Abstract
Brain ischemia is a common acute injury resulting from impaired blood flow to the brain. Translation of effective drug candidates from experimental models to patients has systematically failed. The use of human induced pluripotent stem cells (iPSC) offers new opportunities to gain translational insights into diseases including brain ischemia. We used a human 3D self-assembling iPSC-derived model (human cortical organoids, hCO) to characterize the effects of ischemia caused by oxygen-glucose deprivation (OGD). hCO exposed to 2 h or 8 h of OGD had neuronal death and impaired neuronal network complexity, measured in whole-mounting microtubule-associated protein 2 (MAP-2) immunostaining. Neuronal vulnerability was reflected by a reduction in MAP-2 mRNA levels, and increased release of neurofilament light chain (NfL) in culture media, proportional to OGD severity. Glial fibrillary acidic protein (GFAP) gene or protein levels did not change in hCO, but their release in medium increased after prolonged OGD. In conclusion, this human 3D iPSC-based in vitro model of brain ischemic injury is characterized by marked neuronal injury reflected by the release of the translational biomarker NfL which is relevant for testing neuroprotective strategies.
Keywords: Human cortical organoids, induced pluripotent stem cells, ischemic injury, neurodegeneration, biomarkers
Introduction
Animal models have been successfully used over the past fifty years for disease modelling in neuroscience both in vitro and in vivo, to decipher a variety of pathological events that are otherwise neglected or untreated. However, the biological/genetic differences among species have hampered the discovery of effective therapies for a large part of neurological and neurodegenerative disorders. The use of human tissues as a source for cell culture models, mainly from autopsies or fetal specimens, has been limited by the scant availability of the primary source and scalability of the primary cultures. The advent of human induced pluripotent stem cell (iPSC) technology in the first decade of the new century introduced a revolutionary paradigm for cell therapy and preclinical studies. 1 iPSC-derived in vitro models proved very useful research tool to help understand the basic biochemical and cellular mechanisms involved in different disorders. The three-dimensional (3D) self-organization of iPSC or neural precursors in brain organoids or spheroids recreates cytoarchitectural complexity and intercellular cross-talks which facilitate cell differentiation and the micro-environmental signalling underpinning brain physiology. 2
Brain organoids from the genetically defined patient’s specific iPSC recapitulate in vitro the main pathogenic mechanisms and hallmarks of neurodegenerative disorders, including Alzheimer’s disease (AD), 3 Parkinson’s disease (PD), 4 amyotrophic lateral sclerosis (ALS), 5 Huntington’s disease (HD), 6 and Rett syndrome. 7
Studies on acute brain injury can also benefit from the use of brain organoids. 3D organoids mirror in vitro the complex spatial and temporal involvement of different cell types and signalling networks in acute brain injury. Thus, there is increasing interest in modelling brain injury abnormalities in human organoids to generate effective translational models.8–10 Recently Pasca and colleagues 11 used this model to explore the effect of hypoxia, demonstrating the reliability of this human cellular platform to study the environmental factors underlying acquired brain injury.
Ischemic stroke is one of the most socio-economically impactful acute brain disorders. 12 It has been extensively studied in in vivo and in vitro models 13,14 that have helped understand many biochemical and molecular mechanisms underpinning its pathophysiology. These studies led to the identification of several candidate pharmacological approaches, but none of these effectively translated into clinical use. Over the last few years therefore, ischemic stroke research has adopted specific strategies for more reliable and enhanced potential translatability. 15 Specific guidelines for the use and reporting of work with animal models have been implemented (i.e., IMPROVE 16 ) and multicentric preclinical studies have been done.17,18 Besides animal models, stroke researchers have introduced and/or improved in vitro approaches to mimic stroke. A typical way to mirror ischemic stroke in vitro is oxygen-glucose deprivation (OGD). However, OGD does not completely reproduce the metabolic imbalance induced when flow is reduced to arrest if applied to overly simplified cellular models. 14
We developed a human cell-based 3D in vitro model to investigate the effect of an ischemic-like injury obtained with OGD. Cortical organoids (hCO), with an intricate interconnection of differentiated neurons and astrocytes in a 3D structure, were obtained from the aggregation of human iPSC-derived neural precursors cells (NPC), and exposed to OGD. We characterized the consequences of two durations of OGD on hCO by protein and gene expression analysis of selected brain markers. We also tested whether neurofilament light chain (NfL) and GFAP proteins, which emerged as biomarkers of neuronal and astroglial injury respectively, can be detected in hCO culture media after OGD and increase with ischemia severity. Our hCO model showed overt neurotoxicity, dependent on OGD duration, with less marked involvement of astrocytes.
Methods
Induced pluripotent stem cells (iPSC)
Episomal iPSC were obtained from Gibco™ (Life Technologies, CA, US, Lot V2.0), under the Johns Hopkins University Institutional Stem Cell Research Oversight Committee approval for collection and use. The iPSC line was cultured and expanded in feeder-free conditions by passaging every 3–5 days when they reached 70%–80% confluence, in a xeno-free cell culture medium formulation (StemMACS™ iPS-Brew XF, Miltenyi Biotec S.r.l.). NPC were obtained from the iPSC by blocking the TGF-β/BMP-dependant SMAD signalling (STEMdiff™ SMADi Induction Kit, STEMCELLS™ Technologies), resulting in efficient neural induction. By day 7 of neural induction, primitive NPC were dissociated with Accutase (Life Technologies), passed through a 100-μm strainer, and plated on Matrigel-coated dishes in an NPC expansion medium (STEMdiff™ Neural Progenitor Medium, STEMCELLS™ Technologies). An additional NPC line obtained from Axol Bioscience Ltd (Cod Ax0019) was also used here to generate the hCO. Samples were from the National Institute on Aging (NIA) Repository Collection (National Institutes of Health, Maryland, USA) and obtained under the Internal Review Board approval and patient informed consent. NPC were cultured and expanded in feeder-free condition by passaging every 6-7 days for up to 15 passages. The hCO used in the whole study were obtained from the two cell lines.
Generation of hCO
NPC from the 7th to the 15th passage were used to generate hCO, as previously reported. 19 Briefly, at 90% confluence NPC were enzymatically detached with Accutase and counted. Then a 105 NPC-single cell suspension was plated in 96-well ultra-low attachment plates and cells were grown in NPC expansion medium for one day under constant gyratory shaking (88 rpm). Within a few hours, the floating NPC folded into spherical structures. The next day the medium was changed to hCO complete medium composed of Neurobasal Plus, 1:50 B27 Plus, 1:100 GlutaMAX (Gibco™, Thermo Fisher Scientific), 20 ng/mL human recombinant BDNF and 20 ng/mL human recombinant GDNF (Miltenyi Biotec srl). Half of the medium was changed every two days during hCO formation. Cell aggregates were maintained at 37 °C, 5% CO2 under constant gyratory shaking (88 rpm) to mature up for a minimum of sixty days before injury.
Experimental design
We ran five independent experiments, each containing 4-10 hCO per condition. Ischemia was reproduced by OGD for 2 or 8 hours. The resulting experimental groups are: no injury (CTRL), OGD 2 h or OGD 8 h. At the time points indicated in Figure 1(a), we measured: 1) hCO mortality by lactate dehydroganase (LDH) and propidium iodide (PI) assays, 2) hCO toxicity by highly sensitive immunoassay for neuronal (NfL) and astroglial (GFAP) biomarkers released in the culture media, 3) hCO morphometric characterization by histological analysis and 4) gene expression analysis by RT-PCR.
