Abstract
Developing in silico models that accurately reflect a whole, functional cell is an ongoing challenge in biology. Current efforts bring together mathematical models, probabilistic models, visual representations, and data to create a multi‐scale description of cellular processes. A realistic whole‐cell model requires imaging data since it provides spatial constraints and other critical cellular characteristics that are still impossible to obtain by calculation alone. This review introduces Soft X‐ray Tomography (SXT) as a powerful imaging technique to visualize and quantify the mesoscopic (~25 nm spatial scale) organelle landscape in whole cells. SXT generates three‐dimensional reconstructions of cellular ultrastructure and provides a measured structural framework for whole‐cell modeling. Combining SXT with data from disparate technologies at varying spatial resolutions provides further biochemical details and constraints for modeling cellular mechanisms. We conclude, based on the results discussed here, that SXT provides a foundational dataset for a broad spectrum of whole‐cell modeling experiments.
Keywords: cell structure, soft X‐ray tomography, subcellular organization, whole‐cell modeling
Abbreviations
- 2D
two‐dimensional
- 3D
three‐dimensional
- CCD
charged‐couple device
- cryoET
cryo‐electron microscopy
- DOF
depth of field
- FBA
flux balance analysis
- FEL
free‐electron laser
- KZP
condenser zone plate
- MZP
micro zone plate
- NCXT
National Center for X‐ray Tomography
- ODE
ordinary differential equation
- PBC
pancreatic beta cell
- PDE
partial differential equation
- SXT
soft X‐ray tomography
- TEM
transmission electron microscopy
- TXM
transmission X‐ray microscope
1. INTRODUCTION
One of the overarching goals of modern cell biology is to ascertain the position of molecules with respect to cell structures as well as any relocations of those molecules during physiological processes. To some extent, this is the conceptual equivalent of having a real‐time updated GPS map for each component of a cell. Such maps, analogous to those on smartphones that make it possible to understand traffic conditions in real time, would enable the prediction of outcomes of extracellular manipulations or details of any cellular function. Achieving this goal will require input from many different technologies, for example, structural imaging tools to visualize and categorize cells and their subcellular organization combined with functional tools to characterize the molecules and track their positions over time. This combination of structural and dynamical techniques provides the framework for accurate whole‐cell modeling and facilitates a deeper understanding of cellular behavior, enabling the prediction of modifications associated with disease states.
To build the maps required for whole‐cell modeling, we need information at multiple scales, from atomic to tens of microns. Significant amounts of information at the atomic scale have been generated using techniques such as X‐ray macromolecular crystallography, cryo‐electron microscopy, and free‐electron laser (FEL)‐based serial diffraction. 1 , 2 , 3 , 4 , 5 , 6 , 7 While the information from these molecular‐level data streams is of immense value across the entire spectrum of biology and medicine, it is important to consider the following statement: “Understanding the structure and dynamics of a particular molecular interaction in isolation (i.e., without cellular context) does not necessarily give us information on how the molecule impacts cell phenotype or behavior.” To determine this, we need a quantitative description of molecular location and behavior in the context of the three‐dimensional (3D) organization of cells. This description could be obtained using a spatio‐temporal model, which could be generated in silico using generalized parameters for a cell. 8 , 9 , 10 However, the accuracy and precision of such models can be greatly increased when the computer‐generated models are based on and calibrated to experimental data obtained from the cells under investigation.
Vast amounts of information about cell structure and dynamics have been, and continue to be, generated using established imaging modalities such as electron and light microscopies. 11 , 12 , 13 , 14 , 15 , 16 In this manuscript, we focus on soft X‐ray tomography (SXT) and how it can play an important role in cell modeling efforts. SXT is a relatively new imaging tool that generates 3D structural information of whole cells at mesoscale resolution (~25 nm). 17 , 18 , 19 , 20 In addition to structural data, SXT provides quantitative information about the density and composition of organic molecules in subcellular compartments. 21 , 22 , 23 , 24 The unique capabilities of SXT were recently used to generate 3D cell models depicting the subcellular architecture of a wide range of different cell types in the near‐native state. 19 , 21 , 25 In this manuscript, we will first discuss the principles and capabilities of imaging with soft X‐rays and then describe recent examples in which SXT has provided pivotal data for modeling a variety of whole‐cell processes.
2. IMAGING CELLS USING SOFT X‐RAY TOMOGRAPHY
SXT generates data‐rich 3D images of whole, hydrated cells up to 18 μm thick 26 with isotropic resolution 20 , 27 , 28 , 29 approaching 10 nm in some circumstances. 18 , 20 Cells are in the near‐native state in that they are cryo‐fixed rather than chemically fixed, which reduces the potential for fixation‐induced artifacts. 30 , 31 , 32 , 33 In addition, cell structures are visualized based on their inherent contrast rather than preferential binding of chemical stains, ruling out staining artifacts and providing a reproducible natural contrast for whole‐cell modeling research. 25 , 33 , 34 Unique to SXT, the contrast of structures is quantitative, reflecting information about the molecular composition of each region of the cell. SXT imaging is also fast in that a tomographic reconstruction of a large eukaryotic cell can be generated in 5–10 min from mounting the specimen on the microscope, enabling analysis of statistically significant numbers of cells. 28 , 29 , 35 SXT data are useful for the development of cell models. It provides a subcellular organizational framework for developing models that explain and predict cell function by incorporating other molecular and dynamical data into this framework. In the following sections, we will briefly discuss the principles of imaging with soft X‐rays that make it possible to examine fully hydrated whole cells and how 3D information about the organic composition of cell structures is obtained.
2.1. Image formation using soft X‐rays
Soft X‐rays are most effective for imaging hydrated cells when used in an absorption contrast microscope. 36 The specimen is illuminated with X‐ray photons at energies that fall within the so‐called “water window” (i.e., between the K‐shell absorption edges of carbon at 282 eV and oxygen at 543 eV) 21 (Figure 1A). As photons in this energy range pass through the specimen, they are absorbed an order of magnitude more strongly by carbon‐ and nitrogen‐containing compounds than by water. For this reason, cell structures dense in carbon—for example, membranes or lipid bodies 39 —absorb the X‐rays more than highly solvated regions, such as vacuoles. 40 Consequently, subtle differences in the biochemical composition of structures produce measurable contrast in soft X‐ray images. Since the absorption adheres to Beer–Lambert's law, image contrast is linear with respect to the species and density of biomolecules in each voxel. As a result, each voxel has an associated Linear Absorption Coefficient (LAC). 41 The greater the concentration of biomolecules in a voxel, the higher the LAC value; conversely, voxels with fewer organic molecules have lower LAC values. The high‐contrast images of unstained cell structures generated with soft X‐ray imaging provide fundamental structural information for creating cell models.
FIGURE 1.
