Significance
Inhibitory exometabolites produced by individual root-derived bacteria have been widely studied in plant protection against soil-borne pathogens. However, the prevalence of exometabolite production in root-associated bacterial communities and their ecological relevance for root microbiota establishment remains elusive. We characterized exometabolite-mediated binary interactions between taxonomically diverse root- and soil-derived bacteria. Using genetic and metabolomics approaches, we identified the antimicrobial 2,4-diacetylphloroglucinol (DAPG) and the iron chelator pyoverdine, which together explain most of the inhibitory activity of the highly antagonistic root-associated Pseudomonas brassicacearum. Microbiota reconstitution experiments with a defined bacterial community revealed their cofunctioning as root competence determinants by specifically affecting root microbiota establishment. In natural environments, these exometabolites likely serve as adaptive traits contributing to pseudomonad pervasiveness throughout the root microbiota.
Keywords: root microbiome, microbe–microbe interactions, synthetic ecology, competition, secondary metabolites
Abstract
Soil-dwelling microbes are the principal inoculum for the root microbiota, but our understanding of microbe–microbe interactions in microbiota establishment remains fragmentary. We tested 39,204 binary interbacterial interactions for inhibitory activities in vitro, allowing us to identify taxonomic signatures in bacterial inhibition profiles. Using genetic and metabolomic approaches, we identified the antimicrobial 2,4-diacetylphloroglucinol (DAPG) and the iron chelator pyoverdine as exometabolites whose combined functions explain most of the inhibitory activity of the strongly antagonistic Pseudomonas brassicacearum R401. Microbiota reconstitution with a core of Arabidopsis thaliana root commensals in the presence of wild-type or mutant strains revealed a root niche-specific cofunction of these exometabolites as root competence determinants and drivers of predictable changes in the root-associated community. In natural environments, both the corresponding biosynthetic operons are enriched in roots, a pattern likely linked to their role as iron sinks, indicating that these cofunctioning exometabolites are adaptive traits contributing to pseudomonad pervasiveness throughout the root microbiota.
Plant root-associated bacterial communities – collectively referred to as the root microbiota – are characterized by reduced taxonomic diversity and class-level dominance of Alpha-, Beta-, and Gammaproteobacteria, Actinobacteria, Flavobacteriia, and Bacilli (1–3). Host genetic determinants underlying root microbiota establishment include carbon-rich photoassimilates (i.e., primary metabolites) that are continuously released by plants in the rhizosphere, acting as key mediators of root microbiota assembly and activity (4–6). It is conceivable that specialized metabolites produced by root commensals also contribute to the establishment of the microbiota, as these molecules may be responsible for cooperative or competitive relationships with other bacterial taxa. Specialized inhibitory metabolites (7–9) are often produced by complex, energetically costly enzymatic mechanisms encoded by biosynthetic gene clusters (BGCs) (10, 11). These molecules display diverse antagonistic activities, including inhibition of prokaryotic cell wall biosynthesis, pore formation in the cell envelope, or inhibition of adenosine triphosphate biosynthesis (12–16). For example, the antimicrobial 2,4-diacetylphloroglucinol (DAPG) from fluorescent Pseudomonas strains plays a dominant role in suppressing the fungal take-all disease that affects the roots of grass and cereal plants and has been demonstrated to inhibit bacterial growth as well (17–23). Furthermore, suppression of tomato infections with pathogenic Ralstonia solanacearum can be achieved by chelating inorganic iron with bacterial siderophores that cannot be taken up by the pathogen, including pyoverdines, while growth-promoting siderophores facilitate infections of this bacterial root pathogen (24, 25). In vitro, pyoverdine from P. chlororaphis YL-1 inhibits gram-positive and gram-negative bacteria in an iron concentration-dependent manner and is required for the suppression of the leaf pathogen Xanthomonas oryzae pv. oryzae in rice (26). However, it remains unknown whether such bacterial exometabolites with different modes of action shape root microbiota assembly and promote strain pervasiveness on roots.
Here, we assessed the prevalence of antagonistic binary interactions within the root microbiota using a collection of Arabidopsis thaliana root-derived commensals (At-Sphere) (1). We identify the antimicrobial DAPG and the high-affinity iron chelator pyoverdine as exometabolites underpinning the activities of the highly antagonistic Pseudomonas brassicacearum R401 and demonstrate their contribution to root microbiota establishment.
Results
Widespread Production of Specialized Exometabolites among Root-Associated Bacteria.
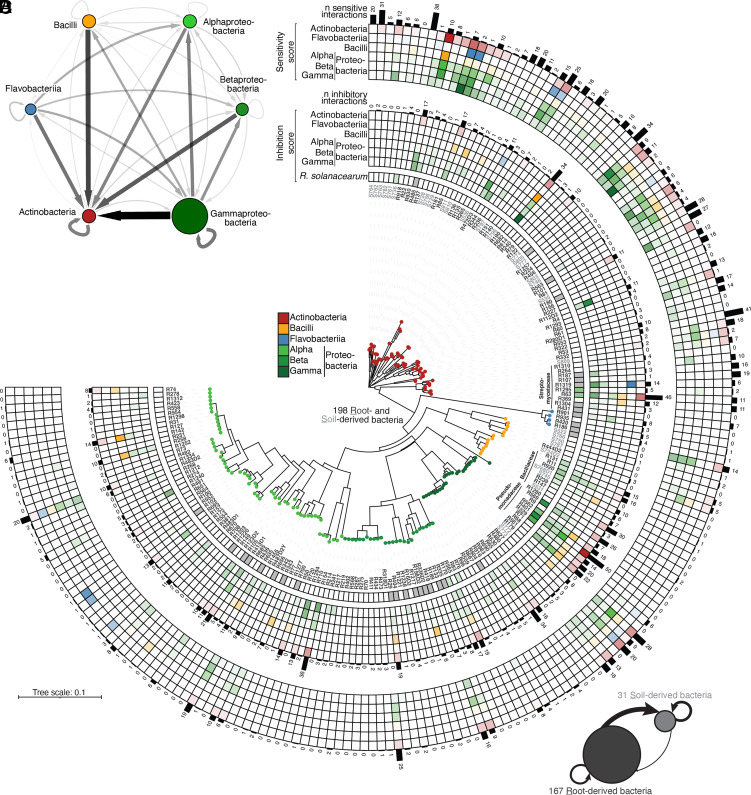
To investigate the prevalence of interbacterial competition mediated by secreted metabolites (hereafter referred to as exometabolites), we used a modified Burkholder plate-based assay (mBA; ref. 27 and SI Appendix, Fig. S1A) and tested 39,204 binary interbacterial interactions, i.e., 198 producer versus 198 target isolates. Independent validation of 7,470 randomly selected interactions (i.e., 19%) revealed 95% reproducibility of interaction phenotypes (SI Appendix, Table S1). We detected an inhibition halo in 1,011 interactions (i.e., 2.6% of tested pairwise interactions involving 66% of the isolates, SI Appendix, Fig. S1B and Table S1), suggesting antibiosis due to specialized exometabolites. Antagonistic interactions were detected between all bacterial classes tested, indicating that the production of exometabolites is common in the At-RSphere culture collection of commensals (1). Actinobacteria isolates were most sensitive to all other classes, especially to Gammaproteobacteria, which showed the highest aggregated frequency and average intensity of inhibitory activities (Fig. 1A). Since we observed taxonomic signals at the class level, we calculated the average halo of inhibition size for all target and producer isolates across all bacterial classes tested (see sensitivity scores in Fig. 1B and inhibition scores in Fig. 1C, see also SI Appendix, Fig. S1B). This revealed that only a few bacteria – mainly belonging to Pseudomonadaceae (R9, R68, R71, R329, R401, R562, and R569) – exhibited broad inhibition of phylogenetically diverse isolates (Fig. 1C). Extensive strain-specific variation in inhibition scores across closely related bacteria was observed, suggesting large standing genetic variation for the production of bacterial exometabolites among root-derived isolates (Fig. 1C). Finally, we observed a 2.7× higher inhibitory activity for root-derived bacteria compared to those originating from soil (P = 0.011; Kruskal–Wallis followed by Dunn’s post-hoc test and Benjamini–Hochberg correction, Fig. 1D), suggesting that the production of exometabolites might be advantageous for bacterial root colonization.
Fig. 1.
