Abstract
Ovulation is an integral part of women’s menstrual cycle and fertility. Understanding the mechanisms of ovulation has broad implications for the treatment of anovulatory diseases and the development of novel contraceptives. Now, few studies have developed effective models that both faithfully recapitulate the hallmarks of ovulation and possess scalability. We established a three-dimensional encapsulated in vitro follicle growth (eIVFG) system that recapitulates folliculogenesis and produces follicles that undergo ovulation in a controlled manner. Here, we determined whether ex vivo ovulation preserves molecular signatures of ovulation and demonstrated its use in discovering novel ovulatory pathways and nonhormonal contraceptive candidates through a high-throughput ovulation screening. Mature murine follicles from eIVFG were induced to ovulate ex vivo using human chorionic gonadotropin and collected at 0, 1, 4, and 8 hours post-induction. Phenotypic analyses confirmed key ovulatory events, including cumulus expansion, oocyte maturation, follicle rupture, and luteinization. Single-follicle RNA-sequencing analysis revealed the preservation of ovulatory genes and dynamic transcriptomic profiles and signaling. Soft clustering identified distinct gene expression patterns and new pathways that may critically regulate ovulation. We further used this ex vivo ovulation system to screen 21 compounds targeting established and newly identified ovulatory pathways. We discovered that proprotein convertases activate gelatinases to sustain follicle rupture and do not regulate luteinization and progesterone secretion. Together, our ex vivo ovulation system preserves molecular signatures of ovulation, presenting a new powerful tool for studying ovulation and anovulatory diseases as well as for establishing a high-throughput ovulation screening to identify novel nonhormonal contraceptives for women.
Keywords: ovulation, RNA-sequencing, soft-clustering, proprotein convertase, nonhormonal contraceptive
Ex vivo ovulation enables novel contraception discovery for women.
Graphical Abstract
Graphical Abstract.
Introduction
Ovulation, an essential step of mammalian reproduction, is the process by which a fully grown ovarian antral follicle ruptures to release a fertilizable egg in response to hormonal cues. Ovulation has significant clinical ramifications for fertility, endocrine cyclicity, and contraception. Approximately 10–15% of reproductive-aged women are infertile, with anovulation as the leading cause and accounting for 22–42% of infertility cases [1]. Defective ovulation is also a key pathological feature of polycystic ovarian syndrome and luteinized unruptured follicle syndrome, two common ovarian conditions that impact numerous adolescent and reproductive-aged women [2, 3]. The popular hormonal birth control pills for women primarily contain progestin, a synthetic progesterone, to block ovulation; however, side effects can be serious, including the development of hormone-related cancers, stroke, depression, irregular periods, and obesity, among others [4, 5]. These side effects can lead to non-use or discontinuation. Thus, there is an urgent need to develop novel nonhormonal contraceptives for women. Central to contraceptive development and understanding ovulatory disease is creating and leveraging biologically faithful ovulatory systems.
In each ovarian cycle, a surge of the luteinizing hormone (LH) from the anterior pituitary stimulates one or multiple preovulatory follicles, depending on the species, to undergo a series of molecular and cellular events, including cumulus cell expansion, oocyte maturation, follicle rupture, and luteinization [6]. As a key stimulus, LH binds to luteinizing hormone/choriogonadotropin receptor (LHCGR) in the granulosa cells of preovulatory follicles to activate several essential signaling molecules to drive ovulation, such as the epidermal growth factor receptor (EGFR) [7], extracellular signal-regulated kinase 1 and 2 (ERK1/2) [8], and progesterone receptor (PGR) [9]. Despite the remarkable advances that have been made in studying these molecules and associated signaling pathways, our understanding of the dynamic and complex process of ovulation is limited due to the difficulty in studying ovulation in vivo. Culture of cumulus-oocyte complexes (COCs) has been used to study ovulation, and this method also has a high potential for large-scale ovulation screening [10]. However, culturing COCs does not adequately replicate follicle rupture and the paracrine communication between mural and cumulus granulosa cells, thereby limiting its use in studying ovulation. Thus, the majority of methods used to date for studying ovulation are not well-suited for scalable drug screening to target anovulatory diseases and identify novel nonhormonal contraceptives for women.
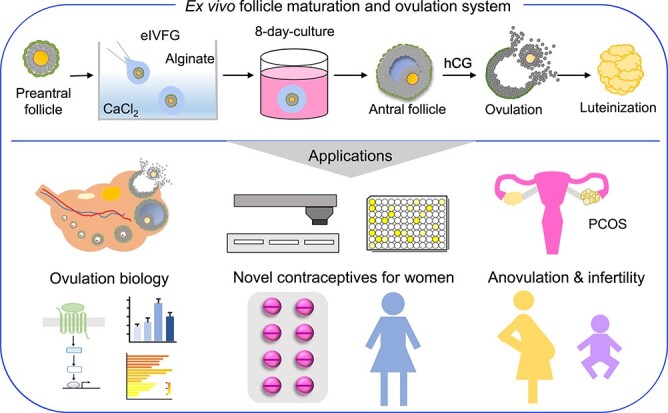
We previously established an alginate hydrogel-based encapsulated in vitro follicle growth (eIVFG) system, which has been used successfully to culture follicles from multiple mammalian species in vitro [11–13]. Alginate encapsulation maintains the three-dimensional (3D) architecture of follicles with the oocyte completely surrounded by companion granulosa cells. This system supports follicle growth from the preantral to antral stages. Mouse follicles derived from eIVFG undergo dynamic changes in gene expression essential for follicle-stimulating hormone (FSH)-dependent follicle maturation, such as genes related to ovarian steroidogenesis and induction of LHCGR in mural granulosa cells [14, 15]. In response to stimulation by human chorionic gonadotropin (hCG), an LH analog, antral follicles derived from eIVFG are able to undergo follicular rupture and release fertilizable eggs arrested at the metaphase of meiosis II [11–13]. These results demonstrate that ovulation can occur outside of the context of the ovary and the systemic environment of the body. In this study, we aimed to further investigate whether these eIVFG-derived follicles preserve the molecular signatures of ovulation in vivo, thus enabling a truly scalable and highly controllable system for studying ovulation biology as well as screening therapeutic and contraceptive candidates.
Results
eIVFG recapitulates key events of follicle development, ovarian steroidogenesis, ovulation, and luteinization
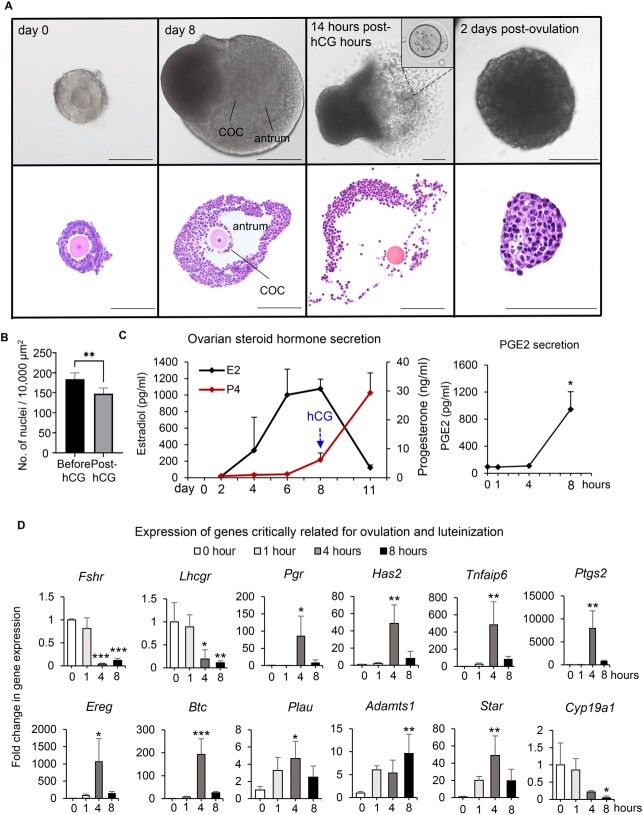
We first cultured immature mouse follicles to determine whether eIVFG preserves structural and hormonal features of folliculogenesis as they occur in vivo. Bright-field and histological follicle images showed that alginate encapsulation maintained the 3D architecture of follicles throughout growth from the secondary stage on day 0 to the antral stage on day 8 (Figure 1A). Antral follicles were characterized by the presence of a fluid-filled cavity and the differentiation of granulosa cells into the mural granulosa cells that form the follicular wall and the cumulus cells that surround the oocyte (Figure 1A). Upon stimulation with hCG on day 8, antral follicles ruptured with expanded COCs (Figure 1A). When ovulated follicles were cultured for an additional 2 days, follicular cells hypertrophied with increased cytoplasmic volume and formed a corpus luteum (CL)-like structure (Figure 1A and B).
Figure 1.
Mouse follicles grown within the eIVFG system recapitulate key events of follicle development, hormone secretion, ovulation, and luteinization. (A) Follicle growth, ovulation, and luteinization within the eIVFG system with representative bright-field follicle images on the top and histological images at the bottom. Follicles were cultured for 8 days to reach maturation; ex vivo ovulation was induced by treating mature antral follicles with 1.5 IU/ml of hCG on day 8 of eIVFG; and ovulated follicles were continuously cultured for 48 hours to allow for luteinization and progesterone secretion. Scale bar: 100 μm. (B) The cytoplasm-nucleus ratios (an indicator of luteinization) of follicular cells after 48-hour culture of ovulated follicles. (C) The concentrations of estradiol (black line) and progesterone (red line) during eIVFG and the concentrations of PGE2 across the time of ex vivo ovulation in the conditioned media. (D) Relative mRNA expression levels of established ovulatory genes examined by RT-qPCR at 0, 1, 4, and 8 hours post-hCG. N = 10–15 follicles in each group. *p < 0.05; **p < 0.01; ***p < 0.001.
Hormonal analysis revealed that follicles grown in the eIVFG system secreted increasing amounts of estradiol, which peaked on day 8 and then markedly decreased after ovulation; in contrast, progesterone concentrations were moderately low before ovulation but greatly increased after ovulation, consistent with the timing of CL formation (Figure 1C). The follicular concentration of PGE2, a key ovulation mediator, has been shown to peak just before ovulation [16, 17]. Our enzyme-linked immunosorbent assay (ELISA) results showed that during ex vivo ovulation, PGE2 concentrations in the conditioned media were nearly non-detectable in the first 4 hours but significantly increased by 8.71-fold from 4 to 8 hours post-hCG (Figure 1C). Collectively, these results demonstrate that eIVFG recapitulates key morphological and hormonal events of follicle growth, differentiation, ovulation, and luteinization.
