Abstract
Although the activator protein‐1 (AP‐1) factor Batf is required for Th17 cell development, its mechanisms of action to underpin the Th17 program are incompletely understood. Here, we find that Batf ensures Th17 cell identity in part by restricting alternative gene programs through its actions to restrain IL‐2 expression and IL‐2‐induced Stat5 activation. This, in turn, limits Stat5‐dependent recruitment of Ets1‐Runx1 factors to Th1‐ and Treg‐cell‐specific gene loci. Thus, in addition to pioneering regulatory elements in Th17‐specific loci, Batf acts indirectly to inhibit the assembly of a Stat5‐Ets1‐Runx1 complex that enhances the transcription of Th1‐ and Treg‐cell‐specific genes. These findings unveil an important role for Stat5‐Ets1‐Runx1 interactions in transcriptional networks that define alternate T cell fates and indicate that Batf plays an indispensable role in both inducing and maintaining the Th17 program through its actions to regulate the competing actions of Stat5‐assembled enhanceosomes that promote Th1‐ and Treg‐cell developmental programs.
Keywords: Batf, Ets1, plasticity, Runx1, Th17
Subject Categories: Chromatin, Transcription & Genomics; Immunology
Batf impairs assembly of a Stat5‐Ets1‐Runx1 complex that promotes Th1 and Treg‐cell developmental programs to maintain Th17 cell identity.
Introduction
The emergence of alternative CD4 T cell fates is controlled by transcriptional networks activated downstream of T cell receptor (TCR) and cytokine signaling inputs. Th17 cells develop from antigen‐activated naïve CD4 T cells in the presence of interleukin‐6 (IL‐6) and transforming growth factor‐β (TGF‐β) (Weaver et al, 2007; Korn et al, 2009). TCR signaling induces the pioneering transcription factors, Batf and Irf4, which act cooperatively to alter chromatin accessibility at Th17‐specifying genomic loci (Ciofani et al, 2012) and, in concert with IL‐6–dependent Stat3, promote the expression of core Th17‐specific genes, including Il17a, Il17f, Il21, Il22, Il23r, and the Th17 master regulator transcription factor, Rorγt (Rorc) (Weaver et al, 2007; Korn et al, 2009; Ciofani et al, 2012). Th17 cells develop in barrier tissues both at homeostasis and in response to pathogen threats. They share overlapping developmental features with peripheral Treg cells (Weaver et al, 2006; Omenetti & Pizarro, 2015), with which they collaborate to maintain barrier function to restrain the commensal microbiota, and to orchestrate host defense against threats from an extracellular pathogen (Mucida & Salek‐Ardakani, 2009; Wang et al, 2014; Omenetti & Pizarro, 2015). However, Th17 cells are also central to immunopathology in multiple immune‐mediated disorders (Weaver et al, 2013). Underlying these deleterious functions of Th17 cells is their propensity to transdifferentiate into Th1‐like cells under inflammatory conditions (Geginat et al, 2016; Harbour et al, 2020; Loos et al, 2020) wherein either IL‐12 or prolonged IL‐23 signaling can promote expression of the Th1 hallmark transcription factor, Tbx21, and abrogate expression of Rorγt (Lexberg et al, 2008; Lee et al, 2009; Mukasa et al, 2010; Morrison et al, 2013). Conversely, ongoing classical IL‐6 signaling appears to be nonredundant in stabilizing and maintaining the Th17 phenotype (Harbour et al, 2020).
Batf expression, while required for early programming of Th17 development (Schraml et al, 2009; Ciofani et al, 2012), is sustained throughout Th17 differentiation and is amplified by IL‐6 signaling (Pham et al, 2019), suggesting that, in addition to pioneering chromatin accessibility at Th17 loci, Batf has other functions in maintaining the Th17 chromatin landscape. In Th17 cells, Batf heterodimerizes with other AP‐1 family proteins, JunB or JunD, to bind DNA (Li et al, 2012b; Murphy et al, 2013). Batf‐containing heterodimers co‐bind with Irf4 at AP‐1‐IRF composite elements, termed AICE motifs (Glasmacher et al, 2012; Li et al, 2012b), to assemble a trimeric complex that is required for Th17‐lineage programming (Ciofani et al, 2012; Glasmacher et al, 2012; Li et al, 2012b). Optimal development of Tfh, Th2, and Th9 cells similarly requires Batf (Schraml et al, 2009; Betz et al, 2010; Jabeen et al, 2013; Sahoo et al, 2015; Kuwahara et al, 2016; Iwata et al, 2017) and given the overlapping gene targets of Batf and Irf4 in these other T cell subsets, Batf and Irf4 likely act cooperatively in these cells as well (Murphy et al, 2013; Huber & Lohoff, 2014). It was recently reported that the principal AP‐1 partner of Batf in Th17 cells is JunB, which, like Batf, is required for optimal Th17 cell development, in part by repressing the expression of genes that control pTreg‐ and Th1‐cell fates (Carr et al, 2017), albeit by mechanisms not fully understood.
In addition to its pioneering function to alleviate chromatin constraints at key gene regulatory elements, Batf has also been shown to reorganize chromatin structure in T cells, at least in part by cooperating with the transcription factor, Ets1, to recruit the architectural trans‐factor CTCF at non‐AICE genomic targets (Pham et al, 2019). Ets1 is a member of a winged helix‐turn‐helix transcription factor family that shares a unique Ets domain and recognizes the core GGAA/T DNA sequence element (Garrett‐Sinha, 2013). To optimally bind DNA, Ets1 requires the cooperation of additional factors to block its autoinhibitory domain, and in T cells, Runx1 is the preferred partner (Garrett‐Sinha, 2013; Kasahara et al, 2017). Together, Ets1 and Runx1 recruit PolII (Cauchy et al, 2016) and CBP/p300 (Kitabayashi et al, 1998; Yang et al, 1998; Hollenhorst et al, 2009) to enhancer elements to promote gene transcription. Ets1 family members have been reported to interact with AP‐1 factors to cooperatively activate transcription in human T cells (Bassuk & Leiden, 1995), and genomic distal regulatory elements that become accessible following TCR stimulation are enriched for AP‐1 and Ets motifs (Bevington et al, 2016). Moreover, in view of the prevailing view that Ets1 may suppress Th17 differentiation (Moisan et al, 2007), and the broad distribution of AP‐1 and Ets1 consensus binding sites across the T cell enhancer landscape, a greater understanding of the interplay of these factors in Th17 development and function is needed.
In this regard, it is interesting that Ets family factors have been reported to bind Stat5 in non‐T cells (Schwaller et al, 2000) and that Ets1 is reported to physically interact with Stat5 in T cells (Rameil et al, 2000). Although the implications of this association in lineage‐specific programming of T cells remain ill‐defined, it is particularly compelling in that IL‐2–dependent activation of Stat5 has been demonstrated to restrain Tfh and Th17 cell development (Laurence et al, 2007; Ballesteros‐Tato et al, 2012), while essential for the development of Foxp3+ regulatory T cells (Ross & Cantrell, 2018) and also supporting the development of Th1 and Th2 cells (Liao et al, 2008, 2011). Although a precise understanding of the role of IL‐2 in modulating effector CD4 T cell development is unclear, mechanisms to limit IL‐2 signaling in order to maintain a balance of Stat3 and Stat5 that is favorable to Th17 programming and phenotype stability appear to be central.
Here, we find that in addition to activating the core genes of the Th17 program, Batf also acts to restrict the expression of genes of the Treg and Th1 programs through its actions to restrain IL‐2‐induced Stat5 activation, thereby limiting Stat5‐dependent recruitment of Ets1‐Runx1 factors to Th1‐ and Treg‐cell‐specific gene loci. Accordingly, Batf acts both to pioneer regulatory elements in Th17‐specific genes and limit assembly of a Stat5‐Ets1‐Runx1 enhanceosome that appears to be required for optimal expression of Th1‐ and Treg‐cell‐specific genes. Our findings define a new role for Batf in T cell differentiation and unveil an important role for Stat5‐Ets1‐Runx1 interactions in transcriptional networks that define alternative T cell developmental fates.
Results
In a transcriptomic survey of WT and Batf‐deficient (Batf KO) T cells polarized under Th17 conditions, diminished expression of Th17 signature genes was observed in cells lacking Batf (Schraml et al, 2009), including Il17a, Il17f, Il21, Maf, Il23r and the hallmark transcription factor, Rorc (Figs 1A and EV1A and B). Accordingly, Gene Ontogeny (GO) analysis indicated that pathways associated with Th17 development including glycolysis, mTORC1 and TGF‐β signaling, and inflammatory responses were enriched in WT Th17 compared with Batf KO cells (Fig 1B; Mangan et al, 2006; Veldhoen et al, 2006; Kurebayashi et al, 2012; Sun et al, 2017). While Batf2 and Batf3 can have compensatory functions in the absence of Batf, Batf2 expression was negligible in WT and KO cells and Batf3 expression was reduced suggesting that the phenotype of the Batf −/− animals was not impacted by these factors (Murphy et al, 2013). Notably, however, a subset of genes related to the interferon response and Foxp3 targets was increased in absence of Batf (Figs 1A and B, and EV1A and B). Consistent with these findings, Batf‐deficient Th17 cells showed increased IFN‐γ and Foxp3 expression with a concomitant decrease in IL‐17a production (Fig 1C and D). IFN‐γ levels were also enhanced in Batf KO Th0 indicating that Batf repression of IFN‐γ is not unique to Th17 cells (Fig EV1D). However, Batf deficiency had no effect on IFN‐γ and Foxp3 expression in cells cultured under iTreg conditions likely due to low Batf expression in WT cells (Fig EV1C). Antibody‐mediated blockade of IFN‐γ completely abrogated IFN‐γ expression in both WT and Batf KO Th0 cells, demonstrating that increased IFN‐γ expression was a cell‐intrinsic result of Batf deficiency in Th17 cultured cells (Fig EV1D). siRNA knock‐down of Batf during Th17 cell differentiation similarly resulted in reduced Rorc and IL‐17a expression and enhanced IFN‐γ and Foxp3 (Fig 1E and F). In addition to Rorc, transcripts encoding other transcription factors that contribute to Th17 development, including Ap‐1 family members (Junb and Fosl2) and Klf2, were also modulated by Batf deficiency (Fig EV1B; Ciofani et al, 2012; Jin et al, 2012; Carr et al, 2017; Downs‐Canner et al, 2017; Hasan et al, 2017). Thus, in agreement with previous reports (Schraml et al, 2009; Ciofani et al, 2012), Batf is required for Th17 differentiation, but it also represses genes associated with Th1‐ and Treg‐cell differentiation.
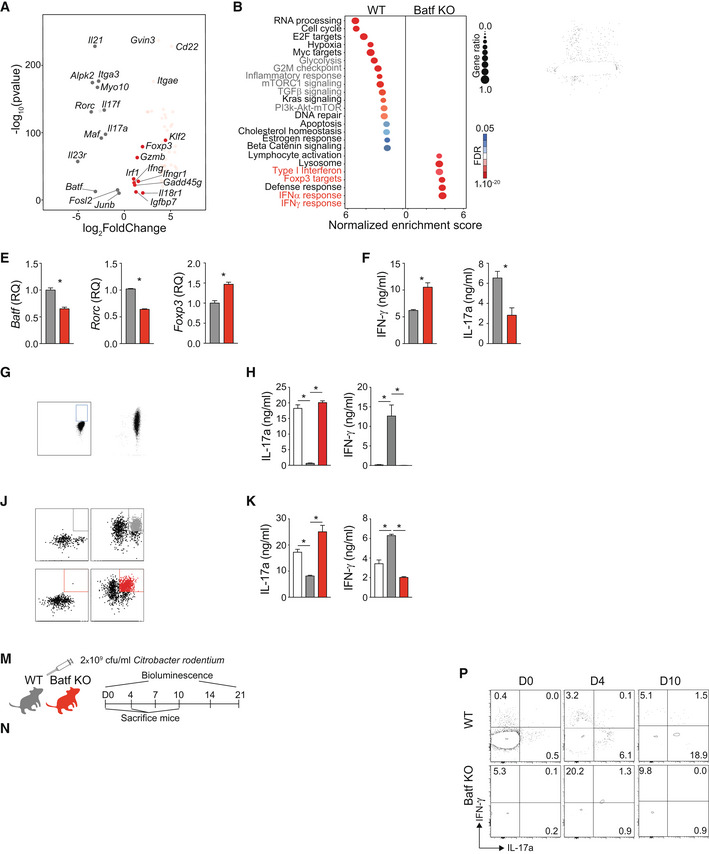
Figure 1. Batf‐deficient CD4 T cells exhibit Th1/Treg‐like phenotypes and fail to protect against Citrobacter rodentium infection.
-
A, BDifferential gene expression in WT versus Batf KO Th17 cells. Naïve WT and Batf KO CD4+CD62Lhi T cells were cultured under Th17‐polarizing conditions for 5 days. Differentiated Th17 cells were reactivated with anti‐CD3 for 6 h, and total RNA was extracted for RNA sequencing. Differential gene expression (fold change > 2, fdr < 0.05) was represented as a volcano plot (A) and subjected to pathway analysis (B).
-
CDay 5 differentiated WT and Batf KO Th17 cells were restimulated with PMA and ionomycin for 5 h, and stained intracellularly for flow cytometric analysis of IFN‐γ, IL‐17a, and Foxp3 expression, represented by plots with percentage ± s.e.m. of positive cells.
-
DHistograms represent Foxp3 expression in resting day 5 WT and Batf KO Th17 and Treg.
-
E, FNaïve WT CD4+CD62Lhi T cells were cultured under Th17‐polarizing conditions. On day 3, cells were transfected with control or siRNA targeting Batf, rested overnight, used for total RNA isolation to assess gene expression (E), or restimulated with anti‐CD3 for 24 h to measure cytokine production by ELISA (F).
-
G–INaïve CD4+CD62Lhi T cells from IL‐17a reporter mice (Il17a eGFP) were cultured under Th17‐polarizing conditions. On day 5, eGFP+/IL‐17a+ cells were sorted and cultured for two additional rounds in the presence of anti‐CD3 alone (TCR(R3)) or Th17‐polarizing cytokines (Th17(R3)) (G). Cells from round 1 (Th17(R1)) and round 3 (TCR(R3) and Th17(R3)) were used to measure cytokine production by ELISA after 24 h of anti‐CD3 stimulation (H) or were stained intracellularly for Batf expression (I).
-
J–LNaïve CD4+CD62Lhi T cells from IL‐17a reporter mice (Il17a eGFP) were cultured under Th17‐polarizing conditions. On day 2, cells were transduced with control (Empty) or retrovirus expressing Batf‐Thy1.1 (Batf). eGFP+/IL‐17a+Thy1.1+ cells, sorted on day 5 and cultured for two additional rounds in the presence of anti‐CD3 alone (J). Thy1.1+ cells from round 1 (Thy1.1(R1)) and round 3 (Thy1.1(R3) and Batf(R3)) were restimulated with 2 μg/ml anti‐CD3 for 24 h to measure cytokine production by ELISA (K) or were stained intracellularly to measure Batf expression (L).
-
M–PSchematic of Citrobacter rodentium (Cr) infection model using WT and Batf KO mice. WT and Batf KO mice were orally infected by gavage with 2 × 109 cfu/ml of Citrobacter rodentium (Cr) (M). Luminescence measurements were obtained (N) with quantification of the signal (O) at the indicated time points. Cr‐infected WT and Batf KO were sacrificed on day 4 and day 10. Cells were isolated from the colon, restimulated with PMA and ionomycin, and stained intracellularly for flow cytometric analysis of cytokine production and Foxp3 expression presented as density plots (P). Data are gated on viable CD4+TCRβ+.
Data information: Data are representative of two independent experiments with similar results (A, B), mean ± s.e.m. of 3–5 independent experiments (C–L), or mean ± s.e.m. of n = 5–6 mice (O) and representative of two independent experiments (M–P). (*P < 0.05; two‐sided t‐test or one‐way ANOVA followed by Tukey's test). RQ, relative quantification.
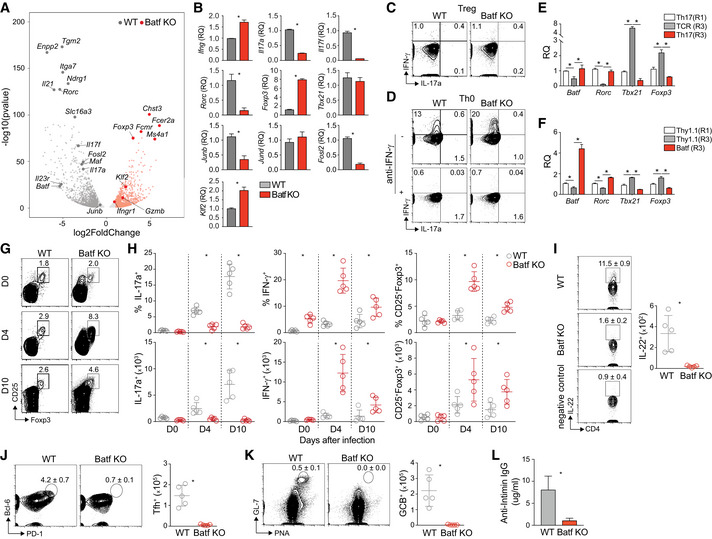
Figure EV1. Batf deficiency skews CD4 T cell development toward Th1/Treg phenotypes resulting in altered response to Citrobacter rodentium infection.
-
ANaïve WT and Batf KO CD4+CD62Lhi T cells were cultured under Th17‐polarizing conditions. On day 5, total RNA was extracted from resting Th17 cells and used for RNA‐seq. Differential gene expression (fold change > 2, fdr < 0.05) was represented as a volcano plot.
-
BNaïve WT and Batf KO CD4+CD62Lhi T cells were cultured under Th17‐polarizing conditions for 5 days. Total RNA was extracted from resting or restimulated (6 h with anti‐CD3) cells and used for gene expression analysis by RT–PCR (resting: Rorc, Foxp3, Tbx21, Junb, Jund, Fosl2, and Klf2; restimulated: Ifng, Il17a, and Il17f).
-
C, DDay 5 of differentiated WT and Batf KO cells cultured under Treg (C) or Th0 conditions (D, cultured with or without anti‐IFN‐γ) were restimulated with PMA and ionomycin for 5 h, and stained intracellularly for IFN‐γ and IL‐17a expression.
