Summary
Salt and drought impair plant osmotic homeostasis and greatly limit plant growth and development. Plants decrease stomatal aperture to reduce water loss and maintain osmotic homeostasis, leading to improved stress tolerance. Herein, we identified the C2H2 transcription factor gene OSMOTIC STRESS INDUCED C 2 H 2 1 (OSIC1) from Populus alba var. pyramidalis to be induced by salt, drought, polyethylene glycol 6000 (PEG6000) and abscisic acid (ABA). Overexpression of OSIC1 conferred transgenic poplar more tolerance to high salinity, drought and PEG6000 treatment by reducing stomatal aperture, while its mutant generated by the CRISPR/Cas9 system showed the opposite phenotype. Furthermore, OSIC1 directly up‐regulates PalCuAOζ in vitro and in vivo, encoding a copper‐containing polyamine oxidase, to enhance H2O2 accumulation in guard cells and thus modulates stomatal closure when stresses occur. Additionally, ABA‐, drought‐ and salt‐induced PalMPK3 phosphorylates OSIC1 to increase its transcriptional activity to PalCuAOζ. This regulation of OSIC1 at the transcriptional and protein levels guarantees rapid stomatal closure when poplar responds to osmotic stress. Our results revealed a novel transcriptional regulatory mechanism of H2O2 production in guard cells mediated by the OSIC1‐PalCuAOζ module. These findings deepen our understanding of how perennial woody plants, like poplar, respond to osmotic stress caused by salt and drought and provide potential targets for breeding.
Keywords: Osmotic stress; OSIC1; PalCuAOζ; Populus alba var. pyramidalis ; stomatal closure; 1,256g
Introduction
Osmotic stress induced by drought and/or high saline impairs the osmotic homeostasis of plant cells, resulting in serious restriction in plant growth and development, or even death, therefore threatening agriculture and forestry production (Boyer, 1982; Pandey and Shukla, 2015). The rapid stomatal closure of plants under osmotic conditions is the most efficient way to save water and thus is indispensable to adapt to and tolerate this stress (Matsuda et al., 2016; Xiong et al., 2002). The dominant regulatory manner of stomatal closure is mediated by abscisic acid (ABA), a key stress‐signalling phytohormone (Gong et al., 2021; Munemasa et al., 2015; Shen et al., 2021). Osmotic stress‐enhanced production of ABA is perceived by the receptors, the PYRABACTIN RESISTANCE (PYR)/PYR1‐LIKE (PYL)/regulatory components of ABA receptor (RCAR) and the co‐receptors clade A type 2C phosphatases (PP2Cs). The formation of the PYR/PYL/RCAR‐ABA‐PP2C complex releases the inhibition of PP2C to downstream targets, thus activates protein kinases, including the SNF1‐related protein kinase 2 family, particularly SnRK2.2/2.3/2.6 (OPEN STOMATA 1, OST1) and GUARD CELL HYDROGEN PEROXIDE‐RESISTANT1 (GHR1) (Hua et al., 2012; Ma et al., 2009; Park et al., 2009). These activated protein kinases phosphorylate and activate downstream ion channels, such as SLOW ANION CHANNEL‐ASSOCIATED1 (SLAC1), SLAH3 (SLAC1 HOMOLOGUE3) and ALUMINUM‐ACTIVATEDMALATE TRANSPORTER 12/QUICKLY ACTIVATED ANIONCHANNEL1 (ALMT12/QUAC1), while inhibiting the inward rectifier potassium channel KAT1, resulting in ion efflux from guard cells (Kim et al., 2010; Qi et al., 2018). This process in turn leads to osmotic water efflux and a decrease in the turgor and volume of the guard cells and stomatal closure (Brandt et al., 2015; Geiger et al., 2009a,b, 2011; Hedrich and Geiger, 2017; Hsu et al., 2021; Imes et al., 2013; Meyer et al., 2010; Sato et al., 2009; Vahisalu et al., 2008).
Abscisic acid‐induced generation and accumulation of the secondary signalling molecules, such as hydrogen peroxide (H2O2), nitric oxide (NO) and hydrogen sulphide (H2S), participate in the regulation of stomatal movement (Hou et al., 2013; Liu and Xue, 2021; Wang et al., 2020; Zhang et al., 2019b). For example, the anion channel SLAC1 of guard cells can also be phosphorylated and activated through apoplastic H2O2 directly activating GHR1(Hua et al., 2012; Sierla et al., 2018), and through intracellular H2O2‐activated mitogen‐activated protein kinase (MAPK) cascades (Singh et al., 2017). Most H2O2 in plants is derived from chloroplasts and mitochondria of leaf cells (Qi et al., 2018). The generation of extracellular H2O2 occurs through ABA‐activated OST1, which can phosphorylate and activate plasma membrane NADPH oxidases/respiratory burst oxidase homologue (RBOH) to produce O2, which is then converted to H2O2 by copper/zinc superoxide dismutase (SOD; Han et al., 2019; Hua et al., 2012; Sirichandra et al., 2009). In addition, copper‐containing polyamine oxidases (CuAOs, EC1.4.3.6) catalyse the oxidation of aliphatic diamines putrescine (Put) and cadaverine (Cad) to generate H2O2 in guard cells (Cona et al., 2006; Rea et al., 2004). In Arabidopsis, a total of 10 CuAO genes have been identified and their activities can be enhanced by ABA (Qu et al., 2014).
The C2H2‐type zinc finger proteins (C2H2‐ZFPs) acting as transcription factors contain conserved hexapeptide residues (QALGGH) in their DNA‐binding domain and play important roles in morphogenesis, transcriptional activation and stress response of plants (Laity et al., 2001; Takatsuji, 1999). ZPT2‐3 in Petunia is up‐regulated by drought and cold stress, and its overexpression confers plants more drought tolerance (Sugano et al., 2003). ZFP252 up‐regulates tolerance to salt and drought by maintaining osmotic homeostasis in rice (Xu et al., 2008). Overexpressing GsZFP1 in Glycine soja results in smaller stomata and impairs ABA‐induced stomatal closure (Luo et al., 2012). In Arabidopsis, ZAT12 is involved in the response to various abiotic stresses (Brumbarova et al., 2016) and links reactive oxide species (ROS, H2O2 is a representative molecule) signalling and iron (Fe) acquisition by interacting with the bHLH transcription factor (Le et al., 2016). PeSTZ1 enhances cold tolerance by regulating PeAPX2 expression (He et al., 2019). ZAT10 in Arabidopsis is significantly induced by cold, salt and osmotic stress. Overexpressing or knockdown of this gene increases the salt tolerance of plants (Mittler et al., 2006; Nguyen et al., 2016).
In this study, we identified that a gene OSMOTIC STRESS INDUCED C 2 H 2 1 (OSIC1) significantly induced by salt, drought, PEG6000 and ABA in Populus alba var. pyramidalis that has high‐stress tolerance (Ma et al., 2018). Overexpressing of this C2H2‐ZFP transcription factor in poplar promoted the number of branches and roots and showed more tolerance to salt and drought stress than the wild‐type (WT) plantlets. However, the CRISPR‐Cas9‐generated osic1 mutant poplars displayed a reduction in salt and drought tolerance. Furthermore, OSIC1 positively regulated stomatal closure of poplars after salt, drought and PEG6000 treatments through its direct regulation of PalCuAOζ expression that increased H2O2 accumulation in the guard cells under these stresses. Finally, we found that PalMPK3 could phosphorylate OSIC1 to promote its transcriptional activation activity to PalCuAOζ. Our findings revealed a novel transcriptional regulation of ABA‐induced H2O2 accumulation under osmotic stress mediated by the OSIC1‐PalCuAOζ regulatory module and highlighted the molecular basis for osmotic stress tolerance in poplar adaptation.
Results
A C 2 H 2 ‐ZFP gene is induced by salt, drought, PEG6000 and ABA in P. alba var. pyramidalis
Our previous RNA‐Seq data showed a gene, pal_pou01649, to be up‐regulated in the salt‐stressed leaves and xylem of P. alba var. pyramidalis compared with the untreated materials, respectively (Yu et al., 2017; Zhang et al., 2021). This gene encodes a typical C2H2‐ZEP transcription factor with two C2H2 domains. In addition, there are two putative phosphorylation sites (P‐Site 1 and P‐Site 2) and a leucine‐rich box (L‐box) that is related to protein interactions (Sakamoto et al., 2000) in this peptide (Figure S1a). Phylogenetic analysis showed that pal_pou01649 grouped with homologues from other poplar species rather than those from Arabidopsis (Figure S1b); therefore, it is not orthologous to these members in Arabidopsis. pal_pou01649 was dominantly expressed in leaves and xylem (Figure 1a) and could also be induced by salt, drought, PEG6000 and exogenous ABA (Figure 1b–e). The spatiotemporal expression pattern of this gene involved osmotic stress‐related cis‐acting elements in its promoter, such as ABA response elements (ABREs) and drought response elements (DREs; Figure S1c). These findings suggested that this gene might be associated with poplar's responses to osmosis‐related stresses. Therefore, we named it OSMOTIC STRESS INDUCED C2H2 1 (OSIC1). Additionally, the subcellular localization of OSIC1 was specifically in the nucleus of poplar mesophyll, which was consistent with the predicted nuclear localization signal (NLS) in OSIC1 (Figure 1f and Figure S1a).
Figure 1.

Identification of the transcription factor OSIC1 in P. alba var. pyramidalis. (a) The tissue expression pattern of OSIC1 in Populus alba var. pyramidalis by qPCR. (b) The relative expression level of OSIC1 in poplar leaves exposed to 0, 100 and 300 mm NaCl solution. (c) The relative expression level of OSIC1 in poplar leaves after 0, 3, 9 and 12 h of drought. Drought stress started when the water content was 70% of the soil. (d) The relative expression level of OSIC1 in poplar leaves after 0, 3, 6, 12 and 24 h of PEG6000 treatment. (e) The relative expression level of OSIC1 in poplar leaves treated with exogenous ABA for 0, 1.5, 3 and 6 h. The error bars represent the SE of the mean fold change for three biological replicates. The poplar UBQ gene was used as the internal reference. Letters above bars represent statistically significant differences between groups (P < 0.05) as determined by one‐way ANOVA Duncan's test. (f) Subcellular localization of OSIC1 in mesophyll cells of P. alba var. pyramidalis leaves. DAPI (blue) was used to indicate the nucleus.
OSIC1 positively regulates the number of branches and roots and enhances tolerance to PEG6000 treatment
To determine the biofunction of OSIC1 in poplar, we generated OSIC1 overexpression lines (OSIC1‐OE‐L1 and L4) and their mutant poplars (osic1‐ko‐L3 and L11) through the CRISPR–Cas9 system (Figure S2). Interestingly, the overexpression poplars showed multi‐branch and multi‐root phenotypes (Figure S3), even the 8‐month‐old plantlets (Figure S3d), whereas the mutant poplars tended to be less roots and branches than the wild‐type (WT) without significances (Figure S3). These results indicate that OSIC1 positively regulates branch and root development in poplar. In addition, to figure out the role of OSIC1 in osmotic stress, we watered the transgenic poplar lines with 25% PEG6000 solution to mimic high osmotic soil condition. After 28 days of treatment, the mature leaves of all plantlets were dried‐out, the OSIC1‐OE poplars showed vibrant apical buds and young leaves, but osic1‐ko mutants' were obviously drooped (Figure S4). Therefore, OSIC1 plays a positive role in tolerance to PEG6000‐caused osmotic stress.
