Abstract
Gastrointestinal tumors have become a worldwide health problem with high morbidity and poor clinical outcomes. Chemotherapy and surgery, the main treatment methods, are still far from meeting the treatment needs of patients, and targeted therapy is in urgent need of development. Recently, emerging evidence suggests that kelch-like (KLHL) proteins play essential roles in maintaining proteostasis and are involved in the progression of various cancers, functioning as adaptors in the E3 ligase complex and promoting the specific degradation of substrates. Therefore, KLHL proteins should be taken into consideration for targeted therapy strategy discovery. This review summarizes the current knowledge of KLHL proteins in gastrointestinal tumors and discusses the potential of KLHL proteins as potential drug targets and prognostic biomarkers.
Keywords: gastrointestinal tumors, KLHL, therapeutic target, prognostic biomarker, substrate
Introduction
Gastrointestinal tumors refer to those occurring in the digestive system, mainly including gastric, liver, colorectal, and pancreatic cancers, according to WHO classification [1]. Because of diagnostic difficulties and high morbidity, treating gastrointestinal tumors remains a difficult problem worldwide. Current treatments mainly include surgical resection and chemotherapy, but their therapeutic benefit implies we are far from an efficient cure [2]. Targeted drugs should achieve better therapeutic effects, but few of them are applied for the treatment of gastrointestinal tumors, highlighting the urgent need for the development of novel drug targets and prognostic biomarkers.
Kelch-like (KLHL) proteins, encoded by Kelch-like (KLHL) gene family members, function as substrate adaptors of the Cul3-scaffold E3 ligase complex and are the key factors that recognize and interact with substrates. KLHL proteins (KLHLs) mediate the ubiquitination of substrates and determine degradation fate and are thereby responsible for maintaining substrate homeostasis. Since substrate homeostasis plays an essential role in a wide range of cellular life activities, the function of KLHLs as key regulators of protein homeostasis under physiological and pathological conditions should be emphasized.
In recent years, an increasing number of studies have revealed that KLHLs are recurrently dysfunctional in gastrointestinal tumors, which is associated with tumor progression, suggesting that aberrant KLHLs generate an imbalance in protein homeostasis. For example, studies revealed that decreased expression of Keap1 (KLHL19) promoted the growth of pancreatic cancer cells [3], and increased KLHL21 expression was closely related to unfavorable outcomes in patients with cholangiocarcinoma [4]. Based on the specificity and importance of KLHLs in regulating substrate ubiquitination and stability, targeted inhibition of KLHL function may provide new insights for gastrointestinal tumor therapy. Therefore, this review will provide an overview of the biological functions and clinical significance of KLHLs in gastrointestinal tumors, attempt to match different KLHLs to drug targets or prognostic biomarkers, and ultimately propose potential targeted strategies for gastrointestinal tumor therapy.
KLHL aberrations in gastrointestinal tumors and their clinical significance
Various aberrations of KLHLs are frequently found in many gastrointestinal tumors, including mutations, deletions, hypermethylation, fusions and aberrant expression (Table 1). These aberrations result in global proteome alterations and proteostasis imbalance, suggesting that KLHLs play crucial roles in gastrointestinal tumors. Several mutations in KLHL19 have been reported in gastrointestinal tumors, e.g., N222S in pancreatic cancer; F280L, Q82H, and G350S in gastric cancer; and Q359X in colorectal cancer [5, 6]. Many of these mutations cause constitutive activation of Nrf2, which is commonly associated with poor prognosis of patients. Surprisingly, mutations at the same site have been observed in various malignancies. For example, G379D has been found in liver and gallbladder cancers [7, 8]. Gene mutations commonly lead to a considerable loss of KLHL activities, followed by dysregulation of downstream proteins and disease development. Additionally, deletions and promoter hypermethylation have been discovered in gastrointestinal tumors. Full-length deletion of KLHL9 was observed in 5.46% of gastric cancers and might partly account for gastric tumorigenesis [9]. Hypermethylation, a vital epigenetic modification, was found in KLHL19 and KLHL35 in colorectal and liver cancers, respectively [10, 11]. Notably, low expression of these KLHLs induced by epigenetic alterations is associated with poor survival. Furthermore, fusion is another common cause of aberrations. The PHOSPHO2-KLHL23 fusion has been reported in both liver and gastric cancers [12, 13], and NR5A2-KLHL29 was detected in colorectal cancer [14]. Clinically, fusions display a tumor-specific pattern and are associated with prognosis.
Table 1.
Aberrations of KLHLs in gastrointestinal tumors.
| KLHLs | Cancer types | Aberrations | Sites | References |
|---|---|---|---|---|
| KLHL4 | READ | Low-expression | N/A | [18] |
| KLHL5 | CRC | High-expression | N/A | [83] |
| STAD | High-expression | N/A | [84] | |
| KLHL6 | GC | High-expression | N/A | [85] |
| KLHL9 | GC | Low-expression | Full-length deletion | [9] |
| KLHL19 | CHOL | Mutation | W554X, C249Y, 543insC Frameshift(codon 181) | [8] |
| CRC | DNA hypermethylation | T330I, V606M, R234W, Q359X | [6, 10, 86] | |
| GBC | Mutation, deletion, high-expression | M1_E441delext-103, D87Y, G379D, S338L, 996delC Frameshift(codon 332) | [8, 87] | |
| GC | Mutation | F280L, Q82H, S233N, C288Y, L281P, G350S | [6] | |
| HCC | Mutation, low-expression | R442_splice, E593X, E542DfsX31, Y396SfsX19, G379D, V271L+, R17Q, D236Y, L342M, N183S, R336Q | [6, 7, 16] | |
| PC | Mutation | N222S, G386R, T142M | [5, 88] | |
| SBC | Mutation | Y490C, R320L, R202H | [89] | |
| KLHL21 | CHOL | High-expression | N/A | [4] |
| HCC | High-expression | N/A | [15] | |
| KLHL22 | CRC | Low-expression | N/A | [19] |
| HB | Mutation | M1L | [90] | |
| KLHL23 | GC | Fusion, high-expression | PHOSPHO2-KLHL23 | [13] |
| HCC | Fusion, low-expression | PHOSPHO2-KLHL23 | [12] | |
| KLHL29 | CRC | Fusion, low-expression | NR5A2-KLHL29 | [14] |
| KLHL30 | HCC | Low-expression | N/A | [17] |
| KLHL32 | PC | Low-expression | N/A | [91] |
| KLHL34 | CRC | N/A | N/A | [92] |
| KLHL35 | HCC | DNA hypermethylation | N/A | [11] |
| KLHL37 | CRC | High-expression | N/A | [20, 93, 94] |
| KLHL39 | CRC | N/A | N/A | [22] |
| ESCA | Low-expression | N/A | [95] |
CHOL cholangiocarcinoma, CRC colorectal cancer, ESCA esophageal cancer, GBC gallbladder cancer, GC gastric cancer, HB hepatoblastoma, HCC hepatocellular carcinoma, N/A not available, PC pancreatic cancer, READ rectum adenocarcinoma, SBC small bowel carcinoma, STAD stomach adenocarcinoma, N/A not available.
Aberrant expression is the most typical type of aberration. For instance, it was reported that KLHL21 was highly expressed in hepatocellular carcinoma [15], whereas KLHL19, KLHL23, and KLHL30 had decreased expression [12, 16, 17]. KLHL4 and KLHL22 were downregulated in colorectal cancer [18, 19], while KLHL37 was upregulated [20]. In addition, we noticed that some KLHLs were important by data analysis, which drew little attention before [21]. For example, the expression levels of KLHL7 and KLHL15 were found to be elevated in esophageal cancer, while the levels of KLHL17 and KLHL36 were decreased in gastric cancer. Various KLHLs, such as KLHL5 and KLHL27, are typically highly expressed in gastrointestinal tumors. Based on their distinct expression profiles, the clinical significance of KLHLs needs to be determined. Reduced KLHL19 expression is related to poor overall survival in liver cancer [16]. A study revealed that higher KLHL21 expression was linked to unfavorable outcomes in cholangiocarcinoma [4]. Downregulation of KLHL39 in colorectal cancer was correlated with higher TNM stage and shorter survival [22]. Additionally, we assessed the expression of the abovementioned KLHLs that had previously garnered little interest in relevant samples. For example, increased expression of KLHL15 was associated with unsatisfactory survival in esophageal cancer, while there was a significant correlation between high KLHL36 expression and good outcomes in pancreatic cancer and cholangiocarcinoma.
