Abstract
Poxviruses are large double-stranded DNA viruses that encode their own DNA replication, transcription, and mRNA biogenesis machinery, which underlies their ability to replicate entirely in the cytoplasm. However, like all other viruses, poxviruses remain dependent on host ribosomes to translate their mRNAs into the viral proteins needed to complete their replication cycle. While earlier studies established a fundamental understanding of how poxviruses wrestle with their hosts for control of translation initiation and elongation factors that guide ribosome recruitment and mRNA decoding, recent work has begun to reveal the extent to which poxviruses directly target the ribosome itself. This review summarizes our current understanding of the regulation of ribosomes and translation in poxvirus infection.
Introduction
Poxviruses have a fascinating medical history and an equally fascinating replication cycle. Members of the poxviridae family include Variola virus, the causative agent of smallpox that killed more people than any other pathogen in history [1]. Smallpox was eradicated using the cross-protection afforded by infection with milder cowpox and Vaccinia (VacV) viruses, ushering in the era of modern vaccines and oncolytics. However, several new and zoonotic poxviruses are emerging as new pandemic threats. Monkeypox, for example, is capable of human-to-human transmission, resulting in outbreaks that are occurring with increasing frequency in regions of Africa, where the virus is naturally endemic, and with global travel, these are extending their reach to other parts of the world [2]. Indeed, the largest known global outbreak of monkeypox is occurring as we write this review.
Besides their medical history, the study of poxvirus replication in cultured cells played a critical role in the discovery of Guanosine-5’-triphosphate 5’-methyl-(GTP) capping, 2’-O-methylation, and 3’ polyadenylation that we now know to occur on most viral and cellular transcripts in order to control both mRNA stability and translation (reviewed in [3]). This is in part because of poxvirus’ remarkable self-sufficiency, encoding not only their own DNA replication and transcription machinery but also mRNA capping, decapping, and polyadenylation factors [1]. Soon after entry into the cell, viral cores begin to transcribe their early genes, which constitute approximately half of the 200 or so total genes encoded by poxviruses [4,5]. Early mRNAs are released into the cytosol where they are translated by the host protein synthesis machinery, with the encoded viral proteins then facilitating disassembly of the viral core and progression of the replication cycle [6,7]. Establishment of infection involves the formation of cytoplasmic ‘mini nuclei’ that are referred to as viral factories (VFs), where DNA replication and transcription of intermediate and late genes occurs. Because of their close kinetics and shared dependence on DNA replication for transcription, intermediate and late genes are often referred to cumulatively as postreplicative genes. While early mRNAs are structurally similar to host transcripts, postreplicative mRNAs have unique features that are tied to a switch in translational control strategies at later stages of infection.
Early infection: nothing too unusual yet
Early poxvirus mRNAs are structurally similar to their host counterparts, including the presence of a 5’-methyl-GTP cap along with a 3’-polyA-tail. Conventional loading of ribosomes onto such mRNAs involves recognition of the 5’ cap by the eukaryotic translation- initiation factor complex, eukaryotic initiation factor (eIF4F) [8,9] (Figure 1). eIF4F recruits the small 40S subunit of the ribosome through a large bridging complex called eIF3. In addition, eIF4F also provides the helicase activity that allows the 40S subunit to scan the mRNA 5’ untranslated region (UTR) in search of the start codon, which is usually a methionine-encoding adenine (A), uracil (U), guanine (G) (AUG). Start codon recognition involves a number of additional eIFs, including the eIF2 complex that carries the initiator Met-tRNA [9]. Once the start codon is recognized, the large 60S subunit of the ribosome then joins and initiates translation of the mRNA open-reading frame (ORF). As the ribosome moves along and reads the ORF, it undergoes large-scale swiveling of the 40S and 60S subunits, known as ratcheting. At the same time, a small number of eukaryotic elongation factors deliver aminoacylated tRNAs and facilitate the elongation phase that synthesizes the encoded protein. Once a stop codon is encountered, eukaryotic-release factors facilitate the recycling of ribosomes for a new round of translation.
Figure 1.
