Abstract
G protein-coupled receptors (GPCRs) within the same subfamily often share high homology in their orthosteric pocket and therefore pose challenges to drug development. The amino acids that form the orthosteric binding pocket for epinephrine and norepinephrine in the β1 and β2 adrenergic receptors (β1AR and β2AR) are identical. Here, to examine the effect of conformational restriction on ligand binding kinetics, we synthesized a constrained form of epinephrine. Surprisingly, the constrained epinephrine exhibits over 100-fold selectivity for the β2AR over the β1AR. We provide evidence that the selectivity may be due to reduced ligand flexibility that enhances the association rate for the β2AR, as well as a less stable binding pocket for constrained epinephrine in the β1AR. The differences in the amino acid sequence of the extracellular vestibule of the β1AR allosterically alter the shape and stability of the binding pocket, resulting in a marked difference in affinity compared to the β2AR. These studies suggest that for receptors containing identical binding pocket residues, the binding selectivity may be influenced in an allosteric manner by surrounding residues, like those of the extracellular loops (ECLs) that form the vestibule. Exploiting these allosteric influences may facilitate the development of more subtype-selective ligands for GPCRs.
Subject terms: X-ray crystallography, Small molecules, G protein-coupled receptors
Constrained catecholamines gain β2AR selectivity. Although the orthosteric pockets are identical in β1AR and β2AR, surrounding residues allosterically modify the pockets and contribute to the β2AR selectivity of the constrained catecholamines.
Introduction
G protein-coupled receptors (GPCRs) are of great interest as therapeutic targets1. One of the major challenges in drug discovery efforts is to minimize off-target side effects, and to achieve greater subtype selectivity of candidate drugs2. Many pharmaceutically important GPCRs have multiple closely related subtypes that fulfill different physiologic roles, but are all activated by the same hormone or neurotransmitter. For example, the nine adrenergic receptor subtypes have different roles in regulating central and sympathetic nervous system functions, but can all be activated by epinephrine (Epi) and norepinephrine (NorEpi), as they share a highly homologous orthosteric binding pocket.
The β1 and β2 adrenergic receptors (ARs) have been among the most extensively studied G protein-coupled receptors (GPCRs) due to their roles in the regulation of cardiac and pulmonary function by the autonomic nervous system. βAR antagonists were among the first GPCR drugs to be developed and β1AR-selective antagonists are used for the treatment of heart failure, arrhythmias, and hypertension3. β2AR-selective agonists have been used in the treatment of asthma and chronic obstructive pulmonary disease4. β1AR and β2AR are equally responsive to Epi, primarily secreted by the adrenal gland, in contrast to NorEpi, which is primarily released from sympathetic nerve terminals, and is approximately 10-fold more selective for the β1AR5.
Previous molecular dynamics (MD) simulations of alprenolol and other ligands binding to the β2AR6 reveal that ligands typically access the orthosteric binding pocket through a portal formed by a narrow cleft, lined by residues in the extracellular vestibule. Recent studies suggest that the preferred pathway may differ for the β1AR and β2AR7 (Fig. 1a). MD simulations suggest that ligand flexibility might be important for navigating through the extracellular vestibule. Ligand flexibility, however, also yields an entropic penalty when the ligand arrives at the orthosteric pocket, where it is constrained to a single or limited set of conformations. The principle sites of flexibility in catecholamines are contributed by two rotatable bonds, one between the catechol ring and the ethanolamine moiety and one within the ethanolamine itself (see Fig. 1b). A key finding revealed in structural studies of Epi-, NorEpi- and isoproterenol (ISO)-bound β-ARs is the selection of one specific set of rotamers of the catecholamines8,9.
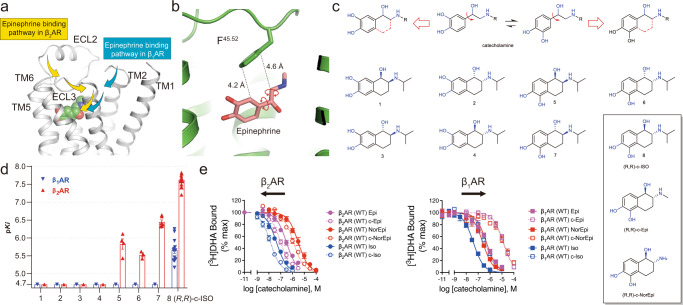
Fig. 1. Design and synthesis of conformationally-constrained catecholamine.
a Epi favors different binding pathways to enter the orthosteric pockets in the β1AR and β2AR7. b There is space between Epi and F45.52 to allow the receptor to accommodate for the conformational restriction of the catecholamine (PDB: 4LDO). c The design and synthesis flow of eight possible conformationally-constrained isoprenaline isomers and the chemical structures of (R,R)-c-Epi and (R,R)-c-NorEpi. d The (R,R)-isomer of c-ISO showed the highest affinity to both the β1AR and β2AR among all the eight possible isomers. Data were given as mean ± SEM of n = 3 (for 1-4,6), n = 4 (for 5), n = 5 (β1AR for 7), n = 6 (β2AR for 7), n = 13 (β2AR for 8), and n = 14 (β1AR for 8) independent experiments. e c-ISO, c-Epi, and c-NorEpi showed β2AR selectivity in a radioligand competition binding assay. Data were given as mean ± SEM of n = 3 (β1AR for Epi), n = 3(β2AR for Epi), n = 3 (β1AR for NorEpi), n = 5(β2AR for NorEpi), n = 3 (β1AR for ISO), n = 3(β2AR for ISO), n = 6 (β1AR for c-Epi), n = 6 (β2AR for c-Epi), n = 5 (β1AR for c-NorEpi), n = 5(β2AR for c-NorEpi), n = 3 (β1AR for cISO), n = 3(β2AR for cISO) independent experiments. Source data are provided as a Source Data file.
In this work, to examine the effect of ligand flexibility and the entropic contribution to catecholamine binding to β1AR and β2AR, we generated conformationally-constrained forms of Epi, NorEpi, and ISO to restrict or constrain the rotatable bonds and characterized their pharmacology and signaling behavior. We show that the constrained catecholamines gain β2AR selectivity over the β1AR despite the orthosteric ligand binding sites are identical in these two receptors. We further show that allosteric effects from surrounding residues of the orthosteric pockets alter the shape of the pockets and contribute to the β2AR selectivity of the constrained catecholamines.
Results
Design and characterization of constrained catecholamines
Two bridging carbons were introduced to constrain the conformation of the catecholamines. Because there are two possible ways to rigidify the compound and each constrained compound has two chiral carbons, each catecholamine will have eight possible constrained isomers (Fig. 1c). Compounds 1–4 would not be expected to fit into the previously reported Epi and NorEpi binding pose; nevertheless, we decided to synthesize and test them for completeness (Supplementary Notes). The synthesis of a subset of similar compounds has previously been described; however, these compounds were not enantiomerically pure, and their pharmacology was not fully characterized10. We synthesized all the eight possible isomers of ISO (Supplementary Notes). Of which, the (R,R) form (compound 8, c-ISO) showed the highest affinity for both the β1AR and the β2AR, but not the αARs, in radioligand competition binding assay (Fig. 1d and Supplementary Table 1). Interestingly, the (R,R)-isomer also showed selectivity towards the β2AR over β1AR (Fig. 1e and Supplementary Table 2). Consequently, we synthesized the constrained (R,R)-isomers of Epi and NorEpi (hereby named c-Epi or c-NorEpi) (Supplementary Notes) and characterized their pharmacological properties side-by-side with the non-constrained forms. Compared to Epi, which has a similar EC50 in G protein activation, adenylyl cyclase activation, and arrestin recruitment for both the β1AR and the β2AR, c-Epi exhibited both an increase in potency at the β2AR and a decrease at the β1AR. On the β2AR, c-Epi has a threefold lower EC50 for G protein activation and adenylyl cyclase stimulation and a 7.5-fold reduction in EC50 in an arrestin recruitment assay, compared to the non-rigidified native hormone (Fig. 2a, Supplementary Fig. 1, Supplementary Tables 2,3). In striking contrast, c-Epi at the β1AR yields a ~14-fold increase in the EC50 in cyclase stimulation and a ~19-fold increase in the EC50 in arrestin recruitment. Radioligand competition binding assays indicate that constrained Epi bound eightfold better on the β2AR but 34-fold worse on the β1AR, compared to unconstrained Epi (Fig. 1e and Supplementary Table 4). Thus, while Epi does not display a preference for either β1AR or β2AR in an arrestin recruitment or radioligand binding assays, c-Epi recruits β-arrestin with an ~600-fold selectivity for β2AR over β1AR. We also note a substantial reduction (~40%) in the Emax for c-Epi-induced arrestin recruitment to β1AR (Fig. 2a and Supplementary Table 3).
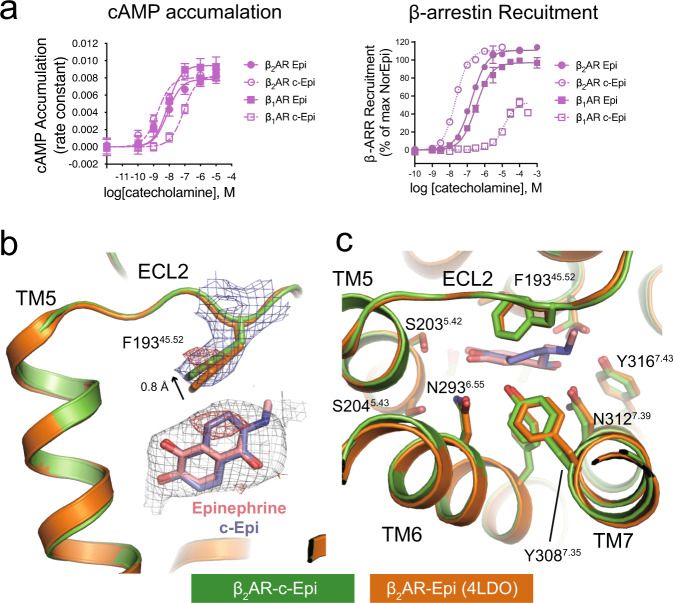
Fig. 2. c-Epi is selective towards the β2AR and binds to the same position as Epi does.
a c-Epi shows β2AR selectivity in both cAMP recruitment and arrestin signaling assays. Data were given as mean ± SEM of n = 6 (β1AR for Epi, arrestin), n = 15 (β1AR for c-Epi, arrestin), n = 17 (β2AR for Epi, arrestin), and n = 21 (β2AR for c-Epi, arrestin) independent experiments. Source data are provided as a Source data file. b c-Epi binds in a nearly identical position as Epi in the β2AR as revealed by the 2fofc density (blue mesh, contoured at 1.0 σ). The extra two carbons of c-Epi compared to Epi as well as the upward movement of F45.52 are revealed by the isomorphous difference map (red mesh, contoured at 3.0 σ). c The binding pocket residues are in similar positions upon Epi or c-Epi binding.
