Abstract
This study compared disease progression of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) in three different models of golden hamsters: aged (≈60 weeks old) wild-type (WT), young (6 weeks old) WT, and adult (14–22 weeks old) hamsters expressing the human-angiotensin-converting enzyme 2 (hACE2) receptor. After intranasal (IN) exposure to the SARS-CoV-2 Washington isolate (WA01/2020), 2-deoxy-2-[fluorine-18]fluoro-D-glucose positron emission tomography with computed tomography (18F-FDG PET/CT) was used to monitor disease progression in near real time and animals were euthanized at pre-determined time points to directly compare imaging findings with other disease parameters associated with coronavirus disease 2019 (COVID-19). Consistent with histopathology, 18F-FDG-PET/CT demonstrated that aged WT hamsters exposed to 105 plaque forming units (PFU) developed more severe and protracted pneumonia than young WT hamsters exposed to the same (or lower) dose or hACE2 hamsters exposed to a uniformly lethal dose of virus. Specifically, aged WT hamsters presented with a severe interstitial pneumonia through 8 d post-exposure (PE), while pulmonary regeneration was observed in young WT hamsters at that time. hACE2 hamsters exposed to 100 or 10 PFU virus presented with a minimal to mild hemorrhagic pneumonia but succumbed to SARS-CoV-2-related meningoencephalitis by 6 d PE, suggesting that this model might allow assessment of SARS-CoV-2 infection on the central nervous system (CNS). Our group is the first to use (18F-FDG) PET/CT to differentiate respiratory disease severity ranging from mild to severe in three COVID-19 hamster models. The non-invasive, serial measure of disease progression provided by PET/CT makes it a valuable tool for animal model characterization.
Keywords: SARS-CoV-2, COVID-19, Hamsters, Computed tomography (CT), 2-Deoxy-2-[fluorine-18]fluoro-D-glucose (18F-FDG) PET/CT, Animal models
1. Introduction
The coronavirus disease 2019 (COVID-19) pandemic has resulted in over 761 million recorded cases and over 6.8 million deaths worldwide (WHO). In response, more than 10 vaccines against the Washington isolate (WA1/2020) of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) have been approved for use in humans (Li et al., 2022), and “boosters” using the spike of variants of concern/interest as an immunogen are already deployed (Link-Gelles et al., 2022). However, genetic variants of concern/interest continue to emerge and challenge the efficacy of vaccine and therapeutic countermeasures. Therefore, more techniques for model characterization could provide a better understanding of the pathophysiology of SARS-CoV-2 infection in the hamster model of COVID-19 for preclinical countermeasure development.
The elderly and individuals with comorbidities, such as diabetes, obesity, and immunodeficiency, are more susceptible to developing severe disease when compared to younger or healthier individuals (Jin et al., 2021; Rydyznski Moderbacher et al., 2020). COVID-19 patients requiring hospitalization frequently develop diffuse alveolar damage that is sometimes fatal (Barton et al., 2020; Borczuk et al., 2020; D'Agnillo et al., 2021; von Stillfried et al., 2022). Therefore, animal models that develop lung injury similar to that seen in COVID-19 patients are needed to study the pathophysiological mechanisms underlying disease progression, identify biomarkers of disease, and provide preclinical evaluation of medical countermeasures.
Wild-type (WT) golden hamsters (Mesocricetus auratus) are naturally susceptible to SARS-CoV-2 infection and are the most frequently used animal model of COVID-19 (Munoz-Fontela et al., 2020; Renn et al., 2021). Previous studies have shown that, despite typically surviving infection, mature (32–34-week-old) hamsters exposed to 105 plaque-forming units (PFU) of SARS-CoV-2 develop more pronounced weight loss, more severe and protracted bronchointerstitial pneumonia, and less robust humoral immune response when compared to young (6-week-old) hamsters (Osterrieder et al., 2020).
In an attempt to model the severe, sometimes fatal, pneumonia seen in a subset of humans with COVID-19, hamsters carrying the human angiotensin-converting enzyme 2 (hACE2) receptor were developed (Golden et al., 2022). Young (6–8-week-old) hACE2 hamsters exposed to a dose of SARS-CoV-2 1000-fold lower than typically administered to WT hamsters showed slowed movement or reduced mobility, weight loss, and high mortality rates (Gilliland et al., 2021). In a similar study with hACE2 hamsters, the virus was detected in the lungs, heart, and brain, with the highest titers found in the lungs (Golden et al., 2022). The virus detected in the brain was associated with a fatal neurotropic meningoencephalitis, which has also been observed in hACE2 mice (Golden et al., 2022; Moreau et al., 2020; Toomer et al., 2022). While the effects of SARS-CoV-2 on the central nervous system (CNS) have not been fully elucidated, a fatal SARS-CoV-2 neurotropic meningoencephalitis has not been reported in the brain tissue of human autopsy patients that succumbed to COVID-19 (Barton et al., 2020; Ho et al., 2022; Khan et al., 2021; Matschke et al., 2020; Stein et al., 2022).
Previously, computed tomography (CT), positron emission tomography with CT (PET/CT), and single-photon emission computerized tomography (SPECT) have been used to evaluate animal models of Middle East respiratory syndrome coronavirus (MERS-CoV) (Chefer et al., 2018; Johnson et al., 2015b, 2016), demonstrating the utility of medical imaging in pre-clinical development. In particular, 2-deoxy-2-[fluorine-18]fluoro-D-glucose (18F-FDG) PET/CT imaging provides detailed spatiotemporal visualization of inflammatory processes deep within the entire body of the hamsters in a longitudinal and non-invasive way. Moreover, semi-quantitative 18F-FDG PET/CT image analysis is less affected by sampling bias, which enables a more comprehensive view of the disease progression.
In this study, a comparison of three established SARS-CoV-2 hamster models representing lung disease severity from mild to moderate to severe was made: 60-week-old (aged) WT, 6-week-old (young) WT, and 14–22-week-old hACE2 (Golden et al., 2022) hamsters exposed to WA01/2020 via the intranasal (IN) route. Disease progression was assessed using body weight, viral load in the upper respiratory tract and lung tissue, whole-body 18F-FDG micro-PET/CT, immunology, and histopathology. Our group successfully differentiated COVID-19 lung disease severity from mild to severe by utilizing 18F-FDG micro-PET/CT in these hamster models. Additionally, we were able to determine the damage to the nasal turbinates, which was supported by histopathology.
2. Materials and methods
2.1. Animals and experiment design
An initial series of natural history of disease studies was followed by a series of PET/CT imaging studies in young and hACE2 hamster models. The clinical, virology, immunology, and histology data were consolidated to increase the sample sizes for statistical purposes.
All hamsters were acclimated for 10–14 d (WT) (or 70 d for hACE2) and individually housed in the IRF-Frederick biosafety level 4 (BSL-4) laboratory, a facility accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC). All hamsters were IN-exposed to either mock inoculum (diluent medium) or to SARS-CoV-2 WA01/2020, diluted in 100 μL of Dulbecco's modified eagle medium (DMEM) and supplemented with 2% heat-inactivated fetal bovine serum (FBS; Sigma-Aldrich, St. Louis, MO, USA). The exposure inoculum was back-titrated real-time to verify the dose received.
Natural history studies were performed in WT young hamsters (LVG hamsters; Charles River Laboratories, Wilmington, MA, USA) after exposure to mock inoculum or 105 (back titer: 1.00^105 PFU) or 103 PFU (back titer: 9.29^102 PFU) of SARS-CoV-2 and in hACE2 hamsters (provided by Dr. Zhongde Wang and Dr. E. Bart Tarbet from Utah State University) after exposure to mock inoculum or 100 (back titer: 1.43^102 PFU), 10 (back titer: 1.51^101 PFU), 1 (back titer: 1.78^100 PFU), or 0.1 PFU (back titer: undetectable) SARS-CoV-2 (Fig. S1A). Body temperatures were assessed using subcutaneously placed transponders in hACE2 hamsters. Body temperature decreases are a reliable indicator of SARS-CoV-2-associated morbidity in the hACE2 model. Body temperatures were assessed in young WT hamsters post-SARS-CoV-2 exposure, but the data is not presented because body temperature did not significantly differ from uninfected controls (Francis et al., 2021; Osterrieder et al., 2020). A natural history of disease study was not performed in aged WT hamsters due to a lack of available 60-week-old hamsters.
The subsequent PET/CT imaging studies were performed in aged, young, and hACE2 hamsters (Fig. S1B). Hamsters were grouped by SARS-CoV-2/mock-inoculum exposure dose and then separated into cohorts based on pre-determined imaging days, followed by serial necropsy days (Fig. S1B). Of 10 aged (≈60-week-old) WT hamsters (HsdHan AURA hamsters; Envigo, Haslett, MI, USA), nine were exposed to a target dose of 105 PFU SARS-CoV-2 (back titer: 9.27^104 PFU) and 1 was mock-exposed (Fig. S1B). The inclusion of an aged hamster group exposed to 103 PFU SARS-CoV-2 was not possible due to the limited availability of 60-week-old hamsters. Young (6-week-old) WT hamsters were exposed to either target dose of 105 PFU (back titer: 5.57^104 PFU) or 103 PFU (back titer: 1.43^103 PFU) with the same group sizes as mock-exposed controls (Fig. S1B). Adult (14–22-week-old) hACE2 hamsters were exposed to lower doses of SARS-CoV-2 due to previously identified higher morbidity and mortality when compared to WT hamsters, and a 10-PFU dose was not included in the imaging study due to uniform mortality observed in the natural history of disease study: 100 PFU (back titer: 5.63^101 PFU), 1 PFU (back titer: undetected), or 0.1 PFU (back titer: undetected) with six mock-exposed controls (natural history and imaged) (Fig. S1B).
Hamsters were weighed and evaluated for clinical signs of disease from the day prior to exposure (−1 d, with exposure designated as day 0) until the end of the study. Pre-exposure PET/CT was performed at 7 ± 3 d in hamster groups included in the serial imaging studies. Clinical disease signs in hACE2 hamsters included the presence of weight loss exceeding 10% of weight relative to day 0, increased body temperatures of 2 °C or more, lethargy, hunched posture, ruffled fur, nasal or ocular discharge, and tachypnea and/or dyspnea. Each of these clinical signs was scored a 0 or 1. Hamsters with cumulative clinical scores greater than 7 or those that demonstrated weight loss equal to or greater than 25% compared to day 0 weight, decreases in body temperature in excess of 3 °C, or unresponsiveness to external stimulus were humanely euthanized. SARS-CoV-2 is not typically lethal in WT hamsters. The clinical signs observed in WT hamsters were weight loss and increased respiratory rate. In addition to daily monitoring for clinical signs of disease, all hamsters were euthanized and necropsied after micro-PET/CT imaging at scheduled or unscheduled (reaching pre-determined endpoint criteria) terminal time points, and samples were collected to assess viral replication and host immune response to infection, as well as for histopathology and SARS-CoV-2 nucleocapsid protein (NP) immunohistochemistry (IHC) to characterize disease progression.
2.2. Virus
The2019-nCoV/USA-WA1-A12/2020 (GenBank #MT020880.1) isolate (Washington isolate; WA01-/2020) of SARS-CoV-2 (Nidovirales: Coronaviridae: Sarbecovirus) was provided by the U.S. Centers for Disease Control and Prevention (CDC; Atlanta, GA, USA) at passage 3 from sample collection. The virus was then passaged once at the Integrated Research Facility at Fort Detrick by inoculating Vero cells E6 (American Type Culture Collection [ATCC], Manassas, VA, USA). The infected cells were incubated for 72 h in DMEM with 4.5 g/L D-glucose, L-glutamine, and 110 mg/L sodium pyruvate (Gibco, Gaithersburg, MD, USA), containing 2% heat-inactivated FBS (SAFC Biosciences, Lenexa, KS, USA) in a humidified atmosphere at 37 °C with 5% carbon dioxide (CO2). The resulting master stock, a passage 4 stock (IRF0394), was quantified by plaque assay using Vero E6 cells with a 2.5% Avicel overlay and stained after 48 h with a 0.2% crystal violet stain (Ricca Chemical, Arlington, TX, USA). Virus stocks were sequenced and compared to the published SARS-CoV-2 Washington isolate genome (GenBank #MT020880.1) to confirm sequence fidelity. Virus stocks were free from adventitious agents, and quality-control assays for endotoxin, mycoplasma, and bacterial contamination were negative.
2.3. Viral load
2.3.1. Oropharyngeal samples collected from swabbing and lavage
Media used for lavage collection (on all WT hamsters) was composed of DMEM with L-glutamine (Gibco, Waltham, MA, USA), 2% vol/vol heat inactivated FBS (Sigma-Aldrich), and 1% vol/vol antibiotic-antimycotic (Gibco). Oropharyngeal samples (for all hACE2 hamsters) were collected using Puritan PurFlock Ultra Sterile Flocked swab (Puritan Medical Products Company LLC, Guilford, ME, USA). The flocked tip was inserted into the oropharyngeal area and then swiped in a circular motion three times. The flocked tip of the swab was placed into a vial containing viral transport media : HBSS, 2%vol/vol heat inactivated FBS (Sigma-Aldrich), 0.2% vol/vol of 250 µg/mL Amphotericin B (Sigma-Aldrich) and 0.2% vol/vol of 50 mg/mL Gentamycin (Gibco); the plastic handle was cut from the tip with clean scissors. Oropharyngeal lavages were performed immediately after euthanasia by instilling 1.0 mL of 2% vol/vol heat inactivated FBS (Sigma-Aldrich) and 1% vol/vol antibiotic-antimycotic (Gibco) in 250-μL increments. Each increment of media was immediately retrieved by pipette and transferred into a sterile plastic vial.
2.3.2. Tissue processing for real-time reverse transcription polymerase chain reaction (RT-qPCR) and plaque assay
Approximately 50 mg of lung tissue was collected in pre-weighed bead beating tubes and maintained at −80 °C prior to processing. Samples for plaque assays and RT-qPCR were reweighed after thawing to calculate the weight of the tested lung tissue. Phosphate-buffered saline (PBS) (Gibco, Grand Island, New York) was added to achieve a 10% suspension for plaque assay, or TRIzol LS (Thermo Fisher Scientific, Waltham, MA, USA) was added to achieve a 10% w/v homogenate for RT-qPCR. Samples were homogenized using an OMNI Bead Ruptor Elite for 3 min and then clarified by centrifugation for 10 min at 2500 rpm.
2.3.3. RT-qPCR
Samples for RNA extraction and downstream RT-qPCR analysis of viral genomic RNA were treated using TRIzol LS (Thermo Fisher Scientific) in accordance with manufacturer's instructions. Briefly, 70 μL of TRIzol-inactivated sample was added to 280 μL of QIAGEN Buffer AVL, containing 3 μg of carrier RNA (polyA, supplied with QIAamp Viral RNA Mini kit [QIAGEN, Germantown, MD, USA]), and extracted using the QIAamp Viral RNA Mini Kit in accordance with manufacturer's instructions. Samples were eluted in 70 μL of Buffer AVE, aliquoted in two equal portions, and stored at −80 °C until performing the assay. SARS-CoV-2 viral RNA targets in the ORF1a gene were then quantified using the CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel protocol using an Applied Biosystems 7500 Fast Dx Real-Time PCR Instrument (Thermo Fisher Scientific) in accordance with the manufacturer's instructions. Cycling conditions were: 50 °C for 5 min, 95 °C for 20 s, 95 °C for 3 s, and 60 °C for 30 s; the last two steps were repeated for 45 cycles. Serially 10-fold-diluted from 9 log10 to 0 log10 viral RNA copies of DNA, comprised of the SARS-CoV-2 RT-qPCR assay target, constructed as gene fragments (gBlocks; Integrated DNA Technologies, Coralville, IA, USA), were used as a standard curve to establish quantifiable cycle threshold (Ct) values. Data were reported as viral RNA copies per mg of tissue. Applied Biosystems 7500 software version 1.4.1 was used to calculate the viral RNA copies in a sample.
2.3.4. Infectious virus burden quantification
Vero E6 cells were plated in 6-well plates at a density of 1 × 106 cells per well to ensure at least 90% confluence the following day. Ten-fold serial dilutions of the tested samples were added in triplicate of 300 μL per well to individual wells, followed by 1 h incubation at 37 °C with 5% CO2, rocking every 15 min. Then, the cells were covered with 2 mL of 2.5% Avicel overlay, diluted (1:1; f/c 2.5%) in 2 × Eagle's minimum essential medium (EMEM; Quality Biological, Gaithersburg, MD, USA), containing 4% FBS and incubated at 37 °C with 5% CO2 for 48 h. The overlay was removed, and the cells were fixed with 0.2% crystal violet in 10% neutral buffered formalin (NBF) for 30 min at ambient temperature. The plates were washed with water and plaques were enumerated. Data were reported as PFU per g of homogenized tissue.