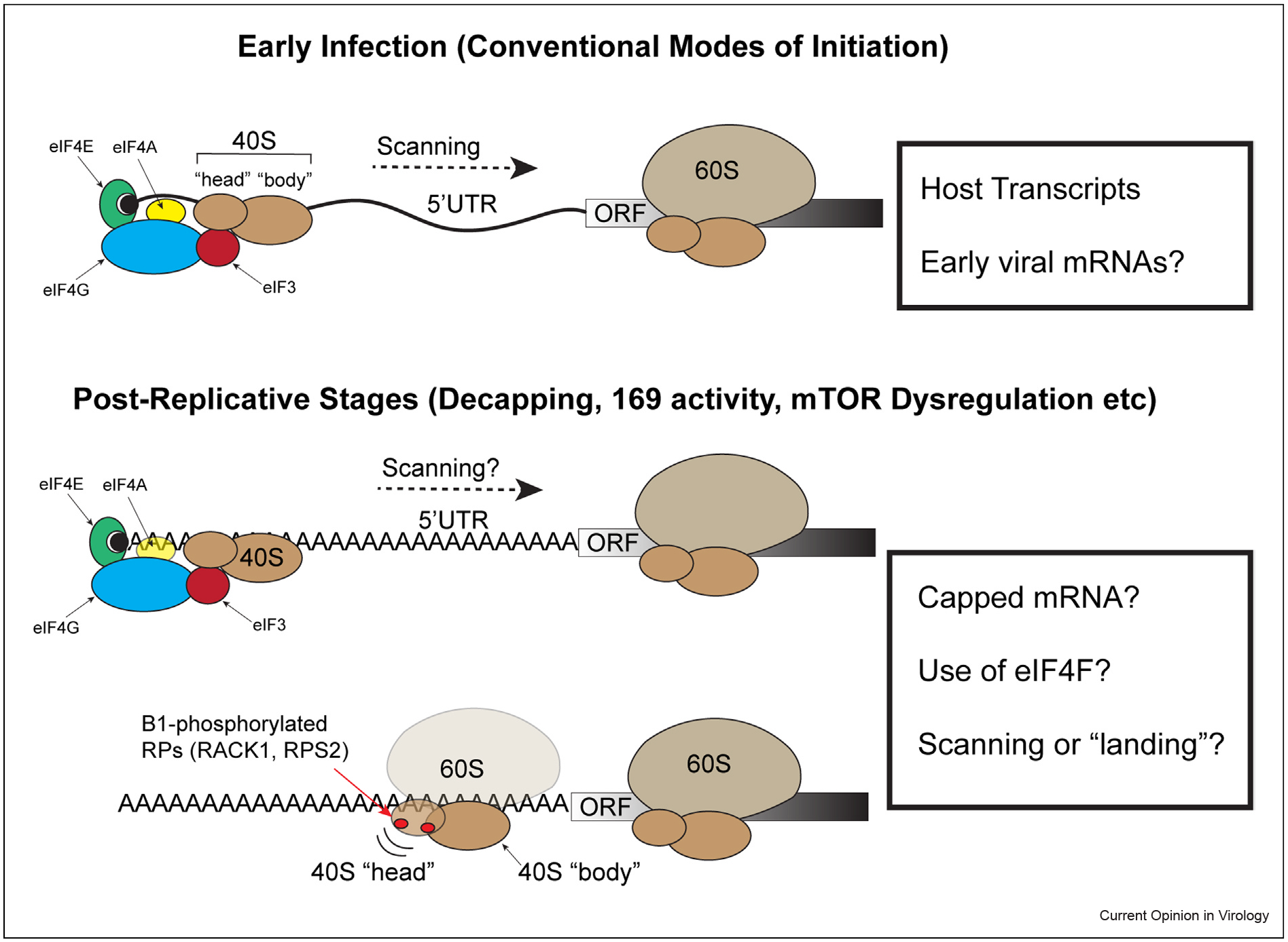
Illustration of key aspects of ribosome recruitment during early and postreplicative stages of poxvirus infection. Top: A typical host or early viral mRNA contains a 5’ 7-methyl-GTP ‘Cap’ (black circle) and an UTR that precedes the ORF. Recruitment of the ribosome to the correct end of the mRNA involves cap recognition by the eIF4E subunit of the eIF4F complex, which further consists of the scaffold protein eIF4G and the RNA helicase eIF4A. The eIF4G scaffold also binds the multi-subunit complex eIF3 that bridges to the 40S ribosomal subunit; the 40S complex contains 33 subunits that form ‘head’ and ‘body’ regions that can swivel as the 40S inspects the mRNA. The helicase activity of eIF4F facilitates 40S scanning of the UTR to find the start codon at which point the 60S ribosomal subunit joins to begin decoding the ORF. The length and structural complexity of the UTR influences both the degree of dependence on eIF4F helicase activity and translation efficiency of different mRNAs. While structurally similar and therefore likely to also require eIF4F and eIF3 activity, it has yet to be directly proven that early viral mRNAs are translated in a cap-dependent manner similar to their host. Bottom: Owing to the presence of viral proteins that drive decapping, mTOR dysregulation, and broader changes to the cell at later stages of infection, it remains unclear whether postreplicative viral mRNAs are capped or use eIFs for initiation, or whether they utilize mixed initiation strategies. The heterogeneous lengths and relatively short nature of the 5’ polyA-leaders found on postreplicative mRNAs also raises questions as to whether scanning is involved. Moreover, unique B1-driven phosphorylation events on small ribosomal proteins, in particular RACK1, alter the swivel motion of the 40S and reduce dependence of certain transcripts on the helicase activity of eIF4F. This likely supports cap-independent recruitment of the ribosome to the unstructured polyA-stretches that directly precede the start codon.
Translation of early poxvirus mRNAs appears to require many of the eIFs that mediate cap-dependent initiation on host mRNAs. Although initial studies suggested that proteolytic cleavage of the eIF4F scaffolding subunit, eIF4G only modestly reduces poxvirus translation [10], these experiments were performed in transformed cell lines that are often translationally hyperactivated and may therefore be less sensitive systems. Subsequent studies using primary normal human fibroblasts and mouse embryo fibroblasts found that inhibition or genetic deletion of the kinase that phosphorylates eIF4E, the cap-binding subunit of eIF4F, reduces VacV spread [11]. Moreover, poxviruses actively stimulate the formation of eIF4F complexes and phosphorylation of eIF4E in primary cells, which is normally associated with increased cap-dependent initiation [11]. Stimulation of eIF4F formation involves activation of mammalian/mechanistic Target of Rapamycin (mTOR), which inactivates small eIF4E-binding proteins that act as repressors of eIF4F assembly [11,12]. mTOR lies downstream of Phosphatidylinositol-3-kinase-AKT (PI3K–Akt) signaling, a pathway that is stimulated very early in infection through viral binding to cell surface receptors during entry [13]. Beyond eIF4F, several subunits of the eIF3 complex that functions in coordinating ribosome recruitment with eIF4F activity are also required for VacV protein synthesis [14]. However, it has yet to be proven that early viral mRNAs directly utilize eIF4F. Complicating our understanding of this, stimulating eIF4E phosphorylation also serves to increase the expression of host proteins that repress interferon production [15]. As such, it is also possible that poxviruses activate and require eIF4F in order to control host responses to infection rather than requiring eIF4F directly for their own translation, or perhaps the virus has coupled both processes. Beyond eIF4F and eIF3, virtually all mRNAs whether they be cellular or viral require eIF2-mediated start codon recognition [8]. This dependency forms the basis of a central line of cellular defense against many viruses, including poxviruses. Owing to transcriptional readthrough, poxviruses produce dsRNA that activates the cellular sensor Protein Kinase R (PKR), which phosphorylates and inactivates eIF2. While this inhibits both host and viral translation, this altruistic suicide quickly limits virus spread. To counter these defenses, poxviruses encode a range of proteins that limit dsRNA production and PKR activity [8,16].
