Abstract
Individuals with neurofibromatosis type 1 develop rat sarcoma virus (RAS)–mitogen-activated protein kinase–mitogen-activated and extracellular signal-regulated kinase (RAS-MAPK-MEK)–driven nerve tumors called neurofibromas. Although MEK inhibitors transiently reduce volumes of most plexiform neurofibromas in mouse models and in neurofibromatosis type 1 (NF1) patients, therapies that increase the efficacy of MEK inhibitors are needed. BI-3406 is a small molecule that prevents Son of Sevenless (SOS)1 interaction with Kirsten rat sarcoma viral oncoprotein (KRAS)-GDP, interfering with the RAS-MAPK cascade upstream of MEK. Single agent SOS1 inhibition had no significant effect in the DhhCre;Nf1fl/fl mouse model of plexiform neurofibroma, but pharmacokinetics (PK)-driven combination of selumetinib with BI-3406 significantly improved tumor parameters. Tumor volumes and neurofibroma cell proliferation, reduced by MEK inhibition, were further reduced by the combination. Neurofibromas are rich in ionized calcium binding adaptor molecule 1 (Iba1)+ macrophages; combination treatment resulted in small and round macrophages, with altered cytokine expression indicative of altered activation. The significant effects of MEK inhibitor plus SOS1 inhibition in this preclinical study suggest potential clinical benefit of dual targeting of the RAS-MAPK pathway in neurofibromas.
SIGNIFICANCE STATEMENT
Interfering with the RAS–mitogen-activated protein kinase (RAS-MAPK) cascade upstream of mitogen activated protein kinase kinase (MEK), together with MEK inhibition, augment effects of MEK inhibition on neurofibroma volume and tumor macrophages in a preclinical model system. This study emphasizes the critical role of the RAS-MAPK pathway in controlling tumor cell proliferation and the tumor microenvironment in benign neurofibromas.
Introduction
Neurofibromatosis type 1 (NF1) is an autosomal dominant cancer predisposition syndrome in which one in 2000–3000 children are born with a mutant NF1 allele and are predisposed to develop benign peripheral nerve sheath tumors called neurofibromas (NFs) (Rasmussen and Friedman, 2000; Uusitalo et al., 2015; Ratner and Miller, 2015). Neurofibromas develop after rare somatic cell loss of the remaining wild-type NF1 allele in a peripheral nerve glial cell, a Schwann cell (SC), or an SC precursor, so that neurofibromas contain SCs with biallelic loss of function at the NF1 locus (Kluwe et al., 1999; Serra et al., 2001; Pemov et al., 2017). Neurofibromas also contain endothelial cells, perineurial cells, fibroblasts, and recruited immune cells (Staser et al., 2012). It is estimated that up to 30% of the cells within plexiform neurofibromas (PNFs) are Iba1+;CD11b+;F4/80+ macrophages (Liao et al., 2018; Fletcher et al., 2019); macrophages restrain tumor development but enhance tumor growth (Prada et al., 2013). About half of NF1 patients develop PNFs in deep nerves (Plotkin et al., 2012). PNFs grow most rapidly in childhood and can grow large enough to cause significant morbidity. Of particular concern are PNF derivatives called malignant peripheral nerve sheath tumors (MPNSTs), which manifest in 8%–15% of NF1 patients coincident with accumulation of genetic changes, often during adolescence or young adulthood (Miettinen et al., 2017).
The NF1 gene product neurofibromin accelerates the hydrolysis of active RAS-GTP to inactive RAS-GDP (Scheffzek and Shivalingaiah, 2019), attenuating its activity. All six RAS proteins [(ras-related protein H (HRAS), ras-related protein N (NRAS), ras-related protein K (KRAS), ras-related protein R (RRAS), ras-related protein R-Ras2 (TC21/RRAS2), and ras-related protein M (MRAS)] are regulated by NF1 (Ohba et al., 2000). Therefore, in cells lacking NF1, RAS-GTP levels are increased and/or sustained. Pathways downstream of RAS-GTP, including the mitogen-activated protein kinase (MAPK) pathway, confer a survival or proliferation advantage and therefore represent promising potential targets for NF1 therapy (Lavoie et al., 2020; Fisher et al., 2021; Widemann et al., 2014). RAS proteins possess intrinsic GTPase activity even in the absence of GAP activity. Binding to guanine-nucleotide exchange factors (GEFs) such as SOS1/SOS2 (Son of Sevenless) facilitates exchange of GDP for GTP, leading to activation of RAS (Hennig et al., 2015; Baltanás et al., 2020). Based on their mode of action in suppressing nucleotide exchange, both SOS1 inhibitors and agents that block downstream signaling, such as mitogen-activated and extracellular signal-regulated kinase (MEK) inhibitors, could address aberrant activation of MAPK pathway signaling and deliver therapeutic benefit to NF1 patients. We therefore postulated that blocking RAS activation with an inhibitor of SOS1 would show therapeutic effects in a murine model of PNF.
In the DhhCre;Nf1fl/fl mouse model of PNF, homozygous deletion of Nf1 occurs in up to half of Schwann cells, causing a highly penetrant phenotype in which mice form neurofibromas adjacent to the spinal cord (Wu et al., 2008). Volumetric (3D) measurements of magnetic resonance imaging (MRI) images can be used to assess the effects of therapeutics on neurofibromas (Wu et al., 2012). Activity of MEK inhibitors in DhhCre;Nf1fl/fl mice preceded phase 1 and phase 2 clinical trials (Jessen et al., 2013; Dombi et al., 2016, Gross et al., 2020), which led to the approval of selumetinib in NF1 patients with plexiform neurofibroma after demonstrating reduction in PN tumor volume in 70% of individuals (Dombi et al., 2016; Gross et al., 2020). However, not all individuals with neurofibromas respond to treatment. Furthermore, despite MEK inhibitor associated side effects, tumor control is dependent on patients remaining on treatment. Therefore, therapies with the potential to increase the response rates and/or efficacy of MEK inhibitors would be of potential clinical utility.
A selective and orally available small-molecule SOS1 inhibitor, BI-3406, was recently described. BI-3406 binds the catalytic domain of SOS1, preventing its interaction with KRAS-GDP and therefore GTP loading. Treatment of cells with BI-3406 reduced the proliferation of KRAS-driven cancer cells. Notably, seven of 14 tested tumor cell lines with NF1 aberrations were sensitive to BI-3406 treatment independent of their KRAS mutation status. Single-agent activity of BI-3406 was observed in KRAS mutant xenograft models, and a deepened response was observed upon combination with MEK inhibition (Hofmann et al., 2021; Hillig et al., 2019). Therefore, we tested whether single-agent or combined SOS1 and MEK inhibition might represent novel approaches in the treatment of neurofibromas. Here we report the results of PK-driven combination of selumetinib with BI-3406 in the DhhCre;Nf1fl/fl mouse model of PNF. Comparable selumetinib levels were observed in mice when administering 10 mg/kg selumetinib monotherapy versus 5 mg/kg in combination with BI-3406. Concentrations of BI-3406 were comparable in groups treated either with monotherapy or combination. Combination therapy resulted in improved efficacy as assessed by change in tumor volume, decreases in cell proliferation, and alterations in the tumor microenvironment.
Materials and Methods
Mouse Husbandry
Mice were housed in temperature- and humidity-controlled facilities on a 12-hour dark/light cycle with free access to food and water. The animal care and use committee of Cincinnati Children’s Hospital Medical Center approved all animal use. The mice were on a largely C57Bl/6 genetic background, with some residual SV129 and FVBN strain background. The Dhh allele was maintained on the male, and mice were interbred to obtain the required genotypes. Mouse genotyping was carried out as previously described (Wu et al., 2008).