OSIC1 positively regulates salt tolerance of poplar
To investigate the roles of OSIC1 in the salt tolerance of poplar, we submerged the leaf discs from the overexpression, WT and mutant poplars into the NaCl solution. The leaf discs of osic1‐ko showed severe necrosis, but those of the OSIC1‐OE plants displayed slight damage compared with the WT (Figure 2a). In addition, the 5‐week‐old, rooted cuttings with different genotypes were transplanted into solid medium with 200 mm NaCl for 7 days, and the WT and osic1‐ko cuttings exhibited severe injury, whereas OSIC1‐OE cuttings were slightly stressful (Figure 2b). Salt stress damaged photosynthetic pigments (Sayyad‐Amin et al., 2016); thus, we examined the contents of total chlorophyll (Ct), chlorophyll a (Ca), chlorophyll b (Cb) and carotenoid (Ck) in these stressed materials, and osic1‐ko showed a significant decrease in these photosynthetic pigments, whereas the overexpression lines had the highest levels compared with WT (Figure 2c). The electrolyte leakage (EL) and malonaldehyde (MDA) contents are indicators of the membrane damage of plant cells (Aghdam et al., 2014), and the levels of EL and MDA in osic1‐ko and OSIC1‐OE plants were up‐regulated and down‐regulated compared with WT, respectively (Figure 2d). In addition, the noninvasive microtest technology (NMT) proved that overexpression of OSIC1 promoted Na+ efflux in the roots of salt‐treated poplar, but which was impaired in the osic1‐ko mutants (Figure S5). These findings suggest that OSIC1 positively modulates the salt tolerance of poplar.
Figure 2.

OSIC1 positively regulates poplar tolerance to salt stress. (a) The phenotype of detached leaf discs from WT, OSIC1‐OE and osic1‐ko poplars before and after salt treatment. (b) The phenotypes of WT, OSIC1‐OE and osic1‐ko rooted cuttings in the solid medium with 200 mm NaCl. (c) The content of photosynthetic pigments in the salt‐stressed poplar cuttings in (b). Ca: content of chlorophyll a; Cb: content of chlorophyll b; Ck: content of carotenoid content; Ct: total chlorophyll content. (d) The measurement of electrolyte leakage (EL) and malondialdehyde (MDA) content in the leaves of WT, OSIC1‐OE and osic1‐ko poplar cuttings exposed to salt stress. (e) The stomatal morphology of WT, OSIC1‐OE and osic1‐ko poplar leaves treated with 0 and 200 mm NaCl, which were observed by a scanning electron microscopy (SEM). (f) The length and width of stomata were measured using the ImageJ software, and area analysis was carried out according to the length and width. The orange, green and blue colour represent WT, OSIC1‐OE and osic1‐ko poplar lines, respectively. These data originated from six independent experiments, and error bars represent the SE of the mean of fold changed from one experiment, and at least 19 stomata of each poplar leaf were measured. All asterisks indicate significant differences: *P < 0.05, **P < 0.01. The ‘ns’ represents no sense.
Because OSIC1 is involved in osmotic stress, we suspected that this gene likely influenced the stomatal phenotype. We examined the stomata of OSIC1‐OE and osic1‐ko poplars under normal conditions, and there were no significant differences in either stomatal morphology or density compared with WT (Figure S6). Moreover, we measured and compared the length, width and area of stoma with or without salt stress, respectively. The stomatal length showed no significant differences across all plants, but the overexpression lines displayed the least width and area after salt treatment, whereas the osic1‐ko plants had the widest and largest stoma (Figure 2e,f). These results indicate that OSIC1 plays a positive role in stomatal closure under salt stress.
OSIC1 positively regulates tolerance to drought stress in poplar
To identify OSIC1's function in drought tolerance, the 2‐month‐old plantlets were used for drought treatment through cutting off the water supply. After 3 days (soil water content: 55%), the osic1‐ko lines showed drooped leaves, but the WT and the overexpression poplars showed no observable changes compared with their status before treatment. After 5 days of treatment, the mutant poplars were severely dehydrated, and the leaf margins of WT plants were dry and crimping. However, OSIC1‐overexpressing poplars exhibited little stressed symptom (Figure 3a). In addition, we measured and compared the relative water content (RWC) of poplar leaves before and after 5 days of drought treatment (soil water content: 25%). The RWC of overexpression poplars after treatment was similar to that before treatment, but the WT and the mutants showed a significantly decreased RWC. The EL and MDA contents also indicated the severe membrane damage in stressed WT and osic1‐ko lines, but OSIC1‐OE poplars seemed to be free from the stress. Proline (Pro) can protect plant cells from osmotic stress, and the OSIC1‐OE poplars had the highest level of Pro content compared with WT and osic1‐ko poplars (Figure 3b). Moreover, we traced the stoma‐related parameters of these poplars after drought treatment, including stomatal conduction, leaf transpiration and net photosynthetic rate. The stomatal conduction and leaf transpiration of OSIC1‐OE displayed a more rapid decline than those of WT and osic1‐ko plants after 5 days of treatment (Figure 3c). These findings indicate that OSIC1 positively modulate stomatal closure under drought stress. Moreover, the net photosynthetic rate of all lines was reduced after 5 days of drought, but the OSIC1‐OE poplars were still significantly higher than those of WT and mutant lines (Figure 3c). These results suggest that OSIC1 positively regulate the drought tolerance of poplar.
Figure 3.

OSIC1 positively regulates drought tolerance. (a) The drought tolerance of different genotypic poplars. Drought stress started when the water content was 85% of the soil, and the poplar status after 0, 3 and 5 days of drought is shown. (b) Quantitation of leaf relative water content (RWC, %), EL, MDA and proline (Pro) content in WT, OSIC1‐OE and osic1‐ko poplar leaves after 0 and 5 days of drought. Error bars represent the SE of the mean fold changes for five biological replicates. All asterisks indicate significant differences: *P < 0.05, **P < 0.01. The ‘ns’ represents no sense. (c) The measurement of stomatal conductance, transpiration rate and net photosynthetic rate of WT, OSIC1‐OE and osic1‐ko poplar leaves after 0 and 5 days of drought. Error bars represent the SE of the mean fold changes from seventy measurements of at least five plantlets for each genotype. All asterisks indicate significant differences: *P < 0.05, **P < 0.01. The ‘ns’ represents no sense.
OSIC1 positively regulates ABA‐mediated stomatal closure
Abscisic acid‐inducible OSIC1 regulates stomatal closure under salt and drought stress, suggesting an association with ABA signalling. To prove this hypothesis, we observed and measured the stomatal aperture of OSIC1‐OE, osic1‐ko and WT poplars treated with 5 μm ABA for 1.5 and 3 h. The length, width and area of the stomata showed no significant differences when the leaves were submerged in the open solution (OS). However, after 1.5 h of ABA application, the width and area of these stomata in overexpression leaves displayed the greatest reduction, which was further reduced after 3 h, and those in the mutants had the largest stomatal area compared with the WT (Figure 4). These findings indicates that OSIC1 plays a positive function in ABA‐mediated stomatal closure.
Figure 4.

OSIC1 promotes ABA‐induced stomatal closure. (a) An observation of ABA‐induced stomatal closure in WT, OSIC1‐OE and osic1‐ko poplar leaves after treatment with an opening solution or 5 μm ABA for 1.5 h and 3 h, respectively. The fifth node leaf of each genotypic plantlet was immersed in a light stoma opening solution (OS) for 2 h. Then, 5 μm ABA was added to the OS to treat the leaves for 1.5 or 3 h. (b) The length and width of stomata were measured using ImageJ software, and stomatal area was calculated according to the length and width. At least 19 stomata of each poplar were measured. Asterisks indicate significant differences: *P < 0.05, **P < 0.01. The ‘ns’ represents no sense.
OSIC1 accelerates H2O2 accumulation in guard cells under salt, PEG6000 and ABA treatment
Abscisic acid induces the accumulation of H2O2 (Shen et al., 2021). We examined H2O2 accumulation in OSIC1‐OE, osic1‐ko and WT poplars before or after exogenous ABA treatment through diaminobenzene (DAB) staining. After 1.5 h of ABA treatment, the OSIC1‐OE leaves turned darkest brown, followed by the WT leaves (Figure S7). This result indicates that OSIC1 enhances ABA‐inducible H2O2. Our quantification of H2O2 content in the leaves also demonstrated that OSIC1 can accelerate H2O2 accumulation under salt and PEG6000 treatments (Figure S8). H2O2 accumulation in guard cells is important for stomatal closure (Qu et al., 2014; Zhang et al., 2019a). We examined the endogenous H2O2 content in the guard cells with or without treatments by salt, PEG6000 and ABA using a H2O2‐sensitive fluorescent dye (Gong et al., 2021). The highest florescence intensity was observed in the OSIC1‐OE lines even though there was no treatment (Figure 5). After three treatments, the florescence intensity was obviously increased in the overexpression lines, followed by the WT, and the osic1‐ko lines showed the weakest florescence (Figure 5). Therefore, OSIC1 positively regulates stress‐induced H2O2 accumulation in guard cells, which is involved in ABA signalling.
Figure 5.
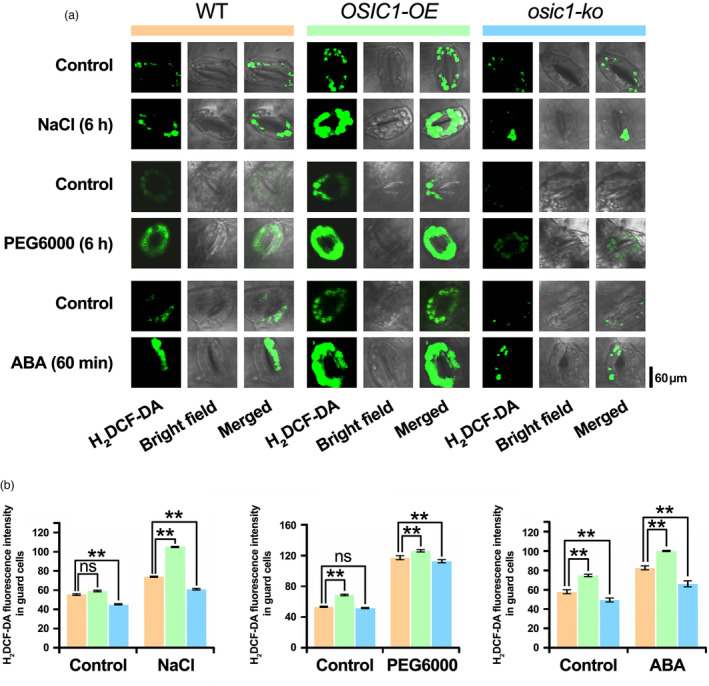
Detection of H2O2 in guard cells under different treatments. (a) Fluorescence images of guard cells from poplar leaves of different genotypes indicated the H2O2 content. The poplar leaves treated with NaCl, PEG6000 and ABA were preloaded with the fluorescent H2O2 indicator dye H2DCFDA. (b) Quantification of the fluorescence intensity in guard cells in (a). Error bars indicate SE (n ≥ 3). Asterisks indicate significant differences: **P < 0.01. The ‘ns’ represents no sense.