-
ENaïve CD4+CD62Lhi T cells from IL‐17a reporter mice (Il17a eGFP) were cultured under Th17‐polarizing conditions. On day 5, eGFP+/IL‐17a+ cells were sorted and cultured for two additional rounds in the presence of anti‐CD3 alone (TCR(R3)) or Th17‐polarizing cytokines (Th17(R3)). Cells from round 1 (Th17(R1)) and round 3 (TCR(R3) and Th17(R3)) were used for total RNA isolation to assess gene expression by RT–PCR (data normalized to Th17(R1)).
-
FNaïve CD4+CD62Lhi T cells from IL‐17a reporter mice (Il17a eGFP) were cultured under Th17‐polarizing conditions. On day 2, cells were transduced with control (Empty) or retrovirus expressing Batf‐Thy1.1 (Batf). eGFP+/IL‐17a+Thy1.1+ cells were sorted on day 5 and cultured for two additional rounds in the presence of anti‐CD3 alone. Thy1.1+ cells from round 1 (Thy1.1(R1)) and round 3 (Thy1.1(R3) and Batf(R3)) were used for total RNA isolation to assess gene expression by RT–PCR (data normalized to Thy1.1(R1)).
-
G, HWT and Batf KO mice were infected by gavage with 2 × 109 cfu/ml Citrobacter rodentium (C.r.). Cells were isolated from the colon, restimulated with PMA and ionomycin, and stained intracellularly for flow cytometric analysis of cytokine production and Foxp3 (G) expression and were presented as percentages of the indicated cell population (H, top) and numbers of positive cells (H, bottom) at the indicated time points.
-
IMice were sacrificed on day 10 postinfection. Cells were isolated from the colon tissue, restimulated with PMA and ionomycin, and intracellularly stained for cytokine production. IL‐22‐producing CD4+ T cells on day 10 postinfection were assessed in the colon of infected WT and Batf KO mice with contour plots showing percentage ± s.e.m. and average of cell numbers. Data are gated on viable TCRβ+. Unstimulated cells were used as negative controls.
-
J–LCells from MLN of WT and Batf KO mice on day 10 postinfection were used for Tfh (CD4+CD44+CXCR5+PD‐1+Bcl6+) (J) and germinal center B cell (B220+PNA+GL‐7+) (K) staining with contour plots showing percentage ± s.e.m. and average of cell numbers. Anti‐Intimin IgG antibody was measured using serum of WT and Batf KO mice on day 21 postinfection (L).
Data information: Data are representative of two independent experiments with similar results (A) or mean ± s.e.m. of three independent experiments (B–F). (*P < 0.05; two‐sided t‐test). RQ, relative quantification. Data are gated on viable CD4+TCRβ+ and mean ± s.e.m. of n = 5 mice and representative of two independent experiments with similar results (G–L) (*P < 0.05; two‐sided t‐test).
Studies by this lab and others have established that Th17 cells are highly plastic and that without sustained IL‐6 and TGF‐β signaling, acquire a Th1‐like signature, similar to what was observed in Batf‐deficient Th17 cells (Lexberg et al, 2008; Lee et al, 2009; Harbour et al, 2020; Fig 1A–L). We therefore postulated that Batf may stabilize the Th17 cell phenotype and thus play an important role in immune homeostasis and Th17‐mediated host defense and disease. To examine the impact of Batf on Th17 phenotype stability, we employed an in vitro T cell culture system to assess Th17 plasticity (Lee et al, 2009) (Fig 1G–L). As Th17 cells stimulated repeatedly in the absence of exogenous cytokines shifted toward a Th1‐like phenotype we noted that, compared to cells maintained with TGF‐β and IL‐6 (Th17 R3), Batf expression decreased significantly by the third round of culture (TCR R3) in cells maintained without exogenous cytokines (Fig 1G–I). This loss of Batf expression correlated with reduced Rorc and IL‐17a, and increased IFN‐γ, Foxp3, and Tbx21, and differed from Th17‐polarized Batf KO cells in which T‐bet expression was unaffected (Fig EV1B and E). This suggested that the maintenance of Batf expression by exogenous cytokines might underpin the Th17 phenotype. In agreement with this, ectopic expression of Batf in WT Th17 cells by retroviral transduction sustained Rorc and IL‐17a expression and limited Tbx21, Foxp3, and IFN‐γ expression (Batf (R3)) compared with cells transduced by empty‐vector virus expressing the reporter alone (Thy1.1; Figs 1J–L and EV1F). Collectively, these data indicate that Batf not only promotes Th17 development but also stabilizes the Th17 program and represses Th1 and regulatory phenotypes by modulating key cytokines and transcription factors.
To extend these findings in vivo, we examined mice infected with the enteric pathogen, Citrobacter rodentium (Cr), which induces a protective Th17 response (Fig 1M–P) (Weaver & Hatton, 2009; Shiomi et al, 2010; Wang et al, 2014; Silberger et al, 2017). In WT mice challenged with a luminescent strain of Cr, bacterial burden sharply declined after 7 days (Fig 1M–O), in concert with the appearance of T cells expressing IL‐17a in the infected colon (Fig 1P). By contrast, Batf‐deficient mice failed to induce IL‐17a+ cells and were unable to control the bacterial load (Fig 1M–P). IL‐22–expressing CD4 T cells (Th22 cells), which are also important in host defense against Cr, were similarly abrogated in the absence of Batf, contributing to the impaired response (Fig EV1I). Consistent with in vitro findings, Batf‐deficient CD4 T cells defaulted primarily to a Th1‐like phenotype in mice challenged with Cr (Fig 1P). Regulatory cells were also expanded, although to a lesser degree (Fig EV1G). Thus, IFN‐γ‐producing and Foxp3‐expressing cells (Figs 1P and EV1G) were significantly increased in Batf KO animals (Figs 1N and O, and EV1H). Batf −/− animals have fewer ILC3s in the colon and are less effective at controlling Cr infection relative to WT mice (Liu et al, 2020). Consistent with our published studies, this is likely due in part to diminished ILC3‐produced IL‐22 (Liu et al, 2020) and we suspect this contributes to overall colitic phenotype in the Batf KO animals during the early phase of the infection (Zindl et al, 2022). In agreement with a requirement for Batf expression in the development of T follicular helper (Tfh) cells and germinal center B (GCB) cells (Betz et al, 2010; Sahoo et al, 2015), Cr‐infected Batf KO mice were profoundly deficient in these responses and, accordingly, exhibited a defective antibody response to the Cr virulence factor intimin, further compounding the impaired host response (Fig EV1J–L). Together, these results indicate that Batf is not only indispensable in driving host‐protective Th17 and Tfh responses to Cr but is also important in tempering the development of Foxp3+ T cells that may dampen the host‐protective Th17/Th22 response. It further suggests that the Th1‐cell response generated is inadequate to control infection.
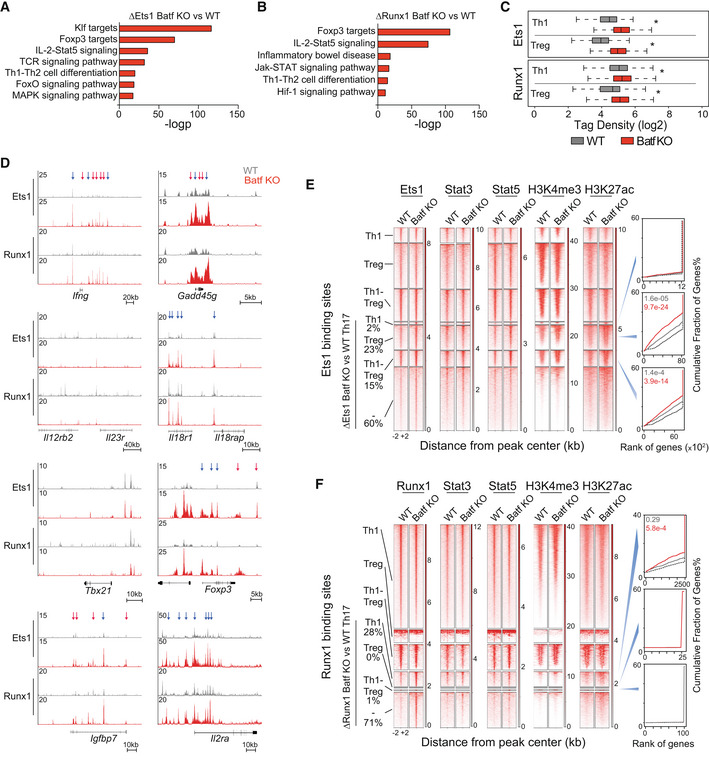
The foregoing studies expose an underappreciated role of Batf in regulating the expression of genes associated with Th1 and Treg phenotypes during Th17 development, with features similar to that recently described for JunB (Carr et al, 2017). Batf functions as a pioneer factor and chromatin modifier in CD4 T cells (Ciofani et al, 2012; Karwacz et al, 2017; Pham et al, 2019). To investigate whether repression of Th1‐ and Treg‐like genes by Batf during Th17 programming might occur via modulation of the chromatin landscape of phenotype‐defining loci, we performed ATAC‐seq and surveyed active (H3K4me3 and H3K27Ac) and repressive (H3K27me3) histone marks in WT versus Batf KO Th17‐polarized cells (Figs 2A and EV2A,B). As predicted (Ciofani et al, 2012), in WT cells subjected to ATAC‐seq, Th17‐specifying loci (e.g., Il23r, Il17a‐f, Rorc, and Ahr) were in an “open” configuration (Fig EV2A) with substantial H3K4me3 and H3K27Ac marks and limited H3K27me3 marks (Fig 2A). By contrast, H3K4me3 marks across the Il17a‐f locus were lost and were greatly diminished at Rorc in absence of Batf, while H3K27me3 generally remained low and little chromatin accessibility was identified by ATAC‐seq (Figs 2A and EV2A). Notably, H3K27me3 was substantially reduced at select Th1‐ and Treg‐associated loci whereas H3K4me3 marks were mostly unaffected by Batf deficiency (Figs 2A and EV2B). Genome‐wide assessment showed that normalized signal intensity of H3K27me3 centered around Batf sites at Th1 and Treg loci (Li et al, 2012b) was significantly reduced in the absence of Batf and was accompanied by increases in H3K4me3 at Treg loci, while H3K4me3 at Th1 loci remained unchanged (Fig EV2C). Though Th17‐specifying gene expression was decreased in Batf KO Th17 (Figs 1A and EV1A; Schraml et al, 2009; Ciofani et al, 2012) recruitment of H3K27me3 to Th17 loci in Batf KO Th17 cells was also reduced, likely due to diminished chromatin accessibility (Ciofani et al, 2012; Karwacz et al, 2017; Pham et al, 2019). Concurrent Batf and H3K27me3 recruitment to the Ifng locus in WT Th17 cells suggests direct repression of IFN‐γ expression and may contribute to Th17 lineage stabilization (Fig 2A). Collectively, these data suggest that in WT Th17 cells Treg and Th1 loci mainly exist in a poised chromatin conformation composed of Batf‐independent permissive marks and Batf‐dependent repressive marks that restrain expression, whereas at Th17‐specifying loci Batf functions primarily as a pioneer factor.
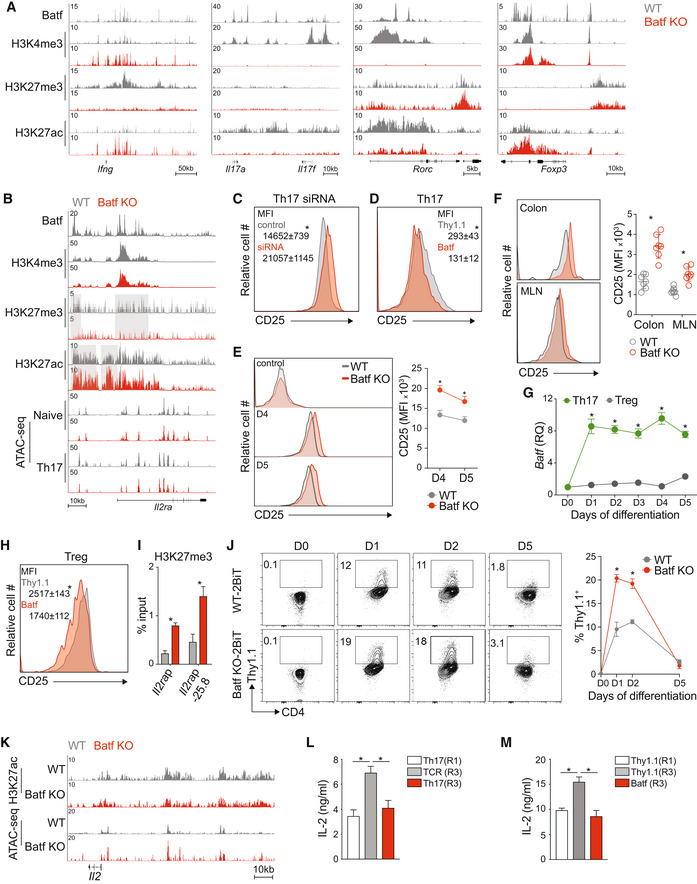
Figure 2. Batf modulates Il2ra and Il2 expression in Th17 cells.
-
A, BNaïve WT and Batf KO CD4+CD62Lhi T cells were cultured under Th17‐polarizing conditions for 5 days. ChIP‐seq for transcription factor (Batf) and histone modifications (H3K4me3, H3K27me3, and H3K27ac) was performed using day 5 WT and Batf KO Th17 cells (A, B). Chromatin accessibility (ATAC‐seq) was performed using naïve or day 2 WT and Batf KO cells cultured under Th17 conditions (data from GSE123209) (B). Tracks visualized using the Integrated Genome Browser (IGB) show the chromatin states of naïve and 48 h Th17 cells compared between WT and Batf KO cells in the Il2ra gene locus (B).
-
CNaïve WT CD4+CD62Lhi T cells were cultured under Th17‐polarizing conditions. On day 3, cells were transfected with control or siRNA targeting Batf. After 24 h, cells were stained for cell surface expression of CD25 and analyzed by flow cytometry (MFI, mean fluorescence intensity).
-
DNaïve WT CD4+CD62Lhi T cells were cultured under Th17‐polarizing conditions. On day 2, cells were transduced with control or retrovirus expressing Batf‐Thy1.1. On day 5 cells were stained for cell surface expression of CD25 and analyzed by flow cytometry with quantified mean fluorescence intensity (MFI). Data are gated on Thy1.1 expression.
-
ENaïve WT and Batf KO CD4+CD62Lhi T cells were cultured under Th17‐polarizing conditions. Naïve (control), day 4, and day 5 of differentiated WT and Batf KO Th17 cells were harvested and stained for cell surface expression of CD25 and analyzed by flow cytometry (MFI, mean fluorescence intensity).
-
FWT and Batf KO mice were orally infected by gavage with 2 × 109 cfu/ml of Citrobacter rodentium (Cr). Cr‐infected WT and Batf KO were sacrificed on day 4. Cells were isolated from mesenteric lymph nodes (MLN) and colonic lamina propria (CLP) and stained for CD25 expression presented as histograms. Data are gated on viable CD4+TCRβ+ cells.
-
GNaïve WT and Batf KO CD4+CD62Lhi T cells were cultured under Th17 or Treg polarizing conditions for 5 days. The kinetics of Batf gene expression was assessed by RT–PCR at the indicated time points (data were normalized to naïve WT cells).
-
H, INaïve WT CD4+CD62Lhi T cells were cultured under iTreg polarizing conditions. On day 2, cells were transduced with control (Thy1.1; Empty) or Batf‐Thy1.1 retrovirus. Thy1.1+ cells were sorted on day 5 and stained for cell surface expression of CD25 and analyzed by flow cytometry (MFI, mean fluorescence intensity) (H) and assessed for H3K27me3 binding at the Il2ra locus by ChIP–qPCR (I).
-
JNaïve WT‐2BiT and Batf KO‐2BiT CD4+CD62Lhi T cells were cultured under Th17‐polarizing conditions. IL‐2‐Thy1.1 production comparing WT‐2BiT and Batf KO‐2BiT cells was measured by flow cytometry at the indicated time points and frequencies of positive cells determined.
-
KTracks visualized using the Integrated Genome Browser (IGB) indicate the chromatin state of the Il2 locus in 5d (H3K27ac) VS 48 h (ATAC) WT and Batf KO Th17 cells.
- L, M
Data information: Data are representative of two independent experiments with similar results (A, B, and K), mean ± s.e.m. of n = 7 mice and representative of two independent experiments (D), or mean ± s.e.m. of 3–5 independent experiments (C, E–J, L and M) (*P < 0.05; two‐sided t‐test).
Figure EV2. Batf modulates chromatin landscape in Th17 cells.
-
AChromatin accessibility (ATAC‐seq) was performed using naïve or day 2 WT and Batf KO cells cultured under Th17 conditions (data from GSE123209). Tracks visualized using the Integrated Genome Browser (IGB) compare chromatin states of WT and Batf KO Th17 in the indicated gene loci.
-
BNaïve CD4+CD62Lhi WT and Batf KO T cells cultured under Th17‐polarizing conditions for 5 days were assessed for binding of Batf, H3K4me3, H3K27me3, and H3K27ac at selected loci.
-
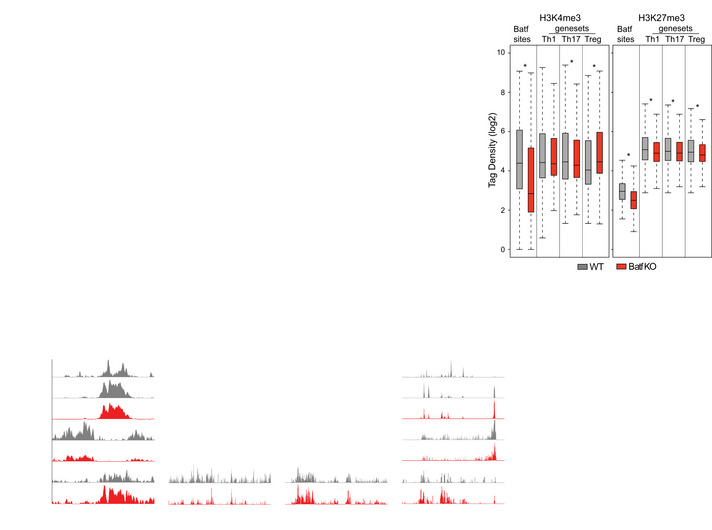
CNaïve WT and Batf KO CD4+CD62Lhi T cells were cultured under Th17‐polarizing conditions for 5 days and ChIP‐seq for transcription factors Batf and histone modification (H3K4me3 and H3K27me3) was performed. Boxplots display normalized tag density of H3K4me3 (left) and H3K27me3 (right) centered around ±1 kb of Batf sites or ± 2.5 kb of peak centers that were annotated to the nearest Th1‐, Th17‐, and Treg‐specific genes (Li et al, 2012b; GSE39756) compared between WT and Batf KO Th17‐polarized cells. Central bands, boxes, and whiskers show median, upper and lower quartile, and maximum and minimum values, respectively.
