Abstract
Background
Diagnostic testing for primary ciliary dyskinesia (PCD) started in 2013 in Palestine. We aimed to describe the diagnostic, genetic and clinical spectrum of the Palestinian PCD population.
Methods
Individuals with symptoms suggestive of PCD were opportunistically considered for diagnostic testing: nasal nitric oxide (nNO) measurement, transmission electron microscopy (TEM) and/or PCD genetic panel or whole-exome testing. Clinical characteristics of those with a positive diagnosis were collected close to testing including forced expiratory volume in 1 s (FEV1) Global Lung Index z-scores and body mass index z-scores.
Results
68 individuals had a definite positive PCD diagnosis, 31 confirmed by genetic and TEM results, 23 by TEM results alone, and 14 by genetic variants alone. 45 individuals from 40 families had 17 clinically actionable variants and four had variants of unknown significance in 14 PCD genes. CCDC39, DNAH11 and DNAAF11 were the most commonly mutated genes. 100% of variants were homozygous. Patients had a median age of 10.0 years at diagnosis, were highly consanguineous (93%) and 100% were of Arabic descent. Clinical features included persistent wet cough (99%), neonatal respiratory distress (84%) and situs inversus (43%). Lung function at diagnosis was already impaired (FEV1 z-score median −1.90 (−5.0–1.32)) and growth was mostly within the normal range (z-score mean −0.36 (−3.03–2.57). 19% individuals had finger clubbing.
Conclusions
Despite limited local resources in Palestine, detailed geno- and phenotyping forms the basis of one of the largest national PCD populations globally. There was notable familial homozygosity within the context of significant population heterogeneity.
Short abstract
Despite limited resources in Palestine, detailed geno- and phenotyping forms the basis of one of the largest national PCD cohorts globally. There is notable familial homozygosity within the context of significant population heterogeneity. https://bit.ly/3HTo96x
Introduction
Primary ciliary dyskinesia (PCD) is a rare heterogeneous disease that can be caused by genetic mutations in over 50 different motile cilia-related genes [1–3]. The associated functional and structural defects of cilia impair mucociliary clearance, causing persistent upper and lower respiratory disease, including daily wet cough and recurrent chest infections, chronic serous otitis media (glue ear) and persistent rhinosinusitis [4]. Pulmonary symptoms often start in the neonatal period and continue throughout life, almost invariably leading to bronchiectasis. Involvement of embryonic nodal cilia causes situs anomalies in approximately 50% of individuals, some with associated congenital heart defects [5]. PCD is estimated to affect about 1 in 7500 people in Europe but is more common in populations where consanguineous marriage is common [6, 7].
Palestine has a highly consanguineous population and many patients in its paediatric pulmonology service have symptoms suggestive of PCD. PCD diagnostic testing is complex and requires specialist equipment and personnel, hence innovative approaches are needed in countries with fewer resources [8]. According to European Respiratory Society (ERS) and American Thoracic Society (ATS) guidelines, a positive diagnosis can only be made in individuals with a supportive clinical history and either confirmatory “hallmark” ultrastructural abnormalities detected by transmission electron microscopy (TEM) or pathogenic mutations in PCD genes that show a consistent inheritance pattern (respective to the recessive, X-linked and dominant subtypes of PCD) [9, 10]. The diagnosis is likely, but not proven, in individuals with a persistent wet cough from infancy, with either situs anomalies or a low nasal nitric oxide (nNO) measurement [11–13].
While great progress has been made in the past decade to understand molecular disease mechanisms and improve diagnosis and treatment, PCD management is still hampered by a lack of fundamental data on epidemiology and clinical course in different settings. To date, a handful of national cohorts have been described and these are almost exclusively based in well-resourced healthcare settings [14]. Most published data are cross-sectional and longitudinal cohorts are now required to extend the knowledge base for PCD.
In this study, we aimed to demonstrate that accurate diagnoses are possible in a resource-limited country through collaboration with specialist centres. We report the diagnostic findings, including the genetic profile and recurrent disease-causing variants in the Palestinian PCD population. We additionally present the spectrum of clinical phenotypes, as well as disease severity close to the time of diagnosis.
Methods
Ethical approval
Ethical approval was provided by Al-Quds University Research Ethics Committee Palestine (Ref. 31/REC/2018), London Bloomsbury Research Ethics Committee, UK (08/H0713/82) and Southampton and South-West Hants Research Ethics Committee (06/Q1702/10) with University of Southampton (ERGO 53155), UK. Participants or parents gave informed consent.
Study design
Patients were evaluated and opportunistically identified for PCD diagnostic testing by the only physician testing patients in Palestine (N.R.) in collaboration with TEM and genetics experts at the University of Southampton (UoS) and University College London. Participants were reported in the study if they had a positive PCD diagnosis based on diagnostic TEM and/or genetic testing [9]. Data were prospectively captured using clinic records and retrospectively added to a study dataset. Key variables for this manuscript are diagnostic results (TEM and nNO), genotype and cross-sectional clinical characteristics at time of diagnosis including lung function (forced expiratory volume in 1 s (FEV1)), nutritional status, and demographics (June 2013–December 2020).
Clinical characteristics
The clinical history, spirometry and anthropometry data were recorded by one clinician (N.R.) at the time of diagnostic testing. Data were retrospectively transferred to a data capture sheet (Excel) with predefined variables. Where necessary, the patient's family was re-contacted following clinic to clarify demographics or symptoms. Spirometry was measured using Smart PFT LabTM (Medical Equipment Europe GmbH, Hammelburg, Germany) according to ATS/ERS guidelines [15] and FEV1 and forced vital capacity (FVC) z-scores were calculated using ERS Global Lung Index (GLI) reference values [16]. Body mass index (BMI) was calculated from clinic measurements of height and weight and z-scores were calculated using the World Health Organization BMI 5–19 years reference values (www.who.int/tools/growth-reference-data-for-5to19-years/indicators/bmi-for-age). In the absence of a suitable reference for adults, we estimated the z-score for adults assuming they were 19 years old (all adults with BMI measurements in the study were aged ≤48 years).
Diagnosis
Diagnostic testing was directed by clinical indication and available resources. nNO measurement was conducted as a screening tool from March 2016 and, if low, then a nasal brushing sample was sent for TEM. Genetic testing was limited by funding and priorities changed over time (e.g. initially prioritising patients with an abnormal TEM [17]). Once confidence at identifying the clinical phenotype and measuring nNO was reached, patients with normal TEM but a highly likely clinical picture and low nNO were included. On occasion, the order of tests differed; for example, if nNO analysis was not possible on the day of nasal brushing it was sometimes measured later. Diagnostic results were transferred to a data capture sheet (Excel) with predefined variables and codes. Many families had several individuals with symptoms, but because of limited resources, in most cases only one person from each family was tested.
