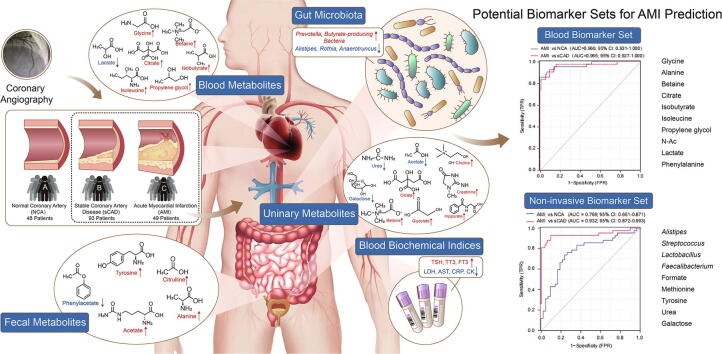
Graphical abstract
Keywords: Acute myocardial infarction (AMI), Stable coronary artery disease (sCAD), Prediction model, Gut microbiota, Metabolite
Highlights
-
•
Multi-omics reveals specific gut microbes and fecal/serum/urinary metabolites are closely related with AMI.
-
•
Combination of gut bacteria and fecal/urinary metabolites provides an effective and non-invasive biomarker set for AMI.
-
•
We provide a potential effective and non-invasive biomarker set for the prediction of AMI, thus contributing to early warning of AMI for patients with sCAD.
Abstract
Introduction
Acute myocardial infarction (AMI) accounts for the majority of deaths caused by coronary artery disease (CAD). Early warning of AMI, especially for patients with stable coronary artery disease (sCAD), is urgently needed. Our previous study showed that alterations in the gut microbiota were correlated with CAD severity.
Objectives
Herein, we tried to discover accurate and convenient biomarkers for AMI by combination of gut microbiota and fecal/blood/urinary metabolomics.
Methods
We recruited 190 volunteers including 93 sCAD patients, 49 AMI patients, and 48 subjects with normal coronary artery (NCA), and measured their blood biochemical parameters, 16S rRNA-based gut microbiota and NMR-based fecal/blood/urinary metabolites. We further selected 20 subjects from each group and analyzed their gut microbiota by whole-metagenome shotgun sequencing.
Results
Multi-omic analyses revealed that AMI patients exhibited specific changes in gut microbiota and serum/urinary/fecal metabolites as compared to subjects with sCAD or NCA. Fourteen bacterial genera and 30 metabolites (11 in feces, 10 in blood, 9 in urine) were closely related to AMI phenotypes and could accurately distinguish AMI patients from sCAD patients. Some species belonging to Alistipes, Streptococcus, Ruminococcus, Lactobacillus and Faecalibacterium were effective to distinguish AMI from sCAD and their predictive ability was confirmed in an independent cohort of CAD patients. We further selected nine indicators including 4 bacterial genera, 3 fecal and 2 urinary metabolites as a noninvasive biomarker set which can distinguish AMI from sCAD with an AUC of 0.932.
Conclusion
Combination of gut microbiota and fecal/urinary metabolites provided a set of potential useful and noninvasive predictive biomarker for AMI from sCAD.
Introduction
Coronary artery disease (CAD) is the leading cause of mortality worldwide [1], accounting for 18.6 million deaths annually in 2019 [2]. Based on the extent of arterial blockage, CAD is classified into two major clinical subtypes, stable coronary artery disease (sCAD) and acute coronary syndromes (ACS) [3]. The sCAD is a clinically stable condition which is mainly caused by reversible, transient episode of blood supply–demand mismatch correlated with myocardial ischemia, whereas ACS manifests as sudden cardiac death (SCD), acute myocardial infarction (AMI), or unstable angina pectoris (UAP) in clinic [4]. Among these, AMI, including ST-segment elevated myocardial infarction (STEMI) and non-ST-segment elevated myocardial infarction (NSTEMI), accounts for the majority of CAD deaths, leading to a considerable health-care cost and societal burden [5]. Although great progress has been achieved, early warning of the risk of AMI remains a big challenge. Therefore, discovery of noninvasive and accurate biomarkers for AMI is urgently needed.
No reliable and effective technique for the early warning of AMI is currently available. Several diagnostic biomarkers for AMI have been identified and used for the clinical diagnosis. The cardiac-specific circulating biomarkers, cardiac troponins I (cTnI) and cardiac troponins T (cTnT), are primary diagnostic markers for AMI [6], but their distinguishing performance for AMI and sCAD is not satisfied because cTnI usually increases in both sCAD and MI patients [7]. Some other diseases are also accompanied by elevation of these two parameters, which may lead to the false prediction of AMI. Recent investigations revealed that exosomal miR-1, miR-1915-3p, miR-4507 and miR-3656, creatinine kinase myocardial band (CK-MB), and creatine kinase (CK) are significantly higher in AMI patients than in sCAD and healthy persons [8], [9], [10], [11], but their prediction accuracy is relatively low. Therefore, more biomarkers with high sensitivity and specificity for AMI are desiderated to nip the occurrence of AMI in the bud.
The gut microbiota and its metabolites are involved in the process of AMI. Animals experiment showed that the families Lachnospiraceae, Syntrophomonadaceae, Eubacteriaceae and Dethiosulfovibrionaceae, and the genera Tissierella and Soehngenia are significantly higher in AMI rats than the SHAM controls [12]. In human beings, AMI patients harbor higher abundance of Megasphaera, Butyricimonas, Acidaminococcus, Desulfovibrio, and decreased Tyzzerella 3, Dialister, [Eubacterium] ventriosum group, Pseudobutyrivibrio and Lachnospiraceae ND3007 than healthy controls [13]. Meanwhile, an elevated plasma level of trimethylamine N-oxide (TMAO), a pro-atherogenic and pro-thrombotic metabolite produced by gut microbiota, is independently associated with risk of major adverse cardiac events in ACS patients [14]. In addition, downstream tryptophan metabolites of the kynurenine pathway are related with an increasing risk of AMI in patients with suspected sCAD [15]. Therefore, the gut microbiota affects the composition and content of blood metabolites in CAD patients [16], thus recognized as potential diagnostic markers of AMI together with serum metabolites.
Our previous study showed that the typical changes of bacterial co-abundance group (CAG) in the different phases of coronary artery disease (CAD) were dominated by Roseburia, Klebsiella, Clostridium IV and Ruminococcaceae [17]. Furthermore, the combination of 24 CAGs and 72 serum metabotypes could effectively distinguish sCAD from ACS with an AUC of 0.897 [17]. However, this prediction model was quite complex and required invasive blood sampling and expensive metabolomic analysis, which heavily limited its broad utilization in clinic settings. Establishment of a simple and, importantly, non-invasive prediction model is in great request.
To discover noninvasive, high-efficiency biomarkers of AMI, we recruited 190 volunteers including 93 sCAD, 49 AMI and 48 NCA subjects. We systemically measured their blood biochemical parameters, 16S rRNA-based gut microbiota and NMR-based fecal/blood/urinary metabolites. We analyzed and compared the profiles of blood biochemical parameters, gut microbial community, and fecal/blood/urinary metabolomics among different groups. Finally, we obtained a noninvasive AMI biomarker set including four gut genera (Alistipes, Streptococcus, Lactobacillus, Faecalibacterium), three fecal metabolites (formate, methionine, tyrosine) and two urinary compounds (urea, galactose), which can accurately distinguish AMI from sCAD with an AUC value of 0.932.
Material and methods
Ethics statement
This study was approved by the Ethics Committee of Fuwai Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College (Beijing, China) in accordance with the ethical standards of the 1964 Declaration of Helsinki and its later amendments (Approval no. 2018–995). Written informed consent was obtained from all participants.
