Abstract
Background
Colorectal cancer (CRC) is a malignant tumor. Recent studies have showed circular RNA (circRNA) participates in the development of CRC. The study was designed to reveal the role of circ_0011385 in CRC progression and underneath mechanism.
Methods
The expression circ_0011385, microRNA-330-3p (miR-330-3p) and myosin VI (MYO6) mRNA were determined by quantitative real-time polymerase chain reaction. Protein expression was detected by Western blot assay. Cell proliferation was investigated by 3-(4,5)-dimethylthiahiazo (-z-y1)-3,5-di-phenytetrazoliumromide (MTT), cell colony formation and flow cytometry assays. Cell apoptosis was demonstrated by flow cytometry analysis. Cell migration and invasion were evaluated by wound-healing assay and transwell invasion assay, respectively. The binding sites between miR-330-3p and circ_0011385 or MYO6 were predicted by CircInteractome or starBase online databases, and identified by dual-luciferase reporter and RNA immunoprecipitation assays.
Results
Circ_0011385 and MYO6 expression were dramatically upregulated, while miR-330-3p expression was downregulated in CRC tissues or cells compared with control groups. Circ_0011385 expression was associated with tumor size, tumor-node-metastasis stage (TNM) stage and lymph node metastasis of CRC patients. Circ_0011385 silencing or MYO6 absence repressed cell proliferation, migration and invasion, whereas induced cell apoptosis in CRC. Additionally, miR-330-3p inhibitor or MYO6 overexpression attenuated the repressive impacts of circ_0011385 silencing on CRC process. Circ_0011385 was associated with miR-330-3p, and miR-330-3p targeted MYO6. Circ_0011385 knockdown inactivated MEK1/2/ERK1/2 signaling pathway by miR-330-3p/MYO6 axis. Furthermore, circ_0011385 knockdown suppressed tumor growth in vivo.
Conclusion
Circ_0011385 regulated CRC process by miR-330-3p/MYO6 axis through MEK1/2/ERK1/2 signaling pathway, providing a novel therapeutic target for CRC.
Keywords: CRC, circ_0011385, miR-330-3p, MYO6
At a glance of commentary
Scientific background on the subject
Colorectal cancer (CRC) is a malignant tumor. Recent studies have shown circular RNA (circRNA) participates in the development of CRC. The study was designed to reveal the role of circ_0011385 in CRC progression and underneath mechanism.
What this study adds to the field
The study showed that circ_0011385 regulated CRC development via miR-330-3p/MYO6 axis through MEK1/2/ERK1/2 signaling pathway. Our findings not only lay a foundation for further revealing the pathogenesis of CRC, but also provide a potential therapeutic target for CRC.
Colorectal cancer (CRC) is the third most aggressive cancer and ranks the second in cancer-related mortality [1]. CRC chiefly affects the elderly, with more than 65% of CRC cases over 65 years old [2]. Researchers have disclosed molecular targeted therapy is a new therapeutic strategy [3,4]. Several treatments based on targeted molecule have emerged, but they are only effective in improving the quality of life of CRC patients, and the long-term survival time of the sufferers is still low [[5], [6], [7]]. Thus, exploring more reliable therapeutic targets is urgent for the therapy of CRC.
Circular RNA (circRNA) is a noncoding RNA and more stable than its linear mRNA [8]. It can modulate gene expression and biological functions by sponging microRNA (miRNA) through acting as a competitive endogenous RNA [9]. Owing to its high expression and stability, circRNA widely participates in the biological activities of cancers, such as cell proliferation, metastasis and apoptosis [10]. In CRC progression, lots of circRNAs are also involved. For example, circ_001680 [11], circ_0136666 [12] and circ_0005576 [13] were reported to function as promoters. Circ-ITGA7 [14], circ_0001946 [15] and circ_0060745 [16] were explained to repress CRC growth. In this study, we found circ_0011385 was overexpressed in CRC tissues and cell lines. However, whether circ_0011385 regulated CRC was unknown.
MiRNAs are a kind of small noncoding RNAs and about 20 nucleotides in length [17]. MiRNAs mainly control mRNA level by targeting their noncoding sequences at post-transcriptional stage [18]. Plentiful researches unveiled that miRNAs were dysregulated in CRC, and regulated CRC evolution. For example, miR-374a-3p [19] and miR-19a [20] were apparently upregulated in CRC cells, and contributed to CRC process. MiR-106b-5p [21] and miR-3622a-3p [22] were lowly expressed in CRC tissues and inhibited CRC growth. In addition, cross-sectional study has revealed miR-330-3p inhibits cell proliferation in CRC [23]. Myosin VI (MYO6) belongs to actin-associated myosin family and also participates in regulating the progression of cancer development [24]. In the study of Liu et al., we found MYO6 facilitated CRC evolution [25]. In this research, we found circ_0011385 possessed the putative binding sites of miR-330-3p, and miR-330-3p also contained the binding sequence of MYO6. Based on the above data, we hypothesized miR-330-3p/MYO6 pathway was responsible for circ_0011385-mediated CRC process.
Thus, mechanism assays were employed to reveal whether miR-330-3p was associated with circ_0011385 or MYO6. Rescue experiments were used to demonstrate the connection among circ_0011385, miR-330-3p and MYO6 in modulating cell proliferation, migration, invasion and apoptosis in CRC.
Materials and methods
Specimen collection and store
Fifty pairs of human CRC tissues and adjacent normal tissues were collected from CRC sufferers from Renmin Hospital of Wuhan University. Obtained specimens were kept at −80 °C in a freezer. All CRC sufferers signed the written informed consents before operation, and this study was allowed by the Ethics Committee of Renmin Hospital of Wuhan University.
Cell purchase and culture
Human normal colonic epithelial cell line NCM460 was obtained from Otwo Biotech (Shenzhen, China). Human CRC cell lines, SW480, SW620, LOVO and HCT116, were purchased from Procell (Wuhan, China). NCM460, SW480 and HCT116 cells were cultivated in Roswell Park Memorial Institute-1640 (RPMI-1640; Procell), SW620 and LOVO cells were grown in Dulbecco's modified Eagle's medium (DMEM; Procell). Medium was supplemented with 10% fetal bovine serum (FBS; Procell) and antibiotics (100 μg/mL penicillin, 100 μg/mL streptomycin) (Procell). Cells were grown at 37 °C in a humid incubator with 5% CO2.
Cell transfection
The small interfering (si) RNAs against circ_0011385 (si-circ_0011385) and MYO6 (si-MYO6), the small hairpin RNA against circ_0011385 (sh-circ_0011385) and control groups (si-NC and sh-NC) were synthesized by GENEWIZ (Suzhou, China). The mimic and inhibitor of miR-330-3p (miR-330-3p and anti-miR-330-3p) and their controls (miR-NC and anti-miR-NC) were provided by GenePharma (Shanghai, China). The overexpression plasmid of MYO6 (MYO6) was built by inserting the complete sequence of MYO6 into pcDNA 3.1 (vector; EK-Bioscience, Shanghai, China). Si-circ_0011385, anti-miR-330-3p, si-MYO6, MYO6 and their controls were used to reveal whether circ_0011385 regulated CRC development by miR-330-3p/MYO6 axis in vitro. MiR-330-3p and miR-NC were transfected into cells to reveal the associated relationships among circ_0011385, miR-330-3p and MYO6. Sh-circ_0011385 and sh-NC were built to disclose the impacts of circ_0011385 silencing on tumor formation in vivo. Cell transfection was performed using Lipofectamine 2000 (Thermo Fisher, Waltham, MA, USA). The synthesized sequences were si-circ_0011385 5′-AGTATAGTGCCAAGGAAAGCA-3′, si-MYO6 5′-CCTAGAAGCCTTTGGAAAT-3′, miR-330-3p 5′-GCAAAGCACACGGCCUGCAGAGA-3′, anti-miR-330-3p 5′-UCUCUGCAGG-CCGUGUGCUUUGC-3′, si-NC 5′-CCTGAATCCGTTAGAGAAT-3′, miR-NC 5′-UUUGUACUACACAAAAGUACUG-3′ and anti-miR-NC 5′-CAGUACUUUUGUGUAGUACAAA-3′.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Tissues and cells were lysed and purified using miRNeasy Mini Kit (Qiagen, Valencia, CA, USA). cDNA was synthesized with FastKing RT Kit (Tiangen, Beijing, China) or MicroRNA RT kit (Thermo Fisher). In order to determine the expression levels of circRNA/miRNA/mRNA, Mx3000P system (Stratagene, Santa Clara, CA, USA) was employed with SuperReal PreMix Color mix (Tiangen). The obtained data were assessed with the 2-ΔΔCt method. U6 and β-actin served as internal references. Relevant primers were displayed in Table 1.
