Significance
The study of how calmodulin (CaM) interacts with ion channels is becoming increasingly important for understanding the molecular basis of several human diseases. Here, we show that it is possible to study a CaM-ion channel complex in its native tissue, with a combination of cryo-electron microscopy and structural proteomics. We studied the interaction of CaM with the rod CNG channel from bovine retina; this interaction remained enigmatic for more than three decades. We show that upon binding to CaM, the channel undergoes structural changes that may also provide insights into other CaM-ion channel complexes.
Keywords: ion channel, structural biology, cryo-EM, proteomics, calmodulin
Abstract
Calmodulin (CaM) regulates many ion channels to control calcium entry into cells, and mutations that alter this interaction are linked to fatal diseases. The structural basis of CaM regulation remains largely unexplored. In retinal photoreceptors, CaM binds to the CNGB subunit of cyclic nucleotide-gated (CNG) channels and, thereby, adjusts the channel’s Cyclic guanosine monophosphate (cGMP) sensitivity in response to changes in ambient light conditions. Here, we provide the structural characterization for CaM regulation of a CNG channel by using a combination of single-particle cryo-electron microscopy and structural proteomics. CaM connects the CNGA and CNGB subunits, resulting in structural changes both in the cytosolic and transmembrane regions of the channel. Cross-linking and limited proteolysis-coupled mass spectrometry mapped the conformational changes induced by CaM in vitro and in the native membrane. We propose that CaM is a constitutive subunit of the rod channel to ensure high sensitivity in dim light. Our mass spectrometry-based approach is generally relevant for studying the effect of CaM on ion channels in tissues of medical interest, where only minute quantities are available.
Cyclic nucleotide-gated (CNG) channels in retinal photoreceptors represent the final target of the light-activated signaling pathway (1, 2). At rest, a small fraction of CNG channels is kept open by an elevated cGMP concentration, and a so-called dark current flows into the cell. Light triggers cGMP hydrolysis by a phosphodiesterase (PDE) (3, 4), in turn CNG channels close, thereby generating membrane hyperpolarization, an electrical signal that is passively transmitted to the synaptic terminal. In rod photoreceptors, 10 to 18% of the dark current is carried by Ca2+ (5, 6). Ca2+ plays a fundamental role in rod photoreceptors: It terminates the light response, adjusts light sensitivity, and restores the dark state. Ca2+ controls several enzymes of the phototransduction cascade via small Ca2+-binding proteins (7–15). One of these regulatory proteins is calmodulin (CaM), which lowers the open probability of the CNG channel (16–19).
CaM consists of two lobes (N- and C-lobe) connected by a highly flexible linker region (20) that allows lobes to orient freely (21). The two lobes display a wide range of Ca2+ affinities (22) and recognize different binding motifs. CaM exploits this intrinsic flexibility to target hundreds of different proteins (23, 24). However, owing to this flexibility, it is notoriously difficult to obtain high-resolution structures of membrane protein/CaM complexes; most structures include only CaM bound to short peptides (25). Although CaM regulates many ion channels, even to the point that CaM is considered a genuine subunit of ion channels (26), the structural underpinnings for CaM modulation are just beginning to emerge (27–30).
From a structural perspective, how CaM modulates the activity of CNG channels in olfactory neurons and photoreceptors remains unknown. By increasing the apparent K1/2 for Cyclic adenosine monophosphate (cAMP) binding up to 50-fold (18, 31), CaM attenuates the activity of olfactory CNG channels, which indicates a need for adaptation in olfactory neurons. By contrast, in rod photoreceptors, the K1/2 shift is maximally twofold (17, 32, 33), which is very small compared to other adaptive changes that occur during light adaptation (34, 35). Moreover, regulation by Ca2+ and Ca2+-dependent factors appears to be different in cones vs. rod channels (36–39). In cones of striped bass, CNG-modulin, which is distinct from CaM, lowers the sensitivity of the cone CNG channel by almost fourfold (39). Thus, the physiological function of CaM in rod and cone photoreceptors and in mammalian vs. lower-vertebrate photoreceptors remains enigmatic.
The rod CNG channel is composed of CNGA1 (40) and CNGB1 (41) subunits in a 3:1 stoichiometry (42–44). CNGA1 and CNGB1 encompass six transmembrane segments (S1–S6), a cytosolic C-linker composed of helices A′ to F′, and a cyclic nucleotide-binding domain (CNBD) composed of helices A to C and a beta-roll. Atomic structures of the rod and cone CNG channels have been resolved in closed and partially open states (45–47). These structures reveal the elements that control the ion passage and gating mechanism, and as well similarities and differences of rod and cone CNG channels (48). In homomeric CNGA1 channels, upon binding of cGMP, the CNBD becomes more compact; consequently, the C-linker and the S6 transmembrane segment move, causing the central hydrophobic gate in the ion pathway to open (49, 50). In heteromeric CNGA/CNGB channels, below the central gate, a single arginine residue in CNGB1 forms an additional gate (45–47), whose specific role is unknown.
A feature not addressed by these recent studies was the structural role of CaM. The rod CNGB1 subunit contains two CaM-binding sites, CaM1 and CaM2. CaM1 is located at the N terminus close to a glutamate-rich disordered region, whereas CaM2 is located C-terminally of the CNBD. The role of the two CaM-binding sites remains enigmatic. Whereas CaM1 is required to increase K1/2 of the rod channel, CaM2 is dispensable although its affinity for CaM is higher as compared to CaM1 (19, 51). The affinity of CaM is exquisitely high (<5 nM CaM) (17, 19, 33, 51), suggesting that CaM is permanently bound to the channel. Remarkably, very low Ca2+ concentrations (40 to 50 nM) can modulate the rod CNG channel (19, 52, 53).
Here, we employ high-resolution cryo-electron microscopy (cryo-EM) in combination with mass spectrometry (MS) to study the structural basis of CaM modulation of the bovine rod CNG channel. Remarkably, our work pioneers the combined use of cross-linking and limited proteolysis-coupled MS to study a minor component in a complex protein mixture. We find that CaM connects the CNGA1 and CNGB1 subunits in the coiled-coil region. The cytosolic and transmembrane regions of the channel adopt a more compact structure in complex with CaM than without. Finally, we map the structural changes upon CaM binding on the channel embedded in its native membrane, and we find that the changes detected by MS are on a par with the cryo-EM structures. These findings constitute a step toward characterizing the structural changes underlying binding of CaM to CNG channels.
Results
Cryo-EM Structure of the Rod CNG Channel Bound to CaM.
