Abstract
PKC epsilon (PKCε) plays important roles in behavioral responses to alcohol and in anxiety-like behavior in rodents, making it a potential drug target for reducing alcohol consumption and anxiety. Identifying signals downstream of PKCε could reveal additional targets and strategies for interfering with PKCε signaling. We used a chemical genetic screen combined with mass spectrometry to identify direct substrates of PKCε in mouse brain and validated findings for 39 of them using peptide arrays and in vitro kinase assays. Prioritizing substrates with several public databases such as LINCS-L1000, STRING, GeneFriends, and GeneMAINA predicted interactions between these putative substrates and PKCε and identified substrates associated with alcohol-related behaviors, actions of benzodiazepines, and chronic stress. The 39 substrates could be broadly classified in three functional categories: cytoskeletal regulation, morphogenesis, and synaptic function. These results provide a list of brain PKCε substrates, many of which are novel, for future investigation to determine the role of PKCε signaling in alcohol responses, anxiety, responses to stress, and other related behaviors.
Graphical Abstract
Highlights
-
•
Identification of direct PKCε substrates in mouse brain with chemical genetics.
-
•
Utilized AS-PKCε and N6-benzyl-ATPγ for covalent capture of substrates.
-
•
Validated direct PKCε substrates using in vitro methods.
-
•
Most substrates associated with cytoskeletal, morphological, or synaptic function.
-
•
Several substrates involved in alcohol-, stress-, and anxiety-related phenotypes.
In Brief
PKC epsilon (PKCε) plays important roles in behavioral responses to alcohol and in anxiety-like behavior, making it a potential drug target for reducing alcohol consumption and anxiety. Here, a chemical genetic screen and mass spectrometry identified 39 direct substrates of PKCε in mouse brain. They were prioritized using several public databases (LINCS-L1000, STRING, GeneFriends, GeneMANIA) to predict interactions between them and PKCε. Many were novel and broadly fell into three functional categories: cytoskeletal regulation, morphogenesis, and synaptic function.
PKC epsilon (PKCε) is a member of the PKC family of serine-threonine kinases, which transduce signals carried by lipid second messengers (1). Studies with KO mice and selective kinase inhibitors have revealed that PKCε plays an important role in behavioral responses to alcohol and in anxiety-related behavior. Compared with WT littermates, PKCε KO (Prkce−/−) mice show markedly reduced ethanol consumption, and a small molecule inhibitor of PKCε reduces alcohol consumption in mice (2, 3, 4). In addition, Prkce transcripts are highly expressed in the brains of in-bred and selected lines of mice that drink high amounts of ethanol (5, 6). Prkce−/− mice also display reduced anxiety-like behavior with reduced levels of the circulating stress hormones ACTH and corticosterone and reduced expression of corticotropin-releasing factor in the central amygdala (7, 8). Therefore, PKCε has become of considerable interest as a central nervous system target for reducing alcohol consumption and anxiety (9, 10, 11).
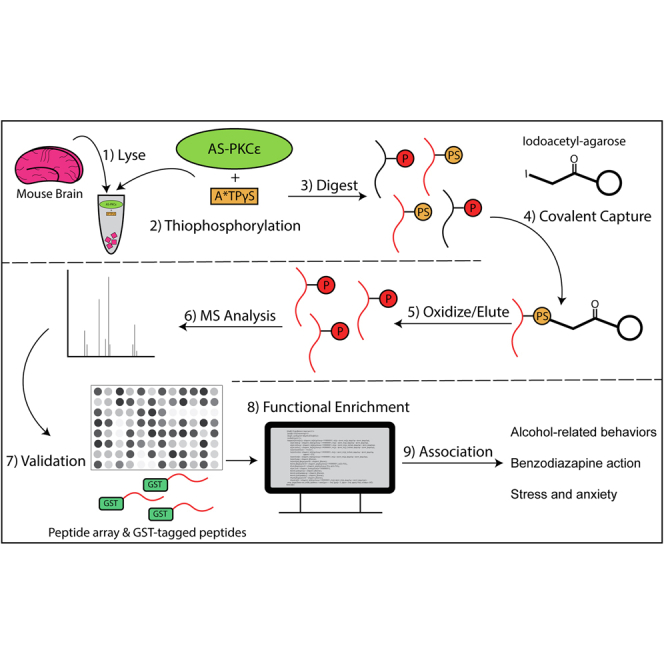
Because PKCε is an attractive drug target, we sought to identify additional proteins in the brain that act within PKCϵ signaling pathways with the assumption that they could be valuable for developing therapeutics. To identify direct PKCϵ substrates, we used a chemical-genetic approach combined with mass spectrometry (12, 13). We identified 39 proteins in mouse forebrain lysates in which we validated sites of phosphorylation using peptide arrays and in vitro kinase assays. Some like neuromodulin and myristoylated alanine-rich PKC substrate (MARCKS) had been previously identified as PKC substrates (14, 15), but most were novel. Bioinformatic analysis revealed involvement of these proteins in cytoskeletal function, morphogenesis, and synaptic function. We prioritized putative substrates based on whether there was existing support for interactions with PKCϵ in publicly available databases, such as LINCS-L1000, STRING, GeneFriends, and GeneMANIA. To formulate hypotheses regarding PKCϵ involvement in alcohol intake and anxiety, we further mined publicly available databases and found that several of these proteins have been previously associated with alcohol-related behaviors, anxiety-related behaviors, or the actions of anxiolytic benzodiazepines. Several are membrane transporters, receptors, or enzymes, which could be considered potential candidates for drug discovery. This database of putative PKCϵ substrates will provide a useful resource for those groups wishing to investigate the function of PKCϵ and potentially could serve as useful biomarkers of dysregulated PKCϵ in disease or for therapeutic development.
Experimental Procedures
Animals
We used male F2 generation WT or Prkce−/− mice on a C57BL/6J x 129S4 background (16). Mice were group-housed in temperature- and humidity-controled rooms with free access to food and water under a 12-h light/dark cycle (lights on at 7:00 AM). Experiments began when mice were 2 to 3 months old. Experiments were approved by the Institutional Animal Care and Use Committee at The University of California San Francisco and The University of Texas at Austin and complied with the ARRIVE guidelines and the National Institutes of Health Guide for the Care and Use of Laboratory Animals (17).
Substrate Labeling
Mice were euthanized with CO2 and decapitated. Their brains were rapidly removed, rinsed in ice-cold PBS, and quickly transferred to ice-cold RIPA buffer (G-Biosciences; Cat No. 786-490) containing 25 mM Tris [2-Amino-2-(hydroxymethyl)propane-1,3-diol]–HCl, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS, pH 7.4 with added Halt Protease Inhibitor Cocktail (Thermo Fisher Scientific; Cat No. 78440). The tissue was homogenized using a glass Dounce tissue grinder, and the homogenate was nutated at 4 °C for 30 min. The homogenate was centrifuged at 4 °C for 5 min at 18,000g, and supernatant proteins were collected and quantified using the Bradford assay method with bovine serum albumin (BSA) as a standard (18). Lysates were diluted to 20 mg/ml of total protein in RIPA buffer.
Proteins in lysates were thiophosphorylated in a reaction buffer containing 20 mM Hepes (pH7.4), 0.1 mM EGTA, 0.03% Triton X-100, 10 mM MgCl2, 1 mM DTT, 1 mM β-mercaptoethanol, 0.59 mM L-⍺-phosphatidylserine (Avanti Polar Lipids; Cat No. 840032), 0.1 mM ATP, 5.0 mM GTP, and 5 mg protein lysate in a final volume of 500 μl in the presence or absence 0.2 μM AS-PKCε (19). The reaction was allowed to proceed for 5 min at 37 °C. Then, N6-benzyl A∗TP-γS (BioLog Life Sci Inst; Cat No. A 060) was added to a final concentration of 0.25 mM, and the reaction was continued for another 10 min at 37 °C. The reaction was stopped by adding EDTA to a final concentration of 500 mM.
Covalent Capture and Analysis of Thiophosphorylated Proteins
Putative substrates were isolated and identified as described, with slight modification (13, 20). Brain lysates were denatured by adding solid urea (60% by volume) and 1M tris(2-carboxyethyl)phosphine to a final concentration of 10 mM and incubating the mixture at 55 °C for 1 h. Proteins were then digested by diluting the urea to 2M by the addition of 100 mM NH4HCO3 (pH 8.0), adding additional tris(2-carboxyethyl)phosphine to 10 mM final concentration and trypsin (Promega; Cat No. VA9000) 1:25 by weight. The reaction solution was adjusted to pH 7.8 using 5% NaOH. Lysates were digested overnight (6–17 h) at 37 °C until the solution was very clear. The solution was then acidified with 0.5% trifluoroacetic acid (TFA) and desalted using a Sep-Pak C18 column (Waters Corp; WAT051910). After the column was washed with 25 ml of 0.1% TFA in water, the peptides were eluted slowly with 0.8 ml 0.1% TFA/70% acetonitrile and then 0.2 ml acetonitrile. The solution containing the desalted peptides was reduced to ∼35 μl using a SpeedVac concentrator. The pH of the peptides was adjusted by adding 40 μl of 200 mM Hepes pH 7.0 with 75 μl acetonitrile and brought to pH 7.0 by the addition of 5% NaOH (approximately 8 μl). The peptide solution was then added to 100 μl iodoacetyl beads (Thermo Scientific; Cat No. 53155) equilibrated with 200 mM Hepes (pH 7.0) and incubated with end-over-end rotation at room temperature in the dark for 12 to 16 h. The beads were then added to small disposable columns, washed with a buffer of H2O, 5M NaCl, 50% acetonitrile, 5% formic acid, and 10 mM DTT, followed by elution with 100 μl and 200 μl (300 μl total) 1 mg/ml oxone (Sigma-Aldrich; Cat No. 228036), desalted, and concentrated using a 10 μl C18 ZipTip (Millipore; Cat No. Z720070) pipette tip eluting into 60 μl total volume.
The resulting phosphopeptides were concentrated to 12 μl on a freezing SpeedVac concentrator, and 5 μl were separated immediately using a 2 to 50% acetonitrile gradient over 30 min (total run time of 53 min) on a nanoACQUITY UPLC System (Waters Corp) reversed phase 75-micron capillary UPLC column on-line to an LTQ Velos Orbitrap mass spectrometer (Thermo Fisher Scientific). Two runs were performed on each sample. In each case, MS1 scans from m/z 350 to 1400 were acquired at a resolution of 30K. In one run, this was followed by HCD fragmentation analysis at a collision energy of 35 and a resolution of 15K of the six most intense multiply charged peaks. In the second run, ETD fragmentation was acquired on the six most intense precursors and fragments were measured in the ion trap.
Peak lists were generated by using PAVA (UCSF in-house software) and searched on Protein Prospector version 5.9.4 (https://prospector.ucsf.edu/prospector/mshome.htm). The database queried consisted of all mouse entries from a Swiss-Prot database downloaded on July 6th 2011 (36,620 sequences). No fixed modifications were considered; variable modifications included oxidation of methionine, phosphorylation of serine and threonine, pyroglutamate formation from peptide N-terminal glutamine, and protein N-terminal acetylation with up to four modifications total allowed per peptide. A precursor mass tolerance of ± 20 ppm was allowed. For HCD, the fragment tolerance was also 20 ppm; for ETD data, a tolerance of 0.6 Da was allowed. Results were thresholded at an expectation value of 0.01. Modification site localization threshold was set to a SLIP score of 6, which corresponds to a 95% localization confidence per spectrum and typically a global false localization rate of around 1%.
Peptide Array
Peptide arrays containing all identified phosphopeptides were generated by the Francis Crick Institute (21). Each array had 184 peptides, each 12-amino-acids in length, derived from the phosphopeptides identified in the covalent capture analysis. The array included control peptides with identified phosphorylation sites and potential off-target serine or threonine residues substituted with alanine residues. Peptide arrays were synthesized using an Intavis ResPepSL Automated Peptide Synthesizer (Intavis Bioanalytical Instruments AG). Arrays were generated on a cellulose membrane through cycles of N(a)-Fmoc amino acid coupling via activation of carboxylic acid groups with diisopropylcarbodiimide in the presence of ethyl cyano(hydroxyamino)acetate (Oxyma Pure) followed by removal of the temporary α-amino protecting group with piperidine. After chain assembly, side chain protection groups were removed by treating membranes with a deprotection cocktail (20 ml 95% trifluoroacetic acid, 3% triisopropylsilane, 2% water for 4h at room temperature) then washing (4× dichloromethane, 4× ethanol, 2× water, 1× ethanol) prior to air drying.
The arrays were incubated overnight at 4 °C in blocking buffer (40 mM Tris–HCl, pH 7.4, 0.02% Triton X-100, 3% w/v BSA) with constant nutation. The next day, the arrays were incubated in wash buffer (40 mM Tris–HCl, pH 7.4, 0.02% Triton X-100) for 10 min, three times at room temperature. Then, the arrays were incubated for 30 min at 37 °C with constant nutation in kinase buffer (40 mM Tris–HCl, pH 7.4, 5 mM MgCl2, 0.2% Triton X-100, 0.5, 1 mg/ml BSA, and 100 μM ATPγS (Abcam; Cat No. ab138911)) containing 2.5 ng/μl PKCε kinase domain. To stop the reaction, EDTA was added to the buffer to a final concentration of 100 mM. The arrays were briefly rinsed with wash buffer before being incubated in wash buffer containing p-nitrobenzyl mesylate (PNBM) (Abcam; Cat No. 138910) added to a final concentration of 2.5 mM. The reaction was allowed to continue overnight at 4 °C. The arrays were then washed with wash buffer for 10 min, three times at room temperature. Afterward, the arrays were incubated for 2h at room temperature with rabbit anti-thiophosphate ester antibody (Abcam; Cat No. ab92570; 1:2000) in blocking buffer, which was composed of TBS-T (10 mM Tris, 150 mM NaCl, 0.05% w/v Tween-20) plus 5% (w/v) nonfat milk. The arrays were then washed with TBS-T for 10 min, three times at room temperature. The arrays were then incubated with donkey anti-rabbit-HRP (Sigma-Aldrich; Cat No. AP182P; 1:5000) for 1 h at room temperature in blocking buffer. Finally, the arrays were washed with TBS-T for 10 min, three times at room temperature. Immunoreactivity was detected by incubating arrays in SuperSignal West Pico PLUS Chemiluminescent Substrate (Thermo Fischer Scientific; Cat No. 34577) for 5 min and imaging them with a ChemiDocMP imager (Bio-Rad). Thiophosphorylation signals were quantified with Image Lab Software 6.1 (Bio-Rad).