Established molecular signatures of in vivo ovulation are conserved ex vivo
To determine whether key genes known to regulate ovulation in vivo undergo similar expression dynamics in our ex vivo ovulation model, we performed single-follicle RT-qPCR on a candidate set of genes using cDNA from follicles at 0, 1, 4, and 8 hours post-hCG. The specific genes, their full names, functions in ovulation, and references are listed in Supplemental Table 1. Overall, the expression profiles of all examined genes in Figure 1D followed similar trends to what has been reported during in vivo ovulation. For example, the gonadotropin receptor genes, Fshr and Lhcgr, had comparable expression levels between 0 and 1 hour but then continuously decreased at 4 and 8 hours. Genes encoding several essential ovulation modulators, including Pgr, EGF-like factors (Ereg and Btc), and cumulus cell expansion regulators (Has2, Tnfaip6, and Ptgs2), exhibited a transient induction at 4 hours. The expression of genes related to proteolysis during ovulation, Plau, and Adamts1 increased and peaked at 4 and 8 hours, respectively. With respect to the ovarian steroidogenesis-related genes, Star increased and peaked at 4 hours and Cyp19a1 continuously decreased during ex vivo ovulation. Together, these results indicate that ex vivo ovulation preserves the dynamic transcriptional changes of well-defined ovulatory genes.
Single-follicle RNA-seq analysis reveals transcriptome-wide temporal changes and conservation of key ovulatory genes and signaling during ex vivo ovulation
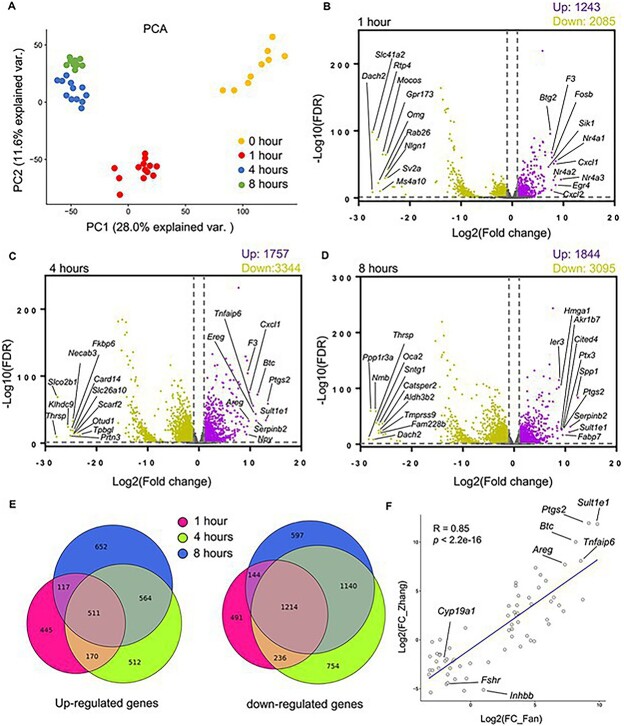
To further characterize the transcriptome during ex vivo ovulation in an unbiased manner, we collected eIVFG-derived follicles at 0, 1, 4, and 8 hours post-hCG and performed single-follicle RNA-seq. High-quality sequencing data were generated from single follicles at each time point, and thousands of genes were detected in ovulatory follicles (Supplemental Figure 1). To understand the global variation in our dataset, we first performed principal component analysis (PCA). PCA separated follicles into distinct clusters based on time post-hCG exposure (Figure 2A), demonstrating that follicles experienced dramatic transcriptomic changes over the 8-hour time period of ex vivo ovulation.
Figure 2.
The transcriptome-wide temporal changes of eIVFG-derived follicles during ex vivo ovulation examined by single-follicle RNA-seq. (A) PCA of the first two principal components for follicles treated with hCG for 0, 1, 4, and 8 hours. N = 9, 13, 12, and 10 follicles at 0, 1, 4, and 8 hours, respectively. (B–D) Volcano plots of all DEGs (FDR < 0.05, fold change >2 or <0.5 compared to follicles at 0 hour). Purple: Upregulated genes; gray: non-significantly altered genes; yellow: downregulated genes. The top 10 upregulated and downregulated DEGs were highlighted in each volcano plot. (E) Comparisons of DEGs at three post-hCG time points (1, 4, and 8 hours vs. 0 hour). (F) Correlation analysis of top 66 LH-ERK1/2 target genes at 4 hours post-hCG injection in the study by Fan et al. in a mouse superovulation model and the same set of genes in the RNA-seq dataset at 4 hours post-hCG in our ex vivo ovulation model, with eight established ovulatory genes highlighted.
We identified thousands of differentially expressed genes (DEGs) (fold change >2 and false discovery rate (FDR) < 0.05) for each time point post-hCG exposure compared to the control follicles at 0 hour, and the top 10 upregulated and downregulated genes at each time point were highlighted in volcano plots (Figure 2B–D). The complete list of all DEGs at these time points is available in our deposited dataset in the Gene Expression Omnibus (GEO186937). There were 1243 upregulated and 2085 downregulated genes at 1 hour post-hCG (Figure 2B). The most upregulated gene was Nr4a3, a gene encoding the nuclear receptor subfamily 4A3 (NR4A3). The other two genes of the NR4A family, Nr4a1 and Nr4a2, were also among the top 10 upregulated genes, suggesting a potential important role in early ovulatory events. At 4 hours, there were 1757 up- and 3344 down-regulated genes (Figure 2C); among the top 10 upregulated genes, 9 of them have been implicated in ovulation previously (Figure 2C and Supplemental Table 1). At 8 hours, 1844 and 3095 genes were upregulated and downregulated, respectively (Figure 2D), with secreted phosphoprotein 1 (Spp1) being the most upregulated gene. Spp1 encodes osteopontin, which regulates cell adhesion, migration, and inflammation, suggesting a possible role in ovulation [18].
We next compared DEGs between each time point to investigate how many genes change their expression consistently across ex vivo ovulation or only uniquely at a particular time point. Interestingly, the expression changes of many genes were unique to a given time point (Figure 2E). For instance, although 511 genes were consistently upregulated at all three times post-hCG, 445, 512, and 652 genes were specifically upregulated at 1, 4, or 8 hours only, respectively (Figure 2E). There were 1214 genes that were consistently downregulated at all three times post-hCG; however, 491, 754, and 597 genes were uniquely downregulated at 1, 4, or 8 hours only, respectively (Figure 2E). Collectively, these results suggest that our RNA-seq data can comprehensively define the temporal transcriptomic signals that drive ovulation.
We further examined whether the DEGs identified in our ex vivo ovulation model are consistent with the gene expression changes observed during ovulation in vivo. Using an in vivo transgenic superovulation mouse model, Fan et al. demonstrated the indispensable role of LH-ERK1/2 signaling in granulosa cells during ovulation and identified many LH-ERK1/2 target genes with microarray analysis [8]. We compared the top 66 differentially expressed LH-ERK1/2 target genes (39 upregulated and 27 downregulated) at 4 hours post-hCG injection identified by Fan et al. and the same set of genes in the RNA-seq dataset at 4 hours post-hCG in our ex vivo ovulation model. Correlation analysis revealed that the majority of LH-ERK1/2 target genes not only showed consistent upregulation or downregulation but also had similar fold changes between the two datasets, with a Pearson correlation coefficient (R) value of 0.85 (Figure 2F with eight ovulatory genes highlighted and Supplemental Table 2).
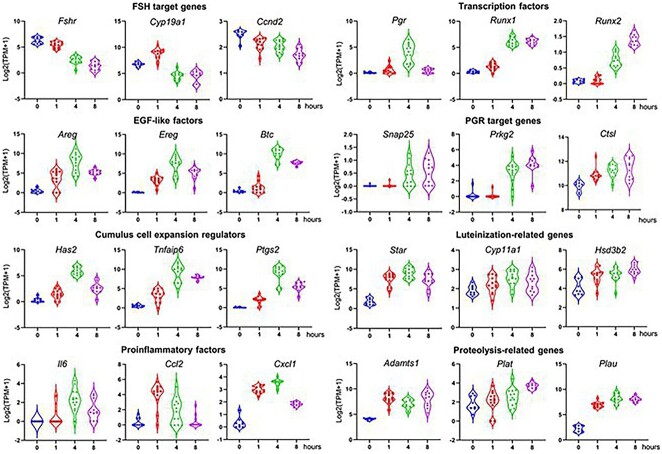
The expression patterns of individual ovulatory genes, including those in Figure 1D and several with literature precedent, were examined across the time-course of ex vivo ovulation (Figure 3). These genes are related to FSH-target genes, EGF-like factors, cumulus expansion, proinflammatory factors, transcriptional factors, PGR-target genes, and genes involved in luteinization and proteolysis. The expression patterns of these ovulatory genes were consistent with the RT-qPCR results (Figure 1D) and also with previous in vivo results (Supplemental Table 1). Given that we identified the conservation of many key ovulatory genes and signaling pathways in our DEGs, and many new ones, these data suggest that our ex vivo ovulation model preserves the transcriptomic features of in vivo ovulation and provides a unique opportunity to define the roles of novel genes and signaling pathways in the temporal events of ovulation.
Figure 3.
mRNA expression (log2[TPM + 1]) of established ovulatory genes in eIVFG-derived follicles at 0, 1, 4, and 8 hours post-hCG examined by single-follicle RNA-seq. N = 9, 13, 12, and 10 follicles at 0, 1, 4, and 8 hours, respectively (TPM: transcripts per million).