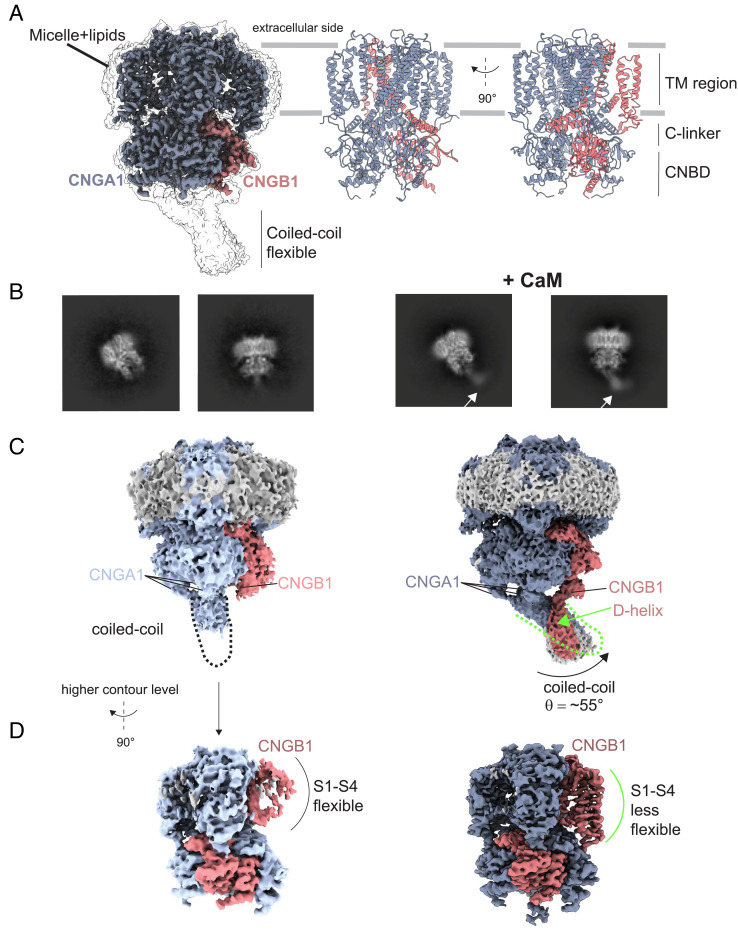
We purified the native CNG channel from rod outer segments of bovine retina (45) (SI Appendix, Fig. S1). Prior to preparation of grids for cryo-EM, we added bovine CaM to the sample at saturating concentration (10 µM). The sample contained 10 mM Ca2+. Single-particle analysis yielded a density map at 2.76 Å (SI Appendix, Figs. S2 and S3) of excellent quality in both the transmembrane and cytosolic regions (Fig. 1A). From this density map, we built an atomic model, which includes CNGA1 residues K151-G615 and CNGB1 residues Q760-L862 and V871-K1203 (Fig. 1A and SI Appendix, Table S1 and Figs. S4 and S5). The two-dimensional (2D) projections of the channel revealed an additional density in the coiled-coil region that is absent in the sample without CaM (Fig. 1B) (45). A side-by-side comparison of the density maps without and with CaM identifies three important differences: First, without CaM, the CNGA1 coiled-coil region, formed by three helices C-terminal of the CNBD is poorly resolved, adopts an orientation perpendicular to the membrane, and it is not making contacts to CNGB1 (Fig. 1C). By contrast, with CaM, the coiled-coil region adopts an angle of ~55° to the axis perpendicular to the membrane. Second, with CaM, the D-helix of CNGB1 contacts the coiled-coil region (Fig. 1C). Finally, the density map of the transmembrane region of CNGB1 is poorly resolved without CaM, whereas it becomes more visible with CaM (Fig. 1D).
Fig. 1.
Comparison of the overall structure of the bovine rod CNG channel without and with CaM. (A) Density map of the channel with CaM shown at two different contour levels. CNGA1 subunits (blue), CNGB1 subunit (pink). The map is shown in transparency at a lower threshold to highlight the density of the micelle and that of the flexible coiled-coil region. (B) 2D class averages of different projections of the channel without (45) (Left) and with CaM added (Right). The presence of additional density within the coiled-coil region is indicated with an arrow. (C and D) Comparison of the density maps at different contour levels without and with CaM. Maps are shown at a lower contour level to appreciate the difference in the orientation of the CNGA1 coiled-coil region and the presence of an additional helix at the C terminus of the CNGB1 subunit named D-helix (indicated by the green arrow). (D) Maps at a higher contour level to show that the transmembrane region of CNGB1 is much better resolved with CaM. Maps are rotated by 90° compared to panel C.
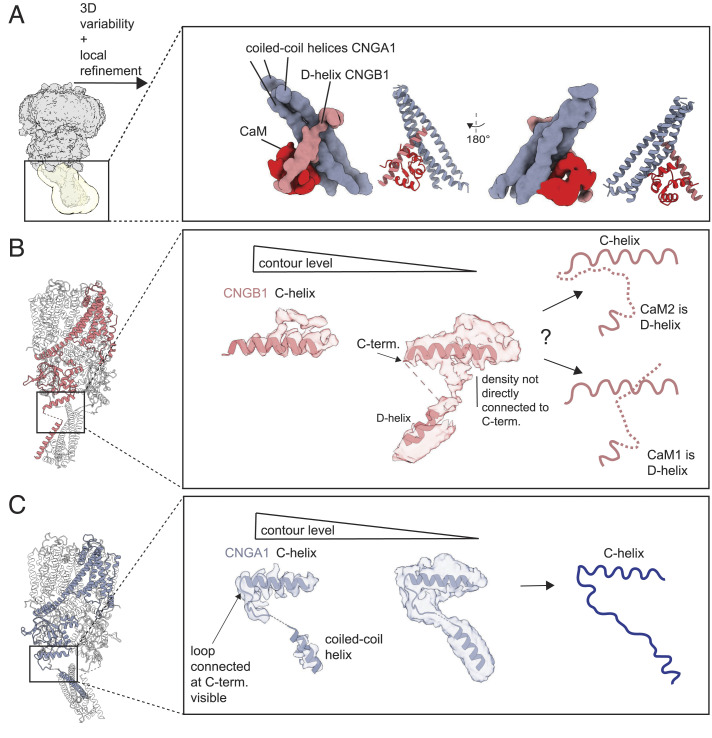
In support of the EM map, three-dimensional (3D) variability analysis (54) shows that the coiled-coil region of CNGA1 and the transmembrane region of CNGB1 are flexible (SI Appendix, Fig. S2). The coiled-coil region switches between two conformations; in one conformation, the interaction between the coiled-coil and the D-helix of CNGB1 is clearly visible, whereas in the other conformation, this interaction is less visible (Movie S1). This pronounced flexibility explains why the coiled-coil region is of low resolution (SI Appendix, Fig. S3). The movement of the coiled-coil region upon binding of CaM is synchronized with the movements in the transmembrane region of CNGB1 (Movie S2). We performed local refinement with a mask that included only the coiled-coil region to improve the density map in this area (SI Appendix, Fig. S6). To reduce heterogeneity, we used a subset of particles isolated from 3D variability analysis, that showed the strongest interaction between CNGA1 and CNGB1 at the coiled-coil region (SI Appendix, Fig. S2). This approach yielded a density map at 4.66 Å (SI Appendix, Fig. S6) that allowed a rigid-body fit of an atomic model containing the CNGA1 coiled-coil helices, the CNGB1 D-helix, and one lobe of CaM (Fig. 2A). This structural information was merged with the high-resolution structure shown in Fig. 1A. In this map, CaM interacts with both CNGA1 and CNGB1. The second lobe of CaM remains not visible probably because it is very flexible. As the connection between the C terminus of the C-helix and the D-helix is difficult to interpret in our density map, it is difficult to determine whether the D-helix encompasses the CaM1 or CaM2 sequence (Fig. 2 C, Center). Two different interpretations are possible: i) the D-helix corresponds to the CaM2 sequence, and the part of the flexible loop between the C- and D-helices is not visible; alternatively, ii) the D-helix corresponds to CaM1, and the visible density near the C-helix is part of the CNGB1 N terminus (Fig. 2 C, Right). By contrast, in CNGA1, the density of the loop sequence that connects the C-helix with the coiled-coil helix is unambiguously resolved (Fig. 2D).
Fig. 2.
Structural details of the coiled-coil region. (A) Left, the density map obtained after nonuniform refinement with a mask around the coiled-coil region in yellow; Right, the density map obtained after local refinement of the coiled-coil region; the three helices of the CNGA1 coiled-coil colored (blue), the CNGB1 D-helix (pink), and CaM (red). A ribbon representation is shown on the side of the density map. (B) Details of the density map obtained after CTF refinement on the entire CNG channel. It illustrates that the density map does not allow to confidently model the connection between C and D helices of CNGB1. Right, two different interpretations are shown: either CaM2 (Upper) or CaM1 (Lower) represents the D-helix. (C) For comparison, the C terminus of the CNGA1 C-helix (chain C is shown) connects the N terminus of the coiled-coil helix unambiguously.