Widespread production of specialized exometabolites among root- and soil-derived bacteria. (A) Inhibitory interaction network based on binary interaction data from a modified Burkholder assay (mBA; n = 198 strains, 39,204 pairwise combinations tested, 1 to 2 biological replicates). Edge width and color depict the aggregated frequency of inhibitions at the class level while node size indicates the mean halo size, measured at 4 d post-inoculation (dpi). Arrows reflect inhibition directionality and intensity. For the complete list of interactions, see SI Appendix, Table S1. (B and C) Phylogenetic tree showing inhibition and sensitivity scores for each strain. The tree was built based on the full-length 16S rRNA gene sequences of 167 root- (dark gray) and 31 soil-derived (light gray) bacteria. All bacteria were reciprocally screened against each other and against Ralstonia solanacearum GMI1000. For each strain, sensitivity scores (average sensitivity of a strain to exometabolites produced by a given bacterial class) (B) and inhibition scores (average exometabolite-dependent inhibition of a given strain to a bacterial class) (C) are shown in the outer and inner heatmaps, respectively. These scores represent the average halo size for a given strain at the class level and were determined by an mBA. The number (“n”) of sensitivities per strain and the number of inhibitory interactions are indicated by black bars. (D) Network showing the aggregated frequency of inhibitions measured for root- and soil-derived bacteria. Node size indicates the mean magnitude of inhibitions from mBA experiments. Arrows reflect inhibition directionality and intensity.
Ralstonia spp. are core members of the root microbiota of healthy Arabidopsis plants in nature (28). We hypothesized that the bacterial root microbiota of A. thaliana contributes to preventing disease in natural environments and tested the inhibitory activities of 167 of the aforementioned bacteria against pathogenic Ralstonia solanacearum GMI1000 (hereafter referred to as Rs; ref. 29 and Fig. 1C). A subset of root- and soil-derived bacteria inhibited the growth of Rs in mBA experiments (10.9% and 10.3%, respectively). These inhibitory activities were mainly manifested by bacteria from three genera: Pseudomonas, Streptomyces, and Bacillus (Fig. 1C).
Genomic Capacity for Specialized Metabolite Production Explains Pronounced Inhibitory Activity.
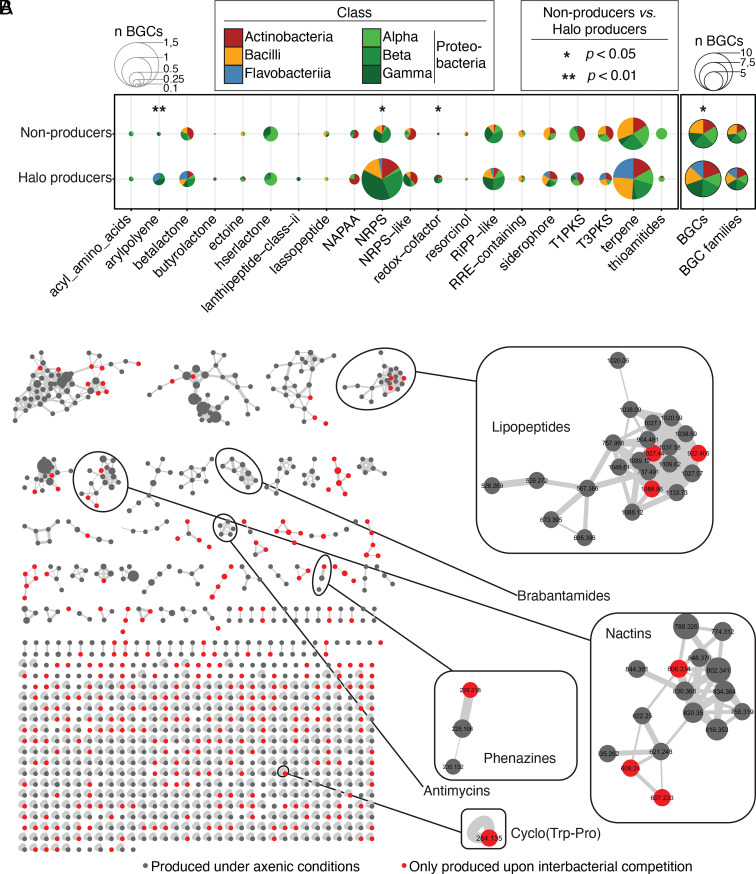
We hypothesized that the ability to produce inhibitory halos is correlated with the genome-encoded potential for the biosynthesis of specialized metabolites. We predicted BGCs for all strains tested in mBA experiments (SI Appendix, Table S2) and examined whether halo producer strains encode more BGCs than nonproducers. The total number of BGCs was significantly increased in the antagonistic isolates (P = 0.0166). BGCs, which are thought to be involved in the biosynthesis of nonribosomal peptides (NRPs), aryl polyene, and redox cofactors, were significantly enriched in halo-producing isolates compared with nonproducers (Fig. 2A), suggesting that these compounds may be important for interbacterial competition.
Fig. 2.
Genomic capacity for specialized metabolite production explains inhibitory activity. (A) Balloon plot depicting the genetic potential for specialized metabolite production. Using antiSMASH 6.0, we predicted BGCs for the genomes of all the analyzed bacteria. On the x-axis, the 20 most abundant BGC families across all the tested bacterial taxa are depicted. Size of pie charts depicts the number of a given BGC averaged across all halo producers or nonproducers (n BGCs; Fig. 1C and SI Appendix, Table S1). Sections of pie charts represent the distribution of BGCs across distinct bacterial classes. Sections are colored by bacterial classes. The total number of BGCs and BGC families is depicted with black outlines. The full dataset can be found in SI Appendix, Table S2. Statistical significance was determined by Kruskal–Wallis followed by Dunn’s post-hoc test and Benjamini–Hochberg adjustment. Significance between halo producer and nonproducers is indicated by black asterisks (∗,∗∗, P < 0.05, and 0.01, respectively). (B) Molecular network of extracts from ten producer strains with broad antagonistic activity, including extracts from inhibition halos with sensitive strains. Media components and nodes found to be produced only by sensitive strains were removed. Of the remainder, 298 nodes (29.5% of the network) were detected only in the interaction zone extracts and are marked in red. Dereplicated molecular families with additional congeners produced upon competition are highlighted.
We next investigated the diversity of bacterial metabolites that could explain the observed inhibitory phenotypes. Metabolites were extracted from individual strains (n = 198) grown on the same agar medium used in mBA experiments with two organic solvents with different polarities, ethyl acetate and methanol, to capture greater chemical diversity. Liquid chromatography-tandem mass spectrometry (UPLC-MS/MS) analysis of the resulting 396 samples yielded ~200,000 mass spectra, which were analyzed using the Global Natural Products Social Molecular Networking (GNPS) workflow (30). Network analysis of the resulting mass spectra revealed 247 families of molecules with similar fragmentation patterns, each representing at least two structurally related analogs (SI Appendix, Fig. S2). While 2,220 nodes were shared between multiple classes, the remaining 1,094 nodes were found to be produced only by individual bacterial classes. Here, Gammaproteobacteria showed the greatest number of class-specific metabolites, indicating that their pronounced inhibitory activity (Fig. 1C) can be explained by the production of an enormous diversity of specialized metabolites.
To determine whether bacteria secrete an increased diversity of specialized metabolites when interacting with competitors (31), we additionally analyzed the metabolomes of 16 inhibitory zones from 10 producer isolates (R63, R71, R68, R342, R401, R562, R569, R690, R920, and R1310) with broad inhibitory activity against three target isolates (R472D3, R480, and R553) using the same UPLC-MS/MS GNPS workflow. Spectra comparison with those of individually cultured strains revealed 298 additional ions detected exclusively in inhibition zones (Fig. 2B). Dereplication of the network revealed that the biosynthesis of polyketides, such as two additional congeners of the nactin antibiotics and peptidic compounds, such as cyclo(Trp-Pro), and congeners of a unique lipopeptide family, were specifically triggered by interactions with other bacterial strains, pointing to a possible role of these molecules in interbacterial competition.
DAPG Contributes to the Inhibitory Activity of P. brassicacearum R401.
