Summary
Despite the established significance of WRKY proteins and phenylpropanoid metabolism in plant immunity, how WRKY proteins modulate aspects of the phenylpropanoid pathway remains undetermined. To understand better the role of WRKY proteins in plant defence, we identified a cotton (Gossypium hirsutum) protein, GhWRKY41, that is, universally and rapidly induced in three disease‐resistant cotton cultivars following inoculation with the plant pathogenic fungus, Verticillium dahliae. We show that overexpression of GhWRKY41 in transgenic cotton and Arabidopsis enhances resistance to V. dahliae, while knock‐down increases cotton more susceptibility to the fungus. GhWRKY41 physically interacts with itself and directly activates its own transcription. A genome‐wide chromatin immunoprecipitation and high‐throughput sequencing (ChIP‐seq), in combination with RNA sequencing (RNA‐seq) analyses, revealed that 43.1% of GhWRKY41‐binding genes were up‐regulated in cotton upon inoculation with V. dahliae, including several phenylpropanoid metabolism master switches, receptor kinases, and disease resistance‐related proteins. We also show that GhWRKY41 homodimer directly activates the expression of GhC4H and Gh4CL, thereby modulating the accumulation of lignin and flavonoids. This finding expands our understanding of WRKY‐WRKY protein interactions and provides important insights into the regulation of the phenylpropanoid pathway in plant immune responses by a WRKY protein.
Keywords: cotton, GhWRKY41, Verticillium dahliae, phenylpropanoid metabolism, Gh4CLGhC4H
Introduction
Phenylpropanoid metabolism is an important secondary metabolic pathway, playing a particularly important role in generating precursors for plant defence against biotic stresses (Dong and Lin, 2021). Lignin biosynthesis is one of the branches of the phenylpropanoid pathway, and the lignin polymer provides much of the mechanical strength of the cell wall which functions as a defensive chemical barrier to limit pathogen colonization (Onohata and Gomi, 2020; Zhao and Dixon, 2011). The deposition of lignin in colonized vessels also prevents the horizontal spread of pathogens to the apoplast and surrounding healthy vessels, compartmentalizing the pathogen at the infection sites and contributing to their elimination (Kashyap et al., 2021). Flavonoids, synthesized through another branch of phenylpropanoid metabolism, are a class of multi‐functional compounds involved in myriad biological processes (Warner et al., 2021). Besides provide protection against UV damage, regulation of auxin transport, and the pigmentation of flowers, many studies have demonstrated roles for flavonoids as an antifungal agent through inhibition of mycelial growth (Gill et al., 2018; Hu et al., 2018a; Warner et al., 2021).
Lignin is a major component of the secondary cell wall (SCW) and its biosynthesis is regulated by a complex transcriptional regulatory network (Ohtani and Demura, 2019). In this hierarchical network, NAC ( NAM, ATAF1/2, CUC2) genes, such as NST1, NST2, SND1, VND6, and VND7, are first‐layer master switches that activate a subset of transcription factors (TFs) and also directly regulate genes that are responsible for the activation of specific processes in the SCW; the second‐layer master switches, such as AtMYB46 and AtMYB83, are direct downstream targets of the above NAC TFs; the third‐layer regulators, including several activators such as MYB58, MYB63, MYB85 and several repressors like MYB4, MYB6, MYB7 and MYB32, directly regulate the expression of lignin biosynthetic genes in the fourth tier (Ohtani and Demura, 2019; Pratyusha and Sarada, 2022). In addition to NAC and MYB TFs widely reported as controlling lignin metabolism, members of other TF families, such as WRKY, AP2/ERF, basic helix–loop–helix (bHLH) and HD‐ZIP, also participate in this process (Du et al., 2015; Guo et al., 2016; Yan et al., 2013; Yang et al., 2016). So far, only WRKY12 and WRKY13 from Arabidopsis have been reported oppositely to regulate the lignification in SCW through acting as direct repressors and activators, respectively, of NST2 (Li et al., 2015; Wang et al., 2010). However, whether other WRKY proteins participate in the lignin metabolism pathway remains unclear.
The flavonoid biosynthetic pathway is regulated by different sets of TFs through distinct mechanisms in a tissue‐ or species‐specific manner (Hassani et al., 2020; Liu et al., 2019; Zhao et al., 2020). Several members of the MYB and bHLH TFs families, together with TRANSPARENT TESTA GLABRA1, can form ternary protein complexes named MBW that have been extensively involved in regulating flavonoid biosynthesis (Xu et al., 2015). Some MYB proteins, such as MYB11, MYB12, and MYB111, act as particular regulators of flavonol biosynthesis (Zhang et al., 2021). To date, few members of the WRKY family have been reported to modulate flavonoid metabolism. For example, WRKY23 regulates the expression of F3'H, which catalyses the conversion of dihydrokaempferol (DHK) to dihydroquercetin (DHQ) (Grunewald et al., 2012). Whether other WRKY members mediate flavonoid metabolism requires further characterization.
WRKY proteins constitute one of the largest TF families (Reboledo et al., 2022). All WRKY factors contain a highly conserved WRKYGQK sequence motif adjacent to a zinc‐finger binding motif (Wani et al., 2021). This domain has a high binding affinity to a W‐box element [TTGAC(C/T)] which has frequently been found in promoter regions of defence‐related genes (Wani et al., 2021). Based on the number and structure of the conserved WRKY zinc‐finger motifs among WRKY proteins, the 75 WRKY members in Arabidopsis can be phylogenetically classified into three Groups (I, II and III), while Group II WRKYs can be further divided into five subgroups (IIa + IIb, IIc, IId + IIe) (Wani et al., 2021). At least four of the seven WRKY subclasses have been demonstrated to participate in protein–protein interactions between WRKY proteins (Chi et al., 2013). In Arabidopsis, three Group IIa WRKY proteins (WRKY18, WRKY40 and WRKY60) interact with each other and with themselves via their leucine zipper motifs present at the N‐terminus of each protein (Abeysinghe et al., 2019; Lahiri et al., 2019). One example is the rice Group IIa WRKY protein, WRKY71, which interacts not only with itself but also with a Group IId WRKY protein, WRKY51 (Xie et al., 2006). Given the number of WRKY‐WRKY protein interactions, studying the complex modes of functional interactions between them has great implications for understanding the regulatory mechanisms mediated by these proteins. To date, very few studies have aimed to address the functional roles of WRKYs through DNA binding and target gene expression analysis.
Nevertheless, tremendous progress have been achieved in discovering and deciphering the roles of WRKYs in plant immunity (Choi et al., 2020; Du et al., 2021; Xiong et al., 2020; Yang et al., 2020), but given a large number of WRKYs and complexity of plant‐pathogen interplay, this understanding is incomplete. In our previous study, a suppression subtractive hybridization complementary DNA library of G. barbadense constructed after inoculation with V. dahliae showed the altered expression of abundant TFs, including large numbers of WRKY genes (Xu et al., 2011). Here, we identified a gene encoding the Group III WRKY protein GhWRKY41, which was clearly up‐regulated and induced by V. dahliae in three disease‐resistant cotton cultivars. GhWRKY41 forms a positive feedback regulatory loop with itself and promotes the accumulation of lignin and a variety of flavonoids by activating the expression of GhC4H and Gh4CL in the phenylpropanoid pathway, and finally enhances cotton resistance to V. dahliae.
Results
Transcription of GhWRKY41 is up‐regulated following V. dahliae infection in disease‐resistant cotton cultivars
To identify defence‐related WRKY in cotton responsive to V. dahliae, two widely planted disease‐resistant cotton cultivars (G. hirsutum cv. 86‐4 and Zhongzhimian2) were selected and inoculated with V. dahliae for RNA‐seq. We first performed BLAST analysis (E‐value <1e‐50) with WRKY genes reported in Arabidopsis to identify all cotton WRKYs, revealing a total of 212 WRKY genes (Table S1). Among these, 71 genes in cv. 86‐4 and 38 in cv. Zhongzhimian2 showed significant transcriptional up‐regulation (fold‐change ≥2) (Figure S1A and Table S1). We previously performed RNA‐seq with the disease‐resistant cultivar G. barbadense cv. Hai7124 after inoculation with V. dahliae (Zhang et al., 2018). We found 115 WRKYs in cv. Hai7124 were significantly induced by V. dahliae (Figure S1A and Table S1). Further analysis indicated that a total of 18 WRKY genes were expressed in all these three disease‐resistant cotton cultivars, and the expression of Gh_A08G2417 showed rapid and universal induction (Figure S1B–D). We thus focused our study on the functional mechanism of the Gh_A08G2417 in possible protection against V. dahliae in cotton. RT‐qPCR was further employed and showed that the expression of Gh_A08G2417 increased in cotton roots post‐inoculation with V. dahliae, and the up‐regulated fold in two resistant cultivars (cv. 86‐4 and cv. Zhongzhimian2) was significantly higher than that in susceptible cultivar (in cv. YZ1) (Figure 1a and Figure S2). And it was ubiquitously expressed in all tissues, with higher transcription levels in the root (Figure 1b).
Figure 1.

The characterization of GhWRKY41. (a–b) Relative expression of GhWRKY41 as determined by reverse transcription quantitative PCR. Expression was normalized to internal reference GhUB7. Mock, untreated control. (a) Expression of GhWRKY41 at different time‐points following inoculation of cotton seedlings (cv. YZ1) at the three‐leaf stage with Verticillium dahliae strain V991. (b) Expression of GhWRKY41 in different tissues of cotton grown under standard conditions. Samples were taken from plants at the three‐leaf stage, and ovules were sampled at 0 and 5 days after pollination. Values represent means ± SD; n = 3. (c) Various truncated coding sequences (CDS) of GhWRKY41 were respectively fused in‐frame to the GAL4 DNA‐binding domain in pGBKT7 and transformed into Y2HGold yeast cells. The transformed cells were plated onto SD/−Trp (growth control) or SD/−Trp + X‐α‐Gal medium. (d) Transcriptional activation activity of GhWRKY41 in cotton protoplasts. 35 S‐GAL4DB and 35 S‐REN were used as the negative and internal controls, respectively. Values represent means ± SD; n = 3. (e) Subcellular localization of GhWRKY41‐GFP fusion proteins in Nicotiana benthamiana leaves. Green fluorescent protein (GFP) represents GFP fluorescence detected; red fluorescent protein (RFP) represents RFP fluorescence detected (AtHY5 encodes a nucleur‐localized protein and was used as control); Merged represents the merged images of bright‐field, GFP and RFP. Significant differences from the mock or empty vector were determined using Student's t test: N.S., not significant; **P < 0.01.
A phylogenetic tree was constructed to assess the evolutionary relationship of the Gh_A08G2417 with WRKY genes from Arabidopsis. Results show that Gh_A08G2417 clusters with WRKY Group III members, of which the closest orthologs of Gh_A08G2417 in Arabidopsis are AtWRKY41 and AtWRKY53 (Figure S3A). Gh_A08G2417 contains a WRKY domain that includes the highly conserved amino acid sequence WRKYGQK and a CX7CX23HX1C zinc‐finger motif (Figure S3B). We thus designated Gh_A08G2417 as GhWRKY41.