OSIC1 causes transcriptome changes in poplar
To determine the influence of ectopic OSIC1 on gene expression in poplar, we performed an RNA‐seq analysis using WT, salt‐stressed WT and nonstressed OSIC1‐OE poplars. Compared with the nonstressed WT, there were 5954 down‐regulated differentially expressed genes (DEGs) and 8056 up‐regulated DEGs in the salt‐stressed WT (Figure 6a and Table S1). In addition, 850 and 990 DEGs were down‐regulated and up‐regulated, respectively, in the nonstressed OSIC1‐OE poplar (Figure 6a and Table S2). Furthermore, we compared these DEGs and found that there were 584 DEGs up‐regulated both by ectopic OSIC1 and salt stress (Figure 6a), and they were mainly annotated to be involved in response to salicylic acid, oxidative stress, hyperosmotic salinity and hydrogen peroxide (Figure 6b, Figure S9a and Table S3). There were 404 DEGs down‐regulated both in OSIC1‐OE and salt‐stressed WT (Figure 6a), and they were associated with defence response, regulation of growth, response to gibberellin and leaf development (Figure 6b, Figure S9b and Table S4). The 73 DEGs which were up‐regulated in OSIC1‐OE but down‐regulated in the stressed WT (Figure 6a) were annotated to be related to salt stress response, flavonoid biosynthetic process, response to sucrose and seed development (Figure 6b, Figure S9c and Table S5). The 103 DEGs with enhanced expression levels in the stressed WT but reduced in OSIC1‐OE (Figure 6a) were implicated in defence response, wounding response, heat response and cold response (Figure 6b, Figure S9d and Table S6). Because OSIC1 plays a positive role in osmotic stress tolerance through H2O2 enhancement, we emphasized 584 co‐up‐regulated DEGs, including CuAOζ, WRKY75, OSMOTIN34, NAC32, RD22, NAC1, CBF4 and CIPK21 (Figure 6b). Therefore, we verified the expression levels of WRKY75, CIPK21, OSMOTIN34, NAC32 and CuAOζ in WT, OSIC1‐OE and osic1‐ko poplars with and without salt stress using a qPCR. All selected candidate genes were significantly up‐regulated in the OSIC1‐OE line (Figure 6c), while OSMOTIN34 and CuAOζ were down‐regulated in the osic1‐ko poplar (Figures 6c and 7a).
Figure 6.

RNA‐seq analysis and comparison of OSIC1‐OE and salt‐stressed WT plants. (a) The Venn diagram indicates the number of differentially expressed genes (DEGs) in OSIC1‐OE, WT and salt‐stressed WT poplars. (b) A heatmap indicates the expression level of the DEGs having similar expression patterns (up‐regulated and down‐regulated), or the DEGs showing opposite expression patterns in OSIC1‐OE and stressed WT poplars. (c) The qPCR verified the expression levels of WRKY75, CIPK21, OSMOTIN34 and NAC32 in WT, OSIC1‐OE and osic1‐ko poplars with or without NaCl treatment. The internal reference was the UBQ gene. The gene‐specific primers used in this qPCR assay are presented in Table S8. The error bars represent the SD of the mean values (n = 3). Letters above bars represent statistically significant differences between groups (P < 0.05) as determined by one‐way ANOVA Duncan's test.
Figure 7.

OSIC1 directly regulates PalCuAOζ expression by binding to the cis‐elements in its promoter. (a) The qPCR verified the expression of PalCuAOζ in WT, OSIC1‐OE and osic1‐ko poplars with or without NaCl treatment. (b) qPCR was used to determine the expression of PalCuAOζ after 0, 1.5, 3 and 6 h of ABA treatment. The internal reference was the UBQ gene. The error bars represent the SD of the mean values (n = 3). Letters above bars represent statistically significant differences between groups (P < 0.05) as determined by one‐way ANOVA Duncan's test. (c) The schemas indicate the distribution of OSIC1 binding elements in the PalCuAOζ promoter. (d) The Y1H assay showed direct binding of OSIC1 to OBE1 and OBE3 in the promoter of PalCuAOζ. The empty vector pGADT7 (AD) with pHIS2‐3 × OBE1 and pHIS2‐3 × OBE3 was used as the negative controls. There was 50 mm 3‐AT used in this assay. This Y1H assay was performed in 5 replicates. (e) Sequences of OBE1 and OBE3 probes, and their mutated probes in the EMSA are shown. These probes were labelled with biotin. (f) The EMSA was performed to identify the binding of MBP‐tagged OSIC1 to OBE1 and OBE3 in vitro.
OSIC1 targets PalCuAOζ
CuAOζ mediates H2O2 accumulation involved in ABA signals in Arabidopsis (Qu et al., 2014). To determine whether PalCuAOζ is the target of OSIC1 that results in the observed H2O2 accumulation before, we first confirmed this PalCuAOζ was also significantly induced by exogenous ABA, salt, PEG6000 and drought treatments (Figure 7b and Figure S10). The analysis of PalCuAOζ's promoter with a 1748‐bp length region showed that there were three potential binding elements of C2H2‐ZEP transcription factors predicted by the JASPAR database. We identified these elements as OSIC1 binding elements (OBEs) are distributed at −1274 bp ~ −1264 bp (OBE1), −864 bp ~ −874 bp (OBE2) and − 587 bp ~ −597 bp (OBE3; Figure 7c). To confirm the binding activity of OSIC1 to these elements, a yeast one‐hybrid assay (Y1H) was performed, and the results showed that OSIC1 could bind to OBE1 and OBE3 (Figure 7d). However, the DNA region containing OBE2 showed strong transcriptional self‐activation activity (Figure S11). Subsequently, we synthesized the biotin‐labelled probes and mutant probes of OBE1 and OBE3 for an electrophoretic mobility shift assay (EMSA; Figure 7e). We found that the (maltose‐binding protein) MBP‐tagged OSIC1 (MBP‐OSIC1) could bind to the labelled probes of OBE1 and OBE3, and this binding could be competitive by the nonlabelled probes (Figure 7f). Moreover, the mutant probes could not be bound by the MBP‐OSIC1 (Figure 7f). These results demonstrated that PalCuAOζ is one of the targets of OSIC1.
OSIC1 positively modulates ABA‐inducible H2O2 accumulation in guard cells via PalCuAOζ
In Vicia faba, CuAO catalyses ABA‐induced H2O2 production in guard cells (Qu et al., 2014). We found that this PalCuAOζ was expressed in the guard cells of poplar (Figure S12). Therefore, we assumed that OSIC1 was mediated by PalCuAOζ to increase H2O2 content of guard cells. To determine this, we generated the PalCuAOζ knockout poplars under the WT (palcuaoζ‐ko) and the OSIC1‐OE background (palcuaoζ‐ko/OSIC1‐OE) by the CRISPR‐Cas9 system, respectively (Figure S13a,b). The width and area of the stoma were enhanced both in palcuaoζ‐ko and palcuaoζ‐ko/OSIC1‐OE poplars compared with the WT, even without ABA application. Furthermore, the decrease in stomatal width and area was significantly less than that of WT after ABA treatment (Figure 8a,b). This result indicated that OSIC1 required PalCuAOζ to regulate ABA‐inducible stomatal closure. Additionally, the H2O2 content in guard cells was examined in these mutant poplars. Knocking out PalCuAOζ led to obviously decreased H2O2 content even if ABA was supplied, which also failed to be restored by overexpressing OSIC1 (Figure 8c,d). Therefore, PalCuAOζ‐ induced H2O2 was required for OSIC1 to modulate stomatal closure and was genetically epistasis to OSIC1.
Figure 8.

OSIC1‐mediated stomatal closure and H2O2 accumulation in guard cells were dependent on PalCuAOζ. (a) Stomatal closure in the leaves of WT, palcuaoζ‐ko and palcuaoζ‐ko/OSIC1‐OE plants treated with ABA. (b) The length, width and area from at least 19 stomata using the ImageJ software. Error bars indicate SE (n ≥ 19). Asterisks indicate significant differences: *P < 0.05, **P < 0.01. The ‘ns’ represents no sense. (c) The H2O2 content in guard cells after ABA treatment was indicated by the H2O2 indicator dye H2DCFDA. (d) The fluorescence intensity in guard cells (c) was calculated. Error bars indicate SE (n ≥ 3). Asterisks indicate significant differences: **P < 0.01.
PalMPK3 interacts with OSIC1 and mediates its phosphorylation
To further elucidate the transcriptional regulation mechanism of OSIC1, yeast two‐hybrid (Y2H) screening was performed to identify the interactional proteins of OSIC1. Among candidate proteins (Table S7), we found a homologue of Arabidopsis MITOGEN‐ACTIVATED PROTEIN KINASE3 (MPK3), which is an environmentally responsive mitogen‐activated protein kinase (MAPK) and its upstream MAPK kinases, MKK4 and MKK5, which are key regulators of stomatal development and patterning (Wang et al., 2007) and are thus named PalMPK3. A point‐to‐point Y2H assay and a co‐immunoprecipitation (Co‐IP) assay further determined this interaction between OSIC1 and PalMPK3 (Figure 9a,b) and we determined this PalMPK3 could be significantly induced by salt, drought and ABA treatment (Figure S14). These results suggest that OSIC1 could be phosphorylated by PalMPK3. To test this hypothesis, we isolated and cloned the PalMPK3 and its kinase PalMKK5 (Figure S15) and purified the recombinant MBP‐tagged OSIC1, PalMPK3 and PalMKK5DD (T218D and S224D), a constitutively active form of PalMKK5 according to its homologue in Arabidopsis (Asai et al., 2002) in Escherichia coli and performed in vitro phosphorylation assays using an anti‐MBP antibody. The results showed that MBP‐OSIC1 was phosphorylated by the active form of MBP‐PalMPK3 by PalMKK5DD, but this phosphorylation did not occur in the absence of PalMPK3 (Figure 9c). Moreover, this phosphorylation of OSIC1 in vivo could be observed when 35S:PalMPK3‐Flag, 35S:PalMKK5 DD and 35S:OSIC1‐GFP constructs were co‐expressed in the tobacco leaves (Figure 9d and Figure S16). The results indicated that PalMPK3 can phosphorylate OSIC1 in vitro and in vivo.
Figure 9.

PalMPK3 interacted with and phosphorylated OSIC1 in vitro and in vivo. (a) The Y2H assay indicated the interaction between PalMPK3 and OSIC1. The AD‐SOS2 with BD‐SOS3 was used as the positive control, while combinations including empty AD with BD, AD with BD‐PalMPK3, AD with BD‐OSIC1, and AD‐OSIC1 with BD were used as negative controls. The 3‐AT concentration in this assay was 30 mm. (b) A Co‐IP assay was performed by co‐expressing Flag‐PalMPK3 and GFP‐OSIC1 in the tobacco leaves. The proteins immunoprecipitated from leaves of tobacco extracts using anti‐Flag agarose beads (IP: anti‐Flag) were analysed by immunoblotting with anti‐GFP antibody (bottom two panels). The protein inputs were assessed by immunoblotting (top two panels). (c) PalMPK3 phosphorylates OSIC1 in vitro. PalMKK5DD, PalMPK3 and OSIC1 were fused with MBP, respectively, and were expressed and purified. The different protein combinations were incubated in the reaction buffer at 28 °C, and then, these mixtures were subjected to a Phos‐tag SDS–PAGE gel. Finally, immunoblotting with anti‐MBP antibody was performed. (d) PalMPK3 phosphorylates OSIC1 in vivo. Flag‐PalMPK3, PalMKK5DD and GFP‐OSIC1 were co‐expressed in tobacco leaves. Total proteins were extracted and subjected to Phos‐tag SDS–PAGE assay and then immunoblotting with anti‐Flag and anti‐GFP antibodies (top panel).