Participants and study design
This is a single-center cross sectional study. We continuously recruited 190 participants under 65 years old with complete information on medical history, clinical and biochemical parameters from Fuwai Hospital between December 2016 and February 2017. The diagnosis was made on the basis of symptoms, laboratory tests, ECG and coronary angiographic results. 190 participants including normal coronary artery (NCA group, n = 48), stable coronary artery disease with the coronary artery stenosis ≥ 70% (sCAD group, n = 93) and acute myocardial infarction (AMI group, n = 49). The criteria for AMI included: 1) symptoms of chest pain at rest (>20 min); 2) ischemic electrocardiographic changes: ST-segment changes and/or T-wave inversions; 3) significant increases in myocardial enzyme levels. For sCAD, the criteria included: 1) chest pain symptoms (<10 min) and electrocardiographic changes only after activity; 2) normal myocardial enzyme level. The coronary angiography was performed on all participants. Plaques or stenosis was not found in age- and sex-matched control subjects. All enrolled participants in the NCA, sCAD and AMI group who were suspected of CAD underwent coronary angiography (CAG)and had no history of unstable angina, myocardial infarction, stroke, cancers, or coronary revascularization. The angiographic data were confirmed independently by two observers in this study. The blood, urine and feces samples were collected the first morning after admission to the hospital. All collected samples were frozen on dry ice within 30 min, and stored in − 80 °C freezers before further analysis.
Nuclear magnetic resonance (NMR) sample collection and preparation
Serum(before the coronary angiography surgery)and urine(early morning urinary)samples were collected and centrifuged at 278 K (5 °C) at 3,000g for 10 min, the supernatants of samples were stored at −80 °C for metabolic profile establishment and statistical analysis. Faeces samples were stored at −80 °C after homogenate with phosphate buffer (0.2 M NaH2PO4/K2HPO4, pH 7.4). Samples were prepared using the previously reported method [18].
NMR spectra acquisition and processing
All NMR spectra were recorded at 298 K (25 °C) using a Bruker Avance 500 MHz spectrometer (1H frequency: 500.13 MHz; Bruker, Germany). The analysis of samples was in conformity with previous study (Supplementary Methods).
NMR multivariate data analysis
Output data were processed with the SIMCA-P + 14.0 software (Umetrics, Sweden) to elucidate patterns in metabolite concentration shifts. Statistical analysis was also conducted with SPSS19.0 (IBM; USA) using the two-tailed Student’s t-test. P-value of<0.05 was considered to be statistically significant between two groups.
Human faecal sample collection and DNA extraction
Fresh feces samples were collected from 190 subjects, and then delivered from Fuwai Hospital to the laboratory in an ice bag using insulating polystyrene foam containers. DNA was extracted using an EZNA™ stool DNA isolation kit (Omega Bio-Tek, VWR, Herlev, Denmark). The DNA was then eluted in 50 µL of elution buffer and stored at −80 °C for further 16S rRNA-based metagenomic analysis Additionally, 60 fecal DNA samples were selected randomly (20 samples from each group) for whole-metagenome shotgun sequencing.
DNA library construction and sequencing
DNA library was constructed using the TruSeq Nano DNA LT Library Preparation Kit (FC-121–4001, Illumina, San Diego, CA, USA). The resulting libraries were sequenced on an Illumina HiSeq 4000 sequencer (Illumina, San Diego, CA, USA). The running mode of metagenomics was paired-end of 150 bp and the running mode of 16S rRNA sequencing was paired-end of 300 bp.
Sequencing data analysis
The sequencing data were analyzed using QIIME 2, and the quality control was conducted using FastQC and MetaPhlAn v2.662 as previous described (Supplementary Methods).
Statistical analysis
The wilcox test were used to analyze the differential clinical indexes for continuous and categorical variables, respectively. Spearman correlations between microbiota, metabolites and clinical indexes were calculated using GraphPad Prism 8.0.1. The visual presentation of multiple omics correlations was performed using the pheatmap package in R.
Results
General characteristics of the study cohort
A total of 190 individuals were recruited in this study which include 93 sCAD and 49 AMI patients, and 48 NCA subjects. Their general demographics are shown in Supplementary Table S1. To discover unique biomarkers for AMI, we systematically analyzed the blood biochemical indices, fecal microbiota, and fecal/blood/urinary metabolomics of each participant. (Supplementary Table S2).
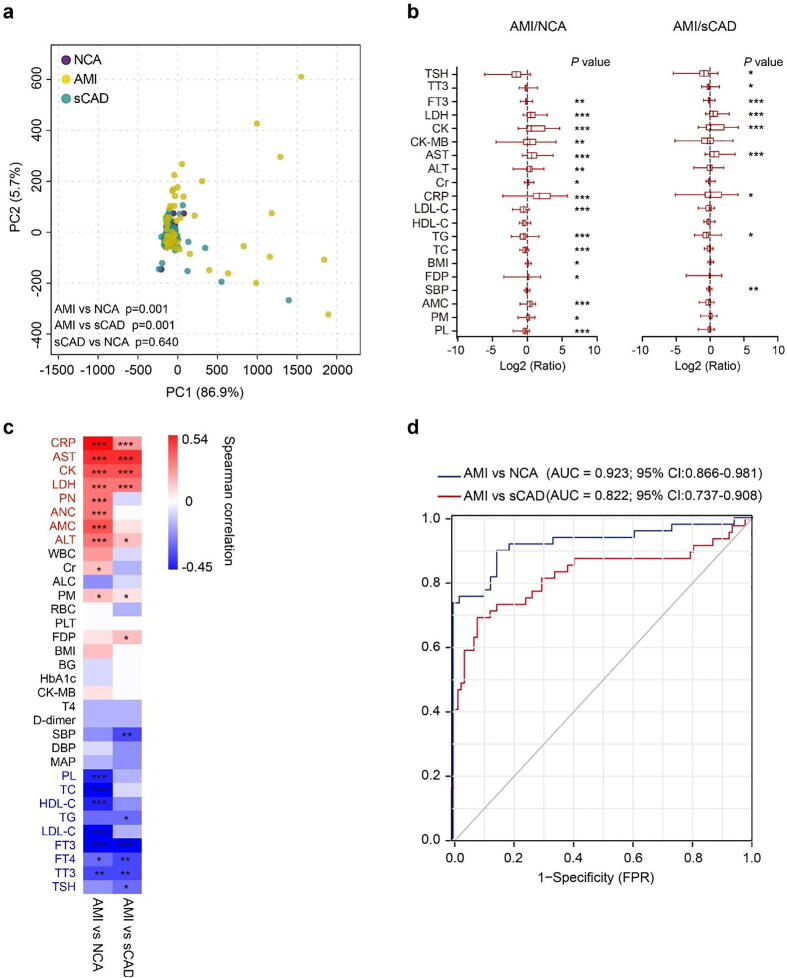
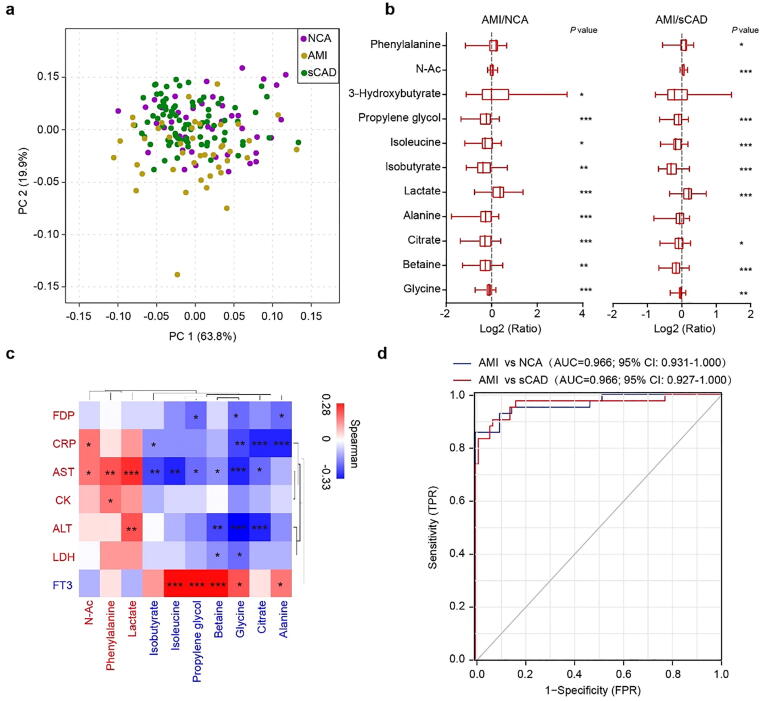
We then performed principal component analysis (PCA), inter-group difference analysis, and correlation analysis with AMI (1 as true and 0 as false) on 33 general blood biochemical indices to find key biomarkers for AMI. PCA diagram displayed significant departure between AMI group and NCA/sCAD groups (Fig. 1a). The serum levels of CRP, AST, CK and LDH in AMI group were significantly higher while FT3 was significantly lower than the other two groups (Fig. 1b, Supplementary Table S2). Among the 33 indices, CRP, CK, AST, ALT and LDH were positive while FT3, TC, LDL-c, HDL-c were negative to AMI occurrence (Fig. 1c). These results suggest that LDH, AST, CRP, CK, ALT, FDP and FT3 may serve as biomarkers to distinguish AMI from NCA/sCAD. Receiver operating characteristic (ROC) curve displayed that combination of these 7 blood indices could distinguish AMI from NCA with an AUC value of 0.923, but its distinguishing ability for AMI and sCAD was relatively weak (AUC = 0.822) (Fig. 1d).
