Summary
The importance of rhizomicrobiome in plant development, nutrition acquisition and stress tolerance is unquestionable. Relevant plant genes corresponding to the above functions also regulate rhizomicrobiome construction. Deciphering the molecular regulatory network of plant‐microbe interactions could substantially contribute to improving crop yield and quality. Here, the plant gene‐related nutrient uptake, biotic and abiotic stress resistance, which may influence the composition and function of microbial communities, are discussed in this review. In turn, the influence of microbes on the expression of functional plant genes, and thereby plant growth and immunity, is also reviewed. Moreover, we have specifically paid attention to techniques and methods used to link plant functional genes and rhizomicrobiome. Finally, we propose to further explore the molecular mechanisms and signalling pathways of microbe‐host gene interactions, which could potentially be used for managing plant health in agricultural systems.
Keywords: microbe‐host gene interactions, microbiome, plant genetic engineering techniques, microbial sequencing techniques, synthetic microbial communities
Introduction
Environment‐friendly production of crops is one of the challenges in agricultural systems to feed growing population (Yashveer et al., 2014). Increasing evidences indicate that the soil rhizomicrobiome benefit plant growth and health and therefore play an important role in dealing with this challenge (Bai et al., 2022; Goh et al., 2013; Mueller and Sachs, 2015; Pieterse et al., 2014; Raza et al., 2021). The host‐associated microbiota inhabits inside various plant tissues and on root surface to access soil nutrients (Bai et al., 2022). Microbial community structure is largely shaped by the host plant and external environment (Dastogeer et al., 2020; Friesen et al., 2011; Raza et al., 2021). When plants are subjected to biotic or abiotic stresses, they can recruit beneficial microorganisms to help them resist these stresses by secreting a range of chemical factors, which is known as the ‘cry for help’ strategy (Bai et al., 2022; Bakker et al., 2018; Carrión et al., 2019; Liu et al., 2020; Liu and Brettell, 2019). To understand the comprehensive ‘cry for help’ strategy of plants, it is vital to unravel the molecular mechanisms of microbiome recruitment in the rhizosphere (Rolfe et al., 2019; Zancarini et al., 2021).
The functions of the plant genes are crucial for understand the signalling cascades that control plant development and stress responses (Depuydt and Vandepoele, 2021). A large number of genes functions have been identified in the model plant Arabidopsis thaliana. Frequently, a single gene or several genes can largely regulate traits such as nutrient uptake, disease resistance, and resistance to abiotic stresses in plants (Liu et al., 2009; Wei et al., 2022; Zhao et al., 2021). These genes were further validated in different crops such as wheat, rice, maize, soybean and sorghum (Li et al., 2018; Liu et al., 2016; Maron et al., 2013; Wei et al., 2022; Yokosho et al., 2011). In recent years, plant functional genes have also been found to play important role in shaping the rhizomicrobiome (Cordovez et al., 2019; Zhang et al., 2019). The discoveries of plant‐microbe interaction at the molecular level provide a new direction for genetic breeding (Kroll et al., 2017; Nerva et al., 2022). Further research on microbes mutually interacting with the host genes is expected to cultivate new germplasm resources (Kroll et al., 2017). Plant functional genes could regulate root phenotypic traits and the secretion of root exudates, such as organic acids and hormones (Kaushal et al., 2021; Wang et al., 2013, 2020; Yu et al., 2021), which have been found to drive microbial community assembly in rhizosphere (Zhalnina et al., 2018). For example, the expression of plant organic acid channel protein genes can promote the production of organic acid, which could recruit beneficial rhizosphere microorganisms by forming stable metal chelate complexes and increasing soil pH (Zhang et al., 2017).
Rhizosphere microorganisms can also confer health advantages to plants by inducing the expression of plant functional genes (Berendsen et al., 2018; Liu et al., 2017). It has been proved that the expression of genes related to nutrient uptake and stress resistance is affected by structural and functional changes in the rhizosphere microbiota. These microbes can regulate the expression of plant functional genes by secreting secondary metabolites, producing volatile compounds and competing for nutrients, thereby directly or indirectly influencing plant growth and development (Hacquard et al., 2017; Hou et al., 2021; Kwak et al., 2018; Netzker et al., 2020; Yuan et al., 2018). Thus, there exists a complex network of interactions between functional plant genes and rhizomicrobiome and both of them play key roles for plant survival and growth (Berendsen et al., 2018; Depuydt and Vandepoele, 2021). However, whether plant genes or rhizobia have a greater influence on plant development is currently being debated, which necessitates particular plant‐microbe contribution algorithms. Exploring this relationship will help us to develop crop varieties with strong adaptability and resistance to various stresses. So far, such studies are only the tip of the iceberg due to the complex mechanism of microbe‐host gene interaction that involve multidisciplinary intersections (Rolfe et al., 2019). Therefore, understanding how plant‐microbe communication is established requires more experimental exploration using cutting‐edge technologies.
Unprecedented technologies facilitate the investigation on the link of plant‐specific genes to rhizosphere microorganisms (Fadiji and Babalola, 2020; Kumar et al., 2019; Levy et al., 2018; Liu et al., 2020; Schaarschmidt et al., 2020; Xu et al., 2018). These technologies include plant‐related CRISPR‐Cas, transgenics and transgenic hairy roots and microbial‐related 16S rRNA sequencing, GeoChip, metagenomics and synthetic communities. Using the ‘top‐down’ and ‘bottom‐up’ designed methods and relevant techniques, great progress has been made in the mechanism of plant functional gene‐microbe interaction (Chi et al., 2018; Ke et al., 2021; Lawson et al., 2019; Xu et al., 2021b). Here, we summarized how plant‐specific genes regulate rhizomicrobiome, and in turn how rhizomicrobiome influence the host genes, as well as the research methods used.
Most microorganisms in nature are unculturable (Kaeberlein et al., 2002). Yet minute and complicated experimental conditions and media combinations have to be considered in the culturable ones, greatly obstructing the development of microbiomics (van Teeseling and Jogler, 2021; Xing et al., 2017). To overcome the shortcomings of pure culture, the microbiome sequencing technology has been developed in recent years (Liu et al., 2012). It became clear that the microbiome could not be fully characterized at the genetic and transcriptional levels (Gao and Chu, 2020). Thus, the development of metaproteomics and metabolomics is particularly necessary for microbial functional studies (White et al., 2016). The unprecedented development of microbiome technologies mentioned above has enabled an in‐depth analysis and understand the composition and function of host‐associated microbiota (Shao et al., 2021).
Plant functional genes regulate rhizosphere microorganisms
Plant‐specific genes influence rhizomicrobiome based on molecular signalling, which is essentially a ‘top‐down’ process. Increasing number of studies examine how plant functional genes affect microbes at the molecular level, such as the structure of rhizomicrobiome selected by cultivars with the same high nutrient use efficiency and stress resistance (Chen et al., 2019; Qiao et al., 2017; Shi et al., 2020). Although some genes explain only a small part of the total variance in the rhizomicrobiome, the microbiome is increasingly considered as an important predictor of plant phenotype (Bai et al., 2022; Ravanbakhsh et al., 2019; Zancarini et al., 2021). In this section, we will specifically address rhizomicrobiome that are likely regulated by plant functional genes regarding nutrient uptake genes, disease and abiotic stress resistance genes.
Nutrient uptake‐related genes affect rhizosphere microbes
Plant growth and development heavily depend on the availability of nutrients that the root system can access to, indicating that plants have to face enormous challenges in extracting nutrients for cellular activities, and any lack of nutrients may decrease the productivity (Morgan and Connolly, 2013; Sukumar et al., 2013). As a result, some plant species have ‘risen to the occasion’ and attempted to recruit soil microorganisms by regulating nutrient uptake genes to enhance defence capability against nutrient deprivation (Millet et al., 2010; Teixeira et al., 2019; Zhang et al., 2019). The main mechanism of nutrient‐uptake‐related‐genes regulating rhizosphere microbes is its capability to increase root surface area, root hairs and lateral roots that are key factors to alter the rhizosphere microbial community (Figure 1a; Contesto et al., 2010; Ditengou et al., 2000; Felten et al., 2009; Hirsch et al., 1997; Yu et al., 2021). For example, the maize (Zea mays) mutant rootless meristem 1 (rum1) is defective in the initiation of embryonic seminal roots and postembryonic lateral roots in primary roots. Rum1 gene is an important checkpoint for auxin‐mediated initiation of lateral and seminal roots in maize, which may participate in the molecular network of root formation by regulating auxin transport in primary roots and auxin perception in primary root pericytes and influencing lateral root formation (Woll et al., 2005). Recently, it has been shown that the rhizosphere bacterial diversity along the root development region of the maize mutant rum1 is significantly reduced compared to the wild type (Yu et al., 2021). This is because the mutant rum1 lacks lateral roots, limiting water and nutrient acquisition during early developmental stages. This suggests that root development‐related genes can control the length and number of lateral roots by mediating a number of keys signalling substances, which in turn affect the microbial composition in the rhizosphere.
Figure 1.
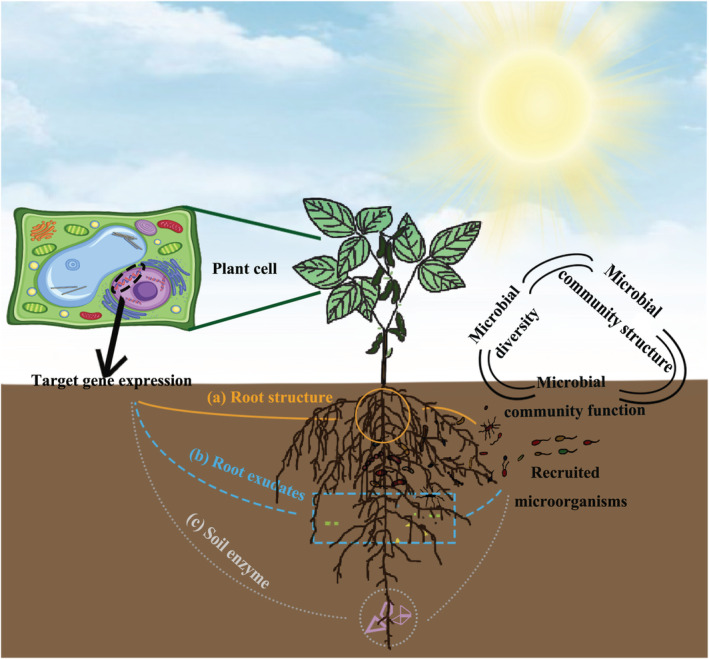
Plant functional genes regulate rhizosphere microbial diversity and function. (a) The development of root morphology (e.g., root length, number of lateral roots) is determined by the relevant plant genes, which usually regulate the synthesis of substances required for root morphology at the genetic and transcriptional levels. Changes in root structure imply differences in the ability of roots to supply nutrients, which affect the assembly of the rhizosphere microbial community to some extent; (b) Plant functional genes influence rhizosphere microbial diversity and structure through the control of root exudates (e.g., phenolics, flavonoids, hormones). (c) The expression of host‐specific genes has a regulatory effect on soil enzyme activity, which in turn is closely related to microorganisms. However, whether plant functional genes can affect microbial diversity, and function by regulating soil enzyme activity remains to be further demonstrated by relevant studies. In short, plant genes can alter the diversity and assembly of rhizosphere microbes. Nevertheless, during these alterations of rhizosphere microorganisms, the functions of the original microbial community may be changed with the newly recruited microorganisms.
A number of genes related to nutrient uptake and transport have been identified, such as ammonium nitrogen, nitrate nitrogen and phosphate root transporters (Wei et al., 2010; Zhu et al., 2016). For instance, several plasma membrane transporters involved in NO3 − have been identified in Arabidopsis and other crops (Hu et al., 2015; Wang et al., 2012). NRT1.1B was found that largely explain the differences in nitrogen utilization efficiency between indica and japonica, which are the two main rice subspecies rice in Asian (Hu et al., 2015). Moreover, NRT1.1 not only transports nitrate but also promotes uptake of the growth promotion hormone from the rhizosphere, which affects lateral root development (Krouk et al., 2010; Léran et al., 2013; Teng et al., 2019). It has been found that wild‐type rice has more rhizosphere microbes involved in the nitrogen cycle compared to the NRT1.1B mutant. NRT1.1B is associated with the relative abundance of root bacteria that harbour key genes in the ammonification process, and these microbes may catalyse the formation of ammonium in the rhizosphere environment and thus affect the acquisition of nitrogen in plants (Zhang et al., 2019). Another example is that Adenosine triphosphate binding cassette (ABC) transporter proteins include a large family have been shown to be involved in membrane transport of endogenous secondary metabolites in plants (Badri et al., 2008; Yazaki, 2005). Some of these members in this family are important in the secretion of antifungal diterpenes and heavy metal detoxification (Brunetti et al., 2015; Jasiński et al., 2001). Studies have shown that Arabidopsis mutants with damaged ABC transporter abcg30 (Atpdr2) increase and decrease the secretion of phenols and sugars, respectively, forming a specific microbial community capable of resisting or degrading phenolic compounds enriched in abcg30 plant secretions and thus reducing rhizobacterial diversity (Badri et al., 2009; Cordovez et al., 2019). Furthermore, karrikin (KAR)/KAR‐like Signals (KLs)/D14L‐ligand‐responsive genes were proved to promote the production of strigolactones and flavonoids, which selectively modify the composition of the rhizomicrobiome (Wang et al., 2020). These studies suggest that some nutrient uptake and transport‐related genes can influence rhizomicrobiome composition by regulating root cell transporter protein activity, secreting root exudates (e.g., secondary metabolites, organic acids, hormones) and thus regulating plant nutrient utilization and altering root environmental conditions (e.g., soil pH, O2 partial pressure, carbon source; Figure 1b; Kaushal et al., 2021; Liu et al., 2022; Wang et al., 2013, 2020; Wen et al., 2022; Yu et al., 2021). Moreover, some root secretions can act as signals to initiate rhizosphere chemical communication recognition processes, thereby influencing microbial‐based crop growth‐defence trade‐off strategies (Chen et al., 2020; Lareen et al., 2016; Vives‐Peris et al., 2020).
There exist some specific mechanisms in legumes by which nutrient uptake‐related genes (e.g., genes that control nodulation and thus increase nitrogen uptake) regulate rhizomicrobiome. This is because symbiotic nitrogen fixation by rhizobia is a mutually beneficial symbiotic process established between legumes and rhizobia through the activation of rhizobia‐induced signalling pathways and the expression of functional genes required for nodule primordium formation (Gao et al., 2021; Yang et al., 2022). Many important genes, including nodule initiation (NIN), nodule requirement (ERN1), Nod factor receptor 5, lotus root histidine kinase 1 and some micro(mi)RNAs, such as MtmiR169a‐MtNFYA1, have been reported to be involved affect nodulation by regulating nodulation signalling pathways or mediating secreting of flavonoid and nitrate (affecting legume rhizobia infection; Combier et al., 2006; Han et al., 2020; Laloum et al., 2014; Lorite et al., 2018; Tsikou et al., 2018). A recent study showed that overexpression of miR169c inhibited nodulation via targeting 3′‐UTR of GmNFYA‐C, while it promoted nodulation when miR169c lost its function (Xu et al., 2021c). In the rhizosphere of prospective host legumes, rhizobia have a close cooperative or competitive relationship with soil microorganisms (Han et al., 2020; Lorite et al., 2018). For instance, exogenous rhizobia can increase the relative abundance of potentially beneficial microorganisms, thus altering the microbial community structure and composition (White et al., 2015; Xu et al., 2020; Zgadzaj et al., 2016; Zhong et al., 2019). Moreover, Bacillus cereus group specifically promotes and suppresses the growth and colonization of Sinorhizobia and Bradyrhizobia, respectively (Han et al., 2020). Therefore, legumes genes can also influence the establishment and modification of the rhizomicrobiome community by mediating rhizobial colonization and nodulation.
