Abstract
Background and Objectives
Coenzyme Q10 (CoQ10) is an important electron carrier and antioxidant. The COQ7 enzyme catalyzes the hydroxylation of 5-demethoxyubiquinone-10 (DMQ10), the second-to-last step in the CoQ10 biosynthesis pathway. We report a consanguineous family presenting with a hereditary motor neuropathy associated with a homozygous c.1A > G p.? variant of COQ7 with abnormal CoQ10 biosynthesis.
Methods
Affected family members underwent clinical assessments that included nerve conduction testing, histologic analysis, and MRI. Pathogenicity of the COQ7 variant was assessed in cultured fibroblasts and skeletal muscle using a combination of immunoblots, respirometry, and quinone analysis.
Results
Three affected siblings, ranging from 12 to 24 years of age, presented with a severe length-dependent motor neuropathy with marked symmetric distal weakness and atrophy with normal sensation. Muscle biopsy of the quadriceps revealed chronic denervation pattern. An MRI examination identified moderate to severe fat infiltration in distal muscles. Exome sequencing demonstrated the homozygous COQ7 c.1A > G p.? variant that is expected to bypass the first 38 amino acid residues at the n-terminus, initiating instead with methionine at position 39. This is predicted to cause the loss of the cleavable mitochondrial targeting sequence and 2 additional amino acids, thereby preventing the incorporation and subsequent folding of COQ7 into the inner mitochondrial membrane. Pathogenicity of the COQ7 variant was demonstrated by diminished COQ7 and CoQ10 levels in muscle and fibroblast samples of affected siblings but not in the father, unaffected sibling, or unrelated controls. In addition, fibroblasts from affected siblings had substantial accumulation of DMQ10, and maximal mitochondrial respiration was impaired in both fibroblasts and muscle.
Discussion
This report describes a new neurologic phenotype of COQ7-related primary CoQ10 deficiency. Novel aspects of the phenotype presented by this family include pure distal motor neuropathy involvement, as well as the lack of upper motor neuron features, cognitive delay, or sensory involvement in comparison with cases of COQ7-related CoQ10 deficiency previously reported in the literature.
Ubiquinone, also known as coenzyme Q10 (CoQ10), is a lipophilic electron carrier, responsible for shuttling electrons from complex I and complex II to complex III in the inner mitochondrial membrane.1,2 The reduced form of CoQ10, ubiquinol, acts as a potent antioxidant in subcellular membranes and participates in many cellular functions including de novo pyrimidine synthesis and sulfide metabolism.1,2 CoQ10 is predominantly synthesized endogenously in association with the matrix side of the inner mitochondrial membrane or obtained in negligible amounts from diet. As such, CoQ10 deficiencies are a group of inherited autosomal recessive mitochondrial diseases that affect the CoQ10 biosynthesis pathway either directly (primary CoQ10 deficiency) or indirectly (secondary CoQ10 deficiency).3 Primary CoQ10 deficiencies have substantial heterogeneity in phenotypic presentation, often presenting as multisystemic disorders including neurologic abnormalities (upper and lower motor neuron disorders and cognitive impairment) and steroid-resistant nephrotic syndrome.1,4,5
Primary CoQ10 deficiency can result from pathogenic variation in COQ7. COQ7 encodes a flavin-dependent monooxygenase, 5-demethoxyubiquinone hydroxylase (COQ7), which catalyzes the hydroxylation of 5-demethoxyubiquinone-10 (DMQ10). There are 7 reported cases of children with primary CoQ10 deficiency resulting from pathogenic COQ7 variation (Table 1). Pathogenicity of these variants has been demonstrated by impaired activity of respiratory chain enzymes,6-9 varying levels of CoQ10 deficiency,6,7,9,10 accumulation of DMQ10, and reduced COQ7 expression.7,9,10 As often observed in CoQ10 deficiency,5 the phenotype includes variable axonal and demyelinating polyneuropathy, developmental delay and regression, and upper motor neuron dysfunction.
Table 1.
Genetically Confirmed Cases of COQ7-Related Primary CoQ10 Deficiency
In this study, we report 3 siblings with a progressive length-dependent motor neuropathy with severe distal atrophy. Exome and genome sequencing identified a homozygous COQ7 variant resulting in CoQ10 deficiency. Unlike the previously reported cases,6-11 the affected siblings have a pure distal motor neuropathy without sensory, cognitive, or visual deficiencies. Because this phenotypic presentation of COQ7-related CoQ10 deficiency differs substantially from those described in earlier case reports,6-11 this report extends the phenotypic spectrum associated with COQ7-related CoQ10 deficiency.
Methods
Patients
We report 3 affected siblings (sibling 1: female aged 24 years, sibling 2: male aged 23 years, and sibling 3: male aged 12 years) referred for progressive distal weakness to the Medical Genetics service for evaluation. The siblings were born to Syrian parents who are first cousins. Next-generation sequencing panels for the Charcot-Marie-Tooth disease, spinal muscle atrophy, distal hereditary motor neuropathy, and mitochondrial DNA (mtDNA) showed negative results.
Standard Protocol Approvals, Registrations, and Patient Consents
The affected siblings and family members were enrolled in the Care4Rare Canada research study12 because of the lack of a molecular diagnosis. Approval of the study design was obtained from the institutional research ethics board (Children's Hospital of Eastern Ontario; #1104E and CTO1577), and free and informed consent was obtained prior to enrollment for all participants.
Clinical Findings in Affected Siblings
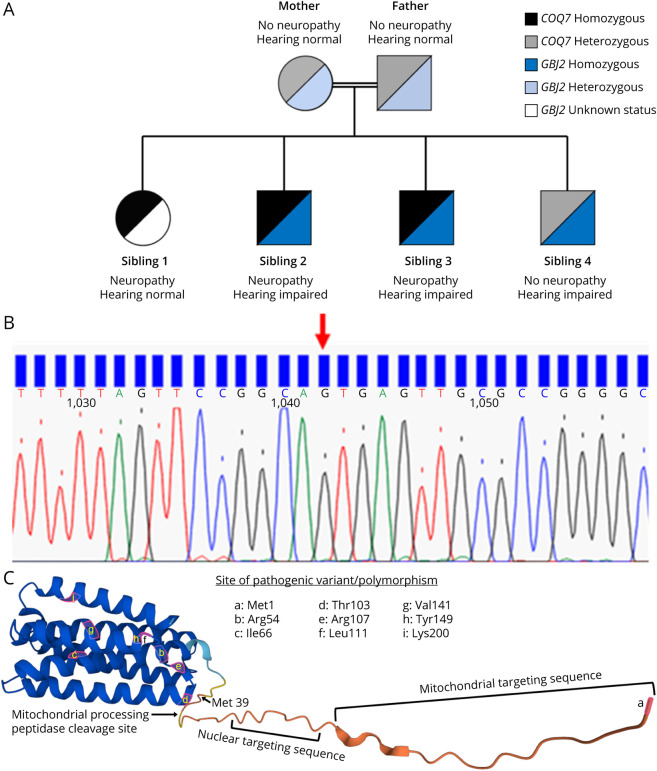
The 3 affected siblings developed distal weakness by 10–14 years of age and muscle atrophy in the distal lower more than upper extremities with preserved sensation (temperature, fine touch, sharp, vibration, and proprioception) and preserved deep tendon reflexes. The parents and unaffected brother (sibling 4 in Figure 1A) had normal strength and no atrophy. Electrodiagnostic testing of all affected siblings revealed reduced distal motor amplitude pattern with preserved sensory responses. Needle EMG studies demonstrated distal chronic reinnervation with no active denervation. In addition, siblings 2, 3, and 4 were deaf since childhood. The deafness did not segregate with motor neuropathy (sibling 1 was not deaf and sibling 4 had no neuropathy) (Figure 1A). Clinical features of the siblings with motor neuropathy and the affected individuals reported in the literature are summarized in Table 1.
