Abstract
Background
Metastatic cholangiocarcinoma (CC), a form of gastrointestinal cancer that originates from the bile ducts, cannot be cured by currently available therapies, and is associated with dismal prognosis. In a previous case report, adoptive transfer of autologous tumor infiltrating lymphocytes (TILs), the majority of which recognized a tumor-specific point mutation, led to a profound and durable cancer regression in a patient with metastatic CC. Thus, more effective treatment for patients with this disease may be developed by using TILs that target cancer-specific mutations, but also other genetic aberrations such as gene fusions. In this context, fusions that involve fibroblast growth factor receptor 2 (FGFR2) and function as oncogenes in a subset of patients with intrahepatic CC (ICC) represent particularly attractive targets for adoptive cell therapy. However, no study to date has explored whether FGFR2 fusions can be recognized by patients’ T cells.
Method
To address whether FGFR2 fusions can be recognized by patients’ T cells, we tested TILs from four patients with FGFR2 fusion-positive ICC for recognition of peptides and minigenes that represented the breakpoint regions of these fusions, which were unique to each of the four patients.
Results
We found that CD4+ TILs from one patient specifically recognized the breakpoint region of a unique FGFR2-TDRD1 (tudor domain-containing 1) fusion, and we isolated a T-cell receptor responsible for its recognition.
Conclusions
This finding suggests that FGFR2 fusion-reactive TILs can be isolated from some patients with metastatic ICC, and thus provides a rationale for future exploration of T cell-based therapy targeting FGFR2 fusions in patients with cancer. Furthermore, it augments the rationale for extending such efforts to other types of solid tumors hallmarked by oncogenic gene fusions.
Keywords: Translational Medical Research; Antigens; Gastrointestinal Neoplasms; Immunotherapy, Adoptive; Lymphocytes, Tumor-Infiltrating
WHAT IS ALREADY KNOWN ON THIS TOPIC
Some gene fusions are immunogenic, that is, they generate unique epitopes that are recognized by autologous T lymphocytes. The immunogenicity of fibroblast growth factor receptor 2 (FGFR2) fusions, which serve as oncogenes in a subset of patients with intrahepatic cholangiocarcinoma, has not yet been explored.
WHAT THIS STUDY ADDS
By finding that a patient with cholangiocarcinoma harbored both a unique FGFR2-TDRD1 (tudor domain-containing 1) fusion and tumor infiltrating T lymphocytes that recognized it in a specific manner, this study demonstrates that FGFR2 fusions can be immunogenic.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
This study establishes a screening protocol for isolating T cells (and T-cell receptors) that may recognize patient-specific FGFR2 fusions. This protocol could be implemented in future design of personalized adoptive T-cell therapy trials for patients with cholangiocarcinoma or other cancers that harbor FGFR2 fusions.
Introduction
Cholangiocarcinoma (CC) is a form of gastrointestinal cancer that originates from the epithelium of either intrahepatic or extrahepatic bile ducts. It accounts for approximately 3% of all gastrointestinal cancers, with reported incidence of one to two cases per 100,000 persons per year in the USA (and much higher incidence in areas with endemic fluke infestations of the biliary tract, for example, Southeast Asia).1 Most cases are diagnosed as locally advanced or metastatic disease, and thus cannot be cured by surgical resection. They are treated with systemic chemotherapy (eg, combination of gemcitabine and cisplatin), but the responses are modest.2 Thus, novel, more effective therapies for this disease are urgently needed.
Approximately 15% of cases of intrahepatic CC (ICC) harbor oncogenic (ie, driver) gene fusions involving the fibroblast growth factor receptor 2 (FGFR2) and one of many diverse partner genes.3–6 Chimeric FGFR2 proteins encoded by many of these fusions retain dimerization motifs contained in the fusion partners, which allows them to undergo spontaneous oligomerization and subsequent autophosphorylation of the FGFR2 kinase domains.3 This activates FGFR2 signaling pathway in a ligand-independent fashion, and ultimately leads to stimulation of cellular pathways that enhance cell proliferation and survival.
Targeting FGFR2 fusions with small molecule inhibitors can induce tumor regressions in up to 36% of patients with fusion-positive ICC who were refractory to systemic chemotherapy. Although this led to Food and Drug Administration-approval of two FGFR inhibitors for treatment of ICC,7 8 the responses to these agents are typically incomplete and short-lived, mainly because of the acquisition of mutations in the FGFR2 kinase domain that confer resistance to the administered drugs.5
Adoptive cell transfer (ACT) is a form of T cell-based immunotherapy that has been effective in treating select patients with metastatic melanoma and metastatic epithelial cancers.9 In one form of ACT, patients receive an infusion of autologous tumor infiltrating lymphocytes (TILs) that are isolated from autologous metastatic tumors and selected for their ability to recognize cancer neoantigens (ie, proteins that are encoded by genes harboring tumor-specific mutations). In another form of ACT, tumor antigen-reactive T-cell receptors (TCRs) are transduced into autologous T cells from the peripheral blood and then infused into the patient.10
Targeting cancer neoantigens resulting from point mutations with either TILs or TCRs was shown to mediate durable regression of metastatic solid tumors.11–15 In one of these studies, a patient with chemotherapy-refractory metastatic ICC was treated twice with autologous CD4+ TILs that targeted a neoantigen derived from a unique point mutation in the putative tumor suppressor ERBB2IP.12 The second treatment, in which approximately 95% of infused TILs recognized the mutant ERBB2IP (vs 25% in the first treatment), resulted in substantial regression of lung and liver metastases, which was augmented by a course of immune checkpoint inhibition, and has been ongoing for more than 72 months following the treatment.
Given this success, the use of ACT to target putative neoantigens arising from FGFR2 fusion breakpoint regions, which do not involve the FGFR2 kinase domain, represents a potential approach to treat patients with metastatic ICC, including those who develop resistance to FGFR inhibitors through acquisition of kinase domain mutations. However, although several studies have demonstrated that T cells from patients with cancer can recognize peptides derived from various gene fusions (eg, BCR-ABL in chronic myeloid leukemia,16 SYT-SSX1 in synovial cell sarcoma,17 MYB-NFIB in adenoid cystic carcinoma,18 or DNAJB1-PRKACA in fibrolamellar hepatocellular carcinoma19), no study to date has shown whether FGFR2 fusions can be recognized by patients’ T cells. Moreover, targeting gene fusions with ACT has remained unexplored.
To test whether T cells can recognize FGFR2 fusions, we focused on a cohort of 12 patients with ICC whose TILs were previously screened for recognition of tumor-specific point mutations under an ACT protocol (NCT01174121). Analysis of the tumor transcriptomes revealed that four of these patients harbored four different in-frame gene fusions involving FGFR2. We then tested whether TILs from these patients could recognize peptides and minigenes corresponding to patient-specific fusion breakpoints. In this study, we describe the isolation and characterization of an FGFR2 fusion-reactive TIL clone from one of these patients.
Materials and methods
Patients
Twelve patients with ICC were consecutively evaluated at the Surgery Branch of the National Cancer Institute (NCI) from 2015 to 2019 for participation in a cell therapy protocol (NCT01174121). All patients provided a written informed consent both for this protocol, and for the accompanying protocol that allowed for use of their samples for research purposes. The information about the patients and the outcomes of the initial neoantigen testing are summarized in online supplemental tables 1,2.
jitc-2022-006303supp008.pdf (241.4KB, pdf)
Whole exome and whole transcriptome sequencing
Genomic DNA and RNA from freshly resected tumors, as well as genomic DNA from the matched normal peripheral blood mononuclear cells (PBMCs), were first extracted using a Maxwell Instrument (Promega, Madison, Wisconsin, USA). Whole exome sequencing (WES) library preparation and identification of tumor-specific point mutations was performed as described previously.20 For whole transcriptome sequencing, complementary DNA (cDNA) sequencing libraries were prepared from total tumor RNA (2 µg per sample) using the Illumina TruSeq Stranded Total RNA Library Prep kit (Illumina, San Diego, California, USA), following the manufacturer’s protocol. Finally, the libraries were subjected to paired-end sequencing on a NextSeq 500 desktop sequencer (Illumina).
Identification of FGFR2 gene fusions from the whole transcriptome sequencing data
Sequencer-generated BCL files were converted to FASTQ files using the bcl2fastq tool in the CASAVA suite (Illumina). Sequencing quality of these samples was evaluated using Fastqc (V.0.11.9) (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/), with subsequent quality trim using Fastp (V.0.20).21 Quality-checked FASTQ files were then processed using FusionCatcher (V.1.20),22 Arriba (V.2.1.0)23 and STAR-fusion (V.1.6.0)24 pipelines, with hg38 human reference genome and default parameters. Results from the three tools were consolidated into a single preliminary table, which provided information such as breakpoint locations for each 5’ and 3’ fusion partner, and quantification of junction reads (reads overlapping each fusion breakpoint) and discordant read pairs (paired reads that mapped to two different partner genes, each on one side of the fusion breakpoint). Transcripts encoding predicted fusions were constructed using in-house designed scripts and by referring to hg38 human genome and GTF downloaded from GATK resource bundle (https://console.cloud.google.com/storage/browser/genomics-public-data/resources/broad/hg38/v0/). In addition to the gene annotation provided by the three tools, annoFuse R package25 was also used to annotate the gene fusions. Finally, the output data was used to extract information about the in-frame FGFR2 fusions, which is summarized in the online supplemental table 3.
