Summary
Tiller number per plant—a cardinal component of ideal plant architecture—affects grain yield potential. Thus, alleles positively affecting tillering must be mined to promote genetic improvement. Here, we report a Tiller Number 1 (TN1) protein harbouring a bromo‐adjacent homology domain and RNA recognition motifs, identified through genome‐wide association study of tiller numbers. Natural variation in TN1 affects its interaction with TIF1 (TN1 interaction factor 1) to affect DWARF14 expression and negatively regulate tiller number in rice. Further analysis of variations in TN1 among indica genotypes according to geographical distribution revealed that low‐tillering varieties with TN1‐hapL are concentrated in Southeast Asia and East Asia, whereas high‐tillering varieties with TN1‐hapH are concentrated in South Asia. Taken together, these results indicate that TN1 is a tillering regulatory factor whose alleles present apparent preferential utilization across geographical regions. Our findings advance the molecular understanding of tiller development.
Keywords: rice, tiller number, BAH domain, GWAS, geographical distribution
Excellent germplasm and gene resources are the material basis of new variety improvement. Breeding practice shows that major breakthroughs in breeding depend on the discovery and utilization of excellent germplasm and gene resources. The author took a genome‐wide association study of 295 germplasm materials and identified a novel gene regulating tiller number in rice. The picture shown are rice cultivars used for GWAS analysis in this study.

Introduction
Rice (Oryza sativa L.) is a global staple food. Thus, rice grain yield must be increased to ensure overall food security. Rice yield dramatically increased following ‘Green Revolution’ in the 1960s, during which semi‐dwarfing and heterosis played significant roles (Wang et al., 2017). Ideal plant architecture and high nitrogen‐use efficiency (NUE) are the current strategies aimed at improving rice yield (Duan et al., 2019; Sun et al., 2014; Wang and Li, 2006). In rice, tillering is relevant to both ideal plant architecture and high NUE (Liu et al., 2021; Wu et al., 2020). Therefore, exploring the mechanisms of tiller development is important in breeding for ideal plant architecture and high NUE.
The currently cloned genes controlling tiller development in rice primarily regulate the initiation or outgrowth of axillary buds. In particular, MONOCULM1 (MOC1), MONOCULM3/TILLERS ABSENT1/STERILE and REDUCED TILLING 1 (MOC3/TAB1/SRT1), LAX PANICLE1 (LAX1), and LAX PANICLE2/ GRAIN NUMBER PER‐PANICLE4 (LAX2/GNP4) induce tiller initiation and regulate tiller number (Li et al., 2003; Lu et al., 2015a; Mjomba et al., 2016; Oikawa and Kyozuka, 2009; Tabuchi et al., 2011; Tanaka et al., 2015; Zhang et al., 2011a). Other genes, specifically those involved in the strigolactone (SL) pathway, such as DWARF3 (D3), DWARF10 (D10), DWARF14/HIGH TILLERING DWARF 2 (D14/HTD2), and DWARF53 (D53), regulate tillering by affecting the growth of axillary buds (Arite et al., 2007, 2009; Gao et al., 2009; Ishikawa et al., 2005; Jiang et al., 2013; Liu et al., 2009; Zhou et al., 2013). D3, D53, and D14 interact with each other under treatment with GR24 (an SL) leading to ubiquitination of D53 by D14 and the SCFD3 complex. Loss‐of‐function mutants d3 and d14 and functional‐gain mutant d53 showed markedly increased tiller numbers (Jiang et al., 2013; Zhou et al., 2013). IDEAL PLANT ARCHITECTURE 1/SOUAMOSA PROMOTER BINDING PROTEIN‐LIKE 14 (IPA1/OsSPL14) promotes panicle branching and contributes to ideal plant architecture (Jiao et al., 2010; Miura et al., 2010). D53 interacts with IPA1 to repress its transcriptional activation, and IPA1 binds the promoter of D53 in feedback regulation under SL‐mediated signal transduction (Song et al., 2017). Rice TEOSINTE BRANCHED 1 (OsTB1) is a negative regulator suppressing axillary bud outgrowth (Minakuchi et al., 2010; Takeda et al., 2003). Furthermore, OsMADS57 negatively regulates D14 expression, although its inhibitory activity is weakened upon interaction with OsTB1 (Guo et al., 2013). IPA1 and CIRCADIAN CLOCK ASSOCIATED 1 (OsCCA1) are the key transcription factors binding the promoter of OsTB1 to restrain tillering, and OsCCA1 acts upstream of D14 and IPA1 to regulate axillary bud outgrowth (Lu et al., 2013; Wang et al., 2020). In addition, NITROGEN‐MEDIATED TILLER GROWTH RESPONSE 5 (NGR5) interacts with Polycomb Repressive Complex 2 (PRC2), influencing the H3K27me3 modification level of D14 and IPA1 to regulate their expression levels (Wu et al., 2020). Overall, there has been extensive research on the regulation of tiller buds.
Typically, bromo‐adjacent homology (BAH) domain‐containing proteins are attached to chromatin‐associated proteins and play important roles in transcriptional regulation and protein–protein interactions (Callebaut et al., 1999; Chambers et al., 2013). Arabidopsis BAH domain proteins EARLY BOLTING IN SHORT DAY (EBS) and SHORT LIFE (SHL) suppress the transcription of flowering genes (Li et al., 2018; Yang et al., 2018). The BAH domain recognizes the histone marks H3 lysine 9 di‐methylation (H3K9me2), H4 lysine 20 di‐methylation (H4K20me2), H3 lysine 27 tri‐methylation (H3K27me3), and unmodified H3K4 (Du et al., 2012; Kuo et al., 2012; Li et al., 2016, 2018; Yang et al., 2018; Zhang et al., 2020). Majority of the RNA‐binding proteins (RBPs) harbour RNA recognition motifs (RRMs) and are primarily involved in post‐transcriptional regulation, such as mRNA splicing, 3′‐end processing, and mRNA stability and decay (Goodarzi et al., 2014; Hogan et al., 2008; Kenan et al., 1991; Roundtree et al., 2017). OsRRM and OsRRMh‐containing RRMs play important roles in plant architecture and cell development in rice endosperm (Chen et al., 2007; Liu and Cai, 2013). Arabidopsis ANTI‐SILENCING 1/INCREASE IN BONSAI METHYLATION 2 (ASI1/IBM2), which contains both a BAH domain and RRMs, interacts with ASI1‐IMMUNOPRECIPITATED PROTEIN 1 (AIPP1) and ENHANCED DOWNY MILDEW 2 (EDM2) to form the AAE complex to facilitate 3′‐distal polyadenylation further, and to stabilize full‐length transcripts. The loss of function of ASI1 alters the expression of heterochromatin‐containing genes with a high level of DNA or histone methylation in the gene body region (Duan et al., 2017; Saze et al., 2013; Wang et al., 2013). Taken together, these reports indicate that proteins containing BAH domains and RRMs play pivotal roles in developmental processes. A recent study showed that OsASI1 (the orthologue of Arabidopsis ASI1 in rice) regulates a nuclear XRN family exonuclease gene, OsXRNL, which further affects the processing of miRNA precursors (You et al., 2021). However, the functions of proteins containing BAH domains and RRMs in the regulation of tiller number remain unknown.
As outlined above, many genes involved in tiller development, including D3, D14, D53, and OsTB1, have been identified through mutant studies. However, due to their unfavourable phenotypes, such mutants have little value in crop improvement. Therefore, favourable alleles/haplotypes affecting tillering must be mined from natural populations. It was recently found that tiller‐regulating genes usually showed high expression in the root at 24:00. Based on this expression pattern and a genome‐wide association study (GWAS), a small auxin‐UP RNA gene, OsSAUR27, was identified in natural populations and has been proven to affect tiller number and plant architecture (Ma et al., 2020).
To this end, in this study, we performed GWAS on tiller number and identified a tillering regulator, Tiller Number 1 (TN1)/OsASI1, containing a BAH domain and an RRM. Its functional variation site (+2163G > A) is located in the first exon. We further showed that TN1 interacts with TIF1 (TN1 interaction factor 1) to positively regulate D14 expression and modulate axillary bud outgrowth, ultimately affecting tiller development. We observed that TN1 has not been under selection during rice domestication, although various haplotypes have been established in different regions. Our results enrich the gene pool for the genetic improvement of tillering in cultivated rice.