Purification and Thiophosphorylation of GST-Tagged Peptides
Some peptides were also analyzed by in vitro thiophosphorylation. Peptide sequences (7–12 amino acids in length) derived by mass spectrometry were utilized to identify corresponding nucleic acid sequences in each gene’s RefSeq file. Confirmed DNA sequences were cloned into pGEX-6p-2 (Sigma-Aldrich; Cat No. GE28-9546-50) at BamH1/XhoI sites in frame with the GST purification tag sequence and transformed into BL21(DE3) Escherichia coli cells for expression.
Following expression and purification with a Pierce GST Spin Purification Kit (Thermo Fisher Scientific; Cat No. 16106), GST-tagged substrate peptides containing the putative phosphorylation site or corresponding alanine mutation were subjected to thiophosphorylation. PKCε kinase domain (2.5 ng/μl) was added to a 40 μl reaction containing kinase buffer (20 mM Hepes [2-[4-(2-Hydroxyethyl)piperazin-1-yl]ethane-1-sulfonic acid], pH 7.4, 10 mM MgCl2, 0.1 mM EGTA, 0.3% Triton-X-100, 1 ng/μl BSA) and 1 mM of ATPγS. Each GST-tagged peptide (2 μg) was added to a seperate reaction. The reaction proceeded for 30 min at 37 °C in a Thermo-Mixer shaking at 1000 RPM and was then stopped by the addition of 10 μl 500 mM EDTA (final concentration: 100 mM). Thiophosphorylated peptides were alkylated by adding 2.5 μl 50 mM PNBM in DMSO (final concentration: 2.5 mM PNBM, 5% DMSO). Samples were incubated for 1h at room temperature with constant nutation, and then 13.1. μL of 5X Laemmli sample buffer was added, and 0.25 μg of each GST-tagged peptide was separated by SDS-PAGE on 12% Tris-Glycine gels. Proteins were then transferred to LF-PVDF membranes, which were blocked for 1 hour in blocking buffer at room temperature. The membranes were then incubated in blocking buffer with a rabbit anti-thiophosphate ester antibody (Abcam; Cat No. ab92570, 1:5000) and mouse anti-GST antibody (Invitrogen; Cat No. MA4-004; 1:5000) overnight at 4 °C. The next day, membranes were washed with TBS-T for 5 min, five times at room temperature. The membranes were then incubated in blocking buffer with goat anti-rabbit StarBright Blue 700 (Bio-Rad; Cat No. 12004162; 1:5000) and goat anti-mouse StarBright Blue 520 (Bio-Rad; Cat No. 12005867; 1:5000) for 1h at room temperature. The membranes were washed with TBS-T for 5 min, six times at room temperature and imaged with a ChemiDocMP imager (Bio-Rad). The thiophosphorylation and GST signal from each substrate peptide was quantified using Image Lab Software 6.1 (Bio-Rad).
Bioinformatics
Prediction of Functional Interactions Between PKCε and Putative Substrates
To determine if evidence exists supporting functional interactions between PKCε and the putative PKCε substrates validated by in vitro kinase assays, we mined publicly available datasets. This analysis included gene expression signatures resulting from PKCε knockdown and overexpression in the NIH L1000 database, protein-protein interaction (PPI) networks in the STRING and GeneMANIA databases, and gene coexpression data in GeneFriends.
LINCS-L1000 (clue.io) is a database that catalogs the effects of pharmacological or genetic perturbation on gene expression in a number of human cell lines (22). We downloaded the differential gene expression signatures for pharmacological and genetic perturbations (LINCS-L1000 level 5 data) from Gene Expression Omnibus (Phase I: GSE92742, Phase II: GSE7013), where each signature is the result of a perturbation, cell line, and time point for which differential expression was assessed. We identified the PKCε subset of L1000 data using the signature id field from the metadata file available on Gene Expression Omnibus. We defined each signature as genes with |z| > 2 as recommended by the Broad Institute (personal communication). To determine which genes are most likely interacting with PKCε, we found the overlapping genes between the putative substrate gene list and the genes that were affected in opposite directions by PKCε knockdown and overexpression. Analyses were implemented in R versions 3.4.4 and 3.5.0.
We determined which putative PKCε substrates are known or are predicted to interact based on information in STRING v11.5 b (string-db.org) (23) and GeneMANIA (genemania.org) (24) databases of known and predicted PPIs. STRING and GeneMANIA databases utilize different sources of primary information regarding known and predicted PPIs, so we performed the analysis with both as a complementary approach. Using STRING, we ran the analysis on PKCε and the 39 validated substrates using low, medium, and high confidence thresholds and Mus musculus as the species. In GeneMANIA, we ran the analysis using the query-dependent weighting method, Mus musculus as the species, max resultant genes set to zero, and max resultant attributes set to 10.
We identified genes that are coexpressed with Prkce using information from GeneFriends v5 (genefriends.org). GeneFriends is a functional genomics tool that can assign putative functions to poorly annotated genes or can identify and rank new candidate genes related to a disease or biological process from a seed list of genes (25, 26). It is based on a gene coexpression map that describes which genes tend to be activated (increased in expression) and deactivated (decreased in expression) simultaneously in a large number of RNA-seq data samples. Because the samples originate from a wide range of conditions, the coexpression map reflects which genes in general tend to be altered simultaneously and are thus under similar transcriptional regulation. We used mouse (Mus musculus), Gene Symbol, and Prkce as the input search terms. Association was defined as the gene being ranked in the top 5% of genes coexpressed with Prkce (which corresponded to the top 1351 correlated genes).
Phosphosite Databases and Amino Acid Motif Logo Creation
We determined if the putative phosphorylation sites are present in two databases of kinase-substrate relationships, PhosphoSitePlus and ChemPhoPro (27, 28). PhosphoSitePlus is a curated posttranslational modification database. The ChemPhoPro database provides predictions regarding target selectivity of 20 kinase inhibitors against 406 kinases using three different cell lines (HL60, MCF7, and NTERA2).
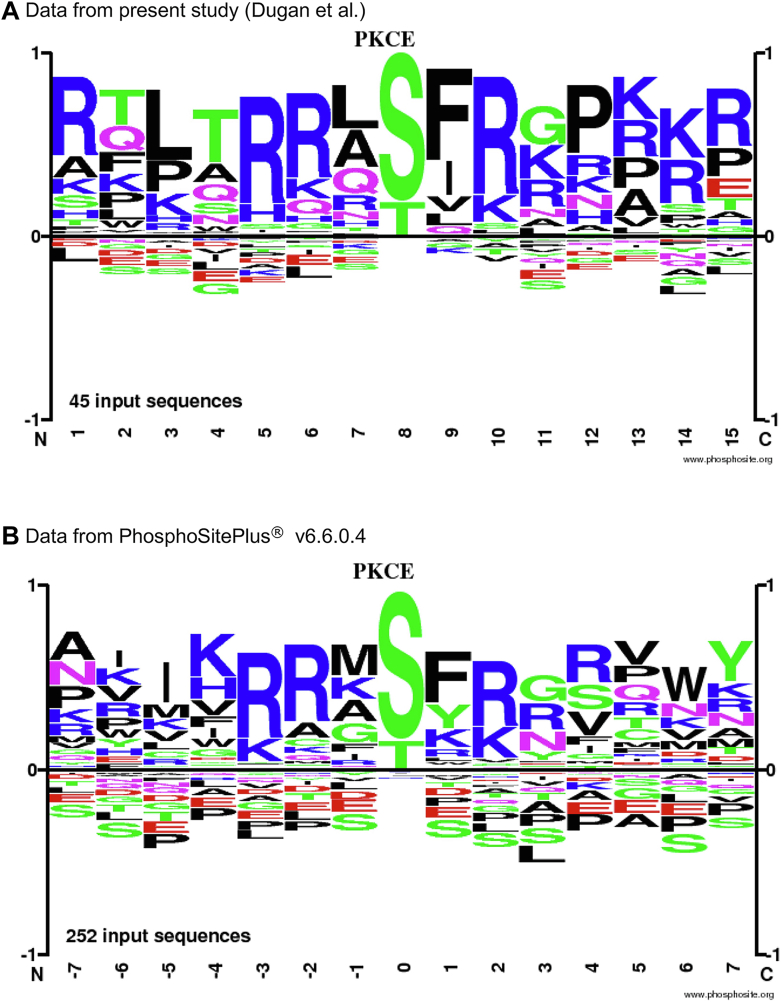
To determine if the sites we identified are in the context of a motif likely to be phosphorylated by PKCε, we generated an amino acid motif logo around the sites of modification identified in the present study ( ± 7 amino acids) using the Sequence Logo tool on PhosphoSitePlus with default settings (27). We compared the amino acid motif logo generated from our data to the amino acid motif logo generated from the sites of modification for PKCε in the PhosphoSitePlus v6.6.0.4 database ( ± 7 AA) and to consensus phosphorylation site motifs for PKCs (29, 30, 31).
Functional Enrichment Analysis
To predict functional associations between PKCε and its putative substrates, we used the functional enrichment analysis tools in STRING. Prkce and each putative substrate gene were queried in STRING to assess the enriched gene ontology (GO) terms broadly separated into three categories: biological processes, molecular functions, and cellular components. Terms were considered significantly enriched if they had FDR-corrected p-value <0.05.
Substrates Related to Alcohol and Stress
To generate hypotheses regarding mechanisms of action for PKCε in alcohol-related behaviors, we compared our putative substrate list to a list of genes related to ethanol-consumption that was curated from mouse gene knockout, gene overexpression, and microarray data (found on the INIA IT-GED website at http://inia.icmb.utexas.edu/).
Because Prkce−/− mice show prolonged sedative and ataxic responses to benzodiazepines and an antianxiety phenotype, we determined whether any putative substrates were also benzodiazepine targets (2, 7). We mined the L1000 database to identify benzodiazepine targets in a manner similar to what we did for the Prkce signatures. We first identified benzodiazepine agonists and antagonists that have been profiled in L1000 and defined each signature as genes with |z| > 2. We then found the overlapping genes between the putative substrate gene list and the genes that were affected in opposite directions by benzodiazepine agonists and antagonists. Analyses were implemented in R versions 3.6.3. Additionally, we compared the prioritized putative substrate list to a list of stress-related genes that was curated from mouse gene expression data after different kinds of stressors including acute, sub-chronic or chronic restraint, cold swim, sub-chronic or chronic variable stress, unpredictable chronic mild stress, chronic social defeat, early life stress, electric footshock (or contextual fear conditioning), and auditory fear conditioning with or without immobilization (found on the Stress Mice portal at http://hpc-bioinformatics.cineca.it/stress_mice/main) (32).
Experimental Procedures and Statistical Rationale
For the thiophosphorylation and covalent capture experiment, four groups of male mice were analyzed in three experiments. The first experiment contained samples from a group of five WT mice. The second experiment had samples from a group of two WT and two Prkce−/− mice. The third experiment contained samples from two WT mice. The resulting phosphopeptides detected in each sample were analyzed in two runs by mass spectrometry using HCD and ETD fragmentation.
We used cellulose peptide arrays containing all phosphopeptides identified in the covalent capture analysis in a thiophosphorylation reaction with the PKCε kinase domain to validate phosphorylation sites. The array included control peptides with identified phosphorylation sites and potential off-target serine or threonine residues substituted with alanine residues. Three peptide arrays were used, and the results were averaged across the three arrays. A peptide was considered validated when its thiophosphorylation signal decreased by more than 50% upon replacement of the putative phosphorylation site residue with alanine.
GST-tagged peptides with and without alanine mutations at candidate phosphorylation sites were analyzed by in vitro thiophosphorylation. Thiophosphorylation signal and GST signal was identified by Western blot analysis in three replicate experiments for each GST-tagged peptide. The ratio of thiophosphorylation signal to GST signal was determined, and the percent change in thiophosphorylation signal was normalized to the GST-tagged peptide without the alanine mutation. Differences in phosphorylation signal between mutant and native peptides were compared by unpaired, two-tailed t-tests and considered significantly different where p< 0.05.
Bioinformatic analysis was conducted on validated substrates to identify PKCε interacting partners, functional associations with PKCε, and their relation to alcohol and stress. Gene signatures were defined as genes with |z| > 2 and terms were considered significantly enriched if the FDR-correct p-value <0.05. Unless otherwise stated, the bioinformatic analyses were conducted in R versions 3.4.4, 3.5.0, or 3.6.3.