-
DNaïve WT CD4+CD62Lhi T cells were cultured under iTreg polarizing conditions. On day 2, cells were transduced with control (Thy1.1; Empty) or Batf‐Thy1.1 retrovirus. Thy1.1+ cells were sorted on day 5 and used for total RNA isolation to assess gene expression by RT–PCR (data normalized to Thy1.1).
Data information: Data are representative of two independent experiments with similar results (A–C), *P < 0.05 (Mann–Whitney test); or mean ± s.e.m. of three independent experiments (D) (*P < 0.05; two‐sided t‐test).
Upon further examination of genome‐wide chromatin accessibility in Batf KO Th17 cells, we noted that the Il2ra locus resembled Th1 and Treg loci, having greatly diminished H3K27me3 with stable H3K4me3 marks (Fig 2B). Increased acetylation was also observed, suggesting heightened Il2ra transcriptional activation in the absence of Batf (Fig 2B). Consistent with this, Th17 cells in which Batf expression was repressed by siRNA transfection showed enhanced CD25 expression compared with control transfectants (Fig 2C). Conversely, the overexpression of Batf substantially decreased Il2ra expression (Fig 2D). Accordingly, CD25 expression was increased on Batf KO Th17 cells, particularly in the later stages of culture on days 4 and 5 (Fig 2E). Consistent with these findings, CD4 T cells isolated from the colon and mesenteric lymph node (MLN) of C.r‐infected Batf KO mice expressed significantly higher levels of CD25 compared with cells from infected WT controls (Fig 2F).
IL‐2 receptor signaling restrains Th17 differentiation (Laurence et al, 2007) but is essential for Treg‐cell development and augments Th1 development (Fujimura et al, 2013; Toomer et al, 2019). The foregoing studies suggested a model in which Batf reinforces the Th17 program, in part by downregulating the high‐affinity IL‐2 receptor, thereby limiting Stat5 activation. Supporting this model, we found that Batf transcript levels were significantly higher in Th17 versus induced Treg cells (Fig 2G). While not essential for Treg development, Batf is required for the differentiation of Ccr7lo tissue‐resident effector Tregs (eTreg) (Vasanthakumar et al, 2015; Hayatsu et al, 2017; Delacher et al, 2020), where it is expressed at substantially higher levels than in Ccr7hi central Treg (cTreg) cells (Hayatsu et al, 2017). Reflecting an enhanced requirement for IL‐2, Ccr7hi cTreg cells have significantly greater CD25 expression than Ccr7lo eTreg cells (Smigiel et al, 2014). In view of this inverse correlation of Batf with CD25 expression and based on our findings in Th17 cells (Fig 2C–E), we examined the ability of Batf to repress the IL‐2 receptor complex in regulatory T cells. The overexpression of Batf in iTreg, as predicted, induced repressive H3K27me3 marks at the Il2ra locus and accordingly, reduced Il2ra and CD25 expression (Figs 2H and I, and EV2D). Moreover, consistent with diminished Il2ra (Burchill et al, 2007), Foxp3 expression was also suppressed by enforced Batf expression, consistent with prior findings (Zhang et al, 2018; Fig 2D). Together, these data implicate Batf as a regulator of Treg and Th17 identity through modulation of IL‐2 receptor expression.
In a previous study, we found that Th17 cells express high levels of IL‐2 (DiToro et al, 2018), a seemingly paradoxical observation given that IL‐2 potently inhibits the Th17 developmental program (Laurence et al, 2007). Because AP‐1 factors contribute to Il2 transcriptional activity (Jain et al, 1995), we tested whether Batf might also control IL‐2 expression as a means to further reinforce Th17 identity. By tracking IL‐2 production in Batf KO and WT mice that had been crossed with the IL‐2 reporter strain, 2BiT (DiToro et al, 2018), we found that early in Th17 differentiation, IL‐2 expression was modestly increased in the absence of Batf (Fig 2J). Accordingly, the Il2 locus was more permissible in the absence of Batf on day 2 but by day 5 was indistinguishable from WT cells (Fig 2K). In a culture model of Th17 plasticity (Lee et al, 2009), as Batf expression diminished over multiple rounds of culture without added cytokines (Fig 1G–L), IL‐2 production increased (Fig 2L). The overexpression of Batf returned IL‐2 production to similar levels as R1 Th17 cells (Fig 2M). Taken together, these results suggest that Batf represses IL‐2 expression early in Th17 development as a mechanism to limit the autocrine upregulation of the IL‐2 receptor (Feng et al, 2014; Li et al, 2014; Hirai et al, 2021).
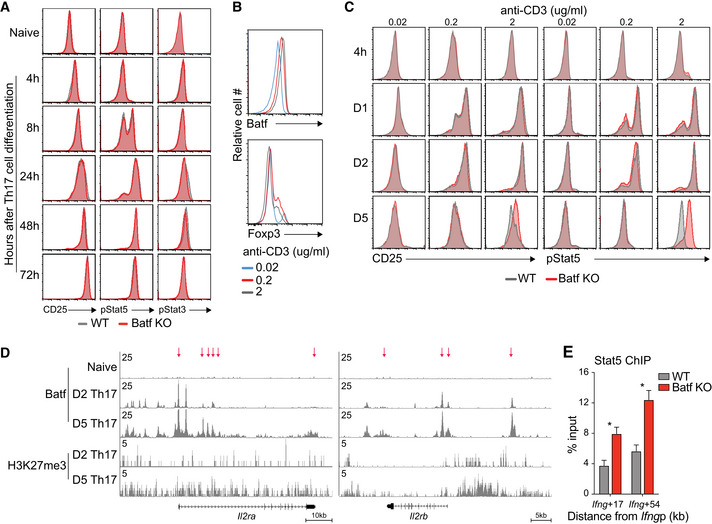
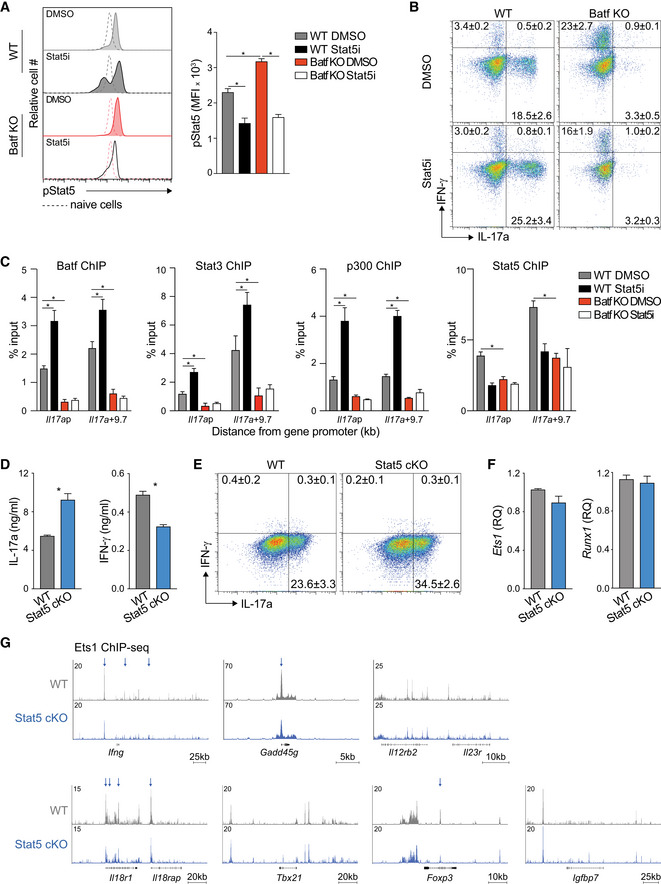
Batf‐dependent CD25 suppression and the associated modulation of pStat5 were most apparent during the later stages of Th17 cell differentiation (Figs 3A and B, and EV3A) and coincided with strong TCR stimulation conditions and heightened levels of Batf (Fig EV3B and C). This suggested that under robust antigenic activation, increased Batf expression underlies CD25 repression. Accordingly, ChIP‐seq analyses revealed greater recruitment of Batf and enhanced H3K27me3 deposition at the Il2ra locus on day 5 compared to earlier in development (Fig EV3D), providing a mechanism by which Batf could limit IL‐2 signaling and stabilize the Th17 phenotype. Similar increases were found for the gene encoding IL‐2Rβ, (Il2rb; Fig EV3D). Collectively, these results indicate that in addition to its pioneering functions, Batf may contribute to Th17 development by restraining IL‐2 production early in differentiation and may reinforce the program by repressing Il2ra expression later in development. Because IL‐2 signaling upregulates CD25 expression (Feng et al, 2014; Li et al, 2014; Hirai et al, 2021), both direct and indirect effects of Batf may be contributory.
Figure 3. Batf mediates Stat5 activation.
-
A, BNaïve WT and Batf KO CD4+CD62Lhi T cells were cultured under Th17‐polarizing conditions. Naïve (control), day 4, and day 5 of differentiated WT and Batf KO Th17‐polarized cells were harvested and stained intracellularly for flow cytometric analysis of phosphorylated Stat3 (pStat3) and phosphorylated Stat5 (pStat5) (A) with quantified mean fluorescence intensity (MFI) (B).
-
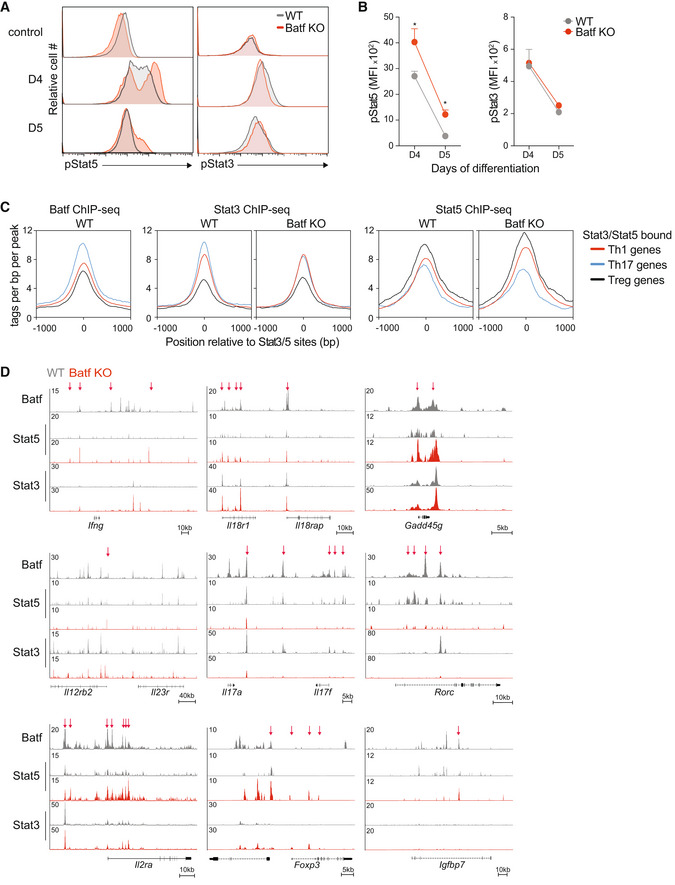
C, DChIP‐seq for Stat3 and Stat5 was performed after 1 h of IL‐6 and IL‐2 stimulation, respectively, using WT and Batf KO cells that were cultured under Th17‐polarizing conditions for 5 days. Histograms show Batf, Stat3, and Stat5 binding occupied around ±1 kb of peak centers of co‐bound Stat3‐Stat5 sites that were annotated near Th1, Th17, or Treg‐specific genes (C). Tracks show Batf, Stat5, and Stat3 ChIP‐seq data compared between WT and Batf KO Th17 in the indicated gene loci (WT, gray; Batf KO, red). Red arrows indicate differential Stat5 binding comparing WT and Batf KO cells (D).
Data information: Data are mean ± s.e.m. of 3–5 independent experiments (A, B) or representative of two independent experiments with similar results (C, D), or (*P < 0.05; two‐sided t‐test).
Figure EV3. Batf regulates the Il2ra gene expression in the late stage of Th17 cell differentiation.
-
A–CNaïve WT and Batf KO CD4+CD62Lhi T cells were cultured under Th17‐polarizing conditions with 2 μg/ml of anti‐CD3 (A) or with increasing amounts of anti‐CD3 (B, C). Expression of CD25 expression, phosphorylated Stat5 (pStat5), and phosphorylated Stat3 (pStat3) were assessed by flow cytometry at the indicated time points, (A, C), or on day 5 (B).
-
DTracks show Batf and H3K27me3 ChIP‐seq data from day 2 (D2) and day 5 (D5) post‐Th17 polarization at the indicated gene loci. Red arrows indicate regions of increased Batf and H3K27me3 recruitment on D5 vs. D2 Th17 cells.
-
ENaïve WT and Batf KO CD4+CD62Lhi T cells were cultured under Th0 conditions. Cells were collected on day 5 and used to assess Stat5 binding at the indicated genomic regions by ChIP–qPCR.
Data information: Data are representative of two independent experiments with similar results (A‐D) or mean ± s.e.m. of 3 independent experiments (E). (*P < 0.05; two‐sided t‐test).
Signaling through the IL‐2 receptor primarily results in the phosphorylation of Stat5 (Ross & Cantrell, 2018). In Th17 cells, IL‐2‐dependent increases in pStat5 can compete with IL‐6‐induced pStat3 for binding at the Il17a‐f locus, resulting in the inhibition of IL‐17 expression (Yang et al, 2011) and suggesting that heightened pStat5 destabilizes the Th17 phenotype by altering the balance of pStat3 and pStat5. Consistent with increased CD25 expression on d4 and d5 Batf KO Th17 cells (Fig 2C), pStat5 was enhanced whereas pStat3 was unaffected (Schraml et al, 2009; Fig 3A,B). These results point to a role for Batf in mediating control of pStat3‐pStat5 occupancy by limiting the availability of pStat5. Thus, we investigated genome‐wide Stat3 and Stat5 recruitment to Th1‐, Th17‐, and Treg‐like gene loci in the presence and absence of Batf, focusing on regions that bind both Stat3 and Stat5 (Fig 3C). As expected, and coinciding with Batf recruitment, Stat3 binding was greatest at Th17 gene loci and accordingly, was markedly impaired in Th17 cells lacking Batf (Fig 3C). By contrast, Stat5 occupancy was highest at chromosomal regions encompassing genes predominantly expressed by Th1 and Treg cells. Batf deficiency further increased Stat5 recruitment to Th1 and Treg loci while Stat3 binding at these locations remained unchanged (Fig 3C). We then interrogated Stat3 and Stat5 recruitment to phenotype‐specifying loci in 5d WT and Batf KO Th17 cells, to more precisely examine the consequence of Batf deficiency on the relative occupancy of these factors (Fig 3D). Consistent with genome‐wide analyses (Fig 3C), we found that while Stat3 binding was largely unperturbed, Stat5 recruitment was increased at Th1 (Ifng, Il18r1‐Il18rap, Gadd45g, Il12rb2) and Treg gene loci (Foxp3 and Igfbp7) in Batf KO cells compared with WT cells (Figs 3D and EV3E). However, while areas of increased Stat3 binding were detected in the Ifng and Il18r1 genomic regions in the absence of Batf, they were accompanied by an overall increase in Stat5 throughout the loci. Moreover, in Batf KO cells, recruitment of both Stat3 and Stat5 to the Il17a‐f and Rorc loci was impaired, reflecting the loss of Batf‐mediated chromatin accessibility (Ciofani et al, 2012; Karwacz et al, 2017; Pham et al, 2019; Fig 3D). Notably, Stat5 occupancy within and upstream of the Il2ra gene was substantially enhanced in Batf‐deficient Th17 cells (Fig 3D). Taken together, these data suggest that by suppressing the expression of the IL‐2 receptor and its Stat5 output, Batf shifts the balance of Stat3 and Stat5 in favor of Stat3 at Th17‐defining loci, thereby promoting gene expression of associated genes.
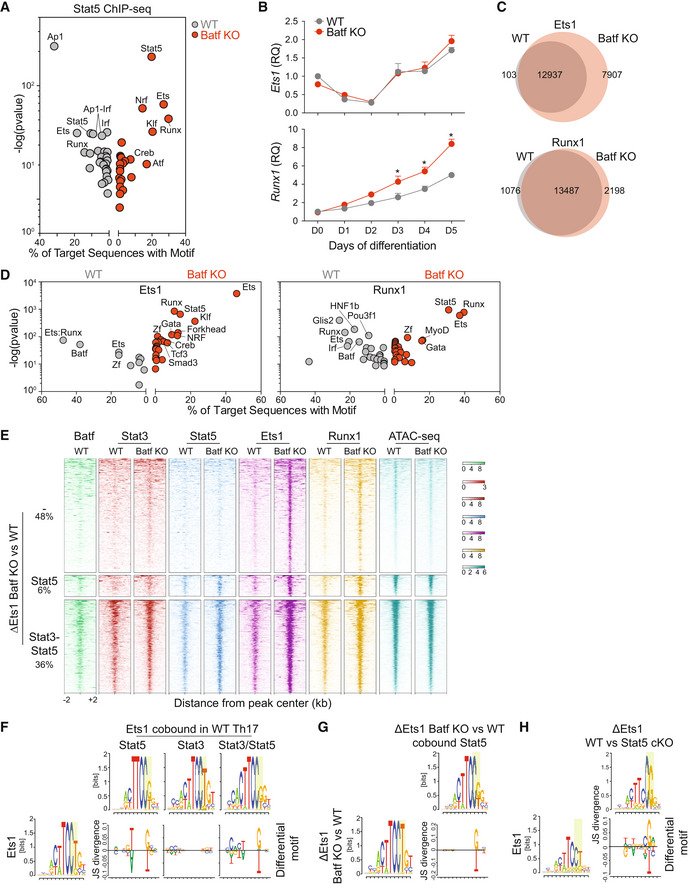
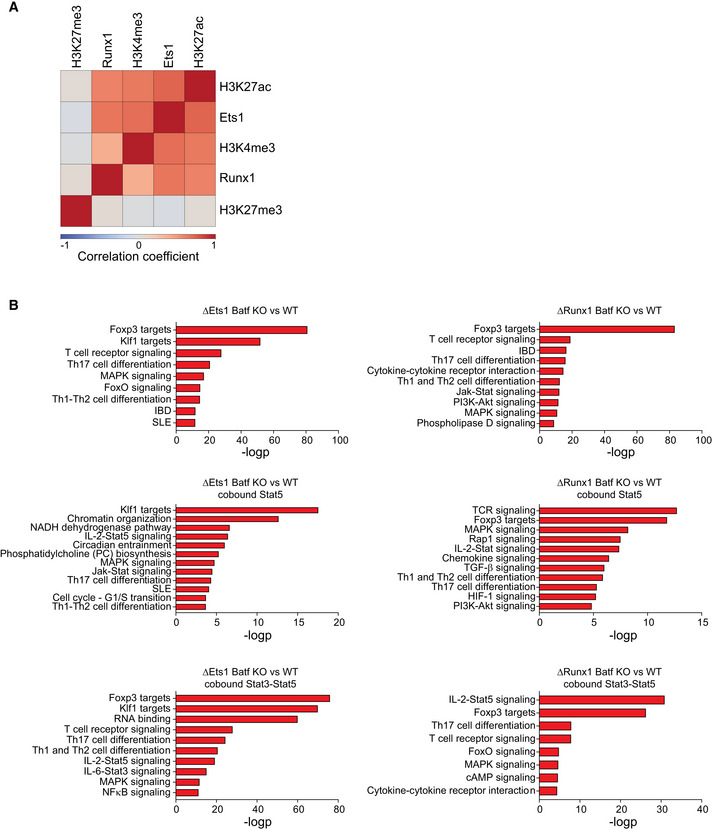
In view of our finding of Batf‐modulated Stat5 recruitment to Th1 and Treg loci in Th17 cells and in light of the ability of Stat5 to interact with several transcription factors to enhance transcription (Able et al, 2017), we sought to identify Stat5‐cooperating partners that might modulate gene expression. Motif analyses of Stat5 peaks in Batf KO compared with WT cells (Fig 4A) revealed that in addition to the expected Stat5 motif, motifs corresponding to Nrf, Klf, Ets, and Runx were substantially enriched (Fig 4A). While Nrf and Klf factors have not been reported to associate with Stat5, both Ets and Runx factors have (Rameil et al, 2000; Ogawa et al, 2008).
