Abstract
Mpox is a zoonotic disease caused by monkeypox virus (MPXV) from the Orthopoxvirus genus. Unprecedented transmission events have led to more than 70 000 cases reported worldwide by October 2022. The change in mpox epidemiology has raised concerns of its ability to establish endemicity beyond its traditional geographical locations. In this review, we discuss the current understanding of mpox virology and viral dynamics that are relevant to mpox diagnostics. A synopsis of the traditional and emerging laboratory technologies useful for MPXV detection and in guiding “elimination” strategies is outlined in this review. Importantly, development in MPXV genomics has rapidly advanced our understanding of the role of viral evolution and adaptation in the current outbreak.
Keywords: diagnostics, genomics, monkeypox, mpox, orthopoxvirus, serology
Abbreviations
- CRISPR
clustered regularly interspaced short palindromic repeats
- LAMP
loop mediated isothermal amplification
- mpox
monkeypox
- MPXV
monkeypox virus
1. INTRODUCTION
Mpox is a zoonotic disease caused by monkeypox virus (MPXV) from the family Poxviridae, Orthopoxvirus genus, which includes variola (VARV), vaccinia (VACV), camelpox, and cowpox (CPXV) viruses. 1 Since its first discovery in 1958 from primate rash samples, periodic outbreaks of mpox have been reported in West African countries and the Congo basin. 1 The reservoir host(s) for MPXVs are uncertain but are thought to be one or more African rodents or other small mammals (e.g., Funisciurus spp., Heliosciurus spp.). 2 Apart from the incidental zoonotic transmission, MPXV can be transmitted between humans through respiratory droplets, direct contact with skin lesions, sexual contact, and through contaminated fomites 3 (Figure 1). Although vertical transmission has not been proven, fetal death has been reported. 4 The majority of mpox cases are self‐limiting, with clade‐dependent case mortality rates of 1%–10% reported in the African continent, with deaths predominantly occurring in children. 5
Figure 1.
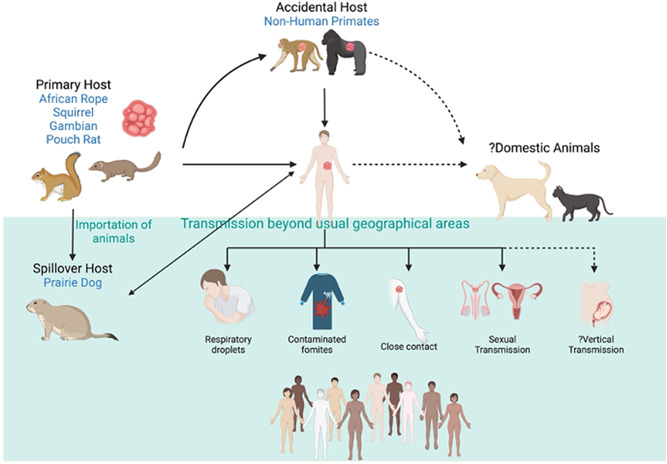
Mpox transmission cycle. Top part of the figure illustrates the traditional transmission routes, in which rodents endemic to the African continent are the primary hosts, with accidental spillover to humans or nonhuman primates through direct contact with lesions. Solid lines indicate established routes of transmission and dotted lines indicate potential routes of transmission. Bottom part of the figure (green background) indicates the most recent human‐to‐human transmissions in the 2022 outbreak (created with Biorender).
In 2003, an outbreak was reported outside the endemic regions involving multiple states in the United States, possibly originating from imported animals. Infection in prairie dogs resulted in the identification of a potential new reservoir for ongoing transmission to humans; however, the outbreak was contained and mpox did not become enzootic in the United States. In mid‐2022, a multicountry spread of mpox across Europe, the Americas, the Western Pacific, countries of the Eastern Mediterranean, and in South East Asia prompted the World Health Organization (WHO) to declare a public health emergency. 6 Unlike previous outbreaks, transmission has been observed predominantly among men who have sex with men (MSM). 7 By October 2022, over 70 000 cases of mpox were reported worldwide. Despite its morbidity, case fatality in this outbreak has been low (mortality rate <1%), with more than half of these deaths occurring in low‐resource settings within the African continent. 6 , 8
The unprecedented global transmission of mpox necessitates a rapid review of diagnostic strategies to interrupt transmission and prevent endemicity. In this review, we summarize the key diagnostic strategies that could be utilized in the prevention and control of MPX.
2. MPOX VIROLOGY
MPXV has a large genome (~200 kb) consists of linear double‐stranded DNA with a covalently closed hairpin on both 5′ and 3′ ends with inverted tandem repeats (ITS). 4 It is the largest virus known to infect humans and has a wide host range and tissue tropism. 9 Many mammalian cell lines are permissive to viral entry. 9 To date, no specific host cell receptor has been identified for viruses within the Orthopoxvirus genus. Viral entry is predominantly mediated through interactions with cell surface ligands (e.g., glycoasaminoglycans, chondroitin sulfate, and heparan sulfate) and membrane fusion. Cytoplasmic replication generates two forms of infectious virions, the intracellular mature virus (IMV) and extracellular enveloped virus (EEV). The external layer of envelope in EEV plays a role in immune evasion and transmission within host, whereas IMV is responsible for transmission between hosts. 10
3. VIRAL DYNAMICS AND SAMPLING STRATEGIES
Assessing viral dynamics (incubation, infectious period, residual DNA shedding) is critical in informing testing strategies for emerging pathogens. Assessment of the incubation period for the current mpox epidemic has been challenging due to multiple exposure events and a change in mode of transmission (i.e., predominantly sexual transmission among MSM). Historically, the incubation period for MPXV, VCV, and VRV have been estimated to be around 12–13 days for droplet or noninvasive transmission routes. 11 , 12 A shorter incubation period of 9 days was estimated for invasive exposures (mucous membrane). 13 A recent report from the Netherlands estimated the incubation period of the current mpox outbreaks to be around 8.5 days based on lognormal distribution. 14
To date, diagnosis of mpox has been largely restricted to testing of symptomatic patients with typical lesions. Multisite studies confirmed that polymerase chain reaction (PCR) testing of skin lesions has the highest yield (clinical sensitivity 91%–100%) 15 , 16 , 17 , 18 , 19 (Figure 2). The clinical sensitivity of upper respiratory specimens (oral swab, nasopharyngeal swab, and saliva) has been reported between 69% and 100% in a number of case reports. 15 , 16 , 17 , 18 Similarly, high sensitivity has been reported for testing of rectal swabs (78%–97%) and seminal fluid (77.8%–100%). 18 , 19 Positive PCR has also been reported for blood (whole blood, plasma, sera), urine, and feces although there are limited data available to estimate their clinical yield accurately. 18 Collectively, these studies suggest that for early detection of mpox (before development of skin lesions), testing at other symptomatic sites (e.g., rectal swabs in proctitis) may be helpful. Variation in the reported sensitivity of oral swabs and blood samples may be explained by the genital transmission in which oropharyngeal involvement is possibly a secondary event and may not occur in restricted genital infection in some cases. 15 To date, viral shedding dynamics in presymptomatic cases are largely unclear. Two studies performed retrospective testing of samples sent for sexually transmitted infection (STI) screening detected MPXV DNA in a small proportion of samples (2%–6.5%), predominantly from anorectal swabs and a few pharyngeal samples. 20 , 21
Figure 2.