KLHLs show distinct clinical significance and characteristic aberrant expression patterns, emphasizing the importance of KLHLs in gastrointestinal tumors. Some aberrations are tightly linked with gastrointestinal tumors, and such aberrations lead to sharply decreased tumor suppression or substantially increased tumor promotion. Decreased tumor suppression may suggest utility as a prognostic biomarker, and increased tumor promotion may suggest utility as a drug target. As adaptors, KLHLs participate in a range of biological processes due to their interactions with various substrates. Aberrations of KLHLs caused by pathological situations lead to substrate dysfunction, eventually resulting in malignant transformation and tumorigenesis. Here, we focus on several representative KLHLs and discuss their regulation of substrates.
The substrates and pathological dysfunction of KLHLs in gastrointestinal tumors
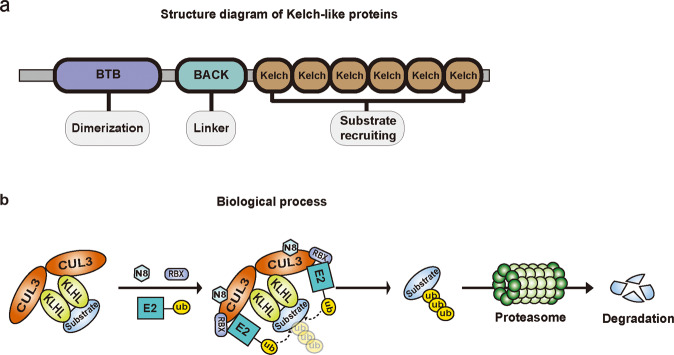
KLHLs are adaptors that interact with a variety of substrates in the E3 enzyme, consisting of three characteristic domains: the bric-a-brac, tramtrack, broad complex (BTB)/poxvirus and zinc finger (POZ) domain, the BACK domain, and the Kelch domain (Fig. 1). The Kelch domain specifically recognizes and interacts with substrates, and the BTB/POZ domain is responsible for binding the scaffold protein Cullin3. Subsequently, neddylated Cullin3 recruits the E2 enzyme and finally promotes the transfer of ubiquitin molecules from E2 onto substrates. Additionally, the BACK domain is a conserved and poorly described region, and some mutations in this domain are reported to be linked to gastrointestinal tumors, e.g., gastric cancer, liver cancer, and cholangiocarcinoma [6, 8].
Fig. 1. The KLHLs structure and the biological process regulated by KLHLs.
a Schematic representation of KLHLs structure. KLHLs consist of BTB/POZ domain, BACK domain and Kelch domain. The BTB/POZ domain is essential for dimerization and contributes to CUL3 interaction. The Kelch domain is comprised of six identical motif repeats and mediates specific substrates recruitment. The BACK domain is served as a linker connecting two domains. b The biological process involved in KLHLs regulation. The KLHLs and CUL3 are both dimerized and form CUL3-KLHL complex to identify substrates. The complex then recruits the E2 enzyme when bound to RBX1 and NEDD8, and jointly transfers ubiquitin onto substrates for subsequent proteasome-mediated degradation.
It is well known that KLHLs perform cellular functions mostly by ubiquitination, during which they specifically interact with substrates. The properties of these substrates bestow distinct roles on KLHLs. If the substrate acts as an oncoprotein, the KLHL will correspondingly play vital anticancer roles. In contrast, if the substrate contributes to tumor suppression, the KLHL will possess pro-cancer functions. Research has identified bona fide substrates of a few KLHLs, while potential substrates that have been documented in the protein interactome as interacting with KLHLs have also been defined, but for such substrates, there is no experimental evidence. Thus, a summary of the substrates and pathological dysfunction of KLHLs in gastrointestinal tumors is imperative to increase the understanding of KLHLs. Here, we provide a review of representative KLHLs (i.e., KLHL7, KLHL15, KLHL19, KLHL22, KLHL23, KLHL27, and KLHL36) and their substrates in gastrointestinal tumors (Fig. 2 and Table 2).
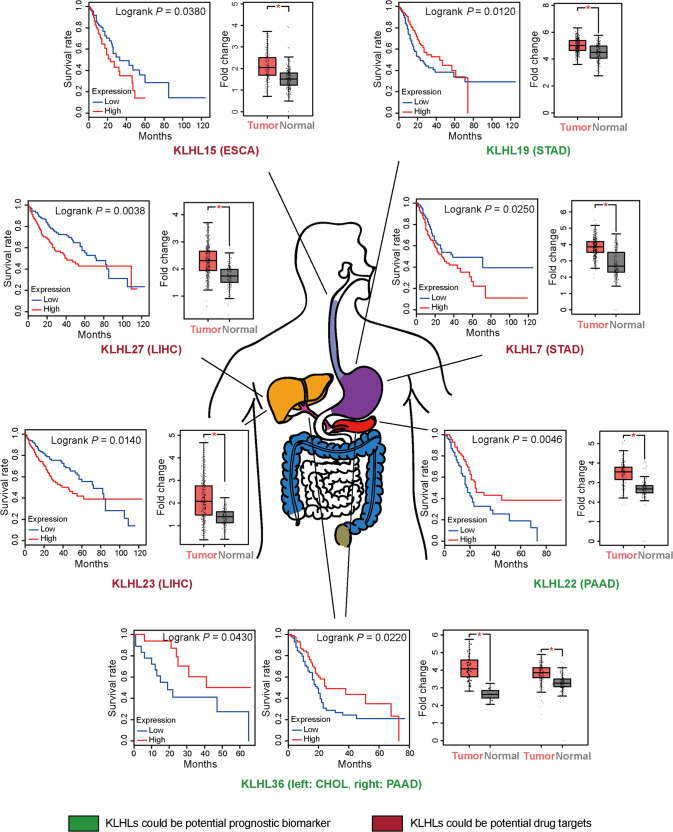
Fig. 2. KLHLs with some properties of drug targets or prognostic biomarkers.
All listed KLHLs are high-expressed in tumor tissues compared to normal tissues. In tumor tissues, KLHLs whose high expression is correlated to low overall survival may match some properties of drug targets. Meanwhile, KLHLs whose high expression is correlated to high overall survival accordingly possibly fulfill some requirements of prognostic biomarkers. KLHLs in red match some properties of drug targets. KLHLs in green match some properties of prognostic biomarkers. CHOL cholangiocarcinoma, ESCA esophageal carcinoma, LIHC liver hepatocellular carcinoma, PAAD pancreatic adenocarcinoma, STAD stomach adenocarcinoma.
Table 2.
Gastrointestinal tumors associated-KLHLs and their interactions.
| KLHLs | Interactions | Cancer types | References | |
|---|---|---|---|---|
| Substrate | Potential substrate | |||
| KLHL7 | TUT1 | N/A | [96] | |
| Fbxl17 | N/A | [97] | ||
| KLHL15 | CtIP | CC, GC, HCC | [36, 37, 39, 98] | |
| PPP2R5B | N/A | [99] | ||
| DCX | N/A | [40] | ||
| DCLK1* | CC, CHOL, ESCA, GC, HCC, PC | [40–42, 44, 45, 47] | ||
| DCLK2 | N/A | [40] | ||
| KLHL19 | Nrf2* | CHOL, CRC, GC, HCC, PC | [23–26, 100] | |
| p62 | CRC, HCC, PC | [27–29, 101] | ||
| IKBKB | CC, GC, HCC | [30–33] | ||
| KLHL22 | DEPDC5 | GISTs, HCC | [48–50] | |
| PLK1* | CC, ESCA, GC, HCC, PC | [51–53, 102–104] | ||
| PD-1* | CRC | [54] | ||
| ADRA1A | N/A | [105] | ||
| NANOG | CRC, ESCA, HCC, PC | [56, 57, 106–108] | ||
| KLHL23 | Actin | N/A | [12] | |
| ANXA1 | CRC, PC | [109–111] | ||
| ANXA7 | HCC | [109, 112] | ||
| BNIP3L | CRC, HCC, PC | [60, 61, 109, 113] | ||
| CDKN1A | CRC, GC, HCC | [63–65, 109] | ||
| TK1 | N/A | [109] | ||
| KLHL27 | HOOK2 | CC, ESCA, GC | [67, 114] | |
| LIG4 | CRC | [68, 115] | ||
| KLHL36 | TRIM55 | HCC | [70, 116] | |
| TRIM63 | N/A | [116] | ||
CC colon cancer, CHOL cholangiocarcinoma, CRC colorectal cancer, ESCA esophageal cancer, GC gastric cancer, GISTs gastrointestinal stromal tumors, HCC hepatocellular carcinoma, N/A not available, PC pancreatic cancer, N/A not available, * substrates have been used as targets in cancer therapy with inhibitors under clinical investigation.
KLHL19
KLHL19, also known as Keap1, is the most thoroughly studied among the KLHL family members. It interacts with its typical substrate, Nrf2, forming the KLHL19-Nrf2 pathway to react against oxidative stress. To date, many proteins have been identified as substrates, and we focused on some representative proteins, namely, Nrf2, p62, and IKBKB.