Subcellular localization and transcriptional activation activity of GhWRKY41
To investigate the transcriptional activation activity of GhWRKY41, various truncated fragments of the coding sequences (CDS) of GhWRKY41 were transformed into the Y2HGold yeast strain. The yeast transformants with the full‐length of GhWRKY41 activated reporter gene expression (conferring α‐galactosidase activity), whereas yeast transformed with the pGBKT7 vector alone did not (Figure 1c). The truncation analysis of the GhWRKY41 CDS showed that all fragments in GhWRKY41 are required for its transcriptional activity (Figure 1c). The dual‐luciferase reporter (DLR) assay show cotton protoplasts harbouring GhWRKY41 increased luciferase activity 15‐fold compared with the negative control (Figure 1d). Confocal microscopy revealed that AtHY5‐RFP and GhWRKY41‐GFP signals predominantly overlapped in the nuclei (Figure 1e). Collectively, these results indicate that GhWRKY41 be a nuclear‐localized protein and possesses transcriptional activity.
GhWRKY41 enhances resistance against V. dahliae in cotton
To investigate the function of GhWRKY41 in cotton defence to V. dahliae, virus‐induced gene silencing (VIGS) was employed to silence GbWRKY41 gene which is highly similar with GhWRKY41. The expression of GbWRKY41 was down‐regulated in TRV:GhWRKY41 plants compared with the control (TRV:00) at 14 days post‐agroinfiltration (Figure 2b). When GbWRKY41 was knocked down, the seedlings exhibited typical disease symptoms at 20 days after inoculation, including chlorosis, wilting and defoliation, whereas the seedlings with TRV:00 grew with mild disease symptoms (Figure 2a,c). Disease indices and rate of diseased plants analysis also support this phenotype (Figure 2d).
Figure 2.

GhWRKY41 is a positive regulator of cotton resistance to Verticillium dahliae. (a) Disease symptoms of cotton seedlings (G. barbadense cv. 7124) after inoculation for 20 days. Seedlings of TRV:00 and TRV:GhWRKY41 were inoculated with V. dahliae and re‐planted in soil with at least 25 plants. (b) Reverse transcription PCR analysis to examine the expression of GbWRKY41 in TRV:00 and TRV:GhWRKY41 plants. The GbUB7 gene was used as a control for expression. (c) V. dahliae hyphal accumulation of in TRV:00 and TRV:GhWRKY41 plant stems. (d) Disease index and rate of diseased plants of TRV:00 and TRV:GhWRKY41 plants after inoculation for 20 days. Values represent means ± SD; n = 3. (e–f) Southern blotting (e) and reverse transcription‐quantitative PCR (RT‐qPCR) analysis (f) to examine the copy number and expression respectively of GhWRKY41 in GhWRKY41‐transgenic cotton lines. The GhUB7 gene was used as the endogenous reference gene. Values represent means ± SD; n = 3. (g) Disease symptoms in wild type (WT), overexpression (OE) and RNAi (Ri) cotton plants after inoculation for 11 days. Seedlings of WT and transgenic lines were inoculated with V. dahliae and re‐planted in soil with at least 25 plants for each line. (h) Disease index of WT and transgenic plants at 5 days after the plants beginning to present disease symptoms. Values represent means ± SD; n = 3. (i) RT‐qPCR analysis of GhWRKY41 in WT and transgenic Arabidopsis lines. The AtACTIN gene was used as the endogenous reference gene. The values are the means ± SD; n = 3. (j) Disease symptoms in WT and overexpressing Arabidopsis lines at 3 weeks after inoculation with V. dahliae. (k) Disease index statistics in WT and transgenic Arabidopsis lines after inoculation with V. dahliae for 3 weeks. Values are the means ± SD; n = 3. All statistical analyses were performed using Student's t test: **, P < 0.01. All assays were repeated three times with similar results.
The transgenic cotton lines were obtained and identified through Southern blotting and Reverse transcription‐quantitative PCR (RT‐qPCR) analysis; two independent GhWRKY41‐overexpression lines (OE‐93 and OE‐112) and two independent RNAi lines (Ri‐228 and Ri‐230) were selected for further study (Figure 2e,f). All the transgenic and WT seedlings were challenged with V. dahliae and showed distinct symptoms. GhWRKY41‐overexpressing plants showed more resistance, while GhWRKY41 knock‐down plants exhibited enhanced susceptibility (Figure 2g). The statistical analyses of the disease index from these experiments were also consistent with the phenotypic disease symptom (Figure 2h). We also obtained two independent GhWRKY41‐overexpressing Arabidopsis lines (OE‐1 and OE‐2) (Figure 2i). Similar to the results in cotton, the GhWRKY41‐overexpressing plants displayed a higher level of resistance compared to WT (Figure 2j,k). Taken together, these findings suggest that the overexpression of GhWRKY41 improved resistance to V. dahliae in both cotton and Arabidopsis, while the knockdown of GhWRKY41 makes cotton more susceptible to the pathogen.
Genome‐wide identification of GhWRKY41 target genes
To further investigate the regulatory roles of GhWRKY41, genome‐wide DNA‐binding sites of GhWRKY41 were surveyed using a ChIP‐seq strategy. We mapped the sequencing reads of ChIP DNA from three biological immunoprecipitated samples to the TM‐1 cotton genome (Zhang et al., 2015) resulting in a total of 2351, 1981 and 2182 peaks, respectively (Figure 3a). Comparative data analysis revealed 1315 overlapping peaks (Figure 3a and Table S2), which were considered as the high‐confidence GhWRKY41‐binding regions, and used for further analysis. The 1315 GhWRKY41 binding peaks were located in the genic regions of 1019 potential target genes (Figure 3d and Table S2). Of these, 39.01% were located in promoter regions (−3 kb to the transcription start site, TSS) (Figure 3b). Of note, the DNA‐binding sites of GhWRKY41 significantly located within the 3‐kb region upstream of the TSS and with the highest distribution in the region adjacent to the TSS (Figure 3c). Among the 1019 GhWRKY41‐binding candidate genes, 439 (43.1%) genes overlap with V. dahliae‐responsive DEGs (Figure 3d and Table S3). Of these, the expression of most of the GhWRKY41‐binding genes was prominently induced by V. dahliae (Figure 3e and Table S3), suggesting that more genes could be up‐regulated directly by GhWRKY41 in cotton resistance against V. dahliae. And some genes were predicted in association with stress/defence responses, and transcriptional regulation and post‐translational modification (Figure 3f).
Figure 3.

Genome‐wide identification and expression profiles of GhWRKY41 target genes in cotton. (a) Chromatin immunoprecipitation and high‐throughput sequencing (ChIP‐Seq) using three biological replicates revealed 1315 high‐confidence GhWRKY41‐binding peaks, which were shared across replicates. (b) Distribution of GhWRKY4‐association sites in different regions of the annotated genes. (c) GhWRKY41‐association sites are highly enriched in the region proximal to the transcriptional start sites (TSS). (d) Venn diagram showing the overlap between genes bound by GhWRKY41 from ChIP‐Seq and the DEGs in cotton (Gossypium hirsutum cv. YZ1) inoculated with Verticillium dahliae by RNA‐Seq. (e) Expression profiles of 439 GhWRKY41‐targeted DEGs in cotton inoculated with V. dahliae at 6, 12 and 24 h. The colour legend indicates normalized gene expression value among genotypes. Clusters indicate up‐regulated genes induced by V. dahliae. The red colour indicates relatively high expression and blue indicates relatively low expression. (f) Twenty up‐regulated DEGs in (e) encode proteins associated with transcriptional regulation, post‐translational modifications and stress/defence response within GhWRKY41 targets in inoculated and uninoculated cotton.
GhWRKY41 binds and activates the expression of itself
GhWRKY41 was identified itself as a potential target gene of GhWRKY41 (Figure 3f). Totally, six W‐box elements were revealed within the promoter of GhWRKY41 (ProGhWRKY41, 2000 bp upstream of ATG codon) (Figure 4a). Yeast one‐hybrid (Y1H) assay was performed and confirmed the ProGhWRKY41‐GhWRKY41 interaction (Figure 4b). And the interaction was confirmed by ChIP‐qPCR (Figure 4c). We further investigated the transcription activation activity of GhWRKY41 by a DLR assay (Figure 4d). The results revealed that GhWRKY41 promoter drives LUC expression weakly without GhWRKY41, but co‐expression of ProGhWRKY41:LUC with GhWRKY41 led to a significant increase in LUC luminescence intensity (Figure 4e). LUC activity increased 6‐fold compared with the control when ProGhWRKY41:LUC and GhWRKY41 were co‐expressed (Figure 4f). These results suggest that GhWRKY41 binds to the promoter of itself and directly activates its transcription.
Figure 4.

GhWRKY41 binds to and directly activates the promoter of itself. (a) Schematic diagrams of the promoter of GhWRKY41. The red rectangles indicate positions of the W‐box elements. P1‐P5 regions show the promoter fragments containing the W‐box elements used for Chromatin immunoprecipitation (ChIP)‐quantitative PCR (qPCR) assay. (b) Yeast one‐hybrid (Y1H) assay shows that growth of yeast cells cotransformed with the ProGhWRKY41‐AbAi and GhWRKY41‐pGADT7 vector, and negative control (ProGhWRKY41‐AbAi + pGADT7 empty vector) on selective medium added without (up) or with (down) aureobasidin A (AbA). (c) ChIP‐qPCR analysis of GhWRKY41 binding affinity to itself own promoter. After normalization with GhUB7, the relative enrichment in no antibody were designed as 1 to normalize that in chromatins immunoprecipitated with anti‐Myc antibody. The values are the means ± SD; n = 3. (d) Schematic diagrams of the effector and reporter constructs used for tobacco transient expression assay. MCS, multiple cloning sites. 35 S and CaMV term are promoter and terminator of CaMV 35 S, respectively. LUC and REN are firefly luciferase and renilla luciferase, respectively. EV represents the emtpy vector and was used as the negative control. (e) Luminescence imaging of transient dual‐luciferase reporter (DLR) assay shows the driven effect of GhWRKY41 to itself. Construct pGreenII‐0800 LUC containing the GhWRKY41 promoter and construct pGreenII 62‐SK with or without the GhWRKY41 coding region were transiently expressed in N. benthamiana leaves (n = 10). (f) Quantification of relevant LUC activities in (e). The average value of fluorescence in EV was set as 1. The values are the means ± SD; n = 6. Statistical analyses in (c) and (f) were performed using Student's t test: **, P < 0.01.