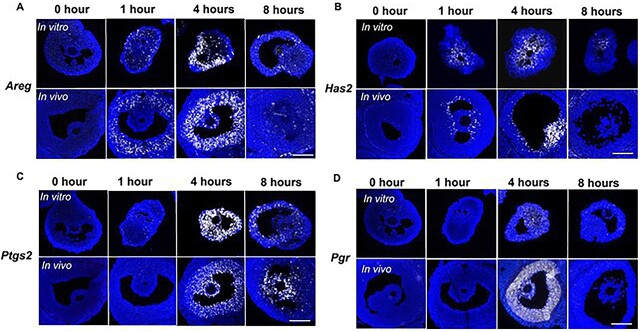
In situ RNA hybridization reveals spatiotemporal expression of ovulatory genes
In situ RNA hybridization was next performed to validate the spatiotemporal expression profiles of four established ovulatory genes: Areg, Has2, Ptgs2, and Pgr. We compared the expression of these genes in follicles at specific time points following ovulation induction both in the ex vivo ovulation model and in mouse ovaries following the induction of superovulation in vivo. The temporal expression patterns of all four genes visualized by in situ RNA hybridization (Figure 4) were consistent with those observed by single-follicle RNA-seq analysis (Figure 3) and RT-qPCR (Figure 1D). Moreover, there was concurrence in the spatiotemporal expression profiles of these genes in follicles following both ex vivo and in vivo ovulation (Figure 4). In response to hCG, Areg was induced in mural granulosa cells at 1 hour (Figure 4A); at 4 hours, Areg was highly expressed in both mural granulosa cells and cumulus cells, and its expression then decreased in both cell populations at 8 hours (Figure 4A). In contrast to Areg, Has2, which regulates cumulus cell expansion, exhibited high expression only in the cumulus cells and the inner layer of mural granulosa cells at 4 hours post-hCG (Figure 4B). Ptgs2, the gene encoding COX-2 (cyclooxygenase-2) for the metabolism of arachidonic acid and synthesis of PGE2 [19], had high expression levels in both cumulus cells and mural granulosa cells at 4 hours post-hCG (Figure 4C). As a nuclear receptor and a transcription factor, PGR induces the transcription of various downstream target genes during ovulation [20–22]. Consistent to the expression pattern of Pgr in vivo, it was transiently induced by hCG at 4 hours in mural granulosa cells during ex vivo ovulation (Figure 4D). These results, coupled with our RT-qPCR and RNA-seq data, further demonstrate that the established molecular signatures of ovulation are maintained spatiotemporally during ex vivo ovulation.
Figure 4.
The spatiotemporal expression of four representative ovulatory genes (A: Areg, B: Has2, C: Ptgs2, and D: Pgr) in ovulatory follicles in both ex vivo and in vivo ovulation models examined by in situ RNA hybridization. N = 3–5 follicles or ovaries in each post-hCG time point and group. Scale bar: 100 μm.
Soft clustering identifies distinct gene expression patterns and novel gene regulatory pathways involved in ovulation
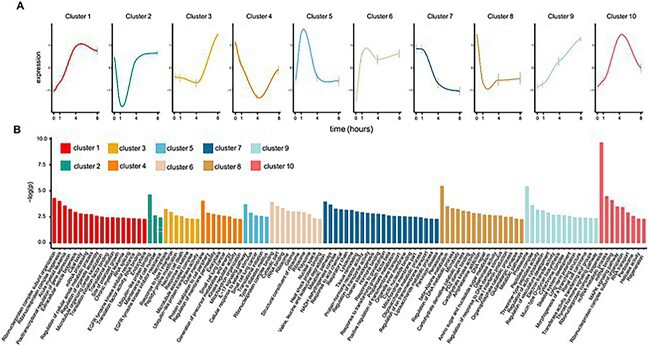
Given that our ex vivo ovulation model well preserves key molecular features of ovulation, we next wanted to identify novel ovulatory genes and signaling. We used soft clustering to cluster all identified DEGs and we identified 10 temporal clusters (Figure 5A), with the specific genes in each cluster listed in Supplemental Table 4. Overall, the expression patterns are dynamic and yield detailed information about the temporal transcriptomic changes during ex vivo ovulation. Several clusters of genes continuously increased (Clusters 1, 3, 6, and 9) or decreased (Clusters 7 and 8) throughout the time-course of ex vivo ovulation, suggesting that the upregulation or downregulation of these genes might critically regulate ovulation. We also identified several clusters that increased (Clusters 5 and 10) or decreased (Clusters 2 and 4) only at 1 or 4 hours, indicating their transient roles in the mid of ovulation. Several clusters identified well-established ovulatory genes, such as Runx1, Lif, Il11, and Hmga1 in Cluster 1, Lhcgr, Fshr, Cyp17a1, and Hsd17b1 in Cluster 7, and Igf1r and Has2 in Cluster 10. The full names and functions of these established ovulatory genes and references are provided in Supplemental Table 1. Soft clustering analysis also allowed us to identify many previously unknown LH/hCG responsive genes in each cluster, suggesting that these genes may also critically control ovulation.
Figure 5.
Soft clustering analysis of DEGs in follicles at 1, 4, and 8 hours post-hCG compared to 0-hour time point. (A) Identified 10 temporal clusters during ex vivo ovulation. (B) Gene Ontology (GO) and KEGG pathway analysis using enriched genes in each identified cluster, with only the top 100 enriched GO terms or altered pathways listed. The full lists of GO and KEGG enrichment results for each cluster are shown in Supplemental Table 5.
To better delineate important ovulatory signaling with high temporal resolution, we next performed GO and KEGG pathway analyses on genes within each cluster (Figure 5B and Supplemental Table 5). These analyses identified both established and new signaling pathways involved in ovulation. In Cluster 1, in which gene expression increased continuously over time, we found enrichment for pathways related to the “mRNA surveillance pathway,” “mTOR signaling pathway,” “RNA transport,” and “JAK-STAT signaling pathway.” In Cluster 7, in which gene expression continuously decreased over time, we found enrichment for “amino acid metabolism,” “hormone-binding,” and “ovarian steroidogenesis.” In Cluster 9, in which gene expression continuously increased over time, we found enrichment for the “proteasome” and several pathways related to “vasculature and tissue remodeling” and “proteolysis.” Angiogenesis [23–26], tissue remodeling [27–29], and proteolysis [30–34] have been proposed to regulate follicle rupture at the late stage of ovulation. For Cluster 10, which contains genes whose expression transiently increased at 4 hours, we found the established “MAPK signaling pathway” and the unknown “Rap1 and Wnt signaling pathways” [8, 35]. Consistent with the concept that ovulation is an inflammatory process [36], both GO and KEGG analyses identified inflammation-related GO terms or signaling pathways, such as the “IL-17 signaling pathway” in Cluster 5 (KEGG), “Type I interferon production” in Clusters 2 and 8 (GO biological process), “NFkB binding” from Cluster 9 (GO molecular function), and “Cytokine receptor activity” from Cluster 10 (GO molecular function). Collectively, these results suggest that ovulation is a dynamic biological process, which involves multiple cellular functions and signaling pathways at precise times. These newly identified genes and pathways may provide us with insights into the mechanisms of ovulation, revealing molecular targets for studying anovulatory diseases and developing novel contraceptives.
The ex vivo ovulation system enables a high-throughput ovulation screening, which identifies proprotein convertase as an essential regulator of follicle rupture
We next explored the utility of this ex vivo ovulation system for a high-throughput ovulation screening to identify nonhormonal contraceptive candidates (Figure 6A). We first tested 10 compounds that target known ovulation mediators, including EGFR, ERK1/2, COX-2, calcium, and MMPs. The compounds’ names, molecular targets, and references are listed in Supplemental Table 6. The majority of these compounds at 10 μM significantly inhibited follicle rupture, and their effects on progesterone secretion were consistent with previous data regarding their roles in luteinization and progesterone secretion (Figure 6B and C). We further screened another 11 compounds that target two new pathways identified in our study (Supplemental Table 6). The first three compounds target proprotein convertases because both Pcsk5, the fifth member of the proprotein convertase family, and proprotein convertase-mediated proteolysis are enriched in Cluster 9 of soft clustering (Figure 5 and Supplemental Table 4). The remaining eight compounds inhibit JAK–STAT signaling enriched in Cluster 1 of soft clustering and KEGG pathway analysis (Figure 5). With respect to the eight JAK inhibitors, the broad JAK inhibitor (JAK inhibitor 1), the selective JAK1 inhibitor (ABT-494), and the selective TYK2 inhibitor (SAR-20347) significantly inhibited follicle rupture but also suppressed the secretion of progesterone (Figure 6B and C). All three compounds targeting proprotein convertases, including proprotein convertase inhibitor (PCI), furin inhibitor I, and furin inhibitor II, greatly inhibited follicle rupture and did not affect progesterone secretion (Figure 6B and C). These results indicate that JAK–STAT signaling critically regulates follicle rupture and also contributes to luteinization and progesterone secretion; however, proprotein convertases control follicle rupture in a steroidogenesis-independent manner.
Figure 6.
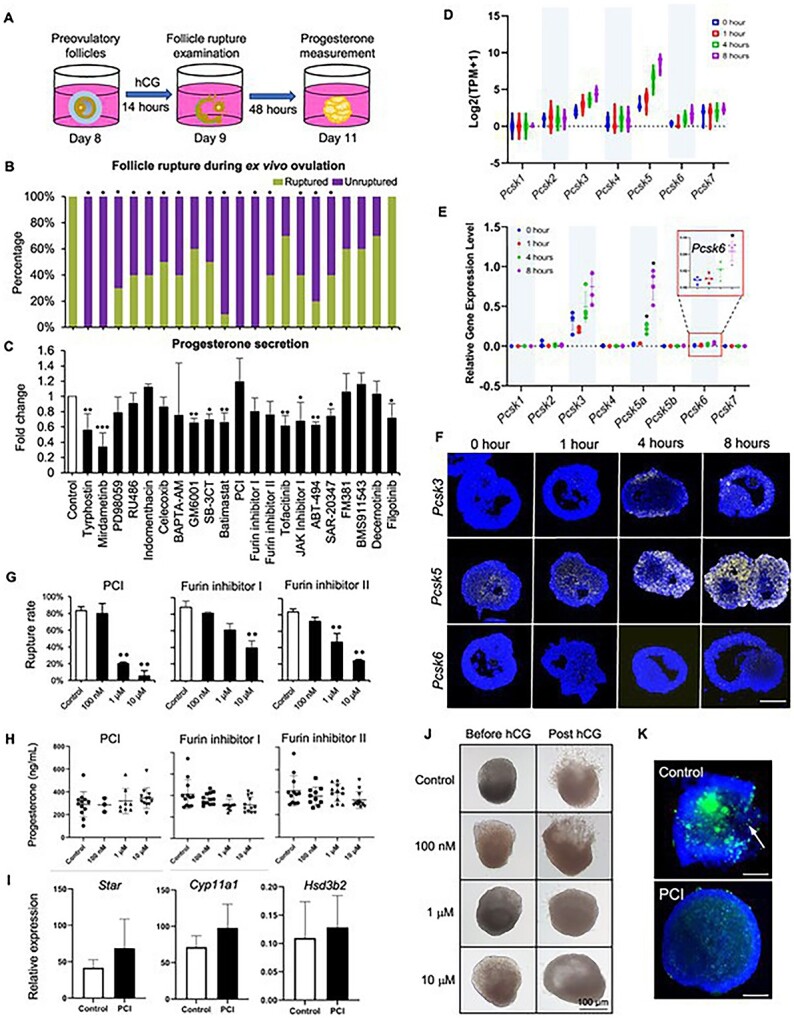
(A) The schematic of a high-throughput ovulation screening. (B) The percentages of ruptured follicles in response to the treatment of vehicle or 21 compounds during ex vivo ovulation. (C) The concentrations of progesterone measured by ELISA after vehicle- or compound-treated follicles were cultured for an extra 48 hours. mRNA expression of all seven PCSK family genes during ex vivo ovulation examined by single-follicle RNA-seq (D) and RT-qPCR (E). (F) mRNA expression of Pcsk3, Pcsk5, and Pcsk6 examined by in situ RNA hybridization in eIVFG-derived follicles at 0, 1, 4, and 8 hours post-hCG during ex vivo ovulation. N = 3–5 follicles at each time point and group. Scale bar: 100 μm. (G) The percentages of ruptured follicles after follicles were treated with 0, 0.1, 1, and 10 μM of proprotein convertase inhibitor (PCI), Furin inhibitor I, or Furin inhibitor II during ex vivo ovulation. (H) The concentrations of progesterone measured by ELISA in the conditioned media after vehicle- or compound-treated follicles were cultured for an extra 48 hours. Error bar represents standard deviation, N = 10–15 follicles per group for follicle rupture and hormone measurement, and three replicates of experiments were performed. (I) mRNA expression of genes related to luteinization at 0 and 4 hours post-hCG after treatment with vehicle or 10 μM PCI. N = 7–8 follicles in each group. Error bar: Standard deviation. (J) Representative images of follicles (before and 14-hour post-hCG treatment) treated with 0, 0.1, 1 and 10 μM of PCI. (K) Representative images of in situ zymography of follicles treated with or without 10 μM PCI at 14 hours post-hCG. N = 4–5 follicles were included in each group. Blue: DAPI; green: fluorescence from degraded DQ-gelatin. Scale bar: 100 μm.
