Abstract
Aging impairs both circadian rhythms and memory, though the relationship between these impairments is not fully understood. Circadian rhythms are largely dictated by clock genes within the body’s central pacemaker, the suprachiasmatic nucleus (SCN), though these genes are also expressed in local clocks throughout the body. As circadian rhythms can directly affect memory performance, one possibility is that memory deficits observed with age are downstream of global circadian rhythm disruptions stemming from the SCN. Here, we demonstrate that expression of clock gene Period1 within a memory-relevant cortical structure, the retrosplenial cortex (RSC), is necessary for incidental learning, and that age-related disruption of Period1 within the RSC—but not necessarily the SCN—contributes to cognitive decline. These data expand the known functions of clock genes beyond maintaining circadian rhythms and suggests that age-associated changes in clock gene expression modulates circadian rhythms and memory performance in a brain region–dependent manner.
Keywords: retrosplenial cortex, aging, Per1, incidental learning, cognitive decline, suprachiasmatic nucleus
1. Introduction
Circadian rhythms regulate many physiological and behavioral processes, including the formation of memory (Davies et al., 1973; Eckel-Mahan et al., 2008). Aging notably impairs both circadian rhythms (Renfrew et al., 1987; Weitzman et al., 1982; Witting et al., 1990) and memory (Craik and McDowd, 1987; Youngjohn and Crook, 1993), though the molecular mechanisms underlying these impairments are not well understood, nor is the relationship between them fully characterized (e.g., if age-related memory impairments are caused by age-related circadian deficits).
In mammals, circadian rhythms are generated chiefly by the body’s central pacemaker, the suprachiasmatic nucleus (SCN), which is found at the base of the hypothalamus and synchronizes satellite oscillators throughout the body. Circadian oscillators, including the SCN, contain molecular clocks largely driven by a transcription-translation feedback loop (TTFL) with four critical components: genes Circadian Locomotor Output Cycles Kaput (Clock) and Brain and Muscle ARNT-like 1 (Bmal1) and gene families Period (Per) and Cryptochrome (Cry). In brief, CLOCK and BMAL heterodimerize and promote transcription of Cry and Per genes. After translation in the cytoplasm, CRY and PER proteins enter the nucleus and inhibit their own transcription, creating a negative feedback loop that lasts approximately 24 hours. The molecular clock within the SCN entrains other oscillators located throughout the rest of the brain and body, however it is worth noting that many of these peripheral oscillators cycle in the absence of SCN input (Yamazaki et al., 2000; Yoo et al., 2004), although many report that the hippocampus does not (Abe et al., 2002; Phan et al., 2011; but see also Chaudhury et al., 2005). Most of the existing research on clock genes focuses on their activity within the SCN, but these genes are also present across both the nervous system (Abe et al., 2002) and other tissues (Plautz et al., 1997). The function of clock genes outside of the TTFL is not yet fully characterized.
Recent reports have suggested that clock genes might operate in satellite brain regions (i.e., outside the SCN) to control local functions, including learning and memory (Snider et al., 2016; Woodruff et al., 2018; reviewed by Smies et al., 2022) and other behavior (McClung et al., 2005; Mukherjee et al., 2010; Spencer et al., 2013) in a brain region–dependent manner. Previous work has demonstrated that one such clock gene, Period1 (Per1) modulates the phosphorylation of cAMP response element binding protein (CREB) in the dorsal hippocampus (DH; Rawashdeh et al., 2016), suggesting a possible role for Per1 in hippocampal memory, given CREB’s known role as a memory modulator (Yin et al., 1994, 1995). We recently demonstrated that bidirectional, local manipulations of Per1 in the DH (Bellfy et al., 2022; Kwapis et al., 2018) and retrosplenial cortex (RSC; Urban et al., 2021) are sufficient to affect spatial memory. Although we reported that Per1 expression in the DH is impaired by aging (Kwapis et al., 2018), no one has yet investigated how Per1 expression in memory-relevant neocortical regions (e.g., the RSC) changes as a result of age and whether these changes are linked to memory performance.
The RSC (Brodmann areas 29 and 30) is a cortical brain structure integral to spatial memory and critically affected by age. This region—located immediately posterior to the corpus callosum in primates and directly dorsal to the DH in rodents—is densely connected with the DH and both the prefrontal and cingulate cortices (Shibata et al., 2004; Wang et al., 2016). Notably, expression of the immediate early gene (IEG) Fos is induced in the RSC following spatial learning (Vann et al., 2000), and lesions of the rodent RSC impair performance in both the Morris water maze (Harker and Whishaw, 2002) and radial arm maze (Vann and Aggleton, 2004). Further studies have suggested that RSC lesions specifically impair the ability to bind complex stimuli together, rather than the act of navigation itself (Nelson et al., 2015). Additionally, the RSC has been demonstrated to play an important role in both the retrieval (Corcoran et al., 2011) and formation (Kwapis et al., 2015; Urban et al., 2021) of contextual fear memory. Both the anterior RSC (aRSC) and the posterior RSC (pRSC) have been implicated in spatial memory, although these subregions may each support different types of information (see Discussion). Interestingly, both contextual fear memory (Moyer Jr. and Brown, 2006) and object location memory (Wimmer et al., 2012) are impaired as a result of aging, but RSC-independent modalities like delay fear conditioning (Trask et al., 2021a) and semantic memory (Wiggs et al., 1998) are resistant to the effects of aging (Craik and Grady, 2002; Moyer Jr. and Brown, 2006; reviewed by Trask and Fournier, 2022). Thus, the RSC is tightly linked to age-related memory deficits, and further investigation may elucidate the link between these memory deficits and age-related disruptions in circadian rhythms.
Here, we report that dysregulation of Per1 within the RSC contributes to age-related memory deficits. We find that although learning induces Per1 expression in the RSC of aging mice, this induction is smaller than in young mice. Additionally, this Per1 induction fluctuates across the day-night cycle in both young and aging animals, although aging induces minor disruptions of this pattern. Furthermore, local downregulation of RSC Per1 in young mice impairs the formation of spatial memory, while local upregulation of Per1 in the RSC of aging mice is sufficient to rescue memory formation. Finally, although learning induces gene expression in the SCN of young but not aging mice, locally downregulating Per1 in the SCN of young mice has no effect on spatial memory. Together, these data indicate that Per1 functions within the RSC to modulate memory, whereas Per1 in the SCN is dispensable for normal memory formation in young mice. Age-related repression of Per1 in the RSC may therefore contribute to age-related spatial memory deficits. Although we did not directly test the role of Per1 as a circadian regulator, these results further elucidate the relationships between clock genes, aging, and learning.
2. Methods
2.1. Subjects
A total of 233 Male (after the removal of outliers) C57BL/6J mice from either Jackson Laboratories or the NIA aging rodent colony (Charles River) were used for all analyses. Mice were housed in a temperature- and humidity-controlled environment with a 12 hr light/dark cycle (lights on at 6:00 AM EST or 7:00 AM DST; off at 6:00 PM EST or 7:00 PM DST) and had ad libitum access to food and water. We used the moment the lights came on as Zeitgeber time (ZT) 0. Thus, ZT5 is 5 hours after the lights came on, and ZT12 is when the lights turned off each evening. Behavioral experiments were conducted between ZT5 and ZT7 unless otherwise specified, as we have previously demonstrated that young mice demonstrate both strong memory for OLM and robust hippocampal Per1 inductions at this timepoint, while aging mice exhibit impairments in both processes at this time (Kwapis et al., 2018). Young mice were between 2 and 3 months of age at the start of experiments while aging mice were between 18 and 20 months of age at the start of experiments. In every experiment, mice were randomly assigned to groups (homecage/trained, Per1 sgRNA/ctrl sgRNA) and all extraneous conditions (objects, boxes, etc.) were appropriately counterbalanced. As we have previously detected an effect of sex on our Per1 manipulations within the RSC (Urban et al., 2021), we chose to only use male mice for this work. All experiments were approved by the Penn State Institutional Animal Care and Use Committee.
2.2. Surgery
Mice were anesthetized with 2% isoflurane (Patterson Veterinary, Greeley CO) dissolved in oxygen. Their heads were shaved and sterilized with betadine (Purdue Products) and 70% ethanol. Ophthalmic ointment (Dechra Veterinary Products, Portland, ME) was applied to the animals’ eyes to prevent desiccation. An incision was made between the eyes with a sterile surgical blade (Aspen Surgical Products, Caledonia, MT) and the skull was dried with 70% ethanol; then, holes were bored with a surgical drill (Foredom Electric Co., Bethel, CT). For the RSC, bilateral infusers connected to injector syringes (Hamilton Co., Reno, NV) were lowered at a rate of 0.2 mm/15 sec to a final depth of 0.75 mm below the skull and allowed to rest for 5 min. Then, 1.0 μL of viral cocktail (see below) was injected per hemisphere at 6 μL/hr. The injectors were allowed to rest for 5 minutes following injection, then were raised by 0.1 mm and allowed to rest for an additional 2 minutes. Finally, the injector was removed at a rate of 0.1mm/15 sec. For the SCN, viruses were infused with a 30-gauge Neuro Hamilton syringe directly mounted to a nanoinjector pump on the stereotaxic apparatus that was lowered at a rate of 0.3mm/15 sec to a final depth of 5.65 mm below the skull, where it was allowed to rest for 5 minutes. A bolus of 1.0 μL virus was injected at 6 μL/hr. Then, the needle rested for 5 minutes before being raised by 0.1mm and resting for an additional 2 minutes. Finally, the needle was removed at a rate of 0.2mm/15 sec and the other hemisphere was injected. Coordinates (from Bregma) were RSC (primarily targeting the anterior RSC): 1.80 mm caudal, 0.45 mm lateral, 0.75 mm ventral. SCN: 0.46 mm caudal, 0.22 mm lateral, 5.65 mm ventral.
