ABSTRACT
Whole-genome sequencing is becoming the gold standard for pathogen characterization and offers considerable advantages for understanding the evolution and dissemination of new determinants of antimicrobial resistance. Despite the benefits of whole-genome sequencing for pathogen characterization, implementation costs and lack of expertise may limit its use by public health laboratories. This article reviews the advantages of whole-genome sequencing for pathogen characterization and the current status of the use of whole-genome sequencing for antimicrobial resistance surveillance in Ecuador. A roadmap is suggested for including whole-genome sequencing for pathogen characterization based on the needs of the health reference institutions through alliances with Ecuadorian universities. Establishing a partnership between public health institutions and academia would be valuable for clinicians, policy-makers, and epidemiologists who could then take reasonable measures in those areas and establish a basis for adapting One Health strategies to tackle antimicrobial resistance in Ecuador.
Keywords: Environmental health surveillance; drug resistance, microbial; whole genome sequencing; Ecuador
RESUMEN
La secuenciación del genoma completo, que está pasando a ser el estándar de referencia para la caracterización de agentes patógenos, ofrece ventajas considerables para comprender la evolución y la diseminación de los nuevos determinantes de la resistencia a los antimicrobianos. Sin embargo, a pesar de los beneficios que genera, los costos de ejecución y la falta de experiencia pueden limitar su uso por parte de los laboratorios de salud pública. En este artículo se evalúan las ventajas de la secuenciación del genoma completo para la caracterización de agentes patógenos y el estado actual del uso de la secuenciación del genoma completo en la vigilancia de la resistencia a los antimicrobianos en Ecuador. Se propone una hoja de ruta para incluir la secuenciación del genoma completo para la caracterización de agentes patógenos según las necesidades de las instituciones de salud de referencia, lo que se haría por medio de alianzas con universidades ecuatorianas. Establecer una asociación entre las instituciones de salud pública y los círculos académicos sería sumamente valioso para los médicos, los responsables de las políticas y los epidemiólogos, que podrían adoptar medidas razonables en sus ámbitos y sentar una base para adaptar las estrategias de “Una salud” a fin de abordar la resistencia a los antimicrobianos en Ecuador.
Palabras clave: Vigilancia sanitaria ambiental, farmacorresistencia microbiana, secuenciación completa del genoma, Ecuador
RESUMO
O sequenciamento do genoma completo está se tornando o padrão ouro para a caracterização de patógenos e oferece vantagens consideráveis para a compreensão da evolução e disseminação de novos determinantes de resistência aos antimicrobianos. Apesar dos benefícios do sequenciamento do genoma completo para a caracterização de patógenos, os custos de implementação e a falta de especialização podem limitar seu uso pelos laboratórios de saúde pública. Este artigo analisa as vantagens do sequenciamento do genoma completo para a caracterização de patógenos e a situação atual do uso desta técnica para a vigilância da resistência aos antimicrobianos no Equador. Sugere-se um roteiro para incluir o sequenciamento de genomas completos para caracterização de patógenos com base nas necessidades das instituições de saúde de referência, por meio de alianças com universidades equatorianas. A criação de uma parceria entre instituições de saúde pública e entidades acadêmicas seria valiosa para clínicos, formuladores de políticas e epidemiologistas, que poderiam, assim, tomar medidas razoáveis nessas áreas e estabelecer uma base para adaptar estratégias de Saúde Única para combater a resistência aos antimicrobianos no Equador.
Palavras-chave: Vigilância sanitária ambiental, resistência microbiana a medicamentos, sequenciamento completo do genoma, Ecuador
In 2015, the World Health Organization (WHO) launched the Global Antimicrobial Resistance and Use Surveillance System (GLASS) to standardize antimicrobial resistance surveillance methods, including guidelines for the collection, analysis, interpretation, and sharing of antimicrobial resistance data on priority pathogens by countries at the clinical level (1). Between 2015 and 2021, 109 countries and territories joined this initiative. However, Ecuador has not yet participated (2).
The GLASS initiative provides advice and recommendations about how to use whole-genome sequencing approaches to complement the phenotypic characterization of priority pathogens. It is expected that this technique will lead to a better understanding of the transmission and inter-relatedness of microbial populations. Today, whole-genome sequencing is an integral part of the first- or second-line methods for surveillance of antimicrobial resistance in many European countries and the USA (3). However, implementing this technology into routine diagnosis and surveillance of pathogens in developing countries is challenging, constraints being implementation costs, infrastructure, capacity and expertise, and availability of reagents (4). Considering these points, the academic sector, including research institutions and universities, in low- and middle-income countries could play an important role in developing and adapting surveillance strategies to optimize the use of resources to fill knowledge gaps and, in this way, utilize the benefits of whole-genome sequencing for the surveillance of pathogens worldwide. This paper aims to review the advantages of whole-genome sequencing for pathogen characterization and the current situation on the use of whole-genome sequencing for antimicrobial resistance surveillance in Ecuador and suggests a roadmap for how to move forward with this technology.