The transition to late stages of replication: … things start to get strange!
As infection progresses to intermediate and late stages, VFs increase in size, shutoff of host translation occurs, and viral translation strategies become more unconventional, while direct targeting of the ribosome itself comes more into play. Several early viral proteins play a role in preparing for this switch as they obviously have to be expressed first to exert their effects. The viral 169 protein causes the accumulation of inactive 80S ribosomes, blocking initiation on a broad range of mRNAs, including the unusual dicistorvirus Internal Ribosome Entry Site (IRES) that initiates translation in the absence of all eIFs including eIF2 [17•]. Protein 169 appears to be excluded from VFs where postreplicative mRNAs are translated [18] and therefore contributes to selective suppression of host translation and antiviral responses [17•]. Whether and how early viral mRNAs are spared, or whether their abundance or kinetics of expression simply evade the effects of Protein 169 is unclear. However, such shutoff strategies are likely to intentionally suppress translation of early viral mRNAs to facilitate the transition to postreplicative phases of infection. Indeed, virally encoded decapping enzymes are also thought to contribute to smoother transitions in phases of virus replication. The decapping protein D9 is expressed early in infection, while the second decapping protein, D10, is expressed as a postreplicative gene. D9 and D10 contribute to both host shutoff and clearance of early viral mRNAs [19,20], as well as clearance of dsRNA to limit PKR activation [21,22•]. Whether and how late viral mRNAs are spared from D9/D10 decapping activity also remains unclear, but there appears to be at least some degree of selective targeting of spliced mRNAs by viral decapping enzymes, which may limit the effects on viral mRNAs that are not spliced [23•]. It has also been suggested that decapping enzymes are excluded from VFs [24]. However, viral transcripts do not appear to be completely spared from decapping and there is growing evidence that postreplicative mRNAs can initiate in a manner that has reduced dependence on cap-binding eIF4E [25•]. Although eIF4E and eIF4G are recruited to VFs where late viral protein synthesis occurs [11,18], in light of more recent findings, it is possible that this recruitment serves to sequester eIFs from the host as part of the poxvirus’ broader shutoff strategy rather than to stimulate viral protein synthesis directly. Adding to this complexity, D9 and in particular the postreplicative decapping protein D10 actually stimulate translation, with more robust effects reported for postreplicate mRNAs that harbor unusual 5’ polyA-leaders [26•]. As discussed below, these and other recent findings have begun to shed light on the functions of these once-enigmatic leaders.
The ability of postreplicate mRNAs to initiate in both cap-dependent and cap-independent manners may give poxviruses the flexibility to replicate under a variety of environmental conditions. Interestingly, as infection progresses, the viral F17 protein targets mTOR in a unique manner. Unlike other viruses that either stimulate or repress mTOR activity by targeting upstream signaling components [8], F17 binds and sequesters Raptor and Rictor, which are key regulatory subunits of the two mTOR complexes mTORC1 and mTORC2, respectively [27,28]. This dysregulates mTORC1–mTORC2 cross-talk to evade cytosolic sensing by cGAS, but also results in cell-type- and cell state-dependent activation or repression of mTOR’s substrates that regulate cap-dependent translation [28]. The ability of postreplicative mRNAs to utilize either mode of initiation likely allows poxviruses to deal with these variable outcomes of mTOR dysregulation in different cell types, along with the production of decapping proteins and the induction of cellular stresses that impair canonical modes of ribosome recruitment.
Another early viral protein, the B1 kinase, phosphorylates a number of ribosomal proteins that then function in this later-stage switch in translation strategies. While it was known for several decades that VacV phosphorylates several ribosomal subunit proteins (RPs) (reviewed in [3]), it was only recently that the precise sites and the functionality of these modifications have been uncovered. By isolating ribosomes from cells infected with a variety of DNA or RNA viruses and conducting mass spectrometry, studies have shown that VacV B1 uniquely phosphorylates residues in the small RP, RPS2, which lies at the mRNA entry channel, and Receptor for Activated C Kinase 1 (RACK1), which lies at the mRNA exit channel [29•,30••] (Figure 1). Both of these phosphorylation events stimulate translation of postreplicative viral mRNAs or reporters harboring polyA-leaders. While the mechanistic basis by which RPS2 phosphorylation regulates translation remains unknown, the manner in which RACK1 is modified by VacV provided unexpected insights into ribosome diversification across species [30••,31] and coincided with a flurry of discoveries in the field of ribosome-mediated quality control (RQC), which also centers around polyA elements [32,33].
Poxviruses and PolyA … perhaps pretending to be protists?
