Abstract
Human papillomavirus (HPV) assays used in cervical cancer screening should be clinically validated according to international criteria. OncoPredict HPV® Screening (SCR) is a partial genotyping multiplex real‐time PCR assay targeting E6/E7 genes of 13 high‐risk (hr) HPVs. OncoPredict HPV® SCR (index assay) identifies HPV‐16 and HPV‐18 separately, 11 other hrHPV in aggregate and includes quality controls for sample adequacy, DNA extraction efficiency and PCR inhibition. 1300 VALGENT‐2 study samples (from women aged 20–60 attending the Scottish cervical cancer screening program) were tested with the index assay and the GP5+/6+ PCR enzyme immunoassay (standard comparator assay). Non‐inferior accuracy detecting cervical intraepithelial neoplasia of grade 2 or worse (CIN2+) of the index versus comparator was verified. Intra‐ and interlaboratory reproducibility of the index was evaluated by overall concordance and Cohen's kappa, using a sub‐population (n = 526). Relative sensitivity and specificity for CIN2+ of the index versus comparator were 1.01 (95% confidence interval [CI]: 0.99–1.03) and 1.02 (95% CI: 1.0–1.04), respectively. Noninferiority p values were all ≤0.05, except for CIN3+ in patients ≥30 years. Excellent intra‐ and interlaboratory reproducibility was shown with concordance >98% and kappas >0.95. OncoPredict HPV® SCR fulfills the three international validation criteria for hrHPV DNA tests in cervical cancer screening.
Keywords: cervical cancer, HPV genotyping, human papillomavirus, OncoPredict HPV® , test validation, VALGENT
1. INTRODUCTION
Human papillomavirus (HPV) infection is the most common sexually transmitted infection globally. It is estimated that more than 85% of individuals will acquire any HPV infection once in their lifetime. 1 Whilst most HPV infections resolve spontaneously without any clinical manifestation within two years, 10% of infections persist and may progress to cervical precancerous lesions. 2 HPVs are classified according to their oncogenic potential, with 12 genotypes considered carcinogenic (Group 1) by the International Agency for Research on Cancer (IARC) and referred to as high‐risk (hr) HPV types (HPV16, HPV18, HPV31, HPV33, HPV35, HPV39, HPV45, HPV51, HPV52, HPV56, HPV58 and HPV59). 3 , 4
These 12 hrHPV types are together responsible for more than 90% of all cervical cancer cases; with HPV 16 possessing the highest oncogenic potential followed by HPV 18, causing together more than 70% of cervical cancers worldwide. 5 , 6 As cervical cancer develops slowly, over approximately 20 years from HPV infection to cancer, screening is an important tool for the early detection and treatment of pre‐cancerous lesions, avoiding progression to invasive disease. Cervical cancer is one of the most successfully preventable neoplasia emphasizing the importance of effective screening. 7 , 8 Strong evidence from large randomized clinical trials has demonstrated a reduction of incidence and mortality from cervical cancer when hrHPV‐based screening is used as compared to screening with conventional cytology. 9 , 10 , 11 As a result, many countries have replaced cytology with HPV testing as a primary screening tool. 12 , 13 , 14 , 15 , 16 This paradigm shift to hrHPV‐based primary screening has been accompanied by the influx and rapid increase of the number of HPV assays on the global market. 17 , 18 Several new HPV molecular assays allowing point of care, manual or high‐throughput testing have been developed, based on various technologies, ranging from immunoassays, PCR and CRISPR‐Cas12a‐based platforms. One of the first HPV assays introduced in cervical cancer screening programs is based on enzyme immunoassay (EIA) technology that detects PCR amplification products. However the majority of current HPV DNA molecular assays are based on real‐time PCR, which is less labor‐intensive and time‐consuming than EIA. More recently droplet digital PCR (ddPCR) has been explored, allowing not only HPV detection but also the accurate and sensitive determination of HPV viral load. 19 Isothermal amplification and CRISPR‐Cas12a technologies have also been evaluated as rapid HPV detection methods, which may prove useful in resources‐constrained settings. 20 , 21 HPV assays selected for cervical cancer screening need to be properly validated in terms of clinical accuracy and reproducibility, according to recognized international guidelines for their use in cervical cancer screening. 22
International criteria for the validation of a new HPV assay require assessment of non‐inferior sensitivity and specificity of the assay compared to a standard comparator test, in addition to achieving high intra and inter‐laboratory reproducibility using well preserved samples that are representative of a screening population. 23
The VALidation of HPV GENotyping Tests (VALGENT) is a collaborative consortium for the comparison and validation of HPV assays that are intended for screening according to the international validation guidelines. 24 Established first in 2012, there have been four iterations of the VALGENT framework to date from different countries in Europe. The VALGENT framework allows evaluation of new HPV assays by providing samples that are representative of a screening population. In this study, we aim to evaluate the clinical performance and reproducibility of the OncoPredict® SCR assay using samples that are nested within an organized screening program from the second installment of the VALGENT framework (VALGENT‐2).