Fig. 1.
The difference of general blood biochemical indices among subjects with NCA, sCAD and AMI. (a) Principal component analysis (PCA) of general blood biochemical indices among three groups. (b) Boxplot of log fold change (AMI/NCA or AMI/sCAD) of 20 blood biochemical indices. (c) Heat map showing the correlation intensity between blood biochemical indices and AMI occurrence. (d) Seven important features (CRP, ALT, AST, LDH, CK, FDP, FT3) to build the prediction model yielded an area under the curve (AUC) based on ROC (receiver operating characteristic) analysis. FPR, false-positive rate; TPR, true positive rate. *p < 0.05, **p < 0.01, ***p < 0.001.
Gut microbiota helps to distinguish AMI patients from sCAD patients
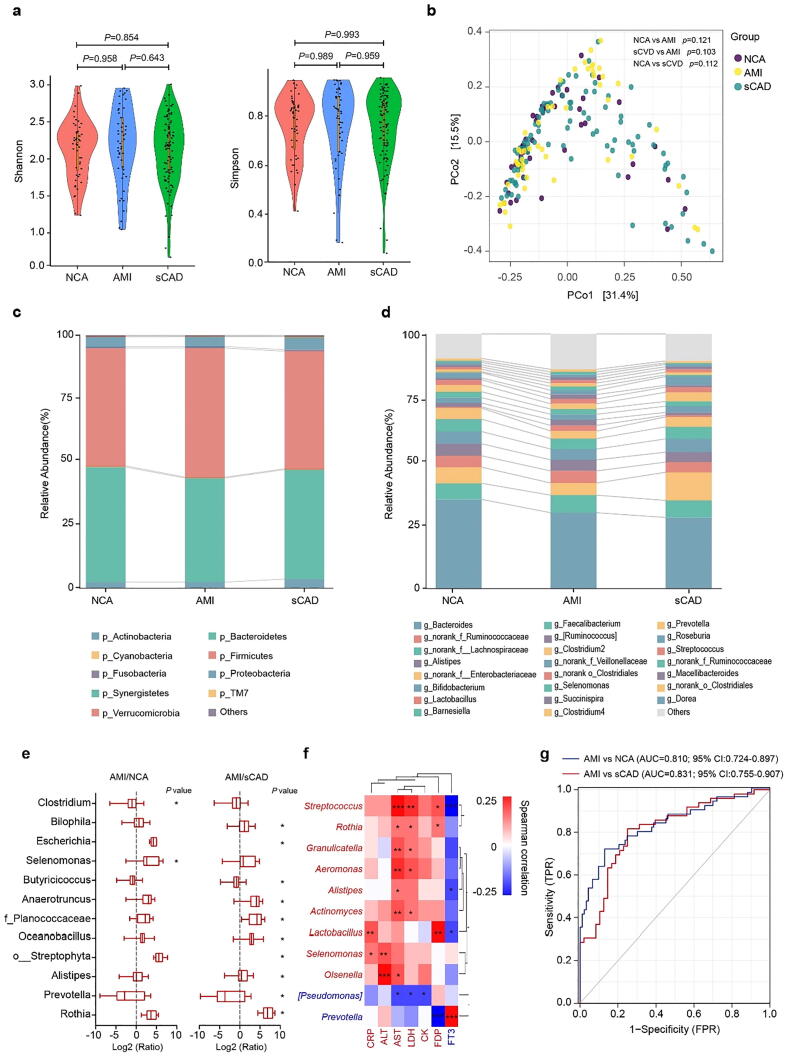
The gut microbiota is closely related to the etiology of heart disease [19], and our previous study showed that the gut microbial composition differed from healthy controls to diverse coronary artery disease subtypes [17]. To find efficient biomarkers that can distinguish AMI from sCAD, we performed 16S rRNA-based metagenomic analysis on fecal samples. Both alpha diversity (Shannon and Simpson indices) (Fig. 2a) and beta diversity (principal coordinate analysis (PCoA)) (Fig. 2b) showed no significant difference among AMI, sCAD and NCA groups, suggesting that the overall structure of the gut microbiota was not obviously changed with the heart disease status. Detailed taxonomic analysis at the phylum and genus levels displayed that the relative abundance of specific taxa was markedly altered among groups (Fig. 2c and d). Nine genera, i.e. Alistipes, Rothia, Oceanobacillus, Anaerotruncus, Selenomonas, Escherichia, Bilophila, unclassified o_Streptophyta and unclassified f_Planococcaceae, were significantly enriched in AMI patients, while Butyricicoccus and Prevotella were more abundant in NCA and sCAD groups (Fig. 2e).
Fig. 2.
Gut microbiota helps to distinguish AMI patients from sCAD patients. (a) Alpha-diversity was assessed by Shannon index and Simpson index. (b) Beta-diversity was assessed by principal coordinate analysis (PCoA) based on bray-curtis distance. (c)Phyla and (d) genera profile of the gut microbiota among three groups. (e) Boxplot of log fold change (AMI/NCA or AMI/sCAD) of 11 genera. (f) Heat map showing the correlation intensity between 11 genera and 7 blood biochemical indices. (g) Fourteen specific genera to build the prediction model yielded an AUC based on ROC analysis. *p < 0.05.
We further correlated the relative abundance of individual genus with the seven key blood biochemical indices to find key bacteria that are closely associated with AMI occurrence. Streptococcus, Alistipes, Rothia, Granulicatella, Lactobacillus, Actinomyces, Anaerotruncus, Selenomonas and Olsenella were positive to AMI risk indicators (CRP, AST, ALT, CK, LDH FDP), while Prevotella and [Pseudomonas] were negative to these indicators (Fig. 2f). In addition, Prevotella was also positive to FT3, an indicator that was negatively associated with AMI occurrence (Fig. 2f). We finally selected 14 genera including Streptococcus, Alistipes, Lactobacillus, Clostridium, Rothia, Oceanobacillus, Butyricicoccus, Selenomonas, Bilophila, [Pseudomonas], Anaerotruncus, Granulicatella, Prevotella and Actinomyces as a gut bacterial biomarker set for AMI. ROC analysis revealed that this gut bacterial set could distinguish AMI from sCAD and NCA with AUC values of 0.831 and 0.810, respectively (Fig. 2g). Although its predictive ability for AMI and NCA was lower than traditional blood biochemical parameters, this gut bacterial set exhibited a slightly higher predicting potential to distinguish AMI from sCAD patients (AUC 0.831 vs 0.822).
