Abstract
The aim of this review was to map the literature assessing associations between maternal or infant immune or gut microbiome biomarkers and child neurodevelopmental outcomes within the first 5 years of life. We conducted a PRISMA‐ScR compliant review of peer‐reviewed, English‐language journal articles. Studies reporting gut microbiome or immune system biomarkers and child neurodevelopmental outcomes prior to 5 years were eligible. Sixty‐nine of 23,495 retrieved studies were included. Of these, 18 reported on the maternal immune system, 40 on the infant immune system, and 13 on the infant gut microbiome. No studies examined the maternal microbiome, and only one study examined biomarkers from both the immune system and the gut microbiome. Additionally, only one study included both maternal and infant biomarkers. Neurodevelopmental outcomes were assessed from 6 days to 5 years. Associations between biomarkers and neurodevelopmental outcomes were largely nonsignificant and small in effect size. While the immune system and gut microbiome are thought to have interactive impacts on the developing brain, there remains a paucity of published studies that report biomarkers from both systems and associations with child development outcomes. Heterogeneity of research designs and methodologies may also contribute to inconsistent findings. Future studies should integrate data across biological systems to generate novel insights into the biological underpinnings of early development.
Keywords: early child development, gut microbiome, immunology, infancy, neurodevelopment, proteomics
1. BACKGROUND
Accumulating evidence spanning the fields of biology, epigenetics, genetics, and neuroscience has identified the critical role of various biological systems in shaping the developing brain (Konkel, 2018, Inguaggiato et al., 2017, Nazzari & Frigerio, 2020, Sordillo et al., 2019). The early developmental period (herein defined as conception through to 5 years following birth) is a critical window where rapid and significant developments to the brain and other biological systems (e.g., the immune system and the gut microbiome) coincide (West, 2002, Borre et al., 2014, Codagnone et al., 2019, Cowan et al., 2020). Studies have described reciprocal and predictive relationships between various biological systems and the developing brain, offering exciting clinical insights into the origins of health and neurodevelopment. Such insights provide novel opportunities for early action that can optimize and protect the developing brain, potentially preventing adverse neurodevelopmental outcomes (Chorna et al., 2020, Nahmias et al., 2019, Puthussery et al., 2018). The characterization of these biological systems has been supported by rapid improvements in the quality and affordability of biological assessments, meaning that highly detailed data are increasingly accessible. This has resulted in the recent proliferation of studies that examine the association between a range of different biomarkers and early neurodevelopmental outcomes (Barbosa et al., 2020, Carlson et al., 2018, Kitano, 2002, Rudolph et al., 2018).
Studies examining the biological underpinnings of early neurodevelopment may use hypothesis‐driven (Rudolph et al., 2018) (including translation of animal models to humans) or exploratory approaches (Bodnar et al., 2018). Historically, these studies have focused on a single biological system—sometimes termed as a single “omics layer” (e.g., the gut microbiome) (Yan et al., 2018). Single‐omics approaches mean that complex interactions between biological systems that likely influence neurodevelopment are rarely examined, though the existence of these complex, “multiomics” interactions is often acknowledged. This knowledge forms the foundation of systems biology approaches to understanding human development (Veenstra, 2021). It is unlikely that the influence of unique biomarkers on the developing brain can be understood until multiomics approaches are integrated into research efforts (Yan et al., 2018). In moving toward a multiomics approach to early neurodevelopment, a sensible place to begin is to consider interactions between biological systems previously identified as important for neurodevelopment, namely, the immune system and the gut microbiome.
Fetal immune programming is initiated in utero, with evidence of responses to a variety of potentially pathogenic and nonpathogenic environmental antigens from the second trimester. There is increasing evidence that the developing fetal brain is influenced by metabolic and immune factors, including maternal immune factors (Estes & McAllister, 2016, Jones et al., 1996). The risk of ill‐health increases when maternal or fetal immune system functioning is compromised, including disruptions to typical neurodevelopmental processes and outcomes (Bilbo & Schwarz, 2009). Maternal, fetal, and infant immune dysregulation in the pre‐ and postnatal periods are associated with pervasive and sustained neurodevelopmental outcomes including neurodevelopmental disorders (e.g., attention deficit hyperactivity disorder, autism), although casual pathways are unclear (Meyer et al., 2011, Instanes et al., 2017, Beversdorf et al., 2018). One proposed mechanistic pathway involves cytokine imbalances, which are thought to disrupt important neurodevelopmental processes (e.g., synaptic pruning and synapse formation) (Nazzari & Frigerio, 2020, Jiang et al., 2018, Ganguli & Chavali, 2021).
The gut microbiome is a complex ecosystem home to trillions of microbes characterized by hundreds of unique species (Tognini, 2017). Connectivity of the brain and the gut microbiome via the microbiota–gut–brain (MGB) axis is well‐established (Borre et al., 2014, Lima‐Ojeda et al., 2017, Warner, 2019, Perez‐Muñoz et al., 2017). This axis provides a channel for the gut microbiome to modulate neurodevelopment via signaling in neuronal, hormonal, and immunological pathways (Warner, 2019, Foster et al., 2017, Cryan & Dinan, 2012, Cryan & de Wit, 2019). Both the gut and the brain develop rapidly in the first years of life, with the gut undergoing swift changes in microbial composition and diversity, under the influence of nutritional and other environmental factors (Bäckhed et al., 2015). These changes play a major role in the postnatal development and maturation of the immune, endocrine, and neurobiological systems (Borre et al., 2014). During this critical period, a healthy gut microbiome helps to support typical neurodevelopment (Foster et al., 2017). Inversely, gut dysbiosis in early life can directly or indirectly compound neurodevelopmental risk (Borre et al., 2014, Lima‐Ojeda et al., 2017). An emerging number of studies have identified an overrepresentation of certain gut microbiota among people with neurodevelopmental and mental health problems, such as autism, anxiety, and schizophrenia (Ho et al., 2020, Iglesias‐Vázquez et al., 2020, Xu et al., 2019, Wang et al., 2011, Malan‐Muller et al., 2018, Szeligowski et al., 2020, Nguyen et al., 2019). Among nonclinical populations, gut microbiome composition is also associated with poorer developmental outcomes in early childhood (Loughman et al., 2020, Jurek et al., 2021, Lacorte et al., 2019). Accumulating evidence suggests that dysbiosis of the gut microbiome may precede the behavioral manifestations of neurodevelopmental impairments (Diaz Heijtz, 2016, Aatsinki et al., 2019, Kelsey et al., 2021, Laue et al., 2020). Although in reverse, neurodevelopmental difficulties may increase the likelihood of gut dysbiosis due to changes in diet (Mayer et al., 2014). It remains unclear how reliable the early microbiome is as an observable indicator for later neurodevelopmental risk. Accordingly, identifying the extent to which gut microbiome composition and function shape neurodevelopment during early life remains a research priority.
Bidirectional relationships between the gut microbiome and immune system are also well‐established in both animal and human models (Tamburini et al., 2016, Hooper et al., 2012, Cerdó et al., 2019, Al Nabhani et al., 2019). For example, studies have demonstrated that immune system dysregulation has consequences for the gut microbiome (Cerdó et al., 2019), and that dysbiosis of the gut microbiome can lead to increased immune system activity (Rothhammer et al., 2016). This highlights the potential indirect effects of the immune system on early neurodevelopment via alterations to the gut microbiome, and vice versa. However, it is not yet clear how specific biomarkers contained within either system interact to jointly shape neurodevelopment. Further, it is also unclear whether the growing recognition for multivariate or “systems‐based” approaches to neurodevelopment has been used to guide empirical examination of the complex interrelationships between biomarkers, biological systems, and neurodevelopment—or whether reductionist approaches (i.e., evaluating how one specific biomarker is predictive of neurodevelopment) continue to represent the rationale for most studies in this field.
1.1. The present review
While there has been a proliferation of studies on the role of the maternal and infant immune system and the gut microbiome in neurodevelopment, synthesis of these findings is needed to provide insight into the state of the evidence. The aim of this systematic scoping review is to map existing literature pertaining to how biomarkers within the human gut microbiome or immune system are associated with neurodevelopmental and related behavioral outcomes within the first 5 years of life. Considering that functioning of the microbiome is also influenced by the immune system (and vice versa) (Cryan & Dinan, 2012, Brestoff & Artis, 2013, van den Berg et al., 2016), we aimed to determine the extent to which these biological systems have been studied together in the context of early neurodevelopment.
Further, we looked beyond dichotomous clinical outcomes (e.g., the presence or absence of disease), and instead examine dimensional indicators of neurodevelopment or behavior. This approach is necessary for the early neurodevelopmental period, as many neurodevelopmental conditions are not recognized until later childhood (Finlay‐Jones et al., 2019). Furthermore, we aimed to provide insight into how these biomarkers are associated with behavioral outcomes in the general population. Providing this comprehensive and up‐to‐date information may help to direct research efforts by identifying promising candidate biomarkers thought to shape the developing brain.
2. MATERIALS AND METHODS
The Joanna Briggs Institute (JBI) framework for conducting scoping reviews was adopted for this study. The results of this scoping review were reported in accordance with the Preferred Reporting Items for Systematic Reviews and Meta‐Analyses extension for Scoping Reviews (PRISMA‐ScR).
2.1. Protocol registration
The protocol for this scoping review was first registered on the Open Science Framework (OSF) (Mancini & Finlay‐Jones, 2021) in September 2020 and last updated April 2021.
2.2. Eligibility criteria
Eligibility for inclusion in this review was based on the population, concept, context, and type of evidence elements reported below.
2.2.1. Population
We included articles that examined human gut microbiome or immune system biomarkers during the first 5 years of life. Biomarkers could be sampled from the child or from their mother during pregnancy. Assessments of neurodevelopment or behavior within the first 5 years of life were also necessary to warrant inclusion in this review. Studies that did not include clearly defined neurodevelopmental or related behavioral assessment within the first 5 years of life were not included.
2.2.2. Concept
We selected articles analyzing each of the following concepts: early human gut microbiome or immune system biomarkers, and neurodevelopment or related behavior within the first 5 years of life. Studies that only examined other biological systems (e.g., the genome or metabolome) or only described neurodevelopment as case–control outcomes were excluded. Studies that stratified subjects into different categories (e.g., “at‐risk” or “not at‐risk”) remained eligible for inclusion provided that dimensional assessments of behavior were reported. Studies that grouped poor neurodevelopmental outcomes with other adverse outcomes (e.g., physical outcomes or mortality) were not eligible for inclusion.
2.2.3. Context
No clinical, cultural, or geographic limits were applied in this review. We also did not limit eligible studies to a particular health context (e.g., preterm birth, or high risk for developmental conditions).
2.2.4. Sources of evidence
Peer‐reviewed studies published (or in preprint) in the English language between January 2010 to April 2021 were eligible for inclusion. Only primary research studies were included to ensure relevance to the research question. Only studies published from 2010 onward were included as our aim was to provide an up‐to‐date review of the extant literature. Meta‐analyses and reviews were used to search for additional studies. Alternative forms of literature (e.g., book chapters, opinion papers, conference proceedings, research protocols, and editorials) were not included.
2.3. Search strategy
Search terms were developed and refined through an iterative process involving experts in human biology, early neurodevelopment, and behavior. Preliminary search terms were piloted and refined to ensure that the final search terms were comprehensive. The final list of search terms (see File S1 for example) was finalized in April 2021. Search terms were adjusted to suit the requirements of each database, which included Embase, Medline, PsycInfo, and Web of Science (see Supporting Information for search syntax). Search results were limited to journal articles published in the English language.
2.4. Study screening and selection
Results were exported into EndNote X9 and deduplicated. One author screened each of the titles and abstracts using the Rayyan (Ouzzani et al., 2016) website interface. A second author screened approximately 30% of titles and abstracts using a machine learning‐enhanced strategy through ASReview (van de Schoot et al., 2021) software. The second reviewer used the program to engage in a conventional title and abstract classification process that is simultaneously used to train a model that actively sorts each remaining study based on relevance. The “next‐most‐relevant” study title and abstract identified by the model are then presented to the reviewer for classification. Each study identified for full‐text extraction was obtained and independently reviewed by two reviewers. This process helps to enhance the accuracy of limited dual review approaches—whereby at least one reviewer will screen a subset of studies (Stoll et al., 2019). Any divergence on whether a study should be included in the review was resolved via discussion between the review team.
2.5. Data charting and presentation of results
A data charting form was jointly developed by several of the study authors to document the relevant details to extract from each included study. This data charting form was updated where appropriate throughout the review process to ensure that relevant information was captured. We extracted data on first author, country, study design, population (including maternal and child characteristics), proportion of children that were male, biomarker type, source, and assessment method (including reference database used for taxonomic profiling in each microbiome study), and neurodevelopmental outcome domain, measure, and age at assessment. The data from each study that was full‐text screened were independently charted by two authors, and then compared. Any divergence on the information included on each study data charting form was resolved through discussion. Due to the heterogeneity across studies, data were summarized narratively.
3. RESULTS
3.1. Literature search
The flow diagram of study selection is presented in Figure 1. The search identified 32,939 articles. Following de‐duplication, 23,495 articles underwent title and abstract screening, which identified 149 articles that met the inclusion criteria based on title or abstract. These 149 studies were captured within the subset of studies screened by the second reviewer (30% of studies deemed as “most relevant” by the software), supporting the efficacy of the machine learning‐enhanced limited dual review approach to article screening (Stoll et al., 2019). The full texts of these articles were independently reviewed by two study authors for eligibility. Eighty studies did not meet inclusion criteria and were rejected with reasons described in Figure 1. In total, 69 studies met inclusion criteria and were retained for this review.
FIGURE 1.
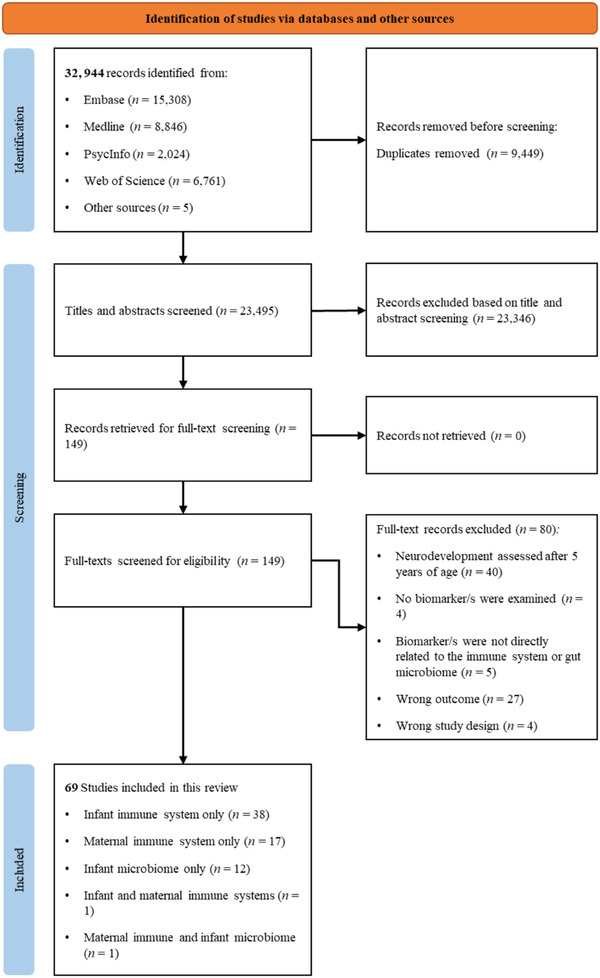
PRISMA flow diagram outlining study selection process
3.2. Population characteristics
Included studies were conducted across 25 countries. Most studies were conducted in the United States (n = 34), followed by Bangladesh (n = 4) and Brazil, China, Ireland, and Ukraine (n = 3 each). In addition to geographical region, study populations differed according to socioeconomic conditions, health status of the mother, and health status of the infant. Most studies recruited community samples, often as part of larger birth cohort studies, or recruiting typically developing samples (n = 22). A large portion of studies (n = 20) recruited samples with infants who were born preterm and/or very low birthweight, or from mothers at increased risk for preterm birth. Several studies (n = 12) included samples that were enriched for different health outcomes, with most of these studies (10 of the 12) comparing these infants/mothers to a control or risk‐free group. Several studies (n = 8) recruited infants based on exposure to a medical event or condition (e.g., infant encephalopathy, alcohol exposure in utero), or from impoverished or rural areas (n = 7). Sample sizes ranged from 26 to 3038 participants. Study details are summarized in Tables 1, 2, 3. We also provide a visual representation of the most common immune infant gut microbiome biomarkers examined for each of the included studies in Tables 4, 5, 6. The aggregated results of associations between the maternal and infant immune system, and infant gut microbiome are reported in Tables 7, 8, 9.
TABLE 1.
Included studies that examine the association between maternal immune system biomarkers and neurodevelopmental outcomes (N = 18)
| Author (year), country | Population context | Covariates | Study design | Final N (% male) | Biomarker(s) | Biosample age | Source | Source analytic method | Outcome measure (domains) | Age at outcome |
|---|---|---|---|---|---|---|---|---|---|---|
| Nazzari et al. (2019), Italy | Healthy | Gestational age, infant sex, maternal prepregnancy BMI, length of heel stick | Cohort | 104 (51.9%) | IL‐6; CRP; sAA | Late pregnancy | Maternal blood and saliva | Quantikine High Sensitivity ELISA kits (R&D Systems Europe); Expanded Range High Sensitivity Cortisol EIA Kit (Salimetrics); α‐Amylase Kinetic Enzyme Assay Kit (Salimetrics) | Behavior observation (regulation; emotional reactivity) | 48–72 h |
| Osborne et al. (2018), UK | Maternal MDD and controls | Maternal prepregnancy BMI | Cohort | 87 (not specified) | IL‐1β; IL‐2; IL‐6; IL‐8; IL‐10; TNF‐α; VEGF; EGF; MCP‐1 | Third trimester | Maternal blood | Sandwich chemiluminescent immunoassay | NBAS (automatic stability; regulation; orientation; motor skills) | 6 days |
| Yan et al. (2020), China | Healthy | Gestational age, birth weight, maternal age, education, spouses’ education, family income, parity, maternal prepregnancy BMI, gestational weight gain, delivery mode, infant sex. | Cohort | 1186 (52.1%) | TNF‐α; IL‐1β; IL‐6; IL‐10; MIP‐1β | Second trimester (16–18 weeks) | Maternal blood | MILLIPLEX MAP Human Cytokine/ChemokineMagnetic Bead Panel 96 Well Plate Assay (Millipore) | CDSC (cognition; language; gross motor skills; fine motor skills; social) | 1 month |
| Nazzari et al. (2020), Italy | Healthy | Gestational age, maternal emotional availability, maternal IQ, infant sex, maternal postnatal depression | Cohort | 104 (51%) | IL‐6; CRP | Mean (SD) = 34.76 (1.12) weeks | Maternal blood | Quantikine ELISA kits (R&D Systems) | BSID‐III (cognition) | 11.96 (1.85) weeks |
| Hunter et al. (2021), USA | Healthy | Maternal education, maternal postpartum depression, anxiety, and stress | Cohort | 127 (51%) | CRP; IL‐6; IL‐8; TNF‐α | 16 weeks | Maternal blood | Beckman‐Coulter high sensitivity assay; R&D Systems high sensitivity assay | IBQ‐R (surgency; negativity; regulation) | 3 months |
| Gustafsson et al. (2019), USA | High ADHD risk | Maternal third trimester depression and stress | Cohort | 68 (60%) | IL‐6; TNF‐α; MCP‐1 | Third trimester | Maternal blood | Enzyme‐linked immunosorbent assay | IBQ‐R + Still face paradigm observation (temperament; regulation) | 6 months |
| Gustafsson et al. (2018), USA | High ADHD risk | Maternal ADHD symptoms, maternal postnatal depression symptoms, infant age | Cohort | 68 (57%) | IL‐6; TNF‐α; MCP‐1; IL‐10 | Third trimester | Maternal blood | Enzyme‐linked immunosorbent assay (R&D Systems); Luminex polystyrene bead‐based multiplex immunoassays (R&D Systems) | IBQ‐R (negative affect; regulation) | 6.8 months (0.8) |
| Bodnar et al. (2018), Ukraine | Alcohol exposure in utero | Maternal marital status, BMI, socioeconomic status, smoking, parity, child sex, and study site | Cohort | 152 (40.9%–68.6%) | 40 cytokines/chemokines (see study for full list) | Second trimester (17–19 weeks) Third trimester (31–34 weeks) | Maternal blood | Meso Scale Discovery V‐PLEX Human Biomarker 40‐Plex kit | BSID‐II (cognition; language; psychomotorv ) | 6 and 12 months |
| Sowell et al. (2018), Ukraine | Alcohol exposed and controls | Maternal age, paternal age, maternal education, SES, prepregnancy BMI, parity, gravidity, MVM use, maternal smoking (cigarettes per day), study site, gestational week at blood draw, child sex, gestational age at birth, birth weight, and birth length | Cohort | 241 (56%) | IL‐1β; TNF‐α; IFN‐γ; GM‐CSF; IL‐2; IL‐4; IL‐6; IL‐8; IL‐10 | Second trimester (19 and 32 weeks) | Maternal blood | Human multiplexing bead immunoassay | BSID‐III (cognition; psychomotor) | 6 and 12 months |
| Freedman et al. (2019), USA | Infection and controls | Infant sex, maternal age, obesity, and depression, maternal age, maternal smoking | Cross‐sectional | 162 (not specified) | CRP; choline | 16 weeks | Maternal blood | Beckman–Coulter high sensitivity assay | IBQ‐R (surgency; negative emotionality; regulation) | 12 months |
| Rasmussen et al. (2019), USA | Healthy | Household income, prepregnancy BMI, obstetric risk, smoking during pregnancy, and infant sex | Cohort | 147 (55.8%) | IL‐6 |
Mean (SD) = 12.6 (2.8) weeks 20.4 (1.5) weeks 30.3 (1.3) weeks |
Maternal blood | Enzyme‐linked immunosorbent assay (eBioscience) | BSID‐III + MRI (cognition; socioemotional development) | 12 months |
| Spann et al. (2018), USA | Adolescent mothers | Sex, postmenstrual age, and scanner upgrade | Cohort | 72 (66.7%) | IL‐6; CRP | Third trimester (34–37 weeks) | Maternal blood | Enzyme‐linked immunosorbent assay; Cobras Integra 400 Plus (Roche Diagnostics) turbidimetric | BSID‐III (cognition) | 14 months |
| Irwin et al. (2019), Seychelles | Healthy | Child sex, maternal age, child age at assessment, number of parents living with child, socioeconomic status | Cohort | 1408 (52.1%) | IL‐1β; IL‐2; IFN‐γ; TNF‐α; IL‐4; IL‐6; IL‐5; IL‐10; CRP; MCP‐1; TARC; VEGF‐D; sFLT‐1 | Third trimester (28 weeks) | Maternal blood | Meso Scale Discovery multiplex assay | BSID‐II (cognition; psychomotor skills); CDI (expressive and receptive language); IBQ‐R (temperament) | 15–25 months |
| Rudolph et al. (2018), USA | Healthy | Not reported | Cohort | 84 (50%) | IL‐6 |
Mean = 12.7 weeks 20.5 weeks 30.4 weeks |
Maternal blood | Enzyme‐linked immunosorbent assay | IBQ + neuroimaging (negative emotionality; working memory) | 24 months |
| Graham et al. (2018), USA | Healthy | Gestational age at birth, age at scan | Cohort | 86 (59.3%) | IL‐6 | First, second, and third trimesters | Maternal blood | Enzyme‐linked immunosorbent assay | Behavioral observation—snack delay task (impulsivity) + neuroimaging | 24 months |
| Sevenoaks et al. (2021), South Africa | Maternal HIV compared to controls | Clinic, maternal smoking during pregnancy, maternal alcohol use during pregnancy, maternal socioeconomic status, maternal BMI at 6 weeks postpartum, infant birth weight, prematurity, infant sex and exclusive breastfeeding (yes/no), maternal HIV disease parameters adjusted for maternal CD4+ during pregnancy, maternal viral load during pregnancy, maternal ART regimen during pregnancy and initiation of ART (before or during pregnancy). | Cohort | 267 (28.8%–39%) | IFN‐γ; IL‐1β; IL‐2; IL‐4; IL‐5; IL‐6; IL‐7; IL‐8; IL‐10; IL‐12p70; IL‐13; TNF‐α; GM‐CSF; NGAL; MMP‐9 | 26 weeks gestation | Maternal blood | Multiplex bead assay; Enzyme‐linked immunosorbent assay | BSID‐III (cognition; language; motor skills) | 24–28 months |
| Monthe‐Dreze et al. (2019), USA | Healthy | Child sex, age at outcome | Cohort | 1246 (51.8%) | CRP | Second trimester | Maternal blood | Enzyme‐linked immunosorbent assay | WRAVMA (intelligence; visual‐motor skills) | 3.2 years |
| Rommel et al. (2020), USA | Healthy | Nutritional supplementation | Cohort | 512 (47.7%) | PGF2‐α | 32.6 weeks | Maternal urine | Gas chromatography; Mass spectrometry | BASC, SRS (Externalizing problems; Internalizing problems; Behavioral Symptoms Index; social problems) | 4–5 years |
Note: Studies are grouped based on age of first neurodevelopmental assessment.
