Graphical abstract
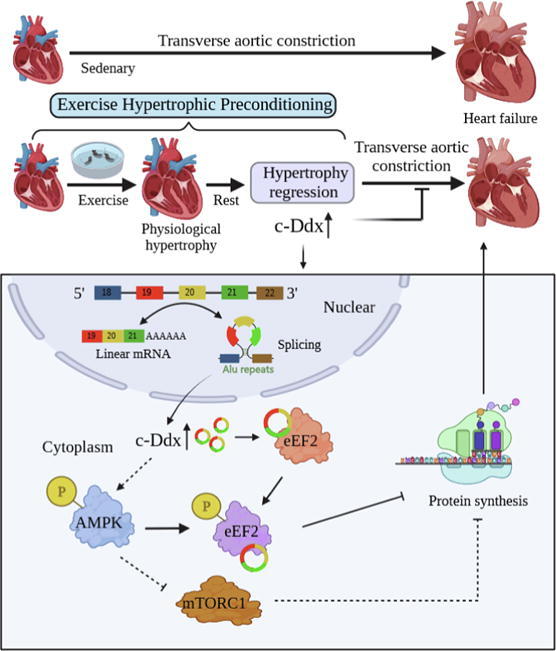
Working model of this study. EHP significantly upregulates circular RNA Ddx60 (c-Ddx) in mouse myocardial tissue. c-Ddx exerts its function by binding eEF2. After binding eEF2, c-Ddx promotes phosphorylation of eEF2 and its upstream AMPK, generating a positive feedback pathway. Phosphorylation of eEF2 prevents protein translational elongation in pathological myocardial hypertrophy, and eventually produces an antihypertrophic effect, while activation of AMPK by c-Ddx/eEF2 complexus would also produce an antihypertrophic role through the well-known signal pathways. Abbreviations: EHP: exercise hypertrophic preconditioning; c-Ddx, circ-Ddx60; eEF2: eukaryotic elongation factor 2; AMPK: AMP-activated protein kinase; mTORC1, mammalian target of rapamycin kinase complex 1. ↑, activation; ⊥, inhibition; ---, pathway confirmed by previous studies. This picture was created with BioRender.com.
Keywords: Myocardial hypertrophy, Exercise hypertrophic preconditioning, Circular RNA, Eukaryotic elongation factor 2
Highlights
-
•
This study is the first attempt to clarify the role of circRNA on exercise hypertrophic precondition (EHP).
-
•
We identified a novel circRNA, named circ-Ddx60, is preferentially expressed in myocardial tissue and significantly up-regulated in response to EHP.
-
•
We confirmed that circ-Ddx60 promotes the formation of antihypertrophic memory of EHP, silencing circ-Ddx60 suppresses the inhibitory effect of EHP on pathological myocardial hypertrophy.
-
•
Circ-Ddx60 silencing inhibits the EHP-enhanced phosphorylation of eEF2 and AMPK.
-
•
This work would provide opportunity to search new therapeutic targets for pathological cardiac hypertrophy.
Abstract
Introduction
We previously reported a phenomenon called exercise hypertrophic preconditioning (EHP), the underlying mechanisms of which need further clarification.
Objectives
We aimed to investigate whether circular RNAs (circRNAs) are involved in EHP.
Methods
CircRNA sequencing of myocardial tissue was performed in male C57BL/6 mice with EHP and sedentary. Bioinformatics analysis and Sanger sequencing were used to screen hub circRNA expression and to detect full-length circRNAs, respectively. Loss-of-function analyses were conducted to assess the effects of circ-Ddx60 (c-Ddx) on EHP. After 21 days of swimming training or resting, mice underwent transverse aortic constriction (TAC) or sham surgery. Echocardiography, invasive hemodynamic measurement and histological analysis were used to evaluate cardiac remodeling and function. The presence of interaction between c-Ddx and proteins was investigated using comprehensive identification of RNA-binding proteins by mass spectrometry (ChIRP-MS).
Results
In this study, we identified a novel circRNA, named c-Ddx that was preferentially expressed in myocardial tissue and significantly up-regulated in EHP mice. Silencing of c-Ddx attenuated the antihypertrophic effect of EHP and worsened heart failure in mice that underwent TAC. ChIRP-MS and molecular docking analysis validated the combination of c-Ddx and eukaryotic elongation factor 2 (eEF2). Mechanistically, c-Ddx silencing inhibited the increase of phosphorylation of eEF2 and its upstream AMP-activated protein kinase (AMPK) induced by EHP.
Conclusions
C-Ddx contributes to the antihypertrophic memory of EHP by binding and activating eEF2, which would provide opportunity to search new therapeutic targets for pathological hypertrophy of heart.
Introduction
Despite the introduction of new therapies for heart failure (HF) such as angiotensin receptor-neprilysin inhibitors and sodium glucose cotransporter 2 inhibitors, HF remains the leading cause of death [1], [2]. Additional preventive and therapeutic approaches to HF are urgently needed. Exercise as a “lifestyle modification” is known to be beneficial for preventing and recovering from HF [3], [4], which may be mediated by triggering beneficial adaptive cellular responses and different molecular mechanisms [5], [6]. Novel therapeutic drug targets for HF may be found by clarifying the molecular mechanisms for the cardiac benefit of exercise. We previously reported a phenomenon termed exercise hypertrophic preconditioning (EHP), by which exercise-induced antihypertrophic memory exists even after physiological hypertrophy has regressed following termination of exercise [7]. Awareness of EHP uncovers potential new therapeutic approaches for treating cardiac hypertrophy and HF, we have demonstrated earlier that the Mhrt779/Brahma-related gene-1 (Brg1)/histone deacetylase 2 (Hdac2)/phosphorylated serine-threonine kinase (p-Akt)/phosphorylated glycogen synthase kinase 3β (p-GSK3β) signal pathway is involved in EHP [7], but other potential mechanisms of EHP remain poorly understood.