2.3. Object Location Memory
OLM was conducted as previously described (Vogel-Ciernia and Wood, 2014). Briefly, mice were handled for 4 days and then habituated to polypropylene arenas (23.0 cm × 30.0 cm × 23.0 cm) for 6 days. Following the last day of habituation, mice underwent viral surgery and were allowed to rest for 72 hours to recover and promote peak HSV expression. After these 72 hours, mice were trained for 10 minutes with two identical objects. Twenty-four hours following training, mice underwent a test session in which one of the objects was moved. The objects were identical in appearance (100 mL beakers filled with hydraulic cement) and differed only in location. For the time course experiment investigating diurnal changes in Per1 expression, OLM was conducted under dim red light, to prevent confounding effects of light-induced expression of Per1. A discrimination index was calculated using the formula DI = (tn − tf) / (tn + tf) * 100%, where tn is the time each mouse spent investigating the object in the novel location and tf is the time each mouse spent investigating the object in the familiar location. Investigation was defined as any time the mouse spent with all 4 paws on the ground, its nose pointed directly at the object, its nose within 1 cm of the object, and not otherwise engaged in some other behavior (e.g., digging, climbing). Recorded videos were scored on a frame-by-frame basis by a blinded experimenter using the GUI of DeepEthogram (Bohnslav et al., 2021). Any animal demonstrating significant preference (| DI | > 20) during training was excluded from subsequent analyses (11 mice across all experiments). Additionally, any animal that spent a total of less than 3 seconds investigating or failed to explore both objects during test was also excluded from subsequent analyses (1 mouse across all experiments).
2.4. Tissue Extraction
Animals were euthanized via cervical dislocation and decapitated with surgical scissors (Fine Science Tools, Foster City, CA). Brains were removed from the skull with rongeurs and a surgical spatula (Fine Science Tools, Foster City, CA) and then flash-frozen in 2-methylbutane (Fisher Scientific, Waltham, MA). Brains were stored at −80°C before being sectioned with a Leica CM150 Cryostat (Leica Biosystems, Wetzlar, Germany). Punches of 500 μm were collected from the RSC or SCN and stored at −80°C. Our punches were specifically targeting the aRSC, but we cannot rule out the inclusion of some pRSC tissue, given the lack of a definite anterior/posterior RSC boundary in mice.
2.5. qPCR
RNA was extracted from punches with RNeasy Mini Kits (Qiagen, Germantown, MD) and cDNA was generated with High-capacity cDNA Reverse Transcription Kits (ThermoFisher, Frederick, MD). PrimeTime primer/probe assays were generated with IDT PrimerQuest Design Tool (IDT, Coralville, IA) and used to quantify expression of Per1, Fos, and Gapdh. Exact sequences: Per1 left primer: 5′-CCTGGAGGAATTGGAGCATATC-3′; Per1 right primer: 5′-CCTGCCTGCTCCGAAATATAG3′; Per1 probe: 5’-6-FAM/AAACCAGGA/Zen/CACCTTCTCTGTGGC/3IABkFQ-3’; Fos left primer: 5′-GGCACTAGAGACGGACAGAT-3′; Fos right primer: 5′-ACAGCCTTTCCTACTACCATTC-3′; Fos probe: 5’-6-FAM/CAGCCGACT/Zen/CCTTCTCCAGCATG/3IABkFQ-3’; Gapdh left primer: 5′-GGAGAAACCTGCCAAGTATGA-3′; Gapdh right primer: 5′-TCCTCAGTGTAGCCCAAGA-3′; Gapdh probe, 5’-HEX/TCAAGAAGG/ZEN/TGGTGAAGCAGGCAT/3IABkFQ/-3’. Analyses and statistics were performed in LightCycler 96 (Roche, Basel, Switzerland) using Roche proprietary algorithms.
2.6. Immunohistochemistry
Immunohistochemistry was performed as previously described (Kwapis et al., 2018). Briefly, 20 μm sections were mounted on slides, fixed with 4% PFA (ThermoFisher, Frederick, MD), permeabilized with 0.01% Triton-X (Fisher Scientific, Waltham, MA), and blocked for 1 hour with 8% normal goat serum (Jackson ImmunoResearch, West Grove, PA). Sections were then incubated overnight with rabbit anti-mCherry (1:500; ab167453; Abcam, Waltham, MA) and chicken anti-GFP (1:250; GFP-1010; Aves Labs, Davis, CA) primary antibodies, followed by a 1-hour incubation with goat anti-rabbit Alexa 555 (1:1000; a21430; ThermoFisher, Frederick, MD) and goat anti-chicken FITC (1:1000; ab6873; Abcam, Waltham, MA) secondary antibodies.
2.7. Viruses
All experiments used neuron-specific herpes simplex viruses (HSVs) to induce local gene expression only in neurons of the RSC or SCN. The CRISPRi virus (HSV-CRISPRi) expresses dCas9-KRAB-MeCp2 under an hSyn promoter followed by an IRES element and then mCherry. The CRISPRa virus (HSV-CRISPRa), similarly, expresses dCas9-VPR under an hSyn promoter followed by an IRES element and then mCherry. The sgRNA viruses (HSV-sgRNA) express the sgRNA under a U6 promoter and GFP under a CMV promoter. Three separate sgRNAs were used, one optimized for targeting Per1 with the CRISPRi system (GAGTTCGACGGCTCCAGAGTA), one optimized for targeting Per1 with the CRISPRa system (GCCCTTGTAAAGCAACCAT; Urban et al., 2021), and a nontargeting control sgRNA used for both systems (GCGAGGTATTCGGCTCCGCG; Lorsch et al., 2019). For each experiment, mice received a 1:1 viral cocktail of one of the CRISPR systems and an appropriate sgRNA (control mice received nontargeting sgRNAs). All viruses were purchased from Dr. Rachael Neve at the Gene Delivery Technology Core of Massachusetts General Hospital.
2.8. Statistics
Data are represented as Mean ± SEM in all figures. For each experiment, datapoints were dropped if they were more than 2 standard deviations away from the mean. Data were analyzed by either unpaired 2-tailed Student’s t-tests (Fig. 3C, 3D, 4B, 4C, 6C, 6D) or 2-way ANOVAs followed by Sidak’s multiple comparison post hoc tests (elsewhere). Data normality was verified by examining residuals, and no significant departures from normality were observed. Sample sizes were based on previous studies (Kwapis et al., 2018; Urban et al., 2021), but no statistical method was used to predetermine sample size. Graphpad Prism 9 (GraphPad Software, San Diego, CA) was used for all analyses.
Figure 3.

Local downregulation of Per1 within the RSC is sufficient to impair memory performance in young mice at ZT5. A. Representative images of HSV spread in the RSC. B. Schematic. Our two-virus HSV-CRIPRi system was infused into the RSC 72 hrs prior to training in the OLM task to downregulate Per1 expression during training. Control animals received the full CRISPRi construct but with a nontargeting sgRNA. C. Control animals show robust memory for the original object location while animals who received the Per1 sgRNA do not (n = 6/group). D. RSC-specific Per1 knockdown has no effect on total object exploration (n = 6/group). Two Per1 downregulation mice were dropped due to demonstrating an object preference during training, and an outlier was identified in the control group and dropped. * denotes p ≤ 0.05, ** denotes p ≤ 0.01, *** denotes p ≤ 0.001, **** denotes p ≤ 0.0001.
Figure 4.

Local upregulation of Per1 within the RSC improves memory performance in aging mice at ZT5. A. Schematic. Our two-virus HSV-CRIPRa system was infused into the RSC 72 hrs prior to training in the OLM task to upregulate Per1 expression during training. Control animals received the full CRISPRa construct but with a nontargeting sgRNA. B. 18-month-old control animals show minimal memory for the original object location while animals who received the Per1 sgRNA demonstrate robust memory (n = 13,10). C. RSC-specific Per1 knockup has no significant effect on total object exploration (n = 13,10). One control mouse was dropped due to insufficient exploration at test, while 6 mice (5 Per1, 1 control) were dropped due to preference during training. Due to this high attrition, this experiment was repeated and data were pooled, as no differences were detected between replications. Additionally, an outlier was identfied in the Per1 group and dropped. * denotes p ≤ 0.05, ** denotes p ≤ 0.01, *** denotes p ≤ 0.001, **** denotes p ≤ 0.0001.
Figure 6.

Local downregulation of Per1 within the SCN has no effect on memory in young mice trained at ZT5. A. Representative images of HSV spread in the SCN. B. Schematic. Our two-virus HSV-CRIPRi system was infused into the SCN 72 hrs prior to training in the OLM task to downregulate Per1 expression during training. Control animals received the full CRISPRi construct but with a nontargeting sgRNA. C. Control animals show robust memory for the original object location, as do animals who received the Per1 sgRNA (n = 5, 5). D. SCN-specific Per1 knockdown has no effect on total object exploration. (n = 5, 5). Two mice from each group were droped due to showing a preference during training. Additionally, an outlier was identified in the Per1 group and dropped. * denotes p ≤ 0.05, ** denotes p ≤ 0.01, *** denotes p ≤ 0.001, **** denotes p ≤ 0.0001.