GhWRKY41 homodimer activates its own transcripts upon V. dahliae infection
To screen possible interactors of GhWRKY41 involved in cotton resistance to V. dahliae, we first carried out a yeast two‐hybrid (Y2H) assay. Since the full‐length of GhWRKY41 protein has strong auto‐activity, the truncated GhWRKY41 with the N‐terminal 190 amino acid residues (GhWRKY41ΔC) was used as bait to screen a Y2H library (Figure 1c). Interestingly, GhWRKY41ΔC was found to be interacted with GhWRKY41 (Figure 5a). Firefly luciferase complementation imaging (LCI) and biomolecular fluorescence complementation (BiFC) assays were conducted to further verify this interaction. As shown in Figure 5b,c, strong Luc fluorescence was detected in N. benthamiana leaves co‐infiltrated with GhWRKY41‐NLuc and GhWRKY41‐CLuc, but no significant Luc fluorescence was observed in the negative controls. Moreover, the co‐expression of GhWRKY41‐NYFP and GhWRKY41‐CYFP led to clear YFP signals in the nucleus while no signal was observed in the negative controls (Figure 5d), suggesting that GhWRKY41 forms a homodimer in vivo. We further performed a Western blotting assay in which GhWRKY41‐Myc fusion protein was expressed in cotton protoplast. The result showed that GhWRKY41‐Myc homodimer and monomer are both present in cotton cells (Figure 5e and Figure S4), suggesting that GhWRKY41 monomer can partly convert into homodimer under normal condition and V. dahliae supernatant treatment. To further verify whether the conversion depends on V. dahliae, we performed a LCI assay in which GhWRKY41‐NLuc and GhWRKY41‐CLuc were co‐expressed in cotton protoplast with different treatments. Under normal condition, GhWRKY41‐NLuc and GhWRKY41‐CLuc showed strong interaction and increased luciferase activity ~10‐fold compared with the negative control (Figure 5f). Furthermore, after treatment with V. dahliae supernatant or chitin solution, luciferase activity of GhWRKY41 protein–protein interaction was significantly increased to 23‐fold ~27‐fold, respectively (Figure 5f), suggesting that both treatments promote the formation of GhWRKY41 homodimer. While after chitinase treatment, the luciferase activity of homodimer significantly decreased (Figure 5f).
Figure 5.
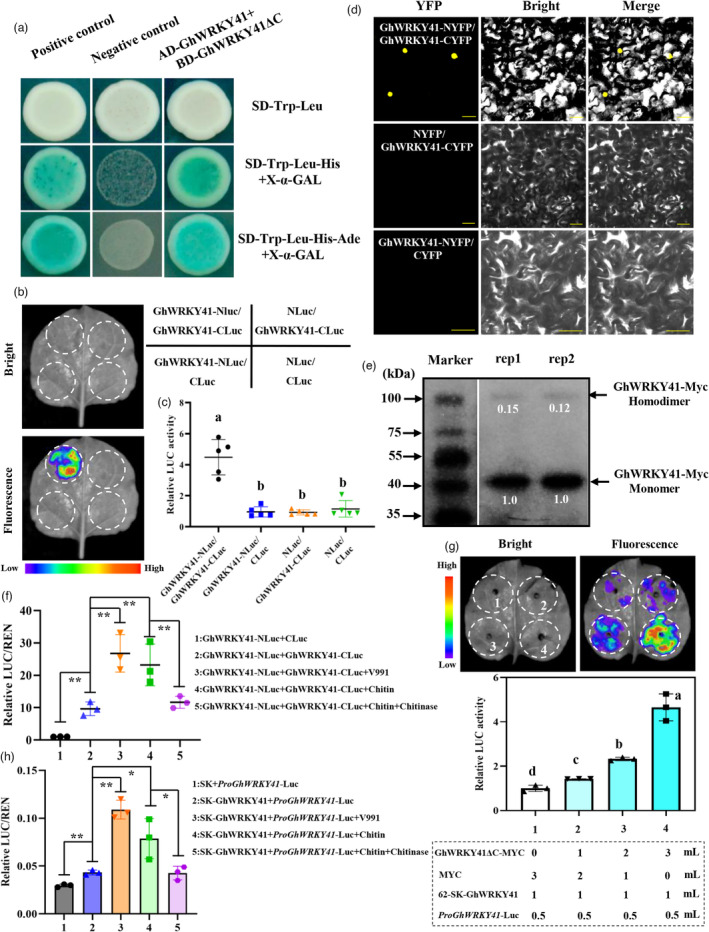
GhWRKY41 interacts with itself to promote its own expression. (a) Yeast two‐hybrid (Y2H) assay to detect interaction between GhWRKY41 and itself. The full‐length GhWRKY41 was fused with the activation domain (GhWRKY41‐pGADT7) and the carboxyl terminus deletion of GhWRKY41 was fused with the binding domain (GhWRKY41ΔC‐pGBKT7). Transformed yeast cells were grown on synthetic dextrose (SD) medium, and the blue colonies on SD‐Trp‐Leu‐His (with 5‐bromo‐4‐chloro‐3‐indolyl‐β‐d‐galactopyranoside acid [X‐α‐Gal]) and SD‐Trp‐Leu‐His‐Ade (with X‐α‐Gal) media indicate positive interactions. (b) A firefly luciferase complementation imaging (LCI) analysis of the interaction between GhWRKY41 and itself. Agrobacterium strains containing the indicated pairs were co‐expressed in N. benthamiana. The luminescent signal was collected at 48 h after infiltration. (c) Quantification of relevant Luc activities in (b). The values are the means ± SD; n = 5. (d) Biomolecular fluorescence complementation (BiFC) assay showing that the interaction between GhWRKY41‐NYFP and GhWRKY41‐CYFP formed a functional yellow fluorescent protein (YFP) in the nuclei. The interactions between GhWRKY41‐NYFP and CYFP, and between GhWRKY41‐CYFP and NYFP, were used as negative controls for the BiFC assay. Merged = merging of YFP and bright. Bars = 20 μm. (e) Western blotting assay showing the presence both of GhWRKY41 homodimer and monomer in cotton protoplast cells. GhWRKY41‐Myc fusion proteins were immunoprecipitated with MYC‐coupled beads, and the immunoblot was probed with anti‐Myc antibodies. (f) LCI assay showing the formation of GhWRKY41 homodimer in cotton protoplast cells was promoted by V. dahliae supernatant and chitin treatment. The values are the means ± SD; n = 3. (g) Luminescence imaging of LCI assay shows the driven effect of GhWRKY41 homodimer to GhWRKY41 promoter. The N. benthamiana leaves (n = 10) expressed 62‐SK‐GhWRKY41 and ProGhWRKY41‐LUC together with GhWRKY41ΔC‐MYC and/or MYC in different ratios as shown in the dotted box. The empty vector containing MYC tag was used as the negative control. The values are the means ± SD; n = 3. (h) DLR assay showing the generated GhWRKY41 homodimer in cotton protoplast cells promoted by V. dahliae supernatant and chitin treatment increased the driven effect to GhWRKY41 promoter. The values are the means ± SD; n = 3. Significant differences in (f–h) were determined using Student's t‐test: *, P < 0.05; **, P < 0.01. In other graphs, different letters indicate significant differences as determined using ANOVA and LSD multiple comparisons (P < 0.05).
We further investigated the relationship between GhWRKY41 homodimer and GhWRKY41 transcripts. Since GhWRKY41ΔC without transcriptional activity could interact with GhWRKY41, the activation effect of the GhWRKY41 homodimer on ProGhWRKY41 was assayed with GhWRKY41 and GhWRKY41ΔC‐Myc. The results showed that the LUC fluorescence gradually increased along with the content of GhWRKY41ΔC‐MYC in N. benthamiana leaves co‐infiltrated with 62‐SK‐GhWRKY41 and ProGhWRKY41‐Luc (Figure 5g). We therefore suggest that GhWRKY41 homodimer and GhWRKY41 transcripts formed a positive regulation loop to amplify its own regulatory activity. We also performed a DLR assay in which SK‐GhWRKY41 and ProGhWRKY41‐Luc were co‐expressed in cotton protoplast with different treatments. The results showed that V. dahliae supernatant and chitin treatments both promote the formation of GhWRKY41 homodimer and significantly increased the transcriptional activity (Figure 5h). While chitinase treatment obviously passivated the effect of chitin treatment for GhWRKY41 to ProGhWRKY41‐Luc (Figure 5h), suggesting that activation effect of the GhWRKY41 homodimer to ProGhWRKY41 depends on V. dahliae and the key pathogen elicitor chitin.
GhWRKY41 promotes lignin and flavonoid synthesis
We identified several putative GhWRKY41‐targeted TFs in the ChIP‐seq data analysis, including GhNST1 and GhSND2 (Figure 3f), which are master switches in the phenylpropanoid pathway (Fang et al., 2020; Zhang et al., 2020; Zhong et al., 2008). Further ChIP‐qPCR analysis revealed that GhWRKY41 directly binds to the promoters of GhNST1 and GhSND2 (Figure S5). Consistent with the expression pattern of GhWRKY41 induced by V. dahliae, the transcripts of GhSND2 were also significantly up‐regulated following V. dahliae inoculation (Figure S6). To investigate whether GhWRKY41 regulates the accumulation of lignin and flavonoids, we performed a histochemical stain assay in GhWRKY41‐transgenic cotton. Phloroglucinol‐HCl staining of stem cross‐sections showed that more lignin was found in GhWRKY41‐overexpression lines, specifically in the xylem and phloem, while less lignin are found in GhWRKY41‐silencing plants compared with WT both under normal growth condition and after inoculation with V. dahliae (Figure 6a). Similarly, NaOH staining suggested that flavonoid accumulation was also increased in overexpressing lines and decreased in RNAi lines, and the infection by V. dahliae significantly increased the flavonoid accumulation (Figure 6b). We further measured the content of lignin and a variety of flavonoids. The results showed that the contents of lignin and several flavonoids, including DHK, DHQ, eriodictyol (Eri), and naringenin (Nar), were significantly higher in GhWRKY41‐overexpressing plants, and lower in RNAi lines, compared with WT (Figure 6c,g), which consistent with the results of histochemical staining. The lignin content in Arabidopsis were also was increased in GhWRKY41‐OE Arabidopsis (Figure S7A), whereas no significant differences in flavonoid accumulation were observed between WT and transgenic Arabidopsis, apart from DHQ (Figure S7B), suggesting that GhWRKY41 may participate in the regulation of the flavonoid pathway in cotton and Arabidopsis in a different way.
Figure 6.

GhWRKY41 promotes lignin and flavonoids accumulation. (a–b) The histochemical analysis to observe the lignin (a) and flavonoids (b) deposition in stem from wild type (WT) and GhWRKY41‐transgenic cotton under normal growth condition and after inoculation with V. dahliae for 7 d. EP: epidermis, Co: cortex, P: phloem, X: xylem, Pi: pith. (c) The determination of the lignin in WT and GhWRKY41 transgenic cotton. The values are the means ± SD; n = 12. (d–g) The content of four flavonoids, including dihydrokaempferol (DHK) (d), dihydroquercetin (DHQ) (e), eriodictyol (Eri) (f) and naringenin (Nar) (g) were measured and analysed in WT and GhWRKY41‐transgenic cotton on HPLC‐MS/MS equipment. Values are the means ± SD; n = 6. Different letters in (c–g) indicate significant differences as determined using ANOVA and LSD multiple comparisons (P < 0.05). (h,i) The fungus was grown for 5 days on the potato dextrose agar (PDA) plate with 0.1 μg/mL each flavonoid (h) and the colony diameter was determined (i). The fluorescence show the signal of green fluorescent protein (GFP) in V. dahliae strain V592 fused GFP. Values are the means ± SD; n = 6. (j) The statistics of fluorescence signal in (h). Values are the means ± SD; n = 3. Statistical analyses in (i–j) were performed using a Student's t test: **P < 0.01.
To test the toxicity of these flavonoids to the V. dahliae strain V592 containing a green fluorescent protein (GFP) reporter gene, we added each flavonoid at different concentrations in potato dextrose agar medium, based on previous reports (Hu et al., 2018a; Ohtani and Demura, 2019). The results showed that all flavonoids could inhibit the growth of V. dahliae with the dramatically reduced colony diameter and GFP fluorescence (Figure 6h,j). There was a clear correlation between the flavonoid concentrations and hyphal growth of V. dahliae (Figure S8). These results suggested that the accumulation of flavonoids in the plants may play essential roles in the defence response to V. dahliae.