Assessment of FGFR2 fusion and HLA gene expression
For FGFR2 fusion gene expression analysis, RNA-sequencing data was first aligned to the hg38 reference genome using STAR aligner.26 Next, soft-clipped and properly-paired reads aligned to the exons harboring the fusion breakpoints were extracted into two separate BAM files using the Samtools/1.15.1.27 Then, the average read depth per base position for the fused and wild type (WT) exons was calculated. These values were scaled to the total sequencing depth in each sample and then divided by the length of the exon to make them comparable between exons across samples. The final exon expression values were reported in Read Base Per KiloBase Per Million.
For the human leukocyte antigen (HLA) gene expression analysis, tumor RNA-sequencing data was analyzed using arcasHLA.28
Validation of predicted FGFR2 fusion sequences in tumor samples
Validation of FGFR2 fusions identified from the whole transcriptome was performed by fusion-specific RT-PCR and Sanger sequencing. First, cDNA synthesis was performed on 0.5 µg total tumor RNA using the SuperScript III First-Strand Synthesis System for RT-PCR (Thermo Fisher Scientific, Waltham, Massachusetts, USA), according to manufacturer’s instructions. Next, fusion-specific amplification primers, listed in the online supplemental table 4, were obtained from Integrated DNA Technologies (Coralville, Iowa, USA). All products were amplified using Hot Start Flex DNA Polymerase (New England Biolabs, Ipswich, Massachusetts, USA) and a touchdown PCR with a final annealing temperature of 61°C and 45 total cycles. A portion of each amplification reaction was subjected to automated gel electrophoresis, using the 4200 TapeStation System (Agilent Technologies, Santa Clara, California, USA). Finally, amplified products were purified using the QIAquick PCR Purification Kit (Qiagen, Gaithersburg, Maryland, USA) and sequenced using Sanger sequencing (Poochon Scientific, Frederick, Maryland, USA).
Generation of tumor infiltrating lymphocytes (TILs)
TILs were cultured from surgically resected tumors following a previously described approach.20 Briefly, tumor tissue was dissected free of hemorrhagic and necrotic areas and cut into approximately 1×1 mm fragments (up to N=24), which were then plated individually in 24-well plates and cultured in 2 mL of RPMI medium supplemented with 2 mM L-glutamine, 25 mM HEPES, 10 µg/mL gentamicin (all from Life Technologies, Carlsbad, California, USA), 10% in-house human AB serum and 6000 IU/mL of interleukin (IL)-2 (Prometheus, San Diego, California, UAA) for 6–8 weeks. Medium was replenished twice weekly; the wells were split in 1:2 fashion when fully confluent and cryopreserved until further use.
Generation of fusion minigenes and peptides
For each FGFR2 fusion, a minigene was designed by linking a nucleotide sequence encoding 150 amino acid-segment of FGFR2 protein that terminated at the fusion breakpoint with a sequence encoding 150 amino acid-segment of the partner protein that initiated at the fusion breakpoint. Control minigenes were designed to encode approximately 300 amino acids mapping to either FGFR2 or to its fusion partners. Control tandem minigenes encoding point mutations previously found to be recognized by TILs from these patients were designed as described previously.20 Next, all the constructs were codon-optimized, synthesized and ligated into a pcRNA2SL vector (GenScript, Piscataway, New Jersey, USA). The plasmids were then in vitro transcribed into RNA using mMESSAGE mMACHINE T7 ULTRA Transcription Kit (Thermo Fisher Scientific, Waltham, Massachusetts, USA). Finally, RNA samples were purified using RNeasy Mini Kit (Qiagen, Germantown, Maryland, USA), quantified by spectrophotometry, and stored at −80°C until further use.
Simultaneously, 26-mer peptides were designed to encode 13-mer amino acid sequence of FGFR2 that terminated upstream of the fusion breakpoint, followed by a 13-mer sequence of the partner protein that initiated downstream of the fusion breakpoint. The 25-mer peptides, each encoding a point mutation previously found to be recognized in TIL screens, were also designed. Next, all peptides were synthesized by GenScript (Piscataway, New Jersey, USA) as lyophilized powder, resuspended in DMSO, and stored at –20°C until use. The amino acid sequences of these peptides, as well as of each minigene used in this study, are shown in the online supplemental tables 3,5.
Generation of antigen presenting cells
Autologous dendritic cells (DCs) or Epstein-Barr virus (EBV)-transformed B cells were used as antigen presenting cells (APCs). DCs were generated from patient leukapheresis samples (ie, PBMCs) using an adherence method. First, PBMCs were plated in tissue culture flasks in AIM V CTS medium (Life Technologies, Carlsbad, California, USA) and incubated at 37°C for 2 hours. Next, following the removal of non-adherent cells, the flasks were washed twice with phosphate-buffered saline (PBS), and then incubated with RPMI medium supplemented with 10% fetal bovine serum (FBS; GE Healthcare Life Sciences, Logan, Utah, USA), 2 mM L-glutamine, 25 mM HEPES and Antibiotic-Antimycotic (all from Life Technologies, Carlsbad, California, USA), as well as with 50 ng/mL granulocyte-macrophage colony-stimulating factor (GM-CSF) and 20 ng/mL IL-4 (both from PeproTech, Rocky Hill, New Jersey, USA). On day 5 or 6, the adherent cells (ie, immature DCs) were harvested using a cell scraper.
To generate EBV-transformed B (EBV-B) cells, 1×108 PBMCs were cultured in 4 mL of complete RPMI medium supplemented with 10% FBS and Antibiotic-Antimycotic, with the addition of 1 mL of B95-8 culture supernatant containing the EBV (American Type Culture Collection, Manassas, Virginia, USA) and 0.5 mg/mL cyclosporine A (Sigma-Aldrich, St. Louis, Missouri, USA). Medium with cyclosporine A was replenished as needed, and the cultures were maintained for approximately 4–6 weeks. After the formation of round colonies (representing immortalized B cells) was observed, the cultures were further expanded in cyclosporine-free medium and cryopreserved until future use.
Assessment of T-cell responses to FGFR2 fusions
Recognition of FGFR2 fusions was assessed by performing overnight co-cultures of T cells and APCs that were either pulsed with relevant peptides or transfected with the relevant minigenes. This was followed by measuring interferon gamma (IFN-γ) production, as well as upregulation of activation marker 4-1BB (CD137) on the surface of T cells by flow cytometry.
For peptide testing, APCs were first incubated with FGFR2 or control peptides (at a final concentration of 10 µM, unless indicated otherwise) for 4–24 hours, and then washed twice prior to the co-culture. For minigene testing, APCs were transfected with minigene RNA using Lipofectamine MessengerMAX (Thermo Fisher Scientific, Waltham, Massachusetts, USA) according to the manufacturer’s instructions. Transfected or peptide-pulsed APCs were rested overnight in the same media that was used to generate these cultures.
Cryopreserved TILs (or TCR-transduced T cells, where applicable) were first rested for 24–48 hours in 50/50 medium supplemented with IL-2 (6000 IU/mL for TILs and 1200 IU/mL for TCR-transduced T cells) and 1 µg/mL DNAse I (STEMCELL Technologies, Vancouver, British Columbia, Canada). The 50/50 medium consisted of equal parts of AIM V CTS medium, supplemented with 5% human AB serum (Valley Biomedical, Winchester, Virginia, USA) and Antibiotic-Antimycotic, and RPMI medium supplemented with 2 mM L-glutamine, 25 mM HEPES, Antibiotic-Antimycotic and 10% human AB serum.
On the day of the co-culture, cells were washed twice to remove excess IL-2 and plated (2×104 cells/well for enzyme-linked immunosorbent assay (ELISpot) and 1×105 cells/well for other assays) with APCs (1×105 cells/well) onto either MultiScreen-IP filter plates (Millipore, Burlington, Massachusetts, USA), which were precoated overnight with IFN-γ detection antibody (isotype 1-D1K; Mabtech, Cincinnati, Ohio, USA) (for ELISpot), or onto regular U-bottom 96-well plates (for other assays). Co-cultures were then incubated overnight at 37°C and 5% CO2. Cell Stimulation Cocktail, a mixture of phorbol 12-myristate 13-acetate and ionomycin, (Affymetrix, San Diego, California, USA) was used as a positive control in 1:1000 v/v ratio.