Circular RNAs (circRNAs) are an enigmatic class of single-stranded RNA with largely unknown functions [8], [9], while most circRNAs are formed by head-to-tail splicing of the exon regions of protein-coding genes, some are derived from intron regions [10]. Prior studies have demonstrated that loss of super-enhancer regulated circNfix or overexpression of circHipk3 can promote cardiac regeneration [11], [12]. It has been reported that certain circRNAs (such as circHRCR and circYAP) attenuate and others (including circSlc8a1 and circ000203) promote pathological cardiac hypertrophy [13], [14], [15], [16]. However, it is unclear whether circRNAs are involved in physiological cardiac hypertrophy and EHP.
Our preliminary findings led us to hypothesize that circ-Ddx60 (c-Ddx) mediated antihypertrophic molecular memory of EHP. This study was designed to characterize c-Ddx, define its contribution to EHP, and clarify the underlying mechanisms.
Materials and methods
The full methods and materials for this study are provided in the supplementary materials and are available upon request. The sequences of primers used in all experiments are shown in Tables S1–S5 (supplemental materials).
Ethics statement
All experiments were performed in accordance with our institutional guidelines for animal research, which conform to the Guide for the Care and Use of Laboratory Animals (National Institutes of Health Publication, 8th Edition, 2011). Approval for this study was granted by the ethics review board at Nanfang Hospital, Southern Medical University (Guangzhou, China). (Approval no. NFYY20190930).
Statistical analysis
Data were summarized by presenting mean ± standard error of the mean. All data analyses were performed using GraphPad Prism version 7.0 (GraphPad Software Inc., San Diego, CA, USA). Statistical differences between 2 groups were evaluated using 2-tailed, unpaired t-tests; comparisons of differences among multiple groups were evaluated by either one-way or two-way (if there were 2 factor levels) analysis of variance, followed by Bonferroni correction for post hoc multiple comparisons. P values<0.05 were considered statistically significant.
Results
Identification of c-Ddx as a novel CircRNA significantly up-regulated by EHP
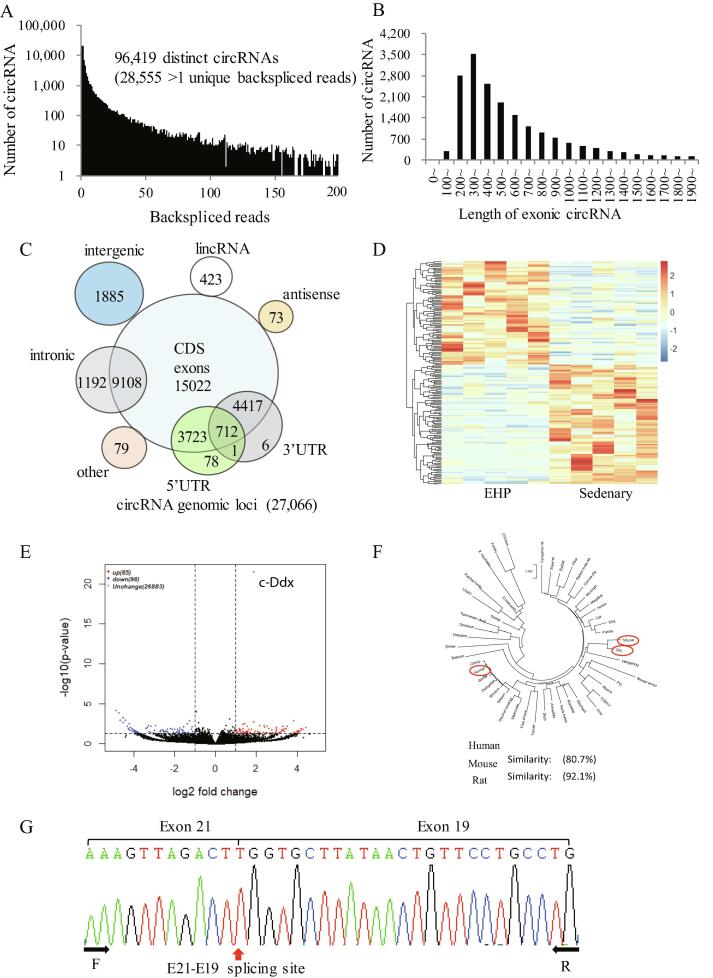
We investigated the potential involvement of circRNAs in EHP (Fig. S1). A total of 96,419 distinct circRNAs were screened, of which 28,555 were supported by 2 unique reads (Fig. 1A). We further screened out 27,066 circRNA molecules that met 6 criteria (Fig. S2A), most of these molecules had exon lengths of 200–500 bps (Fig. 1B). Venn diagrams show the overlap between circRNAs across samples (Fig. S2B) and across circRNA genomic loci of origin (Fig. 1C). We noted that 183 circRNAs were differentially expressed (Filter by |Fold Change|>1 and p<0.05) between the EHP group and the sedentary group (Fig. 1D and E). Of the 85 up-regulated circRNAs, we selected a novel circRNA with a relative high level at baseline and the smallest statistical p value that we named it c-Ddx according to its mother gene Ddx60. ALU repeats are known to promote exon circularization of circRNAs, output of the RepeatMasker program performed on the mm10 DDX60 gene (chr8:61979256–61983943) was detected by the UCSC Genome Browser (https://genome.ucsc.edu) (Fig. S2C). C-Ddx was transcribed from exons 19 to 21 of the DDX60 gene, and the mouse ortholog of c-Ddx exhibited 80.7% and 92.1% nucleotide identity with its human and rat counterpart respectively (Fig. 1F). The full-length sequence of c-Ddx was determined by Sanger sequencing (Fig. 1G). Using an improved bioinformatics analysis method, we also determined that the full length of c-Ddx can be constructed from Illumina paired-end sequencing reads, from which 1 end of each paired-end read contains the back-splice junction. The resulting consensus sequence was comprised of three exons without introns (Fig. S2D).
Fig. 1.