2.4. In vivo micro-PET/CT imaging
To evaluate the SARS-CoV-2-induced pathological changes over time noninvasively, serial micro-PET/CT imaging studies were performed on an MRS PET/CT 120 preclinical scanner (MR Solutions, Guildford, Surrey, UK) at −7±3, 2, 5, and 8 d post-exposure (PE). During each imaging day, each animal was anesthetized using isoflurane and injected intramuscularly in the thigh with 18F-FDG (Cardinal Health, Beltsville, MD, USA). A respiratory-gated high-resolution chest CT (for radiological evaluation), a whole-body helical CT (to generate an attenuation map for PET reconstruction), and a 10-min static micro-PET image (at 60 min after 18F-FDG injection) were acquired sequentially and then reconstructed. CT images were read by a board-certified radiologist. All images were reviewed, and data were measured using MIM v.7.0 (MIM Software, Cleveland, OH, USA).
2.4.1. CT data quantification
The CT abnormalities were first evaluated in the lungs by an experienced radiologist using a qualitative CT consolidation scoring system of 0–5 with 0.5 increments, adapted from human cases and based on lung involvement (Pan et al., 2020). Qualitatively, the pulmonary abnormalities seen in hamsters had a similar spectrum of distribution, morphology, and duration to that described in patients with mild to moderate COVID-19; abnormalities included multifocal, bilateral ground-glass opacities (GGOs), infiltrates, consolidation, air bronchograms, and dilated airways (Hani et al., 2020).
Lobar scores were summed to generate total lung scores. CT scores ranged 0–5 for each lobe (right upper, right middle, right lower, right accessory, and left). Lobes were scored as: 0 = no disease involvement, 1 < 5%, 2 = 5–24%, 3 = 25–49%, 4 = 50–74%, 5 > 74%, with a maximum total lung score of 25.
Percent change in the volume of lung hyperdensity (PCLH) was performed to quantify CT data by first segmenting the lung field in the CT scan using a region growing implementation (Johnson et al., 2015a). Lung volumes were generated for baseline scans and all subsequent CT scans for each hamster. A histogram analysis on the baseline scan for each animal determined a density threshold value (Hounsfield Unit [HU]) for which 5% of voxels were above this threshold. This threshold was applied to all scans performed post-exposure, and the volume of tissue with the HU value greater than this value was compared as a percent change to the volume at baseline. In addition, this hyperdense volume was normalized to the total lung volume and represented as a percent of total lung volume.
2.4.2. PET data quantification
To quantify PET data, whole-body CT and micro-PET scans for a given imaging session were co-registered. Regions of interest (ROIs) were placed manually on the PET images, and location was determined on the co-registered CT images. These regions included specific lung abnormalities, when present, as well as left and right lungs in which no specific abnormalities were present. The lung abnormality with the highest 18F-FDG uptake was first identified by visual inspection of the entire lung by a board-certified radiologist. Then, an ROI was placed, and 18F-FDG maximum standardized uptake value (SUVmax) was measured from the lung abnormality. To minimize the inter-animal variations, another ROI was placed and the mean SUV (SUVmean) was measured in a normal area of the lungs where no specific abnormalities were present. The ratio between the abnormal SUVmax and normal SUVmean (normalized lung abnormality SUVmax) was calculated for each animal. The averaged normalized lung abnormality SUVmax values of the groups were graphed longitudinally. Quantitative analysis was correlated with a qualitative evaluation of CT and micro-PET/CT lung pathology over time, performed by a board-certified radiologist.
To analyze the 18F-FDG uptake in the nasal tissues, a 2.5-mm spherical volume of interest was manually placed on the PET image using the CT as anatomical reference. The SUVmean was measured. To reflect the longitudinal change of the 18F-FDG uptake in the nasal tissues and nasopharyngeal structures, the ratio between the SUVmean of different days post-exposure and that of baseline was calculated (normalized nasal SUVmean). The averaged normalized nasal SUVmean of groups were graphed longitudinally.
2.5. Immunology assessment
2.5.1. Serum spike immunoglobulin G (IgG) enzyme-linked immunosorbent assay (ELISA)
ELISA plates were coated with 50 μL (0.1 μg per well) of SARS-CoV-2 spike protein S1 domain (Sino Biological, Wayne, PA, USA) in PBS and placed on a microplate overnight at 4 °C, followed by washing with PBS with Tween 20 (PBS-T) and blocking in 5% non-fat dry milk (LabScientific, Danvers, MA, USA). The ELISA plates were washed again with PBS-T, and diluted serum samples were serially diluted 2-fold (1:100–1:12,800). After washing, S1 domain-specific IgG antibodies were detected using horseradish peroxidase (HRP) conjugated anti-hamster antibodies (Jackson ImmunoResearch, Laboratories, Inc, West Grove, Pennsylvania, USA) and 3,3′,5,5′-Tetramethylbenzidine (TMB) substrate (Thermo Fisher Scientific) (Malherbe et al., 2021). Reciprocal endpoint titers were determined in Prism software, version 9 (GraphPad, La Jolla, CA, USA), using a sigmoidal 4-parameter logistic fit curve. Endpoint titers were calculated at the point where the curve crossed the ELISA cutoff value. Data are presented as the mean and standard deviation of two independent ELISA runs.
2.5.2. Fluorescence neutralization assay
The fluorescence reduction neutralization assay (FRNA) method for measuring neutralizing antibodies was developed previously (Bennett et al., 2021). Briefly, Vero E6 cells were seeded at 3 x 104 in 100 μL DMEM+10% heat-inactivated FBS in 96-well Operetta plates (Greiner Bio-One, Monroe, NC, USA). The following day, a series of 6-point dilutions of 1:2 were performed in duplicate, starting with a dilution of 1:40–1:960. SARS-CoV-2 virus stock was diluted in serum-free media at a multiplicity of infection (MOI) of 0.5. The mixture of serum and virus were incubated for 1 h at 37 °C with 5% CO2. Following incubation, 100 μL per well of each virus/serum mixture were transferred to the Operetta plates with cells. The virus/serum mixtures were incubated for 24 h at 37 °C with 5% CO2. Subsequently, plates were fixed with 20% NBF at 4 °C for 24 h. Plates were then washed and blocked with 3% bovine serum albumin (BSA) in 1 × PBS for 30 min on a rocker.
The plates were stained with primary antibody, a SARS-CoV NP-specific rabbit monoclonal antibody (Sino Biological), at 1:8000 in blocking buffer at ambient temperature for 60 min on a rocker. The plates were washed and stained with secondary antibody, goat α-rabbit IgG (H + L), Alexa Fluor 594 Conjugate (Thermo Fisher Scientific, Waltham, MA, USA) at 1:2500 and incubated at ambient temperature for 30 min on a rocker in the dark. Finally, plates were read on an Operetta High-Content Analysis System (PerkinElmer, Waltham, MA, USA) with at least four fields of view of >1000 cells each. Data were analyzed using Harmony software (PerkinElmer). Half-maximal neutralization titers (NT50) were calculated by averaging the fluorescence intensity in virus control wells.
2.5.3. Cytokine/chemokine profile
Lung homogenates were analyzed for cytokine and chemokine changes by RT-qPCR assay (Chan et al., 2020). Nine cytokines and chemokines were evaluated: interleukin 2 (IL2), IL6, IL10, interferon gamma (IFNG), tumor necrosis factor alpha (TNFA), transforming growth factor beta 1 (TGFB1), chemokines C–C motif ligand 20 (CCL20), CCL22, and interferon gamma-induced protein 10 (IP-10). Chemokine/cytokine profiling was performed on lung tissues from virus-exposed and mock-exposed hamsters by RT-qPCR. Tissue samples for RNA extraction were processed as above, followed by total RNA extraction. RNA was then purified with a PureLink RNA Mini Kit (Thermo Fisher Scientific, Carlsbad, CA, USA) in accordance with manufacturer's instructions. TaqPath 1-Step Multiplex Master Mix (Thermo Fisher Scientific, Frederick, MD, USA) was used to generate complementary DNA and quantified using Taqman primers and probes (Integrated DNA Technologies) using the Applied Biosystems 7500 Fast Dx Real-Time PCR Instrument in accordance with the manufacturer's instructions. The Ct value of each gene was normalized to the internal reference gene (hamster RPL-18), and the comparative Ct (ΔΔCt) method was used to calculate changes in chemokine and cytokine gene expression. The data were further normalized and graphed, with the mock-exposed animals serving as the calibrator sample for each respective day post-exposure. Results were graphed using GraphPad Prism 9.3.a.
2.6. Histopathology
All hamsters that did not succumb to disease were humanely euthanized by terminal exsanguination under deep isoflurane anesthesia and necropsied at scheduled time points. Animals that reached pre-determined clinical endpoint criteria following infection were similarly euthanized. The lungs, trachea, nasal turbinates, brain, olfactory bulb, heart, spleen, liver, kidneys, adrenal gland, eyes, ovaries/testes, axillary lymph nodes, inguinal lymph nodes, mesenteric lymph nodes, bronchial lymph nodes, submandibular salivary glands, stomach, duodenum, jejunum, ileum, and colon were collected either in 10% NBF for histopathology and routinely processed for histopathology and IHC or frozen to assess viral load by plaque assay and RT-qPCR.
2.6.1. Microscopic examination
Hematoxylin and eosin-stained sections were semiquantitatively scored 0–4 by a pathologist as follows: 0, tissue within normal limits; 1, histologic changes that barely exceeded normal limits; 2, easily identified histologic changes of limited severity; 3, prominent histologic changes that may have caused organ dysfunction but were not of maximal severity; or 4, overwhelming histologic changes likely to cause significant organ dysfunction.
2.6.2. IHC for SARS-CoV-2
SARS-CoV-2 IHC was performed with rabbit-polyclonal anti-SARS-CoV/SARS-CoV-2 nucleocapsid primary antibody (1:6500 dilution; Sino Biological, Wayne, PA, USA) and followed by a biotinylated donkey anti-rabbit IgG secondary antibody (1:1500 dilution; Jackson ImmunoResearch Laboratories, Inc). The biotinylated target was detected by ABC-AP kit (Vector Labs, Newark, CA, USA) using the ImmPACT Vector Red substrate and counterstained with hematoxylin. Stains were semiquantitatively scored for staining intensity and distribution as follows: 0, no staining; 1, weakly positive (fewer than 5 cells per section) and/or low intensity staining; 2, moderately positive (5–20 cells per section) and/or medium intensity staining; 3, strongly positive (more than 20 cells per section) and/or high intensity staining.
2.7. Statistical analyses
All data were analyzed with GraphPad Prism 9.3.1. Survival data were analyzed using Kaplan–Meier survival curves compared by the log-rank (Mantel–Cox) test, followed by pairwise comparison using the Gehan–Breslow–Wilcoxon test. Body weight loss was analyzed using mixed-effects repeated measures model with Tukey's post-test multiple comparisons. Statistical analyses of clinical (RT-qPCR and plaque assay), radiological, and pathology scores were performed by one-way analysis of variance (ANOVA) with Tukey multiple comparison to determine significant differences among groups. Two-way ANOVA and then Tukey's test were used to analyze SARS-CoV-2 S1 ELISA, neutralization antibody, and cytokine/chemokine profiles. Pearson correlation tests were performed on endpoints of interest; p < 0.05 was considered statistically significant.
3. Results
3.1. Clinical presentations
3.1.1. WT hamsters (aged and young) survived high dose exposure of SARS-CoV-2 (up to 105 PFU); hACE2 hamsters showed uniform lethality with 10 PFU SARS-CoV-2 exposure
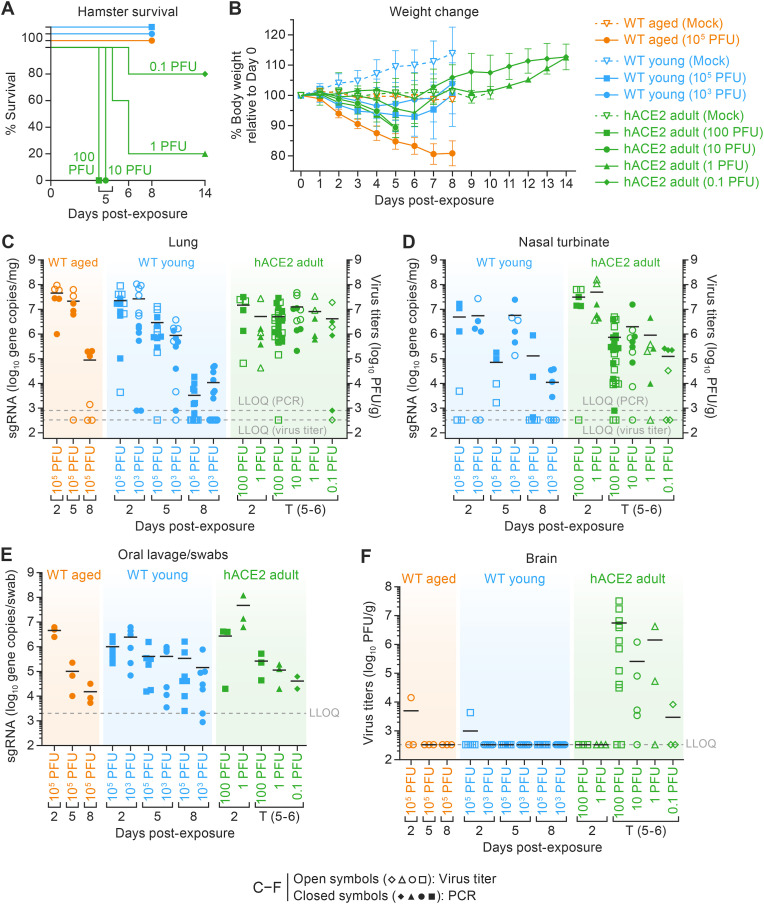
Aside from hamsters serially euthanized at pre-scheduled time points post-exposure (Fig. S1), all WT hamsters exposed to 105 (aged and young) or 103 (young) SARS-CoV-2 (or mock inoculum) survived to their scheduled study endpoints of 2, 5, 8, 10 or 14 d PE. Survival of hACE2 hamsters was dose-dependent; those exposed to either 100 or 10 PFU showed uniform lethality, met pre-defined endpoint criteria, and were euthanized or succumbed to viral meningoencephalitis by 6 d PE (Fig. 1 A and Fig. S1). Four of five hACE2 hamsters exposed to 1 PFU SARS-CoV-2 succumbed by 6 d PE. One of five hACE2 hamsters exposed to 0.1 PFU succumbed at 6 d PE (Fig. 1A and Fig. S1). The three mock-exposed control hACE2 hamsters survived to the end of the study (10 d PE).
Fig. 1.
Clinical signs of disease survival, body weight changes, and viral loads in all three hamster models.
A. Survival of hamsters exposed to SARS-CoV-2 via intranasal inoculation. All WT hamsters survived virus exposure over the course of 8 d. Survival of hACE2 hamsters in a 10–14-d natural history study correlated with exposure dose. Significant differences were seen as p = 0.0013 (**). B. Individual relative body weight changes (weight loss in WT aged 105 PFU in comparison to WT young 103, 105, hACE2 100, 10, 1, and 0.1 PFU; p < 0.0001***) over the duration of the disease. Data are presented as the mean percentage of the weight relative to 0 d (±SD). Statistical significance for average body weight was analyzed across the 8–14-d time course using a mixed-effects repeated measures model with Tukey's post-test multiple comparisons. C-F Virus loads were determined from C. RNA (per mg homogenized right cranial lung lobe) and infectious virus titers (per g homogenized right cranial lung lobe) (WT aged 105 PFU in comparison to WT young 105 PFU; p < 0.03*) and (D) nasal turbinates (E) Viral RNA from oropharyngeal samples collected by swabbing or lavage (per swab or per mL, respectively) to detect the virus shredding over the course of the experiment (F) Infectious virus titers in brain obtained at euthanasia. Horizontal lines show the mean. Points indicate data from individual hamsters. Solid symbols indicate viral RNA; open symbols indicate virus titers; dotted lines indicate the limit of detection; points indicate data from individual hamsters. One-way ANOVA with Tukey multiple comparison was performed in GraphPad Prism 9.3.1. p < 0.033 is *, p < 0.002 is **, and p < 0.001 is ***. SARS-CoV-2, severe acute respiratory syndrome coronavirus-2; WT, wild-type; hACE2, human angiotensin-converting enzyme 2; SEM, standard error of the mean; SD, standard deviation; ANOVA, analysis of variance; LLOQ, lower limit of quantitation; PCR, polymerase chain reaction.
3.1.2. Aged WT hamsters showed significantly more body weight loss when compared to young WT or hACE2 hamsters after exposure to SARS-CoV-2
As expected, mock-exposed young WT hamsters gained weight throughout the experiment. Mock-exposed adult hACE2 and aged WT hamsters showed no weight change throughout the experiment (Fig. 1B). In WT hamsters, signs of disease presented within 3 d PE and included a significant reduction in body weight (Fig. 1B) with an increase in respiratory rate. Decreases in body weight were observed on 2 d PE in all virus-exposed hamsters when compared to mock-exposed hamsters (Fig. 1B). Aged WT hamsters showed the most body weight loss when compared to young WT hamsters and hACE2 hamsters exposed to SARS-CoV-2. At 8 d PE, a recovery in body weight was observed in all surviving hamster groups; however, body weight gains were marginal in aged WT hamsters compared to young WT hamsters (Fig. 1B).