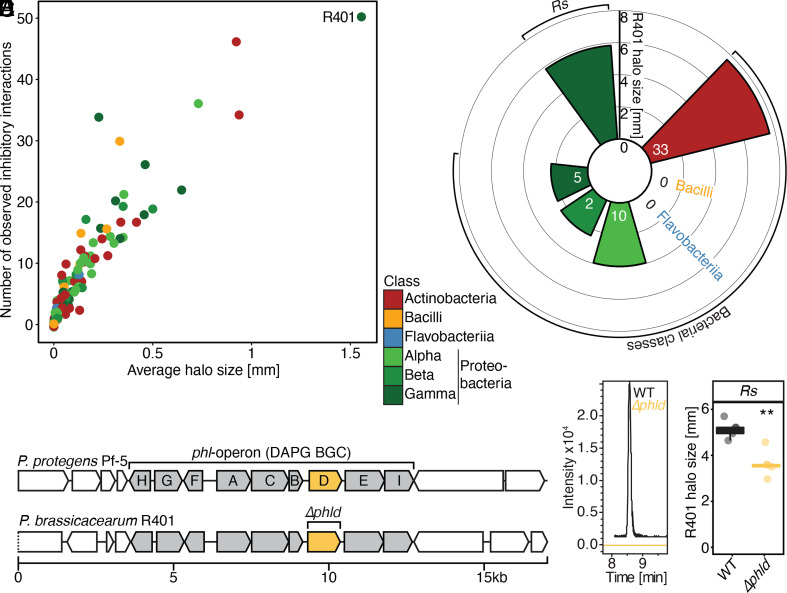
P. brassicacearum R401 showed the greatest number of inhibitory interactions (>17× greater than average) and the largest average halo size (>10× greater than average; Fig. 3A). Except for Bacilli and Flavobacteria, 50 isolates belonging to all other tested bacterial classes were sensitive to the inhibitory activity of R401, particularly Actinobacteria (Fig. 3B), indicating the production of exometabolites with broad spectrum inhibitory activity. antiSMASH-based analysis of a resequenced, circular R401 genome predicted 16 BGCs (SI Appendix, Fig. S3 and Table S2), one of which exactly matches the phl operon of Pseudomonas protegens Pf-5, which has been shown to encode the enzymatic machinery for the production of the Pseudomonas-specific polyketide (DAPG; Fig. 3C) (32). Since no other genome from our culture collection harbors the phl operon (SI Appendix, Fig. S3), this points to a role for DAPG in mediating the inhibitory activity of R401. We generated a marker-free deletion mutant of the key biosynthetic gene phlD in R401 (Fig. 3C). In mBA experiments, the R401 Δphld mutant was significantly—yet only partly—impaired in its antagonistic activity (Fig. 3E), retaining 71% of its inhibitory activity toward Rs. As DAPG production was completely lacking in the Δphld mutant (Fig. 3D), this suggests that DAPG alone is insufficient to explain the full inhibitory activity of R401 toward Rs.
Fig. 3.
DAPG contributes to the inhibitory activity of P. brassicacearum R401. (A) Projection of the number of observed inhibitory activities (out of 198) as a function of the mean halo size for each producer strain as measured in mBA experiments. The most antagonistic strain (P. brassicacearum R401) is highlighted. (B) Mean halo size of R401 against target bacterial classes or R. solanacearum (Rs) are indicated by bar height. Numbers indicate the number of observed inhibitory interactions of R401 per target bacterial class as measured in mBA. (C) Schematic overview of the phl operon that encodes all 2,4-Diacetylphloroglucinol (DAPG) biosynthetic genes in P. protegens Pf-5 and R401. Genes within the BGC are colored in gray, the gene encoding the initial anabolic enzyme PhlD is highlighted in yellow. (D) Extracted ion chromatograms for R401 DAPG (EICs: 211.0601 m/z ± 0.01 [M+H]+) of the WT and mutant extracts, confirming complete lack of DAPG production in the tested mutant. (E) Halo production of R401 WT and Δphld using Rs as target bacterium as measured in mBA. Statistical significance was determined by Kruskal—Wallis followed by Dunn’s post-hoc test and Benjamini—Hochberg adjustment. Significance compared to WT is indicated by black asterisks (∗∗, P < 0.01; n = 5).
DAPG and Pyoverdine Act Additively to Inhibit Taxonomically Distinct Root Microbiota Members.
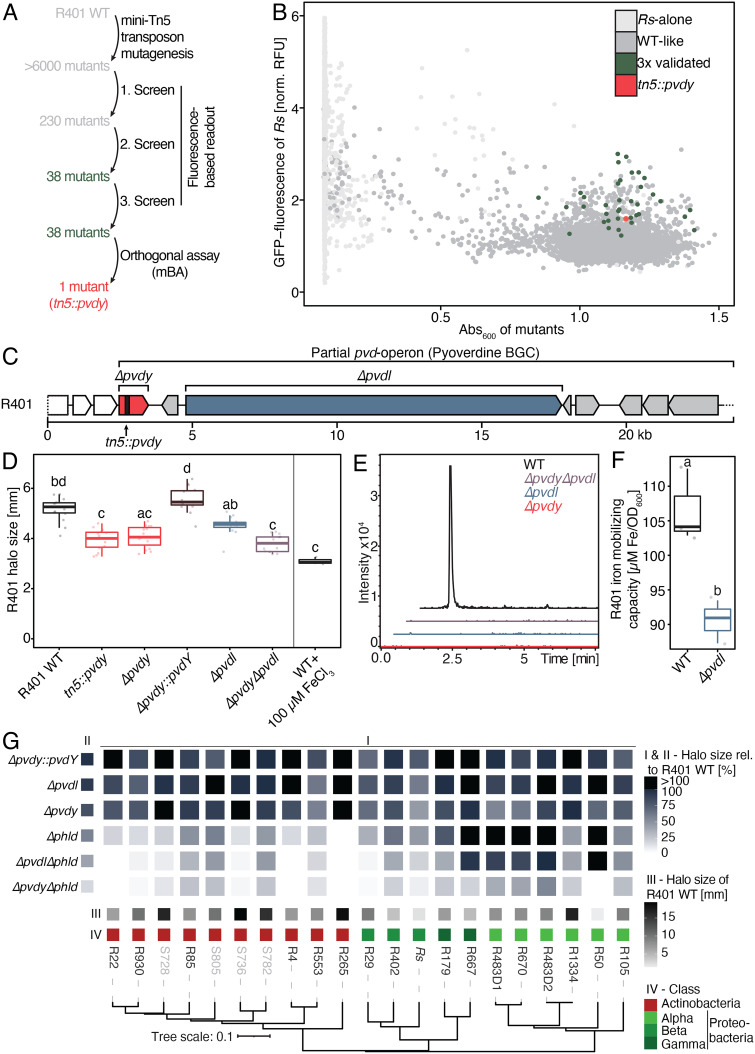
We performed a forward genetic screen to reveal additional determinants mediating the residual inhibitory activity of R401 Δphld. First, we generated a R401 mini-Tn5 transposon library with >6,000 insertion mutants. We adopted a fluorescence-based bacterial co-culture system in liquid medium to test all the R401 insertion mutants individually for their ability to suppress growth of a GFP-expressing Rs GMI1000 strain (Rs GMI1600; ref. 33). Of the 230 candidates identified in the primary screen, 38 R401 Tn5 mutants were robustly impaired in Rs suppression after two independent rounds of validation (Fig. 4 A and B and SI Appendix, Table S3). One of these candidate R401 mutants was also significantly impaired (on average 26.6%) in its inhibitory activity against Rs in mBA experiments on solid agar, suggesting that this mutant is impaired in the production of an inhibitory exometabolite (SI Appendix, Table S3). This R401 mutant carries the Tn5 transposon insertion within the gene of a putative acyltransferase (pvdY) involved in the biosynthesis of the siderophore pyoverdine in Pseudomonas aeruginosa (34). We validated the contribution of R401 pvdY to pyoverdine biosynthesis and Rs inhibition by generating an independent pvdY deletion mutant, Δpvdy, and a R401 deletion mutant of the gene encoding NRP synthetase pvdL, located downstream of pvdY (Δpvdl, Fig. 4C). In mBA experiments with Rs GMI1000, the R401 Δpvdy strain phenocopied the R401 tn5::pvdy mutant and the mutant phenotype was complemented by the expression of pvdY under its native promoter in the R401 Δpvdy background (Δpvdy::pvdY). R401 Δpvdl exhibited a slightly weaker impairment of halo production compared to R401 Δpvdy. It is likely that R401 PvdY is involved in hydroxyornithine acetylation—a component of R401 pyoverdine—while PvdL is involved in the initial amino acid condensation, forming the basis for the peptidic backbone of pyoverdines. Acetyl hydroxyornithine could be involved in the biosynthesis of a third, unknown exometabolite by R401, explaining the difference between both pyoverdine mutants observed in mBA experiments. We also generated a R401 double deletion strain, Δpvdy Δpvdl, which is impaired in its inhibitory activity against pathogenic Rs to a similar extent as the R401 Δpvdy single mutant (Fig. 4D). Using mass spectrometry, we confirmed that all generated R401 pyoverdine mutants had lost their ability to produce pyoverdine (R401 pyoverdine has the sequence: Glu-Q-Lys-AcOHOrn-Ala-Gly-Ser-Ser-OHAsp-Thr, with Q being the fluorophore moiety; Fig. 4E).