Nasal nitric oxide
Patients who were developmentally able (e.g. aged >5 years) were screened using a portable electrochemical nNO analyser (Niox Mino; March 2016–April 2019; Niox Vero September 2019–December 2020). Measurements used tidal breathing with the pursed lips method whenever possible and the open-mouth method in younger children. The measurements were taken when patients were free of acute infection, at a sampling flow of 0.3 L·min−1. A low nNO is not in itself confirmatory of PCD [9] and symptomatic individuals with levels <77 nL·min−1 were considered highly likely to have PCD and were not included in this study unless genetics or TEM confirmed the diagnosis.
Transmission electron microscopy
For patients with a strong clinical suspicion based on signs and symptoms [13], ciliated respiratory epithelium was sampled for TEM by brushing the nasal mucosa using a cytology brush. Samples were chemically fixed in glutaraldehyde and sent to the UoS for quantitative analysis. Using a Tecnai Spirit 12 TEM at ×26 500 minimum magnification, 100–300 cilia in transverse section per sample were scored for assessment of axonemal structure and representative images captured. Using Better Experimental Approaches to Treat (BEAT)-PCD criteria, a positive diagnosis of PCD was confirmed by identification of outer dynein arm defect, inner and outer dynein arm defects, or microtubular disorganisation with inner dynein arm defect [17]. Defects which did not fulfil the criteria for a positive diagnosis (e.g. transposition defect; mislocalisation of basal bodies with few or no cilia) were recorded, but these participants were not considered to have a definite diagnosis unless they had genetic confirmation [9].
Genetic testing and analysis
Blood samples were obtained from probands, their parents and their affected and unaffected siblings where possible. DNA was isolated from peripheral blood lymphocytes by standard procedures, then subject to next-generation sequencing (NGS) either using a targeted gene panel as previously described [18] or whole-exome sequencing (WES). For WES, exome libraries (Agilent SureSelect Human All Exon V6 kit, Agilent Technologies) were paired-end 150-bp sequenced on an Illumina HiSeq2500 by Novogene (UK) Co Ltd. Bioinformatic analyses were performed as described previously for the gene panel [18]. FASTQ files from WES were aligned to the human reference genome (GRCh38/hg38) using BWA-MEM2 [19]. Variant calling was performed according to the GATK 4.1.4.1 best practice pipeline [20] and variant annotation was done using Annovar [21]. Variant prioritisation and filtration were performed as previously described [18]. All variants were reported according to Human Genome Variation Society recommendations [22] and mapped to GRCh38. Variant pathogenicity annotation was performed according to American College of Medical Genetics and Genomics and the Association for Molecular Pathology (ACMG-AMP) variant interpretation guidelines [23]. Variant classification information was also extracted from the Database of Chromosomal Imbalance and Phenotype in Humans using Ensembl Resources (DECIPHER) and ClinVar, a database providing aggregated variant interpretation data collected from clinical and research testing laboratories, expert panels and other groups [24, 25]. Sanger sequencing was used to confirm all variants prioritised using NGS and assure correct segregation within family members. In three families, targeted Sanger sequencing was also used on a candidate variant basis for genetic diagnosis to identify recurrent CCDC39 and DNAAF11 mutations in this population shared with patients with similar TEM findings (supplementary table 1).
Data management and statistical analyses
Data were cleaned and coded in Excel using predefined coding and then imported into SAS9.4 (SAS Institute, Cary, NC) to be formatted and analysed. Descriptive statistics were performed in SAS9.4 using the PROC FREQ, PROC UNIVARIATE and PROC SQL functions. Continuous data were described as mean with standard deviation (sd) or median with interquartile range and categorical data were described as proportions. Data were tidied using Tidyverse packages. For visualisation, plots were created with the ggplot2 package using the ggplot function with more colours added using the RColorBrewer package in R-studio 3.6 (R-Studio team, Boston, MA). Genotype–phenotype clustering was visualised with a Circos plot using the RCircos package. Genetic data from 13 patients were included in a previous publication [18].
Results
464 patients with a clinical history suggestive of PCD were opportunistically investigated. nNO was measured in 350, TEM analysed in 183 and genetic screening in 82 (supplementary figure 1), following which 68 had a positive diagnosis according to ERS guidelines [9]. 23 individuals had a positive diagnosis based on TEM alone, 14 on pathogenic biallelic mutations alone (only recessive forms of PCD were identified), with 31 having both genetic and TEM diagnostic confirmation.
Of 34 individuals with ‘hallmark’ diagnostic TEM who proceeded to genetic testing, 31 (91%) had causative variants in known PCD genes; no variants in known PCD genes were found in the other three, but putative candidate genes with variants of interest are currently being investigated. Of those with normal TEM, 21 proceeded to genetic testing of whom 11 had a genetic cause found (52%) (DNAH11, DRC1, RSPH4A and RSPH9). The genetic and confirmatory familial segregation analysis data are summarised in supplementary table 1.
Palestinian PCD population clinical characteristics (n=68)
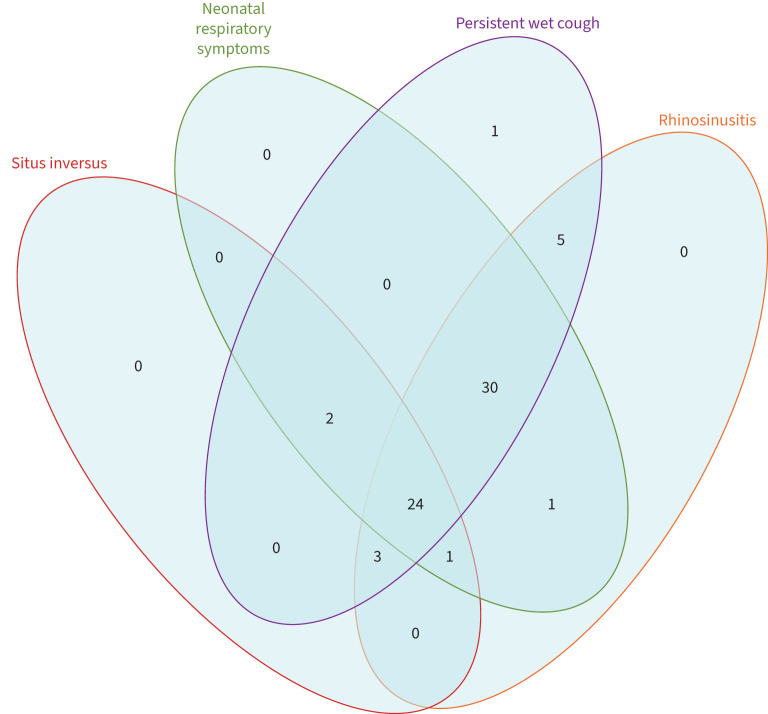
The mean age at diagnosis was 10.0 years (range 3 months–40 years), 39.7% were female and 42.6% had situs inversus (table 1). Individuals were almost all from consanguineous backgrounds (92.6%) and many had symptomatic family members. The reporting of symptoms including neonatal respiratory distress (83.8%), persistent wet cough (98.5%), persistent rhinosinusitis (94.1%) and middle ear disease (73.5%) was similar to previous reports [4], but finger clubbing (19.1%) was reportedly higher than expected (0/65; unpublished Southampton Children's PCD Clinic, UK). Whilst the vast majority of patients reported a combination of several symptoms (36% had chronic cough, rhinosinusitis, neonatal respiratory distress and situs inversus; 45% had chronic cough, rhinosinusitis and neonatal respiratory distress with normal situs), several patients reported fewer symptoms, highlighting the heterogeneity of the syndrome (figure 1).
