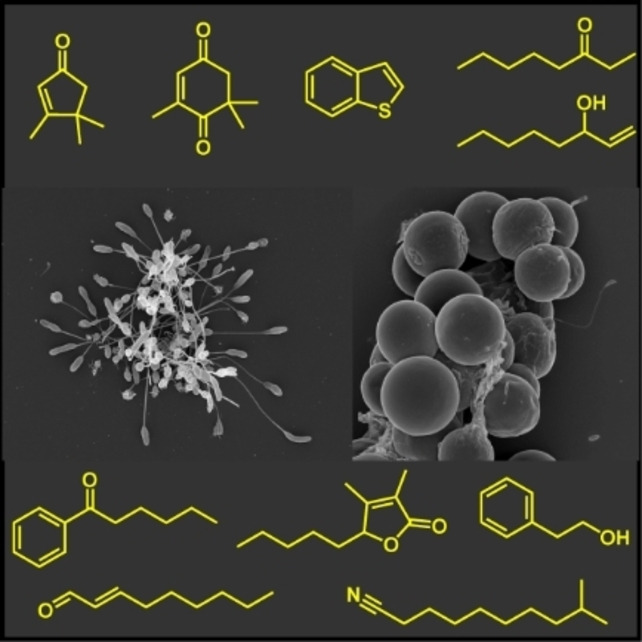
Abstract
Volatiles released by the apicomplexan alga Chromera velia CCAP1602/1 and their associated bacteria have been investigated. A metagenome analysis allowed the identification of the most abundant heterotrophic bacteria of the phycosphere, but the isolation of additional strains showed that metagenomics underestimated the complexity of the algal microbiome, However, a culture‐independent approach revealed the presence of a planctomycete that likely represents a novel bacterial family. We analysed algal and bacterial volatiles by open‐system‐stripping analysis (OSSA) on Tenax TA desorption tubes, followed by thermodesorption, cryofocusing and GC‐MS‐analysis. The analyses of the alga and the abundant bacterial strains Sphingopyxis litoris A01A‐101, Algihabitans albus A01A‐324, “Coraliitalea coralii” A01A‐333 and Litoreibacter sp. A01A‐347 revealed sulfur‐ and nitrogen‐containing compounds, ketones, alcohols, aldehydes, aromatic compounds, amides and one lactone, as well as the typical algal products, apocarotenoids. The compounds were identified by gas chromatographic retention indices, comparison of mass spectra and syntheses of reference compounds. A major algal metabolite was 3,4,4‐trimethylcyclopent‐2‐en‐1‐one, an apocarotenoid indicating the presence of carotenoids related to capsanthin, not reported from algae so far. A low overlap in volatiles bouquets between C. velia and the bacteria was found, and the xenic algal culture almost exclusively released algal components.
Keywords: apocarotenoids, GC-MS, marine bacteria, metagenomics, volatile organic compounds
Passing the smell test: The apicomplexan alga Chromera velia harbours a range of bacteria that have been identified by metagenome analysis. Analysis of the volatile organic compounds in the C. velia and individual bacterial isolates showed an almost absent overlap between the released compounds, thus indicating a low contribution of the bacteria to the odour of this algae, which is dominated by apocarotenoids.
Introduction
Chromera velia CCAP 1602/1, an apicomplexan alga associated with corals, was isolated in 2001 from the scleractinian coral Plesiastrea versipora from Sydney Harbour by Moore et al. [1] This unicellular eukaryotic phototroph is the type species of the phylum Chromerida within the alveolates. The latter represents a eukaryotic superensemble comprising three main phyla of protists, the dinoflagellates, ciliates and apicomplexans. [2] Apicomplexa consist mostly of parasites like the blood parasites piroplasms, coccidians and hemosporidians. These can cause malaria (several Plasmodium species), toxoplasmosis (Toxoplasma gondii) and veterinary coccidiosis (Eimeria). Phylogenetic analyses revealed a closer relationship of C. velia to the apicomplexan parasites than to the dinoflagellates. [1] The complex plastids of chromerids and dinoflagellates encode a form II Rubisco for CO2 fixation, but it is still unclear if they originated from a single endosymbiosis in a common ancestor or two independent more recent acquisitions.[ 3 , 4 ] Due to its phylogenetic position, C. velia is a key species for the understanding of apicomplexan evolution and could be used in medical research on antimalarial drugs.[ 1 , 5 ] This connecting link in apicomplexan evolution has been elected from the German Society for Plant Sciences to be the alga of the year 2020. Interestingly, C. velia lacks chlorophyll c, a typical photosynthesis pigment occurring in diatoms, brown algae and dinoflagellates. The diagnostic algal pigments are chlorophyll a, the carotenoids violaxanthin and an isomer of isofucoxanthin as major components, whereas β‐carotene was found as a minor component. [1] Carotenoids and xanthophylls are known to be produced by algae and are essential for their life cycle.[ 6 , 7 ] They are required for light‐harvesting and also act as scavengers of reactive oxygen species (ROS) thus protecting algal cells from damage by free radicals. [6]
The fatty acid composition of this alga has been investigated by Tomčala et al.. [8] The most abundant fatty acids were palmitic and stearic acids. Monounsaturated fatty acids comprised mainly vaccenic and oleic acids, while major polyunsaturated fatty acids included arachidonic acid, eicosapentaenoic acid (C20:5n‐3), linoleic acid, and dihomo‐γ‐linolenic acid. [8]
Besides these compounds no other constituents of C. velia were reported. We were interested in their release of small volatile organic compounds (VOCs), because such compounds might serve as chemical cues mediating interactions with various other organisms or even between algal cells.[ 9 , 10 ] VOCs can also indicate various physiological states, as algae can be affected by environmental effects such as light and temperature as well as nutrition or abiotic stress. [10] The original C. velia isolate CCAP 1602/1 is non‐axenic and comprises an uncharacterised set of associated heterotrophic bacteria that are living from the exudates of the phototrophic host. However, the composition of the specific bacterial community of Chromera is not known and has never been studied in chromerid algae. Metagenomics of non‐axenic unicellular phototrophs provide the promising perspective to establish genomes of associated heterotrophic bacteria that are difficult to cultivate, as previously shown for cyanobacteria, diatoms and dinoflagellates.[ 11 , 12 , 13 ] To gain a comprehensive insight into the hidden prokaryotic diversity in the xenic culture of Chromera, we collected bacterial isolates from the alga and investigated their abundance in the phycosphere with metagenome binning. Furthermore, we were interested in the identity of the compounds released as VOCs by Chromera and selected co‐occurring bacteria. Chromerid volatiles were investigated for the first time to investigate whether the VOCs are released form the alga or the bacteria and to get insight into the formation of the compounds in these microorganisms.
Results and Discussion
Bacterial isolates
Sixteen different bacterial strains of the classes Alphaproteobacteria, Gammaproteobacteria and Flavobacteriia were isolated from the phycosphere of the xenic alga C. velia CCAP 1602/1 (Table 1). Their classification is based on the analysis of the diagnostic 16S‐rRNA gene that is still serving as a valuable marker for a rapid taxonomic assessment (Table 1 and Table S1 in the Supporting Information). The 16S‐rDNA identities with the closest validly described type strain are ranging between 93.0 % and 100.0 % observed for Salinisphaera sp. A01A‐316 and Pyruvatibacter mobilis A01A‐348, respectively. According to the generally accepted threshold of 98.7 % for the delineation of different species, [14] at least three isolates, the alphaproteobacteria Litoreibacter sp. A01A‐347 (Roseobacteraceae, 96.1 %), Pseudooceanicola sp. A01An‐413 (Roseobacteraceae, 97.7%) and the gammaproteobacterium Salinisphaera sp. A01A‐316 (Salinisphaeraceae, 93.0 %) represent new species. Based on the rather conservative 16S rRNA gene threshold of 94.5 % [15] the latter might even represent the first strain of a novel genus.
Table 1.
Bacterial isolates from C. velia CCAP 1602/1. Strains used for headspace analysis are indicated in bold characters. The abundance of the bacteria in the phycosphere was calculated from the coverage information (= genome equivalents) of the metagenome assembled genomes (MAGs). For a more complete description see Table S1. A, Alphaproteobacteria; G, Gammaproteobacteria; F, Flavobacteriia, n.d., not determined.