Fig. 4.
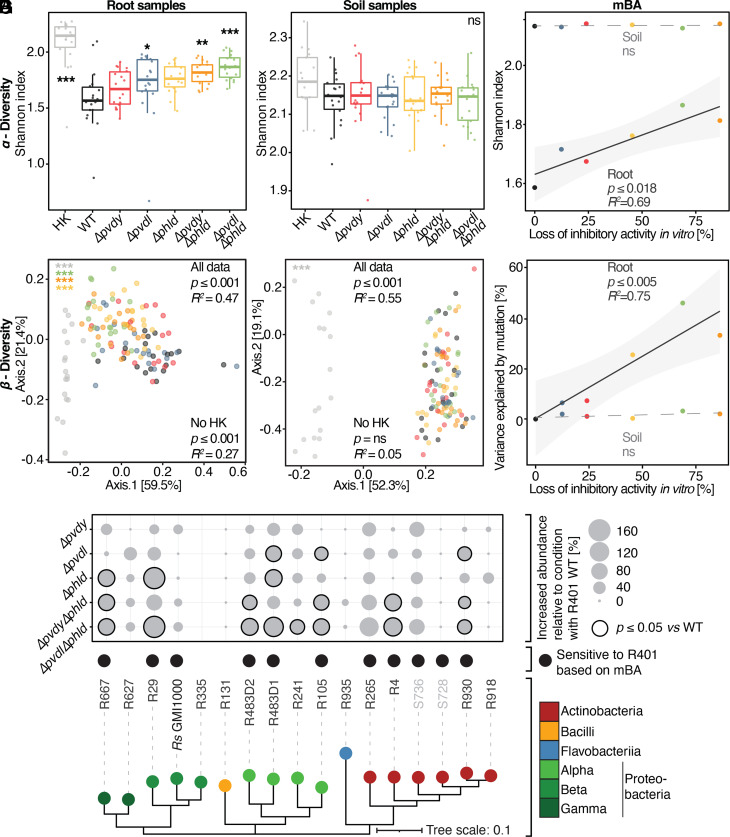
DAPG and pyoverdine act additively to inhibit taxonomically distinct root microbiota members. (A) Schematic overview of the mini-Tn5 transposon mutant screen workflow in R401. (B) Effect of >6,000 R401 transposon mutants on the growth of GFP-expressing R. solanacearum (Rs GMI1600) as a function of the absorption at 600 nm of axenically grown mutants. Rs-alone indicates reaction vessels without R401 mutants. All other points correspond to individual R401 mutants that have been colored based on their phenotype in this screen. (C) Schematic overview of the genomic context of the fragmented pvd-operon which encodes a fraction of pyoverdine biosynthetic genes of R401. Genes within the BGC are colored in gray, pvdY and pvdL are highlighted in red and blue, respectively. Transposon integration sites are highlighted by vertical black bars. (D) Halo production of R401 WT and four different mutants that are impaired in the production of pyoverdine (tn5::pvdy, Δpvdy, Δpvdl, and ΔpvdyΔpvdl) as measured in mBA. Mutant names and colors are depicted as in panel C. Δpvdy::pvdY is a complementation line of Δpvdy. Halo production of R401 WT strains after medium supplementation with 100 µM FeCl3. Rs was used as a target strain. Halo size measurements were taken after 3 d of interaction. Letters indicate statistically significant differences as determined by Kruskal—Wallis followed by Dunn’s post-hoc test and Benjamini—Hochberg adjustment with P < 0.05 (n = 15). (E) Extracted ion chromatograms for the R401 pyoverdine (EICs: 622.2764 m/z ± 0.1 [M+2H]2+, corresponding to the most abundant peak linked to R401 pyoverdines) of the WT and mutant extracts, confirming complete lack of production in all the tested mutants. (F) Ferric iron-mobilizing activity of R401 WT and Δpvdl. Letters indicate statistically significant differences as determined by Kruskal—Wallis followed by Dunn’s post-hoc test and Benjamini–Hochberg adjustment with P < 0.05 (n = 3). (G) Heatmap depicting a halo of mBA screen of R401 WT and single and double mutants that are impaired in DAPG (Δphld) and/or pyoverdine (Δpvdl or Δpvdy) production. Δpvdy::pvdY is a Δpvdy complementation line. A taxonomically diverse set of root- and soil-derived bacteria (comprising Rs) has been used as target bacteria. All target bacteria are sensitive to R401 (SI Appendix, Table S1). Halo sizes have been normalized to the respective WT halo sizes. Average, relative halo sizes are depicted in I. (II) shows the average thereof across all the tested strains, while (III) shows the average absolute halo size of R401 WT on a given target strain; n = 5. (IV) is a phylogenetic tree based on v5v7 16S rRNA genes, depicting strain taxonomy at the class level.
All tested Pseudomonas isolates in the At-RSphere contain pyoverdine biosynthetic genes; however, for R9, no characteristic pyoverdine fluorescence was detected (SI Appendix, Fig. S3), potentially explaining its low inhibitory activity in mBAs (Fig. 1C). Therefore, we hypothesized that pyoverdines contribute more broadly to explaining the unusually high inhibitory activity of the Pseudomonadaceae detected in the mBA experiments and isolated pyoverdine mutants from another Pseudomonas root commensal in our collection. We generated a mini-Tn5 mutant library of approximately 6,000 insertion mutants of pyoverdine-producing Pseudomonas fluorescens R569 and assayed about 2,000 mutants for loss of pyoverdine-mediated fluorescence by fluorimetry (SI Appendix, Fig. S4A). Characterization of the Tn5 integration sites revealed one mutant in each of the R401 pvdY and pvdL homologs (SI Appendix, Table S3 and Fig. S4 B and C). Unlike the partial loss of inhibitory activity of the R401 Δpvdy and Δpvdl single and Δpvdy Δpvdl double mutants against Rs, the R569 Δpvdy and Δpvdl single mutants both showed a complete loss of inhibitory activity to Rs in mBA experiments (SI Appendix, Fig. S4D). Although the genomes of root commensals R401 and R569 are assigned to different Pseudomonas sublineages (Fig. 1B) and the arrangement of genes in the pvd operon is different (Fig. 4C and SI Appendix, Fig. S4C), it is likely that the corresponding pyoverdines are directly responsible for the observed inhibitory activity. Our findings indicate that pyoverdine might either be the sole or one of the several exometabolites produced by root commensals that limit the growth of Rs. Since pyoverdines function as iron chelators, we tested whether their inhibitory activity can be explained by interbacterial competition for iron by supplementing mBA agar medium with excess ferric iron (100 µM FeCl3). This resulted in significantly impaired Rs inhibition when confronted with WT R401 or undetectable inhibition when confronted with WT R569, thus phenocopying results obtained with the corresponding strain-specific pyoverdine mutants (Fig. 4D and SI Appendix, Fig. S4D). Given that the mutants also exhibit drastically reduced iron mobilization capacity compared with the corresponding WT isolates (Fig. 4F and SI Appendix, Fig. S4E), we conclude that their inhibitory activities against root commensals and pathogenic Rs are likely mediated through their iron chelator function. However, we cannot exclude a possible additional pyoverdine activity under limited iron conditions.
To test whether these two classes of compounds account for the full inhibitory activity of R401, we generated two R401 double pyoverdine and DAPG mutants, Δpvdy Δphld and Δpvdl Δphld, and conducted mBA assays using Rs and a representative set of the aforementioned commensals that we previously observed to be sensitive to R401 (SI Appendix, Table S1). The double pyoverdine and DAPG mutants showed severely reduced inhibitory activity against Rs, and all other tested isolates compared to the single mutants, suggesting a cumulative impact of the two metabolites. Although the general sensitivity patterns were again isolate specific, Actinobacteria were typically inhibited by DAPG alone. DAPG and pyoverdine collectively explained >70% of R401 inhibitory activity, but a residual halo of inhibition was still observed, pointing to at least a third—yet to be defined—exometabolite (Fig. 4G). Beyond the known inhibitory activities of DAPG and pyoverdine, our results provide evidence that metabolites with distinct modes of action jointly act to inhibit a taxonomically broad range of bacteria.
DAPG and Pyoverdine Modulate Root Microbiota Assembly and Restrict Bacterial Diversity.