Specific gut bacterial species effectively distinguish AMI from sCAD
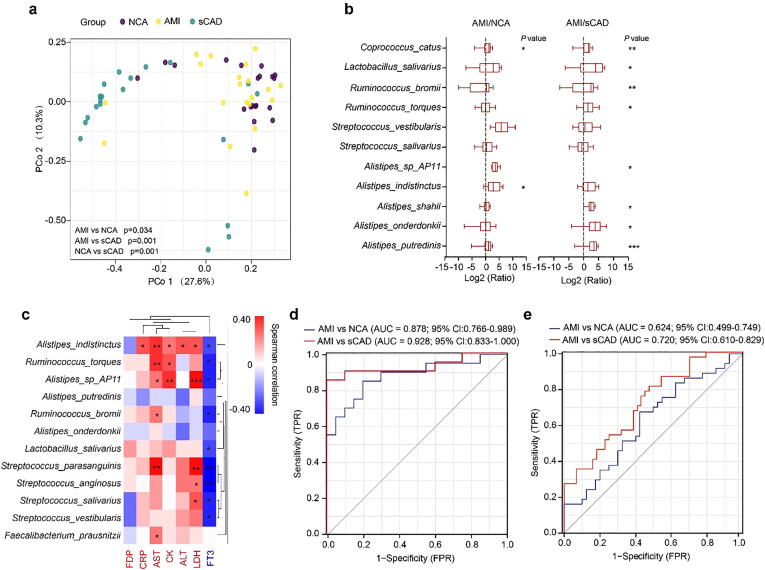
To further identify specific gut bacteria that can accurately predict AMI, we randomly selected 60 individuals from this cohort (20 participants from each group) and analyzed their gut microbiota by whole-metagenome shotgun sequencing. PCoA analysis revealed that AMI patients were significant separated from sCAD and NCA participants, although there were overlaps between AMI and NCA groups (Fig. 3a). The species belonging to Alistipes (A. putredinis, A. onderdonkii, A. shahii, A. indistinctus and Alistipes sp_AP11), Streptococcus (S. salivarius and S. vestibularis), Ruminococcus (R. torques and R. bromii) and Lactobacillus (L. salivarius) were remarkably enriched in AMI patients over sCAD and NCA subjects (Fig. 3b). Correlation analysis between individual species and blood biochemical biomarkers uncovered that A. indistinctus, R. torques, S. parasanguinis, Faecalibacterium prausnitzii, R. bromii, S. salivarius, L. salivarius, S. vestibularis Alistipes sp_AP11 and S. anginosus were positively associated with at least one key AMI risk indicators (Fig. 3c). Collectively, we selected 9 bacterial species including A. shahii, A. indistinctus, Alistipes sp_AP11, S. salivarius, S. vestibularis, R. torques, R. bromii, L. salivarius and F. prausnitzii as an AMI characteristic gut species set. ROC curve showed this gut species set could distinguish AMI patients from sCAD/NCA subjects with AUC values of 0.928 and 0.878, respectively (Fig. 3d).
Fig. 3.
Specific species could effectively distinguish AMI and sCAD patients. (a) PCoA analysis of gut microbiota among three groups. (b) Boxplot of log fold change (AMI/NCA or AMI/sCAD) of 11 species. (c) Heat map showing the correlation intensity between 12 species and 7 blood biochemical indices. (d) Nine specific species to build the prediction model yielded an AUC based on ROC analysis. (e) Six specific species to build the prediction model yielded an AUC based on ROC analysis. *p < 0.05, **p < 0.01, ***p < 0.001.
To validate the predictive ability of this gut species set for AMI, we distinguished AMI from sCAD/NCA subjects using this species set for an independent cohort which was enrolled in a different medical center and included 44 sCAD, 37 MI and 44 NCA participants [17]. In the gut microbiota data of that study, 6 out of the 9 characteristic gut species (A. shahii, A. indistinctus, S. vestibularis, R. bromii, L. salivarius and F. prausnitzii) were detected. We took these six species as a predict set to distinguish AMI from sCAD/NCA which showed an AUC value of 0.720 and 0.624, respectively (Fig. 3e). Although it showed some potential to distinguish AMI from sCAD, the accuracy of this gut species set was largely decreased in this independent cohort, which might be partially due to the undetected key species (Alistipes sp_AP11, S. salivarius and R. torques) and indicated that more types of classifiers are needed to improve the prediction accuracy.
Combination of gut microbiota and fecal metabolites provides a better separation of AMI from sCAD
The low prediction ability of gut microbiota for AMI may be due to the weak linkage between bacterial taxonomy and their actual functions. Several studies have reported that the metabolites of the gut microbiota are closely related to the occurrence of CAD [17]. Therefore, we analyzed the fecal metabolomics to find more effective predictors that could distinguish AMI from sCAD (Supplementary Fig. S1).
As gut bacteria were intrinsically linked with fecal metabolites, we hypothesized that combination of gut microbiota and fecal metabolites might enhance the prediction performance. Correlation between individual gut genus and fecal butyrate content revealed that the most fecal butyrate-related genera were shown in Supplementary table S3.
Blood metabolomics analysis of AMI and sCAD patients
Our previous research revealed that the blood metabolite modules detected by HPLC-MS exhibited potential diagnostic value for differentiating patients with different CAD subtypes [17]. To get more convenient biomarkers for AMI, we analyzed the blood metabolomics of each participant by NMR methods. A total of 33 serum metabolites were detected. Although no significant difference presented in the overall blood metabolites among three groups (Fig. 4a), the AMI patients showed significantly decreased level of glycine, betaine, citrate, alanine, isobutyrate, isoleucine and propylene glycol, whereas the lactate and 3-hydroxybutyrate were significantly raised (Fig. 4b). Correlation analysis of serum metabolites with key blood biochemical biomarkers revealed that N-Ac, lactate and phenylalanine were positive to AMI risk indicators (CRP, AST, ALT, CK, LDH, FDP) and negative to FT3, while glycine, betaine, citrate, alanine, isobutyrate, isoleucine and propylene glycol were negative to AMI risk indicators (Fig. 4c). We selected 10 blood metabolites (glycine, alanine, betaine, citrate, isobutyrate, isoleucine, propylene glycol, N-Ac, lactate and phenylalanine) as blood AMI biomarker metabolites, which could distinguish AMI from sCAD or NCA with an AUC value of 0.966 for both (Fig. 4d).
Fig. 4.
Blood metabolomics analysis of AMI and sCAD patients. (a) PCA analysis of blood metabolites among three groups. (b) Boxplot of log fold change (AMI/NCA or AMI/sCAD) of 8 blood metabolites. (c) Heat map showing the correlation intensity between 10 blood metabolites and 7 blood biochemical indices. (d) Ten specific blood metabolites to build the prediction model yielded an AUC based on ROC analysis. *p < 0.05, **p < 0.01, ***p < 0.001.
Urine metabolomics analysis of AMI and sCAD patients
We also analyzed the NMR-based urine metabolomics for each participant (Supplementary Fig. S2).