Phosphorylation of OSIC1 enhances its transcriptional activation activity
Phosphorylation can activate or repress the transcriptional activation activity of transcription factors to target genes (Jiang et al., 2019; Yang and Wang, 2017). To investigate the influence of phosphorylated OSIC1 on its transcriptional activity, we generated an artificial promoter using three repeats of OBE1‐OBE2‐OBE3 (3 × OBE1/2/3) followed by a 35S minimal promoter to drive a luciferase (LUC) encoding gene (Figure 10a). This construct was co‐expressed in tobacco leaves with OSIC1, a nonphosphorylated OSIC1 AA (S8A and S229A), and an OSIC1 DD (S8D and S229D) that mimics constitutive phosphorylation (Figure 10a). Subsequently, a dual‐LUC assay indicated that co‐expressed OSIC1 AA showed the lowest fluorescence intensity, and the OSIC1 DD displayed more intensity than that of OSIC1 (Figure 10b,c), indicating that the phosphorylation can enhance the transcriptional activation activity of OSIC1. Interestingly, OSIC1 DD co‐expressed with 200 mm NaCl exhibited the highest intensity among all samples (Figure 10b,c). Additionally, another dual‐LUC assay indicated that the OSIC1 co‐expressing PalMPK3 and PalMKK5 DD indeed significantly increased LUC intensity; furthermore, this LUC intensity could be further elevated as the increase in NaCl concentration increased (Figure 10d,e). These results suggest that OSIC1 has other phosphorylation sites that are phosphorylated by other salt stress‐induced protein kinases.
Figure 10.

PalMPK3‐mediated phosphorylation of OSIC1 increases its DNA‐binding ability. (a) The effector and reporter structures of the luciferase assay. (b) The luciferase assay verifies the transcriptional activity of OSIC1AA, OSIC1 and OSIC1DD to the PalCuAOζ promoter in tobacco. (c) Fluorescence intensity of the luciferase assay (b) using ImageJ software. Error bars indicate SE (n ≥ 3). Asterisks indicate significant differences: *P < 0.05, **P < 0.01. (d) The luciferase assay determined that OSIC1 phosphorylated by PalMPK3 and PalMKK5DD or NaCl treatment enhanced its transcriptional activity to the PalCuAOζ promoter in tobacco leaves. (e) Fluorescence intensity analysis of the luciferase assay (d) using ImageJ software. Error bars indicate SE (n ≥ 3). Asterisks indicate significant differences: *P < 0.05, **P < 0.01.
Discussion
High salinity and drought stress lead to osmotic stress (Zhu, 2002). Plants evolve fine‐tuned stomatal movement to balance gas exchange and water loss when these stresses occur (Verma et al., 2019). Abscisic acid and ROS signals are the dominant strategies to mediate rapid stomatal closure (Devireddy et al., 2018; Postiglione and Muday, 2020). However, how these two signals are integrated and orchestrated is still elusive in perennial woody plants, such as P. alba var. pyramidalis, which adapts well to the arid and semiarid lands of northern China (Ma et al., 2018). In this study, we found that a C2H2‐ZFP transcription factor‐encoding gene, OSIC1, in this poplar species substantiated its biofunction in stress‐induced stomatal closure. First, OSIC1 is dominantly expressed in leaves and xylem and is significantly induced by salt, drought, PEG6000 and ABA. Second, in transgenic poplars, OSIC1 plays a positive role in poplar tolerance to salt and drought stresses and causes comprehensive transcriptome reprogramming. Third, OSIC1 accelerates H2O2 accumulation by up‐regulating PalCuAOζ, resulting in stomatal closure. Finally, OSIC1 can be phosphorylated by PalMPK3 to enhance its transcriptional activity. Therefore, we proposed a novel working model of the response to osmotic stress in poplar (Figure 11): under normal conditions, the mRNA abundance of PalMPK3 and OSIC1 is sustained at a low level, and their encoding products are inactivated; thus, the transcription of PalCuAOζ is not massively activated. Therefore, the H2O2 content is insufficient to drive stomatal closure. When the poplar is subject to osmotic stress caused by salt or drought, ABA accumulates. The accumulated ABA induces PalMPK3 and OSIC1 expression and, moreover, activates PalMKK4/5, which further phosphorylates PalMPK3. Subsequently, OSIC1 is phosphorylated by activated PalMPK3. This phosphorylated OSIC1 binds to the promoter of PalCuAOζ's and up‐regulates its expression. PalCuAOζ catalyses the generation of more H2O2 that mediates stomatal closure and reduces water loss.
Figure 11.

Working model of OSIC1 in regulating stomatal closure under salt and drought stress. Under normal conditions, OSIC1, PalMPK3 and PalCuAOζ are expressed at a lower level and the stomata are open. Osmotic stress caused by salt and drought accelerates ABA accumulation, which up‐regulates OSIC1, PalMPK3 and PalCuAOζ expression levels and mediates phosphorylation of OSIC1 by the PalMKK5–PalMPK3 cascade. The phosphorylated OSIC1 can bind to the PalCuAOζ promoter and activate its transcription. PalCuAOζ then promotes the decomposition of polyamines (PAs) to produce H2O2 under stress. The H2O2 acts as a signal molecule to regulate stomatal closure to enhance poplar tolerance to osmotic stress.
ROS act as signalling molecules that function in stomatal closure when plants respond to stresses (Shen et al., 2021; Zhang et al., 2019b). Its generation necessarily relies on the ABA signal and thus activated RBOHD and RBOHF (Chater et al., 2015). Additionally, the CuAOs play an important role as a source of H2O2 and are up‐regulated by ABA (Qu et al., 2014; Tavladoraki et al., 2016). In this study, we found that OSIC1 mediated the transcription of PalCuAOζ, therefore accelerating H2O2 accumulation in stomata when stresses occur (Figures 5 and 7). This process is involved in ABA signals because OSIC1 can be induced by ABA (Figure 1b–e), and ABA‐induced H2O2 accumulation is significantly decreased in the osic1‐ko poplars (Figure 5 and Figure S7). In Arabidopsis, MPK3/MPK6, MKK4/MKK5 and the MAPKKK YODA (YDA) form a MAPK pathway that modulates stomatal development and patterning (Wang et al., 2007) and regulates stomatal immunity interdependent with ABA signals (Su et al., 2017). In poplar, salt‐induced PalMPK3 (Figure S14b) and its encoding product interact with and phosphorylates OSIC1 (Figure 9) to enhance its transcriptional activity to PalCuAOζ (Figure 10). Additionally, we also noticed that the stomatal density and the morphology are independent on OSIC1's expressions (Figure S6), suggesting that the OSIC1 is not responsible for MAPK pathway‐mediated stomatal development. Additionally, whether OSIC1 regulates stomatal immunity requires further investigation. Overall, the revealed regulation mechanism of OSIC1‐PalCuAOζ‐H2O2 at transcriptional and protein level is another pathway of ABA‐associated H2O2 signal mediating stomatal closure under stresses.
ROS are also inherent to regulating plant development. For example, the apoplastic ROS generated by two redundant RBOH isoforms, RBOHH and RBOHJ, are indispensable for proper pollen tube growth (Boisson‐Dernier et al., 2013; Kaya et al., 2014; Lassig et al., 2014). The formation of the Casparian strip within the endodermis is dependent on the precisely localized ROS production that is determined by recruitment of RBOHF to the site of lignin deposition (Lee et al., 2013). The apoplastic ROS produced by RBOHD and RBOHF also regulates the development of the secondary cell wall architecture when cell wall damage is precepted (Denness et al., 2011; Hamann et al., 2009). Stimulation of apoplastic ROS production leads to an influx of extracellular Ca2+, which is required for cell elongation. For example, RBOH isoform C (RBOHC), with its upstream RHO‐RELATED PROTEIN FROM PLANTS2, a GTPase, controls the ROS production in this process because of the deficiency of root hair elongation in rbohc mutants (Foreman et al., 2003; Jones et al., 2007). In addition, the WRKY75 transcription factor represses a catalase (CAT)‐encoding gene to inhibit H2O2 degradation, resulting in H2O2 accumulation, and thus accelerating leaf senescence in Arabidopsis (Guo et al., 2017). Its homologue in poplar regulates the development of adventitious roots and lateral buds by modulating H2O2 content (Wang et al., 2022; Zhang et al., 2022). In addition, apoplastic CuAOs are crucial players in cell wall modifications, growth regulation, programmed cell death, root xylem differentiation, fruit ripening, etc. (Tavladoraki et al., 2016). Herein, we found that OSIC1‐overexpressing poplars exhibited more branches and roots (Figure S3), which might result from the OSIC1's positive role in H2O2 accumulation through PalCuAOζ (Figures 5 and 8c and Figure S7). Therefore, the phenotype of branches and roots might be controlled by the OSIC1‐PalCuAOζ module. In addition, PalWRKY75 was also up‐regulated by ectopic OSIC1 (Figure 6c and Figure S9), indicating that OSIC1 can simultaneously increase H2O2 generation and reduce its scavenging. These results provide another example of ROS function in plant development and a potential target for breeding to screen poplar germplasms with higher osmotic tolerance and more biomass.
Experimental procedures
Plant materials and transformation
The P. alba var. pyramidalis saplings used in this study were cultivated in a greenhouse with a temperature of 25 °C, a 16‐h light/8‐h dark cycle and a relative humidity of 60% (Ma et al., 2018). Agrobacterium‐mediated transformation of leaf discs was used to generate transgenic P. alba var. pyramidalis plants (Bai et al., 2020; Niu et al., 2021). The poplar leaves were cut into pieces and placed on MS medium [containing 0.025 mg/L 6‐benzylaminopurine (6‐BA), 0.5 mg/L thidiazuron (TDZ) and 100 μmol/L acetosyringone] to induce callus formation and then in MS medium [containing 0.1 mg/L 6‐benzylaminopurine (6‐BA) and 1.0 mg/L indole‐3‐butyric acid (IBA)] to induce shoot regeneration. The transgenic plants were identified by PCR to detect the HPTII gene with the gene‐specific primers (Table S8). Transient transformation was performed in tobacco P. alba var. pyramidalis leaves according to previous methods (Ma et al., 2018; Niu et al., 2021).
Stress treatments
For salt treatment of the detached leaves, these leaves were cut into discs with a diameter of 1 cm and submerged in the MS solution with or without 200 mm NaCl for 10 days. In addition, the 1‐month‐old sterile plantlets were transplanted into the solid MS medium with or without 200 mm NaCl for 7 days. For drought treatment, the 50‐day‐old plantlets in the greenhouse were selected for drought treatment. The origin of drought is when the water content of the soil is 85%. These plantlets were evaluated for drought tolerance after 3 and 5 days of treatment. For ABA treatment, the 50‐day‐old plantlets in the greenhouse were sprayed with 150 μm ABA (He et al., 2018). For PEG6000 treatment, 50‐day‐old plantlets were watered with 25% PEG6000 solution twice a week, and after 28 days of treatment, the phenotypes were observed.