Perhaps more interesting is the complete switch in β1AR-β2AR selectivity of NorEpi upon constraining the catecholamine. In β-arrestin recruitment assays NorEpi has ~16-fold selectivity for β1AR over β2AR, whereas c-NorEpi displays a 52-fold selectivity for β2AR over β1AR (Supplementary Fig. 2 and Supplementary Table 3). For isoproterenol, where the EC50 for β1AR and β2AR are nearly identical (19 and 16 nM, respectively) and thus, no subtype selectivity. Constrained ISO, however, displays greater than a 70-fold selectivity for β2AR afforded by a greater than a 100-fold decrease in the potency at β1AR for recruiting β-arrestin (Supplementary Fig. 2 and Supplementary Table 3). Radioligand binding analysis suggests that this dramatic increase in β2AR selectivity was contributed by a tenfold decrease in affinity for β1AR, with a ~3-fold improvement in β2AR affinity. (Fig. 1e and Supplementary Table 4).
An additional side note is that neither c-NorEpi, c-Epi or c-ISO displayed any appreciable activity for the α1AR or α2AR subtypes, suggesting that constraining the catecholamines garners the agonists β2AR-selective over all adrenergic receptors (Supplementary Fig. 3 and Supplementary Table 2).
Structure determination of the β2AR–c-Epi complex
What is most striking about the enhanced potency of these constrained catecholamines on β2AR is the marked decrease in potency at the β1AR. In an attempt to understand the structural basis for this subtype selectivity, we solved the crystal structures of the β2AR in complex with c-Epi and c-ISO at 3.2 and 3.4 Å resolution, respectively. The structures were obtained with a previously described nanobody Nb6B9 and a β2AR construct with T4 lysozyme fused to the N-terminus (T4L-β2AR)8. Nb6B9 binds to the intracellular surface of the β2AR, the G protein binding site, and stabilizes its active conformation.
The c-Epi binding site is clearly revealed by a simulated omit map suggesting that c-Epi and Epi bind in nearly identical positions (Fig. 2b). The bridging carbons that constrain Epi in c-Epi are clearly revealed by an isomorphous difference map between the β2AR–c-Epi data and the β2AR-Epi data (Fig. 2b, red mesh). When comparing the β2AR-Epi structure and β2AR-c-Epi structure, all the orthosteric pocket residues are in similar positions (Fig. 2c). The F19345.52 side chain is displaced slightly upward (~0.8 Å) in the c-Epi bound structure compared to the Epi bound structure, supported by the 2fofc map and isomorphous difference map between the β2AR-c-Epi and β2AR-Epi data (Fig. 2b). Similar upward movement of F19345.52 is observed in the β2AR-c-ISO structure (Supplementary Fig. 4). Even though the β2AR-c-Epi and β2AR-Epi structures are remarkably similar (RMSD of 0.3 Å for all Cα atoms), structure analysis suggests that c-Epi binding provides additional stabilization of ECL3 and the C-terminal end of ECL2, as revealed by a reduction in normalized b-factors (Supplementary Fig. 5).
As previously mentioned, all residues that form the orthosteric pocket (as defined within 4 Å of the ligand) of Epi are conserved between the β2AR and the β1AR. One residue that appears within 3.5–4.5 Å away from bridging carbons of c-Epi is Y3087.35. This residue is phenylalanine in the β1AR (F3597.35), suggesting that the Y3087.35’s hydroxyl moiety could account for the affinity difference between β2AR and the β1AR. Substituting Y3087.35 to phenylalanine in β2AR, however, exhibited only a modest decrease in c-Epi affinity (Supplementary Fig. 6). Likewise, the β1AR-F3597.35Y mutant displayed an almost identical affinity for c-Epi compared to the wild-type β1AR. Taken together, these data suggest that the amino acid difference of Y/F7.35 is not the key determinant of selectivity.
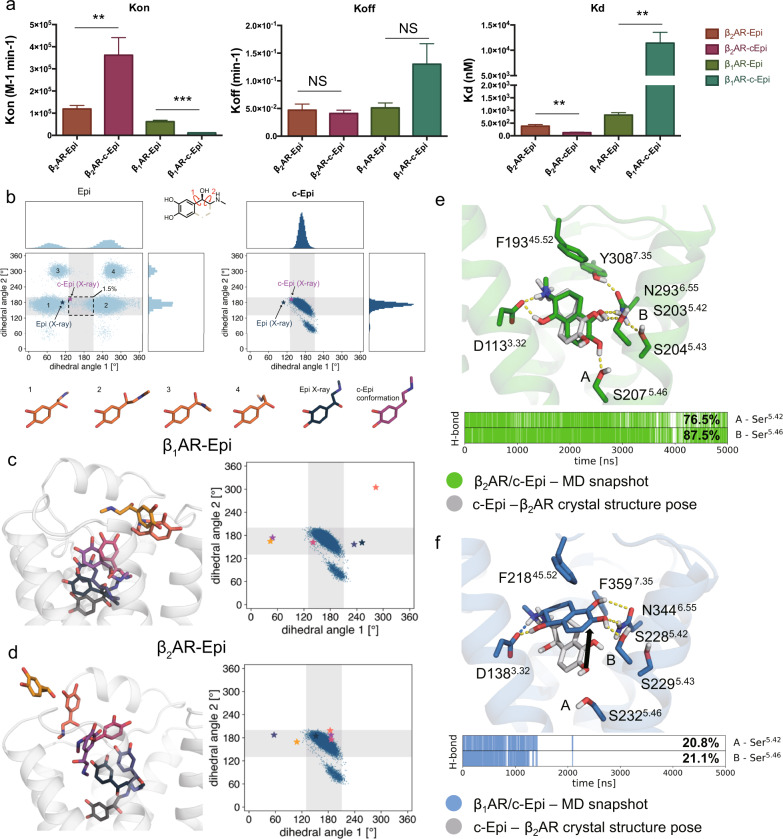
The differences in affinity observed for constrained and non-constrained catecholamines could be due to differences in agonist on-rates, off-rates, or both. We, therefore, analyzed the kinetics of c-Epi and c-NorEpi binding to both β1AR and β2AR. c-Epi exhibits a ~3-fold faster association rate with the β2AR than Epi, suggesting that conformational restriction facilitates ligand binding, probably due to entropic advantages and perhaps influenced by the increased hydrophobicity offered by the addition of the cyclic ring (Fig. 3a). Indeed, MD simulations suggest that Epi adopts a wide spectrum of conformations in the solution that rarely match its bioactive conformation, the conformation observed in the active state structure of the β2AR bound to Epi (Fig. 2b). In contrast, c-Epi appears to exist almost exclusively in the bioactive conformation (Fig. 3b). The entropic gain that results from the conformational restriction could account for the faster association rate of c-Epi compared to Epi with the β2AR. c-NorEpi also displays a modest ~2-fold faster association rate than NorEpi to the β2AR, suggesting a common mechanism for differences in the association rate (Supplementary Fig. 7). The dissociation rates of both c-Epi and c-NorEpi from β2AR were not statistically different compared to Epi or NorEpi, respectively (Fig. 3a and Supplementary Fig. 7).
Fig. 3. The selectivity of c-Epi toward the β2AR reflects differences in the binding pathways and less stable orthosteric binding pocket interactions in the β1AR.
a Binding kinetics studies of Epi and c-Epi towards the β1AR and β2AR. Statistic analysis were performed using two-way ANOVA analysis. (*P < 0.05, **P < 0.005, ***P < 0.0005). Data were given as mean ± SEM of n = 5 (β2AR for Epi), n = 5 (β2AR for c-Epi), n = 3 (β1AR for Epi), n = 3 (β1AR for c-Epi), and independent experiments. Source data are provided as a Source data file. b MD simulations suggest that Epi adopts a wide spectrum of conformations in the solution that rarely matches its bioactive conformation, while the conformational restriction limits the catecholamine in c-Epi to the bioactive conformation. The values of two dihedral angles of the aliphatic part of Epi adopted in simulations are plotted against each other. The stars indicate the values for Epi and c-Epi as present in the β2AR crystal structures. The dashed rectangle is a rough measure for the conformational space occupied by c-Epi in solution. The percentage value indicates the frequency by which Epi entered this space. c, d Epi adopts different conformations during the binding process to the β1AR (c) and β2AR (d)33. The conformational restriction of c-Epi is incompatible with this proposed path for β1AR, but is less so for β2AR. The different poses of Epi represent a potential path of binding suggested by metadynamics simulations. The plot displays the dihedral angles adopted by c-Epi in solution as described for b. The stars show the dihedral adopted by Epi in the stages of association shown in the same color on the respective left side. e c-Epi maintains its crystallographic pose in the β2AR in simulations. A representative frame of the simulations is displayed in green and the crystallographic pose of c-Epi in gray. f In β1AR-c-Epi does not maintain the expected bioactive conformation in simulations. A representative frame of the simulations is displayed in blue and the crystallographic pose of c-Epi in the β2AR in gray. e, f The trace plot, indicated the presence of the canonical hydrogen bonds to Ser5.42 and Ser5.46. The trace is displayed for one representative trajectory. The percentage value is the mean over six simulations, 5 µs each.
