Abstract
Background and Aims
The coronavirus disease 2019 (COVID‐19) has brought serious threats to public health worldwide. Nasopharyngeal, nasal swabs, and saliva specimens are used to detect severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2). However, limited data are available on the performance of less invasive nasal swab for testing COVID‐19. This study aimed to compare the diagnostic performance of nasal swabs with nasopharyngeal swabs using real‐time reverse transcription polymerase chain reaction (RT‐PCR) considering viral load, onset of symptoms, and disease severity.
Methods
A total of 449 suspected COVIDCOVID‐19 individuals were recruited. Both nasopharyngeal and nasal swabs were collected from the same individual. Viral RNA was extracted and tested by real‐time RT‐PCR. Metadata were collected using structured questionnaire and analyzed by SPSS and MedCalc software.
Results
The overall sensitivity of the nasopharyngeal swab was 96.6%, and the nasal swab was 83.4%. The sensitivity of nasal swabs was more than 97.7% for low and moderate C t values. Moreover, the performance of nasal swab was very high (>87%) for hospitalized patients and at the later stage >7 days of onset of symptoms.
Conclusion
Less invasive nasal swab sampling with adequate sensitivity can be used as an alternative to nasopharyngeal swabs for the detection of SARS‐CoV‐2 by real‐time RT‐PCR.
Keywords: COVID‐19, nasal swab, nasopharyngeal swab, real time RT‐PCR, SARS‐CoV‐2, sensitivity
1. INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) spread rapidly worldwide, causing more than 649 million confirmed cases and 6.6 million death till November 2022 (Worldometer, November 2022). Considering the high transmissibility of SARS‐CoV‐2 and the financial toll on healthcare systems, early and precise detection is critical to its control. 1 Many asymptomatic SARS‐CoV‐2 patients, before being identified as symptomatic, have interacted with healthy people due to the lack of appropriate detection assays. 2
Several sampling methods and commercially available kits are used to detect SARS‐CoV‐2. Nasopharyngeal swab is used as the gold standard recommended by World Health Organization (WHO). 3 , 4 However, specimen collection from nasopharynges can be uncomfortable and relatively invasive procedure and may cause sneezing, coughing, and even bleeding. This may also produce aerosols which pose a risk of infection to healthcare personnel. In contrast, nasal swab, saliva, and oropharyngeal (throat) swabs are used as alternatives. 5 , 6 , 7 , 8 Especially nasal swab has been used as a preferred specimen for detecting many respiratory viral RNA, as it can avoid the uncomfortable sampling procedure. 9 , 10 , 11 Additionally, it can be self‐collected which is quicker, more bearable, and reduce the use of stringent personal protective equipment (PPE). As a result, self‐collected nasal swab may encourage patients to provide specimen and can help in early detection and prevent transmission. 12 , 13 Thus, nasal swab is more suitable than nasopharyngeal sampling for mass screening. 14 , 15
The literature review and meta‐analyses currently showed that nasal swab had parallel or acceptable sensitivities compared with nasopharyngeal swab 6 with some inconsistent results. 12 , 13 , 16 , 17 , 18 , 19 Most of the studies were based on the limited number of specimens 19 , 20 , 21 focused on particular groups, especially adults (above 18 years), 13 , 22 used nasal swabs in combination with other types, 13 , 17 or symptomatic cases only. 23 Moreover, the performance of nasal swabs in these studies was dependent on collection media and procedure. 6 In this study, we report a comparative analysis of the overall performance of nasal swab and nasopharyngeal swab based on viral load, diseases severity, hospitalization, and incubation periods of SARS‐CoV‐2.
2. MATERIALS AND METHODS
2.1. Ethical clearance
This study was approved by the icddr,b Institutional Review Board (PR‐21065). We obtained informed consent from all the participants before collecting specimen and data. Proper biosafety and biosecurity protocols were maintained during specimen collection and transportation.
2.2. Participant enrollment and specimen collection
A total of 449 COVID‐19 suspected individuals were recruited from February 6 to March 7, 2022. The following enrollment criteria were set for the study participants: (i) all age groups, and either sex; (ii) acute onset of fever or cough or any three or more of the presented signs and symptoms recommended by WHO for COVID‐19 testing; (iii) both hospitalized and community people; (iv) both symptomatic and asymptomatic cases who visited the health facility. Even though no infants or babies attended our clinic for COVID‐19 testing in the present study. The patients were enrolled at different time points after infections and we categorized them into three arms based on the onset of illness at sample collection: 0–3 days, 4–7 days, or > 7 days of onset of symptoms.