Results
PKCε Substrate Identification
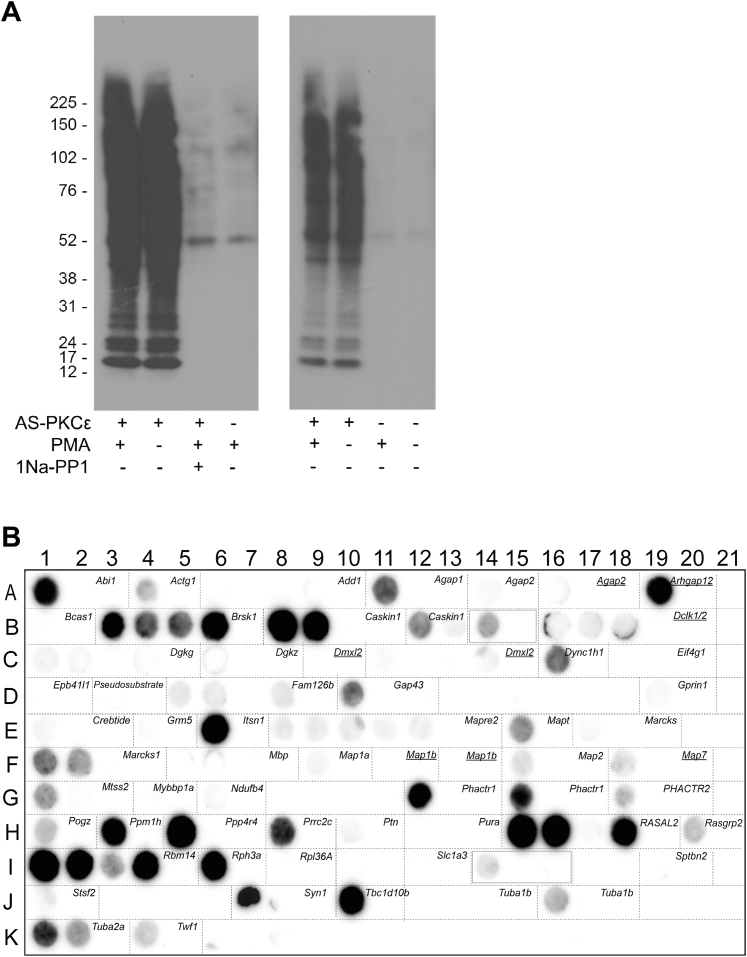
To identify PKCε substrates, we used AS-PKCε, which contains an M486A mutation that allows it to use bulky ATP analogs like N6-benzyl-ATP as phosphate donors, while native kinases cannot generally use these ATP analogs (19). To enrich for labeled substrates, we used N6-benzyl-ATP-γS instead of N6-benzyl-ATP to transfer a thiophosphate group to substrates and produce a highly specific tag that can be covalently purified from complex mixtures of proteins and subsequently analyzed using mass spectrometry (13). To determine whether thiophosphorylation in the lysates was specific for AS-PKCε, we incubated forebrain lysates from WT mice with AS-PKCε and N6-benzyl-ATP-γS in the presence or absence of a PKC activator, phorbol 12-myristate, 13-acetate (0.2 μM) and a specific AS-PKCε inhibitor, 1-napthyl-PP1 (20 μM) (19). Thiophosphorylation was similar with or without addition of phorbol 12-myristate, 13-acetate but drastically reduced in samples incubated with 1-napthyl-PP1 or without AS-PKCε (Fig. 1A).
Fig. 1.
AS-PKCε thiophosphorylation and PKCε substrate peptide array. Whole brain mouse tissue lysates were incubated in a kinase reaction with N6-benzyl ATP-γS in the presence or absence of AS-PKCε, PMA, or 1NA-PP1. A, lysates probed for thiophosphorylation with anti-thiophospho antibody. B, average of three peptide array chemiluminescent images. Images of each individual array are available as supplemental Fig. S3. The position on the peptide array is outlined for each putative substrate with its corresponding gene name in the top right corner of each outlined box. Boxes with underlined gene names show peptides that were only identified in one experiment. Two boxes with capitalized gene names were identified in the human Swiss-Prot database but did not have an equivalent in the mouse Swiss-Prot (see supplemental Table S8). These were excluded from the bioinformatics analyses. Boxes outlined in blue designate irrelevant peptides put on arrays by mistake. Empty, unlabeled boxes did not have a peptide present. PKCε, PKC epsilon; PMA, phorbol 12-myristate, 13-acetate.
We then conducted three experiments using mouse forebrain lysates from four groups of male C57BL/6J x 129S4 mice and identified 53 proteins with their sites of phosphorylation that were detected in at least two of the experiments in samples in which AS-PKCε was present (supplemental Table S1). All peptides with their associated gene symbols and UniProt accession numbers, m/z values, posttranslational modifications, retention times, scores, and associated fragmentation method are listed in supplemental Table S2. A total of 1575 phosphopeptide spectra were detected with 90% found in samples containing AS-PKCε (supplemental Table S3).
HCD and ECD fragmentation yielded 86 peptides in common, but the HCD method detected far more peptides than the ECD method (supplemental Table S4). We included peptides detected by either fragmentation method in subsequent analyses. In one experiment, we compared WT and Prkce−/− samples to see if using knockout tissue would increase the yield of phosphopeptides. Surprisingly, however, we found fewer peptides in samples from Prkce−/− mice, and 9 of the 19 peptides unique to Prkce−/− samples were detected in other experiments that used only WT tissue (supplemental Table S5), suggesting that most putative PKCε substrates could be detected using WT tissue.
Substrate Validation
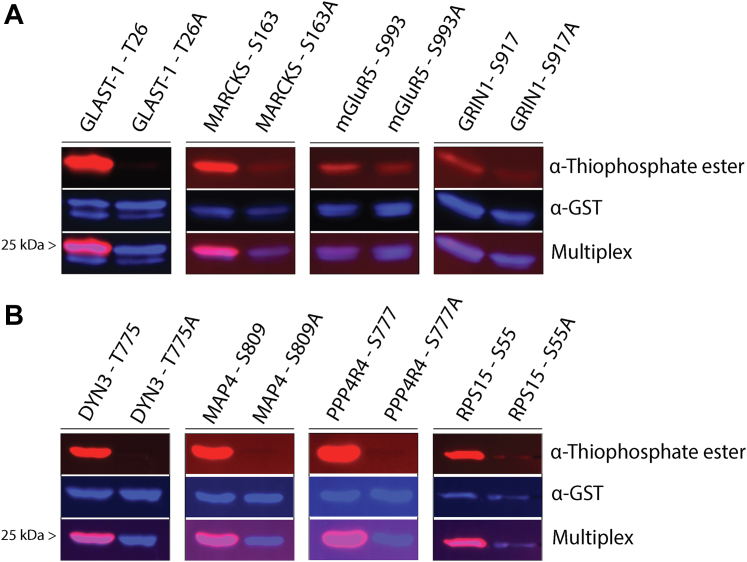
We next confirmed putative PKCε substrates identified from our covalent capture experiments using a peptide array with native peptides and mutant peptides containing alanine residues at identified phosphorylation sites (Fig. 1B and supplemental Table S6). MARCKS is a known substrate of PKCε, but its peptide array signal was very low (14). To further investigate its validity, we generated GST-tagged substrate peptides (7–12 amino acids in length) of MARCKS and five other proteins that were borderline or below the cutoff criteria for validation by the peptide array. These included G protein–regulated inducer of neurite outgrowth 1 (GRIN1), metabotropic glutamate receptor 5 (mGluR5), diacylglycerol kinase gamma (DGK-gamma), diacylglycerol kinase zeta (DGK-zeta), and excitatory amino acid transporter 1 (GLAST-1). We also generated GST-tagged substrate peptides (12 amino acids in length) for seven putative PKCε substrates identified in our covalent capture experiments that were mistakenly excluded from the peptide array. These included dynamin-3, microtubule-associated protein 4 (MAP-4), myelin basic protein, myotubularin, serin/threonine-protein phosphatase 4 regulatory subunit 4 (PPP4R4), and two peptides corresponding to 40S ribosomal protein S15 (RPS15). We subjected these peptides and their corresponding alanine mutant controls to in vitro thiophosphorylation and detection by Western blot analysis. From these substrate candidates, we confirmed that PKCε phosphorylates peptides corresponding to GRIN1, mGluR5, MARCKS, GLAST-1, Dynamin-3, MAP-4, PPP4R4, and RPS15 at S55 (Fig. 2), but not DGK-gamma, DGK-zeta, myelin basic protein, Myotubularin, or RPS15 at S138 (supplemental Fig. S1). The percent change in each thiophosphorylation signal along with the statistical analysis are available in supplemental Figure S2. Therefore, using the peptide array and Western blot analysis of GST-tagged peptides, we confirmed phosphorylation sites in 39 of the putative substrates (Table 1).
Fig. 2.
Representative western blots of validated GST-tagged PKCε substrate peptides. GST-tagged PKCε substrate peptides (7–12 amino acids in length) and alanine controls were subjected to in vitro thiophosphorylation. Each peptide is shown with its site of phosphorylation and corresponding alanine control peptide. A, four of the six putative substrate peptides that were borderline to the cutoff criteria on the peptide array. B, four of the seven putative substrate peptides that were mistakenly absent from the peptide array. A and B, (Top) signal from α-thiophosphate ester antibody representing level of thiophosphorylation for each peptide. (Middle) Signal from α-GST antibody representing total peptide in reaction. (Bottom) Multiplex image of α-thiophosphate ester and α-GST signals. Each validated peptide has a significant increase in thiophosphorylation signal compared with their corresponding alanine control across three experiments as measured by an unpaired two-tailed t test (see supplemental Fig. S2). PKCε, PKC epsilon.
Table 1.
Putative PKCε substrates from mass spectrometry and confirmed by peptide array or Western blot analysis
 |
Existing Support for Functional Interactions with PKCϵ in Publicly Available Databases
We investigated whether there was evidence from previous research that supported interactions between PKCε and these 39 validated substrates. To do this, we mined publicly available databases for relevant information including the following: (1) gene expression signatures resulting from PKCε knockdown and overexpression (L1000/CMap database), (2) PPI networks (STRING and GeneMania databases), and (3) gene coexpression data (GeneFriends database). Out of the 39 putative substrates, gene expression levels of 33 were measured or inferred in the L1000 database after manipulation of PKCε. Genes for three of these substrates were affected in an opposing manner by PKCε knockdown and overexpression (Table 2): Mapre2, Rbm14, and Tubb2a.
Table 2.
Bioinformatics analyses of putative PKCε substrates
| Gene name | PKCε targets |
Alcohol |
Benzo |
Stress |
|||
|---|---|---|---|---|---|---|---|
| L1000 | PPI |
CoEx GeneFriends (Pearson corr. Coeff.) |
INIA IT GED | L1000 | Stress mice | ||
| STRINGdb | GeneMANIA | ||||||
| Mapre2 | X | X | X | X (0.61) | X | X | |
| Tubb2a | X | X | X (0.59) | X | X | ||
| Gap43 | X | X | X (0.54) | X | X | X | |
| Mapt | X | X | X (0.57) | X | X | ||
| Syn1 | X | X | X (0.87) | X | X | ||
| Agap2 | X (med) | X | X (0.82) | X | X | ||
| Rph3a | X | X (0.77) | X | X | |||
| Caskin1 | X | X (0.51) | X | ||||
| Grm5 | X | X (0.87) | X | ||||
| Map2 | X | X (0.76) | X | ||||
| Brsk1 | X | X (0.72) | |||||
| Rasgrp2 | X | X | X | X | |||
| Itsn1 | X | X | X | ||||
| Phactr1 | X (0.84) | X | X | X | |||
| Dync1h1 | X | X | X | ||||
| Marcks | X (high) | X | X | X | |||
| Pogz | X | X | X | ||||
| Actg1 | X | X | X | ||||
| Slc1a3 | X (0.60) | X | |||||
| Gprin1 | X (0.70) | X | |||||
| Rbm14 | X | X | |||||
| Map1a | X (0.78) | X | |||||
| Fam126b | X | X | |||||
| Mtss2 | X | X (0.46) | X | X | |||
| Ppp4r4 | X | X | X | ||||
| Marcksl1 | X (high) | X | X | ||||
| Ndufb4 | X | X | |||||
| Prrc2c | X | X | |||||
| Srsf2 | X | X | |||||
| Ptn | X | X | |||||
| Agap1 | X (0.45) | X | |||||
| Tbc1d10b | X | ||||||
| Tuba1b | X | ||||||
| Twf1 | X | ||||||
| Ppm1h | X | ||||||
| Abi1 | |||||||
| Dnm3 | X (0.60) | X | |||||
| Map4 | X (0.42) | X | |||||
| Rps15 | X | ||||||
The STRING PPI network for PKCε and the 39 substrates is depicted in Figure 3 (medium confidence threshold) with the nodes colored by “synapse,” “cytoskeleton organization,” and “actin binding” annotations, which were highly enriched Gene Ontology terms in the network (supplemental Table S7). We found that 17 of the putative substrates were already noted to interact with PKCε at a low confidence threshold, three at a medium confidence threshold, and two at a high confidence level (Table 2). The PPI networks at each confidence level contained significantly more interactions than expected for a random set of proteins of the same size. Such enrichment suggests that the proteins are biologically connected as a group.
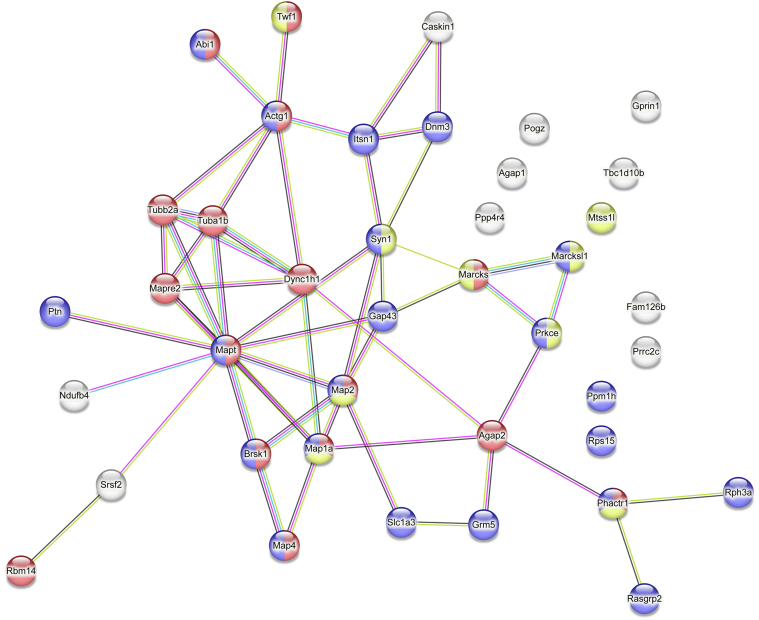
Fig. 3.
