Viruses, one of the most agriculturally important groups of plant pathogens, account for 47% of all emerging plant infectious diseases (Anderson et al., 2004). Viruses with relatively simple genomes and encoding only a few proteins are highly dependent on host factors (or susceptibility [S] factors) to complete their lifecycles. The modification or mutation of S factors might cause the loss of susceptibility, often resulting in recessive resistance. The eukaryotic translation initiation factor 4 E (eIF4E) is a well‐characterized S gene for many potyviruses of the family Potyviridae in dicots, which encodes a cap‐binding protein that binds to methylated guanine triggering assembly of the protein translation initiation complex. The eIF4E has become a major target for engineering viral resistance using the CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats/CRISPR‐associated protein 9) genome‐editing technology. Knocking out eIF4E was shown to result in resistance in dicots such as melon (Pechar et al., 2022) and monocots such as barley (Hoffie et al., 2021).
Wheat yellow mosaic disease caused by Wheat yellow mosaic virus (WYMV) or Chinese wheat mosaic virus (CWMV) in East Asia, and Wheat spindle streak mosaic virus (WSSMV) in Europe and North America, results in severe yield losses in hexaploid wheat. WYMV and WSSMV belong to the genus Bymovirus of the family Potyviridae and are transmitted by the soil‐borne plasmodiophorid Polymyxa graminis. In diploid barley, the bymoviruses Barley yellow mosaic virus (BaYMV) and Barley mild mosaic virus (BaMMV) cause yellow mosaic disease, and most resistance loci are represented by recessive alleles (Jiang et al., 2020). By contrast, 13 resistance genes and/or quantitative trait loci (QTL) have been mapped in wheat, and all are inherited in a dominant manner. However, to date, no resistance genes against viruses have been isolated by map‐based cloning in wheat.
This study reported CRISPR/Cas9‐guided genome editing of wheat orthologous of the barley susceptibility factor gene HveIF4E. In barley, natural polymorphisms in HveIF4E are the causal agent of several resistance alleles (Jiang et al., 2020), and HveIF4E‐edited plants also showed BaMMV resistance (Hoffie et al., 2021). Three TaeIF4E homoeologous genes were identified by BLAST against the reference genome sequence of wheat (IWGSC RefSeq v1.1; Figure S1a). We selected a 20‐bp target sequence for a single guide RNA (sgRNA) upstream of the protospacer‐adjacent motif (PAM) within the conserved region in these three homoeoalleles (Figure 1a and Figure S1a,b) and transformed the expression cassette into wheat cultivar (cv.) ‘Fielder’. Out of 65 independent regenerated plants (T0), eight genome‐editing events were detected (Table S1). Seven of eight edits were single base insertions or deletions of 1 or 2 bp leading to frameshift mutations. There were three, three, and two plants showing edits at the target site in subgenomes 3A, 3B, and 3D, respectively (Table S1). We conducted two rounds of cross‐pollination and marker‐assisted selection to pyramid the TaeIF4E mutations in all three subgenomes in a single hybrid plant, followed by genotyping 318 F2 segregants (Figure S1b–e). Single (aaBBDD, AAbbDD and AABBdd), double (aabbDD, aaBBdd and AAbbdd) and triple (aabbdd) mutants in all three TaeIF4E homoeoalleles were obtained. The CRISPR/Cas9 cassette was present in all mutants (Figure S1f). We predicted and analysed five potential off‐target effects, and no mutations were detected in mutants (Table S2), indicating off‐targets unlikely occurred.
Figure 1.