Nrf2, named nuclear factor erythroid 2-related factor, is a key substrate of KLHL19. Normally, KLHL19 transmits ubiquitin to Nrf2 for degradation. Upon exposure to reactive oxygen species (ROS), KLHL19 is inhibited by conformational changes, thereby stabilizing Nrf2 and eventually leading to downstream antioxidant gene activation. Nrf2 is presumably an oncoprotein that is constitutively and widely activated for protection in gastrointestinal tumors. In a mutant K-ras/p53 mouse model, KLHL19 deletion accelerated cholangiocarcinoma formation alongside Nrf2 activation [23]. Enhanced KLHL19-mediated ubiquitination of Nrf2 triggered ROS-induced cell death in gastric cancer [24]. Knockdown of Nrf2 led to sorafenib-induced ferroptotic events, while KLHL19 knockdown contributed to ferroptosis resistance in liver cancer cells [25]. In addition, some Nrf2 mutations were reported to lead to aberrant regulation of the KLHL19-Nrf2 pathway to protect cancer cells [26].
p62, also called SQSTM1, has emerged as a multifunctional signal hub. p62 interacts with KLHL19 via competitive binding, causing Nrf2 activation [27]. Autophagy is another essential process impacted by p62. In a xenograft mouse model, KLHL19 displayed a higher affinity for phosphorylated p62 for subsequent autophagy-mediated degradation, which significantly facilitated liver tumor growth [28]. KLHL19 accumulation along with p62 reduction generated an inflammatory and senescent phenotype, thereby promoting cancer development in an orthotopic pancreatic cancer model [29].
IKBKB is another recognized substrate. KLHL19 facilitated IKBKB ubiquitination to act as a tumor suppressor while IKBKB deficiency accordingly impeded inflammation-induced intestinal carcinogenesis in a mouse model [30, 31]. Consistently, the pro-cancer ability of IKBKB was demonstrated in a gastric cancer model [32]. Additionally, research revealed that increased KLHL19 and decreased IKBKB contributed to the improvement of hepatic steatosis, whereas IKBKB facilitated lipogenesis and hepatocellular carcinogenesis in vivo [33, 34].
KLHL15
KLHL15 was first identified in silico, and its mRNA is universally expressed in diverse tissues. KLHL15 is reported to be related to agenesis of the corpus callosum [35]. Nevertheless, more studies of KLHL15 to determine its biological mechanisms are underway. Based on the published literature, CtIP, PPP2R5B, DCX, DCLK1, and DCLK2 have all been identified as substrates of KLHL15.
CtIP, also known as RBBP8, functions in genome maintenance, especially in DNA double-strand break repair. Due to its unique function, CtIP is commonly dysregulated in gastrointestinal tumors. For example, CtIP was upregulated in HBV-related liver cancer cells [36]. Knockdown of CtIP slowed the gastric cancer cell growth rate, while inhibition of its ubiquitination by KLHL15 led to chemoresistance and cancer development [37, 38]. Additionally, depletion of CtIP decreased metastasis in an orthotopic colon cancer model [39]. PPP2R5B is a regulatory subunit of PP2A, but its role in gastrointestinal tumors remains unknown. DCX, DCLK1, and DCLK2 were found to be bona fide substrates in a study of neuronal dendritogenesis [40]. DCX and DCLK2 are both rarely studied in gastrointestinal tumors. In contrast, increased expression of DCLK1 has been universally detected in numerous gastrointestinal tumors, e.g., esophageal, colon, and gastric cancers, while its ubiquitination regulator, KLHL15, is expressed at low levels in gastrointestinal tumors [41–43]. A DCLK1-selective inhibitor exhibited significant antitumor effects against cholangiocarcinoma cells in vitro [44]. Recently, numerous studies have illustrated the relationship between DCLK1 and stemness. DCLK1 is recognized as a marker of cancer stem cells in many gastrointestinal tumors [45, 46]. For example, in one study, when injuries and inflammation occurred, DCLK1-positive cells acquired carcinogenic abilities, inducing oncogenic transformation in the pancreas [47].
KLHL22
KLHL22 plays significant roles in tumorigenesis, mitosis, and immune homeostasis. One study revealed that KLHL22 was downregulated in colorectal cancer and attenuated cell invasion, migration, and proliferation in vitro [19]. It has been reported that DEPDC5, PLK1, and PD-1 are substrates of KLHL22, while ADRA1A and NANOG are potential substrates.
DEPDC5, a crucial and conserved subunit of GATOR1, undergoes ubiquitin linkage mediated by KLHL22. It has been reported that DEPDC5 deficiency promotes the growth of gastrointestinal stromal tumor xenografts in nude mice, while its stabilization resulting from KLHL22 depletion hinders tumor progression [48, 49]. Moreover, knockdown of DEPDC5 potentiated ROS resistance in liver cancer, which was associated with poor outcomes [50]. PLK1, a regulator of mitosis, is regulated in localization instead of protein stability [51]. A selective inhibitor of PLK1 had anticancer effects in esophageal cancer both in vivo and in vitro [52]. Overexpression of PLK1 hindered liver cell death, while knockdown reversed this effect [53]. Furthermore, PD-1 is a novel substrate of KLHL22 and prevents excessive inhibition of T cells [54]. PD-1 is normally expressed on the surface of T cells, and inhibition of PD-1 to upregulate the T-cell response and improve the microenvironment is viewed as a promising strategy for cancer therapy. Furthermore, several inhibitors targeting PD-1, such as nivolumab and pembrolizumab, are approved for the treatment of some gastrointestinal tumors and display good efficacy [55].
ADRA1A is a G protein-coupled receptor that senses catecholamines. NANOG is considered a biomarker of cancer stem cells that regulates self-renewal and maintains stemness. In colorectal cancer tumor-bearing mice, NANOG knockdown led to a significant decrease in the tumor growth [56]. Similarly, degradation of NANOG weakened the pancreatic cancer phenotype in vitro [57].
KLHL23
KLHL23 is an actin-binding protein. A study demonstrated that KLHL23 hindered the epithelial-mesenchymal transition of liver cancer cells by impeding actin cytoskeleton remodeling [12]. According to the study, actin was found to be the only definite substrate, while the other 5 proteins, namely, ANXA1, ANXA7, BNIP3L, CDKN1A, and TK1, were identified as potential substrates.
ANXA1 and ANXA7 both belong to the annexin superfamily and can promote or suppress gastrointestinal tumors [58, 59]. BNIP3L, also known as NIX, profoundly impacts mitophagy. In a xenograft model, BNIP3L-dependent mitophagy upregulated glycolysis, thereby promoting the cancer stemness phenotype of liver cancer cells [60]. In addition, BNIP3L deficiency markedly retarded tumor development and augmented survival in a pancreatic cancer mouse model [61]. CDKN1A, famously known as p21, is described as an essential regulator of the cell cycle and DNA damage repair. CDKN1A plays dual roles in cancer depending on cell type [62]. Increased expression of CDKN1A impeded colorectal tumor growth [63]. CDKN1A inhibition was reported to promote cancer progression in a gastric cancer mouse model [64]. However, CDKN1A sustained regeneration in liver injury and promoted hepatocarcinogenesis in vivo and in vitro [65]. Research on TK1 in gastrointestinal tumors has rarely been conducted.
KLHL27
KLHL27 was first characterized as a human homolog of mouse intracisternal a particle-promoted placenta. Moreover, KLHL27 is highly expressed in breast cancer [66]. The literature shows that KLHL27 has no substrate except actin, but it has 2 potential substrates, namely, Hook2 and Lig4.
Hook2 is essential for mitosis and cytokinesis. Given its increased serum levels, Hook2 hypothetically encodes a tumor antigen and is regarded as a potential diagnostic biomarker for esophageal, gastric, and colon cancers [67]. Lig4 is a DNA ligase that is indispensable in the nonhomologous end-joining repair pathway. Consistently, Lig4 knockdown decreased radioresistance in colorectal cancer cells [68].
KLHL36
KLHL36 has rarely been described to date. It has been reported that KLHL36 expression can be elevated by TNFα [69]. Based on the literature, Trim55, and Trim63, also known as Murf2 and Murf1, respectively, are potential substrates.
Trim55 and Trim63 can function as E3 ligases. High expression of Trim55 hampered the migration and invasion of liver cancer cells in vitro [70]. Trim63 is involved in cancer cachexia, but gastrointestinal-related research is limited.
KLHL7
KLHL7 is mostly reported to be associated with genetic diseases. TUT1, also known as Star-PAP, has been identified as the only known substrate to date implicated in U6 snRNA maturation. Fbxl17 has been identified as a potential substrate that sustains cancer cell proliferation by ubiquitination. However, the impacts of these proteins on gastrointestinal tumors remain unexplored.
