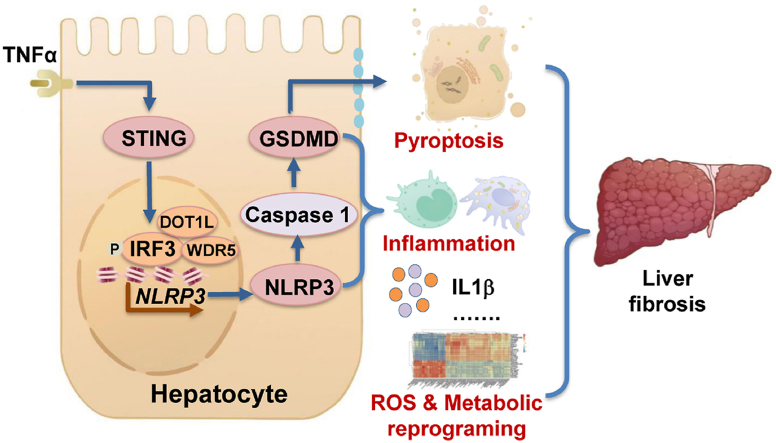
Abstract
The activation of stimulator of interferon genes (STING) and NOD-like receptor protein 3 (NLRP3) inflammasome-mediated pyroptosis signaling pathways represent two distinct central mechanisms in liver disease. However, the interconnections between these two pathways and the epigenetic regulation of the STING-NLRP3 axis in hepatocyte pyroptosis during liver fibrosis remain unknown. STING and NLRP3 inflammasome signaling pathways are activated in fibrotic livers but are suppressed by Sting knockout. Sting knockout ameliorated hepatic pyroptosis, inflammation, and fibrosis. In vitro, STING induces pyroptosis in primary murine hepatocytes by activating the NLRP3 inflammasome. H3K4-specific histone methyltransferase WD repeat-containing protein 5 (WDR5) and DOT1-like histone H3K79 methyltransferase (DOT1L) are identified to regulate NLRP3 expression in STING-overexpressing AML12 hepatocytes. WDR5/DOT1L-mediated histone methylation enhances interferon regulatory transcription factor 3 (IRF3) binding to the Nlrp3 promoter and promotes STING-induced Nlrp3 transcription in hepatocytes. Moreover, hepatocyte-specific Nlrp3 deletion and downstream Gasdermin D (Gsdmd) knockout attenuate hepatic pyroptosis, inflammation, and fibrosis. RNA-sequencing and metabolomics analysis in murine livers and primary hepatocytes show that oxidative stress and metabolic reprogramming might participate in NLRP3-mediated hepatocyte pyroptosis and liver fibrosis. The STING-NLRP3-GSDMD axis inhibition suppresses hepatic ROS generation. In conclusion, this study describes a novel epigenetic mechanism by which the STING-WDR5/DOT1L/IRF3-NLRP3 signaling pathway enhances hepatocyte pyroptosis and hepatic inflammation in liver fibrosis.
Keywords: Liver cirrhosis, GSDMD, IRF3, Histone methylation, Oxidative stress, Metabolic reprogramming
Graphical abstract
Highlights
-
•
A new role for STING in inducing hepatocyte pyroptosis by activating the NLRP3 inflammasome in hepatocytes was discovered.
-
•
A novel epigenetic mechanism by which STING enhanced NLRP3 expression by increasing histone methylation was demonstrated.
-
•
Hepatocyte pyroptosis might promote liver fibrosis via disturbing hepatic metabolic reprogramming and oxidative stress.
Abbreviations
- αSMA
α-smooth muscle actin
- ASC
apoptosis-associated speck like protein
- ALT
alanine aminotransferase
- AST
aspartate aminotransferase
- BDL
bile duct ligation
- CCl4
carbon tetrachloride
- CCL5
C–C motif chemokine ligand 5
- ChIP
chromatin immunoprecipitation
- cl-Caspase 1
cleaved caspase 1
- cl-GSDMD
cleaved GSDMD
- Co-IP
co-immunoprecipitation
- CYP2E1
cytochromeP450 2E1
- DEGs
differentially expressed genes
- DHE
dihydroethidium
- DOT1L
DOT1-like histone H3K79 methyltransferase
- GSDMD
gasdermin D
- H&E
hematoxylin and eosin
- HPLC
high-performance liquid chromatography
- HSCs
hepatic stellate cells
- IF
immunofluorescence
- IFN I
type I interferon
- IHC
immunohistochemistry
- IL
interleukin
- IRF3
interferon regulatory transcription factor 3
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LPS
lipopolysaccharide
- NLRP3
NOD-like receptor protein 3
- PI
propidium iodide
- p-IRF3
phosphorylated IRF3
- p-p65
phosphorylated NFκB-p65
- qPCR
quantitative real-time polymerase chain reaction
- ROS
reactive oxygen species
- SEM
scanning electron microscopy
- STING
stimulator of interferon genes
- TAA
thioacetamide
- TCA
the citric acid
- TEM
transmission electron microscopy
- TNFα
tumor necrosis factor α
- WB
Western blot
- WDR5
WD repeat-containing protein 5
- 4HNE
4-hydroxynonenal
1. Introduction
Hepatocytes, the major parenchymal cells in the liver, play crucial roles in protein synthesis, metabolism, detoxification, and immune regulation. Continuous stimulation of etiologic factors and oxidative stress leads to hepatocyte death [1,2]. In turn, dying hepatocytes release proinflammatory and profibrogenic cytokines to activate hepatic stellate cells (HSCs), leading to their activation, the deposition of extracellular matrix, and ultimately liver cirrhosis [3,4]. Liver cirrhosis is a common end-stage chronic liver disease [5,6]. During chronic liver diseases developing into liver cirrhosis, hepatocyte death results in abnormal liver function, decompensation, liver failure, and even death of the patients [1,7,8]. As hepatocytes are the central metabolic cells for glucose, lipids, and protein, hepatocyte death also leads to metabolic reprogramming, which refers to cells altering their metabolism to support the increased energy demand [9,10]. Thus, hepatocyte death represents a major event in the initiation and development of hepatic inflammation and liver cirrhosis. However, how aberrant hepatocyte death and metabolic reprogramming are induced in response to liver injury is incompletely understood.
Previous studies emphasized the crucial role of apoptosis and necrosis during liver cirrhosis. Pyroptosis is a recently described programmed cell death characterized by nuclear pyknosis, DNA breakage, cell membrane rupture and membranous vesicle formation, and the release of reactive oxygen species (ROS) and inflammatory cytokines [11]. While different from other types of cell death, recent evidence suggests that pyroptosis may initiate local inflammation via releasing inflammatory cytokines and recruiting immune cells, thus playing a pivotal role in the development of chronic liver diseases [8,12]. Pyroptosis is induced by apoptosis-speck-like protein containing a caspase recruitment domain (ASC), NOD-like receptors, and Caspase 1 protein components containing inflammasome activation, such as the most typical NOD-like receptor protein 3 (NLRP3) inflammasome [13]. The NLRP3 inflammasome, which is a complex of NLRP3, ASC and Caspase 1, can be activated by bacterial, viral, and endogenous damage-associated molecular patterns [11]. The activated NLRP3 inflammasome can either amplify the inflammatory response by promoting mature interleukin (IL)-1β and IL18 secretion or induce pyroptosis by cleaving gasdermin D (GSDMD) to form pyroptotic membrane pores [11,14]. Although pyroptosis was first found in macrophages, increasing evidence indicates that NLRP3 inflammasome-mediated hepatocyte pyroptosis is involved in the development of metabolic-associated fatty liver disease and alcoholic hepatitis [7,15,16]. However, the role and underlying mechanism of NLRP3 inflammasome-mediated hepatocyte pyroptosis in liver cirrhosis remain unclear.
The stimulator of interferon genes (STING) signaling pathway, part of the innate immune system activated by cytosolic DNA [17], has been shown to be involved in various liver diseases [18,19]. The STING signaling pathway could cause priming and activation of the NLRP3 inflammasome in immune cells [20]. However, it remains unknown whether STING is also involved in regulating NLRP3 inflammasome-mediated hepatocyte pyroptosis in the context of liver cirrhosis. Upon cytosolic DNA stimulation, STING activates the interferon regulatory transcription factor 3 (IRF3) and NFκB signaling pathways. This leads to the production of ROS and proinflammatory cytokines, such as type I interferon (IFN I) and tumor necrosis factor α (TNFα) [21]. STING can also regulate epigenetic modifications such as histone methylation, which further affect the transcription of inflammatory genes [22,23]. It seems that the activation of STING and NLRP3 inflammasome-mediated pyroptosis signaling pathways represent two distinct central mechanisms in liver disease. However, the interconnections between STING and NLRP3 signaling pathways and the epigenetic regulation of the STING-NLRP3 axis in hepatocyte pyroptosis during liver fibrosis remain unknown.
This study aimed to verify whether blockage of hepatocyte pyroptosis by inhibiting the STING-NLRP3 signaling pathway could attenuate liver fibrosis and elucidate the underlying mechanisms that involve oxidative stress and metabolic reprogramming. We described a novel role and epigenetic mechanism by which the STING-IRF3/WD repeat-containing protein 5 (WDR5)/DOT1-like histone H3K79 methyltransferase (DOT1L)-NLRP3 signaling pathway enhances hepatocyte pyroptosis and hepatic inflammation in liver fibrosis.
2. Materials and methods
2.1. Animals
The animal procedures were approved by the Animal Use and Care Committee of West China Hospital, Sichuan University (#2017005A) and were conducted according to the regulations set by West China Hospital, Sichuan University. Liver fibrosis was induced by thioacetamide (TAA), carbon tetrachloride (CCl4), or bile duct ligation (BDL).