TABLE 1.
Demographics and clinical characteristics of participants in the primary ciliary dyskinesia (PCD) cohort, i.e. those with definite diagnosis of PCD based on transmission electron microscopy and/or genetic testing
| Clinical variables | Clinical outcomes |
| Age at diagnosis | Median 10.0 years; range 3 months–40.0 years |
| Female | 27 (39.7%) |
| Situs inversus | 29 (42.6%) |
| Consanguinity (n=60) | 65 (92.6%) |
| Congenital heart disease# (n=67) | 8 (11.9%) |
| Neonatal respiratory symptoms (n=67) | 57 (85.1%) |
| Persistent wet cough | 57 (83.8%) |
| Persistent rhinosinusitis | 64 (94.1%) |
| Conductive hearing loss and/or otitis media with effusion (glue ear) | 50 (73.5%) |
| Finger clubbing (n=67) | 13 (19.4%) |
| nNO (nL·min−1) (n=36) | Median 14.5; range 5–62.5 |
| FEV1 z-score (n=28) | Median −1.90 (−5.0–1.32) |
| FVC z-score (n=28) | Median −1.72 (−5.64–0.57) |
| BMI z-score n=28 | Median −0.36 (−3.03–2.57) |
n=68 unless otherwise stated. BMI: body mass index; FEV1: forced expiratory volume in 1 s; FVC: forced vital capacity; nNO: nasal nitric oxide.
#Atrial septal defect n=2; right isomerism with complex cardiac defects n=1; situs ambiguous with complex cardiac defects n=1; subaortic membrane n=1; ventricular septal defect n=3.
FIGURE 1.
Venn diagram representing the variability of symptoms characteristic of primary ciliary dyskinesia (n=67; insufficient data for one member of cohort): situs inversus, neonatal respiratory distress, chronic wet cough and rhinosinusitis.
nNO was <77 nL·min−1 in 100% of tested individuals with a positive diagnosis. There was considerable variability of FEV1, FVC and BMI z-scores, but median values for lung function were already below average around the time of diagnosis (table 1).
Genetic causes of PCD in the Palestinian population
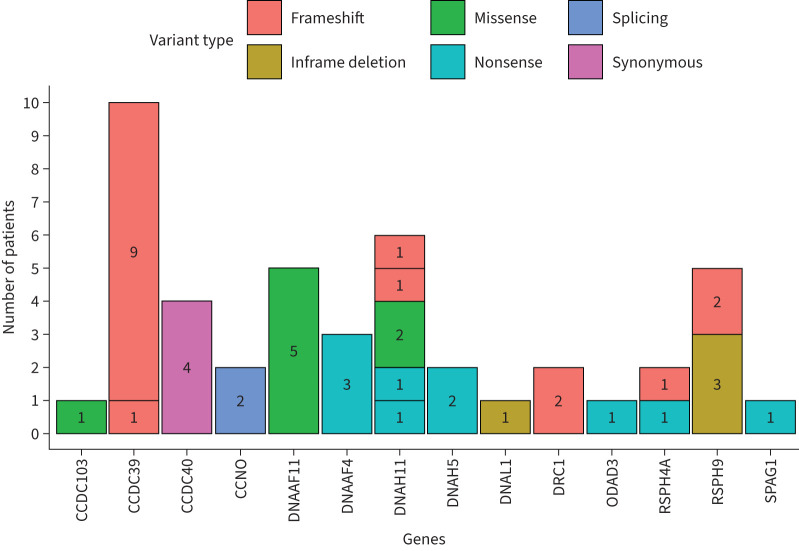
Genetic screening was performed on 82 patients from 63 families. This identified a total of 17 different causal variants and an additional four variants of unknown significance (VUS) in 14 known PCD genes, according to ACMG-AMP variant classification guidelines [23] (table 2, figure 2, supplementary table 1). Of the four variants classed as VUS, there were several lines of evidence to support pathogenicity but all fell below the “likely pathogenic” threshold, except for one well reported variant CCDC103 c.104G>C; p.(Arg35Pro), which is likely pathogenic if PP4 evidence is applied (phenotype/biochemical/immuno-fluorescence staining specificity) (table 2). Eight of the pathogenic/likely pathogenic variants identified in this study were specific to the Palestinian population, not having been reported before in ClinVar (table 2). A genetic diagnosis in agreement with confirmation through proband sequencing and familial segregation analysis to identify biallelic variants meeting ACMG-AMP pathogenic/likely pathogenic criteria [23] resulted in a definitive genetic cause being found in a total of 45 patients: 39 affected individuals from 35 families; additionally, VUS in four known PCD genes consistent with TEM defects were identified in a further six individuals from five families (table 2). One variant, CCDC40 c.48A>G carried in a family with four affected children, differed between its ClinVar score (‘likely benign’) and ACMG score of ‘likely pathogenic', as it is predicted to create a synonymous early missense change, p.(Gly16Gly), but ACMG predicts a likely splicing defect (table 2). Overall, the genetic screening analysis yielded a positive diagnosis in 45 patients from 40 families, providing a positive genetic diagnosis in 62% families and 55% of patients, who all carried homozygous variants in known PCD genes. The other 37 patients screened did not carry indicative variants in known PCD genes, but a number of variants in candidate ciliary genes were identified that are being further investigated.
TABLE 2.