3. Results
3.1. Learning induces Per1 and Fos expression in the RSC of young and aging mice in a circadian-dependent fashion
First, we sought to compare learning-induced expression of Per1 between the young and aging RSC. Here, young (8-week-old) and aging (18-month-old) mice were either trained in OLM or left in their homecage at ZT5 and then sacrificed an hour later to examine Per1 expression via qPCR. Homecage control mice were handled and habituated to the context as part of the OLM protocol but remained in their homecage on the day of training and were sacrificed in a counterbalanced manner with trained animals (Fig. 1A). We chose these ages as we have previously demonstrated that spatial learning upregulates Per1 in the DH of 8-week-old but not 18-month-old mice around ZT5 (Kwapis et al., 2018). Furthermore, mice exhibit clear deficits in OLM at 18-months (Wimmer et al., 2012); thus, this is an ideal age to model age-related memory impairments. Based on our results in the DH, we anticipated that learning would fail to increase expression of Per1 in the RSC of aging mice. We found that while learning did induce Per1 in the aging RSC, the magnitude of this induction was smaller than in young animals (Fig. 1B; 2-way ANOVA: effect of training: F(1,15) = 49.90, p < 0.0001; effect of age: F(1,15) = 4.272, p = 0.0565; interaction: F(1,15) = 5.553, p = 0.0325). Additionally, learning induces expression of the immediate early gene Fos in both the young and aging RSC (Fig. 1C; 2-way ANOVA: effect of training: F(1,15) = 80.39, p < 0.0001; effect of age: F(1,15) = 2.094, p = 0.1684; interaction: F(1,15) = 2.310, p = 0.1493). We investigated the expression of Per1 and Fos in the SCN of these mice and found that learning induces Per1 expression in the SCN of young but not aging mice (Fig. 1D; 2-way ANOVA: effect of training: F(1,15) = 16.12, p = 0.0011; effect of age: F(1,15) = 0.02490, p = 0.8767; interaction: F(1,15) = 10.71, p = 0.0051) while Fos expression is induced within the SCN regardless of age (Fig. 1E; 2-way ANOVA: effect of training: F(1,15) = 38.65, p < 0.0001; effect of age: F(1,15) = 0.0009, p = 0.9762; interaction: F(1,15) = 0.00164, p = 0.9682). Given Per1’s known role as a clock gene and the documented relationship between circadian rhythms and memory, we next investigated how this learning-induced Per1 expression was modulated by time-of-day in both young and aging animals.
Figure 1.
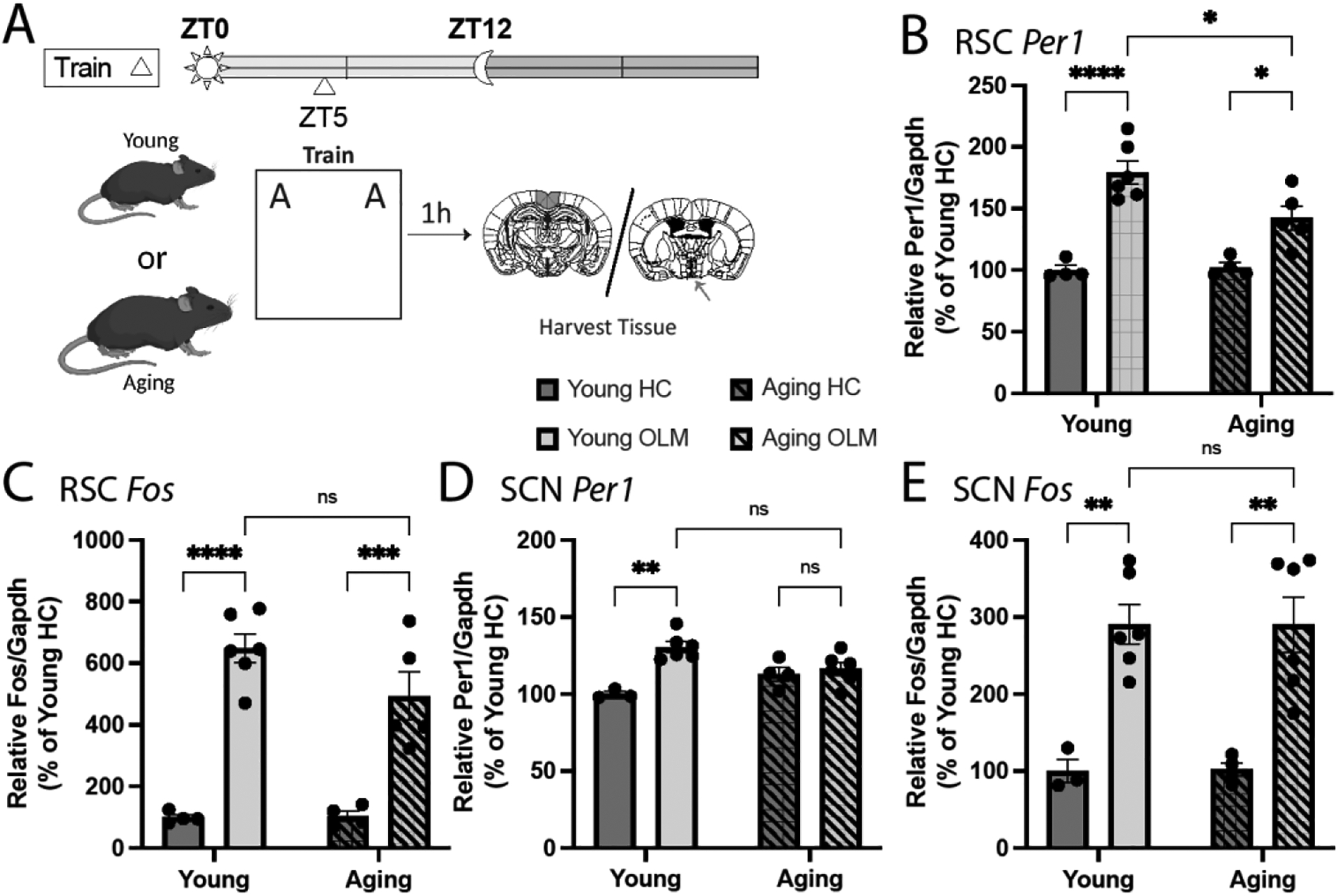
Training in OLM at ZT5 induces changes in gene expression in the RSC and SCN. A. Schematic. Young and aging mice were trained in OLM at ZT5 and sacrificed (along with homecage controls) 1 hr later. Expression of Per1 and Fos in the RSC and SCN was analyzed via qPCR. B. Per1 expression was induced in the RSC following learning, although this induction was attenuated in aging mice (n = 4–6). C. Fos expression was induced in the RSC following learning in both young and aging mice (n = 4–6). D. Per1 expression was induced in the SCN following learning only in young mice (n = 3–6). E. Fos expression was induced in the SCN following learning in both young and aging mice (n = 3–6). Outliers were identified in the young trained RSC Per1 data and in the aging trained RSC Fos data (1 each). * denotes p ≤ 0.05, ** denotes p ≤ 0.01, *** denotes p ≤ 0.001, **** denotes p ≤ 0.0001.
To investigate the interplay between learning, time-of-day, and Per1 expression in the aging RSC,18-month-old C57BL/6J mice were either trained in OLM or left in their homecage at six different timepoints (Zeitgeber Times: ZT1, ZT5, ZT9, ZT13, ZT17, ZT21), sacrificed an hour later, and then retrosplenial Per1 and Fos expression was quantified with qPCR (Fig. 2A). This experiment was set up identically to a previous experiment we ran in young mice that observed robust learning-induced increases in Per1 within the RSC during the day but not the night (Urban et al., 2021). Here, we also report that OLM training induces Per1 expression in the aging RSC in a circadian-dependent manner (Fig. 2B; 2-way ANOVA: effect of training: F(1,68) = 336.9, p < 0.0001; effect of time-of-day: F(5,68) = 17.08, p < 0.0001; interaction: F(5.68) = 3.156, p = 0.0128). Additionally, when we compared the Per1 induction in the aging RSC with what we have previously published in the young RSC (Urban et al., 2021), we find minimal differences between the relative magnitude of this Per1 induction in young and aging animals, with a significant difference only detected at ZT17 (Fig. 2C; 2-way ANOVA: effect of time-of-day: F(5,66) = 9.946, p < 0.0001; effect of age: F(1,66) = 3.651, p = 0.0604; interaction: F(5.66) = 2.691, p = 0.0283). Notably, both young and aging animals express more learning-induced Per1 during the day relative to night (Supplementary Fig. 1A; 2-way ANOVA: effect of day/night: F(1,74) = 5.138, p = 0.0263; effect of age: F(1,74) = 31.01, p < 0.0001; interaction: F(1,74) = 0.05706, p = 0.8119) identical to the pattern observed in memory performance (Bellfy et al., 2022; Chaudhury and Colwell, 2002). We also investigated Fos expression in the same mice and found that learning induces Fos expression in both young (Fig. 2D; 2-way ANOVA: effect of training: F(1,64) = 336.7, p < 0.0001; effect of time-of-day: F(5,64) = 18.98, p < 0.0001; interaction: F(1.64) = 14.28, p < 0.0001) and aging animals (Fig. 2E; 2-way ANOVA: effect of training: F(1,69) = 244.6, p < 0.0001; effect of time-of-day: F(5,69) = 2.065, p = 0.0803; interaction: F(1.69) = 1.729, p = 0.1395), also in a circadian-dependent fashion (Fig. 2F; 2-way ANOVA: effect of time-of-day: F(5,64) = 19.37, p < 0.0001; effect of age: F(1,64) = 25.23, p < 0.0001; interaction: F(5.64) = 16.02, p < 0.0001), though Fos expression was only greater during the day than the night in aging mice (Supplementary Fig. 1B; 2-way ANOVA: effect of day/night: F(1,72) = 8.156, p = 0.0056; effect of age: F(1,72) = 24.63, p < 0.0001; interaction: F(1,72) = 6.98, p = 0.0101). These results suggest that while circadian patterns of learning-induced changes in gene expression are largely maintained in the aging RSC, the absolute amount of learning-induced Per1 is dampened with age.
Figure 2.