The protein–protein interaction network analysis using STRING database. A search of the STRING database with PKCε and the 39 validated, putative PKCε substrates resulted in a network with 40 nodes and 52 edges, with a PPI enrichment value of <1.0e-16. Node color represents functional annotations from Gene Ontology resource: Blue = Synapse; Red = Cytoskeleton Organization, Yellow = Actin Binding. PKCε, PKC epsilon; PPI, protein–protein interaction.
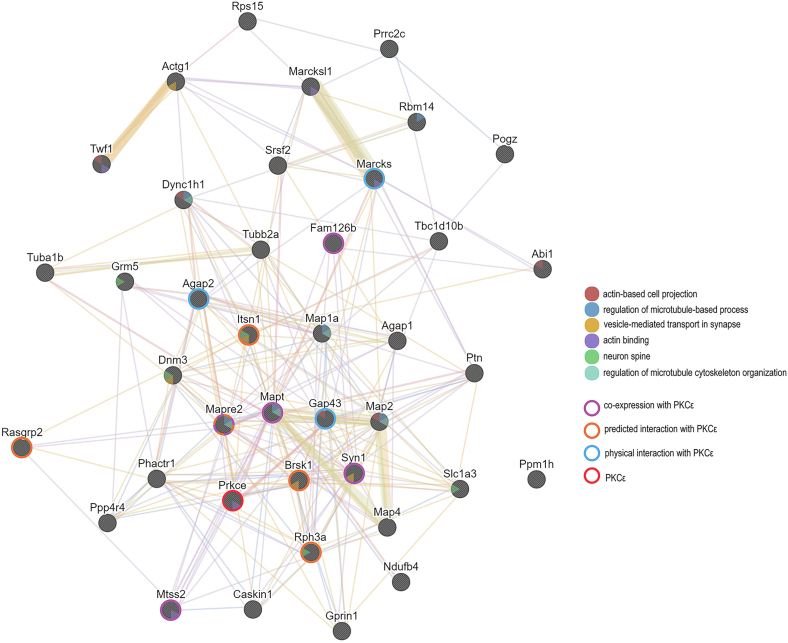
The GeneMANIA analysis also produced a functional association network of PKCε and its putative substrate genes. This network revealed that three identified substrates are already known to physically interact with PKCε, five were predicted to have interactions with PKCε, and five were coexpressed with PKCε (Fig. 4). GeneMANIA identified both MARCKS and neuromodulin as having physical interactions with PKCε, and both are known substrates of PKCε (14, 15). GeneMANIA did not find any putative substrate genes that are colocalized or share protein domains with PKCε. However, several substrates shared protein domains or were colocalized with each other.
Fig. 4.
GeneMANIA network of PKCε and putative substrates. GeneMANIA connected PKCε and the validated, putative substrates by coexpression, predicted interaction, physical interaction, colocalization, and shared protein domain data. Genes associated with the synaptic vesicle and cytoskeletal related functions are colored within each circle. Genes that have coexpression, predicted interactions, or physical interactions with PKCε are denoted with a colored border around each circle. The position of PKCε in the network is highlighted with a red circle border. PKCε, PKC epsilon.
Finally, we queried the GeneFriends database to find genes with expression patterns that are correlated with PKCε, reasoning that such genes could be acting in similar pathways and be potentially interacting. We found 19 genes that are coexpressed with PKCε (Table 2).
Taken together, genes for 29 of the 39 putative PKCε substrates were identified by at least one of these approaches (Table 2), and genes for 12 were identified in at least two of these combined approaches: Mapre2, Tubb2a, Gap43, Mapt, Syn1, Agap2, Rph3a, Caskin1, Grm5, Map2, Brsk1, and Mtss2 (Table 2). These results provide functional evidence linking PKCε and these 29 candidate substrates. The remaining 10 of the substrates we validated in the peptide array and Western blot analyses have no previously known association with PKCε in these four databases and represent potentially novel PKCε substrates (Table 2).
Phosphosite Databases
To determine if the sites we identified are in the context of a motif likely to be phosphorylated by PKCε, we compared the amino acid motif logo generated from our data to the amino acid motif logo generated from the sites of modification for PKCε in the PhosphoSitePlus v6.6.0.4 database ( ± 7 AA) (27). The motifs look strikingly similar for the amino acids at positions −3 and −2 amino-terminal to the phosphorylated site and amino acids +1, +2, and +3 carboxyl-terminal to the phosphorylated site. The motif is also consistent with consensus phosphorylation site motifs for PKCs that include (R/K)X(S/T), (R/K)(R/K)X(S/T), (R/K)XX(S/T), (R/K)X(S/T)(R/K), and (R/K)XX(S/T)X(R/K) but begins to diverge for more distal amino acids (Fig. 5) (29, 30, 31). None of the phosphorylation sites for PKCε in PhosphoSitePlus were identified in our study. However, 34 of the sites we detected were present in PhosphoSitePlus.
Fig. 5.
Amino acid motif surrounding the putative PKCε phosphorylation sites. Plot of the amino acid motif around the 45 identified sites of modification ( ± 7 AA) in this study (A) and for the 252 sites of modification ( ± 7 AA) for PKCε in PhosphoSitePlus v6.6.0.4 (B). Plots were created using the Sequence Logo tool on PhosphoSitePlus with default settings. PKCε, PKC epsilon.
The ChemPhoPro database contains predictions of kinase substrates based on an algorithm developed by Hijazi et al. (28). There were no substrates nominated from the prediction algorithm as possible PKCε substrates. The ChemPhoPro database also includes 83 previously reported substrates sourced from UniProt, one of which was identified in our screen (MARCKS S163). Nine of the sites we detected were present in ChemPhoPro, all of which were also in the PhosphoSitePlus database.
Functional Annotation of PKCε Putative Substrates and Relationship to PKCε CNS Phenotypes
We searched for GO terms that were enriched among the putative PKCε substrate genes using STRING. supplemental Table S7 contains the full results of the GO term enrichment analysis. Synaptic and cytoskeleton functions were highly enriched in the GO terms. This is consistent with previous results published for the synaptic proteome, as 32 of the putative PKCε substrates have been identified as synaptic proteins (33, 34). This is also consistent with previous PKCε literature, demonstrating a role for PKCε in cytoskeletal function, morphogenesis, and the release of synaptic vesicles (9, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47).
Since Prkce−/− mutant mice show decreased alcohol consumption, reduced anxiety-like behavior, and an accentuated response to benzodiazepines, we next focused on identifying substrates that might be involved in these phenotypes (2, 3, 7). For substrates that might regulate alcohol consumption, we queried a database containing ethanol-related genes (INIA IT-GED; http://inia.icmb.utexas.edu/) using our validated substrate gene list. We found that 35 out of the 39 substrate genes have been previously linked to alcohol consumption or other alcohol-related phenotypes (Table 2). The remaining substrate genes were not found in any IT-GED datasets.
To investigate putative PKCε substrates that might regulate anxiety-like behavior or response to benzodiazepines, we identified which PKCε substrates were also benzodiazepine targets based on gene expression signatures in the L1000 database. We identified seven of the substrate genes in this search: Ptn, Ppm1h, Mapt, Gap43, Phactr1, Rph3a, Mtss2. We also compared the putative PKCε substrate genes to genes related to various types of stress (http://hpc-bioinformatics.cineca.it/stress_mice/) and found that 18 out of 39 substrate genes have been linked to stress in that the transcript abundance of the genes is altered following exposure to experimental stress protocols like social defeat stress, fear conditioning, or restraint stress. Two of these genes, Phactr1 and Gap43, were notable because they have been linked to alcohol, stress, benzodiazepine response and have evidence supporting their interaction or coexpression with Prkce from our bioinformatics analyses.
Discussion
Here, we report using a chemical-genetic approach to identify PKCε substrates and their phosphorylation sites from mouse brain. We applied bioinformatic analyses to identify potential novel drug targets for the treatment of excessive alcohol consumption and anxiety. As validation of our method, we identified several known substrates of PKCε, including neuromodulin and MARCKS, confirmed their phosphorylation by PKCε in vitro using peptide arrays or Western blot analysis of GST-fusion proteins and performed motif analysis on the identified phosphorylation sites showing consistency with consensus PKC phosphorylation motifs. We also identified several proteins that have not been previously identified as PKCε substrates. Several are implicated in cytoskeletal regulation, morphogenesis, and synaptic vesicle release.
To our knowledge, ours is the first screen for direct substrates of PKCε in brain. However, there have been two studies that have analyzed the phosphoproteome after PKCε manipulation. One examined protein phosphorylation in the liver of rats fed a high fat diet after treatment with a PKCε-targeted antisense oligonucleotide (48). Two of the phosphorylated proteins they identified were seen in our study, MAP-4 and serine/arginine-rich splicing factor 2. MAP-4 phosphorylation was increased, while phosphorylation of serine/arginine-rich splicing factor 2 was reduced in a PKCε-dependent manner, independent of the high fat diet treatment. However, the sites of phosphorylation in both proteins did not match the sites identified in our present study. Moreover, their approach did not measure direct substrates of PKCε, only how PKCε expression changes the phosphoproteome of the rat liver following a high fat diet. A second study examined protein phosphorylation in cultured mouse astrocytes expressing a constitutively active form of PKCε (49). None of the differentially phosphorylated proteins were identified in our current study, and they also did not measure direct kinase substrates. In addition, they used cultured astrocytes which can have properties different from those that reside in brain. Our comprehensive list of direct substrates is thus unique and could be valuable for groups investigating the function of PKCε and for developing small molecule modulators of PKCε signaling in the context of central nervous system disease.
Other groups have examined the phosphoproteome in animal models of excessive ethanol consumption. One examined the dorsal striatum in ethanol naïve, high, and low alcohol-preferring mice (50). Seven phosphoproteins that were more abundant in the dorsal striatum of high alcohol–preferring mice were also identified in our study, although none of the phosphorylation sites were the same. These seven proteins were Microtubule-associated protein tau (Tau), MAP-1A, BRSK1, MARCKS-related protein, MARCKS, neuromodulin, and MAP2. Ten phosphoproteins that were significantly more abundant in the dorsal striatum of low alcohol–preferring mice were also identified in our study. These were Dynamin-3, Tau, neuromodulin, RAS guanyl-releasing protein 2, Phosphatase and actin regulator 1, MAP-1A, MARCKS, PPP4R4, AGAP-1, and Serine/arginine-rich splicing factor 2. Interestingly, the phosphorylation sites on neuromodulin (S41), MARCKS (S163), and PPP4R4 (S777) that were more abundant in the low alcohol–preferring mice are the same phosphorylation sites we identified. This overlap in identified proteins provides further evidence that these proteins regulate alcohol-related behaviors, especially considering that all but two of these proteins (MAP-1A and BRSK1) were also found in the INIA IT-GED microarray database.
A second study investigated the effect of adolescent corticosterone exposure on adult ethanol and sucrose self-administration and on the phosphoproteome of the adult rat amygdala (51). Adolescent corticosterone exposure increased several phosphoproteins and among those most altered that we also found in our study were MAP2, Tau, and MAP-1A. However, the sites of phosphorylation identified were different from those in our study. Nevertheless, these findings further implicate several substrates of PKCε as being involved in responses to alcohol and stress.
PKCε Substrates in Neurotransmitter Release and Synaptic Regulation
PKCε is present in presynaptic terminals and plays a role in vesicle release (9, 52). Glutamate exocytosis from hippocampal granule cells in guinea pigs depends on the actin-binding domain of PKCε (35). In the rat calyx of Held, activation of PKCε causes its translocation to the presynaptic membrane (37). Activity-dependent potentiation of large dense-core vesicle exocytosis in chromaffin cells is regulated by PKCε and MARCKS through modulation of vesicle translocation (39). In rat brain slices, selective activation of PKCε with α,β-DCP-LA stimulates the release of glutamate from the hippocampus, dopamine from the striatum, and serotonin from the hypothalamus (42). Additionally, PKCε has been implicated in acetylcholine release at the rat neuromuscular junction (44).
PKCε regulation of neurotransmitter release and synaptic function may be particularly important for behavioral responses to alcohol. We previously found that ethanol-induced enhancement of GABAergic transmission in the central amygdala is PKCε dependent (41). PKCε inhibitors block alcohol-induced GABA release in this region and when administered systemically, they reduce alcohol consumption in mice (4). Therefore, PKCε regulation of neurotransmission in the central amygdala may be related to its role in promoting alcohol consumption, suggesting that PKCε substrates involved in neurotransmitter release could be drug targets for reducing alcohol consumption. The substrates involved in neurotransmitter release with at least one line of supporting evidence from our bioinformatic analyses include calcium/calmodulin-dependent serine protein kinase (CASK) Interacting Protein 1 (Caskin-1), Synapsin-1, Brain-Specific Serine/Threonine-Protein Kinase (BRSK1), Rabphilin-3A, and Intersectin-1. None of these proteins have been previously identified as PKCε substrates. Of particular interest are Caskin-1, Synapsin-1, Rabphilin-3A, and Intersectin-1, since there is some evidence that they are altered by chronic exposure to alcohol and stress and therefore might influence alcohol consumption and anxiety.