In contrast, the association rates at β1AR were significantly slower for c-Epi (~5-fold) and even slower for c-NorEpi (~15-fold) compared to the non-constrained catecholamines. The slower on-rate of c-Epi and c-NorEpi suggests that the portal to the orthosteric site on β1AR may be less compatible with the constrained catecholamines. The faster on-rate kinetics on β2AR are consistent with metadynamics simulations sampling of possible ligand binding pathways. While Epi adopts various compatible conformations during the process of binding to β1AR, c-Epi does not (Fig. 3c). This is in contrast to β2AR, where c-Epi’s limited conformations appear more compatible during the process of binding to the orthosteric site (Fig. 3d). This may contribute to the slower on-rate of c-Epi compared to Epi for the β1AR.
c-Epi or c-NorEpi also have a 2–3-fold increased dissociation rate for the β1AR compared to Epi or NorEpi (Fig. 3a and Supplementary Fig. 7), although this difference did not reach statistical significance, it is consistent with MD simulations of c-Epi binding to the orthosteric site of β1AR. Hydrogen bonding of the meta and para-hydroxyl on the catechol ring of catecholamines to two Ser residues on TM5 are critical for binding and efficacy on all catecholamine receptors. MD simulations show that c-Epi is much less stable in the binding pocket of the β1AR, as revealed by a large conformational shift of the ligand in simulations and a loss of H-bonding with TM5 (Fig. 3e, f). The canonical H bond between meta-hydroxyl of c-Epi and Ser5.42, as well as the H bond between para-hydroxyl of c-Epi and Ser5.46 occur 76.5 and 87.5% of the time between c-Epi and the β2AR, while the same H bonds only occur 20.8 and 21.1% of the time between c-Epi and the β1AR (Fig. 3e, f). Thus, the lower affinity of c-Epi or c-NorEpi for the β1AR compared to the β2AR is thus likely due to a combined effect of ligand entropy, less compatible conformations during the binding process, and reduced stability of the ligand binding pocket.
It is important to note that the binding site modeled from crystallographic data reflects the binding site of the active conformation, which is stabilized by the G protein-mimicking nanobody. Differences in access to the orthosteric site may likely change in the active conformation compared to the inactive state. Data from pharmacological studies suggest that access to (association rate) or escape from (dissociation rate) the β2AR orthosteric site differs significantly in the inactive and active states11.
The role of the vestibule formed by the ECLs
The MD simulations on alprenolol binding to the β2AR show transient interactions within the vestibule before ligand entry in the orthosteric site6. This was also observed in simulations following ligand binding to the muscarinic M3 and M4 acetylcholine receptor12. The vestibule on β2AR, located directly above the orthosteric site, is composed of residues contributed by the ECLs. We previously demonstrated that replacing the ECLs of β2AR with that of β1AR could confer a complete switch to high-affinity NorEpi binding by accelerating the association rate7. With the exception of Y3087.35 (F3596.58 in the β1AR), the residues within orthosteric site of β2AR and β1AR that coordinate Epi binding are identical. As noted above we observed very little differences in Y6.58 or F6.58 in c-Epi binding. The conserved aromatic residue F45.52 which contributes toward the formation of a lid over the orthosteric pocket drew our attention. We and others have shown that residue 45.52 affects the association and dissociation rates of ligands and arrestin signaling in other class A GPCRs13,14. Our structures reveal that F19345.52 on β2AR needs to move up by ~0.8 Å in order to accommodate the two carbons that rigidifies c-Epi. We previously showed that F45.52, with contributions from different surrounding residues, has significant effects on NorEpi or Epi affinity7. The F45.52A mutation has a larger effect in reducing Epi and NorEpi’s affinity for the β1AR than β2AR (~250-fold increase in Ki values for Epi and NorEpi for the β1AR, but only a 50-fold increase for Epi and 3-fold for NorEpi for the β2AR)7. In contrast, the same mutation has a larger effect in reducing c-Epi affinity for the β2AR than β1AR (Supplementary Fig. 8).
Analysis of the pharmacological properties of the constrained catecholamines using β1AR-β2AR chimeras, where the extracellular vestibules (consisting of the ECLs as well as the extracellular ends of TMs) of β1AR and β2AR were exchanged (Supplementary Fig. 9)7, strongly suggests that residues within the vestibule surrounding the orthosteric pocket confer the majority of subtype selectivity of the c-Epi and c-NorEpi. As summarized in Supplemental Table 5 and Supplementary Fig. 10, replacing the extracellular vestibule of the β2AR with that of β1AR (β2ARin/β1ARout) eliminates the enhanced on-rate and accelerates the off-rates of the constrained catecholamines yielding rate constants comparable to wild-type β1AR. Likewise, replacing the extracellular vestibule of β1AR with that of β2AR (β1ARin/β2ARout) results in a minimal change in on-rates and reduced off-rates yielding rate constants comparable to wild-type β2AR. In agreement with the binding kinetics data, replacing the extracellular vestibule also results in the change of EC50 for all three constrained catecholamines in the arrestin recruitment assay (Supplementary Fig. 11).
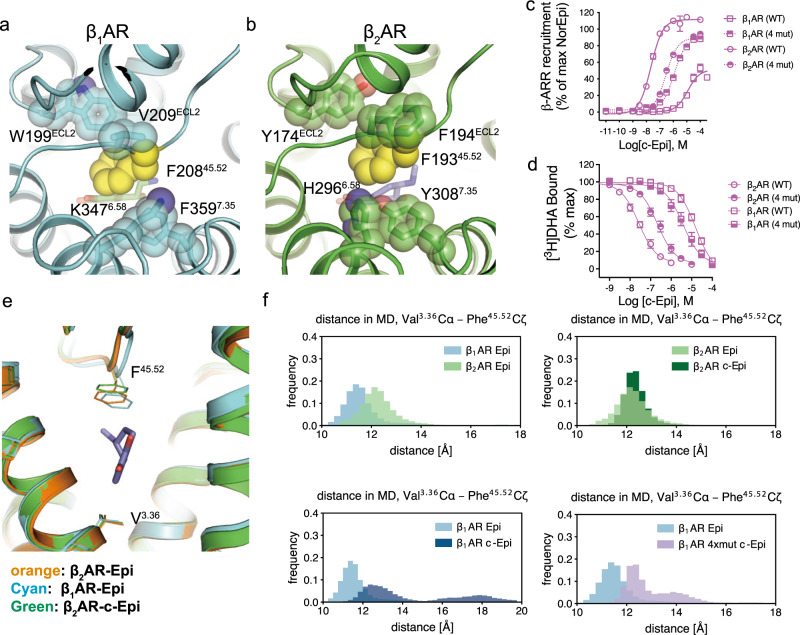
In an attempt to pinpoint key residues within the extracellular vestibule that may contribute to the subtype selectivity offered by the constrained catecholamines, we performed mutagenesis studies to determine the role of key residues that surround F45.52, but are not directly involved in agonist binding. Out of the residues within a 5 Å distance to F45.52, four are different between the β1AR and β2AR. These are W199ECL2, V209ECL2, K3476.58, and F3597.35 in the β1AR (Fig. 4a), and Y174ECL2, F194ECL2, H2966.58, and Y3087.35 in the β2AR (Fig. 4b). Single point substitutions of each of the four residues on β2AR with those of β1AR decreased c-Epi affinity, with F194ECL2V having the largest effect. The reverse mutation in β1AR, V219ECL2F, is the only substitution out of the four that displays slightly increased c-Epi affinity. (Supplementary Fig. 12). Of note, previous studies showed that this pair of mutations (β1AR-V219ECL2F and β2AR-F194ECL2V) had little effect on NorEpi affinity7, suggesting that the selectivity mechanism of c-Epi for the β2AR is different from that of NorEpi for the β1AR.
Fig. 4. Mutating four residues around the F45.52 affect c-Epi affinities in the β1AR and β2AR.
a, b F45.52 (yellow spheres) is conserved in the β1AR and β2AR, but is surrounded by differing residues. The different surrounding residues are W199ECL2, V209ECL2, K3476.58, and F3597.35 in the β1AR (a) and Y174ECL2, F194ECL2, H2966.58, and Y3087.35 in the β2AR (b). c, d Mutating the four residues surrounding F45.52 reduced c-Epi affinity to the β2AR and increased c-Epi affinity to the β1AR, in a β-arrestin recruitment assay (c) and in a competition binding assay (d). Data were given as mean ± SEM of n = 7 (β1AR (4 mut), β2AR (4 mut), arrestin), n = 15 (β1AR, arrestin), and n = 21 (β2AR, arrestin), n = 6 (β1AR, β2AR, β1AR (4 mut), β2AR (4 mut),DHA binding) independent experiments. Source data are provided as a Source data file. e F45.52 has a slightly different conformation in the β2AR-c-Epi, β2AR-Epi, and β1AR-Epi structures, and as a result, the distance between V3.36 and F45.52 is longer in the β2AR-c-Epi structure than in the β2AR-Epi structure, while the distance is the shortest in the β1AR-Epi structure. f The distribution for the distance between the Cα atom of V3.36 and the Cζ atom of F45.52 in MD simulations are displayed for the different simulation conditions. The distribution is shown in light blue for β1AR-Epi, dark blue for β1AR-c-Epi, light green for β2AR-Epi, dark green for β2AR-c-Epi and purple for β1AR_4mut-c-Epi. Cζ of F45.52 is located deeper in the pocket for β1AR-Epi compared to β2AR-Epi, and β1AR-c-Epi shows a similar distribution as β1AR-Epi. The deeper position in β1AR cannot accommodate for c-Epi and leads to a distortion of the F45.52 conformation and, therefore, to higher distances. For β1AR_4mut-c-Epi, distances at a β2AR-like level are observed, suggesting that the β2AR-like surrounding of F45.52 leads to conformations of F45.52 that are located less deep in the pocket as for β1AR-Epi.
