Abstract
Human activities have caused the exchange of species among different parts of the world. When introduced species become naturalized and invasive, they may cause great negative impacts on the environment and human societies, and pose significant threats to biodiversity and ecosystem structure. Knowledge on phylogenetic relatedness between native and non-native species and among non-native species at different stages of species invasion may help for better understanding the drivers of species invasion. Here, I analyze a comprehensive data set including both native and non-native angiosperm species in China to determine phylogenetic relatedness of introduced species across a full invasion continuum (from introduction through naturalization to invasion). This study found that (1) introduced plants are a phylogenetically clustered subset of overall (i.e. native plus non-native) angiosperm flora, (2) naturalized plants are a phylogenetically clustered subset of introduced plants, and (3) invasive plants are a phylogenetically clustered subset of naturalized plants. These patterns hold regardless of spatial scales examined (i.e. national versus provincial scale) and whether basal- or tip-weighted metric of phylogenetic relatedness is considered. These findings are consistent with Darwin's preadaptation hypothesis.
Keywords: Angiosperm, Community assembly, Darwin's preadaptation hypothesis, Exotic species, Introduced species, Phylogenetic dispersion
Highlights
-
•
A comprehensive data set is analyzed to determine phylogenetic relatedness of introduced plants across a full invasion continuum in China.
-
•
Introduced, naturalized, and invasive species are phylogenetically clustered subsets of their respective species pools.
-
•
These patterns hold regardless of spatial scales and evolutionary depths examined.
1. Introduction
Human accidental or intentional transportation of species out of their native geographic ranges occurs across the world (Thuiller et al., 2010). The relocation of organisms has bridged natural dispersal barriers and opened novel opportunities for species to expand into new regions beyond their native ranges (Thuiller et al., 2010; Fristoe et al., 2021). For vascular plants alone, more than 13,000 species have been transported outside their native ranges and have successfully established self-sustaining populations (van Kleunen et al., 2015) without direct human intervention (Richardson et al., 2000; Pyšek et al., 2004; Blackburn et al., 2011). Of these naturalized plant species, some have spread widely and caused great negative impacts on the environment and human societies (Blackburn et al., 2011; Lambertini et al., 2011; Simberloff et al., 2013). These invasive species have posed significant threats to biodiversity and ecosystem structure and functioning (Mack et al., 2000).
For a non-native species to become invasive, it has to progress along a continuum with three stages (i.e. introduction, naturalization, invasion), which is called the introduction−naturalization−invasion (INI) continuum (Blackburn et al., 2011; Richardson and Pyšek, 2012; Divíšek et al., 2018), by passing a number of dispersal barriers and environmental and biotic filters (Richardson et al., 2000; Miller et al., 2017). Understanding the drivers of the success of transitions from one stage to another across the INI continuum has become a major objective in ecology and biogeography (Omer et al., 2022).
Previous studies have shown that the underlying drivers of species invasion can substantially differ between different invasion stages. For example, socio-economic drivers are deemed crucial for introduction and naturalization (Williamson, 2006) while invasiveness is assumed to depend more on species’ traits (Pyšek et al., 2009) and the interaction of non-native species with native species and the physical environment of the recipient region (Essl et al., 2019). Many of the characteristics attributed to successful invasion of plants, such as growth forms, pollination and dispersal types, are strongly associated with particular taxa (Lambdon, 2008). However, for many plant groups, the ecological features that favor invasion success are poorly understood (Gallien et al., 2019). Although key correlates of invasion success have been identified for certain species, measuring relevant functional traits for a large number of species is often impractical (Gallien et al., 2019). Because species with shared ancestry are ecologically more similar to each other than to distantly related species (Cavender-Bares et al., 2009), and previous studies have shown that many functional traits are phylogenetically conserved (Donoghue, 2008; Ackerly, 2009), phylogenies can be used to overcome the problem of missing information on traits.
Phylogenetic relatedness among species could be used as a proxy of similarity in functional traits among the species (Webb, 2000; Zhang et al., 2011; Krishna et al., 2021). Several studies have examined changes in phylogenetic relatedness among species in plant communities as a result of the introduction of non-native plants, but nearly all of these studies focus on only one or two of the three stages of the INI continuum. For example, Lososová et al. (2015), Sandel and Tsirogiannis (2016), Qian et al. (2022a), Qian and Sandel (2022), and Park et al. (2020) have investigated the effect of the introduction of non-native plants on phylogenetic relatedness of plants in the recipient communities, and Zhang et al. (2021) and Qian et al. (2022b) have investigated changes in phylogenetic relatedness in the naturalization−invasion transition of the INI continuum. To my knowledge, Omer et al. (2022) is the only study that has simultaneously investigated all the three stages of the INI continuum. Their study focused on angiosperm species in South Africa, and they concluded that species are more likely to be introduced for cultivation if they are distantly related to the native flora, the probability for cultivated species to become naturalized species is higher for species closely related to the native flora, and the probability for naturalized species to become invasive is higher if they are distantly related to the native flora. These findings suggest that introduced non-native species would be a phylogenetically over-dispersed subset of species with respect to the pool of all species in the recipient community (i.e., including both native and non-native species), naturalized non-native species would be a phylogenetically clustered subset of species with respect to the pool of all non-native species in the recipient community (i.e., including both naturalized and non-naturalized species), and invasive non-native species would be a phylogenetically over-dispersed subset of species with respect to the pool of naturalized non-native species in the recipient community (i.e., including only naturalized species). Whether these apply to other regions across the world needs to be tested.