Identification of mousec-Ddx, a novel circRNA most significantly up-regulated by exercise hypertrophic preconditioning. (A), Distinct circRNAs identified through unique backsplice reads; (B), Distribution of exon lengths of circRNAs; (C), Venn diagram of circRNA genomic loci of origin; (D), Heatmap analysis of sequencing data for differentially expressed circRNAs with |log2 [fold change]| >1 and P < 0.05; (E), Volcano plot of differentially expressed circRNAs; (F), Similarity of DdX60 sequences from exon 19–21 among human, mouse and rat, calculated based on pairwise alignment; (G), Verification of the full sequence of c-Ddx by Sanger sequence analysis. Abbreviations: circRNA, circular RNA; c-Ddx, circ-Ddx60;
Characterization of c-Ddx
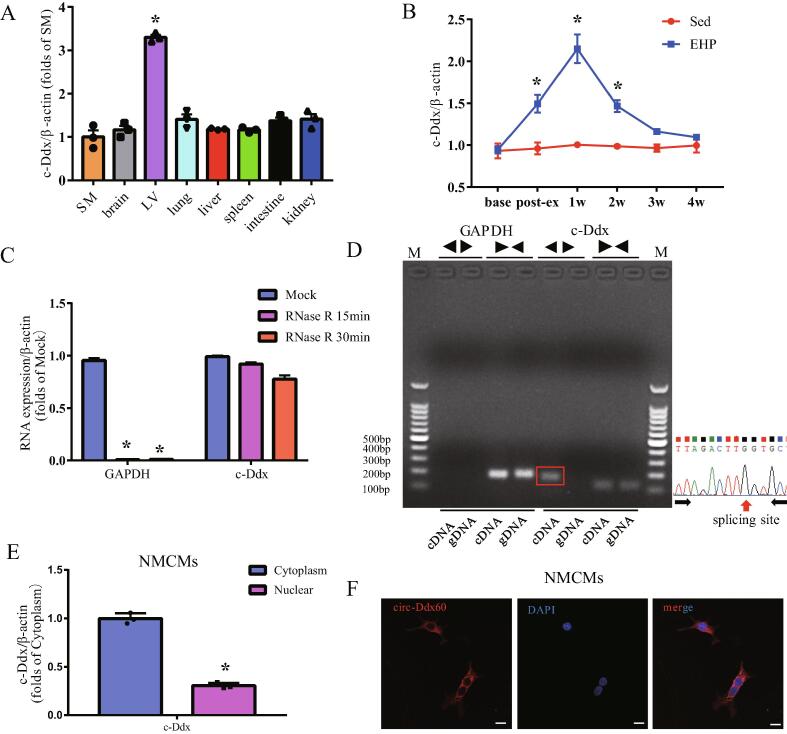
Expression of c-Ddx was significantly more abundant in left ventricular tissue than in other tissues as shown by quantitative PCR (Fig. 2A). We investigated the expression level of c-Ddx in left ventricular tissue among various time points of exercise in mice. After 21 days of exercise training, the c-Ddx levels were significantly increased relative to the Sedenary group. The level of c-Ddx continued to increase after the termination of exercise and peaked at one week, then gradually decreased with time, and approached the level of the Sed group after 4 weeks (Fig. 2B). We further investigated the expression of c-Ddx in the sedentary, EHP and transverse aortic constriction (TAC) groups of mice. The c-Ddx level was higher in the EHP mice than in the sedentary group. At 5 weeks after TAC, c-Ddx was significantly lower than in the sedentary group (Fig. S3). We then investigated the stability and localization of c-Ddx in neonatal mouse cardiomyocytes (NMCMs). Despite digestion with RNase R exonuclease treatment for 15 or 30 min, the enrichment of c-Ddx was apparent, whereas the abundance of linear glyceraldehyde-3-phosphate dehydrogenase (GAPDH) decreased, indicating this RNA species is circular in form (Fig. 2C). Divergent primer amplification of c-Ddx in cDNA but not in genomic DNA confirmed that c-Ddx doesn't arise from genomic rearrangement (Fig. 2D). Quantitative PCR analysis of nuclear and cytoplasmic c-Ddx (Fig. 2E) and fluorescence in situ hybridization for c-Ddx (Fig. 2F) demonstrated that c-Ddx localized preferentially in the cytoplasm rather than in the nucleus, while U6 used as control for nuclear fraction almost localized in nucleus (Fig. S4A-S4B).
Fig. 2.
The properties ofmousec-Ddx. (A), Abundance of c-Ddx expression determined by quantitative PCR in various adult mouse tissues,*P < 0.05 vs. SM, n = 3 per group; (B), A time course of myocardial expression levels of c-Ddx in response to swimming training or sedentary life in mice, swimming was terminated at 3 weeks, *P < 0.05 vs. Sed group, n = 6 per group at each time point; (C), Abundances of c-Ddx and glyceraldehyde-3-phosphate dehydrogenase (GADPH) in adult mouse myocardial tissue treated with RNase R, *P < 0.05 vs. Mock group, n = 3 per group; (D), Divergent primers amplify c-Ddx of adult mouse heart in complementary DNA but not in genomic DNA, divergent and convergent primers are indicated by the directions of the arrows; (E), Abundance of nuclear and cytoplasmic c-Ddx in NMCMs, *P < 0.05 vs. Cytoplasm group, n = 3 per group; (F), RNA fluorescence in situ hybridization for c-Ddx in NMCMs (bar = 5 µm); Abbreviations: c-Ddx, circ-Ddx60; EHP, exercise hypertrophic preconditioning; NMCMs, neonatal mouse cardiomyocytes; Sed, sedentary; SM, skeletal muscle; LV, left ventricular tissue;
Silencing c-Ddx attenuates the inhibitory effect of EHP on pathological cardiac hypertrophy
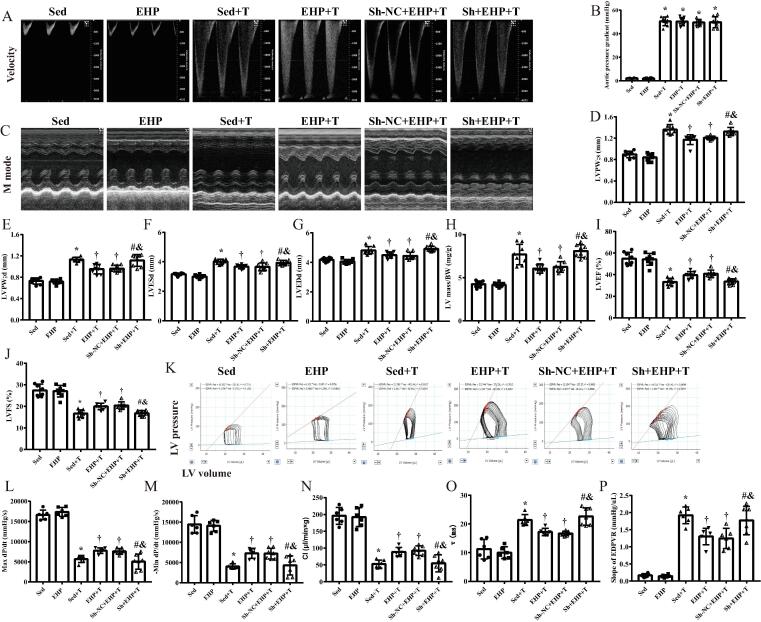