Poxvirus’ use of polyA-leaders is unusual because their mammalian hosts almost exclusively limit adenosine homopolymers to the 3’ UTR in the form of the polyA-tail. The polyA-tail plays roles in mRNA stability and in stimulating cap-dependent translation initiation by binding PolyA-binding proteins [8,9]. PolyA-stretches also play a third, crucial role in alerting the cell to aberrant translation events or improperly processed transcripts. For example, if an mRNA is accidentally internally polyadenylated, or if a stop codon is missing or skipped by a ribosomal frameshift and decoding continues through to the natural polyA-tail, the ribosome begins to slide and ultimately stalls on the adenosine stretches. Trailing ribosomes then collide with stalled leading ribosomes, which triggers regulatory monoubiquitination of Phosphatidylinositol-3-kinase-AKT (RPSs), including RPS10 and RPS20, by the E3 ligase ZNF598 [34–36]. This in turn elicits the broader RQC response whereby the stalled ribosomes are split and recycled back to their 40S and 60S subunits, and the aberrant mRNA and peptide are targeted for destruction [32,33]. In the absence of RQC regulatory factors such as ZNF598, collided ribosomes persist and ultimately move through the polyA stretch, resulting in its decoding rather than RQC activation. In terms of their roles in viral infection, ZNF598 independently regulates Retinoic acid-inducible gene (RIG-I)-mediated interferon production [37,38•], but by using interferon-defective cell lines, a specific role for ZNF598 in poxvirus protein synthesis was identified [38•]. This earlier study used a limited analysis of intermediate and late proteins and proposed that beyond RQC, ZNF598 and monoubiquitination of RPS20 might also be involved in regulating translation of 5’ polyA-mRNAs [38•]. However, a subsequent proteomic study showed that ZNF598 and RPS20 monoubiquitination are more broadly required for both early and late VacV proteins [39•]. Interestingly, although sustained throughout infection, maximal ubiquitination of RPS20 is reached within 1 h of infection, long before early viral proteins are detectable [39•]. How ZNF598 is activated so rapidly remains unknown, but based on ZNF598 insufficiency, it appears that overall, poxvirus protein synthesis may benefit indirectly from cellular RQC activity that recycles stalled ribosomes in order to sustain a sufficient pool of active ribosomes for viral protein synthesis [38•,39•] (Figure 2). Curiously, in cells expressing RQC reporter constructs wherein either a 60-nucleotide control sequence or a polyA linker are present between green fluorescent protein (GFP) and red fluorescent protein cherry Fluorescent Protein (ChFP/RFP), infection increases the relative amounts of RFP to GFP made by polyA-stall reporters at late (16 h) timepoints [39•]. Normally RQC activity reduces RFP expression on these polyA-stall reporters, and restoration of RFP expression is indicative of polyA-readthrough due to reduced ZNF598 or RQC activity (Figure 2a–c). As such, the late-stage increase in RFP expression appears to be kinetically and functionally distinct from the early activation of ZNF598-mediated stall resolution and ribosome recycling that should reduce RFP production. The authors suggested that VacV may sequester ZNF598 from the host and therefore also from these reporters, while utilizing it for viral protein synthesis [39•]. While this is a valid hypothesis that remains to be tested, phosphorylation of RACK1 and broader changes in modes of translation at late stages of infection may offer an alternative explanation for this more unusual readout from these polyA reporters.
Figure 2.
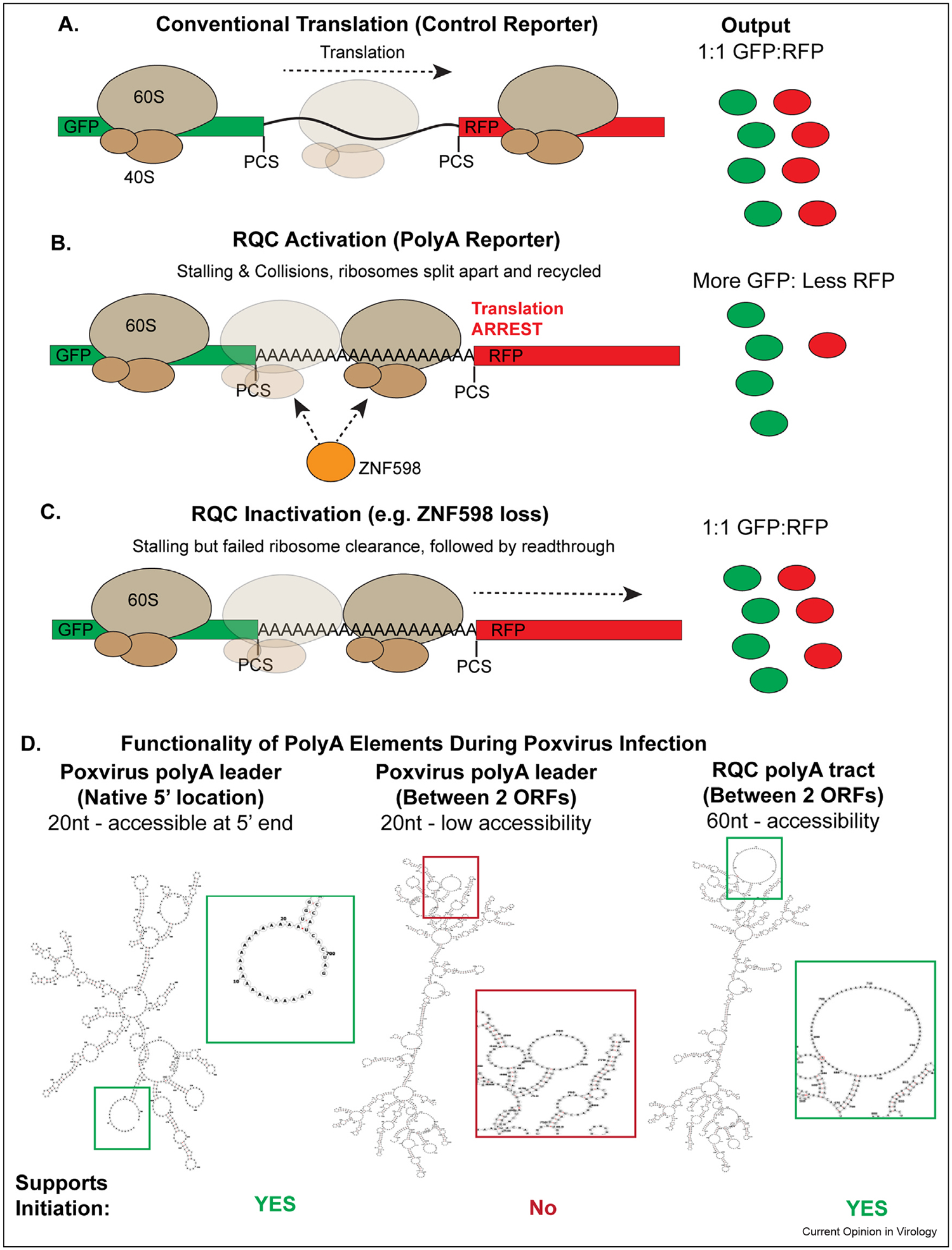
A model for RQC and polyA element activities during poxvirus infection. (a–c) RQC reporters consist of GFP and RFP (or mCherry) separated by a nucleotide test sequence. In addition, GFP and RFP are flanked by protease cleavage sites (PCS) so that GFP or RFP production are not interdependent and GFP can still be made when stalling and RQC occur. (a) Conventional translation of a reporter with a control linker sequence produces equal GFP and RFP. (b) The presence of a 60-nt polyA linker sequence causes ribosomes to stall and collide. ZNF598 senses collisions and through monoubiquitylation, triggers release of the stalled ribosomes to be recycled. As a result, cells make GFP but very little RFP due to the termination of translation of the reporter on the polyA sequence. (c) In the absence of RQC activity, such as with loss of ZNF598, ribosomes stall but eventually move through the polyA tract to continue translation of the internally polyadenylated mRNA. As a result, levels of GFP and RFP normalize to those of control reporters. While it remains unclear how RQC activity is stimulated during early poxvirus infection, the recycling of stalled