Detection of IFN-γ and other cytokines produced by T cells
Secretion of IFN-γ from T cells was measured by either ELISpot or by IFN-γ electrochemiluminescence-based assay. For IFN-γ ELISpot, cell suspensions were first transferred from the MultiScreen-IP filter plates to separate U-bottom 96-well plates, which were set aside for flow cytometric analysis. MultiScreen-IP filter plates were then washed with 0.05% Tween 20 in PBS and incubated with a biotinylated detection antibody (Mabtech, Cincinnati, Ohio, USA) for 2 hours at room temperature. Next, the plates were washed again and incubated with streptavidin-ALP (Mabtech) for 1 hour. Then, they were washed with PBS and incubated with KPL BCIP/NBT substrate solution (SeraCare Life Sciences, Milford, Massachusetts, USA) for 10–15 min at room temperature. Finally, after the plates were washed with tap water and dried, they were scanned and analyzed using the ImmunoSpot plate reader (Cellular Technologies Limited, Shaker Heights, Ohio, USA). T cells from co-cultures that had at least 50 IFN-γ spots, with ≥2-fold increase over the controls, were deemed to have significantly increased IFN-γ production.
For IFN-γ electrochemiluminescence-based assay, supernatants from regular co-culture plates were assayed using the IFN-γ U-PLEX kit (Meso Scale Diagnostics, Rockville, Maryland, USA), according to the manufacturer’s instructions. The plates were read on MESO Sector S600 plate reader (Meso Scale Diagnostics) and analyzed using the accompanying software.
For analysis of other cytokines, supernatants from overnight TIL co-cultures were analyzed using a customized U-PLEX kit (also from Meso Scale Diagnostics), according to the manufacturer’s instructions. The assay was customized to allow simultaneous detection of IFN-γ, tumor necrosis factor alpha, IL-2, GM-CSF, and granzyme B.
Flow cytometry
For all experiments, the U-bottom plates with cell suspensions were washed twice using PBS with 0.5% FBS. The cell pellets were then stained with antibodies diluted in PBS/0.5% FBS in 1:50 V/V ratio at 4°C for 30 min. The following antibodies were used: CD3 (clone SK7), CD4 (clone SK3), CD8 (clone SK1) and CD137 (4-1BB, clone 4B4-1) (all from BD Biosciences, San Jose, California, USA). Flow cytometric analysis was performed on FACS Canto II cell analyzer (BD Biosciences). Cell sorting was performed using the SH800 sorter (Sony Biotechnology, San Jose, California, USA). Data was analyzed using FlowJo V.10.2 software (Tree Star, Ashland, Oregon, USA).
Identification and synthesis of TCRs
To identify TCR sequences responsible for fusion recognition, single-cell TCR sequencing was performed using the Takara SMARTer Human scTCR a/b Profiling Kit – 96 (Takara Bio USA, San Jose, California, USA), according to the manufacturer’s instructions. Briefly, single cells from TIL population that upregulated 4-1BB and downregulated CD3 in response to the relevant peptide were sorted into wells of a 96-well plate and subjected to cDNA synthesis and amplification using SMART technology, which included incorporation of cellular barcoding. Next, cDNA corresponding to TCRA and TCRB transcripts was further amplified and prepared for sequencing, which was performed on a MiSeq instrument with paired-end 2×300 bp reads using the MiSeq Reagent Kit v3 (600 cycle) (Illumina, San Diego, California, USA). Read extraction and clonality counts were determined by the MiXCR software package (V.2.1.12) (MiLaboratory, Russia).
To synthesize the identified TCR, TCRα/β constant regions were replaced with modified mouse TCRα/β constant regions to enhance TCR pairing and surface expression.29–31 TCRα and TCRβ chains were linked with a furin SGSG P2A linker, and then synthesized and cloned into an MSGV retroviral vector.32
Generation of TCR-transduced T cells
On day 1, HEK 293 cells expressing viral proteins GAG and POL (293GP cells) were plated (1×106/well) into poly-D-lysine-coated 6-well plates (Corning, Tewksbury, Massachusetts, USA). On day 2, each well was transfected with 2 µg pMSGV8-TCR and 1 µg of pRD114 using Lipofectamine 2000 Transfection Reagent (Life Technologies, Carlsbad, California, USA), following the manufacturer’s instructions. Simultaneously, ~1–3 x 108 healthy donor PBMCs, obtained via leukapheresis, were stimulated with 50 ng/mL anti-CD3 (clone OKT-3; eBioscience, San Diego, California, USA) and 1200 IU/mL of IL-2 in a tissue culture flask. On day 3, retrovirus-containing supernatants from 293GP cells were harvested and spinoculated onto non-tissue culture-treated 6-well plates coated with RetroNectin (Takara Bio USA, Mountain View, California, USA), at 2000×g for 2 hours at room temperature. Spinoculated plates were then seeded with donor PBMCs (2×106 cells/well), centrifuged at 1000×g for 10 min and incubated overnight at 37°C. Transduced cells were then cultured in complete 50/50 medium supplemented with 1200 IU/mL of IL-2 for additional 5 days. Next, transduction efficiency was assessed by staining the cells with an anti-mouse TCRβ chain antibody (clone H57-597; Invitrogen, Waltham, Massachusetts, USA) and performing flow cytometry. The cells were then cryopreserved until further use.
Determination of HLA restriction element and assessment of FGFR2-TDRD1 minigene recognition
Relevant HLA alleles identified from tumor WES were synthesized and cloned into pcDNA3.1 vector (GeneOracle, Santa Clara, California, USA). Next, COS-7 cells were plated in a flat-bottom 96-well plate (2.5×104 cells/well) and co-transfected the following day with combinations of plasmids encoding individual major histocompatibility complex (MHC) class II molecules (150 ng/well each) using Lipofectamine 2000 transfection reagent (0.5 µl/well). The next day, transfected cells were pulsed with FGFR2-TDRD1 26-mer peptide for 2 hours, washed twice in complete RPMI, and co-cultured overnight with TCR-transduced T cells (1×105/well) from two unrelated and non-HLA matched healthy donors. IFN-γ production from these cells was assessed by ELISpot, as described above.
To assess the recognition of the FGFR2-TDRD1 minigene, COS-7 cells (2.5×104 cells/well) were co-transfected (using Lipofectamine 2000 transfection reagent) in a flat-bottom 96-well plate with pcDNA3.1 plasmids encoding HLA-DRA1*01:01 and HLA-DRB1*04:04 (50 ng each per well) and a pcDNA3.1 plasmid encoding either the control TDRD1, control FGFR2 or the FGFR2-TDRD1 protein (100 ng per well). The following day, TCR-transduced T-cells were added; IFN-γ production was analyzed after the overnight co-culturing using IFN-γ U-PLEX, as described above.
Immunohistochemistry
Tumor sections were cut onto glass slides and allowed to dry overnight at room temperature. Immunohistochemistry (IHC) staining was performed using the Leica BOND RX automated stainer (Leica Biosystems, Buffalo Grove, Illinois, USA). Slides were deparaffinized and subjected to antigen retrieval (ER2, 20 min). The following antibodies were used: MHC-I (clone HC10, dil. 1:1000; in-house) and HLA-DR (clone TAL.1B5, dil. 1:200; Dako, Carpinteria, California, USA). Whole slide imaging of IHC slides was performed using a Hamamatsu NanoZoomer S60 Digital slide scanner C13210-01 (Hamamatsu, Bridgewater, New Jersey, USA).
Statistical analysis
Statistical analyses were performed on GraphPad Prism V.7.0 software (GraphPad Software, La Jolla, California, USA). When applicable, data were expressed as mean±SD.
Results
Identification of FGFR2 fusions in patients with ICC
The study cohort consisted of 12 patients with metastatic ICC whose tumors had been subjected to both whole exome and RNA sequencing and whose TILs were previously screened for recognition of tumor-specific point mutations. Patent and tumor characteristics, and the results of previous TIL screening for recognition of point mutations are summarized in the online supplemental tables 1 and 2, respectively.
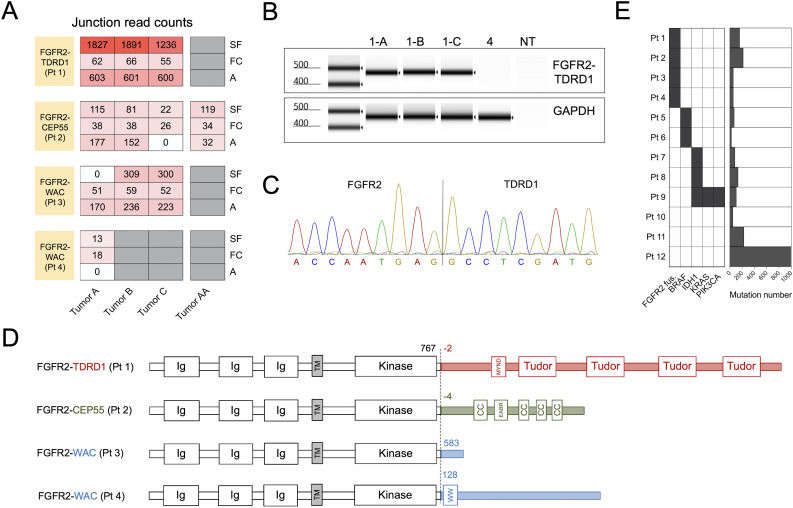
The analysis of tumor transcriptomes, performed using three different fusion prediction algorithms, revealed that tumors from 4 out of 12 patients harbored in-frame gene fusions involving FGFR2 as a 5’ partner (figure 1A and online supplemental table 3). The 3’ partners were tudor domain-containing 1 (TDRD1) in patient 1, centrosomal protein of 55 kDa (CEP55) in patient 2, and WW domain-containing adapter protein with coiled-coil (WAC) in patients 3 and 4. Transcripts that spanned the fusion breakpoints, termed ‘junction reads’, were detected by at least one fusion caller in all the tumors that were resected from each of these patients (figure 1A). Similar result was observed for paired reads that mapped to different sides of the fusion breakpoint, termed ‘discordant read pairs’ (online supplemental figure 1A). Among the four patients, the estimated transcript levels mapping to the FGFR2-TDRD1 breakpoint in patient 1 were significantly higher than the levels of other fusion breakpoints in patients 2–4 (online supplemental figure 1B).