Trained nurses and medical technologists collected nasopharyngeal and nasal swabs from same individuals following the standard sample collection protocol and placed in a separate tube of 1 mL viral transport media for real‐time reverse transcription polymerase chain reaction (RT‐PCR). Then the specimens were stored at the refrigerator for 2–8°C until tested.
2.3. Real‐time RT‐PCR
Viral RNA was extracted from nasopharyngeal swabs and nasal swab using Qiagen miniElute Viral RNA extraction kit. COVID‐19 detection was performed by using a WHO‐recommended, semi‐quantitative, probe‐based real‐time RT‐PCR assay. In brief, we prepared the real‐time RT‐PCR reaction mixtures using iTaq™ Universal Probes and One‐Step Reaction Mix (Bio‐Rad Laboratories, Inc.) in CFX96 Touch™ Real‐time PCR Detection System (Bio‐Rad Laboratories, Inc.). In this PCR system, two genes (RdRp and N genes) of SARS‐CoV‐2 were targeted and tested according to the protocol suggested by the Chinese Center for Disease Control and Prevention (China CDC) and WHO. 24 , 25
2.4. Data analysis
If either nasopharyngeal or nasal swab was positive by real‐time RT‐PCR (Ct<37), the specimen was regarded as true positive. Data were analyzed using SPSS version 22.0. In addition, we calculated the sensitivity, specificity, and accuracy with a 95% confidence interval (95% CI) through online based‐software MedCalc (https://www.medcalc.org/calc/diagnostic_test.php). The sensitivity of nasopharyngeal swab or nasal swab is the ability of detecting true positive individuals who had confirmed SARS‐CoV‐2 using real‐time PCR (positive either in nasopharyngeal swab or nasal swab).
The specificity of nasopharyngeal swab or nasal swab is the ability of detecting the negative individuals who were confirmed negative for SARS‐CoV‐2 using real‐time PCR (negative for both nasopharyngeal swab and nasal swab).
The formula was: Diagnostic sensitivity = a/(a + c) × 100 (%) (95% CI), specificity = d/(b + d) × 100 (%) (95% CI) and accuracy = a + d/(a + b + c + d) × 100 (%) (95% CI); where a, true positive (TP); b, false positive (FP); c, false‐negative (FN); and d, true negative (TN).
3. RESULTS
3.1. Metadata analysis of the study participants
Among 449 suspected cases, (230 hospitalized, 219 nonhospitalized), 279 were male and 170 female and their average age was 41.19 ± 18.25 years. The highest number of the participants were within 18–30 years (n = 125) followed by 31–40 years (n = 100). Regarding the onset of symptoms, 83, 180, and 168 individuals submitted their specimens within 0–3 days, 4–7 days, and more than 7 days, respectively. Considering different clinical features of the suspected cases, cough (66%) was manifested as the highest followed by runny nose (62.8%), headache (39.4%), and fever (38.5%). Moreover, 124 individuals had comorbid conditions and 18 were asymptomatic (Table 1).
Table 1.
Demographic and clinical data.