Abbreviations: ADHD, attention deficit hyperactivity disorder; BASC, Behavior Assessment System for Children; BMI, Body Mass Index; BSID‐II, Bayley Scales of Infant Development—Second Edition; BSID‐III, Bayley Scales of Infant Development—Third Edition; B CDI, Communicative Development Inventory; CDSC, Child Developmental Scale of China; HIV, human immunodeficiency virus; IBQ‐R, Infant Behavior Questionnaire—Revised; MDD, Major Depressive Disorder; MRI, magnetic resonance imaging; NBAS, Neonatal Behavioral Assessment Scale; SRS, Social Responsiveness Scale; TD, typically developing; WRAVMA, Wide Range Assessment of Visual Motor Abilities.
TABLE 2.
Included studies that examine the association between infant immune system biomarkers and neurodevelopmental outcomes (N = 40)
| Author (year), country | Population context | Covariates | Study design | Final N (% male) | Biomarker(s) | Source | Source analytic method | Biosample age | Outcome measure (domains) | Age at outcome |
|---|---|---|---|---|---|---|---|---|---|---|
| Ghassabian et al. (2018), USA | Typically developing (TD) | Gestational age, birth order, maternal age and level of education, maternal smoking during pregnancy, and maternal depressive symptoms | Cohort | 3038 (52%) | bFGF; IL‐8; IL‐1ra; IL‐1α; MCP‐1, MIP‐1α; MIP‐1β; VEGF; CCL21; CCL27; IL‐16; IL‐20; MCP‐2; MIP‐1d; CCL12; CCL‐17; sICAM‐1; sVCAM‐1; Cathepsin D; MPO; PDGF‐AA; PAI‐1; NCAM; CRP | Newborn dried blood spot | Milliplex Panels II and III (Millipore) | 2–3 days | ASQ; (cognitive, motor, communication, social) | 4–36 months |
| Nist, Pickler, et al. (2020), USA | Very preterm | PMA at birth, DOL of blood collection for cytokine analysis, and infant age at the time of neurobehavioral/neurodevelopmental assessment | Cohort | 62 (60%) | IL‐1RA; IL‐6; IL‐10; IL‐8; MCP‐1; GCSF; GMCSF | Infant blood | Bio‐Plex multiplex assay | 0–3 weeks postbirth | BSID‐III; (cognition, language, motor skills); NAPI (irritability, alertness) | 6 months |
| Liu & Feng (2010), China | HIE | Not reported | Cohort | 52 (not specified) | IL‐1β; IL‐8; TNF‐α | Child blood | Human multiplexing bead immunoassays | 0–1 week | DDST‐II (gross motor skills; fine motor skills; language, social skills) | 6 and 12 months |
| Varner et al. (2015), USA | CP or NDD | Gestational age at birth, maternal education level, and exposure to magnesium sulfate (MgSO4) | Case–control | 615 (not specified) | IL‐1β; IL‐8; TNF‐α | Umbilical cord blood | Enzyme‐linked immunosorbent assay | Birth | BSID‐II (cognition; motor skills) | 6, 12, and 24 months |
| Camargos et al. (2017), Brazil | Overweight, obese or normal‐weight infants | Age and gender of child | Cross‐sectional | 50 | sTNFR1; sTNFR2; MCP‐1; IL‐8; IP‐10; RANTES; MIG | Infant blood | Blood plasma, serum erythrocyte lysate | 6–24 months | BSID‐III (cognition; motor skills) | 6–24 months |
| Jensen et al. (2019), Bangladesh | Living in poverty | Gestational age | Cohort | 122 (not specified) | CRP | Infant blood | Enzyme‐linked immunosorbent assays (Immuniidiagnostik) | 4–12 months | MSEL (gross motor skills; fine motor skills); WPPSI (visual reception; receptive language; expressive language; intelligence) |
6, 27, and 36 months (MSEL) 60 months (WPPSI) |
| Xie et al. (2019), Bangladesh | Living in poverty, high risk for chronic systemic inflammation PROVIDE and CRYPTO cohort | Height‐for‐age Z‐scores | Cohort | 260 (not specified) | CRP | Infant blood | Not specified | 18–104 weeks | MSEL (cognition); WPPSI‐III (intelligence); Face‐oddball task (event‐related potential) |
6 and 36 months (ERP) 27 months (MSEL) 48 months (WPPSI‐III) |
| Magalhaes et al. (2017), Brazil | Poor motor development compared to control | Not reported. | Cohort | 40 (55%) | IL‐1β; IL‐6; IL‐8; IL‐10; IL‐12p70; TNF‐α; MCP‐1; RANTES; IP‐10; MIG | Infant urine, infant blood | Cytometric bead assay (BD Biosciences, California) Human Inflammatory Kit (CBA) Human Chemokine Kit (CBA) | 0–3 weeks | TIMP (posture motor coordination) | 48 h, 72 h, and 3 weeks after birth |
| Nist, Shoben, et al. (2020), USA | Very preterm | Length of stay in NICU | Cohort | 68 (66.2%) | IL‐1β; IL‐4; IL‐6; IL‐8; IL‐10; IL‐17A; TNF‐α; MCP‐1 | Infant blood | Bio‐plex multiplex assay | 0–35 weeks | NAPI (motor development and vigor alertness and orientation) | 35 weeks |
| Jiang et al. (2014), Bangladesh | Living in poverty | Sex, monthly family income, maternal education, febrile illness, and LAZ at 12 months | Cohort | 127 (58.3%) | IL‐1β; IL‐6; TNF‐α; IL‐4; IL‐10 | Infant blood | Human Bio‐Plex Pro Assays (Bio‐Plex 200 Platform) | 6 months | BSID‐III (cognition; language; motor skills) | 12 months |
| Benavides et al. (2022), USA | Very preterm, ELBW | Sex, birth weight, GA at birth, day of age, and pretransfusion hemoglobin within 24 h of assessment | Cohort | 71 (data on BSID‐II for 26) (39.4%) | IL‐1β; IL‐2; IL‐4; IL‐6; IL‐8; IL‐10; IL‐12; IL‐13; IL‐17; IL‐18; IFN‐γ; IP‐10; MCP‐1; IFN‐γ; TNF‐α; TNF‐β; sICAM‐1; sVCAM‐1; VEGF; TPO | Infant blood | V‐PLEX Plus Pro‐inflammatory Panel 1 Human Kit (Multiplex); V‐PLEX Plus Vascular Injury Panel 2 Human Kit (Multiplex); U‐PLEX Biomarker Group 1 (hu) Assays (Multiplex) | 22–29 weeks | BSID‐III (cognition; language; motor skills) | 12 months |
| Etheredge et al. (2018), Tanzania | Non‐stunted from low‐income country | Assessor, study baseline, length‐for‐age, weight‐for‐age, exclusive breastfeeding at 6 weeks and 6 months, any respiratory infection or diagnosis, anemia, iron deficiency, and inflammation | RCT | 107 (49.5%) | CRP | Infant blood | Immunoturbidimetric assay (Roche Diagnostics) | 6 weeks, 6 months, 12 months | BSID‐III (cognition; language; motor skills) | 15 months |
| Chalak et al. (2014), USA | HIE | Not reported | Cohort | 27 (57%−70%) | Il‐1; Il‐6; IL‐8; VEGF; TNF‐α; INF‐γ | Umbilical blood | Enzyme‐linked immunosorbent assay | 6–24, 48, 72, and 78 h | BSID‐III (cognitive; language motor) | 15–18 months |
| Dietrick et al. (2020), Ireland | NE compared to control | Gestational age and sex | Cohort | 185 (40%−57%) | IL‐6; IL‐8; IL‐10; VEGF | Infant blood and cerebrospinal fluid | Custom multiplex enzyme‐linked immunosorbent assay | 0–1 week postbirth | BSID‐III (cognition; language; motor skills) | 15–30 months |
| Lee et al. (2021), South Korea | Very preterm and/or VLBW | Postmenstrual age and gestational age | Cohort | 94 (41%–62%) | IL‐1β; IL‐6; IL‐8; TNF‐α; CRP | Infant blood and cerebrospinal fluid | Enzyme‐linked immunosorbent assay (Cloud‐Clone Corp, Texas) Quantikine (R&D systems, Minnesota) | <35 weeks gestational age | BSID‐III (cognition; language; motor skills social–emotional development; adaptive behavior) | 18 months |
| Rose et al. (2016), USA | Very preterm, VLBW | Not reported | Cohort | 92 (not specified) | CRP | Infant blood | Not specified | Within 14 days postbirth | BSID‐III (cognition; language; motor skills) | 18–22 months |
| Carlo et al. (2011), USA | ELBW | Race and gestational age | Cohort | 755 (41.6%) | IL‐1β; IL‐8; TNF‐α; RANTES; IL‐2 | Umbilical cord blood and infant whole blood spots | Multiplex Luminex assay | 0–3 weeks | BSID‐II (cognition; motor skills) | 18–22 months |
| Sweetman et al. (2021), Ireland | NE compared to control | Not reported | Cohort | 94 (not specified) | GM‐CSF; IL‐8 | Infant blood and cerebrospinal fluid | Human Ultra‐Sensitive IL‐8; Human Ultra‐Sensitive GM‐CSF | 1, 3, and 7 days | BSID‐III; MRI (cognition; language; motor skills) | 18–24 months |
| Dilli et al. (2013) | VLBW, with or without sepsis | Not reported | Cohort | 40 (50%) | IL‐6; CRP; CD64 | Infant blood | Tinaquant CRP (Latex) high sensitive immunoturbidimetric assay for CRP; chemiluminescent sequential immunometric assay for IL‐6 Flow cytometry for CD64 | First 7 days after birth | BSID‐II (cognition; language; motor skills) | 18–24 months |
| Jiang et al. (2017), Bangladesh | Living in poverty PROVIDE | Sex, family income, education, length‐for‐age | Cohort | 422 (51.7%) | TNF‐α; IL‐1β; IL‐6; IL‐10; CRP | Infant blood | Human Bio‐Plex Pro Assays Enzyme‐linked immunosorbent assay | 18 weeks postbirth for all biomarkers beside CRP. CRP assessed at 6, 18, 40, 53, and 104 weeks | BSID‐III (cognition; language; social–emotional development) | 78 and 104 weeks |
| Silveira & Procianoy (2011), Brazil | Very preterm, VLBW, high risk for sepsis | Not reported | Cohort | 62 (not specified) | IL‐1β; IL‐6; IL‐8; IL‐10; TNF‐α | Infant blood | Human cytokine lincoplex kit (Linco Research) | 24–32 weeks gestational age | BSID‐II (cognition; motor skils) | 22–24 months |
| Rodríguez‐Trujillo et al. (2019), Spain | Preterm | Not reported | Cohort | 98 (59.5%) | IL‐6 | Amniotic fluid | Enzyme‐linked immunosorbent assay | Birth | ASQ‐III (communication; fine motor skills; gross motor skills; problem solving; personal–social development) | 23.5 months |
| Leviton et al. (2013), USA | Extremely preterm ELGAN | Gestational age, sex | Cohort | 805 (51.7%) | CRP; SAA; MPO; IL‐1β; IL‐6; IL‐6R; TNF‐α; TNF‐R1; TNF‐R2; IL‐8; MCP‐1; MCP‐4; MIP‐1β; RANTES; I‐TAC; ICAM‐1; ICAM‐3; VCAM‐1; E‐SEL; MMP‐1; MMP‐9; VEGF; VEGF‐R1; VEGF‐R2; IGFBP‐1 | Infant blood spots | Meso Scale Discovery electrochemiluminescence multiplex platform Sector Imager 2400 | 0–1 week | BSID‐II (cognition) | 24 months |
| Leviton et al. (2016), USA | Extremely preterm ELGAN | Birthweight Z‐score | Cohort | 750 (not specified) | CRP; SAA; MPO; IL‐1β; IL‐6; IL‐6R; TNF‐α; TNF‐R2; IL‐8; RANTES; ICAM‐1; MMP‐9; VEGF; VEGF‐R2; TSH; EPO | Infant blood spots | Meso Scale Discovery electrochemiluminescence | 3–4 weeks | BSID‐II (cognition; motor skills) | 24 months |
| O'Shea et al. (2012), USA | Extremely preterm ELGAN | Gestational age | Cohort | 939 (53%) | CRP; SAA; MPO; IL‐1β; IL‐6; IL‐6R; TNF‐α; TNF‐R1; TNF‐R2; IL‐8; MCP‐1; MCP‐4; MIP‐1β; RANTES; I‐TAC; ICAM‐1; ICAM‐3; VCAM‐1; E‐SEL; MMP‐1; MMP‐9; VEGF; VEGF‐R1; VEGF‐R2; IGFBP‐1 | Infant blood | Meso Scale Discovery multiplex platform and Sector Imager 2400 (MSD, Maryland) | 0–2 weeks | BSID‐II (cognition; language; motor skills) | 24 months |
| O'Shea et al. (2013), USA | Extremely preterm ELGAN | Gestational age | Cohort | 800 (not specified) | CRP; SAA; MPO; IL‐1β; IL‐6; IL‐6R; TNF‐α; TNF‐R1; TNF‐R2; IL‐8; MCP‐1; MCP‐4; MIP‐1β; RANTES; I‐TAC; ICAM‐1; ICAM‐3; VCAM‐1; E‐SEL; MMP‐1; MMP‐9; VEGF; VEGF‐R1; VEGF‐R2; IGFBP‐1 | Infant blood | Multiplex assays (electrochemiluminescence multiplex detection system) | 0–2 weeks | BSID‐II, VABS (cognition; motor skills) | 24 months |
| O'Shea et al. (2014), USA | Extremely preterm ELGAN | Gestational age, birth weight Z‐score, first pregnancy, and multifetal pregnancy | Cohort | 600 (not specified) | CRP; SAA; MPO; IL‐1β; IL‐6; IL‐6R; TNF‐α; TNF‐R1; TNF‐R2; IL‐8; MCP‐1; MCP‐4; MIP‐1β; RANTES; I‐TAC; ICAM‐1; ICAM‐3; VCAM‐1; E‐SEL; MMP‐1; MMP‐9; VEGF; VEGF‐R1; VEGF‐R2; IGFBP‐1 | Infant blood | Multiplex assays (electrochemiluminescence multiplex detection system) | 0–2 weeks | BSID‐II (cognition); CBCL (Behavior emotional reactivity) | 24 months |
| van den Berg et al. (2016), Netherlands | Very preterm | Gestational age, birth weight, sex, and one or more serious neonatal infections | Case–control | 79 (59.21%) | IL‐1β; Il‐2; IL‐4; IL‐6; IL‐8; IL‐10; IL‐17; IFN‐ γ; TNF‐α | Blood samples | Fluorescent bead‐based multiplex immunoassay (Luminex xMAP technology), and cytokine assay kits were purchased from Bio‐rad (Hercules, California) | 48 h postbirth; preintervention 7 days; 14 days postnatal | BSID‐II/III (mental developmental index, psychomotor index) | 2 years |
| Sevenoaks et al. (2021), South Africa | Maternal HIV compared to control | Maternal sociodemographic and lifestyle factors (adjusted for clinic, maternal smoking during pregnancy, maternal alcohol use during pregnancy, maternal socioeconomic status, maternal BMI at 6 weeks postpartum), infant health (adjusted for birth weight, prematurity, infant sex and exclusive breastfeeding [yes/no]), maternal HIV disease parameters (adjusted for maternal CD4+ during pregnancy, maternal viral load during pregnancy, maternal ART regimen during pregnancy and initiation of ART [before or during pregnancy]) | Cohort | 267 (28.8%–39%) | IFN‐γ; IL‐1β; IL‐2; IL‐4; IL‐5; IL‐6; IL‐7; IL‐8; IL‐10; IL‐12p70; IL‐13; TNF‐α; GM‐CSF; NGAL; MMP‐9 | Infant blood | Multiplex bead assay; Enzyme‐linked immunosorbent assay | 6–10 weeks; 24–28 months | BSID‐III (cognition; language; motor skills) | 24–28 months |
| Lodha et al. (2010), Canada | NEC | Not reported | Cohort | 40 (57.5%) | TNF‐α; IL‐6; IL‐8 | Infant blood | DPC Immulite system | 2–3 weeks | BSID‐II (cognition; motor skills) | 24–28 months |
| Krakowiak et al. (2017), USA | ASD compared to control | Age at bloodspot collection, years between sample collection and elution, birth month, gestational age | Case–control | 303 (59%–88%) | IL‐1β; IL‐2; IL‐4; IL‐5; IL‐6; IL‐10; IL‐12; IL‐13; IFN‐γ; TNF‐α; IL‐8; MCP‐1; MIP‐1α; MIP‐1β; IP‐10; RANTES | Infant blood | Luminex multiplex Bio‐Plex Precision Pro Human Cytokine Assays MILLIPLEX human cytokine kits | Birth | MSEL (cognitive development); VABS (personal‐social skills); ABC (maladaptive behavior) | 24–60 months |
| Ashwood et al. (2011), USA | ASD compared to control | Child's age at blood draw and gender, diagnostic group | Case–control | 223 (80%–86.6%) | GM‐CSF; IFN‐γ; IL‐1β; IL‐2; IL‐4; IL‐5; IL‐6; IL‐8; IL‐10; IL‐12(p40); IL‐13; TNF‐α | Child blood | Human multiplexing bead immunoassays | 24–60 months (not specified) | SCQ (ASD symptoms); VABS (adaptive behavior); MSEL (cognition); ABC (maladaptive behavior) | 24–60 months (not specified) |
| Voltas et al. (2017), Spain | TD | Socioeconomic status, breastfeeding, gestational age, mother's age (score), GHQ mother, exposure to tobacco during pregnancy, bottle fed iron fortified, BMI at 30 months | Cohort | 51 (49.02%) | Il‐1β; IL‐4; IL‐6 | Infant blood | Multiplex assay (FlowCytomix) | 6–12 months | BSID‐II (cognition; motor skills); CBCL (behavior; emotional reactivity) | 30 months |
| Kinjo et al. (2011), Japan | VLBW | APGAR scores | Cohort | 29 (55%) | IL‐1β; IL‐6; IL‐8; IL‐10; TNF‐α; IL‐12p70; CXCL9; CXCL10; MCP‐1 | Infant blood | Cytometric bead array (Becton Dickinson) | Birth 4 weeks | KSPD (Postural motor; cognition; adaptive behavior; language; social skills) | 36 months |
| Ahearne et al. (2017), Ireland | HIE | Not reported | Cohort | 33 (66.6%) | IL‐6; IL‐16 | Umbilical cord blood | Luminex assays | Birth | BSID‐III (cognition; language; motor skills) | 36–42 months |
| Abraham et al. (2021), (Israel) | RD | SES, parents and children age and gender | Cohort | 47 (55.3%) | s‐IgA | Child saliva | Enzyme‐linked immunosorbent assay | 36–55 months | Behavior observation (regulation; negative emotionality) | 40.41 months (4.82) |
| Blok et al. (2011), Netherlands | Very preterm | Not reported | Cohort | 105 (46%) | IL‐6; IL‐8; TNF‐α | Infant blood | Enzyme‐linked immunosorbent assay | 0–5 days | GMDS (personal–social; hearing and speech; locomotor; eye‐hand coordination; performance; practical reasoning) | 3.5 years |
| Sorokin et al. (2014), USA | Mothers at risk for preterm birth | Gestational age at delivery, treatment group | Cohort | 465 (not specified) | IL‐6; CRP; MPO | Umbilical cord blood | Enzyme‐linked immunosorbent assay | Birth | BSID‐II (mental development; psychomotor development) | 36–42 months |
| Kyriklaki et al. (2019), Greece | Typically developing | Prepregnancy BMI, smoking early in pregnancy, parity, birth weight, preterm birth, BMI at the age of 4 and passive smoking of the child at 4 years of age | Cross‐sectional (nested) | 642 (52%) | IL‐1β; IL‐6; IL‐8; IL‐10; IL‐17; TNF‐α; MIP‐1α; IFN‐γ | Child blood | Milliplex Map human high sensitivity T cell magnetic bead panel (Millipore, MA) | 4 years | MSCA (cognition; memory; verbal; motor skills; quantitative; perceptual) | 4 years |
| von Ehrenstein et al. (2012), USA | Typically developing | Birth weight, gender, race/ethnicity, and smoking during pregnancy | Cohort | 369 (51.5%) | IL‐4; IL‐10; IL‐12p70; TNF‐α; IFN‐γ | Umbilical cord blood | Linco Immunoassay; Luminex 100 IS (Millipore) | Birth | WPPSI‐R (verbal intelligence; performance intelligence) | 5 years |
Abbreviations: ABC, Aberrant Behavior Checklist; ASQ, Ages & Stages Questionnaire; BSID‐II, Bayley Scales of Infant Development—Second Edition; BSID‐III, Bayley Scales of Infant Development—Third Edition; CBCL, Child Behavior Checklist; DDST, Denver Developmental Screening Test; GMDS, Griffiths Mental Developmental Scales; KSPD, Kyoto Scale of Psychological Development; MRI, magnetic resonance imaging; MSCA, McCarthy Scales of Children's Abilities; MSEL, Mullen Scales of Early Learning; NAPI, Neurobehavioral Assessment of the Preterm Infant, Revised Second Edition; TIMP, Test of Infant Motor Performance; VABS, Vineland Adaptive Behavior Scale; WPPSI, Wechsler Preschool and Primary Scale of Intelligence.