While the individual single substitutions of the β1AR (W199ECL2Y, K3476.58H, and F3597.35Y) do not increase the receptor’s affinity for c-Epi, a combination of all these three mutations together with the V219ECL2F (β1AR-4mut) showed an increased c-Epi affinity qualitatively similar to the β1ARin/β2ARout chimera (Fig. 4c, d, Supplementary Fig. 10, and Table 2). The results suggest that cooperation between different residues within the vestibule is required in order to establish a high-affinity c-Epi binding pocket in the β1AR. Combining all four mutations in the β2AR (β2AR-4mut) shows decreased c-Epi affinity compared to wild-type β2AR and is quantitively similar to the β2ARin/β1ARout chimera (Supplementary Fig. 11 and Table 2). Interestingly, the reduced affinity of c-Epi on β2AR-4mut was not as large as the single substitution β2AR-F194ECL2V, highlighting the complex behavior of the allosteric interactions between residues. We further examined the effects of the four-residue substitutions on c-NorEpi and c-ISO. In arrestin recruitment assays, the β2AR-4mut displays decreased c-NorEpi and c-ISO potency compared to the β2AR, while β1AR-4mut shows increased c-NorEpi and c-ISO potency compared to the β1AR (Supplementary Fig. 13 and Supplementary Table 2).
To investigate the effect of the four-residue mutations on F45.52 conformation and dynamics, we simulated the binding of c-Epi to β1AR-4mut and compared the results with the MD simulations of Epi or c-Epi binding to wild-type β1AR and β2AR. As previously mentioned, c-Epi is not stable in the orthosteric pocket of β1AR and the canonical H bonds between the catecholamine and TM5 are seldom observed in the simulations. With β1AR-4mut, however, the c-Epi binding pose is more stable where canonical H bonds occur 43.9% of the time for the meta-hydroxyl and Ser5.42 and 55.4% of the time for para-hydroxyl and Ser5.46 (Supplementary Fig. 14a), all in good agreement with the functional data.
As observed in the crystal structure, the upward 0.8 Å movement of the F19345.52 side chain is required in order to accommodate the extra two carbon atoms in c-Epi. In the β2AR-Epi structure, the distance between the Cα atom of V1173.36, which is located at the bottom of the orthosteric site, and the Cζ atom of F19345.52 is 11 Å. In contrast, this V3.36/Cα – F45.52/Cζ distance is 11.7 Å in the β2AR-c-Epi structure and 10.6 Å in the β1AR-Epi structure. Similarly, MD simulations suggest that the V3.36/Cα – F45.52/Cζ distance is slightly longer in the β2AR-c-Epi complex compared to the β2AR-Epi complex, and slightly shorter in the β1AR-Epi complex compared to the β2AR-Epi complex (Fig. 4e, f). This small difference does not appear to have a major effect on the affinity of the smaller Epi, whereas the addition of the two carbons in c-Epi, requires the additional space created by the upward movement of F19345.52. It is likely that the allosteric effects of the residues surrounding F45.52 in β1AR do not permit its upward movement to the same extent as in β2AR without a rotamer change. MD simulations suggest that the side chain of F45.52 in the β1AR needs to flip away in order to avoid a clash with the c-Epi. This would require more flexibility of F45.52 and the surrounding region of the ECL2 in the β1AR-c-Epi simulation compared to the β2AR-c-Epi. Substituting the four surrounding residues, as with β1AR-4mut, results in reduced flexibility of F45.52 and ECL2 (Supplementary Fig. 14b), displaying a similar V3.36/Cα – F45.52/Cζ distance distribution as observed in the β2AR-c-Epi structure (Fig. 4e, f).
Of note, the F45.52 that is crucial for high-affinity binding of the β2AR to c-Epi is not conserved as Phe, but as branched-chain amino acids (leucine, isoleucine, valine) in the α1AR or α2AR subtypes (Supplementary Fig. 15a). Comparison with the α2BAR structure (PDB code: 6K41)15 suggests the sidechain of leucine may clash with c-Epi (Supplementary Fig. 15b). Binding studies also suggest F45.52V mutation decreases c-Epi affinity by ~100-fold in the β2AR (Supplementary Fig. 8b). The difference in position 45.52 may explain the lack of activity of constrained catecholamine to the αAR subtypes.
Signaling and signal bias
In agreement with MD simulations and radioligand binding assays, the constrained catecholamines display enhanced potency on β2AR in adenylyl cyclase assays while decreasing their potency through β1AR stimulation (Fig. 2a). Similar enhanced effects of c-Epi and c-NorEpi were observed in β-arrestin recruitment, ie. enhanced potency on β2AR and diminished potency at β1AR, but with their magnitude of differences appear to be magnified in comparison to their responses in cyclase assays. Moreover, the diminished potency of the constrained catecholamines in β1AR-promoted β-arrestin recruitment assays (19- and 25-fold decreased potency of c-Epi and c-NorEpi, respectively) is accompanied by a decreased efficacy (~50% decreased efficacy of both c-Epi and c-NorEpi). It is likely that the β1AR, known to be a relatively poor arrestin-2 recruiter16 for catecholamines, exhibits an even poorer response to constrained catecholamines due to a combination of suboptimal binding at the hormone-binding site, as described above, and the relative instability of the vestibule and ECLs associated with F45.52. Thus, c-Epi and c-NorEpi confer a combined effect of lowering activity at β1AR while improving potency at β2AR, yielding an overall improvement in β2AR-β1AR selectivity.
To compare the relative effectiveness of the constrained catecholamines in cAMP and β-arrestin assays, we analyzed the dose-response relationships using an Operational Model of Bias, proposed by ref. 17. In brief, we first calculated the “transduction coefficient” of each ligand in cAMP and arrestin recruitment assays. The transduction coefficient is defined as log(τ/Ka), where τ represents the efficacy of the agonist and Ka represents the dissociation constant of the ligand (Supplementary Table 6a). Then the relative effectiveness (RE) between the two agonists were calculated as the inverse logarithm of Δlog(τ/Ka) between these two ligands. An RE >1 implies that a new agonist, relative to a control agonist, displays superior pharmacological properties based on efficacy and affinity. The bias factors, or the comparison between the RE of the two agonists in two separate signaling assays, was calculated as the inverse logarithm of ΔΔlog(τ/Ka) of the same pair of agonists in two different signaling pathways (Supplementary Table 6b). In cAMP assays with β2AR, c-Epi (RE~4), and c-NorEpi (RE ~20), compared to Epi and NorEpi, respectively, were both much higher than on β1AR (Supplemental Table 6). In β-arrestin recruitment assays, both c-Epi and c-NorEpi appeared dramatically more effective (c-Epi: RE~21, c-NorEpi: RE ~65) compared to Epi and NorEpi, respectively. Taken together, these data suggest that both c-Epi (bias factor ~5.4) and c-NorEpi (bias factor ~3.2) on β2AR display a signaling bias toward β-arrestin recruitment over adenylyl cyclase activity. It is important to note that although c-NorEpi displays a larger bias factor of ~ 51 on β1AR, the relative effectiveness (RE) for c-NorEpi on β1AR was ~2400-fold lower than NorEpi in cyclase assays and ~47-fold lower in β-arrestin recruitment assays. Interestingly, constraining isoproterenol had a major impact on β-arrestin recruitment through β1AR (100-fold decrease in potency compared to ISO) with little enhancing effect on β-arrestin recruitment by β2AR. This was accompanied by only modest effects on β1AR potency in cAMP assays and subtle effects on β2AR-mediated efficacy. Taken together, these data suggest that constraining isoproterenol appears as a β1AR-selective diminution, yielding a large effect on the relative effectiveness and hence biasing the signaling away from β-arrestin recruitment.
Discussion
A common goal in drug development is to target specific receptor subtypes and avoid off-target binding events that often lead to adverse side effects. However, distinguishing specific receptor subtypes that natively bind the same hormone has been a major challenge in drug discovery. Indeed, advances in the structural biology of GPCRs have provided some insight into strategies to identify subtype-selective compounds. We previously reported the development of selective antagonists for the M3 muscarinic acetylcholine receptor and an orexin 1 receptor-selective antagonist based on single amino acid differences between these subtypes (M3AChR and M2AChR or OX1R and OX2R, respectively)18,19. In this study, we further show that even for GPCR subtypes like β1AR and β2AR that share identical orthosteric pockets, the shape and stability of the orthosteric pocket can be influenced by surrounding residues resulting in marked differences in ligand affinity.
Previous studies by ref. 20 revealed that entropy plays an important role in agonist binding to the turkey β1AR. The authors found that isoproterenol binding to turkey red blood cell membranes was highly dependent on temperature, with higher affinity observed at lower temperatures. Antagonist binding was unaffected by temperature. The authors concluded that agonist binding is associated with an unfavorable decrease in entropy. In this current study, we designed constrained catecholamines to determine the effect of reducing the entropic penalty of binding. Of the eight possible constrained enantiomers of iso (Fig. 1c), the (R,R)-enantiomer bound to the β2AR with the highest affinity. The constrained catecholamines c-NorEpi and c-Epi exhibited a faster association rate for binding to the β2AR compared to the non-constrained agonists (Fig. 3a and Supplementary Fig. 7), consistent with a reduced entropic penalty upon binding. However, we were surprised to find that conformationally-constrained catecholamine exhibited a high degree of β2AR selectivity, with a marked decrease in affinity for the β1AR as well as α1ARs and α2ARs (Supplementary Fig. 3). This selectivity for β2AR over β1AR was not due to differences in the residues that directly interact with the ligand in the orthosteric binding site. Binding kinetics studies and MD simulations suggest that the difference in affinity is due to a combined effect of the loss of flexibility needed to access the orthosteric pocket of the β1AR, and a less stable binding pose of c-Epi in the β1AR. When comparing the β1AR and β2AR, we found that the position of F45.52, common to both receptors, is slightly displaced in the β1AR relative to the β2AR. This difference in the orientation of F45.52 (a smaller V3.36/Cα – F45.52/Cζ distance), which leads to a change in the shape of the binding pocket and accounting for the apparent subtype selectivity, can largely be attributed to as few as four aromatic amino acids in β2AR ECLs that surround F45.52.