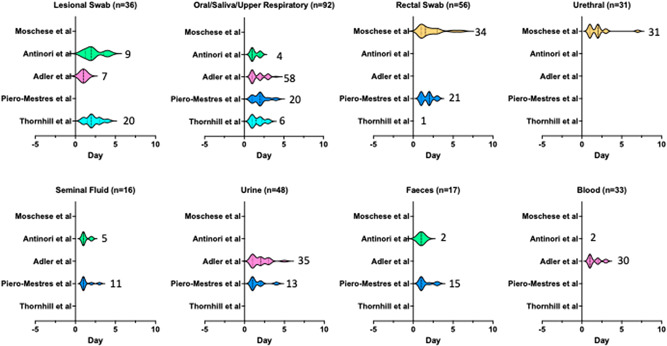
Schematic representation of mpox DNA shedding studies. Y axis indicates Monkeypox virus (MPXV) DNA dynamic studies from 2022 outbreaks which examined the association between polymerase chain reaction (PCR) positivity and symptom onset. Each violin plot represents the distribution of samples tested positive for MPXV from multiple patients over time, in relation to symptom onset (Day 0). Numbers next to the Violin plots indicate the number of PCR‐positive samples and the width of the plots correspond to the duration of DNA shedding. Solid lines within the Violin plots signify median day from symptom onset.
One major limitation of published data to date is that studies are largely restricted to early disease sampling. In the early symptomatic period, most patients would have positive MPXV PCR from many body sites (Figure 2). Prolonged shedding of more than 21 days has been reported in oral, nasopharyngeal, rectal swabs, skin lesions, and urine but whether this reflects chronicity of infection or residual DNA shedding is still unclear. 17 Presence of other STI pathogens is also very common in the 2022 outbreak and detection of an alternative pathogen should not be used to exclude mpox infection. 22 , 23 , 24
4. NUCLEIC ACID DETECTION
Most of the available molecular diagnostic assays for MPXV are based on real‐time quantitative PCR (qPCR), with a few Loop mediated isothermal amplification (LAMP)‐based assays in development. 25 , 26 , 27 A summary of molecular assays and platforms (commercially available and in development) is provided in Supporting Information: Table S1. Published primers and probes were designed around the conserved regions of the central coding region (E9L‐NVAR within DNA polymerase gene, B6R within the envelope protein gene and F3L within the ORF) and the ITS region (G2R within the tumor necrosis gene). 28 , 29 A large number of commercial reagent kits are now available for Orthopoxvirus or MPXV detection with similar analytical sensitivity and specificity, although information on clinical performance and sample type evaluation is limited 25 , 30 (Table S1). Multiplexing Orthopoxvirus and MPXV detection has the advantage of differentiating natural infection from disseminated VACV in countries using replication‐competent VACV vaccine (e.g., ACAM2000, Dryvax) in ring vaccination of close contacts. In addition, these assays can be readily applied using existing qPCR platforms for high‐throughput testing, without going through extensive assay validation to meet regulatory requirements. Clade‐specific assays in the format of multiplexed PCR and LAMP had also been described. 31 , 32 These assays designed their primers and probes to detect MPXV genomic regions (F3L and G2R) with a few nucleotide differences between Clades I, IIa, and IIb. Given the rapid accumulation of mutations in the 2022 outbreak, ongoing evaluation of primer and probe dropout should be considered if adopting this approach. Post‐COVID‐19, many laboratories in endemic and nonendemic countries are now equipped with modular or all‐in‐one fully automated platforms (e.g., Hologic Panther Fusion, Abbott Alinity M) (Table S1), which allow random access. 33 Some of these platforms also permit the use of laboratory‐developed tests. The current gap in testing, however, is the limited number of rapid molecular point‐of‐care tests (POCT) that can be deployed to testing clinics and remote regions in mpox endemic countries.
In resource‐limited settings, LAMP‐based assay is an attractive tool, where access to thermocycler, stable electricity supply, and temperature control is challenging. Despite the ease of use, many traditional LAMP‐based assays still require a laboratory set up and suffers from complex primer and probe design. Advancements in isothermal technologies such as combining the use of recombinase polymerase amplification (RPA) with clustered regularly interspaced short palindromic repeats (CRISPR)‐based technologies allow both simplification of procedures with improvement in assay sensitivity. 27 , 34 , 35 A recent report of combining an RPA‐CRISPR‐cas12a technology with lateral flow assay (LFA) read out shows promises in adapting this technology as a true POCT. 36
5. POCT
A number of rapid antigen and antibody tests have been developed in the format of LFAs for a range of specimen types (serum, plasma, lesion fluid, oropharyngeal swab). 25 To date, both the analytical and clinical performance of these assays are unclear. In particular, the utility of RAT from nonlesional sites with lower viral loads requires rigorous assessment. The extent and implication of false positive and false negative results should also be carefully considered when used, particularly if the goal of testing is to detect every case as part of a public health “elimination” strategy.