2. MATERIALS AND METHODS
2.1. Study population and VALGENT‐2 panel
The VALGENT‐2 18 iteration was conducted in Scotland. Scotland has a national organized cervical screening program with high uptake of around 70%. At the time of sample collection in 2012, women were then screened by cytology at three‐year intervals from the age of 20 up to 60 years old. The study population for VALGENT‐2 included 1300 samples of which 1000 were from consecutive samples collated from the routinely screened population (screening population) and 300 samples were cytologically abnormal samples (enrichment population). All samples for the VALGENT‐2 panel were collated at one of the National Health Service (NHS) cytopathology laboratories that supported the cervical screening program in the storage medium PreservCyt liquid (Hologic). The panel was collated in August 2012. The median age of the women contributing the specimens was 38 years (range 19–68 years).
2.2. Testing of samples with index assay
OncoPredict HPV® SCR assay (Hiantis) is a partial genotyping qualitative real‐time PCR assay targeting E6 and E7 DNA sequences of 13 hrHPV types. It allows separate genotyping for HPV‐16 and HPV‐18 and detection of 11 other hrHPV types (HPV −31, −33, −35, −39, −45, −51, −52, −56, −58, −59, and −68) in aggregate. The multiplex real‐time PCR assay includes two reaction wells. The first well is the sample's quality control tube (QC) which permits evaluation of: (i) sample's cellularity by a C‐C Motif Chemokine Receptor 5 (CCR5) gene through a quantitative assay; (ii) an exogenous control gene target which is added to the sample prior to nucleic acid extraction, to assess DNA recovery; (iii) an amplification control target included in the real‐time PCR mix to determine potential PCR inhibition. The second well allows screening for the presence or absence of HPV‐16, HPV‐18 and the 11 hrHPV types. This tube also includes the amplification control to verify for PCR inhibition. The CCR5 DNA control dilutions required to prepare the standard curve for target quantification are included in the OncoPredict HPV® kit.
Nucleic acid extraction and HPV testing with OncoPredict HPV® SCR assay were performed at the University of Milano‐Bicocca from May to June 2021, using a Fluent 480 (Tecan) automated workstation. DNA extraction was performed by means of Quick‐DNA/RNA MagBead (Zymo) from 400 µl of sample starting volume. Ten microliters of exogenous control was added to the sample before nucleic acids extraction to evaluate the efficiency of the extraction procedure. The extracted DNA was eluted in a final 100 µl volume into a clean 96‐well plate. OncoPredict HPV® SCR was performed according to the manufacturer's protocol. Briefly, the automated liquid handler dispensed 10 µl of mastermix QC and SCR, respectively into two reaction wells, and subsequently 5 µl of sample's DNA extract added to each well, resulting in a 15 µl total PCR reaction volume. Negative and positive controls provided by the manufacturer were included in each PCR run. PCR was carried out using a CFX384 Touch Real‐Time PCR Detection System (Bio‐Rad). Samples were considered adequate for the analysis if (i) containing at least 400 cells/reaction; (ii) extraction efficiency ≥10%; (iii) amplification control cycle threshold (Ct) ≤30. Samples were defined as HPV positive if the Ct was ≤40 for the HPV channels.
2.3. Testing of samples with standard comparator test
GP5+/6+ PCR enzyme immunoassay (hereinafter referred to as GP‐EIA [Diassay) 25 was the standard comparator test used for the VALGENT‐2 samples. GP‐EIA is an enzyme immunoassay‐based method able to detect HPV DNA, after PCR amplification of the targeted DNA sequences. It detects an aggregated presence of 14 hrHPV types (HPV −16, −18, −31, −33, −35, −39, −45, −51, −52, −56, −58, −59, −66, and −68) and an aggregated presence of six low‐risk HPV types (HPV −6, −11, −40, −42, −43, and −44). The GP‐EIA, together with the Hybrid Capture 2 (HC2) 26 assay, is one of the clinically validated assays currently recommended as standard comparator test, given substantial evidence of their clinical performance coming from large randomized controlled trials. 5 , 27 GP‐EIA was the comparator assay against which the index assay OncoPredict HPV® SCR was evaluated. As the GP‐EIA has no genotyping capability, the LMNX genotyping kit HPV GP hr (GP5+/6+LMNX Diassay) (hereinafter referred to as “LMNX Diassay” [Diassay BV—previous version marketed as the digene HPV Genotyping LQ Test by Qiagen]) 23 was used to compare type‐specific HPV genotyping. The LMNX Diassay includes individual resolution of 18 HPV types, including the same 14 hrHPV types as in GP‐EIA in addition to four additional probable/possible high‐risk (pHR)‐HPV types (IARC Group 2 A/B carcinogens) (HPV −26, −53, −73, and −82) and an internal control for a human DNA target to evaluate the quality of the specimen. 22 Testing of samples with the GP‐EIA and LMNX Diassay was performed starting from 500 µl of the original samples at DDL Diagnostic Laboratory during April to May 2013 and April to September 2013. 23 All samples have since been stored at −80°C in the Scottish HPV Archive, a biobank to support HPV research at the University of Edinburgh, Scotland (https://shine.ed.ac.uk/research-updates/scottish-hpv-archive). They were retrieved from −80°C and dispatched with a dedicated courier for cold storage to University of Milano‐Bicocca for testing with the index assay.