Figure 1. Genetic Testing.
(A) Pedigree depicting the segregation of COQ7 and GBJ2 variants. (B) Sequencing chromatogram shows the c.1A > G variant (NM_016138.5) detected in COQ7 in the muscle biopsy sample from sibling 2. (C) Native human COQ7 protein as predicted using AlphaFold.36,37 With the loss of the start codon, COQ7 will putatively initiate instead at methionine 39, resulting in the loss of the cleavable n-terminal presequence (containing the mitochondrial and nuclear targeting sequences),29 as well as the amino acids at positions 37 and 38. The amino acids highlighted in pink correspond to pathogenic variants in published cases of COQ7-related CoQ10 deficiency. Panel C is used with permission from DeepMind, the creators of AlphaFold. CoQ10 = coenzyme Q10.
MRI
Whole-body muscle MRI with coronal T1W1 sequences of the thorax and abdomen and axial T1W1 and short tau inversion recovery sequences of the thorax, abdomen, and upper and lower limbs were performed on a 1.5T Siemens MRI.
Identification of Rare Variants by Exome Sequencing
For duo-exome sequencing (sibling 1 and sibling 2), exonic DNA was selected using the Agilent SureSelect 50 Mb (V5) All Exon Kit following manufacturer's instructions and sequenced on an Illumina HiSeq 2,500. Read alignment, variant calling, and annotation were performed as per the Care4Rare in-house pipeline as previously described for FORGE and Care4Rare Canada projects (github.com/ccmbioinfo/crg2).12-15 As per Care4Rare quality requirements, more than 95% of the Consensus Coding Sequence (CCDS) bases were covered by at least 10 reads, and more than 90% of CCDS bases were covered by at least 20 reads for each sample. Variants were disregarded if they were present at >1% in gnomAD or seen in more than 5 samples from our in-house database (approximately 2,000 exomes previously sequenced at the McGill University and Genome Quebec Innovation Center). PCR and Sanger sequencing were used to validate the variants identified by genome-wide sequencing. Variants present in ≥0.1% minor allele frequency in gnomAD were excluded.
Sequencing of PCR Amplicons
Total RNA was extracted from muscle biopsy from sibling 2 using TRIzol reagent (Thermo Fisher Scientific) and was reverse transcribed by using qScript cDNA Supermix (Quanta Biosciences) according to instructions from the manufacturers. N-terminal part of the coding region of COQ7 was amplified by using Phusion High-Fidelity DNA Polymerase (New England BioLabs), and the primer pairs used were as follows: 5′-AGTCCGAGCCAAGGGCAC-3′ and 5′- TTCAGTGTCGCCAACTGTCC-3’. PCR products were directly used for sequencing (McGill University and Génome Québec Innovation Center).
Tissue Sampling
Full-thickness skin samples were obtained from the 3 affected siblings, an unaffected sibling (sibling 4), and the father using standard punch biopsy technique. In addition, an open muscle biopsy of the left vastus lateralis of sibling 2 was performed using a standard protocol. Hematoxylin and eosin, Gomori trichrome, and oil red O staining were performed to search for ragged red fibers and detection of abnormal lipid deposits. Stains for nicotinamide adenine dinucleotide (NADH), succinic dehydrogenase, cytochrome oxidase, and myosin ATPase (at pH 4.3, 4.7, and 10.4) were performed to identify fibers with respiratory chain abnormalities. Transmission electron microscopy was performed by Eastern Ontario Regional Laboratory Pathology Laboratory according to standard techniques. A portion of the sampled vastus lateralis was snap frozen in liquid nitrogen and stored at −80°C for later analyses, and 1 portion was placed in ice-cold biopsy preservation solution for mitochondrial respiration analysis (biopsy preservation solution, pH 7.1; 5.77 mM Na2ATP, 7 10 ethylene glycol tetraacetic acid-CaEGTA buffer [0.1 µM free Ca2+], 6.56 mM MgCl2-6H2O, 20 mM taurine, 60 mM K-lactobionate, 15 mM phosphocreatine, 20 mM imidazole, 0.5 mM dithiothreitol, and 50 mM 2-(N-morpholino)ethanesulfonic acid). Samples of muscle were also obtained from wild-type and COQ7-knockout mice and an unrelated human donor.
Cell Cultures
Skin-derived fibroblasts were maintained according to 1 of 2 protocols. In the first protocol, fibroblasts were maintained at 37°C in Dulbecco modified Eagle medium (DMEM; #319-005-CL, Wisent) supplemented with 10% fetal bovine serum (#090–150, Wisent), 1x GlutaMAX (Gibco, 35050-061), 1% antibiotic-antimycotic (#450-115-EL, Wisent), and 5% CO2. In the second protocol, fibroblasts were maintained at 37°C in DMEM (Gibco, 11995-065) supplemented with 10% fetal bovine serum, 1x GlutaMAX (Gibco, 35050-061), 1% penicillin-streptomycin solution (Gibco, 15140-122), and 5% CO2. Immunoblots, quinone analyses, and viability assessments were performed in cells with passage numbers between 8 and 20. Cell viability was assessed in standard glucose media and in media containing 10 mM galactose and no glucose, which stimulates oxidative phosphorylation. Cell viability was determined using the resazurin reduction assay with 0.15 mg/mL resazurin solution at 10% v/v of the total culture volume incubated for 2 hours.
Analysis of Fibroblast Cellular Bioenergetics
The Seahorse XFe96 Analyzer (Agilent) was used to measure oxygen consumption rates and extracellular acidification rates (ECAR). Fibroblasts were seeded at 15,000 cells/well 24 hours prior to analysis. The following day, cell culture media was replaced with phenol red-free, sodium bicarbonate–free Seahorse XF medium, pH 7.4 (DMEM, 25 mM d-glucose) supplemented with 4 mM L-glutamine, 1 mM Na-pyruvate 30 minutes prior to loading into the XF Analyzer. Following measurements of basal respiration, fibroblasts were treated with sequential injections of oligomycin (1 μM), carbonyl cyanide p-trifluoromethoxyphenyl hydrazine (FCCP) (1 μM), antimycin A (0.5 μM) with rotenone (0.5 μM), and monensin (10 μM). After completion of the assay, Seahorse XF medium was removed, and cells were washed with PBS, followed by lysis with radioimmunoprecipitation lysis buffer (0.5M Tris-HCl, pH 7.4, 1.5M NaCl, 2.5% deoxycholic acid, 10% NP-40, and 10 mM EDTA). The amount of protein per well was determined by the BCA assay using the Pierce BCA Protein Assay Kit (Thermofisher, 23225). Data were analyzed using the XF software (Seahorse Biosciences), and bioenergetic capacity and fuel flexibility were investigated by graphing glycolytic and oxidative ATP production, as previously described.16
Enzyme Activities
Enzyme activities for citrate synthase (CS) and complex I (CI) were determined as previously described17 using the BioTek Synergy Mx Microplate Reader (BioTek Instruments, Inc., Winooski, VT). For CS activity, fibroblasts were lysed in 20 mM hypotonic potassium phosphate buffer (pH 7.5) using an 18-gauge needle and centrifuged at 14,000g for 10 minutes at 4°C. CS activity was determined by measuring absorbance at 412 nm in 50 mM Tris-HCl (pH 8.0) with 0.2 mM DTNB, 0.1 mM acetyl-coA, and 0.25 mM oxaloacetate.