Results
Candidate gene analysis of locus qTN2
In a panel of 295 cultivated rice accessions, tiller number per plant ranged from 3.7 to 19.0 (Dataset S1). Association analysis using a mixed linear model (MLM) with the top three principal components (PCs) as the fixed effects (Figure S1) revealed 13 quantitative trait loci (QTLs) associated with tiller number, which were named qTN1–qTN13 (Dataset S2). As qTN2 showed the strongest signal in GWAS (Figure 1a and Figure S2a), candidate genes in a 122 kb interval of qTN2 were screened and 12 genes were identified (Figure 1b, Dataset S3). Expression data from RiceXPro website (Sato et al., 2011) indicated that five of these genes produced no transcripts at any growth stage. Thus, we analysed haplotypes of the remaining seven genes and found no significant differences in the expression levels of these genes among the different haplotype varieties (Figure 1d, Figures S2d and S3). Of these, only LOC_Os01g42460 and LOC_Os01g42490 possess significant non‐synonymous SNPs in the functional encoding regions. The expression analysis showed that LOC_Os01g42460 in roots at 24:00 during vegetative growth was significantly higher than that of LOC_Os01g42490 (Figure S2b). Additionally, the expression level of LOC_Os01g42460 at the base of tiller buds at 45 days after transplanting was significantly higher than that of LOC_Os01g42490 (Figure S2c). These results prompted us to focus further on LOC_Os01g42460, which we named TN1.
Figure 1.

GWAS of tiller number to identify TN1. (a) Manhattan plot of GWAS results. Red dots represent QTLs. (b) Regional Manhattan plot of qTN2 and pairwise LD analysis. Significant SNPs (−log10(P) ≥ 4) are presented as red dots. Green and red boxes indicate annotated genes, triangles denote QTLs, and dots represent SNPs. (c) TN1‐based association mapping. Red dots indicate SNPs (−log10(P) ≥ 4) in the 2000 bp promoter and 7467 bp genomic sequence. (d) Haplotypes (hap) of TN1 among 279 accessions; major and minor alleles are indicated in yellow and green respectively. Data are presented as mean ± SE. Different letters indicate significant differences at P < 0.05 according to two‐tailed Student's t‐test.
Using six significant SNP variants above the suggestive significance threshold of GWAS in the promoter and protein coding regions, we identified four TN1 haplotypes (hap1, hap2, hap3, and hap4) in the indica subpopulation and one haplotype (hap2) in the japonica subpopulation (Figure 1c,d). In the indica subpopulation, tiller numbers in hap1, hap2, and hap3 accessions were similar, being significantly higher than in hap4. This implied that the SNP (+2163G > A) in the coding region causing a mutation at the 83rd amino acid (arginine/glutamine) of the BAH domain may be the functional variation. Meanwhile, a previous study showed that a mis‐sense substitution in BAHEBS decreases the binding activity of EBS, thereby affecting flowering in Arabidopsis (Li et al., 2018; Pineiro et al., 2003). Then, we tested the promoter activities of three haplotypes (hap1, hap2, and hap4) in rice protoplasts but noted no significant differences (Figure S2e,f). Meanwhile, there were no obvious differences in the expression level of TN1 in indica accessions containing hap1, hap2 (high tiller), and hap4 (low tiller) (Figure S2d). Collectively, these results suggest that the non‐synonymous SNP variant (+2163G > A) of LOC_Os01g42460 is vital for the tiller development and is the most probable candidate gene.
TN1 negatively regulates tiller formation in rice
TN1 encodes a protein with 502 amino acids (aa) and contains a BAH domain (amino acid 28–145) and an RRM (amino acid 340–416). To identify whether TN1/LOC_Os01g42460 regulates tiller number, we knocked out the BAH domain or RRM in separate lines. Mutant tn1‐1 carried a 6 bp deletion in the first exon, resulting in a 3 amino acid change in the BAH domain compared with the wild‐type Nipponbare (NIP). Mutants tn1‐2 and tn1‐3 carried a 4 bp deletion and a 1 bp insertion, respectively, in the sixth exon of TN1; both mutations were located in the RRM and caused frameshifts (Figure 2a and Figure S4a,b). Mutant tn1‐4 carried a thymine (T) insertion in the BAH domain, causing premature termination (Figure S4b). Tiller number in tn1‐1 and tn1‐4 increased by 29.8% and 31.7% compared with that in NIP, while the number in tn1‐2 and tn1‐3 was not significantly different from that in NIP (Figure 2b–e and Figure S4c). Thus, the BAH domain might play an important role in tillering regulation. Further, we introduced the coding sequence (CDS) of TN1 driven by the CaMV35S promoter into NIP to generate constitutive TN1‐overexpression lines. Tiller number in TN1‐OE1 and TN1‐OE2 was significantly reduced compared with that in NIP (Figure 2f–h). Moreover, the initiation of tillering buds was not affected in either tn1‐1 or TN1‐OE (Figure S5). Taken together, these results indicate that TN1 negatively controls tillering growth to affect tiller number.
Figure 2.

Phenotypic characterization of TN1‐transgenic plants. (a) Sequences of CRISPR‐knockout lines. (b) Phenotypes of NIP and tn1‐1 lines at the reproductive stage. Scale bar = 20 cm. (c) Phenotypes of NIP and tn1‐2 and tn1‐3 lines at the reproductive stage. Scale bar = 20 cm. (d) Tiller number per plant of NIP and tn1‐1 lines. (e) Tiller number per plant of NIP and tn1‐2 and tn1‐3 lines. (f) Phenotype of NIP and TN1‐OE lines at the reproductive stage. Scale bar = 20 cm. (g) Expression level of TN1 in NIP versus TN1‐OE lines. Data are presented as mean ± SD. (n = 3 biologically independent samples). (h) Tiller number per plant of NIP and TN1‐OE lines. (i) Seed setting rate of NIP, tn1‐1, tn1‐2, tn1‐3, and TN1‐OE lines. (j) Heading date of NIP, tn1‐1, tn1‐2, tn1‐3, and TN1‐OE lines. In (d–j), P‐values were determined using two‐tailed Student's t‐tests. **P < 0.01. Data are presented as mean ± SD (n = 15).
Expression patterns of TN1
To determine the subcellular localization of TN1, we transformed the 35 S:TN1‐GFP vector into rice protoplasts and tobacco leaves. The results showed that TN1 is a nuclear protein (Figure 3a and Figure S6). Analysis of dynamic TN1 expression in different tissues (Figure S7a,b) revealed high expression in tiller buds at 30 and 60 days after transplanting (DT_30 and DT_60 respectively). In addition, we generated ProTN1:GUS transgenic plants and detected strong GUS activity in tiller buds (Figure 3b,e). These results are consistent with the function of TN1 in regulating tiller growth.
Figure 3.
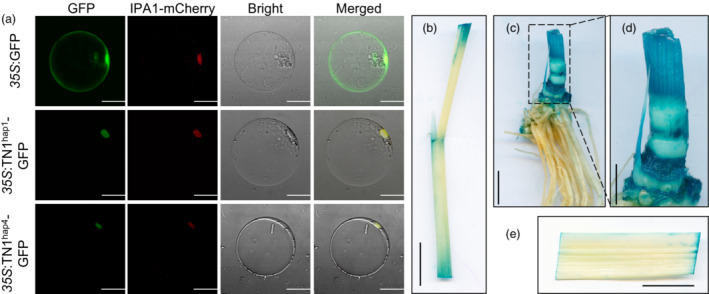
Expression pattern of TN1. (a) Subcellular localization of TN1‐GFP fusion protein in rice protoplasts of different haplotypes. Scale bar = 20 μm. (b, c, e) GUS staining of tissues (leaf sheath, tiller, and leaf respectively) in ProTN1:GUS‐transgenic plants at 30 days after transplanting. Scale bar = 2 cm. (d) Enlarged version of dashed box in (c). Scale bar = 1 cm.
Tn1 recognizes the H3K27me3 mark and interacts with TIF1
We blasted the TN1 protein and noted that it is highly homologous to Arabidopsis ASI1/IBM2 (Figure 4a). Given that both these proteins harbour a BAH domain and this domain can recognize a series of histone marks, we performed a histone pull‐down assay and found that TN1 could bind the H3K27me3 mark (Figure 4b). H3K27me3 is a histone mark that suppress gene expression; thus, TN1 may recognize the H3K27me3 mark of downstream genes to regulate their expression.
Figure 4.

TN1 interacts with TIF1. (a) Phylogenetic analysis of the TN1 protein in 14 species. Monocots and dicots are indicated in orange and green respectively. (b) TN1 recognizes the H3K27me3 peptide in pull‐down assay. (c) Yeast two‐hybrid assay showing interactions of different genotypes of TN1 with TIF1. (d) Luciferase complementation image (LCI) assay in Nicotiana Benthamian leaves. The fluorescence intensity of TN1hap1 and TIF1 was set as 1, and the assay was repeated eight times. (e) Bimolecular fluorescence complementation (BiFC) assay in rice protoplasts. Scale bar = 10 μm. (f) TN1 directly interacts with TIF1 in Co‐IP assay. Black arrow indicates TIF1‐Myc.