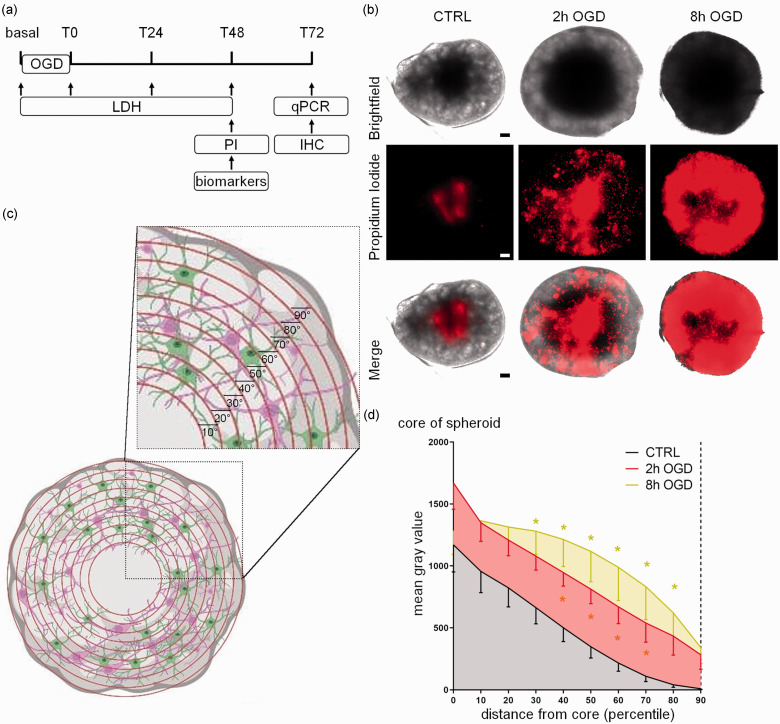
Figure 1.
Experimental design and effects of OGD on propidium iodide distribution in hCO. (a) Schedule of experiments including cell death assays (LDH release and PI fluorescent intensity), biomarker measures, gene profiling (real-time PCR) and immunohistochemistry (IHC). The basal line indicates the initial condition before OGD, usually referred to in the text as Tpre (b) Representative images of hCO exposed to OGD for 2 or 8 hours and marked with PI (red; scale bar 15 µm). (c) Representative illustration of the segmentation process for the quantification of PI in hCO that generates 10 concentric areas (percentiles) numbered in ascending order from the center (core) to the periphery and (d) PI fluorescence quantification in untreated (CTRL), 2 h and 8 h OGD-treated hCO. Data are shown as mean grey values – SEM (n 13–15 over four experimental replications) and analyzed by two-way RM ANOVA followed by Tukey’s multiple comparisons. *p < 0.05 vs CTRL. Panel (c) has been created in BioRender.com.
Ischemic injury
OGD was induced as previously described. 20 The complete culture medium was removed, hCO were washed three times with PBS and transferred into a temperature‐controlled (37 °C) hypoxic chamber (H35 Hypoxystation, Don Whitley Scientific) at O2 0.1%, CO2 5%, N2 95%. In the hypoxic chamber the PBS was replaced with deoxygenated glucose‐free Neurobasal A medium (Gibco™, Thermo Fisher Scientific). After OGD, hCO were returned to a normoxic incubator and medium was replaced with normal medium. Control hCO were maintained in a normoxic incubator and medium changes were done with hCO complete medium.
Lactate dehydrogenase assay
Conditioned media of hCO was collected at different time-points, as reported in the experimental design: before OGD (Tpre), at the end of the OGD period (T0), after 24 h of recovery (T24) and after 48 h of recovery (T48). To evaluate cell death in hCO, the lactate dehydrogenase (LDH) activity released in the conditioned media was measured with a commercial bioluminescent plate-based assay (LDH-Glo™ Cytotoxicity Assay, Promega, Madison, WI). Luminescence was measured with a multimode plate reader (Tecan, Infinite M200) after 30 min of incubation with single determinations per well. LDH level differences from Tpre values were compared and normalized to uninjured conditions.
Propidium iodide incorporation
At the end of the recovery period (48 h), hCO were incubated with propidium iodide (PI; Merck KGaA) diluted 1:200 in hCO complete medium for 30 mins. Then PI solution was removed, and images were acquired with an optical microscope equipped with CellR software (Olympus IX81). Serial focal planes (30 μm size) on the Z axis were acquired in two different channels to obtain the brightfield and the fluorescent (Texas red filter) images. Analyses were then done with Fiji software (https://imagej.net/Fiji/Downloads) as follows: based on the brightfield images, the hCO surface was devided into ten concentric regions equidistant from the periphery to the central core (calculated as 40% of the hCO surface), thus dividing the hCO surface into 10 percentiles (Figure 1(c)). The mean grey values from the PI fluorescent signal were then measured for each percentile and plotted on a graph.
Biomarker measurements
The release of NfL and GFAP in the culture media at T48 was measured by single molecule array (simoa) immunoassay (Quanterix, Billerica, MA) to assess neuronal and astrocytic damage respectively. Analyses were run using commercial simoa assays on a SR-X Analyzer: NfL advantage assay (catalog #103400) and GFAP discovery assay (catalog #102336). Samples were diluted 1:200 in diluent buffer. A single lot of reagents was used for all samples. All samples were examined twice.
Immunohistochemistry
hCO were fixed with 4% formaldheyde for 1 h at room temperature, followed by dehydration with different methanol concentrations. Permeabilization was then achieved using 3% BSA, 0.5% Triton X-100 in PBS for 2 h. hCO were then incubated with the primary antibodies GFAP (mouse; Millipore) and MAP-2 (rabbit; Merk Life Science Srl) diluted 1:200 in blocking solution (5%BSA, 0.1% Triton X-100 in PBS) at 37 °C for 24 h in agitation (50 rpm) . The secondary antibodies anti-mouse 488 and anti-rabbit 647 (Tebu-Bio Srl) diluted 1:250 in blocking solution were then incubated for a further 24 h with agitation at 37 °C. An additional 2 h incubation with HOECHST 33258 (Merck Life Science S.r.l.) diluted 1:200 in PBS to identify cell nuclei followed when needed. hCO were then dehydrated sequentially in 50% and 100% methanol solutions and exposed to Visikol HISTO-M™ (Merck KGaA) solution for 30 minutes in Ibidi, μ-Slide 2 Well Co-Culture plates (Ibidi GMBH) for tissue clearing before image acquisition.