TABLE 3.
Included studies that examine the association between infant gut microbiome biomarkers and neurodevelopmental outcomes (N = 13)
| Author (year), country | Population context | Covariates | Study design | Final N (% male) | Biomarker(s) | Biosample age | Source | Source analytic method | Outcome measure (domains) | Age at outcome |
|---|---|---|---|---|---|---|---|---|---|---|
| Kelsey et al. (2021), USA | TD | Birthweight, income, breastfeeding, gestational age, head circumference | Cross‐sectional | 63 (58.73%) |
16S rRNA 1. Alpha diversity: Shannon Index and Chao1 2. Virulence Factors |
9–56 days post‐natal | Diaper stool via collection tube by parents | Shotgun DNA gene sequencing Database: Custom built from NCBI, incl draft/complete genomes of Bacteria, Archaea, Fungi, Viruses, Protozoa | IBQ‐R (negative emotionality; regulation/orienting; surgency/positive emotionality); FNIRS (functional brain connectivity) | 25 days |
| Aatsinki et al. (2019), Finland | TD | Sex, mode of delivery, gestational age, infant age during sampling, antibiotic treatments, breastfeeding status at 2.5 months of age | Prospective cohort | 301 (53%) |
1. 16S rRNA 2. Alpha diversity 3. Alpha richness 4. Observed species (profile, genera, OTUs): V. dispar: Enterobacteriaceae, Clostridium neonatale; Bacteroides: Bacteroides fragilis and other species; Bifidobacterium/Enterobacteriaceae, Erwinia, Streptococcus |
2.5 months postnatal | Diaper stool via collection tube by parents | 16S ribosomal RNA gene sequencing with MiSeq (Illumina) Database: GreenGenes | IBQ‐R (negative emotionality; regulation/orienting; surgency/positive emotionality) | 6 months |
| Aatsinki et al. (2022), Finland | Elevated prenatal symptoms of depression and anxiety | Maternal age and education, BMI, prenatal SSRI/SNRI use, prenatal end‐of‐pregnancy depressive symptoms, infant gestational age at birth, birth length and height, age at fecal sampling, breastfeeding status, infant antibiotics intake, mode of delivery | Secondary prospective cohort within a case–control | 131 (53%) |
16S rRNA 1. Alpha diversity 2. Observed taxa: Bifidobacterium, Clostridium |
2.5 months postnatal | Diaper stool via collection tube by parents | 16S ribosomal RNA gene sequencing with MiSeq (Illumina) Database: GreenGenes | Stimulus‐based assessment of attention to emotional faces | 8 months |
| Carlson et al. (2018), USA | TD | Breastfeeding at time of sample collection, paternal ethnicity, mode of delivery | Prospective cohort | 89 (55.1%) |
16S rRNA 1. Alpha diversity 2. Specific Taxa 3. Richness |
1 year | Diaper by parents |
16S rRNA via Illumina MiSeq Database: Not specified |
MSEL (gross motor skills; fine motor skills; visual reception; expressive and receptive language) | 1 year |
| Rozé et al. (2020), France | Preterm | Gestational age, maternal age, maternal country of birth, maternal education, birth weight Z‐score, cesarean delivery, and individual therapeutics | Cohort | 577 (52.5%) |
16S rRNA 1. Alpha diversity: Shannon Index 2. Observed taxa: Enterococcus, Staphylococcus, Escherichia, Shigella |
Median age of 23 days postbirth | Diaper stool at −80°C by researchers |
16S ribosomal RNA gene sequencing with MiSeq (Illumina) Database: RDP |
ASQ (overall neurodevelopment) | 2 years |
| Loughman et al. (2020), Australia | TD | Gestational age, mode of birth, antibiotic use during labor, breastfeeding at 4 weeks, number of siblings, household pet ownership, sex of child, age of child at time of developmental assessment | Prospective cohort | 201 (52.7%) |
16S rRNA 1. Alpha diversity 2. Beta diversity |
1, 6, and 12 months postnatal | Diaper by parents |
16S RNA via Illumina MiSeq Database: SILVA |
CBCL (internalizing problems; externalizing problems; total problems) | 2 years |
| Loughman et al. (2021), Australia | Colic | Mode of birth, sex, birth weight, gestational age, infant feeding type, infant age at baseline, antibiotic or probiotic use in 24 h prior to sample collection, baseline crying/fussing time, maternal postpartum depression at baseline and 1 month, maternal education level | Prospective cohort | 118 (53%) |
16S rRNA 1. Alpha diversity 2. Beta diversity |
7.4 weeks | Diaper by parents |
16S rRNA via Illumina MiSeq platform, using 2 × 300 bp paired‐end sequencing, Database: Not specified |
CBCL (internalizing problems; externalizing problems; total problems) | 2 years |
| Gao et al. (2019), USA | TD | Sex, maternal education, paternal age, paternal ethnicity, twin status, postnatal age at scan, income | Prospective cohort | 39 (61.5%) |
16S rRNA 1. Alpha diversity: Shannon, Chao1, Faith's Phylogenetic 2. Observed species |
1 year | Diaper by parents |
16S rRNA via Illumina MiSeq Database: Not specified |
MSEL (gross motor skills, fine motor skills; visual reception; expressive and receptive language) | 2 years |
| van den Berg et al. (2016), Netherlands | Very preterm | Gestational age, birth weight, sex, and one or more serious neonatal infections | Case–control | 79 (59.21%) | Bifidobacteria count |
48 h postnatal Preintervention 7 days 14 days postnatal |
Stool | Bacterial cells | BSID‐II/III (cognition; motor skills) | 2 years |
| Christian et al. (2015), USA | TD | Temperament characteristics, surgency/extraversion, sociability, high‐intensity pleasure and activity level, eating behavior | Prospective cohort | 77 (53.25%) |
16S rRNA 1. Alpha diversity 2. Beta diversity |
18–27 months postnatal | Diaper via collection tube by parents |
16S rRNA via Roche 454 FLX Titanium Database: GreenGenes |
ECBQ (negative affectivity; surgency/extraversion; effortful control) | 18–27 months |
| Rothenberg et al. (2021), China | Living rurally | Child had diarrhea or vomited in the previous 12 months, types of food consumed in previous 24 h, weight, height, weight‐for‐height | Cross section within a prospective cohort | 46 (55%) |
16S rRNA 1. Specific Taxa |
36 months | Sterile cotton swab by parents |
16S rRNA Illumina MiSeq 2 × 300 bp paired‐end sequencing Database: RDP |
BSID‐II (cognition; motor skills) | 36 months |
| Sordillo et al. (2019), USA | Asthma intervention | Age at ASQ‐3 assessment, cholecalciferol (vitamin D) treatment group, clinical site, mode of delivery, child's sex, and antibiotic administration in the first days of life, gestational age, maternal age, marital status, education level, family income, infant race/ethnicity, breastfeeding status 6 months after birth | Prospective cohort | 309 (55%) |
16S rRNA 1. Taxa Abundance 2. Specific Taxa |
3–6 months | Diaper via collection tube by parents |
16S rRNA via Roche 454 FLX Titanium Database: RDP |
ASQ‐3 (motor skills; problem solving; communication; social skills) | 3 years |
| Laue et al. (2020), USA | TD | Maternal self‐reported smoking during pregnancy, early exclusive breastfeeding, delivery mode, peripartum antibiotics, gestational age, age at SRS‐2, maternal education, marital status, maternal age, paternal age, and child sex | Prospective cohort | 140 (51.6%) |
16S rRNA 1. Alpha diversity: Shannon Diversity Index 2. Specific Taxa |
6 weeks, 1 year, 2 years, and 3 years postnatal | Study‐provided diaper or Receptacle tube by parents (depending on age) |
16S rRNA via Illumina MiSeq and Shotgun sequencing via Illumina NextSeq Database: GreenGenes (16S rRNA) MetaPhlAn (Shotgun) |
SRS‐2 (awareness; cognition; communication; motivation; restricted interests/repetitive behavior) | 3 years |
Abbreviations: ASQ, Ages & Stages Questionnaire; BSID‐III, Bayley Scales of Infant Development—Third Edition; CBCL, Child Behavior Checklist; ECBQ, Early Childhood Behavior Questionnaire; FNIRS, Functional Near Infrared Spectroscopy; IBQ‐R, Infant Behavior Questionnaire—Revised; MSEL, Mullen Scales of Early Learning; OTU, Operational Taxonomic Unit; SRS‐2, Social Responsiveness Scale 2; TD, typically developing.
TABLE 4.
Maternal immune system biomarkers measured across the retained studies (N = 18)
| Maternal immune biomarkers | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Study authors | Final N | IL‐6 | TNF‐α | CRP | IL‐10 | IL‐1β | IL‐8 | MCP‐1 | IL‐4 | IL‐2 | IFN‐γ | IL‐5 | TARC | VEGF | VEGF‐D | GM‐CSF | IL‐12p70 | IL‐13 | MIP‐1β | IL‐7 | sFLT‐1 |
| Bodnar et al. (2018) a | 152 | S– | S↓ | S↓ | S↓ | S– | S– | S– | S– | S↓ | S– | S– | S– | S– | S– | S– | S– | S– | S– | S– | |
| Irwin et al. (2019) b | 1408 | C↓M↓L↑E↕ | C↓M↓ L↕ E↓ | C↑M↑ L↓E↓ | C↓M↑ L↓E↓ | C↑M↓ L↕E↑ | C↑M↑ L↓E↕ | C↓M↓ L↓E↓ | C↓M↓ L↕E↑ | C↑M↑L↕E↓ | C↓M↓ L↕E↓ | C↑M↑ L↑E↑ | C↑M↑ L↑E↑ | C↓M↓ L↓E↓ | |||||||
| Sowell et al. (2018) c | 241 | C↕M↕ | C↕M↕ | C↑M↑ | C↑M↑ | C↕M↕ | C↑M↑ | C↕M↕ | C↑M↑ | ||||||||||||
| Spann et al. (2018) | 72 | C↑ | C↑ | ||||||||||||||||||
| Rasmussen et al. (2019) | 147 | C↓E– | |||||||||||||||||||
| Nazzari et al. (2020) | 104 | C↓ | C↓ | ||||||||||||||||||
| Yan et al. (2020) | 1186 | C↓L↓ M↓E↓ | C↓L↓ M↓E↓ | C↓L↓ M↓E↓ | C↓L↓ M↓E↓ | C↓L↓M↓E↓ | |||||||||||||||
| Rudolph et al. (2018) | 84 | C↓ | |||||||||||||||||||
| Freedman et al. (2019) | 162 | E↓ | |||||||||||||||||||
| Gustafsson et al. (2019) b | 68 | E↓ | E↓ | E↓ | |||||||||||||||||
| Gustaffson et al. (2018) b | 68 | E↓ | E↓ | E↓ | E↓ | ||||||||||||||||
| Hunter et al. (2021) | 127 | E↓ | E– | E↑ | E– | ||||||||||||||||
| Osborne et al. (2018) | 87 | L↓M↑E↓ | L↓M↑E↓ | L↓M↑E↓ | L–M–E– | L–M–E– | L–M–E– | L↓M↑E↓ | |||||||||||||
| Monthe‐Dreze et al. (2019) | 1246 | C↓ | |||||||||||||||||||
| Graham et al. (2018) | 86 | E↓ | |||||||||||||||||||
| Nazzari et al. (2019) | 104 | E↓ | E↓ | ||||||||||||||||||
| Sevenoaks et al. (2021) | 267 | C↓L↓M↑ | C↓L↓M↓ | C↓L↓M↓ | C↓L↓M↓ | C↓L↓M↓ | C↑L↓M↓ | C↓L↓M↓ | C↓L↓M↓ | C↓L↓M↓ | C↑L↓M↑ | C↓L↓M↓ | C↓L↓M↓ | C↓L↓M↓ | |||||||
| Rommel et al. (2020) | 512 | ||||||||||||||||||||
Note: Only biomarkers that were examined in two or more of the included studies are reported in this table. A complete list of all biomarkers examined across these studies is provided in the Supporting Information. Biomarkers not included in this table but measured in one of these studies are EGF, bFGF, VEGF‐C, Eotaxin, Eotaxin‐3, IL‐16, MCP‐4, TNF‐β, GMCSF, IP‐10, MDC, sICAM‐1, sVCAM‐1, IL‐1α, IL‐12, IL‐15, HNP1‐3, MIP‐1α, NGAL, MMP‐9, SAA, sAA Choline, and PGF2‐α. Rommel et al. (2020) measured one biomarker (PGF2‐α) not listed in the table above. Associations depicted in red denote at least one statistically significant association between biomarker and neurodevelopmental outcome reported by study authors. C, cognitive development; L, language/communication development; M, motor development; A, adaptive behavior; E, social–emotional development; S, neurodevelopmental outcome composed of several domains. ↓ = associated with poorer neurodevelopmental outcome. ↑ = associated with better neurodevelopmental outcome. ↕ = mixed (positive and negative) association with better outcome across specific domains of subtypes neurodevelopmental outcome (e.g., vocabulary produced, or vocabulary understood as subtypes of language/communication development), timing of the biosample (e.g., sampled at multiple points in time), or source of biosample (e.g., blood and cerebrospinal fluid). – = direction and/or significance of association with neurodevelopmental outcome not reported.
Study examined cytokine “networks” composed of specific biomarkers.
Study examined latent “inflammatory” or “anti‐inflammatory” profiles, with specific biomarkers identified as items driven by the latent construct.
Study also examined a ratio of inflammatory to anti‐inflammatory biomarkers.
TABLE 5.
Infant immune system biomarkers measured across the retained studies (N = 40)
| Infant immune biomarker | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Study authors | Final N | IL‐6 | IL‐8 | TNF‐α | IL‐1β | CRP | IL‐10 | MCP‐1 | IL‐4 | RANTES | VEGF | MIP‐1β | IFN‐γ | MPO | IP‐10 | IL‐2 | IL‐12p70 | IL‐6R | SAA | MCP‐4 | MMP‐9 |
| Jiang et al. (2017) | 422 | C–L↓M–E– | C–L↓M–E– | C–L↓M–E– | C↓L↓M–E↓ | C↓L↓M↓E↓ | C–L↓M–E– | ||||||||||||||
| Jiang et al. (2014) | 127 | C↓L↓M↓ | C↑L↑M↓ | C↑L↑M↓ | C↑L↓M↓ | C↑L↑M↑ | |||||||||||||||
| Ahearne et al. (2017) | 33 | C–L–M– | |||||||||||||||||||
| Dietrick et al. (2020) | 185 | C↓M↓L↓ | C↓M↓L↓ | C↓M↓L↓ | C↓M↑L↓ | ||||||||||||||||
| Camargos et al. (2017) | 50 | C↑M– | C–M– | C–M↓ | C–M– | ||||||||||||||||
| Chalak et al. (2014) | 27 | S↓ | S↓ | S↓ | S↓ | S↓ | |||||||||||||||
| Lee et al. (2021) | 94 | C–L–M–A–S– | C↕L↕M↕A↕S↕ | C↓L↓M↓A↓S↕ | C↓L↓M↓A↓S↓ | C↓L↓M↓A↓S↓ | |||||||||||||||
| Etheredge et al. (2018) | 107 | C↑M↓L↑ | |||||||||||||||||||
| Benavides et al. (2022) | 71 | C– | C↓ | C– | |||||||||||||||||
| Nist, Shoben, et al. (2020) a | 62 | C↓M– | C↓M– | C–M↓ | C–M– | C–M– | C–M– | ||||||||||||||
| Carlo et al. (2011) | 755 | C↑M↓ | C↑M↑ | C↑M↑ | C↑M↑ | C↓M↑ | |||||||||||||||
| Varner et al. (2014) | 615 | C↑M↑ | C↓M↓ | C↑M↑ | |||||||||||||||||
| Silveira & Procianoy (2011) | 62 | C↓M↓ | C↓M↓ | C↓M↓ | C↓M↓ | C↓M↓ | |||||||||||||||
| Díllí et al. (2013) | 40 | C↓M– | C↓M– | ||||||||||||||||||
| Lodha et al. (2010) | 40 | C–M– | C–M– | C–M– | |||||||||||||||||
| Leviton et al. (2013) | 805 | C↓ | C↓ | C↓ | C↓ | C↓ | C↓ | C↓ | C↓ | C↓ | C↓ | C↓ | C↓ | C↓ | C↓ | ||||||
| Leviton et al. (2016) | 750 | C↓ | C↓ | C↓ | C↕ | C↓ | C↑ | ||||||||||||||
| O'Shea et al. (2012) | 939 | C↓M↓ | C↓M↓ | C↓ M↓ | C↕ M↓ | C↓ M↓ | C↑M↕ | C↑M↕ | C↑M↕ | C↓M↕ | C↕ M↓ | C↕M↕ | C↓M↓ | C↕M↕ | |||||||
| O'Shea et al. (2013) | 800 | C↓ | C↓ | C↓ | C↓ | C↓ | C↓ | C↓ | C↓ | C↓ | C↓ | C↓ | C↓ | C↓ | C↓ | ||||||
| O'Shea et al. (2014) | 600 | C↓E↕ | C↓E↕ | C↓E↕ | C↓E↕ | C↓E↕ | C↓E↕ | C↓E↕ | C↓E↕ | C↓E↑ | C↓E↕ | C↓E↕ | C↓E↕ | C↓E↓ | C↓E↕ | ||||||
| Voltas et al. (2017) | 51 | C↓L↓M↓E↓ | C↓L↓M↓E↓ | C↓L↓M↓E↓ | |||||||||||||||||
| Ghassabian et al. (2018) | 3038 | S– | S– | S– | S– | S↑ | S– | ||||||||||||||
| Rodriguez‐Trujillo et al. (2019) | 98 | S↓ | |||||||||||||||||||
| Xie et al (2019) | 260 | C↓ | |||||||||||||||||||
| Krakowiak et al. (2017) | 303 | C–A–L– | C–A–L– | C–A–L– | C–A–L↑ | C–A–L– | C–A–L– | L↓ | C–A–L– | C–A–L– | C–A–L– | C–A–L– | C–A–L– | ||||||||
| Jensen et al. (2019) | 122 | C↓L↓M↓ | |||||||||||||||||||
| Ashwood et al. (2011) | 223 | C–A– | C↓A↓ | C–A– | C–A– | C–A– | C–A– | C–A– | C–A– | ||||||||||||
| Liu & Feng (2010) | 52 | S↓ | S↓ | S↓ | |||||||||||||||||
| Blok et al. (2011) | 105 | S– | S– | S– | |||||||||||||||||
| Kinjo et al. (2011) | 29 | C↓L–M– | C↓L↓M↓ | C–L–M– | C↓L–M↓ | C–L–M– | C–L–M– | C–L–M– | |||||||||||||
| Kyriklaki et al. (2019) b | 642 | C↑M↑ | C↓M↓ | C↓M↓ | C↕M↓ | C↑M↑ | |||||||||||||||
| Nist, Pickler, et al. (2020) | 68 | C–L–M– | C↑L–M– | C–L–M– | C–L–M– | C–L–M– | C–L–M– | C–L–M– | |||||||||||||
| Magalhaes et al. (2017) | 40 | M↑ | M↑ | M↑ | M↕ | M↓ | M↓ | M↓ | M↑ | M↓ | |||||||||||
| Von Ehrenstein et al. (2012) | 369 | C↓ | C↕ | C↕ | C↕ | C↓ | |||||||||||||||
| Sorokin et al. (2014) | 465 | C–M– | C–M– | C–M– | |||||||||||||||||
| Rose et al. (2016) | 92 | C↓M↓L↓ | |||||||||||||||||||
| Van Den Berg et al. (2016) | 79 | C– | C– | C– | C↓M– | C– M– | C –M↓ | C– M↓ | C– M↓ | ||||||||||||
| Sevenoaks et al. (2021) | 267 | C↓L↓M↓ | C↓L↓M↓ | C↓L↓M↓ | C↓L↓M↓ | C↓L↓M↓ | C↓L↓M↓ | C↓L↓M↓ | C↓L↓M↓ | C↓L↓M↓ | |||||||||||
| Sweetman et al. (2021) | 94 | C↓M↓L↓ | |||||||||||||||||||
| Abraham et al. (2021) | 47 | ||||||||||||||||||||
Note: Only biomarkers that were examined in five or more studies are reported in this table. A complete list of all biomarkers examined across these studies is provided in the Supporting Information. Biomarkers not included in this table but measured in these studies are noted here. Biomarkers investigated in four studies are GMCSF, ICAM‐1, MIP‐1α, TNF‐R1, TNF‐R2, I‐TAC, VCAM‐1, E‐SEL, MMP‐1, VEGF‐R1, VEGF‐R2, and IGFBP‐1. Biomarkers investigated in three studies are IL‐16, ICAM‐3, IL‐5, IL‐1α, and IL‐13. Biomarkers investigated in two studies are MIG, TNF‐β, IL‐17, Eotaxin, TARC, sVCAM‐1, sICAM‐1, and IL‐7. Biomarkers investigated in only one study are sTNFR1, sTNFR2, IL‐1, IL‐1‐RA, GCSF, CD64, CXCL9, CXCL10, IL‐17A, Adiponectin, s‐IgA, IL‐12, FGF, IL‐1ra, 6Ckine, CTACK, IL‐20, MCP‐2, MIP‐1d, SDF‐1, Cathepsin D, PDGF‐AA, PAI‐1, NCAM, CD26E, Eotaxin‐3, bFGF, MDC, sFlt‐1 , VEGF‐C, PIGF, IL‐15, VEGF‐D, TIE‐1, IL‐17α , and NGAL. Associations depicted in red denote at least one statistically significant association between biomarker and neurodevelopmental outcome reported by study authors. C, cognitive development; L, language/communication development; M, motor development; A, adaptive behavior; E, social–emotional development; S, neurodevelopmental outcome composed of several domains. ↓ = associated with poorer neurodevelopmental outcome. ↑ = associated with better neurodevelopmental outcome. ↕ = mixed (positive and negative) association with better outcome across specific domains of subtypes neurodevelopmental outcome (e.g., vocabulary produced, or vocabulary understood as subtypes of language/communication development), timing of the biosample (e.g., sampled at multiple points in time), or source of biosample (e.g., blood and cerebrospinal fluid). – = direction and/or significance of association with neurodevelopmental outcome not reported.