Phylogenetic analysis, sequence alignment and promoter analysis
The OSIC1 gene sequence was obtained from P. alba var. pyramidalis genome (Zhang et al., 2021) and was verified by PCR using gene‐specific primers. The sequences of OSIC1 homologues from other species were retrieved and downloaded from JGI (https://phytozome.jgi.doe.gov/pz/portal.html) and NCBI (https://www.ncbi.nlm.nih.gov/). Amino acid sequence alignments of C2H2 proteins were performed using MEGA v.6.0.6 and GeneDoc software. The phylogenetic tree of the C2H2 domain based on the maximum likelihood method was constructed using RAxML software with 1000 bootstrap replicates. The cis‐elements in the promoter of OSIC1 were predicted using online software (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
Net Na+ flux analysis in roots
The net Na+ was measured at Xu‐Yue Research Institute (Beijing, China) using NMT (NMT150 series; Younger USA LLC, Amherst, MA) and imFluxes V2.0 software (Younger USA LLC, Amherst, MA). Root samples of WT, OSIC‐OE and osic1 plantlets were excised, immediately rinsed with redistilled water, and equilibrated for 30 min in a basic solution [KCl (0.1 mm), CaCl2 (0.1 mm), MgCl2·6H2O (0.1 mm), NaCl (0.5 mm), MES (0.3 mm), Na2SO4 (0.2 mm), pH = 6.5]. Thereafter, before adding salt, a stable Na+ flux was recorded in the apical region (approximately 300 μm from the root tip) for 8 min. Salt shock (100 mm NaCl) was performed by adding NaCl stock solution (1 M). Monitor the transient ion flux for another 30 min. Due to the diffusion effect of inventory additions, flux data collected in the first 2–3 min were not included in the analysis. For each treatment, NaCl‐altered Na+ kinetics was recorded from at least six individual plants.
Nucleic acid extraction and quantitative real‐time PCR (qPCR) analysis
Total RNA was extracted from poplars with a Plant RNA Extraction Kit TRIzol® Reagent (Invitrogen, America, 15 596‐026), and 1 μg of total RNA was used to generate complementary DNA (cDNA) using a PrimeScript™ RT reagent Kit with gDNA Eraser (Perfect Real Time) kit (TaKaRa, Japan). Gene expression levels were analysed by qPCR with TB Green Premix Ex Tag II (TaKaRa, Japan). Ubiquitin (UBQ) was used as reference gene to calculate the relative levels of gene expression. The qPCR program was as follows: 95 °C for 30 s; 40 cycles of 95 °C for 5 s, 60 °C for 35 s; 95 °C for 1 min, 55 °C for 30 s, 95 °C for 30 s. All the primers used are listed in Table S8. In addition, the genomic DNA for promoter cloning, transgenic and mutant identification was extracted by a CTAB method (Rogers and Bendich, 1985).
RNA‐seq analysis
Nontreated 1‐month‐old OSIC1‐OE and WT plantlets, together with the salt‐stressed WT (200 mm NaCl for 3 days), were selected for RNA‐seq. These RNA‐Seq data were generated at Annoroad Gene Technology (Beijing, China) with an Illumina HiSeq2000 system (Illumina, San Diego, CA). After processing, we used the Hisat2 algorithm to map the clean reads to the P. alba var. pyramidalis reference genome (Zhang et al., 2021). Differentially expressed genes (DEGs) were identified by edgeR (Robinson et al., 2010), while Venny 2.1 software was used to create a Venn diagram of DEGs (https://bioinfogp.cnb.csic.es/tools/venny/index.html). The DEG heatmaps were generated using EHBIO gene technology resources (http://www.ehbio.com/Cloud_Platform/front/#/). The relative expression levels were determined by qPCR analysis.
Vector construction
For the overexpression of OSIC1, OSIC1 was isolated and cloned by polymerase chain reaction (PCR) with gene‐specific primers using complementary DNA (cDNA) (Table S8). PCR was performed with PrimeSTAR® HS DNA polymerase (Takara, China) in a total volume of 25 μL at 98 °C for 2 s, 38 cycles of 98 °C for 10 s, 55 °C for 5 s and 72 °C for 40 s, and a final extension of 72 °C for 10 min. The PCR product was ligated into the pCXSN vector (Chen et al., 2009), to generate a 35S promoter‐driven OSIC1. For the OSIC1 promoter, the 1748‐bp promoter fragment of OSIC1 was isolated and cloned from poplar genomic DNA by PCR using specific primers. This DNA fragment followed and drove a GFP reporter gene. This construct could be introduced into tobacco leaves using Agrobacterium‐mediated transient transformation. For OSIC1 and PalCuAOζ loss‐of‐function mutants (osic1‐ko and palcuaoζ‐ko), the targets were synthesized by BGI (Beijing, China). The targets were cloned into the entry vector pYLgRNA and then assembled into the destination vector pYLCRISPR/Cas9 according to a previous method (Fan et al., 2015; Ma et al., 2018). The mutated poplars were verified by PCR using HPTII gene‐specific primers (Table S8).
Physiological and biochemical analysis of poplars
To measure the photosynthetic pigment content, 0.1 g of fresh poplar leaves were submerged and homogenized with 80% acetone (v/v). Add 2 mL 80% acetone, transfer the homogenate into a centrifuge tube, and wash the mortar with an appropriate amount of 80% acetone. All liquid was collected into a centrifuge tube, diluted to 10 mL with 80% acetone and then centrifuged at 1,456g. This process was carried out at room temperature in the dark. The supernatant was taken for 2 mL to measure the absorbance at 663 nm, 645 nm and 440 nm wavelengths, respectively, and calculate the chlorophyll a (Ca), chlorophyll b (Cb), carotenoid concentration (Ck) and total chlorophyll content (Ct) according to the formula: Ca = 12.72 × D663–2.59 × D645; Cb = 22.88 × D645–4.67 × D663; Ck = 4.7 × D440–0.27 × (Ca + Cb); Ct = Ca + Cb. Two millilitres of 80% acetone were used as a reference.
The electrolyte leakage (EL) was measured as described previously (Dahro et al., 2016). The leaf blade at the eighth node was cut into circular leaf discs with a diameter of 1 cm and immersed in 30 mL of deionized distilled water. A tube containing the same volume of water served as the control. These tubes were shaken at 20 rpm at room temperature for 1 h, and then, the initial conductivities of the sample (E1) and blank (CK1) were measured by a conductivity metre. After that, the tubes were boiled at 121 °C for 20 min and cooled to room temperature, and then, the conductivity (E2 and CK2) was measured again. The EL was represented by relative conductance (E) calculated using the formula E (%) = (E1 − CK1)/(E2 − CK2) × 100%.
The malondialdehyde (MDA) content was determined using a kit (Comin Biotechnology, China) according to the manufacturer's protocol (www.cominbio.com). Fresh leaves (0.1 g) were homogenized with 1 mL of extract buffer in an ice bath and then centrifuged at 8000 g for 10 min at 4 °C. Next, 0.2 mL supernatant was transferred to a new tube and mixed immediately with 0.6 mL of Reagent 1. The reaction mixture was heated with a boiling water bath for 30 min and centrifuged at 10000 g for 20 min at 4 °C after cooling on ice. The absorbance of 1 mL of the supernatant was measured at 532 nm and 600 nm wavelength and calculated using the following formula MDA content (nmol/g fresh weight) = 25.8 × (A 532 − A 600)/W, W: sample mass (g). At least five biological replicates were performed.
Proline (Pr) content was determined using a kit (Comin Biotechnology, China) according to the manufacturer's protocol (www.cominbio.com). Fresh leaf (0.1 g) was homogenized with 1 mL of extract buffer in an ice bath. After that, it was shaken at 90 °C for 10 min and then centrifuged at 10000 g for 10 min at 25 °C. Next, 0.5 mL supernatant was transferred to a new tube and mixed immediately with 0.5 mL Reagent I and 0.5 mL reagent II. The mixture was heated with a boiling water bath for 30 min and shaken every 10 min. After cooling, 1 mL reagent III was added to the mixture and shaken for 30 s, until liquid separation. one millilitre of the upper liquid layer was pipetted to measure its absorbance at 520 nm wavelength. The Pro content was calculated using the formula Pro content (nmol/g fresh weight) = 19.2 × (A 520 + 0.0021)/W, W: sample mass (g). At least five biological replicates were performed.
The methods of measurement of the relative water content (RWC) were described (He et al., 2018). The leaves at the eighth node were detached and used for RWC measurements. The leaf FW (fresh leaves were weighed), leaf turgid weight (TW, leaves were measured after submergence in water for 8 h) and leaf dry weight (DW, leaves were measured after drying at 80 °C for 72 h) were measured. The RWC was calculated as (FW − DW)/(TW − DW) × 100%. At least five biological replicates were performed.
The methods used for the analysis of the stomatal movement were described (He et al., 2018). Stomatal conductance (Gs), transpiration (Tr) and net photosynthetic rate (Pn) were measured using the LI‐COR 6800 portable photosynthesis analysis system. At least ten plants of each line were measured.
Stomatal morphology observation
The leaf at the eighth node from 2‐month‐old poplar was fixed immediately with liquid nitrogen for 30 s. Stomatal images were collected using a Hitachi S‐3400 N scanning electron microscope. More than 100 guard cells from each sample were measured. For ABA‐induced stomatal closure, the leaves were submerged in a stomata‐opening solution (OS) containing 0.01 M KCl, 0.1 M CaCl2 and 0.01 mm MES‐KOH for 0.5 h in the dark and 2.0 h in the light, and then, 5 μm ABA was added. The stomata were fixed with liquid nitrogen after 0, 1.5 and 3 h of ABA treatment. The stomatal images were collected using a Hitachi S‐3400 N scanning electron microscope. More than 19 guard cells from each sample were measured. The stomatal area was calculated as length × width ×3.14 × 1/4 (μm2) (Wang et al., 2018).
The detection of H2O2 in leaves and guard cells
The H2O2 content in guard cells was indicated by H2DCFDA (2′,7′‐dichlorodihydrofluorescein diacetate; AbMole) as a H2O2 indicator as described previously (Gong et al., 2021). The leaves were detached from 2‐month‐old WT, OSIC1‐OE, osic1‐ko, palcuaoζ‐ko and palcuaoζ‐ko/OSIC1‐OE poplars and submerged in 50 μm H2DCFDA solution. After incubation for 25 min in the dark at 25 °C, these leaves were washed with ddH2O to remove excess dye. These leaves were submerged in an OS for 4 h and then transferred into 100 mm NaCl solution for 6 h, 7.5% PEG6000 solution for 6 h and 50 mm ABA for 1 h at room temperature. Confocal microscopy observations and images were performed (FV3000; Olympus, Japan) with a 40 × objective at 488 nm wavelength. The ImageJ software was used to calculate the fluorescence of guard cells. The H2O2 accumulation in leaves was indicated by histochemical staining using 3,3′‐diaminobenzidine‐4‐HCl (DAB). Leaves treated with salt or ABA were harvested and submerged in DAB solution and removed the photosynthetic pigments as described previously (He et al., 2018).
Yeast one‐hybrid (Y1H) assays
The JASPAR database was used to analyse the OSIC1 binding sites of the 1750‐bp PalCuAOζ promoter region (He et al., 2019) and assigned them to OBE1, OBE2 and OBE3. These binding sites were repeated three times and then fused with the pHIS2 vector to generate the pHIS2‐3 × OBE1, pHIS2‐3 × OBE2 and pHIS2‐3 × OBE3 constructs, respectively. The CDS fragment of OSIC1 was ligated to pGADT7 to generate the AD‐OSIC1 construct. These constructs were cointroduced into the Y187 yeast strain according to the manufacturer's instructions (Matchmaker Gold Y1H Library Screening System; Clontech, CA), and the transformants were grown on the synthetic dropout (SD) medium lacking tryptophan (Trp, W) and leucine (Leu, L). These transformants were screened on the SD medium lacking Trp, Leu and histidine (His, H) and with or without 50 mm 3‐amino‐1,2,4‐triazole (3‐AT). The empty pGADT7 and pHIS2‐3 × OBEs constructs were used as negative controls.
