Significance
Cell-free DNA (cfDNA) is one of the most promising biomarkers for cancer detection. In the era of precision medicine, an approach that exploits our burgeoning knowledge of the cancer genome and epigenome for the detection of cfDNA would provide unprecedented utility. We thereby have developed an approach (MethylSaferSeqS) that simultaneously evaluates for both genetic and epigenetic alterations in the same DNA molecules. In this proof-of-principle study, we demonstrate the ability of MethylSaferSeqS to detect the expected mutations, copy number changes, and methylation in cfDNA from cancer patients and healthy individuals. MethylSaferSeqS is a versatile method that is readily adaptable to existing sequencing and experimental strategies to simultaneously detect genetic and epigenetic features of the same DNA molecules.
Keywords: biomarker, cfDNA, methylation, mutation, copy number alteration
Abstract
The analysis of cell-free DNA (cfDNA) from plasma offers great promise for the earlier detection of cancer. At present, changes in DNA sequence, methylation, or copy number are the most sensitive ways to detect the presence of cancer. To further increase the sensitivity of such assays with limited amounts of sample, it would be useful to be able to evaluate the same template molecules for all these changes. Here, we report an approach, called MethylSaferSeqS, that achieves this goal, and can be applied to any standard library preparation method suitable for massively parallel sequencing. The innovative step was to copy both strands of each DNA-barcoded molecule with a primer that allows the subsequent separation of the original strands (retaining their 5-methylcytosine residues) from the copied strands (in which the 5-methylcytosine residues are replaced with unmodified cytosine residues). The epigenetic and genetic alterations present in the DNA molecules can then be obtained from the original and copied strands, respectively. We applied this approach to plasma from 265 individuals, including 198 with cancers of the pancreas, ovary, lung, and colon, and found the expected patterns of mutations, copy number alterations, and methylation. Furthermore, we could determine which original template DNA molecules were methylated and/or mutated. MethylSaferSeqS should be useful for addressing a variety of questions relating genetics and epigenetics.
Detecting cancers at a stage when they can still be cured is one of the major objectives of current cancer research. Assays based on the detection of genetic or epigenetic alterations of DNA, chromosome copy number changes, or changes in the abundance of specific proteins or RNA molecules are being explored for this purpose. Among them, changes in DNA sequence are the most sensitive and specific, with the capacity to identify one altered DNA allele of a given gene mixed with more than 100,000 normal alleles of the same gene (1). To detect such rare alleles requires the purification of DNA from multiple milliliters (mL) of plasma, as 1 mL plasma contains an average of only ~1,000 to 2,000 alleles of each gene. Past studies have shown that increases in sensitivity can be obtained by combining assays for DNA alterations and changes in protein abundance (2, 3). It would be ideal if epigenetic changes could also be assessed in the same cell-free DNA (cfDNA) molecules used to detect genetic changes, thereby reducing the total volume of plasma required from the patient.
The most commonly used method for investigating the epigenetic status of DNA employs bisulfite-mediated deamination to convert unmethylated deoxycytidine residues to deoxyuridine (4–6). Unfortunately, bisulfite treatment makes it impossible to detect the type of mutation that is most common in cancers, i.e., C to T transitions (7). We here report a technique called MethylSaferSeqS that employs bisulfite-mediated deamination but allows detection of any type of genetic or epigenetic change in DNA, including C to T transitions, in the same DNA template.
Results
Overview of MethylSaferSeqS.
The most sensitive way to detect rare mutations is through duplex sequencing, i.e., the independent determination of the sequence of both strands of DNA. Errors during DNA purification, library preparation, or sequencing are much more likely to alter a single base, rather than both bases at that position in complementary (Watson and Crick) DNA strands. Thus, duplex sequencing can exponentially reduce error rates. Several methods for duplex sequencing have been described (1, 8–13). MethylSaferSeqS can be applied to any of these library preparation methods using the following approach:
-
1.
Copy both strands (Watson and Crick) of each template DNA molecules after adapter ligation.
-
2.
Separate the original DNA strands from the copied strands.
-
3.
Prepare two libraries, one from the original DNA strands (a) and one from the copied strands (b):
-
a.
Deaminate unmodified cytosine residues in the original strands using bisulfite, PCR-amplify, and assess epigenetic changes;
-
b.
PCR-amplify the copied strands and assess genetic changes.
-
a.
Though these steps are simple in principle and involve only minimal changes to conventional library preparation methods, performing them in a way that preserves the majority of the initial DNA template molecules for both genetic and epigenetic analysis is challenging. The final, optimized protocol is summarized in Fig. 1, with further details described below and in Materials and Methods.
Fig. 1.
Overview of MethylSaferSeqS. Following ligation of Y-adaptors to each double-stranded DNA molecule during standard library construction, each original strand is copied with a primer that targets the P7 sequence. The primer contains a dual biotin modification and a deoxyuridine (dU) base at the 5′ end. The 5′ biotins allow the copied strand (still hybridized to the original strand) to bind to streptavidin beads. After binding, the original strands are separated from the beads via heat denaturation. The remaining, copied strand is then removed from the beads via cleavage of the deoxyuridine residue at the 5′ end of the primer. This process yields two libraries: one from the copied strands (Left) used for genetic analysis, and the other from original strands (Right) used for epigenetic analysis. UID: Unique identifier. USER: Uracil DNA glycosylase and the DNA glycosylase-lyase Endonuclease VIII.
Principles of MethylSaferSeqS.
The first principle involves modifications to adaptor sequences to ensure compatibility with standard library preparation and sequencing methods. The adapters appended to the template molecules must be modified such that bisulfite-mediated deamination does not change the adapter sequences in a way that would make them incompatible with other components of standard, commercially available library generation, and sequencing processes. Our preferred change was to substitute cytosine (C) residues with 5-methylcytosines (5-meC’s) in the adapter sequences. Adapters could also be changed in other ways for the same purpose, such as using only A, T, and G’s without any C’s.
The second principle is embodied in the statement “every molecule is sacred” (14). Because abnormal genetic or epigenetic changes are expected to be very rare in certain cfDNA samples, avoiding losses of these alleles during library preparation is critical. The SaferSeqS library preparation method yields a higher fraction of template molecules than most other library preparation methods (1), and was therefore used as the base for the MethylSaferSeqS protocol. SaferSeqS also facilitates duplex sequencing, which is key to the reliable detection of rare mutations. Though the base workflow described here for MethylSaferSeqS is that used for SaferSeqS, the protocol can be easily adapted to any library preparation method.
Step 1: Copy template molecules.