We conducted root microbiota reconstitution experiments with germ-free A. thaliana Col-0 in Flowpots to study the contribution of DAPG and pyoverdine to root microbiota structure and to limiting the growth of Rs (35). We tested heat-killed (HK) or live WT R401, R401 Δpvdy, and Δpvdl single as well as Δpvdy Δphld and Δpvdl Δphld double mutants on a phylogenetically diverse synthetic community (SynCom) that comprises 18 bacterial isolates (SI Appendix, Fig. S5A and Table S1), in both “soil” (peat matrix) and root compartments. After 3 weeks of plant—microbe cocultivation, DNA was extracted from soil and root samples and was subjected to 16S rRNA amplicon sequencing at isolate-specific resolution. Shoot fresh weight did not differ between conditions, and no wilting was observed (SI Appendix, Fig. S5 B and C), indicating that Rs was unable to cause disease on A. thaliana, possibly due to the presence of the SynCom. This is consistent with very low Rs relative abundances found in root samples (SI Appendix, Fig. S5D) and a previous report that direct immersion of A. thaliana roots with a very high Rs inoculum of 108 cells/mL was needed to induce wilting symptoms in the crucifer (36).
The addition of live, WT R401 to the SynCom significantly reduced bacterial alpha diversity (Shannon index) compared to the (HK) R401 condition in the root compartment (P < 0.001). This major impact on the community is gradually lost when R401 exometabolite mutants were co-inoculated with the SynCom (Fig. 5A). Importantly, no such effects were observed in the soil compartment (Fig. 5B). Bacterial beta diversity (Bray—Curtis dissimilarity) was also drastically affected by WT R401 inoculation, explaining most of the variation and resulting in a clear separation along axis 1 (HK versus WT, P < 0.001; R2 = 0.63). All mutant samples fall in between these extremes and follow a clear trajectory, WT > single mutants > double mutants > HK, suggesting that DAPG and pyoverdine have a cumulative influence on bacterial community structure in the root compartment (Fig. 5D). This is likely due to direct exometabolite activities, as the production of either metabolite did not affect plant phenotypes (SI Appendix, Fig. S5C). Furthermore, in silico depletion of R401 16S rRNA sequence reads leads to similar changes in beta diversity with even higher significance levels in some cases (Δpvdl versus WT; P = 0.002; R2 = 0.14; SI Appendix, Fig. S5E), indicating that, irrespective of R401 abundance, the bacterial community is altered by DAPG and pyoverdine production. While in the soil compartment, inoculation of WT R401 leads to a similar shift as in the root compartment (HK versus WT, P < 0.001; R2 = 0.66), genetic depletion of either DAPG or pyoverdine biosynthetic abilities alone or together does not alter community structure (Fig. 5E), which indicates niche-specific activity of these exometabolites in the root compartment. The impact of R401 on community structure was still significant upon in silico depletion of R401 16S rRNA sequences (HK versus WT, P < 0.001; R2 = 0.35; SI Appendix, Fig. S5F), indicating that R401 uses other, unknown mechanisms to influence community structure in soil.
Fig. 5.
DAPG and pyoverdine modulate root microbiota assembly and restrict bacterial diversity. (A–G) Community profiling of an 18-member bacterial SynCom comprising either heat-killed R401 [HK, (WT) background], live R401 WT, or corresponding mutants that are impaired in the production of DAPG (Δphld), pyoverdine (Δpvdy, Δpvdl), or both (Δpvdy Δphld, Δpvdl Δphld). SynComs and A. thaliana Col-0 seeds were co-inoculated into the peat-based gnotobiotic Flowpot system. At 21 dpi, roots and soil samples were taken. Data from three full-factorial experiments with six biological replicates each. (A and B) Alpha diversity (Shannon index) of root (A) and soil (B) samples in response to R401 or its mutants. Statistical significance was determined by Kruskal–Wallis followed by Dunn’s post-hoc test and Benjamini–Hochberg adjustment. Significance compared to WT is indicated by black asterisks (∗,∗∗,∗∗∗, P < 0.05, 0.01, and 0.001, respectively; “ns”: not significant; n = 18). (D and E) PCoA based on Bray–Curtis community dissimilarities between samples in root (D) and soil (E) in response to R401 or its mutants. PERMANOVA-derived P-values are represented as asterisks (∗∗∗, P < 0.001; n = 18), colored by the respective condition. PERMANOVA analysis on the full dataset before (all data) or after (no HK) in silico depletion of HK samples is indicated in black; R2 represents the variance explained by R401 genotype. (C and F) Regression analysis of loss of inhibitory activity of R401 mutants in an mBA (Fig. 4G) and Shannon indices (C; data from A and B) or variance explained by R401 (F; data from D and E). P-values and R2 derive from a linear model and have been computed for soil and root samples separately; ns, not significant. Confidence intervals are depicted in light gray. (G) Balloon plot depicting the increase in relative abundance of SynCom members at the root relative to the condition in which R401 WT has been inoculated. Statistical significance was determined by Kruskal–Wallis followed by Dunn’s post-hoc test and Benjamini–Hochberg adjustment. Significance compared to WT in non-normalized dataset is indicated by black circles, indicating P < 0.05 (n = 18). Susceptibility toward R401 WT in the halo assay is depicted as black spheres. Black spheres indicate sensitivity toward R401, while colored spheres represent the respective bacterial class.
To determine whether the mBA results obtained in vitro have physiological relevance in planta, we inspected relative isolate abundances across conditions, reasoning that isolates insensitive to at least one of the R401 exometabolites in vitro would not benefit from genetic disruption of either BGC in a community context in the root compartment. This analysis revealed that only DAPG- and/or pyoverdine-sensitive isolates benefited from disruption of the corresponding BGCs present in R401 (Fig. 5G), suggesting that binary interaction data in vitro can explain the impact on individual commensal isolates in a community context in the root compartment. This observation prompted us to test whether mBA data could predict community-scale effects as well as effects on single isolates. We computed the average reduction in inhibitory activity for each R401 mutant (Fig. 4G) and tested whether the lack of competitiveness could inform changes in alpha- and beta-diversity. Linear regression analyses revealed that binary interaction data largely explained the observed effect size in the root compartment for both alpha- and beta-diversity indices but had no predictive power in the soil compartment (Fig. 5 C and F, respectively). In conclusion, two exometabolites produced by a single isolate have large effects on key ecological indices of a taxonomically diverse SynCom and can be linked to pairwise in vitro interaction experiments.
DAPG and Pyoverdine Act as Root Competence Determinants in a Community Context.
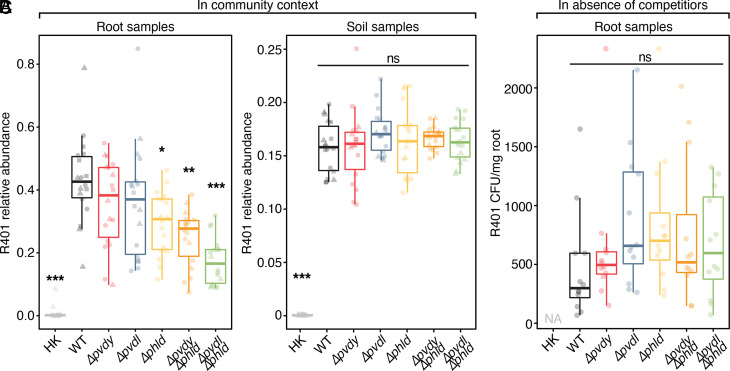
To examine whether R401-induced modulation of bacterial assembly and diversity promoted R401 competitiveness, we determined the relative abundance of R401 in SynCom samples collected from root and soil compartments. In both root and soil samples, the 16S rRNA reads of live R401 by far exceeded the barely detectable HK R401 reads, indicating that live R401 inoculum proliferates in both compartments under all conditions (Fig. 6 A and B). However, R401 accumulated in association with roots at >2x higher abundance than in the soil compartment. R401 accumulation in the root compartment was gradually reduced in the SynCom context: Single or double mutations of DAPG and pyoverdine biosynthetic genes were sufficient to reduce the R401 abundance by up to approximately 39% compared to WT R401 (Fig. 6A). Importantly, the capacity of these deletion mutants to colonize the soil compartment remained unaffected (Fig. 6B). We also investigated the root colonization capacity of WT and all the five R401 deletion strains in mono-associations on axenic A. thaliana plants in an agar-based system (37) and found no significant differences in live R401 cell counts (Fig. 6C). Neither the growth of R401 DAPG or pyoverdine mutants was impaired in axenic culture media (SI Appendix, Fig. S6 B and C). Similarly, colonization experiments with WT R401 or the Δpvdy Δphld double mutant in the Flowpot system revealed no difference in root colonization capacity in the absence of bacterial competitors (SI Appendix, Fig. S6A). Thus, DAPG and pyoverdine cofunction as R401 root competence determinants specifically in competition with other members of this SynCom in the root compartment.
