Abstract
Human Endogenous Retroviruses (HERVs) are viral sequences integrated into the human genome, resulting from the infection of human germ‐line cells by ancient exogenous retroviruses. Despite losing their replication and retrotransposition abilities, HERVs appear to have been co‐opted in human physiological functions while their aberrant expression is linked to human disease. The role of HERVs in multiple malignancies has been demonstrated, however, the extent to which HERV activation and expression participate in the development of cancer is not yet fully comprehended. In this review article, we discuss the presumed role of HERVs in carcinogenesis and their promising diagnostic and prognostic implications. Additionally, we explore recent data on the HERVs in cancer therapeutics, either through the manipulation of their expression, to induce antitumor innate immunity responses or as cancer immunotherapy targets. Finally, more precise and higher resolution high‐throughput sequencing approaches will further elucidate HERV participation in human physiological and pathological processes.
Keywords: cancer, evolution, HERVs in oncogenesis, HERV‐K (HML‐2), human endogenous retrovirus
1. INTRODUCTION
The role of infectious agents and most dominantly viruses is well‐established in the development of human malignancy. In fact, an estimated 12% of human cancers are linked to viral infections on a global scale, with this estimation even higher for the developing world. 1 Despite viruses being neither a necessary nor a sufficient condition for the initiation of the carcinogenic process, several mechanisms that lead to the “hallmarks of cancer,” including uncontrollable cell proliferation and immortalization, evasion of the host's growth‐inhibitory and immunity mechanisms, modification in energy metabolism, cellular invasion and metastasis as well as angiogenesis, can be virally facilitated. 2
Human Endogenous Retroviruses (HERVs) are viral sequences integrated into the human genome, resulting from multiple invasion events in human germ‐line cells by ancient exogenous retroviruses. 3 After these elements have been endogenized and reached fixation in the human genome, HERVs are vertically transmitted via Mendelian inheritance and are found scattered throughout the mammalian genome 3 (Figure 1). The only HERV family to integrate the human genome after the human‐chimpanzee divergence is HERV‐K (HML‐2), with these integrations occurring about 500 000 years ago. 4 Unlike endogenous retroviruses of other mammalian species that are capable of replication and reinfection, and have a clear oncogenic potential, 5 HERVs (with the probable exception of HERV‐K [HML‐2]) have become replication and retrotransposition defective, and their transcription is strictly regulated by epigenetic mechanisms. 6 Some HERVs seem to have been co‐opted in the hosts' physiological functions and their exaptation offered evolutionary advantages to the host, with HERV products present in healthy tissues 7 and aberrant HERV expression observed in human inflammation, autoimmunity, and malignancy. 8
Figure 1.
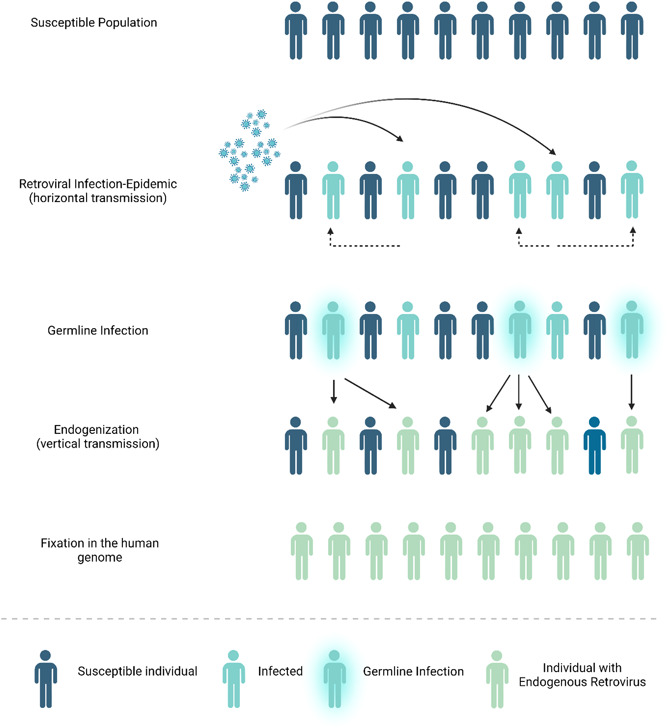
HERV life cycle. HERVS are viral sequences integrated into the human genome, resulting from multiple invasion events of ancient exogenous retroviruses, after their horizontal transmission and the occurrence of retroviral epidemics. Endogenization occurred after the infection of germ‐line cells and subsequent vertical transmission, and these elements either became extinct or reached fixation in the human genome, HERVs are vertically transmitted via Mendelian inheritance and are found scattered throughout the host's genome.
HERVs, human endogenous retroviruses.
These render HERVs important compartments in the shaping of human evolution, homeostasis preservation, and disease development. Despite these implications, their exact functions and roles in cancer development remain largely a terra‐incognita and a promising field of research regarding their pathophysiology in malignancy development and their potential utilization as therapeutic targets.
In this review, we aim to summarize recent data on the potential role of endogenous retroviruses in the development of cancer in humans. First, we describe their structure and their main expression control mechanisms, along with the physiological functions of their co‐opted products and the presumed HERV oncogenic mechanisms. Then, we describe their role in various human malignancies, and the involved mechanisms and discuss their potential functions as diagnostic and prognostic biomarkers in human cancer as well as their role in cancer therapeutics
2. HERVS: STRUCTURE AND EPIGENETIC CONTROL, CO‐OPTION, AND TUMORIGENIC IMPLICATIONS
The original structure of a HERV resembles the one of a simple replicable retrovirus. Long terminal repeats (LTRs) flank the proviral sequences which contain gag, pro, pol, and env genes, essential for its survival and preservation. LTRs serve a dual function, both as mediators for the proper retroviral integration and as regulatory elements, with promoter and enhancer function. 9 Accumulation of deleterious mutations, either nonsense or causing frameshifts, and recombination events leading to the creation of solitary LTRs, have eliminated the coding capacity of the majority of HERV sequences, while multiple transcription factors and epigenetic mechanisms control their expression. 6 DNA methylation and histone modifications, methylation, and deacetylation, are the main epigenetic mechanisms that regulate the expression of HERVs. 10 Interestingly, the age of the LTR integration is determining for the type of epigenetic control these elements are under, with evolutionarily young LTRs, with high CpG density being regulated through DNA methylation by DNA methyltransferases (DNMTs). 10 On the other hand, intermediate‐age LTRs are controlled epigenetically by histone lysine methyltransferases, and different LTRs appear to show tropism for particular methyltransferases. 10 Interestingly, evolutionarily old LTRs appear to be controlled epigenetically by histone lysine methyltransferases, while a major factor in their transcriptional inactivation is the accumulation of non‐sense mutations in their sequences. 10
HERVs have been preserved in the human genome and some of their products have been co‐opted for the survival of their hosts. The most striking example of HERV exaptation is the importance of syncytins in the successful placentation and the maternal immune tolerance to the fetus. 11 , 12 Syncytin‐1 and Syncytin‐2, the Envelope proteins of HERV‐W and HERV‐FRD respectively, are highly fusogenic glycoproteins that participate in the formation of syncytia and are responsible for the fusion of the villous trophoblast to the syncytiotrophoblast during early pregnancy. 13 Furthermore, Syncytins contribute to the protection of the developing fetus by inducing maternal immunosuppression during gestation, leading to the evasion of the paternal antigens from the maternal immune system 14 and more specifically, the highly preserved immunosuppressive domain of Syncytin‐2 is a potent immunosuppressive factor that contributes to this effect. 14
No conclusive findings point to HERVs as oncogenic, but numerous studies have demonstrated multiple mechanisms by which HERVs and their products participate in the development of the hallmarks of cancer.
The fusogenic properties of HERV envelope proteins, most prominently the syncytins, can promote mitotic effects, cellular invasion, and metastasis when they are expressed in an inappropriate setting. More specifically, Syncytin‐1 was found to enhance mitosis and the metastatic dynamics of hepatocellular carcinoma, 15 while its fusogenic features with cell‐cell adhesion increased the invasiveness in endometrial carcinoma 16 , 17 (Figure 2A). Furthermore, the immunosuppressive domain of Syncytin‐2 exerts its effect through the activation of mitogen‐activated protein kinase (MAP kinase) and causes T‐helper 1 (Th1) immunosuppression with the inhibition of Th1‐related cytokines. 18 These potent immunosuppressive properties of Syncytin‐2 have been linked to immune evasion and immunological tolerance of tumors. 18
Figure 2.
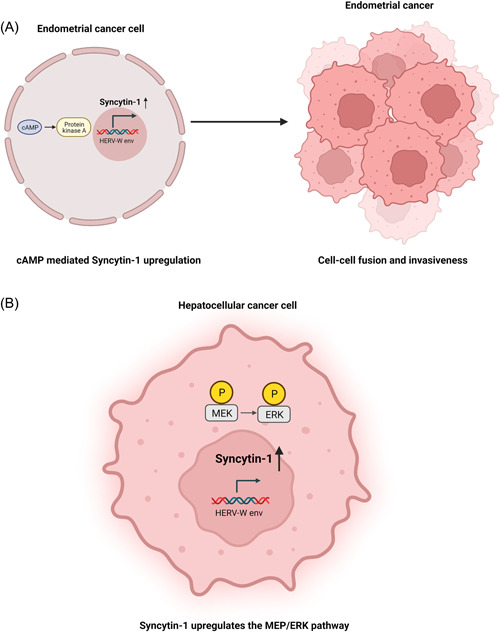
Oncogenic mechanisms related to Syncytin 1 expression: the examples of endometrial and hepatocellular carcinoma. (A) Upregulation of Syncytin‐1 mediated by the cAMP pathway leads to increased cellular fusion and thus increased invasiveness in endometrial carcinoma. (B) Syncytin‐1 leads to the development of malignant features in hepatocellular carcinoma cells through the activation of the MEK/ERK pathway and has been correlated to doxorubicin resistance. cAMP, cyclic adenosine monophosphate; ERK, extracellular‐signal regulated kinase; MEK, mitogen‐activated protein kinase. Created with BioRender.com.