Combination of gut bacterial and fecal/urinary metabolites provides an effective and non-invasive biomarker set for AMI
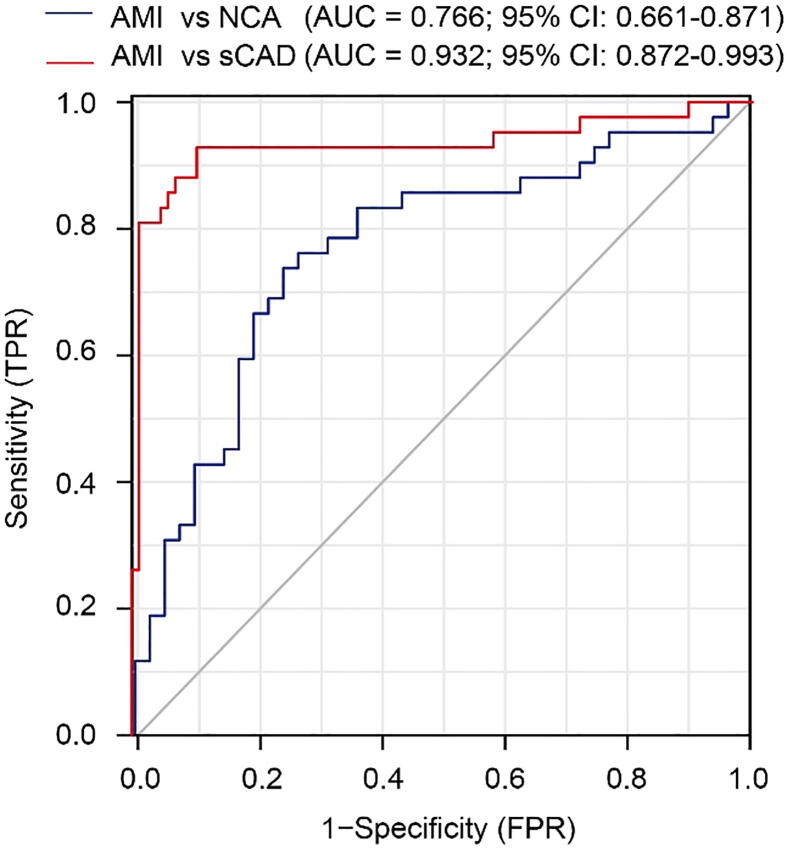
Although the present and previous studies demonstrated that the blood metabolites were effective to distinguish AMI from other CAD subtypes [17], [20], the intrinsic invasiveness largely restricted its clinical application. Based on our systemic measurements of gut microbiota and blood/fecal/urinary metabolomics, we set up a set of non-invasive biomarkers that may effectively and conveniently distinguish AMI patients from sCAD patients. This non-invasive biomarker set contained four gut bacterial genera (Alistipes, Streptococcus, Lactobacillus and Faecalibacterium), three fecal metabolites (formate, methionine and tyrosine) and two urine metabolites (urea and galactose). The ROC curve showed this non-invasive biomarker set distinguished AMI from NCA with an AUC of 0.766 and distinguished AMI from sCAD with an AUC as high as 0.932 (Fig. 5), revealing a promising utility for prediction of AMI from sCAD patients.
Fig. 5.
Combination of gut bacterial and fecal/urinary metabolites provides an effective and non-invasive biomarker set for AMI. A non-invasive biomarker set consisted of four gut bacterial genera (Alistipes, Streptococcus, Lactobacillus and Faecalibacterium), three fecal metabolites (formate, methionine and tyrosine) and two urine metabolites (urea and galactose) was established.
Relationship between the gut microbiota and serum/fecal metabolites in AMI
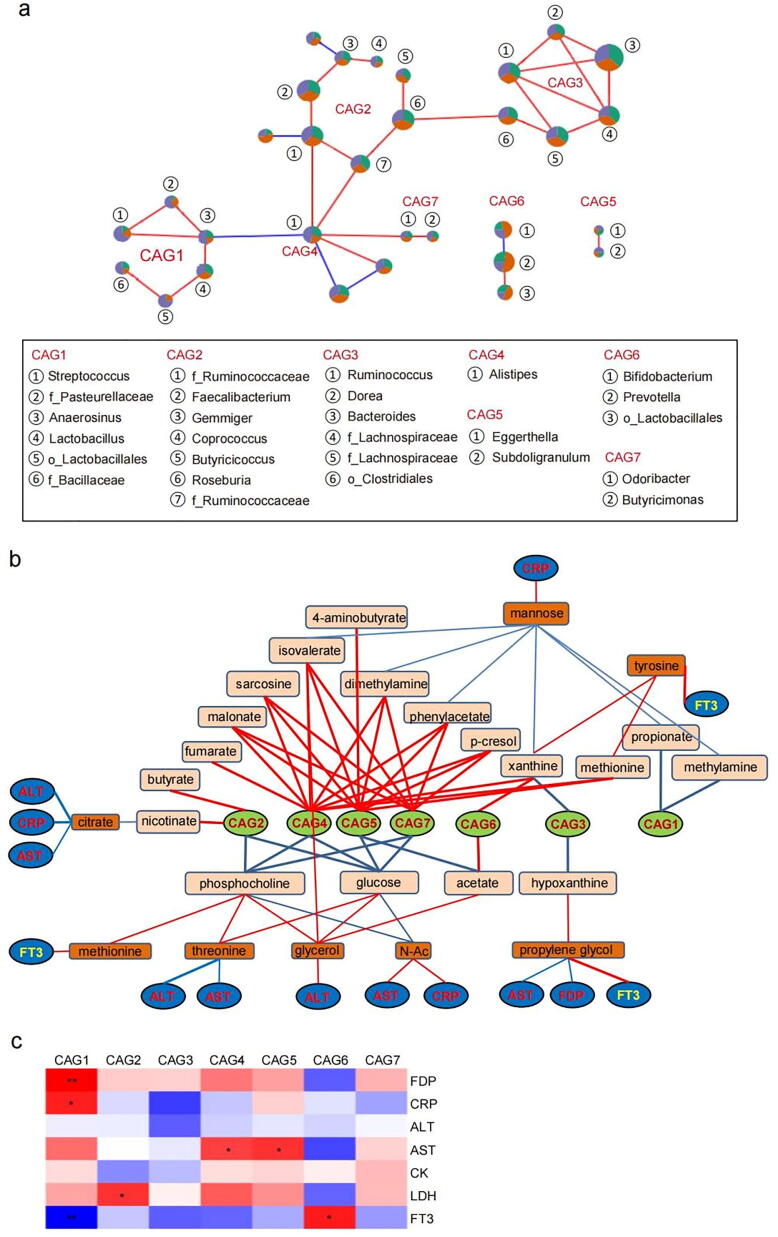
We subsequently correlated the gut microbiota with fecal/blood metabolites to further explore the characteristics of microbiota in patients with AMI. As bacteria act as functional groups (guilds) in the gut ecosystem [21], we constructed a co-abundance network based on SparCC correlation coefficients and clustered the gut genera into 7 closely related CAGs (r > 0.3, p < 0.01) (Fig. 6a). The CAG2 was mainly composed of potential butyrate-producers such as Roseburia, Butyricicoccus and Faecalibacterium, while CAG4 was solely consisted of Alistipes. Of these, CAG1, CAG4, CAG5 and CAG7 was increased in AMI patients compared with the sCAD and NCA subjects, while CAG6 was significantly decreased in AMI patients compared with sCAD patients.
Fig. 6.
Relationship between the gut microbiota and serum/fecal metabolites in AMI. (a) The co-abundance network based on SparCC correlation coefficients. (b) The correlation between CAGs and serum/fecal metabolites. (c) Correlation between CAGs and key blood AMI indices.
Given an FDR of 5%, the 7 CAGs were significantly correlated with 18 fecal metabolites, as analyzed by Spearman correlation coefficients (Fig. 6b). CAG2, mainly consisted of butyrate-producers, was positive to fecal butyrate and nicotinate, partially validating this correlation analysis. CAG4, CAG5 and CAG7, enriched in AMI group, were positively correlated with multiple fecal metabolites, such as malonate, sarcosine, isovalerate, dimethylamine, phenylacetate, methionine and p-cresol, but negatively correlated with glucose and o-phosphocholine. CAG1 was negative to methylamine and propionate, CAG3 was negative to hypoxanthine and xanthine, while CAG6 was positive to xanthine and acetate (Fig. 6b).