2.4. Reproducibility
To assess the intra‐ and interlaboratory reproducibility of the OncoPredict HPV® SCR, a subset of 526 samples were randomly selected by the Unit of Cancer Epidemiology, Sciensano from the initial VALGENT‐2 panel, which included 30% hrHPV positive samples as foreseen by the validation guidelines. 16 In particular the reproducibility panel contained 157 hrHPV positive samples and 369 hrHPV negative samples, based on the results of VALGENT‐2 standard comparator test GP‐EIA. Residual DNA extracts following initial testing for the selected 526 samples were retested twice at the University of Milano‐Bicocca (Laboratory 1) to assess intra‐laboratory reproducibility and subsequently tested once at PTP (Parco Tecnologico Padano) (Laboratory 2) to assess inter‐laboratory reproducibility. A flowchart depicting the sample flow and testing procedures conducted in this study is displayed in Figure 1.
Figure 1.
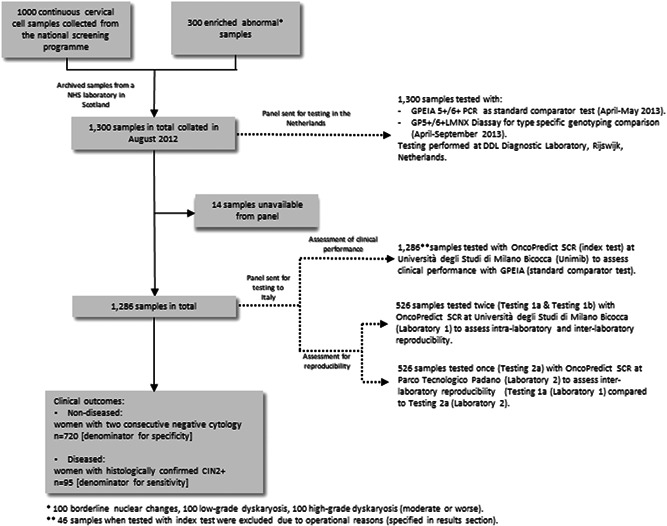
Flowchart of samples and testing of the OncoPredict HPV® SCR assay using the VALGENT‐2 panel
2.5. Clinical outcomes
Classification of cytological and histological outcomes was based on the reporting guidelines by the British Society for Clinical Cytopathology. 24 According to the screening program guidance in Scotland, women with abnormal cytological results were referred to colposcopy and biopsies were performed where clinically indicated. For this study, outcomes for clinical sensitivity were determined when patients received a histologically confirmed diagnosis of cervical intraepithelial neoplasia grade 2 or worse (CIN2+). Outcomes for clinical specificity or the nondiseased group estimates were determined when women had two repeated negative cytology samples across two screening rounds. In this case, women who received two negative cytology results enrolled in the Scottish screening program over the span of 3 years were considered the non‐diseased group.
2.6. Statistical analyses
The absolute clinical sensitivity for CIN2+ and specificity for ≤CIN1 of the OncoPredict HPV® SCR were calculated with 95% CI in all women and women aged 30 and older. 16 Non‐inferior accuracy statistics compared to the standard comparator test (GP‐EIA) as proposed by Tang et al. 28 were assessed applying the benchmarks of 0.90 for relative sensitivity and 0.98 for relative specificity. McNemar (McN) test was applied to determine any differences between matched proportions.
Intra‐reproducibility of the OncoPredict HPV® SCR testing was assessed by the University of Milano‐Bicocca laboratory, whereas the inter‐reproducibility assessment included the initial testing at the University of Milano‐Bicocca and subsequent testing at the PTP laboratory (Parco Tecnologico Padano). The reproducibility was expressed as the overall percentage agreement which is the ratio of the number of concordant results (positive on both assays + negative on both assays) overall test results. The reproducibility validation criterion was considered as fulfilled when the left 95% confidence interval (CI) bound for hrHPV concordance exceeded 87% and the kappa > 0.5. 16
Additionally, genotype‐specific concordance was assessed between the OncoPredict HPV® SCR and LMNX Diassay. The concordance between OncoPredict HPV® SCR and LMNX Diassay was assessed for HPV‐16 and HPV‐18 and aggregate hrHPVs using Kappa statistics. Cohen's kappa (as defined by Fleiss 29 ) were categorized as following levels of agreement between the two assays: (1.00 ≥ K > 0.80): excellent; (0.80 ≥ K > 0.60): good; (0.60 ≥ _K > 0.40): moderate; (0.40 ≥ _K > 0.20): fair; (0.20 ≥ K > 0.00): poor.
Ninety‐five percent exact CIs were calculated (95% CI) for all proportions. The CI around relative sensitivity and specificity took the paired design of comparisons into account. The level of statistical significance was set at 0.05. Statistical analyses were performed with STATA version 16.
2.7. Ethical approval statement
Ethical approval was obtained for the study. As the cervical samples are anonymized residual samples surplus to diagnostic requirements which have been donated to the Scottish HPV Archive which comes under the governance of the National Research Scotland Lothian Bioresource (East of Scotland Research Ethics Service Ref 15 ‐ES‐0094 and 20/ES/0061). Samples were provided for this study following approval from the Scottish HPV Archive steering committee (HPV Archive application Ref‐0060).