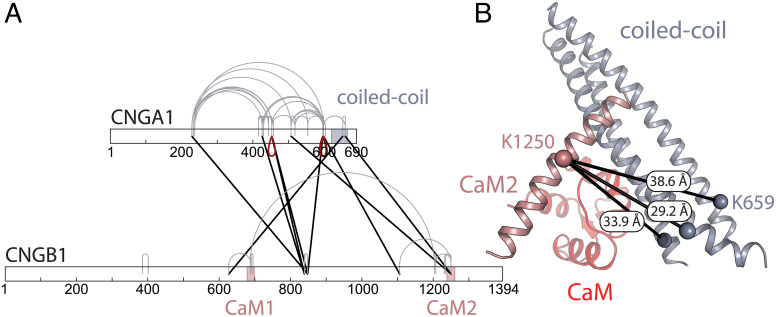
To determine which part of the CNGB1 sequence forms the D-helix, we performed cross-linking-mass spectrometry (XL-MS) on the purified CNG channel in the presence of saturating concentrations of CaM (10 µM) and Ca2+ (9 mM). We used the amine-reactive cross-linker disuccinimidyl suberate (DSS) and digested the sample with the endoproteinase Lys-C and trypsin (55). We identified three different cross-links between CNGA1 subunits (Fig. 3A, red drops); this is expected for a channel that encompasses three CNGA1 subunits (Fig. 3B). The cross-linking sites are found on residues K453 of the C-linker domains of two CNGA1 subunits and residues K596 or K598 of the CNBDs of two CNGA1 subunits (SI Appendix, Fig. S7). Considering the distance between the cross-linked lysine residues, the links between K596xK596 and K598xK598 can be explained by the channel’s protein-forming dimers (45). Importantly, we find a cross-link between residue K659 of the coiled-coil helices of CNGA1 and K1250 on the CaM2 domain of CNGB1. The measured distances between these two residues in our model of the coiled-coil region with CaM2 are 38.6 Å, 33.9 Å, and 29.2 Å for CNGA1 chains A, B, and C, respectively. The cross-link between chain C and CaM2 matches the expected Cα cross-link distance of DSS (26 to 30 Å) (56), suggesting that this is the link we observe in our data. These results imply that the D-helix of CNGB1 corresponds to the CaM2 sequence. Additionally, we find two cross-links between CaM2 and the CNBD of both CNGA1 and CNGB1: CNGB1 K1250 × CNGA1 K507 and CNGB1 K1250 × CNGB1 K1106: These cross-links reflect the increased flexibility of the coiled-coil region that is in contact with CaM2 (Movies S1 and S2). Importantly, we also detect a cross-link between K687 in CaM1 and K1203 in the C-helix of the CNBD of CNGB1; although the two sites are far apart in the linear polypeptide, this result suggests that, in the 3D-fold, CaM1 is close to CaM2. Although the distance between K1203 and the D-helix in our model would also be below 30 Å, we do not observe any cross-links between CaM1 and the coiled-coil domain, further supporting the notion that the D-helix corresponds to CaM2. Furthermore, K627 of CNGB1 (not visible in the map because it is located on the flexible N terminus) cross-links with K652 of the CNGA1 coiled-coil region. This indicates that the N terminus of CNGB1 is in close proximity to the CNBD and coiled-coil region, suggesting that CaM1 might be bound to the other lobe of CaM that is not visible in our density map.
Fig. 3.
Cross-linking mass spectrometry on the purified rod CNG channel. (A) Cross-linking map of CNGA1 and CNGB1. Cross-links that can be unequivocally attributed to links between adjacent CNGA1 subunits in the same channel protein (or different channel proteins in a dimer) are highlighted as red drops; cross-links either within a single subunit (CNGA1 or CNGB1) or, alternatively, between the same subunits (CNGA1xCNGA1 or CNGB1xCNGB1) are indicated in gray; cross-links between different subunits (CNGA1xCNGB1) are indicated in black. The coiled-coil region and the two CaM-binding sites are highlighted by blue and pink areas, respectively. (B) Model of the coiled-coil region including CaM2 and the CaM C-lobe. The identified cross-link between CaM2 (pink) and the coiled-coil region (blue) (CNGB1 K1250 x CNGA1 K659) is indicated in black, with the measured distances for CNGA1 chain A, B, and C. CaM is depicted in light red.
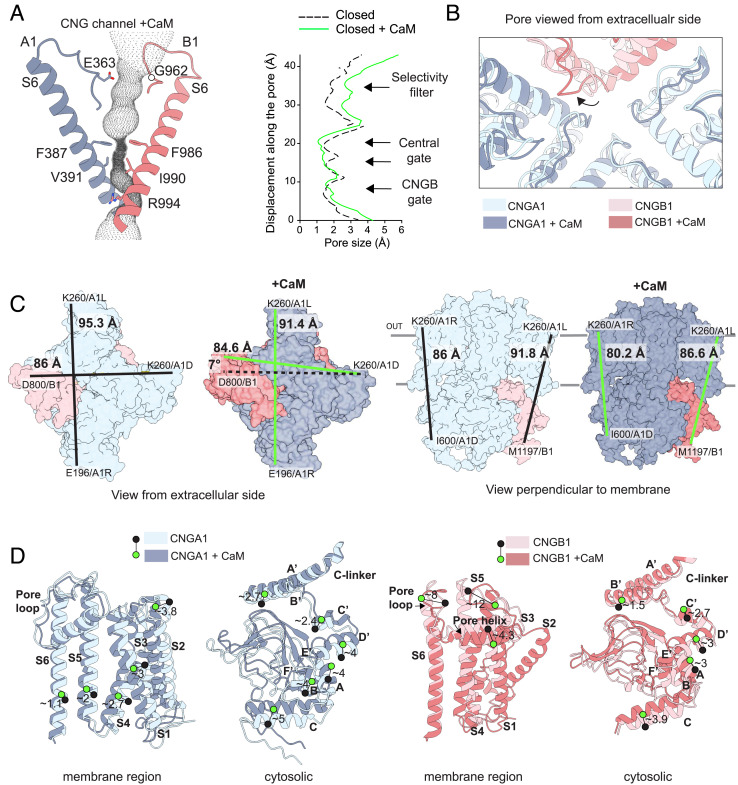
We further compared the structures of the channel without and with CaM, to ask whether the binding of CaM to the coiled-coil region causes structural changes in the cytosolic region (composed of the CNBD and C-linker) and the transmembrane region (Fig. 4). The channel bound to CaM is, as expected, in the closed state. Residues F387 and V391 of CNGA1, and residues F986 and I990 of CNGB1, represent the central hydrophobic gate; their side chains are oriented toward the center of the pore (Fig. 4A and SI Appendix, Fig. S8). Residue R994 of CNGB1, which forms a second gate below the central gate, is also oriented toward the pore. In the presence of CaM, the constriction of the pore at the central gate and at the CNGB1 gate is not altered, with the constriction at the central gate being only slightly more pronounced as compared to the structure without CaM (Fig. 4B). However, in the presence of CaM, the ion-conduction pathway is moderately wider at the level of the selectivity filter. Here, the filter loop of CNGB1, located between the S5-S6 pore helix and the S6 segment, is oriented away from the pore. By contrast, the pore loops of CNGA1 do not change orientation in the two structures (Fig. 4B). The physiological significance of a wider selectivity filter remains to be understood. Between the pore loops and the central gate, we observed a density that may correspond to trapped water/divalent ion(s) (SI Appendix, Fig. S8); this density is also present without CaM (45). To distinguish the three CNGA1 subunits, we refer to them as A1L, A1D, and A1R based on their left, diagonal, and right position, respectively, relative to the CNGB1 subunit (46).