Yeast two‐hybrid (Y2H) assay
The Y2H screening was performed according to the Matchmaker GAL4 Two‐hybrid system 3 & Libraries User Manual (TaKaRa). The cDNA library of P. alba var. pyramidalis was customized using the Clontech kit (TaKaRa). The CDS of OSIC1 was ligated onto the pGBKT7 vector to generate the BD‐OSIC1 construct as the bait. The bait plasmid was first introduced into the AH109 yeast strain, and then, the transformants were transformed with the cDNA library according to the manufacturer's manual (Clontech). These transformed yeasts were screened on the SD lacking W, L and H, containing 30 mm 3‐AT. These screened yeast clones were subjected to PCR with primers (T7: TAATACGACTCACTATAGGGC; 3′AD AGATGGTGCACATGCACAG), and then, the PCR products were identified by sequencing.
For point‐to‐point Y2H, the CDSs of OSIC1 and PalMPK3 were ligated into pGADT7 and pGBKT7, respectively. These constructs were cointroduced into the AH109 yeast strain and the interaction was screened on SD medium lacking W, L and H, containing 30 mm 3‐AT. The vector combinations AD + BD, AD + BD‐PalMPK3, AD + BD‐OSIC1 and AD‐OSIC1 + BD were used as the negative controls. The combination AD‐SOS2 (GenBank: XM_011019470.1) + BD‐SOS3 (GenBank: XM_011016039.1) served as the positive control. α‐Gal was used to stain the positive transformants.
Electrophoretic mobility shift assay (EMSA)
The recombinant protein MBP‐OSIC1 was purified and used in an EMSA. Biotin‐labelled and unlabelled promoter fragments of PalCuAOζ were synthesized by BGI (Beijing, China) as probes and cold probes, respectively (Table S8). A LightShift® Chemiluminescent EMSA Kit was used in the subsequent EMSA experiments (20 148, Thermo Fisher Scientific), as detailed in the instructions (www.thermofisher.com).
Luciferase (LUC) assay
The CDSs of OSIC1, OSIC1 AA , OSIC1 DD , PalMPK3 and PalMKK5 DD were cloned and driven by a 35S promoter. The OBE1, OBE2 and OBE3 were tandem and then repeated three times. This artificial DNA fragment was followed by a 35S mini promoter to drive a LUC gene. The constructs were introduced into tobacco leaves through an Agrobacterium‐mediated transient transformation. Tobacco leaves were injected with Agrobacterium tumefaciens for 48 h, and the reaction substrate fluorescein was sprayed onto the surface of the injected leaves. The fluorescence intensity was detected by a living plant CCD imaging system.
Co‐immunoprecipitation (Co‐IP) assay
The constructs 35S:Flag‐PalMPK3 and 35S:GFP‐OSIC1 were co‐expressed in tobacco leaves by an Agrobacterium‐mediated transient transformation (Liu and Xue, 2021). After 72 h, the infected leaves were ground in liquid nitrogen, and 1 g powder was mixed in a new tube with 2 mL IP buffer (Tris–HCl 20 mm; NaCl 150 mm; EDTA 1 mm; Glycerol 10%; TritonX‐100 0.3%; PMSF 1 mm; 1× Cocktail 10 μL). Centrifuge the mixture at 1,465g, 4 °C for 10 min. Then, 200 μL supernatant mixed with 5 × protein loading buffer was pipetted as the input sample. The other extract was incubated with Flag affinity beads (ab1240; Abcam, Cambridge, UK) for 2 h at 4 °C. The beads were collected and washed three times with IP buffer containing 0.1% TritonX‐100. Finally, these beads were mixed with 5 × protein loading buffer and incubated at 95 °C for 10 min. IP products and the input samples were separated by 12% sodium dodecyl sulphate‐polyacrylamide gel electrophoresis (SDS–PAGE), and the target protein was detected by immunoblotting using anti‐Flag (1 : 1000, Abcam ab32) or anti‐GFP (1 : 1000, Abcam ab32) antibodies.
Phosphorylation assay
For protein expression, the constructed pMAL‐C2X‐OSIC1, pMAL‐C2X‐MPK3 and pMAL‐C2X‐MKK5DD vectors were introduced into E. coli strain BL21 (DE3). When the OD600 reached 0.5, the isopropyl β‐D‐1‐thiogalactopyranoside (IPTG; 1 mm) was added and shaken at 16 °C overnight to induce protein expression.
MBP fusion proteins were purified with maltose Sepharose resin (NEB, USA). A two‐step reaction was performed for the in vitro phosphorylation assay. First, MBP‐PalMKK5DD was incubated with MBP‐PalMPK3 in 20 μL of reaction buffer (200 mm Tris–HCl (pH 7.5), 100 mm MgCl2, 10 mm DTT, 1 mm ATP and 10 mm MnCl2) at 28 °C for 60 min. Second, 15 μL of the solution from reaction I was incubated with MBP‐OSIC1 in 40 μL of the reaction buffer. The reaction was stopped by adding SDS loading buffer. The protein gel was separated by SDS–PAGE or 50 μm Phos‐tag™ SDS–PAGE using 12% (w/v) acrylamide (304–93 521, WAKO, Beijing, China; https://labchem‐wako.fujifilm.com). The phosphorylated proteins were visualized by immunoblotting using an anti‐MBP antibody. For in vivo phosphorylation, co‐expressing 35S:Flag‐PalMPK3 and 35S::GFP‐OSIC1 in tobacco leaves, protein extraction and immunoblotting were performed using the same protocols as Co‐IP. The proteins were separated by SDS–PAGE or 50 μm Phos‐tag™ SDS–PAGE using 12% (w/v) acrylamide, and the phosphorylated proteins were visualized by immunoblotting.
Statistical analyses
The data from each group are presented as the means ± standard errors from three independent experiments. The physiological measurement results and the degree of stomatal opening and closing were statistically analysed by one‐way analysis of variance using SPSS 17.0 software (SPSS, Inc., Chicago, IL).
Conflicts of interest
The authors declare no competing interests.
Author contributions
D.W. designed the experiments. Q.B., Z.N., C.G. and Q.C. performed the experiments. Y.J., Q.B., J.B., J.L. and M.Z. contributed to the data analyses. D.W., Y.J., Q.B. and J.L wrote the manuscript.
Supporting information
Figure S1 Identification of the transcription factor OSIC1 in Populus alba var. pyramidalis.
Figure S2 Generation and identification of OSIC1‐OE and osic1‐ko poplar.
Figure S3 Growth status of OSIC1‐OE and osic1‐ko poplars.
Figure S4 Tolerance of different genotypic poplars to osmotic stress.
Figure S5 Net Na+ flux in roots of WT, OSIC1‐OE and osic1‐ko plants.
Figure S6 Stomata density of WT, OSIC1‐OE and osic1‐ko leaves.
Figure S7 DAB staining was performed to indicate H2O2 content in WT, OSIC1‐OE and osic1‐ko leaves before and after ABA treatment. The ABA concentration is 150 μm.
Figure S8 Quantification of H2O2 content.
Figure S9 GO enrichment of the DEGs.
Figure S10 Relative expression level of PalCuAOζ with different treatment.
Figure S11 Y1H assay showed the binding activity of OSIC1 to the OBE2 motif in the PalCuAOζ promoter.
Figure S12 PalCuAOζ expresses in guard cells of poplar.
Figure S13 Mutant identification of palcuaoζ‐ko and palcuaoζ‐ko/OSIC1‐OE generated by CRISPR/Cas9 system.
Figure S14 Identification of PalMPK3 in Populus alba var. pyramidalis.
Figure S15 Identification of PalMKK5 in Populus alba var. pyramidalis.
Figure S16 Expression of GFP‐tagged OSIC1 and Flag‐tagged PalMPK3 co‐expressed in tobacco leaves was detected by an immunoblotting using anti‐GFP and anti‐Flag antibodies.
Table S1 DEGs in salt‐stressed WT compared with WT in normal condition.
Table S2 DEGs in OSIC1 overexpression line compared with WT.
Table S3 Five hundred eighty‐four DEGs up‐regulated both in OSIC1‐OE and salt‐stressed WT.
Table S4 Four hundred four DEGs down‐regulated both in OSIC1‐OE and salt‐stressed WT.
Table S5 Seventy‐three DEGs up‐regulated in OSIC1‐OE but down‐regulated in salt‐stressed WT.
Table S6 One hundred three DEGs down‐regulated in OSIC1‐OE but up‐regulated in salt‐stressed WT.
Table S7 Proteins potentially interacting with OSIC1 according to Y2H screening assay
Table S8 Primers used in this study.
Acknowledgements
We thank Dr. Huanhuan Liu (Sichuan University) for the suggestions on experiments and gifts to related vectors. This research was supported by grants from the National Natural Science Foundation of China (No. 31870580), the National Key Research and Development Plan of China (2021YFD2200202) and the Supercomputing Center of Lanzhou University. The Core Facility of the School of Life Sciences, Lanzhou University, provided us with the qRT‐PCR facilities, SEM, CCD chemiluminescence system and fluorescence stereomicroscope.
Contributor Information
Yuanzhong Jiang, Email: wandsh@lzu.edu.cn, Email: jyz88623@126.com.
Dongshi Wan, Email: wandsh@lzu.edu.cn.