Given the profound involvement of proprotein convertases in the proteolytic activation of MMPs and subsequent tissue remodeling [37, 38], which have been implicated in ovulation, we performed more in-depth investigations to determine whether the proprotein convertase-mediated proteolytic activation may be a target for a nonhormonal contraceptive that only blocks ovulation without affecting steroidogenesis. RNA-seq results showed that, in addition to Pcsk5, the expression of another two genes from the same family, Pcsk3 and Pcsk6, also increased across ex vivo ovulation (Figure 6D), which was confirmed by the results of RT-qPCR (Figure 6E). There are two isoforms of Pcsk5: Pcsk5A and Pcsk5B, with Pcsk5A but not Pcsk5B highly induced by hCG (Figure 6E). In situ RNA hybridization further revealed that these Pcsk genes were primarily induced in the granulosa cells of ovulatory follicles during ex vivo ovulation (Figure 6F).
We next tested three proprotein convertase inhibitors by including a broader concentration range. All three inhibitors inhibited follicle rupture in a dose-dependent manner (Figure 6G), with IC50s at 0.48 μM, 1.15 μM, and 0.81 μM, respectively, calculated by a four-parameter logistic regression model. Follicles treated with all three inhibitors at all concentrations secreted comparable progesterone levels relative to controls (Figure 6H). The dispensable role of proprotein convertases in luteinization and progesterone secretion was further confirmed by RT-qPCR results, demonstrating that 10 μM PCI did not affect the expression of luteinization-related genes, including Star, Cyp11a1, and Hsd3b2, at 14 hours post-hCG (Figure 6I). Representative images of follicles before and after hCG stimulation in both vehicle- and PCI- treated groups are shown in Figure 6J.
Proprotein convertases activate gelatinases to regulate various physiological processes and diseases [38–40]. In situ zymography was next performed to examine the activation of gelatinases during ex vivo ovulation. At 14 hours post-hCG, follicles had remarkably increased fluorescent signals in ovulatory follicles (Figure 6K), indicating increased gelatinase activities at the late stages of ovulation. However, the hCG-induced gelatinase activation was greatly inhibited by the co-treatment with PCI (Figure 6K), suggesting that the inhibited gelatinase activation and defective ECM remodeling result in failed follicle rupture. In summary, these results indicate that proprotein convertases critically regulate the rupture of ovulatory follicles but do not contribute to luteinization and progesterone secretion, which make them attractive as potential targets for the development of nonhormonal female contraceptives.
Discussion
While LH is the established physiological stimulus that triggers ovulation, the complete molecular events of ovulation remain poorly defined due to limited research models. Many systems have been developed to assess ovulatory processes and contraceptive development including non-mammalian models [41]. The method of eIVFG has been successfully used to culture follicles of multiple species [42–44]. Follicles grown from eIVFG preserve key structural and molecular features of folliculogenesis [14, 15]. Here, we further demonstrated that ovulation, the culminating follicular event, is preserved in eIVFG-derived follicles in a variety of aspects, including morphological, hormonal, and gene regulatory pathway changes; we also demonstrated the potential applications of this ex vivo ovulation model in discovering novel ovulatory pathways and identifying female nonhormonal contraceptives through a high-throughput ovulation screening.
Our study demonstrated that upon hCG stimulation, mature follicles from eIVFG recapitulated key biological events of ovulation, including cumulus cell expansion, resumption of oocyte meiosis, follicle rupture, and subsequent luteinization and progesterone secretion. Mechanistically, the LH surge activates several intrafollicular gene regulatory pathways to drive ovulation. Some ovulatory genes are transiently induced at 1 or 4 hours and decline after, and many other genes continuously increase through the late stages of ovulation [45]. In terms of previously known ovulatory genes listed in Supplemental Table 1 and LH target genes in Supplemental Table 2, the results of our RT-qPCR, RNA-seq, in situ hybridization, and correlation analyses revealed that 63 out of 66 genes had consistent changes between in vivo and ex vivo ovulation models, such as the transcriptional reduction of FSH-target genes (Fshr, Cyp19a1, and Ccnd2) and the induction of EGF-like factors (Areg, Ereg, and Btc) and luteinization-related genes (Star, Cyp11a1, and Hsd3b2). These results indicate that eIVFG-based ex vivo ovulation preserves the morphological and hormonal features of ovulation as well as key ovulatory genes. For the three genes that did not correlate (Nos3, Dusp8, and Tnfsf11), they all have smaller fold changes within the range of −2 to 2 in both in vivo and our ex vivo data sets. Based on the criteria (≤ −2 or ≥2) we used to define DEGs, the fold changes of these genes are insignificant. In addition, it is challenging to directly compare the RNA-seq and microarray data due to their different technical backgrounds and quantification methods. Thus, it is more important that the patterns match rather than the absolute values.
Ovulation is an inflammatory process [46]. Our RNA-seq analysis showed that many inflammatory genes were induced during ex vivo ovulation, such as genes encoding cytokines (Il6, Il7, Il11, Il33, and Cxcl1, 2, 3, 4, 5, 10). These inflammatory factors produced from granulosa and theca cells have been postulated to recruit circulating leukocytes to weaken the basal lamina underlying the theca cell layers and also the ovarian surface epithelium (OSE), which enables the rupture of both ovulatory follicle and OSE [47]. Although there were no immune cells and OSE in our ex vivo ovulation system, the induction of inflammatory regulators suggests that eIVFG-derived follicles preserve follicle-intrinsic inflammatory responses upon the stimulation of hCG. PGE2 is another important inflammatory regulator during ovulation [48, 49]. Two prostaglandin synthases regulate the metabolism of arachidonic acid to generate PGE2: COX-1 and COX-2, with their gene names as Ptgs1 and Ptgs2, respectively. COX-1 is primarily expressed in theca cells, and COX-2 is highly induced in granulosa cells following the LH surge [50, 51]. Our study consistently revealed that Ptgs2 but not Ptgs1 was rapidly induced by hCG in granulosa cells. Indeed, Ptgs2 is the most up-regulated gene by hCG at 4 hours, with a fold change of 11,536. The transcriptional induction of Ptgs2 is consistent with the ELISA data that there was a remarkably increased secretion of PGE2 at 8 hours post-hCG. The JAK–STAT signaling was enriched in the cluster 1 of soft clustering analysis, suggesting the temporal expression of genes related to the JAK/STAT signaling which has been implicated in cumulus expansion by inducing the transcription of Has2, Tnfaip6, and Ptx3 [52]. Our ovulation screening results showed that JAK–STAT signaling also critically contributes to the rupture of ovulatory follicles and luteinization.
While inflammation is central to ovulation, it is not fully understood how and when inflammatory pathways, particularly the induction of inflammatory factors in granulosa and theca cells, are initiated in response to the LH surge. Our soft clustering analysis revealed the enrichment of “IL-17 signaling pathway” from Cluster 5 and “Cytokine receptor activity” from Cluster 10, suggesting that some inflammatory pathways are activated as earlier as 1 hour and then turned down/off afterwards. For example, Nr4a1, Nr4a2, and Nr4a3 were among the top 10 upregulated genes at 1 hour post-hCG. These orphan nuclear receptors have been found to turn on immediately by multiple stressors and regulate the homeostasis of inflammation [53]. The transient activation of these inflammatory genes and pathways is indeed consistent with a recent study finding that the PGR-dependent induction of NFkBIA (NFkB inhibitor α, an NFkB inhibitor family member) prevents the hyperinflammatory damage at the late stages of ovulation [54]. In addition to Nfkbia, our RNA-seq data revealed that several other NFkB inhibitor genes were also highly induced by hCG (Nfkbib, Nfkbid, Nfkbie, and Nfkbiz, Supplemental Figure 2), suggesting the redundant roles of these NFkB inhibitors during ovulation. Together, these results demonstrate the preservation of inflammatory responses during ex vivo ovulation; in addition, the newly identified inflammatory genes and pathways may also critically govern ovulation.
Proteolysis critically regulates ovulation through remodeling ECM and breaking down follicular connective tissues [36]. Three major proteolytic systems have been found to regulate proteolysis during ovulation, including the enzyme family of a disintegrin and metalloproteinase with thrombospondin motifs (ADASTS), plasminogen activator, and matrix metalloproteinases (MMPs) [36]. ADAMTS1, the first member of the ADAMTS family, is essential for ovulation as mice lacking Adamts1 had anovulation possibly due to failed cleavage of versican [55]. Our RNA-seq data showed that Adamts1 was induced as early as 1 hour. Plasminogen activators, including both urokinase-type (PLAU) and tissue-type (PLAT), have been reported to cleave plasminogen to form the active plasmin to degrade ECM components during ovulation [56, 57]. We found that Plau was highly induced (28-fold) at 1 hour and then further, respectively, increased by 67- and 51-fold at 4 and 8 hours post-hCG; however, Plat remained the same at 0, 1, and 4 hours and then increased for 2.8-fold at 8 hours. This is consistent with Fan et al.’s previously reported work, showing that Plau but not Plat was greatly induced in granulosa cells at 4 hours in an in vivo mouse superovulation model [8]. The PLAU/PLAT-mediated activation of plasmin has been speculated to cleave and activate another group of proteolytic system, MMPs, which further degrade multiple ECM components of ovulatory follicles [58, 59]. However, mice with single deletion of Plat or Plau have normal fertility and the dual deletion only resulted in a 26% decrease in ovulation rate [57], indicating that other proteolytic systems may also contribute to the activation of MMPs and subsequent follicle rupture during ovulation.