Caskin-1 is a synaptic scaffolding protein that binds to calcium/calmodulin-dependent serine protein kinase and may link CASK to downstream intracellular effectors by binding the cytoplasmic tails of neurexins and other cell-surface proteins (53). Behavioral tests of Caskin1 KO mice suggest that Caskin-1 contributes to a wide spectrum of nervous system functions, including gait, nociception, memory formation, dendritic spine morphology, and the stress response (54, 55). Caskin1 transcripts are reduced in cortical astrocytes from humans with alcohol use disorder, in frontal cortex of C57BL/6J mice after chronic, every-other-day ethanol consumption, and in mouse cortical astrocytes after chronic intermittent ethanol exposure (56, 57, 58). Moreover, Caskin1 was a hub gene in an astrocyte-specific gene network that was downregulated by chronic intermittent ethanol treatment, suggesting that Caskin-1 in astrocytes could be a system regulator of the response to chronic ethanol consumption (58).
Synapsin-1 is the first identified synaptic vesicle protein and the most abundant phosphoprotein found at synapses (59, 60, 61). It is primarily expressed in neurons and is composed of a short N-terminal sequence (A-domain), a short linker (B-domain), a large conserved central sequence (C-domain), and a proline-rich C-terminal region (D-domain) (62). The A-domain of Synapsin-1 attaches to synaptic vesicles and can be phosphorylated by cAMP-dependent PKA and calcium/calmodulin-dependent protein kinase 1 (CaM kinase I) at Ser-9 (63, 64). Phosphorylation of the A-domain causes Synapsin-1 to dissociate from synaptic vesicles (65). The D-domain of Synapsin-1 can be phosphorylated by calcium/calmodulin-dependent protein kinase II (CaM kinase II) at Ser-566 and Ser-603 (62, 63, 66, 67). Phosphorylation at these sites is implicated in the regulation of synaptic vesicle mobilization and neurotransmitter release (68, 69). These sites are close to the putative PKCε phosphorylation site identified in this study, Ser-568. This suggests a role for PKCε in regulating synaptic vesicle release via Synapsin-1. Synapsin-1 is involved in various aspects of the synaptic vesicle cycle and plays an important role in the synchronization of GABA release in interneuron populations (70). Chronic ethanol exposure increases the clustering of Synapsin-1 in hippocampal neurons, and Synapsin-1 phosphorylation is increased in the brains of mice after an acute exposure to ethanol (71, 72). Syn1 gene expression is reduced in the cerebellum of C57BL/6J mice after one session of “drinking-in-the-dark”, which is a behavioral model of alcohol binge drinking (5). Syn1 expression is in contrast upregulated in the frontal cortex of postmortem human brains from individuals with a history of alcohol dependence (73).
In addition to ethanol responses, Synapsin-1 has also been implicated in the transcriptional response to chronic social defeat stress (CSDS) in mice. The CSDS model is a mouse model of resilience and susceptibility to psychosocial stress (74, 75). There appears to be a strong genetic component to CSDS-induced social avoidance, as different inbred strains of mice show varying proportions of resilient and susceptible mice (76). Syn1 transcript levels are reduced in the ventral hippocampus and bed nucleus of the stria terminalis of both DBA/2 and C57BL/6 mice following CSDS (76, 77). However, Syn1 transcript levels are higher in the nucleus accumbens of C57BL/6J mice, which are relatively resilient to CSDS (78). Moreover, the transcriptional response in the nucleus accumbens was associated with antidepressant responses to ketamine and imipramine, suggesting the transcriptional changes in the nucleus accumbens could be critical for stress responses (78).
Rabphilin-3A is a vesicle-associated protein involved in synaptic vesicle trafficking and release. Rabphilin-3A was first identified as a protein that interacts with the GTP-bound form of RAB3A and localizes with synaptic vesicles (79, 80, 81). Rabphilin-3A is phosphorylated by both PKA and CaM kinase II, but the role these events play in the regulation of synaptic vesicles is unclear (82, 83). Expression of Rph3a is reduced in the dorsal and ventral striatum of C57BL/6J mice after a single drinking-in-the-dark procedure, and an SNP in Rph3A has been associated with alcohol consumption in Korean men (5, 84).
Intersectin-1 is a scaffold protein involved in multiple cellular signaling pathways, regulation of the synaptic vesicle cycle, neuronal migration, and synaptic plasticity (85, 86, 87). There are two main isoforms of Intersectin-1 that have differential tissue expression. The short isoform is expressed ubiquitously, and the long isoform is expressed specifically in neurons (88, 89). In the brain, Intersectin-1 regulates dendritic spine development in hippocampal neurons and is an important mediator of fast neurotransmission (90, 91). A role for Intersectin-1 in fast neurotransmission involves its coordination of Synapsin-1 localization and the clearance of SNARE complexes at presynaptic release sites (92, 93). Itsn1 transcripts are reduced in the basolateral amygdala, nucleus accumbens shell, and nucleus accumbens core of ethanol-naïve High Drinking in the Dark (HDID-1) male mice (6). These mice have been selectively bred from heterogenous stock HS/Npt mice for binge-like drinking to high blood alcohol levels (94, 95). This finding suggests a role for Itsn1 in predisposition to binge drinking.
PKCε Substrates Involved in Cytoskeletal Regulation and Morphogenesis
Our PKCε substrate screen revealed several genes associated with cytoskeletal function and morphogenesis. A role for PKCε in cytoskeletal regulation is consistent with the presence of an actin-binding domain in PKCε that is unique among the PKC family and its previously described role in cytoskeletal regulation in non-neuronal cells (9, 35, 36, 38, 40, 45). Among our list of validated substrates, we found several microtubule-associated proteins to be putative PKCε substrates, including microtubule associated protein RP/EB family member 2 (EB2), microtubule associated protein tau, microtubule associated protein 2 (MAP2), and tubulin beta 2a.
EB2 has previously been associated with congenital skin disorders and was recently directly linked to human cranial neural crest migration (96, 97, 98). However, the function of EB2 in the mature brain has yet to be characterized. An SNP in Mapre2 has been associated with alcohol dependence (99). Mapre2 transcript levels are higher in the brains of alcohol-preferring rodents that in nonalcohol-preferring lines (100). Mapre2 expression is also linked to stress as it is expressed at lower levels in the hippocampus after CSDS in C57BL/6 mice (76).
Tau is abundant in neurons, plays a primary role in the assembly and stabilization of microtubules, and is key to the pathogenesis of several neurodegenerative disorders known as tauopathies (101, 102, 103). The binding of Tau to microtubules is negatively regulated by phosphorylation at specific sites by MAP2 kinase (104). PKCε can phosphorylate the microtubule-binding domains of Tau in vitro, but no studies have identified Thr-694 as a site of PKCε phosphorylation (105, 106, 107). Whether Thr-694 phosphorylation occurs in brain is not known. There is evidence that alcohol consumption may exacerbate tauopathies, and a large multiomics analysis found increased MAPT expression associated with increased alcohol consumption in humans (108, 109, 110).
MAP2 is another microtubule-associated protein that is enriched in neuronal dendrites and plays functional roles in microtubule stabilization, bundling, and proper spacing in dendrites and axons (111, 112, 113, 114, 115). Phosphorylation by PKA, PKC, and MARK1 regulate the association of MAP2 with microtubules (116). These kinases phosphorylate the microtubule-binding domain of MAP2 and cause it to dissociate from microtubules (117, 118, 119, 120, 121, 122, 123). Map2 expression is reduced following chronic alcohol consumption and is increased during withdrawal in specific rat brain regions (124). Map2 mRNA is also increased in the extended amygdala of ethanol-naïve HDID-1 mice compared with HS/Npt mice from which they were selected (125).
Tubulin beta-2A chain is a member of the tubulin family of proteins that co-assemble into microtubules. The peptide we identified is also present in Tubulin beta-2B, 4A, 4B, and five chain proteins. Microtubules are highly dynamic and provide cellular support, allow for intracellular trafficking, and are critical for neuronal migration during development (126, 127). Mutations in tubulin genes can cause brain malformations known as tubulinopathies (128). While no mutations in these genes have been linked to alcohol consumption, tubulin proteins overall are reduced, while expression of TUBB2B, TUBB4A, and TUBB4B genes is increased, in brain tissue from humans with alcohol use disorder (73, 129, 130). Rodent models have also revealed alcohol-related changes in brain tubulin gene expression and protein levels. Tubulin beta-2A chain is increased in the nucleus accumbens of male C57BL/6J mice stably consuming ∼15 g/kg/day with continuous voluntary access to 20% alcohol, and Tubb2a expression is increased in the extended amygdala of ethanol-naïve HDID-1 mice compared with HS/Npt mice (125, 131). However, Tubb4b expression is decreased in the prefrontal cortex of female C57BL/6J mice after intermittent access to 20% alcohol, and Tubb5 expression is decreased in the nucleus accumbens shell and bed nucleus of the stria terminalis of ethanol-naïve HDID-1 mice compared with HS/Mpt mice (125, 132). There is also differential expression of Tubb2b and Tubb4b in the brains of mice genetically predisposed to elevated alcohol consumption, and Tubb5 expression is differentially expressed in mice after a drinking-in-the-dark procedure (5, 100). In addition, after chronic social defeat stress, Tubb2a, Tubb2b, and Tubb4b expression is increased in the hippocampus of male DBA2/J and C57BL/6J mice, while Tubb4a and Tubb4b expression is increased in the nucleus accumbens of male C57BL/6J mice (76, 78). Therefore, several members of the tubulin family of proteins are responsive to ethanol and are differentially expressed in rodents selectively bred to consume high levels of ethanol. Taken together, these findings suggest a mechanistic link between PKCε, cytoskeletal regulation, and alcohol consumption.
Summary
In summary, we identified 39 putative PKCε substrates, most of them novel, using a chemical genetic screen. Bioinformatics analyses showed that many are important for neurotransmitter release and microtubule function. Transcripts for several substrates are altered by alcohol exposure and chronic stress. Therefore, this investigation provides a substantial list of candidates for further investigation into the role of PKCε in alcohol consumption and responses to stress.
There are caveats to our results that deserve consideration. First, production of whole brain lysates eliminates the normal compartmentalization of proteins within cells and can lead to kinase-substrate interactions that do not normally occur under physiological conditions. Therefore, it is likely that some of the proteins we identified are not true PKCε substrates in intact tissues.
A second caveat involves the method used to capture thiophosphates which also captures peptides with cysteines. These form thioether linkages that are stable to oxidation and are retained on the iodoacetyl beads. In silico analysis of trypsin digests of the human proteome estimates that about 24% of the peptides contain cysteine and therefore would be missed by the method we used (12).
Third, although many proteins were thiophosphorylated in the bulk samples (Fig. 1A), we only identified peptides that were specifically present in samples incubated with AS-PKCε and were detected in at least two experiments. This stringent analysis decreased false positives but may have resulted in false negatives for those phosphoproteins of low abundance or with some abundance in samples incubated without AS-PKCε. Indeed, when we inadvertently included five peptides that appeared in only one of the three experiments, we found that two of these (peptides associated with genes Arhgap12 and Map7) were validated by the peptide array. We did not include these two in the bioinformatics analyses.
Some putative substrates confirmed by Western blot analysis of phosphorylated GST fusion proteins, mGluR5 and GLAST-1, were not detected on the peptide array. This may have occurred because anchoring of some peptide sequences to the array membrane may interfere with kinase-substrate interactions that more readily occur in solution. Therefore, while peptide arrays are efficient at screening for substrates, they may miss some substrates of importance. Follow-up solution-based kinase assays can be used to detect these proteins.
Finally, our studies did not determine the functional significance of the identified phosphorylation sites. Future work will be needed to investigate the importance of these phosphorylation sites for protein and cellular function and for animal behavior.
Data Availability
The authors declare that the main data supporting the findings of this study are available within the article and its Supplemental materials. Raw mass spectrometry data has been submitted to MassIVE (massive.ucsd.edu) with the identifier MSV000089659. Annotated spectra for all results can be viewed using MSViewer (133) at https://msviewer.ucsf.edu/prospector/cgi-bin/msform.cgi?form=msviewer using the following search keys:
NH620 ETD data: os9qgidggu
NH620 HCD data: z2slyneu87
NH623 ETD data: ccxvkteg0x
NH623 HCD data: zhb7mdgn08
NH625 ETD data: 38eph2azey
NH625 HCD data: dmolm7wdix
Supplemental data
This article contains supplemental data (134).
Conflicts of Interest
The authors with the exception of K. M. S. and N. T. H declare that there are no competing interests associated with the manuscript. K. M. S. is an inventor on patents covering the analog sensitive and analog specific kinase engineering owned by Princeton University. K. M. S. has consulting agreements for the following companies, which involve monetary and/or stock compensation: Revolution Medicines, Black Diamond Therapeutics, BridGene Biosciences, Denali Therapeutics, Dice Molecules, eFFECTOR Therapeutics, Erasca, Genentech/Roche, Janssen Pharmaceuticals, Kumquat Biosciences, Kura Oncology, Mitokinin Inc, Nested, Type6 Therapeutics, Venthera, Wellspring Biosciences (Araxes Pharma), Turning Point, Ikena, Initial Therapeutics, Vevo, and BioTheryX. N. T. H owns shares and is an employee of Mitokinin Inc.
Acknowledgments
We would like to acknowledge and thank Dai-Fei Wu PhD for assistance in performing thiophosphorylation and covalent capture experiments.
