Summary
Plant‐specialized metabolism is complex, with frequent examples of highly branched biosynthetic pathways, and shared chemical intermediates. As such, many plant‐specialized metabolic networks are poorly characterized.
The N‐methyl Δ1‐pyrrolinium cation is a simple pyrrolidine alkaloid and precursor of pharmacologically important tropane alkaloids. Silencing of pyrrolidine ketide synthase (AbPyKS) in the roots of Atropa belladonna (Deadly Nightshade) reduces tropane alkaloid abundance and causes high N‐methyl Δ1‐pyrrolinium cation accumulation. The consequences of this metabolic shift on alkaloid metabolism are unknown. In this study, we utilized discovery metabolomics coupled with AbPyKS silencing to reveal major changes in the root alkaloid metabolome of A. belladonna.
We discovered and annotated almost 40 pyrrolidine alkaloids that increase when AbPyKS activity is reduced. Suppression of phenyllactate biosynthesis, combined with metabolic engineering in planta, and chemical synthesis indicates several of these pyrrolidines share a core structure formed through the nonenzymatic Mannich‐like decarboxylative condensation of the N‐methyl Δ1‐pyrrolinium cation with 2‐O‐malonylphenyllactate. Decoration of this core scaffold through hydroxylation and glycosylation leads to mono‐ and dipyrrolidine alkaloid diversity.
This study reveals the previously unknown complexity of the A. belladonna root metabolome and creates a foundation for future investigation into the biosynthesis, function, and potential utility of these novel alkaloids.
Keywords: Atropa belladonna, metabolomics, nonenzymatic catalysis, pyrrolidine alkaloids, Solanaceae, specialized metabolism, tropane alkaloids
Introduction
Plants synthesize complex mixtures of specialized metabolites of various chemical classes that facilitate reproduction and adaptation to abiotic and biotic challenges (Tewksbury & Nabhan, 2001; Weinhold & Baldwin, 2011; A. C. Huang et al., 2019; Voges et al., 2019; Fiesel et al., 2022; Roy et al., 2022). The bioactive properties of plant‐specialized metabolites have led to the cooption of plants and their chemical components for use as pharmaceuticals, flavorings, fragrances, and agrochemicals. In turn, this has driven intense interest toward purifying bioactive components from plant extracts, elucidating their structures, resolving their underlying mechanisms of biosynthesis, and developing metabolic engineering strategies for sustainable or increased production (Westfall et al., 2012; Galanie et al., 2015; Srinivasan & Smolke, 2020; Mitreiter & Gigolashvili, 2021; Fiesel et al., 2022).
Alkaloids are a large and diverse class of specialized metabolites commonly defined by the presence of a nitrogen atom within a heterocyclic ring (Ziegler & Facchini, 2008). Pyrrolidine alkaloids possess a core structural signature comprised of a heterocyclic ring containing a single nitrogen atom and four carbon atoms (Fig. 1). As such, the pyrrolidine core represents one of the simplest alkaloid structures, and its formation denotes a case of convergent evolution as pyrrolidine alkaloids occur in plants, bacteria, fungi, and animals, and are formed through distinct biosynthetic routes (Tsuda et al., 2005; Kim et al., 2016; Touchard et al., 2016; Zheng et al., 2017; Lin et al., 2020). Structural diversity of pyrrolidines can occur through simple chemical decorations such as hydroxylation or the addition of aliphatic and aromatic groups (Fig. 1). However, the pyrrolidine ring can also be incorporated into more structurally complex multi‐ring metabolites, including flavoalkaloids, sulfated guaiane sesquiterpenoids, and Emiline (Shen et al., 2013; Blair et al., 2017; Wu et al., 2018). Pyrrolidine alkaloids possess varied bioactivities, and there is considerable interest in their use as pharmacologically and agriculturally active lead compounds (Prandi & Occhiato, 2019; Li Petri et al., 2021; Basha et al., 2022).
Fig. 1.
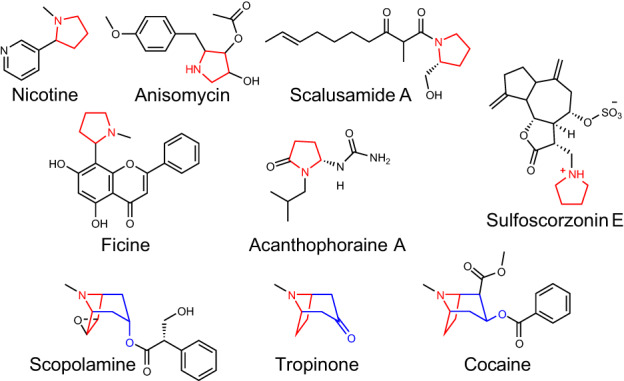
Examples of pyrrolidine and tropane alkaloid diversity formed through distinct biosynthetic routes. The pyrrolidine core is colored red. The first ring of the tropane core is a pyrrolidine highlighted in red, while the remaining portion of the tropane core is highlighted in blue.
Tropane alkaloids are a pharmacologically important subgroup of pyrrolidine alkaloids that occur in phylogenetically distinct plant families, including the Solanaceae (Kim et al., 2016). Tropanes possess a core 8‐azabicyclo[3.2.1]octane core structure, and the nitrogen‐containing ring of this bicyclic structure is derived from the N‐methyl‐Δ1‐pyrrolinium cation, a simple pyrrolidine building block (Fig. 1). The N‐methyl‐Δ1‐pyrrolinium cation is also the source of the heterocyclic ring(s) of simple pyrrolidines such as hygrine (which co‐occurs in tropane producing plants, Fig. 2) and also forms one of the rings of nicotine in Nicotiana spp. (Leete, 1990; Humphrey & O'Hagan, 2001). In vivo labeling and subsequent enzyme characterization revealed that the N‐methyl‐Δ1‐pyrrolinium cation is derived from ornithine through four distinct steps (Fig. 2): (1) the decarboxylation of ornithine to putrescine, catalyzed by ornithine decarboxylase; (2) methylation of putrescine to N‐methylputrescine by putrescine methyltransferase; (3) oxidation of N‐methylputrescine to 4‐methylaminobutanal by methylputrescine oxidase; and (4) spontaneous, nonenzymatic cyclization of 4‐methylaminobutanal to form the N‐methyl‐Δ1‐pyrrolinium cation (Hibi et al., 1994; Humphrey & O'Hagan, 2001; Heim et al., 2007; Katoh et al., 2007; Kim et al., 2016).
Fig. 2.
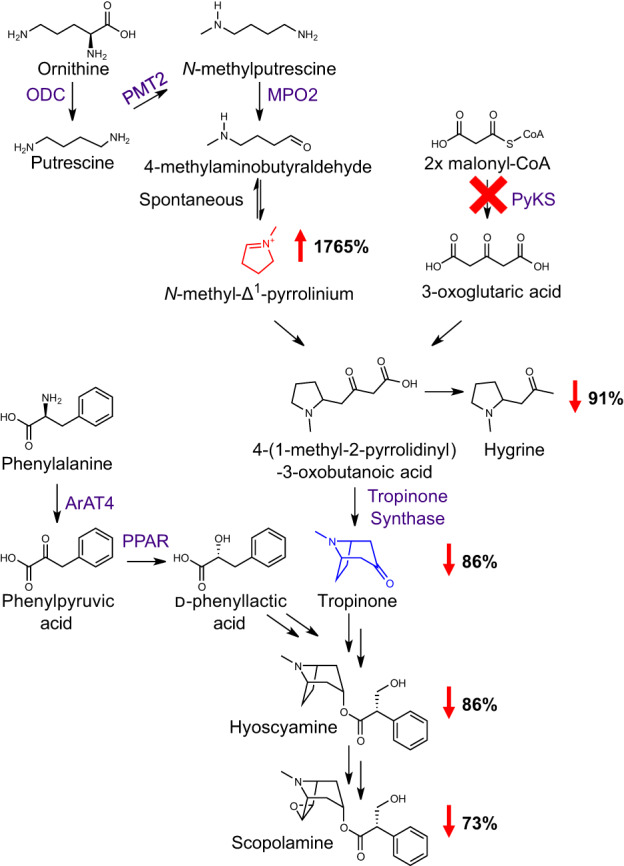
Effect of silencing PyKS on tropane metabolism in Atropa belladonna roots. Virus‐induced gene silencing of a root‐expressed polyketide synthase (PyKS) in A. belladonna roots, indicated by the red ‘X’, disrupts tropane alkaloid biosynthesis and causes an accumulation of the N‐methyl‐Δ1‐pyrrolinium ion shown in red. The upward‐pointing red arrow reflects the percentage increase in the N‐methyl‐Δ1‐pyrrolinium ion in AbPyKS‐silenced lines compared with TRV2 empty vector controls. Downward red arrows represent the percentage decrease in the pyrrolidine alkaloid hygrine and tropane alkaloids in AbPyKS‐silenced lines compared with TRV2 empty vector controls. Data are adapted from Bedewitz et al. (2018). For simplicity, an abbreviated tropane pathway is shown and omitted steps are indicated by double arrows. Tropinone, the first tropane alkaloid in the pathway with the characteristic bicyclic ring structure of all tropanes, is shown in blue. Enzyme abbreviations are as follows; ArAT4, phenylalanine:4‐hydroxyphenylpyruvate aminotransferase; MPO2, methylputrescine oxidase; ODC, ornithine decarboxylase; PMT2, putrescine methyltransferase; PPAR, phenylpyruvic acid reductase.
The biosynthesis of tropane alkaloids is well studied in the Solanaceae family, particularly in Atropa belladonna (Deadly Nightshade), where the availability of a multi‐tissue transcriptome assembly helped elucidate the biosynthetic pathway to hyoscyamine and scopolamine, two medicinally important tropanes (Bedewitz et al., 2014, 2018; Qiu et al., 2018, 2020; Srinivasan & Smolke, 2020). In the Solanaceae, the formation of tropinone is a key step in tropane alkaloid biosynthesis as it represents the first metabolite in the pathway with the characteristic 8‐azabicyclo[3.2.1]octane core skeleton shared by all tropanes (Fig. 2). The type III polyketide synthase, pyrrolidine ketide synthase (AbPyKS), is required for tropinone biosynthesis, likely through provision of 3‐oxoglutarate, which undergoes a nonenzymatic Mannich‐like decarboxylative condensation with the N‐methyl Δ1‐pyrrolinium cation to form 4‐(1‐methyl‐2‐pyrrolidinyl)‐3‐oxobutanoate (Bedewitz et al., 2018; J.‐P. Huang et al., 2019; Hasebe et al., 2021). This diketide metabolite is not only spontaneously decarboxylated to form the simple pyrrolidine hygrine, but also undergoes P450‐mediated cyclization to yield tropinone (Bedewitz et al., 2018). Thus, tropane and pyrrolidine alkaloid metabolism are intrinsically linked, and the resulting metabolic diversity is determined by a combination of enzyme‐catalyzed and nonenzymatic reactions.