In summary, accumulating experimental evidence has established that KLHLs promote or suppress gastrointestinal tumors in a substrate-dependent manner. If a substrate is a protumor factor, the KLHL suppresses tumor development by promoting substrate degradation and therefore could match properties of a biomarker of good prognosis. On the other hand, if a substrate is a tumor suppressor, the KLHL will restrain the antitumor functions of the substrate and promote tumor growth, suggesting that the KLHL might conform to the characteristics of a drug target. Therefore, based on the existing reports and database results, KLHL7, KLHL15, KLHL23, and KLHL27 could be potential candidates of drug targets, whereas KLHL19, KLHL22, and KLHL36 meet certain criteria of prognostic biomarkers.
Strategies to target KLHLs for gastrointestinal tumor therapeutics
Targeted therapies are emerging for gastrointestinal tumors and have shown tangible benefits with fewer side effects and higher efficacy than chemotherapy and radiotherapy. Targeting KLHLs is regarded as a feasible and promising approach due to their crucial roles in gastrointestinal tumors. As adaptors of E3 enzymes, KLHLs are involved in multiple cellular activities at key points, and their dysfunction often leads to tumorigenesis. Furthermore, KLHL inhibition strategies must consider the specificity of individual KLHLs. Due to the high expression of certain KLHLs in gastrointestinal tumors and the specificity of interactions between KLHLs and substrates, targeting KLHLs will not impair intracellular biological processes in normal tissues. Instead, inhibitors can be concentrated in tumor cells and specifically repress them, tremendously minimizing unfavorable side effects. More importantly, KLHLs are targetable, and several strategies are proposed here.
Protein‒protein interaction (PPI) inhibitors, whether small molecules or peptides, have been designed to hinder interactions by binding to KLHLs. Different KLHLs possess unique Kelch domains that interact with specific substrates, offering a structural basis for designing PPI inhibitors with specificity that disrupts the KLHL-substrate interaction. To date, many studies have reported small molecule inhibitors. The K67 derivative 5d possessed potent inhibitory activity and attenuated chemoresistance in liver cancer, as demonstrated by cell experiments [71]. Another PPI inhibitor, esculetin, hindered proliferation and induced apoptosis of pancreatic cancer cells in vitro [72]. Furthermore, peptide inhibitors, such as cyclic peptide 3, are continually emerging [73]. Apart from traditional druggable sites, novel sites are constantly being uncovered to accelerate drug discovery. A study showed that dimethyl fumarate (DMF) bound to the top region of the KLHL19 Kelch domain instead of the bottom region, to which Nrf2 bound [74], facilitating the development of new potent selective drugs. Targeting the CUL3-KLHL interface is also a theoretical strategy. According to one study, disruption of the interaction between a hydrophobic groove of KLHL11 and the CUL3 N-terminal extension led to sharply decreased affinity [75], suggesting a promising target for KLHL11 inhibition. A designed stapled peptide was also reported to efficiently mimic the CUL3-BTB interaction [76], providing the possibilities for the subsequent development of inhibitors to disrupt the CUL3-KLHL interaction.
Allosteric inhibitors are commonly defined as inhibitors that bind to nonactive sites of enzymes, subsequently triggering conformational changes and ultimately altering enzyme activities. Taking KLHL19 as an example, Cys151, Cys273, and Cys288 are the major sensor residues among its 27 cysteines, which all sense ROS and electrophilic chemicals [77]. In the presence of oxidative stress, the conformation of KLHL19 changes as a consequence of modification, subsequently preventing Nrf2 degradation. Inhibitors of cysteines are mainly electrophiles that inactivate KLHL19, and one of them, DMF, has been approved for the treatment of relapsing multiple sclerosis. Furthermore, the potency of some inhibitors has been confirmed to impede gastrointestinal tumor growth. TCE 31 reduced the formation of preneoplastic foci in the livers of aflatoxin B1-treated mice [78]. Moreover, sulforaphane triggered apoptosis of colon cancer cells by G2/M phase arrest in vitro [79].
Promoting KLHL degradation appears to be a direct and effective method to hamper KLHL functions. To date, proteolysis-targeting chimeras (PROTACs) have been developed as a main tool to target proteins of interest (POIs) for degradation by hijacking the ubiquitination system. A PROTAC is composed of three parts. One ligand binds to the POI, another binds to the E3 ligase, and a linker connects the ligands in the middle. PROTACs should be designed to target unique regions of KLHLs that enable formation of distinct structures and binding pockets, ensuring specific binding with KLHLs. A study reported that a PROTAC targeting KLHL19 successfully triggered the degradation of Tau [80], suggesting that KLHLs could be targeted by PROTACs; this result indicated that PROTACs could be designed to inactivate KLHLs by degradation if one ligand was targeted for KLHLs and another ligand recruited an E3 ligase. A number of PROTACs have shown strong effects in gastrointestinal tumors [81]. In addition, research found that in Drosophila, KLHL19 and KLHL18 could be self-degraded by ubiquitination in a proteasome-independent manner [82]. Although the detailed mechanism of KLHL self-ubiquitination is unclear, it offers potential novel insights into targeting KLHLs for degradation, which could ultimately be translated into new drugs.
Discussion
In this review, literature searches and sample analyses were performed to identify KLHL aberrations in gastrointestinal tumors and their clinical value. Then, based on their significance, we focused on several representative KLHLs, i.e., KLHL7, KLHL15, KLHL19, KLHL22, KLHL23, KLHL27, and KLHL36, and provided a systematic summary of their substrates and pathological dysfunction in gastrointestinal tumors to provide novel insights. Furthermore, possible therapeutic strategies to inhibit aberrant KLHLs were proposed. Our review highlights a number of intracellular roles of KLHLs in gastrointestinal tumors and thereby provides a theoretical basis for KLHL inhibitor design.
Gastrointestinal tumors are highly heterogeneous. Even if the clinical and pathologic features are similar, the eventual outcomes are different. Thus, biomarkers of gastrointestinal tumors are of vital importance and urgently needed. Many studies have revealed that some KLHLs are related to prognosis in gastrointestinal tumors. Moreover, the majority of KLHLs are located in the cytoplasm and thereby can be easily detected. However, many KLHLs are analyzed based on immunohistochemistry, which cannot be easily performed in the clinic. The accuracy, sensitivity, robustness, etc., of such assays must all be determined and verified. This review provides potential biomarkers for the diagnosis of gastrointestinal tumors.
KLHLs are potential ideal therapeutic targets for treating gastrointestinal tumors for the following reasons: (1) The first is the effectiveness of intervention targets (KLHLs), and studies have revealed that biological methods, such as knockdown, of KLHLs has a therapeutic effect in gastrointestinal tumors. (2) The second is the targetability of KLHLs. At present, PPI inhibitors, allosteric inhibitors and PROTACs are all technically well developed, and substantial evidence shows that some inhibitors derived from the above strategies show inhibitory effects on KLHLs. (3) The third reason concerns specificity. The specificity of the KLHL protein structure (the unique substrate binding pocket) and the specificity of inhibitor targeting strategies (PPIs and PROTACs) both ensure the specificity of the targeted inhibition of KLHLs. Apart from inhibitors of KLHLs with tumor-promoting function, activators of tumor suppressor KLHLs are another promising option. However, activators that can rescue KLHL functions have not been studied in gastrointestinal tumors, and their technical feasibility remains to be further evaluated and demonstrated. Furthermore, current knowledge about KLHLs in gastrointestinal tumors is still limited. For example, not all of the substrate proteins mentioned in this review fit the characteristics of definite ubiquitinated substrates of KLHLs, and to expand the list of substrates, several interacting proteins were defined as potential substrates though sufficient experimental evidence is lacking. Therefore, further research should focus on substrate identification and biological function clarification. Only significant progress in target validation can facilitate the development of KLHL inhibitors. Furthermore, inhibitor development still faces some challenges: (1) the moderate efficacy of PPI inhibitors should be improved, (2) some allosteric inhibitors of KLHLs will need to be identified without knowledge of the allosteric site, and (3) the good druggability of PROTAC molecules should be ensured on the premise of ensuring effectiveness.
In conclusion, we described KLHL aberrations and their practical significance in gastrointestinal tumors, outlined the biological functions of representative KLHLs and their substrates under pathological conditions in gastrointestinal tumors, and finally offered feasible strategies for therapies. KLHLs are important in cells because of their extraordinary abilities. KLHLs have substrate-dependent antitumor and protumor effects in practice. KLHL7, KLHL15, KLHL23, and KLHL27 could be potential candidates of drug targets. The availability of many inhibitors makes it possible to develop drugs targeting KLHLs in gastrointestinal tumors. In addition, KLHL19, KLHL22, and KLHL36 could be potential targets for the identification of prognostic biomarkers in gastrointestinal tumors. This review provides guidelines for a comprehensive understanding of the biological functions of KLHLs and offers references for the feasible development of inhibitors targeting KLHLs in gastrointestinal tumors.
