Abstract
Adipose tissue inflammation is a driving factor for the development of obesity‐associated metabolic disturbances, and a role of adipose tissue T cells in initiating the pro‐inflammatory signaling is emerging. However, data on human adipose tissue T cells in obesity are limited, reflected by the lack of phenotypic markers to define tissue‐resident T cell subsets. In this study, we performed a deep characterization of T cells in blood and adipose tissue depots using multicolor flow cytometry and RNA sequencing. We identified distinct subsets of T cells associated with obesity expressing the activation markers, CD26 and CCR5, and obesity‐specific genes that are potentially engaged in activating pro‐inflammatory pathway, including ceramide signaling, autophagy, and IL‐6 signaling. These findings increase our knowledge on the heterogeneity of T cells in adipose tissue and on subsets that may play a role in obesity‐related pathogenesis.
Keywords: adipose tissue, multicolor flow cytometry, obesity, obesity‐specific genes, T cells
By an in‐depth profiling of human adipose tissue we identified distinct subsets of T cells associated with obesity. The findings increase of knowledge on heterogeneity of T cells in adipose tissue and on subsets that may play a role in obesity‐related pathogenesis.
Introduction
Chronic, low‐grade inflammation originating in adipose tissue and reflected in the circulation represents a key mechanism for obesity‐induced insulin resistance. Emerging evidence from experimental model systems suggests early accumulation and immunological influence of T cells in adipose tissue inflammation [1, 2]. CD4+ and CD8+ T cells are important components of the adaptive immune system and can be classified into distinct functional subsets based on variation in expression of surface receptors, transcription factors, and cytokine secretion profile. CD4+ T cells exist as immunosuppressive regulatory T cells (Tregs), T helper effector cells (Th1, Th2, Th17), and T follicular helper (Tfh) cells, whereas CD8+ T cells differentiate into memory and cytotoxic T lymphocytes upon activation with high killing capacity through granule release [3, 4]. Antigen‐experienced CD4+ and CD8+ T cells, such as central and effector memory cells (TCM and TEM, respectively), take part in surveilling circulation, lymph nodes, and peripheral tissues. In tissues, TEM can further differentiate into tissue‐resident memory T (TRM) cells upon antigenic stimulation that permanently reside in tissues through adaptations and interactions with other tissue‐resident cells [5].
In mice, lean adipose tissue is enriched with anti‐inflammatory T cells, whereas obesity induces a shift towards Treg depletion and accumulation of pro‐inflammatory CD4+ T cells and cytotoxic CD8+ T cells, predominantly in the visceral adipose tissue (VAT) compared with the subcutaneous adipose tissue (SAT) [1, 2, 6]. In humans, similar observations have been reported by some [7, 8] but not by others [9, 10], underscoring the need for confirmative studies. Phenotypically, TRM cells express canonical tissue residency markers, such as CD69, CD49a, and CD103, and as well as tissue‐specific signature markers reflecting adaptations to the microenvironment [5]. However, such markers defining T cells residing in human adipose tissue are not well characterized, limiting our current understanding of how T cell subsets may be involved in obesity‐related metabolic disturbances. Comparing the phenotypic markers of T cells between blood and adipose tissue from lean subject and people with obesity may improve our understanding of the tissue‐specific adaptations to obesity. Studies report that weight loss effectively reduces the levels of pro‐inflammatory T cells [11], but the exact effects of body weight changes on T cell composition and downstream signaling pathways are poorly understood. Here, we performed multicolor flow cytometry and RNA sequencing on T cells from different cohorts of patients with obesity and from lean controls to identify protein and gene expression signatures in blood and tissue. We show that T cells in blood and adipose tissue display distinct patterns and identify unique subpopulations of T cells residing in adipose tissue that may have a potential role in the pathogenesis of obesity.
Results and discussion
Broad receptor profiling identifies distinct T cell proteins in blood and adipose tissue
As an initial approach to identify adipose tissue‐specific T cell surface proteins, we performed a surface proteomics analysis using the LEGENDScreenTM immunoprofiling kit on available CD3+ T cells in blood and SAT (Fig. 1A, cohort 1). Several proteins were expressed at various levels on the T cells in blood and adipose tissue (Fig. 1B and C), and we also identified commonly expressed proteins between the two compartments (Fig. 1C and Fig. S1). Among the differentially expressed proteins, most were expressed by a higher proportion of blood T cells, such as CD81, CD99, CD100, CD162, CD205, and CD229, whereas CCR10 and CD279 (PD‐1) were higher on SAT T cells (Fig. 1B and C). CCR10 has been reported to regulate the maintenance and function of skin‐resident T cells [12], but a functional role on adipose tissue T cells has not yet been described. The inhibitory protein, PD‐1, is expressed on activated T cells to mediate T cell exhaustion and has been described as a tissue‐resident T cell marker in human pancreas [13]. Interestingly, its ligand, PD‐L1, was recently linked to reduced inflammation in human adipose tissue [14]. The comparison of LEGENDScreenTM proteins between blood and SAT is potentially a powerful method to reveal tissue‐specific T cell markers. However, our analysis did not detect established tissue residency markers, such as CD69 and CD103, as tissue‐specific, which indicated a limitation of this approach. This unexpected finding may, at least in part, be explained by the fact that, due to the high number of cells required in the surface proteome analysis, each donor only provided sufficient cells for a limited number of proteins, and each protein was only measured once, making the results sensitive to both technical and biological variation. The unmatched blood‐ and adipose tissue‐derived donors may also have impacted the findings.
