Significance
Amino acid substitutions within the cervid prion protein (PrP) can decrease susceptibility to chronic wasting disease, generally with more prominent effects in homozygous animals. Using novel gene-targeted mouse models expressing S138N reindeer/caribou PrP, we demonstrate subclinical infection with prion seeding activity in spleen and fecal prion shedding in heterozygous 138SN and homozygous 138NN mice. A lower percentage of heterozygous 138SN-PrP than homozygous 138NN-PrP expressing mice harbored seeding-efficient prions in tissues. This is caused by dominant-negative interference of the PrP variants occurring only if they are coexpressed. Our findings are relevant to inform conservation efforts for caribou, an endangered species in North America. Furthermore, our study provides new mechanistic insights into genetic resistance and dominant-negative interference of conversion-competent PrP variants.
Keywords: prion, chronic wasting disease, prion protein polymorphism, pmca, gene-targeted mouse model
Abstract
Prions are proteinaceous infectious particles that replicate by structural conversion of the host-encoded cellular prion protein (PrPC), causing fatal neurodegenerative diseases in mammals. Species-specific amino acid substitutions (AAS) arising from single nucleotide polymorphisms within the prion protein gene (Prnp) modulate prion disease pathogenesis, and, in several instances, reduce susceptibility of homo- or heterozygous AAS carriers to prion infection. However, a mechanistic understanding of their protective effects against clinical disease is missing. We generated gene-targeted mouse infection models of chronic wasting disease (CWD), a highly contagious prion disease of cervids. These mice express wild-type deer or PrPC harboring the S138N substitution homo- or heterozygously, a polymorphism found exclusively in reindeer (Rangifer tarandus spp.) and fallow deer (Dama dama). The wild-type deer PrP-expressing model recapitulated CWD pathogenesis including fecal shedding. Encoding at least one 138N allele prevented clinical CWD, accumulation of protease-resistant PrP (PrPres) and abnormal PrP deposits in the brain tissue. However, prion seeding activity was detected in spleens, brains, and feces of these mice, suggesting subclinical infection accompanied by prion shedding. 138N-PrPC was less efficiently converted to PrPres in vitro than wild-type deer (138SS) PrPC. Heterozygous coexpression of wild-type deer and 138N-PrPC resulted in dominant-negative inhibition and progressively diminished prion conversion over serial rounds of protein misfolding cyclic amplification. Our study indicates that heterozygosity at a polymorphic Prnp codon can confer the highest protection against clinical CWD and highlights the potential role of subclinical carriers in CWD transmission.
Prion diseases are invariably fatal neurodegenerative disorders of humans, e.g., Creutzfeldt–Jakob disease (CJD), and animals, such as scrapie in sheep and goat or bovine spongiform encephalopathy (BSE) in cattle (1). They are caused by infectious proteins termed prions, which consist of PrPSc, a misfolded form of the cellular prion protein (PrPC) that aggregates and accumulates in the brains of affected individuals (2). Chronic wasting disease (CWD) is a highly contagious prion disease of cervids. First described in mule deer in Colorado in the late 1960s (3, 4), it is now found in free-ranging and domesticated cervids in 30 US states, four Canadian provinces, South Korea, and Nordic countries in Europe (5–8). Infected animals accumulate prions in lymphoreticular and other peripheral tissues, e.g., skeletal muscle, and shed infectious prions in saliva, urine and feces, contributing to direct and environmental transmission and rapidly increasing geographic distribution of CWD (9–12). This threatens endangered cervid species such as caribou (Rangifer tarandus spp.) and poses a potential risk to humans, as recent studies indicate that the cervid-to-human transmission barrier might not be impermeable (13, 14). Therefore, understanding factors reducing susceptibility to limit transmission of CWD is of critical importance. Single nucleotide polymorphisms (SNPs) within the prion protein gene (Prnp) that result in nonsynonymous amino acid substitutions (AAS) are known to affect susceptibility to prion infection, such as the M129V polymorphism in humans (15). In cervids, the G96S and Q95H AAS found in white-tailed deer (WTD) (Odocoileus virginianus), S225F in mule deer (Odocoileus hemionus), and M132L found in elk (Cervus canadensis) cause prolonged CWD incubation periods and incomplete attack rates (16–19). In wild populations, cervids expressing these allelic variants were less likely found positive for CWD than those expressing wild-type PrPC (20, 21). However, not all cervid PrP AAS delay CWD pathogenesis, such as those found in red deer (Cervus elaphus) and moose (Alces sp.), and some were shown to result in the emergence of new prion strains (22).
An AAS from serine to asparagine at codon 138 of the cervid Prnp has been reported exclusively in fallow deer (Dama dama) and reindeer/caribou (Rangifer tarandus spp.; 23–24). Fallow deer only encode asparagine at this codon (138NN) and are resistant to peripheral prion infection (23), but susceptible to intracerebral (i.c.) prion inoculation, albeit with prolonged disease incubation periods (25). The majority of reindeer/caribou are homozygous for serine at Prnp codon 138 (138SS; wild-type), with 138SN and 138NN genotypes present in North American populations (24, 26, 27). Interestingly, results from two different CWD infection studies in reindeer are not straightforward (28, 29). One study reported that homozygous 138NN reindeer succumb to disease faster than their wild-type and heterozygous counterparts upon i.c., but not peripheral, infection (28). Another study reported that wild-type reindeer were susceptible to CWD, while the only 138SN reindeer in the study did not develop clinical prion disease upon oral exposure at the time the study was terminated (29), indicating reduced susceptibility. However, only small numbers of 138NN and/or 138SN reindeer were included in these studies and inoculating sufficient numbers of each genotype with a panel of well-characterized CWD strains appears not feasible. Testing different strains is important as genotypes associated with resistance to CWD infection can be permissive to less prevalent CWD strains (18, 30).
To overcome these limitations and systematically study the impact of the S138N substitution on CWD pathogenesis, fecal shedding, and prion conversion, we generated gene-targeted mice expressing wild-type deer (138SS), or the 138SN and 138NN caribou Prnp genotypes, by replacing the mouse Prnp protein coding sequence. We infected them i.c. or intraperitoneally (i.p.) with a range of North American CWD isolates from reindeer and WTD, originating from animals with different genotypes and containing diverse CWD strains (18, 29–31). Mice expressing the wild-type deer PrP (Prnp.Cer.Wt) all succumbed to CWD, regardless of isolate and infection route, and recapitulated the PrPSc distribution in lymphatic organs and fecal shedding observed in cervids. Meanwhile, their 138SN (Prnp.Cer.138SN) and 138NN (Prnp.Cer.138NN) counterparts did not develop clinical disease. However, prion-seeding activity was detectable in the brain, spleen, and feces, indicating subclinical infection and potential for contagiousness. Intriguingly, seeding activity was more abundant in the spleens than in the brains of these mice, even in i.c. inoculated animals, but more limited in Prnp.Cer.138SN mice than in the other genotypes. Using in vitro protein misfolding cyclic amplification (PMCA) assays we demonstrated that the conversion efficiency of PrP substrate only from Prnp.Cer.138SN brain homogenates (BH) decreased over subsequent rounds in serial PMCA. This effect was not observed when we combined Prnp.Cer.Wt and Prnp.Cer.138NN mouse BH at an equal ratio (1:1) as substrate, directly indicating conversion-limiting interactions of PrP variants only when coexpressed in cis, i.e., within the same cells.
Caribou in North America are categorized as endangered and attempts at breeding programs are underway (32, 33). Our findings will be valuable information for caribou preservation programs and provide novel insights into conversion-limiting effects exerted upon coexpression of Prnp allelic variants.
Results
Generation and Characterization of Gene-Targeted Mouse Models.
We used the murine C57BL/6-derived embryonic stem cell line Bruce 4 and a CRISPR/Cas9-based gene targeting strategy to replace (knock-in) the coding sequence for mouse PrP with that for 138S or 138N cervid PrP. From these cells, we generated two novel mouse lines homozygous for 138S (Prnp.Cer.Wt) or 138N (Prnp.Cer.138NN) PrP, which we crossed to generate heterozygous offspring (Prnp.Cer.138SN). We confirmed the Prnp sequences of the mouse lines as well as mRNA and protein levels (SI Appendix, Fig. S1 A–C). We found no significant differences in total PrPC and Prnp (mRNA) expression levels in the brains of Prnp.Cer.Wt, Prnp.Cer.138NN, and C57BL/6N control mice (SI Appendix, Fig. S1B). We also confirmed correct cellular processing of PrPC by de-glycosylation using PNGase-F and Endo-H enzymes to confirm the presence of complex N-linked glycans (SI Appendix, Fig. S1C).