For assessment of CI activity, mitochondrial-enriched fractions were prepared by lysing cells in 10 mM ice-cold hypotonic Tris buffer (pH 7.6) with a 22-guage needle. The cell lysate was mixed with 1.5 M sucrose solution and subsequently centrifuged at 600g for 10 minutes at 4°C. The supernatant was collected and centrifuged at 14,000g for 10 minutes at 4°C. The mitochondrial pellet was resuspended in 10 mM ice-cold hypotonic Tris buffer (pH 7.6) and subjected to 2 flash-freeze cycles to improve rotenone sensitivity. CI activity was determined by measuring absorbance at 340 nm in 50 mM potassium phosphate buffer (pH 7.5), 3 mg/mL fatty acid–free bovine serum albumin, 300 μM KCN, 100 μM NADH, and 60 μM ubiquinone. Enzyme activity was calculated using the extinction coefficient (ε = 13.6 mmol−1 cm−1 for CS; 6.2 mmol−1 cm−1 for CI) and expressed per microgram of protein.
Immunoblotting
As previously performed,9 protein content of whole-cell lysates of biopsied muscle tissue (from sibling 2) and cultured fibroblast samples was determined using the BCA assay (Thermo Fisher Scientific), and standard immunoblotting techniques were used to probe COQ7 expression in samples (75 μg of total protein). Immunoblots were performed once. Porin was probed as a loading control. Decaprenyl-diphosphate synthase subunit 2 (PDSS2), the first enzyme involved in COQ biosynthesis, was probed to determine whether there were other effects on the COQ biosynthesis pathway in the affected siblings. Primary antibodies were rabbit polyclonal anti-COQ7 (1:1,000 dilution; Proteintech Group Inc, Chicago, IL), anti-VDAC1/Porin (1:1,000 dilution; Cell Signaling Technology, Danvers, MA), and anti-PDSS2 (1:2000 dilution; Proteintech Group Inc, Chicago, IL). Flash-frozen skeletal muscle obtained from a 31-year-old male volunteer (ZenBio, Inc., NC) was used as a wild-type control.
Mitochondrial Supercomplex Analysis
Vastus lateralis samples were assessed for ETC supercomplexes (SCs) by blue native PAGE (BN‐PAGE), as previously described.18 The following primary antibodies were used: CI (NADH dehydrogenase [ubiquinone] 1α subcomplex subunit 9, mitochondrial [NDUFA9]) (459100; Thermo Fisher Scientific, Waltham, MA), complex II (CII; Fp) succinate dehydrogenase complex, subunit A, flavoprotein variant (459200; Thermo Fisher Scientific), complex III (CIII; ubiquinol‐cytochrome c reductase core protein II) (Ab14745; MitoSciences [Abcam, Cambridge, United Kingdom]), complex IV (CIV; subunit I) (459600; Thermo Fisher Scientific), and complex V (CV; ATP synthase subunit a, mitochondrial) (Ab14748; MitoSciences). ETC SCs were analyzed based on their banding pattern, as previously confirmed by 2D‐BN‐PAGE.19
Quantitation of CoQ10
Quinone extraction and quantitation by high-performance liquid chromatography (HPLC) were conducted as previously described10 using 2 biological replicates. In brief, cells were lysed in a RIPA buffer (Tris-HCl, pH 7.5, 1% NP-40, 0.5% deoxycholate, 10 mM EDTA, and 150 mM NaCl) and extracted with a mixture of 28.5% ethanol and 71.5% hexane (vol/vol) for 2 minutes by vigorously vortexing. After centrifugation, the upper organic layer was transferred to a new tube, and hexane was evaporated by drying in a SpeedVac concentrator (Thermo Fisher Scientific) and kept at −80°C. The left residual was finally redissolved in a mixture of methanol and ethanol (7:3, vol/vol) before injection into HPLC (a Agilent 1,260 Infinity LC system). During the HPLC, the samples were separated on a reverse-phase C18 column (2.1 × 50 mm, 1.8 μm, Agilent m), eluted with mixture of 70% methanol and 30% ethanol (vol/vol) at 1.8 mL/minute and detected at 275 nm. Peak identities and CoQ10 amounts were established by comparison with the profile obtained from CoQ10 standard (Sigma-Aldrich). Protein content in the quinone extracts was determined by the BCA assay (Thermo Fisher Scientific) and used to normalize quinone levels. The muscle sample of sibling 1 was also run after being spiked with a 65-ng CoQ10 standard to further verify the CoQ10 peak identity; this experiment was performed once.
High-Resolution Respirometry of Vastus Lateralis Muscle
High-resolution respirometry was conducted on saponin-permeabilized vastus lateralis muscle from sibling 2 and healthy controls using an Oxygraph-2k system with an attached fluorometer (Oroboros, Innsbruck, Austria).
The Oxygraph-2k units were calibrated, and all measurements were performed in duplicate at 37°C in 2 mL of mitochondrial respiration media (MiR05:110 mM sucrose, 60 mM K-lactobionate, 20 mM N-2-hydroxyethylpiperazine-N-2-ethane sulfonic acid, 20 mM taurine, 10 mM KH2PO4, 3 mM MgCl2, 0.5 mM ethylene glycol tetraacetic acid, 1 g/L bovine serum albumin, pH 7.1).
As previously described,20 the assay protocol consisted of consecutive additions of 2 mM malate, 5 mM pyruvate, 10 mM glutamate (CI leak), 5 mM ADP (CI OXPHOS), 10 mM succinate (CI + II OXPHOS), 2.5 µM oligomycin (CI + II Leak), 0.5 µM titrations of FCCP (Max respiration), 2.5 µM antimycin A (nonmitochondrial respiration), 0.5 mM N,N,N′,N′-Tetramethyl-p-phenylenediamine dihydrochloride, 2 mM sodium l-ascorbate (CIV activity), and 100 mM sodium azide (CIV inhibitor). Values were corrected to nonmitochondrial oxygen consumption (i.e., that in the presence of antimycin A), and CIV was corrected to sodium azide.
Statistical Analysis
Group comparisons of fibroblast respirometry were made using 2-tailed Student t tests for independent samples. Group comparisons of quinone levels in fibroblasts were made using 2-way analysis of variance, followed by Dunnett multiple comparison tests. Statistical significance was defined at p < 0.05.
Data Availability
Data are available from the corresponding author on reasonable request.