Proprotein convertases belong to another important class of proteases, and they have been shown to regulate ovarian functions by activating enzymes, peptide hormones, and growth factors through cleaving protein precursors. PCSK3 or Furin is highly induced in granulosa cells of ovulatory follicles in rats [37, 60], and Furin inhibitor blocks the activation of MMP2 to decrease rat ovulation [37]. The deletion of Furin in oocytes causes infertility in mice due to the developmental arrest of early secondary follicles [61]. PCSK5 has been reported to activate oocyte-specific proteins of GDF9 and BMP15 [62, 63] and inhibin secreted from granulosa cells of maturing follicles [64]. However, it is unknown whether other proprotein convertases also activate MMPs during ovulation. Our study revealed that hCG continuously increased the expression of multiple proprotein convertases, including Pcsk3, 5, and 6. The treatment of all three tested proprotein convertase inhibitors blocked follicle rupture, which is likely through the deactivation of gelatinases. More importantly, none of these inhibitors disrupted progesterone secretion, suggesting that this protease family and associated regulatory pathways are attractive druggable targets for developing nonhormonal female contraceptives. Because all three tested inhibitors non-selectively inhibit proprotein convertases, in vitro and/or in vivo loss-of-function models are needed to determine whether the specific Pcsk isoforms have distinct or redundant functions in regulating follicle rupture. Indeed, compared to Pcsk3 and Pcsk6, Pcsk5a has the highest expression levels and also the greatest induction at 8 hours post-hCG, suggesting a prominent role in this process. Tissue specificity is critical for viable drug targets; based on the Genotype-Tissue Expression (GTEx) Portal database (gtexportal.org) that houses tissue-specific gene expression in various human tissues, Pcsk5 has the most abundant expression in the ovaries compared to all other tissues (e.g. uterus, breast, heart, and liver; Supplemental Figure 3). Thus, whether a selective PCSK5 inhibitor, particularly for the isoform of PCSK5A, may block ovulation without off-target effects warrants further investigations.
When examining the temporal expression pattern of a large gene set, such as RNA-seq results, the expression of many genes is not binary but rather exhibits complex dynamics [65]. We thus used the soft clustering to identify distinct gene expression patterns across the time-course of ovulation. Although genes in the same cluster are not necessarily regulated by the same mechanism, we can gain clues regarding when certain genes or regulatory pathways are activated or deactivated. For example, follicle maturation-related genes, such as Fshr and Cyp17a1, were enriched in Cluster 7, which contains continuously decreased genes; in contrast, inflammatory factors (e.g. Lif, Tnf, Cxcl5, and Ccl7) and other known ovulatory regulators (Runx1, Has2, and Crebbp) were enriched in Clusters 1, 5, and 10 that consist of transiently induced genes at 1 or 4 hours, the early or middle stage of ovulation. In addition, soft clustering also enables us to identify novel ovulatory pathways in an unbiased way. For example, “proteasome” was enriched from Cluster 9 having genes continuously increased during ex vivo ovulation. Proteasome cooperates with ubiquitin to regulate proteolysis in eukaryotic cells [66]. Though there is little research regarding the role of proteasome in ovulation, one study from Teeli et al. reported the possible functions of proteasome-ubiquitin system in luteinization [67]. As luteinization occurs after ovulation, it is not surprising that the proteasome pathway was enriched in a cluster containing continuously upregulated genes. Also, “ubiquitin-like protein binding” and “ubiquitin-like protein transferase activity” were enriched in Cluster 2 and Cluster 3, respectively, both of which consisted of genes that were first downregulated but then upregulated during ex vivo ovulation.
One limitation of the ex vivo ovulation system is that there are no recruited immune cells and OSE, which are integral parts of ovulation in vivo. The identified gene regulatory pathways are restricted to the events taking place in granulosa and theca cell layers during ovulation. In future studies, we aim to co-culture follicles with immune cells and ovarian epithelial cells to reconstitute a more complex and in vivo-like ovary organoid. Another option is to use the emerging microfluidic technology, such as EVATAR™ we created [68], to culture ovarian explants that contain all critical components of the ovary. In addition, the majority of identified DEGs are granulosa cell related genes. Two of our recently published studies demonstrate that eIVFG well preserves both somatic theca and granulosa cells as well as the follicle-enclosed oocytes [15, 69]. In future studies, single-cell RNA-seq can be used to differentiate individual follicular cell types and also their transcriptional dynamics during ex vivo ovulation.
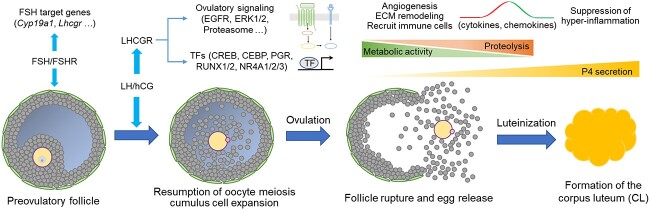
Based on the results of RNA-seq and soft clustering and others, we speculate the temporal molecular events underpinning ovulation, including several new gene regulatory pathways identified in our study (Figure 7): (1) FSH-target genes and signaling are firstly turned down in response to the LH surge; (2) LH stimulates transcriptional factors (PGR, CEBP, RUNX1/2, and NR4A) and pathways (EGF, MAPKs, and newly identified pathways, e.g. proteosome) to trigger the complex molecular events of ovulation; (3) inflammation factors, such as cytokines, chemokines, and other newly identified inflammatory factors, are induced at early to mid-stages of ovulation to support immune cell attraction, ECM remodeling, and angiogenesis; (4) the inflammatory responses are then suppressed to avoid hyperinflammatory damage through the PGR-dependent NFKB inhibitors; (5) the metabolic activities continuously decrease across the time of ovulation; (6) the proteolytic activities continuously increase to facilitate remodeling of ECM and reorganization of follicular cell layers, which enable the ultimate follicle rupture and ovulation; and (7) the follicular somatic cells luteinize to secrete increasing levels of progesterone to regulate early ovulatory events (e.g. follicle rupture) and eventually form the CL to underpin downstream uterine receptivity and embryo implantation.
Figure 7.
Speculated spatiotemporal molecular events underpinning mammalian ovulation.
In conclusion, we demonstrated that follicles derived from the eIVFG system preserve mammalian ovulation phenotypically, hormonally, and mechanistically. Our transcriptomic analyses identify novel genes and signaling pathways that may critically govern ovulation, and we show direct application for the discovery of novel nonhormonal contraceptive candidates. This ex vivo ovulation model system is a robust tool for studying ovulation biology, which can be used to further develop contraceptives and study anovulatory diseases.
Materials and methods
Animals
Adult CD-1 male and female mice were purchased and used to establish a breeding colony (Envigo, Indianapolis, IN). Mice were housed in polypropylene cages in the animal facilities of Northwestern University or Rutgers University. In both places, mice were kept under a temperature-, humidity-, and light-controlled barrier facilities (12-hour light/12-hour dark) and were provided with food and water ad libitum. All animal procedures were approved by the Northwestern University and Rutgers University Institutional Animal Care and Use Committees (IACUC).
Follicle isolation and eIVFG
Mouse secondary follicles were isolated from 16-day-old CD-1 female mice using insulin gauge needles (BD Biosciences, Franklin Lakes, NJ) in L15 media (Invitrogen, Carlsbad, CA) supplemented with 1% fetal bovine serum (FBS; Peak Serum, Inc., Wellington, CO). Follicles were classified based on morphology and size (150–180 μm diameter). Follicles with intact morphology were encapsulated in 0.5% (w/v) alginate (Sigma-Aldrich, St. Louis, MO) for the preservation of the 3D structure of follicles throughout the gonadotropin-dependent folliculogenesis as previously described [11]. Briefly, follicles suspended within 5 μl alginate hydrogel beads were immersed in calcium solution containing 50 mM CaCl2 (Thermo Fisher Scientific, Waltham, MA) and 140 mM NaCl (Thermo Fisher Scientific) for 2 minutes to allow alginate polymers to crosslink and form an alginate bead. To ensure a high yield of preovulatory follicles for the following ex vivo ovulation assay, multi-layered secondary follicles were selected to be cultured. A single multi-layered secondary follicle was encapsulated in each alginate bead. Follicles within alginate beads were cultured in 96-well plates, with each well containing one bead and 100 μl growth media. The growth media consisted of 50% αMEM Glutamax (Thermo Fisher Scientific) and 50% F-12 Glutamax (Thermo Fisher Scientific) supplemented with 3 mg/ml bovine serum albumin (BSA; Sigma-Aldrich), 10 mIU/ml recombinant follicle stimulating hormone (rFSH; gifted by Organon, Jersey City, NJ, USA), 1 mg/ml bovine fetuin (Sigma-Aldrich), and 5 μg/ml insulin-transferrin-selenium (ITS, Sigma-Aldrich). Follicles were cultured for 8 days at 37°C in a humidified environment of 5% CO2 in air. Half of the growth media was replaced every other day and the conditioned media was stored at −20°C for enzyme-linked immunosorbent assay (ELISA). Cultured follicles were imaged at each media change using an Olympus CKX53 Inverted Microscope with 10× and 2× objectives (Olympus Corporation, Hachioji-shi, Tokyo, Japan). On days 0 and 8 of eIVFG or 14 hours and 2 days post-ovulation inductions, five to seven follicles were collected and fixed in 3.8% paraformaldehyde (PFA; Thermo Fisher Scientific) with 0.1% Triton X (Cat. # X100, Sigma-Aldrich) at 37°C for 1 hour for histology.