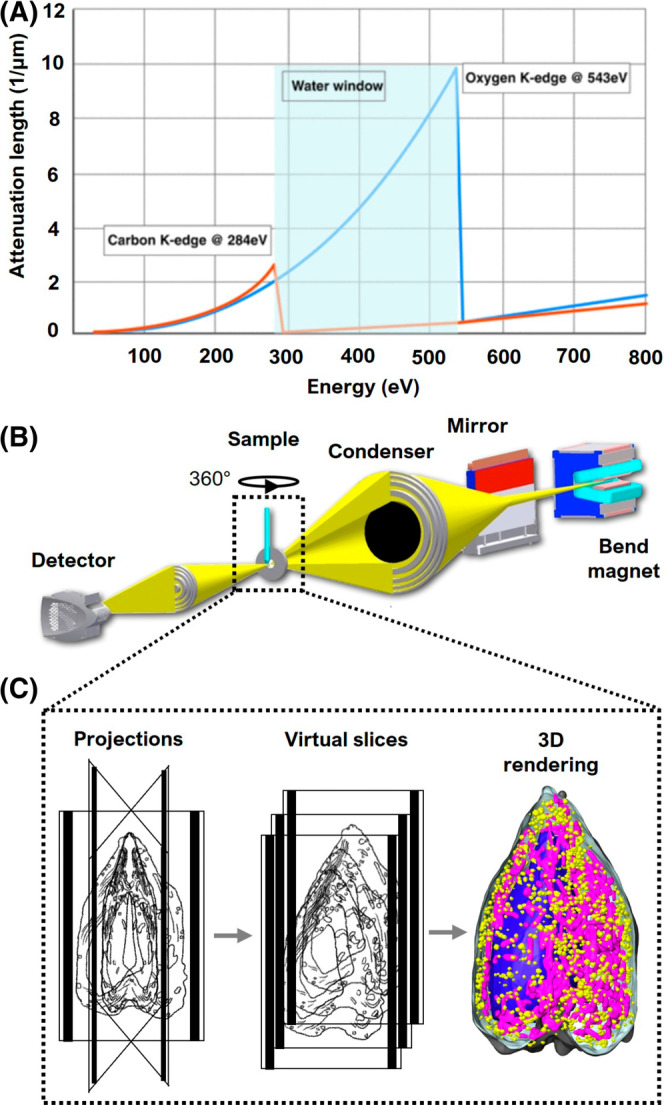
Overview of imaging cells with SXT. (A) “Water window” (light blue): region of the spectrum where the X‐ray absorption is less attenuated by water, 23 giving rise to natural contrast of cell structures in SXT. (B) Schematic representation of XM‐2, the soft X‐ray microscope located at Advanced Light Source, Lawrence Berkeley National Laboratory. Details about the microscope can be found on the National Center for X‐ray Tomography website (https://ncxt.org/) and in previous articles 37 ; (C) Specimen acquisition: (Left) single projections at incremental degrees of rotation are collected, aligned around the rotation axis, and reconstructed; (Center) virtual sections, or orthoslices, of the reconstructed tomograms; (Right) 3D volume of the reconstructed cell with select organelles segmented and color‐coded (reconstructed data adapted from White et al., 2020 38 ).
In addition to visualizing cell structures based on their inherent contrast, soft X‐rays have sufficient energy to penetrate whole cells up to ~18 μm in diameter. 26 , 42 , 43 This eliminates the need for sectioning and places soft X‐ray imaging in a unique position as a structural imaging technology. Traditional plastic section transmission electron microscopy (TEM) is restricted to imaging stained specimens that have been embedded in plastic and sectioned (typically 80–100 nm thick). 44 , 45 Cryo‐electron microscopy images native‐state cells, but is limited to specimens in the range of 100–200 nm thick because of the scattering of electrons in thick specimens. 11 , 12 , 13 , 46 In contrast, soft X‐ray microscopy can image an entire cell in the absence of chemical fixatives and stains. 27 , 32 , 36 This is highly advantageous but, for the case of a single image, is accompanied by the complexity of interpreting the data. Each image collected with soft X‐ray microscopy is a two‐dimensional (2D) projection of a large 3D object, and each projection image contains numerous superimposed structures in a complex, difficult to interpret image. To fully retrieve the information obtainable from soft X‐ray imaging, tomographic procedures must be used.
2.2. Soft X‐ray tomography: From 2D to 3D
Tomographic methods have long been used by the medical community to non‐invasively examine human internal anatomy. A series of 2D projection images is collected from multiple angles around the patient, with each image containing numerous superimposed structures seen from a slightly different angle. 47 Using a mathematical algorithm, such as filtered back projection 48 or iterative reconstruction, 49 these 2‐D images are converted into a tomogram—a 3‐D volumetric representation of the internal human anatomy. We use a very similar approach in SXT, with notable differences being that we use lower energy X‐rays and rotate the specimen around a single axis rather than rotate the X‐ray source and camera, 29 , 36 , 37 , 50 the preferred method for collecting a series of 2‐D images in medical tomography (Figure 1C).
Generation of a high‐quality tomographic reconstruction requires that the projection image dataset is collected at small, uniform, angular increments with the entire specimen in each field of view. Since each pixel of a projection image includes the information contained in a line integral of the X‐ray absorption along the X‐ray path through the specimen, rotation of the specimen through 180° is necessary to collect a complete projection image dataset. Such a “full rotation” dataset results in an isotropic resolution reconstructed volume, that is, the voxels of the reconstructed volume have the same size in all directions. 51 Specimen rotation through 180°, often reported as ±90° data collection, is the conventional approach used by the National Center for X‐ray Tomography (NCXT; https://ncxt.org), where the soft X‐ray microscope, XM‐2, is equipped with a full‐rotation cryo‐stage designed to accommodate cells mounted in a thin‐walled glass capillary (Figure 1B). 22 , 37 Microscopes at other synchrotrons are equipped with tilt stages that typically accommodate planar specimens. 52 , 53 , 54 With those stages, tomograms are reconstructed from specimens that can only be imaged between ±60–70° of tilt angle, due to physical limitations of the microscope as well as the nature of an extended specimen grown on a flat substrate. 50 As these stages are tilted, the sample progressively increases in thickness, becoming ~3x thicker at a 70° tilt. 55 In other words, a specimen imaged at 0° must be ≤5 μm thick because it reaches the limit of soft X‐ray penetration (~15 μm) at 70°. The limited rotation range means that information contained in ~±20° of the specimen (angles between ±70° to ±90°) is missing from the reconstruction; this leads to what is known as a “missing wedge” artifact, as seen in data obtained using cryo‐electron tomography (cryo‐ET). 11 , 29 These artifacts include distorted cell structures, which require added image processing and interpolation of information to restore their shapes. Full‐rotation tomography, in contrast, avoids those distortions and uncertainties.
Production of a high‐quality tomogram also requires collecting a large number of images of the same specimen and, consequently, significant exposure to potentially damaging X‐ray radiation. In order to prevent damaging the specimen during imaging, cells are first cryo‐immobilized using well‐known rapid freezing methods, such as gravity‐driven plunge freezing or high‐pressure freezing. 28 Freezing cells for imaging also immobilize molecules to prevent movement artifacts during data collection. At the NCXT, we use a custom‐made freezing apparatus that allows setting the rate at which the cell is plunged into the liquid nitrogen‐cooled propane (Figure 2A). This allows us to freeze large cells faster than gravity‐driven instruments and assures that the entire cell is well frozen, and molecules remain in the same position. Although the increased pressure occasionally causes a slight distortion of cells due to the force of the cryogen stream pushing the cell upward, it does not interfere with data interpretation in most instances. 38 , 56 Our ability to plunge freeze cells 10–20 μm thick enables us to obtain well‐frozen thick cells while mitigating radiation damage of the specimen. 22
FIGURE 2.
