Abstract
The Hickeliinae (Poaceae: Bambusoideae) is an ecologically and economically significant subtribe of tropical bamboos restricted to Madagascar, Comoros, Reunion Island, and a small part of continental Africa (Tanzania). Because these bamboos rarely flower, field identification is challenging, and inferring the evolutionary history of Hickeliinae from herbarium specimens is even more so. Molecular phylogenetic work is critical to understanding this group of bamboos. Here, comparative analysis of 22 newly sequenced plastid genomes showed that members of all genera of Hickeliinae share evolutionarily conserved plastome structures. We also determined that Hickeliinae plastome sequences are informative for phylogenetic reconstructions. Phylogenetic analysis showed that all genera of Hickeliinae are monophyletic, except for Nastus, which is paraphyletic and forms two distant clades. The type species of Nastus (Clade II) is endemic to Reunion Island and is not closely related to other sampled species of Nastus endemic to Madagascar (Clade VI). Clade VI (Malagasy Nastus) is sister to the Sokinochloa + Hitchcockella clade (Clade V), and both clades have a clumping habit with short-necked pachymorph rhizomes. The monotypic Decaryochloa is remarkable in having the longest floret in Bambuseae and forms a distinct Clade IV. Clade III, which has the highest generic diversity, consists of Cathariostachys, Perrierbambus, Sirochloa, and Valiha, which are also morphologically diverse. This work provides significant resources for further genetic and phylogenomic studies of Hickeliinae, an understudied subtribe of bamboo.
Keywords: Bamboo, Hickeliinae, Madagascar, Paleotropical, Phylogenomics, Plastome
Highlights
-
•
Hickeliinae is a Bambusoideae subtribe endemic to Madagascar, Comoros, La Reunion and Tanzania.
-
•
Our study provides a comprehensive understanding of Hickeliinae phylogeny at the generic level based on plastome data.
-
•
Molecular phylogeny indicates that all genera of Hickeliinae are monophyletic except for Nastus Juss.
1. Introduction
The Bambusoideae (bamboos) is one of 12 currently recognized subfamilies of the Poaceae (grasses) unambiguously supported by recent analyses (Bouchenak-Khelladi et al., 2008; GPWG II 2012; Wu and Ge, 2012; Saarela et al., 2018). Bamboos, which naturally occur on all continents except Europe and Antarctica (Li, 1998; Ohrnberger, 1999; Bamboo Phylogeny Group et al., 2012; Soreng et al., 2022), currently total 1642 species in 115 genera (Vorontsova et al., 2016), although these numbers continue to change with new combinations and discoveries (e.g., Haevermans et al., 2020; Tong et al., 2020; Zhang et al., 2020; Ruiz-Sanchez et al., 2021; Ye et al., 2021b). Three tribes, i.e., Arundinarieae (temperate woody bamboos), Bambuseae (tropical woody bamboos) and Olyreae (herbaceous bamboos), are widely recognized. The tribe Bambuseae is further divided into the paleotropical woody bamboos and the neotropical woody bamboos (Sungkaew et al., 2009; Triplett et al., 2014; Kellogg, 2015; Saarela et al., 2018; Guo et al., 2019). The paleotropical woody bamboos have, in turn, been divided into eight subtribes, one of which is named the Hickeliinae A. Camus (Camus, 1924; Dransfield 1994, 1997, 1998, 2000, 2002, 2003, 2016).
All native woody bamboo species of Madagascar fall into two subtribes, the Hickeliinae (Bambuseae) and the Thamnocalaminae (Arundinarieae) (Zhou et al., 2017; Zhang et al., 2017, 2020), except for Schizostachyum perrieri A. Camus (Vorontsova et al., 2016). However, Schizostachyum Nees is a genus of the Melocalanninae distributed in Southeast Asia, extending to New Guinea, Melanesia and Polynesia. The identity of S. perrieri is questionable (Zhou et al., 2022) and probably belongs to Oldeania Stapleton judging from the type specimen deposited at the Muséum National d’Histoire Naturelle. Dransfield (2000) inferred that it may be conspecific with Arundinaria madagascariensis A. Camus which has been transferred into Oldeania (Zhang et al., 2017). After transferring 11 Malesian species of Nastus Juss. to Chloothamnus Buse (subtribe Racemobambosinae Stapleton), Ruhooglandia S. Dransf. & K.M. Wong (subtribal position uncertain) and Widjajachloa K.M. Wong & S. Dransf. (subtribe Racemobambosinae) (Widjaja and Wong, 2016; Wong and Dransfield, 2016), Hickeliinae currently includes 9 genera and 32 species: Cathariostachys S. Dransf. (2 spp.), Decaryochloa A. Camus (1 sp.), Hickelia A. Camus (4 spp.), Hitchcockella A. Camus (1 sp.), Nastus (12 spp.), Perrierbambus A. Camus (2 spp.), Sirochloa S. Dransf. (1 sp.), Sokinochloa S. Dransf. (7 spp.), and Valiha S. Dransf. (2 spp.) (Dransfield, 2016; Vorontsova et al., 2016). The heterogeneity and revision of Nastus s.l. have also been revealed and supported by plastid phylogeny (Chokthaweepanich, 2014; Zhou et al., 2017). All Hickeliinae species, except Nastus borbonicus J. F. Gmel. (Reunion Island) and Hickelia africana S. Dransf. (Tanzania), are endemic to Madagascar, with Sirochloa parvifolia S. Dransf. extending to Comoros. The majority of species in the Hickeliinae are found on the eastern escarpment, where remnants of wet rainforest still exist and a variety of ecological niches are found. Hickelia is found in montane forests (Dransfield, 1994), whereas Sirochloa and Valiha are found in coastal areas at lower elevations (Dransfield, 1998, 2002). Perrierbambus grows in the drier vegetation in the west of Madagascar (Vorontsova et al., 2018). Nastus, the largest genus, is abundant in forested areas and forest margins (Dransfield, 2003). Sokinochloa, which is the most recently described and revised genus, is also found across Madagascar’s forests and their margins (Dransfield, 2016). Madagascar’s fauna, particularly lemurs, and its people depend on these bamboos (Dransfield, 2003; King et al., 2013). Despite the common occurrence of Hickeliinae species and their close relationships with economically significant invasive and edible species (Canavan et al., 2016), the phylogenetic relationships among Hickeliinae genera are poorly understood.