ribosomes driven by RQC factors likely ensures a sufficient pool of active ribosomes to support the high levels of viral protein synthesis that occurs. (d) A model for differences in how polyA sequences and reporters behave in poxvirus-infected cells. Note that poxvirus polyA-leaders average 10–40 nts and their functionality as an IRES has been performed using representative 20-nt elements. By contrast, RQC reporters use 60-nt polyA-stretches to induce ribosome stalling. RNAfold (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi.) was used to model the effects of these fundamental differences in polyA lengths, using GFP as a model ORF. Boxed regions zoom in on the modeled polyA sequence for each panel. Left: Native poxvirus leaders are short but are located at the free 5’ end of the mRNA, making them readily accessible to ‘landing’ by poxvirus-modified ribosomes. Middle: The 20-nt poxvirus leader modeled between two GFP ORFs forms a small and likely poorly accessible region for ribosomes attempting to land internally. They therefore likely lack IRES functionality in bicistronic reporter assays. Right: The 60-nt polyA element used in RQC studies forms a large unstructured element that is likely more accessible to poxvirus-modified ribosomes to randomly initiate internally as well as normally at the 5’UTR, sustaining higher levels of RFP than GFP expression. While different reporter setups are used in different studies, the simplest explanation for the different functionalities observed to date is accessibility of the polyA tracts based on their sizes and locations.
RACK1 is a multifunctional protein that directly regulates translation, mediates signaling to and from the ribosome, and also plays a role in RQC [40]. Although RACK1 has been implicated in facilitating ZNF598-mediated modifications to RPs [34], RACK1 phosphorylation occurs at later stages of poxvirus infection and RACK1 appears to be specifically required for postreplicative viral protein synthesis [30••,41••,42•]. RACK1 has also been shown to be required for c-Jun N-terminal kinase (JNK) and p38 activation in response to ribosome collisions, which occur as part of RQC and the broader ribotoxic stress response (RSR) [33,41••,43]. While poxvirus infection also activates JNK and p38, again unlike RQC and RPS20 ubiquitination, this occurs later in infection and requires DNA replication for maximum activation [44]. This activation has been linked to host responses to infection [45,46], and using genetic knockouts of RACK1, it was found that unlike RSR signaling, RACK1 is not essential for JNK activation in poxvirus-infected cells [42•]. Cumulatively, this suggests that RACK1’s roles in stress signaling and RQC are not a predominant aspect of its role in infection, which seems more specific to postreplicative mRNA translation.
Beyond signaling, RACK1 also directly regulates translation and has been implicated in selective translation of specific classes of mRNAs, notably including initiation on RNA virus mRNAs that contain cap-independent IRES elements [47–49]. Although poxvirus polyA-leaders do not have bona fide IRES activity, in vitro studies have shown that they reduce the dependency of mRNAs on eIFs, likely due to their lack of structural complexity [50]. However, the activity of polyA-leaders is greatly enhanced in poxvirus-infected cells and this enhancer activity requires RACK1 and the viral B1 kinase that phosphorylates RACK1 [25•,30••]. Intriguingly, 5’ polyA-leaders are not natural in mammalian cells but they are utilized by protists and dicot plants [31]. Moreover, poxviruses produce polyA-leaders in a nontemplated fashion that involves slippage of the viral RNA polymerase, but why they do this remained enigmatic for several decades (reviewed in [3]). The poxvirus B1 kinase phosphorylates Serine 278 within a variable linker that lies between conserved β-propeller blades in RACK1 [30••]. Extensive phylogenetic comparisons revealed that this linker region contains negatively charged residues in protists and dicot plants but not in mammals, and expression of either poxvirus S278E-phosphomimetic RACK1 or chimeric human RACK1 with a dicot plant linker (which is also common in protists) enhances translation of polyA-leaders similarly [30••,31]. This suggests that upon infection, the poxvirus B1 kinase converts their mammalian host’s ribosomes into a plant/protist-like state to maximize translation of their non-native 5’ polyA-mRNAs. While there are no plant poxviruses, there are suggestions that mammalian poxviruses originated from a protist ancestor [51]. This raises the intriguing hypothesis that ancestral poxviruses evolved to produce polyA-leaders in a host that naturally utilized similar leaders, but upon jumping to mammals, poxviruses evolved to recreate this optimal environment through phosphorylation of mammalian RACK1.
