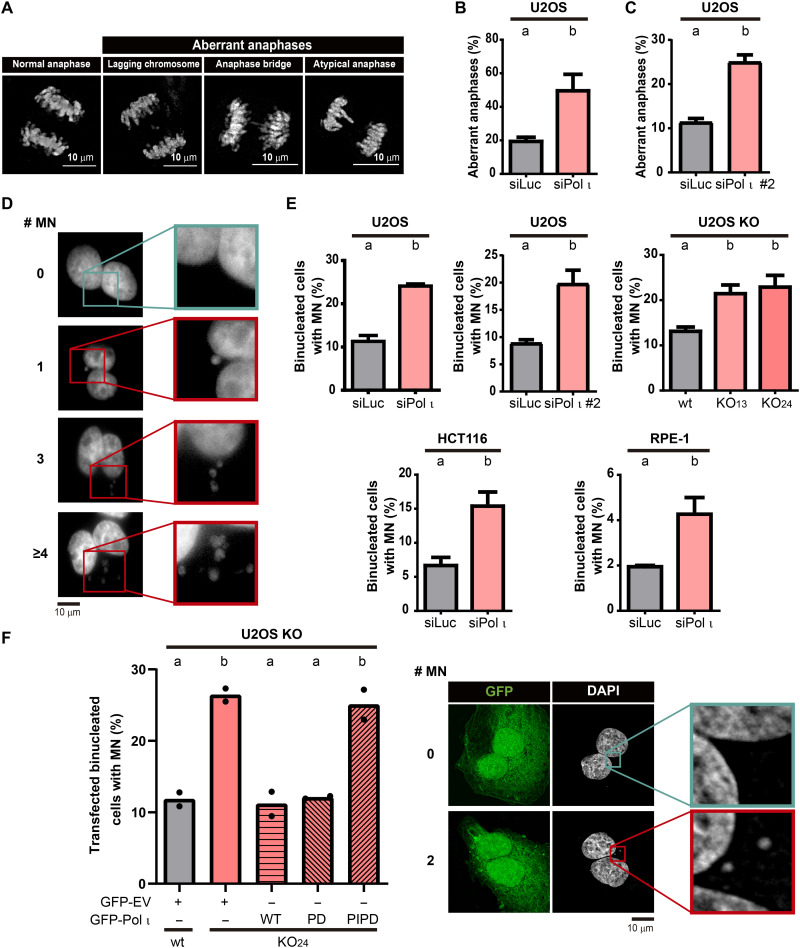
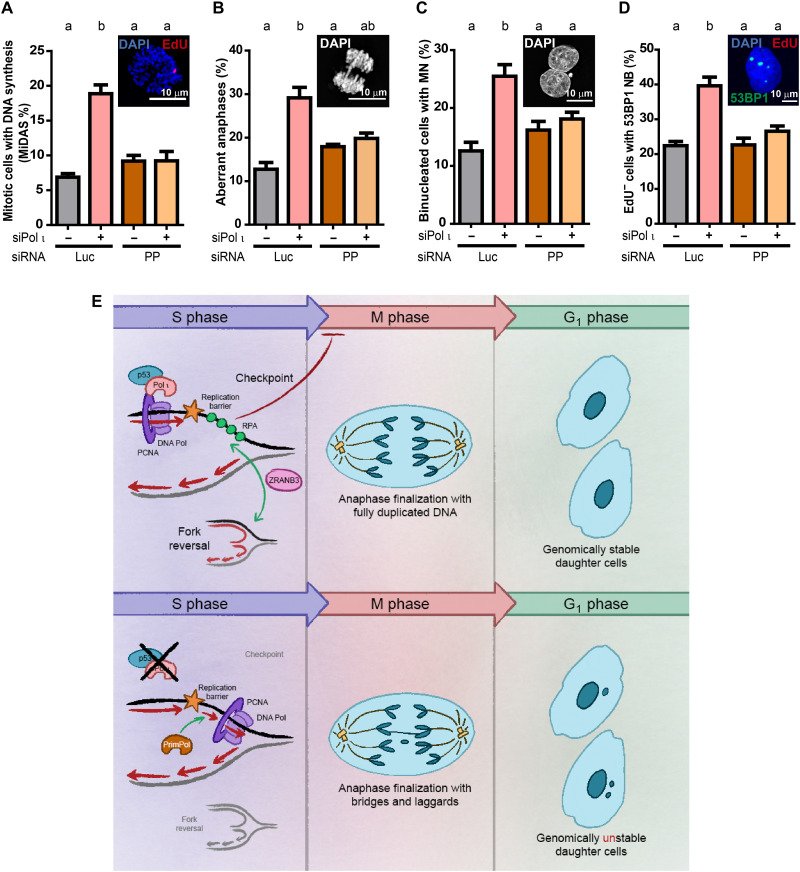
We speculated that the peculiar combination of changes that follow Pol ι depletion (i.e., ranging from DNA replication parameters including increases of both nascent DNA track lengths and origin firing frequency to the defective accumulation of checkpoint and other DDR signals) could prompt a premature entrance to G2 and M phase with unfinished DNA duplication. A way to test this hypothesis is to measure mitotic DNA synthesis (MiDAS), which should increase if DNA replication is not completed in S phase (37). RAD52-mediated MiDAS levels increased in Pol ι–depleted cells (fig. S8, A to C). We also found increased levels of other mitotic defects, such as accumulation of anaphase aberrations (Fig. 5, A to C) and micronuclei (38) in different cell lines depleted or knocked out for Pol ι (Fig. 5, D and E). When testing the effect of exogenous Pol ι, the wild-type and PD but not the PCNA-binding [PCNA-interacting domain dead (PIPD)] mutant were able to rescue Pol ι KO, thus implying that the up-regulation of chromosome instability and replication stress (Figs. 1H and 5F) are triggered by a common mechanism promoted by Pol ι depletion. Also, the proportion of G1 cells with 53BP1 nuclear bodies, which are associated with DNA regions with under-replicated DNA in G1 (39), increased in Pol ι–depleted cells (fig. S8, D to F). In contrast to MiDAS, anaphase aberrations, micronuclei, and 53BP1 nuclear bodies observed after Pol ι depletion were up-regulated independently of RAD52 expression (fig. S8, G to I), implying that chromosome instability induced by Pol ι depletion may result from unresolved S-phase intermediates, which are not processed by MiDAS in M phase.
Fig. 5. Pol ι depletion causes the accumulation of chromosomal instability.
(A) Representative images of normal and aberrant anaphases showing, e.g., lagging chromosomes, anaphase bridges, and atypical anaphases. (B) Percentage (mean ± SD) of aberrant anaphases in U2OS cells transfected with siLuc and siPol ι. At least 40 anaphases per sample were analyzed in three independent experiments (statistics: Student’s t test). (C) Percentage (mean ± SD) of aberrant anaphases in U2OS cells transfected with siLuc and siPol ι #2. At least 50 anaphases per sample were analyzed in three independent experiments (statistics: Student’s t test). (D) Representative images of binucleated cells without (teal frame) and with (red frame) increasing number of micronuclei. (E) Percentage (mean ± SD) of binucleated cells with micronuclei in U2OS, RPE-1, and HCT116 cells transfected with the indicated siRNAs, and in U2OS control (wt) and Pol ι KO clones. At least 150 binucleated cells per sample were analyzed in three independent experiments (statistics: Student’s t test for U2OS, HCT116, and RPE-1 and one-way ANOVA with Tukey post-test for U2OS KO). (F) Left: Percentage of binucleated cells with micronuclei in control (wt) and Pol ι KO U2OS cells transfected with GFP-EV, GFP–Pol ι WT, PD, and PIPD mutants. At least 200 binucleated cells per sample were analyzed. Dots represent the individual values of two independent experiments (statistics: one-way ANOVA and Tukey post-test). Right: Representative images of transfected binucleated cells without (teal frame) and with (red frame) micronuclei.