To clarify the molecular mechanism of TN1, we cloned the homologous proteins of AIPP1 and EDM2 in rice. LOC_Os07g15270 and LOC_Os08g24946 shared the highest homology with AIPP1 and EDM2 (Figure S8a) and were named TN1 interaction factor 1 (TIF1) and OsEDM2 respectively. Yeast two‐hybrid (Y2H) assays showed that TN1, TIF1, and OsEDM2 did not exhibit transactivation activity (Figure S9a). TN1hap4 showed a stronger interaction with TIF1 than TN1hap1 in vitro (Figure 4c). Likewise, these differences in interaction intensity were observed in luciferase complementation image (LCI) assays (Figure 4d). In addition, subcellular localization assay revealed that TIF1 is a nuclear protein (Figure S8b). Therefore, bimolecular fluorescence complementation (BiFC) assay was performed in rice protoplasts and tobacco leaves, together with co‐immunoprecipitation (Co‐IP) assays, which further confirmed the interaction between TN1 and TIF1 in vivo (Figure 4e,f and Figure S8c). However, both TN1 and TIF1 did not interact with OsEDM2 in yeast, and yeast three‐hybrid (Y3H) assays showed no complex formation between TN1, TIF1, and OsEDM2 (Figure S9a–c).
TIF1 is a negative regulator of tiller number in rice
TIF1 showed a similar expression pattern to TN1, with high expression in tiller buds at 30 days after transplanting (Figure 5a). To investigate the function of TIF1, we knocked out TIF1 with CRISPR–Cas9 using two target sites located in the first exon (Figure 5b) and obtained two homozygous mutant lines, namely tif1‐1 and tif1‐2. The former harboured a 73 bp deletion between the two target sites, and the latter carried a 2 bp deletion at the first site and a 1 bp insertion at the second site, with both causing frameshifts that resulted in early translation termination (Figure 5b). Similar to that in tn1‐1, tiller number was significantly increased in tif1‐1 and tif1‐2 compared with that in NIP (Figure 5c,d). These results indicate that TIF1 and TN1 co‐regulate tiller formation in rice.
Figure 5.

TIF1 negatively regulates tiller number in rice. (a) Expression levels of TN1 in tillering tissues. Data are presented as mean ± SD (n = 3 independent biological samples). DT30_Ab, DT45_Ab, DT60_Ab, ST_1, ST_2, and ST_3 represent developmental stages described in Materials and Methods. (b) Mutation targets of tif1‐1 and tif1‐2. (c) Phenotypes of NIP and tif1‐CRISPR lines at the reproductive stage. Scale bar = 20 cm. (d) Tiller number per plant of NIP and tif1‐CRISPR lines. P‐values were determined using two‐tailed Student's t‐tests. **P < 0.01. Data are presented as mean ± SD (n = 15).
TN1 and TIF1 affect D14 expression to regulate tiller development
In Arabidopsis, ASI1 regulates polyadenylation to ensure the transcription of downstream target genes (Saze et al., 2013; Wang et al., 2013). To identify genes downstream of TN1, we conducted RNA‐Seq analysis and determined differentially expressed genes (DEGs) at the base of axillary buds between NIP and tn1‐1 (Dataset S4). A total of 2938 genes were down‐regulated [log2(tn1‐1/NIP) ≤ −1, P < 0.05] and 2840 genes were up‐regulated [log2(tn1‐1/NIP) ≥ 1, P < 0.05] in tn1‐1 plants (Figure 6a). Gene ontology (GO) analysis revealed that 5778 specific DEGs were enriched in fundamental biological processes (Figure S10a). The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis showed that many DEGs were enriched in the mRNA surveillance pathway (Figure S10b). These results indicate that TN1 might also be required for the expression of intragenic heterochromatin‐containing genes.
Figure 6.

Both TN1 and TIF1 regulate D14 expression. (a) Volcano map of differentially expressed genes between NIP and tn1‐1. (b) Heat map showing the expression of tillering‐related genes in NIP and tn1‐1. (c) Relative expression levels of D14, OsCCA1, and D3 in NIP, tn1‐1, TN1‐OE, and tif1 lines. (d) Phenotypes of tn1‐1 and D14‐OE/tn1‐1 lines at the reproductive stage. Scale bar = 10 cm. (e, f) D14 expression and tiller number in tn1‐1 and D14‐OE/tn1‐1 lines at the reproductive stage. (g) Model of TN1 and TIF1 functioning synergistically to regulate tiller number by affecting D14 transcripts. Red dotted lines indicate transcript abundance. Dotted circles indicate dysfunctionality. Solid circles indicate full function. Broken circles indicate loss of function. Red crosses indicated inhibition. Data in (c) and (e) are presented as mean ± SD of three biologically independent samples.
Next, the expression levels of seven cloned genes controlling tiller development, namely DWARF AND LOW‐TILLERING/DLT (Tong et al., 2009), OsPIN2 (Chen et al., 2012), OsPIN5b (Lu et al., 2015b), D14, D3, OsTB1, and OsCCA1, were altered in tn1‐1 compared with those in NIP (Figure 6b). The expression patterns of these genes in tn1‐1 were consistent with their functions in promoting tiller formation. Next, we assessed the expression levels of these genes in tn1‐1 and TN1‐OE lines using qRT‐PCR and found that the expression of OsCCA1, D3, and D14 was significantly decreased in tn1‐1 but increased in TN1‐OE1 and TN1‐OE2 compared with that in NIP (Figure 6c and Figure S11a–d). These results indicate that TN1 positively regulates the expression of OsCCA1, D3, and D14. OsCCA1, D3, and D14 are involved in the same pathway controlling tiller development (Wang et al., 2020). These results prompted us to investigate whether TN1 acts upstream of the OsCCA1–D3–D14 module. In addition, we found the expression level of TIF1 remained unaffected in tn1‐1 or TN1‐OE (Figure S11e).
Given that D14 expression is related to the H3K27me3 modification level, we considered that TN1 may recognize the H3K27me3 histone mark of D14 to affect its expression. We next generated D14‐overexpression lines in the tn1‐1 background and found that the positive overexpression lines developed less tillers than tn1‐1 (Figure 6d–f). Moreover, the expression levels of D14, D3, and OsCCA1 were significantly decreased in the tif1‐1 and tif1‐2 mutants compared with those in NIP (Figure 6c), suggesting that TIF1 also affects the expression of these genes to regulate tiller number. Collectively, these results indicate that TN1 and TIF1 regulate tiller number by affecting D14 transcription (Figure 6g).
Natural variations in the TN1 gene
A diverse panel of 2154 cultivated and 221 wild rice accessions (Huang et al., 2012; Wang et al., 2018) was used to investigate variations in TN1 in various rice germplasms. Based on the SNP variation +2163G > A, the accessions were divided into two haplotypes, namely TN1‐hapH (SNP_G) and TN1‐hapL (SNP_A). Phylogenetic analysis revealed that Tn1‐hapH (SNP_G) and Tn1‐hapL (SNP_A) were present in both cultivated and wild rice, indicating that differentiation of TN1 originated in wild rice. TN1‐hapL was present only in the indica subgroup, while TN1‐hapH was present both in indica and japonica subgroups (Figure 7a,b). Haplotype network analysis showed TN1‐hapL and TN1‐hapH were similarly distributed in cultivated and wild rice accessions (Figure S12). We next analysed the nucleotide diversity and Tajima's D value of eight subpopulations (Or‐I, Or‐II, Or‐III, Aus, Aro, Ind, GJ‐tmp, and GJ‐trp) and observed no significant differences in Tajima's D value (Table S1), indicating that there has been no selective divergence of TN1 haplotypes. Investigation of the geographical distribution of TN1 alleles revealed that TN1‐hapL accessions are mainly concentrated in Southeast and East Asia, whereas most TN1‐hapH accessions are concentrated in South Asia (Figure 7c). These results indicate apparent preferential utilization of TN1 alleles across geographical regions. We constructed a near‐isogenic line (NIL), NIL‐TN1 CH1230, with TN1‐hapH from CH1230 in a 93–11 genetic background (Figure S13). NIL‐TN1 CH1230 developed more tillers and showed an earlier heading date than NIL‐TN1 93‐11 (Figure 7d), while there were no significant differences in plant height, spikelet number per panicle, and 1000‐grain weight. The results indicate that the TN1‐hapH allele could be used to promote more tillers. However, the frequency of the TN1‐hapL allele was increased in improved varieties (IMPs) compared with that in landraces (LAN), indicating the utilization of TN1‐hapL in modern rice varieties (Figure 7f).
Figure 7.