3. RESULTS
3.1. Study population
The demographics and cytopathological results of the VALGENT‐2 population have been described previously. 18 Of the 1300 samples from VALGENT‐2, only 1286 residual aliquots were available for the present study. Of these, a further 46 (25 from the screening and 21 from the enrichment population) were excluded, 17 due to an insufficient starting volume (<400 µl, required for testing with OncoPredict HPV® SCR) and 29 (2.3%) of invalid samples due to either low sample cellularity (six samples with <400 cells/reaction) or reduced nucleic acid extraction efficiency (23 with nucleic acid recovery <10%). In total, there were 1240 samples with valid OncoPredict HPV® SCR results. Initial testing of the 1300 samples with GP‐EIA excluded two samples due to operational issues. Therefore, the final number of matched samples between GP‐EIA and OncoPredict HPV® SCR was n = 1239. From the screening population, 720 women had two consecutive negative cytology results and were used as the nondiseased group whereas 95 patients had a diagnosis of CIN2+ and were used as the diseased group. From the diseased group, 50 patients had a diagnosis of CIN3+. The hrHPV prevalence in the Scottish screening population was 16.2% (95% CI = 13.9%–18.7%) when assessed with OncoPredict HPV® SCR and 17.3% (95% CI = 14.9%–19.8%) when assessed with GP‐EIA (Table 1).
Table 1.
hrHPV positivity within the VALGENT‐2 screening population assessed with OncoPredict HPV® SCR assay and GP5+/6+‐PCR‐EIA
| Number screened | Number hrHPV+ | % hrHPV+ | |
|---|---|---|---|
| OncoPredict HPV SCR | 968 | 157 | 16.2% |
| GP5+/6+‐PCR‐EIA | 968 | 167 | 17.3% |
Abbreviations: HPV, human papillomavirus; hrHPV, high‐risk HPV.
3.2. Clinical performance of OncoPredict HPV® SCR assay
In the total study population, OncoPredict HPV® SCR detected 90/95 of CIN2+ and 49/50 of CIN3+ cases corresponding to a sensitivity of 94.7% (95% CI = 93.2%–96.3%) and 98.0 (95% CI = 97.0–99.0), respectively. In the nondiseased group, 659/720 samples were OncoPredict HPV® SCR negative resulting in a specificity for ≤CIN1 of 91.5% (95% CI = 89.6%–93.5%). In patients aged 30 years or older, the absolute sensitivity for CIN2+ and CIN3+ was 92.9% (95% CI = 90.9%–94.8%) and 95.5% (95% CI = 93.8%–97.1%), respectively. Specificity for ≤CIN1 of OncoPredict HPV® SCR was slightly higher in women aged 30 years and older compared to the total study population and was 93.3% (95% CI = 91.4–95.2). The accuracy estimates of OncoPredict HPV® SCR and GP‐EIA 5+/6+ are shown in Table 2.
Table 2.
Absolute sensitivity and specificity of the OncoPredict HPV® SCR compared to GP‐EIA 5+/6+ in the total study population and women aged ≥30 years
| Parameter | Outcome | n/N a | OncoPredict HPV® SCR, % (95% CI) | n/N a | GP‐EIA 5+/6+, % (95% CI) |
|---|---|---|---|---|---|
| Total study population | |||||
| Sensitivity | CIN2+ | 90/95 | 94.7 (93.2–96.3) | 89/95 | 93.7 (92.0–95.4) |
| Sensitivity | CIN3+ | 49/50 | 98.0 (97.0–99.0) | 49/50 | 98.0 (97.0–99.0) |
| Specificity | ≤CIN1 | 659/720 | 91.5 (89.6–93.5) | 649/720 | 90.1 (88.1–92.2) |
| Women ≥ 30 years | |||||
| Sensitivity | CIN2+ | 39/42 | 92.9 (90.–94.8) | 39/42 | 92.9 (90.9–94.8) |
| Sensitivity | CIN3+ | 21/22 | 95.5 (93.8–97.1) | 21/22 | 95.5 (93.8–97.1) |
| Specificity | ≤CIN1 | 569/610 | 93.3 (91.4–95.2) | 563/610 | 92.3 (90.3–94.3) |
Note: For more details, see Supporting Information: Table 1.
Abbreviation: HPV, human papillomavirus.
For sensitivity, n is number of hrHPV+ among N women with disease (CIN2 + or CIN3 + ). For specificity, n is number of hrHPV‐ among N women with two consecutive negative cytology results (i.e., non‐diseased group [≤CIN1]).
Relative sensitivities for CIN2+ and CIN3+ and relative specificity for ≤CIN1 of OncoPredict HPV® SCR versus GP‐EIA 5+/6+ are shown in Table 3. The relative sensitivity for CIN2+ in the total study population was 1.01 (95% CI = 0.99–1.03) and 1.00 (95% CI = 0.95–1.05) in patients aged 30 years and older. The relative specificity for ≤CIN1 was 1.02 (95% CI = 1.00–1.04) and 1.01 (95% CI = 0.99–1.03) in all women and in women aged 30 years or older, respectively. OncoPredict HPV® SCR demonstrated noninferior sensitivity for CIN2+ and CIN3+ and noninferior specificity for ≤CIN1 compared to GP‐EIA 5+/6+ (p ni = 0.001 and p ni = 0.0004, respectively) in the total study population and also in the population of patients of 30 years and older (p ni always >0.05, except for the sensitivity for CIN3+ among patients aged 30 years and older). However, the relative sensitivity for CIN3+ in this latter population was the same (21/22 for OncoPredict HPV® SCR and 21/22 for GP‐EIA) and the p McN was 1.00.