Study examined latent “inflammatory” or “anti‐inflammatory” profiles, with specific biomarkers identified as items driven by the latent construct.
Study also examined a ratio of inflammatory to anti‐inflammatory biomarkers.
TABLE 6.
Infant gut microbiome biomarkers measured across the retained studies (N = 13)
| Study | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Infant gut microbiome biomarker | Rozé et al. (2020) | Aatsinki et al. (2022) | Aatsinki et al. (2019) | Kelsey et al. (2021) | Laue et al. (2020) | Loughman et al. (2020) | Loughman et al. (2021) | Christian et al. (2015) | Rothenburg et al. (2021) | Carlson et al. (2018) | Gao et al. (2019) | Sordillo et al. (2019) a | Van den Berg et al. (2016) |
| Final N | 577 | 131 | 301 | 63 | 140 | 201 | 118 | 77 | 46 | 89 | 39 | 309 | 79 |
| Alpha diversity | |||||||||||||
| Shannon–Wiener | S– | T– | T↑ | T↓ | E↓ | E↓ | E↓ | T↕ | M–C– | C↓ | C– | C↓L↓M↓E↓ | |
| Simpson | E↓ | E↓ | |||||||||||
| Chao1 | T– | T– | T↑ | E↓ | E↓ | C↓ | C– | ||||||
| Phylogenetic | T↑ | M–C– | C↓ | C– | |||||||||
| Other(s) | E↓ | M–C– | C–M– | ||||||||||
| Gene ontology | |||||||||||||
| Virulence factors | T↑ | ||||||||||||
| Resistome genes | T– | ||||||||||||
| Observed taxa | S– | T– | T– | T– | E↑ | E↓ | E↓ | M–C– | C↓ | C– | |||
| Taxa count | E↑ | C–M– | |||||||||||
| Beta diversity | |||||||||||||
| Unifrac (Weighted/Unweighted) | E↑ | E↓ | E↓ | T↑ | M–C– | ||||||||
| Aquificae | |||||||||||||
| Thermovibrio guaymasensis | T– | ||||||||||||
| Firmicutes | S↓ | ||||||||||||
| Anaerococcus | |||||||||||||
| Anaerostipes | M–C– | ||||||||||||
| Blautia | M–C– | ||||||||||||
| Blautia producta | E↓ | ||||||||||||
| Butyricicoccus pullicaecorum | |||||||||||||
| Catenibacterium | |||||||||||||
| Cellulosibacter | M–C– | ||||||||||||
| Clostridiales | C–L↓M–E– | ||||||||||||
| Clostridium | T– | T– | E↓ | M–C– | C–L–M–E– | ||||||||
| Clostridium perfringens | T– | ||||||||||||
| Clostridium disporicum | T– | ||||||||||||
| Clostridium sensu stricto | S– | ||||||||||||
| Coprobacillus | |||||||||||||
| Coprococcus | E↓ | M–C– | |||||||||||
| Dialister | T↑ | ||||||||||||
| Enterococcus | |||||||||||||
| Enterococcus faecalis | S– | T– | |||||||||||
| Eubacterium | |||||||||||||
| Eubacterium hallii | |||||||||||||
| Eubacterium dolichum | |||||||||||||
| Faecalibacterium | M–C– | C– | |||||||||||
| Flavonifractor | M–C– | ||||||||||||
| Flavonifractor plautii | E↓ | ||||||||||||
| Fusicatenibacter | M–C– | ||||||||||||
| Gemmiger | M–C– | ||||||||||||
| Holdemania | |||||||||||||
| Lactobacillus | E↓ | ||||||||||||
| Lachnospiraceae | T– | E↓ | E↓ | M–C– | C–L–M–E– | ||||||||
| Lactococcus | |||||||||||||
| Megasphaera | |||||||||||||
| Megamonas | M–C– | ||||||||||||
| Oscillibacter | M–C– | ||||||||||||
| Phascolarctobacterium | M–C– | ||||||||||||
| Peptoniphilus | |||||||||||||
| Roseburia | |||||||||||||
| Roseburia peoriensis | T– | ||||||||||||
| Ruminococcaceae | T↑ | C– | |||||||||||
| Ruminococcus | M–C– | ||||||||||||
| Ruminococcus gnavus | E↓ | E↓ | |||||||||||
| Ruminococcus torques | E↓ | ||||||||||||
| Bacterium_6_1_63FAA | E↓ | ||||||||||||
| Sarcina | |||||||||||||
| Sporacetigenium | |||||||||||||
| Streptococcus | T↑ | M–C– | |||||||||||
| Streptococcus salivarius | T– | ||||||||||||
| Streptococcus vestibularis | T– | ||||||||||||
| Staphylococcus | S– | E↓ | |||||||||||
| Weissella | |||||||||||||
| Veillonella | C–L–M–E– | ||||||||||||
| Bacteroidetes | |||||||||||||
| Alistipes | M–C– | ||||||||||||
| Alloprevotella | |||||||||||||
| Bacteroides | T↓ | M–C– | C– | C–L–M–E– | |||||||||
| Bacteroides fragilis | T– | ||||||||||||
| Bacteroides caccae | T– | ||||||||||||
| Porphyromonas | |||||||||||||
| Parabacteroides | T↑ | M–C– | |||||||||||
| Prevotella | E↓ | ||||||||||||
| Rikenellaceae | T↑ | ||||||||||||
| Actinobacteria | |||||||||||||
| Actinomyces | E↑ | ||||||||||||
| Adlercreutzia equolifaciens | E↓ | ||||||||||||
| Bifidobacterium | T↑ | E↓ | E↕ | M–C– | C–L–M–E– | C↑M– | |||||||
| Bifidobacterium catenulatum | T– | ||||||||||||
| Bifidobacterium dentium | T– | ||||||||||||
| Bifidobacterium pseudocatenulatum | T– | ||||||||||||
| Collinsella | M–C– | ||||||||||||
| Rothia | E↓ | ||||||||||||
| Proteobacteria | S↑ | ||||||||||||
| Campylobacter | |||||||||||||
| Enterobacteriaceae | T↑ | C–L–M–E– | |||||||||||
| Enterobacter aerogenes | S– | ||||||||||||
| Erwinia | T↓ | ||||||||||||
| Escherichia | S– | C–L–M–E– | |||||||||||
| Escherichia coli | S– | T– | |||||||||||
| Escherichia shigella | M–C– | ||||||||||||
| Klebsiella | E↓ | ||||||||||||
| Shigella | S– | ||||||||||||
| Succinivibrio | |||||||||||||
| Sutterella | |||||||||||||
| Campylobacter | |||||||||||||
| Verrucomicrobia | |||||||||||||
| Akkermansia | S– | ||||||||||||
| Coriobacteriaceae | |||||||||||||
| Atopobium | S– | T↓ | |||||||||||
Note: Associations depicted in red denote at least one statistically significant association between biomarker and neurodevelopmental outcome reported by study authors. Biomarker names are derived from the naming conventions used in each of the included studies. However, it is important to recognize that certain phyla were renamed as part of the NCBI Taxonomy to include phylum rank in taxonomic names. C, cognitive development; L, language/communication development; M, motor development; A, adaptive behavior; E, social–emotional development; S, neurodevelopmental outcome composed of several domains; T, Temperament. ↓ = associated with poorer neurodevelopmental outcome. ↑ = associated with better neurodevelopmental outcome. ↕ = mixed (positive and negative) association with better outcome across specific domains of subtypes neurodevelopmental outcome (e.g., vocabulary produced, or vocabulary understood as subtypes of language/communication development), timing of the biosample (e.g., sampled at multiple points in time), or source of biosample (e.g., blood and cerebrospinal fluid). – = direction and/or significance of association with neurodevelopmental outcome not reported.
Study identified clustered or grouped specific phylum.
TABLE 7.
Summary of maternal immune system biomarkers outcomes across neurodevelopment domains (N = 18)
| IL‐6 | TNF‐α | CRP | IL‐10 | IL‐1β | IL‐8 | MCP‐1 | IL‐4 | IL‐2 | IFN‐γ | IL‐5 | TARC | VEGF | VEGF‐D | GM‐CSF | IL‐12p70 | IL‐13 | MIP‐1β | IL‐7 | sFLT‐1 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| # Studies | 15 | 10 | 8 | 7 | 6 | 5 | 4 | 4 | 4 | 3 | 3 | 2 | 2 | 2 | 3 | 2 | 2 | 2 | 2 | 2 |
| C | 1↕, 1↑, 6↓ | 1↕, 3↓ | 2↑, 2↓ | 1↑, 3↓ | 2↑, 2↓ | 1↕, 1↓ | 1↑ | 2↑, 1↓ | 1↕, 2↓ | 1↑, 1↓ | 2↓ | 1↑ | 1↑ | 2↑ | 1↓ | 1↓ | 1↓ | 1↓ | 1↓ | |
| L | 1↑, 3↓ | 1↕, 3↓ | 1↓ | 4↓ | 1↕, 2↓, 1– | 1↓, 1– | 1↓ | 2↓ | 1↕, 1↓, 1– | 1↕, 1↓ | 1↕, 1↓ | 1↑ | 1↓ | 1↑ | 1↓ | 1↓ | 1↓ | 1↓ | 1↓ | 1↓ |
| M | 1↕, 2↑, 2↓ | 1↕, 1↑, 3↓ | 1↑ | 3↑, 2↓ | 1↑, 3↓, 1– | 1↕, 1↓, 1– | 1↑ | 1↑, 2↓ | 1↕, 2↓, 1– | 1↑, 1↓ | 2↓ | 1↑ | 1↑ | 1↑ | 2↑ | 1↓ | 1↓ | 1↓ | 1↓ | 1↓ |
| E | 1↕, 7↓, 1– | 5↓, 1– | 1↑, 3↓ | 4↓ | 1↑, 1↓, 1– | 2– | 1↕, 2↓ | 1↓ | 1↑, 1– | 1↓ | 1↓ | 1↑ | 1↓ | 1↑ | 1↓ | 1↓ | ||||
| S | 1– | 1↓ | 1↓ | 1↓ | 1– | 1– | 1– | 1– | 1↓ | 1– | 1– | 1– | 1– | 1– | 1– | 1– | 1– | 1– | 1– |
Note: C, cognitive domain; L, language domain; M, motor domain; E, emotional domain; S, social domain.
TABLE 8.
Summary of infant immune system biomarkers outcomes across neurodevelopment domains and the reported direction of findings (N = 40)
| IL‐6 | IL‐8 | TNF‐α | IL‐1β | CRP | IL‐10 | MCP‐1 | IL‐4 | RANTES | VEGF | MIP‐1β | IFN‐γ | MPO | IP‐10 | IL‐2 | IL‐12p70 | IL‐6R | SAA | MCP‐4 | MMP‐9 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| # Studies | 28 | 26 | 25 | 21 | 14 | 14 | 12 | 9 | 8 | 7 | 6 | 6 | 6 | 4 | 5 | 4 | 5 | 4 | 4 | 4 |
| C | 1↑, 13↓, 10– | 1↕, 4↑, 12↓, 4– | 2↑, 11↓, 8– | 3↕, 3↑, 10↓, 3– | 1↑, 11↓, 1– | 1↕, 2↑, 3↓, 7– | 1↑, 4↓, 5– | 1↕, 1↑, 2↓, 3– | 2↑, 3↓, 2– | 1↑, 4↓ | 4↓, 1– | 1↕, 1↓, 3– | 1↕, 3↓, 1– | 3– | 2↓, 3– | 2↓, 1– | 1↕, 1↑, 3↓ | 4↓ | 1↕, 3↓ | 4↓ |
| L | 5↓, 5– | 1↕, 4↓, 2– | 1↑, 3↓, 3– | 2↑, 4↓, 3– | 1↑, 4↓ | 4↓, 3– | 3– | 1↑, 3↓, 1– | 1– | 1↓ | 1– | 1↓, 1– | 1– | 1↓, 1– | 1↓, 1– | 1↓ | ||||
| M | 2↑, 6↓, 9– | 1↕, 2↑, 7↓, 5– | 2↑, 8↓, 4– | 1↕, 2↑, 8↓, 3– | 6↓, 2– | 1↑, 5↓, 5– | 1↕, 1↓, 4– | 1↑, 3↓, 2– | 1↕, 1↑, 2↓ | 1↕, 1↑ | 1↕ | 2↓ | 1↓, 1– | 1↑, 1– | 1↑, 2↓ | 2↓, 1– | 1↕ | 1↓ | 1↕ | 1↓ |
| A | 3– | 1↕, 1↓, 1– | 1↓, 2– | 1↓, 2– | 1↓ | 2– | 1– | 1– | 1– | 1– | 2– | 1– | 2– | |||||||
| E | 1↕, 1↓, 1– | 1↕, 1– | 1↕, 1– | 1↕, 2↓ | 1↕, 1↓ | 1– | 1↕ | 1↓ | 1↕ | 1↕ | 1↑ | 1↕ | 1↕ | 1↕ | 1↓ | 1↕ | ||||
| S | 2↓, 2– | 1↕, 2↓, 2– | 1↕, 2↓, 1– | 2↓ | 1↓, 1– | 1– | 1↓, 1– | 1↑ | 1↓ | 1– |
Note: C, cognitive domain; L, language domain; M, motor domain; A, adaptive behavior domain; E, emotional domain; S, social domain.
TABLE 9.
Summary of infant gut microbiome biomarkers outcomes across neurodevelopmental domains (N = 13)
| Infant gut microbiome biomarker genus | Alpha diversity | Beta diversity | Aquificae | Firmicutes | Bacteroidetes | Actinobacteria | Proteobacteria | Verrucomicrobia | Coriobacteriaceae |
|---|---|---|---|---|---|---|---|---|---|
| # Studies | 42 | 5 | 1 | 51 | 11 | 14 | 13 | 1 | 2 |
| C | 5↓, 10– | 1– | 21– | 5– | 1↑, 3– | 3– | |||
| L | 1↓ | 1↓, 3– | 1– | 1– | 2– | ||||
| M | 1↓, 6– | 1– | 19– | 4– | 4– | 3– | |||
| E | 2↑, 11↓ | 1↑, 2↓ | 12↓, 4– | 1↓, 1– | 1↕, 1↑, 3↓, 1– | 1↓, 2– | |||
| S | 2– | 1↓, 3– | 1↑, 4– | 1– | 1– | ||||
| T | 5↑, 2↓, 7– | 1↑ | 1– | 3↑, 9– | 2↑, 1↓, 2– | 1↑, 3– | 1↑, 1↓, 1– | 1↓ |
Note: C, cognitive domain; L, language domain; M, motor domain; A, adaptive behavior domain; E, emotional domain; S, social domain; T, temperament.
3.3. Biomarkers examined in the included studies
The 69 included studies were organized into groups based on the source (mother, infant, or both) and type of biomarker (immune system, gut microbiome, or both). Only one study examined biomarkers from both maternal and child samples (Sevenoaks et al., 2021), and only one study examined biomarkers pertaining to both the immune system and the gut microbiome (Tamburini et al., 2016). Sixty‐eight of the 69 eligible studies measured immune system and/or gut microbiome markers from either the mother or infant. No studies examined the maternal gut microbiome. There were 81 unique immune biomarkers investigated across the included studies. The maximum number of immune biomarkers assayed in one study was 40, while the most frequently assayed immune biomarkers across all studies was IL‐6, then TNF‐α. It should be noted that while studies can assay for specific immune biomarkers, microbiome studies rarely studied a select number of species.
Of these 69 included studies, 18 studies provided their first (or only) measure of biomarkers during pregnancy, 25 studies provided their first (or only) measure of biomarkers within the first month of life, 14 studies provided their first (or only) measure of biomarkers between the first and sixth month of life, seven studies provided their first (or only) measure of biomarkers between 6 and 12 months, and five studies provided their first (or only) measure of biomarkers between 12 and 60 months. Most (n = 56) of the included studies featured only one period of biomarker assessment; 13 studies assessed biomarkers at two or more timepoints (see Tables 1, 2, 3).
3.3.1. Maternal immune system
Eighteen studies examined maternal immune system biomarkers (see Table 1). There were 44 unique biomarkers examined across these 18 studies. Each study measured between one and 35 unique maternal immune system biomarkers. The most frequently examined maternal immune system biomarkers were cytokines IL‐6 (n = 15 studies), TNF‐α (n = 9 studies), and CRP (n = 8 studies). These biomarkers for each study are presented as part of Tables 1 and 4. Maternal immune functioning was most frequently assessed through blood samples (i.e., plasma, serum, or whole blood samples; n = 17 studies). Across studies, samples were most frequently analyzed using a form of immunoassay or gas chromatography.
3.3.2. Infant immune system
There were 40 studies that examined infant immune biomarkers (see Table 2). Each study measured between one and 38 unique immune system biomarkers. The most frequently examined immune system biomarkers were cytokines IL‐6 (n = 28 studies), IL‐8 (n = 26 studies), TNF‐α (n = 25 studies), and IL‐1β (n = 21 studies). Biomarkers examined in each study are presented as part of Tables 2 and 5. Infant immune functioning was most frequently assessed through infant blood samples (plasma, serum, or whole blood samples; n = 27 studies), or cerebrospinal fluid and saliva samples (n = 4 studies). Six studies included multiple biological sources to observe immune system functioning. These samples were most frequently analyzed using a form of immunoassay or gas chromatography.
3.3.3. Infant gut microbiome
Thirteen studies examined the link between the gut microbiome and neurodevelopmental outcomes in the first 5 years of life (see Tables 3 and 6). These studies identified differences in gut microbial composition (n = 1) and alpha diversity (n = 10) and in subsequent analysis identified 96 genera/species‐specific group differences. These changes were found in the phyla of Firmicutes (n = 49), Proteobacteria (n = 12), Bacteroidetes (n = 9), Actinobacteria (n = 8), Verrucomicrobia (n = 1), and Aquificae (n = 1).