Pathogenicity classifications for all identified variants in known primary ciliary dyskinesia (PCD) genes
| Study ID | Gene | RefSeq transcript | Variant (all homozygous) | Genomic location (GRCh38) | Exon/total | LOF | NMD (DECIPHER) | ACMG/AMP evidence | ACMG/AMP classification | ClinVar classification | Recorded in previous publications/similar population |
| 0-1, 0-2, 0-8, 0-9, 0-11, 0-20, 0-21, 0-25, 0-31 | CCDC39 | NM_181426.1 | c.1871_1872del; p.(Ile624Lysfs*3) | 3: 180641995_180641996del | 13/20 | Yes | Yes | PVS1_very strong PM2_moderate PM3_moderate |
Pathogenic | Pathogenic | One case from Saudi exome project [26] |
| 0-35 | CCDC39 | NM_181426.1 | c.2190del; p.(Glu731Asnfs*31) | 3: 180619334del | 16/20 | Yes | Yes | PVS1_very strong PM2_moderate PM3_moderate |
Pathogenic | Pathogenic | Algerian/Tunisian founder effect [27, 39] |
| 0-6 (4 aff) | CCDC40 | NM_017950.3 | c.48A>G; p.(Gly16Gly) | 17: 80038141A>G | 2/20 | No | na | PM2_moderate PM3_supporting PP1_strong |
Likely pathogenic (may create splice enhancer) | Likely benign | No |
| 0-36, 0-39 | CCNO | NM_021147.5 | c.381+5G>C | 5: 55233138C>G | 1/3 | No | No | PM2_moderate PM3_moderate PP1_moderate PP3_supporting PP4_supporting |
Likely pathogenic | Absent | No |
| 0-16, 0-30, 0-40 | DNAAF4 (DYX1C1) | NM_130810.3 | c.384_390del; p.(Tyr128*) | 15: 55491138_55491144del | 5/10 | Yes | Yes | PVS1_very strong PM2_moderate PM3_moderate |
Pathogenic | Likely pathogenic | No |
| 0-4, 0-15, 0-18, 0-26, 0-42 | DNAAF11 (LRRC6) | NM_012472.4 | c.436G>C; p.(Asp146His) | 8: 132632957C>G | 5/12 | No | na | PS3_strong PM3_moderate PP1_strong |
Pathogenic | Pathogenic | Two unrelated Palestinian families [28, 40] |
| 0-23 (2 aff) | DNAH5 | NM_001369.2 | c.10050G>A; p.(Trp3350*) | 5: 13766027C>T | 59/79 | Yes | Yes | PVS1_very strong PM2_moderate PM3_supporting |
Pathogenic | Absent | No |
| 0-24 | DNAH11 | NM_001277115.1 | c.6727C>T; p.(Arg2243*) | 7: 21710596C>T | 41/82 | Yes | Yes | PVS1_very strong PM3_moderate |
Pathogenic | Pathogenic/likely pathogenic | [41] |
| 0-7 | DNAH11 | NM_001277115.1 | c.12646G>T; p.(Glu4216*) | 7: 21892563G>T | 77/82 | Yes | Yes | PVS1_very strong PM2_moderate PM3_moderate |
Pathogenic | Absent | No |
| 0-22 | DNAH11 | NM_001277115.1 | c.13240dup; p.(Thr4414Asnfs*34) | 7: 21900057dup | 81/82 | Yes | Yes | PVS1_very strong PM2_moderate PM3_moderate |
Pathogenic | Pathogenic | No |
| 0-28 | DNAH11 | NM_001277115.1 | c.13436_13440dup; p.(Tyr4481Leufs*7) | 7: 21901139_21901143dup | 82/82 | Yes | No | PVS1_moderate PM2_moderate PM3_moderate |
Likely Pathogenic | Absent | No |
| 0-14, 0-19 | DRC1 | NM_145038.5 | c.1521_1524del; p.(Glu508Alafs*4) | 2: 26450009_26450012del | 12/17 | Yes | Yes | PVS1_strong PM2_moderate PM3_moderate |
Likely Pathogenic | Absent | No |
| 0-5 | ODAD3 (CCDC151) | NM_145045.4 | c.850C>T; p.(Gln284*) | 19: 11426257G>A | 7/13 | Yes | Yes | PVS1_strong PM2_moderate PM3_supporting |
Likely Pathogenic | Absent | No |
| 0-29 | RSPH4A | NM_001010892.3 | c.367del; p.(Pro123Leufs*44) | 6: 116616991del | 1/6 | Yes | Yes | PVS1_very strong PM2_moderate PM3_supporting |
Pathogenic | Absent | No |
| 0-37 | RSPH4A | NM_001010892.3 | c.72G>A; p.(Trp24*) | 6: 116616695G>A | 1/6 | Yes | Yes | PVS1_very strong PM2_moderate PM3_supporting |
Pathogenic | Absent | No |
| 0-27, 0-32, 0-33 | RSPH9 | NM_152732.4 | c.800_802del; p.(Lys268del) | 6: 43670922_43670924del | 5/5 | No | na | PS3_strong PM2_supporting PM3_moderate PM4_supporting PP1_strong |
Pathogenic | Pathogenic | Two Bedouin families; one Saudi family [29, 30, 42] |
| 0-34 | SPAG1 | NM_172218.3 | c.742C>T; p.(Arg248*) | 8: 100187160C>T | 8/19 | Yes | Yes | PVS1_very strong PM2_moderate PM3_supporting PP1_supporting |
Pathogenic | Absent | No |
| 0-3 | CCDC103 | NM_213607.3 | c.104G>C; p.(Arg35Pro) | 17: 44901102G>C | 2/4 | No | na | PM2_moderate PM3_moderate PP3_supporting PP4_supporting |
VUS (likely pathogenic) | VUS | No |
| 0-10, 0-12 | DNAH11 | NM_001277115.1 | c.563T>C; p.Met188Thr | 7:21558869T>C | 3/82 | No | na | PM2_moderate PM3_moderate PP1_supporting |
VUS | Absent | No |
| 0-17 | DNAL1 | NM_031427.4 | c.285_287del; p.(Glu97del) | 14: 73687283_73687285del | 6/8 | No | No | PM2_moderate PM3_supporting PM4_supporting |
VUS | VUS | No |
| 0-13 (**aff+aff father) | RSPH9 | NM_152732.4 | c.760del; p.(Arg254Alafs*76) | 6: 43670878del | 5/5 | Yes | No | PVS1_moderate PM2_moderate PM3_supporting |
VUS | Absent | No |
Study ID indicates family number and each family has a single affected child with a positive genetic diagnosis, except where ‘aff’ is shown, indicating that there is more than one affected child in the family (number of affected individuals is shown in these cases). Family 0-13 is the only family with an affected parent in addition to affected children. Database of Chromosomal Imbalance and Phenotype in Humans using Ensembl Resources (DECIPHER), American College of Medical Genetics and Genomics and the Association for Molecular Pathology (ACMG-AMP) and ClinVar classifications indicate likelihood that variants cause protein loss of function, transcript nonsense-mediated decay (NMD), in addition to showing their predicted pathogenic, likely pathogenic or variants of unknown significance (VUS) status. LOF: loss of function; na: not applicable (i.e. cannot be done for missense variants).