Most of the shortcomings of antigen detection assays can be overcome with recent development in nucleic acid POCTs, such as the CRISPR‐based LFA for SARS‐CoV‐2 detection. This technology utilizes isothermal amplification and fluorescein labeling of target nucleic acid followed by capturing with Biotin‐labeled CRISPR‐Cas ribonucleoprotein complex. Secondary antibody capture of this complex allows visualization on LFA strip. Analytical sensitivity of this assay can rival conventional real‐time PCR for a number of viruses. 37 , 38 , 39 , 40 Modification of CRISPR‐Cas enzymes to capture DNA can be readily performed for MPXV detection.
6. VIRAL CULTURE AND PHENOTYPIC DRUG SUSCEPTIBILITY TESTING
MPXV culture should be performed in a high containment laboratory (PC3/BSL3) and VACV vaccine is generally recommended for laboratory staff performing this work. MPXV can be readily cultured in a number of human (HEK293, HeLa, A549, MRC‐5), nonhuman primate (NHP) (Vero, Vero E6, MK2, MA104, RMK), and mammalian (RK13)‐derived cell lines. 41 , 42 , 43 Routine liquid transport media (VTM, UTM) are generally suitable, whereas swabs containing semisolid media or additives (e.g., gel, charcoal) could potentially be cytotoxic to cells. Extensive cytopathic effects (CPE) can be observed in as early as 2–3 days postinoculation, often with widespread cellular detachment and degeneration. A more distinct CPE pattern with infectious foci is seen when using cell culture passaged virus. When using primary inoculum from lesional samples, MPXV CPE appears to be more extensive than herpesviruses (e.g., HSV and VZV). 44 MPXV can also be cultured from nonlesional sites (oral, pharyngeal, rectal) but with higher risks of bacterial contamination.
Although viral culture may not be used routinely in the diagnostic testing of MPXV, it is an essential part of laboratory outbreak response, particularly in the generation of positive control material for NAAT and serological assay development. Viral culture has also been used in public health studies as a surrogate for putative infectivity, especially in understanding the viral infectivity period in body fluids (e.g., seminal fluid) 45 and the significance of fomite transmission. 46 One correlation study suggests that qPCR C t of ≥35 or ≥DNA 4300 copies/ml corresponds with non‐ or marginal infectivity. 47 Due to a lack of standardization of MPXV qPCR, this finding may not be directly generalizable to other laboratories. 29 Other important applications of MPXV culture include phenotypic susceptibility testing for antivirals, assessment of population immunity through neutralization assays, and assessment of cellular/host tropism in identifying the potential organ or animal reservoir. 9
Phenotypic drug susceptibility assays performed for in vitro assessment of antiviral susceptibility to Orthopoxviruses are mostly in the format of plaque reduction assay. 48 For VARV, resistance to Cidofovir and Brincidofovir has been attributed to mutations in the DNA polymerase gene (F8L—A314T and A684V), conferring an increase of at least fivefold in EC50. 49 These mutations result in morphological alterations to cidofovir‐triphosphate binding sites. As A314 and A684 amino acid sequence is highly conserved within the genus, mutations in these regions are likely to result in resistance to all Orthopoxviruses. Similarly, Tecovirimat (ST‐246) resistance can be assessed using plaque reduction assays and in vitro resistance has been mapped to VP13, a protein responsible for viral transportation. 50 , 51 , 52 A number of VP37 amino acid substitutions have been linked to high‐level resistance in cell culture (≥10‐fold increase in EC50) although clinical data is lacking. 53
7. SEROLOGICAL ASSAYS
Serological assessment of mpox infection is useful in a number of settings, for example, identification of self‐attenuated infection, assessing population seroprevalence to determine asymptomatic infection, and assessment of population immunity. However, mpox serology is not extensively utilized in diagnostic laboratories due to lack of commercial assays. There are two major challenges in mpox serological development, (1) limited availability of MPXV antigens/inactivated viral particles and (2) serological cross‐reactivity between Orthopoxviruses. 54 Despite this, a number of in‐house assays (e.g., immunofluorescence assay [IFA], enzyme‐linked immunosorbent assay [ELISA], hemagglutination inhibition assay [HAI]) have been developed over the years to meet the demand for surveillance and to differentiate vaccination from natural infection (Table 1). Previous serodynamic studies using ELISA suggest that IgM may persists for weeks to months, whereas IgG persists for years. 56 , 62 , 64
Table 1.
Orthopoxvirus serological assays and applications
| Assay | Complexity | Scalability | Application | References | ||
|---|---|---|---|---|---|---|
| Assess recent versus distant infection | Differentiate natural infection versus immunization | Assessment of humoral immunity | ||||
| IFA | ++ | +++ | Yes | No | No | [55] |
| ELISA | + | ++++ | Yes | No | No | [56] |
| Post‐adsorption ELISA | ++ | +++ | Yes | Yes | No | [57] |
| Peptide‐based ELISA (IgG) | ++ | + | No | Yes | No | [58] |
| Neutralization assay | +++ | + | No | No | Yes | [59] |
| HAI | + | +++ | Yes | No | No | [60] |
| Postadsorption gel precipitation | ++ | ++ | No | Yes | No | [61] |
| Complement fixation | + | +++ | Yes | No | No | [54] |
| Postadsorption raiodimmunoassay | ++ | ++ | No | Yes | No | [62] |
| Western blot | ++ | + | No | Yes | No | [57] |
| Cross neutralization | ++ | ++ | No | Yes | Yes | [63] |
Abbreviations: ELISA, enzyme‐linked immunosorbent assay; HAI, hemagglutination inhibition assay; IFA, immunofluorescence assay.