Three small intervention c-Ddx (si) were designed according to the back-splice junction of c-Ddx. The silencing efficiency was tested in NMCMs (Fig. S5A). Si-1 sequences were used to construct short hairpin of c-Ddx (Sh-c-Ddx) carried by adeno-associated virus type 9 (AAV9). Four weeks after in situ injection of AAV9 carrying plasmids of Sh-c-Ddx in the mouse myocardium, the transfection efficiency was confirmed by GPF fluorescence (Fig. S5B). Treatment with Sh-c-Ddx for 4 weeks resulted in an approximately half down-regulation of c-Ddx (Fig. S5C). Then mice were conducted to swimming exercise for 21 days. After rest for 1 week, mice subjected to TAC surgery. Quality control of TAC surgery was evaluated by both Doppler blood flow velocity at the banding site (Fig. 3A) and the aortic pressure gradient calculated from the flow velocity (Fig. 3B). The aortic pressure gradient (PG) is an indicator of LV pressure overload which can be calculated from the Doppler velocity of blood flow (V) across the aortic stenosis: PG = 4 × V2. Five weeks after TAC, echocardiography indicated that the EHP group had significantly decreased LV posterior wall thickness and LV diameter and significantly increased left ventricular ejection fraction and left ventricular fractional shortening compared with the sedentary group, these changes were blocked by pretreatment with Sh-c-Ddx (Fig. 3C-J). Pressure-volume loop measurements indicated that the EHP group had a significantly increased LV dp/dt max, dp/dt min and cardiac index (CI) and significantly decreased τ and slope of end-diastolic pressure volume relationship (EDPVR) compared with the sedentary group; pretreatment with Sh-c-Ddx canceled the hemodynamic improvements attributed to EHP (Fig. 3K-P).
Fig. 3.
Effect of silencing of c-Ddx on the antihypertrophic effect of EHPin mice. (A), Doppler flow velocity at the banding site of aortic arch in mice at 3 d after surgery; (B), Aortic pressure gradient calculated from Doppler flow velocity, *P < 0.01 vs. Sed group; (C), Representative M-mode echocardiography of LV; (D), LV posterior wall systolic thickness; (E), LV posterior wall diastolic thickness; (F), LV end-systolic diameter; (G), LV end-diastolic diameter; (H), Echocardiographic LV mass/BW; (I), LV ejection fraction; (J), LV fractional shortening; n = 8 in each groups. (K), Representative LV pressure–volume loops at vena cava during occlusion; (L), Maximum rising rate of LV pressure (dp/dt max); (M), Maximum descending rate of LV pressure (dp/dt min); (N), Cardiac index; (O), Exponential LV relaxation time constant (τ); (P), End-diastolic pressure volume relationship as a measure of LV diastolic stiffness. For panels K-P, n = 6–7 in each group (a few animals died of procedure failure due to rupture of the artery). *P < 0.05 vs. Sed group, †P < 0.05 vs. Sed + T group, &P < 0.05 vs. EHP + T group, #P < 0.05 vs. Sh-NC + EHP + T group. Abbreviations: c-Ddx, circ-Ddx60; EHP, exercise hypertrophic preconditioning; LV, left ventricle; Sed, sedentary group; Sh, short hairpin of circ-Ddx60; T, transverse aortic constriction; NC, negative control; BW, body weight (g).
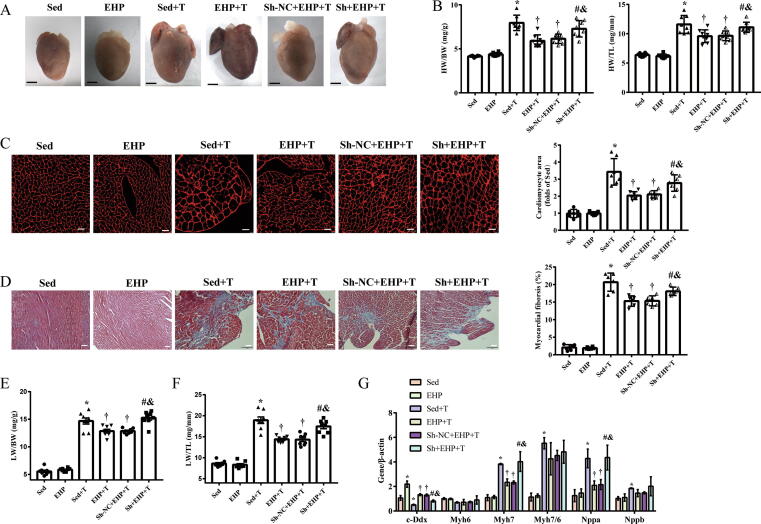
EHP significantly attenuated TAC-induced myocardial hypertrophy and fibrosis, as reflected by lower heart weight/body weight and heart weight/tibia length ratios (Fig. 4A-B), smaller cardiomyocyte area (Fig. 4C), less myocardial fibrosis (Fig. 4D), smaller lung weight/body weight and lung weight/tibia length ratios (Fig. 4E-F), and lower expression of fatal natriuretic peptide type A (NPPA) and myosin heavy chain 7 (MYH7) genes (Fig. 4G). These changes were blocked by pretreatment with Sh-c-Ddx (Fig. 4G).
