Abstract
The high morbidity and mortality associated with SARS-CoV-2 infection, the etiological agent of COVID-19, has had a major impact on global public health. Significant progress has been made in the development of an array of vaccines and biologics, however, the emergence of SARS-CoV-2 variants and breakthrough infections are an ongoing major concern. Furthermore, there is an existing paucity of small-molecule host and virus-directed therapeutics and prophylactics that can be used to counter the spread of SARS-CoV-2, and any emerging and re-emerging coronaviruses. We describe herein our efforts to address this urgent need by focusing on the structure-guided design of potent broad-spectrum inhibitors of SARS-CoV-2 3C-like protease (3CLpro or Main protease), an enzyme essential for viral replication. The inhibitors exploit the directional effects associated with the presence of a gem-dimethyl group that allow the inhibitors to optimally interact with the S4 subsite of the enzyme. Several compounds were found to potently inhibit SARS-CoV-2 and MERS-CoV 3CL proteases in biochemical and cell-based assays. Specifically, the EC50 values of aldehyde 1c and its corresponding bisulfite adduct 1d against SARS-CoV-2 were found to be 12 and 10 nM, respectively, and their CC50 values were >50 μM. Furthermore, deuteration of these compounds yielded compounds 2c/2d with EC50 values 11 and 12 nM, respectively. Replacement of the aldehyde warhead with a nitrile (CN) or an α-ketoamide warhead or its corresponding bisulfite adduct yielded compounds 1g, 1eand1f with EC50 values 60, 50 and 70 nM, respectively. High-resolution cocrystal structures have identified the structural determinants associated with the binding of the inhibitors to the active site of the enzyme and, furthermore, have illuminated the mechanism of action of the inhibitors. Overall, the high Safety Index (SI) (SI=CC50/EC50) displayed by these compounds suggests that they are well-suited to conducting further preclinical studies.
Keywords: SARS-CoV-2, MERS-CoV, Coronavirus, 3-Chymotrypsin-like protease (3CLpro), Broad-spectrum inhibitors, gem-dimethyl effect
Graphical abstract
1. Introduction
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) is an enveloped, positive-sense single-stranded β-coronavirus belonging to the family Coronaviridae [1,2]. Infections by SARS-CoV-2 continue to have a major impact on public health worldwide [3] and the emergence of multiple variants [[4], [5], [6]], the declining effectiveness of vaccines and biologics [[7], [8], [9]], and the currently limited number of available small-molecule direct-acting antivirals (DAAs) that can be deployed against the virus as therapeutics and prophylactics, have collectively hampered efforts to halt the ongoing spread of infection and coronavirus disease 2019 (COVID-19). Thus, there is currently an urgent need for the discovery and development of host and virus-targeted direct-acting antivirals to complement and enhance the armamentarium of SARS-CoV-2 therapeutics, which currently includes only PaxlovidR (a combination of the 3CLpro inhibitor nirmatrelvir and ritonavir, a CYP450 3A4 inhibitor which suppresses the metabolism of nirmaltrevir), and the viral RNA-dependent RNA-polymerase inhibitors Molnupiravir and Remdesivir. The availability of a portfolio of direct-acting antivirals that act by different mechanisms of action would be highly advantageous and make possible the optimal use of combination therapy, creating a high barrier for the emergence of resistance [[10], [11], [12], [13]].
SARS-CoV-2 3CL protease (SARS-CoV-2 3CLpro or Main protease, Mpro) and papain-like protease (PLpro) are the two cysteine proteases encoded by the SARS-CoV-2 genome. These play an essential role in viral replication by cleaving the viral genome-encoded polyproteins pp1a and pp1ab to generate nonstructural and structural proteins that are essential for viral replication [1]. High-resolution X-ray crystal structures and biochemical studies have established that SARS-CoV-2 3CLpro is a 33 kDa homodimer with a catalytic Cys145-His [41] dyad [14,15]. Furthermore, the protease has a malleable and extended active site with a substrate specificity for a -R-U-Leu-Gln-X sequence, where X is a small amino acid such as Ser or Gly, R is a small hydrophobic amino acid and U can be Thr, Val, Arg or Lys, corresponding to amino acid residues -P4-P3-P2-P1-P1’- [16,17]. Recent crystallographic studies have illuminated further the molecular determinants associated with the interaction of SARS-CoV-23CLpro with its substrates [[18], [19], [20]]. The protease displays a strong preference for a P1 Gln (or Gln mimic) at the S1 subsite and no human homologous protease is known to have the same primary substrate specificity. Furthermore, the plasticity of the S2 pocket enables it to accommodate an array of unnatural hydrophobic amino acids [21,22]. Importantly, the substrate binding cleft is fairly conserved among coronaviruses, suggesting that the design of broad-spectrum DAAs may be feasible [23]. The protease is a validated target for the development of SARS-CoV-2 therapeutics [[24], [25], [26], [27]] and has been the subject of intense investigations by us [28] and others [29]. We report herein the structure-guided design, synthesis, structure-activity relationships, and biochemical, structural, and antiviral activity studies of potent, broad-spectrum inhibitors of SARS-CoV-2 3CLpro that exploit the conformational control and other beneficent attributes of the gem-dimethyl moiety to access new chemical space and enhance enzyme-inhibitor interactions in the S3-S4 active site domains of the protease.
2. Results and discussion
Inhibitor design. The gem-dimethyl moiety has been widely used in drug design and optimization campaigns.30,28(d) The introduction of a gem-dimethyl group frequently increases target engagement by enabling an inhibitor to assume an entropically favorable bioactive conformation, resulting in enhanced potency. Guided by high-resolution cocrystal structures, we envisaged constructing an inhibitor (represented by general structure (I), Table 1 ) capable of exploiting the directional control afforded by the gem-dimethyl group to orient a phenyl or substituted phenyl group toward the malleable and hydrophobic S4 pocket of the protease and a tethered P2-P1 recognition element (a Leu-Gln surrogate fragment) that is congruent with the known substrate specificity of the protease capable of engaging in multiple H-bonding and hydrophobic interactions. It was also anticipated that oxidation of the thioether to generate a new stereocenter would allow optimal stereochemical control and furnish a new locus for H-bonding interactions. The warhead (Z) was chosen to be an aldehyde or latent aldehyde bisulfite adduct, however, for comparative purposes, we also interrogated the effect on potency of an α-ketoamide [15,31,32] or masked α-ketoamide [33], and nitrile [24,34] warheads, as well as the effect of deuteration, on potency [35].
Table 1.
IC50 and CC50 values of compounds 1 c-g and 2c/d-17c/d against SARS-CoV-2 3CLpro and MERS-CoV 3CLpro, and EC50 values of selected compounds against SARS-CoV-2.
General Structure (I) of inhibitors for 1 c-g, 2c/d – 7c/d, 10c/d – 16c/d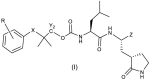 | ||||||||
|---|---|---|---|---|---|---|---|---|
| Compound Code | R | X | Y | Z | IC50 (μM)a |
EC50 (μM)a,b |
CC50 (μM) | |
| SARS-CoV-2 | MERS-CoV | SARS-CoV-2 | ||||||
| 1c | H | S | H | -CHO | 0.07 ± 0.02 | 0.08 ± 0.02 | 0.012 ± 0.01 | >50 |
| 1d | -CH(OH)SO3Na | 0.09 ± 0.01 | 0.09 ± 0.02 | 0.010 ± 0.02 | >50 | |||
| 1e | 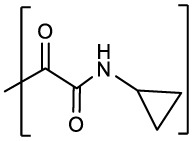 |
0.21 ± 0.02 | 0.04 ± 0.01 | 0.05 ± 0.03 | >50 | |||
| 1f | 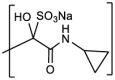 |
0.45 ± 0.05 | 0.18 ± 0.06 | 0.07 ± 0.05 | >50 | |||
| 1g | -CN | 0.49 ± 0.05 | 0.41 ± 0.08 | 0.06 ± 0.06 | >50 | |||
| 2c | H | S | D | -CHO | 0.20 ± 0.02 | 0.06 ± 0.01 | 0.011 ± 0.02 | >50 |
| 2d | -CH(OH)SO3Na | 0.31 ± 0.03 | 0.04 ± 0.01 | 0.012 ± 0.03 | >50 | |||
| 3cd | H | 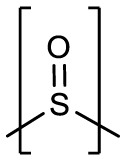 |
H | -CHO | 0.33 ± 0.01 | 0.19 ± 0.07 | NT | >50 |
| 3dd | -CH(OH)SO3Na | 0.35 ± 0.10 | 0.25 ± 0.09 | NT | >50 | |||
| 4c | H | 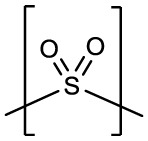 |
H | -CHO | 0.19 ± 0.05 | 0.26 ± 0.01 | NT | >50 |
| 4d | -CH(OH)SO3Na | 0.20 ± 0.01 | 0.28 ± 0.01 | NT | >50 | |||
| 5c | m-F | S | H | -CHO | 0.14 ± 0.02 | 0.10 ± 0.02 | 0.009 ± 0.02 | >50 |
| 5d | -CH(OH)SO3Na | 0.13 ± 0.01 | 0.13 ± 0.03 | 0.010 ± 0.02 | >50 | |||
| 6c | p-F | S | H | -CHO | 0.17 ± 0.03 | 0.19 ± 0.07 | NT | >50 |
| 6d | -CH(OH)SO3Na | 0.27 ± 0.02 | 0.15 ± 0.01 | NT | >50 | |||
| 7c | m-Cl | S | H | -CHO | 0.15 ± 0.01 | 0.11 ± 0.04 | 0.010 ± 0.01 | >50 |
| 7d | -CH(OH)SO3Na | 0.17 ± 0.03 | 0.10 ± 0.02 | 0.011 ± 0.01 | >50 | |||
| 8c |  |
-CHO | 0.32 ± 0.03 | 0.14 ± 0.04 | NT | >50 | ||
| 8d | -CH(OH)SO3Na | 0.31 ± 0.02 | 0.14 ± 0.05 | NT | >50 | |||
| 9c | 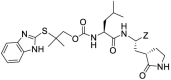 |
-CHO | 0.30 ± 0.02 | 0.12 ± 0.04 | NT | >50 | ||
| 9d | -CH(OH)SO3Na | 0.31 ± 0.09 | 0.21 ± 0.10 | NT | >50 | |||
| 10c | H | O | H | -CHO | 0.31 ± 0.05 | 0.04 ± 0.01 | NT | >50 |
| 10d | -CH(OH)SO3Na | 0.24 ± 0.05 | 0.02 ± 0.02 | NT | >50 | |||
| 11c | p-Cl | O | H | -CHO | 0.46 ± 0.11 | 0.02 ± 0.01 | NT | >50 |
| 11d | -CH(OH)SO3Na | 0.34 ± 0.03 | 0.11 ± 0.05 | NT | >50 | |||
| 12c | p-Cl | O | D | -CHO | 0.17 ± 0.05 | 0.04 ± 0.01 | 0.015 ± 0.05 | >50 |
| 12d | -CH(OH)SO3Na | 0.17 ± 0.02 | 0.02 ± 0.02 | 0.012 ± 0.03 | >50 | |||
| 13c | m-Cl | O | H | -CHO | 0.19 ± 0.02 | 0.02 ± 0.02 | 0.013 ± 0.01 | >50 |
| 13d | -CH(OH)SO3Na | 0.21 ± 0.03 | 0.02 ± 0.02 | 0.014 ± 0.01 | >50 | |||
| 14c | m-Cl | O | D | -CHO | 0.30 ± 0.08 | 0.04 ± 0.03 | NT | >50 |
| 14d | -CH(OH)SO3Na | 0.28 ± 0.07 | 0.07 ± 0.02 | NT | >50 | |||
| 15c | p-F | O | H | -CHO | 0.22 ± 0.03 | 0.03 ± 0.02 | NT | >50 |
| 15d | -CH(OH)SO3Na | 0.23 ± 0.01 | 0.04 ± 0.02 | NT | >50 | |||
| 16c | m-F | O | H | -CHO | 0.20 ± 0.01 | 0.05 ± 0.02 | NT | >50 |
| 16d | -CH(OH)SO3Na | 0.12 ± 0.02 | 0.04 ± 0.02 | NT | >50 | |||
| 17cd | 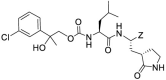 |
-CHO | 0.29 ± 0.02 | 0.12 ± 0.01 | 0.025 ± 0.07 | >50 | ||
| 17dd | -CH(OH)SO3Na | 0.32 ± 0.12 | 0.10 ± 0.01 | 0.025 ± 0.01 | >50 | |||
| GC373e | 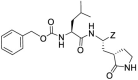 |
-CHO | 0.38 ± 0.09 | 0.28 ± 0.05 | 0.031 ± 0.02 | >100 | ||
| GC376e | -CH(OH)SO3Na | 0.41 ± 0.07 | 0.25 ± 0.10 | 0.027 ± 0.01 | >100 | |||
Chemistry. The synthetic routines employed in the synthesis of precursor alcohols 1–17 are illustrated in Schemes S1 and S2 and entailed the reaction of a substituted or non-substituted phenol, thiophenol or heterocyclic thiol, with ethyl 2-bromo-2-methylpropionate or 2-bromoisobutyric acid to yield the alkylated product [[36], [37], [38]] which was reduced to alcohols 1, 5–7,9 using LiAlH4 or alcohols 10–16 using carbonyl diimidazole/NaBH4 or NaBD4 respectively (Scheme S1) [39]. In some cases (alcohol 2) the ester was first hydrolyzed to the acid with aqueous LiOH/aqueous THF followed by reduction of the acid using carbonyl diimidazole/NaBD4. Alcohols 3–4 were synthesized by controlled oxidation of alcohol 1 using m-chloroperoxybenzoic acid (Scheme S1) while alcohol 17 was synthesized via an epoxide intermediate as shown in Scheme S2 [40,41]. Alcohol 8 was commercially available. Some of the generated alcohols (1–9, 17) were reacted with leucine methyl ester isocyanate, followed by ester hydrolysis and peptide coupling with a Gln surrogate methyl ester to yield dipeptidyl esters (1-9b, 17b) which were reduced with LiBH4 and subsequently oxidized with Dess-Martin periodinane reagent to obtain aldehyde inhibitors 1-9c and 17c (Scheme 1 ). The rest of the alcohols (10–16) were subsequently treated with disuccinimidyl carbonate [42] and the activated mixed esters were reacted with a Leu-Gln surrogate amino alcohol to yield the dipeptidyl alcohols (10b-16b), which were then oxidized to the corresponding aldehydes (10c-16c) with Dess-Martin periodinane reagent (Scheme 2 ). Compounds 1-17c and 1e were transformed into the bisulfite salts (1-17d and 1f) by treatment with sodium bisulfite as described previously (Scheme 1, Scheme 2) [28,43]. α-Ketoamide 1e was prepared by reacting aldehyde 1c with cyclopropyl isonitrile followed by oxidation with Dess-Martin periodinane reagent, while nitrile inhibitor 1g was obtained by converting dipeptidyl ester 1b to the primary amide, followed by dehydration of the amide (Scheme 1) [24].
Scheme 1.
General Synthesis of Inhibitors 1-9c/d, 1e-g and 17c/d.
Scheme 2.
General Synthesis of Inhibitors 10-16c/d.
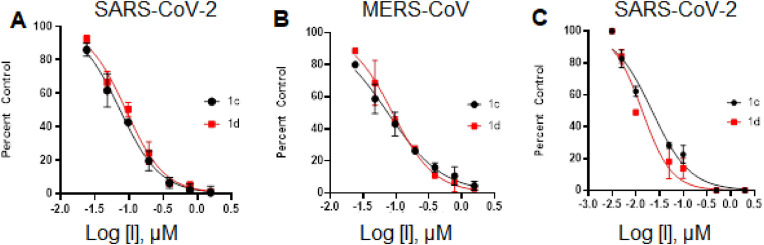
Biochemical studies. The inhibitory activity of compounds 1c-g and 2c/d-17c/d in biochemical assays toward SARS-CoV-2 and MERS-CoV 3CLpro (b), (d), as well as the cytotoxicity of the compounds [28i], were determined and the results are listed in Table 1. A select number of compounds were tested in a cell-based assay against SARS-CoV-2,28h,44 as described in the experimental section. The IC50 values, EC50 values for a select number of inhibitors, and the CC50 values in 293T cells are summarized in Table 1 and are the average of at least three determinations. All tested compounds were active against both SARS-CoV-2 and MERS-CoV 3CLpro and the IC50 values ranged between 70-490 nM and 20–410 nM for SARS-CoV-2 and MERS-CoV 3CL proteases, respectively. The EC50 values of compounds 1c/d, 2c/d, 5c/d, 7c/d, 12c/d and 13c/13d against SARS-CoV-2 3CLpro were found to be in the 9–15 nM range and the CC50 values were >50. The Safety Indices (defined as SI=CC50/EC50) for the aldehyde and bisulfite adducts were very high and ranged between 3333 and 5556. For comparative purposes, the IC50 and EC50 values of GC376 and its precursor aldehyde GC373 are also listed in Table 1. The EC50 values of α-ketoamide 1e and corresponding bisulfite adduct 1f were determined to be 50 and 70 nM, respectively. The EC50 of nitrile inhibitor 1g was found to be 60 nM. Representative dose-dependent inhibition curves in enzyme and cell-based assays are shown in Fig. 1 . A select number of compounds were found to inhibit feline coronavirus (feline infectious peritonitis virus, FIPV, an alpha coronavirus), and EC50s were ranged between 0.06 and 1.32 μM. The EC50 values of aldehyde 1c, α-ketoamide 1e and nitrile inhibitor 1g of against FIPV were 0.08, 0.71 and 1.32 μM, respectively (Table 2 ). The selectivity of representative compounds against a panel of human proteases, including thrombin, chymotrypsin, neutrophil elastase, cathepsin D and cathepsin G was also examined. The compounds did not show any inhibitory activity against these proteases up to 50 μM (Table 2). However, the selected compounds inhibited cathepsin B and cathepsin L with calculated IC50s in the range 0.8–2.5 μM and 0.02–0.04 μM, respectively (Table 2).
Fig. 1.
Inhibition plots of compounds 1c and 1d in enzyme assay (panel A and B) and cell-based SARS-CoV-2 replicon assay (panel C).
Table 2.
EC50 or IC50 values of selected compounds against feline coronavirus (FIPV) in cell culture or human proteases, including thrombin, chymotrypsin, neutrophil elastase, cathepsin B (Cat B), cathepsin D (Cat D), cathepsin G (Cat G) and cathepsin L (Cat L) in enzyme assays, respectively.
| Compound | EC50 (μM)a |
IC50 (μM)a |
||||||
|---|---|---|---|---|---|---|---|---|
| FIPV | Thrombin | Chymo-trypsin | Neutrophil elastase | Cat B | Cat D | Cat G | Cat L | |
| 1c | 0.08 ± 0.01 | >50 | >50 | >50 | 1.9 ± 0.9 | >50 | >50 | 0.03 ± 0.01 |
| 1e | 0.71 ± 0.14 | >50 | >50 | >50 | 2.0 ± 0.4 | >50 | >50 | 0.04 ± 0.02 |
| 1g | 1.32 ± 0.82 | >50 | >50 | >50 | 2.6 ± 0.1 | >50 | >50 | 0.03 ± 0.01 |
| 2c | 0.10 ± 0.03 | >50 | >50 | >50 | 2.0 ± 0.4 | >50 | >50 | 0.03 ± 0.02 |
| 7c | 0.19 ± 0.04 | >50 | >50 | >50 | 1.4 ± 0.4 | >50 | >50 | 0.03 ± 0.01 |
| 12c | 0.10 ± 0.02 | >50 | >50 | >50 | 0.8 ± 0.4 | >50 | >50 | 0.04 ± 0.01 |
| 13c | 0.06 ± 0.02 | >50 | >50 | >50 | 0.9 ± 0.3 | >50 | >50 | 0.02 ± 0.01 |
| 15c | 0.07 ± 0.02 | >50 | >50 | >50 | 1.9 ± 0.9 | >50 | >50 | 0.03 ± 0.02 |
Mean ± SD for IC50 and EC50 determined as described in the experimental section.