Phylogenetic origins and utilization of TN1 in breeding. (a) Phylogenetic analysis of TN1 in a natural rice population. Colour of the outer circle refers to eight ecological groups, including Or‐I, Or‐II, Or‐III, Ind (XI‐1A, XI‐1B, XI‐2, and XI‐3), temperate japonica (GJ‐tmp), tropical japonica (GJ‐trp), Aus, and aromatic (Aro). Inner solid colours indicate the two haplotypes: TN1‐hapH and TN1‐hapL. (b) Haplotype analysis of the functional SNPs of TN1. Colours correspond to the inner branch in (a). (c) Frequencies of TN1‐hapH and TN1‐hapL in ecotypes XI‐1A, XI‐2, and XI‐3. (d‐e) Phenotypes of NIL‐TN1 93‐11 and NIL‐TN1 CH1230 at the reproductive stage. (f) Frequency of alleles in improved varieties (IMP) and landraces (LAN). (g) Combined haplotype analysis of TN1‐TIF1 in the indica subpopulation.
We further analysed TIF1 and found three haplotypes, namely TIF1‐Hap1, TIF1‐Hap2, and TIF1‐Hap3. TIF1‐Hap1 and TIF1‐Hap2 were present in both indica and japonica subgroups, while TIF1‐Hap3 was present only in the indica subgroups (Figure S14). Accessions containing TIF1‐Hap3 developed less tillers than accessions containing the other two haplotypes. We further conducted a joint haplotype analysis of TN1 and TIF1. A panel of 264 accessions was divided into five haplotypes, H1–H5. H1, carrying TN1‐hapH and TIF1‐Hap1 exhibited the highest tiller number. Therefore, it can be used as an elite haplotype in breeding (Figure 7g). Furthermore, the expression level of D14 in H1 and H2 varieties was lower than that in H3 and H4 varieties, indicating that the different combinations of TN1 and TIF1 in germplasm affect D14 expression to regulate tiller number (Figure S15).
Discussion
TN1 is a homologue of Arabidopsis ASI1/IBM2, harbouring a BAH domain and an RRM. OsASI1 has been reported to modulate miRNA and pollen development in rice (You et al., 2021). In this study, TN1, a novel allele was identified from rice germplasm participating in tiller development. Based on the phenotypes of transgenic materials (Figure 2b,c and Figure S4c), we further discovered a novel function of the BAH domain and found that TN1 is involved in multiple pathways. From our results, we proposed a mechanism in which TN1 interacts with TIF1 to affect D14 expression for regulating tiller number in rice. Furthermore, we noted natural variation in TN1 (TN1‐hapH and TN1‐hapL) according to geographical distribution, highlighting its potential use in breeding.
Here, the BAH domain and RRM were separately knocked out using CRISPR–Cas9 to explore their respective functions. Proteins containing the BAH domain, such as EBS, SHL, and AIPP3, have been shown to regulate flowering time in Arabidopsis (Li et al., 2018; Yang et al., 2018; Zhang et al., 2020). In this study, the tn1‐1 line with a 6 bp deletion in the BAH domain developed more tillers, whereas the tn1‐2 and tn1‐3 lines were similar to WT in terms of tiller number. In tn1‐4, mutation in the BAH domain resulted in early translation termination, and these lines presented more tillers than NIP. These results indicate that the BAH domain is involved in tiller development. MEIOSIS ARRESTED AT LEPTOTENE2 (MEL2) is an RRM protein participating in the transition to meiosis at the proper timing. The mutation of MEL2 causes pollen abortion and decreased seed setting rate (Miyazaki et al., 2015; Nonomura et al., 2011; Zhao et al., 2021). The tn1‐2, tn1‐3 (Figure 2i), and tn1‐4 (Figure S4c) lines lacking RRM presented a descending fertility phenotype compared with NIP. Although tn1‐1 harboured a complete RRM, its seed setting rate decreased to 47%. In addition, tn1‐1 presented a low seed setting rate, mainly because of delayed heading date (Figure 2j). Rice plants with this phenotype suffer chilling damage during the grain filling stage in Beijing. Meanwhile, tn1‐4 harboured abnormal BAH domain and exhibited a late heading date (Figure S4c), while tn1‐2 and tn1‐3 with intact BAH domains showed normal flowering time (Figure 2j). Therefore, the BAH domain also affects heading date. However, there were no differences between TN1‐OE and NIP in terms of heading date and seed setting rate (Figure 2i,j).
Previous studies have reported that ASI1 interacts with EDM2 in Arabidopsis through an AIPP1 bridge protein, which is involved in post‐transcriptional regulation. These proteins form a complex to ensure correct transcription of genes with heterochromatin in their introns (Duan et al., 2017; Saze et al., 2013; Wang et al., 2013). However, in this study, examination of interactions among TN1, TIF1, and OsEDM2 showed that only TN1 and TIF1 interacted with each other to control tiller number. This result differs from the tripartite interaction reported in Arabidopsis (Duan et al., 2017), indicating that the molecular mechanism of TN1 varies across species or that TN1 may form a complex with other proteins allied to AIPP1 or EDM2.
The rice loss‐of‐function mutant d14 exhibited increased tiller number, whereas D14‐overexpressing plants were phenotypically similar to the WT plants (Zhao et al., 2014). Tiller number in tn1‐1 was similar to that in d14. Overexpression of full‐length D14 in tn1‐1 (D14‐OE/tn1‐1) decreased tiller number in tn1‐1, indicating that D14 acts downstream of TN1 (Figure 6g). Furthermore, the expression level of D14 was lower in the tif1 mutant (Figure 6c). From these results, we proposed a model in which the functional loss of either TN1 or TIF1 decreases the amount of D14 transcripts to regulate tiller number. In addition, we tested the expression level of D14 in multiple germplasms and noted that H1, H2, H3, and H4 with different genotype combinations of TN1 and TIF1 showed different D14 transcript accumulation (Figure S16), as TN1hap1 and TN1hap4 formed different interactions with TIF1 (Figure 4c,d). These results indicate that TN1 and TIF1 coordinately regulate D14 expression to affect tiller number. This model is similar to the mechanism where OsMADS57 interacts with TB1 to modulate rice tilling via D14 expression (Guo et al., 2013). We also tested the expression of TN1, D3, and OsCCA1 in tif1 lines and found that TN1 and D3 transcript levels were decreased in both tif1 lines, while OsCCA1 transcript level was decreased only in tif1‐2 (Figure 6c, Figure S11f). The results indicate that TIF1 not only interacts with TN1 but also affects its expression. However, the mechanism underlying TIF1‐mediated regulation of TN1 warrants further research in the future.
ASI1 is involved in polyadenylation to control the position of poly A in Arabidopsis. Although we found no alternative 3′‐splicing of D14 in tn1‐1 lines (Dataset S5), a recent study has shown that the interaction of NGR5 with the PRC2 complex alters the H3K27me3 modification level in the D14 genomic region to regulate its expression (Wu et al., 2020). In a previous study, variations of the BAH domain were shown to affect binding capability to histone peptides (Li et al., 2018). We also found TN1 recognizes H3K27me3 (Figure 4f). Meanwhile, two types of TN1 protein, namely TN1hap1 and TN1hap4, exhibited differential interaction capability with TIF1 (Figure 4b), suggesting that variations of the BAH domain affect its biological function. Therefore, we speculate that TIF1 interacts with TN1hap1 and TN1hap4 differently, leading to the differential recognition of H3K27me3 by TN1 to affect D14 expression for controlling tiller number.
Tillering is a complex phenotype, and different approaches are used to improve tiller utilization. Yield per plant can be increased by increasing the number of effective tillers. However, tiller number decreases with increasing plant density, albeit with little effect on grain yield per unit area (Huang et al., 2016). Evidently, both TN1‐hapL and TN1‐hapH have been utilized in rice domestication, albeit with different origins. TN1‐hapL varieties are mainly present in Southeast Asia and East Asia, whereas most TN1‐hapH varieties are present in South Asia. The geographical distribution of the TN1 allele is similar to that of OsTCP19 (Liu et al., 2021). Varieties with high‐ and low‐tillering haplotypes are more abundant in nitrogen‐poor and nitrogen‐rich regions respectively. We also found TN1 expression was induced by nitrogen (Figure S16). Therefore, TN1‐hapH may be responsive to nitrogen fertilization. In recent times, an ideal plant architecture with fewer tillers has been advocated in China and by the International Rice Research Institute in The Philippines (Jiao et al., 2010; Khush, 1995; Virk et al., 2004). The high proportion of improved varieties with TN1‐hapL may be attributed to the advocated breeding strategy. Indica rice is classified into two major genetic subgroups, indica I (IndI) and indica II (IndII), and yield heterosis in superior 3‐line hybrid rice cultivars is achieved through crosses between IndI and IndII accessions, such as the cross between Zhenshan97 (IndI) and Minghui63 (IndII) (Xie et al., 2015; Lv et al., 2020). In this study, Zhenshan97 presented TN1‐hapL, while Minghui63 presented TN1‐hapH, indicating that TN1 may also contribute to heterosis.