Most of the published differentiation assays employ preadsorption or cross‐adsorption with VACV and/or MPXV antigens (Table 1). Postadsorption ELISA is performed using plasma samples, separately preadsorbed with equivalent amounts of high titer, inactivated VACV, and MPXV cell lysate (6 × 108 plaque forming unit equivalents/ml). A differential fold change of anti‐MPXV IgG >2.5 between post‐MPXV and post‐VACV adsorptions generally correlates with mpox infection (with or without vaccination). A lack of fold difference is seen mainly in VACV‐immune individuals but also in a small proportion of individuals immune to both viruses. 43 Differential Western nlot described by Dubois et al. 57 detects three MPX‐specific protein bands, 39, 124, and 148 kDa proteins. When analyzed individually, 39 kDa reactivity has the highest sensitivity for primary and secondary infections (100% and 75%, respectively) but with a reduced specificity of 70%–80%. 124 and 148 kDa bands have high specificity but with a significantly compromised sensitivity. Overall, differential Western nlot may provide additional serological confirmation but with restricted diagnostic utility as a standalone test due to limited scalability and complexity in interpretation.
Although the mechanism underlying cross‐protective immunity to MPXV from VACV immunization is complex, there is evidence from human and NHP challenge studies that polyclonal neutralizing antibodies (Nab) play an important role. 65 , 66 , 67 , 68 Most commonly used Nab assays to Orthopoxviruses are based on plaque reduction neutralization test (PRNT) against IMV, as Nab assays using EEV are more challenging to develop. 69 Higher throughput PRNT assays using recombinant VACV expressing reporter genes (β‐galactosidase or green fluorescent protein) can be used to assess Nab to various Orthopoxviruses. 70 At present, there is a lack of human correlation studies to inform the level of Nab that correlates with clinical protection. Evidence from small animal lethal infection studies suggests that the presence of mixed antibodies and diversity of target epitope towards both EV and MV correlate best with protection. 71
8. TRANSMISSION ELECTRON MICROSCOPY (TEM)
The use of TEM in the differential diagnosis of herpesvirus and poxvirus infection dates back to 1947, during a smallpox outbreak in the United States. 72 Indeed, the direct examination of vesicular fluid using negative contrast TEM remains a current and appropriate methodology for the initial rapid identification of poxvirus virions in clinical samples. 73 Using modern TEM protocols and equipment, it is possible to confirm MPXV infection from high titer samples, such as vesicle fluids, in under 30 min, rivaling turn‐around times utilizing rapid PCR methods. Rapid TEM negative‐contrast examinations of vesicular material can be of use to differentiate members of the genus Orthopoxvirus from the more commonly encountered members of the genus Parapoxvirus (e.g., Orf virus) or genus Molluscipoxvirus (e.g., Molluscum contagiosum virus).
Given the biohazard‐related considerations associated with the handling of suspected or confirmed MPXV samples and derivatives, there are limitations regarding the extent to which TEM examination can progress. For instance, cryogenic electron microscopy protocols that have been used to successfully examine the three‐dimensional structure of related Orthopoxviruses such as VACV, 74 are complicated by risk‐group three associated handling requirements of high‐titer preparations, necessary for such procedures. Traditional TEM protocols, such as negative‐contrast staining of fluid samples or culture supernatants (Figure 3D) and thin‐sectioning of tissues (Figure 3A,B) are more amenable to TEM observation of morphology and morphogenesis, as the requirement for inactivation using glutaraldehyde and/or paraformaldehyde is easily integrated into routine TEM examination protocols. Samples prepared using routine thin‐sectioning protocols are also suitable for the examination of thick‐sections (≥150 nm) using TEM tomography; this permits the three‐dimensional reconstruction of virus particles and associated cellular ultrastructure (Figure 3C).
Figure 3.
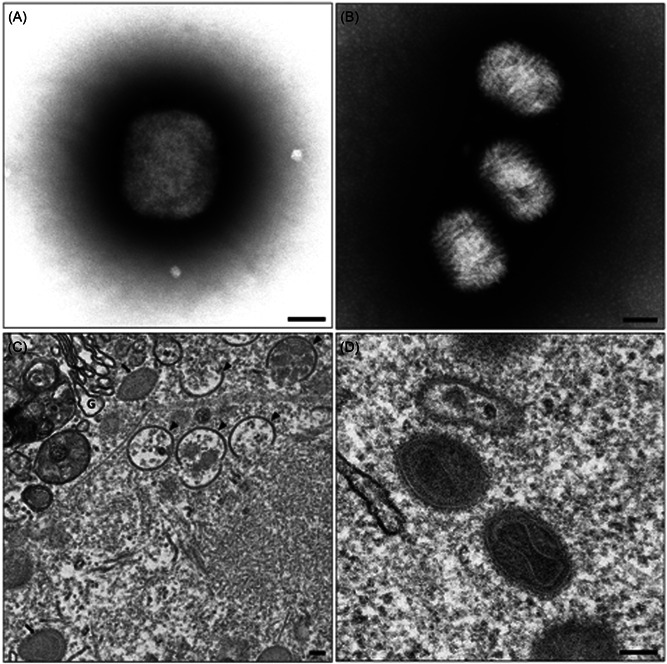
Transmission electron micrographs provided by Dr. Jason Roberts. Negative‐stained Monkeypox virus (MPXV) positive cell culture supernatant, showing an approximately 250 nm × 300 nm brick‐like particle indicative of the genus Orthopoxvirus, (B) Negative‐stained molluscum contagiosum positive material showing multiple 190 nm × 260 nm ovoid particles with a woven appearance of surface tubules characteristic of the genus Molluscipoxvirus, (C) cytoplasmic region of an MPXV infected Vero cell showing a typical virus factory or virosome, intracellular mature virus particles (black arrows) and intermediate crescent stage immature virus particles (black arrowheads) are visible, G = Golgi body, (D) multiple intracellular mature MPXV particles can be seen with clearly defined dumbbell‐shaped core and striated palisade layer between inner and outer membranes. Scale bar = 100 nm.