Figure 1.
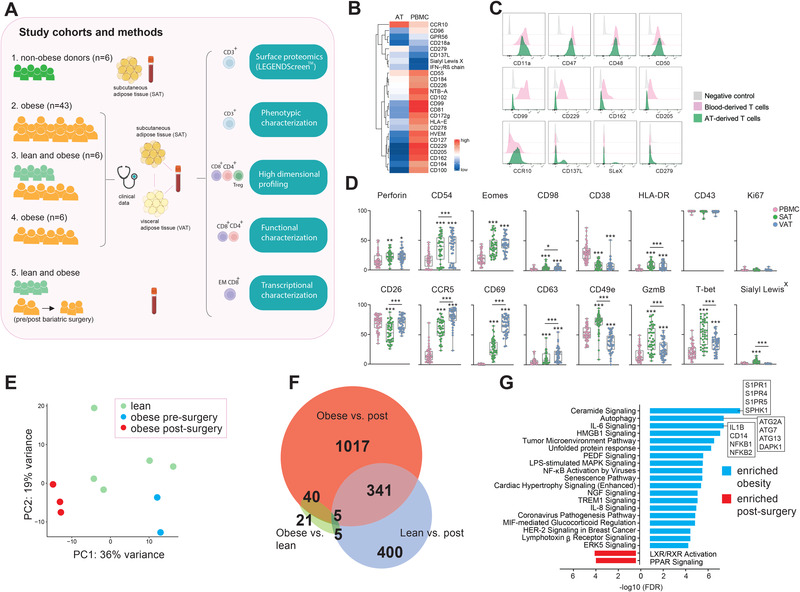
Broad characterization of T cells (A) Overview of study cohorts and methods. EM, effector memory. Pink line indicates paired samples. The figure is partly created with BioRender.com (B) Heatmap depicting percentage of CD3+ T cells expressing the individual proteins in subcutaneous adipose tissue (SAT) and peripheral blood (PBMC) (C) Flow cytometry histograms of modal protein expression on T cells in SAT and PBMC as well as isotype control. Histograms display proteins highly expressed in both compartments (upper), highest in PBMC (middle), or highest in SAT (bottom). sLeX, Sialyl LewisX (D) Box‐and‐whisker plots showing percentage of positive CD3+ T cells expressing the individual proteins in PBMC, SAT and visceral adipose tissue (VAT) from individuals with obesity (n = 43). Line represents median and the whiskers represent the min and max value. The Wilcoxon matched‐paired sign rank test was used for comparison between groups. (*) indicates significant difference relative to blood unless otherwise specified. **p < 0.01, ***p < 0.001. (E) PCA plot of 500 most variable genes in CD8+ effector memory T (TEM) cells from blood of lean subjects (n = 5), and individuals with obesity before (n = 2) and one year after (n = 3) bariatric surgery. (F) Venn diagram showing differentially expressed genes (DEGs) between the three patient groups. (G) Ingenuity Pathway Analysis (IPA) of top 20 out of 69 canonical pathways enriched in individuals with obesity before (blue) and after (red) bariatric surgery. Pathways ordered by FDR. A selection of genes is listed for the top 3 obesity‐enriched pathways.
Therefore, in a second approach, we applied multicolor flow cytometry to investigate the expression of a selection of both surface‐ and intracellular lymphocyte proteins on CD3+ T cells in matched blood, SAT and VAT samples from 43 patients with obesity class III (Fig. 1A, cohort 2). The experimental design did not allow for inclusion of CD4 and CD8, thus limiting the current analysis to CD3+ T cells only. In contrast to LEGENDScreenTM, most of the analyzed proteins were enriched on the T cells in adipose tissue compared with in blood, including CD69 (Fig. 1D and Fig. S2). Moreover, significant differences in expression pattern were observed between SAT and VAT, demonstrating depot‐specific protein signatures (Fig. 1D). In particular, CD49e, granzyme B, and T‐bet were identified as SAT‐specific proteins, whereas VAT T cells displayed increased levels of CD69, CD26 (DPP4), CD63, and CCR5, proteins related to T cell activation [15, 16]. This points to more activated T cells in VAT than in SAT, potentially reflecting adaptations to a stressed VAT microenvironment. Due to the low T cell resolution in this analysis, we cannot conclude whether the observed differences reflect altered phenotypes on the cell per.se or altered proportion of T cell subsets.
Bariatric surgery induces transcriptional changes in circulating CD8+ T cells
To understand how weight loss affects T cells, we next performed RNA sequencing on circulating CD8+ TEM cells, a subset that traffic between circulation and inflamed tissues and has been found altered in mice with diet‐induced obesity [17]. The T cells were obtained from peripheral blood of individuals with obesity, paired before (obese) and one year after (post) bariatric surgery and from lean controls (lean) (Fig. 1A, cohort 5). Using principal component analysis (PCA) of the 500 most changed genes, we observed that the three groups clustered separately, reflecting transcriptional heterogeneity (Fig. 1E). The number of differentially expressed genes (DEGs) was highest between obese and post (1403) compared with the number of DEGs between lean and obese (71) (Fig. 1F, Fig. S3A, and Table S1). Moreover, only 40 DEGs were both affected by obesity and normalized to lean state after surgery (Fig. S3B). Together, these data suggest that the circulating CD8+ TEM cell transcriptome is affected by weight loss, but the transcriptional changes cannot be explained solely by the difference in body weight. It is, however, important to acknowledge the strength of using paired (post vs obese) versus unpaired (lean vs obese) samples that eliminate interindividual variation as a confounder, which may explain some of the discrepancies between the lean and post group. Additionally, some of the differences may be explained by dynamic changes that occur during weight loss that may vary more than the static differences between lean and obese state.