|
|
Organism |
Strain] |
Abundance |
16S rDNA Identity (type strain) [%] |
Taxonomy |
Accession no. (16S‐rRNA) |
|---|---|---|---|---|---|---|
|
1 |
Pelagibius litoralis |
A01A‐306 |
145.9x |
98.95 |
A‐Hyphomicrobiales |
|
|
2 |
Algihabitans albus |
A01A‐324 |
59.4x |
99.58 |
A‐Rhodospirillales |
|
|
3 |
“Coraliitalea coralii” |
A01A‐333 |
33.3x |
98.76 |
F‐Flavobacteriales |
|
|
4 |
Sphingopyxis litoris |
A01A‐101 |
14.3x |
99.86 |
A‐Sphingomonadales |
|
|
5 |
Oceanicaulis alexandrii |
A01A‐340 |
12.4x |
99.65 |
A‐Maricaulales |
|
|
6 |
Litoreibacter sp. |
A01A‐347 |
10.5x |
96.06 |
A‐Rhodobacterales |
|
|
7 |
Roseovarius mucosus |
A01A‐006 |
n.d. |
99.77 |
A‐Rhodobacterales |
|
|
8 |
Sulfifobacter porphyrae |
A01A‐216 |
n.d. |
99.47 |
A‐Rhodobacterales |
|
|
9 |
Pseudooceanicola sp. |
A01An‐413 |
n.d. |
97.69 |
A‐Rhodobacterales |
|
|
10 |
Ponticoccus alexandrii |
A01An‐410 |
n.d. |
99.79 |
A‐Rhodobacterales |
|
|
11 |
Marivita cryptomonadis |
A01An‐414 |
n.d. |
100.00 |
A‐Rhodobacterales |
|
|
12 |
Pseudosulfitobacter pseudonitzschiae |
A01A‐217 |
n.d. |
99.22 |
A‐Rhodobacterales |
|
|
13 |
Stappia indica |
A01A‐202 |
n.d. |
99.16 |
A‐Hyphomicrobiales |
|
|
14 |
Porphyrobacter sanguineus |
A01A‐314 |
n.d. |
99.93 |
A‐Sphingomonadales |
|
|
15 |
Pyruvatibacter mobilis |
A01A‐348 |
n.d. |
100.00 |
A‐Hyphomicrobiales |
|
|
16 |
Salinisphaera bacterium |
A01A‐316 |
n.d. |
93.55 |
G‐Salinisphaerales |
The abundance of 14/16 isolated Alphaproteobacteria either represent their dominance on the surface of C. velia or a cultivation bias that was investigated by metagenome analyses (see below). Our alphaproteobacterial isolates are representing a broad range of families including Rhodovibrionaceae, Rhodospirillaceae, Sphingomonadaceae, Maricaulaceae, Stappiaceae, Erythrobacteraceae and Parvibaculaceae, but the dominant lineage is the family Roseobacteraceae with seven strains. [16] The isolation of roseobacters from the genera Litoreibacter, Roseovarius, Sulfitobacter, Pseudooceanicola, Ponticoccus, Marivita and Pseudosulfitobacter clearly shows that the phycosphere is a hotspot for this ecologically important generalist lineage of marine bacteria. [17] This conclusion is in agreement with the isolation of well‐studied taxa such as Marinovum algicola DG898 with more than a dozen extrachromosomal replicons or the model organism Dinoroseobacter shibae DFL 12 from cultures of non‐axenic dinoflagellates. [18] D. shibae and Phaeobacter inhibens have also been detected in algal blooms of the coccolithophore Emiliania huxleyi. [19]
Metagenomic binning
The metagenome of C. velia was analysed with a bioinformatic pipeline that was initially developed for non‐axenic cyanobacteria. [12] At nearly 200 Mbp, the genome of the eukaryotic alga is more than 40 times larger than the genomes of cyanobacteria typically ranging between 3 and 5 Mbp in size. This unfavourable host to heterotroph ratio poses a challenge for metagenomic binning. Accordingly, we used a comparably high Illumina sequencing depth to establish a sufficient amount of DNA sequences for the metagenomic binning pipeline (3.4+2.9 Gbp of raw data; BioProject: PRJNA822780; Accession: SRX14906536, SRX14773631). Furthermore, we were curious about the role of “contaminating” eukaryotic DNA from the alga, because the binning algorithms were developed for the establishment of metagenome assembled genomes (MAGs) from prokaryotes. Fortunately, our strategy was successful and metagenomic binning with MaxBin, MetaBAT and Concoct resulted in 12, 13 and 170 bins, respectively (Table S2). However, many of them do not represent authentic bacterial MAGs, which is, for example, documented for 154/170 comparably small Concoct bins that exhibit a calculated genomic completeness of 0.0 %. Nevertheless, all three programs binned very large portions of the eukaryotic C. velia genome and a prime example is the MetaBAT “bin.5” with a size of 163.4 Mbp comprising more than 80 % of the algal genome. Their contradictory CheckM‐based “marker lineage” classification as root, Archaea or Bacteria reflects the prokaryotic default options. Authentic bacterial bins have a completeness of at least 50 %, but binning of the same assembly with three different methods provided largely diverging results. MaxBin, MetaBAT and Concoct have individual strengths and weaknesses, [12] but the most accurate binning results for the associated heterotrophic bacteria of C. velia were again obtained with the dereplication, aggregation and scoring strategy of DAS Tool [20] (Table S2).
The DAS Tool approach resulted in eight bacterial bins with a 10‐ to 150‐fold genomic coverage (C. velia: 52x), which is illustrated in the word cloud in Figure 1A. We established isolates of the five abundant Alphaproteobacteria that are highlighted in blue and the red flavobacterium “C. coralii” A01A‐333, but could neither isolate the planctomycete of maxbin.007 nor the uncharacterised alphaproteobacterium of bin 65. Both MAGs represent partial bins with a completeness of only 50 %, and they are lacking the 16S‐rRNA gene, which reflects a typical binning problem of short read Illumina sequences.[ 12 , 21 ] Furthermore, the alphaproteobacterial bin even lacked the diagnostic RpoB protein. However, the missing 16S rDNA gene of the planctomycete could be identified in an incomplete bin of the MaxBin analysis (maxbin.011; 26.6 % completeness). The comparably low RpoB identity of 73.2 % of the planctomycete MAG with the next genome‐sequenced relative was indicative of a very distinct phylogenetic positioning. This conclusion was confirmed by the nearly complete 16S‐rRNA gene with a size of 1494 nt that showed a sequence identity of only 86.5 % with the closest related type strain Poriferisphera corsica DSM 103958. [22] This value exactly matches the threshold for the delineation of different bacterial families, [15] which proposes that the uncultivated planctomycete from the phycosphere of C. velia CCAP 1602/1 might represent a novel family in the planctomycete order Phycisphaerales. Planctomycetes are quite abundant on the surface of macroalgae [23] and Phycisphaerales MAGs have recently been identified in the phycosphere of the dinoflagellates Gambierdiscus carolinianus and Gambierdiscus caribaeus. [13]
Figure 1.
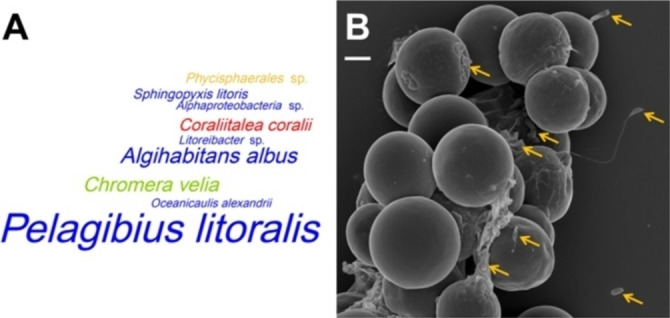
Distribution of the most abundant heterotrophic bacteria in the phycosphere of C. velia CCAP 1602/1. A) Word cloud of metagenomic bacterial bins with a completeness of >50 %. The size of the taxon names corresponds to the genome coverage of the respective bins; the coverage of the eukaryotic algae was inferred from partial bins (Table S2). The following colour code was used to distinguish between the most abundant lineages on phylum/class level: green‐Chromerida, blue‐Alphaproteobacteria, red‐Flavobacteriia, yellow‐Planctomycetes. B) Scanning electron microscopy of C. velia; scale bar: 2 μm. Some rod‐shaped, coccoid and stalked bacteria are highlighted by orange arrows (photo courtesy of Manfred Rohde).
The comparison between isolation and metagenomics clearly shows that both strategies provided complementary insights into the composition of Chromera's microbiome. We were able to isolate the three most abundant bacteria of the phycosphere, that is, P. litoralis A01A‐306 (Rhodovibrionaceae), A. albus A01A‐234 (Rhodospirillaceae) and “C. coralii” A01A‐333 (Flavobacteriaceae). These heterotrophs dominate the microbial flora on the algal surface comprising a potpourri of rod‐shaped, coccoid and stalked bacteria (Figure 1B). One impressive example on the right hand side of the scanning electron micrograph is a bacterium with a strikingly long and curved stalk, which likely represents the low abundant strain P. mobilis A01A‐348. However, the sequencing depth of the metagenome was not sufficient for the recovery of ten isolated bacteria including five of six Roseobacteraceae (Tables 1 and S2), which shows that the complexity of the microbiome would be underestimated exclusively based on metagenome analyses. Deep sequencing of non‐axenic cyanobacteria recently showed that the abundance of associated heterotrophic bacteria might differ in three orders of magnitude. [12] Nevertheless, the current study clearly showed the benefit of the metagenomic approach regardless of the optimizable sequencing depth. The planctomycete MAG illustrates that non‐axenic algae might be treasure troves of a hidden microbial diversity. The analysis of already cultured consortia has an ultimate advantage over environmental metagenome studies, as fastidious bacteria can be isolated afterwards.
Taxon sampling for chemical profiling
Entries 1–6 in Table 1 represent the most abundant bacterial isolates, assigned based on 16S rDNA amplicon sequencing results. As bacterial VOCs often serve distinct functions, [9] we tried to characterise the VOCs released by the six most abundant strains via headspace analysis. A. albus A01A‐324, “C. coralii” A01A‐333, S. litoris A01A‐101 and Litoreibacter sp. A01A‐347 grew readily in culture, while the most abundant organism, P. litoralis A01A‐306, as well as O. alexandrii A01A‐340, grew very slow and were therefore not included into the VOC analyses. A comparative analysis of the released VOCs was performed to identify the source of specific volatiles in the algae‐bacteria system, whose results are discussed in the next sections.
Volatile analysis
The identification of VOCs from bacteria is well established and can be performed using varying sensitive headspace methods, followed by GC‐MS. [24] In contrast, this approach is difficult for algae, mostly because the very slow growth rate results in comparatively lower emission of compounds during a given time period. While standard algal culture conditions for C. velia CCAP1602/1 were unsuccessful in generating enough material for analysis, growth conditions were optimized by the company CellDeg GmbH (Berlin, Germany; https://celldeg.com/) in HD100 cultivators with a defined light regime and external CO2 supply, which resulted in much higher growth rates and a markedly higher cell density.
For analysis, the headspace of such liquid cultures was drawn with a small pump over a Tenax TA desorption tube (open‐system‐stripping analysis, OSSA), followed by thermodesorption, cryofocussing, and GC‐MS‐analysis. The thermodesorption time and temperature had to be optimized to high sensitivity to detect the still low amounts of volatiles released. C. velia CCAP1602/1 and the four culturable dominant associated bacteria were investigated, with two or three biological replicates, respectively. The results for C. velia are shown in Table 2, while the bacterial results are shown in Tables S3–S6.