WHOLE-GENOME SEQUENCING FOR PATHOGEN RESEARCH
The costs associated with whole-genome sequencing are decreasing rapidly, enabling broader and more affordable access to this new technology in most countries (5). Whole-genome sequencing is also becoming the gold standard for pathogen characterization at the research level. Based on a PubMed search with the term “Whole-genome sequencing of bacterial pathogens,” published papers have increased substantially from 93 in 2012 to 767 in 2021. In addition, the number of biosample bacterial sequences deposited at the National Center for Biotechnology Information database was 2 042 430 up to May 2022 (6). Among the many benefits of whole-genome sequencing, this technology characterizes bacteria by identifying the species, serotype, genotype, prophage, resistance and virulence genes, genome annotation, and phylogeny, all within a single laboratory workflow (7). Despite these advantages for identifying antimicrobial resistance genotypes, some enhancements must be developed to improve their use as a tool for clinical decision-making (8). Even though whole-genome sequencing is not a substitute for phenotypic methods for detecting antimicrobial resistance for public health purposes, the many advantages of this technology are crucial for understanding the evolution and dissemination of new determinants of antimicrobial resistance (9).
Nowadays, several bioinformatic tools that do not require strong programming skills are available. For antimicrobial resistance surveillance, the online services of the Center for Genomic Epidemiology allow the typing, phenotyping, and elucidation of phylogeny in an easy interface (10). Other bioinformatics platforms that have a large number of different tools and workflows for characterization of antimicrobial resistance linked to epidemiological surveillance have been developed by the Centre for Genomic Pathogen Surveillance of the United Kingdom of Great Britain and Northern Ireland (Pathogenwatch, Microreact, Data-flo, and Epicollect5) (11). This software allows the prediction of antimicrobial resistance genotypes, geospatial analysis of dissemination of antimicrobial resistance, evaluation of potential hotspots of resistance, and visualization of antimicrobial resistance patterns in specific geographical locations. Despite the advantages of whole-genome sequencing for pathogen characterization, several challenges associated with costs and lack of expertise may limit its use by public health laboratories (12).
ANTIMICROBIAL RESISTANCE SURVEILLANCE AND ONE HEALTH IN ECUADOR
In Ecuador, the surveillance of bacterial pathogens and antimicrobial resistance in the clinical context is mainly carried out by the Reference Laboratory of Antimicrobial Resistance of the National Institute of Public Health (Instituto Nacional de Investigación en Salud Pública), using both conventional and molecular techniques (13). This laboratory serves as a support for the country’s hospital network in the characterization of human pathogens. In addition, the laboratory collaborates with Ecuadorian universities by complementing specific analyses for characterization of antimicrobial resistance using both phenotypic and genotypic methods, which allows enhanced pathogen surveillance (14, 15). The laboratories of the Agency for Phytosanitary and Zoosanitary Regulation and Control (Agencia de Regulación y Control Fito y Zoosanitario AGROCALIDAD) are the reference laboratory in Ecuador for pathogen characterization in the animal sector. This institution analyzes and detects plant and veterinary pathogens using molecular and conventional techniques (16). The laboratories do not currently implement whole-genome sequencing as part of their surveillance procedures. In addition, no official analysis has been done using whole-genome sequencing by public authorities investigating antimicrobial resistance in the environment.
Implementing whole-genome sequencing techniques in Ecuador to characterize antimicrobial resistance in pathogens has been mainly the prerogative of the universities, often in collaboration with the National Institute of Public Health and foreign research institutions. For instance, the research group UNIETAR of the Central University of Ecuador maintains a close partnership with the New York State Department of Agriculture and Markets (USA) and McGill University (Canada). Together they have worked on the characterization by whole-genome sequencing (Illumina®) of zoonotic pathogens (Salmonella enterica serovar Infantis and Escherichia coli) from the poultry sector in northern Ecuador (17). This collaboration has also been extended to other universities. For example, the research group UTA-RAM-One Health at the Technical University of Ambato (central Ecuador) used whole-genome sequencing with Illumina® sequencing to analyze strains of S. Infantis and S. enterica serovar Kentucky circulating in the layer poultry farms in this geographical area (18). This group is also investigating the dissemination of the mobile colistin resistance gene (mcr-1) in E. coli from environmental sources in collaboration with the US Food and Drug Administration, the National Institute of Public Health, the Catalan Institute for Water Research (Spain), and the Structured Operational Research and Training Initiative (SORT-IT), again using Illumina® sequencing (19). Another institution that uses whole-genome sequencing to characterize E. coli and Salmonella from animal, human, food, and environmental sources is the University San Francisco of Quito, which uses Illumina® and Oxford Nanopore Sequencing. In addition, this institution works in collaboration with the University of Minnesota (USA) (20). The study results from these university collaborations have been primarily published in journals classified as Q1 according to the ranking Scimago Scopus, which includes the most important scientific journals worldwide.