3.4. Neurodevelopmental and behavioral outcomes
Children's neurodevelopment was most frequently assessed via behavior‐rating scales used to measure a range of functional outcomes, including cognition, fine and gross motor skills, communication and language skills, social skills, memory, intelligence, adaptive functioning, and temperament. The most frequently used neurodevelopmental assessment was the Bayley Scales of Infant Development (BSID) (n = 32), followed by the Infant Behavior Questionnaire (n = 8), Mullen Scales of Early Learning (n = 7), Ages and Stages Questionnaire (n = 5), and Weschler Preschool and Primary Scale of Intelligence (n = 3). The remaining methods of neurodevelopmental assessment include other forms of behavioral rating scales, or lab‐based observational assessments. In addition, a small number of studies also included additional physiological measures (e.g., fMRI). Most studies used only one measure of neurodevelopment. The measures used to assess neurodevelopmental outcomes are presented as part of Tables 1, 2, 3.
The age of neurodevelopmental assessment ranged from 3 to 60 months, with approximately 72% of studies having one assessment between 12 and 36 months. Neurodevelopmental outcomes were assessed at a single timepoint in 61 studies, four studies included two timepoints, two studies included three timepoints, and two studies performed neurodevelopmental assessments at four timepoints.
3.5. Associations between biomarkers with neurodevelopmental and/or behavioral outcomes
Few studies integrated biological information across mothers and their infants or measured both the immune system and gut microbiome. Therefore, for greater interpretability, the included studies were grouped and described according to the source (i.e., mother and/or infant) and system (i.e., immune system and/or microbiome). Across each of the 69 included studies, we observed substantial heterogeneity across most study components. Specifically, studies were highly varied with respect to the research design enlisted, sample population recruited, the timing and analytic procedures used to quantify biomarkers, and the timing and methods of neurodevelopmental assessment. A key implication of this variability across the included studies was the identification of mixed results and subsequent conclusions derived about how the immune system and gut microbiome were associated with early dimensions of neurodevelopment. No single biomarker(s) were robustly and consistently associated with neurodevelopmental outcomes (e.g., cognition, language, motor development, social–emotional development, or adaptive behavior).
3.5.1. Maternal immune biomarkers and associations with neurodevelopment
Of the 18 studies that quantified associations between the maternal immune system and infant neurodevelopment, four studies measured maternal immune activity in trimester two, and 10 studies in trimester three. Only four studies included assessment across trimesters (Rudolph et al., 2018, Bodnar et al., 2018, Graham et al., 2018, Rasmussen et al., 2019). These studies enlisted a methodology whereby IL‐6 concentrations obtained over trimesters were averaged to yield an approximate average of infant exposure to IL‐6 over pregnancy. While multiple assessments are more resource intensive, they offer critical insights regarding infant's exposure to inflammation—particularly because IL‐6 concentrations may have natural variation on any given day, or as a response to an atypical event during pregnancy (e.g., a recent period of increased stress, or recovering from a brief illness). However, it does appear that IL‐6 concentrations over pregnancy (in studies that included assessment over multiple trimesters) are relatively consistent, which is exemplified in one study that demonstrated strong correlations (r = .55–.68; or 30%–46% shared variance) between IL‐6 levels at the first, second, and third trimesters (Rudolph et al., 2018). Although strong by conventional standards, it does remain important to acknowledge that somewhere between 54% and 70% of the variability in IL‐6 concentrations over time is not explained by earlier levels, thus underscoring the need for multiple assessments during pregnancy, where possible. Importantly, while demonstrated for IL‐6, less is known about the stability of different immune system biomarkers over these trimesters in the context of the current review (i.e., associations with neurodevelopment).
The most frequently examined immune biomarkers were those most typically defined as pro‐inflammatory (i.e., IL‐6, TNF‐α, and CRP). IL‐6 was measured in 15 of the 18 studies; TNF‐α and CRP were measured in 10 and eight of the included studies, respectively. While the heterogeneity across these studies limits our ability to make clear inferences about the significance of any specific biomarkers or co‐expression profiles in shaping specific neurodevelopmental outcomes during early life, some preliminary trends were identified. These are displayed in Table 7. For example, maternal IL‐6 was inversely associated with offspring cognitive outcomes in six out of eight studies, language (three out of four studies), and emotional development (seven out of nine studies). Maternal TNF‐α was inversely associated with offspring cognition and language (three out of four studies, each), motor development (three out of five studies), and emotional development (five out of six studies). Most of the studies to identify inverse associations between maternal inflammatory biomarkers and infant neurodevelopmental outcomes involved healthy mothers, although studies that also enlisted clinical maternal samples (e.g., maternal HIV, alcohol exposure in utero, or at risk for a psychiatric condition) also yielded similar directional findings. These findings are broadly suggestive of the adverse neurodevelopmental consequences associated with increased levels of maternal inflammation. Interestingly, the only study to find a positive association between maternal IL‐6 and CRP with infant cognitive development was unable to fully account for these findings, though the authors speculated that two possible causes may be that IL‐6 is a part of both pro‐inflammatory and anti‐inflammatory pathways, or that infants may mobilize adaptive neurodevelopmental responses when exposed to maternal immune activation (e.g., exposure to elevated but not clinical or sustained inflammation may induce resilience responses in later life) (Spann et al., 2018). However, most studies focused on a smaller number of inflammatory biomarkers, meaning less is known about other inflammatory biomarkers, anti‐inflammatory biomarkers, and their co‐expression.
With many of the included studies statistically underpowered to detect statistically significant effects that are small‐to‐medium in size, it was perhaps unsurprising to note that most included studies—while providing an indication of directional associations—did not reach statistical significance. Among these maternal immune studies, three studies did include large sample sizes (>1000), which may provide insights not attenuated by sample size (Irwin et al., 2019, Monthe‐Dreze et al., 2019, Yan et al., 2020). Even among these large‐scale studies, many of the observed effects were small and/or nonsignificant. For example, the largest study (N = 1453) found some inverse associations between specific biomarkers (i.e., IL‐1β, IL‐10) with neurodevelopmental outcomes, while others were positively associated (i.e., TARC, sFlt‐1, VEGF‐D), though the most frequently examined biomarkers (e.g., IL‐6, TNF‐α, and CRP) were not significantly associated with neurodevelopmental outcomes (Irwin et al., 2019).
3.5.2. Infant immune biomarkers and associations with neurodevelopment
Among the 40 studies examining associations between infant immune biomarkers and neurodevelopmental outcomes, pro‐inflammatory biomarkers were the most extensively evaluated across studies. Measures of IL‐6, IL‐8, TNF‐α, and IL‐1β were each assessed in over half of the 40 included studies, while CRP and IL‐10 were each assessed in 14 studies. Cognitive and motor domains, followed by language domains, were the most frequently assessed outcomes in studies examining infant immune biomarkers. Associations between infant immune biomarkers and adaptive, emotional, or social domains were assessed with less frequency. One of the broad findings was that biomarkers generally recognized as pro‐inflammatory (i.e., IL‐6, IL‐8, TNF‐α, CRP, and IL‐1β) were more frequently found to be inversely associated with cognitive development. For example, Table 5 demonstrates that IL‐6 was inversely associated with better cognitive outcomes in 13 of the 24 studies where it was assessed.
TNF‐α was inversely associated with cognitive development in 11 of the 21 studies (two studies showing a positive association, eight nonsignificant associations), while IL‐8 was inversely associated with cognitive outcomes in 12 of the 21 studies where it was measured (four studies showing a positive association, four nonsignificant, and one mixed result). For the neurodevelopmental domain of motor development, inverse associations were most observed for IL‐8 (seven of 15 studies), TNF‐α and IL‐1β (eight of 14 studies, each), and CRP (six of eight studies). For language, five of the 10 studies measuring associations between IL‐6 and language found evidence for inverse associations, while the remaining five found no significant linear association; however, most studies that included measures of IL‐8, TNF‐α, IL‐1β, CRP, and IL‐10 more frequently reported inverse associations with language. The variability in strength and direction of associations between biomarkers and neurodevelopmental outcomes was present across the sample source (cord blood, serum, plasma, etc.) and method of quantifying the biomarkers contained within these samples. Based on the reviewed studies, no specific sampling or analytic strategy resulted in consistent or more pronounced associations between specific biomarkers and neurodevelopmental outcomes in children.
Inferences about the role of less‐frequently examined biomarkers are more challenging to extrapolate, yet may warrant further investigation. For example, each study measuring SAA and MMP‐9 (four studies, each) reported inverse associations with children's cognitive development. Similarly, four of the five studies assessing VEGF and MIP‐1β also reported inverse associations with cognitive outcomes.
Consistent with the other included studies, most studies measuring infant immune biomarkers and neurodevelopment enlisted single‐timepoint assessments of the immune system, constraining the ability to make inferences on periods of heightened sensitivity (or resilience) to inflammation. Among the included studies, immune biomarkers were measured in some studies as early as birth (e.g., from cord blood) and as late as 4 years of age in others—a period spanning significant developmental changes for children. Multiple assessments of immune biomarkers over time were achieved in a smaller number of these included studies, though variation across these studies with respect to the context, timing, method, biomarkers, and outcomes examined limits our ability to draw comparisons across studies. Taken broadly, the current results suggest that certain immune biomarkers measured during early life are often found to be inversely associated with development in the cognitive, motor, and language domains, though the current heterogeneity of available studies constrains our ability to synthesize these findings beyond observing trends.
3.5.3. Infant gut microbiome and associations with neurodevelopment
Table 6 provides a summary of gut microbiome studies. There were high levels of variability among the 13 studies that examined the association between the infant gut microbiome and neurodevelopment. Table 6 provides the information regarding the specific composition (i.e., alpha and/or beta diversity) or phylum of the gut microbiome investigated within each study, and the reported strength and direction of association with neurodevelopmental domains. Regarding compositional measures, alpha diversity described the amount of microbiome diversity within a single subject, while beta diversity described the amount of microbiome diversity between multiple subjects. All 13 studies provided at least one measure of alpha diversity and five of these studies provided a measure of beta diversity. Several of the included studies were able to report on associations at the phylum level (e.g., Aquificae, Firmicutes, Bacteroidetes, Actinobacteria, Proteobacteria, Verrucomicrobia, Coriobacteriaceae) and at the genera level (Table 9). The strengths of the reported associations between specific genera and neurodevelopmental outcomes were largely comparable with the strength of the associations between the broader measures of overall gut microbiome diversity (i.e., alpha and beta diversity indices).
The descriptions of studies reported in Table 3 provide key information for each study, including the age at which the gut microbiome was assessed. Changes to the composition of the gut microbiome over early life are expected as infants transition out of a mostly milk‐based diet and are introduced to new foods. For example, one study demonstrated nonsignificant associations between 1‐ and 6‐month gut microbiome composition with behavioral outcomes at 24 months, though the composition of infant gut microbiomes at 12 months was associated with behavioral outcomes at 24 months (Loughman et al., 2020). Although the variability in gut microbiome composition across these two timepoints was not compared statistically, these findings suggest that the early infant gut microbiome may be more weakly associated with later neurodevelopment, potentially attributable to the greater homogeneity of gut microbiomes across infants (e.g., as most infants will be breastfed). However, as these infants are acclimated to different foods and other sources over time, important variability in the gut microbiome may then contribute to neurodevelopmental differences. For example, one of the included studies established relationships between gut microbiome at 12 months and later neurodevelopment—though earlier assessments were not obtained (Laue et al., 2020).
All but two studies accounted for mode of delivery, either adjusting for this variable as a covariate in their models or comparing associations between the gut microbiome with neurodevelopmental outcomes between different delivery methods (i.e., natural birth or caesarean birth). One study obtained very early assessment of the infant microbiome (i.e., within the first 4 weeks of life), two studies assessed the microbiome of infants at 4 weeks, four studies performed assessments between 8 and 12 weeks, one longitudinal study assessed the gut microbiome at two periods (3–6 months, and again at 36 months), three studies at 12 months, and two studies assessed the gut microbiome between 18 and 36 months. Most of these studies assessed the infant microbiome at a single timepoint. Neurodevelopmental outcomes were assessed as early as 25 days of age, and as late as 36 months of age, which also has similar implications for interpretation.
Taking study variability into account, some very broad results can be extrapolated. First, like the immune biomarkers, no measure of the gut microbiome was consistently associated with neurodevelopmental outcomes among the included studies. Weak or nonsignificant associations were observed with greater frequency compared to those studies demonstrating large or significant associations. Two studies reported inverse associations between alpha diversity and children's cognitive development, though the overall pattern of findings yielded inconclusive results. A similar pattern of findings was also observed for children's language and motor development. Where significant associations with neurodevelopmental outcomes were identified, these were most frequently observed with the domain of social–emotional development (present in four of the 13 studies). However, it remains important to examine the specific details of each study given the variability in which they were conducted.
4. DISCUSSION
The aim of this review was to summarize existing studies that examined associations between gut microbiome or immune system biomarkers and neurodevelopmental outcomes spanning the first 5 years of life. The 69 included studies enlisted a diverse range of populations and methodologies to evaluate 81 unique biomarkers that had substantial variability in their association with neurodevelopmental outcomes. Our results broadly support the presence of associations between certain biomarkers and later neurodevelopmental outcomes, yet the strength and direction of these associations are heterogeneous. The presence of these associations highlights the potential to generate insights into early biological processes associated with later neurodevelopmental risk. However, the ability to synthesize evidence to identify consistent findings is diminished by the substantial variability in study populations and methodologies, and a tendency to provide a “snapshot” of biomarkers from either maternal or infant sources. Specifically, the method and timing in which biomarkers were measured, the type and age in which neurodevelopmental outcomes were assessed, and the statistical procedures used to test hypotheses differed greatly between studies, making findings difficult to compare.
One important observation of the current literature is that it is dominated by studies that measured biomarkers at a single timepoint. This represents a major limitation as the ability to distinguish changes to the gut or immune function over time is missed, which likely have different implications for subsequent associations with later neurodevelopmental outcome. Longitudinal studies with repeated biomarker assessments are required to establish baseline immune activation and identify the impacts of chronic and acute inflammation. Such studies would also allow us to determine whether there are developmental windows of greater vulnerability when such perturbations may have differential effects. For example, in utero events may be expected to be different from postnatal events, though these were seldom included within the available studies. Longitudinal studies with repeated assessments are critical to understanding the potential sequential or cumulative impacts of disruptions to immune or gut microbiome function at different timepoints.
4.1. Immune system and gut microbiome biomarkers remain poorly integrated in understanding early neurodevelopment
Interactions between the immune system and gut microbiome are often described in the broader human biology literature (Pronovost & Hsiao, 2019). However, studies that have tested the potential modulating associations between these systems are not well‐represented, especially in humans. In the current review, only one study was identified that examined both immune and gut microbiome biomarkers; in this study, only independent associations between immune and gut microbiome biomarkers with later neurodevelopment were reported, meaning that potential interactions between biomarkers across these systems were not investigated (van den Berg et al., 2016). This is a critical gap in the research given the implications of gut–immune interactions for neurodevelopment. For example, the neurodevelopmental implications of gut microbiome dysbiosis may vary according to how an individual's immune system responds to this challenge. A healthy immune system may respond to these challenges adaptively and therefore reduce the likelihood of significant neurodevelopmental detriment. Inversely, a dysregulated immune system may compound the impact of gut microbiome dysbiosis, as well as induce further neurodevelopmental risk via the production of harmful pro‐inflammatory response. Further research in this area may help to generate novel insights and further opportunities for intervention, such as new methods of promoting healthy immune functioning via microbiome‐oriented interventions that can deliver downstream benefits to neurodevelopmental functioning. However, we acknowledge that the development of methods to meaningfully integrate information from both the immune system and microbiome embodies the much broader challenge in adopting multiomics approaches to understanding the development of health and disease (Pinu et al., 2019).
4.2. Interactions between biomarkers may offer further insight into early neurodevelopmental processes
While none of the included studies examined interactions between biomarkers between the immune system and gut microbiome, there were a small number of studies that did examine interactions between biomarkers within either system. These methods are particularly valuable, as many of the methods used to quantify specific biomarkers can assess multiple markers at the same time. However, many of these studies were based on “snapshots” (i.e., assessments at a single timepoint), which have noted limitations given the variable nature of immune and gut microbiome activity. Nevertheless, these exploratory studies offer potentially novel insights into how the interactions between biomarkers may be a superior predictor of neurodevelopmental outcome compared to biomarkers in isolation.
For example, one large scale (n = 634) cross‐sectional study examined the individual associations between seven pro‐inflammatory biomarkers (e.g., IL‐6, TNF‐α, IL‐1β) and one anti‐inflammatory biomarker (IL‐10) with several cognitive and motor outcomes in infants at 4 years of age (Kyriklaki et al., 2019). Consistent with findings from other studies included in this review, the associations between individual biomarkers and cognitive outcomes were weak and nonsignificant. However, a higher ratio of TNF‐α to IL‐10 was a strong and significant predictor of poorer motor development and several domains of cognitive development, including memory, executive function, and overall cognitive ability (Kyriklaki et al., 2019). Additional support for the interaction between maternal immune biomarkers was provided by another study (Sowell et al., 2018), where elevated ratios of TNF‐α to IL‐10, and IL‐6 to IL‐10, in late pregnancy predicted poorer neurodevelopment (cognitive and motor development) at 12 months of age, though individual levels of TNF‐α, IL‐10, and IL‐6 were not associated with neurodevelopment (Sowell et al., 2018). One explanation for why the ratio of pro‐inflammatory to anti‐inflammatory markers may be a better predictor of neurodevelopmental outcomes is that the effects of inflammation may be greater if there is a diminished counterregulatory anti‐inflammatory response. While crude, previous research has described how this “imbalance” may give rise to poor neurodevelopment, including the onset of neurodevelopmental disorders (Jiang et al., 2018). Consequently, direct associations between specific inflammatory biomarkers (e.g., IL‐6 or TNF‐α) may be moderated by levels of anti‐inflammatory biomarkers (e.g., IL‐10). This notion may also help to partially explain the somewhat inconsistent pattern of associations between specific biomarkers and early neurodevelopmental outcomes, and why many of the direct associations between specific biomarkers and neurodevelopmental outcomes are often small. Therefore, further examination of the interaction between immune system biomarkers may represent an important next step in this field of research. Our review identified only two studies that included ratios of inflammatory to anti‐inflammatory markers. However, 18 other studies included in this review measured IL‐10, IL‐6, and/or TNF‐α. The existing availability of these data creates opportunities for the exploration of interactions between inflammatory and anti‐inflammatory biomarkers on early neurodevelopment.
While measures of microbial diversity were common, comparisons between studies, or even within the same study, can prove challenging. Data‐driven approaches to identify various microbial “clusters” can help to classify subjects within a study; however, the replicability of these clusters is generally limited due to well‐established variations in infant microbiota attributed to genetic, environmental, and geographic reasons. Further, while the prospective association between early microbiome and later neurodevelopment was a focus of this review, the potential for reverse (i.e., neurodevelopment impacting the microbiome) or bidirectional relationships should not be dismissed. However, none of the studies identified in the current review tested bidirectional associations.
4.3. Direct associations between biomarkers and neurodevelopment were usually small
Broadly, the results of the 69 studies included in this review highlighted that direct associations between specific biomarkers and early neurodevelopmental outcomes were often small, or small‐to‐moderate in size. Many of these associations failed to reach statistical significance, with most studies enlisting traditional null hypothesis significance testing approaches that use the conventional p < .05 level to determine significance. Furthermore, while there was a trend toward increased inflammatory activity being associated with poorer neurodevelopmental outcomes, the reverse relationship was reported in some studies. These findings may reflect the lack of meaningful associations between immune and gut microbiome development and neurodevelopmental outcomes, in addition to more traditional methodological reasons (e.g., measurement error). They may also be explained by several other factors. For example, Spann et al. (2018) proposed that the positive association between inflammatory markers and better cognitive outcomes observed in their study may represent an adaptive “resilience” response that makes the brain less vulnerable to future challenges.
It is also important to consider that the longitudinal nature of most of the included studies provided an assessment of early immune and/or gut microbiome functioning and an assessment of neurodevelopment in later life. These assessment periods could be years apart, allowing greater opportunities for the effects of extraneous factors (biological, environmental, or genetic) to additionally influence neurodevelopment. Therefore, it is not surprising to see stronger correlations observed in cross‐sectional studies when compared to longitudinal studies of increasing duration between assessment periods. Additionally, as noted in a previous review of studies examining associations between biomarkers and neurodevelopment (Nazzari & Frigerio, 2020), the use of inadequate statistical analyses pervades this area of research. Many studies included in this review likely lacked the statistical power necessary to detect statistically significant effects, meaning that potentially important associations are missed. This may be further problematic in studies where interpretations and directions for future research focus on those biomarkers with statistically significant associations with neurodevelopment.
A generic recommendation that researchers ensure sample sizes are sufficiently sized is only partially useful given the often‐nuanced samples of interest and resources needed to collect both biological and neurodevelopmental information. Consequently, an additional recommendation is for researchers to consider complementary approaches to perform and interpret research using smaller sample sizes. One method may be to place greater emphasis on the strength of an observed effect, as traditional approaches to statistical significance (i.e., null hypothesis significance testing) are highly influenced by sample size. The strength of nonsignificant associations between biomarkers and neurodevelopmental outcomes is not always reported, and subsequently represents a limitation among existing studies. Among our included studies, it was unsurprising to see many of the included studies failing to reach statistical significance. Similarly, many of the included studies with large sample sizes (e.g., 500+ participants) often reported significant associations with neurodevelopmental outcomes, irrespective of the size of the observed effect (Leviton et al., 2016, Leviton et al., 2013, Leviton et al., 2019, O'Shea et al., 2012, O'Shea et al., 2014). Awareness of the influence of sample size on statistical significance is critical in ensuring that the developments in the field are not solely guided by the results of large studies where even trivial associations may be of statistical significance.