Figure 1.
Tumors from four patients with metastatic intrahepatic cholangiocarcinoma (ICC) harbored in-frame FGFR2 gene fusions. (A) Patients’ metastases were surgically resected and subjected to whole transcriptome sequencing. Three different computational algorithms (‘A’, Arriba; ‘FC’, FusionCatcher; and ‘SF’, STAR-Fusion) were used to identify in-frame gene fusions involving the FGFR2. The heatmap depicts junction read counts (ie, reads spanning fusion breakpoints) reported by each fusion caller (rows) for all the tumor samples from four FGFR2 fusion-positive patients (columns). For each patient, all the tumors were resected at the same time, except for patient 2, who had a resection prior to the administration of adoptive T cell therapy targeting ERBBI2PE805G mutation (Tumor AA), and a repeat resection after the therapy (Tumors A–C). (B) RT-PCR was performed on tumor cDNA using primers that flanked the breakpoints of each FGFR2 fusion. Result of automated electrophoresis of the FGFR2-TDRD1 amplification reaction from patient 1’s tumors (1-A, 1-B and 1-C) is depicted. cDNA from patient 4’s tumor was used as a negative control, along with the reaction with no added template (NT). Primers for the wild type human GAPDH were used to generate a positive control. (C) Amplified products from (B) were purified and subjected to Sanger sequencing. The chromatogram depicts the sequencing result for tumor A from patient 1 (same sequence was detected in tumors B and C). (D) Breakpoint coordinates of each FGFR2 fusion transcript were used to reconstruct the sequences of FGFR2 fusion proteins. The dashed line indicates the fusion breakpoints; the numbers next to it indicate the amino acid position at which wild type FGFR2 protein terminates (black) or at which the partner protein starts (colored). The names of the protein domains are provided in the boxes. CC, coiled-coil domain; Ig, immunoglobulin-like domain; MYND (myeloid, Nervy, and DEAF-1)-type zinc finger domain; TM, transmembrane domain. Note that FGFR2 and partner proteins are not depicted on the same scale for clarity purposes. (E) The left panel depicts distribution of FGFR2 fusions and driver (hotspot) point mutations in the study cohort. The hotspot mutations include BRAFD594N (patient 5), BRAFV600E (patient 6), IDH1R132C (patients 7–9), and KRASG12V and PIK3CAH1047R (patient 9). The right panel depicts the overall number of tumor-specific mutations (including single nucleotide variants and insertions/deletions) in the same cohort. cDNA, complementary DNA; FGFR2, fibroblast growth factor receptor 2; TDRD1, tudor domain-containing 1.
jitc-2022-006303supp001.pdf (8.9MB, pdf)
To validate the results of the initial fusion calling, RT-PCR was performed on tumor cDNA using primers that flanked the breakpoints of each fusion (online supplemental table 4). RT-PCR with primers flanking the FGFR2-TDRD1 breakpoint amplified this fusion from all three tumors resected from patient 1 (figure 1B). Subsequent sequencing of the amplification product revealed a complete match to the sequence that was predicted from the whole transcriptome analysis (figure 1C). RT-PCR and subsequent amplification product sequencing performed on available RNA samples from patients 2 to 4 yielded the same results (online supplemental figure 1B–D).
The analysis of these fusion sequences revealed that FGFR2 portions of transcripts from all four fusions terminated at the identical breakpoint, which mapped to a sequence of the FGFR2 gene that encoded glutamic acid at position 767 (E767) in the C-terminal portion of the protein (figure 1D and online supplemental figure 2), as expected based on the previous reports.3 4 All the partner genes were located on the same chromosome as the FGFR2 gene (chromosome 10) but were on the opposite strand (online supplemental figure 2), indicating that these fusions were generated by intrachromosomal inversions. The 5’ ends of transcript portions that corresponded to TDRD1 (patient 1) and CEP55 genes (patient 2) mapped to the 5’ untranslated region of each gene. This indicated that the resulting fusion proteins contained additional two or four amino acids, respectively, before the start of the canonical sequence of each of the fusion partners (figure 1D). In contrast, transcript portions of WAC gene from patients 3 and 4 mapped to different regions within the WAC protein.
jitc-2022-006303supp002.pdf (9.8MB, pdf)
The analysis of tumor-specific point mutations in our 12-patient cohort (figure 1E and online supplemental figure 3) revealed no significant difference in the average tumor mutation burden between the FGFR2 fusion-positive and negative patients (124.8 vs 221.1 mutations, p=0.61), nor in the number of point mutations that were recognized by TILs in the previous screens (1.75 vs 1.17 neoantigens per patient, p=0.53). None of the driver point mutations known to occur in ICC were detected in fusion-positive patients (figure 1E), consistent with the previous reports.4 5
jitc-2022-006303supp003.pdf (1.4MB, pdf)
TILs from patient 1 specifically recognized a peptide spanning the FGFR2-TDRD1 fusion breakpoint
Patient 1, a 45-year-old female with chemotherapy-refractory metastatic ICC, underwent resection of three separate lung metastases at the NCI. The IHC analysis of these specimens revealed diffuse and strong MHC class I expression on tumor cells, while the MHC class II expression, which was also detectable by RNA sequencing, was restricted to larger infiltrating cells (resembling DCs or macrophages) and rare nests of tumor cells (online supplemental figure 4). In a previous study, TILs isolated from these tumors were screened for recognition of 152 tumor-specific mutations identified by the WES, which led to identification of a CD4+ TIL clone that specifically recognized a point mutation in the ras suppressor protein 1 gene (RSU1P150T) (online supplemental table 2).
jitc-2022-006303supp004.pdf (15MB, pdf)
To test whether TILs from patient 1 could recognize the FGFR2-TDRD1 fusion specific to this patients’ tumors, they were co-cultured with autologous DCs that were either pulsed with a 26-mer peptide or were transfected with the minigene representing the FGFR2-TDRD1 breakpoint region. Both peptide and minigene approaches were used to maximize the chance of capturing antigen responses from CD8+ TILs, which were previously found to occur more frequently in response to minigenes, and from CD4+ TILs, which were found to occur more frequently in response to mutant peptides.20 Both approaches relied on autologous DCs, which presumably shared MHC class I and II molecules with the tumor cells, thus circumventing the need to use MHC binding prediction algorithms, which are known to miss some bona fide cancer antigens.9
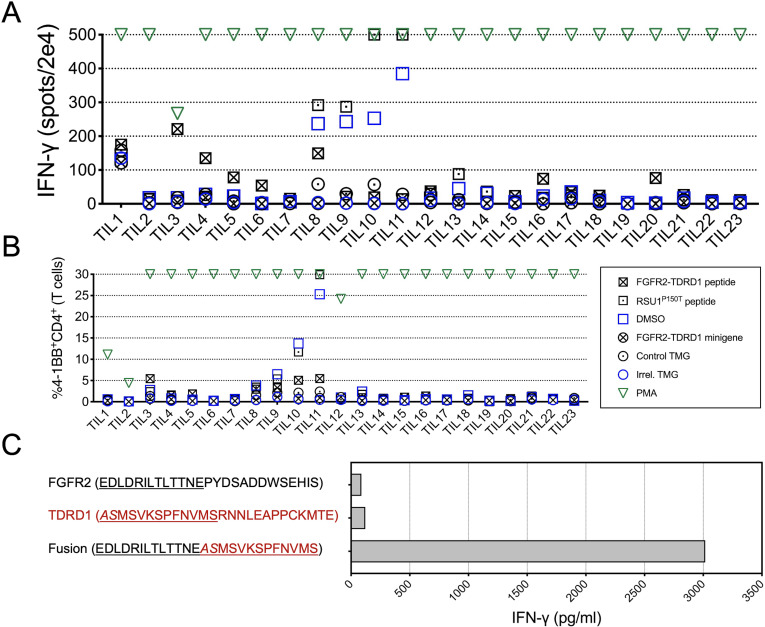
As indicated in figure 2A, several TIL cultures (No 3, 4, 5, 6, 16 and 20) exhibited significantly increased IFN-γ production in response to the FGFR2-TDRD1 peptide. Simultaneous flow cytometric analysis (figure 2B) revealed that CD4+ cells from TIL3 culture also upregulated the activation marker 4-1BB (CD137). Such upregulation was not detected in any other TIL cultures that exhibited increased IFN-γ production, suggesting that the fraction of fusion-reactive cells in these cultures was low. There was no significant 4-1BB upregulation on any of the CD8+ TILs (online supplemental figure 5A).