Results
Genetic Testing
Genetic testing for the Charcot-Marie-Tooth disease, spinal muscle atrophy, distal hereditary motor neuropathy, and mtDNA panel was performed on a clinical basis and showed negative results for a pathogenic variant. As such, the family was enrolled in research and duo-exome sequencing was performed on sibling 1 and sibling 2 and identified a shared homozygous variant of uncertain significance in COQ7 (NM_01638.5) c.1A > G p (Figure 1C). Affected sibling 3 also shared the homozygous, variant of uncertain significance in COQ7. The parents and sibling 4 were heterozygous carriers for the COQ7 variant. The COQ7 variant was absent in gnomAD, and in silico prediction modeling predicted pathogenicity. This variant is the substitution of guanine for adenine at nucleotide position 1 and is predicted to result in a loss of the initiating methionine. Sequencing of PCR amplicons of COQ7 cDNA from muscle biopsy in sibling 2 also indicated a c.1A > G substitution (Figure 1B). The putative impact on the COQ7 protein is shown in Figure 1C.
In addition, for the 3 siblings who had congenital deafness (siblings 2, 3, and 4), genetic testing also revealed a homozygous GJB2 variant: c.35delG; p.(Gly12Valfs*2) (NM_004004.5), which has previously been reported as pathogenic and causing autosomal recessive deafness in multiple families.21,22 The parents were found to be heterozygous carriers of the GJB2 variant (Figure 1A).
Whole-Body MRI
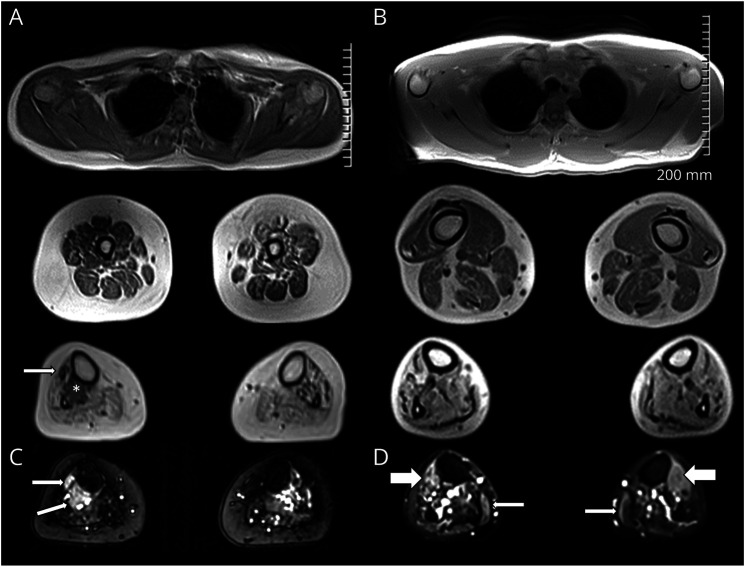
Consistent with the distal atrophy observed clinically, muscle MRI of siblings 1 and 2 demonstrated normal proximal musculature with significant distal muscle wasting and moderate to severe fat infiltration starting the level of the distal third of the thigh and most pronounced in the calves bilaterally (Figure 2).
Figure 2. Distal Denervation Atrophy on Muscle MRI and Muscle Biopsy.
MRI axial images T1 Dixon 2 point in-phase images (A, B) and short tau inversion recovery images (C, D) of patient 1 (sister, left) and patient 2 (older brother, right). Upper extremity musculature is normal (A top, B top). In the distal third of the thighs, there is bilateral fat infiltration of variable degree, ranging from mild to severe. The findings are more diffuse and advanced in the sister, involving all muscle compartments (A middle). In the brother (B middle), the anterior compartment involvement is less evident. Within the calves (A bottom, B bottom), there is bilateral severe fatty atrophy of most muscles, particularly in the soleus and gastrocnemius. In the sister (A bottom), there is more evident asymmetric sparing of the tibialis posterior (*) and tibialis anterior (arrow), which are remarkably spared on the right side. short tau inversion recovery images demonstrate edema-like signal changes of spared muscle fibers. In the sister (C), in the tibialis posterior/tibialis anterior (arrows). In the brother (D), in the anterior muscle compartments (thick arrows) and the gastrocnemius (thin arrows).
Muscle Pathology
Muscle biopsy of left vastus lateralis of sibling 2 demonstrated neurogenic atrophy with wide variation in fiber size and shape due to the presence of scattered angular, atrophic fibers, occasionally in groups, as well as abundant pyknotic nuclear clumps. There were no ragged red fibers or cytochrome oxidase negative fibers. Myofibrillar ATPase staining revealed group atrophy and fiber type grouping (eFigure 1, links.lww.com/NXG/A572).
CoQ10 Analysis
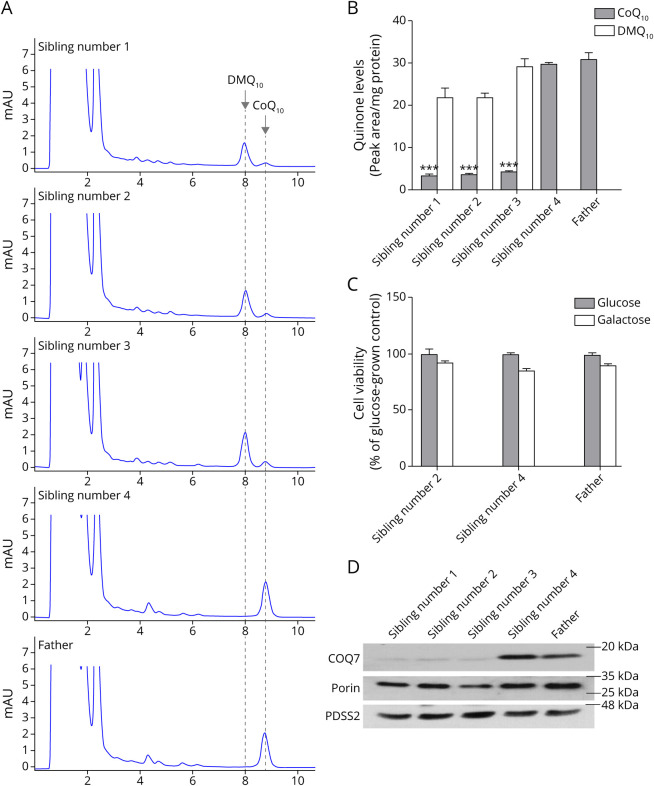
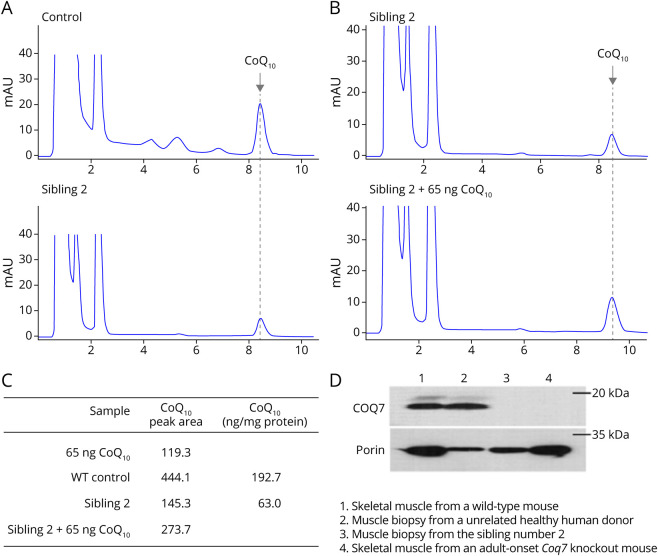
HPLC analysis yielded no detectable DMQ10 (the substrate of COQ7) in cultured skin fibroblasts from the father or sibling 4, whereas a significant amount of DMQ10 was detected in siblings 1, 2, and 3 (Figure 3A, B). On average, cultured fibroblasts from siblings 1, 2, and 3 had 87% lower CoQ10 than their father and unaffected sibling (Figure 3, A and B). Muscle CoQ10 in sibling 2 was 67% lower than muscle CoQ10 of the unrelated human donor (63 ng/mg protein vs 192.7 ng/mg protein) (Figure 4, A–C). However, no DMQ10 peak was seen in the muscle of sibling 2 (Figure 4, A and B).