Entropic gains with the constrained catecholamines enhanced β2AR affinity with a simultaneous decrease in binding affinity for β1AR, observable in ligand binding and G protein coupling assays. What was unanticipated was the greater magnitude of differences in responses in β-arrestin recruitment where the constrained catecholamines appear to have a diminutive effect on β1AR, while enhancing the potency at β2AR, in the case of c-Epi and c-NorEpi. Thus, constraining NorEpi completely switched the β1AR-β2AR selectivity from β1AR-selective NorEpi responses to β2AR-selective arrestin recruitment with c-NorEpi. The mechanism for the magnitude differences in β-arrestin recruitment compared to G protein or cyclase assays, ie. enhancement for β2AR and diminution with β1AR, is unclear but potentially related to one clear structural difference in the catecholamine-bound β-ARs versus the constrained counterparts. Constraining the catecholamines through the addition of two carbon atoms and the formation of a second cyclic ring, impacts F45.52 in both β1AR and β2AR, altering the structures of the ECLs and vestibule located above the orthosteric, hormone-binding site. Accordingly, c-Epi binding to β1AR would require significant changes in the conformation of F45.52 and ECL2, whereas c-Epi binding to β2AR appears to stabilize F45.52 and ECL2 in a conformation that favors arrestin recruitment. Indeed, previous studies on the serotonin 5-HT2B receptor suggest that ligands that influence the structure of ECLs appear to influence G protein and arrestin bias13,14. Structural data on β2AR-arrestin complexes stabilized with c-Epi or c-NorEpi might provide some insight into how the ECLs may be involved in arrestin recruitment and hence help to delineate the basis for the observed effect on signaling bias.
Orthosteric pockets being allosterically modified by surrounding residues, such as those in the ECLs, are unlikely to be a unique case for adrenergic receptors. This study suggests that it may be possible to develop subtype-selective drugs for other GPCRs by carefully exploring the dynamics of the orthosteric pocket differences between subtypes. In this regard, high-resolution structures and MD simulations provide valuable information to guide drug development.
Methods
Protein expression and purification
A previously reported human β2AR – T4 lysozyme fusion construct was used in this study8. A FLAG epitope was fused to the N-terminus. T4 lysozyme was connected to the β2AR at position D291.28 with two alanine residues. The flexible ICL3 (S236-K263) was removed and the C-terminus of the receptor was truncated at position K348. The resulting T4L-β2AR construct was expressed in Sf9 insect cells with BestBac expression systems. Purification of T4L-β2AR and Nb6B9 were performed according to the methods described previously8. Briefly, Nb6B9 was purified by nickel affinity chromatography followed by size exclusion chromatography. T4L-β2AR was solubilized with dodecylmaltoside and purified by M1-FLAG affinity chromatography followed by alprenolol functional chromatography. The elution of the alprenolol column was loaded to the M1-FLAG affinity column for ligand exchange to c-Epi and detergent exchange to L-MNG. The purified T4L-β2AR was incubated with Nb6B9 overnight with a 1:1.5 molar ratio. The excess Nb6B9 was removed by a final size exclusion chromatography with the buffer containing 20 mM HEPES, pH 7.5, 100 mM NaCl, 0.01% MNG, 0.001% CHS, and 100 μM c-Epi. The purified T4L-β2AR-Nb6B9-c-Epi was concentrated at 40 mg/mL and aliquoted.
Crystallization
The T4L-β2AR-Nb6B9-c-Epi complex was reconstituted into the lipidic cubic phase with a 1:1 mass ratio of protein to lipid as previously reported21. The lipid stock consisted of a 10:1 mass ratio of 7.7 MAG with cholesterol. Crystals were grown using 30–100 nL drops with 1 μL of precipitant solution using a GryphonLCP robot. The crystal condition consisted of 26–31% PEG400, 100 mM MES, pH 6.2–6.7, 75–125 mM ammonium phosphate dibasic, and 1 mM c-Epi. Crystals grew after 1–3 days and were harvested for data collection.
Data collection and structure determination
The diffraction data were collected at SPring-8 beamline BL32XU. The micro-focused beam with 10 μm × 15 μm size and 1.0 Å wavelength was used for automatic data collection22. For each crystal, a 10° dataset was collected with 0.1° oscillation per frame. Automatic data processing was performed by KAMO23. Fifty-eight crystals were merged to generate the final 3.1 Å T4L-β2AR-Nb6B9-c-Epi dataset and 43 crystals were merged to generate the final 3.4 Å T4L-β2AR-Nb6B9-c-ISO dataset. The structure of the T4L-β2AR-Nb6B9-c-Epi complex was solved by molecular replacement method with PHENIX24 and T4L-β2AR-Nb6B9-Epinephrine structure (PDE code: 4LDO) as the search model. Structure refinement was carried out with PHENIX and COOT. Molprobity25 was used to validate the final structure. The statistics for data collection and structure refinement were summarized in Supplementary Table 7. The structure figures were prepared using PyMol (The PyMOL Molecular Graphics System, Schrodinger, LLC).
The isomorphous difference map was calculated using FFT26 in CCP427. In brief, the T4L-β2AR-Nb6B9-c-Epi data and the T4L-β2AR-Nb6B9-Epi data were set to the same scale and a Fo-Fo difference map was calculated by subtracting the T4L-β2AR-Nb6B9-Epi from the T4L-β2AR-Nb6B9-c-Epi data.
Normalized b-factor was calculated by dividing the b-factor of each residue by the overall b-factor of the receptor. To visualize the b-factor change, the residue is colored blue if the normalized b-factor is smaller in the β2AR-Nb6B9-c-Epi structure than in the β2AR-Nb6B9-Epi structure, and colored in red if the normalized b-factor is larger in the β2AR-Nb6B9-c-Epi structure than in the β2AR-Nb6B9-Epi structure. The darkness of blue or red color correlates with how large the normalized b-factor difference is between the two structures.
Radioligand binding
The wild-type and mutated human β2AR and β1AR constructs were cloned into pFastbac vector and expressed in Sf9 cells with a Bac-to-Bac expression system. The cell membrane was isolated and resuspended with a binding buffer consisting of 20 mM HEPES, pH 7.5, and 100 mM NaCl. For competition binding assays, the diluted membrane was incubated for 2 h with various concentrations of cold ligands and 2 nM [3H]DHA in the cold binding buffer containing 0.5% BSA to a final volume of 500 μL. The membrane filtration was performed with Brandel 48-well harvester and the collected filter papers and membranes were incubated with OptiPhase HiFafe 3 liquid scintillation cocktail. The Microbeta2 scintillation counter was used for radioactivity counting. The competition binding curves were fitted by GraphPad Prism 6.0 (GraphPad LLC, CA).
Cloning
The human β1AR, β2AR, α1A, α1B, β1AR_mut4, β2AR_mut4, and the murine α2A receptor were fused to the PK1 sequence, the α2B and α2C receptor to ARMS2-PK2 and all cloned to pCMV (DiscoverX, Eurofins) for β-arrestin-2 recruitment assays, respectively, using polymerase chain reaction and Gibson Assembly (New England Biolabs)28. Sequence integrity was verified by DNA sequencing (Eurofins Genomics). The β1AR/β2AR chimeras were generated by switching the N-terminus of the receptors as well as 55 residues between W1.31 and the C-terminus of the receptor. The detailed sequences were shown in Supplementary Figure 9, which is modified from a previous publication from our group7.
Radioligand binding assay with membranes from HEK cells
Binding affinities towards the human β1AR and β2AR were determined as described previously29,30. In brief, membranes were prepared from HEK293T cells transiently transfected with the cDNA for β1AR and β2AR (obtained from the cDNA resource center, www.cdna.org). Receptor densities (Bmax value) and specific binding affinities (KD value) for the radioligand [³H]CGP12,177 (specific activity 51 Ci/mmol, PerkinElmer, Rodgau, Germany) were determined as 4.3 ± 1.1 pmol/mg protein and 0.125 ± 0.032 nM for β1AR and 2.7 ± 0.4 pmol/mg protein and 0.080 ± 0.011 nM for β2AR, respectively. Competition binding experiments were performed by incubating membranes in binding buffer (25 mM HEPES, 5 mM MgCl2, 1 mM EDTA, and 0.006% bovine serum albumin at pH 7.4) at a final protein concentration of 2–6 µg/well, together with the radioligand (final concentration 0.2–0.3 nM) and varying concentrations of the competing ligands for 60 min at 37 °C. Non-specific binding was determined in the presence of unlabeled CGP12,177 at a final concentration of 10 µM. Protein concentration was established using the method of ref. 31.
The resulting competition curves were analyzed by nonlinear regression using the algorithms implemented in PRISM 8.0 (GraphPad Software, San Diego, CA) to provide an IC50 value, which was subsequently transformed into a Ki value employing the equation of Cheng and Prusoff 32. Mean Ki values (±SEM) were derived from 3 to 14 experiments, each performed in triplicates.
