Abstract
Background
Rapid and efficient strategies are needed to discover neutralizing antibodies (nAbs) from B cells derived from virus-infected patients.
Methods
Here, we report a high-throughput single-B-cell cloning method for high-throughput isolation of nAbs targeting diverse epitopes on the SARS-CoV-2-RBD (receptor binding domain) from convalescent COVID-19 patients. This method is simple, fast and highly efficient in generating SARS-CoV-2-neutralizing antibodies from COVID-19 patients’ B cells.
Results
Using this method, we have developed multiple nAbs against distinct SARS-CoV-2-RBD epitopes. CryoEM and crystallography revealed precisely how they bind RBD. In live virus assay, these nAbs are effective in blocking viral entry to the host cells.
Conclusion
This simple and efficient method may be useful in developing human therapeutic antibodies for other diseases and next pandemic.
Statement of Significance: We described a single-B-cell cloning method on how to rapidly and efficiently discover SARS-CoV-2-neutralizing antibodies; this method is potentially useful for discovering therapeutic antibodies for many diseases.
INTRODUCTION
COVID-19 pandemics caused by the coronavirus SARS-CoV-2 has rapidly and continuously spread around the world, and caused a huge health and economic crisis. In particular, the occurrence of the Omicron variant of SARS-CoV-2 has raised grave alarm because they are spreading fast [1]. Therefore, there is an urgent need to develop new therapies against the Omicron variant as well as new technologies that allow fast development of new therapies. SARS-CoV-2 encodes a spike (S) glycoprotein on the surface, which has two functional subunits S1 and S2. The S1 subunit contains receptor-binding domain (RBD), which binds to human endogenous angiotensin converting enzyme 2 (ACE2) receptor [2]. Like SARS-CoV, the SARS-CoV-2 enters human host cells through binding of RBD to ACE2 [3–5]. Neutralizing monoclonal antibodies (nAbs) against RBD have been the major effective therapy approved by regulatory agencies (such as US FDA) to treat SARS-CoV-2 infections. One approach to generate nAbs is to clone nAbs from single B cells in patients who had been infected with COVID-19 convalescent patients. Therapeutic proteins may be induce anti-drug (proteins or mAbs) antibodies (ADAs) in human patients, which may alter mAb efficacy and even lead to unwanted side effects. Cloning of nAbs from convalescent patients is not only more efficient (take less time and effort) but also more likely to generate nAbs with significantly less ADA (anti-drug antibodies).
Single-B-cell cloning and single-B-cell sequencing had been used to discover SARS-CoV-2 nAbs from convalescent COVID-19 patients [6–15]. Single-B-cell cloning technique involves isolating B cells from patients, sorting and cloning genes of the nAbs in single B cells, and expression of the cloned genes in mammalian cells. However, since the functionality of the mAbs can only be verified after cloning and expression, these methods are costly and time-consuming. Here, we report the development of neutralizing antibodies against SARS-CoV-2 including the Omicron variant, using a high-throughput method. This method involves isolation of potent neutralizing antibodies from a large number of single B cells derived from convalescent COVID-19 patients, using a high-throughput cloning method combined with a highly sensitive HTRF (Homogeneous Time Resolved Fluorescence) RBD-ACE2 blocking assay. More than a thousand paired heavy chain and light chain (PCR products) were transfected (without DNA purification) and functional screening was performed for candidate mAbs selection. Subsequently, potent neutralizing mAbs were expressed and purified for the analyses, including viral neutralization and structure characterization. With this method, we could detect thousands of mAbs’ functional characteristics without construction of thousands expression recombinant plasmids. We isolated multiple nAbs that target diverse epitopes on virus spike protein. A5–10 retained potent neutralizing activity against the Delta variant and A34–2 retained potent neutralizing activity against the Omicron variant. A5–10 and A34–2 were selected for clinical development, and A5–10 has now been submitted for IND (Investigational New Drug) approval. This simple and efficient approach could also be useful in isolating therapeutic antibodies against other infectious virus and next pandemic.