2.1. X-ray crystallography studies
X-ray crystallography and screening were iteratively used to guide the synthesis of analogues and optimize potency. Furthermore, the high-resolution cocrystal structures were used to gain insight and understanding into the molecular interactions associated with binding and to unravel the mechanism of action of the compounds.
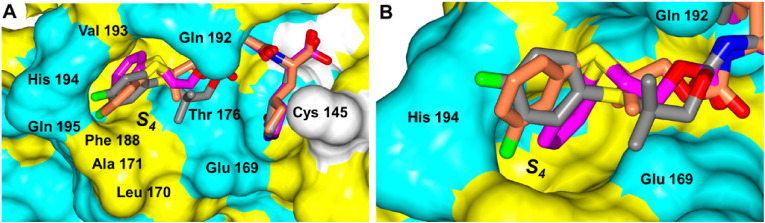
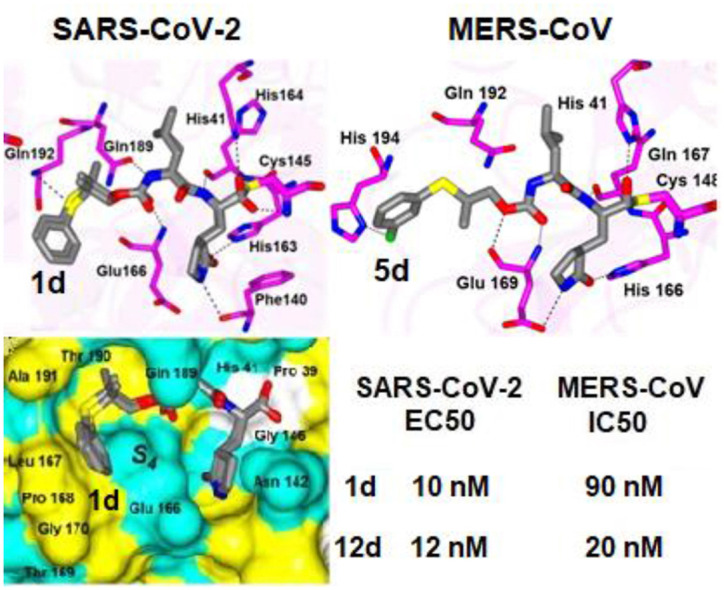
Structural Studies of MERS-CoV 3CLpro. The crystal structures of MERS-CoV 3CLpro were obtained in complex with inhibitors 3d, 5d, 6d and 8d. The inhibitors that contain a sulfide group, 5d, 6d and 8d, were first analyzed. In particular, 5d and 6d which both contain a gem-dimethyl group and only differ by the position of the fluorine atom on the aryl ring adopt similar binding modes (Fig. 2 A-D). Both compounds form the typically observed hydrogen bonds to the protein including His166 and Glu169, except the contact to Gln 192 is absent for 5d. However, 5d forms a new contact between the m-fluoro atom of the inhibitor and a side chain nitrogen atom of His 194. This places the gem-dimethyl group and m-fluoroaryl ring of 5d deep into the S4 subsite (Fig. 3 ). Conversely, 6d which contains a p-fluoroaryl ring is positioned over the top of the His 194 residue in the S4 subsite and the gem-dimethyl group is directed away from S4 pocket (Fig. 3). Interestingly, 8d which does not contain a gem-dimethyl group and lacks substituents on the aryl ring is positioned the furthest into the S4 pocket relative to 5d and 6d and shown in Fig. 3.
Fig. 2.
Binding mode of inhibitors 5d (A-B), 6d (C-D) and 8d (E-F) with MERS-CoV 3CLpro. A, C, E) Fo-Fc Polder omit map (green mesh) contoured at 3σ. B, D, E) Hydrogen bond interactions indicated by the dashed lines. PDB IDs: 5d (8E6C), 6d (8E6D), 8d (8E6E).
Fig. 3.
Comparison of 5d (coral), 6d (gray) and 8d (magenta) in the MERS-CoV 3CLpro active site. A) Surface representation showing the orientation of the superimposed inhibitors. Neighboring residues are colored yellow (nonpolar), cyan (polar), and white (weakly polar). B) Zoomed-in view showing the aryl rings in the S4 pocket. PDB IDs: 5d (8E6C), 6d (8E6D), 8d (8E6E).
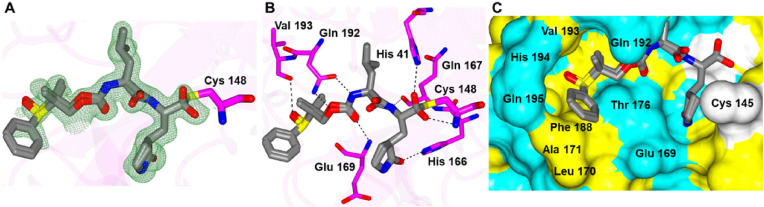
Inhibitor 3d, generated as a mixture of diastereomers, differs from the aforementioned compounds in that a gem-dimethyl group is present along with a sulfoxide group. This compound forms the typical hydrogen bonds to the protein but also contains a new contact between the sulfoxide and the backbone carbonyl group of Val 194 (Fig. 4 A and B). This results in the positioning of the gem-dimethyl group into a cleft in the S4 pocket formed between Gln 192 and Val 193 (Fig. 4C).
Fig. 4.
Binding mode of 3d with MERS-CoV 3CLpro. A) Fo-Fc Polder omit map (green mesh) contoured at 3σ. B) Hydrogen bond interactions. C) Surface representation showing the orientation of 3d in the MERS-CoV 3CLpro binding pocket. Neighboring residues are colored yellow (nonpolar), cyan (polar), and white (weakly polar). PDB IDs: 3d (8E6B).
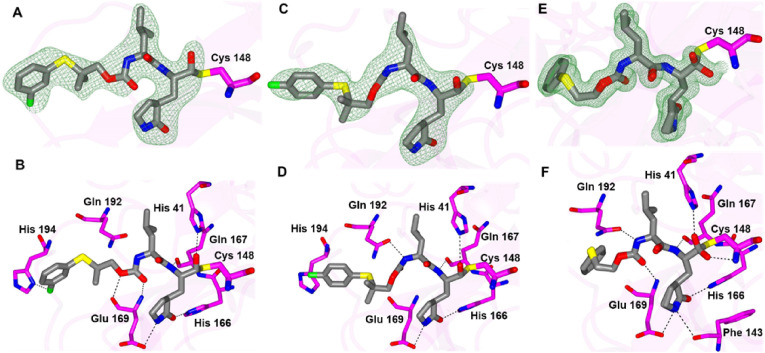
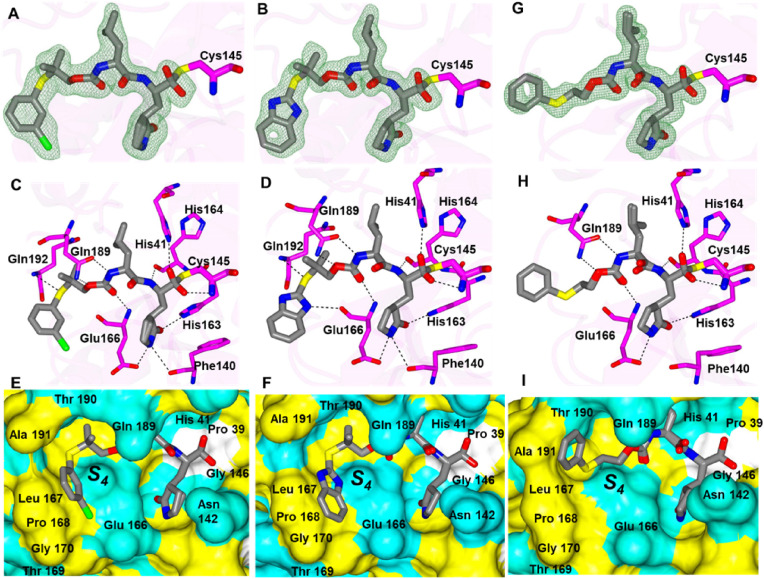
Structural Studies of SARS-CoV-2 3CLpro. The crystal structures of several inhibitors in complex with SARS-CoV-2 3CLpro were obtained for the sulfur containing compounds 1d, 1f, 1g, 3d, 4d, 7d, 8d and 9d, and the oxygen bridging compounds 11d, 15d, 16d and 17d.
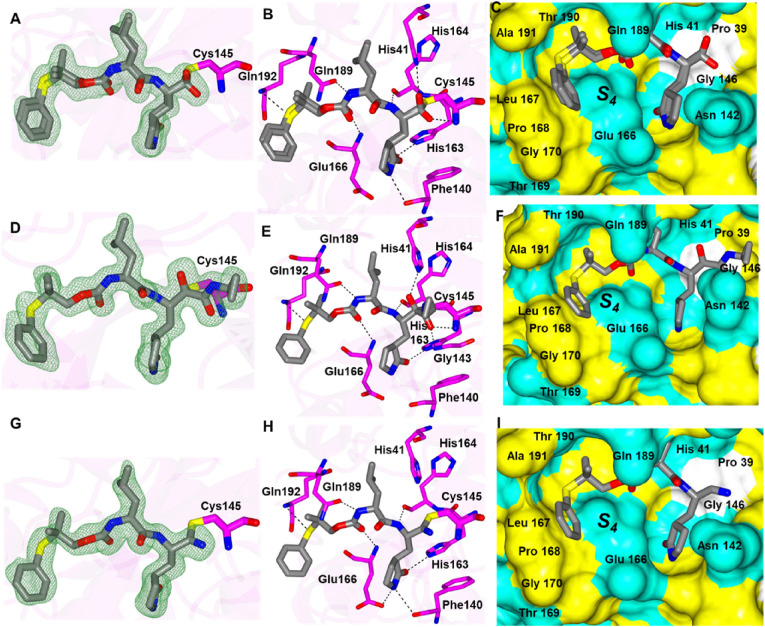
Inhibitors 1d, 1f, 1g each contain a gem-dimethyl phenylthio-group and differ in their aldehyde bisulfite, cyclopropyl ketoamide bisulfite and cyano warheads, respectively. Each of these compounds forms the typically observed H-bonding interactions (Fig. 5 ) except for the bifurcated hydrogen bond from the N-atom of the glutamine surrogate with Glu 166 and Phe 140 that was not observed for 1f. The sulfur atom of each inhibitor forms a contact with the backbone N-atom of Gln 192. This positions the phenyl group against the hydrophobic ridge of the S4 subsite and orients the gem-dimethyl group within the S4 pocket.
Fig. 5.
Binding modes of 1d (A-C), 1f (D-F) and 1g (G-I) with SARS-CoV-2 3CLpro A/D/G) Fo-Fc omit map (green mesh) contoured at 3σ. B/E/H) Hydrogen bond interactions are indicated by dashed lines. C/F/I) Surface representation showing the orientation of the aryl rings of the inhibitor in the S4 subsite of SARS2 3CLpro. Neighboring residues are colored yellow (nonpolar), cyan (polar), and white (weakly polar). PDB IDs: 1d (8F44), 1f (8F45) and 1g (8F46).
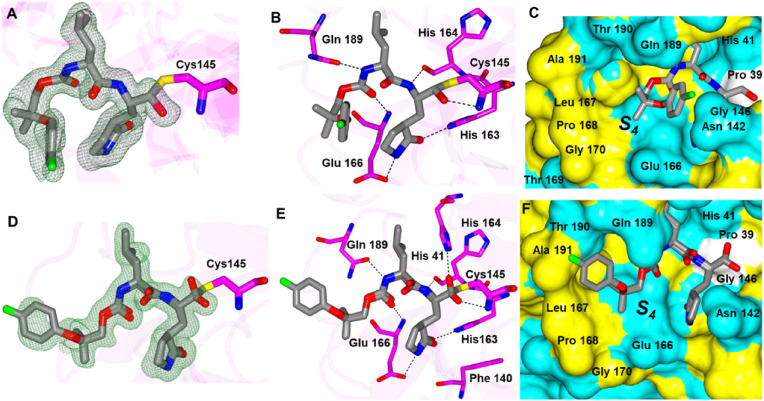
Compounds 3d and 4d contain the sulfoxide (SO) and sulfone (SO2) groups, respectively, which provides an additional polar group(s) to interact with SARS-CoV-2 3CLpro (Fig. 6 A/D). As such, along with the typical hydrogen bond interactions, 3d forms an additional contact between the sulfoxide and the N-atom of Gln 189 which positions the gem-dimethyl group towards the S4 pocket (Fig. 6B and C). Conversely, the sulfone group of 4d forms new contacts with the backbone atoms of SARS-CoV-2 3CLpro Thr 190 and Glu 166 carbonyl oxygens and the N-atom of Gln 192. This results in an approximate 90° rotation of the aryl ring and the gem-dimethyl group of 4d relative to 3d as shown in Fig. 6E-F which is positioned near Leu 167 and Pro 168 of the hydrophobic ridge.
Fig. 6.
Binding modes of 3d (A-C) and 4d (D-F) with SARS-CoV-2 3CLpro A/D) Fo-Fc omit map (green mesh) contoured at 3σ. B/E) Hydrogen bond interactions are indicated by dashed lines. C/F) Surface representation showing the orientation of the aryl rings of the inhibitor in the S4 subsite of SARS2 3CLpro. Neighboring residues are colored yellow (nonpolar), cyan (polar), and white (weakly polar). PDB IDs: 3d (8E5X) and 4d (8E5Z).
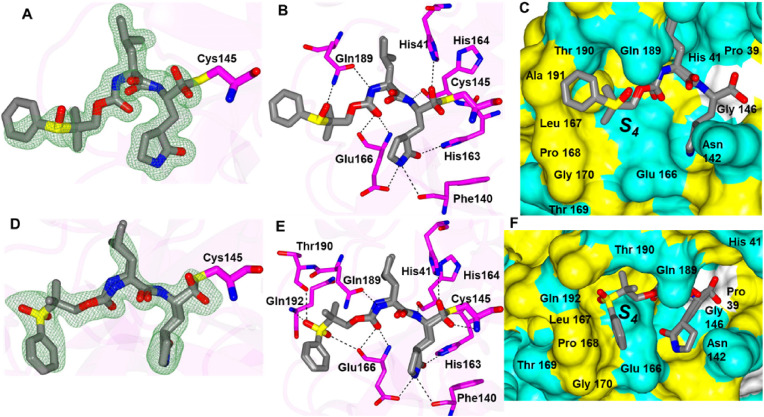
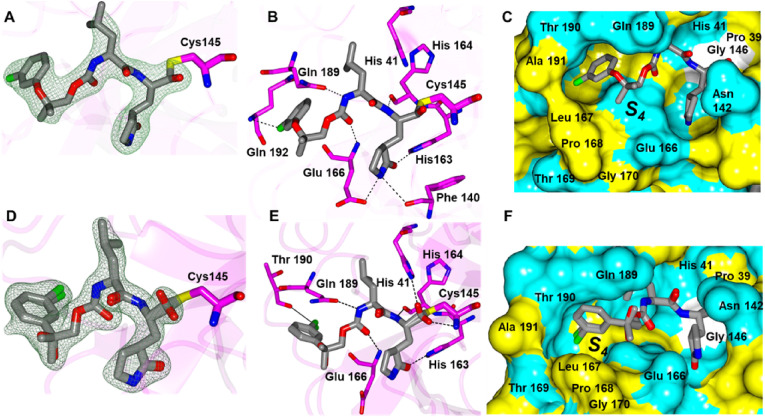
Inhibitors 7d, 8d and 9d all contain sulfide groups with an adjacent gem-dimethyl except for 8d. Interestingly, 7d and 9d adopt similar binding modes although they have different aryl groups at the terminal ends (Fig. 7 A-D). Both compounds contain an additional contact between the sulfur atom of the inhibitor and the backbone N-atom of Gln 192 and the aryl groups are positioned near the hydrophobic ridge in the S4 subsite (Fig. 7E and F). Compound 9d also harbors an additional contact observed between the benzimidazole nitrogen and the backbone carbonyl of Gln 166. This orients the gem-dimethyl groups of each inhibitor in a cleft formed by Gln 189, Thr 190 and Ala 191 in the S4 pocket. Inhibitor 8d possesses a different binding mode that lacks a contact between the sulfur atom and Gln 192 and the terminal aryl ring is positioned on top of Ala 191 (Fig. 7G-I). As such, it appears the presence of the gem-dimethyl groups in 7d and 9d may serve to orient these inhibitors in the S4 subsite and facilitate the contacts with Gln 192. This seems to be the case as well for 4d described above.
Fig. 7.
Binding modes of 7d (A/C/E), 9d (B/D/F) and 8d (G/H/I) with SARS-CoV-2 3CLpro. A/B/G) Fo-Fc omit map (green mesh) contoured at 3σ. C/D/H) Hydrogen bond interactions are indicated by dashed lines. Surface representations (E/F/I) showing the orientation of the aryl rings of the inhibitor in the S4 subsite of SARS2 3CLpro. Neighboring residues are colored yellow (nonpolar), cyan (polar), and white (weakly polar). PDB IDs: 7d (8E61), 8d (8E63), 9d (8E64).
Compounds 11d, 15d, 16d and 17d all contain an oxo linker adjacent to the gem-dimethyl group, except for 17d which contains a methyl and hydroxyl. Comparison of 11d and 15d, which only differ in that each contains a p-chloroaryl and p-fluoroaryl substituent respectively, revealed quite interesting differences. The p-chloroaryl of 11d is directed towards the surface away from the S4 subsite whereas the p-fluoroaryl group of 15d is positioned on top of the hydrophobic ridge near Ala 191 S4 subsite and the gem-dimethyl group is directed toward the interior of the S4 pocket (Fig. 8 ). This observation was unexpected given the small difference between the two compounds. However, further inspection of the crystal packing of the SARS-CoV-2 3CLpro structure with 11d, which is a different crystal form from the other structures (Table S1), revealed that a helix (Gln 244-Thr 257) from a symmetry related protein molecule is in close proximity to the active site (Fig. S1). As such, these symmetry related residues would clash with the p-chlorophenyl group of 11d if they were to adopt a binding mode similar to 15d. Therefore, we urge caution in using this monoclinic C crystal form, that was observed for 11d, when analyzing inhibitor binding modes for SARS-CoV-2 3CLpro by X-ray crystallography.
Fig. 8.