Learning-induced expression of Per1 and Fos within the RSC is modulated by both time-of-day and aging. A. Schematic. Young and aging mice were trained in OLM at 6 different timepoints across the day-night cycle and then sacrified an hour later to examine gene expression in the RSC. B. Per1 expression in the aging RSC normalized to the mean of the ZT1 HC group (n = 5–7). C. Relative amount of Per1 induction for young and aging mice. Here, each timepoint’s OLM values are normalized to the mean of the corresponding HC group (e.g., young ZT5 OLM mice are each normalized to the mean of the young ZT5 HC group; n= 5–7). Note that the young data presented here was previously published in the Neurobiology of Learning and Memory in 2021 as part of a different analysis (Urban et al., 2021). D. Fos expression in the young RSC normalized to the mean of the ZT1 HC group (n = 5–7). E. Fos expression in the aging RSC normalized to the mean of the ZT1 HC group (n = 5–8). F. Relative amount of Fos induction for young and aging mice. Here, each timepoint’s OLM values are normalized to the mean of the corresponding HC group (n = 5–8). Outliers were identified in the Aging RSC Fos ZT13 HC group, ZT13 OLM group, and ZT9 OLM group (1 each). * denotes p ≤ 0.05, ** denotes p ≤ 0.01, *** denotes p ≤ 0.001, **** denotes p ≤ 0.0001.
3.2. Local downregulation of Per1 expression in the RSC of young mice impairs the formation of spatial memory
Next, we investigated if direct modulation of Per1 expression within the RSC can affect memory performance on the OLM task. Given the differences we observed in Per1 expression between young and aging animals (Fig. 1B), we thought that age-related dysregulation (i.e., reduction) of Per1 expression in the RSC might contribute to known age-related impairments in OLM (Wimmer et al., 2012). Additionally, previous work from our group has demonstrated that local reduction of Per1 expression in the RSC can attenuate the formation of a contextual fear memory in young mice (Urban et al., 2021). Thus, we reasoned that locally downregulating Per1 expression in the RSC of young mice during memory formation could impair subsequent test performance in the OLM task.
To locally downregulate Per1 within the RSC, we used HSV-CRISPRi as previously described (Urban et al., 2021). In brief, the CRISPRi system contains a deactivated Cas9 protein (dCas9) fused to two transcriptional repressors (Krüppel associated box [KRAB] and methyl CpG binding protein 2 [MeCP2]). This construct is directed by a single guide RNA (sgRNA) to a gene of interest—here, Per1. When the dCas9 binds as specified by the sgRNA, the attached transcriptional repressors downregulate the targeted gene (Larson et al., 2013). Given the known role of the circadian rhythm in memory performance and the potential off-target effects of systemic Per1 manipulation, we used herpes simplex viruses (HSVs) to manipulate Per1 expression only in RSC neurons. Additionally, since HSV expression peaks only 3 days following injection and then rapidly declines (Neve et al., 2005), this method allows for high temporal control. Given the role of gene expression in memory consolidation, we timed our viral manipulations so that peak viral expression (and, therefore, maximal inhibition of Per1) would occur at the time of OLM training. This HSV-CRISPRi system has been previously validated to reduce Per1 expression in both cultured cells and the mouse RSC (Urban et al., 2021).
To investigate the effect of RSC-specific Per1 downregulation on the formation of spatial memory, young mice were first handled and habituated at ZT5 (when memory for OLM is typically best (Bellfy et al., 2022). Then, they were injected with the HSV-CRISPRi system with a Per1-targeting sgRNA, while control animals received the same CRISPRi construct but with a nontargeting sgRNA, controlling for any effect of viral surgery or CRISPRi expression. Following surgery, mice were permitted to rest for 72 hrs (allowing HSV expression to peak; Neve et al., 2005) before being trained in OLM at ZT5. Twenty-four hours after training, mice were tested in OLM (Fig. 3B). We found that local downregulation of Per1 expression in the RSC of young mice impairs spatial memory, with Per1 knockdown mice showing significantly lower DIs at test relative to control animals (Fig. 3C; 2-tailed t-test: t10 = 2.451, p = 0.0342) without affecting total object exploration (Fig. 3D; 2-tailed t-test: t10 = 0.1044, p = 0.9189) or total distance moved (Supplementary Fig. 2A; 2-way repeated measures ANOVA: effect of day: F(4.214,42.14) = 57.17, p < 0.0001; effect of virus: F(1,10) = 3.741, p = 0.0819; interaction: F(7,70) = 1.605, p = 0.1482; effect of subject: F(10,70) = 7.628, p < 0.0001). These results demonstrate for the first time that Per1 expression within the RSC is necessary for incidental spatial memory, confirming what we had previously shown in contextual fear conditioning (Urban et al., 2021). Additionally, these results, together with our other published work (Bellfy et al., 2022; Kwapis et al., 2018), demonstrate that Per1 modulates a single learning task (OLM) in multiple brain regions—the RSC and the DH.
3.3. Local upregulation of Per1 expression in the RSC of aging mice restores the formation of spatial memory
In a complementary experiment, we examined whether local upregulation of Per1 in the RSC of aging mice is sufficient to rescue memory performance in OLM. Here, we used HSV-CRISPRa, as previously described (Urban et al., 2021), to locally increase Per1 expression within the RSC of aging mice. The CRISPRa system contains a dCas9 fused to three transcriptional activation domains (VP64, p65, and Rta; collectively known as VPR) that drive transcription of a gene target (here, Per1) specified by an sgRNA (Chavez et al., 2015). As with our Per1 CRISPRi system, the CRISPRa construct is downstream of an hSyn promoter and validated to upregulate Per1 expression in both cultured hippocampal neurons and the mouse RSC (Urban et al., 2021).
To elucidate how RSC-specific Per1 upregulation affects spatial memory, we performed OLM at ZT5 with aging mice, a timepoint at which we have previously observed age-related memory impairments in 18-month-old mice (Kwapis et al., 2019, 2018). The animals were handled and habituated before receiving retrosplenial injections of the HSV-CRISPRa system with a Per1-targeting sgRNA or nontargeting sgRNA (control animals). Three days after injection, the mice were trained in OLM and then tested the following day, both at ZT5 (Fig. 4A). We found that local Per1 upregulation within the RSC is sufficient to improve OLM in aging mice (Fig 4B; 2-tailed t-test: t21 = 2.562, p = 0.0182). As with the Per1 knockdown, this manipulation does not affect the total exploration of the two OLM objects during test (Fig. 4C; 2-tailed t-test: t21 = 1.992, p = 0.0595) or total distance traveled (Supplementary Fig. 2B; 2-way repeated measures ANOVA: effect of day: F(3.353,73.77) = 91.97, p < 0.0001; effect of virus: F(1,22) = 0.02730, p = 0.8703; interaction: F(7,154) = 0.389, p = 0.9077; effect of subject: F(22,154) = 7.155, p < 0.0001). Together with the previous experiment, these results reveal that bidirectional manipulation of Per1 expression in the RSC is sufficient to modulate memory performance.
3.4. Spatial learning induces gene expression in the SCN of young and aging mice
Although our results thus far have clearly demonstrated that Per1 in the RSC is critical for proper spatial memory formation, an open question remains as to the SCN’s role in memory, with various reports describing conflicting effects of SCN lesion on memory performance (Cain et al., 2012; Fernandez et al., 2014; Mulder et al., 2014; Phan et al., 2011; Shimizu et al., 2016; Stephan and Kovacevic, 1978; briefly reviewed by Ruby, 2021). Thus far, no one has specifically investigated how local Per1 expression in the brain’s central pacemaker might control memory performance. We decided to examine how OLM training affects Per1 expression in the SCN and test whether Per1’s control over spatial memory is exclusive to memory-relevant brain regions.
First, we examined learning-induced changes in Per1 and Fos expression in the SCN (Fig. 5A) using tissue from the young and aging mice presented in Figure 2. Interestingly, we found here that training in OLM modestly increases Per1 expression within the SCN in both young (Fig. 5B; 2-way ANOVA: effect of training: F(1,66) = 27.63, p < 0.0001; effect of time-of-day: F(5,66) = 2.754, p = 0.0255; interaction: F(5.66) = 0.8846, p = 0.4965) and aging mice (Fig. 5C; 2-way ANOVA: effect of time-of-day: F(5,81) = 3.772, p = 0.0040; effect of training: F(1,81) = 18.42, p < 0.0001; interaction: F(5.81) = 0.2808, p = 0.9224), though this Per1 induction is consistent across the day-night cycle and not modulated by age (Fig. 5D; 2-way ANOVA: effect of time-of-day: F(5,74) = 0.5483, p = 0.7391; effect of age: F(1,74) = 2.866, p = 0.0947; interaction: F(5.74) = 1.932, p = 0.0991). Although we observed a main effect of training on Per1 expression in aging animals, none of the post hoc tests revealed any pairwise differences between trained and homecage groups at each timepoint, consistent with what we reported in Fig. 1D. Additionally, there was no gross day/night differences of magnitude of Per1 induction in either young or aging animals (Supplementary Fig. 1C; 2-way ANOVA: effect of day/night: F(1,82) = 3.216, p = 0.0766; effect of age: F(1,82) = 0.1375, p = 0.7118; interaction: F(1,82) = 1.119, p = 0.2932) Training in OLM also induces Fos expression in the SCN in young (Fig. 5E; 2-way ANOVA: effect of training: F(1,58) = 154.6, p < 0.0001; effect of time-of-day: F(5,58) = 0.4096, p = 0.8402; interaction: F(5.58) = 0.6998, p = 0.6258) and aging mice (Fig. 5F; 2-way ANOVA: effect of training: F(1,79) = 182.0, p < 0.0001; effect of time-of-day: F(5,79) = 2.951, p = 0.0171; interaction: F(5.79) = 0.3958, p = 0.0030). Unlike with Per1, OLM-induced Fos expression in the SCN was modulated by time-of-day and varied with age (Fig. 5G; 2-way ANOVA: effect of time-of-day: F(5,67) = 6.835, p < 0.0001; effect of age: F(1,67) = 0.01917, p = 0.8903; interaction: F(5.67) = 3.758, p = 0.0047). Additionally, there was only a significant difference of induced Fos expression between day and night in aging animals (Supplementary Fig. 1D; 2-way ANOVA: effect of day/night: F(1,75) < 0.001, p = 0.9979; effect of age: F(1,75) = 23.77, p < 0.0001; interaction: F(1,75) = 2.300, p = 0.1336; post hoc Sidak’s test: p <0.001), although for young mice this difference approached significance (post hoc Sidak’s test: p= 0.0573).