China is rich not only for its native flora but also for its non-native flora. According to Lin et al. (2022), China has been introduced with over 13,400 species of vascular plants. Here, I investigate phylogenetic relatedness of non-native angiosperm species at each of the three stages across the INI continuum for China as a whole and for those province-level regions that have large numbers of introduced plant species.
2. Materials and methods
China was divided into 28 province-level regions (Fig. S1). Special administrative regions and municipalities were combined with their respective neighboring provinces, as in previous studies (e.g. Qian, 2007; Qian et al., 2022a); specifically, Beijing and Tianjin were combined with Hebei Province, Chongqing with Sichuan Province, Hong Kong, Macau, and Shenzhen with Guangdong Province, and Shanghai with Zhejiang Province. Angiosperm species lists for the regions, and data on species nativity status (i.e. native versus exotic) and invasion stage of non-native species (introduction, naturalization, invasion) were obtained from the Flora of China (http://www.efloras.org/index.aspx; Wu et al., 1994–2013), A Dataset on Catalogue of Alien Plants in China (http://doi.org/10.57760/sciencedb.01711; Lin et al., 2022), and Hao and Ma (2023). Because the main goal of this study is to test the conclusions of Omer et al. (2022), and because non-native species in their study included only angiosperms introduced to Southern Africa for cultivation, non-native species in the present study also included only those that were originally introduced to China for cultivation. However, because 99% of the non-native plant species in China were introduced for cultivation (Lin et al., 2022), the conclusion of this study should hold for all introduced plant species in China. Botanical nomenclature was standardized according to the World Flora Online (WFO, www.worldfloraonline.org), using the package U.Taxonstand (Zhang and Qian, 2023). Infraspecific taxa were combined with their parental species. As a result, a total of 41,949 angiosperm species were included in this study, of which 12,902, 1061, and 182 were introduced, naturalized, and invasive, respectively. The invasive species were a subset of the naturalized species, which were in turn a subset of the introduced species. The number of introduced angiosperm species in a province-level region varied from 185 to 5619. Including regions with a relatively small number of introduced species in an analysis may bias the result of the analysis. Therefore, for regional-level analyses, I only included those regions that had at least 1500 introduced angiosperm species. Consequently, eight province-level regions were included in regional-level analyses (Fig. S1). Non-native species in a particular province-level region do not include those species that are non-native to the region but native to other part of China.
The package V.PhyloMaker2 (build.nodes.1; Jin and Qian, 2019, 2022) was used to generate a phylogeny for the 41,949 angiosperm species, based on an updated and expanded version of the dated megaphylogeny GBOTB reported by Smith and Brown (2018) as a backbone phylogeny (i.e., GBOTB.extended.TPL.tre; Jin and Qian, 2022). All families in my data set were resolved in the megaphylogeny. I added the genera and species in my data set that were absent from the megaphylogeny to their respective families and genera using V.PhyloMaker2 software (Scenario 3; Jin and Qian, 2022), which sets branch lengths of added taxa in a family by placing the nodes evenly between dated nodes and terminals within the family and placing a missing species at the mid-point of the branch length of its genus. I pruned the megaphylogeny to generate a phylogenetic tree retaining only the 41,949 angiosperm species present in China. Qian and Jin (2021) showed that, for the phylogenetic metrics used here, values derived from a fully-resolved species level tree are nearly identical to those from a tree resolved only at genus level. Because the vast majority of the genera and a large number of species in my data set were resolved in the phylogeny, the phylogenetic metrics derived from the phylogeny are expected to be robust (Qian and Jin, 2021). Smaller phylogenetic trees were extracted from the phylogenetic tree for various analyses (see below for details).
Net relatedness index (NRI) and nearest taxon index (NTI) (Webb, 2000; Webb et al., 2002) were used to measure phylogenetic relatedness among species. These two metrics are among the best metrics measuring phylogenetic relatedness and have been commonly used in studies on community phylogenetics, including studies on naturalized species and invasive species (e.g. Zhang et al., 2021; Qian and Sandel, 2022). Positive values of NRI and NTI indicate that species within assemblages are more closely related than expected for a random draw from the species pool. These metrics were calculated using the package PhyloMeasures (Tsirogiannis and Sandel, 2016). NRI represents phylogenetic relatedness based on the average phylogenetic distance of all species in an assemblage whereas NTI quantifies phylogenetic relatedness by incorporating only the distances of the closest relative (Webb et al., 2002; Cadotte et al., 2018). Therefore, using NRI and NTI simultaneously in this study allows exploring patterns of species assembly at both deep and shallow evolutionary histories.