GhWRKY41 activates expression of GhC4H and Gh4CL
Based on the results that GhWRKY41 could promote accumulation of lignin and flavonoids, we determined expression levels of phenylalanine ammonia‐lyase (GhPAL), cinnamate 4‐hydroxylase (GhC4H), and cinnamate 4‐hydroxylase (Gh4CL), which collectively referred to as the general phenylpropanoid pathway (GPP) for the synthesis of lignin, flavonoids, and other derivatives (Liu et al., 2015). It was found that the expression of both GhC4H and Gh4CL, but not GhPAL, was up‐regulated in GhWRKY41‐overexpression lines, while transcripts of all genes were reduced significantly in RNAi plants (Figure 7a). Four W‐box elements were detected in promoters of GhC4H and two in Gh4CL (Figure 7b). The results of ChIP‐qPCR assay showed that the W‐box region of the GhC4H and Gh4CL promoters were significantly enriched when compared with the mock, which indicated that GhWRKY41 binds to GhC4H and Gh4CL promoters in vivo (Figure 7c). We also conducted a DLR assays in tobacco and cotton protoplasts. Comparing with the empty vector control, GhWRKY41 dramatically increased the firefly LUC reporter expression (Figure 7e,f), in agreement with the expression of GhC4H and Gh4CL levels in the GhWRKY41‐overexpressing plants, indicating that GhWRKY41 directly activates GhC4H and Gh4CL. To investigate whether GhWRKY41 homodimer regulates the expression of GhC4H and Gh4CL, we performed DLR dose response assays. Consistent with the self‐regulation of the GhWRKY41 via homodimer, the intensity of Luc fluorescence also gradually increased along with an increase of injection volume of GhWRKY41ΔC in N. benthamiana leaves co‐infiltrated with 62‐SK‐GhWRKY41 and ProGhC4H‐LUC/ProGh4CL‐LUC together with GhWRKY41ΔC‐MYC or MYC (Figure 7g,h).
Figure 7.

GhWRKY41 directly activates the GhC4H and Gh4CL. (a) RT‐qPCR analysis of relative expression of GhPAL, GhC4H and Gh4CL from general phenylpropanoid pathway in GhWRKY41‐transgenic cotton. Expression was normalized to the internal reference gene GhUB7, and values are relative to those in the WT, which were set as 1. Data are means (±SD), n = 3. (b) Schematic diagrams of the promoters of GhC4H and Gh4CL. The yellow and orange triangles indicate positions of the W‐box elements. P1‐P5 regions show the promoter fragments used for ChIP‐qPCR primers design. (c) ChIP‐qPCR analysis of GhWRKY41 binding affinity to the promoters of GhC4H and Gh4CL. After normalization with GhUB7, the relative enrichment in no antibody were designed as 1 to normalize that in chromatins immunoprecipitated with anti‐Myc antibody. The values are the means ± SD; n = 3. (d) Schematic diagrams of the effector and reporter constructs used for tobacco transient expression assay. EV represents the empty vector and was used as the negative control. (e) Luminescence imaging of transient dual‐luciferase reporter (DLR) assay shows the driven effect of GhWRKY41 to GhC4H and Gh4CL, respectively. Construct pGreenII‐0800 LUC containing the ProGhC4H or ProGh4CL and construct pGreenII 62‐SK with or without the GhWRKY41 coding region were transiently expressed in N. benthamiana leaves (n = 10). (f) The LUC/REN activity ratios of the GhWRKY41 protein to ProGhC4H‐LUC or ProGh4CL‐LUC in cotton protoplast. The average value of fluorescence in EV was set as 1. The values are the means ± SD; n = 3. (g,h) Luminescence imaging of LCI assay shows the driven effect of GhWRKY41 homo‐complex to the ProGhC4H (g) and ProGh4CL (h), respectively. The N. benthamiana leaves (n = 10) expressed 62‐SK‐GhWRKY41 and ProGhC4H‐LUC (g) or ProGh4CL‐LUC (h) together with GhWRKY41ΔC‐MYC and/or MYC in different ratios. The values are the means ± SD; n = 3. Statistical analyses in (a), (c) and (f) were performed using Student's t test: *, P < 0.05; **, P < 0.01. Different letters in (g) and (h) indicate significant differences as determined using ANOVA and LSD multiple comparisons (P < 0.05).
Discussion
GhWRKY41 is a defence‐responsive regulator of the phenylpropanoid pathway
In the last decade, increasing and compelling evidence indicated the roles of WRKYs in immune responses in various plants, including rice, Arabidopsis, cotton, and so on (Abeysinghe et al., 2019; Choi et al., 2020; Du et al., 2021; Xiong et al., 2020). In particular, so far, multiple WRKY family members in cotton have been demonstrated to participate in the defence response of cotton to verticillium wilt (Li et al., 2014; Xiong et al., 2019, 2020). For example, GhWRKY70D13 and GbWRKY1 negatively regulate cotton's resistance to V. dahliae through repressing jasmonic acid (JA) and ethylene signalling pathway (Li et al., 2014; Xiong et al., 2020), and the silencing of GhWRKY70 in cotton lead to increased resistance to V. dahliae which may depends on the coordination effect of salicylic acid and JA signalling (Xiong et al., 2019). These findings indicated that WRKY proteins mediate the disease response of cotton through regulation of different metabolic pathway. In addition, a series of physiological, morphological and molecular studies indicated that the phenylpropanoid metabolism is one of the most important secondary metabolic pathways involved in plant defence against V. dahliae (Hu et al., 2018a; Luo et al., 2021; Xu et al., 2011;). Both resistant and susceptible cotton cultivars show increased lignin deposition after V. dahliae infection, but the resistant plants have the denser xylem with a higher lignin content (Xu et al., 2011). A cotton laccase, GhLac1, modulates broad‐spectrum biotic stress tolerance by mediating the metabolic flux between the monolignol and flavonoid pathways (Hu et al., 2018a). A spontaneous cotton mutant with red coloration contains abundant flavonoids and shows antifungal activity to V. dahliae with reduced pathogen colonization (Long et al., 2019). In our study, GhWRKY41 rapidly and universally responds V. dahliae and directly activates the expression of genes of GPP involved in the biosynthesis of monolignol and flavonoids, and simultaneously promotes accumulation of lignin and a range of flavonoids, leading to enhanced cotton resistance to V. dahliae (Figure 8).
Figure 8.

Schematic model illustrating the proposed roles of GhWRKY41 in plant defence against V. dahliae. GhWRKY41 is rapidly induced by V. dahliae and subsequently forms homodimer with itself, which not only directly activates GhWRKY41 itself to amplify the activation effect, but also binds and activates the expression of GhC4H and Gh4CL from the general phenylpropanoid pathway to promote the accumulation of lignin and flavonoids, thus leading to improved cotton resistance to V. dahliae.
Numerous regulators of lignin and flavonoid biosynthesis have been identified and characterized, and the key TFs belong mainly to the MYB and NAC families (Liu et al., 2015; Xu et al., 2015; Zhang et al., 2020; Zhao and Dixon, 2011; Zhong et al., 2008). The NAC‐MYB‐based gene regulatory network model is widely considered to underpin lignin biosynthesis (Ohtani and Demura, 2019). Studies indicate that a few members of other TF families likely participate in this hierarchical network. E2Fc, a member of the E2F TF family, is a key upstream regulator of the first‐layer master switches VND6, VND7 and of SCW biosynthesis genes (Taylor‐Teeples et al., 2015). WRKY12 in Arabidopsis also binds the promoter of the first‐layer master gene NST2 and leads to its repression (Wang et al., 2010). Our study identified a novel positive regulator of lignin deposition, GhWRKY41, which directly acts on the fourth‐layer monolignol biosynthetic genes GhC4H and Gh4CL (Figure 7), suggesting that GhWRKY41 is a third‐layer regulator in the hierarchical network. It is worth noting that in our ChIP‐seq results, we identified two master factors GhNST1 and GhSND2 (Figure 3 and Figure S2), whose homologous genes in Arabidopsis reside in first‐ and second‐layers, respectively, of the regulatory network (Hussey et al., 2011; Zhang et al., 2020; Zhong et al., 2008). Further studies are necessary to define the detailed mechanisms by which GhWRKY41 directly regulates GhNST1 and GhSND2.
GhWRKY41 forms a positive regulation loop with itself
WRKY proteins often interact with other proteins to carry out biological functions (Chi et al., 2013; Li et al., 2022; Viana et al., 2018). A growing body of evidence also point to WRKY‐WRKY protein–protein interactions (Chi et al., 2013). Three Group IIa WRKY (WRKY18, WRKY40 and WRKY60) from Arabidopsis interact individually with themselves and with each other (Xu et al., 2006). Additionally, Y2H assays revealed that WRKY40 and WRKY60 interact with WRKY36 (Group IId) and WRKY38 (Group III) (Consortium, 2011). The formation of heterodimers or homodimers between WRKY TFs have also been found in several other species, such as rice and apple (Dong et al., 2020; Viana et al., 2018). In our study, the identified Group III WRKY member GhWRKY41 interacts physically with itself, and moreover, GhWRKY41 homodimer and monomer are both present in cotton cells (Figure 5), suggesting that GhWRKY41 monomer can partly convert into homodimer to exert its biofunction in plant. Very recently, we observed that GhWRKY41 could also interact with another Group III protein, GhWRKY30, to form heterodimers (unpublished data), expanding our understanding of interactions between Group III WRKY members. Additional studies are necessary to confirm whether similar molecular mechanisms exist among different WRKY members in the regulation of phenylpropanoid metabolism and plant immunity.
The presence of the W‐box in the promoters of multiple WRKY suggests auto‐regulatory or cross‐regulatory properties of these TFs, whereby WRKYs regulate the expression of each other (Viana et al., 2018). This also implies regulation by internal feedback loops possibly autoregulate their expression (Eulgem et al. 2000). For instance, AtWRKY53 is not only an interaction partner of AtWRKY18, but each is also an upstream regulator, and downstream target of the other (Potschin et al., 2014). ChIP‐seq assay showed GhWRKY41 can target itself, and we demonstrated that GhWRKY41 could autoactivate its own expression (Figure 4). Further study found that GhWRKY41 homodimer could amplify the activation effect of GhWRKY41 on itself, and moreover, the activation effect was promoted by V. dahliae supernate and chitin (Figure 5). We therefore speculate that upon V. dahliae infection, GhWRKY41 forms a positive feedback loop with itself to exert its regulatory function in the cotton immune response (Figure 8). It is worth noting that many other WRKY members, such as Gh_D05G1968, Gh_A11G2016, Gh_A06G0879 and Gh_D04G1122 were identified to act as candidate targets of GhWRKY41 (Table S2), but whether these WRKYs are directly regulated by GhWRKY41 remains to be determined.