Table 2.
VOCs identified in headspace extracts of C. velia CCAP 1602/1.
|
Compound |
RI (exp)[a] |
RI (lit)[b] |
Identification[c] |
Replicate 1 |
Replicate 2 |
|---|---|---|---|---|---|
|
dimethyl disulfide (28) |
763 |
761 |
ms, ri |
xx |
xx |
|
4‐hydroxy‐4‐methyl‐2‐pentanone (95) |
838 |
839 |
ms, ri |
xx |
xxx |
|
hexanenitrile (39) |
878 |
879 |
ms, ri |
xx |
xx |
|
nitric acid pentyl ester (51) |
923 |
905 |
ms, ri |
xx |
x |
|
dimethyl trisulfide (29) |
965 |
965 |
ms, ri |
xx |
xx |
|
oct‐1‐en‐3‐ol (23) |
980 |
980 |
ms, ri |
xxx |
xx |
|
octan‐3‐one (70) |
985 |
985 |
ms, ri |
xxx |
xx |
|
benzoxazole (16) |
1016 |
1067 |
ms, ri |
xx |
xx |
|
3,4,4‐trimethylcyclopent‐2‐en‐1‐one (1) |
1049 |
1050 |
ms, ri, syn |
xx |
xx |
|
(E)‐oct‐2‐en‐1‐ol (22) |
1070 |
1070 |
ms, ri, syn |
xx |
|
|
nonan‐2‐one (53) |
1092 |
1092 |
ms, ri |
xx |
xx |
|
2‐methylbenzofuran (17) |
1100 |
1109 |
ms, ri |
xx |
|
|
2‐hydroxy‐2,6,6‐trimethylcyclohexan‐1‐one (2) |
1107 |
1109 |
ms, ri, syn |
xx |
xx |
|
4‐oxoisophorone (3) |
1143 |
1142 |
ms, ri |
xxx |
xxx |
|
(E)‐non‐2‐enal (25) |
1160 |
1160 |
ms, ri |
xx |
|
|
2,6,6‐trimethyl‐1,4‐cyclohexanedione (4) |
1168 |
1169 |
ms, ri |
xx |
xx |
|
2,3‐benzothiophene (18) |
1189 |
1189 |
ms, ri |
xx |
xx |
|
3,5,5‐trimethylcyclohex‐3‐en‐1‐yl acetate (5) |
1194 |
1192 |
ms, ri, syn |
xx |
xx |
|
1,2,3‐benzothiadiazol (21) |
1249 |
|
ms |
xx |
xx |
|
1‐phenylbutan‐1‐one (12) |
1252 |
1252 |
ms, ri |
xx |
xx |
|
(E)‐dec‐2‐enal (26) |
1262 |
1262 |
ms, ri |
xx |
|
|
1‐(2‐hydroxyphenyl)propan‐1‐one (14) |
1262 |
1262 |
ms, ri, syn |
xx |
xx |
|
3‐methylbenzo[b]thiophene (19) |
1307 |
1306 |
ms, ri, syn |
xx |
xx |
|
9‐methyldecanenitrile (42) |
1351 |
|
ms |
xx |
|
|
unknown compound M [196] |
1486 |
|
|
xxx |
xxx |
|
(E)‐β‐ionone (6) |
1487 |
1486 |
ms, ri |
xx |
xx |
|
dihydrobovolide (27) |
1520 |
1525 |
ms, ri, syn |
xx |
xx |
|
unknown compound M [180] |
1526 |
|
|
xx |
xx |
|
dihydroactinidiolide (7) |
1531 |
1532 |
ms, ri |
xx |
xx |
|
2,2′,5,5′‐tetramethyl‐1,1′‐biphenyl‐Isomer |
1681 |
|
ms |
xx |
xx |
|
dibenzo[b,d]thiophene (20) |
1750 |
1742 |
ms, ri |
xx |
x |
[a] The retention indices are averaged values of the measurements of all used replicates. [b] Retention indices were from NIST Chemistry WebBook or our own database. [c] The compounds were identified by comparison of the mass spectrum to a database spectrum (ms), comparison of the retention index to a published retention index on the same or similar GC fused silica capillary column (ri) or comparison to a synthetic or commercially available reference compound (syn). exp=experimental. lit=literature. The amounts of the compounds are given as 0–2 % (x), 2–20 % (xx), 20–100 % (xxx) relative to the largest peak area in the total ion chromatogram.
The GC‐MS analysis of the algal headspace (Figure 2) revealed about 300 compounds, most of them being background components from the growth medium. This was shown by control experiments with unocullated medium serving as control. Nevertheless, due to the high sensitivity of the headspace method used and good chromatographic separation, 31 compounds specific for C. velia were detected. The volatile profile comprised mainly sulfur‐ and nitrogen‐containing compounds, ketones, alcohols, aromatic compounds, one lactone as well as apocarotenoids.
Figure 2.
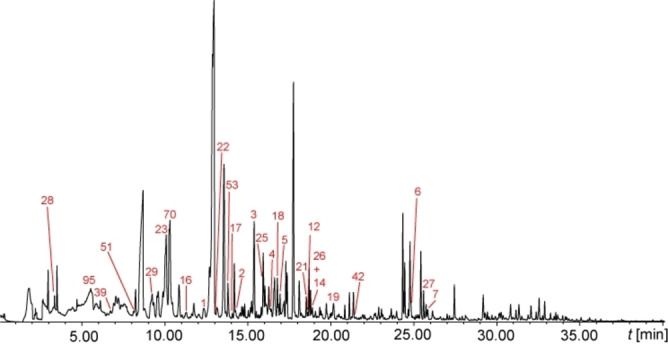
Total ion chromatogram of the headspace of C. velia CCAP1602/1. The numbers in the chromatograms refer to the compound numbers found in Table 2. Unnumbered peaks are medium constituents.
Similarly, bacteria were grown in MB medium, which was also used as negative control (Figure S1). S. litoris A01A‐101 released 27 compounds, containing mainly saturated and unsaturated ketones, alkyl‐substituted pyridines, nitriles and amides. A. albus A01A‐324 showed only nine VOCs, including mostly amides and ketones, whereas “C. coralii” A01A‐333 released the largest number of VOCs of all investigated strains. These 69 compounds included predominantly ketones and minor compounds such as alcohols, amides, aromatic, sulfur‐ and nitrogen‐containing VOCs. Litoreibacter sp. A01A‐347 released 30 VOCs comprising mostly saturated and unsaturated ketones, as well as aromatic compounds. There was minimal overlap of identical released volatiles of the analysed bacteria and the alga, but some of the amides, aromatic compounds and ketones were released by more than one bacterial strain. These results are also evident form the Venn diagram (Figure S2). In the following sections the identified compounds will be described in detail.
Apocarotenoids
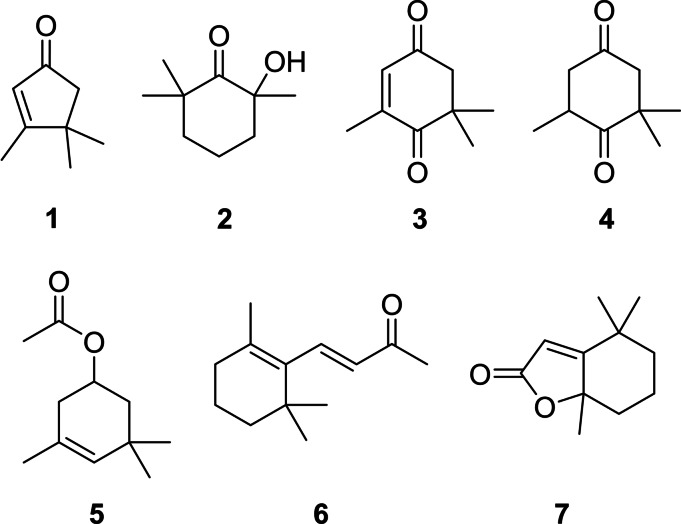
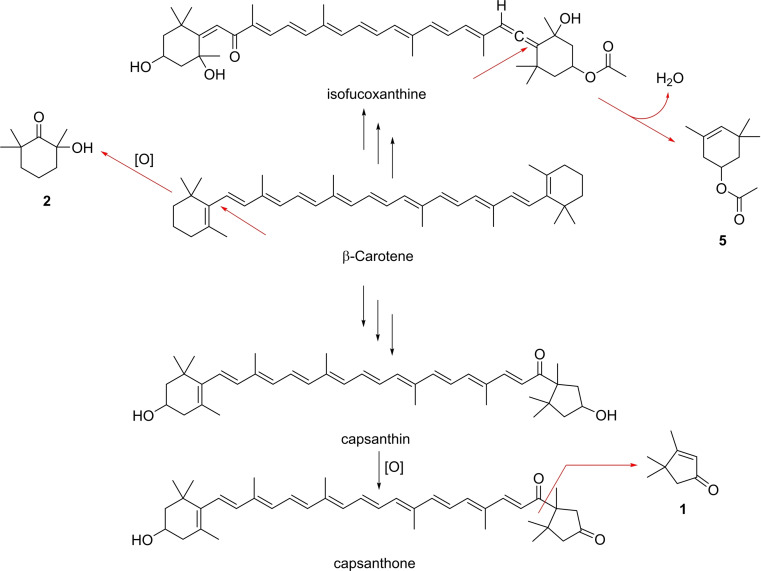
One of the most important groups of C. velia volatiles constitute apocarotenoids, small carotenoid degradation products (Figure 3). These compounds may arise from carotenoids by oxidative stress or ROS in the algal cells. The C13‐norisoprenoids dihydroactinidiolide (7) and (E)‐β‐ionone (6) were released in low concentrations. Both are typical degradation products of β‐carotene [25] and were found previously e. g. in extracts of ulvophycean green algae Ulva prolifera, Ulva linza and Monostroma nitidum[ 25 , 26 ] as well as VOCs released from different black sea red algae [27] or the dinoflagellate Prorocentrum cordatum. [28] The C9‐norisoprenoid 4‐oxoisophorone (3), one of the major compounds of C. velia, is a violaxanthin and zeaxanthin derived degradation product. [25] It is described as a volatile released by P. cordatum [28] and different marine and freshwater algae, [29] but is also a constituent of the essential oils of the macroalgae Cystoseira tamariscifolia, Sargassum muticum and Ulva lactuca.[ 25 , 30 ] The last species also released the C9‐norisoprenoid 2,6,6‐trimethyl‐1,4‐cyclohexanedione (4). [30] 3,4,4‐Trimethylcyclopent‐2‐en‐1‐one (1) is of special interest because it is a degradation product of the keto carotenoids capsanthin and capsanthone, the main carotenoids in ripe fruits of the red bell pepper Capsicum annuum.[ 25 , 31 ] Capsanthin/one and 1 have not been reported before from algae or bacteria. This occurrence suggests that β‐carotene is converted into capsanthin/one or another carotenoid with the capsanthin head group during carotenogenesis. A cleavage of capsanthone to β‐citraurin leads to the formation of 1 as has been suggested for Capsicum annuum (Scheme 1). [25]
Figure 3.