2.2. Ethical approval of human liver tissue collection
All research was conducted in accordance with both the Declaration of Helsinki and Istanbul. The study was approved by the Ethical Committee of West China Hospital and registered in the Chinese Clinical Trial Registry (ChiCTR2200063108). Written informed consent forms were received from all patients.
2.3. Statistical analysis
All data are presented as the mean ± standard deviation and were analyzed by GraphPad software (version 8, GraphPad Software Inc., La Jolla, CA, USA). A t-test and one-way ANOVA followed by Bonferroni post-hoc test were used to analyze the data. p < 0.05 was considered statistically significant.
The Supporting Materials include induction of liver fibrosis, human liver tissue collection, hematoxylin and eosin staining (H&E), immunofluorescence (IF), immunohistochemistry (IHC), transmission electron microscopy (TEM), scanning electron microscopy (SEM), serum biochemistry, Milliplex mouse multiplex assay, primary murine hepatocyte isolation, primary murine HSC (mHSC) isolation, cell culture and treatments, determination of hepatocyte pyroptosis, Primary mHSCs and hepatocyte co-culture, RNA-sequencing and data analysis, metabolomics, quantitative RT‒PCR (qPCR), Western blot (WB), co-immunoprecipitation (co-IP), histone-modifying enzyme compound screening system, and chromatin immunoprecipitation (ChIP), cytochromeP450 2E1 (CYP2E1) activity assay, measurement of hepatic O−2 contents and oxidative stress.
3. Results
3.1. STING knockout and pharmacological inhibition reduce liver fibrosis and hepatic inflammation
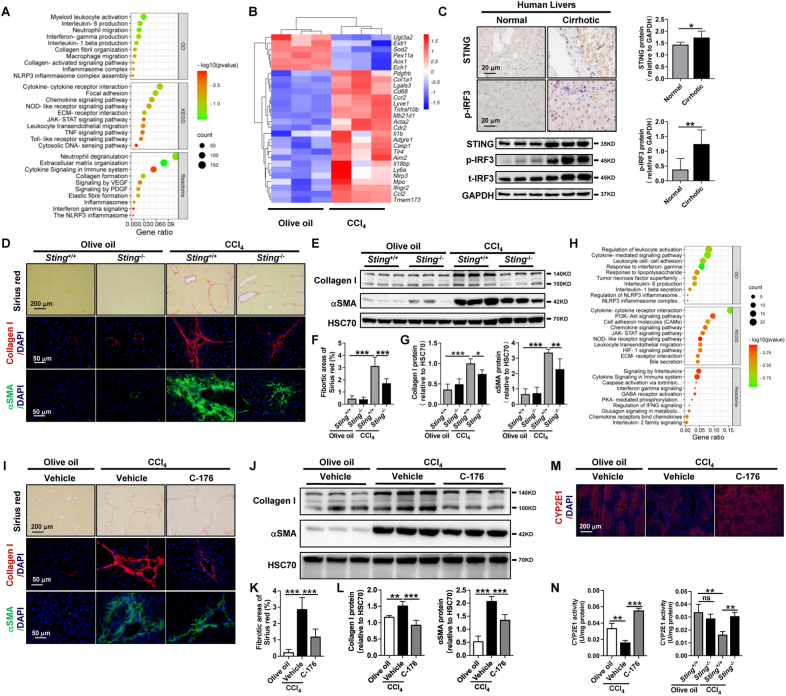
To explore the genes and signaling pathways involved in liver fibrosis, bulk RNA sequencing of livers from a CCl4-induced murine model was performed. The ontology analysis of upregulated differentially expressed genes (DEGs) indicated that the cytosolic DNA-sensing pathway and NLRP3 inflammasome signaling pathway were significantly activated in fibrotic livers (Fig. 1A). The main molecule of the cytosolic DNA-sensing pathway is STING [17]. As shown by the heatmap of key DEGs, the mRNA levels of Sting (Tmem173), Nlrp3, Il1β, and Caspase 1 were significantly increased in fibrotic livers compared to control livers (Fig. 1B). Furthermore, compared to healthy livers, hepatocytes in fibrotic livers exhibited increased STING protein expression in murine and human livers (Fig. 1C, Supporting Figs. S1A–E). IRF3 and NFκB are crucial downstream transcription factors of the STING signaling pathway [17]. Hepatocyte nuclei were positive for both phosphorylated IRF3 (p-IRF3) and phosphorylated NFκB-p65 (p-p65) (Fig. 1C, Supporting Figs. S1A, C, F). However, only p-IRF3 was significantly increased in murine and human cirrhotic livers compared to the respective controls (Fig. 1C, Supporting Figs. S1A–G). These results indicate that the STING signaling pathway is associated with the development of liver fibrosis.
Fig. 1.
STING knockout and pharmacological inhibition attenuate liver fibrosis and hepatic inflammation
(A-B) Wild-type mice were injected with olive oil or CCl4 for 6 weeks. Enriched signaling pathway analysis of upregulated (CCl4vs. olive oil) DEGs (A) and the heatmap of target DEGs (B) from RNA-sequencing were analyzed (n = 3/group). (C) Seven normal human livers and 11 cirrhotic human livers were collected. The distribution and protein levels of STING, t-IRF3, and p-IRF3 were analyzed by IHC or WB. (D–G)Sting+/+ and Sting−/− mice were injected with olive oil or CCl4 for 6 weeks (n = 6/group). Liver fibrosis was analyzed by Sirius red staining, IF, and WB for collagen I and αSMA. (H) RNA-sequencing was performed in CCl4-treated Sting+/+ and Sting−/− mice. Enriched pathway analysis of downregulated DEGs (Sting−/−vs. Sting+/+) was performed (n = 3/group). (I–L) Wild-type mice were treated with either olive oil or CCl4 for 6 weeks in addition to either vehicle or STING inhibitor C-176 (20 mg/kg, n = 6/group). Liver fibrosis was analyzed by Sirius red staining, IF, and WB for collagen I and αSMA. (M–N) The protein levels and activity of CYP2E1 were detected by IF (M) and activity kit (N). *p < 0.05, **p < 0.01, ***p < 0.001. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Next, to study the involvement of STING in liver fibrosis, we utilized Sting knockout mice. Sting knockout was confirmed by WB (Supporting Fig. S2A). In control Sting+/+ mice, CCl4 insult led to liver fibrosis compared to olive oil-treated mice as determined by Sirius red, IF, and WB of collagen I and α-smooth muscle actin (αSMA, Fig. 1D–G). However, CCl4-mediated liver fibrosis was significantly reduced in Sting−/− mice (Fig. 1D–G). We previously demonstrated that liver fibrosis was accompanied by increased hepatic inflammation, and the major infiltrated immune cells were macrophages and neutrophils [24]. Consistently, the expression of macrophage (Cd68, Ccr2, Lgals3, Adgre1) and neutrophil (Mpo) markers was significantly increased in fibrotic livers compared to controls (Fig. 1B). Thus, we next explored the role of STING in hepatic inflammation. There was an increase in immune cell infiltration as evidenced by H&E staining, increased MPO-positive neutrophils and F4/80-positive macrophages in CCl4-treated Sting+/+ mice compared to olive oil-treated Sting+/+ mice (Supporting Fig. S3A). Nonetheless, neutrophil and macrophage infiltration was significantly attenuated in Sting knockout mice (Supporting Fig. S3A).
To understand the proinflammatory role of STING, RNA-sequencing was performed in livers from CCl4-treated Sting+/+ and Sting−/− mice. IFNγ secretion, NLRP3 inflammasome, IL1β secretion, cytokine‒cytokine receptor interaction, and leukocyte activation/adhesion signaling pathways were significantly downregulated in Sting−/− mice (Fig. 1H).
The involvement of STING in liver fibrosis and hepatic inflammation was further confirmed using the STING pharmacological inhibition by C-176. CCl4-mediated liver fibrosis was significantly decreased in mice treated with C-176, as determined by Sirius red staining as well as collagen I and αSMA protein levels (Fig. 1I-L). The increased immune cell infiltration in the CCl4-induced fibrosis model was significantly attenuated in mice treated with C-176, as evidenced by H&E staining, IHC of MPO-positive neutrophils, and F4/80-positive macrophages (Supporting Fig. S3B). CYP2E1 is essential to the metabolic function of the liver [25]. CYP2E1 is mainly expressed in hepatocytes (Fig. 1M). The reduced CYP2E1 protein level in wild-type mice induced by CCl4 was restored after C-176 treatment (Fig. 1M). Comparable hepatic protein level and activity of CYP2E1 were observed in Sting+/+ and Sting−/− mice treated with olive oil (Fig. 1N, Supporting Fig. S3C). Additionally, a decreased CYP2E1 activity was induced by CCl4 treatment when compared to olive oil treatment in Sting+/+ mice (Fig. 1N). However, the decreased CYP2E1 activity induced by CCl4 was significantly attenuated by Sting knockout and C-176 treatment (Fig. 1N). In summary, STING promotes the development of liver fibrosis and hepatic inflammation in mice.
3.2. NLRP3 inflammasome activation and hepatocyte pyroptosis persist in murine and human cirrhotic livers
There are two groups of inflammasomes in terms of receptors, with NLRP3 inflammasome being the most typical inflammasome [26]. Of all inflammasomes, the NLRP3 inflammasome signaling pathway was activated in fibrotic livers and downregulated after Sting knockout (Fig. 1A–B, H). The NLRP3 inflammasome, an NLRP3/ASC/Caspase 1 protein complex that cleaves GSDMD or IL1β/IL18, plays a pivotal role in pyroptosis and inflammation [14]. Whether the NLRP3 inflammasome and hepatocyte pyroptosis are activated in liver fibrosis remains controversial. In the TAA-induced murine fibrotic liver and human cirrhotic liver, the crucial pyroptotic molecules NLRP3, cleaved Caspase 1 (cl-Caspase 1), and cleaved GSDMD (cl-GSDMD) determined by WB and/or IHC were significantly increased, and NLRP3 and cl-Caspase 1 were mainly expressed in hepatocytes (Supporting Figs. S4A–D). Moreover, pyroptotic hepatocytes characterized by ruptured membranes and the formation of membrane vesicles were observed by H&E staining and TEM in murine fibrotic livers (Supporting Fig. S4E). In summary, NLRP3 inflammasome activation and hepatocyte pyroptosis are present in murine and human fibrotic livers.