Funding and additional information
This work was supported by NIH Grant AA013588 and funds provided by the M. June and J. Virgil Waggoner Chair in Molecular Biology to R. O. M., a University of Texas at Austin Bruce-Jones Fellowship awarded to M. P. D., an NIH F32 AA028148 award to L. B. F, a Miriam and Sheldon G. Adelson Medical Research Foundation grant awarded to A. L. B., and an NIH Alcohol Research Training Grant T32 AA007471 to L. B. F. P. J. P. was supported by the Francis Crick Institute, which receives its core funding from Cancer Research U.K. (FC001130), the U.K. Medical Research Council (FC001130), and the Wellcome Trust (FC001130). N. T. H. and K. M. S. acknowledge support by the Howard Hughes Medical Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author contributions
M. P. D. and L. B. F. validation; M. P. D. and L. B. F. formal analysis; M. P. D. and L. B. F. writing–original draft; N. T. H., R. J. C., and P. J. P. investigation; A. L. B. and R. O. M. supervision; K. M. S. and R. O. M. conceptualization; K. M. S., P. J. P., and R. O. M. writing–review and editing; R. O. M. funding acquisition.
Supplemental Data
References
- 1.Battaini F., Mochly-Rosen D. Happy birthday protein kinase C: past, present and future of a superfamily. Pharmacol. Res. 2007;55:461–466. doi: 10.1016/j.phrs.2007.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hodge C.W., Mehmert K.K., Kelley S.P., McMahon T., Haywood A., Olive M.F., et al. Supersensitivity to allosteric GABA A receptor modulators and alcohol in mice lacking PKCε. Nat. Neurosci. 1999;2:997–1002. doi: 10.1038/14795. [DOI] [PubMed] [Google Scholar]
- 3.Olive M.F., Mehmert K.K., Messing R.O., Hodge C.W. Reduced operant ethanol self-administration and in vivo mesolimbic dopamine responses to ethanol inPKCε-deficient mice. Eur. J. Neurosci. 2000;12:4131–4140. doi: 10.1046/j.1460-9568.2000.00297.x. [DOI] [PubMed] [Google Scholar]
- 4.Blasio A., Wang J., Wang D., Varodayan F.P., Pomrenze M.B., Miller J., et al. Novel small-molecule inhibitors of protein kinase C epsilon reduce ethanol consumption in mice. Biol. Psychiatry. 2018;84:193–201. doi: 10.1016/j.biopsych.2017.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mulligan M.K., Rhodes J.S., Crabbe J.C., Mayfield R.D., Harris R.A., Ponomarev I. Molecular profiles of drinking alcohol to intoxication in C57BL/6J mice. Alcohol. Clin. Exp. Res. 2011;35:659–670. doi: 10.1111/j.1530-0277.2010.01384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferguson L.B., Ozburn A.R., Ponomarev I., Metten P., Reilly M., Crabbe J.C., et al. Genome-wide expression profiles drive discovery of novel compounds that reduce binge drinking in mice. Neuropsychopharmacology. 2018;43:1257–1266. doi: 10.1038/npp.2017.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hodge C.W., Raber J., McMahon T., Walter H., Sanchez-Perez A.M., Olive M.F., et al. Decreased anxiety-like behavior, reduced stress hormones, and neurosteroid supersensitivity in mice lacking protein kinase Cε. J. Clin. Invest. 2002;110:1003–1010. doi: 10.1172/JCI15903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lesscher H.M.B., McMahon T., Lasek A.W., Chou W.-H., Connolly J., Kharazia V., et al. Amygdala protein kinase C epsilon regulates corticotropin-releasing factor and anxiety-like behavior. Genes Brain Behav. 2008;7:323–333. doi: 10.1111/j.1601-183X.2007.00356.x. [DOI] [PubMed] [Google Scholar]
- 9.Shirai Y., Adachi N., Saito N. Protein kinase cε: Function in neurons. FEBS J. 2008;275:3988–3994. doi: 10.1111/j.1742-4658.2008.06556.x. [DOI] [PubMed] [Google Scholar]
- 10.Newton P.M., Messing R.O. The substrates and binding partners of protein kinase Cε. Biochem. J. 2010;427:189–196. doi: 10.1042/BJ20091302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kandasamy R., Price T.J. In: Pain Control. Schaible H.-G., editor. Springer; Berlin, Heidelberg: 2015. The pharmacology of nociceptor priming. (15–37). [Google Scholar]
- 12.Blethrow J.D., Glavy J.S., Morgan D.O., Shokat K.M. Covalent capture of kinase-specific phosphopeptides reveals Cdk1-cyclin B substrates. Proc. Natl. Acad. Sci. U. S. A. 2008;105:1442–1447. doi: 10.1073/pnas.0708966105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hertz N.T., Wang B.T., Allen J.J., Zhang C., Dar A.C., Burlingame A.L., et al. Chemical genetic approach for kinase-substrate mapping by covalent capture of thiophosphopeptides and analysis by mass spectrometry. Curr. Protoc. Chem. Biol. 2010;2:15–36. doi: 10.1002/9780470559277.ch090201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Überall F., Giselbrecht S., Hellbert K., Fresser F., Bauer B., Gschwendt M., et al. Conventional PKC-α, novel PKC-ε and PKC-θ, but not atypical PKC-λ are MARCKS kinases in intact NIH 3T3 fibroblasts. J. Biol. Chem. 1997;272:4072–4078. doi: 10.1074/jbc.272.7.4072. [DOI] [PubMed] [Google Scholar]
- 15.Oehrlein S.A., Parker P.J., Herget T. Phosphorylation of GAP-43 (growth-associated protein of 43 kDa) by conventional, novel and atypical isotypes of the protein kinase C gene family: Differences between oligopeptide and polypeptide phosphorylation. Biochem. J. 1996;317:219–224. doi: 10.1042/bj3170219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khasar S.G., Lin Y.-H., Martin A., Dadgar J., McMahon T., Wang D., et al. A novel nociceptor signaling pathway revealed in protein kinase C ε mutant mice. Neuron. 1999;24:253–260. doi: 10.1016/s0896-6273(00)80837-5. [DOI] [PubMed] [Google Scholar]
- 17.Sert N. P. du, Hurst V., Ahluwalia A., Alam S., Avey M.T., Baker M., et al. The ARRIVE guidelines 2.0: updated guidelines for reporting animal research. PLoS Biol. 2020;18 doi: 10.1371/journal.pbio.3000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 19.Qi Z.-H., Song M., Wallace M.J., Wang D., Newton P.M., McMahon T., et al. Protein kinase cε regulates γ-aminobutyrate type A receptor sensitivity to ethanol and benzodiazepines through phosphorylation of γ2 subunits. J. Biol. Chem. 2007;282:33052–33063. doi: 10.1074/jbc.M707233200. [DOI] [PubMed] [Google Scholar]
- 20.Ultanir S.K., Hertz N.T., Li G., Ge W.-P., Burlingame A.L., Pleasure S.J., et al. Chemical genetic identification of NDR1/2 kinase substrates AAK1 and Rabin8 uncovers their roles in dendrite arborization and spine development. Neuron. 2012;73:1127–1142. doi: 10.1016/j.neuron.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hilpert K., Winkler D.F., Hancock R.E. Cellulose-bound peptide arrays: preparation and applications. Biotechnol. Genet. Eng. Rev. 2007;24:31–106. doi: 10.1080/02648725.2007.10648093. [DOI] [PubMed] [Google Scholar]
- 22.Subramanian A., Narayan R., Corsello S.M., Peck D.D., Natoli T.E., Lu X., et al. A next generation connectivity map: l1000 platform and the first 1,000,000 profiles. Cell. 2017;171:1437–1452.e17. doi: 10.1016/j.cell.2017.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucl. Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Warde-Farley D., Donaldson S.L., Comes O., Zuberi K., Badrawi R., Chao P., et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucl. Acids Res. 2010;38:W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Dam S., Cordeiro R., Craig T., van Dam J., Wood S.H., de Magalhães J.P. GeneFriends: an online co-expression analysis tool to identify novel gene targets for aging and complex diseases. BMC Genomics. 2012;13:535. doi: 10.1186/1471-2164-13-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Dam S., Craig T., de Magalhães J.P. GeneFriends: A human RNA-seq-based gene and transcript co-expression database. Nucl. Acids Res. 2015;43:D1124–D1132. doi: 10.1093/nar/gku1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hornbeck P.V., Zhang B., Murray B., Kornhauser J.M., Latham V., Skrzypek E. PhosphoSitePlus, 2014: Mutations, PTMs and recalibrations. Nucl.Acids Res. 2015;43:D512–D520. doi: 10.1093/nar/gku1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hijazi M., Smith R., Rajeeve V., Bessant C., Cutillas P.R. Reconstructing kinase network topologies from phosphoproteomics data reveals cancer-associated rewiring. Nat. Biotechnol. 2020;38:493–502. doi: 10.1038/s41587-019-0391-9. [DOI] [PubMed] [Google Scholar]
- 29.Kreegipuu A., Blom N., Brunak S., Järv J. Statistical analysis of protein kinase specificity determinants. FEBS Lett. 1998;430:45–50. doi: 10.1016/s0014-5793(98)00503-1. [DOI] [PubMed] [Google Scholar]
- 30.Nishikawa K., Toker A., Johannes F.-J., Songyang Z., Cantley L.C. Determination of the specific substrate sequence motifs of protein kinase C isozymes. J. Biol. Chem. 1997;272:952–960. doi: 10.1074/jbc.272.2.952. [DOI] [PubMed] [Google Scholar]
- 31.Pearson R.B., Kemp B.E. Methods in Enzymology. Academic Press; 1991. [3] protein kinase phosphorylation site sequences and consensus specificity motifs: tabulations; pp. 62–81. [DOI] [PubMed] [Google Scholar]
- 32.Flati T., Gioiosa S., Chillemi G., Mele A., Oliverio A., Mannironi C., et al. A gene expression atlas for different kinds of stress in the mouse brain. Sci. Data. 2020;7:437. doi: 10.1038/s41597-020-00772-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trinidad J.C., Specht C.G., Thalhammer A., Schoepfer R., Burlingame A.L. Comprehensive identification of phosphorylation sites in postsynaptic density preparations. Mol. Cell. Proteomics. 2006;5:914–922. doi: 10.1074/mcp.T500041-MCP200. [DOI] [PubMed] [Google Scholar]
- 34.Taoufiq Z., Ninov M., Villar-Briones A., Wang H.-Y., Sasaki T., Roy M.C., et al. Hidden proteome of synaptic vesicles in the mammalian brain. Proc. Natl. Acad. Sci. U. S. A. 2020;117:33586–33596. doi: 10.1073/pnas.2011870117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prekeris R., Mayhew M.W., Cooper J.B., Terrian D.M. Identification and localization of an actin-binding motif that is unique to the epsilon isoform of protein kinase C and participates in the regulation of synaptic function. J. Cell Biol. 1996;132:77–90. doi: 10.1083/jcb.132.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prekeris R., Hernandez R.M., Mayhew M.W., White M.K., Terrian D.M. Molecular analysis of the interactions between protein kinase C-ε and filamentous actin. J. Biol. Chem. 1998;273:26790–26798. doi: 10.1074/jbc.273.41.26790. [DOI] [PubMed] [Google Scholar]
- 37.Saitoh N., Hori T., Takahashi T. Activation of the epsilon isoform of protein kinase C in the mammalian nerve terminal. Proc. Natl. Acad. Sci. U. S. A. 2001;98:14017–14021. doi: 10.1073/pnas.241333598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Akita Y. Protein kinase C-ε (PKC-ε): its unique structure and function. J. Biochem. 2002;132:847–852. doi: 10.1093/oxfordjournals.jbchem.a003296. [DOI] [PubMed] [Google Scholar]
- 39.Park Y.-S., Hur E.-M., Choi B.-H., Kwak E., Jun D.-J., Park S.-J., et al. Involvement of protein kinase C-ε in activity-dependent potentiation of large dense-core vesicle exocytosis in chromaffin cells. J. Neurosci. 2006;26:8999–9005. doi: 10.1523/JNEUROSCI.2828-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Akita Y. Protein kinase cε: multiple roles in the function of, and signaling mediated by, the cytoskeleton. FEBS J. 2008;275:3995–4004. doi: 10.1111/j.1742-4658.2008.06557.x. [DOI] [PubMed] [Google Scholar]
- 41.Bajo M., Cruz M.T., Siggins G.R., Messing R., Roberto M. Protein kinase C epsilon mediation of CRF- and ethanol-induced GABA release in central amygdala. Proc. Natl. Acad. Sci. U. S. A. 2008;105:8410–8415. doi: 10.1073/pnas.0802302105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shimizu T., Kanno T., Tanaka A., Nishizaki T. α, β-DCP-LA selectively activates PKC-ε and stimulates neurotransmitter release with the highest potency among 4 Diastereomers. Cell. Physiol. Biochem. 2011;27:149–158. doi: 10.1159/000325217. [DOI] [PubMed] [Google Scholar]
- 43.Chen Y., Tian Q. The role of protein kinase C epsilon in neural signal transduction and neurogenic diseases. Front. Med. 2011;5:70. doi: 10.1007/s11684-011-0119-9. [DOI] [PubMed] [Google Scholar]