The addition of this step represents the major deviation from conventional library preparations. In conventional library preparations, once adapters are ligated to the DNA templates, the library is amplified using PCR to obtain sufficient DNA for target enrichment or for other purposes. Once amplification is performed, however, all covalent modifications of the DNA templates, such as methylation or hydroxymethylation, are lost and replaced by unmodified DNA residues during the copying process. In MethylSaferSeqS, the two DNA strands are heat denatured as in a typical PCR step, but then each strand is copied with a single primer containing a dual biotin modification and a deoxyuridine (dU) at its 5′ end (described in Step 2 below). Because only one rather than two primers per reaction is used for this step, exponential amplification does not occur. Following this copying, there are n+1 strands rather than 2n strands (n = the number of PCR cycles performed): the original DNA strand (containing covalent modifications such as methylation) and n copied strands (devoid of such modifications).
During the initial development of the MethylSaferSeqS protocol, we used only one round of denaturation and copying with the biotinylated, dU-containing primer. In later stages, we used two or three sequential rounds of denaturing and copying, always with a single primer rather than two primers to avoid exponential amplification. The reason for using more than one cycle was to increase yield, given the importance of recovering as many molecules as possible. With three cycles, essentially all of the template molecules that were successfully recovered in the base protocol (SaferSeqS) were also recovered with MethylSaferSeqS, as assessed by duplex sequencing (Fig. 2). With one and two cycles, the recoveries were 37% and 81%, respectively.
Fig. 2.
Duplex recovery of MethylSaferSeqS compared to standard library construction approach (SaferSeqS) and the number of original molecules. The number of distinct molecules present in MethylSaferSeqS and SaferSeqS was assessed through duplex sequencing after target enrichment through amplification of 48 genomic regions of interest. Each distinct molecule is tagged with a UID, which allows for precise quantification of the number of initial DNA molecules. A duplex UID is defined as an UID that was found in both the Watson and Crick strands of the molecule. (A) The average duplex recoveries were 77% and 70% for MethylSaferSeqS (LA) and SaferSeqS libraries, respectively, compared to the total number of initial DNA molecules. (B) Within the MethylSaferSeqS libraries, similar numbers of distinct molecules were partitioned for methylation analysis prior to bisulfite treatment (pre-LM) and mutation detection (LA). The length of the error bars is the SEM.
Step 2: Separating the copied strands from the original strands.
The primer used for copying contains a dual biotin modification and a dU at the 5′ end as described above. The 5′ dual biotin modification allows the copied strand (still hybridized to the original strand) to bind to streptavidin beads and ensures that this interaction is not disrupted by heat. After binding, the original strands are separated from the beads and bound copied strands via heat denaturation. Lastly, the remaining, copied strand is removed from the beads by cleavage at the dU residue, using Uracil DNA glycosylase and DNA glycosylase-lyase Endonuclease VIII. The removal of DNA templates from beads was done because we found that for subsequent library construction, amplification in solution was more efficient than amplification on beads.
Many variations of Steps 1 and 2 were evaluated during the development of MethylSaferSeqS. These included singly biotinylated rather than doubly biotinylated primers for copying the original template, various numbers of deoxyuridines (from zero to six) at various positions in the primers, various conditions for binding to streptavidin beads, temperatures, ethylenediaminetetracetic acid concentrations, and pH for separating the original strands and copied strands from the beads. Though all variations recovered the desired molecules to some degree, the detailed protocol described in Methods performed best of those tested.
Step 3a: Use bisulfite to deaminate the unmethylated cytosines in the original strands.
Bisulfite-mediated deamination results in cleavage of DNA (15–17). Longer reaction times and higher temperatures in the presence of bisulfite result in higher conversion rates of unmethylated cytosine to uracil, but lower DNA recovery. Given that many of the aberrantly methylated molecules in plasma from early-stage cancer patients are expected to be present at low frequencies, we attempted to find conditions that would lead to reasonable DNA yields while maintaining an acceptable conversion efficiency. Through trial and error, we found that bisulfite treatment at 50 °C for 3 h yielded results that were optimal for our purposes. This preserved the majority (~70%) of the input template molecules with 80% of unmethylated C’s converted to U’s. This conversion efficiency was adequate for the regions of interest to us – i.e., those containing relatively abundant cytosine-phosphate-guanine (CpG) sites. Greater (>98%) conversion of the unmethylated C’s could be achieved by exposure to bisulfite at 50 °C for 16 h, but this resulted in loss of half of the original DNA template molecules when assessed by the Methods described in Performance Evaluation 2 below.
The amplification of the bisulfite-treated library was performed as in Step 3b below with one important difference: the primers for PCR were shorter, truncated at the 3′ end after a guanine-cytosine (GC) dyad. In the copied molecules, all C’s would be converted to T’s following bisulfite treatment, including those in the primer-binding sites. Thus, only the original molecules that have methylated adaptors would retain homology to primer sequences and would be selectively amplified during PCR. The reason for the primer change from the original library amplification primers was to minimize the effects of any unintended copied strands that end up in the libraries of the original DNA strands during Step 2. There are two conceivable ways for copied strands to be carried over into the library of original molecules. First, although only one primer was used in Step 2, any contaminating adapter-derived sequences from prior ligation steps could act as primers to produce nonbiotinylated copies of the original strand. Because these copies would no longer be methylated at CpG sites, they could artificially decrease the apparent fraction of methylated CpG sites in libraries from original template strands. Second, although the use of dual rather than single biotin should prevent dissociation of biotinylated copied strands from streptavidin during heating (18), any small fraction of copied strands that also come off the streptavidin beads during heat denaturation would contaminate the library with original molecules. Of several primers tested to specifically amplify the original molecules following bisulfite conversion, the one employed in the final protocol (SI Appendix, Table S1; Materials and Methods) performed best in preserving the expected methylation patterns, as described in Performance Evaluation 5 below.
Step 3b: PCR-amplify the copied strands.
This step was performed identically to that of the basic SaferSeqS protocol (1) after the copied molecules are cleaved from the streptavidin beads described in Step 2. In this step, primers targeting the adaptor sequences at the ends of each DNA fragment are used for amplification (SI Appendix, Table S1).
At the end of these three steps, the procedure yields two libraries: one from copied molecules for assessment of genetic alterations such as mutations and copy number changes, named “LA” (Step 3b), and one from original molecules for assessment of DNA methylation, named “LM” (Step 3a).
Performance Evaluation #1: What Fraction of the Original Template Molecules Are Represented in the Libraries?