Fig. 4.
Comparison of the rod CNG channel structures without and with CaM. (A) Details of the pore tunnel formed by CNGA1 (blue) and CNGB1 (pink) with the calculated cavity volume in dotted gray. Right, pore radius along the central pore axis in the closed state calculated with HOLE (57) without CaM (dashed, black) (PDB: 7O4H) and with CaM (continuous, green). (B) View from the extracellular side down into the pore of the two superimposed structures showing the pore loops and the ion-conduction pathway. The shift in position of the CNGB1 pore loop is indicated with a black arrow. (C) Left, view from the extracellular side without and with CaM represented as colored surface. Distances between residues are indicated with lines in black (no CaM), or green (with CaM). Right, side view from the axis perpendicular to the membrane. (D) Left, details of the displacement between pairs of residues in transmembrane segments or the cytosolic region for CNGA1 (chain C) without (light blue) and with CaM (dark blue); Right, same analysis for the CNGB1 subunit. Green and black circles indicate the position of C-alpha atoms for the same amino acid in the two structures. Distances are indicated in Angstroms (Å).
Overall, the channel conformations without and with CaM are different (Fig. 4C). Specifically, the channel structure undergoes a compression of ~4 Å estimated by two measures: the distance between the two opposing CNGA1 subunits on the extracellular side (K260 on A1L and E196 on A1R) and also ~4 Å between two K260 residues on A1R and A1L subunits. In the CNGA1 subunits, the compactness of the transmembrane region is more pronounced for S1, S2, and S3, with an average distance between the transmembrane helices of ~3.5 Å, and less for segments S4, S5, and S6 with an average distance of ~2 Å (Fig. 4D). Between CNGB1 and A1D, the distance remains similar, but the transmembrane region of CNGB1 undergoes a small clockwise movement of ~7° that does not occur in CNGA1 (Fig. 4C). A more rigorous comparison of the transmembrane region of CNGB1 with and without CaM is not possible because, without CaM, the structure of this region is not well resolved (45) (Fig. 1C). However, the regions of CNGB1 that are resolved in both maps display large changes (Fig. 4D). For example, the part of S5 of CNGB1 facing the extracellular side of the membrane is displaced by ~12 Å as compared to the structure without CaM. Likewise, the CNGB1 pore helix moves toward the intracellular side by ~4 Å (Fig. 4D). It is possible that the more compact structure of the transmembrane region may stabilize the membrane segments in CNGB1 and for this reason, they may be less flexible than without CaM.
In the presence of CaM, the cytosolic region of both CNGA1 and CNGB1 is compressed by ~5 to 6 Å toward the membrane (Fig. 4 C and D). The compactness is particularly enhanced for the C-helices of the CNBD of CNGA1: The C-helices move toward the membrane by ~5 Å, and the distance between carbon chains is reduced, whereas the helices of the C-linker are displaced by only ~2.5 Å, with the smaller changes observed for helices A′ and B′ (Fig. 4D). Considering that CaM, owing to its high affinity, is constitutively bound to the channel, we hypothesize that this compact structure is the principal state.
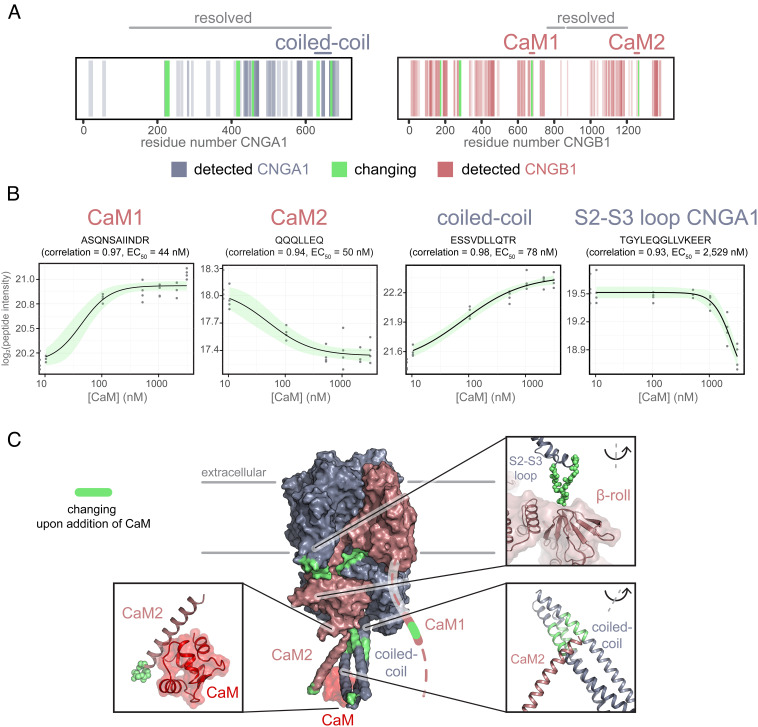
The structures produced by cryo-EM have been obtained with a channel protein embedded in a detergent/lipid mixture. To test whether the structural changes induced by CaM can be also detected in native membranes, we performed limited proteolysis-coupled-mass spectrometry (LiP-MS) (58, 59) on suspensions of rod outer segment (ROS) membranes. This technique has previously been used to study protein–protein interactions through measurements of changes in protease accessibility of complex systems (60). Membranes were washed with EDTA to remove endogenous CaM and were resuspended in a buffer containing 1 mM Ca2+. Samples were incubated with CaM at concentrations ranging between 0 and 3 µM. After incubation, the samples were digested with proteinase K; after 5 min, the reaction was quenched. Four parameter dose–response curves were fitted for each identified peptide; the correlation between the dose–response model we fit onto our data and the measured data was calculated (61). To keep the false-discovery rate low, only peptides with a Pearson’s correlation coefficient > 0.85 were considered significantly changing in their protease accessibility (SI Appendix, Fig. S9). A similar cutoff has recently been shown to be applicable for LiP-MS experiments on membrane suspensions (60). In our experiment, the sequence coverage of CNGA1 and CNGB1 was 44.6% and 55.4%, respectively. While the coverage includes most of the cytosolic part of both subunits, we did not identify peptides covering the transmembrane domains (Fig. 5A), which is to be expected due to the high hydrophobicity of these sequences (60, 62). We identified 12 peptides with significant correlations of the measured signal to the dose–response curves fit to the data (seven peptides on CNGA1 and five on CNGB1). We grouped these peptides according to their EC50 values. Peptides are clustered in three main groups (SI Appendix, Table S2): The first group encompasses peptides with EC50 values around 50 nM, and their amino acid sequences correspond to the CaM1 and CaM2 sites (Fig. 5B, panel 1 and 2). This indicates that the analysis faithfully identifies the CaM-binding sites and indicates that these regions are best protected from proteolysis due to the high affinity for CaM. Interestingly, these peptides show different directionalities, which could be related to their position within the binding motifs or differences in their binding modes. Next, we find peptides with EC50 values between 50 and 550 nM, corresponding to the CNGA1 coiled-coil region that, in the EM structure, is in contact with CaM and CaM2 (e.g., peptide in Fig. 5B, panel 3). Thus, the formation of a ternary complex between CaM, CNGA1, and CNGB1 is identified under native conditions and supports the conclusions derived from the cryo-EM structure. Finally, we find peptides with EC50 values >2,000 nM in CNGA1 sequences involved in intersubunit interactions (Fig. 5B, panel 4). These sites are likely not directly involved in the interaction with CaM, and we hypothesize that they could be a result of secondary effects caused by the complex background the experiment is performed in. The changing sites include the S2-S3 loop (residues 219 to 232, Fig. 5 C, Top Left zoom-in) and the C′-D′ helices of CNGA1 (residues 454 to 460) that interact with the β-roll of the CNBD of CNGB1. We identify additional changes on the A′ helix of the C-linker of CNGA1 (residues 412 to 422, Fig. 5 C, Top Left zoom-in) that is at contact distance to the B′-helix of the C-linker of CNGB1. Lastly, we find changes in the CNGB1 N terminus (residues 169-177 and 274-287), indicating an additional CaM-induced change in accessibility.