I calculated several sets of NRI and NTI for China as a whole and for each province-level region using tailor-made species pools. First, NRI and NTI for each group of non-native species (introduced, naturalized, invasive) for China as a whole and for each province-level region were calculated. For this set of NRI and NTI, species pool for NRI and NTI of introduced species included all (i.e., native and introduced) species; species pool for NRI and NTI of naturalized species included introduced species only; species pool for NRI and NTI of invasive species included naturalized species only. Second, I calculated NRI and NTI for introduced and native species separately with respect to species pools including both introduced and native species, for naturalized and non-naturalized introduced species separately with respect to species pools including both naturalized and non-naturalized introduced species, and for invasive and non-invasive naturalized species with respect to species pools including both invasive and non-invasive naturalized species. Third, I calculated NRI and NTI for invasive species with respect to the pool of introduced species. Species pools for different plant groups in each province-level region included only respective species (i.e., all, introduced, naturalized) in the province-level region. Fourth, I calculated NRI and NTI for naturalized species in each province-level region with respect to the species pool including all naturalized species in China, and for invasive species in each province-level region with respect to the species pool including all invasive species in China.
An observed value of NRI or NTI was compared to a null expectation generated from 999 random draws of equal species richness (i.e., being equal to the number of introduced, naturalized or invasive species in the raw assemblage under consideration) from the species pool. For normally distributed data, significance at p-value <0.05 is equivalent to NRI or NTI >1.96 (i.e., significantly more phylogenetic clustering), or < −1.96 (i.e., significantly more phylogenetic over-dispersion) (Hortal et al., 2011).
To assess the relationships between phylogenetic relatedness and climatic conditions, I obtained mean annual temperature and annual precipitation climate data from the CHELSA climate database (boi1 and bio12, respectively; https://chelsa-climate.org/bioclim). The mean value of each of the two climate variables was calculated for each province-level region using 30-arc-second resolution data. I also investigated the relationship between phylogenetic relatedness and latitude. Pearson's correlation analysis was conducted to assess the strength of each of the relationships.
3. Results
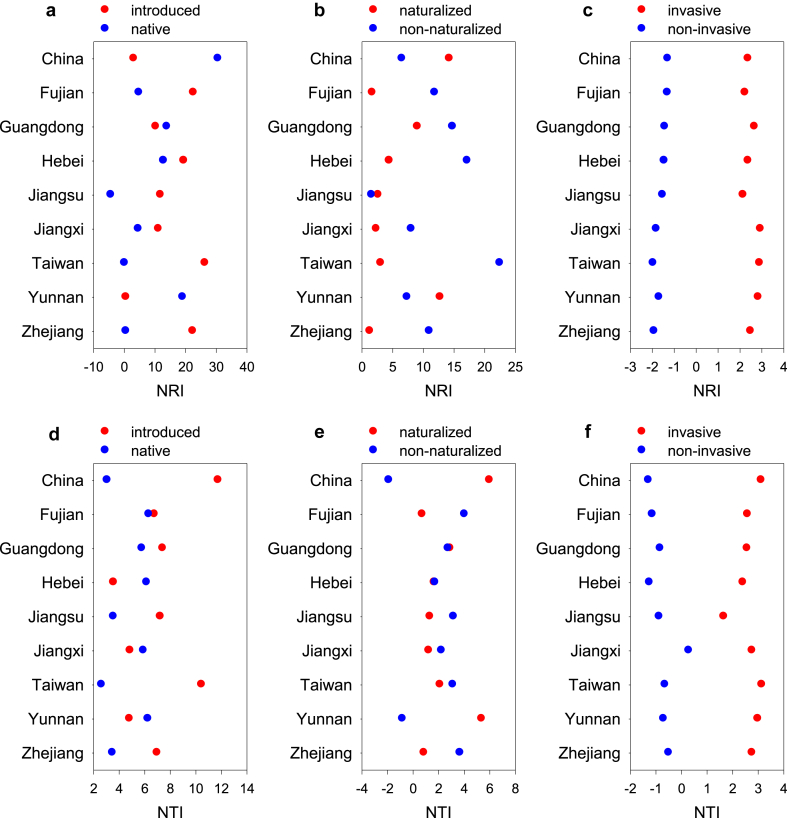
When China as a whole was considered, NRI for introduced species was much smaller than that for native species when they were calculated using the species pool including both introduced and native species in China (Fig. 1a), indicating that introduced species were phylogenetically over-dispersed relative to native species; however, the opposite pattern was observed when NTI was considered (Fig. 1d). Naturalized introduced species were more phylogenetically clustered than non-naturalized introduced species with respect to the species pool including all introduced species in China, and invasive naturalized species were also more phylogenetically clustered than non-invasive naturalized with respect to the species pool including all naturalized species in China, regardless of whether NRI or NTI was considered (Fig. 1). For province-level regions, when NRI and NTI were considered collectively, introduced species were phylogenetically more clustered than native species in 11 out of 18 cases, naturalized introduced species were phylogenetically more over-dispersed than non-naturalized introduced species in 11 out of 18 cases, and invasive naturalized species were phylogenetically more clustered than non-invasive naturalized species in all the 18 cases (Fig. 1).
Fig. 1.
NRI and NTI for native versus introduced species (a, d), for naturalized versus non-naturalized introduced species (b, e), and for invasive versus non-invasive naturalized species (c, f) for China as a whole and for the eight selected province-level regions. Species pools used in calculating NRI and NTI included all (introduced plus native) species in panels a and d, all introduced (naturalized plus non-naturalized) species in panels b and e, and all naturalized (invasive plus non-invasive) species in panels c and f. Species pools for different plant groups in each province-level region included only respective species in the region.