An Ingenuity Pathways Analysis (IPA) of DEGs before and after surgery identified 69 pathways significantly activated in T cells at either one of the time points, in which most were enriched before rather than after surgery (Fig. 1G; Table S2). On top, we found genes related to ceramide signaling, autophagy, and IL‐6 signaling. These pathways are well known to be dysregulated in obesity and metabolic diseases, and it was interesting that they were also changed in the T cells themselves, suggesting a more pro‐inflammatory CD8+ TEM cell in obesity. Our findings coincide with other studies demonstrating the importance of these pathways for T cell responses in obesity [18, 19]. Moreover, the activation of the anti‐inflammatory pathways LXR/RXR activation and PPAR signaling [20] after bariatric surgery suggests a switch back to an anti‐inflammatory state following weight loss (Fig. 1G). Together, we identified genes and related signaling pathways that may be relevant for the pro‐inflammatory T cell responses in obesity. However, whether the identified genes are drivers of T cell‐induced inflammation warrants further investigation. Moreover, using paired blood and adipose tissue samples may reveal whether changes in the circulation are also reflected in the adipose tissue.
Elevated levels of CD8+ but not CD4+ T cells in adipose tissue from people with obesity
Having established that CD3+ T cells in VAT are phenotypically distinct from those in SAT and that CD8+ TEM cells are modified in circulation upon weight loss, we next performed multicolor flow cytometry to compare the abundance and phenotype of T cells in paired blood, SAT, and VAT samples from individuals with obesity and in paired blood and VAT samples from lean subjects diagnosed with intraductal papillary mucinous neoplasms (IPMN; a premalignant pancreatic lesion found in otherwise healthy individuals) (Table 1, cohort 3). Since obesity is known to promote changes in the relative balance of pro‐ and anti‐inflammatory T cells, we characterized CD4+ T cells, Tregs, and CD8+ T cells (Fig. 2A). The frequencies of T cell subsets in blood and VAT did not differ significantly between the two patient groups, but a trend toward less CD4+ T cells and Tregs and more CD8+ T cells was observed in adipose tissue of the individuals with obesity (Fig. 2B). This is similar to previous findings, although CD4+ T cell numbers have been found elevated in obesity [7]. While no significant differences were found by us, it should be noted that the lean group had considerably higher median age than the obese group (Table 1). Similar to obesity, aging affects adipose tissue T cells and increases the levels of CD4+ and CD8+ T cells [21], suggesting that the differences in T cell frequencies would possibly have reached significance if the groups were of similar age.
Table 1.
Clinical and biochemical characteristics of patient cohorts
| Cohort 2 | Cohort 3 | Cohort 4 | ||
|---|---|---|---|---|
| Obese | Lean | Obese | Obese | |
| Subjects | 43 | 6 | 6 | 6 |
| Gender (females) | 69.8% (30/43) | 50% (3/6) | 100% | 66.7% (4/6) |
| Age (years) | 40 (19‐65) | 72 (61‐84) | 44 (22‐61) | 44 (23‐55) |
| T2D | 9/43 | 0/6 | 1/6 | 0 |
| BMI (kg/m2) | 38.8 (33.1‐51.8) | 22.5 (18.3‐23.7) | 42.6 (35.1‐48.5) | 40.7 (36.9‐47.5) |
| Fasting glucose (mmol/L) | 5.6 (4.4‐11.4) | 6.7 (5.4‐10) | 4.9 (4.6‐6.8) | 5.5 (5.3‐6.1) |
| HbA1c (mmol/mol) | 36 (29‐95) | 38 (28‐67) | 36 (32‐49) | 36 (34‐39) |
| CRP (mg/L) | 5 (1‐16) | 1 (1‐30) | 2.5 (1‐21) | 4 (1‐20) |
| Insulin (mIU/L) | 18.3 (5.2‐63.5) | 16.3 (4.7‐30.7) | 16.8 (6.6‐32.4) | |
| HOMA‐IR | 4.7 (1.1‐18.3) | 3.5 (1‐9.3) | 4.0 (1.6‐7.9) | |
| C‐peptide (mmol/L) | 1.1 (0.3‐2.9) | 0.9 (0.4‐1.6) | ||
| Total cholesterol (mmol/L) | 4.6 (2.5‐8.3) | 4.1 (3.1‐7.0) | 1.0 (0.7‐1.7) | |
| LDL (mmol/L) | 3.1 (1.1‐6.1) | 2.5 (1.6‐4.8) | 2.5 (1.8‐4.3) | |
| HDL (mmol/L) | 1.1 (0.6‐2.0) | 1.1 (0.9‐1.8) | 1.3 (0.8‐1.6) | |
| TAG (mmol/L) | 1.6 (0.5‐6.9) | 1.2 (0.9‐1.5) | 1.4 (1‐0‐3.3) | |
Data are given as median (range). All circulating parameters are measured from fasting blood samples. F, females; T2D, type 2 diabetes; BMI, body mass index; CRP, C‐reactive protein; HOMA‐IR, homeostatic model assessment of insulin resistance index; LDL, low‐density lipoprotein cholesterol; HDL, high‐density lipoprotein cholesterol; TAG; triacylglycerol.