Figure 4. Batf‐dependent Stat5 activation modulates Ets1 and Runx1 binding in Th17 cells.
-
AStat5 ChIP‐seq was performed after 1 h of IL‐2 stimulation using WT and Batf KO cells that were cultured under Th17‐polarizing conditions for 5 days. De novo motif enrichment analysis was performed using differential Stat5‐binding sites compared between WT and Batf KO Th17 cells (fold change >, fdr < 0.05).
-
BNaïve WT and Batf KO CD4+CD62Lhi T cells were cultured under Th17‐polarizing conditions for 5 days. The kinetics of Ets1 and Runx1 gene expression was assessed by RT–PCR in the indicated time points (data normalized to naïve WT cells).
-
C, DNaïve WT and Batf KO CD4+CD62Lhi T cells were cultured under Th17‐polarizing conditions for 5 days and Ets1 and Runx1 ChIP‐seq were performed. Venn diagrams displaying differential Ets1 and Runx1 binding compared between WT and Batf KO Th17 cells (C, fold change > 2, fdr < 0.05), and de novo motif enrichment analysis (D) are shown.
-
EHeatmaps show Batf, Stat3, Stat5, Ets1, and Runx1 binding and chromatin accessibility (ATAC‐seq, data from GSE123209) at regions within ±2 kb from peak centers of enriched Ets1 peaks identified in Batf KO compared with WT cells (ΔEts1 Batf KO vs. WT) with the percentage of regions that co‐bound either Stat5 or Stat3‐Stat5 composite sites indicated.
-
F–HChIP‐seq for Stat3 and Stat5 was performed after 1 h of IL‐6 and IL‐2 stimulation, respectively, using day 5 WT Th17 cells. ChIP‐seq for Ets1 was performed using WT, Batf KO, or Stat5 cKO cells that were cultured under Th17‐polarizing conditions for 5 days. Comparison of Ets1 motif in unique Ets1 peaks and Ets1 peaks co‐bound by Stat3, Stat5, or both Stat3 and Stat5 in WT Th17 cells (F). Comparison of Ets1 motif in the enriched Ets1 peaks in Batf KO cells compared with WT cells (ΔEts1 Batf KO vs. WT) with or without co‐bound by Stat5 (G). Comparison of Ets1 motif in unique Ets1 peaks and enriched Ets1 peaks in Stat5 cKO cells compared with WT cells (ΔEts1 Stat5 cKO vs. WT) (H).
Data information: Data are mean ± s.e.m. of three independent experiments (B), or representative of two independent experiments with similar results (A, C, D–H). (*P < 0.05; two‐sided t‐test). RQ, relative quantification.
Ets1 and Runx1 play pivotal and complex roles in T helper cell development (Komine et al, 2003; Moisan et al, 2007; Ono et al, 2007; Mouly et al, 2010; Strempel et al, 2010; Lazarevic et al, 2011; Nguyen et al, 2012; Li et al, 2012a). Ets1 cooperates with T‐bet during Th1 development and, in Tregs, acts to demethylate Foxp3 (Grenningloh et al, 2005; Mouly et al, 2010). Optimal IL‐2 production is dependent on Ets1 (Grenningloh et al, 2005; Tsao et al, 2013) and it is thought that Ets1 suppresses Th17 cell differentiation largely via enhanced IL‐2 expression (Moisan et al, 2007). Conversely, the overexpression of Runx1 inhibits IL‐2 expression (Wong et al, 2011) and promotes Th17 cell development by enhancing Rorc expression (Zhang et al, 2008a). Runx1 also reinforces Treg‐cell differentiation by maintaining Foxp3 expression (Ono et al, 2007; Rudra et al, 2009). Though Ets1 and Runx1 have independent T helper subset‐specific functions, the majority of Runx1‐bound regions colocalize with Ets1 at active enhancer elements genome‐wide, due in part to Runx1's direct interaction with and abrogation of the activity of the autoinhibitory module of Ets1, thereby facilitating Ets1 binding to DNA (Hollenhorst et al, 2009; Garrett‐Sinha, 2013). Together Ets1 and Runx1 cooperate with the transactivator CBP (CREB‐binding protein; Hollenhorst et al, 2009) and p300 (Jayaraman et al, 1999; Iwatani et al, 2010) to promote gene expression. Thus, these data suggest that in the absence of Batf, Stat5 cooperates with Ets1, Runx1, and chromatin modifiers to direct gene activation at Th1 and Treg loci in Th17 cells.
To examine how Ets1 and Runx1 function to enhance the expression of Th1 and Treg genes in Batf KO Th17 cells, we first assessed the effects of Batf deficiency on the expression of both factors during Th17 development (Fig 4B). Ets1 was highly expressed in naïve CD4 T cells and transiently downregulated following Th17 polarization, similar to that observed in Th1 and Th2 cells (Grenningloh et al, 2005). Expression was restored by day 3 and continued to increase, reaching its highest level on day 5, and notably was unaffected by Batf deficiency. Naïve CD4 T cells also express high levels of Runx1 (Komine et al, 2003), but in contrast to Ets1, expression steadily increased during Th17 differentiation and was enhanced in the absence of Batf (Fig 4B). Based on the high levels of Ets1 and Runx1 on day 5 of Th17 cultures, we expected that Ets1 and Runx1 would be widely distributed genome‐wide; indeed, we found both Ets1 and Runx1 were recruited to approximately 13,000 genomic regions (Fig 4C). Ets1‐ and Runx1‐bound regions were also positively correlated with H3K4me3 and H3K27ac histone marks, thereby associating these factors with gene activation in late‐stage Th17 cells (Fig EV4A). Ets1 binding was increased in the absence of Batf, and Runx1 occupancy was also increased, albeit to a lesser degree than Ets1 (Fig 4C). Motif analyses indicated that regions bound uniquely by Ets1 (7907) or Runx1 (2198) in Batf KO cells were highly enriched for Stat5 while AP‐1 consensus sites were notably absent (Fig 4D). These data implicated a functional cooperation of Stat5, Ets1, and Runx1 that together promote Th1‐ and Treg‐like gene expression. Moreover, they suggested that assembly of this enhanceosome may not be inhibited by Batf binding directly but rather by Batf modulation of Il2ra expression that resulted in diminished Stat5 levels.
Figure EV4. Ets1 and Runx1 occupancy associates with active histone modification marks and Foxp3 targets and IL‐2–Stat5 pathway in the absence of Batf.
-
ANaïve WT CD4+CD62Lhi T cells were cultured under Th17‐polarizing conditions for 5 days and recovered cells used to perform ChIP‐seq using antibodies to Batf, Ets1, Runx1, H3K4me3, H3K27me3, and H3K27ac. Stat3 and Stat5 ChIP‐seq were performed after IL‐6 and IL‐2 stimulation, respectively, using day 5 WT Th17 cells. Heatmap shows Pearson correlation in transcription factor binding (Ets1 and Runx1) and chromatin states (H3K4me3, H3K27me3, and H3K27ac) in WT Th17 cells.
-
BNaïve WT and Batf KO CD4+CD62Lhi T cells were cultured under Th17‐polarizing conditions for 5 days and recovered cells used to perform ChIP‐seq using antibodies to Ets1 and Runx1. Stat3 and Stat5 ChIP‐seq were performed after IL‐6 and IL‐2 stimulation, respectively. Differential enriched Ets1 (ΔEts1 Batf KO/WT) and Runx1 (ΔRunx1 Batf KO/WT) peaks in Batf KO compared with WT cells that co‐bound either Stat5 or Stat3‐Stat5 sites were used for genomic annotation analysis.
To further examine the interplay of Stat5, Ets1, and Runx in Th17 cells genome‐wide, we interrogated the regions that uniquely bound Ets1 in the absence of Batf. Of the 7,907 Ets‐bound Batf KO unique peaks, approximately 42% colocalized with either Stat5 motifs or Stat3‐Stat5 composite sites and were associated with increased Stat5 binding in Batf KO cells compared with WT cells (Fig 4E). Though pStat3 expression was unaffected by Batf deficiency (Schraml et al, 2009; Fig 3A and B) increased Stat3 recruitment to the unique Ets1‐bound elements was also observed, indicating that enhanced Stat5 binding does not prevent Stat3 recruitment and suggesting a cooperative interaction of Stat3 with Stat5 at these regions. Approximately half of the unique Ets1‐bound peaks in Batf KO were Stat3‐Stat5–independent, likely reflecting increased Runx1 expression that functions to promote Ets1 DNA binding. Of note, Batf deficiency had little effect on chromatin accessibility at Th1 and Treg loci (Fig EV2A). Accordingly, the accessibility of Ets1‐bound unique regions was similarly unaltered demonstrating that increased Ets1 binding was not associated with greater accessibility (Fig 4E). Thus, Stat3‐Stat5 enrichment at unique regions of Ets1 binding in the absence of Batf supports a mechanism whereby Stat5 is central to activating Th1 and Tregs genes through association with Ets1 and Runx1.
Stat5 physically interacts with the autoinhibitory domain of Ets1 to release it from a closed conformation, thereby augmenting the DNA‐binding activity of Ets1 (Rameil et al, 2000; Dittmer, 2003; Ogawa et al, 2008). Stat5 may also increase the efficiency of Ets1 binding to DNA by modulating DNA methylation via an association with TET2 (Stephens & Poon, 2016; Ma et al, 2018). In view of the reported Stat5 functions that may promote Ets1 binding, we examined the DNA sequence of Ets1‐bound regions in the presence or absence of co‐bound Stat3‐Stat5 (Fig 4F–H). We found that without Stat5, Ets1 preferentially bound to the core motif TTCCT whereas TTCCG, containing a CpG motif, was more prevalent when co‐occupied with Stat5 (Fig 4H). TTCCG was similarly enriched at the unique Batf KO Ets1‐Stat5 co‐bound sites (Fig 4G) and at regions of diminished Ets1 binding in Stat5‐deficient Th17 cells (Fig 4G and H). These results suggest that Stat5 assists Ets1 binding, particularly at sites where DNA methylation may inhibit Ets1 binding.
As expected, given the increased recruitment of Stat5 to Ets1‐ and Runx1‐unique peaks in Batf KO Th17 cells (ΔEts1 Batf KO vs. WT Th17 and ΔRunx1 Batf KO vs. WT Th17 cells; Fig 4E), genomic annotation analysis identified loci associated with IL‐2‐Stat5 signaling (Fig 5A and B). Corresponding to enriched forkhead factor motifs identified in all Ets1‐bound regions in Batf KO Th17 cells (Fig 4D), genomic targets of Foxp3 were also identified in the unique peaks, even in the absence of Stat3‐Stat5 co‐binding, (Figs 5A and B, and EV4B). This supports the previously described role of Ets1 and Runx1 to facilitate Foxp3 binding to Treg enhancers (Samstein et al, 2012). Together, these analyses imply that recruitment of Ets1 and Runx1 to Th1 and Treg loci is upregulated in the absence of Batf in Th17 cells and indeed, normalized tag density of Ets1 and Runx1 binding at the genomic regions annotated near Th1‐ and Treg‐specific genes was significantly increased (Fig 5C). More precise analyses of Th1‐ and Treg‐specific loci revealed that in the absence of Batf, Ets1, and Runx1 occupancy were heightened and correlated with Stat5 binding (Fig 5D, indicated by blue arrows).
Figure 5. Functional cooperation between Ets1, Runx1, and Stat5 in regulating gene expression in Th17 cells.
-
A–F(A) Naïve WT and Batf KO CD4+CD62Lhi T cells were cultured under Th17‐polarizing conditions for 5 days and recovered to perform ChIP‐seq using antibodies to Batf, Ets1, Runx1, H3K4me3, H3K27me3, and H3K27ac. Stat3 and Stat5 ChIP‐seq were performed after IL‐6 and IL‐2 stimulation, respectively, using day 5 WT and Batf KO Th17 cells. WT Th1 or Treg cells were used to perform ChIP‐seq using antibodies to Ets1 and Runx1. Differentially enriched Ets1 and Runx1 peaks in Batf KO vs. WT Th17‐polarized cells (ΔEts1 Batf KO vs. WT, ΔRunx1 Batf KO vs. WT) were used for genomic annotation analysis (A, B). Boxplots show normalized tag density of Ets1 and Runx1 surrounding ±1 kb of peak centers, annotated to the nearest Th1‐ and Treg‐specific genes comparing WT and Batf KO Th17‐polarized cells. Central bands, boxes, and whiskers represent the median, upper quartile, and lower quartile and maximum and minimum values, respectively (C). Tracks show Ets1 and Runx1 ChIP‐seq data from WT and Batf KO Th17‐polarized cells at the indicated gene loci (D; WT, gray; Batf KO, red). Arrows indicate differential Ets1 and Runx1 binding with (red) or without (blue) the presence of Batf binding comparing WT and Batf KO cells. Ets1 (E) or Runx1 (F) peaks from WT Th1, Treg, or both (Th1‐Treg) were integrated with enriched Ets1 or Runx1 peaks in Batf KO compared with WT Th17 cells (Fig 4C) to separate peaks into unique and overlapping clusters. Heatmaps show Stat3, Stat5, H3K4me3, H3K27ac, Ets1 (E), and Runx1 (F) occupying ±2 kb of peak centers of Ets1‐ (E) or Runx1‐bound (F) sites in WT Th1, Treg, or both (Th1‐Treg) that overlap with uniquely enriched Ets1‐ (E) or Runx1‐ bound (F) sites from Batf KO compared with WT Th17 cells (Fig 4C—ΔEts1 Batf KO vs. WT, E; ΔRunx1 Batf KO vs. WT, F) along with the percentage of co‐bound and unique peaks. Histograms represent a rank of genes based on the regulatory potential score of Ets1 (E, right panels) or Runx1 (F, right panels) binding from each cluster to the differentially expressed genes in WT (gray) vs. Batf KO (red) Th17‐polarized cells derived from the integration of ChIP‐seq and RNA‐seq data using BETA algorithm.
Data information: Data are representative of two independent experiments with similar results. *P < 0.05 (Mann–Whitney test).
Collectively, the foregoing studies point to the Stat5‐facilitated assembly of a Th1/Treg‐like transcription‐enhancing module that in Th17 cells is kept in check by Batf‐mediated Stat5 suppression. Because Ets1, Runx1, and Stat5 each play important roles in Th1 and Treg development and function (Garrett‐Sinha, 2013; Jones et al, 2020), we hypothesized that cooperation of these factors promotes phenotype‐specifying gene expression in Th1 and Treg cells. To explore this, we performed Ets1 and Runx1 ChIP‐seq on WT Th1 and Treg cells, integrating bound regions with Th17 Ets1‐ and Runx1‐occupied peaks, including Batf KO unique peaks (Fig 5E and F). We found that in Th1 and Treg cells, Ets1 and Runx1 were recruited normally—regardless of Batf deficiency. Regions that bound Ets1 only in the absence of Batf in Th17 (Fig 4C) were also shared in Th1 and Treg cells and, importantly, were associated with increased Stat5 binding, upregulated active histone marks H3K4me3 and H3K27ac levels, and correlated with genes that were more highly expressed in Batf KO Th17 (Fig 5E). Batf deficiency resulted in substantially fewer peaks of unique Runx1 binding than Ets1 binding, and these regions, mainly shared with Th1 cells, were also enriched for Stat5, H3K4me3, and H3K27ac binding and corresponded to genes with increased expression in the absence of Batf (Fig 5F). Together these results indicate that the Stat5‐Ets1‐Runx1 enhanceosome identified at Th1 and Treg loci in Batf KO Th17 cells is similarly employed by Th1 and Treg cells to promote the expression of lineage‐specific genes.