Confocal microscopy and digital image analysis
Confocal microscopy was done on a Nikon A1 confocal scan unit, managed by NIS elements software. hCO were imaged at laser excitation of 405 (for nuclei), 488 (for GFAP) and 640 nm (for MAP-2) with sequential scanning mode to avoid bleed-through effects. Overview large field acquisition was obtained with a 20 × 0.5 NA, using 10% image overlapping to allow stitching. Each image had a pixel size of 0.6 µm and was acquired over a 200–250 µm z-axis (step size of 2.6 µm). Fields of view for GFAP and MAP-2 quantification were acquired with a 40 × 0.75 NA objective using Nyquist zoom. Each image was 209 × 209 × 20 µm, had a pixel size of 0.2 µm and a step size of 0.9 µm. Four 40× fields of view – representing ≈ 20% of the total hCO were positioned referring to the overview image, as depicted in Figure 3(b). The fields of view were randomly positioned so to sample the hCO evenly around its core, in an area showing OGD-induced damage according to the PI distribution. The volume occupied by GFAP or MAP-2 positive voxels was quantified by building isosurface renderings using Imaris (Bitplane, CH). We set a smoothing of 0.40 µm (double the pixel size), 1 µm for maximum sphere inscribed in object, and applied an autothreshold gray value cut-off. Volumes were normalized over the total assessed hCO volume.
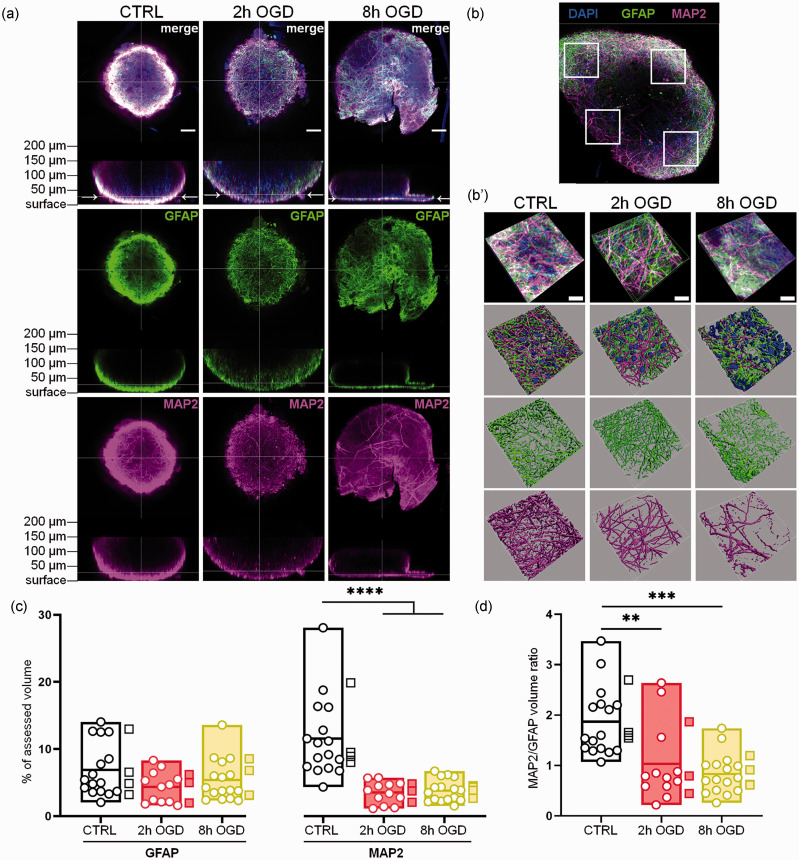
Figure 3.
Neuron and astrocyte immunostaining analysis in whole-mounted hCO. Whole-mount immunostaining for MAP2 and GFAP was done in control, 2 h or 8h OGD-treated hCO and imaged at T72. (a) Overview images of GFAP (green) and MAP2 (magenta) fluorescent signal distribution in whole hCO, showing xy and xz focal planes. Arrows in the xz focal planes indicate the focal level where most of the fluorescent signal was observed based on light penetration. Nuclei are stained in blue by Hoechst 33258. Scale bars 50 µm. (b) Typical distribution of the four randomly selected areas (white squares) used for analysis at 40x magnification. (b′) Representative 40x images of GFAP and MAP2 signals in the selected areas and their 3D reconstruction by isosurfaces. Scale bars 20 µm. (c) Quantification of GFAP and MAP2 signal volumes reported as percentages of the total hCO volume analyzed and (d) Ratio of MAP2 to GFAP signal volumes. Data shown as floating min-to-max bars with line at mean. Round symbols indicate individual 40x fields of view (n = 12–16) from 3-4 hCO per group (mean value per hCO indicated by the square symbols). Two-way RM ANOVA followed by Sidak’s multiple comparisons, ****p< 0.0001 (c) or Kruskall-Wallis test, **p < 0.01, ***p < 0.001 (d).
MAP-2 network analysis was done using an originally developed ImageJ algorithm to segment and skeletonize the signal. Briefly, the background was normalized imposing a minimum gray value of 1200, then the image was corrected with a median filter with a radius of 5 and binarized over the z stack by the Li method. 21 An extended focus image of the binary image was obtained and the skeletonize function was applied. Then the ‘Analyze Skeleton 2D/3D’ ImageJ plugin was applied with no endpoint pruning. Representative images were finally processed with GIMP.
Gene expression analysis
To characterize hCO maturation total RNA was extracted from NPC and from hCO after 14 or 60 days in vitro (DIV). To assess OGD effects on hCO, samples were collected three days after injury, immediately frozen on dry ice and stored at −80°C until analysis. Total RNA was extracted with a Quiagen kit or Ribospin II kit (GeneAll). Samples were treated with DNase (Life Technologies) and reverse-transcribed with random hexamer primers using Multi-Scribe Reverse Transcriptase (Life Technologies). Real-time reverse transcription PCR was done with b-actin as housekeeping gene. Relative gene expression was determined by the ΔΔCt method using mean Ct from three replicates per sample. Data are expressed as the log 2-fold difference over the NPC or CTRL groups. Genes and primer sequences are listed in Table 1.
Table 1.
Genes and primer sequences used for real-time PCR.