RESULTS
High-throughput isolation of functional mAbs from single B cells
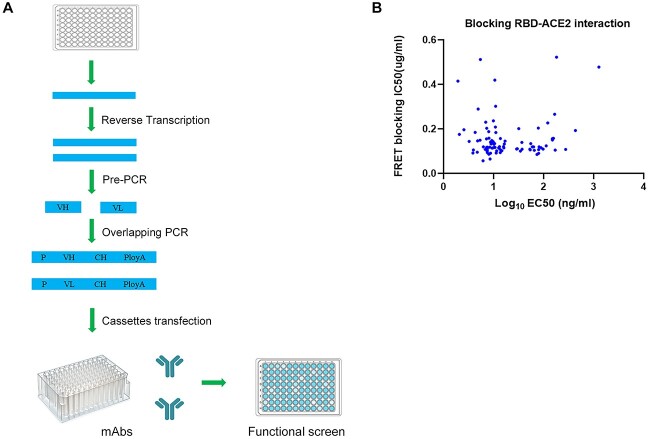
nAbs are capable of blocking the viral entry into human host cells by interacting with their primary target SARS-CoV-2 spike protein. Not all antibodies bind RBD with high affinity, and not all high-affinity antibodies can block viral entry. To obtain a large number of high affinity nAbs against diverse epitopes on the antigen RBD, we obtained blood samples from 19 convalescent patients from several hospitals in China. To isolate memory B cells that produce SARS-CoV-2 specific nAbs, peripheral blood mononuclear cells (PBMCs) from convalescent patients were stained with a fluorescently labeled SARS-CoV-2 S protein and sorted by fluorescence activated cell sorting with gating for an CD27 + IgM − IgG + Spike + population (Fig. S1). Technologies include single-B-cell cloning, memory B-cell immortalization and B-cell activation have been previously used to isolate human mAbs from memory B cells [16]. Transcriptionally active PCR has been used to rapidly generate monoclonal antibodies directly from plasma cells [17]. We developed a high-throughput single-B-cell cloning method for human mAbs discovery based on transcriptionally active PCR (Fig. 1A). There are three steps involved in this method. First, reverse transcription and pre-PCR to obtain heavy-chain and light-chain variable regions (VH and VL) from human single B cells. Second, overlapping PCR to generate expression cassettes with promoter, VH and VL fragments, constant regions and poly A. Third, transient expression of the mAb-containing cassettes into CHO cells. Supernatants of the transfected cells were evaluated for the RBD specific binding activities by ELISA. Since these reactions were carried out using RNAs from a single B cell in a single well, natural pairing of VH:VL was ensured. Direct transfection of paired VH:VL cassettes eliminated the need for individually constructing and expressing antibody vectors, and therefore dramatically increased the efficiency of antibody cloning. With this method, we sorted 1800 memory B cells from COVID-19 convalescent patients into 96-well plates in a single-cell manner and 1089 pairs of VH/VL were successfully cloned. The VH-VL cassettes from single memory B cells were directly transfected into CHO cells to produce mAbs.
Figure 1.
Strategy of cloning functional mAbs from single B cells. (A) Schematic diagram of the method for high-throughput cloning of antibody genes from single B cells. VH and VL gene fragments were amplified via Pre-PCR from single B cells’ cDNA. Then, variable region genes, promoter DNA and constant region gene were combined and amplified to produce VH-VL cassettes. The VH-VL cassettes were directly transfected into CHO cell line to express mAbs. Anti-S protein antibodies were identified by ELISA of the supernatants of the transfected cells. (B) MAbs were evaluated for binding and blocking to target by ELISA and HTRF. MAbs blocking ACE2-RBD interaction would cause a reduction in HTRF signal. Apparent binding EC50s are reported as ng/ml and blocking IC50s are reported as μg/ml.
To quickly identify RBD neutralizing antibodies, we used the RBD-ACE2 blocking assay (Supplementary Fig. S2) to screen for antibodies that can block ACE2 and RBD binding. In total, 93 RBD-binding antibodies blocked ACE2-RBD effectively (IC50 < 0.5 μg/ml) with high binding affinity (Fig. 1B). Thus, we succeeded in isolating 93 potent RBD-ACE2 blocking mAbs using this simple approach.