3.3. STING induces hepatocyte pyroptosis by activating the NLRP3 inflammasome
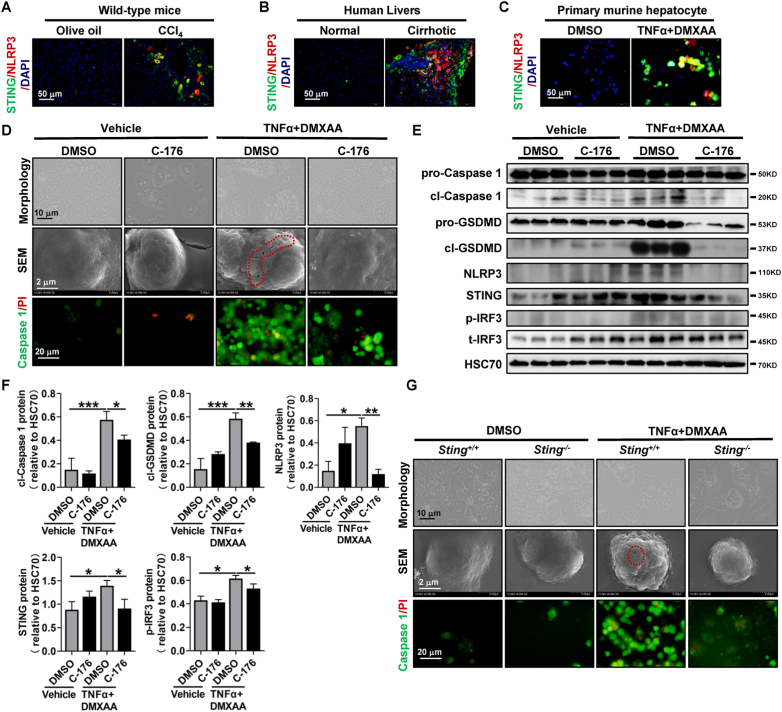
Next, we sought to understand whether STING is involved in NLRP3-mediated pyroptosis. In vivo, STING and NLRP3 were coexpressed in hepatocytes around fibrotic septa of the murine and human cirrhotic liver, as determined by colocalization of STING and NLRP3 (Fig. 2A and B). Additionally, hepatocyte pyroptosis was significantly reduced in Sting knockout mice, as determined by IHC of cl-Caspase 1 (Supporting Fig. S2B).
Fig. 2.
STING induces hepatocyte pyroptosis by activating the NLRP3 inflammasome
(A) Wild-type mice were injected with olive oil or CCl4 for 6 weeks. Colocalization of STING and NLRP3 was assessed by IF. (B) IF was used to analyze the colocalization of STING and NLRP3 in human cirrhotic livers. (C–F) Primary hepatocytes isolated from wild-type mice were treated with DMSO or the STING inhibitor C-176 for 2 h in addition to either vehicle or TNFα+DMXAA for an additional 6 h. Colocalization of STING and NLRP3 was determined by IF (C). Morphological changes, SEM, and colocalization of Caspase 1/PI were performed (D). The protein levels of STING, p-IRF3, cl-Caspase 1, and cl-GSDMD were determined by WB (E), and the quantitative data are shown (F). (G) Primary hepatocytes isolated from Sting+/+ and Sting−/− mice were treated with vehicle or TNFα+DMXAA for 6 h. The morphological changes, SEM, and colocalization of Caspase 1/PI were analyzed. n = 3/group, *p < 0.05, **p < 0.01, ***p < 0.001.
In vitro, TNFα and lipopolysaccharide (LPS) are commonly used stimulators of pyroptosis [27]. However, TNFα (25, 50, 100 ng/mL) or LPS (1, 5, 10 μg/mL) alone could not induce primary murine hepatocyte pyroptosis as determined by cell morphology, Caspase 1/PI colocalization, and WB of NLRP3, cl-Caspase 1 and cl-GSDMD (Supporting Figs. S5A–D). To mimic the microenvironment of liver fibrosis, the TNFα plus STING agonist DMXAA (TNFα+DMXAA) was utilized to induce hepatocyte pyroptosis in murine primary hepatocytes. TNFα+DMXAA increased STING and NLRP3 expression and colocalization in primary murine hepatocytes (Fig. 2C). Meanwhile, TNFα+DMXAA treatment caused a typical pyroptotic cell morphology in primary murine hepatocytes, such as cell shrinkage, membrane rupture, and membrane blebbing, compared to the controls (Fig. 2D). Moreover, TNFα+DMXAA stimulation increased the double-positive cells of activated Caspase 1 and propidium iodide (PI, Fig. 2D). The protein levels of the NLRP3 inflammasome complex, such as NLRP3, cl-Caspase 1, and cl-GSDMD, were also significantly enhanced after TNFα+DMXAA stimulation (Fig. 2E and F). However, the pyroptotic cell morphology was significantly reversed by the STING inhibitor C-176 in primary murine hepatocytes (Fig. 2D). Additionally, the increase in the NLRP3 inflammasome and pyroptotic cell markers NLRP3, cl-Caspase 1, and cl-GSDMD induced by TNFα+DMXAA was also significantly decreased by the STING inhibitor C-176 in primary murine hepatocytes (Fig. 2E and F). Furthermore, as determined by the increased protein levels of STING and p-IRF3, TNFα+DMXAA treatment induced the activation of the STING-IRF3 signaling pathway in primary murine hepatocytes (Fig. 2E and F). However, the enhanced protein levels of STING and p-IRF3 caused by TNFα+DMXAA were markedly decreased by the STING inhibitor C-176 (Fig. 2E and F). Consistently, Sting knockout reduced the aforementioned pyroptotic changes, such as pyroptotic cell morphology and double-positive cells of activated Caspase 1/PI in primary murine hepatocytes (Fig. 2G). Moreover, TNFα stimulation in Sting-overexpressing AML12 cells also induced hepatocyte pyroptosis, as determined by morphological changes and increased mRNA and protein levels of NLRP3 and cl-Caspase 1 (Supporting Figs. S6A–D). In summary, STING induces hepatocyte pyroptosis via activating the NLRP3 inflammasome.
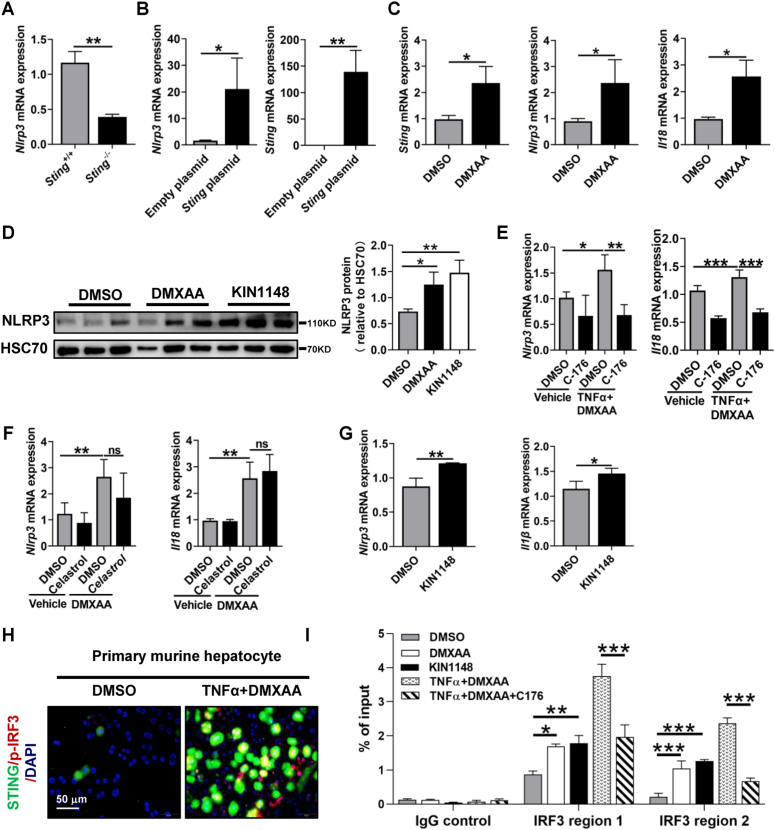
3.4. STING promotes IRF3 recruitment to the Nlrp3 promoter region in hepatocytes
Given that STING and IRF3 were activated in liver fibrosis, we next examined whether the STING-IRF3 signaling pathway regulates NLRP3 expression in primary murine hepatocytes. In primary murine hepatocytes, Sting knockout significantly decreased the mRNA level of Nlrp3 (Fig. 3A). In contrast, Sting overexpression upregulated the mRNA and protein levels of NLRP3 in AML12 hepatocytes (Fig. 3B, Supporting Figs. S6C–D). Additionally, the STING agonist DMXAA enhanced the mRNA levels of Sting, Nlrp3, Il18, Caspase 1, and p65 (Fig. 3C, Supporting Fig. S6E) and increased the protein level of NLRP3 in primary murine hepatocytes (Fig. 3D). However, the elevated mRNA levels of Nlrp3 and Il18 induced by TNFα+DMXAA were reduced by the STING inhibitor C-176 but not by the NFκB inhibitor celastrol (Fig. 3E and F). Furthermore, as described in Fig. 2E and F, the increased protein level of NLRP3 caused by TNFα+DMXAA was significantly abrogated by the STING inhibitor C-176. In line with these data, NLRP3 mRNA and protein levels and Il1β mRNA expression were enhanced by the IRF3 agonist KIN1148 (Fig. 3D, G).