Table 3.
Relative sensitivity and specificity of the OncoPredict HPV® SCR compared to GP 5+/6+ PCR‐EIA
| Comparison | Outcome | Relative accuracy (95% CI) | p Mcn | p n.inf |
|---|---|---|---|---|
| Total study population | ||||
| Relative sensitivity | CIN2+ | 1.01 (0.99–1.03) | 1.000 | <0.0005 |
| Relative sensitivity | CIN3+ | 1.00 (0.96–1.03) | 1.000 | 0.0098 |
| Relative specificity | ≤CIN1 | 1.02 (1.00–1.04) | 0.164 | 0.0004 |
| Women ≥ 30 years | ||||
| Relative sensitivity | CIN2+ | 1.00 (0.95–1.05) | 1.000 | 0.0187 |
| Relative sensitivity | CIN3+ | 1.00 (0.92–1.09) | 1.000 | 0.0633 |
| Relative specificity | ≤CIN1 | 1.01 (0.99–1.03) | 0.377 | 0.0021 |
Abbreviation: HPV, human papillomavirus.
3.3. Intra‐ and interlaboratory reproducibility of the OncoPredict HPV® SCR assay
Of the 526 samples that were selected for the reproducibility assessment, three samples were excluded due to low sample cellularity (<400 cells/reaction). Results for the intralaboratory assessment showed 136 and 381 samples that were concordant for hrHPV positivity and negativity, whereas six sample pairs were discordant leaving an excellent overall agreement of 98.9% (95% CI = 97.5%–99.6%) based on a kappa value of 0.971 (95% CI = 0.947–0.994). The interlaboratory analysis showed 132 concordantly hrHPV positive and 381 concordantly hrHPV negative results whereas 10 sample pairs were discordant: excellent overall agreement of 98.1% (95% CI = 96.5%–99.1%) based on a kappa value of 0.951 (95% CI = 0.920–0.981). Results of the reproducibility analyses are presented in Table 4.
Table 4.
Intralaboratory and interlaboratory reproducibility of OncoPredict HPV® SCR
| First analysis in Unimib | ||||
|---|---|---|---|---|
| Intralaboratory analysis a | ||||
| Second analysis in Unimib | Positive | Negative | Total | |
| Positive | 136 | 1 | 137 | |
| Negative | 5 | 381 | 376 | |
| Total | 141 | 382 | 523 | |
| Interlaboratory analysis b | ||||
| Second analysis in PTP | Positive | Negative | Total | |
| Positive | 132 | 1 | 133 | |
| Negative | 9 | 381 | 390 | |
| Total | 141 | 832 | 523 | |
Abbreviation: HPV, human papillomavirus.
Overall HPV test agreement: 98.9 (95% CI 97.5%–99.6%); Kappa: 0.971 (95% CI 0.947–0.994)—indicating an excellent intralaboratory agreement.
Overall HPV test agreement: 98.1 (95% CI 96.5%–99.1%); Kappa: 0.951 (95% CI 0.920–0.981)—indicating an excellent inter‐laboratory agreement.
3.4. Type‐specific HPV prevalence of OncoPredict HPV® SCR and GP5+/6+ LMNX
Genotyping agreement when compared to the GP5+/6+ LMNX in the total study population was excellent for HPV16 genotyping (general agreement of 99.1% [95% CI: 98%–100%], a kappa value of 0.94), good for HPV 18 genotyping (general agreement of 98.8% [95% CI: 98%–99%], a kappa value of 0.77), and excellent for the pooled identification of other hrHPV types (general agreement of 95.6% [95% CI: 94%–97%], a kappa value of 0.87) (Table 5).
Table 5.
hrHPV genotyping agreement in the total study population between OncoPredict HPV® SCR and GP5+/6+ PCR‐LMNX
| HPV type | −/−a | +/+a | −/+a | +/−a | General agreement (95% CI) | Kappa (95% CI) |
|---|---|---|---|---|---|---|
| HPV 16 | 1135 | 93 | 1 | 10 | 99% (98%–100%) | 0.94 (0.90–0.98) |
| HPV 18 | 1198 | 26 | 2 | 13 | 99% (98%– 99%) | 0.77 (0.66–0.88) |
| Other hrHPVb | 948 | 236 | 37 | 18 | 96% (94%–97%) | 0.87 (0.83–0.90) |
Note: Color legend (adapted from Landis & Koch 31 ) for the levels of agreement: dark green (1.00 ≥ K > 0.80): excellent; light green (0.80 ≥ K > 0.60): good; yellow (0.60 ≥ _K > 0.40): moderate; orange (0.40 ≥ _ K > 0.20): fair; red (0.20 ≥ K > 0.00): poor.