As seen in young WT hamsters, adult hACE2 hamsters showed weight loss (Fig. 1B) that was dose-dependent. Exposure to 100 PFU and 10 PFU resulted in a similar average of 11% body weight loss, those exposed to 1 PFU showed 5%, and those exposed to 0.1 PFU showed 1% body weight loss at 5 d PE. An increase in respiratory rate and a significant decrease in body temperature (up to 4 °C compared to baseline), with ruffled fur, lethargy, and snuffling were also seen in hamsters prior to reaching endpoint criteria.
3.2. Viral load
3.2.1. SARS-CoV-2 load peaked in the lungs at 2 d PE in all three models but persisted in hACE2 hamsters
To determine the tropism of SARS-CoV-2, viral RNA concentrations were measured by RT-qPCR for the envelope gene (E) and/or viral titers were assessed in cell culture from oropharyngeal swab/lavage samples, nasal turbinate tissue, and lung tissue at 2, 5, and 8 d PE. Virus was not detectable in tissues sampled at 10 or 14 d PE (Fig. 1C–G). Lung viral burden peaked in all three models at 2 d PE (Fig. 1C). At 5 d PE, viral burden in the lungs decreased by 1 log10 in young WT hamsters exposed to 105 PFU and by nearly 2 log10 in those exposed to 103 PFU. By 8 d PE, viral burden in the lung tissue of young WT hamsters exposed to 105 PFU decreased further, by nearly 2.5 log10, and in those exposed to 103 PFU viral burden decreased by 2 log10, when compared to 5 d PE (Fig. 1C). In contrast, and independent of dose, hACE2 hamsters maintained high average viral burdens in the lungs throughout the observation period (Fig. 1D).
3.2.2. Viral burden decreases temporally in nasal turbinates and oropharyngeal area
Viral burden in the nasal turbinates of young WT and adult hACE2 hamsters gradually decreased from 2 d PE over time and was barely detectable by 8 d PE (Fig. 1D). Viral RNA amplified from oropharyngeal samples taken via swab or lavage from aged WT hamsters demonstrated a high initial viral burden of nearly 7 log10 gene copies per sample, which dropped to 4 log10 gene copies per swab by 8 d PE (Fig. 1E). hACE2 hamsters in the 100- and 10-PFU exposure groups had high viral RNA burdens at 2 d PE that decreased by 2 log10 at 5–6 d PE (Fig. 1E). Regrettably, nasal turbinate samples were not collected in aged WT hamsters.
3.2.3. SARS-CoV-2 is neuroinvasive in hACE2 hamsters
Low titers of virus were found in the brain tissue of aged WT and young WT hamsters exposed to 105 PFU at 2 d PE (Fig. 1F), but at no other time points, and no evidence of neuroinvasion was found histopathologically. In contrast, infectious SARS-CoV-2 was detected in the brain tissue from hACE2 hamsters in the 100- and 1-PFU exposure groups until the 5–6 d PE time point, when they all succumbed to disease. As expected, hACE2 hamsters exposed to100 PFU had the highest average viral titers in the brain, but, interestingly, the average viral titers in the 10-PFU-exposed group that succumbed to disease were slightly lower than those exposed to 1 PFU that succumbed. The hACE2 hamsters exposed to 0.1 PFU had the lowest average viral titers in the brain (Fig. 1F). No virus was detected in the brain tissue of hACE2 hamsters exposed to 1 or 0.1 PFU virus that survived to pre-determined study endpoints.
3.3. 18F-FDG PET and CT Imaging
3.3.1. Chest CT quantified by lung consolidation scores and analyzed by PCLH
After qualitative review by radiologists, semi-quantitative CT scoring and quantitative evaluation using PCLH were performed (Chefer et al., 2018).
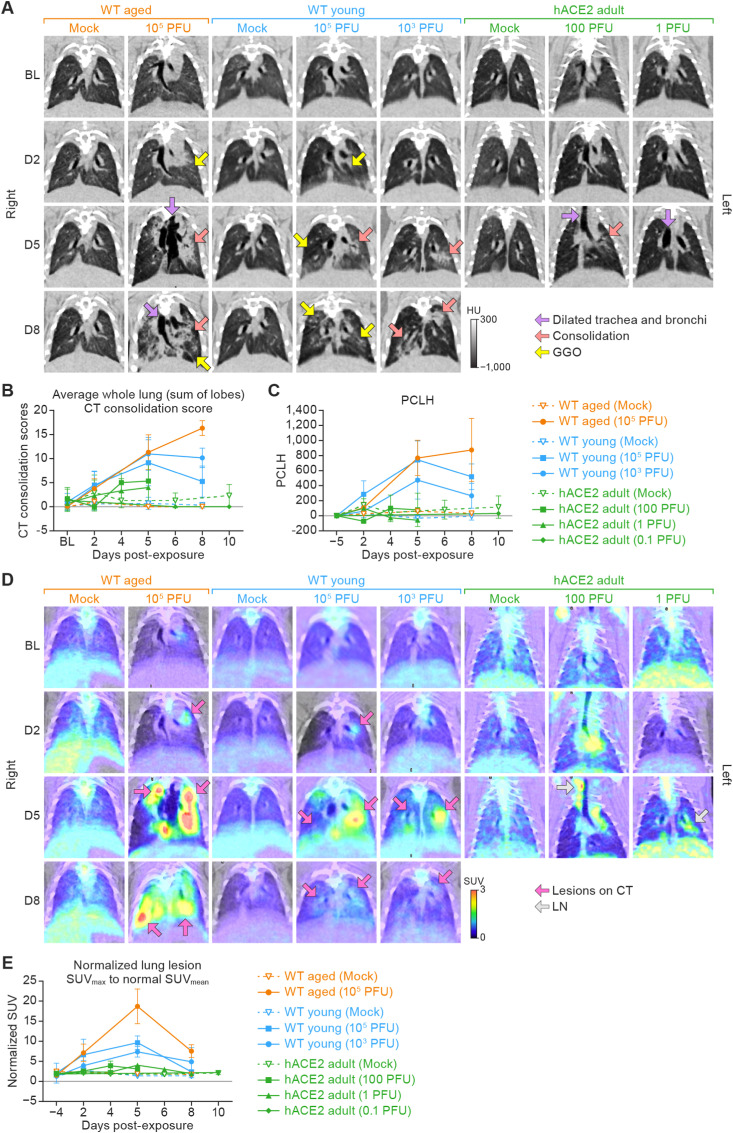
Aged WT (105 PFU) hamsters showed moderate multifocal bilateral GGOs, linear opacities, small foci of consolidation. In all four right lobes, pulmonary abnormalities worsened until 8 d PE (n = 3), as indicated by increasing consolidation scores (Fig. 2 A, red arrows, and Fig. 2B). By comparison, the mean scores for left-lobe CT consolidation scores showed that the pulmonary abnormalities peaked at 5 d PE (4.43 ± 1.13) and started to resolve at 8 d PE (3.75 ± 1.26). In one aged WT hamster, the pneumonia observed in the left lobe at 2 d resolved at 5 d, but the total lung CT consolidation score showed no improvement due to ongoing pneumonia in the right lobes. In three of six hamsters, a mild (one hamster) or severe, pneumomediastinum (two hamsters) that extended into the neck or left hilum was seen at 5 d PE (both underwent scheduled euthanized on 5 d). The observed differences in disease between right and left lung lobes are likely attributable to the anatomy of the hamster respiratory tract wherein the right main bronchus is larger than the left (Kennedy et al., 1978) which would facilitate increased virus deposition in the right lung lobe. Overall, the pulmonary abnormalities in aged WT hamsters first observed at 2 d worsened until 8 d (study end), as evidenced by progressive increases in whole-lung consolidation scores, which were 0.11 ± 0.33 (baseline), 3.78 ± 2.05 (2 d), 11.33 ± 3.62 (5 d), and 16.33 ± 1.53 (8 d) (Fig. 2B) and the quantitative PCLH were 0, 108.52 ± 73.10, 769.359 ± 235.75, and 875.77 ± 419.07 (Fig. 2C) at the same time points. Mock-exposed aged WT hamsters had whole-lung consolidation scores and PCLH values of 0 throughout the study.
Fig. 2.
Micro-PET/CT imaging of the lungs from three models of virus-exposed hamsters over an 8–10-d period.
A. Representative chest micro-PET/CT images in the coronal plane. Columns from left to right show lung images from groups of aged WT hamsters (exposed to mock inoculum and 105 PFU virus), young WT hamsters (exposed to mock inoculum, 105 PFU virus, and 103 PFU virus), and adult hACE2 hamsters (exposed to mock inoculum, 100 PFU virus, and 1 PFU virus). All hamsters were exposed to mock inoculum or SARS-CoV-2 intranasally. Rows from top to bottom are images show disease progress over time (baseline to 8 d). The scale bar indicates radiodensity value (in HU). B. Semi-quantitative average of the whole lung (sum of lobes) CT consolidation score (±SD) from three hamster models. C. Quantitative lung CT: average of PCLH (±SD) from three hamster models. D.18F-FDG PET images show 18F-FDG-avid areas, which generally correlated with CT lung consolidation. (PET images show the same lungs from panels in A.) The color bar indicates SUV. E. Quantitation of 18F-FDG uptake in lungs. Normalized SUVmax was calculated to reflect the longitudinal change of the 18F-FDG uptake in lungs at the different days post-exposure. Data are presented as the average of the normalized lung SUVmax (±SD). Imaging data were analyzed using GraphPad Prism 9.3.1. Two-way ANOVA with Tukey multiple comparison test was used to determine statistical significance. Micro-PET, micro-positron emission tomography; CT, computed tomography; WT, wild-type; hACE2, human angiotensin-converting enzyme 2; SARS-CoV-2, severe acute respiratory syndrome coronavirus-2; HU, Hounsfield Unit; SD, standard deviation; PCLH, percent change in lung hyperdense volume; 18F-FDG, 2-deoxy-2-[fluorine-18]fluoro-D-glucose; SUV, standardized uptake value; GGOs, ground-glass opacities; ANOVA, analysis of variance; BL, baseline; D2, day 2; D5, day 5; D8, day 8; LN, lymph node.
All virus-exposed young WT hamsters showed CT lung abnormalities at 2 d PE. As seen in aged WT hamsters, findings included moderate multifocal bilateral GGOs (Fig. 2A, yellow arrow), linear opacities, small foci of consolidation (Fig. 2A, red arrow). Infiltrations and multiple bilateral areas of consolidation were mostly present in the left lobe of all virus-exposed hamsters (Fig. 2A).
The radiological abnormalities in the lungs peaked at 5 d PE and increased with increasing exposure dose, with a mean CT consolidation score of 9.14 ± 4.78 and PCLH of 472.60 ± 249.91 for the hamsters exposed to 103 PFU SARS-CoV-2 and a mean CT consolidation score of 11.0 ± 3.41 and PCLH of 744.52 ± 251.24 for the hamsters exposed to 105 PFU virus. Lung disease improved in both young WT hamster dose groups at 8 d PE (Fig. 2A, yellow and red arrows), supported by decreases in whole-lung consolidation scores (Fig. 2B) and PCLH (Fig. 2C).
In contrast to WT hamsters, the lungs from SARS-CoV-2-exposed hACE2 hamsters at all exposure doses showed minor abnormalities at 2 d PE that worsened at 4 or 5 d but remained mild (Fig. 2A). CT consolidation scores were markedly less severe than those measured in either group of WT hamsters at 5 d (Fig. 2B), but disease biased towards the left lobe was still observed. The peak whole-lung CT consolidation score at 5 d was 5.33 and 4.00 for hACE2 hamsters exposed to 100 and 1 PFU, respectively (Fig. 2B). Similarly, the PCLH values for hACE2 hamsters at all exposure doses were far lower than those for WT hamsters (Fig. 2C). CT consolidation scores and PCLH values for the 0.1 PFU hACE2 group suggested no difference when compared to mock-exposed hACE2 hamsters. CT abnormalities at later time points could not be assessed for the hACE2 hamsters exposed to 100 or 10 PFU virus, because they succumbed to neurotropic meningoencephalitis at 5 or 6 d PE (Fig. 1A). hACE2 hamsters that were exposed to mock inoculum showed no significant CT abnormalities.
3.3.2. 18F-FDG uptake in lungs
SUV is a unitless measure of radioactive uptake within a defined volume of interest and is used to measure metabolic activity or pooling of radiotracers, such as 18F-FDG. Using micro-PET/CT, the normalized SUVmax only increased slightly in the 100- and 1-PFU dose groups but remained no greater than 4 throughout the study (Fig. 2D and E).
The average normalized lung SUVmax was 1.53 ± 0.23 (n = 9) in aged WT hamsters at baseline. At 2 d PE, 18F-FDG-avid lung lesions were evident in all aged WT hamsters, with a significantly increased normalized lung SUVmax of 7.13 ± 2.18 (n = 9). Pulmonary inflammation developed rapidly 2–5 d PE, manifested by an average normalized lung SUVmax of 18.7 ± 4.33 (n = 6) and then began to decline by 8 d PE to near the values measured at 2 d, with an average normalized lung SUVmax that decreased to 7.57 ± 1.60 (Fig. 2D and E).
The average normalized SUVmax for young WT hamsters exposed to 103 PFU was 1.31 ± 0.18 (n = 9) at baseline. A significant increase was observed at 2 d PE (3.93 ± 1.28, n = 9), with further increase observed at 5 d (7.38 ± 1.30, n = 3) in all young hamsters when compared to baseline. At 8 d PE, a mixed response was observed, with one hamster showing decreased normalized lung SUVmax (2.43 at 8 d versus 8.55 at 5 d), while the another showed a continued increase (7.43 at 8 d versus 1.58 at 2 d). In young WT hamsters exposed to 105 PFU, the normalized lung SUVmax showed a similar trend as seen in young WT hamsters exposed to 103 PFU, which was 1.29 ± 0.25 at baseline, increased at 2 d PE (6.08 ± 3.03, n = 4), peaked at 5 d (10.11 ± 1.18, n = 5), and decreased at 8 d (2.59 ± 0.46, n = 4) (Fig. 2D and E).
For the hACE2 hamsters, the normalized SUVmax only increased slightly in the 100- and 1-PFU exposure dose groups but remained no greater than 4 throughout the study (Fig. 2E). The normalized SUVmax for the 10-PFU dose group did not appreciably increase throughout the course of the experiment. SUVmax remained below 3 in all mock-exposed, hACE2 hamsters throughout the study (Fig. 2D and E). The normalized SUVmax for the 0.1-PFU dose group did not appreciably increase above that of the mock-exposed animals throughout the course of the study. Mock-exposed hACE2 hamsters showed no significant CT lung abnormalities, and the normalized lung SUVmax remained below 3 in all mock-exposed hACE2 hamsters throughout the study (Fig. 2E).
To summarize the imaging findings in the lungs, 18F-FDG PET/CT demonstrated that aged WT hamsters develop more severe and protracted pneumonia and hACE2 hamsters develop mild pneumonia after SARS-CoV-2 exposure.
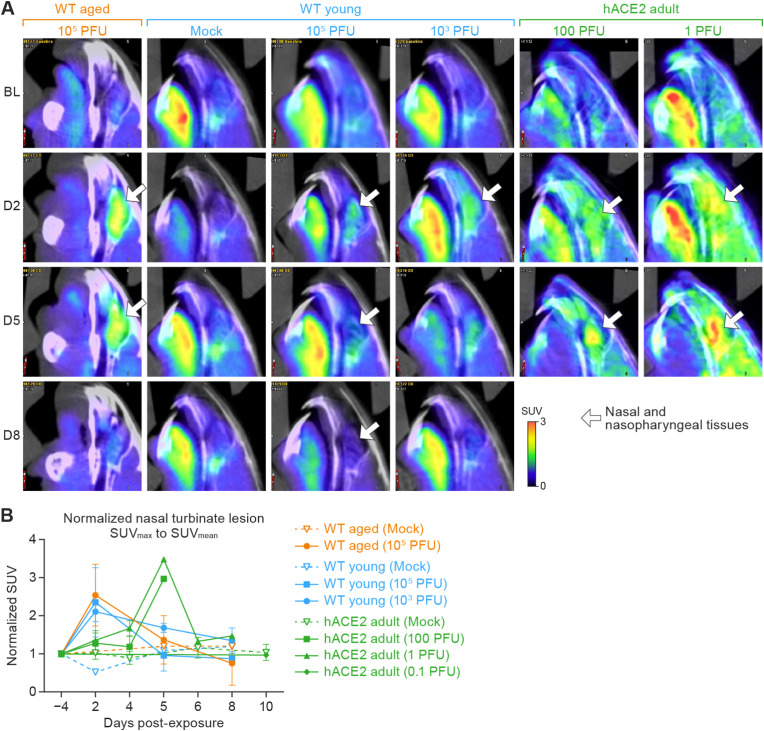
3.3.3. 18F-FDG PET of nasal tissues and nasopharyngeal structures
Increased 18F-FDG uptake in the posterior nasal soft tissues was observed at 2 d PE in aged WT hamsters exposed to 105 PFU, young WT hamsters exposed to 105 PFU, and young WT hamsters exposed to 103 PFU (Fig. 3 A, white arrows). The SUVmean then decreased 5–8 d PE. Accordingly, the normalized nasal and nasopharyngeal tissue SUVmean, compared to baseline as ratios were 2.54 ± 0.81, 2.36 ± 0..26, and 2.11 ± 0.91 for the aged WT (105 PFU), young WT (105 PFU), and young WT (103 PFU) hamsters, respectively (Fig. 3A and B). The normalized nasal and nasopharyngeal tissue SUVmean subsequently decreased to baseline levels at 5 d and remained stable through 8 d.