Fig. 5.
LiP-MS identifies conformational changes of CNGA1/CNGB1 in the native membrane upon addition of CaM. (A) Barcode plots indicating identified peptides on the protein sequences of CNGA1 and CNGB1 (blue and pink). Significantly changing peptides (r > 0.85) are highlighted in green. Regions resolved in the cryo-EM structure are labeled (“resolved”) in gray on top of the barcode plot. The coiled-coil domain, CaM1, and CaM2 are indicated similarly on top of the barcode plots in their respective protein colors (blue and pink). (B) Example peptides with significant correlations on CaM1, CaM2, the coiled-coil domain, and at the interface of CNGA1 and CNGB1. Pearson’s r (correlation) and EC50 values are listed in the titles. Gray dots indicate the measured log2-transformed peptide intensity for each experimental replicate (experiment was conducted in quadruplicates). As controls, peptide intensities without CaM are shown as half-dots on the y axis. The black line indicates the four-parameter dose–response curve modeled onto the data points (log-logarithmic model). The green-shaded area depicts the 95% CI of the fitted model (upper and lower limits). Dose–response curves were fit using the R package drc implemented in the R package protti. (C) Protease accessibility changes (green) mapped onto the protein structure. The nonresolved N terminus of CNGB1 with CaM1 is indicated as a dashed line. Some of the significantly changing regions are shown in zoom-ins, with changing regions highlighted in green or shown as green spheres to aid visibility. CaM is shown in red.
Discussion
CaM modulates the activity of many ion channels (25, 26). Mutations in humans that impair CaM or interactions with CaM can lead to fatal outcomes (63, 64). Structural information on how CaM interacts with ion channels may be key to develop therapies. However, it remains challenging to obtain atomic structures of CaM bound to cellular partners, and especially to membrane proteins. This is often due to the intrinsic flexibility of CaM (21) and flexible or disordered regions in target proteins (25).
In retinal rods, Ca2+ feedback on the phototransduction pathway is pivotal for adjusting light sensitivity (11, 52), but Ca2+ levels may be also crucial in detecting single-photon events. Ca2+/CaM lowers the EC50 of the rod CNG channels for cGMP <twofold (17, 32, 33). This modulation is very light as compared to other Ca2+ feedback mechanisms that target Rh inactivation, regulation of cGMP synthesis, and hydrolysis by PDE (65). The EC50 for CNG channel regulation by Ca2+ is about 44 nM, whereas resting [Ca2+]i is 250 to 500 nM. The local [Ca2+] at the inner mouth of the highly Ca2+-permeable CNG channel may be even higher. The exquisitely high affinity for CaM binding (K1/2 < 5 nM) and the low EC50 for the action of Ca2+ on channel activity suggest that CaM represents a genuine, constitutive channel subunit and, in addition to adjusting the cGMP sensitivity, may serve a structural role. CaM interacts with CaM1 and CaM2 sites at nanomolar concentrations (19). The role of the two sites remains elusive. Removal of CaM2 did not affect CaM modulation, indicating that binding of CaM to CaM1 in the unstructured N terminus is important for modulating the channel’s activity (19). To date, structural information about how CaM interacts with the rod CNG channel is limited to NMR structures of CaM bound to CaM1 and CaM2 peptides (66) (SI Appendix, Fig. S10). In olfactory neurons, Ca2+-free apo-CaM is also permanently associated with the CNG channel and is poised to lower the cAMP sensitivity at higher Ca2+ concentrations (67).
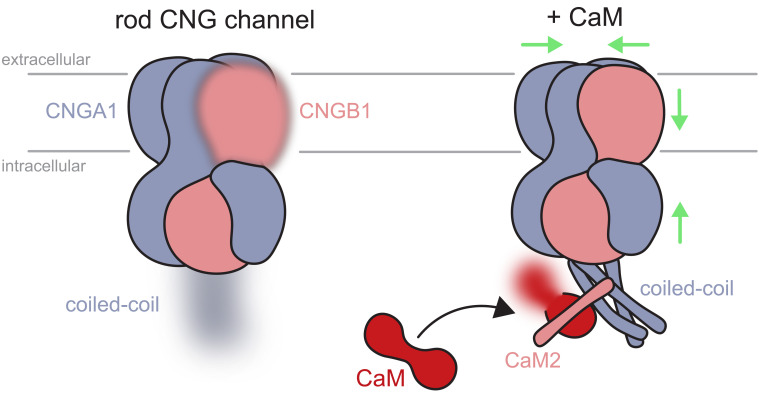
Here, we provide insights into the structural changes caused by binding of CaM to the native rod CNG channel. We find that CaM distinctively alters the structure of the rod CNG channel in several aspects. CaM creates a ternary complex formed by CNGA1-CaM-CNGB1; the CNGA1 coiled-coil region adopts an angle of ~55° as compared to the orientation of the coiled-coil region in the structure without CaM. Moreover, CaM triggers a contraction of the channel notably in the cytosolic and the membrane regions. This suggests that binding of CaM near the coiled-coil region is propagated through the entire polypeptide to adopt a different conformation (Fig. 6). Thus, a local binding event affects the channel conformation globally. It is plausible that the compact ternary complex is energetically more stable, which may raise the energy barrier between closed and open states. In olfactory channels, CaM binding is also conferred by two IQ-type sites on CNGB1b, a shorter splice form of rod CNGB1a, which also harbors CaM1 and CaM2 (68), and the CNGA4 subunit (67). The shift of EC50 for cAMP by CaM is much larger in the olfactory CNG channel (67) than for cGMP in the rod channel. Further insight into the specific functions of CaM1 and CaM2 regulation of the ligand sensitivity of CNG channels is expected to come from structures of the holo state of the complex between CaM and olfactory or cone CNG channel subunits.
Fig. 6.
Schematic representation of the structural changes taking place upon binding of CaM on the bovine rod CNG channel. The rod CNG channel is composed of three CNGA1 (blue) and one CNGB1 subunits (pink). The C terminus of the three CNGA1 subunits forms a coiled-coil structure that is flexible and thus not well resolved in the absence of CaM; for the same reason, the transmembrane region of CNGB1 is only partially resolved (Left, fuzzy parts). CaM creates a ternary complex between the CNGA1 coiled-coil helices (blue) and the CNGB1 CaM2-binding site located on the D-helix (helix in pink), which becomes visible in the coiled-coil region (Right). This binding event produces a contraction of the channel, as indicated by the green arrows. The transmembrane region of CNGB1 and the coiled-coil region become more visible in the EM density map in the presence of CaM. One lobe of CaM (red) is not well resolved (fuzzy part).