PrimPol causes unleashed elongation of DNA replication and chromosome instability in Pol ι–depleted cells
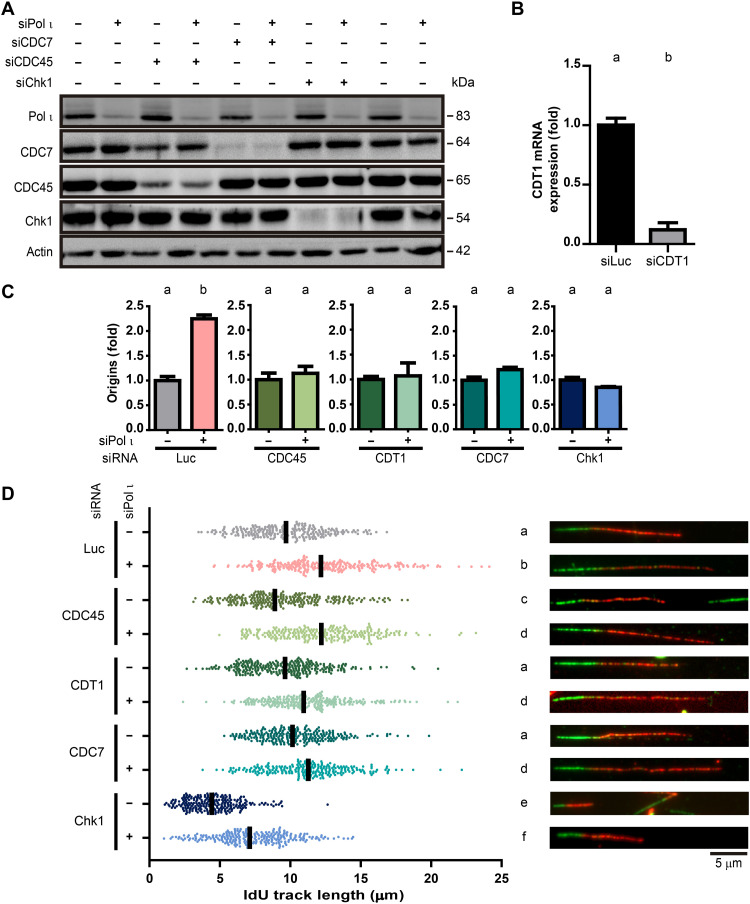
We reasoned that the RAD52-independent chromosome instability in Pol ι–depleted cells was triggered by unleashed replication elongation and/or origin firing. To determine whether these alterations in the replication program could trigger chromosome instability, we first tested if Pol ι affected both replication parameters independently. Therefore, we prevented changes in origin firing and tested if Pol ι depletion could still augment the length of replication tracks. To do so, we depleted different regulators of origin firing in the context of Pol ι down-regulation. Specifically, we down-modulated factors involved in the licensing or firing of origins (siCDC45, siCDT1, or siCDC7) (40) or their regulators (siChk1) (Fig. 6, A and B) (24). As expected, siPol ι failed to modulate origin firing in the context of the abovementioned siRNAs (Fig. 6C and fig. S9). Notably, in all backgrounds tested, Pol ι depletion caused nascent track lengthening (Fig. 6D). We concluded that fork elongation is directly regulated by Pol ι in a manner that is not dependent on the changes in origin firing frequency.
Fig. 6. Pol ι depletion accelerates nascent DNA replication independently of excess origin firing.
(A) Western blot of Pol ι, CDC7, CDC45, and Chk1 levels in U2OS samples transfected with the indicated siRNAs. Actin was used as a loading control. (B) qRT-PCR revealing CDT1 mRNA levels normalized to GAPDH in U2OS samples transfected with siLuc and siCDT1. Error bars represent the SD of two technical replicates of one representative experiment out of two independent ones (statistics: Student’s t test). (C) Fold changes (mean ± SD) in origin firing in samples from U2OS cells transfected with the indicated siRNAs. The frequency of origin firing was calculated as the relative number of origins [(red-green-red + red only fibers)/total fibers]. At least 300 fibers per condition were analyzed in three independent experiments (statistics: Student’s t test). (D) IdU track length quantification from the same samples used in (C). At least a total of 240 fibers were analyzed in three independent experiments. Median is shown in black as in all track length quantifications in this article (statistics: Kruskal-Wallis and Dunn post-test).
By carefully looking at our initial screening results (Fig. 1C), we found that Pol ι depletion reduced the pan-nuclear γH2AX accumulation in all backgrounds tested except for PrimPol-depleted samples. Given that PrimPol promotes DNA replication elongation (5, 7, 41), we hypothesized that PrimPol could trigger excess nascent DNA elongation in Pol ι–depleted samples. We combined siPol ι and siPrimPol and observed that PrimPol depletion abrogated unleashed nascent DNA synthesis by Pol ι knockdown (Fig. 7A). Confirming the functional association between p53 and Pol ι, PrimPol depletion also reverted long nascent tracks in p53-depleted samples (fig. S10A). Increased origin firing resulting from Pol ι depletion was also attenuated by PrimPol depletion (Fig. 7B and fig. S10B), suggesting that enhanced origin firing could result from reduced replication stress signaling.
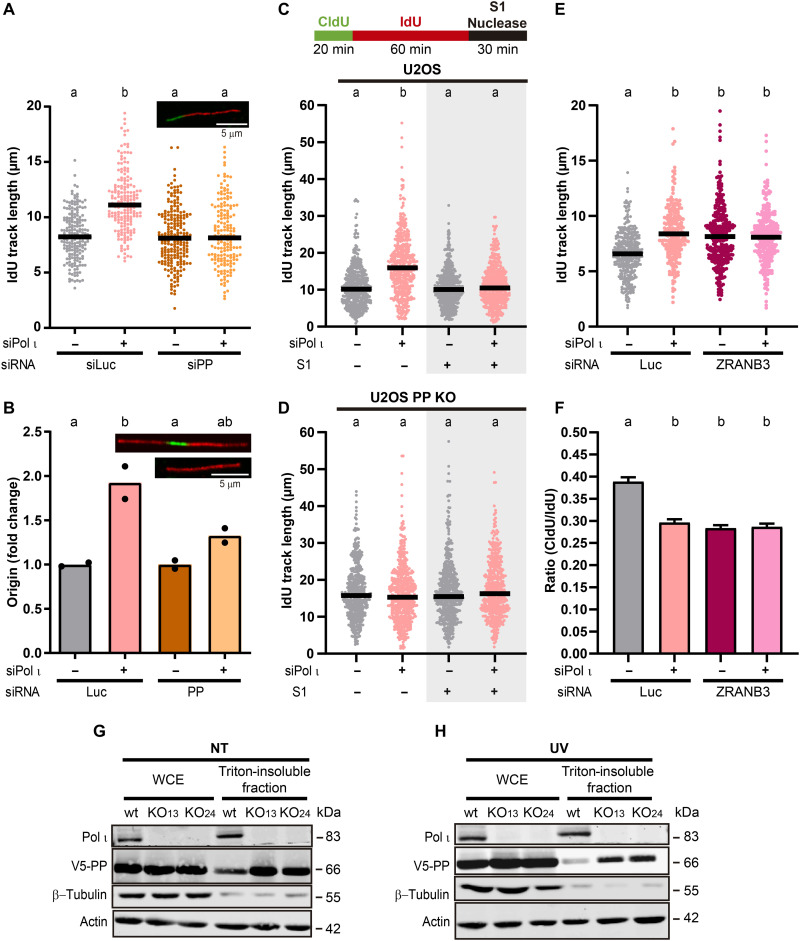
Fig. 7. DNA damage tolerance after Pol ι depletion depends on PrimPol-dependent repriming.
(A) IdU track lengths from U2OS cells transfected with the indicated siRNAs. At least 180 fibers per condition were analyzed in two independent experiments (statistics: Kruskal-Wallis and Dunn post-test). (B) Fold changes of origin firing from samples in (A). At least 200 fibers per condition were analyzed. Dots represent the individual values in two independent experiments (statistics: one-way ANOVA and Tukey post-test). (C) Top: Representation of the fiber protocol used when adding the S1 nuclease. Bottom: IdU track length quantification from U2OS cells transfected with the indicated siRNAs, with (gray background) and without S1 treatment. Three hundred fibers per condition were analyzed in three independent experiments (statistics: Kruskal-Wallis and Dunn post-test). (D) IdU track length from U2OS PrimPol KO cells transfected with the indicated siRNAs, with (gray background) and without S1 treatment. At least 300 fibers per condition were analyzed in three independent experiments (statistics: Kruskal-Wallis and Dunn post-test). (E) IdU track length from U2OS cells transfected with the indicated siRNAs (#3 for Pol ι) and treated with 1 μM CDDP for the last 30 min. At least 240 fibers were analyzed per condition in two experiments (statistics: Kruskal-Wallis and Dunn post-test). (F) Average (mean ± SD) CldU/IdU ratios of U2OS cells from samples in (E) (statistics: one-way ANOVA and Tukey post-test). (G) Western blots of whole-cell extracts and insoluble fractions from control (wt) and Pol ι KO U2OS cells transfected with V5-PP. Tubulin was used to validate the extraction protocol and actin as a loading control. V5 and actin were developed on the same membrane, while two independent membranes were used for Pol ι and tubulin. (H) Western blots of whole-cell extracts and insoluble fractions of control (wt) and Pol ι KO U2OS cells transfected with V5-PP, after 4 hours of UV irradiation (40 J/m2) processed as in (G).
Given the crucial role of PrimPol in repriming (5–7), we wondered if the lengthening in track lengths after Pol ι depletion is the result of discontinuous nascent DNA elongation. To assess for the presence of postreplicative ssDNA gaps, we treated samples with the ssDNA-specific S1 nuclease before DNA fiber spreading as previously described by us (42, 43). We found that S1 treatment leads to shorter tracts upon Pol ι depletion, indicating the presence of gaps (Fig. 7C and fig. S10C). Moreover, in PrimPol KO cells, in agreement with our data using siRNA, Pol ι depletion failed to unleash DNA replication forks (Fig. 7D and fig. S10D). In addition, S1 treatment in this background did not cause any tract shortening (Fig. 7D). Together, these data point to PrimPol-dependent repriming after Pol ι depletion and consequent accumulation of postreplicative ssDNA gaps.
Several recent studies (43–47) reported a competition between PrimPol-mediated repriming and fork reversal. The depletion of the fork reversal promoting factor ZRANB3 (zinc finger RANBP2-type containing 3) (48) and of Pol ι have a similar and epistatic impact on the length of nascent DNA tracks (Fig. 7, E and F, and fig. S10E), suggesting that both Pol ι and ZRANB3 oppose PrimPol-driven events. In agreement with this notion, we observed that the levels of PrimPol in the triton-insoluble fraction are higher in Pol ι–depleted samples than in samples expressing Pol ι (Fig. 7, G and H).