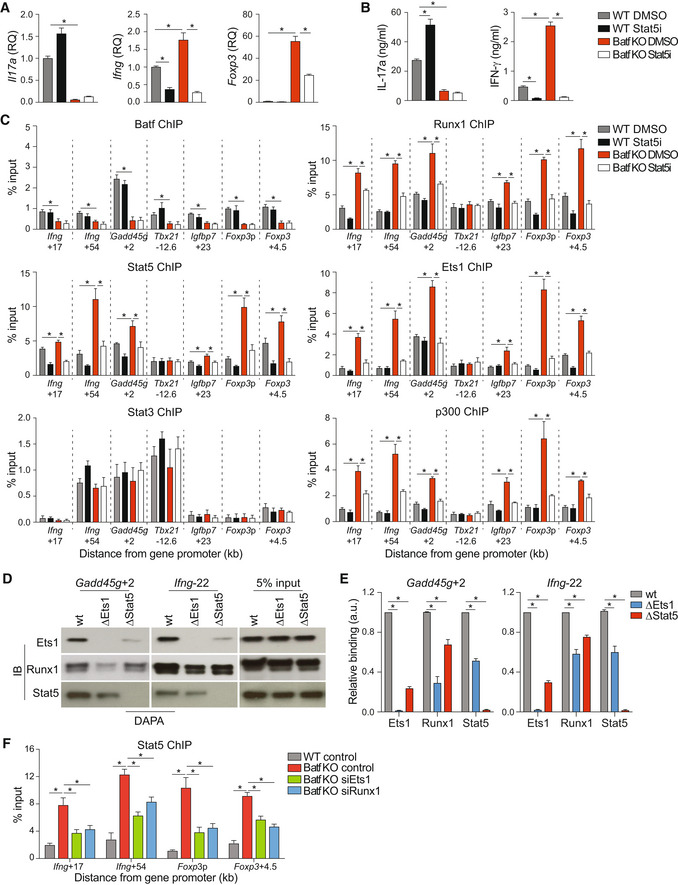
To determine whether increased expression of Th1‐ and Treg‐target genes in Batf KO Th17 cells was dependent on Stat5‐mediated assembly of the Ets1‐Runx1 complex, we pharmacologically repressed Stat5 activation. We first validated that chemical inhibition of Stat5 activation in WT and Batf KO Th17 cells phenocopied Stat5‐deficient cells (Figs 6A and B, and EV5A, B and D–G). We then treated cells with the Stat5 inhibitor (Stat5i) and analyzed lineage‐defining gene expression, cytokine production, and factor recruitment under conditions of diminished Stat5 (Figs 6A–C and EV5A–C). Consistent with previous studies (Laurence et al, 2007), Stat5 inhibition in WT Th17 cells promoted enhanced binding of Batf, Stat3, and p300 at the Il17a locus, resulting in increased IL‐17a production (Figs 6A and B, and EV5C). In line with our previous data (Figs 1A, C and D, and EV1A and B), Ifng/IFN‐γ and Foxp3 expression were increased in Batf KO Th17 cells but were dramatically reduced by inhibiting Stat5 activation—in the case of Ifng/IFN‐γ, to levels lower than in WT Th17 cells (Fig 6A and B). Diminished expression coincided with the loss of Ets1, Runx1, Stat5, and p300 binding at the Ifng and Foxp3 loci and was also observed at loci encoding the Th1 gene, Gadd45g, and the Treg gene, Igfbp7 (Fig 6C). Ets1, Runx1, Stat5, and p300 did not bind appreciably to the Tbx21 locus regardless of Batf deficiency or Stat5 activation status, in line with unaltered Tbx21 expression in the absence of Batf in Th17 cells (Figs 6C and EV1B).
Figure 6. Stat5‐mediated assembly of Ets1/Runx1 enhanceosome.
-
A–CNaïve WT and Batf KO CD4+CD62Lhi T cells cultured under Th17‐polarizing conditions were treated with or without 1 μM Stat5 inhibitor. On day 5, cells were harvested and used for RNA extraction to assess gene expression (A; data were normalized to WT DMSO; Foxp3, unstimulated; Il17a and Ifng, 6 h 2 μg/ml anti‐CD3 restimulation) or restimulated with 2 μg/ml anti‐CD3 for 24 h to assess cytokine production by ELISA (B). WT and Batf KO Th17‐polarized cells were collected on day 5 of culture and used to perform ChIP using antibodies to Stat5, Batf, Ets1, Runx1, Stat3, and p300 (C).
-
D, ENuclear extracts from WT Th17 cells were incubated with biotinylated oligonucleotides containing Ets1‐Runx1‐Stat5‐specific binding sites at the indicated genomic regions (wild‐type or mutant [Δ] as indicated). Immunoblots of precipitated proteins (D), with densitometry measurements (E).
-
FNaïve WT and Batf KO CD4+CD62Lhi T cells were cultured under Th17‐polarizing conditions. On day 3, cells were transfected with control or siRNA targeting Ets1 or Runx1, cultured for an additional 24 h without restimulation, and used to assess Stat5 binding at the indicated genomic regions by ChIP–qPCR.
Data information: Data are mean ± s.e.m. of four independent experiments. (*P < 0.05; two‐sided t‐test or one‐way ANOVA followed by Tukey's test). a.u., arbitrary unit.
Figure EV5. Stat5 inhibition enhances Batf, Stat3, and p300 binding to the Il17a locus.
-
A–CNaïve WT and Batf KO CD4+CD62Lhi T cells were cultured under Th17‐polarizing conditions with or without the addition of 1 μM Stat5 inhibitor. On day 3, cells were harvested and stained intracellularly for flow cytometric analysis of phosphorylated Stat5 (pStat5) to validate the effect of Stat5 inhibitor (A). Day 5 of differentiated WT and Batf KO cells were restimulated with PMA and ionomycin for 5 h and stained intracellularly for IFN‐γ and IL‐17a production represented by plots with percentage ± s.e.m. of positive cells (B). On day 5, cells were harvested and used to perform ChIP–qPCR using antibodies to Batf, Stat3, p300, and Stat5 (C).
-
D–GNaïve WT and Stat5 cKO CD4+CD62Lhi T cells were cultured under Th17‐polarizing conditions. Differentiated Th17 cells were reactivated with anti‐CD3 for 24 h to measure cytokine production by ELISA (D), restimulated with PMA and ionomycin for 5 h, and stained intracellularly for IFN‐γ and IL‐17a production represented by plots with percentage ± s.e.m. of positive cells (E), used for total RNA isolation to assess gene expression by RT–PCR (F), or used to performed ChIP‐seq to assess Ets1 binding at the indicated gene loci (G). Blue arrows indicate regions of increased Ets1 recruitment on WT vs. Stat5 cKO cells.
Data information: Data are mean ± s.e.m. of three independent experiments (A–F) or representative of two independent experiments with similar results (G) (*P < 0.05; two‐sided t‐test or one‐way ANOVA followed by Tukey's test).
To confirm that Stat5 recruitment promoted the formation of the Ets1/Runx1 enhancer complexes at Th1‐ and Treg‐like loci, we performed a DNA‐affinity precipitation assay (DAPA) (Fig 6D and E) using nuclear extracts from d5 Th17 cells and DNA probes encompassing sites of cooperative Ets1, Runx1, and Stat5 recruitment (Figs 1C and D, and 5C–F). Consistent with our ChIP‐seq studies, Ets1, Runx1, and Stat5 co‐bound DNA probes corresponding to distal sequences in the Gadd45g and Ifng loci. Mutation of the Stat5 site within the probe sequence abrograted Stat5 binding and significantly reduced binding of Ets1 and Runx1, with Ets1 being particularly affected in accord with a role for Stat5 in enhancing Ets1 DNA binding (Rameil et al, 2000; Dittmer, 2003; Ogawa et al, 2008; Fig 6D and E). Mutation of the Ets1 site greatly reduced Runx1 binding, consistent with cooperative Ets1‐Runx1 binding (Hollenhorst et al, 2009; Garrett‐Sinha, 2013) but also diminished Stat5 binding, albeit to a lesser degree. Consistent with these findings, Stat5 recruitment to the Ifng and Foxp3 loci in Batf KO cells was significantly reduced when Ets1 and Runx1 expression was reduced by siRNA targeting (Fig 6F). Collectively, these studies indicate that cooperative binding of Stat5, Ets1, and Runx1 is required for the assembly of an enhancer complex that forms at Th1 and Treg loci in Batf‐deficient Th17 cells.
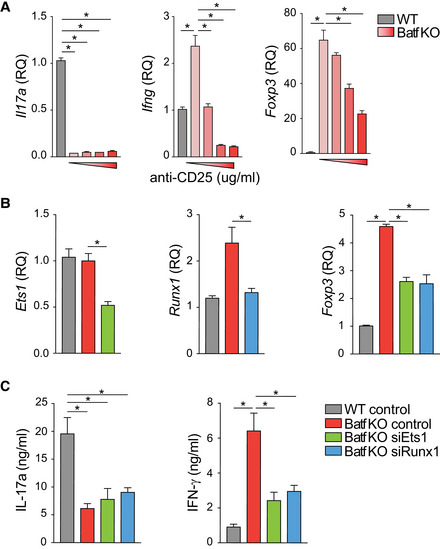
In an extension of these studies, we found that CD25 blockade resulted in reduced Ifng and Foxp3 expression in a dose‐dependent manner (Fig EV6A), demonstrating that reduced Stat5 output of the IL‐2R impaired Ifng and Foxp3 expression in Th17 cells. Notably, we found that Stat3 occupancy was not diminished at sites of increased Stat5 binding where Ets1 uniquely bound in Batf KO Th17 cells (Fig 4E), nor was Stat3 occupancy increased at Th1‐ and Treg‐specific gene loci when Stat5 activation was inhibited (Fig 6C). Moreover, Batf recruitment was not affected by Stat5 inhibition (Fig 6C) suggesting that Batf and Stat3 do not directly compete with Stat5 for binding at these sites. Importantly, the upregulation of Stat5 resulting from Batf deficiency in Th17 cells led to increased Stat5 occupancy and increased loading of Ets1‐Runx1‐p300 complexes at Th1 and Treg loci. The fact that both Ets1 and Runx1 were required for the observed increases in expression of Ifng and Foxp3 in the context of Batf deficiency was established by targeted siRNA knock‐downs (Fig EV6B and C), demonstrating that impaired expression of either factor led to comparably diminished gene expression. Collectively, these data indicate that optimal assembly of an activating Ets1‐Runx1‐p300 complex at Th1 and Treg gene loci in Th17 cells is dependent on Stat5 co‐binding; availability of Stat5 is central to the formation of this complex, irrespective of Stat3 status.
Figure EV6. Ets1 and Runx1 regulate Ifng/IFN‐γ and Foxp3 expression in Batf‐deficient Th17‐polarized cells.
-
ANaïve Batf KO T cells were cultured under Th17‐polarizing conditions with the addition of 0, 1, 5, or 10 μg/ml anti‐CD25 antibody (red bars). Untreated WT cells (gray bars) served as a control. On day 5, cells were harvested for total RNA isolation to assess gene expression by RT–PCR (Foxp3, no restimulation; Ifng and Il17a, restimulated with 2 μg/ml anti‐CD3 for 6 h). Data are normalized to untreated WT Th17 cells.
-
B, CNaïve WT and Batf KO CD4+CD62Lhi T cells were cultured under Th17‐polarizing conditions. On day 3, cells were transfected with control or siRNA targeting Ets1 or Runx1, and cultured an additional 24 h without restimulation prior to total RNA isolation to assess gene expression (B; data normalized to WT control), or with the addition of 2 μg/ml anti‐CD3 for measurement of cytokine production by ELISA (C).
Data information: Data are mean ± s.e.m. of three independent experiments. (*P < 0.05; one‐way ANOVA followed by Tukey's test).
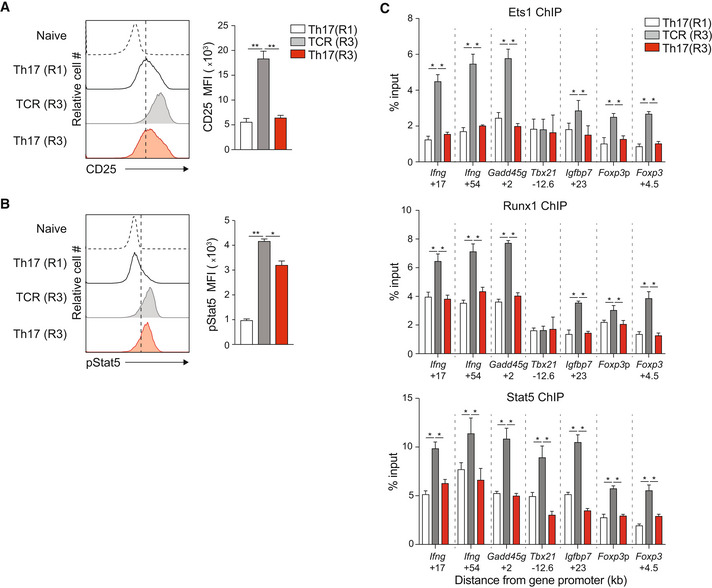
In a culture model of Th17 plasticity (Lee et al, 2009), Th17 cells maintained without IL‐6 and TGF‐β lose expression of Batf and transdifferentiate to a Th1‐like phenotype (Fig 1G–L). Consistent with our previous experiments, diminished Batf expression correlated with increased CD25 and pStat5 expression in this model (Fig 7A and B), and Stat5, Ets1, and Runx1 co‐binding to Th1 and Treg loci were significantly upregulated in transdifferentiated cells (TCR R3) (Fig 7C). Thus, without signals to maintain high Batf expression, the Th17 program is not maintained, driven in part by enhanced Ets1, Runx1, and Stat5 binding at key Th1 and Treg genomic targets. Collectively, these experiments support a model in which sustained Batf expression reinforces Th17 stability by repressing IL‐2R signaling that, in turn, limits Stat5‐dependent transcriptional activity at Th1 and Treg loci. Moreover, these studies establish that Stat5 contributes to the assembly of an Ets1‐Runx1‐p300 enhanceosome that is critical for optimal Th1 and Treg gene expression and that modulation of the Stat3‐Stat5 balance supports T helper phenotype identity by controlling the development of this enhancer complex.
Figure 7. Batf regulates IL‐2–Stat5 signaling in Th17 cells under long‐term culture conditions.
-
A–CNaïve CD4+CD62Lhi T cells from IL‐17a reporter mice (Il17a eGFP) were cultured under Th17‐polarizing conditions. On day 5, eGFP+/IL‐17a+ cells were sorted (Th17(R1)) and cultured for two additional rounds in the presence of anti‐CD3 alone (TCR(R3)) or Th17‐polarizing cytokines (Th17(R3)). Cells from round 1 (Th17(R1)) and round 3 (TCR(R3) and Th17(R3)) were assessed for surface CD25 expression (A) and intracellular pStat5 expression (B) by flow cytometry (A, B), and Ets1, Runx1, and Stat5 binding at the indicated genomic regions by ChIP–qPCR (C).
Data information: Data are mean ± s.e.m. of three independent experiments. (*P < 0.05; **P < 0.01; one‐way ANOVA followed by Tukey's test).
Discussion
An important facet of the Th17 cell program is its intrinsic instability; the early development of Th17 cells shares overlapping features with that of Treg cells and Th17 cells are prone to transdifferentiation into Th1‐like cells (Lee et al, 2009; Ahern et al, 2010; Mukasa et al, 2010; Harbour et al, 2015). The transcriptional network underpinning the Th17 developmental program is critically dependent on Batf (Schraml et al, 2009), which has a nonredundant role in activating the core genes of developing Th17 cells. Studies herein, and recent studies by the Ciofani group (Murphy et al, 2013; Carr et al, 2017), highlight additional roles for Batf and one of its heterodimeric partners, JunB, respectively, in repressing alternate gene expression programs to restrain the Treg and Th1 pathways in order to establish and maintain the Th17 program. Moreover, here we identify a novel mechanism by which Batf represses alternate gene programs: Batf acts via its downmodulation of components of the IL‐2 receptor, thereby limiting IL‐2R Stat5 output and restraining the assembly of a Stat5‐Ets1‐Runx1 enhanceosome that promotes Th1 and Treg‐specifying gene expression. Thus, although Batf function in Th17 programming is tied to IL‐6–induced Stat3 signaling that is required for activation of core Th17 genes (Zhou et al, 2007; Durant et al, 2010; Yang et al, 2011; Harbour et al, 2020), the current study elucidates a role for Batf in repressing IL‐2–induced Stat5 signaling that would otherwise activate Treg‐ and Th1‐cell gene loci, as well as antagonize Stat3 binding at some Th17 gene loci (e.g., the extended Il17a‐Il17f locus; Laurence et al, 2007; Yang et al, 2011). In view of our recent finding of a requirement for ongoing classical IL‐6 signaling to maintain the Th17 program (Harbour et al, 2020), elevated Batf expression driven by persistent IL‐6–dependent Stat3 signaling in mature Th17 cells appears to be essential to both initiate and sustain the metastable Th17 program, at least in part by the blunting assembly of the Stat5‐Ets1‐Runx1 enhanceosome.
In studies leading to the discovery of Th17 cells, a breakthrough was the finding that IFN‐γ–induced Stat1 signaling, which is integral to the differentiation of Th1 cells, prevented the development of Th17 cells (Harrington et al, 2005; Weaver, 2020). Similarly, it was found that IL‐4–induced Stat6 signaling blocked Th17 development (Harrington et al, 2005), and subsequently, IL‐2–induced Stat5 signaling repressed the development of Th17 cells (Yang et al, 2011). Finally, it was found that the transdifferentiation of Th17 cells into Th1‐like cells was mediated by the Stat4 output of either IL‐23 or IL‐12 receptors, leading to the notion of plasticity in the Th17 program (Lee et al, 2009; Ahern et al, 2010; Mukasa et al, 2010; Harbour et al, 2015). Indeed, since the early studies that defined CD4 T cell subsets, a hierarchy of CD4 signals that simultaneously activate—and suppress—alternate developmental programs has come to be appreciated (Murphy & Stockinger, 2010), as have differing intrinsic stabilities of each program. For each program, the transcriptional networks integrated with different Stat signals that emanate from distinct cytokine receptors have exerted both reinforcing and antagonizing effects. In the case of Th17 cell differentiation, core activating TFs include Stat3, Batf, Irf4, and JunB (Chen et al, 2006; Schraml et al, 2009; Ciofani et al, 2012; Li et al, 2012b; Carr et al, 2017), whereas Stat1, Stat5, Stat4 and Stat6 and FosL (Laurence et al, 2007; Villarino et al, 2010; Ciofani et al, 2012; Glosson‐Byers et al, 2014; Guenova et al, 2015) are each either inhibitory or deviating and act at overlapping or distinct stages of Th17 development. Findings in the current study advance our understanding of the latter, adding to Batf's central role in activating the Th17 program and its actions to repress alternate programming of Treg and Th1 genes, largely through restraining IL‐2‐induced Stat5.