Figure 2.

High dimensional profiling of T cell subsets in adipose tissue associated with obesity (A) Gating strategy to identify T cell subsets. Plots showing one representative visceral adipose tissue (VAT) sample. (B) Percentages of CD4+ and CD8+ T cells out of total, live T cells and Tregs out of CD4+ T cells in peripheral blood (PBMC) and VAT of lean subjects (n = 6) and PBMC, VAT and subcutaneous adipose tissue (SAT) of individuals with obesity (n = 6). (C) Box‐and‐whisker plots showing percentage of CD4+ and CD8+ T cells expressing the indicated proteins in blood and adipose tissue from the same individuals. Line represents median and whiskers represent min and max value. The Wilcoxon matched‐paired sign rank test was used for comparison between paired samples and The Mann‐Whitney U test was used for comparison between the lean subjects and individuals with obesity. (*) indicates significant difference between blood and adipose tissue in the same patient group, and (#) indicates significant difference between lean and obese for the indicated tissue compartment. */# p < 0.05, **/## p < 0.01. (D) UMAP plot of T cells. CD3+ T cells from VAT and blood from lean subjects and individuals with obesity were gated, barcoded, and down sampled. Colors indicate the origin of cells. (E) UMAP plots showing expression intensities of 25 phenotypic markers. (F) Twenty‐seven identified PhenoGraph clusters and donut plot visualizing the fractions of each cluster out of the total T cell population. (G) Stacked bars showing proportions of lean‐ and obese‐derived (top) and blood‐ and VAT‐derived (bottom) T cells within each cluster. (H) Selected PhenoGraph clusters displayed over UMAP embeddings showing obese‐, lean‐, VAT‐, and blood‐enriched clusters. (I) Heatmap displaying Z‐score transformed median fluorescence intensity (MFI) expression values for each of the proteins within the 27 clusters. Color scale is determined for each column separately, based on the lowest and highest Z‐score value for that protein. The heatmap was clustered using the ward.D cluster method. Lineage marker annotations are based on expression of CD4, CD8 or double negative (DN), CCR7, and CD45RA (J) Representative histograms showing modal expression intensities of indicated proteins in clusters that are obesity‐enriched (#26 and #20), shared (#18) or lean‐enriched (#14, #17).
People with obesity had higher frequencies of CD8+ T cells and lower frequencies of CD4+ T cells in both adipose tissue depots (only significant in SAT) compared with blood (Figure 2B). Interestingly, no significant differences were found between the two compartments among the lean subjects, indicating that the tissue‐specific differences in T cell subset distribution are more pronounced in the obese state. Moreover, the CD4+ and CD8+ TEM cells (CCR7‐, CD45RA‐; see Fig. S4 for details) were highest in patients with obesity, and higher in the adipose tissue compared with blood. This corroborates recent findings of increased memory T cell number in adipose tissue of obese mice [22] and may indicate a higher presence of T cell‐activating signals in adipose tissue in obesity leading to the generation of TRM. Such environmental cues may include local persisting antigens, inflammatory signals, and nutrients, such as free fatty acids [23]. Sex hormones may also regulate the differentiation and maintenance of TRM, as recently shown for VAT‐resident Tregs [24]. Comparing TRM in men and women would be interesting, but should be the scope of future studies, since in our cohort, patients with obesity consisted only of women.
Differences in protein expression on T cell subsets in lean and obese visceral adipose tissue
To explore how obesity affects the phenotype of T cell subsets, we investigated expression levels of the tissue‐specific proteins identified from the CD3+ T cell phenotyping described above in combination with other recognized T cell markers. Due to low cell recovery from several patient samples, the Treg phenotype is not discussed here but can be found in Fig. S5. As expected, the CD4+ and CD8+ T cells varied in their expression of phenotypic markers (Fig. 2C). The CD4+ T cells generally expressed higher levels of CD127 and CCR7, whereas the CD8+ T cells had higher expression of perforin and granzyme B, supporting their cytotoxic functions [4]. As expected, the tissue residency markers, CD69, CD49a, and CD103, were upregulated by both subsets in adipose tissue compared with blood, a finding evident in both patient groups. In particular, CD69 expression was highest among the TEM cells (Supplementary Fig. 4B and C), in line with this subset comprising of TRM cells [5]. VAT‐specific differences in T cell protein expression were also found between the people with obesity and lean subjects (Fig. 2C). In this depot, the CD4+ and CD8+ T cells from the obese group expressed higher levels of perforin, Eomes, CCR5, and CD69 compared with the lean group, although only significant for perforin on the CD4+ T cells and Eomes and CCR5 on the CD8+ T cells. Given the effector and regulatory functions of these proteins, obesity seems to be associated with more functionally active T cells in VAT. In contrast, the lean subjects displayed significantly elevated levels of CD38 and CD39 on the CD4+ T cells and CD39, CCR7, and CD127 on the CD8+ T cells (Fig. 2C). Interestingly, the metabolic protein, CD39, has been linked to both anti‐inflammatory regulatory functions and pro‐inflammatory memory functions in inflamed tissue [25] and was also identified as a marker of pro‐inflammatory Th17 cells in VAT [26]. Accordingly, whether the observed downregulation of this protein in obese VAT reflects a more pro‐inflammatory T cell phenotype requires further investigation. In summary, we found that obesity is associated with alterations in CD4+ and CD8+ T cell protein profile in VAT. The identified proteins may define phenotypically distinct subtypes of T cells that play a role in the inflammatory signaling linking obesity and metabolic dysfunction.