Fig. 4.
Effect of silencing of c-Ddx on myocardial hypertrophy, cardiac fibrosis and pulmonary congestion in EHP mice with pressure overload. (A), Representative mouse hearts (scale bar = 2 mm); (B), HW:BW ratio and HW:TL ratio; (C), Representative myocardial cross-sections stained with wheat germ agglutinin (scale bar = 25 µm) and quantization of cardiomyocyte cross-sectional area; (D), Representative myocardial cross-sections stained with masson trichrome (scale bar = 50 µm) and quantization of myocardial fibrosis; (E), LW:BW ratio; (F), LW:TL ratio; (G), Silencing of c-Ddx blocked the EHP-induced downregulation of Myh7 and ANP. For all data panels, n = 8 in each groups. *P < 0.05 vs. Sed group, †P < 0.05 vs. Sed + T group, &P < 0.05 vs. EHP + T group, #P < 0.05 vs. Sh-NC + EHP + T group. Abbreviations: c-Ddx, circ-Ddx60; EHP, exercise hypertrophic preconditioning; BW, body weight (g); HW, heart weight (mg); LW, lung weight (mg); TL, tibial length (mm); Sed, sedentary group; EHP, exercise hypertrophic preconditioning; Sh, short hairpin of c-Ddx; T, transverse aortic constriction. NC, negative control.
c-Ddx activates eEF2 and AMPK to attenuate cardiac hypertrophy
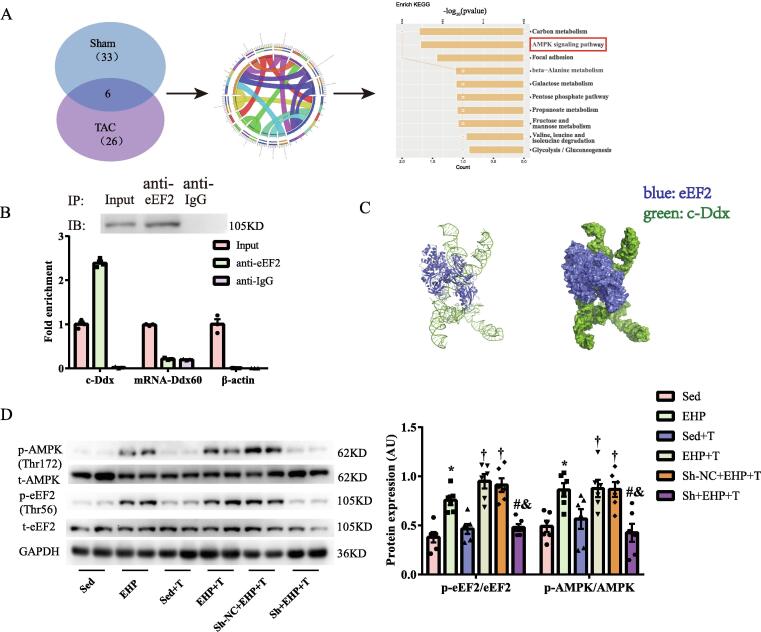
We performed comprehensive identification of RNA-binding proteins by mass spectrometry (ChIRP-MS) to clarify the interaction between c-Ddx and proteins (Fig. S6). The ChIRP-MS procedures are presented in Fig. S6A. The quality control report confirmed that the products of the probes could be used for subsequent mass spectrometry (Table S5, Fig. S6B-S6C). Mass spectrometry identified 182 proteins capable of binding c-Ddx. Filtered by fold change > 1.5 (ie, fold change relative to a linear RNA control) and unique peptide number ≥ 2, the number of proteins specifically enriched in the ChIRP lysate ranged from 33 among Sham mice to 26 among TAC mice (Tables S6-7). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis showed that most of these proteins are involved in cardiomyopathy and metabolism (Fig. S6D-S6E).
We believe that proteins that bind with c-Ddx in both groups have higher specificity. Six proteins (Flnc, Ttn, Pfkm, Aldh6a1, Vcl, eEF2) were coincidentally enriched in the ChIRP lysate of mouse myocardial tissue in both the sham and TAC groups. Functions of these proteins were predicted using KEGG pathway enrichment analysis (Fig. 5A), we focused on eukaryotic elongation factor 2 (eEF2), a protein involved in the adenosine 5′-monophosphate–activated protein kinase (AMPK)-mediated signal pathway for cardiac antihypertrophy. Using RNA immunoprecipitation (RIP)-Western blot, we validated that c-Ddx binds to eEF2 (Fig. 5B). Molecular-docking analysis indicated that c-Ddx is folded to form a DNA-like double helix structure, which makes the RNA have high stability. The folded c-Ddx- wraps the eFF2 protein and fits the protein very well, which is conducive to the formation of a stable complex between RNA and protein (Fig. 5C). We noted that EHP (c-Ddx up-regulated) increased the protein expression of phosphorylated AMPK (p-AMPK) and phosphorylated eEF2 (p-eEF2) in pressure-overloaded mice, however this change was blocked by pretreatment with Sh-c-Ddx (Fig. 5D). To further explore the effect of c-Ddx on the activation of eEF2 and AMPK, we constructed adenovirus (Ad) overexpressing mouse c-Ddx without affecting the expression of its parent gene. We found that c-Ddx overexpression significantly upregulated p-eEF2 and p-AMPK proteins (Fig. S7A-S7B), and markedly attenuated PE-induced hypertrophy in cultured cardiomyocytes (Fig. S7C).
Fig. 5.