We then tested if the choice of PrimPol-dependent repriming tolerance events is the cause of the reduced replication stress signaling and the augmented MiDAS and chromosome instability observed after Pol ι depletion. The percentage of cells with a high number of RPA foci after CDDP treatment was restored to similar levels as in control samples when treated with a combination of siPol ι/siPrimPol (fig. S11, A and B). This result suggests that repriming and checkpoint activation are associated in Pol ι–depleted cells, and they are simultaneously restored to control levels when PrimPol is depleted. In line with a restoration of the DNA replication program in siPol ι/siPrimPol–transfected samples, the excess of MiDAS events was lost in these samples (Fig. 8A). Moreover, chromosome instability, detected by anaphase aberrations, micronuclei, and 53BP1 nuclear bodies in G1, was impaired in Pol ι /PrimPol–depleted samples (Fig. 8, B to D, and fig. S11C). Together, these results demonstrate that, in the absence of Pol ι, the dysregulated participation of PrimPol in nascent DNA elongation uncouples DNA replication and checkpoint activation, triggering chromosome instability in M and in the following G1 phase. Pol ι and PrimPol knockdown shows only mild effects on cell survival (fig. S11, D and E), compared with Pol η knockdown as a reference. Together, these data indicate that Pol ι and PrimPol might have a more prominent role in the maintenance of genomic stability than in cell survival.
Fig. 8. Elimination of PrimPol-mediated repriming prevents the increase in chromosome instability in Pol ι–depleted samples.
(A) Percentage (mean ± SD) of mitotic cells actively synthesizing DNA by MiDAS. At least 100 mitotic cells per sample were analyzed in three independent experiments (statistics: one-way ANOVA and Tukey post-test). (B) Percentage (mean ± SD) of aberrant anaphases/total anaphases in U2OS cells transfected with the indicated siRNAs. 30 to 60 anaphases per sample were analyzed in three independent experiments (statistics: one-way ANOVA and Tukey post-test). (C) Percentage (mean ± SD) of binucleated cells with micronuclei (MN). At least 150 binucleated cells per sample were analyzed in three independent experiments (statistics: one-way ANOVA and Tukey post-test). (D) Percentage (mean ± SD) of EdU− nuclei with one to five 53BP1 nuclear bodies (NB). Three hundred EdU− nuclei per sample were analyzed in three independent experiments (statistics: one-way ANOVA and Tukey post-test). (E) Top: When Pol ι is expressed, a PrimPol-independent nascent DNA synthesis promotes the accumulation of optimal checkpoint-initiating RPA-coated ssDNA and the ZRANB3-mediated reversion of forks. In that context, checkpoint activation, nascent DNA elongation, and origin firing are coordinated so that cells fully duplicate DNA before chromosome separation in anaphase. In that way, Pol ι contributes to the maintenance of genomic stability. Bottom: When Pol ι is depleted or knocked out, the accumulation of RPA-coated ssDNA is low. The limited basal levels of checkpoint activation are accompanied by a more rapid PrimPol-dependent nascent DNA synthesis and increased frequency of origin firing. The execution of such a faster replication program fails to accomplish complete duplication of DNA, impairing the separation of DNA in daughter cells and giving rise to chromosome instability.
DISCUSSION
The existence of a DNA damage tolerance pathway choice that involves a competition between PrimPol-mediated repriming and fork reversal has been proposed in several recent studies (43, 44, 46, 47), although the underlying mechanism remains elusive. Here, we show that Pol ι regulates such a pathway choice, as it decelerates DNA replication epistatically with ZRANB3 and antagonizes PrimPol-dependent repriming. This mechanism discriminates Pol ι from other TLS polymerases, as it enhances replication stress for optimal checkpoint activation, ultimately preventing chromosome instability. Our findings unravel opposed roles of Pol ι and PrimPol in maintaining the genomic stability of cells.
The replication program is regulated by Pol ι in a TLS-independent manner
Pol ι is a polymerase from the Y family so far mostly considered to serve a backup role in TLS (30). However, this study demonstrates a prominent contribution of Pol ι to DNA replication, which is unrelated to the canonical TLS program. Classically, TLS impairment is associated with reduced and more spread out replication track lengths [summarized in table 1 of (19)]. Pol ι loss causes the opposite effect. Specifically, in untreated Pol ι–depleted samples, tracks are longer than those of control samples, and after CDDP treatment, there is no detectable fork stalling. Moreover, the recruitment of other Y family polymerases to replication factories as well as PCNA ubiquitination were unaltered in Pol ι–depleted samples. Collectively and together with our previous analysis of DNA fiber dynamics (10), these observations indicate that canonical TLS is not the primary function of Pol ι, at least in cells expressing the tumor suppressor p53. Such conclusions, however, cannot be generalized, as we have recently observed that Pol ι may differentially affect track lengths depending on the differentiation status of primary human and ovarian cancer cells, suggesting context dependency (12).
While the contribution of Pol ι to TLS may be minor, this does not discard a TLS-independent contribution of Pol ι to DNA replication. Pol ι modulates DNA replication dynamics on both undamaged and damaged DNA templates. Specifically, our results indicate that the role of Pol ι is to ensure the slowdown of replication forks at replication barriers of different origins, ensuring optimal levels of checkpoint activation. Our initial screening revealed that Pol ι is a specialized DNA polymerase with the ability to augment, rather than reduce, γH2AX phosphorylation, implying that replication stress is not prevented but amplified when Pol ι participates in DNA replication. Such a Pol ι–induced signaling contributes to the completion of DNA replication within S phase. Our results suggest that, counterintuitively, optimal checkpoint activation observed in control samples (fig. S4, C and D) requires a certain degree of fork slowdown, which is granted by Pol ι.
Pol ι promotes checkpoint activation
We have previously reported an interaction between Pol ι, p53, and PCNA at sites of nascent DNA synthesis that positions the exonuclease activity of p53 at these sites (10, 12). On the basis of these and further findings involving a transcription-independent and exonuclease-dependent function of p53 (49), we suggested that by combining its DNA polymerase activity with the exonuclease activity of p53, Pol ι promotes idling events at ongoing replication forks (10). Such cycles of incorporation and removal of nucleotides (50, 51) keep the enzyme at a fixed position, preventing the progression of the replisome. Our work presented here demonstrates that the PCNA-binding domain of Pol ι, but not its polymerase domain, is required to prevent aberrant DNA elongation. While such a finding does not rule out the need for idling events by Pol ι (or perhaps other DNA polymerases), they suggest that the primary role of Pol ι at replication forks is to promote the recruitment of p53 (and perhaps other factors) to PCNA. The regulation of the PCNA interactome by Pol ι may influence different aspects of the DNA damage tolerance pathway choice, including transient reversal of forks and the extent of ssDNA accumulation before tolerance occurs. Such events synergize to maximize ATR-Chk1 and downstream target activation. In the absence of Pol ι, the reduced speed of nascent DNA synthesis at replication barriers may underlie reduced RPA accumulation at replication forks, which prevents optimal checkpoint activation (model in Fig. 8E). ATR and Chk1 activation defects were previously shown to unleash origin firing (24). Hence, the combined acceleration of the replication program and the defective checkpoint activation may favor a premature S to G2 transition, very likely triggering the M phase defects that follow Pol ι depletion. Together, these observations unravel an unanticipated contribution of Pol ι to the successful completion of the replication program, which is needed for a timely S-G2-M transition.
Pol ι cooperates with p53 and ZRANB3 and prevents PrimPol participation in nascent DNA synthesis
PrimPol is a primase in human cells with the ability to start DNA chains with deoxynucleotide triphosphates (dNTPs) (7). In vitro, PrimPol has both primase and TLS activity (5–7). However, it is unclear whether the TLS activity of PrimPol has any physiologically relevant function in vivo (41). PrimPol’s active site cannot accommodate bulky DNA adducts (52). Moreover, a recent report suggests that PrimPol can perform DNA strand displacement, which is not associated with TLS events (53). Such observations support the notion that PrimPol mostly acts as a repriming enzyme (5). Accordingly, PrimPol-mediated repriming is required to transverse interstrand cross-links in the leading strand template (54) and can circumvent transcriptional conflicts caused by DNA:RNA hybrids or R-loops, as demonstrated in avian DT40 cells (55). In agreement with this notion, our data demonstrate that the role of PrimPol in a replication adaptation program after Pol ι loss is related to repriming rather than TLS (19, 56). Our findings that PrimPol promotes nascent DNA elongation after depletion of Pol ι or p53 imply that it compensates for a defect in a bypass mechanism other than TLS, albeit at the expense of accumulating postreplicative gaps.
Multiple pieces of evidence demonstrated that template switching and fork reversal events are pathways alternative to PrimPol-dependent DNA synthesis. For example, we have shown that cells depleted of RAD51 and exposed to UVC display excessive fork elongation dependent on PrimPol (28). Moreover, both SMARCAL1 down-regulation and PrimPol overexpression promote DNA elongation mediated by PrimPol (46). In our hands, limiting nascent DNA elongation by Pol ι is epistatic with p53 and ZRANB3. Moreover, the loss of Pol ι or p53 promotes PrimPol-mediated nascent DNA elongation. This result suggests that PrimPol-mediated DNA replication is facilitated by the loss of the concerted actions of Pol ι, p53, and ZRANB3. We, therefore, propose that Pol ι, in concert with p53, plays a central role in the pathway choice between fork reversal and repriming.