Fig. 3.
STING promotes IRF3 recruitment to the Nlrp3 promoter region in hepatocytes
(A) Primary hepatocytes were isolated from Sting+/+ and Sting−/− mice, and the mRNA level of Nlrp3 was then quantified by qPCR. (B) AML12 hepatocytes were transfected with empty plasmid or Sting overexpression plasmid (OE), and the mRNA level of Nlrp3 was determined by qPCR. (C–D) Primary hepatocytes isolated from wild-type mice were treated with the STING agonist DMXAA (50 μg/mL) or IRF3 agonist KIN1148 (10 μg/mL) for 6 h qPCR was applied to evaluate the mRNA levels of Sting, Nlrp3, and Il18 (C), and the protein levels of NLRP3 were assessed by WB (D). (E–F) Primary hepatocytes isolated from wild-type mice were treated with DMSO, the STING inhibitor C-176 (2 μM, E) or the NFκB inhibitor celastrol (50 nM, F) for 2 h in addition to either vehicle or TNFα (25 ng/mL)+DMXAA (50 μg/mL) for an additional 6 h. The mRNA levels of Nlrp3 and Il18 were analyzed by qPCR. (G) Primary hepatocytes isolated from wild-type mice were treated with the IRF3 agonist KIN1148 (10 μg/mL) for 6 h qPCR was applied to evaluate the mRNA levels of Sting, Nlrp3, and Il18. (H) Primary hepatocytes isolated from wild-type mice were treated with DMSO or TNFα (25 ng/mL)+DMXAA (50 μg/mL) for 6 h, and then colocalization of STING and p-IRF3 was determined by IF. (I) Primary hepatocytes isolated from wild-type mice were treated with DMSO, DMXAA, KIN1148, TNFα+DMXAA or TNFα+DMXAA + C-176 for 6 h. Then, p-IRF3 levels at Nlrp3 promoter regions were evaluated by ChIP for p-IRF3 followed by qPCR. n = 3/group, ns: not significant, *p < 0.05, **p < 0.01, ***p < 0.001.
STING activates inflammatory gene expression by recruiting the transcription factor IRF3 [17]. The above results indicate that both STING and IRF3 can regulate NLRP3. We next verified whether STING can increase Nlrp3 transcription by recruiting IRF3. TNFα+DMXAA increased the expression and colocalization of STING and IRF3 in primary murine hepatocytes (Fig. 3H). By using the IRF3 ChIP-sequencing dataset from dendritic cells (GSE125340) [28], we found 2 IRF3 binding regions on the promoter of Nlrp3 (Supporting Fig. S7). ChIP-qPCR was then applied to confirm the binding of IRF3 to the Nlrp3 promoter. As expected, the STING agonist DMXAA, IRF3 agonist KIN1148, and TNFα+DMXAA increased the binding of IRF3 to both promoter regions of Nlrp3, which was abrogated by the STING inhibitor C-176 (Fig. 3I). In summary, the STING-IRF3 but not the STING-NFκB signaling pathway promotes NLRP3 expression in hepatocytes.
3.5. STING increases IRF3 recruitment at Nlrp3 promoter regions by promoting histone methylation via WDR5/DOT1L in AML12 hepatocytes
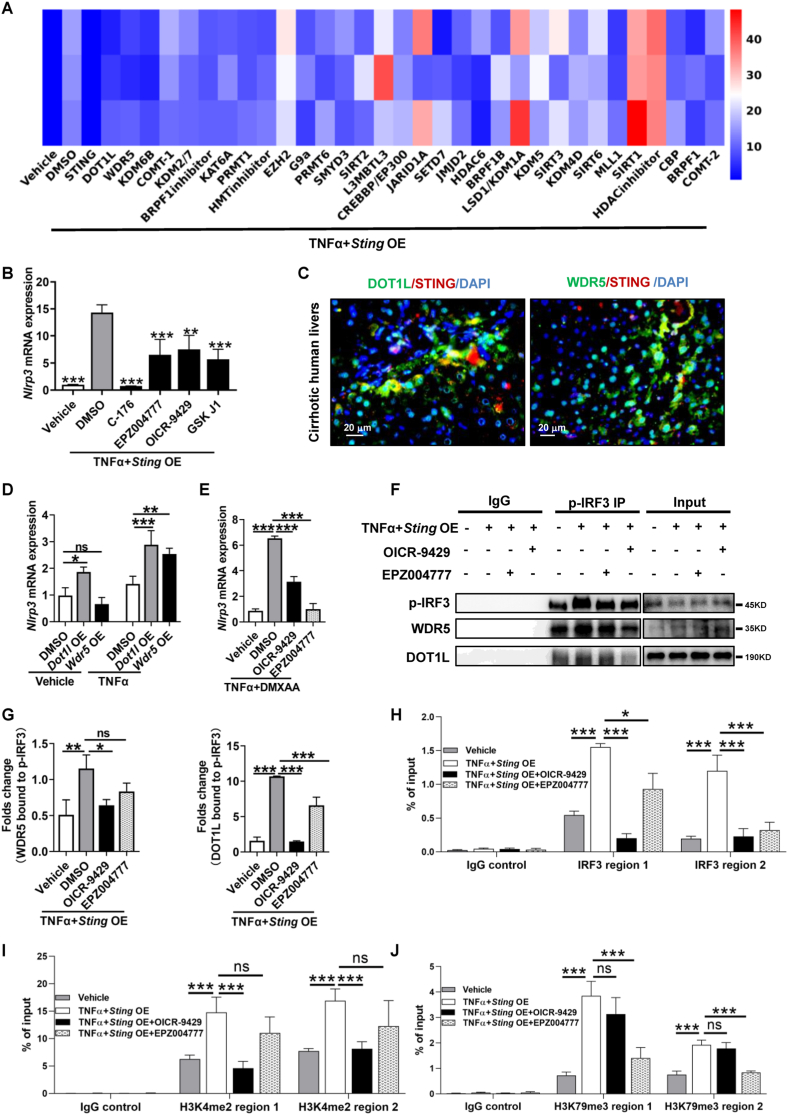
STING enhances the transcriptional activity of inflammatory genes by promoting histone modifications to recruit transcription factors [22,23]. To explain how STING leads to IRF3 recruitment, a histone-modifying enzyme compound system with 32 targets was applied in TNFα-treated AML12 hepatocytes overexpressing Sting (Fig. 4A). The H3K4-specific histone methyltransferase WDR5 and H3K79 methyltransferase DOT1L inhibitors suppressed TNFα plus Sting overexpression-mediated Nlrp3 upregulation (Fig. 4A and B). Of all the histone-modifying enzymes identified by the system, WDR5 and DOT1L colocalized with STING in human cirrhotic livers (Fig. 4C). Both WDR5 and DOT1L overexpression increased Nlrp3 transcription in vehicle- and TNFα-treated AML12 hepatocytes (Fig. 4D). Moreover, Nlrp3 upregulation by TNFα+DMXAA was significantly abrogated by the WDR5 and DOT1L inhibitors OICR-9429 and EPZ004777, respectively (Fig. 4E). To examine whether WDR5 and DOT1L form a transcription activator complex with p-IRF3 for Nlrp3 gene transcription, Co-IP was utilized. TNFα plus Sting overexpression insult promoted p-IRF3 binding to WDR5 and DOT1L (Fig. 4F and G). These interactions were suppressed by the WDR5 and DOT1L inhibitors OICR-9429 and EPZ004777, respectively (Fig. 4F and G). H3K4me2 and H3K79me3 are active histone marks [23,29]. Furthermore, ChIP of p-IRF3, H3K4me2, or H3K79me3 demonstrated that p-IRF3 and histone methylation were increased at the promoter regions of Nlrp3 upon TNFα plus Sting overexpression, which were abolished by either OICR-9429 or EPZ004777 (Fig. 4H–J). In summary, we have established the interconnection between STING and NLRP3 signaling pathways and the epigenetic regulation of the STING-NLRP3 axis in hepatocyte pyroptosis. These results suggest that STING increases IRF3 recruitment to Nlrp3 promoter regions through WDR5/DOT1L-mediated histone methylation.
Fig. 4.
STING increases IRF3 recruitment at Nlrp3 promoter regions by promoting histone methylation via WDR5/DOT1L in AML12 hepatocytes
(A-B) A selected histone-modifying enzyme compound system with 32 targets was applied to test Nlrp3 inhibition in TNFα+Sting-overexpressing AML12 hepatocytes. The heatmap of Nlrp3 expression was analyzed by qPCR (A). Inhibitors that suppress Nlrp3 upregulation are shown (B). (C) IF was used to analyze the colocalization of STING and DOT1L or WDR5 in human cirrhotic livers. (D) AML12 hepatocytes transfected with the Dot1l or Wdr5 overexpression plasmid were treated with vehicle, TNFα (25 ng/mL) for 6 h. The mRNA level of Nlrp3 was analyzed by qPCR. (E) AML12 hepatocytes were treated with DMSO, the DOT1L inhibitor EPZ004777 (7 μM) or the WDR5 inhibitor OICR-9429 (7 μM) for 2 h in addition to either vehicle or TNFα (25 ng/mL)+DMXAA (50 μg/mL) for an additional 6 h. The mRNA level of Nlrp3 was analyzed by qPCR. (F–G) AML12 hepatocytes transfected with the Sting overexpression plasmid were treated with vehicle, TNFα plus OICR-9429 or EPZ004777 for 6 h. Then, cell lysates of nuclear fractionation were subjected to co-IP against p-IRF3, and p-IRF3, WDR5, and DOT1L were then evaluated by WB. (H–J) AML12 hepatocytes transfected with the Sting overexpression plasmid were treated with vehicle, TNFα and TNFα plus OICR-9429 or EPZ004777 for 6 h. Then, p-IRF3 (H), H3K4me2 (I), and H3K79me3 (J) levels at Nlrp3 promoter regions were evaluated by ChIP‒qPCR. n = 3/group. ns: not significant, *p < 0.05, **p < 0.01, ***p < 0.001.