Because each original template molecule has a unique molecular barcode [unique identifier (UID)], we could estimate the fraction of template molecules recovered in the LA and LM libraries through amplification of the libraries with specific primers for genomic regions of interest. We used 48 regions that were commonly mutated in cancers as previously described (1) to evaluate libraries generated from the plasma DNA of seven healthy individuals (SI Appendix, Table S2). This procedure uses a hemi-nested PCR to enrich the regions of interest. Because evaluation of both strands of the same molecules is required for the most sensitive and accurate detection of mutations, the recovery of duplex UIDs was used as a metric for the efficiency of library construction. Each of the cfDNA samples was divided into two equal aliquots: one aliquot was used to construct MethylSaferSeqS libraries and the other to construct standard SaferSeqS libraries. As shown in Fig. 2A, the number of duplex UIDs recovered in the LA form of MethylSaferSeqS libraries was comparable to that in the standard SaferSeqS libraries. The average recovery of duplex UIDs for the MethylSaferSeqS LA libraries was 78% [interquartile range (IQR): 68 to 89%] of the initial DNA template molecules in the cfDNA. This recovery represents a lower bound because it does not take into account losses of template molecules during the hemi-nested PCR enrichment step used for mutation analysis.
To estimate the fraction of templates recovered during preparation of the LM libraries, we generated “pre-LM libraries” from the DNA strands eluted from streptavidin beads after heat denaturation but before bisulfite conversion in Step 2. In parallel, we amplified the DNA from the copied strands which remained bound to the beads (i.e., the LA libraries) using the same primers. The duplex UIDs recovered from the pre-LM libraries averaged 97% (IQR 94 to 100%) of those recovered in the LA libraries (Fig. 2B).
We did not evaluate the fraction of templates remaining after bisulfite conversion in the LM libraries using this hemi-nested PCR-based approach. After bisulfite treatment, the Watson and Crick strands are no longer complementary, making primer design targeting the same region of both strands to accurately estimate duplex recovery difficult, and duplexes were not required for the methylation experiments described below.
Performance Evaluation #2: Are the Same Template Molecules Represented in the LA and LM Libraries?
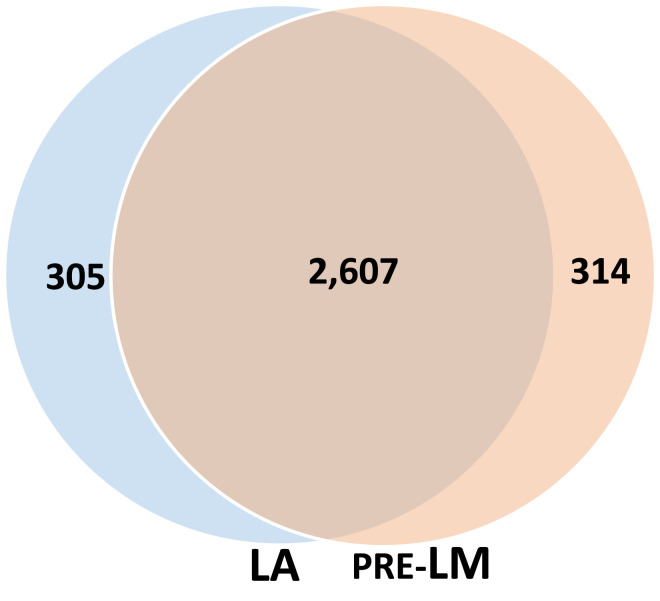
As the majority of the original plasma template molecules were present in both the LA and pre-LM libraries (Fig. 2), there must be considerable overlap between the template molecules in these two libraries based on arithmetic. To quantify the degree of overlap, we performed experiments similar to those described above, but evaluated the fraction of molecules that were shared between the two libraries as determined by their common UIDs (Fig. 3) rather than by estimation of the total number of template molecules recovered in the libraries.
Fig. 3.
DNA molecules represented in both MethylSaferSeqS LA and pre-LM libraries. Venn diagram depicting the number of DNA molecules represented in each library, as estimated by the number of common duplex UIDs.
Analogous to the experiment depicted in Fig. 2B, we first evaluated the shared molecules in the pre-LM libraries made prior to bisulfite-mediated deamination. An average of 2,921 UIDs (IQR: 2,885 to 2,957) were identified in the pre-LM libraries made from the original template strands and 2,912 UIDs (IQR: 2,834 to 2,991) in the LA libraries made from the copied template strands. On an average, 2,607 UIDs (IQR: 2,553 to 2,662) were present in both the pre-LM and LA libraries, which comprise about 90% of each library. Conversely, an average of 314 UIDs (10.7%, IQR 10.2 to 11.2%) were present only in the pre-LM libraries, while 305 (10.5%, IQR: 9.9 to 11%) were present only in the LA libraries (Fig. 3).
Performance Evaluation #3: Can MethylSaferSeqS Libraries Be Used to Detect the Mutations Observed in Standard Libraries?
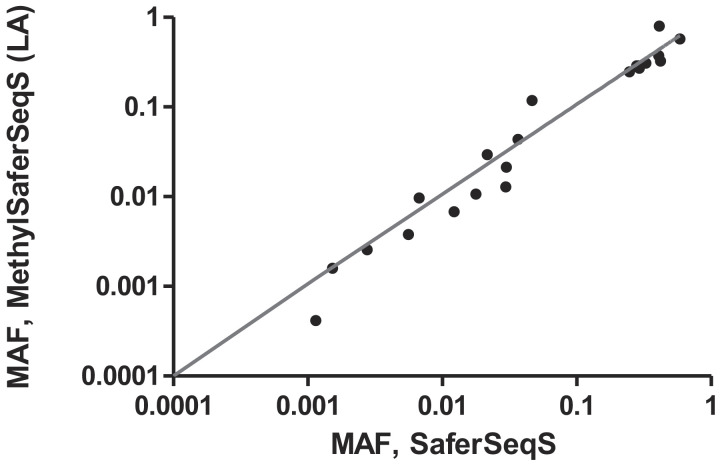
Cell-free plasma DNA samples from 19 patients with advanced colorectal cancer (CRC) (SI Appendix, Table S2) were divided into two equal aliquots. One aliquot was used to construct MethylSaferSeqS libraries and the other to construct standard SaferSeqS libraries. These libraries were then queried for somatic mutations in 48 genomic regions commonly mutated in these cancers, as described for Performance Test 1. A total of 35 somatic mutations were observed and, of these, 22 (63%) were observed in both the MethylSaferSeqS and SaferSeqS libraries. The mutant allele frequencies (MAFs) of the mutations found in both libraries were highly concordant (Fig. 4; Pearson’s r = 0.9, P < 1 × 10−9), with an average MAF of 17.6% (range 0.04 to 79.6%). The remaining 13 (37%) mutations were found in only one of the two libraries (five in SaferSeqS only, and eight in MethylSaferSeqS only) at much lower MAF (average 0.16%; range 0.04 to 0.69%), and all of these were in the stochastic realm as each library generated was from a separate ~5-mL aliquot of plasma.