Pleiotropic functions of GhWRKY41 and its orthologs
Accumulating evidence indicated that some WRKY exert myriad regulatory roles in multiple processes (Schluttenhofer and Yuan, 2015; Viana et al., 2018). In the last decade, two homologues of GhWRKY41 in Arabidopsis, AtWRKY41 and AtWRKY53, have been reported that mediate diverse physiological processes. For example, AtWRKY41 is induced by the flagellin, which constitutively expresses the PR5 gene, but suppresses the methyl jasmonate‐induced PDF1.2 gene (Higashi et al., 2008). AtWRKY53 interacts with the histone deacetylase HDA9 to antagonistically regulate salt tolerance in plants (Zheng et al., 2020). AtWRKY53 protein is not only degraded by E3 ubiquitin ligase or repressed by H3K4 demethylase, but also directly regulates several TFs from other families to modulate senescence (Miao and Zentgraf, 2010; Zentgraf and Doll, 2019). Recently, GhWRKY27 (the same protein as GhWRKY41 described in this manuscript) has also been identified as a positive regulator of leaf senescence, elevating expression of senescence‐associated genes (SAGs) and leading to early senescence in Arabidopsis (Gu et al., 2019). We also identified several SAGs and many genes involved in photosynthesis and chloroplast formation in our ChIP‐seq data (Figure 3 and Table S2), implying that GhWRKY41 may also mediate plant senescence in cotton. In addition to the diverse physiological processes mentioned above, AtWRKY53 also regulates plant responsive to pathogens (Hu et al., 2012; Miao and Zentgraf, 2007). In Arabidopsis, SA can influence AtWRKY53 DNA‐binding activity, representing a link to pathogen responses (Dong et al., 2003). WRKY46 can coordinate with WRKY70 and WRKY53 to ensure basal resistance against pathogen Pseudomonas syringae (Hu et al., 2012). AtWRKY53 is therefore tightly regulated through different mechanisms, and thus may represent an important node in which signals converge for senescence, biotic and abiotic stress responses. GhWRKY41, identified in the current work, positively regulates resistance to V. dahliae through promoting deposition of lignin and flavonoids in cotton tissue (Figures 2 and 6). Approximately 50% of GhWRKY41‐targeted genes significantly up‐regulated in cotton inoculated with V. dahliae included those with defence‐related functions such as PR proteins, disease‐related proteins, AOS, MPK2, and so on (Figure 4 and Table S2), with important roles in plant immunity (Liu et al., 2022; Pollmann et al., 2019; Ren et al., 2020; Wang et al., 2019), and indicating the essential role of GhWRKY41 in a defence response of cotton against V. dahliae. We have also found that GhWRKY41 mediates salt stress tolerance via promoting chlorophyll biosynthesis and photosynthesis (unpublished data). Considering the multiple functions of GhWRKY41 in diverse physiological processes, GhWRKY41 may be a valuable candidate gene for broad‐spectrum resistance breeding in cotton.
Materials and methods
Plant materials
Wild type (Gossypium hirsutum cv. YZ1) and GhWRKY41‐transgenic cotton generated from cv. YZ1 were planted in a field in Wuhan, Hubei province, China for propagation. The seeds of cotton were grown in permeant vermiculite with Hoagland's solution and five‐day‐old seedlings were subsequently transplanted to Hoagland's solution for further growth under long‐day conditions (16 h/8 h light/dark, 25–28 °C, and 60% humidity). The seeds of tobacco (Nicotiana benthamiana) were sown in soil, and the seedlings were grown in a greenhouse maintained under same growth conditions with cotton. Wild type (Col‐0) and GhWRKY41‐overexpressing Arabidopsis were grown in permeant vermiculite Hoagland's solution at 20 °C under long‐day conditions (16 h/8 h light/dark and 60% humidity).
Plant transformation
The full‐length of GhWRKY41 was inserted into pK2GW7.0 using BP and LR Clonase enzyme mixes (Invitrogen), and the overexpression vector was introduced into Agrobacterium tumefaciens (strain GV3101), which was used to transform Arabidopsis (Hao et al., 2012) and cotton (Li et al., 2019; Wang et al., 2020) as previously described, respectively. The fragment from C‐terminal to 3’ UTR region of GhWRKY41 was inserted into pHellsgate4 through BP recombination reactions, and the RNAi vector was introduced into GV3101 to transform cotton. The primers used in this study are listed in Table S4.
Pathogen cultivation, plant inoculation and disease assay
Pathogen cultivation, including V. dahliae strain V991 or V592 (harbouring the GFP reporter gene) (Wang et al., 2021), was performed as described previously (Xiao et al., 2021b). The seedlings of YZ1 were grown in sterilized MS medium. The V. dahliae strain V991 was cultured at 25 °C for 4 days in 100 mL Czapek liquid medium supplemented with the root sections of seven‐day‐old YZ1 seedlings, and the culture was initially filtered by double gauze then centrifuged at 5000 g for 15 min to obtain supernatant. The culture supernatants were further filtered by passage through a 0.22‐μm filter (Millipore Express PES Membrane) and concentrated to 1.5 mL using vacuum freeze dryer for next use. For two cotton cultivars (G. hirsutum cv. YZ1 and G. barbadense cv. 7124) for resistance identification to V. dahliae, the roots of at least 25 seedlings at 2–4 leaf stage were inoculated with a spore suspension (106 spores/mL) by root dipping in the spore suspension for 2 min, and then re‐planted in soil. For two cotton cultivars (G. hirsutum cv. 86‐4 and Zhongzhimian2) used in the RNA‐seq analysis, the roots of cotton seedlings at three‐leaf stage from nine plants were inoculated and harvested at 6, 12 and 48 h for RNA extraction. For Arabidopsis, three‐week‐old seedlings with vermiculite were gently poured out when the vermiculite is slightly dry to avoid excessive damage to the young roots, and the almost complete roots were directly soaked into the spore suspensions for 3 min. In all V. dahliae inoculation assays, an equal volume of bidistilled water was used as a mock treatment (control). The measurements of the disease index were recorded using at least 20 cotton and Arabidopsis plants per treatment with three biological repeats according to previously described methods (Xu et al., 2014).
RT‐qPCR and Southern blotting
The total RNA of cotton roots and whole Arabidopsis seedlings were extracted using RNA Extraction Kit (Tiangen Biotech, China) according to the manufacturer's protocol. RT‐qPCR were performed on a 7500 Real Time PCR system in a 15 μL reaction volume as described previously (Xiao et al., 2021b). Genomic DNA of transgenic cotton seedlings was extracted from young leaves using the plant genomic DNA kit DP305 (Tiangen Biotech). As described previously (Hu et al., 2018a), a quantity of 15 μg of genomic DNA was digested with HindIII (New England Biolabs) for 3 days, and Southern blotting was carried out using the DIG‐High Prime DNA Labeling and Detection Starter Kit II (Roche) according to the manufacturer's instructions.
RNA sequencing and data analysis
For RNA‐seq analysis, total RNAs were extracted from the roots of cotton (G. hirsutum cv. 86‐4 and Zhongzhimian2) using an RNA Extraction Kit (Tiangen Biotech). The RNA‐seq analysis was performed by the Beijing Novogene Bioinformatics Institute (China). Differentially expressed genes were identified using the filter criteria P ≤ 0.05 and fold‐change ≥2.
VIGS assay
The pTRV1 and pTRV2 vectors, and G. barbadense cv. 7124 were used for the VIGS assay (Gao et al., 2013). The fragment from C‐terminal to 3’ UTR region of GhWRKY41 was inserted into pTRV2 using ClonExpress II One Step Cloning Kit (Vazyme). The primers used in this study are listed in Table S4. All vectors were separately introduced into A. tumefaciens strain GV3101. All Agrobacterium cultures were adjusted to OD600 = 0.8 and A. tumefaciens samples with different TRV vectors were mixed in equal volumes and agro‐infiltrated into cotton cotyledons by vacuum infiltration as previously described (Gao et al., 2013).
Transcription activation assays
The full‐length of GhWRKY41 was inserted into the 35 S‐GAL4DB vector as an effector, and the empty 5 × GAL4‐LUC vector was used as a reporter. The dual‐luciferase reporter (DLR) assays in cotton protoplasts were performed as described previously (Min et al., 2015). The various truncated CDS of GhWRKY41 were inserted into the vector pGBKT7 and subsequently introduced into yeast strain Y2HGold, respectively. The positive yeast transformants were transferred to SD/−Trp (with or without X‐α‐Gal) and grown at 30 °C for 3–4 days. The primers used in this study are listed in Table S4.
Subcellular localization
The full‐length of GhWRYK41 was inserted into pGWB743 vector fused at the C‐terminus with GFP, and the 35 S‐GFP vector was used as a control. An Arabidopsis TF AtHY5, which localizes in the cell nucleus was constructed into vector fused a RFP reporter gene (Dong et al., 2021). The three vectors were separately introduced into Agrobacterium tumefaciens strain GV3101 and N. benthamiana leaves were injected with the bacterial suspension (OD600 = 0.5). After 2–3 days, injected leaves were cut to observe the fluorescence of GFP and RFP signals using confocal microscopy.
Chromatin immunoprecipitation (ChIP) assay
The full‐length of GhWRKY41 was inserted into the pGWB417 vector that was fused with a 4 × Myc tag at the C‐terminus to obtain the GhWRKY41‐Myc vector. The cotton protoplasts were transfected with 9 μg GhWRKY41‐Myc plasmid by PEG 4000 transformation. The transformed protoplasts were cultured at 25 °C in the dark for 20 h and collected for ChIP assay. According to previously described methods (Wang et al., 2016), samples were frozen in liquid nitrogen and subsequently the chromatin was isolated and ultrasonicated. Anti‐Myc monoclonal antibodies (Abcam, UK) were used to immunoprecipitate the protein‐DNA complex, and the precipitated DNA was recovered. The precipitated DNA was sequenced using the Illumina HiSeq2000 platform. Sequencing reads (200 bp) were mapped to the G. hirsutum cv. TM‐1 reference genome (https://cottonfgd.org/) (Zhang et al., 2015). MACS (Model‐based Analysis of ChIP‐Seq) version 2.1.0 was used to implement a peak‐finding algorithm to identify regions of IP enrichment against the background (Feng et al., 2012). A q‐value threshold of enrichment of 0.05 was used for all data sets. The chromosome, peak width, significance level, and peak summit number per peak distributions were all displayed. For ChIP‐qPCR assays, the isolated ChIP DNA was used as a template for PCR amplification. Enrichment folds of GhWRKY41‐bound DNA fragments were calculated by comparing chromatin samples immunoprecipitated with anti‐Myc antibody to that in the negative controls (no antibody).
Yeast two‐hybrid (Y2H) assay
Y2H assays were performed using the Matchmaker Gold Yeast Two‐Hybrid System (Clontech, CA) according to the manufacturer's instructions. The N‐terminal 190 amino acid residues of GhWRKY41 (GhWRKY41ΔC) were inserted into pGBKT7 and introduced into the yeast strain Y2HGold to generate BD‐GhWRKY41ΔC bait. The primers used are listed in Table S4.The library that we used for this experiment was a cotton cDNA library prepared from cotton roots under infection conditions with V. dahliae using Match‐maker Library Construction and Screening Kits (Clontech, CA). To confirm the interaction between GhWRKY41 and itself, the full‐length of GhWRKY41 was inserted into pGADT7 to generate GhWRKY41‐AD and introduced into yeast strain Y187. The positive clones of the bait and prey were mixed and mated in 30 °C shaker for 24 h, followed cultured on SD‐Leu‐Trp, SD‐Leu‐Trp‐His (with X‐α‐Gal, Coolaber, Beijing, China) and SD‐Leu‐Trp‐His‐Ade (with X‐α‐Gal) medium.