Apocarotenoids of C. velia.
Scheme 1.
Possible pathway of the formation of the apocarotenoids 1, 2, and 5.
The hydroxy ketone 2 and ester 5 represent further apocarotenoids. Compound 2 was reported from the headspace of U. prolifera and U. linza sampled in Japan [26] and from P. cordatum. [28] Ketol 2 could arise from oxidative cleavage of β‐carotene and hydroxylation (Scheme 1). As discussed, isofucoxanthine is produced by C. velia. [8] 3,5,5‐Trimethylcyclohex‐3‐en‐1‐yl acetate (5) could be generated from the cleavage of the allene double bond and dehydration. To our knowledge, compound 5 has not been reported from algae or bacteria so far. The apocarotenoids represent a specific group of compounds for C. velia and were not released by the investigated bacterial cultures.
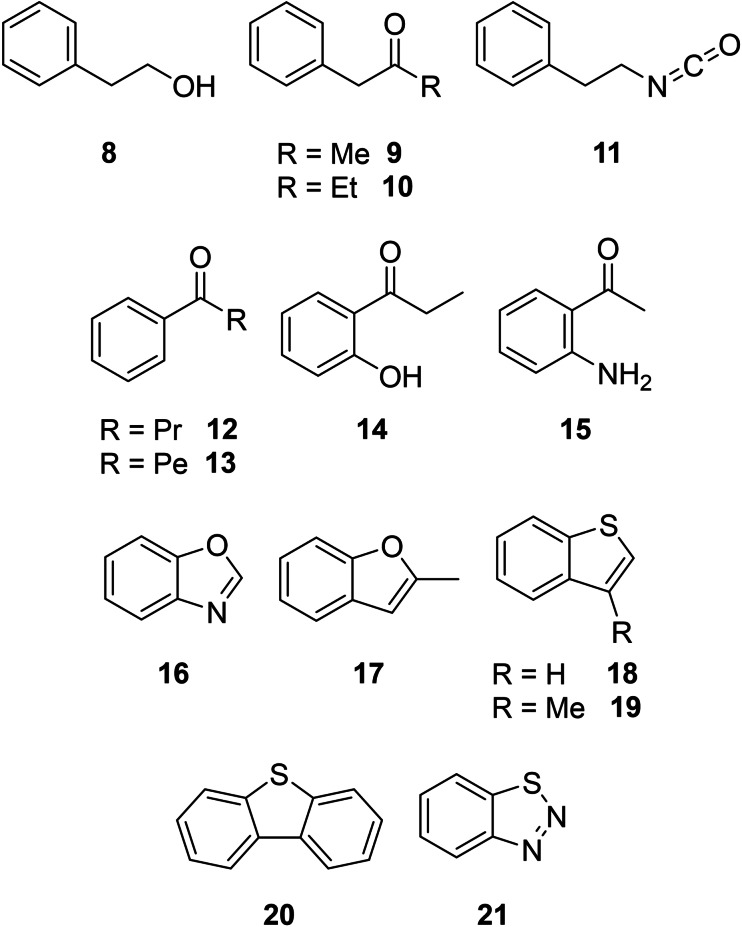
Aromatic compounds
Another important VOC class were oxygenated or heteroatom‐bearing aromatic compounds (Figure 4). C. velia CCAP 1602/1 emitted them as minor secondary metabolites, whereas individual cultures of the bacteria “C. coralii” A01A‐333 and Litoreibacter sp. A01A‐347 produced these compounds in larger amounts. 2‐Phenylethanol (8), a typical widespread aromatic compound in bacteria,[ 32 , 33 , 34 ] was the most abundant compound of “C. coralii“ A01A‐333. Larger amounts of 1‐phenylpropan‐2‐one (9) occurred in “C. coralii” and Litoreibacter sp., whereas its homologue 1‐phenylbutan‐2‐one (10) was specific to the latter strain. Both compounds were identified earlier in actinobacteria. [35] β‐Phenylethyl isocyanate (11) was identifed from “C. coralii” A01A‐333, but has not been reported from bacteria or algae so far. 1‐Phenylbutan‐1‐one (12) and benzoxazole (16) occurred only in C. velia and are known from Pseudomonas aeruginosa PA14, [36] but 12 has not been reported from algae before. The homologue of 12, 1‐phenylhexan‐1‐one (13), occurred only in the headspace extracts of Litoreibacter sp., whereas 1‐(2‐hydroxyphenyl)propan‐1‐one (14) was specific for C. velia. 2‐Aminoacetophenone (15) was identified in “C. coralii” and is a common bacterial product,[ 32 , 35 ] serving as quorum‐sensing signal in Pseudomonas. [37] The bi‐ and tricyclic aromatic compounds 16–21 occurred in C. velia. 2‐Methylbenzofuran (17) was identified in only one analytical replicate; it is associated with serious taste and odour problems in lakes in China resulting from cyanobacterial blooms. [38] 2,3‐Benzothiophene (18) showed highly toxic effects on nitrifying bacteria and the freshwater green alga Pseudokirchneriella subcapitata at a 4–6 mg/L concentration [39] and is a volatile metabolite of the pathogen Clostridium difficile. [40] While 3‐methylbenzo[b]thiophene (19) and dibenzo[b,d]thiophene (20) were not previously described from bacteria or algae, the latter is ecotoxic to algae and daphnids. [41] 1,2,3‐Benzothiadiazol (21) was tentatively identified, as it was only confirmed by its mass spectrum, but no synthetic material was at hand for comparison. Although 21 has also not been described as a natural volatile so far, it functions as a plant protecting agent, inducing systemic resistance in cotton plants against the pathogenic fungus Macrophomina phaseolina. [42] In bean, cucumber and tomato it is also inducing a dose‐dependent resistance against the grey mould caused by Botrytis cinerea. [43]
Figure 4.
Aromatic compounds of C. velia CCAP 1602/1 and isolated bacteria from the phycosphere.
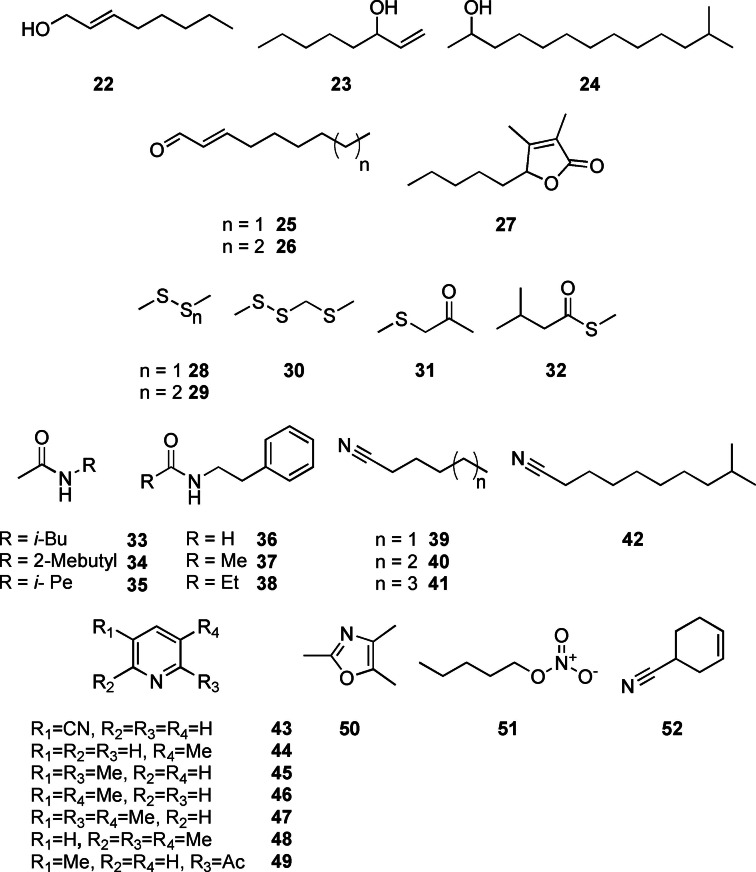
Aliphatic compounds
The detected aliphatic compounds comprised alcohols, aldehydes, amides, sulfur‐ and nitrogen containing compounds as well as one lactone (Figure 5). (E)‐Oct‐2‐en‐1‐ol (22) and oct‐1‐en‐3‐ol (23) were both algal compounds, while 12‐methyltridecan‐2‐ol (24) occurred in “C. coralii” A01A‐333. Alcohol 22 is known from several actinomycetes, [35] but it has not been reported from algae so far. The widespread oct‐1‐en‐3‐ol (23) is a typical fungal volatile [44] but has also been reported from the intertidal red macroalga Pyropia haitanensis, in which this alcohol induces synthesis of other signalling compounds. [45] To our knowledge, 12‐methyltridecan‐2‐ol (24) has not been described from algae or bacteria, but related methyl‐branched alcohols are typical products released from branched fatty acid metabolism in bacteria.[ 32 , 46 ]
Figure 5.