Based on the presence of a 292nt long deletion at the pol‐env gene region or its lack, HERV‐K (HML‐2) is further divided into type I and II viruses, with their main features being the production of Env alternative transcripts Np9 and Rec respectively. 19 These alternativeenv splicing products, with not yet known physiological functions have been proposed as oncoproteins and have been found both in malignant and normal tissues. 20 Np9 can promote cell expansion, growth and survival by triggering multiple molecular pathways in leukemia stem/progenitor cells including the Wnt/β‐catenin, c‐myc/AKT, ras‐ERK, and Notch1 signaling path pathways leading to increased cell stemness in cancer. 21 , 22 Rec and Np9 exercise tumorigenic effects in spermatogonial stem cells, by leading to cellular proliferation and survival, as they interact with the Promyelocytic Leukemia Zinc Finger (PLZF) transcription repressor and prevent the repression of the MYC proto‐oncogene (NCBI Gene ID: 4609). 23 A summary of the mechanisms by which HML‐2 facilitates the development of the hallmarks of cancer and the progression of human malignancy can be found in Figure 3.
Figure 3.
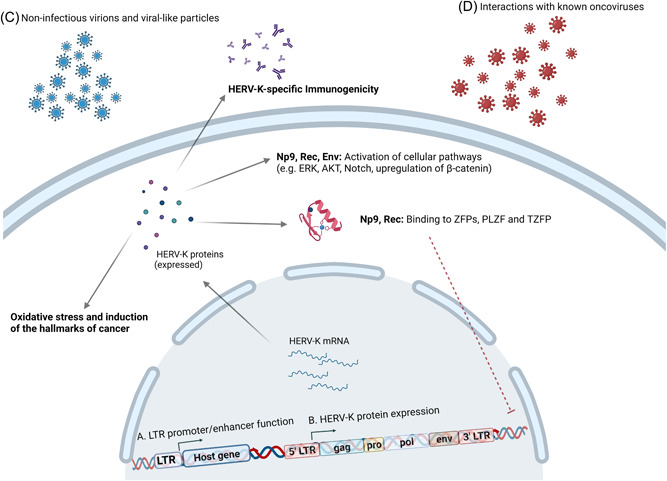
HERV‐K related oncogenesis. (A) LTRs through their promoter and enhancer functions modulate the expression of host genes and thus participate in human carcinogenesis. (B) HERV‐K protein expression appears to exert oncogenic functions including the induction of the hallmarks of cancer and increased cellular oxidative stress, the activation of cancer‐related molecular pathways like the ERK, AKT, and Notch pathway, and the upregulation of β‐catenin. Furthermore, Np9 and Rec seem to bind to ZFPs (PLZF and TZFP) and prevent them from repressing the c‐myc proto‐oncogene expression. Finally, HERV‐K specific immunogenicity has been correlated to multiple malignancies. (C) Presence of HERV‐K noninfectious virions and viral‐like particles have been correlated to human malignancy, most strikingly in the case of teratocarcinoma. (D) Transactivation of HERVs induced by known onco‐virus and intra‐viral interactions between HERV‐K and onco‐viruses have also been linked to the development of the hallmarks of cancer in multiple virally mediated cancers.
ERK, extracellular‐signal regulated kinase; HERV, human endogenous retrovirus; LTR, long terminal repeat; PLZF, promyelocytic leukemia zinc finger; TZFP, testicular zinc finger protein; ZFP, zinc finger protein. Created with BioRender.com.
Moreover, HERV LTRs participate in human carcinogenesis through the modification of the expression of host genes, through their promoter and enhancer functions. “Global hypomethylation” patterns are characteristic in malignant cells and are speculated to be vital for the promotion of the hallmarks of cancer, as they lead to the inappropriate transcriptomic release of multiple genomic sites, enabling HERV activation. 24 Two HERV‐K (HML‐2) proviruses (3q12.3 and 11p15.4) have been recognized to be more active as promoters in tumorigenic human mammary epithelial cells, and the polymorphic RORA binding site, with its more active form present in almost half of the population, on the 5'LTR of the polymorphic 11p5.4 HERV‐K (HML‐2) provirus was found to play a central role in carcinogenesis. 25
Reprogramming of mature cells towards stemness expression patterns that lead to features, such as pluripotency and self‐renewal potential, is essential in the maintenance of tumors and the heterogeneity observed among their cellular populations. 26 Such malignant cell populations, Cancer Stem Cells (CSCs), are often held responsible for increased tumor aggressiveness and treatment resistance. 26
HERV‐H, the most abundant HERV in the human genome, is highly activated during the early stages of embryonic development and is postulated to be able to swift mature cells towards pluripotency. 27 HERV‐H LTRs, and most prominently LTR7, are activated by multiple reprogramming transcription factors, including the Octamer‐binding Transcription Factor 4 (OCT4), Sex Determining Region Y‐box 2 (SOX2), and Krüppel‐like Factor 4 (KLF4/OSK) and their activation is ceased upon programmed differentiation. 27 In addition, stemness induction has also been correlated to the HERV‐K (HML‐2) Env protein, mediated by the upregulation of the mammalian target of rapamycin (mTOR) pathway and epigenetic modifications mediated by lysophosphatidylcholine acyltransferase (LPCAT1). 28 Additionally, Env inhibition has been linked to downregulation of the OCT‐4 pathway and thus reduction of stemness features and increased terminal differentiation towards neural cells. 28 Finally, a novel full‐length HERV envelope protein, HEMO [human endogenous MER34 (medium‐reiteration‐frequency‐family‐34) Open Reading Frame (ORF)] has been recognized as an effector of pluripotency and stemness, and is actively excreted in the circulation of pregnant women, as well as highly expressed in multiple tumors. 29
3. HERV PARTICIPATION IN MALIGNANCIES AND CLINICAL IMPLICATIONS
In this part of our manuscript, we aim to discuss recent data on the participation of HERVs in specific human malignancies, examine the mechanisms that are employed per case, and review the implication of HERVs in these tumors regarding clinal aspects of the disease, such as tumor behavior, prognosis, and potential therapeutic targets.
3.1. Breast cancer
The potential pathogenic role of HERVs, and more importantly HERV‐K (HML‐2), in the development of breast cancer is one of the most extensively studied in malignancy concerning transposable elements. HERV‐K (HML‐2) mRNA and antibodies against its proteins, most importantly Rec, are elevated in serum and HERV‐K (HML‐2) env expression is described in breast cancer tissue of patients with in situ and stage I ductal carcinoma and was particularly increased in patients that developed metastatic disease, suggesting potential prognostic implications. 30
Interestingly, knockdown of HERV‐K (HML‐2) env in breast cancer cells led to reduced expression of oncogenic proteins, by affecting the Ras/p‐RSK/p‐ERK pathways, while its restoration leads to the re‐establishment of the aforementioned pathways, the downregulation of p53 proapoptotic protein and the upregulation of the Transforming Growth Factor‐β1 (TGF‐β1), a crucial protein for epithelial‐to‐mesenchymal transition. 31 Contrary to the healthy epithelium, stimulation of T‐47D (RRID:CVCL_0553) breast cancer cell‐line with female sex hormones leads to the activation of HERV‐K env and pol genes through their respective hormonal nuclear receptors. 32 Also, the octamer motif, a transcription factor binding site, located in the LTR, through its interaction with these hormonal signals modulates the promoter/enhancer function of the LTR5H with regard to downstream genes. 32 This effect of estradiol and progesterone on LTR5H behavior is mediated through the interaction of mainly the Progesterone Receptor and the OCT4 motif in the LTR region. 32 Since hormonal stimulation is a well‐established contributor in breast cancer, its tumorigenic effect could be HERV‐mediated at least to some extent. Additionally, HERV‐K Env is highly expressed in basal‐like invasive ductal carcinoma and HERV‐K expression was correlated to the presence of the wild‐type H‐Ras protein upregulation, which promotes cell proliferation, and inactivation of the Retinoblastoma protein through phosphorylation. 33 Finally, a long‐noncoding RNA (lncRNA) of endogenous retroviral origin that includes the complete LTR70 and sequences from LTR56 and MER67C, named TROJAN (GenBank: AK124454), is highly expressed in Triple Negative Breast Cancer (TNBC), one particularly aggressive and challenging to treat type of breast cancer. 34 TROJAN increases the proliferative and metastatic potential in TNBC and its effects were reversed upon the deletion of the TROJAN sequence, as well as after the administration of antisense oligonucleotides in mice. 34 TROJAN exercises its malignant potential by binding to the antitumor Zinc Finger‐MYND‐type‐containing 8 protein and leads to its degradation through ubiquitination. 34 In the case of basal‐like ductal carcinoma and TNBR, HERVs seem appealing therapeutic targets, since both are aggressive malignancies with limited options, due to the lack of estradiol, progesterone and Human Epidermal Growth Factor 2 (HER2) receptors on both of these tumor types.