- 44.Obis T., Besalduch N., Hurtado E., Nadal L., Santafe M.M., Garcia N., et al. The novel protein kinase C epsilon isoform at the adult neuromuscular synapse: Location, regulation by synaptic activity-dependent muscle contraction through TrkB signaling and coupling to ACh release. Mol. Brain. 2015;8:8. doi: 10.1186/s13041-015-0098-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sen A., Hongpaisan J., Wang D., Nelson T.J., Alkon D.L. Protein kinase cε (PKCε) promotes synaptogenesis through membrane accumulation of the postsynaptic density protein PSD-95. J. Biol. Chem. 2016;291:16462–16476. doi: 10.1074/jbc.M116.730440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schaffer T.B., Smith J.E., Cook E.K., Phan T., Margolis S.S. PKCε inhibits neuronal dendritic spine development through dual phosphorylation of Ephexin5. Cell Rep. 2018;25:2470–2483.e8. doi: 10.1016/j.celrep.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cooke M., Baker M.J., Kazanietz M.G., Casado-Medrano V. PKCε regulates Rho GTPases and actin cytoskeleton reorganization in non-small cell lung cancer cells. Small GTPases. 2021;12:202–208. doi: 10.1080/21541248.2019.1684785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gassaway B.M., Petersen M.C., Surovtseva Y.V., Barber K.W., Sheetz J.B., Aerni H.R., et al. PKCε contributes to lipid-induced insulin resistance through cross talk with p70S6K and through previously unknown regulators of insulin signaling. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E8996–E9005. doi: 10.1073/pnas.1804379115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Burgos M., Fradejas N., Calvo S., Kang S.U., Tranque P., Lubec G. A proteomic analysis of PKCε targets in astrocytes: Implications for astrogliosis. Amino Acids. 2011;40:641–651. doi: 10.1007/s00726-010-0691-3. [DOI] [PubMed] [Google Scholar]
- 50.Grecco G.G., Haggerty D.L., Doud E.H., Fritz B.M., Yin F., Hoffman H., et al. A multi-omic analysis of the dorsal striatum in an animal model of divergent genetic risk for alcohol use disorder. J. Neurochem. 2021;157:1013–1031. doi: 10.1111/jnc.15226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bertholomey M.L., Stone K., Lam T.T., Bang S., Wu W., Nairn A.C., et al. Phosphoproteomic analysis of the amygdala response to adolescent glucocorticoid exposure reveals g-protein coupled receptor kinase 2 as a target for reducing motivation for alcohol. Proteomes. 2018;6:41. doi: 10.3390/proteomes6040041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saito N., Itouji A., Totani Y., Osawa I., Koide H., Fujisawa N., et al. Cellular and intracellular localization of ε-subspecies of protein kinase C in the rat brain; presynaptic localization of the ε-subspecies. Brain Res. 1993;607:241–248. doi: 10.1016/0006-8993(93)91512-q. [DOI] [PubMed] [Google Scholar]
- 53.Tabuchi K., Biederer T., Butz S., Südhof T.C. CASK participates in alternative tripartite complexes in which mint 1 competes for binding with Caskin 1, a novel CASK-binding protein. J. Neurosci. 2002;22:4264–4273. doi: 10.1523/JNEUROSCI.22-11-04264.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Katano T., Takao K., Abe M., Yamazaki M., Watanabe M., Miyakawa T., et al. Distribution of Caskin1 protein and phenotypic characterization of its knockout mice using a comprehensive behavioral test battery. Mol. Brain. 2018;11:63. doi: 10.1186/s13041-018-0407-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bencsik N., Pusztai S., Borbély S., Fekete A., Dülk M., Kis V., et al. Dendritic spine morphology and memory formation depend on postsynaptic Caskin proteins. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-53317-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brenner E., Tiwari G.R., Kapoor M., Liu Y., Brock A., Mayfield R.D. Single cell transcriptome profiling of the human alcohol-dependent brain. Hum. Mol. Genet. 2020;29:1144–1153. doi: 10.1093/hmg/ddaa038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Osterndorff-Kahanek E.A., Becker H.C., Lopez M.F., Farris S.P., Tiwari G.R., Nunez Y.O., et al. Chronic ethanol exposure produces time- and brain region-dependent changes in gene coexpression networks. PLoS One. 2015;10 doi: 10.1371/journal.pone.0121522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Erickson E.K., Grantham E.K., Warden A.S., Harris R.A. Neuroimmune signaling in alcohol use disorder. Pharmacol. Biochem. Behav. 2019;177:34–60. doi: 10.1016/j.pbb.2018.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.De Camilli P., Harris S.M., Huttner W.B., Greengard P. Synapsin I (Protein I), a nerve terminal-specific phosphoprotein. II. Its specific association with synaptic vesicles demonstrated by immunocytochemistry in agarose-embedded synaptosomes. J. Cell Biol. 1983;96:1355–1373. doi: 10.1083/jcb.96.5.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dolphin A.C., Greengard P. Serotonin stimulates phosphorylation of Protein I in the facial motor nucleus of rat brain. Nature. 1981;289:76–79. doi: 10.1038/289076a0. [DOI] [PubMed] [Google Scholar]
- 61.Huttner W.B., Schiebler W., Greengard P., De Camilli P. Synapsin I (protein I), a nerve terminal-specific phosphoprotein. III. Its association with synaptic vesicles studied in a highly purified synaptic vesicle preparation. J. Cell Biol. 1983;96:1374–1388. doi: 10.1083/jcb.96.5.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sudhof T.C., Czernik A.J., Kao H.T., Takei K., Johnston P.A., Horiuchi A., et al. Synapsins: Mosaics of shared and individual domains in a family of synaptic vesicle phosphoproteins. Science. 1989;245:1474–1480. doi: 10.1126/science.2506642. [DOI] [PubMed] [Google Scholar]
- 63.Huttner W.B., DeGennaro L.J., Greengard P. Differential phosphorylation of multiple sites in purified protein I by cyclic AMP-dependent and calcium-dependent protein kinases. J. Biol. Chem. 1981;256:1482–1488. [PubMed] [Google Scholar]
- 64.Kennedy M.B., Greengard P. Two calcium/calmodulin-dependent protein kinases, which are highly concentrated in brain, phosphorylate protein I at distinct sites. Proc. Natl. Acad. Sci. U. S. A. 1981;78:1293–1297. doi: 10.1073/pnas.78.2.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hosaka M., Hammer R.E., Südhof T.C. A phospho-switch controls the dynamic association of synapsins with synaptic vesicles. Neuron. 1999;24:377–387. doi: 10.1016/s0896-6273(00)80851-x. [DOI] [PubMed] [Google Scholar]
- 66.Huttner W.B., Greengard P. Multiple phosphorylation sites in protein I and their differential regulation by cyclic AMP and calcium. Proc. Natl. Acad. Sci. U. S. A. 1979;76:5402–5406. doi: 10.1073/pnas.76.10.5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Czernik A.J., Pang D.T., Greengard P. Amino acid sequences surrounding the cAMP-dependent and calcium/calmodulin-dependent phosphorylation sites in rat and bovine synapsin I. Proc. Natl. Acad. Sci. U. S. A. 1987;84:7518–7522. doi: 10.1073/pnas.84.21.7518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chi P., Greengard P., Ryan T.A. Synaptic vesicle mobilization is regulated by distinct synapsin I phosphorylation pathways at different frequencies. Neuron. 2003;38:69–78. doi: 10.1016/s0896-6273(03)00151-x. [DOI] [PubMed] [Google Scholar]
- 69.Llinás R., Gruner J.A., Sugimori M., McGuinness T.L., Greengard P. Regulation by synapsin I and Ca(2+)-calmodulin-dependent protein kinase II of the transmitter release in squid giant synapse. J. Physiol. 1991;436:257–282. doi: 10.1113/jphysiol.1991.sp018549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Forte N., Binda F., Contestabile A., Benfenati F., Baldelli P. Synapsin I synchronizes GABA release in distinct interneuron subpopulations. Cereb. Cortex. 2020;30:1393–1406. doi: 10.1093/cercor/bhz174. [DOI] [PubMed] [Google Scholar]
- 71.Carpenter-Hyland E.P., Woodward J.J., Chandler L.J. Chronic ethanol induces synaptic but not extrasynaptic targeting of NMDA receptors. J. Neurosci. 2004;24:7859–7868. doi: 10.1523/JNEUROSCI.1902-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Conti A.C., Jr J.W.M., Moulder K.L., Jiang X., Dave B.A., Mennerick S., et al. Adenylyl cyclases 1 and 8 initiate a presynaptic homeostatic response to ethanol treatment. PLoS One. 2009;4 doi: 10.1371/journal.pone.0005697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ponomarev I., Wang S., Zhang L., Harris R.A., Mayfield R.D. Gene coexpression networks in human brain identify epigenetic modifications in alcohol dependence. J. Neurosci. 2012;32:1884–1897. doi: 10.1523/JNEUROSCI.3136-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Krishnan V., Han M.-H., Graham D.L., Berton O., Renthal W., Russo S.J., et al. Molecular adaptations underlying susceptibility and resistance to social defeat in brain reward regions. Cell. 2007;131:391–404. doi: 10.1016/j.cell.2007.09.018. [DOI] [PubMed] [Google Scholar]
- 75.Hammels C., Pishva E., De Vry J., van den Hove D.L.A., Prickaerts J., van Winkel R., et al. Defeat stress in rodents: from behavior to molecules. Neurosci. Biobehav. Rev. 2015;59:111–140. doi: 10.1016/j.neubiorev.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 76.Laine M.A., Trontti K., Misiewicz Z., Sokolowska E., Kulesskaya N., Heikkinen A., et al. Genetic control of myelin plasticity after chronic psychosocial stress. eNeuro. 2018;5 doi: 10.1523/ENEURO.0166-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Misiewicz Z., Iurato S., Kulesskaya N., Salminen L., Rodrigues L., Maccarrone G., et al. Multi-omics analysis identifies mitochondrial pathways associated with anxiety-related behavior. PLoS Genet. 2019;15 doi: 10.1371/journal.pgen.1008358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bagot R.C., Cates H.M., Purushothaman I., Vialou V., Heller E.A., Yieh L., et al. Ketamine and imipramine reverse transcriptional signatures of susceptibility and induce resilience-specific gene expression profiles. Biol. Psychiatry. 2017;81:285–295. doi: 10.1016/j.biopsych.2016.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shirataki H., Kaibuchi K., Yamaguchi T., Wada K., Horiuchi H., Takai Y. A possible target protein for smg-25A/rab3A small GTP-binding protein. J. Biol. Chem. 1992;267:10946–10949. [PubMed] [Google Scholar]
- 80.Shirataki H., Kaibuchi K., Sakoda T., Kishida S., Yamaguchi T., Wada K., et al. Rabphilin-3A, a putative target protein for smg p25A/rab3A p25 small GTP-binding protein related to synaptotagmin. Mol. Cell. Biol. 1993;13:2061–2068. doi: 10.1128/mcb.13.4.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mizoguchi A., Yano Y., Hamaguchi H., Yanagida H., Ide C., Zahraoui A., et al. Localization of rabphilin-3A on the synaptic vesicle. Biochem. Biophys. Res. Commun. 1994;202:1235–1243. doi: 10.1006/bbrc.1994.2063. [DOI] [PubMed] [Google Scholar]
- 82.Numata S., Shirataki H., Hagi S., Yamamoto T., Takai Y. Phosphorylation of rabphilin-3A, a putative target protein for Rab3A, by cyclic AMP-dependent protein kinase. Biochem. Biophys. Res. Commun. 1994;203:1927–1934. doi: 10.1006/bbrc.1994.2413. [DOI] [PubMed] [Google Scholar]
- 83.Kato M., Sasaki T., Imazumi K., Takahashi K., Araki K., Shirataki H., et al. Phosphorylation of rabphilin-3A by calmodulin-dependent protein kinase II. Biochem. Biophys. Res. Commun. 1994;205:1776–1784. doi: 10.1006/bbrc.1994.2875. [DOI] [PubMed] [Google Scholar]
- 84.Baik I., Cho N.H., Kim S.H., Han B.-G., Shin C. Genome-wide association studies identify genetic loci related to alcohol consumption in Korean men. Am. J. Clin. Nutr. 2011;93:809–816. doi: 10.3945/ajcn.110.001776. [DOI] [PubMed] [Google Scholar]
- 85.Hunter M.P., Russo A., O’Bryan J.P. Emerging roles for intersectin (ITSN) in regulating signaling and disease pathways. Int. J. Mol. Sci. 2013;14:7829–7852. doi: 10.3390/ijms14047829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pechstein A., Shupliakov O., Haucke V. Intersectin 1: a versatile actor in the synaptic vesicle cycle. Biochem. Soc. Trans. 2010;38:181–186. doi: 10.1042/BST0380181. [DOI] [PubMed] [Google Scholar]
- 87.Jakob B., Kochlamazashvili G., Jäpel M., Gauhar A., Bock H.H., Maritzen T., et al. Intersectin 1 is a component of the Reelin pathway to regulate neuronal migration and synaptic plasticity in the hippocampus. Proc. Natl. Acad. Sci. U. S. A. 2017;114:5533–5538. doi: 10.1073/pnas.1704447114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pucharcos C., Casas C., Nadal M., Estivill X., de la Luna S. The human intersectin genes and their spliced variants are differentially expressed. Biochim. Biophys. Acta. 2001;1521:1–11. doi: 10.1016/s0167-4781(01)00276-7. [DOI] [PubMed] [Google Scholar]
- 89.Ma Y.J., Okamoto M., Gu F., Obata K., Matsuyama T., Desaki J., et al. Neuronal distribution of EHSH1/intersectin: molecular linker between clathrin-mediated endocytosis and signaling pathways. J. Neurosci. Res. 2003;71:468–477. doi: 10.1002/jnr.10500. [DOI] [PubMed] [Google Scholar]