The fecal metabolites were further correlated with the blood metabolites. Eight blood metabolites, i.e. mannose, tyrosine, citrate, methionine, threonine, glycerol, N-Ac, propylene glycol, were closely related with above fecal metabolites (p < 0.05 but FDR > 0.05). Among these, blood mannose was negatively related with fecal contents of isovalerate, dimethylamine, phenylacetate, xanthine, propionate and methylamine, and it was positive to AMI risk factor CRP. Blood citrate was negative to fecal nicotinate and it was also negative to AMI risk factors CRP, ALT and AST. Blood threonine was positive to fecal phosphocholine and glucose and it was negative to ALT and AST. In contrast, blood N-Ac was negative to fecal phosphocholine and glucose and positive to ALT and AST. Blood propylene glycol was positive to fecal hypoxanthine and it was negative to AMI risk factors AST and FDP but positive to FT3 (Fig. 6b).
Correlation analysis between CAGs and various blood AMI risky factors revealed that CAG1 was positive to FDP and CRP and negative to FT3, CAG5 were positive AST, while CAG7 was positive to FT3 (Fig. 6c). Systemic correlation among CAGs, fecal and blood metabolites showed that the positive relationship between CAG1 and CRP might be mediated by fecal propionate, methylamine and blood mannose. The positive correlation between CAG4/5 with AST might be mediated by fecal phosphocholine, glucose and blood threonine, N-Ac. CAG6 (Bifidobacterium, Prevotella and unclassified_o_Lactobacillales) was negative to AMI risky factors (FDP, CRP, ALT, AST, LDH) and positive to FT3, eventually negative to AMI occurrence. However, its correlation with AMI-related factors cannot be linked by fecal and blood metabolites (Fig. 6b and c).
Discussion
The pathophysiology of CAD is atherosclerotic plagues in coronary arteries coupled with or driven by inflammatory reactions [22], and the lesions of vulnerable atherosclerotic plagues are responsible for the clinical conversion from sCAD to deadly AMI [23]. Current clinical examinations are detecting cardiac ischemia or coronary artery stenosis, and do not directly optimize preventative strategies to the CAD patients who would suffer an AMI. Furthermore, the “gold standard” for the diagnosis of CAD is still rested with coronary angiography, which is invasive and along with lethal side effects [24]. Therefore, it still remains a challenge for the accurate prediction and timely intervention of coronary atherosclerosis patients to prevent the occurrence of AMI. This comprehensive analysis based on blood biochemical indicators, gut microbiota, and blood, urinary and fecal metabolomics data uncovered that the gut microbiota is closely related to the occurrence of AMI, which will facilitate clinical diagnosis of AMI. At the same time, we identified a simple, non-invasive prediction model that distinguished AMI from sCAD accurately, thus greatly facilitating patients with AMI and doctors to monitor the risk of MI recurrence at any time. Although the clinical predictive efficacy of this model still needs to be verified among a larger population, it still has great clinical value.
Regular testing of pathological indicators in sCAD patients is clinically needed to assess the progression and risk of AMI. The pathological indicators current used are as follows. (1) Myocardial injury factors, including creatine kinase (CK), lactate dehydrogenase (LDH), cardiac troponin I (cTnI) and myoglobin, are significantly higher in AMI patients than in normal individuals [25]. Among above factors, LDH and CK levels are closely related with AMI occurrence with the AUC value more than 0.9 [26]. (2) The coagulation-related parameters such as FDP and fibrin D-dimer, which are correlated with a greater risk of MI/CHD (coronary heart disease) death [27]. (3) Thyroid hormones, T3 and FT3, are important to evaluate cardiac function during AMI. The TSH was also positively related with cardiac ejection fraction [28]. (4) The other indicators AST and AST/ALT ratio are considerably associated with the mortality of AMI, among which AST/ALT ratio is a strong and independent predictor for long-term mortality after AMI [29]. Additionally, a diagnostic model including AST and CK is established to differentiate AMI patients from AP patients with an AUC value of 0.975 [26]. The inflammatory biomarkers CRP is also powerful to predict MI death [27]. The complex of MI occurrence and less specificity of above various factors urged us to find more markers to improve the accuracy of prediction. In present study, seven factors including CRP, AST, ALT, CK, LDH, FDP and FT3 were proved to be closely related to the occurrence of AMI. Combination of them distinguished NCA from AMI with the AUC value of 0.923, but the effectiveness of distinguishing sCAD from AMI is relatively weak with AUC value of 0.822. These may be due to the similar pathological basis of both diseases, causing the reduced specificity of blood biochemical factors. Therefore, new factors or models are urgently needed to improve the accuracy of the prediction for AMI.
The serum metabolites can comprehensively reflect the state of MI, and therefore possess a better ability to predict AMI. Study revealed that high level of serum dimethylglycine was related with the mortality in CHD patients, thus improving risk prediction of CHD [30]. Additionally, serum level of choline was also significantly lower in patients with a history of AMI than those without AMI history [31]. Other studies revealed that lactate level is related to the development and prognosis of AMI [32]. Although the serum metabolites can well predict the AMI occurrence or recurrence, it needs to perform specialized serum metabolomics analysis, which is expensive and needs long time, as well as difficult to be widely used in clinical detection. Our study simplified serum metabolites related to AMI occurrence and found 10 serum metabolites including glycine, betaine, citrate, alanine, isobutyrate, isoleucine, propylene glycol, N-Ac, lactate and phenylalanine accurately distinguished AMI from NCD and sCAD with AUC value of both 0.966.
The gut microbiota is closely related to the occurrence and progression of MI. Our previous study showed that the composition of gut microbiota varies greatly in patients with different stages of CAD [17]. The changes of relative abundance of genera Treponema 2, Rikenellaceae RC9 group, Prevotellaceae UCG-003, and Bacteroides may contribute to the pathogenesis of AMI [33]. In this study, Streptococcus and Alistipes were significantly increased in patients with AMI. Existing study showed that Streptococcus oralis caused infective endocarditis and aortic valves, then developed into AMI manifested by chest pain and dyspnoea with cardiovascular collapse [34]. Meanwhile, the genus of Alistipes can produce trimethylamine-N-oxide (TMAO), thus promote the development of MI [35]. Additionally, bacteria such as Butyricicoccus [36], Prevotella [37] were also involved in the development of MI. In our study, combination of 14 genera including Streptococcus, Alistipes, Butyricicoccus, Prevotella et al. can distinguish AMI from sCAD with an AUC of 0.831. More interestingly, specific species of Streptococcus and Alistipes possessed the ability to distinguish AMI from sCAD with the AUC value up to 0.963. Further verification for this prediction model in another publicly available population data showed that it distinguished AMI from sCAD with AUC reached to 0.738. Although this value is not as high as 0.963 in present study, it is higher than their original AUC value of 0.695 calculated by 72 blood metabolites and 24 CAGs. Therefore, our specific species still possess a strong ability to distinguish AMI from sCAD in different populations, and have high predictive values for AMI recurrence.
Since the bacterial metabolites are more closely related to their function. We therefore further examined the relationship between fecal metabolome and AMI occurrence, and then correlated fecal metabolites with gut microbiota. We found methionine significantly increased in AMI patients, which was in accordance with existing study shown that the methionine metabolite homocysteine (Hcy) linked to atherothrombosis [38] and may cause the development of MI. Additionally, phenylalanine and tyrosine levels were also significantly correlated with increased risk of CAD [39]. In present study, 11 fecal metabolites set better distinguished AMI from sCAD with AUC value up to 0.831. Combination fecal metabolites and gut microbiota enhanced the predictive ability with the AUC value reached to 0.961. Thus, gut microbiota and fecal metabolites are closely related to the occurrence of AMI and can be used as predictive indicators.