| Metadata | Participants, n = 449 |
|---|---|
| Sex (n, %) | |
| Male | 279 (62) |
| Female | 170 (38) |
| Age group (years) (n, %) | |
| <5 | 1 (0.2) |
| 5–17 | 26 (5.8) |
| 18–30 | 125 (27.8) |
| 31–40 | 100 (22.3) |
| 41–50 | 62 (13.8) |
| 51–60 | 56 (12.5) |
| >60 | 79 (17.6) |
| Agea | 41.19 ± 18.25 |
| Clinical features | |
| Fever (n, %) | 173 (38.5) |
| Cough (n, %) | 310 (66) |
| Runny nose (n, %) | 282 (62.8) |
| Sore throat (n, %) | 73 (16.3) |
| Shortness of breath (n, %) | 75 (16.7) |
| Chills (n, %) | 16 (3.6) |
| Vomiting (n, %) | 35 (7.8) |
| Nausea (n, %) | 37 (8.2) |
| Diarrhea (n, %) | 20 (4.5) |
| Altered smell (n, %) | 27 (6) |
| Headache (n, %) | 177 (39.42) |
| Conjunctivitis (n, %) | 4 (0.9) |
| Muscle aches (n, %) | 105 (23.4) |
| Joint aches (n, %) | 58 (12.9) |
| Loss of appetite (n, %) | 96 (21.4) |
| Altered consciousness (n, %) | 3 (0.7) |
| Onset of symptoms | |
| 0–3 days | 83 (18.5) |
| 4–7 days | 180 (40.1) |
| >7 days | 168 (37.4) |
| No symptoms | 18 (4.0) |
| Hospitalization | |
| Yes | 230 (70) |
| No | 219 (30) |
| Comorbidities (n, %) | |
| Yes | 124 (27.6) |
| No | 325 (62.4) |
| Body temperature (⁰C)a | 36.39 ± 0.58 |
| Respiratory rate (/minutes)a | 19.49 ± 1.76 |
| Pulse rate (/minutes)a | 82.25 ± 9.88 |
| Systolic blood pressure (mm/Hg)a | 115.45 ± 11.11 |
| Diastolic blood pressure (mm/Hg)a | 75.88 ± 8.10 |
| Oxygen saturation (%)a | 98.10 ± 1.23 |
Note: Comorbidity includes obesity, cancer, diabetes, asthma, heart diseases, lung diseases (nonasthma), liver diseases, kidney diseases.
Abbreviation: n, study participant.
Results presented as mean ± SD.
3.2. Diagnostic performance between nasopharyngeal and nasal swab
The diagnostic performance between nasopharyngeal and nasal swab is shown in Figure 1 and Supporting Information: Table S1. We compared the sensitivity, specificity, and accuracy of nasopharyngeal and nasal swabs by testing through the gold standard method real‐time RT‐PCR. The overall sensitivity of nasopharyngeal and nasal swabs was 96.6% and 83.4%, respectively (Figure 1). The specificity for both nasopharyngeal and nasal swabs was 100% and the accuracy was 98.4% and 92.4%, respectively (Supporting Information: Table S1).
Figure 1.
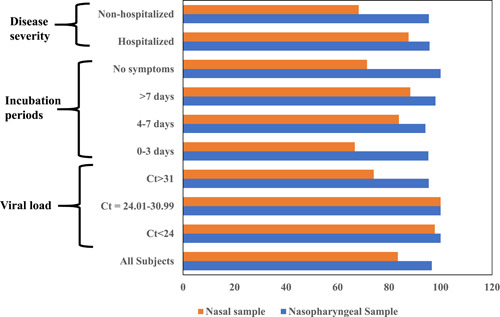
Comparison of sensitivity of nasopharyngeal and nasal swabs. Sensitivity was calculated by, True positive/(True positive+ False negative) through MedCalc Software.
The performance of nasopharyngeal and nasal swabs was analyzed based on real‐time PCR‐generated C t values. The sensitivity of nasopharyngeal swabs, in case of low C t <24, was 100% while nasal swabs showed 97.7%. Interestingly, the sensitivity was 100% for both of nasopharyngeal and nasal swabs with moderate C t values, 24–31. Although sensitivity for both specimens was declined with high C t values, >31, the nasopharyngeal swabs showed significantly higher (95.4%) performance compared with nasal swabs (74%) (Figure 1).
We also evaluated the sensitivity based on the onset of symptoms. Participants with 0–3 days of symptoms showed 95.2% sensitivity for nasopharyngeal swabs and 67% for nasal swabs. In case of 4–7 days of symptoms, nasopharyngeal and nasal swabs showed the sensitivity 94.1% and 83.8%, respectively. We observed the highest sensitivity among the group with >7 days of symptoms; 98% for nasopharyngeal swab, and 88.2% for nasal swab. In case of asymptomatic individuals, the sensitivity of nasopharyngeal swabs was 100% while only 71.4% for nasal swabs (Figure 1).
The sensitivity of nasopharyngeal swabs was almost similar for both of the hospitalized and nonhospitalized patients (96%). Whereas, the sensitivity of nasal swabs was found higher in severe cases for example, hospitalized patients (87.6%) compared with the nonhospitalized patients (68.2%) (Figure 1).