Sequence analysis of RBD-ACE2 blocking mAbs
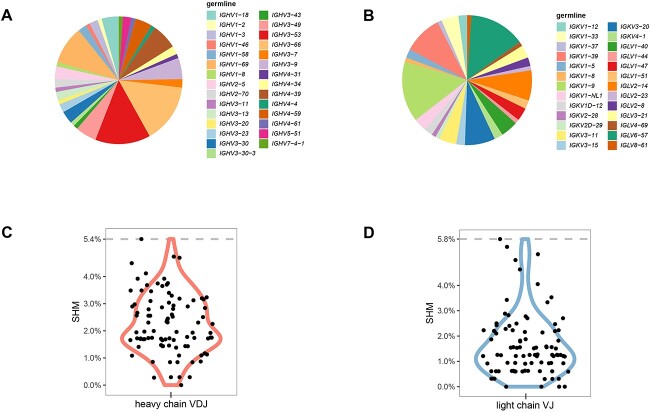
To assess the diversity of 93 RBD-ACE2 blocking mAbs, we analyzed sequences of the 93 RBD-ACE2 blocking antibodies. We found that these antibodies have a broad use of heavy chain and light chain genes at germline levels (Fig. 2A and B). IGHV3–53 has been shown to be the most frequently used germline gene for targeting SARS-CoV-2 RBD [18]. Among heavy-chain germline genes of the 93 antibodies, IGHV3–53, IGHV3–66 and IGHV1–69 were most frequently observed. Germline genes are capable of generating potent SARS-CoV-2 nAbs with limited mutations [14]. Consistent with this notion, almost all 93 RBD-ACE2 blocking antibodies were somatically mutated, with low level of somatic hypermutation (SHM) in VH and VL (Fig. 2C and D). Similarly, neutralizing antibodies to MERS-CoV were also somatically mutated with low SHM [19]. These results suggest that the human germline gene can respond effectively to coronavirus with little mutation. Together, our sequence analysis confirmed that potential nAbs isolated by the high-throughput single-B-cell cloning method we developed exhibited antibody diversity at the germline genes level.
Figure 2.
RBD-ACE2 blocking mAbs sequence analysis. (A) Usage of VH germline genes. The area ratio of each part in the pie chart represents the ratio of the VH germline. (B) Usage of VL germline genes. The area ratio of each part in the pie chart represents the ratio of the VL germline. (C) Violin plot showing somatic hypermutation of heavy chain. Each dot represents the median SHM percentage of mAb’s heavy chain. (D) Violin plot showing somatic hypermutation of light chain. Each dot represents the median SHM percentage of mAb’s light chain.
Virus neutralization
Among the 93 antibodies, we selected 14 antibodies based on their diversity of germline genes. We determined the binding constant (Ka) and dissociation constant (Kd) to RBD of these nAbs by Biolayer Interferometry (Supplementary Table S1). The binding affinities of the nAbs (KD) were derived by KD=Kd/Ka. Next, we used a replication-defective lentivirus expressing SARS-CoV-2 S to evaluate the abilities of the nAbs to block SARS-CoV-2 pseudovirus infection. The neutralization profiles of these 14 mAbs are shown in Fig. 3A. Based on their KDs and IC50 in the pseudovirus assay, we selected nine nAbs to measure their capabilities to inhibit live SARS-CoV-2 using plaque reduction neutralization tests (PRNT) (Fig. 3B). Consistent with their abilities against SARS-CoV-2 pseudovirus infection, the nine nAbs were highly potent in blocking the live virus, with IC50 values ranging from 0.03 to 0.55 μg/ml. To roughly map the binding epitopes on the RBD that the nAbs bind, we established competition experiments based on HTRF. Based on the amount of competition, epitopes binning was performed for the nine nAbs. The percentage of competition for each pair of nAbs was measured by competition experiments (Supplementary Fig. S3). These analyses revealed potential some nAbs that are not competitive and target to several epitopes on RBD.
Figure 3.