9. MPOX GENOMICS
A number of methods can be employed in the direct sequencing of clinical samples containing MPXV, including PCR amplicon, hybridization‐probe capture, and metagenomics sequencing, largely due to the high viral load presence in cutaneous lesions. 75 , 76 , 77 Although the yield may not differ between methods in high viral load samples, PCR amplicon sequencing has the advantage of cost‐effectiveness and better results (genomic coverage) for low viral load samples (e.g., nonlesional body sites). 77 However, it is yet unclear of the extent and frequency of primer dropout in amplicon sequencing in the current outbreak. Currently, there is no head‐to‐head comparison of hybridization capture and PCR amplicon approach. Nanopore sequencing is an appealing platform due to its portability and potential adaptation to field testing; however, additional consideration in using data polishing tools (Canu, Medaka, etc.) to correct indels should be considered due to issues with base‐calling in homopolymer‐rich regions. 76 In addition, due to the presence of repetitive and hypervariable regions in the ITS, masking of the first 1500 and 7000 bp of the genome is often performed to prevent any read mapping error. 78 Open source bioinformatics workflow (masking, pairwise alignments to reference sequence, phylogenetic reconstruction, clade and lineage assignments) is available through Nextclade. 79
The 2022 outbreak has also prompted a nonstigmatizing reclassification of MPXV genomics into Clade I (previous Congo Basin), IIa (West African), and IIb (2022 outbreak), which is endorsed by WHO. 80 , 81 As a characterization of the MPXV genomics for the current outbreak is underway, early studies suggest evolutionary changes to MPXV genome may potentially influence transmissibility and outbreak projections. 82 , 83 Divergence analyses of the 2022 outbreak virus (Lineage B.1) showed approximately 50 SNPs difference from its ancestral lineage (lineage A.1) from virus isolated in 2018 and 2019 (Nigeria, United Kingdom, Israel, and Singapore). 82 A significant increase in the previously established substitution rates (background of 1–2 SNPs/genome/year) 84 and the preferential mutational pattern (GA > AA, TC > TT) are suggestive of viral adaptation, possibly through genome editing by APOBEC3A. 83 , 85 , 86 While it is unclear if these changes were driven by human or animal adaption, additional phenotypic studies are required to characterize the transmissibility of these mutational changes. The utility of genomics in case‐linkage and informing diagnostics and therapeutics require further research.
10. CURRENT GAPS AND FUTURE DIRECTIONS
As the evidence for viral adaptation is emerging, there is a high possibility of mpox establishing endemicity beyond its historical geographical locations. Improving accessibility to testing is therefore critical in worldwide eradication efforts. A recent survey conducted in Europe showed that most European countries have effective mpox outbreak management and testing capabilities and a robust surveillance system. 87 In the African continent, PCR testing capability has also improved since the COVID‐19 pandemic, but sequencing service is only available in seven countries. 33 Other factors that hinder an effective outbreak response (e.g., accessibility to PCR in remote areas, training, surveillance system, vaccines, and therapeutics) continue to pose a challenge for the eradication of mpox in the endemic regions. 33
To date, PCR remains the cornerstone of mpox testing but the use of more accessible, lower sensitivity assays (e.g., rapid antigen tests) should also be considered if PCR testing is not available due to capacity or geographical restrictions. Further effort of assay standardization is equally important and may help increase the applicability of data generated from culture correlation studies. Advances in technologies and innovative approaches developed during SARS‐CoV‐2 pandemics, for example, CRISPR‐based POCT, microfluidics, Virolens system, biosensors, and wastewater surveillance could be customized to MPXV testing. 88 , 89 Finally, the incorporation of both genotypic and phenotypic assays for viral characterization is critical for an adaptive public health response to evolving viral epidemiology.
AUTHOR CONTRIBUTIONS
Chuan Kok Lim, Eike Steinig, Mona L. Taouk, and Deborah Ann Williamson planned and determined the scope of the manuscript. Chuan Kok Lim, Jason Roberts, Michael Moso, Kwee Chin Liew, Eike Steinig, and Eloise Williams wrote the manuscript. Chuan Kok Lim and Jason Roberts generated figures and tables for the manuscript. Thomas Tran, Mona L. Taouk, Eike Steinig, Leon Caly, Eloise Williams, and Deborah Ann Williamson reviewed the manuscript.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supporting information
Supplementary information.
ACKNOWLEDGMENTS
The authors acknowledge the work performed by VIDRL staff, particularly the data generated by the Viral Identification, Serology and Translational Diagnostics Laboratories. The authors also acknowledge the contribution of clinical staff from Melbourne Sexual Health Clinic and the patients affected by mpox. Open access publishing facilitated The University of Melbourne, Australia as part of the Wiley ‐ The University of University of Melbourne, Australia agreement via the Council of Australian University Librarians.
Lim CK, Roberts J, Moso M, et al. Mpox diagnostics: review of current and emerging technologies. J Med Virol. 2023;95:e28429. 10.1002/jmv.28429
DATA AVAILABILITY STATEMENT
The data that supports the findings of this study are available in the supplementary material of this article.