Identification of proteins bound by c-Ddx and the influence of c-Ddx on the activity of eEF2 and AMPK in mice. (A), Gene ontology analysis of 6 functional proteins bound by c-Ddx in Sham and TAC groups; (B), Verification of the combination of eEF2 and c-Ddx by RNA binding protein immunoprecipitation; (C), Verification of the combination of eEF2 and c-Ddx by molecular docking analysis. The binding score of RNA to eFF2 protein was −325.38 kcal/mol. Right picture represented the electrostatic surface of eEF2 and c-Ddx. (D), The Effects of c-Ddx kncokdown on EHP-induced phosphorylation of eEF2 and AMPK. *P < 0.05 vs. Sed group, †P < 0.05 vs. Sed + T group, &P < 0.05 vs. EHP + T group, #P < 0.05 vs. Sh-NC + EHP + T group. Abbreviations: c-Ddx, circ-Ddx60; AMPK, adenosine 5′-monophosphate activated protein kinase; eEF2, eukaryotic elongation factor 2; Sed, sedentary group; EHP, exercise hypertrophic preconditioning; NC, negative control; Sh, short hairpin of c-Ddx; TAC, transverse aortic constriction.
The RIP-Western blot and IP-Western blot showed that c-Ddx did not bind AMPK and no combination between eEF2 and AMPK (Fig. S8A-S8B). After silencing eEF2 (Fig. S9A), we found that c-Ddx-induced phosphorylation of eEF2 and AMPK was significantly blocked in neonatal rat cardiomyocytes (Fig. S9B). When co-cultured with dorsomorphin (Compound C, CC), an AMPK chemical inhibitor, c-Ddx induced phosphorylation of AMPK but not eEF2 in cardiomyocytes was abolished (Fig. S10A).
Discussion
Previous studies have found former athletes are likely to live longer than nonathletic counterparts and leisure time physical activity is associated with reduced risk of cardiovascular disease and all-cause mortality. These findings suggest beneficial memory of exercise can be sustained [17], [18], although the specific mechanisms of this effect are poorly understood. Our recent report also demonstrated that people with a habit of optimal exercise dose (vigorous physical activity group) had the best risk reduction effect of chronic HF (OR 0.443, 95% CI: 0.286–0.658) [19]. Several investigations have confirmed that myocardial hypertrophic precondition is cardioprotective which manifested by its inhibitory effect on pathological cardiac hypertrophy and myocardial ischemia as well as its protection on HF induced by cardiac hypertrophy and myocardial ischemia [7], [20], [21], [22], [23].
CircRNA can act as an endogenous miRNA sponge, interact with functional proteins, regulate gene transcription or even have coding potential [24], [25]. While investigators have attempted to clarify the role of circRNAs in pathological cardiac hypertrophy, the influence of circRNAs on physiological cardiac hypertrophy and EHP remains unclear. Due to their unique circular structure, circRNAs are more stable than linear RNA resulting in resistance to exonuclease degradation and the capacity to accumulate in non-proliferating cells such as cardiomyocytes [26], [27]. The function of circRNAs is inconsistent with their associated linear isoforms, and often is expressed in a tissue-specific and disease-specific manner [10], [28]. In this study, we identified and characterized a novel circRNA named c-Ddx, and clarified its contribution to the antihypertrophic effect of EHP.
We found that silencing c-Ddx in EHP mice, which lead to c-Ddx levels closer to sedentary, significantly inhibited the antihypertrophic effect of EHP on pathological myocardial hypertrophy. These findings suggest that the antihypertrophic effect of EHP is mediated through up-regulation of c-Ddx.
To clarify the potential mechanisms by which c-Ddx mediates the antihypertrophic effect of EHP, we performed ChIRP-MS and found that c-Ddx could bind eEF2 in myocardial tissues of both sham and TAC mice. eEF2 is a GTPase capable of translocating peptidyl-transfer RNA from the A-site to the P-site of the ribosome, thereby promoting translation elongation of protein [29]. We noted that EHP up-regulated the phosphorylation of eEF2 and AMPK proteins in TAC mice but these protective effects were blocked by pre-treatment of silencing c-Ddx, which suggests that c-Ddx may be a mechanism for the antihypertrophic memory of EHP. We did not detect a combination between c-Ddx and AMPK. It is well known that the AMPK/eEF2 pathway is closely associated with myocardial hypertrophy [30], [31]. Phosphorylation of eEF2 inhibits the activity of eEF2 leading to suppression of myocardial hypertrophy [31]. AMPK activation has been shown to inhibit cardiac hypertrophy by reducing O-GlcNAcylation of proteins [32] or affecting downstream targets such as the mammalian target of rapamycin/p70 ribosomal S6 protein kinase [33]. It seems interesting that Compound C blocked c-Ddx-induced AMPK activation but not eEF2 activation, suggesting AMPK does not severe as an upstream of eEF2 in the presence of a combination of c-Ddx and eEF2. It is likely that AMPK activation by c-Ddx may exert its antihypertrophic role through other signal pathways.
Many other factors-mediated mechanisms contribute to the antihypertrophic effect of EHP. We previously reported that the long noncoding RNA Mhrt779 contributes to the antihypertrophic effect of EHP by inhibiting the Brg1/Hdac2/Akt/GSK3β pathway [7]. Interestingly, the promoter of eEF2 has been shown to bind Brg1 [34], eEF2 can bind Hdac5 [35], and Hdac5 has been demonstrated to interact with Hdac2 [36]. These findings suggest that c-Ddx may exert an antihypertrophic effect through the eEF2/Brg1/Hdac2/5/Akt/GSK3β pathway. Since many proteins bind c-Ddx, further studies are needed to clarify whether additional mechanisms are involved in the antihypertrophic memory of c-Ddx.
There are some limitations in this study which need further investigation. (1) It is completely unknown whether c-Ddx also exerts antihypertrophic role in patients with pathological hypertrophy. (2) Due to the gender difference existing in cardiovascular disease, our conclusions from male mice are not necessary appropriate for female mice. (3) We only elucidate the role of c-Ddx in cardiac hypertrophy by affecting the activity of eEF2 and AMPK, but did not further analyze its effect on metabolic pathway by acting on AMPK.