Propagation of CWD Prions in Gene-Targeted Mouse Models.
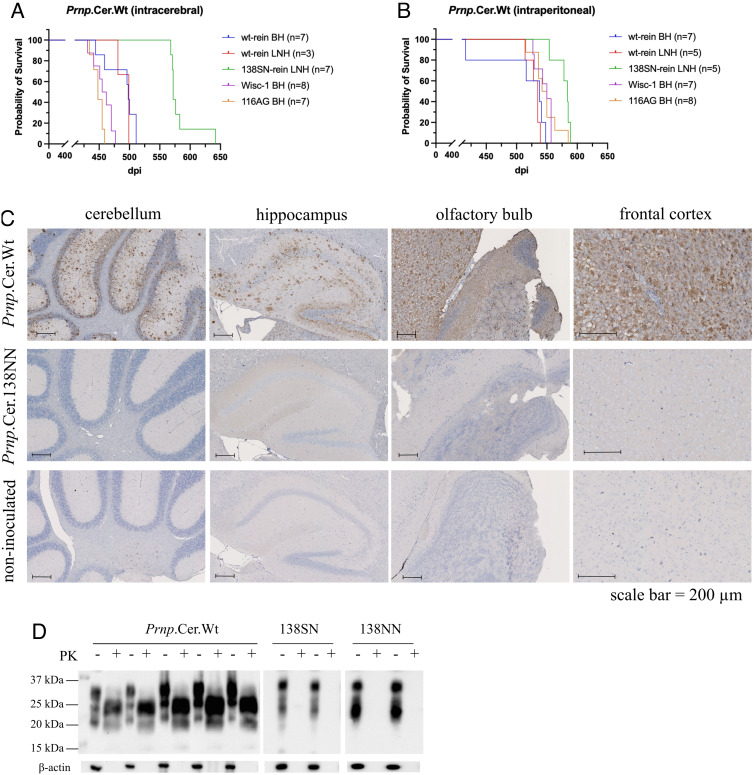
We used well-characterized BH from CWD-positive WTD with the wild-type Prnp genotype harboring the Wisc-1 CWD strain (18, 31), from WTD heterozygous for the A116G polymorphism containing 116AG prions (30, 31), and from wild-type reindeer (29), as well as CWD-positive lymph node homogenates (LNH) from wild-type and 138SN reindeer (29), for i.c. and i.p. inoculation into Prnp.Cer.Wt, Prnp.Cer.138NN, and Prnp.Cer.138SN gene-targeted mice. Of note, the 138SN reindeer LNH was derived from an orally infected reindeer not showing clinical signs when euthanized (29). Therefore, prior to inoculation, we confirmed the presence of prions in the reindeer tissue homogenates by real-time quaking-induced conversion (RT-QuIC) and compared the amounts of seeding activity by endpoint dilution. The LNHs, even though they were derived from a clinical (wild-type) and a nonclinical (138SN) reindeer, respectively, harbored comparable levels of seeding activity with a dilution endpoint of 2 × 10−3, while seeding activity in the wild-type reindeer BH was 100-fold higher (SI Appendix, Fig. S2). All inoculated Prnp.Cer.Wt mice developed terminal prion disease (Fig. 1 A and B and Table 1). Their brains were positive for abnormal PrP deposition in immunohistochemistry (IHC) (Fig. 1C and SI Appendix, Fig. S3) and proteinase K-resistant PrPSc (PrPres) in western blot (Fig. 1D), confirming the susceptibility of this mouse line to both i.c. and i.p. (peripheral) CWD infection.
Fig. 1.
Propagation of white-tailed deer and reindeer CWD prions in gene-targeted mice. Survival times were measured in number of days postinoculation (dpi). Gene-targeted Prnp.Cer.Wt mice were inoculated (A) intracerebrally and (B) intraperitoneally. Number of mice per group included in the survival graph analyses are indicated in brackets. Survival graphs were generated in Graphpad Prism v9. The x axis represents dpi, and the y axis represents the percentage of mice surviving. (C) Representative immunohistochemistry (IHC) showing abnormal PrP deposits in brain sections of Prnp.Cer.Wt (511 dpi) and Prnp.Cer.138NN (616 dpi; subclinical) mice inoculated intracerebrally with wild-type reindeer BH. A non-inoculated Prnp.Cer.Wt mouse brain was used as negative control. IHC figures from left to right: cerebellum, hippocampus, olfactory bulb, and frontal cortex. (D) Representative western blots for PrPres detection in brains of terminal Prnp.Cer.Wt mice (488.4 ± 26.3 dpi) and subclinical Prnp.Cer.138SN and NN mice (≥700 dpi) inoculated intracerebrally with wild-type reindeer BH. Samples were digested with 50 µg/mL proteinase K for 1 h and PrPres was detected using the 4H11 anti-PrP antibody (1:500). Beta-actin (β-actin) was used as a loading control. PK: proteinase K; kDa: kilodalton; BH: brain homogenate; LNH: lymph node homogenate; wt: wild-type; rein: reindeer; dpi: days post-inoculation.
Table 1.
Susceptibility of knock-in mice expressing wild-type deer (138SS; 116AA), 138SN and 138NN PrPC to reindeer or white-tailed deer CWD isolates following intracerebral (i.c.) or intraperitoneal (i.p.) inoculation.
| Average survival times ± SD in days (N1/Ntotal) | |||||
|---|---|---|---|---|---|
| Inoculum | Tissue | roi | Prnp.Cer.Wt | Prnp.Cer.138SN* | Prnp.Cer.138NN* |
| Reindeer CWD (NA) | |||||
| wild-type (138SS) | Brain | i.c. | 488.4 ± 26.3 (7/7) | 514 (0/2) | 594 (0/3) |
| ID #31-47 | 714 (0/3) | 702 (0/2) | |||
| i.p. | 532.8 ± 13.5 (6/6) | 529 (0/2) | 588 (0/3) | ||
| 715 (0/4) | 702 (0/2) | ||||
| RPLN | i.c. | 493 ± 10.4 (3/3) | 514 (0/2) | 509, 523 (0/4) | |
| 718 (0/3) | 701 (0/4) | ||||
| i.p. | 530.2 ± 9.9 (5/6) | 529 (0/2) | 581 (0/3) | ||
| 711 (0/3) | 755 (0/2) | ||||
| 138SN | RPLN | i.c. | 583.4 ± 26.3 (7/7)† | 558, 581 (0/3) | 606 (0/3) |
| ID #17 | 710 (0/4) | 702 (0/3) | |||
| i.p. | 578 ± 14 (5/5) | 578 (0/2) | 435-611 (0/4) | ||
| 709 (0/4) | 701 (0/1) | ||||
| White-tailed deer CWD (NA) | |||||
| wild-type (116AA; 138SS) | Brain | i.c. | 457 ± 15.7 (8/8) | ni | 561, 593 (0/4) |
| ID #1277; Wisc-1 strain | 700, 705 (0/2) | ||||
| i.p. | 545.7 ± 13.1 (7/7) | ni | 585 (0/3) | ||
| 701 (0/3) | |||||
| 116AG | Brain | i.c. | 448.6 ± 8.8 (7/7) | ni | 581, 612 (0/4) |
| ID N/A; 116AG strain | 705 (0/2) | ||||
| i.p. | 547.1 ± 20.7 (8/8) | ni | 513 (0/3) | ||
| 701 (0/2) | |||||
SD: standard deviation; N1: number of clinical mice; Ntotal: number of mice analyzed per group; CWD: chronic wasting disease; NA: North America; roi: route of inoculation; RPLN: retropharyngeal lymph node; i.c.: intracerebral; i.p.: intraperitoneal; ni: not inoculated; N/A: not applicable.
*Prnp.Cer.138SN and Prnp.Cer.138NN samples are further divided into two subgroups each i.e., mice euthanized around the latest survival time point of their terminal Prnp.Cer.Wt counterparts and mice euthanized upon our experimental endpoint at >700 dpi, corresponding to top and bottom rows within each group, respectively.
†One mouse had a longer survival period (642 dpi) than the rest (< 590 dpi).