Fig. 3.
18F-FDG uptake determined by tracking of the posterior nasal soft tissues using micro-PET/CT images.
A. Representative 18F-FDG micro-PET/CT images from groups of WT aged hamsters (105 PFU), young hamsters (mock, 105 PFU, and 103 PFU) and adult hACE2 hamsters (100 and 1 PFU) exposed to SARS-CoV-2 intranasally with different doses (columns from left to right) in the sagittal plane show metabolic activities located in the regions of interests (nasal tissues; white arrow) for each indicated study day (rows from top to bottom). B. Quantitation of 18F-FDG uptake in nasal cavities. Normalized nasal SUVmean was calculated to reflect the longitudinal change of the 18F-FDG uptake in the nasal and nasopharyngeal structures at the different days post-exposure. Data are presented as the mean percentage of the normalized nasal SUVmean (±SD). Imaging data were analyzed using GraphPad Prism 9.3.1. Two-way ANOVA with Tukey multiple comparison test was used to determine statistical significance. 18F-FDG, 2-deoxy-2-[fluorine-18]fluoro-D-glucose; micro-PET, micro-positron emission tomography; CT, computed tomography; WT, wild-type; hACE2, human angiotensin-converting enzyme 2; SARS-CoV-2, severe acute respiratory syndrome coronavirus-2; SUV, standardized uptake value; SD, standard deviation; ANOVA, analysis of variance; BL, baseline; D2, day 2; D5, day 5; D8, day 8.
In the hACE2 hamsters exposed to 100 PFU, the normalized nasal and nasopharyngeal tissue SUVmean showed an increasing trend until euthanasia or succumbing to infection at 5 d. The SUVmean in the nasal and nasopharyngeal tissue of the hACE2 hamster exposed to 1 PFU that survived past 6 d was highest at 5 d, decreased at 6 d, and remained stable through 8 d (Fig. 3A and B). All mock-exposed WT and hACE2 hamsters showed stable 18F-FDG uptake with normalized SUVmean around 1.0 in the nasal and nasopharyngeal areas throughout the study.
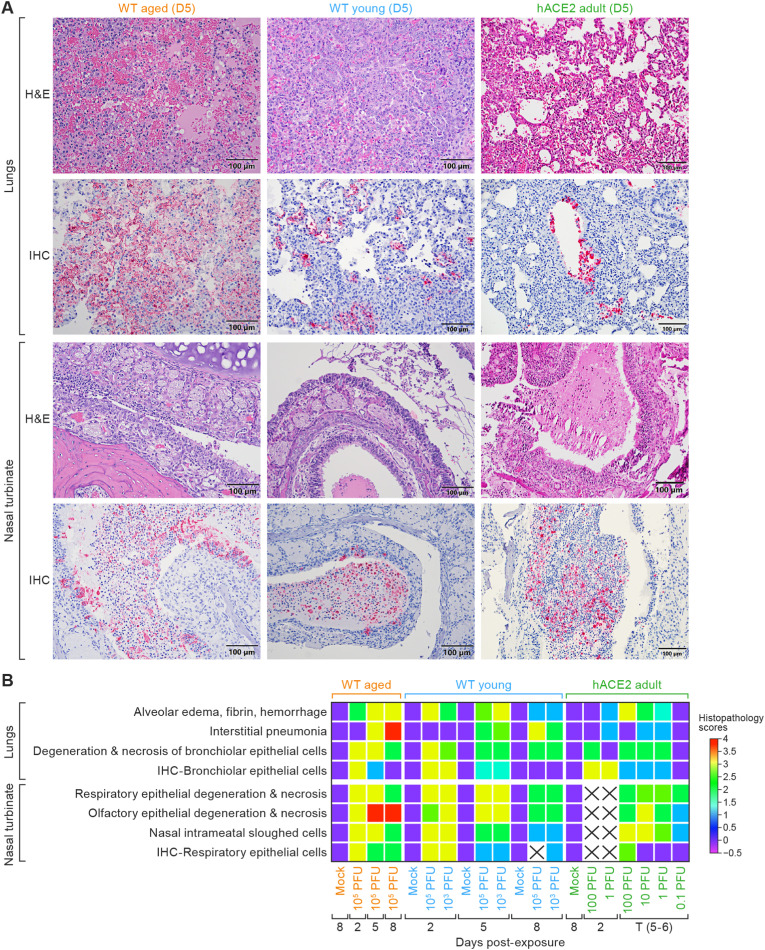
3.4. Lung histopathology corresponded with CT and micro-PET/CT findings
3.4.1. Histopathology in lungs demonstrate aged WT hamsters develop more severe and protracted pneumonia and hACE2 hamsters develop mild pneumonia following SARS-CoV-2 exposure
Histopathology in the lung tissue of aged WT, young WT, and hACE2 hamsters strongly correlated with CT and PET/CT findings. In aged WT hamsters at 2 d PE a mild to moderate necrohemorrhagic interstitial pneumonia with pulmonary edema and fibrin exudation, epithelial cell degeneration and necrosis, and vascular leakage with leukocyte migration across the walls of pulmonary veins was seen (Fig. 4 B and Table 1 ). Semiquantitative interstitial pneumonia scores (Fig. 4B, left columns) did not differ from young WT hamsters (Fig. 4B, middle columns) at this time point. However, by 5 d, aged WT hamsters showed more severe intra-alveolar hemorrhage but less interstitial pneumonia (Fig. 4A, upper-left panel) when compared to young WT hamsters (Fig. 4A, upper middle panel). IHC showed that the intensity and number of SARS-CoV-2-positive cells in aged WT hamsters (Fig. 4A, lower-left panel) was higher than that observed in young WT hamsters (Fig. 4A, lower middle panel) at 5 d. By 8 d PE, aged WT hamsters showed an ongoing, multifocal to coalescing, moderate to severe necrohemorrhagic interstitial pneumonia with a severe alveolar hemorrhage, fibrin, and edema when compared to young hamsters (Fig. 4B and Table 1). IHC for SARS-CoV-2 was scant but positive in the lung tissue from all three aged hamsters at 8 d PE (Fig. 4B, left columns, and Table 1).
Fig. 4.
Histopathologic changes in the lungs and nasal turbinates.
A. Histopathology (row 1: lungs; row 3: nasal turbinates) and immunochemistry (row 2: lungs; row 4: nasal turbinates) from aged WT, young WT, and hACE2 hamsters at 5 d (WT 105 PFU and hACE2 100 PFU, respectively). At 5 d PE, aged WT hamsters showed a hemorrhagic interstitial pneumonia (upper-left panel) that lacked the robust, adenomatous, type II pneumocyte hyperplasia (upper-middle panel) seen in young hamster's lungs, while hACE2 hamsters exposed to 100 PFU of virus showed only a mild interstitial pneumonia at the same post-exposure time point. B. A heat map of histopathology scores in the lungs and nasal turbinates by day post-exposure and SARS-CoV-2 exposure dose in the three hamster models. An “X” indicates insufficient sample collection for comparison. Data were analyzed using GraphPad Prism 9.3.1, Two-way ANOVA with Tukey multiple comparison test. (WT aged 105 PFU in comparison to hACE2 100 PFU, degeneration necrosis and interstitial pneumonia p < 0.05*). WT, wild-type; hACE2, human angiotensin-converting enzyme 2; PE, post-exposure; IHC, immunohistochemistry; SARS-CoV-2, severe acute respiratory syndrome coronavirus-2; NP, nucleocapsid protein; ANOVA, analysis of variance; D5, day 5; H&E, hematoxylin and eosin.
Table 1.
Histopathology findings in respiratory track of hamsters exposed to SARS-CoV-2 at each day post-exposure.
| Day 2 |
Day 5 |
Day 8 |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Strain/Age | HE | IHC | ISH | HE | IHC | ISH | HE | IHC | ISH |
| Lung | |||||||||
| Aged WT | Similar lesions as seen in young WT hamsters | +++ 3/3 |
+++ 3/3 |
As same as 2 d, but more severe necrohemorrhagic and lesser interstitial pneumonia | +++ 3/3 |
+++ 3/3 |
As same as 5 d, still predominated by necrohemorrhagic pneumonia | ++ 3/3 |
+ 1/3* - 2/3 |
| Young WT | Multifocal, mild to moderate, necrohemorrhagic pneumonia, bronchiolar intraluminal hemorrhagic exudates and cell debris, and vascular leakage and leukocyte migration in pulmonary veins | +++ 6/6 |
++ 1/6 +++ 5/6 |
Multifocal, mild to moderate mixture of hemorrhagic and interstitial (glandular pattern of hypertrophic and hyperplastic type II pneumocytes) pneumonia, bronchiolar epithelial hypertrophy and hyperplasia forming folds with syncytial cells |
+ 6/6 |
+ 2/6 +++ 1/6 - 3/6 |
Early alveolar and bronchiolar repair characterized by alveoli and bronchioles lined by hyperplastic, flattened type II pneumocytes or cuboidal epithelial cells, respectively, and no or small numbers of intra-alveolar macrophages | + 1/6* |
- 6/6 |
| hACE2 adult | No significant lesions (1/3). Minimal (1/3) to mild (1/3), multifocal, lymphohistiocytic bronchointerstitial pneumonia | +++ 3/3 |
<25% 3/3 |
Multifocal, minimal (2/3) to mild (1/3) hemorrhagic and interstitial pneumonia | +++ 3/3 |
<25% 3/3 |
ND | ND | ND |
| Nasal Turbinates | |||||||||
| WT aged | Similar lesions | +++ 3/3 |
+++ 3/3 |
As same as 2 d, predominated by epithelial cell degeneration and necrosis | +++ 3/3 |
+++ 3/3 |
As same as 5 d, still presented with multifocal, mild (1/3) to severe (2/3) olfactory epithelial degeneration and necrosis | + 1/3 +++ 2/3 |
+ 1/3** |
| WT young | Multifocal, acute rhinitis characterized epithelial (respiratory, olfactory, and submucosal glandular) degeneration and necrosis with loss of cilia, inflammatory infiltrates of mucosa and submucosa, large amount/numbers of hemorrhagic exudates, sloughed epithelial cells, and degenerate neutrophils. | +++ 6/6 |
+++ 6/6 |
Multifocal, moderate to severe respiratory epithelial hypertrophy and hyperplasia or squamous metaplasia, olfactory epithelia attenuation by irregular hyperplastic epithelial cells, and lesser exudate in the nasal cavities. | + 6/6** |
++ 2/6 ++ 2/6 |
Respiratory epithelium was partially repaired either by normal pseudostratified or metaplastic squamous epithelia. The olfactory epithelium was partially repaired by multiple irregular layers of elongated, basophilic, hyperplastic epithelial cells. | − (6/6)** |
− 6/6** |
| hACE2 adult | Mild (1/3) to moderate (2/3), multifocal to locally extensive, neutrophilic and lymphohistiocytic rhinitis | +++ 3/3 |
ND | Minimal (1/3) to mild (1/3) to moderate (1/3), multifocal to locally extensive, neutrophilic rhinitis | +++ 1/3 |
ND | ND | ND | ND |
Abbreviations: WT, wild-type; hACE2, human angiotensin-converting enzyme 2; HE, hematoxylin; IHC, immunohistochemistry; ISH, in situ hybridization; ND, not done.
Notes: +: weakly positive; ++: moderately positive; +++: strongly positive. * focal, <5 positive cells in a young hamster; ** Rare intraluminal cell debris was weakly positive in some hamsters.
The histopathological findings in the lungs of young WT hamsters at 2 d PE were similar to those seen in aged WT hamsters (Fig. 4B and Table 1). Interestingly, there were no significant differences in the histopathologic lesions in the lungs between the 105 and 103 PFU exposure doses in young WT hamsters. Young WT hamsters from both exposure dose groups showed multifocal weak to strong SARS-CoV-2 antigen positivity in bronchiolar epithelial cells, type II pneumocytes, macrophages, and luminal-sloughed cell debris (Table 1). Young hamsters from both exposure dose groups showed multifocal weak to strong SARS-CoV-2 antigen positivity in bronchiolar epithelial cells, type II pneumocytes, macrophages, and luminal-sloughed cell debris (Table 1). At 5 d PE, young WT hamsters showed a robust multifocal glandular pattern (adenomatosis) of interstitial pneumonia characterized by densely packed hypertrophic and hyperplastic type II pneumocytes admixed with less hemorrhage and fewer inflammatory cells (Fig. 4A, upper-middle panels, and Table 1). By 8 d, young WT hamsters showed signs of alveolar and bronchiolar epithelial regeneration and repair (Fig. 4B) that was not evident in aged WT hamsters. Alveoli were lined by more flattened type II pneumocytes, the alveolar spaces were air-filled or contained small numbers of macrophages. IHC for SARS-CoV-2 was positive in only one of three young WT hamsters at 8 d (Table 1).
Histopathology in the lung tissue of hACE2 hamsters exposed to 100 PFU SARS-CoV-2 showed only a mild to moderate, multifocal, lymphohistiocytic bronchointerstitial pneumonia (Fig. 4A, upper-right panel, and Table 1) and the severity of pneumonia lessened in an exposure-dose-dependent manner. Semiquantitative histopathology scores of the pneumonia showed that the pneumonia in hACE2 hamsters was significantly milder than that seen in both aged and young WT hamsters (Fig. 4B, right columns). IHC for SARS-CoV-2 NP in the lung tissue was positive in all hACE2 hamsters at 2 d except for 1 exposed to 100 PFU (Fig. 4A, lower-right panel) and staining intensity and distribution decreased by 5 d PE and by exposure dose (Fig. 4B, right columns and Table 1). hACE2 hamsters that survived to scheduled study endpoints (1 exposed to 1 PFU and five of eight exposed to 0.1 PFU) were grossly and histopathologically within normal limits and showed no immunohistochemical positivity for SARS-CoV-2 NP.
3.4.2. Histopathology in the nasal turbinates showed persistent rhinitis in aged WT hamsters exposed to SARS-CoV-2 but resolution in young WT hamsters exposed to the same virus dose
The rhinitis seen at 2 d PE was similar in both young and aged WT hamsters and characterized by a multifocal acute, exudative rhinitis, with infiltration of large numbers of mixed inflammatory cells and mild to moderate epithelial (respiratory, olfactory, and submucosal glandular) degeneration and necrosis with loss of cilia (Fig. 4A, upper and middle panels, and Table 1). By 5 d, young WT hamsters showed multiple layers of reactive, hyperplastic mucosal olfactory epithelial cells with squamous metaplasia (Fig. 4A, upper-middle panel). Similar, but milder olfactory epithelial regeneration was observed in aged WT hamsters (Fig. 4A, upper-left panel). IHC for SARS-CoV-2 NP showed a small number of positive epithelial cells (both respiratory and olfactory) in young WT hamsters (Fig. 4A, lower middle panel, and Table 1). In contrast, all three aged WT hamsters showed strong SARS-CoV-2 antigen positivity in epithelial cells (respiratory, olfactory, and submucosal glandular; Fig. 4A, lower-left panel and Table 1). By 8 d, the respiratory and olfactory epithelium from young WT hamsters continued to show evidence of repair (Fig. 4B, middle columns and Table 1). By 8 d, IHC for SARS-CoV-2 was negative in the in all young WT hamsters (Table 1) In contrast, all three aged WT hamsters showed strong SARS-CoV-2 NP antigen positivity in epithelial cells (respiratory, olfactory, and submucosal glandular).
At 2 d hACE2 hamsters exposed to 100 PFU of SARS-CoV-2 and necropsied showed a mild to moderate, locally extensive, necrotizing, exudative neutrophilic and lymphohistiocytic rhinitis with multifocal erosion and ulceration that decreased in severity by exposure dose. By 5 d PE, the rhinitis was similar but with evidence of reactive mucosal epithelial cell hyperplasia (Fig. 4A, upper-right panel, Fig. 4B, right columns and Table 1).
IHC for SARS-CoV-2 NP at 2 d in hACE2 hamsters showed strong, multifocal to locally extensive positivity in the respiratory epithelium and sustentacular cells. By 5 d, IHC positivity was more variable (14 of 17 showed IHC positivity for SARS-CoV-2; Fig. 4A, lower-right panel). The single hACE2 hamster exposed to 1 PFU of virus that survived to the scheduled study endpoint at 14 d showed no significant histopathologic lesions in the nasal turbinates and no IHC positivity for SARS-CoV-2 NP (Fig. 4B, right columns). Detailed histopathologic descriptions are shown in Table 1.
Of the tissues collected at the time of necropsy, histopathologic lesions were limited to the lung and nasal cavity in all hamsters, and the lung, nasal cavity, and brain, and heart in hACE2 hamsters. No significant lesions or IHC positivity for SARS-CoV-2 NP were seen in other tissues examined.