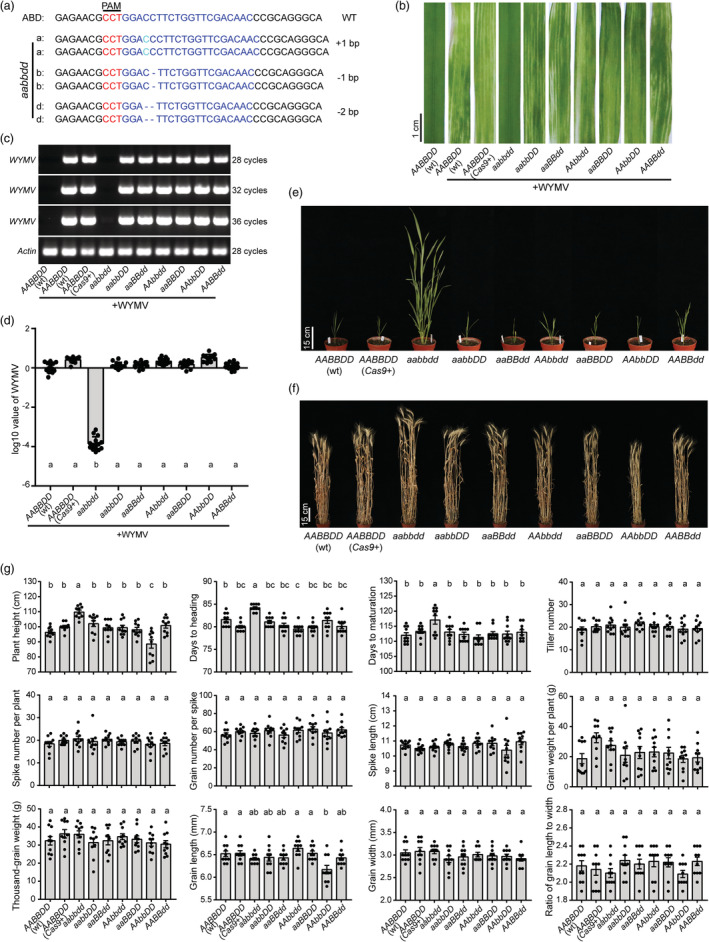
TaeIF4E‐edited triple mutant improves WYMV resistance without yield penalty in wheat. (a) Mutations in the TaeIF4E nucleotide sequence in triple mutants. The sgRNA and the protospacer‐adjacent motif (PAM) are highlighted in blue and red, respectively. WT, wild type. (b) The symptoms of the triple, double and single mutants, the wild type (wt, non‐transformed ‘Fielder’) and the mock control (harbouring the Cas9 cassette, without detected edits) at 6 weeks postinoculation with WYMV. The accumulation of WYMV was determined by RT‐PCR (c) and RT‐qPCR (d). The wheat Actin gene served as the endogenous control. In (d), black dots represent samples (n = 14). (e) Phenotypes of the TaeIF4E‐edited mutant lines at jointing stage under greenhouse conditions. Eight‐week‐old WYMV‐inoculated plants were transplanted to soil and grown in the greenhouse conditions until seed harvest. (f) Phenotype of TaEIF4E‐edited mutants at the maturation stage in garden experiment. (g) Statistics for 12 agronomic traits. Ten plants per genotype were analysed. Black dots represent individual measurements. Statistical significance was determined using Tukey's post hoc test (P = 0.05). Error bars represent standard deviation (SD).
We mechanically inoculated nine homozygous lines, representing edited mutants, non‐edited plants (AABBDD/Cas9+) and wild‐type ‘Fielder’ with WYMV‐infected wheat leaves or mock (buffer). The mock‐inoculated plants stayed green and grew normally (Figure S2a,b). However, upon WYMV infection, only the triple‐edited plants remained green, resembling wild‐type plants without infection, whereas the other plants were susceptible to WYMV with yellow discoloration (Figure 1b). By using reverse transcription (RT)‐PCR and quantitative RT‐PCR, viral RNA accumulation was hardly (ca. 10−4) detected in triple‐edited plants, whereas abundant viral RNA accumulated in wild‐type plants and other mutants (Figure 1c,d). We transplanted 8‐week‐old plants that had been inoculated with WYMV or mock to the normal greenhouse conditions. Whereas triple‐edited plants that were inoculated with WYMV grew normally, plants of the other genotypes showed severely stunted growth (Figure 1e), fewer spikes and lower seed setting (Figure S3a). Both mock‐inoculated TaeIF4E‐edited mutants and wild‐type plants were well developed (Figure S3b). In addition, we inoculated these genotypes with CWMV, another soil‐borne P. graminis‐transmitted virus that belongs to the genus Furovirus of the Virgaviridae family. All the TaeIF4E‐edited mutants were susceptible to CWMV, like wild‐type ‘Fielder’, with yellow chlorotic streaking or stripes on the leaves (Figure S4a). We observed no significant differences in viral accumulation among lines (Figure S4b,c). These results demonstrate that TaeIF4E is a host factor in the host compatibility of WYMV but not CWMV.
We further recorded 12 agronomic traits under normal growth conditions in garden experiment. The TaeIF4E‐aabbdd mutant showed an elevated plant height, as these plants were ca. 10 cm taller than the other lines (Figure 1f,g), as well as delayed heading (ca. 4 days) and maturity (ca. 5 days; Figure 1g and Figure S5). Except the single mutant TaeIF4E‐AAbbDD that showed a significant difference in plant height and grain length, no significant differences were detected for the remaining traits (Figure 1g). The simultaneous targeting of the three TaeIF4E homoeoalleles resulted in pleiotropic changes in plant height and heading/maturity, but without yield penalty.