Mice inoculated i.c. with the WTD-116AG isolate had the shortest survival time at 448.6 ± 8.8 dpi, followed by those inoculated i.c. with WTD Wisc-1 prions at 457.1 ± 15.7 dpi (Fig. 1A and Table 1). The corresponding i.p.-inoculated groups reached terminal disease ~98.5 and ~88.6 d later, respectively (545.7 ± 13.1 and 547.1 ± 20.7 dpi, respectively; Fig. 1B and Table 1). Mice inoculated i.c with the wild-type reindeer BH and LNH reached terminal endpoints at 488.4 ± 26.3 dpi and 493 ± 10.4 dpi, respectively (Fig. 1A and Table 1). Those inoculated i.p. came down at 511.8 ± 54.9 dpi and 530.2 ± 9.9 dpi, respectively (Fig. 1B and Table 1). In contrast to all other inocula, groups inoculated i.c and i.p with the 138SN reindeer LNH presented no significant difference, with survival times at 583.4 ± 26.3 dpi and 578.0 ± 14.0 dpi, respectively (Fig. 1 A and B and Table 1). None of the inoculated Prnp.Cer.138SN and Prnp.Cer.138NN mice developed clinical disease, showed accumulation of PrPres, or had detectable PrP deposits at an experimental endpoint of ≥700 dpi (Fig. 1 C and D and Table 1).
Prion Seeding Activity in Spleens and Brains of Infected Prnp.Cer.138SN and Prnp.Cer.138NN Mice.
To determine whether nonclinical Prnp.Cer.138SN and Prnp.Cer.138NN mice inoculated with the CWD isolates accumulated prions in their brains and spleens, we euthanized a subset of mice (n = 1 to 2 per group) around the latest survival time point of their Prnp.Cer.Wt counterparts. The remaining mice (n = 2 to 5 per group) were euthanized at a predetermined experimental endpoint of ≥700 dpi (Table 1), in the absence of clinical signs.
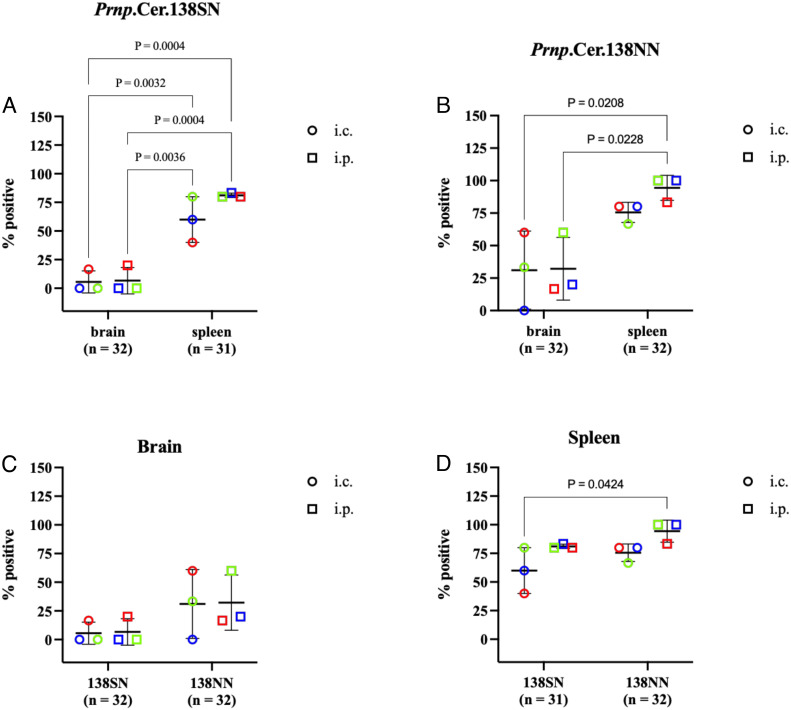
We detected seeding activity by RT-QuIC in 71% of Prnp.Cer.138SN (n = 31) and 88.5% of Prnp.Cer.138NN (n = 52) mouse spleens tested (Fig. 2 A and B and Table 2). Sixty percent of Prnp.Cer.138SN mouse spleens tested positive for seeding activity when mice were inoculated i.c., compared to 81.3% when inoculated i.p (Fig. 2A and Table 2). Similarly, 80.8% of Prnp.Cer.138NN mouse spleens had seeding activity when mice were inoculated i.c., compared to 96.2% in i.p.-inoculated groups (Fig. 2B and Table 2). There were no significant differences in the percentage of spleens with detectable seeding activity between Prnp.Cer.138SN and Prnp.Cer.138NN mice, except between spleens of i.c.-inoculated Prnp.Cer.138SN and i.p.-inoculated Prnp.Cer.138NN mice, which is not a meaningful comparison (Fig. 2D). Ninety-eight percent (97.9%) of Prnp.Cer.Wt mouse spleens tested positive for seeding activity; only one mouse, inoculated i.p. with the 138SN reindeer LNH, had no detectable seeding activity out of the 48 mouse spleens tested (Table 2). Abnormal PrP deposition was also detected in Prnp.Cer.Wt, 138SN and 138NN mouse spleens (SI Appendix, Fig. S4).
Fig. 2.
Percentage of spleens and brains positive in RT-QuIC in subclinical (A) Prnp.Cer.138SN mice and (B) Prnp.Cer.138NN mice inoculated with wild-type reindeer BH (blue) or LNH (green), or 138SN reindeer LNH (red). Comparison of (C) brains and (D) spleens between the two mouse lines. Each data point represents the percentage of positive samples out of the total number of samples analyzed per group (n = 3 to 8 per group; grouped by reindeer isolate and route of inoculation). Samples were considered positive when a minimum of 2 out of 4 replicates crossed the threshold relative fluorescence unit (RFU) at any seed dilution (refer to Table 2). The y axis represents the percentage of positive samples and the x axis the different tissues (A and B) or mouse lines (C and D). Groups were compared using a two-way ANOVA followed by Sidak’s multiple comparison test. Only statistically significant comparisons are shown (P value < 0.05). Statistical analyses and graphs were generated in Graphpad Prism v9. i.c.: intracerebral; i.p.: intraperitoneal.
Table 2.
Percentage of brains and spleens positive for prion-seeding activity in RT-QuIC per the total number of samples tested within each inoculation group
| Percentage (%) of positive samples per RT-QuIC seed dilution | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Prnp.Cer.Wt | Prnp.Cer.138SN* | Prnp.Cer.138NN* | |||||||||||||||
| Spleen | Brain | Spleen | Brain | Spleen | |||||||||||||
| Inoculum | Tissue | roi | −1 | −2 | −3 | −1 | −2 | −3 | −1 | −2 | −3 | −1 | −2 | −3 | −1 | −2 | −3 |
| Reindeer CWD (NA) | |||||||||||||||||
| WT (138SS) | Brain | i.c. | 100% | 100% | 50% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 100% | 67% | 33% |
| ID #31-47 | 0% | 0% | 0% | 100% | 67% | 67% | 0% | 0% | 0% | 50% | 50% | 0% | |||||
| i.p. | 80% | 100% | 60% | 0% | 0% | 0% | 50% | 50% | 0% | 33% | 33% | 0% | 100% | 33% | 0% | ||
| 0% | 0% | 0% | 100% | 75% | 50% | 0% | 0% | 0% | 50% | 100% | 50% | ||||||
| RPLN | i.c. | 100% | 100% | 33% | 0% | 0% | 0% | 50% | 0% | 0% | 0% | 33% | 0% | 100% | 33% | 0% | |
| 0% | 0% | 0% | 100% | 67% | 33% | 0% | 33% | 0% | 33% | 33% | 0% | ||||||
| i.p. | 80% | 100% | 80% | 0% | 0% | 0% | 50% | 0% | 0% | 33% | 100% | 33% | 100% | 33% | 33% | ||
| 0% | 0% | 0% | 100% | 33% | 100% | 0% | 0% | 0% | 100% | 50% | 0% | ||||||
| 138SN | RPLN | i.c. | 100% | 83% | 67% | 0% | 33% | 0% | 100% | 50% | 0% | 0% | 100% | 33% | 67% | 33% | 67% |
| ID #17 | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 50% | 0% | |||||
| i.p. | 67% | 67% | 33% | 0% | 50% | 0% | 100% | 100% | 0% | 0% | 0% | 0% | 100% | 75% | 50% | ||
| 0% | 0% | 33% | 67% | 33% | 33% | 0% | 50% | 0% | 0% | 0% | 0% | ||||||
| WTD CWD (NA) | |||||||||||||||||
| WT (116AA; 138SS) | Brain | i.c. | 100% | 100% | 67% | ni | ni | ni | ni | ni | ni | 0% | 0% | 0% | 67% | 67% | 0% |
| ID #1277; Wisc-1 strain | 0% | 50% | 0% | 50% | 0% | 0% | |||||||||||
| i.p. | 100% | 100% | 40% | ni | ni | ni | ni | ni | ni | 0% | 0% | 0% | 100% | 100% | 67% | ||
| 0% | 0% | 50% | 100% | 100% | 50% | ||||||||||||
| 116AG | Brain | i.c. | 80% | 100% | 40% | ni | ni | ni | ni | ni | ni | 0% | 0% | 0% | 33% | 100% | 33% |
| ID N/A; 116AG strain | 0% | 50% | 50% | 100% | 100% | 100% | |||||||||||
| i.p. | 67% | 100% | 100% | ni | ni | ni | ni | ni | ni | 0% | 0% | 0% | 67% | 100% | 100% | ||
| 0% | 0% | 0% | 100% | 100% | 50% | ||||||||||||
CWD: chronic wasting disease; NA: North America; roi: route of inoculation; RPLN: retropharyngeal lymph node; i.c.: intracerebral; i.p.: intraperitoneal; ni: not inoculated; N/A: not applicable.