Materials and methods
Plant material
A panel of 295 Oryza sativa accessions, including 117 accessions from the core collection (Zhang et al., 2011b) and 178 accessions from the International Rice Molecular Breeding Program (Wang et al., 2018; Yu et al., 2003), was planted in the Shangzhuang Experimental Farm of China Agricultural University in Beijing, China (39°9′ N, 116°3′ E). Five plants in the middle row were randomly selected from each accession to determine tiller statistics. The phenotypes of all materials used for GWAS are listed in Dataset S1. To construct near isogenic lines (NILs) of TN1, we developed pairs of molecular markers located in the TN1 genome region (Dataset S6) and produced a BC3F2 NILs using marker selection from the donor variety CH1230 (hap1) to recurrent parent 93–11 (hap4).
Vector construction and genetic transformation
A 20 bp specific sequence from the first exon of the TN1 CDS was selected and integrated into SK‐gRNA to construct the TN1‐CRISPR vector, as described previously (Wang et al., 2015). A protospacer adjacent motif sequence from the RRM was integrated into pHUE411 to construct the TN1‐RRM‐CRISPR vector (Xing et al., 2014). The TN1 CDS without the stop codon was integrated into pSuper1300‐GFP to construct the TN1 overexpression vector. A 2.6 kb promoter fragment upstream of TN1 was fused to pMDC162 to construct a ProTN1:GUS vector. The above constructs were transformed into Agrobacterium tumefaciens strain EHA105. The tn1‐1, tn1‐2, tn1‐3, and tn1‐4 transgenic lines were obtained through Agrobacterium‐mediated infection of NIP (Oryza sativa L. subsp. japonica) callus. The D14 CDS without the stop codon was integrated into pMDC32 to construct the D14‐OE vector, which was transformed into tn1‐1 line to obtain D14‐OE/tn1‐1 lines. D14‐OE/tn1‐1 lines were planted in Sanya, the other transgenic lines in Beijing during May to October.
GWAS, QTL gene annotation, and haplotype analysis
Sequencing data were derived from the 3000 Rice Genome Project. Association analysis using MLM was performed in TASSEL 5.0. After 1000 permutation analyses, we adopted a threshold of P = 10−4 at a genome‐wide level. The standard of delimiting QTL followed previously described methods (Huang et al., 2010). A QTL was defined as a region containing least three clustered significant SNPs within a distance of <170 kb between one another. Non‐synonymous SNPs were separated from all SNPs identified in the 295 accessions based on information on MSU‐RGAP 7.0. Haplotype division was based on significant SNPs (above the suggestive significance threshold of association) concurrently located in the 2 kb promoter and exon of genes.
qRT‐PCR
Total RNA was extracted from different plant tissues using TRIzol reagent (Invitrogen, Carlsbad) and treated with RNase‐free DNase I. cDNA was generated from 1 μg RNA using M‐MLV reverse transcriptase (Takara, Osaka, Japan). qRT‐PCR was performed using TB Premix Ex Taq II with ROX Reference Dye II (Takara, #RR820A) on the Applied Biosystems 7500 Fast Real‐Time PCR System. Rice ubiqutin1 was used as the internal control (Dataset S6). Relative gene expression level was analysed using the comparative critical threshold (▵▵Ct) method (Livak and Schmittgen, 2001). RNA for expression pattern analysis was extracted from the base of tiller buds at 30, 45, and 60 days after transplanting (DT_30, DT_45, and DT_60 respectively) and the base of first, second, and third internode at 75 days after transplanting (ST_1, ST_2, and ST_3 respectively).
Subcellular localization
Different haplotypes of TN1 CDS were cloned and used to generate 35 S:TN1‐hap1‐GFP and 35 S:TN1‐hap4‐GFP vectors for subcellular localization assays. All isolated plasmids were transformed into rice protoplasts using polyethylene glycol‐mediated transformation. Fluorescence was examined under a two‐photon laser confocal microscope (Zeiss LSM880) after 16 h of growth in the dark.
GUS staining
The solution buffer for GUS staining contained 50 mm Na2HPO4, 10 mm Na2EDTA, 0.5 mm K3Fe (CN)6, 0.5 mm K4Fe (CN)6, 0.1% Trition X‐100, and 1 mg/mL 1,5‐bromo‐4‐chloro‐3‐indolyl β‐D‐glucuronic acid. Different tissues of ProTN1:GUS transgenic plants were dipped into GUS staining buffer at 37°C for 12 h and then decolorized with 95% alcohol to remove chlorophyll.
BiFC assays
The TN1 and TIF1 CDSs were cloned from NIP and integrated into plasmids pUC‐SPYNE/pSPYNE‐35 S and pUC‐SPYCE/pSPYCE‐35 S respectively. TN1‐nYFP and TIF1‐cYFP plasmids were co‐transfected into rice protoplasts, and mixed Agrobacterium suspension containing the TN1‐nYFP and TIF1‐cYFP plasmids was injected into Nicotiana benthamiana (tobacco) leaves. YFP signals in the rice protoplasts and tobacco leaves was observed after 16 and 48 h under a confocal laser scanning microscope.
Y2H and Y3H assays
Y2H assays were performed according to the manufacturer's recommendations (Clontech). The integrated vectors pGBKT7‐TN1 and pGADT7‐TIF1 were co‐transformed into yeast strain Y2HGold. The yeast cells were plated onto SD‐L/T (lacking Leu and Trp) and SD‐L/T/H/A (lacking Leu, Trp, His, and Ade) to assess interactions. Y3H assays were performed as previously described (Zhang et al., 2018). The selective media SD‐L/T/M (lacking Leu, Trp, and Met) and SD‐L/T/M/H (lacking Leu, Trp, Met, and His) were used to assess interactions. OD of yeast suspension was set to the same value as that before the test to determine interaction intensity.
LCI assay
The TN1 and TIF1 CDSs were fused to the pCAMBIA1300‐nLUC and pCAMBIA1300‐cLUC vectors respectively. Then, the TN1‐nLUC and TIF1‐cLUC vectors were transfected into Agrobacterium tumefaciens (strain EHA105) and co‐transformed into tobacco leaves. After incubation for 48 h, the leaves were sprayed with 1 mM beetle luciferin solution (Promega, E1605), and LUC signals were observed with a cooled CCD imaging instrument (Vilber, Fusion FX7 EDGE).
Dual‐luciferase assay
According to haplotype analysis in the indica subpopulation, we cloned 2.6 kb promoter fragments of TN1 from CH1230 (hap1) and 93–11 (hap4) and inserted them into a pGreenII0800‐LUC vector to generate proTN1‐hap1‐LUC and proTN1‐hap4‐LUC. All vectors were transformed into rice protoplasts. The activities of firefly luciferase (LUC) and Renilla luciferase (REN) were examined using the Dual‐Luciferase Reporter Assay System kit (Promega, E1910). LUC/REN results were used to measure the activities of different promoters, and an empty pGreenII0800‐LUC vector was used as the control. Four replicates were set for the analysis of each vector.
Co‐IP assays
Co‐IP assays were performed as previously described (Zhang et al., 2017, 2020). HA‐TN1 and TIF1‐Myc were expressed separately as well as jointly in tobacco leaves. Proteins were extracted from the tobacco leaves and detected with primary anti‐Myc (Sigma, M4439) and anti‐HA (Sigma, H3663), followed by secondary goat anti‐mouse IgG (Light chain specific) (Easybio, BE0105).
RNA‐Seq
RNA samples with three biological replicates were collected from the bases of axillary buds in NIP and tn1‐1 lines at 55 days after transplanting. Sample extraction and Illumina sequencing were performed by the Beijing Genome Institute (Wuhan). Clean reads were mapped to the rice genome (MSU‐RGAP 7.0) using Bowtie2 (v2.2.5). Gene expression levels were calculated based on RSEM. A volcano map of differentially expressed genes between the wildtype (NIP) and tn1‐1 was plotted using Hiplot (https://hiplot.com.cn/). A heat map was plotted using TBtools (https://github.com/CJ‐Chen/TBtools). All changes in gene expression are presented in Dataset S4.
Nucleotide diversity and selection analysis
A 7.5 kb genomic sequence of TN1 plus a 2 kb upstream region was used for analysis. Rice groups classified in the 3 K project were examined using DnaSP 5.10.
Evolutionary analysis of TN1 and its genome sequence
Homologous protein sequences of TN1 and TIF1 were downloaded from UniProt (https://www.uniprot.org/blast/). A neighbour‐joining tree (1000 replications of bootstrap tests) containing 14 species was constructed using MEGA 6.0. The EvolView tool (Zhang et al., 2012) was used to visualize the evolutionary tree.
A panel of 2154 cultivated and 221 wild rice varieties was used to construct a phylogenetic tree of the TN1 gene sequence according to previously described methods (Li et al., 2021). A minimum spanning tree of haplotypes using the same data was calculated following a previously described method (Guo et al., 2020).