4. DISCUSSION
In this study, we compared the diagnostic performance of nasopharyngeal and nasal swabs for the COVID‐19 detection using real‐time PCR results. Although, nasopharyngeal swab was found to be more sensitive, nasal swabs could also be used as an alternative specimen type for symptomatic and severe cases with sensitivity and specificity >80%.
The performance of nasal swabs was analyzed considering different factors such as C t value, the onset of symptoms, and disease severity. We observed that the sensitivity of the nasopharyngeal and nasal swabs was almost similar in lower to moderate C t values due to high to medium viral load while the sensitivity of nasal swabs was declined to a greater extent with higher C t value than nasopharyngeal swab. Different studies showed similar performance (75%–100%) of nasal swab. 20 , 22 , 26 , 27 , 28 It is likely that the nasopharynx consumes the highest SARS‐CoV‐2 viral load. Several studies explained that the patients tested with high C t >31 value had lower viral load and noninfectious. 29 , 30 So, the low sensitivity in high C t value will not affect the transmissibility of live SARS‐CoV‐2 which is observed in our study. Based on the onset of symptoms, nasal swabs showed lower performance at the earlier stage (less than 3 days of onset of symptoms) while nasopharyngeal swabs showed higher performance all through the disease progression. Similarly, the performance of nasal swab was very low in an asymptomatic patient (73.4%).
We also observed that sensitivity and specificity of nasal swabs were higher among hospitalized patient rather than nonhospitalized patient that was similar with other studies. 21 , 31 Most of the hospitalized patients had a severe illness and more chance of spreading the virus into different parts of the body. Therefore, more viruses were detected in the nasal swabs of hospitalized patients with higher sensitivity. In contrast, nonhospitalized patients had mild symptoms and the virus might be localized in the respiratory tract only. Therefore, the sensitivity of nasal swab in nonhospitalized patients was lower than in hospitalized ones. As it is difficult to collect nasopharyngeal swabs from hospitalized, especially intensive care unit (ICU)‐admitted patients with auxiliary oxygen supply, nasal swab could be a right choice. Another important point from our finding is that, if the participants with low viral load are excluded from the study, the sensitivity of the nasopharyngeal and nasal swabs are almost similar in all conditions (data not shown).
Nasal swab has definite advantages over nasopharyngeal swab, such as more comfortable, less invasive, and easy for self‐collection. In addition, nasal swab collection does not require trained medical staff and minimize the risk of transmission. 32 , 33 , 34 , 35 , 36 , 37 Thus, nasal swab sampling may encourage more people for COVID‐19 testing and reduce the transmissibility of the diseases.
The study had one limitation. We could not enroll an equal number of patients in different groups such as very few participants were enrolled in 0–3 days of symptoms onset. It might have some effects on the sensitivity of nasal swab.
In conclusion, nasal swabs can be an easy and potential alternative sample source for SARS‐CoV‐2 detection which is less invasive and have adequate sensitivity.
AUTHOR CONTRIBUTIONS
Kamrun Nahar: Data curation; investigation; writing—original draft. Mst Noorjahan Begum: Investigation; validation; writing—review & editing. Selim Reza Tony: Formal analysis; writing—review & editing. Mohammad Jubair: Data curation. Md. Abir Hossain: Data curation. Yeasir Karim: Investigation. Abdullah Al. Faisal: Investigation. Mohammad Enayet Hossain: Writing—review & editing. Mohammed Ziaur Rahman: Writing—review & editing. Mustafizur Rahman: Conceptualization; funding acquisition; methodology; project administration; supervision; writing—review & editing.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
TRANSPARENCY STATEMENT
The lead author Mustafizur Rahman affirms that this manuscript is an honest, accurate, and transparent account of the study being reported; that no important aspects of the study have been omitted; and that any discrepancies from the study as planned (and, if relevant, registered) have been explained.
Supporting information
Supporting information.
ACKNOWLEDGMENTS
The study was funded by the Global affairs Canada approved by the icddr,b institutional review board (PR‐21065). icddr,b is also appreciative to the core donors; the Government of Bangladesh, Canada, Sweden, and the UK for providing core/unrestricted support.