Table 1.
Primer sequences used in qRT-PCR in this research.
| Gene | Sequences |
|---|---|
| circ_0011385 sense | 5′-GACAACAATGAGCCCTACA-3′ |
| circ_0011385 anti-sense | 5′-AGCCGAATTGGTCTTGAGAA-3′ |
| EIF3I sense | 5′-TAACCACAACCTCCACCA-3′ |
| EIF3I anti-sense | 5′-GCTGTAGCTCTTGCCATCA-3′ |
| miR-330-3p sense | 5′-ACACTCCAGCTGGGGCAAAGCACACGGCCTG-3′ |
| miR-330-3p anti-sense | 5′-TGGTGTCGTGGAGTCG-3′ |
| miR-361-3p sense | 5′- ACACTCCAGCTGGGTCCCCCAGGTGTGATTC-3′ |
| miR-361-3p anti-sense | 5′-TGGTGTCGTGGAGTCG-3′ |
| miR-526b-5p sense | 5′-GTCTCTTGAGGGAAGCACT-3′ |
| miR-526b-5p anti-sense | 5′-GTGCAGGGTCCGAGGT-3′ |
| miR-149-5p sense | 5′-ACACTCCAGCTGGGTCTGGCTCCGTGTCTTC-3′ |
| miR-149-5p anti-sense | 5′-TGGTGTCGTGGAGTCG-3′ |
| miR-616-3p sense | 5′-ACACTCCAGCTGGGAGTCATTGGAGGGTTT-3′ |
| miR-616-3p anti-sense | 5′-TGGTGTCGTGGAGTCG-3′ |
| MYO6 sense | 5′-TTCCGCATCCCATTCATC-3′ |
| MYO6 anti-sense | 5′-TCGTCCTTACCAGCCACA-3′ |
| β-actin sense | 5′-CACCATTGGCAATGAGCGGTTC-3′ |
| β-actin anti-sense | 5′-AGGTCTTTGCGGATGTCCACGT-3′ |
| U6 sense | 5′-CTCGCTTCGGCAGCACA-3′ |
| U6 anti-sense | 5′-AACGCTTCACGAATTTGCGT-3′ |
RNase R resistance analysis of circ_0011385 and actinomycin D treatment assays
For RNase R resistance analysis of circ_0011385 assay, HCT116 and SW480 cells were collected and RNA was extracted. RNase R (Geneseed, Guangzhou, China) was incubated with RNA at 37 °C for 30 min. After that, RNeasy MinElute Cleaning Kit (Qiagen) was used to purify RNA. For actinomycin D treatment assay, the collected CRC cells were incubated with actinomycin D (Amresco, Solon, OH, USA) (2 mg/L) for 0 h, 6 h, 12 h, 18 h and 24 h. Finally, the expression of circ_0011385 was determined by qRT-PCR. EIF3I mRNA acted as a control.
3-(4,5)-dimethylthiahiazo (-z-y1)-3,5-di-phenytetrazoliumromide (MTT) assay
HCT116 and SW480 cells were seeded in 96-well plates for 14 h. The cells were transfected according to the defined purposes, and cultivated for 0 h, 24 h, 48 h and 72 h, respectively. Following that, MTT solution (Beyotime, Shanghai, China) was utilized to incubate cells about 3 h. And produced formazan was dissolved with dimethyl sulfoxide (Millipore, Bradford, MA, USA). The output of wavelength at 570 nm was detected with microplate reader (Thermo Fisher).
Cell colony formation assay
The colony-forming ability of CRC cells was investigated by cell colony formation assay. Shortly, cells were suspended in RPMI-1640 (Procell) and then added in 6-well plates. Cells were grown about 2 weeks and RPMI-1640 was replaced every 2–3 days. Proliferative colonies were immobilized using paraformaldehyde (Beyotime) and dyed by crystal violet (Beyotime). The colony-forming ability was determined by counting cell numbers. A colony was deemed when cell numbers over 50.
Flow cytometry analysis
The apoptosis and cell cycle of HCT116 and SW480 cells were demonstrated by cell apoptosis and cell cycle detection kits (Beyotime), respectively. Shortly, cells were digested using trypsin (Thermo Fisher) and collected by centrifuging at 1000 g for 4 min. Cells were mixed using 70% ethanol (Millipore) at 4 °C overnight (only in cell cycle assay) and then suspended in phosphate buffer solution (PBS; Thermo Fisher). After that, Annexin V-fluorescein isothiocyanate (Annexin V-FITC; Beyotime) and propidium iodide (PI; Beyotime) were used to incubate cells in cell apoptosis assay; dye buffer (Beyotime), PI solution (Beyotime) and RNase A (Beyotime) were chosen to incubate cells in cell cycle assay. Samples were assessed using flow cytometry (Thermo Fisher).
Wound-healing assay
Cells were seeded in 6-well plates, and washed using PBS (Thermo Fisher) when the confluence of cells reached about 100%, and cell wounds were created. Following that, cells were cultured in FBS-free RPMI-1640 (Procell) for 24 h. The migratory ability of CRC cells was determined by assessing the width of the wounds under microscope (Nikon, Tokyo, Japan) with 100 × magnification.
Transwell invasion assay
In order to demonstrate the invasive capacity of CRC cells, transwell chambers with Matrigel (Corning, Madison, New York, USA) were employed. Briefly, HCT116 and SW480 cells were grown in the upper chambers supplemented with FBS-free RPMI-1640 (Procell). In the lower chambers, RPMI-1640 (Procell) containing 12% FBS (Procell) was added. Twenty-four hours later, cells were washed and incubated with methanol (Beyotime) and crystal violet (Beyotime). Results were determined by figuring up the number of cells in the lower chambers with microscope (Nikon) at a 100(×) magnification.
Western blot assay
RIPA buffer (Beyotime) was employed to lyse collected tissues and cells. The lysates were mixed with loading buffer (Thermo Fisher) and loaded on 12% bis-tris-acrylamide gel (Thermo Fisher). Following that, protein bands were electrotransferred onto polyvinylidene fluoride membranes (PVDF; Millipore) and immersed in fat-free milk (Solarbio, Beijing, China). Then, the membranes were incubated with anti-mitogen-activated protein kinase's kinase 1 (anti-MEK1/2; 1:1000; Affinity, Nanjing, China), anti-phosphorylated MEK1/2 (anti-p-MEK1/2; 1:2000; Affinity), anti-extracellular signal-regulated kinase 1/2 (anti-ERK1/2; 1:3000; Affinity), anti-p-ERK1/2 (1:1000; Affinity), anti-cyclin dependent kinase 1 (anti-CDK1) (1:1500; Affinity), anti-Cyclin D1 (1:1000; CST, Boston, MA, USA), anti-MYO6 (1:2500; Affinity) and anti-β-actin (1:15,000; Affinity), respectively. At 12 h after incubation, PVDF membranes were washed and incubated with secondary antibody (1:2000; CST). Finally, protein bands were presented by eyoECL Plus Kit (Beyotime). β-actin was utilized as a reference.
