Abstract
Introduction
The molecular transformation of the human preaccess vein after arteriovenous fistula (AVF) creation is poorly understood. This limits our ability to design efficacious therapies to improve maturation outcomes.
Methods
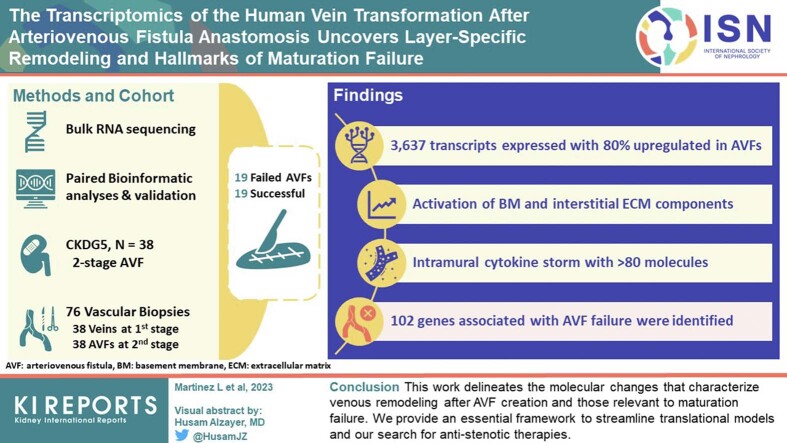
Bulk RNA sequencing (RNA-seq) followed by paired bioinformatic analyses and validation assays were performed in 76 longitudinal vascular biopsies (veins and AVFs) from 38 patients with stage 5 chronic kidney disease or end-stage kidney disease undergoing surgeries for 2-stage AVF creation (19 matured, 19 failed).
Results
A total of 3637 transcripts were differentially expressed between veins and AVFs independent of maturation outcomes, with 80% upregulated in fistulas. The postoperative transcriptome demonstrated transcriptional activation of basement membrane and interstitial extracellular matrix (ECM) components, including preexisting and novel collagens, proteoglycans, hemostasis factors, and angiogenesis regulators. A postoperative intramural cytokine storm involved >80 chemokines, interleukins, and growth factors. Postoperative changes in ECM expression were differentially distributed in the AVF wall, with proteoglycans and fibrillar collagens predominantly found in the intima and media, respectively. Interestingly, upregulated matrisome genes were enough to make a crude separation of AVFs that failed from those with successful maturation. We identified 102 differentially expressed genes (DEGs) in association with AVF maturation failure, including upregulation of network collagen VIII in medial smooth muscle cells (SMCs) and downregulation of endothelial-predominant transcripts and ECM regulators.
Conclusion
This work delineates the molecular changes that characterize venous remodeling after AVF creation and those relevant to maturation failure. We provide an essential framework to streamline translational models and our search for antistenotic therapies.
Keywords: arteriovenous fistula, extracellular matrix, maturation, transcriptomics
Graphical abstract
The first few weeks after AVF creation unravel a highly unpredictable remodeling process that results in a mature vascular access with flow >600 ml/min1 or a failed fistula with significant stenoses due to excessive wall fibrosis and aggressive intimal hyperplasia.2 The odds of these scenarios have remained around 50:50 for more than 50 years.3, 4, 5 The mechanistic landscape determining maturation or failure has been based so far on animal models, extrapolations from arterial biology, and computer modeling with almost no confirmation in human cohorts. In fact, the questions of how a human preaccess vein transforms into a fistula at the molecular level and what determines maturation success during this transformation remain unanswered.
The main trigger for the transformation of the vein to the AVF is the rush of highly oxygenated blood from the artery after anastomosis.6 Initial distention of the vein alleviates the elevated wall shear stress while driving further increases in luminal diameter and quick remodeling of the wall to withstand circumferential wall stress and secure hemostasis.6 Regardless of the maturation outcome, all fistulas increase in cross-sectional diameter and wall thickness through extensive reorganization of the ECM as well as selective cell survival and proliferation.2,7, 8, 9 In animal models, these processes are orchestrated by mechanisms involving transforming growth factor β (TGFβ) signaling, heme oxygenase 1, interleukin 6, metalloproteinases, and monocyte chemoattractant protein 1, among others.10, 11, 12, 13 Nonetheless, whether these mechanistic paradigms apply to the remodeling of a human preaccess vein to a fistula remains unknown for lack of sufficient human-based studies.
We recently applied RNA-seq to preaccess veins from a small cohort of patients with 2-stage AVF to find molecular signatures that predisposed for AVF failure.14 We showed increased expression of proinflammatory genes in association with nonmaturation.14 In addition, microarray-based analyses of 4 preaccess veins and 6 AVFs by independent groups highlighted the role of TGFβ signaling, inflammatory cytokines, and INHBA and NR4A2-regulated nuclear factor-κB activity in the adaptation of the vein to arterial circulation.15,16 While the emphasis of our previous work was on the preintervention transcriptomic profile, we now apply RNA-seq and paired bioinformatic analyses to vein and AVF tissues from a larger cohort to discover gene expression changes underlying the postoperative transformation. This method also controls for other factors contributing to transcriptional variability. We illustrate for the first time the functional changes required for proper AVF remodeling after arterial anastomosis and those that are relevant for maturation or failure.
Methods
Detailed methods are described in the Supplementary Methods section.
Study Design
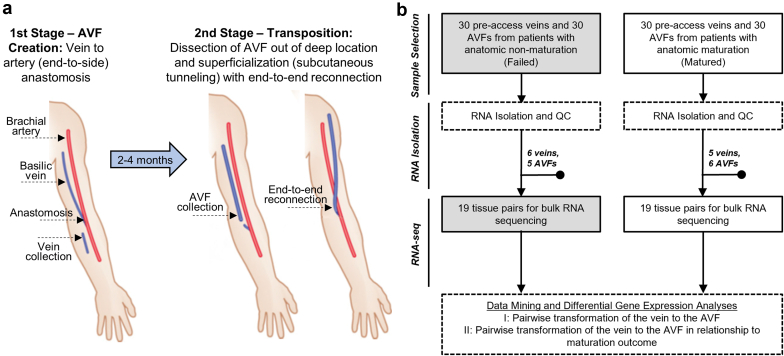
Longitudinal vascular biopsies (preaccess veins and juxta-anastomotic AVF segments) were collected from 60 participants ≥21 years of age with stage 5 chronic kidney disease or end-stage kidney disease and scheduled for 2-stage AVF creation surgery at Jackson Memorial Hospital and the University of Miami Hospital from November 2014 to November 2018 as part of an actively enrolling biorepository. These included 30 patients with successful AVF maturation and 30 with maturation failure (Figure 1a and b), selected from the repository such that both groups had similar representation of baseline characteristics (age, sex, ethnicity, comorbidities, and previous vascular access history) and followed predefined inclusion or exclusion criteria (Supplementary Methods). The AVF specimen was collected approximately 2 cm from the initial anastomosis if the length of the transected AVF was long enough to allow its transposition (Figure 1a). Vein and AVF samples 1 to 5 mm in length were submerged in RNAlater (QIAGEN, Germantown, MD) for RNA isolation. In 19 patients from each group, both the preaccess vein and AVF passed RNA quality control requirements and were selected for bulk RNA-seq (Figure 1b). If enough tissue was available, a 1- to 2-mm cross-section was fixed in 10% neutral formalin (Sigma-Aldrich, St. Louis, MO) for paraffin embedding and sectioning. The study was performed according to the ethical principles of the Declaration of Helsinki and regulatory requirements at both institutions. The ethics committee and Institutional Review Board at the University of Miami approved the study. Preaccess veins from 19 patients were previously analyzed in Martinez et al.14
Figure 1.
Study design and sample collection. (a) Flow diagram of surgical procedures and sample collection. Reprinted (with minor changes) with permission from Martinez et al.2 (b) Total number of veins and AVFs collected for RNA isolation per maturation outcome and number of patients selected for bulk RNA sequencing after applying RNA quality control inclusion criteria (both preaccess vein and AVF sample with RNA integrity score > 5). AVF, arteriovenous fistula; QC, quality control.