Nahar K, Begum MN, Tony SR, et al. Nasal swab as an alternative specimen for the detection of severe acute respiratory syndrome coronavirus 2. Health Sci Rep. 2023;6:e1213. 10.1002/hsr2.1213
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Gable P,Huang JY, Gilbert SE, et al. A comparison of less invasive severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) diagnostic specimens in nursing home Residents—Arkansas, June–August 2020. Clin Infect Dis. 2021;73:S58‐S64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ji T, Liu Z, Wang G, et al. Detection of COVID‐19: a review of the current literature and future perspectives. Biosens Bioelectron. 2020;166:112455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. World Health Organization . Laboratory Testing for Coronavirus Disease 2019 (COVID‐19) in Suspected Human Cases. World Health Organization; 2020a:1‐7. [Google Scholar]
- 4. Zou L, Ruan F, Huang M, et al. SARS‐CoV‐2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382:1177‐1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hanson KE, Caliendo AM, Arias CA, et al. The infectious diseases society of america guidelines on the diagnosis of COVID‐19: molecular diagnostic testing. Clin Infect Dis. 2021:ciab048. 10.1093/cid/ciab048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lee RA, Herigon JC, Benedetti A, Pollock NR, Denkinger CM. Performance of saliva, oropharyngeal swabs, and nasal swabs for SARS‐CoV‐2 molecular detection: a systematic review and meta‐analysis. J Clin Microbiol. 2021;59:e02881‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Griesemer SB, Van Slyke G, Ehrbar D, et al. Evaluation of specimen types and saliva stabilization solutions for SARS‐CoV‐2 testing. J Clin Microbiol. 2021;59:e01418‐e01420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Callahan C, Lee RA, Lee GR, Zulauf K, Kirby JE, Arnaout R. Nasal swab performance by collection timing, procedure, and method of transport for patients with SARS‐CoV‐2. J Clin Microbiol. 2021;59:e0056921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Seaman CP, Tran LTT, Cowling BJ, Sullivan SG. Self‐collected compared with professional‐collected swabbing in the diagnosis of influenza in symptomatic individuals: a meta‐analysis and assessment of validity. J Clin Virol. 2019;118:28‐35. [DOI] [PubMed] [Google Scholar]
- 10. Haussig JM, Targosz A, Engelhart S, et al. Feasibility study for the use of self‐collected nasal swabs to identify pathogens among participants of a population‐based surveillance system for acute respiratory infections (GrippeWeb‐Plus)‐Germany, 2016. Influenza Other Respir Viruses. 2019;13:319‐330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Frazee BW, Rodríguez‐Hoces de la Guardia A, Alter H, et al. Accuracy and discomfort of different types of intranasal specimen collection methods for molecular influenza testing in emergency department patients. Ann Emerg Med. 2018;71:509‐517. [DOI] [PubMed] [Google Scholar]
- 12. To KKW, Tsang OTY, Leung WS, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS‐CoV‐2: an observational cohort study. Lancet Infect Dis. 2020;20:565‐574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Berenger BM, Fonseca K, Schneider AR, et al. Sensitivity of nasopharyngeal, nasal and throat swab for the detection of SARS‐CoV‐2. MedRxiv. 2020. [Google Scholar]
- 14. Nagura‐Ikeda M, Imai K, Tabata S, et al. Clinical evaluation of self‐collected saliva by quantitative reverse transcription‐PCR (RT‐qPCR), direct RT‐qPCR, reverse transcription‐loop‐mediated isothermal amplification, and a rapid antigen test to diagnose COVID‐19. J Clin Microbiol. 2020;58:e01438‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Iwasaki S, Fujisawa S, Nakakubo S, et al. Comparison of SARS‐CoV‐2 detection in nasopharyngeal swab and saliva. J Infect. 2020;81:e145‐e147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. LeBlanc JJ, Heinstein C, MacDonald J, Pettipas J, Hatchette TF, Patriquin G. A combined oropharyngeal/nares swab is a suitable alternative to nasopharyngeal swabs for the detection of SARS‐CoV‐2. J Clin Virol. 2020;128:104442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wehrhahn MC, Robson J, Brown S, et al. Self‐collection: an appropriate alternative during the SARS‐CoV‐2 pandemic. J Clin Virol. 