Fig. 6.
DAPG and pyoverdine act as root competence determinants in a community context. (A and B) Relative abundance of R401 WT or mutants in root (A) and soil (B) samples in competition with 18-member SynCom, as shown in Fig. 5; n = 18. Statistical significance was determined by Kruskal—Wallis followed by Dunn’s post-hoc test and Benjamini—Hochberg adjustment. Significance compared to WT is indicated by black asterisks (∗,∗∗,∗∗∗, P < 0.05, 0.01, and 0.001, respectively; ns, not significant). (C) Colonization capability of R401 or its mutants in mono-associations on axenically grown A. thaliana Col-0 roots. Plants were grown on ½ MS-agar plates for 14 d. Colony-forming units have been determined and normalized to root fresh weight; n = 12. Statistical significance was determined by ANOVA followed by Tukey’s HSD test. No significant differences were detected as indicated by “ns.”
To assess the prevalence of Pseudomonas strains capable of producing DAPG and pyoverdine in association with plants when grown in natural soils, we analyzed the genomes of several commensal culture collections of 1,567 isolates from roots or leaves of A. thaliana or roots of the legume Lotus japonicus (1, 6, 38, 39). These plants had been grown in the same or different soils on different continents, and our genome analysis revealed increased abundance of DAPG and pyoverdine BGCs in root-derived Pseudomonas isolates (SI Appendix, Fig. S6 D and E). These results not only support our conclusions obtained with a defined core commensal community and gnotobiotic A. thaliana but also suggest a broader yet root-specific role of these two exometabolites in natural environments beyond the model crucifer.
Discussion
Our identification of a subset of specialized metabolites, primarily polyketides, and NRPs, that are specifically produced upon competition sensing, suggests that interbacterial competition can activate cryptic BGCs in the A. thaliana root microbiota. A related study using an A. thaliana phyllosphere bacterial culture collection also demonstrated widespread production of specialized metabolites and identified 725 binary inhibitory interactions in vitro, with only 3.6% of 224 strains mediating most of these interactions (40). The A. thaliana leaf and root microbiota overlap extensively at higher taxonomic ranks, and Pseudomonas species are core members of both communities (1). However, the comparison between root- and leaf-derived Pseudomonas genomes collected from natural environments reveals complete or partial niche specificity, respectively, for Pseudomonas production of DAPG and pyoverdine in the root compartment (SI Appendix, Fig. S6D). Furthermore, the total number of BGCs is significantly higher (approximately 1.6×) in root-derived Pseudomonas isolates (SI Appendix, Fig. S6E). Likewise, the number of antagonistic interbacterial interactions within the root microbiota significantly exceeds those between abundant soil-derived bacteria (Fig. 1D). These findings could be explained by the fact that natural, unplanted soils and leaves represent oligotrophic habitats (4, 41), whereas the rhizoplane is a microenvironment with a continuous supply of root exudation-derived nonstructural carbohydrates to support bacterial proliferation (42). It is likely that the large number of antagonistic interactions we found between root microbiota members in vitro is also influenced by the agar medium, which is rich in nonstructural organic carbon to mimic the nutrient-rich rhizoplane, thereby inducing the costly biosynthesis of exometabolites.
We provided genetic evidence for a root niche-specific cofunction of the exometabolites DAPG and pyoverdine as root competence determinants of commensal R401 that scales at the community level by influencing alpha- and beta-diversity indices. R401 also proliferates in the soil compartment, but its growth there and its influence on the structure of the bacterial soil community are independent of these exometabolites (Figs. 5 and 6 A and B). These observations, together with the fact that plant roots are a major sink for the uptake of rhizospheric mineral iron (43–45), imply that the public good iron becomes rate limiting in the root compartment. This could explain why the high-affinity iron chelator pyoverdine and antimicrobial DAPG cofunction and maximize R401 growth at the expense of its commensal competitors in the root niche, despite their different modes of action. Consistent with this model, the expression of core biosynthetic genes involved in the production of both DAPG and pyoverdine is induced under iron-limiting conditions in rhizospheric P. fluorescens and human pathogenic P. aeruginosa PAO1 (46, 47). The indistinguishable root colonization capacity of WT R401, pyoverdine, or DAPG single, or double mutants in mono-associations with A. thaliana is also consistent with our model and further supports the specific role of these exometabolites in interspecies competition during root microbiota establishment. Likewise, P. fluorescens C7R12 pyoverdine mutants were unaffected in rhizosphere competence under axenic conditions (48). All the seven A. thaliana root-derived Pseudomonas spp. strains in the At-RSphere culture collection have the genetic capacity to produce pyoverdines (SI Appendix, Fig. S3). Together with the essential function of pyoverdines produced by multiple Pseudomonas root commensals in limiting the growth of pathogenic Rs (Fig. 4D, SI Appendix, Fig. S4D, and ref. 25), this suggests a widespread role for these siderophores in determining Pseudomonas competitiveness in the root microbiota, even in soils that contain replete bioavailable inorganic iron for plant growth (1, 24, 25, 43). Thus, it is possible that in natural soils, regulated secretion of pyoverdines is an adaptive trait of the genus Pseudomonas to simultaneously compete against the root iron sink and enable pervasiveness of the Pseudomonadaceae lineage via interbacterial competition (2, 3, 49).
Our genetic data show that a sequential reduction in the diversity of specialized exometabolites in one species is sufficient to increase alpha-diversity indices and significantly alter the beta diversity of a synthetic root microbiota, thereby establishing a causal link between within-species genetic diversity and interspecies diversity changes. This suggests that in the root microenvironment, DAPG and pyoverdine produced by WT R401 cofunction to locally inhibit the growth of multiple SynCom members, which in turn increases the abundance of the producer R401. Our results obtained with the SynCom in coculture with gnotobiotic A. thaliana grown in peat matrix bear striking similarity to the changes in alpha diversity reported for this plant when grown in natural soils, where bacterial alpha diversity in bulk soil, rhizosphere, and root compartments gradually decreases toward the root (2, 3, 28). This suggests that exometabolite-mediated antagonistic interactions of commensals underpin at least part of the reduction of alpha diversity consistently observed in natural environments at the soil—root interface.
Given the prevalence of operons predicted to encode pyoverdines among root-derived Pseudomonas spp. isolates in the At-RSphere culture collection (ref. 1 and SI Appendix, Fig. S3), we consider the moderately complex 18-member SynCom employed here as necessary to overcome genetic redundancy, at least for pyoverdine production and probably also for the DAPG exometabolite in the root microbiota, when A. thaliana plants are grown in natural soils (SI Appendix, Table S2 and refs. 32, 50). It remains to be tested whether genetic depletion of individual bacterial exometabolites can have similarly striking effects on highly complex natural microbial communities, as the assessment of microbial antagonism in vitro is often not transferable to field conditions (51–53). However, DAPG- and phenazine-overproducing mutant strains of Pseudomonas putida WCS358r were shown to differentially shape root-associated fungal communities of field-grown wheat over an experimental period of up to 139 d compared to the WCS358r WT strain (54, 55). This suggests that bacterial exometabolites—such as DAPG—can have long-lasting effects on the structure of complex microbial communities. The approach described here to remove genetic redundancy by using annotated genomes of cultured microbiota members may be more generally applicable to explore community functions of other microbial genetic determinants with SynComs in gnotobiotic plant growth systems. Taken together, our study suggests that high-throughput binary interaction experiments, combined with genome mining for BGCs of root microbiota culture collections, can be applied to identify strains with broad-spectrum antagonistic activities that are likely robust root colonizers. This might have relevance for future interventions in the root microbiota with rational biologicals that confer beneficial traits on the host, including indirect pathogen protection and mineral nutrition.
Materials and Methods
Detailed descriptions of all utilized methods and data analysis workflows can be found in SI Appendix.
Screen for Antagonistic Interbacterial Interactions.
For all mBA experiments, bacterial strains were cultured axenically. Target strains were embedded in molten 25% tryptic soy agar (TSA) and producer strains were then dropped out on top. After up to 96 h of cultivation, pictures were taken and halos were quantified.
Metabolomic Analyses.
Untargeted metabolomic analysis was either conducted with axenically grown strains or on the mBA inhibition zones of ten strains (R63, R68, R71, R342, R401, R562, R569, R690, R920, and R1310), each tested against three target strains (R472D3, R480, and R553).