Biochemical and structural studies have also shed light on how RACK1 phosphorylation regulates ribosome function and translation. Through structure modeling and binding assays using chimeric linkers with various charge organizations, it became evident that single or spaced negative charge increases electrostatic forces in the head domain of the 40S ribosome but with minimal impact on the overall affinity of RACK1 for the ribosome [31]. By contrast, while clustered charge organizations retain the ability to enhance polyA-leader mRNA translation, this comes at a cost of reduced affinity for the ribosome. With most species that encode polyA-leaders using either single or spaced RACK1 linker charge organizations [31], the introduction of a single phosphate into the shorter human RACK1 linker demonstrates the exquisite precision with which poxviruses mimic other species to optimize electrostatic forces in the 40S head domain. Recent studies using RACK1-knockout cells rescued with either a wildtype or poxvirus S278E-phosphomimetic RACK1 further revealed that these electrostatic forces alter the swivel motion of the 40S head domain, an effect that is also observed upon 40S binding to type-III/-IV IRES elements found in some RNA viruses [41••]. Moreover, in cells expressing poxvirus phosphomimetic RACK1 subsets of cellular mRNAs become resistant to various translation inhibitors, including those targeting eIF4A, the helicase subunit of eIF4F that is critical to cap-dependent translation of cellular mRNAs under normal conditions [41••]. This suggests that in the absence of direct IRES-like activity of polyA-leaders themselves [25•,50], phosphorylation of the RACK1 linker performs the 40S remodeling function that is often associated with IRES elements. These findings also show that while RACK1 phosphorylation selectively enhances translation of certain transcripts, it is not repressive and therefore likely does not contribute to virus-induced host shutoff. Moreover, its enhancer activity is not exclusive to polyA-leaders. Instead, RACK1 phosphorylation likely supports a less complex mode of ribosome recruitment to mRNAs with unstructured 5’ UTRs that include 5’ polyA-leaders, akin to bacterial ribosome landing on Shine–Delgarno sequences. This is in line with reports that despite host shutoff, some host mRNAs are translationally upregulated during infection and these too have shorter, less structured 5’ UTRs [52]. Intriguingly, in testing GFP-polyA-RFP reporters that are used to study RQC and stalling described above, studies showed that GFP production declined, while RFP production was either sustained or increased specifically in cells expressing phosphomimetic RACK1 [41••]. This is the opposite to what happens when ribosomes stall and produce less RFP, or when readthrough occurs upon ZNF598 depletion where GFP:RFP ratios normalize (Figure 2a–c). This suggests that phosphomimetic RACK1 may bypass GFP and support cap-independent internal initiation on the long polyA sequences in these reporters to produce RFP [41••]. As discussed above in relation to ZNF598 and RQC, recent studies in infected cells using the same reporters showed a similar increase in RFP production that alters RFP:GFP ratios specifically at late stages of infection [39•]. While this was interpreted as a reduction in RQC activity, it is also possible that this reflects the switch to alternative cap-independent initiation on polyA elements that occurs at postreplicative stages of infection, or perhaps a combination of both events. While shorter polyA elements found on the free 5’ end of viral mRNAs do not have inherent IRES activity when inserted into bicistronic reporters [25•], it is quite possible that the longer 60-nucleotide polyA elements used in RQC reporters do, at least when coupled with phosphorylated RACK1 that allows the ribosome to adopt this landing behavior (Figure 2d).
Concluding remarks
We now have a considerable understanding of how poxviruses target the host protein synthesis machinery, including ribosomes themselves in order to control translation at various stages of their replication. However, many outstanding questions remain to be addressed. Although likely, it is still uncertain whether early viral mRNAs truly require eIF4F directly or whether viral manipulation of and requirements for eIF4F activity center more around control of host responses to infection. It would also be interesting to determine how RQC is activated so early in infection and precisely how factors such as ZNF598 impact both early and postreplicative phases of viral protein synthesis. While we now have a better understanding of how poxviruses both produce their unusual 5’ polyA-leaders and regulate translation of postreplicative mRNAs, there are still many avenues to explore in terms of the finer mechanistic details and broader factors involved. Beyond the fascinating cell biology that underlies poxvirus replication and the continually surprising insights they provide into fundamental aspects of cell biology, the recent outbreak of monkeypox underscores the importance of poxvirus research.
Acknowledgements
This work was supported by funding from the National Institutes of Health, USA to D.W. [Grant number R01 AI127456]. We apologize to authors whose work could not be cited due to reference limitations.
Footnotes
Conflict of interest statement
The authors declare no conflicts of interest associated with this work.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest.