Figure 3. Depleted CoQ10 and COQ7 in Cultured Fibroblasts.
(A) HPLC chromatograms of quinone extracts from human skin fibroblasts. The cells from the 3 affected siblings show a significant loss of CoQ10 and accumulation of the biosynthetic precursor DMQ10. (B) Quinone quantification. Values are shown as mean ± standard error of the mean (SEM) (n = 2 biological replicates). ***p < 0.001 compared with the CoQ10 level in the father's cells (2-way analysis of variance, followed by Dunnett multiple comparison tests). (C) Cell viability after a 4-day culture in galactose medium. Values are shown as mean ± SEM (n = 6). (D) Western blot analysis of expression of COQ7. In the skin fibroblasts from the 3 affected siblings, there is a severe reduction in COQ7 expression, in comparison with the cells from the carrier sibling and their carrier father. The level of the COQ biosynthetic enzyme PDSS2 is unchanged. The mitochondrial outer membrane protein, porin, was used as a loading control. CoQ10 = coenzyme Q10; HPLC = high-performance liquid chromatography.
Figure 4. Depleted CoQ10 and COQ7 in Skeletal Muscle.
(A) HPLC chromatograms of quinone extracts from muscle biopsy samples. Flash-frozen skeletal muscle obtained from a 31-year-old male volunteer was used as a control. (B) HPLC chromatograms of quinone extracted from 1.2 mg of muscle biopsy sample from sibling 2. One sample was spiked with 65 ng of CoQ10 standard before HPLC injection. The result show that the area of the peak of interest is greater in the CoQ10-spiked sample, providing further confirmation of the CoQ10 peak identity and allowing quantitation of CoQ10. (C) Quantitation indicated that the muscle of sibling 2 has 67% less CoQ10 than the control muscle. (D) Western blot analysis of expression of COQ7. Compared with the control, there is a severely diminished level of COQ7 in the muscle sample from sibling 2. Skeletal muscles from an adult-onset COQ7-knockout38 or a wild-type mouse were used for verification of the COQ7 band identity. The mitochondrial outer membrane protein, porin, was used as a loading control. Western blots were cropped to show only relevant bands. CoQ10 = coenzyme Q10; HPLC = high-performance liquid chromatography.
Immunoblot Analysis
The affected siblings had severely diminished levels of COQ7 in fibroblasts (Figure 3D), with COQ7 to porin ratios 85% ± 7% lower in affected siblings than their carrier father and sibling 4. The molecular weights of the truncated and wild-type proteins were comparable. The level of the COQ biosynthetic enzyme PDSS2 was unchanged in all 3 affected siblings. No COQ7 was detected by Western blot analysis in the muscle biopsy from sibling 2 (Figure 4D). No comparisons of COQ7 protein expression were made between heterozygous carriers and controls.
Mitochondrial Function Is Impaired in Skeletal Muscle From Sibling 2
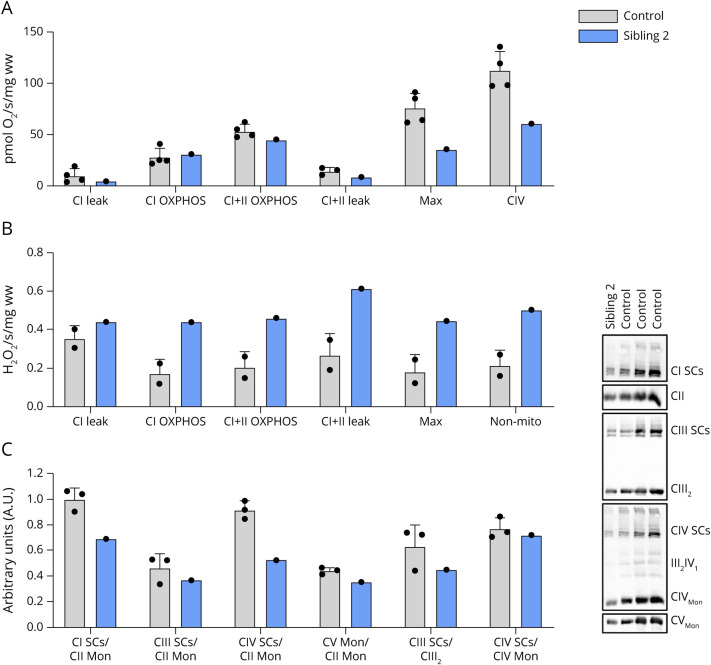
High-resolution respirometry analysis of biopsied vastus lateralis muscle from sibling 2 revealed minimal differences in respiratory flux for CI leak, CI OXPHOS, and CI + CII OXPHOS compared with that of vastus lateralis obtained from unrelated controls. However, FCCP-induced maximal mitochondrial respiration and CIV activity were lower in vastus lateralis muscle from sibling 2 compared with those in muscle from unrelated controls (Figure 5A). Simultaneous fluorometric analysis demonstrated higher H2O2 emissions in muscle from sibling 2 compared with those in muscle from healthy controls (Figure 5B). Because mitochondrial SCs are known to enhance electron flow and decrease reactive oxygen species (ROS), we therefore assessed SC content in vastus lateralis from sibling 2. BN-PAGE analysis of skeletal muscle mitochondrial SC content demonstrated decreased content of complexes I and IV containing SCs in sibling 2, whereas levels of complex III containing SCs were in range of controls (Figure 5C).
Figure 5. Increased H2O2 Emissions and Decreased Mitochondrial Respiration in Skeletal Muscle From Sibling 2.
(A) High-resolution respiratory flux per mg of saponin-permeabilized vastus lateralis muscle from COQ7-affected sibling 2 and unrelated controls. FCCP-induced maximal respiration and CIV activity were lower in the COQ7-affected muscle. (B) Simultaneous fluorometric quantification of H2O2 emissions was higher in vastus lateralis from affected sibling 2. (C) BN-PAGE analysis revealed a decrease in CI and CIV mitochondrial supercomplexes in vastus lateralis muscle of affected sibling 2. BN‐PAGE = blue native PAGE.