Selumetinib and SOS1 Inhibitor Dosing
Male and female DhhCre;Nf1fl/fl mice (n = 16 males and n = 20 females) did not differ significantly in tumor volume. Thus, in volumetric measurements of MRI scans we found insignificant differences between males and female (at 5 months, P = 0.63; at 7 months, P = 0.27; at 9 months, P = 0.21). In addition, the average change in tumor volume in tumor-bearing male and female mice between 7 months and 9 months was indistinguishable. Thus, we randomly used mice of both sexes in each treatment group. Mice were treated with selumetinib (AZD6244; Selleck Chemicals, LLC, Houston, TX) in 0.5% [w/v] methyl cellulose solution E-50 solution with 0.2% [v/v] polysorbate 80 [Tween-80] twice daily via oral gavage. The SOS1 inhibitor BI-3406 was provided by Boehringer Ingelheim and was administered at 50 mg/kg suspended in 0.5% Natrosol (hydroxyethylcellulose; 250HX) twice daily via oral gavage. For combinations of 5 or 10 mg/kg selumetinib + 50 mg/kg SOS1i (Combo), the drugs were made at 2× concentration and combined prior dosing. Each mouse received the same volume (10 ml/kg) for either single agent, vehicle, or the combinations. The duration of dosing is provided in the text for each experiment. Institutional Animal Care and Use Committee (IACUC) regulations mandated euthanasia if 20% decrease in body weight occurred.
Cardiac Puncture/Tumor Storage and Fixation
Four hours after final dose administration, mice were placed into an isofluorane-filled chamber until breathing ceased. Blood was collected into a 1.5-ml EDTA tube after cardiac puncture and then placed on ice for 30–60 minutes. Plasma was extracted after centrifugation at 4°C for 10 minutes. at 13,000 g. Paraspinal tumors were resected after cardiac puncture, and half of the tumors from each mouse were either flash frozen in liquid nitrogen for future use or placed in 4% paraformaldehyde for 1 hour at room temperature for histologic processing. Time from cardiac puncture to tumor removal was under 10 minutes for each mouse.
MRI and Neurofibroma Volumetric Measurement
A 7T Bruker BioSpec (Ettlingen, Germany) MRI was used to collect images in anesthetized DhhCre;Nf1fl/fl mice at 5, 7, and 9 months of age for neurofibroma volumetric measurements, as described previously for this model system (Wu et al., 2012; Jousma et al., 2015).
MRI, Statistical Analysis
Neurofibroma volumes for each mouse at each time point (5, 7, and 9 months) were compared using Fisher’s exact test. Volumetric changes were subsequently compared using log transformed tumor volume data with a random effects model using the Statistical Analysis System (SAS) mixed procedure (Wu et al., 2012).
Pharmacokinetic Analysis
All compounds were quantified in mouse plasma using BIB1355BS as internal standard and high-performance liquid chromatography tandem mass spectrometry (HPLC-MS/MS) (AB SCIEX QTRAP 6500+ Triple Quad with ESI-Probe, positive mode, 1-μl injection volume). Plasma proteins were removed from the samples by protein precipitation with acetonitrile. All compounds were separated by a gradient mobile phase (A: 5 mM ammonium acetate in water; B: 0.1% formic acid in acetonitrile) with a C-18 column (XBridge BEH C18 2.5-μm 2.1 × 50 mm). The Q1/Q3s (in positive mode) for selumetinib and BI-3406 were 457.2/301.1 and 463.3/206.1, respectively. The lower limit of quantification for BI-3406 was 1 nM using 1 μl of plasma. The lower limit of quantification for selumetinib was 0.5 ng/ml using 20 μl of plasma. Pharmacokinetic parameters including area under the curve (AUC) and the elimination rate constant (kel) were estimated (from average data) using noncompartmental methods with add-ins on Excel. Selumetinib and SOS1 plasma concentration were analyzed using a two-way repeated measures ANOVA.
Immunohistochemistry
Tumors were fixed for 1 hour in 4% paraformaldehyde, and then paraffin was embedded. Blocks were sectioned at 4-μm thickness and then baked at 60°C for 1 hour for air drying. We deparaffinized, hydrated, and transferred sections to 0.1 M citrate buffer (pH 6.0) for antigen retrieval. Sections were quenched with 3% H2O2 for 10 minutes, rinsed in PBS, and blocked using serum of appropriate species (10% serum with 0.3% Triton-X-100 in PBS) for 1 hour. After blocking, paraffin sections were incubated overnight in one of the following primary antibodies: Rabbit Anti-Ki67 (1:300; 12202; Cell Signaling, Danvers, MA), rabbit anti-Iba1 (1:3000; 019-19741; Wako Chemicals, Richmond, VA), anti-cyclin D1 (1:300; 9661S; Cell Signaling) and rabbit anti-cleaved caspase 3 (1:300; 9661S; Cell Signaling). After rinsing in PBS, sections were incubated in appropriate biotinylated secondary antibody (1:200; Vector Laboratories). Sections were rinsed after secondary antibodies, incubated in ABC (PK-S100; Vector Laboratories) for 1 hour, rinsed, and then incubated in DAB (3,3′-diaminobenzidine; SK-1400; Vector Laboratories). After DAB and rinses, sections were counterstained with Harris Hematoxylin (Poly Scientific R&D), rinsed in water, dehydrated in alcohol, cleared in xylene, and cover glassed using Histomount (Life Technologies). We acquired microscopic images with OpenLab software suites on a Zeiss Axiovert 200. For cell counting, three fields in each of three sections per animal from designated numbers of animals per group were analyzed. Cells were counted using Image J software. Statistical comparisons were conducted using Student’s t test one-way ANOVA; when normality and equal variance were not sufficient, a one-way ANOVA on Ranks was used.
Macrophage Morphometric Analysis
Analysis was carried out on ionized calcium binding adaptor molecule 1 (Iba1)-positive cells in paraffin sections. Images were acquired using a Nikon brightfield microscope with 40× magnification (objective: Nikon Plan APO Lambda 40×/0.95 DIC N2). Fifty to one hundred nonoverlapping cells per tumor were selected at random from the tumor center moving outward. Cropped images containing selected cells were processed to obtain a binary mask, and pixels from background and neighboring cells were removed to ensure a single continuous set of pixels. Each cell was processed with the original image as reference to avoid bias. To quantify changes in cell complexity, four morphologic parameters were measured using Fiji (ImageJ): area was measured as the total number of pixels present in the filled shape of the cell image, perimeter was based on the single outline cell shape as the number of pixels, circularity was calculated as (4π × cell area)/(cell perimeter)2, and roundness was calculated as (4A)/(πM2) (A = cell area; M = the longest axis of an ellipse fit to each cell). K-means clustering was used to define subpopulations of Iba1+ cells. Second, we performed Sholl analysis for each cell to quantify cell complexity; we quantified the number of Iba1+ cell branches intersecting concentric circles originating from the soma and spaced 0.05 μm apart.
Generation Characterization of Bone Marrow–Derived Macrophages
Bone marrow–derived macrophages (BMDMs) were generated from 6- to 9-month-old tumor-bearing (Nf1fl/fl; DhhCre+) mice. Autologous serum was derived after centrifugation of whole blood from the same tumor-bearing animals. Femurs and tibiae were dissected after euthanasia and flushed with Opti-MEM + GlutaMAX (Fisher). Cells were mechanically dissociated and filtered through a 40-micron cell strainer and then plated at 2 × 106 cells per well in 24-well plates. Nonadherent cells were washed away with warmed media after 2 hours to purify monocytes. Cells were maintained in Opti-MEM+ GlutaMAX plus 1% autologous serum and 1× Pen/Strep solution (GIBCO) for 6 days to derive mature BMDMs. We incubated BMDMs in serum-free Opti-MEM + GlutaMAX for 48 hours prior to treatment with PD0325901 (1μM), BI-3406 (1μM), a combination of the two inhibitors, or vehicle (DMSO) for 4 hours. Cells were then washed twice with warm PBS, and total cellular RNA was isolated via TRIzol reagent (Fisher). RNA was converted to cDNA using the High-Capacity cDNA Reverse Transcription Kit (Fisher). Real-time quantitative reverse-transcription polymerase chain reaction (qRT-PCR) of cytokine mRNAs was performed on the Bio-Rad CFS OPUS 96 wit Power SYBR Green Master Mix using cells from two tumor-bearing mice, each in three independent experiments; each qRT-PCR reaction was performed in duplicate. Expression of each gene was normalized to β-actin. Mouse primers are shown in Supplemental Table 1.