FIGURE 2.
Range of homozygous gene variants identified in 45 genetically diagnosed Palestine primary ciliary dyskinesia patients. Per gene, each different block represents a different variant, as listed in table 2. Numbers within the blocks indicate the number of patients per mutation.
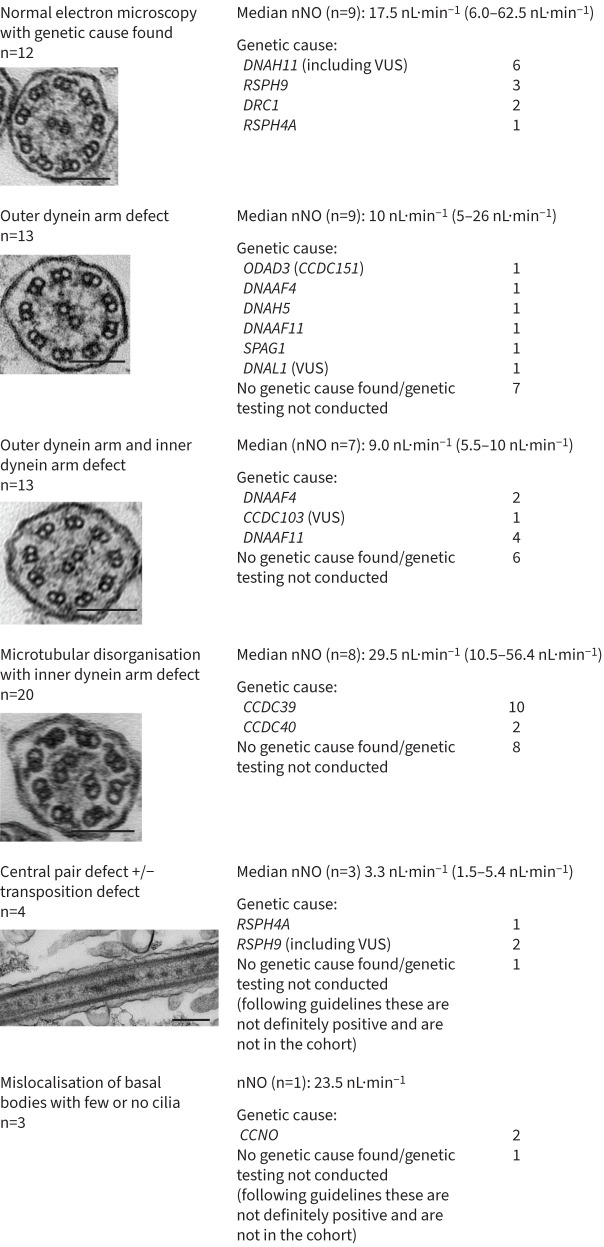
TEM findings correlated with genotype (figure 3, supplementary table 1). A remarkable 100% of genetic diagnoses involved homozygous variants, yet the homozygous variants were found in 12 different genes (14 genes including VUS results, figure 2). This reflects a high degree of genetic heterogeneity, alongside family allelic homogeneity reflecting likely shared parental ancestry within families. It is also notable that seven variants were recurrent within the population, i.e. detected in two or more families, suggesting a shared ancestral mutation/founder effect: CCDC39 c.1871_1872del; p.(Ile624Lysfs*3), CCNO c.381+5G>C, DNAAF4 c.384_390del; p.(Tyr128*), DNAAF11 c.436G>C; p.(Asp146His), DRC1 c.1521_1524del; p.(Glu508Alafs*4) and RSPH9 c.800–802del; p.(Lys268del) as well as a DNAH11 VUS p.(Met188Thr) (figure 2, table 2, supplementary table 1). This hypothesis is supported by previous reports showing that three of these recurrent variants were carried in PCD patients of Arabic Bedouin and Palestinian origin [26–31] (table 2).
FIGURE 3.
Diagnostic outcomes in the Palestinian primary ciliary dyskinesia (PCD) population. Nasal nitric oxide (nNO) and PCD genes with pathogenic mutations are reported according to transmission electron microscopy (TEM) findings. Two patients with a genetic diagnosis did not have TEM conducted (RSPH9 and DNAH5). VUS: variants of unknown significance. All scale bars=200 nm.
For the two most commonly occurring mutations, nine families carrying the same homozygous CCDC39 c.1871_1872del variants and five carrying homozygous DNAAF11 c.436G>C together account for 35% of overall diagnoses alone. The variants DNAAF4 c.384_390del and RSPH9 c.800–802del found in six families account for another 15%. These four variants alone therefore cause significant levels of disease amongst the tested Palestinian families and highlight that, in less resourced settings, identification of commonly occurring disease alleles can allow for highly targeted genetic diagnosis. As illustrated in this study, it was possible to use allele-specific PCR for diagnostics in three families carrying CCDC39 c.1871_1872del and DNAAF11c.436G>C mutations (supplementary table 1).
Despite commonly recurrent mutations, the number of genetically confirmed patients was insufficient to identify meaningful genotype–phenotype correlations amongst the population (supplementary figure 2). Even individuals with the same variant and some from the same family have differing clinical presentations.
Discussion
This is the first report of PCD diagnostic findings, including genetic variants, within the Palestinian population. In 8 years, we successfully established a diagnostic process, resulting in a large clinical population of 68 individuals with confirmed PCD from Palestine. A further 57 are highly likely to have the diagnosis based on situs inversus with sinopulmonary symptoms and/or low nNO levels and many more symptomatic family members will now undergo targeted screening. The data includes clinical manifestations, genotype and axoneme ultrastructure. This work highlights the importance of international collaboration for rare diseases like PCD, particularly where the diagnosis is complex and requires expertise and equipment that is only available in specialist centres. Networks such as BEAT-PCD and European Reference Network (ERN) for the lung (ERN-LUNG) support the diagnosis and management of PCD by supporting local physicians.
As expected, the clinical characteristics were similar to those previously described, including neonatal respiratory distress, persistent wet cough from infancy and middle ear disease [4, 13]. Interestingly, when we first started diagnosing patients, we had a predominance of patients with laterality defects, but the incidence of situs inversus in the clinic population is now only 42.6%, presumably reflecting the improved detection by other features.