REFERENCES
- 1. Brown K, Leggat PA. Human monkeypox: current state of knowledge and implications for the future. Trop Med Infect Dis. 2016;1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Spickler AR 2022. Accessed September 19, 2022. http://www.cfsph.iastate.edu/DiseaseInfo/factsheets.php
- 3. Monkeypox: Background information. UK Health Security Agency, Public Health England; 2022. [Google Scholar]
- 4. Family: Poxviridae, Chapter Version: ICTV Ninth Report. Accessed September 19, 2022. https://ictv.global/report_9th/dsDNA/poxviridae [Google Scholar]
- 5. Bunge EM, Hoet B, Chen L, et al. The changing epidemiology of human monkeypox‐A potential threat? A systematic review. PLoS Neglected Trop Dis. 2022;16:e0010141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. WHO. Multi‐Country Outbreak of Monkeypox, External Situation Report #5‐7 September. Vol 2022. WHO; 2022. [Google Scholar]
- 7. Liu X, Zhu Z, He Y, et al. Monkeypox claims new victims: the outbreak in men who have sex with men. Infect Dis Poverty. 2022;11:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mathieu ES, Dattani F, Ritchie S, Roser HM. Monkeypox. Our World in Data. 2022. Accessed September 19, 2022. OurWorldInData.org. Global Health. [Google Scholar]
- 9. McFadden G. Poxvirus tropism. Nat Rev Microbiol. 2005;3:201‐213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Keasey S, Pugh C, Tikhonov A, et al. Proteomic basis of the antibody response to monkeypox virus infection examined in cynomolgus macaques and a comparison to human smallpox vaccination. PLoS One. 2010;5:e15547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nishiura H. Determination of the appropriate quarantine period following smallpox exposure: an objective approach using the incubation period distribution. Int J Hyg Environ Health. 2009;212:97‐104. [DOI] [PubMed] [Google Scholar]
- 12. Eichner M. Transmission potential of smallpox: estimates based on detailed data from an outbreak. Am J Epidemiol. 2003;158:110‐117. [DOI] [PubMed] [Google Scholar]
- 13. Reynolds MG, Yorita KL, Kuehnert MJ, et al. Clinical manifestations of human monkeypox influenced by route of infection. J Infect Dis. 2006;194:773‐780. [DOI] [PubMed] [Google Scholar]
- 14. Miura F, van Ewijk CE, Backer JA, et al. Estimated incubation period for monkeypox cases confirmed in the Netherlands, May 2022. Euro Surveill. 2022;27(24):2200448. 10.2807/1560-7917.ES.2022.27.24.2200448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Thornhill JP, Barkati S, Walmsley S, et al. Monkeypox virus infection in humans across 16 countries—April‐June 2022. N Engl J Med. 2022;387:679‐691. [DOI] [PubMed] [Google Scholar]
- 16. Peiro‐Mestres A, Fuertes I, Camprubi‐Ferrer D, et al. Frequent detection of monkeypox virus DNA in saliva, semen, and other clinical samples from 12 patients, Barcelona, Spain, May to June 2022. Euro Surveill. 2022;27(28):2200503. 10.2807/1560-7917.ES.2022.27.28.2200503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Adler H, Gould S, Hine P, et al. Clinical features and management of human monkeypox: a retrospective observational study in the UK. Lancet Infect Dis. 2022;22:1153‐1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Antinori A, Mazzotta V, Vita S, et al. Epidemiological, clinical and virological characteristics of four cases of monkeypox support transmission through sexual contact, Italy, May 2022. Euro Surveill. 2022;27(22):2200421. 10.2807/1560-7917.ES.2022.27.22.2200421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Moschese D, Pozza G, Mileto D, et al. Isolation of viable monkeypox virus from anal and urethral swabs, Italy, May to July 2022. Euro Surveill. 2022;27(36):2200675. 10.2807/1560-7917.ES.2022.27.36.2200675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ferre VM, Bachelard A, Zaidi M, et al. Detection of monkeypox virus in anorectal swabs from asymptomatic men who have sex with men in a sexually transmitted infection screening program in Paris, France. Ann Intern Med. 2022;175(10):1491-1492. 10.7326/M22-2183 [DOI] [PubMed] [Google Scholar]
- 21. De Baetselier I, Van Dijck C, Kenyon C, et al. Retrospective detection of asymptomatic monkeypox virus infections among male sexual health clinic attendees in Belgium. Nature Med. 2022;28:2288‐2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bížová B, Veselý D, Trojánek M, Rob F. Coinfection of syphilis and monkeypox in HIV positive man in Prague, Czech Republic. Travel Med Infect Dis. 2022;49:102368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Heskin J, Belfield A, Milne C, et al. Transmission of monkeypox virus through sexual contact—a novel route of infection. J Infect. 2022;85:334‐363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hammerschlag Y, MacLeod G, Papadakis G, et al. Monkeypox infection presenting as genital rash, Australia, May 2022. Euro Surveill. 2022;27(22):2200411. 10.2807/1560-7917.ES.2022.27.22.2200411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. FindDx . 2022. Accessed October 16, 2022. https://www.finddx.org/mpx-test-directory/
- 26. Iizuka I, Saijo M, Shiota T, et al. Loop‐mediated isothermal amplification‐based diagnostic assay for monkeypox virus infections. J Med Virol. 2009;81:1102‐1108. [DOI] [PubMed] [Google Scholar]
- 27. Davi SD, Kissenkötter J, Faye M, et al. Recombinase polymerase amplification assay for rapid detection of Monkeypox virus. Diagn Microbiol Infect Dis. 2019;95:41‐45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dabrowski PW, Radonić A, Kurth A, Nitsche A. Genome‐wide comparison of cowpox viruses reveals a new clade related to Variola virus. PLoS One. 2013;8:e79953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Huggett JF, French D, O'Sullivan DM, et al. Monkeypox: another test for PCR. Euro Surveill. 27(32):2200497. 10.2807/1560-7917.ES.2022.27.32.2200497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Michel J, Targosz A, Rinner T, et al. Evaluation of 11 commercially available PCR kits for the detection of monkeypox virus DNA, Berlin, July to September 2022. Euro Surveill. 2022;27(45):pii=2200816. 10.2807/1560-7917.ES.2022.27.45.2200816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huo S, Chen Y, Lu R, et al. Development of two multiplex real‐time PCR assays for simultaneous detection and differentiation of monkeypox virus IIa, IIb, and I clades and the B.1 lineage. Biosaf Health. 