Fig. 4.
Concordance of mutations and their MAFs identified in cfDNA by MethylSaferSeqS and SaferSeqS. Equal aliquots (~5 mL) of plasma from 19 patients with Stage IV CRCs were made into MethylSaferSeqS and SaferSeqS libraries. The MethylSaferSeqS LA and SaferSeqS libraries were assessed for mutations in 48 commonly mutated genomic regions. Twenty-two mutations were identified in both libraries. The mutant allele fraction (MAF) in the MethylSaferSeqS vs. SaferSeqS libraries for these mutations is shown (axes are on logarithmic base 10 scale, range: 0.01 to 100%).
In addition to the 19 patients for whom paired conventional SaferSeqS and MethylSaferSeqS libraries were available, we analyzed MethylSaferSeqS LA libraries from another 246 patients with the same panel of 48 amplicons. In sum, we evaluated MethylSaferSeqS LA libraries from 265 patients, including 198 patients with cancers of the colon, ovaries, pancreas, or lung, as well as 67 healthy individuals, to determine whether the fraction of samples with somatic mutations detectable in the cfDNA conformed to those expected from prior studies. Of the 198 cancer patients, 121 (61%) patients had at least one detectable somatic mutation (Table 1). The mutations and their MAFs detected are listed in SI Appendix, Table S3. As expected, higher fractions of patients with later-stage cancers (79% of patients with Stage III or IV cancers) than in early-stage cancers (49% of patients with Stage I or II cancers, P < 1 × 10−11, Binomial test) had detectable mutations. The mutant allele fractions observed were also considerably higher in patients with later-stage cancers than early-stage cancers (6.6 vs. 1.5%, P < 0.005, Student’s t test). In the 67 samples from healthy individuals, only one (1.5%) had a detectable mutation in their cfDNA at a low MAF (0.25%; SI Appendix, Table S3). This particular mutation (GNAS p.R201H) is commonly observed in individuals with clonal hematopoiesis of indeterminate potential (CHIP) (19).
Table 1.
Fraction of patients with detectable mutations in cfDNA using MethylSaferSeqS
| Cancer type/stage | # Patients total | # Patients detected (% of total) | Avg MAF (range), % |
|---|---|---|---|
| Lung (total) | 32 | 25 (78%) | 2.21% (0.03 to 26.02%) |
| III | 22 | 20 (91%) | 2.72% (0.03 to 26.02%) |
| II | 4 | 2 (50%) | 0.14% (0.13 to 0.14%) |
| I | 6 | 3 (50%) | 0.14% (0.09 to 0.19%) |
| Ovarian (total) | 37 | 25 (68%) | 3.61% (0.07 to 24.19%) |
| IV | 2 | 2 (100%) | 1.05% (0.35 to 1.74%) |
| III | 17 | 12 (71%) | 2.98% (0.07 to 17.12%) |
| II | 5 | 5 (100%) | 7.04% (0.15 to 24.19%) |
| I | 5 | 2 (40%) | 1.10% (0.12 to 2.08%) |
| NA | 8 | 4 (50%) | 3.77% (0.10 to 8.17%) |
| Pancreas (total) | 102 | 49 (48%) | 0.86% (0.06 to 9.12%) |
| IV | 5 | 5 (100%) | 0.45% (0.26 to 0.66%) |
| III | 6 | 1 (17%) | 0.87% (0.87 to 0.87%) |
| II | 75 | 37 (49%) | 1.00% (0.07 to 9.12%) |
| I | 16 | 6 (38%) | 0.28% (0.06 to 0.93%) |
| CRC (total) | 27 | 22 (81%) | 14.16% (0.07 to 58.59%) |
| IV | 26 | 22 (85%) | 14.16% (0.07 to 58.59%) |
| II | 1 | 0 (0%) | 0.00% (0.00 to 0.00%) |
| Healthy control (total) | 67 | 1 (1%) | 0.25% (0.25 to 0.25%) |
Performance Evaluation #4: Can MethylSaferSeqS Libraries Be Used to Detect Copy Number Alterations Observed in Standard Libraries?
For the analysis of copy number alterations, we used the cfDNA from 17 patients with CRC (SI Appendix, Table S2), from which an aliquot was used to construct MethylSaferSeqS libraries and another to construct standard SaferSeqS libraries, as described above. The libraries were PCR-amplified using primers suitable for loading on an Illumina instrument (SI Appendix, Table S1), followed by whole-genome sequencing (WGS) to ~0.75×. Nearly identical changes in specific chromosomal regions, as well as breakpoints, were observed in both the SaferSeqS and MethylSaferSeqS LA libraries (Fig. 5 and SI Appendix, Table S4). There were 122 regions with a copy number gain or loss identified in MethylSaferSeqS libraries, of which, 112 (92%) were found with the same change (copy number gain or loss) in the corresponding SaferSeqS libraries. Conversely, there were 132 regions with copy number gain or loss identified in the SaferSeqS libraries, of which 114 (86%) changes were also found in the MethylSaferSeqS libraries. When more stringent criteria were applied to exclude borderline regions (Materials and Methods), nearly all remaining calls (98%, or 120 of 122) in the SaferSeqS and MethylSaferSeqS libraries were concordant. No copy number alterations were observed in the SaferSeqS and MethylSaferSeqS libraries derived from cfDNA of the healthy control (Fig. 5).
Fig. 5.
Concordance of copy number alterations identified in cfDNA by MethylSaferSeqS and SaferSeqS. MethylSaferSeqS (LA) and SaferSeqS libraries were constructed from equal aliquots of plasma from patients with advanced CRC and healthy controls for identification of copy number changes. Representative regions are shown for three patients with CRC and a healthy control. Y-axis represents the number of bins (each bin represents a 500 KB genomic region).
Performance Evaluation #5: Can MethylSaferSeqS Libraries Reliably Detect DNA Methylation?