*Prnp.Cer.138SN and NN samples are further divided into two subgroups each i.e., mice euthanized around the latest survival time point of their terminal Prnp.Cer.Wt counterpart and mice euthanized at >700 dpi, corresponding to top and bottom rows of each group, respectively. −1, −2, −3 refers to 2 × 10−1, 2 × 10−2, 2 × 10−3 RT-QuIC seed dilutions, respectively.
When we investigated the brains of Prnp.Cer.138SN (n = 32) and Prnp.Cer.138NN (n = 52) mice for seeding activity in RT-QuIC, we found that only 6.3% of Prnp.Cer.138SN brains tested positive, while 25% of Prnp.Cer.138NN brains tested positive, regardless of inoculum and inoculation route (Fig. 2C and Table 2). However, there was no significant difference in brain seeding activity between i.c.- and i.p.-inoculated animals within and between the two mouse lines (Fig. 2C). We also compared the percentage of RT-QuIC-positive spleens and brains within both mouse lines and found that the number of brains with seeding activity was significantly lower in general when compared to their respective spleens, regardless of inoculum and inoculation route (Fig. 2 A and B). We also detected seeding activity in the spleens of Prnp.Cer.138NN mice much earlier than the brains (~300 dpi), even in i.c.-inoculated animals.
Gene-Targeted Mice Shed Prions in their Feces.
We collected fecal samples from inoculated Prnp.Cer.Wt, Prnp.Cer.138SN and Prnp.Cer.138NN mice upon their terminal or experimental endpoints and tested them for prion seeding activity by RT-QuIC. Out of the 26 Prnp.Cer.Wt samples tested, 16 were positive (61.54%), while 4 out of 8 (50%) of the Prnp.Cer.138SN and only 4 out of 24 (16.67%) of the Prnp.Cer.138NN mice fecal samples were positive. These results show that Prnp.Cer.Wt mice had the highest fecal prion shedding, followed by 138SN and then 138NN mice. These results provide evidence of fecal prion shedding in our gene-targeted mouse models inoculated with CWD prions, regardless of whether they were clinical (Prnp.Cer.Wt) or subclinical (Prnp.Cer.138SN and Prnp.Cer.138NN). Representative fecal RT-QuIC graphs are shown in SI Appendix, Fig. S5.
138NN-PrPC Was Less Efficiently Converted to PrPres In Vitro.
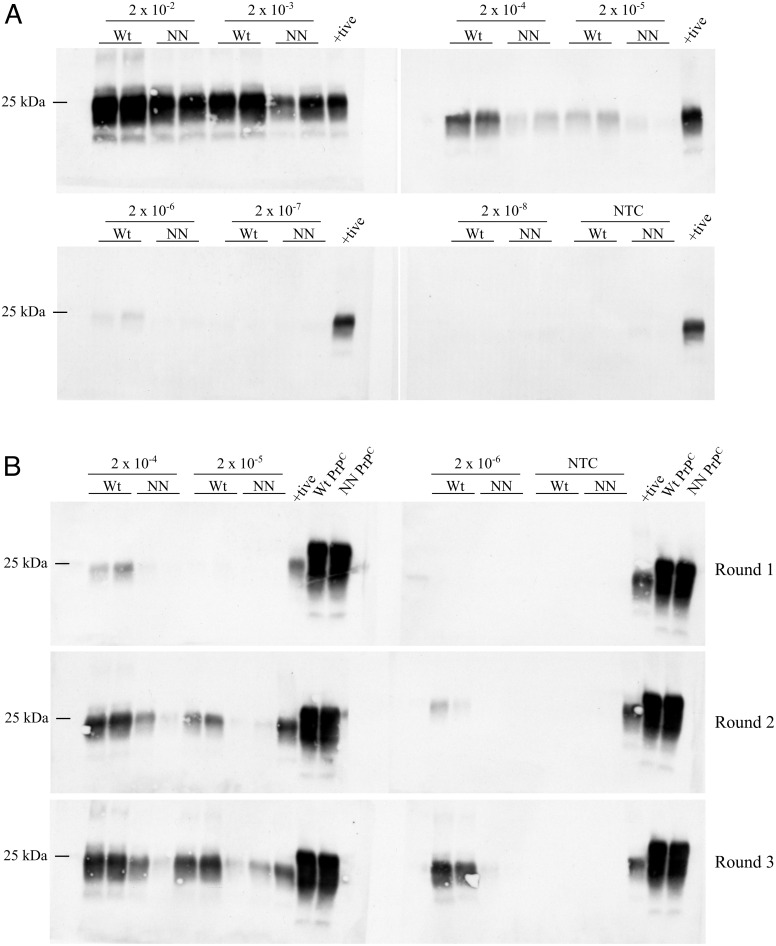
To assess whether the partially protective effect of the S138N AAS is related to an impaired ability to convert into PrPSc, we compared the PrPC to PrPres conversion efficiency of wild-type (138SS) vs. 138NN PrPC in PMCA. We used BH from non-inoculated Prnp.Cer.Wt and Prnp.Cer.138NN mice as substrates and seeded PMCA reactions with 2 × 10−2 to 2 × 10−8 dilutions of 10% (w/v) CWD-positive wild-type reindeer BH, which had the highest seeding activity in RT-QuIC (SI Appendix, Fig. S2). We found that 138NN-PrPC was not as efficiently converted into PrPres as wild-type deer (138SS) PrPC. The 138NN-PrPC substrate was not able to amplify reactions seeded with a 2 × 10−6 BH dilution in round one of PMCA (Fig. 3A), in line with previous in vitro replication studies (34, 35). Next, we performed serial PMCA of reactions seeded with the 2 × 10−4 to 2 × 10−6 BH dilutions to determine whether amplification efficiency increases. Serial PMCA also abolishes a potential species barrier effect as PrPSc newly generated in round one used as seed in subsequent rounds consists of the respective substrate PrPC variant. By round 3, we detected low amounts of PrPres in reactions using the 138NN-PrPC substrate at the 2 × 10−6 seed dilution (Fig. 3B), indicating increased amplification capacity of the 138NN-PrPC over serial PMCA, but still with lower efficiency compared to 138SS-PrPC.
Fig. 3.
Representative western blots showing PrPres levels from PMCA reactions using 10% (w/v) BH from non-inoculated Prnp.Cer.Wt and 138NN knock-in mice (wild-type deer and 138NN brain-derived PrPC) as substrates, seeded with a 10% (w/v) CWD-infected wild-type reindeer BH. (A) One round of PMCA with 2 × 10−2 to 2 × 10−8 seed dilutions. (B) Serial PMCA up to three rounds with 2 × 10−4 to 2 × 10−6 seed dilutions. Positive control for the western blots was a 263K hamster-seeded PMCA reaction with hamster PrPC substrate from a non-inoculated 10% hamster BH. PMCA samples were digested for 1 h with 50 µg/mL proteinase K at 37 °C and positive control with 150 µg/mL proteinase K at 55 °C. PrPres was detected with the 4H11 antibody (1:500). Wt: Prnp.Cer.Wt substrate; NN: Prnp.Cer.138NN substrate; +tive: positive control; NTC: no seed control; kDa: kilodalton. Each experiment was repeated at least three times.