Macrophage Polarization Assay
Whole bone marrow monocytes were collected as above but from wild-type mice. We cultured equal numbers of monocytes and Nf1–/– Schwann cells for 48 hours; Schwann cells were isolated from the embryonic day 13.5 dorsal root ganglia (DRG) of Nf1–/– mutant embryos (Ratner et al., 2006). Coculture was carried out in Opti-MEM + GlutaMAX. Cells were detached using Accutase cell detachment solution for 5 minutes at 37°C (StemCell Technologies) and then washed twice with 1 ml of cold PBS by centrifugation. Subsequent steps were performed on ice. Cells were then stained with a live-dead exclusion marker (LIVE/DEAD Blue; Fisher) per manufacturer’s recommended protocol; washed with 1 ml of Flow Buffer (BioLegend); and then resuspended in Flow Buffer with Brilliant Buffer (1:200; BD Biosciences), True-Stain Monocyte Blocker (1:200; BioLegend), CD16/CD32 rat anti-mouse 2.4G2 block (1 μM; BD Biosciences), and primary antibodies (Supplemental Table 2) for 30 minutes. After incubation, cells were fixed with 2% freshly prepared paraformaldehyde in PBS (30 minutes at room temperature), and samples were run on a Cytek Aurora spectral analyzer within 24 hours. Single color controls and unstained controls were used to unmix samples.
Results
Dose Adjustment of Selumetinib To Maintain Selumetinib Concentration in Combination with BI-3406 Due to Drug-Drug Interaction
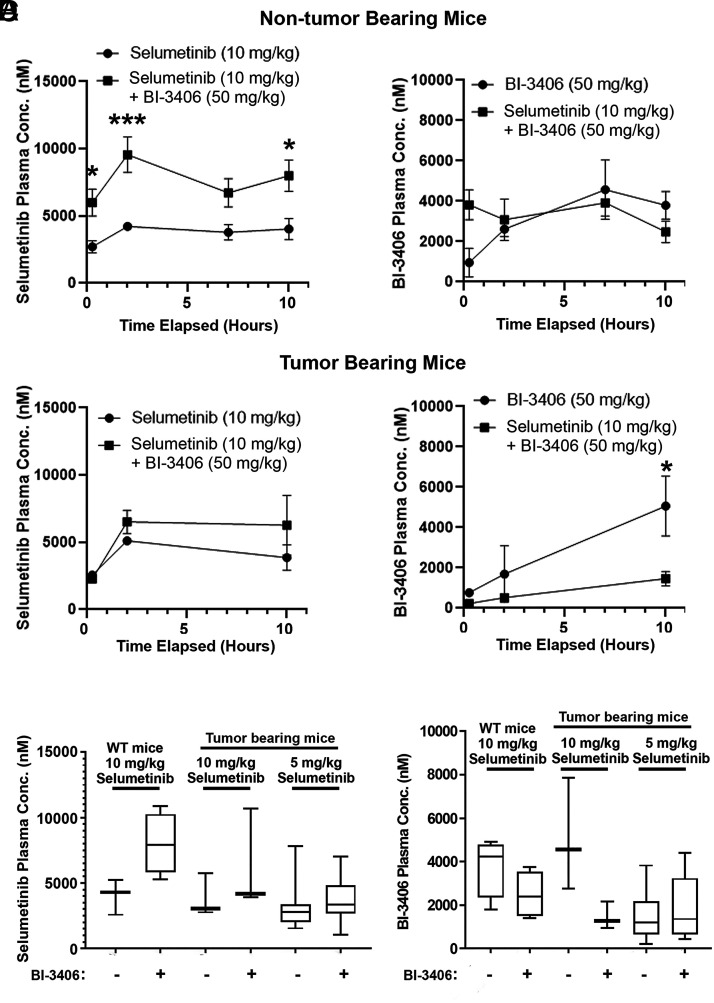
Drug plasma concentrations of selumetinib and BI-3406 (SOS1 inhibitor) were measured after administration alone or in combination to assess the potential of drug-drug interactions in combination-treated mice. The plasma concentration of selumetinib was increased in two experiments in combination with BI-3406. In the first experiment using non–tumor-bearing mice the increase was significant (Fig. 1A, left; two-way repeated measures ANOVA; overall difference of means = 3876.88 nM;P = 0.005), and in the second experiment with DhhCre;Nf1fl/fl tumor-bearing mice a trend to increased exposures was observed (nonsignificant, Fig. 1B, left; two-way repeated measures ANOVA). Given the apparent drug-drug interaction between the two compounds, in subsequent efficacy studies the dose of selumetinib was reduced in the combination group from 10 mg/kg to 5 mg/kg to achieve similar exposures for both monotherapy and combination treatments in the efficacy trial. At the end of the efficacy (daily dosing for 58 days) experiment, plasma was collected 4 hours after the last dose and exposures were determined and compared with results at 4 hours from the two prior tests (Fig. 1C). Comparable selumetinib levels were observed in mice when comparing 10 mg/kg monotherapy versus 5 mg/kg in combination with BI-3406 (Fig. 1C). The plasma concentration of BI-3406 was not statistically different when administered alone or in combination with selumetinib in non–tumor-bearing mice (Fig. 1A, right). In tumor-bearing mice, plasma levels of BI-3406 were reduced in combination with selumetinib (Fig. 1B, right). The dose of BI-3406 was not adjusted in combination as the observed reduction was only significant at one time point and there was no trend in the same direction in non–tumor-bearing mice (Fig. 1A, right). In the efficacy experiment (daily dosing for 58 days), concentrations of BI-3406 were comparable in groups treated either with monotherapy or combination (Fig. 1D).
Fig. 1.
PK analysis of selumetinib and the SOS1 inhibitor BI-3406. (A) and (B). Plasma concentrations (nM) of selumetinib (10 mg/kg) and BI-3406 (50 mg/kg) were measured over 10 hours. Two doses were separated by 6 hours. The first PK measurement was at 15 minutes. After the first dose, a second dose was administered at 6 hours. Plasma concentration of 10 mg/kg selumetinib (left panels) and 50 mg/kg BI-3406 (right panels) in non–tumor-bearing mice [e.g., wild-type littermates of tumor-bearing mice with genotypes Nf1fl/fl or Nf1fl/+ (A)] and tumor-bearing mice (B). Plasma concentrations of 10 mg/kg selumetinib (C) and 50 mg/kg BI-3406 (D) compared from three experiments: tumor-bearing mice, non–tumor-bearing mice, and after 2 months of dosing, each using selumetinib at the designated concentration of 5 or 10 mg/kg, each at 4 hours after a final dose of designated drugs.