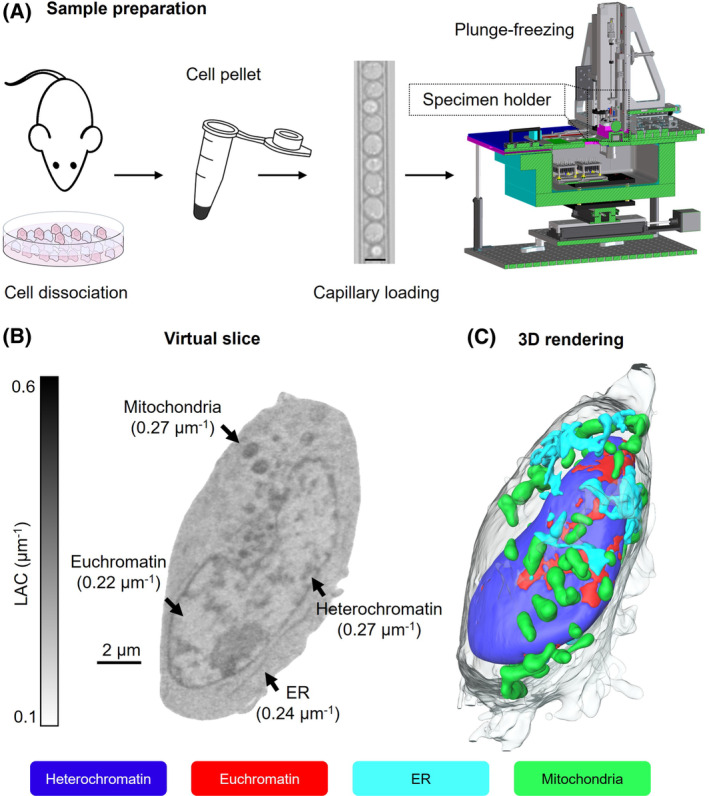
SXT sample preparation and data analysis. (A) Cells are dissociated either from tissues or culture, harvested, and loaded in thin‐walled (100–200 nm) glass capillaries. The diameter of the capillary is variable and based on cell size (scale bar, 8 μm). 39 Each capillary is plunge‐frozen in liquid nitrogen‐cooled liquid propane using a custom‐made freezing apparatus, then stored in liquid nitrogen until data collection. 22 , 28 (B) Orthoslice of a human lymphoblastoid cell, with LAC values shown between 0.1–0.6 μm−1; the average values for each organelle are reported in brackets. (C) 3D rendering of the segmented cell seen in (B), showing two major nuclear components (euchromatin‐red, and heterochromatin‐blue) seen at the nuclear surface, mitochondria (green), and endoplasmic reticulum (cyan). Scale bar 2 μm.
2.3. SXT instrumentation
Biological SXT is typically carried out using a full‐field transmission X‐ray microscope (TXM). The majority of such microscopes are located at synchrotrons around the world. 30 , 57 Although the principles of soft X‐ray imaging are the same, the design of each of these microscopes varies slightly. We will focus on the design of the microscope located in the U.S.A. since it was used to collect the data shown in this manuscript and is the only instrument producing full‐rotation tomographic datasets. This X‐ray microscope, known as XM‐2, was designed and constructed by the National Center for X‐ray Tomography for the sole purpose of imaging biological specimens (Figure 1B). It is located at beamline 2.1.2 of the Advanced Light Source synchrotron (located at Lawrence Berkeley National Laboratory). The optical design is similar to that of a light microscope, except that it uses Fresnel zone plates as lenses. 21 , 58 The condenser lens (traditionally called KZP, German for Condenser Zone Plate) focuses the beam onto the specimen and the objective lens (usually called the MZP for Micro Zone Plate) magnifies the image onto the CCD detector. The KZP also functions as an X‐ray monochromator. 37 The focal length of the KZP is inversely proportional to the focused wavelength; thus, moving the KZP will alter the energy used to illuminate the specimen. This allows us to select the desired “water window” X‐rays, typically 520 eV. The microscope is also equipped with a device that makes it possible to readily change the MZP. 37 , 59 The resolution of an MZP is inversely proportional to its depth of field (DOF); in other words, the better the resolution of the MZP, the shallower the DOF, and the smaller the size of specimen that will be entirely in focus. 37 , 59 Having the ability to rapidly change MZPs makes it possible to readily switch from imaging cells as small as bacteria to larger eukaryotic cells. 59 As a result, XM‐2 typically collects data using MZPs ranging from those achieving 35 nm to 80 nm resolution. 56 , 59 , 60 Algorithms and other technologies that will enable imaging larger specimens with better resolution zone plates are under development. 26 , 51 In the interim, the flexibility of rapidly switching optics assures that all cells imaged, no matter the size, are focused throughout and yield tomograms containing a wealth of quantitative information ripe for mining.
2.4. Data interpretation and analyses
Soft X‐ray tomograms contain a vast array of organelles, membraneless structures such as chromatin and nucleoli, as well as inter‐organelle cytoplasmic molecules (Figure 2B). There is no single approach to interpreting these complex datasets. One of the first things many investigators do is to “segment” structures, in other words, isolate a specific region from the other cell contents based on size, shape, and/or location in the cell (Figure 2C). The segmented data can then be used to determine numbers, sizes, shapes, and volumes of the organelles. With SXT data, there is another valuable measurement that aids in identification and analyses—the LAC values associated with each structure (Figure 2B). The LAC values can be used to assist segmentation approaches. They also can be extremely valuable for a variety of stand‐alone projects that require knowledge about the molecular density of, and/or around, structures.