Aliphatic and nitrogen containing compounds for C. velia and associated bacteria.
The unsaturated aldehydes (E)‐oct‐2‐enal and (E)‐non‐2‐enal (25, 26) were only found in the algal extracts. They are, as the alcohols 22 and 23, products of oxidative degradation of polyunsaturated fatty acids, often formed in response to abiotic or biotic stress. [47] Especially polyunsaturated aldehydes are formed when diatoms such as Thalassiosira rotula are exposed to stress due to cell disruption. [48] Aldehydes 25 and 26 could result from an oxidative cleavage of unsaturated fatty acids, for example, arachidonic acid present in C. velia, as described for the brown alga Laminaria angustata. [49] Both aldehydes are common in algae, for example, in the green algal seaweed Capsosiphon fulvescens. [50]
Amides were specific to the investigated bacteria. The aliphatic amides N‐isobutylacetamide (33), N‐2‐methylbutylacetamide (34) and N‐isopentylacetamide (35) were detected in some of the strains in low amounts. These amides were occasionally reported from bacteria (35),[ 51 , 52 ] including marine actinomycete Salinispora pacifica. [53] Similarly, phenylethylamides 36–38 occurred in “C. coralii” A01A‐333 in low amounts, whereas N‐(2‐phenylethyl)acetamide (37) was the only compound released by S. litoris A01A‐101. All amides were previously reported from macroalgae‐associated Roseobacteraceae, [54] and also other bacteria. [55]
The only lactone identified was dihydrobovolide (27) released by C. velia. It is known from the essential oil of several plants like Sonchus arvensis [56] and also from the thallus culture of the marine green alga Ulva pertusa. [57] This compound is probably an oxidative degradation product of furan fatty acids that are produced by many algae. [58]
Several sulfur compounds were released from the alga, but more importantly from the flavobacterium “C. coralii” A01A‐333. Dimethyl disulfide (28) and dimethyl trisulfide (29) are known as volatiles derived from bacteria and marine algae and are discussed in several reviews.[ 32 , 59 ] 2,3,5‐Trithiahexane (30), 4‐thia‐2‐pentanone (31) and S‐methyl 3‐methylbutanethioate (32) were also produced by “C. coralii”. Compound 30 is a photolysis product of 28. [60] The only thioester 32 is known from several streptomycetes. [35] Methylthioketone 31 occurred as a sulfur volatile when Microbacterium foliorum is co‐cultivated with the yeast Debaryomyces hansenii. [61]
The sphingomonad S. litoris A01A‐101 and C. velia released a number of nitrogen containing VOCs. Hexanenitrile (39) was detected in both replicates, whereas 9‐methyldecanenitrile (42) occurred only once in C. velia. Nitrile 39 and its homologues heptanenitrile (40) and octanenitrile (41) occurred in S. litoris as minor compounds. Aliphatic nitriles represent a neglected class of microbial volatiles and have only been reported from the gammaproteobacterium Pseudomonas veronii and the actinomycete Micromonospora echinospora. [62] They constitute a new algal volatile class. 3‐Pyridinecarbonitrile (43) was identified in “C. coralii”, being reported from the actinomycete Thermomonospora curvata earlier. [35] The alkylpyridines 3‐methylpyridine (44), 2,5‐ (45) and 3,5‐dimethylpyridine (46), as well as 2,3,5‐ (47) and 2,3,6‐trimethylpyridine (48) were released by S. litoris, while 2‐acetyl‐5‐methylpyridine (49), was only tentatively identified due to lack of a synthetic standard. To our knowledge, this pyridine and 44 have not been reported as a volatiles from bacteria or algae. The other pyridines are constituents of refined oils from various algae and bacteria.[ 63 , 64 ] The pyridine 47 was detected in P. aeruginosa isolates [65] and the green alga Chlamydomonas sp. [66] 2,4,5‐Trimethyloxazol (50) was present in “C. coralii” and previously reported from Bacillus bacteria. [67] The unusual nitric acid pentyl ester (51) occurred in very low concentrations in both replicates of C. velia. It is not known from algae, but was first identified in fresh ripe tomatoes. [68] 4‐Cyanocyclohexene (52) was detected in Litoreibacter sp. during our study. Like 45, it has been reported from the pyrolysis oil from the haptophycean microalga Isochrysis. [63]
Ketones
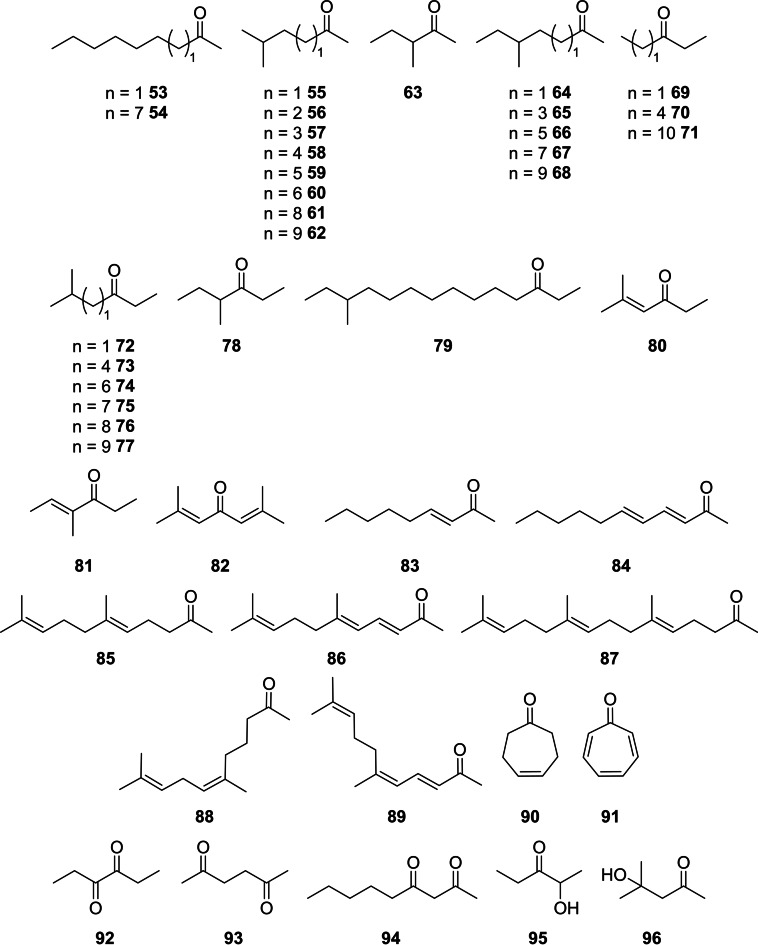
Although ketones certainly mostly qualify as aliphatic compounds, they are treated here separately due to their large structural variety found within the investigated microorganisms. Ketones were the most frequently produced volatile compounds released from “C. coralii” A01A‐333, whereas only a few were produced by the alga and the other strains (Figure 6). Flavobacteria are known for their abundant ketone formation. [46] These ketones appeared predominantly in high concentrations and comprised saturated and unsaturated linear ketones, cyclic ketones, hydroxy‐ or diketones. They are products of the fatty acid biosynthetic pathway and their occurrence has been reviewed.[ 32 , 33 ] Methyl‐branched methyl ketones and ethyl ketones were dominating and their identity was confirmed by calculation of their gas chromatographic retention indices according to an empirical method. [46] The methyl ketones showed C5 to C15 chains, usually with an iso‐ or anteiso‐methyl substituent (53–68).
Figure 6.
Ketones identified in the headspace of C. velia and associated bacteria.
11‐Methyltridecan‐2‐one (67) was the most abundant volatile in “C. coralii” A01A‐333. All these ketones have been reported from different bacterial strains.[ 46 , 52 , 69 , 70 , 71 , 72 ] Because of their widespread occurrence, such methyl ketones have been classified as common bacterial volatiles. [9] 5‐Methylhexan‐2‐one (55) was emitted also from Litoreibacter sp., while 3‐methylpentan‐2‐one (63) occurred in S. litoris, also known from various other bacteria.[ 70 , 72 , 73 ] Simple methyl ketones with an odd number of carbon atoms like nonan‐2‐one (53), released from the alga, and pentadecan‐2‐one (54), found in “C. coralii”, are usually derived from even‐numbered β‐keto acids by decarboxylation. [32] Nonan‐2‐one (53) is known as a volatile released by the green alga Cladophora vagabunda, [71] whereas 54 was identified in the brown alga Caulocystis cephalornithos and the red alga Hypnea musciformis. [74] Ethyl ketones (69–79) comprised similarly mostly iso‐branched, but also anteiso as well as unbranched compounds, predominantly produced by “C. coralii”. The short chain ethyl ketones 72 and 78 occurred in S. litoris A01A‐101. 13‐Methyltetradecan‐3‐one (77) and its corresponding anteiso‐isomer 79 are known from the Cytophaga‐Flavobacterium‐Bacteroides group. [46] 5‐Methylhexan‐3‐one (72) is produced by diverse bacteria,[ 52 , 70 , 75 ] whereas its isomer 78 was reported only tentatively. [76] The ethyl ketones 73–76 have not been reported from bacteria to our knowledge. Unbranched pentan‐3‐one (69) appeared in S. litoris, octan‐3‐one (70) in C. velia and tetradecan‐3‐one (71) in “C. coralii”. Compound 69 was reported from the heterokont microalga Mycrochloropsis salina [77] and also appeared in the aqueous fraction from hydrothermal liquefaction of different fresh‐ and seawater algae, [78] but is also known from several bacteria.[ 70 , 79 ] Ketone 70 occurs in actinomycetes and cyanobacteria[ 32 , 80 ] as well as in the red alga Palmaria palmata, [81] while 71 was found as a volatile of bacterial strains isolated from the rhizosphere of lemon plants. [82]
Several unsaturated ketones were emitted from the bacteria as well. (E)‐4‐Methylhex‐4‐en‐3‐one (81), phorone (82) and 5‐methylhex‐4‐en‐3‐one (80) were present in the headspace extract of S. litoris and (E)‐3‐nonen‐2‐one (83) and (3E,5E)‐undeca‐3,5‐dien‐2‐one (84) in the headspace of Litoreibacter sp. as minor compounds. Ketone 80, only confirmed by MS, was earlier reported from Myxococcus xanthus [75] and Paenisporosarcina sp. [70] and 81 was reported from Streptomyces afghaniensis. [83] (E)‐Non‐3‐en‐2‐one (83) is known from Lactobacillus helveticus, [84] whereas 82 and 84 have not been reported from bacteria.