Combination Drug Treatment Is Tolerable, Reduces Neurofibroma Extracellular Signal-Regulated Kinase Phosphorylation, and Leads to Reduced Tumor Cell Proliferation
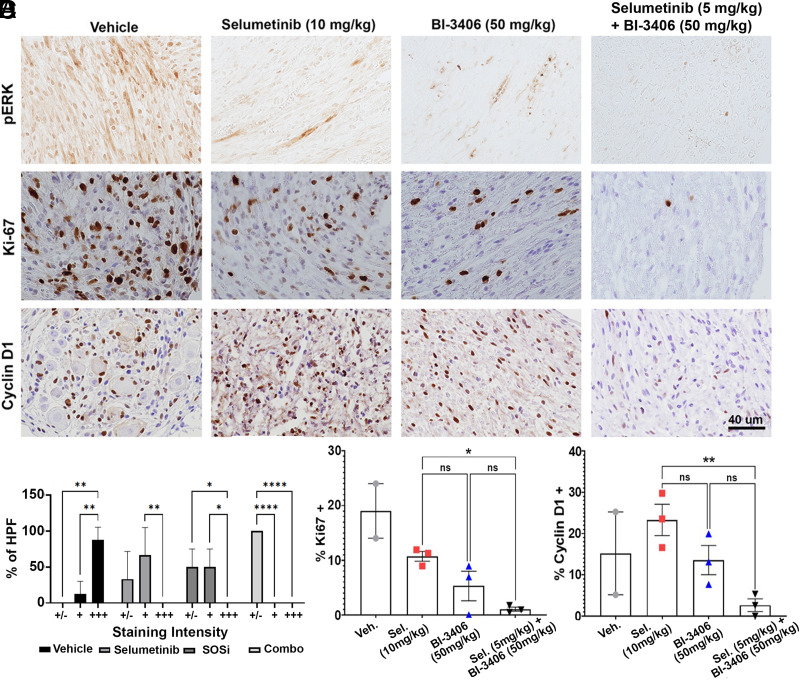
Seven-month-old tumor-bearing mice were treated for 15 days with either vehicle, selumetinib, BI-3406, or a combination of selumetinib and BI-3406 at the doses described above. A small decrease in the weight of mice treated with selumetinib in monotherapy was observed, but the effect was transient, and no mice showed >10% weight loss over the experimental period (Supplemental Fig. 1). Extracellular signal-regulated kinase phosphorylation (P-ERK) was assessed as a surrogate marker for RAS-MAPK pathway activation in tumor sections made 4 hours after final dosing of drug(s) (Fig. 2A). P-ERK staining decreased in all treatment groups when compared with vehicle-treated mice. The pronounced reduction was observed in combination groups (Fig. 2A). Figure 2A also shows representative tissue sections stained with either anti-Ki67 or anti-cyclin D1 to monitor the effect of the treatment on cell proliferation. On average, cell proliferation (Ki67+ cells) in selumetinib-treated tumors (10.7% ± 0.9%) decreased versus vehicle-treated tumors (18%), consistent with results of previous studies of MEK inhibitors in the DhhCre;Nf1fl/fl mouse model and clinical trials in patients (Jessen et al., 2013; Dombi et al., 2016, Gross et al., 2020). The percentage of Ki67+ cells in tumors was also reduced after single-agent treatment with BI-3406 (5.3% ± 2.7%) and combination treatment (1.0% ± 0.4%). Ki67+ cells were statistically reduced in the selumetinib/BI-3406–treated mice compared with selumetinib alone (P = 0.029; Fig. 2C). Quantification of cyclin D1+ cells similarly showed a significant reduction of the percentage of cyclin D1+ in the selumetinib/BI-3406–treated group compared with selumetinib as a single agent (P = 0.007; Fig. 2D). A trend, but no significant enhanced effect, was observed comparing the selumetinib/BI-3406 combination with either drug alone. Also, as is common in this model, one of three vehicle-treated mice died before the end of this biomarker study, limiting the statistical comparison with vehicle.
Fig. 2.
Combination drug treatment reduces neurofibroma P-ERK and tumor cell proliferation at 14 days of dosing. Representative images from IHC staining for P-ERK, cyclin D1, and Ki67 (A); quantification of P-ERK staining intensity (B); analysis in at least five high-powered fields per section per mouse; number of mice as shown for (C) and (D). Intensity is graded from least (+/−) to most (+++). Quantification of the percentage of positive cells in three sections per mouse >1000 cells were counted per condition; each symbol represents the average of data from a different mouse for Ki-67 (C). Quantification of the percent of cells expressing or cyclin D1 (D). For vehicle, n = 2 mice; for selumetinib, n = 3; for BI-3406, n = 3; for combination, n = 3.
In this model, levels of apoptosis in untreated and vehicle-treated tumors is under 1%; Mitogen activated protein kinase kinase inhibitor (MEKi) administration does not enhance apoptosis (Jessen et al., 2013). No increase in markers of apop-tosis (CC3+ cleaved caspase 3 positive cells) was observed upon treatment of neurofibromas with either drug alone or in the combination (not shown).
Efficacy of Selumetinib and Selumetinib Plus SOS1 Inhibition in Neurofibroma
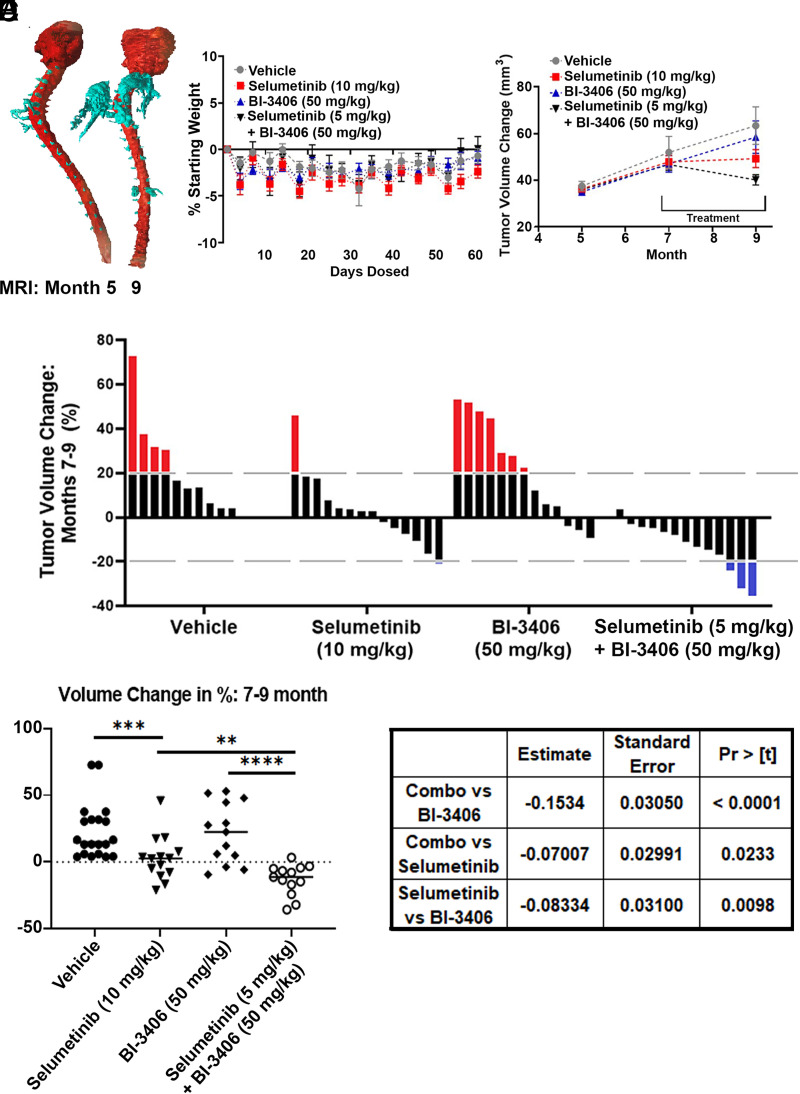
Tumor-bearing mice were treated with vehicle or drug(s) for 58 days to determine if SOS1 inhibitors show single agent or combination effects on neurofibroma growth. MRI scans from a representative vehicle treated, tumor-bearing mouse at 5 and 9 months of age was transformed into a finite element model (FEM) to for illustrative purposes, to show enlarging tumors primarily around the cervical enlargement (Fig. 3A). No mouse in any treatment group showed a decrease in body weight of more than 10% compared with their starting body weight during long-term continuous dosing (Fig. 3B).
Fig. 3.
Drug efficacy results after 58 days of dosing. (A) Finite element model (FEM) of a representative vehicle-treated mouse depicting tumor growth at 5 months and 9 months. Areas of spinal nerve/dorsal root ganglia and tumor are shown in turquoise; brain and spinal cord are shown in red. (B) Body weight measurements show no significant decrease in body weight. (C) Average tumor volume change per mouse, measured via volumetric MRI analysis. (D) Waterfall plot shows the percent change in tumor volume in individual mice between 7 and 9 months. Change in tumor volume for each mouse is shown as a single bar. Dotted lines show 20% change.