Diagnostic morphological characters of subtribe Hickeliinae include an erect culm in the lower part with a scrambling or climbing upper part, nodes that dip downward with an extravaginal branch, determinate inflorescence (spike or capitate) with one floret in the spikelet, and pachymorph rhizomes (Vorontsova et al., 2018). Woody bamboos are famous for their rare and often unpredictable flowering (Janzen, 1976; Li et al., 2006; Ma et al., 2017), and the Hickeliinae is no exception (Dransfield, 2000). Consequently, collecting flowering material for identification and classification is challenging. Molecular data and plastome sequences, in particular, have provided invaluable information for resolving the phylogenetic relationships of bamboos and grasses (Zhou et al., 2017; Saarela et al., 2018). However, molecular approaches have yet to clarify phylogenetic relationships in Hickeliinae. Although one study used five chloroplast markers to confirm the monophyly of the subtribe, these markers lacked sufficient informative sites to resolve phylogenetic relationships among genera (Hackel et al., 2018). One potential approach to resolving complex evolutionary relationships such as those in the subtribe Hickeliinae is the use of comparative plastome analysis (Shaw et al., 2007; Moore et al., 2010; Park et al., 2017; Li et al., 2021). Plastomes, which have a circular quadripartite structure containing a large single-copy (LSC) region and a small single-copy (SSC) region separated by two inverted repeats (IRs) (Palmer, 1991), show a relatively high degree of conservatism in size, structure, and gene content across land plants (Wicke et al., 2011). In the past ten years, plastid phylogenomic analyses have been used to address evolutionary questions and have been widely applied in the Bambusoideae to resolve relationships at the subfamilial, tribal, and subtribal levels (Zhang et al., 2011; Wu and Ge, 2012; Ma et al., 2014; Wysocki et al., 2015; Ye et al., 2021a).
Here, we used genome skimming to acquire whole plastid genomes of 22 representative species of the subtribe Hickeliinae. Our aim was to (1) determine whether Hickeliinae plastome sequences are structurally similar to those of other Bambusoideae; (2) assess whether plastome sequences are sufficiently informative to resolve evolutionary relationships within this subtribe; (3) confirm whether Hickeliinae genera are monophyletic. We hope to contribute to a valuable phylogenetic framework for future taxonomic and ecological studies of bamboos in Madagascar.
2. Materials and methods
2.1. Plant material and taxon sampling
We sampled 22 species, covering all genera and about 69% of the known species of Hickeliinae. All species sampled here are endemic to Malagasy except Nastus borbonicus (from Reunion Island) and Hickelia africana (from Tanzania). The samples included in this study were collected during targeted field trips in Madagascar in 2017 and 2018. Fresh leaves and other plant materials were collected from the eastern humid forest (Andasibe and Maromizaha) and northeastern mountains (Marojejy) to the western drier areas (Ambanja) of Madagascar. These localities were chosen because of previous collections of the Hickeliinae and their rich biodiversity. During the field survey, we collected flowering material of three bamboo species: Hickelia perrieri (A. Camus) S. Dransf., Nastus tsaratananensis A. Camus and Nastus perrieri A. Camus. This material represents the first modern collection of native flowering bamboos from the montane forest bamboo diversity hotpot on Madagascar’s highest mountain, Tsaratanana. Fresh leaves were silica-gel-dried and stored for DNA extraction. Voucher specimens were deposited at herbaria in Parc Botanique et Zoologique de Tsimbazaza (TAN), Kunming Institute of Botany, Chinese Academy of Sciences (KUN), Royal Botanic Gardens, Kew (K), and Muséum National d'Histoire Naturelle (P). Some experimental materials were sampled from specimens deposited at K and P. Data on collection provenance and voucher specimens are shown in Table 1.
Table 1.
Voucher information and provenance of species sampled in this study. An additional reference sequence originally assembled by Wysocki et al. (2015) was also included: KJ870994, based on Dransfield 1349 collected from Ambatofitorahana in Madagascar.
| Species | Country of origin | Silica gel or herbarium specimens used (S/H) | Voucher | Accession number |
|---|---|---|---|---|
| Hickelia madagascariensis (reference) | Madagascar | H | S. Dransfield 1349 | KJ870994 |
| Cathariostachys capitata | Madagascar | H | S. Dransfield 1064 | GWHBHNA01000000 |
| C. madagascariensis | Madagascar | S | R. A. Rakotonasolo 015 | GWHBHMZ01000000 |
| Decaryochloa diadelpha | Madagascar | S | R. A. Rakotonasolo 016 | GWHBHMY01000000 |
| Hickelia africana | Tanzania | H | S. Bidgood 221 | GWHBHMX01000000 |
| H. madagascariensis | Madagascar | H | S. Dransfield 1290 | GWHBHMW01000000 |
| H. perrieri | Madagascar | H | R. A. Rakotonasolo 071 | GWHBHMV01000000 |
| Hitchcockella baronii | Madagascar | H | D. Ravelonarivo & T. Augustin 3430 | GWHBHMU01000000 |
| Nastus aristatus | Madagascar | S | R. A. Rakotonasolo 022 | GWHBHMT01000000 |
| N. borbonicus | Reunion Island | S | T. Hubert s.n. | GWHBHMS01000000 |
| N. elongatus | Madagascar | S | R. A. Rakotonasolo 024 | GWHBHMR01000000 |
| N. emirnensis | Madagascar | H | S. Dransfield 1119 | GWHBHMQ01000000 |
| N. humbertianus | Madagascar | H | H. Humbert 13668 | GWHBHMP01000000 |
| N. perrieri | Madagascar | S | R. A. Rakotonasolo 032 | GWHBHMO01000000 |
| N. tsaratananensis | Madagascar | S | H. Perrier 16170 | GWHBHMN01000000 |
| Perrierbambus madagascariensis | Madagascar | H | L. Nusbaumer 1954 | GWHBHMM01000000 |
| Sirochloa parvifolia | Madagascar | S | M. S. Vorontsova 1769 | GWHBHML01000000 |
| Sokinochloa australis | Madagascar | H | S. Dransfield 1506 | GWHBHMK01000000 |
| S. brachyclada | Madagascar | S | R. A. Rakotonasolo 028 | GWHBHMJ01000000 |
| S. chapelieri | Madagascar | H | D. K. Edelman 152 | GWHBHMI01000000 |
| S. chiataniae | Madagascar | H | Chia Tan 099 | GWHBHMH01000000 |
| S. viguieri | Madagascar | H | J. Dransfield 7749 | GWHBHMG01000000 |
| Valiha diffusa | Madagascar | H | R. A. Rakotonasolo 014 | GWHBHMF01000000 |
2.2. DNA extraction and sequencing
Genomic DNA was extracted from leaf tissue and isolated using the TIANGEN DNAsecure Plant Kit following the manufacturer’s instructions, or from herbarium specimens following Zeng et al. (2018). The Illumina Hiseq platform was used to sequence paired-end reads. Library preparation and sequencing were carried out at the Laboratory of Molecular Biology, the Germplasm Bank of Wild Species (Kunming, China) and BGI Genomics Co., Ltd. (Shenzhen, China).