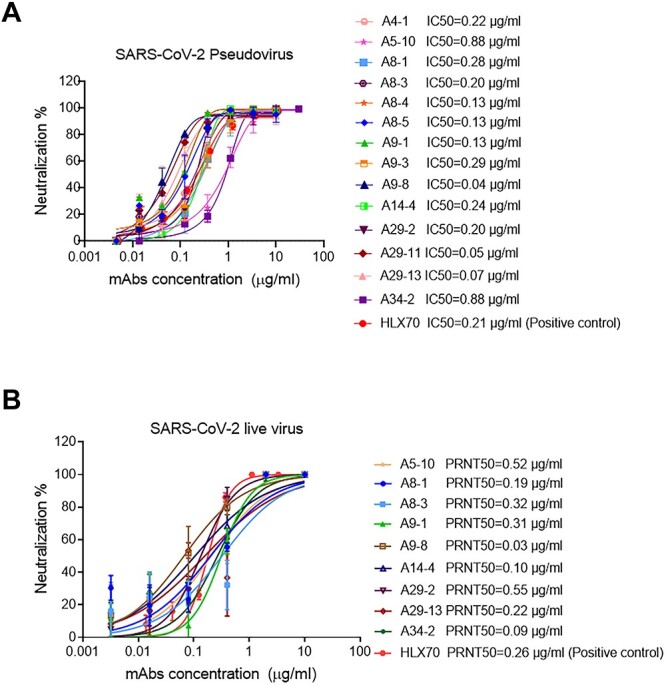
SARS-CoV-2 neutralizing mAbs. (A) Neutralization activities of nAbs against SARS-CoV-2 pseudovirus. SARS-CoV-2 pseudovirus were incubated with 3-fold serial dilutions of nAbs, with the highest concentration set to 10 μg/ml. The same experiments were done twice independently. IC50 were determined by a four-parameter logistic model. (B) Neutralization profiles of the nine purified nAbs against SARS-CoV-2 live virus. nAbs with 5-fold serial dilutions were incubated with the same volume of approximately 100 PFU of SARS-CoV-2, with the highest concentration set to 10 μg/ml. The mixtures were then added to the Vero cell monolayers. The same experiments were done twice independently. The PRNT50 values were determined using non-linear regression analysis. The positive control antibody is HLX70, and US Food and Drug Administration has approved the application for a phase I clinical trial with HLX70.
Structural basis for neutralization
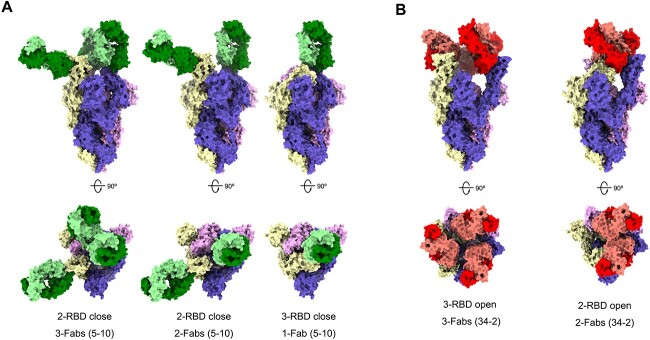
To gain insights into the structural basis of the epitopes, we selected three antibodies to solve the structure of RBD antibodies or Spike trimer antibodies. The analyses of competition revealed nAbs A8–1, A5–10 and A34–2 may target to different epitopes on RBD (Supplementary Fig. S3). We solved the crystal structure of RBD-A8–1-Fab complex by X-ray crystallography (Supplementary Fig. S4 and Supplementary Tables S2 and S3). The structure of RBD-A8–1-Fab complex was superimposed with hACE2-RBD complex (PDB: 6M0J) [20], and analysis revealed that about 78% of the residues constituting the epitope of A8–1 are within the RBM (receptor binding motif) directly binding with ACE2, indicating that A8–1 competes directly with RBD in binding to hACE2 (Supplementary Fig. S4). Each SARS-CoV-2 RBD could exist in two distinct configurations referred to as close and open state [3]. Neutralizing antibodies may recognize a specific RBD configuration in the S trimer [21–23]. To determine which states of RBD in the S trimer that A5–10 and A34–2 could interact with, we produced cryo-EM reconstructions of A5–10 Fab and A34–2 Fab in complex with the S trimer of SARS-CoV-2 (Supplementary Fig. S5 and Supplementary Table S4). Three main states were observed in A5–10 Fab-S trimer particle, a 3-Fab-bound complex with two RBDs closed (i.e. two A5–10 Fab bound to two closed state RBDs and one A5–10 Fab bound to an open state RBD) at 6.1 Å resolution, a 2-Fab-bound complex with two RBDs closed at 4.8 Å resolution, and a 1-Fab-bound complex with three RBDs closed at 4.7 Å resolution (Fig. 4A). Thus, nAb A5–10 could interact with RBD in both close form and open form. Two main states were observed in A34–2 Fab-S trimer particles, a 3-Fab-bound complex with three RBDs open at 5.3 Å resolution, and a 2-Fab-bound complex with two RBDs open at 5.3 Å resolution (Fig. 4B). The nAb A34–2 could therefore interact only with open state RBD. Altogether, cryo-EM reconstructions confirmed that the nAbs we discovered could not only target to diverse epitopes but also recognize different RBD states.
Figure 4.