Stat5a and Stat5b are induced by receptors for multiple cytokines and growth factors, including those using the common γ (cγ) receptor subunit (Wei et al, 2008). Of the cγ cytokines, IL‐2 has the broadest effects across different T cell subsets. IL‐2 signaling through Stat5 plays a central role in promoting Th1 and Th2 differentiation from naïve CD4 T cells by enhancing the expression of Il12rb2 and Il4ra, respectively (Liao et al, 2008, 2011), and is essential for the development of Foxp3+ regulatory T cells (Ross & Cantrell, 2018). Stat5 has been shown to activate transcription by recruiting p300/CBP (Wingelhofer et al, 2018) and, in Tregs, through the recruitment of DNA demethylases TET1/2 that are required to stabilize Foxp3 expression (Yang et al, 2015). By contrast, the repression of Th17 cell development by IL‐2 signaling has been shown to be mediated by Stat5 antagonism of Stat3 binding at the Il17a‐Il17f locus, diminishing Stat3‐induced permissive histone marks and limiting Il17a and Il17f transcription (Laurence et al, 2007; Yang et al, 2011). IL‐2 similarly antagonizes Tfh cell development (Ballesteros‐Tato et al, 2012). Our finding that Stat5 contributes to the recruitment of Ets1‐Runx1 complexes at bivalent sites in Th1 and Treg lineage genes identifies a mechanism through which Stat5 signaling downstream of the IL‐2 receptor may impact multiple T lineages, whether to enforce or antagonize transcriptional networks in each.
With respect to Th17 programming, which is inhibited by IL‐2 receptor signaling, the role of Batf in antagonizing the signal output of the IL‐2 receptor appears to be key. In this regard, it is notable that we find overlapping recruitment of Batf, Stat5, Ets1, and Runx1 across the Il2ra gene locus, where the overexpression of Batf limited expression of IL‐2Rα (CD25) while deficiency of Batf enhanced the binding of Stat5, Ets1, and Runx1 to these shared target sequences and increased IL‐2Rα expression. Not only does this implicate a role for the modulation of assembly of the Stat5‐Ets1‐Runx1 enhanceosome as a central mechanism for the regulation of IL‐2 signaling in T cell development, as an expression of inducible components of the IL‐2 receptor complex, particularly IL‐2Rα (CD25), is upregulated in a positive feedback loop downstream of IL‐2‐induced Stat5 signaling, it also suggests that Batf may act directly at the Il2ra locus to antagonize assembly of the Stat5‐Ets1‐Runx1 enhanceosome as a mechanism to limit Stat5 signaling in Th17 cells. While Batf was originally described as a transcriptional repressor (Murphy et al, 2013) and has been shown to induce a repressed chromatin configuration at the Foxp3 locus (Zhang et al, 2018) it is unclear how Batf mediates repression of IL‐2 receptor signaling. However, our finding that Batf binding to the Il2ra locus coincident with increased H3K27me3 suggests that its recruitment of inhibitory histone marks may be contributory. A similar mechanism appears to be operative at the Il2rb locus. The direct interaction of Batf with the polycomb repressive complex 2 (PRC2) has not been described, although Batf, in cooperation with IRF4 can enhance the expression of the PRC2 component, Ezh2, which catalyzes the addition of methyl groups to histone H3 at lysine 27 (Ochiai et al, 2018). Although further studies will be needed to understand whether, and how, Batf may directly interfere with Il2ra and Il2rb transcription, our findings indicate that Batf interrupts the positive feedback loop that reinforces IL‐2 signaling and thus Stat5 output as a mechanism to block IL‐2‐mediated repression of Th17 development.
Through interrogation of genomic regions of increased Stat5 occupancy in Batf KO Th17 cells, we identified high concordance with the binding of Ets1 and Runx1. The genomic association of Stat5, Ets1, and Runx1 has been described (Rameil et al, 2000; Dittmer, 2003; Ogawa et al, 2008; Schmidl et al, 2014), although not in the context of Th17 development. Moreover, Ets1 has been found to associate with T‐bet (Grenningloh et al, 2005), Gata3 (Wang et al, 2006; Strempel et al, 2010), and Foxp3 (Mouly et al, 2010) to fine‐tune subset‐specific enhancer activity to promote Th1, Th2, and Treg development, respectively. Accessible enhancers with coincident Foxp3, Ets1, Runx1, and Stat5 motifs are enriched in regulatory T cells while enhancers that are associated with increased Ets1, Runx1, and Stat5 binding (Schmidl et al, 2014) are enriched in other T cell subsets, suggesting that Stat5 plays a central role in Ets1‐Runx1 enhancer activity in both regulatory and effector T cells. Because Th17 cells express all of the components of this enhancer complex and the chromatin landscape surrounding Th1 and Treg loci is largely accessible, it is not surprising that a mechanism evolved to manage the aberrant expression of non‐Th17 phenotype genes. The physical interaction of Ets1 with numerous factors promotes its binding to DNA and enhances gene expression (Hollenhorst et al, 2011), and Ets1 and Runx1 cooperatively bind to DNA at composite motifs that are highly enriched at loci encoding genes involved in T cell activation and development. Together they recruit CBP to enhance transcriptional activation (Hollenhorst et al, 2009). Stat5 can directly interact with both Ets1 and Runx1 and also enhances transcriptional activity by interacting with p300 and CBP (Rameil et al, 2000; Ogawa et al, 2008; Able et al, 2017). The ability of Stat5 to recruit both Ets1 and Runx1 along with each factor's intrinsic capacity to interact with CBP and p300 would appear to amplify the independent activities of each factor in the composite enhanceosome. As Stat5 is not thought to be a pioneer factor, it may also act to promote Ets1‐Runx1 binding at sites with relatively low affinity for the two factors alone, although this will require further study.
Notably, we found that Stat3 bound to many of the same regions bound by Stat5, Ets1, and Runx1 in Th1 and Treg gene loci, as well as the Il2ra and Il2rb loci, raising the possibility that there might be competitive binding between these factors akin to what has been observed at Th17 core genes, such as the Il17a‐Il17f locus. However, while Stat5 availability was inhibited by Batf's downmodulation of the IL‐2 receptor, Stat3 binding at these non‐Th17 loci was unaffected by inhibition of Stat5—or deficiency of Batf—indicating that Stat5 and Stat3 do not compete for binding at these sites, and establishing that Batf is not required to pioneer these sites for Stat3 (or Stat5) binding. Similarly, pharmacological inhibition of Stat5 signaling did not increase Batf binding to these targets. Thus, in contrast to core Th17 gene loci, where Batf is required to pioneer sites that can bind both Stat3 and Stat5 as competitors, its major function at these alternative sites is uncoupled from regulation of Stat3 binding or competitive interference for Stat5 binding; the actions of Batf at these sites would appear to be largely dependent on its indirect control of Stat5 availability rather than promoting Stat3 competition or limiting Stat5 binding.
Collectively, published results and findings herein support a model in which Batf activates the core Th17 program by pioneering sites for Stat3 (and Rorγt) binding while restraining IL‐2–dependent Stat5 signaling that would otherwise compete for Stat3 binding at these sites and limit Stat5‐dependent assembly of Ets1‐Runx1 complexes that promote the activation of Th1 and Treg genes that are independent of Batf pioneering function or Stat3 antagonism. In essence, heightened Batf expression induced by IL‐6–driven Stat3 signaling early in Th17 commitment regulates the Stat3‐Stat5 balance in favor of Stat3 at dual‐responsive elements and against Stat5 at sites of Ets1‐Runx1 binding. Through this mechanism, Stat3 signaling begets increased Batf expression that favors Th17 programming by limiting Stat5 availability for Treg‐ and Th1‐cell programs, and persistence of Stat3 signaling is required to sustain increased Batf that sustains the Th17 program. In this regard, it is noteworthy that while both Ets1 and Runx1 are expressed by naïve T cells, and although the Ets1 transcript undergoes transient downregulation early in Th17 cells—which may act to further limit IL‐2 production and signaling, it recovers and increases thereafter such that transcripts of both Ets1 and Runx1 are substantially elevated late in Th17 cell development. While it is currently unclear why Ets1 and Runx1 expressions rise in late‐stage Th17 cells, our data suggest that this may further destabilize the Th17 program and contribute to the propensity of Th17 cells to undergo transdifferentiation into Th1 or even Treg cells. Future studies that examine the modulation of Ets1 and Runx1 during this late developmental window should be informative, as should efforts to better define factors that control the amplitude and timing of IL‐2 receptor signaling during this period.
In view of the universal induction of Batf expression downstream of TCR signaling, the suppression of inducible components of the IL‐2 receptor by Batf may seem counterintuitive for non‐Th17 effector developmental programs, such as Treg and Th1, which are critically dependent on, or amplified by, IL‐2 signaling, respectively. However, while increased CD25 expression has been reported in Batf‐deficient Treg cells (Wheaton & Ciofani, 2020) and CD8 T cells (Kurachi et al, 2014), suggesting that repression of IL‐2–induced Stat5 may be a common feature of Batf actions across T cell programs, the reduced levels of Batf induced in developing non‐Th17 cells would appear insufficient to deter their IL‐2–dependent programming, consistent with a gradient effect of Batf's actions to impair Il2ra transcription. Moreover, the greatest effect of Batf on restraining CD25 expression appeared to be later in the arc of CD25 expression kinetics, where it may be less impactful for non‐Th17 cells. In accord with this, Batf deficiency targeted to innate lymphoid cells also resulted in skewing toward a Th1‐like phenotype, upregulated transcription of all three IL‐2R chains, and enriched Ets1 and Runx1 motifs at Batf‐dependent loci in ILC3s (Wu et al, 2022), suggesting a conserved role for Batf‐dependent regulation of a Stat5‐Ets1‐Runx1 enhancesome in type 3 immune cells.
Our findings that a Th1‐Treg gene enhancer module regulated by a Stat5‐Ets1‐Runx1 enhanceosome is actively repressed in Th17 cells may have implications for novel therapeutics to treat Th17‐mediated diseases. Small molecule inhibitors that inhibit Ets1‐ and Runx1‐DNA interactions (Illendula et al, 2016; Currie et al, 2018) have been developed, and the use of these agents and others to restrain assembly and/or function of the Stat5‐Ets1‐Runx1 enhanceosome could potentially dampen transdifferentiation of protective Th17 cells into more pathogenic Th1‐like cells to achieve beneficial therapeutic effects. Although a potential complication of this approach is the possibility that suppression of Ets1 and Runx1 may impair the development and function of regulatory T cells, thereby limiting efficacy, attenuation of the development of pathogenic Th1‐like cells while sparing nonpathogenic Th17 cells holds sufficient promise that additional studies to explore the modulation of this pathway for therapeutic ends are warranted.
Materials and Methods
Experimental model and subject details mice
C57BL/6, Stat5 fl/fl, and Il17a gfp were purchased from The Jackson Laboratory (Bar Harbor, ME, USA) and Batf −/− (Batf KO) were previously described (Schraml et al, 2009). Cd4‐Cre were purchased from Taconic and bred to Stat5 fl/fl to generate Stat5 fl/fl Cd4‐Cre mice (Stat5 cKO). Littermates were used as wild‐type (WT) control mice. Mice were maintained under specific pathogen‐free conditions. All experiments were performed with the approval of the University of Alabama Institutional Animal Care and Use Committee.
Cell culture conditions
Naïve CD4+CD62Lhi T cells were isolated from the spleen and lymph nodes using a MACS isolation system (Miltenyi Biotec) or sorted by flow cytometry for CD4+CD44lowCD62Lhigh naïve cell population. Naïve CD4 T cells were activated with plate‐bound anti‐CD3 (2 μg/ml, 145‐2C11) and soluble anti‐CD28 (37.51, 0.5 μg/ml) with additional cytokines and antibodies to generate Th1 cells (5 ng/ml IL‐12; 50 U/ml human IL‐2; and 10 μg/ml anti‐IL‐4, 11B11), Th17 cells (20 ng/ml IL‐6; 5 ng/ml human TGF‐β; 10 ng/ml IL‐1β, 10 U/ml human IL‐2 on d3; 10 μg/ml anti‐IL‐4, 11B11; and 10 μg/ml anti‐IFN‐γ, XMG), and Treg cells (2.5 ng/ml human TGF‐β; and anti‐IL‐4, 11B11). Cells were expanded in the fresh medium after 3 days with half concentration (Th17) of the original cytokines or additional 50 U/ml human IL‐2 in Treg culture. In long‐term culture conditions, IL‐17a‐producing cells were sorted based on GFP marker. Cells were plated at 0.5 × 106 cells/ml and activated with 1 μg/ml plate‐bound anti‐CD3 alone or with the addition of Th17 cytokines (IL‐6 and TGF‐β). Recombinant cytokines and antibodies are from BD Biosciences, Bio X Cell, or R&D. In some experiments, Stat5 inhibitor (Millipore, Stat5 Inhibitor III, Pimozide) or anti‐mouse CD25 (BioLegend, PC61) were added to the culture on day 0, and cells were analyzed on day 5.
Retroviral expression vectors and retroviral transduction
Retrovirus expressing Thy1.1 and Batf‐Thy1.1 were previously described (Jabeen et al, 2013). Retroviral stocks were made by calcium phosphate transfection of Platinum‐E retroviral packaging cell line (Cell BioLabs). Naive CD4+CD62Lhi were cultured under Th17 culture conditions. On day 2, cells were transduced with Thy1.1 or Batf‐Thy1.1 retroviruses by centrifugation at 2,000 rpm at 25°C for 1 h in the presence of 8 μg/ml polybrene. After spin infection, the viral supernatant was replaced with the former culture supernatant. Cells were expanded on day 3, harvested on day 5, and used for long‐term culture experiments.
Transfection of siRNA
siRNAs targeting Batf, Ets1, and Runx1 were purchased from Santa Cruz Biotechnology. Naïve CD4 T cells were cultured under Th17 conditions and transfected with siRNA on day 3 using Amaxa Nucleofector kit (Lonza). Transfected cells were rested overnight with 50 U/ml hIL‐2, and restimulated with anti‐CD3 for 24 h for gene expression and cytokine production analyses.
Citrobacter rodentium (Cr) infection
6–8 weeks WT and Batf KO mice were gavaged with 1‐2 × 109 cfu of Citrobacter rodentium strain DBS100 (ATCC 51459). Mice were sacrificed on day 4 and day 10 postinfection. Cells were isolated from mesenteric lymph nodes (MLN) and colon tissues, restimulated with PMA and ionomycin, and intracellularly stained for cytokines and transcription factors. Serum was collected on the day postinfection and used for antibody titer measurement by ELISA. For bioluminescence imaging, WT and Batf KO mice were infected with 1–2 × 109 cfu of the bioluminescent C.r. strain ICC180, shaved, and imaged with an IVIS 100 Imaging System (Xenogen, Inc.) as described previously (Basu et al, 2012).
Cell sorting and FACS
Cells were stained with fluorochrome‐conjugated surface antibodies and sorted using a BD FACS Aria. Sorted cells were used for total RNA isolation to assess gene expression by RT–PCR. For surface staining, cells were stained with fluorochrome‐conjugated antibodies for 30 min at 4°C, washed with 2% BSA/PBS, and fixed with 2% formaldehyde for 10 min at room temperature before analysis. Using a Foxp3/Transcription factor staining kit (eBioscience), intracellular staining for cytokines and transcription factors was performed by stimulating cells with 50 ng/ml PMA and 500 ng/ml ionomycin, adding 3 μM monensin (Sigma Aldrich) 2 h poststimulation followed by FACs analysis at 5 h. pStat3 and pStat5 were analyzed by FACS using cells fixed with 1.5% formaldehyde for 10 min at room temperature, permeabilized using 100% ice‐cold methanol for 10 min at 4°C, and stained with fluorochrome‐conjugated antibodies for 30 min at room temperature.
ELISA
Blood was spun at 10,000 rpm for 10 min, and serum was collected. 96‐well NUNC MaxiSorp plates were coated with Intimin 385β protein and incubated overnight at 4°C. Capture IgG (Southern Biotech) was used as standard control. Plates were washed twice in PBS‐0.05% Tween (PBST), blocked at room temperature (RT) with 1.5% BSA‐PBS, and washed twice with PBST. Unlabeled IgG standards (Southern Biotech) and serum diluted in PBS with 0.2% BSA were added, incubated for 2 h at RT, and washed with PBST. HRP‐conjugated goat anti‐mouse IgG (Southern Biotech) was added and incubated for 2 h at RT. Plates were washed with PBST and developed with TMB Substrate (Life Technologies) OD values were obtained using a VERSAmax tunable microplate reader, and data were analyzed using SoftMax Pro. Antigen‐specific Ig concentrations were calculated from the linear portion of the standard curve.
Gene expression by RT–PCR and RNA‐seq
Total RNA was isolated using Trizol reagent (Invitrogen Life Technologies) and reversed transcribed to make cDNA using iScript cDNA Synthesis Kit (Bio‐Rad). Gene expression was assessed by RT–PCR and normalized to housekeeping gene expression (β2‐microglobulin, β2m) (Table EV1). The relative gene expression was calculated by the change‐in‐threshold (−ΔΔC T) method. For RNA‐seq, total RNA was used to prepare the library for sequencing using TruSeq RNA library prep kit (Illumina). RNA‐seq libraries were sequenced with single‐end 50 bp using Illumina HiSeq 2500.
RNA‐seq analysis
Raw sequencing data were mapped to the mm9 genome using Tophat2. Mapped reads were filtered by samtools, counted, and normalized using HOMER feature analyzeRNA.pl. Differential expressed genes were generated using DESeq2. Heatmap of differential expressed genes were created using Heatmapper (Babicki et al, 2016). Differential expressed genes were subjected to geneset enrichment analysis (GSEA) (Subramanian et al, 2005).
Chromatin immunoprecipitation (ChIP) and ChIP‐seq
Cells (10 × 106) were cross‐linked for 10 min with 1% formaldehyde, quenched with 0.125 M Glycine for 5 min, washed with PBS, resuspended in lysis buffer (5 mM Pipes, 85 mM KCl, 0.5% NP‐40 with protease inhibitor), homogenized by running cells through an 18‐gauge needle, and pelleted by centrifugation. Nuclei were resuspended in RIPA buffer (1X PBS, 1% NP‐40, 0.5% sodium deoxycholate, 0.1% SDS, and protease inhibitor) and sonicated to a size range of 200–500 bp using a Diagenode Bioruptor. Cells were pelleted and, resuspended in RIPA buffer. Antibodies (rabbit anti‐Batf [Schraml et al, 2009], rabbit anti‐Ets1 [abcam, ab124282], rabbit anti‐Runx1 [abcam, ab23980], rabbit anti‐p300 [Santa Cruz Biotech, sc‐585] rabbit anti‐H3K27me3 [Millipore, 07‐449], rabbit anti‐H3K4me3 [Millipore, 07‐473], rabbit anti‐H3K27ac [abcam, ab4729], rabbit anti‐Stat3 [Millipore, 06‐596], rabbit anti‐Stat5 [R&D Systems, AF2168], and normal rabbit IgG [Millipore, 12–370]) were coupled to Dynabeads M‐280 Sheep anti‐rabbit IgG (Thermo Fisher Scientific) at 4°C with rotation overnight then washed 3 times with PBS/BSA solution. Antibody‐bound beads were added to sonicated chromatin, incubated at 4°C overnight, washed with 5 times LiCl buffer (100 mM Tris–pH 7.5, 500 mM LiCl, 1% NP‐40, 1% sodium deoxycholate) and once with TE buffer, resuspended in elution buffer (1% SDS and 0.1 M NaHCO3), and incubated at 65°C overnight. Immunoprecipitated DNA purified using MinElute columns (Qiagen) was analyzed by RT–PCR (Table EV1). ChIP data were analyzed using the percent input method and adjusted to IgG control background.