The genotyping revealed a high diagnostic rate and a fascinating genetic architecture for this population. Remarkably, families from this small geographical isolate show extensive genetic heterogeneity, since 12 different PCD genes were found to carry 17 different causal mutations amongst the affected families. A further four VUS of unproven diagnostic status, except for one that may be causal if PP4 applied from the ACMG classification system, were detected in four of the known PCD genes. Just as striking, however, is the finding that all genetic diagnoses involved homozygous variants in the affected individuals, with both parents in each individual family being heterozygotes, carrying a single copy of the same mutant allele. There was one case of a family with an affected father carrying the same homozygous variants as their affected child, but this relates to a VUS of less certain causation (RSPH9 c.760del). We found a number of recurrent mutations affecting two or more families, carried in homozygous state in a total of 26 PCD families. Most prominent of the recurrent mutations were CCDC39 c.1871_1872del and DNAAF11 c.436G>C. This reflects common ancestry and direct consanguinity amongst Palestinian marriages, with cousin marriages being common [32]. Notably, homozygous variants were also found in the children of the few parents who were not knowingly related, reflecting a high population endogamy.
The literature on PCD from the Arab countries has mostly been limited to a few case reports and case series [33]. A study of 33 families from Egypt reported genetic heterogeneity with variants in 13 genes [34], but although the responsible genes were similar to the ones described in this study, the variants were unique to either Palestinian or Egyptian families. A study from the United Arab Emirates identified a number of unique deleterious variants, but also a large number of variants of unknown significance in PCD-related genes [35]. In a cohort from Tunisia (34 families), CCDC39 was the most common mutant gene (44.1%) similar to our population, followed by DNAH5 (11.8%), HYDIN (8.8%) and TTC25 (5.9%) [36]. In Saudi Arabia the most commonly mutated gene was RSPH9 among 18 cases, which included the RSPH9 c.800-802del mutation also reported here [37].
A study from Israel included predominantly Arab individuals and reported mutations in DNAH11 [38] to be the most common cause of PCD. A study of five children from two unrelated consanguineous Palestinian families revealed novel mutations in DNAAF11/LRRC6 (c.436G>C), which also affected families in this study [28].
There are four main providers of healthcare in Palestine, namely the Ministry of Health responsible for the public health system, the United Nations Relief and Works Agency for Palestinian Refugees (UNRWA), non-governmental and civil society organisations (like the Palestinian Red Crescent), and the private health sector. Although there have been significant healthcare improvements in recent years, the system still lacks sufficient infrastructure to serve the growing population. According to the Palestinian Central Bureau of Statistics (PCBS) (www.pcbs.gov.ps), in 2018 the average number of doctors per 1000 population was 2.09 and hospital beds 1.33 per 1000 population, which is considerably less than most developed countries. The healthcare system is closely connected to and affected by social, economic and political situations. Therefore, it faces challenges due to many years of restrictions and blockade. In 2017, the PCBS reported that 29.2% of Palestinians were living below the poverty level, about four times higher in Gaza (53%) than in the West Bank (13.9%) (www.pcbs.gov.ps). This makes it challenging for many Palestinians to access quality healthcare. The diagnostic testing conducted during this study was dependent on a number of small research grants with support from collaborators in UK. Moving forward, the PCD service in Palestine is not funded by the public health system and therefore patients will have to either pay out of their own pocket or purchase expensive private insurance to cover the costs of clinic visits and tests.
In addition, the blockade on the West Bank and Gaza and the restrictions on free movement make it difficult for many Palestinians to have immediate access to healthcare. Patients with chronic medical conditions like PCD, who should ideally have regular monitoring, often seek medical attention only when they are ill and cannot afford routine monitoring such as sputum microbiology, imaging and pulmonary function testing. These challenges were significant obstacles for a standardised approach to PCD diagnostic testing in Palestine in this study. The choice and order of the tests depended on the resources available at the time and the access to clinics. For example, in Gaza, it was easier for local physicians to ship blood samples for genetic testing rather than patients travelling from Gaza to the West Bank for nNO and ciliary biopsy.
Moreover, the lack of a specialised multidisciplinary PCD centre in Palestine is another barrier to providing PCD care for patients. Diagnostic services are fractioned and potentially inconsistent due to nNO measurements and nasal brushings being conducted at four different locations (Jerusalem, Bethlehem, Hebron and Ramallah).
Identifying the causative mutations in the Palestinian population can allow targeted mutation testing in future cases that could be traced back to one of the known families in this study. Such testing can be done locally at a cheap cost, thus saving resources and avoiding many of the other cumbersome tests if a diagnosis could be reached. However, such an approach should be interpreted with caution since it may be good to “rule in” but not “rule out” PCD. In the future, it may even be possible to develop a population-specific mutation panel that could be utilised for a newborn screening programme.
In summary, we report the first detailed study of the genetics and clinical features of PCD in the Palestinian population. The cohort of individuals with PCD was rapidly formed within an isolated population with limited diagnostic resources and continues to grow. There is a large number of families and individuals with symptoms who are still awaiting investigation and it is likely that the prevalence of PCD in Palestine will be higher than reported in international literature. We have highlighted the significant genetic heterogeneity and the remarkable finding of 100% homozygous variants.
Supplementary material
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary material 00714-2022.supplement (407.2KB, pdf)
Footnotes
This article has an editorial commentary: https://doi.org/10.1183/23120541.00122-2023
Provenance: Submitted article, peer reviewed.
Support statement: N. Rumman was the recipient of a European Respiratory Society (ERS) Fellowship (STRTF 2014-6816). AAIR Charity (charity number 1129698) funded the whole-exome sequencing and Circassia loaned two Niox Mino analysers for this study. The National PCD Diagnostic Service at University Hospital Southampton is commissioned and funded by NHS England. Some authors are participants of BEAT-PCD: Better Evidence to Advance Therapeutic options for PCD (COST Action BM 1407; ERS Clinical Research Collaboration). BEAT-PCD COST Action funded J.S. Lucas to visit N. Rumman to support the new PCD Service and development of this cohort in Palestine. N. Rumman was funded by an Academy of Medical Sciences Daniel Turnberg Travel Fellowship to visit H.M. Mitchison for completion of the genetic data. H.M. Mitchison acknowledges funding from Great Ormond Street Children's Charity, NIHR Biomedical Research Centre at Great Ormond Street Hospital, the British Council Newton-Mosharafa Fund and Ministry of Higher Education in Egypt. Some authors participate in European Reference Network for Rare Respiratory Diseases (ERN-LUNG) project identifier number 739546. J.S. Lucas chaired the ERS Task Force for the development of a practice guideline for diagnosis of PCD (ERS TF-2014-04) and ERS Task Force for “Nasal nitric oxide measurement in children for the diagnosis of primary ciliary dyskinesis: a technical standard”. M.R. Fassad is supported by a Wellcome Trust Collaborative Award in Science (210585/Z/18/Z). Funding information for this article has been deposited with the Crossref Funder Registry.