4.4. Associations may be indirect
The present review constrained the definition of neurodevelopment to assessments that rely on observable behaviors. While a strength of many of the included studies was the use of gold standard, clinician‐administered assessments of neurodevelopment (i.e., the Bayley Scales), it is important to acknowledge that these assessments represent the behavioral manifestation that is driven in part by the function of the infant brain. The sequence in which changes to the immune system or gut microbiome trigger structural or functional changes to brain activity that then manifest in behavioral atypicality remains comparatively underexamined. For example, one of the included studies provided evidence of an indirect effect of maternal IL‐6 in pregnancy on infant cognition at 12 months of age via alterations to frontolimbic white matter (Rasmussen et al., 2019), while another study demonstrated how alpha diversity in the gut microbiome could predict changes to the amygdala that then manifest in observable behavioral changes (Gao et al., 2019). Although there were few studies that included objective measures of the immune system and/or gut microbiome, objective indicators of brain functioning (e.g., fMRI), and a neurodevelopmental assessment, the available evidence is consistent with the well‐established notion that behavioral assessments of neurodevelopment are driven by—but not a perfect representation of—the structure and function of the brain (Rasmussen et al., 2019, Gao et al., 2019, Spann et al., 2018). Future research that further explores relationships between these biomarkers and both structural and behavioral indicators of neurodevelopment will help to better explain how biological systems are linked.
4.5. Associations between biomarkers and neurodevelopmental outcomes are varied
Perhaps unsurprisingly, the heterogeneity of included studies was matched with mixed findings regarding the strength and direction of associations between observed biomarkers and neurodevelopmental outcomes. To provide an illustrative example, one study reported a positive association between levels of maternal CRP in the third trimester and cognitive development assessed by the BSID‐III at 12 weeks postbirth (Nazzari et al., 2020), while another reported a negative association between third‐trimester CRP and cognition assessed by the BSID at 14 months postbirth (Gao et al., 2019). A multitude of plausible (and not mutually exclusive) reasons may explain such discrepancies, including but not limited to the analytic method used to quantify CRP, the timing of the neurodevelopmental assessment, individual variation, geographic context, clinical characteristics of the sample, or differences in the quantitative methods employed. This example is symbolic of the broader findings described in the narrative and tabular summaries of the included findings, where the strength and direction of associations between biomarkers and neurodevelopment were varied—even among the most frequently examined biomarkers (e.g., IL‐6, CRP, alpha diversity). While the size and scope of the studies retained for this scoping review do not enable explanations for these variations to be clearly identified, the current findings underscore the need for future work to focus on capturing and interrogating genetic, environmental, and temporal influences on the associations between specific biomarkers and later developmental outcomes, as well as on capturing interactions between biomarkers. Such lines of enquiry are perhaps best supported by large‐scale, collaborative studies with standardized protocols for data capture, analysis, and reporting.
4.6. Neurodevelopmental outcomes are diverse
Our scoping review characterized the existing literature as one of significant heterogeneity. However, a high degree of consistency in the operationalization of neurodevelopmental outcome in the first 5 years of life was examined. Specifically, most of the included studies enlisted the BSID. This clinician‐administered instrument provides an indication of a child's development in the domains of motor skills, cognition, and language. The measure is widely used, is available in multiple languages, and typically demonstrates favorable psychometric properties and is the “gold standard” measure of children's neurodevelopment during this developmental period. However, it is important to consider that neurodevelopment continues to be variable into later development, meaning that BSID scores in early life are not deterministic of neurodevelopment in later childhood (Luttikhuizen dos Santos et al., 2013). Moreover, other potentially important dimensions of neurodevelopment (e.g., regulation or negative emotionality) may be underrepresented. Consequently, previous evidence suggesting that the immune system and gut microbiome may shape other dimensions related to neurodevelopment, such as temperament, emotionality, attachment, and social responsiveness, will also require further investigation. These outcomes may be particularly relevant when considering the role of systems biology in providing insight into risk and resilience for childhood mental illness.
4.7. Strengths and limitations of the current review
Preregistration of the scoping review protocol and adherence to best‐practice scoping methodology helped to increase rigor of the current review. A further strength of this review was the comprehensiveness of the literature search, with 23,495 individual titles and abstracts screened for eligibility. Understandably, an inevitability of this thorough search is that the time‐intensive nature of this process means that a smaller number of additional studies may be published between the date of the search and the publication of this manuscript. A limitation of this review was the exclusion of unpublished literature and studies in the non‐English language. While the studies we identified in this review included a mix of statistically significant and nonsignificant associations between specific biomarkers and early neurodevelopment, the potential for publication bias may still impact the broadband associations that these studies point toward. Assessments of study quality and publication bias are seldom included as part of the scoping review process but may serve to complement future evidence synthesis studies in future studies that may enlist a narrower scope that permits a meta‐analysis of findings; systematic review and meta‐analysis of the included studies were not appropriate due to the heterogeneity of included studies and purpose of this scoping review.
4.8. Future recommendations
Well‐established associations between the human immune system and gut microbiome have not yet been comprehensively investigated in the context of early neurodevelopment. It currently remains unclear how the interactions between these two systems may modulate brain development during this particularly sensitive neurodevelopmental window. Studies including measures of immune and gut microbiome biomarkers will enable the identification of critical interactions between these systems and subsequent impact on neurodevelopmental outcomes. This also represents an important step toward adopting a “systems” approach to neurodevelopment, whereby the combined role of biological systems (and their interactions) is already widely described, but seldom quantified in the context of early, transdiagnostic indicators of neurodevelopment (e.g., cognition, language, and motor development). Multiple observations spanning the perinatal period will help researchers overcome potential error and limitations associated with using a single observation of biological functioning—a common approach identified in many of the studies included in this review. Important and individualized biological changes during pregnancy and the early developmental periods are not captured by studies that provide only a “snapshot” of biological functioning at a single point in time but are critical in guiding effective intervention strategies.
5. CONCLUSION
The early neurodevelopmental implications of interactions between the immune system and gut microbiome are not yet well‐understood. The dynamic nature of the maternal and infant immune system and microbiome is also underrepresented due to a few large‐scale, longitudinal studies that integrate biological systems and sources. Studies that begin to evaluate the interaction between biological systems over the early developmental period are necessary to advance our understanding of the developing brain, and to ultimately develop interventions that provide a novel pathway for promoting developmental resilience (Tillisch et al., 2013, Steenbergen et al., 2015, Singh et al., 2014, Kang et al., 2017). Findings within this emerging field have already started to identify candidate biomarkers that are relevant to overall brain development, and in the onset of specific neurodevelopmental conditions (Nazzari & Frigerio, 2020, Parsons et al., 2020, Lima‐Ojeda et al., 2017). Much of this direction has been guided by the results of animal studies. However, the benefits of harnessing these biomarkers to aid in the very early identification of neurodevelopmental risk in humans has not yet been fully actualized (Nahmias et al., 2019, Puthussery et al., 2018). As the human body is populated by a vast number of unique biomarkers that share complex interactions with other biomarkers within—and between—biological systems, the identification of specific biomarkers that are robustly and meaningfully associated with early neurodevelopment represents an important, but challenging, scientific undertaking.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supporting information
Supplementary File 1 Search Terms and Results from Embase (Ovid Interface) Database Search
Supplement Material
ACKNOWLEDGMENTS
This manuscript was prepared with support from (1) the Western Australian Child Health Research Fund and (2) the Raine Medical Research Foundation awarded to Amy Finlay‐Jones.
Open access publishing facilitated by Curtin University, as part of the Wiley ‐ Curtin University agreement via the Council of Australian University Librarians.
Mancini, V. O. , Brook, J. , Hernandez, C. , Strickland, D. , Christophersen, C. T. , D'Vaz, N. , Silva, D. , Prescott, S. , Callaghan, B. , Downs, J. , & Finlay‐Jones, A. (2023). Associations between the human immune system and gut microbiome with neurodevelopment in the first 5 years of life: A systematic scoping review. Developmental Psychobiology, 65, e22360. 10.1002/dev.22360
DATA AVAILABILITY STATEMENT
All files associated with this review are available at Open Science Framework https://osf.io/nk72p/.
Index
| Immune biomarkers | Inflammatory | |
|---|---|---|
| CD64 | Cluster of Differentiation 64 | |
| CRP | C‐reactive protein | pro |
| E‐SEL | E‐selectin | |
| GCSF | Granulocyte colony‐stimulating factor | |
| GDNF | Glial cell line‐derived neurotrophic factor | |
| GM‐CSF | Granulocyte‐macrophage colony‐stimulating factor | |
| ICAM‐1 | Intercellular adhesion molecule 1 | |
| ICAM‐3 | Intercellular adhesion molecule 3 | |
| IFN‐γ | Interferon gamma | anti |
| IGFBP‐1 | Insulin‐like growth factor‐binding protein 1 | |
| IL‐1 | Interleukin‐1 | |
| IL‐10 | Interleukin‐10 | anti |
| IL‐12 | Interleukin‐12 | |
| IL‐12p70 | Interleukin‐12p70 | |
| IL‐13 | Interleukin‐13 | |
| IL‐16 | Interleukin‐16 | |
| IL‐17 / IL‐17A | Interleukin‐17 | |
| IL‐17α | Interleukin‐17α | |
| IL‐18 | Interleukin‐18 | |
| IL‐1RA | Interleukin‐1 receptor antagonist | |
| IL‐1β | Interleukin‐1β | pro |
| IL‐2 | Interleukin‐2 | anti |
| IL‐4 | Interleukin‐4 | anti |
| IL‐5 | Interleukin‐5 | anti |
| IL‐6 | Interleukin‐6 | pro |
| IL‐6R | Interleukin‐6 receptor | |
| IL‐7 | Interleukin‐7 | |
| IL‐8 | Interleukin‐8 | |
| IL‐9 | Interleukin‐9 | |
| IL12p40 | Interleukin‐12p40 | |
| IP‐10 | Interferon gamma‐induced protein 10 | |
| M‐CSF | Macrophage colony‐stimulating factor | |
| MCP‐1 | Monocyte chemoattractant protein‐1 | pro |
| MCP‐4 | Monocyte chemoattractant protein‐4 | |
| MIG | Monokine induced by IFN‐γ | |
| MIP‐1α | Macrophage inflammatory protein‐1α | |
| MIP‐1β | Macrophage inflammatory protein‐1β | |
| MMP‐9 | Matrix metallopeptidase 9 | |
| MPO | Myeloperoxidase | |
| NGAL | neutrophil gelatinase‐associated lipocalin | |
| PAI‐1 | Plasminogen activator inhibitor‐1 | |
| PDGF‐AA | Platelet‐derived growth factor‐AA | |
| PGF2α | Prostaglandin F2α | |
| RANTES | Regulated upon Activation, Normal T Cell Expressed and Presumably Secreted | |
| s‐IgA | Salivary Immunoglobulin A | |
| SAA/sAA | Serum amyloid A | |
| SDF‐1 | Stromal cell‐derived factor 1 | |
| sFLT‐1 | Soluble fms‐like tyrosine kinase‐1 | pro |
| sICAM‐1 | Soluble intercellular adhesion molecule‐1 | |
| sTNFR1 | Soluble tumor necrosis factor receptor‐1 | |
| sTNFR2 | Soluble tumor necrosis factor receptor‐2 | |
| sVCAM‐1 | Soluble vascular cell adhesion molecule‐1 | |
| TARC | Thymus and activation‐regulated chemokine | |
| TNF‐R1 | Tumor necrosis factor receptor 1 | |
| TNF‐R2 | Tumor necrosis factor receptor 2 | |
| TNF‐α | Tumor necrosis factor α | |
| TNF‐β | Tumor necrosis factor β | |
| TPO | Thyroid peroxidase | |
| VEGF | Vascular endothelial growth factor | |
| VEGF‐D | Vascular endothelial growth factor‐D | |
| VEGF‐R1 | Vascular endothelial growth factor receptor 1 | |
| VEGF‐R2 | Vascular endothelial growth factor receptor 2 | |
| Neurodevelopment measures | ||
| ABC | Aberrant Behavior Checklist | |
| ADHD | Attention Deficit Hyperactivity Disorder | |
| ASD | Autism Spectrum Disorder | |
| ASQ | Ages and Stages Questionnaire (several editions) | |
| BASC | Behavior Assessment System for Children (several editions) | |
| BMI | Body Mass Index | |
| BSID | Bayley Scales of Infant Development (II and III Editions) | |
| CBCL | Child Behavior Checklist | |
| CDI | Communicative Development Inventory | |
| CDSC | Child Developmental Scale of China | |
| CI | Confidence Interval | |
| CP | cerebral palsy | |
| CRYPTO | Burden of Cryptosporidiosis | |
| DDST | Denver Developmental Screening Test | |
| ECBQ | Early Childhood Behavior Questionnaire | |
| ELBW | extremely low birth weight | |
| ELGAN | Extremely Low Gestational Age Newborn cohort study | |
| FNIRS | Functional Near Infrared Spectroscopy | |
| GMDS | Griffiths Mental Developmental Scales | |
| HIE | hypoxic‐ischemic encephalopathy | |
| HIV | Human immunodeficiency Virus | |
| IBQ / IBQ‐R | Infant Behavior Questionnaire (‐Revised) | |
| IQ | Intelligence Quotient | |
| KSPD | Kyoto Scale of Psychological Development | |
| MDD | Major Depressive Disorder | |
| MDI | Mental Development Index of the BSID | |
| (f)MRI | (functional) Magnetic Resonance Imaging | |
| MSCA | McCarthy Scales of Children's Abilities | |
| MSEL | Mullen Scales of Early Learning | |
| NAPI | Neurobehavioral Assessment of the Preterm Infant, Revised Second Edition | |
| NBAS | Neonatal Behavioral Assessment Scale | |
| NDD | neurodevelopmental disability | |
| NE | neonatal encephalopathy (?) | |
| NEC | necrotizing enterocolitis | |
| NHST | null hypothesis significance testing | |
| PDI | Psychomotor Development Index of the BSID | |
| PRAM‐D | Psychiatry Research and Motherhood ‐ Depression study | |
| PROVIDE | Performance of Rotavirus and Oral Polio Vaccines in Developing Countries cohort study | |
| SDI | Shannon Diversity Index | |
| SRS | Social Responsiveness Scale (several editions) | |
| TD | Typically Developing | |
| TIMP | Test of Infant Motor Performance | |
| VABS | Vineland Adaptive Behavior Scale | |
| VLBW | very low birth weight | |
| WPPSI / WPPSI‐R | Weschler Preschool and Primary Scale of Intelligence (‐Revised) | |
| WRAVMA | Wide Range Assessment of Visual Motor Abilities | |
REFERENCES
- Aatsinki, A. K. , Kataja, E. L. , Munukka, E. , Lahti, L. , Keskitalo, A. , Korja, R. , Nolvi, S. , Häikiö, T. , Tarro, S. , Karlsson, H. , & Karlsson, L. (2022). Infant fecal microbiota composition and attention to emotional faces. Emotion, 22(6), 1159–1170. [DOI] [PubMed] [Google Scholar]
- Aatsinki, A.‐K. , Lahti, L. , Uusitupa, H.‐M. , Munukka, E. , Keskitalo, A. , Nolvi, S. , O'Mahony, S. , Pietilä, S. , Elo, L. L. , Eerola, E. , Karlsson, H. , & Karlsson, L. (2019). Gut microbiota composition is associated with temperament traits in infants. Brain, Behavior, and Immunity, 80, 849–858. [DOI] [PubMed] [Google Scholar]
- Aboussaleh, Y. , Farsi, M. , El Hioui, M. , & Ahami, A. O. T. (2012). Anemia and obesity coexist among women of reproductive age in north west of Morocco. Mediterranean Journal of Nutrition and Metabolism, 5(3), 213–217. [Google Scholar]
- Abraham, A. , Zagoory‐Sharon, O. , & Feldman, R. (2021). Early maternal and paternal caregiving moderates the links between preschoolers' reactivity and regulation and maturation of the HPA‐immune axis. Developmental Psychobiology, 63, 1482–1498. [DOI] [PubMed] [Google Scholar]
- Ahearne, C. E. , Chang, R. Y. , Walsh, B. H. , Boylan, G. B. , & Murray, D. M. (2017). Cord blood IL‐16 is associated with 3‐year neurodevelopmental outcomes in perinatal asphyxia and Hypoxic‐ischaemic encephalopathy. Developmental Neuroscience, 39(1–4), 59–65. [DOI] [PubMed] [Google Scholar]
- Al Nabhani, Z. , Dulauroy, S. , Marques, R. , Cousu, C. , Al Bounny, S. , Déjardin, F. , Sparwasser, T. , Bérard, M. , Cerf‐Bensussan, N. , & Eberl, G. (2019). A weaning reaction to microbiota is required for resistance to immunopathologies in the adult. Immunity, 50(5), 1276.e5–1288.e5. [DOI] [PubMed] [Google Scholar]
- Ashwood, P. , Krakowiak, P. , Hertz‐Picciotto, I. , Hansen, R. , Pessah, I. , & Van de Water, J. (2011). Elevated plasma cytokines in autism spectrum disorders provide evidence of immune dysfunction and are associated with impaired behavioral outcome. Brain, Behavior, and Immunity, 25(1), 40–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckhed, F. , Roswall, J. , Peng, Y. , Feng, Q. , Jia, H. , Kovatcheva‐Datchary, P. , Li, Y. , Xia, Y. , Xie, H. , Zhong, H. , Khan, M. T. , Zhang, J. , Li, J. , Xiao, L. , Al‐Aama, J. , Zhang, D. , Lee, Y. S. , Kotowska, D. , Colding, C. , … Wang, J. (2015). Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host & Microbe, 17(6), 852–852. [DOI] [PubMed] [Google Scholar]
- Barbosa, S. , Khalfallah, O. , Forhan, A. , Galera, C. , Heude, B. , Glaichenhaus, N. , & Davidovic, L. (2020). Serum cytokines associated with behavior: A cross‐sectional study in 5‐year‐old children. Brain, Behavior, and Immunity, 87, 377–387. [DOI] [PubMed] [Google Scholar]
- Benavides, A. , Bell, E. F. , Georgieff, M. K. , Josephson, C. D. , Stowell, S. R. , Feldman, H. A. , Nalbant, D. , Tereshchenko, A. , Sola‐Visner, M. , & Nopoulos, P. (2022). Sex‐specific cytokine responses and neurocognitive outcome after blood transfusions in preterm infants. Pediatric Research, 91(4), 947–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beversdorf, D. Q. , Stevens, H. E. , & Jones, K. L. (2018). Prenatal stress, maternal immune dysregulation, and their association with autism spectrum disorders. Current Psychiatry Reports, 20(9), 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilbo, S. , & Schwarz, J. (2009). Early‐life programming of later‐life brain and behavior: A critical role for the immune system. Frontiers in Behavioral Neuroscience, 3, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blok, C. A. , Krediet, T. G. , Kavelaars, A. , Koopman‐Esseboom, C. , Vreman, H. J. , & van Bel, F. (2011). Early end‐tidal carbon monoxide levels and neurodevelopmental outcome at 3 years 6 months of age in preterm infants. Developmental Medicine and Child Neurology, 53(12), 1113–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodnar, T. S. , Raineki, C. , Wertelecki, W. , Yevtushok, L. , Plotka, L. , Zymak‐Zakutnya, N. , Honerkamp‐Smith, G. , Wells, A. , Rolland, M. , Woodward, T. S. , Coles, C. D. , Kable, J. A. , Chambers, C. D. , Weinberg, J. , & Collaborative Initiative on Fetal Alcohol Spectrum Disorders (CIFASD) . (2018). Altered maternal immune networks are associated with adverse child neurodevelopment: Impact of alcohol consumption during pregnancy. Brain, Behavior, and Immunity, 73, 205–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borre, Y. E. , O'Keeffe, G. W. , Clarke, G. , Stanton, C. , Dinan, T. G. , & Cryan, J. F. (2014). Microbiota and neurodevelopmental windows: Implications for brain disorders. Trends in Molecular Medicine, 20(9), 509–518. [DOI] [PubMed] [Google Scholar]