Understanding of the tropane and pyrrolidine alkaloid network is incomplete. Silencing of AbPyKS in the roots of A. belladonna reduces tropane alkaloid biosynthesis and is accompanied by a dramatic increase in the abundance of the electrophilic N‐methyl Δ1‐pyrrolinium cation (Bedewitz et al., 2018; Fig. 2). The consequences of this metabolic shift on the tropane and pyrrolidine alkaloid network are unknown. In this study, we utilize high‐resolution mass spectrometry‐based metabolomics coupled with multivariate statistical analyses to define the metabolic consequences of AbPyKS silencing. This discovery metabolomics approach revealed dramatic changes to the tropane and pyrrolidine alkaloid network that were not revealed through earlier targeted metabolite analyses (Bedewitz et al., 2018). In particular, we report the existence of a previously unidentified pyrrolidine alkaloid subnetwork that shows increased abundance due to AbPyKS silencing. Multiple metabolites within this subnetwork share a core structure derived through nonenzymatic Mannich‐like decarboxylative condensation between the N‐methyl Δ1‐pyrrolinium cation and 2‐O‐malonylphenyllactate. Further metabolic complexity within this subnetwork occurs through decorations of this core structure through multiple glycosylations and hydroxylations, as well as additional nonenzymatic reactions to yield dipyrrolidines. Overall, this study uncovers previously unidentified metabolic connectivity between tropane and pyrrolidine alkaloid metabolism and highlights the importance of nonenzymatic reactions and catalytic promiscuity in generating specialized metabolite diversity.
Materials and Methods
Virus‐induced gene silencing and metabolite extractions
Root metabolite extracts from AbPyKS‐silenced and the corresponding TRV2 control Atropa belladonna L. plants, described previously in Bedewitz et al. (2018), were utilized for the discovery metabolomics experiments described later. A DNA construct targeting AbPPAR (Qiu et al., 2018) for virus‐induced gene silencing (VIGS) was assembled in the TRV2‐LIC vector (Dong et al., 2007) using primers listed in Supporting Information Table S1. Virus‐induced gene silencing of AbPPAR was performed as described in Bedewitz et al. (2018), and roots were harvested from AbPPAR‐silenced (n = 22 plants) and TRV2 controls (n = 24 plants) at 4 wk postinfiltration. Metabolite extractions from AbPPAR and the corresponding TRV2 controls were performed as described previously in Bedewitz et al. (2018) except that samples were incubated at room temperature during the extraction process. Gene silencing was validated by quantitative RT‐PCR by measuring transcript abundances in six silenced and six control plants based on their median phenyllactic acid (PLA) levels as measured by LC–MS. RNA extractions and quantitative RT‐PCR were performed as described in Bedewitz et al. (2018) using the primer sets listed in Table S1.
Discovery metabolomics and metabolite annotation
For discovery metabolite profiling of AbPyKS‐silenced roots, metabolite extracts were diluted 1 : 50 in acetonitrile and analyzed on a Waters Xevo G2‐XS Q‐TOF mass spectrometer (Waters Corp., Milford, MA, USA) equipped with a Shimadzu LC‐20AD high‐performance liquid chromatography (HPLC) system (Shimadzu Scientific Instruments, Columbia, MD, USA) and an Ascentis® Express F5 column (2.1 × 100 mm with 2.7 μm particle size) at an oven temperature of 50°C. A 20‐min hydrophilic interaction liquid chromatography gradient in Table S2 was used for HPLC separation with a sample injection volume of 10 μl and a flow rate of 0.3 ml min−1. Mass spectra were captured using electrospray ionization in positive‐ion mode with continuum acquisition without dynamic range extension and an acquisition mass range of m/z 50–1200 in sensitivity mode. Leucine enkephalin was used as lock mass, and telmisartan was added to the extraction solvent as an internal standard at a concentration of 0.25 μM. The scan time was 0.3 s, and the capillary and cone voltages were 3.00 kV and 40 V, respectively. Cone and desolvation gas flow rates were 20 and 500 l h−1, respectively, and the source and desolvation temperatures were 100°C and 350°C, respectively. The high‐energy spectra were generated using a collision voltage ramp of 15–60 V with argon as the collision gas. The diluted samples were pooled, and a pooled sample was analyzed after eight samples to facilitate the retention alignment of peaks throughout the experiment. A solvent blank sample followed the pooled sample to help assign background impurities. The order of sample analysis was randomized to minimize batch effects. AbPPAR‐silenced samples were profiled similarly except data were selectively extracted for targeted analysis of metabolites of interest.
The raw mass spectra from AbPyKS‐silenced roots were imported into Progenesis QI v.2.4 software (Nonlinear Dynamics Ltd, Newcastle, UK) for further processing. The data were processed using the parameters found in Table S3. Peaks were aligned using the first pooled sample as an alignment reference, peaks were deisotoped, and adduct signals from the adducts listed in Table S3 were combined. Peak picking with an absolute ion intensity threshold of five revealed 6291 features (including signals from multiple adducts of the same metabolite) for 5817 ions. Peaks were normalized to the internal standard and filtered. Filtering of features removed 4053 ions with levels highest in blank samples, 255 ions highest in a pooled fraction of all samples, 105 ions with relative mass defect (RMD) values above 1100 ppm (filtered because metabolites with RMD values above 1100 were compound classes not of interest (i.e. inorganics)), and 274 ions outside of a 0.98‐ to‐13.6‐min retention time window. Peaks found in the pooled and blank controls but not the experimental samples were removed from the analysis as these are likely contaminants. Peaks outside of 0.98‐ to 13.60‐min retention time range were removed as well as metabolites with RMD values over 1100 because most pyrrolidine and tropane alkaloids elute between 1.0 and 13.6 min and have mass defects between 300 and 900 ppm. RMD values were calculated as follows (Ekanayaka et al., 2015).
The processed data were imported into EZInfo v.3.0.2 software (Umetrics, Umeå, Sweden) for statistical analysis. Data were scaled using Pareto scaling to reduce the influence of metabolite abundance on the statistical model. Orthogonal projections to latent structures discriminant analysis (OPLS‐DA) was used to identify the ions responsible for the chemotypic differences between genotypes. The S‐plot generated from the OPLS‐DA model was used to identify ions of interest for downstream analyses. Metabolites with P(corr) values over +0.6 and below −0.6 were sorted by the P(1) values because this value correlates with ion abundance. The three most abundant metabolites in each MS survey scan were selected for data‐dependent tandem mass spectrometry for putative metabolite identification.
Tandem mass spectrometry (MS/MS) experiments were performed using a Waters Xevo G2‐XS Q‐TOF mass spectrometer equipped with a Shimadzu LC‐20AD HPLC system and an Ascentis® Express F5 column (2.1 × 100 mm with 2.7 μm particle size) with the same chromatographic separation parameters described in the untargeted metabolomics study. Tandem mass spectra were acquired using electrospray ionization in positive mode with an acquisition mass range of m/z 50–1200 using the Fast DDA TOF function in sensitivity mode. The source parameters were as described in the untargeted experiment excluding a change in cone voltage to 90 V and desolvation gas flow to 350 l h−1. The survey and MS/MS functions were collected with scan times of 0.5 s using centroided data acquisition. The intensity threshold was set to 5000 for the survey scan with a switchback threshold of 106. The MS/MS spectra used argon as a collision gas with a collision voltage ramp from 10 to 60 V. Several MS/MS experiments were performed with these parameters with altered exclusion lists ((1) the exclusion of ion masses below m/z 300, (2) below m/z 900, and (3) above m/z 310 and experiments with inclusion mass windows from (4) m/z 310–600 and (5) m/z 600–900) on metabolite extracts from both TRV2‐ and PyKS‐silenced samples. Negative mode tandem mass spectrometry experiments were performed with the same HPLC conditions, but the source parameters were modified as follows. The scan time was 0.5 s, and the capillary and cone voltage were 3.15 kV and 60 V, respectively. Cone and desolvation gas flow rates were 6 and 600 l h−1, respectively. The MS/MS functions were collected with scan times of 0.5 s with centroided data formats for mass m/z 290.14 collecting masses between m/z 50 and 700. The MS/MS spectra used argon as a collision gas with a collision voltage ramp from 10 to 60 V. Both AbPPAR‐ and AbPyKS‐silenced metabolite extractions were profiled undiluted in negative mode using reverse‐phase chromatography to measure PLA and 2‐O‐malonylphenyllactate (Table S4).
Metabolite purification and structural characterization
Detailed methods describing the purification and structural characterization of the P1 and P2 (m/z 292.15) metabolites and 2‐O‐malonylphenyllactate are provided in Methods S1, and Tables S5 and S6.
Production of P1 and P2 (m/z 292.15) metabolites through transient expression in N. benthamiana
Nicotiana benthamiana Domin seeds were treated with 10% w/v trisodium orthophosphate for 5 min, rinsed with distilled water, and sown into four‐inch square pots of Redi‐Earth. Seedlings were thinned to one plant per pot following emergence and grown for 6–8 wk in a growth room under fluorescent lights (145 μmol m−2 s−1) at 23°C with a 16‐h : 8‐h, light : dark photoperiod. Constructs of full‐length open reading frames for AbPMT2 and AbMPO2 in the pEAQ‐DEST1 vector were described previously (Bedewitz et al., 2018). Full‐length open reading frames of AbArAT4 and AbPPAR were amplified from plasmid DNA, cloned into the pEAQ‐HT vector (Sainsbury et al., 2009) using In‐Fusion™ cloning (Takara Bio Inc., Shiga, Japan), and transformed into A. tumefaciens strain LBA4404. Agrobacterium tumefaciens culture preparation and N. benthamiana infiltrations proceeded as described in Bedewitz et al. (2018) except that infiltrated leaves were harvested 5 d following infiltration. Metabolites were extracted from c. 100 mg of powdered leaf tissue in 1 ml of extraction solvent (20% methanol, 0.1% formic acid, and 0.25 μM telmisartan in water) using gentle rocking on an orbital shaker for 3 h at room temperature.