Conclusion
This study is a first attempt clarifying the effect of a circRNA on EHP. We identified a novel circRNA, named c-Ddx that was preferentially expressed in myocardial tissue and significantly up-regulated in EHP mice. We confirmed that c-Ddx promotes antihypertrophic memory of EHP, evidenced by our finding that silencing c-Ddx suppresses the inhibitory effect of EHP on pathological myocardial hypertrophy and heart failure. Furthermore, we found that EHP activated while silencing c-Ddx inhibited the phosphorylated of eEF2 and AMPK in mice underwent TAC, which suggests c-Ddx contributes to the antihypertrophic memory of EHP by binding and activating eEF2 (Graphical abstract). These findings would provide opportunity to search new therapeutic targets for pathological cardiac hypertrophy.
Compliance with Ethics Requirements
All experiments were performed in accordance with our institutional guidelines for animal research, which conform to the Guide for the Care and Use of Laboratory Animals (National Institutes of Health Publication, 8th Edition, 2011). Approval for this study was granted by the ethics review board at Nanfang Hospital, Southern Medical University (Guangzhou, China). (Approval no. NFYY20190930).
No human study included in this study.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgement
This work was supported by grants from the Joint Funds of the National Natural Science Foundation of China (U1908205 to Y. L.), the Municipal Planning Projects of Scientific Technology of Guangzhou (201804020083 to Y. L.), the Key Program of Natural Science Foundation of Guangdong Province (2018B0303110008 to Y. L.).
Data availability
The datasets used and analyzed supporting the findings of this study are available in this paper or the Supplementary Materials. Any other raw data generated or analyzed in this study can be obtained upon reasonable request from the corresponding authors.
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2022.06.005.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.van der Meer P., Gaggin H.K., Dec G.W. Acc/aha versus esc guidelines on heart failure: Jacc guideline comparison. J Am Coll Cardiol. 2019;73:2756–2768. doi: 10.1016/j.jacc.2019.03.478. [DOI] [PubMed] [Google Scholar]
- 2.Seferovic P.M., Ponikowski P., Anker S.D., Bauersachs J., Chioncel O., Cleland J.G.F., et al. Clinical practice update on heart failure 2019: Pharmacotherapy, procedures, devices and patient management. An expert consensus meeting report of the heart failure association of the european society of cardiology. Eur J Heart Fail. 2019;21(10):1169–1186. doi: 10.1002/ejhf.1531. [DOI] [PubMed] [Google Scholar]
- 3.Cattadori G., Segurini C., Picozzi A., Padeletti L., Anzà C. Exercise and heart failure: an update. ESC Heart Fail. 2018;5:222–232. doi: 10.1002/ehf2.12225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma S., Liao Y. Noncoding rnas in exercise-induced cardio-protection for chronic heart failure. EBioMedicine. 2019;46:532–540. doi: 10.1016/j.ebiom.2019.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu G., Zhang X., Gao F. The epigenetic landscape of exercise in cardiac health and disease. J Sport Health Sci. 2021;10:648–659. doi: 10.1016/j.jshs.2020.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fiuza-Luces C., Santos-Lozano A., Joyner M., Carrera-Bastos P., Picazo O., Zugaza J.L., et al. Exercise benefits in cardiovascular disease: Beyond attenuation of traditional risk factors. Nat Rev Cardiol. 2018;15(12):731–743. doi: 10.1038/s41569-018-0065-1. [DOI] [PubMed] [Google Scholar]
- 7.Lin H., Zhu Y., Zheng C., Hu D., Ma S., Chen L., et al. Antihypertrophic memory after regression of exercise-induced physiological myocardial hypertrophy is mediated by the long noncoding rna mhrt779. Circulation. 2021;143(23):2277–2292. doi: 10.1161/CIRCULATIONAHA.120.047000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., et al. Circular rnas are a large class of animal rnas with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 9.He A.T., Liu J., Li F., Yang B.B. Targeting circular rnas as a therapeutic approach: current strategies and challenges. Signal Transduct Target Ther. 2021;6:185. doi: 10.1038/s41392-021-00569-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kristensen L.S., Andersen M.S., Stagsted L.V.W., Ebbesen K.K., Hansen T.B., Kjems J. The biogenesis, biology and characterization of circular rnas. Nat Rev Genet. 2019;20:675–691. doi: 10.1038/s41576-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 11.Huang S., Li X., Zheng H., Si X., Li B., Wei G., et al. Loss of super-enhancer-regulated circrna nfix induces cardiac regeneration after myocardial infarction in adult mice. Circulation. 2019;139(25):2857–2876. doi: 10.1161/CIRCULATIONAHA.118.038361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Si X., Zheng H., Wei G., Li M., Li W., Wang H., et al. Circrna hipk3 induces cardiac regeneration after myocardial infarction in mice by binding to notch1 and mir-133a. Mol Ther Nucleic Acids. 2020;21:636–655. doi: 10.1016/j.omtn.2020.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang K., Long B., Liu F., Wang J.-X., Liu C.-Y., Zhao B., et al. A circular rna protects the heart from pathological hypertrophy and heart failure by targeting mir-223. Eur Heart J. 2016;37(33):2602–2611. doi: 10.1093/eurheartj/ehv713. [DOI] [PubMed] [Google Scholar]
- 14.Wu N., Xu J., Du W.W., Li X., Awan F.M., Li F., et al. Yap circular rna, circyap, attenuates cardiac fibrosis via binding with tropomyosin-4 and gamma-actin decreasing actin polymerization. Mol Ther. 2021;29(3):1138–1150. doi: 10.1016/j.ymthe.2020.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lim T.B., Aliwarga E., Luu T.D.A., Li Y.P., Ng S.L., Annadoray L., et al. Targeting the highly abundant circular rna circslc8a1 in cardiomyocytes attenuates pressure overload induced hypertrophy. Cardiovasc Res. 2019;115(14):1998–2007. doi: 10.1093/cvr/cvz130. [DOI] [PubMed] [Google Scholar]