In agreement with Pol ι promoting DNA damage tolerance events that antagonize repriming, we also found that Pol ι depletion facilitates PrimPol recruitment to chromatin. PrimPol loading seems primarily mediated by its interaction with the ssDNA binding protein RPA (5, 57, 58) and occurs only when RPA levels on ssDNA are not high, at least in vitro (59, 60). As PrimPol cannot displace RPA from ssDNA (59, 61), it is possible that the reduced RPA levels observed in Pol ι–depleted cells [this report and (10)] may facilitate PrimPol loading to replicating DNA, which in turn causes a DNA replication program entailing M phase aberrations. Perhaps less anticipated is the inverse association between PrimPol activity and checkpoint activation observed by others after UV irradiation in PrimPol knockdown samples (6) and herein by us (in the context of Pol ι–depleted conditions). Such results may suggest that the location (at forks and behind) and/or the size of ssDNA tracks may be relevant to the efficiency of checkpoint activation.
Pol ι–mediated suppression of PrimPol-dependent nascent DNA synthesis protects the genomic stability of cells
While several reports have explored the association of nascent DNA gap formation and cell death (62), here, we report that the changes in replication dynamics caused by the excess participation of PrimPol in DNA synthesis in S phase cause an accumulation of under-replicated DNA in M phase and trigger chromosome instability. The under-replicated DNA that accumulates after Pol ι KO or knockdown can be processed by RAD52-dependent MiDAS. DNA substrates for MiDAS were reported to be either stalled or progressing replication forks that aberrantly entered the M phase before the completion of DNA synthesis (37, 39). The accumulation of such aberrant replication intermediates is compatible with the concept of an impaired checkpoint after Pol ι depletion. While MiDAS is up-regulated after Pol ι knockdown in a PrimPol-dependent manner, MiDAS impairment via RAD52 depletion does not affect the accumulation of chromosome aberrations and micronuclei, suggesting that chromosome instability affects DNA regions that are not engaged in MiDAS. MiDAS was reported to be inefficient in other scenarios [e.g., after Chk1 depletion (63)], and it might as well be inefficient after Pol ι knockdown. Therefore, in the context of Pol ι knockdown, regions of the genome that failed to initiate replication or properly complete replication in the S phase and do not enter MiDAS in M phase may trigger the formation of anaphase bridges, micronuclei, and 53BP1-labeled scars in the next G1. These results highlight that the changes in the replication program caused by Pol ι loss and PrimPol-dependent gap formation do not induce replication stress markers in S phase but trigger devastating consequences in M phase that MiDAS cannot prevent. G1-specific 53BP1 nuclear bodies were previously observed in genetic backgrounds that accumulate PrimPol-dependent gaps, but only when the repair of these gaps by gap-filling mechanisms was impaired (46). The difference between these results and the findings reported here opens the possibility that Pol ι might also be involved in gap filling.
While the pathways leading to cell death and chromosome instability are conceived as mechanistically entangled (38), the PrimPol-dependent chromosome instability observed after Pol ι depletion correlates with only a modest augmentation in cell death. Such results are very intriguing as Pol ι and PrimPol may be part of a limited set of factors that selectively regulate only one of these variables.
MATERIALS AND METHODS
Cell culture and reagents
U2OS [American Type Culture Collection (ATCC)] (RRID: CVCL_0042), U2OS PrimPol KO (43), RPE-1-hTERT (ATCC), and HCT116 (gifts from B. Vogelstein, Johns Hopkins University, Baltimore, MD) were grown in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen) with 10% fetal bovine serum (Natocor). UVC irradiation was performed as described previously (27). CDDP and hydroxyurea (HU) were purchased from Sigma-Aldrich (#P4394 and #127-07-1, respectively). All cell lines were maintained in a humidified, 5% CO2 incubator and passaged as needed. Cell lines were regularly checked for mycoplasma contamination. The p53 and p21 status of all cell lines was checked, and none of the cell lines used are in the list of commonly misidentified cell lines maintained by the International Cell Line Authentication Committee.
CRISPR-Cas9 KO U2OS cell lines
The single-guide RNAs (sgRNAs) used to KO DNA polymerase ι were designed using Benchling [Biology Software, 2017, retrieved from (64)]. The oligos used to clone Pol ι sgRNA into pX459 (Addgene) were the following: forward: CACCGATCCAGAGTCATAGTACATG; reverse: aaacCATGTACTATGACTCTGGATC.
U2OS cells were then transfected with pX459-IOTA-sgRNA using Lipofectamine 3000 (Thermo Fisher Scientific) and selected with puromycin (1 μg/ml) for 48 hours to enrich for positive transfectants. Live cells were sorted by flow cytometry (FACSAria Fusion C, BD Biosciences), and single cells were seeded into 96-well plates and allowed to grow for 2 weeks. Individually expanded clones were screened for loss of protein expression by immunoblotting with Pol ι–specific antibody (Abcam, catalog no. ab228783).
siRNAs and vector expression plasmids
Transfections of siRNAs and plasmid expression vectors were performed using jetPRIME (Polyplus) following the manufacturer’s instructions. Except for survival assays, cells were harvested 48 or 72 hours after transfection. GFP–Pol η was a gift from A. Lehmann (Genome Damage and Stability Centre, School of Life Sciences, University of Sussex, Falmer, Brighton, UK) (29); GFP–Pol ι was provided by R. Woodgate [Laboratory of Genomic Integrity, National Institute of Child Health and Human Development, National Institutes of Health (NIH), Bethesda, MD, USA]; and GFP-REV1 was donated by E. Friedberg (Department of Pathology, UT Southwestern Medical Center, Dallas, TX, USA). V5-PP was a gift from J. Méndez (Spanish National Cancer Research Centre, Madrid, Spain).
GFP–Pol ι point mutations that eliminate its catalytic function (PD: D34A/D126A/E127A) and its ability to bind PCNA (PIPD: Y426A/Y427A) were obtained by site-directed mutagenesis by Gene Universal and cloned into the original GFP–Pol ι vector (backbone pEGFP-C3) using restriction enzymes XhoI and EcoRI. The sequences of these mutants are described in the Supplementary Materials.
siRNAs were purchased from Dharmacon, Eurofins Genomics, and Qiagen: siLuc: 5′-CGUACGCGGAAUACUUCGA-3′ (65); siPol ι: 5′-AACAGUCUUAUUACCUGAAAGUU-3′; siPol ι #2: 5′-GAAGUAAAUUCUGGCACAA-3′; siPol ι #3 Smart Pool: 5′-AAGCCUCAUACAGUGAGAUUA-3′, 5′-UUCGGAUUAGCGGUUUAUUAA-3′, 5′-GCGGUUUAUUAAGCUCUUCUA-3′, and 5′-ACCGGGAACAUCAGGCUUUAA-3′; siPP: 5′-GAGGAAACCGUUGUCCUCAGUGUAU-3′ (28); siPol κ: 5′-AAGAUUAUGAAGCCCAUCCAA-3′; siPol η: 5′-CUGGUUGUGAGCAUUCGUGUA-3′; siREV3 Smart Pool: 5′-GAGAGUACCUCCAGAUUUA-3′, 5′-GCUAAUCCCUCUAUAGUUA-3′, and 5′-CAAAGAUGCUGCUACAUUA-3′; siREV1: 5′-AAAAGCAUCAAAGCUGGACGACU-3′; siCDC45: 5′-GCAAGACAAGAUCACUCAA-3′; siCDT1: 5′-GCGCAAGUUGGCCAGAUC-3′ (66); siCDC7: 5′-GCAGUCAAAGACUGUGGAU-3′ (67); siChk1: 5′-GAAGCAGUCGCAGUGAAGA-3′ (65); siZRANB3 Smart Pool: 5′-CCGGAUUCACAUCUAUACUAA-3′, 5′-UGCCAGUGUAUGACACCUUAA-3′, 5′-CAGACUCGCAAUUAUGUUUCA-3′, and 5′-CCCACUUGCCAGACUAAGCAA-3′; sip53: 5′-GACUCCAGUGGUAAUCUA-3′; siRAD52: 5′-GGAGUGACUCAAGAAUUA-3′ (63).