Dual-luciferase reporter assay
In order to build the wild-type (WT) and mutant (MUT) luciferase reporter plasmids, the binding sites of miR-330-3p in circ_0011385 and 3′-untranslated regions (3′UTR) of MYO6 were predicted by CircInteractome or starBase online databases. The wild-type plasmids (circ_0011385-WT and MYO6 3′UTR-WT) and mutant plasmids (circ_0011385-MUT and MYO6 3′UTR-MUT) were constructed by Geneseed Co., Ltd. Conducted plasmids were transfected with miR-330-3p or miR-NC using DharmaFECT 4 (Thermo Fisher) according to manufacturer's instructions. Luciferase activities were assessed by Dual-Lucy Assay Kit (Solarbio) with Renilla luciferase activity as a control.
RNA immunoprecipitation (RIP) assay
Whether circ_0011385 and MYO6 were directly associated with miR-330-3p was confirmed by RIP assay. In brief, cells were collected and lysed using RIP lysis buffer (Millipore). The lysates were incubated with magnetic beads (Millipore) coated with anti-argonaute2 (anti-Ago2; Abcam, Cambridge, UK) or anti-immunoglobulin G (anti-IgG; Abcam), respectively. Twenty-four hours later, magnetic beads were washed, and the expression of circ_0011385, miR-330-3p and MYO6 was determined through qRT-PCR assay.
In vivo assay
Six-week-old male BALB/c nude mice were provided by Vital River Laboratories (Beijing, China) and fed in pathogen-free environment. Nude mice were randomly divided into 2 groups (N = 6, respectively). 5 × 106 SW480 cells transfected with sh-NC or sh-circ_0011385 were severally suspended in 200 μL PBS (Thermo Fisher), which were then inoculated into mice. One week later, tumor volume was measured every 7 days. Five weeks later, all mice were killed and tumor weight was detected. In addition, whether circ_0011385 silencing affected the expression levels of miR-330-3p and MYO6 was revealed using the harvested neoplasms. The Animal Care and Use Committee of Renmin Hospital of Wuhan University allowed this research.
Statistical analysis
All data were obtained based on three independent duplicate tests, and assessed by GraphPad Prism. Results were presented as means ± standard deviations (SD). Significant differences were compared by Spearman's rank correlation coefficient test in assessing the linear relationships among circ_0011385, miR-330-3p and MYO6. Significant differences were compared by Student's t-tests, Wilcoxon signed-rank test or One-way analysis of variance (ANOVA). Log-rank test was employed to compare the groups between low and high circ_0011385 expression. Independent prognostic factors related to overall survival were analyzed by univariate and multivariate overall survival analyses using Cox's proportional hazard regression model. Statistical significance was deemed when P value less than 0.05.
Results
Circ_0011385 expression was dramatically upregulated in CRC tissues and cells
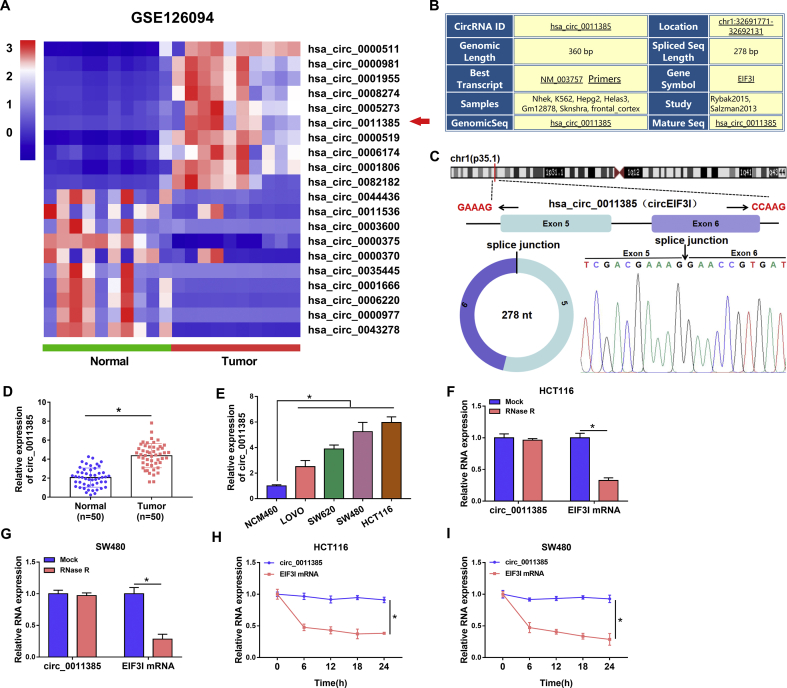
In order to seek differently expressed circRNAs in CRC, GEO dataset was employed. GSE126094 dataset showed that circ_0011385 expression was dramatically upregulated in CRC tissues compared with control groups (Fig. 1A). Also, we found that circ_0011385 expression was associated with tumor size, tumor-node-metastasis stage (TNM) stage and lymph node metastasis of CRC patients (Table S1). The results from the multivariate analysis indicated that circ_0011385 expression was an independent predictor for overall survival (Table S2). We further found circ_0011385 was located in chromosome1 (chr1):32691771–32692131 and its spliced sequence was 278 base pairs (bp) in length (Fig. 1B). Also, Fig. 1C showed circ_0011385 was composed of exons 5 and 6 of the EIF3I, and confirmed the back-splicing of circ_0011385 by Sanger sequencing. Subsequently, qRT-PCR results presented circ_0011385 expression was apparently upregulated in CRC tissues and LOVO, SW620, SW480 and HCT116 cells relative to normal tissues and NCM460 cells, respectively (Fig. 1D and E). Furthermore, results showed circ_0011385 had no obvious change in expression after RNase R or actinomycin D treatment, whereas the expression of its linear form, EIF3I mRNA, was notably decreased in HCT116 and SW480 cells (Fig. 1F–I), suggesting circ_0011385 was a circular RNA. The above data demonstrated that circ_0011385 might participate in regulating CRC process.
Fig. 1.
Circ_0011385 expression was upregulated in CRC tissues and cells. (A) The differently expressed circRNAs were analyzed by GSE126094 dataset in CRC and normal tissues. (B) CircBase online database was employed to present the genetic messages of circ_0011385. (C) The presence of circ_0011385 was confirmed by qRT-PCR, followed by Sanger sequencing. (D and E) QRT-PCR was conducted to detect the expression level of circ_0011385 in 50 pairs of CRC, paracancerous normal tissues and NCM460, LOVO, SW620, SW480, HCT116 cells. (F and G) RNase R resistance analysis of circ_0011385 assay was carried out to illustrate circ_0011385 was a circular RNA. (H and I) The expression levels of circ_0011385 and EIF3I mRNA were determined by qRT-PCR at 0 h, 6 h, 12 h, 18 h and 24 h, respectively, after actinomycin D treatment in HCT116 and SW480 cells. ∗p < 0.05.