- Brestoff, J. R. , & Artis, D. (2013). Commensal bacteria at the interface of host metabolism and the immune system. Nature Immunology, 14(7), 676–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargos, A. C. R. , Mendonca, V. A. , Oliveira, K. S. C. , de Andrade, C. A. , Leite, H. R. , da Fonseca, S. F. , Vieira, E. L. , Teixeira, A. L. Jr. , & Lacerda, A. C. (2017). Association between obesity‐related biomarkers and cognitive and motor development in infants. Behavioural Brain Research, 325, 12–16. [DOI] [PubMed] [Google Scholar]
- Carlo, W. A. , McDonald, S. A. , Tyson, J. E. , Stoll, B. J. , Ehrenkranz, R. A. , Shankaran, S. , Goldberg, R. N. , Das, A. , Schendel, D. , Thorsen, P. , Skogstrand, K. , Hougaard, D. M. , Oh, W. , Laptook, A. R. , Duara, S. , Fanaroff, A. A. , Donovan, E. F. , Korones, S. B. , Stevenson, D. K. , … Eunice Kennedy Shriver National Institute of Child Health and Human Development Neonatal Research Network . (2011). Cytokines and neurodevelopmental outcomes in extremely low birth weight infants. Journal of Pediatrics, 159(6), 919.e3–925.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson, A. L. , Xia, K. , Azcarate‐Peril, M. A. , Goldman, B. D. , Ahn, M. , Styner, M. A. , Thompson, A. L. , Geng, X. , & Gilmore, J. H. (2018). Infant gut microbiome associated with cognitive development. Biological Psychiatry, 83(2), 148–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson, A. L. , Xia, K. , Azcarate‐Peril, M. A. , Goldman, B. D. , Ahn, M. , Styner, M. A. , Thompson, A. L. , Geng, X. , Gilmore, J. H. , & Knickmeyer, R. C. (2018). Infant gut microbiome associated with cognitive development. Biological Psychiatry, 83(2), 148–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerdó, T. , Diéguez, E. , & Campoy, C. (2019). Early nutrition and gut microbiome: Interrelationship between bacterial metabolism, immune system, brain structure, and neurodevelopment. American Journal of Physiology‐Endocrinology and Metabolism, 317(4), E617–E630. [DOI] [PubMed] [Google Scholar]
- Chalak, L. F. , Sanchez, P. J. , Adams‐Huet, B. , Laptook, A. R. , Heyne, R. J. , & Rosenfeld, C. R. (2014). Biomarkers for severity of neonatal hypoxic‐ischemic encephalopathy and outcomes in newborns receiving hypothermia therapy. Journal of Pediatrics, 164(3), 468–474.e461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chorna, O. , Cioni, G. , & Guzzetta, A. (2020). Principles of early intervention. In: Gallagher A, Bulteau C, Cohen D, Michaud JL, eds. Handbook of clinical neurology . Vol, 174. Elsevier, 333–341. [DOI] [PubMed] [Google Scholar]
- Christian, L. M. , Galley, J. D. , Hade, E. M. , Schoppe‐Sullivan, S. , Kamp Dush, C. , & Bailey, M. T. (2015). Gut microbiome composition is associated with temperament during early childhood. Brain, Behavior, and Immunity, 45, 118–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codagnone, M. G. , Spichak, S. , O'Mahony, S. M. , O'Leary, O. F. , Clarke, G. , Stanton, C. , Dinan, T. G. , & Cryan, J. F. (2019). Programming bugs: Microbiota and the developmental origins of brain health and disease. Biological Psychiatry, 85(2), 150–163. [DOI] [PubMed] [Google Scholar]
- Cowan, C. S. M. , Dinan, T. G. , & Cryan, J. F. (2020). Annual Research Review: Critical windows – The microbiota–gut–brain axis in neurocognitive development. Journal of Child Psychology and Psychiatry, 61(3), 353–371. [DOI] [PubMed] [Google Scholar]
- Cryan, J. F. , & de Wit, H. (2019). The gut microbiome in psychopharmacology and psychiatry. Psychopharmacology, 236(5), 1407–1409. [DOI] [PubMed] [Google Scholar]
- Cryan, J. F. , & Dinan, T. G. (2012). Mind‐altering microorganisms: The impact of the gut microbiota on brain and behaviour. Nature Reviews Neuroscience, 13(10), 701–712. [DOI] [PubMed] [Google Scholar]
- Diaz Heijtz, R. (2016). Fetal, neonatal, and infant microbiome: Perturbations and subsequent effects on brain development and behavior. Seminars in Fetal and Neonatal Medicine, 21(6), 410–417. [DOI] [PubMed] [Google Scholar]
- Dietrick, B. , Molloy, E. , Massaro, A. N. , Strickland, T. , Zhu, J. , Slevin, M. , Donoghue, V. , Sweetman, D. , Kelly, L. , O'Dea, M. , McGowan, M. , Vezina, G. , Glass, P. , Vaidya, D. , Brooks, S. , Northington, F. , & Everett, A. D. (2020). Plasma and cerebrospinal fluid candidate biomarkers of neonatal encephalopathy severity and neurodevelopmental outcomes. Journal of Pediatrics, 226, 71–79.e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díllí, D. , Eras, Z. , Dilmen, U. , & Sakrucu, E. D. (2013). Neurodevelopmental evaluation of very low birth weight infants with sepsis at 18 to 24 months' corrected age. Indian Pediatrics, 50(3), 327–330. [DOI] [PubMed] [Google Scholar]
- Estes, M. L. , & McAllister, A. K. (2016). Maternal immune activation: Implications for neuropsychiatric disorders. Science, 353(6301), 772–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etheredge, A. J. , Manji, K. , Kellogg, M. , Tran, H. , Liu, E. , McDonald, C. M. , Kisenge, R. , Aboud, S. , Fawzi, W. , Bellinger, D. , Gewirtz, A. T. , & Duggan, C. P. (2018). Markers of environmental enteric dysfunction are associated with neurodevelopmental outcomes in Tanzanian children. Journal of Pediatric Gastroenterology and Nutrition, 66(6), 953–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay‐Jones, A. , Varcin, K. , Leonard, H. , Bosco, A. , Alvares, G. , & Downs, J. (2019). Very early identification and intervention for infants at risk of neurodevelopmental disorders: A transdiagnostic approach. Child Development Perspectives, 13(2), 97–103. [Google Scholar]
- Foster, J. A. , Rinaman, L. , & Cryan, J. F. (2017). Stress & the gut‐brain axis: Regulation by the microbiome. Neurobiology of Stress, 7, 124–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman, R. , Hunter, S. K. , Law, A. J. , Wagner, B. D. , D'Alessandro, A. , Christians, U. , Noonan, K. , Wyrwa, A. , & Hoffman, M. C. (2019). Higher gestational choline levels in maternal infection are protective for infant brain development. Journal of Pediatrics, 208, 198–206.e192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguli, S. , & Chavali, P. L. (2021). Intrauterine viral infections: Impact of inflammation on fetal neurodevelopment. Frontiers in Neuroscience, 15(1509), 771557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, W. , Salzwedel, A. , Carlson, A. , Xia, K. , Azcarate‐Peril, M. A. , Styner, M. A. , Thompson, A. L. , Geng, X. , Goldman, B. D. , Gilmore, J. H. , & Knickmeyer, R. C. (2019). Gut microbiome and brain functional connectivity in infants—A preliminary study focusing on the amygdala. Psychopharmacology, 236(5), 1641–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghassabian, A. , Sundaram, R. , Chahal, N. , McLain, A. C. , Bell, E. M. , Lawrence, D. A. , Gilman, S. E. , & Yeung, E. H. (2018). Concentrations of immune marker in newborn dried blood spots and early childhood development: Results from the Upstate KIDS Study. Paediatric and Perinatal Epidemiology, 32(4), 337–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham, A. M. , Rasmussen, J. M. , Rudolph, M. D. , Heim, C. M. , Gilmore, J. H. , Styner, M. , Potkin, S. G. , Entringer, S. , Wadhwa, P. D. , Fair, D. A. , & Buss, C. (2018). Maternal systemic interleukin‐6 during pregnancy is associated with newborn amygdala phenotypes and subsequent behavior at 2 years of age. Biological Psychiatry, 83(2), 109–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson, H. C. , Holton, K. F. , Anderson, A. N. , Nousen, E. K. , Sullivan, C. A. , Loftis, J. M. , Nigg, J. T. , & Sullivan, E. L. (2019). Increased maternal prenatal adiposity, inflammation, and lower omega‐3 fatty acid levels influence child negative affect. Frontiers in Neuroscience, 13, 1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson, H. C. , Sullivan, E. L. , Nousen, E. K. , Sullivan, C. A. , Huang, E. , Rincon, M. , Nigg, J. T. , & Loftis, J. M. (2018). Maternal prenatal depression predicts infant negative affect via maternal inflammatory cytokine levels. Brain, Behavior, and Immunity, 73, 470–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, L. K. H. , Tong, V. J. W. , Syn, N. , Nagarajan, N. , Tham, E. H. , Tay, S. K. , Shorey, S. , Tambyah, P. A. , & Law, E. C. N. (2020). Gut microbiota changes in children with autism spectrum disorder: A systematic review. Gut Pathogens, 12(1), 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper, L. V. , Littman, D. R. , & Macpherson, A. J. (2012). Interactions between the microbiota and the immune system. Science, 336(6086), 1268–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter, S. K. , Hoffman, M. C. , D'Alessandro, A. , Noonan, K. , Wyrwa, A. , Freedman, R. , & Law, A. J. (2021). Male fetus susceptibility to maternal inflammation: C‐reactive protein and brain development. Psychological Medicine, 51(3), 450–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias‐Vázquez, L. , Van Ginkel Riba, G. , Arija, V. , & Canals, J. (2020). Composition of gut microbiota in children with autism spectrum disorder: A systematic review and Meta‐analysis. Nutrients, 12(3), 792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inguaggiato, E. , Sgandurra, G. , & Cioni, G. (2017). Brain plasticity and early development: Implications for early intervention in neurodevelopmental disorders. Neuropsychiatrie de l'Enfance et de l'Adolescence, 65(5), 299–306. [Google Scholar]
- Instanes, J. T. , Halmøy, A. , Engeland, A. , Haavik, J. , Furu, K. , & Klungsøyr, K. (2017). Attention‐deficit/hyperactivity disorder in offspring of mothers with inflammatory and immune system diseases. Biological Psychiatry, 81(5), 452–459. [DOI] [PubMed] [Google Scholar]
- Irwin, J. L. , McSorley, E. M. , Yeates, A. J. , Mulhern, M. S. , Strain, J. J. , Watson, G. E. , Grzesik, K. , Thurston, S. W. , Love, T. M. , Smith, T. H. , Broberg, K. , Shamlaye, C. F. , Myers, G. J. , Davidson, P. W. , & van Wijngaarden, E. (2019). Maternal immune markers during pregnancy and child neurodevelopmental outcomes at age 20 months in the Seychelles Child Development Study. Journal of Neuroimmunology, 335, 577023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, S. K. G. , Tofail, F. , Haque, R. , Petri, W. A. , & Nelson, C. A. (2019). Child development in the context of biological and psychosocial hazards among poor families in Bangladesh. PLoS ONE, 14(5), e0215304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, N. M. , Cowan, M. , Moonah, S. N. , & Petri, W. A. Jr (2018). The impact of systemic inflammation on neurodevelopment. Trends in Molecular Medicine, 24(9), 794–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, N. M. , Tofail, F. , Ma, J. Z. , Haque, R. , Kirkpatrick, B. , Nelson, C. A. , & Petri, W. A. (2017). Early life inflammation and neurodevelopmental outcome in Bangladeshi infants growing up in adversity. American Journal of Tropical Medicine and Hygiene, 97(3), 974–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, N. M. , Tofail, F. , Moonah, S. N. , Scharf, R. J. , Taniuchi, M. , Ma, J. Z. , Hamadani, J. D. , Gurley, E. S. , Houpt, E. R. , Azziz‐Baumgartner, E. , Haque, R. , & Petri, W. A. Jr (2014). Febrile illness and pro‐inflammatory cytokines are associated with lower neurodevelopmental scores in Bangladeshi infants living in poverty. BMC Pediatrics, 14, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, A. C. , Miles, E. A. , Warner, J. O. , Colwell, B. M. , Bryant, T. N. , & Warner, J. A. (1996). Fetal peripheral blood mononuclear cell proliferative responses to mitogenic and allergenic stimuli during gestation. Pediatric Allergy and Immunology, 7(3), 109–116. [DOI] [PubMed] [Google Scholar]
- Jurek, L. , Sevil, M. , Jay, A. , Schröder, C. , Baghdadli, A. , Héry‐Arnaud, G. , & Geoffray, M. M. (2021). Is there a dysbiosis in individuals with a neurodevelopmental disorder compared to controls over the course of development? A systematic review. European Child & Adolescent Psychiatry, 30(11), 1671–1694. [DOI] [PubMed] [Google Scholar]
- Kang, D.‐W. , Adams, J. B. , Gregory, A. C. , Borody, T. , Chittick, L. , Fasano, A. , Khoruts, A. , Geis, E. , Maldonado, J. , McDonough‐Means, S. , Pollard, E. L. , Roux, S. , Sadowsky, M. J. , Lipson, K. S. , Sullivan, M. B. , Caporaso, J. G. , & Krajmalnik‐Brown, R. (2017). Microbiota transfer therapy alters gut ecosystem and improves gastrointestinal and autism symptoms: An open‐label study. Microbiome, 5(1), 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsey, C. M. , Prescott, S. , McCulloch, J. A. , Trinchieri, G. , Valladares, T. L. , Dreisbach, C. , Alhusen, J. , & Grossmann, T. (2021). Gut microbiota composition is associated with newborn functional brain connectivity and behavioral temperament. Brain, Behavior, and Immunity, 91, 472–486. [DOI] [PubMed] [Google Scholar]
- Kinjo, T. , Ohga, S. , Ochiai, M. , Honjo, S. , Tanaka, T. , Takahata, Y. , Ihara, K. , & Hara, T. (2011). Serum chemokine levels and developmental outcome in preterm infants. Early Human Development, 87(6), 439–443. [DOI] [PubMed] [Google Scholar]
- Kitano, H. (2002). Systems biology: A brief overview. science, 295(5560), 1662–1664. [DOI] [PubMed] [Google Scholar]
- Konkel, L. (2018). The brain before birth: Using fMRI to explore the secrets of fetal neurodevelopment. Environmental Health Perspectives, 126(11), 112001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krakowiak, P. , Goines, P. E. , Tancredi, D. J. , Ashwood, P. , Hansen, R. L. , Hertz‐Picciotto, I. , & Van de Water, J. (2017). Neonatal cytokine profiles associated with autism spectrum disorder. Biological Psychiatry, 81(5), 442–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriklaki, A. , Margetaki, K. , Kampouri, M. , Koutra, K. , Bitsios, P. , Chalkiadaki, G. , Dermitzaki, E. , Venihaki, M. , Sarri, K. , Anousaki, D. , Kogevinas, M. , & Chatzi, L. (2019). Association between high levels of inflammatory markers and cognitive outcomes at 4years of age: The Rhea mother‐child cohort study, Crete, Greece. Cytokine, 117, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacorte, E. , Gervasi, G. , Bacigalupo, I. , Vanacore, N. , Raucci, U. , & Parisi, P. (2019). A systematic review of the microbiome in children with neurodevelopmental disorders. Frontiers in Neurology, 10, 727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laue, H. E. , Korrick, S. A. , Baker, E. R. , Karagas, M. R. , & Madan, J. C. (2020). Prospective associations of the infant gut microbiome and microbial function with social behaviors related to autism at age 3 years. Scientific Reports, 10(1), 15515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laue, H. E. , Korrick, S. A. , Baker, E. R. , Karagas, M. R. , & Madan, J. C. (2020). Prospective associations of the infant gut microbiome and microbial function with social behaviors related to autism at age 3 years. Scientific Reports, 10(1), 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, E. S. , Kim, E. K. , Shin, S. , Choi, Y. H. , Jung, Y. H. , Kim, S. Y. , Koh, J. W. , Choi, E. K. , Cheon, J. E. , & Kim, H. S. (2021). Factors associated with neurodevelopment in preterm infants with systematic inflammation. BMC Pediatrics, 21, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leviton, A. , Allred, E. N. , Fichorova, R. N. , Kuban, K. C. K. , Michael O'Shea, T. , & Dammann, O. (2016). Systemic inflammation on postnatal days 21 and 28 and indicators of brain dysfunction 2 years later among children born before the 28th week of gestation. Early Human Development, 93, 25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leviton, A. , Fichorova, R. N. , O'Shea, T. M. , Kuban, K. , Paneth, N. , Dammann, O. , Allred, E. N. , & ELGAN Study Investigators . (2013). Two‐hit model of brain damage in the very preterm newborn: Small for gestational age and postnatal systemic inflammation. Pediatric Research, 73(3), 362–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leviton, A. , Joseph, R. M. , Fichorova, R. N. , Allred, E. N. , Gerry Taylor, H. , Michael O'Shea, T. , & Dammann, O. (2019). Executive dysfunction early postnatal biomarkers among children born extremely preterm. Journal of Neuroimmune Pharmacology, 14(2), 188–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima‐Ojeda, J. M. , Rupprecht, R. , & Baghai, T. C. (2017). “I am I and my bacterial circumstances”: Linking gut microbiome, neurodevelopment, and depression. Frontiers in Psychiatry, 8, 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. , & Feng, Z. C. (2010). Increased umbilical cord plasma interleukin‐1 beta levels was correlated with adverse outcomes of neonatal hypoxic‐ischemic encephalopathy. Journal of Tropical Pediatrics, 56(3), 178–182. [DOI] [PubMed] [Google Scholar]
- Lodha, A. , Asztalos, E. , & Moore, A. (2010). Cytokine levels in neonatal necrotizing enterocolitis and long‐term growth and neurodevelopment. Acta Paediatrica, International Journal of Paediatrics, 99(3), 338–343. [DOI] [PubMed] [Google Scholar]
- Loughman, A. , Ponsonby, A. L. , O'Hely, M. , Symeonides, C. , Collier, F. , Tang, M. L. K. , Carlin, J. , Ranganathan, S. , Allen, K. , Pezic, A. , Saffery, R. , Jacka, F. , Harrison, L. C. , Sly, P. D. , Vuillermin, P. , & BIS Investigator Group . (2020). Gut microbiota composition during infancy and subsequent behavioural outcomes. EBioMedicine, 52, 102640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughman, A. , Quinn, T. , Nation, M. L. , Reichelt, A. , Moore, R. J. , Van, T. T. H. , Sung, V. , & Tang, M. L. K. (2021). Infant microbiota in colic: Predictive associations with problem crying and subsequent child behavior. Journal of Developmental Origins of Health and Disease, 12(2), 260–270. [DOI] [PubMed] [Google Scholar]
- Luttikhuizen dos Santos, E. S. , de Kieviet, J. F. , Königs, M. , van Elburg, R. M. , & Oosterlaan, J. (2013). Predictive value of the Bayley scales of infant development on development of very preterm/very low birth weight children: A meta‐analysis. Early Human Development, 89(7), 487–496. [DOI] [PubMed] [Google Scholar]
- Magalhaes, R. C. , Moreira, J. M. , Vieira, E. L. M. , Rocha, N. P. , Miranda, D. M. , & Simoes, E. (2017). Urinary levels of IL‐1beta and GDNF in preterm neonates as potential biomarkers of motor development: A prospective study. Mediators of Inflammation, 2017, 8201423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malan‐Muller, S. , Valles‐Colomer, M. , Raes, J. , Lowry, C. A. , Seedat, S. , & Hemmings, S. M. (2018). The gut microbiome and mental health: Implications for anxiety‐and trauma‐related disorders. Omics: A Journal of Integrative Biology, 22(2), 90–107. [DOI] [PubMed] [Google Scholar]
- Mancini, V. O. , & Finlay‐Jones, A. (2021, June 30). The role of the microbiome and immune systems on human development in the first five years of life: A scoping review. 10.17605/OSF.IO/NK72P [DOI]
- Mayer, E. A. , Padua, D. , & Tillisch, K. (2014). Altered brain‐gut axis in autism: Comorbidity or causative mechanisms? BioEssays, 36(10), 933–939. [DOI] [PubMed] [Google Scholar]
- Meyer, U. , Feldon, J. , & Dammann, O. (2011). Schizophrenia and autism: Both shared and disorder‐specific pathogenesis via perinatal inflammation? Pediatric Research, 69(8), 26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monthe‐Dreze, C. , Rifas‐Shiman, S. L. , Gold, D. R. , Oken, E. , & Sen, S. (2019). Maternal obesity and offspring cognition: The role of inflammation. Pediatric Research, 85(6), 799–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahmias, A. S. , Pellecchia, M. , Stahmer, A. C. , & Mandell, D. S. (2019). Effectiveness of community‐based early intervention for children with autism spectrum disorder: A meta‐analysis. Journal of Child Psychology and Psychiatry, 60(11), 1200–1209. [DOI] [PubMed] [Google Scholar]
- Nazzari, S. , Fearon, P. , Rice, F. , Ciceri, F. , Molteni, M. , & Frigerio, A. (2020). Neuroendocrine and immune markers of maternal stress during pregnancy and infant cognitive development. Developmental Psychobiology, 62(8), 1100–1110. [DOI] [PubMed] [Google Scholar]
- Nazzari, S. , Fearon, P. , Rice, F. , Dottori, N. , Ciceri, F. , Molteni, M. , & Frigerio, A. (2019). Beyond the HPA‐axis: Exploring maternal prenatal influences on birth outcomes and stress reactivity. Psychoneuroendocrinology, 101, 253–262. [DOI] [PubMed] [Google Scholar]
- Nazzari, S. , & Frigerio, A. (2020). The programming role of maternal antenatal inflammation on infants’ early neurodevelopment: A review of human studies: Special section on “Translational and Neuroscience Studies in Affective Disorders” Section Editor, Maria Nobile MD, PhD. Journal of Affective Disorders, 263, 739–746. [DOI] [PubMed] [Google Scholar]