3.6. Hepatocyte NLRP3 deficiency and NLRP3 pharmacological inhibition ameliorate hepatic inflammation and liver fibrosis
The above results have uncovered the molecular mechanism of STING in NLRP3 regulation. However, the role of NLRP3 in hepatocyte pyroptosis and liver fibrosis remains controversial and is the focus of this section. In vitro, TNFα plus NLRP3 agonist nigericin treatment induced the pyroptotic phenotype, increased Caspase 1/PI colocalization, and enhanced protein levels of cl-Caspase 1 and cl-GSDMD in primary murine hepatocytes, and these pyroptotic changes were significantly attenuated by the NLRP3 inhibitor MCC950 (Supporting Figs. S8A–C). Activated HSCs are the major source of hepatic myofibroblasts and ECM in liver fibrosis [30]. To test the role of hepatocyte pyroptosis in the activation of HSCs, a transwell assay was setup by plating primary murine hepatocytes into the top well with corresponding treatments and primary murine HSCs into the bottom of the transwell apparatus. Pyroptoic hepatocytes induced by TNFα plus STING agonist DMXAA increased HSC activation, as determined by IF of αSMA in HSCs, which was reversed when STING inhibitor C-176 or NLRP3 inhibitor MCC950 was added in hepatocytes (Supporting Figs. S9A–B).
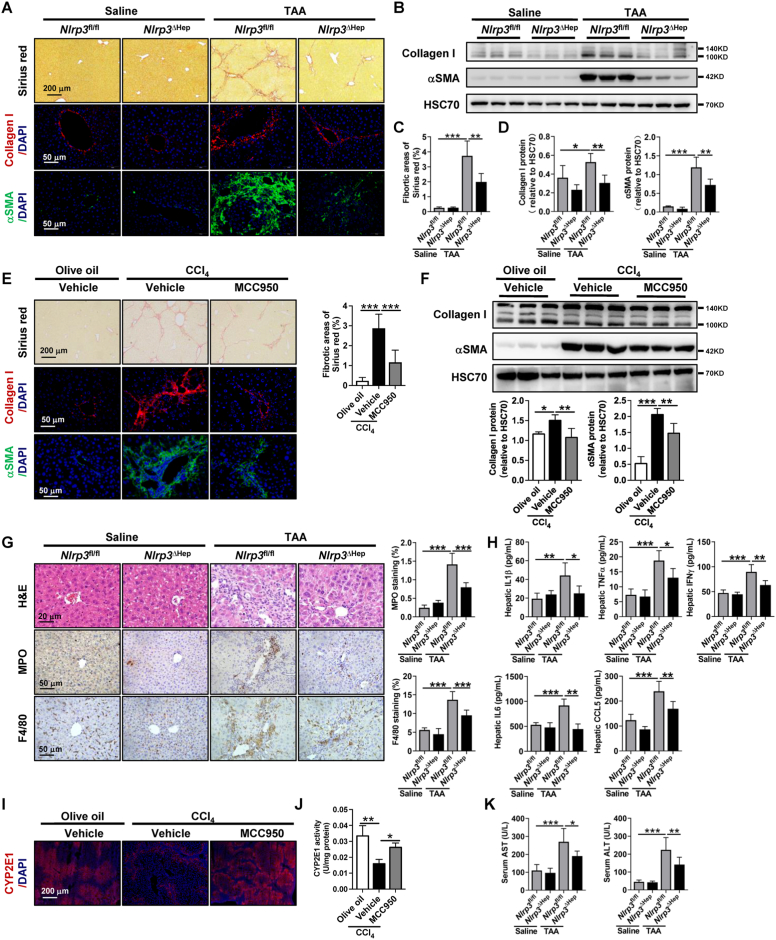
In vivo, we next investigated the role of hepatocyte-specific Nlrp3 deletion and NLRP3 pharmacological inhibition by MCC950 in the development of liver fibrosis in TAA or CCl4 treatment or BDL surgery murine models. Hepatocyte-specific Nlrp3 deletion mice (Nlrp3△Hep) were generated by crossing Nlrp3flox mice with Alb-Cre mice and were confirmed by IHC of NLPR3 (Supporting Fig. S10A). As expected, pyroptotic hepatocytes induced by TAA were also significantly abrogated in Nlrp3△Hep mice, as indicated by IHC of cl-Caspase 1 (Supporting Fig. S10B). TAA or CCl4 treatment or BDL surgery led to increased liver fibrosis in control mice compared to vehicle or sham, as determined by Sirius red, IHC, and WB of collagen I and αSMA (Fig. 5A–F, Supporting Figs. S11A–D). However, liver fibrosis was significantly reduced in TAA- or BDL-treated Nlrp3△Hep mice compared to Nlrp3fl/fl mice (Fig. 5A–D, Supporting Figs. S11A–D). Similarly, liver fibrosis induced by CCl4 treatment was also significantly attenuated by NLRP3 inhibitor MCC950 (Fig. 5E and F).
Fig. 5.
Hepatocyte NLRP3 deficiency and NLRP3 pharmacological inhibition ameliorate hepatic inflammation and liver fibrosis
(A-D)Nlrp3fl/fl and Nlrp3ΔHep mice were injected with normal saline or TAA for 8 weeks. Liver fibrosis was analyzed by Sirius red staining, IF, and WB for collagen I and αSMA. (E–F) Wild-type mice were treated with either olive oil or CCl4 for 6 weeks in addition to either vehicle or NLRP3 inhibitor MCC950 (20 mg/kg, n = 6/group). Liver fibrosis was analyzed by Sirius red staining, IF, and WB for collagen I and αSMA. (G) Hepatic inflammation was analyzed by H&E staining, IHC for the neutrophil marker MPO, and the macrophage marker F4/80, and the quantitative data are shown. (H) Hepatic IL1β, TNFα, IFNγ, IL6, and CCL5 levels were quantified by the Milliplex mouse multiplex assay. (I–J) The protein levels and activity of CYP2E1 were detected by IF (I) and activity kit (J). (K) Liver function was analyzed by serum AST and ALT. n = 6/group. *p < 0.05, **p < 0.01, ***p < 0.001. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Unlike other types of cell death, pyroptosis can initiate local inflammation by releasing inflammatory cytokines and recruiting immune cells [8,12]. We next explored the impact of NLRP3 on hepatic inflammation. TAA or CCl4 treatment or BDL surgery on Nlrp3fl/fl or wild-type mice increased immune cell infiltration compared to that in the respective controls, as evidenced by H&E staining, IHC of MPO-positive neutrophils, and F4/80-positive macrophages (Fig. 5G, Supporting Figs. S11E–F, Supporting Fig. S12A). Consistently, the increase in hepatic inflammation in TAA treatment or BDL surgery was further confirmed by the enhanced hepatic cytokines and chemokines, including IL1β, TNFα, IFNγ, IL6, and C–C motif chemokine ligand 5 (CCL5, Fig. 5H, Supporting Fig. S11G). This increase in immune cell infiltration, hepatic cytokines and chemokines in the TAA- or BDL-induced fibrosis models were significantly attenuated in Nlrp3△Hep mice (Fig. 5G and H, Supporting Figs. S11E–G). The increase in immune cell infiltration induced by CCl4 treatment was also significantly ameliorated by NLRP3 pharmacological inhibition with MCC950 (Supporting Fig. S12A). Regarding different liver fibrosis models, the decreased activity and protein level of CYP2E1 induced by CCl4 were enhanced by MCC950 (Fig. 5I and J). Moreover, a similar hepatic protein level and activity of CYP2E1 were observed in Nlrp3fl/fl and Nlrp3△Hep mice treated with saline (Supporting Figs. S12B–D). In comparison, the decreased protein level of CYP2E1 induced by TAA treatment was also abrogated in Nlrp3△Hep mice (Supporting Figs. S12B–C). As defined by increased serum ALT and AST, liver injury was ameliorated in Nlrp3△Hep mice of TAA- or BDL-induced fibrosis models (Fig. 5K, Supporting Fig. S11H). Additionally, the increased total bilirubin in BDL surgery mice was also abrogated in Nlrp3△Hep mice (Supporting Fig. S11H). In summary, NLRP3 inhibition attenuates hepatocyte pyroptosis, and hepatocyte-specific Nlrp3 deletion reduces liver fibrosis, hepatic inflammation, and liver injury.
3.7. Inhibition of pyroptosis by gsdmd knockout attenuates liver fibrosis
NLPR3-dependent GSDMD cleavage is crucial to promoting cell pyroptosis [31]. To investigate the pro-fibrotic effect of pyroptosis, NLRP3 downstream Gsdmd knockout mice were subjected to TAA or BDL. Gsdmd knockout was confirmed by WB (Supporting Fig. S10C). By quantifying cl-Caspase 1, TAA-mediated hepatocyte pyroptosis was significantly attenuated in Gsdmd−/− mice (Supporting Fig. S10D). Compared to Gsdmd+/+ mice, TAA- or BDL-mediated liver fibrosis was significantly decreased in Gsdmd−/− mice, as determined by Sirius red staining as well as collagen I and αSMA protein levels (Supporting Figs. S13A–D, Supporting Figs. S14A–D). GSDMD cleaves the cell membrane of pyroptotic cells to release cell content and inflammatory factors [11]. We next confirmed that Gsdmd knockout inhibited hepatic inflammation and liver injury. The increased immune cell infiltration, hepatic cytokines and chemokines (IL1β, TNFα, IFNγ, IL6, and CCL5), as well as serum ALT, AST, and total bilirubin in the TAA- or BDL-induced fibrosis models were significantly attenuated in Gsdmd knockout mice (Supporting Figs. S13E–H, Supporting Figs. S14E–H). A comparable hepatic protein level of CYP2E1 was observed in Gsdmd+/+ and Gsdmd−/− mice treated with saline (Supporting Fig. S13I). However, the reduced protein level of CYP2E1 induced by TAA treatment was restored in Gsdmd−/− mice (Supporting Fig. S13I). In summary, inhibition of pyroptosis by Gsdmd knockout attenuates liver fibrosis, hepatic inflammation, and liver injury.