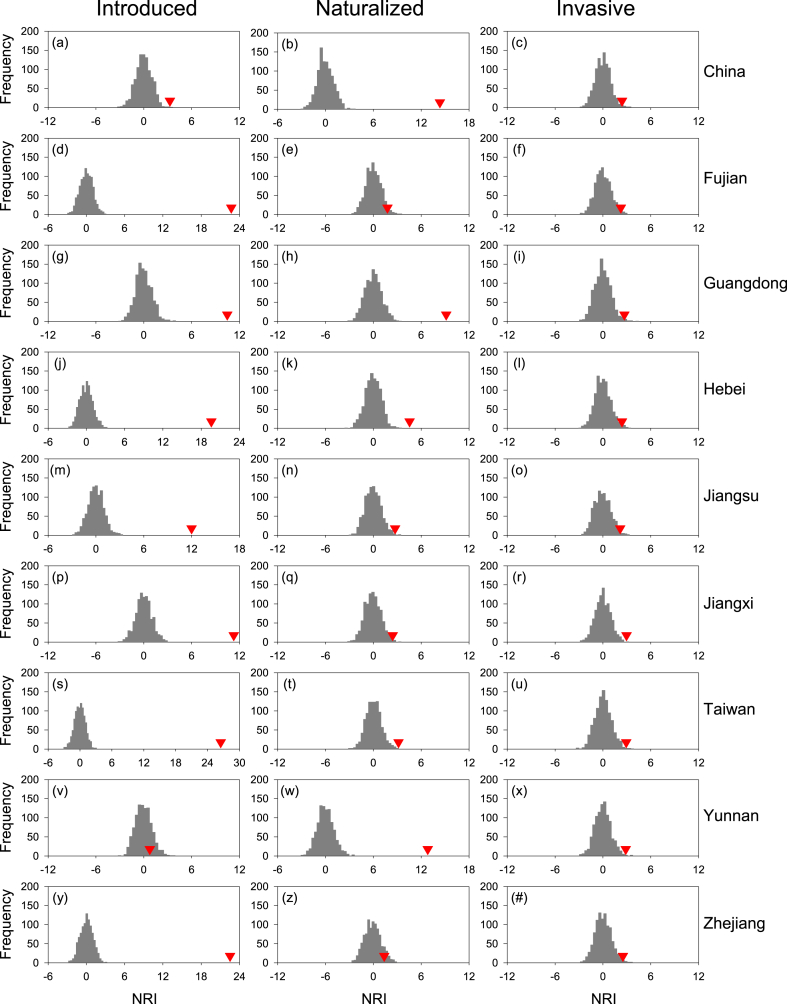
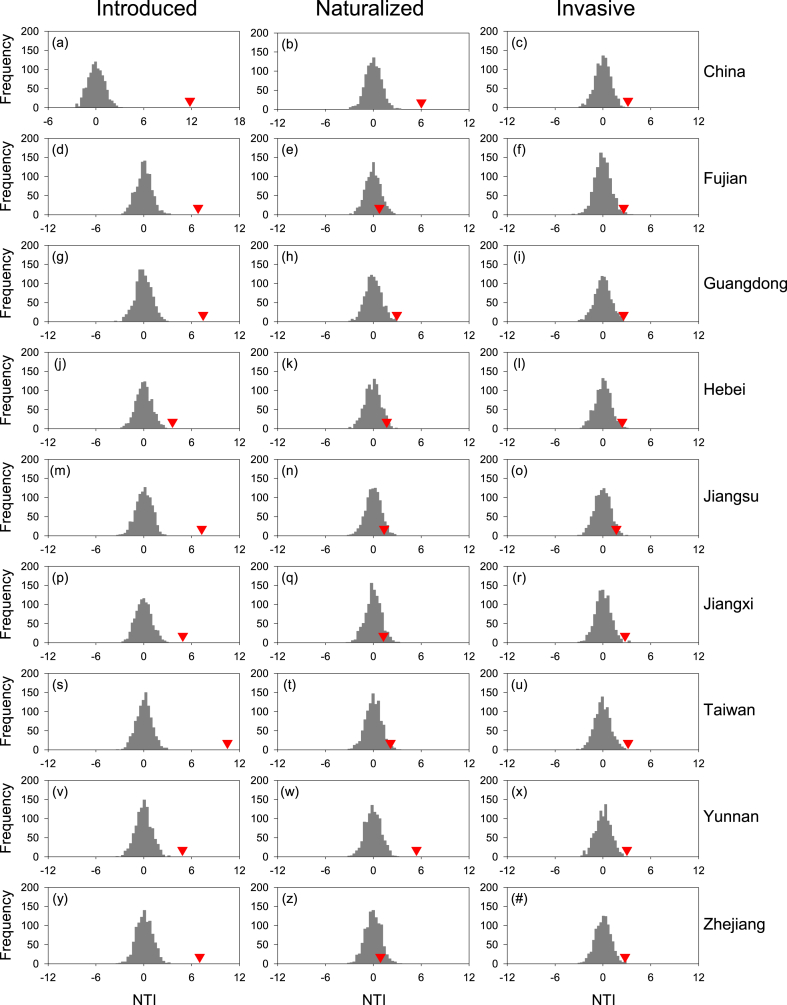
For China as a whole, values of NRI and NTI were positive for introduced angiosperm species with respect to overall (native plus introduced) angiosperm species, for naturalized angiosperm species with respect to introduced angiosperm species, and for invasive angiosperm species with respect to naturalized angiosperm species (Fig. 2, Fig. 3). NRI was smaller than NTI for introduced and invasive species (3.28 versus 11.78 and 2.40 versus 3.14, respectively), and was greater than NTI for naturalized species (14.34 versus 6.02). Because all the values of NRI and NTI were greater than 1.96, introduced, naturalized, and invasive angiosperm species were significantly (p < 0.05) clustered with respect to their species pools. Invasive species were also significantly clustered with respect to the pool of all introduced species in China (NRI and NTI = 8.87 and 3.85, respectively).