DLR assays
The DLR system assays were performed in N. benthamiana leaves and cotton protoplast as described previously (Xiao et al., 2021a). The promoters were inserted into pGreenII 0800‐LUC serving as reporters and the full‐length of GhWRKY41 was inserted into pGreenII 62‐SK serving as effector. The primers used are listed in Table S4. The cotton protoplasts were transfected with a mixture of 6 μg of effector plasmid, 6 μg of reporter plasmid, and 0.5 μg of internal control 35 S‐REN plasmid by polyethylene glycol (PEG) transformation. For V. dahliae supernatant and chitin treatment in cotton protoplasts, the 1 mL culture solution of transformed protoplasts added 20 μL ddH2O, 20 μL V. dahliae supernatant, 20 μL 10 mg/mL chitin (sigma), 20 μL chitin+0.5 mU chitinase (sigma), respectively. After 20 h at 25°C under dark conditions, the luciferase assay was conducted using the DLR assay system (Promega). For DLR dose response assays in N. benthamiana, GhWRKY41ΔC was inserted into the vector pGWB417 to generate the constructs GhWRKY41ΔC‐MYC containing C‐terminal tagged MYC protein, while the empty vector containing MYC tag was used as negative control. All of the plasmids were transformed individually into A. tumefaciens strain GV3101 for transient expression. The concentration of A. tumefaciens was adjusted to OD600 = 2.0 and mixed at respective ratios of 0.5:1:0:3, 0.5:1:1:2, 0.5:1:2:1 and 0.5:1:3:0 (pGreenII 0800‐LUC: pGreenII 62‐SK: MYC: GhWRKY41ΔC‐MYC), and all pairs of the suspensions were adjusted to the same OD600 = 0.6 prior to coinfiltration into N. benthamiana leaves.
Firefly luciferase complementation imaging (LCI) assay
The full‐length of GhWRKY41 was inserted into the pCAMBIA‐NLuc and pCAMBIA‐CLuc vectors to obtain GhWRKY41‐NLuc and GhWRKY41‐CLuc, respectively. The primers used are listed in Table S4. All vectors were transformed into N. benthamiana plants via the A. tumefaciens strain GV3101. Equal amounts of Agrobacterium cultures containing CLuc and NLuc constructs were mixed, and then co‐infiltrated into N. benthamiana leaves. The infiltrated leaves were analysed for relative Luc activity 48–72 h after infiltration using a low‐light cooled charge‐coupled device camera (Night owl LB985, Germany). Quantitative analysis was performed using the IndiGo software (Berthold Technologies, Germany).
GhWRKY41‐NLuc, GhWRKY41‐CLuc and 35 S‐REN plasmid were co‐transfected into cotton protoplasts to detect the interaction of GhWRKY41 with itself. 1 mL culture solution of transformed protoplasts added 20 μL ddH2O, 20 μL V. dahliae supernatant, 20 μL 10 mg/mL chitin (sigma), 20 μL chitin+0.5 mU chitinase (sigma), respectively. The protoplasts were cultured at 25°C in the dark for 20 h and collected to measure LUC and REN value as described previously (Xiao et al., 2021a).
Biomolecular fluorescence complementation (BiFC) assay
To generate the BiFC constructs, full‐length of GhWRKY41 was inserted into pS1301‐NYFP or pS1301‐CYFP vectors (Yuan et al., 2010). GhWRKY41‐NYFP and empty pS1301‐CYFP vectors, and GhWRKY41‐CYFP and empty pS1301‐NYFP vectors were used as negative controls for the BiFC assays. All vectors were transformed into N. benthamiana plants via the A. tumefaciens GV3101. Fluorescence signals in leaf epidermal cells were observed by confocal microscopy (Olympus FV1200).
Yeast one‐hybrid (Y1H) assay
Y1H assays were performed to confirm the interactions between GhWRKY41 and the promoters of its putative targets using the Matchmaker™ Gold Yeast One‐Hybrid Library Screening System (Clontech). The full‐length of GhWRKY41 was inserted into the pGADT7 vector as the prey, and the promoters were inserted into the pAbAi vector used to generate the bait. Both fusion constructs were transformed to the Y1HGold strain and cultured on SD/−Leu/‐Ura screening medium. Then, the interactions between GhWRKY41 and the promoter fragments were detected on SD/−Leu/‐Ura medium with 100 ng/mL aureobasidin A (AbA).
Immunoblot analysis
To examine the protein expression of GhWRKY41‐Myc in cotton protoplast, the GhWRKY41‐Myc homodimer and monomer immunoprecipitated by the Anti‐Myc monoclonal antibody (from “ChIP assay”) was used to perform Western blotting assay. Total proteins were extracted from the cotton protoplasts in extraction buffer (50 mm Tris–HCl, pH 8.0, 150 mm KCl, 1 mm EDTA, 0.5% Triton X‐100, 1 mm DTT, 1 mm PMSF, and a 1× protease inhibitor cocktail tablet [Roche]) and immunoblotted according to a previous method (Hu et al., 2018b).
Histochemical staining and determination of lignin and flavonoid content
Histochemical staining of hand‐cut cross sections from cotton cotyledonary basal nodes were used to visualize lignin and flavonoids deposition. Lignin was visualized using Wiesner reagent (Xu et al., 2011). For the histochemical staining of flavonoids, sections were cut at the same position and visualized using 5% (w/v) NaOH (Hu et al., 2018a). Cotton roots and Arabidopsis stem samples were dried in a lyophilizer and ground to a fine powder. The 5 mg powder was determined using the lignin‐thioglycolic acid reaction according to a previous study (Bubna et al., 2011). The lignin content was shown as the weight percentage of dry weight. To determinate the endogenous concentrations of DHK, DHQ, Eri and Nar in cotton and Arabidopsis, 0.15 g cotton root and Arabidopsis plant was extracted with 300 μL 80% (v/v) methanol containing acetonitrile/water/phosphoric acid (80:20:0.1, v/v/v) after shaking at 4 °C for 12 h for the detection of flavonoids. The extract was analysed on LC–MS or HPLC equipment for flavonoid detection according to previously studies (Tan et al., 2013).
Effects of diverse flavonoids on pathogens growth
To test the toxicity of diverse flavonoids (Sigma‐Aldrich) to V. dahliae strain V592, 1 mg/mL DHK, DHQ, Nar and Eri were prepared with 80% (v/v) methanol. Each flavonoid was individually added into 100 mL PDA medium, and the final concentrations were adjusted to 0, 0.1, 0.5, 1, 2, 5, 10, 20 and 50 μg/mL. The added total volume with 80% (v/v) methanol in medium was same. The diameters of V. dahliae colonies in PDA medium were measured after growth at 25°C in an incubator for ~5 days. The GFP bioluminescence of V. dahliae strain V592 was observed and measured using low‐light cooled charge‐coupled device camera (Nighowl LB985, Germany).
Accession numbers
Arabidopsis genes sequence involved in this work can be found in the TAIR (The Arabidopsis Information Resource, http://www.Arabidopsis.org/), accession numbers as following: AtACTIN (AT3G18780), AtWRKY41 (AT4G11070), AtWRKY53 (AT4G23810). Cotton genes sequence information can be obtained from the CottonFGD database (https://cottonfgd.org/), accession numbers as following: GhUB7 (DQ116441), GhWRKY41 (Gh_A08G2417), GhC4H (Gh_D11G2286), Gh4CL (Gh_A05G1188), GhNST1 (Gh_A08G0961), GhSND2 (Gh_A06G2028).
Conflict of interest
The authors declare no competing interests.
Author contributions
L.Z., S.X. A.A and Q.H. conceived and designed the project and the experiments. S.X. performed experiments for the gene biofunction assays and wrote the manuscript draft; Y.M. help to perform DLR and LCI assay; Z.Y., H.S., S.L. and X.Z help to perform ChIP assay and data analysis; W.W. and Y.Y. help to plant transgenic cotton; L.Z., S.J.K, K.L, J.K. and X.Z. revised the manuscript. All the authors discussed the results and the conception of the article.
Funding
This work was supported by funding from National Natural Science Foundation of China (32230076), Hubei Hongshan Laboratory Foundation (2021hszd006).
Supporting information
Figure S1 The identification of WRKY genes by RNA‐seq in three cotton cultivars infected with V. dahliae.
Figure S2 Expression of GhWRKY41 at different time‐points following inoculation of two cotton cultivar (86‐4 and Zhongzhimian 2) at the three‐leaf stage with Verticillium dahliae strain V991.
Figure S3 Phylogenetic tree analysis and amino acid sequence alignment of GhWRKY41.
Figure S4 Western blotting assay in cotton protoplast cells showing that V. dahliae supernatant treatment promotes the conversion of the GhWRKY41 monomer to the homodimer.
Figure S5 ChIP‐qPCR analysis of GhWRKY41 binding affinity to the promoter of GhSND2 and GhNST1.
Figure S6 Heat map of expression of GhSND2 in two cotton cultivars infected with V. dahliae.
Figure S7 The determination of lignin and flavonoids in WT and GhWRKY41‐overexpressing Arabidopsis.
Figure S8 Negative relationship between flavonoid concentration and hyphal growth.
Table S1 The counts and FPKM of all WRKYs in RNA‐seq derived from G. hirsutum (cv. 86‐4 and Zhongzhimian 2) and G. barbadense (cv. Hai7124) roots infected with V. dahliae.
Table S2 The 1315 overlapping peaks of three biological replicates in ChIP‐seq data and 1019 potential target genes of GhWRKY41.
Table S3 DEGs identified in cotton (G. hirsutum cv. YZ1) by RNA‐seq following inoculation with V. dahliae.
Table S4 Primers used in this study.
Acknowledgements
We are indebted to Huazhi Song and Dandan Yue (National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University) for assistance with the laser scanning confocal microscope and to Hongbo Liu (National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University) for flavonoids quantification.
Contributor Information
Alifu Aierxi, Email: alip68@126.com.
Longfu Zhu, Email: lfzhu@mail.hzau.edu.cn.
Data availability statement
The data that support the findings of this study are available in the supporting information of this article.