The combined strategy for analyzing circulating clones focuses on epidemiological connections between the origin of multidrug-resistant strains and the occurrence of these isolates in different sources in Ecuador. For instance, research on bacteria that harbor mcr-1 has mainly focused on animal and environmental sources (21). Based on previous studies and the deposited sequences available at the National Center for Biotechnological Information database, a core genome multilocus sequence typing (cgMLST) analysis has been carried out, as shown by the minimum spanning tree method using the BacWGSTdb 2.0 repository (22). cgMLST phylogeny offers an accurate discriminatory capacity compared with other conventional phylogenetic methods. However, because of the limited number of isolates, it is currently not possible to find an epidemiological connection between the isolates given the complex diversity of the lineages, the scarcity of clinical isolates, and the bias about the origin of the samples, which are limited to Pichincha, Cotopaxi, and Tungurahua provinces (Figure 1). Comprehensive information about the characteristics of the samples is included in Table 1, including the antibiotic resistance genes detected in silico using BacWGSTdb 2.0.
FIGURE 1. Minimum spanning tree of the core genome multilocus sequence type (cgMLST) allelic profiles of mcr-1 positive Escherichia. coli isolates from Ecuador with whole-genome sequencing.
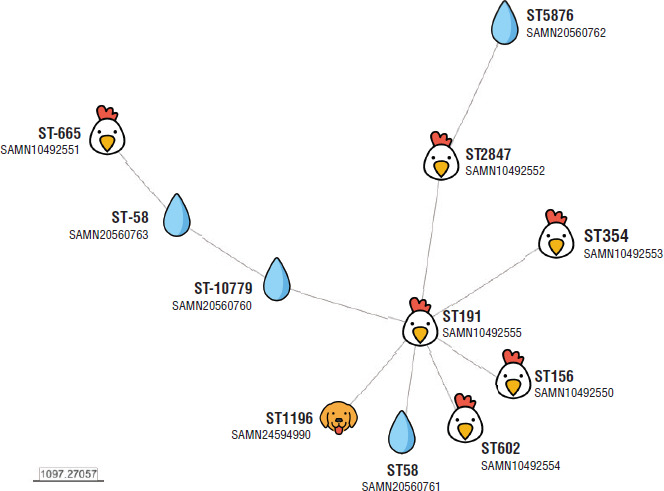
Notes: Data retrieved from at the National Center for Biotechnology Information database until June 2022.
The origin of the isolates (poultry, dog and environment) is represented using free icons (getemoji.com).
The multilocus sequence type (Warwick scheme) and the biosample number are shown.
Source: Prepared by authors based on information retrieved from the BacWGST 2.0 repository (22).
TABLE 1. Genomic and general characteristics of the Escherichia coli harboring mcr-1 gene whole-genome sequences from Ecuadorian samples.
|
ID NCBI |
Strain name |
MLST (classic Warwick) |
Clonal complex |
Sampling point |
Year |
Sample |
Antibiotic resistance genes detected (ResFinder database) |
|
|---|---|---|---|---|---|---|---|---|
|
Latitude |
Longitude |
|||||||
|
SAMN10492550 |
1CT 86A |
156 |
156 |
–0,18 |
–78,46 |
2014 |
Poultry |
aac(3)-VIa, aac(6′)-Ib, aadA1, aph(3″)-Ib, bla CTX-M-2 , bla TEM-1B , catA1, catB3, dfrA1, dfrA14, mcr-1.1, mdf(A), sul1, sul2, tet(B) |
|
SAMN10492551 |
1CT 109B |
665 |
None |
–0,18 |
–78,46 |
2014 |
Poultry |
aac(3)-VIa, aph(3″)-Ib, aph(6)-Id ,bla CTX-M-140 , bla TEM-1B , catA1, dfrA1, erm(B), mcr- 1.1, mdf(A), mph(B), sul1, sul2, tet(A) |
|
SAMN10492552 |
1CT 136A |
2847 |
None |
–0,18 |
–78,46 |
2014 |
Poultry |
aac(3)-IV, aadA1, aph(3″)-Ib, aph(4)-Ia, aph(6)-Id, bla CTX-M-65 , bla KPC-3 , dfrA17, floR, fosA3, mcr-1.1, mdf(A), sul2, tet(A) |
|
SAMN10492553 |
1CT 22A |
354 |
354 |
–0,18 |
–78,46 |
2014 |
Poultry |
aac(3)-IId, aph(3″)-Ib, aph(3′)-Ia, aph(6)-Id, bla CMY-2 , bla TEM-1B , dfrA17, floR, fosA, mcr-1.1, mdf(A), sul2 |
|
SAMN10492554 |
1CT 160A |
602 |
446 |
–0,18 |
–78,46 |
2014 |
Poultry |
aac(3)-IV, aph(4)-Ia, aph(6)-Id, bla CTX-M-65 , bla TEM-1B , catA1, dfrA17, floR, fosA3, fosA7, mcr-1.1, mdf(A), oqxA, oqxB, tet(B) |
|
SAMN10492555 |
1CT 188B |
191 |
None |
–0,18 |
–78,46 |
2014 |
Poultry |
aadA1, aadA2, bla CMY-2 , cmlA1, dfrA1, floR, mcr-1.1, mdf(A), qnrB19, sul3, tet(A) |
|
SAMN20560760 |