Author contributions: N. Rumman and J.S. Lucas planned the diagnostic pathway and development of a PCD cohort, and C.L. Jackson taught N. Rumman to take and process diagnostic samples during an ERS Fellowship visit to UoS. N. Rumman collected clinical data, nasal brushings and DNA samples; N. Rumman holds the ethical approvals for the Palestinian cohort. N. Rumman performed all the nasal NO testing. Electron microscopy was analysed by P. Goggin, R. Doherty and J. Thompson. Genetic samples were analysed by M.R. Fassad, G.M. Freke, M.A. Mohamed, M.P. Patel, R. Pengelly, G. Wheway and H.M. Mitchison. The data analysis plan was developed by J.S. Lucas and B. Rubbo. Genetic data analysis was performed by M.R. Fassad, N.S. Thomas, G. Wheway and H.M. Mitchison. Data cleaning was led by C. Driessens and J. Chopra. Clinical data analyses were undertaken by C. Driessens and J.S. Lucas. B. Fashho conducted spirometry. N. Abdelrahman, A. Adwan, R. Abu Nema, A. Qaaqour, A. Hasballah and M. Albakri identified and referred patients for PCD diagnostic testing to N. Rumman. H.M. Mitchison and J.S. Lucas take responsibility for the data and analyses.
Conflict of interest: N. Rumman received support for the present manuscript from two NIOX MINO devices from Circassia (previously called Aerocrine) to perform nasal nitric oxide testing and AAIR Charity funded whole-exome sequencing testing, and support for attending meetings and/or travel from the European Respiratory Society for a short term research training fellowship (3 months at Southampton University) (STRTF 2014-6816), outside the submitted work. M.R. Fassad received support for the present manuscript from a Wellcome Trust Collaborative Award in Science (210585/Z/18/Z). C. Driessens received support for the present manuscript from NHS England: work for this manuscript was performed while working for the National PCD Diagnostic Service at University Hospital Southampton; this service is commissioned and funded by NHS England. R. Pengelly received support for the present manuscript from Asthma Allergy and Inflammation Research. G. Wheway received support for the present manuscript from Asthma Allergy and Inflammation Research Trust; grants or contracts from UKRI COVID-19 Agile Response Fund, Wessex Medical Research/Rosetrees Trust PhD studentship, and Asthma Allergy and Inflammation Research Trust, outside the submitted work; stock or stock options for Illumina Inc., outside the submitted work; and receipt of equipment, materials, medical writing, gifts or other services from Synthego, outside the submitted work. H.M. Mitchison received support for the present manuscript from Great Ormond Street Children's Charity, NIHR Biomedical Research Centre at Great Ormond Street Hospital, and British Council Newton-Mosharafa Fund and Ministry of Higher Education in Egypt. J.S. Lucas received grants or contracts from NIHR, AAIR Charity, and NHS England, outside the submitted work. The remaining authors have nothing to disclose.
References
- 1.Lucas JS, Davis SD, Omran H, et al. Primary ciliary dyskinesia in the genomics age. Lancet Respir Med 2020; 8: 202–216. doi: 10.1016/S2213-2600(19)30374-1 [DOI] [PubMed] [Google Scholar]
- 2.Wallmeier J, Nielsen KG, Kuehni CE, et al. Motile ciliopathies. Nat Rev Dis Primers 2020; 6: 77. doi: 10.1038/s41572-020-0209-6 [DOI] [PubMed] [Google Scholar]
- 3.Legendre M, Zaragosi LE, Mitchison HM. Motile cilia and airway disease. Semin Cell Dev Biol 2021; 110: 19–33. doi: 10.1016/j.semcdb.2020.11.007 [DOI] [PubMed] [Google Scholar]
- 4.Goutaki M, Meier AB, Halbeisen FS, et al. Clinical manifestations in primary ciliary dyskinesia: systematic review and meta-analysis. Eur Respir J 2016; 48: 1081–1095. doi: 10.1183/13993003.00736-2016 [DOI] [PubMed] [Google Scholar]
- 5.Best S, Shoemark A, Rubbo B, et al. Risk factors for situs defects and congenital heart disease in primary ciliary dyskinesia. Thorax 2019; 74: 203–205. doi: 10.1136/thoraxjnl-2018-212104 [DOI] [PubMed] [Google Scholar]
- 6.O'Callaghan C, Chetcuti P, Moya E. High prevalence of primary ciliary dyskinesia in a British Asian population. Arch Dis Child 2010; 95: 51–52. doi: 10.1136/adc.2009.158493 [DOI] [PubMed] [Google Scholar]
- 7.Hannah WB, Seifert BA, Truty R, et al. The global prevalence and ethnic heterogeneity of primary ciliary dyskinesia gene variants: a genetic database analysis. Lancet Respir Med 2022; 10: 459–468. doi: 10.1016/S2213-2600(21)00453-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rumman N, Jackson C, Collins S, et al. Diagnosis of primary ciliary dyskinesia: potential options for resource-limited countries. Eur Respir Rev 2017; 26: 160058. doi: 10.1183/16000617.0058-2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lucas JS, Barbato A, Collins SA, et al. European Respiratory Society guidelines for the diagnosis of primary ciliary dyskinesia. Eur Res J 2017; 49: 1601090. doi: 10.1183/13993003.01090-2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shapiro AJ, Davis SD, Polineni D, et al. Diagnosis of primary ciliary dyskinesia. an official American Thoracic Society clinical practice guideline. Am J Respir Crit Care Med 2018; 197: e24–e39. doi: 10.1164/rccm.201805-0819ST [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leigh MW, Hazucha MJ, Chawla KK, et al. Standardizing nasal nitric oxide measurement as a test for primary ciliary dyskinesia. Ann Am Thorac Soc 2013; 10: 574–581. doi: 10.1513/AnnalsATS.201305-110OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collins SA, Gove K, Walker W, et al. Nasal nitric oxide screening for primary ciliary dyskinesia: systematic review and meta-analysis. Eur Respir J 2014; 44: 1589–1599. doi: 10.1183/09031936.00088614 [DOI] [PubMed] [Google Scholar]
- 13.Behan L, Dimitrov BD, Kuehni CE, et al. PICADAR: a diagnostic predictive tool for primary ciliary dyskinesia. Eur Respir J 2016; 47: 1103–1112. doi: 10.1183/13993003.01551-2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ardura-Garcia C, Goutaki M, Carr SB, et al. Registries and collaborative studies for primary ciliary dyskinesia in Europe. ERJ Open Res 2020; 6: 00005-2020. doi: 10.1183/23120541.00005-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller MR, Hankinson J, Brusasco V, et al. Standardisation of spirometry. Eur Respir J 2005; 26: 319–338. doi: 10.1183/09031936.05.00034805 [DOI] [PubMed] [Google Scholar]