Overall, these studies suggest that nutrient‐related genes can directly or indirectly influence the structure of rhizosphere microbial communities by altering root structure morphology, influencing plant nutrient use efficiency, regulating nodule colonization, and thus altering the rhizosphere microenvironment. These mechanisms can provide information for molecular breeding strategies to improve nutrient utilization and thus productivity in crops.
Disease resistance genes affect rhizosphere microbes
Plant growth in variable environment is threatened by various biotic stresses such as pathogen, and gradually domesticate the corresponding resistance mechanisms (Bakker et al., 2018; Chen et al., 2020; Li et al., 2021; Liu et al., 2020, 2021; Song et al., 2021). When plants are invaded by pathogens, disease resistance genes will be activated, which in turn trigger plant‐specific molecular immune recognition systems (Teixeira et al., 2019). Previous studies have shown that the cell membrane receptor protein kinase FERONIA (FER) can regulate microbe‐associated molecular patterns (MAMP)‐induced reactive oxygen species (ROS) burst and basal ROS levels in roots through the small G protein (ROP2), which is a positive regulator of plasma membrane NADPH oxidase (Duan et al., 2010; Stegmann et al., 2017). After genetic analysis of different gene mutants, researchers found that ROP2‐mediated basal level ROS regulation was essential for growth regulation of Pseudomonas interrogans (Bergonci et al., 2014; Haruta et al., 2014; Li et al., 2016; Wang et al., 2020; Zhu et al., 2020). Researchers found that the fer‐8 mutant reduced basal levels of ROS in the root system after pathogen invasion and lacked NADPH oxidase mutants showed elevated rhizosphere Pseudomonas (Song et al., 2021), suggesting that plants may mediate the plant immune system through the RALF‐FER signalling pathway or affect the release of specific secretions that increase Pseudomonas populations to resist pathogen invasion (Figure 1b; Berendsen et al., 2018; Liu et al., 2017; Rudrappa et al., 2008). As the most representative plant secondary metabolites, coumarins, benzoxazinoids and triterpenes play a pivotal role in improving plant disease resistance (Koprivova and Kopriva, 2022). Recently, it was found that two Multidrug and Toxic Compound Extrusion (MATE) transporter proteins (CmMATE1 and ClMATE1) involved in the transport of their respective cucurbitacins (a triterpenoid unique to Cucurbitaceae). They further showed that the transport of cucurbitacin B from melon roots into the soil regulates the rhizosphere microbiome by selectively enriching two bacterial genera, Enterobacter and Bacillus, leading to strong resistance to the soil‐borne wilt fungus Fusarium oxysporum (Zhong et al., 2022; Zhou et al., 2016). Together, these studies demonstrate that plants' disease resistance genes can recruit beneficial microorganisms or alter microbial community structure by activating the plant immune system or regulating the synthesis of several key metabolite in plants. These research efforts pave the way for the use of the rhizosphere microbiome to improve resistance to soil‐borne diseases.
Abiotic stress resistance genes affect rhizosphere microbes
In addition to biotic stresses, plant‐microbial symbiotic organisms are also subjected to many abiotic stresses (Zhang et al., 2020; Zhu, 2016), such as nutrient deficiency (i.e., nitrogen, iron and phosphorus) and high heavy metal (e.g., aluminium, cadmium and lead) stresses (Castrillo et al., 2017; Fang et al., 2020; Finkel et al., 2019; Harbort et al., 2020; López‐Arredondo et al., 2014; Ma, 2007; von Uexküll and Mutert, 1995). The Arabidopsis root‐specific R2R3‐type MYB transcription factor MYB72 has become an important component of the induced systemic resistance (ISR) episode (Van der Ent et al., 2008). In addition to its role in ISR, MYB72 is also induced in Arabidopsis roots under growth conditions of iron limitation and distorted iron uptake (Buckhout et al., 2009; Colangelo and Guerinot, 2004; van de Mortel et al., 2008). It was strongly demonstrated that the transcription factor MYB72 and MYB72‐controlled β‐glucosidase BGLU42 act key roles in regulating the beneficial rhizobacteria‐ISR and iron‐uptake responses, by regulating coumarin exudation to inhibit soil‐borne fungal pathogens and promote the growth of growth‐promoting and ISR‐inducing rhizobacteria (Stringlis et al., 2018). Rhizomicrobiome therefore have become a ‘secret weapon’ for plants to seize scarce soil iron resources, providing new ideas to regulate soil iron mobilization and activation, and promoting the widespread application of the plant functional gene‐rhizosphere microorganism model in crop resistance to abiotic stresses (Stringlis et al., 2018).
The plant MATE family transports a wide range of substrates such as organic acids, phytohormones and secondary metabolites (Magalhaes et al., 2007; Seo et al., 2012). The functions of many MATE transporter proteins have been illustrated in plants (Takanashi et al., 2014), including the transport of secondary metabolites such as alkaloids (Shoji et al., 2009), disease resistance regulation (Nawrath et al., 2002; Sun et al., 2011), iron translocation (Durrett et al., 2007; Yokosho et al., 2009) and Al detoxification (Wu et al., 2014). MATE transporter proteins are also present and involved in Aluminium (Al) resistance and tolerance in crops such as rice, maize, soybean and sorghum (Liu et al., 2016; Maron et al., 2013; Yokosho et al., 2011). When soybean was exposed to high Al stress, the expression of GmMATE58 and GmMATE1 genes increased, promoting the secretion of substances such as malic acid, oxalic acid and phenolic compounds (Chen and Liao, 2016; Li et al., 2018; Liu et al., 2016; Zhou et al., 2018), which can recruit beneficial microorganisms to enhance soybean to resist Al toxicity. In particular, the recruited microorganisms, such as Burkholderia, can improve the solubility of phosphorus in the soil and undergo denitrification, thus improving soybean tolerance to Al toxicity (Lian et al., 2019). Noteworthy, not only the normal or overexpression of genes but also the loss of plant functional gene can affect the host resistance to various stresses. Our recent research found that the rice could influence rhizosphere microorganisms by changing plant metabolites, such as salicin, arbutin, glycolic acid phosphate, after loss of the function of sst (seedling salt tolerant) gene and then assist host to resist salt stress (Lian et al., 2020). These studies illustrated that, like nutrient‐related genes, abiotic stress resistance genes can regulate rhizobia by inducing systemic stress resistance and regulating specific metabolites.
The expression of host‐specific genes also has a regulatory effect on soil enzyme activities (Figure 1c; Chen et al., 2011; Fließbach et al., 2012). This mainly attributes to that soil enzymes are mainly derived from exudates of plant root (Guan et al., 1986) and metabolites of microorganisms (Zimmermann and Frey, 2002). The amount and functions of microorganisms were closely related to the activities of soil enzymes (Durán et al., 2018; Velmourougane and Blaise, 2017), including urease, sucrase and cellulase. The transgenic AFPCHI disease‐resistant sugar beet was found to have increased urease, dehydrogenase, protease, catalase, pronase, acid and alkaline phosphatase activities through greenhouse trials (Bezirganoglu and Uysal, 2017). However, it has been reported that tobacco planted with trans‐antimicrobial protein gene and trans‐null plasmid gene had some inhibitory effects on peroxidase and urease activities in purple soil at specific periods (Wang et al., 2013). It remains to be further verified that plant disease resistance genes may affect the assembly of rhizosphere microorganisms by regulating the activity of soil enzymes.
Mechanisms of genes regulating the rhizosphere microbes
According to the description above, plant functional genes affect rhizomicrobiome mainly by regulating root structure morphology, plant nutrient use efficiency, rhizobial colonization, secondary metabolites and hormones, and activating the plant immune system, which in turn affect the rhizosphere microenvironment or directly signal to microorganisms (Egamberdieva et al., 2017; Eichmann et al., 2021; López‐Ráez et al., 2017). However, these mechanisms often act interactively in plants. For example, phytohormones can influence root structural morphology, plant‐dependent defence processes and root exudate secretion (Eichmann et al., 2021; Fu et al., 2021). Growth hormone regulates Arabidopsis root development mainly through the growth hormone synthesis pathway and the polar transport carrier pathway. Deletion of the growth hormone synthesis genes rty (rooty) and sur (super root) can lead to excessive endogenous IAA synthesis, resulting in a high number of lateral roots (Boerjan et al., 1995). Ethylene promotes Arabidopsis root hair growth by regulating the activity of EIN3/EIL1 and RHD6/RSL1 transcriptional complexes (Feng et al., 2017). Moreover, plant immune system can be divided into two layers, and hormonal signals are essential for both layers (Aerts et al., 2021). In the first layer, plants are damaged, recognize microorganisms/pathogens and release small molecule damage‐associated molecular patterns that trigger immune signals leading to pattern‐triggered immunity (PTI; Dangl et al., 2013; Erb and Reymond, 2019). Gene expression in PTI immunity is almost always influenced by interactions between sectors (Hillmer et al., 2017). In the second layer, pathogens secrete variable effectors that hinder PTI by inhibiting defence hormones, and resistant plants recognize the effectors, triggering effector immunity (ETI; Han and Kahmann, 2019). In ETI, all divisions can (partially) take over the response if one of them is inactive (Tsuda et al., 2009). However, in the actual defence process, there is a complex crosstalk between molecular pathways of different hormones and this crosstalk is critical and complex for adjusting plant growth and development and thus affecting microorganisms (Aerts et al., 2021). Furthermore, hormones can mediate the secretion of root exudates. It has been shown that disruption of ET signalling pathways leads to differences in the composition of root exudates, including smaller amounts of esculetin, gallic acid, L‐fucose, eicosapentaenoic acid, and higher amounts of β‐aldehyde, and that these root exudate metabolites can affect the assembly and function of bacterial taxa (Fu et al., 2021).
Effects of rhizosphere microbes on plant functional genes
Rhizosphere microorganisms can decompose soil organic matter into inorganic nutrients for plants and their physiological metabolic activities can also improve soil quality (Bhatti et al., 2017; Li et al., 2022; Li and Gong, 2021; Mishra et al., 2017; Wang et al., 2018). Rhizosphere microorganisms increase nutrient availability to plants, promote plant development and achieve the microbial ecological service function in farmland ecosystems. However, obtaining genetic varieties with high nutrient utilization and cross‐stress resistance is the fundamental way to improve the yield and quality of farmland crops (Ali et al., 2018; Anwar and Kim, 2020). Therefore, uncovering the mechanisms of ‘bottom‐up’ regulation of plant functional genes by rhizosphere microbes is of great importance for agricultural production. In this section, the microbial effects on the expression of functional genes related to plant growth promotion, flowering, immune regulation and stress tolerance were reviewed.
Effects of rhizosphere microbes on plant growth and development genes
For decades, researches on beneficial plant‐microbe interactions have formed a strong molecular framework (Lugtenberg and Kamilova, 2009; Saleem et al., 2019). Microbes can up‐ or down‐regulate the genes related to plant nutrient absorption by immobilizing nitrogen and to secondary metabolites, thus promoting or inhibiting plant growth (Figure 2). The main microorganisms associated with nitrogen fixation in the rhizosphere of specific plants include Klebsiella, Paenibacillus and Azospirillum (Grady et al., 2016; Mehnaz et al., 2007; Ryu et al., 2020). Wang et al. (2021) pointed out that specific relationships exist between given host genetics and associated microbes. This is further supported by the results from synthetic communities (SynComs) application that can systemically regulate the transcription of genes involved in multiple facets of growth and nutrient metabolism, especially auxin responses and nutrient signalling pathways. The expression of nitrate transporter genes and nitrate reductase genes were down‐regulated in plants treated with SynComs, which improved the biological fixation of N2, thereby inhibiting the direct absorption of nitrogen by roots and related metabolic pathways (Wang et al., 2021). Many phosphorus starvation (PSR) genes, phosphate transport and metabolism genes are also activated by SynComs (Wang et al., 2021; Wu et al., 2013). This indicates that microorganisms not only contribute to phosphorus release from insoluble forms but also activate the PSR signalling system, thereby enhancing the absorption of environmental phosphorus and promoting inter‐tissue phosphorus recycling (Desbrosses and Stougaard, 2011; Li et al., 2019; Wu et al., 2013; Zhong et al., 2019). Additionally, GO analysis showed that auxin responsive genes were abundant among differentially expressed genes affected by SynComs application (Hinsinger et al., 2011; Wang et al., 2021). Recruited rhizosphere microbes can produce phytohormones to regulate plant flowering signalling pathways (Rodriguez et al., 2019). A current focus is to decipher the link between rhizosphere microorganisms and plant functional genes on flowering time (Figure 2). For one thing, microorganisms can produce IAA from tryptophan (Trp; Duca et al., 2014; Molina et al., 2018; Patten et al., 2013; Treesubsuntorn et al., 2018). One of the abundant rhizosphere microorganisms, Arthrobacter, has been reported to have the ability to produce IAA, which is beneficial for plant growth (Li et al., 2018). IAA was the direct driver that down‐regulated the expression of genes involved in flowering, which delays the flowering time (Lu et al., 2018; Mai et al., 2011). It was also found that the selectively enriched rhizosphere microorganism regulated the flowering time by affecting the available nitrogen content in autoclaved potting‐mix soils (Ishioka et al., 1991; Panke‐Buisse et al., 2015). Notably, microbes also synthesize and emit many volatile organic compounds (VOCs) to perturb host flowering time (Hung et al., 2013; Sánchez‐López et al., 2016b). VOCs are low molecular weight (<300 Dalton) molecules that are easily dispersed by air and water due to high vapour pressure and low boiling point (Bitas et al., 2013; Schmidt et al., 2016; Schulz and Dickschat, 2007). The appearance of floral buds in VOCs treated ahk2/4 and ahk3/4 plants occurred 3–4 days before non‐treated Arabidopsis. In contrast, VOCs did not exert any significant effect on the time of floral bud appearance in both ahk2/3 and 35S:CKX1 plants (ahk2/4, ahk3/4 and ahk2/3 are CK signalling mutants; 35S:CKX1 plants are CK oxidase/dehydrogenase1 over‐expressing plants). These findings provide strong evidence that VOCs‐promoted early flowering involves suppression of NO action through the scavenging of NO molecules by CKs (Riefler et al., 2006; Sánchez‐López et al., 2016a; Werner et al., 2008). Together, microbes synthesize a multitude of nutrients and hormones, that affect the expression of plant flowering‐related genes, and may directly or indirectly regulate plant flowering time (De‐la‐Peña and Loyola‐Vargas, 2014).
Figure 2.
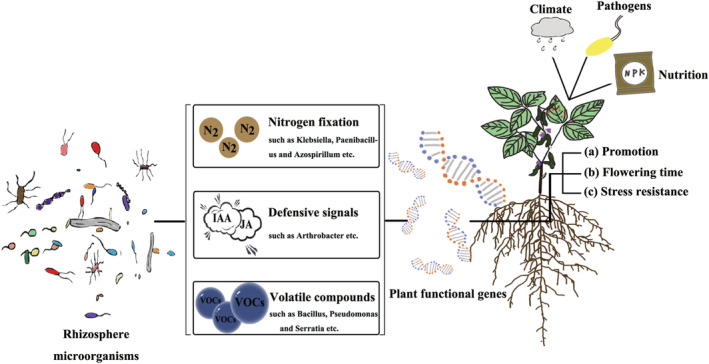
Rhizosphere microbes affect the plant functional genes related to growth and development, flowering time and stress resistance. (a) Microbes improved the biological fixation of N2. Microorganisms alter the content of various material components of the nitrogen cycle by stimulating the nitrogen cycle genes in the host, which in turn compensates for the material elements that the host is deprived of. In this way, microorganisms promote the nutrient uptake of the host and enable it to thrive. In addition to perceiving root secretions from plants, microbes themselves are capable of secondary metabolism to secrete specific substances, such as volatile compounds and hormones. Among them, volatile compounds have been shown to be direct drivers of plant growth hormone synthesis genes and photosynthesis genes. With the addition of plant growth hormone and photosynthesis, the growth of plants is affected. (b) Rhizosphere microorganisms directly or indirectly regulate the flowering time of plants. On the one hand, microbes secrete IAA to directly regulate flowering genes to influence flowering time; on the other hand, microbes indirectly influence flowering time by constraining plant nutrient requirements and secreting volatile compounds. (c) Under biotic and abiotic stresses, microorganisms influence the expression of defence genes (e.g., climate, pathogens, and nutrient deprivation), which in turn activate defence signalling pathways.