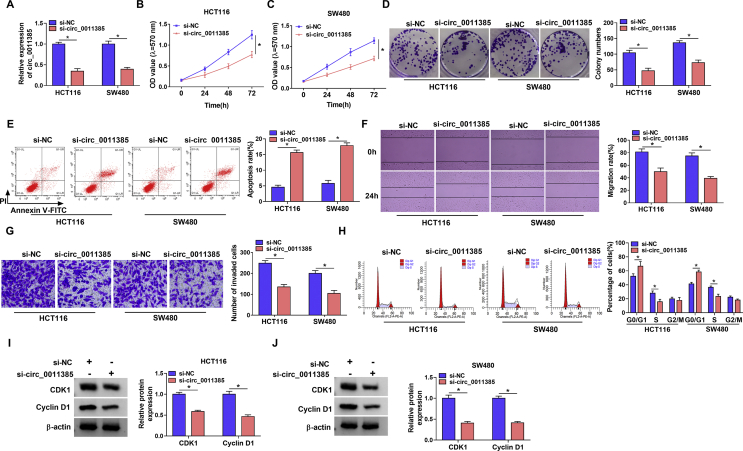
Circ_0011385 silencing repressed cell proliferation, migration and invasion, while promoted cell apoptosis and cell cycle arrest in CRC
Based on the above results, the impacts of circ_0011385 knockdown on CRC process were explored. To validate this, the small interfering RNA targeting circ_0011385 was transfected into HCT116 and SW480 cells, and its silencing efficiency was firstly detected. Data showed circ_0011385 expression was dramatically downregulated, while the expression of its parent, EIF3I, had little change in si-circ_0011385 group as compared to their expression in si-NC group in HCT116 and SW480 cells (Fig. 2A and Fig. S1), suggesting si-circ_0011385 was successfully synthesized. Subsequently, MTT and cell colony formation assays displayed circ_0011385 silencing inhibited the viability and colony-forming ability of HCT116 and SW480 cells (Fig. 2B–D). On the contrary, circ_0011385 absence induced cell apoptosis in HCT116 and SW480 cells (Fig. 2E). Additionally, the migration and invasion of HCT116 and SW480 cells were repressed after si-circ_0011385 transfection (Fig. 2F and G). The impact of circ_0011385 silencing on cell cycle of HCT116 and SW480 cells was further revealed. Results showed circ_0011385 knockdown induced cell cycle arrest at G0/G1 phase (Fig. 2H), further demonstrating that circ_0011385 silencing repressed the proliferation of HCT116 and SW480 cells. Meanwhile, Western blot analysis showed circ_0011385 repression downregulated the protein levels of CDK1 and Cyclin D1 (Fig. 2I and J), suggesting circ_0011385 silencing repressed cell proliferation at molecular level. These data demonstrated circ_0011385 acted as an oncogene in CRC process.
Fig. 2.
Circ_0011385 knockdown inhibited CRC progression. (A) The interfering efficiency of si-circ_0011385 was determined by qRT-PCR in HCT116 and SW480 cells. (B–D) The impact of circ_0011385 silencing on the proliferation of HCT116 and SW480 cells was presented by MTT and cell colony formation assays. (E and H) Flow cytometry analysis was used to determine the influences of circ_0011385 downregulation on the apoptosis and cell cycle of HCT116 and SW480 cells. (F and G) The impacts of circ_0011385 absence on the migration and invasion of HCT116 and SW480 cells were investigated by wound-healing and transwell invasion assays. (I and J) Western blot was performed to determine the effects of circ_0011385 silencing on the protein expression of CDK1 and Cyclin D1 in HCT116 and SW480 cells. ∗p < 0.05.
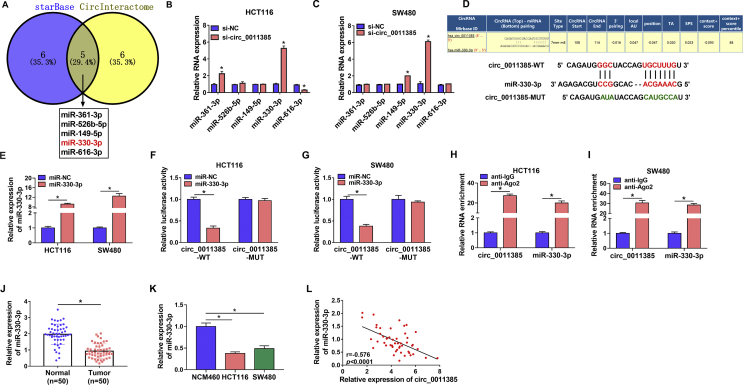
Circ_0011385 functioned as a sponge of miR-330-3p in CRC cells
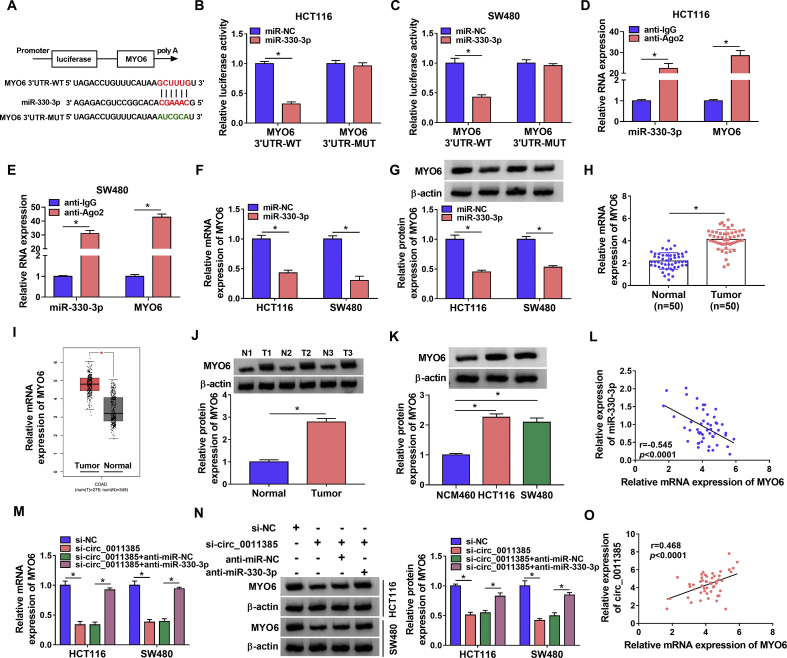
In order to reveal the regulatory mechanism of circ_0011385 in CRC progression, we predicted circ_0011385-associated miRNA(s) by starBase and CircInteractome online databases. Results presented that 5 miRNAs (miR-361-3p, miR-526b-5p, miR-149-5p, miR-330-3p and miR-616-3p) containing putative binding sites of circ_0011385 were simultaneously predicted by both starBase and CircInteractome online databases (Fig. 3A). QRT-PCR analysis further revealed only miR-330-3p expression was dramatically upregulated by si-circ_0011385 in both HCT116 and SW480 cells (Fig. 3B and C). Thus, miR-330-3p was chosen as a target miRNA of circ_0011385. And CircInteractome online database showed that the binding sites of circ_0011385 for miR-330-3p (Fig. 3D). Subsequently, qRT-PCR data also showed miR-330-3p expression was greatly increased after miR-330-3p mimic transfection (Fig. 3E), implicating miR-330-3p mimic was effective in increasing miR-330-3p expression. Additionally, dual-luciferase reporter assay displayed that relative luciferase activity was dramatically repressed in circ_0011385-WT + miR-330-3p group, but not in circ_0011385-MUT + miR-330-3p group (Fig. 3F and G). Meanwhile, RIP assay demonstrated the amount of circ_0011385 and miR-330-3p was apparently enriched by anti-Ago2 in HCT116 and SW480 cells compared with them in anti-IgG group (Fig. 3H and I). The study also found miR-330-3p expression was notably downregulated in CRC tissues (N = 50) and HCT116 and SW480 cells in contrast to adjacent normal tissues (N = 50) and NCM460 cells, respectively (Fig. 3J and K). Furthermore, we found miR-330-3p expression was negatively related to circ_0011385 expression (Fig. 3L). These evidences proved circ_0011385 was associated with miR-330-3p in CRC cells.