References
- Abeysinghe, J. , Lam, K. and Ng, D. (2019) Differential regulation and interaction of homoeologous WRKY18 and WRKY40 in Arabidopsis allotetraploids and biotic stress responses. Plant J. 97, 352–367. [DOI] [PubMed] [Google Scholar]
- Bubna, G.A. , Lima, R.B. , Zanardo, D.Y. , Dos Santos, W.D. , Ferrarese, M.deL and Ferrarese-Filho, O. (2011) Exogenous caffeic acid inhibits the growth and enhances the lignification of the roots of soybean (Glycine max). J Plant Physiol. 168, 1627–1633. [DOI] [PubMed] [Google Scholar]
- Chi, Y. , Yang, Y. , Zhou, Y. , Zhou, J. , Fan, B. , Yu, J.‐Q. and Chen, Z. (2013) Protein–protein interactions in the regulation of WRKY transcription factors. Mol. Plant 6, 287–300. [DOI] [PubMed] [Google Scholar]
- Choi, N. , Im, J. , Lee, E. , Lee, J. , Choi, C. , Park, S. and Hwang, D. (2020) WRKY10 transcriptional regulatory cascades in rice are involved in basal defense and Xa1‐mediated resistance. J. Exp. Bot. 71, 3735–3748. [DOI] [PubMed] [Google Scholar]
- Consortium, AIM . (2011) Evidence for network evolution in an "Arabidopsis" interactome map. Science, 333, 601–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, N. and Lin, H. (2021) Contribution of phenylpropanoid metabolism to plant development and plant‐environment interactions. J. Integr.Plant Biol. 63, 180–209. [DOI] [PubMed] [Google Scholar]
- Dong, J. , Chen, C. and Chen, Z. (2003) Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol. Biol. 51, 21–37. [DOI] [PubMed] [Google Scholar]
- Dong, Q. , Zheng, W. , Duan, D. , Huang, D. , Wang, Q. , Liu, C. , Li, C. et al. (2020) MdWRKY30, a group IIa WRKY gene from apple, confers tolerance to salinity and osmotic stresses in transgenic apple callus and Arabidopsis seedlings. Plant Sci. 299, 110611. [DOI] [PubMed] [Google Scholar]
- Dong, H. , Hu, C. , Liu, C. , Wang, J. , Zhou, Y. and Yu, J. (2021) ELONGATED HYPOCOTYL 5 mediates blue light‐induced starch degradation in tomato. J. Exp. Bot. 72, 2627–2641. [DOI] [PubMed] [Google Scholar]
- Du, Q. , Avci, U. , Li, S. , Gallego‐Giraldo, L. , Pattathil, S. , Qi, L. , Hahn, M. et al. (2015) Activation of miR165b represses AtHB15 expression and induces pith secondary wall development in Arabidopsis . Plant J. 83, 388–400. [DOI] [PubMed] [Google Scholar]
- Du, D. , Zhang, C. , Xing, Y. , Lu, X. , Cai, L. , Yun, H. , Zhang, Q. et al. (2021) The CC‐NB‐LRR OsRLR1 mediates rice disease resistance through interaction with OsWRKY19. Plant Biotechnol. J. 19, 1052–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem, T. , Rushton, P. , Robatzek, S. and Somssich, I. (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. [DOI] [PubMed] [Google Scholar]
- Fang, S. , Shang, X. , Yao, Y. , Li, W. and Guo, W. (2020) NST‐ and SND‐subgroup NAC proteins coordinately act to regulate secondary cell wall formation in cotton. Plant Sci. 301, 110657. [DOI] [PubMed] [Google Scholar]
- Feng, J. , Liu, T. , Qin, B. , Zhang, Y. and Liu, X. (2012) Identifying ChIP‐seq enrichment using MACS. Nature Protoc. 7, 1728–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, W. , Long, L. , Zhu, L.F. , Xu, L. , Gao, W.H. , Sun, L.Q. , Liu, L.L. et al. (2013) Proteomic and virus‐induced gene silencing (VIGS) Analyses reveal that gossypol, brassinosteroids, and jasmonic acid contribute to the resistance of cotton to Verticillium dahliae. Mol. Cell. Proteomics 12, 3690–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill, U. , Uppalapati, S. , Gallego‐Giraldo, L. , Ishiga, Y. , Dixon, R. and Mysore, K. (2018) Metabolic flux towards the (iso)flavonoid pathway in lignin modified alfalfa lines induces resistance against Fusarium oxysporum f. sp. medicaginis. Plant Cell Environ. 41, 1997–2007. [DOI] [PubMed] [Google Scholar]
- Grunewald, W. , DeSmet, I. , Lewis, D.R. , Löfke, C. , Jansen, L. , Goeminne, G. , VandenBossche, R. et al. (2012) Transcription factor WRKY23 assists auxin distribution patterns during Arabidopsis root development through local control on flavonol biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 109, 1554–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, L. , Dou, L. , Guo, Y. , Wang, H. , Li, L. , Wang, C. , Ma, L. et al. (2019) The WRKY transcription factor GhWRKY27 coordinates the senescence regulatory pathway in upland cotton (Gossypium hirsutum L.). BMC Plant Biol. 19, 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, W. , Jin, L. , Miao, Y. , He, X. , Hu, Q. , Guo, K. , Zhu, L. et al. (2016) An ethylene response‐related factor, GbERF1‐like, from Gossypium barbadense improves resistance to Verticillium dahliae via activating lignin synthesis. Plant Mol. Biol. 91, 305–318. [DOI] [PubMed] [Google Scholar]
- Hao, J. , Tu, L. , Hu, H. , Tan, J. , Deng, F. , Tang, W. , Nie, Y. et al. (2012) GbTCP, a cotton TCP transcription factor, confers fibre elongation and root hair development by a complex regulating system. J. Exp. Bot. 63, 6267–6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassani, D. , Fu, X. , Shen, Q. , Khalid, M. , Rose, J.K.C. and Tang, K. (2020) Parallel transcriptional regulation of artemisinin and flavonoid biosynthesis. Trends Plant Sci. 25, 466–476. [DOI] [PubMed] [Google Scholar]
- Higashi, K. , Ishiga, Y. , Inagaki, Y. , Toyoda, K. , Shiraishi, T. and Ichinose, Y. (2008) Modulation of defense signal transduction by flagellin‐induced WRKY41 transcription factor in Arabidopsis thaliana . Mol. Genet. Genomics 279, 303–312. [DOI] [PubMed] [Google Scholar]
- Hu, Y. , Dong, Q. and Yu, D. (2012) Arabidopsis WRKY46 coordinates with WRKY70 and WRKY53 in basal resistance against pathogen Pseudomonas syringae. Plant Sci. 185‐186, 288–297. [DOI] [PubMed] [Google Scholar]
- Hu, Q. , Min, L. , Yang, X. , Jin, S. , Zhang, L. , Li, Y. , Ma, Y. et al. (2018a) Laccase GhLac1 modulates broad‐spectrum biotic stress tolerance via manipulating phenylpropanoid pathway and jasmonic acid synthesis. Plant Physiol. 176, 1808–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Q. , Zhu, L. , Zhang, X. , Guan, Q. , Xiao, S. , Min, L. and Zhang, X. (2018b) GhCPK33 negatively regulates defense against Verticillium dahliae by phosphorylating GhOPR3. Plant Physiol. 178, 876–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussey, S. , Mizrachi, E. , Spokevicius, A. , Bossinger, G. , Berger, D. and Myburg, A. (2011) SND2, a NAC transcription factor gene, regulates genes involved in secondary cell wall development in Arabidopsis fibres and increases fibre cell area in Eucalyptus. BMC Plant Biol. 11, 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashyap, A. , Planas‐Marquès, M. , Capellades, M. , Valls, M. and Coll, N. (2021) Blocking intruders: inducible physico‐chemical barriers against plant vascular wilt pathogens. J. Exp. Bot. 72, 184–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahiri, A. , Venkatasubramani, P. and Datta, A. (2019) Bayesian modeling of plant drought resistance pathway. BMC Plant Biol. 19, 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C. , He, X. , Luo, X. , Xu, L. , Liu, L. , Min, L. , Jin, L. et al. (2014) Cotton WRKY1 mediates the plant defense‐to‐development transition during infection of cotton by Verticillium dahliae by activating JASMONATE ZIM‐DOMAIN1 expression. Plant Physiol. 166, 2179–2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W. , Tian, Z. and Yu, D. (2015) WRKY13 acts in stem development in Arabidopsis thaliana . Plant Sci. 236, 205–213. [DOI] [PubMed] [Google Scholar]
- Li, J. , Wang, M. , Li, Y. , Zhang, Q. , Lindsey, K. , Daniell, H. , Jin, S. et al. (2019) Multi‐omics analyses reveal epigenomics basis for cotton somatic embryogenesis through successive regeneration acclimation process. Plant Biotechnol. J. 17, 435–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. , Li, Y. , Dang, M. , Li, S. , Chen, S. , Liu, R. , Zhang, Z. et al. (2022) Jasmonate‐responsive transcription factors NnWRKY70a and NnWRKY70b positively regulate benzylisoquinoline alkaloid biosynthesis in lotus (Nelumbo nucifera). Front. Plant Sci. 13, 862915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. , Osbourn, A. and Ma, P. (2015) MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol. Plant 8, 689–708. [DOI] [PubMed] [Google Scholar]
- Liu, M. , Zhang, C. , Duan, L. , Luan, Q. , Li, J. , Yang, A. , Qi, X. et al. (2019) CsMYB60 is a key regulator of flavonols and proanthocyanidans that determine the colour of fruit spines in cucumber. J. Exp. Bot. 70, 69–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, T. , Chen, T. , Kan, J. , Yao, Y. , Guo, D. , Yang, Y. , Ling, X. et al. (2022) The GhMYB36 transcription factor confers resistance to biotic and abiotic stress by enhancing PR1 gene expression in plants. Plant Biotechnol. J. 20, 722–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, L. , Liu, J. , Gao, Y. , Xu, F. , Zhao, J. , Li, B. and Gao, W. (2019) Flavonoid accumulation in spontaneous cotton mutant results in red coloration and enhanced disease resistance. Plant Physiol. Biochem. 143, 40–49. [DOI] [PubMed] [Google Scholar]
- Luo, X. , Li, Z. , Xiao, S. , Ye, Z. , Nie, X. , Zhang, X. , Kong, J. et al. (2021) Phosphate deficiency enhances cotton resistance to Verticillium dahliae through activating jasmonic acid biosynthesis and phenylpropanoid pathway. Plant Sci. 302, 110724. [DOI] [PubMed] [Google Scholar]
- Miao, Y. and Zentgraf, U. (2007) The antagonist function of Arabidopsis WRKY53 and ESR/ESP in leaf senescence is modulated by the jasmonic and salicylic acid equilibrium. Plant Cell 19, 819–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao, Y. and Zentgraf, U. (2010) A HECT E3 ubiquitin ligase negatively regulates Arabidopsis leaf senescence through degradation of the transcription factor WRKY53. Plant J. 63, 179–188. [DOI] [PubMed] [Google Scholar]
- Min, L. , Hu, Q. , Li, Y. , Xu, J. , Ma, Y. , Zhu, L. , Yang, X. et al. (2015) LEAFY COTYLEDON1‐CASEIN KINASE I‐TCP15‐PHYTOCHROME INTERACTING FACTOR4 network regulates somatic embryogenesis by regulating auxin homeostasis. Plant Physiol. 169, 2805–2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtani, M. and Demura, T. (2019) The quest for transcriptional hubs of lignin biosynthesis: beyond the NAC‐MYB‐gene regulatory network model. Curr.Opin Biotechnol. 56, 82–87. [DOI] [PubMed] [Google Scholar]
- Onohata, T. and Gomi, K. (2020) Overexpression of jasmonate‐responsive OsbHLH034 in rice results in the induction of bacterial blight resistance via an increase in lignin biosynthesis. Plant Cell Rep. 39, 1175–1184. [DOI] [PubMed] [Google Scholar]