Segmentation is typically carried out using commonly available image‐processing software packages. However, even with aids such as built‐in segmentation tools, the task of segmenting remains the rate‐limiting step in SXT data analysis. The reconstruction of a large mammalian cell can take anywhere from days or even weeks of effort by an experienced researcher. Consequently, significant efforts are being put toward the application of advanced image processing techniques for automating and de‐skilling this process. 61 , 62 , 63 , 64 , 65 , 66 , 67
2.5. Examples of cells imaged using SXT
SXT is capable of imaging a wide variety of cells of varying sizes and shapes. The one qualification, as previously discussed, is that soft X‐ray tomography is typically used to analyze single cells since soft X‐rays are unable to penetrate cells thicker than 15 μm in diameter. This includes specimens in which one dimension alone exceeds that thickness, such as spindle‐shaped cells or tubular cells 50 μm or more in length. There are many examples demonstrating the power of imaging single cells with SXT to address a broad spectrum of research questions, including studies of chromatin compaction, organelle‐organelle interactions, and studies of regional LAC values to model molecular movements. 56 , 68 , 69 , 70 In this manuscript, we highlight five different cell types, as seen in Figure 3. The top panel (Figure 3A) shows an example of nuclei obtained from mouse olfactory epithelial cells (multipotent stem cell, neuronal progenitor, and mature olfactory sensory neuron). 70 In this study, the investigators quantified the reorganization of chromatin during neurogenesis and showed that there is an increase in the percent heterochromatin during differentiation. A similar study conducted during hematopoiesis in cells obtained from mice also showed increased chromatin compaction during blood cell differentiation (not shown). 74 To demonstrate the power of SXT in studies testing drug efficacy, investigators examined the stages of red blood cell sickling over time (Figure 3B) and determined the point at which the drug was no longer capable of reversing the sickling process. 75 SXT is also a powerful tool for monitoring changes in response to a variety of exogenous treatments. This was powerfully demonstrated in algae (Figure 3C), where there was a dramatic increase in both the volume and location of lipids in response to the addition of glucose, followed by a complete reversal of that organization upon the removal of glucose. 71 Smaller cells such as the yeast, S. cerevisea, were characterized at different stages of the cell cycle, showing the reorganization and duplication of cell organelles (Figure 3D) 72 and chromatin compaction and DNA‐protein interactions were quantified during both bacterial growth and environmental adaptation in E. coli (Figure 3E). 73 , 76 Other manuscripts, not highlighted here, show the power of SXT to quantify and metric cell phenotypes, to determine statistically significant differences between healthy and diseased cells, to quantify subcellular differences between wild‐type cells and genetically modified cells and between cells exposed to different environments. 29 , 77 , 78 , 79 Thus, SXT data can be used as the framework for cell models that represent experimental or environmental conditions, bringing reality (in terms of experimental observations) and phenotypic accuracy to whole‐cell models.
FIGURE 3.
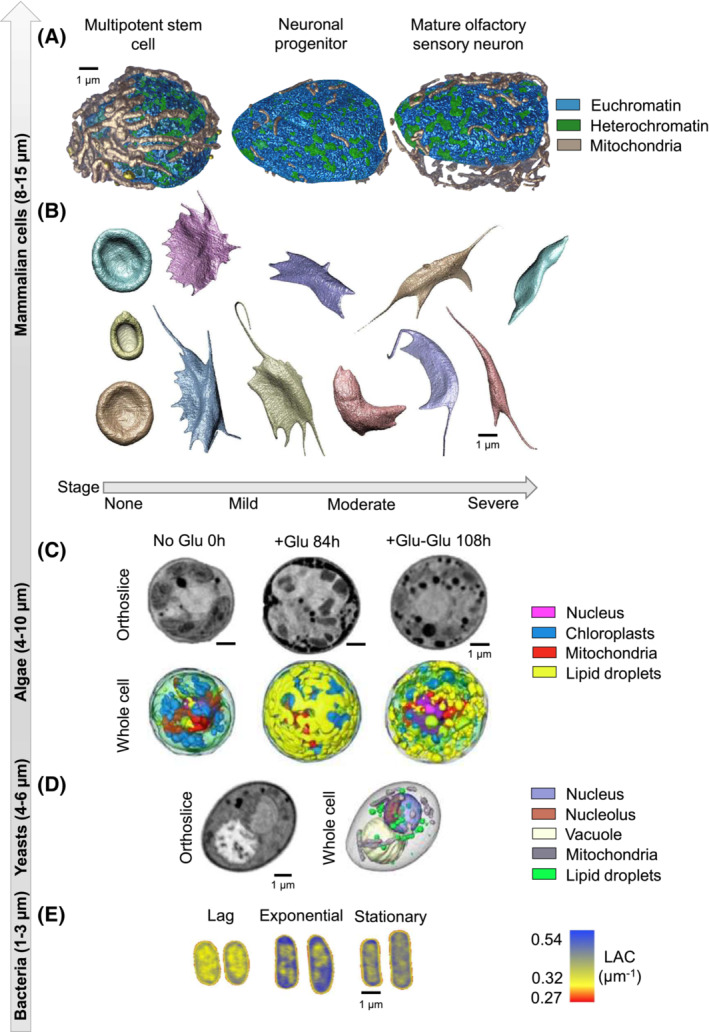
Different cell types visualized by SXT. (A) Changes in nuclear organization of mouse olfactory epithelial cells (multi‐potent stem cell, neuronal progenitor, mature olfactory sensory neuron). 70 (B) Morphological changes in sickled blood cells at different stages of the disease. 57 (C) Changes in the ultrastructure and organization of the green alga, Chromochloris zofingiensis, after different timepoints after glucose stimulation and reversal 71 ; (D) organization of Saccharomyces cerevisiae 72 ; and (E) alterations in nucleoid compaction in Escherichia coli 73 at different stages of cell growth (lag, exponential, and stationary). Scale bar 1 μm.
3. WHOLE‐CELL COMPUTATIONAL MODELS
Our goal in this article is to demonstrate that SXT can provide a measured structural framework for a whole‐cell computational model—a 3D map upon which the activities of genes and gene products can be assessed during the complex biological processes of an entire cell. Although there is a significant body of literature that provides a comprehensive overview of whole‐cell modeling (e.g., see review by Singla et al., 2018 for overview 80 ), it is more extensive than we can discuss in this manuscript. However, given our goals of establishing how SXT data can function as a valuable contribution to this endeavor, we will briefly summarize a wide range of different modeling approaches and the type of predictive understanding they aim to enable.
Models based on mathematical formalisms, such as ordinary differential equations (ODEs), partial differential equations (PDEs), Boolean networks, and flux balance analysis (FBA), have been used to model cell functions. 81 , 82 , 83 Constraint‐based methods have been used to integrate biochemical, genetic, and genomic data into a mathematical framework that enables a mechanistic description of metabolic physiology. 82 There are many other tools currently available for the integration of different cell data, for example: (1) E‐cell, a modeling tool for genetic and biochemical processes 84 ; (2) V‐cell, a platform based on a set of ordinary or partial equations that simulates molecular mechanisms based on restraints derived by imaging the cellular localization of molecules in the cell (https://vcell.org) 85 ; and (3) M‐cell, a 3D reaction–diffusion model using Monte Carlo methods (https://mcell.org). 86 Both V‐cell and M‐cell combine information about molecular spatial distributions with mathematical descriptions of the interaction of different molecular species, providing details about molecular dynamics in the cell. A good overview of general approaches to whole‐cell modeling can be found at the Human Whole‐Cell Modeling Project (https://www.wholecell.org). 87
Another effort that combines atomic data with the cell's 3D landscape derived from experimental data is the models produced by using the software cellPack (http://cellPACK.org/). This software combines biological building blocks, such as molecules, at atomic resolution with cellular structural components at the mesoscale (e.g., organelles), using an algorithm that optimizes the molecular packing in restraint compartments. 10 , 88
3.1. Examples of SXT data used for whole‐cell modeling
Modeling methods can define the most likely path taken by a targeted biochemical component. An elegant example of this principle was reported by Isaacson and colleagues. 89 , 90 The authors used LAC values of chromatin obtained from SXT tomograms to model the time required for proteins to find specific DNA‐binding sites in the nucleus. Simulating the motion of the protein using a Markovian continuous‐time random walk revealed that proteins could find their DNA target faster than would be possible by diffusion alone.