Fig. 3.
Circ_0011385 bound to miR-330-3p in CRC cells. (A) Circ_0011385-associated miRNAs were predicted by starBase and CircInteractome online databases, and there were 5 miRNAs that were simultaneously predicted by the two online databases. (B and C) The impacts of circ_0011385 silencing on the expression levels of miR-361-3p, miR-526b-5p, miR-149-5p, miR-330-3p and miR-616-3p were determined by qRT-PCR in HCT116 and SW480 cells. (D) The schema showed the supposed binding sites between circ_0011385 and miR-330-3p and the mutated sequence of circ_0011385. (E) The efficiency of miR-330-3p overexpression was revealed by qRT-PCR in HCT116 and SW480 cells. (F–I) Dual-luciferase reporter assay and RIP assay were performed to demonstrate that circ_0011385 was associated with miR-330-3p in HCT116 and SW480 cells. (J and K) QRT-PCR was employed to detect miR-330-3p expression in CRC tissues (N = 50), adjacent normal tissues (N = 50) and NCM460, HCT116, SW480 cells. (L) Spearman correlation analysis was employed to disclose the linear relationship between miR-330-3p expression and circ_0011385 expression in CRC tissues. ∗p < 0.05.
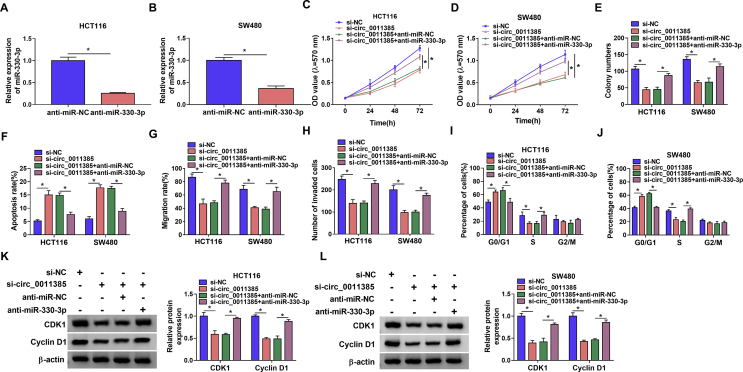
Circ_0011385 regulated CRC evolution by sponging miR-330-3p
Our data had presented that circ_0011385 acted as a sponge of miR-330-3p, whether circ_0011385 modulated CRC process by sponging miR-330-3p was further explored. Data firstly exhibited miR-330-3p inhibitor was effective in knocking down miR-330-3p expression in HCT116 and SW480 cells (Fig. 4A and B). Subsequently, results showed miR-330-3p inhibitor attenuated the repressive effects of circ_0011385 silencing on the viability and colony-forming ability of HCT116 and SW480 cells (Fig. 4C–E). As expected, miR-330-3p inhibitor also restored the promoting impact of circ_0011385 silencing on the apoptosis of HCT116 and SW480 cells (Fig. 4F). Additionally, miR-330-3p inhibitor could impair the inhibitory effects of circ_0011385 silencing on cell migration and invasion (Fig. 4G and H), and the promoting impact of that on cell cycle arrest in HCT116 and SW480 cells (Fig. 4I and J). Furthermore, circ_0011385 absence-mediated effects on the protein expression of CDK1 and Cyclin D1 were restrained by anti-miR-330-3p (Fig. 4K and L). Thus, these results confirmed that circ_0011385 modulated CRC process by binding to miR-330-3p.
Fig. 4.
Circ_0011385 silencing suppressed CRC development by interacting with miR-330-3p. (A and B) The effect of miR-330-3p inhibitor on miR-330-3p expression was determined by qRT-PCR in HCT116 and SW480 cells. (C–E) The impacts between circ_0011385 silencing and miR-330-3p inhibitor on the proliferation of HCT116 and SW480 cells were unveiled by MTT assay and cell colony formation assay. (F, I and J) Flow cytometry analysis was utilized to illustrate the effects between circ_0011385 silencing and miR-330-3p inhibitor on the apoptosis and cell cycle of HCT116 and SW480 cells. (G and H) Wound-healing and transwell assays were performed to reveal the influences between circ_0011385 absence and miR-330-3p inhibitor on the migration and invasion of HCT116 and SW480 cells, respectively. (K and L) Western blot assay was conducted to determine the influences between circ_0011385 downregulation and miR-330-3p inhibitor on the protein expression of CDK1 and Cyclin D1 in HCT116 and SW480 cells. ∗p < 0.05.
Circ_0011385 silencing decreased MYO6 expression via sponging miR-330-3p in HCT116 and SW480 cells
In order to unveil the mechanism of miR-330-3p in regulating CRC development, the study predicted its target gene by starBase online database. Result showed MYO6 possessed the supposed binding sites of miR-330-3p (Fig. 5A). Subsequently, dual-luciferase reporter assay presented relative luciferase activity was obviously suppressed in MYO6 3′UTR-WT + miR-330-3p group, but the relative luciferase activity had no apparent variation in MYO6 3′UTR-MUT + miR-330-3p group (Fig. 5B and C), showing miR-330-3p bound to MYO6. RIP assay also showed anti-Ago2 could apparently enrich miR-330-3p and MYO6 as significantly high expression of miR-330-3p and MYO6 in anti-Ago2 group when compared with them in anti-IgG group (Fig. 5D and E). Additionally, it was found that MYO6 mRNA and protein levels were notably downregulated by miR-330-3p mimic (Fig. 5F and G). Subsequent data showed MYO6 mRNA and protein expression were apparently upregulated in CRC tissues or HCT116 and SW480 cells compared to control groups (Fig. 5H, J and K). TCGA dataset further revealed MYO6 mRNA expression was significantly upregulated in CRC tissues as compared to normal tissues (Fig. 5I). Spearman correlation analysis demonstrated that miR-330-3p expression was negatively correlated with MYO6 expression (Fig. 5L). The above findings suggested miR-330-3p was directly associated with MYO6 in HCT116 and SW480 cells.
Fig. 5.
Circ_0011385 regulated MYO6 expression via binding to miR-330-3p. (A) The schema presented the putative binding sites between miR-330-3p and MYO6, and the mutated sites of MYO6. (B–E) Dual-luciferase reporter assay and RIP assay were conducted to demonstrate miR-330-3p was associated with MYO6 in HCT116 and SW480 cells. (F and G) The effects of miR-330-3p mimic on the mRNA and protein expression of MYO6 were determined by qRT-PCR and Western blot, respectively, in HCT116 and SW480 cells. (H) QRT-PCR was utilized to detect the mRNA expression of MYO6 in 50 pairs of CRC tissues and paracancerous normal tissues. (I) TCGA dataset was employed to predict the mRNA expression of MYO6 in CRC tissues (N = 275) and normal tissues (N = 349). (J and K) Western blot was performed to detect the protein expression of MYO6 in 50 pairs of CRC and paracancerous normal tissues and NCM460, HCT116 and SW480 cells. (L and O) Spearman correlation analysis was conducted to determine the linear relationships between MYO6 mRNA expression and miR-330-3p expression or circ_0011385 expression in CRC tissues. (M and N) The impacts between circ_0011385 silencing and miR-330-3p inhibitor on the mRNA and protein levels of MYO6 were revealed by qRT-PCR and Western blot analysis, respectively, in HCT116 and SW480 cells. ∗p < 0.05.