Figure 2.
TILs from patient 1 specifically recognized FGFR2-TDRD1 fusion. (A) TILs (n=23) were co-cultured with autologous DCs that were pulsed with FGFR2-TDRD1 peptide or transfected with FGFR2-TDRD1 minigene. Results of IFN-γ ELISpot are depicted. A tandem minigene (TMG) encoding several point mutations from another patient (Irrel. TMG) and DMSO were used as negative controls for minigene and peptide testing, respectively. A 25-mer peptide representing a point mutation in the RSU1 gene (RSU1P150T), previously identified as a neoantigen, as well as a TMG incorporating this mutation (Control TMG), were used as positive controls. PMA/ionomycin (PMA) was used as a non-specific positive control. (B) Same co-cultures as in (A) were analyzed by flow cytometry for expression of surface activation marker 4-1BB (CD137). The graph depicts the frequency of 41BB+CD4+ cells among all live T cells (a composite of CD8+ and CD4+ cells). (C) TIL3 cells were co-cultured with DCs pulsed with FGFR2-TDRD1 peptide or the control TDRD1 and FGFR2 peptides. IFN-γ production was measured the following day using an IFN-γ electrochemiluminescence assay. DCs, dendritic cells; DMSO, dimethylsulfoxide; ELISpot, enzyme-linked immunosorbent assay; FGFR2, fibroblast growth factor receptor 2; IFN-γ, interferon gamma;PMA, phorbol 12-myristate 13-acetate; TDRD1, tudor domain-containing 1; TIL, tumor infiltrating lymphocyte.
jitc-2022-006303supp005.pdf (2.6MB, pdf)
To determine whether FGFR2-TDRD1 fusion was recognized in a specific manner, TIL3 cells were co-cultured with DCs pulsed with the fusion peptide, as well as corresponding control FGFR2 and TDRD1 peptides. As indicated in figure 2C, only the fusion peptide elicited IFN-γ production from TIL3 cells. This response was also associated with increased production of IL-2 and GM-CSF, and a more modest increase in granzyme B (online supplemental figure 5B), indicating a cytokine secretion pattern mostly consistent with that of type 1 helper CD4+ T cells.
TILs from patients 2, 3 and 4 exhibited no significant IFN-γ production or 4-1BB upregulation following overnight co-cultures with DCs that were pulsed with peptides or transfected with minigenes corresponding to patient-specific FGFR2 fusions (online supplemental figure 6).
jitc-2022-006303supp006.pdf (5.4MB, pdf)
A TCR expressed by CD4+ TILs from patient 1 specifically recognized the FGFR2-TDRD1 fusion
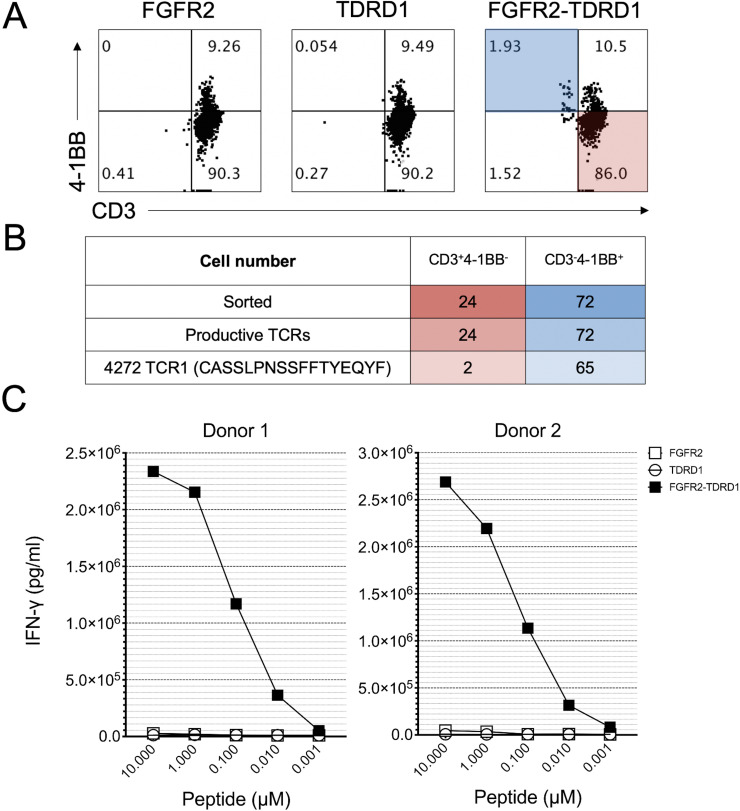
To isolate a TCR responsible for recognition of the FGFR2-TDRD1 fusion, TIL3 cells were co-cultured overnight with autologous DCs pulsed with either fusion or the control peptides. The following day, flow cytometric analysis revealed that approximately 2% of CD4+ T lymphocytes were activated (ie, exhibited simultaneous downregulation of CD3 and upregulation of 4-1BB) when co-cultured with the fusion peptide, in comparison to <0.1% cells in corresponding co-culture controls (figure 3A). These activated CD4+ T cells were then sorted and subjected to single-cell TCR sequencing, which identified a single TCR that was significantly enriched in activated versus non-activated cells (figure 3B).
Figure 3.
TCR derived from TIL3 cells specifically recognized FGFR2-TDRD1 fusion peptide. (A) TIL3 cells from patient 1 were co-cultured with autologous DCs that were pulsed with FGFR2-TDRD1 or FGFR2 and TDRD1 wild type peptides. Dot plots depict the flow cytometric analysis that was performed the following day. Data was gated on live CD4+ T cells. Cells in the shaded areas (CD3−4-1BB+ in blue and CD3+4-1BB− in red) were subjected to single-cell sorting and TCR sequencing. (B) Results of single-cell TCR sequencing. The table indicates the number of cells that were sorted from CD3−4-1BB+ and CD3+4-1BB− fractions (top row), along with the number of cells that exhibited any productive CDR3b sequence (middle row) or a CDR3b sequence corresponding to the TCR1 (bottom row). (C) PBMCs from two healthy donors were transduced with TCR1 and co-cultured with patient 1’s B cells that were pulsed with decreasing concentrations of FGFR2-TDRD1, or control FGFR2 and TDRD1 peptides. The following day, IFN-γ production was measured using IFN-γ electrochemiluminescence assay. Data represents average reads from duplicate co-culture wells; error bars represent SD. Calculated half maximal effective concentrations (EC50s) for IFN-γ production were 0.089 µM for Donor 1 and 0.189 µM for Donor 2. A representative of two independent experiments is shown; all experiments showed comparable EC50 values. DCs, dendritic cells; FGFR2, fibroblast growth factor receptor 2; IFN-γ, interferon gamma; PBMCs, peripheral blood mononuclear cells; TCRs, T-cell receptors; TDRD1, tudor domain-containing 1; TIL, tumor infiltrating lymphocyte.
To test the specificity and the affinity of this TCR, its sequence was cloned into an MSGV plasmid and transduced into PBMCs from two unrelated (non-HLA-matched) healthy donors with greater than 50% efficiency (online supplemental figure 7A). These TCR-transduced T cells were then co-cultured with patient 1’s B cells that were pulsed with either FGFR2-TDRD1 or the control peptides. The analysis of IFN-γ production performed on the following day demonstrated that TCR-transduced T cells from both donors recognized the fusion peptide in a dose-dependent manner, whereas only baseline reactivity was observed in response to control peptides (figure 3C).