Histone peptide pull‐down assay
A part of TN1 CDS (amino acid 1–190) was integrated into pGEX‐6P‐1 vector, which was transformed into E. coli BL21 to purify the TN1‐BAH protein; an empty pGEX‐6P‐1 vector was used as the control. A pull‐down assay was performed as previously described (Li et al., 2018; Zhang et al., 2017, 2020). Next, 15 μL of streptavidin beads (NEB, S1421S) and 2 μg of H3K27me3 histone peptide (ANASPEC, AS‐64367) were incubated in a binding buffer for 1 h at 4°C. After washing three to five times with a binding buffer to remove free peptides, 2–3 μg of purified TN1‐BAH‐GST or GST was added to 800 μL of binding buffer with a mixture of beads and H3K27me3 peptides and gently agitated two to three times. Then, 100 μL of the mixture was absorbed as the input sample. The remaining 700 μL of buffer was induced at 4°C for 3 h. The protein–bead mixture was washed five times with a binding buffer before immunoblotting using an anti‐GST antibody (Abcam, ab92, 1:2000). Western blots were observed with a cooled CCD imaging instrument (Vilber, Fusion FX7 EDGE).
Primers
Primers used in the study are listed in Dataset S6.
Author contributions
Q. Z, Z. Z, and H. Z designed the research. Q. Z and J. X performed data analysis. Q. Z performed most of experiments. X. Z, Q. M, T. Y, S. Z, M. L, and L. L performed part of the experiments. Y. L, Y. P, D. L, J. L, and Z. L provided technical assistance. Q. Z wrote the article. N.U. Khan assisted with revisions for the article. H. Z, Z. Z, and Z. L conceived and supervised the writing.
Conflict of interest
The authors declare that they have no competing interests.
Supporting information
Dataset S1 List of the 295 varieties used in this study.
Dataset S2 The QTLs associated with tiller numbers.
Dataset S3 The candidate gene in qTN2.
Dataset S4 The differentially expressed genes between NIP and tn1‐1.
Dataset S5 The alternative 3′ splicing between NIP and tn1‐1 lines.
Dataset S6 Primers used in this study.
Figure S1 Population structure of 295 rice accessions.
Figure S2 Functional variation analysis of TN1.
Figure S3 Haplotype and expression analysis of candidate gene in qTN2.
Figure S4 Target sites in tn1‐2, tn1‐3, and tn1‐4 lines and the phenotype of tn1‐4.
Figure S5 Comparison of tiller buds among the NIP, tn1‐1, and TN1‐OE lines.
Figure S6 Subcellular localization of TN1‐GFP fusion protein in tobacco leaves.
Figure S7 Expression pattern of TN1 in different tissues of NIP, as determined by qRT‐PCR.
Figure S8 Phylogenetic analysis of TIF1 and OsEDM2 proteins and relationship of TN1 and TIF1 in tobacco leaves.
Figure S9 Interaction among TN1, TIF1, and OsEDM2.
Figure S10 Enrichment analysis of 5778 DEGs.
Figure S11 Expression levels of DLT, OsTB1, OsPIN2, OsPIN5b, and TIF1 in NIP, tn1‐1, and TN1‐OE lines and expression level of TN1 in tif1 lines.
Figure S12 Evolutionary pattern of TN1 based on minimum spanning.
Figure S13 Genome constitution of NIL‐TN1 CH1230.
Figure S14 Haplotype analysis of TIF1.
Figure S15 Expression levels of D14 among different combination of TN1 and TIF1 haplotypes in natural germplasms.
Figure S16 TN1 is induced by nitrogen treatment.
Table S1 Nucleotide diversity and selection analysis of TN1.
Acknowledgements
We thank Robert A. McIntosh (University of Sydney) for critical reading and suggested revisions for the article. Our confocal observation and western signal detection were performed at the CAB Public Instrument Platform of Chinese Agricultural University and we thank Chen Sun for her help. This work was supported by grants from the National Natural Science Foundation of China (32172030, 31971922, 32272123 and 32072036), the project of the Administrative Bureau of Sanya Yazhou Bay Science and Technology City (SYND‐2022‐29, B21HJ0508), the Ministry of Science and Technology of the People's Republic of China (2016YFD0100803‐2), and the open project of GKLRGB‐SKLAB union laboratory, Guangxi Academy of Agricultural Sciences (2018‐15‐Z06‐KF10, 2022‐36‐Z01‐KF03).
Contributor Information
Hongliang Zhang, Email: zhangl@cau.edu.cn.
Zhanying Zhang, Email: zhangzhanying@cau.edu.cn.
References
- Arite, T. , Iwata, H. , Ohshima, K. , Maekawa, M. , Nakajima, M. , Kojima, M. , Sakakibara, H. et al. (2007) DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. Plant J. 51, 1019–1029. [DOI] [PubMed] [Google Scholar]
- Arite, T. , Umehara, M. , Ishikawa, S. , Hanada, A. , Maekawa, M. , Yamaguchi, S. and Kyozuka, J. (2009) d14, a strigolactone‐Insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol. 50, 1416–1424. [DOI] [PubMed] [Google Scholar]
- Callebaut, I. , Courvalin, J.C. and Mornon, J.P. (1999) The BAH (bromo‐adjacent homology) domain: a link between DNA methylation, replication and transcriptional regulation. FEBS Lett. 446, 189–193. [DOI] [PubMed] [Google Scholar]
- Chambers, A. , Pearl, L. , Oliver, A. and Downs, J. (2013) The BAH domain of Rsc2 is a histone H3 binding domain. Nucleic Acids Res. 41, 9168–9182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S. , Wang, Z. and Cai, X. (2007) OsRRM, a Spen‐like rice gene expressed specifically in the endosperm. Cell Res. 17, 713–721. [DOI] [PubMed] [Google Scholar]
- Chen, Y. , Fan, X. , Song, W. , Zhang, Y. and Xu, G. (2012) Over‐expression of OsPIN2 leads to increased tiller numbers, angle and shorter plant height through suppression of OsLAZY1 . Plant Biotechnol. J. 10, 139–149. [DOI] [PubMed] [Google Scholar]
- Du, J. , Zhong, X. , Bernatavichute, Y.V. , Stroud, H. , Feng, S. , Caro, E. , Vashisht, A.A. et al. (2012) Dual binding of chromomethylase domains to H3K9me2‐containing nucleosomes directs DNA methylation in plants. Cell 151, 167–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, C. , Wang, X. , Zhang, L. , Xiong, X. , Zhang, Z. , Tang, K. , Pan, L. et al. (2017) A protein complex regulates RNA processing of intronic heterochromatin‐containing genes in Arabidopsis . Proc. Natl. Acad. Sci. U. S. A. 114, E7377–E7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, E. , Wang, Y. , Li, X. , Lin, Q. , Zhang, T. , Wang, Y. , Zhou, C. et al. (2019) OsSHI1 regulates plant architecture through modulating the transcriptional activity of IPA1 in rice. Plant Cell 31, 1026–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, Z. , Qian, Q. , Liu, X. , Yan, M. , Feng, Q. , Dong, G. , Liu, J. et al. (2009) Dwarf 88, a novel putative esterase gene affecting architecture of rice plant. Plant Mol. Biol. 71, 265–276. [DOI] [PubMed] [Google Scholar]
- Goodarzi, H. , Zhang, S. , Buss, C.G. , Fish, L. , Tavazoie, S. and Tavazoie, S.F. (2014) Metastasis‐suppressor transcript destabilization through TARBP2 binding of mRNA hairpins. Nature 513, 256–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, S. , Xu, Y. , Liu, H. , Mao, Z. , Zhang, C. , Ma, Y. , Zhang, Q. et al. (2013) The interaction between OsMADS57 and OsTB1 modulates rice tillering via DWARF14 . Nat. Commun. 4, 1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, H. , Zeng, Y. , Li, J. , Ma, X. , Zhang, Z. , Lou, Q. , Li, J. et al. (2020) Differentiation, evolution and utilization of natural alleles for cold adaptability at the reproductive stage in rice. Plant Biotechnol. J. 18, 2491–2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan, D.J. , Riordan, D.P. , Gerber, A.P. , Herschlag, D. and Brown, P.O. (2008) Diverse RNA‐binding proteins interact with functionally related sets of RNAs, suggesting an extensive regulatory system. PLoS Biol. 6, 2297–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, X. , Wei, X. , Sang, T. , Zhao, Q. , Feng, Q. , Zhao, Y. , Li, C. et al. (2010) Genome‐wide association studies of 14 agronomic traits in rice landraces. Nat. Genet. 42, 961–976. [DOI] [PubMed] [Google Scholar]