UTA20 |
10779 |
None |
–1,19 |
–78,60 |
2018 |
Environment |
aadA1, aadA2, bla CTX-M-55 , cmlA1, dfrA1, fosA3, mcr-1.1, mdf(A), sul3, tet(B) |
|
SAMN20560761 |
UTA30 |
58 |
155 |
–0,94 |
–78,61 |
2018 |
Environment |
aac(3)-IV, aph(3″)-Ib, aph(3′)-Ia, aph(4)-Ia, aph(6)-Id, bla CTX-M-65 , bla TEM-1B , dfrA5, floR, fosA3, mcr-1.1, mdf(A), qnrB19, sul2, tet(A) |
|
SAMN20560762 |
UTA144 |
5876 |
None |
–0,94 |
–78,61 |
2018 |
Environment |
aac(3)-IV, aph(3″)-Ib, aph(4)-Ia, aph(6)-Id, bla CTX-M-65 , bla TEM-1B , floR, mcr- 1.1, mdf(A), tet(B) |
|
SAMN20560763 |
UTA175 |
10 |
10 |
–0,95 |
–78,61 |
2018 |
Environment |
aac(3)-IIa, aadA1, aadA2, aph(3″)-Ib, aph(6)-Id, bla CTX-M-15 , bla TEM-1B , cmlA1, dfrA14, floR, mcr-1.1, mdf(A), qnrS1, sul2, sul3, tet(A), tet(M) |
|
SAMN24594990 |
UTA069 |
1196 |
1196 |
–1,27 |
–78,64 |
2018 |
Dog |
aadA1, aadA2, bla CTX-M-55 , cmlA1, dfrA12, fosA3, mcr-1.1, mdf(A), sul3, tet(A) |
MLST, multilocus sequence type.
Note: Available at the National Center for Biotechnology Information database until June 2022 (22).
Source: Prepared by authors based on information retrieved from the National Center for Biotechnology Information database (6) and BacWGST 2.0 repository (22).
WHOLE-GENOME SEQUENCING FOR PATHOGEN CHARACTERIZATION
Pathogen surveillance using whole-genome sequencing could enhance our understanding of the dissemination and evolution of antimicrobial resistance worldwide. This information would offer unprecedented opportunities to detect outbreaks, track microbial sources, detect reservoirs of antimicrobial resistance, and undertake clinical analysis. As noted above, some Ecuadorian universities are capable of performing whole-genome sequencing analysis, including both sequencing and bioinformatic analysis. Additionally, most universities and the antimicrobial resistance reference laboratories maintain solid collaborative networks between each other, as illustrated by their publication records. Although access to bioinformatics expertise and competence for routine whole-genome sequencing data analysis is still limited in Ecuador, the public entities (National Institute of Public Health, AGROCALIDAD, and Ministry of Environment) could enhance collaboration with the universities that possess the skills and expertise to complement the genomic characterization.
The intrinsic characteristics of the health reference institutions and the Ecuadorian universities make it necessary to define suitable frameworks for using whole-genome sequencing surveillance (Figure 2) and establish specific responsibilities for each institution. The National Planification for the Prevention and Control of Antimicrobial Resistance (2019–2023), headed by the Ministry of Health, needs to be updated and include collaboration with academia for the surveillance of the critical pathogens. Guidelines are needed to define the participation of the ministries of health, environment, and agriculture and representation from the universities. These guidelines can be further developed to map the current situation, outline the strengths and weaknesses of both public health institutions and universities, and agree on the strategies that would combine and complement their roles in surveillance of antimicrobial resistance.
FIGURE 2. Tasks to be undertaken for implementation of WGS surveillance of antimicrobial resistance in Ecuador.
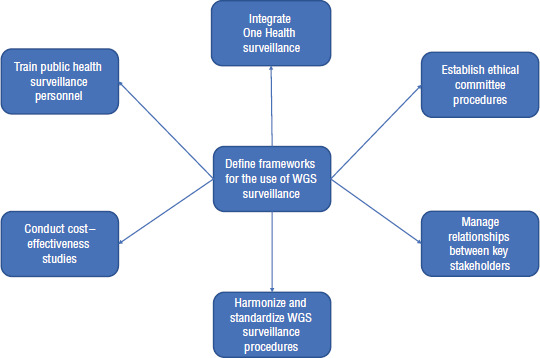
WGS, whole-genome sequencing.