2022;4(6):392-398. 10.1016/j.bsheal.2022.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mao L, Ying J, Selekon B, et al. Development and characterization of recombinase‐based isothermal amplification assays (RPA/RAA) for the rapid detection of monkeypox virus. Viruses. 2022;14:2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Moyo E, Musuka G, Murewanhema G, Moyo P, Dzinamarira T. Monkeypox outbreak: a perspective on Africa's diagnostic and containment capacity. Int J Infect Dis. 2022;123:127‐130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Piepenburg O, Williams CH, Stemple DL, Armes NA. DNA detection using recombination proteins. PLoS Biol. 2006;4:e204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Glökler J, Lim TS, Ida J, Frohme M. Isothermal amplifications—a comprehensive review on current methods. Crit Rev Biochem Mol Biol. 2021;56:543‐586. [DOI] [PubMed] [Google Scholar]
- 36. Zhao FWP, Wang H, Liu S, et al. CRISPR/Cas12a‐mediated ultrasensitive and on‐site monkeypox viral testing. medRxiv. 2022. 10.1101/2022.10.10.22280931 [DOI] [PubMed] [Google Scholar]
- 37. Yuan T, Mukama O, Li Z, et al. A rapid and sensitive CRISPR/Cas12a based lateral flow biosensor for the detection of Epstein‐Barr virus. Analyst (Lond). 2020;145:6388‐6394. [DOI] [PubMed] [Google Scholar]
- 38. Soh JH, Balleza E, Abdul Rahim MN, et al. CRISPR‐based systems for sensitive and rapid on‐site COVID‐19 diagnostics. Trends Biotechnol. 2022;40:1346‐1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Broughton JP, Deng X, Yu G, et al. CRISPR‐Cas12‐based detection of SARS‐CoV‐2. Nature Biotechnol. 2020;38:870‐874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ali Z, Sánchez E, Tehseen M, et al. Bio‐SCAN: a CRISPR/dCas9‐based lateral flow assay for rapid, specific, and sensitive detection of SARS‐CoV‐2. ACS Synth Biol. 2022;11:406‐419. [DOI] [PubMed] [Google Scholar]
- 41. Gong Q, Wang C, Chuai X, Chiu S. Monkeypox virus: a re‐emergent threat to humans. Virol Sin. 2022;37:477‐482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Realegeno S, Puschnik AS, Kumar A, et al. Monkeypox virus host factor screen using haploid cells identifies essential role of GARP complex in extracellular virus formation. J Virol. 2017;91:e00011‐e00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cho CT, Wenner HA. In vitro growth characteristics of monkeypox virus. Exp Biol Med. 1972;139:916‐920. [DOI] [PubMed] [Google Scholar]
- 44. Mazzotta V, Mondi A, Carletti F, et al. Ocular involvement in monkeypox: description of an unusual presentation during the current outbreak. J Infect. 2022;85:573‐607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lapa D, Carletti F, Mazzotta V, et al. Monkeypox virus isolation from a semen sample collected in the early phase of infection in a patient with prolonged seminal viral shedding. Lancet Infect Dis. 2022;22:1267‐1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pfeiffer JA, Collingwood A, Rider LE, et al. High‐contact object and surface contamination in a household of persons with monkeypox virus infection—Utah, June 2022. MMWR Morb Mortal Wkly Rep. 2022;71:1092‐1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Paran N, Yahalom‐Ronen Y, Shifman O, et al. Monkeypox DNA levels correlate with virus infectivity in clinical samples, Israel, 2022. Euro Surveill. 2022;27(35):2200636. 10.2807/1560-7917.ES.2022.27.35.2200636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Andrei G, Gammon DB, Fiten P, et al. Cidofovir resistance in vaccinia virus is linked to diminished virulence in mice. J Virol. 2006;80:9391‐9401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kannan SR, Sachdev S, Reddy AS, et al. Mutations in the monkeypox virus replication complex: potential contributing factors to the 2022 outbreak. J Autoimmun. 2022;133:102928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Grosenbach DW, Jordan R, Hruby DE. Development of the small‐molecule antiviral ST‐246 as a smallpox therapeutic. Future Virol. 2011;6:653‐671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Duraffour S, Lorenzo MM, Zöller G, et al. ST‐246 is a key antiviral to inhibit the viral F13L phospholipase, one of the essential proteins for orthopoxvirus wrapping. J Antimicrob Chemother. 2015;70:1367‐1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Berhanu A, Prigge JT, Silvera PM, Honeychurch KM, Hruby DE, Grosenbach DW. Treatment with the smallpox antiviral tecovirimat (ST‐246) alone or in combination with ACAM2000 vaccination is effective as a postsymptomatic therapy for monkeypox virus infection. Antimicrob Agents Chemother. 2015;59:4296‐4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. US FDA. FDA Monkeypox Response. US FDA; 2002. https://www.fda.gov [Google Scholar]
- 54. Esposito JJ, Obijeski JF, Nakano JH. Serological relatedness of monkeypox, variola, and vaccinia viruses. J Med Virol. 1977;1:35‐47. [DOI] [PubMed] [Google Scholar]
- 55. Gispen R, Huisman J, Brand‐Saathof B, Hekker AC. Immunofluorescence test for persistent poxvirus antibodies. Arch Gesamte Virusforsch. 1974;44:391‐395. [DOI] [PubMed] [Google Scholar]
- 56. Karem KL, Reynolds M, Braden Z, et al. Characterization of acute‐phase humoral immunity to monkeypox: use of immunoglobulin M enzyme‐linked immunosorbent assay for detection of monkeypox infection during the 2003 North American outbreak. Clin Vaccine Immunol. 2005;12:867‐872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dubois ME, Slifka MK. Retrospective analysis of monkeypox infection. Emerging Infect Dis. 2008;14:592‐599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Dubois ME, Hammarlund E, Slifka MK. Optimization of peptide‐based ELISA for serological diagnostics: a retrospective study of human monkeypox infection. Vector Borne Zoonotic Dis. 2012;12:400‐409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kramski M, Drozd A, Lichtfuss GF, Dabrowski PW, Ellerbrok H. Rapid detection of anti‐vaccinia virus neutralizing antibodies. Virol J. 2011;8:139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Jezek Z, Nakano JH, Arita I, Mutombo M, Szczeniowski M, Dunn C. Serological survey for human monkeypox infections in a selected population in Zaire. J Trop Med Hyg. 1987;90:31‐38. [PubMed] [Google Scholar]