Ex vivo ovulation and follicle rupture assay
On day 8 of eIVFG, mature antral follicles with a diameter of 300–350 μm were removed from alginate beads by incubation in L15 media containing 1% FBS and 10 IU/ml alginate lyase from Flavobacterium multivorum (Sigma-Aldrich) at 37°C for 20 minutes. Follicles were then transferred to the maturation media and ovulation was induced by incubation in the maturation media containing 50% αMEM Glutamax and 50% F-12 Glutamax supplemented with 10%FBS, 1.5 IU/ml hCG (Sigma-Aldrich), 10 ng/ml epidermal growth factor (EGF; BD Biosciences), and 10 mIU/ml rFSH. At 0, 1, 4, and 8 hours post-hCG treatment, follicles were collected and washed in 1X phosphate-buffered saline (PBS; Invitrogen) three times and then either: [1] snap-frozen for single-follicle reverse transcription-quantitative polymerase chain reaction (RT-qPCR), [2] transferred into the RLT buffer (Qiagen, Hilden, Germany) and snap-frozen for single-follicle RNA sequencing, or [3] fixed in 3.8% PFA with 0.1% Triton X at 37°C for 1 hour for histology or in situ RNA hybridization. For the follicle rupture assay, follicles were incubated in the maturation media with or without the presence of tested compounds at the concentration of 10 μM (Supplemental Table 6) for 14 hours. For each compound tested, 10 follicles were included per group. Follicles in which one side of the follicular wall was breached with the extrusion of COCs were defined as “ruptured” follicles, and follicles with an intact follicular wall were defined as “unruptured” follicles. After examining the follicle rupture success, the remaining somatic cells were cultured in the maturation media for another 48 hours to induce luteinization. The conditioned media was collected after 48 hours and stored at −20°C for ELISA.
Measuring concentrations of hormones in conditioned media
The concentrations of estradiol and progesterone secreted in the culture media during follicular growth and luteinization, respectively, were measured using ovarian steroid hormone ELISA kits (Calbiotech, Spring Valley, CA). The concentration of prostaglandin E2 (PGE2) during in vitro ovulation was measured using the PGE2 ELISA Kit (Abcam, Cambridge, UK). All assays were performed according to the manufacturer’s instructions with five biological replicates.
RNA extraction and target gene quantification by RT-qPCR
Follicles collected at 0, 1, 4, and 8 hours post-hCG treatment were used to examine the expression of ovulation-related genes by RT-qPCR. Total RNA of each follicle was extracted by using the PicoPure RNA isolation kit (Thermo Fisher Scientific). Total RNA was then reversely transcribed into cDNA by using the Superscript III First-Strand Synthesis System with random hexamer primers (Invitrogen) and stored at −80°C. RT-qPCR was performed in a 96-well plate by using the Power SYBR Green PCR Master Mix (Thermo Fisher Scientific) in a StepOnePlus Real-Time PCR system (Thermo Fisher Scientific). RT-qPCR thermocycler was programmed for 10 min at 95°C, followed by 40 cycles of 15 seconds at 95°C and 40 seconds at 60°C, and ended with a melting curve stage to determine the specificity of primers. The primer sequences (5′ to 3′) were listed in Supplemental Table 3 and the relative gene expression level of each gene was calculated by 2^(Ct reference – Ct target).
Single-follicle RNA-seq data generation
Libraries for RNA-seq were generated from single follicles as previously described for single cells work flow but with less amplification cycles [70]. Briefly, we first performed a cell counting experiment to confirm that single follicles had similar amounts of ~12 500 follicular cells per follicle, which ensures that cell counts and hence RNA content are comparable between 0, 1, 4, and 8 hours post-hCG. Smart-Seq2 cDNA synthesis was performed on isolated follicle RNA with Maxima H Minus reverse transcriptase (ThermoFisher). Whole transcriptome amplified (WTA) product was purified with AMPure XP beads (Beckman Coulter), quantified with Quanit-iT PicoGreen dsDNA Assay (ThermoFisher), normalized, and used for preparing paired-end sequencing libraries with Nextera XT (Illumina, #FC-131) according to the manufacturer’s instructions. Libraries were pooled equally and sequenced on a NextSeq500/550 (Illumina) using a 75 cycle v2 sequencing kit.
Analysis of transcriptomics data
Following sequencing, BCL files were converted to merged, demultiplexed FASTQs. FASTQs were checked for size to ensure a comparable sequencing depth across all samples. These were then mapped to the GRCM38 genome using HISAT2 [71] and quality control metrics were extracted using publicly available workflows on Terra (https://portal.firecloud.org/?return=terra#methods/GP-TAG/SS2_scRNA_pipeline/32). After alignment, count data were batch-corrected using ComBat-seq [72]. Batch-corrected matrices and converted Transcripts Per Million matrices were used for subsequent analyses. Count matrices were input to Partek Flow Software (Version 10.0) where PCA was performed and DESeq2 tool embedded in the software was applied for differential expression.
Fuzzy clustering of transcriptomics data
We used custom scripts to perform fuzzy clustering with MFuzz [65, 73]. First, gene symbols were checked with the HUGO Gene Nomenclature Committee (HGNC) and filtered to include valid symbols and protein-coding genes. We then merged differential expression results across each pairwise comparison (post-hCG 0 hour versus 1 hour, 0 hour versus 4 hours, and 0 hour versus 8 hours) and any genes with a 1.5-fold change and padj < 0.05 were flagged. We then used standard MFuzz workflows to cluster DEGs at 1, 4, and 8 hours post-ovulation induction compared to 0 hour, yielding 10 different clusters. Genes with membership >0.6 were flagged for each cluster and used as input for pathway analyses. Gene ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis were performed on cluster-associated genes using WebGestalt [74].
Mouse ovary sample collection
23-day-old CD-1 female mice were treated with 5 IU pregnant mare serum gonadotropin (PMSG; Calbiochem, San Diego, CA) via intraperitoneal (IP) injection to stimulate follicle maturation. At 46 hours, PMSG-treated mice were intraperitoneally injected with 5 IU hCG (Sigma-Aldrich) to induce ovulation. Mice were then sacrificed to collect ovaries at 0, 1, 4, and 8 hours, which are consistent with the time points examined for ex vivo ovulation. Ovaries were fixed in 10% neutral buffered formalin (EMD, Burlington, MA) at 4°C overnight for histology and in situ RNA hybridization.
Histology and in situ RNA hybridization
Fixed follicles from eIVFG or ovaries from in vivo mouse superovulation model were paraffin-embedded and serially sectioned at 5 μm. Selected sections were stained with hematoxylin and eosin (H&E, ThermoFisher Scientific) for histology. To examine the spatiotemporal expression patterns of genes involved in ovulation, in situ RNA hybridization was performed on histological sections from both cultured follicles and ovaries using the RNAscope Multiplex Fluorescent Detection Kit V2 and HybEZ Hybridization System (Advanced Cell Diagnostics, Inc., Newark, CA) according to the manufacturer’s instructions. Briefly, follicle or ovarian tissue sections were pretreated with heat, H2O2, and protease prior to hybridization with target gene probes. An HRP-based signal amplification system was then hybridized to the target probes followed by the fluorescent dye labeling. Tissue sections were mounted with Vectashield antifade mounting medium with DAPI (Maravai LifeSciences, San Diego, CA) and imaged with a confocal microscope (Leica, Wetzlar, Germany).
Proprotein convertase inhibitor treatment
Antral follicles grown from eIVFG were pre-incubated with 0, 0.01, 0.1, 1, and 10 μM proprotein convertase inhibitor (PCI, Sigma-Aldrich), or Furin inhibitor I (Sigma-Aldrich), or Furin inhibitor II (Sigma-Aldrich) in growth media for 2 hours at 37°C in 5% CO2 in air and then transferred into maturation media with the same concentrations of PCI for 0 and 4 hours for single-follicle RT-qPCR or 14 hours for the follicle rupture assay. After follicle rupture examination, follicles were pooled together by a group of 10, dissociated in 500 μl Accutase (ThermoFisher Scientific) at 37°C for 15 min, stained with Trypan blue 0.4% (Invitrogen) at a 1:1 ratio, and live cell number was counted by Countess 3 (Invitrogen). Live cell percentages were 95% and 93% in the control and PCI-treated groups, respectively. Ruptured follicles were also cultured in maturation media without FSH for an additional 48 hours for luteinization. Conditioned media was collected for progesterone measurement using the ELISA kit.
In situ zymography
Follicles on day 8 eIVFG were removed out of alginate beads then incubated in growth media with 10 μM PCI or dimethyl sulfoxide (DMSO) as a vehicle control for 2 hours, then transferred to maturation media containing 100 μg/ml fluorescent-conjugated DQ gelatin (Invitrogen) with the same concentration of PCI and incubated at 37°C in a humidified atmosphere of 5% CO2 in air for 14 hours. After incubation, follicles were fixed with 3.8% PFA at 37°C for 1 hour, then stained with DAPI to visualize nuclei and mounted for visualization. Fluorescent images of DQ gelatin were obtained using EVOS cell imaging system (Thermo Fisher Scientific) at an excitation wavelength of 495 nm and a detection wavelength of 515 nm.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 9.0 (GraphPad Software Inc., San Diego, CA, USA). Data were expressed as the mean ± standard deviation (SD). When comparing gene expressions at different time points and conducting dose response experiments including hormone measurement, one-way analysis of variance followed by Dunnett’s multiple comparison test were used to analyze the statistical significance between control and treatment groups. Otherwise, Welch’s t test was applied. For follicle rupture assay, Yates’ chi-square test was used to compare the percentages of ruptured follicles. A p-value <0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We are grateful for the insightful suggestions from and discussions with all team members of the Ovarian Contraceptive Discovery Initiative (OCDI) and Dr. Stephen Ward and Dr. Daniel Goldberg from the Bill & Melinda Gates Foundation.
Footnotes
† Funding Support: This work was supported, in whole or in part, by the Bill & Melinda Gates Foundation [INV-003385] (FED, SX, BAG, and AKS) and [OPP1200269] (TKW and FED). Under the grant conditions of the Foundation, a Creative Commons Attribution 4.0 Generic License has already been assigned to the Author Accepted Manuscript version that might arise from this submission. This work was also supported by the National Institutes of Health (NIH) K01ES030014 (SX) and R01ES032144 (SX and QZ).
Contributor Information
Jiyang Zhang, Department of Pharmacology and Toxicology, Ernest Mario School of Pharmacy, Environmental and Occupational Health Sciences Institute (EOHSI), Rutgers University, Piscataway, NJ, USA; Department of Obstetrics and Gynecology, Feinberg School of Medicine, Northwestern University, Chicago, IL, USA.
Brittany A Goods, Thayer School of Engineering, Dartmouth College, Hanover, NH, USA.
Pawat Pattarawat, Department of Pharmacology and Toxicology, Ernest Mario School of Pharmacy, Environmental and Occupational Health Sciences Institute (EOHSI), Rutgers University, Piscataway, NJ, USA.