High‐dimensional analysis identifies obesity‐enriched T cell clusters with unique protein signatures
For a higher resolution of phenotypic diversity, we employed the non‐linear dimensionality reduction technique, Uniform Manifold Approximation Projection (UMAP), on the flow cytometry data to identify protein signatures on a single‐cell level, potentially revealing additional CD4+ and CD8+ T cell subpopulations associated with obesity. As expected, the expression intensities of CD4 and CD8 proteins defined two distinct clusters in the UMAP plot (Fig. 2D and E; Fig. S6). Both clusters contained smaller subclusters of cells deriving from obese VAT exclusively, confirming our previous assumptions that obese VAT contains phenotypically distinct T cell populations. Indeed, these VAT‐specific CD4+ and CD8+ T cell subclusters displayed increased expression levels of CD26, CD69, and CCR5, suggesting that the obesity‐induced activation of T cells at this site is only evident for specific subpopulations (Fig. 2E). In line with their role as leukocyte activation markers, CD26 and CD69 are previously found expressed on activated T cells in adipose tissue of individuals with obesity [10, 27]. CD26 induces T cell co‐stimulatory signals and activation by specifically interacting with its ligand, adenosine deaminase [16]. The chemokine receptor, CCR5, directs cells to the site of inflammation and has been found upregulated on T cells in obese VAT previously [28, 29]. Together, the upregulation of CD26, CD69, and CCR5 may reflect enhanced migration and interaction with target cells in the adipose tissue to promote T cell activation.
To further explore the phenotypic heterogeneity of the UMAP plot clusters, we performed PhenoGraph clustering and identified 27 clusters of T cells with different relative sizes of the total T cell population (Fig. 2F and Fig. S7). Each cluster also varied in the proportion of cells originating from lean subjects and individuals with obesity, and from blood and VAT (Fig. 2G). In line with the manual gating, the most extreme lean‐enriched (#12, #14, #15, #17) and obesity‐enriched (#20, #21 #23, #25, #26) clusters contained almost exclusively VAT‐derived T cells (Fig. 2G and H). When summarizing normalized protein expression levels between the identified clusters along with annotations of lineage markers, CD4+ and CD8+ T cells defined each set of the lean‐ and obesity‐enriched VAT clusters, all displaying memory phenotypes (Fig. 2I). Combined with relative high levels of CD49a and CD69, these clusters appeared to comprise VAT TRM cells (Fig. 2I). While CCR5 was expressed in all five obesity‐enriched clusters, Eomes levels were highest in CD8+ cluster #20, and CD26 levels were highest in CD4+ clusters #25 and #26 (Figure 2I, J). Thus, despite sharing phenotypic features, the unique protein signatures within the individual clusters point to the existence of additional subpopulations of CD4+ and CD8+ TRM cells in obese VAT. Further sub‐gating of CD4+ Th cells was not possible in our data and future studies should investigate how these proteins are expressed on various Th subsets, such as Th1, Th2, and Th17 cells.
The elevated expression levels of the T cell activation protein, CD26, in the two most obesity‐enriched CD4+ T cell clusters indicated that these cells might display enhanced effector functions. In line with this notion, we measured stress‐induced responses of adipose tissue T cells from a separate cohort of individuals with obesity (Table 1, cohort 4) and observed that CD4+CD26+ T cells in adipose tissue displayed a trend toward increased pro‐inflammatory IFN‐γ and TNF production compared with the CD4+CD26− subset (Fig. S8). Protein expression also varied between the two subsets, with the entire population of CD4+CD26+ T cells expressing the co‐stimulatory marker, CD28, confirming their activation status [30] and lacking granzyme B expression, indicating poor cytotoxicity (Fig. S9). This is in line with Th cells representing the cytokine‐producing cells with low capacity to kill, and our data indicate that these signature functions are mainly confined to the CD26+ Th cells. However, validation in larger cohorts is required in addition to assessing how the subset markers relate to metabolic dysfunction.
Taken together, we identified protein signatures of obesity‐associated CD4+ and CD8+ TRM cells in adipose tissue. While some of these proteins have been found expressed on human adipose tissue T cells previously, the high dimensional analysis allowed us to measure co‐expression patterns, highlighting the advantages of integrating 29‐parameters flow cytometry with dimensionality reduction techniques. Recent advances in single cell RNA sequencing have led to the identification of T cell subsets in adipose tissue of mice [31] and humans [32] defined by distinct cluster marker genes. However, we are not aware of other studies performing deep‐phenotyping of human adipose tissue T cell to the same extent as shown by us. It should be noted that the high dimensional clustering analysis included only VAT samples from the individuals with obesity to allow direct comparison with the lean counterparts, and whether the identified clusters also exist in obese and lean SAT remains to be determined.