jitc-2022-006303supp007.pdf (2.6MB, pdf)
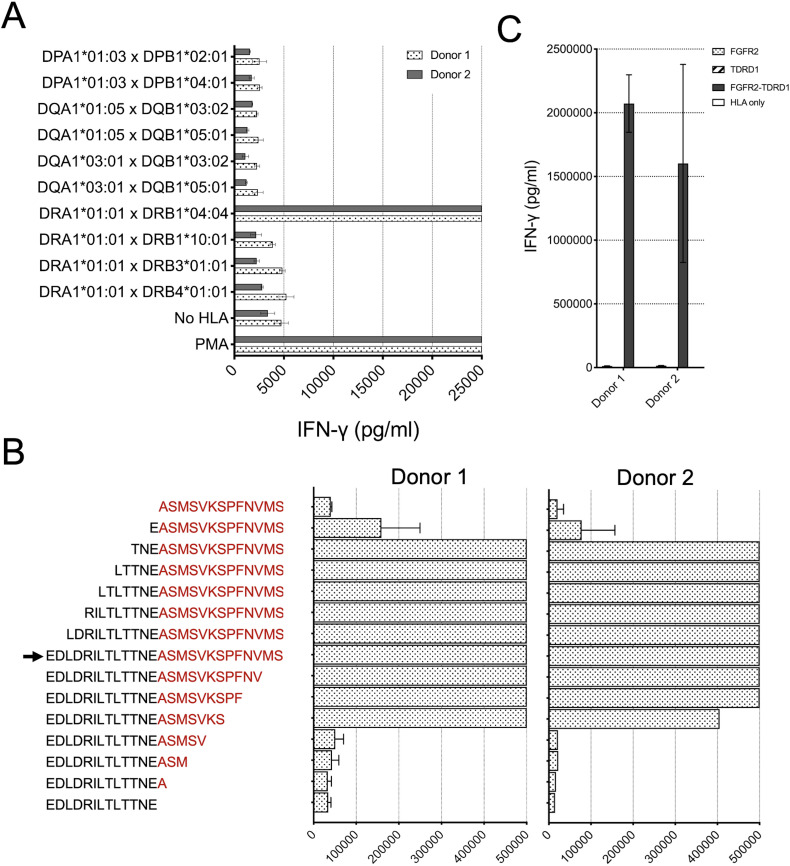
To determine the MHC restriction element for the fusion-reactive TCR, COS-7 cells were transfected with pairs of plasmids encoding all the MHC class II molecules identified by WES in the tumors from patient 1. Next, transfected cells were pulsed for 2 hours with the FGFR2-TDRD1 peptide, followed by an overnight co-culture with TCR-transduced T cells. As indicated in figure 4A, IFN-γ production was detected only when TCR-transduced T cells were co-cultured in presence of HLA-DRA1*01:01 and HLA-DRB1*04:04, thus identifying this MHC-II pair as the restriction element for the fusion-reactive TCR.
Figure 4.
TCR1 demonstrated HLA-DRB1*04:04-restricted recognition of FGFR2-TDRD1 peptide, which could be derived from the breakpoint region in the FGFR2-TDRD1 minigene. (A) TCR1-transduced T cells from two healthy donors were co-cultured with COS-7 cells, which were first transfected with pairs of plasmids encoding all MHC class II molecules detected in patient 1’s tumors, and then pulsed with the FGFR2-TDRD1 peptide. The following day, IFN-γ production was measured using an IFN-γ electrochemiluminescence assay. Mock-transfected COS-7 cells (No HLA) were used as a negative control; PMA/ionomycin (PMA) was used as a positive control. (B) Same TCR1-transduced T cells were co-cultured with patient 1’s B cells that were pulsed with peptides derived from the FGFR2-TDRD1 26-mer (arrowhead) by serial truncations of the FGFR2 (black) or TDRD1 portion of the fusion (red). IFN-γ production was measured the following day. (C) TCR1-transduced T cells were co-cultured with COS-7 cells that were transfected with plasmids encoding HLA-DRA1*01:01 and DRB1*04:04 molecules and a plasmid encoding either the breakpoint region of the FGFR2-TDRD1 protein, or the corresponding portions of normal FGFR2 or TDRD1 proteins. IFN-γ production was measured via electrochemiluminescence. COS-7 cells transfected only with the HLA plasmids were used as a negative control. For all experiments in this figure, bar graphs represent average reads from duplicate co-culture wells; error bars represent SD. FGFR2, fibroblast growth factor receptor 2; HLA, human leukocyte antigen; IFN-γ, interferon gamma; MHC, major histocompatibility complex; PMA, phorbol 12-myristate 13-acetate; TCR, T-cell receptor; TDRD1, tudor domain-containing 1.
To delineate the minimal epitope that mediated FGFR2-TDRD1 recognition, TCR-transduced T cells were co-cultured with patient 1’s B cells pulsed with serial two-amino acid truncations of fusion 26-mer peptide. As indicated in figure 4B, the loss of the second and third amino acids in the N-terminal direction of the fusion breakpoint, and the loss of sixth and seventh amino acid in the C-terminal direction of the breakpoint (TNEASMSVKS) resulted in significantly decreased IFN-γ production, suggesting that the TNEASMSVKS 10-mer peptide, or potentially one of the 9-mers contained in its sequence, was likely the minimal epitope required for fusion recognition. Interestingly, in a computational analysis using NetMHCIIpan-4.0,33 the best predicted HLA-DRB1*04:04 binder that overlapped with this minimal epitope had a predicted affinity rank of 63% among all the peptides that spanned the FGFR2-TDRD1 breakpoint and that were predicted to bind to any of the patient’s MHC-II molecules (online supplemental figure 7B). Thus, the epitope discovered in our study would have probably been missed if an MHC-II binding prediction algorithm were used to prioritize shorter peptides for TIL screening. This highlights the advantage of bypassing current MHC-II binding predictions by using longer, 25-mer peptides (and corresponding minigenes) presented by the autologous APCs for detection of MHC-II neoepitopes.
Finally, to assess whether cells bearing the FGFR2-TDRD1 gene can form a peptide presented on the cell surface MHC-II molecules, TCR-transduced T cells were co-cultured with COS-7 cells transfected with DNA expression plasmids encoding HLA-DRA1*01:01, HLA-DRB1*04:04, and either a 300-mer portion of the FGFR2-TDRD1 protein, centered around the fusion breakpoint, or corresponding FGFR2 and TDRD1 control proteins. Expression of the HLA and fusion molecules was confirmed by flow cytometry and by RT-PCR, respectively (online supplemental figure 7CD). As indicated in figure 4C, IFN-γ production was detected in presence of the fusion gene, but not the control genes, indicating that the fusion gene could indeed be transcribed, translated, and intracellularly processed into a peptide (epitope) recognized by T cells.
Discussion
In this study, we analyzed whole transcriptome data derived from tumors of 12 patients with chemotherapy-refractory metastatic ICC. We found that four of them harbored fusions involving FGFR2, each with a unique breakpoint sequence. We then tested their TILs for recognition of peptides and minigenes corresponding to breakpoint regions of these fusions. We found that CD4+ TILs from patient 1 specifically recognized a peptide spanning the breakpoint of the FGFR2-TDRD1 protein. Furthermore, we isolated an HLA-DRB1*04:04-restricted TCR from the reactive TILs and confirmed that it specifically recognized the fusion, but not the control FGFR2 or TDRD1 peptides or minigenes.
More than 100 FGFR2 fusions with different partner genes have been reported in patients with ICC.34 Although some of them are shared among patients (eg, FGFR2-BICC, which can be detected in approximately one-third of fusion-positive ICC cases), most fusions have unique partner genes or exhibit unique breakpoint sequences, and so require a personalized sequencing-based approach for detection.5 8 Consistently, FGFR2-TDRD1 and FGFR2-CEP55 fusions described here have not yet been reported in ICC but were reported in other cancers, although with different breakpoint sequences.35–37 Thus, the epitope encoded by the FGFR2-TDRD1 breakpoint appears unique to patient 1. Conversely, an FGFR2-WAC fusion seemingly matching the breakpoint detected in patient 4 (but not in patient 3) has been previously reported in metastatic ICC.38 39
All FGFR2 fusions reported here included partner genes that were located on the same chromosome as the FGFR2 (chromosome 10), but on the opposite DNA strand. This suggested that the mechanism of fusion formation was intrachromosomal inversion, which is consistent with a previous finding that more than 50% of FGFR2 fusions in ICC arise from intrachromosomal arrangements.5 All four fusion partners reported here were transcribed in the same reading frame as the FGFR2, and, except for WAC from patient 3, retained domains with known or presumed dimerization motifs (figure 1D).40–42 This is consistent with a proposed model ascribing the oncogenic potential of FGFR2 fusion proteins to their ability to self-aggregate and thus initiate the FGFR2 signaling cascade in a ligand-independent fashion.3
Our study provides evidence that TILs from ICC can specifically recognize an FGFR2 fusion and can be used to isolate a TCR that mediates this recognition. Due to its small size, it cannot be used to accurately determine the proportion of all patients with ICC who harbor FGFR2 fusion-reactive TILs, nor how many responses are mediated by CD8+ versus CD4+ cells. Rather, it provides a rationale and a blueprint to accomplish this by screening TILs from a larger, prospective cohort of patients with fusion-positive ICC.
The only recognized fusion in our study, FGFR2-TDRD1, exhibited significantly higher expression levels in the tumors from patient 1 than the non-immunogenic fusions in other patients. Although this suggested that the expression level may influence fusion recognition, as was previously demonstrated for point mutations in gastrointestinal cancers,20 the correlation between the FGFR2 fusion expression and immunogenicity could be established only by testing T cells from a larger number of patients with ICC. Expanding the screening efforts could also help determine whether current or prior FGFR inhibitor treatment can influence recognition of FGFR2 fusions, and thus inform the optimal timing for isolation of fusion-reactive T cells.
Our study could not address whether TILs or T cells transduced with the MHC class II-restricted, FGFR2-TDRD1-reactive TCR described here would be of clinical benefit if used for the ACT while patient 1 was still alive. Because of the lack of preserved tumor material, we could not test whether these T cells could recognize and destroy autologous tumor cells or patient-derived xenografts, nor whether the immunogenic peptide could be eluted from the tumor MHC-II molecules. Likewise, we could not test whether low MHC class II expression, which was demonstrated in the tumors of patient 1 by IHC, could be enhanced after the fusion-reactive T cells produced IFN-γ in response to encountering the immunogenic peptide on the surface of antigen presenting cells in the tumor.