Binding mode of 11d (A-C) and 15d (D-F) with SARS-CoV-2 3CLpro. A/D) Fo-Fc omit map (green mesh) contoured at 3σ. B/E) Hydrogen bond interactions are indicated by dashed lines. Surface representations (C/F) showing the orientation of the aryl rings of the inhibitor in the S4 subsite of SARS-CoV-2 3CLpro. Neighboring residues are colored yellow (nonpolar), cyan (polar), and white (weakly polar). PDB IDs: 11d (8E65), 15d (8E68).
Comparison of inhibitors 16d and 17d, which contain m-halogen substituents, revealed quite similar binding modes. Along with the typical hydrogen bonds to the protein, a contact with the backbone N-atom of Gln 192 and the m-fluoro group of the inhibitor was observed for 16d as shown in Fig. 9 A-C. Similarly, the m-chloro atom of 17d forms a contact with the backbone carbonyl of Thr 190 (Fig. 9D-F). The m-haloaryl rings for both compounds are positioned deep within the pocket of the S4 subsite but the rings are rotated approximately 90o relative to one another to accommodate interactions with these different residues. By comparison, 15d which contains a p-fluorophenyl group is moved away from the S4 pocket relative to 16d and 17d as shown in Fig. 10 .
Fig. 9.
Binding mode of 16d (A–C) and 17d (D–F) with SARS-CoV-2 3CLpro. A/D) Fo-Fc omit map (green mesh) contoured at 3σ. B/E) Hydrogen bond interactions are indicated by dashed lines. Surface representations (C/F) showing the orientation of the aryl rings of the inhibitor in the S4 subsite of SARS-CoV-2 3CLpro. Neighboring residues are colored yellow (nonpolar), cyan (polar), and white (weakly polar). PDB IDs: 16d (8E69), 17d (8E6A).
Fig. 10.
Superposition of the SARS-CoV-2 3CLpro structures in complex with 16d (gray), 17d (gold) and 15d (coral). A) Rendering showing the differences in the inhibitors in the S4 subsite. B) Surface representations showing the position of the inhibitors in the S4 pocket with neighboring residues are colored yellow (nonpolar), cyan (polar), and white (weakly polar). PDB IDs: 15d (8E68), 16d (8E69), 17d (8E6A).
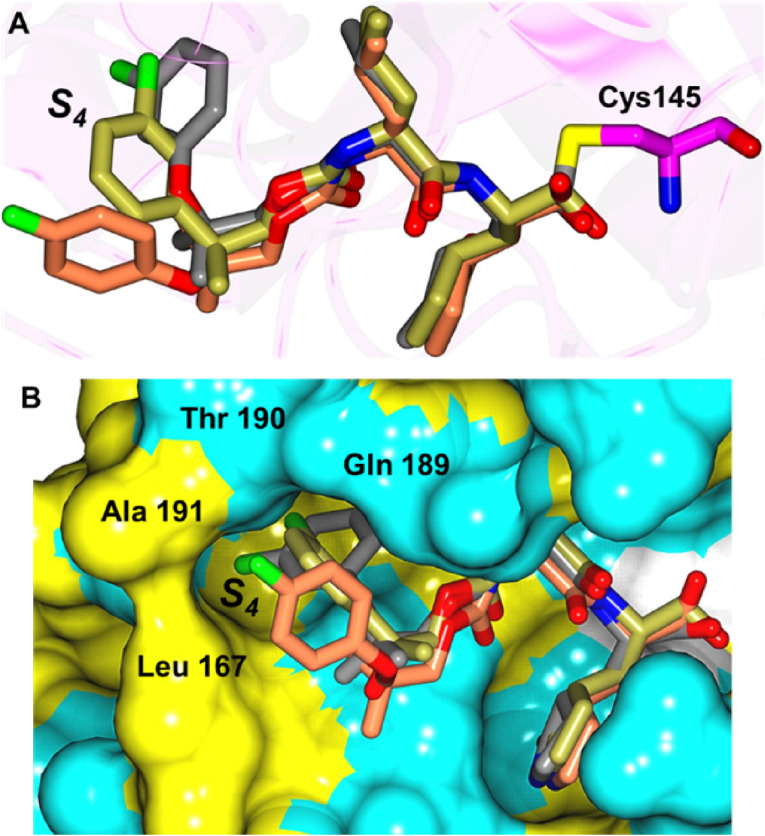
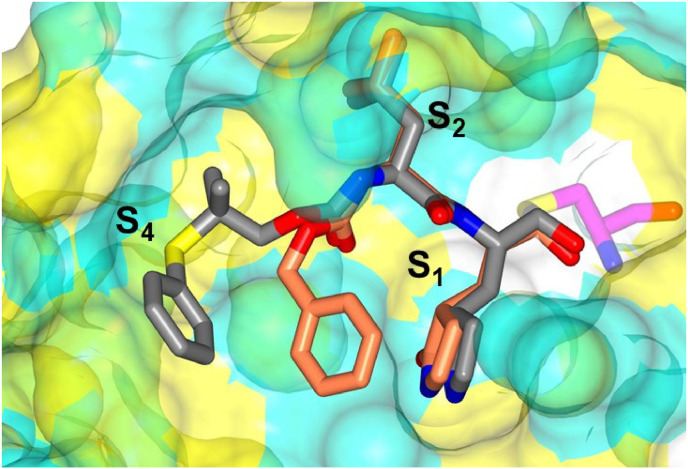
In comparing the potency of compounds 1c/d to the potency of GC373 (aldehyde) and GC376 (bisulfite adduct), the greater potency of the former arises from the ability of the gem-dimethyl group to direct the aryl group into an extended conformation, thereby engaging in favorable binding interactions with the S4 pocket, while the hairpin “closed” conformation assumed by the phenyl group in GC373/GC376 cannot engage in such interactions (Fig. 11 ).
Fig. 11.
Superposition of inhibitor GC376 (coral) bound to SARS-CoV-2 3CLpro (PDB 7K0G) superimposed with compound 1d (gray). Surface representation of SARS-CoV-2 3CLpro with neighboring residues are colored yellow (nonpolar), cyan (polar), and white (weakly polar).
Structure-Activity Relationships. Accessing new chemical space using conformational bias has been a generally fruitful strategy in drug design [30]. In the studies described herein, high-resolution cocrystal structures and the directional control afforded by the gem-dimethyl element were used in the design of a novel series of inhibitors of SARS-CoV-2 and MERS-CoV-2 3CL proteases. Overall, the results listed in Table 1 clearly demonstrate that the generated compounds potently inhibit both SARS-CoV-2 3CLpro and MERS-CoV 3CLpro, are devoid of cytotoxicity and, furthermore, several compounds display a broad spectrum of activity. For example, compounds 1c/1d (Table 1) are nanomolar inhibitors of the two proteases in biochemical and cell-based assays and are well-suited to conducting animal studies in SARS-CoV-2 and MERS-CoV mouse models of infection.28(a,d) When the sulfur is replaced by oxygen, inhibitor 10c is 2-fold more potent against MERS-CoV 3CLpro and more than 4-fold less active against SARS-CoV-2 3CLpro. In general, the replacement of sulfur by oxygen yielded compounds that were less potent against SARS-CoV-2 3CLpro (compare compounds 10c/d through 16c/d vs 1c/d and 5c/d through 7c/d) but more potent against MERS-CoV 3CLpro (Table 1, compounds 1c/d vs 10c/d; 5c/d vs 16c/d; 6c/d vs 15c/d; and 7c/d vs 13c/d). The beneficial effect of the gem-dimethyl group is clearly evident by comparing the potency of compound 1c with that of compound 8c, which is 4.6-fold and nearly 2-fold higher against SARS-CoV-2 3CLpro and MERS-CoV 3CLpro, respectively. Replacement of the phenyl ring in 1c with a benzimidazole ring (9c) resulted in >4-fold and 1.5-fold reduction in potency against SARS-CoV-2 3CLpro and MERS-CoV 3CLpro, respectively. Oxidation of the thioether in 1c/d generated the corresponding sulfoxides as a mixture of diastereomers which, along with the corresponding sulfones, were found to be less potent than the precursor sulfides (1c/d versus 3c/3d or 4c/d). Potency was near invariant to ring substitution in the X = O series (11c/d through 16c/d) and diminished in the X = S series of compounds (1c/d versus 5c/5d through 7c/d). The replacement of the aldehyde/aldehyde bisulfite adduct warheads with an α-ketoamide/α-ketoamide bisulfite adduct yielded compounds with decreased potency (compounds 1c/d versus compounds 1e/f). Importantly, the EC50 values of 1e/1f against SARS-CoV-2 were nearly equipotent (50 and 70 nM, respectively), rendering the α-ketoamide bisulfite adducts amenable to multiparameter optimization of their PK characteristics through further derivatization [45]. The potency of nitrile (Z = CN) 1g was comparable to the α-ketoamide/α-ketoamide bisulfite adduct compounds 1e/1f and equipotent against SARS-CoV-2 and MERS-CoV 3CLpro, respectively. However, the EC50 value of 1g was 5 or 6-fold higher than those of compounds 1c/1d. Deuterium substitution frequently improves the PK characteristics of drugs and, although not investigated here, a preliminary assessment of the impact of deuteration on potency was made by synthesizing several deuterated inhibitors. The EC50 values of 1c/d and the corresponding deuterated compound (2c/d) against SARS-CoV-2 3CLpro remained the same. In comparing the IC50 values of nondeuterated and deuterated pairs (Table 1, 11c/d versus 12c/d and 13c/d versus 14c/d), there was ∼2-fold increase in potency in the former pair and no change in potency in the latter pair against SARS-CoV-2 3CLpro. The IC50 values of compounds 13c/d and 14c/d remained in the nanomolar range, however, there was a 2 and 3.5-fold decrease in potency against MERS-CoV 3CLpro. There was a 1.5-fold loss in potency against SARS-CoV-2 3CLpro upon replacement of one of the methyl groups in gem-dimethyl with a polar group (OH) (Table 1, compounds 17c/d). Selected 3CLpro inhibitors showed potent inhibition against cathepsin L and moderate inhibitory activity against cathepsin B (Table 2). Interestingly, aldehyde 1c, α-ketoamide 1e, and nitrile inhibitor 1g showed similar potency against both cathepsins B and L (Table 2), which is in contrast to the variable inhibitory effects displayed against the 3CLpros (Table 1). In a previous report, we demonstrated that 3CLpro inhibitors with dual effects on cathepsin L did not inhibit the entry of SARS-CoV-2 in cells expressing both ACE2 and TMPRSS2 using pseudotyped virus assay with virus spike protein [28i]. Similarly, we found that the selected compounds used in the present studies did not inhibit the entry of SARS-CoV-2 in cells expressing both ACE2 and TMPRSS2 (data not shown).
3. Conclusions
The emergence of SARS-CoV-2, the etiological agent of COVID-19, and the severe impact of the resulting pandemic on public health worldwide, has accelerated efforts to develop effective countermeasures to combat the spread of the virus. The rapid development of vaccines and biologics to address the emergence of variants of concern and ongoing studies to better define SARS-CoV-2 biology and pathogenesis, have greatly ameliorated the impact of the virus. There is, however, an urgent need to expand the arsenal of small-molecule host and virus-targeted direct-acting antivirals as therapeutics and prophylactics. In this report, we describe the design of inhibitors of SARS-CoV-2 3CLpro that optimally utilizes structural information, and the directional and conformational effects of the gem-dimethyl group. The generated inhibitors were profiled using structure-activity relationships, and biochemical and structural studies. These studies enabled the identification of compounds that display potent activity against both SARS-CoV-2 3CLpro and MERS-CoV 3CLpro. A select number of inhibitors were also evaluated in cell-based assays, and their evaluation in relevant mouse models of infection is envisaged. High-resolution cocrystal structures confirmed the mechanism of action with the catalytic cysteine (Cys145) covalently bound to the carbonyl carbon of the inhibitors and provided valuable insights regarding the primary structural determinants associated with binding.
4. Experimental section
General. Reagents and dry solvents were purchased from various chemical suppliers (Sigma-Aldrich, Acros Organics, Chem-Impex, TCI America, Oakwood chemical, Cambridge Isotopes, AK scientific, Alpha Aesar and Fisher) and were used as obtained. Silica gel (230–450 mesh) used for flash chromatography was purchased from Sorbent Technologies (Atlanta, GA). Normal phase chromatography was performed on a Teledyne ISCO CombiFlash system using RediSep normal phase silica cartridges (35–70 μm particle size range). Thin layer chromatography was performed using Analtech silica gel plates. Visualization was accomplished using UV light and/or iodine. NMR spectra were recorded in CDCl3 or DMSO‑d 6 using a Varian XL-400 spectrometer. The purity of the final compounds was found to be ≥ 95%, as determined by absolute qNMR analysis (supplemental information) using a Bruker AV III 500 NMR spectrometer equipped with a CPDUL CRYOprobe and CASE autosampler (the University of Kansas Nuclear Magnetic Resonance Laboratory). Dimethyl sulfone TraceCERT® was used as the internal calibrant. High resolution mass spectrometry (HRMS) was performed at the Wichita State University Mass Spectrometry lab using an Orbitrap Velos Pro mass spectrometer (Thermosphere, Waltham, MA) equipped with an electrospray ion source.
Synthesis of compounds 1-9b, 17b (Scheme 1). General procedure. A solution of alcohol (1 eq/20 mmol) in dry acetonitrile (10 mL/g alcohol) was treated with triethylamine (2 eq/mmol), followed by the amino acid methyl ester isocyanate (1 eq). The resulting reaction mixture was refluxed for 2 h and then allowed to cool to room temperature. The solution was concentrated, and the residue was redissolved in ethyl acetate (20 mL/g alcohol). The organic layer was washed with 5% HCl (2 × 10 mL/g alcohol) and brine (10 mL/g alcohol). The organic layer was dried over anhydrous sodium sulfate, filtered, and concentrated to yield a crude oil. Purification of the crude oil by flash chromatography yielded the carbamate ester as a colorless oil.
A solution of the carbamate ester (1 eq) in THF (10 mL/g ester) was treated with 1 M aqueous LiOH (4 eq). The reaction mixture was stirred for 3 h at room temperature. The solvent was removed by rotovap and the solution was acidified to pH ∼2 using 5% aqueous hydrochloric acid. The aqueous layer was extracted with ethyl acetate (3 × 10 mL/g ester) and the combined organic layers were dried over anhydrous sodium sulfate, filtered, and concentrated to yield the carbamate acid.
1,1′-Carbonyldiimidazole (1.2 eq) was added to a solution of carbamate acid (1 eq) in anhydrous acetonitrile (7 mL/g carbamate acid) and stirred for 30 min at room temperature. In a separate round botttom flask, a solution of glutamine surrogate HCl (1 eq) in anhydrous acetonitrile (7 mL/g carbamate acid) was cooled to 0−5 °C, treated with TEA (4 eq) and stirred for 30 min, whereupon the two solutions were mixed and stirred overnight. The solvent was removed, and the residue was redissolved in ethyl acetate (30 ml/g carbamate acid). The organic layers were washed with saturated sodium bicarbonate (2 × 10 mL/g carbamate acid) and brine (10 mL/g carbamate acid), dried over anhydrous Na2SO4, filtered, and concentrated in vacuo. The resulting crude product was purified by flash chromatography (silica gel/ethyl acetate/hexane) to yield dipeptidyl esters 1-9b, 17b.
Synthesis of compounds 10-16b (Scheme 2). General procedure. To a solution of alcohol (1 eq) in anhydrous acetonitrile (10 mL/g alcohol) was added N,N′-disuccinimidyl carbonate (1.2 eq) and TEA (3.0 eq), and the reaction mixture was stirred for 4 h at room temperature. The solvent was removed in vacuo and the residue was dissolved in ethyl acetate (40 mL/g alcohol). The organic phase was washed with saturated aqueous NaHCO3 (2 × 20 mL/g alcohol), followed by brine (20 mL). The organic layers were combined and dried over anhydrous Na2SO4, filtered and concentrated in vacuo to yield the mixed carbonate which was used in the next step without further purification.
To a solution of Leu-Gln surrogate amino alcohol a (1.0 eq) in dry methylene chloride (10 mL/g of amino alcohol) was added TEA (1.5 eq) and the reaction mixture was stirred for 20 min at room temperature (solution 1). In a separate flask, the mixed carbonate was dissolved in dry methylene chloride (10 mL/g of carbonate) (solution 2). Solution 1 was added to solution 2 and the reaction mixture was stirred for 3 h at room temperature. Methylene chloride was added to the organic phase (40 mL/g of carbonate) and then washed with saturated aqueous NaHCO3 (2 × 20 mL/g alcohol), followed by brine (20 mL). The organic phase was dried over anhydrous Na2SO4, filtered and concentrated in vacuo. The resultant crude product was purified by flash chromatography (silica gel/hexane/ethyl acetate) to yield dipeptidyl alcohols 10-16b as a white solid.
Synthesis of compounds 1-17c (Scheme 1, Scheme 2). General procedure. For compounds 1-9b, 17b (dipeptidyl esters), the dipeptidyl alcohols were synthesized as follows: lithium borohydride (2 M in THF, 3 eq) was added to a solution of dipeptidyl ester (1 eq) in anhydrous THF (10 mL/g ester), stirred for 30 min, followed by dropwise addition of anhydrous methanol (10 mL/g ester), and stirred at room temperature overnight. The reaction mixture was concentrated, acidified with 5% aqueous hydrochloric acid and extracted with ethyl acetate (3 × 10 mL/g ester). The combined organic layers were washed with brine (10 mL/g ester), dried over anhydrous sodium sulfate, filtered, and concentrated to yield a crude off-white solid, which was purified by flash chromatography to yield the dipeptidyl alcohol as a white solid.
Dipeptidyl alcohols 10-16b and dipeptidyl alcohols derived from above 1-9b, 17b were used in the next step.
To a solution of the dipeptidyl alcohol (1 eq) in anhydrous dichloromethane (300 mL/g dipeptidyl alcohol) kept at 0–5 °C under a N2 atmosphere was added Dess-Martin periodinane reagent (3 eq) and the reaction mixture was stirred for 3 h at 15–20 °C. The organic phase was washed with 10% aq Na2S2O3 (2 × 100 mL/g dipeptidyl alcohol), followed by saturated aqueous NaHCO3 (2 × 100 mL/g dipeptidyl alcohol), distilled water (2 × 100 mL/g dipeptidyl alcohol), and brine (100 mL). The organic phase was dried over anhydrous Na2SO4, filtered and concentrated in vacuo. The resulting crude product was purified by flash chromatography (silica gel/hexane/ethyl acetate) to yield aldehydes 1-17c.
Synthesis of compounds 1-17d (Scheme 1, Scheme 2). General procedure. To a solution of dipeptidyl aldehyde c (1 eq) in ethyl acetate (10 mL/g of dipeptidyl aldehyde) was added absolute ethanol (5 mL/g of dipeptidyl aldehyde) with stirring, followed by a solution of sodium bisulfite (1 eq) in water (1 mL/g of dipeptidyl aldehyde). The reaction mixture was stirred for 3 h at 50 ± 5 °C. The reaction mixture was allowed to cool to room temperature and then vacuum filtered. The solid was thoroughly washed with absolute ethanol and the filtrate was dried over anhydrous sodium sulfate, filtered, and concentrated to yield a white solid. The white solid was stirred with dry ethyl ether (3 × 10 mL/g of dipeptidyl aldehyde), followed by careful removal of the solvent using a pipette and dried using a vacuum pump for 2 h to yield dipeptidyl bisulfite adduct d as a white solid.