The Alber Lab used probabilistic modeling approaches to show that large centromere clusters play a key role in overall chromosome positioning in the nucleus and in stabilizing specific chromatin interactions. 69 They first used Hi‐C data analyses of human B‐cell nuclei to identify possible packing patterns of chromosomes and the location of their centromeres. They then matched and confirmed the predicted centromere positions from Hi‐C with the locations previously identified in X‐ray tomograms based on their high LAC values.
Soft X‐ray tomograms were also used to model molecular diffusion, evaluating the contribution of cytoplasmic organelles as barriers to the molecular motion. 91 The investigators modeled the time required for a protein to diffuse from the plasma membrane to the nucleus in the presence or absence of specific organelles and compared the results with diffusion through an “empty” cytosol. 91 The study suggests that strong intracellular signal inactivation will produce sharper and more robust signaling from cell membrane to the nucleus. Mathematical models were then used to describe and quantify basic chemical signals, and to estimate the arrival times at the nuclear membrane of proteins that are activated at the cell membrane and diffuse throughout the crowded cytosol. The evaluation of an organelle barrier in the calculations was enabled using the reconstructed volumes obtained by SXT.
Single‐cell parasites such as Toxoplasma gondii can undergo several motile motifs, including exotic helical gliding. The Dunn lab used a theoretical approach to evaluate the role of self‐organized F‐actin in motion generation and used X‐ray tomograms to understand how the impact of several F‐actin patterns at the molecular level would be reflected at the cellular scale. 92 They adapted an initial theoretical model of actin self‐reorganization to the physical shape of the parasite acquired using SXT. Depending on the directional properties of the actin filaments, they justified complex motion motifs, inducing helical gliding of the parasite. This example presents an elegant application of SXT to a multiscale model, depicting the role of volumetric imaging techniques in informing theoretical models based on molecular features.
The combination of probabilistic models, visual representations, mathematical models, and experimental data models is being used by the Pancreatic Beta Cell Consortium (https://www.pbcconsortium.org/) with the goal of designing a comprehensive whole‐cell model of a pancreatic β cell (PBC). 80 SXT is contributing to this model by providing the structural organization of statistically significant populations of cells before stimulation (control) and after manipulation. The Bayesian metamodeling by Raveh et al. 93 integrates several multiscale and temporal models and serves as a tool to contextualize the original input models. The metamodel augments accuracy and also improves precision and completeness by resolving conflicts among the original input models. The Goodsell lab 24 contributed to the knowledge of the multiscale system by integrating proteomic and structural data at the molecular and cellular scale to provide a direct description of insulin secretory granules at different stages of maturation. These investigators took advantage of the different LAC values of insulin granules in the X‐ray tomograms to quantify the concentration of proteins and lipids in the experimental data of an insulin granule. Thus, based on the LAC values, they were able to simulate the composition of three different pools of insulin granules at different stages of maturation and identify their location in the cell. In addition, the SXT data enabled them to determine which of three proposed proteomes were responsible for the maturation process.
SXT data contribute to the design of models. These data can provide mesoscale details of multiple landscapes of organelle interactions and connectivity, 56 determine morphological alterations of the overall ultrastructure of cells 38 and integrate ‐omic (proteomic, metabolic, lipidomic) data by providing information about the spatial enrichment of metabolites under specific stimuli (Figure 4).
FIGURE 4.
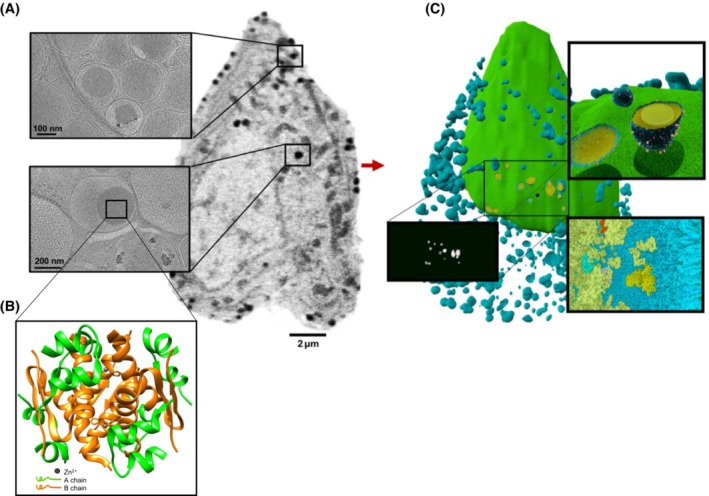
Data integration for a comprehensive whole‐cell model of pancreatic beta cells (from White et al., 2020 38 ©, some rights reserved; exclusive licensee AAAS; distributed under a CC BY‐NC 4.0 License (http://creativecommons.org/licenses/by‐nc/4.0/). (A) Imaging data from SXT 38 and cryo‐ET 94 of INS‐1 E rat insulinoma pancreatic beta cells showing details of information at different spatial scales. (B) Structural features of the insulin crystal of cells (insulin structural model 95 ; PDB ID: 2OM0). (C) Model of the cell produced in CellPack (http://cellPACK.org/), based on information from (A) and (B).
4. CONCLUSIONS
A comprehensive model of a whole cell must be based on both theoretical and experimental input obtained from a broad range of methods and technologies. Mapping this diverse body of information onto cellular structures requires 3D images of cells at multiple different time points and under a variety of physiological conditions. We have shown here that SXT can provide the structural framework for such models. Each voxel in an X‐ray tomogram contains information such as the chemical composition and molecular density, which allows it to be classified as part of a specific organelle or other subcellular feature. Since the absorption of X‐rays by cell structures is quantifiable, SXT can also detect changes in the biochemical composition of that structure. It is also possible to incorporate molecular information into the model by overlaying data from fluorescence microscopy onto the X‐ray data from the same cell. 39 , 68 Additional details of specific structures seen in SXT can be discerned by correlating images of those structures with cryoET data (Figure 4). Incorporating these data into the whole‐cell model will generate a much more precise framework for analyzing and predicting cell function.
Currently, SXT requires the use of a soft X‐ray microscope located at a synchrotron light source. This limits access to the technique, with demand outstripping available hours of experimental access. However, tremendous advances are being made in developing “tabletop” soft X‐ray sources 96 , 97 (such as the SiriusXT source 98 ; https://siriusxt.com), slated to become commercially available soon. As their name suggests, these new sources are sized to fit in a typical academic or industrial laboratory. Widening the availability of SXT will almost certainly lead to an explosion in applying the technique, particularly as major experimental input for whole‐cell modeling.
AUTHOR CONTRIBUTIONS
Carolyn A. Larabell designed the research. Valentina Loconte, Jian‐Hua Chen, and Bieke Vanslembrouck prepared the samples. Valentina Loconte and Jian‐Hua Chen collected and analyzed the data. Axel A. Ekman reconstructed the data and performed post‐processing analysis. Valentina Loconte analyzed the data. Gerry McDermott and Mark A. Le Gros provided support during the data analysis. Mark A. Le Gros provided support during the data acquisition. Valentina Loconte, Gerry McDermott, Mark A. Le Gros, and Carolyn A. Larabell wrote the paper with the contribution of all co‐authors.