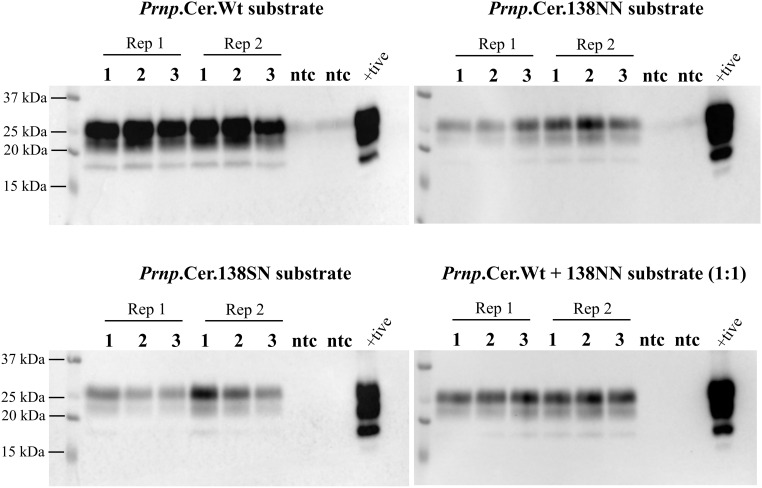
Heterozygous 138SN-PrPC Fails to Sustain Prion Propagation In Vitro.
We then performed serial PMCA using 138SN-PrPC from non-inoculated Prnp.Cer.138SN mouse BH (10% w/v) as substrate in comparison with the Prnp.Cer.Wt and Prnp.Cer.138NN substrates (138SS- and 138NN-PrPC). In addition, we included a control substrate consisting of Prnp.Cer.Wt and Prnp.Cer.138NN BH (10% w/v) mixed at a 1:1 ratio. Serial PMCA reactions were performed, in which the first round was seeded with a 2 × 10−3 dilution of a 10% CWD-positive Prnp.Cer.Wt mouse BH. Interestingly, the 138SN-PrPC substrate from heterozygous Prnp.Cer.138SN mouse brains showed a progressive reduction in its ability to convert into PrPres over three rounds of PMCA (Fig. 4), unlike the 138SS- and 138NN-PrPC substrates, which gained amplification efficiency (Figs. 3B and 4). However, this reduction of amplification efficiency over subsequent PMCA rounds was not observed in reactions with the 1:1 (equal ratios) 138SS- and 138NN-PrPC substrate concoction, suggesting a dominant-negative interference in prion conversion only if the two PrP variants are co-expressed in the same individual (Fig. 4).
Fig. 4.
Representative western blot for n = 3 independent experiments showing PrPres levels from PMCA rounds 1 to 3. Reactions were seeded with a 10% (w/v) mCWD-positive Prnp.Cer.Wt BH at the 2 × 10−3 dilution and passaged 1:10 into the next round. Positive control for the western blot was the BH used to seed the experiments. Samples were digested for 1 h with 50 µg/mL proteinase K at 37 °C. PrPres was detected with the 4H11 antibody (1:500). Rep 1 and 2: replicate 1 and 2; +tive: positive control; ntc: no seed control; kDa: kilodalton.
Discussion
Using novel gene-targeted mouse models expressing wild-type deer (138SS) and 138NN PrP variants, we were able to recapitulate CWD pathogenesis, including fecal prion shedding, without the confounding effects of random integration transgenics, including overexpression, unpredictable and improper spatial expression patterns, and off-target effects (36). Most importantly, we generated mice heterozygous at codon 138 (138SN), allowing us to simulate CWD pathogenesis in heterozygous (138SN) reindeer/caribou. Here we show, for the first time, that heterozygosity for a protective PrP amino acid substitution exhibits more prominent effects than homozygosity in hindering CWD prion propagation only if both allelic variants are co-expressed in “cis” in the brain.
In alignment with other gene-targeted mouse models expressing deer or elk PrPC (37), our Prnp.Cer.Wt mice were susceptible to i.c. and peripheral infection with all North American CWD strains (18, 30, 31) tested here. We also demonstrate that our gene-targeted mice recapitulate CWD prion fecal shedding, with the highest percentage of positive feces found in Prnp.Cer.Wt mice. The variability of shedding between individual mice within the same experimental group is in line with findings in fecal samples collected from different deer at the same time point postinfection, and for the same deer at different time points (38).
Both the Prnp.Cer.138SN and Prnp.Cer.138NN mouse lines failed to develop clinical signs of CWD up to ~2 y postinoculation upon infection with all the CWD isolates tested here, including the 138SN reindeer LNH, which should be able to propagate in the absence of a transmission barrier in the Prnp.Cer.138SN line. However, the majority of Prnp.Cer.138NN and Prnp.Cer.138SN mice were subclinically infected. Prion seeding activity was mostly confined to their spleens, reminiscent of subclinical infection in cases of scrapie and variant CJD (39, 40), with minimum levels of seeding activity in the brains. Interestingly, we observed the preference for spleen replication not only in i.p.-inoculated mice, but also in i.c.-inoculated animals. This suggests that either 1) prions colonized the spleen through anterograde transport or 2) that low concentrations of the inoculum escaped the brain through blood vessels during the inoculation process (41). In either scenario, PrPSc was able to replicate more efficiently in the spleen than the brain tissue environment, possibly due to the presence of spleen-specific cofactors of conversion, or lower PrPC expression levels, resulting in an overall less restrictive prion replication in the spleen, as observed previously (42, 43). The less restrictive replication in the spleen can also explain the similar survival times of Prnp.Cer.Wt mice inoculated i.c. and i.p. with the 138SN reindeer LNH. 138SN LNH prions may not efficiently replicate in the brain even upon i.c. inoculation, but rather colonize the spleen first, resulting in a subset of prions with a wild-type PrP-adapted conformer and their subsequent retrograde transport to the brain, followed by a more efficient replication. Alternatively, the S138N substitution results in a PrPC conformation more amenable to prion conversion in the spleen environment. Initial spleen replication is followed by selection and/or adaptation of prions with the ability to enter and replicate in the brain, giving rise to a concept of PrPC primary structure as a determinant of prion tissue tropism. However, transport and entry into the lymphoid system is not necessarily governed by prion strain (42, 44), once again highlighting the permissibility of the spleen to most, if not all, prions. Thus, we cannot exclude the possibility of a very protracted pathogenesis with old age-related death preceding prion neuroinvasion in our Prnp.Cer.138NN and SN mice.
We were intrigued to find more Prnp.Cer.138NN mouse brains positive in RT-QuIC than Prnp.Cer.138SN, suggesting less prion replication in the brains of heterozygous animals. In cervid Prnp polymorphisms investigated to date, homozygous carriers of the minor alleles appear to be less susceptible to CWD infection than heterozygous individuals (16, 45, 46). This is different from the M129V polymorphism in humans, with 129M homozygotes being the most, and heterozygotes the least, susceptible to sporadic and acquired prion disease (15, 47, 48).
Prnp.Cer.138SN mouse brains also proved to be the least efficient substrate for generating prions in single-round and serial PMCA. Serial PMCA mimics passaging and adaptation and/or prion strain selection in mice (49, 50), and compensates for potential transmission barrier effects between the wild-type reindeer seed and the 138NN- or 138SN-PrPC substrate. While amplification of PrPres using 138NN-PrPC substrate improved by the third round of serial PMCA, passaging reactions with the 138SN-PrPC substrate resulted in reduced PrPres levels. This phenomenon was previously observed in serial PMCA using deer-96SS PrPC substrate (51). However, in our case, the heterozygous PrP, and not the homozygous 138NN-PrPC, presented this outcome, suggesting dominant-negative inhibition, since both 138SS- and 138NN-PrPC alone are competent to sustain and increase prion replication.
Dominant-negative inhibition related to the heterozygous presence of protective PrP polymorphisms can be explained by dimer formation of polymorphic PrP with the conversion-competent wild-type PrP, rendering it nonconvertible, or by competitive binding of cofactors required for conversion (52, 53). Another possible explanation is the “heterozygous inhibition” mechanism suggested by Hizume et al., whereby amyloid formation is delayed in individuals expressing two different PrPC variants that are both conversion-competent, due to a mismatch of PrPSc molecules which need to “stack” on top of each other to form an amyloid, thus acting as amyloid formation “decelerators” to each other (54).