Average tumor volumes and waterfall plot showing tumor volume changes for individual mice treated with each agent are shown in Fig. 3, C and D. Tumor volume, as measured by repeated MRI, increased in vehicle treated mice (months 7–9) and decreased during treatment in some selumetinib-treated mice (6/14), as expected based on previous results (Jessen et al., 2013; Jousma et al., 2015; Dombi et al., 2016). At the end of the study, tumor size decreased in only three BI-3406 treated mice (3/13), and tumor volume was decreased in all but one mouse treated with the combination of selumetinib plus BI-3406 (12/13). (Fig. 3D). Compared with the control group, a significantly higher number of mice showed decreased tumor size after treatment with either (P < 0.0002) combination or selumetinib monotherapy treated groups (P = 0.001; one-sided nonparametric Mann Whitney-Wilcoxon U test, adjusted for multiple comparison according to Bonferroni Holm). No difference was observed in the BI-3406 treated group when compared with vehicle (P = 0.3891). When only the treatment groups are compared, the percent volume change of the tumor in the selumetinib/BI-3406 treated group was significantly greater when compared with either BI-3406 (P = 0.0002) or selumetinib monotherapy (P = 0.0033; one-sided nonparametric Mann Whitney-Wilcoxon U test, adjusted for multiple comparison according to Bonferroni Holm). Of note, treatment alone or in combination resulted in retardation of tumor growth rather than regression, as judged by comparing tumor volumes in mice at the end of the experiment to those measured in 5-month-old mice, when therapy was initiated (Fig. 3, C and D; statistical analysis shown in Fig. 3E).
Mixed models analysis allows a more detailed analysis of volume change, as it accounts for the known heterogeneity in neurofibroma growth in the DhhCre;Nf1fl/fl mouse model. Heterogeneity in change in tumor size is observed both across a group and in longitudinal measurements for each mouse (Wu et al., 2012). Random coefficient model analysis results agreed with the tumor shrinkage analysis: tumor shrank during the treatment period (months 7–9) in the selumetinib/BI-3406 combination treated and the selumetinib treated mice (P < .0001, P < .0001) but not in the BI-3406 treated or the vehicle treated group (P = 0.1819, P = 0.3607). The shrinkage observed in the combination treated group was statistically larger than that compared with either single agent treated group; P < .0001 and P = 0.0233 when compared with the BI-3406 and selumetinib treated groups, respectively. Results of this statistical analysis are shown in Fig. 3F.
Long-Term Treatment Shows Sustained Reductions in Ki-67 and Cyclin D1
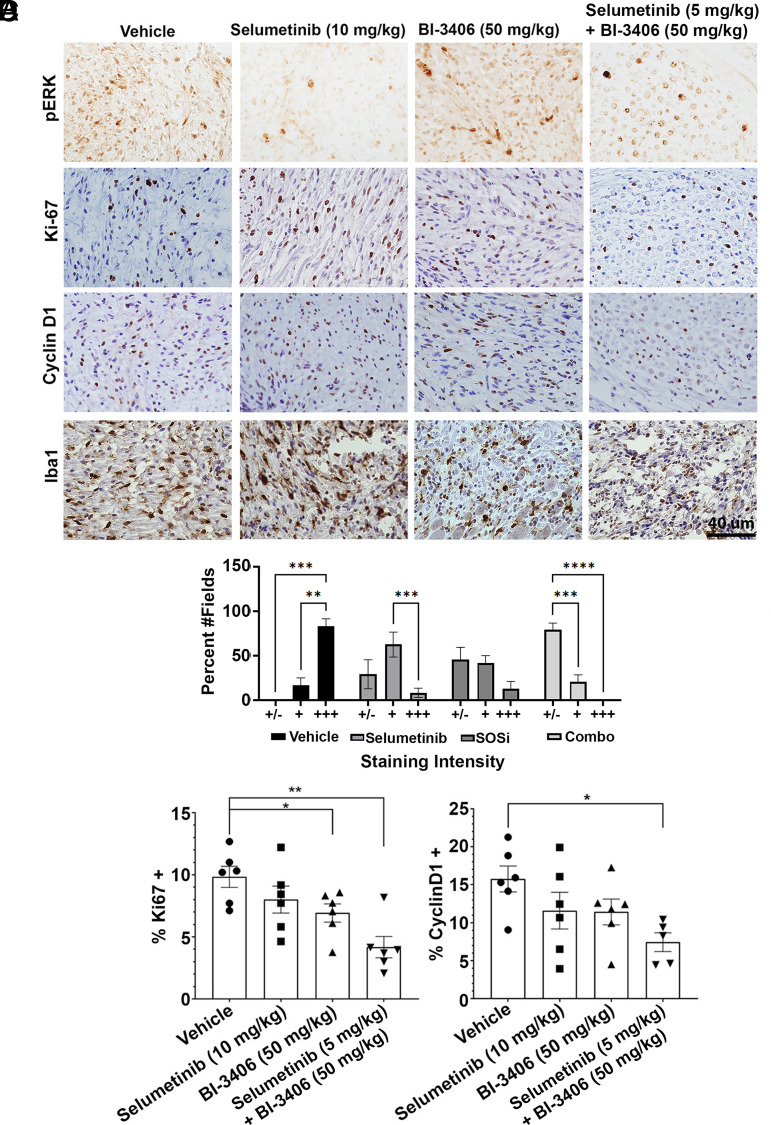
Four hours after administration of final dose of drug(s) in the efficacy study, tissue was collected, fixed, and embedded for immunohistochemical assessment (Fig. 4A). Immunohistochemistry (IHC) analysis demonstrated P-ERK reduction in the selumetinib and selumetinib/B-3406–treated groups compared with the vehicle. In contrast, BI-3406 exposure did not reduce P-ERK at this time point even though a reduction in cell proliferation was sustained for the 8 weeks of dosing in the combination. For example, Ki67+ cells represented 9.8% ± 0.9% of cells in vehicle-treated samples and 4.2% ± 0.9% in the combination. A one-way ANOVA showed significant decreases in cell proliferation, as assessed by Ki67+ positive cells, upon treatment with a combination of both drugs compared with vehicle (P < 0.001) or to selumetinib alone (P = 0.032) (Fig. 4B). Similar results were observed upon analysis of the cell cycle marker cyclin D1 in the combination treatment group compared with the vehicle (P = 0.037; Fig. 4C).
Fig. 4.
Two-month treatment results in sustained reductions in cell proliferation. IHC images and analysis. (A) Representative images of P-ERK, Ki-67, cyclin D1, and Iba1 stained sections. Analysis of P-ERK staining intensity (B). Quantitative analysis of the percentage of cells that are Ki67+ (C) or cyclin D1+ (D). Quantification was as for Fig. 2. Reduced proliferation in the combination treatment is more than in the selumetinib-treated samples.
Although treatment did not reduce Iba1+ macrophage number, a change in Iba1+ cell morphology was observed (Supplemental Fig. 2A). Given the large numbers of macrophages in neurofibromas and the observation that macrophage morphology changes with macrophage function (McWhorter et al., 2013), we analyzed macrophage shape in more detail (Fig. 5).
Fig. 5.