Terpenoids (85–89) comprised (E)‐geranylacetone (85), one of the major components of S. litoris, (Z)‐geranylacetone (88), (3E,5E)‐ and (3E,5Z)‐6,10‐dimethylundeca‐3,5,9‐trien‐2‐one (86 and 89), as well as farnesyl acetone (87). Terpenoids are common volatiles produced from bacteria and their distribution has been discussed in several reviews.[ 32 , 34 ] Cyclic ketones cyclohept‐4‐en‐1‐one (90) and tropone (91) were both present in “C. coralii”, while the latter one was also found in A. albus. Both 90 and 91 were reported from diverse actinomycetes[ 35 , 83 , 85 ] and marine Roseobacteraceae.[ 83 , 86 ] As tropone formation has been shown to proceed according to the phenylacetate pathway, [87] it seems likely that 90 could also be derived from this biosynthetic route.
The diketones hexane‐2,5‐dione (93) and nonane‐2,4‐dione (94) occurred in Litoreibacter sp., whereas hexane‐3,4‐dione (92) was found in “C. coralii”. Diketones have previously been reported from bacteria.[ 32 , 34 ] However, compounds 92–94 are not known from bacteria or algae so far, but 2,3‐diketones and the related acyloins are typical fermentation products of bacteria.[ 32 , 34 ] Diacetone alcohol (96) was released from C. velia and is known from the red algae Corallina mediterranea and Laurencia coronopus, [27] from diverse soil bacteria, [88] as well as from the marine actinomycete Salinispora tropica. [85] The acyloin 95 of “C. coralii” is a widespread volatile of bacteria. [35] It is a biosynthetic precursor of alkylated pyrazines. [89]
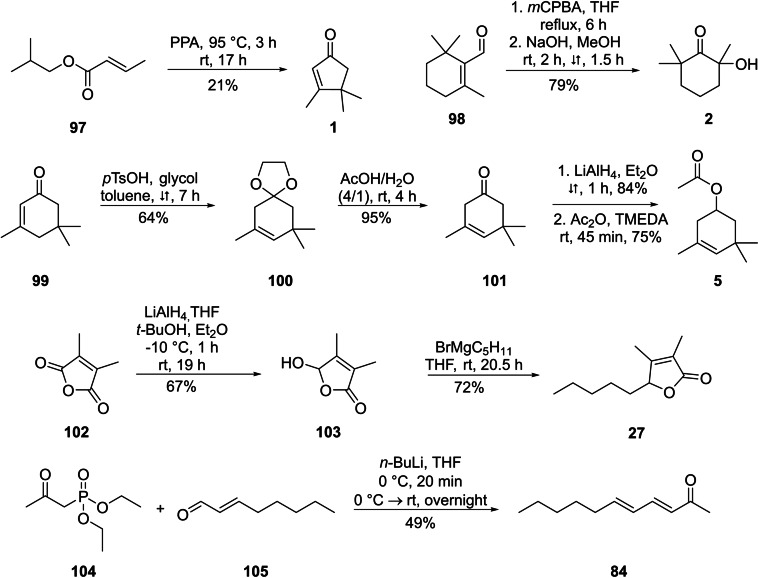
Syntheses of reference compounds
Although mass spectra of most of the identified compounds were available in various data bases or our in‐house MS library, or were obtained from commercial samples, several of the structures proposed from the mass spectra needed to be confirmed by synthetic reference compounds, notably 1, 2, 5, 27, and 84. Therefore, these compounds were synthesised (Scheme 2). Compound 1 was obtained by a nazarov‐cyclisation of isobutyl (E)‐but‐2‐enoate (97) with polyphosphoric acid (PPA). [90] For the synthesis of 2‐hydroxy‐2,6,6‐trimethylcyclohexan‐1‐one (2), Baeyer‐Villiger oxidation of β‐cyclocitral (98) followed by hydrolysis with methanolic sodium hydroxide solution yielded compound 2. [91] The acetate 5 was obtained over a four‐steps‐synthesis starting from α‐isophorone (99). Conversion into ketal 100 with glycol and acid catalysis induced a rearrangement of the double bond into the β,γ‐position. Compound 100 was deprotected in the presence of water and acetic acid to give 101. [92] A reduction of the carbonyl group with LiAlH4 to the β,γ‐unsaturated alcohol [93] and a final esterification with acetic acid anhydride yielded ester 5. [94] Dihydrobovolide (3,4‐dimethylpent‐5‐ylfuran‐2(5H)‐one, 27) was obtained by reduction of 3,4‐dimethylfuran‐2,5‐dione (102) with LiAlH4 to 5‐hydroxy‐3,4‐dimethylfuran‐2(5H)‐one (103), [95] followed by Grignard reaction of 103 with pentylmagnesium bromide to give the lactone 27. [96] The di‐unsaturated ketone (3E,5E)‐undeca‐3,5‐dien‐2‐one (84) was obtained by a Horner‐Wadsworth‐Emmons reaction of phosphonate 104 and (E)‐oct‐2‐enal (105). [97] The mass spectra and the retention indices of these compounds were compared with those of the unknown compounds, confirming in all cases the proposed structures (see the Supporting Information).
Scheme 2.
Syntheses of reference compounds 1, 2, 5, 27 and 84.
Comparative analysis
As is evident from the preceding paragraphs, there was little overlap in emitted compounds between the non‐axenic alga and some dominant bacteria isolated from its phycosphere. Accordingly, there seems to be a comparably low influence of these bacteria on the VOC composition. Although the bacteria are capable to produce various volatiles and largely different odour bouquets, they are not observed in the algal cultures.
The bouquet of C. velia largely consists of compounds that are formed by oxidative degradation of larger compounds. This is indicated by the apocarotenoids 1–7 derived from carotenoids, but also by chain cleavage products of unsaturated fatty acids such as 23, 25, 26, 53, and 70, as well as 27, a degradation product of furane fatty acids. The most abundant volatiles were 4‐oxoisophorone (3), oct‐1‐en‐3‐ol (23), a known signalling compound, octan‐3‐one (70) and (E)‐non‐2‐enal (25). Nevertheless, several unique compounds were also released, including the mostly undescribed aromatic sulfur compounds 18–21, the nitriles 39 and 42, and the nitric acid ester 51. Furthermore, abundant sulfides 28 and 29 were emitted, which were surprisingly not detected in the analysed bacteria, although they constitute very common volatiles of other bacteria.[ 9 , 32 , 33 , 34 , 35 ]
The analysis of the four bacteria did not show much similarity with respect to the emitted compounds. While A. albus A01A‐324 produced mostly amides, but only few other compounds, the other Alphaproteobacteria emitted more components. The VOCs of S. litoris A01A‐101 also comprised amides, but moreover contained various methylpyridines, nitriles and short ketones. Such ketones were also present in Litoreibacter sp. A01A‐347, but a considerable amount of compounds remained unidentified. Finally, “C. coralii” A01A‐333 represents from a chemical point of view a typical member of the flavobacteria, comprising the compound classes that were previously reported for other isolates. [46] Aliphatic methyl‐ and ethyl ketones were predominant, but also including compounds known to be used as bacterial signals, such as 15. These largely different bouquets might indicate different physiology, maybe indicative of different physiological niches filled by the bacteria when grown together with C. velia.
Conclusions
To the best of our knowledge, we have performed the first metagenome study of a non‐axenic chromerid alga. Our combined binning strategy, including the use of DAS‐Tool, allowed us to establish authentic bins of heterotrophic bacteria from the phycosphere of Chromera velia CCAP 1602/1. The establishment of authentic RpoB and 16S‐rRNA genes from the planctomycete reflects the advantage of metagenome analyses of low‐complexity microbial communities.
For the first time, the volatiles released during the growth of the apicomplexan alga C. velia have been analysed. Although only low amounts of VOCs are released, a sensitive headspace analysis allowed structural characterization of the emitted volatiles. Primary compounds are the degradation products of fatty acids and especially carotenoids, which provide the first hints for the presence of carotenoids beyond isofucoxanthin in this alga that are yet unknown. Furthermore, this study revealed the release of other unique compounds not known from other algae.
The VOC bouquet of the bacteria shows no similarity to that of the alga. At first sight this indicates that the bacteria have no direct influence on the bouquet of volatiles of non‐axenic algal cultures. However, due to the use of different media for the cultivation of the photoautotrophic alga and the heterotrophic bacteria, further analyses are needed to confirm this prediction. Several of the volatiles reported here, especially some nitriles, are reported for the first time from microorganisms.
Conflict of interest
The authors declare no conflict of interest.
1.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Acknowledgments
We thank Jessica Grube, Vanessa Stiller, Orsola Päuker and the CellDeg GmbH for technical support of this work. We also thank the two anonymous reviewers for their constructive criticism. The research was funded by the Deutsche Forschungsgemeinschaft (DFG) Schu984/12‐3 and Project‐ID 34509606‐TRR 51. Open Access funding enabled and organized by Projekt DEAL.
Koteska D., Marter P., Huang S., Pradella S., Petersen J., Schulz S., ChemBioChem 2023, 24, e202200530.
Data Availability Statement
The data that support the findings of this study are available in the supplementary material of this article.