The binding kinetics assay
The binding kinetics assays were performed as previously described in ref. 7. In brief, off-rate measurements in membranes containing the target receptors (the human β1AR, β2AR as well as the β1ARin/β2ARout and β2ARin/β1ARout chimeras) were pre-incubated with 0.1–0.5 nM [3H]DHA for 1 h at RT. [3H]DHA dissociation rates were initiated with the addition of excess propranolol (50 μM) and samples were subjected to rapid filtration at various times. Association rates were determined by incubating membranes with [3H]DHA (ranging from 0.1–0.5 nM concentration) in the absence or presence of three different concentrations of (3, 10, and 30 μM NorEpi for β2AR; 0.3, 1, and 3 μM NorEpi for β1AR; 3, 10, and 30 μM NorEpi for β1ARin/β2ARout; 0.3, 1, and 3 μM NorEpi for β2ARin/β1ARout; 1, 3, and 10 μM c-NorEpi on β2AR; 3, 10, and 30 μM c-NorEpi for β1AR; 1, 3, and 10 μM c-NorEpi on β1AR and β2AR; 3, 10, and 30 μM c-NorEpi for β1ARin/β2ARout; 1, 3, and 10 μM c-NorEpi on β2ARin/β1ARout; 0.3, 1, and 3 μM Epi for β2AR; 1, 3, and 10 μM Epi on β1AR; 0.3, 1, and 3 μM Epi for β2ARin/β1ARout; 3, 10, and 30 μM Epi for β1ARin/β2ARout; 3, 10, and 30 μM; c-Epi for β1AR; 0.1, 0.3, and 1 μM; c-Epi for β2AR; 3, 10, and 30 μM c-Epi for β2ARin/β1ARout; 3, 10, and 30 μM c-Epi for β1ARin/β2ARout) the catecholamine. Aliquots were removed at various times and subjected to rapid filtration. Membranes containing bound [3H]DHA were harvested by filtering through GF/C UnifilterTM (Perkin Elmer) and counted on a Top CountTM (Perkin Elmer) scintillation counter. Binding kinetics were calculated using the Kinetics of Competitive Binding fit in GraphPad Prism 6.0 (GraphPad LLC, CA). The kinetics parameters of [3H]DHA used in the analysis were derived from a previous study7.
β-Arrestin-2 recruitment and G protein IP-one assay
Determination of receptor-stimulated β-arrestin-2 recruitment was performed applying the PathHunter assay (DiscoverX, Birmingham, UK), which is based on the measurement of fragment complementation of β-galactosidase as described in ref. 33. In detail, HEK293T cells stably expressing the enzyme acceptor (EA) tagged β-arrestin-2 fusion protein were transfected with the cDNA for β1AR, β2AR, β1AR_mut4, β2AR_mut4, α1A, α1B, or α2A receptor each fused to the ProLink-PK1 fragment for enzyme complementation and transferred into 384 well microplates. α2B and α2C receptors fused to the ProLink-ARMS2-PK2 fragment were treated analogously. Measurement started by incubating cells with agonist for 90 min (β1AR, β2AR, β2AR_mut4, α2B, or α2C, respectively), 180 min (β1AR_mut4, α1B, or α2A, respectively), or 300 min (α1A), respectively. Chemoluminescence was monitored with a Clariostar plate reader (BMG, Ortenberg, Germany) and analyzed by normalizing the raw data relative to basal activity (0%) and the maximum effect of Norepi (100%). Four to 21 repeats in duplicate were analyzed by applying the algorithms for four-parameter nonlinear regression implemented in Prism 8.0 (GraphPad LLC, CA) to get dose-response curves representing EC50 and Emax value. Bias calculations were performed as described by ref. 17.
The determination of receptor-mediated G protein signaling by β1AR and β2AR was performed by applying an IP accumulation assay (IP-One HTRF®, Cisbio, Codolet, France) according to the manufacturer’s protocol and in analogy to previously described protocols18. In brief, HEK 293 T cells were co-transfected with the cDNA for β1AR or β2AR and the hybrid G protein Gαqs (Gαq protein with the last five amino acids at the C-terminus replaced by the corresponding sequence of Gαs (gift from The J. David Gladstone Institutes, San Francisco, CA), respectively and transferred into 384 well microplates. Cells were incubated with an agonist for 120 min and accumulation of the second messenger was stopped by adding detection reagents (IP1-d2 conjugate and Anti-IP1cryptate TB conjugate). After 60 min, TR-FRET was measured with a Clariostar plate reader. FRET-signals from four to 14 repeats in duplicates were normalized to vehicle (0%) and the maximum effect of Norepi (100%) and analyzed to get EC50 and Emax values.
cAMP accumulation assay
Initial cAMP assays on β2AR were performed using clonal selected HEK293 ΔGNAS cell line34, stably expressing the cAMP biosensor pink flamido35 (encoded on a pcDNA4.0/TO/Zeocin plasmid, generously provided by Dr. Jin Zhang, UCSD). Briefly, cells were treated with 4 μg/mL doxycycline to induce overexpression of the cAMP biosensor 48 h prior to measurement of cAMP. Cells were gently harvested, washed in Hanks’ Balanced Salt Solution (HBSS, Sigma), and seeded onto clear bottom poly-d-lysine coated, black 96-well polystyrene assay plate (Costar) at ~2 × 106 cells per well. Cells were treated with a dilution series of catecholamine (10−4–10−10 M) in HBSS buffer with 20 mM HEPES, pH 7.8, 600 μM 3-Isobutyl-1-methylxanthine (IBMX), and 3 mM ascorbic acid. Real-time fluorescence measurements were collected immediately following agonist application: excitation 535 nm, emission 612 nm, integration time 40 µs, bottom read, a kinetic interval of 13 s, and a delay of 10 ms using a fluorescence plate reader (BioTek, Winooski, VT). The cAMP accumulation was monitored for 17.5 min. The fluorescence emission data were fitted to a single exponential to obtain rate constants (from t = 5 s to t = 305 s) for each agonist concentration using Prizm (GraphPad LLC, CA). Rate constants were plotted as a function of catecholamine concentration and fitted to a logistics curve using Prism.
Subsequent cAMP assays on β1AR and β2AR were performed in Sf9 cells. Briefly, cells were infected with baculoviruses for the cAMP biosensor pink flamido, together with either β1AR or β2AR. Twenty-four hours following infection, cells were gently harvested, washed in HBSS, and seeded onto clear bottom poly-d-lysine coated, black 96-well polystyrene assay plate (Costar) at ~1 × 106 cells per well. Cells were treated with a dilution series of catecholamine (10−4–10−10 M) in HBSS buffer with 20 mM HEPES, pH 7.8, 600 μM IBMX, and 3 mM ascorbic acid. Real-time fluorescence measurements were collected immediately following agonist application: excitation 535 nm, emission 612 nm, integration time 40 µs, bottom read, a kinetic interval of 13 s, and a delay of 10 ms using a fluorescence plate reader (SpectraMax M5, Molecular Devices, CA). The cAMP accumulation was monitored for 10 min. The fluorescence emission data were fitted to a single exponential to obtain rate constants (from t = 5 s to t = 305 s) for each agonist concentration using Prizm (GraphPad LLC, CA). Rate constants were plotted as a function of catecholamine concentration and fitted to a logistics curve using Prism. All statistical analysis for EC50 and Emax, as well as relative effectiveness and bias, was performed using Prism. Relative effectiveness and Bias calculations were performed as described by ref. 17.
[35S]GTPγS binding assays
Membranes were prepared from Sf9 cells expressing Gαsβ1γ2 and β2AR. Membranes (~15 μg) were pretreated with GDP (final assay concentration of 1 μM) in a GTPγS assay buffer (20 mM HEPES, pH 7.4, 100 mM NaCl, 10 mM MgCl2, and 1 mM ascorbic acid) and different concentrations of agonist (Epi, c-Epi, NorEpi, c-NorEpi, Iso or c-Iso) for 10 min at room temperature before adding [35S]GTPγS (for a final concentration of 0.1 nM). The assay was incubated at 30 oC for 30 min before stopping by rapid filtration through GF/B Unifilter plates (Whatman) and washing with ice-cold assay buffer. Filter plates were dried before adding Microscint 0™ and counting bound [35S]GTPγS (Perkin Elmer) using a TopCount™ (Perkin Elmer). Data were analyzed using Prizm 6.0 (GraphPad, LLC, CA). Figures show the combined results from three separate experiments performed in duplicate.
Unbiased simulations of receptor-ligand complexes
Simulations of β2AR were based on the c-Epi bound crystal structure described in this manuscript and the epinephrine-bound crystal structure (PDB entry 4LDO)8. Simulations of β1AR were based on a BI-167107 bound crystal structure of β1AR (PDB entry 7BU7)7. For the simulations of β1AR, BI-167107 was replaced with either Epi or c-Epi by structurally aligning the crystal structures β1AR and the respective β2AR and transferring the coordinates of the ligands. Mutations to β1AR were introduced utilizing Maestro (Schrödinger, LCC, New York, NY, 2018) while selecting the rotamers with the highest probability and at the same time, resembling the respective conformation in the c-Epi bound β2AR crystal structure.
Coordinates were prepared by removing the nanobody and the T4L fusion protein. Only crystal waters within or close to the receptor were retained. Prime (Schrödinger, LCC, New York, NY, 2018) was used to model missing side chains. Hydrogen atoms were added, and the protein chain termini were capped with the neutral acetyl and methyl amide groups.
Except for D2.50, E3.41, and D3.49, all titratable residues were left in their dominant protonation state at pH 7.0. Previous studies suggested that residues D2.50 and D3.49 for β2AR are protonated in the active state36,37, and residue E3.41 directly contacts the lipid interface and, therefore, will also exist predominantly in its protonated state38,39. We thus protonated these three residues in our simulations. Epi and c-Epi were protonated at the secondary amine allowing the formation of the canonical salt bridge to D3.32 conserved in aminergic GPCRs.
The prepared protein structures were then aligned to the Orientation of Proteins in Membranes (OPM)40 structure of active β2AR (PDB entry 3SN6) and internal water was added utilizing Dowser41. Each complex was inserted into a pre-equilibrated bilayer of palmitoyl-oleoyl-phosphatidylcholine (POPC) lipids using Dabble, a simulation preparation software (Betz, R. Dabble (v.2.6.3). 10.5281/zenodo.836914 (2017)). Sodium and chloride ions were added to neutralize each system at a concentration of approximately 150 mM. The final box dimensions were approximately 85 × 75 × 85 Å3.