Anatomic AVF maturation failure was defined retrospectively as an AVF that never achieved a luminal diameter ≥6 mm, which required a shorter transposition, graft extension, or ligation instead of the standard transposition during second-stage surgery.7 The anatomic maturation definition is a simplified version of the “rule of sixes,” specifically suited to evaluate the biology of early vascular remodeling before the confounding effects of cannulation.
RNA-Seq and Differential Gene Expression Analysis
Preparation and sequencing of RNA libraries was carried out in the John P. Hussman Institute for Human Genomics, Center for Genome Technology. DEGs between preaccess veins and AVFs were identified in DESeq217 using a paired algorithm accounting for the time of tissue collection (vein vs. AVF) and the patient origin of each set of paired samples. The raw RNA-seq data are accessible in NCBI Gene Expression Omnibus through the GEO accession numbers GSE119296 and GSE220796 (Supplementary Table S1).
Cytokine Multiplex Assays
Tissue protein lysates normalized to 400 μg/ml were profiled using the Human Cytokine/Chemokine 48-Plex Discovery Assay (catalog #HD48) and the TGFβ 3-Plex Discovery Assay (#TGFB1-3) by Eve Technologies Corporation (Calgary, AB, Canada).
Histology and Immunohistochemistry Analyses
Tissue sections were stained with Masson’s trichrome (Polysciences, catalog #25088-1) and Alcian blue (Abcam, #ab150662) for gross histomorphometric analysis. For immunohistochemistry , specific proteins were detected using the primary antibodies listed in the Supplementary Methods.
Statistical Analysis
Statistical analyses of morphometry and protein validation data were performed using GraphPad Prism 8.4.0 (San Diego, CA).
Results
Study Design and Characteristics of the Study Cohort
Longitudinal gene expression changes were assessed by comparing preaccess veins obtained at the time of access creation (first-stage surgery) with juxta-anastomotic AVF samples procured at the time of AVF superficialization (second-stage surgery) from 38 individuals (Figure 1a and b). The time between first-stage and second-stage surgeries was 81 ± 30 days (mean ± SD). Both the maturation and failure groups included 19 participants (Figure 1b) and were similar in terms of demographic composition and clinical characteristics (Table 1).
Table 1.
Baseline characteristics of the paired transcriptomics discovery cohort
| Characteristic | Matured (n = 19) | Failed (n = 19) |
|---|---|---|
| Demographics | ||
| Age, yr | 54 ± 17 | 59 ± 13 |
| Female, n (%) | 10 (53) | 9 (47) |
| Race/ethnicity, n (%) | ||
| Black | 9 (47) | 10 (53) |
| Hispanic | 6 (32) | 7 (37) |
| White | 4 (21) | 2 (11) |
| Comorbidities | ||
| Hypertension, n (%) | 19 (100) | 18 (95) |
| Diabetes, n (%) | 9 (47) | 14 (74) |
| ASCVD, n (%) | 6 (32) | 8 (42) |
| CHF, n (%) | 3 (16) | 3 (16) |
| AVF features/history | ||
| Brachiobasilic, n (%) | 17 (89) | 16 (84) |
| Left arm, n (%) | 16 (84) | 17 (89) |
| Vein diameter, mm | 4.0 [4.0–4.0] | 4.0 [4.0–4.0] |
| AVF diameter, mm | 7.0 [6.0–7.5] | 4.5 [4.0–5.0] |
| Time intervala, d | 78 ± 29 | 85 ± 32 |
| Stage-5 predialysis, n (%) | 2 (11) | 5 (26) |
| Previous AVF, n (%) | 3 (16) | 4 (21) |
ASCVD, atherosclerotic cardiovascular disease; AVF, arteriovenous fistula; CHF, congestive heart failure.
Continuous variables are expressed as mean ± SD or median (interquartile range).
Time interval between first-stage and second-stage surgeries.
Transcriptome-Wide Changes During the Vein to AVF Transformation
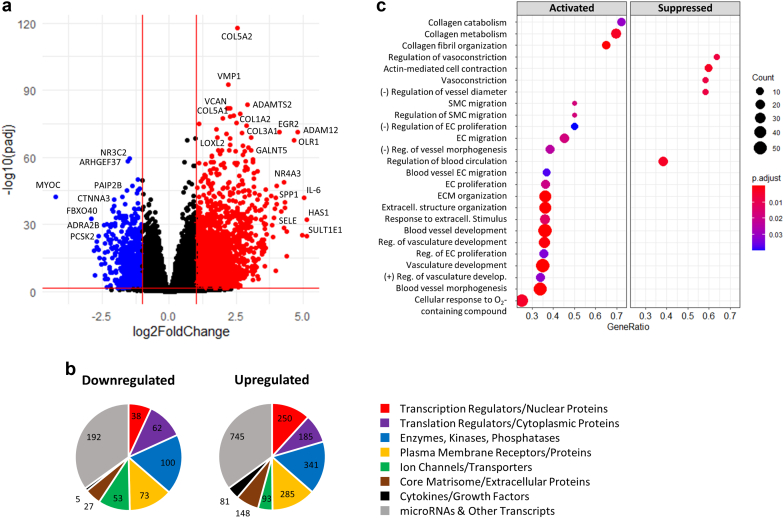
A total of 3637 transcripts were differentially expressed between veins and AVFs in paired analyses (absolute log2 fold change [FC] ≥ 1, false discovery rate [FDR] < 0.05; Figure 2a, Supplementary Figure S1, Supplementary Data File S1). Eighty percent of these transcripts were upregulated after anastomosis (clusters 1 and 2 in Supplementary Figure S1A and C), whereas the remaining 20% had lower transcriptional activity in fistulas (cluster 3). Clusters 1 and 3 best reflected the transformation between veins and AVFs and included the highest-expressing genes and low proportion of pseudogenes and other nonfunctional transcripts (Supplementary Figure S1C and D). Among 2678 DEGs with identified functions in digital databases,18 there was a similar distribution of gene functions between downregulated and upregulated DEGs in AVFs. Exceptions included a higher representation of transcription regulators or nuclear proteins (12% vs. 7%), more cytokines or growth factors (4% vs. 1%), and lower proportion of ion channels and transporters (4% vs. 10%) in upregulated versus downregulated genes, respectively (Figure 2b).
Figure 2.
DEGs and functional pathways in the vein to AVF transformation. (a) Volcano plot representation of 3637 DEGs in AVFs compared with their corresponding preaccess veins (pairwise comparisons). Changes in gene expression are presented as log2(fold change) in AVFs with respect to veins. Red dots indicate upregulated genes in AVFs (log2 fold change ≥ 1, false discovery rate < 0.05), and blue dots are genes downregulated in fistulas (log2 fold change ≤ −1, false discovery rate < 0.05). (b) Distribution of functions in 2678 DEGs with functional classifications available. (c) Pathway enrichment analyses of DEGs indicating activated and suppressed biological processes during the vein to AVF transformation. Biological processes are organized by gene ratio on the x-axis (also known as enrichment ratio), which is defined as the ratio of DEGs annotated in a term (count) to the total number of genes in this process in genome-wide annotation packages. DEG, Differentially expressed gene; EC, endothelial cell; ECM, extracellular matrix; SMC, smooth muscle cell.
Gene set enrichment analyses of DEGs revealed transcriptional activation of genes related to vasculature development, ECM remodeling, SMC migration, and endothelial cell (EC) proliferation and migration (Figure 2c, Supplementary Figure S2A; see Supplementary Data File S2 for full list of pathways). Vasculature development pathways involved upregulation of basement membrane and interstitial ECM components (e.g., collagens, proteoglycans, and glycoproteins), cell adhesion molecules (e.g., CDH2), hemostasis factors (e.g., SERPINE1), multiple cytokines, and angiogenesis regulators (e.g., SEMA4A, ESM1, PTGS2, and SPARC) (Supplementary Figure S2). Fibrillar and nonfibrillar collagens, proteoglycans, and ECM-modifying enzymes also formed part of gene sets involved in ECM organization and collagen fibril formation.