| Gene | Primer forward | Primer reverse |
|---|---|---|
| β-ACTIN | ACTCTTCCAGCCTTCCTTC | ATCTCCTTCTGCATCCTGTC |
| BDNF | AGAAAGTTCGGCCCAATGAA | GAGCATCACCCTGGACGTGTA |
| DCX | TATGCGCCGAAGCAAGTCTCCA | CATCCAAGGACAGAGGCAGGTA |
| GAD1 | GGGTTCGCACAGGTCATCC | CCGTTGATGTCAGCCATTCTC |
| GFAP | AGGAGAACCGGATCACCATT | TTGTGATTTTCCCCGTCTTT |
| GLAST | GTGGACTGGTTCCTGGATCG | TCGTGACAAGTGCTCCACAA |
| GLT1 | GACTTAGAAGTGGTTGCTAGGC | AGGGGTTTTTCCTGGTCACTAC |
| GRIN1 | CCTCATCTCCAGCCAGGTCT | TGGGAGTGAAGTGGTCGTTG |
| MAP2 | TCCAAATGTGGCTCTCTGAA | TAGCTTGGGCCTTTTCTTTG |
| NESTIN | GATCGCTCAGGTCCTGGAAG | TCCAGGAGGGTCCTGTACG |
| NeuN | ACGATCGTAGAGGGACGGAA | TTTAGCTTCCAGCCGTTGGT |
| S100β | GAAFAAATCCGAACYGAAGGAGC | TCCTGGAAGTCACATTCGCCGT |
| SOX1 | GCTGACACCAGACTTGGGTTT | CCCCTCGAGCAAAGAAAACG |
| SOX2 | CATGAAGGAGCACCCGGATT | CTCGCCATGCTATTGCCG |
| SYP | TGCAGAACAAGTACCGAGAG | CTGTCTCCTTAAACACGAACC |
| Tuj1 | GATCGGGGCCAAGTTCTGG | GCCTCGTTGTAGTAGACGCT |
| VEGFA | CGAGGGCCTGGAGTGTGT | CCGCATAATCTGCATGGTGAT |
Electrophysiological recordings
Cortical organoids were transferred from the incubator to the recording chamber and held in place with the custom-made anchor. We took whole-cell path-clamp recordings at room temperature in an extracellular solution containing 130 mM NaCl, 3.5 mM KCl, 1.2 mM NaH2PO4, 1.3 mM MgCl2, 25 mM NaHCO3, 11 mM D-glucose, 2 mM CaCl2, constantly bubbled with 95% O2 and 5% CO2. Patch electrodes (5–10 MΩ resistance when filled) contained the following: 140 mM CsCl, 4 mM NaCl, 1 mM MgCl2, 10 mM HEPES, 2 mM Mg-ATP and 0.4 mM Mg-GTP. Cells were visualized using an upright Olympus microscope (BX51 WI) with infrared differential interference contrast optics and a water immersion 40x objective. We used a MultiClamp 700B amplifier and Digidata 1440 A digitizer and Pclamp10 software for data acquisition and stimulus delivery. Signals were filtered at 3 Hz using a Bessel filter and digitized at 10 kHz. Clampfit 10 software was used for data analysis. Evoked action potentials were recorded in current-clamp mode using a series of injected currents from −60 pA to 160 pA in 20 pA steps. Whole-cell currents were recorded in voltage-clamp mode with a basal holding potential of −60 mV followed by stimulating pulses from −80 mV to 40 mV with step size 20 mV.
Statistical analysis
Data are reported as mean and standard deviation (SD) from at least three independent experiments and statistically analyzed applying mixed models as detailed below. Group sizes were established to ensure a minimum power of 0.80 to detect moderate effect sizes of 0.2 to 0.25 between controls and other groups at a two-sided α of 0.05. For statistical analyses we used GraphPad Prism (GraphPad Software Inc., USA, version 9.2.0). A parametric unpaired t-test and One-way ANOVA with Tukey's post-analysis or One-way Welch-corrected ANOVA followed by Dunnett’s T3 test were used to compare the differences between two or more groups. Bartlett’s test was used to test the equality of variances. D'Agostino & Pearson omnibus normality test and relative QQ plots were used to assess the assumption of normality. Two-way Anova and Kruskall-Wallis or Sidak’s multiple comparisons tests were used for comparisons between two categorical variables and one quantitative dependent variable. P values lower than 0.05 was considered statistically significant.
Results
Consistently with previous findings, 19 a time-dependent increase was observed in the levels of genes related to glial cells. An increase in differentiated neuron-related genes was detected at 14 DIV and persisted up to 60 DIV, together with a decline in immature neuron and stem cell patterns from 14 to 60 DIV (Figure S1). Recordings of action potentials and inward/outward currents by patch clamp in 60 DIV hCO showed the presence of responsive neurons (Figure S2). 60 DIV hCO were used for all subsequent experiments.
OGD induced cytotoxicity in hCO
OGD effects on cell death were investigated by imaging analysis after hCO exposure to PI at T48 of re-oxygenation. OGD changed the PI distribution in hCO (Figure 1(b)), as shown by the radial increase of the PI signal around the core. We therefore analysed the spatial changes in PI by applying concentric fields extending from the hCO core. Compared with CTRL, we did not find any significant differences in the PI intensity induced by OGD in the core (Figure 1(d)), where a physiological necrotic area is still expected in untreated conditions. However, there was a significant increase in PI signal from the 40th to the 70th percentile for the 2 h OGD and from the 30th to the 80th percentile for the 8 h OGD (p < 0.05 vs CTRL for all, Figure 1(d)), indicating a correlation between OGD duration and injury severity.
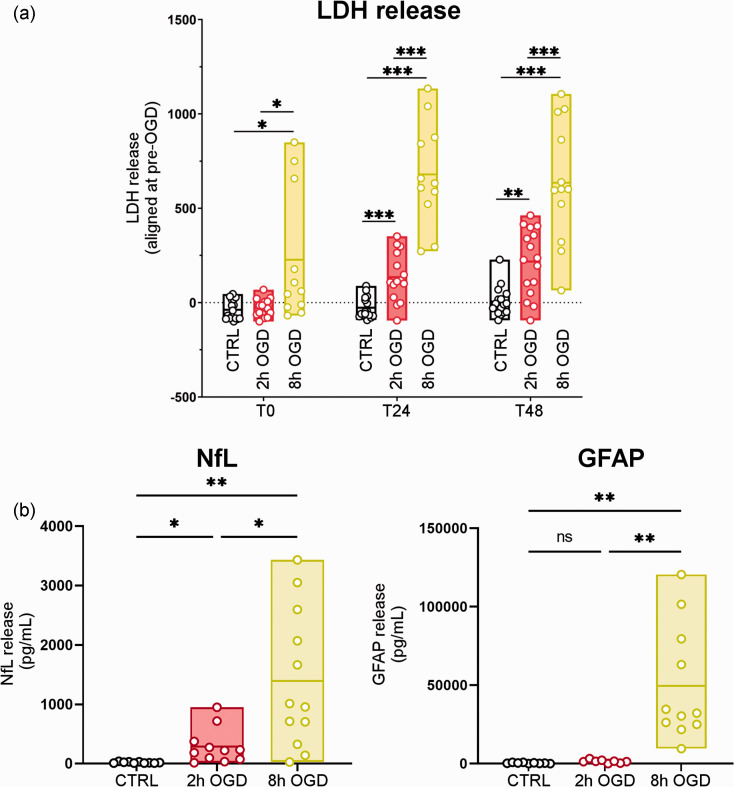
CTRL hCO showed no changes in LDH release over time (Figure 2(a)). 2 h OGD induced a time-dependent increase in LDH release with significantly higher values than CTRL at T24 (p < 0.001) and T48 (p < 0.01) of re-oxygenation. 8 h OGD further exacerbated this condition, with LDH release higher than CTRL (p < 0.05) and 2 h OGD (p < 0.05) already detectable at T0, that persisted at T24 and T48 (p < 0.001). While 2 h OGD showed increased LDH release over time (T0 vs T24: p < 0.001; T24 vs T48: p < 0.05), 8 h OGD reached a plateau at T24 (T0 vs T24: p < 0.001) and LDH remained high at T48 (T24 vs T48: n.s.). These data indicate that injury severity is proportional to OGD duration.
Figure 2.
Injury-dependent cell death and biomarker release in hCO after OGD. (a) LDH release was quantified in conditioned media collected before (pre-OGD), immediately after (T0), and 24 h (T24) and 48 h (T48) after treatment in control (CTRL), 2 h or 8 h OGD conditions and (b) Conditioned media were collected 48 h after OGD. NfL and GFAP concentrations in the media were recorded by simoa immunoassay. Data are shown as floating min-to-max bars with line at mean. Symbols indicate individual hCO (n = 11–16) over four experimental replications. LDH levels were compared with one-way ANOVA and Tukey’s multiple comparisons test, *p < 0.05, ** p < 0.01, ***p < 0.001. NfL and GFAP data had unequal variances as determined by Bartlett’s test and were therefore analyzed by one-way Welch-corrected ANOVA followed by Dunnett’s T3 multiple comparisons test, *p < 0.05, **p < 0.01.