Intriguingly, the structural changes induced by cGMP binding result in a similar contraction of the cytosolic region of the homomeric TAX-4 and human CNGA1 channels (49, 50). However, the cGMP-induced changes translate in a dilation of the channel pore, which is not observed upon CaM binding: The channel pore in the ternary complex is only slightly narrower in some parts than the channel without CaM. Another change in the pore region involves the ion selectivity filter: With CaM, the pore loop of CNGB1 is differently oriented and this feature possibly contributes to make the pore moderately wider in this region. As we observed previously in the structure without CaM (45), the resolution of the filter loops is lower than the average map resolution, and side chains on the loops are at times difficult to model (SI Appendix, Fig. S8). The conserved glutamate (E363) on CNGA1 (69–71) does not coordinate any ion, but density that may correspond to trapped ion(s) in the pore is found in both structures above the closed hydrophobic central gate (SI Appendix, Fig. S8). It remains to be understood how the filter loop of CNGB1 shifts away from the pore and whether its movement is truly related to the structural changes observed within the CNGB1 pore helix.
The flexibility of the coiled-coil region limits the resolution of the cryo-EM map in this part of the channel. Similar to our map, the density map of the human CNG channel between the C- and D-helices of CNGB1 (46) is also difficult to interpret (72). The D-helix of the human rod CNG channel adopts the same orientation as in our map (SI Appendix, Fig. S11), thus supporting the notion that the human structures contain CaM, as previously proposed (72). Because flexibility limits our interpretation of the EM maps, we employed XL-MS to identify the sequence of CNGB1 to which CaM binds. The results suggest that this sequence corresponds to the CaM2 site. Moreover, we can infer that changes in the coiled-coil region are induced by the CaM2 domain of CNGB1. The coiled-coil domain ties together the CNGA1 subunits at their C-terminal end and controls the 3A:1B stoichiometry. We propose that the Ca2+/CaM-mediated interaction between CNGB1 CaM2 and the coiled-coil region formed by three CNGA1 subunits represents an allosteric mechanism to synchronously control three A subunits by a single B subunit. It remains to be fully understood whether a single CaM molecule binds to both CaM1 and CaM2 simultaneously, because one lobe of CaM remains flexible and is not visible in the cryo-EM structure. The CaM1 site may serve an additional structural function. CNGB1 carries a unique N-terminal domain that is as large as the CNGA1 subunit. This domain, called glutamic acid-rich part (GARP), is structurally disordered due to long stretches of Glu residues (41, 73). Of note, CNGA1 is lacking a GARP domain and CaM-binding sites. The GARP domain makes contact across a small cytosolic space with proteins in the rim region of photoreceptor disks (74, 75). The CaM1 site is adjacent to the GARP domain. Ca2+ ions entering via CNG channels will pass along and interact electrostatically with this negatively charged, “polymeric” string. CaM1/CaM may serve as a Ca2+ sensor that probes the electrostatics of this link and relays it to the CNG channel protein.
XL-MS provided evidence for the presence of a dimeric CNG channel species. Whether this conformation has functional relevance or is an artifact of the purification with detergent remains to be addressed. Moreover, we applied LiP-MS to the channel embedded in its native membrane. LiP-MS shows that CaM causes accessibility changes in the regions involved in the interaction with CaM1, CaM2, and CNGA1 coiled-coil, but also regions of CNGA1 that are at contact distance to CNGB1, including the C-linker and CNBD. One key advantage of our setup is that it requires minute amounts of material, making it suitable to study membrane protein interactions in tissues of medical interest. Our work shows the feasibility of this approach to characterize proteins in rod photoreceptor membranes that are much less abundant than rhodopsin and likely not amenable to other complementary methods such as native mass spectrometry (76). We propose that the combined information obtained by XL-MS and LiP-MS will as well become particularly important for studying functional domains on flexible protein regions that remain difficult to visualize by cryo-EM.
A few other components of the phototransduction cascade carry CaM-binding sites, including the Na+/Ca2+-K+ exchanger (77), which interacts with the channel (78). Whether CaM orchestrates Ca2+-dependent interactions between different components of the signaling pathway remains to be addressed. Our approach opens avenues for addressing how intrinsic flexibility and CaM binding is translated into the modulation of basic properties of CNG channels in visual and olfactory neurons.
Materials and Methods
Materials and methods are expanded in SI Appendix, which include a detailed description of the experimental approach used for carrying out sample preparation, cryo-EM, image processing, atomic model building, and all mass-spectrometry experiments. Specifically, the following paragraphs are contained in SI Appendix: Sample preparation and cryo-EM, single-particle processing and model building, structure analysis, LiP-MS sample preparation, limited proteolysis, tryptic digest, C18 clean-up, LC-MS/MS, data analysis, XL-MS chemical cross-linking, tryptic digest, C18 clean-up, LC-MS/MS, data analysis, and data visualization of mass spectrometry results.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We are indebted to Miroslav Peterek at the Scientific Center for Optical and Electron Microscopy (ScopeM) of ETH Zurich, who provided support during acquisition of the datasets on Titan Krios. We are also grateful to Jonas Mühle and Oliver Miguel Tejero at Paul Scherrer Institute (PSI) for their assistance with the production of rod outer segment (ROS) membranes. We are thankful to our colleagues at PSI for maintenance of the computing cluster “Merlin 6.” J.M. received funding from Holcim Stiftung (Holderbank, Switzerland), Promedica Stiftung (#1461/M), Swiss NSF (SNSF) (#19082), and Novartis Stiftung for Biomedical Research (#20C198). V.M.K. received funding from SNSF (grant #184951). G.F.X.S. acknowledges SNSF grants #173335 and #192760.
Author contributions
D.C.A.B., D.S., A.L., P.P., G.F.X.S., U.B.K., V.M.K., and J.M. designed research; D.C.A.B., D.S., and J.M. performed research; V.M.K., A.L. and P.P. contributed new reagents/analytic tools; D.C.A.B., D.S., M.J.R., U.B.K., and J.M. analyzed data; and D.S., U.B.K., and J.M. wrote the paper.
Competing interests
G.F.X.S. is a co-founder and scientific advisor of the companies leadXpro AG and InterAx Biotech AG.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
Atomic coordinates, and density maps, have been deposited at the wwPDB under accession numbers PDB: 8BX7, EMD-16311, and EMD-16313. Limited proteolysis-coupled-mass spectrometry data are deposited on PRIDE under the accession number PXD038640, and the cross-linking mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD038640. The R code used for data analysis is available on GitHub (https://github.com/dschust-r/CNG_LiP_MS) and PRIDE.