Abbreviations: HPV, human papillomavirus; hrHPV, high‐risk HPV.
−/− both tests are concordantly negative; +/+ both tests are concordantly positive; −/+ OncoPredict HPV® SCR negative, GP5+/6+ LMNX positive; +/− OncoPredict HPV® SCR positive, GP5+/6+ LMNX negative.
Other hrHPV includes the aggregate of HPV types 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 68 for OncoPredict HPV® SCR whilst GP5+/6+ LMNX also includes the additional HPV types: −66, −26, −53, −73, and −82.
4. DISCUSSION
In this study, the clinical performance of OncoPredict HPV® SCR was assessed, in combination with an automated preanalytical workflow for nucleic acid extraction and PCR plate preparation, using VALGENT‐2 framework samples collected as part of a routine cervical screening program, according to international clinical validation criteria. hrHPV DNA testing with OncoPredict HPV® SCR assay has shown non‐inferior accuracy for the detection of cervical precancerous lesions and a level of intra‐ and inter‐laboratory reproducibility that meets the international validation criteria for cervical cancer screening.
The strong evidence of the effectiveness of HPV‐based screening in the last decade has seen a sudden increase in the number of HPV assays in the market. A recent inventory estimated that over 250 distinct HPV assays are currently available in the market but over 90% have not been clinically validated according to international validation guidelines for use in cervical cancer screening. 13 It is essential that new HPV assays will be clinically validated using samples that are representative of the screening context. OncoPredict HPV® SCR is a novel HPV assay with partial genotyping capability that enables the detection of 13 hrHPV types in a single reaction well, with separate identification of HPV‐16, HPV‐18, and 11 other pooled hrHPVs. The OncoPredict HPV® SCR assay also includes a unique quality control assessment of both sample adequacy and sample processing in a second reaction well. This feature enables an evaluation of the quality of sample collection and technical issues that may occur during the end to end process of testing. In particular, cellularity is evaluated by the quantification of the human CCR5 gene, present in a constant copy number/cell, allowing to control for the quality of sample collection. The amplification of the human target is evaluated in a separate reaction well from that set up for the detection of HPV positivity, so avoiding any potential interference due to competition of the amplification reactions. Moreover spiking the specimen with an exogenous control target before DNA extraction provides insight into yield and recovery of DNA in this preanalytical phase, allowing the possibility for laboratories to use OncoPredict HPV® SCR assay on their own extraction systems according to local available infrastructure/liquid handling platforms. Furthermore, potential PCR inhibition is assessed by means of an amplification control target included in both QC and SCR mastermixes. This extended quality control assessment of OncoPredict HPV® SCR assay allows a reduction of potential “false negative” results due to inadequate sample collection, particularly important in the case of self‐collected samples, and can highlight technical issues related to analytical (PCR) steps. This allows for more accurate troubleshooting and mitigation.
Although the VALGENT‐2 panel was collated back in 2012, this study shows that the performance of OncoPredict HPV® SCR was not affected by specimen storage over approximately 10 years. This study showed only 0.5% (6/1286) of samples invalid due to inadequate cellularity, possibly pointing to changes in the number of cells usable for HPV testing. While further exploration of a sample's storage life and its effects on the quality of HPV testing is justifiable, it is notable that valid samples that were tested with OncoPredict HPV® SCR did not show any reduction of sensitivity and specificity when compared to the GP‐EIA. A linkage of baseline data with the Scottish Pathology registry beyond the VALGENT2 follow‐up period is planned when all aliquots will be exhausted. This will identify cases with CIN2+ not yet discovered within the VALGENT observation period.
As HPV‐16 and HPV‐18 are linked to over 70% of cervical cancer cases worldwide, 5 , 6 limited genotyping assays which delineate HPV‐16 and HPV‐18 are often used to triage positive hrHPV samples. The use of assays with limited genotyping offers high‐throughput and quick turnover time when used in a screening context.
In conclusion, OncoPredict HPV® SCR shows non‐inferior clinical accuracy to that of the standard comparator test and sufficient interand intralaboratory reproducibility using the stringent international validation criteria evaluated through the VALGENT‐2 framework. This means that OncoPredict HPV® SCR may be added to the list of hrHPV assays considered as validated for screening. 30
AUTHOR CONTRIBUTIONS
Sharonjit K. Dhillon and Marc Arbyn have contributed to the study design and were involved in the practical coordination of the study. Clementina E. Cocuzza, Marianna Martinelli, Chiara Giubbi, Ruth C. Njoku, Ramya Bhatia, and Kate Cuschieri were involved in data collection and/or samples testing. Sharonjit K. Dhillon and Marc Arbyn have analyzed the data. Sharonjit K. Dhillon, Clementina E. Cocuzza, Pui Yan Jenny Chung, and Marc Arbyn have written the manuscript. All authors reviewed and/or edited the manuscript. All co‐authors approved the final manuscript and its submission to this journal.