Metabolites were detected by the exact masses and coretention with metabolites in AbPyKS‐silenced plants using a Waters Xevo G2‐XS Q‐TOF mass spectrometer equipped with a Shimadzu LC‐20AD HPLC system and an Ascentis® C18 column (2.1 × 100 mm with 2.7 μm particle size) at an oven temperature of 45°C and reverse‐phase chromatographic separation shown in Table S4. Centroided data were collected using electrospray ionization in positive‐ion mode with a scan time of 0.1 s. Spectra were acquired in sensitivity mode with a mass range of m/z 50–1200. Capillary and cone voltage were set to 3 kV and 90 V, respectively. The cone and desolvation gas flows were 20 and 350 l h−1, and the source and desolvation temperatures were 100 and 350°C, respectively. Quasi‐simultaneous acquisition of low‐ and high‐collision energy mass spectra (MSE) was acquired, using leucine enkephalin as lock mass. The high‐energy spectra were acquired using a collision energy ramp from 20 to 80 V with argon as the collision gas.
Chemical synthesis
The N‐methyl‐Δ1‐pyrrolinium chloride was synthesized as described previously (Ma et al., 2020). Nonenzymatic synthesis of P1 and P2 ([M+H]+ m/z 292.15) together with P4 and P5 ([M+H]+ m/z 375.23) metabolites was achieved by combining 1 mM of N‐methyl‐Δ1‐pyrrolinium chloride with 1 mM 2‐O‐malonylphenyllactate purified from A. belladonna. Assays were performed in 25 mM potassium phosphate buffer pH 8.0 with a 100 μl reaction volume producing a final pH of 7.3. The reactions were incubated for 1 h at 30°C and then quenched by adding 25 μl of 5% formic acid containing 2 μM telmisartan (final concentration = 1% formic acid, 0.4 μM telmisartan). Control reactions lacking N‐methyl‐Δ1‐pyrrolinium chloride were included along with an N‐methyl‐Δ1‐pyrrolinium chloride control. Replicates (n = 3) were included for each reaction. Metabolites were detected as described previously for transient assays in N. benthamiana.
Results
Discovery metabolomics reveals an increase in pyrrolidine alkaloids in AbPyKS ‐silenced lines
In order to investigate alternate fates of the N‐methyl‐Δ1‐pyrrolinium cation in A. belladonna roots, we used discovery LC–MS‐based metabolomics to profile previously characterized AbPyKS‐silenced plants relative to the TRV2 controls (Bedewitz et al., 2018). After filtering of detected LC–MS signals (see the Materials and Methods section), 1130 ions remained and were used for orthogonal partial least squares discriminant analysis (also referred to as orthogonal projection to latent structures; OPLS‐DA) with AbPyKS and TRV2 controls selected as the treatment groups. This analysis revealed R2Y and Q2 values of 99% and 98% of the overall model (Fig. 3). The S‐plot generated from this model contained 154 and 257 signals with P(corr) values below and above the −0.6 and 0.6 thresholds, respectively (Table S7). In total, 62 ions were sufficiently abundant to enable targeted peak integration and tandem mass spectrometry experiments for structural annotation, and all yielded q‐values, a measure of false discovery rate, < 0.001 (Tables S8, S9).
Fig. 3.
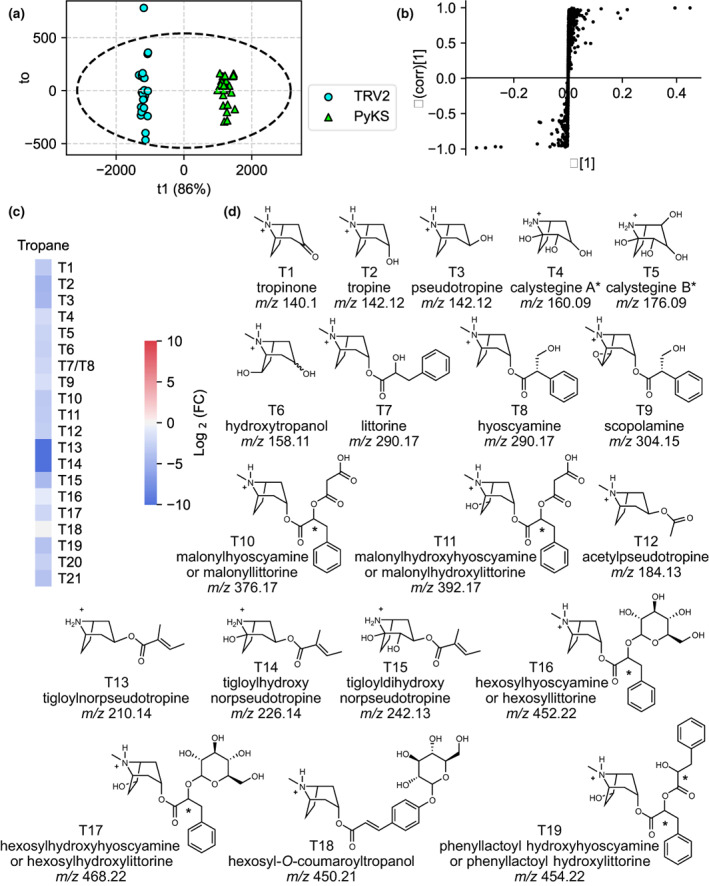
Discovery metabolite profiling of PyKS‐silenced lines reveals a decrease in tropane alkaloids in Atropa belladonna roots. (a) The orthogonal projections to latent structures discriminant analysis (OPLS‐DA) scores plot showing the separation of TRV2 control and PyKS‐silenced plants. The first component (T1 or predictive component shown on the x‐axis), the genotypic component, explains 86% of the variation between the samples. Each point represents a single metabolite extraction taken from a TRV2 (blue) or PyKS (green) plant. Within‐group variance (to or the orthogonal component) is shown on the y‐axis, and Hotelling's T2 (95%) ellipse is shown with the dashed ellipse. (b) The OPLS‐DA S‐plot reveals the ions that contribute to the differences between TRV2 control and PyKS‐silenced lines. Each point represents a signal from the metabolite profiles of the TRV2‐ and PyKS‐silenced plants. Positive x‐ and y‐values indicate that these signals are more abundant in PyKS‐silenced plants compared with the TRV2 controls while negative values indicate that these signals are more abundant in TRV2 control plants compared with PyKS‐silenced plants. X‐values approaching axes extremes (−1 or 0.5) designate that those signals greatly contribute to the model differences, whereas y‐values approaching axes extremes (−1 or 1) are indicative of increased confidence. (c) Change in tropane alkaloid abundance in PyKS‐silenced plants compared with TRV2 controls (log2(FC) refers to log2(fold change)). Red indicates the corresponding metabolite is increased in abundance in PyKS‐silenced plants relative to TRV2 controls, while blue represents metabolites that are decreased. (d) Structural annotations of quantified tropane alkaloids. Compounds are numbered as they appear in the text and annotations are based on tandem mass spectrometry fragmentation (Supporting Information Table S8). Metabolite structures (T1–T19) are displayed in the [M+H]+ form. m/z corresponds to the mass‐to‐charge ratio. An asterisk indicates an ambiguous structural annotation due to unresolved stereochemistry or the presence of isomers.
As expected, based on prior targeted metabolite analysis (Bedewitz et al., 2018), several of the ions that decreased in AbPyKS‐silenced plants are known tropane alkaloids, including scopolamine, constitutional isomers hyoscyamine/littorine, tropine, pseudotropine, and tropinone. Tandem mass spectrometry experiments evaluating ions of decreased relative abundance in AbPyKS‐silenced plants provided useful information for assigning metabolites to structural classes. For example, metabolites yielding protonated fragment ions m/z 142.09, 128.08, 158.08, and 110.1 were consistent with those typically observed for nortropane alkaloids, whereas fragment ions m/z 142.12, 140.11, and 124.11 suggested structures related to N‐methylated tropane alkaloids (Fig. S1; Table S8). Several of these tropane and nortropane alkaloids were identified previously in A. belladonna roots, including the tropanols and calystegines (T1–T6), tropane aromatic esters (T7–T9), tropane aromatic esters with acylations (T10 and T11), and tropane and nortropane aliphatic esters (T12–T15; Fig. 3; Bedewitz et al., 2018; Sadre et al., 2022). Six tropane alkaloids annotated here appear to be novel, including T16–T21 displaying [M+H]+ at m/z 452.23, 468.22, 450.21, 454.22, 306.17, and 386.16 (Table S8; Fig. 3). The tandem mass spectra for three of these compounds (T16–T18) exhibited a neutral loss of 162.057 Da, yielding fragment ions m/z 290.17, 306.17, and 288.16, suggesting that these compounds are hexosides of littorine or hyoscyamine, hydroxylittorine or hydroxyhyoscyamine, and coumaroyltropine, respectively. The T19 metabolite exhibited a neutral mass loss of 166.06 Da to yield m/z 288.16, consistent with the loss of PLA to generate dehydro‐littorine or dehydro‐hyoscyamine. Consequently, this metabolite was annotated as phenyllactoyl hydroxyhyoscyamine or phenyllactoyl hydroxylittorine. These annotations allowed us to propose a pathway in which the major aromatic tropane esters littorine and hyoscyamine are decorated in A. belladonna roots with malonate, hexose, and phenyllactoyl groups (Fig. S2). Two remaining metabolites, T20 and T21, lack putative annotations (Table S8). Although the exact mass of T20 [M+H]+, m/z 306.1699 corresponds to C17H24NO4 +, consistent with hydroxyhyoscyamine and hydroxylittorine, the fragment ions m/z 200.13 and 154.12 are inconsistent with this annotation as these metabolites yield m/z 158.12 and m/z 140.12 fragment ions (Schlesinger et al., 2021).
Tandem mass spectrometry of ions increased in AbPyKS‐silenced plants revealed that 36 metabolites gave rise to an m/z 84.08 fragment ion (C5H10N+) consistent with metabolites that contain a methylpyrrolidine moiety (Fig. 4; Table S9). In contrast to these novel pyrrolidines, hygrine (P3) abundance decreased as the disruption of 3‐oxoglutarate biosynthesis prevented the formation of 4‐(1‐methyl‐2‐pyrrolidinyl)‐3‐oxobutanoic acid and its subsequent decarboxylation to hygrine (Bedewitz et al., 2018). We focused on the structural annotation of the most abundant pyrrolidine alkaloids including P1 and P2 (m/z 292.15), P4 and P5 (m/z 375.23), P6 (m/z 144.1), P7 (m/z 227.17), P8 and P9 (m/z 308.15), and P10 (m/z 454.2); ions correspond to [M+H]+. The measured, exact masses of the novel pyrrolidines were used to assign the molecular formula of each ion. The odd‐nitrogen rule was used to predict that each [M+H]+ ion contained at least one nitrogen, except P4 and P5 (the m/z 375.23 isomers) presumably containing two nitrogen atoms (Table S9). Fragmentation of P1, P2, P4, and P5 revealed that fragment ions m/z 144.1 and 209.16 coincided with the neutral losses of 148.05 and 166.06 Da, respectively (Fig. 5a,b). These neutral losses are consistent with cinnamoyl and phenyllactoyl‐like moieties. To confirm this assessment, isomers P1 and P2 were analyzed in negative mode ([M−H]−, m/z 290.14), yielding fragment ions m/z 165.055 and 147.0446 analogous to those generated in positive‐ion mode (Table S10).