Fibroblast Mitochondrial Function
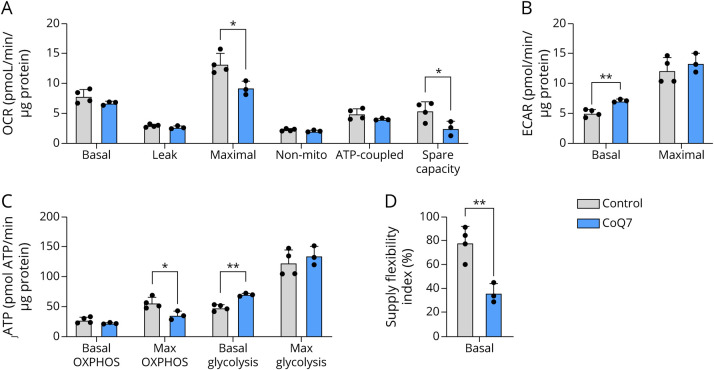
Basal oxygen consumption and oligomycin-induced leak respiration were preserved in cultured fibroblasts from affected siblings. Consistent with the impaired maximal respiration observed in vastus lateralis from sibling 2, cultured fibroblasts from affected siblings demonstrated similar defects in maximal respiration compared with fibroblasts from unrelated controls (p < 0.05; Figure 6A and eFigure 2A, links.lww.com/NXG/A572). Spare respiratory capacity was lower in fibroblasts from affected siblings, reflecting impaired ability to respond to increased energetic demands. The basal ECAR was higher in fibroblasts from affected siblings (p < 0.01; Figure 6B and S2B). Maximal glycolytic rate achieved after the addition of monensin was not different in affected siblings compared with that in control fibroblasts. Using the Mookerjee method16 to assess bioenergetic capacity and metabolic flexibility, fibroblasts from affected siblings exhibited a higher glycolytic index (eFigure 2C, links.lww.com/NXG/A572), demonstrating a bioenergetic profile favoring glycolysis and the Warburg effect (p < 0.001; Figure 6C). When assessing the supply flexibility index of the fibroblasts (the ability to change the source of ATP supply between glycolytic and oxidative pathways in response to changes in ATP demand), fibroblasts from affected siblings displayed impaired capacity to switch between ATP production pathways (p < 0.01; Figure 6D). Complex I activity was in range of controls (eFigure 2D, links.lww.com/NXG/A572). Mitochondrial content, as assessed by CS activity, was not different in fibroblasts from affected siblings relative to controls (eFigure 2E, links.lww.com/NXG/A572). Despite the observed impairment of mitochondrial respiratory function, the skin fibroblasts from sibling 2 showed similar viability in galactose medium, compared with that of sibling 4 and father with wild-type level of CoQ10 (Figure 3C).
Figure 6. Seahorse Analysis on Cultured Skin Fibroblasts.
(A) Maximal respiration and spare respiratory capacity are decreased in fibroblasts from affected siblings (COQ7) compared with their carrier father and sibling (control). (B) Basal extracellular acidification rate, a proxy measure of glycolysis, is higher in fibroblasts from affected siblings. (C) The rate of ATP formation from OXPHOS and glycolysis. (D) The supply flexibility index. All values are presented as mean values ± SD. Comparisons between groups were as determined using a 2-tailed Student t test for independent samples. *p < 0.05, **p < 0.01.
Discussion
Previously reported cases of COQ7-related CoQ10 deficiency have presented with a global demyelinating and axonal sensorimotor polyneuropathy, developmental delay, and upper motor neuron features.6-11 We presented 3 siblings with pathogenic variation in COQ7 and a pure motor length–dependent neuropathy with no clinical or electrophysiologic evidence of sensory nerve involvement and no evidence of demyelination. The siblings had no developmental delay or cognitive impairment and no upper motor neuron features. Pure motor neuropathies are rare, and the clinical presentation of the family reported in this study represents a distinct phenotype from previously reported cases of COQ7-related CoQ10 deficiency. A caveat to the clinical phenotype described here is that other features may develop as the siblings get older and their condition progresses. While COQ7 has been associated with hearing loss in past reports,6,7,10 hearing loss did not segregate with COQ7 variants in this report. In this family, deafness was explained by the pathogenic homozygous c35delG p.(GLy12Valfs*2) variants in GJB2. This GBJ2 variant is a well-established cause of nonsyndromic hearing loss.21,22
In the absence of the COQ7 start codon, the c.1A > G p.? variant is expected to truncate the translated protein by 38 amino acid residues at the n-terminus, initiating instead with methionine at position 39.23 The resulting translated protein is expected to be approximately 17.5% smaller than the typical protein (i.e., 179 vs 217 amino acids) in length, lacking the n-terminal mitochondrial targeting presequence that enables import of COQ7 into mitochondria and 2 subsequent amino acids. The presequence is normally cleaved within the mitochondria, accounting for the similar molecular weights observed in the immunoblot. The primary biochemical consequence of CoQ10 deficiency is impaired electron flow that results in decreased mitochondrial respiration. In the cases presented in this study, COQ7 was severely depleted and DMQ10 accumulated in cultured skin fibroblasts from the affected siblings, consistent with the notion that CoQ10 biosynthesis is intact excepting the last 2 steps. In contrast to the results from cultured fibroblasts, the muscle from sibling 2 did not accumulate DMQ10 despite the lack of detectable COQ7 and a 67% reduction in CoQ10. To explain the absence of DMQ10 but the presence of CoQ10 in muscle, we speculate that in a differentiated tissue such as muscle, compared with quickly dividing fibroblasts, the overall CoQ synthesis pathway is so severely depressed that insufficient DMQ10 is produced to allow for detection. By contrast, the muscle tissue might respond to low CoQ10 production by slowing CoQ10 breakdown enough to produce a measurable peak. The fibroblasts and permeabilized skeletal muscle from individuals homozygous for the c.1A > G p.? COQ7 variant displayed decreased maximal respiration, whereas resting respiration in the cultured fibroblasts and respiration with CI and CI + II substrates in permeabilized skeletal muscle were preserved. The preserved substrate-driven respiration contrasts that of the c.422T > A p.(Val141Glu) COQ7 variant where respiration in the presence of CI and CI + II substrates was impaired in primary patient fibroblasts,7 as well as the patient fibroblasts with the c.308C > T p.(Thr103Met) and c.332T > C p.(Leu111Pro) COQ7 variant that did not exhibit lower maximal respiration.10 The diverse mitochondrial phenotypes observed in these different variants likely contribute to the heterogeneity in phenotypic presentations of COQ7 deficiency.
The severe fat infiltration observed in the affected siblings is similar to reports that involve myopathic presentations of CoQ10 deficiency,24,25 in which excessive lipid droplets are commonly observed in histologic examinations of skeletal muscle. Lipid accumulation likely results from decreased fatty acid usage in mitochondrial oxidative processes and increased reliance on glycolytic metabolism. Consistent with this notion, COQ7-knockout mouse embryonic fibroblasts have increased glycolytic capacity and fail to survive when cultured in galactose media.26 In line with these observations, the COQ7 fibroblasts from the affected siblings presented in this study exhibited a bioenergetic profile favoring glycolysis and an impaired ability to increase oxidative metabolism in response to increased energy demands. Fibroblasts from affected siblings were found to be viable in galactose. Cells lacking COQ7 are capable of mitochondrial respiration mediated by DMQ at low levels27; however, COQ7 deficiency is associated with the activation of the mammalian target of rapamycin (mTOR)/hypoxia-inducible factor 1-alpha (HIF-1α) and ROS/HIF-1α signaling pathways that promote aerobic glycolysis.28
In addition to the mitochondrial targeting sequence, a nuclear targeting sequence between amino acids 11 and 29 in COQ7 has been reported.29 While the existence of nuclear-located COQ7 is controversial,30 nuclear-located COQ7 reportedly functions independent from the mitochondrial form and plays a specific role in mediating ROS metabolism and mitochondrial stress responses.29 Human cells lacking nuclear COQ7 have increased ROS emissions and are more susceptible to ROS-induced cell death.29 Moreover, Caenorhabditis elegans worms null in clk-1, the C elegans homolog of COQ7, have increased expression of genes involved in the mitochondrial unfolded protein response, which is ameliorated in the presence of nuclear-located COQ7.29 In context of the patients presented in this study, the increased H2O2 emission observed in the permeabilized skeletal muscle of sibling 2 may be attributable to the lack of nuclear-located COQ7. Moreover, the decreased assembly of mitochondrial SCs in the muscle from sibling 2 may contribute to enhanced ROS production in COQ7 patients.