Acknowledgements
This work was supported by the State Key Program of the National Natural Science Foundation of China (No. 81830107) and the Fundamental Research Funds for the Central Universities (No. 2021XZZX037).
Competing interests
The authors declare no competing interests.
Contributor Information
Sen-feng Xiang, Email: sxiang@zju.edu.cn.
Qiao-jun He, Email: qiaojunhe@zju.edu.cn.
Mei-dan Ying, Email: mying@zju.edu.cn.
References
- 1.Nagtegaal ID, Odze RD, Klimstra D, Paradis V, Rugge M, Schirmacher P, et al. The 2019 WHO classification of tumours of the digestive system. Histopathology. 2020;76:182–8. doi: 10.1111/his.13975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vincent A, Herman J, Schulick R, Hruban RH, Goggins M. Pancreatic cancer. Lancet. 2011;378:607–20. doi: 10.1016/S0140-6736(10)62307-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abu-Alainin W, Gana T, Liloglou T, Olayanju A, Barrera LN, Ferguson R, et al. UHRF1 regulation of the Keap1-Nrf2 pathway in pancreatic cancer contributes to oncogenesis. J Pathol. 2016;238:423–33. doi: 10.1002/path.4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen J, Song W, Du Y, Li Z, Xuan Z, Zhao L, et al. Inhibition of KLHL21 prevents cholangiocarcinoma progression through regulating cell proliferation and motility, arresting cell cycle and reducing Erk activation. Biochem Biophys Res Commun. 2018;499:433–40. doi: 10.1016/j.bbrc.2018.03.152. [DOI] [PubMed] [Google Scholar]
- 5.Valero V, 3rd, Saunders TJ, He J, Weiss MJ, Cameron JL, Dholakia A, et al. Reliable detection of somatic mutations in fine needle aspirates of pancreatic cancer with next-generation sequencing: implications for surgical management. Ann Surg. 2016;263:153–61. doi: 10.1097/SLA.0000000000001156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoo NJ, Kim HR, Kim YR, An CH, Lee SH. Somatic mutations of the KEAP1 gene in common solid cancers. Histopathology. 2012;60:943–52. doi: 10.1111/j.1365-2559.2012.04178.x. [DOI] [PubMed] [Google Scholar]
- 7.Schulze K, Imbeaud S, Letouze E, Alexandrov LB, Calderaro J, Rebouissou S, et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet. 2015;47:505–11. doi: 10.1038/ng.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shibata T, Kokubu A, Gotoh M, Ojima H, Ohta T, Yamamoto M, et al. Genetic alteration of Keap1 confers constitutive Nrf2 activation and resistance to chemotherapy in gallbladder cancer. Gastroenterology. 2008;135:1358–68. doi: 10.1053/j.gastro.2008.06.082. [DOI] [PubMed] [Google Scholar]
- 9.Lee B, Yoon K, Lee S, Kang JM, Kim J, Shim SH, et al. Homozygous deletions at 3p22, 5p14, 6q15, and 9p21 result in aberrant expression of tumor suppressor genes in gastric cancer. Genes Chromosomes Cancer. 2015;54:142–55. doi: 10.1002/gcc.22226. [DOI] [PubMed] [Google Scholar]
- 10.Hanada N, Takahata T, Zhou Q, Ye X, Sun R, Itoh J, et al. Methylation of the KEAP1 gene promoter region in human colorectal cancer. BMC Cancer. 2012;12:66. doi: 10.1186/1471-2407-12-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shitani M, Sasaki S, Akutsu N, Takagi H, Suzuki H, Nojima M, et al. Genome-wide analysis of DNA methylation identifies novel cancer-related genes in hepatocellular carcinoma. Tumour Biol. 2012;33:1307–17. doi: 10.1007/s13277-012-0378-3. [DOI] [PubMed] [Google Scholar]
- 12.Peng JM, Bera R, Chiou CY, Yu MC, Chen TC, Chen CW, et al. Actin cytoskeleton remodeling drives epithelial-mesenchymal transition for hepatoma invasion and metastasis in mice. Hepatology. 2018;67:2226–43. doi: 10.1002/hep.29678. [DOI] [PubMed] [Google Scholar]
- 13.Choi ES, Lee H, Lee CH, Goh SH. Overexpression of KLHL23 protein from read-through transcription of PHOSPHO2-KLHL23 in gastric cancer increases cell proliferation. FEBS Open Bio. 2016;6:1155–64. doi: 10.1002/2211-5463.12136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun Z, Ke X, Salzberg SL, Kim D, Antonescu V, Cheng Y, et al. The novel fusion transcript NR5A2-KLHL29FT is generated by an insertion at the KLHL29 locus. Cancer. 2017;123:1507–15. doi: 10.1002/cncr.30510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi L, Zhang W, Zou F, Mei L, Wu G, Teng Y. KLHL21, a novel gene that contributes to the progression of hepatocellular carcinoma. BMC Cancer. 2016;16:815. doi: 10.1186/s12885-016-2851-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J, Yu Y, Ji T, Ma R, Chen M, Li G, et al. Clinical implication of Keap1 and phosphorylated Nrf2 expression in hepatocellular carcinoma. Cancer Med. 2016;5:2678–87. doi: 10.1002/cam4.788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kong W, Gao M, Jin Y, Huang W, Huang Z, Xie Z. Prognostic model of patients with liver cancer based on tumor stem cell content and immune process. Aging (Albany NY) 2020;12:16555–78. doi: 10.18632/aging.103832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang W, Li G, Wang Z, Zhou L, Yin X, Yang T, et al. A ten-N(6)-methyladenosine (m(6)A)-modified gene signature based on a risk score system predicts patient prognosis in rectum adenocarcinoma. Front Oncol. 2020;10:567931. doi: 10.3389/fonc.2020.567931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song Y, Yuan H, Wang J, Wu Y, Xiao Y, Mao S. KLHL22 regulates the EMT and proliferation in colorectal cancer cells in part via the Wnt/beta-Catenin signaling pathway. Cancer Manag Res. 2020;12:3981–93. doi: 10.2147/CMAR.S252232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cui Y, Yang J, Bai Y, Li Q, Yao Y, Liu C, et al. ENC1 facilitates colorectal carcinoma tumorigenesis and metastasis via JAK2/STAT5/AKT axis-mediated epithelial mesenchymal transition and stemness. Front Cell Dev Biol. 2021;9:616887. doi: 10.3389/fcell.2021.616887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen HY, Hu JY, Chen TH, Lin YC, Liu X, Lin MY, et al. KLHL39 suppresses colon cancer metastasis by blocking KLHL20-mediated PML and DAPK ubiquitination. Oncogene. 2015;34:5141–51. doi: 10.1038/onc.2014.435. [DOI] [PubMed] [Google Scholar]
- 23.Nabeshima T, Hamada S, Taguchi K, Tanaka Y, Matsumoto R, Yamamoto M, et al. Keap1 deletion accelerates mutant K-ras/p53-driven cholangiocarcinoma. Am J Physiol Gastrointest Liver Physiol. 2020;318:G419–G27. doi: 10.1152/ajpgi.00296.2019. [DOI] [PubMed] [Google Scholar]
- 24.Liu JZ, Hu YL, Feng Y, Jiang Y, Guo YB, Liu YF, et al. BDH2 triggers ROS-induced cell death and autophagy by promoting Nrf2 ubiquitination in gastric cancer. J Exp Clin Cancer Res. 2020;39:123. doi: 10.1186/s13046-020-01620-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun X, Ou Z, Chen R, Niu X, Chen D, Kang R, et al. Activation of the p62-Keap1-NRF2 pathway protects against ferroptosis in hepatocellular carcinoma cells. Hepatology. 2016;63:173–84. doi: 10.1002/hep.28251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qin JJ, Cheng XD, Zhang J, Zhang WD. Dual roles and therapeutic potential of Keap1-Nrf2 pathway in pancreatic cancer: a systematic review. Cell Commun Signal. 2019;17:121. doi: 10.1186/s12964-019-0435-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee Y, Chou TF, Pittman SK, Keith AL, Razani B, Weihl CC. Keap1/Cullin3 modulates p62/SQSTM1 activity via UBA domain ubiquitination. Cell Rep. 2017;19:188–202. doi: 10.1016/j.celrep.2017.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ichimura Y, Waguri S, Sou YS, Kageyama S, Hasegawa J, Ishimura R, et al. Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol Cell. 2013;51:618–31. doi: 10.1016/j.molcel.