VOCs released from rhizomicrobiome can also improve multiple functions in ecosystems, such as plant growth and development (Gutiérrez‐Luna et al., 2010; Kanchiswamy et al., 2015; Ortíz‐Castro et al., 2009; Schulz‐Bohm et al., 2018). A variety of bacteria or fungi have been identified to produce VOCs, such as Bacillus, Pseudomonas and Serratia spp. (Hassani et al., 2018; Plyuta et al., 2016; Xie et al., 2020). Sun et al. (2020) found that treatment with F.luteovirens VOCs reduces primary root growth by aggravating auxin accumulation through the repression of the abundance of auxin efflux carrier PIN‐FORMED 2 (PIN2) protein, whereas it increases the lateral root number of A. thaliana seedlings. In addition to modulating root system architecture, treatment with F. luteovirens VOCs markedly increased aboveground growth. The transcriptomic and metabolomic analyses further supported the idea that F. luteovirens VOCs regulate plant growth and development by inducing up‐ or down‐regulation of genes related to carbon/nitrogen metabolism and antioxidant defence (Sun et al., 2020). Overall, these findings suggest that rhizosphere microbes can regulate plant growth‐related gene expression by mobilizing nutrients, altering plant nutrient use efficiency and producing hormones such as IAA and volatile compounds.
Effects of rhizosphere microorganisms on plant resistance genes
As sessile organisms, plants have to cope with various biotic and abiotic stress for long‐term domestication (Zhu, 2016). In this process, corresponding resistance genes and mechanisms have evolved continuously to resist adverse environmental conditions (Chong et al., 2019; Frantzeskakis et al., 2020). Recently, a paradigm shift in the life sciences has emerged, in which microbial communities are viewed as core drivers of tolerance mechanisms (Cordovez et al., 2019). Beneficial microbes were recruited to build defence signalling pathways (Figure 2), which are based on the interaction of plants, pathogenic bacteria and rhizomicrobiome in response to biotic stress, that is likely a survival strategy conserved across the plant kingdom (Liu et al., 2019; Liu and Brettell, 2019). Durum wheat infected by the fungal pathogen Fusarium pseudograminearum (Fp) leads to an enrichment of the beneficial bacterium Stenotrophomonas rhizophila (SR80) in the rhizosphere. As an early warning factor, the assembled SR80 was able to promote the large‐scale expression of pathogenesis‐related (PR) genes involved in the salicylic acid and jasmonic acid signalling pathways, enhancing host immunity against crown rot disease (Liu et al., 2016, 2021). Another case is that VOCs produced by Fusarium culmorum stimulated the production of flavonoid sodorifen VOCs of bacterium Serratia plymuthica, which induced the expression of associated defence genes in Arabidopsis (Raza et al., 2021; Schmidt et al., 2017).
Under abiotic stresses, the effect of rhizomicrobiome on plant genes might be more comprehensible. As for low phosphorus availability, the phosphorous starvation response induced by microbial invasion can promote the expression of master transcriptional regulator PhR1 to alter orthophosphate (Pi) metabolism in plants. PHR1 directly activates microbiome‐enhanced response to phosphate limitation while repressing microbially driven plant immune system. It suggests that microbes have changed the transcription level of defence genes to enhance the immunity of plants (Castrillo et al., 2017). Furthermore, rhizomicrobiome may change more defensive pathways to mitigate abiotic stress. Inoculation of Arbuscular Mycorrhizal Fungi (AMF) under drought stress condition has been found to be able to induce the expression of 1‐pyrrolin‐5‐carboxylic acid synthase (P5CS) gene. P5CS is a key enzyme involved in the proline synthesis, which can promote cell water retention, thus improving the ability of plants to resist osmotic stress (Hu et al., 1992; Ruiz‐Lozano et al., 2006). At the same time, the AMF also enhance plant resistance to drought stress by regulating 9‐cis‐epoxycarotenoid dioxygenase (NCED) gene expression. NCED is an important enzyme in controlling abscisic acid (ABA) metabolism which catalyses the oxidative cleavage of epoxy carotenoids into xanthoxins (Chauffour et al., 2019; Taylor et al., 2000). As a whole, the evidence above suggests that microbes can regulate the expression of plant genes through multiple pathways, such as secreting hormones, producing VOCs, enhancing the plant immune system, and building defence signalling pathways, to help the host adapt to various stresses. These studies shed light on the important role of plant‐associated rhizosphere microbiota for plant functions and broaden multiple ideas for manipulating host growth through microbial intervention.
Research methods of linking plant functional genes to rhizosphere microorganisms
The link of plant functional gene to rhizomicrobiome is complicated. Not only the ‘functional genes‐downstream, genes‐metabolite or other signalling substances‐microbe’ pathway need to be considered but also factors such as inter‐microbe, environments and plant residues (Chen et al., 2019; Edwards et al., 2018; Gao, Han, et al., 2019; Gao, Karlsson, et al., 2019; Geddes et al., 2019; Pascale et al., 2020; Trivedi et al., 2021; Trubl et al., 2018). These signalling substances, such as hormones and small RNA, are frequently exchanged between the host and the microorganism, triggering structural and functional changes on both sides (Huang et al., 2019; Middleton et al., 2021; Yang et al., 2016). The desired substantive function of microorganisms leads to healthy plant growth. The new findings suggest that the ultimate outcome of host health may depend on not only the exchange of substance between host and microbes but also the signalling and metabolic interactions among microbiome members (Durán et al., 2018; Finkel et al., 2019; Harbort et al., 2020; Xu et al., 2021b). Thus, understanding plant gene‐microbe interactions may require examining these relationships at the level of host and microbial functional capacity, activity, and molecular exchange (Xu et al., 2021b). However, a larger reason for the slow progress in our understanding of the functionality of plant gene‐microbe interactions may lie on our choice of methods and tools. How to integratively combine host‐centric molecular technologies, such as CRISPR/Cas system (e.g., Cas9 and Cas12a), gene silencing (e.g., RNA interference) and gene overexpression, with microbial‐centric histological sequencing technologies, such as amplicon sequencing, shotgun metagenomics, metatranscriptomics and metabolomics, are important to unlock complex mechanisms of plant gene‐microbe interactions (Figure 3; Fitzpatrick et al., 2020).
Figure 3.
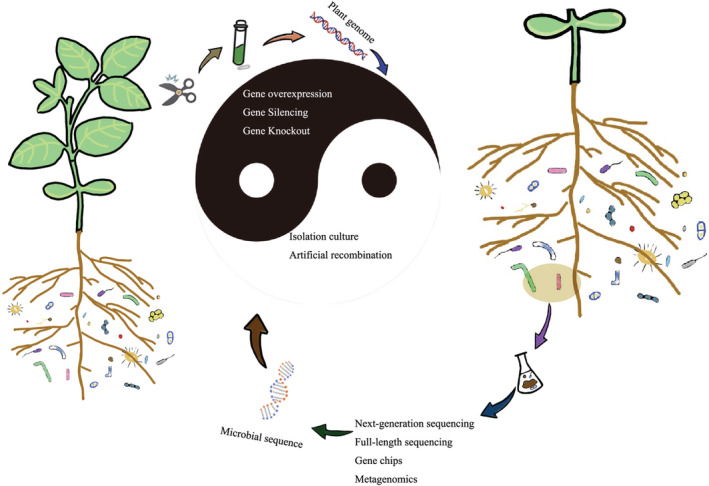
Connecting plant and microbes relegated technologies. Plant genomic data obtained from plant tissues and microbial sequence data obtained by various microbial sequencing techniques were used for plant functional gene and rhizosphere microbial function probes. The integration of plant gene‐centred and rhizosphere microbial function‐centred technologies is the key to promote the study of plant gene and rhizosphere microbial interactions.
Genome‐Wide Association Studies (GWAS) well attach phytomics to microbiomics and demonstrate that host genomics does influence the composition of the microbiome (Trivedi et al., 2021). GWAS were previously used to study the phyllosphere microbiome with quantitative methods to map microbiomes as phenotypes (Horton et al., 2014; Oyserman et al., 2022; Roman‐Reyna et al., 2019; Wallace et al., 2018) and are increasingly focusing on the rhizosphere microbiomes of plants such as Arabidopsis, sorghum, barley, maize and tomato (Bergelson et al., 2019; Escudero‐Martinez et al., 2022; Oyserman et al., 2018, 2022; Wagner et al., 2020). Compared to phyllosphere, the rhizosphere has proven to be the most promising part for unravelling the genetic power of the host microbiome (Deng et al., 2021). This may be owing to the high complexity of the rhizosphere microbiome and its strong colonization ability (Bano et al., 2021; Rico et al., 2014; Schlechter et al., 2019). GWAS break down the wall of the tripartite relationship between plant genotype phenotype‐microbiome, thus identifying the link between plant phenotype and microbiome function (Horton et al., 2014; Vorholt et al., 2017). Therefore, the abundance and community structure of the rhizosphere microbiome can be used to infer the relevant genetic loci and plant genes (Deng et al., 2021; Escudero‐Martinez et al., 2022). The inferred plant genes can be validated by artificially modified mutants to elucidate the potential host genetic causes of microbiome changes (Schäfer et al., 2022; Wagner et al., 2020). Even if candidate genes for microbial recruitment are identified, there are difficulties in reproducing and validating them (Zancarini et al., 2021). Conversely, host genotype data can also predict the composition of microorganisms, which determines whether there is inter‐ and intra‐species variation in the microbiome in the host being tested (Deng et al., 2021; Fitzpatrick et al., 2018; Walters et al., 2018). GWAS of plant microbiome associations promote a comprehensive understanding of the host molecular mechanisms of microbiome assembly and lay the foundation for microbiome characterization to be implemented into breeding programs. Notably, heritable rhizosphere microbes showed strong overlap in different host genotypes (Deng et al., 2021). This fraction may be the few pivotal microorganisms that have stabilized on the species domestication and temporal evolutionary scales and subsequently regulate the proliferation of other members of the community (Brachi et al., 2022). Furthermore, since environmental conditions are a major component of variability and plant genotypes can explain only a few microbial variations, GWAS need to develop more comprehensive models to reveal the effects of genotype, microbiome, environment and their interactions on plant phenotypes (Zancarini et al., 2021).
For the microbial sequencing, the widely used technology is still second‐generation sequencing, which has undoubtedly enabled researchers to gain a broad understanding of the structure of microbial community (Metzker, 2010; Niedringhaus et al., 2011). Increasingly, studies are also focusing on the activity and function of microbes by incorporating metatranscriptomics and macrogenomics, gene chips and viral omics, with the potential for more advanced approaches to be developed later (McDonald et al., 2022; Wang et al., 2019; Xu et al., 2021a; Zaramela et al., 2021). Each of the technology mentioned above has its own advantages and disadvantages (Table 1), and researchers can choose the appropriate method according to the needs as well as the funding budget. It is worth noting that the gene impact on rhizomicrobiome is often accomplished through metabolism or small molecule signalling substances. Therefore, metabolomics and the detection of the transfer of small molecule signalling substances (i.e., miRNAs) becomes particularly important in this reciprocal process (Middleton et al., 2021; Pang et al., 2021). Therefore, data integration approaches are essential to unravel the relationship between plant functional gene and microbes. Pang et al. (2021) reviewed the statistical approaches developed for integration of plant metabolites and microbiome data, and Zancarini et al. (2021) reviewed statistical approaches for integrating plant omics with microbiome data. It is certain that rapidly developing multi‐omics combinatorial analyses may further elucidate mechanisms of interaction between genes and microbiomes.
Table 1.
Commonly used technologies on plant functional gene and microbiome sequencing technologies
| Techniques | Advantages | Disadvantages | References |
|---|---|---|---|
| Gene overexpression |
Amplify gene function Commonly used for resistance to stress |
Lethal effects | Dalman et al. (2017), Huo et al. (2021), Prabhu et al. (2017), Shalmani et al. (2021), Truong et al. (2021), Yang et al. (2017) |
| Gene knockout |
High knockdown efficiency simple operation multiple targets can be edited simultaneously |
Severe off‐target property Prone to mutations |
Ding et al. (2013), Gao et al. (2021), Irie et al. (2015) |
| Gene silencing |
TGS:Genetic stability PTGS:High specificity Reproducibility easily manipulated |
TGS:Mutants may have defects PTGS:positional effects transient incomplete gene knockouts |
Ashfaq et al. (2020), Huang et al. (2021), Matzke and Mosher (2014), Sigman and Slotkin (2016), Tan et al. (2020) |
| Next‐generation sequencing |
High throughput low cost high efficiency |
Low resolution difficult to detect low‐abundance microorganisms at the genus level PCR preference |
Metzker (2010), Niedringhaus et al. (2011), Shendure and Ji (2008) |
| Full‐length sequencing |
No PCR amplification required with ultra‐long sequencing read length high precision no GC preference |
High cost high error rate (15%–40%) low throughput |
Gui‐Feng and Hai‐Yan (2020), Singer et al. (2016), Xu et al. (2019) |
| Gene chips |
High density Rapid real‐time detection automation |
Too much information on hybridization slow processing and analysis of hybridization data specificity and sensitivity of hybridization need to be improved preparation of high‐quality nucleic acid samples needs to be improved |
Gabig and Wegrzyn (2001), Yang and Chen (2000) |
| Metagenomic |
Allows access to large numbers of assembled genomes (MAGs) improves resolution of microbial community analysis Allows access to more diverse compositional and functional data |
Expensive and host genes are vulnerable to contamination | Liu et al. (2021), New and Brito (2020) |
A large amount of data is available based on the above methods to support the linkage between plant genes and microbial structure and function, but the validations of microbial functions are still lacking. Recently, artificial recombination has been increasingly used to validate microbial functions (Durán et al., 2018; Zhuang et al., 2021). We believe that the rapid development of technologies such as the high‐throughput partitioning methods developed by Zhang et al. (2021) and computer‐guided synthesis of artificial colonies could assist to explore the plant gene and microbe interaction. Specifically, there was a significant difference in root bacterial community composition between root diffusion barrier genotypes of Arabidopsis and wild type after inoculation with artificial bacterial communities (Salas‐González et al., 2021). These results suggest that the endothelial diffusion component of the Arabidopsis root system regulates the conformation of the microbial community (Zhou et al., 2020). Furthermore, the synthetic population repressed the transcriptional response to ABA (cluster C2) by comparing differentially expressed genes in wild‐type plants and mutant myb36‐2 roots, leading to the speculation that the microbiome regulates suberization and lignification through ABA‐dependent pathways (Barberon et al., 2016; Salas‐González et al., 2021). It is clear that synthetic biology is establishing transboundary links, as well as establishing molecular links between plant nutrition and defence (Berendsen et al., 2018; Castrillo et al., 2017; Liu et al., 2020; Zhou et al., 2012). However, recent developments in synthetic communities have ignored fundamental issues in microbiome studies, namely standardization and reproducibility (Zengler et al., 2019). Little standardization in the culture systems is used to study complex microbial communities. Researchers typically use single strains or simple co‐cultures, which are often poor models for complex and metabolically diverse microbial communities in nature (Ruby, 2008). While these methods can provide a better understanding of the native microbiome, they lack reproducibility (even within a single laboratory) in the absence of microbial inoculate that are stable over time.