References
- 1. Moore R. B., Oborník M., Janouškovec J., Chrudimský T., Vancová M., Green D. H., Wright S. W., Davies N. W., Bolch C. J. S., Heimann K., et al., Nature 2008, 451, 959. [DOI] [PubMed] [Google Scholar]
- 2. Adl S. M., Simpson A. G. B., Lane C. E., Lukeš J., Bass D., Bowser S. S., Brown M. W., Burki F., Dunthorn M., Hampl V., et al., J. Eukaryotic Microbiol. 2012, 59, 429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Janouskovec J., Horák A., Oborník M., Lukes J., Keeling P. J., Proc. Natl. Acad. Sci. USA 2010, 107, 10949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Petersen J., Ludewig A.-K., Michael V., Bunk B., Jarek M., Baurain D., Brinkmann H., Genome Biol. Evol. 2014, 6, 666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Weatherby K., Carter D., Adv. Appl. Microbiol. 2013, 85, 119. [DOI] [PubMed] [Google Scholar]
- 6. Novoveská L., Ross M. E., Stanley M. S., Pradelles R., Wasiolek V., Sassi J.-F., Mar. Drugs 2019, 17, 640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jin E.-S., Polle J. E., Lee H.-K., Hyun S.-M., Chang M., J. Microbiol. Biotechnol. 2003, 13, 165. [Google Scholar]
- 8. Tomčala A., Michálek J., Schneedorferová I., Füssy Z., Gruber A., Vancová M., Oborník M., Biomol. Eng. 2020, 10, 1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Weisskopf L., Schulz S., Garbeva P., Nat. Rev. Microbiol. 2021, 19, 391. [DOI] [PubMed] [Google Scholar]
- 10. Zuo Z., Front. Microbiol. 2019, 10, 491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.
- 11a. Cornet L., Bertrand A. R., Hanikenne M., Javaux E. J., Wilmotte A., Baurain D., Microb. Genomics 2018, 4, e000212, DOI 10.1099/mgen.0.000212; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11b. Di Costanzo F., Di Dato V., van Zyl L. J., Cutignano A., Esposito F., Trindade M., Romano G., Int. J. Mol. Sci. 2021, 22, 13199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Marter P., Huang S., Brinkmann H., Pradella S., Jarek M., Rohde M., Bunk B., Petersen J., Genes 2021, 12, 389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rambo I. M., Dombrowski N., Constant L., Erdner D., Baker B. J., Environ. Microbiol. 2020, 22, 1764. [DOI] [PubMed] [Google Scholar]
- 14. Stackebrandt E., Ebers J., Microbiol. Today 2006, 8, 6. [Google Scholar]
- 15. Yarza P., Yilmaz P., Pruesse E., Glöckner F. O., Ludwig W., Schleifer K.-H., Whitman W. B., Euzéby J., Amann R., Rosselló-Móra R., Nat. Rev. Microbiol. 2014, 12, 635. [DOI] [PubMed] [Google Scholar]
- 16. Liang K. Y. H., Orata F. D., Boucher Y. F., Case R. J., Front. Microbiol. 2021, 12, 683109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.
- 17a. Newton R. J., Griffin L. E., Bowles K. M., Meile C., Gifford S., Givens C. E., Howard E. C., King E., Oakley C. A., Reisch C. R., et al., ISME J. 2010, 4, 784; [DOI] [PubMed] [Google Scholar]
- 17b. Simon M., Scheuner C., Meier-Kolthoff J. P., Brinkhoff T., Wagner-Döbler I., Ulbrich M., Klenk H.-P., Schomburg D., Petersen J., Göker M., ISME J. 2017, 11, 1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.
- 18a. Frank O., Göker M., Pradella S., Petersen J., Environ. Microbiol. 2015, 17, 4019; [DOI] [PubMed] [Google Scholar]
- 18b. Wagner-Döbler I., Ballhausen B., Berger M., Brinkhoff T., Buchholz I., Bunk B., Cypionka H., Daniel R., Drepper T., Gerdts G., et al., ISME J. 2010, 4, 61. [DOI] [PubMed] [Google Scholar]
- 19. Segev E., Wyche T. P., Kim K. H., Petersen J., Ellebrandt C., Vlamakis H., Barteneva N., Paulson J. N., Chai L., Clardy J., et al., eLife 2016, 5, e17473, DOI: 10.7554/eLife.17473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sieber C. M. K., Probst A. J., Sharrar A., Thomas B. C., Hess M., Tringe S. G., Banfield J. F., Nat. Microbiol. 2018, 3, 836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hiseni P., Snipen L., Wilson R. C., Furu K., Rudi K., Front. Microbiol. 2021, 12, 822301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kallscheuer N., Wiegand S., Kohn T., Boedeker C., Jeske O., Rast P., Müller R.-W., Brümmer F., Heuer A., Jetten M. S. M., et al., Front. Microbiol. 2020, 11, 602250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.
- 23a. Lage O. M., Bondoso J., Front. Microbiol. 2014, 5, 267; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23b. Wiegand S., Jogler M., Boedeker C., Pinto D., Vollmers J., Rivas-Marín E., Kohn T., Peeters S. H., Heuer A., Rast P., et al., Nat. Microbiol. 2020, 5, 126.31740763 [Google Scholar]
- 24. Dickschat J. S., Nat. Prod. Rep. 2014, 31, 838. [DOI] [PubMed] [Google Scholar]
- 25. Krammer G. E., Werkhoff P., Sommer H., Schmidt C. O., Gatfield I., Bertram H.-J. in ACS Symposium Series (Eds.: Winterhalter P., Rouseff R. L.), American Chemical Society, Washington, DC, 2001, pp. 206–219. [Google Scholar]
- 26. Yamamoto M., Baldermann S., Yoshikawa K., Fujita A., Mase N., Watanabe N., Sci. World J. 2014, 2014, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kamenarska Z., Ivanova A., Stancheva R., Stoyneva M., Stefanov K., Dimitrova-Konaklieva S., Popov S., Bot. Mar. 2006, 49, 47, DOI 10.1515/BOT.2006.006. [DOI] [Google Scholar]
- 28. Koteska D., Sanchez Garcia S., Wagner-Döbler I., Schulz S., Mar. Drugs 2022, 20, 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Moran L., Bou G., Aldai N., Ciardi M., Morillas-España A., Sánchez-Zurano A., Barron L. J. R., Lafarga T., Sci. Rep. 2022, 12, 3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. El Amrani Zerrifi S., El Khalloufi F., Mugani R., El Mahdi R., Kasrati A., Soulaimani B., Barros L., Ferreira I. C. F. R., Amaral J. S., Finimundy T. C., et al., Toxin Rev. 2020, 12, 527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.
- 31a. Deli J., Molnár P., Matus Z., Tóth G., J. Agric. Food Chem. 2001, 49, 1517; [DOI] [PubMed] [Google Scholar]
- 31b. Philip T., Francis F. J., J. Food Sci. 1971, 36, 96. [Google Scholar]
- 32. Schulz S., Dickschat J. S., Nat. Prod. Rep. 2007, 24, 814. [DOI] [PubMed] [Google Scholar]
- 33. Schulz S., Biwer P., Harig T., Koteska D., Schlawis C. in Comprehensive Natural Products III (Eds.: Liu H.-W., P. Begley T.), Elsevier, 2020, pp. 161–178. [Google Scholar]
- 34. Schulz S., Schlawis C., Koteska D., Harig T., Biwer P. in Bacterial Volatile Compounds as Mediators of Airborne Interactions (Eds.: Ryu C.-M., Weisskopf L., Piechulla B.), Springer, Singapore, 2020, pp. 93–121. [Google Scholar]
- 35. Citron C. A., Rabe P., Dickschat J. S., J. Nat. Prod. 2012, 75, 1765. [DOI] [PubMed] [Google Scholar]
- 36. Bean H. D., Dimandja J.-M. D., Hill J. E., J. Chromatogr. B 2012, 901, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kesarwani M., Hazan R., He J., Que Y., Apidianakis Y., Lesic B., Xiao G., Dekimpe V., Milot S., Deziel E., et al., PLoS Pathog. 2011, 7, e1002192, DOI 10.1371/journal.ppat.1002192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang R., Qi F., Liu C., Zhang Y., Wang Y., Song Z., Kumirska J., Sun D., Ecotoxicol. Environ. Saf. 2019, 181, 499. [DOI] [PubMed] [Google Scholar]
- 39. Eilersen A. M., Arvin E., Henze M., Water Sci. Technol. 2004, 50, 277. [PubMed] [Google Scholar]
- 40. Rees C. A., Shen A., Hill J. E., J. Chromatogr. B 2016, 1039, 8. [DOI] [PubMed] [Google Scholar]
- 41. Eisentraeger A., Brinkmann C., Hollert H., Sagner A., Tiehm A., Neuwoehner J., Environ. Toxicol. Chem. 2008, 27, 1590. [DOI] [PubMed] [Google Scholar]
- 42. Adrees H., Haider M. S., Anjum T., Akram W., Crop Prot. 2019, 115, 75. [Google Scholar]
- 43. Azami-Sardooei Z., Seifi H. S., de Vleesschauwer D., Höfte M., Australas. Plant Pathol. 2013, 42, 485. [Google Scholar]
- 44. Schnürer J., Olsson J., Börjesson T., Fungal Genet. Biol. 1999, 27, 209. [DOI] [PubMed] [Google Scholar]
- 45. Qian F., Luo Q., Yang R., Zhu Z., Chen H., Yan X., J. Appl. Phycol. 2015, 27, 621. [Google Scholar]
- 46. Dickschat J. S., Helmke E., Schulz S., Chem. Biodiversity 2005, 2, 318. [DOI] [PubMed] [Google Scholar]
- 47. Creelman R. A., Mulpuri R., The Arabidopsis Book 2002, 1, e0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.