In agreement with the development of an enlarged vessel, suppressed pathways included those related to cell contraction, vasoconstriction, and regulation of vessel diameter and blood circulation (Figure 2c). Inhibition of these processes was explained by downregulation of various adrenergic receptors, intermediate filament proteins (e.g., DES), voltage-gated potassium channels, and genes involved in regulation of intracellular calcium (Supplementary Figure S2). Upregulation of mechanosensitive TRPV4 and LRRC8A channels (Supplementary Data File S1) may also illustrate a multigenic mechanistic apparatus in control of flow-induced vasodilation.19,20
Transcriptional Regulation of the Vein to AVF Transformation
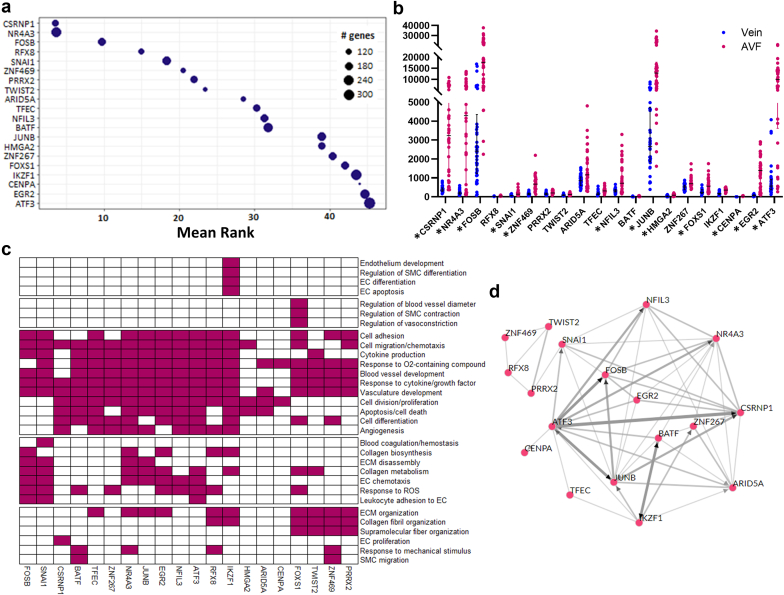
The increased proportion of transcriptional regulators among upregulated DEGs prompted us to search for potential transcriptional drivers of the vein to AVF transformation. For an unbiased prediction of transcription factors (TFs) responsible for DEGs between veins and AVFs, a transcription factor enrichment analysis21 was performed using highly expressed DEGs (1170 with base mean >50) as putative targets. The 20 best-ranked TFs are presented in Figure 3a (full list in Supplementary Data File S3), 12 of which were among the list of DEGs and 7 others had higher expression in AVFs than veins (FDR < 0.05) but did not make the cutoff of log2FC ≥ 1 (log2FC ranging from 0.40 in PRRX2 to 0.97 in IKZF1). Pathway analyses of target DEGs controlled by these factors identified specific vascular functions in which these TFs may play a role. For example, highly upregulated TFs such as CSRNP1, NR4A3, JUNB, EGR2, and ATF3 seemed to control mostly cellular functions; NR4A3, JUNB, SNAI1, and FOSB played specific roles in ECM disassembly, whereas ZNF469 and FOXS1 regulated ECM organization (both), blood vessel diameter (FOXS1), and response to mechanical stimulus (ZNF469) (Figure 3c). The transcription factor enrichment analysis algorithm also supports coexpression and/or direct transcriptional regulation of several of these TFs by other factors in this set (Figure 3d).
Figure 3.
Transcriptional regulation of the vein to AVF transformation. (a) TF enrichment analysis indicating the 20 best-ranked TFs in the transcriptional regulation of the vein to AVF transformation. This analysis ranks TFs based on the number of predicted targets in the input data set compared with the total number of targets experimentally identified by chromatin immunoprecipitation sequencing. (b) RNA expression in the vein and AVF of the 20 best TFs predicted in (a). The y-axis presents normalized counts, and the asterisk symbol indicates TFs that are differentially expressed during the vein to AVF transformation (log2 fold change ≥ 1, false discovery rate < 0.05). (c) Prediction of biological processes in which the above TFs participate based on pathway analyses of their corresponding gene targets. (d) Coregulatory networks of the above TFs indicating coexpression of TFs in chromatin immunoprecipitation sequencing libraries (gray lines) or direct transcriptional regulation by an indicated factor (pointed arrows). AVF, arteriovenous fistula; EC, endothelial cell; ECM, extracellular matrix; ROS, reactive oxygen species; SMC, smooth muscle cell; TF, transcription factor.
MicroRNAs contribute to posttranscriptional regulation by inhibiting translation and destabilizing mRNA. In addition to the above TFs, multiple differentially expressed microRNAs were identified as participants of vasculature development processes (Supplementary Figure S2A, Supplementary Data File S2).
ECM Remodeling During the Vein to AVF Transformation
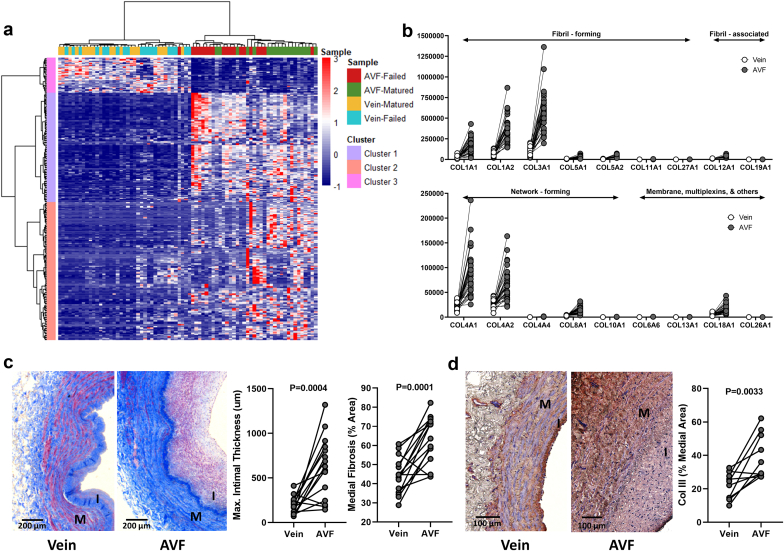
Considering the essential role of ECM remodeling in AVF development and maturation outcomes and that any differences in ECM genes between AVFs and preaccess veins likely reflect stable transcriptional signatures of this early remodeling period, we then focused our prepost comparisons on “matrisome” gene expression, the ensemble of genes encoding for core ECM and ECM-associated proteins. Using as reference the MatrisomeDB annotation database22 and a summary of proteoglycan biosynthetic or modifying enzymes,23 a total of 285 matrisome genes were identified in the overall set of DEGs between veins and AVFs (Figure 4a, Supplementary Data File S4). More than 87% of these matrisome DEGs were upregulated in AVFs compared with those in preaccess veins (Figure 4a, Supplementary Figure S3A) and included some of the genes with highest FC in genome-wide differential expression analyses (Figure 2a). Interestingly, upregulated matrisome genes (clusters 1 and 2) in Figure 4a were enough to make a crude separation of AVFs that failed from those with successful maturation.
Figure 4.
Extracellular matrix remodeling during the vein to AVF transformation. (a) Heatmap of 285 differentially expressed “matrisome” genes (absolute log2 fold change ≥ 1, false discovery rate < 0.05) in AVFs compared with their corresponding preaccess veins. Clusters 1 and 2 are upregulated in AVFs with respect to veins, whereas cluster 3 is downregulated in fistulas. (b) Expression levels and magnitude of fold change in upregulated collagen genes during the vein to AVF transformation. Y-axes indicate normalized counts. False discovery rate < 0.05 for all paired comparisons. (c) Representative trichrome stainings and quantification of morphometry in veins and AVF tissue sections. (d) Immunohistochemistry of collagen III and quantification in veins and AVF tissue sections. Values in (c) and (d) were compared using paired t-tests. AVF, arteriovenous fistula; I, intima; M, media.