The care needs to be taken when using metagenomes to detect endophytes in leaves, stems and roots, as cross‐contamination of host genes and microbial genes can occur (Liu et al., 2021). To solve this problem, the host genome needs to be identified to remove the host gene contamination in subsequent analyses (Marotz et al., 2018; Song and Xie, 2020). This has certainly limited much of the research from being carried out in some crops. In addition, choosing the appropriate time to sample is a challenge as there is often little or no a priori knowledge of when host and microbiome responses occur and how their interactions change as the plant grows (Xiong et al., 2021). As some of the techniques involved above are expensive, we therefore recommend that researchers select multiple sampling time points and use suitable techniques to explore the dynamic processes of plant gene and microbial interactions in focal periods using a multi‐omics approach. In addition, as soil type strongly influences microorganisms (Bai et al., 2017; Girvan et al., 2003; Pershina et al., 2018), we suggest that experiments should be verified in different soil types, so as to find general patterns of gene regulation of microorganisms.
Summary and outlook
Microbial communities with manipulated plant functional gene expression have great potential for bioengineering, agricultural and environmental remediation. There is clear evidence that functional plant genes and rhizomicrobiome can interact with each other. However, more fundamental studies are needed to decrypt the ‘on or off’ of functional genes in plant‐microbe communication. There is no shortage of emerging technologies and methodological approaches that can be used to further explore the molecular mechanisms and signalling pathways of microbe‐host gene interactions. However, it is still a long way to construct a complete network of plant functional genes and rhizosphere microbes, with a plethora of outstanding questions: (i) How to find plant genes that can regulate rhizomicrobiome on a large scale? GWAS may be a very good approach, considering that GWAS could be used to identify specific gene‐regulated microorganisms; (ii) To what extent and how long can plant genes play a role in shaping the rhizospheric microbiota and its associated functions? Conversely, microorganisms to specific genes? (iii) Are there other signals, such as miRNA, for the plant and microbe interaction besides metabolites? This knowledge will enable us to reshape the microbiome through genetic engineering, or to regulate the functional genes of plants through microbes, ultimately optimizing plant growth.
Conflict of interest
The authors declare that they have no competing interests.
Author contributions
Tengxiang Lian, Jian Jin and Hai Nian designed the content and structure of the review. Qi Liu, Tengxiang Lian, Lang Cheng wrote the main manuscript. Qi Liu, Lang Cheng prepared Figures 1, 2, 3 and Table 1. The authors read and approved the final version of the manuscript.
Acknowledgments
The authors gratefully acknowledge all members for helpful conversations on this topic. The authors also acknowledge support from the National Natural Science Foundation of China (Grant No. 32170115), Double First‐class Discipline Promotion Project (Grant No. 2021B10564001), and Guangzhou Science and Technology Innovation Development Funding (Grant No. 202102020068).
Contributor Information
Jian Jin, Email: jinjian29@hotmail.com.
Tengxiang Lian, Email: liantx@scau.edu.cn.
References
- Aerts, N. , Pereira Mendes, M. and Van Wees, S.C.M. (2021) Multiple levels of crosstalk in hormone networks regulating plant defense. Plant J. 105, 489–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali, J. , Jewel, Z.A. , Mahender, A. , Anandan, A. , Hernandez, J. and Li, Z. (2018) Molecular genetics and breeding for nutrient use efficiency in rice. Int. J. Mol. Sci. 19, 1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anwar, A. and Kim, J.K. (2020) Transgenic breeding approaches for improving abiotic stress tolerance: recent progress and future perspectives. Int. J. Mol. Sci. 21, 2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashfaq, M.A. , Kumar, V.D. , Reddy, P.S.S. , Kumar, C.H.A. , Kumar, K.S. , Rao, N.N. , Tarakeswari, M. et al. (2020) Post‐transcriptional gene silencing: basic concepts and applications. J. Biosci. 45, 128. [PubMed] [Google Scholar]
- Badri, D.V. , Loyola‐Vargas, V.M. , Broeckling, C.D. , De‐la‐Peña, C. , Jasinski, M. , Santelia, D. , Martinoia, E. et al. (2008) Altered profile of secondary metabolites in the root exudates of Arabidopsis ATP‐binding cassette transporter mutants. Plant Physiol. 146, 762–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badri, D.V. , Quintana, N. , El Kassis, E.G. , Kim, H.K. , Choi, Y.H. , Sugiyama, A. , Verpoorte, R. et al. (2009) An ABC transporter mutation alters root exudation of phytochemicals that provoke an overhaul of natural soil microbiota. Plant Physiol. 151, 2006–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai, B. , Liu, W. , Qiu, X. , Zhang, J. , Zhang, J. and Bai, Y. (2022) The root microbiome: community assembly and its contributions to plant fitness. J. Integr. Plant Biol. 64, 230–243. [DOI] [PubMed] [Google Scholar]
- Bai, R. , Wang, J.T. , Deng, Y. , He, J.Z. , Feng, K. and Zhang, L.M. (2017) Microbial community and functional structure significantly varied among distinct types of paddy soils but responded differently along gradients of soil depth layers. Front. Microbiol. 8, 945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker, P.A.H.M. , Pieterse, C.M.J. , de Jonge, R. and Berendsen, R.L. (2018) The soil‐borne legacy. Cell, 172, 1178–1180. [DOI] [PubMed] [Google Scholar]
- Bano, S. , Wu, X. and Zhang, X. (2021) Towards sustainable agriculture: rhizosphere microbiome engineering. Appl. Microbiol. Biotechnol. 105, 7141–7160. [DOI] [PubMed] [Google Scholar]
- Barberon, M. , Vermeer, J.E. , De Bellis, D. , Wang, P. , Naseer, S. , Andersen, T.G. , Humbel, B.M. et al. (2016) Adaptation of root function by nutrient‐induced plasticity of endodermal differentiation. Cell, 164, 447–459. [DOI] [PubMed] [Google Scholar]
- Berendsen, R.L. , Vismans, G. , Yu, K. , Song, Y. , de Jonge, R. , Burgman, W.P. , Burmølle, M. et al. (2018) Disease‐induced assemblage of a plant‐beneficial bacterial consortium. ISME J. 12(6), 1496–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergelson, J. , Mittelstrass, J. and Horton, M.W. (2019) Characterizing both bacteria and fungi improves understanding of the Arabidopsis root microbiome. Sci. Rep. 9, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergonci, T. , Ribeiro, B. , Ceciliato, P.H. , Guerrero‐Abad, J.C. , Silva‐Filho, M.C. and Moura, D.S. (2014) Arabidopsis thaliana RALF1 opposes brassinosteroid effects on root cell elongation and lateral root formation. J. Exp. Bot. 65, 2219–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezirganoglu, I. and Uysal, P. (2017) Impact of transgenic AFPCHI (Cucumis melo L. Silver Light) fungal resistance melon on soil microbial communities and enzyme activities. J. Plant Biotechnol. 44, 156–163. [Google Scholar]
- Bhatti, A.A. , Haq, S. and Bhat, R.A. (2017) Actinomycetes benefaction role in soil and plant health. Microb. Pathog. 111, 458–467. [DOI] [PubMed] [Google Scholar]
- Bitas, V. , Kim, H.S. , Bennett, J.W. and Kang, S. (2013) Sniffing on microbes: diverse roles of microbial volatile organic compounds in plant health. Mol. Plant Microbe Interact. 26, 835–843. [DOI] [PubMed] [Google Scholar]
- Boerjan, W. , Cervera, M.T. , Delarue, M. , Beeckman, T. , Dewitte, W. , Bellini, C. , Caboche, M. et al. (1995) Superroot, a recessive mutation in Arabidopsis, confers auxin overproduction. Plant Cell, 7, 1405–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachi, B. , Filiault, D. , Whitehurst, H. , Darme, P. , Le Gars, P. , Le Mentec, M. , Morton, T.C. et al. (2022) Plant genetic effects on microbial hubs impact host fitness in repeated field trials. Proc. Natl Acad. Sci. USA, 119, e2201285119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunetti, P. , Zanella, L. , De Paolis, A. , Di Litta, D. , Cecchetti, V. , Falasca, G. , Barbieri, M. et al. (2015) Cadmium‐inducible expression of the ABC‐type transporter AtABCC3 increases phytochelatin‐mediated cadmium tolerance in Arabidopsis . J. Exp. Bot. 66, 3815–3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckhout, T.J. , Yang, T.J. and Schmidt, W. (2009) Early iron‐deficiency‐induced transcriptional changes in Arabidopsis roots as revealed by microarray analyses. BMC Genomics, 10, 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrión, V.J. , Perez‐Jaramillo, J. , Cordovez, V. , Tracanna, V. , de Hollander, M. , Ruiz‐Buck, D. , Mendes, L.W. et al. (2019) Pathogen‐induced activation of disease‐suppressive functions in the endophytic root microbiome. Science, 366, 606–612. [DOI] [PubMed] [Google Scholar]
- Castrillo, G. , Teixeira, P.J. , Paredes, S.H. , Law, T.F. , de Lorenzo, L. , Feltcher, M.E. , Finkel, O.M. et al. (2017) Root microbiota drive direct integration of phosphate stress and immunity. Nature, 543, 513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauffour, F. , Bailly, M. , Perreau, F. , Cueff, G. , Suzuki, H. , Collet, B. , Frey, A. et al. (2019) Multi‐omics analysis reveals sequential roles for ABA during seed maturation. Plant Physiol. 180, 1198–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. , Bonkowski, M. , Shen, Y. , Griffiths, B.S. , Jiang, Y. , Wang, X. and Sun, B. (2020) Root ethylene mediates rhizosphere microbial community reconstruction when chemically detecting cyanide produced by neighbouring plants. Microbiome, 8, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Z.H. , Chen, L.J. , Zhang, Y.L. and Wu, Z.J. (2011) Microbial properties, enzyme activities and the persistence of exogenous proteins in soil under consecutive cultivation of transgenic cottons (Gossypium hirsutum L.). Plant Soil Environ. 57, 67–74. [Google Scholar]
- Chen, Q. , Jiang, T. , Liu, Y.X. , Liu, H. , Zhao, T. , Liu, Z. , Gan, X. et al. (2019) Recently duplicated sesterterpene (C25) gene clusters in Arabidopsis thaliana modulate root microbiota. Sci. China Life Sci. 62, 947–958. [DOI] [PubMed] [Google Scholar]
- Chen, Z.C. and Liao, H. (2016) Organic acid anions: an effective defensive weapon for plants against aluminum toxicity and phosphorus deficiency in acidic soils. J. Genet. Genomics, 43, 631–638. [DOI] [PubMed] [Google Scholar]
- Chen, S. , Waghmode, T.R. , Sun, R. , Kuramae, E.E. , Hu, C. and Liu, B. (2019) Root‐associated microbiomes of wheat under the combined effect of plant development and nitrogen fertilization. Microbiome, 7, 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi, H. , Wang, X. , Shao, Y. , Qin, Y. , Deng, Z. , Wang, L. and Chen, S. (2018) Engineering and modification of microbial chassis for systems and synthetic biology. Synth. Syst. Biotechnol. 4, 25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong, G.L. , Foo, M.H. , Lin, W.D. , Wong, M.M. and Verslues, P.E. (2019) Highly ABA‐induced 1 (HAI1)‐Interacting protein HIN1 and drought acclimation‐enhanced splicing efficiency at intron retention sites. Proc. Natl Acad. Sci. USA, 116, 22376–22385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colangelo, E.P. and Guerinot, M.L. (2004) The essential basic helix‐loop‐helix protein FIT1 is required for the iron deficiency response. Plant Cell, 16(12), 3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combier, J.P. , Frugier, F. , de Billy, F. , Boualem, A. , El‐Yahyaoui, F. , Moreau, S. , Vernié, T. et al. (2006) MtHAP2‐1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula . Genes Dev. 20, 3084–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contesto, C. , Milesi, S. , Mantelin, S. , Zancarini, A. , Desbrosses, G. , Varoquaux, F. , Bellini, C. et al. (2010) The auxin‐signaling pathway is required for the lateral root response of Arabidopsis to the rhizobacterium Phyllobacterium brassicacearum . Planta, 232, 1455–1470. [DOI] [PubMed] [Google Scholar]
- Cordovez, V. , Dini‐Andreote, F. , Carrión, V.J. and Raaijmakers, J.M. (2019) Ecology and evolution of plant microbiomes. Annu. Rev. Microbiol. 73, 69–88. [DOI] [PubMed] [Google Scholar]
- Dalman, K. , Wind, J.J. , Nemesio‐Gorriz, M. , Hammerbacher, A. , Lundén, K. , Ezcurra, I. and Elfstrand, M. (2017) Overexpression of PaNAC03, a stress induced NAC gene family transcription factor in Norway spruce leads to reduced flavonol biosynthesis and aberrant embryo development. BMC Plant Biol. 17, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl, J.L. , Horvath, D.M. and Staskawicz, B.J. (2013) Pivoting the plant immune system from dissection to deployment. Science, 341, 746–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dastogeer, K.M.G. , Tumpa, F.H. , Sultana, A. , Akter, M.A. and Chakraborty, A. (2020) Plant microbiome—an account of the factors that shape community composition and diversity. Curr. Plant Biol. 23, 100161. [Google Scholar]
- De‐la‐Peña, C. and Loyola‐Vargas, V.M. (2014) Biotic interactions in the rhizosphere: a diverse cooperative enterprise for plant productivity. Plant Physiol. 166, 701–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, X. , Zhang, N. , Shen, Z. , Zhu, C. , Liu, H. , Xu, Z. , Li, R. et al. (2021) Soil microbiome manipulation triggers direct and possible indirect suppression against Ralstonia solanacearum and Fusarium oxysporum . NPJ Biofilms Microbiomes. 7, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depuydt, T. and Vandepoele, K. (2021) Multi‐omics network‐based functional annotation of unknown Arabidopsis genes. Plant J. 108, 1193–1212. [DOI] [PubMed] [Google Scholar]
- Desbrosses, G.J. and Stougaard, J. (2011) Root nodulation: a paradigm for how plant‐microbe symbiosis influences host developmental pathways. Cell Host Microbe. 10, 348–358. [DOI] [PubMed] [Google Scholar]