Detection of R401 DAPG and Pyoverdine.
Using metabolite analyses, R401 DAPG and pyoverdine were detected in WT extracts and lack of metabolite production was confirmed in the respective mutants.
BGC Prediction Using antiSMASH.
antiSMASH 6.0 (56) was used to predict BGCs for all the tested strains.
Mutant Generation.
Using homologous recombination, marker-free targeted R401 knockout mutants were generated lacking DAPG and/or pyoverdine biosynthetic genes.
Establishment of mini-Tn5 Transposon Mutant Collections in R401 and R569.
R401 and R569 were transformed with pUTmTn5Km2 (57), carrying a mini-Tn5 transposon, and individual colonies were then picked into 96-well plates.
mini-Tn5 Transposon Mutant Screen for Loss of R401-Mediated Growth Inhibition of Rs GMI1600.
R401 mini-Tn5 transposon mutants were evaluated in parallel for their WT-like growth and loss of inhibitory activity against GFP-expressing Rs GMI1600.
mini-Tn5 Transposon Mutant Screen for Lack of Pyoverdine Fluorescence of R569.
Axenically grown R569 mini-Tn5 transposon mutants were evaluated for their lack of fluorescence characteristic of pyoverdine at λexcitation = 395 nm and λemission = 470 nm.
Identification of mini-Tn5 Transposon Integration Sites in the Genomes of R401 and R569.
The chromosomal mini-Tn5 transposon integration sites in the R401 or R569 genomes were determined similarly as described before (57).
Complementation of R401 Δpvdy.
Complementation of R401 Δpvdy was conducted by expressing the coding region of R401 pvdY with its native 5′ regulatory sequences from the low-copy pSEVA22l plasmid (58) in the Δpvdy mutant background.
In vitro Iron Mobilization Assay.
The capability of R401 and R569 mutants to solubilize inaccessible ferric iron was tested using a previously described photometric assay (59).
Validation of Bacterial Growth Rates.
The growth of each R401 mutant and wild type was assessed by continuously measuring the OD600 of actively growing bacterial cultures.
Microbiota Reconstitution in the Gnotobiotic Flowpot System.
Flowpots were assembled according to ref. 35 with minor modifications. A final bacterial OD600 of 0.0025 was inoculated in ½ MS, and five surface-sterilized A. thaliana Col-0 seeds per Flowpot were sown. After 21 dpi, the fresh weight of shoots was determined, and the roots and peat matrix were harvested for bacterial community profiling.
Monoassociation Experiment of R401 on A. thaliana Seedlings.
Colony-forming units of WT R401 or its mutants colonizing A. thaliana seedlings grown in ½ MS agar were determined as described (37).
DNA Isolation.
DNA was isolated from A. thaliana roots and Flowpot peat using a modified high-throughput version of the FastDNA SPIN kit for Soil (MP Biomedicals).
Library Preparation for Bacterial 16S rRNA Gene Profiling.
The v5v7 variable regions of the bacterial 16S rRNA gene were amplified from DNA template derived from roots and Flowpot peat. The resulting amplicons were tagged with sample-specific barcodes using a dual-indexing approach. Illumina paired-end sequencing was then performed in-house with the MiSeq benchtop sequencer.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
This research was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy—EXC-number 2048/1—project 390686111, a European Research Council advanced grant (ROOTMICROBIOTA) to P.S.-L., as well as funds to P.S.-L. from the Max Planck Society. Furthermore, this work was supported by funds to S.H. from a European Research Council starting grant (MICRORULES 758003) and the Max Planck Society, as well as the Cluster of Excellence on Plant Sciences and the Priority Programme: Deconstruction and Reconstruction of the Plant Microbiota (SPP DECRyPT 2125; project P.S.-L.: SCHU 799/8-1; project S.H.: HA 8169/2-2), both funded by the DFG. Work in the Schäberle lab was supported by the German Federal Ministry of Education and Research (BMBF). L.W. was funded by the China Scholarship Council (CSC No. 201908080177). K.S. and Y.I. were funded through JP22H00364. We thank the Max Planck-Genome-Centre Cologne for advising and performing the resequencing of the R401 genome and Jose Flores-Uribe for his support in the genome assembly and annotation. We thank Daniel Machado for his constructive feedback on the analyses. We thank Jeff Dangl for his critical and constructive comments on our manuscript. We thank Brigitte Pickel and Dieter Becker for their support with experiments. Thanks to Neysan Donnelly for editing the manuscript and Saurabh Pophaly for support with data submission.
Author contributions
F.G., M.A.H., A.J.F., K.S., T.F.S., S.H., and P.S.-L. designed research; F.G., M.A.H., M.C., M.M., Y.I., N.B., A.J.F., L.W., J.O., K.-W.M., C.J.H., H.W., and T.F.S. performed research; F.G., M.A.H., M.C., M.M., P.Z., N.B., T.T., H.W., S.M., and R.G.-O. analyzed data; and F.G., T.F.S., S.H., and P.S.-L. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
Reviewers: G.C., University of Nottingham; and A.O., John Innes Centre.
Contributor Information
Stéphane Hacquard, Email: hacquard@mpipz.mpg.de.
Paul Schulze-Lefert, Email: schlef@mpipz.mpg.de.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Bai Y., Functional overlap of the arabidopsis leaf and root microbiota. Nature 528, 364–369 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Bulgarelli D., Revealing structure and assembly cues for arabidopsis root-inhabiting bacterial microbiota. Nature 488, 91–95 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Lundberg D. S., Defining the core arabidopsis thaliana root microbiome. Nature 488, 86–90 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eilers K. G., Lauber C. L., Knight R., Fierer N., Shifts in bacterial community structure associated with inputs of low molecular weight carbon compounds to soil. Soil Biol. Biochem. 42, 896–903 (2010). [Google Scholar]
- 5.Hu L., Root exudate metabolites drive plant-soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nat. Commun. 16, 2738 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wippel K., Host preference and invasiveness of commensal bacteria in the lotus and arabidopsis root microbiota. Nat. Microbiol. 6, 1150–1162 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clough S. E., Jousset A., Elphinstone J. G., Friman V. P., Combining in vitro and in vivo screening to identify efficient Pseudomonas biocontrol strains against the phytopathogenic bacterium ralstonia solanacearum. Microbiologyopen 11, e1283 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dimkić I., Janakiev T., Petrović M., Degrassi G., Fira D., Plant-associated bacillus and pseudomonas antimicrobial activities in plant disease suppression via biological control mechanisms - A review. Physiol. Mol. Plant Pathol. 117, 101754 (2022). [Google Scholar]
- 9.Fira D., Dimkić I., Berić T., Lozo J., Stanković S., Biological control of plant pathogens by bacillus species. J. Biotechnol. 285, 44–55 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Charlop-Powers Z., Owen J. G., Reddy B. V. B., Ternei M. A., Brady S. F., Chemical-biogeographic survey of secondary metabolism in soil. Proc. Natl. Acad. Sci. U.S.A. 111, 3757–3762 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crits-Christoph A., Diamond S., Butterfield C. N., Thomas B. C., Banfield J. F., Novel soil bacteria possess diverse genes for secondary metabolite biosynthesis. Nature 558, 440–444 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Dimkic I., et al. , The profile and antimicrobial activity of bacillus lipopeptide extracts of five potential biocontrol strains. Front. Microbiol. 8, 925 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Falardeau J., Wise C., Novitsky L., Avis T. J., Ecological and mechanistic insights into the direct and indirect antimicrobial properties of bacillus subtilis lipopeptides on plant pathogens. J. Chem. Ecol. 39, 869–878 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Jasim B., Sreelakshmi K. S., Mathew J., Radhakrishnan E. K., , Surfactin Iturin, and fengycin biosynthesis by endophytic bacillus sp. from bacopa monnieri. Microbial. Ecol. 72, 106–119 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Kwak Y. S., et al. , Saccharomyces cerevisiae genome-wide mutant screen for sensitivity to 2,4-diacetylphloroglucinol, an antibiotic produced by pseudomonas fluorescens. Appl. Environ. Microbiol. 77, 1770–1776 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Makovitzki A., Avrahami D., Shai Y., Ultrashort antibacterial and antifungal lipopeptides. Proc. Natl. Acad. Sci. U.S.A. 103, 15997–16002 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Isnansetyo A., Cui L., Hiramatsu K., Kamei Y., Antibacterial activity of 2,4-diacetylphloroglucinol produced by pseudomonas sp. AMSN isolated from a marine alga, against vancomycin-resistant staphylococcus aureus. Int. J. Antimicrob. Agents 22, 545–547 (2003). [DOI] [PubMed] [Google Scholar]