Synthesis of compound 1e (Scheme 1). Acetic acid (1.1 eq) and cyclopropyl isonitrile were added to a solution of 1c (1 eq) in anhydrous ethyl acetate (18 mL/g aldehyde), stirred at room temperature overnight, whereupon ethyl acetate was removed on the rotovap. The residue was redissolved in methanol (15 mL/g aldehyde). A solution of potassium carbonate (2.5 eq) in water (10 ml/g K2CO3) was added and stirred for 2 h at room temperature. The reaction mixture was concentrated and extracted with ethyl acetate (3 × 20 mL/g aldehyde). The combined organic layers were washed with 5% aqueous hydrochloric acid (20 mL/g aldehyde), brine (20 mL/g aldehyde), dried over anhydrous sodium sulfate, filtered, and concentrated to yield a crude off-white solid, which was purified by flash chromatography to yield the α-OH amide intermediate.
To a solution of α-OH amide intermediate (1 eq) in anhydrous dichloromethane (300 mL/g dipeptidyl alcohol) kept at 0–5 °C under a N2 atmosphere was added Dess-Martin periodinane reagent (3 eq) and the reaction mixture was stirred for 3 h at 15–20 °C. The organic phase was washed with 10% aq Na2S2O3 (2 × 100 mL/g dipeptidyl alcohol), followed by saturated aqueous NaHCO3 (2 × 100 mL/g dipeptidyl alcohol), distilled water (2 × 100 mL/g dipeptidyl alcohol), and brine (100 mL). The organic phase was dried over anhydrous Na2SO4, filtered and concentrated in vacuo. The resulting crude product was purified by flash chromatography (silica gel/hexane/ethyl acetate) to yield α-ketoamide 1e.
Synthesis of compounds 1f (Schemes 1). To a solution of 1e (1 eq) in ethyl acetate (10 mL/g of dipeptidyl aldehyde) was added absolute ethanol (5 mL/g of dipeptidyl aldehyde) with stirring, followed by a solution of sodium bisulfite (1 eq) in water (1 mL/g of dipeptidyl aldehyde). The reaction mixture was stirred for 3 h at 50 ± 5 °C. The reaction mixture was allowed to cool to room temperature and then vacuum filtered. The solid was thoroughly washed with absolute ethanol and the filtrate was dried over anhydrous sodium sulfate, filtered, and concentrated to yield a white solid. The white solid was stirred with dry ethyl ether (3 × 10 mL/g of dipeptidyl aldehyde), followed by careful removal of the solvent using a pipette and dried using a vacuum pump for 2 h to yield dipeptidyl bisulfite adduct d as a white solid.
Synthesis of compounds 1g (Scheme 1). 7 M Ammonia in methanol (150 eq) was added to dipeptidyl ester 1b (1 eq) in anhydrous methanol (4 mL/g ester), stirred for 6 h, followed by addition of 7 M Ammonia in methanol (50 eq), and stirred at room temperature overnight. 7 M Ammonia in methanol (50 eq) were added consecutively on day 2 and day 3 while stirring. On day 4, the reaction mixture was concentrated in vacuo to obtain dipeptidyl amide as a white solid.
Dipeptidyl amide (1 eq) and 1H-imidazole (3 eq) were dissolved in a 1:1 mixture of anhydrous dichloromethane and anhydrous pyridine. Phosphorus oxychloride (6 eq) was added dropwise at −35 °C in a dry ice-acetonitrile bath and stirred for 4 h. Aqueous 5% hydrochloric acid (10%, 80 ml/g amide) was added, and solution was allowed to warm up to room temperature over 1 h while stirring. The reaction mixture was extracted with methylene chloride (3 × 20 mL/g ester). The combined organic layers were washed with brine (20 mL/g ester), dried over anhydrous sodium sulfate, filtered, and concentrated to yield a crude off-white solid, which was purified by flash chromatography to yield 1g as a white solid.
4.1. Characterization of compounds
2-Methyl-2-(phenylthio)propyl ((S)-4-methyl-1-oxo-1-(((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)amino)pentan-2-yl)carbamate (1c). Yield (90%).1H NMR (400 MHz, cdcl3) δ 9.49 (s, 1H), 8.44–8.40 (m, 1H), 7.57–7.50 (m, 2H), 7.40–7.30 (m, 2H), 7.26–7.24 (m, 1H), 5.78 (s, 1H), 5.29 (d, J = 9.5 Hz, 1H), 4.37–4.23 (m, 2H), 3.98 (s, 2H), 3.40–3.28 (m, 2H), 2.00–1.81 (m, 2H), 1.78–1.65 (m, 3H), 1.62–1.49 (m, 3H), 1.25 (s, 6H), 0.98 (d, J = 6.1 Hz, 6H). 13C NMR (126 MHz, DMSO) δ 200.84, 178.24, 173.14, 155.92, 137.28, 130.40, 129.28, 128.89, 69.73, 56.25, 53.01, 47.69, 40.58, 39.41, 37.21, 29.26, 27.27, 25.31, 25.28, 24.21, 22.98, 21.41. HRMS m/z: [M+Na]+ Calculated for C24H35N3NaO5S 500.2195; Found 500.2185.
Sodium (2S)-1-hydroxy-2-((S)-4-methyl-2-(((((1S,2S)-2-phenylcyclopropyl)methoxy)carbonyl)amino)pentanamido)-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonate (1d). Yield (39%). 1H NMR (400 MHz, dmso) δ 7.68–7.27 (m, 14H), 5.41 (d, J = 6.1 Hz, 1H), 5.30 (d, J = 6.1 Hz, 1H), 4.29–3.59 (m, 10H), 3.18–2.90 (m, 4H), 2.23–1.85 (m, 6H), 1.80–1.32 (m, 10H), 1.26–1.03 (m, 12H), 0.93–0.78 (m, 12H). 13C NMR (126 MHz, DMSO) δ 179.25, 179.05, 171.95, 171.85, 155.94, 155.85, 137.28, 130.44, 129.25, 128.91, 84.55, 83.80, 69.73, 53.49, 53.34, 48.97, 48.45, 47.71, 40.90, 40.77, 40.40, 39.40, 37.74, 37.64, 32.09, 29.76, 27.54, 27.34, 25.33, 24.25, 24.19, 23.19, 23.11, 22.92, 21.67, 21.43, 21.29. HRMS m/z: [M+Na]+ Calculated for C24H36N3Na2O8S2 604.1740; Found 604.1721.
2-Methyl-2-(phenylthio)propyl ((S)-1-(((S)-4-(cyclopropylamino)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl)butan-2-yl)amino)-4-methyl-1-oxopentan-2-yl)carbamate (1e). Yield (64%). 1H NMR (400 MHz, cdcl3) δ 8.40 (d, J = 5.8 Hz, 1H), 7.52 (d, J = 6.6 Hz, 2H), 7.40–7.27 (m, 3H), 7.09 (s, 1H), 6.68 (s, 1H), 5.48 (d, J = 8.8 Hz, 1H), 5.29–5.15 (m, 1H), 4.44–4.27 (m, 1H), 4.02–3.88 (m, 2H), 3.39–3.24 (m, 2H), 2.82–2.71 (m, 1H), 2.62–2.28 (m, 2H), 2.10–1.87 (m, 3H), 1.80–1.58 (m, 2H), 1.57–1.44 (m, 1H), 1.23 (d, J = 3.6 Hz, 6H), 0.95 (dd, J = 6.5, 4.3 Hz, 6H), 0.88–0.76 (m, 2H), 0.68–0.51 (m, 2H). 13C NMR (126 MHz, DMSO) δ 196.43, 178.05, 172.78, 162.13, 155.81, 137.29, 130.41, 129.28, 128.89, 69.75, 52.73, 52.18, 47.67, 40.69, 37.72, 31.13, 27.18, 25.32, 25.29, 24.10, 22.97, 22.45, 21.52, 5.44, 5.39. HRMS m/z: [M+Na]+ Calculated for C28H40N4NaO6S 583.2567; Found 583.2535.
Sodium (3S)-1-(cyclopropylamino)-2-hydroxy-3-((S)-4-methyl-2-(((2-methyl-2-(phenylthio)propoxy)carbonyl)amino)pentanamido)-1-oxo-4-((S)-2-oxopyrrolidin-3-yl)butane-2-sulfonate (1f). Yield (81%). 1H NMR (400 MHz, dmso) δ 8.82–8.18 (m, 2H), 7.75–7.17 (m, 16H), 5.52–5.34 (m, 1H), 5.10–4.93 (m, 1H), 4.64–3.35 (m, 8H), 3.24–2.65 (m, 4H), 2.40–1.98 (m, 4H), 1.98–1.30 (m, 11H), 1.30–1.09 (m, 12H), 1.09–0.76 (m, 16H), 0.69–0.33 (m, 4H). 13C NMR (126 MHz, DMSO) δ 179.25, 178.04, 172.78, 170.52, 155.82, 137.29, 131.44, 130.41, 129.28, 128.90, 128.73, 125.99, 89.94, 69.71, 58.79, 53.16, 47.68, 40.60, 37.72, 31.11, 27.50, 27.18, 25.33, 24.10, 23.27, 23.09, 22.97, 22.59, 22.47, 21.39, 21.13, 5.96, 5.54, 5.44, 5.40. HRMS m/z: [M+Na]+ Calculated for C28H41N4Na2O9S2 687.2111. Found 687.2267.
2-Methyl-2-(phenylthio)propyl ((S)-1-(((S)-1-cyano-2-((S)-2-oxopyrrolidin-3-yl)ethyl)amino)-4-methyl-1-oxopentan-2-yl)carbamate (1g). Yield (78%). 1H NMR (400 MHz, cdcl3) δ 8.37 (d, J = 6.8 Hz, 1H), 7.52 (d, J = 6.7 Hz, 2H), 7.41–7.25 (m, 3H), 6.74 (s, 1H), 5.58 (d, J = 8.8 Hz, 1H), 4.88–4.75 (m, 1H), 4.37–4.20 (m, 1H), 4.03–3.88 (m, 2H), 3.40–3.24 (m, 2H), 2.54–2.28 (m, 3H), 2.00–1.91 (m, 1H), 1.90–1.75 (m, 1H), 1.75–1.47 (m, 3H), 1.28–1.21 (m, 6H), 0.95 (dd, J = 6.4, 4.0 Hz, 6H). 13C NMR (126 MHz, DMSO) δ 177.51, 172.62, 155.98, 137.29, 130.40, 129.28, 128.89, 119.58, 69.81, 52.94, 47.66, 40.15, 39.48, 38.22, 37.03, 33.40, 26.99, 25.32, 24.20, 22.94, 21.34. HRMS m/z: [M+Na]+ Calculated for C24H34N4NaO4S 497.2199; Found 497.2175.
2-Methyl-2-(phenylthio)propyl-1,1-d2 ((S)-4-methyl-1-oxo-1-(((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)amino)pentan-2-yl)carbamate (2c). Yield (43%). 1H NMR (400 MHz, cdcl3) δ 9.58 (s, 0H), 9.49 (s, 1H), 8.37 (s, 1H), 7.57–7.48 (m, 2H), 7.40–7.30 (m, 3H), 5.76 (s, 1H), 5.31–5.19 (m, 1H), 4.32 (d, J = 8.8 Hz, 1H), 3.43–3.24 (m, 2H), 2.53–2.27 (m, 1H), 1.99–1.81 (m, 2H), 1.74–1.50 (m, 6H), 1.32–1.18 (m, 6H), 1.02–0.85 (m, 6H). 13C NMR (126 MHz, DMSO) δ 200.85, 178.25, 173.15, 155.94, 137.27, 130.41, 129.28, 128.90, 59.75, 56.26, 53.02, 47.54, 40.58, 39.42, 37.21, 29.27, 27.28, 25.26, 24.22, 23.11, 22.98, 21.42. HRMS m/z: [M+Na]+ Calculated for C24H33D2N3NaO5S 502.2321; Found 502.2295.
Sodium (2S)-1-hydroxy-2-((S)-4-methyl-2-(((2-methyl-2-(phenylthio)propoxy-1,1-d2)carbonyl)amino)pentanamido)-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonate (2d). Yield (77%). 1H NMR (400 MHz, dmso) δ 7.64–7.30 (m, 14H), 5.42 (s, 1H), 5.31 (s, 1H), 4.25–3.76 (m, 4H), 3.20–2.90 (m, 4H), 2.34–1.79 (m, 6H), 1.75–1.29 (m, 10H), 1.26–1.09 (m, 12H), 0.97 (d, J = 9.6 Hz, 2H), 0.91–0.76 (m, 12H). 13C NMR (126 MHz, DMSO) δ 179.32, 179.11, 178.32, 172.19, 171.97, 170.05, 156.00, 155.91, 137.30, 130.45, 130.29, 129.41, 129.29, 129.01, 128.94, 128.76, 126.01, 84.62, 83.84, 75.56, 61.30, 56.08, 54.55, 53.63, 53.49, 53.37, 48.86, 48.45, 47.57, 47.28, 43.14, 40.79, 40.41, 39.46, 39.02, 37.66, 37.39, 27.56, 27.37, 27.27, 25.30, 24.28, 24.22, 23.86, 23.23, 23.15, 23.11, 21.46, 21.32, 21.17, 18.58, 15.15. HRMS m/z: [M+Na]+ Calculated for C24H34D2N3Na2O8S2 606.1865; Found 606.1838.
2-Methyl-2-(phenylsulfinyl)propyl ((S)-4-methyl-1-oxo-1-(((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)amino)pentan-2-yl)carbamate (3c). Yield (46%). 1H NMR (400 MHz, cdcl3) δ 9.50 (s, 2H), 9.00 (s, 1H), 8.59 (s, 1H), 7.66–7.56 (m, 4H), 7.56–7.47 (m, 6H), 6.79 (s, 1H), 6.32 (s, 1H), 5.56 (d, J = 8.3 Hz, 1H), 5.41 (d, J = 8.4 Hz, 1H), 4.44–3.95 (m, 7H), 3.42–3.26 (m, 4H), 2.57–2.45 (m, 2H), 2.45–2.34 (m, 2H), 2.11–1.47 (m, 13H), 1.14 (s, 12H), 1.00–0.89 (m, 12H). 13C NMR (126 MHz, DMSO) δ 200.84, 178.29, 173.11, 173.05, 155.67, 155.60, 139.09, 139.04, 131.45, 128.73, 126.01, 125.97, 66.37, 66.26, 58.76, 56.34, 53.14, 53.04, 40.65, 40.51, 39.44, 37.27, 29.29, 29.27, 27.31, 24.25, 24.13, 23.16, 22.99, 21.58, 21.43, 17.09, 16.95, 16.80, 16.67. HRMS m/z: [M+Na]+ Calculated for C24H35N3NaO6S 516.2145; Found 516.2140.
Sodium (2S)-1-hydroxy-2-((2S)-4-methyl-2-(((2-methyl-2-(phenylsulfinyl)propoxy)carbonyl)amino)pentanamido)-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonate (3d). Yield (81%). 1H NMR (400 MHz, dmso) δ 7.79–7.26 (m, 16H), 5.53 (dd, J = 15.6, 6.2 Hz, 1H), 5.37 (dd, J = 15.6, 5.9 Hz, 1H), 4.26–3.82 (m, 8H), 3.19–2.91 (m, 4H), 2.31–1.84 (m, 6H), 1.80–1.32 (m, 10H), 1.12–0.98 (m, 12H), 0.93–0.80 (m, 12H). 13C NMR (126 MHz, DMSO) δ 179.28, 179.09, 171.97, 171.94, 171.88, 171.79, 155.70, 155.63, 155.52, 139.10, 139.02, 131.47, 131.39, 131.32, 128.75, 126.11, 126.05, 125.99, 84.55, 84.51, 83.82, 83.76, 66.27, 66.22, 58.86, 58.82, 58.77, 53.72, 53.56, 53.50, 53.40, 49.04, 48.97, 48.58, 48.51, 40.81, 40.65, 40.38, 40.33, 39.44, 37.82, 37.78, 37.71, 32.21, 31.98, 29.99, 29.73, 27.55, 27.37, 24.29, 24.25, 23.20, 23.13, 21.45, 21.30, 17.10, 17.04, 17.01, 16.95, 16.76, 16.70, 15.19. HRMS m/z: [M+Na]+ Calculated for C24H36N3Na2O9S2 620.1689; Found 620.1677.
2-Methyl-2-(phenylsulfonyl)propyl ((S)-4-methyl-1-oxo-1-(((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)amino)pentan-2-yl)carbamate (4c). Yield (23%). 1H NMR (400 MHz, cdcl3) δ 9.48 (s, 1H), 8.49 (d, J = 5.7 Hz, 1H), 7.90 (d, J = 7.6 Hz, 2H), 7.68 (t, J = 7.5 Hz, 1H), 7.58 (t, J = 7.6 Hz, 2H), 6.05 (s, 1H), 5.29 (d, J = 8.8 Hz, 1H), 4.39–4.25 (m, 2H), 3.41–3.28 (m, 2H), 2.56–2.31 (m, 2H), 1.92–1.65 (m, 7H), 1.58–1.45 (m, 1H), 1.39–1.29 (m, 6H), 1.01–0.86 (m, 6H). 13C NMR (126 MHz, DMSO) δ 200.80, 178.25, 172.92, 155.39, 155.30, 135.34, 134.26, 129.93, 129.25, 65.31, 62.21, 56.29, 53.02, 40.54, 39.42, 37.23, 29.25, 27.29, 24.15, 22.96, 21.38, 18.67, 18.64. HRMS m/z: [M+Na]+ Calculated for C24H35N3NaO7S 532.2094; Found 532.2078.
Sodium (2S)-1-hydroxy-2-((S)-4-methyl-2-(((2-methyl-2-(phenylsulfonyl)propoxy)carbonyl)amino)pentanamido)-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonate (4d). Yield (56%). 1H NMR (400 MHz, dmso) δ 7.88–7.75 (m, 6H), 7.71–7.13 (m, 9H), 5.37 (d, J = 6.3 Hz, 1H), 5.26 (d, J = 5.8 Hz, 1H), 4.23–4.02 (m, 4H), 4.01–3.77 (m, 4H), 3.19–2.94 (m, 4H), 2.27–1.80 (m, 6H), 1.78–1.32 (m, 10H), 1.31–1.16 (m, 12H), 1.10–1.06 (m, 1H), 0.90–0.76 (m, 12H). 13C NMR (126 MHz, DMSO) δ 200.84, 179.26, 179.06, 171.80, 171.69, 155.43, 155.32, 135.44, 135.38, 134.29, 134.25, 129.95, 129.28, 84.53, 83.78, 65.35, 64.94, 62.30, 62.23, 53.59, 53.38, 49.01, 48.51, 40.72, 40.30, 39.43, 37.78, 37.68, 27.55, 27.37, 24.20, 24.14, 23.18, 23.10, 21.41, 21.25, 18.74, 18.57, 15.19. HRMS m/z: [M+Na]+ Calculated for C24H36N3Na2O10S2 636.1638; Found 636.1627.