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 29.Shao C, Tu C, Cheng X, Xu Z, Wang X, Shen J, et al. Inflammatory and senescent phenotype of pancreatic stellate cells induced by Sqstm1 downregulation facilitates pancreatic cancer progression. Int J Biol Sci. 2019;15:1020–9. doi: 10.7150/ijbs.27825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee DF, Kuo HP, Liu M, Chou CK, Xia W, Du Y, et al. KEAP1 E3 ligase-mediated downregulation of NF-kappaB signaling by targeting IKKbeta. Mol Cell. 2009;36:131–40. doi: 10.1016/j.molcel.2009.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koliaraki V, Pasparakis M, Kollias G. IKKbeta in intestinal mesenchymal cells promotes initiation of colitis-associated cancer. J Exp Med. 2015;212:2235–51. doi: 10.1084/jem.20150542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sakamoto K, Hikiba Y, Nakagawa H, Hayakawa Y, Yanai A, Akanuma M, et al. Inhibitor of kappaB kinase beta regulates gastric carcinogenesis via interleukin-1alpha expression. Gastroenterology. 2010;139:226–38. doi: 10.1053/j.gastro.2010.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gu L, Zhu Y, Lin X, Lu B, Zhou X, Zhou F, et al. The IKKbeta-USP30-ACLY axis controls lipogenesis and tumorigenesis. Hepatology. 2021;73:160–74. doi: 10.1002/hep.31249. [DOI] [PubMed] [Google Scholar]
- 34.Wang C, Hu NH, Yu LY, Gong LH, Dai XY, Peng C, et al. 2,3,5,4’-Tetrahydroxystilbence-2-O-beta-D-glucoside attenuates hepatic steatosis via IKKbeta/NF-kappaB and Keap1-Nrf2 pathways in larval zebrafish. Biomed Pharmacother. 2020;127:110138. doi: 10.1016/j.biopha.2020.110138. [DOI] [PubMed] [Google Scholar]
- 35.Schumann M, Hofmann A, Krutzke SK, Hilger AC, Marsch F, Stienen D, et al. Array-based molecular karyotyping in fetuses with isolated brain malformations identifies disease-causing CNVs. J Neurodev Disord. 2016;8:11. doi: 10.1186/s11689-016-9144-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang D, Liu H, Lin J, Ye D. Hepatitis B virus infection dampens CtIP expression in Hepatoma cell. J Cancer. 2018;9:1182–7. doi: 10.7150/jca.23649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu Y, Chen L, Zhao G, Li H, Guo Q, Zhu S, et al. RBBP8/CtIP suppresses P21 expression by interacting with CtBP and BRCA1 in gastric cancer. Oncogene. 2020;39:1273–89. doi: 10.1038/s41388-019-1060-7. [DOI] [PubMed] [Google Scholar]
- 38.Zhang C, Zhou B, Gu F, Liu H, Wu H, Yao F, et al. Micropeptide PACMP inhibition elicits synthetic lethal effects by decreasing CtIP and poly(ADP-ribosyl)ation. Mol Cell. 2022;82:1297–312. doi: 10.1016/j.molcel.2022.01.020. [DOI] [PubMed] [Google Scholar]
- 39.Ren J, Wu Y, Wang Y, Zhao Y, Li Y, Hao S, et al. CtIP suppresses primary microRNA maturation and promotes metastasis of colon cancer cells in a xenograft mouse model. J Biol Chem. 2021;296:100707. doi: 10.1016/j.jbc.2021.100707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song J, Merrill RA, Usachev AY, Strack S, The X-linked intellectual disability gene product and E3 ubiquitin ligase KLHL15 degrades doublecortin proteins to constrain neuronal dendritogenesis. J Biol Chem. 2020;296:100082. [DOI] [PMC free article] [PubMed]
- 41.Wu X, Qu D, Weygant N, Peng J, Houchen CW, Cancer stem cell marker DCLK1 correlates with tumorigenic immune infiltrates in the colon and gastric adenocarcinoma microenvironments. Cancers. 2020;12:274. [DOI] [PMC free article] [PubMed]
- 42.Vega KJ, May R, Sureban SM, Lightfoot SA, Qu D, Reed A, et al. Identification of the putative intestinal stem cell marker doublecortin and CaM kinase-like-1 in Barrett’s esophagus and esophageal adenocarcinoma. J Gastroenterol Hepatol. 2012;27:773–80. doi: 10.1111/j.1440-1746.2011.06928.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen Y, Zhou H, Wang Z, Huang Z, Wang J, Zheng M, et al. Integrated analysis of ceRNA network and tumor-infiltrating immune cells in esophageal cancer. Biosci Rep. 2021;41:BSR20203804. [DOI] [PMC free article] [PubMed]
- 44.Lorenzo N, Sabina DM, Guido C, Ilaria Grazia Z, Samira S, Valeria A, et al. DCLK1, a putative stem cell marker in human cholangiocarcinoma. Hepatology. 2021;73:144–59. doi: 10.1002/hep.31571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ali N, Allam H, May R, Sureban SM, Bronze MS, Bader T, et al. Hepatitis C virus-induced cancer stem cell-like signatures in cell culture and murine tumor xenografts. J Virol. 2011;85:12292–303. doi: 10.1128/JVI.05920-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakanishi Y, Seno H, Fukuoka A, Ueo T, Yamaga Y, Maruno T, et al. Dclk1 distinguishes between tumor and normal stem cells in the intestine. Nat Genet. 2013;45:98–103. doi: 10.1038/ng.2481. [DOI] [PubMed] [Google Scholar]
- 47.Westphalen CB, Takemoto Y, Tanaka T, Macchini M, Jiang Z, Renz BW, et al. Dclk1 defines quiescent pancreatic progenitors that promote injury-induced regeneration and tumorigenesis. Cell Stem Cell. 2016;18:441–55. doi: 10.1016/j.stem.2016.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen J, Ou Y, Yang Y, Li W, Xu Y, Xie Y, et al. KLHL22 activates amino-acid-dependent mTORC1 signalling to promote tumorigenesis and ageing. Nature. 2018;557:585–9. doi: 10.1038/s41586-018-0128-9. [DOI] [PubMed] [Google Scholar]
- 49.Pang Y, Xie F, Cao H, Wang C, Zhu M, Liu X, et al. Mutational inactivation of mTORC1 repressor gene DEPDC5 in human gastrointestinal stromal tumors. Proc Natl Acad Sci USA. 2019;116:22746–53. doi: 10.1073/pnas.1914542116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mizuno Y, Shimada S, Akiyama Y, Watanabe S, Aida T, Ogawa K, et al. DEPDC5 deficiency contributes to resistance to leucine starvation via p62 accumulation in hepatocellular carcinoma. Sci Rep. 2018;8:106. doi: 10.1038/s41598-017-18323-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beck J, Maerki S, Posch M, Metzger T, Persaud A, Scheel H, et al. Ubiquitylation-dependent localization of PLK1 in mitosis. Nat Cell Biol. 2013;15:430–9. doi: 10.1038/ncb2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu M, Wang Y, Yang D, Gong Y, Rao F, Liu R, et al. A PLK1 kinase inhibitor enhances the chemosensitivity of cisplatin by inducing pyroptosis in oesophageal squamous cell carcinoma. EBioMedicine. 2019;41:244–55. doi: 10.1016/j.ebiom.2019.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Komatsu S, Takenobu H, Ozaki T, Ando K, Koida N, Suenaga Y, et al. Plk1 regulates liver tumor cell death by phosphorylation of TAp63. Oncogene. 2009;28:3631–41. doi: 10.1038/onc.2009.216. [DOI] [PubMed] [Google Scholar]
- 54.Zhou XA, Zhou J, Zhao L, Yu G, Zhan J, Shi C, et al. KLHL22 maintains PD-1 homeostasis and prevents excessive T cell suppression. Proc Natl Acad Sci USA. 2020;117:28239–50. doi: 10.1073/pnas.2004570117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ramos-Casals M, Brahmer JR, Callahan MK, Flores-Chavez A, Keegan N, Khamashta MA, et al. Immune-related adverse events of checkpoint inhibitors. Nat Rev Dis Prim. 2020;6:38. doi: 10.1038/s41572-020-0160-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang C, Zhao Y, Yang Y, Zhong C, Ji T, Duan J, et al. RNAi mediated silencing of Nanog expression suppresses the growth of human colorectal cancer stem cells. Biochem Biophys Res Commun. 2021;534:254–60. doi: 10.1016/j.bbrc.2020.11.101. [DOI] [PubMed] [Google Scholar]
- 57.Tan P, Xu Y, Du Y, Wu L, Guo B, Huang S, et al. SPOP suppresses pancreatic cancer progression by promoting the degradation of NANOG. Cell Death Dis. 2019;10:794. doi: 10.1038/s41419-019-2017-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Foo SL, Yap G, Cui J, Lim LHK. Annexin-A1—A blessing or a curse in cancer? Trends Mol Med. 2019;25:315–27. doi: 10.1016/j.molmed.2019.02.004. [DOI] [PubMed] [Google Scholar]