Among the upregulated collagen genes, the ones with the highest magnitude of FC and expression levels (normalized counts) were the fibrillar collagens I and III, network-forming collagens IV and VIII, and multiplexin collagen XVIII, which is also considered a heparan sulfate proteoglycan (Figure 3b). Collagens I and III are the main collagenous components of the interstitial ECM, whereas the hexagonal networks of collagens IV and VIII provide an anchoring scaffold and mechanical strength to the basement membrane.24 Increased expression of collagen V was also noted and that of collagen XII, which is induced by excess mechanical stress and further stabilizes fibrillar collagens.25,26 Increased expression of genes involved in collagen processing and metabolism suggests that organization of the ECM is an active process in the AVF. These include SERPINH1, an essential procollagen chaperone in the endoplasmic reticulum,27 various endoplasmic reticulum–resident prolyl and lysyl hydroxylases (P3H1, P3H4, P4HA3, and PLOD1), which generate stable triple helices,28 procollagen C-proteinases and N-proteinases (BMP1, ADAMTS2), procollagen C-proteinase enhancer (PCOLCE), and multiple metalloproteinases (Supplementary Figure S3B). Overall, the widespread upregulation of collagen genes in AVFs corresponded to a significant increase in medial fibrosis and fibrillar collagen deposition in the media, as exemplified by collagen III, the most abundant and one of the most upregulated after anastomosis (Figure 4c and d).
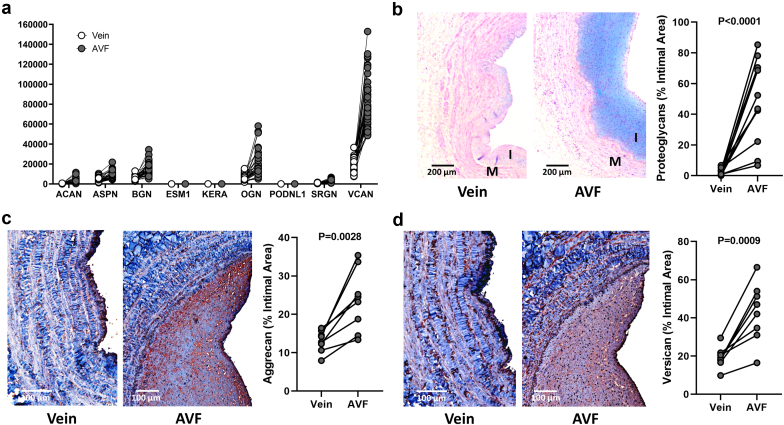
Increased expression of collagens and processing factors was paralleled by notable ECM glycoproteins (FN1, ELN, laminins, fibrillins, thrombospondins, SPARC, CTGF, CYR61, etc.) and cross-linking enzymes (LOX and LOXL2) with functions in ECM remodeling and wall stability (Supplementary Figure S3B). Five proteoglycans stood out in terms of FC and magnitude of expression in AVFs compared with those in preaccess veins, including ACAN, ASPN, BGN, OGN, and VCAN (Figure 5a). There was also significant upregulation of proteoglycan biosynthetic or modifying enzymes such as HAS1, HAS2, B3GNT5, CHST2, and GALNT5. (Supplementary Figure S3B). Interestingly, increased collagen deposition in AVFs predominantly affected the medial layer, whereas postoperative intimal expansion was characterized by overt proteoglycan biosynthesis (Figure 5), as confirmed by a significant increase in glycosaminoglycan-targeted (Alcian blue) and core protein–targeted staining (Figure 5b–d).
Figure 5.
Intimal remodeling during the vein to AVF transformation. (a) Expression levels and magnitude of fold change in upregulated proteoglycan genes during the vein to AVF transformation. The y-axis indicates normalized counts. False discovery rate < 0.05 for all paired comparisons. (b) Representative Alcian blue stainings and quantification in veins and AVF tissue sections. (c,d) Immunohistochemistry of aggrecan (c) and versican (d) and quantification in veins and AVF tissue sections. Values in (b) to (d) were compared using paired t-tests. AVF, arteriovenous fistula; I, intima; M, media.
Secreted Factors and Inflammation During the Vein to AVF Transformation
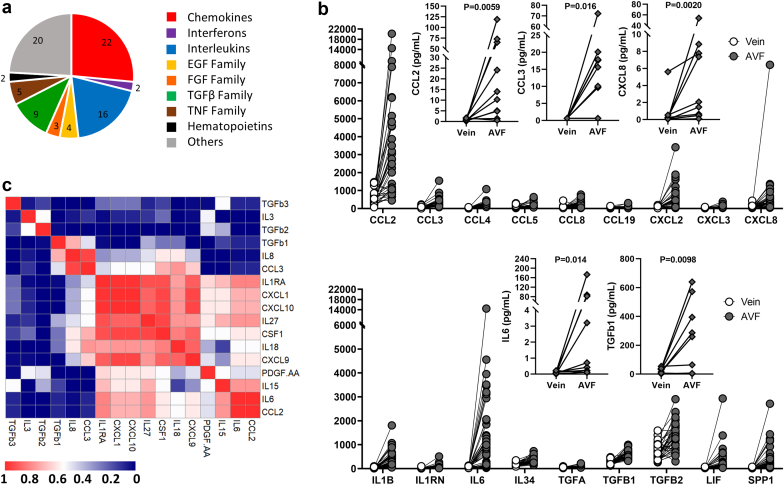
Many secreted factors are integral constituents of the ECM because they bind to glycosaminoglycans and ECM protein domains and act as autocrine or paracrine signals that regulate matrisome gene expression. Of the 91 secreted factors differentially expressed between AVFs and preaccess veins, 90% were upregulated in fistula tissues (Figure 6a, Supplementary Figure S3A). Chemokines with the highest fold increase from veins to AVFs were MCP-1 (CCL2), MIP-1α (CCL3), CXCL2, and interleukin-8 (CXCL8) (Figure 6b). Other factors that stood out in terms of FC and magnitude of expression included IL1B, IL6, TGFβ1 and TGFβ2, LIF, and SPP1, which is also a glycoprotein. Upregulation of many of these secretable factors after anastomosis was confirmed at the protein level using multiplex assays (Figure 6b) and were found localized throughout the AVF wall, including the intima and media (Supplementary Figure S3C). Interestingly, interleukins and chemokines in AVFs correlated for the most part at the protein level, whereas the concentrations of TGFβ1 and TGFβ2 correlated neither with these factors nor with each other (Figure 6c).
Figure 6.
Inflammation during the vein to AVF transformation. (a) Family distribution of upregulated secretable factors during the vein to AVF transformation. (b) RNA expression levels (dots) and magnitude of fold change in upregulated secretable factors. Y-axes indicate normalized counts. False discovery rate < 0.05 for all paired comparisons. Inset plots present protein quantifications (diamonds) of select factors as determined by multiplex assays. Protein levels were compared using Wilcoxon matched-pairs signed-rank tests. (c) Spearman correlation of upregulated secretable factors for which protein levels were available. AVF, arteriovenous fistula; EGF, epidermal growth factor; FGF, fibroblast growth factor; TGF, transforming growth factor; TNF, tumor necrosis factor.