Fig. 2.
NRI (red triangle) for introduced (left), naturalized (middle), and invasive (right) angiosperm species for China as a whole and for each of the eight selected province-level regions, with respect to their respective national and provincial species pools. Species pool for introduced species included all (i.e., native and introduced) species; species pool for naturalized species included introduced species only; species pool for invasive species included naturalized species only. Species pools for different plant groups in each province-level region included only respective species (i.e., all, introduced, naturalized) in the region. Each histogram represents the frequency of NRI values derived from 999 null assemblages randomly drawn from the species pool.
Fig. 3.
NTI (red triangle) for introduced (left), naturalized (middle), and invasive (right) angiosperm species for China as a whole and for each of the eight selected province-level regions, with respect to their respective national and provincial species pools. Species pool for introduced species included all (i.e., native and introduced) species; species pool for naturalized species included introduced species only; species pool for invasive species included naturalized species only. Species pools for different plant groups in each province-level region included only respective species (i.e., all, introduced, naturalized) in the region. Each histogram represents the frequency of NTI values derived from 999 null assemblages randomly drawn from the species pool.
When the eight selected province-level regions were considered separately, values of NRI and NTI were positive in all cases, i.e., for introduced, naturalized, and invasive angiosperm species with respect to their respective regional species pools (Fig. 2, Fig. 3). Of the 48 values of NRI and NTI (i.e., 8 regions by 3 types of non-native species by 2 phylogenetic metrics), 39 (81%) were significant (p < 0.05; Fig. 2, Fig. 3). NRI was greater than NTI for introduced species in seven of the eight regions (i.e., all except for Yunnan), for naturalized species in all the eight regions, and for three of the eight regions (i.e., Guangdong, Jiangxi, and Jiangsu). The mean of the 24 values of NRI was significantly greater than that of NTI (7.69 versus 3.75; paired t-test, p < 0.05). When NRI and NTI were calculated based on species pool including all introduced species in each region, they were positive in all the 16 cases, 13 (81%) of which were significant (Table S1).
NRI and NTI for regional plant assemblages were negatively correlated with mean annual temperature and annual precipitation and positively correlated with latitude, regardless of whether naturalized species in each region were compared with all introduced species in China or invasive species in each region were compared with all naturalized species in China (Table 1). This indicates that phylogenetic relatedness of both naturalized and invasive species increased with increasing climate stressfulness.
Table 1.
Correlation coefficients for the relationships of phylogenetic relatedness metrics (NRI, NTI) with mean annual temperature (MAT), annual precipitation (AP), and latitude (LAT) for naturalized and invasive species in the eight selected province-level regions in China. NRI and NTI for naturalized species in each region were derived from the species pool including all introduced (naturalized plus non-naturalized) species in China; similarly, NRI and NTI for invasive species in each region were derived from the species pool including all naturalized (invasive plus non-invasive) species in China.
| Metric | MAT | AP | LAT |
|---|---|---|---|
| NRI (naturalized) | −0.290 | −0.314 | 0.299 |
| NTI (naturalized) | −0.533 | −0.413 | 0.461 |
| NRI (invasive) | −0.125 | −0.145 | 0.143 |
| NTI (invasive) | −0.562 | −0.178 | 0.263 |
4. Discussion
This study found that (1) introduced plants for cultivation are a phylogenetically clustered subset of overall (i.e., native plus introduced) angiosperm flora, (2) naturalized plants are a phylogenetically clustered subset of introduced plants for cultivation, and (3) invasive plants are a phylogenetically clustered subset of naturalized plants. In other word, changes in phylogenetic relatedness across the INI continuum (i.e., from introduction through naturalization to invasion) are in the same direction (i.e., towards more phylogenetic clustering). These three patterns hold regardless of spatial scales examined (i.e., national versus provincial scale) and whether basal-weighted or tip-weighted metric of phylogenetic relatedness is considered, as shown in Fig. 2, Fig. 3. To my knowledge, this study is the first to investigate patterns of phylogenetic relatedness of non-native angiosperms across a full INI continuum by comparing phylogenetic relatedness of non-native species at different invasion stages (i.e., introduction, naturalization, invasion) with their respective species pools within sampling areas. Omer et al. (2022) conducted a study investigating phylogenetic relatedness of non-native species at different invasion stages for angiosperms in South Africa; however, their study did not investigate phylogenetic relatedness of non-native species at each of invasion stages with respect to the pool of species for each study area; instead, their study compared introduced and naturalized angiosperm species to the pool of global angiosperm species that do not belong to the native flora of Southern Africa. Because the conclusions of their study are relevant to those of the present study, I discuss them below.