- 59.Guo C, Liu S, Greenaway F, Sun MZ. Potential role of annexin A7 in cancers. Clin Chim Acta. 2013;423:83–9. doi: 10.1016/j.cca.2013.04.018. [DOI] [PubMed] [Google Scholar]
- 60.Chen YY, Wang WH, Che L, Lan Y, Zhang LY, Zhan DL, et al. BNIP3L-dependent Mitophagy promotes HBx-induced cancer stemness of hepatocellular carcinoma cells via glycolysis metabolism reprogramming. Cancers 2020;12:655. [DOI] [PMC free article] [PubMed]
- 61.Humpton TJ, Alagesan B, DeNicola GM, Lu D, Yordanov GN, Leonhardt CS, et al. Oncogenic KRAS induces NIX-mediated mitophagy to promote pancreatic cancer. Cancer Discov. 2019;9:1268–87. doi: 10.1158/2159-8290.CD-18-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Georgakilas AG, Martin OA, Bonner WM. p21: A two-faced genome guardian. Trends Mol Med. 2017;23:310–9. doi: 10.1016/j.molmed.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 63.Zhang S, Yu C, Yang X, Hong H, Lu J, Hu W, et al. N-myc downstream-regulated gene 1 inhibits the proliferation of colorectal cancer through emulative antagonizing NEDD4-mediated ubiquitylation of p21. J Exp Clin Cancer Res. 2019;38:490. [DOI] [PMC free article] [PubMed]
- 64.Chen J, Li GQ, Zhang L, Tang M, Cao X, Xu GL, et al. Complement C5a/C5aR pathway potentiates the pathogenesis of gastric cancer by down-regulating p21 expression. Cancer Lett. 2018;412:30–6. doi: 10.1016/j.canlet.2017.10.003. [DOI] [PubMed] [Google Scholar]
- 65.Marhenke S, Buitrago-Molina LE, Endig J, Orlik J, Schweitzer N, Klett S, et al. p21 promotes sustained liver regeneration and hepatocarcinogenesis in chronic cholestatic liver injury. Gut. 2014;63:1501–12. doi: 10.1136/gutjnl-2013-304829. [DOI] [PubMed] [Google Scholar]
- 66.Govindaraj V, Yaduvanshi NS, Krishnamachar H, Rao AJ. Expression of thyroid-stimulating hormone receptor, octamer-binding transcription factor 4, and intracisternal A particle-promoted polypeptide in human breast cancer tissues. Horm Mol Biol Clin Investig. 2012;9:173–8. doi: 10.1515/hmbci-2011-0130. [DOI] [PubMed] [Google Scholar]
- 67.Kobayashi S, Hiwasa T, Arasawa T, Kagaya A, Ishii S, Shimada H, et al. Identification of specific and common diagnostic antibody markers for gastrointestinal cancers by SEREX screening using testis cDNA phage library. Oncotarget. 2018;9:18559–69. doi: 10.18632/oncotarget.24963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jun S, Jung Y-S, Suh HN, Wang W, Kim MJ, Oh YS, et al. LIG4 mediates Wnt signalling-induced radioresistance. Nat Commun. 2016;7:10994. [DOI] [PMC free article] [PubMed]
- 69.Bauer D, Mazzio E, Soliman KFA. Whole transcriptomic analysis of Apigenin on TNFalpha Immuno-activated MDA-MB-231 breast cancer cells. Cancer Genom Proteom. 2019;16:421–31. doi: 10.21873/cgp.20146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li X, Huang L, Gao W. Overexpression of Tripartite Motif containing 55 (TRIM55) inhibits migration and invasion of Hepatocellular Carcinoma (HCC) cells via epithelial-mesenchymal transition and matrix metalloproteinase-2 (MMP2) Med Sci Monit. 2019;25:771–7. doi: 10.12659/MSM.910984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yasuda D, Ohe T, Takahashi K, Imamura R, Kojima H, Okabe T, et al. Inhibitors of the protein–protein interaction between phosphorylated p62 and Keap1 attenuate chemoresistance in a human hepatocellular carcinoma cell line. Free Radic Res. 2020;54:859–71. doi: 10.1080/10715762.2020.1732955. [DOI] [PubMed] [Google Scholar]
- 72.Arora R, Sawney S, Saini V, Steffi C, Tiwari M, Saluja D. Esculetin induces antiproliferative and apoptotic response in pancreatic cancer cells by directly binding to KEAP1. Mol Cancer. 2016;15:64. doi: 10.1186/s12943-016-0550-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lu MC, Jiao Q, Liu T, Tan SJ, Zhou HS, You QD, et al. Discovery of a head-to-tail cyclic peptide as the Keap1-Nrf2 protein-protein interaction inhibitor with high cell potency. Eur J Med Chem. 2018;143:1578–89. doi: 10.1016/j.ejmech.2017.10.052. [DOI] [PubMed] [Google Scholar]
- 74.Unni S, Deshmukh P, Krishnappa G, Kommu P, Padmanabhan B. Structural insights into the multiple binding modes of dimethyl fumarate (DMF) and its analogs to the Kelch domain of Keap1. FEBS J. 2021;288:1599–613. doi: 10.1111/febs.15485. [DOI] [PubMed] [Google Scholar]
- 75.Canning P, Cooper CDO, Krojer T, Murray JW, Pike ACW, Chaikuad A, et al. Structural basis for Cul3 protein assembly with the BTB-Kelch family of E3 ubiquitin ligases. J Biol Chem. 2013;288:7803–14. doi: 10.1074/jbc.M112.437996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.de Paola I, Pirone L, Palmieri M, Balasco N, Esposito L, Russo L, et al. Cullin3-BTB interface: a novel target for stapled peptides. PLoS One. 2015;10:e0121149. doi: 10.1371/journal.pone.0121149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lu MC, Ji JA, Jiang ZY, You QD. The Keap1-Nrf2-ARE pathway as a potential preventive and therapeutic target: an update. Med Res Rev. 2016;36:924–63. doi: 10.1002/med.21396. [DOI] [PubMed] [Google Scholar]
- 78.Honda T, Yoshizawa H, Sundararajan C, David E, Lajoie MJ, Favaloro FG, Jr., et al. Tricyclic compounds containing nonenolizable cyano enones. A novel class of highly potent anti-inflammatory and cytoprotective agents. J Med Chem. 2011;54:1762–78. doi: 10.1021/jm101445p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu KC, Shih TY, Kuo CL, Ma YS, Yang JL, Wu PP, et al. Sulforaphane induces cell death through G2/M phase arrest and triggers apoptosis in HCT 116 human colon cancer cells. Am J Chin Med. 2016;44:1289–310. doi: 10.1142/S0192415X16500725. [DOI] [PubMed] [Google Scholar]
- 80.Lu M, Liu T, Jiao Q, Ji J, Tao M, Liu Y, et al. Discovery of a Keap1-dependent peptide PROTAC to knockdown Tau by ubiquitination-proteasome degradation pathway. Eur J Med Chem. 2018;146:251–9. doi: 10.1016/j.ejmech.2018.01.063. [DOI] [PubMed] [Google Scholar]
- 81.Nieto-Jimenez C, Morafraile EC, Alonso-Moreno C, Ocana A. Clinical considerations for the design of PROTACs in cancer. Mol Cancer. 2022;21:67. doi: 10.1186/s12943-022-01535-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhou Z, Xu C, Chen P, Liu C, Pang S, Yao X, et al. Stability of HIB-Cul3 E3 ligase adaptor HIB is regulated by self-degradation and availability of its substrates. Sci Rep. 2015;5:12709. doi: 10.1038/srep12709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim SC, Hong CW, Jang SG, Kim YA, Yoo BC, Shin YK, et al. Establishment and characterization of paired primary and peritoneal seeding human colorectal cancer cell lines: identification of genes that mediate metastatic potential. Transl Oncol. 2018;11:1232–43. doi: 10.1016/j.tranon.2018.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wu Q, Yin G, Lei J, Tian J, Lan A, Liu S. KLHL5 is a prognostic-related biomarker and correlated with immune infiltrates in gastric cancer. Front Mol Biosci. 2020;7:599110. doi: 10.3389/fmolb.2020.599110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Deng J, Guo J, Ma G, Zhang H, Sun D, Hou Y, et al. Prognostic value of the cancer oncogene Kelch-like 6 in gastric cancer. Br J Surg. 2017;104:1847–56. doi: 10.1002/bjs.10628. [DOI] [PubMed] [Google Scholar]
- 86.Gao L, Yuan F, Che G, Xiao X, Nie X, Wang Y, et al. Epigenetic modifications but not genetic polymorphisms regulate KEAP1 expression in colorectal cancer. J Cell Biochem. 2019;120:12311–20. doi: 10.1002/jcb.28495. [DOI] [PubMed] [Google Scholar]