Yingzheng Wang, Department of Pharmacology and Toxicology, Ernest Mario School of Pharmacy, Environmental and Occupational Health Sciences Institute (EOHSI), Rutgers University, Piscataway, NJ, USA.
Tessa Haining, The Ragon Institute of MGH, MIT and Harvard, Cambridge, MA, USA.
Qiang Zhang, Gangarosa Department of Environmental Health, Rollins School of Public Health, Emory University, Atlanta, GA, USA.
Alex K Shalek, The Ragon Institute of MGH, MIT and Harvard, Cambridge, MA, USA; The Institute for Medical Science and Engineering, Department of Chemistry, Koch Institute for Integrative Cancer Research, MIT, Cambridge, MA, USA; Broad Institute of MIT and Harvard, Cambridge, MA, USA.
Francesca E Duncan, Department of Obstetrics and Gynecology, Feinberg School of Medicine, Northwestern University, Chicago, IL, USA.
Teresa K Woodruff, Department of Obstetrics and Gynecology, Michigan State University, East Lansing, MI, USA.
Shuo Xiao, Department of Pharmacology and Toxicology, Ernest Mario School of Pharmacy, Environmental and Occupational Health Sciences Institute (EOHSI), Rutgers University, Piscataway, NJ, USA.
Author contributions
JZ and BAG contributed to the experimental design, data analysis, bioinformatics, and manuscript writing. PP, YW, TH, and QZ contributed to the data collection, analysis, and manuscript writing. TKW, SX, FED, and AKS contributed to the experimental design, data analysis and interpretation, manuscript writing, and approval.
Conflict of interest
AKS reports compensation for consulting and/or SAB membership from Merck, Honeycomb Biotechnologies, Cellarity, Repertoire Immune Medicines, Ochre Bio, Third Rock Ventures, Hovione, Relation Therapeutics, FL82, and Dahlia Biosciences. BAG reports compensation for consulting for FL82.
Data and materials availability
All custom scripts used to analyze RNA-seq data in this manuscript are publicly available (https://github.com/bagoods/OvulationTimeCourse). Associated RNA-seq data can be found at the Gene Expression Omnibus (GEO186837).
References
- 1. Stephen GM, Corson L. The national regional advisory council practice survey for 2000. Fertil Steril 2002; 77:448–455. [DOI] [PubMed] [Google Scholar]
- 2. LeMaire GS. The luteinized unruptured follicle syndrome: anovulation in disguise. J Obstet Gynecol Neonatal Nurs 1987; 16:116–120. [DOI] [PubMed] [Google Scholar]
- 3. Luciano AA, Lanzone A, Goverde AJ. Management of female infertility from hormonal causes. Int J Gynaecol Obstet 2013; 123:S9–S17. [DOI] [PubMed] [Google Scholar]
- 4. Casado-Espada NM, de Alarcon R, de la Iglesia-Larrad JI, Bote-Bonaechea B, Montejo AL. Hormonal contraceptives, female sexual dysfunction, and managing strategies: a review. J Clin Med 2019; 8:908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Britton LE, Alspaugh A, Greene MZ, McLemore MR. CE: an evidence-based update on contraception. Am J Nurs 2020; 120:22–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Richards JS, Ascoli M. Endocrine, paracrine, and autocrine Signaling pathways that regulate ovulation. Trends Endocrinol Metab 2018; 29:313–325. [DOI] [PubMed] [Google Scholar]
- 7. Hsieh M, Lee D, Panigone S, Horner K, Chen R, Theologis A, Lee DC, Threadgill DW, Conti M. Luteinizing hormone-dependent activation of the epidermal growth factor network is essential for ovulation. Mol Cell Biol 2007; 27:1914–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fan HY, Liu Z, Shimada M, Sterneck E, Johnson PF, Hedrick SM, Richards JAS. MAPK3/1 (ERK1/2) in ovarian granulosa cells are essential for female fertility. Science 2009; 324:938–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lydon JP, DeMayo FJ, Funk CR, Mani SK, Hughes AR, Montgomery CA, Shyamala G, Conneely OM, O'Malley BW. Mice lacking progesterone receptor exhibit pleiotropic reproductive abnormalities. Genes Dev 1995; 9:2266–2278. [DOI] [PubMed] [Google Scholar]
- 10. D'Alessandris C et al. Control of mouse cumulus cell-oocyte complex integrity before and after ovulation: plasminogen activator synthesis and matrix degradation. Endocrinology 2001; 142:3033–3040. [DOI] [PubMed] [Google Scholar]
- 11. Xu M, Kreeger PK, Shea LD, Woodruff TK. Tissue-engineered follicles produce live, fertile offspring. Tissue Eng 2006; 12:2739–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xiao S, Duncan FE, Bai L, Nguyen CT, Shea LD, Woodruff TK. Size-specific follicle selection improves mouse oocyte reproductive outcomes. Reproduction 2015; 150:183–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xiao S, Zhang J, Romero MM, Smith KN, Shea LD, Woodruff TK. In vitro follicle growth supports human oocyte meiotic maturation. Sci Rep-Uk 2015; 5:17323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Skory RM, Bernabé BP, Galdones E, Broadbelt LJ, Shea LD, Woodruff TK. Microarray analysis identifies COMP as the most differentially regulated transcript throughout in vitro follicle growth. Mol Reprod Dev 2013; 80:132–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang Y, Drake RS, Russo DD, Pattarawat P, Zhang Q, Zelinski MB, Shalek AK, Goods BA, Xiao S. Vitrification preserves murine ovarian follicular cell transcriptome in a 3D encapsulated in vitro follicle growth system. Biol Reprod 2021; 105:1378–1380. 10.1093/biolre/ioab185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Markosyan N, Duffy DM. Prostaglandin E2 acts via multiple receptors to regulate plasminogen-dependent proteolysis in the primate periovulatory follicle. Endocrinology 2009; 150:435–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim SO, Harris SM, Duffy DM. Prostaglandin E2 (EP) receptors mediate PGE2-specific events in ovulation and luteinization within primate ovarian follicles. Endocrinology 2014; 155:1466–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lund SA, Giachelli CM, Scatena M. The role of osteopontin in inflammatory processes. J Cell Commun Signal 2009; 3:311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sirois J, Sayasith K, Brown KA, Stock AE, Bouchard N, Doré M. Cyclooxygenase-2 and its role in ovulation: a 2004 account. Hum Reprod Update 2004; 10:373–385. [DOI] [PubMed] [Google Scholar]
- 20. Shimada M, Yanai Y, Okazaki T, Yamashita Y, Sriraman V, Wilson MC, Richards JAS. Synaptosomal-associated protein 25 gene expression is hormonally regulated during ovulation and is involved in cytokine/chemokine exocytosis from granulosa cells. Mol Endocrinol 2007; 21:2487–2502. [DOI] [PubMed] [Google Scholar]
- 21. Jo M, Curry TE Jr. Luteinizing hormone-induced RUNX1 regulates the expression of genes in granulosa cells of rat periovulatory follicles. Mol Endocrinol 2006; 20:2156–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sriraman V, Rudd MD, Lohmann SM, Mulders SM, Richards JS. Cyclic guanosine 5′-monophosphate-dependent protein kinase II is induced by luteinizing hormone and progesterone receptor-dependent mechanisms in granulosa cells and cumulus oocyte complexes of ovulating follicles. Mol Endocrinol 2006; 20:348–361. [DOI] [PubMed] [Google Scholar]
- 23. Hazzard TM, Molskness TA, Chaffin CL, Stouffer RL. Vascular endothelial growth factor (VEGF) and angiopoietin regulation by gonadotrophin and steroids in macaque granulosa cells during the peri-ovulatory interval. Mol Hum Reprod 1999; 5:1115–1121. [DOI] [PubMed] [Google Scholar]
- 24. Hazzard TM, Xu F, Stouffer RL. Injection of soluble vascular endothelial growth factor receptor 1 into the preovulatory follicle disrupts ovulation and subsequent luteal function in rhesus monkeys. Biol Reprod 2002; 67:1305–1312. [DOI] [PubMed] [Google Scholar]
- 25. Wulff C, Wilson H, Wiegand SJ, Rudge JS, Fraser HM. Prevention of thecal angiogenesis, antral follicular growth, and ovulation in the primate by treatment with vascular endothelial growth factor Trap R1R2. Endocrinology 2002; 143:2797–2807. [DOI] [PubMed] [Google Scholar]
- 26. Gutman G, Barak V, Maslovitz S, Amit A, Lessing JB, Geva E. Regulation of vascular endothelial growth factor-A and its soluble receptor sFlt-1 by luteinizing hormone in vivo: implication for ovarian follicle angiogenesis. Fertil Steril 2008; 89:922–926. [DOI] [PubMed] [Google Scholar]
- 27. Yang WL, Godwin AK, Xu XX. Tumor necrosis factor-alpha-induced matrix proteolytic enzyme production and basement membrane remodeling by human ovarian surface epithelial cells: molecular basis linking ovulation and cancer risk. Cancer Res 2004; 64:1534–1540. [DOI] [PubMed] [Google Scholar]
- 28. Lind AK, Weijdegård B, Dahm-Kähler P, Mölne J, Sundfeldt K, Brännström M. Collagens in the human ovary and their changes in the perifollicular stroma during ovulation. Acta Obstet Gynecol Scand 2006; 85:1476–1484. [DOI] [PubMed] [Google Scholar]
- 29. Irving-Rodgers HF, Hummitzsch K, Murdiyarso LS, Bonner WM, Sado Y, Ninomiya Y, Couchman JR, Sorokin LM, Rodgers RJ. Dynamics of extracellular matrix in ovarian follicles and corpora lutea of mice. Cell Tissue Res 2010; 339:613–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Brannstrom M, Woessner JF Jr, Koos RD, Sear CH, LeMaire WJ. Inhibitors of mammalian tissue collagenase and metalloproteinases suppress ovulation in the perfused rat ovary. Endocrinology 1988; 122:1715–1721. [DOI] [PubMed] [Google Scholar]
- 31. Hagglund AC, Ny A, Leonardsson G, Ny T. Regulation and localization of matrix metalloproteinases and tissue inhibitors of metalloproteinases in the mouse ovary during gonadotropin-induced ovulation. Endocrinology 1999; 140:4351–4358. [DOI] [PubMed] [Google Scholar]
- 32. Curry TE Jr, Song L, Wheeler SE. Cellular localization of gelatinases and tissue inhibitors of metalloproteinases during follicular growth, ovulation, and early luteal formation in the rat. Biol Reprod 2001; 65:855–865. [DOI] [PubMed] [Google Scholar]
- 33. Ny T, Wahlberg P, Brandstrom IJ. Matrix remodeling in the ovary: regulation and functional role of the plasminogen activator and matrix metalloproteinase systems. Mol Cell Endocrinol 2002; 187:29–38. [DOI] [PubMed] [Google Scholar]