To further these earlier studies, we performed serial PMCA reactions using a mixture of equal amounts of 138SS- and 138NN-PrPC substrates to mimic the heterozygous state, but did not observe a loss of prion amplification, demonstrating that the inhibitory mechanism only occurs when the wild-type (138S) and 138N PrPC alleles are expressed in the same cell. It also indicates that the interaction is strong and not affected by sonication or detergents present in the conversion buffer. These findings support the “molecular heterosis” effect proposed by Tahiri-Alaoui et al., where the time required to form amyloids was significantly extended only in preformed heterozygous soluble β-oligomers (human PrP M129V), when compared to homogenous soluble β-oligomer preparations (129M or V), or a mix of homogenously preformed soluble β-oligomer preparations at equal ratios (55). In this scenario, if heterozygous 138SN animals harbor preformed 138SN soluble β-oligomers, this entity will “stabilize” the 138S and 138N monomers, and since soluble β-oligomers are unable to form β-sheet rich multimers without reverting to its monomeric form, amyloid formation thereby decelerates, as observed in our 138SN-PrPC PMCA reactions. Based on our preferred hypothesis that PrPC conversion in the spleen is more efficient than in the brain, we further propose that the molecular heterosis effect is less pronounced in the spleen, because of lower PrPC expression levels (41) and thus a less likelihood of intermolecular PrP interactions.
To conclude, our study demonstrates that CWD-infected animals harboring S138N PrP might be “silent spreaders” of CWD prions and highlights the importance of lymphatic tissues in the detection of CWD, particularly in caribou, even in the absence of clinical manifestation. It is important to keep in mind that even protective genotypes may be permissive to certain minor or newly emerging CWD strains. Our results provide new mechanistic insights into dominant-negative inhibition of prion conversion, the tissue specificity of this effect, and suggests that PrPC primary structure is a determinant for tissue-specific prion replication.
Materials and Methods
Ethics Statement.
Animal experiments were performed under the strict guidelines of the Canadian Council for Animal Care and were approved by the University of Calgary Health Sciences Animal Care Committee under protocols AC 18-0047 and AC 22-0015.
Generation of Gene-Targeted Mouse Lines.
Bruce 4 C57BL/6 embryonic stem cells (ESCs), a gift from Branko Zevnik (CECAD, University of Cologne), were cultured and engineered as described previously (56), with the exception that in the provided reference different cell lines and targeting vectors were used. Cervid Prnp targeting constructs were generated by ligating respective Prnp open reading frame (ORF) variants (Prnp.Cer.Wt and Prnp.Cer.138NN) between EagI and ClaI sites of the intermediate vector pWJPrP101 (56) containing homology regions and a removable neomycin selection cassette. Maps of targeting vectors are available at https://tinyurl.com/mryb5fy7. Cas9 vector used for double-strand break generation in Prnp gene is available from Addgene (plasmid #78621; ref. 56).
Gene-edited ESCs were injected into 8-cell embryos of CD1-Elite white mice (Charles River) and placed in the uterus of pseudo-pregnant females of the same mouse line. Resulting chimera offspring were bred to albino C57BL/6J mice to obtain hemizygous pups, which were then bred to “FLP (flippase)-deleter” mice on a C57BL/6J background (strain #009086, JAX) to remove the frt-flanked neomycin resistance cassette required for ESC selection. The “FLP-ed” mice were back-crossed twice to C57BL/6N mice, genotyped to confirm removal of the Flp recombinase gene, and bred to homozygosity for the wild-type deer Prnp (Prnp.Cer.Wt) or 138NN caribou Prnp (Prnp.Cer.138NN). Heterozygous F1 offspring expressing 138SN caribou Prnp (Prnp.Cer.138SN) was obtained by crossing the two homozygous lines.
Reverse-Transcriptase Quantitative PCR (RT-qPCR).
RNA extraction was performed on frozen mouse brains using the TRIzol method with an Omni™ homogenizer and stored at −80 °C until further use. Complementary DNA (cDNA) was synthesized using 2 µg of extracted mRNA, 3 µg of random primers, 0.5 mM dNTPs, 1× strand buffer, 1 mM DDT, 1 µL reverse transcriptase (ThermoFisher) and molecular grade dH2O. The resulting cDNA was used as a template for RT-qPCR reactions using primers that detect the cervid Prnp ORF, i.e., cerKI-Prnp-F (5′- GCA TCG GTG GCA GGA CT -3′) and cerKI-Prnp-R (5′- GAT CCA GCT GCC TAT GTG GC - 3′). Actin was also amplified with primers actin-F (5′- CTC AGG AGG AGC AAT GAT CTT GAT -3′) and actin-R (5′-TAC CAC CAT GTA CCC AGG CA -3′) as the housekeeping gene to normalize Ct values. Each reaction was setup with 10 µL of 2× SBYR™ Green MasterMix (ThermoFisher), 0.5 M forward primer, 0.5 M reverse primer, 5 µL cDNA and molecular grade dH2O for a total of 20 µL reaction volume. Values were calculated as the fold-difference (2−∆∆Ct) compared to wild-type C57BL/6N mice after normalizing to actin Ct values.
Mouse Inoculations, Monitoring, and Euthanasia.
WTD CWD isolates Wisc-1 and 116AG, previously described (18, 30, 31), were stored as 20% (w/v) BH in 1× phosphate-buffered saline (PBS) at −80 °C. Wild-type reindeer brain and wild-type and 138SN reindeer lymph nodes from experimentally-inoculated animals (29) were homogenized as 10% (w/v) homogenates in 1× PBS using a gentleMACS™ Dissociator (Miltenyi Biotec) and tested for their prion seeding activities in RT-QuIC (SI Appendix, Fig. S2). Eight mice per group were inoculated between 6 and 8 wk of age. Mice were inoculated i.c. with 20 µL of 1% BH or LNH using a 0.6 × 4 mm steel needle (Unimed, Switzerland), or i.p. with 100 µL of 5% BH or LNH using a 27G needle (BD) under anesthesia using isoflurane as described previously (30).
Mice were monitored weekly for signs of clinical prion disease and monitored daily at the onset of at least one confirmatory sign as described previously (30) and were euthanized by isoflurane overdose followed by cardiac perfusion when they reached terminal prion disease. Survival time was defined as the number of days from the date of inoculation to the date of euthanasia upon terminal prion disease, also referred to as days post-inoculation (dpi). Time-point euthanasia was performed on several nonclinical mice around the survival time (dpi) of their prion-sick counterparts. All mice experiments were terminated at a predetermined experimental endpoint of ≥700 dpi.
Mouse Tissue Collection and Processing.
Brain, spleen, and feces were collected upon euthanasia and stored at −80 °C or in formalin for biochemical analyses or IHC, respectively. Frozen tissues were homogenized in sterile 1× PBS either as 10% or 20% (w/v) homogenates in FastPrep-24™ Lysing D Matrix tubes with ceramic beads (MP Biomedicals) using the FastPrep-24™ Tissue Homogenizer (MP Biomedicals). Tissue homogenates were aliquoted and stored at −80 °C until further use.
Fecal samples were homogenized as previously described (57), except for using the FastPrep-24™ Tissue Homogenizer. Briefly, fecal samples were suspended in fecal extraction buffer (20 mM sodium phosphate pH 7.1, 130 mM NaCl, 0.05% Tween 20, 1 mM PMSF and 1× cOmplete™ Protease Inhibitor, EDTA-free [Roche]) at a final concentration of 10 % (w/v), homogenized, incubated on a rocking platform for ~1 h, centrifuged to remove insoluble debris, distributed into 200 to 250 µL aliquots, and stored at −80 °C until further use.
Real-Time Quaking-Induced Conversion (RT-QuIC).
Recombinant prion protein (rPrP) with the full-length mouse prion protein sequence (aa 23-231) was prepared as previously described (57). The RT-QuIC reaction cocktail contained 10 µg of rPrP, 10 µM Thioflavin-T (ThT), 170 mM NaCl, 1× PBS (contains 130 mM NaCl), 1 mM EDTA and H2O. Ninety-eight microliters of the RT-QuIC cocktail was added into each well of a 96-well plate (Thermo Fisher Scientific, #165305) followed by 2 µL of seed. The seeds used were 10% (w/v) BH or LNH diluted in a buffer containing 0.1% SDS and 1× N2 media supplement, with dilutions ranging from undiluted to 2 × 10−6 dilutions and set up in four replicates. Plates were sealed with a sealing tape, placed into a BMG Labtech FLUOstar™ plate reader and set for shaking and incubation intervals of 1-min shaking (700 rpm) and 1-min rest. The readings were plotted as the average relative fluorescence unit (RFU) against time in hours (h). Samples were determined positive when two out of four replicates crossed the threshold, i.e., the average RFU of the negative control replicate wells plus five times its SD.