LM libraries underwent bisulfite-mediated deamination as described in Step 3a above, with an average conversion efficiency of 77% (IQR: 75 to 79%). We evaluated the LM libraries in two ways. First, we assessed the contribution of various cell types to cfDNA from healthy individuals. We performed whole-genome sequencing to an average depth of ~7 haploid genome equivalents. We then used 5,612 differentially methylated regions (DMR) that were characteristic of given cell types, as previously described (20). This allowed us to define the contributions from the liver, lungs, colon, small intestines, pancreas, adrenal glands, esophagus, heart, brain, T cells, B cells, and neutrophils using quadratic programming in 31 healthy individuals (SI Appendix, Table S5). In the MethylSaferSeqS libraries, the fraction of template molecules derived from each of the twelve cell types analyzed was similar to expected (SI Appendix, Fig. S1). For example, leukocytes were by far the most predominant contributors, with average contributions of 68.6% (IQR: 67.2-73.3%), consistent with prior findings (20, 21). Among leukocyte subsets, neutrophils were the greatest contributor, with average contributions of 43.8% (IQR: 40.5 to 48.9%). The average neutrophil-to-lymphocyte ratio was 1.8 (IQR: 1.6 to 2.1), consistent with the expected ~2:1 ratio in the circulation of healthy individuals (22). Normal liver was the highest contributor among nonhematopoietic cells, with an average contribution of 8.2% (IQR: 5 to 7.8%), also consistent with prior findings (20). There was greater-than-expected fraction of cfDNA (7.5%, IQR 6.4 to 8.6%) identified as derived from the brain. This higher-than-expected fraction has been observed previously (21), which has been attributed to an artifact of deconvolution algorithms and, less likely, turnover of peripheral neurons.
Second, LM libraries from the cfDNA of cancer patients were PCR-amplified using the primers described in Step 3a that were specific for the original molecules after bisulfite conversion (SI Appendix, Table S1). We then performed whole-genome sequencing to an average depth of ~4.1 haploid genome equivalents. To illustrate that MethylSaferSeqS could detect cancer-specific methylation, the data from 11 genomic regions that have previously been reported to be hypermethylated in cancers are recorded in SI Appendix, Table S6 (23–28). The average size of the regions was 114 bp (range 93 to 132 bp), containing an average of 12.6 CpG sites per region (range 7 to 19 bp). This provided an opportunity to discover methylation differences in cfDNA samples in which sufficient amounts of DNA were derived from neoplastic cells. By rough estimation, each marker could be covered in at most 52 distinct DNA molecules (average of 12.6 CpG’s × average coverage of 4.1 per CpG). Thus, we applied the panel to plasma samples with mutation MAF > 2%, as determined in Performance Evaluation #3, assuming at least one mutant DNA molecule was captured by the panel.
Fractional methylation was calculated for each marker as the ratio of the total number of unconverted cytosines in CpG sites to the total number of CpG sites (Materials and Methods). The fractional methylation was considerably higher in the cfDNA of the 29 cancer patients who had sufficient neoplastic content (MAF > 2%) than that in the cfDNA samples from the 67 healthy individuals used as controls (34% vs. 5%, P < 1 × 10−5, Student’s t test; SI Appendix, Table S7). Moreover, there was a correlation between the mutation MAF found in LA libraries and the fractional methylation in these samples (Pearson’s r = 0.9, P < 1 × 10−9; SI Appendix, Fig. S2 and Table S7). Finally, although the markers in our panel were chosen to be cancer specific rather than tissue specific, some markers appeared to be specific for certain cancers. For example, the ones reported to be altered in CRCs were more often methylated in the cfDNA from patients with CRCs than those in patients with other types of cancers or in healthy individuals (Fig. 6).
Fig. 6.
Fraction of methylation identified in cfDNA samples using markers that are hypermethylated in CRCs. MethylSaferSeqS LM libraries from plasma samples of 29 patients with colorectal (CRC), ovarian, pancreatic, and lung cancers that had sufficient neoplastic content (>2% mutant allele frequency based on mutation assay using LA libraries) as well as 67 healthy controls were subjected to whole-genome sequencing. The fractions of methylation in four representative markers that are commonly hypermethylated in CRCs are shown.
Finally, we wished to determine whether we could reliably access the methylation status of mutated DNA fragments present in the plasma. For this purpose, we examined two patient samples with mutations that occurred in a CpG dyad that affected the methylation status of the strands (Fig. 7). Because the Watson and Crick strands of the original molecules lose complementarity after bisulfite conversion, we used two sets of PCR primers (one for each strand) that reflected the conversion of C’s to T’s in the LM libraries (SI Appendix, Table S1). For example, in the plasma from patient INDI 995, a patient with Stage IB ovarian clear cell adenocarcinoma, a PIK3CA p.H1047R mutation was found in 3.1% in the LA library. This mutation creates a CpG site (CAT → CGT) in the mutant molecules. To assess whether the mutant molecules found in LA molecules were methylated in the LM libraries, we amplified the LM libraries with primers in SI Appendix, Table S1. Overall, 409 UIDs were found in both LM and LA libraries. Of those, 11 (2.7%) contained the PIK3CA p.H1047R mutation. The lower fraction of mutants detected (2.7% vs. 3.1%) is likely due to the distance of primers from the mutation of interest, such that some very short amplified DNA molecules (of the 409) did not span the mutation. As expected, a high fraction of mutant UIDs (82% of 11) were methylated in the CGT trimer, compared to only 1.3% of the 398 C residues in the analogous CAT trimer in the wild-type UIDs, presumably due to incomplete bisulfite conversion (Fig. 7).
Fig. 7.
Determination of methylation status in wild-type and mutant DNA fragments in cfDNA. Two patient samples had mutations identified in the MethylSaferSeqS LA libraries that are expected to alter the methylation status of mutant molecules in the LM libraries. In INDI 955, the PIK3CA p.H1047R mutation was found at 3.1% of the UIDs in the MethylSaferSeqS LA library. This mutation creates a CpG site in mutant molecules. In INDIA 133, the TP53 p.R248W mutation was found in 9.3% of the UIDs in the MethylSaferSeqS LA library. This mutation erases a CpG site in mutant molecules. The expected methylation status of the wild-type and mutant molecules is shown for each patient, along with the fraction of methylated UIDs found for the wild-type and mutant molecules. MAF = mutant allele fraction.
Similarly, in another patient, INDIA 1333, who had Stage IIB pancreatic adenocarcinoma, a TP53 p.R248W mutation was found in the LA libraries at 9.3% MAF. This mutation occurred at a CpG site (CCG → CCA). As above, we performed targeted enrichment for this mutant position in the LM libraries with the primers listed in SI Appendix, Table S1. There were 900 UIDs found in both LA and LM libraries. Of those, 65 UIDs (7.2%) contained the TP53 p.R248W mutation. A high fraction of wild-type UIDs (98% of 835 UIDs) were methylated in the CCG trimer. As expected, only 3.1% of the 65 C residues in the analogous CCA trimer in the mutant UIDs appeared to be methylated, presumably due to incomplete bisulfite conversion (Fig. 7).