We used the CHARMM36m42–46 parameter set for protein molecules, lipid molecules, and salt ions and the CHARMM TIP3P model for water. Parameters for Epi and c-Epi were generated using the CHARMM General Force Field (CGenFF)47–49 with the ParamChem server (paramchem.org) version 1.0.0.
We performed the simulations using the CUDA version of PMEMD (particle-mesh Ewald molecular dynamics) in AMBER1650. We performed six independent simulations under each condition. Each simulation was equilibrated independently as follows: Three rounds of minimization were performed, each consisting of 500 iterations of steepest descent minimization, followed by 500 iterations of conjugate gradient descent minimization, with harmonic restraints of 10.0, 5.0, and 1.0 kcal mol−1 Å−2 placed on the protein and lipids, respectively. Systems were heated from 0 to 100 K in the NVT ensemble over 12.5 ps and then from 100 to 310 K in the NPT ensemble over 125 ps, using 10.0 kcal mol−1 Å−2 harmonic restraints applied to lipid and protein-heavy atoms. Systems were then equilibrated at 310 K in the NPT ensemble at 1 bar, with harmonic restraints on all protein-heavy atoms for 10 ns. Starting at 5.0 kcal mol−1 Å−2, the restraints were reduced in a stepwise fashion by 1.0 kcal mol−1 Å−2 every 2 ns. This was followed by additional 20 ns of equilibration with again harmonic restraints on all protein-heavy atoms. Starting at 1.0 kcal mol−1 Å−2, the restraints were reduced in a stepwise fashion by 0.1 kcal mol−1 Å−2 every 2 ns. Production simulations were performed in the NPT ensemble at 310 K and 1 bar, using a Langevin thermostat for temperature coupling and a Monte Carlo barostat for pressure coupling. These simulations used a 4 fs time step with hydrogen mass repartitioning51. Bond lengths to hydrogen atoms were constrained using SHAKE52. Periodic boundary conditions were applied. Non-bonded interactions were cut off at 9.0 Å, and long-range electrostatic interactions were computed using particle-mesh Ewald (PME)53 with an Ewald coefficient of approximately 0.31 Å and an interpolation order of four. The fast Fourier transform (FFT) grid size was chosen such that the width of a grid cell was ~1 Å.
During production simulations, all residues within 5 Å of the G protein interface at β2AR were restrained to the initial structure using 5.0 kcal mol−1 Å−2 harmonic restraints applied to non-hydrogen atoms. Using such restraints instead of the intracellular binding partner reduces the overall system size, enabling faster simulation, while ensuring that the receptor maintains an active conformation throughout the simulation.
Trajectory snapshots were saved every 200 ps during production simulations. Analysis of the trajectories was performed using Visual Molecular Dynamics (VMD)54, CPPTRAJ55, and GetContacts (https://getcontacts.github.io/). Root-mean-square fluctuations (rmsf) were calculated to an average structure omitting the first 500 ns of each simulation trajectory to avoid including any initial relaxation or equilibration of the system. Trajectories were aligned to the initial crystal structure on all transmembrane helix alpha carbons. For each simulation condition, an average structure was generated by considering trajectory snapshots from all simulations under that condition. The rmfs for each alpha carbon was then calculated for each trajectory under that condition relative to this average position using CPPTRAJ. Visualization was performed using the PyMOL Molecular Graphics System, Version 2.1.1 (Schrödinger, LLC). Plots were created using Matplotlib 3.0.256.
Unbiased simulations of ligands in solution
Parameter topology and coordinate files were built up using the tleap module of AMBER1857. For the simulations, the general AMBER force field (GAFF)58 was used for Epi and c-Epi. The ligands were solvated in a truncated octahedron with a minimum solute-to-wall distance of 25 Å. The systems were neutralized with one chloride ion. The TIP3P water model59 was applied. Parameters for Epi and c-Epi were assigned using antechamber57. The structure of Epi and c-Epi were optimized using Gaussian 1660 at the B3LYP/6-31 G(d) level of theory and charges were calculated at HF/6-31 G(d) level of theory. Subsequently, atom point charges were assigned according to the RESP procedure61. A formal charge of +1 was assigned to Epi and c-Epi.
We performed the simulations using the CUDA version of PMEMD in AMBER1857. Each simulation system was energy minimized 2500 iterations of steepest descent minimization, followed by 7500 iterations of conjugate gradient descent minimization and equilibrated in the NVT ensemble at 310 K for 1 ns followed by the NPT ensemble for 1 ns with harmonic restraints of 10.0 kcal mol–1 on the ligand. In the NVT ensemble, the Langevin thermostat was used. In the NPT ensemble, the Monte Carlo barostat was applied. The system was further equilibrated for 12 ns with restraints on the ligand atoms. Here, the restraints were reduced every 3 ns in a stepwise fashion to be 5.0, 1.0, 0.5, and 0.1 kcal mol−1, respectively. Productive simulations were performed using periodic boundary conditions and a time step of 2 fs. Bond lengths to hydrogen atoms were constrained using SHAKE52. Long-range electrostatic interactions were computed using the PME53 method. Non-bonded interactions were cut off at 8.0 Å.
Trajectory snapshots were saved every 50 ps during production simulations. Trajectories were visualized using VMD54 and cluster based on the ligand atoms using the CPPTRAJ55 module of AMBER18. Visualization was performed using PyMOL Molecular Graphics System, Version 2.1.1 (Schrödinger, LLC). Plots were created using Matplotlib 3.0.256.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We acknowledge the computing resources and support provided by the Erlangen Regional Computing Center (RRZE) and support provided by the Radioisotope Laboratory, Center of Biomedical Analysis, Tsinghua University. This work was supported by the Tsinghua-Peking Center for Life Sciences, Beijing Frontier Research Center for Biological Structure, and Beijing Advanced Innovation Center for Structural Biology, Tsinghua University (X.X. and X.L.), by National Natural Science Foundation of China (Grant 32122041 to X.L. and Grant 32100968 to X.X.), China Postdoctoral Science Foundation 2021M691809 (X.X.), the DFG grant GRK 1910 (P.G., J.S., and J.K.), and National Institute of General Medical Sciences grants GM106990 (B.K.K., P.G., A.S., and R.K.S.), GM083118 (R.K.S.), and R01GM127359 (R.O.D.). B.K.K. is a Chan Zuckerberg Biohub investigator and an Einstein BIH Visiting Fellow.
Source data
Author contributions
X.X. and X.L. performed protein expression, purification, crystallization, structure determination, and refinement. J.S., A.S., and L.M. performed the design, chemical synthesis, and analytical characterization of compounds. J.K. and A.J.V. performed and analyzed MD simulations supervised by R.O.D. M.J.C. and D.M. characterized the pharmacology properties of βARs and mutants supervised by R.K.S. and H.H. and P.G. X.X. performed the mutagenesis studies and radioligand binding of βAR mutants supervised by X.L. and B.K.K. K.H. performed automatic data collection and processing. The manuscript was written by X.L., R.K.S., and B.K.K., with input and suggestions from X.X., H.H., J.K., and P.G. All authors contributed to the editing of the manuscript. R.K.S., P.G., and B.K.K. initiated the project, coordinated the experiments, and supervised the overall research.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work.
Data availability
The coordinates and structural factors of T4L-β2AR/NB6B9/c-Epi and T4L-β2AR/NB6B9/c-ISO structures have been deposited into Protein Data Bank under the accession code 7XKA (T4L-β2AR/NB6B9/c-Epi structure) and 7XK9 (T4L-β2AR/NB6B9/c-ISO structure), respectively. Source data are provided with this paper.
Competing interests
B.K.K. is a co-founder of and consultant for ConfometRx, Inc. The remaining authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
5/24/2023
A Correction to this paper has been published: 10.1038/s41467-023-38820-y
Contributor Information
Brian K. Kobilka, Email: kobilka@stanford.edu
Roger K. Sunahara, Email: rsunahara@health.ucsd.edu
Xiangyu Liu, Email: liu_xy@mail.tsinghua.edu.cn.
Peter Gmeiner, Email: peter.gmeiner@fau.de.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-023-37808-y.