Precisely why the c.1A > G p.? COQ7 variant affects distal motor but not sensory function is not known, but metabolic disturbances and oxidative stress associated with CoQ10 deficiency should be considered potential pathomechanisms.31 Selective degeneration of motor neurons is associated with endoplasmic reticulum stress, impaired mitochondria fusion, altered transport of mitochondria, and mitochondrial dysfunction in distal hereditary motor neuropathies.32,33 Mitochondrial oxidative stress has been also been implicated in the distal motor axonopathy seen in patients with amyotrophic lateral sclerosis (ALS) and in animal models of ALS,34 a condition that presents with limited or subtle sensory involvement.35 Thus, the metabolic dysfunction observed in the fibroblasts and biopsied muscle of the affected patients combined with the elevated H2O2 emission observed in the biopsy with the observed neuropathy likely contribute to the distal motor phenotype. The hypothesis that motor axon viability is challenged by mitochondrial oxidative stress in COQ7 neurologic disease should be explored in future studies.
CoQ10 deficiencies are frequently treated with a high-dose oral supplementation of CoQ10 or CoQ10 precursor molecules that bypass the relevant genetic alteration, with 2,4-dihydroxybensoic acid as a notable example for COQ7-related CoQ10 deficiency.5-7,9-11 However, the efficacy and safety of 2,4-dihydroxybensoic acid has recently been called into question.9 Responses to oral supplementation are variable and may be tissue dependent.5-7,10,11 CoQ10 treatment has not resulted in a long-term improvement in COQ7 phenotypes,6,7,10,11 but advances in methods to solubilize CoQ10 may lead to better outcomes in the future.9 No experiments to rescue metabolic function of cultured fibroblasts from the affected siblings were performed in this study.
In conclusion, we present the first report of pure distal hereditary motor neuropathy associated with a homozygous pathogenic variant in the gene encoding the ubiquinone biosynthesis protein COQ7. This clinical presentation extends the phenotypic spectrum of COQ7 neurologic disease, which has been previously reported with axonal and demyelinating polyneuropathy, spastic paraparesis, and cognitive delay. This study also highlights the importance of COQ7 for motor nerve function. Further functional studies are needed to improve the understanding of the underlying pathomechanism(s) and the potential overlap with other motor neuropathies.
Acknowledgment
The authors are grateful to the family featured in this study for their participation and for consenting to the publication of this report.
Glossary
- ALS
amyotrophic lateral sclerosis
- BN‐PAGE
blue native PAGE
- CI
complex I
- CoQ10
coenzyme Q10
- CS
citrate synthase
- DMEM
Dulbecco modified Eagle medium
- DMQ10
5-demethoxyubiquinone-10
- ECAR
extracellular acidification rates
- FCCP
carbonyl cyanide p-trifluoromethoxyphenyl hydrazine
- HPLC
high-performance liquid chromatography
- mtDNA
mitochondrial DNA
- NADH
nicotinamide adenine dinucleotide
- PDSS2
Decaprenyl-diphosphate synthase subunit 2
- ROS
reactive oxygen species
- SCs
supercomplexes
Appendix 1. Authors
Appendix 2. Coinvestigators
Study Funding
This study was supported by the Physician Services Incorporated and, in part, by the Care4Rare Canada Consortium funded by Genome Canada and the Ontario Genomics Institute (OGI-147), the Canadian Institutes of Health Research, Ontario Research Fund, Genome Alberta, Genome British Columbia, Genome Quebec, and Children's Hospital of Eastern Ontario Foundation.
Disclosure
I.C. Smith is supported by the Eric Poulin ALS Translational Research Fund. A. Breiner is supported by the ALS Eric Poulin Research Chair. H. Lochmüller receives support from the Canadian Institutes of Health Research (Foundation Grant FDN-167281), the Canadian Institutes of Health Research and Muscular Dystrophy Canada (Network Catalyst Grant for NMD4C), the Canada Foundation for Innovation (CFI-JELF 38412), and the Canada Research Chairs program (Canada Research Chair in Neuromuscular Genomics and Health, 950–232279). K.M. Boycott is supported by a CIHR Foundation Grant (FDN-154279) and a Tier 1 Canada Research Chair in Rare Disease Precision Health. M.-E. Harper is supported by a University of Ottawa Research Chair and a Canadian Institutes of Health Research foundation grant (FDN-143278). J. Warman-Chardon is supported by a Department of Medicine Clinical Research Chair and Physician Services Incorporated, Muscular Dystrophy Canada, and Canadian Institutes of Health Research grants. Full disclosure form information provided by the authors is available with the full text of this article at Neurology.org/NG.