2-((3-fluorophenyl)thio)-2-methylpropyl ((S)-4-methyl-1-oxo-1-(((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)amino)pentan-2-yl)carbamate (5c). Yield (91%). 1H NMR (400 MHz, cdcl3) δ 9.49 (s, 1H), 8.39 (d, J = 5.9 Hz, 1H), 7.35–7.22 (m, 3H), 7.12–7.03 (m, 1H), 6.25 (s, 1H), 5.38 (d, J = 8.6 Hz, 1H), 4.44–4.17 (m, 2H), 4.03–3.92 (m, 2H), 3.41–3.23 (m, 2H), 2.54–2.31 (m, 2H), 2.10–1.51 (m, 6H), 1.26 (s, 6H), 0.98 (d, J = 6.3 Hz, 6H). 13C NMR (126 MHz, DMSO) δ 200.84, 178.26, 173.15, 162.56, 160.59, 155.91, 155.81, 133.35, 132.74, 132.68, 130.60, 130.53, 123.54, 123.38, 116.50, 116.34, 69.54, 56.29, 53.02, 48.45, 48.42, 40.62, 39.42, 37.23, 29.28, 27.28, 25.33, 24.22, 24.15, 22.99, 21.40, 21.37. HRMS m/z: [M+Na]+ Calculated for C24H34FN3NaO5S 518.2101; Found 518.2092.
Sodium (2S)-2-((S)-2-(((2-((3-fluorophenyl)thio)-2-methylpropoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonate (5d). Yield (55%). 1H NMR (400 MHz, dmso) δ 7.71–7.21 (m, 14H), 5.50 (d, J = 6.1 Hz, 1H), 5.37 (d, J = 5.9 Hz, 1H), 4.08–3.59 (m, 8H), 3.16–2.92 (m, 4H), 2.25–1.78 (m, 5H), 1.72–1.30 (m, 9H), 1.27–1.14 (m, 12H), 1.11–1.02 (m, 2H), 0.91–0.79 (m, 12H). 13C NMR (126 MHz, DMSO) δ 179.26, 179.06, 178.94, 172.26, 172.02, 171.87, 162.55, 160.59, 155.93, 155.84, 155.81, 133.38, 132.77, 132.71, 130.67, 130.60, 123.51, 123.35, 116.49, 116.33, 84.56, 83.81, 69.59, 69.47, 53.50, 53.34, 48.94, 48.55, 48.44, 40.86, 40.80, 40.42, 39.42, 37.75, 37.65, 37.48, 32.46, 32.15, 29.74, 27.66, 27.53, 27.34, 25.36, 24.25, 24.22, 24.19, 23.21, 23.13, 23.03, 21.50, 21.42, 21.29. HRMS m/z: [M+Na]+ Calculated for C24H35FN3Na2O8S2 622.1645; Found 622.1636.
2-((4-Fluorophenyl)thio)-2-methylpropyl ((S)-4-methyl-1-oxo-1-(((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)amino)pentan-2-yl)carbamate (6c). Yield (91%). 1H NMR (400 MHz, cdcl3) δ 9.49 (s, 1H), 8.36 (d, J = 5.9 Hz, 1H), 7.50 (t, J = 6.8 Hz, 2H), 7.01 (t, J = 8.7 Hz, 2H), 6.18 (s, 1H), 5.36 (d, J = 8.7 Hz, 1H), 4.40–4.16 (m, 2H), 4.03–3.87 (m, 2H), 3.41–3.26 (m, 2H), 2.55–2.30 (m, 2H), 2.08–1.47 (m, 6H), 1.23 (s, 6H), 0.97 (d, J = 6.4 Hz, 6H). 13C NMR (126 MHz, DMSO) δ 200.83, 178.26, 173.16, 163.90, 161.93, 155.92, 139.58, 139.51, 126.24, 126.21, 116.03, 115.86, 69.59, 56.29, 53.02, 47.84, 40.59, 39.42, 37.23, 29.29, 27.28, 25.23, 25.18, 24.22, 22.98, 21.41, 21.36. HRMS m/z: [M+Na]+ Calculated for C24H34FN3NaO5S 518.2101; Found 518.2098.
Sodium (2S)-2-((S)-2-(((2-((4-fluorophenyl)thio)-2-methylpropoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonate (6d). Yield (92%). 1H NMR (400 MHz, dmso) δ 7.90–6.89 (m, 14H), 5.56 (s, 1H), 5.39 (s, 1H), 4.28–3.72 (m, 8H), 3.20–2.93 (m, 4H), 2.37–2.01 (m, 4H), 1.99–1.77 (m, 2H), 1.75–1.30 (m, 10H), 1.25–1.12 (m, 12H), 0.94–0.79 (m, 12H). 13C NMR (126 MHz, DMSO) δ 178.98, 178.85, 172.29, 172.04, 163.91, 161.95, 155.95, 155.89, 139.59, 139.52, 126.25, 116.04, 115.87, 69.62, 69.55, 56.32, 56.06, 53.39, 53.35, 53.07, 47.88, 47.85, 47.80, 40.89, 40.84, 40.77, 40.61, 39.46, 37.50, 37.45, 37.30, 37.25, 29.30, 27.68, 27.65, 27.37, 27.30, 25.24, 25.19, 24.24, 23.12, 23.06, 22.99, 22.93, 21.68, 21.51, 21.42, 21.38, 15.18, 15.13, 15.10. HRMS m/z: [M+Na]+ Calculated for C24H35FN3Na2O8S2 622.1645; Found 622.1639.
2-((3-Chlorophenyl)thio)-2-methylpropyl ((S)-4-methyl-1-oxo-1-(((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)amino)pentan-2-yl)carbamate (7c). Yield (58%). 1H NMR (400 MHz, cdcl3) δ 9.57 (s, 0H), 9.48 (s, 1H), 8.38 (s, 1H), 7.54 (s, 1H), 7.42 (d, J = 7.5 Hz, 1H), 7.37–7.31 (m, 1H), 7.27 (d, J = 6.5 Hz, 1H), 6.08 (s, 1H), 5.33 (s, 1H), 4.31 (s, 2H), 3.98 (s, 2H), 3.42–3.28 (m, 2H), 2.57–2.30 (m, 2H), 2.03–1.82 (m, 2H), 1.79–1.63 (m, 2H), 1.62–1.39 (m, 2H), 1.26–1.24 (m, 6H), 0.97 (d, J = 5.1 Hz, 6H). 13C NMR (126 MHz, DMSO) δ 200.84, 178.25, 173.13, 155.88, 136.26, 135.84, 133.07, 132.70, 130.55, 129.39, 69.50, 56.28, 53.01, 48.45, 40.63, 39.42, 37.22, 29.27, 27.28, 25.30, 24.22, 22.99, 21.42. HRMS m/z: [M+Na]+ Calculated for C24H34ClN3NaO5S 534.1806; Found 534.1791.
Sodium (2S)-2-((S)-2-(((2-((3-chlorophenyl)thio)-2-methylpropoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonate (7d) Yield (80%). 1H NMR (400 MHz, dmso) δ 7.68–7.28 (m, 14H), 5.38 (d, J = 6.0 Hz, 1H), 5.28 (s, 1H), 4.08–3.67 (m, 8H), 3.18–2.92 (m, 4H), 2.24–1.95 (m, 4H), 1.92–1.30 (m, 10H), 1.25–1.15 (m, 12H), 1.11–1.02 (m, 2H), 0.91–0.80 (m, 12H). 13C NMR (126 MHz, DMSO) δ 179.25, 179.05, 171.96, 171.84, 155.90, 155.81, 136.23, 135.85, 133.06, 132.75, 130.63, 129.36, 84.54, 83.80, 69.55, 53.50, 53.34, 48.96, 48.46, 40.81, 40.43, 39.41, 37.75, 37.65, 27.53, 27.34, 25.34, 24.25, 24.19, 23.21, 23.13, 21.44, 21.30. HRMS m/z: [M+Na]+ Calculated for C24H35ClN3Na2O8S2 638.1350; Found 638.1341.
2-(Phenylthio)ethyl ((S)-4-methyl-1-oxo-1-(((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)amino)pentan-2-yl)carbamate (8c). Yield (44%). 1H NMR (400 MHz, cdcl3) δ 9.55 (s, 0H), 9.47 (s, 1H), 8.37 (s, 1H), 7.41–7.32 (m, 2H), 7.32–7.24 (m, 2H), 7.23–7.15 (m, 1H), 6.20 (s, 0H), 5.32 (d, J = 8.5 Hz, 1H), 4.57–4.13 (m, 4H), 3.44–3.25 (m, 2H), 3.21–3.06 (m, 2H), 2.94–2.68 (m, 1H), 2.61–2.23 (m, 2H), 2.03–1.79 (m, 3H), 1.79–1.63 (m, 2H), 1.61–1.48 (m, 1H), 0.96 (d, J = 5.8 Hz, 6H). 13C NMR (126 MHz, DMSO) δ 200.89, 178.32, 173.10, 155.85, 135.35, 129.15, 128.21, 128.15, 125.95, 125.92, 62.26, 56.31, 53.05, 40.59, 39.46, 37.25, 31.26, 29.27, 27.31, 24.21, 22.96, 21.52. HRMS m/z: [M+Na]+ Calculated for C22H31N3NaO5S 472.1882; Found 472.1878.
Sodium (2S)-1-hydroxy-2-((S)-4-methyl-2-(((2-(phenylthio)ethoxy)carbonyl)amino)pentanamido)-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonate (8d). Yield (81%). 1H NMR (400 MHz, dmso) δ 7.60–7.19 (m, 14H), 6.08 (d, J = 6.8 Hz, 1H), 5.46–5.24 (m, 1H), 4.49–3.58 (m, 10H), 3.24–2.96 (m, 8H), 2.30–1.94 (m, 4H), 1.88–1.28 (m, 10H), 1.09–1.01 (m, 2H), 0.92–0.78 (m, 12H). 13C NMR (126 MHz, DMSO) δ 179.26, 179.08, 171.96, 171.85, 155.79, 135.36, 129.15, 128.16, 125.95, 125.89, 84.49, 83.76, 62.25, 62.21, 53.61, 53.46, 49.00, 48.54, 40.72, 40.33, 39.44, 37.78, 37.68, 31.24, 31.13, 31.04, 27.54, 27.37, 24.25, 24.20, 23.17, 23.08, 21.50, 21.34. HRMS m/z: [M+Na]+ Calculated for C22H32N3Na2O8S2 576.1427; Found 576.1418.
2-((1H-benzo[d]imidazole-2-yl)thio)-2-methylpropyl ((S)-4-methyl-1-oxo-1-(((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)amino)pentan-2-yl)carbamate (9c). Yield (17%). 1H NMR (400 MHz, cdcl3) δ 9.67 (d, J = 8.7 Hz, 1H), 9.57 (s, 0H), 9.47 (s, 1H), 8.52 (d, J = 5.4 Hz, 1H), 8.24–8.17 (m, 1H), 7.75–7.67 (m, 1H), 7.44–7.34 (m, 2H), 5.99 (d, J = 15.3 Hz, 1H), 5.25 (d, J = 8.5 Hz, 1H), 4.54–4.18 (m, 4H), 3.41–3.22 (m, 2H), 2.52–2.26 (m, 2H), 1.99–1.83 (m, 2H), 1.79–1.62 (m, 4H), 1.60–1.49 (m, 6H), 1.01–0.87 (m, 6H). 13C NMR (126 MHz, DMSO) δ 200.85, 178.29, 173.14, 163.05, 145.55, 142.87, 125.35, 125.13, 122.56, 121.56, 119.08, 118.39, 110.90, 70.12, 59.76, 56.32, 53.01, 50.38, 40.67, 39.43, 37.25, 29.26, 27.30, 25.38, 25.17, 24.22, 22.97, 21.47. HRMS m/z: [M+Na]+ Calculated for C25H36N5O5S 518.2437; Found 518.2428.
Sodium (2S)-2-((S)-2-(((2-((1H-benzo[d]imidazole-2-yl)thio)-2-methylpropoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonate (9d). Yield (68%). 1H NMR (400 MHz, dmso) δ 8.14–8.07 (m, 2H), 7.76–7.70 (m, 2H), 7.67–7.59 (m, 2H), 7.57–7.48 (m, 3H), 7.46–7.36 (m, 2H), 7.26–7.15 (m, 3H), 7.09–7.04 (m, 1H), 4.37–3.95 (m, 8H), 3.14–2.97 (m, 4H), 2.35–2.19 (m, 2H), 2.16–2.05 (m, 2H), 1.95–1.81 (m, 2H), 1.70–1.40 (m, 13H), 1.40–1.27 (m, 12H), 0.92–0.78 (m, 12H). HRMS m/z: [M+Na]+ Calculated for C25H36N5Na2O8S2 644.1801; Found 644.1787.
2-Methyl-2-phenoxypropyl ((S)-4-methyl-1-oxo-1-(((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)amino)pentan-2-yl)carbamate (10c). Yield (35%). 1H NMR (400 MHz, dmso) δ 9.41 (d, J = 7.5 Hz, 1H), 8.48 (d, J = 7.6 Hz, 1H), 7.67–7.41 (m, 2H), 7.33–7.24 (m, 2H), 7.09 (t, J = 7.4 Hz, 1H), 7.01 (d, J = 6.8 Hz, 2H), 5.76–5.65 (m, 1H), 4.28–4.07 (m, 1H), 4.00–3.87 (m, 2H), 3.19–2.96 (m, 2H), 2.35–2.06 (m, 2H), 1.94–1.77 (m, 1H), 1.75–1.34 (m, 5H), 1.23 (s, 6H), 0.96–0.81 (m, 6H). 13C NMR (126 MHz, DMSO) δ 200.86, 178.27, 173.22, 156.04, 154.27, 129.06, 124.05, 123.61, 78.68, 68.58, 56.28, 53.05, 40.65, 39.41, 37.24, 29.30, 27.29, 24.24, 23.58, 23.13, 23.00, 21.42. HRMS m/z: [M+Na]+ Calculated for C24H35N3NaO6 484.2424; Found 484.2401.
Sodium (2S)-1-hydroxy-2-((S)-4-methyl-2-(((2-methyl-2-phenoxypropoxy)carbonyl)amino)pentanamido)-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonate (10d). Yield (65%). 1H NMR (400 MHz, dmso) δ 7.71–7.38 (m, 5H), 7.34–7.24 (m, 4H), 7.13–7.06 (m, 2H), 7.05–6.93 (m, 4H), 6.10 (d, J = 7.4 Hz, 1H), 5.89 (d, J = 6.2 Hz, 0H), 5.58 (d, J = 6.2 Hz, 1H), 5.47 (d, J = 6.1 Hz, 0H), 5.43 (d, J = 5.9 Hz, 1H), 4.49–3.58 (m, 10H), 3.18–2.95 (m, 4H), 2.27–1.77 (m, 6H), 1.72–1.31 (m, 9H), 1.28–1.18 (m, 12H), 1.12–1.04 (m, 2H), 0.94–0.80 (m, 12H). 13C NMR (126 MHz, DMSO) δ 179.32, 179.11, 172.17, 172.00, 156.10, 156.02, 154.30, 129.10, 124.08, 123.61, 84.61, 83.85, 78.71, 68.61, 53.54, 53.41, 48.91, 48.48, 40.81, 40.45, 39.45, 37.79, 37.69, 32.27, 29.80, 27.56, 27.37, 24.30, 24.24, 23.62, 23.24, 23.16, 21.45, 21.33. HRMS m/z: [M+Na]+ Calculated for C24H36N3Na2O9S 588.1968; Found 588.1951.
2-(4-Chlorophenoxy)-2-methylpropyl ((S)-4-methyl-1-oxo-1-(((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)amino)pentan-2-yl)carbamate (11c). Yield (83%). 1H NMR (400 MHz, dmso) δ 9.40 (d, J = 6.4 Hz, 1H), 8.48 (d, J = 7.6 Hz, 1H), 7.63 (s, 1H), 7.52 (d, J = 8.1 Hz, 1H), 7.32 (d, J = 8.8 Hz, 2H), 7.04 (d, J = 8.7 Hz, 2H), 4.26–4.13 (m, 1H), 4.13–4.03 (m, 1H), 3.96 (s, 2H), 3.19–3.00 (m, 2H), 2.34–2.06 (m, 2H), 1.93–1.84 (m, 1H), 1.73–1.38 (m, 5H), 1.23 (s, 6H), 0.94–0.81 (m, 6H). 13C NMR (126 MHz, DMSO) δ 200.85, 178.28, 173.21, 155.98, 153.25, 128.96, 127.64, 125.74, 79.38, 68.44, 56.30, 53.05, 40.65, 39.42, 37.25, 29.31, 27.29, 24.24, 23.42, 23.00, 21.41. HRMS m/z: [M+Na]+ Calculated for C24H34ClN3NaO6 518.2034; Found 518.2028.
Sodium (2S)-2-((S)-2-(((2-(4-chlorophenoxy)-2-methylpropoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonate (11d). Yield (20%). 1H NMR (400 MHz, dmso) δ 7.71–7.45 (m, 4H), 7.37–7.29 (m, 4H), 7.10–6.97 (m, 4H), 5.47 (d, J = 6.2 Hz, 1H), 5.31 (d, J = 5.9 Hz, 1H), 4.28–3.82 (m, 10H), 3.20–2.96 (m, 4H), 2.31–1.87 (m, 6H), 1.79–1.37 (m, 10H), 1.23 (s, 12H), 0.91–0.81 (m, 12H). 13C NMR (126 MHz, DMSO) δ 179.25, 179.07, 172.00, 171.93, 156.02, 155.94, 153.24, 153.21, 129.00, 128.98, 127.61, 125.89, 125.79, 125.74, 83.77, 79.37, 79.33, 68.40, 68.34, 53.58, 53.40, 48.99, 48.52, 40.78, 40.44, 39.41, 37.78, 37.68, 32.08, 27.54, 27.35, 24.28, 24.23, 23.52, 23.42, 23.20, 23.13, 21.41, 21.29. HRMS m/z: [M+Na]+ Calculated for C24H35ClN3Na2O9S 622.1578; Found 622.1558.
2-(4-Chlorophenoxy)-2-methylpropyl-1,1-d2 ((S)-4-methyl-1-oxo-1-(((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)amino)pentan-2-yl)carbamate (12c). Yield (79%). 1H NMR (400 MHz, dmso) δ 9.40 (d, J = 7.1 Hz, 1H), 8.48 (d, J = 7.6 Hz, 1H), 7.63 (s, 1H), 7.52 (d, J = 8.1 Hz, 1H), 7.36–7.29 (m, 2H), 7.04 (d, J = 8.8 Hz, 2H), 4.25–4.17 (m, 1H), 4.12–4.04 (m, 1H), 3.20–3.02 (m, 2H), 2.33–2.07 (m, 2H), 1.95–1.84 (m, 1H), 1.74–1.37 (m, 5H), 1.23 (s, 6H), 0.94–0.83 (m, 6H). 13C NMR (126 MHz, DMSO) δ 200.84, 178.28, 173.22, 155.99, 153.26, 128.96, 127.64, 125.72, 79.27, 56.30, 53.05, 40.65, 39.43, 37.25, 29.32, 27.29, 24.24, 23.37, 23.00, 21.41. HRMS m/z: [M+Na]+ Calculated for C24H32D2ClN3NaO6 520.2160; Found 520.2147.