- 16.Li H., Xu J.-D., Fang X.-H., Zhu J.-N., Yang J., Pan R., et al. Circular rna circrna_000203 aggravates cardiac hypertrophy via suppressing mir-26b-5p and mir-140-3p binding to gata4. Cardiovasc Res. 2020;116(7):1323–1334. doi: 10.1093/cvr/cvz215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kettunen J.A., Kujala U.M., Kaprio J., Bäckmand H., Peltonen M., Eriksson J.G., et al. All-cause and disease-specific mortality among male, former elite athletes: an average 50-year follow-up. Br J Sports Med. 2015;49(13):893–897. doi: 10.1136/bjsports-2013-093347. [DOI] [PubMed] [Google Scholar]
- 18.Holtermann A., Schnohr P., Nordestgaard B.G., Marott J.L. The physical activity paradox in cardiovascular disease and all-cause mortality: the contemporary copenhagen general population study with 104 046 adults. Eur Heart J. 2021;42:1499–1511. doi: 10.1093/eurheartj/ehab087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen K., Sha T., Zhu Y., Ma S., Chen L., Liao W., et al. Optimal dose of physical exercise for preventing cardiac and renal dysfunction, data from nhanes survey. Eur J Prev Cardiol. 2022 doi: 10.1093/eurjpc/zwac096. [DOI] [PubMed] [Google Scholar]
- 20.Wei X., Wu B., Zhao J., Zeng Z., Xuan W., Cao S., et al. Myocardial hypertrophic preconditioning attenuates cardiomyocyte hypertrophy and slows progression to heart failure through upregulation of s100a8/a9. Circulation. 2015;131(17):1506–1517. doi: 10.1161/CIRCULATIONAHA.114.013789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma L.-L., Ding Z.-W., Yin P.-P., Wu J., Hu K., Sun A.-J., et al. Hypertrophic preconditioning cardioprotection after myocardial ischaemia/reperfusion injury involves aldh2-dependent metabolism modulation. Redox Biol. 2021;43:101960. doi: 10.1016/j.redox.2021.101960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma L.-L., Kong F.-J., Ma Y.-J., Guo J.-J., Wang S.-J., Dong Z., et al. Hypertrophic preconditioning attenuates post-myocardial infarction injury through deacetylation of isocitrate dehydrogenase 2. Acta Pharmacol Sin. 2021;42(12):2004–2015. doi: 10.1038/s41401-021-00699-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu J., Lu J., Huang J., You J., Ding Z., Ma L., et al. Variations in energy metabolism precede alterations in cardiac structure and function in hypertrophic preconditioning. Front Cardiovasc Med. 2020;7 doi: 10.3389/fcvm.2020.602100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song H.K., Hong S.-E., Kim T., Kim D.H., Schönbach C. Deep rna sequencing reveals novel cardiac transcriptomic signatures for physiological and pathological hypertrophy. PLoS ONE. 2012;7(4) doi: 10.1371/journal.pone.0035552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang L., Wang J., Li G., Xiao J. Non-coding rnas in physiological cardiac hypertrophy. Adv Exp Med Biol. 2020;1229:149–161. doi: 10.1007/978-981-15-1671-9_8. [DOI] [PubMed] [Google Scholar]
- 26.Chen L.L. The biogenesis and emerging roles of circular rnas. Nat Rev Mol Cell Biol. 2016;17:205–211. doi: 10.1038/nrm.2015.32. [DOI] [PubMed] [Google Scholar]
- 27.Jeck W.R., Sorrentino J.A., Wang K., Slevin M.K., Burd C.E., Liu J., et al. Circular rnas are abundant, conserved, and associated with alu repeats. RNA. 2013;19(2):141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li X., Yang L., Chen L.L. The biogenesis, functions, and challenges of circular rnas. Mol Cell. 2018;71:428–442. doi: 10.1016/j.molcel.2018.06.034. [DOI] [PubMed] [Google Scholar]
- 29.Yamada S., Kamata T., Nawa H., Sekijima T., Takei N. Ampk activation, eef2 inactivation, and reduced protein synthesis in the cerebral cortex of hibernating chipmunks. Sci Rep. 2019;9:11904. doi: 10.1038/s41598-019-48172-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kameshima S., Okada M., Yamawaki H. Eukaryotic elongation factor 2 (eef2) kinase/eef2 plays protective roles against glucose deprivation-induced cell death in h9c2 cardiomyoblasts. Apoptosis. 2019;24:359–368. doi: 10.1007/s10495-019-01525-z. [DOI] [PubMed] [Google Scholar]
- 31.Kameshima S., Okada M., Ikeda S., Watanabe Y., Yamawaki H. Coordination of changes in expression and phosphorylation of eukaryotic elongation factor 2 (eef2) and eef2 kinase in hypertrophied cardiomyocytes. Biochem Biophys Rep. 2016;7:218–224. doi: 10.1016/j.bbrep.2016.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gélinas R., Mailleux F., Dontaine J., Bultot L., Demeulder B., Ginion A., et al. Ampk activation counteracts cardiac hypertrophy by reducing o-glcnacylation. Nat Commun. 2018;9(1) doi: 10.1038/s41467-017-02795-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan J., Yan J.-Y., Wang Y.-X., Ling Y.-N., Song X.-D., Wang S.-Y., et al. Spermidine-enhanced autophagic flux improves cardiac dysfunction following myocardial infarction by targeting the ampk/mtor signalling pathway. Br J Pharmacol. 2019 doi: 10.1111/bph.14706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang G., Balamotis M.A., Stevens J.L., Yamaguchi Y., Handa H., Berk A.J. Mediator requirement for both recruitment and postrecruitment steps in transcription initiation. Mol Cell. 2005;17:683–694. doi: 10.1016/j.molcel.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 35.Federspiel J.D., Greco T.M., Lum K.K., Cristea I.M. Hdac4 interactions in huntington's disease viewed through the prism of multiomics. Mol Cell Proteomics. 2019;18:S92–s113. doi: 10.1074/mcp.RA118.001253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eom G.H., Nam Y.S., Oh J.G., Choe N., Min H.-K., Yoo E.-K., et al. Regulation of acetylation of histone deacetylase 2 by p300/cbp-associated factor/histone deacetylase 5 in the development of cardiac hypertrophy. Circ Res. 2014;114(7):1133–1143. doi: 10.1161/CIRCRESAHA.114.303429. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and analyzed supporting the findings of this study are available in this paper or the Supplementary Materials. Any other raw data generated or analyzed in this study can be obtained upon reasonable request from the corresponding authors.