References
- 1.Alcazar-Fabra M, Rodriguez-Sanchez F, Trevisson E, Brea-Calvo G. Primary Coenzyme Q deficiencies: a literature review and online platform of clinical features to uncover genotype-phenotype correlations. Free Radic Biol Med. 2021;167:141-180. doi: 10.1016/j.freeradbiomed.2021.02.046. [DOI] [PubMed] [Google Scholar]
- 2.Santos-Ocana C, Cascajo MV, Alcazar-Fabra M, et al. . Cellular models for primary CoQ deficiency pathogenesis study. Int J Mol Sci. 2021;22(19):10211. doi: 10.3390/ijms221910211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Quinzii CM, Hirano M. Primary and secondary CoQ(10) deficiencies in humans. Biofactors. 2011;37(5):361-365. doi: 10.1002/biof.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quinzii CM, Emmanuele V, Hirano M. Clinical presentations of coenzyme q10 deficiency syndrome. Mol Syndromol. 2014;5(3-4):141-146. doi: 10.1159/000360490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alcazar-Fabra M, Trevisson E, Brea-Calvo G. Clinical syndromes associated with Coenzyme Q10 deficiency. Essays Biochem. 2018;62(3):377-398. doi: 10.1042/ebc20170107. [DOI] [PubMed] [Google Scholar]
- 6.Kwong AK, Chiu AT, Tsang MH, et al. . A fatal case of COQ7-associated primary coenzyme Q(10) deficiency. JIMD Rep. 2019;47(1):23-29. doi: 10.1002/jmd2.12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freyer C, Stranneheim H, Naess K, et al. . Rescue of primary ubiquinone deficiency due to a novel COQ7 defect using 2, 4-dihydroxybensoic acid. J Med Genet. 2015;52(11):779-783. doi: 10.1136/jmedgenet-2015-102986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Theunissen TEJ, Nguyen M, Kamps R, et al. . Whole exome sequencing is the preferred strategy to identify the genetic defect in patients with a probable or possible mitochondrial cause. Front Genet. 2018;9:400. doi: 10.3389/fgene.2018.00400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y, Gumus E, Hekimi S. A novel COQ7 mutation causing primarily neuromuscular pathology and its treatment options. Mol Genet Metab Rep. 2022;31:100877. doi: 10.1016/j.ymgmr.2022.100877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y, Smith C, Parboosingh JS, Khan A, Innes M, Hekimi S. Pathogenicity of two COQ7 mutations and responses to 2, 4-dihydroxybenzoate bypass treatment. J Cell Mol Med. 2017;21(10):2329-2343. doi: 10.1111/jcmm.13154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hashemi SS, Zare-Abdollahi D, Bakhshandeh MK, et al. . Clinical spectrum in multiple families with primary COQ 10 deficiency. Am J Med Genet A. 2021;185(2):440-452. doi: 10.1002/ajmg.a.61983. [DOI] [PubMed] [Google Scholar]
- 12.Beaulieu CL, Majewski J, Schwartzentruber J, et al. . FORGE Canada Consortium: outcomes of a 2-year national rare-disease gene-discovery project. Am J Hum Genet. 2014;94(6):809-817. doi: 10.1016/j.ajhg.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754-1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McKenna A, Hanna M, Banks E, et al. . The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297-1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mookerjee SA, Gerencser AA, Nicholls DG, Brand MD. Quantifying intracellular rates of glycolytic and oxidative ATP production and consumption using extracellular flux measurements. J Biol Chem. 2017;292(17):7189-7207. doi: 10.1074/jbc.m116.774471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spinazzi M, Casarin A, Pertegato V, Salviati L, Angelini C. Assessment of mitochondrial respiratory chain enzymatic activities on tissues and cultured cells. Nat Protoc. 2012;7(6):1235-1246. doi: 10.1038/nprot.2012.058. [DOI] [PubMed] [Google Scholar]
- 18.Pileggi CA, Blondin DP, Hooks BG, et al. . Exercise training enhances muscle mitochondrial metabolism in diet-resistant obesity. EBioMedicine. 2022;83:104192. doi: 10.1016/j.ebiom.2022.104192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Acin-Perez R, Fernandez-Silva P, Peleato ML, Perez-Martos A, Enriquez JA. Respiratory active mitochondrial supercomplexes. Mol Cell. 2008;32(4):529-539. doi: 10.1016/j.molcel.2008.10.021. [DOI] [PubMed] [Google Scholar]
- 20.Pesta D, Gnaiger E. High-resolution respirometry: OXPHOS protocols for human cells and permeabilized fibers from small biopsies of human muscle. Methods Mol Biol. 2012;810:25-58. doi: 10.1007/978-1-61779-382-0_3. [DOI] [PubMed] [Google Scholar]
- 21.Bolz H, Schade G, Ehmer S, Kothe C, Hess M, Gal A. Phenotypic variability of non-syndromic hearing loss in patients heterozygous for both c.35delG of GJB2 and the 342-kb deletion involving GJB6. Hearing Res. 2004;188(1-2):42-46. doi: 10.1016/s0378-5955(03)00346-0. [DOI] [PubMed] [Google Scholar]
- 22.Azadegan-Dehkordi F, Ahmadi R, Koohiyan M, Hashemzadeh-Chaleshtori M. Update of spectrum c.35delG and c.-23+1G>A mutations on the GJB2 gene in individuals with autosomal recessive nonsyndromic hearing loss. Ann Hum Genet. 2019;83:1-10. doi: 10.1111/ahg.12284. [DOI] [PubMed] [Google Scholar]
- 23.Asaumi S, Kuroyanagi H, Seki N, Shirasawa T. Orthologues of the Caenorhabditis elegans longevity gene clk-1 in mouse and human. Genomics. 1999;58(3):293-301. doi: 10.1006/geno.1999.5838. [DOI] [PubMed] [Google Scholar]
- 24.Horvath R, Schneiderat P, Schoser BG, et al. . Coenzyme Q10 deficiency and isolated myopathy. Neurology. 2006;66(2):253-255. doi: 10.1212/01.wnl.0000194241.35115.7c. [DOI] [PubMed] [Google Scholar]
- 25.Trevisson E, DiMauro S, Navas P, Salviati L. Coenzyme Q deficiency in muscle. Curr Opin Neurol. 2011;24(5):449-456. doi: 10.1097/wco.0b013e32834ab528. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, Hekimi S. Mitochondrial respiration without ubiquinone biosynthesis. Hum Mol Genet. 2013;22(23):4768-4783. doi: 10.1093/hmg/ddt330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Hekimi S. Minimal mitochondrial respiration is required to prevent cell death by inhibition of mTOR signaling in CoQ-deficient cells. Cell Death Discov. 2021;7(1):201. doi: 10.1038/s41420-021-00591-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gu R, Zhang F, Chen G, et al. . Clk1 deficiency promotes neuroinflammation and subsequent dopaminergic cell death through regulation of microglial metabolic reprogramming. Brain Behav Immun. 2017;60:206-219. doi: 10.1016/j.bbi.2016.10.018. [DOI] [PubMed] [Google Scholar]
- 29.Monaghan RM, Barnes RG, Fisher K, et al. . A nuclear role for the respiratory enzyme CLK-1 in regulating mitochondrial stress responses and longevity. Nat Cell Biol. 2015;17(6):782-792. doi: 10.1038/ncb3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu JL, Yee C, Wang Y, Hekimi S. A single biochemical activity underlies the pleiotropy of the aging-related protein CLK-1. Sci Rep. 2017;7(1):859. doi: 10.1038/s41598-017-00754-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quinzii CM, Lopez LC, Gilkerson RW, et al. . Reactive oxygen species, oxidative stress, and cell death correlate with level of CoQ10 deficiency. FASEB J. 2010;24(10):3733-3743. doi: 10.1096/fj.09-152728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharma G, Pfeffer G, Shutt TE. Genetic neuropathy due to impairments in mitochondrial dynamics. Biology (Basel). 2021;10(4):268. doi: 10.3390/biology10040268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gregianin E, Pallafacchina G, Zanin S, et al. . Loss-of-function mutations in the SIGMAR1 gene cause distal hereditary motor neuropathy by impairing ER-mitochondria tethering and Ca2+ signalling. Hum Mol Genet. 2016;25(17):3741-3753. doi: 10.1093/hmg/ddw220. [DOI] [PubMed] [Google Scholar]
- 34.Moloney EB, de Winter F, Verhaagen J. ALS as a distal axonopathy: molecular mechanisms affecting neuromuscular junction stability in the presymptomatic stages of the disease. Front Neurosci. 2014;8:252. doi: 10.3389/fnins.2014.00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tao QQ, Wei Q, Wu ZY. Sensory nerve disturbance in amyotrophic lateral sclerosis. Life Sci. 2018;203:242-245. doi: 10.1016/j.lfs.2018.04.052. [DOI] [PubMed] [Google Scholar]
- 36.Jumper J, Evans R, Pritzel A, et al. . Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596(7873):583-589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Varadi M, Anyango S, Deshpande M, et al. . AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022;50(D1):D439-D444. doi: 10.1093/nar/gkab1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y, Oxer D, Hekimi S. Mitochondrial function and lifespan of mice with controlled ubiquinone biosynthesis. Nat Commun. 2015;6(1):6393. doi: 10.1038/ncomms7393. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are available from the corresponding author on reasonable request.