Fig. 4.
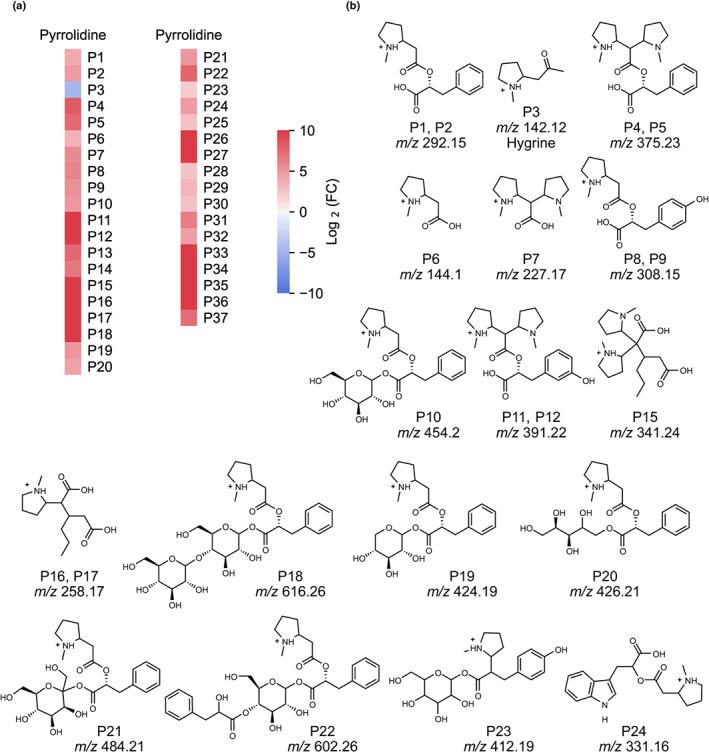
Discovery metabolite profiling of PyKS‐silenced lines reveals an increase in pyrrolidine alkaloids in Atropa belladonna roots. (a) Change in pyrrolidine alkaloid abundance in PyKS‐silenced plants compared with TRV2 controls (log2(FC) refers to log2(fold change)). Red indicates the corresponding metabolite is increased in abundance in PyKS‐silenced plants relative to TRV2 controls, while blue represents metabolites that are decreased. (b) Structural annotations of quantified pyrrolidine alkaloids. Compounds are numbered as they appear in the text and annotations are based on tandem mass spectrometry fragmentation (Supporting Information Table S9). Metabolite structures (P1–P24) are displayed in the [M+H]+ form. The stereochemistries of the sugar moieties of P10 and P18 through P23 as well as the positions of the hydroxyl groups on the aromatic ring of P8, P9, P11, P12, and P23 are unresolved. m/z corresponds to the mass‐to‐charge ratio.
Fig. 5.
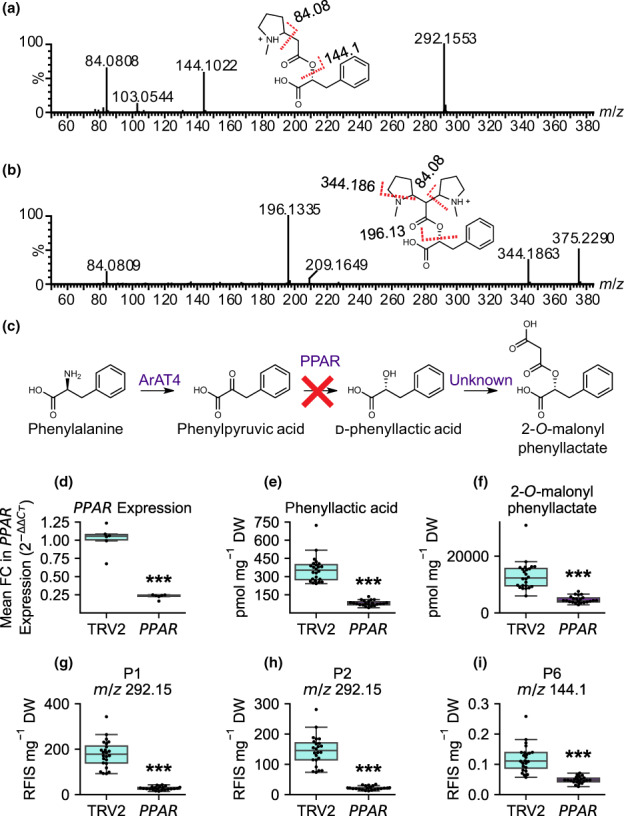
Phenyllactic acid (PLA) is incorporated into pyrrolidine alkaloid metabolites. Liquid chromatography with tandem mass spectrometry spectra obtained for the (a) P1 (m/z 292.15) and (b) P4 (m/z 375.23) metabolites collected in positive mode with electrospray ionization. The x‐axis indicates the mass‐to‐charge ratio (m/z) of the fragment ions, whereas the y‐axis indicates percent abundance. Red‐dashed lines indicate fragmentation of the molecule at that location with the numerical value of the fragment yielded indicated above the dashed line. A loss of 31 Da in (b) (producing the m/z 344 and m/z 196 fragments) indicates a neutral loss of methylamine. Fragmentation to m/z 144.1 and m/z 196.13 in panels (a) and (b) represents a neutral mass loss of PLA and a neutral mass loss of PLA and methylamine, respectively. (c) The pathway for 2‐O‐malonylphenyllactate formation in Atropa belladonna with the red ‘X’ indicating silencing of PPAR. (d) Mean fold change (FC) in PPAR expression in TRV2 control (n = 6) and PPAR‐silenced (n = 6) A. belladonna plants with the six median PLA levels for each genotype. Genotypes are shown on the x‐axis and the mean FC in PPAR expression is shown on the y‐axis calculated as mean . (e–i) Metabolite abundances of PLA, 2‐O‐malonylphenyllactate, P1, P2, and P6 in TRV2 (n = 24; left, cyan) and PPAR (n = 22; right, purple). (d–i). Box and whisker plots are displayed with horizontal lines indicating upper extreme, upper quartile, median, lower quartile, and lower extreme in order from top to bottom. The dots on the box and whisker plots represent individual samples. Statistical significance is indicated as determined by Welch's t‐test: ***, P < 0.001.
We annotated P6 (C7H14NO2, as the neutral species) as N‐methylpyrrolidin‐2‐yl acetic acid based on exact mass and the subsequent fragmentation of P6 to m/z 84.08 and P7 as 2,2‐di(N‐methylpyrrolidin‐2‐yl)acetic acid with an extra pyrrolidinyl attached to carbon‐2 of acetic acid. P6 occurred both as a neutral fragment of P1, P2, P8, P9, and P10 and as an intact independent metabolite; however, whether this metabolite is formed independently through a Mannich‐like decarboxylative condensation or through hydrolysis of another metabolite is unknown. The predicted molecular formulas of P4 and P5, P8 and P9, and P10 are consistent with the molecular formula of the P1 and P2 with the addition of an N‐methylpyrrolidinyl, hydroxyl, and hexosyl moiety, respectively. Fragmentation of P10 revealed the neutral loss of 162.057, yielding m/z 292.15 consistent with the loss of a hexose moiety. MS/MS spectra of P8, P9, and P10 exhibited fragment ions at m/z 144.1 and 84.08 matching the fragmentation pattern of P1 and P2 isomers. The fragmentation of P8 and P9 to m/z 144.1 suggests that the additional hydroxyl group is located on the cinnamoyl/phenyllactoyl‐like moiety. However, P4 and P5 differ in their fragmentation pattern, likely due to the predicted dipyrrolidinyl moiety (but the exact attachment of the pyrrolidines and their stereochemistry is unknown). Several metabolites detected as odd mass [M+H]+ ions increased in AbPyKS‐silenced lines. P11 and P12 (m/z 391.22), P13 and P14 (m/z 453.2), and P15 (m/z 341.24) were annotated as dipyrrolidines due to fragmentation patterns similar to P4 and P5, including the formation of fragment ions m/z 227.17, 209.16, and 196.13 (Table S9). P15 shares a neutral loss of 114.07 Da with the protonated P16 and P17 (m/z 258.17) monopyrrolidines, suggesting that this metabolite is the dipyrrolidine analog of P16 and P17. However, the identity of the remaining portion of the molecule requires further investigation. The exact mass of P11 and P12 suggests that these metabolites are derived from P4 and P5 but contain an additional oxygen atom. The shared m/z 277.17, 209.16, and 196.13 fragments correspond to the dipyrrolidinylacetic acid found in the P4 and P5 isomers, indicating that the additional hydroxyl group is on the PLA portion of the molecule (Table S9).
Further investigation of tandem mass spectra of compounds with elevated concentrations in AbPyKS‐silenced lines revealed a collection of metabolites yielding fragment ions of m/z 292.15, 144.1, and 84.08. The neutral losses (162.06, 132.05, 134.06, and 192.07 Da) suggest that several of these metabolites possess sugar moieties, including P18 (m/z 616.26), P19 (m/z 424.19), P20 (m/z 426.21), and P21 (m/z 484.21). The exact masses in combination with the tandem mass spectra support the putative annotation of P18–P21 as esterified derivatives of P1/P2 with the addition of dihexose, pentose, reduced pentose, and heptose to the carboxylic acid group, respectively (Table S9). Additionally, P22 (m/z 602.26) fragments and loses 166.06 Da, consistent with a neutral loss of PLA, and fragments further to yield m/z 292.15 after the loss of 144.047 Da supporting the structural annotation as a P1 or P2 metabolite with the addition of a hexosyl and an additional phenyllactoyl moiety. The mass spectrum of P23 (m/z 412.19) is consistent with the addition of N‐methylpyrrolidine to a cinnamic acid hexosyl ester, although there are structural ambiguities when trying to identify the acid portion of the molecule. Although much of the pyrrolidine diversity in A. belladonna root is seemingly derived from pyrrolidines containing PLA, the concentrations of several other pyrrolidine alkaloids increased in AbPyKS‐silenced plants. They were identified as the protonated cations of P16 and P17 (m/z 258.17), P24 (m/z 331.16), P25 (m/z 314.13), and P26 (m/z 370.13) containing alternative and unknown substituents (Table S9).