Maturation Failure: When the Vein to AVF Transformation Goes Wrong
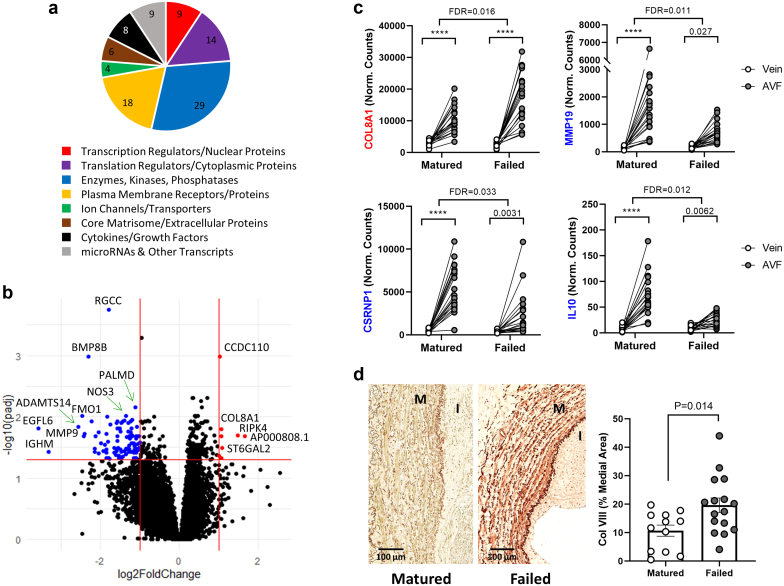
Twenty-eight AVFs of those analyzed by RNA-seq had enough tissue available for histology and immunohistochemistry (12 mature and 16 failures). The percentage of medial fibrosis was higher in AVFs that failed than in those that matured (68.01 ± 12.66% vs. 57.99 ± 9.69%, P = 0.036), whereas maximal intimal thickness was similar between outcomes (992.0 ± 606.0 vs. 751.7 ± 614.3 μm, P = 0.32, respectively). Next, we sought to identify the changes in gene expression during the vein to AVF transformation that were relevant to maturation failure. Paired differential gene expression analyses conditional to maturation outcomes identified 102 DEGs in association with AVF failure (absolute log2FC ≥ 1, FDR < 0.05; Figure 7a and b, Supplementary Data File S5). Half (51) of these DEGs increased in expression from the vein to the AVF irrespective of maturation outcomes (Supplementary Data Files S1 and S5) but either increased significantly more in AVFs that failed (COL8A1, RIPK4, ST6GAL2, and AP000808.1) or did not increase to the same extent (47 of 51; e.g., IL10, MMP19, and CSRNP1) compared with those in successful maturation (Figure 7c). Four additional DEGs decreased from the vein to the AVF irrespective of maturation outcomes and significantly more so with maturation failure (S100B, GFRA3, L1CAM, and ST6GALNAC1). The remaining 47 failure-associated DEGs were detected for the first time in these outcome-dependent analyses and not in the general vein to AVF comparisons because of punctuated differences between outcomes (4 upregulated [TRMT9B, CCDC110, INHBA-AS1, and AL160153.1] and 43 downregulated with failure [e.g., NOS3, ARG2, and HYAL2]; Supplementary Figure S4).
Figure 7.
Differentially expressed genes (DEGs) during the vein to AVF transformation in association with maturation failure. (a) Distribution of functions in 97 of 102 failure-associated DEGs during the vein to AVF transformation. The functional classification of 5 DEGs has not been determined. (b) Volcano plot representation of differential gene expression analysis between AVFs and their corresponding preaccess veins conditional to maturation failure. Log2(fold change) on the x-axis represents how much more (or less) a gene changed in tissue pairs that failed versus those that matured. Red dots indicate upregulated genes with failure compared with successful maturation (log2 fold change ≥ 1, false discovery rate [FDR] < 0.05), and blue dots are genes downregulated with failure (log2 fold change ≤ −1, FDR < 0.05). (c,d) Examples of DEGs during the vein to AVF transformation in association with maturation failure. COL8A1 is upregulated, whereas MMP19, CSRNP1, and IL10 are downregulated with failure. Values above brackets indicate the FDR for the corresponding comparison. ∗∗∗∗FDR < 0.001. Immunohistochemistry of collagen VIII and quantification in AVFs that matured or failed. Values were compared using an unpaired t-test. AVF, arteriovenous fistula.
Ninety-seven failure-associated DEGs had functions identified in digital databases (Figure 7a),18 and more than one-fourth (28 of 102) formed part of the matrisome, including core matrisome or ECM proteins, secretable factors, multiple proteoglycan biosynthetic or modifying enzymes, and enzymatic ECM regulators (Supplementary Data File S4). This agrees with previous associations of adverse ECM remodeling with maturation failure.2,29 Interestingly, the only collagen gene upregulated with failure was COL8A1, which was confirmed by immunohistochemistry (Figure 7d). Increased deposition of collagen VIII occurred predominantly in the media, in contrast with its initial identification as a component of the endothelial basement membrane30 but in agreement with reports of injured SMCs and the media of developing vessels.31, 32, 33 Another profibrotic factor upregulated in AVFs that failed was INHBA-AS1, which is associated with increased fibrillar collagen deposition in hypertrophic scar formation.34
The glycosaminoglycan biosynthetic enzyme ST6GAL2 was also upregulated during the vein to AVF transformation in AVFs that failed compared with those that matured, whereas other proteoglycan-modifying enzymes (HYAL2 and HPSE), metalloproteinases (MMP9, MMP19, ADAMTS9, and ADAMTS14), and at least 2 anti-inflammatory factors (IL1RN and IL10) had decreased expression in cases of failure (Supplementary Data File S4, Supplementary Figure S4). Bone morphogenetic protein 2, which antagonizes TGFβ signaling,35 and SERTAD1, a transcriptional coactivator of BMP2-induced Smad1 signaling,36 were also downregulated with maturation failure. Altogether, these and the above changes suggest a dysregulation of ECM remodeling in maturation failure.
Finally, at least 17 endothelium-predominant genes were downregulated during the vein to AVF transformation in AVFs that failed compared with those that matured. These included NOS3, ARG2, ICAM2, SELP, SELL, APOLD1, PALMD, and SRGN, among others (Supplementary Figure S4, Supplementary Data File S5). This downregulation may be related to lower levels of TFs that regulate endothelial genes (e.g., CSRNP1, SOX17, ZNF331, FOSL1, and MYC) in AVFs that failed (Figure 7c, Supplementary Figures S4 and S5, Supplementary Data File S6), or the antiangiogenic effect of vastatin, a collagen VIII–derived matrikine.37 In support of the latter, COL8A1 expression in AVFs negatively correlated with RNA levels of NOS3 and PALMD (Supplementary Figure S4B). Downregulation of EC markers may also indicate a higher degree of endothelial dysfunction in maturation failure.
Discussion
The AVF created in 2 stages is one of the few clinical scenarios that allows the longitudinal analysis of human veins to study the mechanism underlying flow-induced venous remodeling. In this work, to our knowledge, we present for the first time the postoperative changes in the transcriptomic landscape of the human vein after anastomosis. We identified genes and regulatory networks that explain the ability of the vessel to dilate and thicken in response to supra-arterial circulation. We provided evidence of layer-specific ECM reorganization in AVFs. Finally, we discovered a small list of genes associated with maturation failure that can be potentially targeted to prevent stenoses. Having longitudinal human data of gene expression changes associated with failure is crucial not only to obtain a clinically relevant understanding of the AVF remodeling process but also to reassess and adapt our experimental models for the accurate design of future therapies.