Neuron and astrocyte injury in hCO after OGD
To quantify the cell-specific damage induced by OGD we then measured the concentrations of markers related to neurons (NfL) or astrocytes (GFAP) released in the conditioned media after OGD. 2 h OGD increased NfL release compared to CTRL (p < 0.05, Figure 2(b)). 8 h OGD further increased neuronal damage, with greater NfL release compared to both CTRL (p < 0.01) and 2 h OGD (p < 0.05; Figure 2(b)). GFAP showed a significant increase in release only for the 8 h OGD condition (p < 0.01 vs CTRL and vs 2 h OGD; Figure 2(b)).
To determine the specific effects of OGD at a cellular level we then did an image analysis for neuronal (MAP2) and astrocytic (GFAP) markers on whole-mounted hCO (Figure 3(a)) at T72, with a 3D reconstruction of the fluorescent signal (Figure 3(b) and (b′)). When comparing the GFAP-positive volumes, no differences between CTRL and OGD hCO were seen (Figure 3(c)). In contrast, compared to CTRL MAP2 was significantly reduced in the hCO exposed to 2 h or 8 h OGD (p < 0.0001; Figure 3(c)). The MAP2/GFAP volume ratio was 2 ± 0.75 in CTRL and was significantly reduced by injury (2 h, p < 0.01 and 8 h, p < 0.001) to levels below one after 8 h OGD (Figure 3(d)).
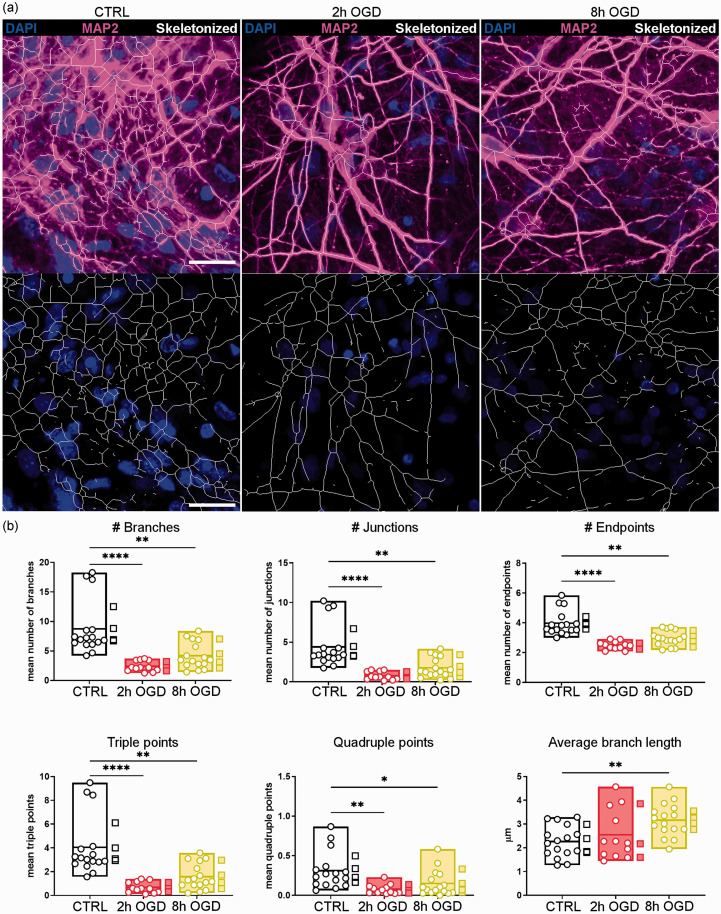
We next analysed different morphometric parameters of MAP2-positive neuron branches to characterize the neuron network pattern in the various conditions (representative images in Figure 4(a)). Both 2 h and 8 h OGD significantly affected the complexity of neuronal branches by reducing the mean number of all the parameters analysed (Figure 4(b)), including the number of branches, junctions, endpoints, triple and quadruple points. The spared MAP2-positive neurons in 8 h OGD had increased branch length (p < 0.01 vs CTRL, Figure 4(b)), consistent with a loss of collateral ramifications. This indicates significant impairment by OGD of the physiological complex neuron arborization and networking.
Figure 4.
OGD impaired the neuron network pattern. Neurons in hCO were immunostained with MAP2 and four 40x fields of view were randomly selected for analysis. (a) Representative images of selected areas in control (CTRL), 2h and 8h OGD-treated hCO (upper panels). The MAP2 fluorescent signal (magenta) was segmented and skeletonized with an originally developed ImageJ algorithm (white traces in the lower panels). Nuclei are in blue. Scale bars 20 µm and (b) Different parameters describing the neuron arborization complexity were calculated by ImageJ’s analyze skeleton plugin: number of branches, junctions, endpoints, junctions with 3 (triple) or 4 (quadruple) branches, and the average branch length. Data shown as floating min-to-max bars with line at mean. Round symbols indicate individual 40x fields of view (n = 12–16) from 3–4 hCO per group (mean value per hCO indicated by the square symbols). One-way ANOVA followed by Tukey’s multiple comparisons for Average branch length, Kruskall-Wallis test for the other parameters, *p < 0.05, **p < 0.01, ****p < 0.0001.
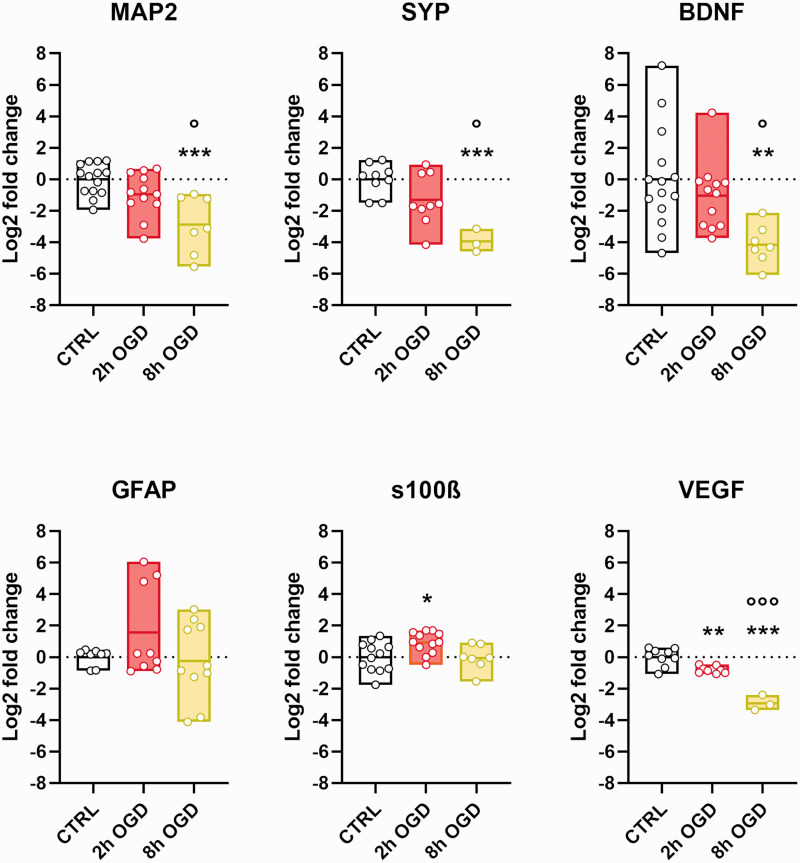
Gene expression analysis at T72 also detected a neurotoxic effect, with MAP2 and SYP neuronal genes, as well as neuronal associated growth factor BDNF showing significant reduction after 8 h OGD and less marked changes after 2 h OGD (p < 0.01 OGD 8 h vs CTRL; p < 0.05 OGD 8 h vs OGD 2 h; Figure 5). When astrocyte-related genes were analysed (GFAP and S100β, Figure 5), differences were negligible between conditions, except for a significant increase, although small, in S100β only for 2 h OGD compared to CTRL (p < 0.05). VEGFA expression, which in our hCO could be ascribed only to astrocytes, showed clear injury-dependent down-regulation (p < 0.01 OGD 2 h vs CTRL, p < 0.001 OGD 8 h vs CTRL, p < 0.001 OGD 2 h vs OGD 8 h).