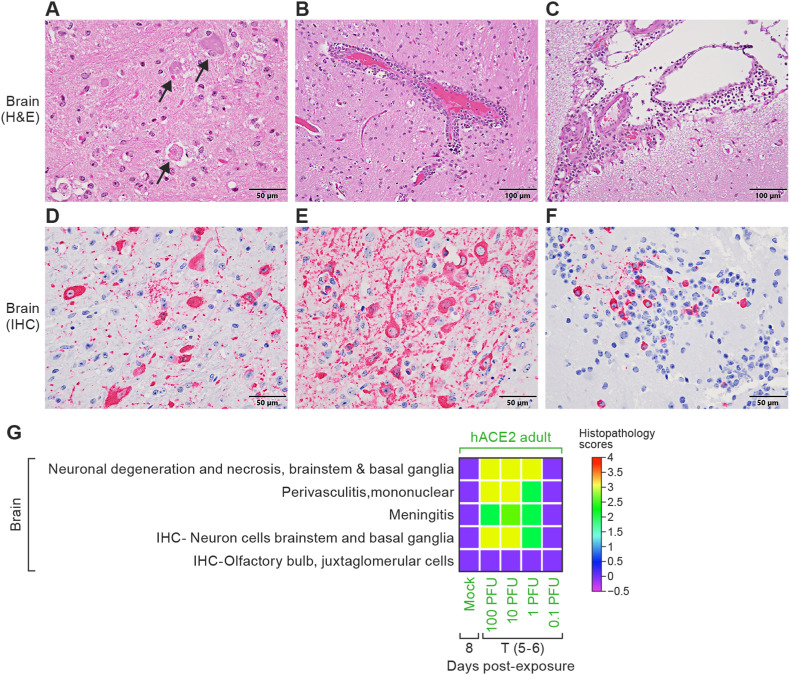
3.4.3. SARS-CoV-2 is neurotropic in hACE2 hamsters but not in WT hamsters
In one of three euthanized hACE2 hamsters, SARS-CoV-2 neurotropism was first observed at 2 d PE with strong IHC SARS-CoV-2 NP cytoplasmic positivity in caudal brainstem neurons, despite negative SARS-CoV-2 titers in the brain tissue at this time point. By 4 d PE, neuronal necrosis was multifocal and moderate (Fig. 5 A). All hACE2 hamsters showed a mild to moderate multifocal to locally extensive perivascular lymphoplasmacytic meningoencephalitis (Fig. 5B and C and 5G) extending from the proximal to distal brainstem with bilaterally symmetrical, strong, widespread cytoplasmic positivity for SARS-CoV-2 in neurons (Fig. 5D–G) with similar histopathologic lesions and IHC SARS-CoV-2 neuronal positivity was seen in hACE2 hamsters exposed to 1 PFU that succumbed between 4–6 d PE (Fig. 5G). The hACE2 hamster exposed to 1 PFU that survived to the scheduled study endpoint (14 d) showed a mild perivascular encephalitis but IHC for SARS-CoV-2 NP negative in the brain tissue examined. Brain tissue from the five hACE2 hamsters exposed to 0.1 PFU virus and mock-exposed hamsters that were euthanized at study end at 14 d PE and mock-exposed controls were histopathologically normal and negative for SARS-CoV-2 antigen (Fig. 5G). Despite low viral titers in the brain tissue from young and aged WT hamsters exposed to 105 PFU virus at 2 d, no histopathologic lesions were seen in the brain and IHC for SARS-CoV-2 was negative in all WT hamsters at all PE time points examined (Fig. 5G).
Fig. 5.
Brain histopathology of hACE2 hamsters.
Histopathology (A–C) and IHC (D–F) of the brain and olfactory bulb from one hACE2 hamster that sucumbed to disease at 5 d. A. Moderate, multifocal, neuronal necrosis of brainstem neurons (black arrows). B. Dense, perivascular lymphoplasmacytic cuffing. C. Lymphoplasmacytic meningitis. D. Strong cytoplasmic positivity in brainstem neurons for SARS-CoV-2 NP antigen. E. Most neurons in the basal ganglia showed strong cytoplasmic positivity for SARS-CoV-2 NP. F. Scattered juxtaglomerular cells within the olfactory bulb showed SARS-CoV-2 NP antigen positivity. G. Correlation heat map of brain histopathology and IHC scores among hACE2 hamsters. Data were analyzed using GraphPad Prism 9.3.1, Two-way ANOVA with Tukey multiple comparison test (at 5 d post-exposure, hACE2 100 PFU versus hACE2 0.1 PFU; p < 0.001 is ***). hACE2, human angiotensin-converting enzyme 2; SARS-CoV-2, severe acute respiratory syndrome coronavirus-2; NP, nucleoprotein; ANOVA, analysis of variance; H&E, hematoxylin and eosin; IHC, immunohistochemistry; T (5–6), terminal.
3.5. Immune response to SARS-CoV-2 exposure
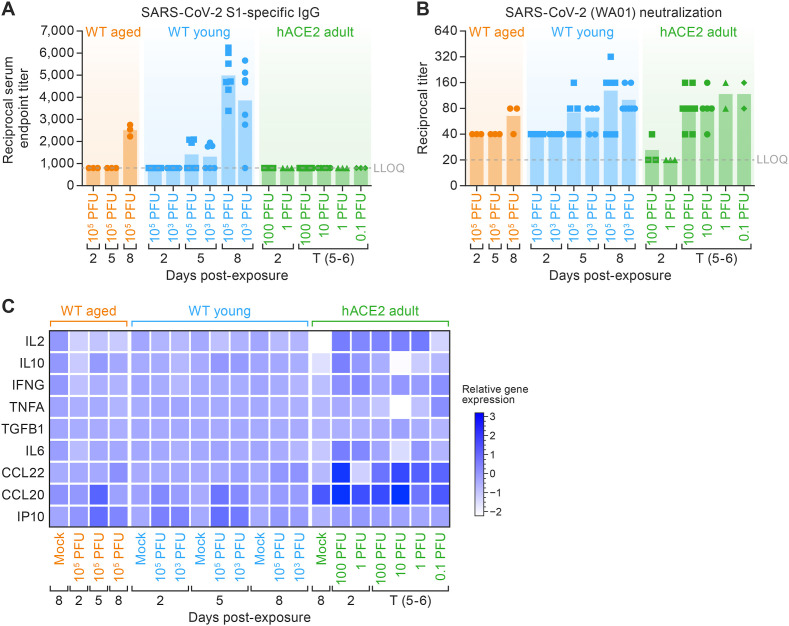
3.5.1. Aged WT hamsters and hACE2 hamsters showed a less robust and delayed humoral immune response when compared to young WT hamsters
An IgG ELISA against the SARS-CoV-2 S1 protein was developed to assess the temporal development and magnitude of the humoral immune response (Fig. 6 A). Young WT hamsters showed an IgG response against the S1 protein at 5 d PE that correlated with exposure dose and was robust by 8 d PE (Fig. 6A). Aged WT hamsters did not develop appreciable IgG titers against the S1 protein until 8 d. At 8 d (endpoint), titers in young WT hamsters were significantly higher than those measured in aged WT hamsters (p < 0.001). No appreciable IgG antibody responses were detected against the S1 protein in hACE2 hamsters by 5 d at any exposure dose (Fig. 6A).
Fig. 6.
Humoral systemic immune response and SARS-CoV-2 acute local immune gene activation.
A. Median of endpoint SARS-CoV-2-specific IgG antibody in serum obtained at the indicated study days and measured by ELISA. (IgG significantly increased in WT young 105 PFU and 103 PFU with respect to WT aged 105 PFU, p < 0.001*** and p < 0.05*). B. Reciprocal live virus neutralization titers in serum obtained at the indicated days post-exposure. Horizontal bars show the median. Points indicate data from individual hamsters. (WT aged 105 PFU in comparison to hACE2 100; p < 0.05*) C. Heat map of cytokine and chemokine protein expression levels in lung homogenates collected from the three hamster models from different SARS-CoV-2 dose groups at the indicated time points. Heatmap are a representation of the log2 fold change of selected hamster chemokine/cytokine RNA levels as determined by RT-qPCR on lung extracts and normalized against RPL18 mRNA levels. (IL2 reduced significantly in WT aged and young 105 PFU in comparison to hACE2 100 and 10 PFU; p = 0.019 is * and p = 0.0003 is ***. IL10 significantly increased WT aged and young; p < 0.0063 is * and p < 0.001 is ***.) SARS-CoV-2, severe acute respiratory syndrome coronavirus-2; IgG, immunoglobulin G; ELISA, enzyme-linked immunosorbent assay; WT, wild-type; hACE2, human angiotensin-converting enzyme 2; RT-qPCR, real-time reverse transcription polymerase chain reaction; mRNA, messenger RNA; IL, interleukin; WA01, Washington isolate; IFNG, interferon gamma; TNFA, tumor necrosis factor alpha; TGFB1, transformation growth factor beta 1; CCL, C–C motif ligand; IP-10, IFNG-induced protein 10 kDa; T (5–6), terminal.
Neutralizing antibody titers in SARS-CoV-2-exposed hamsters was quantified using an in vitro live-virus neutralization assay (Bennett et al., 2021) to determine if viral clearance was due to the action of neutralizing antibodies. We observed a strong correlation between S1 IgG antibody responses and neutralization endpoint titers among aged and young hamsters. By 5 d PE, young WT hamsters and hACE2 hamsters exhibited an appreciable level of neutralizing antibody, which increased further by 8 d PE in young WT hamsters. These findings suggested that neutralizing antibody capacity of young hamsters significantly outpaced the timing and extent of SARS-CoV-2 neutralizing capacity in hACE2 and aged hamsters (Fig. 6B).
3.5.2. Chemokine profiles in the lungs were significantly elevated at early (2 d PE) and late (5 d PE) time points in hACE2 hamsters relative to WT hamsters
Lung tissues from aged and young WT hamsters were examined by RT-qPCR for cytokine gene-expression profiles. In the lungs of aged WT hamsters, an early trend (2 d PE) of reduced expression of numerous cytokine transcripts associated with an active inflammatory response (IL2, IL6, IL10, and IFNG) was seen (Fig. 6C, left columns) relative to mock-exposed controls; however, this early trend was absent from young WT hamsters (Fig. 6C, middle columns). Additionally, young and aged WT hamsters exhibited enhanced local gene expression of the chemokines CCL20 and IP-10, although this occurred early in the young hamsters (at 2 d) and later in aged hamsters (at 5 d). By 8 d PE, the overall gene-expression profiles in the lungs of both young and aged WT hamsters had largely returned to baseline—excluding trending increases in CCL20, CCL22, and IP-10 in young hamsters and CCL22 and IP-10 in aged hamsters.
Interestingly, the transcripts for IL2, IL6, IL10, and IFNG were elevated early in hACE2 hamsters (Fig. 6C, right columns), and these increases occurred in a dose-dependent manner. TNF expression remained comparable to baseline in WT hamsters, though 0.1-PFU dose hACE2 adult show elevated level of TNF (p < 0.05). TGF expression was found to be WT aged > WT young > hACE2 adult hamster at 2 d (p < 0.05).
At baseline, CCL20 expression was higher in the hACE2 mock-exposed group when compared to WT hamsters, and CCL20 expression was significantly higher in hACE2 hamsters when compared to WT hamsters (p < 0.05) at all post-exposure time points. As noted, IP-10 expression did not increase in the hACE2 hamsters at any time point and infectious dose. CCL22 expression is comparable in WT young and WT aged hamster at 5 d PE. The order was hACE2 adult > WT young > WT aged (p < 0.001).
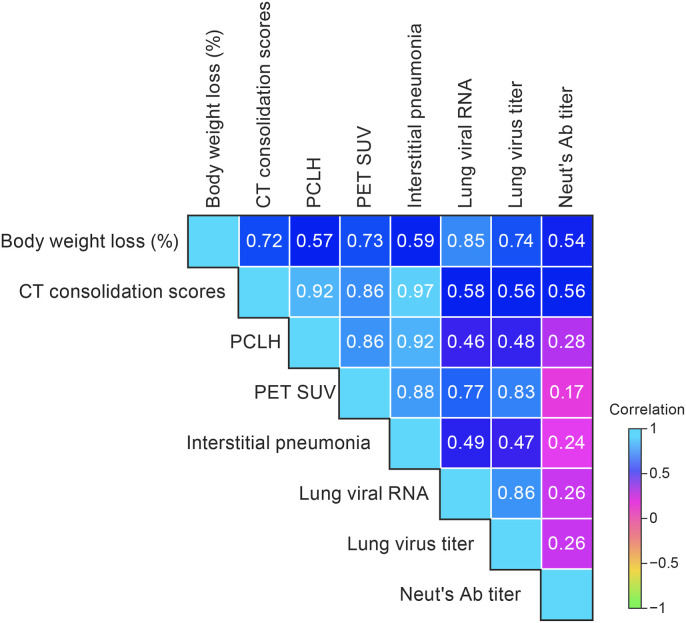
3.6. In vivo imaging assessments of pulmonary disease strongly correlated with disease outcomes including body weight loss, viral load, and semiquantitative histopathology interstitial pneumonia scores
Pearson correlation analyses were performed on data collected at 5 d PE (peak disease) to determine if there were correlations between in vivo micro-PET/CT imaging, lung histopathology, and clinical outcomes. Pearson correlation coefficients were calculated for body weight loss, CT whole-lung consolidation scores, PCLH, normalized SUVmax, lung histopathology (interstitial pneumonia), and viral load in lung tissues (RT-qPCR and infectious virus titers) as well as neutralizing antibody titers (Fig. 7 ). The strongest positive correlations were seen between CT whole-lung consolidation scores and interstitial pneumonia histopathology scores (0.97), CT whole-lung consolidation scores and PCLH (0.92), CT whole-lung consolidation scores and PET SUVmax scores (0.86). We found that percent body weight loss was highly correlated with whole-lung CT scores (0.72) and with lung viral load (RT-qPCR, 0.85; plaque assay, 0.74). Viral load, measured by RT-qPCR and plaque assay, correlated well (0.86); and both correlated well with PET SUV scores (0.77 and 0.83, respectively). SARS-CoV-2 neutralizing antibody titers did not correlate well as with other outcomes (range 0.17–0.56; Fig. 7).
Fig. 7.
Correlation heatmap of imaging, pathology, and clinical data.
Spearman correlations of lung imaging (CT consolidation score, PCLH, and normalized SUVmax), histopathology scores (interstitial pneumonia), clinical signs of disease (weight loss, viral shedding, lung titers) and neutralizing antibody titers at 5 d (Spearman correlation analysis, p < 0.05; significant correlations are shown in red). CT, computed tomography; PCLH, percent change in lung hyperdense volume; SUV, standardized uptake value; PET, positron emission tomography; Ab, antibody.
4. Discussion
As of this writing, no animal model recapitulates every feature of severe COVID-19-associated pneumonia; most models show a mild to moderate disease course. While WT hamsters are naturally susceptible to SARS-CoV-2 infection and consistently develop an exposure-dose-dependent rhinitis and bronchointerstitial pneumonia following IN exposure (Rosenke et al., 2020; Sia et al., 2020), they fail to develop the severe diffuse alveolar damage with hyaline membrane formation and thrombosis often seen in patients requiring hospitalization (Barton et al., 2020; von Stillfried et al., 2022). Disease severity also differs among SARS-CoV-2 variants, presenting a further obstacle to model development (Carroll et al., 2022; Cochin et al., 2022; Halfmann et al., 2022; Su et al., 2022; Toomer et al., 2022; Yuan et al., 2022). Early in the pandemic, advanced age became a well-known risk factor for developing severe COVID-19 and led to investigation of aged hamsters as a potentially improved model of severe disease. While the pathophysiologic mechanisms underlying higher morbidity and mortality seen in the elderly with COVID-19 are complex and not completely understood (Bartleson et al., 2021), the data reported here support the previous literature (Osterrieder et al., 2020), showing that aged hamsters would be an improved, although imperfect, model of severe COVID-19 in the elderly when compared to either young WT or hACE2 hamsters.
No studies have characterized and evaluated the aged WT, young WT, and hACE2 hamster models in parallel using 18F-FDG PET/CT. The goal of this study was to use 18F-FDG micro-PET/CT to further characterize and compare the disease progression and severity of SARS-CoV-2 in aged (60-week-old) WT and young (5–6-week-old) WT hamsters to the less-well-studied adult (22-24-week-old) hACE2 (Golden et al., 2022) hamster model using commonly reported measures of disease severity including body weights, viral burden in the upper respiratory tract and lungs, and serial histopathology. In humans, chest CT is considered to be among the most accurate diagnostic modalities of SARS-CoV-2 infection (Benameur et al., 2021). Accordingly, we utilized serial high-resolution respiratory-gated chest CT and whole-body 18F-FDG micro-PET/CT to assess the morphological and metabolic changes during disease progression and regression, quantitatively and non-invasively.
In this study, we utilized a semi-automated lung-segmentation tool aided by artificial intelligence and a quantitative radiodensity-based parameter (PCLH) for reliable and unbiased analyses of chest CTs in SARS-CoV-2-exposed hamsters. Respiratory-gated chest CTs were evaluated by an experienced radiologist. We found the whole-lung CT consolidation scores and PCLH to be sensitive and specific methods to detect and follow the progression of SARS-CoV-2-induced lung lesions in the hamster model.