2020;128:104417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Basu A, Zinger T, Inglima K, et al. Performance of abbott ID now COVID‐19 rapid nucleic acid amplification test using nasopharyngeal swabs transported in viral transport media and dry nasal swabs in a New York city academic institution. J Clin Microbiol. 2020;58:e01136‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kojima N, Turner F, Slepnev V, et al. Self‐collected oral fluid and nasal swabs demonstrate comparable sensitivity to clinician collected nasopharyngeal swabs for coronavirus disease 2019 detection. Clin Infect Dis. 2021;73:e3106‐e3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Péré H, Podglajen I, Wack M, et al. Nasal swab sampling for SARS‐CoV‐2: a convenient alternative in times of nasopharyngeal swab shortage. J Clin Microbiol. 2020;58(6):e00721‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pinninti S, Trieu C, Pati SK, et al. Comparing nasopharyngeal and midturbinate nasal swab testing for the identification of severe acute respiratory syndrome coronavirus 2. Clin Infect Dis. 2021;72:1253‐1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Callahan C, Lee RA, Lee GR, Zulauf K, Kirby JE, Arnaout R. Nasal‐Swab Testing Misses Patients with Low SARS‐CoV‐2 Viral Loads. medRxiv. 2020:06.12.20128736. 10.1101/2020.06.12.20128736 [DOI] [Google Scholar]
- 23. Harrington A, Cox B, Snowdon J, et al. Comparison of abbott ID now and Abbott m2000 methods for the detection of SARS‐CoV‐2 from nasopharyngeal and nasal swabs from symptomatic patients. J Clin Microbiol. 2020;58:e00798‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. CDC . China CDC Primers and Probes For Detection 2019‐nCoV. CDC; 2020. [Google Scholar]
- 25. World Health Organization . WHO In‐House Assays for 2019‐ Novel Coronavirus (2019‐nCoV) Real‐Time rRT‐PCR. WHO; 2020b. [Google Scholar]
- 26. Tu YP, Jennings R, Hart B, et al. Swabs collected by patients or health care workers for SARS‐CoV‐2 testing. N Engl J Med. 2020;383:494‐496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Berenger BM, Conly JM, Fonseca K, et al. Saliva collected in universal transport media is an effective, simple and high‐volume amenable method to detect SARS‐CoV‐2. Clin Microbiol Infect. 2021;27:656‐657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Arnaout R, Lee RA, Lee GR, et al. The limit of detection matters: the case for benchmarking severe acute respiratory syndrome coronavirus 2 testing. Clin Infect Dis. 2021;73:e3042‐e3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. La Scola B, Le Bideau M, Andreani J, et al. Viral RNA load as determined by cell culture as a management tool for discharge of SARS‐CoV‐2 patients from infectious disease wards. Eur J Clin Microbiol Infect Dis. 2020;39:1059‐1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li P, Fu JB, Li KF, et al. Transmission of COVID‐19 in the terminal stages of the incubation period: a familial cluster. Int J Infect Dis. 2020;96:452‐453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jamal AJ, Mozafarihashjin M, Coomes E, et al. Sensitivity of nasopharyngeal swabs and saliva for the detection of severe acute respiratory syndrome coronavirus 2. Clin Infect Dis. 2021;72:1064‐1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. He X, Lau EHY, Wu P, et al. Temporal dynamics in viral shedding and transmissibility of COVID‐19. Nature Med. 2020;26:672‐675. [DOI] [PubMed] [Google Scholar]
- 33. Huang CG, Lee KM, Hsiao MJ, et al. Culture‐based virus isolation to evaluate potential infectivity of clinical specimens tested for COVID‐19. J Clin Microbiol. 2020;58:e01068‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jefferson T, Spencer EA, Brassey J, Heneghan C. Viral cultures for coronavirus disease 2019 infectivity assessment: a systematic review. Clin Infect Dis. 2021;73:e3884‐e3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wölfel R, Corman VM, Guggemos W, et al. Virological assessment of hospitalized patients with COVID‐2019. Nature. 2020;581:465‐469. [DOI] [PubMed] [Google Scholar]
- 36. Yamada S, Fukushi S, Kinoshita H, et al. Assessment of SARS‐CoV‐2 infectivity of upper respiratory specimens from COVID‐19 patients by virus isolation using VeroE6/TMPRSS2 cells. BMJ Open Respir Res. 2021;8:e000830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yang J, Zheng Y, Gou X, et al. Prevalence of comorbidities and its effects in patients infected with SARS‐CoV‐2: a systematic review and meta‐analysis. Int J Infect Dis. 2020;94:91‐95. 10.1016/j.ijid.2020.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