References
- Asai, T. , Tena, G. , Plotnikova, J. , Willmann, M.R. , Chiu, W.L. , Gomez‐Gomez, L. , Boller, T. et al. (2002) Alleviation of postharvest chilling injury of tomato fruit by salicylic acid treatment. Nature 415, 977–983.11875555 [Google Scholar]
- Aghdam, M.S. , Asghari, M. , Khorsandi, O. and Mohayeji, M. (2014) MAP kinase signalling cascade in Arabidopsis innate immunity. J. Food Sci. Technol. 51, 2815–2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai, Q. , Duan, B. , Ma, J. , Fen, Y. , Sun, S. , Long, Q. , Lv, J. et al. (2020) Coexpression of PalbHLH1 and PalMYB90 genes from Populus alba enhances pathogen resistance in poplar by increasing the flavonoid content. Front. Plant Sci. 10, 1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisson‐Dernier, A. , Lituiev, D.S. , Nestorova, A. , Franck, C.M. , Thirugnanarajah, S. and Grossniklaus, U. (2013) ANXUR receptor‐like kinases coordinate cell wall integrity with growth at the pollen tube tip via NADPH oxidases. PLoS Biol. 11, e1001719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer, J.S. (1982) Plant productivity and environment. Science, 218, 443–448. [DOI] [PubMed] [Google Scholar]
- Brandt, B. , Munemasa, S. , Wang, C. , Nguyen, D. , Yong, T. , Yang, P.G. , Poretsky, E. et al. (2015) Calcium specificity signaling mechanisms in abscisic acid signal transduction in Arabidopsis guard cells. Elife, 4, e03599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumbarova, T. , Le, C.T. , Ivanov, R. and Bauer, P. (2016) Regulation of ZAT12 protein stability: the role of hydrogen peroxide. Plant Signal. Behav. 11, e1137408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chater, C. , Peng, K. , Movahedi, M. , Dunn, J.A. , Walker, H.J. , Liang, Y.K. , McLachlan, D.H. et al. (2015) Elevated CO2‐induced responses in stomata require ABA and ABA signaling. Curr. Biol. 25, 2709–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S. , Songkumarn, P. , Liu, J. and Wang, G.‐L. (2009) A versatile zero background T‐vector system for gene cloning and functional genomics. Plant Physiol. 150, 1111–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cona, A. , Rea, G. , Angelini, R. , Federico, R. and Tavladoraki, P. (2006) Functions of amine oxidases in plant development and defence. Trends Plant Sci. 11, 80–88. [DOI] [PubMed] [Google Scholar]
- Dahro, B. , Wang, F. , Peng, T. and Liu, J.H. (2016) PtrA/NINV, an alkaline/neutral invertase gene of Poncirus trifoliata, confers enhanced tolerance to multiple abiotic stresses by modulating ROS levels and maintaining photosynthetic efficiency. BMC Plant Biol. 16, 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denness, L. , McKenna, J.F. , Segonzac, C. , Wormit, A. , Madhou, P. , Bennett, M. , Mansfield, J. et al. (2011) Cell wall damage‐induced lignin biosynthesis is regulated by a reactive oxygen species‐ and jasmonic acid‐dependent process in Arabidopsis. Plant Physiol. 156, 1364–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devireddy, A.R. , Zandalinas, S.I. , Gómez‐Cadenas, A. , Blumwald, E. and Mittler, R. (2018) Coordinating the overall stomatal response of plants: Rapid leaf‐to‐leaf communication during light stress. Sci. Signal. 11, eaam9514. [DOI] [PubMed] [Google Scholar]
- Fan, D. , Liu, T. , Li, C. , Jiao, B. , Li, S. , Hou, Y. and Luo, K. (2015) Efficient CRISPR/Cas9‐mediated targeted mutagenesis in Populus in the first generation. Sci. Rep. 5, 12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foreman, J. , Demidchik, V. , Bothwell, J.H.F. , Mylona, P. , Miedema, H. , Torres, M.A. and Dolan, L. (2003) Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature, 422, 442–446. [DOI] [PubMed] [Google Scholar]
- Geiger, D. , Scherzer, S. , Mumm, P. , Stange, A. , Marten, I. , Bauer, H. , Ache, P. et al. (2009a) Activity of guard cell anion channel SLAC1 is controlled by drought‐stress signaling kinase‐phosphatase pair. Proc. Natl. Acad. Sci. 106, 21425–21430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger, D. , Scherzer, S. , Mumm, P. , Stange, A. , Marten, I. , Bauer, H. and Hedrich, R. (2009b) Activity of guard cell anion channel SLAC1 is controlled by drought‐stress signaling kinase‐phosphatase pair. Proc. Natl. Acad. Sci. 106, 21425–21430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger, D. , Maierhofer, T. , Al‐Rasheid, K.A. , Scherzer, S. , Mumm, P. , Liese, A. , Ache, P. et al. (2011) Stomatal closure by fast abscisic acid signaling is mediated by the guard cell anion channel SLAH3 and the receptor RCAR1. Sci. Signal. 4, ra32‐ra32. [DOI] [PubMed] [Google Scholar]
- Gong, L. , Liu, X.‐D. , Zeng, Y.‐Y. , Tian, X.‐Q. , Li, Y.‐L. , Turner, N.C. and Fang, X.‐W. (2021) Stomatal morphology and physiology explain varied sensitivity to abscisic acid across vascular plant lineages. Plant Physiol. 186, 782–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, P. , Li, Z. , Huang, P. , Li, B. , Fang, S. , Chu, J. and Guo, H. (2017) A tripartite amplification loop involving the transcription factor WRKY75, salicylic acid, and reactive oxygen species accelerates leaf senescence. Plant Cell, 29, 2854–2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann, T. , Bennett, M. , Mansfield, J. and Somerville, C. (2009) Identification of cell‐wall stress as a hexose‐dependent and osmosensitive regulator of plant responses. Plant J. 57, 1015–1026. [DOI] [PubMed] [Google Scholar]
- Han, J.P. , Köster, P. , Drerup, M.M. , Scholz, M. , Li, S. , Edel, K.H. , Hashimoto, K. et al. (2019) Fine‐tuning of RBOHF activity is achieved by differential phosphorylation and Ca2+ binding. New Phytol. 221, 1935–1949. [DOI] [PubMed] [Google Scholar]
- He, F. , Wang, H.L. , Li, H.G. , Su, Y. , Li, S. , Yang, Y. , Feng, C.H. et al. (2018) PeCHYR1, a ubiquitin E3 ligase from Populus euphratica, enhances drought tolerance via ABA‐induced stomatal closure by ROS production in Populus . Plant Biotechnol. J. 16, 1514–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, F. , Li, H.G. , Wang, J.J. , Su, Y. , Wang, H.L. , Feng, C.H. , Yang, Y. et al. (2019) PeSTZ1, a C2H2‐type zinc finger transcription factor from Populus euphratica, enhances freezing tolerance through modulation of ROS scavenging by directly regulating PeAPX2 . Plant Biotechnol. J. 17, 2169–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrich, R. and Geiger, D. (2017) Biology of SLAC 1‐type anion channels–from nutrient uptake to stomatal closure. New Phytol. 216, 46–61. [DOI] [PubMed] [Google Scholar]
- Hou, Z. , Wang, L. , Liu, J. , Hou, L. and Liu, X. (2013) Hydrogen sulfide regulates ethylene‐induced stomatal closure in Arabidopsis thaliana . J. Integr. Plant Biol. 55, 277–289. [DOI] [PubMed] [Google Scholar]
- Hsu, P.K. , Dubeaux, G. , Takahashi, Y. and Schroeder, J.I. (2021) Signaling mechanisms in abscisic acid‐mediated stomatal closure. Plant J. 105, 307–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua, D. , Wang, C. , He, J. , Liao, H. , Duan, Y. , Zhu, Z. , Guo, Y. et al. (2012) A plasma membrane receptor kinase, GHR1, mediates abscisic acid‐and hydrogen peroxide‐regulated stomatal movement in Arabidopsis. Plant Cell, 24, 2546–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imes, D. , Mumm, P. , Böhm, J. , Al‐Rasheid, K.A.S. , Marten, I. , Geiger, D. and Hedrich, R. (2013) Open stomata 1 (OST 1) kinase controls R–type anion channel QUAC 1 in Arabidopsis guard cells. Plant J. 74, 372–382. [DOI] [PubMed] [Google Scholar]
- Jiang, H. , Tang, B. , Xie, Z. , Nolan, T. , Ye, H. , Song, G.Y. , Walley, J. et al. (2019) GSK3pen stomata 1 (OST) 1osphorylates RD26 to potentiate drought signaling in Arabidopsis. Plant J. 100, 923–937. [DOI] [PubMed] [Google Scholar]
- Jones, M.A. , Raymond, M.J. , Yang, Z. and Smirnoff, N. (2007) NADPH oxidase‐dependent reactive oxygen species formation required for root hair growth depends on ROP GTPase. J. Exp. Bot. 58, 1261–1270. [DOI] [PubMed] [Google Scholar]
- Kaya, H. , Nakajima, R. , Iwano, M. , Kanaoka, M.M. , Kimura, S. , Takeda, S. , Kawarazaki, T. et al. (2014) Ca2+‐activated reactive oxygen species production by Arabidopsis RbohH and RbohJ is essential for proper pollen tube tip growth. Plant Cell, 26, 1069–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, T.‐H. , Böhmer, M. , Hu, H. , Nishimura, N. and Schroeder, J.I. (2010) Guard cell signal transduction network: advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu. Rev. Plant Biol. 61, 561–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laity, J.H. , Lee, B.M. and Wright, P.E. (2001) Zinc finger proteins: new insights into structural and functional diversity. Curr. Opin. Struct. Biol. 11, 39–46. [DOI] [PubMed] [Google Scholar]
- Lassig, R. , Gutermuth, T. , Bey, T.D. , Konrad, K.R. and Romeis, T. (2014) Pollen tube NAD(P)H oxidases act as a speed control to dampen growth rate oscillations during polarized cell growth. Plant J. 78, 94–106. [DOI] [PubMed] [Google Scholar]
- Le, C.T. , Brumbarova, T. , Ivanov, R. , Stoof, C. , Weber, E. , Mohrbacher, J. , Fink‐Straube, C. et al. (2016) ZINC FINGER OF ARABIDOPSIS THALIANA12 (ZAT12) interacts with FER‐LIKE IRON DEFICIENCY‐INDUCED TRANSCRIPTION FACTOR (FIT) linking iron deficiency and oxidative stress responses. Plant Physiol. 170, 540–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y. , Rubio, M.C. , Alassimone, J. and Geldner, N. (2013) A mechanism for localized lignin deposition in the endodermis. Cell, 153, 402–412. [DOI] [PubMed] [Google Scholar]
- Liu, H. and Xue, S. (2021) Interplay between hydrogen sulfide and other signaling molecules in the regulation of guard cell signaling and abiotic/biotic stress response. Plant Commun. 2, 100179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, X. , Cui, N. , Zhu, Y. , Cao, L. , Zhai, H. , Cai, H. , Ji, W. et al. (2012) Over‐expression of GsZFP1, an ABA‐responsive C2H2‐type zinc finger protein lacking a QALGGH motif, reduces ABA sensitivity and decreases stomata size. J. Plant Physiol. 169, 1192–1202. [DOI] [PubMed] [Google Scholar]
- Ma, Y. , Szostkiewicz, I. , Korte, A. , Moes, D. , Yang, Y. , Christmann, A. and Grill, E. (2009) Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science, 324, 1064–1068. [DOI] [PubMed] [Google Scholar]
- Ma, J. , Wan, D. , Duan, B. , Bai, X. , Bai, Q. , Chen, N. and Ma, T. (2018) Genome sequence and genetic transformation of a widely distributed and cultivated poplar. Plant Biotechnol. J. 17, 451–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda, S. , Takano, S. , Sato, M. , Furukawa, K. , Nagasawa, H. , Yoshikawa, S. , Kasuga, J. et al. (2016) Rice stomatal closure requires guard cell plasma membrane ATP‐binding cassette transporter RCN1/OsABCG5. Mol. Plant, 9, 417–427. [DOI] [PubMed] [Google Scholar]
- Meyer, S. , Mumm, P. , Imes, D. , Endler, A. , Weder, B. , Al‐Rasheid, K.A.S. , Geiger, D. et al. (2010) AtALMT12 represents an R‐type anion channel required for stomatal movement in Arabidopsis guard cells. Plant J. 63, 1054–1062. [DOI] [PubMed] [Google Scholar]