The first objective of this study was to characterize the coordinated molecular changes evoked by overflow in the structure and composition of the AVF wall to understand physiological remodeling. We demonstrated increased expression of mechanosensitive channels, stretch or flow-induced collagens, proteoglycans, cytokines, and ECM-modifying enzymes, along with downregulation of genes involved in vasoconstriction after anastomosis. Mechanosensation of increased flow and volume induces wall remodeling and thickening to withstand circumferential wall stress and secure hemostasis.6,38 Mechanosensitive channels such as TRPV4 activate ECM remodeling by inducing matrix synthesis, MMP activity, and cytokine/growth factor signaling, including TGFβ.39 Increased synthesis of fibrillar collagens I and III provides tensile strength and integrity to the AVF wall.40 Concurrently, the significant increase in network-forming collagen IV relates to its crucial role in stabilizing the basement membrane while allowing wall stretching.24,40 Collagen types V, VIII, XII, and XVIII were barely transcribed in the preaccess vein but are significantly upregulated in AVFs to further stabilize basement membranes and regulate fibril formation.24, 25, 26,41,42
Another hallmark of this study was the finding that proteoglycans significantly accumulate in the intima after AVF creation. Proteoglycans, particularly heparan sulfate, hyaluronan, and sialic acid–modified proteins, play an important function in shear-induced mechanotransduction because they help increase nitric oxide production,43, 44, 45, 46, 47 although this effect is debated in larger vessels.48 Fluid shear stress stimulates hyaluronan synthesis by ECs.49 However, the localization of proteoglycans and Alcian blue staining throughout the entire intima supports their production by neointimal cells more so than ECs. This characteristic of the intima has also been observed in animal models of restenosis and vein grafts50,51 and speaks to the uniqueness of this layer as sponge and/or reservoir of secretable factors, regulator of immune cell infiltration, and potential biomechanical support to resist hemodynamic compression. Animal models have shown an increase in hyaluronic acid during AVF remodeling and a role for the hyaluronic acid receptor CD44 in promoting inflammation.52 The upregulation of ST6GAL2 and downregulation of HYAL2, HPSE, and ST6GALNAC1 in AVFs that failed suggest a disbalance in glycosaminoglycan structure and composition compared with mature vessels, although the exact mechanisms remain to be identified. Moreover, whether intimal proteoglycans are a requirement for intimal cell migration and survival or other characteristics of this layer is a critical question for future studies, with profound implications for therapies aiming to control intimal expansion.
As a final objective of this study, to our knowledge, we have now discovered for the first time the association between changes in gene expression after anastomosis and AVF failure. Surprisingly, only a small number of DEGs seems to be modifying the remodeling process to determine maturation or failure. Among the few upregulated genes with failure was COL8A1, whose accumulation was further confirmed surrounding SMCs in the media. Similar to collagen IV, the postoperative deposition of collagen type VIII may contribute to wall stability.24 However, collagen VIII increases collagen I and III deposition and TGFβ signaling under volume overload conditions53 and may be related to vascular stiffness and ECM rigidity in AVFs that fail. Collagen VIII also promotes SMC migration,54 suggesting a potential role in intimal hyperplasia. Although we did not observe differences in fibrillar collagens at the RNA level between AVFs that matured and those that failed, it is possible that these changes were detectable early in remodeling and correspond to an increase in medial fibrosis with maturation failure as demonstrated previously.2 Expression of collagen VIII by SMCs is induced by injury mechanisms and/or increases in flow as observed in animal models of AVF and balloon injury.12,31,33 Importantly, unlike collagen IV, global Col8a1/Col8a2 knockout animals are viable and present normal vascular development,55 which suggests that this is a targetable collagen to improve AVF maturation.
Interestingly, multiple endothelial-predominant genes are downregulated with AVF failure compared with successful maturation. These differences may represent changes in EC phenotypes caused by downregulation of important EC-active TFs with failure. Alternatively, it may represent endothelial dysfunction or deficient postoperative luminal reendothelialization because there are no significant differences in intramural neovascularization between maturation outcomes.56 Of note, reduced expression of PALMD with failure may impair alignment of EC nuclei with respect to flow, increasing mechanical stress and risk of stenosis.57 The endothelial proteoglycan SRGN is another mechanosensitive gene that promotes nitric oxide synthesis in response to abnormal wall shear stress58 and is downregulated with AVF failure. Altogether, these examples may illustrate deficiencies in mechanosensitive adaptations in AVF nonmaturation. It is difficult to predict nitric oxide availability in these human samples as both NOS3 and ARG2 are downregulated with failure. However, NOS3 overexpression and perivascular nitric oxide delivery have shown promising results in animal AVF models.59, 60, 61
The higher number of individuals and application of paired bioinformatics in the present work proved a powerful approach to find postoperative changes related to outcomes while controlling for other factors contributing to transcriptional variability. These analyses lay the groundwork for future omics studies to create a more comprehensive molecular framework of the AVF transformation. The combination of these data with proteomics and single-cell analyses will help identify specific cell phenotypes and intercellular and extracellular regulatory networks that determine the proper adaptation of the vein after AVF creation. These findings will also allow a better interrogation of existing animal models and an improved retrotranslational approach (bedside-bench) for the discovery of novel therapies. We will be able to generate new genetically modified models to establish the causality of these molecular mechanisms and help close the gap between experimental AVFs and clinically relevant remodeling processes.
The limitations of this study include the single-center setting and the exclusion of forearm AVFs. Assessment of the postoperative juxta-anastomotic transcriptomic profile at the time of superficialization may also preclude the detection of gene expression changes occurring early after anastomosis or in other regions of the AVF that may be relevant to maturation or failure. Despite these limitations, our work provides a clinically relevant framework of the molecular transformation from the vein to the AVF to be further dissected in future mechanistic studies. The new data highlight the potential role of collagen VIII as a new ECM regulator of postoperative venous remodeling. Considering the antiangiogenic properties of this collagen, perivascular interventions targeting collagen VIII at the time of AVF creation may be a feasible strategy to improve outward remodeling and even endothelial function, without significantly compromising wall strength or increasing the risk of aneurysms.
Disclosure
All the authors declared no competing interests.
Acknowledgments
This study was supported by the National Institutes of Health grants R01-DK098511 to LHS and RIV-P, R01-DK121227 and R01-DK132888 to RIV-P, K08-HL151747 to LM, and the VA Merit Award IBX004658 to RIV-P.
Footnotes
Supplementary Methods.
Figure S1. Summary of transcriptomic changes during the vein to AVF transformation.
Figure S2. Pathways involved in the vein to AVF transformation.
Figure S3. Matrisome expression changes during the vein to AVF transformation.
Figure S4. Gene expression changes associated with AVF maturation failure.
Figure S5. Transcriptional regulation of genes associated with maturation failure.
Table S1. Accession numbers of raw sequencing data in the NCBI Gene Expression Omnibus repository.
Data File S1. Differential gene expression analysis of 57,505 transcripts between veins and AVFs (paired analysis).
Data File S2. Gene set enrichment analysis of 3637 DEGs between veins and AVFs.
Data File S3. Transcription factor enrichment analysis of putative factors regulating DEGs between veins and AVFs.
Data File S4. Summary and functional classification of matrisome genes differentially expressed between veins and AVFs and in association with failure.
Data File S5. Differential gene expression analysis of 17,992 transcripts between veins and AVFs in association with failure (paired analysis).
Data File S6. Transcription factor enrichment analysis of putative factors regulating DEGs in association with failure.
Supplementary Material
Supplementary Methods.
Figure S1. Summary of transcriptomic changes during the vein to AVF transformation.
Figure S2. Pathways involved in the vein to AVF transformation.
Figure S3. Matrisome expression changes during the vein to AVF transformation.
Figure S4. Gene expression changes associated with AVF maturation failure.
Figure S5. Transcriptional regulation of genes associated with maturation failure.
Table S1. Accession numbers of raw sequencing data in the NCBI Gene Expression Omnibus repository.
Data File S1. Differential gene expression analysis of 57,505 transcripts between veins and AVFs (paired analysis).
Data File S2. Gene set enrichment analysis of 3637 DEGs between veins and AVFs.
Data File S3. Transcription factor enrichment analysis of putative factors regulating DEGs between veins and AVFs.
Data File S4. Summary and functional classification of matrisome genes differentially expressed between veins and AVFs and in association with failure.
Data File S5. Differential gene expression analysis of 17,992 transcripts between veins and AVFs in association with failure (paired analysis).
Data File S6. Transcription factor enrichment analysis of putative factors regulating DEGs in association with failure.