- Huang, X. , Kurata, N. , Wei, X. , Wang, Z.‐X. , Wang, A. , Zhao, Q. , Zhao, Y. et al. (2012) A map of rice genome variation reveals the origin of cultivated rice. Nature 490, 497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, X. , Yang, S. , Gong, J. , Zhao, Q. , Feng, Q. , Zhan, Q. , Zhao, Y. et al. (2016) Genomic architecture of heterosis for yield traits in rice. Nature 537, 629–633. [DOI] [PubMed] [Google Scholar]
- Ishikawa, S. , Maekawa, M. , Arite, T. , Onishi, K. , Takamure, I. and Kyozuka, J. (2005) Suppression of tiller bud activity in tillering dwarf mutants of rice. Plant Cell Physiol. 46, 79–86. [DOI] [PubMed] [Google Scholar]
- Jiang, L. , Liu, X. , Xiong, G. , Liu, H. , Chen, F. , Wang, L. , Meng, X. et al. (2013) DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 504, 401–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao, Y. , Wang, Y. , Xue, D. , Wang, J. , Yan, M. , Liu, G. , Dong, G. et al. (2010) Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 42, 541–544. [DOI] [PubMed] [Google Scholar]
- Kenan, D.J. , Query, C.C. and Keene, J.D. (1991) RNA recognition: towards identifying determinants of specificity. Trends Biochem. Sci. 16, 214–220. [DOI] [PubMed] [Google Scholar]
- Khush, G.S. (1995) Breaking the yield frontier of rice. GeoJournal 35, 329–332. [Google Scholar]
- Kuo, A.J. , Song, J. , Cheung, P. , Ishibe‐Murakami, S. , Yamazoe, S. , Chen, J.K. , Patel, D.J. et al. (2012) The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier‐Gorlin syndrome. Nature 484, 115–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. , Qian, Q. , Fu, Z. , Wang, Y. , Xiong, G. , Zeng, D. , Wang, X. et al. (2003) Control of tillering in rice. Nature 422, 618–621. [DOI] [PubMed] [Google Scholar]
- Li, S. , Yang, Z. , Du, X. , Liu, R. , Wilkinson, A.W. , Gozani, O. , Jacobsen, S.E. et al. (2016) Structural basis for the unique multivalent readout of unmodified H3 tail by Arabidopsis ORC1b BAH‐PHD cassette. Structure 24, 486–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z. , Fu, X. , Wang, Y. , Liu, R. and He, Y. (2018) Polycomb‐mediated gene silencing by the BAH‐EMF1 complex in plants. Nat. Genet. 50, 1254–1261. [DOI] [PubMed] [Google Scholar]
- Li, J. , Zeng, Y. , Pan, Y. , Zhou, L. , Zhang, Z. , Guo, H. , Lou, Q. et al. (2021) Stepwise selection of natural variations at CTB2 and CTB4a improves cold adaptation during domestication of japonica rice. New Phytol. 231, 1056–1072. [DOI] [PubMed] [Google Scholar]
- Liu, D. and Cai, X. (2013) OsRRMh, a Spen‐like gene, plays an important role during the vegetative to reproductive transition in rice. J. Integr. Plant Biol. 55, 876–887. [DOI] [PubMed] [Google Scholar]
- Liu, W. , Wu, C. , Fu, Y. , Hu, G. , Si, H. , Zhu, L. , Luan, W. et al. (2009) Identification and characterization of HTD2: a novel gene negatively regulating tiller bud outgrowth in rice. Planta 230, 649–658. [DOI] [PubMed] [Google Scholar]
- Liu, Y. , Wang, H. , Jiang, Z. , Wang, W. , Xu, R. , Wang, Q. , Zhang, Z. et al. (2021) Genomic basis of geographical adaptation to soil nitrogen in rice. Nature 590, 600–605. [DOI] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lu, Z. , Yu, H. , Xiong, G. , Wang, J. , Jiao, Y. , Liu, G. , Jing, Y. et al. (2013) Genome‐wide binding analysis of the transcription activator IDEAL PLANT ARCHITECTURE1 reveals a complex network regulating rice plant architecture. Plant Cell 25, 3743–3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, Z. , Shao, G. , Xiong, J. , Jiao, Y. , Wang, J. , Liu, G. , Meng, X. et al. (2015a) MONOCULM 3, an ortholog of WUSCHEL in rice, is required for tiller bud formation. J. Genet. Genomics 42, 71–78. [DOI] [PubMed] [Google Scholar]
- Lu, G. , Coneva, V. , Casaretto, J.A. , Ying, S. , Mahmood, K. , Liu, F. , Nambara, E. et al. (2015b) OsPIN5b modulates rice (Oryza sativa) plant architecture and yield by changing auxin homeostasis, transport and distribution. Plant J. 83, 913–925. [DOI] [PubMed] [Google Scholar]
- Lv, Q. , Li, W. , Sun, Z. , Ouyang, N. , Jing, X. , He, Q. , Wu, J. et al. (2020) Resequencing of 1,143 indica rice accessions reveals important genetic variations and different heterosis patterns. Nat. Commun. 11, 4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, X. , Li, F. , Zhang, Q. , Wang, X. , Guo, H. , Xie, J. , Zhu, X. et al. (2020) Genetic architecture to cause dynamic change in tiller and panicle numbers revealed by genome‐wide association study and transcriptome profile in rice. Plant J. 104, 1603–1616. [DOI] [PubMed] [Google Scholar]
- Minakuchi, K. , Kameoka, H. , Yasuno, N. , Umehara, M. , Luo, L. , Kobayashi, K. , Hanada, A. et al. (2010) FINE CULM1 (FC1) works downstream of strigolactones to inhibit the outgrowth of axillary buds in rice. Plant Cell Physiol. 51, 1127–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura, K. , Ikeda, M. , Matsubara, A. , Song, X.‐J. , Ito, M. , Asano, K. , Matsuoka, M. et al. (2010) OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 42, 545–549. [DOI] [PubMed] [Google Scholar]
- Miyazaki, S. , Sato, Y. , Asano, T. , Nagamura, Y. and Nonomura, K.I. (2015) Rice MEL2, the RNA recognition motif (RRM) protein, binds in vitro to meiosis‐expressed genes containing U‐rich RNA consensus sequences in the 3'‐UTR. Plant Mol. Biol. 89, 293–307. [DOI] [PubMed] [Google Scholar]
- Mjomba, F.M. , Zheng, Y. , Liu, H. , Tang, W. , Hong, Z. , Wang, F. and Wu, W. (2016) Homeobox is pivotal for OsWUS controlling tiller development and female fertility in rice. G3‐Genes Genomes Genet 6, 2013–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonomura, K.I. , Eiguchi, M. , Nakano, M. , Takashima, K. , Komeda, N. , Fukuchi, S. , Miyazaki, S. et al. (2011) A novel RNA‐recognition‐motif protein is required for premeiotic G1/S‐phase transition in rice (Oryza sativa L.). PLoS Genet. 7, e1001265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa, T. and Kyozuka, J. (2009) Two‐step regulation of LAX PANICLE1 protein accumulation in axillary meristem formation in rice. Plant Cell 21, 1095–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineiro, M. , Gomez‐Mena, C. , Schaffer, R. , Martinez‐Zapater, J.M. and Coupland, G. (2003) Early bolting in short days is related to chromatin remodeling factors and regulates flowering in Arabidopsis by repressing FT . Plant Cell 15, 1552–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roundtree, I.A. , Evans, M.E. , Pan, T. and He, C. (2017) Dynamic RNA modifications in gene expression regulation. Cell 169, 1187–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, Y. , Antonio, B.A. , Namiki, N. , Takehisa, H. , Minami, H. , Kamatsuki, K. , Sugimoto, K. et al. (2011) RiceXPro: a platform for monitoring gene expression in japonica rice grown under natural field conditions. Nucleic Acids Res. 39, D1141–D1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saze, H. , Kitayama, J. , Takashima, K. , Miura, S. , Harukawa, Y. , Ito, T. and Kakutani, T. (2013) Mechanism for full‐length RNA processing of Arabidopsis genes containing intragenic heterochromatin. Nat. Commun. 4, 2301. [DOI] [PubMed] [Google Scholar]
- Song, X. , Lu, Z. , Yu, H. , Shao, G. , Xiong, J. , Meng, X. , Jing, Y. et al. (2017) IPA1 functions as a downstream transcription factor repressed by D53 in strigolactone signaling in rice. Cell Res. 27, 1128–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, H. , Qian, Q. , Wu, K. , Luo, J. , Wang, S. , Zhang, C. , Ma, Y. et al. (2014) Heterotrimeric G proteins regulate nitrogen‐use efficiency in rice. Nat. Genet. 46, 652–656. [DOI] [PubMed] [Google Scholar]