Figure 5.

Learning-induced expression of Per1 and Fos within the SCN is modulated by both time-of-day and aging. A. Schematic. Young and aging mice were trained in OLM at 6 different timepoints across the day-night cycle and then sacrified an hour later to examine gene expression in the SCN. B. Per1 expression in the young SCN normalized to the mean of the ZT1 HC group (n = 5–8). C. Per1 expression in the aging SCN normalized to the mean of the ZT1 HC group (n = 7–8). D. Relative amount of Per1 induction for young and aging mice. Here, each timepoint’s OLM values are normalized to the mean of the corresponding HC group (e.g., young ZT5 OLM mice are normalized to the mean of the young ZT5 HC group; n = 5–8). E. Fos expression in the young SCN normalized to the mean of the young ZT1 HC group; n = 5–7). F. Fos expression in the aging SCN normalized to the mean of the aging ZT1 HC group; n = 7–8). G. Relative amount of Fos induction for young and aging mice. Here, each timepoint’s OLM values are normalized to the mean of the corresponding HC group (n = 5–7). An outlier was identified in the Aging SCN Fos ZT17 OLM group and removed. * denotes p ≤ 0.05, ** denotes p ≤ 0.01, *** denotes p ≤ 0.001, **** denotes p ≤ 0.0001.
3.5. Local downregulation of Per1 expression in the SCN of young mice has no effect on the formation of spatial memory
Although we found that learning induces Per1 expression in both the RSC and the SCN, we observed that the RSC Per1 induction was more tightly linked to memory performance than that of the SCN—that is, learning-induced RSC Per1 and memory performance (Bellfy et al., 2022) both peak during the day. Thus, we reasoned that Per1 activity in the RSC is more likely to support memory formation and sought to rule out the effect of SCN Per1 on memory performance. To do so, we used the same HSV-CRISPRi construct described previously to locally downregulate Per1 within the SCN in young mice. As in the RSC CRISPRi experiment, mice were handled and habituated to the OLM chambers, administered HSV-CRISPRi, allowed to rest for 72 hours, and then trained and tested in OLM (Fig. 6B). Control animals received HSV-CRISPRi but with a nontargeting RNA, controlling for any effects of viral surgery or CRISPRi expression. We found that downregulation of Per1 within the SCN has no effect on the formation of spatial memory (Fig 6C; 2-tailed t-test: t8 = 0.3976, p = 0.7013), on total object exploration (Fig 6D; 2-tailed t-test: t8 = 0.1425, p = 0.8902), or distance travelled (Supplementary Fig. 2C; 2-way repeated measures ANOVA: effect of day: F(2.387,19.09) = 31.00, p < 0.0001; effect of virus: F(1,8) = 1.554, p = 0.2478; interaction: F(7,56) = 1.961, p = 0.0769; effect of subject: F(8,56) = 6.627, p < 0.0001). These data support our hypothesis that the previously observed memory effects can be specifically attributed to Per1 activity within the RSC.
4. Discussion
Here, we investigated the relationship between brain region–specific Per1 expression, spatial memory, and aging. Our results reveal that Per1 expression in the RSC is affected by aging and necessary for the formation of long-term spatial memory in OLM. We find that targeted downregulation of Per1 within the RSC of young animals impairs learning, while upregulation of Per1 within the RSC of aging mice improves learning. Finally, although learning induces Per1 expression in the SCN of young but not aging mice, locally downregulating Per1 expression in the SCN of young mice does not affect the formation of spatial memory. Together, these findings expand the known role of Per1 as a memory-relevant gene in neocortical and subcortical regions and further demonstrate the diverse roles clock genes play outside of the SCN. Additionally, these data support the idea that age-related dysregulation of clock genes (namely, Per1) contributes to multiple symptoms of aging (circadian disruptions and memory deficits) in a brain region–dependent fashion.
Per1 has long been known to play an important role in learning and memory, but many previous studies (Abarca et al., 2002; Jilg et al., 2010; Rawashdeh et al., 2014) have relied on global Per1 knockout mice, making it difficult to discern if the observed changes in learning are a result of decreased Per1 activity within the SCN or within memory-relevant structures like the DH and RSC. Previous work from our group (Bellfy et al., 2022; Kwapis et al., 2018; Urban et al., 2021) has demonstrated that targeted manipulations of Per1 expression in just the DH and RSC are sufficient to modulate memory formation, and this work builds on those studies by investigating how RSC Per1 is changed by aging. We have previously identified that local upregulation of Per1 within the DH is sufficient to rescue age-related deficits in OLM (Kwapis et al., 2018), so it is not surprising that similar trends are observed in the RSC. Interestingly, we previously reported that DH Per1 is not induced by learning in aging animals (Kwapis et al., 2018), but here we found that Per1 is induced by learning within the RSC of aging mice (Fig. 1). These results suggest that some critical threshold of Per1 expression might be necessary to consolidate memory, and that while learning does induce some Per1 expression within the aging RSC, this induction is insufficient to reach levels necessary for proper memory formation.
It is also possible that circadian fluctuations in retrosplenial Per1 may contribute to circadian fluctuations in memory performance. Notably, both learning-induced RSC Per1 expression (Urban et al., 2021) and long-term memory performance (Bellfy et al., 2022; Chaudhury and Colwell, 2002; Eckel-Mahan et al., 2008) peak during the day in young animals, supporting the hypothesis that Per1 drives memory formation in a circadian fashion. However, we have also previously revealed that for context fear conditioning, locally increasing Per1 expression within the RSC during the subjective night does not restore memory to day-time levels, and overexpression of Per1 in the RSC during the day can impair memory performance (Urban et al., 2021). These data suggest that different mechanisms may contribute to memory formation during the day and night, and that increased Per1 does not always improve memory. One explanation for this discrepancy is that age-related changes in memory performance and day-night changes in memory performance rely on different molecular mechanisms, such that that age-related impairments can be rescued by Per1 upregulation while night-related impairments cannot. Alternatively, Per1 may be modulating contextual fear memory and OLM via different mechanisms. These differently motivated paradigms rely on different brain structures (e.g., the amygdala is engaged by fear conditioning but not OLM) and its possible that the mnemonic effects of Per1 are circuit-specific. Future studies should test whether overexpression of Per1 during the day can also negatively affect OLM performance, and whether age-related impairments in fear conditioning are restored by RSC Per1 overexpression.
The RSC is believed to be functionally graded along its anterior-posterior axis. Prevailing theories of RSC function characterize the anterior portion (the aRSC) as more important for encoding “what” information while the posterior portion (pRSC) is more responsible for tracking “where” information (Neave et al., 1994; Trask et al., 2021b; Vann et al., 2003; Vann and Aggleton, 2004). Since OLM is a spatial memory paradigm (the only difference between the objects is their position in the arena), one might expect the pRSC to be more heavily involved than the aRSC. Nonetheless, previous work (de Landeta et al., 2020) has revealed a clear role for the aRSC in OLM and other spatial tasks (Kwapis et al., 2015). Additionally, we have previously identified Per1 in the aRSC as important for contextual fear conditioning, so we elected to investigate Per1’s role in the aRSC during OLM. Our viral manipulations and punches were specifically targeting the aRSC (see Methods), but the boundary between anterior and posterior RSC is not clearly defined in mice and, as such, we cannot conclusively determine that our injection was isolated to the anterior portion. Thus, we have simply referred to our target region as the RSC throughout.
Intriguingly, we did find that learning affects Per1 and Fos expression within the SCN, although Per1 expression here is not modulated by time-of-day and is uniform across the day and night (Fig. 5; Supplemental Fig. 1). We also demonstrate here, for the first time, that targeted downregulation of Per1 within the SCN does not affect the formation of spatial memory, consistent with prior work suggesting that SCN ablation fails to impair memory (Mulder et al., 2014; Stephan and Kovacevic, 1978; though it is worth noting there are other reports to the contrary: Phan et al., 2011; Shimizu et al., 2016). We did not perform activity monitoring on these animals to see if this manipulation affected their circadian rhythm, though the lack of detectable differences in OLM performance suggests that any possible circadian rhythm changes are insufficient to affect memory performance. Furthermore, even Per1−/− mice demonstrate largely intact circadian rhythms, suggesting this manipulation would minimally affect their circadian rhythms, if at all (Cermakian et al., 2001). Finally, these animals were housed on a 12 hr light/dark cycle, so their diurnal rhythms would have been entrained to the zeitgeber of the vivarium lights, and this entrainment is also intact in Per1−/− mice (Cermakian et al., 2001).