- 1.Moss B: Poxviridae: the viruses and their replication. In Fields Virology. Edited by Knipe DM, Howley PM. Lippincott Williams & Wilkins; 2007:2849–2883. [Google Scholar]
- 2.Simpson K, et al. : Human monkeypox - after 40 years, an unintended consequence of smallpox eradication. Vaccine 2020, 38:5077–5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meade N, DiGiuseppe S, Walsh D: Translational control during poxvirus infection. Wiley Inter Rev RNA 2019, 10:e1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang Z, et al. : Expression profiling of the intermediate and late stages of poxvirus replication. J Virol 2011, 85:9899–9908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang Z, et al. : Simultaneous high-resolution analysis of vaccinia virus and host cell transcriptomes by deep RNA sequencing. Proc Natl Acad Sci USA 2010, 107:11513–11518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kilcher S, et al. : siRNA screen of early poxvirus genes identifies the AAA+ ATPase D5 as the virus genome-uncoating factor. Cell Host Microbe 2014, 15:103–112. [DOI] [PubMed] [Google Scholar]
- 7.Liu B, et al. : Identification of poxvirus genome uncoating and DNA replication factors with mutually redundant roles. J Virol 2018, 92:e02152–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jan E, Mohr I, Walsh D: A cap-to-tail guide to mRNA translation strategies in virus-infected cells. Annu Rev Virol 2016, 3:283–307. [DOI] [PubMed] [Google Scholar]
- 9.Hershey JWB, Sonenberg N, Mathews MB: Principles of translational control. Cold Spring Harb Perspect Biol 2019, 11:a011528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mulder J, et al. : Vaccinia virus protein synthesis has a low requirement for the intact translation initiation factor eIF4F, the cap-binding complex, within infected cells. J Virol 1998, 72:8813–8819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walsh D, et al. : Eukaryotic translation initiation factor 4F architectural alterations accompany translation initiation factor redistribution in poxvirus-infected cells. Mol Cell Biol 2008, 28:2648–2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zaborowska I, Walsh D: PI3K signaling regulates rapamycin-insensitive translation initiation complex formation in vaccinia virus-infected cells. J Virol 2009, 83:3988–3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soares JA, et al. : Activation of the PI3K/Akt pathway early during vaccinia and cowpox virus infections is required for both host survival and viral replication. J Virol 2009, 83:6883–6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walsh D, Mohr I: Coupling 40S ribosome recruitment to modification of a cap-binding initiation factor by eIF3 subunit e. Genes Dev 2014, 28:835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herdy B, et al. : Translational control of the activation of transcription factor NF-kappaB and production of type I interferon by phosphorylation of the translation factor eIF4E. Nat Immunol 2012, 13:543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rothenburg S, Brennan G: Species-specific host-virus interactions: implications for viral host range and virulence. Trends Microbiol 2020, 28:46–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.•.Strnadova P, et al. : Inhibition of translation initiation by protein 169: a vaccinia virus strategy to suppress innate and adaptive immunity and alter virus virulence. PLoS Pathog 2015, 11:e1005151. [DOI] [PMC free article] [PubMed] [Google Scholar]; Revealed an unusual strategy to target and inactivate ribosomes to counter host responses to infection.
- 18.Katsafanas GC, Moss B: Colocalization of transcription and translation within cytoplasmic poxvirus factories coordinates viral expression and subjugates host functions. Cell Host Microbe 2007, 2:221–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parrish S, Resch W, Moss B: Vaccinia virus D10 protein has mRNA decapping activity, providing a mechanism for control of host and viral gene expression. Proc Natl Acad Sci USA 2007, 104:2139–2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parrish S, Moss B: Characterization of a second vaccinia virus mRNA-decapping enzyme conserved in poxviruses. J Virol 2007, 81:12973–12978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu SW, et al. : Poxvirus decapping enzymes enhance virulence by preventing the accumulation of dsRNA and the induction of innate antiviral responses. Cell Host Microbe 2015, 17:320–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.•.Burgess HM, Mohr I: Cellular 5’−3’ mRNA exonuclease Xrn1 controls double-stranded RNA accumulation and anti-viral responses. Cell Host Microbe 2015, 17:332–344. [DOI] [PMC free article] [PubMed] [Google Scholar]; Together with Reference 23, revealed that poxvirus decapping enzymes function to control the accumulation of dsRNA that activates PKR responses.
- 23.•.Ly M, et al. : Vaccinia virus D10 has broad decapping activity that is regulated by mRNA splicing. PLoS Pathog 2022, 18:e1010099. [DOI] [PMC free article] [PubMed] [Google Scholar]; Together with Reference 22, revealed that poxvirus decapping enzymes function to control the accumulation of dsRNA that activates PKR responses.
- 24.Cao S, et al. : A poxvirus decapping enzyme colocalizes with mitochondria to regulate RNA metabolism and translation and promote viral replication. mBio (3) 2022, 13:e00300–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.•.Dhungel P, Cao S, Yang Z: The 5’-poly(A) leader of poxvirus mRNA confers a translational advantage that can be achieved in cells with impaired cap-dependent translation. PLoS Pathog 2017, 13:e1006602. [DOI] [PMC free article] [PubMed] [Google Scholar]; Together with Reference 30, revealed that poxvirus infection activates translation of 5’ polyA-mRNAs with reduced dependence on host initiation factors.
- 26.•.Cantu F, et al. : Poxvirus-encoded decapping enzymes promote selective translation of viral mRNAs. PLoS Pathog 2020, 16:e1008926. [DOI] [PMC free article] [PubMed] [Google Scholar]; Revealed that poxvirus decapping enzymes enhance translation of 5’ polyA-mRNAs.