Finally, HERVs were shown to be potential targets for therapeutic interventions against cancer. Released HERV expression via decreased H3K27me3 histone modification indicates responsiveness to trastuzumab, a monoclonal antibody against HER2, administered in HER‐positive malignancies, and resistance to trastuzumab was correlated to elevated levels of this histone modification. 35 This effect is attributed to the type‐I interferon (IFN) rich tumor environment resulting from the presence of HERVs. 35 Reversal of HERV silencing due to increased H3K27me3 has been proposed as a strategy to circumvent trastuzumab resistance and reduce immune evasion in HER‐positive breast cancer, utilizing antiviral responses against HERVs. 35 Additionally, chimeric antigen receptor (CAR) T‐cell immunotherapy appears to have promising applications in targeting HERV expression on tumors. More specifically, CAR T‐cells targeting HERV‐K Env have been shown to trigger robust inflammatory responses against breast cancer cells and not MCF‐10A (RRID:CVCL_0598) healthy breast cells in cell cultures, while in xenograft models in mice, CAR T‐cells against HERV‐K Env led to significantly reduced breast tumor size and metastatic potential which was correlated with increased p53 expression and reduced p‐ERK and Ras pathway induction. 36
3.2. Teratocarcinoma and testicular cancer
Along with the implication of HERVs in mammary carcinomas, one of the first correlations between cancer and HERVs regarded teratocarcinoma, as HERV‐K (HML‐2) mRNA, proteins, noninfectious virions were identified and HERV‐K (HML‐2) is capable to trigger immune responses in teratocarcinoma tissues. 37 In Tera‐1 (RRID:CVCL_2776) cell line, a distinct proviral profile has been reported with the prevalence of human‐specific evolutionarily young proviruses, forming noninfectious virions, and the transcription of HERV‐K (HML‐2) proviruses correlated to the promoter activity of their corresponding LTRs, with younger LTR5H demonstrating the highest activity. 37 Additionally, gag transcripts displayed the highest abundancy, and interestingly, the majority of the expressed proviruses in this cell line belonged to type I HERV‐K (HML‐2). 37 Similarly, in the NCCIT (RRID:CVCL_1451) teratocarcinoma cells, Np9 contributes to the survival and migration potential of cells and its Np9 loss with CRISPR/Cas manipulation, although not sufficient to eliminate these characteristics, significantly reduced the metastatic potential of these cells and led to their increased vulnerability to stressors like cellular starvation and chemotherapeutics. 19 Hence, targeting Np9 could be a useful adjuvant treatment to classical pharmacological approaches.
Multiple HERV families have been involved in the development of testicular carcinomas. Testicular Zinc Finger Protein (TZFP) is a protein of little understood function, which appears to be a transcriptional regulator during spermatogenesis, and represses the Androgen Receptor (AR) (NCBI Gene ID: 367) and C‐myc oncoprotein. 38 HERV‐K (HML‐2) accessory protein Rec binds to TZFP and leads to the release of AR and MYC gene expression in testicular cancer, while HERV‐K (HML‐2) full‐length Rec‐transfected mice were prone to in situ testicular cancer. 38 Activation of HERV‐W loci mediated by LTR hypomethylation has also been described in testicular cancer. 39
On the contrary, HERV‐9 LTR12 is a promoter for the expression of a proapoptotic isoform of p63 protein (GTAp63) in male germ‐line cells and is silenced in testicular tumors, which led to the assumption of the evolutionary preservation of such integration to ensure the host's fitness and reproductive ability. 40 Interestingly, Histone deacetylase (HDAC) inhibitors, were found at least partially to owe their antitumor proapoptotic effects in testicular cancer to the upregulation of GTAp63 by reestablishing the transcription of HERV‐9 LTR12 promoter function. 40 Finally, in a study where patients with multiple types of germ‐cell tumors were included persistence of antibodies against HERV‐K Gag and Env after the administration of chemotherapy may be predictive of residual or metastatic disease. 41
3.3. Epithelial ovarian cancer
HERV activity was correlated to Epithelial ovarian cancer (EOC), as increased levels of HERV‐K Env are described in ovarian cancer tissue compared to healthy ovarian tissues from the same patients and benign ovarian tumors, and increased reverse transcriptase activity was noted in these tissues and the plasma of the patients. 42 Additionally, HERV‐K hypomethylation was correlated to resistance to platinum and reduced progression‐free and overall survival in patients with ovarian clear cell carcinoma (OCCC). 43
Most importantly, HERV‐K Env is immunogenic in EOC patients, as elevated levels of HERV‐K Env‐specific antibodies were found in this patient group and HERV‐K Env can elicit adaptive immune responses leading to increased specificity T‐cells against EOC tissue. 42 An ovarian cancer prognosis score was designed, based on the expression of multiple HERV families, and increased scores are indicative of a favorable prognosis, even in the case of high‐grade malignancy. 44 In fact, HERV expression provokes increased infiltration of cytotoxic CD8 + PD1 + T‐cells, and acts preventatively against the immune evasion of malignant cells, and based on this observation it was postulated that at least to some extent the DNA‐methylation manipulation through DNMT‐inhibitors may be useful in antitumor immune activation. 44
3.4. Endometrial carcinoma
HERVs and more particularly their Env proteins seem to play a role in the progression and invasiveness of endometrial carcinoma. Differential expression of multiple HERV env genes has been shown in endometrial cancer and Syncytin‐1 and ‐2 expression increased with advanced endometrial cancer stage, while both syncytins and ERV‐3 expression demonstrated specificity for glandular carcinoma histology. 45 Syncytin‐1 expression is positively correlated to biomarkers of endometrial cancer including the Estrogen Receptor and proliferation biomarker Ki‐67, as well as overall survival. 17 Finally, Syncytin‐1 induces cell‐cell adhesion, cellular proliferation, epithelial‐to‐mesenchymal transition, invasion and migration in endometrial cancer. 16 , 17
3.5. Prostate cancer
HERVs both at an mRNA and protein level have been proposed as diagnostic and prognostic biomarkers in prostate cancer. HERV‐K Gag protein demonstrates increased expression, mediated both by the demethylation of its promoter and also by hormonal activation via androgen stimulation in prostate cancer cell lines. 46 Also, Gag upregulation appears to be specific for malignant prostate tissue, compared to benign neoplasia and normal prostate tissues, 46 , 47 and has been suggested as a useful immunohistochemistry biomarker for prostate cancer diagnosis. 47 Furthermore, specific antibodies against immunogenic epitopes of the HERV‐K and HERV‐H Env proteins have been recognized, providing further potential diagnostic biomarkers. 48 Interestingly, increases in the specific antibodies targeting HERV‐K Gag protein in prostate cancer patient plasma are correlated to increased prostate cancer mortality, possibly due to increased antigenic stimulation in the frames of increased disease burden. 46
3.6. Urinary tract carcinoma
Aberrant expression of HERVs has also been described in urinary tract malignancies. Syncytin‐1 upregulation is more frequently encountered in tissues of bladder urothelial cancer compared to healthy tissues of the affected organ, while Syncytin‐1 overexpressing tumors were more aggressive. 49 This elevation in Syncytin‐1 expression was correlated to increased HERV‐W 3' LTR promoter activity due to point mutations in its sequences. 49 Hence, the detection of these 3' LTR mutations of HERV‐W could serve as prognostic indicators of more aggressive disease behavior.