References
- 1.Robbin M.L., Greene T., Cheung A.K., et al. Arteriovenous fistula development in the first 6 weeks after creation. Radiology. 2016;279:620–629. doi: 10.1148/radiol.2015150385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martinez L., Duque J.C., Tabbara M., et al. Fibrotic venous remodeling and nonmaturation of arteriovenous fistulas. J Am Soc Nephrol. 2018;29:1030–1040. doi: 10.1681/ASN.2017050559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee T., Qian J.Z., Zhang Y., Thamer M., Allon M. Long-term outcomes of arteriovenous fistulas with unassisted versus assisted maturation: a retrospective National Hemodialysis Cohort Study. J Am Soc Nephrol. 2019;30:2209–2218. doi: 10.1681/ASN.2019030318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huijbregts H.J., Bots M.L., Wittens C.H., Schrama Y.C., Moll F.L., Blankestijn P.J. Hemodialysis arteriovenous fistula patency revisited: results of a prospective, multicenter initiative. Clin J Am Soc Nephrol. 2008;3:714–719. doi: 10.2215/CJN.02950707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robbin M.L., Greene T., Allon M., et al. Prediction of arteriovenous fistula clinical maturation from postoperative ultrasound measurements: findings from the hemodialysis fistula maturation study. J Am Soc Nephrol. 2018;29:2735–2744. doi: 10.1681/ASN.2017111225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dixon B.S. Why don’t fistulas mature? Kidney Int. 2006;70:1413–1422. doi: 10.1038/sj.ki.5001747. [DOI] [PubMed] [Google Scholar]
- 7.Tabbara M., Duque J.C., Martinez L., et al. Pre-existing and postoperative intimal hyperplasia and arteriovenous fistula outcomes. Am J Kidney Dis. 2016;68:455–464. doi: 10.1053/j.ajkd.2016.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan J.S., Campos B., Wang Y., et al. Proliferation patterns in a pig model of AV fistula stenosis: can we translate biology into novel therapies? Semin Dial. 2014;27:626–632. doi: 10.1111/sdi.12240. [DOI] [PubMed] [Google Scholar]
- 9.de Graaf R., Dammers R., Vainas T., Hoeks A.P., Tordoir J.H. Detection of cell-cycle regulators in failed arteriovenous fistulas for haemodialysis. Nephrol Dial Transplant. 2003;18:814–818. doi: 10.1093/ndt/gfg033. [DOI] [PubMed] [Google Scholar]
- 10.Hu H., Lee S.R., Bai H., et al. TGFbeta (transforming growth factor-beta)-activated kinase 1 regulates arteriovenous fistula maturation. Arterioscler Thromb Vasc Biol. 2020;40:e203–e213. doi: 10.1161/ATVBAHA.119.313848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Juncos J.P., Tracz M.J., Croatt A.J., et al. Genetic deficiency of heme oxygenase-1 impairs functionality and form of an arteriovenous fistula in the mouse. Kidney Int. 2008;74:47–51. doi: 10.1038/ki.2008.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hall M.R., Yamamoto K., Protack C.D., et al. Temporal regulation of venous extracellular matrix components during arteriovenous fistula maturation. J Vasc Access. 2015;16:93–106. doi: 10.5301/jva.5000290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nguyen M., Thankam F.G., Agrawal D.K. Sterile inflammation in the pathogenesis of maturation failure of arteriovenous fistula. J Mol Med (Berl) 2021;99:729–741. doi: 10.1007/s00109-021-02056-4. [DOI] [PubMed] [Google Scholar]
- 14.Martinez L., Tabbara M., Duque J.C., et al. Transcriptomics of human arteriovenous fistula failure: genes associated with nonmaturation. Am J Kidney Dis. 2019;74:73–81. doi: 10.1053/j.ajkd.2018.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jie K., Feng W., Boxiang Z., et al. Identification of pathways and key genes in venous remodeling after arteriovenous fistula by bioinformatics analysis. Front Physiol. 2020;11 doi: 10.3389/fphys.2020.565240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hashimoto Y., Okamoto A., Saitoh H., et al. Gene expression changes in venous segment of overflow arteriovenous fistula. Int J Nephrol. 2013;2013 doi: 10.1155/2013/980923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.QIAGEN ingenuity pathway analysis. QIAGEN. https://digitalinsights.qiagen.com/IPA
- 19.Gao F., Wang D.H. Hypotension induced by activation of the transient receptor potential vanilloid 4 channels: role of Ca2+-activated K+ channels and sensory nerves. J Hypertens. 2010;28:102–110. doi: 10.1097/HJH.0b013e328332b865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alghanem A.F., Abello J., Maurer J.M., et al. The SWELL1-LRRC8 complex regulates endothelial AKT-eNOS signaling and vascular function. eLife. 2021;10 doi: 10.7554/eLife.61313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keenan A.B., Torre D., Lachmann A., et al. ChEA3: transcription factor enrichment analysis by orthogonal omics integration. Nucleic Acids Res. 2019;47:W212–W224. doi: 10.1093/nar/gkz446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shao X., Taha I.N., Clauser K.R., Gao Y.T., Naba A. MatrisomeDB: the ECM-protein knowledge database. Nucleic Acids Res. 2020;48:D1136–D1144. doi: 10.1093/nar/gkz849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gottschalk J., Elling L. Current state on the enzymatic synthesis of glycosaminoglycans. Curr Opin Chem Biol. 2021;61:71–80. doi: 10.1016/j.cbpa.2020.09.008. [DOI] [PubMed] [Google Scholar]
- 24.Ricard-Blum S. The collagen family. Cold Spring Harb Perspect Biol. 2011;3:a004978. doi: 10.1101/cshperspect.a004978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fluck M., Giraud M.N., Tunc V., Chiquet M. Tensile stress-dependent collagen XII and fibronectin production by fibroblasts requires separate pathways. Biochim Biophys Acta. 2003;1593:239–248. doi: 10.1016/s0167-4889(02)00394-4. [DOI] [PubMed] [Google Scholar]
- 26.Chiquet M., Birk D.E., Bonnemann C.G., Koch M. Collagen XII: protecting bone and muscle integrity by organizing collagen fibrils. Int J Biochem Cell Biol. 2014;53:51–54. doi: 10.1016/j.biocel.2014.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koide T., Nagata K. Collagen biosynthesis. Top Curr Chem. 2005;247:85–114. doi: 10.1007/b103820. [DOI] [Google Scholar]
- 28.Myllyharju J. Intracellular post-translational modifications of collagens. Top Curr Chem. 2005;247:115–147. doi: 10.1007/b103821. [DOI] [Google Scholar]
- 29.Hernandez D.R., Applewhite B., Martinez L., et al. Inhibition of lysyl oxidase with beta-aminopropionitrile improves venous adaptation after arteriovenous fistula creation. Kidney360. 2021;2:270–278. doi: 10.34067/kid.0005012020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sage H., Trueb B., Bornstein P. Biosynthetic and structural properties of endothelial cell type VIII collagen. J Biol Chem. 1983;258:13391–13401. doi: 10.1016/S0021-9258(17)44129-9. [DOI] [PubMed] [Google Scholar]
- 31.Bao H., Li Z.T., Xu L.H., et al. Platelet-derived extracellular vesicles increase Col8a1 secretion and vascular stiffness in intimal injury. Front Cell Dev Biol. 2021;9 doi: 10.3389/fcell.2021.641763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iruela-Arispe M.L., Sage E.H. Expression of type VIII collagen during morphogenesis of the chicken and mouse heart. Dev Biol. 1991;144:107–118. doi: 10.1016/0012-1606(91)90483-j. [DOI] [PubMed] [Google Scholar]
- 33.Sibinga N.E., Foster L.C., Hsieh C.M., et al. Collagen VIII is expressed by vascular smooth muscle cells in response to vascular injury. Circ Res. 1997;80:532–541. doi: 10.1161/01.res.80.4.532. [DOI] [PubMed] [Google Scholar]