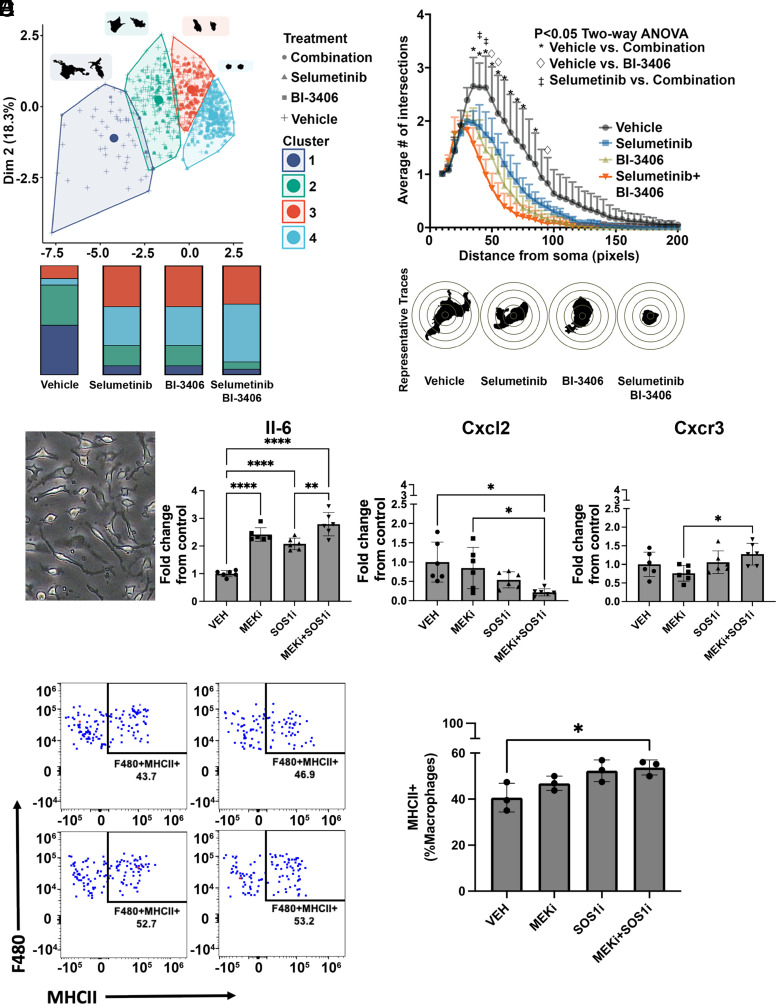
Treatment with selumetinib in combination with BI-3406 alters Iba1+ macrophage shape. (A, left) Scatter plot showing clusters (n = 4) of Iba1+ macrophages, defined via K-means cluster analysis based on cell size and cell shape characteristics. Each dot represents data from a single cell in samples of PNF treated with vehicle, selumetinib, BI-3406, or selumetinib and BI-3406. Examples of two cells with characteristic morphology for each cluster are shown in black (A, right). Bar graphs showing the frequency of cells in each cluster for each treatment (B, right). Sholl plot indicates a significant loss of arborization in Iba1+ cells, traced from animals that received selumetinib in combination with BI-3406 when compared with either vehicle-treated (*) or selumetinib-treated (•) animals, (P < 0.05, two-way ANOVA, n = 4, 50–100 cells per animal) (B, left). Traces of cells representative of each treatment condition. (C) Representative phase contrast image of mouse bone marrow–derived macrophages collected from tumor-bearing mice and maintained in defined serum-free medium. (D) Changes in mRNA expression of cytokines after 4-hour treatment with vehicle, selumetinib, BI-3406, or selumetinib + BI-3406. Graphs show significant increases in IL6 and reduced levels of Cxcl2 after combination treatments. Macrophage analysis by Flow cytometry (E and F). Representative plots (G) and quantification (H) of MHCII expression in F480+ macrophages grown for 48 hours with Nf1−/− Schwann cells.
Macrophages Change Morphology after Selumetinib/SOS1 Inhibition
Changes in the tumor microenvironment were analyzed after treatment with selumetinib and BI-3406 alone or in combination. Alterations in macrophage shape is directly associated with a change in cytokine expression and activation profile in vitro (Rodell et al., 2018) and indirectly with changes in cytokine expression and patient survival in vivo (Donadon et al., 2020). To identify alterations in macrophage morphology, individually traced Iba1+ macrophages were assessed by size (area and perimeter) and cell complexity (circularity and roundness). Macrophages were separated into four K-means clusters using shape characteristics so that each cluster contained cells of a different shape (Supplemental Fig. 2, B and C); quantification showed that treatment with selumetinib, BI-3406, or a combination of the two drugs increased the frequency of Iba1+ cells in cluster 4, containing small, round macrophages, versus vehicle-treated mice (Fig. 5A). We next performed Sholl analysis, a more sensitive analysis of Iba1+ cell morphology, in which the number of cell processes that cross concentrically placed rings are counted for each cell (McWhorter et al., 2013). Selumetinib in combination with BI-3406 resulted in a significant loss of Iba1+ cell arborization, with lesser effects in other treatment groups. Results are quantified in Fig. 5B, which also shows representative single cells indicative of a profound change in Iba1+ cells in the tumor microenvironment.
To determine if treatment with BI-3406, MEK inhibitor, or the combination directly modulates macrophages, we treated mature macrophages from tumor-bearing mice (Fig. 5C). Cells were treated for 4 hours, and then changes in gene expression were assessed using qRT-PCR. Combination treatment resulted in a significant decrease of Cxcl2 gene expression (a cytokine known to polarize macrophages toward an anti-inflammatory phenotype) and a significant increase in interleukin 6 (IL-6), known to polarize macrophages toward a proinflammatory phenotype (Fig. 5D). We also observed a slight but significant increase in Cx3cr1 compared with MEK inhibitor treatment; this receptor is known to modulate macrophage inflammatory response (Fig. 5D). Increased levels of major histocompatibility complex class II (MHCII) mark macrophage activation; notably, normal nerve macrophages express MHCII (Ydens et al., 2020). Therefore, we also tested if combination treatment alters macrophage activation using MHCII as a readout. Macrophages were identified by multiparametric flow cytometry, as shown in Supplemental Fig. 3. We cocultured wild-type monocytes with Nf1−/− Schwann cells to mimic tumor-bearing mice. In this setting, the combination treatment resulted in a significant 25% increase in macrophages (CD45+;CD11b+;F480+;Ly6C-;Ly6G-) with high MHCII expression (Fig. 5, E and F).
Discussion
MEK inhibitor treatment results in reduction in tumor growth in the DhhCre;Nf1fl/fl neurofibroma model (Jessen et al., 2013). Evidence of activity in 70% of individuals with PNF resulted in FDA approval of selumetinib in children and young adults with NF1 (Dombi et al., 2016;). Notably, despite clinically meaningful and durable improvements (lasting >1 year) and the fact that drug resistance was not noted, some patients had to discontinue treatment due to toxicities common to MEK inhibitors, shrinkage was only partial, and dose reduction resulted in progression in five out of six patients (Gross et al., 2020). Here we tested the hypothesis that RAS/MAPK pathway inhibition using a novel SOS1 inhibitor could potentiate the activity of MEKi in neurofibroma (Kessler et al., 2021). Although the robust effect of selumetinib makes it difficult to discern additional effects, we report significant improvement in several tumor parameters upon combination with SOS1 inhibition.
In published studies using KRAS mutant tumor models, monotherapy with BI-3406 resulted in a modest antiproliferative effect but showed strong synergy in combination with MEK inhibitors, presumably by blocking feedback release mechanisms (Hofmann et al., 2021). In the DhhCre;Nf1fl/fl mouse model, monotherapy with BI-3406 (Fig. 3, C and D) resulted in no beneficial efficacy. However, an enhanced effect was observed upon combination with an exposure adjusted dose of the MEK inhibitor selumetinib. Combination of these agents might increase the durability of the response by attenuating resistance mechanisms mediated by negative feedback signaling or upstream receptor tyrosine kinase (RTK) signaling (Kun et al., 2021). Further, preclinical studies are needed to test this hypothesis. The beneficial efficacy in the combination correlated with reduced proliferation and cell cycling as assessed by cyclin D1 and Ki67 staining. It is possible that use of an MEK inhibitor with longer circulation half-life (e.g., trametinib or PD-0325901) would enhance the observed combination effects. Like selumetinib, these MEK inhibitors have undergone clinical testing in individuals with PNF (Weiss et al., 2021), and combination studies could be carried out to assess this hypothesis.