2.3. Plastid genome assembly and annotation
Plastid genomes were assembled using the GetOrganelle toolkit (Jin et al., 2020). We used Bandage v.8.0 (Wick et al., 2015) to visualize newly assembled plastome structure. Hickelia madagascariensis A. Camus was retrieved from GenBank (accession number: KJ_870994) and used as the reference plastome (Wysocki et al., 2015). Annotation was conducted in Geneious v.9.1.4 using MAFFT (Kearse et al., 2012) to align plastome sequences; annotations were transferred from the reference plastome. MAFFT was also used to align all plastomes. Annotations and alignments were checked manually. Gene maps of circular complete plastomes were drawn using OGDRAW (Lohse et al., 2013). Geneious v.9.1.4 was used to inspect the boundaries between LSC, IRa, SSC, IRb and the GC content for each species. Whole plastid sequence data have been deposited in the Genome Warehouse in National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences/China National Center for Bioinformation, which are publicly accessible at https://ngdc.cncb.ac.cn/gwh.
2.4. Comparative genomic and phylogenetic analysis
Using Hickelia madagascariensis (KJ_870994) as a reference, we compared the plastomes of the species sampled in this study. The assembled plastomes of Hickeliinae were aligned and plotted using the mVista program with LAGAN mode (Frazer et al., 2004). The rearrangement analysis of all plastomes was performed using Mauve alignment with default values for all parameters (Darling et al., 2004). The borders between LSC, IRa, SSC and IRb were investigated using IRscope (Amiryousefi et al., 2018).
To determine the phylogenetic relationships of the species sampled and test the support for the relationships within the subtribe, maximum likelihood (ML) was used to reconstruct a phylogenetic tree using whole plastome sequences of all sampled species in RAxML v.8.2.10 with the GTR + G + I model and 1000 rapid bootstrap replicates (Stamatakis, 2014). Bayesian inference (BI) was carried out in MrBayes v.3.2.6 (Ronquist et al., 2012). Markov chain Monte Carlo (MCMC) chains were run for 2 × 10,000,000 generations. Both analyses were performed on CIPRES Science Gateway (Miller et al., 2010). To root the tree, three species of Bambusa Schreb. were retrieved from GenBank and used as outgroup taxa (B. teres Buch.-Ham. ex Munro: NC_050751.1, B. oliveriana Gamble: MK679792.1 and B. vulgaris Schrad. ex J.C.Wendl: NC_050780.1).
3. Results
3.1. Plastome organizations and features in the subtribe Hickeliinae
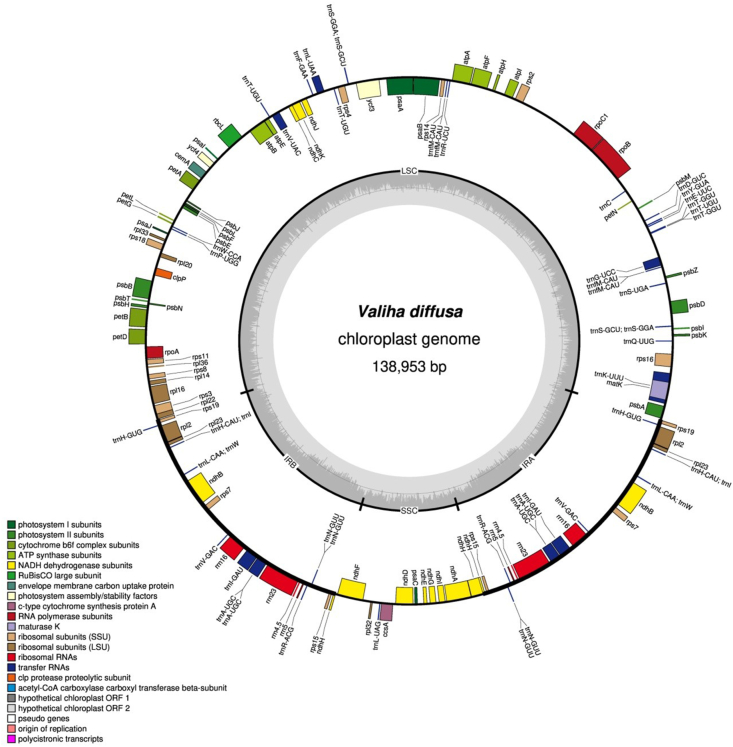
The plastomes of all species showed a typical quadripartite structure (Fig. 1). Data on plastome size, length, and GC content of LSC, SSC and IR are shown in Table 2. All plastomes shared a common structure, consisting of two IR (21,773–21,804 bp) regions separated by an LSC (81,925–83,018 bp) region and an SSC (12,485–12,863 bp) region. The SSC region had the lowest GC content (33.1%–33.4%) followed by the LSC region (36.9%–37%), while that of the IR regions (44.2%) was constant and the highest. All plastomes contained the same 128 genes. A total of 110 unique genes were found, and 18 genes were duplicated in the IR regions (Table 3). The 110 unique genes comprise 77 protein-coding genes, 29 tRNA genes and 4 rRNA genes.
Fig. 1.
Plastome map of Valiha diffusa. Genes inside the circle are transcribed clockwise; genes outside the circle are transcribed counter-clockwise. The light gray inner circle corresponds to AT content, and the dark gray to GC content. Genes belonging to different functional groups are shown in different colors.
Table 2.