Source: Prepared by the authors during the Structured Operational Research and Training Initiative (SORT-IT) course.
An excellent starting point could be the analysis of pathogens considered priority 1 by WHO (23) (carbapenem-resistant Acinetobacter baumanii, carbapenem-resistant Pseudomonas aeruginosa, nd carbapenem-resistant, extended spectrum beta-lactamase-producing Enterobacterales) and isolates that harbor emerging antibiotic resistance genes to last-resort antibiotics (mobile colistin resistance and mobile tigecycline resistance genes) detected in the hospital network of laboratories of the National Institute of Public Health. This information would be valuable for clinicians, policy-makers, and epidemiologists, who could then take reasonable measures in those areas and establish a basis for adapting One Health strategies to tackle antimicrobial resistance in Ecuador.
CONCLUSION
We hope that this opinion paper will encourage the public health authorities in Ecuador to work more closely with academia in order for this body to play an active role in the future National Planification for the Prevention and Control of Antimicrobial Resistance, based on their experience with genomic analysis and bioinformatics, and their involvement in the One Health approach. In addition, we outline a roadmap for including whole-genome sequencing in antimicrobial resistance surveillance in Ecuador that would allow the characterization of outbreaks, assessment of reservoirs, and dissemination routes of pathogens of critical interest.
Disclaimer.
The authors hold sole responsibility for the views expressed in the manuscript, which may not necessarily reflect the opinion or policy of the Revista Panamericana de Salud Pública / Pan American Journal of Public Health and/or those of the Pan American Health Organization.
Acknowledgements.
This article was developed through SORT-IT, a global partnership coordinated by the WHO Special Programme for Research and Training in Tropical Diseases (TDR), United Nations Children’s Fund, United Nations Development Programme, World Bank. The specific SORT IT program that led to this research article included an implementation partnership of: TDR and the Pan American Health Organization, and WHO country offices of Colombia and Ecuador; Ministry of Health and Social Protection, Colombia; Food and Agriculture Organization, Freetown, Sierra Leone; Sustainable Health Systems, Freetown, Sierra Leone; Tuberculosis Research and Prevention Center Non-Governmental Organization, Yerevan, Armenia; International Union Against Tuberculosis and Lung Diseases, Paris, France and South-East Asia offices, India; Institute of Tropical Medicine, Antwerp, Belgium; Damien Foundation, Brussels, Belgium; Indian Council of Medical Research, National Institute of Epidemiology, New Delhi, India; Jawaharlal Institute of Postgraduate Medical Education & Research, Pondicherry, India; GMERS Medical College Gotri, Vadodara, Gujarat, India; India Medical College Baroda, Gujarat, India; Sri Manakula Vinayagar Medical College, Madagadipet, India; Public Health, Ontario, Canada; Universidade Federal de Ciencias de Saude de Porto Alegre, Porto Alegre, Brazil; Universidade de Brasilia, Brasilia, Brazil; Universidad de Concepcion, Concepcion, Chile; Universidad de los Andes, Bogata, Colombia; Universidad Pontificia Bolivariana, Medellin, Colombia; Universidad Pedagógica y Tecnológica de Colombia, Tunja, Columbia; Universidad Central del Ecuador, Quito, Ecuador; California State University of Fullerton, Fullerton, USA; and Universidad Autónoma de Yucatán, Merida, Mexico.
Funding Statement
The SORT IT antimicrobial resistance programme is funded by the National Institute of Health Research, Department of Health & Social Care of the United Kingdom and supported by implementing partners.
Footnotes
Author contributions.
WC-C and ADH conceived the original idea. WC-C, NO-G, TS, C-AG-D, CB-C, MSR, and ADH collaborated in the protocol design. WC-C, ADH, NO-G, TS, C-AG-D, CB-C, and MSR obtained approval of the work from the Ethics Advisory Groups of PAHO and The Union. ADH and MSR supervised the entire work. WC-C wrote the manuscript. All the authors contributed to manuscript revision and editing, and read and approved the final version.
Funding.
The SORT IT antimicrobial resistance programme is funded by the National Institute of Health Research, Department of Health & Social Care of the United Kingdom and supported by implementing partners.
Conflicts of interest.
None declared.
REFERENCES
- 1.Global antimicrobial resistance surveillance system: manual for early implementation. Geneva: World Health Organization; 2015. [Google Scholar]; Global antimicrobial resistance surveillance system: manual for early implementation. Geneva: World Health Organization; 2015.
- 2.GLASS: the detection and reporting of colistin resistance. Second edition. Geneva: World Health Organization; 2021. [Google Scholar]; GLASS: the detection and reporting of colistin resistance. Second edition. Geneva: World Health Organization; 2021.