Targeting of class II antigens by ACT has proven clinically efficacious in patients with several cancer types.12 43 44 However, unlike in the case of targeting MHC class I antigens, the question whether therapeutic success of class II antigen targeting requires direct recognition of tumor cells (vs recognition of tumor-infiltrating APCs) remains unresolved. For instance, studies that used murine tumor models found that adoptively transferred tumor-specific CD4+ T cells were able to completely eradicate established, MHC-II deficient tumors, and that tumor rejection was dependent on T cell-mediated activation of intratumoral macrophages.45 46 Thus, because the mechanism underlying successful MHC-II targeting in humans is not yet fully understood, the lack of evidence of autologous tumor-cell recognition by the TCR reported in this study would have not necessarily precluded an attempt to use it for patient treatment.
Because of the dismal prognosis associated with the ICC treated with currently available therapies, including the small molecule FGFR inhibitors, and because of the previous success of ACT targeting a point mutation in a patient with metastatic ICC, targeting FGFR2 fusions with T cells should be explored in an attempt to improve clinical outcomes for patients with this disease. Moreover, fusion-directed ACT may bypass toxicities associated with inhibition of vital cellular pathways such as the FGFR signaling cascade, which in some cellular contexts may exhibit tumor-suppressive properties.47
The personalized screening approach for discovery of fusion-reactive T cells described in this study could be used for future design of personalized clinical trials for patients with ICC or other cancers that harbor FGFR2 fusions.3 These trials, in which putative MHC class I or II-restricted TILs or TCR-transduced T cells recognizing different FGFR2 fusions would be infused to the patients, could be conducted similarly to (or even as a part of) the ongoing personalized ACT trials in which tumor-specific point mutations are targeted by TILs (NCT01174121) or TCRs (NCT04102436). Importantly, because most FGFR2 fusion breakpoints in cholangiocarcinoma are patient-specific, most of the discovered fusion-reactive TILs or TCRs would not be beneficial for treatment of other patients. This is exemplified by the discovery of the FGFR2-TDRD1-reactive TCR from patient 1, which targeted a fusion sequence that was not reported in other cases of ICC.
Several oncogenic (ie, driver) fusions have been identified in malignancies other than ICC, including the FGFR3-TACC fusions in glioblastoma, and fusions involving ALK and NTRK1 or NTRK2 in other types of solid cancers.48 Targeting these fusions with TILs and TCRs may be akin to targeting driver point mutations, which is considered beneficial for several reasons, including the high expression levels needed to sustain the malignant phenotype, as well as their relatively homogenous distribution among cancer cells.9 49 The benefit of targeting driver mutations is illustrated by clinical studies that demonstrated profound cancer regressions after targeting driver KRASG12D mutations in patients with metastatic colorectal and pancreatic cancer by either KRASG12D-reactive TILs14 and the TCR-transduced T cells,15 respectively.
The benefit of targeting driver fusions may be even more pronounced than targeting driver point mutations because the oncogenic fusions may occur in absence of other oncogenes.50 This increases tumor dependence on the fusion expression, and thus potentially minimizes the ability of the tumor to evade the immune attack through fusion loss or downregulation. Such dependency is illustrated by the outcomes of studies in which different oncogenic fusions were targeted with pharmacological therapies. For instance, complete and sustained cancer regressions were seen when targeting BCR-ABL fusion in patients with chronic myeloid leukemia with tyrosine kinase inhibitor imatinib,51 or when targeting EML4-ALK fusions in patients with lung adenocarcinoma with several ALK inhibitors.52 However, these therapies either do not exhibit curative potential, or are unavailable for most patients with fusion-driven solid cancers.
In conclusion, we demonstrated that an FGFR2-TDRD1 fusion from a patient with ICC elicited a TCR-mediated, MHC class II-restricted recognition by autologous TILs. This proof-of-principle study provides evidence that TILs from a patient with an aggressive metastatic cancer can specifically recognize a peptide derived from an FGFR2 fusion, thereby providing the further rationale for future studies of adoptively transferred, fusion-reactive T cells in the treatment of patients with fusion-driven solid tumors.
Acknowledgments
This work used the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov).
Footnotes
Contributors: BSW and SS: designed and conducted experiments, analyzed data and wrote manuscript. VH, BG, SN, JJG, TDP and YL: provided resources and analyses for the study and edited the manuscript. DG, PR and SAR: supervised research and edited the manuscript. VL: study guarantor; conceptualized the project, designed experiments, supervised research and wrote and edited the manuscript.
Funding: This study was funded by the Intramural Research Program (award number N/A) of the National Cancer Institute, National Institutes of Health, US Department of Health and Human Services.
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
All data relevant to the study are included in the article or uploaded as supplementary information.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
Patients were enrolled on a protocol approved by the NCI Institutional Review Board (reference #03C0277; NCT00068003). They had provided an informed consent for all subsequent analyses of their banked blood and tumor samples, which were performed in the current study. Participants gave informed consent to participate in the study before taking part.
References
- 1.Patel N, Benipal B. Incidence of cholangiocarcinoma in the USA from 2001 to 2015: A US cancer statistics analysis of 50 states. Cureus 2019;11:e3962. 10.7759/cureus.3962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kam AE, Masood A, Shroff RT. Current and emerging therapies for advanced biliary tract cancers. Lancet Gastroenterol Hepatol 2021;6:956–69. 10.1016/S2468-1253(21)00171-0 [DOI] [PubMed] [Google Scholar]
- 3.Wu Y-M, Su F, Kalyana-Sundaram S, et al. Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov 2013;3:636–47. 10.1158/2159-8290.CD-13-0050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arai Y, Totoki Y, Hosoda F, et al. Fibroblast growth factor receptor 2 tyrosine kinase fusions define a unique molecular subtype of cholangiocarcinoma. Hepatology 2014;59:1427–34. 10.1002/hep.26890 [DOI] [PubMed] [Google Scholar]
- 5.Silverman IM, Hollebecque A, Friboulet L, et al. Clinicogenomic analysis of fgfr2-rearranged cholangiocarcinoma identifies correlates of response and mechanisms of resistance to pemigatinib. Cancer Discov 2021;11:326–39. 10.1158/2159-8290.CD-20-0766 [DOI] [PubMed] [Google Scholar]
- 6.Lowery MA, Ptashkin R, Jordan E, et al. Comprehensive molecular profiling of intrahepatic and extrahepatic cholangiocarcinomas: potential targets for intervention. Clin Cancer Res 2018;24:4154–61. 10.1158/1078-0432.CCR-18-0078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abou-Alfa GK, Sahai V, Hollebecque A, et al. Pemigatinib for previously treated, locally advanced or metastatic cholangiocarcinoma: a multicentre, open-label, phase 2 study. Lancet Oncol 2020;21:671–84. 10.1016/S1470-2045(20)30109-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Javle MM, Roychowdhury S, Kelley RK, et al. Final results from a phase II study of infigratinib (BGJ398), an FGFR-selective tyrosine kinase inhibitor, in patients with previously treated advanced cholangiocarcinoma harboring an fgfr2 gene fusion or rearrangement. JCO 2021;39:265. 10.1200/JCO.2021.39.3_suppl.265 [DOI] [Google Scholar]
- 9.Leko V, Rosenberg SA. Identifying and targeting human tumor antigens for T cell-based immunotherapy of solid tumors. Cancer Cell 2020;38:454–72. 10.1016/j.ccell.2020.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosenberg SA, Restifo NP. Adoptive cell transfer as personalized immunotherapy for human cancer. Science 2015;348:62–8. 10.1126/science.aaa4967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levi ST, Copeland AR, Nah S, et al. Neoantigen identification and response to adoptive cell transfer in anti-PD-1 naïve and experienced patients with metastatic melanoma. Clin Cancer Res 2022;28:3042–52. 10.1158/1078-0432.CCR-21-4499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tran E, Turcotte S, Gros A, et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science 2014;344:641–5. 10.1126/science.1251102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zacharakis N, Chinnasamy H, Black M, et al. Immune recognition of somatic mutations leading to complete durable regression in metastatic breast cancer. Nat Med 2018;24:724–30. 10.1038/s41591-018-0040-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tran E, Robbins PF, Lu Y-C, et al. T-cell transfer therapy targeting mutant KRAS in cancer. N Engl J Med 2016;375:2255–62. 10.1056/NEJMoa1609279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leidner R, Sanjuan Silva N, Huang H, et al. Neoantigen T-cell receptor gene therapy in pancreatic cancer. N Engl J Med 2022;386:2112–9. 10.1056/NEJMoa2119662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yotnda P, Firat H, Garcia-Pons F, et al. Cytotoxic T cell response against the chimeric p210 BCR-ABL protein in patients with chronic myelogenous leukemia. J Clin Invest 1998;101:2290–6. 10.1172/JCI488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sato Y, Nabeta Y, Tsukahara T, et al. Detection and induction of ctls specific for SYT-SSX-derived peptides in HLA-A24(+) patients with synovial sarcoma. J Immunol 2002;169:1611–8. 10.4049/jimmunol.169.3.1611 [DOI] [PubMed] [Google Scholar]