- 90.Thomas S., Ritter B., Verbich D., Sanson C., Bourbonnière L., McKinney R.A., et al. Intersectin regulates dendritic spine development and somatodendritic endocytosis but not synaptic vesicle recycling in hippocampal neurons. J. Biol. Chem. 2009;284:12410–12419. doi: 10.1074/jbc.M809746200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sakaba T., Kononenko N.L., Bacetic J., Pechstein A., Schmoranzer J., Yao L., et al. Fast neurotransmitter release regulated by the endocytic scaffold intersectin. Proc. Natl. Acad. Sci. U. S. A. 2013;110:8266–8271. doi: 10.1073/pnas.1219234110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gerth F., Jäpel M., Pechstein A., Kochlamazashvili G., Lehmann M., Puchkov D., et al. Intersectin associates with synapsin and regulates its nanoscale localization and function. Proc. Natl. Acad. Sci. U. S. A. 2017;114:12057–12062. doi: 10.1073/pnas.1715341114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jäpel M., Gerth F., Sakaba T., Bacetic J., Yao L., Koo S.-J., et al. Intersectin-Mediated clearance of SNARE complexes is required for fast neurotransmission. Cell Rep. 2020;30:409–420.e6. doi: 10.1016/j.celrep.2019.12.035. [DOI] [PubMed] [Google Scholar]
- 94.Thiele T.E., Navarro M. Drinking in the dark” (DID) procedures: a model of binge-like ethanol drinking in non-dependent mice. Alcohol. 2014;48:235–241. doi: 10.1016/j.alcohol.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Crabbe J.C., Metten P., Rhodes J.S., Yu C.-H., Brown L.L., Phillips T.J., et al. A line of mice selected for high blood ethanol concentrations shows drinking in the dark to intoxication. Biol. Psychiatry. 2009;65:662–670. doi: 10.1016/j.biopsych.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Isrie M., Breuss M., Tian G., Hansen A.H., Cristofoli F., Morandell J., et al. Mutations in either TUBB or MAPRE2 cause circumferential skin creases kunze type. Am. J. Hum. Genet. 2015;97:790–800. doi: 10.1016/j.ajhg.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Berkun L., Slae M., Mor-Shaked H., Koplewitz B., Eventov-Friedman S., Harel T. Homozygous variants in MAPRE2 and CDON in individual with skin folds, growth delay, retinal coloboma, and pyloric stenosis. Am. J. Med. Genet. A. 2019;179:2454–2458. doi: 10.1002/ajmg.a.61355. [DOI] [PubMed] [Google Scholar]
- 98.Thues C., Valadas J.S., Deaulmerie L., Geens A., Chouhan A.K., Duran-Romaña R., et al. MAPRE2 mutations result in altered human cranial neural crest migration, underlying craniofacial malformations in CSC-KT syndrome. Sci. Rep. 2021;11:4976. doi: 10.1038/s41598-021-83771-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Edenberg H.J., Koller D.L., Xuei X., Wetherill L., McClintick J.N., Almasy L., et al. Genome-wide association study of alcohol dependence implicates a region on chromosome 11. Alcohol. Clin. Exp. Res. 2010;34:840–852. doi: 10.1111/j.1530-0277.2010.01156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mulligan M.K., Ponomarev I., Hitzemann R.J., Belknap J.K., Tabakoff B., Harris R.A., et al. Toward understanding the genetics of alcohol drinking through transcriptome meta-analysis. Proc. Natl. Acad. Sci. U. S. A. 2006;103:6368–6373. doi: 10.1073/pnas.0510188103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Goedert M., Jakes R. Expression of separate isoforms of human tau protein: Correlation with the tau pattern in brain and effects on tubulin polymerization. EMBO J. 1990;9:4225–4230. doi: 10.1002/j.1460-2075.1990.tb07870.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhang C.-C., Xing A., Tan M.-S., Tan L., Yu J.-T. The role of MAPT in neurodegenerative diseases: genetics, mechanisms and therapy. Mol. Neurobiol. 2016;53:4893–4904. doi: 10.1007/s12035-015-9415-8. [DOI] [PubMed] [Google Scholar]
- 103.Strang K.H., Golde T.E., Giasson B.I. MAPT mutations, tauopathy, and mechanisms of neurodegeneration. Lab. Invest. 2019;99:912–928. doi: 10.1038/s41374-019-0197-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Drechsel D.N., Hyman A.A., Cobb M.H., Kirschner M.W. Modulation of the dynamic instability of tubulin assembly by the microtubule-associated protein tau. Mol. Biol. Cell. 1992;3:1141–1154. doi: 10.1091/mbc.3.10.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Boyce J.J., Shea T.B. Phosphorylation events mediated by protein kinase Cα and ε participate in regulation of tau steady-state levels and generation of certain “Alzheimer-like” phospho-epitopes. Int. J. Dev. Neurosci. 1997;15:295–307. doi: 10.1016/s0736-5748(97)00010-5. [DOI] [PubMed] [Google Scholar]
- 106.Shea T.B., Ekinci F.J. In: Molecular and Cellular Mechanisms of Neuronal Plasticity: Basic and Clinical Implications. Ehrlich Y.H., editor. Springer US; Boston, MA: 1998. Influence of phospholipids and sequential kinase activities on tau in Vitro. (181–201). [DOI] [PubMed] [Google Scholar]
- 107.Taniguchi T., Kawamata T., Mukai H., Hasegawa H., Isagawa T., Yasuda M., et al. Phosphorylation of tau is regulated by PKN. J. Biol. Chem. 2001;276:10025–10031. doi: 10.1074/jbc.M007427200. [DOI] [PubMed] [Google Scholar]
- 108.Kapoor M., Chao M.J., Johnson E.C., Novikova G., Lai D., Meyers J.L., et al. Multi-omics integration analysis identifies novel genes for alcoholism with potential overlap with neurodegenerative diseases. Nat. Commun. 2021;12:5071. doi: 10.1038/s41467-021-25392-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hoffman J.L., Faccidomo S., Kim M., Taylor S.M., Agoglia A.E., May A.M., et al. In: International Review of Neurobiology. Deak T., Savage L.M., editors. Academic Press; 2019. Chapter Five - alcohol drinking exacerbates neural and behavioral pathology in the 3xTg-AD mouse model of Alzheimer’s disease; pp. 169–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Peng B., Yang Q., B Joshi R., Liu Y., Akbar M., Song B.-J., et al. Role of alcohol drinking in Alzheimer’s Disease, Parkinson’s Disease, and Amyotrophic Lateral Sclerosis. Int. J. Mol. Sci. 2020;21:2316. doi: 10.3390/ijms21072316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Murphy D.B., Vallee R.B., Borisy G.G. Identity and polymerization-stimulatory activity of the nontubulin proteins associated with microtubules. Biochemistry. 1977;16:2598–2605. doi: 10.1021/bi00631a004. [DOI] [PubMed] [Google Scholar]
- 112.Vallee R.B., Borisy G.G. Removal of the projections from cytoplasmic microtubules in vitro by digestion with trypsin. J. Biol. Chem. 1977;252:377–382. [PubMed] [Google Scholar]
- 113.Gamblin T.C., Nachmanoff K., Halpain S., Williams R.C. Recombinant microtubule-associated protein 2c reduces the dynamic instability of individual microtubules. Biochemistry. 1996;35:12576–12586. doi: 10.1021/bi961135d. [DOI] [PubMed] [Google Scholar]
- 114.Chen J., Kanai Y., Cowan N.J., Hirokawa N. Projection domains of MAP2 and tau determine spacings between microtubules in dendrites and axons. Nature. 1992;360:674–677. doi: 10.1038/360674a0. [DOI] [PubMed] [Google Scholar]
- 115.Cunningham C.C., Leclerc N., Flanagan L.A., Lu M., Janmey P.A., Kosik K.S. Microtubule-associated protein 2c reorganizes both microtubules and microfilaments into distinct cytological structures in an actin-binding protein-280–deficient melanoma cell line. J. Cell Biol. 1997;136:845–857. doi: 10.1083/jcb.136.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ramkumar A., Jong B.Y., Ori-McKenney K.M. ReMAPping the microtubule landscape: how phosphorylation dictates the activities of microtubule-associated proteins. Dev. Dyn. 2018;247:138–155. doi: 10.1002/dvdy.24599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rubino H.M., Dammerman M., Shafit-Zagardo B., Erlichman J. Localization and characterization of the binding site for the regulatory subunit of type II cAMP-dependent protein kinase on MAP2. Neuron. 1989;3:631–638. doi: 10.1016/0896-6273(89)90273-0. [DOI] [PubMed] [Google Scholar]
- 118.Itoh T.J., Hisanaga S., Hosoi T., Kishimoto T., Hotani H. Phosphorylation states of microtubule-associated protein 2 (MAP2) determine the regulatory role of MAP2 in microtubule dynamics. Biochemistry. 1997;36:12574–12582. doi: 10.1021/bi962606z. [DOI] [PubMed] [Google Scholar]
- 119.Ainsztein A.M., Purich D.L. Stimulation of tubulin polymerization by MAP-2. Control by protein kinase C-mediated phosphorylation at specific sites in the microtubule-binding region. J. Biol. Chem. 1994;269:28465–28471. [PubMed] [Google Scholar]
- 120.Illenberger S., Drewes G., Trinczek B., Biernat J., Meyer H.E., Olmsted J.B., et al. Phosphorylation of microtubule-associated proteins MAP2 and map4 by the protein kinase p110mark: Phosphorylation sites and regulation of microtubule dynamics. J. Biol. Chem. 1996;271:10834–10843. doi: 10.1074/jbc.271.18.10834. [DOI] [PubMed] [Google Scholar]
- 121.Drewes G., Ebneth A., Preuss U., Mandelkow E.-M., Mandelkow E. MARK, a novel family of protein kinases that phosphorylate microtubule-associated proteins and trigger microtubule disruption. Cell. 1997;89:297–308. doi: 10.1016/s0092-8674(00)80208-1. [DOI] [PubMed] [Google Scholar]
- 122.Ebneth A., Drewes G., Mandelkow E.-M., Mandelkow E. Phosphorylation of MAP2c and MAP4 by MARK kinases leads to the destabilization of microtubules in cells. Cell Motil. 1999;44:209–224. doi: 10.1002/(SICI)1097-0169(199911)44:3<209::AID-CM6>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 123.Ozer R.S., Halpain S. Phosphorylation-dependent localization of microtubule-associated protein MAP2c to the actin cytoskeleton. Mol. Biol. Cell. 2000;11:3573–3587. doi: 10.1091/mbc.11.10.3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Putzke J., De Beun R., Schreiber R., De Vry J., Tölle T.R., Zieglgänsberger W., et al. Long-term alcohol self-administration and alcohol withdrawal differentially modulate microtubule-associated protein 2 (MAP2) gene expression in the rat brain. Mol. Brain Res. 1998;62:196–205. doi: 10.1016/s0169-328x(98)00253-8. [DOI] [PubMed] [Google Scholar]
- 125.Ferguson L.B., Zhang L., Kircher D., Wang S., Mayfield R.D., Crabbe J.C., et al. Dissecting brain networks underlying alcohol binge drinking using a systems genomics approach. Mol. Neurobiol. 2019;56:2791–2810. doi: 10.1007/s12035-018-1252-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Feng Y., Walsh C.A. Protein–Protein interactions, cytoskeletal regulation and neuronal migration. Nat. Rev. Neurosci. 2001;2:408–416. doi: 10.1038/35077559. [DOI] [PubMed] [Google Scholar]
- 127.Guzik B.W., Goldstein L.S. Microtubule-dependent transport in neurons: steps towards an understanding of regulation, function and dysfunction. Curr. Opin. Cell Biol. 2004;16:443–450. doi: 10.1016/j.ceb.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 128.Romaniello R., Arrigoni F., Fry A.E., Bassi M.T., Rees M.I., Borgatti R., et al. Tubulin genes and malformations of cortical development. Eur. J. Med. Genet. 2018;61:744–754. doi: 10.1016/j.ejmg.2018.07.012. [DOI] [PubMed] [Google Scholar]
- 129.Labisso W.L., Raulin A.-C., Nwidu L.L., Kocon A., Wayne D., Erdozain A.M., et al. The loss of α- and β-tubulin proteins are a pathological hallmark of chronic alcohol consumption and natural brain ageing. Brain Sci. 2018;8:175. doi: 10.3390/brainsci8090175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Enculescu C., Kerr E.D., Yeo K.Y.B., Schenk G., Fortes M.R.S., Schulz B.L. Proteomics reveals profound metabolic changes in the alcohol use disorder brain. ACS Chem. Neurosci. 2019;10:2364–2373. doi: 10.1021/acschemneuro.8b00660. [DOI] [PubMed] [Google Scholar]
- 131.Faccidomo S., Swaim K.S., Saunders B.L., Santanam T.S., Taylor S.M., Kim M., et al. Mining the nucleus accumbens proteome for novel targets of alcohol self-administration in male C57BL/6J mice. Psychopharmacology (Berl.) 2018;235:1681–1696. doi: 10.1007/s00213-018-4870-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Osterndorff-Kahanek E., Ponomarev I., Blednov Y.A., Harris R.A. Gene expression in brain and liver produced by three different regimens of alcohol consumption in mice: comparison with immune activation. PLoS One. 2013;8 doi: 10.1371/journal.pone.0059870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Baker P.R., Chalkley R.J. MS-viewer: a web-based spectral viewer for proteomics results. Mol. Cell. Proteomics. 2014;13:1392. doi: 10.1074/mcp.O113.037200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.The UniProt Consortium UniProt: the universal protein knowledgebase in 2021. Nucl. Acids Res. 2021;49:D480–D489. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that the main data supporting the findings of this study are available within the article and its Supplemental materials. Raw mass spectrometry data has been submitted to MassIVE (massive.ucsd.edu) with the identifier MSV000089659. Annotated spectra for all results can be viewed using MSViewer (133) at https://msviewer.ucsf.edu/prospector/cgi-bin/msform.cgi?form=msviewer using the following search keys:
NH620 ETD data: os9qgidggu
NH620 HCD data: z2slyneu87
NH623 ETD data: ccxvkteg0x
NH623 HCD data: zhb7mdgn08
NH625 ETD data: 38eph2azey
NH625 HCD data: dmolm7wdix