Discussion
In the era of precision medicine, it would be useful to develop tools that make use of our burgeoning knowledge of the cancer genome and epigenome. Here, we described a method, MethylSaferSeqS, that can detect genetic and epigenetic changes in the same DNA molecules. Using plasma samples from cancer patients, we showed that the expected mutations, copy number changes, and methylation can be detected with MethylSaferSeqS. With simple modifications, MethylSaferSeqS can be adapted to any library preparation method compatible with next-generation sequencing, including those that support duplex sequencing, and can do so without greatly compromising the efficiency of molecule recovery.
There are a variety of potential applications for MethylSaferSeqS. One obvious application is to incorporate tumor-specific epigenetic and genetic markers for cancer detection. For this purpose, MethylSaferSeqS allows for concurrent construction of the libraries for epigenetic and genetic assessments, alleviating the need to split the sample for each assay and maximizing sensitivity. Furthermore, methylation markers can be tissue specific as well as cancer specific, such that they essentially provide fingerprints that can be used to trace the tissue of origin. Finally, MethylSaferSeqS libraries can be used to study other features related to cancer, such as cfDNA fragmentomics, in the context of the genetic and epigenetic status of each DNA molecule.
There are several limitations to our study. First, we did not distinguish somatic mutations that were derived from neoplastic cells from those derived from hematopoietic cells. Thus, we did not exclude mutations arising from CHIP. In a diagnostic test, those mutations can be excluded by studying the DNA from matched white blood cells. This was not within the scope of our study, as our goal was to demonstrate that MethylSaferSeqS can reliably detect somatic mutations regardless of the source. However, given only one of the 67 healthy individuals (1.5%) had a detectable mutation, we do not believe that CHIP was a major source of mutations reported in our study.
Another limitation in our study was the loss of DNA during bisulfite treatment, resulting in relatively low number of DNA molecules queried for methylation in LM libraries. Even with our modified protocol and reduced conversion time, a substantial fraction of DNA (~30%) was degraded during bisulfite treatment. To increase recovery further would require gentler treatment conditions that result in a lower conversion efficiency, which could lead to overestimation of the fraction of methylated molecules. In our study, the lower recovery did not affect our ability to interpret shallow whole-genome sequencing data. It also did not impact our ability to evaluate methylation with hemi-nested PCR, which can be used to assess orders of magnitude more genome fragments than WGS at equivalent cost. Furthermore, the optimal genomic regions to distinguish plasma DNA samples from patients with and without cancers are those that contain multiple CpG sites. An 80% bisulfite conversion rate is suitable for evaluation of such regions because the probability that multiple CpG sites within a region are not bisulfite converted is exponentially less than that for a single site. For the evaluation of a single CpG site, however, an 80% conversion rate would not be sufficient to make definitive conclusions. For such purposes, MethylSaferSeqS can be readily adapted to incorporate bisulfite-free methods for generating LM libraries that result in less DNA degradation while maintaining higher conversion efficiencies (29–31).
Finally, we did not attempt to show that the concurrent evaluation of methylation, mutation, and copy number changes in the same DNA molecules could add sensitivity to existing detection methods. But with this demonstration of a versatile method that is readily adaptable to existing sequencing and experimental strategies for the simultaneous detection of these features, our study sets the stage for a large prospective trial that can evaluate MethylSaferSeqS for various clinical applications.
Materials and Methods
Plasma Collection and DNA Collection.
This study was approved by the Institutional Review Boards for Human Research at Johns Hopkins Hospital (Institutional Review Board Numbers: IRB00075499 and NA_00090530). All the participants provided a written informed consent in accordance with the principles of the Declaration of Helsinki. DNA was purified from ~5 mL plasma using a BioChain cfDNA Extraction kit (Cat # K5011625).
Library Preparation.
Libraries were prepared as previously described (1) with the following modifications: Custom 3′ and 5′ adaptors containing 5-methylcytosines rather than unmethylated cytosines were used (SI Appendix, Table S1) during ligation. The adaptor sequences otherwise are identical to what has been used for SaferSeqS. Following ligation, both strands of the templates were copied with a single, deoxyuridine-containing, dual-biotinylated primer at 5 uM targeting the 3′ adaptor (SI Appendix, Table S1) for three PCR cycles with KAPA HiFi HotStart Uracil + ReadyMix Kit (Roche Diagnostics; cat # 7959052001). The resultant molecules (one original strand bound to a complementary copied strand from the last PCR cycle, and two additional copied strands from the first two PCR cycles) were bound to Streptavidin MyOne Dynabeads T1 (ThermoFisher; cat # 65601). Beads and bound DNA were heated to 75 °C in 22.5 μL water for 2 min to separate the original strands (now in the supernatant) from the copied strands that remain attached to the beads. The beads were then incubated again in 22.5 μL water at 75 °C for 2 min, and the DNA in the supernatant combined with the DNA in the first supernatant (total of 45 μL).
For preparation of the LM library, the DNA from the supernatant was deaminated with bisulfite using reagents from the EZ DNA Methylation Kit (Zymo Research; cat # D5004) per manufacturer’s protocol with the following modifications: After denaturation, 100 μL CT Conversion Reagent was added to each well and incubated in the dark for 3 h at 50 °C, followed by a 10-min hold at 4 °C. The conversion reaction was then added to the Silicon-A-Binding plate and mixed with 600 μL M-Binding buffer. The plate was centrifuged at 2,800 g for 5 min, and each column was subsequently washed with 500 μL M-Wash buffer. After washing, 200 μL M-Desulphonation buffer was added to each well and incubated at room temperature for 15 min. The plate was then centrifuged at 2,800 g for 5 min and washed twice with 600 μL then 400 μL M-Wash buffer. The DNA was finally eluted twice in 21 μL M-elution buffer each time for a total volume of ~42 μL and amplified as below.
For preparation of the LA library, the copied strands bound to streptavidin beads were released by treatment with USER (Uracil-Specific Excision Reagent) enzyme (New England Biolabs; cat # M5505S). USER contains a mixture of Uracil DNA glycosylase and the DNA glycosylase-lyase Endonuclease VIII that in combination cleaves at the deoxyuridine base in the primer (Fig. 1). The Dynabeads with bound copied molecules were incubated with 3 μL USER and 37 μL Elution Buffer (Qiagen, Cat 19086) in a 40 μL reaction for 60 min at 37 °C. Following incubation, the cleaved DNA strands were separated from the beads using a magnet, collected in the supernatant, and added to PCR reactions as described below.