CONFLICT OF INTEREST
C. E. C. is a minority shareholder of Hiantis Srl and received research grants and/or gratis consumables from Beckton Dickinson, Copan Italia, Seegene, Novosanis and Fujirebio. C. E. C. has received speaker honoraria and/or travel funds from Seegene, Beckton Dickinson, Copan Italia. C. E. C. is a minority shareholder of Hiantis Srl. K. C. and R. B.'s institution has received research funding or gratis consumables to support research from the following commercial entities in the last 3 years: Cepheid, Roche, Abbott, Euroimmun, GeneFirst, SelfScreen, Hiantis, Seegene and Hologic.
Supporting information
Supporting information.
ACKNOWLEDGMENTS
The authors would like to thank Giulia Bassanini from Parco Tecnologico Padano (PTP), Lodi, Italy for her support in the inter‐laboratory reproducibility testing. S. K. D. and M. A.'s institution has received support from the Horizon 2020 Framework Programme for Research and Innovation of the European Commission, through the RISCC Network (Grant No. 847845) and from the VALGENT framework, as explained in the VALGENT protocol (Arbyn et al., J ClinVirol 2016; 76 (S1): S14‐S21). S. K. D., C. E. C., P. Y. J. C., K. C., and M. A. received support from the SME Instrument Phase 2 Project: HPV OncoPredict [Grant agreement ID: 806551].
Dhillon SK, Cocuzza CE, Chung PYJ, et al. Evaluation of the clinical performance of OncoPredict HPV® SCR assay within the VALGENT‐2 framework. J Med Virol. 2022;95:e28417. 10.1002/jmv.28417
DATA AVAILABILITY STATEMENT
No data were deposited in a repository.
REFERENCES
- 1. International Agency forResearch on Cancer . Cervical Cancer Screening. IARC Handbooks of Cancer Prevention . Vol 18. IARC Publications. https://publications.iarc.fr/604 [Google Scholar]
- 2. Martin CM, O'Leary JJ. Histology of cervical intraepithelial neoplasia and the role of biomarkers. Best Pract Res Clin Obstet Gynaecol. 2011;25(5):605‐615. 10.1016/j.bpobgyn.2011.04.005 [DOI] [PubMed] [Google Scholar]
- 3. International Agency for Research on Cancer . Human Papillomaviruses. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. Volume 90. Vol 90. IARC Press; 2007. [Google Scholar]
- 4. Bouvard V, Baan R, Straif K, et al. A review of human carcinogens—Part B: biological agents. Lancet Oncol. 2009;10(4):321‐322. 10.1016/s1470-2045(09)70096-8 [DOI] [PubMed] [Google Scholar]
- 5. Arbyn M, Ronco G, Anttila A, et al. Evidence regarding human papillomavirus testing in secondary prevention of cervical cancer. Vaccine. 2012;30(suppl 5):F88‐F99. 10.1016/j.vaccine.2012.06.095 [DOI] [PubMed] [Google Scholar]
- 6. Ronco G, Giorgi‐Rossi P, Carozzi F, et al. Efficacy of human papillomavirus testing for the detection of invasive cervical cancers and cervical intraepithelial neoplasia: a randomised controlled trial. Lancet Oncol. 2010;11(3):249‐257. 10.1016/S1470-2045(09)70360-2 [DOI] [PubMed] [Google Scholar]
- 7. Sankaranarayanan R, Nene BM, Shastri SS, et al. HPV screening for cervical cancer in rural India. N Engl J Med. 2009;360(14):1385‐1394. 10.1056/NEJMoa0808516 [DOI] [PubMed] [Google Scholar]
- 8. Hillemanns P, Friese K, Dannecker C, et al. Prevention of Cervical Cancer: Guideline of the DGGG and the DKG (S3 Level, AWMF Register Number 015/027OL, December 2017)—Part 1 with introduction, screening and the pathology of cervical dysplasia. Geburtshilfe Frauenheilkd. 2019;79(2):148‐159. 10.1055/a-0818-5440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Maver PJ, Poljak M. Primary HPV‐based cervical cancer screening in Europe: implementation status, challenges, and future plans. Clin Microbiol Infect. 2020;26(5):579‐583. 10.1016/j.cmi.2019.09.006 [DOI] [PubMed] [Google Scholar]
- 10. Zeferino L, Bastos J, Vale D, et al. Guidelines for HPV‐DNA testing for cervical cancer screening in Brazil. Rev Bras Ginecol Obstet. 2018;40(6):360‐368. 10.1055/s-0038-1657754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huh WK, Ault KA, Chelmow D, et al. Use of primary high‐risk human papillomavirus testing for cervical cancer screening: interim clinical guidance. Obstet Gynecol. 2015;125(2):330‐337. 10.1097/AOG.0000000000000669 [DOI] [PubMed] [Google Scholar]
- 12. Cheung LC, Egemen D, Chen X, et al. 2019 ASCCP risk‐based management consensus guidelines: methods for risk estimation, recommended management, and validation. J Low Genit Tract Dis. 2020;24(2):90‐101. 10.1097/LGT.0000000000000528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Poljak M, Oštrbenk Valenčak A, Gimpelj Domjanič G, Xu L, Arbyn M. Commercially available molecular tests for human papillomaviruses: a global overview. Clin Microbiol Infect. 2020;26(9):1144‐1150. 10.1016/j.cmi.2020.03.033 [DOI] [PubMed] [Google Scholar]