Suppression of phenyllactate biosynthesis disrupts pyrrolidine alkaloid accumulation in A. belladonna roots
Tandem mass spectrometry experiments suggested that several of the metabolite features with the highest OPLS‐DA P(corr) values lose a cinnamoyl‐like moiety (Fig. 5a,b; Table S9). Tandem mass spectra collected in negative‐ion mode also reveal that P1 and P2 (in their [M−H]− form, m/z 290.14) fragments to form m/z 165.05, a mass consistent with deprotonated PLA (Table S10). To investigate whether the P1 and P2 metabolites and their derivatives are PLA‐derived, we used VIGS to silence AbPPAR, a gene encoding phenylpyruvic acid reductase (PPAR; Fig. 5c), which catalyzes the conversion of phenylpyruvate to PLA in the pathway leading to scopolamine formation (Qiu et al., 2018). Silencing of AbPPAR resulted in a 79% decrease in transcript abundance relative to TRV2 plants. Targeted metabolite analysis showed that PLA is reduced in PPAR‐silenced plants relative to TRV2 control plants (Fig. 5d–i). The novel pyrrolidines, P1, P2, and P6, are reduced in PPAR‐silenced plants supporting the derivation of P1 and P2 from PLA and the subsequent hydrolysis of either of these isomers to form P6 (Figs 4, 5). These data do not support the hypothesis that P6 is formed independently of P1 and P2. The levels of P4 and P5 could not be quantified in PPAR‐silenced plants as they were below the limits of detection in these extracts. However, P8 and P10 were reduced in PPAR‐silenced plants (Fig. S3). P9 is a lower abundance isomer and was below the limit of detection in this experiment.
We were able to confirm the structural annotation derived from MS/MS by purifying the two major P1 and P2 isomers from bulk‐silenced AbPyKS root tissue and performing 1D‐ and 2D‐NMR experiments. The NMR experiments confirmed that P1 and P2 are 2‐(2‐(1‐methylpyrrolidin‐2‐yl)acetoxy)‐3‐phenylpropanoic acid diastereomers (Tables S11, S12; Dataset S1). Note that protons on or adjacent to the pyrrolidinyl ring of each isomer are reported (Tables S11, S12; Dataset S1) and shown (Dataset S1. NMR spectra a and h) as multiplets without coupling constants as they were unresolved. This is likely the result of high‐speed oscillations including nitrogen inversion within the five‐membered ring that influence HCCH dihedral angles and broaden NMR signals, a common phenomenon observed in pyrrolidine NMR (Steimbach et al., 2021).
We hypothesized that P1 and P2 are formed via a Mannich‐like decarboxylative condensation of 2‐O‐malonylphenyllactate with the N‐methyl‐Δ1‐pyrrolinium cation due to the attachment of N‐methylpyrrolidine moieties to 2‐O‐acetylphenyllactate (Fig. 6; Tables S11, S12; Dataset S1). Indeed, 2‐O‐malonylphenyllactate was annotated in A. belladonna root extracts based on its exact mass (m/z 251.07 [M−H]−) and silencing of PPAR resulted in a reduction in this metabolite (Fig. 5f). Purification and NMR analysis confirmed this putative assignment (Table S13). Furthermore, in agreement with the role of 2‐O‐malonylphenyllactate in the formation of P1 and P2, the abundance of this metabolite decreased in AbPyKS‐silenced plants while the levels of PLA were unaltered (Fig. S4).
Fig. 6.
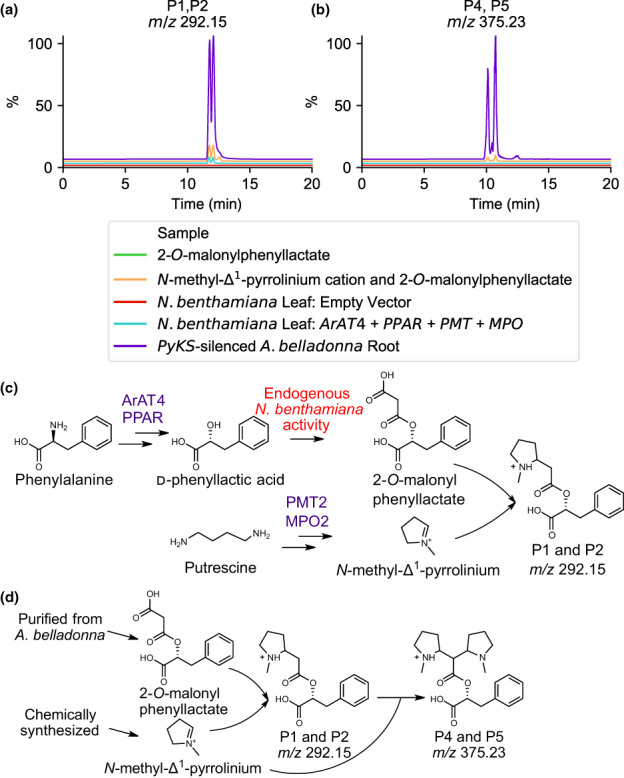
In planta and nonenzymatic formation of pyrrolidine alkaloids. Extracted ion chromatogram of (a) P1 and P2 (m/z 292.15) and (b) P4 and P5 (m/z 375.23) showing nonenzymatic formation (orange) and in Nicotiana benthamiana (cyan) in comparison with an PyKS‐silenced Atropa belladonna root sample (purple). Assay experimental design for pyrrolidine alkaloid formation in (c) N. benthamiana leaf and (d) nonenzymatic synthesis. Metabolite structures are displayed in the [M+H]+ form for P1, P2, P4, and P5. Enzyme abbreviations are as follows; ArAT4, phenylalanine:4‐hydroxyphenylpyruvate aminotransferase; MPO2, methylputrescine oxidase; PMT2, putrescine methyltransferase; PPAR, phenylpyruvic acid reductase. m/z corresponds to the mass‐to‐charge ratio.
Pyrrolidine alkaloid diversity driven by cation electrophilicity
To explore the formation of P1 and P2, we sought to produce these metabolites in N. benthamiana by using transient expression of tropane alkaloid pathway genes to produce PLA and the N‐methyl‐Δ1‐pyrrolinium cation. When AbArAT4 and AbPPAR are transiently expressed in N. benthamiana, PLA is produced and subsequently malonylated by unknown endogenous N. benthamiana enzymes to produce appreciable levels of the 2‐O‐malonylphenyllactate, more than 10‐fold greater than PLA, as determined using an authentic standard purified from A. belladonna (Fig. S5). We demonstrated previously that transient expression of AbPMT2 and AbMPO2 in N. benthamiana leaves leads to the formation of the N‐methyl‐Δ1‐pyrrolinium cation (Bedewitz et al., 2018). Transient expression of AbArAT4, AbPPAR, AbPMT2, and AbMPO2 resulted in the production of P1 and P2 as determined by coretention with the metabolites in a PyKS‐silenced A. belladonna root sample and tandem mass spectrometry (Fig. 6; Table S14). Metabolites P6, P8, P9, P10, P18, P20, and P22 were also formed in this experiment, suggesting the presence of promiscuous hydroxylases, glycosyltransferases, transferases, and esterases in N. benthamiana that catalyze their formation (Fig. S6). Although an m/z 424.19 metabolite is produced in N. benthamiana plants transiently expressing ArAT4, PPAR, PMT, and MPO, this metabolite does not co‐elute with P19 (with the same protonated m/z 424.19 molecular ion) that accumulates in A. belladonna PyKS‐silenced root suggesting that these metabolites are distinct isomers. The P4 and P5 isomers were not detected in these N. benthamiana leaf extracts. We next sought to determine whether P1 and P2 could form nonenzymatically. We synthesized N‐methyl‐Δ1‐pyrrolinium chloride and used 2‐O‐malonylphenyllactate purified from A. belladonna to assay P1 and P2 formation in the absence of enzyme. The reaction was incubated for 1 h at 30°C following the addition of 1 mM of each reactant. The addition of these two products resulted in the formation of P1 and P2, as well as P4 and P5 that matched the AbPyKS‐silenced products in exact mass, retention time, and tandem mass spectrometry fragmentation pattern (Fig. 6; Table S14).
Discussion
Atropa belladonna has emerged as a model species for exploring the biosynthesis of the medicinally important tropane alkaloids hyoscyamine and scopolamine. As such, metabolite profiling in A. belladonna has mainly focused on the targeted analysis of the scopolamine pathway and its associated intermediates (Bedewitz et al., 2014, 2018; Qiu et al., 2018, 2020). Consequently, there remains an overall lack of understanding of the tropane‐related metabolic network. To improve this knowledge gap, we utilized discovery metabolomics based on high‐resolution mass spectrometry and multivariate statistical analysis to determine the metabolic consequences of silencing AbPyKS, a key enzyme in tropane alkaloid biosynthesis. This approach revealed substantial changes to the A. belladonna root metabolome, including the discovery of previously unreported tropane and pyrrolidine alkaloids and the identification of a pyrrolidine alkaloid metabolic subnetwork comprised of P1 and P2 derivatives formed by the nonenzymatic Mannich‐like condensation of the N‐methyl‐Δ1‐pyrrolinium cation and 2‐O‐malonylphenyllactate. As such, this study illustrates the potential of combining discovery metabolomics with targeted genetic perturbations for expanding the understanding of plant‐specialized metabolism.
Nonenzymatic reactions play key roles in metabolism because of the energy spent to prevent cellular damage by reactive metabolites and the importance of nonenzymatic reactions in the evolution of enzymes and metabolic pathways (Keller et al., 2015; Lerma‐Ortiz et al., 2016). For example, nonenzymatic glycolysis and the pentose phosphate pathway‐like reactions occur in Fe(II)‐rich conditions, an environment potentially provided by Archean oceans (Keller et al., 2014). Recently, the nonenzymatic formation of isoprene, an abundant volatile synthesized by isoprene synthase (IspS) in plants and bacteria, was demonstrated using Mn2+ (Oku et al., 2022). The existence of nonenzymatic versions of these pathways in certain conditions (i.e. availability of divalent cations) supports the hypothesis that pathway enzymes evolve to regulate and support these nonenzymatic reactions and eventually allow them to occur in the absence of the original catalysts (Keller et al., 2014). Nonenzymatic reactions also occur in plant‐specialized metabolism, including striking examples that lead to the formation of betalain pigments in Caryophyllales and nesocodin, an alkaloid responsible for the blood‐red pigmentation of floral nectar in Nesocodon mauritianus (Campanulaceae) and Jaltomata herrerae (Solanaceae; Schliemann et al., 1999; Roy et al., 2022). These latter two examples involve the formation of iminium ion intermediates through the condensation of a primary amine with an aldehyde.