Figure 5.
OGD effect on neuron and astrocyte gene expression. Total RNA was extracted from control (CTRL), 2 h and 8 h OGD-treated hCO. Real-time PCR was done with β-actin as housekeeping gene. Relative gene expression for MAP2, synaptophysin (SYP), BDNF, GFAP and S100β and VEGFA is expressed as the log 2-fold difference over the CTRL group. Data shown as floating min-to-max bars with line at mean. Symbols indicate individual hCO (n = 12–14 for CTRL and 2 h OGD and n = 3–10 for 8 h OGD) over three experimental replications. One-way ANOVA followed by Tukey’s multiple comparisons test, *p < 0.05, **p < 0.01, ***p < 0.001 vs CTRL, ***p < 0.001; °°°p < 0.001 vs OGD 2 h.
Discussion
We developed and characterized an ischemic brain injury model consisting of 3D self-assembling hCO from iPSC of human origin and exposed to OGD. The model presents massive and persistent OGD-induced cell death with overt neuronal damage, as revealed by histological and gene expression analyses, as well as by the biomarker release in culture media. These pathological features were proportional to the length of the ischemic stimulus, indicating more severe damage with longer OGD duration.
Human-derived iPSC have been used over the last few years to generate cell models with the aim of improving the translational impact of preclinical research studies by overcoming the genetic and phenotypic differences between species. Cell cultures serve as reliable tools for the study of stroke-associated pathogenic mechanisms and drug testing. Compared to 2D cell cultures, 3D cultures mimic the in vivo tissue microenvironment better in terms of cell-cell and cell-extracellular matrix (ECM) interactions, differentiation and maturation rates, neuron electrophysiological properties and nutrient requirements.22–25 We used a protocol able to generate, in 8 weeks of in vitro culture, hCO that reproduce neuronal-glial interactions and connectivity, 19 and provide new data on the sensitivity to ischemia of human neurons grown in a 3D microenvironment by showing clear alterations in the dendritic branching pattern and neuronal morphology induced by OGD in whole-mounted hCO.
iPSC have been used to model pathological genetic conditions resulting in stroke, such as the Marfan syndrome, 26 cerebral autosomal dominant arteriopathy,27,28 and cerebral cavernous malformation. 29 However, the use of iPSC-derived brain organoids to recapitulate ischemia and metabolic failure by depleting energy substrates in the culture system is in its infancy. Heme-induced cell apoptosis and structural changes were described by Harbuzariu et al. 9 in cortical organoids. Hypoxia has been shown to impair the expression of dorsal brain genes, oligodendrocytes, and neuronal progenitors in developing brain organoids, 10 and to have different effects on neural cells, with outer radial glia and immature neurons more vulnerable to hypoxic injury compared to neural precursors.30,31 The vulnerability of differentiating cells to OGD was also confirmed recently by Van Breedam and colleagues, who developed a luminescent human iPSC-derived neurospheroid model that allows real-time measurement of neural viability under OGD. 32 By this approach the authors demonstrated that OGD-mediated injury was reversible in younger (1-week-old), but not in more mature (4-week-old) neurospheroids, suggesting the need for longer neurospheroid maturation for in vitro stroke research. Paşca et al. identified impairments in neuron progenitor distribution in specific areas of brain spheroids cultured in hypoxic conditions, 11 but no toxic effects on differentiated neurons were reported.
Here we adopted the model by Pamies et al., 19 which showed critical elements of neuronal function, including synaptogenesis and neuron-to-neuron and neuronal-glial interactions, and describe a clear neuronal susceptibility to ischemia with cell death and neurotoxicity proportional to the duration of metabolic substrate depletion in hCO characterized by differentiated neurons and astrocytes. 3D spheroids constitutively include cells in different states, i.e., proliferating, quiescent, apoptotic, hypoxic, and necrotic cells. The outer layers receive sufficient oxygen and nutrients, allowing cell differentiation, while a necrotic core builds up inside the organoid owing to lack of oxygen and nutrient diffusion. 2 When analyzing the radial distribution of PI cell incorporation in concentric areas of hCO, we found specific OGD-mediated toxicity in the outer layers, but not in the core, confirming different responses of the distinct hCO layers to OGD. We therefore restricted the analysis of neuronal dendritic branching patterns and astrogliosis to hCO outer layers. We found cell-specific impairments such as reduced neuron arborization and neuron/astrocyte ratio proportional to the severity of the injury. Accordingly, there was an injury severity-dependent reduction of genes associated with neuronal structure/function (MAP2, SYP) and trophic support (BDNF).
Our study demonstrates the alteration of neurite development and networking organization under ischemic conditions with the NfL increase in culture media indicating ischemic severity-dependency. NfL is a neuronal cytoskeletal element, vital for maintaining cellular integrity and axonal transport. When neuronal injury and axonal membrane disruption occur, NfL is released into the interstitial fluid and ultimately ends in the cerebrospinal fluid (CSF) and blood. 33 In ischemic stroke patients NfL serum levels are elevated and correlate with infarct volume and recurrent ischemic lesions, 34 predicting long term functional disability and higher all-cause mortality. 35 Importantly, NfL has proved valid as a pharmacodynamic biomarker in monitoring treatment response after acute or chronic neurological disorders.36–38
Different preclinical models of ischemia show an increase in the immunoreactivity of NfL in the ischemic areas,39,40 likely originating from the abundance of NfL degradation products detected in the ischemic tissue. 40 Data from in vivo models of proteopathic neurodegenerative lesions and of traumatic brain injury show increased circulating NfL levels.41,42 However, no data are available from preclinical ischemia models. Here we report a severity-dependent NfL release after ischemia, offering a solid parameter to test the efficacy of neuroprotective approaches for future interventional studies.