Another important finding of our study is that the urine metabolome is extremely associated with AMI occurrence. Nine urine metabolites accurately distinguished AMI from sCAD with AUC value up to 0.935. Even just urea alone can accurately distinguish AMI from sCAD. Previous studies showed that the prediction model consisting of age, CRP, eGFR, myoglobin, and urea can predict cardiac death with an AUC of 77.5%[29]. Moreover, urea can be quantified accurately and quickly using a general urea kit in routine tests, so it is of great clinical value. Of course, this finding still needs to be supported by more and larger clinical research data.
AMI usually requires lifelong medication and lifelong monitoring. Current clinical tests need patients to provide their blood samples, which are not suitable for frequent and long-term monitoring. To address this problem, the non-invasive, convenient and accurate monitoring indicators are clinically needed. In this study, four bacterial genera, two urine metabolites, and three fecal metabolites were used to establish model to predict AMI recurrence. The predictive model confers each parameter a weight coefficient based on training data and gives a prediction value for each person. A cut-off value (i.e. 0.5) can be set to determine whether a subject is in dangerous AMI prophase. Although it does not require all parameters to be detected in a special person, we recommend, based on our experience, to detect least 6 of them to guarantee the accuracy of the prediction. For more convenience, feces are also enough to predict AMI based on gut microbiota and/or fecal metabolites. This model is non-invasive, easy to collect and test, and suitable to design as kits to monitor the recurrence of AMI in a convenient manner. It is quite suitable for regular monitor of AMI, especially for patients having difficulties to visit hospital/doctor frequently. Although its efficacy needs to be confirmed by more clinical trials, a new idea is proposed for predicting the recurrence of AMI through non-invasive testing methods such as gut microbiota, urine and fecal metabolites to facilitate the daily monitoring of AMI.
Conclusion
In conclusion, our study and further analysis showed that blood metabolites work better for the early warning of AMI, which consistent with the results of our previous research. However, detection for blood metabolites is an invasive manner. To address this problem and gain high accurate of prediction for AMI, we found combinations of bacterial genera, fecal metabolites and urine metabolites are not only a good prediction model for early warning of AMI recurrence, but also a method with convenient detection merit. Furthermore, we found the specific combinations of species of gut microbiota that are equally good for early warning of AMI.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
We thank all study participants, research staffs, and students who participated in this work.
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in the repository of the Genome Sequence Archive Sequence Database of National Genomics Data Center (https://bigd.big.ac.cn/) under the accession number CRA002142.
Funding sources
This work was supported by National Natural Science Foundation of China (82073837, 81973217), Disciplines Construction Project [201920200807], Chinese Academy of Medical Sciences Innovation Fund for Medical Sciences [CIFMS 2021-I2M-1-028], Chinese Academy of Medical Sciences Innovation Fund for Medical Sciences [CIFMS 2016-I2M-1–009], Fundamental Research Funds for the Central Universities (3332021103), National Natural Science Foundation of China(82030124, 81873041).
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2022.06.008.
Contributor Information
Chaoran Dong, Email: dongcr@imm.ac.cn.
Yinghong Wang, Email: wyh@imm.ac.cn.
Feng Gao, Email: gaofengardota@sina.com.
Qifeng Liu, Email: lqf@imm.ac.cn.
Jianxun Liu, Email: liujx0324@sina.com.cn.
Yida Tang, Email: tang_yida@163.com.
Shuyang Zhang, Email: shuyangzhang103@nrdrs.org.
Chongming Wu, Email: chomingwu@163.com.
Haibo Zhu, Email: zhuhaibo@imm.ac.cn.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Libby P., Theroux P. Pathophysiology of coronary artery disease. Circulation. 2005;111(25):3481–3488. doi: 10.1161/CIRCULATIONAHA.105.537878. [DOI] [PubMed] [Google Scholar]
- 2.McCaffrey T.A., Toma I., Yang Z., Katz R., Reiner J., Mazhari R., et al. Rna sequencing of blood in coronary artery disease: Involvement of regulatory t cell imbalance. BMC Med Genomics. 2021;14(1) doi: 10.1186/s12920-021-01062-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Piepoli M.F., Hoes A.W., Agewall S., Albus C., Brotons C., Catapano A.L., et al. 2016 European guidelines on cardiovascular disease prevention in clinical practice. Kardiol Pol. 2016;74(9):821–936. doi: 10.5603/KP.2016.0120. [DOI] [PubMed] [Google Scholar]
- 4.Ahmadi A., Argulian E., Leipsic J., Newby D.E., Narula J. From subclinical atherosclerosis to plaque progression and acute coronary events: Jacc state-of-the-art review. J Am Coll Cardiol. 2019;74(12):1608–1617. doi: 10.1016/j.jacc.2019.08.012. [DOI] [PubMed] [Google Scholar]
- 5.Virani S.S., Alonso A., Benjamin E.J., Bittencourt M.S., Callaway C.W., Carson A.P., et al. Heart disease and stroke statistics-2020 update: a report from the american heart association. Circulation. 2020;141(9) doi: 10.1161/CIR.0000000000000757. [DOI] [PubMed] [Google Scholar]
- 6.Farmakis D., Mueller C., Apple F.S. High-sensitivity cardiac troponin assays for cardiovascular risk stratification in the general population. Eur Heart J. 2020;41:4050–4056. doi: 10.1093/eurheartj/ehaa083. [DOI] [PubMed] [Google Scholar]
- 7.Jeremias A., Kleiman N.S., Nassif D., Hsieh W.-H., Pencina M., Maresh K., et al. Prevalence and prognostic significance of preprocedural cardiac troponin elevation among patients with stable coronary artery disease undergoing percutaneous coronary intervention: results from the evaluation of drug eluting stents and ischemic events registry. Circulation. 2008;118(6):632–638. doi: 10.1161/CIRCULATIONAHA.107.752428. [DOI] [PubMed] [Google Scholar]
- 8.Chaitman B.R., Alexander K.P., Cyr D.D., Berger J.S., Reynolds H.R., Bangalore S., et al. Myocardial infarction in the ischemia trial: Impact of different definitions on incidence, prognosis, and treatment comparisons. Circulation. 2021;143(8):790–804. doi: 10.1161/CIRCULATIONAHA.120.047987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O′Sullivan J.F., Neylon A., McGorrian C., Blake G.J. Mirna-93-5p and other mirnas as predictors of coronary artery disease and stemi. Int J Cardiol. 2016;224:310–316. doi: 10.1016/j.ijcard.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 10.Su J., Li J., Yu Q., Wang J., Li X., Yang J., et al. Exosomal mirnas as potential biomarkers for acute myocardial infarction. IUBMB Life. 2020;72(3):384–400. doi: 10.1002/iub.2189. [DOI] [PubMed] [Google Scholar]
- 11.Ren X., Ellis B.W., Ronan G., Blood S.R., DeShetler C., Senapati S., et al. A multiplexed ion-exchange membrane-based mirna (mix center dot mir) detection platform for rapid diagnosis of myocardial infarction. Lab Chip. 2021;21 doi: 10.1039/d1lc00685a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu Z.-X., Li S.-F., Chen H., Song J.-X., Gao Y.-F., Zhang F., et al. The changes of gut microbiota after acute myocardial infarction in rats. PLoS ONE. 2017;12(7):e0180717. doi: 10.1371/journal.pone.0180717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han Y., Gong Z., Sun G., Xu J., Qi C., Sun W., et al. Dysbiosis of gut microbiota in patients with acute myocardial infarction. Front Microbiol. 2021;12 doi: 10.3389/fmicb.2021.680101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li X.S., Obeid S., Klingenberg R., Gencer B., Mach F., Raber L., et al. Gut microbiota-dependent trimethylamine n-oxide in acute coronary syndromes: a prognostic marker for incident cardiovascular events beyond traditional risk factors. Eur Heart J. 2017;38:814–824. doi: 10.1093/eurheartj/ehw582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pedersen E.R., Tuseth N., Eussen S.J.P.M., Ueland P.M., Strand E., Svingen G.F.T., et al. Associations of plasma kynurenines with risk of acute myocardial infarction in patients with stable angina pectoris. Arterioscler Thromb Vasc Biol. 2015;35(2):455–462. doi: 10.1161/ATVBAHA.114.304674. [DOI] [PubMed] [Google Scholar]