An increasing number of reports underline the role of HERV expression in clear‐cell Renal Cell Carcinoma (ccRCC) with important therapeutic implications, as HERV‐E‐derived antigens appear to be promising targets for immunotherapy. Increased expression of potentially immunogenic HERVs was correlated to gene expression patterns indicative of immune check‐point activation, especially in the case of ccRCC, and these HERVs include members of the HERV‐K family, ERV‐3‐2 and HERV‐E. 50 , 51 Three HERV‐E transcripts were identified in ccRCC tissues in contrast to healthy renal tissue and other types of tumors, one of which is found to encode the whole sequence of the env gene and evidence is suggestive of HERV‐E‐derived peptides on the surface of ccRCC cells, that appear capable to elicit potent CD8 + T‐cell responses. 51 Interestingly, the expression of HERV‐4700 (HERVERI‐gammaretrovirus), which was ccRCC‐specific, demonstrates high translational potential and data suggest the presence of anti‐HERV‐4700‐specific CD8 + T cells. 52 Additionally, the presence of HERV‐4700 epitopes on ccRCC cells was predictive of response to anti‐PD1 treatments, 52 but this was not verified in all cohorts. 53 Nevertheless, the implication of HERVs in immune responses in the setting of malignancy and their potential role as therapeutic targets is a promising study field that warrants further elucidation. Notably, decitabine, a DNA demethylating agent, facilitates responses to immune checkpoint blockade in kidney cancer cell lines by releasing the expression of transposable elements, including HERV‐4700. 54 This was postulated to enhance viral mimicry on cancer cells, leading to innate immunity antiviral responses and this pro‐inflammatory cytokine microenvironment leads to specific T‐cellular responses in the treated cells. 54
Finally, recent data suggest that HERV‐K Env is upregulated on the surface of ccRCC cells in comparison to healthy kidney tissue and higher levels of HERV‐K Env in ccRCC cells were correlated to better survival outcomes in patients with ccRCC, indicating the potential function of HERV‐K Env as an immunogenic tumor‐antigen. 55
3.7. Gastrointestinal cancer
Aberrant HERV expression, and more particularly deregulation of the expression of the HERV‐K viral genes env, np9, rec, and gag has been recognized both in gastric and colon cancer. 56
The majority of the available evidence on HERV participation in gastrointestinal malignancies regards colorectal cancer. Upregulation of HERV‐H transcripts including HERV‐H noncoding spliced transcripts is present in colorectal cancer samples and increased HERV‐H activity was shown in malignant regions, in contrast to healthy adjacent tissues. 57 Interestingly, young HERV‐H integrations were the most intensely upregulated loci throughout the stages of colon cancer. 58 HERV‐H expression was correlated with lymph node involvement and microsatellite instability, a phenomenon characterized by impaired mismatch mutation repair mechanisms, leading to tumor hypermutability and indicative of higher tumor immunogenicity, lymphocyte tumor infiltration and better prognosis. 58 Additionally, HERV‐Kenv expression has been linked to tumorigenic characteristics in the DLD‐1 (RRID:CVCL_0248) cell line and its knockout was linked to the reduction of the proliferative and metastatic potential of these cells, while evidence suggests that the tumorigenic properties of the HERV‐K env gene are linked to increased oxidative stress burden. 59 However, these findings were not confirmed in colon cancer tissues. 60 , 61
HERV LTRs demonstrated increased hypomethylation in colon cancer tissues compared to healthy colon, leading to HERV‐H, HERV‐K, and HERV‐P transcriptional release. 60 Interestingly, right‐sided localization of colon cancer correlated to significantly increased HERV‐P and HERV‐R expression compared to left‐sided tumors, indicating a different HERV‐mediated malignancy profile, compatible with the different molecular patterns observed per colon cancer localization. 60 Increased transcription of HERV‐W‐1, HERV‐FRD, ERV‐3‐1 (a completeenv sequence belonging to the HERV‐R family) and HERV‐V‐1 has been shown after the induction of resistance to etoposide in colon cancer cells. 62 Moreover, increased Syncytin‐1 expression was linked to shortened overall survival in rectal, but did not demonstrate the same effect in epithelial cancers of the colon.
Taken together, all these data indicate an important role of HERV expression in colon cancer development, expansion and treatment resistance and possibly HERVs may serve as targets of ancillary treatment to enhance sensitivity to common chemotherapeutics, in the cases where the prognosis is guarded.
3.8. Hepatocellular carcinoma
HERV expression is implicated mainly in disease progression and the prognosis of Hepatocellular Carcinoma (HCC). Syncytin‐1 is upregulated in HCC tissues, while higher expression levels are independently correlated with higher tumor burden and worse outcomes. 15 Syncytin‐1 enhances the malignant dynamics of HCC through the phosphorylation of the mitogen‐activated protein kinase (MEK)/ERK pathway and its expression predisposes to doxorubicin resistance 15 (Figure 2B). Additionally, HERV‐K (HML‐2) expression levels have been suggested as a novel prognostic biomarker for HCC overall survival, with poorer outcomes in HCC patients with increased HERV‐K (HML‐2) transcription in HCC tissues. 63
Intriguingly, HERV transactivation by Hepatitis B and C Viruses (HBV and HCV, respectively) is another potential manner in which HERVs take part in HCC development. Notably, higher levels of Syncytin‐1 are found in HCC tissues of Hepatitis B surface antigen positive individuals compared to negative ones 15 and Hepatitis B virus oncoprotein X (HbX) leads to increased levels of spliced and non‐spliced transcripts of syncytin‐1 gene and increased concentrations of Syncytin‐1 protein in Hep‐G2 (RRID:CVCL_0027) cells. 64 Finally, HCV upregulates the transcription of HERV‐H and HERV‐Kpol genes compared to controls, which remain elevated even after successful treatment and viral clearance. 65 This finding offers a plausible explanation for the maintainance of increased HCC rates even after successful HCV clearance. 66
3.9. Pancreatic cancer
Evidence on HERV implication in pancreatic cancer is limited, but it suggests that increased activation of full‐length HERV‐K Env is present in pancreatic tissues and cancer cell lines. 67 Additionally, increased reverse transcriptase activity was observed, and viral‐like particles were excreted from pancreatic cancer cell lines. 67 HERV‐K was shown to be immunogenic in pancreatic cancer patients as they demonstrated increased antibody titers against HERV‐K Env and Np9. 67 HERV‐K Env appears to facilitate cell proliferation and metastasis. 67 Interestingly, in pancreatic cancer cells, one novel splice variant of the HERV‐K env gene was identified (GenBank: AY037928.1). 67
3.10. Lung cancer
Metagenomic analysis of the virome in the blood and nonsmall cell lung cancer tissues has revealed an abundant HERV presence. 68 Significantly increased levels of HERV‐K, HERV‐H, HERV‐P, and HERV‐R env mRNAs have been recognized in lung cancer tissues compared to healthy controls. 69 Adenocarcinoma tissues demonstrated the most prominent increases compared to other lung cancer histological types, and HERV‐K, HERV‐H, and HERV‐P levels were distinctive for lung adenocarcinoma compared to squamous and small‐cell lung cancer. 69
Aside from the potential of HERVs as diagnostic and differentiating biomarkers in lung cancer, MER4 has been proposed as a biomarker of responsiveness to immune check‐point treatment in non‐small cell lung cancer, as increased expression of MER4 was associated with better response to these treatments and higher immune infiltration in these tumors, leading to lower proliferation and malignant potential. 70
3.11. Glioblastoma multiforme
HERV contribution to inflammation and degeneration in the central nervous system (CNS) has long been recognized. Recent data suggest that HERVs may serve as a valuable treatment target of Glioblastoma Multiforme (GBM), an especially challenging to treat malignancy and the most common CNS malignancy in adults. In fact, LTRs of multiple HERV families, in their vast majority belonging to the ERV‐1 superfamily, demonstrate an upregulation in the GBM compared to healthy brain tissues, in a specific manner, which renders these elements promising targets. 71 Interestingly, HERV‐K, the family with the widest implication in human pathology, demonstrated the greatest downregulation in GBM tissues, 71 and these findings are in line with previous results, where HERV‐K was not found to be a suitable biomarker and target for GBM. 72
Additionally, multiple HERV families were found dysregulated in the GBM highly proliferative A‐172 (RRID:CVCL_0131) cell line, including the upregulated HERV‐K (HML‐6) and ERV3 superfamily and the downregulated ERV1 and PRIMA superfamilies. 73 Most strikingly, significant hypomethylation at the HERV‐K (HML‐6) locus 19q43b was correlated with this increase in HERV‐K (HML‐6) expression. 73 Interestingly, HERV‐K (HML‐6) contains an intact ORF for a small Env protein ERVK3‐1, increased expression of which was recognized in the A‐172 cell line and GBM tissue samples and its presence was correlated with reduced survival, independent of the presence isocitrate dehydrogenases (IDH) mutations in GBM tissues, 73 a significant prognostic biomarker in GBM. 74 Thus, HERV‐K (HML‐6) and mainly ERVK3‐1 expression may serve as a prognosticator and treatment target for GBM.
3.12. Melanoma
Overexpression of HERV‐K (HML‐2) has been associated with the development of melanoma and HERV‐K (HML‐2) has been implied as a potential diagnostic and therapeutic target. More specifically, upregulated transcription of HERV‐K (HML‐2) loci has been shown in melanoma cells compared to normal melanocytes and these loci often include ORFs for the expression of HERV‐K (HML‐2) Gag, Env, Np9, and Rec retroviral proteins. 75 Reverse transcription of spliced HERV‐K (HML‐2) transcripts has also been shown in malignant cells, either employing the L1 machinery of Long Interspersed Nuclear Elements (LINEs) or the traditional retroviral reverse transcription. 75 ERK‐6 locus on 7p22.1 [HERV‐K(HML‐2.HOM)] presents particular interest and is a promising diagnostic biomarker since its transcription was found to be melanoma‐specific and lack thereof characterized healthy melanocytes. 75 Transcriptional balance of the HERV‐K (HML‐2) proteins seems to be an important regulator between the proliferative and the invasive state of melanoma, and the Melanocyte Inducing Transcription Factor (MIFT) plays an important part probably through its binding to the HERV‐K (HML‐2) LTR5H. 76 In fact, the binding of MIFT to the LTR5H leads to the upregulation of Rec expression and enhances the proliferative behavior in melanoma cells, while at the same time Rec knock‐down cells demonstrated a reduced transcription of MIFT, upregulation of Env and Gag expression, increased epithelial‐to‐mesenchymal transition and development of invasive behavior and metastatic features. 76 Additionally, significant Env presence was found in melanoma tissue samples compared to other normal control tissues and CAR T‐cells targeting HERV‐K Env demonstrated promising results in a mouse xenograft model. 77
Transcriptomic release of transposable elements in melanoma tissues has also been demonstrated with the decreased levels of CpG methylation levels of HERV‐K and LINE‐1 elements in melanoma cells compared to healthy melanocytes of non‐malignant nevi. 78 Intriguingly, significant correlations occurred only between the levels of CpG methylation levels of HERV‐K elements and clinical parameters. More specifically, shorter relapse‐free survival and worse disease prognosis were linked to lower methylation levels and higher expression of HERV‐K elements, independently of age and gender. 78 In agreement with these findings, HERV‐K expression appears to be non‐redundant for the development of the CD133 antigen on melanoma cells in melanoma cell lines, a marker tied to stemness features in melanoma cells, including increased proliferation, metastatic characteristics, and drug resistance. 79
3.13. Leukemia
The participation of HERVs in the development and progression of leukemias has been suggested by various studies. In Acute Myeloid Leukemia (AML), ERVs demonstrate enhancer function for genes related to cellular proliferation and growth. In fact, ERV deregulated expression, mostly for LTR2 elements was shown to modify the expression of adjacent genes, such as Zinc Finger Protein 611 (ZFP611) and apolipoprotein C1/2/4‐2 (APOC1/2/4‐2) and E (APOE). 80 Most strikingly, APOC1 and APOE expression was correlated to cellular death resistance in AML and to adverse prognosis with lower overall survival rates. 80 Deletion of LTR2 elements leads to reduced expression of the aforementioned genes, consequently, leading to cellular proliferation restriction and increased apoptosis of malignant cells. 80 Thus, these elements could be important targets for gene expression manipulation in the therapeutics of cancer.