Sodium (2S)-2-((S)-2-(((2-(4-chlorophenoxy)-2-methylpropoxy-1,1-d2)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonate (12d). Yield (50%). 1H NMR (400 MHz, dmso) δ 7.69–7.44 (m, 5H), 7.37–7.28 (m, 4H), 7.10–6.95 (m, 4H), 5.46 (d, J = 6.2 Hz, 1H), 5.30 (d, J = 5.9 Hz, 1H), 4.28–3.79 (m, 5H), 3.17–2.96 (m, 4H), 2.25–1.90 (m, 5H), 1.79–1.33 (m, 10H), 1.27–1.18 (m, 12H), 1.11–1.03 (m, 1H), 0.92–0.79 (m, 12H). 13C NMR (126 MHz, DMSO) δ 179.26, 179.07, 172.06, 171.95, 156.04, 155.96, 153.26, 153.22, 129.01, 127.61, 125.87, 125.78, 84.53, 83.78, 79.27, 79.23, 53.58, 53.40, 48.97, 48.52, 40.78, 40.44, 39.42, 37.79, 37.69, 32.12, 29.93, 27.54, 27.35, 24.28, 24.23, 23.48, 23.39, 23.21, 23.13, 21.42, 21.29. HRMS m/z: [M+Na]+ Calculated for C24H33D2ClN3Na2O9S 624.1704; Found 624.1683.
2-(3-Chlorophenoxy)-2-methylpropyl ((S)-4-methyl-1-oxo-1-(((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)amino)pentan-2-yl)carbamate (13c). Yield (24%). 1H NMR (400 MHz, dmso) δ 9.40 (d, J = 6.9 Hz, 1H), 8.48 (d, J = 7.7 Hz, 1H), 7.63 (s, 1H), 7.57–7.42 (m, 1H), 7.32 (t, J = 8.1 Hz, 1H), 7.20–7.13 (m, 1H), 7.10–6.94 (m, 2H), 4.29–3.86 (m, 4H), 3.21–2.98 (m, 2H), 2.36–2.07 (m, 2H), 1.96–1.83 (m, 1H), 1.79–1.36 (m, 5H), 1.25 (d, J = 3.7 Hz, 6H), 0.95–0.82 (m, 6H). 13C NMR (126 MHz, DMSO) δ 200.85, 178.28, 173.20, 155.97, 155.48, 133.05, 130.43, 123.95, 123.70, 122.65, 79.78, 68.41, 56.31, 53.03, 40.70, 39.43, 37.26, 29.31, 27.30, 24.23, 23.42, 23.34, 23.01, 21.39. HRMS m/z: [M+Na]+ Calculated for C24H34ClN3NaO6 518.2034; Found 518.2017.
Sodium (2S)-2-((S)-2-(((2-(3-chlorophenoxy)-2-methylpropoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonate (13d). Yield (33%). 1H NMR (400 MHz, dmso) δ 7.68–7.38 (m, 5H), 7.31 (t, J = 8.1 Hz, 2H), 7.20–7.12 (m, 2H), 7.06 (s, 2H), 7.00 (d, J = 8.4 Hz, 2H), 6.07 (d, J = 7.0 Hz, 1H), 5.71 (dd, J = 10.5, 6.0 Hz, 1H), 4.47–3.54 (m, 8H), 3.20–2.93 (m, 4H), 2.20 (d, J = 54.8 Hz, 3H), 1.95–1.77 (m, 1H), 1.76–1.32 (m, 9H), 1.29–1.20 (m, 12H), 1.12–1.02 (m, 4H), 0.94–0.79 (m, 12H). 13C NMR (126 MHz, DMSO) δ 179.28, 179.08, 172.06, 171.93, 156.00, 155.92, 155.49, 133.02, 130.49, 123.93, 123.68, 122.67, 84.56, 79.80, 68.44, 53.53, 53.38, 48.97, 48.50, 40.83, 40.46, 39.43, 39.08, 37.78, 37.68, 29.82, 27.54, 27.35, 24.27, 24.20, 23.46, 23.36, 23.22, 23.14, 21.42, 21.30. HRMS m/z: [M+Na]+ Calculated for C24H35ClN3Na2O9S 622.1578; Found 622.1557.
2-(3-Chlorophenoxy)-2-methylpropyl-1,1-d2 ((S)-4-methyl-1-oxo-1-(((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)amino)pentan-2-yl)carbamate (14c). Yield (51%). 1H NMR (400 MHz, dmso) δ 9.40 (d, J = 6.9 Hz, 1H), 8.48 (d, J = 7.6 Hz, 1H), 7.63 (s, 1H), 7.57–7.47 (m, 1H), 7.32 (t, J = 8.1 Hz, 1H), 7.20–7.13 (m, 1H), 7.10–6.92 (m, 2H), 5.71 (dd, J = 10.4, 6.0 Hz, 1H), 4.27–4.16 (m, 1H), 4.15–4.06 (m, 1H), 3.20–2.98 (m, 2H), 2.33–2.05 (m, 1H), 1.95–1.79 (m, 1H), 1.77–1.36 (m, 5H), 1.25 (d, J = 3.6 Hz, 6H), 0.94–0.83 (m, 6H). 13C NMR (126 MHz, DMSO) δ 200.85, 178.29, 173.21, 155.99, 155.49, 133.06, 130.44, 123.94, 123.70, 122.63, 79.68, 56.32, 53.04, 40.70, 39.44, 37.27, 29.31, 27.30, 24.24, 23.38, 23.29, 23.06, 23.02, 21.42. HRMS m/z: [M+Na]+ Calculated for C24H32D2ClN3NaO6 520.2160; Found 520.2147.
Sodium (2S)-2-((S)-2-(((2-(3-chlorophenoxy)-2-methylpropoxy-1,1-d2)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonate (14d). Yield (47%). 1H NMR (400 MHz, dmso) δ 7.66–7.40 (m, 4H), 7.32 (t, J = 8.0 Hz, 2H), 7.20–7.13 (m, 2H), 7.09–6.95 (m, 4H), 6.09 (d, J = 7.1 Hz, 1H), 4.47–4.33 (m, 1H), 4.09–3.59 (m, 4H), 3.48–3.42 (m, 1H), 3.16–2.96 (m, 3H), 2.28–1.78 (m, 5H), 1.74–1.34 (m, 9H), 1.30–1.20 (m, 12H), 1.13–1.07 (m, 4H), 0.92–0.78 (m, 12H). 13C NMR (126 MHz, DMSO) δ 178.28, 178.23, 173.21, 173.13, 155.99, 155.49, 133.06, 133.01, 130.48, 130.44, 123.93, 123.70, 122.72, 122.63, 96.75, 96.23, 84.55, 79.68, 61.39, 61.17, 56.32, 56.04, 53.33, 53.04, 50.96, 50.69, 40.69, 39.43, 37.69, 37.56, 37.31, 37.27, 30.61, 29.91, 29.52, 29.41, 29.31, 28.26, 27.64, 27.55, 27.30, 24.24, 24.20, 23.38, 23.01, 21.41, 21.39. HRMS m/z: [M+Na]+ Calculated for C24H33D2ClN3Na2O9S 624.1704; Found 624.1683.
2-(4-Fluorophenoxy)-2-methylpropyl ((S)-4-methyl-1-oxo-1-(((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)amino)pentan-2-yl)carbamate (15c). Yield (46%). 1H NMR (400 MHz, dmso) δ 9.40 (d, J = 7.2 Hz, 1H), 8.48 (d, J = 7.6 Hz, 1H), 7.63 (s, 1H), 7.56–7.40 (m, 1H), 7.14–6.94 (m, 4H), 4.28–4.16 (m, 1H), 4.12–4.03 (m, 1H), 3.94 (s, 2H), 3.20–2.97 (m, 2H), 2.33–2.06 (m, 2H), 1.95–1.84 (m, 1H), 1.74–1.33 (m, 5H), 1.21 (s, 6H), 0.94–0.82 (m, 6H). 13C NMR (126 MHz, DMSO) δ 200.85, 178.28, 173.22, 159.43, 157.52, 156.01, 150.36, 125.75, 125.68, 115.57, 115.39, 78.95, 68.34, 56.30, 53.05, 40.65, 39.42, 37.25, 29.31, 27.29, 24.24, 23.44, 23.40, 23.00, 21.41. HRMS m/z: [M+Na]+ Calculated for C24H35FN3O6 480.2510; Found 480.2493.
Sodium (2S)-2-((S)-2-(((2-(4-fluorophenoxy)-2-methylpropoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonate (15d). Yield (25%). 1H NMR (400 MHz, dmso) δ 7.66–7.30 (m, 5H), 7.16–6.83 (m, 8H), 5.43 (d, J = 6.1 Hz, 1H), 5.28 (d, J = 5.8 Hz, 1H), 4.28–3.71 (m, 9H), 3.17–2.87 (m, 4H), 2.26–1.89 (m, 5H), 1.81–1.31 (m, 10H), 1.26–1.02 (m, 13H), 0.94–0.65 (m, 12H). 13C NMR (126 MHz, DMSO) δ 179.28, 179.09, 172.05, 171.96, 159.44, 157.53, 156.06, 155.98, 150.37, 125.89, 125.82, 125.74, 125.70, 115.62, 115.44, 84.55, 83.79, 78.96, 78.91, 68.32, 68.26, 53.58, 53.41, 48.99, 48.53, 40.80, 40.46, 39.42, 37.80, 37.70, 32.13, 29.92, 29.30, 27.55, 27.36, 27.29, 24.29, 24.24, 23.55, 23.45, 23.21, 23.13, 23.11, 21.43, 21.31. HRMS m/z: [M+Na]+ Calculated for C24H35FN3Na2O9S 606.1874; Found 606.1856.
2-(3-Fluorophenoxy)-2-methylpropyl ((S)-4-methyl-1-oxo-1-(((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)amino)pentan-2-yl)carbamate (16c). Yield (43%). 1H NMR (400 MHz, dmso) δ 9.40 (d, J = 7.1 Hz, 1H), 8.48 (d, J = 7.6 Hz, 1H), 7.63 (s, 1H), 7.57–7.42 (m, 1H), 7.37–7.26 (m, 1H), 6.99–6.78 (m, 3H), 4.30–3.87 (m, 4H), 3.19–2.98 (m, 2H), 2.34–2.06 (m, 2H), 1.95–1.82 (m, 1H), 1.77–1.37 (m, 5H), 1.26 (d, J = 2.7 Hz, 6H), 0.94–0.82 (m, 6H). 13C NMR (126 MHz, DMSO) δ 200.85, 178.28, 173.20, 163.30, 161.35, 155.96, 155.87, 130.16, 130.08, 119.91, 111.18, 111.00, 110.50, 110.33, 79.66, 68.44, 56.30, 53.03, 40.68, 39.42, 37.26, 29.30, 27.29, 24.23, 23.44, 23.36, 23.00, 21.40. HRMS m/z: [M+Na]+ Calculated for C24H34FN3NaO6 502.2330; Found 502.2314.
Sodium (2S)-2-((S)-2-(((2-(3-fluorophenoxy)-2-methylpropoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonate (16d). Yield (47%). 1H NMR (400 MHz, dmso) δ 7.64–7.25 (m, 7H), 6.95–6.73 (m, 5H), 5.39 (d, J = 6.0 Hz, 1H), 5.28 (d, J = 5.9 Hz, 1H), 4.28–3.72 (m, 10H), 3.19–2.90 (m, 4H), 2.26–1.89 (m, 5H), 1.80–1.35 (m, 11H), 1.30–1.11 (m, 12H), 0.94–0.70 (m, 12H). 13C NMR (126 MHz, DMSO) δ 179.29, 179.09, 172.08, 171.95, 163.29, 161.35, 156.02, 155.98, 155.94, 155.89, 130.24, 130.16, 119.99, 119.94, 111.16, 110.98, 110.47, 110.31, 84.58, 83.83, 79.69, 68.49, 53.54, 53.38, 48.96, 48.49, 40.82, 40.44, 39.43, 37.78, 37.68, 32.22, 29.81, 27.54, 27.35, 24.27, 24.21, 23.48, 23.38, 23.21, 23.13, 21.42, 21.29. HRMS m/z: [M+Na]+ Calculated for C24H35FN3Na2O9S 606.1874; Found 606.1853.
2-(3-Chlorophenyl)-2-hydroxypropyl ((S)-4-methyl-1-oxo-1-(((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)amino)pentan-2-yl)carbamate (17c). Yield (70%). 1H NMR (400 MHz, cdcl3) δ 9.46–9.41 (m, 2H), 9.08 (s, 1H), 8.90 (s, 1H), 7.52 (s, 2H), 7.37–7.27 (m, 4H), 7.26–7.20 (m, 2H), 6.08–6.00 (m, 2H), 5.24 (d, J = 8.6 Hz, 1H), 5.06 (d, J = 8.9 Hz, 1H), 4.73–4.40 (m, 3H), 4.38–4.21 (m, 3H), 4.18–3.96 (m, 2H), 3.46–3.33 (m, 4H), 2.57–2.37 (m, 4H), 1.98–1.61 (m, 12H), 1.55 (s, 3H), 1.47 (s, 3H), 1.01–0.85 (m, 12H). 13C NMR (126 MHz, DMSO) δ 200.86, 178.33, 173.18, 155.99, 149.00, 148.92, 132.73, 129.65, 126.49, 125.48, 124.27, 72.22, 72.02, 71.15, 56.35, 52.98, 40.74, 39.48, 37.28, 29.29, 27.32, 26.40, 24.17, 23.01, 21.45. HRMS m/z: [M+Na]+ Calculated for C23H32ClN3NaO6 504.1878; Found 504.1857.
Sodium (2S)-2-((2S)-2-(((2-(3-chlorophenyl)-2-hydroxypropoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonate.
Chemical Formula: C23H33ClN3NaO9S (17d). Yield (68%). 1H NMR (400 MHz, dmso) δ 7.74–7.13 (m, 14H), 6.10 (d, J = 7.0 Hz, 1H), 5.66 (d, J = 5.9 Hz, 1H), 5.50–5.39 (m, 2H), 4.20–3.86 (m, 8H), 3.19–2.99 (m, 4H), 2.23–1.70 (m, 6H), 1.61–1.34 (m, 14H), 0.89–0.68 (m, 12H). 13C NMR (126 MHz, DMSO) δ 179.31, 179.13, 172.21, 171.98, 156.06, 156.00, 149.06, 148.98, 132.74, 132.71, 129.69, 126.50, 126.46, 125.48, 124.31, 84.53, 83.80, 72.29, 72.07, 71.13, 64.96, 56.08, 53.54, 53.38, 48.93, 48.55, 40.92, 40.88, 40.47, 37.82, 37.72, 32.10, 32.04, 29.86, 27.53, 27.36, 26.67, 26.50, 26.40, 26.36, 26.29, 24.21, 24.18, 23.25, 23.15, 21.47, 21.31, 18.59. HRMS m/z: [M+Na]+ Calculated for C23H33ClN3Na2O9S 608.1422; Found 608.1401.
4.2. Cloning and expression of 3CL proteases
The codon-optimized cDNA of full length of 3CLpro of SARS-CoV-2 (GenBank number MN908947.3) fused with sequences encoding 6 histidine at the N-terminal was synthesized by Integrated DNA (Coralville, IA). The synthesized gene was subcloned into the pET-28a(+) vector. The expression and purification of SARS-CoV-2 3CLpro was conducted following a standard procedure described previously [28b]. The expression and purification of the 3CL protease of MERS-CoV performed by standard methods described previously by our lab [28d].
4.3. Biochemical FRET assays
Briefly, a stock solution of an inhibitor was prepared in DMSO and diluted in assay buffer comprised of 20 mM HEPES buffer, pH 8, containing NaCl (200 mM), EDTA (0.4 mM), glycerol (60%), and 6 mM dithiothreitol (DTT). SARS-CoV-2 3CLpro (or MERS-CoV 3CLpro) was mixed with serial dilutions of inhibitors 1c-g and 2c/d-17c/d or with DMSO in 25 μL of assay buffer and incubated at 37 °C for 1 h, followed by the addition of 25 μL of assay buffer containing substrate (FAM-SAVLQ/SG-QXL®520, AnaSpec, Fremont, CA). The substrate was derived from the cleavage sites on the viral polyproteins of SARS-CoV (or MERS-CoV). Fluorescence readings were obtained using an excitation wavelength of 480 nm and an emission wavelength of 520 nm on a fluorescence microplate reader (FLx800; Biotec, Winoosk, VT) 1 h following the addition of substrate. Relative fluorescence units (RFU) were determined by subtracting background values (substrate-containing well without protease) from the raw fluorescence values, as described previously [28b]. The dose-dependent FRET inhibition curves were fitted with a variable slope using GraphPad Prism software (GraphPad, La Jolla, CA) in order to determine the IC50 values of the compounds.
4.4. Cell-based inhibition assays
To assess antiviral effects of selected compounds (dissolved in DMSO) in cell culture, the SARS-CoV-2 replicon system with pSMART-T7-scv2-replicon (pSMART® BAC V2.0 Vector Containing the SARS-CoV-2, Wuhan-Hu-1 Non-Infectious Replicon) was used [44]. The clone was obtained from BEI Resources and experiments were performed in a BSL-2 setting. The synthetic SARS-CoV-2 replicon RNA was prepared from the pSMART-T7-scv2-replicon as described [28h], and the Neon Electroporation system (ThermoFisher, Chicago, IL) was used for the RNA electroporation to 293T cells. After the electroporation, cells were incubated with DMSO (0.1%) or each compound at 2, 0.5, 0.1 and 0.02 μM for 30 h, and luciferase activities were measured for antiviral effects. The EC50s of eight selected compounds, including 1c, 1e, 1g, 2c, 7c, 12c, 13c and 15c, were examined against feline coronavirus (FIPV) for their broad-spectrum inhibitory effects against coronaviruses belonging to a different genus. The Crandell-Rees Feline Kidney (CRFK) cells were infected with FIPV-1146 (ATCC, Manassas, VA) and various concentrations of each compound were added to cell medium. The EC50 values were determined as described previously [28g]. The dose-dependent inhibition curve for each compound was prepared and the 50% effective concentration (EC50) values were determined by GraphPad Prism software using a variable slope (GraphPad, La Jolla, CA).
4.5. Measurement of in vitro cytotoxicity (nonspecific cytotoxic effects)
Confluent cells grown in 96-well plates were incubated with various concentrations (1–100 μM) of each compound for 72 h. Cell cytotoxicity was measured in 293T cells by a CytoTox 96 nonradioactive cytotoxicity assay kit (Promega, Madison, WI), and the CC50 values were calculated using a variable slope by GraphPad Prism software. The in vitro Safety Index was calculated by dividing the CC50 by the EC50.
Effects of selected compounds against human proteases. The IC50s of eight selected compounds, including 1c, 1e, 1g, 2c, 7c, 12c, 13c and 15c, were examined against a panel of human proteases, including thrombin (AnaSpec, Fremont, CA), alpha-chymotrypsin (Sigma-Aldrich, St Louis, MO), human neutrophil elastase (Sigma-Aldrich), cathepsin B (Abcam, Eugene, OR), cathepsin D (Abcam), cathepsin G (Novus Biological, Englewood, CO) and cathepsin L (AnaSpec) in enzyme assays following manufacturer's procedures.