3.8. Hepatocyte NLRP3 deficiency attenuates liver fibrosis via metabolic reprogramming
The abovementioned results verified that blocking hepatocyte pyroptosis by NLRP3 and GSDMD deficiency can abrogate liver fibrosis. We next clarified how hepatocyte pyroptosis deficiency attenuates liver fibrosis using transcriptome and metabolomics. In vitro, primary murine hepatocytes treated with TNFα plus NLRP3 agonist nigericin and TNFα plus nigericin plus MCC950 were analyzed by RNA-sequencing (Supporting Fig. S8D). The DEG ontology analysis showed that 209 signaling pathways were significantly enriched, with the top being oxidative phosphorylation, ATP metabolism, ROS metabolism, metabolism of RNA, and the citric acid (TCA) cycle (Supporting Fig. S8D).
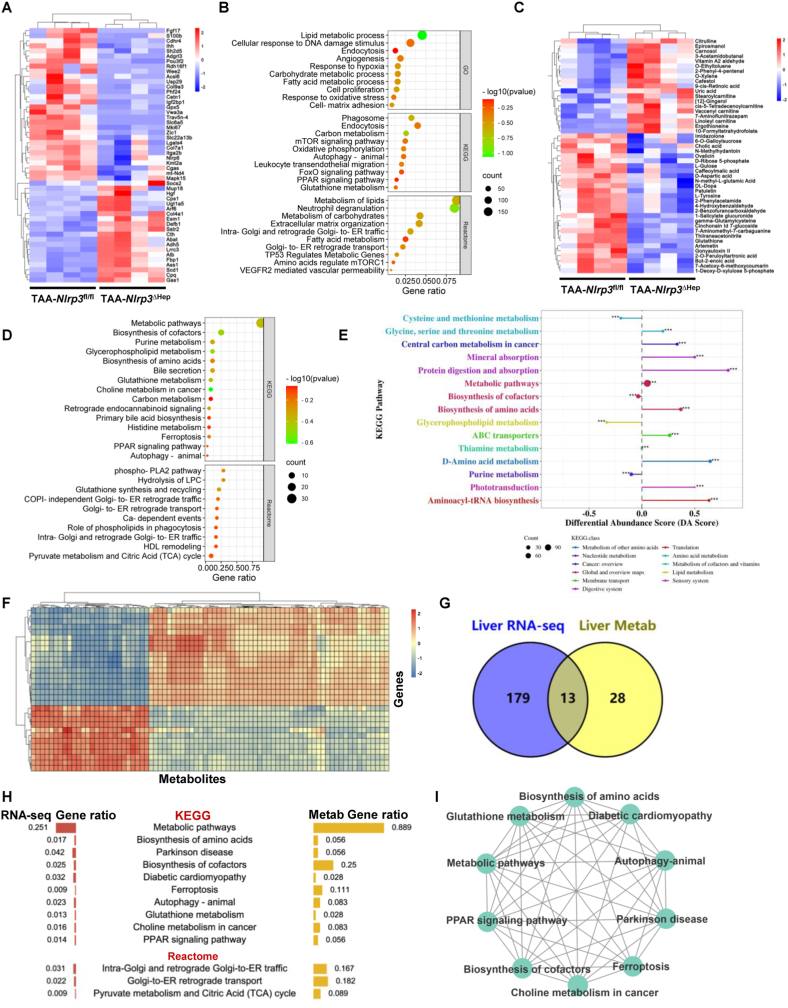
In vivo, livers from TAA-treated Nlrp3fl/fl or Nlrp3△Hep mice were analyzed by RNA sequencing. The ECM (Col9a3, Col7a1, Itga2b, Esm1), cell proliferation (Mki67, Mapk15, Ihh), and metabolism (Fgf17, Acsl6, Gpx5, mt-Nd4, Socs2)-related genes were significantly downregulated in Nlrp3△Hep mice (Fig. 6A). Conversely, liver protective genes, such as Hgf, Alb, and Col4a1, were upregulated in Nlrp3△Hep mice (Fig. 6A). Metabolism of lipids, carbon, and glutathione, response to oxidative stress, oxidative phosphorylation, cell proliferation, cell-matrix adhesion, leukocyte transendothelial migration, and neutrophil degranulation were significantly enriched in DEG ontology analysis (Fig. 6B).
Fig. 6.
Hepatocyte-specific Nlrp3 deletion attenuates liver fibrosis via metabolic reprogramming
(A-E) RNA-sequencing and metabolomics of livers from Nlrp3fl/fl and Nlrp3ΔHep mice injected with TAA for 8 weeks (n = 4/group). RNA-sequencing was performed to show the heatmap (A) and the enriched signaling pathways (B) of DEGs. Ultra HPLC-based metabolomics of livers was applied to reveal the heatmap (C), the enriched signaling pathway (D), and the KEGG metabolic pathway-based differential abundance analysis (E) of differential metabolites. (F–I) Integrative multiomics analysis of the transcriptome and metabolomics. Hierarchical clustering for canonical correlation analysis of the mRNAs and metabolomics (F). Venn diagram (G) and ontology analysis (H) displaying the shared signaling pathways between the transcriptome and metabolomics. Functional enrichment analysis was then performed to gain further insights into the KEGG of DEGs and metabolites (I).
Hepatocytes are the central metabolic cells for glucose, lipids, and protein. Hepatocyte death also leads to metabolic reprogramming [9,10]. As metabolism-related pathways were enriched in DEG ontology analysis of NLRP3 inhibition both in vitro (Supporting Fig. S8D) and in vivo (Fig. 6B), ultra-high-performance liquid chromatography (HPLC)-based metabolomics was utilized for livers from TAA-treated Nlrp3fl/fl or Nlrp3△Hep mice. We identified 104 differential metabolites, 79 Kyoto Encyclopedia of Genes and Genomes (KEGG) signaling pathways, and 11 Reactome signaling pathways, with the top metabolites being the metabolism of purine, glycerophospholipid, glutathione, choline, carbon, and histidine (Fig. 6C and D). Consistently, KEGG metabolic pathway-based differential abundance analysis in TAA-treated Nlrp3fl/fl mice and Nlrp3△Hep mice also confirmed the dysregulation of the abovementioned metabolic pathways (Fig. 6E). To explore the potential relationships between the transcriptome and metabolomics, the comparison of the transcriptome with the metabolomics exhibited significant correlations between mRNAs and metabolites (Fig. 6F). Afterward, the 13 common signaling pathways between the transcriptome and metabolomics were selected (Fig. 6G). Metabolic pathways, such as biosynthesis of amino acids, autophagy, glutathione metabolism, choline metabolism, the PPAR signaling pathway, pyruvate metabolism and the TCA cycle, were shared between the transcriptome and metabolomics (Fig. 6H). Functional enrichment analysis of these 10 KEGG pathways was performed to gain further insights into the biological functions and interactions of DEGs and metabolites. The results showed that terms associated with these metabolic pathways interplay with each other (Fig. 6I). The above results indicated that alteration of these metabolic pathways, which lead to metabolic reprogramming and oxidative stress, may contribute to NLRP3-mediated reduction of pyroptosis and liver fibrosis. However, detailed mechanisms remain to be elucidated and will be the focus of further investigations. In summary, hepatocyte NLRP3 deficiency might ameliorate liver fibrosis by abrogating hepatic metabolic reprogramming.
3.9. STING-NLRP3-GSDMD axis inhibition suppresses hepatic ROS generation
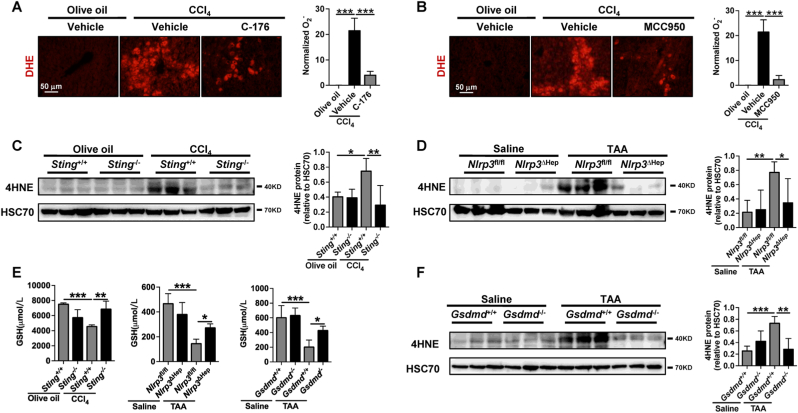
As ROS metabolism and response to oxidative stress were enriched during NLRP3-mediated hepatocyte pyroptosis (Fig. 6B, Supporting Fig. S8D), the role of STING-NLRP3-GSDMD axis inhibition on the suppression of ROS generation was then determined. Compared to the healthy controls, the ROS levels were significantly increased in TAA- or CCl4-induced murine fibrotic livers, as determined by dihydroethidium (DHE) assay, WB of 4-hydroxynonenal (4HNE), and GSH concentration (Fig. 7A–F). However, the DHE levels were significantly decreased after treatment with STING inhibitor C-176 and NLRP3 inhibitor MCC950 in CCl4-induced fibrotic livers (Fig. 7A and B). The increased hepatic 4HNE and GSH induced by TAA were also significantly abrogated in Sting−/−, Nlrp3△Hep, and Gsdmd−/− mice (Fig. 7C–F). In summary, the STING-NLRP3-GSDMD axis inhibition suppresses hepatic ROS generation.