- 18.W. T. Julian, A. v. Vasilchenko, D. D. Shpindyuk, D. v. Poshvina, A. S. Vasilchenko, Bacterial-derived plant protection metabolite 2,4-diacetylphloroglucinol: Effects on bacterial cells at inhibitory and subinhibitory concentrations. Biomolecules 11, 13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keel C., et al. , Suppression of root diseases by pseudomonas fluorescens CHA0: Importance of the bacterial secondary metabolite 2,4-diacetylphloroglucinol. Mol. Plant Microbe Interact. 5, 4–13 (1992). [Google Scholar]
- 20.Lanteigne C., Gadkar V. J., Wallon T., Novinscak A., Filion M., Production of DAPG and HCN by pseudomonas sp. LBUM300 contributes to the biological control of bacterial canker of tomato. Phytopathology 102, 967–973 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Raaijmakers J. M., Weller D. M., Natural plant protection by 2,4-diacetylphloroglucinol-producing pseudomonas spp. Take-all decline soils. Mol. Plant Microbe Interact. 11, 144–152 (1998). [Google Scholar]
- 22.Zhou H., Improving biocontrol activity of pseudomonas fluorescens through chromosomal integration of 2,4-diacetylphloroglucinol biosynthesis genes. Chinese Sci. Bull. 50, 775–781 (2005). [Google Scholar]
- 23.Zhou T. T., et al. , phlF− mutant of Pseudomonas fluorescens J2 improved 2,4-DAPG biosynthesis and biocontrol efficacy against tomato bacterial wilt. Biol. Control 78, 1–8 (2014). [Google Scholar]
- 24.Gu S., et al. , Siderophore-mediated interactions determine the disease suppressiveness of microbial consortia. mSystems 5, e00811-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gu S., et al. , Competition for iron drives phytopathogen control by natural rhizosphere microbiomes. Nat. Microbiol. 5, 1002 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y., et al. , Pyoverdines are essential for the antibacterial activity of pseudomonas chlororaphis YL-1 under low-iron conditions. Appl. Environ. Microbiol. 87, 1–17 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burkholder P. R., Pfister R. M., Leitz F. H., Production of a pyrrole antibiotic by a marine bacterium. Appl. Microbiol. 14, 649 (1966). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thiergart T., et al. , Root microbiota assembly and adaptive differentiation among european arabidopsis populations. Nat. Ecol. Evol. 4, 122–131 (2020). [DOI] [PubMed] [Google Scholar]
- 29.Boucher A., Transposon mutagenesis of pseudomonas solanacearum: Isolation of Tn5induced avirulent mutants. Microbiology 131, 2449–2457 (1985). [Google Scholar]
- 30.Wang M., et al. , Sharing and community curation of mass spectrometry data with global natural products social molecular networking. Nat. Biotechnol. 34, 828–837 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cornforth D. M., Foster K. R., Competition sensing: The social side of bacterial stress responses. Nat. Rev. Microbiol. 11, 285–293 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Keel C., et al. , Conservation of the 2,4-diacetylphloroglucinol biosynthesis locus among fluorescent pseudomonas strains from diverse geographic locations. Appl. Environ. Microbiol. 62, 552–563 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aldon D., Brito B., Boucher C., Genin S., A bacterial sensor of plant cell contact controls the transcriptional induction of ralstonia solanacearum pathogenicity genes. EMBO J. 19, 2304 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lamont I. L., Martin L. W., Sims T., Scott A., Wallace M., Characterization of a gene encoding an acetylase required for pyoverdine synthesis in pseudomonas aeruginosa. J. Bacteriol. 188, 3149–3152 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kremer J. M., Peat-based gnotobiotic plant growth systems for arabidopsis microbiome research. Nat. Protoc. 16, 2450–2470 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deslandes L., et al. , Genetic characterization of RRS1, a recessive locus in arabidopsis thaliana that confers resistance to the bacterial soilborne pathogen ralstonia solanacearum. Mol. Plant Microbe Interact. 11, 659–667 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Ma K. W., Coordination of microbe–host homeostasis by crosstalk with plant innate immunity. Nat. Plants 7, 814–825 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karasov T. L., et al. , Arabidopsis thaliana and pseudomonas pathogens exhibit stable associations over evolutionary timescales. Cell Host Microbe 24, 168–179.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levy A., Genomic features of bacterial adaptation to plants. Nat. Genetics 50, 138–150 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Helfrich E. J. N., Bipartite interactions, antibiotic production and biosynthetic potential of the arabidopsis leaf microbiome. Nat. Microbiol. 3, 909–919 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vorholt J. A., Microbial life in the phyllosphere. Nat. Rev. Microbiol. 10, 828–840 (2012). [DOI] [PubMed] [Google Scholar]
- 42.F. el Z. Haichar, T. Heulin, J. P. Guyonnet, W. Achouak, Stable isotope probing of carbon flow in the plant holobiont. Curr. Opin. Biotechnol. 41, 9–13 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Harbort C. J., et al. , Root-secreted coumarins and the microbiota interact to improve iron nutrition in arabidopsis. Cell Host Microbe 28, 825–837.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kobayashi T., Nishizawa N. K., Iron uptake, translocation, and regulation in higher plants. Annu. Rev. Plant Biol. 63, 131–152 (2012). [DOI] [PubMed] [Google Scholar]
- 45.Loper J. E., Lindow S. E., A biological sensor for iron available to bacteria in their habitats on plant surfaces. Appl. Environ. Microbiol. 60, 1934–1941 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lim C. K., Hassan K. A., Tetu S. G., Loper J. E., Paulsen I. T., The effect of iron limitation on the transcriptome and proteome of pseudomonas fluorescens Pf-5. PLoS One 7, e39139 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Palma M., Worgall S., Quadri L. E. N., Transcriptome analysis of the pseudomonas aeruginosa response to iron. Arch. Microbiol. 180, 374–379 (2003). [DOI] [PubMed] [Google Scholar]
- 48.Mirleau P., et al. , Fitness in soil and rhizosphere of pseudomonas fluorescens C7R12 compared with a C7R12 mutant affected in pyoverdine synthesis and uptake. FEMS Microbiol. Ecol. 34, 35–44 (2000). [DOI] [PubMed] [Google Scholar]
- 49.Hacquard S., et al. , Microbiota and host nutrition across plant and animal kingdoms. Cell Host Microbe 17, 603–616 (2015). [DOI] [PubMed] [Google Scholar]
- 50.Tracanna V., et al. , Dissecting disease-suppressive rhizosphere microbiomes by functional amplicon sequencing and 10× metagenomics. mSystems 6, e0111620 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Knudsen I. M. B., et al. , Selection of biological control agents for controlling soil and seed-borne diseases in the field. Eur. J. Plant Pathol. 103, 775–784 (1997). [Google Scholar]
- 52.Shtienberg D., Elad Y., Incorporation of weather forecasting in integrated. Biological-chemical management of botrytis cinerea. Phytopathology 87, 332–340 (2007). [DOI] [PubMed] [Google Scholar]
- 53.Whitaker B. K., Bakker M. G., Bacterial endophyte antagonism toward a fungal pathogen in vitro does not predict protection in live plant tissue. FEMS Microbiol. Ecol. 95, fiy237 (2019). [DOI] [PubMed] [Google Scholar]
- 54.Bakker P. A. H. M., et al. , Effects of pseudomonas putida modified to produce phenazine-1-carboxylic acid and 2,4-diacetylphloroglucinol on the microflora of field grown wheat. Antonie Van Leeuwenhoek 81, 617–624 (2002). [DOI] [PubMed] [Google Scholar]
- 55.Glandorf D. C. M., et al. , Effect of genetically modified pseudomonas putida WCS358r on the fungal rhizosphere microflora of field-grown wheat. Appl. Environ. Microbiol. 67, 3371–3378 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.K. Blin et al. , antiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 49, W29–W35 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Merrell D. S., Hava D. L., Camilli A., Identification of novel factors involved in colonization and acid tolerance of vibrio cholerae. Mol. Microbiol. 43, 1471–1491 (2002). [DOI] [PubMed] [Google Scholar]
- 58.Martínez-García E., et al. , SEVA 3.0: An update of the standard european vector architecture for enabling portability of genetic constructs among diverse bacterial hosts. Nucleic Acids Res. 48, D1164–D1170 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Arora N. K., Verma M., Modified microplate method for rapid and efficient estimation of siderophore produced by bacteria. 3 Biotech 7, 381 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and/or SI Appendix.