Structure of the plastomes analyzed in this study.
| Taxon name | Plastome genome size (bp) | LSC length (bp) | GC content of LSC (%) | IR length (bp) | GC content of IR (%) | SSC length (bp) | GC content of SSC (%) |
|---|---|---|---|---|---|---|---|
| Hickelia madagascariensis (reference) | 138,276 | 81,925 | 37 | 21,804 | 44.2 | 12,743 | 33.1 |
| Cathariostachys capitata | 138,606 | 82,550 | 36.9 | 21,782 | 44.2 | 12,492 | 33.3 |
| C. madagascariensis | 138,462 | 82,425 | 37 | 21,776 | 44.2 | 12,485 | 33.3 |
| Decaryochloa diadelpha | 139,073 | 82,762 | 37 | 21,782 | 44.2 | 12,747 | 33.2 |
| Hickelia africana | 138,290 | 81,966 | 37 | 21,790 | 44.2 | 12,744 | 33.1 |
| H. madagascariensis | 138,284 | 81,936 | 37 | 21,803 | 44.2 | 12,742 | 33.1 |
| H. perrieri | 138,250 | 81,941 | 37 | 21,788 | 44.2 | 12,733 | 33.2 |
| Hitchcockella baronii | 139,167 | 82,779 | 36.9 | 21,786 | 44.2 | 12,616 | 33.1 |
| Nastus aristatus | 139,380 | 82,961 | 37 | 21,778 | 44.2 | 12,863 | 33.1 |
| N. borbonicus | 138,953 | 82,619 | 36.9 | 21,787 | 44.2 | 12,760 | 33.2 |
| N. elongatus | 139,287 | 82,973 | 37 | 21,775 | 44.2 | 12,764 | 33.2 |
| N. emirnensis | 139,291 | 82,962 | 37 | 21,774 | 44.2 | 12,781 | 33.2 |
| N. humbertianus | 139,291 | 82,962 | 37 | 21,774 | 44.2 | 12,781 | 33.2 |
| N. perrieri | 139,328 | 82,946 | 37 | 21,776 | 44.2 | 12,830 | 33.3 |
| N. tsaratananensis | 139,272 | 82,950 | 37 | 21,774 | 44.2 | 12,774 | 33.3 |
| Perrierbambus madagascariensis | 139,134 | 82,827 | 36.9 | 21,786 | 44.2 | 12,735 | 33.1 |
| Sirochloa parvifolia | 139,346 | 83,018 | 36.9 | 21,782 | 44.2 | 12,764 | 33.1 |
| Sokinochloa australis | 138,258 | 81,958 | 37 | 21,773 | 44.2 | 12,754 | 33.3 |
| S. brachyclada | 138,286 | 82,001 | 37 | 21,779 | 44.2 | 12,727 | 33.3 |
| S. chapelieri | 139,149 | 82,847 | 37 | 21,782 | 44.2 | 12,738 | 33.2 |
| S. chiataniae | 139,045 | 82,749 | 37 | 21,781 | 44.2 | 12,734 | 33.3 |
| S. viguieri | 139,147 | 82,801 | 37 | 21,782 | 44.2 | 12,782 | 33.2 |
| Valiha diffusa | 138,937 | 82,862 | 37 | 21,775 | 44.2 | 12,525 | 33.4 |
Table 3.
Gene content in the Hickeliinae plastomes.
| Category | Group of gene | Name of gene | |||
|---|---|---|---|---|---|
| Self-replication | Ribosomal RNA genes | rrn4.5a | rrn5a | rrn16a | rrn23a |
| Transfer RNA genes | trnA-UGCa | trnC-GCA | trnD-GUC | trnE-UUC | |
| trnF-GAA | trnfM-CAU∗ | trnG-UCC | trnH-CAU | ||
| trnH-GUGa | trnI-GAUa | trnK-UUU | trnL-CAA | ||
| trnL-UAA | trnL-UAG | trnN-GUUa | trnP-UGG | ||
| trnQ-UUG | trnR-ACGa | trnR-UCU | trnS-GCU | ||
| trnS-GGA | trnS-UGA | trnT-GGU | trnT-UGU | ||
| trnV-GACa | trnV-UAC | trnW-CCA | trnY-GUA | ||
| Small subunit of ribosome | rps2 | rps3 | rps4 | rps7a | |
| rps8 | rps11 | rps12a,b | rps14 | ||
| rps15a | rps16 | rps18 | rps19a | ||
| Large subunit of ribosome | rpl2a | rpl14 | rpl16 | rpl20 | |
| rpl22 | rpl23a | rpl32 | rpl33 | ||
| rpl36 | |||||
| RNA polymerase subunit | rpoA | rpoB | rpoC1 | rpoC2 | |
| Photosynthesis | Subunits of photosystem I | psaA | psaB | psaC | psaI |
| psaJ | |||||
| Subunits of photosystem II | psbA | psbB | psbC | psbD | |
| psbE | psbF | psbH | psbI | ||
| psbJ | psbK | psbL | psbM | ||
| psbN | psbT | psbZ | |||
| Subunits of cytochrome | petA | petB | petD | petG | |
| petL | petN | ||||
| Subunits of ATP synthetase | atpA | atpB | atpE | atpF | |
| atpH | atpI | ||||
| Large subunit of rubisco | rbcL | ||||
| Subunits of NADH | ndhA | ndhBa | ndhC | ndhD | |
| ndhE | ndhF | ndhG | ndhHa | ||
| ndhI | ndhJ | ndhK | |||
| Other gene | Translation initiation factor | infA | |||
| Maturase | matK | ||||
| Envelope membrane protein | cemA | ||||
| C-type cytochrome synthesis gene | ccsA | ||||
| Protease | clpP | ||||
| Unknown function | Conserved open reading frames | ycf3 | ycf4 | ||
Two gene copies in IRs.
Gene divided into two independent transcription units.
Gene containing one intron.
3.2. Structural variations of the plastome
The structure of all plastomes in this study was relatively conserved, except for in the trnG-trnT region (Fig. S1). Plastome size ranged from 138,250 bp (Hickelia perrieri) to 139,380 bp (Nastus aristatus A. Camus). Although gene organization did not show evidence of rearrangement from the examination with Mauve (Fig. S2), the length of three plastome regions varied between species. In addition, the LSC and SSC regions were less conserved than the IR region. We also noted variations at the four boundaries between the single-copy region and the IR region shown by the results of IRscope (Fig. S3). ndhH was the only gene that spans the junction of the single-copy region and IR region in all species. ndhF exclusively spans the JSA junction of Sokinochloa viguieri (A. Camus) S. Dransf. and Nastus aristatus.