For the amplification of LM libraries, ~42 μL purified DNA after bisulfite conversion was added to PCR reactions performed with KAPA HiFi HotStart Uracil +ReadyMix Kit (Roche Diagnostics; Cat # 7959052001) with primers at a final concentration of 5 μM (SI Appendix, Table S1) in 100 μL reactions. For the amplification of LA libraries, 40 μL DNA that was cleaved from the streptavidin beads was added to PCR reactions performed with KAPA HiFi HotStart +ReadyMix Kit (Roche Diagnostics; cat # K2601) with primers at a final concentration of 5 μM (SI Appendix, Table S1) in 100 μL reactions. All libraries were amplified for eight cycles as follows: 98 °C for 45 s, then eight cycles of 98 °C for 15 s to denature, 60 °C for 30 s to anneal, and 72 °C for 30 s to extend. The amplified DNA from each reaction was purified using 180 μL SPRISelect beads (Beckman Coulter) and eluted using 100 μL Elution Buffer (Qiagen, cat # 19086). For WGS of either the LA or LM libraries, 100 ng library DNA was amplified in 50 μL reactions in Ultra Q5 with primers at 2 μM (SI Appendix, Table S1) for seven cycles with the following conditions: 98 °C for 30 s, then seven cycles of 98 °C for 10 s to denature, and 65 °C for 75 s to anneal and extend. For amplification of specific regions of the genome, anchored hemi-nested PCR for LM libraries was done using the primers described in SI Appendix, Table S1. For amplification of the 48 regions in LA performed, primers were used as previously described (1).
Sequencing and Analysis.
Barcoded libraries were sequenced using 100 bp paired-end runs (200 cycles) on a NovaSeq 6000 to an average depth of 75 million read pairs per sample. Mutation analysis was performed after demultiplexing, and grouping reads into duplex families was carried out as previously described (1). For methylation analysis, Bisulfite Sequence MAPping program (BSMAP) was used to align each paired-end read to the bisulfite-converted hg19 genome, and the fractional methylation was calculated with BSMAP’s methratio.py script (32). The intercept function in BEDtools (33) was then used to identify the fraction of methylation in the targeted regions (SI Appendix, Table S6). The conversion efficiency for each sample was estimated from regions that are unmethylated such as in the 14 unmethylated bases in the UIDs (SI Appendix, Table S7). For comparison of UIDs in the LA and LM libraries, all the 14 bases in UIDs were presumed to undergo complete conversion during bisulfite treatment. The methylation fraction for each region was calculated from the total number of unconverted cytosines in CpG’s to the total number of CpGs per region:
where n represents the total number of CpG sites in the region, i represents a particular CpG site, Nc,i represents the number of unconverted C’s in the CpG site, and Nt,i represents the number of converted C to T’s in the CpG site.
This fraction was then adjusted to obtain values above the nonconverted background and to account for minor differences in conversion efficiency across experiment as follows:
Only regions with more than 10 reads are included in SI Appendix, Table S7. For cfDNA tissue deconvolution in plasma from healthy individuals using quadratic programming, the methods as previously described (20) were used with a total of 5,653 markers. For the identification of copy number changes, the reads from WGS of MethylSaferSeqS LA libraries and SaferSeqS libraries were processed using ichorCNA algorithm (34). As more stringent criteria, we required logR ratio > 0.16 (for copy number gain) or < −0.16 (for copy number loss) to filter out borderline regions, as well as number of bins per region >20 (equivalent of 20 × 500,000 basepairs, or 10 MB) to ensure that sufficiently large number of markers were used for mapping. Mutations are identified in the SaferSeqS and MethylSaferSeqS libraries using the following criteria: # mutant UIDs (Supercalimutants) >1, family count within each UID family >1, mutations that were either inactivating in tumor suppressors or present with >5 counts in the Catalogue Of Somatic Mutations In Cancer Database (35). All sequencing data will be deposited in European Bioinformatics Institute EGAS00001006839 (36).
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank the individuals who participated in this study for their courage and generosity. We also thank Dr. S. Markowitz for insightful comments and review of this manuscript. We are grateful to C. Blair and K. Judge for expert technical and administrative assistance and to E. Cook for illustrative assistance. This work was supported by The Lustgarten Foundation for Pancreatic Cancer Research, The Virginia and 297 D.K. Ludwig Fund for Cancer Research, The Conrad N. Hilton Foundation, The Sol Goldman Charitable Foundation, Burroughs Wellcome Fund Career Award for Medical Scientists, the Commonwealth Foundation, NCATS 1R21TR004059, NINDS R21NS113016, and National Cancer Institute grants/NIH grants (R37 CA230400, U01 CA200469, U01 CA62924, U01 CA06973, U01 CA230691, T32 GM136577, and T32 CA009071).
Author contributions
Y.W., C.D, J.D.C., A.M., C.B., N.P., K.W.K., and B.V. designed research; Y.W., C.D., J.D.C., A.M., S.C., N.S., M.P., J.P., L.D., N.N., J.C.D., M.S., M.Z., C.B., N.P., K.W.K., and B.V. performed research; L.T.H.-P., B.N.H.T., T.S.T., T.V.N., C.B., N.P., K.W.K., and B.V. contributed new reagents/analytic tools; Y.W., C.D., J.D.C., A.M., S.C., C.B., N.P., K.W.K., and B.V. analyzed data; and Y.W. and B.V. wrote the paper.
Competing interests
B.V., K.W.K., and N.P. are founders of Thrive Earlier Detection, an Exact Sciences Company. K.W.K., N.P., Y.W., and C.D. are consultants to Thrive Earlier Detection. B.V., K.W.K., N.P., and C.D. hold equity in Exact Sciences. B.V., K.W.K., and N.P. are founders of or consultants to and own equity in ManaT Bio., Haystack Oncology, Neophore, CAGE Pharma, and Personal Genome Diagnostics. J.D.C. is a founder and holds equity in Haystack Oncology. N.P. is consultant to Vidium. B.V. is a consultant to and holds equity in Catalio Capital Management. C.B. is a consultant to Depuy-Synthes, Bionaut Labs, Haystack Oncology, and Galectin Therapeutics. C.B. is a co-founder of OrisDx. The companies named above, as well as other companies, have licensed previously described technologies related to the work described in this paper from Johns Hopkins University. B.V., K.W.K., N.P., Y.W., A.M., J.D.C., and C.D. are inventors on some of these technologies. Licenses to these technologies are or will be associated with equity or royalty payments to the inventors as well as to Johns Hopkins University. Patent applications on the work described in this paper may be filed by Johns Hopkins University. The terms of all these arrangements are being managed by Johns Hopkins University in accordance with its conflict of interest policies.