Additionally, while not conventionally used as a primary modality, 18F-FDG PET, first developed and utilized by our group in a SARS-CoV-2 hamster model, is able to identify inflammation earlier and with greater sensitivity than anatomic imaging modalities used in patients with COVID-19 (Bai et al., 2021; Jacobi et al., 2020). Typical CT manifestations, such as GGOs with or without superimposed consolidation, are appreciated in peripheral, basilar, and posterior predominant patterns with increased metabolic activity on 18F-FDG PET imaging (Jacobi et al., 2020). High 18F-FDG uptake at the sites of infection and inflammation are associated with inflammatory responses, such as proinflammatory cytokines and migration of inflammatory cells, focal hyperemia, and increased vascular permeability (Eibschutz et al., 2022). It is notable that the aged WT hamsters that had the highest normalized lung SUVmax at 5 d PE also had the most significant histopathological findings, including bilateral hemorrhagic interstitial pneumonia, multifocal alveolar edema, and fibrin deposition, as well as infiltrations of mixed inflammatory cells (lymphocytes, plasma cells, macrophages, and/or neutroophils) within the lung interstitium. Activated inflammatory cells are characterized by high-energy consumption and upregulated glucose transporter expression and therefore transport more of the glucose analog 18F-FDG into their cytoplasm. Consequently, this study demonstrates the direct correlation between the 18F-FDG uptake and the metabolic activity of cells at sites of inflammation.
The changes in the normalized lung SUVmax correlated well with whole-lung CT consolidation scores and PCLH in all groups up to 5 d PE (Fig. 7). At 8 d PE, the aged WT hamsters (105 PFU) had decreased metabolic activity in the lungs, while the whole-lung CT score and PCLH remained elevated. However, our quantitative micro-PET analysis used normalized lung SUVmax, which characterized the signal intensity of the 18F-FDG-avid lesions but not the volume affected. A combination of hypermetabolic volume and SUVmax has been developed to classify inflammation in patients with COVID-19, with a low level of inflammation defined by a hypermetabolic volume less than 50 mL and SUVmax less than 7, and any values above those levels considered a high level of inflammation (Dietz et al., 2021). We may adapt this method for use in the hamster COVID-19 model for future imaging studies to improve our ability to quantify the extent of inflammation in the lungs during the course of infection.
Viral burden in the lungs also correlated with the severity of the pneumonia in WT hamsters, peaking in aged and young WT hamsters at 2 d PE and decreasing at 5 and 8 d. This pattern correlated with the intensity and distribution of the IHC staining in the lungs for SARS-CoV-2 NP. Viral burden remained higher in aged WT hamsters compared to young WT hamsters, consistent with the more protracted pulmonary disease course observed in WT hamsters and similar to findings of a previous study in aged hamsters by Selvaraj et al. which, however, reported fatal disease at 5–8 d PE in 56–86-week-old WT hamsters exposed IN to 105 tissue culture infectious dose (TCID50; 5–7x104 PFU virus) (Selvaraj et al., 2021). We did not observe fatal disease using a 105 PFU dose of the same SARS-CoV-2 variant administered IN 60-week-old WT hamsters, nor did a study in aged (32–34-week-old) hamsters reported by Osterrieder et al. using a 105 PFU IN exposure dose of the SARS-CoV-2 München (BetaCoV/Germany/BavPat1/2020) isolate. These three studies raise important questions regarding what defines an “aged” hamster in the context of the hamster model of COVID-19 and further enforces the importance of uniformity and replicability. The sample size of aged hamsters in our study (n = 10) was limited by the scarcity of aged hamsters available from commercial vendors.
Paradoxically, and despite the later temporal development and milder pneumonia observed in hACE2 hamsters, SARS-CoV-2 titers remained high in hACE2 hamsters through 5–6 d PE at all exposure doses, and infection was uniformly fatal at this time point in hamsters exposed to 100 or 10 PFU. The later temporal development and more mild pneumonia in hACE2 hamsters when compared to WT hamsters may be explained by the 1000 and 10,000 lower exposure doses necessitated by the fatal neurotropic meningoencephalitis induced by the Washington variant at 4–5 d PE in this model (Golden et al., 2022). However, recent studies using the Omicron variant B.1.1.529 (BA.1) at doses of 1000 PFU yielded only 25% mortality in hACE2 hamsters (Halfmann et al., 2022) and at doses of 4.5–6.3 × 106 PFU (9x106 TCID50) yielded no mortality in hACE2 mice (Toomer et al., 2022). However, the brain was not examined for evidence of neurotropism in either of these studies. This is important because Seehusen et al. evaluated the propensity for neurotropism of several SARS-CoV-2 variants in hACE2 mice, including Omicron B.1.1.529, and at a dose of 103 showed no evidence of neurotropism and minimal pulmonary disease (Seehusen et al., 2022). The variability in neurotropism of different SARS-CoV-2 variants in hACE2 rodent models makes it imperative that future studies utilizing these models to evaluate SARS-CoV-2 pathogenesis, pathogenicity/virulence, and/or to evaluate medical countermeasures include evaluation of the brain to assess the impact of neurotropism/meningoencephalitis on the disease course and survivorship. In our hands, using the WA1/2020 isolate, the hACE2 hamster does not recapitulate the pathogenesis of COVID-19 in humans.
Our data support the understanding that the hACE-2 hamster model has many of the same drawbacks as the K-18 and murine models of SARS-CoV-2 infection (Gruber et al., 2021). The K-18 hACE2 hamster model and murine models develop a rapidly fatal neuropathological phenotype following exposure to the WA1/2020 variant (Golden et al., 2022; Gruber et al., 2021; Moreau et al., 2020). However, recent studies using hACE2 mice have shown that SARS-CoV-2 neurotropism is variant-specific (Seehusen et al., 2022). It is not known if this variability in neurotropism of SARS-CoV-2 is seen in the hACE2 hamster model.
We compare the immunological responses to SARS-CoV-2 infection between WT and hACE2 hamsters which might explain the difference in viral titer. Hamsters had high levels of SARS-2 specific IgG which would naturally decrease levels of free virus in tissues. IgG titers against S1 in serum samples from young WT hamsters were detected at 5 d PE and were approximately 1.5- to 2-fold higher in young WT hamsters when compared to aged WT hamsters at 8 d, when IgG was first detected. Temporally similar humoral immune responses have been reported in clinical cases (Luo et al., 2020; Lynch et al., 2021), and these findings are consistent with the more severe and protracted pneumonia seen in WT hamsters.
Neutralizing antibody titers followed the same dose-dependent pattern in WT hamsters. Young WT hamsters exposed to 105 PFU developed higher neutralizing antibody titers than those exposed to 103 PFU and higher neutralizing antibody titers than aged WT hamsters exposed to the same 105 PFU virus dose. In contrast, no SARS-CoV-2 IgG titers were detected in hACE2 hamsters at any post-exposure time point. hACE2 hamsters did develop neutralizing antibody titers by 5–6 d PE, but these neutralizing antibody titers had an inverse relationship to exposure dose. Neutralizing antibody titers at 5–6 d were higher in hACE2 hamsters exposed to 1 and 0.1 PFU virus than those exposed to 100 or 10 PFU virus. The failure of hACE2 hamsters to develop comparative anti-SARS-CoV-2 IgG titers at 4–6 d is likely due to the lower exposure doses (100, 10, 1, or 0.1 PFU), with rapid neuroinvasion resulting in earlier death following exposure to 100 and 10 PFU, and may also be the result of more mild pulmonary disease seen at lower exposure doses of 1 and 0.1 PFU. The development of higher neutralizing antibody titers at lower exposure doses in hACE2 hamsters is the opposite of what was seen in young WT hamsters exposed to 103 and 105 virus and warrants further investigation, as neutralizing antibodies have been shown to protect humans from clinically relevant disease (Khoury et al., 2021).
We assessed several key cytokines and chemokines that have been used to correlate disease progression, or as predictors of outcome, in humans in these three models to determine if effector cell responses reflect what is seen in humans with COVID-19. IL2, IL6, and IL10 are markers of disease severity and mortality (Azar et al., 2020; Wong et al., 2019). Through the secretion of IL10 and TGFB1, T cells suppress inflammation and restore homeostasis (Wong et al., 2019). Studies in humans have found that impairment of cognitive function in recovered patients is associated with underlying inflammatory conditions (Zhou et al., 2020). Increased expression of both IL2 and IL10 has been associated with patients requiring critical care at the hospital for COVID-19 (Abers et al., 2021).
The increases in IL2 and IL10 in hACE2 hamsters exposed to 100, 10, and 1 PFU are consistent with the higher mortality seen in hACE2 hamsters at all exposure doses when compared to WT hamsters (all survived); however, hACE2 hamsters that succumbed following virus exposure showed only a minimal to mild pneumonia compared the severe pneumonia seen in cases with fatal outcome in the clinic. Exposure to SARS-CoV-2 resulted in inverse responses in IL2 expression in WT hamsters when compared to hACE2 hamsters. Young WT hamsters exposed to 105 PFU virus had significantly lower IL2 concentrations in serum when compared to mock-exposed controls at 2–8 d PE, and IL2 concentrations did not differ in aged WT hamsters. IL10 was increased in hACE2 hamsters at 2 d PE but did not differ from mock-exposed controls at 5 d or at any post-exposure time point in WT hamsters. These IL2 and IL10 responses may accurately reflect the WT hamster as a consistent survivor model of COVID-19 but they do not explain the dysregulated IL2 and IL10 immune responses observed in hACE2 hamsters with only mild pulmonary disease.
An inverse association between CCL22 expression and COVID-19 severity has been observed in humans (Abers et al., 2021); however, CCL22 expression was higher in hACE2 hamsters at 2 d PE to the end of the study, except the hACE2 hamsters exposed to 1 PFU (at 2 d). Increases in CCL20 expression have been considered a predictor for severe COVID-19 (Brunet-Ratnasingham et al., 2021; Wilson et al., 2022), but not mild or moderate COVID-19; however, in hACE2 hamsters CCL20 was higher at 2 d to the end of the study when compared to mock-exposed controls, despite only developing a minimal to mild pneumonia.
The induction of IP-10 is a robust indicator of viral respiratory infections (Cheemarla et al., 2021), and expression of IP-10 in the lungs would be expected to correlate with a local enhancement of anti-viral effector T cells and natural killer cells expressing the cognate receptor CXCR3. Increases in IP-10 have been reported in COVID-19 patients in all severity groups (Brunet-Ratnasingham et al., 2021). Consistent with this finding in humans, both the young and aged WT hamsters had a significant induction in IP-10, but this change was strikingly absent from the hACE2 animals, indicating a delay in at least one portion of the local innate and adaptive cellular response in the hACE2 hamsters.
Several studies have documented that the golden hamster provides a better model, as the similarity to humans with regard to disease symptoms, pathogenesis, and immune responses is greater than mouse models. It has been demonstrated that human cytokines, including granulocyte-macrophage colony-stimulating factor (GM-CSF) and IL12 are fully functional in hamster models but not in mouse models. Hamster models are used for other diseases also, such as those caused by Nipah, Ebola, yellow fever, and West Nile viruses (Miao et al., 2019). Frer et al. (2022) showed that both influenza A virus and SARS-CoV-2 induce a systemic antiviral response; only the latter results in a sustained inflammatory pathology that extends well beyond clearance of the primary infection (Frere et al., 2022).
In our study, TNF expression was found to be hACE2>WT young > WT aged. However, in a study done by Francis et al. (2021), hamsters exposed with SARS-CoV-2 isolate Canada/ON/VIDO-01/2020 induced a higher level of TNF expression in lungs at an early stage of infection (Francis et al., 2021). Our TGFB expression profile was, in order, WT aged > WT young > hACE2 adult. Similar results were reported by Oishi et al. (2022); older animals demonstrated elevated suppressor T cells and neutrophils in the respiratory tract, correlating with an increase in TGFB and IL17 induction (Oishi et al., 2022). Together, these data support the posit that diminished immunity is one of the underlying causes of age-related morbidity. Bednash et al., 2021 reported elevated expression of IFNG early in hamsters exposed with WA01/2020. In our study, IFNG expression was found to be, in order, hACE2 adult > WT young > WT aged (Bednash et al., 2022).
Golden hamsters are the best model to test antiviral countermeasures and vaccine for SARS-COV-2. Early acute proinflammatory response associated with increased production of IL6, IL2, CXCL10, CCL22, CCL20, and IFNG is similar irrespective of age, sex and variants. There is difference in late adaptive response.
Incorporating 18F-FDG micro-PET/CT into these experiments supports the utility of medical imaging in animal-model development. The serial imaging data correlated to the histopathology of respiratory-tract disease and demonstrated onset, progression, and regression in all three models. Lung disease was evident in the WT animals, and 18F-FDG-avid areas overlapped with lesions identified by CT. Scoring of the lesions and changes in 18F-FDG uptake also corresponded with histopathology suggesting that serial imaging can be used to monitor disease progression and can be expanded to evaluate countermeasures similar to what was done for MERS-CoV (Johnson et al., 2015b). Furthermore, the imaging identified commonalities between the models in that all three hamster models developed a dose-associated hemorrhagic, exudative, and necrotizing rhinitis, supported by engorgement of the posterior nasal soft tissues observed on CT and by increased 18F-FDG uptake in the nasal and nasopharyngeal structures.
Our group is the first to compare the three SARS-CoV-2 (WA-01/2020) hamster models with escalating doses, incorporation of medical imaging, and time-matched histopathology. We conclude that the hACE2 hamster model exposed to the SARS-CoV-2 Washington isolate IN showed the least clinical relevance to COVID-19, while the respiratory disease in aged WT hamsters exposed to 105 PFU of virus most closely recapitulates COVID-19. The aged WT hamster would be an improved, more stringent, preclinical model for the study of SARS-CoV-2 pathogenesis and countermeasure development. This work will provide useful information for scientific community's future studies on medical countermeasure for emerging SARS-CoV-2 variants of concern and could be broadly applied to other viral pathogens that could emerge.
Funding
This work was supported in part through Laulima Government Solutions, LLC, prime contract with the National Institutes of Health (NIH) National Institute of Allergy and Infectious Diseases (NIAID) [contract number HHSN272201800013C]. Y.C., D.L.P, K.C., H.W., S.D., D.X.L., I.M.F., C.B., J.S., R.A., A.S., J.L., N.S., R.H., N.I., T.B., D.D.L., L.M.H., R.B., D.R., J.W., J.R.K., and M.R.H. performed this work as employees of Laulima Government Solutions, LLC. J.L. and A.W.H. performed this work as employees of Tunnell Government Services (TGS), a subcontractor of Laulima Government Solutions, LLC under Contract No. HHSN272201800013C. This work was also supported in part with federal funds from the NIH National Cancer Institute (NCI) [contract number 75N91019D00024, Task Order No. 75N91019F00130]. J.S. was supported by the Clinical Monitoring Research Program Directorate, Frederick National Laboratory for Cancer Research, sponsored by NCI. Support was also provided by a contract between the Utah State University and the NIH NIAID Division of Microbiology and Infectious Diseases (DMID) Virology Branch [contract number HHSN272201700041I, task order 75N93020F00001]. This work was also supported by the NIH NIAID Division of Intramural Research.
The views and conclusions contained in this document are those of the authors and should not be interpreted as necessarily representing the official policies, either expressed or implied, of the U.S. Department of Health and Human Services or of the institutions and companies affiliated with the authors, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
The animal study protocol was reviewed and approved by the NIH NIAID DCR IRF-Frederick Animal Care and Use Committee in compliance with all applicable federal regulations governing the protection of animals and research.
Author contributions
Y.C., R.F.J., M.R.H., L.E.H. and C.S.S. contributed to the study conception and design.
J.L., D.L.P., K.C., H.W., S.D., D.X.L., J.S., C.B., J.S., I.M.F., R.A., A.S., J.L., N.S., R.H., N.I., R.B., T.B., D.D.L., A.W.H., L.M.H., R.B., D.R., J.R.K., and M.C.S. contributed to study performance, and sample and data collection.
Y.C., R.F.J., D.L.P., M.R.H., J.L., H.W., I.M.F., D.X.L., R.A., A.S., J.S., J.L., D.B., J.J., C.B., D.A.H, J.R.K., J.W., and K.D.Y. contributed to data analyses, interpretation, and writing.
All authors read and approved the final manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank all the staff of the National Institutes of Health (NIH) National Institute of Allergy and Infectious Diseases (NIAID) Division of Clinical Research (DCR) Integrated Research Facility at Fort Detrick (IRF-Frederick) who supported this study, in particular, Kyra Hadley, Zachary Hubble, Collin Waters, Abel Ordonez, Joseph Hischak, Erin Kollins, Nicholas Vaughan, Bryan Aristy, Sarah Dowling, Becky Reeder, Oscar Rojas, Marcelo Castro, Philip Sayre, Deepa Varghese, Sean Bartlinski, Gregory Kocher, Elena N. Postnikova, Robin Gross, Shuiqing Yu, Lindsay Marron, Steve Mazur, Shalamar Georgia, Erika Maynor, Rebecca Bernbaum, Blake Davis, Anthony Marketon, Nejra Isic, Anya Crane, and Travis K. Warren.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.antiviral.2023.105605.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Study design—timeline/activities and experimental groups/cohorts
A. Natural history of disease studies were performed to characterize the disease pathogenesis in aged WT, young WT, and hACE2 hamsters. Hamsters were acclimated and individually housed. Hamsters were grouped by exposure dose and then separated into cohorts based on pre-determined imaging/serial euthanasia days and exposed to either mock inoculum (diluent medium) or to SARS-CoV-2. B. Subsequent micro-PET/CT imaging studies were performed in aged WT, young WT, and hACE2 hamsters. Hamsters were grouped by exposure dose and then separated into cohorts based on pre-determined imaging days followed by serial euthanasia. Hamsters were weighed and evaluated for clinical signs of disease from the −1 d until the end of the study. PET/CT was performed on 7 ± 3 d for hamsters in the “imaged” groups. All hamsters were euthanized and necropsied after PET/CT imaging at scheduled or unscheduled terminal time points, and samples were collected. Sample sizes of each group/cohort are shown. WT, wild type; T, terminal; NH, natural history; PET/CT, positron emission tomography with computed tomography; hACE2, human angiotensin-converting enzyme 2; D2, Day 2; D4, Day 4; D5, Day 5; D6, Day 6; D8, Day 8; D10, Day 10; D14, Day 14.