For fecal homogenates (FH), samples were subjected to sodium phosphotungstic acid (Na-PTA) precipitation as previously described (11, 57) before testing in RT-QuIC. Briefly, samples were incubated with 10% sarkosyl, followed by Na-PTA stock solution, and centrifuged at maximum speed. The resulting pellet was resuspended at a 1:10 ratio of the original volume in RT-QuIC seed dilution buffer, further diluted up to 2 × 10−2, followed by RT-QuIC reaction setup as described (57).
Protein Misfolding Amplification Assay (PMCA).
PMCA was performed using protocols by Pritzkow et al. (58) and Paul et al. (59) with modifications. Briefly, brains from non-inoculated Prnp.Cer.Wt, Prnp.Cer.138NN and Prnp.Cer.138SN mice were prepared as 10% (w/v) BH using a Potter–Elvehjem PTFE pestle and glass tube (Sigma-Aldrich, #P7984) in cold PMCA conversion buffer containing 4 mM EDTA, 1% Triton X-100 and 1 tablet cOmplete™ Protease Inhibitor Mini (Roche) in 1× PBS and adjusted to a pH of 7.4. Samples were centrifuged at 13,000 rpm for 1 min to remove insoluble debris. The homogenates were aliquoted in sterile 1.5-mL tubes and stored at −80 °C until further use.
Ninety µL of substrate was added to either 0.5-mL tubes (Eppendorf, #30124537) with 20 ± 2 mg of glass beads (Roth, #A554.1, 0.75-1 mm ø) or 0.2-mL tubes (ThermoFisher, #AB0337) with three PTFE balls (McMaster-Carr, #9660K12, 3/32 in ø). Ten percent (w/v) BH from CWD-positive wild-type reindeer or Prnp.Cer.Wt mouse were diluted in 10-fold serial dilutions ranging from 2 × 10−1 to 2 × 10−7 in PMCA substrate. 10 µL of each seed dilution was added, with a final seed concentration of 2 × 10−2 to 2 × 10−8 in each reaction tube. All reactions were prepared in duplicate. Tubes were sealed with parafilm, decontaminated in a 0.3% NaOH + 0.2% SDS bath or 2.5 % bleach for 5 min, rinsed with H2O, and centrifuged briefly before they were placed in a tube rack inside a microplate sonicator horn assembly (431MPXH and Q700, QSonica) connected to a circulating water bath (CC304-B, Huber). The sonicator was set for 30 s of sonication at 375 to 395 W followed by 29.5 min rest and run for 48 h for a total of 96 sonication-rest cycles, corresponding to one round of PMCA. For serial PMCA (sPMCA), 10 µL of sample was transferred into 90 µL of fresh PMCA substrate and run for another 48 h with the same settings. This process was repeated to obtain three rounds of sPMCA.
Proteinase K (PK) Digestion, SDS–PAGE, and Western Blotting.
Mouse tissue homogenates.
Twenty percent (w/v) BHs were diluted at a 1:1 ratio in 2× lysis buffer (100 mM Tris-HCl pH 7.5, 100 mM NaCl, 10 mM EDTA, 1% [v/v] Triton-X, 1% [w/v] sodium deoxycholate, H2O) to make a 10% (w/v) BH. Twenty microliters of each sample was digested with 50 µg/mL PK (Sigma) for 1 h at 37 °C and prepared for SDS-PAGE by boiling for 10 min at 95 °C in sample buffer. Twenty microliters of the final +PK sample volume was loaded onto a 12.5% SDS-PAGE gel. For total PrP, or -PK, samples were not digested with PK and 5 µL was loaded onto the gel.
PMCA products.
PMCA samples were digested with 50 µg/mL PK in a solution containing 1% (w/v) sarkosyl and 0.06% (w/v) SDS in PBS or in 1× lysis buffer for 1 h at 37 °C under constant shaking at 450 rpm. Sample loading buffer was added to each tube and the samples were boiled for 10 min. Aliquots of the final sample solution were loaded onto a 12% Mini-PROTEAN® TGX™ pre-cast SDS-polyacrylamide gel (BioRad) or 12.5% SDS–PAGE gels. Proteins were transferred to polyvinylidene difluoride (PVDF) membranes and probed with the 4H11 anti-PrP antibody at a 1:500 dilution followed by a secondary goat-antimouse HRP-conjugated antibody at a 1:5,000 dilution. Membranes were developed using an HRP substrate (Immobilon Forte) and imaged using the Bio-Rad Chemidoc™ MP Imaging System and/or exposed to X-ray films to visualize the protein bands.
Immunohistochemistry for Detection of Abnormal PrP Deposits.
Brains or spleens fixed in 10% buffered formalin were dehydrated and embedded in paraffin. IHC was performed as previously described (18). Briefly, sagittal brain sections (4.5 to 6 µm) were mounted on positively charged slides and deparaffinized, followed by autoclaving in antigen retrieval buffer. Sections were then incubated for 30 min in 98% formic acid and treated with 4 M guanidine thiocyanate for 2 h. The detection of abnormal PrP deposits was achieved by exposure of the sections to BAR224 anti-PrP primary antibody (1:2,000; Cayman), followed by incubation with a biotinylated secondary anti-mouse antibody (DAKO™ ARK), horseradish peroxidase-streptavidin conjugate, chromogen substrate, and hematoxylin counterstain.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
This study was supported by funds from the Natural Sciences and Engineering Research Council, the Margaret Gunn Endowment for Animal Research, Parks Canada Agency, and Genome Canada. S.G. is supported by the Canada Research Chair program. Portions of the paper were developed from the thesis of M.I.A. We would like to thank Cameron Fielding and Kenichi Ito at the Clara Christie Centre for Mouse Genomics (CCCMG) facility at the University of Calgary for their services in generating and genotyping our gene-targeted mouse models, and staff from the CCCMG and the Prion-Virology Animal Facility for excellent animal care. We would also like to express our gratitude to Ronald Shikiya and Jason Bartz (Creighton University) for their generous advice in setting up our PMCA equipment and reactions.