- 87.Pandey A, Stawiski EW, Durinck S, Gowda H, Goldstein LD, Barbhuiya MA, et al. Integrated genomic analysis reveals mutated ELF3 as a potential gallbladder cancer vaccine candidate. Nat Commun. 2020;11:4225. doi: 10.1038/s41467-020-17880-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.DeNicola GM, Karreth FA, Humpton TJ, Gopinathan A, Wei C, Frese K, et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475:106–9. doi: 10.1038/nature10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Quaas A, Heydt C, Waldschmidt D, Alakus H, Zander T, Goeser T, et al. Alterations in ERBB2 and BRCA and microsatellite instability as new personalized treatment options in small bowel carcinoma. BMC Gastroenterol. 2019;19:21. doi: 10.1186/s12876-019-0942-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jia D, Dong R, Jing Y, Xu D, Wang Q, Chen L, et al. Exome sequencing of hepatoblastoma reveals novel mutations and cancer genes in the Wnt pathway and ubiquitin ligase complex. Hepatology. 2014;60:1686–96. doi: 10.1002/hep.27243. [DOI] [PubMed] [Google Scholar]
- 91.Xu D, Wang Y, Liu X, Zhou K, Wu J, Chen J, et al. Development and clinical validation of a novel 9-gene prognostic model based on multi-omics in pancreatic adenocarcinoma. Pharmacol Res. 2021;164:105370. doi: 10.1016/j.phrs.2020.105370. [DOI] [PubMed] [Google Scholar]
- 92.Ha YJ, Kim CW, Roh SA, Cho DH, Park JL, Kim SY, et al. Epigenetic regulation of KLHL34 predictive of pathologic response to preoperative chemoradiation therapy in rectal cancer patients. Int J Radiat Oncol Biol Phys. 2015;91:650–8. doi: 10.1016/j.ijrobp.2014.11.013. [DOI] [PubMed] [Google Scholar]
- 93.Uddin MN, Li M, Wang X. Identification of transcriptional signatures of colon tumor stroma by a meta-analysis. J Oncol. 2019;2019:8752862. doi: 10.1155/2019/8752862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Garcia-Bilbao A, Armananzas R, Ispizua Z, Calvo B, Alonso-Varona A, Inza I, et al. Identification of a biomarker panel for colorectal cancer diagnosis. BMC Cancer. 2012;12:43. doi: 10.1186/1471-2407-12-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang Y, Cheng J, Xie D, Ding X, Hou H, Chen X, et al. NS1-binding protein radiosensitizes esophageal squamous cell carcinoma by transcriptionally suppressing c-Myc. Cancer Commun. 2018;38:33. doi: 10.1186/s40880-018-0307-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kim J, Tsuruta F, Okajima T, Yano S, Sato B, Chiba T. KLHL7 promotes TUT1 ubiquitination associated with nucleolar integrity: Implications for retinitis pigmentosa. Biochem Biophys Res Commun. 2017;494:220–6. doi: 10.1016/j.bbrc.2017.10.049. [DOI] [PubMed] [Google Scholar]
- 97.Mason B, Flach S, Teixeira FR, Manzano Garcia R, Rueda OM, Abraham JE, et al. Fbxl17 is rearranged in breast cancer and loss of its activity leads to increased global O-GlcNAcylation. Cell Mol Life Sci. 2020;77:2605–20. doi: 10.1007/s00018-019-03306-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ferretti LP, Himmels SF, Trenner A, Walker C, von Aesch C, Eggenschwiler A, et al. Cullin3-KLHL15 ubiquitin ligase mediates CtIP protein turnover to fine-tune DNA-end resection. Nat Commun. 2016;7:12628. doi: 10.1038/ncomms12628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Oberg EA, Nifoussi SK, Gingras AC, Strack S. Selective proteasomal degradation of the B’beta subunit of protein phosphatase 2A by the E3 ubiquitin ligase adaptor Kelch-like 15. J Biol Chem. 2012;287:43378–89. doi: 10.1074/jbc.M112.420281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang Q, Zhang ZY, Du H, Li SZ, Tu R, Jia YF, et al. DUB3 deubiquitinates and stabilizes NRF2 in chemotherapy resistance of colorectal cancer. Cell Death Differ. 2019;26:2300–13. doi: 10.1038/s41418-019-0303-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ren F, Shu G, Liu G, Liu D, Zhou J, Yuan L, et al. Knockdown of p62/sequestosome 1 attenuates autophagy and inhibits colorectal cancer cell growth. Mol Cell Biochem. 2014;385:95–102. doi: 10.1007/s11010-013-1818-0. [DOI] [PubMed] [Google Scholar]
- 102.Raab M, Sanhaji M, Matthess Y, Horlin A, Lorenz I, Dotsch C, et al. PLK1 has tumor-suppressive potential in APC-truncated colon cancer cells. Nat Commun. 2018;9:1106. doi: 10.1038/s41467-018-03494-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chen Z, Chai Y, Zhao T, Li P, Zhao L, He F, et al. Effect of PLK1 inhibition on cisplatin-resistant gastric cancer cells. J Cell Physiol. 2019;234:5904–14. doi: 10.1002/jcp.26777. [DOI] [PubMed] [Google Scholar]
- 104.Zhang C, Sun X, Ren Y, Lou Y, Zhou J, Liu M, et al. Validation of Polo-like kinase 1 as a therapeutic target in pancreatic cancer cells. Cancer Biol Ther. 2012;13:1214–20. doi: 10.4161/cbt.21412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sokolina K, Kittanakom S, Snider J, Kotlyar M, Maurice P, Gandia J, et al. Systematic protein-protein interaction mapping for clinically relevant human GPCRs. Mol Syst Biol. 2017;13:918. doi: 10.15252/msb.20167430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang X, Jin J, Wan F, Zhao L, Chu H, Chen C, et al. AMPK promotes SPOP-mediated NANOG degradation to regulate prostate cancer cell stemness. Dev Cell. 2019;48:345–60. doi: 10.1016/j.devcel.2018.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Deng L, Xiang X, Yang F, Xiao D, Liu K, Chen Z, et al. Functional evidence that the self-renewal gene NANOG regulates esophageal squamous cancer development. Biochem Biophys Res Commun. 2017;490:161–8. doi: 10.1016/j.bbrc.2017.06.016. [DOI] [PubMed] [Google Scholar]
- 108.Cao J, Zhao M, Liu J, Zhang X, Pei Y, Wang J, et al. RACK1 promotes self-renewal and chemoresistance of cancer stem cells in human hepatocellular carcinoma through stabilizing Nanog. Theranostics. 2019;9:811–28. doi: 10.7150/thno.29271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Vinayagam A, Stelzl U, Foulle R, Plassmann S, Zenkner M, Timm J, et al. A directed protein interaction network for investigating intracellular signal transduction. Sci Signal. 2011;4:rs8. doi: 10.1126/scisignal.2001699. [DOI] [PubMed] [Google Scholar]
- 110.Hagihara T, Kondo J, Endo H, Ohue M, Sakai Y, Inoue M. Hydrodynamic stress stimulates growth of cell clusters via the ANXA1/PI3K/AKT axis in colorectal cancer. Sci Rep. 2019;9:20027. doi: 10.1038/s41598-019-56739-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pessolano E, Belvedere R, Bizzarro V, Franco P, Marco I, Porta A, et al. Annexin A1 may induce pancreatic cancer progression as a key player of extracellular vesicles effects as evidenced in the in vitro MIA PaCa-2 Model System. Int J Mol Sci. 2018;19:3878. [DOI] [PMC free article] [PubMed]
- 112.Ibrahim MM, Sun MZ, Huang Y, Jun M, Jin Y, Yue D, et al. Down-regulation of ANXA7 decreases metastatic potential of human hepatocellular carcinoma cells in vitro. Biomed Pharmacother. 2013;67:285–91. doi: 10.1016/j.biopha.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 113.Yan C, Luo L, Guo CY, Goto S, Urata Y, Shao JH, et al. Doxorubicin-induced mitophagy contributes to drug resistance in cancer stem cells from HCT8 human colorectal cancer cells. Cancer Lett. 2017;388:34–42. doi: 10.1016/j.canlet.2016.11.018. [DOI] [PubMed] [Google Scholar]
- 114.Luck K, Kim DK, Lambourne L, Spirohn K, Begg BE, Bian W, et al. A reference map of the human binary protein interactome. Nature. 2020;580:402–8. doi: 10.1038/s41586-020-2188-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Woods NT, Mesquita RD, Sweet M, Carvalho MA, Li X, Liu Y, et al. Charting the landscape of tandem BRCT domain-mediated protein interactions. Sci Signal. 2012;5:rs6. doi: 10.1126/scisignal.2002255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Nowak M, Suenkel B, Porras P, Migotti R, Schmidt F, Kny M, et al. DCAF8, a novel MuRF1 interaction partner, promotes muscle atrophy. J Cell Sci. 2019;132:jcs233395. [DOI] [PubMed]