Cryo-EM structures of the SARS-CoV-2 S trimer in complex with Fabs. All the structures were generated by rigid-body docking of the coordinates of S trimer subunits (PDB: 6vsb). The Fab-RBD-complexes reconstructed into the cryo-EM density maps and shown from both the side view (up) and top view (down). (A) Structures of S trimer in complex with A5–10 have three distinctive conformations. Left: only one RBD rotating up but each RBD binding with a Fab 5–10. Middle: two RBDs (one up and one down) binding with two Fabs and one RBD free. Right: three RBDs in ‘all-closed’ with only one accessible to Fab 5–10. (B) Structures of S trimer in complex with A34–2 revealed two conformations—one with RBDs in the ‘all-up’ configurations, each being recognized by a Fab (left); another with one RBD in down and two RBDs in up and only the ones in up binding with Fab 34–2.
Neutralizing of SARS-CoV-2 variants by antibodies
SARS-CoV-2 spike protein mutations are being discovered continuously [24, 25]. The SARS-CoV-2 variants including the Beta variant (B.1.351), Delta variant (B.1.617.2) and the emerging Omicron variant (B.1.1.529) significantly alter the abilities of nAbs to neutralize SARS-CoV-2 infection [26–29]. Indeed, several neutralizing antibodies drugs including bamlanivimab, REGN10933/REGN10987, AZD1061/ AZD8895 and BRII-196 were significantly reduced by the Delta variant or the Omicron variant [30–34]. To assess the impact of mutations on nAbs A8–1, A5–10 and A34–2, we tested the 50% neutralization dose (ND50) of these antibodies to neutralize the SARS-CoV-2 variants live virus. The Beta variant reduced the neutralization capability of A8–1 (Fig. 5A). From the atomic Fab-RBD structures, it is clear that the binding epitope of mAb A8–1 includes K417 and N501 (Fig. S4 and Supplementary Table S3), which are the main mutations occurring in the Beta variant. The Omicron variant contains 15 mutations on the RBD, including G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y and Y505H [35]. For nAb A5–10, the N440K in Omicron broke down a hydrogen bond formed between A5–10 and WT RBD. For nAb A8–1, the mutations of Q498R, Y505H and Q493R broke down two, two and one hydrogen bonds, respectively, that were present in the complex of A8–1 and WT RBD, and R493 even clashed with V101. These changes resulted in the largely decreased neutralizing activities of A5–10 and A8–1 against Omicron. However, the evasion of Omicron against A34–2 is not effective, since the only mutation of N417 that is present on the epitope of A34–2 can form the same hydron bond with E31 as was shown with K417 in the WT RBD, which formed an hydrogen bond with E31 (Fig. 5B). We also tested the ability of nAbs A8–1, A5–10 and A34–2 to neutralize the pseudoparticle variants (VOCs, BA.2, BA.2.12.1, BA.3, BA.4/5). The neutralization potency of A8–1, A5–10 and A34–2 are greatly reduced by Omicron sublineages BA.2, BA.2.12.1, BA.3, BA.4/5 (Supplementary Table S5). Together, these analyses suggest that Omicron sublineages would escape most neutralizing antibodies against wild-type SARS-CoV-2.
Figure 5.
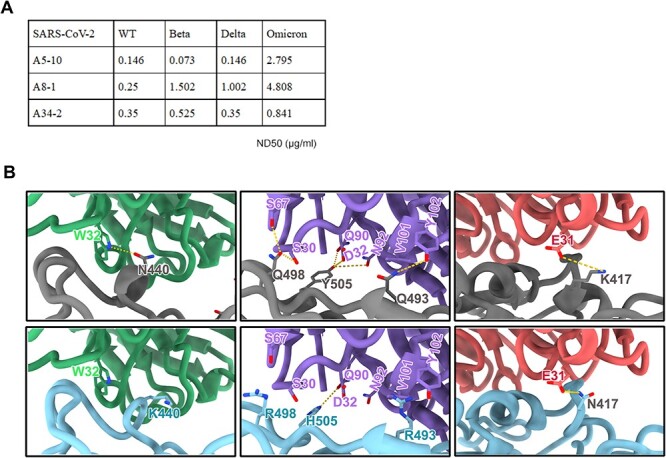
Neutralizing activity of nAbs against SARS-CoV-2 variants. (A) Neutralizing activities of nAbs A5–10, A8–1 and A34–2 against SARS-CoV-2 and variants. The ND50 of nAbs for each variant are shown in the table. ND50 values indicated the dose of each mAb for 50% neutralization of viral infection. (B) The structural insights into the immune evasion of A5–10 (left), A8–1 (middle) and A34–2 (right) by Omicron. The structures of A5–10, A8–1 and A34–2 in complex with WT-RBD (top) were superimposed with the structure of Omicron RBD (PDB code: 7WED) (bottom) with main mutations responsible for immune invasion marked out. Antibody A5–10, A8–1 and A34–2, WT-RBD and Omicron-RBD were colored in blue, purple, red, grey and cyan, respectively.