Supporting Information
References
- 1.Matulef K., Zagotta W. N., Cyclic nucleotide-gated ion channels. Annu. Rev. Cell Dev. Biol. 19, 23–44 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Kaupp U. B., Seifert R., Cyclic nucleotide-gated ion channels. Physiol. Rev. 82, 769–824 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Cote R. H., Photoreceptor phosphodiesterase (PDE6): Activation and inactivation mechanisms during visual transduction in rods and cones. Pflugers Arch. 473, 1377–1391 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baehr W., Devlin M. J., Applebury M. L., Isolation and characterization of cGMP phosphodiesterase from bovine rod outer segments. J. Biol. Chem. 254, 11669–11677 (1979). [PubMed] [Google Scholar]
- 5.Nakatani K., Yau K. W., Calcium and magnesium fluxes across the plasma membrane of the toad rod outer segment. J. Physiol. 395, 695–729 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yau K.-W., Nakatani K., Light-suppressible, cyclic GMP-sensitive conductance in the plasma membrane of a truncated rod outer segment. Nature 317, 252–255 (1985). [DOI] [PubMed] [Google Scholar]
- 7.Palczewski K., et al. , Molecular cloning and characterization of retinal photoreceptor guanylyl cyclase-activating protein. Neuron 13, 395–404 (1994). [DOI] [PubMed] [Google Scholar]
- 8.Palczewski K., Polans A. S., Baehr W., Ames J. B., Ca(2+)-binding proteins in the retina: Structure, function, and the etiology of human visual diseases. Bioessays 22, 337–350 (2000). [DOI] [PubMed] [Google Scholar]
- 9.Peshenko I. V., Dizhoor A. M., Guanylyl cyclase-activating Proteins (GCAPs) are Ca2+/Mg 2+ Sensors: Implications for photoreceptor guanylyl cyclase (RetGC) regulation in mammalian photoreceptors. J. Biol. Chem. 279, 16903–16906 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Burns M. E., Mendez A., Chen J., Baylor D. A., Dynamics of cyclic GMP synthesis in retinal rods. Neuron 36, 81–91 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Reingruber J., Holcman D., Fain G. L., How rods respond to single photons: Key adaptations of a G-protein cascade that enable vision at the physical limit of perception. BioEssays 37, 1243–1252 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao Y., et al. , Structure of the visual signaling complex between transducin and phosphodiesterase 6. Mol. Cell 80, 237–245 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turunen T., Koskelainen A., Functional modulation of phosphodiesterase-6 by calcium in mouse rod photoreceptors. Sci. Rep. 11, 8938 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zang J., Neuhauss S. C. F., The binding properties and physiological functions of recoverin. Front. Mol. Neurosci. 11, 473 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hurley J. B., Recoverin, a calcium-binding protein in photoreceptors. Behav. Brain Sci. 18, 497–498 (1995). [Google Scholar]
- 16.Chen J., et al. , Channel modulation and the mechanism of light adaptation in mouse rods. J. Neurosci. 30, 16232–16240 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsu Y.-T., Molday R. S., Modulation of the cGMP-gated channel of rod photoreceptor cells by calmodulin. Nature 361, 76–79 (1993). [DOI] [PubMed] [Google Scholar]
- 18.Chen T., Yau K., Direct modulation by Ca2+ – calmodulin of cyclic nucleotide-activated channel of rat olfactory receptor neurons. Nature 368, 545–548 (1994). [DOI] [PubMed] [Google Scholar]
- 19.Weitz D., et al. , Calmodulin controls the rod photoreceptor CNG channel through an unconventional binding site in the N-terminus of the β-subunit. EMBO J. 17, 2273–2284 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Babu Y. S., Bugg C. E., Cook W. J., Structure of calmodulin refined at 2.2 Å resolution. J. Mol. Biol. 204, 191–204 (1988). [DOI] [PubMed] [Google Scholar]
- 21.Barbato G., Ikura M., Kay L. E., Pastor R. W., Bax A., Backbone dynamics of calmodulin studied by 15N relaxation using inverse detected two-dimensional NMR spectroscopy: The central helix is flexible. Biochemistry 31, 5269–5278 (1992). [DOI] [PubMed] [Google Scholar]
- 22.Potter J. D., Strang-Brown P., Walker P. L., Iida S., Ca2+ binding to calmodulin. Methods Enzymol. 102, 135–143 (1983). [DOI] [PubMed] [Google Scholar]
- 23.Vetter S. W., Leclerc E., Novel aspects of calmodulin target recognition and activation. Eur. J. Biochem. 270, 404–414 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Yap K. L., et al. , Calmodulin target database. J. Struct. Funct. Genomics 1, 8–14 (2000). [DOI] [PubMed] [Google Scholar]
- 25.Kovalevskaya N. V., et al. , Structural analysis of calmodulin binding to ion channels demonstrates the role of its plasticity in regulation. Pflugers Arch. Eur. J. Physiol. 465, 1507–1519 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Saimi Y., Kung C., Calmodulin as an ion channel subunit. Annu. Rev. Physiol. 64, 289–311 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Zhou L., Liu H., Zhao Q., Wu J., Yan Z., Architecture of the human NALCN channelosome. Cell Discov. 8, 33 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun J., MacKinnon R., Cryo-EM structure of a KCNQ1/CaM complex reveals insights into congenital long QT syndrome. Cell 169, 1042–1050.e9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee C. H., MacKinnon R., Activation mechanism of a human SK-calmodulin channel complex elucidated by cryo-EM structures. Science (80-) 360, 508–513 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kschonsak M., et al. , Structural architecture of the human NALCN channelosome. Nature 603, 180–186 (2022). [DOI] [PubMed] [Google Scholar]
- 31.Balasubramanian S., Lynch J. W., Barry P. H., Calcium-dependent modulation of the agonist affinity of the mammalian olfactory cyclic nucleotide-gated channel by calmodulin and a novel endogenous factor. J. Membr. Biol. 152, 13–23 (1996). [DOI] [PubMed] [Google Scholar]
- 32.Gordon S. E., Downing-Park J., Zimmerman A. L., Modulation of the cGMP-gated ion channel in frog rods by calmodulin and an endogenous inhibitory factor. J. Physiol. 486, 533–546 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bauer P. J., Cyclic GMP-gated channels of bovine rod photoreceptors: affinity, density and stoichiometry of Ca2+-calmodulin binding sites. J. Physiol. 494, 675–685 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koutalos Y., Yau K.-W., Regulation of sensitivity in vertebrate rod photoreceptors by calcium. Trends Neurosci. 19, 73–81 (1996). [DOI] [PubMed] [Google Scholar]
- 35.Govardovskii V. I., Calvert P. D., Arshavsky V. Y., Photoreceptor light adaptation. Untangling desensitization and sensitization. J. Gen. Physiol. 116, 791–794 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hackos D. H., Korenbrot J. I., Calcium modulation of ligand affinity in the cyclic GMP-gated ion channels of cone photoreceptors. J. Gen. Physiol. 110, 515–528 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rebrik T. I., Korenbrot J. I., In intact cone photoreceptors, a Ca2+-dependent, diffusible factor modulates the cGMP-gated ion channels differently than in rods. J. Gen. Physiol. 112, 537–548 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rebrik T. I., Korenbrot J. I., In intact mammalian photoreceptors, Ca2+-dependent modulation of cGMP-gated ion channels is detectable in cones but not in rods. J. Gen. Physiol. 123, 63–75 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rebrik T. I., Botchkina I., Arshavsky V. Y., Craft C. M., Korenbrot J. I., CNG-modulin: A novel Ca-dependent modulator of ligand sensitivity in cone photoreceptor CGMP-gated ion channels. J. Neurosci. 32, 3142–3153 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaupp U. B., et al. , Primary structure and functional expression from complementary DNA of the rod photoreceptor cyclic GMP-gated channel. Nature 342, 762–766 (1989). [DOI] [PubMed] [Google Scholar]
- 41.Körschen H. G., et al. , A 240 kDa protein represents the complete β subunit of the cyclic nucleotide-gated channel from rod photoreceptor. Neuron 15, 627–636 (1995). [DOI] [PubMed] [Google Scholar]
- 42.Weitz D., Ficek N., Kremmer E., Bauer P. J., Kaupp U. B., Subunit stoichiometry of the CNG channel of rod photoreceptors. Neuron 36, 881–889 (2002). [DOI] [PubMed] [Google Scholar]