Footnotes
Reviewers: C.C., University of Cambridge; and J.-P.I., Coriell Institute for Medical Research.
Contributor Information
Yuxuan Wang, Email: yuxuan.wang@jhmi.edu.
Bert Vogelstein, Email: vogelbe@jhmi.edu.
Data, Materials, and Software Availability
Sequencing data have been deposited in European Genome-phenome Archive (EGA) with accession no. EGAS00001006839 (36).
Supporting Information
References
- 1.Cohen J. D., et al. , Detection of low-frequency DNA variants by targeted sequencing of the Watson and Crick strands. Nat. Biotechnol. 39, 1220–1227 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cohen J. D., et al. , Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 359, 926–930 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Douville C., et al. , Assessing aneuploidy with repetitive element sequencing. Proc. Natl. Acad. Sci. U.S.A. 117, 4858–4863 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frommer M., et al. , A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. U.S.A. 89, 1827–1831 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayatsu H., Wataya Y., Kai K., Iida S., Reaction of sodium bisulfite with uracil, cytosine, and their derivatives. Biochemistry 9, 2858–2865 (1970). [DOI] [PubMed] [Google Scholar]
- 6.Shapiro R., DiFate V., Welcher M., Deamination of cytosine derivatives by bisulfite. Mechanism of the reaction. J. Am. Chem. Soc. 96, 906–912 (1974). [DOI] [PubMed] [Google Scholar]
- 7.Alexandrov L. B., et al. , The repertoire of mutational signatures in human cancer. Nature 578, 94–101 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoang M. L., et al. , Genome-wide quantification of rare somatic mutations in normal human tissues using massively parallel sequencing. Proc. Natl. Acad. Sci. U.S.A. 113, 9846–9851 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmitt M. W., et al. , Detection of ultra-rare mutations by next-generation sequencing. Proc. Natl. Acad. Sci. U.S.A. 109, 14508–14513 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abascal F., et al. , Somatic mutation landscapes at single-molecule resolution. Nature 593, 405–410 (2021). [DOI] [PubMed] [Google Scholar]
- 11.Ueda S., et al. , A quantification method of somatic mutations in normal tissues and their accumulation in pediatric patients with chemotherapy. Proc. Natl. Acad. Sci. U.S.A. 119, e2123241119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kinde I., Wu J., Papadopoulos N., Kinzler K. W., Vogelstein B., Detection and quantification of rare mutations with massively parallel sequencing. Proc. Natl. Acad. Sci. U.S.A. 108, 9530–9535 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Newman A. M., et al. , Integrated digital error suppression for improved detection of circulating tumor DNA. Nat. Biotechnol. 34, 547–555 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Braun R. E., Every sperm is sacred–or is it? Nat. Genet. 18, 202–204 (1998). [DOI] [PubMed] [Google Scholar]
- 15.Olova N., et al. , Comparison of whole-genome bisulfite sequencing library preparation strategies identifies sources of biases affecting DNA methylation data. Genome Biol. 19, 33 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanaka K., Okamoto A., Degradation of DNA by bisulfite treatment. Bioorg Med. Chem. Lett. 17, 1912–1915 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Kint S., De Spiegelaere W., De Kesel J., Vandekerckhove L., Van Criekinge W., Evaluation of bisulfite kits for DNA methylation profiling in terms of DNA fragmentation and DNA recovery using digital PCR. PLoS One 13, e0199091 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bearden S., Wang F., Hall A. R., Simple and efficient room-temperature release of biotinylated nucleic acids from streptavidin and its application to selective molecular detection. Anal Chem. 91, 7996–8001 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steensma D. P., et al. , Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood 126, 9–16 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun K., et al. , Plasma DNA tissue mapping by genome-wide methylation sequencing for noninvasive prenatal, cancer, and transplantation assessments. Proc. Natl. Acad. Sci. U.S.A. 112, E5503–5512 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moss J., et al. , Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease. Nat. Commun. 9, 5068 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song M., Graubard B. I., Rabkin C. S., Engels E. A., Neutrophil-to-lymphocyte ratio and mortality in the United States general population. Sci. Rep. 11, 464 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li M., et al. , Sensitive digital quantification of DNA methylation in clinical samples. Nat. Biotechnol. 27, 858–863 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park J. K., et al. , The role of quantitative NPTX2 hypermethylation as a novel serum diagnostic marker in pancreatic cancer. Pancreas 41, 95–101 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Wei B., et al. , A panel of DNA methylation biomarkers for detection and improving diagnostic efficiency of lung cancer. Sci. Rep. 11, 16782 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ying L., et al. , Methylation-based cell-free DNA signature for early detection of pancreatic cancer. Pancreas 50, 1267–1273 (2021). [DOI] [PubMed] [Google Scholar]
- 27.Yi J. M., et al. , Novel methylation biomarker panel for the early detection of pancreatic cancer. Clin. Cancer Res. 19, 6544–6555 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patai A. V., et al. , Comprehensive DNA methylation analysis reveals a common ten-gene methylation signature in colorectal adenomas and carcinomas. PLoS One 10, e0133836 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Y., et al. , Bisulfite-free direct detection of 5-methylcytosine and 5-hydroxymethylcytosine at base resolution. Nat. Biotechnol. 37, 424–429 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Schutsky E. K., et al. , Nondestructive, base-resolution sequencing of 5-hydroxymethylcytosine using a DNA deaminase. Nat. Biotechnol. 36, 1083–1090 (2018), 10.1038/nbt.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vaisvila R., et al. , Enzymatic methyl sequencing detects DNA methylation at single-base resolution from picograms of DNA. Genome Res. 31, 1280–1289 (2021), 10.1101/gr.266551.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xi Y., Li W., BSMAP: Whole genome bisulfite sequence MAPping program. BMC Bioinform. 10, 232 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quinlan A. R., Hall I. M., BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adalsteinsson V. A., et al. , Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat. Commun. 8, 1324 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tate J. G., et al. , COSMIC: The catalogue of somatic mutations in cancer. Nucleic Acids Res. 47, D941–D947 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y., et al. , Detection of rare mutations, copy number variation, and DNA methylation in the same template DNA molecules. European Genome-Phenome Archive. https://ega-archive.org/studies/EGAS00001006839. Deposited 3 December 2022.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
Sequencing data have been deposited in European Genome-phenome Archive (EGA) with accession no. EGAS00001006839 (36).