- 14. Arbyn M, Snijders PJF, Meijer CJLM, et al. Which high‐risk HPV assays fulfil criteria for use in primary cervical cancer screening? Clin Microbiol Infect. 2015;21(9):817‐826. 10.1016/j.cmi.2015.04.015 [DOI] [PubMed] [Google Scholar]
- 15. Meijer CJLM, Berkhof H, Heideman DaM, Hesselink AT, Snijders PJF. Validation of high‐risk HPV tests for primary cervical screening. J Clin Virol. 2009;46(suppl 3):S1‐S4. 10.1016/S1386-6532(09)00540-X [DOI] [PubMed] [Google Scholar]
- 16. Meijer CJLM, Berkhof J, Castle PE, et al. Guidelines for human papillomavirus DNA test requirements for primary cervical cancer screening in women 30 years and older. Int J Cancer. 2009;124(3):516‐520. 10.1002/ijc.24010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Arbyn M, Depuydt C, Benoy I, et al. VALGENT: a protocol for clinical validation of human papillomavirus assays. J Clin Virol. 2016;76(suppl 1):S14‐S21. 10.1016/j.jcv.2015.09.014 [DOI] [PubMed] [Google Scholar]
- 18. Cuschieri K, Geraets DT, Moore C, Quint W, Duvall E, Arbyn M. Clinical and analytical performance of the onclarity HPV assay using the VALGENT framework. J Clin Microbiol. 2015;53(10):3272‐3279. 10.1128/JCM.01366-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rotondo JC, Oton‐Gonzalez L, Mazziotta C, et al. Simultaneous detection and viral DNA load quantification of different human papillomavirus types in clinical specimens by the high analytical droplet digital PCR method. Front Microbiol. 2020;11:591452. 10.3389/fmicb.2020.591452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gong J, Zhang G, Wang W, et al. A simple and rapid diagnostic method for 13 types of high‐risk human papillomavirus (HR‐HPV) detection using CRISPR‐Cas12a technology. Sci Rep. 2021;11(1):12800. 10.1038/s41598-021-92329-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang W, Du H, Huang X, et al. Evaluation of an isothermal amplification HPV detection assay for primary cervical cancer screening. Infect Agent Cancer. 2020;15:65. 10.1186/s13027-020-00328-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Geraets DT, Heideman DaM, de Koning MNC, et al. High‐throughput genotyping of high‐risk HPV by the digene HPV genotyping LQ test using GP5+/6+‐PCR and xMAP technology. J Clin Virol. 2009;46(suppl 3):S21‐S26. 10.1016/S1386-6532(09)70297-5 [DOI] [PubMed] [Google Scholar]
- 23. Geraets DT, Cuschieri K, de Koning MNC, et al. Clinical evaluation of a GP5+/6+‐based luminex assay having full high‐risk human papillomavirus genotyping capability and an internal control. J Clin Microbiol. 2014;52(11):3996‐4002. 10.1128/JCM.01962-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dudding N, Sutton J. BSCC terminology conference, koilocytosis and mild dyskaryosis. Cytopathology. 2002;13(6):379‐381. 10.1046/j.1365-2303.2002.00448_1.x [DOI] [PubMed] [Google Scholar]
- 25. Jacobs MV, Snijders PJ, van den Brule AJ, Helmerhorst TJ, Meijer CJ, Walboomers JM. A general primer GP5+/GP6(+)‐mediated PCR‐enzyme immunoassay method for rapid detection of 14 high‐risk and 6 low‐risk human papillomavirus genotypes in cervical scrapings. J Clin Microbiol. 1997;35(3):791‐795. 10.1128/jcm.35.3.791-795.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hesselink AT, van den Brule AJC, Brink AATP, et al. Comparison of hybrid capture 2 with in situ hybridization for the detection of high‐risk human papillomavirus in liquid‐based cervical samples. Cancer. 2004;102(1):11‐18. 10.1002/cncr.11904 [DOI] [PubMed] [Google Scholar]
- 27. Ronco G, Dillner J, Elfström KM, et al. Efficacy of HPV‐based screening for prevention of invasive cervical cancer: follow‐up of four European randomised controlled trials. Lancet. 2014;383(9916):524‐532. 10.1016/S0140-6736(13)62218-7 [DOI] [PubMed] [Google Scholar]
- 28. Tang NS, Tang ML, Chan ISF. On tests of equivalence via non‐unity relative risk for matched‐pair design. Stat Med. 2003;22(8):1217‐1233. 10.1002/sim.1213 [DOI] [PubMed] [Google Scholar]
- 29. Fleiss JL, Cohen J. The equivalence of weighted kappa and the intraclass correlation coefficient as measures of reliability. Educ Psychol Meas. 1973;33(3):613‐619. 10.1177/001316447303300309 [DOI] [Google Scholar]
- 30. Arbyn M, Simon M, Peeters E, et al. 2020 list of human papillomavirus assays suitable for primary cervical cancer screening. Clin Microbiol Infect. 2021;27(8):1083‐1095. 10.1016/j.cmi.2021.04.031 [DOI] [PubMed] [Google Scholar]
- 31. Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33(1):159‐174. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.
Data Availability Statement
No data were deposited in a repository.