When focusing on astroglial changes we detected an increase of GFAP in the media, suggesting astrocyte injury 43 and/or reactive astrogliosis. 44 However, we did not detect any changes in the GFAP immunostained volume or gene expression of structural genes (GFAP and S100β) within the organoid after OGD. Astrocytes are an inflammatory population that activates after acute brain injury by showing hypertrophysm and by packing up to form the gliotic scar at the boundaries of a lesion. Thus, reactive astrocytes might have been expected in our model in response to OGD. However, the OGD model induces a global, not a focal lesion around which a gliotic scar could form. We may hypothesize that both astrocyte injury and activation occurred in our model, and the effects were balanced enough to be adequately identified by our analysis of GFAP immunoreactivity and gene expression. Nevertheless, we detected a clear injury-dependent down-regulation of VEGFA, suggesting dysfunction in astrocyte reaction. Vascularized organoids now offer the possibility to explore the interplay between astrocyte activation and vasculature 45,46 and enable future investigation on the role of VEGFA in vascular remodeling.
Besides technical reasons, we should also mention that our hCO may not mirror all the inflammatory consequences of brain ischemia. Indeed, brain components with close interactions with astrocytes, like endothelial cells and microglia, are not present in the hCO. We provide evidence here for differentiated and electrophysiologically active neurons, and model optimization will aim at obtaining fully mature neural cells and thorough functional characterization.
Nevertheless, our model represents a step forward in basic stroke research, specifically by offering a valuable tool to study neuronal damage. Neurons are grown in three-dimensional self-assembled structures, which is a prerequisite for closer in vitro mirroring of their physiology.47,48 The translational biomarker of neuronal injury NfL is measurable in culture medium and proved sensitive to OGD duration and severe consequences. The model is based on human- iPSC, allowing future studies of precision medicine as iPSC may be obtained from selected donors by non-invasive sampling, for instance using urine cells. 49 This will enable us to better consider some genetic-based comorbidities that may affect stroke outcomes.
Another important implication of our study is that it fits the requirement for a valid complimentary method to animal studies. While animal studies cannot be abandoned in stroke research, a reliable in vitro benchmark to screen candidate therapeutic approaches would help reduce their use.
In conclusion, we provide a reliable in vitro model of brain ischemia in a 3 D complex brain environment, namely cortical organoids. OGD reproduced key features of in vivo pathology, affecting neuron survival and networking. The neurodegenerative effects are proportional to the duration of OGD, monitored by clinically relevant biomarkers of brain injury. This evidence paves the way for prospective interventional studies with direct translational implications.
Supplemental Material
Supplemental material, sj-pdf-1-jcb-10.1177_0271678X231152023 for Neural cortical organoids from self-assembling human iPSC as a model to investigate neurotoxicity in brain ischemia by Massimiliano De Paola, Francesca Pischiutta, Davide Comolli, Alessandro Mariani, Joe Kelk, Ilaria Lisi, Milica Cerovic, Stefano Fumagalli, Gianluigi Forloni, Elisa R Zanier in Journal of Cerebral Blood Flow & Metabolism
Acknowledgements
The authors acknowledge Francesca Marrulli for her contribution in culture handling and live imaging acquisition.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was funded by the Italian Ministry of Health – Rete IRCCS delle Neuroscienze e della Neuroriabilitazione (RIN) [Project code RCR‐2021‐23671214, cup code D42C21000600001]; the European Union´s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant (ENTRAIN) [agreement No. 813294]; Fondazione Regionale per la Ricerca Biomedica, Regione Lombardia (VASCO) [project ID 1739635].
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Authors’ contributions
MDP and EZ conceived the study and were assisted by SF and FP in the design and execution of the experiments. DC, AM, JPK and IL contributed to the experiment itself and data analysis. MDP, EZ, SF and FP were the main contributors to writing the manuscript, with comments from all authors. MC contributed to the model characterization. GF supervised the study and the manuscript writing. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The study was conducted according to the ethical principles of the Declaration of Helsinki. Cell lines used in this study were purchased from independent companies. Data and information from the donors were properly anonymised and informed consent was obtained at the time of sample collection. Cell lines were approved for use by The Johns Hopkins University Institutional Stem Cell Research Oversight Committee and the National Institute on Aging (NIA) Repository Collection (National Institutes of Health, Maryland, USA).
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in Zenodo repository (https://zenodo.org/deposit/7228188).
ORCID iDs
Massimiliano De Paola https://orcid.org/0000-0003-2818-9039
Elisa R Zanier https://orcid.org/0000-0002-3011-8718
Supplemental material
Supplemental material for this article is available online.
References
- 1.Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007; 131: 861–872. [DOI] [PubMed] [Google Scholar]
- 2.Qian X, Song H, Ming G.Brain organoids: advances, applications and challenges. Development 2019; 146: dev166074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raja WK, Mungenast AE, Lin Y-T, et al. Self-organizing 3D human neural tissue derived from induced pluripotent stem cells recapitulate Alzheimer’s disease phenotypes. PLoS ONE 2016; 11: e0161969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim H, Park HJ, Choi H, et al. Modeling G2019S-LRRK2 sporadic Parkinson’s disease in 3D midbrain organoids. Stem Cell Reports 2019; 12: 518–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pereira JD, DuBreuil DM, Devlin A-C, et al. Human sensorimotor organoids derived from healthy and amyotrophic lateral sclerosis stem cells form neuromuscular junctions. Nat Commun 2021; 12: 4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conforti P, Besusso D, Bocchi VD, et al. Faulty neuronal determination and cell polarization are reverted by modulating HD early phenotypes. Proc Natl Acad Sci USA 2018; 115: E762–E771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gomes AR, Fernandes TG, Vaz SH, et al. Modeling Rett syndrome with human patient-specific forebrain organoids. Front Cell Dev Biol 2020; 8: 610427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramirez S, Mukherjee A, Sepulveda S, et al. Modeling traumatic brain injury in human cerebral organoids. Cells 2021; 10: 2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harbuzariu A, Pitts S, Cespedes JC, et al. Modelling heme-mediated brain injury associated with cerebral malaria in human brain cortical organoids. Sci Rep 2019; 9: 19162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boisvert EM, Means RE, Michaud M, et al. Minocycline mitigates the effect of neonatal hypoxic insult on human brain organoids. Cell Death Dis 2019; 10: 325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pașca AM, Park J-Y, Shin H-W, et al. Human 3D cellular model of hypoxic brain injury of prematurity. Nat Med 2019; 25: 784–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feigin VL, Stark BA, Johnson CO, et al. Global, regional, and national burden of stroke and its risk factors, 1990–2019: a systematic analysis for the global burden of disease study 2019. The Lancet Neurology 2021; 20: 795–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kleinschnitz C, Fluri F, Schuhmann M.Animal models of ischemic stroke and their application in clinical research. Drug Des Devel Ther 2015; 9: 3445–3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holloway PM, Gavins FNE.Modeling ischemic stroke in vitro: status quo and future perspectives. Stroke 2016; 47: 561–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dirnagl U.Thomas willis lecture: is translational stroke research broken, and if so, how can we fix it? Stroke 2016; 47: 2148–2153. [DOI] [PubMed] [Google Scholar]