FUNDING INFORMATION
The National Center for X‐ray Tomography was supported by NIH NIGMS (P41GM103445 and P30GM138441) and the DOE's Office of Biological and Environmental Research (DE‐AC02‐5CH11231). The Center is located at the Advanced Light Source, a U.S. DOE Office of Science User Facility under contract no. DE‐AC02‐05CH11231. CAL, VL and BV were supported by NSF 1902936.
DISCLOSURES
The authors declare no competing interest.
ACKNOWLEDGMENTS
The authors thank Kate L. White at University of Southern California and all the members of the Pancreatic Beta Cell Consortium for the constructive discussion.
Loconte V, Chen J‐H, Vanslembrouck B, et al. Soft X‐ray tomograms provide a structural basis for whole‐cell modeling. The FASEB Journal. 2023;37:e22681. doi: 10.1096/fj.202200253R
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.
REFERENCES
- 1. Patwardhan A, Ashton A, Brandt R, et al. A 3D cellular context for the macromolecular world. Nat Struct Mol Biol. 2014;21(10):841‐845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schmidt M. Time‐resolved macromolecular crystallography at pulsed X‐ray sources. Int J Mol Sci. 2019;20(6):1401. doi: 10.3390/ijms20061401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dickerson JL, Garman EF. The potential benefits of using higher X‐ray energies for macromolecular crystallography. J Synchrotron Radiat. 2019;26:922‐930. [DOI] [PubMed] [Google Scholar]
- 4. Wolff AM, Young ID, Sierra RG, et al. Comparing serial X‐ray crystallography and microcrystal electron diffraction (MicroED) as methods for routine structure determination from small macromolecular crystals. IUCrJ. 2020;7:306‐323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nakane T, Kotecha A, Sente A, et al. Single‐particle cryo‐EM at atomic resolution. Nature. 2020;587(7832):152‐156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pandey S, Bean R, Sato T, et al. Time‐resolved serial femtosecond crystallography at the European XFEL. Nat Methods. 2020;17(1):73‐78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sobolev E, Zolotarev S, Giewekemeyer K, et al. Megahertz single‐particle imaging at the European XFEL. Commun Phys. 2020;3(1):1‐11. [Google Scholar]
- 8. Karr JR, Sanghvi JC, MacKlin DN, et al. A whole‐cell computational model predicts phenotype from genotype. Cell. 2012;150(2):389‐401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fava E, Dehghany J, Ouwendijk J, et al. Novel standards in the measurement of rat insulin granules combining electron microscopy, high‐content image analysis and in silico modelling. Diabetologia. 2012;55(4):1013‐1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Johnson GT, Goodsell DS, Autin L, Forli S, Sanner MF, Olson AJ. 3D molecular models of whole HIV‐1 virions generated with cellPACK. Faraday Discuss. 2014;169:23‐44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lučić V, Rigort A, Baumeister W. Cryo‐electron tomography: the challenge of doing structural biology in situ. J Cell Biol. 2013;202(3):407‐419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mahamid J, Pfeffer S, Schaffer M, et al. Visualizing the molecular sociology at the HeLa cell nuclear periphery. Science. 2016;351(6276):969‐972. [DOI] [PubMed] [Google Scholar]
- 13. Buckley G, Gervinskas G, Taveneau C, Venugopal H, Whisstock JC, de Marco A. Automated cryo‐lamella preparation for high‐throughput in‐situ structural biology. J Struct Biol. 2020;210(2):107488. [DOI] [PubMed] [Google Scholar]
- 14. Valm AM, Cohen S, Legant WR, et al. Applying systems‐level spectral imaging and analysis to reveal the organelle interactome. Nature. 2017;546(7656):162‐167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Guo Y, Li D, Zhang S, et al. Visualizing intracellular organelle and cytoskeletal interactions at nanoscale resolution on millisecond timescales. Cell. 2018;175(5):1430‐1442.e17. [DOI] [PubMed] [Google Scholar]
- 16. Zhang J, Campbell RE, Ting AY, Tsien RY. Creating new fluorescent probes for cell biology. Nat Rev Mol Cell Biol. 2002;3(12):906‐918. [DOI] [PubMed] [Google Scholar]
- 17. Denbeaux G, Anderson E, Chao W, et al. Soft X‐ray microscopy to 25 nm with applications to biology and magnetic materials. Nucl Instrum Methods Phys Res, Sect A. 2001;467‐468(1):841‐844. [Google Scholar]
- 18. Attwood D, Chao W, Anderson EH, et al. Imaging at high spatial resolution: soft X‐ray microscopy to 15 nm. J Biomed Nanotechnol. 2006;2(2):75‐78. [Google Scholar]
- 19. Weiss D, Schneider G, Vogt S, et al. Tomographic imaging of biological specimens with the cryo transmission X‐ray microscope. Nucl Instrum Methods Phys Res A. 2001;467(Part 2):1308‐1311. [Google Scholar]
- 20. Chao W, Harteneck BD, Liddle JA, Anderson EH, Attwood DT. Soft X‐ray microscopy at a spatial resolution better than 15 nm. Nature. 2005;435(7046):1210‐1213. [DOI] [PubMed] [Google Scholar]
- 21. Attwood D. Soft X‐Rays and Extreme Ultraviolet Radiation: Principles and Applications. Cambridge University Press; 2007. [Google Scholar]
- 22. Parkinson DY, Epperly LR, McDermott G, Le Gros MA, Boudreau RM, Larabell CA. Nanoimaging cells using soft X‐ray tomography. Methods Mol Biol. 2013;950:457‐481. [DOI] [PubMed] [Google Scholar]
- 23. LeGros MA, Chen J‐H, Do M, McDermott G, Larabell CA. Putting molecules in the picture: using correlated light microscopy and soft X‐ray tomography to study cells. In: Jaeschke EJ, Khan S, Schneider JR, Hastings JB, eds. Synchrotron Light Sources and Free‐Electron Lasers: Accelerator Physics, Instrumentation and Science Applications. Springer Nature Switzerland AG 2020; 2016:1367‐1391. [Google Scholar]
- 24. Autin L, Barbaro BA, Jewett AI, et al. Integrative structural modelling and visualisation of a cellular organelle. QRB Discov. 2022;3:e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Weiss D, Schneider G, Niemann B, Guttmann P, Rudolph D, Schmahl G. Computed tomography of cryogenic biological specimens based on X‐ray microscopic images. Ultramicroscopy. 2000;84(3–4):185‐197. [DOI] [PubMed] [Google Scholar]
- 26. Ekman A, Chen J‐H, Vanslembrouck B, Larabell CA, Le Gros MA, Weinhardt V. Extending of imaging volume in soft x‐ray tomography. bioRxiv. 2022:491437. doi: 10.1101/2022.05.11.491437 [DOI] [Google Scholar]
- 27. Sakdinawat A, Attwood D. Nanoscale X‐ray imaging. Nat Photonics. 2010;4(12):840‐848. [Google Scholar]