- 34.Yang Y., Xiao C., Liu K., Song L., Zhang Y., Dong B. Silencing of long noncoding INHBA antisense RNA1 suppresses proliferation, migration, and extracellular matrix deposition in human hypertrophic scar fibroblasts via regulating microRNA-141-3p/myeloid cell leukemia 1 axis. Bioengineered. 2021;12:1663–1675. doi: 10.1080/21655979.2021.1919013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Calvier L., Chouvarine P., Legchenko E., et al. PPARgamma links BMP2 and TGFbeta1 pathways in vascular smooth muscle cells, regulating cell proliferation and glucose metabolism. Cell Metab. 2017;25:1118–1134.e7. doi: 10.1016/j.cmet.2017.03.011. [DOI] [PubMed] [Google Scholar]
- 36.Peng Y., Zhao S., Song L., Wang M., Jiao K. Sertad1 encodes a novel transcriptional co-activator of SMAD1 in mouse embryonic hearts. Biochem Biophys Res Commun. 2013;441:751–756. doi: 10.1016/j.bbrc.2013.10.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shen Z., Yao C., Wang Z., et al. Vastatin, an endogenous antiangiogenesis polypeptide that is lost in hepatocellular carcinoma, effectively inhibits tumor metastasis. Mol Ther. 2016;24:1358–1368. doi: 10.1038/mt.2016.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen Y.S., Lu M.J., Huang H.S., Ma M.C. Mechanosensitive transient receptor potential vanilloid type 1 channels contribute to vascular remodeling of rat fistula veins. J Vasc Surg. 2010;52:1310–1320. doi: 10.1016/j.jvs.2010.05.095. [DOI] [PubMed] [Google Scholar]
- 39.Ji C., McCulloch C.A. TRPV4 integrates matrix mechanosensing with Ca(2+) signaling to regulate extracellular matrix remodeling. FEBS J. 2021;288:5867–5887. doi: 10.1111/febs.15665. [DOI] [PubMed] [Google Scholar]
- 40.Tang V.W. Collagen, stiffness, and adhesion: the evolutionary basis of vertebrate mechanobiology. Mol Biol Cell. 2020;31:1823–1834. doi: 10.1091/mbc.E19-12-0709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miosge N., Simniok T., Sprysch P., Herken R. The collagen type XVIII endostatin domain is co-localized with perlecan in basement membranes in vivo. J Histochem Cytochem. 2003;51:285–296. doi: 10.1177/002215540305100303. [DOI] [PubMed] [Google Scholar]
- 42.Chanut-Delalande H., Fichard A., Bernocco S., Garrone R., Hulmes D.J., Ruggiero F. Control of heterotypic fibril formation by collagen V is determined by chain stoichiometry. J Biol Chem. 2001;276:24352–24359. doi: 10.1074/jbc.m101182200. [DOI] [PubMed] [Google Scholar]
- 43.Pohl U., Herlan K., Huang A., Bassenge E. EDRF-mediated shear-induced dilation opposes myogenic vasoconstriction in small rabbit arteries. Am J Physiol. 1991;261:H2016–H2023. doi: 10.1152/ajpheart.1991.261.6.H2016. [DOI] [PubMed] [Google Scholar]
- 44.Hecker M., Mulsch A., Bassenge E., Busse R. Vasoconstriction and increased flow: two principal mechanisms of shear stress-dependent endothelial autacoid release. Am J Physiol. 1993;265:H828–H833. doi: 10.1152/ajpheart.1993.265.3.H828. [DOI] [PubMed] [Google Scholar]
- 45.Bartosch A.M.W., Mathews R., Mahmoud M.M., Cancel L.M., Haq Z.S., Tarbell J.M. Heparan sulfate proteoglycan glypican-1 and PECAM-1 cooperate in shear-induced endothelial nitric oxide production. Sci Rep. 2021;11 doi: 10.1038/s41598-021-90941-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mochizuki S., Vink H., Hiramatsu O., et al. Role of hyaluronic acid glycosaminoglycans in shear-induced endothelium-derived nitric oxide release. Am J Physiol Heart Circ Physiol. 2003;285:H722–H726. doi: 10.1152/ajpheart.00691.2002. [DOI] [PubMed] [Google Scholar]
- 47.Florian J.A., Kosky J.R., Ainslie K., Pang Z., Dull R.O., Tarbell J.M. Heparan sulfate proteoglycan is a mechanosensor on endothelial cells. Circ Res. 2003;93:e136–e142. doi: 10.1161/01.RES.0000101744.47866.D5. [DOI] [PubMed] [Google Scholar]
- 48.Ruane-O’Hora T., Ahmeda A., Markos F. The vascular glycocalyx is not a mechanosensor in conduit arteries in the anesthetized pig. PeerJ. 2020;8 doi: 10.7717/peerj.8725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gouverneur M., Spaan J.A., Pannekoek H., Fontijn R.D., Vink H. Fluid shear stress stimulates incorporation of hyaluronan into endothelial cell glycocalyx. Am J Physiol Heart Circ Physiol. 2006;290 doi: 10.1152/ajpheart.00592.2005. H458-H452. [DOI] [PubMed] [Google Scholar]
- 50.Zhang W.D., Bai H.Z., Sawa Y., et al. Association of smooth muscle cell phenotypic modulation with extracellular matrix alterations during neointima formation in rabbit vein grafts. J Vasc Surg. 1999;30:169–183. doi: 10.1016/s0741-5214(99)70189-8. [DOI] [PubMed] [Google Scholar]
- 51.Wight T.N., Merrilees M.J. Proteoglycans in atherosclerosis and restenosis: key roles for versican. Circ Res. 2004;94:1158–1167. doi: 10.1161/01.RES.0000126921.29919.51. [DOI] [PubMed] [Google Scholar]
- 52.Kuwahara G., Hashimoto T., Tsuneki M., et al. CD44 promotes inflammation and extracellular matrix production during arteriovenous fistula maturation. Arterioscler Thromb Vasc Biol. 2017;37:1147–1156. doi: 10.1161/ATVBAHA.117.309385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Skrbic B., Engebretsen K.V., Strand M.E., et al. Lack of collagen VIII reduces fibrosis and promotes early mortality and cardiac dilatation in pressure overload in mice. Cardiovasc Res. 2015;106:32–42. doi: 10.1093/cvr/cvv041. [DOI] [PubMed] [Google Scholar]
- 54.Hou G., Mulholland D., Gronska M.A., Bendeck M.P. Type VIII collagen stimulates smooth muscle cell migration and matrix metalloproteinase synthesis after arterial injury. Am J Pathol. 2000;156:467–476. doi: 10.1016/S0002-9440(10)64751-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hopfer U., Fukai N., Hopfer H., et al. Targeted disruption of Col8a1 and Col8a2 genes in mice leads to anterior segment abnormalities in the eye. FASEB J. 2005;19:1232–1244. doi: 10.1096/fj.04-3019com. [DOI] [PubMed] [Google Scholar]
- 56.Duque J.C., Martinez L., Tabbara M., et al. Vascularization of the arteriovenous fistula wall and association with maturation outcomes. J Vasc Access. 2020;21:161–168. doi: 10.1177/1129729819863584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sainz-Jaspeado M., Smith R.O., Plunde O., et al. Palmdelphin regulates nuclear resilience to mechanical stress in the endothelium. Circulation. 2021;144:1629–1645. doi: 10.1161/CIRCULATIONAHA.121.054182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ma Q., Gu W., Li T., et al. SRGN, a new identified shear-stress-responsive gene in endothelial cells. Mol Cell Biochem. 2020;474:15–26. doi: 10.1007/s11010-020-03830-7. [DOI] [PubMed] [Google Scholar]
- 59.Pike D., Shiu Y.T., Cho Y.F., et al. The effect of endothelial nitric oxide synthase on the hemodynamics and wall mechanics in murine arteriovenous fistulas. Sci Rep. 2019;9:4299. doi: 10.1038/s41598-019-40683-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Somarathna M., Hwang P.T., Millican R.C., et al. Nitric oxide releasing nanomatrix gel treatment inhibits venous intimal hyperplasia and improves vascular remodeling in a rodent arteriovenous fistula. Biomaterials. 2022;280 doi: 10.1016/j.biomaterials.2021.121254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Falzon I., Northrup H., Guo L., Totenhagen J., Lee T., Shiu Y.T. The geometry of arteriovenous fistulas using endothelial nitric oxide synthase mouse models. Kidney360. 2020;1:925–935. doi: 10.34067/kid.0001832020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.