3.3. Plastid phylogenomic analyses
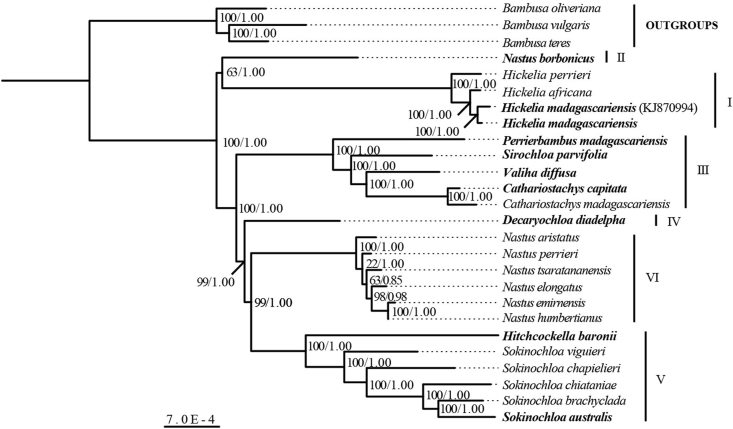
Our analyses showed that the subtribe Hickeliinae consists of six clades. The topologies reconstructed by BI and ML analyses were largely congruent (Fig. 2). The topologies from the BI and ML analyses were either the same, or ML analyses yielded less resolved or less supported trees than BI analysis. Analyses of complete plastome data resolved the phylogenetic relationships within Hickeliinae with high statistical support on most nodes (MLBS >90, PP = 1.00). In our tree, Nastus borbonicus from Reunion Island is sister to Hickelia with relatively low bootstrap support. The tree is compatible with a basal trichotomy for the three main clades: Clade II/Clade I/and Clade III + IV + V + VI.
Fig. 2.
ML tree based on whole plastome sequences using three species of Bambusa as outgroup taxa. Numbers near nodes indicate maximum likelihood bootstrap (MLBS)/Bayesian inference posterior probability (PP). Species names in bold are types of genera.
Major clades are well supported with strong statistical support (Fig. 2). Clade I consists of all sampled species of Hickelia (H. africana, H. perrieri and H. madagascariensis). Clade II contains only one species, Nastus borbonicus, which is endemic to Reunion Island. Clade III consists of five species from three oligotypic genera and one monotypic genus, i.e., Perrierbambus madagascariensis A. Camus, Sirochloa parvifolia, Valiha diffusa S. Dransf., Cathariostachys madagascariensis (A. Camus) S. Dransf. and Cathariostachys capitata (Kunth) S. Dransf. Clade IV consists of the monotypic genus Decaryochloa. Clade V consists of Sokinochloa and the monotypic Hitchcockella. Clade VI consists of Malagasy species of Nastus (N. aristatus, N. elongatus A. Camus, N. emirnensis A. Camus, N. perrieri, N. humbertianus A. Camus and N. tsaratananensis). All sampled Malagasy species of Nastus (Clade VI) are in another clade of the Hickeliinae with strong bootstrap support, whereas Nastus borbonicus from Reunion Island forms a basal clade of Hickeliinae sister to Hickelia with high posterior probability and low bootstrap support.
4. Discussion
The accessibility of plant plastome sequences has increased swiftly over the last decade. However, plastome data from species of Hickeliinae has remained scarce. We present here the most comprehensively sampled data set of plastid genomes and an explicit phylogeny for the neglected bamboo subtribe Hickeliinae. In total, 22 plastome sequences were newly generated covering all recognized genera of Hickeliinae.
4.1. Plastome structure is conserved in Hickeliinae and similar to that of other Bambusoideae subtribes
The structure and length of all the plastomes analyzed in our study are conserved. The plastome structures, gene contents and gene orders of Hickeliinae are consistent with that of other bamboo species (Zhang et al., 2011; Ma et al., 2014; Wysocki et al., 2015). These similarities indicate that the plastome structure of Hickeliinae is largely conserved following patterns across the Bambusoideae.
4.2. Monophyly of the Hickeliinae and phylogenetic relationships among main clades
Our phylogenetic analysis based on plastome data confirmed that, except for Nastus, all currently-recognized genera of Hickeliinae are monophyletic. These genera had previously been delimited based on morphology. In our phylogenetic tree, Clade II consists of Nastus borbonicus, which is the type species and only species of Nastus distributed outside Madagascar. Morphologically, Nastus borbonicus is characterized by its erect culms and pachymorph rhizomes with long necks (Dransfield, 2000; Bosser and Renvoize, 2018).
Clade I consists of species of Hickelia, which was previously suggested as an early diverged genus in Hickeliinae (Chokthaweepanich, 2014; Zhou et al., 2017; Hackel et al., 2018). Hickelia is found in Malagasy dry highland forests with one species in wet montane forests of Tanzania (Dransfield, 1994). The culm of it is erect at the base and then scrambles over nearby vegetation at the upper part (Dransfield, 1994). Based on the order of divergence in Clade II, Hickelia is most likely native to Madagascar and subsequently dispersed to Africa.
Clade III is highly heterogeneous, including four genera (i.e., Cathariostachys, Perrierbambus, Sirochloa, and Valiha) of plants characterized by having culms that are erect on the lower part. Plants of these four genera grow at low to mid-elevations, distributed from montane forests to relatively drier coastal forests. Sirochloa has an open clumping habit, whereas the other three genera grow scattered. The culms of Perrierbambus and Sirochloa are solid, whereas those of the other two are hollow. The inflorescence types are capitate, paniculate, spicate, and racemose in Cathariostachys, Perrierbambus, Sirochloa, and Valiha, respectively (Dransfield, 1998, 2000, 2002, 2003).
Members of Clades IV–VI are wet forest climbers that grow in mid-elevation habitats (Dransfield, 1997, 2016; Vorontsova et al., 2018). The monotypic genus Decaryochloa (Clade IV), which is remarkable in having the longest floret in tropical woody bamboos (Bambuseae), is sister to Clades V and VI, the most speciose clades. The climbing bamboos of Malagasy Nastus (Clade VI) and Sokinochloa (Clade V) have the highest species diversity in the subtribe (Dransfield, 2016). Species in these two clades are similar in having pachymorph rhizomes with short necks that lead to their clumping habit. However, the capitate inflorescence of Sokinochloa is significantly different from that of the other two genera, which are paniculate or racemose. The wet forest climber Hitchcokella is one of the rarest and most enigmatic members of the Hickeliinae and is much in need of further study (Vorontsova et al., 2018).
Our work shows that plastome sequences are sufficiently informative to resolve the phylogeny of Hickeliinae. In addition, our Hickeliinae phylogeny is the most explicit and robust framework to date for this bamboo subtribe. Our phylogeny updates the most recent study involving Hickeliinae, which included a sampling of 15 species from all genera of Hickeliinae and used 5 plastid markers to infer phylogeny (Hackel et al., 2018). Our analyses shared two samples with theirs: One is the plastome sequence of Hickelia madagascariensis (Dransfield 1349 from Ambatofitorahana in Madagascar) previously sequenced by Wysocki et al. (2015) and used here as the reference sequence. Another is a herbarium sample of the rare species Hitchcockella baronii A. Camus (Ravelonarivo and Augustin 3430 from Marojejy in Madagascar, http://legacy.tropicos.org/Specimen/100323444), which Hackel et al. (2018) received as a silica specimen from the Missouri Botanical Garden DNA Bank. Because the previous study focused on a larger scale (i.e., grasses in Madagascar) and only several markers were used, posterior probabilities for most inner nodes of Hickeliinae were considerably low and prevent further discussion.