- 16.Percie Du Sert N, Alfieri A, Allan SM, et al. The IMPROVE guidelines (ischaemia models: procedural refinements of in vivo experiments). J Cereb Blood Flow Metab 2017; 37: 3488–3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Orset C, Haelewyn B, Allan SM, et al. Efficacy of alteplase in a mouse model of acute ischemic stroke: a retrospective pooled analysis. Stroke 2016; 47: 1312–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Llovera G, Hofmann K, Roth S, et al. Results of a preclinical randomized controlled multicenter trial (pRCT): anti-CD49d treatment for acute brain ischemia. Sci Transl Med 2015; 7: 299ra121. [DOI] [PubMed] [Google Scholar]
- 19.Pamies D, Barreras P, Block K, et al. A human brain microphysiological system derived from induced pluripotent stem cells to study neurological diseases and toxicity. Altex 2017; 34: 362–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pischiutta F, Brunelli L, Romele P, et al. Protection of brain injury by amniotic mesenchymal stromal cell-secreted metabolites. Crit Care Med 2016; 44: e1118–e1131. [DOI] [PubMed] [Google Scholar]
- 21.Li CH, Tam PKS.An iterative algorithm for minimum cross entropy thresholding. Pattern Recogn Lett 1998; 19: 771–776. [Google Scholar]
- 22.Duval K, Grover H, Han L-H, et al. Modeling physiological events in 2D vs. 3D cell culture. Physiology (Bethesda) 2017; 32: 266–277. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 23.Hopkins AM, DeSimone E, Chwalek K, et al. 3D in vitro modeling of the central nervous system. Prog Neurobiol 2015; 125: 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edmondson R, Broglie JJ, Adcock AF, et al. Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors. ASSAY Drug Dev Technol 2014; 12: 207–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Antoni D, Burckel H, Josset E, et al. Three-dimensional cell culture: a breakthrough in vivo. Int J Mol Sci 2015; 16: 5517–5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Granata A, Serrano F, Bernard WG, et al. An iPSC-derived vascular model of marfan syndrome identifies key mediators of smooth muscle cell death. Nat Genet 2017; 49: 97–109. [DOI] [PubMed] [Google Scholar]
- 27.Kelleher J, Dickinson A, Cain S, et al. Patient-specific iPSC model of a genetic vascular dementia syndrome reveals failure of mural cells to stabilize capillary structures. Stem Cell Reports 2019; 13: 817–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ling C, Liu Z, Song M, et al. Modeling CADASIL vascular pathologies with patient-derived induced pluripotent stem cells. Protein Cell 2019; 10: 249–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beltran AA, Molina SG, Marquez A, et al. Generation of an induced pluripotent stem cell line (UNCCi002-A) from a healthy donor using a non-integration system to study cerebral cavernous malformation (CCM). Stem Cell Res 2021; 54: 102421. [DOI] [PubMed] [Google Scholar]
- 30.Daviaud N, Chevalier C, Friedel RH, et al. Distinct vulnerability and resilience of human neuroprogenitor subtypes in cerebral organoid model of prenatal hypoxic injury. Front Cell Neurosci 2019; 13: 336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim MS, Kim D-H, Kang HK, et al. Modeling of hypoxic brain injury through 3D human neural organoids. Cells 2021; 10: 234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Breedam E, Nijak A, Buyle-Huybrecht T, et al. Luminescent human iPSC-derived neurospheroids enable modeling of neurotoxicity after oxygen–glucose deprivation. Neurotherapeutics 2022; 19: 550–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nielsen HH, Soares CB, Høgedal SS, et al. Acute neurofilament light chain plasma levels correlate with stroke severity and clinical outcome in ischemic stroke patients. Front Neurol 2020; 11: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tiedt S, Duering M, Barro C, et al. Serum neurofilament light a biomarker of neuroaxonal injury after ischemic stroke. Neurology 2018; 91: e1338–e1347. [DOI] [PubMed] [Google Scholar]
- 35.Gendron TF, Badi MK, Heckman MG, et al. Plasma neurofilament light predicts mortality in patients with stroke. Sci Transl Med 2020; 12: 19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuhle J, Gaiottino J, Leppert D, et al. Serum neurofilament light chain is a biomarker of human spinal cord injury severity and outcome. J Neurol Neurosurg Psychiatry 2015; 86: 273–279. [DOI] [PubMed] [Google Scholar]
- 37.Scott G, Zetterberg H, Jolly A, et al. Minocycline reduces chronic microglial activation after brain trauma but increases neurodegeneration. Brain 2018; 141: 459–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuhle J, Kropshofer H, Haering DA, et al. Blood neurofilament light chain as a biomarker of MS disease activity and treatment response. Neurology 2019; 92: e1007–e1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mages B, Aleithe S, Altmann S, et al. Impaired neurofilament integrity and neuronal morphology in different models of focal cerebral ischemia and human stroke tissue. Front Cell Neurosci 2018; 12: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mages B, Fuhs T, Aleithe S, et al. The cytoskeletal elements MAP2 and NF-L show substantial alterations in different stroke models while elevated serum levels highlight especially MAP2 as a sensitive biomarker in stroke patients. Mol Neurobiol 2021; 58: 4051–4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bacioglu M, Maia LF, Preische O, Erratum et al. Neurofilament light chain in blood and CSF as marker of disease progression in mouse models and in neurodegenerative diseases. (Neuron 2016; 91: 494–496. [DOI] [PubMed] [Google Scholar]
- 42.O’Brien WT, Pham L, Brady RD, et al. Temporal profile and utility of serum neurofilament light in a rat model of mild traumatic brain injury. Exp Neurol 2021; 341: 113698. [DOI] [PubMed] [Google Scholar]
- 43.Yue JK, Yuh EL, Korley FK, et al. Association between plasma GFAP concentrations and MRI abnormalities in patients with CT-negative traumatic brain injury in the TRACK-TBI cohort: a prospective multicentre study. Lancet Neurol 2019; 18: 953–961. [DOI] [PubMed] [Google Scholar]
- 44.Yang Z, Wang KKW.Glial fibrillary acidic protein: from intermediate filament assembly and gliosis to neurobiomarker. Trends Neurosci 2015; 38: 364–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun X-Y, Ju X-C, Li Y, et al. Generation of vascularized brain organoids to study neurovascular interactions. eLife 2022; 11: e76707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cakir B, Xiang Y, Tanaka Y, et al. Engineering of human brain organoids with a functional vascular-like system. Nat Methods 2019; 16: 1169–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ko E, Poon MLS, Park E, et al. Engineering 3D cortical spheroids for an in vitro ischemic stroke model. ACS Biomater Sci Eng 2021; 7: 3845–3860. [DOI] [PubMed] [Google Scholar]
- 48.Song G, Zhao M, Chen H, et al. The application of brain organoid technology in stroke research: challenges and prospects. Front Cell Neurosci 2021; 15: 646921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bouma MJ, Arendzen CH, Mummery CL, et al. Reprogramming urine‐derived cells using commercially available self‐replicative RNA and a single electroporation. Current Protocols in Stem Cell Biology 2020; 55: e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-jcb-10.1177_0271678X231152023 for Neural cortical organoids from self-assembling human iPSC as a model to investigate neurotoxicity in brain ischemia by Massimiliano De Paola, Francesca Pischiutta, Davide Comolli, Alessandro Mariani, Joe Kelk, Ilaria Lisi, Milica Cerovic, Stefano Fumagalli, Gianluigi Forloni, Elisa R Zanier in Journal of Cerebral Blood Flow & Metabolism
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in Zenodo repository (https://zenodo.org/deposit/7228188).