In the Solanaceae, oxidation of N‐methylputrescine to N‐methylaminobutanal by methylputrescine oxidase leads to spontaneous intramolecular condensation and formation of the N‐methyl‐Δ1‐pyrrolinium cation (Heim et al., 2007; Katoh et al., 2007; Lichman, 2021). This mechanism is postulated to have evolved from polyamine catabolism, where putrescine is converted to 4‐aminobutanal before subsequent reactions to yield succinate (Naconsie et al., 2014). Analogous reactions occur in ring formation of piperidine and pyrrolizidine alkaloids (Sun et al., 2012; Zakaria et al., 2022). Iminium cations are potent electrophiles, and 4‐(1‐methyl‐2‐pyrrolidinyl)‐3‐oxobutanoic acid, an intermediate in tropane alkaloid biosynthesis, can be formed through a Mannich‐like decarboxylative condensation between the N‐methyl‐Δ1‐pyrrolinium cation and 3‐oxoglutaric acid, while cuscohygrine is formed through the same mechanism by condensation of 4‐(1‐methyl‐2‐pyrrolidinyl)‐3‐oxobutanoic acid with a second N‐methyl‐Δ1‐pyrrolinium cation (Bedewitz et al., 2018; J.‐P. Huang et al., 2019). Silencing of AbPyKS in A. belladonna root leads to the substantial accumulation of the N‐methyl‐Δ1‐pyrrolinium cation and reduced flux toward tropane alkaloids (Fig. 2; Bedewitz et al., 2018). This reduced tropane accumulation ensures that there is a large pool of PLA in A. belladonna roots, the majority of which is malonylated by an uncharacterized malonyltransferase (Fig. 5). This malonate group serves as a nucleophile that reacts spontaneously with the electrophilic N‐methyl‐Δ1‐pyrrolinium cation to yield P1 and P2, and with a second N‐methyl‐Δ1‐pyrrolinium cation to generate P4 and P5 (Fig. 6; Table S14). The difference in the relative accumulation of P1, P2, P4, and P5 in AbPyKS‐silenced root tissue compared with N. benthamiana leaf and in chemical assays (Fig. 6), is possibly attributed to the time scales over which these experiments were conducted. For example, samples from A. belladonna VIGS experiments were harvested 4 wk postinfiltration, providing a considerable time for metabolite accumulation to occur. By contrast, samples were harvested after 5 d for transient expression assays in N. benthamiana leaf and after only an hour in chemical reactivity assays. While we propose that P1, P2, P4, and P5 are formed nonenzymatically in planta, we cannot completely exclude the possibility that enzymes contribute to their formation in A. belladonna root and N. benthamiana leaf.
The identification of the P1 and P2 isomers and their derivatives reveals alternate fates for PLA in A. belladonna roots beyond its well‐defined role in the biosynthesis of principal aromatic tropane esters littorine, hyoscyamine, and scopolamine (Robins et al., 1994). However, we cannot exclude the possibility that pyrrolidine alkaloids with hydroxyl groups on the aromatic ring (P8, P9, P11, P12, and P23) are derived from 4‐hydroxyphenyllactate rather than PLA. For example, the promiscuous activities of AbArAT4 and AbPPAR could yield 4‐hydroxyphenylpyruvate and 4‐hydroxyphenyllactate from tyrosine, respectively, and the latter could be malonated before condensation with the N‐methyl‐Δ1‐pyrrolinium cation. AbArAT4 can catalyze the transfer of the amino group from tyrosine to phenylpyruvate to yield phenylalanine and 4‐hydroxyphenylpyruvate although this reaction is 250‐fold less catalytically efficient than when phenylalanine is used as the amino donor substrate (Bedewitz et al., 2014). Similarly, AbPPAR is c. 10‐fold less catalytically efficient when 4‐hydroxyphenylpyruvate is used as a substrate rather than phenylpyruvate (Qiu et al., 2018). Phenyllactic acid is a broad‐spectrum antimicrobial agent that has attracted considerable interest from the food, animal feed, and pharmaceutical industries (Dieuleveux et al., 1998; Rajanikar et al., 2021). The range of roles of PLA in plants remains undefined, but this study reveals that a large PLA pool exists in A. belladonna roots that is channeled into diverse metabolites (Figs 4, 6). Future efforts could focus on examining the potential bioactivities of these novel metabolites, both in terms of biomedical or agricultural applications, but also with respect to their potential influence on the rhizosphere.
In total, c. 400 ions met our statistical threshold for altered abundance in AbPyKS‐silenced roots relative to controls (Table S7). Of these, we were able to annotate c. 60 tropane and pyrrolidine metabolites, including many that are previously unreported (Tables S8, S9). However, most of the metabolites remain unannotated, illustrating the challenges of metabolite annotation and indicating that our understanding of root metabolism remains incomplete. For example, c. 40 metabolites were annotated as simple pyrrolidines based on their ability to yield m/z 84 fragments characteristic of the N‐methyl‐Δ1‐pyrrolinium cation (Table S9). Based on the proposed formation of P1 and P2 isomers through Mannich‐like decarboxylative condensation, we hypothesize that all these simple pyrrolidines are formed through similar reactions with nucleophilic intermediate metabolites. However, except for the 15 metabolites derived from the condensation of the N‐methyl‐Δ1‐pyrrolinium cation with 2‐O‐malonylphenyllactate, the identities of the corresponding nucleophiles that give rise to the remaining metabolites are yet to be determined.
The propensity of the N‐methyl‐Δ1‐pyrrolinium cation to react and form products with different nucleophiles is advantageous for the synthesis of diverse pyrrolidines and could represent a strategy for the generation of scaffold molecules for subsequent modification through metabolic engineering or chemical synthesis. Indeed, we engineered the production of P1 and P2 in N. benthamiana through transient expression of AbPMT2, AbMPO2, AbArAT4, and AbPPAR while taking advantage of a promiscuous endogenous malonyltransferase that forms 2‐O‐malonylphenyllactate (Fig. 6). However, the electrophilicity of the N‐methyl‐Δ1‐pyrrolinium cation could also present challenges for engineering the production of specific pyrrolidines due to the formation of unwanted by‐products or damage to nucleophilic macromolecules or host metabolites (Lerma‐Ortiz et al., 2016). For example, although metabolic engineering of the N‐methyl‐Δ1‐pyrrolinium cation and tropane alkaloids has been achieved in microbial systems, metabolite analysis of the engineered strains is thus far limited to the desired target molecules and the potential formation of off‐target metabolites was not considered (Ping et al., 2019a,b; Srinivasan & Smolke, 2019, 2020).
Metabolite profiling of PyKS‐silenced material revealed a subnetwork of metabolites putatively assigned by MS/MS experiments as P1, P2, P4, and P5 core structures, with sugar or hydroxyl group additions. These metabolites and their modified derivatives also accumulate in nonsilenced A. belladonna roots, albeit at a considerably lower percentage of the total ion chromatogram compared with tropane alkaloids. The pathway model (Fig. 7) hypothesizes that these additions are catalyzed by enzymes involved in specialized metabolism such as UDP‐glycosyltransferases – enzyme classes notorious for their promiscuity (Louveau & Osbourn, 2019). Interestingly, the same decorations occur in the P1 and P2 derivatives produced via metabolic engineering in N. benthamiana leaves (Fig. S6), indicating that the corresponding enzyme activities are conserved in different Solanaceae genera (Atropa and Nicotiana) and in distinct tissues (roots and leaves). Similarly, we identified several glycosylated and malonylated forms of tropane aromatic esters that decreased in PyKS‐silenced plants (Fig. S2; Table S8). Such modifications increase the complexity of the alkaloid metabolome and may act in the detoxification of molecules that would otherwise be harmful to the cell, or possibly serve as forms that are more readily transported (Louveau & Osbourn, 2019; Zhang et al., 2022). The identification of these modified alkaloids paves the way for future characterization of the enzymes that catalyze their formation.
Fig. 7.
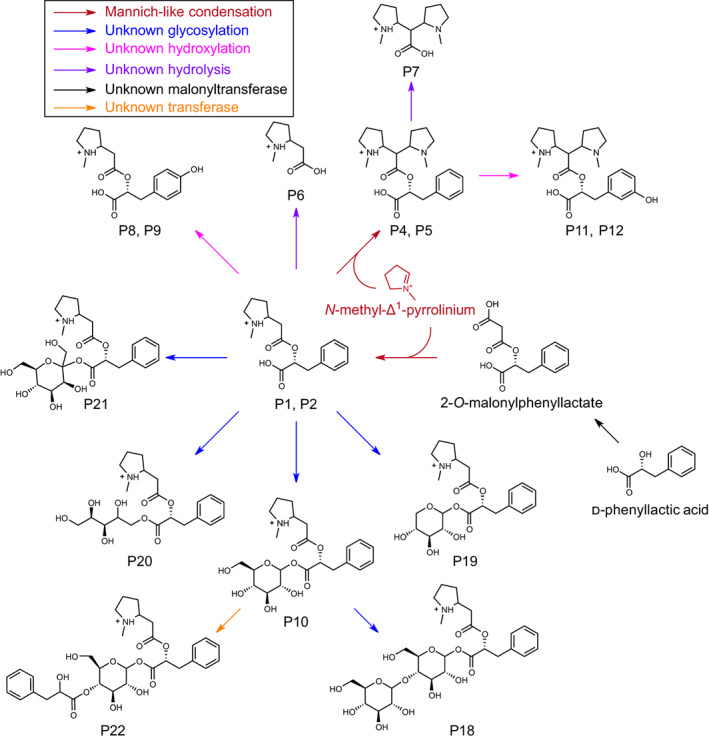
Proposed pyrrolidine alkaloid subnetwork in Atropa belladonna root. 2‐O‐malonylphenyllactate is formed from d‐phenyllactic acid through catalysis by an unknown acyltransferase. 2‐O‐malonyl phenyllactate undergoes Mannich‐like decarboxylative condensation with the N‐methyl‐Δ1‐pyrrolinium ion to form the m/z 292.15 metabolite (P1 and P2). The m/z 292.15 metabolite is subsequently modified through addition of a second N‐methyl‐Δ1‐pyrrolinium ion to yield the m/z 375.23 metabolite (P4 and P5). These scaffold molecules then undergo further modifications by unknown enzymes leading to increased pyrrolidine alkaloid diversity. For simplicity, only single isomers of each metabolite are shown. Structures inferred from MS/MS fragmentation (Supporting Information Table S9). Metabolite structures are displayed in the [M+H]+ form (P1, P2, P4–P12, and P18–P22). m/z corresponds to the mass‐to‐charge ratio.
Competing interests
None declared.