For ChIP‐seq, immunoprecipitated DNA was used to prepare libraries using NEBNext Ultra II DNA library prep kit and NEBNext Multiplex Oligos for Illumina (NEB). ChIP‐seq libraries were sequenced with single‐end 50 bp using Illumina HiSeq 2,500.
ChIP‐seq analysis
Raw sequencing data were mapped to mm9 genome using Bowtie1 (Langmead et al, 2009) with options ‐m 1 k 1. ChIP‐seq peaks were called using MACS2 (Zhang et al, 2008b) with P‐value cutoff at 1 × 10−5. Tag counts were normalized to 10 million tags and calculated using annotatePeaks function of HOMER (Heinz et al, 2010). Heatmaps were visualized using deepTools (Ramírez et al, 2016). Motif analysis was performed using findMotifsGenome function of HOMER. ChIP‐seq peaks were annotated to mm9 genome using annotatePeaks function of HOMER. Gene ontology analysis of associated peaks was performed using annotatePeaks function of HOMER with option ‐go. ChIP‐seq tracks were normalized to 10 million tags and viewed using the Integrated Genome Brower (IGB) (Nicol et al, 2009). The log2 transformed normalized read data were used to generate box plots (indicate interquartile range with whiskers ±1.5 times) using R. Binding and Expression Target Analysis (BETA) (Wang et al, 2013) was used to integrate ChIP‐seq of transcription factors with differential gene expression. Difflogo (Nettling et al, 2015) was used to visualize pair‐wise differences between the motifs.
DNA‐affinity precipitation assay (DAPA)
Differentiated T cells were pelleted and washed with 1× PBS, and the following buffers were added sequentially and incubated on ice: lysis buffer (10 mM Tris–HCl pH 7.4, 10 mM NaCl, and 0.3% MgCl2) for 10 min, 20% NP‐40 for 10 min, cells were pelleted, nuclear lysis buffer (50 mM Tris–HCl pH 7.4, 150 mM NaCl, and 0.5% NP‐40) for 20 min. Cells were pelleted, and the supernatant was collected as a nuclear extract. Oligonucleotides (biotinylated at 5′) containing wild‐type (WT), mutated Ets1 (ΔEts1), or Stat5 (ΔStat5)‐binding sites (underlined) were:
Ifng‐22 WT
5′‐ACAGGAAGGAGATGGGAAGTCAGATCAAAGCTGGCTTAACACCTCTGTCCTG‐3′;
Ifng‐22 ΔEts1
5′‐ACAAAAAGGAGATGGGAAGTCAGATCAAAGCTGGCTTAACACCTCTGTCCTG‐3′;
Ifng‐22 ΔStat5
5′‐ACAGGAAGGAGATAAACCGTCAGATCAAAGCTGGCTTAACACCTCTGTCCTG‐3′;
Gadd45g + 2 WT 5′‐GCAGGTGTGACTCAGCAAGCAGCCTTCCAGTGAAAGGAGGG‐3′;
Gadd45g + 2 ΔEts
5′‐GCAGGTGTGACTCAGCAAGCAGAAAACCAGTGAAAGGAGGG‐3′;
Gadd45g + 2 ΔStat5
5′‐GCAGGTGTGACTCAGCAAGCAGCCTTCCAGTGAAAAAAGGG‐3′.
Biotinylated oligonucleotides were incubated with streptavidin‐agarose beads for 30 min at 4°C and washed with pull‐down buffer (25 mM HEPES, 15 mM NaCl, 0.5 mM DTT, 0.5% NP‐40, 0.1 mM EDTA pH 7.5, and 10% glycerol). Nuclear extracts were added and incubated at 4°C for 2 h. The complex was washed with pull‐down buffer, eluted at 100°C for 5 min in Laemmli's sample buffer, separated using 10% SDS–PAGE gel, and immunoblotted with primary antibodies (rabbit anti‐Ets1 [abcam, ab124282], rabbit anti‐Runx1 [abcam, ab23980], and rabbit anti‐Stat5 [R&D Systems, AF2168]).
Statistical analysis
Data are mean ± s.e.m. Statistical analyses were calculated using Prism (GraphPad Software) or R. P‐values < 0.05 is considered as statistical significance.
Author contributions
Duy Pham: Conceptualization; data curation; formal analysis; investigation; methodology; writing – original draft; writing – review and editing. Daniel J Silberger: Data curation; formal analysis; investigation; methodology. Kim N Nguyen: Data curation; investigation; methodology. Min Gao: Data curation; formal analysis; validation; visualization; methodology. Casey T Weaver: Conceptualization; data curation; formal analysis; supervision; funding acquisition; writing – original draft; project administration; writing – review and editing. Robin D Hatton: Conceptualization; data curation; formal analysis; supervision; funding acquisition; validation; visualization; methodology; writing – review and editing.
Disclosure and competing interests statement
The authors declare that they have no conflict of interest.
Supporting information
Expanded View Figures PDF
Table EV1
PDF+
Acknowledgements
The authors are grateful to members of the Weaver laboratory for their helpful comments and suggestions. We thank B. Dale, A. Smith, E. Higginbotham, and H. Turner for technical assistance. We gratefully acknowledge the UAB Epitope Recognition and Immunoreagent Core Facility (NIH P30 AR048311) for antibody preparations and the CFAR Basic and Translational Sciences Core Facility (NIH P30 AI27667) for FACS sorting. This work was supported by NIH R01 DK115172 to RDH and CTW, T32 AI007051‐37 to DP and DJS, and UAB Institutional Funds to CTW.
The EMBO Journal (2023) 42: e109803
Contributor Information
Casey T Weaver, Email: cweaver@uab.edu.
Robin D Hatton, Email: robinhatton@uabmc.edu.
Data availability
RNA‐seq and ChIP‐seq have been deposited in the NCBI Gene Expression Omnibus with the accession number GSE167420 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE167420).
References
- Able A, Burrell J, Stephens J (2017) STAT5‐interacting proteins: a synopsis of proteins that regulate STAT5 activity. Biology 6: 20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahern PP, Schiering C, Buonocore S, McGeachy MJ, Cua DJ, Maloy KJ, Powrie F (2010) Interleukin‐23 drives intestinal inflammation through direct activity on T cells. Immunity 33: 279–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babicki S, Arndt D, Marcu A, Liang Y, Grant JR, Maciejewski A, Wishart DS (2016) Heatmapper: web‐enabled heat mapping for all. Nucleic Acids Res 44: W147–W153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesteros‐Tato A, León B, Graf BA, Moquin A, Adams PS, Lund FE, Randall TD (2012) Interleukin‐2 inhibits germinal center formation by limiting T follicular helper cell differentiation. Immunity 36: 847–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassuk AG, Leiden JM (1995) A direct physical association between ETS and AP‐1 transcription factors in normal human T cells. Immunity 3: 223–237 [DOI] [PubMed] [Google Scholar]
- Basu R, O'Quinn DB, Silberger DJ, Schoeb TR, Fouser L, Ouyang W, Hatton RD, Weaver CT (2012) Th22 cells are an important source of IL‐22 for host protection against enteropathogenic bacteria. Immunity 37: 1061–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz BC, Jordan‐Williams KL, Wang C, Kang SG, Liao J, Logan MR, Kim CH, Taparowsky EJ (2010) Batf coordinates multiple aspects of B and T cell function required for normal antibody responses. J Exp Med 207: 933–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevington SL, Cauchy P, Piper J, Bertrand E, Lalli N, Jarvis RC, Gilding LN, Ott S, Bonifer C, Cockerill PN (2016) Inducible chromatin priming is associated with the establishment of immunological memory in T cells. EMBO J 35: 515–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burchill MA, Yang J, Vogtenhuber C, Blazar BR, Farrar MA (2007) IL‐2 receptor beta‐dependent STAT5 activation is required for the development of Foxp3+ regulatory T cells. J Immunol 178: 280–290 [DOI] [PubMed] [Google Scholar]
- Carr TM, Wheaton JD, Houtz GM, Ciofani M (2017) JunB promotes Th17 cell identity and restrains alternative CD4+ T‐cell programs during inflammation. Nat Commun 8: 301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cauchy P, Maqbool MA, Zacarias‐Cabeza J, Vanhille L, Koch F, Fenouil R, Gut M, Gut I, Santana MA, Griffon A et al (2016) Dynamic recruitment of Ets1 to both nucleosome‐occupied and ‐depleted enhancer regions mediates a transcriptional program switch during early T‐cell differentiation. Nucleic Acids Res 44: 3567–3585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Laurence A, Kanno Y, Pacher‐Zavisin M, Zhu B‐M, Tato C, Yoshimura A, Hennighausen L, O'Shea JJ (2006) Selective regulatory function of Socs3 in the formation of IL‐17‐secreting T cells. Proc Natl Acad Sci USA 103: 8137–8142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciofani M, Madar A, Galan C, Sellars M, Mace K, Pauli F, Agarwal A, Huang W, Parkhurst CN, Muratet M et al (2012) A validated regulatory network for Th17 cell specification. Cell 151: 289–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie SL, Warner SL, Vankayalapati H, Liu X, Sharma S, Bearss DJ, Graves BJ (2018) Development of high‐throughput screening assays for inhibitors of ETS transcription factors. SLAS Discov 24: 77–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delacher M, Imbusch CD, Hotz‐Wagenblatt A, Mallm J‐P, Bauer K, Simon M, Riegel D, Rendeiro AF, Bittner S, Sanderink L et al (2020) Precursors for nonlymphoid‐tissue Treg cells reside in secondary lymphoid organs and are programmed by the transcription factor BATF. Immunity 52: 295–312.e11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiToro D, Winstead CJ, Pham D, Witte S, Andargachew R, Singer JR, Wilson CG, Zindl CL, Luther RJ, Silberger DJ et al (2018) Differential IL‐2 expression defines developmental fates of follicular versus nonfollicular helper T cells. Science 361: eaao2933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmer J (2003) The biology of the Ets1 proto‐oncogene. Mol Cancer 2: 29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs‐Canner S, Berkey S, Delgoffe GM, Edwards RP, Curiel T, Odunsi K, Bartlett DL, Obermajer N (2017) Suppressive IL‐17A+Foxp3+ and ex‐Th17 IL‐17AnegFoxp3+ Treg cells are a source of tumour‐associated Treg cells. Nat Commun 8: 14649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durant L, Watford WT, Ramos HL, Laurence A, Vahedi G, Wei L, Takahashi H, Sun H‐W, Kanno Y, Powrie F et al (2010) Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis. Immunity 32: 605–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Arvey A, Chinen T, van der Veeken J, Gasteiger G, Rudensky AY (2014) Control of the inheritance of regulatory T cell identity by a cis element in the Foxp3 locus. Cell 158: 749–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura K, Oyamada A, Iwamoto Y, Yoshikai Y, Yamada H (2013) CD4 T cell‐intrinsic IL‐2 signaling differentially affects Th1 and Th17 development. J Leukoc Biol 94: 271–279 [DOI] [PubMed] [Google Scholar]
- Garrett‐Sinha LA (2013) Review of Ets1 structure, function, and roles in immunity. Cell Mol Life Sci 70: 3375–3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geginat J, Paroni M, Kastirr I, Larghi P, Pagani M, Abrignani S (2016) Reverse plasticity: TGF‐β and IL‐6 induce Th1‐to‐Th17‐cell transdifferentiation in the gut. Eur J Immunol 46: 2306–2310 [DOI] [PubMed] [Google Scholar]
- Glasmacher E, Agrawal S, Chang AB, Murphy TL, Zeng W, Vander Lugt B, Khan AA, Ciofani M, Spooner CJ, Rutz S et al (2012) A genomic regulatory element that directs assembly and function of immune‐specific AP‐1‐IRF complexes. Science 338: 975–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glosson‐Byers NL, Sehra S, Kaplan MH (2014) STAT4 is required for IL‐23 responsiveness in Th17 memory cells and NKT cells. JAK‐STAT 3: e955393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenningloh R, Kang BY, Ho I‐C (2005) Ets‐1, a functional cofactor of T‐bet, is essential for Th1 inflammatory responses. J Exp Med 201: 615–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenova E, Skabytska Y, Hoetzenecker W, Weindl G, Sauer K, Tham M, Kim K‐W, Park J‐H, Seo JH, Ignatova D et al (2015) IL‐4 abrogates T(H)17 cell‐mediated inflammation by selective silencing of IL‐23 in antigen‐presenting cells. Proc Natl Acad Sci USA 112: 2163–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbour SN, Maynard CL, Zindl CL, Schoeb TR, Weaver CT (2015) Th17 cells give rise to Th1 cells that are required for the pathogenesis of colitis. Proc Natl Acad Sci USA 112: 7061–7066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbour SN, DiToro DF, Witte SJ, Zindl CL, Gao M, Schoeb TR, Jones GW, Jones SA, Hatton RD, Weaver CT (2020) TH17 cells require ongoing classic IL‐6 receptor signaling to retain transcriptional and functional identity. Sci Immunol 5: eaaw2262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington LE, Hatton RD, Mangan PR, Turner H, Murphy TL, Murphy KM, Weaver CT (2005) Interleukin 17‐producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat Immunol 6: 1123–1132 [DOI] [PubMed] [Google Scholar]
- Hasan Z, Koizumi S‐I, Sasaki D, Yamada H, Arakaki N, Fujihara Y, Okitsu S, Shirahata H, Ishikawa H (2017) JunB is essential for IL‐23‐dependent pathogenicity of Th17 cells. Nat Commun 8: 15628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayatsu N, Miyao T, Tachibana M, Murakami R, Kimura A, Kato T, Kawakami E, Endo TA, Setoguchi R, Watarai H et al (2017) Analyses of a mutant Foxp3 allele reveal BATF as a critical transcription factor in the differentiation and accumulation of tissue regulatory T cells. Immunity 47: 268–283.e9 [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK (2010) Simple combinations of lineage‐determining transcription factors prime cis‐regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai T, Ramos TL, Lin P‐Y, Simonetta F, Su LL, Picton LK, Baker J, Lin J‐X, Li P, Seo K et al (2021) Selective expansion of regulatory T cells using an orthogonal IL‐2/IL‐2 receptor system facilitates transplantation tolerance. J Clin Invest 131: e139991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenhorst PC, Chandler KJ, Poulsen RL, Johnson WE, Speck NA, Graves BJ (2009) DNA specificity determinants associate with distinct transcription factor functions. PLoS Genet 5: e1000778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenhorst PC, McIntosh LP, Graves BJ (2011) Genomic and biochemical insights into the specificity of ETS transcription factors. Annu Rev Biochem 80: 437–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber M, Lohoff M (2014) IRF4 at the crossroads of effector T‐cell fate decision. Eur J Immunol 44: 1886–1895 [DOI] [PubMed] [Google Scholar]
- Illendula A, Gilmour J, Grembecka J, Tirumala VSS, Boulton A, Kuntimaddi A, Schmidt C, Wang L, Pulikkan JA, Zong H et al (2016) Small molecule inhibitor of CBFβ‐RUNX binding for RUNX transcription factor driven cancers. EBioMedicine 8: 117–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata A, Durai V, Tussiwand R, Briseño CG, Wu X, Grajales‐Reyes GE, Egawa T, Murphy TL, Murphy KM (2017) Quality of TCR signaling determined by differential affinities of enhancers for the composite BATF‐IRF4 transcription factor complex. Nat Immunol 18: 563–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwatani K, Fujimoto T, Ito T (2010) Cyclin D1 blocks the anti‐proliferative function of RUNX3 by interfering with RUNX3‐p300 interaction. Biochem Biophys Res Commun 400: 426–431 [DOI] [PubMed] [Google Scholar]
- Jabeen R, Goswami R, Awe O, Kulkarni A, Nguyen ET, Attenasio A, Walsh D, Olson MR, Kim MH, Tepper RS et al (2013) Th9 cell development requires a BATF‐regulated transcriptional network. J Clin Invest 123: 4641–4653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain J, Loh C, Rao A (1995) Transcriptional regulation of the IL‐2 gene. Curr Opin Immunol 7: 333–342 [DOI] [PubMed] [Google Scholar]
- Jayaraman G, Srinivas R, Duggan C, Ferreira E, Swaminathan S, Somasundaram K, Williams J, Hauser C, Kurkinen M, Dhar R et al (1999) p300/cAMP‐responsive element‐binding protein interactions with ets‐1 and ets‐2 in the transcriptional activation of the human stromelysin promoter. J Biol Chem 274: 17342–17352 [DOI] [PubMed] [Google Scholar]
- Jin H, Burns EJ, Nadeau M, Yeste A, Kumar D, Rangachari M, Zhu C, Xiao S, Seavitt J, Georgopoulos K et al (2012) Aiolos promotes TH17 differentiation by directly silencing Il2 expression. Nat Immunol 13: 770–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DM, Read KA, Oestreich KJ (2020) Dynamic roles for IL‐2‐STAT5 signaling in effector and regulatory CD4+ T cell populations. J Immunol 205: 1721–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karwacz K, Miraldi ER, Pokrovskii M, Madi A, Yosef N, Wortman I, Chen X, Watters A, Carriero N, Awasthi A et al (2017) Critical role of IRF1 and BATF in forming chromatin landscape during type 1 regulatory cell differentiation. Nat Immunol 18: 412–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara K, Shiina M, Fukuda I, Ogata K, Nakamura H (2017) Molecular mechanisms of cooperative binding of transcription factors Runx1‐CBFβ‐Ets1 on the TCRα gene enhancer. PLoS ONE 12: e0172654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitabayashi I, Yokoyama A, Shimizu K, Ohki M (1998) Interaction and functional cooperation of the leukemia‐associated factors AML1 and p300 in myeloid cell differentiation. EMBO J 17: 2994–3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komine O, Hayashi K, Natsume W, Watanabe T, Seki Y, Seki N, Yagi R, Sukzuki W, Tamauchi H, Hozumi K et al (2003) The Runx1 transcription factor inhibits the differentiation of naive CD4+ T cells into the Th2 lineage by repressing GATA3Expression. J Exp Med 198: 51–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn T, Bettelli E, Oukka M, Kuchroo VK (2009) IL‐17 and Th17 cells. Annu Rev Immunol 27: 485–517 [DOI] [PubMed] [Google Scholar]