Crystallization and data Collection. Purified MERS 3CLpro and SARS-CoV-2 3CLpro were prepared as described previously and concentrated between 10 and 12 mg/mL in 100 mM NaCl, 20 mM Tris pH 8.0 for crystallization screening. All crystallization experiments were setup using an NT8 drop-setting robot (Formulatrix Inc.) and UVXPO MRC (Molecular Dimensions) sitting drop vapor diffusion plates at 18 °C 100 nL of protein and 100 nL crystallization solution were dispensed and equilibrated against 50 μL of the latter. 100 mM stock solutions of the inhibitors were prepared in DMSO and complexes were prepared by mixing 1 μL of the ligand with 49 μL of the 3CLpros to give a 2 mM ligand concentration. The samples were incubated on ice for 1 h prior to screening. Crystals were typically obtained in 1–2 days from the following conditions and were cryoprotected as described.
SARS-CoV-2 3CLpro. Crystals were obtained from the PACT screen (Molecular Dimensions) unless otherwise noted. All samples were frozen in a cryoprotectant solution consisting of 80% (v/v) crystallant and 20% (v/v) PEG 200 except for 7d in which the crystallization solution served as the cryoprotectant. The crystallization conditions are as follows: 1g-condition C4 (25% (w/v) PEG 1500, 100 mM PCTP pH 7.0), 3d and 9d-condition G4 (20% (w/v) PEG 3350, 100 mM Bis-Trs Propane pH 7.5, 200 mM KSCN). 4d-condition B5 (25% (w/v) PEG 1500, 100 MIB pH 8.0). 7d-condition E6 (30% (w/v) PEG 550 MME, 100 mM Bis-Tris pH 6.5, 50 mM calcium chloride). 8d-condition B5 (25% (w/v) PEG 1500, 100 mM MIB pH 8.0). 15d-condition F10 (20% (w/v) PEG 3350, 100 mM Bis-Tris propane pH 6.5, 20 mM sodium/potassium phosphate). 16d-condition F10 (20% (w/v) PEG 3350, 100 mM Bis-Tris propane pH 6.5, 20 mM sodium/potassium phosphate). 17d-condition B7 (20% (w/v) PEG 6000, 100 mM MES pH 6.0, 200 mM NaCl). 11d- Proplex screen (Molecular Dimensions) condition F6 (15% (w/v) PEG 20000, 100 mM Hepes pH 7.0). 1d- Index HT Screen (Hampton Research) condition F2 (20% (w/v) PEG 2000 MME, 100 mM Tris pH 8.5, 200 mM Trimethylamine N-oxide dihydrate). 1f- 28% (w/v) PEG 2000 MME, 100 mM Bis-Tris pH 6.5.
MERS-CoV 3CLpro. Crystals were obtained from the Index HT screen (Hampton Research). 3d- Condition H12 (30% (w/v) PEG 2000 MME, 0.15 M Potassium bromide). Cryoprotectant: Fresh drop of crystallization solution. 5d- Condition H1 (25% (w/v) PEG 3350, 100 mM Tris pH 8.5, 200 mM magnesium chloride). Cryoprotectant 1:1 (v/v) crystallization solution and 50% (w/v) PEG 3350. 6d- Condition F11 (25% (w/v) PEG 3350, 100 mM Bis-Tris pH 6.5, 200 mM sodium chloride). Cryoprotectant: Fresh drop of crystallization solution. 8d- Condition D7 (25% (w/v) PEG 3350, 100 mM Bis-Tris pH 6.5). Cryoprotectant 1:1 (v/v) crystallization solution and 50% (w/v) PEG 3350.
Structure Solution and Refinement. X-ray diffraction data were collected at the National Synchrotron Light Source-II (NSLS-II) NYX beamline 19-ID and the Advanced Photon Source (APS) IMCA-CAT beamline 17ID. Diffraction data for SARS-CoV-2 3CLpro in complex with 1f, 3d and 9d were collected in-house with a Bruker D8 Venture sealed tube microfocus (IμS 3.0, Incoatec) diffractometer equipped with Helios multilayer optics and a Photon III C14 pixel array detector. Intensities were integrated using XDS [46,47] via Autoproc [48] and the Laue class analysis and data scaling were performed with Aimless [49]. Structure solution was conducted by molecular replacement with Phaser [50] using a previously determined inhibitor bound structures of MERS-CoV 3CLpro (5WKK) and SARS-CoV-2 3CLpro (PDB 6XMK) as the search models. Structure refinement and manual model building were conducted with Phenix [51] and Coot [52], respectively. Disordered side chains were truncated to the point for which electron density could be observed. Structure validation was conducted with Molprobity [53] and figures were prepared using the CCP4MG package [54]. Crystallographic data are provided in Table S1 and S2. [[55], [56], [57], [58], [59], [60]].
Author contributions
All authors have approved the final article. Study design: W.C.G., K-O.C., Y.K, and S.L., experiments: C.S. D., M.J.M., A.Z., H.N.N., T.K.M., H.A.T., A.J.M., A.C., L.L., K.P.B., article preparation: W.C.G., K-O.C., Y.K., S.L., C.S.D.
Funding
This research was supported by the National Institutes of Health [grants R01 AI130092 and AI161085 to K.O.C.]); the National Institute of General Medical Sciences [P30GM110761]; the companies of the Industrial Macromolecular Crystallography Association; the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences [contract no. DE-AC02-06CH11357]; the U.S. Department of Energy, Office of Science [contract No. DE-SC0012704]; the Office of the Director of the National Institutes of Health [S10OD030394]. the National Institutes of Health [shared Instrumentation grant #S10RR024664]; the National Science Foundation [major research instrumentation Award # 1625923]; the National Institutes of Health, National Institute of General Medical Sciences [P30GM133893]; the Department of Energy, Office of Biological and Environmental Research [KP1605010]. The funding sources had no involvement in study design, the collection, analysis and interpretation of data, the writing of the report, and in the decision to submit the article for publication.
Accession codes
Coordinates and structure factors for complexes with the following with inhibitors were deposited to the Worldwide Protein Databank (wwPDB) with the accession codes: MERS-CoV 3CLpro complexes (3d) (8E6B), (5d) (8E6C), (6d) (8E6D) and (8d) (8E6E). SARS-CoV-2 3CLpro complexes 1d (8F44), 1f (8F45), 1g (8F46), 3d (8E5X), 4d (8E5Z), 7d (8E61), 8d (8E63), 9d (8E64), 11d (8E65), 15d (8E68), 16d (8E69) and 17d (8E6A).
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The following reagent was obtained through BEI Resources, NIAID, NIH: pSMART® BAC V2.0 Vector Containing the SARS-Related Coronavirus 2, Wuhan-Hu-1 Non-Infectious Replicon, NR-54972.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ejmech.2023.115376.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Supporting Information is available free of charge and contains synthesis of alcohol inputs, 1H NMR, 13C NMR and HRMS spectra, absolute qNMR data, superposition of the SARS-CoV-2 3CLpro structures in complex with 11d and 15d, and crystallographic data for MERS 3CLpro and SARS-CoV-2 3CLpro inhibitor complexes.
Data availability
Data will be made available on request.
References
- 1.Perlman S., Masters P.S., Coronaviridae . In: Fields Virology: Emerging Viruses. Howley P.M., Knipe D.M., Whelan S., editors. Vol. 1. 2020. The viruses and their replication; pp. 410–448. (Wolters Kluwer: Wichita). [Google Scholar]
- 2.Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.COVID-19 Map - Johns Hopkins Coronavirus Resource Center. Total Cases: 659,945,888 Infections and 6,688,724 Deaths (accessed December 30, 2022).
- 4.Tao K., Tzou P.L., Nouhin J., Gupta R.K., de Oliveira T., Pond S.L.K., Fera D., Shafer R.W. The biological and clinical significance of emerging SARS-CoV-2 variants. Nat. Rev. Genet. 2021;22:757–773. doi: 10.1038/s41576-021-00408-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abdool Karim S.S., de Oliveira T. New SARS-CoV-2 variants – clinical, public health, and vaccine implications. N. Engl. J. Med. 2021;384:1866–1868. doi: 10.1056/NEJMc2100362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shuai H., Chan J.F.-W., Hu B., Chai Y., Yuen T.T.-T., Yin F., Huang X., Yoon C., Hu J.-C., Liu H., Shi J., Liu Y., Zhu T., Zhang J., Hou Y., Wang Y., Lu L., Cai B.-Z., Huang J.-D., To K.K.-W., Yuen K.-Y., Chu H. Attenuated replication and pathogenicity of SARS-CoV-2B.11.529 omicron. Nature. 2022;603:693. doi: 10.1038/s41586-0232-04442-5. [DOI] [PubMed] [Google Scholar]
- 7.Moore J.P., Offit P.A. SARS-CoV-2 vaccines and the growing threat of viral variants. JAMA. 2021;325:821–822. doi: 10.1001/jama.2021.1114. [DOI] [PubMed] [Google Scholar]
- 8.Creech C.B., Walker S.C., Samuels R.J. SARS-CoV-2 vaccines. JAMA. 2021;325(13):1318–1320. doi: 10.1001/jama.2021.3199. [DOI] [PubMed] [Google Scholar]
- 9.Kumar S., Chandele A., Sharma A. Current status of therapeutic monoclonal antibodies against SARS-CoV-2. PLoS Pathog. 2021;17(9) doi: 10.1371/journal.ppat.1009885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghosh A.K., Mishevich J.L., Mesecar A.D., Mitsuya H. Recent drug development and medicinal approaches for the treatment of SARS coronavirus and Covid‐19. ChemMedChem. 2022;17 doi: 10.1002/cmdc.202200440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jonsson C.B., Golden J.E., Meibohm B. Time to “Mind the Gap” in novel small molecule drug discovery for direct-acting antivirals for SARS-CoV-2. Curr. Op. Virol. 2021;50:1–7. doi: 10.1016/j.coviro.2021.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gil C., Ginex T., Maestro I., Nozal V., Barrado-Gil L., Cuesta-Geijo M.A., Urquiza J., Ramírez D., Alonso C., Campillo N.E., Martinez A. COVID-19: drug targets and potential treatments. J. Med. Chem. 2020;63:12359–12386. doi: 10.1021/acs.jmedchem.0c00606. [DOI] [PubMed] [Google Scholar]
- 13.Sharma A., Tiwari S., Deb M.K., Marty J.L. Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2): a global pandemic and treatments strategies. Int. J. Antimicrob. Agents. 2020;56(2) doi: 10.1016/j.ijantimicag.2020.106054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C. Structure of M pro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 15.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rut, W.; Groborz, K.; Zhang, L.; Sun, X.; Zmudzinski, M.; Hilgenfeld, R.; Drag, M.; Substrate specificity profiling of SARS-CoV-2 Mpro protease provides basis for anti-COVID-19 drug design. bioRxiv preprint. 10.1101/2020.03.07.981928. (accessed March 7, 2020). [DOI]
- 17.The nomenclature used is that of Schechter I and Berger A. Schechter, I. On the size of the active site in proteases. I. Papain. Biochem. Biophys. Res. Commun. 1967;425(3):497–502. doi: 10.1016/j.bbrc.2012.08.015. where the residues on the N-terminus side of the peptide bond that is cleaved are designated as P1-Pn and those on the C-terminus side are designated P1’-Pn’. The corresponding active site subsites are designated S1-Sn and S1’-Sn’. S1 is the primary substrate specificity subsite and P1-P1’ is the scissile bond. [DOI] [PubMed] [Google Scholar]
- 18.Zhao Y., Zhu Y., Liu X., Jin Z., Duan Y., Zhang Q., Wu C., Feng L., Du X., Zhao J., Shao M., Zhang B., Yang X., Wu L., Ji X., Guddat L.W., Yang K., Rao Z., Yang H. Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2. Proc. Natl. Acad. Sci. U.S.A. 2022;119 doi: 10.1073/pnas.211714211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee J., Kenward C., Worrall L.J., Vuckovic M., Gentile F., Ton A.-T., Ng M., Cherkasov A., Strynadka N.C.J., Poetzel M. X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation. Nat. Commun. 2022;13:5196. doi: 10.1038/s41467-022-32854-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.MacDonald E.A., Frey G., Namchuk M.N., Harrison S.C., Hinshaw S.M., Windsor I.W. Recognition of divergent viral substrates by the SARS-CoV-2 Main protease. ACS Infect. Dis. 2021;7:2591–2595. doi: 10.1021/acsinfecdis.1c00237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kneller D.W., Phillips G., O'Neill H.M., Jedrzejczak R., Stols L., Langan P., Joachimiak A., Coates L., Kovalevsky A. Structural plasticity of SARS-CoV-2 3CL Mpro active site cavity revealed by room temperature X-ray crystallography. Nat. Commun. 2020;11:3202. doi: 10.1038/s41467-020-16954-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deshmukh M.G., Ippolito J.A., Zhang C.-H., Stone E.A., Reilly R.A., Miller S.J., Jorgensen W.L. Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease. Structure. 2021;29:823–833. doi: 10.1016/j.str.2021.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hegyi A., Ziebuhr J. Conservation of substrate specificities among coronavirus main proteases. J. Gen. Virol. 2002;83:595–599. doi: 10.1099/0022-1317-83-3-595. [DOI] [PubMed] [Google Scholar]
- 24.Owen D.R., Allerton C.M.N., Anderson A.S., Aschenbrenner L., Avery L., Berritt S., Boras B., Cardin R.D., Carlo A., Coffman K.J., Dantonio A., Di L., Eng H., Ferre R., Gajiwala K.S., Gibson S.A., Greasley S.E., Hurst B.L., Kadar E.P., Kalgutkar A.S., Lee J.C., Lee J., Liu W., Mason S.W., Noell S., Novak R., Obach R.S., Ogilvie K., Patel N.C., Pettersson M., Rai D.K., Reese M.R., Sammons M.F., Sathish J.G., Singh S.P., Steppan C.M., Stewart A.E., Tuttle J.B., Updyke L., Verhoest P.R., Wei L., Yang Q., Zhu Y. An oral SARS-CoV-2 Mpro inhibitor clinical candidate for the treatment of COVID-19. Science. 2021;374(6575):1586–1593. doi: 10.1126/science.abl4784. [DOI] [PubMed] [Google Scholar]
- 25.Boras B., Jones R.M., Anson B.J., Arenson D., Aschenbrenner L., Bakowski M.A., Beutler N., Binder J., Chen E., Eng E., Hammond H., Hammond J., Haupt R.E., Hoffman R., Kadar E.P., Kania R., Kimoto E., Kirkpatrick M.G., Lanyon L., Lendy E.K., Lillis J.R., Logue J., Luthra S.A., Ma C., Mason S.W., McGrath M.E., Noell S., Obach R.S., O'Brien M.N., O'Connor R., Ogilvie K., Owen D., Pettersson M., Reese M.R., Rogers T.F., Rosales R., Rossulek M.I., Sathish J.G., Shirai N., Steppan C., Ticehurst M., Updyke L.W., Weston S., Zhu Y., White K.M., Garcia-Sastre A., Wang J., Chatterjee A.K., Mesecar A.D., Frieman M.B., Anderson A.S., Allerton C. Preclinical characterization of an intravenous coronavirus 3CL protease inhibitor for the potential treatment of COVID19. Nat. Commun. 2021;12:6055. doi: 10.1038/s41467-021-26239-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quan B.-X., Shuai H., Xia A.-J., Hou Y., Zeng R., Liu X.-L., Lin G.-F., Quiao J.-X., Li W.-P., Wang F.-L., Wang K., Zhou R.-J., Yuen T.T.-T., Chen M.-X., Yoon, Wu M., Zhang S.-Y., Huang C., Wang Y.-F., Yang W., Tian C., Li W.-M., Wei Y.-Q., Yuen K.-Y., Chan J.F.-W., Lei J., Chu H., Yang S. An orally available Mpro inhibitor is effective against wild-type SARS-CoV-2 and variants including omicron. Nat. Microbiol. 2022;7:716–725. doi: 10.1038/s41564-022-01119-7. [DOI] [PubMed] [Google Scholar]
- 27.Gao K., Wang R., Chen J., Tepe J.J., Huang F., Wei G.-W. Perspectives on SARS-CoV-2 main protease inhibitors. J. Med. Chem. 2021;64:16922–16955. doi: 10.1021/acs.jmedchem.1c00409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.(a) Dampalla C.S., Zheng J., Perera K.D., Wong L.-Y.R., Meyerholz D.K., Nguyen H.N., Kashipathy M.M., Battaile K.P., Lovell S., Kim Y., Perlman S., Groutas W.C., Chang K.-O. Post-infection treatment with a protease inhibitor increases survival of mice with a fatal SARS-CoV-2 infection. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2101555118. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Dampalla C.S., Kim Y., Bickmeier N., Rathnayake A.D., Nguyen H.N., Zheng J., Kashipathy M.M., Baird M.A., Battaile K.P., Lovell S., Perlman S., Chang K.O., Groutas W.C. Structure-guided design of conformationally-constrained cyclohexane inhibitors of severe acute respiratory syndrome coronavirus-2 3CL protease. J. Med. Chem. 2021;64:10047–10058. doi: 10.1021/acs.jmedchem.1c00319. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Dampalla C.S., Rathnayake A.D., Perera K.D., Jesri A.-R.M., Nguyen H.N., Miller M.J., Thurman H.A., Zheng J., Kashipathy M.M., Battaille K.P., Lovell S., Perlman S., Kim Y., Groutas W.C., Chang K.O. Structure-guided design of potent inhibitors of SARS-CoV-2 3CL protease: structural, biochemical, and cell-based studies. J. Med. Chem. 2021;64:17846–17865. doi: 10.1021/acs.jmedchem.1c01037. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Rathnayake A.D., Zheng J., Kim Y., Perera K.D., Mackin S., Meyerholz D.K., Kashipathy M.M., Battaile K.P., Lovell S., Perlman S. 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV–infected mice. Science Transl. Méd. 2020;12 doi: 10.1126/scitranslmed.abc5332. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Pedersen N.C., Kim Y., Liu H., Galasiti Kankanamalage A.C., Eckstrand C., Groutas W.C., Bannasch M., Meadows J.M., Chang K.-O. Efficacy of a 3C-like protease inhibitor in treating various forms of acquired feline infectious peritonitis. J. Feline Med. Surg. 2018;20:378–392. doi: 10.1177/1098612X17729626. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Kim Y., Liu H., Galasiti Kankanamalage A.C., Weerasekara S., Hua D.H., Groutas W.C., Chang K.-O., Pedersen N.C. Reversal of the progression of fatal coronavirus infection in cats by a broad-spectrum coronavirus protease inhibitor. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005531. [DOI] [PMC free article] [PubMed] [Google Scholar]; (g) Kim Y., Lovell S., Tiew K.