- 28. Chen J‐H, Vanslembrouck B, Loconte V, et al. A protocol for full‐rotation soft X‐ray tomography of single cells. STAR Protoc. 2022;3(1):101176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Loconte V, Chen J‐H, Cortese M, et al. Using soft X‐ray tomography for rapid whole‐cell quantitative imaging of SARS‐CoV‐2‐infected cells. Cell Rep Methods. 2021;1(7):100117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Weinhardt V, Chen JH, Ekman A, McDermott G, Le Gros MA, Larabell C. Imaging cell morphology and physiology using X‐rays. Biochem Soc Trans. 2019;47:489‐508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kirz J, Jacobsen C, Howells M. Soft‐X‐ray microscopes and their biological applications. Q Rev Biophys. 1995;28(1):33‐130. [DOI] [PubMed] [Google Scholar]
- 32. Larabell C, McDermott G, LeGros M. 3D imaging of cells with soft x‐rays. In: Abstracts of Papers of the American Chemical Society, Vol. 251. American Chemical Society; 2016. [Google Scholar]
- 33. Schneider G, Guttmann P, Heim S, et al. Three‐dimensional cellular ultrastructure resolved by X‐ray microscopy. Nat Methods. 2010;7(12):985‐987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McDermott G, Fox DM, Epperly L, et al. Visualizing and quantifying cell phenotype using soft X‐ray tomography. Bioessays. 2012;34(4):320‐327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Larabell CA, Le Gros MA. X‐ray tomography generates 3‐D reconstructions of the yeast, Saccharomyces cerevisiae, at 60‐nm resolution. Mol Biol Cell. 2004;15(3):957‐962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Larabell CA, Nugent KA. Imaging cellular architecture with X‐rays. Curr Opin Struc Biol. 2010;20(5):623‐631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Le Gros MA, McDermott G, Cinquin BP, et al. Biological soft X‐ray tomography on beamline 2.1 at the advanced light source. J Synchrotron Radiat. 2014;21(6):1370‐1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. White KL, Singla J, Loconte V, et al. Visualizing subcellular rearrangements in intact β cells using soft X‐ray tomography. Sci Adv. 2020;6(50):1‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Smith EA, McDermott G, Do M, et al. Quantitatively imaging chromosomes by correlated cryo‐fluorescence and soft x‐ray tomographies. Biophys J. 2014;107(8):1988‐1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Uchida M, Sun Y, McDermott G, et al. Identification of an NADH‐dependent 5‐hydroxymethylfurfural‐reducing alcohol dehydrogenase in Saccharomyces cerevisiae . Yeast. 2008;25:191‐198. [DOI] [PubMed] [Google Scholar]
- 41. Do M, Isaacson SA, McDermott G, Le Gros MA, Larabell CA. Imaging and characterizing cells using tomography. Arch Biochem Biophys. 2015;581:111‐121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Duke E, Dent K, Razi M, Collinson LM. Biological applications of cryo‐soft X‐ray tomography. J Microsc. 2014;255(2):65‐70. [DOI] [PubMed] [Google Scholar]
- 43. Chen HY, Chiang DML, Lin ZJ, et al. Nanoimaging granule dynamics and subcellular structures in activated mast cells using soft X‐ray tomography. Sci Rep. 2016;6:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Winey M, Meehl JB, O'Toole ET, Giddings TH. Conventional transmission electron microscopy. Mol Biol Cell. 2014;25(3):319‐323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kornfeld J, Denk W. Progress and remaining challenges in high‐throughput volume electron microscopy. Curr Opin Neurobiol. 2018;50:261‐267. [DOI] [PubMed] [Google Scholar]
- 46. Graham BJ, Hildebrand DGC, Kuan AT, et al. High‐throughput transmission electron microscopy with automated serial sectioning. bioRxiv. 2019:657346. doi: 10.1101/657346 [DOI] [Google Scholar]
- 47. Goldman LW. Principles of CT and CT technology. J Nucl Med Technol. 2007;35(3):115‐128. [DOI] [PubMed] [Google Scholar]
- 48. Guenter L, Wolfgang HH. theoretical framework for filtered back projection in tomosynthesis. ProcSPIE. 1998;3338:1127‐1137. [Google Scholar]
- 49. Willemink MJ, de Jong PA, Leiner T, et al. Iterative reconstruction techniques for computed tomography part 1: technical principles. Eur Radiol. 2013;23(6):1623‐1631. [DOI] [PubMed] [Google Scholar]
- 50. Schneider G, Anderson E, Vogt S, et al. Computed tomography of cryogenic cells. Surf Rev Lett. 2002;9(1):177‐183. [Google Scholar]
- 51. Parkinson DY, Knoechel C, Yang C, Larabell CA, Le Gros MA. Automatic alignment and reconstruction of images for soft X‐ray tomography. J Struct Biol. 2012;177(2):259‐266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sorrentino A, Nicolas J, Valcarcel R, et al. MISTRAL: a transmission soft X‐ray microscopy beamline for cryo nano‐tomography of biological samples and magnetic domains imaging. J Synchrotron Radiat. 2015;22(4):1112‐1117. [DOI] [PubMed] [Google Scholar]
- 53. Carzaniga R, Domart M‐C, Collinson LM, Duke E. Cryo‐soft X‐ray tomography: a journey into the world of the native‐state cell. Protoplasma. 2014;251(2):449‐458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Groen J, Conesa JJ, Valcárcel R, Pereiro E. The cellular landscape by cryo soft X‐ray tomography. Biophys Rev. 2019;11(4):611‐619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cinquin BP, Do M, McDermott G, et al. Putting molecules in their place. J Cell Biochem. 2014;115(2):209‐216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Loconte V, Singla J, Li A, et al. Soft X‐ray tomography to map and quantify organelle interactions at the mesoscale. Structure. 2022;30:1‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Harkiolaki M, Darrow MC, Spink MC, Kosior E, Dent K, Duke E. Cryo‐soft X‐ray tomography: using soft X‐rays to explore the ultrastructure of whole cells. Emerg Top Life Sci. 2018;2(1):81‐92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Schmahl G. X‐ray microscopy. Nucl Instrum Methods Phys Res. 1983;208(1):361‐365. [Google Scholar]
- 59. Weinhardt V, Chen JH, Ekman AA, et al. Switchable resolution in soft X‐ray tomography of single cells. PLoS One. 2020;15(1):1‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Loconte V, White KL. The use of soft X‐ray tomography to explore mitochondrial structure and function. Mol Metab. 2021;57:101421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gibson E, Giganti F, Hu Y, et al. Automatic multi‐organ segmentation on abdominal CT with dense V‐networks. IEEE Trans Med Imaging. 2018;37(8):1822‐1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Dima AA, Elliott JT, Filliben JJ, et al. Comparison of segmentation algorithms for fluorescence microscopy images of cells. Cytometry A. 2011;79(7):545‐559. [DOI] [PubMed] [Google Scholar]
- 63. Ekman A, Chen J‐H, Dermott GM, Le Gros MA, Larabell C. Task based semantic segmentation of soft X‐ray CT images using 3D convolutional neural networks. Microsc Microanal. 2020;26(S2):3152‐3154. [Google Scholar]
- 64. Francis JP, Wang H, White K, Syeda‐Mahmood T, Stevens R. Neural Network Segmentation of Cell Ultrastructure Using Incomplete Annotation, 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), 3‐7 April 2020, 2020: 1183–1187.