4.3. Paraphyly of the genus Nastus
The genus Nastus was originally published by Jussieu (1789) based on Nastus borbonicus as the type species. It is distributed in Madagascar with only the type species on Reunion Island. However, the result of plastid phylogenomic analysis resolved Nastus as polyphyletic and further confirmed the results of Zhou et al. (2017). The central point of incongruence in Nastus with Hackel et al. (2018) is in the placement of Nastus borbonicus: Hackel et al. (2018) analyzed ndhF and rps16 originally sequenced by Kelchner and Bamboo Phylogeny Group (2013), which is sister to three Malagasy Nastus but without statistical support. Our phylogeny rejects the current delimitation of Nastus and strongly suggests that it may now encompass two distinct genera. In addition to their allopatric nature, species currently treated in Nastus from Madagascar can be distinguished from Nastus borbonicus by a few morphological characteristics. All Nastus species from Madagascar, described by A. Camus (1925–1953), have determinate inflorescences with one-floret spikelets, which match the inflorescence structure of Nastus borbonicus from Reunion Island. The spikelet consists of five transitional glumes, one floret, and a rachilla extension usually with rudimentary floret. However, they differ from Nastus borbonicus in having slender culms that are erect with dropping tips (N. elongatus) or erect at lower parts with scrambling upper parts. Moreover, the species from Madagascar have a single branch bud at each node, which is borne below the supranodal ridge where the sheath scar curves downward, whereas Nastus borbonicus has multiple branch buds at each node which are borne above the sheath scar which is not curved. Malagasy Nastus species have a clumping habit with commonly short necks in the rhizome. However, the rhizomes have long necks in Nastus borbonicus, so that it does not form a clump. The fact that Jussieu based the genus on Nastus borbonicus, recovered here as its own lineage, suggests that other species of Nastus will need to be accommodated in a different, probably new genus. Further phylogenetic studies with broader sampling across its distribution range and evidence from nuclear genes are needed to clarify relationships between Malagasy species of Nastus and the type species, Nastus borbonicus.
5. Conclusion
This study shows that the plastome structure of Hickeliinae is largely conserved, with characteristics that are similar to those of other Bambusoideae species. Our plastid phylogenomic analysis provides a robust framework of Hickeliinae and confirms previous delimitation of genera based on morphological characters, although further phylogenetic analyses based on nuclear genes and morphological characters are still needed. Furthermore, our phylogeny suggests that the currently-defined genus Nastus is paraphyletic and might be divided into two different genera. However, given the reticulate evolutionary history of woody bamboos, this needs to be verified by nuclear data (Guo et al., 2019, 2021). We, therefore, recommend a taxonomic review of the genus Nastus on the basis of comprehensive phylogenomic study. Our work has demonstrated that comparative plastome analysis provides insight into the phylogeny of Hickeliinae and using complete plastomes can resolve the complex phylogenetic relationships. Our work also highlights the need for an evaluation of the conservation status of each species of the Malagasy Hickeliinae across all distributions and in various ecological niches, which is indispensable for protecting and sustainable utilizing Malagasy bamboos.
Author contributions
RAR conducted field survey and phylogenomic analysis, and drafted the manuscript; TH, MSV, HR and SD assisted in field sampling and herbarium work; MSV, TH and SD provided comments and revised the draft; MYZ and DZL conceived the study and wrote the paper. All authors contributed to the paper and gave final approval.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank the curators of the herbarium of the Royal Botanic Gardens, Kew (Richmond, UK), the staff of the Kew Madagascar Conservation Center (Antananarivo), curators of the Muséum National d'Histoire Naturelle (Paris), and staff of the Parc Botanique et Zoologique de Tsimbazaza (Antananarivo) for their help in fieldwork and all Malagasy authorities who granted the research permits. The authors sincerely thank the Germplasm Bank of Wild Species for bioinformatics and DNA analysis. This study was supported by the National Natural Science Foundation of China (31670396 and 32120103003), the Program of Science and Technology Talents Training of Yunnan Province, China (2017HA014), and the Large-scale Scientific Facilities of the CAS (2017–LSFGBOWS–02). Special thanks to the University of Chinese Academy of Sciences for the fellowship (CAS-TWAS 2016) to the first author, who worked on this study as part of his doctoral studies.
Footnotes
Peer review under responsibility of Editorial Office of Plant Diversity.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2022.06.001.
Contributor Information
Meng-Yuan Zhou, Email: zhoumengyuan@mail.kib.ac.cn.