Given that SCN-specific Per1 downregulation does not affect learning in young animals (Fig. 6), we suspect our observed changes in SCN gene expression are unlikely to be directly linked to learning per se. However, we did not directly test whether targeted upregulation in aging mice rescues long-term memory in OLM, although given that SCN-specific Per1 manipulation had no effect on learning in young mice, we would not expect this to be the case. Additionally, there was no effect of time-of-day on induced Per1 expression within the SCN (Fig. 5B, 5C; Supplemental Fig. 1), suggesting Per1 here is not linking memory strength with time-of-day. However, exposure to a zeitgeber (e.g., a light pulse) induces pronounced expression of Per1 (Shigeyoshi et al., 1997) and Fos (Kornhauser et al., 1990) within the SCN, and it is conceivable that our OLM paradigm constitutes a zeitgeber for trained animals, as these animals have undergone 7 consecutive days of behavior occurring at precisely the same time each day. Further, since training was conducted under dim red light and mice were housed and transported in light-protected conditions, this Per1 induction cannot be attributed to light exposure. It is worth noting, however, that we detected day/night differences in learning-induced Fos expression (Supplemental Fig. 1) in the SCN of only aging animals, though the values for young animals approached significance. This is consistent with the long-known fact that the SCNs of both nocturnal and diurnal mammals tend to exhibit greater neuronal activity during the subjective day (Inouye and Kawamura, 1979). Interestingly, we only detected day/night differences in the SCN Fos expression of trained animals, with minimal differences between homecage groups (Fig. 5).
This study does not investigate the specific mechanism by which Per1 is downregulated in the aging brain. However, a previous report from our group demonstrated that age-related deficits in hippocampal Per1 expression were largely due to the repressive histone deacetylase HDAC3 (Kwapis et al., 2018). Thus, it is reasonable that the same HDAC3-induced repressive chromatin structure observed in the aging DH may also be present in the aging RSC and preventing adequate Per1 expression following a learning event. Although we did detect an attenuated Per1 induction in aging animals in our initial experiment (Fig. 1), our subsequent time course experiment suggests that Per1 is induced to comparable levels between the young and aging RSC (Fig. 2). Given the fact the initial experiment was the only one to directly compare young and aging mRNA levels, we believe this best represents the true relationship between learning-induced Per1 and Fos induction in the young and aging RSC. That is, it seems that learning induces some Per1 expression in the aging RSC, but not to the level seen in young animals (Fig. 1). The time course experiments (Fig. 2 and Fig. 5) were ran separately as young and aging cohorts, and then each trained mouse Per1 value was divided by the average of its ZT-matched homecage group to produce a normalized induction value. Thus, it is possible that abnormally low homecage expression of Per1 or Fos might exaggerate the apparent magnitude of induction seen in Figure 2.
Additionally, in this study, we did not investigate which aspect of memory (i.e., acquisition, consolidation, or retrieval) was affected by Per1 manipulation. Although we timed our HSV manipulations to ensure maximal virus expression during OLM training, there was likely still expression of our constructs during the test session 24 hours later. Thus, while these data suggest Per1 most likely influences memory acquisition or consolidation, we cannot conclusively rule out an effect of Per1 on memory retrieval. However, another recent report from our group suggests that diurnal changes in memory performance can be specifically attributed to memory consolidation (Bellfy et al., 2022). Given the relationship between time-of-day, memory performance, and RSC Per1 expression, it is most likely that Per1 specifically affects memory consolidation. Additionally, in the DH, PER1 is known to function by shuttling p90RSK into the nucleus to phosphorylate cAMP response element binding protein (CREB; Rawashdeh et al., 2016) Given CREB’s known role in memory consolidation (Yin et al., 1995, 1994), this further suggests that Per1 is also necessary for memory consolidation, possibly by controlling the allocation of individual neurons to a newly formed memory trace or engram (Zhou et al., 2009).
A notable weakness of this study is the decision to only include male subjects. Previous work from our group has noted that Per1 modulates memory formation within the RSC in a sex-dependent manner (Urban et al., 2021). Therefore, we decided to split subsequent investigations into male and female cohorts to minimize sample sizes. Male mice were investigated first (this study) as their behavior was more readily affected by our Per1 manipulations (Urban et al., 2021). We are currently investigating these effects in female mice in a separate series of experiments.
It has been difficult to decouple the relationship between the circadian rhythm, learning, and aging. These results expand what is known about the function of clock genes beyond the molecular clock, how the expression of these genes impacts memory formation, and how this expression is modulated by aging. Previous work has argued that age-related memory impairments are, at least partially, caused by circadian rhythm disruption stemming from the SCN (Antoniadis et al., 2000; Pang et al., 2006). In this older model (Fig. 7A), aging would result in dysregulation of the expression of Per1 and other clock genes within the SCN, thereby disrupting circadian rhythm and subsequently affecting the downstream process of learning. However, the data presented here, in combination with previous work from our group (Bellfy et al., 2022; Kwapis et al., 2018; Urban et al., 2021) and elsewhere (Rawashdeh et al., 2014, 2016) support a model (Fig. 7B) in which age-related deficits in memory performance can be attributed to local disruption of Per1 in memory-relevant brain regions (i.e., the RSC and the DH), while disruption of Per1 within the SCN contributes to circadian dysfunction. This second model is further supported by other work (Kwapis et al., 2018) from our group suggesting these Per1 disruptions stem from local epigenetic changes (i.e., excessive histone deacetylase activity in the DH) rather than disrupted communication with the SCN (an argument further supported by the negative results in Fig. 6 here). This has broad implications for clinicians interested in chronotherapy aimed at rescuing memory performance and circadian rhythms in aging populations, as therapies intended to restore circadian rhythm globally may not remedy the underlying local disruption of Per1 within the DH, RSC, and other memory-relevant brain regions.
Figure 7.

Two possible models explaining the interaction between age and its associated circadian disruptions and memory impairments. A. Model in which aging predominately disrupts the expression of Per1 (and other clock genes) within the SCN, leading to circadian dysregulation. Then, an impaired circadian rhythm negatively affects the downstream processes of learning and results in memory deficits. B. Model in which the local dysregulation of clock genes within various memory-relevant brain regions is directly contributing to age-related memory impairments, as supported by data here and elsewhere (Kwapis et al., 2018).
Supplementary Material
Supplemental Figure 1. Learning-induced changes in Per1 within the RSC and Fos within the SCN are greater during the day than at night. A. Direct comparison between day (ZT1, ZT5, and ZT9) and night (ZT13, ZT17, ZT21) RSC Per1 induction in young and aging mice. B. Direct comparison between day and night RSC Fos induction in young and aging mice. C. Direct comparison between day and night SCN Per1 induction in young and aging mice. D. Direct comparison between day and night SCN Fos induction in young and aging mice. These are additional analyses of data already appearing in Fig. 2 and Fig. 5. Note that the young RSC Per1 data presented here was previously published in the Neurobiology of Learning and Memory in 2021 as part of a different analysis (Urban et al., 2021).
Supplemental Figure 2. No HSV treatments had any effect on total distance traveled in any of the behavioral experiments. A. Total distance traveled each day for mice in the young RSC Per1 downregulation experiment (Fig. 3). B. Total distance traveled each day for mice in the aging RSC Per1 upregulation experiment (Fig. 4). C. Total distance traveled each day for mice in the young SCN Per1 downregulation experiment (Fig. 6).
Highlights.
Learning induces Per1 expression within the RSC of young and aging mice
This RSC Per1 induction rhythmically cycles in young and aging mice
Bidirectional RSC Per1 manipulations affect spatial memory
Learning also induces Per1 within the SCN of young (but not aging) mice
Per1 knockdown in the SCN of young mice has no effect on memory
Acknowledgements
Figures were made partially with BioRender. We thank Dr. Rachael Neve for help designing and packaging all HSV vectors used here. We also thank Gavyn Partlow for designing applications used to streamline behavioral scoring.
Funding
This work was funded by NIA grants R00AG056595, R21AG068444, and R01AG074041; Whitehall Foundation grant 2020-05-06; and AFAR Junior Faculty grant A21105; all to JLK.
Abbreviations:
- BMAL
brain and muscle ARNT-like
- CLOCK
Circadian Locomotor Output Cycles Kaput
- CREB
cAMP response element binding protein
- Cry
Cryptochrome
- DH
dorsal hippocampus
- HSV
herpes simplex virus
- IEG
immediate early gene
- OLM
object location memory
- Per1
Period1
- RSC
retrosplenial cortex
- SCN
suprachiasmatic nucleus
- sgRNA
single guide RNA
- TTFL
transcription-translation feedback loop
- ZT
zeitgeber time
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations of interest
none
Data Availability
The data presented here are available from the corresponding author upon reasonable request.