Additionally, the expression of ERV‐3‐1 is upregulated in the blood and bone marrow cells of patients with AML, and this upregulation was more prominent in the myeloid lineage compared to the monocytic one. 81 More importantly, the presence of the ERV‐3‐1 protein was confirmed on the surface of leukemic cells of all myeloid‐phenotype AML patients and the majority of monocytic‐phenotype patients in immunohistochemical studies. 81 ERV‐3‐1 upregulated expression was correlated to chromosome 8 trisomy leukemic genotype, however, this correlation was not significant in the immunohistochemical study for the identification of the ERV‐3‐1 protein on leukemic cells. 81 Deregulated HERV expression patterns are recognized in multiple AML differentiation types. Significant upregulation of the expression of HERV‐E and concurrent downregulation of ERV‐3‐1 were found in AML with erythroid and megakaryocytic differentiation, while significant downregulation of HERV‐K (HML‐2) was observed in myeloid and monocytic AML cells. 82 These findings suggest that a more detailed analysis of the HERV signatures in each AML phenotype may aid as a biomarker in cases where diagnostic dilemmas are imposed.
HERV participation has also been described in Chronic lymphocytic leukemia (CLL). Increased transcription of the HERV‐K np9 and gag genes was found in B‐cell CLL patients, with the increases in the gag gene expression being more modest and not reaching statistical significance. 83 Nevertheless, such implications of HERV‐K have long been discussed in CLL, as Peripheral Blood Mononuclear Cells (PBMCs) of CLL patients demonstrated HERV‐K‐10‐likegag gene upregulation compared to those from healthy donors 84 and both of these genes are postulated to have oncogenic capacity and may be promising treatment targets.
Interestingly, despite the positive correlations between the level of HERV expression and increased age in humans 85 and the limited number of studies on HERV expression in pediatric diseases, associations between HERV expression and leukemias have also been demonstrated in pediatric populations. Higher HERV‐K expression was found in pediatric leukemia patients compared to healthy age‐matched controls, especially regarding the upregulation of the pol gene expression in Acute Lymphoblastic Leukemia (ALL) patients. 86 Additionally to ALL, other study groups have demonstrated the role of the expression of the HERV‐K env in pediatric myeloid malignancies. 87
3.14. Lymphomas
Hodgkin Lymphoma (HL), a mature B‐cell malignancy, has been correlated to the downregulation in HL cells of ERV‐3, a HERV with speculated antitumor properties such as reduced cell growth and expansion. 88 Interestingly, HL cells treatment with the histone deacetylation inhibitor, vorinostat, and hypoxia mimetic agents leads to increases in the expression of ERV‐3 accompanied by increased cellular apoptosis. 88 Additionally, HL is characterized by the ectopic expression of the Colony Stimulating Factor‐1 Receptor (CSF1‐R) on B‐cells, which leads to the CSF1/CSF1‐R binding and consequently tumor growth. 89 The LTR of the “mammalian apparent LTR retrotransposon” (MaLR) THE1 family acts as a promoter for this ectopic expression of CSF1‐R on B‐cells in HL and similar effects have been shown in aplastic large cell lymphoma, possibly through the demethylation of THE1 LTR. 89
Regarding non‐Hodgkin Lymphomas, Diffuse Large B‐cell Lymphoma, the most common malignancy of this type, demonstrates upregulation of the gag gene with relatively intact ORFs, belonging to the HERV‐V family (gagV1), which is a promising biomarker. 90 Conversely, Mycosis Fungoides (MF), the most common histological type of primary cutaneous T‐cell lymphoma, demonstrates significantly increased transcription levels of HERV‐W sequences compared to healthy skin tissues of the same patient, and mainly stemming from HERV‐W loci on chromosome 6q21 and 7q21.2. 91 MF skin immunohistochemistry studies revealed the presence of syncytin‐1 encoded by the HERV‐W locus at chromosome 7q21.2, and its presence was recognized as specific on the surface of malignant T‐cells. 91 Interestingly, lack of Syncytin‐1 expression was found on non‐malignant T‐cells located in the skin due to local inflammation due to Lichen Ruber Planus, indicating that its expression is not indicative of a general inflammation state but is rather lymphoma‐specific. 91
In the case of the participation of HERVs in lymphoma development, we should also consider the interaction between HERVs and known oncogenic viruses implicated in the development of lymphomas, notably Human T‐lymphotropic Virus‐1 (HTLV‐1) and Epstein‐Barr Virus (EBV). One of the main oncogenic mechanisms by which HTLV‐1 infection leads to Adult T cell lymphoma is through the oncogenic function of the Tax protein. 92 Interestingly, among other genomic sites, the transcription of which is modified by Tax, Tax modulates the expression of HERV‐W LTRs in Jurkat cells (RRID:CVCL_0065), possibly via the cAMP response element‐binding protein (CREB) and the NF‐κB pathways. 92 It is reasonable to assume that, among other mechanisms, this deregulation of HERV elements mediates the modified gene expression that characterizes the development of malignancy in HTLV‐1 infection. 92 Similarly, EBV infection is a key player in the development of B‐cell lymphomas and most importantly, the Diffuse Large B‐cell Lymphoma. 93 Activation of LTR promoter and enhancers is shown in EBV‐transformed primary B‐cells, and this retroelement activation has been attributed to both DNA methylation modification by the EBV infection, and also, to the binding of EBV‐derived transcription factors, such as EBV Nuclear Antigen 2 (EBNA2). 93
3.15. Multiple myeloma
Currently, evidence on the role and the expression patterns of HERVs in multiple myeloma is limited. Increased expression of HERV‐Fc has been described in multiple myeloma and plasma‐cell leukemia compared to controls. 82
Intriguingly, two endogenous retroviral loci, one belonging to the HERV‐F1 family located on chromosome X and the other belonging to the HERV‐K family, on chromosome 1, appear to act synergistically in the development and progression of multiple myeloma, through either complementation or even through recombination. 94 In fact, the presence of almost complete gag and env genes of HERV‐Fc1 locus and the pol gene of HERV‐K could potentially lead to the creation of a recombinant replication capable virus, which can contribute positively to the development of plasma‐cell dyscrasias and, thus, multiple myeloma. 94
3.16. Kaposi's sarcoma and primary effusion lymphoma
Despite the distinct histological origins and pathological features of Kaposi's sarcoma (KS) and Primary Effusion Lymphoma (PEL), we study these tumors together due to their aetiological association with Kaposi's Sarcoma Herpesvirus (KSHV) and we describe the role of the HERV‐KSHV interactions in the pathogenetic pathways of these cancers.
Recent in vitro and in vivo data suggest that de novo infection and latency with KSHV, mediated by the Latency‐associated nuclear antigen (LANA, ORF73) and viral FADD‐like interferon converting enzyme inhibitory protein (vFLIP, ORF71), lead to the transactivation of HERV‐K. 95 This endogenous retroviral transactivation appears to be mediated both by intracellular pathways, including the MAPK and the NF‐κB pathway as well as by transcription factors binding at HERV‐K LTRs, like transcription factor Sp1. 95 Furthermore, HERV‐K Env has been linked to the development of invasive characteristics in primary Human Umbilical Vein Endothelial Cells (HUVEC) and its knockdown reversed this effect, while the expression of the Vascular Endothelial Growth Factor (VEGF) and its receptor VEGFR1 was also diminished. 95 Finally, Np9 seems to be the most important endogenous retroviral oncogenic protein participating in the development of KS, as KSHV de novo infection induces a significant increase in np9 transcription in HUVEC, Np9 was found to upregulate oncogenic pathways such as AKT, ERK and Notch‐1 pathways and increase β‐catenin expression, and Np9 expression was significantly increased in KS samples compared to normal skin tissue and latently infected Telomerase‐immortalized Endothelial Cells (TIVE). 95
Regarding the role of HERV‐KSHV interactions in the development of PEL, recent data suggest the dysregulation of multiple HERV families. 96 Strikingly, similarly to KS, HERV‐K expression was found upregulated in PEL cells and PBMCs from KSHV infected HIV‐positive persons compared to KSHV uninfected HIV‐positive ones. 95
4. CONCLUSION
The role of HERVs in multiple malignancies has been demonstrated and even though the extent to which HERV activation and expression affect the development of cancer in humans is not yet totally comprehended, it is evident that HERVs seem to be important players in human disease and not just evolutionary fossils as they have been once perceived. Multiple studies have shown their promising perspectives as diagnostic and prognostic biomarkers in various cancer types. Aside from the role of the expression of HERV proteins during the early stages of cancer development and throughout the disease course, where these proteins serve a presumably carcinogenic role, recent advances in the field of cancer therapeutics shed light on a novel and promising role of HERV protein expression, this time as potent immune response triggers. This is suggested by various studies where significant antitumor results are demonstrated, either after the manipulation of HERV expression, leading to viral mimicry phenomena and subsequent induction of innate immunity responses, or after the generation of HERV protein‐specific CAR T‐cells, as HERV‐targeting cancer immunotherapy.