In these studies, we confirmed that selumetinib as a monotherapy causes a transient reduction in P-ERK levels in a mouse model (Dombi et al., 2016). In agreement with the sensitivity observed in a subset of NF1 mutant cancer cell lines (Hofmann et al., 2021), we observed that BI-3406 similarly reduces levels of P-ERK in neurofibroma tumors in vivo after 2 weeks of dosing. This data supports the concept that neurofibromas rely on SOS1 as a guanine-nucleotide exchange factor (GEF) for RAS. However, at the end of a 2-month treatment period, P-ERK was only marginally reduced by BI-3406 as a single agent, suggesting that cells adapt to overcome long-term exposure to this inhibitor. BI-3406 only targets SOS1 but not its paralog SOS2. It will be interesting to analyze in subsequent studies if, in neurofibromatosis cells, upregulation of alternative exchange factors such as SOS2 occurs.
Activation of the MAPK pathway results in negative feedback mechanisms that act through SOS1. In the presence of pathway inhibitors, these regulatory pathways are released so that targeting SOS1 makes sense as a combination partner for MAPK inhibitors such as MEK inhibitors. Indeed, SOS1 inhibitors have been shown to enhance the efficacy of MEK inhibitors, resulting in a stronger pathway modulation compared with both single agents (Hofmann et al., 2021). This described effect likely explains the combination effects that we observed on MAPK pathway modulation and efficacy after treatment with selumetinib plus BI-3406 in the DhhCre;Nf1fl/fl mouse model. Additional analysis of RAS-MAPK signaling in short- and long-term treatment groups would be useful, including analysis of downstream effector protein mRNAs (e.g., Dusp6, Spry 2, Spry 4). Dusp6 mRNA was reduced 20-fold 6 hours after dosing the MEK inhibitor PD-0325901 in the neurofibroma model (Jessen et al., 2013).
We analyzed the effect of treatment on macrophages because macrophage infiltration is observed in nerve before neurofibroma formation and macrophages are the dominant immune cells in mouse and human neurofibromas (Prada et al., 2013; Liao et al., 2018; Fletcher et al., 2019). Macrophages become permissive for tumor growth once tumors are established (Prada et al., 2013). Treatment with selumetinib, BI-3406, or a combination of the two drugs resulted in striking changes in tumor macrophage morphology, a change to smaller cells with fewer processes that correlates with macrophage activation in other settings (McWhorter et al., 2013; Dewhurst et al., 2017). For example, blocking SOS1 reduced levels of Src family or Abl tyrosine kinase activation in macrophages and diminished macrophage migration (Baruzzi et al., 2015). In our mouse model, the observed effects are at least in part cell autonomous to macrophages. Treating macrophages with BI-3406 and MEK inhibitor uniquely decreased CXCL2 [chemokine (C-X-C motif) ligand 2] expression. This change, together with increases in IL6 and MHCII, is predicted to alter tumor macrophage phenotype so that macrophages are less tumor supporting.
We have shown that efficacy of the combination of SOS1 and MEK inhibition is greater compared with single-agent MEK inhibition and correlates with an enhanced antiproliferative effect and changes in the tumor microenvironment. The combination with BI-3406 could allow a reduction in the required dose of selumetinib and potentially prevent some MEK inhibitor–induced side effects. Reducing toxicity and enhanced efficacy is important because tumor regrowth is observed in NF1 patients who undergo MEK inhibitor dose reductions due to adverse side effects. The combination of an MEKi with an SOS1 in this preclinical study holds the promise of increased clinical benefit that needs to be investigated in a phase 1 clinical trial.
Acknowledgments
The authors thank Katherine A. Chaney for assistance with dosing animals. The authors also thank Oezlem Yuece Petronczki for critical review of the manuscript.
Abbreviations
- BMDM
bone marrow–derived macrophage
- Iba1
ionized calcium binding adaptor molecule 1
- IHC
immunohistochemistry
- IL6
interleukin 6
- KRAS
ras-related protein K
- MAPK
mitogen-activated protein kinase
- MEK
mitogen-activated and extracellular signal-regulated kinase
- MHCII
major histocompatibility complex class II
- MRI
magnetic resonance imaging
- NF1
neurofibromatosis type 1
- P-ERK
extracellular signal-regulated kinase phosphorylation
- PK
pharmacokinetics
- PNF
plexiform neurofibroma
- qRT-PCR
real-time quantitative reverse-transcription polymerase chain reaction
- SC
Schwann cell
- SOS
Son of Sevenless
Authorship Contributions
Participated in research design: Ahmari, Arnhof, Boehmelt, Düchs, Long, Maier, Hofmann, Ratner.
Conducted experiments: Jackson, Ahmari, Wu, Rizvi. Fugate, Dombi, Arnhof.
Contributed new reagents or analytic tools: Arnhof, Trapani.
Performed data analysis: Kim, Arnhof.
Wrote or contributed to the writing of the manuscript: Jackson, Ahmari, Wu, Rizvi, Fugate, Kim, Dombi, Arnhof, Boehmelt, Düchs, Long, Maier, Trapani, Hofmann, Ratner.
Footnotes
This work was funded in part by Boehringer Ingelheim and by National Institutes of Health National Institute of Neurological Disorders and Stroke [Grant R01-NS28840] (to N.R.).
H.A., G.B., M.J.D., C.J.L., U.M., F.T., and M.H.H. are full-time employees at affiliations of Boehringer Ingelheim. Revolution Medicines funds some of N.R.’s research, unrelated to this manuscript.
 This article has supplemental material available at jpet.aspetjournals.org.
This article has supplemental material available at jpet.aspetjournals.org.