- Ding, W.T. , Zhang, G.C. and Liu, J.J. (2013) 3′ Truncation of the GPD1 promoter in Saccharomyces cerevisiae for improved ethanol yield and productivity. Appl. Environ. Microbiol. 79, 3273–3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditengou, F.A. , Béguiristain, T. and Lapeyrie, F. (2000) Root hair elongation is inhibited by hypaphorine, the indole alkaloid from the ectomycorrhizal fungus Pisolithus tinctorius, and restored by indole‐3‐acetic acid. Planta, 211, 722–728. [DOI] [PubMed] [Google Scholar]
- Duan, Q. , Kita, D. , Li, C. , Cheung, A.Y. and Wu, H.M. (2010) FERONIA receptor‐like kinase regulates RHO GTPase signaling of root hair development. Proc. Natl Acad. Sci. USA, 107, 17821–17826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duca, D. , Lorv, J. , Patten, C.L. , Rose, D. and Glick, B.R. (2014) Indole‐3‐acetic acid in plant‐microbe interactions. Antonie Van Leeuwenhoek, 106, 85–125. [DOI] [PubMed] [Google Scholar]
- Durán, P. , Thiergart, T. , Garrido‐Oter, R. , Agler, M. , Kemen, E. , Schulze‐Lefert, P. and Hacquard, S. (2018) Microbial interkingdom interactions in roots promote Arabidopsis survival. Cell, 175, 973–983.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrett, T.P. , Gassmann, W. and Rogers, E.E. (2007) The FRD3‐mediated efflux of citrate into the root vasculature is necessary for efficient iron translocation. Plant Physiol. 144, 197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, J.A. , Santos‐Medellín, C.M. , Liechty, Z.S. , Nguyen, B. , Lurie, E. , Eason, S. , Phillips, G. et al. (2018) Compositional shifts in root‐associated bacterial and archaeal microbiota track the plant life cycle in field‐grown rice. PLoS Biol. 16, e2003862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egamberdieva, D. , Wirth, S.J. , Alqarawi, A.A. , Abd Allah, E.F. and Hashem, A. (2017) Phytohormones and beneficial microbes: essential components for plants to balance stress and fitness. Front Microbiol. 8, 2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichmann, R. , Richards, L. and Schäfer, P. (2021) Hormones as go‐betweens in plant microbiome assembly. Plant J. 105, 518–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erb, M. and Reymond, P. (2019) Molecular interactions between plants and insect herbivores. Annu. Rev. Plant Biol. 70, 527–557. [DOI] [PubMed] [Google Scholar]
- Escudero‐Martinez, C. , Coulter, M. , Alegria Terrazas, R. , Foito, A. , Kapadia, R. , Pietrangelo, L. , Maver, M. et al. (2022) Identifying plant genes shaping microbiota composition in the barley rhizosphere. Nat. Commun. 13, 3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadiji, A.E. and Babalola, O.O. (2020) Metagenomics methods for the study of plant‐associated microbial communities: a review. J. Microbiol. Methods, 170, 105860. [DOI] [PubMed] [Google Scholar]
- Fang, Q. , Zhang, J. , Zhang, Y. , Fan, N. , van den Burg, H.A. and Huang, C.F. (2020) Regulation of aluminum resistance in Arabidopsis involves the SUMOylation of the zinc finger transcription factor STOP1. Plant Cell, 32, 3921–3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felten, J. , Kohler, A. , Morin, E. , Bhalerao, R.P. , Palme, K. , Martin, F. , Ditengou, F.A. et al. (2009) The ectomycorrhizal fungus Laccaria bicolor stimulates lateral root formation in poplar and Arabidopsis through auxin transport and signaling. Plant Physiol. 151, 1991–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, Y. , Xu, P. , Li, B. , Li, P. , Wen, X. , An, F. , Gong, Y. et al. (2017) Ethylene promotes root hair growth through coordinated EIN3/EIL1 and RHD6/RSL1 activity in Arabidopsis . Proc. Natl Acad. Sci. USA, 114, 13834–13839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel, O.M. , Salas‐González, I. , Castrillo, G. , Spaepen, S. , Law, T.F. , Teixeira, P.J.P.L. , Jones, C.D. et al. (2019) The effects of soil phosphorus content on plant microbiota are driven by the plant phosphate starvation response. PLoS Biol. 17, e3000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick, C.R. , Copeland, J. , Wang, P.W. , Guttman, D.S. , Kotanen, P.M. and Johnson, M.T.J. (2018) Assembly and ecological function of the root microbiome across angiosperm plant species. Proc. Natl Acad. Sci. USA, 115, E1157–E1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick, C.R. , Salas‐González, I. , Conway, J.M. , Finkel, O.M. , Gilbert, S. , Russ, D. , Teixeira, P.J.P.L. et al. (2020) The plant microbiome: from ecology to reductionism and beyond. Annu. Rev. Microbiol. 74, 81–100. [DOI] [PubMed] [Google Scholar]
- Fließbach, A. , Messmer, M. , Nietlispach, B. , Infante, V. and Mäder, P. (2012) Effects of conventionally bred and Bacillus thuringiensis (Bt) maize varieties on soil microbial biomass and activity. Biol. Fertil. Soils, 48, 315–324. [Google Scholar]
- Frantzeskakis, L. , Di Pietro, A. , Rep, M. , Schirawski, J. , Wu, C.H. and Panstruga, R. (2020) Rapid evolution in plant‐microbe interactions—a molecular genomics perspective. New Phytol. 225, 1134–1142. [DOI] [PubMed] [Google Scholar]
- Friesen, M.L. , Porter, S.S. , Stark, S.C. , von Wettberg, E.J. , Sachs, J.L. and Martinez‐Romero, E. (2011) Microbially mediated plant functional traits. Annu. Rev. Ecol. Evol. Syst. 42, 23–46. [Google Scholar]
- Fu, R. , Feng, H. , Dini‐Andreote, F. , Wang, Z. , Bo, C. , Cao, W. , Yang, K. et al. (2021) Modulation of the tomato rhizosphere microbiome via changes in root exudation mediated by the ethylene receptor NR. Microorganisms, 9, 2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabig, M. and Wegrzyn, G. (2001) An introduction to DNA chips: principles, technology, applications and analysis. Acta Biochim Pol. 48, 615–622. [PubMed] [Google Scholar]
- Gao, C. (2021) Genome engineering for crop improvement and future agriculture. Cell, 184, 1621–1635. [DOI] [PubMed] [Google Scholar]
- Gao, G.F. and Chu, H.Y. (2020) Techniques and methods of microbiomics and their applications. J. Plant Ecol. 44, 395–408. [Google Scholar]
- Gao, Z. , Han, M. , Hu, Y. , Li, Z. , Liu, C. , Wang, X. , Tian, Q. et al. (2019) Effects of continuous cropping of sweet potato on the fungal community structure in rhizospheric soil. Front. Microbiol. 10, 2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, Z. , Karlsson, I. , Geisen, S. , Kowalchuk, G. and Jousset, A. (2019) Protists: puppet masters of the rhizosphere microbiome. Trends Plant Sci. 24, 165–176. [DOI] [PubMed] [Google Scholar]
- Gao, J.P. , Xu, P. , Wang, M. , Zhang, X. , Yang, J. , Zhou, Y. , Murray, J.D. et al. (2021) Nod factor receptor complex phosphorylates GmGEF2 to stimulate ROP signaling during nodulation. Curr. Biol. 31, 3538–3550.e5. [DOI] [PubMed] [Google Scholar]
- Geddes, B.A. , Paramasivan, P. , Joffrin, A. , Thompson, A.L. , Christensen, K. , Jorrin, B. , Brett, P. et al. (2019) Engineering transkingdom signalling in plants to control gene expression in rhizosphere bacteria. Nat. Commun. 10, 3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girvan, M.S. , Bullimore, J. , Pretty, J.N. , Osborn, A.M. and Ball, A.S. (2003) Soil type is the primary determinant of the composition of the total and active bacterial communities in arable soils. Appl. Environ. Microbiol. 69(3), 1800–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh, C.H. , Veliz Vallejos, D.F. , Nicotra, A.B. and Mathesius, U. (2013) The impact of beneficial plant‐associated microbes on plant phenotypic plasticity. J. Chem. Ecol. 39, 826–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grady, E.N. , MacDonald, J. , Liu, L. , Richman, A. and Yuan, Z.C. (2016) Current knowledge and perspectives of Paenibacillus: a review. Microb. Cell Fact. 15, 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, S.Y. , Zhang, D. and Zhang, Z. (1986) Soil Enzyme and Its Research Methods, pp. 274–297. Beijing: Chinese Agricultural Press. [Google Scholar]
- Gutiérrez‐Luna, F.M. , López‐Bucio, J. , Altamirano‐Hernández, J. , Valencia‐Cantero, E. , Reyes de la Cruz, H. and Macías‐Rodríguez, L. (2010) Plant growth‐promoting rhizobacteria modulate root‐system architecture in Arabidopsis thaliana through volatile organic compound emission. Symbiosis, 51, 75–83. [Google Scholar]
- Hacquard, S. , Spaepen, S. , Garrido‐Oter, R. and Schulze‐Lefert, P. (2017) Interplay between innate immunity and the plant microbiota. Annu. Rev. Phytopathol. 55, 565–589. [DOI] [PubMed] [Google Scholar]
- Han, X. and Kahmann, R. (2019) Manipulation of phytohormone pathways by effectors of filamentous plant pathogens. Front. Plant Sci. 10, 822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, Q. , Ma, Q. , Chen, Y. , Tian, B. , Xu, L. , Bai, Y. , Chen, W. et al. (2020) Variation in rhizosphere microbial communities and its association with the symbiotic efficiency of rhizobia in soybean. ISME J. 14(8), 1915–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbort, C.J. , Hashimoto, M. , Inoue, H. , Niu, Y. , Guan, R. , Rombolà, A.D. , Kopriva, S. et al. (2020) Root‐secreted coumarins and the microbiota interact to improve iron nutrition in Arabidopsis . Cell Host Microbe, 28, 825–837.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta, M. , Sabat, G. , Stecker, K. , Minkoff, B.B. and Sussman, M.R. (2014) A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science, 343, 408–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassani, M.A. , Durán, P. and Hacquard, S. (2018) Microbial interactions within the plant holobiont. Microbiome, 6, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillmer, R.A. , Tsuda, K. , Rallapalli, G. , Asai, S. , Truman, W. , Papke, M.D. , Sakakibara, H. et al. (2017) The highly buffered Arabidopsis immune signaling network conceals the functions of its components. PLoS Genet. 13, e1006639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinsinger, P. , Betencourt, E. , Bernard, L. , Brauman, A. , Plassard, C. , Shen, J. , Tang, X. et al. (2011) P for two, sharing a scarce resource: soil phosphorus acquisition in the rhizosphere of intercropped species. Plant Physiol. 156, 1078–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch, A.M. , Fang, Y. , Asad, S. and Kapulnik, Y. (1997) The role of phytohormones in plant‐microbe symbioses. Plant Soil. 194, 171–184. [Google Scholar]
- Horton, M.W. , Bodenhausen, N. , Beilsmith, K. , Meng, D. , Muegge, B.D. , Subramanian, S. , Vetter, M.M. et al. (2014) Genome‐wide association study of Arabidopsis thaliana leaf microbial community. Nat. Commun. 5, 5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, S. , Thiergart, T. , Vannier, N. , Mesny, F. , Ziegler, J. , Pickel, B. and Hacquard, S. (2021) A microbiota‐root‐shoot circuit favours Arabidopsis growth over defence under suboptimal light. Nat. Plants, 7, 1078–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, C.A. , Delauney, A.J. and Verma, D.P. (1992) A bifunctional enzyme (delta 1‐pyrroline‐5‐carboxylate synthetase) catalyzes the first two steps in proline biosynthesis in plants. Proc. Natl Acad. Sci. USA, 89, 9354–9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, B. , Wang, W. , Ou, S. , Tang, J. , Li, H. , Che, R. , Zhang, Z. et al. (2015) Variation in NRT1.1B contributes to nitrate‐use divergence between rice subspecies. Nat. Genet. 47, 834–838. [DOI] [PubMed] [Google Scholar]
- Huang, C.Y. , Wang, H. , Hu, P. , Hamby, R. and Jin, H. (2019) Small RNAs—big players in plant‐microbe interactions. Cell Host Microbe, 26, 173–182. [DOI] [PubMed] [Google Scholar]
- Huang, S. , Yan, Y. , Su, F. , Huang, X. , Xia, D. , Jiang, X. , Dong, Y. et al. (2021) Research progress in gene editing technology. Front. Biosci. 26, 916–927. [DOI] [PubMed] [Google Scholar]
- Hung, R. , Lee, S. and Bennett, J.W. (2013) Arabidopsis thaliana as a model system for testing the effect of Trichoderma volatile organic compounds. Fungal Ecol. 6, 19–26. [Google Scholar]
- Huo, T. , Wang, C.T. , Yu, T.F. , Wang, D.M. , Li, M. , Zhao, D. , Li, X.T. et al. (2021) Overexpression of ZmWRKY65 transcription factor from maize confers stress resistances in transgenic Arabidopsis . Sci. Rep. 11, 4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie, N. , Weinberger, L. , Tang, W.W. , Kobayashi, T. , Viukov, S. , Manor, Y.S. , Dietmann, S. et al. (2015) SOX17 is a critical specifier of human primordial germ cell fate. Cell, 160, 253–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishioka, N. , Tanimoto, S. and Harada, H. (1991) Roles of nitrogen and carbohydrate in floral‐bud formation in pharbitis apex cultures. J. Plant Physiol. 138, 573–576. [Google Scholar]
- Jasiński, M. , Stukkens, Y. , Degand, H. , Purnelle, B. , Marchand‐Brynaert, J. and Boutry, M. (2001) A plant plasma membrane ATP binding cassette‐type transporter is involved in antifungal terpenoid secretion. Plant Cell, 13, 1095–1107. [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein, T. , Lewis, K. and Epstein, S.S. (2002) Isolating “uncultivable” microorganisms in pure culture in a simulated natural environment. Science, 296, 1127–1129. [DOI] [PubMed] [Google Scholar]
- Kanchiswamy, C.N. , Malnoy, M. and Maffei, M.E. (2015) Chemical diversity of microbial volatiles and their potential for plant growth and productivity. Front. Plant Sci. 6, 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushal, R. , Peng, L. , Singh, S.K. , Zhang, M. , Zhang, X. , Vílchez, J.I. , Wang, Z. et al. (2021) Dicer‐like proteins influence Arabidopsis root microbiota independent of RNA‐directed DNA methylation. Microbiome, 9, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke, J. , Wang, B. and Yoshikuni, Y. (2021) Microbiome engineering: synthetic biology of plant‐associated microbiomes in sustainable agriculture. Trends Biotechnol. 39, 244–261. [DOI] [PubMed] [Google Scholar]
- Koprivova, A. and Kopriva, S. (2022) Plant secondary metabolites altering root microbiome composition and function. Curr. Opin. Plant Biol. 67, 102227. [DOI] [PubMed] [Google Scholar]
- Kroll, S. , Agler, M.T. and Kemen, E. (2017) Genomic dissection of host‐microbe and microbe‐microbe interactions for advanced plant breeding. Curr. Opin. Plant Biol. 36, 71–78. [DOI] [PubMed] [Google Scholar]
- Krouk, G. , Lacombe, B. , Bielach, A. , Perrine‐Walker, F. , Malinska, K. , Mounier, E. , Hoyerova, K. et al. (2010) Nitrate‐regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev Cell. 18, 927–937. [DOI] [PubMed] [Google Scholar]