Data availability
Data will be made available on request.
References
- Abers M.S., Delmonte O.M., Ricotta E.E., Fintzi J., Fink D.L., de Jesus A.A.A., Zarember K.A., Alehashemi S., Oikonomou V., Desai J.V., Canna S.W., Shakoory B., Dobbs K., Imberti L., Sottini A., Quiros-Roldan E., Castelli F., Rossi C., Brugnoni D., Biondi A., Bettini L.R., D'Angio’ M., Bonfanti P., Castagnoli R., Montagna D., Licari A., Marseglia G.L., Gliniewicz E.F., Shaw E., Kahle D.E., Rastegar A.T., Stack M., Myint-Hpu K., Levinson S.L., DiNubile M.J., Chertow D.W., Burbelo P.D., Cohen J.I., Calvo K.R., Tsang J.S., Su H.C., Gallin J.I., Kuhns D.B., Goldbach-Mansky R., Lionakis M.S., Notarangelo L.D. An immune-based biomarker signature is associated with mortality in COVID-19 patients. JCI Insight. 2021;6 doi: 10.1172/jci.insight.144455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azar M.M., Shin J.J., Kang I., Landry M. Diagnosis of SARS-CoV-2 infection in the setting of the cytokine release syndrome. Expert Rev. Mol. Diagn. 2020;20:1087–1097. doi: 10.1080/14737159.2020.1830760. [DOI] [PubMed] [Google Scholar]
- Bai Y., Xu J., Chen L., Fu C., Kang Y., Zhang W., Fakhri G.E., Gu J., Shao F., Wang M. Inflammatory response in lungs and extrapulmonary sites detected by [(18)F] fluorodeoxyglucose PET/CT in convalescing COVID-19 patients tested negative for coronavirus. Eur. J. Nucl. Med. Mol. Imag. 2021;48:2531–2542. doi: 10.1007/s00259-020-05083-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartleson J.M., Radenkovic D., Covarrubias A.J., Furman D., Winer D.A., Verdin E. SARS-CoV-2, COVID-19 and the aging immune system. Nature Aging. 2021;1:769–782. doi: 10.1038/s43587-021-00114-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton L.M., Duval E.J., Stroberg E., Ghosh S., Mukhopadhyay S. COVID-19 autopsies, Oklahoma, USA. Am. J. Clin. Pathol. 2020;153:725–733. doi: 10.1093/ajcp/aqaa062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednash J.S., Kagan V.E., Englert J.A., Farkas D., Tyurina Y.Y., Tyurin V.A., Samovich S.N., Farkas L., Elhance A., Johns F., Lee H., Cheng L., Majumdar A., Jones D., Mejia O.R., Ruane-Foster M., Londino J.D., Mallampalli R.K., Robinson R.T. Syrian hamsters as a model of lung injury with SARS-CoV-2 infection: pathologic, physiologic, and detailed molecular profiling. Transl. Res. 2022;240:1–16. doi: 10.1016/j.trsl.2021.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benameur N., Mahmoudi R., Zaid S., Arous Y., Hmida B., Bedoui M.H. SARS-CoV-2 diagnosis using medical imaging techniques and artificial intelligence: a review. Clin. Imag. 2021;76:6–14. doi: 10.1016/j.clinimag.2021.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett R.S., Postnikova E.N., Liang J., Gross R., Mazur S., Dixit S., Kocher G., Yu S., Georgia-Clark S., Gerhardt D., Cai Y., Marron L., Lukin V.V., Holbrook M.R. Scalable, micro-neutralization assay for assessment of SARS-CoV-2 (COVID-19) virus-neutralizing antibodies in human clinical samples. Viruses. 2021;13 doi: 10.3390/v13050893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borczuk A.C., Salvatore S.P., Seshan S.V., Patel S.S., Bussel J.B., Mostyka M., Elsoukkary S., He B., Del Vecchio C., Fortarezza F., Pezzuto F., Navalesi P., Crisanti A., Fowkes M.E., Bryce C.H., Calabrese F., Beasley M.B. COVID-19 pulmonary pathology: a multi-institutional autopsy cohort from Italy and New York City. Mod. Pathol. 2020;33:2156–2168. doi: 10.1038/s41379-020-00661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet-Ratnasingham E., Anand S.P., Gantner P., Dyachenko A., Moquin-Beaudry G., Brassard N., Beaudoin-Bussières G., Pagliuzza A., Gasser R., Benlarbi M., Point F., Prévost J., Laumaea A., Niessl J., Nayrac M., Sannier G., Orban C., Messier-Peet M., Butler-Laporte G., Morrison D.R., Zhou S., Nakanishi T., Boutin M., Descôteaux-Dinelle J., Gendron-Lepage G., Goyette G., Bourassa C., Medjahed H., Laurent L., Rébillard R.M., Richard J., Dubé M., Fromentin R., Arbour N., Prat A., Larochelle C., Durand M., Richards J.B., Chassé M., Tétreault M., Chomont N., Finzi A., Kaufmann D.E. Integrated immunovirological profiling validates plasma SARS-CoV-2 RNA as an early predictor of COVID-19 mortality. Sci. Adv. 2021;7 doi: 10.1126/sciadv.abj5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll T., Fox D., van Doremalen N., Ball E., Morris M.K., Sotomayor-Gonzalez A., Servellita V., Rustagi A., Yinda C.K., Fritts L., Port J.R., Ma Z.M., Holbrook M.G., Schulz J., Blish C.A., Hanson C., Chiu C.Y., Munster V., Stanley S., Miller C.J. The B.1.427/1.429 (epsilon) SARS-CoV-2 variants are more virulent than ancestral B.1 (614G) in Syrian hamsters. PLoS Pathog. 2022;18 doi: 10.1371/journal.ppat.1009914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F., Zhang A.J., Yuan S., Poon V.K., Chan C.C., Lee A.C., Chan W.M., Fan Z., Tsoi H.W., Wen L., Liang R., Cao J., Chen Y., Tang K., Luo C., Cai J.P., Kok K.H., Chu H., Chan K.H., Sridhar S., Chen Z., Chen H., To K.K., Yuen K.Y. Simulation of the clinical and pathological manifestations of coronavirus disease 2019 (COVID-19) in a golden Syrian hamster model: implications for disease pathogenesis and transmissibility. Clin. Infect. Dis. 2020;71:2428–2446. doi: 10.1093/cid/ciaa325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheemarla N.R., Watkins T.A., Mihaylova V.T., Wang B., Zhao D., Wang G., Landry M.L., Foxman E.F. Dynamic innate immune response determines susceptibility to SARS-CoV-2 infection and early replication kinetics. J. Exp. Med. 2021;218 doi: 10.1084/jem.20210583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chefer S., Seidel J., Cockrell A.S., Yount B., Solomon J., Hagen K.R., Liu D.X., Huzella L.M., Kumar M.R., Postnikova E., Bohannon J.K., Lackemeyer M.G., Cooper K., Endlich-Frazier A., Sharma H., Thomasson D., Bartos C., Sayre P.J., Sims A., Dyall J., Holbrook M.R., Jahrling P.B., Baric R.S., Johnson R.F. The human sodium iodide symporter as a reporter gene for studying Middle East respiratory syndrome coronavirus pathogenesis. mSphere. 2018;3 doi: 10.1128/mSphere.00540-18. e00540-00518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochin M., Luciani L., Touret F., Driouich J.S., Petit P.R., Moureau G., Baronti C., Laprie C., Thirion L., Maes P., Boudewijns R., Neyts J., de Lamballerie X., Nougairede A. The SARS-CoV-2 Alpha variant exhibits comparable fitness to the D614G strain in a Syrian hamster model. Commun Biol. 2022;5:225. doi: 10.1038/s42003-022-03171-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Agnillo F., Walters K.-A., Xiao Y., Sheng Z.-M., Scherler K., Park J., Gygli S., Rosas L.A., Sadtler K., Kalish H., Blatti C.A., Zhu R., Gatzke L., Bushell C., Memoli M.J., O'Day S.J., Fischer T.D., Hammond T.C., Lee R.C., Cash J.C., Powers M.E., O'Keefe G.E., Butnor K.J., Rapkiewicz A.V., Travis W.D., Layne S.P., Kash J.C., Taubenberger J.K. Lung epithelial and endothelial damage, loss of tissue repair, inhibition of fibrinolysis, and cellular senescence in fatal COVID-19. Sci. Transl. Med. 2021;13 doi: 10.1126/scitranslmed.abj7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz M., Chironi G., Claessens Y.E., Farhad R.L., Rouquette I., Serrano B., Nataf V., Hugonnet F., Paulmier B., Berthier F., Keita-Perse O., Giammarile F., Perrin C., Faraggi M., Group M. COVID-19 pneumonia: relationship between inflammation assessed by whole-body FDG PET/CT and short-term clinical outcome. Eur. J. Nucl. Med. Mol. Imag. 2021;48:260–268. doi: 10.1007/s00259-020-04968-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eibschutz L.S., Rabiee B., Asadollahi S., Gupta A., Assadi M., Alavi A., Gholamrezanezhad A. FDG-PET/CT of COVID-19 and other lung infections. Semin. Nucl. Med. 2022;52:61–70. doi: 10.1053/j.semnuclmed.2021.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis M.E., Goncin U., Kroeker A., Swan C., Ralph R., Lu Y., Etzioni A.L., Falzarano D., Gerdts V., Machtaler S., Kindrachuk J., Kelvin A.A. SARS-CoV-2 infection in the Syrian hamster model causes inflammation as well as type I interferon dysregulation in both respiratory and non-respiratory tissues including the heart and kidney. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frere J.J., Serafini R.A., Pryce K.D., Zazhytska M., Oishi K., Golynker I., Panis M., Zimering J., Horiuchi S., Hoagland D.A., Møller R., Ruiz A., Kodra A., Overdevest J.B., Canoll P.D., Borczuk A.C., Chandar V., Bram Y., Schwartz R., Lomvardas S., Zachariou V., tenOever B.R. SARS-CoV-2 infection in hamsters and humans results in lasting and unique systemic perturbations after recovery. Sci. Transl. Med. 2022;14 doi: 10.1126/scitranslmed.abq3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilliland T., Liu Y., Li R., Dunn M., Cottle E., Terada Y., Ryckman Z., Alcorn M., Vasilatos S., Lundy J., Larson D., Wu H., Luke T., Bausch C., Egland K., Sullivan E., Wang Z., Klimstra W.B. Protection of human ACE2 transgenic Syrian hamsters from SARS CoV-2 variants by human polyclonal IgG from hyper-immunized transchromosomic bovines. bioRxiv. 2021 doi: 10.1101/2021.07.26.453840. 2021.2007.2026. [DOI] [Google Scholar]
- Golden J.W., Li R., Cline C.R., Zeng X., Mucker E.M., Fuentes-Lao A.J., Spik K.W., Williams J.A., Twenhafel N., Davis N., Moore J.L., Stevens S., Blue E., Garrison A.R., Larson D.D., Stewart R., Kunzler M., Liu Y., Wang Z., Hooper J.W. Hamsters expressing human angiotensin-converting enzyme 2 develop severe disease following exposure to SARS-CoV-2. mBio. 2022;13 doi: 10.1128/mbio.02906-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber A.D., Firsching T.C., Trimpert J., Dietert K. Hamster models of COVID-19 pneumonia reviewed: how human can they be? Vet. Pathol. 2021 doi: 10.1177/03009858211057197. 3009858211057197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann, P.J., Iida, S., Iwatsuki-Horimoto, K., Maemura, T., Kiso, M., Scheaffer, S.M., Darling, T.L., Joshi, A., Loeber, S., Singh, G., Foster, S.L., Ying, B., Case, J.B., Chong, Z., Whitener, B., Moliva, J., Floyd, K., Ujie, M., Nakajima, N., Ito, M., Wright, R., Uraki, R., Warang, P., Gagne, M., Li, R., Sakai-Tagawa, Y., Liu, Y., Larson, D., Osorio, J.E., Hernandez-Ortiz, J.P., Henry, A.R., Ciuoderis, K., Florek, K.R., Patel, M., Odle, A., Wong, L.-Y.R., Bateman, A.C., Wang, Z., Edara, V.-V., Chong, Z., Franks, J., Jeevan, T., Fabrizio, T., DeBeauchamp, J., Kercher, L., Seiler, P., Gonzalez-Reiche, A.S., Sordillo, E.M., Chang, L.A., van Bakel, H., Simon, V., Alburquerque, B., Alshammary, H., Amoako, A.A., Aslam, S., Banu, R., Cognigni, C., Espinoza-Moraga, M., Farrugia, K., van de Guchte, A., Khalil, Z., Laporte, M., Mena, I., Paniz-Mondolfi, A.E., Polanco, J., Rooker, A., Sominsky, L.A., Douek, D.C., Sullivan, N.J., Thackray, L.B., Ueki, H., Yamayoshi, S., Imai, M., Perlman, S., Webby, R.J., Seder, R.A., Suthar, M.S., García-Sastre, A., Schotsaert, M., Suzuki, T., Boon, A.C.M., Diamond, M.S., Kawaoka, Y. & Consortium Mount Sinai Pathogen Surveillance study, g., 2022. SARS-CoV-2 Omicron virus causes attenuated disease in mice and hamsters. Nature 603, 687-692. 10.1038/s41586-022-04441-6. [DOI] [PMC free article] [PubMed]
- Hani C., Trieu N.H., Saab I., Dangeard S., Bennani S., Chassagnon G., Revel M.P. COVID-19 pneumonia: a review of typical CT findings and differential diagnosis. Diagn Interv Imaging. 2020;101:263–268. doi: 10.1016/j.diii.2020.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho C.-Y., Salimian M., Hegert J., O'Brien J., Choi S.G., Ames H., Morris M., Papadimitriou J.C., Mininni J., Niehaus P., Burke A., Canbeldek L., Jacobs J., LaRocque A., Patel K., Rice K., Li L., Johnson R., LeFevre A., Blanchard T., Shaver C.M., Moyer A., Drachenberg C. Postmortem assessment of olfactory tissue degeneration and microvasculopathy in patients with COVID-19. JAMA Neurol. 2022 doi: 10.1001/jamaneurol.2022.0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobi A., Chung M., Bernheim A., Eber C. Portable chest X-ray in coronavirus disease-19 (COVID-19): a pictorial review. Clin. Imag. 2020;64:35–42. doi: 10.1016/j.clinimag.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J., Agarwala N., Kundu P., Harvey B., Zhang Y., Wallace E., Chatterjee N. Individual and community-level risk for COVID-19 mortality in the United States. Nat. Med. 2021;27:264–269. doi: 10.1038/s41591-020-01191-8. [DOI] [PubMed] [Google Scholar]
- Johnson R.F., Bagci U., Keith L., Tang X., Mollura D.J., Zeitlin L., Qin J., Huzella L., Bartos C.J., Bohorova N., Bohorov O., Goodman C., Kim D.H., Paulty M.H., Velasco J., Whaley K.J., Johnson J.C., Pettitt J., Ork B.L., Solomon J., Oberlander N., Zhu Q., Sun J., Holbrook M.R., Olinger G.G., Baric R.S., Hensley L.E., Jahrling P.B., Marasco W.A. 3B11-N, a monoclonal antibody against MERS-CoV, reduces lung pathology in rhesus monkeys following intratracheal inoculation of MERS-CoV Jordan-n3/2012. Virology. 2016;490:49–58. doi: 10.1016/j.virol.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R.F., Hammoud D.A., Lackemeyer M.G., Yellayi S., Solomon J., Bohannon J.K., Janosko K.B., Jett C., Cooper K., Blaney J.E., Jahrling P.B. Small particle aerosol inoculation of cowpox Brighton Red in rhesus monkeys results in a severe respiratory disease. Virology. 2015;481:124–135. doi: 10.1016/j.virol.2015.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R.F., Via L.E., Kumar M.R., Cornish J.P., Yellayi S., Huzella L., Postnikova E., Oberlander N., Bartos C., Ork B.L., Mazur S., Allan C., Holbrook M.R., Solomon J., Johnson J.C., Pickel J., Hensley L.E., Jahrling P.B. Intratracheal exposure of common marmosets to MERS-CoV Jordan-n3/2012 or MERS-CoV EMC/2012 isolates does not result in lethal disease. Virology. 2015;485:422–430. doi: 10.1016/j.virol.2015.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy A.R., Desrosiers A., Terzaghi M., Little J.B. Morphometric and histological analysis of the lungs of Syrian golden hamsters. J. Anat. 1978;125:527–553. [PMC free article] [PubMed] [Google Scholar]