- 3.Stevens EL, Carleton HA, Beal J, Tillman GE, Lindsey RL, Lauer AC, et al. The use of whole-genome sequencing by the Federal Interagency Collaboration for Genomics for Food and Feed Safety in the United States. J Food Prot. 2022;85(5):755–772. doi: 10.4315/JFP-21-437. [DOI] [PubMed] [Google Scholar]; Stevens EL, Carleton HA, Beal J, Tillman GE, Lindsey RL, Lauer AC, et al. The use of whole-genome sequencing by the Federal Interagency Collaboration for Genomics for Food and Feed Safety in the United States. J Food Prot. 2022;85(5):755–72. 10.4315/JFP-21-437 [DOI] [PubMed]
- 4.Alleweldt F, Kara Ş, Best K, Aarestrup FM, Beer M, Bestebroer TM, et al. Economic evaluation of whole genome sequencing for pathogen identification and surveillance – results of case studies in Europe and the Americas 2016 to 2019. Euro Surveill. 2021;26(9):1900606. doi: 10.2807/1560-7917.ES.2021.26.9.1900606. [DOI] [PMC free article] [PubMed] [Google Scholar]; Alleweldt F, Kara Ş, Best K, Aarestrup FM, Beer M, Bestebroer TM, et al. Economic evaluation of whole genome sequencing for pathogen identification and surveillance – results of case studies in Europe and the Americas 2016 to 2019. Euro Surveill. 2021;26(9):1900606. 10.2807/1560-7917.ES.2021.26.9.1900606 [DOI] [PMC free article] [PubMed]
- 5.Quainoo S, Coolen JPM, van Hijum SAFT, Huynen MA, Melchers WJG, van Schaik W, et al. Whole-genome sequencing of bacterial pathogens: the future of nosocomial outbreak analysis. Clin Microbiol Rev. 2017;30(4):1015–1063. doi: 10.1128/CMR.00016-17. [DOI] [PMC free article] [PubMed] [Google Scholar]; Quainoo S, Coolen JPM, van Hijum SAFT, Huynen MA, Melchers WJG, van Schaik W, et al. Whole-genome sequencing of bacterial pathogens: the future of nosocomial outbreak analysis. Clin Microbiol Rev. 2017;30(4):1015–63. 10.1128/CMR.00016-17. [DOI] [PMC free article] [PubMed]
- 6.Genome. Bethesda: National Library of Medicine; 2022. [cited 2022 Jun 7]. Internet. Available from: https://www.ncbi.nlm.nih.gov/genome/ [Google Scholar]; Genome [Internet]. Bethesda: National Library of Medicine; 2022 [cited 2022 Jun 7]. Available from: https://www.ncbi.nlm.nih.gov/genome/
- 7.Kekre M, Arevalo SA, Valencia MF, Lagrada ML, Macaranas PKV, Nagaraj G, et al. Integrating scalable genome sequencing into microbiology laboratories for routine antimicrobial resistance surveillance. Clin Infect Dis. 2021;73(Suppl 4):S258–S266. doi: 10.1093/cid/ciab796. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kekre M, Arevalo SA, Valencia MF, Lagrada ML, Macaranas PKV, Nagaraj G, et al. Integrating scalable genome sequencing into microbiology laboratories for routine antimicrobial resistance surveillance. Clin Infect Dis. 2021;73(Suppl 4):S258–66. 10.1093/cid/ciab796 [DOI] [PMC free article] [PubMed]
- 8.Ellington MJ, Ekelund O, Aarestrup FM, Canton R, Doumith M, Giske C, et al. The role of whole genome sequencing in antimicrobial susceptibility testing of bacteria: report from the EUCAST Subcommittee. Clin Microbiol Infect. 2017;23(1):2–22. doi: 10.1016/j.cmi.2016.11.012. [DOI] [PubMed] [Google Scholar]; Ellington MJ, Ekelund O, Aarestrup FM, Canton R, Doumith M, Giske C, et al. The role of whole genome sequencing in antimicrobial susceptibility testing of bacteria: report from the EUCAST Subcommittee. Clin Microbiol Infect. 2017;23(1):2–22. 10.1016/j.cmi.2016.11.012 [DOI] [PubMed]
- 9.Doyle RM, O’Sullivan DM, Aller SD, Bruchmann S, Clark T, Pelegrin AC, et al. Discordant bioinformatic predictions of antimicrobial resistance from whole-genome sequencing data of bacterial isolates: an inter-laboratory study. Microb Genoms. 2020;6(2):e000335. doi: 10.1099/mgen.0.000335. [DOI] [PMC free article] [PubMed] [Google Scholar]; Doyle RM, O’Sullivan DM, Aller SD, Bruchmann S, Clark T, Pelegrin AC, et al. Discordant bioinformatic predictions of antimicrobial resistance from whole-genome sequencing data of bacterial isolates: an inter-laboratory study. Microb Genoms. 2020;6(2):e000335. 10.1099/mgen.0.000335 [DOI] [PMC free article] [PubMed]