- Nguyen, T. T. , Kosciolek, T. , Maldonado, Y. , Daly, R. E. , Martin, A. S. , McDonald, D. , Knight, R. , & Jeste, D. V. (2019). Differences in gut microbiome composition between persons with chronic schizophrenia and healthy comparison subjects. Schizophrenia Research, 204, 23–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nist, M. D. , Pickler, R. H. , Harrison, T. M. , Steward, D. K. , & Shoben, A. B. (2020). Inflammatory predictors of neurobehavior in very preterm infants. Early Human Development, 147, 105078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nist, M. D. , Shoben, A. B. , & Pickler, R. H. (2020). Early inflammatory measures and neurodevelopmental outcomes in preterm infants. Nursing Research, 69(5S Suppl 1), S11–S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne, S. , Biaggi, A. , Chua, T. E. , Du Preez, A. , Hazelgrove, K. , Nikkheslat, N. , Previti, G. , Zunszain, P. A. , Conroy, S. , & Pariante, C. M. (2018). Antenatal depression programs cortisol stress reactivity in offspring through increased maternal inflammation and cortisol in pregnancy: The Psychiatry Research and Motherhood ‐ Depression (PRAM‐D) study. Psychoneuroendocrinology, 98, 211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Shea, T. M. , Allred, E. N. , Kuban, K. C. K. , Dammann, O. , Paneth, N. , Fichorova, R. , Hirtz, D. , Leviton, A. , & Extremely Low Gestational Age Newborn (ELGAN) Study Investigators . (2012). Elevated concentrations of inflammation‐related proteins in postnatal blood predict severe developmental delay at 2 years of age in extremely preterm infants. Journal of Pediatrics, 160(3), 395–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Shea, T. M. , Joseph, R. M. , Kuban, K. C. , Allred, E. N. , Ware, J. , Coster, T. , Fichorova, R. N. , Dammann, O. , Leviton, A. , & ELGAN Study Investigators . (2014). Elevated blood levels of inflammation‐related proteins are associated with an attention problem at age 24 mo in extremely preterm infants. Pediatric Research, 75(6), 781–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Shea, T. M. , Shah, B. , Allred, E. N. , Fichorova, R. N. , Kuban, K. C. K. , Dammann, O. , Leviton, A. , & ELGAN Study Investigators . (2013). Inflammation‐initiating illnesses, inflammation‐related proteins, and cognitive impairment in extremely preterm infants. Brain, Behavior, and Immunity, 29, 104–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouzzani, M. , Hammady, H. , Fedorowicz, Z. , & Elmagarmid, A. (2016). Rayyan ‐ A web and mobile app for systematic reviews. Systematic Reviews, 5, 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons, E. , Claud, K. , & Petrof, E. O. (2020). The infant microbiome and implications for central nervous system development. Progress in Molecular Biology and Translational Science, 171, 1–13. [DOI] [PubMed] [Google Scholar]
- Perez‐Muñoz, M. E. , Arrieta, M.‐C. , Ramer‐Tait, A. E. , & Walter, J. (2017). A critical assessment of the “sterile womb” and “in utero colonization” hypotheses: Implications for research on the pioneer infant microbiome. Microbiome, 5(1), 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinu, F. R. , Beale, D. J. , Paten, A. M. , Kouremenos, K. , Swarup, S. , Schirra, H. J. , & Wishart, D. (2019). Systems biology and multi‐omics integration: Viewpoints from the metabolomics research community. Metabolites, 9(4), 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pronovost, G. N. , & Hsiao, E. Y. (2019). Perinatal interactions between the microbiome, immunity, and neurodevelopment. Immunity, 50(1), 18–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthussery, S. , Chutiyami, M. , Tseng, P.‐C. , Kilby, L. , & Kapadia, J. (2018). Effectiveness of early intervention programs for parents of preterm infants: A meta‐review of systematic reviews. BMC Pediatrics, 18(1), 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen, J. M. , Graham, A. M. , Entringer, S. , Gilmore, J. H. , Styner, M. , Fair, D. A. , Wadhwa, P. D. , & Buss, C. (2019). Maternal interleukin‐6 concentration during pregnancy is associated with variation in frontolimbic white matter and cognitive development in early life. Neuroimage, 185, 825–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez‐Trujillo, A. , Rios, J. , Angeles, M. A. , Posadas, D. E. , Murillo, C. , Rueda, C. , Botet, F. , Bosch, J. , Vergara, A. , Gratacós, E. , Palacio, M. , & Cobo, T. (2019). Influence of perinatal inflammation on the neurodevelopmental outcome of premature infants. Journal of Maternal‐Fetal and Neonatal Medicine, 32(7), 1069–1077. [DOI] [PubMed] [Google Scholar]
- Rommel, A.‐S. , Milne, G. L. , Barrett, E. S. , Bush, N. R. , Nguyen, R. , Sathyanarayana, S. , Swan, S. H. , & Ferguson, K. K. (2020). Associations between urinary biomarkers of oxidative stress in the third trimester of pregnancy and behavioral outcomes in the child at 4 years of age. Brain, Behavior, and Immunity, 90, 272–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, J. , Vassar, R. , Cahill‐Rowley, K. , Hintz, S. R. , & Stevenson, D. K. (2016). Neonatal biomarkers of inflammation: Correlates of early neurodevelopment and gait in very‐low‐birth‐weight preterm children. American Journal of Perinatology, 33(1), 71–78. [DOI] [PubMed] [Google Scholar]
- Rothenberg, S. E. , Chen, Q. , Shen, J. , Nong, Y. , Nong, H. , Trinh, E. P. , Biasini, F. J. , Liu, J. , Zeng, X. , Zou, Y. , Ouyang, F. , & Korrick, S. A. (2021). Neurodevelopment correlates with gut microbiota in a cross‐sectional analysis of children at 3 years of age in rural China. Scientific Reports, 11(1), 7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothhammer, V. , Mascanfroni, I. D. , Bunse, L. , Takenaka, M. C. , Kenison, J. E. , Mayo, L. , Chao, C. C. , Patel, B. , Yan, R. , Blain, M. , Alvarez, J. I. , Kébir, H. , Anandasabapathy, N. , Izquierdo, G. , Jung, S. , Obholzer, N. , Pochet, N. , Clish, C. B. , Prinz, M. , … Quintana, F. J. (2016). Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nature Medicine, 22(6), 586–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozé, J.‐C. , Ancel, P.‐Y. , Marchand‐Martin, L. , Rousseau, C. , Montassier, E. , Monot, C. , Le Roux, K. , Butin, M. , Resche‐Rigon, M. , Aires, J. , Neu, J. , Lepage, P. , Butel, M.‐J. , & EPIFLORE Study Group . (2020). Assessment of neonatal intensive care unit practices and preterm newborn gut microbiota and 2‐Year neurodevelopmental outcomes. JAMA Network Open, 3(9), e2018119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph, M. D. , Graham, A. M. , Feczko, E. , Miranda‐Dominguez, O. , Rasmussen, J. M. , Nardos, R. , Entringer, S. , Wadhwa, P. D. , Buss, C. , & Fair, D. A. (2018). Maternal IL‐6 during pregnancy can be estimated from newborn brain connectivity and predicts future working memory in offspring. Nature Neuroscience, 21(5), 765–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevenoaks, T. , Wedderburn, C. J. , Donald, K. A. , Barnett, W. , Zar, H. J. , Stein, D. J. , & Naudé, P. J. W. (2021). Association of maternal and infant inflammation with neurodevelopment in HIV‐exposed uninfected children in a South African birth cohort. Brain, Behavior, and Immunity, 91, 65–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silveira, R. C. , & Procianoy, R. S. (2011). High plasma cytokine levels, white matter injury and neurodevelopment of high risk preterm infants: Assessment at two years. Early Human Development, 87(6), 433–437. [DOI] [PubMed] [Google Scholar]
- Singh, K. , Connors, S. L. , Macklin, E. A. , Smith, K. D. , Fahey, J. W. , Talalay, P. , & Zimmerman, A. W. (2014). Sulforaphane treatment of autism spectrum disorder (ASD). Proceedings of the National Academy of Sciences of the United States of America, 111(43), 15550–15555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sordillo, J. E. , Korrick, S. , Laranjo, N. , Carey, V. , Weinstock, G. M. , Gold, D. R. , O'Connor, G. , Sandel, M. , Bacharier, L. B. , Beigelman, A. , Zeiger, R. , Litonjua, A. A. , & Weiss, S. T. (2019). Association of the infant gut microbiome with early childhood neurodevelopmental outcomes: An ancillary study to the VDAART randomized clinical trial. JAMA Network Open, 2(3), e190905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin, Y. , Romero, R. , Mele, L. , Iams, J. D. , Peaceman, A. M. , Leveno, K. J. , Harper, M. , Caritis, S. N. , Mercer, B. M. , Thorp, J. M. , O'Sullivan, M. J. , Ramin, S. M. , Carpenter, M. W. , Rouse, D. J. , & Sibai, B. (2014). Umbilical cord serum interleukin‐6, C‐reactive protein, and myeloperoxidase concentrations at birth and association with neonatal morbidities and long‐term neurodevelopmental outcomes. American Journal of Perinatology, 31(8), 717–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowell, K. D. , Uriu‐Adams, J. Y. , Van de Water, J. , Chambers, C. D. , Coles, C. D. , Kable, J. A. , Yevtushok, L. , Zymak‐Zakutnya, N. , Wertelecki, W. , Keen, C. L. , & Collaborative Initiative on Fetal Alcohol Spectrum Disorders (CIFASD) . (2018). Implications of altered maternal cytokine concentrations on infant outcomes in children with prenatal alcohol exposure. Alcohol, 68, 49–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spann, M. N. , Monk, C. , Scheinost, D. , & Peterson, B. S. (2018). Maternal immune activation during the third trimester is associated with neonatal functional connectivity of the salience network and fetal to toddler behavior. The Journal of Neuroscience, 38(11), 2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spann, M. N. , Monk, C. , Scheinost, D. , & Peterson, B. S. (2018). Maternal immune activation during the third trimester is associated with neonatal functional connectivity of the salience network and fetal to toddler behavior. Journal of Neuroscience, 38(11), 2877–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenbergen, L. , Sellaro, R. , van Hemert, S. , Bosch, J. A. , & Colzato, L. S. (2015). A randomized controlled trial to test the effect of multispecies probiotics on cognitive reactivity to sad mood. Brain, Behavior, and Immunity, 48, 258–264. [DOI] [PubMed] [Google Scholar]
- Stoll, C. R. T. , Izadi, S. , Fowler, S. , Green, P. , Suls, J. , & Colditz, G. A. (2019). The value of a second reviewer for study selection in systematic reviews. Research Synthesis Methods, 10(4), 539–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetman, D. U. , Strickland, T. , Melo, A. M. , Kelly, L. A. , Onwuneme, C. , Watson, W. R. , Murphy, J. F. A. , Slevin, M. , Donoghue, V. , O'Neill, A. , & Molloy, E. J. (2021). Neonatal encephalopathy is associated with altered IL‐8 and GM‐CSF which correlates with outcomes. Frontiers in Pediatrics, 8, 556216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szeligowski, T. , Yun, A. L. , Lennox, B. R. , & Burnet, P. W. (2020). The gut microbiome and schizophrenia: The current state of the field and clinical applications. Frontiers in Psychiatry, 11, 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamburini, S. , Shen, N. , Wu, H. C. , & Clemente, J. C. (2016). The microbiome in early life: Implications for health outcomes. Nature Medicine, 22(7), 713–722. [DOI] [PubMed] [Google Scholar]
- Tillisch, K. , Labus, J. , Kilpatrick, L. , Jiang, Z. , Stains, J. , Ebrat, B. , Guyonnet, D. , Legrain‐Raspaud, S. , Trotin, B. , Naliboff, B. , & Mayer, E. A. (2013). Consumption of fermented milk product with probiotic modulates brain activity. Gastroenterology, 144(7), 1394–1401e1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tognini, P. (2017). Gut microbiota: A potential regulator of neurodevelopment. Frontiers in Cellular Neuroscience, 11, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg, J. P. , Westerbeek, E. A. M. , Broring‐Starre, T. , Garssen, J. , & Van Elburg, R. M. (2016). Neurodevelopment of preterm infants at 24 months after neonatal supplementation of a prebiotic mix: A randomized trial. Journal of Pediatric Gastroenterology and Nutrition, 63(2), 270–276. [DOI] [PubMed] [Google Scholar]
- van de Schoot, R. , de Bruin, J. , Schram, R. , Zahedi, P. , de Boer, J. , Weijdema, F. , Kramer, B. , Huijts, M. , Hoogerwerf, M. , Ferdinands, G. , Harkema, A. , Willemsen, J. , Ma, Y. , Fang, Q. , Hindriks, S. , Tummers, L. , & Oberski, D. L. (2021). An open source machine learning framework for efficient and transparent systematic reviews. Nature Machine Intelligence, 3(2), 125–133. [Google Scholar]
- Varner, M. W. , Marshall, N. E. , Rouse, D. J. , Jablonski, K. A. , Leveno, K. J. , Reddy, U. M. , Mercer, B. M. , Iams, J. D. , Wapner, R. J. , Sorokin, Y. , Thorp, J. M. , Malone, F. D. , Carpenter, M. , O'Sullivan, M. J. , Peaceman, A. M. , Hankins, G. D. , Dudley, D. J. , Caritis, S. N. , & The Eunice Kennedy Shriver National Institute of Child Health Human Development Maternal‐Fetal Medicine Units Network . (2015). The association of cord serum cytokines with neurodevelopmental outcomes. American Journal of Perinatology, 30(2), 115–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenstra, T. D. (2021). Omics in systems biology: Current progress and future outlook. Proteomics, 21(3‐4), 2000235. [DOI] [PubMed] [Google Scholar]
- Voltas, N. , Arija, V. , Hernandez‐Martinez, C. , Jimenez‐Feijoo, R. , Ferre, N. , & Canals, J. (2017). Are there early inflammatory biomarkers that affect neurodevelopment in infancy? Journal of Neuroimmunology, 305, 42–50. [DOI] [PubMed] [Google Scholar]
- von Ehrenstein, O. S. , Neta, G. I. , Andrews, W. , Goldenberg, R. , Goepfert, A. , & Zhang, J. (2012). Child intellectual development in relation to cytokine levels in umbilical cord blood. American Journal of Epidemiology, 175(11), 1191–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L. , Christophersen, C. T. , Sorich, M. J. , Gerber, J. P. , Angley, M. T. , & Conlon, M. A. (2011). Low relative abundances of the mucolytic bacterium Akkermansia muciniphila and Bifidobacterium spp. in feces of children with autism. Applied and Environmental Microbiology, 77(18), 6718–6721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner, B. B. (2019). The contribution of the gut microbiome to neurodevelopment and neuropsychiatric disorders. Pediatric Research, 85(2), 216–224. [DOI] [PubMed] [Google Scholar]
- West, L. J. (2002). Defining critical windows in the development of the human immune system. Human & Experimental Toxicology, 21(9‐10), 499–505. [DOI] [PubMed] [Google Scholar]
- Xie, W. , Kumar, S. , Kakon, S. H. , Haque, R. , Petri, W. A. , & Nelson, C. A. (2019). Chronic inflammation is associated with neural responses to faces in Bangladeshi children. NeuroImage, 202, 116110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, M. , Xu, X. , Li, J. , & Li, F. (2019). Association between gut microbiota and autism spectrum disorder: A systematic review and Meta‐analysis. Frontiers in Psychiatry, 10, 473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, J. , Risacher, S. L. , Shen, L. , & Saykin, A. J. (2018). Network approaches to systems biology analysis of complex disease: Integrative methods for multi‐omics data. Briefings in Bioinformatics, 19(6), 1370–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, J. , Zhu, Y. , Cao, L. J. , Liu, Y. Y. , Zheng, Y. Z. , Li, W. , & Huang, G. W. (2020). Effects of maternal folic acid supplementation during pregnancy on infant neurodevelopment at 1 month of age: A birth cohort study in China. European Journal of Nutrition, 59(4), 1345–1356. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary File 1 Search Terms and Results from Embase (Ovid Interface) Database Search
Supplement Material
Data Availability Statement
All files associated with this review are available at Open Science Framework https://osf.io/nk72p/.
Index
| Immune biomarkers | Inflammatory | |
|---|---|---|
| CD64 | Cluster of Differentiation 64 | |
| CRP | C‐reactive protein | pro |
| E‐SEL | E‐selectin | |
| GCSF | Granulocyte colony‐stimulating factor | |
| GDNF | Glial cell line‐derived neurotrophic factor | |
| GM‐CSF | Granulocyte‐macrophage colony‐stimulating factor | |
| ICAM‐1 | Intercellular adhesion molecule 1 | |
| ICAM‐3 | Intercellular adhesion molecule 3 | |
| IFN‐γ | Interferon gamma | anti |
| IGFBP‐1 | Insulin‐like growth factor‐binding protein 1 | |
| IL‐1 | Interleukin‐1 | |
| IL‐10 | Interleukin‐10 | anti |
| IL‐12 | Interleukin‐12 | |
| IL‐12p70 | Interleukin‐12p70 | |
| IL‐13 | Interleukin‐13 | |
| IL‐16 | Interleukin‐16 | |
| IL‐17 / IL‐17A | Interleukin‐17 | |
| IL‐17α | Interleukin‐17α | |
| IL‐18 | Interleukin‐18 | |
| IL‐1RA | Interleukin‐1 receptor antagonist | |
| IL‐1β | Interleukin‐1β | pro |
| IL‐2 | Interleukin‐2 | anti |
| IL‐4 | Interleukin‐4 | anti |
| IL‐5 | Interleukin‐5 | anti |
| IL‐6 | Interleukin‐6 | pro |
| IL‐6R | Interleukin‐6 receptor | |
| IL‐7 | Interleukin‐7 | |
| IL‐8 | Interleukin‐8 | |
| IL‐9 | Interleukin‐9 | |
| IL12p40 | Interleukin‐12p40 | |
| IP‐10 | Interferon gamma‐induced protein 10 | |
| M‐CSF | Macrophage colony‐stimulating factor | |
| MCP‐1 | Monocyte chemoattractant protein‐1 | pro |
| MCP‐4 | Monocyte chemoattractant protein‐4 | |
| MIG | Monokine induced by IFN‐γ | |
| MIP‐1α | Macrophage inflammatory protein‐1α | |
| MIP‐1β | Macrophage inflammatory protein‐1β | |
| MMP‐9 | Matrix metallopeptidase 9 | |
| MPO | Myeloperoxidase | |
| NGAL | neutrophil gelatinase‐associated lipocalin | |
| PAI‐1 | Plasminogen activator inhibitor‐1 | |
| PDGF‐AA | Platelet‐derived growth factor‐AA | |
| PGF2α | Prostaglandin F2α | |
| RANTES | Regulated upon Activation, Normal T Cell Expressed and Presumably Secreted | |
| s‐IgA | Salivary Immunoglobulin A | |
| SAA/sAA | Serum amyloid A | |
| SDF‐1 | Stromal cell‐derived factor 1 | |
| sFLT‐1 | Soluble fms‐like tyrosine kinase‐1 | pro |
| sICAM‐1 | Soluble intercellular adhesion molecule‐1 | |
| sTNFR1 | Soluble tumor necrosis factor receptor‐1 | |
| sTNFR2 | Soluble tumor necrosis factor receptor‐2 | |
| sVCAM‐1 | Soluble vascular cell adhesion molecule‐1 | |
| TARC | Thymus and activation‐regulated chemokine | |
| TNF‐R1 | Tumor necrosis factor receptor 1 | |
| TNF‐R2 | Tumor necrosis factor receptor 2 | |
| TNF‐α | Tumor necrosis factor α | |
| TNF‐β | Tumor necrosis factor β | |
| TPO | Thyroid peroxidase | |
| VEGF | Vascular endothelial growth factor | |
| VEGF‐D | Vascular endothelial growth factor‐D | |
| VEGF‐R1 | Vascular endothelial growth factor receptor 1 | |
| VEGF‐R2 | Vascular endothelial growth factor receptor 2 | |
| Neurodevelopment measures | ||
| ABC | Aberrant Behavior Checklist | |
| ADHD | Attention Deficit Hyperactivity Disorder | |
| ASD | Autism Spectrum Disorder | |
| ASQ | Ages and Stages Questionnaire (several editions) | |
| BASC | Behavior Assessment System for Children (several editions) | |
| BMI | Body Mass Index | |
| BSID | Bayley Scales of Infant Development (II and III Editions) | |
| CBCL | Child Behavior Checklist | |
| CDI | Communicative Development Inventory | |
| CDSC | Child Developmental Scale of China | |
| CI | Confidence Interval | |
| CP | cerebral palsy | |
| CRYPTO | Burden of Cryptosporidiosis | |
| DDST | Denver Developmental Screening Test | |
| ECBQ | Early Childhood Behavior Questionnaire | |
| ELBW | extremely low birth weight | |
| ELGAN | Extremely Low Gestational Age Newborn cohort study | |
| FNIRS | Functional Near Infrared Spectroscopy | |
| GMDS | Griffiths Mental Developmental Scales | |
| HIE | hypoxic‐ischemic encephalopathy | |
| HIV | Human immunodeficiency Virus | |
| IBQ / IBQ‐R | Infant Behavior Questionnaire (‐Revised) | |
| IQ | Intelligence Quotient | |
| KSPD | Kyoto Scale of Psychological Development | |
| MDD | Major Depressive Disorder | |
| MDI | Mental Development Index of the BSID | |
| (f)MRI | (functional) Magnetic Resonance Imaging | |
| MSCA | McCarthy Scales of Children's Abilities | |
| MSEL | Mullen Scales of Early Learning | |
| NAPI | Neurobehavioral Assessment of the Preterm Infant, Revised Second Edition | |
| NBAS | Neonatal Behavioral Assessment Scale | |
| NDD | neurodevelopmental disability | |
| NE | neonatal encephalopathy (?) | |
| NEC | necrotizing enterocolitis | |
| NHST | null hypothesis significance testing | |
| PDI | Psychomotor Development Index of the BSID | |
| PRAM‐D | Psychiatry Research and Motherhood ‐ Depression study | |
| PROVIDE | Performance of Rotavirus and Oral Polio Vaccines in Developing Countries cohort study | |
| SDI | Shannon Diversity Index | |
| SRS | Social Responsiveness Scale (several editions) | |
| TD | Typically Developing | |
| TIMP | Test of Infant Motor Performance | |
| VABS | Vineland Adaptive Behavior Scale | |
| VLBW | very low birth weight | |
| WPPSI / WPPSI‐R | Weschler Preschool and Primary Scale of Intelligence (‐Revised) | |
| WRAVMA | Wide Range Assessment of Visual Motor Abilities | |