- Khan M., Yoo S.-J., Clijsters M., Backaert W., Vanstapel A., Speleman K., Lietaer C., Choi S., Hether T.D., Marcelis L., Nam A., Pan L., Reeves J.W., Van Bulck P., Zhou H., Bourgeois M., Debaveye Y., De Munter P., Gunst J., Jorissen M., Lagrou K., Lorent N., Neyrinck A., Peetermans M., Thal D.R., Vandenbriele C., Wauters J., Mombaerts P., Van Gerven L. Visualizing in deceased COVID-19 patients how SARS-CoV-2 attacks the respiratory and olfactory mucosae but spares the olfactory bulb. Cell. 2021;184:5932–5949. doi: 10.1016/j.cell.2021.10.027. e5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury D.S., Cromer D., Reynaldi A., Schlub T.E., Wheatley A.K., Juno J.A., Subbarao K., Kent S.J., Triccas J.A., Davenport M.P. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021;27:1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- Li M., Wang H., Tian L., Pang Z., Yang Q., Huang T., Fan J., Song L., Tong Y., Fan H. COVID-19 vaccine development: milestones, lessons and prospects. Signal Transduct. Targeted Ther. 2022;7:146. doi: 10.1038/s41392-022-00996-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link-Gelles R., Ciesla A.A., Fleming-Dutra K.E., Smith Z.R., Britton A., Wiegand R.E., Miller J.D., Accorsi E.K., Schrag S.J., Verani J.R., Shang N., Derado G., Pilishvili T. Effectiveness of bivalent mRNA vaccines in preventing symptomatic SARS-CoV-2 infection - increasing community access to testing Program, United States, september-november 2022. MMWR Morb. Mortal. Wkly. Rep. 2022;71:1526–1530. doi: 10.15585/mmwr.mm7148e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y.R., Chakraborty I., Yun C., Wu A.H.B., Lynch K.L. Kinetics of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) antibody avidity maturation and association with disease severity. Clin. Infect. Dis. 2020;73:e3095–e3097. doi: 10.1093/cid/ciaa1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch K.L., Whitman J.D., Lacanienta N.P., Beckerdite E.W., Kastner S.A., Shy B.R., Goldgof G.M., Levine A.G., Bapat S.P., Stramer S.L., Esensten J.H., Hightower A.W., Bern C., Wu A.H.B. Magnitude and kinetics of anti–severe acute respiratory syndrome coronavirus 2 antibody responses and their relationship to disease severity. Clin. Infect. Dis. 2021;72:301–308. doi: 10.1093/cid/ciaa979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malherbe D.C., Kurup D., Wirblich C., Ronk A.J., Mire C., Kuzmina N., Shaik N., Periasamy S., Hyde M.A., Williams J.M., Shi P.-Y., Schnell M.J., Bukreyev A. A single dose of replication-competent VSV-vectored vaccine expressing SARS-CoV-2 S1 protects against virus replication in a hamster model of severe COVID-19. npj Vaccines. 2021;6:91. doi: 10.1038/s41541-021-00352-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matschke J., Lütgehetmann M., Hagel C., Sperhake J.P., Schröder A.S., Edler C., Mushumba H., Fitzek A., Allweiss L., Dandri M., Dottermusch M., Heinemann A., Pfefferle S., Schwabenland M., Sumner Magruder D., Bonn S., Prinz M., Gerloff C., Püschel K., Krasemann S., Aepfelbacher M., Glatzel M. Neuropathology of patients with COVID-19 in Germany: a post-mortem case series. Lancet Neurol. 2020;19:919–929. doi: 10.1016/S1474-4422(20)30308-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao J., Chard L.S., Wang Z., Wang Y. Syrian hamster as an animal model for the study on infectious diseases. Front. Immunol. 2019;10:2329. doi: 10.3389/fimmu.2019.02329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau G.B., Burgess S.L., Sturek J.M., Donlan A.N., Petri W.A., Mann B.J. Evaluation of K18-hACE2 mice as a model of SARS-CoV-2 infection. Am. J. Trop. Med. Hyg. 2020;103:1215–1219. doi: 10.4269/ajtmh.20-0762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Fontela C., Dowling W.E., Funnell S.G.P., Gsell P.S., Riveros-Balta A.X., Albrecht R.A., Andersen H., Baric R.S., Carroll M.W., Cavaleri M., Qin C., Crozier I., Dallmeier K., de Waal L., de Wit E., Delang L., Dohm E., Duprex W.P., Falzarano D., Finch C.L., Frieman M.B., Graham B.S., Gralinski L.E., Guilfoyle K., Haagmans B.L., Hamilton G.A., Hartman A.L., Herfst S., Kaptein S.J.F., Klimstra W.B., Knezevic I., Krause P.R., Kuhn J.H., Le Grand R., Lewis M.G., Liu W.C., Maisonnasse P., McElroy A.K., Munster V., Oreshkova N., Rasmussen A.L., Rocha-Pereira J., Rockx B., Rodriguez E., Rogers T.F., Salguero F.J., Schotsaert M., Stittelaar K.J., Thibaut H.J., Tseng C.T., Vergara-Alert J., Beer M., Brasel T., Chan J.F.W., Garcia-Sastre A., Neyts J., Perlman S., Reed D.S., Richt J.A., Roy C.J., Segales J., Vasan S.S., Henao-Restrepo A.M., Barouch D.H. Animal models for COVID-19. Nature. 2020;586:509–515. doi: 10.1038/s41586-020-2787-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi K., Horiuchi S., Frere J., Schwartz R.E., tenOever B.R. A diminished immune response underlies age-related SARS-CoV-2 pathologies. Cell Rep. 2022;39 doi: 10.1016/j.celrep.2022.111002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterrieder N., Bertzbach L.D., Dietert K., Abdelgawad A., Vladimirova D., Kunec D., Hoffmann D., Beer M., Gruber A.D., Trimpert J. Age-dependent progression of SARS-CoV-2 infection in Syrian hamsters. Viruses. 2020;12 doi: 10.3390/v12070779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan F., Ye T., Sun P., Gui S., Liang B., Li L., Zheng D., Wang J., Hesketh R.L., Yang L., Zheng C. Time course of lung changes at chest CT during recovery from coronavirus disease 2019 (COVID-19) Radiology. 2020;295:715–721. doi: 10.1148/radiol.2020200370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renn M., Bartok E., Zillinger T., Hartmann G., Behrendt R. Animal models of SARS-CoV-2 and COVID-19 for the development of prophylactic and therapeutic interventions. Pharmacol. Ther. 2021;228 doi: 10.1016/j.pharmthera.2021.107931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenke K., Meade-White K., Letko M., Clancy C., Hansen F., Liu Y., Okumura A., Tang-Huau T.L., Li R., Saturday G., Feldmann F., Scott D., Wang Z., Munster V., Jarvis M.A., Feldmann H. Defining the Syrian hamster as a highly susceptible preclinical model for SARS-CoV-2 infection. Emerg. Microb. Infect. 2020;9:2673–2684. doi: 10.1080/22221751.2020.1858177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydyznski Moderbacher C., Ramirez S.I., Dan J.M., Grifoni A., Hastie K.M., Weiskopf D., Belanger S., Abbott R.K., Kim C., Choi J., Kato Y., Crotty E.G., Kim C., Rawlings S.A., Mateus J., Tse L.P.V., Frazier A., Baric R., Peters B., Greenbaum J., Ollmann Saphire E., Smith D.M., Sette A., Crotty S. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell. 2020;183:996. doi: 10.1016/j.cell.2020.09.038. 1012.e1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seehusen F., Clark J.J., Sharma P., Bentley E.G., Kirby A., Subramaniam K., Wunderlin-Giuliani S., Hughes G.L., Patterson E.I., Michael B.D., Owen A., Hiscox J.A., Stewart J.P., Kipar A. Neuroinvasion and neurotropism by SARS-CoV-2 variants in the K18-hACE2 mouse. Viruses. 2022;14:1020. doi: 10.3390/v14051020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvaraj P., Lien C.Z., Liu S., Stauft C.B., Nunez I.A., Hernandez M., Nimako E., Ortega M.A., Starost M.F., Dennis J.U., Wang T.T. SARS-CoV-2 infection induces protective immunity and limits transmission in Syrian hamsters. Life Sci. Alliance. 2021;4 doi: 10.26508/lsa.202000886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia S.F., Yan L.M., Chin A.W.H., Fung K., Choy K.T., Wong A.Y.L., Kaewpreedee P., Perera R., Poon L.L.M., Nicholls J.M., Peiris M., Yen H.L. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature. 2020;583:834–838. doi: 10.1038/s41586-020-2342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein S.R., Ramelli S.C., Grazioli A., Chung J.-Y., Singh M., Yinda C.K., Winkler C.W., Sun J., Dickey J.M., Ylaya K., Ko S.H., Platt A.P., Burbelo P.D., Quezado M., Pittaluga S., Purcell M., Munster V.J., Belinky F., Ramos-Benitez M.J., Boritz E.A., Lach I.A., Herr D.L., Rabin J., Saharia K.K., Madathil R.J., Tabatabai A., Soherwardi S., McCurdy M.T., Babyak A.L., Perez Valencia L.J., Curran S.J., Richert M.E., Young W.J., Young S.P., Gasmi B., Sampaio De Melo M., Desar S., Tadros S., Nasir N., Jin X., Rajan S., Dikoglu E., Ozkaya N., Smith G., Emanuel E.R., Kelsall B.L., Olivera J.A., Blawas M., Star R.A., Hays N., Singireddy S., Wu J., Raja K., Curto R., Chung J.E., Borth A.J., Bowers K.A., Weichold A.M., Minor P.A., Moshref M.A.N., Kelly E.E., Sajadi M.M., Scalea T.M., Tran D., Dahi S., Deatrick K.B., Krause E.M., Herrold J.A., Hochberg E.S., Cornachione C.R., Levine A.R., Richards J.E., Elder J., Burke A.P., Mazzeffi M.A., Christenson R.H., Chancer Z.A., Abdulmahdi M., Sopha S., Goldberg T., Sangwan Y., Sudano K., Blume D., Radin B., Arnouk M., Eagan J.W., Palermo R., Harris A.D., Pohida T., Garmendia-Cedillos M., Dold G., Saglio E., Pham P., Peterson K.E., Cohen J.I., de Wit E., Vannella K.M., Hewitt S.M., Kleiner D.E., Chertow D.S., Consortium N.C.-A. SARS-CoV-2 infection and persistence in the human body and brain at autopsy. Nature. 2022;612:758–763. doi: 10.1038/s41586-022-05542-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su W., Choy K.T., Gu H., Sia S.F., Cheng K.M., Nizami S.I.N., Krishnan P., Ng Y.M., Chang L.D.J., Liu Y., Cheng S.M., Peiris M., Poon L.L., Nicholls J.M., Yen H.L. Omicron BA.1 and BA.2 sub-lineages show reduced pathogenicity and transmission potential than the early SARS-CoV-2 D614G variant in Syrian hamsters. J. Infect. Dis. 2022 doi: 10.1093/infdis/jiac276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toomer G., Burns W., Garcia L., Henry G., Biancofiori A., George A., Duffy C., Chu J., Sides M., Muñoz M., Garcia K., Nikolai-Yogerst A., Peng X., Westfall L., Baker R. Characterization of three variants of SARS-CoV-2 in vivo shows host-dependent pathogenicity in hamsters, while not in K18-hACE2 mice. Viruses. 2022;14 doi: 10.3390/v14112584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Stillfried S., Bülow R.D., Röhrig R., Boor P., Böcker J., Schmidt J., Tholen P., Majeed R., Wienströer J., Weis J., Bremer J., Knüchel R., Breitbach A., Cacchi C., Freeborn B., Wucherpfennig S., Spring O., Braun G., Römmele C., Märkl B., Claus R., Dhillon C., Schaller T., Sipos E., Hirschbühl K., Wittmann M., Kling E., Kröncke T., Heppner F.L., Meinhardt J., Radbruch H., Streit S., Horst D., Elezkurtaj S., Quaas A., Göbel H., Hansen T., Titze U., Lorenzen J., Reuter T., Woloszyn J., Baretton G., Hilsenbeck J., Meinhardt M., Pablik J., Sommer L., Holotiuk O., Meinel M., Mahlke N., Esposito I., Crudele G., Seidl M., Amann K.U., Coras R., Hartmann A., Eichhorn P., Haller F., Lange F., Schmid K.W., Ingenwerth M., Rawitzer J., Theegarten D., Birngruber C.G., Wild P., Gradhand E., Smith K., Werner M., Schilling O., Acker T., Gattenlöhner S., Stadelmann C., Metz I., Franz J., Stork L., Thomas C., Zechel S., Ströbel P., Wickenhauser C., Fathke C., Harder A., Ondruschka B., Dietz E., Edler C., Fitzek A., Fröb D., Heinemann A., Heinrich F., Klein A., Kniep I., Lohner L., Möbius D., Püschel K., Schädler J., Schröder A.-S., Sperhake J.-P., Aepfelbacher M., Fischer N., Lütgehetmann M., Pfefferle S., Glatzel M., Krasemann S., Matschke J., Jonigk D., Werlein C., Schirmacher P., Domke L.M., Hartmann L., Klein I.M., Schwab C., Röcken C., Friemann J., Langer D., Roth W., Strobl S., Rudelius M., Stock K.F., Weichert W., Delbridge C., Kasajima A., Kuhn P.-H., Slotta-Huspenina J., Weirich G., Barth P., Wardelmann E., Evert K., Büttner A., Manhart J., Nigbur S., Bittmann I., Fend F., Bösmüller H., Granai M., Klingel K., Warm V., Steinestel K., Umathum V.G., Rosenwald A., Kurz F., Vogt N. First report from the German COVID-19 autopsy registry. The Lancet Regional Health - Europe. 2022;15 doi: 10.1016/j.lanepe.2022.100330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO, WHO Coronavirus (COVID-19) Dashboard. https://covid19.who.int/.
- Wilson J.C., Kealy D., James S.R., Plowman T., Newling K., Jagger C., Filbey K., Mann E.R., Konkel J.E., Menon M., Knight S.B., Simpson A., Prihartadi A., Forshaw G., Todd N., Yates D.R.A., Grainger J.R., Hussell T., Kaye P.M., Signoret N., Lagos D. Integrated miRNA/cytokine/chemokine profiling reveals severity-associated step changes and principal correlates of fatality in COVID-19. iScience. 2022;25 doi: 10.1016/j.isci.2021.103672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J.J.M., Leong J.Y., Lee J.H., Albani S., Yeo J.G. Insights into the immuno-pathogenesis of acute respiratory distress syndrome. Ann. Transl. Med. 2019;7:504. doi: 10.21037/atm.2019.09.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S., Ye Z.W., Liang R., Tang K., Zhang A.J., Lu G., Ong C.P., Man Poon V.K., Chan C.C., Mok B.W., Qin Z., Xie Y., Chu A.W., Chan W.M., Ip J.D., Sun H., Tsang J.O., Yuen T.T., Chik K.K., Chan C.C., Cai J.P., Luo C., Lu L., Yip C.C., Chu H., To K.K., Chen H., Jin D.Y., Yuen K.Y., Chan J.F. Pathogenicity, transmissibility, and fitness of SARS-CoV-2 Omicron in Syrian hamsters. Science. 2022;377:428–433. doi: 10.1126/science.abn8939. [DOI] [PubMed] [Google Scholar]
- Zhou H., Lu S., Chen J., Wei N., Wang D., Lyu H., Shi C., Hu S. The landscape of cognitive function in recovered COVID-19 patients. J. Psychiatr. Res. 2020;129:98–102. doi: 10.1016/j.jpsychires.2020.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Study design—timeline/activities and experimental groups/cohorts
A. Natural history of disease studies were performed to characterize the disease pathogenesis in aged WT, young WT, and hACE2 hamsters. Hamsters were acclimated and individually housed. Hamsters were grouped by exposure dose and then separated into cohorts based on pre-determined imaging/serial euthanasia days and exposed to either mock inoculum (diluent medium) or to SARS-CoV-2. B. Subsequent micro-PET/CT imaging studies were performed in aged WT, young WT, and hACE2 hamsters. Hamsters were grouped by exposure dose and then separated into cohorts based on pre-determined imaging days followed by serial euthanasia. Hamsters were weighed and evaluated for clinical signs of disease from the −1 d until the end of the study. PET/CT was performed on 7 ± 3 d for hamsters in the “imaged” groups. All hamsters were euthanized and necropsied after PET/CT imaging at scheduled or unscheduled terminal time points, and samples were collected. Sample sizes of each group/cohort are shown. WT, wild type; T, terminal; NH, natural history; PET/CT, positron emission tomography with computed tomography; hACE2, human angiotensin-converting enzyme 2; D2, Day 2; D4, Day 4; D5, Day 5; D6, Day 6; D8, Day 8; D10, Day 10; D14, Day 14.
Data Availability Statement
Data will be made available on request.