- 16.Quanjer PH, Stanojevic S, Cole TJ, et al. Multi-ethnic reference values for spirometry for the 3–95-yr age range: the global lung function 2012 equations. Eur Respir J 2012; 40: 1324–1343. doi: 10.1183/09031936.00080312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shoemark A, Boon M, Brochhausen C, et al. International consensus guideline for reporting transmission electron microscopy results in the diagnosis of primary ciliary dyskinesia (BEAT PCD TEM criteria). Eur Respir J 2020; 55: 1900725. doi: 10.1183/13993003.00725-2019 [DOI] [PubMed] [Google Scholar]
- 18.Fassad MR, Patel MP, Shoemark A, et al. Clinical utility of NGS diagnosis and disease stratification in a multiethnic primary ciliary dyskinesia cohort. J Med Genet 2020; 57: 322–330. doi: 10.1136/jmedgenet-2019-106501 [DOI] [PubMed] [Google Scholar]
- 19.Vasimuddin M, Misra S, Li H, et al. Efficient architecture-aware acceleration of BWA-MEM for multicore systems. IEEE International Parallel and Distributed Processing Symposium (IPDPS), Rio de Janeiro, Brazil, 2019. pp. 314–324. doi: 10.1109/IPDPS.2019.00041 [DOI] [Google Scholar]
- 20.McKenna A, Hanna M, Banks E, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010; 20: 1297–1303. doi: 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010; 38: e164. doi: 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.den Dunnen JT, Dalgleish R, Maglott DR, et al. HGVS recommendations for the description of sequence variants: 2016 update. Hum Mutat 2016; 37: 564–569. doi: 10.1002/humu.22981 [DOI] [PubMed] [Google Scholar]
- 23.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015; 17: 405–424. doi: 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Firth HV, Richards SM, Bevan AP, et al. DECIPHER: database of chromosomal imbalance and phenotype in humans using ensembl resources. Am J Hum Genet 2009; 84: 524–533. doi: 10.1016/j.ajhg.2009.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Landrum MJ, Lee JM, Benson M, et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res 2018; 46: D1062–D10d7. doi: 10.1093/nar/gkx1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Monies D, Abouelhoda M, Assoum M, et al. Lessons learned from large-scale, first-tier clinical exome sequencing in a highly consanguineous population. Am J Hum Genet 2019; 104: 1182–1201. doi: 10.1016/j.ajhg.2019.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merveille AC, Davis EE, Becker-Heck A, et al. CCDC39 is required for assembly of inner dynein arms and the dynein regulatory complex and for normal ciliary motility in humans and dogs. Nat Genet 2011; 43: 72–78. doi: 10.1038/ng.726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horani A, Ferkol TW, Shoseyov D, et al. LRRC6 mutation causes primary ciliary dyskinesia with dynein arm defects. PLoS One 2013; 8: e59436. doi: 10.1371/journal.pone.0059436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Castleman VH, Romio L, Chodhari R, et al. Mutations in radial spoke head protein genes RSPH9 and RSPH4A cause primary ciliary dyskinesia with central-microtubular-pair abnormalities. Am J Hum Genet 2009; 84: 197–209. doi: 10.1016/j.ajhg.2009.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reish O, Slatkin M, Chapman-Shimshoni D, et al. Founder mutation(s) in the RSPH9 gene leading to primary ciliary dyskinesia in two inbred Bedouin families. Ann Hum Genet 2010; 74: 117–125. doi: 10.1111/j.1469-1809.2009.00559.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alsaadi MM, Gaunt TR, Boustred CR, et al. From a single whole exome read to notions of clinical screening: primary ciliary dyskinesia and RSPH9 p.Lys268del in the Arabian Peninsula. Ann Hum Genet 2012; 76: 211–220. doi: 10.1111/j.1469-1809.2012.00704.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sirdah MM. Consanguinity profile in the Gaza Strip of Palestine: large-scale community-based study. Eur J Med Genet 2014; 57: 90–94. doi: 10.1016/j.ejmg.2014.01.003 [DOI] [PubMed] [Google Scholar]
- 33.Hammoudeh S, Gadelhak W, Janahi IA. Primary ciliary dyskinesia among Arabs: where do we go from here? Paediatr Respir Rev 2019; 29: 19–22. doi: 10.1016/j.prrv.2018.09.002 [DOI] [PubMed] [Google Scholar]
- 34.Fassad MR, Shoman WI, Morsy H, et al. Clinical and genetic spectrum in 33 Egyptian families with suspected primary ciliary dyskinesia. Clin Genet 2020; 97: 509–515. doi: 10.1111/cge.13661 [DOI] [PubMed] [Google Scholar]
- 35.Alsamri MT, Alabdouli A, Iram D, et al. A study on the genetics of primary ciliary dyskinesia. J Clin Med 2021; 10: 5102. doi: 10.3390/jcm10215102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mani R, Belkacem S, Soua Z, et al. Primary ciliary dyskinesia gene contribution in Tunisia: identification of a major Mediterranean allele. Hum Mutat 2020; 41: 115–121. doi: 10.1002/humu.23905 [DOI] [PubMed] [Google Scholar]
- 37.Alzaid M, Al-Mobaireek K, Almannai M, et al. Clinical and molecular characteristics of primary ciliary dyskinesia: a tertiary care centre experience. Int J Pediatr Adolesc Med 2021; 8: 258–263. doi: 10.1016/j.ijpam.2021.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gileles-Hillel A, Mor-Shaked H, Shoseyov D, et al. Whole-exome sequencing accuracy in the diagnosis of primary ciliary dyskinesia. ERJ Open Res 2020; 6: 00213-2020. doi: 10.1183/23120541.00213-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boaretto F, Snijders D, Salvoro C, et al. Diagnosis of primary ciliary dyskinesia by a targeted next-generation sequencing panel: molecular and clinical findings in Italian patients. J Mol Diagn 2016; 18: 912–922. doi: 10.1016/j.jmoldx.2016.07.002 [DOI] [PubMed] [Google Scholar]
- 40.Zariwala MA, Gee HY, Kurkowiak M, et al. ZMYND10 is mutated in primary ciliary dyskinesia and interacts with LRRC6. Am J Hum Genet 2013; 93: 336–345. doi: 10.1016/j.ajhg.2013.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lai M, Pifferi M, Bush A, et al. Gene editing of DNAH11 restores normal cilia motility in primary ciliary dyskinesia. J Med Genet. 2016; 53: 242–249. doi: 10.1136/jmedgenet-2015-103539 [DOI] [PubMed] [Google Scholar]
- 42.Alsaadi MM, Erzurumluoglu AM, Rodriguez S, et al. Nonsense mutation in coiled-coil domain containing 151 gene (CCDC151) causes primary ciliary dyskinesia. Hum Mutat 2014; 35: 1446–1448. doi: 10.1002/humu.22698 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary material 00714-2022.supplement (407.2KB, pdf)