Fig. 7.
STING-NLRP3-GSDMD axis inhibition suppresses hepatic ROS generation
(A-B) Wild-type mice were treated with either olive oil or CCl4 for 6 weeks in addition to either vehicle STING inhibitor C-176 (20 mg/kg) or NLRP3 inhibitor MCC950 (20 mg/kg). Hepatic O− was analyzed by DHE. (C–D)Sting+/+ and Sting−/− mice were injected with olive oil or CCl4 for 6 weeks. Nlrp3fl/fl and Nlrp3ΔHep mice were injected with normal saline or TAA for 8 weeks. The protein level of 4HNE was analyzed by WB. (E) The levels of GSH in the liver were quantified. (F)Gsdmd+/+ and Gsdmd−/− mice were injected with normal saline or TAA for 8 weeks. The protein level of 4HNE was analyzed by WB. n = 6/group. *p < 0.05, **p < 0.01, ***p < 0.001.
4. Discussion
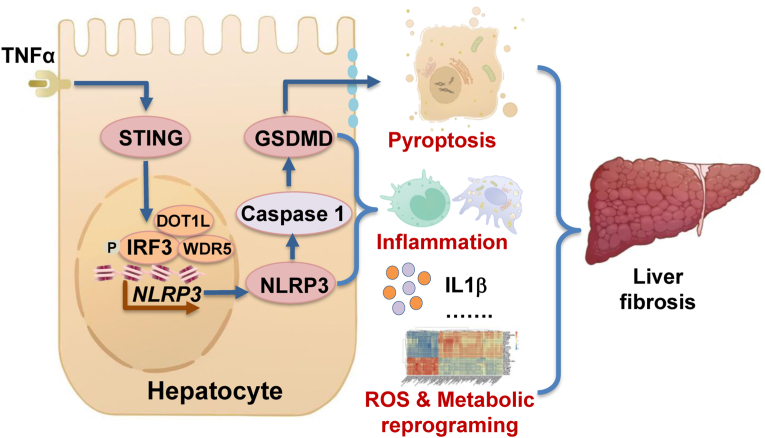
Hepatocyte pyroptosis results in the dysfunctional hepatocytes, abnormal liver function, and eventually liver fibrosis [1,7,8]. Thus, it is crucial to protect hepatocytes against pyroptosis. The activation of STING and NLRP3 inflammasome-mediated pyroptosis signaling pathways represents two distinct central mechanisms in liver disease [18,19]. However, the interconnections between these two pathways and the epigenetic regulation of the STING-NLRP3 axis in hepatocyte pyroptosis during liver fibrosis remain unknown. We discovered a new role for STING in inducing hepatocyte pyroptosis by activating the NLRP3 inflammasome in hepatocytes. We demonstrated a new epigenetic mechanism by which STING enhanced NLRP3 expression by increasing histone methylation at Nlrp3 promoter regions. Furthermore, we found new therapeutic targets in which Sting knockout, hepatocyte-specific Nlrp3 deletion, or downstream Gsdmd knockout ameliorated pyroptosis, hepatic inflammation, and liver fibrosis. Hepatic metabolic reprogramming and oxidative stress might also contribute to the anti-fibrotic effect of hepatocyte-specific Nlrp3 deletion. Collectively, this study describes a novel role and epigenetic mechanism by which the STING-IRF3/WDR5/DOT1L-NLRP3 signaling pathway enhances hepatocyte pyroptosis and hepatic inflammation in liver fibrosis (Fig. 8).
Fig. 8.
Proposed mechanisms
STING increases NLPR3 and NLRP3 inflammasome-mediated hepatocyte pyroptosis during liver injury via epigenetic regulation of the IRF3/WDR5/DOT1L transcription activator complex. STING-NLRP3 pathway leads to liver fibrosis by increasing hepatocyte pyroptosis, hepatic inflammation, and metabolic reprogramming.
Most research on pyroptosis focuses on monocytes and macrophages in liver diseases, and a recent study demonstrated that hepatocytes are resistant to inflammasome-activated pyroptosis [27]. However, hepatocytes can undergo pyroptosis and perpetuate inflammasome-driven fibrogenesis [7]. Consistent with these findings, the present study confirmed the existence of hepatocyte pyroptosis in human and murine fibrotic livers and that STING can induce pyroptosis in primary murine hepatocytes. Pyroptosis is mainly caused by NLRP3 inflammasome activation, which cleaves GSDMD to form pyroptotic membrane pores [8,26]. In the liver, hepatocyte pyroptosis contributes to myofibroblast activation [7,32], and the space of dead hepatocytes is gradually replaced by myofibroblasts, ultimately leading to liver fibrosis [4,8]. In our study, Sting knockout, hepatocyte-specific Nlrp3 deletion, or Gsdmd knockout reduced pyroptosis, hepatic inflammation, and liver fibrosis. Additionally, co-culture of HSC with pyroptotic hepatocytes increased HSC activation, and HSC activation was reversed when hepatocyte pyroptosis was abrogated by STING or NLRP3 inhibitor. The above information supports the concept that blocking hepatocyte pyroptosis via STING/NLRP3/GSDMD inhibition has therapeutic potential to attenuate liver fibrosis.
Hepatic metabolic reprogramming and oxidative stress are associated with hepatic fibrosis in both humans and rodents [33,34]. Pharmacologic treatment of hepatic metabolism and oxidative stress-related molecules reduces fibrosis in a murine model [34]. Consistently, we found that hepatic metabolism reprogramming and oxidative stress might contribute to the anti-fibrotic effect of hepatocyte-specific Nlrp3 deletion. Metabolomic factors released from apoptotic cells might influence the surrounding tissue microenvironment and induce oxidative stress [35,36]. In this study, STING-NLRP3-GSDMD axis inhibition reduces hepatocyte pyroptosis and hepatic ROS generation in murine liver fibrosis. Hepatocyte pyroptosis might also contribute to liver fibrosis by releasing metabolites and ROS generation. Increased CYP2E1 activity is one of the hepatic enzymes contributing to ROS generation in alcohol- or lipid-induced liver injuries [25]. However, TAA and CCl4 are suicide substrates of CYP2E1, leading to decreased CYP2E1 activity [37,38]. Additionally, increased IFN-γ can also suppresses native CYP2E1 promoter activity and may decrease CYP2E1 expression [39]. Thus, the protein level and/or activity of CYP2E1 were consistently decreased in the current CCl4-and TAA-induced liver fibrosis model, and were restored by STING-NLRP3-GSDMD axis inhibition. Unlike alcohol- or lipid-induced liver fibrosis, other molecules other than CYP2E1 might contribute to ROS generation. Further mechanistic studies should be performed to clarify how the hepatocyte STING-NLRP3-GSDMD axis changes the metabolic signaling pathway and ROS generation.
Although NLRP3 is crucial in inflammasome activation, inflammation, and pyroptosis, the mechanism of NLRP3 transcription has not been fully illustrated. Generally, the expression of NLRP3 is initially stimulated by LPS, cytosolic DNA, TNFα and other endogenous molecules via activation of the NFκB signaling pathway [11,12]. Other signaling pathways, such as the Hippo-YAP signaling pathway [40] and histone deacetylase 2 (HDAC2) [41], are also involved in NLRP3 regulation. Despite our increasing knowledge of NLRP3 regulation in other diseases, the transcriptional regulation of NLRP3 in liver cirrhosis remains elusive. STING can enhance the transcriptional activity of inflammatory genes by changing histone modifications to recruit transcription factors [22,23]. Here, STING enhanced NLRP3 expression and NLPR3 inflammasome-mediated hepatocyte pyroptosis by increasing histone methylation at Nlrp3 promoter regions via WDR5 and DOT1L and recruiting IRF3 at Nlrp3 promoter regions. Moreover, the STING-IRF3 signaling pathway also participates in NLRP3 inflammasome-mediated hepatocyte pyroptosis. The current study established a fundamental effect of the STING-IRF3/WDR5/DOT1L-NLRP3 signaling pathway in hepatocyte pyroptosis and liver fibrosis.
In conclusion, STING increases NLPR3 and NLRP3 inflammasome-mediated hepatocyte pyroptosis via epigenetic regulation of the IRF3/WDR5/DOT1L transcription activator complex. Inhibition of the STING-NLRP3-GSDMD signaling pathway protects hepatocytes from pyroptosis and attenuates hepatic inflammation and liver fibrosis. These results shed light on the anti-pyroptosis strategy as a potential therapeutic candidate for patients with liver cirrhosis.
Author contribution
JG, CT, and EK conceived and supervised the study; YX, CZ, YT, BL, TL, EL, CG and WD performed the experiments; CZ, YX, YT, and YG analyzed the data; YX, EK, CT, and JG wrote the manuscript with input from all the authors.
Grant support
This work was supported by the National Natural Science Fund of China (82170623, 82170625, U1702281, 81873584, 82000613, and 82000574), the National Key R&D Program of China (2017YFA0205404), Sichuan Science and Technology Program (2020YJ0084 and 2021YFS0147), and the 135 projects for disciplines of excellence of West China Hospital, Sichuan University (ZYGD18004).
Declaration of competing interest
All authors declare no conflict of interest.
Acknowledgment
The authors thank Dr. Vijay H. Shah from Mayo Clinic for his supervision.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2023.102691.
Contributor Information
Yang Xiao, Email: xiaoyangsg@foxmail.com.
Chong Zhao, Email: zhaochong.suzee@qq.com.
Yang Tai, Email: tyscu@foxmail.com.
Bei Li, Email: libei@scu.edu.cn.
Tian Lan, Email: lantian-scu@foxmail.com.
Enjiang Lai, Email: EnjiangLai@foxmail.com.