- Tabuchi, H. , Zhang, Y. , Hattori, S. , Omae, M. , Shimizu‐Sato, S. , Oikawa, T. , Qian, Q. et al. (2011) LAX PANICLE2 of rice encodes a novel nuclear protein and regulates the formation of axillary meristems. Plant Cell 23, 3276–3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda, T. , Suwa, Y. , Suzuki, M. , Kitano, H. , Ueguchi‐Tanaka, M. , Ashikari, M. , Matsuoka, M. et al. (2003) The OsTB1 gene negatively regulates lateral branching in rice. Plant J. 33, 513–520. [DOI] [PubMed] [Google Scholar]
- Tanaka, W. , Ohmori, Y. , Ushijima, T. , Matsusaka, H. , Matsushita, T. , Kumamaru, T. , Kawano, S. et al. (2015) Axillary meristem formation in rice requires the WUSCHEL ortholog TILLERS ABSENT1 . Plant Cell 27, 1173–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong, H. , Jin, Y. , Liu, W. , Li, F. , Fang, J. , Yin, Y. , Qian, Q. et al. (2009) DWARF AND LOW‐TILLERING, a new member of the GRAS family, plays positive roles in brassinosteroid signaling in rice. Plant J. 58, 803–816. [DOI] [PubMed] [Google Scholar]
- Virk, P.S. , Khush, G.S. and Peng, S. (2004) Breeding to enhance yield potential of rice at IRRI: the ideotype approach. Int. Rice Res. Notes. 29, S1–S9. [Google Scholar]
- Wang, Y. and Li, J. (2006) Genes controlling plant architecture. Curr. Opin. Biotechnol. 17, 123–129. [DOI] [PubMed] [Google Scholar]
- Wang, X. , Duan, C.‐G. , Tang, K. , Wang, B. , Zhang, H. , Lei, M. , Lu, K. et al. (2013) RNA‐binding protein regulates plant DNA methylation by controlling mRNA processing at the intronic heterochromatin‐containing gene IBM1 . Proc. Natl. Acad. Sci. U. S. A. 110, 15467–15472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C. , Shen, L. , Fu, Y. , Yan, C. and Wang, K. (2015) A simple CRISPR/Cas9 system for multiplex genome editing in rice. J. Genet. Genomics 42, 703–706. [DOI] [PubMed] [Google Scholar]
- Wang, S. , Wu, K. , Qian, Q. , Liu, Q. , Li, Q. , Pan, Y. , Ye, Y. et al. (2017) Non‐canonical regulation of SPL transcription factors by a human OTUB1‐like deubiquitinase defines a new plant type rice associated with higher grain yield. Cell Res. 27, 1142–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W. , Mauleon, R. , Hu, Z. , Chebotarov, D. , Tai, S. , Wu, Z. , Li, M. et al. (2018) Genomic variation in 3,010 diverse accessions of Asian cultivated rice. Nature 557, 43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, F. , Han, T. , Song, Q. , Ye, W. , Song, X. , Chu, J. , Li, J. et al. (2020) The rice circadian clock regulates tiller growth and panicle development through strigolactone signaling and sugar sensing. Plant Cell 32, 3124–3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, K. , Wang, S. , Song, W. , Zhang, J. , Wang, Y. , Liu, Q. , Yu, J. et al. (2020) Enhanced sustainable green revolution yield via nitrogen‐responsive chromatin modulation in rice. Science 367, eaaz2046. [DOI] [PubMed] [Google Scholar]
- Xie, W. , Wang, G. , Yuan, M. , Yao, W. , Lyu, K. , Zhao, H. , Yang, M. et al. (2015) Breeding signatures of rice improvement revealed by a genomic variation map from a large germplasm collection. Proc. Natl. Acad. Sci. U. S. A. 112, E5411–E5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing, H.‐L. , Dong, L. , Wang, Z.‐P. , Zhang, H.‐Y. , Han, C.‐Y. , Liu, B. , Wang, X.‐C. et al. (2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 14, 327–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z. , Qian, S. , Scheid, R.N. , Lu, L. , Chen, X. , Liu, R. , Du, X. et al. (2018) EBS is a bivalent histone reader that regulates floral phase transition in Arabidopsis . Nat. Genet. 50, 1247–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You, L. , Lin, J. , Xu, H. , Chen, C. , Chen, J. , Zhang, J. , Zhang, J. et al. (2021) Intragenic heterochromatin‐mediated alternative polyadenylation modulates miRNA and pollen development in rice. New Phytol. 232, 835–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, S. , Xu, W. , Vijayakumar, C.H. , Ali, J. , Fu, B. , Xu, J. , Jiang, Y. et al. (2003) Molecular diversity and multilocus organization of the parental lines used in the International Rice Molecular Breeding Program. Theor. Appl. Genet. 108, 131–140. [DOI] [PubMed] [Google Scholar]
- Zhang, Z. , Li, J. , Yao, G. , Zhang, H. , Dou, H. , Shi, H. and Sun, X. (2011a) Fine mapping and cloning of the grain number per‐panicle gene (Gnp4) on chromosome 4 in rice (Oryza sativa L.). Agric. Sci. China 10, 1825–1833. [Google Scholar]
- Zhang, H. , Zhang, D. , Wang, M. , Sun, J. , Qi, Y. , Li, J. , Wei, X. et al. (2011b) A core collection and mini core collection of Oryza sativa L. in China. Theor. Appl. Genet. 122, 49–61. [DOI] [PubMed] [Google Scholar]
- Zhang, H. , Gao, S. , Lercher, M.J. , Hu, S. and Chen, W.‐H. (2012) EvolView, an online tool for visualizing, annotating and managing phylogenetic trees. Nucleic Acids Res. 40, W569–W572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z. , Li, J. , Pan, Y. , Li, J. , Zhou, L. , Shi, H. , Zeng, Y. et al. (2017) Natural variation in CTB4a enhances rice adaptation to cold habitats. Nat. Commun. 8, 14788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z. , Li, J. , Tang, Z. , Sun, X. , Zhang, H. , Yu, J. , Yao, G. et al. (2018) Gnp4/LAX2, a RAWUL protein, interferes with the OsIAA3‐OsARF25 interaction to regulate grain length via the auxin signaling pathway in rice. J. Exp. Bot. 69, 4723–4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y.‐Z. , Yuan, J. , Zhang, L. , Chen, C. , Wang, Y. , Zhang, G. , Peng, L. et al. (2020) Coupling of H3K27me3 recognition with transcriptional repression through the BAH‐PHD‐CPL2 complex in Arabidopsis . Nat. Commun. 11, 6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J. , Wang, T. , Wang, M. , Liu, Y. , Yuan, S. , Gao, Y. , Yin, L. et al. (2014) DWARF3 participates in an SCF complex and associates with DWARF14 to suppress rice shoot branching. Plant Cell Physiol. 55, 1096–1109. [DOI] [PubMed] [Google Scholar]
- Zhao, T. , Ren, L. , Zhao, Y. , You, H. , Zhou, Y. , Tang, D. , Du, G. et al. (2021) Reproductive cells and peripheral parietal cells collaboratively participate in meiotic fate acquisition in rice anthers. Plant J. 108, 661–671. [DOI] [PubMed] [Google Scholar]
- Zhou, F. , Lin, Q. , Zhu, L. , Ren, Y. , Zhou, K. , Shabek, N. , Wu, F. et al. (2013) D14‐SCFD3‐dependent degradation of D53 regulates strigolactone signalling. Nature 504, 406–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Dataset S1 List of the 295 varieties used in this study.
Dataset S2 The QTLs associated with tiller numbers.
Dataset S3 The candidate gene in qTN2.
Dataset S4 The differentially expressed genes between NIP and tn1‐1.
Dataset S5 The alternative 3′ splicing between NIP and tn1‐1 lines.
Dataset S6 Primers used in this study.
Figure S1 Population structure of 295 rice accessions.
Figure S2 Functional variation analysis of TN1.
Figure S3 Haplotype and expression analysis of candidate gene in qTN2.
Figure S4 Target sites in tn1‐2, tn1‐3, and tn1‐4 lines and the phenotype of tn1‐4.
Figure S5 Comparison of tiller buds among the NIP, tn1‐1, and TN1‐OE lines.
Figure S6 Subcellular localization of TN1‐GFP fusion protein in tobacco leaves.
Figure S7 Expression pattern of TN1 in different tissues of NIP, as determined by qRT‐PCR.
Figure S8 Phylogenetic analysis of TIF1 and OsEDM2 proteins and relationship of TN1 and TIF1 in tobacco leaves.
Figure S9 Interaction among TN1, TIF1, and OsEDM2.
Figure S10 Enrichment analysis of 5778 DEGs.
Figure S11 Expression levels of DLT, OsTB1, OsPIN2, OsPIN5b, and TIF1 in NIP, tn1‐1, and TN1‐OE lines and expression level of TN1 in tif1 lines.
Figure S12 Evolutionary pattern of TN1 based on minimum spanning.
Figure S13 Genome constitution of NIL‐TN1 CH1230.
Figure S14 Haplotype analysis of TIF1.
Figure S15 Expression levels of D14 among different combination of TN1 and TIF1 haplotypes in natural germplasms.
Figure S16 TN1 is induced by nitrogen treatment.
Table S1 Nucleotide diversity and selection analysis of TN1.