References
- 1.Sriram K, Insel PA. G protein-coupled receptors as targets for approved drugs: how many targets and how many drugs. Mol. Pharm. 2018;93:251–258. doi: 10.1124/mol.117.111062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan HCS, Li Y, Dahoun T, Vogel H, Yuan S. New binding sites, new opportunities for GPCR drug discovery. Trends Biochem. Sci. 2019;44:312–330. doi: 10.1016/j.tibs.2018.11.011. [DOI] [PubMed] [Google Scholar]
- 3.Ogrodowczyk M, Dettlaff K, Jelinska A. Beta-blockers: current state of knowledge and perspectives. Mini Rev. Med. Chem. 2016;16:40–54. doi: 10.2174/1389557515666151016125948. [DOI] [PubMed] [Google Scholar]
- 4.Billington CK, Penn RB, Hall IP. β(2) agonists. Handb. Exp. Pharm. 2017;237:23–40. doi: 10.1007/164_2016_64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Minneman KP, Pittman RN, Molinoff PB. Beta-adrenergic receptor subtypes: properties, distribution, and regulation. Annu. Rev. Neurosci. 1981;4:419–461. doi: 10.1146/annurev.ne.04.030181.002223. [DOI] [PubMed] [Google Scholar]
- 6.Dror RO, et al. Pathway and mechanism of drug binding to G-protein-coupled receptors. Proc. Natl Acad. Sci. USA. 2011;108:13118–13123. doi: 10.1073/pnas.1104614108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu X, et al. Binding pathway determines norepinephrine selectivity for the human beta1AR over beta2AR. Cell Res. 2021;31:569–579. doi: 10.1038/s41422-020-00424-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ring AM, et al. Adrenaline-activated structure of beta2-adrenoceptor stabilized by an engineered nanobody. Nature. 2013;502:575–579. doi: 10.1038/nature12572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Warne T, Edwards PC, Dore AS, Leslie AGW, Tate CG. Molecular basis for high-affinity agonist binding in GPCRs. Science. 2019;364:775–778. doi: 10.1126/science.aau5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishikawa M, et al. Selective beta-adrenoceptor activities of tetrahydronaphthalene derivatives. Life Sci. 1975;16:305–314. doi: 10.1016/0024-3205(75)90029-6. [DOI] [PubMed] [Google Scholar]
- 11.DeVree BT, et al. Allosteric coupling from G protein to the agonist-binding pocket in GPCRs. Nature. 2016;535:182–186. doi: 10.1038/nature18324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kenakin T, Strachan RT. PAM-antagonists: a better way to block pathological receptor signaling. Trends Pharm. Sci. 2018;39:748–765. doi: 10.1016/j.tips.2018.05.001. [DOI] [PubMed] [Google Scholar]
- 13.Wacker D, et al. Crystal structure of an LSD-bound human serotonin receptor. Cell. 2017;168:377. doi: 10.1016/j.cell.2016.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCorvy JD, et al. Structure-inspired design of beta-arrestin-biased ligands for aminergic GPCRs. Nat. Chem. Biol. 2018;14:126–134. doi: 10.1038/nchembio.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuan D, et al. Activation of the alpha2B adrenoceptor by the sedative sympatholytic dexmedetomidine. Nat. Chem. Biol. 2020;16:507–512. doi: 10.1038/s41589-020-0492-2. [DOI] [PubMed] [Google Scholar]
- 16.Shiina T, Kawasaki A, Nagao T, Kurose H. Interaction with beta-arrestin determines the difference in internalization behavor between beta1- and beta2-adrenergic receptors. J. Biol. Chem. 2000;275:29082–29090. doi: 10.1074/jbc.M909757199. [DOI] [PubMed] [Google Scholar]
- 17.van der Westhuizen ET, Breton B, Christopoulos A, Bouvier M. Quantification of ligand bias for clinically relevant beta2-adrenergic receptor ligands: implications for drug taxonomy. Mol. Pharm. 2014;85:492–509. doi: 10.1124/mol.113.088880. [DOI] [PubMed] [Google Scholar]
- 18.Liu H, et al. Structure-guided development of selective M3 muscarinic acetylcholine receptor antagonists. Proc. Natl Acad. Sci. USA. 2018;115:12046–12050. doi: 10.1073/pnas.1813988115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hellmann J, et al. Structure-based development of a subtype-selective orexin 1 receptor antagonist. Proc. Natl Acad. Sci. USA. 2020;117:18059–18067. doi: 10.1073/pnas.2002704117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weiland GA, Minneman KP, Molinoff PB. Fundamental difference between the molecular interactions of agonists and antagonists with the beta-adrenergic receptor. Nature. 1979;281:114–117. doi: 10.1038/281114a0. [DOI] [PubMed] [Google Scholar]
- 21.Caffrey M, Cherezov V. Crystallizing membrane proteins using lipidic mesophases. Nat. Protoc. 2009;4:706–731. doi: 10.1038/nprot.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirata K, et al. ZOO: an automatic data-collection system for high-throughput structure analysis in protein microcrystallography. Acta Crystallogr. D. Struct. Biol. 2019;75:138–150. doi: 10.1107/S2059798318017795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamashita K, Hirata K, Yamamoto M. KAMO: towards automated data processing for microcrystals. Acta Crystallogr. D. Struct. Biol. 2018;74:441–449. doi: 10.1107/S2059798318004576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D. Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. Sect. D. Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ten Eyck LF. Fast Fourier transform calculation of electron density maps. Methods Enzymol. 1985;115:324–337. doi: 10.1016/0076-6879(85)15024-X. [DOI] [PubMed] [Google Scholar]
- 27.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. Sect. D., Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 29.Stanek M, et al. Hybridization of beta-adrenergic agonists and antagonists confers G protein bias. J. Med Chem. 2019;62:5111–5131. doi: 10.1021/acs.jmedchem.9b00349. [DOI] [PubMed] [Google Scholar]
- 30.Hubner H, et al. Structure-guided development of heterodimer-selective GPCR ligands. Nat. Commun. 2016;7:12298. doi: 10.1038/ncomms12298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951;193:265–275. doi: 10.1016/S0021-9258(19)52451-6. [DOI] [PubMed] [Google Scholar]
- 32.Cheng Y, Prusoff WH. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharm. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 33.Liu X, et al. An allosteric modulator binds to a conformational hub in the beta2 adrenergic receptor. Nat. Chem. Biol. 2020;16:749–755. doi: 10.1038/s41589-020-0549-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stallaert W, et al. Purinergic receptor transactivation by the beta2-adrenergic receptor increases intracellular Ca(2+) in nonexcitable cells. Mol. Pharm. 2017;91:533–544. doi: 10.1124/mol.116.106419. [DOI] [PubMed] [Google Scholar]
- 35.Harada K, et al. Red fluorescent protein-based cAMP indicator applicable to optogenetics and in vivo imaging. Sci. Rep. 2017;7:7351. doi: 10.1038/s41598-017-07820-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghanouni P, et al. The effect of pH on beta(2) adrenoceptor function. Evidence for protonation-dependent activation. J. Biol. Chem. 2000;275:3121–3127. doi: 10.1074/jbc.275.5.3121. [DOI] [PubMed] [Google Scholar]
- 37.Ranganathan A, Dror RO, Carlsson J. Insights into the role of Asp79(2.50) in beta2 adrenergic receptor activation from molecular dynamics simulations. Biochemistry. 2014;53:7283–7296. doi: 10.1021/bi5008723. [DOI] [PubMed] [Google Scholar]
- 38.Dror RO, et al. Identification of two distinct inactive conformations of the beta2-adrenergic receptor reconciles structural and biochemical observations. Proc. Natl Acad. Sci. USA. 2009;106:4689–4694. doi: 10.1073/pnas.0811065106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosenbaum DM, et al. Structure and function of an irreversible agonist-beta(2) adrenoceptor complex. Nature. 2011;469:236–240. doi: 10.1038/nature09665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lomize MA, Lomize AL, Pogozheva ID, Mosberg HI. OPM: orientations of proteins in membranes database. Bioinformatics. 2006;22:623–625. doi: 10.1093/bioinformatics/btk023. [DOI] [PubMed] [Google Scholar]
- 41.Zhang L, Hermans J. Hydrophilicity of cavities in proteins. Proteins. 1996;24:433–438. doi: 10.1002/(SICI)1097-0134(199604)24:4<433::AID-PROT3>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 42.Best RB, Mittal J, Feig M, MacKerell AD., Jr. Inclusion of many-body effects in the additive CHARMM protein CMAP potential results in enhanced cooperativity of α-helix and β-hairpin formation. Biophys. J. 2012;103:1045–1051. doi: 10.1016/j.bpj.2012.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Best RB, et al. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ(1) and χ(2) dihedral angles. J. Chem. Theory Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang J, MacKerell AD., Jr. CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J. Comput. Chem. 2013;34:2135–2145. doi: 10.1002/jcc.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klauda JB, et al. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang J, et al. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods. 2017;14:71–73. doi: 10.1038/nmeth.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vanommeslaeghe K, et al. CHARMM general force field: a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vanommeslaeghe K, Raman EP, MacKerell AD., Jr. Automation of the CHARMM general force field (CGenFF) II: assignment of bonded parameters and partial atomic charges. J. Chem. Inf. Model. 2012;52:3155–3168. doi: 10.1021/ci3003649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vanommeslaeghe K, MacKerell AD., Jr. Automation of the CHARMM general force field (CGenFF) I: bond perception and atom typing. J. Chem. Inf. Model. 2012;52:3144–3154. doi: 10.1021/ci300363c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Case, D. A. et al. AMBER 2016. University of California, San Francisco (2016).
- 51.Hopkins CW, Le Grand S, Walker RC, Roitberg AE. Long-time-step molecular dynamics through hydrogen mass repartitioning. J. Chem. Theory Comput. 2015;11:1864–1874. doi: 10.1021/ct5010406. [DOI] [PubMed] [Google Scholar]
- 52.Ryckaert J-P, Ciccotti G, Berendsen HJC. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. doi: 10.1016/0021-9991(77)90098-5. [DOI] [Google Scholar]
- 53.Darden T, York D, Pedersen L. Particle mesh Ewald - an N.Log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. doi: 10.1063/1.464397. [DOI] [Google Scholar]
- 54.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 55.Roe DR, Cheatham TE., 3rd PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013;9:3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 56.Hunter JD. Matplotlib: a 2D graphics environment. Comput. Sci. Eng. 2007;9:90–95. doi: 10.1109/MCSE.2007.55. [DOI] [Google Scholar]
- 57.Case, D. A. et al. AMBER 2018. University of California, San Francisco (2018).
- 58.Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J. Comput. Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 59.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. doi: 10.1063/1.445869. [DOI] [Google Scholar]
- 60.Frisch, M. J. et al. Gaussian, Inc., Gaussian 16. Revision C.01, Wallingford CT (2016).
- 61.Bayly CI, Cieplak P, Cornell W, Kollman PA. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model. J. Phys. Chem. 1993;97:10269–10280. doi: 10.1021/j100142a004. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The coordinates and structural factors of T4L-β2AR/NB6B9/c-Epi and T4L-β2AR/NB6B9/c-ISO structures have been deposited into Protein Data Bank under the accession code 7XKA (T4L-β2AR/NB6B9/c-Epi structure) and 7XK9 (T4L-β2AR/NB6B9/c-ISO structure), respectively. Source data are provided with this paper.