References
- Abarca C, Albrecht U, Spanagel R, 2002. Cocaine sensitization and reward are under the influence of circadian genes and rhythm. Proceedings of the National Academy of Sciences 99, 9026–9030. 10.1073/pnas.142039099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe M, Herzog ED, Yamazaki S, Straume M, Tei H, Sakaki Y, Menaker M, Block GD, 2002. Circadian Rhythms in Isolated Brain Regions. J Neurosci 22, 350–356. 10.1523/JNEUROSCI.22-01-00350.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniadis EA, Ko CH, M.R., McDonald RJ, 2000. Circadian rhythms, aging and memory. Behavioural Brain Research 111, 25–37. 10.1016/S0166-4328(00)00145-5 [DOI] [PubMed] [Google Scholar]
- Bellfy L, Smies CW, Bernhardt AR, Bodinayake KK, Sebastian A, Stuart EM, Wright DS, Lo C-Y, Murakami S, Boyd HM, von Abo MJ, Albert I, Kwapis JL, 2022. The clock gene Per1 expression may exert diurnal control over hippocampal memory consolidation. bioRxiv 2022.10.11.511798. 10.1101/2022.10.11.511798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnslav JP, Wimalasena NK, Clausing KJ, Dai YY, Yarmolinsky DA, Cruz T, Kashlan AD, Chiappe ME, Orefice LL, Woolf CJ, Harvey CD, 2021. DeepEthogram, a machine learning pipeline for supervised behavior classification from raw pixels. eLife 10, e63377. 10.7554/eLife.63377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cain SW, Chalmers JA, Ralph MR, 2012. Circadian modulation of passive avoidance is not eliminated in arrhythmic hamsters with suprachiasmatic nucleus lesions. Behavioural Brain Research 230, 288–290. 10.1016/j.bbr.2012.02.022 [DOI] [PubMed] [Google Scholar]
- Cermakian N, Monaco L, Pando MP, Dierich A, Sassone-Corsi P, 2001. Altered behavioral rhythms and clock gene expression in mice with a targeted mutation in the Period1 gene. EMBO J 20, 3967–3974. 10.1093/emboj/20.15.3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury D, Colwell CS, 2002. Circadian modulation of learning and memory in fear-conditioned mice. Behavioural Brain Research 133, 95–108. 10.1016/S0166-4328(01)00471-5 [DOI] [PubMed] [Google Scholar]
- Chaudhury D, Wang LM, Colwell CS, 2005. Circadian Regulation of Hippocampal Long-Term Potentiation. J Biol Rhythms 20, 225–236. 10.1177/0748730405276352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez A, Scheiman J, Vora S, Pruitt BW, Tuttle M, Iyer E, Lin S, Kiani S, Guzman CD, Wiegand DJ, Ter-Ovanesyan D, Braff JL, Davidsohn N, Housden BE, Perrimon N, Weiss R, Aach J, Collins JJ, Church GM, 2015. Highly-efficient Cas9-mediated transcriptional programming. Nat Methods 12, 326–328. 10.1038/nmeth.3312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran KA, Donnan MD, Tronson NC, Guzmán YF, Gao C, Jovasevic V, Guedea AL, Radulovic J, 2011. NMDA Receptors in Retrosplenial Cortex Are Necessary for Retrieval of Recent and Remote Context Fear Memory. J Neurosci 31, 11655–11659. 10.1523/JNEUROSCI.2107-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craik FIM, Grady CL, 2002. Aging, Memory, and Frontal Lobe Functioning, in: Stuss DT, Knight RT (Eds.), Principles of Frontal Lobe Function. Oxford University Press, p. 0. 10.1093/acprof:oso/9780195134971.003.0031 [DOI] [Google Scholar]
- Craik FIM, McDowd JM, 1987. Age differences in recall and recognition. Journal of Experimental Psychology: Learning, Memory, and Cognition 13, 474–479. 10.1037/0278-7393.13.3.474 [DOI] [Google Scholar]
- Davies JA, Navaratnam V, Redfern PH, 1973. A 24-hour rhythm in passive-avoidance behaviour in rats. Psychopharmacologia 32, 211–214. 10.1007/BF00428692 [DOI] [PubMed] [Google Scholar]
- de Landeta AB, Pereyra M, Medina JH, Katche C, 2020. Anterior retrosplenial cortex is required for long-term object recognition memory. Sci Rep 10, 4002. 10.1038/s41598-020-60937-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckel-Mahan KL, Phan T, Han S, Wang H, Chan GC-K, Scheiner ZS, Storm DR, 2008. Circadian oscillation of hippocampal MAPK activity and cAMP: implications for memory persistence. Nat Neurosci 11, 1074–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez F, Lu D, Ha P, Costacurta P, Chavez R, Heller HC, Ruby NF, 2014. Dysrhythmia in the suprachiasmatic nucleus inhibits memory processing. Science 346, 854–857. 10.1126/science.1259652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harker KT, Whishaw IQ, 2002. Impaired Spatial Performance in Rats with Retrosplenial Lesions: Importance of the Spatial Problem and the Rat Strain in Identifying Lesion Effects in a Swimming Pool. J. Neurosci 22, 1155–1164. 10.1523/JNEUROSCI.22-03-01155.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye ST, Kawamura H, 1979. Persistence of circadian rhythmicity in a mammalian hypothalamic “island” containing the suprachiasmatic nucleus. Proc Natl Acad Sci U S A 76, 5962–5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jilg A, Lesny S, Peruzki N, Schwegler H, Selbach O, Dehghani F, Stehle JH, 2010. Temporal dynamics of mouse hippocampal clock gene expression support memory processing. Hippocampus 20, 377–388. 10.1002/hipo.20637 [DOI] [PubMed] [Google Scholar]
- Kornhauser JM, Nelson DE, Mayo KE, Takahashi JS, 1990. Photic and circadian regulation of c-fos gene expression in the hamster suprachiasmatic nucleus. Neuron 5, 127–134. 10.1016/0896-6273(90)90303-W [DOI] [PubMed] [Google Scholar]
- Kwapis JL, Alaghband Y, Kramár EA, López AJ, Vogel Ciernia A, White AO, Shu G, Rhee D, Michael CM, Montellier E, Liu Y, Magnan CN, Chen S, Sassone-Corsi P, Baldi P, Matheos DP, Wood MA, 2018. Epigenetic regulation of the circadian gene Per1 contributes to age-related changes in hippocampal memory. Nat Commun 9, 3323. 10.1038/s41467-018-05868-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwapis JL, Alaghband Y, López AJ, Long JM, Li X, Shu G, Bodinayake KK, Matheos DP, Rapp PR, Wood MA, 2019. HDAC3-Mediated Repression of the Nr4a Family Contributes to Age-Related Impairments in Long-Term Memory. J Neurosci 39, 4999–5009. 10.1523/JNEUROSCI.2799-18.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwapis JL, Jarome TJ, Lee JL, Helmstetter FJ, 2015. The retrosplenial cortex is involved in the formation of memory for context and trace fear conditioning. Neurobiol Learn Mem 123, 110–116. 10.1016/j.nlm.2015.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson MH, Gilbert LA, Wang X, Lim WA, Weissman JS, Qi LS, 2013. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat Protoc 8, 2180–2196. 10.1038/nprot.2013.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorsch ZS, Hamilton PJ, Ramakrishnan A, Parise EM, Salery M, Wright WJ, Lepack AE, Mews P, Issler O, McKenzie A, Zhou X, Parise LF, Pirpinias ST, Ortiz Torres I, Kronman HG, Montgomery SE, Loh Y-HE, Labonté B, Conkey A, Symonds AE, Neve RL, Turecki G, Maze I, Dong Y, Zhang B, Shen L, Bagot RC, Nestler EJ, 2019. Stress resilience is promoted by a Zfp189-driven transcriptional network in prefrontal cortex. Nat Neurosci 22, 1413–1423. 10.1038/s41593-019-0462-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung CA, Sidiropoulou K, Vitaterna M, Takahashi JS, White FJ, Cooper DC, Nestler EJ, 2005. Regulation of dopaminergic transmission and cocaine reward by the Clock gene. Proceedings of the National Academy of Sciences 102, 9377–9381. 10.1073/pnas.0503584102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer JR Jr., Brown TH, 2006. Impaired trace and contextual fear conditioning in aged rats. Behavioral Neuroscience 120, 612. 10.1037/0735-7044.120.3.612 [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Coque L, Cao J-L, Kumar J, Chakravarty S, Asaithamby A, Graham A, Gordon E, Enwright JF, DiLeone RJ, Birnbaum SG, Cooper DC, McClung CA, 2010. Knockdown of Clock in the Ventral Tegmental Area Through RNA Interference Results in a Mixed State of Mania and Depression-Like Behavior. Biological Psychiatry, Animal Models for Mania and Melancholia 68, 503–511. 10.1016/j.biopsych.2010.04.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder CK, Papantoniou C, Gerkema MP, Van Der Zee EA, 2014. Neither the SCN nor the adrenals are required for circadian time-place learning in mice. Chronobiol Int 31, 1075–1092. 10.3109/07420528.2014.944975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neave N, Lloyd S, Sahgal A, Aggleton JP, 1994. Lack of effect of lesions in the anterior cingulate cortex and retrosplenial cortex on certain tests of spatial memory in the rat. Behavioural Brain Research 65, 89–101. 10.1016/0166-4328(94)90077-9 [DOI] [PubMed] [Google Scholar]
- Nelson AJD, Hindley EL, Pearce JM, Vann SD, Aggleton JP, 2015. The effect of retrosplenial cortex lesions in rats on incidental and active spatial learning. Front Behav Neurosci 9, 11. 10.3389/fnbeh.2015.00011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neve RL, Neve KA, Nestler EJ, Carlezon WA, 2005. Use of herpes virus amplicon vectors to study brain disorders. BioTechniques 39, 381–391. 10.2144/05393PS01 [DOI] [PubMed] [Google Scholar]
- Pang KCH, Miller JP, Fortress A, McAuley JD, 2006. Age-related disruptions of circadian rhythm and memory in the senescence-accelerated mouse (SAMP8). AGE 28, 283–296 (2006). 10.1007/s11357-006-9013-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan TH, Chan GC-K, Sindreu CB, Eckel-Mahan KL, Storm DR, 2011. The Diurnal Oscillation of MAP (Mitogen-Activated Protein) Kinase and Adenylyl Cyclase Activities in the Hippocampus Depends on the Suprachiasmatic Nucleus. J Neurosci 31, 10640–10647. 10.1523/JNEUROSCI.6535-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plautz JD, Kaneko M, Hall JC, Kay SA, 1997. Independent Photoreceptive Circadian Clocks Throughout Drosophila. Science 278, 1632–1635. 10.1126/science.278.5343.1632 [DOI] [PubMed] [Google Scholar]