References
- Baltanás FC, Zarich N, Rojas-Cabañeros JM, Santos E (2020) SOS GEFs in health and disease. Biochim Biophys Acta Rev Cancer 1874:188445. [DOI] [PubMed] [Google Scholar]
- Baruzzi A, Remelli S, Lorenzetto E, Sega M, Chignola R, Berton G (2015) Sos1 regulates macrophage podosome assembly and macrophage invasive capacity. J Immunol 195:4900–4912. [DOI] [PubMed] [Google Scholar]
- Dewhurst JA, Lea S, Hardaker E, Dungwa JV, Ravi AK, Singh D (2017) Characterisation of lung macrophage subpopulations in COPD patients and controls. Sci Rep 7:7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombi EBaldwin AMarcus LJFisher MJWeiss BKim AWhitcomb PMartin SAschbacher-Smith LERizvi TA, et al. (2016) Activity of selumetinib in neurofibromatosis type 1-related plexiform neurofibromas. N Engl J Med 375:2550–2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donadon MTorzilli GCortese NSoldani CDi Tommaso LFranceschini BCarriero RBarbagallo MRigamonti AAnselmo A, et al. (2020) Macrophage morphology correlates with single-cell diversity and prognosis in colorectal liver metastasis. J Exp Med 217:e20191847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher MJShih CSRhodes SDArmstrong AEWolters PLDombi EZhang CAngus SPJohnson GLPacker RJ, et al. ; Neurofibromatosis Clinical Trials Consortium (2021) Cabozantinib for neurofibromatosis type 1-related plexiform neurofibromas: a phase 2 trial. Nat Med 27:165–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher JS, Springer MG, Choi K, Jousma E, Rizvi TA, Dombi E, Kim MO, Wu J, Ratner N (2019) STAT3 inhibition reduces macrophage number and tumor growth in neurofibroma. Oncogene 38:2876–2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross AMWolters PLDombi EBaldwin AWhitcomb PFisher MJWeiss BKim ABornhorst MShah AC, et al. (2020) Selumetinib in children with inoperable plexiform neurofibromas. N Engl J Med 382:1430–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig A, Markwart R, Esparza-Franco MA, Ladds G, Rubio I (2015) Ras activation revisited: role of GEF and GAP systems. Biol Chem 396:831–848. [DOI] [PubMed] [Google Scholar]
- Hillig RCSautier BSchroeder JMoosmayer DHilpmann AStegmann CMWerbeck NDBriem HBoemer UWeiske J, et al. (2019) Discovery of potent SOS1 inhibitors that block RAS activation via disruption of the RAS-SOS1 interaction. Proc Natl Acad Sci USA 116:2551–2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann MHGmachl MRamharter JSavarese FGerlach DMarszalek JRSanderson MPKessler DTrapani FArnhof H, et al. (2021) BI-3406, a potent and selective SOS1-KRAS interaction inhibitor, is effective in KRAS-driven cancers through combined MEK inhibition. Cancer Discov 11:142–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessen WJMiller SJJousma EWu JRizvi TABrundage MEEaves DWidemann BKim MODombi E, et al. (2013) MEK inhibition exhibits efficacy in human and mouse neurofibromatosis tumors. J Clin Invest 123:340–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jousma ERizvi TAWu JJanhofer DDombi EDunn RSKim MOMasters ARJones DRCripe TP, et al. (2015) Preclinical assessments of the MEK inhibitor PD-0325901 in a mouse model of neurofibromatosis type 1. Pediatr Blood Cancer 62:1709–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler DGerlach DKraut NMcConnell DB. . Targeting Son of Sevenless 1: the pacemaker of KRAS.2021. Curr Opin Chem Biol 62:109–118. [DOI] [PubMed] [Google Scholar]
- Kluwe L, Friedrich R, Mautner VF (1999) Loss of NF1 allele in Schwann cells but not in fibroblasts derived from an NF1-associated neurofibroma. Genes Chromosomes Cancer 24:283–285. [DOI] [PubMed] [Google Scholar]
- Kun E, Tsang YTM, Ng CW, Gershenson DM, Wong KK (2021) MEK inhibitor resistance mechanisms and recent developments in combination trials. Cancer Treat Rev 92:102137. [DOI] [PubMed] [Google Scholar]
- Lavoie H, Gagnon J, Therrien M (2020) ERK signalling: a master regulator of cell behaviour, life and fate. Nat Rev Mol Cell Biol 21:607–632. [DOI] [PubMed] [Google Scholar]
- Liao C-P, Booker RC, Brosseau J-P, Chen Z, Mo JTchegnon E, Wang Y, Clapp DW, and Le LQ, (2018) Contributions of inflammation and tumor microenvironment to neurofibroma tumorigenesis. J Clin Invest 128:2848–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWhorter FY, Wang T, Nguyen P, Chung T, Liu WF (2013) Modulation of macrophage phenotype by cell shape. Proc Natl Acad Sci USA 110:17253–17258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miettinen MMAntonescu CRFletcher CDMKim ALazar AJQuezado MMReilly KMStemmer-Rachamimov AStewart DRViskochil D, et al. (2017) Histopathologic evaluation of atypical neurofibromatous tumors and their transformation into malignant peripheral nerve sheath tumor in patients with neurofibromatosis 1-a consensus overview. Hum Pathol 67:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba Y, Mochizuki N, Yamashita S, Chan AM, Schrader JW, Hattori S, Nagashima K, Matsuda M (2000) Regulatory proteins of R-Ras, TC21/R-Ras2, and M-Ras/R-Ras3. J Biol Chem 275:20020–20026. [DOI] [PubMed] [Google Scholar]
- Pemov ALi HPatidar RHansen NFSindiri SHartley SWWei JSElkahloun AChandrasekharappa SCBoland JF, et al. ; NISC Comparative Sequencing Program; NCI DCEG Cancer Genomics Research Laboratory (2017) The primacy of NF1 loss as the driver of tumorigenesis in neurofibromatosis type 1-associated plexiform neurofibromas. Oncogene 36:3168–3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkin SRBredella MACai WKassarjian AHarris GJEsparza SMerker VLMunn LLMuzikansky AAskenazi M, et al. (2012) Quantitative assessment of whole-body tumor burden in adult patients with neurofibromatosis. PLoS One 7:e35711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prada CEJousma ERizvi TAWu JDunn RSMayes DACancelas JADombi EKim MOWest BL, et al. (2013) Neurofibroma-associated macrophages play roles in tumor growth and response to pharmacological inhibition. Acta Neuropathol 125:159–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen SA, Friedman JM (2000) NF1 gene and neurofibromatosis 1. Am J Epidemiol 151:33–40. [DOI] [PubMed] [Google Scholar]
- Ratner N, Miller SJ (2015) A RASopathy gene commonly mutated in cancer: the neurofibromatosis type 1 tumour suppressor. Nat Rev Cancer 15:290–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratner N, Williams JP, Kordich JJ, Kim HA (2006) Schwann cell preparation from single mouse embryos: analyses of neurofibromin function in Schwann cells. Methods Enzymol 407:22–33. [DOI] [PubMed] [Google Scholar]
- Rodell CB, Arlauckas SP, Cuccarese MF, Garris CS, Li R, Ahmed MS, Kohler RH, Pittet MJ, Weissleder R (2018) TLR7/8-agonist-loaded nanoparticles promote the polarization of tumour-associated macrophages to enhance cancer immunotherapy. Nat Biomed Eng 2:578–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffzek K, Shivalingaiah G (2019) Ras-specific GTPase-activating proteins-structures, mechanisms, and interactions. Cold Spring Harb Perspect Med 9:a031500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra E, Rosenbaum T, Nadal M, Winner U, Ars E, Estivill X, Lázaro C (2001) Mitotic recombination effects homozygosity for NF1 germline mutations in neurofibromas. Nat Genet 28:294–296. [DOI] [PubMed] [Google Scholar]
- Staser K, Yang FC, Clapp DW (2012) Pathogenesis of plexiform neurofibroma: tumor-stromal/hematopoietic interactions in tumor progression. Annu Rev Pathol 7:469–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uusitalo E, Leppävirta J, Koffert A, Suominen S, Vahtera J, Vahlberg T, Pöyhönen M, Peltonen J, Peltonen S (2015) Incidence and mortality of neurofibromatosis: a total population study in Finland. J Invest Dermatol 135:904–906. [DOI] [PubMed] [Google Scholar]
- Weiss BDWolters PLPlotkin SRWidemann BCTonsgard JHBlakeley JAllen JCSchorry EKorf BRobison NJ, et al. (2021) NF106: a Neurofibromatosis Clinical Trials Consortium phase II trial of the MEK inhibitor mirdametinib (PD-0325901) in adolescents and adults with NF1-related plexiform neurofibromas. J Clin Oncol 39:797–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widemann BCDombi EGillespie AWolters PLBelasco JGoldman SKorf BRSolomon JMartin SSalzer W, et al. (2014) Phase 2 randomized, flexible crossover, double-blinded, placebo-controlled trial of the farnesyltransferase inhibitor tipifarnib in children and young adults with neurofibromatosis type 1 and progressive plexiform neurofibromas. Neuro-oncol 16:707–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Dombi E, Jousma E, Scott Dunn R, Lindquist D, Schnell BM, Kim MO, Kim A, Widemann BC, Cripe TP, Ratner N (2012) Preclincial testing of sorafenib and RAD001 in the Nf(flox/flox); DhhCre mouse model of plexiform neurofibroma using magnetic resonance imaging. Pediatr Blood Cancer 58:173–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Williams JP, Rizvi TA, Kordich JJ, Witte D, Meijer D, Stemmer-Rachamimov AO, Cancelas JA, Ratner N (2008) Plexiform and dermal neurofibromas and pigmentation are caused by Nf1 loss in desert hedgehog-expressing cells. Cancer Cell 13:105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ydens EAmann LAsselbergh BScott CLMartens LSichien DMossad OBlank TPrijck SDLow D, et al. (2020) Profiling peripheral nerve macrophages reveals two macrophage subsets with distinct localization, transcriptome and response to injury. Nat Neurosci 23:676–689. [DOI] [PMC free article] [PubMed] [Google Scholar]