- Kumar, K.R. , Cowley, M.J. and Davis, R.L. (2019) Next‐generation sequencing and emerging technologies. Semin. Thromb. Hemost. 45, 661–673. [DOI] [PubMed] [Google Scholar]
- Kwak, M.J. , Kong, H.G. , Choi, K. , Kwon, S.K. , Song, J.Y. , Lee, J. , Lee, P.A. et al. (2018) Rhizosphere microbiome structure alters to enable wilt resistance in tomato. Nat. Biotechnol. 36, 1100–1109. [DOI] [PubMed] [Google Scholar]
- Laloum, T. , Baudin, M. , Frances, L. , Lepage, A. , Billault‐Penneteau, B. , Cerri, M.R. , Ariel, F. et al. (2014) Two CCAAT‐box‐binding transcription factors redundantly regulate early steps of the legume‐rhizobia endosymbiosis. Plant J. 79, 757–768. [DOI] [PubMed] [Google Scholar]
- Lareen, A. , Burton, F. and Schäfer, P. (2016) Plant root‐microbe communication in shaping root microbiomes. Plant Mol. Biol. 90, 575–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson, C.E. , Harcombe, W.R. , Hatzenpichler, R. , Lindemann, S.R. , Löffler, F.E. , O'Malley, M.A. , García Martín, H. et al. (2019) Common principles and best practices for engineering microbiomes. Nat. Rev. Microbiol. 17, 725–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léran, S. , Muños, S. , Brachet, C. , Tillard, P. , Gojon, A. and Lacombe, B. (2013) Arabidopsis NRT1.1 is a bidirectional transporter involved in root‐to‐shoot nitrate translocation. Mol. Plant. 6, 1984–1987. [DOI] [PubMed] [Google Scholar]
- Levy, A. , Conway, J.M. , Dangl, J.L. and Woyke, T. (2018) Elucidating bacterial gene functions in the plant microbiome. Cell Host Microbe, 24, 475–485. [DOI] [PubMed] [Google Scholar]
- Li, Y. and Gong, X. (2021) Effects of dissolved organic matter on the bioavailability of heavy metals during microbial dissimilatory iron reduction: a review. Rev. Environ. Contam. Toxicol. 257, 69–92. [DOI] [PubMed] [Google Scholar]
- Li, M. , Guo, R. , Yu, F. , Chen, X. , Zhao, H. , Li, H. and Wu, J. (2018) Indole‐3‐Acetic acid biosynthesis pathways in the plant‐beneficial bacterium arthrobacter pascens ZZ21. Int. J. Mol. Sci. 19, 443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X.Z. , Qin, S.M. , Chen, Z.J. , Zhang, J. , Yao, L.G. , Li, N. , Pang, F.H. et al. (2022) Polyamine‐producing bacteria regulated the community structure of rhizosphere bacteria and reduced the absorption of Cd in wheat. Huan Jing Ke Xue, 43, 1031–1039. [DOI] [PubMed] [Google Scholar]
- Li, G.Z. , Wang, Z.Q. , Yokosho, K. , Ding, B. , Fan, W. , Gong, Q.Q. , Li, G.X. et al. (2018) Transcription factor WRKY22 promotes aluminum tolerance via activation of OsFRDL4 expression and enhancement of citrate secretion in rice (Oryza sativa). New Phytol. 219, 149–162. [DOI] [PubMed] [Google Scholar]
- Li, C. , Wu, H.M. and Cheung, A.Y. (2016) FERONIA and her pals: functions and mechanisms. Plant Physiol. 171, 2379–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. , Zhang, C. , Qu, X. , Luo, Z. , Lu, S. , Kuzyakov, Y. , Alharbi, H.A. et al. (2021) Microbial communities and functions in the rhizosphere of disease‐resistant and susceptible camellia spp. Front. Microbiol. 12, 732905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Zhang, Y. , Wang, Q.X. , Wang, T.T. , Cao, X.L. , Zhao, Z.X. , Zhao, S.L. et al. (2018) Resistance to powdery MILDEW8.1 boosts pattern‐triggered immunity against multiple pathogens in Arabidopsis and rice. Plant Biotechnol. J. 16, 428–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C. , Zhou, J. , Wang, X. and Liao, H. (2019) A purple acid phosphatase, GmPAP33, participates in arbuscule degeneration during arbuscular mycorrhizal symbiosis in soybean. Plant Cell Environ. 42, 2015–2027. [DOI] [PubMed] [Google Scholar]
- Lian, T. , Huang, Y. , Xie, X. , Huo, X. , Shahid, M.Q. , Tian, L. , Lan, T. et al. (2020) Rice SST variation shapes the rhizosphere bacterial community, conferring tolerance to salt stress through regulating soil metabolites. mSystems, 5, e00721‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian, T.X. , Ma, Q. , Shi, Q. , Cai, Z. , Zhang, Y. , Cheng, Y. and Nian, H. (2019) High aluminum stress drives different rhizosphere soil enzyme activities and bacterial community structure between aluminum‐tolerant and aluminum‐sensitive soybean genotypes. Plant Soil, 440, 409–425. [Google Scholar]
- Liu, H. and Brettell, L.E. (2019) Plant defense by VOC‐induced microbial priming. Trends Plant Sci. 24, 187–189. [DOI] [PubMed] [Google Scholar]
- Liu, H. , Brettell, L.E. , Qiu, Z. and Singh, B.K. (2020) Microbiome‐mediated stress resistance in plants. Trends Plant Sci. 25, 733–743. [DOI] [PubMed] [Google Scholar]
- Liu, H. , Carvalhais, L.C. , Kazan, K. and Schenk, P.M. (2016) Development of marker genes for jasmonic acid signaling in shoots and roots of wheat. Plant Signal Behav. 11, e1176654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Chen, L. , Wu, G. , Feng, H. , Zhang, G. , Shen, Q. and Zhang, R. (2017) Identification of root‐secreted compounds involved in the communication between cucumber, the beneficial Bacillus amyloliquefaciens, and the soil‐borne pathogen Fusarium oxysporum . Mol. Plant Microbe Interact. 30, 53–62. [DOI] [PubMed] [Google Scholar]
- Liu, Y. , Evans, S.E. , Friesen, M.L. and Tiemann, L.K. (2022) Root exudates shift how N mineralization and N fixation contribute to the plant‐available N supply in low fertility soils. Soil Biol. Biochem. 165, 108541. [Google Scholar]
- Liu, S. , Hao, H. , Lu, X. , Zhao, X. , Wang, Y. , Zhang, Y. , Xie, Z. et al. (2017) Transcriptome profiling of genes involved in induced systemic salt tolerance conferred by Bacillus amyloliquefaciens FZB42 in Arabidopsis thaliana . Sci. Rep. 7, 10795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H. , Li, J. , Carvalhais, L.C. , Percy, C.D. , Prakash Verma, J. , Schenk, P.M. and Singh, B.K. (2021) Evidence for the plant recruitment of beneficial microbes to suppress soil‐borne pathogens. New Phytol. 229, 2873–2885. [DOI] [PubMed] [Google Scholar]
- Liu, J. , Li, Y. , Wang, W. , Gai, J. and Li, Y. (2016) Genome‐wide analysis of MATE transporters and expression patterns of a subgroup of MATE genes in response to aluminum toxicity in soybean. BMC Genomics, 17, 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H. , Macdonald, C.A. , Cook, J. , Anderson, I.C. and Singh, B.K. (2019) An ecological loop: host microbiomes across multitrophic interactions. Trends Ecol. Evol. 34, 1118–1130. [DOI] [PubMed] [Google Scholar]
- Liu, J. , Magalhaes, J.V. , Shaff, J. and Kochian, L.V. (2009) Aluminum‐activated citrate and malate transporters from the MATE and ALMT families function independently to confer Arabidopsis aluminum tolerance. Plant J. 57, 389–399. [DOI] [PubMed] [Google Scholar]
- Liu, F. , Mao, J. , Kong, W. , Hua, Q. , Feng, Y. , Bashir, R. and Lu, T. (2020) Interaction variability shapes succession of synthetic microbial ecosystems. Nat. Commun. 11, 309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y.X. , Qin, Y. , Chen, T. , Lu, M. , Qian, X. , Guo, X. and Bai, Y. (2021) A practical guide to amplicon and metagenomic analysis of microbiome data. Protein Cell, 12, 315–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, G.H. , Ye, Z.F. and Wu, W.Z. (2012) Culture‐dependent and culture‐independent approaches to studying soil microbial diversity. Acta Ecologica Sinica, 32, 4421–4433. [Google Scholar]
- López‐Arredondo, D.L. , Leyva‐González, M.A. , González‐Morales, S.I. , López‐Bucio, J. and Herrera‐Estrella, L. (2014) Phosphate nutrition: improving low‐phosphate tolerance in crops. Annu. Rev. Plant Biol. 65, 95–123. [DOI] [PubMed] [Google Scholar]
- López‐Ráez, J.A. , Shirasu, K. and Foo, E. (2017) Strigolactones in plant interactions with beneficial and detrimental organisms: the yin and yang. Trends Plant Sci. 22, 527–537. [DOI] [PubMed] [Google Scholar]
- Lorite, M.J. , Estrella, M.J. , Escaray, F.J. , Sannazzaro, A. , Videira, E. , Castro, I.M. , Monza, J. et al. (2018) The rhizobia‐lotus symbioses: deeply specific and widely diverse. Front. Microbiol. 9, 2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, T. , Ke, M. , Lavoie, M. , Jin, Y. , Fan, X. , Zhang, Z. , Fu, Z. et al. (2018) Rhizosphere microorganisms can influence the timing of plant flowering. Microbiome, 6, 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugtenberg, B. and Kamilova, F. (2009) Plant‐growth‐promoting rhizobacteria. Annu. Rev. Microbiol. 63, 541–556. [DOI] [PubMed] [Google Scholar]
- Ma, J.F. (2007) Syndrome of aluminum toxicity and diversity of aluminum resistance in higher plants. Int. Rev. Cytol. 264, 225–252. [DOI] [PubMed] [Google Scholar]
- Magalhaes, J.V. , Liu, J. , Guimarães, C.T. , Lana, U.G. , Alves, V.M. , Wang, Y.H. , Schaffert, R.E. et al. (2007) A gene in the multidrug and toxic compound extrusion (MATE) family confers aluminum tolerance in sorghum. Nat. Genet. 39, 1156–1161. [DOI] [PubMed] [Google Scholar]
- Mai, Y.X. , Wang, L. and Yang, H.Q. (2011) A gain‐of‐function mutation in IAA7/AXR2 confers late flowering under short‐day light in Arabidopsis . J. Integr. Plant Biol. 53, 480–492. [DOI] [PubMed] [Google Scholar]
- Maron, L.G. , Guimarães, C.T. , Kirst, M. , Albert, P.S. , Birchler, J.A. , Bradbury, P.J. , Buckler, E.S. et al. (2013) Aluminum tolerance in maize is associated with higher MATE1 gene copy number. Proc. Natl Acad. Sci. USA, 110, 5241–5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marotz, C.A. , Sanders, J.G. , Zuniga, C. , Zaramela, L.S. , Knight, R. and Zengler, K. (2018) Improving saliva shotgun metagenomics by chemical host DNA depletion. Microbiome, 6, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke, M.A. and Mosher, R.A. (2014) RNA‐directed DNA methylation: an epigenetic pathway of increasing complexity. Nat. Rev. Genet. 15, 394–408. [DOI] [PubMed] [Google Scholar]
- McDonald, T.R. , Rizvi, M.F. , Ruiter, B.L. , Roy, R. , Reinders, A. and Ward, J.M. (2022) Posttranslational regulation of transporters important for symbiotic interactions. Plant Physiol. 188, 941–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehnaz, S. , Weselowski, B. and Lazarovits, G. (2007) Azospirillum canadense sp. nov., a nitrogen‐fixing bacterium isolated from corn rhizosphere. Int. J. Syst. Evol. Microbiol. 57, 620–624. [DOI] [PubMed] [Google Scholar]
- Metzker, M.L. (2010) Sequencing technologies—the next generation. Nat. Rev. Genet. 11, 31–46. [DOI] [PubMed] [Google Scholar]
- Middleton, H. , Yergeau, É. , Monard, C. , Combier, J.P. and El Amrani, A. (2021) Rhizospheric plant‐microbe interactions: miRNAs as a key mediator. Trends Plant Sci. 26, 132–141. [DOI] [PubMed] [Google Scholar]
- Millet, Y.A. , Danna, C.H. , Clay, N.K. , Songnuan, W. , Simon, M.D. , Werck‐Reichhart, D. and Ausubel, F.M. (2010) Innate immune responses activated in Arabidopsis roots by microbe‐associated molecular patterns. Plant Cell, 22, 973–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra, J. , Singh, R. and Arora, N.K. (2017) Alleviation of heavy metal stress in plants and remediation of soil by rhizosphere microorganisms. Front. Microbiol. 8, 1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina, R. , Rivera, D. , Mora, V. , López, G. , Rosas, S. , Spaepen, S. , Vanderleyden, J. et al. (2018) Regulation of IAA biosynthesis in azospirillum brasilense under environmental stress conditions. Curr. Microbiol. 75, 1408–1418. [DOI] [PubMed] [Google Scholar]
- Morgan, J.B. and Connolly, E.L. (2013) Plant‐soil interactions: nutrient uptake. Nat. Educ. Knowl. 4, 2. [Google Scholar]
- van de Mortel, J.E. , Schat, H. , Moerland, P.D. , Loren, V. , van Themaat, E. , van der Ent, S. , Blankestijn, H. et al. (2008) Expression differences for genes involved in lignin, glutathione and sulphate metabolism in response to cadmium in Arabidopsis thaliana and the related Zn/Cd‐hyperaccumulator Thlaspi caerulescens . Plant Cell Environ. 31, 301–324. [DOI] [PubMed] [Google Scholar]
- Mueller, U.G. and Sachs, J.L. (2015) Engineering microbiomes to improve plant and animal health. Trends Microbiol. 23, 606–617. [DOI] [PubMed] [Google Scholar]
- Nawrath, C. , Heck, S. , Parinthawong, N. and Métraux, J.P. (2002) EDS5, an essential component of salicylic acid‐dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. Plant Cell, 14, 275–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nerva, L. , Sandrini, M. , Moffa, L. , Velasco, R. , Balestrini, R. and Chitarra, W. (2022) Breeding toward improved ecological plant‐microbiome interactions. Trends Plant Sci. 27, 1134–1143. [DOI] [PubMed] [Google Scholar]
- Netzker, T. , Shepherdson, E.M.F. , Zambri, M.P. and Elliot, M.A. (2020) Bacterial volatile compounds: functions in communication, cooperation, and competition. Annu. Rev. Microbiol. 74, 409–430. [DOI] [PubMed] [Google Scholar]
- New, F.N. and Brito, I.L. (2020) What is metagenomics teaching us, and what is missed? Annu. Rev. Microbiol. 74, 117–135. [DOI] [PubMed] [Google Scholar]
- Niedringhaus, T.P. , Milanova, D. , Kerby, M.B. , Snyder, M.P. and Barron, A.E. (2011) Landscape of next‐generation sequencing technologies. Anal. Chem. 83, 4327–4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortíz‐Castro, R. , Contreras‐Cornejo, H.A. , Macías‐Rodríguez, L. and López‐Bucio, J. (2009) The role of microbial signals in plant growth and development. Plant Signal. Behav. 4, 701–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyserman, B.O. , Flores, S.S. , Griffioen, T. , Pan, X. , van der Wijk, E. , Pronk, L. , Lokhorst, W. et al. (2022) Disentangling the genetic basis of rhizosphere microbiome assembly in tomato. Nat. Commun. 13, 3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyserman, B.O. , Medema, M.H. and Raaijmakers, J.M. (2018) Road MAPs to engineer host microbiomes. Curr. Opin. Microbiol. 43, 46–54. [DOI] [PubMed] [Google Scholar]