Given the targeted relationships among circ_0011385, miR-330-3p and MYO6, whether circ_0011385 could modulate MYO6 expression via sponging miR-330-3p was revealed. Results displayed circ_0011385 silencing obviously downregulated the mRNA and protein levels of MYO6, whereas these effects were restored after miR-330-3p inhibitor transfection (Fig. 5M and N). Also, it was found MYO6 mRNA expression was positively related to circ_0011385 expression (Fig. 5O). These results demonstrated circ_0011385 controlled MYO6 expression by sponging miR-330-3p in HCT116 and SW480 cells.
Circ_0011385 regulated CRC cell malignancy by interacting with MYO6 through MEK1/2/ERK1/2 pathway
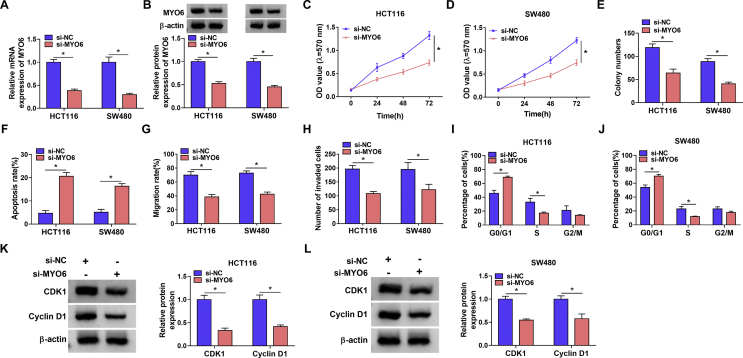
The impacts of MYO6 silencing on CRC development were continued to be demonstrated. Results firstly showed si-MYO6 was effective in downregulating MYO6 expression based on the apparently low mRNA and protein levels of MYO6 in HCT116 and SW480 cells transfected with si-MYO6 (Fig. 6A and B). Subsequently, MTT and cell colony formation assays demonstrated MYO6 absence repressed cell viability and colony-forming ability, respectively, in HCT116 and SW480 cells (Fig. 6C–E). Flow cytometry analysis displayed that MYO6 silencing promoted cell apoptosis (Fig. 6F). Additionally, low MYO6 expression repressed cell migration and invasion, and induced cell cycle arrest at G0/G1 phase in HCT116 and SW480 cells (Fig. 6G–J). The expression levels of cell cycle-related proteins, CDK1 and Cyclin D1, were significantly downregulated after MYO6 silencing in HCT116 and SW480 cells (Fig. 6K and L). These results illustrated MYO6 silencing suppressed CRC development. Based on the above result, the study further investigated whether MYO6 overexpression could rescue circ_0011385 silencing-mediated action. Fig. S2A and B showed the efficiency of MYO6 overexpression in both HCT116 and SW480 cells. Subsequent data displayed that the repressing effects of circ_0011385 knockdown on cell proliferation, migration and invasion were overturned after MYO6 overexpression (Fig. S2C, D, E, G, H, K and L). Also, the increased apoptosis and cell cycle arrest by circ_0011385 knockdown were relieved by ectopic MYO6 expression (Fig. S2F, I and J). Importantly, we found that circ_0011385 depletion inactivated MEK1/2/ERK1/2 signaling pathway, which was attenuated by miR-330-3p downregulation or MYO6 overexpression (Fig. S3A–D). As shown in Fig. S3E and F, MYO6 knockdown also blocked MEK1/2/ERK1/2 pathway. Taken together, circ_0011385 regulated CRC cell malignancy by interacting with MYO6 through MEK1/2/ERK1/2 pathway.
Fig. 6.
MYO6 silencing suppressed CRC progression. (A and B) The impacts of MYO6 silencing on the mRNA and protein levels of MYO6 were determined by qRT-PCR and Western blot, respectively, in HCT116 and SW480 cells. (C–E) MTT and cell colony formation assays were employed to determine the effect of MYO6 knockdown on the proliferation of HCT116 and SW480 cells. (F, I and J) Flow cytometry analysis was conducted to disclose the influences of MYO6 absence on the apoptosis and cell cycle of HCT116 and SW480 cells. (G and H) Wound-healing and transwell invasion assays were performed to reveal the effects of MYO6 silencing on the migration and invasion of HCT116 and SW480 cells, respectively. (K and L) The influences of MYO6 knockdown on the protein expression of CDK1 and Cyclin D1 were presented by Western blot analysis in HCT116 and SW480 cells. ∗P < 0.05.
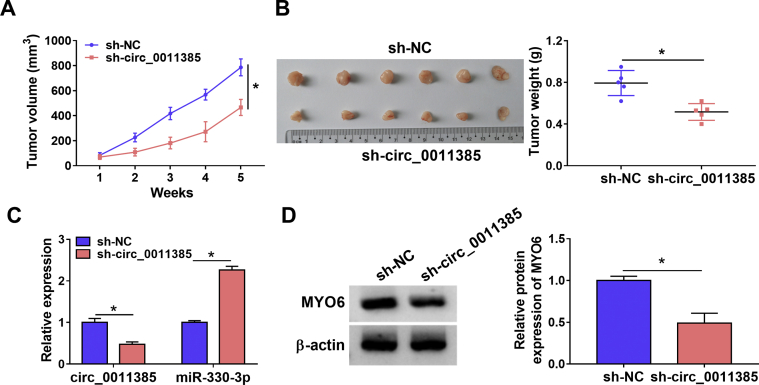
Circ_0011385 knockdown inhibited tumor growth in vivo
The impacts of circ_0011385 silencing on tumor growth in vivo were further revealed. Results presented that the volume and weight of tumors were obviously smaller or lighter in sh-circ_0011385 group than in sh-NC group (Fig. 7A and B), illustrating circ_0011385 silencing could suppress tumor formation in vivo. Additionally, it was found circ_0011385 expression was dramatically low after sh-circ_0011385 transfection (Fig. 7C), meaning sh-circ_0011385 was successfully built. Furthermore, data disclosed circ_0011385 knockdown could upregulate miR-330-3p expression and downregulate MYO6 protein expression in vivo (Fig. 7C and D). These findings demonstrated circ_0011385 silencing inhibited tumor growth by upregulating miR-330-3p expression and downregulating MYO6 expression in vivo.
Fig. 7.
Circ_0011385 knockdown repressed tumor growth in vivo. (A and B) The impacts of circ_0011385 silencing on the volume and weight of tumors were determined in vivo. (C) QRT-PCR was employed to determine the expression levels of circ_0011385 and miR-330-3p after circ_0011385 silencing in the forming tumors from sh-NC and sh-circ_0011385 group. (D) The impact of sh-circ_0011385 on the protein expression of MYO6 was revealed by Western blot in the forming tumors from sh-NC and sh-circ_0011385 group. ∗p < 0.05.
Discussion
CRC is posing a heavy burden to human health because of its strong proliferative and metastatic capacities [26]. CircRNA-miRNA-mRNA network is revealed to play important roles in revealing the pathogenesis of human diseases [27,28]. In CRC, similar mechanisms have been disclosed. For example, circ_0053277 contributed to CRC evolution by miR-2467-3p/matrix metallopeptidase 14 (MMP14) axis [29]. Circ_0136666 facilitated cell proliferation and invasion by regulating miR-136/SH2B adaptor protein 1 (SH2B1) signaling pathway in CRC [30]. Herein, we found circ_0011385 silencing repressed CRC development by miR-330-3p/MYO6 axis.