Finally, future studies with the use of more precise and higher resolution approaches will elucidate the exact role of HERVs and transposable elements in general in human physiological and pathological processes, such as carcinogenesis.
AUTHOR CONTRIBUTIONS
Konstantina Kitsou: Conceptualization, writing – original draft preparation, writing‐ reviewing and editing. Pagona Lagiou: Conceptualization, writing – reviewing and editing. Gkikas Magiorkinis: Conceptualization, writing – reviewing and editing, supervision. All authors contributed to the article and approved the submitted version.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ACKNOWLEDGEMENT
Κ. Κ. is supported through a donation of SYN‐ENOSIS, the Greek Shipowners' Social Welfare Company, to GM.
Kitsou K, Lagiou P, Magiorkinis G. Human endogenous retroviruses in cancer: oncogenesis mechanisms and clinical implications. J Med Virol. 2022;e28350. 10.1002/jmv.28350
DATA AVAILABILITY STATEMENT
Not applicable.
REFERENCES
- 1. Kitsou K, Iliopoulou M, Spoulou V, Lagiou P, Magiorkinis G. Viral causality of human cancer and potential roles of human endogenous retroviruses in the multi‐omics era: an evolutionary epidemiology review. Front Oncol. 2021;11:4511. 10.3389/FONC.2021.687631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discovery. 2022;12(1):31‐46. 10.1158/2159-8290.CD-21-1059 [DOI] [PubMed] [Google Scholar]
- 3. Li Y, Zhang G, Cui J. Origin and deep evolution of human endogenous retroviruses in Pan‐Primates. Viruses. 2022;14(7):1370. 10.3390/V14071370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Burn A, Roy F, Freeman M, Coffin JM. Widespread expression of the ancient HERV‐K (HML‐2) provirus group in normal human tissues. PLoS Biol. 2022;20(10):e3001826. 10.1371/JOURNAL.PBIO.3001826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mager DL, Stoye JP. Mammalian endogenous retroviruses. Microbiol Spectr. 2015;3(1). 10.1128/MICROBIOLSPEC.MDNA3-0009-2014 [DOI] [PubMed] [Google Scholar]
- 6. Geis FK, Goff SP. Silencing and transcriptional regulation of endogenous retroviruses: an overview. Viruses. 2020;12(8):884. 10.3390/v12080884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chuong EB. The placenta goes viral: retroviruses control gene expression in pregnancy. PLoS Biol. 2018;16(10):e3000028. 10.1371/journal.pbio.3000028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mao J, Zhang Q, Cong YS. Human endogenous retroviruses in development and disease. Comput Struct Biotechnol J. 2021;19:5978‐5986. 10.1016/J.CSBJ.2021.10.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ito J, Sugimoto R, Nakaoka H, et al. Systematic identification and characterization of regulatory elements derived from human endogenous retroviruses. PLoS Genet. 2017;13(7):e1006883. 10.1371/journal.pgen.1006883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ohtani H, Liu M, Zhou W, Liang G, Jones PA. Switching roles for DNA and histone methylation depend on evolutionary ages of human endogenous retroviruses. Genome Res. 2018;28(8):1147‐1157. 10.1101/GR.234229.118/-/DC1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liang CY, Wang LJ, Chen CP, Chen LF, Chen YH, Chen H. GCM1 regulation of the expression of syncytin 2 and its cognate receptor MFSD2A in human placenta. Biol Reprod. 2010;83(3):387‐395. 10.1095/BIOLREPROD.110.083915 [DOI] [PubMed] [Google Scholar]
- 12. Zhu H, Peng B, Klausen C, et al. NPFF increases fusogenic proteins syncytin 1 and syncytin 2 via GCM1 in first trimester primary human cytotrophoblast cells. FASEB J. 2020;34(7):9419‐9432. 10.1096/FJ.201902978R [DOI] [PubMed] [Google Scholar]
- 13. Denner J. Expression and function of endogenous retroviruses in the placenta. APMIS. 2016;124(1‐2):31‐43. 10.1111/APM.12474 [DOI] [PubMed] [Google Scholar]
- 14. Mangeney M, Renard M, Schlecht‐Louf G, et al. Placental syncytins: genetic disjunction between the fusogenic and immunosuppressive activity of retroviral envelope proteins. Proc Natl Acad Sci. 2007;104(51):20534‐20539. 10.1073/PNAS.0707873105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhou Y, Liu L, Liu Y, et al. Implication of human endogenous retrovirus W family envelope in hepatocellular carcinoma promotes MEK/ERK‐mediated metastatic invasiveness and doxorubicin resistance. Cell Death Discov. 2021;7(1):177. 10.1038/s41420-021-00562-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Strick R, Ackermann S, Langbein M, et al. Proliferation and cell‐cell fusion of endometrial carcinoma are induced by the human endogenous retroviral Syncytin‐1 and regulated by TGF‐β. J Mol Med. 2006;85(1):23‐38. 10.1007/S00109-006-0104-Y/FIGURES/9 [DOI] [PubMed] [Google Scholar]
- 17. Liu C, Xu J, Wen F, et al. Upregulation of syncytin‐1 promotes invasion and metastasis by activating epithelial‐mesenchymal transition‐related pathway in endometrial carcinoma. Onco Targets Ther. 2019;12:31‐40. 10.2147/OTT.S191041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lokossou AG, Toudic C, Nguyen PT, et al. Endogenous retrovirus‐encoded Syncytin‐2 contributes to exosome‐mediated immunosuppression of T cells. Biol Reprod. 2019;102(1):185‐198. 10.1093/BIOLRE/IOZ124 [DOI] [PubMed] [Google Scholar]
- 19. Chan SM, Sapir T, Park S‐S, et al. The HERV‐K accessory protein Np9 controls viability and migration of teratocarcinoma cells. PLoS One. 2019;14(2):e0212970. 10.1371/journal.pone.0212970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schmitt K, Heyne K, Roemer K, Meese E, Mayer J. HERV‐K(HML‐2) rec and np9 transcripts not restricted to disease but present in many normal human tissues. Mob DNA. 2015;6(1):4. 10.1186/S13100-015-0035-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen T, Meng Z, Gan Y, et al. The viral oncogene Np9 acts as a critical molecular switch for co‐activating β‐catenin, ERK, Akt and Notch1 and promoting the growth of human leukemia stem/progenitor cells. Leukemia.2013;27(7):1469‐1478. 10.1038/leu.2013.8 [DOI] [PubMed] [Google Scholar]
- 22. Shang S, Hua F, Hu ZW. The regulation of β‐catenin activity and function in cancer: therapeutic opportunities. Oncotarget. 2017;8(20):33972‐33989. 10.18632/ONCOTARGET.15687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Denne M, Sauter M, Armbruester V, Licht JD, Roemer K, Mueller‐Lantzsch N. Physical and functional interactions of human endogenous retrovirus proteins Np9 and Rec with the promyelocytic leukemia zinc finger protein. J Virol. 2007;81(11):5607‐5616. 10.1128/JVI.02771-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Babaian A, Mager DL. Endogenous retroviral promoter exaptation in human cancer. Mob DNA. 2016;7(1):24. 10.1186/S13100-016-0080-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Montesion M, Williams ZH, Subramanian RP, Kuperwasser C, Coffin JM. Promoter expression of HERV‐K (HML‐2) provirus‐derived sequences is related to LTR sequence variation and polymorphic transcription factor binding sites. Retrovirology. 2018;15(1):57. 10.1186/S12977-018-0441-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hepburn AC, Steele RE, Veeratterapillay R, et al. The induction of core pluripotency master regulators in cancers defines poor clinical outcomes and treatment resistance. Oncogene.2019;38(22):4412‐4424. 10.1038/s41388-019-0712-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ohnuki M, Tanabe K, Sutou K, et al. Dynamic regulation of human endogenous retroviruses mediates factor‐induced reprogramming and differentiation potential. Proc Natl Acad Sci. 2014;111(34):12426‐12431. 10.1073/pnas.1413299111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang T, Medynets M, Johnson KR, et al. Regulation of stem cell function and neuronal differentiation by HERV‐K via mTOR pathway. Proc Natl Acad Sci. 2020;117(30):17842‐17853. 10.1073/PNAS.2002427117/-/DCSUPPLEMENTAL [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Heidmann O, Béguin A, Paternina J, et al. HEMO, an ancestral endogenous retroviral envelope protein shed in the blood of pregnant women and expressed in pluripotent stem cells and tumors. Proc Natl Acad Sci. 2017;114(32):E6642‐E6651. 10.1073/PNAS.1702204114/-/DCSUPPLEMENTAL [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang‐Johanning F, Li M, Esteva FJ, et al. Human endogenous retrovirus type K antibodies and mRNA as serum biomarkers of early‐stage breast cancer. Int J Cancer. 2014;134(3):587‐595. 10.1002/ijc.28389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhou F, Li M, Wei Y, et al. Activation of HERV‐K Env protein is essential for tumorigenesis and metastasis of breast cancer cells. Oncotarget. 2016;7(51):84093‐84117. 10.18632/ONCOTARGET.11455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nguyen TD, Davis J, Eugenio RA, Liu Y. Female sex hormones activate human endogenous retrovirus type K through the OCT4 transcription factor in T47D breast cancer cells. AIDS Res Hum Retroviruses. 2019;35(3):348‐356. 10.1089/AID.2018.0173 [DOI] [PubMed] [Google Scholar]