- 48a. Lavrentyev P., Franzè G., Pierson J., Stoecker D., Mar. Drugs 2015, 13, 2834; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48b. Barofsky A., Pohnert G., Org. Lett. 2007, 9, 1017. [DOI] [PubMed] [Google Scholar]
- 49. Boonprab K., Matsui K., Akakabe Y., Yoshida M., Yotsukura N., Chirapart A., Kajiwara T., J. Appl. Phycol. 2006, 18, 409. [Google Scholar]
- 50. Sun S.-M., Chung G.-H., Shin T.-S., J. Appl. Phycol. 2012, 24, 1003. [Google Scholar]
- 51.
- 51a. El-Mehalawy A. A., Int. J. Agric. Biol. 2005, 7, 188; [Google Scholar]
- 51b. Kowalewska J., Zelazowska H., Babuchowski A., Hammond E. G., Glatz B. A., Ross F., J. Dairy Sci. 1985, 68, 2165. [Google Scholar]
- 52. Dickschat J. S., Bode H. B., Wenzel S. C., Müller R., Schulz S., ChemBioChem 2005, 6, 2023. [DOI] [PubMed] [Google Scholar]
- 53. Harig T., Schlawis C., Ziesche L., Pohlner M., Engelen B., Schulz S., J. Nat. Prod. 2017, 80, 3289. [DOI] [PubMed] [Google Scholar]
- 54. Ziesche L., Bruns H., Dogs M., Wolter L., Mann F., Wagner-Döbler I., Brinkhoff T., Schulz S., ChemBioChem 2015, 16, 2094. [DOI] [PubMed] [Google Scholar]
- 55. Böröczky K., Laatsch H., Wagner-Döbler I., Stritzke K., Schulz S., Chem. Biodiversity 2006, 3, 622. [DOI] [PubMed] [Google Scholar]
- 56. Radulović N., Blagojević P., Palić R., Nat. Prod. Commun. 2009, 4, 405. [PubMed] [Google Scholar]
- 57. Fujimura T., Kawai T., Shiga M., Kajiwara T., Hatanaka A., Phytochemistry 1990, 29, 745. [Google Scholar]
- 58. Spiteller G., Lipids 2005, 40, 755. [DOI] [PubMed] [Google Scholar]
- 59.
- 59a. Stefels J., J. Sea Res. 2000, 43, 183; [Google Scholar]
- 59b. Blunden G., Prog. Phycol. Res. 1986, 4, 39. [Google Scholar]
- 60. Buttery R. G., Seifert R. M., J. Agric. Food Chem. 1977, 25, 434.838989 [Google Scholar]
- 61. Deetae P., Spinnler H.-E., Bonnarme P., Helinck S., Appl. Microbiol. Biotechnol. 2009, 82, 169. [DOI] [PubMed] [Google Scholar]
- 62. Montes Vidal D., von Rymon-Lipinski A.-L., Ravella S., Groenhagen U., Herrmann J., Zaburannyi N., Zarbin P. H. G., Varadarajan A. R., Ahrens C. H., Weisskopf L., et al., Angew. Chem. Int. Ed. 2017, 56, 4342. [DOI] [PubMed] [Google Scholar]
- 63. Wang X., Zhao B., Tang X., Yang X., Bioresour. Technol. 2015, 179, 58. [DOI] [PubMed] [Google Scholar]
- 64.
- 64a. Kawale H. D., Kishore N., Energy 2019, 178, 344; [Google Scholar]
- 64b. Patzelt D. J., Hindersin S., Elsayed S., Boukis N., Kerner M., Hanelt D., J. Appl. Phycol. 2015, 27, 2239; [Google Scholar]
- 64c. Li J., Wang G., Chen M., Li J., Yang Y., Zhu Q., Jiang X., Wang Z., Liu H., Bioresour. Technol. 2014, 169, 110. [DOI] [PubMed] [Google Scholar]
- 65. Bean H. D., Rees C. A., Hill J. E., J. Breath Res. 2016, 10, 47102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhang B., Wang L., Li R., Rahman Q. M., Shahbazi A., Energ. Fuego At. 2017, 31, 12223. [Google Scholar]
- 67. Robacker D. C., Martinez A. J., Garcia J. A., Bartelt R. J., Fla. Entomol. 1998, 81, 497. [Google Scholar]
- 68. Buttery R. G., Takeoka G. R., J. Agric. Food Chem. 2004, 52, 6264. [DOI] [PubMed] [Google Scholar]
- 69.
- 69a. Dickschat J. S., Martens T., Brinkhoff T., Simon M., Schulz S., Chem. Biodiversity 2005, 2, 837; [DOI] [PubMed] [Google Scholar]
- 69b. Weise T., Kai M., Gummesson A., Troeger A., von Reuß S., Piepenborn S., Kosterka F., Sklorz M., Zimmermann R., Francke W., et al., Beilstein J. Org. Chem. 2012, 8, 579; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69c. DeMilo A. B., Lee C.-J., Moreno D. S., Martinez A. J., J. Agric. Food Chem. 1996, 44, 607. [Google Scholar]
- 70. Wang E., Liu X., Si Z., Li X., Bi J., Dong W., Chen M., Wang S., Zhang J., Song A., et al., Agriculture 2021, 11, 368. [Google Scholar]
- 71. Elenkov I., Georgieva T., Hadjieva P., Dimitrova-Konaklieva S., Popov S., Phytochemistry 1995, 38, 457. [Google Scholar]
- 72. Garbeva P., Hordijk C., Gerards S., de Boer W., FEMS Microbiol. Ecol. 2014, 87, 639. [DOI] [PubMed] [Google Scholar]
- 73. Jollivet N., Bzenger M.-C., Vayssier Y., Belin J.-M., Appl. Microbiol. Biot. 1992, 36, 790. [Google Scholar]
- 74.
- 74a. Amico V., Biondi D., Cunsolo F., Ruberto G., J. Nat. Prod. 1990, 53, 1379; [Google Scholar]
- 74b. Rafiquzzaman S. M., Ahmad M. U., Lee J. M., Kim E.-Y., Kim Y.-O., Kim D.-G., Kong I.-S., J. Food Process. Preserv. 2016, 40, 1074. [Google Scholar]
- 75. Dickschat J. S., Wenzel S. C., Bode H. B., Müller R., Schulz S., ChemBioChem 2004, 5, 778. [DOI] [PubMed] [Google Scholar]
- 76. Wilkins K., Chemosphere 1996, 32, 1427. [Google Scholar]
- 77. Fisher C. L., Lane P. D., Russell M., Maddalena R., Lane T. W., Metabolites 2020, 10, 361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Maddi B., Panisko E., Wietsma T., Lemmon T., Swita M., Albrecht K., Howe D., Biomass Bioenergy 2016, 93, 122. [Google Scholar]
- 79.
- 79a. Jia S., Li Y., Zhuang S., Sun X., Zhang L., Shi J., Hong H., Luo Y., Food Microbiol. 2019, 84, 103248; [DOI] [PubMed] [Google Scholar]
- 79b. Antolak H., Jeleń H., Otlewska A., Kręgiel D., Food Res. Int. 2019, 121, 379. [DOI] [PubMed] [Google Scholar]
- 80. Le Pape M.-A., Grua-Priol J., Prost C., Demaimay M., J. Agric. Food Chem. 2004, 52, 550. [DOI] [PubMed] [Google Scholar]
- 81. Höckelmann C., Jüttner F., Water Sci. Technol. 2004, 49, 47. [PubMed] [Google Scholar]
- 82. Gutiérrez-Luna F. M., López-Bucio J., Altamirano-Hernández J., Valencia-Cantero E., de La Cruz H. R., Macías-Rodríguez L., Symbiosis 2010, 51, 75. [Google Scholar]
- 83. Citron C. A., Barra L., Wink J., Dickschat J. S., Org. Biomol. Chem. 2015, 13, 2673. [DOI] [PubMed] [Google Scholar]
- 84. Montanari C., Sado Kamdem S. L., Serrazanetti D. I., Vannini L., Guerzoni M. E., J. Appl. Microbiol. 2013, 115, 1388. [DOI] [PubMed] [Google Scholar]
- 85. Groenhagen U., Leandrini De Oliveira A. L., Fielding E., Moore B. S., Schulz S., ChemBioChem 2016, 17, 1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Thiel V., Brinkhoff T., Dickschat J. S., Wickel S., Grunenberg J., Wagner-Döbler I., Simon M., Schulz S., Org. Biomol. Chem. 2010, 8, 234. [DOI] [PubMed] [Google Scholar]
- 87. Teufel R., Gantert C., Voss M., Eisenreich W., Haehnel W., Fuchs G., J. Biol. Chem. 2011, 286, 11021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Al-Daghari D. S. S., Al-Mahmooli I. H., Al-Sadi A. M., Al-Sabahi J. N., Velazhahan R., Indian Phytopathol. 2020, 73, 771. [Google Scholar]
- 89. Dickschat J. S., Wickel S., Bolten C. J., Nawrath T., Schulz S., Wittmann C., Eur. J. Org. Chem. 2010, 2010, 2687. [Google Scholar]
- 90. Vacas S., Navarro I., Marzo J., Navarro-Llopis V., Primo J., J. Agric. Food Chem. 2019, 67, 9441. [DOI] [PubMed] [Google Scholar]
- 91. Subbaraju G. V., Manhas M. S., Bose A. K., Synthesis 1992, 1992, 816. [Google Scholar]
- 92. Babler J. H., Malek N. C., Coghlan M. J., J. Org. Chem. 1978, 43, 1821. [Google Scholar]
- 93. Rosini G., Ballini R., Zanotti V., Tetrahedron 1983, 39, 1085. [Google Scholar]
- 94. Kadam S. T., Lee H., Kim S. S., B. Kor. Chem. Soc. 2009, 30, 1071. [Google Scholar]
- 95. Schobert R., Barnickel B., Synthesis 2009, 2778. [Google Scholar]
- 96. Surmont R., Verniest G., de Kimpe N., J. Org. Chem. 2010, 75, 5750. [DOI] [PubMed] [Google Scholar]
- 97. Gao Z., Fletcher S. P., Chem. Commun. 2018, 54, 3601. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Data Availability Statement
The data that support the findings of this study are available in the supplementary material of this article.