De-Zhu Li, Email: dzl@mail.kib.ac.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Multimedia component 1
References
- Amiryousefi A., Hyovonen J., Poczai P. IRscope: an online program to visualise the junction sites of chloroplast genomes. Bioinformatics. 2018;34:3030–3031. doi: 10.1093/bioinformatics/bty220. [DOI] [PubMed] [Google Scholar]
- Bamboo Phylogeny Group (BPG) In: Proc. of the 9th World Bamboo Congress. Gielis J., Potters G., editors. The World Bamboo Organization; Antwerp, Belgium: 2012. An updated tribal and subtribal classification for the Bambusoideae (Poaceae) pp. 3–27. [Google Scholar]
- Bosser J., Renvoize S.A. In: Flore des Mascareignes. La Réunion, Maurice, Rodrigues. 203: Graminées. Autrey J.C., Bosser J., Ferguson I.K., editors. IRD éditions; Marseille, France: 2018. pp. 16–18. [Google Scholar]
- Bouchenak-Khelladi Y., Salamin N., Savolainen V., et al. Large multi-gene phylogenetic trees of the grasses (Poaceae): progress towards complete tribal and generic level sampling. Mol. Phylogenet. Evol. 2008;47:488–505. doi: 10.1016/j.ympev.2008.01.035. [DOI] [PubMed] [Google Scholar]
- Camus A. Genres nouveaux de Bambusées malgaches. Compt. Rend. Hebd. Séances Acad. Sci. 1924;179:478–480. [Google Scholar]
- Canavan S., Richardson D.M., Visser V., et al. The global distribution of bamboos: assessing correlates of introduction and invasion. AoB Plants. 2016;9:plw078. doi: 10.1093/aobpla/plw078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chokthaweepanich H. Dissertation, Iowa State University; Ames, Iowa, U.S.A: 2014. Phylogenetics and Evolution of the Paleotropical Woody Bamboos (Poaceae: Bambusoideae: Bambuseae) [Google Scholar]
- Darling A.C., Mau B., Blattner F.R., et al. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dransfield S. The genus Hickelia (Gramineae–Bambusoideae) Kew Bull. 1994;49:429–443. [Google Scholar]
- Dransfield S. Notes on the genus Decaryochloa (Gramineae–Bambusoideae) from Madagascar. Kew Bull. 1997;52:593–600. [Google Scholar]
- Dransfield S. Valiha and Cathariostachys, two new bamboo genera (Gramineae–Bambusoideae) from Madagascar. Kew Bull. 1998;53:375–397. [Google Scholar]
- Dransfield S. In: Grasses Systematics and Evolution. Jacobs S.W.L., Everett J., editors. CSIRO Publishing; Melbourne: 2000. Woody bamboos (Gramineae–Bambusoideae) of Madagascar; pp. 43–50. [Google Scholar]
- Dransfield S. Sirochloa, a new bamboo genus from Madagascar (Poaceae–Bambusoideae) Kew Bull. 2002;57:963–970. [Google Scholar]
- Dransfield S. In: The Natural History of Madagascar. Goodman S.M., Benstead J.P., editors. University of Chicago Press; Chicago: 2003. Poaceae, Bambuseae, bamboos; pp. 467–471. [Google Scholar]
- Dransfield S. Sokinochloa, a new bamboo genus (Poaceae-Bambusoideae) from Madagascar. Kew Bull. 2016;71:40. [Google Scholar]
- Frazer K.A., Pachter L., Poliakov A., et al. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GPWG II (Grass Phylogeny Working Group II) New grass phylogeny resolves deep evolutionary relationships and discovers C4 origins. New Phytol. 2012;193:304–312. doi: 10.1111/j.1469-8137.2011.03972.x. [DOI] [PubMed] [Google Scholar]
- Guo Z.H., Ma P.F., Yang G.Q., et al. Genome sequences provide insights into the reticulate origin and unique traits of woody bamboos. Mol. Plant. 2019;12:1353–1365. doi: 10.1016/j.molp.2019.05.009. [DOI] [PubMed] [Google Scholar]
- Guo C., Ma P.F., Yang G.Q., et al. Parallel ddRAD and genome skimming analyses reveal a radiative and reticulate evolutionary history of the temperate bamboos. Syst. Biol. 2021;70:756–773. doi: 10.1093/sysbio/syaa076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackel J., Vorontsova M.S., Nanjarisoa O.P., et al. Grass diversification in Madagascar: in situ radiation of two large C3 shades clades and support for a Miocene to Pliocene origin of C4 grassy biomes. J. Biogeogr. 2018;45:750–761. [Google Scholar]
- Haevermans T., Mantuano D., Zhou M.Y., et al. Discovery of the first succulent bamboo (Poaceae, Bambusoideae) in a new genus from Laos’ karst areas, with a unique adaptation to seasonal drought. PhytoKeys. 2020;156:125–137. doi: 10.3897/phytokeys.156.51636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janzen D.H. Why bamboos wait so long to flower. Annu. Rev. Ecol. Syst. 1976;7:374–391. [Google Scholar]
- Jin J.J., Yu W.B., Yang J.B., et al. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020;21 doi: 10.1186/s13059-020-02154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jussieu A.H.L. Viduam Herssant and Theophilum Barrois; Paris, France: 1789. Genera Plantarum. [Google Scholar]
- Kearse M., Moir R., Wilson A., et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelchner S.A., Bamboo Phylogeny Group Higher level phylogenetic relationships within the bamboos (Poaceae: Bambusoideae) based on five plastid markers. Mol. Phylogenet. Evol. 2013;67:404–413. doi: 10.1016/j.ympev.2013.02.005. [DOI] [PubMed] [Google Scholar]
- Kellogg E.A. In: Kubitzki K., editor. vol. 13. Springer; Heidelberg, Germany: 2015. Flowering plants. Monocots: Poaceae. (The Families and Genera of Vascular Plants Series). [Google Scholar]
- King T., Randrianarimanana H.L., Rakotonirina L.H., et al. Large-culmed bamboos in Madagascar: distribution and field identification of the primary food sources of the critically endangered greater bamboo lemur Prolemur simus. Primate Conserv. 2013;27:33–53. [Google Scholar]
- Li D.Z. In: Bamboo – Conservation, Diversity, Ecogeography, Germplasm Resource Utilization and Taxonomy: Proceedings of Training Course Cum Workshop, 10–17 May 1998, Kunming and Xishuangbanna. Rao A.N., Ramanatha Rao V., editors. IPGRI-APO; Yunnan, China. Serdang, Malaysia: 1998. Taxonomy and biogeography of the Bambuseae (Gramineae:Bambusoideae) pp. 235–247. [Google Scholar]
- Li D.Z., Wang Z.P., Zhu Z.D., et al. In: Flora of China. Wu Z.Y., Raven P.H., Hong D.Y., editors. Beijing: Science Press; St. Louis: Missouri Botanical Garden Press; 2006. Bambuseae. [Google Scholar]
- Li H.T., Luo Y., Gan L., et al. Plastid phylogenomic insights into relationships of all flowering plant families. BMC Biology. 2021;19:1–13. doi: 10.1186/s12915-021-01166-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M., Drechsel O., Kahlau S., et al. OrganellarGenomeDRAW-a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 2013;41:W575–W581. doi: 10.1093/nar/gkt289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma P.F., Zhang Y.X., Zeng C.X., et al. Chloroplast phylogenomic analyses resolve deep-level relationships of an intractable bamboo tribe Arundinarieae (Poaceae) Syst. Biol. 2014;63:933–950. doi: 10.1093/sysbio/syu054. [DOI] [PubMed] [Google Scholar]