- 33. Johanning GL, Malouf GG, Zheng X, et al. Expression of human endogenous retrovirus‐K is strongly associated with the basal‐like breast cancer phenotype. Sci Rep. 2017;7:41960. 10.1038/srep41960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jin X, Xu XE, Jiang YZ, et al. The endogenous retrovirus‐derived long noncoding RNA TROJAN promotes triple‐negative breast cancer progression via ZMYND8 degradation. Sci Adv. 2019;5(3):9820. 10.1126/SCIADV.AAT9820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hirukawa A, Singh S, Wang J, et al. Reduction of global H3K27me 3 enhances HER2/ErbB2 targeted therapy. Cell Rep. 2019;29(2):249‐257. 10.1016/J.CELREP.2019.08.105 [DOI] [PubMed] [Google Scholar]
- 36. Zhou F, Krishnamurthy J, Wei Y, et al. Chimeric antigen receptor T cells targeting HERV‐K inhibit breast cancer and its metastasis through downregulation of Ras. Oncoimmunology. 2015;4(11):e1047582. 10.1080/2162402X.2015.1047582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bhardwaj N, Montesion M, Roy F, Coffin J. Differential expression of HERV‐K (HML‐2) proviruses in cells and virions of the teratocarcinoma cell line Tera‐1. Viruses. 2015;7(3):939‐968. 10.3390/V7030939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kaufmann S, Sauter M, Schmitt M, et al. Human endogenous retrovirus protein Rec interacts with the testicular zinc‐finger protein and androgen receptor. J Gen Virol. 2010;91(Pt 6):1494‐1502. 10.1099/VIR.0.014241-0 [DOI] [PubMed] [Google Scholar]
- 39. Gimenez J, Montgiraud C, Pichon JP, et al. Custom human endogenous retroviruses dedicated microarray identifies self‐induced HERV‐W family elements reactivated in testicular cancer upon methylation control. Nucleic Acids Res. 2010;38(7):2229‐2246. 10.1093/NAR/GKP1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Beyer U, Krönung SK, Leha A, Walter L, Dobbelstein M. Comprehensive identification of genes driven by ERV9‐LTRs reveals TNFRSF10B as a re‐activatable mediator of testicular cancer cell death. Cell Death Differ. 2016;23(1):64‐75. 10.1038/CDD.2015.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kleiman A, Senyuta N, Tryakin A, et al. HERV‐K(HML‐2) GAG/ENV antibodies as indicator for therapy effect in patients with germ cell tumors. Int J Cancer. 2004;110(3):459‐461. 10.1002/IJC.11649 [DOI] [PubMed] [Google Scholar]
- 42. Rycaj K, Plummer JB, Yin B, et al. Cytotoxicity of human endogenous retrovirus K‐specific T cells toward autologous ovarian cancer cells. Clin Cancer Res. 2015;21(2):471‐483. 10.1158/1078-0432.CCR-14-0388 [DOI] [PubMed] [Google Scholar]
- 43. Iramaneerat K, Rattanatunyong P, Khemapech N, Triratanachat S, Mutirangura A. HERV‐K hypomethylation in ovarian clear cell carcinoma is associated with a poor prognosis and platinum resistance. Int J Gynecol Cancer. 2011;21(1):51‐57. 10.1097/IGC.0B013E3182021C1A [DOI] [PubMed] [Google Scholar]
- 44. Natoli M, Gallon J, Lu H, et al. Transcriptional analysis of multiple ovarian cancer cohorts reveals prognostic and immunomodulatory consequences of ERV expression. J Immunother Cancer. 2021;9(1):e001519. 10.1136/jitc-2020-001519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Strissel PL, Ruebner M, Thiel F, et al. Reactivation of codogenic endogenous retroviral (ERV) envelope genes in human endometrial carcinoma and prestages: emergence of new molecular targets. Oncotarget. 2012;3(10):1204‐1219. 10.18632/ONCOTARGET.679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Reis BS, Jungbluth AA, Frosina D, et al. Prostate cancer progression correlates with increased humoral immune response to a human endogenous retrovirus GAG protein. Clin Cancer Res. 2013;19(22):6112‐6125. 10.1158/1078-0432.CCR-12-3580 [DOI] [PubMed] [Google Scholar]
- 47. Rezaei SD, Hayward JA, Norden S, et al. HERV‐K Gag RNA and protein levels are elevated in malignant regions of the prostate in males with prostate cancer. Viruses. 2021;13(3):449. 10.3390/V13030449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Manca MA, Solinas T, Simula ER, et al. HERV‐K and HERV‐H env proteins induce a humoral response in prostate cancer patients. Pathogens. 2022;11(1):95. 10.3390/PATHOGENS11010095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yu H, Liu T, Zhao Z, et al. Mutations in 3′‐long terminal repeat of HERV‐W family in chromosome 7 upregulate syncytin‐1 expression in urothelial cell carcinoma of the bladder through interacting with c‐Myb. Oncogene. 2014;33(30):3947‐3958. 10.1038/ONC.2013.366 [DOI] [PubMed] [Google Scholar]
- 50. Panda A, de Cubas AA, Stein M, et al. Endogenous retrovirus expression is associated with response to immune checkpoint pathway in clear cell renal cell carcinoma. JCI Insight. 2018;3(16):e121522. 10.1172/jci.insight.121522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cherkasova E, Scrivani C, Doh S, et al. Detection of an immunogenic HERV‐E envelope with selective expression in clear cell kidney cancer. Cancer Res. 2016;76(8):2177‐2185. 10.1158/0008-5472.CAN-15-3139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Smith CC, Beckermann KE, Bortone DS, et al. Endogenous retroviral signatures predict immunotherapy response in clear cell renal cell carcinoma. J Clin Invest. 2018;128(11):4804‐4820. 10.1172/JCI121476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Au L, Hatipoglu E, Robert de Massy M, et al. Determinants of anti‐PD‐1 response and resistance in clear cell renal cell carcinoma. Cancer Cell. 2021;39(11):1497‐1518. 10.1016/J.CCELL.2021.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. de Cubas AA, Dunker W, Zaninovich A, et al. DNA hypomethylation promotes transposable element expression and activation of immune signaling in renal cell cancer. JCI Insight. 2020;5(11):e137569. 10.1172/JCI.INSIGHT.137569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Weyerer V, Strissel PL, Stöhr C, et al. Endogenous retroviral–K envelope is a novel tumor antigen and prognostic indicator of renal cell carcinoma. Front Oncol. 2021;11:11. 10.3389/fonc.2021.657187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tavakolian S, Goudarzi H, Lak E, Faghihloo E. The evaluation of HERV‐K env, np9, rec, gag expression in normal, polyp and cancerous tissues of gastric and colon. Future Virol. 2020;14(12):805‐812. [Google Scholar]
- 57. Liang Q, Xu Z, Xu R, Wu L, Zheng S. Expression patterns of non‐coding spliced transcripts from human endogenous retrovirus HERV‐H elements in colon cancer. PLoS One. 2012;7(1):e29950. 10.1371/JOURNAL.PONE.0029950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pérot P, Mullins CS, Naville M, et al. Expression of young HERV‐H loci in the course of colorectal carcinoma and correlation with molecular subtypes. Oncotarget. 2015;6(37):40095‐40111. 10.18632/oncotarget.5539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ko EJ, Ock MS, Choi YH, et al. Human endogenous retrovirus (Herv)‐K env gene knockout affects tumorigenic characteristics of nupr1 gene in dld‐1 colorectal cancer cells. Int J Mol Sci. 2021;22(8):3941. 10.3390/IJMS22083941/S1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Dolci M, Favero C, Tarantini L, et al. Human endogenous retroviruses env gene expression and long terminal repeat methylation in colorectal cancer patients. Med Microbiol Immunol. 2020;209(2):189‐199. 10.1007/s00430-020-00662-6 [DOI] [PubMed] [Google Scholar]
- 61. Dolci M, Favero C, Toumi W, et al. Human endogenous retroviruses long terminal repeat methylation, transcription, and protein expression in human colon cancer. Front Oncol. 2020;10:569015. 10.3389/FONC.2020.569015/FULL [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Díaz‐Carballo D, Acikelli AH, Klein J, et al. Therapeutic potential of antiviral drugs targeting chemorefractory colorectal adenocarcinoma cells overexpressing endogenous retroviral elements. J Exp Clin Cancer Res. 2015;34(1):81. 10.1186/s13046-015-0199-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ma W, Hong Z, Liu H, et al. Human endogenous retroviruses‐K (HML‐2) expression is correlated with prognosis and progress of hepatocellular carcinoma. BioMed Res Int. 2016;2016:1‐9. 10.1155/2016/8201642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Liu C, Liu L, Wang X, Liu Y, Wang M, Zhu F. HBV X protein induces overexpression of HERV‐W env through NF‐κB in HepG2 cells. Virus Genes. 2017;53(6):797‐806. 10.1007/s11262-017-1479-2 [DOI] [PubMed] [Google Scholar]