-C., Mandadapu S.R., Alliston K.R., Battaile K.P., Groutas W.C., Chang K.-O. Broad-spectrum antivirals against 3C or 3C-like proteases of picornaviruses, noroviruses, and coronaviruses. J. Virol. 2012;86:11754–11762. doi: 10.1128/JVI.01348-12. [DOI] [PMC free article] [PubMed] [Google Scholar]; (h) Dampalla C.S., Rathnayake A.D., Galasiti Kankanamalage A.C., Kim Y., Perera K.D., Nguyen H.N., Miller M.J., Madden T.K., Picard H.R., Thurman H.A., Kishipathy M.M., Liu L., Battaille K.P., Lovell S., Chang K.O., Groutas W.C. Structure-guided design of potent spirocyclic inhibitors of severe acute respiratory syndrome coronavirus-2 3C-like protease. J. Med. Chem. 2022;65:7818–7832. doi: 10.1021/acs.jmedchem.2c00224. [DOI] [PMC free article] [PubMed] [Google Scholar]; (i) Dampalla C.S., Nguyen H.N., Rathnayake A.D., Kim Y., Perera K.D., Madden T.K., Thurman H.A., Machen A.J., Kashipathy M.M., Liu L., Battaile K.P., Lovell S., Chang K.-O., Groutas W.C. Broad-spectrum cyclopropane-based inhibitors of coronavirus 3C-like proteases: biochemical, structural, and virological studies. Pharmacol. Transl. Sci. 2022;6:181–194. doi: 10.1021/acspytsci.2c00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.(a) Sacco M.D., Ma C., Lagarias P., Gao A., Townsend J.A., Meng X., Dube P., Zhang X., Hu Y., Kitamura N., Hurst B., Tarbet B., Marty M.T., Kolocouris A., Xiang Y., Chen Y., Wang J. Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against Mpro and cathepsin L. Sci. Adv. 2020;6 doi: 10.1126/sciadv.abe0751. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Dai W., Jochmans D., Xie H., Yang H., Li J., Su H., Chang D., Wang J., Peng J., Zhu L., Nian Y., Hildenfeld R., Jiang H., Chen K., Zhang L., Xu Y., Neyts J., Liu H. Design, synthesis, and biological evaluation of peptidomimetic aldehydes as broad-spectrum inhibitors against enterovirus and SARS-CoV-2. J. Med. Chem. 2022;65:2794–2808. doi: 10.1021/acs.jmedchem.0c02258. [DOI] [PubMed] [Google Scholar]; (c) Dai W., Zhang B., Jiang X.-M., Su H., Li J., Zhao Y., Xie X., Jin Z., Peng J., Liu F. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020;368:1331–1335. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Qiao J., Li, Zeng R., Liu F.-L., Luo R.-H., Huang C., Wang Y.-F., Zhang J., Quan B., Shen C., Mao X., Liu X., Sun W., Yang W., Ni X., Wang K., Xu L., Duan Z.-L., Zou Q.-C., Zhang H.-L., Qu W., Peng Y.-H., Li M.-H., Yang R.-C., Liu X., You J., Zhou Y., Yao R., Li W.-P., Liu J.-M., Chen P., Liu Y., Lin G.-F., Yang X., Zou J., Li L., Hu Y., Lu G.-W., Li W.-M., Wei Y.-Q., Zheng Y.-T., Lei J., Yang S. SARS-CoV-2 Mpro inhibitors with antiviral activity in a transgenic mouse model. Science. 2021;371:1374–1378. doi: 10.1126/science.abf1611. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Bai B., Belovodskiy A., Hena M., Kandadai A.S., Joyce M.A., Saffran H.A., Shields J.A., Khan M.B., Arutyunova E., Lu J., Bajwa S.K., Hockman D., Fischer C., Lamer T., Vuong W., van Belkum M.J., Gu Z., Lin F., Du Y., Xu J., Rahim M., Young H.S., Vederas J.C., Tyrrell L.D., Lemieux J., Nieman J.A. Peptidomimetic α-acyloxymethylketone warheads with six-membered lactam P1 glutamine mimic: SARS-CoV-2 3CL protease inhibition, coronavirus antiviral activity, and in vitro biological stability. J. Med. Chem. 2022;65:2905–2925. doi: 10.1021/acs.jmedchem.1c00616. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Ghosh A.K., Raghavaih J., Shahabi D., Yadav M., Anson B.J., Lendy E.K., Hattori S., Higashi-Kuwata N., Mesecar A.D. Indole chloropyridinyl ester-derived SARS-CoV-2 3CLpro inhibitors: enzyme inhibition, antiviral efficacy, structure-activity relationship and X-ray structural studies. J. Med. Chem. 2021;64:14702–14714. doi: 10.1021/acs.jmedchem.1c01214. [DOI] [PMC free article] [PubMed] [Google Scholar]; (g) Konno S., Kobayashi K., Senda M., Funai Y., Tamai I., Schakel L., Sakata K., Pillaiyar T., Taguchi A., Taniguchi A., Gutschow M., Muller C.E., Takeuchi K., Hirohama M., Kawaguchi A., Kojima M., Senda T., Shirasaka Y., Kamitani W., Hayashi Y. 3CL protease inhibitors with an electrophilic acylketone moiety as anti-SARS-CoV-2 agents. J. Med. Chem. 2022;65:2926–2939. doi: 10.1021/acs.jmedchem.1c00665. [DOI] [PubMed] [Google Scholar]; (h) Unoh Y., Uehara S., Nakahara K., Nobor H., Yamatsu Y., Yamamoto S., Maruyama Y., Taoda Y., Kasamatsu K., Suto T., Kouki K., Nakahashi A., Kawashima S., Sanaki T., Toba S., Uemura K., Mizutare T., Ando S., Sasaki M., Orba Y., Sawa H., Sato A., Sato T., Kato T., Tachibana Y. Discovery of S-127622, a noncovalent oral SARS-CoV-2 3CL protease inhibitor clinical candidate for treating COVID-19. J. Med. Chem. 2022;65:6499–6512. doi: 10.1021/acs.jmedchem.2c00117. [DOI] [PMC free article] [PubMed] [Google Scholar]; (i) Han S.H., Goins C.M., Arya T., Shin W.-J., Maw J., Hooper A., Sonawane D.P., Porter M.R., Bannister B.E., Crouch, Lindsey A.A., Lakatos G., Marttinez S.R., Alvarado J., Akers W.S., Wang N.S., Jung J.U., Macdonald J.D., Stauffer S.R. Structure-based optimization of ML300-derived, noncovalent inhibitors targeting the severe acute respiratory syndrome coronavirus 3CL protease (SARS-CoV-3CLpro) J. Med. Chem. 2022;65:2880–2904. doi: 10.1021/acs.jmedchem.1c00598. [DOI] [PMC free article] [PubMed] [Google Scholar]; (j) Breidenbach J., Lemke C., Pillaiyar T., Schakel L., Al Hamwi G., Diett M., Gedschold R., Geiger N., Lopez V., Mirza S., Namasivayam V., Schiedel A.C., Sylvester K., Thimm D., Vielmuth C., Vu L.P., Zyulina M., Bodem J., Gutschow M., Muller C.E. Targeting the main protease of SARS-CoV-2: from the establishment of a high throughput screening to the design of tailored inhibitors. Angew. Chem. Int. Ed. 2021;60:2–9. doi: 10.1002/anie.202016961. [DOI] [PMC free article] [PubMed] [Google Scholar]; (k) Kitamura N., Sacco M.D., Ma C., Hu Y., Townsend J.A., Meng X., Zhang F., Zhang X., Ba M., Szeto T., Kukuljac A., Marty M.T., Schultz D., Cherry S., Xiang Y., Chen Y., Wang J. Expedited approach toward the rational design of noncovalent SARS-CoV-2 main protease inhibitors. J. Med. Chem. 2022;65:2848–2865. doi: 10.1021/acs.jmedchem.1c00509. [DOI] [PMC free article] [PubMed] [Google Scholar]; (l) Xia Z., Sacco M., Hu Y., Ma C., Meng X., Zhang F., Szeto T., Xiang Y., Chen Y., Wang J. Rational design of hybrid SARS-CoV-2 main protease inhibitors guided by the superimposed cocrystal structures with the peptidomimetic inhibitors GC-376, Telaprevir, and Boceprevir. ACS Pharmacol. Transl. Sci. 2021;4:1408–1421. doi: 10.1021/acsptsci.1c00099. [DOI] [PMC free article] [PubMed] [Google Scholar]; (m) Malla T.R., Brewitz L., Muntean D.-G., Aslam H., Owen C.D., Salah E., Tumber A., Lukacik P., Strain-Damerell C., Mikolajek H., Walsh M.A., Schofield C.J. Penicillin derivatives inhibit SARS-CoV-2 main protease by reaction with its nucleophilic cysteine. J. Med. Chem. 2022;65:7682–7696. doi: 10.1021/acs.jmedchem.1c02214. [DOI] [PMC free article] [PubMed] [Google Scholar]; (n) Ma C., Xia Z., Sacco M.D., Hu Y., Townsend A., Meng X., Choza J., Tan H., Jang J., Gongora M.V., Zhang X., Zhang F., Xiang Y., Marty M.T., Chen Y., Wang J. Discovery of di- and trihaloacetamides as covalent SARS-CoV-2 main protease inhibitors with high target specificity. J. Am. Chem. Soc. 2021;143:20697–20709. doi: 10.1021/jacs.1c08060. [DOI] [PMC free article] [PubMed] [Google Scholar]; (o) Huff S., Kummetha I.R., Tiwari S.K., Huante M.B., Clark A.E., Wang S., Bray W., Smith D., Carlin A.F., Endsley M., Rana T.M. Discovery and mechanism of SARS-CoV-2 main protease inhibitors. J. Med. Chem. 2022;65:2866–2879. doi: 10.1021/acs.jmedchem.1c00566. [DOI] [PMC free article] [PubMed] [Google Scholar]; (p) Ashhurst A.S., Tang A.H., Fajtova P., Yoon M.C., Aggarwal A., Bedding M.J., Stoye A., Beretta L., Pwee D., Drelich A., Skinner D., Li L., Meek T.D., McKerrow J.H., Hook V., Tseng C.-T., Larance M., Turville S., Gerwick W.H., O'Donoghue A.J., Payne R.J. Potent anti-SARS-CoV-2 activity by the natural product gallinamide A and analogues via inhibition of cathepsin L. J. Med. Chem. 2022;65:2956–2970. doi: 10.1021/acs.jmedchem.1c01494. [DOI] [PMC free article] [PubMed] [Google Scholar]; (q) Mondal S., Chen Y., Lockbaum G.J., Sen S., Chaudhuri S., Reyes A.C., Lee J.M., Kaur A.N., Sultana N., Cameron M.D., Shaffer S.A., Schiffer C.A., Fitzgerald K.A., Thomson P.R. Dual inhibitors of main protease (Mpro) and cathepsin L as potent antivirals against SARS-CoV-2. J. Am. Chem. Soc. 2022;144:21035–21045. doi: 10.1021/jacs.2c04626. [DOI] [PMC free article] [PubMed] [Google Scholar]; (r) Ma X.R., Alugubelli Y.R., Ma Y., Vatansever E.C., Scott D.A., Qiao Y., Yu G., Xu S., Liu W.R. MPI8 is potent against SARS-CoV-2 by inhibiting dually and selectively the SARS-CoV-2 main protease and the host cathepsin L. ChemMedChem. 2022;17 doi: 10.1002/cmdc.202100456. [DOI] [PMC free article] [PubMed] [Google Scholar]; (s) Pillaiyar T., Flury P., Kruger N., Su H., Schakel L., Da Silva E.B., Eppler O., Kronenberger T., Nie T., Luedtke S., Rocha C., Sylvester K., Petry M.R.I., McKerrow J.H., Poso A., Pohlmann S., Gutschow M., O'Donoghue A.J., Xu Y., Muller C.E., Laufer S.A. Small-molecule thioesters as SARS-CoV-2 main protease inhibitors: enzyme inhibition, structure-activity relationships, antiviral activity, and X-ray structure determination. J. Med. Chem. 2022;65:9376–9395. doi: 10.1021/acs.jmedchem.2c00636. [DOI] [PubMed] [Google Scholar]; (t) Gao S., Song L., Claff T., Woodson M., Sylvester K., Jing L., Weibe R.H., Cheng Y., Strater N., Schakel L., Gutschow M., Ye B., Yang M., Zhang T., Kang D., Toth K., Tavis J., Tollefson A.E., Muller C.E., Zhan P., Liu X. Discovery and crystallographic studies of nonpeptidic piperazine derivatives as covalent SARS-CoV-2 main protease inhibitors. J. Med. Chem. 2022;65:16902–16917. doi: 10.1021/acs.jmedchem.2c01716. [DOI] [PubMed] [Google Scholar]
- 30.Talele T.T. Natural products-inspired use of the gem-dimethyl group in medicinal chemistry. J. Med. Chem. 2018;61:2166–2210. doi: 10.1021/acs.jmedchem.7b00315. [DOI] [PubMed] [Google Scholar]
- 31.Robello M., Barresi E., Baglini E., Salerno S., Taliani S., Da Settimo F. The alpha ketoamide moiety as a privileged motif in medicinal chemistry: current insights and emerging opportunities. J. Med. Chem. 2021;64:3508–3545. doi: 10.1021/acs.jmedchem.0c01808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cooper M.S., Zhang L., Ibrahim M., Zhang K., Sun X., Roske J., Gohl M., Bronstrup M., Cowell J.K., Sauerhering L., Becker S., Vangeel L., Jochmans D., Neyts J., Rox K., March G.P., Maple H.J., Hilgenfgeld R. Diastereomeric resolution yields highly potent inhibitor of SARS-CoV-2 main protease. J. Med. Chem. 2022;65:13328–13342. doi: 10.1021/acs.jmedchem.2c01131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mandadapu S.R., Gunnam M.R., Tiew K.C., Uy R.A., Prior A.M., Alliston K.R., Hua D.H., Kim Y., Chang K.O., Groutas W.C. Inhibition of norovirus 3Cl protease by bisulfite adducts of transition state inhibitors. Bioorg. Med. Chem. Lett. 2013;23:62–65. doi: 10.1016/j.bmcl.2012.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vankadara S., Dawson M.D., Fong J.Y., Oh Q.Y., Ang Q.A., Liu B., Chang H.Y., Koh J., Tan Q.W., Joy J., Chia C.S.B. A warhead substitution study on the coronavirus main protease inhibitor nirmaltrevir. Med. Chem. Lett. 2022;13:1345–1350. doi: 10.1021/acsmedchemlett.2c00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pirali T., Serafini M., Cargnin S., Genazzani A.A. Applications of deuterium in medicinal chemistry. J. Med. Chem. 2019;62:5276–5297. doi: 10.1021/acs.jmedchem.8b01808. [DOI] [PubMed] [Google Scholar]
- 36.Davis R.D., Fitzgerald R.N., Guo J. Improved method for the synthesis of 2-methyl-2-aryloxypropanoic derivatives. Synthesis. 2004;12:1959–1962. doi: 10.1055/s-2004-829171. [DOI] [Google Scholar]
- 37.Ammazzalorso A., Amoroso R., Baraldi M., Bettoni G., Braghiroli D., De Filippis B., Giampietro L., Tricca M.L., Vezzalini F. Synthesis and antiplatelet activity of thioaryloxyacids analogues of clofibric acid. Eur. J. Med. Chem. 2005;40:918–921. doi: 10.1016/j.ejmech.2005.03.022. [DOI] [PubMed] [Google Scholar]
- 38.Giampietro L., Ammazzalorso A., Giancristofaro A., Lannutti F., Bettonni G., De Filippis B., Fantacuzzi M., Maccallini C., Petruzelli M., Morgano A., Moschetta A., Amoroso R. Synthesis and biological evaluation of 2-heteroarylthioalkanoic acid analogues of clofibric acid as peroxisome proliferator-activated receptor α agonists. J. Med. Chem. 2009;52:6224–6332. doi: 10.1021/jm900878u. [DOI] [PubMed] [Google Scholar]
- 39.Kim H.-O., Kahn M. The synthesis of aminoazole analogs of lysine and arginine: the Mitsunobu reaction with lysinol and argininol. Synlett. 1999;8:1239–1240. doi: 10.1055/s-1999-2819. [DOI] [Google Scholar]
- 40.Corey E.J., Chaykovsky M. Methylenecyclohexane oxide. Org. Synth. 1969;49:78–80. doi: 10.15227/orgsyn.049.0078. [DOI] [Google Scholar]
- 41.Fan R.-H., Hou X.-L. Tetrabutylammonium bisulfate: a new effective catalyst for the hydrolysis of aziridines or epoxides. Org. Biomol. Chem. 2003;1:1565–1567. doi: 10.1039/B301081C. [DOI] [PubMed] [Google Scholar]
- 42.Ghosh A.K., Duong T.T., McKee S.P., Thompson W.J.N. N’-Disuccinimidyl carbonate: a useful reagent for alkoxycarbonylation of amines. Tetrahedron Lett. 1992;33:2781–2784. doi: 10.1016/S0040-4039(00)78856-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kjell D.P., Slattery B.J., Semo M.J. A novel, nonaqueous method for regeneration of aldehydes from bisulfite adducts. J. Org. Chem. 1999;64:5722–5724. doi: 10.1021/jo990543v. [DOI] [PubMed] [Google Scholar]
- 44.He X., Quan S., Xu M., Rodriguez S., Goh S.L., Wei J., Fridman A., Koeplinger K.A., Carroll S.S., Grobler J.A., Espeseth A.S., Olsen D.B., Hazudas D.J., Wang D. Generation of SARS-CoV-2 reporter replicon for high-throughput antiviral screening and testing. Proc. Natl. Acad. Sci. U.S.A. 2021;118 doi: 10.1073/pnas.2025866118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Galasiti Kankanamalage A.C., Kim Y., Rathnayake A.D., Alliston K.R., Butler M.M., Cardinale S.C., Bowlin T.L., Groutas W.C., Chang K.O. Design, synthesis and evaluation of novel prodrugs of transition state inhibitors of norovirus 3CL protease. J. Med. Chem. 2017;60:6239–6248. doi: 10.1021/acs.jmedchem.7b00497. [DOI] [PubMed] [Google Scholar]
- 46.Kabsch W. Automatic indexing of rotation diffraction patterns. J. Appl. Crystallogr. 1988;21:67–72. doi: 10.1107/S0021889887009737. [DOI] [Google Scholar]
- 47.Kabsch W. XDS. Acta Crystallogr. D. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vonrhein C., Flensburg C., Keller P., Sharff A., Smart O., Paciorek W., Womack T., Bricogne G. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2011;67:293–302. doi: 10.1107/S0907444911007773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Evans P.R. An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2011;67:282–292. doi: 10.1107/S090744491003982X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.-W., Kapral G.J., Grosse-Kunstleve R.W. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen V.B., Arendall W.B., Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Potterton L., McNicholas S., Krissinel E., Gruber J., Cowtan K., Emsley P., Murshudov G.N., Cohen S., Perrakis A., Noble M. Developments in the CCP4 molecular-graphics project. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2004;60:2288–2294. doi: 10.1107/S0907444904023716. [DOI] [PubMed] [Google Scholar]
- 55.Evans P.R. An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr. D Biol. Crystallogr. 2011;67:282–292. doi: 10.1107/S090744491003982X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Evans P. Scaling and assessment of data quality. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 57.Diederichs K., Karplus P.A. Improved R-factors for diffraction data analysis in macromolecular crystallography. Nat. Struct. Biol. 1997;4:269–275. doi: 10.1038/nsb0497-269. [DOI] [PubMed] [Google Scholar]
- 58.Weiss M.S. Global indicators of X-ray data quality. J. Appl. Crystallogr. 2001;34:130–135. doi: 10.1107/S0021889800018227. [DOI] [Google Scholar]
- 59.Karplus P.A., Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Evans P. Biochemistry. Resolving some old problems in protein crystallography. Science. 2012;336:986–987. doi: 10.1126/science.1222162. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information is available free of charge and contains synthesis of alcohol inputs, 1H NMR, 13C NMR and HRMS spectra, absolute qNMR data, superposition of the SARS-CoV-2 3CLpro structures in complex with 11d and 15d, and crystallographic data for MERS 3CLpro and SARS-CoV-2 3CLpro inhibitor complexes.
Data Availability Statement
Data will be made available on request.