- Ma P.F., Vorontsova M.S., Nanjarisoa O.P., et al. Negative correlation between rates of molecular evolution and flowering cycles in temperate woody bamboos revealed by plastid phylogenomics. BMC Plant Biol. 2017;17:1–15. doi: 10.1186/s12870-017-1199-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M.A., Pfeiffer W., Schwartz T. Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, Louisiana, 14 Nov 2010. IEEE; Piscataway: 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees; pp. 45–52. [Google Scholar]
- Moore M.J., Soltis P.S., Bell C.D., et al. Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proc. Natl. Acad. Sci. U.S.A. 2010;107:4623–4628. doi: 10.1073/pnas.0907801107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohrnberger D. Elsevier; the Netherlands: 1999. The Bamboos of the World: Annotated Nomenclature and Literature of the Species and the Higher and Lower Taxa. [Google Scholar]
- Palmer J.D. In: The Molecular Biology of Plastids. Bogorad L., Vasil I.K., editors. Academic Press; San Diego: 1991. Plastid chromosomes: structure and evolution; pp. 5–53. [Google Scholar]
- Park I., Kim W.J., Yang S., et al. The complete chloroplast genome sequence of Aconitum coreanum and Aconitum carmichaelii and comparative analysis with other Aconitum species. PLoS One. 2017;12 doi: 10.1371/journal.pone.0184257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F., Teslenko M., van der Mark P., et al. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Sanchez E., Tyrrell C.D., Londono X., et al. Diversity, distribution, and classification of Neotropical woody bamboos (Poaceae: Bambusoideae) in the 21st century. Bot. Sci. 2021;99:198–228. [Google Scholar]
- Saarela J.M., Burke S.V., Wysocki W.P., et al. A 250 plastome phylogeny of the grass family (Poaceae): topological support under different data partitions. PeerJ. 2018;6:e4299. doi: 10.7717/peerj.4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw J., Lickey E.B., Schilling E.E., et al. Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: the tortoise and the hare III. Am. J. Bot. 2007;94:275–288. doi: 10.3732/ajb.94.3.275. [DOI] [PubMed] [Google Scholar]
- Soreng R.J., Peterson P.M., Zuloaga F.O., et al. A worldwide phylogenetic classification of the Poaceae (Gramineae) III: an update. J. Syst. Evol. 2022;60:476–521. [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sungkaew S., Stapleton C., Salamin N., et al. Non-monophyly of the woody bamboos (Bambuseae; Poaceae): a multi-gene region phylogenetic analysis of Bambusoideae. J. Plant Res. 2009;122:95–108. doi: 10.1007/s10265-008-0192-6. [DOI] [PubMed] [Google Scholar]
- Tong Y.H., Zheng X., Zhang Y.Y., et al. Khoonmengia honbaensis, a new genus and species of temperate bamboo (Poaceae, Bambusoideae) from central-southern Vietnam. PhytoKeys. 2020;138:163–177. doi: 10.3897/phytokeys.138.39512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triplett J.K., Clark L.G., Fisher A.E., et al. Independent allopolyploidization events preceded speciation in the temperate and tropical woody bamboos. New Phytol. 2014;204:66–73. doi: 10.1111/nph.12988. [DOI] [PubMed] [Google Scholar]
- Vorontsova M.S., Clark L.G., Dransfield J., et al. World Checklist of Bamboos and Rattans. INBAR; Beijing: 2016. [Google Scholar]
- Vorontsova M.S., Dransfield S., Renvoize S.A., et al. Royal Botanic Gardens; Kew: 2018. Identification Guide to Grasses and Bamboos in Madagascar. [Google Scholar]
- Wick R.R., Schultz M.B., Zobel J., et al. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 2015;31:3350–3352. doi: 10.1093/bioinformatics/btv383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicke S., Schneeweiss G.M., Pamphilis C.W., et al. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol. Biol. 2011;76:273–297. doi: 10.1007/s11103-011-9762-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widjaja E.A., Wong K.M. New combinations in Chloothamnus (Poaceae: Bambusoideae), a genus of malesian bamboos formerly confused with Nastus. Sandakania. 2016;22:37–40. [Google Scholar]
- Wong K.M., Dransfield S. Ruhooglandia and Widjajachloa, two new genera of malesian bamboos (Poaceae: Bambusoideae) and their distinction from Nastus and Chloothamnus. Sandakania. 2016;22:1–9. [Google Scholar]
- Wu Z.Q., Ge S. The phylogeny of the BEP Clade in grasses revisited: evidence from the whole-genome sequences of chloroplasts. Mol. Phylogenet. Evol. 2012;62:573–578. doi: 10.1016/j.ympev.2011.10.019. [DOI] [PubMed] [Google Scholar]
- Wysocki W.P., Clark L.G., Attigala L., et al. Evolution of the bamboos (Bambusoideae: Poaceae): a full plastome phylogenomic analysis. BMC Evol. Biol. 2015;15:50. doi: 10.1186/s12862-015-0321-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X.Y., Ma P.F., Guo C., et al. Phylogenomics of Fargesia and Yushania reveals a history of reticulate evolution. J. Syst. Evol. 2021;59:1183–1197. [Google Scholar]
- Ye X.Y., Zhang Y.X., Li D.Z. Two new species of Yushania (Poaceae: Bambusoideae) from South China, with a taxonomic revision of related species. Plant Divers. 2021;43:492–501. doi: 10.1016/j.pld.2021.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C.X., Hollingsworth P.M., Yang J., et al. Genome skimming herbarium specimens for DNA barcoding and phylogenomics. Plant Methods. 2018;14:43. doi: 10.1186/s13007-018-0300-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.J., Ma P.F., Li D.Z. High-throughput sequencing of six bamboo chloroplast genomes: Phylogenetic implications for temperate woody bamboos (Poaceae: Bambusoideae) PLoS One. 2011;6 doi: 10.1371/journal.pone.0020596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.X., Ma P.F., Havermans T., et al. In search of the phylogenetic affinity of the temperate woody bamboos from Madagascar, with description of a new species (Bambusoideae, Poaceae) J. Syst. Evol. 2017;55:453–465. [Google Scholar]
- Zhang Y.X., Guo C., Li D.Z. A new subtribal classification of Arundinarieae (Poaceae, Bambusoideae) with the description of a new genus. Plant Divers. 2020;42:127–134. doi: 10.1016/j.pld.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M.Y., Zhang Y.X., Haevermans T., et al. Towards a complete generic-level plastid phylogeny of the paleotropical woody bamboos (Poaceae: Bambusoideae) Taxon. 2017;66:539–553. [Google Scholar]
- Zhou M.Y., Liu J.X., Ma P.F., et al. Plastid phylogenomics shed lights on intergeneric relationships and spatiotemporal evolutionary history of Melocanninae (Poaceae: Bambusoideae) J. Syst. Evol. 2022;60:640–652. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multimedia component 1