This study found that NRI and NTI for introduced species in China were significantly greater than 1.96 with respect to the species pool including both introduced and native species in China. This suggests that species are more likely to be introduced for cultivation if they are closely related to the native flora of China. This appears to be contrary to the conclusion of Omer et al. (2022) that non-native species distantly related to native species were more likely to be introduced for cultivation in South Africa. However, their conclusion is drawn based on the comparison of introduced species in South Africa with those species in the global species pool that have not been introduced into South Africa, and the model that only explained a small amount (< 10%) of the variation, as Omer et al. (2022) noted. My study confirms the notion that the species that have been intentionally introduced are not a random selection from the global flora but possess certain characteristics that make them of interest for cultivation, because humans might preferentially introduce plant species with characteristics that could provide economic and social benefits or ecosystem services that are not provided by species of the native flora (Omer et al., 2022). Many introduced plant species for cultivation may belong to a relatively few major clades (e.g., families or orders), causing phylogenetic clustering. For example, of the 246 families of the introduced angiosperms in this study, 71% of the 12,902 introduced species belong to the top 10% of the families, and 14% of the introduced angiosperm species occur in only three families (Orchidaceae, Cactaceae, and Fabaceae); this proportion is much higher than the proportion (56%) of the species in the top 10% of the 309 families in the overall angiosperm flora of China.
My study found that naturalized plants are a phylogenetically clustered subset of introduced plants for cultivation in China. This finding is consistent with the observation of Omer et al. (2022) for Southern Africa that those introduced angiosperm species that are more closely related to native species were more likely to naturalize, which is expected to increase phylogenetic relatedness (clustering) among species in a community. This pattern of increasing phylogenetic relatedness among species in the recipient community of naturalized species has been broadly documented for both plants (Qian et al., 2022a; Qian and Sandel, 2022) and animals (e.g. Xu et al., 2022). The observed pattern is consistent with Darwin's preadaptation hypothesis (Qian and Sandel, 2022). Specifically, Darwin proposed that introduced species may be more likely to naturalize if they are closely related to natives in the recipient communities, a hypothesis that is based on assuming similar ecological niches among closely related species, and focuses on the influence of environmental filtering, rather than competitive exclusion. The presence of a closely related native species in a region suggests that environmental conditions are also suitable for that non-native species, so should be a positive indicator of likely naturalization success.
This study found that invasive plant species are a phylogenetically clustered subset of naturalized plant species, consistent to Darwin's preadaptation hypothesis. However, this finding appears to be contrary to the finding of Omer et al. (2022) that naturalized plant species most distantly related to the natives are more likely to become invasive in South Africa, causing phylogenetic relatedness to decrease. Unlike the present study which focused on invasive angiosperm species of plants introduced to China for cultivation, Qian et al. (2022b) conducted a study including all invasive angiosperm species of introduced plants; they found that invasive plants are a phylogenetically clustered subset of naturalized plants, which is consistent with the finding of the present study. In addition, they found that more harmful invasive species are more phylogenetically clustered with respect to species pools of naturalized plants, compared to less harmful invasive plants, in China.
This study showed that when species in the pool of introduced species are assembled into regional naturalized species assemblages, species in naturalized communities tend to be phylogenetically more clustered in more stressful climatic conditions (lower temperature and precipitation). Similarly, when species in the pool of naturalized species are assembled into regional invasive species assemblages, species in invasive communities also tend to be phylogenetically more clustered in more stressful climatic conditions. These patterns are broadly consistent with those reported in previous studies for native angiosperms in China (e.g. Lu et al., 2018; Qian et al., 2019). The negative relationships of phylogenetic relatedness of naturalized species with temperature and precipitation observed in this study are also consistent with those reported in previous studies for China (Qian et al., 2022b). However, the negative relationships of phylogenetic relatedness of invasive species with temperature and precipitation observed in this study tend to be inconsistent, in some cases, with those reported by Qian et al. (2022a), who investigated a different set of invasive species and a different set of regions in China. If the negative relationships of phylogenetic relatedness with temperature and precipitation are general for native, naturalized, and invasive angiosperms, this would suggest that the assembly of plants in these three groups from species pools into regional communities is likely driven by the same environment-filtering mechanisms, such as phylogenetic niche conservatism (Donoghue, 2008; Qian et al., 2022a, b).
In sum, in this study, I analyzed a comprehensive data set to determine phylogenetic relatedness of non-native angiosperm species at different stages of the introduction−naturalization−invasion continuum. I found that invasive plants are a phylogenetically clustered subset of naturalized plants, which are a phylogenetically clustered subset of introduced plants for cultivation, which in turn are a phylogenetically clustered subset of overall (i.e. native plus introduced) angiosperm flora in China. These patterns, all of which are consistent with Darwin's preadaptation hypothesis, hold true regardless of whether national or regional floras are considered and whether deep or shallow evolutionary history is considered. Future studies should test whether the patterns observed in this study can be observed in other regions across the world, and whether the patterns observed for plant species apply to animal species.
Author contributions
H.Q. designed research, analyzed data, and wrote the paper.
Declaration of competing interest
The author declares no conflict of interest.
Acknowledgements
I am grateful to anonymous reviewers for their constructive comments on the manuscript.
Footnotes
Peer review under responsibility of Editorial Office of Plant Diversity.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2022.387.