- Kurachi M, Barnitz RA, Yosef N, Odorizzi PM (2014) The transcription factor BATF operates as an essential differentiation checkpoint in early effector CD8+ T cells. Nat Immunol 15: 373–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurebayashi Y, Nagai S, Ikejiri A, Ohtani M, Ichiyama K, Baba Y, Yamada T, Egami S, Hoshii T, Hirao A et al (2012) PI3K‐Akt‐mTORC1‐S6K1/2 axis controls Th17 differentiation by regulating Gfi1 expression and nuclear translocation of RORγ. Cell Rep 1: 360–373 [DOI] [PubMed] [Google Scholar]
- Kuwahara M, Ise W, Ochi M, Suzuki J, Kometani K, Maruyama S, Izumoto M, Matsumoto A, Takemori N, Takemori A et al (2016) Bach2–Batf interactions control Th2‐type immune response by regulating the IL‐4 amplification loop. Nat Commun 7: 12596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory‐efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurence A, Tato CM, Davidson TS, Kanno Y, Chen Z, Yao Z, Blank RB, Meylan F, Siegel R, Hennighausen L et al (2007) Interleukin‐2 signaling via STAT5 constrains T helper 17 cell generation. Immunity 26: 371–381 [DOI] [PubMed] [Google Scholar]
- Lazarevic V, Chen X, Shim J‐H, Hwang E‐S, Jang E, Bolm AN, Oukka M, Kuchroo VK, Glimcher LH (2011) T‐bet represses T(H)17 differentiation by preventing Runx1‐mediated activation of the gene encoding RORγt. Nat Immunol 12: 96–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YK, Turner H, Maynard CL, Oliver JR, Chen D, Elson CO, Weaver CT (2009) Late developmental plasticity in the T helper 17 lineage. Immunity 30: 92–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lexberg MH, Taubner A, Förster A, Albrecht I, Richter A, Kamradt T, Radbruch A, Chang H‐D (2008) Th memory for interleukin‐17 expression is stable in vivo . Eur J Immunol 38: 2654–2664 [DOI] [PubMed] [Google Scholar]
- Li L, Patsoukis N, Petkova V, Boussiotis VA (2012a) Runx1 and Runx3 are involved in the generation and function of highly suppressive IL‐17‐producing T regulatory cells. PLoS ONE 7: e45115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Spolski R, Liao W, Wang L, Murphy TL, Murphy KM, Leonard WJ (2012b) BATF‐JUN is critical for IRF4‐mediated transcription in T cells. Nature 490: 543–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Liang Y, LeBlanc M, Benner C, Zheng Y (2014) Function of a Foxp3 cis‐element in protecting regulatory T cell identity. Cell 158: 734–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao W, Schones DE, Oh J, Cui Y, Cui K, Roh T‐Y, Zhao K, Leonard WJ (2008) Priming for T helper type 2 differentiation by interleukin 2‐mediated induction of interleukin 4 receptor alpha‐chain expression. Nat Immunol 9: 1288–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao W, Lin J‐X, Wang L, Li P, Leonard WJ (2011) Modulation of cytokine receptors by IL‐2 broadly regulates differentiation into helper T cell lineages. Nat Immunol 12: 551–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Kim MH, Friesen L, Kim CH (2020) BATF regulates innate lymphoid cell hematopoiesis and homeostasis. Sci Immunol 5: eaaz8154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loos J, Schmaul S, Noll TM, Paterka M, Schillner M, Löffel JT, Zipp F, Bittner S (2020) Functional characteristics of Th1, Th17, and ex‐Th17 cells in EAE revealed by intravital two‐photon microscopy. J Neuroinflammation 17: 357–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Gao W, Sun X, Wang W (2018) STAT5 and TET2 cooperate to regulate FOXP3‐TSDR demethylation in CD4+ T cells of patients with colorectal cancer. J Immunol Res 2018: 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan PR, Harrington LE, O'Quinn DB, Helms WS, Bullard DC, Elson CO, Hatton RD, Wahl SM, Schoeb TR, Weaver CT (2006) Transforming growth factor‐beta induces development of the T(H)17 lineage. Nature 441: 231–234 [DOI] [PubMed] [Google Scholar]
- Moisan J, Grenningloh R, Bettelli E, Oukka M, Ho I‐C (2007) Ets‐1 is a negative regulator of Th17 differentiation. J Exp Med 204: 2825–2835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison PJ, Bending D, Fouser LA, Wright JF, Stockinger B, Cooke A, Kullberg MC (2013) Th17‐cell plasticity in helicobacter hepaticus‐induced intestinal inflammation. Mucosal Immunol 6: 1143–1156 [DOI] [PubMed] [Google Scholar]
- Mouly E, Chemin K, Nguyen HV, Chopin M, Mesnard L, Leite‐de‐Moraes M, Burlen‐defranoux O, Bandeira A, Bories J‐C (2010) The Ets‐1 transcription factor controls the development and function of natural regulatory T cells. J Exp Med 207: 2113–2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucida D, Salek‐Ardakani S (2009) Regulation of TH17 cells in the mucosal surfaces. J Allergy Clin Immunol 123: 997–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukasa R, Balasubramani A, Lee YK, Whitley SK, Weaver BT, Shibata Y, Crawford GE, Hatton RD, Weaver CT (2010) Epigenetic instability of cytokine and transcription factor gene loci underlies plasticity of the T helper 17 cell lineage. Immunity 32: 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy KM, Stockinger B (2010) Effector T cell plasticity: flexibility in the face of changing circumstances. Nat Immunol 11: 674–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy TL, Tussiwand R, Murphy KM (2013) Specificity through cooperation: BATF‐IRF interactions control immune‐regulatory networks. Nat Rev Immunol 13: 499–509 [DOI] [PubMed] [Google Scholar]
- Nettling M, Treutler H, Grau J, Keilwagen J, Posch S, Grosse I (2015) DiffLogo: a comparative visualization of sequence motifs. BMC Bioinformatics 16: 387–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HV, Mouly E, Chemin K, Luinaud R, Despres R, Fermand JP, Arnulf B, Bories JC (2012) The Ets‐1 transcription factor is required for Stat1‐mediated T‐bet expression and IgG2a class switching in mouse B cells. Blood 119: 4174–4181 [DOI] [PubMed] [Google Scholar]
- Nicol JW, Helt GA, Blanchard SG, Raja A, Loraine AE (2009) The integrated genome browser: free software for distribution and exploration of genome‐scale datasets. Bioinformatics 25: 2730–2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochiai K, Kondo H, Okamura Y, Shima H, Kurokochi Y, Kimura K, Funayama R, Nagashima T, Nakayama K, Yui K et al (2018) Zinc finger‐IRF composite elements bound by Ikaros/IRF4 complexes function as gene repression in plasma cell. Blood Adv 2: 883–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa S, Satake M, Ikuta K (2008) Physical and functional interactions between STAT5 and Runx transcription factors. J Biochem 143: 695–709 [DOI] [PubMed] [Google Scholar]
- Omenetti S, Pizarro TT (2015) The Treg/Th17 Axis: a dynamic balance regulated by the Gut microbiome. Front Immunol 6: 639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono M, Yaguchi H, Ohkura N, Kitabayashi I, Nagamura Y, Nomura T, Miyachi Y, Tsukada T, Sakaguchi S (2007) Foxp3 controls regulatory T‐cell function by interacting with AML1/Runx1. Nature 446: 685–689 [DOI] [PubMed] [Google Scholar]
- Pham D, Moseley CE, Gao M, Savic D, Winstead CJ, Sun M, Kee BL, Myers RM, Weaver CT, Hatton RD (2019) Batf pioneers the reorganization of chromatin in developing effector T cells via Ets1‐dependent recruitment of Ctcf. Cell Rep 29: 1203–1220.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rameil P, Lécine P, Ghysdael J, Gouilleux F, Kahn‐Perlès B, Imbert J (2000) IL‐2 and long‐term T cell activation induce physical and functional interaction between STAT5 and ETS transcription factors in human T cells. Oncogene 19: 2086–2097 [DOI] [PubMed] [Google Scholar]
- Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, Manke T (2016) deepTools2: a next generation web server for deep‐sequencing data analysis. Nucleic Acids Res 44: W160–W165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross SH, Cantrell DA (2018) Signaling and function of Interleukin‐2 in T lymphocytes. Annu Rev Immunol 36: 411–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudra D, Egawa T, Chong MMW, Treuting P, Littman DR, Rudensky AY (2009) Runx‐CBFβ complexes control expression of the transcription factor Foxp3 in regulatory T cells. Nat Immunol 10: 1170–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahoo A, Alekseev A, Tanaka K, Obertas L, Lerman B, Haymaker C, Clise‐Dwyer K, McMurray JS, Nurieva R (2015) Batf is important for IL‐4 expression in T follicular helper cells. Nat Commun 6: 7997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samstein RM, Samstein RM, Arvey A, Arvey A, Josefowicz SZ, Josefowicz SZ, Peng X, Peng X, Reynolds A, Reynolds A et al (2012) Foxp3 exploits a pre‐existent enhancer landscape for regulatory T cell lineage specification. Cell 151: 153–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidl C, Hansmann L, Lassmann T, Balwierz PJ, Kawaji H, Itoh M, Kawai J, Nagao‐Sato S, Suzuki H, Andreesen R et al (2014) The enhancer and promoter landscape of human regulatory and conventional T‐cell subpopulations. Blood 123: e68–e78 [DOI] [PubMed] [Google Scholar]
- Schraml BU, Hildner K, Ise W, Lee W‐L, Smith WAE, Solomon B, Sahota G, Sim J, Mukasa R, Cemerski S et al (2009) The AP‐1 transcription factor Batf controls TH17 differentiation. Nature 460: 405–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwaller J, Parganas E, Wang D, Cain D, Aster JC, Williams IR, Lee CK, Gerthner R, Kitamura T, Frantsve J et al (2000) Stat5 is essential for the myelo‐ and lymphoproliferative disease induced by TEL/JAK2. Mol Cell 6: 693–704 [DOI] [PubMed] [Google Scholar]
- Shiomi H, Masuda A, Nishiumi S, Nishida M, Takagawa T, Shiomi Y, Kutsumi H, Blumberg RS, Azuma T, Yoshida M (2010) Gamma interferon produced by antigen‐specific CD4+ T cells regulates the mucosal immune responses to Citrobacter rodentium infection. Infect Immun 78: 2653–2666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberger DJ, Zindl CL, Weaver CT (2017) Citrobacter rodentium: a model enteropathogen for understanding the interplay of innate and adaptive components of type 3 immunity. Mucosal Immunol 10: 1108–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smigiel KS, Richards E, Srivastava S, Thomas KR, Dudda JC, Klonowski KD, Campbell DJ (2014) CCR7 provides localized access to IL‐2 and defines homeostatically distinct regulatory T cell subsets. J Exp Med 211: 121–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens DC, Poon GMK (2016) Differential sensitivity to methylated DNA by ETS‐family transcription factors is intrinsically encoded in their DNA‐binding domains. Nucleic Acids Res 44: 8671–8681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strempel JM, Grenningloh R, Ho I‐C, Vercelli D (2010) Phylogenetic and functional analysis identifies Ets‐1 as a novel regulator of the Th2 cytokine gene locus. J Immunol 184: 1309–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES et al (2005) Gene set enrichment analysis: a knowledge‐based approach for interpreting genome‐wide expression profiles. Proc Natl Acad Sci USA 102: 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Fu J, Zhou Y (2017) Metabolism controls the balance of Th17/T‐regulatory cells. Front Immunol 8: 1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toomer KH, Lui JB, Altman NH, Ban Y, Chen X, Malek TR (2019) Essential and non‐overlapping IL‐2Rα‐dependent processes for thymic development and peripheral homeostasis of regulatory T cells. Nat Commun 10: 1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao H‐W, Tai T‐S, Tseng W, Chang H‐H, Grenningloh R, Miaw S‐C, Ho I‐C (2013) Ets‐1 facilitates nuclear entry of NFAT proteins and their recruitment to the IL‐2 promoter. Proc Natl Acad Sci USA 110: 15776–15781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasanthakumar A, Moro K, Xin A, Liao Y, Gloury R, Kawamoto S, Fagarasan S, Mielke LA, Afshar‐Sterle S, Masters SL et al (2015) The transcriptional regulators IRF4, BATF and IL‐33 orchestrate development and maintenance of adipose tissue–resident regulatory T cells. Nat Immunol 16: 276–285 [DOI] [PubMed] [Google Scholar]
- Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, Stockinger B (2006) TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL‐17‐producing T cells. Immunity 24: 179–189 [DOI] [PubMed] [Google Scholar]
- Villarino AV, Gallo E, Abbas AK (2010) STAT1‐activating cytokines limit Th17 responses through both T‐bet‐dependent and ‐independent mechanisms. J Immunol 185: 6461–6471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Shannon MF, Young IG (2006) A role for Ets1, synergizing with AP‐1 and GATA‐3 in the regulation of IL‐5 transcription in mouse Th2 lymphocytes. Int Immunol 18: 313–323 [DOI] [PubMed] [Google Scholar]
- Wang S, Sun H, Ma J, Zang C, Wang C, Wang J, Tang Q, Meyer CA, Zhang Y, Liu XS (2013) Target analysis by integration of transcriptome and ChIP‐seq data with BETA. Nat Protoc 8: 2502–2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Friedrich C, Hagemann SC, Korte WH, Goharani N, Cording S, Eberl G, Sparwasser T, Lochner M (2014) Regulatory T cells promote a protective Th17‐associated immune response to intestinal bacterial infection with C. rodentium. Mucosal Immunol 7: 1290–1301 [DOI] [PubMed] [Google Scholar]
- Weaver CT (2020) One road to the TH17 pathway: how TH1 led to TH17 (and vice versa), and first came last. Nat Immunol 21: 819–821 [DOI] [PubMed] [Google Scholar]
- Weaver CT, Hatton RD (2009) Interplay between the TH17 and TReg cell lineages: a (co‐)evolutionary perspective. Nat Rev Immunol 9: 883–889 [DOI] [PubMed] [Google Scholar]
- Weaver CT, Harrington LE, Mangan PR, Gavrieli M, Murphy KM (2006) Th17: an effector CD4 T cell lineage with regulatory T cell ties. Immunity 24: 677–688 [DOI] [PubMed] [Google Scholar]
- Weaver CT, Hatton RD, Mangan PR (2007) IL‐17 family cytokines and the expanding diversity of effector T cell lineages. Annu Rev Immunol 25: 821–852 [DOI] [PubMed] [Google Scholar]
- Weaver CT, Elson CO, Fouser LA, Kolls JK (2013) The Th17 pathway and inflammatory diseases of the intestines, lungs, and skin. Annu Rev Pathol Mech Dis 8: 477–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, Laurence A, O'Shea JJ (2008) New insights into the roles of Stat5a/b and Stat3 in T cell development and differentiation. Semin Cell Dev Biol 19: 394–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheaton JD, Ciofani M (2020) JunB controls intestinal effector programs in regulatory T cells. Front Immunol 11: 444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingelhofer B, Neubauer HA, Valent P, Han X, Constantinescu SN, Gunning PT, Müller M, Moriggl R (2018) Implications of STAT3 and STAT5 signaling on gene regulation and chromatin remodeling in hematopoietic cancer. Leukemia 32: 1713–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong WF, Kurokawa M, Satake M, Kohu K (2011) Down‐regulation of Runx1 expression by TCR signal involves an autoregulatory mechanism and contributes to IL‐2 production. J Biol Chem 286: 11110–11118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Khatun A, Kasmani MY, Chen Y, Zheng S, Atkinson S, Nguyen C, Burns R, Taparowsky EJ, Salzman NH et al (2022) Group 3 innate lymphoid cells require BATF to regulate gut homeostasis in mice. J Exp Med 219: 11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Shapiro LH, Rivera M, Kumar A, Brindle PK (1998) A role for CREB binding protein and p300 transcriptional coactivators in Ets‐1 transactivation functions. Mol Cell Biol 18: 2218–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X‐P, Ghoreschi K, Steward‐Tharp SM, Rodriguez‐Canales J, Zhu J, Grainger JR, Hirahara K, Sun H‐W, Wei L, Vahedi G et al (2011) Opposing regulation of the locus encoding IL‐17 through direct, reciprocal actions of STAT3 and STAT5. Nat Immunol 12: 247–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R, Qu C, Zhou Y, Konkel JE, Shi S, Liu Y, Chen C, Liu S, Liu D, Chen Y et al (2015) Hydrogen sulfide promotes Tet1‐ and Tet2‐mediated Foxp3 demethylation to drive regulatory T cell differentiation and maintain immune homeostasis. Immunity 43: 251–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Meng G, Strober W (2008a) Interactions among the transcription factors Runx1, RORgammat and Foxp3 regulate the differentiation of interleukin 17‐producing T cells. Nat Immunol 9: 1297–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W et al (2008b) Model‐based analysis of ChIP‐Seq (MACS). Genome Biol 9: R137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Xiao X, Lan P, Li J, Dou Y, Chen W, Ishii N, Chen S, Xia B, Chen K et al (2018) OX40 Costimulation inhibits Foxp3 expression and Treg induction via BATF3‐dependent and independent mechanisms. Cell Rep 24: 607–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Ivanov II, Spolski R, Min R, Shenderov K, Egawa T, Levy DE, Leonard WJ, Littman DR (2007) IL‐6 programs TH‐17 cell differentiation by promoting sequential engagement of the IL‐21 and IL‐23 pathways. Nat Immunol 8: 967–974 [DOI] [PubMed] [Google Scholar]
- Zindl CL, Witte SJ, Laufer VA, Gao M, Yue Z, Janowski KM, Cai B, Frey BF, Silberger DJ, Harbour SN et al (2022) A nonredundant role for T cell‐derived interleukin 22 in antibacterial defense of colonic crypts. Immunity 55: 494–511.e11 [DOI] [PMC free article] [PubMed] [Google Scholar]