Concluding remarks
Targeting T cells to regulate adipose tissue inflammation may represent a potential therapeutic strategy for treating obesity‐related complications. However, such strategy requires detailed characterization of T cells residing in adipose tissue, which is largely missing in the studies that addresses these issues, with only few phenotypic markers available to describe human adipose tissue T cell subsets. Future pharmacological strategies to reduce inflammatory signaling must also take into account that some pro‐inflammatory signaling in the adipose tissue is in fact beneficial as they are necessary for the regulation of energy homeostasis [33]. This warrants a detailed understanding of phenotypic heterogeneity of T cells residing in adipose tissue and how they contribute to these essential processes. By employing high‐resolution flow cytometry and transcriptomic analysis, we here characterized T cells in blood and adipose tissue from several independent cohorts and identified obesity‐associated CD4+ and CD8+ T cell subtypes in VAT defined by distinct protein expression patterns. Given the pathological role of VAT in obesity, our findings provide framework for future studies to investigate their functional relevance in obesity‐related metabolic disturbances.
Material and methods
Characteristics of patients and controls
Blood and adipose tissue samples were obtained from different cohorts of individuals with obesity and from lean subjects (cohorts and available clinical and biochemical data are presented in Fig. 1 and Table 1). Written consents were obtained from all patients, and the study was approved by the Regional Committees for Medical and Health Research Ethics (REK 2015/2343 and 2010/502) in Norway and by the local ethics committee in Stockholm, Sweden (2013/2285‐31/3, 2017/589‐32, 2014/979‐31/1 and 2015/1688‐32/1).
Sample preparation and isolation of immune cells
Adipose tissue biopsies were collected in Krebs Ringer Phosphate (KRP) buffer, followed by enzymatic digestion to isolate the stromal vascular cells (SVC) as previously described [34]. SVC was then freshly stained (proteome screening) or cryopreserved in freezing media until flow cytometry experiments were performed. Peripheral blood mononuclear cells (PBMC) were isolated from heparin blood using density gradient medium as previously described [34].
Surface proteome screening
LegendScreenTM Human PE Kit (Biolegend) was applied on freshly isolated SVC from liposuction and PBMC from donors to screen the T cells for 315 surface proteins. To separate the cells by the tissue of origin, blood‐ and adipose tissue immune cells were CD45 barcoded before subjected to the kit plates. The samples were acquired on the BD Fortessa instrument and the obtained data was analyzed using FlowJo v10.
Flow cytometry
Frozen PBMC and SVC were stained with extra‐and intracellular antibodies for 29‐color flow cytometry and acquired on a BD LSR Symphony containing 5 lasers (355, 405, 488, 561, and 637 nm). Data were analyzed in FlowJo V10 with the Downsample, Uniform Manifold Approximation and Projection (UMAP), and PhenoGraph plugins.
RNA sequencing
Five hundred picogram total RNA was used as input to prepare cDNA using SMART‐Seq v4 Ultra Low Input RNA Kit for Sequencing (Takara Bio. Cat. No. 634898). The cDNA quality was examined on Agilent TapeStation system using a High Sensitivity D5000 ScreenTape (Agilent, Cat. No. 5067–5592). One nanogram cDNA was used for library preparation using Nextera XT DNA Library Preparation Kit (Illumina, Cat. Nos. FC‐131‐1024 & FC‐131‐1096). The yield and quality of the amplified libraries were analyzed using Qubit by Thermo Fisher and the Agilent Tapestation. The indexed cDNA libraries were normalized and combined, and the pools were sequenced on the Illumina Novaseq S4 with 300‐cycles paired end sequencing.
Statistics
Extracted FlowJo data were analyzed using Prism version 9.2.0 (GraphPad). D'Agostino & Pearson omnibus normality test was used to determine normality of the data. The Wilcoxon matched‐pairs test was used for comparison between paired samples and the Mann–Whitney U‐test was used for comparison between different patient groups. A p‐value < 0.05 was considered statistically significant.
More details on the methods are provided in Supporting Information.
Conflict of interest
The authors declare no commercial or financial conflict of interest.
Ethics approval statement for human studies
The studies involve human participants and have been approved by the Regional Committees for Medical and Health Research Ethics (REK) in Norway and by the local ethics committee in Stockholm, Sweden.
Patient consent statement
The patients/participants provided their written informed consent to participate in this study.
Author contributions
M.E.H., M.C., G.M., J.F., and N.K.B. were associated with conceptualization and methodology. M.E.H., M.C., J.F., and N.K.B. performed analysis and drafted the manuscript. M.E.H., M.C., K.S., N.S., D.S., L.L.A., and A.P. contributed to experimental data. I.D.H. and E.S. contributed to clinical data. C.B., H.N., J.H., and J.K.H. acquired patient sample. All authors revised the final manuscript.
Peer review
The peer review history for this article is available at https://publons.com/publon/10.1002/eji.202249990
Abbreviations
- DEG
differentially Expressed Gene
- EM
effector memory
- PBMC
peripheral blood mononuclear cell
- SAT
subcutaneous adipose tissue
- Treg
regulatory T cell
- UMAP
uniform manifold approximation and projection
- VAT
visceral adipose tissue
Supporting information
Supporting Information
Acknowledgements
The flow cytometry was performed at the Flow Cytometry Core Facility, Department of Clinical Science, University of Bergen and at the Center for Infectious Medicine, Department of Medicine Huddinge, Karolinska Institutet, Karolinska University Hospital. We also would like to thank the core facility at Novum, BEA, Bioinformatics and Expression Analysis, which is supported by the board of research at the Karolinska Institute and the research committee at the Karolinska hospital. The computations and data handling were enabled by resources provided by the Swedish National Infrastructure for Computing (SNIC) at UPPMAX partially funded by the Swedish Research Council through grant agreement no. 2018–05973. This work was funded by the Western Norway Regional Health Authority (Helse Vest RHF), Swedish Research Council, the Swedish Cancer Society, the Swedish Foundation for Strategic Research, Knut and Alice Wallenberg Foundation, the Novo Nordisk Foundation, the Center for Innovative Medicine at Karolinska Institutet, the Stockholm County Council, and Karolinska Institutet.