- 61. Esposito JJ, Obijeski JF, Nakano JH. The virion and soluble antigen proteins of variola, monkeypox, and vaccinia viruses. J Med Virol. 1977;1:95‐110. [DOI] [PubMed] [Google Scholar]
- 62. Hammarlund E, Lewis MW, Carter SV, et al. Multiple diagnostic techniques identify previously vaccinated individuals with protective immunity against monkeypox. Nature Med. 2005;11:1005‐1011. [DOI] [PubMed] [Google Scholar]
- 63. Baxby D. The surface antigens of orthopoxviruses detected by cross‐neutralization tests on cross‐absorbed antisera. J Gen Virol. 1982;58:251‐262. [DOI] [PubMed] [Google Scholar]
- 64. Taub DD, Ershler WB, Janowski M, et al. Immunity from smallpox vaccine persists for decades: a longitudinal study. Am J Med. 2008;121:1058‐1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kempe CH, Bowles C, Meiklejohn G, et al. The use of vaccinia hyperimmune gamma‐globulin in the prophylaxis of smallpox. Bull World Health Organ. 1961;25:41‐48. [PMC free article] [PubMed] [Google Scholar]
- 66. Belyakov IM, Earl P, Dzutsev A, et al. Shared modes of protection against poxvirus infection by attenuated and conventional smallpox vaccine viruses. Proc Natl Acad Sci USA. 2003;100:9458‐9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Edghill‐Smith Y, Golding H, Manischewitz J, et al. Smallpox vaccine‐induced antibodies are necessary and sufficient for protection against monkeypox virus. Nature Med. 2005;11:740‐747. [DOI] [PubMed] [Google Scholar]
- 68. Gilchuk I, Gilchuk P, Sapparapu G, et al. Cross‐Neutralizing and protective human antibody specificities to poxvirus infections. Cell. 2016;167:684‐694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Vanderplasschen A, Hollinshead M, Smith GL. Antibodies against vaccinia virus do not neutralize extracellular enveloped virus but prevent virus release from infected cells and comet formation. J Gen Virol. 1997;78(Pt 8):2041‐2048. [DOI] [PubMed] [Google Scholar]
- 70. Gates I, Olson V, Smith S, Patel N, Damon I, Karem K. Development of a high‐content orthopoxvirus infectivity and neutralization assays. PLoS One. 2015;10:e0138836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Moss B. Smallpox vaccines: targets of protective immunity. Immunol Rev. 2011;239:8‐26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Nagler FPO, Rake G. The use of the electron microscope in diagnosis of variola, vaccinia, and varicella. J Bacteriol. 1948;55:45‐51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Gelderblom H. Specimen collection for electron microscopy. Emerging Infect Dis. 2000;6:433‐434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Cyrklaff M, Risco C, Fernández JJ, et al. Cryo‐electron tomography of vaccinia virus. Proc Natl Acad Sci USA. 2005;102:2772‐2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Chen N, Li G, Liszewski MK, et al. Virulence differences between monkeypox virus isolates from West Africa and the Congo basin. Virology. 2005;340:46‐63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Vandenbogaert M, Kwasiborski A, Gonofio E, et al. Nanopore sequencing of a monkeypox virus strain isolated from a pustular lesion in the Central African Republic. Sci Rep. 2022;12:10768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Chen NFG, Gagne L, Doucette M, et al. Monkeypox virus multiplexed PCR amplicon sequencing (PrimalSeq). protocols. 2022. Accessed September 19, 2022. https://www.protocols.io/view/monkeypox-virus-multiplexed-pcr-amplicon-sequencin-5qpvob1nbl4o/v1?version_wwarning=no
- 78. Gigante CM, Korber B, Seabolt MH, et al. Multiple lineages of monkeypox virus detected in the United States, 2021‐2022. Science. 2022;378:560‐565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nextclade. Monkeypox. Vol 2022. Github. Accessed October 19, 2022. https://github.com/nextstrain/monkeypox
- 80. Happi C, Adetifa I, Mbala P, et al. Urgent need for a non‐discriminatory and non‐stigmatizing nomenclature for monkeypox virus. PLoS Biol. 2022;20(8):e3001769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. WHO. Monkeypox: Experts Give Virus Variants New Names. WHO; 2022. [Google Scholar]
- 82. Isidro J, Borges V, Pinto M, et al. Phylogenomic characterization and signs of microevolution in the 2022 multi‐country outbreak of monkeypox virus. Nature Med. 2022;28:1569‐1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. O'Toole AR, Rambaut A. Initial observations about putative APOBEC3 deaminase editing driving short‐term evolution of MPXV since 2017. Virological. 2022. https://virological.org/t/initial-observations-about-putative-apobec3-deaminase-editing-driving-short-term-evolution-of-mpxv-since-2017.830 [Google Scholar]
- 84. Firth C, Kitchen A, Shapiro B, Suchard MA, Holmes EC, Rambaut A. Using time‐structured data to estimate evolutionary rates of double‐stranded DNA viruses. Mol Biol Evol. 2010;27:2038‐2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Crystal M, Gigante MP, Ali Ruprecht, et al. Genomic deletions and rearrangements in monkeypox virus from the 2022 outbreak, USA. bioRxiv. 2022. 10.1101/2022.09.16.508251 [DOI] [Google Scholar]
- 86. Gigante CrystalM, B, K , Seabolt MatthewH, Wilkins Kimberly, et al. Multiple lineages of Monkeypox virus detected in the United States, 2021‐ 2022. Science. 2022;378:6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Grothe JH, Cornely OA, Salmanton‐García J, et al. Monkeypox diagnostic and treatment capacity at epidemic onset: a VACCELERATE online survey. J Infect Public Health. 2022;15:1043‐1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. de Jonge EF, Peterse CM, Koelewijn JM, et al. The detection of monkeypox virus DNA in wastewater samples in the Netherlands. Sci Total Environ. 2022;852:158265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Tiwari A, Adhikari S, Kaya D, et al. Monkeypox outbreak: wastewater and environmental surveillance perspective. Sci Total Environ. 2023;856:159166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information.
Data Availability Statement
The data that supports the findings of this study are available in the supplementary material of this article.