- 18.Yang W, Lee K-W, Srivastava RM, et al. Immunogenic neoantigens derived from gene fusions stimulate T cell responses. Nat Med 2019;25:767–75. 10.1038/s41591-019-0434-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirk AM, Chou C-H, Crawford JC, et al. Abstract 1383: characterization of CD8 T cell responses to dnajb1-PRKACA fusion neoantigens in fibrolamellar carcinoma. Cancer Res 2022;82:1383. 10.1158/1538-7445.AM2022-1383 [DOI] [Google Scholar]
- 20.Parkhurst MR, Robbins PF, Tran E, et al. Unique neoantigens arise from somatic mutations in patients with gastrointestinal cancers. Cancer Discov 2019;9:1022–35. 10.1158/2159-8290.CD-18-1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen S, Zhou Y, Chen Y, et al. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018;34:i884–90. 10.1093/bioinformatics/bty560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nicorici D, Satalan M, Edgren H, et al. FusionCatcher - a tool for finding somatic fusion genes in paired-end RNA-sequencing data. Bioinformatics [Preprint]. 10.1101/011650 [DOI] [Google Scholar]
- 23.Uhrig S, Ellermann J, Walther T, et al. Accurate and efficient detection of gene fusions from RNA sequencing data. Genome Res 2021;31:448–60. 10.1101/gr.257246.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haas BJ, Dobin A, Stransky N, et al. STAR-fusion: fast and accurate fusion transcript detection from RNA-seq. Bioinformatics [Preprint]. 10.1101/120295 [DOI] [Google Scholar]
- 25.Gaonkar KS, Marini F, Rathi KS, et al. AnnoFuse: an R package to annotate, prioritize, and interactively explore putative oncogenic RNA fusions. BMC Bioinformatics 2020;21:577. 10.1186/s12859-020-03922-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, Handsaker B, Wysoker A, et al. The sequence alignment/map format and samtools. Bioinformatics 2009;25:2078–9. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orenbuch R, Filip I, Comito D, et al. ArcasHLA: high-resolution HLA typing from rnaseq. Bioinformatics 2020;36:33–40. 10.1093/bioinformatics/btz474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cohen CJ, Zhao Y, Zheng Z, et al. Enhanced antitumor activity of murine-human hybrid T-cell receptor (TCR) in human lymphocytes is associated with improved pairing and TCR/CD3 stability. Cancer Res 2006;66:8878–86. 10.1158/0008-5472.CAN-06-1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cohen CJ, Li YF, El-Gamil M, et al. Enhanced antitumor activity of T cells engineered to express T-cell receptors with a second disulfide bond. Cancer Res 2007;67:3898–903. 10.1158/0008-5472.CAN-06-3986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haga-Friedman A, Horovitz-Fried M, Cohen CJ. Incorporation of transmembrane hydrophobic mutations in the TCR enhance its surface expression and T cell functional avidity. J Immunol 2012;188:5538–46. 10.4049/jimmunol.1103020 [DOI] [PubMed] [Google Scholar]
- 32.Wargo JA, Robbins PF, Li Y, et al. Recognition of NY-ESO-1+ tumor cells by engineered lymphocytes is enhanced by improved vector design and epigenetic modulation of tumor antigen expression. Cancer Immunol Immunother 2009;58:383–94. 10.1007/s00262-008-0562-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reynisson B, Alvarez B, Paul S, et al. NetMHCpan-4.1 and netmhciipan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res 2020;48:W449–54. 10.1093/nar/gkaa379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Milind M J, Karthikeyan M, Rachna T S, et al. n.d. Profiling of 3,634 cholangiocarcinomas (CCA) to identify genomic alterations (GA), tumor mutational burden (TMB), and genomic loss of heterozygosity (gloh). J Clin Oncol;15_suppl:4087. 10.1200/JCO.2019.37.15_suppl.4087 [DOI] [Google Scholar]
- 35.Gu W, Yang J, Wang Y, et al. Comprehensive identification of fgfr1-4 alterations in 5 557 chinese patients with solid tumors by next-generation sequencing. Am J Cancer Res 2021;11:3893–906. [PMC free article] [PubMed] [Google Scholar]
- 36.Basturk O, Berger MF, Yamaguchi H, et al. Pancreatic intraductal tubulopapillary neoplasm is genetically distinct from intraductal papillary mucinous neoplasm and ductal adenocarcinoma. Mod Pathol 2017;30:1760–72. 10.1038/modpathol.2017.60 [DOI] [PubMed] [Google Scholar]
- 37.Meissner T, Amallraja A, Mark A, et al. Abstract P1-05-22: the value of RNA-seq for the detection of clinically actionable targets in breast cancer - A small cohort analysis. Cancer Res 2017;77:1–05. 10.1158/1538-7445.SABCS16-P1-05-22 [DOI] [Google Scholar]
- 38.Sarinya K, Apinya J, Jing Quan L, et al. Lack of targetable FGFR2 fusions in endemic fluke-associated cholangiocarcinoma. JCO Glob Oncol 2020;6:GO.20.00030. 10.1200/GO.20.00030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jusakul A, Cutcutache I, Yong CH, et al. Whole-genome and epigenomic landscapes of etiologically distinct subtypes of cholangiocarcinoma. Cancer Discov 2017;7:1116–35. 10.1158/2159-8290.CD-17-0368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chuma S, Hiyoshi M, Yamamoto A, et al. Mouse tudor repeat-1 (MTR-1) is a novel component of chromatoid bodies/nuages in male germ cells and forms a complex with snrnps. Mech Dev 2003;120:979–90. 10.1016/s0925-4773(03)00181-3 [DOI] [PubMed] [Google Scholar]
- 41.Zhao W, Seki A, Fang G. Cep55, a microtubule-bundling protein, associates with centralspindlin to control the midbody integrity and cell abscission during cytokinesis. Mol Biol Cell 2006;17:3881–96. 10.1091/mbc.e06-01-0015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu GM, Arnaout MA. WAC, a novel WW domain-containing adapter with a coiled-coil region, is colocalized with splicing factor SC35. Genomics 2002;79:87–94. 10.1006/geno.2001.6684 [DOI] [PubMed] [Google Scholar]
- 43.Lu Y-C, Parker LL, Lu T, et al. Treatment of patients with metastatic cancer using a major histocompatibility complex class II-restricted T-cell receptor targeting the cancer germline antigen MAGE-A3. J Clin Oncol 2017;35:3322–9. 10.1200/JCO.2017.74.5463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hunder NN, Wallen H, Cao J, et al. Treatment of metastatic melanoma with autologous CD4+ T cells against NY-ESO-1. N Engl J Med 2008;358:2698–703. 10.1056/NEJMoa0800251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haabeth OAW, Fauskanger M, Manzke M, et al. CD4+ T-cell-mediated rejection of MHC class II-positive tumor cells is dependent on antigen secretion and indirect presentation on host apcs. Cancer Res 2018;78:4573–85. 10.1158/0008-5472.CAN-17-2426 [DOI] [PubMed] [Google Scholar]
- 46.Muranski P, Restifo NP. Adoptive immunotherapy of cancer using CD4(+) T cells. Curr Opin Immunol 2009;21:200–8. 10.1016/j.coi.2009.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer 2010;10:116–29. 10.1038/nrc2780 [DOI] [PubMed] [Google Scholar]
- 48.Schram AM, Chang MT, Jonsson P, et al. Fusions in solid tumours: diagnostic strategies, targeted therapy, and acquired resistance. Nat Rev Clin Oncol 2017;14:735–48. 10.1038/nrclinonc.2017.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reiter JG, Makohon-Moore AP, Gerold JM, et al. Minimal functional driver gene heterogeneity among untreated metastases. Science 2018;361:1033–7. 10.1126/science.aat7171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao Q, Liang W-W, Foltz SM, et al. Driver fusions and their implications in the development and treatment of human cancers. Cell Rep 2018;23:227–38. 10.1016/j.celrep.2018.03.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hochhaus A, Larson RA, Guilhot F, et al. Long-term outcomes of imatinib treatment for chronic myeloid leukemia. N Engl J Med 2017;376:917–27. 10.1056/NEJMoa1609324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gerber DE, Minna JD. ALK inhibition for non-small cell lung cancer: from discovery to therapy in record time. Cancer Cell 2010;18:548–51. 10.1016/j.ccr.2010.11.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2022-006303supp008.pdf (241.4KB, pdf)
jitc-2022-006303supp001.pdf (8.9MB, pdf)
jitc-2022-006303supp002.pdf (9.8MB, pdf)
jitc-2022-006303supp003.pdf (1.4MB, pdf)
jitc-2022-006303supp004.pdf (15MB, pdf)
jitc-2022-006303supp005.pdf (2.6MB, pdf)
jitc-2022-006303supp006.pdf (5.4MB, pdf)
jitc-2022-006303supp007.pdf (2.6MB, pdf)
Data Availability Statement
All data relevant to the study are included in the article or uploaded as supplementary information.