- 27.Meade N, et al. : Poxviruses evade cytosolic sensing through disruption of an mTORC1-mTORC2 regulatory circuit. Cell 2018, 174:1143–1157 e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meade N, et al. : mTOR dysregulation by vaccinia virus F17 controls multiple processes with varying roles in infection. J Virol 2019, 93:e00784–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.•.DiGiuseppe S, et al. : Proteomic and mechanistic dissection of the poxvirus-customized ribosome. J Cell Sci 2020, 134:jcs246603. [DOI] [PMC free article] [PubMed] [Google Scholar]; Using mass spectrometry, this study identified unique phosphorylation events on ribosomal proteins in poxvirus-infected cells and uncovered a role for RPS2 phosphorylation.
- 30.••.Jha S, et al. : Trans-kingdom mimicry underlies ribosome customization by a poxvirus kinase. Nature 2017, 546:651–655. [DOI] [PMC free article] [PubMed] [Google Scholar]; Together with Reference 25, revealed how poxviruses stimulate translation of mRNAs with 5’ polyA leaders, including through phosphorylation of RACK1.
- 31.Rollins MG, et al. : RACK1 evolved species-specific multifunctionality in translational control through sequence plasticity within a loop domain. J Cell Sci 2019, 132:jcs228908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Joazeiro CAP: Mechanisms and functions of ribosome-associated protein quality control. Nat Rev Mol Cell Biol 2019, 20:368–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.D’Orazio KN, Green R: Ribosome states signal RNA quality control. Mol Cell 2021, 81:1372–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sundaramoorthy E, et al. : ZNF598 and RACK1 regulate mammalian ribosome-associated quality control function by mediating regulatory 40S ribosomal ubiquitylation. Mol Cell 2017, 65:751–760 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garzia A, et al. : The E3 ubiquitin ligase and RNA-binding protein ZNF598 orchestrates ribosome quality control of premature polyadenylated mRNAs. Nat Commun 2017, 8:16056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Juszkiewicz S, Hegde RS: Initiation of quality control during poly (A) translation requires site-specific ribosome ubiquitination. Mol Cell 2017, 65:743–750 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang G, et al. : Attenuation of the innate immune response against viral infection due to ZNF598-promoted binding of FAT10 to RIG-I. Cell Rep 2019, 28:1961–1970 e4. [DOI] [PubMed] [Google Scholar]
- 38.•.DiGiuseppe S, et al. : ZNF598 plays distinct roles in interferon-stimulated gene expression and poxvirus protein synthesis. Cell Rep 2018, 23:1249–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]; Together with reference 39, this study revealed the importance of ZNF598 to poxvirus protein synthesis.
- 39.•.Sundaramoorthy E, et al. : Ribosome quality control activity potentiates vaccinia virus protein synthesis during infection. J Cell Sci 2021, 134:jcs257188. [DOI] [PMC free article] [PubMed] [Google Scholar]; Together with reference 38, this study confirmed the importance of ZNF598 to poxvirus protein synthesis. However, through mass spectrometry this study also revealed that ZNF598 does not just regulate translation of 5’ polyA-mRNAs but is, instead, broadly required for poxvirus translation.
- 40.Adams DR, Ron D, Kiely PA: RACK1, A multifaceted scaffolding protein: structure and function. Cell Commun Signal 2011, 9:1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.••.Rollins MG, et al. : Negative charge in the RACK1 loop broadens the translational capacity of the human ribosome. Cell Rep 2021, 36:109663. [DOI] [PMC free article] [PubMed] [Google Scholar]; Using phosphomimetics of poxvirus-modified RACK1, this studied revealed structural changes in the ribosome including altered swivel motion in the 40S head domain that selectively regulate translation.
- 42.•.Park C, Walsh D: RACK1 regulates poxvirus protein synthesis independently of its role in ribosome-based stress signaling. J Virol 2022, 96:e0109322. [DOI] [PMC free article] [PubMed] [Google Scholar]; This studied revealed that RACK1’s role in ribosome-based JNK stress signaling is not directly involved in regulating poxvirus protein sythesis.
- 43.Kim TS, et al. : JNK activation induced by ribotoxic stress is initiated from 80S monosomes but not polysomes. BMB Rep 2019, 52:502–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pereira AC, et al. : A vaccinia virus-driven interplay between the MKK4/7-JNK1/2 pathway and cytoskeleton reorganization. J Virol 2012, 86:172–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang P, et al. : Protein kinase PKR-dependent activation of mitogen-activated protein kinases occurs through mitochondrial adapter IPS-1 and is antagonized by vaccinia virus E3L. J Virol 2009, 83:5718–5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taghavi N, Samuel CE: Protein kinase PKR catalytic activity is required for the PKR-dependent activation of mitogen-activated protein kinases and amplification of interferon beta induction following virus infection. Virology 2012, 427:208–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.LaFontaine E, et al. : Ribosomal protein RACK1 enhances translation of poliovirus and other viral IRESs. Virology 2020, 545:53–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Majzoub K, et al. : RACK1 controls IRES-mediated translation of viruses. Cell 2014, 159:1086–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu Y, et al. : Mass spectrometric analysis of the human 40S ribosomal subunit: native and HCV IRES-bound complexes. Protein Sci 2005, 14:1438–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shirokikh NE, Spirin AS: Poly(A) leader of eukaryotic mRNA bypasses the dependence of translation on initiation factors. Proc Natl Acad Sci USA 2008, 105:10738–10743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koonin EV, Yutin N: Evolution of the large nucleocytoplasmic DNA viruses of eukaryotes and convergent origins of viral gigantism. Adv Virus Res 2019, 103:167–202. [DOI] [PubMed] [Google Scholar]
- 52.Dai A, et al. : Ribosome profiling reveals translational upregulation of cellular oxidative phosphorylation mRNAs during vaccinia virus-induced host shutoff. J Virol 2017, 91:e01858–16. [DOI] [PMC free article] [PubMed] [Google Scholar]