- 10.Center for Genomic Epidemiology. Copenhagen: Technical University of Denmark; 2011. [cited 2022 Oct 14]. Internet. Available from: https://cge.food.dtu.dk/ [Google Scholar]; Center for Genomic Epidemiology [Internet]. Copenhagen: Technical University of Denmark; 2011 [cited 2022 Oct 14]. Available from: https://cge.food.dtu.dk/
- 11.Centre for Genomic Pathogen Surveillance. Hinxton: Our Software; 2015. [cited 2022 Oct 14]. Internet. Available from: https://www.pathogensurveillance.net/our-software/ [Google Scholar]; Centre for Genomic Pathogen Surveillance [Internet]. Hinxton; Our Software; 2015 [cited 2022 Oct 14]. Available from: https://www.pathogensurveillance.net/our-software/
- 12.Revez J, Espinosa L, Albiger B, Leitmeyer KC, Struelens MJ. Survey on the use of whole-genome sequencing for infectious diseases surveillance: rapid expansion of European national capacities, 2015–2016. Front Public Heal. 2017;5:347. doi: 10.3389/fpubh.2017.00347. [DOI] [PMC free article] [PubMed] [Google Scholar]; Revez J, Espinosa L, Albiger B, Leitmeyer KC, Struelens MJ. Survey on the use of whole-genome sequencing for infectious diseases surveillance: rapid expansion of European national capacities, 2015–2016. Front Public Heal. 2017;5:347. 10.3389/fpubh.2017.00347 [DOI] [PMC free article] [PubMed]
- 13.Instituto Nacional de Investigación en Salud Pública . Centro de Resistencia a los Antimicrobianos |CRN–RAMI. Quito: Instituto Nacional de Investigación en Salud Pública; 2022. [cited 2022 Jun 1]. Internet. Available from: http://www.investigacionsalud.gob.ec/webs/ram/ [Google Scholar]; Instituto Nacional de Investigación en Salud Pública. Centro de Resistencia a los Antimicrobianos |CRN–RAMI [Internet].Quito: Instituto Nacional de Investigación en Salud Pública 2022 [cited 2022 Jun 1]. Available from: http://www.investigacionsalud.gob.ec/webs/ram/
- 14.Albán MV, Tamayo R, Villavicencio FX, Núñez EJ, Zurita J, Sevillano G, et al. Canines with different pathologies as carriers of different lineages of Escherichia coli harboring mcr-1 and clinically relevant β-lactamases in central Ecuador. J Glob Antimicrob Resist. 2020;22:182–183. doi: 10.1016/j.jgar.2020.05.017. [DOI] [PubMed] [Google Scholar]; Albán MV, Tamayo R, Villavicencio FX, Núñez EJ, Zurita J, Sevillano G, et al. Canines with different pathologies as carriers of different lineages of Escherichia coli harboring mcr-1 and clinically relevant β-lactamases in central Ecuador. J Glob Antimicrob Resist. 2020;22:182–3. 10.1016/j.jgar.2020.05.017 [DOI] [PubMed]
- 15.Sánchez-Salazar E, Gudiño ME, Sevillano G, Zurita J, Guerrero-López R, Jaramillo K, et al. Antibiotic resistance of Salmonella strains from layer poultry farms in central Ecuador. J Appl Microbiol. 2020;128(5):1347–1354. doi: 10.1111/jam.14562. [DOI] [PubMed] [Google Scholar]; Sánchez-Salazar E, Gudiño ME, Sevillano G, Zurita J, Guerrero-López R, Jaramillo K, et al. Antibiotic resistance of Salmonella strains from layer poultry farms in central Ecuador. J Appl Microbiol. 2020;128(5):1347–54. 10.1111/jam.14562 [DOI] [PubMed]
- 16.Agrocalidad Ecuador . Dirección de Diagnóstico Animal. Quito: Ministry of Agriculture, Livestock, Aquaculture and Fisheries; 2022. [cited 2022 Jun 7]. Internet. Available from: https://www.agrocalidad.gob.ec/?page_id=38762. [Google Scholar]; Agrocalidad Ecuador. Dirección de Diagnóstico Animal [Internet]. Quito: Ministry of Agriculture, Livestock, Aquaculture and Fisheries; 2022 [cited 2022 Jun 7]. Available from: https://www.agrocalidad.gob.ec/?page_id=38762
- 17.Burnett E, Ishida M, de Janon S, Naushad S, Duceppe MO, Gao R, et al. Whole-genome sequencing reveals the presence of the blaCTX-M-65 gene in extended-spectrum β-lactamase-producing and multi-drug-resistant clones of Salmonella serovar Infantis isolated from broiler chicken environments in the Galapagos Islands. Antibiotics (Basel) 2021;10(3):267. doi: 10.3390/antibiotics10030267. [DOI] [PMC free article] [PubMed] [Google Scholar]; Burnett E, Ishida M, de Janon S, Naushad S, Duceppe MO, Gao R, et al. Whole-genome sequencing reveals the presence of the blaCTX-M-65 gene in extended-spectrum β-lactamase-producing and multi-drug-resistant clones of Salmonella serovar Infantis isolated from broiler chicken environments in the Galapagos Islands. Antibiotics (Basel). 2021;10(3):267. 10.3390/antibiotics10030267 [DOI] [PMC free article] [PubMed]