- 34. Peluffo MC, Murphy MJ, Baughman ST, Stouffer RL, Hennebold JD. Systematic analysis of protease gene expression in the rhesus macaque ovulatory follicle: metalloproteinase involvement in follicle rupture. Endocrinology 2011; 152:3963–3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Su YQ, Wigglesworth K, Pendola FL, O'Brien MJ, Eppig JJ. Mitogen-activated protein kinase activity in cumulus cells is essential for gonadotropin-induced oocyte meiotic resumption and cumulus expansion in the mouse. Endocrinology 2002; 143:2221–2232. [DOI] [PubMed] [Google Scholar]
- 36. Duffy DM, Ko C, Jo M, Brannstrom M, Curry TE. Ovulation: parallels with inflammatory processes. Endocr Rev 2019; 40:369–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kelty BP, Curry TE Jr. Ovarian furin (proprotein convertase subtilisin/kexin type3): expression, localization, and potential role in ovulation in the rat. Biol Reprod 2010; 83:147–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yana I, Weiss SJ. Regulation of membrane type-1 matrix metalloproteinase activation by proprotein convertases. Mol Biol Cell 2000; 11:2387–2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stawowy P, Meyborg H, Stibenz D, Stawowy ŃBP, Roser M, Thanabalasingam U, Veinot JP, Chrétien M, Seidah NG, Fleck E, Graf K. Furin-like proprotein convertases are central regulators of the membrane type matrix metalloproteinase-pro-matrix metalloproteinase-2 proteolytic cascade in atherosclerosis. Circulation 2005; 111:2820–2827. [DOI] [PubMed] [Google Scholar]
- 40. Remacle AG, Rozanov DV, Fugere M, Day R, Strongin AY. Furin regulates the intracellular activation and the uptake rate of cell surface-associated MT1-MMP. Oncogene 2006; 25:5648–5655. [DOI] [PubMed] [Google Scholar]
- 41. Jiang K, Zhang J, Huang Y, Wang Y, Xiao S, Hadden MK, Woodruff TK, Sun J. A platform utilizing drosophila ovulation for nonhormonal contraceptive screening. Proc Natl Acad Sci U S A 2021; 118:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Brito IR, Lima IMT, Xu M, Shea LD, Woodruff TK, Figueiredo JR. Three-dimensional systems for in vitro follicular culture: overview of alginate-based matrices. Reprod Fertil Dev 2014; 26:915–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zubizarreta ME, Xiao S. Bioengineering models of female reproduction. Biodes Manuf 2020; 3:237–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Simon LE, Kumar TR, Duncan FE. In vitro ovarian follicle growth: a comprehensive analysis of key protocol variables. Biol Reprod 2020; 103:455–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hernandez-Gonzalez I, Gonzalez-Robayna I, Shimada M, Wayne CM, Ochsner SA, White L, Richards JAS. Gene expression profiles of cumulus cell oocyte complexes during ovulation reveal cumulus cells express neuronal and immune-related genes: does this expand their role in the ovulation process? Mol Endocrinol 2006; 20:1300–1321. [DOI] [PubMed] [Google Scholar]
- 46. Espey LL. Ovulation as an inflammatory reaction--a hypothesis. Biol Reprod 1980; 22:73–106. [DOI] [PubMed] [Google Scholar]
- 47. Ushigoe K, Irahara M, Fukumochi M, Kamada M, Aono T. Production and regulation of cytokine-induced neutrophil chemoattractant in rat ovulation. Biol Reprod 2000; 63:121–126. [DOI] [PubMed] [Google Scholar]
- 48. LeMaire WJ, Leidner R, Marsh JM. Pre and post ovulatory changes in the concentration of prostaglandins in rat Graafian follicles. Prostaglandins 1975; 9:221–229. [DOI] [PubMed] [Google Scholar]
- 49. Kim SO, Duffy DM. Mapping PTGERs to the ovulatory follicle: regional responses to the ovulatory PGE2 signal. Biol Reprod 2016; 95:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sirois J, Levy LO, Simmons DL, Richards JS. Characterization and hormonal regulation of the promoter of the rat prostaglandin endoperoxide synthase 2 gene in granulosa cells. Identification of functional and protein-binding regions. J Biol Chem 1993; 268:12199–12206. [PubMed] [Google Scholar]
- 51. Sirois J. Induction of prostaglandin endoperoxide synthase-2 by human chorionic gonadotropin in bovine preovulatory follicles in vivo. Endocrinology 1994; 135:841–848. [DOI] [PubMed] [Google Scholar]
- 52. Liu Z, de Matos DG, Fan HY, Shimada M, Palmer S, Richards JAS. Interleukin-6: an autocrine regulator of the mouse cumulus cell-oocyte complex expansion process. Endocrinology 2009; 150:3360–3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Safe S, Jin UH, Morpurgo B, Abudayyeh A, Singh M, Tjalkens RB. Nuclear receptor 4A (NR4A) family - orphans no more. J Steroid Biochem Mol Biol 2016; 157:48–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Park CJ, Lin PC, Zhou S, Barakat R, Bashir ST, Choi JM, Cacioppo JA, Oakley OR, Duffy DM, Lydon JP, Ko CMJ. Progesterone receptor serves the ovary as a trigger of ovulation and a terminator of inflammation. Cell Rep 2020; 31:107496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Brown HM, Dunning KR, Robker RL, Boerboom D, Pritchard M, Lane M, Russell DL. ADAMTS1 cleavage of versican mediates essential structural remodeling of the ovarian follicle and cumulus-oocyte matrix during ovulation in mice. Biol Reprod 2010; 83:549–557. [DOI] [PubMed] [Google Scholar]
- 56. Peng XR, Hsueh AJ, Ny T. Transient and cell-specific expression of tissue-type plasminogen activator and plasminogen-activator-inhibitor type 1 results in controlled and directed proteolysis during gonadotropin-induced ovulation. Eur J Biochem 1993; 214:147–156. [DOI] [PubMed] [Google Scholar]
- 57. Leonardsson G, Peng XR, Liu K, Nordström L, Carmeliet P, Mulligan R, Collen D, Ny T. Ovulation efficiency is reduced in mice that lack plasminogen activator gene function: functional redundancy among physiological plasminogen activators. Proc Natl Acad Sci U S A 1995; 92:12446–12450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Curry TE Jr, Osteen KG. The matrix metalloproteinase system: changes, regulation, and impact throughout the ovarian and uterine reproductive cycle. Endocr Rev 2003; 24:428–465. [DOI] [PubMed] [Google Scholar]
- 59. Baramova EN, Bajou K, Remacle A, L'Hoir C, Krell HW, Weidle UH, Noel A, Foidart JM. Involvement of PA/plasmin system in the processing of pro-MMP-9 and in the second step of pro-MMP-2 activation. FEBS Lett 1997; 405:157–162. [DOI] [PubMed] [Google Scholar]
- 60. Bae JA, Park HJ, Seo YM, Roh J, Hsueh AJW, Chun SY. Hormonal regulation of proprotein convertase subtilisin/kexin type 5 expression during ovarian follicle development in the rat. Mol Cell Endocrinol 2008; 289:29–37. [DOI] [PubMed] [Google Scholar]
- 61. Meng TG, Hu MW, Ma XS, Huang L, Liang QX, Yuan Y, Hou Y, Wang H, Schatten H, Wang ZB, Sun QY. Oocyte-specific deletion of furin leads to female infertility by causing early secondary follicle arrest in mice. Cell Death Dis 2017; 8:e2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mottershead DG, Pulkki MM, Muggalla P, Pasternack A, Tolonen M, Myllymaa S, Korchynskyi O, Nishi Y, Yanase T, Lun S, Juengel JL, Laitinen M et al. Characterization of recombinant human growth differentiation factor-9 signaling in ovarian granulosa cells. Mol Cell Endocrinol 2008; 283:58–67. [DOI] [PubMed] [Google Scholar]
- 63. Hashimoto O, Moore RK, Shimasaki S. Posttranslational processing of mouse and human BMP-15: potential implication in the determination of ovulation quota. Proc Natl Acad Sci U S A 2005; 102:5426–5431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Antenos M, Lei L, Xu M, Malipatil A, Kiesewetter S, Woodruff TK. Role of PCSK5 expression in mouse ovarian follicle development: identification of the inhibin alpha- and beta-subunits as candidate substrates. PLoS One 2011; 6:e17348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kumar L, Futschik ME. Mfuzz: a software package for soft clustering of microarray data. Bioinformation 2007; 2:5–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tanaka K. The proteasome: overview of structure and functions. Proc Jpn Acad Ser B Phys Biol Sci 2009; 85:12–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Teeli AS, Leszczyński P, Krishnaswamy N, Ogawa H, Tsuchiya M, Śmiech M, Skarzynski D, Taniguchi H. Possible mechanisms for maintenance and regression of corpus luteum through the ubiquitin-proteasome and autophagy system regulated by transcriptional factors. Front Endocrinol (Lausanne) 2019; 10:748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Xiao S, Coppeta JR, Rogers HB, Isenberg BC, Zhu J, Olalekan SA, McKinnon KE, Dokic D, Rashedi AS, Haisenleder DJ, Malpani SS, Arnold-Murray CA et al. A microfluidic culture model of the human reproductive tract and 28-day menstrual cycle. Nat Commun 2017; 8:14584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Babayev E, Xu M, Shea LD, Woodruff TK, Duncan FE. Follicle isolation methods reveal plasticity of granulosa cell steroidogenic capacity during mouse in vitro follicle growth. Mol Hum Reprod 2022; 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Trombetta JJ, Gennert D, Lu D, Satija R, Shalek AK, Regev A. Preparation of single-cell RNA-Seq libraries for next generation sequencing. Curr Protoc Mol Biol 2014; 107:21–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 2019; 37:907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zhang Y, Parmigiani G, Johnson WE. ComBat-seq: batch effect adjustment for RNA-seq count data. NAR Genom Bioinform 2020; 2:lqaa078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Futschik ME, Carlisle B. Noise-robust soft clustering of gene expression time-course data. J Bioinforma Comput Biol 2005; 3:965–988. [DOI] [PubMed] [Google Scholar]
- 74. Liao Y, Wang J, Jaehnig EJ, Shi Z, Zhang B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res 2019; 47:W199–W205. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.