Author contributions
M.I.A., L.K., W.J., and S.G. designed research; M.I.A., L.K., D.Z., S.H., C.L., and S.C.C. performed research; L.K., G.M., D.M., M.B., and W.J. contributed new reagents/analytic tools; M.I.A. analyzed data; and M.I.A. and S.G. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Zabel M. D., Reid C., A brief history of prions. Pathog. Dis. 73, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prusiner S. B., Novel proteinaceous infectious particles cause scrapie. Science 216, 136–144 (1982). [DOI] [PubMed] [Google Scholar]
- 3.Williams E. S., Young S., Chronic wasting disease of captive mule deer: A spongiform encephalopathy. J. Wildl. Dis. 16, 89–98 (1980). [DOI] [PubMed] [Google Scholar]
- 4.Williams E. S., Young S., Spongiform encephalopathy of rocky mountain elk. J. Wild. Dis. 18, 465–471 (1982). [DOI] [PubMed] [Google Scholar]
- 5.Benestad S. L., Mitchell G., Simmons M., Ytrehus B., Vikøren T., First case of chronic wasting disease in Europe in a Norwegian free-ranging reindeer. Vet. Res. 47, 88 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pirisinu L., et al. , Novel type of chronic wasting disease detected in Moose (Alces alces), Norway. Emerg. Infect. Dis. 24, 2210–2218 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sohn H.-J., et al. , A case of chronic wasting disease in an elk imported to Korea from Canada. J. Vet. Med. Sci. 64, 855–858 (2002). [DOI] [PubMed] [Google Scholar]
- 8.Richards B. J., Chronic wasting disease distribution in the United States by state and county (2022). 10.5066/P9HQKKFO. 22 October 2022. [DOI]
- 9.Angers R. C., et al. , Prions in skeletal muscles of deer with chronic wasting disease. Science 311, 1117 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Haley N. J., Seelig D. M., Zabel M. D., Telling G. C., Hoover E. A., Detection of CWD prions in urine and saliva of deer by transgenic mouse bioassay. PLoS One 4, e4848 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng Y. C., et al. , Early and non-invasive detection of chronic wasting disease prions in elk feces by real-time quaking induced conversion. PLoS One 11, e0166187 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davenport K. A., et al. , PrPC expression and prion seeding activity in the alimentary tract and lymphoid tissue of deer. PLoS One 12, e0183927 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hannaoui S., et al. , Transmission of cervid prions to humanized mice demonstrates the zoonotic potential of CWD. Acta Neuropathol. 144, 767–784 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Z., et al. , Generation of human chronic wasting disease in transgenic mice. Acta Neuropathol. Commun. 9, 158 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palmer M. S., Dryden A. J., Hughes J. T., Collinge J., Homozygous prion protein genotype predisposes to sporadic Creutzfeldt-Jakob disease. Nature 352, 340–342 (1991). [DOI] [PubMed] [Google Scholar]
- 16.Moore S. J., et al. , Pathologic and biochemical characterization of PrPSc from elk with PRNP polymorphisms at codon 132 after experimental infection with the chronic wasting disease agent. BMC Vet. Res. 14, 80 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson C., et al. , Prion protein polymorphisms in white-tailed deer influence susceptibility to chronic wasting disease. J. Gen. Virol. 87, 2109–2114 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Velásquez C. D., et al. , Deer prion proteins modulate the emergence and adaptation of chronic wasting disease strains. J. Virol. 89, 12362–12373 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fox K. A., Jewell J. E., Williams E. S., Miller M. W., Patterns of PrPCWD accumulation during the course of chronic wasting disease infection in orally inoculated mule deer (Odocoileus hemionus). J. Gen. Virol. 87, 3451–3461 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Jewell J. E., Conner M. M., Wolfe L. L., Miller M. W., Williams E. S., Low frequency of PrP genotype 225SF among free-ranging mule deer (Odocoileus hemionus) with chronic wasting disease. J. Gen. Virol. 86, 2127–2134 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Haley N. J., et al. , Estimating relative CWD susceptibility and disease progression in farmed white-tailed deer with rare PRNP alleles. PLoS One 14, e0224342 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arifin M. I., et al. , Cervid prion protein polymorphisms: Role in chronic wasting disease pathogenesis. Int. J. Mol. Sci. 22, 2271 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rhyan J. C., et al. , Failure of fallow deer (Dama dama) to develop chronic wasting disease when exposed to a contaminated environment and infected mule deer (Odocoileus hemionus). J. Wildl. Dis. 47, 739–744 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Arifin M. I., et al. , Large-scale prion protein genotyping in Canadian caribou populations and potential impact on chronic wasting disease susceptibility. Mol. Ecol. 29, 3830-3840 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamir A. N., et al. , Experimental transmission of chronic wasting disease (CWD) from elk and white-tailed deer to fallow deer by intracerebral route: Final report. Can. J. Vet. Res. 75, 152–156 (2011). [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng Y. C., Musiani M., Cavedon M., Gilch S., High prevalence of prion protein genotype associated with resistance to chronic wasting disease in one Alberta woodland caribou population. Prion 11, 136–142 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Happ G. M., Huson H. J., Beckmen K. B., Kennedy L. J., Prion protein genes in caribou from Alaska. J. Wildl. Dis. 43, 224–228 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Moore S. J., et al. , Horizontal transmission of chronic wasting disease in reindeer. Emerg. Infect. Dis. 22, 2142–2145 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitchell G. B., et al. , Experimental oral transmission of chronic wasting disease to reindeer (Rangifer tarandus tarandus). PLoS One 7, e39055 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hannaoui S., et al. , New and distinct chronic wasting disease strains associated with cervid polymorphism at codon 116 of the Prnp gene. PLoS Pathog. 17, e1009795 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hannaoui S., et al. , Destabilizing polymorphism in cervid prion protein hydrophobic core determines prion conformation and conversion efficiency. PLoS Pathog. 13, e1006553 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bauduin S., McIntire E., St-Laurent M.-H., Cumming S. G., Compensatory conservation measures for an endangered caribou population under climate change. Sci. Rep. 8, 16438 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Serrouya R., et al. , Saving endangered species using adaptive management. Proc. Natl. Acad. Sci. U.S.A. 116, 6181–6186 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haley N. J., et al. , Estimating chronic wasting disease susceptibility in cervids using real-time quaking-induced conversion. J. Gen. Virol. 98, 2882–2892 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raymond G. J., et al. , Evidence of a molecular barrier limiting susceptibility of humans, cattle and sheep to chronic wasting disease. EMBO J. 19, 4425–4430 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jackson W. S., Kaczmarczyk L., Astonishing advances in mouse genetic tools for biomedical research. Swiss Med. Wkly. 145, w14186 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Bian J., et al. , Primary structural differences at residue 226 of deer and elk PrP dictate selection of distinct CWD prion strains in gene-targeted mice. Proc. Natl. Acad. Sci. U.S.A. 116, 12478–12487 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tamgüney G., et al. , Asymptomatic deer excrete infectious prions in faeces. Nature 461, 529–532 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Collis S. C., Kimberlin R. H., Long-term persistence of scrapie infection in mouse spleens in the absence of clinical disease. FEMS Microbiol. Lett. 29, 111–114 (1985). [Google Scholar]
- 40.Bishop M. T., et al. , Prion infectivity in the spleen of a PRNP heterozygous individual with subclinical variant Creutzfeldt-Jakob disease. Brain 136, 1139–1145 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beringue V., et al. , Facilitated cross-species transmission of prions in extraneural tissue. Science 335, 472–475 (2012). [DOI] [PubMed] [Google Scholar]
- 42.Wagner K., et al. , Tissue-specific biochemical differences between chronic wasting disease prions isolated from free-ranging white-tailed deer (Odocoileus virginianus). J. Biol. Chem. 298, 101834 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Béringue V., et al. , Host prion protein expression levels impact prion tropism for the spleen. PLoS Pathog. 16, e1008283 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shikiya R. A., et al. , PrPSc formation and clearance as determinants of prion tropism. PLoS Pathog. 13, e1006298 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnson C. J., et al. , Prion protein polymorphisms affect chronic wasting disease progression. PLoS One 6, e17450 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wolfe L. L., Fox K. A., Miller M. W., “Atypical” chronic wasting disease in PRNP genotype 225FF mule deer. J. Wildl. Dis. 50, 660–665 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Kobayashi A., et al. , Deciphering the pathogenesis of sporadic Creutzfeldt-Jakob disease with codon 129 M/V and type 2 abnormal prion protein. Acta Neuropathol. Commun. 1, 74 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Asante E. A., et al. , Dissociation of pathological and molecular phenotype of variant Creutzfeldt-Jakob disease in transgenic human prion protein 129 heterozygous mice. Proc. Natl. Acad. Sci. U.S.A. 103, 10759–10764 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Castilla J., et al. , Crossing the species barrier by PrPSc replication in vitro generates unique infectious prions. Cell 134, 757–768 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vanni I., et al. , In vitro replication highlights the mutability of prions. Prion 8, 154–160 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Otero A., Duque Velásquez C., Aiken J., McKenzie D., White-tailed deer S96 prion protein does not support stable in vitro propagation of most common CWD strains. Sci. Rep. 11, 11193 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kaneko K., et al. , Evidence for protein X binding to a discontinuous epitope on the cellular prion protein during scrapie prion propagation. Proc. Natl. Acad. Sci. U.S.A. 94, 10069–10074 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Perrier V., et al. , Dominant-negative inhibition of prion replication in transgenic mice. Proc. Natl. Acad. Sci. U.S.A. 99, 13079–13084 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hizume M., et al. , Human prion protein (PrP) 219K is converted to PrPSc but shows heterozygous inhibition in variant Creutzfeldt-Jakob disease infection*. J. Biol. Chem. 284, 3603–3609 (2009). [DOI] [PubMed] [Google Scholar]
- 55.Tahiri-Alaoui A., Sim V. L., Caughey B., James W., Molecular heterosis of prion protein β-oligomers: A potential mechanism of human resistance to disease*. J. Biol. Chem. 281, 34171–34178 (2006). [DOI] [PubMed] [Google Scholar]
- 56.Kaczmarczyk L., Mende Y., Zevnik B., Jackson W. S., Manipulating the prion protein gene sequence and expression levels with CRISPR/Cas9. PLoS One 11, e0154604 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cheng Y. C., et al. , Real-time quaking-induced conversion assay for detection of CWD prions in fecal material. J. Vis. Exp 127, e56373 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pritzkow S., et al. , Quantitative detection and biological propagation of scrapie seeding activity in vitro facilitate use of prions as model pathogens for disinfection. PLoS One 6, e20384 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Paul L., Kirsch P., Thomzig A., Thöne-Reineke C., Beekes M., Practical approaches for refinement and reduction of animal experiments with bank voles in prion research. Berliner und Münchener Tierärztliche Wochenschrift 131, 359–367 (2018). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and/or SI Appendix.