Data availability statement
The original contributions presented in the study are included in the article and Supporting Information. Further inquiries can be directed to the corresponding author.
References
- 1. Nishimura, S. , Manabe, I. , Nagasaki, M. , Eto, K. , Yamashita, H. , Ohsugi, M. , Otsu, M. et al., CD8+ effector T cells contribute to macrophage recruitment and adipose tissue inflammation in obesity. Nat. Med. 2009. 15: 914–920. [DOI] [PubMed] [Google Scholar]
- 2. Kintscher, U. , Hartge, M. , Hess, K. , Foryst‐Ludwig, A. , Clemenz, M. , Wabitsch, M. , Fischer‐Posovszky, P. et al., T‐lymphocyte infiltration in visceral adipose tissue: A primary event in adipose tissue inflammation and the development of obesity‐mediated insulin resistance. Arterioscler. Thromb. Vasc. Biol. 2008. 28: 1304–1310. [DOI] [PubMed] [Google Scholar]
- 3. Zhu, X. and Zhu, J. , CD4 T Helper Cell Subsets and Related Human Immunological Disorders. Int. J. Mol. Sci. 2020. 21: 8011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhang, N. and Bevan, M. J. , CD8+ T Cells: Foot Soldiers of the Immune System. Immunity. 2011. 35: 161–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mueller, S. N. and Mackay, L. K. , Tissue‐resident memory T cells: local specialists in immune defence. Nat. Rev. Immunol. 2015. 16: 79–89. [DOI] [PubMed] [Google Scholar]
- 6. Feuerer, M. , Herrero, L. , Cipolletta, D. , Naaz, A. , Wong, J. , Nayer, A. , Lee, J. et al., Lean, but not obese, fat is enriched for a unique population of regulatory T cells that affect metabolic parameters. 2009. [DOI] [PMC free article] [PubMed]
- 7. Duffaut, C. , Zakaroff‐Girard, A. , Bourlier, V. , Decaunes, P. , Maumus, M. , Chiotasso, P. , Sengenès, C. et al., Interplay between human adipocytes and T lymphocytes in obesity: CCL20 as an adipochemokine and T lymphocytes as lipogenic modulators. Arterioscler. Thromb. Vasc. Biol. 2009. 29: 1608–1614. [DOI] [PubMed] [Google Scholar]
- 8. Fabbrini, E. , Cella, M. , McCartney, S. A. , Fuchs, A. , Abumrad, N. A. , Pietka, T. A. , Chen, Z. et al., Association between specific adipose tissue CD4+ T‐cell populations and insulin resistance in obese individuals. Gastroenterology. 2013. 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Verboven, K. , Wouters, K. , Gaens, K. , Hansen, D. , Bijnen, M. , Wetzels, S. , Stehouwer, C. D. et al., Abdominal subcutaneous and visceral adipocyte size, lipolysis and inflammation relate to insulin resistance in male obese humans. Sci Reports 2018. 8: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Travers, R. L. , Motta, A. C. , Betts, J. A. , Bouloumié, A. and Thompson, D. , The impact of adiposity on adipose tissue‐resident lymphocyte activation in humans. Int. J. Obes. 2015. 39: 762–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fathy, S. M. and Morshed, G. , Clinical research Peripheral blood lymphocyte subsets (CD4+, CD8+ T cells), leptin level and weight loss after laparoscopic greater curvature plication in morbidly obese patients. Arch. Med. Sci. 2014. 10: 886–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xia, M. , Hu, S. , Fu, Y. , Jin, W. , Yi, Q. , Matsui, Y. , Yang, J. et al., CCR10 regulates balanced maintenance and function of resident regulatory and effector T cells to promote immune homeostasis in the skin. J. Allergy Clin. Immunol. 2014. 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Weisberg, S. P. , Carpenter, D. J. , Chait, M. , Dogra, P. , Gartrell‐Corrado, R. D. , Chen, A. X. , Campbell, S. et al., Tissue‐Resident Memory T Cells Mediate Immune Homeostasis in the Human Pancreas through the PD‐1/PD‐L1 Pathway. Cell Rep. 2019. 29: 3916–3932.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schwartz, C. , Schmidt, V. , Deinzer, A. , Hawerkamp, H. C. , Hams, E. , Bayerlein, J. , Röger, O. et al., Innate PD‐L1 limits T cell‐mediated adipose tissue inflammation and ameliorates diet‐induced obesity. Sci. Transl. Med. 2022. 14. [DOI] [PubMed] [Google Scholar]
- 15. Woodward Davis, A. S. , Roozen, H. N. , Dufort, M. J. , DeBerg, H. A. , Delaney, M. A. , Mair, F. , Erickson, J. R. et al., The human tissue‐resident CCR5+ T cell compartment maintains protective and functional properties during inflammation. Sci. Transl. Med. 2019. 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ohnuma, K. , Dang, N. H. and Morimoto, C. , Revisiting an old acquaintance: CD26 and its molecular mechanisms in T cell function. Trends Immunol. 2008. 29: 295–301. [DOI] [PubMed] [Google Scholar]