Collectively, our results demonstrate that CRISPR/Cas9‐guided knockout of TaeIF4E improved WYMV resistance in hexaploid wheat. This is identical in principal to our finding that the simultaneous editing of TaPDIL5‐1 homoeoalleles conferred complete resistance against WYMV (Kan et al., 2022). The triple‐mutated TaeIF4E (this study) or TaPDIL5‐1 (Kan et al., 2022) plants were resistant against WYMV, whereas the single or double‐mutants were susceptible, demonstrating the functional redundancy of the homoeoalleles of the host factor genes and their ability to compensate for each other to facilitate WYMV infection. These results explain the recessive resistance to bymoviruses that has often been detected in diploid barley but not in polyploid wheat (Jiang et al., 2020). Importantly, we detected no yield penalty in the TaeIF4E triple‐edited mutant. This observation is different from diploid barley, in which yield reduction was observed (Hoffie et al., 2021) and in diploid tomato, with severe dwarfing or growth defects (Gauffier et al., 2016) as well as diploid melon, with male sterility (Pechar et al., 2022). We speculate that there are unknown mechanisms that complement with the eIF4E deficiency in polyploidy wheat. Our results highlight the potential of engineering virus resistance via genome editing of eIF4E in polyploid species such as hexaploid wheat.
Conflict of interest
A patent application related to this work has been filed.
Author contributions
PY designed the research. JK, YC, CC, PY, CJ, SC and ZH performed the experiments and data analysis. PY and JK wrote the manuscript. All authors read and approved the final manuscript.
Supporting information
Table S1. Editing events in 65 regenerated T0 plants, as determined by Sanger sequencing of plasmids derived from PCR products.
Table S2. Analysis of potential off‐target effects in edited mutants.
Table S3. PCR primers used in this study.
Figure S1. Generation of TaeIF4E‐edited mutants by genome editing and marker‐assisted allele stacking.
Figure S2. Phenotypes of the TaeIF4E‐edited mutant lines following mock inoculation under greenhouse conditions.
Figure S3. Phenotypes of WYMV‐inoculated and mock‐inoculated plants at the maturation stage under greenhouse conditions.
Figure S4. TaeIF4E‐edited triple mutant in common wheat shows no resistance to CWMV.
Figure S5. Triple knockout TaeIF4E mutant shows delayed maturation.
Acknowledgements
The authors thank Drs. Xingguo Ye and Ke Wang (Chinese Academy of Agricultural Sciences) for assistance on wheat transformation. This work was funded by the National Natural Science Foundation of China (32071997; 32001547), the Fundamental Research Funds for Central Non‐Profit of CAAS (Y2022XK25) and Independent Innovation of Agricultural Science and Technology in Jiangsu Province (CX(22)3149).
References
- Anderson, P.K. , Cunningham, A.A. , Patel, N.G. , Morales, F.J. , Epstein, P.R. and Daszak, P. (2004) Emerging infectious diseases of plants: pathogen pollution, climate change and agrotechnology drivers. Trends. Ecol. Evol. 19, 535–544. [DOI] [PubMed] [Google Scholar]
- Gauffier, C. , Lebaron, C. , Moretti, A. , Constant, C. , Moquet, F. , Bonnet, G. , Caranta, C. et al. (2016) A TILLING approach to generate broad‐spectrum resistance to potyviruses in tomato is hampered by eIF4E gene redundancy. Plant. J. 85, 717–729. [DOI] [PubMed] [Google Scholar]
- Hoffie, R.E. , Otto, I. , Perovic, D. , Budhagatapalli, N. , Habekuss, A. , Ordon, F. and Kumlehn, J. (2021) Targeted knockout of eukaryotic translation initiation factor 4 E confers Bymovirus resistance in winter barley. Front. Genome. Ed. 3, 784233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, C.C C., Kan, J.H. , Ordon, F. , Perovic, D. and Yang, P. (2020) Bymovirus‐induced yellow mosaic diseases in barley and wheat: viruses, genetic resistances and functional aspects. Theor. Appl. Genet. 133, 1623–1640. [DOI] [PubMed] [Google Scholar]
- Kan, J.H. , Cai, Y. , Cheng, C.Y. , Jiang, C.C. , Jin, Y.L. and Yang, P. (2022) Simultaneous editing of host factor gene TaPDIL5‐1 homoeoalleles confers wheat yellow mosaic virus resistance in hexaploid wheat. New. Phytol. 234, 340–344. [DOI] [PubMed] [Google Scholar]
- Pechar, G.S. , Donaire, L. , Gosalvez, B. , García‐Almodovar, C. , Sánchez‐Pina, M.A. , Truniger, V. and Aranda, M.A. (2022) Editing melon eIF4E associates with virus resistance and male sterility. Plant. Biotechnol. J. 20, 2006–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Editing events in 65 regenerated T0 plants, as determined by Sanger sequencing of plasmids derived from PCR products.
Table S2. Analysis of potential off‐target effects in edited mutants.
Table S3. PCR primers used in this study.
Figure S1. Generation of TaeIF4E‐edited mutants by genome editing and marker‐assisted allele stacking.
Figure S2. Phenotypes of the TaeIF4E‐edited mutant lines following mock inoculation under greenhouse conditions.
Figure S3. Phenotypes of WYMV‐inoculated and mock‐inoculated plants at the maturation stage under greenhouse conditions.
Figure S4. TaeIF4E‐edited triple mutant in common wheat shows no resistance to CWMV.
Figure S5. Triple knockout TaeIF4E mutant shows delayed maturation.