- Pang, Z. , Chen, J. , Wang, T. , Gao, C. , Li, Z. , Guo, L. , Xu, J. et al. (2021) Linking plant secondary metabolites and plant microbiomes: a review. Front. Plant Sci. 12, 621276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panke‐Buisse, K. , Poole, A.C. , Goodrich, J.K. , Ley, R.E. and Kao‐Kniffin, J. (2015) Selection on soil microbiomes reveals reproducible impacts on plant function. ISME J. 9, 980–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascale, A. , Proietti, S. , Pantelides, I.S. and Stringlis, I.A. (2020) Modulation of the root microbiome by plant molecules: the basis for targeted disease suppression and plant growth promotion. Front. Plant Sci. 10, 1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patten, C.L. , Blakney, A.J. and Coulson, T.J. (2013) Activity, distribution and function of indole‐3‐acetic acid biosynthetic pathways in bacteria. Crit. Rev. Microbiol. 39, 395–415. [DOI] [PubMed] [Google Scholar]
- Pershina, E.V. , Ivanova, E.A. , Korvigo, I.O. , Chirak, E.L. , Sergaliev, N.H. , Abakumov, E.V. , Provorov, N.A. et al. (2018) Investigation of the core microbiome in main soil types from the East European plain. Sci. Total Environ. 631‐632, 1421–1430. [DOI] [PubMed] [Google Scholar]
- Pieterse, C.M. , Zamioudis, C. , Berendsen, R.L. , Weller, D.M. , Van Wees, S.C. and Bakker, P.A. (2014) Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 52, 347–375. [DOI] [PubMed] [Google Scholar]
- Plyuta, V. , Lipasova, V. , Popova, A. , Koksharova, O. , Kuznetsov, A. , Szegedi, E. , Chernin, L. et al. (2016) Influence of volatile organic compounds emitted by Pseudomonas and Serratia strains on Agrobacterium tumefaciens biofilms. APMIS, 124, 586–594. [DOI] [PubMed] [Google Scholar]
- Prabhu, A.A. , Boro, B. , Bharali, B. , Chakraborty, S. and Dasu, V.V. (2017) Gene and process level modulation to overcome the bottlenecks of recombinant proteins expression in Pichia pastoris . Curr. Pharm. Biotechnol. 18(15), 1200–1223. [DOI] [PubMed] [Google Scholar]
- Qiao, Q. , Wang, F. , Zhang, J. , Chen, Y. , Zhang, C. , Liu, G. , Zhang, H. et al. (2017) The variation in the rhizosphere microbiome of cotton with soil type, genotype and developmental stage. Sci. Rep. 7, 3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravanbakhsh, M. , Kowalchuk, G.A. and Jousset, A. (2019) Root‐associated microorganisms reprogram plant life history along the growth‐stress resistance tradeoff. ISME J. 13, 3093–3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raza, W. , Wei, Z. , Jousset, A. , Shen, Q. and Friman, V.P. (2021) Extended plant metarhizobiome: understanding volatile organic compound signaling in plant‐microbe metapopulation networks. mSystems, 6, e0084921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rico, L. , Ogaya, R. , Terradas, J. and Peñuelas, J. (2014) Community structures of N2 ‐fixing bacteria associated with the phyllosphere of a Holm oak forest and their response to drought. Plant Biol. 16, 586–593. [DOI] [PubMed] [Google Scholar]
- Riefler, M. , Novak, O. , Strnad, M. and Schmülling, T. (2006) Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell, 18, 40–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez, P.A. , Rothballer, M. , Chowdhury, S.P. , Nussbaumer, T. , Gutjahr, C. and Falter‐Braun, P. (2019) Systems biology of plant‐microbiome interactions. Mol. Plant. 12, 804–821. [DOI] [PubMed] [Google Scholar]
- Rolfe, S.A. , Griffiths, J. and Ton, J. (2019) Crying out for help with root exudates: adaptive mechanisms by which stressed plants assemble health‐promoting soil microbiomes. Curr. Opin. Microbiol. 49, 73–82. [DOI] [PubMed] [Google Scholar]
- Roman‐Reyna, V. , Pinili, D. , Borja, F.N. , Quibod, I.L. , Groen, S. , Mulyaningsih, E. et al. (2019) The rice leaf microbiome has a conserved community structure controlled by complex host–microbe interaction. bioRxiv. [Google Scholar]
- Ruby, E.G. (2008) Symbiotic conversations are revealed under genetic interrogation. Nat. Rev. Microbiol. 6, 752–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudrappa, T. , Czymmek, K.J. , Paré, P.W. and Bais, H.P. (2008) Root‐secreted malic acid recruits beneficial soil bacteria. Plant Physiol. 148, 1547–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz‐Lozano, J.M. , Porcel, R. and Aroca, R. (2006) Does the enhanced tolerance of arbuscular mycorrhizal plants to water deficit involve modulation of drought‐induced plant genes? New Phytol. 171, 693–698. [DOI] [PubMed] [Google Scholar]
- Ryu, M.H. , Zhang, J. , Toth, T. , Khokhani, D. , Geddes, B.A. , Mus, F. , Garcia‐Costas, A. et al. (2020) Control of nitrogen fixation in bacteria that associate with cereals. Nat. Microbiol. 5, 314–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas‐González, I. , Reyt, G. , Flis, P. , Custódio, V. , Gopaulchan, D. , Bakhoum, N. , Dew, T.P. et al. (2021) Coordination between microbiota and root endodermis supports plant mineral nutrient homeostasis. Science, 371, eabd0695. [DOI] [PubMed] [Google Scholar]
- Saleem, M. , Hu, J. and Jousset, A. (2019) More than the sum of its parts: microbiome biodiversity as a driver of plant growth and soil health. Annu. Rev. Ecol. Evol. Syst. 50, 145–168. [Google Scholar]
- Sánchez‐López, Á.M. , Bahaji, A. , De Diego, N. , Baslam, M. , Li, J. , Muñoz, F.J. , Almagro, G. et al. (2016a) Arabidopsis responds to Alternaria alternata volatiles by triggering plastid phosphoglucose isomerase‐independent mechanisms. Plant Physiol. 172, 1989–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez‐López, Á.M. , Baslam, M. , De Diego, N. , Muñoz, F.J. , Bahaji, A. , Almagro, G. , Ricarte‐Bermejo, A. et al. (2016b) Volatile compounds emitted by diverse phytopathogenic microorganisms promote plant growth and flowering through cytokinin action. Plant Cell Environ. 39, 2592–2608. [DOI] [PubMed] [Google Scholar]
- Schaarschmidt, S. , Fischer, A. , Lawas, L.M.F. , Alam, R. , Septiningsih, E.M. , Bailey‐Serres, J. , Jagadish, S.V.K. et al. (2020) Utilizing PacBio Iso‐Seq for novel transcript and gene discovery of abiotic stress responses in Oryza sativa L. Int. J. Mol. Sci. 21, 8148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer, M. , Vogel, C.M. , Bortfeld‐Miller, M. , Mittelviefhaus, M. and Vorholt, J.A. (2022) Mapping phyllosphere microbiota interactions in planta to establish genotype‐phenotype relationships. Nat. Microbiol. 7, 856–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlechter, R.O. , Miebach, M. and Remus‐Emsermann, M.N.P. (2019) Driving factors of epiphytic bacterial communities: a review. J. Adv. Res. 19, 57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, R. , Etalo, D.W. , de Jager, V. , Gerards, S. , Zweers, H. , de Boer, W. and Garbeva, P. (2016) Microbial small talk: volatiles in fungal‐bacterial interactions. Front. Microbiol. 6, 1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, R. , Jager, V. , Zühlke, D. , Wolff, C. , Bernhardt, J. , Cankar, K. , Beekwilder, J. et al. (2017) Fungal volatile compounds induce production of the secondary metabolite Sodorifen in Serratia plymuthica PRI‐2C. Sci. Rep. 7, 862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz, S. and Dickschat, J.S. (2007) Bacterial volatiles: the smell of small organisms. Nat. Prod. Rep. 24, 814–842. [DOI] [PubMed] [Google Scholar]
- Schulz‐Bohm, K. , Gerards, S. , Hundscheid, M. , Melenhorst, J. , de Boer, W. and Garbeva, P. (2018) Calling from distance: attraction of soil bacteria by plant root volatiles. ISME J. 12, 1252–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, P.J. , Park, J. , Park, M.J. , Kim, Y.S. , Kim, S.G. , Jung, J.H. and Park, C.M. (2012) A Golgi‐localized MATE transporter mediates iron homoeostasis under osmotic stress in Arabidopsis . Biochem. J. 442, 551–561. [DOI] [PubMed] [Google Scholar]
- Shalmani, A. , Ullah, U. , Muhammad, I. , Zhang, D. , Sharif, R. , Jia, P. , Saleem, N. et al. (2021) The TAZ domain‐containing proteins play important role in the heavy metals stress biology in plants. Environ. Res. 197, 111030. [DOI] [PubMed] [Google Scholar]
- Shao, Q.Y. , Dong, C.B. , Han, Y.F. and Liang, Z.Q. (2021) Research progress in the rhizosphere microbiome of plants. J. Plant Nutr. Fertilizer, 27, 144–152. [Google Scholar]
- Shendure, J. and Ji, H. (2008) Next‐generation DNA sequencing. Nat. Biotechnol. 26, 1135–1145. [DOI] [PubMed] [Google Scholar]
- Shi, Q. , Liu, Y. , Shi, A. , Cai, Z. , Nian, H. , Hartmann, M. and Lian, T. (2020) Rhizosphere soil fungal communities of aluminum‐tolerant and ‐sensitive soybean genotypes respond differently to aluminum stress in an acid soil. Front. Microbiol. 11, 1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji, T. , Inai, K. , Yazaki, Y. , Sato, Y. , Takase, H. , Shitan, N. , Yazaki, K. et al. (2009) Multidrug and toxic compound extrusion‐type transporters implicated in vacuolar sequestration of nicotine in tobacco roots. Plant Physiol. 149, 708–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigman, M.J. and Slotkin, R.K. (2016) The first rule of plant transposable element silencing: location, location, location. Plant Cell, 28, 304–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer, E. , Andreopoulos, B. , Bowers, R.M. , Lee, J. , Deshpande, S. , Chiniquy, J. , Ciobanu, D. et al. (2016) Next generation sequencing data of a defined microbial mock community. Sci. Data, 3, 160081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, L. and Xie, K. (2020) Engineering CRISPR/Cas9 to mitigate abundant host contamination for 16S rRNA gene‐based amplicon sequencing. Microbiome, 8, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, Y. , Wilson, A.J. , Zhang, X.C. , Thoms, D. , Sohrabi, R. , Song, S. , Geissmann, Q. et al. (2021) FERONIA restricts Pseudomonas in the rhizosphere microbiome via regulation of reactive oxygen species. Nat. Plants, 7, 644–654. [DOI] [PubMed] [Google Scholar]
- Stegmann, M. , Monaghan, J. , Smakowska‐Luzan, E. , Rovenich, H. , Lehner, A. , Holton, N. , Belkhadir, Y. et al. (2017) The receptor kinase FER is a RALF‐regulated scaffold controlling plant immune signaling. Science, 355, 287–289. [DOI] [PubMed] [Google Scholar]
- Stringlis, I.A. , Yu, K. , Feussner, K. , de Jonge, R. , Van Bentum, S. , Van Verk, M.C. , Berendsen, R.L. et al. (2018) MYB72‐dependent coumarin exudation shapes root microbiome assembly to promote plant health. Proc. Natl Acad. Sci. USA, 115, E5213–E5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukumar, P. , Legué, V. , Vayssières, A. , Martin, F. , Tuskan, G.A. and Kalluri, U.C. (2013) Involvement of auxin pathways in modulating root architecture during beneficial plant‐microorganism interactions. Plant Cell Environ. 36, 909–919. [DOI] [PubMed] [Google Scholar]
- Sun, L. , Cao, M. , Liu, F. et al. (2020) The volatile organic compounds of Floccularia luteovirens modulate plant growth and metabolism in Arabidopsis thaliana . Plant Soil, 456, 207–221. [Google Scholar]
- Sun, X. , Gilroy, E.M. , Chini, A. , Nurmberg, P.L. , Hein, I. , Lacomme, C. , Birch, P.R. et al. (2011) ADS1 encodes a MATE‐transporter that negatively regulates plant disease resistance. New Phytol. 192, 471–482. [DOI] [PubMed] [Google Scholar]
- Takanashi, K. , Shitan, N. and Yazaki, K. (2014) The multidrug and toxic compound extrusion (MATE) family in plants. Plant Biotechnol. 31, 417–430. [Google Scholar]
- Tan, H. , Li, B. and Guo, H. (2020) The diversity of post‐transcriptional gene silencing mediated by small silencing RNAs in plants. Essays Biochem. 64, 919–930. [DOI] [PubMed] [Google Scholar]
- Taylor, I.B. , Burbidge, A. and Thompson, A.J. (2000) Control of abscisic acid synthesis. J. Exp. Bot. 51, 1563–1574. [DOI] [PubMed] [Google Scholar]
- Teixeira, P.J.P. , Colaianni, N.R. , Fitzpatrick, C.R. and Dangl, J.L. (2019) Beyond pathogens: microbiota interactions with the plant immune system. Curr. Opin. Microbiol. 49, 7–17. [DOI] [PubMed] [Google Scholar]
- Teng, Y. , Liang, Y. , Wang, M. , Mai, H. and Ke, L. (2019) Nitrate transporter 1.1 is involved in regulating flowering time via transcriptional regulation of FLOWERING LOCUS C in Arabidopsis thaliana . Plant Sci. 284, 30–36. [DOI] [PubMed] [Google Scholar]
- Treesubsuntorn, C. , Dhurakit, P. , Khaksar, G. and Thiravetyan, P. (2018) Effect of microorganisms on reducing cadmium uptake and toxicity in rice (Oryza sativa L.). Environ. Sci. Pollut. Res. Int. 25, 25690–25701. [DOI] [PubMed] [Google Scholar]
- Trivedi, P. , Leach, J.E. , Tringe, S.G. , Sa, T. and Singh, B.K. (2021) Author correction: plant‐microbiome interactions: from community assembly to plant health. Nat. Rev. Microbiol. 19, 72. [DOI] [PubMed] [Google Scholar]
- Trubl, G. , Jang, H.B. , Roux, S. , Emerson, J.B. , Solonenko, N. , Vik, D.R. , Solden, L. et al. (2018) Soil viruses are underexplored players in ecosystem carbon processing. mSystems, 3, e00076–e00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong, H.A. , Lee, S. , Trịnh, C.S. , Lee, W.J. , Chung, E.H. , Hong, S.W. and Lee, H. (2021) Overexpression of the HDA15 gene confers resistance to salt stress by the induction of NCED3, an ABA biosynthesis enzyme. Front. Plant Sci. 12, 640443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsikou, D. , Yan, Z. , Holt, D.B. , Abel, N.B. , Reid, D.E. , Madsen, L.H. , Bhasin, H. et al. (2018) Systemic control of legume susceptibility to rhizobial infection by a mobile microRNA. Science, 362, 233–236. [DOI] [PubMed] [Google Scholar]
- Tsuda, K. , Sato, M. , Stoddard, T. , Glazebrook, J. and Katagiri, F. (2009) Network properties of robust immunity in plants. PLoS Genet. 5, e1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Ent, S. , Verhagen, B.W. , Van Doorn, R. , Bakker, D. , Verlaan, M.G. , Pel, M.J. , Joosten, R.G. et al. (2008) MYB72 is required in early signaling steps of rhizobacteria‐induced systemic resistance in Arabidopsis . Plant Physiol. 146, 1293–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Teeseling, M.C.F. and Jogler, C. (2021) Cultivation of elusive microbes unearthed exciting biology. Nat. Commun. 12, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velmourougane, K. and Blaise, D. (2017) Impact of transgenic Bt cotton on soil health. CAB Rev. 12, 1–8. [Google Scholar]
- Vives‐Peris, V. , de Ollas, C. , Gómez‐Cadenas, A. and Pérez‐Clemente, R.M. (2020) Root exudates: from plant to rhizosphere and beyond. Plant Cell Rep. 39, 3–17. [DOI] [PubMed] [Google Scholar]
- von Uexküll, H.R. and Mutert, E. (1995) Global extent, development and economic impact of acid soils. Plant Soil. 171, 1–15. [Google Scholar]