Author contributions
HMP and CSB conceived the study and wrote the article. HMP, ADJ and CSB designed the research. MAB generated the AbPyKS‐silenced plant material and first noted the existence of the m/z 292.15 isomers. HMP, JMG and MAB contributed to the development and analysis of AbPPAR‐silenced plant material. HMP performed UHPLC/MS, multivariate statistical and UHPLC/MS/MS analyses and performed chemical synthesis assays and subsequent data analyses. HMP, MAC and ADJ annotated metabolites. HMP and JMG quantified metabolites. MAB, CSB and HMP generated plant expression constructs performed transient expression analysis in N. benthamiana. MAB and SMH purified and performed NMR experiments to resolve the structure of 2‐O‐malonylphenyllactate. HMP and MAC purified and performed NMR experiments to resolve the structures of the P1 and P2 (m/z 292.15) isomers. SMH and MAC interpreted NMR spectra. ADJ reviewed the NMR interpretations. HMP and KDW synthesized the N‐methyl‐Δ1‐pyrrolinium chloride. ADJ, KDW and MAC edited the manuscript. All authors read and approved the manuscript.
Supporting information
Dataset S1 NMR spectra of pyrrolidine metabolites P1 and P2.
Fig. S1 Tandem mass spectra and proposed fragmentation of representative tropane and nortropane alkaloids.
Fig. S2 Proposed subnetwork of decorated aromatic tropane esters in Atropa belladonna root.
Fig. S3 Modified pyrrolidine alkaloids are decreased in phenylpyruvic acid reductase (PPAR)‐silenced Atropa belladonna roots.
Fig. S4 Abundance of phenyllactic acid and 2‐O‐malonylphenyllactate in PyKS‐silenced Atropa belladonna roots.
Fig. S5 Phenyllactic acid production through transient expression in Nicotiana benthamiana.
Fig. S6 Conserved enzymatic activity results in pyrrolidine alkaloid modifications in Nicotiana benthamiana.
Methods S1 Metabolite purification and structural characterization.
Table S1 Oligonucleotide primers used in the study.
Table S2 UPLC mobile phase gradient utilized for LC‐TOF‐MS analyses of pyrrolidine and tropane alkaloids.
Table S3 Metadata describing Progenesis and EZInfo processing of the AbPyKS vs TRV2 metabolomics experiment.
Table S4 UPLC mobile phase gradient utilized for LC‐TOF‐MS metabolite analysis.
Table S5 LC mobile phase gradient utilized for semi‐preparative separation of metabolite extracts of bulk PyKS‐silenced Atropa belladonna.
Table S6 LC isocratic mobile phase conditions utilized for semi‐preparative separation of P1 and P2 (m/z 292.15) isomers.
Table S7 List of ions from the S‐plot of the orthogonal projections to latent structures discriminant analysis analysis of PykS‐silenced Atropa belladonna roots compared with TRV2 empty vector controls.
Table S8 Liquid chromatography coupled with tandem mass spectrometry fragmentation of positive ions [M+H]+ that are decreased in PyKS‐silenced Atropa belladonna roots compared with TRV2 empty vector controls.
Table S9 Liquid chromatography coupled with tandem mass spectrometry fragmentation of positive ions [M+H]+ that are increased in PyKS‐silenced Atropa belladonna roots compared with TRV2 empty vector controls.
Table S10 Liquid chromatography coupled with tandem mass spectrometry fragmentation of m/z 292.15 (P1 and P2) isomers in negative mode [M−H]− (m/z 290.14).
Table S11 NMR chemical shift data for m/z 292.15 isomer 1 (P1).
Table S12 NMR chemical shift data for m/z 292.15 isomer 2 (P2).
Table S13 NMR chemical shift data for 2‐O‐malonylphenyllactate.
Table S14 Liquid chromatography coupled with tandem mass spectrometry fragmentation of m/z 292.15 (P1 and P2) and 375.23 (P4 and P5) isomers formed in Nicotiana benthamiana and in chemical assays.
Please note: Wiley is not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Acknowledgements
We acknowledge the support of the Max T. Rogers NMR Facility at Michigan State University for their assistance in collecting NMR spectra and thank Professor George Lomonossoff (John Innes Centre) for providing the pEAQ vectors. This research was supported by the National Science Foundation awards IOS‐1546617 and MCB‐1714093. MAB was supported in part by a research assistantship from the Plant Breeding, Genetics and Biotechnology Graduate Program at Michigan State University. CSB and ADJ are supported in part by Michigan AgBioResearch and through USDA National Institute of Food and Agriculture, Hatch project numbers MICL02552 and MICL02474. This publication was made possible by a Plant Biotechnology for Health and Sustainability predoctoral training award to HMP from Grant Number T32‐GM110523 from the National Institute of General Medical Sciences of the National Institutes of Health. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIGMS or NIH.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- Basha NJ, Basavarajaiah SM, Shyamsunder K. 2022. Therapeutic potential of pyrrole and pyrrolidine analogs: an update. Molecular Diversity 26: 2915–2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedewitz MA, Gongora‐Castillo E, Uebler JB, Gonzales‐Vigil E, Wiegert‐Rininger KE, Childs KL, Hamilton JP, Vaillancourt B, Yeo YS, Chappell J et al. 2014. A root‐expressed l‐phenylalanine:4‐hydroxyphenylpyruvate aminotransferase is required for tropane alkaloid biosynthesis in Atropa belladonna . Plant Cell 26: 3745–3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedewitz MA, Jones AD, D'Auria JC, Barry CS. 2018. Tropinone synthesis via an atypical polyketide synthase and P450‐mediated cyclization. Nature Communications 9: 5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair LM, Calvert MB, Sperry J. 2017. Flavoalkaloids‐isolation, biological activity, and total synthesis. The Alkaloids: Chemistry and Biology 77: 85–115. [DOI] [PubMed] [Google Scholar]
- Dieuleveux V, Lemarinier S, Gueguen M. 1998. Antimicrobial spectrum and target site of D‐3‐phenyllactic acid. International Journal of Food Microbiology 40: 177–183. [DOI] [PubMed] [Google Scholar]
- Dong Y, Burch‐Smith TM, Liu Y, Mamillapalli P, Dinesh‐Kumar SP. 2007. A ligation‐independent cloning tobacco rattle virus vector for high‐throughput virus‐induced gene silencing identifies roles for NbMADS4‐1 and ‐2 in floral development. Plant Physiology 145: 1161–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekanayaka EAP, Celiz MD, Jones AD. 2015. Relative mass defect filtering of mass spectra: a path to discovery of plant specialized metabolites. Plant Physiology 167: 1221–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiesel PD, Parks HM, Last RL, Barry CS. 2022. Fruity, sticky, stinky, spicy, bitter, addictive, and deadly: evolutionary signatures of metabolic complexity in the Solanaceae. Natural Product Reports 39: 1438–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanie S, Thodey K, Trenchard IJ, Interrante MF, Smolke CD. 2015. Complete biosynthesis of opioids in yeast. Science 349: 1095–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasebe F, Yuba H, Hashimoto T, Saito K, Funa N, Shoji T. 2021. CRISPR/Cas9‐mediated disruption of the PYRROLIDINE KETIDE SYNTHASE gene reduces the accumulation of tropane alkaloids in Atropa belladonna hairy roots. Bioscience Biotechnology and Biochemistry 85: 2404–2409. [DOI] [PubMed] [Google Scholar]
- Heim WG, Sykes KA, Hildreth SB, Sun J, Lu RH, Jelesko JG. 2007. Cloning and characterization of a Nicotiana tabacum methylputrescine oxidase transcript. Phytochemistry 68: 454–463. [DOI] [PubMed] [Google Scholar]
- Hibi N, Higashiguchi S, Hashimoto T, Yamada Y. 1994. Gene‐expression in tobacco low‐nicotine mutants. Plant Cell 6: 723–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang AC, Jiang T, Liu YX, Bai YC, Reed J, Qu BY, Goossens A, Nutzmann HW, Bai Y, Osbourn A. 2019. A specialized metabolic network selectively modulates Arabidopsis root microbiota. Science 364: eaau6389. [DOI] [PubMed] [Google Scholar]
- Huang J‐P, Fang C, Ma X, Wang L, Yang J, Luo J, Yan Y, Zhang Y, Huang S‐X. 2019. Tropane alkaloids biosynthesis involves an unusual type III polyketide synthase and non‐enzymatic condensation. Nature Communications 10: 4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey AJ, O'Hagan D. 2001. Tropane alkaloid biosynthesis. A century old problem unresolved. Natural Product Reports 18: 494–502. [DOI] [PubMed] [Google Scholar]
- Katoh A, Shoji T, Hashimoto T. 2007. Molecular cloning of N‐methylputrescine oxidase from tobacco. Plant and Cell Physiology 48: 550–554. [DOI] [PubMed] [Google Scholar]
- Keller MA, Piedrafita G, Ralser M. 2015. The widespread role of non‐enzymatic reactions in cellular metabolism. Current Opinion in Biotechnology 34: 153–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller MA, Turchyn AV, Ralser M. 2014. Non‐enzymatic glycolysis and pentose phosphate pathway‐like reactions in a plausible Archean ocean. Molecular Systems Biology 10: 725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N, Estrada O, Chavez B Jr, Stewart C, D'Auria JC. 2016. Tropane and granatane alkaloid biosynthesis: a systematic analysis. Molecules 21: 1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leete E. 1990. Recent developments in the biosynthesis of the tropane alkaloids. Planta Medica 56: 339–352. [DOI] [PubMed] [Google Scholar]
- Lerma‐Ortiz C, Jeffryes JG, Cooper AJL, Niehaus TD, Thamm AMK, Frelin O, Aunins T, Fiehn O, de Crecy‐Lagard V, Henry CS et al. 2016. ‘Nothing of chemistry disappears in biology’: the Top 30 damage‐prone endogenous metabolites. Biochemical Society Transactions 44: 961–971. [DOI] [PubMed] [Google Scholar]
- Li Petri G, Raimondi MV, Spano V, Holl R, Barraja P, Montalbano A. 2021. Pyrrolidine in drug discovery: a versatile scaffold for novel biologically active compounds. Topics in Current Chemistry 379: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichman BR. 2021. The scaffold‐forming steps of plant alkaloid biosynthesis. Natural Product Reports 38: 103–129. [DOI] [PubMed] [Google Scholar]
- Lin X, Ai W, Li M, Zhou X, Liao S, Wang J, Liu J, Yang B, Liu Y. 2020. Collacyclumines A‐D from the endophytic fungus Colletotrichum salsolae SCSIO 41021 isolated from the mangrove Kandelia candel . Phytochemistry 171: 112237. [DOI] [PubMed] [Google Scholar]