- Rawashdeh O, Jilg A, Jedlicka P, Slawska J, Thomas L, Saade A, Schwarzacher SW, Stehle JH, 2014. PERIOD1 coordinates hippocampal rhythms and memory processing with daytime. Hippocampus 24, 712–723. 10.1002/hipo.22262 [DOI] [PubMed] [Google Scholar]
- Rawashdeh O, Jilg A, Maronde E, Fahrenkrug J, Stehle JH, 2016. Period1 gates the circadian modulation of memory-relevant signaling in mouse hippocampus by regulating the nuclear shuttling of the CREB kinase pP90RSK. J. Neurochem 138, 731–745. 10.1111/jnc.13689 [DOI] [PubMed] [Google Scholar]
- Renfrew JW, Pettigrew KD, Rapoport SI, 1987. Motor activity and sleep duration as a function of age in healthy men. Physiology & Behavior 41, 627–634. 10.1016/0031-9384(87)90321-0 [DOI] [PubMed] [Google Scholar]
- Ruby NF, 2021. Suppression of Circadian Timing and Its Impact on the Hippocampus. Front Neurosci 15, 642376. 10.3389/fnins.2021.642376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata H, Kondo S, Naito J, 2004. Organization of retrosplenial cortical projections to the anterior cingulate, motor, and prefrontal cortices in the rat. Neuroscience Research 49, 1–11. 10.1016/j.neures.2004.01.005 [DOI] [PubMed] [Google Scholar]
- Shigeyoshi Y, Taguchi K, Yamamoto S, Takekida S, Yan L, Tei H, Moriya T, Shibata S, Loros JJ, Dunlap JC, Okamura H, 1997. Light-Induced Resetting of a Mammalian Circadian Clock Is Associated with Rapid Induction of the mPer1 Transcript. Cell 91, 1043–1053. 10.1016/S0092-8674(00)80494-8 [DOI] [PubMed] [Google Scholar]
- Shimizu K, Kobayashi Y, Nakatsuji E, Yamazaki M, Shimba S, Sakimura K, Fukada Y, 2016. SCOP/PHLPP1β mediates circadian regulation of long-term recognition memory. Nat Commun 7, 12926. 10.1038/ncomms12926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smies CW, Bodinayake KK, Kwapis JL, 2022. Time to learn: The role of the molecular circadian clock in learning and memory. Neurobiology of Learning and Memory 193, 107651. 10.1016/j.nlm.2022.107651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider KH, Dziema H, Aten S, Loeser J, Norona FE, Hoyt K, Obrietan K, 2016. Modulation of learning and memory by the targeted deletion of the circadian clock gene Bmal1 in forebrain circuits. Behav Brain Res 308, 222–235. 10.1016/j.bbr.2016.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer S, Falcon E, Kumar J, Krishnan V, Mukherjee S, Birnbaum SG, McClung CA, 2013. Circadian genes Period 1 and Period 2 in the nucleus accumbens regulate anxiety-related behavior. Eur J Neurosci 37, 242–250. 10.1111/ejn.12010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan FK, Kovacevic NS, 1978. Multiple retention deficit in passive avoidance in rats is eliminated by suprachiasmatic lesions. Behavioral Biology 22, 456–462. 10.1016/S0091-6773(78)92565-8 [DOI] [PubMed] [Google Scholar]
- Trask S, Ferrara NC, Grisales K, Helmstetter FJ, 2021a. Optogenetic inhibition of either the anterior or posterior retrosplenial cortex disrupts retrieval of a trace, but not delay, fear memory. Neurobiology of Learning and Memory 185, 107530. 10.1016/j.nlm.2021.107530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trask S, Fournier DI, 2022. Examining a role for the retrosplenial cortex in age-related memory impairment. Neurobiology of Learning and Memory 189, 107601. 10.1016/j.nlm.2022.107601 [DOI] [PubMed] [Google Scholar]
- Trask S, Pullins SE, Ferrara NC, Helmstetter FJ, 2021b. The anterior retrosplenial cortex encodes event-related information and the posterior retrosplenial cortex encodes context-related information during memory formation. Neuropsychopharmacology 46, 1386–1392. 10.1038/s41386-021-00959-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban MW, Lo C, Bodinayake KK, Brunswick CA, Murakami S, Heimann AC, Kwapis JL, 2021. The circadian clock gene Per1 modulates context fear memory formation within the retrosplenial cortex in a sex-specific manner. Neurobiology of Learning and Memory 185, 107535. 10.1016/j.nlm.2021.107535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vann SD, Aggleton JP, 2004. Testing the importance of the retrosplenial guidance system: effects of different sized retrosplenial cortex lesions on heading direction and spatial working memory. Behavioural Brain Research 155, 97–108. 10.1016/j.bbr.2004.04.005 [DOI] [PubMed] [Google Scholar]
- Vann SD, Brown MW, Aggleton JP, 2000. Fos expression in the rostral thalamic nuclei and associated cortical regions in response to different spatial memory tests. Neuroscience 101, 983–991. 10.1016/S0306-4522(00)00288-8 [DOI] [PubMed] [Google Scholar]
- Vann SD, Kristina Wilton LA, Muir JL, Aggleton JP, 2003. Testing the importance of the caudal retrosplenial cortex for spatial memory in rats. Behavioural Brain Research 140, 107–118. 10.1016/S0166-4328(02)00274-7 [DOI] [PubMed] [Google Scholar]
- Vogel‐Ciernia A, Wood MA, 2014. Examining Object Location and Object Recognition Memory in Mice. Current Protocols in Neuroscience 69. 10.1002/0471142301.ns0831s69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Nie B, Duan S, Zhu H, Liu H, Shan B, 2016. Functionally Brain Network Connected to the Retrosplenial Cortex of Rats Revealed by 7T fMRI. PLoS One 11, e0146535. 10.1371/journal.pone.0146535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzman ED, Moline ML, Czeisler CA, Zimmerman JC, 1982. Chronobiology of aging: Temperature, sleep-wake rhythms and entrainment. Neurobiology of Aging 3, 299–309. 10.1016/0197-4580(82)90018-5 [DOI] [PubMed] [Google Scholar]
- Wiggs CL, Weisberg J, Martin A, 1998. Neural correlates of semantic and episodic memory retrieval. Neuropsychologia 37, 103–118. 10.1016/S0028-3932(98)00044-X [DOI] [PubMed] [Google Scholar]
- Wimmer M, Hernandez P, Blackwell J, Abel T, 2012. Aging impairs hippocampus-dependent long-term memory for object location in mice. Neurobiol Aging 33, 2220–2224. 10.1016/j.neurobiolaging.2011.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witting W, Kwa IH, Eikelenboom P, Mirmiran M, Swaab DF, 1990. Alterations in the circadian rest-activity rhythm in aging and Alzheimer’s disease. Biological Psychiatry 27, 563–572. 10.1016/0006-3223(90)90523-5 [DOI] [PubMed] [Google Scholar]
- Woodruff ER, Chun LE, Hinds LR, Varra NM, Tirado D, Morton SJ, McClung CA, Spencer RL, 2018. Coordination between Prefrontal Cortex Clock Gene Expression and Corticosterone Contributes to Enhanced Conditioned Fear Extinction Recall. eNeuro 5, ENEURO.0455–18.2018. 10.1523/ENEURO.0455-18.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki S, Numano R, Abe M, Hida A, Takahashi R, Ueda M, Block GD, Sakaki Y, Menaker M, Tei H, 2000. Resetting Central and Peripheral Circadian Oscillators in Transgenic Rats. Science 288, 682–685. 10.1126/science.288.5466.682 [DOI] [PubMed] [Google Scholar]
- Yin JCP, Del Vecchio M, Zhou H, Tully T, 1995. CREB as a Memory Modulator: induced expression of a dCREB2 activator isoform enhances long-term memory in drosophila. Cell 81, 107–115. 10.1016/0092-8674(95)90375-5 [DOI] [PubMed] [Google Scholar]
- Yin JCP, Wallach JS, Del Vecchio M, Wilder EL, Zhou H, Quinn WG, Tully T, 1994. Induction of a dominant negative CREB transgene specifically blocks long-term memory in Drosophila. Cell 79, 49–58. 10.1016/0092-8674(94)90399-9 [DOI] [PubMed] [Google Scholar]
- Yoo S-H, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong H-K, Oh WJ, Yoo OJ, Menaker M, Takahashi JS, 2004. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A 101, 5339–5346. 10.1073/pnas.0308709101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngjohn JR, Crook TH, 1993. Learning, forgetting, and retrieval of everyday material across the adult life span. Journal of Clinical and Experimental Neuropsychology 15, 447–460. 10.1080/01688639308402570 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Won J, Karlsson MG, Zhou M, Rogerson T, J. B, Neve R, Poirazi P, Silva AJ, 2009. CREB regulates excitability and the allocation of memory to subsets of neurons in the amygdala. Nat Neurosci 12, 1438–1443. 10.1038/nn.2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Learning-induced changes in Per1 within the RSC and Fos within the SCN are greater during the day than at night. A. Direct comparison between day (ZT1, ZT5, and ZT9) and night (ZT13, ZT17, ZT21) RSC Per1 induction in young and aging mice. B. Direct comparison between day and night RSC Fos induction in young and aging mice. C. Direct comparison between day and night SCN Per1 induction in young and aging mice. D. Direct comparison between day and night SCN Fos induction in young and aging mice. These are additional analyses of data already appearing in Fig. 2 and Fig. 5. Note that the young RSC Per1 data presented here was previously published in the Neurobiology of Learning and Memory in 2021 as part of a different analysis (Urban et al., 2021).
Supplemental Figure 2. No HSV treatments had any effect on total distance traveled in any of the behavioral experiments. A. Total distance traveled each day for mice in the young RSC Per1 downregulation experiment (Fig. 3). B. Total distance traveled each day for mice in the aging RSC Per1 upregulation experiment (Fig. 4). C. Total distance traveled each day for mice in the young SCN Per1 downregulation experiment (Fig. 6).
Data Availability Statement
The data presented here are available from the corresponding author upon reasonable request.