Appendix ASupplementary data
The following is the Supplementary data to this article:
1
References
- Ackerly D. Conservatism and diversification of plant functional traits: evolutionary rates versus phylogenetic signal. Proc. Natl. Acad. Sci. U.S.A. 2009;106:19699–19706. doi: 10.1073/pnas.0901635106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn T.M., Pyšek P., Bacher S., et al. A proposed unified framework for biological invasions. Trends Ecol. Evol. 2011;26:333–339. doi: 10.1016/j.tree.2011.03.023. [DOI] [PubMed] [Google Scholar]
- Cadotte M.W., Campbell S.E., Li S.P., et al. Preadaptation and naturalization of nonnative species: Darwin's two fundamental insights into species invasion. Annu. Rev. Plant Biol. 2018;69:661–684. doi: 10.1146/annurev-arplant-042817-040339. [DOI] [PubMed] [Google Scholar]
- Cavender-Bares J., Kozak K.H., Fine P.V.A., et al. The merging of community ecology and phylogenetic biology. Ecol. Lett. 2009;12:693–715. doi: 10.1111/j.1461-0248.2009.01314.x. [DOI] [PubMed] [Google Scholar]
- Divíšek J., Chytrý M., Beckage B., et al. Similarity of introduced plant species to native ones facilitates naturalization, but differences enhance invasion success. Nat. Commun. 2018;9:4631. doi: 10.1038/s41467-018-06995-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoghue M.J. A phylogenetic perspective on the distribution of plant diversity. Proc. Natl. Acad. Sci. U.S.A. 2008;105:11549–11555. doi: 10.1073/pnas.0801962105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essl F., Dawson W., Kreft H., et al. Drivers of the relative richness of naturalized and invasive plant species on Earth. AoB Plants. 2019;11:plz051. doi: 10.1093/aobpla/plz051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fristoe T.S., Milan C., Dawson W., et al. Dimensions of invasiveness: links between abundance, geographic range size and habitat breadth in Europe's alien and native floras. Proc. Natl. Acad. Sci. U.S.A. 2021;118 doi: 10.1073/pnas.2021173118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallien L., Thornhill A.H., Zurell D., et al. Global predictors of alien plant establishment success: combining niche and trait proxies. Proc. R. Soc. B-Biol. Sci. 2019;286 doi: 10.1098/rspb.2018.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Q., Ma J.-S. Invasive alien plants in China: an update. Plant Divers. 2023;45:117–121. doi: 10.1016/j.pld.2022.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hortal J., Diniz-Filho J.A.F., Bini L.M., et al. Ice age climate, evolutionary constraints and diversity patterns of European dung beetles. Ecol. Lett. 2011;14:741–748. doi: 10.1111/j.1461-0248.2011.01634.x. [DOI] [PubMed] [Google Scholar]
- Jin Y., Qian H. V.PhyloMaker: an R package that can generate very large phylogenies for vascular plants. Ecography. 2019;42:1353–1359. doi: 10.1016/j.pld.2022.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y., Qian H. V.PhyloMaker2: an updated and enlarged R package that can generate very large phylogenies for vascular plants. Plant Divers. 2022;44:335–339. doi: 10.1016/j.pld.2022.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna M., Winternitz J., Garkoti S.C., et al. Functional leaf traits indicate phylogenetic signals in forests across an elevational gradient in the central Himalaya. J. Plant Res. 2021;134:753–764. doi: 10.1007/s10265-021-01289-1. [DOI] [PubMed] [Google Scholar]
- Lambdon P.W. Is invasiveness a legacy of evolution? Phylogenetic patterns in the alien flora of Mediterranean islands. J. Ecol. 2008;96:46–57. [Google Scholar]
- Lambertini M., Leape J., Marton-Lefevre J., et al. Invasives: a major conservation threat. Science. 2011;333:404–405. doi: 10.1126/science.333.6041.404-b. [DOI] [PubMed] [Google Scholar]
- Lin Q., Xiao C., Ma J. A dataset on catalogue of alien plants in China. Biodivers. Sci. 2022;30 [Google Scholar]
- Lososová Z., de Bello F., Chytrý M., et al. Alien plants invade more phylogenetically clustered community types and cause even stronger clustering. Global Ecol. Biogeogr. 2015;24:786–794. [Google Scholar]
- Lu L.-M., Mao L.-F., Yang T., et al. Evolutionary history of the angiosperm flora of China. Nature. 2018;554:234–238. doi: 10.1038/nature25485. [DOI] [PubMed] [Google Scholar]
- Mack R.N., Simberloff D., Lonsdale W.M., et al. Biotic invasions: causes, epidemiology, global consequences, and control. Ecol. Appl. 2000;10:689–710. [Google Scholar]
- Miller J.T., Hui C., Thornhill A.H., et al. Is invasion success of Australian trees mediated by their native biogeography, phylogenetic history, or both? AoB Plants. 2017;9:plw80. doi: 10.1093/aobpla/plw080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omer A., Fristoe T., Yang Q., et al. The role of phylogenetic relatedness on alien plant success depends on the stage of invasion. Nat. Plants. 2022;8:906–914. doi: 10.1038/s41477-022-01216-9. [DOI] [PubMed] [Google Scholar]