In this research, in order to find differently expressed circRNAs in CRC, GEO dataset was employed. Results showed that circ_0011385 was dramatically upregulated in CRC tissues, implicating circ_0011385 might be involved in CRC evolution. Previous researches have revealed circ_0011385 contributes to thyroid cancer process by sponging miR-361-3p [31] and may be enrolled in the regulation of bladder carcinogenesis growth [32]. In this paper, we found circ_0011385 promoted CRC progression for the first time. Our findings presented circ_0011385 was overexpressed in CRC specimens and cell lines, and associated with tumor size, TNM stage and lymph node metastasis of CRC patients. Circ_0011385 absence restrained cell proliferative, migratory and invasive capacities, but upregulated cell apoptotic rate in CRC. Meanwhile, in vivo assay explained circ_0011385 downregulation inhibited tumor volume and weight, suggesting circ_0011385 could suppress CRC growth in vivo. The above data illustrated circ_0011385 served as an oncogene in CRC development.
For seeking circ_0011385-associated miRNAs, CircInteractome and starBase online databases were performed. Results showed miR-330-3p possessed the supposed targeting sequence of circ_0011385. Further experiments proved circ_0011385 absorbed miR-330-3p. Existing studies indicated miR-330-3p accelerated the evolution of lung cancer [33] and breast cancer [34], whereas restrained the progression of gastric cancer [35] and glioma [36]. In CRC, Huang et al. explained miR-330-3p expression was low in CRC tumors and cell lines; its downregulation facilitated cell proliferation and metastasis, while inhibited cell apoptosis [37], implicating miR-330-3p served as an anti-oncogene in CRC evolution. Similarly, we also observed obviously low expression of miR-330-3p in CRC specimens and cell lines. Additionally, miR-330-3p inhibitor impaired the impacts of circ_0011385 repression on CRC growth, meaning miR-330-3p hindered CRC progression. Our findings were consistent with the published data [37]. In the meantime, our data also suggested circ_0011385 could modulate CRC evolution through absorbing miR-330-3p.
Subsequently, we found miR-330-3p interacted with MYO6 based on the result predicted by starBase online database and identified by dual-luciferase reporter and RIP assays. It was found that MYO6 expression was greatly high in CRC tissues and cells. As expected, loss-of-function experiments further revealed MYO6 silencing suppressed CRC growth. Also, MYO6 overexpression remitted the repressing impacts of circ_0011385 knockdown on CRC cell malignancy. Previous research has explained that MYO6 facilitates cell proliferative and metastatic abilities and hinders cell apoptosis in CRC [38], which is in line with our results. Meanwhile, Spearman correlation analysis showed circ_0011385 was positively related to MYO6 in expression. Rescue experiments presented miR-330-3p inhibitor impaired the repressive impact of circ_0011385 silencing on MYO6 expression, which suggested circ_0011385 could modulate MYO6 expression through absorbing miR-330-3p. Yu and his colleagues have ascertained MYO6 knockdown restrained prostate cancer progression by decreasing ERK1/2 phosphorylation [39]. The current study also showed that MYO6 silencing blocked the activation of MEK1/2/ERK1/2 signaling pathway revealed, and that circ_0011385 modulated MEK1/2/ERK1/2 signaling pathway by miR-330-3p/MYO6 axis.
Excited data have showed that MYO6 regulates prostate cancer cell malignancy by mediating the expression of thioredoxin-interacting protein 1 (TXNIP), a tumor suppressor gene [40], and whether the molecular mechanism behind MYO6 modulating CRC progression was attributed to TXNIP needed to be further explored.
All in all, circ_0011385 was overexpressed in CRC specimens and cell lines. Its silencing repressed CRC growth in vivo and in vitro. Circ_0011385 modulated MYO6 expression via sponging miR-330-3p. Additionally, miR-330-3p absence attenuated the influences of circ_0011385 knockdown on CRC growth. Circ_0011385 silencing inactivated MEK1/2/ERK1/2 signaling pathway by affecting miR-330-3p/MYO6 axis. Collectively, circ_0011385 regulated CRC development via miR-330-3p/MYO6 axis through MEK1/2/ERK1/2 signaling pathway. Our findings not only lay a foundation for further revealing the pathogenesis of CRC, but also provide a potential therapeutic target for CRC.
Funding
This work was supported by the National Natural Science Foundation of China (No. U1604175) and National Key R&D Program of China (2018YFC1311300) and Independent Research Project of Wuhan University (2042019kf0103) and Beijing Kangmeng Charity Foundation (2020HX0026).
Conflicts of interest
The authors have nothing to disclose.
Footnotes
Peer review under responsibility of Chang Gung University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bj.2022.01.007.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Kim J.H. Chemotherapy for colorectal cancer in the elderly. World J Gastroenterol. 2015;21(17):5158–5166. doi: 10.3748/wjg.v21.i17.5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen W.Z., Jiang J.X., Yu X.Y., Xia W.J., Yu P.X., Wang K., et al. Endothelial cells in colorectal cancer. World J Gastrointest Oncol. 2019;11(11):946–956. doi: 10.4251/wjgo.v11.i11.946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang L.C., Fan C.W., Tseng W.K., Hua C.C. Associations between the Nrf2/Keap1 pathway and mitochondrial functions in colorectal cancer are affected by metastasis. Cancer Biomark. 2020;27(2):163–171. doi: 10.3233/CBM-190828. [DOI] [PubMed] [Google Scholar]
- 5.Sur D., Cainap C., Burz C., Havasi A., Chis I.C., Vlad C., et al. The role of miRNA -31-3p and miR-31-5p in the anti-EGFR treatment efficacy of wild-type K-RAS metastatic colorectal cancer. Is it really the next best thing in miRNAs? J BUON. 2019;24(5):1739–1746. [PubMed] [Google Scholar]
- 6.Morabito A., De Maio E., Di Maio M., Normanno N., Perrone F. Tyrosine kinase inhibitors of vascular endothelial growth factor receptors in clinical trials: current status and future directions. Oncologist. 2006;11(7):753–764. doi: 10.1634/theoncologist.11-7-753. [DOI] [PubMed] [Google Scholar]
- 7.Jung W.B., Shin J.Y., Suh B.J. The short-term outcome and safety of laparoscopic colorectal cancer resection in very elderly patients. Korean J Gastroenterol. 2017;69(5):291–297. doi: 10.4166/kjg.2017.69.5.291. Korean. [DOI] [PubMed] [Google Scholar]