- 16.Zhang L., Wei T.-T., Li Y., Li J., Fan Y., Huang F.-Q., et al. Functional metabolomics characterizes a key role for -acetylneuraminic acid in coronary artery diseases. Circulation. 2018;137:1374–1390. doi: 10.1161/CIRCULATIONAHA.117.031139. [DOI] [PubMed] [Google Scholar]
- 17.Liu H., Chen X.i., Hu X., Niu H., Tian R., Wang H., et al. Alterations in the gut microbiome and metabolism with coronary artery disease severity. Microbiome. 2019;7(1) doi: 10.1186/s40168-019-0683-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang C.-Y., Yang K.-M., Yang L., Miao Z.-X., Wang Y.-H., Zhu H.-b., et al. A (1)h nmr-based metabonomic investigation of time-related metabolic trajectories of the plasma, urine and liver extracts of hyperlipidemic hamsters. PLoS ONE. 2013;8(6):e66786. doi: 10.1371/journal.pone.0066786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang W.H.W., Hazen S.L. The gut microbiome and its role in cardiovascular diseases. Circulation. 2017;135(11):1008–1010. doi: 10.1161/CIRCULATIONAHA.116.024251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kazemian N., Mahmoudi M., Halperin F., Wu J.C., Pakpour S. Gut microbiota and cardiovascular disease: opportunities and challenges. Microbiome. 2020;8:36. doi: 10.1186/s40168-020-00821-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu G., Zhao N., Zhang C., Lam Y.Y., Zhao L. Guild-based analysis for understanding gut microbiome in human health and diseases. Genome Med. 2021;13:22. doi: 10.1186/s13073-021-00840-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansson G.K., Hermansson A. The immune system in atherosclerosis. Nat Immunol. 2011;12(3):204–212. doi: 10.1038/ni.2001. [DOI] [PubMed] [Google Scholar]
- 23.Bentzon J.F., Otsuka F., Virmani R., Falk E. Mechanisms of plaque formation and rupture. Circ Res. 2014;114(12):1852–1866. doi: 10.1161/CIRCRESAHA.114.302721. [DOI] [PubMed] [Google Scholar]
- 24.Stefanini G.G., Windecker S. Can coronary computed tomography angiography replace invasive angiography? Coronary computed tomography angiography cannot replace invasive angiography. Circulation. 2015;131(4):418–426. doi: 10.1161/CIRCULATIONAHA.114.008148. [DOI] [PubMed] [Google Scholar]
- 25.Kruse J.M., Enghard P., Schröder T., Hasper D., Kühnle Y., Jörres A., et al. Weak diagnostic performance of troponin, creatine kinase and creatine kinase-mb to diagnose or exclude myocardial infarction after successful resuscitation. Int J Cardiol. 2014;173(2):216–221. doi: 10.1016/j.ijcard.2014.02.033. [DOI] [PubMed] [Google Scholar]
- 26.Shi L.Y., Han Y.S., Chen J., Li Z.B., Li J.C., Jiang T.T. Screening and identification of potential protein biomarkers for the early diagnosis of acute myocardial infarction. Ann Transl Med. 2021;9:743. doi: 10.21037/atm-20-7891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eapen D.J., Manocha P., Patel R.S., Hammadah M., Veledar E., Wassel C., et al. Aggregate risk score based on markers of inflammation, cell stress, and coagulation is an independent predictor of adverse cardiovascular outcomes. J Am Coll Cardiol. 2013;62(4):329–337. doi: 10.1016/j.jacc.2013.03.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.She J., Feng J., Deng Y., Sun L., Wu Y., Guo M., et al. Correlation of triiodothyronine level with in-hospital cardiac function and long-term prognosis in patients with acute myocardial infarction. Dis Markers. 2018;2018:1–8. doi: 10.1155/2018/5236267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baars T., Sowa J.-P., Neumann U., Hendricks S., Jinawy M., Kälsch J., et al. Liver parameters as part of a non-invasive model for prediction of all-cause mortality after myocardial infarction. Arch Med Sci: AMS. 2020;16(1):71–80. doi: 10.5114/aoms.2018.75678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Svingen G.FT., Schartum-Hansen H., Ueland P.M., Pedersen E.R., Seifert R., Ebbing M., et al. Elevated plasma dimethylglycine is a risk marker of mortality in patients with coronary heart disease. Eur J Prev Cardiol. 2015;22(6):743–752. doi: 10.1177/2047487314529351. [DOI] [PubMed] [Google Scholar]
- 31.Mueller D.M., Allenspach M., Othman A., Saely C.H., Muendlein A., Vonbank A., et al. Plasma levels of trimethylamine-n-oxide are confounded by impaired kidney function and poor metabolic control. Atherosclerosis. 2015;243(2):638–644. doi: 10.1016/j.atherosclerosis.2015.10.091. [DOI] [PubMed] [Google Scholar]
- 32.Wu W., Ziemann M., Huynh K., She G., Pang Z.-D., Zhang Y., et al. Activation of hippo signaling pathway mediates mitochondria dysfunction and dilated cardiomyopathy in mice. Theranostics. 2021;11(18):8993–9008. doi: 10.7150/thno.62302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun L., Jia H., Li J., Yu M., Yang Y., Tian D., et al. Cecal gut microbiota and metabolites might contribute to the severity of acute myocardial ischemia by impacting the intestinal permeability, oxidative stress, and energy metabolism. Front Microbiol. 2019;10:1745. doi: 10.3389/fmicb.2019.01745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sugi K., Nakano S., Fukasawa Y., Maruyama R., Tanno J., Senbonmatsu T., et al. Percutaneous coronary intervention for septic emboli in the left main trunk as a complication of infective endocarditis. Heart Lung Circ. 2015;24(11):e176–e179. doi: 10.1016/j.hlc.2015.06.819. [DOI] [PubMed] [Google Scholar]
- 35.Yang T., Qu H., Song X., Liu Q., Yang X., Xu J., et al. Luhong granules prevent ventricular remodelling after myocardial infarction by reducing the metabolites tmao and lps of the intestinal flora. Evid Based Complement Alternat Med. 2019;2019:1–10. doi: 10.1155/2019/8937427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen Y., Peng L., Shi S., Guo G., Wen H. Boeravinone b alleviates gut dysbiosis during myocardial infarction-induced cardiotoxicity in rats. J Cell Mol Med. 2021;25(13):6403–6416. doi: 10.1111/jcmm.16620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sardu C., Consiglia Trotta M., Santella B., D'Onofrio N., Barbieri M., Rizzo M.R., et al. Microbiota thrombus colonization may influence athero-thrombosis in hyperglycemic patients with st segment elevation myocardialinfarction (stemi). Marianella study. Diabetes Res Clin Pract. 2021;173:108670. doi: 10.1016/j.diabres.2021.108670. [DOI] [PubMed] [Google Scholar]
- 38.Kaye A.D., Jeha G.M., Pham A.D., Fuller M.C., Lerner Z.I., Sibley G.T., et al. Folic acid supplementation in patients with elevated homocysteine levels. Adv Ther. 2020;37(10):4149–4164. doi: 10.1007/s12325-020-01474-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jauhiainen R., Vangipurapu J., Laakso A., Kuulasmaa T., Kuusisto J., Laakso M. The association of nine amino acids with cardiovascular events in finnish men in a 12-year follow-up study. J Clin Endocrinol Metab. 2021 doi: 10.1210/clinem/dgab562. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the repository of the Genome Sequence Archive Sequence Database of National Genomics Data Center (https://bigd.big.ac.cn/) under the accession number CRA002142.