- Vorholt, J.A. , Vogel, C. , Carlström, C.I. and Müller, D.B. (2017) Establishing causality: opportunities of synthetic communities for plant microbiome research. Cell Host Microbe, 22, 142–155. [DOI] [PubMed] [Google Scholar]
- Wagner, M.R. , Roberts, J.H. , Balint‐Kurti, P. and Holland, J.B. (2020) Heterosis of leaf and rhizosphere microbiomes in field‐grown maize. New Phytol. 228, 1055–1069. [DOI] [PubMed] [Google Scholar]
- Wallace, J. , Kremling, K.A. , Kovar, L. and Buckler, E. (2018) Quantitative genetics of the maize leaf microbiome. Phytobiomes, 2, 208–224. [Google Scholar]
- Walters, W.A. , Jin, Z. , Youngblut, N. , Wallace, J.G. , Sutter, J. , Zhang, W. , González‐Peña, A. et al. (2018) Large‐scale replicated field study of maize rhizosphere identifies heritable microbes. Proc. Natl Acad. Sci. USA, 115, 7368–7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, B. , Shen, H. , Yang, X. , Guo, T. , Zhang, B. and Yan, W. (2013) Effects of chitinase‐transgenic (McChit1) tobacco on the rhizospheric microflora and enzyme activities of the purple soil. Plant Soil Environ. 59, 241–246. [Google Scholar]
- Wang, C. , Li, Y. , Li, M. , Zhang, K. , Ma, W. , Zheng, L. , Xu, H. et al. (2021) Functional assembly of root‐associated microbial consortia improves nutrient efficiency and yield in soybean. J. Integr. Plant Biol. 63, 1021–1035. [DOI] [PubMed] [Google Scholar]
- Wang, L. , Xu, Q. , Yu, H. , Ma, H. , Li, X. , Yang, J. , Chu, J. et al. (2020) Strigolactone and karrikin signaling pathways elicit ubiquitination and proteolysis of SMXL2 to regulate hypocotyl elongation in Arabidopsis . Plant Cell, 32, 2251–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L. , Yang, T. , Lin, Q. , Wang, B. , Li, X. , Luan, S. and Yu, F. (2020) Receptor kinase FERONIA regulates flowering time in Arabidopsis . BMC Plant Biol. 20, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. , Wang, C. , Sui, J. , Liu, Z. , Li, Q. , Ji, C. , Song, X. et al. (2018) Isolation and characterization of phosphofungi, and screening of their plant growth‐promoting activities. AMB Express. 8, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. , Wei, Z. , Yang, K. , Wang, J. , Jousset, A. , Xu, Y. , Shen, Q. et al. (2019) Phage combination therapies for bacterial wilt disease in tomato. Nat. Biotechnol. 37, 1513–1520. [DOI] [PubMed] [Google Scholar]
- Wang, Y.Y. , Hsu, P.K. and Tsay, Y.F. (2012) Uptake, allocation and signaling of nitrate. Trends Plant Sci. 17, 458–467. [DOI] [PubMed] [Google Scholar]
- Wei, M.S. , Wei, F.X. , Su, M.L. , Xue, Q.Z. and Gang, Q.D. (2010) Responses of two rice cultivars differing in seedling‐stage nitrogen use efficiency to growth under low‐nitrogen conditions. Plant Soil, 326, 291–302. [Google Scholar]
- Wei, S. , Li, X. , Lu, Z. , Zhang, H. , Ye, X. , Zhou, Y. , Li, J. et al. (2022) A transcriptional regulator that boosts grain yields and shortens the growth duration of rice. Science, 377, 386–396. [DOI] [PubMed] [Google Scholar]
- Wen, T. , Hui, Y.G. , Dan, H.W. , Yuan, J. , Qing, N.G. , Hao, X.P. , Sheng, S.F. et al. (2022) Root exudate chemistry affects soil carbon mobilization via microbial community reassembly. Fundam. Res. 2, 697–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner, T. , Holst, K. , Pörs, Y. , Guivarc'h, A. , Mustroph, A. , Chriqui, D. , Grimm, B. et al. (2008) Cytokinin deficiency causes distinct changes of sink and source parameters in tobacco shoots and roots. J. Exp. Bot. 59, 2659–2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, L.J. , Jothibasu, K. , Reese, R.N. , Brözel, V.S. and Subramanian, S. (2015) Spatio temporal influence of isoflavonoids on bacterial diversity in the soybean rhizosphere. Mol. Plant Microbe Interact. 28, 22–29. [DOI] [PubMed] [Google Scholar]
- White, R.A. , Callister, S.J. , Moore, R.J. , Baker, E.S. and Jansson, J.K. (2016) The past, present and future of microbiome analyses. Nat. Protoc. 11, 2049–2053. [Google Scholar]
- Woll, K. , Borsuk, L.A. , Stransky, H. , Nettleton, D. , Schnable, P.S. and Hochholdinger, F. (2005) Isolation, characterization, and pericycle‐specific transcriptome analyses of the novel maize lateral and seminal root initiation mutant rum1. Plant Physiol. 139, 1255–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, P. , Shou, H. , Xu, G. and Lian, X. (2013) Improvement of phosphorus efficiency in rice on the basis of understanding phosphate signaling and homeostasis. Curr. Opin. Plant Biol. 16, 205–212. [DOI] [PubMed] [Google Scholar]
- Wu, X. , Li, R. , Shi, J. , Wang, J. , Sun, Q. , Zhang, H. , Xing, Y. et al. (2014) Brassica oleracea MATE encodes a citrate transporter and enhances aluminum tolerance in Arabidopsis thaliana . Plant Cell Physiol. 55, 1426–1436. [DOI] [PubMed] [Google Scholar]
- Xie, S. , Liu, J. , Gu, S. , Chen, X. , Jiang, H. and Ding, T. (2020) Antifungal activity of volatile compounds produced by endophytic Bacillus subtilis DZSY21 against Curvularia lunata . Ann. Microbiol. 70, 1–10. [Google Scholar]
- Xing, L. , Zhao, S.G. , Zheng, N. , Li, S.L. and Wang, J.Q. (2017) Advance in isolation and culture techniques of uncultured microbes: a review. Microbiol. China, 44, 3053–3066. [Google Scholar]
- Xiong, C. , Singh, B.K. , He, J.Z. , Han, Y.L. , Li, P.P. , Wan, L.H. , Meng, G.Z. et al. (2021) Plant developmental stage drives the differentiation in ecological role of the maize microbiome. Microbiome, 9, 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, H. , Li, Y. , Zhang, K. , Li, M. , Fu, S. , Tian, Y. , Qin, T. et al. (2021a) miR169c‐NFYA‐C‐ENOD40 modulates nitrogen inhibitory effects in soybean nodulation. New Phytol. 229, 3377–3392. [DOI] [PubMed] [Google Scholar]
- Xu, H. , Yang, Y. , Tian, Y. , Xu, R. , Zhong, Y. and Liao, H. (2020) Rhizobium inoculation drives the shifting of rhizosphere fungal community in a host genotype dependent manner. Front. Microbiol. 10, 3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, L. , Dong, Z. , Chiniquy, D. , Pierroz, G. , Deng, S. , Gao, C. , Diamond, S. et al. (2021b) Genome‐resolved metagenomics reveals role of iron metabolism in drought‐induced rhizosphere microbiome dynamics. Nat. Commun. 12, 3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, L. , Naylor, D. , Dong, Z. , Simmons, T. , Pierroz, G. , Hixson, K.K. , Kim, Y.M. et al. (2018) Drought delays development of the sorghum root microbiome and enriches for monoderm bacteria. Proc. Natl Acad. Sci. USA, 115, E4284–E4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, L. , Pierroz, G. , Wipf, H.M. , Gao, C. , Taylor, J.W. , Lemaux, P.G. and Coleman‐Derr, D. (2021c) Holo‐omics for deciphering plant‐microbiome interactions. Microbiome, 9, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y. , Ma, Y. , Hu, X. and Wang, J. (2019) Analysis of prospective microbiology research using third‐generation sequencing technology. Biodiv. Sci. 27, 534–542. [Google Scholar]
- Yang, J. , Lan, L. , Jin, Y. , Yu, N. , Wang, D. and Wang, E. (2022) Mechanisms underlying legume‐rhizobium symbioses. J. Integr. Plant Biol. 64, 244–267. [DOI] [PubMed] [Google Scholar]
- Yang, J.S. and Chen, S.S. (2000) Microarray technology and its application. Biotechnol. Adv. 18, 35–46. [DOI] [PubMed] [Google Scholar]
- Yang, S.Y. , Zhou, X.L. , Chen, Z.Y. and Chen, S.Y. (2017) Technical overview of the validation of gene function in plants. J. Anhui Agric. Sci. 45, 136–140. [Google Scholar]
- Yang, Y. , Gao, K.X. , Yan, W.U. and Liu, X.G. (2016) Indole‐3‐acetic acid‐mediated cross‐kingdom signalling involved in plant‐bacteria interactions. Biotechnol. Bull. 32, 14–21. [Google Scholar]
- Yashveer, S. , Singh, V. , Kaswan, V. , Kaushik, A. and Tokas, J. (2014) Green biotechnology, nanotechnology and bio‐fortification: perspectives on novel environment‐friendly crop improvement strategies. Biotechnol. Genet. Eng. Rev. 30, 113–126. [DOI] [PubMed] [Google Scholar]
- Yazaki, K. (2005) Transporters of secondary metabolites. Curr. Opin. Plant Biol. 8, 301–307. [DOI] [PubMed] [Google Scholar]
- Yokosho, K. , Yamaji, N. and Ma, J.F. (2011) An Al‐inducible MATE gene is involved in external detoxification of Al in rice. Plant J. 68, 1061–1069. [DOI] [PubMed] [Google Scholar]
- Yokosho, K. , Yamaji, N. , Ueno, D. , Mitani, N. and Ma, J.F. (2009) OsFRDL1 is a citrate transporter required for efficient translocation of iron in rice. Plant Physiol. 149, 297–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, P. , He, X. , Baer, M. , Beirinckx, S. , Tian, T. , Moya, Y.A.T. , Zhang, X. et al. (2021) Plant flavones enrich rhizosphere Oxalobacteraceae to improve maize performance under nitrogen deprivation. Nat. Plants, 7, 481–499. [DOI] [PubMed] [Google Scholar]
- Yuan, J. , Raza, W. and Shen, Q. (2018) Root exudates dominate the colonization of pathogen and plant growth‐promoting rhizobacteria. In Root Biology Soil Biology( Giri, B. , Prasad, R. and Varma, A. , eds), pp. 167–180. Cham: Springer International Publishing. [Google Scholar]
- Zancarini, A. , Westerhuis, J.A. , Smilde, A.K. and Bouwmeester, H.J. (2021) Integration of omics data to unravel root microbiome recruitment. Curr. Opin. Biotechnol. 70, 255–261. [DOI] [PubMed] [Google Scholar]
- Zaramela, L.S. , Moyne, O. , Kumar, M. , Zuniga, C. , Tibocha‐Bonilla, J.D. and Zengler, K. (2021) The sum is greater than the parts: exploiting microbial communities to achieve complex functions. Curr. Opin. Biotechnol. 67, 149–157. [DOI] [PubMed] [Google Scholar]
- Zengler, K. , Hofmockel, K. , Baliga, N.S. , Behie, S.W. , Bernstein, H.C. , Brown, J.B. , Dinneny, J.R. et al. (2019) EcoFABs: advancing microbiome science through standardized fabricated ecosystems. Nat. Methods, 16, 567–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zgadzaj, R. , Garrido‐Oter, R. , Jensen, D.B. , Koprivova, A. , Schulze‐Lefert, P. and Radutoiu, S. (2016) Root nodule symbiosis in Lotus japonicus drives the establishment of distinctive rhizosphere, root, and nodule bacterial communities. Proc. Natl Acad. Sci. USA, 113, E7996–E8005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhalnina, K. , Louie, K.B. , Hao, Z. , Mansoori, N. , da Rocha, U.N. , Shi, S. , Cho, H. et al. (2018) Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat. Microbiol. 3, 470–480. [DOI] [PubMed] [Google Scholar]
- Zhang, H. , Zhao, Y. and Zhu, J.K. (2020) Thriving under stress: how plants balance growth and the stress response. Dev. Cell, 55, 529–543. [DOI] [PubMed] [Google Scholar]
- Zhang, J. , Liu, Y.X. , Zhang, N. , Hu, B. , Jin, T. , Xu, H. , Qin, Y. et al. (2019) NRT1.1B is associated with root microbiota composition and nitrogen use in field‐grown rice. Nat. Biotechnol. 37, 676–684. [DOI] [PubMed] [Google Scholar]
- Zhang, J. , Liu, Y.X. , Guo, X. , Qin, Y. , Garrido-Oter, R. , Schulze-Lefert, P. and Bai, Y. (2021) High-throughput cultivation and identification of bacteria from the plant root microbiota. Nat Protoc. 16, 988–1012. [DOI] [PubMed] [Google Scholar]
- Zhang, Y.N. , Zhang, Z.Y. and Chen, Q. (2017) Research progress in regulation mechanism for organic acid transporter gene expression in plants under aluminum stress. MPB, 15, 4899–4904. [Google Scholar]
- Zhao, Z.X. , Feng, Q. , Liu, P.Q. , He, X.R. , Zhao, J.H. , Xu, Y.J. , Zhang, L.L. et al. (2021) RPW8.1 enhances the ethylene‐signaling pathway to feedback‐attenuate its mediated cell death and disease resistance in Arabidopsis . New Phytol. 229, 516–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, Y. , Xun, W. , Wang, X. , Tian, S. , Zhang, Y. , Li, D. , Zhou, Y. et al. (2022) Root‐secreted bitter triterpene modulates the rhizosphere microbiota to improve plant fitness. Nat. Plants, 8, 887–896. [DOI] [PubMed] [Google Scholar]
- Zhong, Y. , Yang, Y. , Liu, P. , Xu, R. , Rensing, C. , Fu, X. and Liao, H. (2019) Genotype and rhizobium inoculation modulate the assembly of soybean rhizobacterial communities. Plant Cell Environ. 42, 2028–2044. [DOI] [PubMed] [Google Scholar]
- Zhou, F. , Emonet, A. , Dénervaud Tendon, V. , Marhavy, P. , Wu, D. , Lahaye, T. and Geldner, N. (2020) Co‐incidence of damage and microbial patterns controls localized immune responses in roots. Cell, 180, 440–453.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, X.H. , Tian, F. , Du, L.P. , Wang, K. , Lin, Z.S. and Ye, X.G. (2012) Advances in molecular biology research of interaction between plants and beneficial microorganisms and their applications in plant improvement. Scientia Agricultura Sinica, 45, 2801–2814. [Google Scholar]
- Zhou, Y. , Ma, Y. , Zeng, J. , Duan, L. , Xue, X. , Wang, H. , Lin, T. et al. (2016) Convergence and divergence of bitterness biosynthesis and regulation in Cucurbitaceae. Nat. Plants, 2, 16183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y. , Yang, Z. , Xu, Y. , Sun, H. , Sun, Z. , Lin, B. , Sun, W. et al. (2018) Soybean NADP‐malic enzyme functions in malate and citrate metabolism and contributes to their efflux under al stress. Front. Plant Sci. 8, 2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, C.Q. , Zhu, X.F. , Hu, A.Y. , Wang, C. , Wang, B. , Dong, X.Y. and Shen, R.F. (2016) Differential effects of nitrogen forms on cell wall phosphorus remobilization are mediated by nitric oxide, pectin content, and phosphate transporter expression. Plant Physiol. 171, 1407–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J.K. (2016) Abiotic stress signaling and responses in plants. Cell, 167, 313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, S. , Estévez, J.M. , Liao, H. , Zhu, Y. , Yang, T. , Li, C. , Wang, Y. et al. (2020) The RALF1‐FERONIA complex phosphorylates eIF4E1 to promote protein synthesis and polar root hair growth. Mol. Plant, 13, 698–716. [DOI] [PubMed] [Google Scholar]
- Zhuang, L. , Li, Y. , Wang, Z. , Yu, Y. , Zhang, N. , Yang, C. , Zeng, Q. et al. (2021) Synthetic community with six Pseudomonas strains screened from garlic rhizosphere microbiome promotes plant growth. Microb. Biotechnol. 14, 488–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, S. and Frey, B. (2002) Soil respiration and microbial properties in an acid forest soil: effects of wood ash. Soil Biol. Biochem. 34, 1727–1737. [Google Scholar]