- 17. Karlsson, E. A. , Sheridan, P. A. and Beck, M. A. , Diet‐induced obesity in mice reduces the maintenance of influenza‐specific CD8+ memory T cells. J. Nutr. 2010;140: 1691–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guerrero‐Ros, I. , Clement, C. C. , Reynolds, C. A. , Patel, B. , Santambrogio, L. , Cuervo, A. M. and Macian, F. , The negative effect of lipid challenge on autophagy inhibits T cell responses. Autophagy. 2020 Feb 1;16: 223–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xu, E. , Pereira, M. M. A. , Karakasilioti, I. , Theurich, S. , Al‐Maarri, M. , Rappl, G. , Waisman, A. et al., Temporal and tissue‐specific requirements for T‐lymphocyte IL‐6 signalling in obesity‐associated inflammation and insulin resistance. Nat. Commun. 2017. 8: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kidani, Y. and Bensinger, S. J. , Liver X receptor and peroxisome proliferator‐activated receptor as integrators of lipid homeostasis and immunity. Immunol. Rev. 2012. 249: 72–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kalathookunnel Antony, A. , Lian, Z. and Wu, H. T Cells in Adipose Tissue in Aging. Front. Immunol. 2018. 9: 2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Misumi, I. , Starmer, J. , Uchimura, T. , Beck, M. A. , Magnuson, T.0 and Whitmire, J. K. , Obesity Expands a Distinct Population of T Cells in Adipose Tissue and Increases Vulnerability to Infection. Cell Rep. 2019. 27: 514–524.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Steinbach, K. , Vincenti, I. and Merkler, D. , Resident‐Memory T Cells in Tissue‐Restricted Immune Responses: For Better or Worse? Front. Immunol. 2018. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vasanthakumar, A. , Chisanga, D. , Blume, J. , Gloury, R. , Britt, K. , Henstridge, D. C. , Zhan, Y. et al., Sex‐specific adipose tissue imprinting of regulatory T cells. Nat 2020 5797800. 2020. 579: 581–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Moncrieffe, H. , Nistala, K. , Kamhieh, Y. , Evans, J. , Eddaoudi, A. , Eaton, S. and Wedderburn, L. R. , High Expression of the Ectonucleotidase CD39 on T Cells from the Inflamed Site Identifies Two Distinct Populations, One Regulatory and One Memory T Cell Population. J. Immunol. 2010. 185: 134–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pandolfi, J. B. , Ferraro, A. A. , Sananez, I. , Gancedo, M. C. , Baz, P. , Billordo, L. A. , Fainboim, L. et al., ATP‐Induced Inflammation Drives Tissue‐Resident Th17 Cells in Metabolically Unhealthy Obesity. J. Immunol. 2016. 196: 3287–3296. [DOI] [PubMed] [Google Scholar]
- 27. Zhong, J. , Rao, X. , Deiuliis, J. , Braunstein, Z. , Narula, V. , Hazey, J. , Mikami, D. et al., A Potential Role for Dendritic Cell/Macrophage‐Expressing DPP4 in Obesity‐Induced Visceral Inflammation. Diabetes. 2013. 62: 149–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huber, J. , Kiefer, F. W. , Zeyda, M. , Ludvik, B. , Silberhumer, G. R. , Prager, G. , Zlabinger, G. J. et al., CC Chemokine and CC Chemokine Receptor Profiles in Visceral and Subcutaneous Adipose Tissue Are Altered in Human Obesity. J. Clin. Endocrinol. Metab. 2008. 93: 3215–3221. [DOI] [PubMed] [Google Scholar]
- 29. Wu, H. , Ghosh, S. , Perrard, X. D. , Feng, L. , Garcia, G. E. , Perrard, J. L. , Sweeney, J. F. et al., T‐cell accumulation and regulated on activation, normal T cell expressed and secreted upregulation in adipose tissue in obesity. Circulation. 2007. 115: 1029–1038. [DOI] [PubMed] [Google Scholar]
- 30. Acuto, O. and Michel, F. , CD28‐mediated co‐stimulation: a quantitative support for TCR signalling. Nat. Rev. Immunol. 2003. 3: 939–951. [DOI] [PubMed] [Google Scholar]
- 31. Emont, M. P. , Jacobs, C. , Essene, A. L. , Pant, D. , Tenen, D. , Colleluori, G. , Di Vincenzo, A. et al., A single‐cell atlas of human and mouse white adipose tissue. Nat 2022 6037903. 2022. 603: 926–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hildreth, A. D. , Ma, F. , Wong, Y. Y. , Sun, R. , Pellegrini, M. and O'Sullivan, T. E. , Single‐cell sequencing of human white adipose tissue identifies new cell states in health and obesity. Nat Immunol. 2021. 22: 639–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wernstedt Asterholm, I. , Tao, C. , Morley, T. S. , Wang, Q. A. , Delgado‐Lopez, F. , Wang, Z V. and Scherer, P. E. , Adipocyte inflammation is essential for healthy adipose tissue expansion and remodeling. Cell Metab. 2014. 20: 103–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Strand, K. , Stiglund, N. , Haugstøyl, M. E. , Kamyab, Z. , Langhelle, V. , Lawrence‐Archer, L. , Busch, C. et al., Subtype‐Specific Surface Proteins on Adipose Tissue Macrophages and Their Association to Obesity‐Induced Insulin Resistance. Front Endocrinol (Lausanne). 2022. 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Data Availability Statement
The original contributions presented in the study are included in the article and Supporting Information. Further inquiries can be directed to the corresponding author.