- 18.Calero-Cáceres W, Villacís J, Ishida M, Burnett E, Vinueza-Burgos C. Whole-genome sequencing of Salmonella enterica serovar Infantis and Kentucky isolates obtained from layer poultry farms in Ecuador. Microbiol Resour Announc. 2020;9(13):e00091–e00020. doi: 10.1128/MRA.00091-20. [DOI] [PMC free article] [PubMed] [Google Scholar]; Calero-Cáceres W, Villacís J, Ishida M, Burnett E, Vinueza-Burgos C. Whole-genome sequencing of Salmonella enterica serovar Infantis and Kentucky isolates obtained from layer poultry farms in Ecuador. Microbiol Resour Announc. 2020;9(13):e00091–20. 10.1128/MRA.00091-20 [DOI] [PMC free article] [PubMed]
- 19.Calero-Cáceres W, Tadesse D, Jaramillo K, Villavicencio X, Mero E, Lalaleo L, et al. Characterization of the genetic structure of mcr-1 gene among Escherichia coli isolates recovered from surface waters and sediments from Ecuador. Pt 2Sci Total Environ. 2022;806:150566. doi: 10.1016/j.scitotenv.2021.150566. [DOI] [PubMed] [Google Scholar]; Calero-Cáceres W, Tadesse D, Jaramillo K, Villavicencio X, Mero E, Lalaleo L, et al. Characterization of the genetic structure of mcr-1 gene among Escherichia coli isolates recovered from surface waters and sediments from Ecuador. Sci Total Environ. 2022;806(Pt 2):150566. 10.1016/j.scitotenv.2021.150566 [DOI] [PubMed]
- 20.Delgado-Blas JF, Ovejero CM, David S, Montero N, Calero-Caceres W, Garcillan-Barcia MP, et al. Population genomics and antimicrobial resistance dynamics of Escherichia coli in wastewater and river environments. Commun Biol. 2021;4(1):457. doi: 10.1038/s42003-021-01949-x. [DOI] [PMC free article] [PubMed] [Google Scholar]; Delgado-Blas JF, Ovejero CM, David S, Montero N, Calero-Caceres W, Garcillan-Barcia MP, et al. Population genomics and antimicrobial resistance dynamics of Escherichia coli in wastewater and river environments. Commun Biol. 2021;4(1):457. 10.1038/s42003-021-01949-x [DOI] [PMC free article] [PubMed]
- 21.Bastidas-Caldes C, Waard JH de, Salgado MS, Villacís MJ, Coral-Almeida M, Yamamoto Y, et al. Worldwide prevalence of mcr-mediated colistin-resistance Escherichia coli in isolates of clinical samples, healthy humans, and livestock-a systematic review and meta-analysis. Pathogens. 2022;11(6):659. doi: 10.3390/pathogens11060659. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bastidas-Caldes C, Waard JH de, Salgado MS, Villacís MJ, Coral-Almeida M, Yamamoto Y, et al. Worldwide prevalence of mcr-mediated colistin-resistance Escherichia coli in isolates of clinical samples, healthy humans, and livestock-a systematic review and meta-analysis. Pathogens. 2022;11(6):659. 10.3390/pathogens11060659 [DOI] [PMC free article] [PubMed]
- 22.Feng Y, Zou S, Chen H, Yu Y, Ruan Z. BacWGSTdb 2.0: a one-stop repository for bacterial whole-genome sequence typing and source tracking. Nucleic Acids Res. 2021;49(D1):D644–D650. doi: 10.1093/nar/gkaa821. [DOI] [PMC free article] [PubMed] [Google Scholar]; Feng Y, Zou S, Chen H, Yu Y, Ruan Z. BacWGSTdb 2.0: a one-stop repository for bacterial whole-genome sequence typing and source tracking. Nucleic Acids Res. 2021;49(D1):D644–D650. 10.1093/nar/gkaa821 [DOI] [PMC free article] [PubMed]
- 23.Prioritization of pathogens to guide discovery, research and development of new antibiotics for drug-resistant bacterial infections, including tuberculosis. World Health Organization; Geneva: World Health Organization; 2017. [cited 2022 Jun 7]. Available from: https://apps.who.int/iris/handle/10665/311820. [Google Scholar]; Prioritization of pathogens to guide discovery, research and development of new antibiotics for drug-resistant bacterial infections, including tuberculosis. World Health Organization. Geneva: World Health Organization; 2017 [cited 2022 Jun 7]. Available from: https://apps.who.int/iris/handle/10665/311820