- Park D.S., Feng X., Maitner B.S., et al. Darwin's naturalization conundrum can be explained by spatial scale. Proc. Natl. Acad. Sci. U.S.A. 2020;117:10904–10910. doi: 10.1073/pnas.1918100117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyšek P., Richardson D.M., Rejmánek M., et al. Alien plants in checklists and floras: towards better communication between taxonomists and ecologists. Taxon. 2004;53:131–143. [Google Scholar]
- Pyšek P., Jaroík V.c., Pergl J., et al. The global invasion success of Central European plants is related to distribution characteristics in their native range and species traits. Divers. Distrib. 2009;15:891–903. [Google Scholar]
- Qian H. Relationships between plant and animal species richness at a regional scale in China. Conserv. Biol. 2007;21:937–944. doi: 10.1111/j.1523-1739.2007.00692.x. [DOI] [PubMed] [Google Scholar]
- Qian H., Deng T., Jin Y., et al. Phylogenetic dispersion and diversity in regional assemblages of seed plants in China. Proc. Natl. Acad. Sci. U.S.A. 2019;116:23192–23201. doi: 10.1073/pnas.1822153116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian H., Jin Y. Are phylogenies resolved at the genus level appropriate for studies on phylogenetic structure of species assemblages? Plant Divers. 2021;43:255–263. doi: 10.1016/j.pld.2020.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian H., Sandel B. Darwin's preadaptation hypothesis and the phylogenetic structure of native and alien regional plant assemblages across North America. Global Ecol. Biogeogr. 2022;31:531–545. [Google Scholar]
- Qian H., Rejmánek M., Qian S. Are invasive species a phylogenetically clustered subset of naturalized species in regional floras? A case study for flowering plants in China. Divers. Distrib. 2022;28:2084–2093. [Google Scholar]
- Qian H., Qian S., Sandel B. Phylogenetic structure of alien and native species in regional plant assemblages across China: testing niche conservatism hypothesis versus niche convergence hypothesis. Global Ecol. Biogeogr. 2022;31:1864–1876. [Google Scholar]
- Richardson D.M., Pyšek P. Naturalization of introduced plants: ecological drivers of biogeographic patterns. New Phytol. 2012;196:383–396. doi: 10.1111/j.1469-8137.2012.04292.x. [DOI] [PubMed] [Google Scholar]
- Richardson D.M., Pyšek P., Rejmánek M., et al. Naturalization and invasion of alien plants: concepts and definitions. Divers. Distrib. 2000;6:93–107. [Google Scholar]
- Sandel B., Tsirogiannis C. Species introductions and the phylogenetic and functional structure of California's grasses. Ecology. 2016;97:472–483. doi: 10.1890/15-0220.1. [DOI] [PubMed] [Google Scholar]
- Simberloff D., Martin J.L., Genovesi P., et al. Impacts of biological invasions: what's what and the way forward. Trends Ecol. Evol. 2013;28:58–66. doi: 10.1016/j.tree.2012.07.013. [DOI] [PubMed] [Google Scholar]
- Smith S.A., Brown J.W. Constructing a broadly inclusive seed plant phylogeny. Am. J. Bot. 2018;105:302–314. doi: 10.1002/ajb2.1019. [DOI] [PubMed] [Google Scholar]
- Thuiller W., Gallien L., Boulangeat I., et al. Resolving Darwin's naturalization conundrum: a quest for evidence. Divers. Distrib. 2010;16:461–475. [Google Scholar]
- Tsirogiannis C., Sandel B. PhyloMeasures: a package for computing phylogenetic biodiversity measures and their statistical moments. Ecography. 2016;39:709–714. [Google Scholar]
- van Kleunen M., Dawson W., Essl F., et al. Global exchange and accumulation of non-native plants. Nature. 2015;525:100–103. doi: 10.1038/nature14910. [DOI] [PubMed] [Google Scholar]
- Webb C.O. Exploring the phylogenetic structure of ecological communities: an example for rain forest trees. Am. Nat. 2000;156:145–155. doi: 10.1086/303378. [DOI] [PubMed] [Google Scholar]
- Webb C.O., Ackerly D.D., McPeek M.A., et al. Phylogenies and community ecology. Annu. Rev. Ecol. Systemat. 2002;33:475–505. [Google Scholar]
- Williamson M. Explaining and predicting the success of invading species at different stages of invasion. Biol. Invasions. 2006;8:1561–1568. [Google Scholar]
- Wu C.-Y., Raven P.H., Hong D.-Y., editors. vols. 2–25. Science Press, Beijing and Missouri Botanical Garden Press; St. Louis: 1994-2013. (Flora of China). [Google Scholar]
- Xu M., Li S.-P., Dick J.T.A., et al. Exotic fishes that are phylogenetically close but functionally distant to native fishes are more likely to establish. Global Change Biol. 2022;28:5683–5694. doi: 10.1111/gcb.16360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A., Hu X., Yao S., et al. Alien, naturalized and invasive plants in China. Plants. 2021;10:2241. doi: 10.3390/plants10112241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Qian H. U.Taxonstand: an R package for standardizing scientific names of plants and animals. Plant Divers. 2023;45:1–5. doi: 10.1016/j.pld.2022.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S.-B., Slik J.W.F., Zhang J.-L., et al. Spatial patterns of wood traits in China are controlled by phylogeny and the environment. Global Ecol. Biogeogr. 2011;20:241–250. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1