Immunostaining and fluorescence detection
For the quantification and immunodetection of specialized Y family polymerases, 53BP1 and γH2AX, cells were fixed in 2 to 4% paraformaldehyde (PFA)/2% sucrose and permeabilized with 0.1% Triton X-100 in phosphate-buffered saline (PBS) as described previously (27, 68). For the detection of the chromatin-bound fraction of RPA, a pre-extraction with ice-cold 0.5% Triton X-100 in CSK buffer [10 mM Pipes (pH 7.5), 100 mM NaCl, 300 mM sucrose, and 3 mM MgCl2] was performed for 5 min before fixation with 2 to 4% PFA/2% sucrose. When detecting CPDs, a denaturing step with 0.07 M NaOH for 4 min was performed after fixation. When using BrdU (10 μM; Sigma-Aldrich, catalog no. B9285), cells were fixed with methanol-acetone and subjected to a denaturing step involving a 30-min incubation with 1.5 N HCl, which exposes the BrdU epitopes enabling their detection with specific antibodies. For the detection of ssDNA under nondenaturing conditions, cells were pulsed and labeled with CldU (10 μM) or BrdU (10 μM) for 36 hours, treated with CDDP (6 μg/ml) for 4 hours, washed and left to recover in cell culture medium for 24 hours, and later fixed with 2 to 4% PFA/2% sucrose. After fixation, samples were not subjected to treatment with denaturing agent. Samples were permeabilized, and regular immunofluorescence was performed. For the visualization of 53BP1 nuclear bodies in 5-ethynyl-2′-deoxyuridine (EdU)–negative cells, cells were pulsed with EdU (10 μM) for 15 min before fixation with 2% PFA/2% sucrose. Samples were permeabilized, EdU staining was carried out using EdU detection kit following the manufacturer’s instructions (Click-iT EdU kit, Invitrogen, catalog no. C10338), and later a regular immunofluorescence was performed for 53BP1 staining. In all cases, blocking was performed overnight in PBS–2% donkey serum (Sigma-Aldrich) before incubation with primary antibodies. Coverslips were then incubated for 1 hour with primary antibodies. Primary antibodies used in this study were the following: anti-53BP1 (Santa Cruz Biotechnology, catalog no. sc-22760, RRID: AB_2256326), anti-γH2AX (Millipore, catalog no. 05-636, RRID: AB_309864), anti–α-BrdU (GE HealthCare, catalog no. RPN 202, RRID: AB_2314032), anti-RPA (Millipore, catalog no. NA18-100UG, RRID: AB_213121), anti-BrdU (Accurate Chemical and Scientific Corporation, catalog no. OBT-0030, RRID: AB_2341179 to detect CldU), anti-BrdU (BD Biosciences, catalog no. 347580, RRID: AB_400326 to detect IdU), and anti-CPDs (MBL International Corporation, D194-1). Secondary anti-mouse/rabbit–conjugated Alexa Fluor 488/546 antibodies were purchased from Invitrogen. GFP-tagged specialized Y polymerases were visualized by GFP autofluorescence. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) (Sigma-Aldrich). Images were obtained with a Zeiss Axioplan microscope, a Zeiss Axio Observer 3 microscope, a Zeiss LSM 510 META confocal microscope, or a Zeiss LSM Pascal confocal microscope. For the quantification of 53BP1 nuclear bodies in EdU-negative cells, since they are described as 2- to 3-μm structures according to (69), small foci (<1.5 μm), with a size typical of S phase, were not counted. For the quantification of RPA foci, the nuclei foci average number was determined for each condition.
Micronuclei assay
Transfected cells and U2OS KO cells were replated at low density. Twenty-four hours after replating, cytochalasin B (4.5 μg/ml; Sigma-Aldrich) was added to the medium, and 40 hours later, cells were fixed with 2 to 4% PFA/2% sucrose for 20 min. DAPI (Sigma-Aldrich) staining served to visualize cellular nuclei. Three hundred binucleated cells per sample were analyzed.
Anaphase aberration assay
Forty-eight hours after siRNA transfection, asynchronous samples were fixed with 2% PFA/2% sucrose for 20 min. DAPI (Sigma-Aldrich) staining was used to visualize anaphases. At least 50 anaphases were measured per sample in each experiment. Z-stacks were acquired with a Zeiss LSM 510 META confocal microscope. Maximum intensity projections were generated using ImageJ software (ImageJ 1.52a).
DNA fiber spreading
DNA fiber spreading was performed exactly as previously described (27, 68, 70).
S1 nuclease DNA fiber assay
DNA fiber spreading with the ssDNA-specific S1 nuclease was performed as previously described (42, 71) with minor modifications. Briefly, exponentially growing transfected U2OS and PRIMPOL KO U2OS cells (43) were pulse-labeled with 30 μM CldU (Millipore Sigma) for 20 min, washed twice with PBS, then pulse-labeled with 250 μM IdU (Millipore Sigma). Cells were then permeabilized with CSK100 [100 mM NaCl, 10 mM Mops (pH 7.2), 3 mM MgCl2, 300 mM sucrose, and 0.5% Triton X-100] for 3 min at room temperature (RT) and incubated in S1 buffer [30 mM sodium acetate, 10 mM zinc acetate, 5% glycerol, and 50 mM NaCl (pH 4.6)] with or without S1 nuclease (20 U/ml; Thermo Fisher Scientific, catalog no. 18001016) for 30 min at 37°C. Next, nuclei were scraped in PBS + 0.1% bovine serum albumin (BSA), pelleted at 4600g for 5 min at 4°C, and resuspended in PBS for a final concentration of 1500 cells/μl. Three microliters of nuclei suspension were lysed with 7 μl of lysis buffer [200 mM tris-HCl (pH 7.5), 50 mM EDTA, and 0.5% SDS] on a positively charged glass slide for 4 min at RT. Slides were then tilted at a 20° to 40° angle to spread DNA. After air drying, DNA was fixed with freshly prepared methanol:acetic acid (3:1) for 5 min and stored at 4°C for at least overnight. For immunostaining of DNA fibers, DNA was denatured with 2.5 M HCl for 1 hour at RT. Slides were then blocked with 5% BSA at 37°C for 1 hour and incubated with mouse anti-BrdU (1:250, BD Biosciences, catalog no. 347580) and rat anti-BrdU (1:100; Abcam, catalog no. ab6326) for 1 hour at 37°C followed by washes with PBS–0.1% Tween 20 and incubation with anti-mouse Alexa Fluor 594 (1:500; Thermo Fisher Scientific, catalog no. A21125) and anti-rat Alexa Fluor 488 (1:300; Thermo Fisher Scientific, catalog no. A11006) for 1 hour at RT. After three washes with PBS, DNA was fixed with 4% PFA for 10 min at RT and washed three times with PBS and three times with PBS–0.1% Tween 20. Last, slides were mounted with ProLong Gold Antifade Reagent (Thermo Fisher Scientific, catalog no. P36930). Images were acquired with the Leica Application Suite X software using SP8 confocal microscope (Leica) with a 63× oil immersion objective. At least 125 to 150 individual fibers were scored per dataset using ImageJ (NIH). Experiments were performed three times independently.
EdU labeling and detection in mitotic cells
Asynchronously growing cells were treated with 10 μM RO-3306 (CDK1i, Merck, catalog no. 217699) for 16 hours to enrich the samples with cells at the G2 phase of the cell cycle. Cells were released from G2 by washing samples three times with PBS. Samples were pulse-labeled with 20 μM EdU for 45 min immediately after CDKi release. EdU was detected using the Click-iT EdU Alexa Fluor 555 Imaging Kit from Life Technologies following the manufacturer’s instructions. DAPI (Sigma-Aldrich) staining served to identify cells that were transiting mitosis. In every single experiment, over 100 metaphases per sample were analyzed. Z-stacks were acquired with a Zeiss LSM 510 Meta confocal microscope. Maximum intensity projections were generated using ImageJ software (ImageJ 1.52a).
Quantitative real-time PCR
Quantitative PCR was conducted exactly as previously described (28, 70). Primer sequences were as follows: GAPDH: 5′-AGCCTCCCGCTTCGCTCTCT-3′ (forward) and 5′-GAGCGATGTGGCTCGGCTGG-3′ (reverse) (63); beta-ACTIN: 5′-CTCGCCTTTGCCGATCC-3′ (forward) and 5′-ATGCCGGAGCCGTTGTC-3′ (reverse); Pol ι: 5′-GTCGTGAGAGTCGTCAGTGC-3′ (forward) and 5′-GCTTGCCAGAGCGTGAAGTA-3′ (reverse); RAD52: 5′-ACAGCGTTTGCCACCAGAA-3′ (forward) and 5′-ATGAGATTCCCAGTTTCCTGT-3′ (reverse); CDT1: 5′-GAACGGCTGCCTGAGCT-3′ (forward) and 5′-CCATTTCCCCAGGGCTCA-3′ (reverse); PrimPol: 5′-TGTGGCTTTGGAGGTTACTGA-3′ (forward) and 5′-TTCTACTGAAGTGCCGATACTGT-3′ (reverse); REV3L: 5′-GACTTATGACCGGCGCTCTG-3′ (forward) and 5′-CCTTTCCGCCCTTCAGGTTC-3′ (reverse).