- 65. Tovo PA, Garazzino S, Daprà V, et al. Chronic HCV infection is associated with overexpression of human endogenous retroviruses that persists after drug‐induced viral clearance. Int J Mol Sci. 2020;21(11):3980. 10.3390/IJMS21113980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Baumert TF, Jühling F, Ono A, Hoshida Y. Hepatitis C‐related hepatocellular carcinoma in the era of new generation antivirals. BMC Med. 2017;15(1):52. 10.1186/S12916-017-0815-7/TABLES/2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Li M, Radvanyi L, Yin B, et al. Correction: downregulation of human endogenous retrovirus type K (HERV‐K) viral env RNA in pancreatic cancer cells decreases cell proliferation and tumor growth. Clin Cancer Res. 2019;25(9):2936. 10.1158/1078-0432.CCR-19-0700 [DOI] [PubMed] [Google Scholar]
- 68. Cai HZ, Zhang H, Yang J, Zeng J, Wang H. Preliminary assessment of viral metagenome from cancer tissue and blood from patients with lung adenocarcinoma. J Med Virol. 2021;93:5126‐5133. 10.1002/jmv.26887 [DOI] [PubMed] [Google Scholar]
- 69. Zare M, Mostafaei S, Ahmadi A, et al. Human endogenous retrovirus env genes: potential blood biomarkers in lung cancer. Microb Pathog. 2018;115:189‐193. 10.1016/J.MICPATH.2017.12.040 [DOI] [PubMed] [Google Scholar]
- 70. Lecuelle J, Favier L, Fraisse C, et al. MER4 endogenous retrovirus correlated with better efficacy of anti‐PD1/PD‐L1 therapy in non‐small cell lung cancer. J Immunother Cancer. 2022;10(3):e004241. 10.1136/JITC-2021-004241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Yuan Z, Yang Y, Zhang N, et al. Human endogenous retroviruses in glioblastoma multiforme. Microorganisms. 2021;9(4):764. 10.3390/MICROORGANISMS9040764/S1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kessler AF, Wiesner M, Denner J, et al. Expression‐analysis of the human endogenous retrovirus HERV‐K in human astrocytic tumors. BMC Res Notes. 2014;7(1):159. 10.1186/1756-0500-7-159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Shah AH, Govindarajan V, Doucet‐O'hare TT, et al. Differential expression of an endogenous retroviral element [HERV‐K(HML‐6)] is associated with reduced survival in glioblastoma patients. Sci Rep.2022;12(1):6902. 10.1038/s41598-022-10914-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kayabolen A, Yilmaz E, Bagci‐Onder T. IDH mutations in glioma: double‐edged sword in clinical applications. Biomedicines. 2021;9(7):799. 10.3390/BIOMEDICINES9070799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Schmitt K, Reichrath J, Roesch A, Meese E, Mayer J. Transcriptional profiling of human endogenous retrovirus group HERV‐K(HML‐2) loci in melanoma. Genome Biol Evol. 2013;5(2):307‐328. 10.1093/gbe/evt010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Singh M, Cai H, Bunse M, Feschotte C, Izsvák Z. Human endogenous retrovirus K Rec forms a regulatory loop with MITF that opposes the progression of melanoma to an invasive stage. Viruses. 2020;12(11):1303. 10.3390/v12111303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Krishnamurthy J, Rabinovich BA, Mi T, et al. Genetic engineering of T cells to target HERV‐K, an ancient retrovirus on melanoma. Clin Cancer Res. 2015;21(14):3241‐3251. 10.1158/1078-0432.CCR-14-3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Cardelli M, Doorn R, Larcher L, et al. Association of HERV‐K and LINE‐1 hypomethylation with reduced disease‐free survival in melanoma patients. Epigenomics. 2020;12(19):1689‐1706. 10.2217/EPI-2020-0127 [DOI] [PubMed] [Google Scholar]
- 79. Argaw‐Denboba A, Balestrieri E, Serafino A, et al. HERV‐K activation is strictly required to sustain CD133+ melanoma cells with stemness features. J Exp Clin Cancer Res. 2017;36(1):20. 10.1186/s13046-016-0485-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Deniz Ö, Ahmed M, Todd CD, Rio‐Machin A, Dawson MA, Branco MR. Endogenous retroviruses are a source of enhancers with oncogenic potential in acute myeloid leukaemia. Nat Commun. 2020;11(1):3506. 10.1038/S41467-020-17206-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Nakagawa S, Kawashima M, Miyatake Y, et al. Expression of ERV3‐1 in leukocytes of acute myelogenous leukemia patients. Gene. 2021;773:145363. 10.1016/j.gene.2020.145363 [DOI] [PubMed] [Google Scholar]
- 82. Engel K, Wieland L, Krüger A, et al. Identification of differentially expressed human endogenous retrovirus families in human leukemia and lymphoma cell lines and stem cells. Front Oncol. 2021;11:11. 10.3389/FONC.2021.637981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Fischer S, Echeverría N, Moratorio G, et al. Human endogenous retrovirus np9 gene is over expressed in chronic lymphocytic leukemia patients. Leuk Res Rep. 2014;3(2):70‐72. 10.1016/J.LRR.2014.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Depil S, Roche C, Dussart P, Prin L. Expression of a human endogenous retrovirus, HERV‐K, in the blood cells of leukemia patients. Leukemia. 2002;16(2):254‐259. 10.1038/sj.leu.2402355 [DOI] [PubMed] [Google Scholar]
- 85. Dolei A, Ibba G, Piu C, Serra C. Expression of HERV genes as possible biomarker and target in neurodegenerative diseases. Int J Mol Sci. 2019;20(15):3706. 10.3390/ijms20153706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Bergallo M, Montanari P, Mareschi K, et al. Expression of the pol gene of human endogenous retroviruses HERV‐K and ‐W in leukemia patients. Arch Virol. 2017;162(12):3639‐3644. 10.1007/S00705-017-3526-7 [DOI] [PubMed] [Google Scholar]
- 87. Januszkiewicz‐Lewandowska D, Nowicka K, Rembowska J, et al. Env gene expression of human endogenous retrovirus‐K and human endogenous retrovirus‐W in childhood acute leukemia cells. Acta Haematol. 2013;129(4):232‐237. 10.1159/000345407 [DOI] [PubMed] [Google Scholar]
- 88. Kewitz S, Staege MS. Expression and regulation of the endogenous retrovirus 3 in Hodgkin's lymphoma cells. Front Oncol. 2013;3:3. 10.3389/FONC.2013.00179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Stacey KJ, Sagulenko V. A clear link between endogenous retroviral LTR activity and Hodgkin's lymphoma. Cell Res. 2010;20(8):869‐871. 10.1038/cr.2010.96 [DOI] [PubMed] [Google Scholar]
- 90. Boso G, Fleck K, Carley S, Liu Q, Buckler‐White A, Kozak CA. The oldest co‐opted gag gene of a human endogenous retrovirus shows placenta‐specific expression and is upregulated in diffuse large B‐Cell lymphomas. Mol Biol Evol. 2021;38(12):5453‐5471. 10.1093/MOLBEV/MSAB245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Maliniemi P, Vincendeau M, Mayer J, et al. Expression of human endogenous retrovirus‐w including syncytin‐1 in cutaneous T‐cell lymphoma. PLoS One. 2013;8(10):e76281. 10.1371/JOURNAL.PONE.0076281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Toufaily C, Landry S, Leib‐Mosch C, Rassart E, Barbeau B. Activation of LTRs from different human endogenous retrovirus (HERV) families by the HTLV‐1 tax protein and T‐cell activators. Viruses. 2011;3(11):2146‐2159. 10.3390/v3112146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Leung A, Trac C, Kato H, et al. LTRs activated by Epstein‐Barr virus–induced transformation of B cells alter the transcriptome. Genome Res. 2018;28(12):1791‐1798. 10.1101/gr.233585.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Schmidt KLM, Vangsted AJ, Hansen B, et al. Synergy of two human endogenous retroviruses in multiple myeloma. Leuk Res. 2015;39(10):1125‐1128. 10.1016/J.LEUKRES.2015.06.014 [DOI] [PubMed] [Google Scholar]
- 95. Dai L, Del Valle L, Miley W, et al. Transactivation of human endogenous retrovirus K (HERV‐K) by KSHV promotes Kaposi's sarcoma development. Oncogene. 2018;37(33):4534‐4545. 10.1038/s41388-018-0282-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Ahuja A, Journo G, Eitan R, Rubin E, Shamay M. High levels of LINE‐1 transposable elements expressed in Kaposi's sarcoma‐associated herpesvirus‐related primary effusion lymphoma. Oncogene. 2020;40(3):536‐550. 10.1038/s41388-020-01549-9 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.