- 8.Qu S., Zhong Y., Shang R., Zhang X., Song W., Kjems J., et al. The emerging landscape of circular RNA in life processes. RNA Biol. 2017;14(8):992–999. doi: 10.1080/15476286.2016.1220473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhong Y., Du Y., Yang X., Mo Y., Fan C., Xiong F., et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. 2018;17(1):79. doi: 10.1186/s12943-018-0827-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao Z.J., Shen J. Circular RNA participates in the carcinogenesis and the malignant behavior of cancer. RNA Biol. 2017;14(5):514–521. doi: 10.1080/15476286.2015.1122162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jian X., He H., Zhu J., Zhang Q., Zheng Z., Liang X., et al. Hsa_circ_001680 affects the proliferation and migration of CRC and mediates its chemoresistance by regulating BMI1 through miR-340. Mol Cancer. 2020;19(1):20. doi: 10.1186/s12943-020-1134-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y., Zang H., Zhang X., Huang G. circ_0136666 facilitates the progression of colorectal cancer via miR-383/CREB1 Axis. Cancer Manag Res. 2020;12:6795–6806. doi: 10.2147/CMAR.S251952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu C., Li S., Hu X. Circ_0005576 promotes malignant progression through miR-874/CDK8 Axis in colorectal cancer. OncoTargets Ther. 2020;13:7793–7805. doi: 10.2147/OTT.S249494. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Yang G., Zhang T., Ye J., Yang J., Chen C., Cai S., et al. Circ-ITGA7 sponges miR-3187-3p to upregulate ASXL1, suppressing colorectal cancer proliferation. Cancer Manag Res. 2019;11:6499–6509. doi: 10.2147/CMAR.S203137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng Z., Li X., Wang H., Geng Y., Cai Y., Tang Y., et al. Dysregulation of CircRNA_0001946 contributes to the proliferation and metastasis of colorectal cancer cells by targeting MicroRNA-135a-5p. Front Genet. 2020;11:357. doi: 10.3389/fgene.2020.00357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu B., Yang N., Liu Y., Kong P., Han M., Li B. Circ_cse1l inhibits colorectal cancer proliferation by binding to eIF4A3. Med Sci Monit. 2020;26 doi: 10.12659/MSM.923876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garzon R., Calin G.A., Croce C.M. MicroRNAs in cancer. Annu Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 18.Bartel D.P. Metazoan MicroRNAs. Cell. 2018;173(1):20–51. doi: 10.1016/j.cell.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Z., Yao H., Wang S., Li G., Gu X. CircTADA2A suppresses the progression of colorectal cancer via miR-374a-3p/KLF14 axis. J Exp Clin Cancer Res. 2020;39(1):160. doi: 10.1186/s13046-020-01642-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y., Liu R., Yang F., Cheng R., Chen X., Cui S., et al. miR-19a promotes colorectal cancer proliferation and migration by targeting TIA1. Mol Cancer. 2017;16(1):53. doi: 10.1186/s12943-017-0625-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhuang M., Zhao S., Jiang Z., Wang S., Sun P., Quan J., et al. MALAT1 sponges miR-106b-5p to promote the invasion and metastasis of colorectal cancer via SLAIN2 enhanced microtubules mobility. EBioMedicine. 2019;41:286–298. doi: 10.1016/j.ebiom.2018.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang S., Sun G., Zhang D., Li Q., Qian H. MiR-3622a-3p acts as a tumor suppressor in colorectal cancer by reducing stemness features and EMT through targeting spalt-like transcription factor 4. Cell Death Dis. 2020;11(7):592. doi: 10.1038/s41419-020-02789-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shirjang S., Mansoori B., Mohammadi A., Shajari N., H G Duijf P., Najafi S., et al. miR-330 regulates colorectal cancer oncogenesis by targeting BACH1. Adv Pharm Bull. 2020;10(3):444–451. doi: 10.34172/apb.2020.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lei C., Du F., Sun L., Li T., Li T., Min Y., et al. miR-143 and miR-145 inhibit gastric cancer cell migration and metastasis by suppressing MYO6. Cell Death Dis. 2017;8(10) doi: 10.1038/cddis.2017.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Y., Li H., Ye X., Ji A., Fu X., Wu H., et al. Hsa_circ_0000231 knockdown inhibits the glycolysis and progression of colorectal cancer cells by regulating miR-502-5p/MYO6 axis. World J Surg Oncol. 2020;18(1):255. doi: 10.1186/s12957-020-02033-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brody H. Colorectal cancer. Nature. 2015;521(7551):S1. doi: 10.1038/521S1a. [DOI] [PubMed] [Google Scholar]
- 27.Xiong D.D., Dang Y.W., Lin P., Wen D.Y., He R.Q., Luo D.Z., et al. A circRNA-miRNA-mRNA network identification for exploring underlying pathogenesis and therapy strategy of hepatocellular carcinoma. J Transl Med. 2018;16(1):220. doi: 10.1186/s12967-018-1593-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rong D., Sun H., Li Z., Liu S., Dong C., Fu K., et al. An emerging function of circRNA-miRNAs-mRNA axis in human diseases. Oncotarget. 2017;8(42):73271–73281. doi: 10.18632/oncotarget.19154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao H., Liu M. Circular RNA hsa_circ_0053277 promotes the development of colorectal cancer by upregulating matrix metallopeptidase 14 via miR-2467-3p sequestration. J Cell Physiol. 2020;235(3):2881–2890. doi: 10.1002/jcp.29193. [DOI] [PubMed] [Google Scholar]
- 30.Jin C., Wang A., Liu L., Wang G., Li G. Hsa_circ_0136666 promotes the proliferation and invasion of colorectal cancer through miR-136/SH2B1 axis. J Cell Physiol. 2019;234(5):7247–7256. doi: 10.1002/jcp.27482. [DOI] [PubMed] [Google Scholar]
- 31.Xia F., Chen Y., Jiang B., Bai N., Li X. Hsa_circ_0011385 accelerates the progression of thyroid cancer by targeting miR-361-3p. Cancer Cell Int. 2020;20:49. doi: 10.1186/s12935-020-1120-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu H.C., Yao J.Q., Yang X., Han J., Wang J.Z., Xu K., et al. Identification of a potentially functional circRNA-miRNA-mRNA regulatory network for investigating pathogenesis and providing possible biomarkers of bladder cancer. Cancer Cell Int. 2020;20:31. doi: 10.1186/s12935-020-1108-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen T., Yang Z., Liu C., Wang L., Yang J., Chen L., et al. Circ_0078767 suppresses non-small-cell lung cancer by protecting RASSF1A expression via sponging miR-330-3p. Cell Prolif. 2019;52(2) doi: 10.1111/cpr.12548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mesci A., Huang X., Taeb S., Jahangiri S., Kim Y., Fokas E., et al. Targeting of CCBE1 by miR-330-3p in human breast cancer promotes metastasis. Br J Cancer. 2017;116(10):1350–1357. doi: 10.1038/bjc.2017.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guan A., Wang H., Li X., Xie H., Wang R., Zhu Y., et al. MiR-330-3p inhibits gastric cancer progression through targeting MSI1. Am J Transl Res. 2016;8:4802–4811. [PMC free article] [PubMed] [Google Scholar]
- 36.Wang H., Liu G., Li T., Wang N., Wu J., Zhi H. MiR-330-3p functions as a tumor suppressor that regulates glioma cell proliferation and migration by targeting CELF1. Arch Med Sci. 2020;16(5):1166–1175. doi: 10.5114/aoms.2020.95027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang Y., Sun H., Ma X., Zeng Y., Pan Y., Yu D., et al. HLA-F-AS1/miR-330-3p/PFN1 axis promotes colorectal cancer progression. Life Sci. 2020;254:117180. doi: 10.1016/j.lfs.2019.117180. [DOI] [PubMed] [Google Scholar]
- 38.Luan Y., Li X., Luan Y., Zhao R., Li Y., Liu L., et al. Circulating lncRNA UCA1 promotes malignancy of colorectal cancer via the miR-143/MYO6 Axis. Mol Ther Nucleic Acids. 2020;19:790–803. doi: 10.1016/j.omtn.2019.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang D., Zhu L., Liao M., Zeng T., Zhuo W., Yang S., et al. MYO6 knockdown inhibits the growth and induces the apoptosis of prostate cancer cells by decreasing the phosphorylation of ERK1/2 and PRAS40. Oncol Rep. 2016;36(3):1285–1292. doi: 10.3892/or.2016.4910. [DOI] [PubMed] [Google Scholar]
- 40.Dunn T.A., Chen S., Faith D.A., Hicks J.L., Platz E.A., Chen Y., et al. A novel role of myosin VI in human prostate cancer. Am J Pathol. 2006;169(5):1843–1854. doi: 10.2353/ajpath.2006.060316. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.