Protein analysis
For Western blot analysis, samples were lysed in Laemmli buffer. When analyzing insoluble protein fraction, samples were treated with ice-cold PBS–0.5% Triton X-100 for 45 s. The soluble fraction was discarded, and the triton-resistant fraction was lysed in Laemmli buffer. Antibodies used were the following: anti-actin (Sigma-Aldrich, catalog no. A2066, RRID: AB_476693), anti-Ku70 (Santa Cruz Biotechnology, catalog no. sc-5309, RRID: AB_628453), anti-GFP (Santa Cruz Biotechnology, catalog no. sc-9996, RRID: AB_627695), anti–Pol ι (Abcam, catalog no. ab228783), anti–phospho-RPAS4/S8 (Bethyl Laboratories, catalog no. A300-245A, RRID: AB_210547), anti–phospho-KAP-1S824 (Bethyl Laboratories, catalog no. A300-767A, RRID: AB_669740), anti–phospho-H2AXS139 (Millipore, catalog no. 05-636, RRID: AB_309864), anti–phospho-ATMS1981(Millipore, catalog no. 05-740, RRID: AB_11214302), anti–phospho-Chk1S345 (Cell Signaling Technology, catalog no. 2348, RRID: AB_331212), anti-CDC7 SPM171 (Santa Cruz Biotechnology, catalog no. sc-56275, RRID: AB_831150), anti-p53 DO-1 (supernatant solution from hybridoma culture) and PAb 1801 (supernatant solution from hybridoma culture), anti–β-tubulin (Cell Signaling Technology, catalog no. 5346, RRID: AB_1950376), anti-PCNA (Santa Cruz Biotechnology, catalog no. sc-56, RRID: AB_628110), anti–ubiquitin-PCNA (Cell Signaling Technology, catalog no. 13439, RRID: AB_2798219), anti-REV1 (Santa Cruz Biotechnology, catalog no. sc-393022, RRID: AB_2885169), anti-REV3L (Abnova, catalog no. H00005980-A01, RRID: AB_875819), anti–Pol κ (Bethyl Laboratories, catalog no. A301-977A, RRID: AB_1548020), anti-PrimPol (rabbit, homemade from J. Méndez laboratory) (5), anti-V5 (Bethyl Laboratories, catalog no. A190-120A, RRID: AB_67586), and anti-ZRANB3 (Thermo Fisher Scientific, catalog no. PA5-97227, RRID: AB_2809029). HRP-conjugated secondary antibodies were from Sigma-Aldrich, and detection was performed with ECL according to the manufacturers’ instructions (Amersham GE HealthCare). Fluorescent secondary antibodies: Invitrogen, anti-rabbit (Thermo Fisher Scientific, catalog no. A32808, RRID: AB_2762837), anti-mouse (Thermo Fisher Scientific, catalog no. A32789, RRID: AB_2762832). Western blot images were acquired with ImageQuant LAS4000 (GE HealthCare ImageQuant, LAS 4000 v 1.0) and Odyssey CLx Imager and processed with ImageJ software (ImageJ 1.52a). Raw Western blot data can be found in figs. S12 and S13.
S-phase exit protocol
Forty-eight hours after siRNA transfection, cells were labeled for 15 min with 20 μM CldU (Fig. 4D, green boxes), washed with PBS, and incubated in fresh medium (DMEM + 10% fetal bovine serum) for different times (2, 4, 6, or 8 hours) (Fig. 4D, gray boxes), before a subsequent incubation with 100 μM IdU (Fig. 4D, red boxes). Cells were washed with PBS and fixed with ice-cold methanol (−20°C) for 20 min and acetone for 40 s. Cells were then denatured with 1.5 N HCl for 30 min, permeabilized with 0.1% Triton X-100 in PBS at 4°C for 15 min, and blocked with donkey serum for 1 hour in a humid chamber. CldU and IdU were detected by immunofluorescence using a rat anti-BrdU antibody (to detect CldU, 1:200) and mouse anti-BrdU antibody (to detect IdU, 1:50) for 1 hour. The secondary antibodies used were Alexa 488 anti-rat and Alexa 546 anti-mouse (diluted 1:200), respectively. DAPI (Sigma-Aldrich) was used to detect nuclei, and images were obtained using a Zeiss Axio Observer 3 microscope. HU (0.75 mM) was used as a control that prevented S-phase exit for the experiment’s total (8 hours) length.
Cell survival assays
Twenty-four hours after transfection, 1500 cells per well were replated in 96-well plates. Twenty-four hours later, cells were treated with either CDDP (1 μg/ml) or UV (7 J/m2), and 48 hours after treatments, cells were fixed with 4% PFA/2% sucrose for 20 min. DAPI staining served to visualize nuclei. INCell 2200 and INCell Analyzer WorkStation were used to image and count nuclei, respectively.
Statistical analysis
All the statistical analysis was performed using GraphPad Prism 8 software and Infostat software. Frequency distributions of DNA track length and ratios were determined with GraphPad Prism 8 software. In non-Gaussian distributions, Mann-Whitney and Kruskal-Wallis tests were used for statistical analyses when comparing two and more than two variables, respectively. In all graphs, different letters indicate groups that are significantly different. Thus, if two samples share the same letter, they are not significantly different, while if two samples do not share any letter, they are significantly different. P < 0.001 or P < 0.05 was considered significant for frequency distribution, and data were shown as the mean of independent experiments, respectively.
Source data
All Western blot membranes used to assemble figures in this article are shown in figs. S12 and S13. Source data from all datasets analyzed during the current study are listed in the Excel file called “ade7997 Source data” in the Supplementary Materials.
Acknowledgments
We would like to thank all members of the Gottifredi, Quinet, and Wiesmüller laboratories for insightful comments and discussions. We also thank R. Woodgate (NIH, USA) for the gift of GFP-Pol ι (72); J. Méndez (CNIO, Spain) for the gifts of V5-PP (8) and anti-PrimPol antibody (5); A. Vindigni (Washington University in St. Louis, USA) for providing U2OS PP KO cells; and A. Gamarnik, G. C. Navarro, and H. M. Pallarés in Fundación Instituto Leloir for reagents and valuable advice for cloning. We thank P. Rodriguez, E. Miglietta, A. H. Rossi, and C. Pascuale for technical support with tissue culture and microscopy.
Funding: This work was supported by Fondo para la Investigación Científica y Tecnológica (FONCyT) PICT2019-2001 to V.G.; Instituto Nacional del Cancer INC-Asistencia Financiera-IV to V.G.; FONCyT PICT2017-0612 to S.F.M.; German Research Foundation, Project A3 in Research Training Group, 1789 “Cellular and Molecular Mechanisms in Aging” and Project B3 in the Collaborative Research Center 1506 “Aging at Interfaces” to L.W.; and INSERM/CNRS ATIP-Avenir grant (project no. C23002LS) and startup package from UMR008/iRCM/IBFJ to A.Q. S.F.M. and V.G. are researchers from the National Council of Scientific and Technological Research (CONICET), L.W. is a professor at the University of Ulm (Germany), and A.Q. is an INSERM researcher (France). A.P.B., S.V.A., S.O.S., M.V.C., and M.B.d.l.V. were supported by fellowships from the National Agency for the Promotion of Science and Technology (ANPCyT) and CONICET; B.A.C. is a member of the International Graduate School in Molecular Medicine Ulm; and T.J. was supported by a CFR fellowship from the Centre d’Energie Atomique (CEA). V.G. was supported by the Alexander von Humboldt Foundation during her extended stay in the Wiesmüller laboratory (Friedrich Wilhelm Bessel Award to V.G.).
Author contributions: V.G. conceived the original project. V.G., S.F.M., L.W., and A.Q. supervised the study. All authors designed, analyzed, and interpreted the experiments. S.F.M., A.P.B., S.V.A., B.A.C., T.J., J.K.S., S.O.S., M.V.C., and M.B.d.l.V. performed the experiments. S.F.M., S.V.A., L.W., and V.G. designed the manuscript and flow of figures. S.F.M. and S.V.A. generated the figures. V.G. wrote the manuscript. S.F.M., S.V.A., A.Q., L.W., and V.G. edited the manuscript, and all authors agreed to its final version and this description of each author’s contributions.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S13
Legend for GFP-Pol ι mutants sequences
Legend for Source data
Other Supplementary Material for this manuscript includes the following:
GFP-Pol ι mutants sequences
Source data
REFERENCES AND NOTES
- 1.S. P. Jackson, J. Bartek, The DNA-damage response in human biology and disease. Nature 461, 1071–1078 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.E. Bournique, M. Dall'Osto, J. S. Hoffmann, V. Bergoglio, Role of specialized DNA polymerases in the limitation of replicative stress and DNA damage transmission. Mutat. Res. 808, 62–73 (2018). [DOI] [PubMed] [Google Scholar]
- 3.J. E. Sale, Translesion DNA synthesis and mutagenesis in eukaryotes. Cold Spring Harb. Perspect. Biol. 5, a012708 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.P. Tonzi, T. T. Huang, Role of Y-family translesion DNA polymerases in replication stress: Implications for new cancer therapeutic targets. DNA Repair 78, 20–26 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.S. Mouron, S. Rodriguez-Acebes, M. I. Martínez-Jiménez, S. García-Gómez, S. Chocrón, L. Blanco, J. Méndez, Repriming of DNA synthesis at stalled replication forks by human PrimPol. Nat. Struct. Mol. Biol. 20, 1383–1389 (2013). [DOI] [PubMed] [Google Scholar]