- 43.Zhong Haining, Molday Laurie, Molday Robert S., Yau King-Wai., The heteromeric cyclic nucleotide-gated channel adops a 3A:1B stoichiometry. Nature 14, 193–198 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng J., Trudeau M. C., Zagotta W. N., Rod cyclic nucleotide-gated channels have a stoichiometry of three CNGA1 subunits and one CNGB1 subunit. Neuron 36, 891–896 (2002). [DOI] [PubMed] [Google Scholar]
- 45.Barret D. C. A., Schertler G. F. X., Benjamin Kaupp U., Marino J., The structure of the native CNGA1/CNGB1 CNG channel from bovine retinal rods. Nat. Struct. Mol. Biol. 29, 32–39 (2022). [DOI] [PubMed] [Google Scholar]
- 46.Xue J., Han Y., Zeng W., Jiang Y., Structural mechanisms of assembly, permeation, gating, and pharmacology of native human rod CNG channel. Neuron 110, 1–10 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheng X., Hu Z., Li H., Yang J., Structure of the human cone photoreceptor cyclic nucleotide-gated channel. Nat. Struct. Mol. Biol. 29, 40–46 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barret D. C. A., Kaupp U. B., Marino J., The structure of cyclic nucleotide-gated channels in rod and cone photoreceptors. Trends Neurosci. 45, 763–776 (2022). 10.1016/j.tins.2022.07.001 [DOI] [PubMed] [Google Scholar]
- 49.Zheng X., et al. , Mechanism of ligand activation of a eukaryotic cyclic nucleotide−gated channel. Nat. Struct. Mol. Biol. 27, 625–634 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xue J., Han Y., Zeng W., Wang Y., Jiang Y., Structural mechanisms of gating and selectivity of human rod CNGA1 channel. Neuron 109, 1302–1313 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grunwald M. E., Yu W., Yu H., Yau K., Identification of a domain on the b-subunit of the rod cGMP-gated cation channel that mediates inhibition by calcium-calmodulin. J. Biol. Chem. 273, 9148–9157 (1998). [DOI] [PubMed] [Google Scholar]
- 52.Nakatani K., Koutalos Y., Yau K. W., Ca2+ modulation of the cGMP-gated channel of bullfrog retinal rod photoreceptors. J. Physiol. 484, 69–76 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sagoo M. S., Lagnado L., The action of cytoplasmic calcium on the cGMP-activated channel in salamander rod photoreceptors. J. Physiol. 497, 309–319 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Punjani A., Fleet D. J., 3D variability analysis: Resolving continuous flexibility and discrete heterogeneity from single particle cryo-EM. J. Struct. Biol. 213, 107702 (2021). [DOI] [PubMed] [Google Scholar]
- 55.Leitner A., Walzthoeni T., Aebersold R., Lysine-specific chemical cross-linking of protein complexes and identification of cross-linking sites using LC-MS/MS and the xQuest/xProphet software pipeline. Nat. Protoc. 9, 120–137 (2014). [DOI] [PubMed] [Google Scholar]
- 56.Merkley E. D., et al. , Distance restraints from crosslinking mass spectrometry: mining a molecular dynamics simulation database to evaluate lysine-lysine distances. Protein Sci. 23, 747–759 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smart O. S., Neduvelil J. G., Wang X., Wallace B. A., Sansom M. S. P., HOLE: A program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 14, 354–360 (1996). [DOI] [PubMed] [Google Scholar]
- 58.Schopper S., et al. , Measuring protein structural changes on a proteome-wide scale using limited proteolysis-coupled mass spectrometry. Nat. Protoc. 12, 2391–2410 (2017). [DOI] [PubMed] [Google Scholar]
- 59.Feng Y., et al. , Global analysis of protein structural changes in complex proteomes. Nat. Biotechnol. 32, 1036–1044 (2014). [DOI] [PubMed] [Google Scholar]
- 60.Holfeld A., et al. , Systematic identification of structure-specific protein-protein interactions. bioRxiv 2023.02.01.522707 (2023), 10.1101/2023.02.01.522707 (Accessed 3 February 2023). [DOI] [PMC free article] [PubMed]
- 61.Piazza I., et al. , A machine learning-based chemoproteomic approach to identify drug targets and binding sites in complex proteomes. Nat. Commun. 11, 4200 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Alfonso-Garrido J., Garcia-Calvo E., Luque-Garcia J. L., Sample preparation strategies for improving the identification of membrane proteins by mass spectrometry. Anal. Bioanal. Chem. 407, 4893–4905 (2015). [DOI] [PubMed] [Google Scholar]
- 63.Crotti L., et al. , Calmodulin mutations associated with recurrent cardiac arrest in infants. Circulation 127, 1009–1017 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Crotti L., et al. , Abstract 14840: International Calmodulinopathy Registry (ICaMR). Circulation 134, A14840–A14840 (2016). [Google Scholar]
- 65.Yiannis K., Yau K.-W., Regulation of sensitivity in verebrate photoreceptors by calcium. Trends Neurosci. 19, 73–81 (1996). [DOI] [PubMed] [Google Scholar]
- 66.Bej A., Ames J. B., NMR structures of calmodulin bound to two separate regulatory sites in the retinal cyclic nucleotide-gated channel. Biochemistry 61, 1955–1965 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bradley J., Bönigk W., Yau K. W., Frings S., Calmodulin permanently associates with rat olfactory CNG channels under native conditions. Nat. Neurosci. 7, 705–710 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bönigk W., et al. , The native rat olfactory cyclic nucleotide-gated channel is composed of three distinct subunits. J. Neurosci. 19, 5332–5347 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Root M. J., MacKinnon R., Identification of an external divalent cation-binding site in the pore of a cGMP-activated channel. Neuron 11, 459–466 (1993). [DOI] [PubMed] [Google Scholar]
- 70.Eismann E., Muller F., Heinemann S. H., Kaupp U. B., A single negative charge within the pore region of a cGMP-gated channel controls rectification, Ca2+ blockage, and ionic selectivity. Proc. Natl. Acad. Sci. U.S.A. 91, 1109–1113 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Seifert R., Eismann E., Ludwig J., Baumann A., Kaupp U. B., Molecular determinants of a Ca2+-binding site in the pore of cyclic nucleotide-gated channels: S5/S6 segments control affinity of intrapore glutamates. EMBO J. 18, 119–130 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Barret D. C. A., Schertler G. F. X., Kaupp U. B., Marino J., Structural basis of the partially open central gate in the human CNGA1/CNGB1 channel explained by additional density for calmodulin in cryo-EM map. J. Struct. Biol. 214, 107828 (2022). [DOI] [PubMed] [Google Scholar]
- 73.Batra-Safferling R., et al. , Glutamic acid-rich proteins of rod photoreceptors are natively unfolded. J. Biol. Chem. 281, 1449–1460 (2006). [DOI] [PubMed] [Google Scholar]
- 74.Körschen H. G., et al. , Interaction of glutamic-acid-rich proteins with the cGMP signalling pathway in rod photoreceptors. Nature 400, 761–766 (1999). [DOI] [PubMed] [Google Scholar]
- 75.Poetsch A., Molday L. L., Molday R. S., The cGMP-gated channel and related glutamic acid-rich proteins interact with Peripherin-2 at the rim region of rod photoreceptor disc membranes. J. Biol. Chem. 276, 48009–48016 (2001). [DOI] [PubMed] [Google Scholar]
- 76.Chen S., et al. , Capturing a rhodopsin receptor signalling cascade across a native membrane. Nature 604, 384–390 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Thibodeau S., Yang W., Sharma S., Lytton J., Calmodulin binds and modulates K(+)-dependent Na(+)/Ca(2+)-exchanger isoform 4, NCKX4. J. Biol. Chem. 296, 100092 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kang K., et al. , Assembly of retinal rod or cone Na+/Ca2+-K+ exchanger oligomers with cGMP-gated channel subunits as probed with heterologously expressed cDNAs. Biochemistry 42, 4593–4600 (2003). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
Atomic coordinates, and density maps, have been deposited at the wwPDB under accession numbers PDB: 8BX7, EMD-16311, and EMD-16313. Limited proteolysis-coupled-mass spectrometry data are deposited on PRIDE under the accession number PXD038640, and the cross-linking mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD038640. The R code used for data analysis is available on GitHub (https://github.com/dschust-r/CNG_LiP_MS) and PRIDE.