Wenting Dai, Email: dai_wenting@qq.com.
Yangkun Guo, Email: yangkunGdr@foxmail.com.
Can Gan, Email: gancan_medical@foxmail.com.
Enis Kostallari, Email: kostallari.enis@mayo.edu, enis.kostallari@gmail.com.
Chengwei Tang, Email: shcqcdmed@163.com.
Jinhang Gao, Email: Gao.jinhang@scu.edu.cn, Gao.jinhang@qq.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
Data availability
Data will be made available on request.
References
- 1.Brenner C., Galluzzi L., Kepp O., Kroemer G. Decoding cell death signals in liver inflammation. J. Hepatol. 2013;59:583–594. doi: 10.1016/j.jhep.2013.03.033. [DOI] [PubMed] [Google Scholar]
- 2.Ibrahim S.H., Hirsova P., Malhi H., Gores G.J. Nonalcoholic steatohepatitis promoting kinases. Semin. Liver Dis. 2020;40:346–357. doi: 10.1055/s-0040-1713115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schuster S., Cabrera D., Arrese M., Feldstein A.E. Triggering and resolution of inflammation in NASH. Nat. Rev. Gastroenterol. Hepatol. 2018;15:349–364. doi: 10.1038/s41575-018-0009-6. [DOI] [PubMed] [Google Scholar]
- 4.Kisseleva T., Brenner D. Molecular and cellular mechanisms of liver fibrosis and its regression. Nat. Rev. Gastroenterol. Hepatol. 2021;18:151–166. doi: 10.1038/s41575-020-00372-7. [DOI] [PubMed] [Google Scholar]
- 5.Asrani S.K., Devarbhavi H., Eaton J., Kamath P.S. Burden of liver diseases in the world. J. Hepatol. 2019;70:151–171. doi: 10.1016/j.jhep.2018.09.014. [DOI] [PubMed] [Google Scholar]
- 6.Han S., et al. Epidemiology of alcohol-associated liver disease. Clin. Liver Dis. 2021;25:483–492. doi: 10.1016/j.cld.2021.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaul S., et al. Hepatocyte pyroptosis and release of inflammasome particles induce stellate cell activation and liver fibrosis. J. Hepatol. 2021;74:156–167. doi: 10.1016/j.jhep.2020.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gautheron J., Gores G.J., Rodrigues C.M.P. Lytic cell death in metabolic liver disease. J. Hepatol. 2020;73:394–408. doi: 10.1016/j.jhep.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delgado M.E., Cárdenas B.I., Farran N., Fernandez M. Metabolic reprogramming of liver fibrosis. Cells. 2021;10 doi: 10.3390/cells10123604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scheidecker B., et al. Induction of in vitro metabolic zonation in primary hepatocytes requires both near-physiological oxygen concentration and flux. Front. Bioeng. Biotechnol. 2020;8:524. doi: 10.3389/fbioe.2020.00524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gan C., Cai Q., Tang C., Gao J. Inflammasomes and pyroptosis of liver cells in liver fibrosis. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.896473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inzaugarat M.E., et al. NLR family pyrin domain-containing 3 inflammasome activation in hepatic stellate cells induces liver fibrosis in mice. Hepatology. 2019;69:845–859. doi: 10.1002/hep.30252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Carvalho Ribeiro M., Szabo G. Role of the inflammasome in liver disease. Annu. Rev. Pathol. 2022;17:345–365. doi: 10.1146/annurev-pathmechdis-032521-102529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szabo G., Petrasek J. Inflammasome activation and function in liver disease. Nat. Rev. Gastroenterol. Hepatol. 2015;12:387–400. doi: 10.1038/nrgastro.2015.94. [DOI] [PubMed] [Google Scholar]
- 15.Wree A., et al. NLRP3 inflammasome activation results in hepatocyte pyroptosis, liver inflammation, and fibrosis in mice. Hepatology. 2014;59:898–910. doi: 10.1002/hep.26592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khanova E., et al. Pyroptosis by caspase11/4-gasdermin-D pathway in alcoholic hepatitis in mice and patients. Hepatology. 2018;67:1737–1753. doi: 10.1002/hep.29645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen R., Du J., Zhu H., Ling Q. The role of cGAS-STING signalling in liver diseases. JHEP Rep. 2021;3 doi: 10.1016/j.jhepr.2021.100324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu Y., et al. STING-mediated inflammation in Kupffer cells contributes to progression of nonalcoholic steatohepatitis. J. Clin. Invest. 2019;129:546–555. doi: 10.1172/JCI121842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo X., et al. Expression of STING is increased in liver tissues from patients with NAFLD and promotes macrophage-mediated hepatic inflammation and fibrosis in mice. Gastroenterology. 2018;155:1971–1984.e1974. doi: 10.1053/j.gastro.2018.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaidt M.M., et al. The DNA inflammasome in human myeloid cells is initiated by a STING-cell death Program upstream of NLRP3. Cell. 2017;171:1110–1124.e1118. doi: 10.1016/j.cell.2017.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang R., Kang R., Tang D. The STING1 network regulates autophagy and cell death. Signal Transduct. Targeted Ther. 2021;6:208. doi: 10.1038/s41392-021-00613-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malik P., et al. NET23/STING promotes chromatin compaction from the nuclear envelope. PLoS One. 2014;9 doi: 10.1371/journal.pone.0111851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cai Q., Gan C., Tang C., Wu H., Gao J. Mechanism and therapeutic opportunities of histone modifications in chronic liver disease. Front. Pharmacol. 2021;12 doi: 10.3389/fphar.2021.784591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao J., et al. Endothelial p300 promotes portal hypertension and hepatic fibrosis through C-C motif chemokine ligand 2-mediated angiocrine signaling. Hepatology. 2021;73:2468–2483. doi: 10.1002/hep.31617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barnes M.A., Roychowdhury S., Nagy L.E. Innate immunity and cell death in alcoholic liver disease: role of cytochrome P4502E1. Redox Biol. 2014;2:929–935. doi: 10.1016/j.redox.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knorr J., Wree A., Feldstein A.E. Pyroptosis in steatohepatitis and liver diseases. J. Mol. Biol. 2021 doi: 10.1016/j.jmb.2021.167271. [DOI] [PubMed] [Google Scholar]
- 27.Sun P., et al. Hepatocytes are resistant to cell death from canonical and non-canonical inflammasome-activated pyroptosis. Cell Mol Gastroenterol Hepatol. 2022;13:739–757. doi: 10.1016/j.jcmgh.2021.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Csumita M., et al. Specific enhancer selection by IRF3, IRF5 and IRF9 is determined by ISRE half-sites, 5' and 3' flanking bases, collaborating transcription factors and the chromatin environment in a combinatorial fashion. Nucleic Acids Res. 2020;48:589–604. doi: 10.1093/nar/gkz1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Habash N.W., Sehrawat T.S., Shah V.H., Cao S. Epigenetics of alcohol-related liver diseases. JHEP reports : innovation in hepatology. 2022;4 doi: 10.1016/j.jhepr.2022.100466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao J., et al. Hepatic stellate cell autophagy inhibits extracellular vesicle release to attenuate liver fibrosis. J. Hepatol. 2020;73:1144–1154. doi: 10.1016/j.jhep.2020.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi J., et al. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526:660–665. doi: 10.1038/nature15514. [DOI] [PubMed] [Google Scholar]
- 32.Wree A., et al. NLRP3 inflammasome driven liver injury and fibrosis: roles of IL-17 and TNF in mice. Hepatology. 2018;67:736–749. doi: 10.1002/hep.29523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khomich O., Ivanov A.V., Bartosch B. Metabolic hallmarks of hepatic stellate cells in liver fibrosis. Cells. 2019;9 doi: 10.3390/cells9010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang J., et al. Molecular profiling reveals a common metabolic signature of tissue fibrosis. Cell reports. Medicine. 2020;1 doi: 10.1016/j.xcrm.2020.100056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Medina C.B., et al. Metabolites released from apoptotic cells act as tissue messengers. Nature. 2020;580:130–135. doi: 10.1038/s41586-020-2121-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patel R.P., Lang J.D., Smith A.B., Crawford J.H. Redox therapeutics in hepatic ischemia reperfusion injury. World J. Hepatol. 2014;6:1–8. doi: 10.4254/wjh.v6.i1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knockaert L., et al. Carbon tetrachloride-mediated lipid peroxidation induces early mitochondrial alterations in mouse liver. Laboratory investigation; a journal of technical methods and pathology. 2012;92:396–410. doi: 10.1038/labinvest.2011.193. [DOI] [PubMed] [Google Scholar]
- 38.Shirato T., Homma T., Lee J., Kurahashi T., Fujii J. Oxidative stress caused by a SOD1 deficiency ameliorates thioacetamide-triggered cell death via CYP2E1 inhibition but stimulates liver steatosis. Arch. Toxicol. 2017;91:1319–1333. doi: 10.1007/s00204-016-1785-9. [DOI] [PubMed] [Google Scholar]
- 39.Qiu L.O., Linder M.W., Antonino-Green D.M., Valdes R., Jr. Suppression of cytochrome P450 2E1 promoter activity by interferon-gamma and loss of response due to the -71G>T nucleotide polymorphism of the CYP2E1*7B allele. J. Pharmacol. Exp. Therapeut. 2004;308:284–288. doi: 10.1124/jpet.103.057208. [DOI] [PubMed] [Google Scholar]
- 40.Wang D., et al. YAP promotes the activation of NLRP3 inflammasome via blocking K27-linked polyubiquitination of NLRP3. Nat. Commun. 2021;12:2674. doi: 10.1038/s41467-021-22987-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Y., et al. Histone deacetylase 2 regulates ULK1 mediated pyroptosis during acute liver failure by the K68 acetylation site. Cell Death Dis. 2021;12:55. doi: 10.1038/s41419-020-03317-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.