- 6.J. Bianchi, S. G. Rudd, S. K. Jozwiakowski, L. J. Bailey, V. Soura, E. Taylor, I. Stevanovic, A. J. Green, T. H. Stracker, H. D. Lindsay, A. J. Doherty, PrimPol bypasses UV photoproducts during eukaryotic chromosomal DNA replication. Mol. Cell 52, 566–573 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.S. Garcia-Gomez, A. Reyes, M. I. Martínez-Jiménez, E. S. Chocrón, S. Mourón, G. Terrados, C. Powell, E. Salido, J. Méndez, I. J. Holt, L. Blanco, PrimPol, an archaic primase/polymerase operating in human cells. Mol. Cell 52, 541–553 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.D. Gonzalez-Acosta, E. Blanco-Romero, P. Ubieto-Capella, K. Mutreja, S. Míguez, S. Llanos, F. García, J. Muñoz, L. Blanco, M. Lopes, J. Méndez, PrimPol-mediated repriming facilitates replication traverse of DNA interstrand crosslinks. EMBO J. 40, e106355 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.A. Vaisman, R. Woodgate, Mysterious and fascinating: DNA polymerase ɩ remains enigmatic 20 years after its discovery. DNA Repair 93, 102914 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.S. Hampp, T. Kiessling, K. Buechle, S. F. Mansilla, J. Thomale, M. Rall, J. Ahn, H. Pospiech, V. Gottifredi, L. Wiesmüller, DNA damage tolerance pathway involving DNA polymerase ι and the tumor suppressor p53 regulates DNA replication fork progression. Proc. Natl. Acad. Sci. U.S.A. 113, E4311–E4319 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.R. Betous, M.-J. Pillaire, L. Pierini, S. van der Laan, B. Recolin, E. Ohl-Séguy, C. Guo, N. Niimi, P. Grúz, T. Nohmi, E. Friedberg, C. Cazaux, D. Maiorano, J.-S. Hoffmann, DNA polymerase κ-dependent DNA synthesis at stalled replication forks is important for CHK1 activation. EMBO J. 32, 2172–2185 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.M. Ihle, S. Biber, I. S. Schroeder, C. Blattner, M. Deniz, G. Damia, V. Gottifredi, L. Wiesmüller, Impact of the interplay between stemness features, p53 and pol iota on replication pathway choices. Nucleic Acids Res. 49, 7457–7475 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Y. Auclair, R. Rouget, J. M. Belisle, S. Costantino, E. A. Drobetsky, Requirement for functional DNA polymerase eta in genome-wide repair of UV-induced DNA damage during S phase. DNA Repair 9, 754–764 (2010). [DOI] [PubMed] [Google Scholar]
- 14.T. Ogi, A. R. Lehmann, The Y-family DNA polymerase kappa (pol kappa) functions in mammalian nucleotide-excision repair. Nat. Cell Biol. 8, 640–642 (2006). [DOI] [PubMed] [Google Scholar]
- 15.T. Ogi, S. Limsirichaikul, R. M. Overmeer, M. Volker, K. Takenaka, R. Cloney, Y. Nakazawa, A. Niimi, Y. Miki, N. G. Jaspers, L. H. F. Mullenders, S. Yamashita, M. I. Fousteri, A. R. Lehmann, Three DNA polymerases, recruited by different mechanisms, carry out NER repair synthesis in human cells. Mol. Cell 37, 714–727 (2010). [DOI] [PubMed] [Google Scholar]
- 16.M. Raschle, P. Knipscheer, M. Enoiu, T. Angelov, J. Sun, J. D. Griffith, T. E. Ellenberger, O. D. Schärer, J. C. Walter, Mechanism of replication-coupled DNA interstrand crosslink repair. Cell 134, 969–980 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.J. McIntyre, Polymerase iota—An odd sibling among Y family polymerases. DNA Repair 86, 102753 (2020). [DOI] [PubMed] [Google Scholar]
- 18.V. Bergoglio, A. S. Boyer, E. Walsh, V. Naim, G. Legube, M. Y. W. T. Lee, L. Rey, F. Rosselli, C. Cazaux, K. A. Eckert, J. S. Hoffmann, DNA synthesis by Pol η promotes fragile site stability by preventing under-replicated DNA in mitosis. J. Cell Biol. 201, 395–408 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.A. P. Bertolin, S. F. Mansilla, V. Gottifredi, The identification of translesion DNA synthesis regulators: Inhibitors in the spotlight. DNA Repair 32, 158–164 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.M. B. Federico, S. O. Siri, N. L. Calzetta, N. S. Paviolo, M. B. de la Vega, J. Martino, M. C. Campana, L. Wiesmüller, V. Gottifredi, Unscheduled MRE11 activity triggers cell death but not chromosome instability in polymerase eta-depleted cells subjected to UV irradiation. Oncogene 39, 3952–3964 (2020). [DOI] [PubMed] [Google Scholar]
- 21.A. V. Makarova, C. Grabow, L. V. Gening, V. Z. Tarantul, T. H. Tahirov, T. Bessho, Y. I. Pavlov, Inaccurate DNA synthesis in cell extracts of yeast producing active human DNA polymerase iota. PLOS ONE 6, e16612 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.L. Haracska, N. Acharya, I. Unk, R. E. Johnson, J. Hurwitz, L. Prakash, S. Prakash, A single domain in human DNA polymerase iota mediates interaction with PCNA: Implications for translesion DNA synthesis. Mol. Cell. Biol. 25, 1183–1190 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.P. Kotsantis, E. Petermann, S. J. Boulton, Mechanisms of oncogene-induced replication stress: Jigsaw falling into place. Cancer Discov. 8, 537–555 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.M. A. Gonzalez Besteiro, V. Gottifredi, The fork and the kinase: A DNA replication tale from a CHK1 perspective. Mutat. Res. Rev. Mutat. Res. 763, 168–180 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.A. A. Goodarzi, A. T. Noon, D. Deckbar, Y. Ziv, Y. Shiloh, M. Löbrich, P. A. Jeggo, ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol. Cell 31, 167–177 (2008). [DOI] [PubMed] [Google Scholar]
- 26.A. K. Ashley, M. Shrivastav, J. Nie, C. Amerin, K. Troksa, J. G. Glanzer, S. Liu, S. O. Opiyo, D. D. Dimitrova, P. le, B. Sishc, S. M. Bailey, G. G. Oakley, J. A. Nickoloff, DNA-PK phosphorylation of RPA32 Ser4/Ser8 regulates replication stress checkpoint activation, fork restart, homologous recombination and mitotic catastrophe. DNA Repair 21, 131–139 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.S. F. Mansilla, G. Soria, M. B. Vallerga, M. Habif, W. Martinez-Lopez, C. Prives, V. Gottifredi, UV-triggered p21 degradation facilitates damaged-DNA replication and preserves genomic stability. Nucleic Acids Res. 41, 6942–6951 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.M. B. Vallerga, S. F. Mansilla, M. B. Federico, A. P. Bertolin, V. Gottifredi, Rad51 recombinase prevents Mre11 nuclease-dependent degradation and excessive PrimPol-mediated elongation of nascent DNA after UV irradiation. Proc. Natl. Acad. Sci. U.S.A. 112, E6624–E6633 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.P. Kannouche, B. C. Broughton, M. Volker, F. Hanaoka, L. H. F. Mullenders, A. R. Lehmann, Domain structure, localization, and function of DNA polymerase eta, defective in xeroderma pigmentosum variant cells. Genes Dev. 15, 158–172 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.J. G. Jansen, P. Temviriyanukul, N. Wit, F. Delbos, C. A. Reynaud, H. Jacobs, N. de Wind, Redundancy of mammalian Y family DNA polymerases in cellular responses to genomic DNA lesions induced by ultraviolet light. Nucleic Acids Res. 42, 11071–11082 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.A. R. Lehmann, A. Niimi, T. Ogi, S. Brown, S. Sabbioneda, J. F. Wing, P. L. Kannouche, C. M. Green, Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair 6, 891–899 (2007). [DOI] [PubMed] [Google Scholar]
- 32.K. N. Choe, G. L. Moldovan, Forging ahead through darkness: PCNA, still the principal conductor at the replication fork. Mol. Cell 65, 380–392 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O. D. Scharer, Nucleotide excision repair in eukaryotes. Cold Spring Harb. Perspect. Biol. 5, a012609 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.S. Biber, H. Pospiech, V. Gottifredi, L. Wiesmuller, Multiple biochemical properties of the p53 molecule contribute to activation of polymerase iota-dependent DNA damage tolerance. Nucleic Acids Res. 48, 12188–12203 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.C. G. Sansam, D. Goins, J. C. Siefert, E. A. Clowdus, C. L. Sansam, Cyclin-dependent kinase regulates the length of S phase through TICRR/TRESLIN phosphorylation. Genes Dev. 29, 555–566 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.A. Maya-Mendoza, P. Moudry, J. M. Merchut-Maya, M. H. Lee, R. Strauss, J. Bartek, High speed of fork progression induces DNA replication stress and genomic instability. Nature 559, 279–284 (2018). [DOI] [PubMed] [Google Scholar]
- 37.S. Minocherhomji, S. Ying, V. A. Bjerregaard, S. Bursomanno, A. Aleliunaite, W. Wu, H. W. Mankouri, H. Shen, Y. Liu, I. D. Hickson, Replication stress activates DNA repair synthesis in mitosis. Nature 528, 286–290 (2015). [DOI] [PubMed] [Google Scholar]
- 38.S. O. Siri, J. Martino, V. Gottifredi, Structural chromosome instability: Types, origins, consequences, and therapeutic opportunities. Cancers 13, 3056 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.A. P. Bertolin, J. S. Hoffmann, V. Gottifredi, Under-replicated DNA: The byproduct of large genomes? Cancers 12, 2764 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.M. Yekezare, B. Gomez-Gonzalez, J. F. Diffley, Controlling DNA replication origins in response to DNA damage—Inhibit globally, activate locally. J. Cell Sci. 126, 1297–1306 (2013). [DOI] [PubMed] [Google Scholar]