- Mittler, R. , Kim, Y. , Song, L. , Coutu, J. , Coutu, A. , Ciftci‐Yilmaz, S. , Lee, H. et al. (2006) Gain‐ and loss‐of‐function mutations in Zat10 enhance the tolerance of plants to abiotic stress. FEBS Lett. 580, 6537–6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munemasa, S. , Hauser, F. , Park, J. , Waadt, R. , Brandt, B. and Schroeder, J.I. (2015) Mechanisms of abscisic acid‐mediated control of stomatal aperture. Curr. Opin. Plant Biol. 28, 154–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, X.C. , Kim, S.H. , Hussain, S. , An, J. , Yoo, Y. , Han, H.J. , Yoo, J.S. et al. (2016) A positive transcription factor in osmotic stress tolerance, ZAT10, is regulated by MAP kinases in Arabidopsis. J. Plant Biol. 59, 55–61. [Google Scholar]
- Niu, Z. , Li, G. , Hu, H. , Lv, J. , Zheng, Q. , Liu, J. and Wan, D. (2021) A gene that underwent adaptive evolution, LAC2 (LACCASE), in Populus euphratica improves drought tolerance by improving water transport capacity. Hort. Res. 8, 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey, V. and Shukla, A. (2015) Acclimation and tolerance strategies of rice under drought stress. Rice Sci. 22, 147–161. [Google Scholar]
- Park, S.‐Y. , Fung, P. , Nishimura, N. , Jensen, D.R. , Fujii, H. , Zhao, Y. , Lumba, S. et al. (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science, 324, 1068–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postiglione, A.E. and Muday, G.K. (2020) The role of ROS homeostasis in aba‐induced guard cell signaling. Front. Plant Sci. 11, 968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, J. , Song, C.P. , Wang, B. , Zhou, J. , Kangasjärvi, J. , Zhu, J.K. and Gong, Z. (2018) Reactive oxygen species signaling and stomatal movement in plant responses to drought stress and pathogen attack. J. Integr. Plant Biol. 60, 805–826. [DOI] [PubMed] [Google Scholar]
- Qu, Y. , An, Z. , Zhuang, B. , Jing, W. , Zhang, Q. and Zhang, W. (2014) Copper amine oxidase and phospholipase D act independently in abscisic acid (ABA)‐induced stomatal closure in Vicia faba and Arabidopsis . J. Plant Res. 127, 533–544. [DOI] [PubMed] [Google Scholar]
- Rea, G. , de Pinto, M.C. , Tavazza, R. , Biondi, S. , Gobbi, V. , Ferrante, P. , De Gara, L. et al. (2004) Ectopic expression of maize polyamine oxidase and pea copper amine oxidase in the cell wall of tobacco plants. Plant Physiol. 134, 1414–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, M.D. , McCarthy, D.J. , and Smyth, G.K. (2010) edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, S.O. and Bendich, A.J. (1985) Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues. Plant Mol. Biol. 5, 69–76. [DOI] [PubMed] [Google Scholar]
- Sakamoto, H. , Araki, T. , Meshi, T. and Iwabuchi, M. (2000) Expression of a subset of the Arabidopsis Cys2 /His2 ‐type zinc‐finger protein gene family under water stress. Genes, 248, 23–32. [DOI] [PubMed] [Google Scholar]
- Sato, A. , Sato, Y. , Fukao, Y. , Fujiwara, M. , Umezawa, T. , Shinozaki, K. , Hibi, T. et al. (2009) Threonine at position 306 of the KAT1 potassium channel is essential for channel activity and is a target site for ABA‐activated SnRK2/OST1/SnRK2. 6 protein kinase. Biochem. J. 424, 439–448. [DOI] [PubMed] [Google Scholar]
- Sayyad‐Amin, P. , Jahansooz, M.‐R. , Borzouei, A. and Ajili, F. (2016) Changes in photosynthetic pigments and chlorophyll‐a fluorescence attributes of sweet‐forage and grain sorghum cultivars under salt stress. J. Biol. Phys. 42, 601–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, C. , Zhang, Y. , Li, Q. , Liu, S. , He, F. , An, Y. , Zhou, Y. et al. (2021) PdGNC confers drought tolerance by mediating stomatal closure resulting from NO and H2O2 production via the direct regulation of PdHXK1 expression in Populus . New Phytol. 230, 1868–1882. [DOI] [PubMed] [Google Scholar]
- Sierla, M. , Hõrak, H. , Overmyer, K. , Waszczak, C. , Yarmolinsky, D. , Maierhofer, T. , Vainonen, J.P. et al. (2018) The receptor‐like pseudokinase GHR1 is required for stomatal closure. Plant Cell, 30, 2813–2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, R. , Parihar, P. , Singh, S. , Mishra, R.K. , Singh, V.P. and Prasad, S.M. (2017) Reactive oxygen species signaling and stomatal movement: Current updates and future perspectives. Redox Biol. 11, 213–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirichandra, C. , Gu, D. , Hu, H.‐C. , Davanture, M. , Lee, S. , Djaoui, M. , Valot, B. et al. (2009) Phosphorylation of the Arabidopsis AtrbohF NADPH oxidase by OST1 protein kinase. FEBS Lett. 583, 2982–2986. [DOI] [PubMed] [Google Scholar]
- Su, J. , Zhang, M. , Zhang, L. , Sun, T. , Liu, Y. , Lukowitz, W. , Xu, J. et al. (2017) Regulation of stomatal immunity by interdependent functions of a pathogen‐responsive MPK3/MPK6 cascade and abscisic acid. Plant Cell, 29, 526–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugano, S. , Kaminaka, H. , Rybka, Z. , Catala, R. , Salinas, J. , Matsui, K. , Ohme‐Takagi, M. et al. (2003) Stress‐responsive zinc finger gene ZPT2‐3 plays a role in drought tolerance in petunia. Plant J. 36, 830–841. [DOI] [PubMed] [Google Scholar]
- Takatsuji, H. (1999) Zinc‐finger proteins: the classical zinc finger emerges in contemporary plant science. Plant Mol. Biol. 39, 1073–1078. [DOI] [PubMed] [Google Scholar]
- Tavladoraki, P. , Cona, A. and Angelini, R. (2016) Copper‐containing amine oxidases and FAD‐dependent polyamine oxidases are key players in plant tissue differentiation and organ development. Front. Plant Sci. 7, 824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahisalu, T. , Kollist, H. , Wang, Y.F. , Nishimura, N. , Chan, W.Y. , Valerio, G. , Lamminmaki, A. et al. (2008) SLAC1 is required for plant guard cell S‐type anion channel function in stomatal signalling. Nature, 452, 487–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma, G. , Srivastava, D. , Tiwari, P. and Chakrabarty, D. (2019) ROS modulation in crop plants under drought stress. In Nitrogen Sulfur Species Plants, pp. 311–336. John Wiley and Sons, Ltd. [Google Scholar]
- Wang, H. , Ngwenyama, N. , Liu, Y. , Walker, J.C. and Zhang, S. (2007) Stomatal development and patterning are regulated by environmentally responsive mitogen‐activated protein kinases in Arabidopsis . Plant Cell, 19, 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W. , Su, M. , Li, H. , Zeng, B. , Chang, Q. and Lai, Z. (2018) Effects of supplemental lighting with different light qualities on growth and secondary metabolite content of Anoectochilus roxburghii . PeerJ, 6, e5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H.Q. , Sun, L.P. , Wang, L.X. , Fang, X.W. , Li, Z.Q. , Zhang, F.F. , Hu, X. et al. (2020) Ethylene mediates salicylic‐acid‐induced stomatal closure by controlling reactive oxygen species and nitric oxide production in Arabidopsis . Plant Sci. 294, 110464. [DOI] [PubMed] [Google Scholar]
- Wang, H. , Pak, S. , Yang, J. , Wu, Y. , Li, W. , Feng, H. , Yang, J. et al. (2022) Two high hierarchical regulators, PuMYB40 and PuWRKY75, control the low phosphorus driven adventitious root formation in Populus ussuriensis . Plant Biotechnol. J. 20, 1561–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, L. , Schumaker, K.S. and Zhu, J.K. (2002) Cell signaling during cold, drought, and salt stress. Plant Cell, 14, S165–S183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, D.Q. , Huang, J. , Guo, S.Q. , Yang, X. , Bao, Y.M. , Tang, H.J. and Zhang, H.S. (2008) Overexpression of a TFIIIA‐type zinc finger protein gene ZFP252 enhances drought and salt tolerance in rice (Oryza sativa L.). FEBS Lett. 582, 1037–1043. [DOI] [PubMed] [Google Scholar]
- Yang, M. and Wang, X. (2017) Multiple ways of BES1/BZR1 degradation to decode distinct developmental and environmental cues in plants. Mol. Plant, 10, 915–917. [DOI] [PubMed] [Google Scholar]
- Yu, L. , Ma, J. , Niu, Z. , Bai, X. , Lei, W. , Shao, X. , Chen, N. et al. (2017) Tissue‐specific transcriptome analysis reveals multiple responses to salt stress in Populus euphratica seedlings. Genes, 8, 372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L. , Shi, X. , Zhang, Y. , Wang, J. , Yang, J. , Ishida, T. , Jiang, W. et al. (2019a) CLE9 peptide‐induced stomatal closure is mediated by abscisic acid, hydrogen peroxide, and nitric oxide in Arabidopsis thaliana . Plant Cell Environ. 42, 1033–1044. [DOI] [PubMed] [Google Scholar]
- Zhang, X. , Liu, L. , Chen, B. , Qin, Z. , Xiao, Y. , Zhang, Y. , Yao, R. et al. (2019b) Progress in understanding the physiological and molecular responses of Populus to salt stress. Int. J. Mol. Sci. 20, 1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L. , Zhao, J. , Bi, H. , Yang, X. , Zhang, Z. , Su, Y. , Li, Z. et al. (2021) Bioinformatic analysis of chromatin organization and biased expression of duplicated genes between two poplars with a common whole‐genome duplication. Hortic. Res. 8, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Yang, X. , Nvsvrot, T. , Huang, L. , Cai, G. , Ding, Y. , Ren, W. et al. (2022) The transcription factor WRKY75 regulates the development of adventitious roots, lateral buds and callus by modulating hydrogen peroxide content in poplar. J. Exp. Bot. 73, 1483–1498. [DOI] [PubMed] [Google Scholar]
- Zhu, J.K. (2002) Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53, 247–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Identification of the transcription factor OSIC1 in Populus alba var. pyramidalis.
Figure S2 Generation and identification of OSIC1‐OE and osic1‐ko poplar.
Figure S3 Growth status of OSIC1‐OE and osic1‐ko poplars.
Figure S4 Tolerance of different genotypic poplars to osmotic stress.
Figure S5 Net Na+ flux in roots of WT, OSIC1‐OE and osic1‐ko plants.
Figure S6 Stomata density of WT, OSIC1‐OE and osic1‐ko leaves.
Figure S7 DAB staining was performed to indicate H2O2 content in WT, OSIC1‐OE and osic1‐ko leaves before and after ABA treatment. The ABA concentration is 150 μm.
Figure S8 Quantification of H2O2 content.
Figure S9 GO enrichment of the DEGs.
Figure S10 Relative expression level of PalCuAOζ with different treatment.
Figure S11 Y1H assay showed the binding activity of OSIC1 to the OBE2 motif in the PalCuAOζ promoter.
Figure S12 PalCuAOζ expresses in guard cells of poplar.
Figure S13 Mutant identification of palcuaoζ‐ko and palcuaoζ‐ko/OSIC1‐OE generated by CRISPR/Cas9 system.
Figure S14 Identification of PalMPK3 in Populus alba var. pyramidalis.
Figure S15 Identification of PalMKK5 in Populus alba var. pyramidalis.
Figure S16 Expression of GFP‐tagged OSIC1 and Flag‐tagged PalMPK3 co‐expressed in tobacco leaves was detected by an immunoblotting using anti‐GFP and anti‐Flag antibodies.
Table S1 DEGs in salt‐stressed WT compared with WT in normal condition.
Table S2 DEGs in OSIC1 overexpression line compared with WT.
Table S3 Five hundred eighty‐four DEGs up‐regulated both in OSIC1‐OE and salt‐stressed WT.
Table S4 Four hundred four DEGs down‐regulated both in OSIC1‐OE and salt‐stressed WT.
Table S5 Seventy‐three DEGs up‐regulated in OSIC1‐OE but down‐regulated in salt‐stressed WT.
Table S6 One hundred three DEGs down‐regulated in OSIC1‐OE but up‐regulated in salt‐stressed WT.
Table S7 Proteins potentially interacting with OSIC1 according to Y2H screening assay
Table S8 Primers used in this study.