- Pollmann, S. , Springer, A. , Rustgi, S. , von Wettstein, D. , Kang, C. , Reinbothe, C. and Reinbothe, S. (2019) Substrate channeling in oxylipin biosynthesis through a protein complex in the plastid envelope of Arabidopsis thaliana . J. Exp. Bot. 70, 1483–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potschin, M. , Schlienger, S. , Bieker, S. and Zentgraf, U. (2014) Senescence networking: WRKY18 is an upstream regulator, a downstream target gene, and a protein interaction partner of WRKY53. J. Plant Growth Regul. 33, 106–118. [Google Scholar]
- Pratyusha, D. and Sarada, D. (2022) MYB transcription factors‐master regulators of phenylpropanoid biosynthesis and diverse developmental and stress responses. Plant Cell Rep. 41, 2245–2260. [DOI] [PubMed] [Google Scholar]
- Reboledo, G. , Agorio, A. and PonceDeLeón, I. (2022) Moss transcription factors regulating development and defense responses to stress. J. Exp. Bot. 73, 4546–4561. [DOI] [PubMed] [Google Scholar]
- Ren, H. , Bai, M. , Sun, J. , Liu, J. , Ren, M. , Dong, Y. , Wang, N. et al. (2020) RcMYB84 and RcMYB123 mediate jasmonate‐induced defense responses against Botrytis cinerea in rose (Rosa chinensis). Plant J. 103, 1839–1849. [DOI] [PubMed] [Google Scholar]
- Schluttenhofer, C. and Yuan, L. (2015) Regulation of specialized metabolism by WRKY transcription factors. Plant Physiol. 167, 295–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, J. , Tu, L. , Deng, F. , Hu, H. , Nie, Y. and Zhang, X. (2013) A genetic and metabolic analysis revealed that cotton fiber cell development was retarded by flavonoid naringenin. Plant Physiol. 162, 86–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor‐Teeples, M. , Lin, L. , de Lucas, M. , Turco, G. , Toal, T.W. , Gaudinier, A. , Young, N.F. et al. (2015) An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature 517, 571–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viana, V. , Busanello, C. , da Maia, L. , Pegoraro, C. and Costa de Oliveira, A. (2018) Activation of rice WRKY transcription factors: an army of stress fighting soldiers? Curr. Opin. Plant Biol. 45, 268–275. [DOI] [PubMed] [Google Scholar]
- Wang, H. , Avci, U. , Nakashima, J. , Hahn, M. , Chen, F. and Dixon, R. (2010) Mutation of WRKY transcription factors initiates pith secondary wall formation and increases stem biomass in dicotyledonous plants. Proc. Natl. Acad. Sci. U. S. A. 107, 22338–22343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M. , Wang, P. , Tu, L. , Zhu, S. , Zhang, L. , Li, Z. , Zhang, Q. et al. (2016) Multi‐omics maps of cotton fibre reveal epigenetic basis for staged single‐cell differentiation. Nucleic Acids Res. 44, 4067–4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, G. , Hu, C. , Zhou, J. , Liu, Y. , Cai, J. , Pan, C. , Wang, Y. et al. (2019) Systemic root‐shoot signaling drives jasmonate‐based root defense against nematodes. Curr. Biol. 29, 3430–3438.e3434. [DOI] [PubMed] [Google Scholar]
- Wang, Q. , Alariqi, M. , Wang, F. , Li, B. , Ding, X. , Rui, H. , Li, Y. et al. (2020) The application of a heat‐inducible CRISPR/Cas12b (C2c1) genome editing system in tetraploid cotton (G. hirsutum) plants. Plant Biotechnol. J. 18, 2436–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H. , Chen, B. , Tian, J. and Kong, Z. (2021) Verticillium dahliae VdBre1 is required for cotton infection by modulating lipid metabolism and secondary metabolites. Environ. Microbiol. 23, 1991–2003. [DOI] [PubMed] [Google Scholar]
- Wani, S. , Anand, S. , Singh, B. , Bohra, A. and Joshi, R. (2021) WRKY transcription factors and plant defense responses: latest discoveries and future prospects. Plant Cell Rep. 40, 1071–1085. [DOI] [PubMed] [Google Scholar]
- Warner, R. , Wu, B. , MacPherson, S. and Lefsrud, M. (2021) A review of strawberry photobiology and fruit flavonoids in controlled environments. Front. Plant Sci. 12, 611893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, S. , Hu, Q. , Shen, J. , Liu, S. , Yang, Z. , Chen, K. , Klosterman, S. et al. (2021a) GhMYB4 downregulates lignin biosynthesis and enhances cotton resistance to Verticillium dahliae . Plant Cell Rep. 40, 735–751. [DOI] [PubMed] [Google Scholar]
- Xiao, S. , Hu, Q. , Zhang, X. , Si, H. , Liu, S. , Chen, L. , Chen, K. et al. (2021b) Orchestration of plant development and defense by indirect crosstalk of salicylic acid and brassinosteorid signaling via transcription factor GhTINY2. J. Exp. Bot. 72, 4721–4743. [DOI] [PubMed] [Google Scholar]
- Xie, Z. , Zhang, Z.‐L. , Zou, X. , Yang, G. , Komatsu, S. and Shen, Q.J. (2006) Interactions of two abscisic‐acid induced WRKY genes in repressing gibberellin signaling in aleurone cells. Plant J. 46, 231–242. [DOI] [PubMed] [Google Scholar]
- Xiong, X. , Sun, S. , Li, Y. , Zhang, X. , Sun, J. and Xue, F. (2019) The cotton WRKY transcription factor GhWRKY70 negatively regulates the defense response against Verticillium dahliae . Crop J. 7, 393–402. [Google Scholar]
- Xiong, X. , Sun, S. , Zhang, X. , Li, Y. , Liu, F. , Zhu, Q. , Xue, F. et al. (2020) GhWRKY70D13 regulates resistance to Verticillium dahliae in cotton through the ethylene and jasmonic acid signaling pathways. Front. Plant Sci. 11, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, X. , Chen, C. , Fan, B. and Chen, Z. (2006) Physical and functional interactions between pathogen‐induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 18, 1310–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, L. , Zhu, L. , Tu, L. , Liu, L. , Yuan, D. , Jin, L. , Long, L. et al. (2011) Lignin metabolism has a central role in the resistance of cotton to the wilt fungus Verticillium dahliae as revealed by RNA‐Seq‐dependent transcriptional analysis and histochemistry. J. Exp. Bot. 62, 5607–5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, L. , Zhang, W. , He, X. , Liu, M. , Zhang, K. , Shaban, M. , Sun, L. et al. (2014) Functional characterization of cotton genes responsive to Verticillium dahliae through bioinformatics and reverse genetics strategies. J. Exp. Bot. 65, 6679–6692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, W. , Dubos, C. and Lepiniec, L. (2015) Transcriptional control of flavonoid biosynthesis by MYB–bHLH–WDR complexes. Trends Plant Sci. 20, 176–185. [DOI] [PubMed] [Google Scholar]
- Yan, L. , Xu, C. , Kang, Y. , Gu, T. , Wang, D. , Zhao, S. and Xia, G. (2013) The heterologous expression in Arabidopsis thaliana of sorghum transcription factor SbbHLH1 downregulates lignin synthesis. J. Exp. Bot. 64, 3021–3032. [DOI] [PubMed] [Google Scholar]
- Yang, L. , Zhao, X. , Yang, F. , Fan, D. , Jiang, Y. and Luo, K. (2016) PtrWRKY19, a novel WRKY transcription factor, contributes to the regulation of pith secondary wall formation in Populus trichocarpa. Sci. Rep. 6, 18643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J. , Wang, Q. , Luo, H. , He, C. and An, B. (2020) HbWRKY40 plays an important role in the regulation of pathogen resistance in Hevea brasiliensis . Plant Cell Rep. 39, 1095–1107. [DOI] [PubMed] [Google Scholar]
- Yuan, M. , Chu, Z. , Li, X. , Xu, C. and Wang, S. (2010) The bacterial pathogen Xanthomonas oryzae overcomes rice defenses by regulating host copper redistribution. Plant Cell. 22, 3164–3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentgraf, U. and Doll, J. (2019) Arabidopsis WRKY53, a node of multi‐layer regulation in the network of senescence. Plants (Basel). 8, 578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, T. , Hu, Y. , Jiang, W. , Fang, L. , Guan, X. , Chen, J. , Zhang, J. et al. (2015) Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM‐1) provides a resource for fiber improvement. Nat. Biotechnol. 33, 531–537. [DOI] [PubMed] [Google Scholar]
- Zhang, L. , Wang, M. , Li, N. , Wang, H. , Qiu, P. , Pei, L. , Xu, Z. et al. (2018) Long noncoding RNAs involve in resistance to Verticillium dahliae, a fungal disease in cotton. Plant Biotechnol. J. 16, 1172–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Q. , Luo, F. , Zhong, Y. , He, J. and Li, L. (2020) Modulation of NAC transcription factor NST1 activity by XYLEM NAC DOMAIN1 regulates secondary cell wall formation in Arabidopsis . J. Exp. Bot. 71, 1449–1458. [DOI] [PubMed] [Google Scholar]
- Zhang, X. , He, Y. , Li, L. , Liu, H. and Hong, G. (2021) Involvement of the R2R3‐MYB transcription factor MYB21 and its homologs in regulating flavonol accumulation in Arabidopsis stamen. J. Exp. Bot. 72, 4319–4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Q. and Dixon, R. (2011) Transcriptional networks for lignin biosynthesis: more complex than we thought? Trends Plant Sci. 16, 227–233. [DOI] [PubMed] [Google Scholar]
- Zhao, R. , Song, X. , Yang, N. , Chen, L. , Xiang, L. , Liu, X. and Zhao, K. (2020) Expression of the subgroup IIIf bHLH transcription factor CpbHLH1 from Chimonanthus praecox (L.) in transgenic model plants inhibits anthocyanin accumulation. Plant Cell Rep. 39, 891–907. [DOI] [PubMed] [Google Scholar]
- Zheng, Y. , Ge, J. , Bao, C. , Chang, W. , Liu, J. , Shao, J. , Liu, X. et al. (2020) Histone deacetylase HDA9 and WRKY53 transcription factor are mutual antagonists in regulation of plant stress response. Mol. Plant 13, 598–611. [DOI] [PubMed] [Google Scholar]
- Zhong, R. , Lee, C. , Zhou, J. , McCarthy, R.L. and Ye, Z.‐H. (2008) A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis . Plant Cell 20, 2763–2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 The identification of WRKY genes by RNA‐seq in three cotton cultivars infected with V. dahliae.
Figure S2 Expression of GhWRKY41 at different time‐points following inoculation of two cotton cultivar (86‐4 and Zhongzhimian 2) at the three‐leaf stage with Verticillium dahliae strain V991.
Figure S3 Phylogenetic tree analysis and amino acid sequence alignment of GhWRKY41.
Figure S4 Western blotting assay in cotton protoplast cells showing that V. dahliae supernatant treatment promotes the conversion of the GhWRKY41 monomer to the homodimer.
Figure S5 ChIP‐qPCR analysis of GhWRKY41 binding affinity to the promoter of GhSND2 and GhNST1.
Figure S6 Heat map of expression of GhSND2 in two cotton cultivars infected with V. dahliae.
Figure S7 The determination of lignin and flavonoids in WT and GhWRKY41‐overexpressing Arabidopsis.
Figure S8 Negative relationship between flavonoid concentration and hyphal growth.
Table S1 The counts and FPKM of all WRKYs in RNA‐seq derived from G. hirsutum (cv. 86‐4 and Zhongzhimian 2) and G. barbadense (cv. Hai7124) roots infected with V. dahliae.
Table S2 The 1315 overlapping peaks of three biological replicates in ChIP‐seq data and 1019 potential target genes of GhWRKY41.
Table S3 DEGs identified in cotton (G. hirsutum cv. YZ1) by RNA‐seq following inoculation with V. dahliae.
Table S4 Primers used in this study.
Data Availability Statement
The data that support the findings of this study are available in the supporting information of this article.