- 41.S. Tirman, E. Cybulla, A. Quinet, A. Meroni, A. Vindigni, PRIMPOL ready, set, reprime! Crit. Rev. Biochem. Mol. Biol. 56, 17–30 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.A. Quinet, D. J. Martins, A. T. Vessoni, D. Biard, A. Sarasin, A. Stary, C. F. M. Menck, Translesion synthesis mechanisms depend on the nature of DNA damage in UV-irradiated human cells. Nucleic Acids Res. 44, 5717–5731 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.A. Quinet, S. Tirman, J. Jackson, S. Šviković, D. Lemaçon, D. Carvajal-Maldonado, D. González-Acosta, A. T. Vessoni, E. Cybulla, M. Wood, S. Tavis, L. F. Z. Batista, J. Méndez, J. E. Sale, A. Vindigni, PRIMPOL-mediated adaptive response suppresses replication fork reversal in BRCA-deficient cells. Mol. Cell 77, 461–474.e9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.G. Bai, C. Kermi, H. Stoy, C. J. Schiltz, J. Bacal, A. M. Zaino, M. K. Hadden, B. F. Eichman, M. Lopes, K. A. Cimprich, HLTF promotes fork reversal, limiting replication stress resistance and preventing multiple mechanisms of unrestrained DNA synthesis. Mol. Cell 78, 1237–1251.e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.K. Jacobs, C. Doerdelmann, J. Krietsch, D. González-Acosta, N. Mathis, S. Kushinsky, E. Guarino, C. Gómez-Escolar, D. Martinez, J. A. Schmid, P. J. Leary, R. Freire, A. R. Ramiro, C. M. Eischen, J. Mendez, M. Lopes, Stress-triggered hematopoietic stem cell proliferation relies on PrimPol-mediated repriming. Mol. Cell 82, 4176–4188.e8 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.S. Tirman, A. Quinet, M. Wood, A. Meroni, E. Cybulla, J. Jackson, S. Pegoraro, A. Simoneau, L. Zou, A. Vindigni, Temporally distinct post-replicative repair mechanisms fill PRIMPOL-dependent ssDNA gaps in human cells. Mol. Cell 81, 4026–4040.e8 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.M.-M. Genois, J.-P. Gagné, T. Yasuhara, J. Jackson, S. Saxena, M.-F. Langelier, I. Ahel, M. T. Bedford, J. M. Pascal, A. Vindigni, G. G. Poirier, L. Zou, CARM1 regulates replication fork speed and stress response by stimulating PARP1. Mol. Cell 81, 784–800 e788 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.M. Vujanovic, J. Krietsch, M. C. Raso, N. Terraneo, R. Zellweger, J. A. Schmid, A. Taglialatela, J.-W. Huang, C. L. Holland, K. Zwicky, R. Herrador, H. Jacobs, D. Cortez, A. Ciccia, L. Penengo, M. Lopes, Replication fork slowing and reversal upon DNA damage require PCNA polyubiquitination and ZRANB3 DNA translocase activity. Mol. Cell 67, 882–890.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.V. Gottifredi, L. Wiesmuller, The tip of an iceberg: Replication-associated functions of the tumor suppressor p53. Cancers 10, 250 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.P. Garg, C. M. Stith, N. Sabouri, E. Johansson, P. M. Burgers, Idling by DNA polymerase delta maintains a ligatable nick during lagging-strand DNA replication. Genes Dev. 18, 2764–2773 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.V. Khare, K. A. Eckert, The proofreading 3′-->5′ exonuclease activity of DNA polymerases: A kinetic barrier to translesion DNA synthesis. Mutat. Res. 510, 45–54 (2002). [DOI] [PubMed] [Google Scholar]
- 52.O. Rechkoblit, Y. K. Gupta, R. Malik, K. R. Rajashankar, R. E. Johnson, L. Prakash, S. Prakash, A. K. Aggarwal, Structure and mechanism of human PrimPol, a DNA polymerase with primase activity. Sci. Adv. 2, e1601317 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.E. O. Boldinova, E. A. Belousova, D. I. Gagarinskaya, E. A. Maltseva, S. N. Khodyreva, O. I. Lavrik, A. V. Makarova, Strand displacement activity of primpol. Int. J. Mol. Sci. 21, 9027 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.E. O. Boldinova, A. V. Yudkina, E. S. Shilkin, D. I. Gagarinskaya, A. G. Baranovskiy, T. H. Tahirov, D. O. Zharkov, A. V. Makarova, Translesion activity of PrimPol on DNA with cisplatin and DNA-protein cross-links. Sci. Rep. 11, 17588 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.S. Šviković, A. Crisp, S. M. Tan-Wong, T. A. Guilliam, A. J. Doherty, N. J. Proudfoot, G. Guilbaud, J. E. Sale, R-loop formation during S phase is restricted by PrimPol-mediated repriming. EMBO J. 38, e99793 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.C. E. Edmunds, L. J. Simpson, J. E. Sale, PCNA ubiquitination and REV1 define temporally distinct mechanisms for controlling translesion synthesis in the avian cell line DT40. Mol. Cell 30, 519–529 (2008). [DOI] [PubMed] [Google Scholar]
- 57.L. Wan, J. Lou, Y. Xia, B. Su, T. Liu, J. Cui, Y. Sun, H. Lou, J. Huang, hPrimpol1/CCDC111 is a human DNA primase-polymerase required for the maintenance of genome integrity. EMBO Rep. 14, 1104–1112 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.T. A. Guilliam, S. K. Jozwiakowski, A. Ehlinger, R. P. Barnes, S. G. Rudd, L. J. Bailey, J. M. Skehel, K. A. Eckert, W. J. Chazin, A. J. Doherty, Human PrimPol is a highly error-prone polymerase regulated by single-stranded DNA binding proteins. Nucleic Acids Res. 43, 1056–1068 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.M. I. Martinez-Jimenez, A. Lahera, L. Blanco, Human PrimPol activity is enhanced by RPA. Sci. Rep. 7, 783 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.T. A. Guilliam, N. C. Brissett, A. Ehlinger, B. A. Keen, P. Kolesar, E. M. Taylor, L. J. Bailey, H. D. Lindsay, W. J. Chazin, A. J. Doherty, Molecular basis for PrimPol recruitment to replication forks by RPA. Nat. Commun. 8, 15222 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.C. Kim, B. F. Paulus, M. S. Wold, Interactions of human replication protein A with oligonucleotides. Biochemistry 33, 14197–14206 (1994). [DOI] [PubMed] [Google Scholar]
- 62.K. Cong, S. B. Cantor, Exploiting replication gaps for cancer therapy. Mol. Cell 82, 2363–2369 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.N. L. Calzetta, M. A. Gonzalez Besteiro, V. Gottifredi, Mus81-Eme1-dependent aberrant processing of DNA replication intermediates in mitosis impairs genome integrity. Sci. Adv. 6, eabc8257 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Biology Software, Cloud-based platform for biotech R&D | Benchling (2017); https://benchling.com.
- 65.J. Speroni, M. B. Federico, S. F. Mansilla, G. Soria, V. Gottifredi, Kinase-independent function of checkpoint kinase 1 (Chk1) in the replication of damaged DNA. Proc. Natl. Acad. Sci. U.S.A. 109, 7344–7349 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.H. Beck, V. Nähse-Kumpf, M. S. Y. Larsen, K. A. O'Hanlon, S. Patzke, C. Holmberg, J. Mejlvang, A. Groth, O. Nielsen, R. G. Syljuåsen, C. S. Sørensen, Cyclin-dependent kinase suppression by WEE1 kinase protects the genome through control of replication initiation and nucleotide consumption. Mol. Cell. Biol. 32, 4226–4236 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.E. Petermann, M. Woodcock, T. Helleday, Chk1 promotes replication fork progression by controlling replication initiation. Proc. Natl. Acad. Sci. U.S.A. 107, 16090–16095 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.S. F. Mansilla, A. P. Bertolin, V. Bergoglio, M. J. Pillaire, M. A. González Besteiro, C. Luzzani, S. G. Miriuka, C. Cazaux, J. S. Hoffmann, V. Gottifredi, Cyclin Kinase-independent role of p21(CDKN1A) in the promotion of nascent DNA elongation in unstressed cells. eLife 5, e18020 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.J. A. Harrigan, R. Belotserkovskaya, J. Coates, D. S. Dimitrova, S. E. Polo, C. R. Bradshaw, P. Fraser, S. P. Jackson, Replication stress induces 53BP1-containing OPT domains in G1 cells. J. Cell Biol. 193, 97–108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.M. A. G. Besteiro, N. L. Calzetta, S. M. Loureiro, M. Habif, R. Bétous, M.-J. Pillaire, A. Maffia, S. Sabbioneda, J.-S. Hoffmann, V. Gottifredi, Chk1 loss creates replication barriers that compromise cell survival independently of excess origin firing. EMBO J. 38, e101284 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.A. Quinet, D. Carvajal-Maldonado, D. Lemacon, A. Vindigni, DNA fiber analysis: Mind the gap! Methods Enzymol. 591, 55–82 (2017). [DOI] [PubMed] [Google Scholar]
- 72.B. S. Plosky, A. E. Vidal, A. R. F. de Henestrosa, M. P. McLenigan, J. P. McDonald, S. Mead, R. Woodgate, Controlling the subcellular localization of DNA polymerases iota and eta via interactions with ubiquitin. EMBO J. 25, 2847–2855 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S13
Legend for GFP-Pol ι mutants sequences
Legend for Source data
GFP-Pol ι mutants sequences
Source data