- Louveau T, Osbourn A. 2019. The sweet side of plant‐specialized metabolism. Cold Spring Harbor Perspectives in Biology 11: a034744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XY, Huang JP, Yang J, Liu XY, Hu XJ, Huang SX. 2020. Concise and efficient syntheses of methyl 4‐(1‐methylpyrrolidin‐2‐yl)‐3‐oxobutanoate and hygrine. Canadian Journal of Chemistry 98: 191–193. [Google Scholar]
- Mitreiter S, Gigolashvili T. 2021. Regulation of glucosinolate biosynthesis. Journal of Experimental Botany 72: 70–91. [DOI] [PubMed] [Google Scholar]
- Naconsie M, Kato K, Shoji T, Hashimoto T. 2014. Molecular evolution of N‐methylputrescine oxidase in tobacco. Plant and Cell Physiology 55: 436–444. [DOI] [PubMed] [Google Scholar]
- Oku H, Mutanda I, Fukuta M, Inafuku M. 2022. Non‐enzymatic formation of isoprene and 2‐methyl‐3‐buten‐2‐ol (2‐MBO) by manganese. Scientific Reports 12: 2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ping Y, Li X, Xu B, Wei W, Wei W, Kai G, Zhou Z, Xiao Y. 2019a. Building microbial hosts for heterologous production of N‐methylpyrrolinium. ACS Synthetic Biology 8: 257–263. [DOI] [PubMed] [Google Scholar]
- Ping Y, Li X, You W, Li G, Yang M, Wei W, Zhou Z, Xiao Y. 2019b. De novo production of the plant‐derived tropine and pseudotropine in yeast. ACS Synthetic Biology 8: 1257–1262. [DOI] [PubMed] [Google Scholar]
- Prandi C, Occhiato EG. 2019. From synthetic control to natural products: a focus on N‐heterocycles. Pest Management Science 75: 2385–2402. [DOI] [PubMed] [Google Scholar]
- Qiu F, Yang C, Yuan L, Xiang D, Lan X, Chen M, Liao Z. 2018. A phenylpyruvic acid reductase is required for biosynthesis of tropane alkaloids. Organic Letters 20: 7807–7810. [DOI] [PubMed] [Google Scholar]
- Qiu F, Zeng J, Wang J, Huang J‐P, Zhou W, Yang C, Lan X, Chen M, Huang S‐X, Kai G et al. 2020. Functional genomics analysis reveals two novel genes required for littorine biosynthesis. New Phytologist 225: 1906–1914. [DOI] [PubMed] [Google Scholar]
- Rajanikar RV, Nataraj BH, Naithani H, Ali SA, Panjagari NR, Behare PV. 2021. Phenyllactic acid: a green compound for food biopreservation. Food Control 128: 108184. [Google Scholar]
- Robins RJ, Woolley JG, Ansarin M, Eagles J, Goodfellow BJ. 1994. Phenyllactic acid but not tropic acid is an intermediate in the biosynthesis of tropane alkaloids in Datura and Brugansia transformed root cultures. Planta 194: 86–94. [Google Scholar]
- Roy R, Moreno N, Brockman SA, Kostanecki A, Zambre A, Holl C, Solhaug EM, Minami A, Snell‐Rood EC, Hampton M et al. 2022. Convergent evolution of a blood‐red nectar pigment in vertebrate‐pollinated flowers. Proceedings of the National Academy of Sciences, USA 119: e2114420119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadre R, Anthony TM, Grabar JM, Bedewitz MA, Jones AD, Barry CS. 2022. Metabolomics-guided discovery of cytochrome P450s involved in pseudotropine-dependent biosynthesis of modified tropane alkaloids. Nature Communications 13: 3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainsbury F, Thuenemann EC, Lomonossoff GP. 2009. pEAQ: versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnology Journal 7: 682–693. [DOI] [PubMed] [Google Scholar]
- Schlesinger D, Salama F, Davidovich Rikanati R, Sertchook R, Tal O, Yahyaa M, Faigenboim A, Ibdah M, Inbar M, Lewinsohn E. 2021. Further insights on the Datura innoxia hyoscyamine 6β‐hydroxylase (DiH6H) based on biochemical characterization and molecular modeling. American Journal of Plant Sciences 12: 53–70. [Google Scholar]
- Schliemann W, Kobayashi N, Strack D. 1999. The decisive step in betaxanthin biosynthesis is a spontaneous reaction. Plant Physiology 119: 1217–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen SM, Shen LG, Zhang J, Li GZ, Li ZY, Pan RL, Si JY. 2013. Emiline, a new alkaloid from the aerial parts of Emilia sonchifolia . Phytochemistry Letters 6: 467–470. [Google Scholar]
- Srinivasan P, Smolke CD. 2019. Engineering a microbial biosynthesis platform for de novo production of tropane alkaloids. Nature Communications 10: 3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan P, Smolke CD. 2020. Biosynthesis of medicinal tropane alkaloids in yeast. Nature 585: 614–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steimbach RR, Tihanyi G, Geraldy MNE, Wzorek A, Miller AK, Klika KD. 2021. Can an intermediate rate of nitrogen inversion affect drug efficacy? Symmetry 13: 1753. [Google Scholar]
- Sun J, Morita H, Chen G, Noguchi H, Abe I. 2012. Molecular cloning and characterization of copper amine oxidase from Huperzia serrata . Bioorganic & Medicinal Chemistry Letters 22: 5784–5790. [DOI] [PubMed] [Google Scholar]
- Tewksbury JJ, Nabhan GP. 2001. Seed dispersal – directed deterrence by capsaicin in chillies. Nature 412: 403–404. [DOI] [PubMed] [Google Scholar]
- Touchard A, Aili SR, Fox EGP, Escoubas P, Orivel J, Nicholson GM, Dejean A. 2016. The biochemical toxin arsenal from ant venoms. Toxins 8: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda M, Sasaki M, Mugishima T, Komatsu K, Sone T, Tanaka M, Mikami Y, Kobayashi J. 2005. Scalusamides A‐C, new pyrrolidine alkaloids from the marine‐derived fungus Penicillium citrinum . Journal of Natural Products 68: 273–276. [DOI] [PubMed] [Google Scholar]
- Voges M, Bai Y, Schulze‐Lefert P, Sattely ES. 2019. Plant‐derived coumarins shape the composition of an Arabidopsis synthetic root microbiome. Proceedings of the National Academy of Sciences, USA 116: 12558–12565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinhold A, Baldwin IT. 2011. Trichome‐derived O‐acyl sugars are a first meal for caterpillars that tags them for predation. Proceedings of the National Academy of Sciences, USA 108: 7855–7859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westfall PJ, Pitera DJ, Lenihan JR, Eng D, Woolard FX, Regentin R, Horning T, Tsuruta H, Melis DJ, Owens A et al. 2012. Production of amorphadiene in yeast, and its conversion to dihydroartemisinic acid, precursor to the antimalarial agent artemisinin. Proceedings of the National Academy of Sciences, USA 109: 111–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q‐X, He X‐F, Jiang C‐X, Zhang W, Shi Z‐N, Li H‐F, Zhu Y. 2018. Two novel bioactive sulfated guaiane sesquiterpenoid salt alkaloids from the aerial parts of Scorzonera divaricata . Fitoterapia 124: 113–119. [DOI] [PubMed] [Google Scholar]
- Zakaria MM, Stegemann T, Sievert C, Kruse LH, Kaltenegger E, Girreser U, Cicek SS, Nimtz M, Ober D. 2022. Insights into polyamine metabolism: homospermidine is double‐oxidized in two discrete steps by a single copper‐containing amine oxidase in pyrrolizidine alkaloid biosynthesis. Plant Cell 34: 2364–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Wang S, Yang J, Kang C, Huang L, Guo L. 2022. Glycosylation of plant secondary metabolites: regulating from chaos to harmony. Environmental and Experimental Botany 194: 104703. [Google Scholar]
- Zheng XQ, Cheng QX, Yao F, Wang XZ, Kong LX, Cao B, Xu M, Lin SJ, Deng ZX, Chooi YH et al. 2017. Biosynthesis of the pyrrolidine protein synthesis inhibitor anisomycin involves novel gene ensemble and cryptic biosynthetic steps. Proceedings of the National Academy of Sciences, USA 114: 4135–4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler J, Facchini PJ. 2008. Alkaloid biosynthesis: metabolism and trafficking. Annual Review of Plant Biology 59: 735–769. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Dataset S1 NMR spectra of pyrrolidine metabolites P1 and P2.
Fig. S1 Tandem mass spectra and proposed fragmentation of representative tropane and nortropane alkaloids.
Fig. S2 Proposed subnetwork of decorated aromatic tropane esters in Atropa belladonna root.
Fig. S3 Modified pyrrolidine alkaloids are decreased in phenylpyruvic acid reductase (PPAR)‐silenced Atropa belladonna roots.
Fig. S4 Abundance of phenyllactic acid and 2‐O‐malonylphenyllactate in PyKS‐silenced Atropa belladonna roots.
Fig. S5 Phenyllactic acid production through transient expression in Nicotiana benthamiana.
Fig. S6 Conserved enzymatic activity results in pyrrolidine alkaloid modifications in Nicotiana benthamiana.
Methods S1 Metabolite purification and structural characterization.
Table S1 Oligonucleotide primers used in the study.
Table S2 UPLC mobile phase gradient utilized for LC‐TOF‐MS analyses of pyrrolidine and tropane alkaloids.
Table S3 Metadata describing Progenesis and EZInfo processing of the AbPyKS vs TRV2 metabolomics experiment.
Table S4 UPLC mobile phase gradient utilized for LC‐TOF‐MS metabolite analysis.
Table S5 LC mobile phase gradient utilized for semi‐preparative separation of metabolite extracts of bulk PyKS‐silenced Atropa belladonna.
Table S6 LC isocratic mobile phase conditions utilized for semi‐preparative separation of P1 and P2 (m/z 292.15) isomers.
Table S7 List of ions from the S‐plot of the orthogonal projections to latent structures discriminant analysis analysis of PykS‐silenced Atropa belladonna roots compared with TRV2 empty vector controls.
Table S8 Liquid chromatography coupled with tandem mass spectrometry fragmentation of positive ions [M+H]+ that are decreased in PyKS‐silenced Atropa belladonna roots compared with TRV2 empty vector controls.
Table S9 Liquid chromatography coupled with tandem mass spectrometry fragmentation of positive ions [M+H]+ that are increased in PyKS‐silenced Atropa belladonna roots compared with TRV2 empty vector controls.
Table S10 Liquid chromatography coupled with tandem mass spectrometry fragmentation of m/z 292.15 (P1 and P2) isomers in negative mode [M−H]− (m/z 290.14).
Table S11 NMR chemical shift data for m/z 292.15 isomer 1 (P1).
Table S12 NMR chemical shift data for m/z 292.15 isomer 2 (P2).
Table S13 NMR chemical shift data for 2‐O‐malonylphenyllactate.
Table S14 Liquid chromatography coupled with tandem mass spectrometry fragmentation of m/z 292.15 (P1 and P2) and 375.23 (P4 and P5) isomers formed in Nicotiana benthamiana and in chemical assays.
Please note: Wiley is not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