- 65. Pelt DM, Sethian JA. A mixed‐scale dense convolutional neural network for image analysis. Proc Natl Acad Sci. 2018;115(2):254‐259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Li A, Zhang X, Singla J, et al. Auto‐segmentation and time‐dependent systematic analysis of mesoscale cellular structure in β‐cells during insulin secretion. PLoS One. 2022;17(3):e0265567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Li A, Zhang S, Loconte V, et al. An intensity‐based post‐processing tool for 3D instance segmentation of organelles in soft X‐ray tomograms. PLoS One. 2022;17(9):e0269887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Elgass KD, Smith EA, LeGros MA, Larabell CA, Ryan MT. Analysis of ER‐mitochondria contacts using correlative fluorescence microscopy and soft X‐ray tomography of mammalian cells. J Cell Sci. 2015;128(15):2795‐2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Tjong H, Li W, Kalhor R, et al. Population‐based 3D genome structure analysis reveals driving forces in spatial genome organization. Proc Natl Acad Sci U S A. 2016;113(12):E1663‐E1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Gros L, Mark A, Clowney EJ, et al. Soft X‐ray tomography reveals gradual chromatin compaction and reorganization during neurogenesis In vivo. Cell Rep. 2016;17(8):2125‐2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Roth MS, Gallaher SD, Westcott DJ, et al. Regulation of oxygenic photosynthesis during trophic transitions in the green alga chromochloris zofingiensis. Plant Cell. 2019;31(3):579‐601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Uchida M, Sun Y, McDermott G, et al. Quantitative analysis of yeast internal architecture using soft X‐ray tomography. Yeast. 2011;28(3):227‐236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hammel M, Amlanjyoti D, Reyes FE, et al. HU multimerization shift controls nucleoid compaction. Sci Adv. 2016;2(7):e1600650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ugarte F, Sousae R, Cinquin B, et al. Progressive chromatin condensation and H3K9 methylation regulate the differentiation of embryonic and hematopoietic stem cells. Stem Cell Reports. 2015;5(5):728‐740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Darrow MC, Zhang YJ, Cinquin BP, et al. Visualizing red blood cell sickling and the effects of inhibition of sphingosine kinase 1 using soft X‐ray tomography. J Cell Sci. 2016;129(18):3511‐3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Remesh SG, Verma SC, Chen JH, et al. Nucleoid remodeling during environmental adaptation is regulated by HU‐dependent DNA bundling. Nat Commun. 2020;11(1):1‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Dasgupta S, Auth T, Gov NS, et al. Membrane‐wrapping contributions to malaria parasite invasion of the human erythrocyte. Biophys J. 2014;107(1):43‐54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Hanssen E, Knoechel C, Dearnley M, et al. Soft X‐ray microscopy analysis of cell volume and hemoglobin content in erythrocytes infected with asexual and sexual stages of Plasmodium falciparum . J Struct Biol. 2012;177(2):224‐232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Myllys M, Ruokolainen V, Aho V, et al. Herpes simplex virus 1 induces egress channels through marginalized host chromatin. Sci Rep. 2016;6(1):28844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Singla J, McClary KM, White KL, Alber F, Sali A, Stevens RC. Opportunities and challenges in building a spatiotemporal multi‐scale model of the human pancreatic β cell. Cell. 2018;173(1):11‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Bordbar A, Monk JM, King ZA, Palsson BO. Constraint‐based models predict metabolic and associated cellular functions. Nat Rev Genet. 2014;15(2):107‐120. [DOI] [PubMed] [Google Scholar]
- 82. Feist AM, Palsson B. The growing scope of applications of genome‐scale metabolic reconstructions using Escherichia coli . Nat Biotechnol. 2008;26(6):659‐667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Gelbach PE, Zheng D, Fraser SE, White KL, Graham NA, Finley SD. Kinetic and data‐driven modeling of pancreatic β‐cell central carbon metabolism and insulin secretion. PLoS Comput Biol. 2022;18(10):e1010555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Tomita M, Hashimoto K, Takahashi K, et al. E‐CELL: software environment for whole‐cell simulation. Bioinformatics. 1999;15(1):72‐84. [DOI] [PubMed] [Google Scholar]
- 85. Schaff JC, Slepchenko BM, Loew LM. Physiological modeling with virtual cell framework. Methods Enzymol. 2000;321:1‐23. [DOI] [PubMed] [Google Scholar]
- 86. Kerr RA, Bartol TM, Kaminsky B, et al. Fast Monte Carlo simulation methods for biological reaction‐diffusion systems in solution and on surfaces. SIAM J Sci Comput. 2008;30(6):3126‐3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Szigeti B, Roth YD, Sekar JAP, Goldberg AP, Pochiraju SC, Karr JR. A blueprint for human whole‐cell modeling. Curr Opin Syst Biol. 2018;7:8‐15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Johnson GT, Autin L, Al‐Alusi M, Goodsell DS, Sanner MF, Olson AJ. CellPACK: a virtual mesoscope to model and visualize structural systems biology. Nat Methods. 2014;12(1):85‐91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Isaacson SA, McQueen DM, Peskin CS. The influence of volume exclusion by chromatin on the time required to find specific DNA binding sites by diffusion. Proc Natl Acad Sci U S A. 2011;108(9):3815‐3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Isaacson SA, Larabell CA, Le Gros MA, McQueen DM, Peskin CS. The influence of spatial variation in chromatin density determined by X‐ray tomograms on the time to find DNA binding sites. Bull Math Biol. 2013;75(11):2093‐2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ma J, Do M, Le Gros MA, et al. Strong intracellular signal inactivation produces sharper and more robust signaling from cell membrane to nucleus. PLoS Comput Biol. 2020;16(11):1‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Hueschen CL, Zarko L‐AS, Chen J‐H, et al. Emergent actin flows explain diverse parasite gliding modes. bioRxiv. 2022:495399. doi: 10.1101/2022.06.08.495399 [DOI] [Google Scholar]
- 93. Raveh B, Sun L, White KL, et al. Bayesian metamodeling of complex biological systems across varying representations. Proc Natl Acad Sci. 2021;118(35):e2104559118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Zhang X, Carter SD, Singla J, et al. Visualizing insulin vesicle neighborhoods in β cells by cryo‐electron tomography. Sci Adv. 2020;6(50):eabc8258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Norrman M, Schluckebier G. Crystallographic characterization of two novel crystal forms of human insulin induced by chaotropic agents and a shift in pH. BMC Struct Biol. 2007;7:1‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Kordel M, Dehlinger A, Seim C, et al. Laboratory water‐window x‐ray microscopy. Optica. 2020;7(6):658‐674. [Google Scholar]
- 97. Kördel M, Fogelqvist E, Carannante V, et al. Biological laboratory X‐ray microscopy. 2019, 28. [DOI] [PMC free article] [PubMed]
- 98. Fahy K, Weinhardt V, Vihinen‐Ranta M, et al. Compact cell imaging device (CoCID) provides insights into the cellular origins of viral infections. J Phys Photonics. 2021;3(3):031002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.