DISCUSSION
Therapeutic antibodies have become a major modality for the treatment of many diseases in virtually all therapeutic areas. Among all therapeutic antibody discovery technologies, antibodies derived from human B cells are highly valuable and sometimes preferred method of discovering therapeutic antibodies, especially during a fast-developing pandemic [36, 37]. New strategies in discovering high-quality neutralizing antibodies from patient derived B cells are desired to effectively treat infectious diseases. In this study, we report a fast and simple method that allows high-throughput isolation of nAbs. This method helped us to identify a large collection of nAbs against SARS-CoV-2 RBD with high affinity and high potency that could simultaneously bind several epitopes on RBD and block the viral entry to the host cells. It is conceivable that this method could be used in developing antibodies to treat other diseases.
A number of different approaches have been used in isolating monoclonal antibodies from human B cells. Single-B-cell cloning has been used to obtain antibody genes from single human B cells with natively paired VH: VL antibodies. This method involves FACS of single B cells, followed by cloning and expression of the cloned nAb genes individually, and therefore is time-consuming and low throughput. Another method is B-cell cloning based on microfluidic technologies. For example, Berkeley Lights’ Beacon optofluidic system is used to distribute thousands of single B cells at one time on a culture chip. The antibody-secreting B cells could be identified by a simple fluorescence detection system, and the antibody genes could be cloned by single-cell cloning [38, 39]. However, in vitro activation and expansion of memory B cells is time-consuming and difficult. The single-B-cell sequencing technique developed recently has outperformed the above methods. Single B cells with antigen (e.g. RBD) specific antibodies on their surface are isolated by FACS sorting, followed by high-throughput single-cell RNA and VDJ sequencing of antibody repertoires using 10X Chromium. Even so, there is still a need for simple, high-throughput, highly efficient method to clone the genes for therapeutic antibodies from single B cells, for infectious as well as non-infectious diseases.
In this study, we used a high-throughput method for isolation of potent nAbs that target diverse epitopes on the RBD of SARS-CoV-2 from convalescent COVID-19 patients. The first part of our method is similar to the ones by others, namely, we enriched S trimer or RBD-specific B cells from convalescent COVID-19 patients. The second part, we coupled single-B-cell VH-VL expression cassettes transfection and RBD-ACE2 blocking assay, and used a simple method for fast and high-throughput isolation and profiling of a large number of mAbs. Given that RNAs from a single B cell were used to carry out the two reactions in a single well (Fig. 1A), natural pairing of VH and VL of IgG was guaranteed. By combining single-cell reverse transcription, pre-PCR and transfection of linear PCR products in a single well, there is no need to use expensive single-cell sequencing instruments or microfluidic device. ELISA assay was used to identify the mAbs, which could bind RBD (Fig. 1B). With this method, we could obtain and screen for thousands of mAbs within 2 weeks from convalescent COVID-19 patients. These mAbs could be used for subsequent functional testing directly.
The diversity of antibodies is the key in developing high-quality neutralizing antibodies. This is achieved by both germline and somatic mutations of the antibody genes. Interestingly, we found that potent neutralizing antibodies isolated by our method have a broad use of heavy-chain and light-chain genes at germline levels, with an excellent diversity. Of the nine nAbs we selected, all were highly potent in blocking live SARS-CoV-2, and can target diverse RBD epitopes. The structures solved and subsequent structural analyses suggest that the nAbs A5–10, A8–1 and A34–2 target to distinct epitopes on RBD and recognize different SARS-CoV-2 S trimer conformations, reminiscence of a lot of published mAbs with high potency and breadth, e.g., XGv347 [40], S309 [41], J08 [42], 10–28 [43] and LY-CoV1404 [44]. These antibodies have been suggested to target more effectively the S trimers, which exert up-to-down dynamic conformations. The diversity of neutralizing antibodies is crucial not only in responding to viral mutations but also in antibody drug discovery in general. This high-throughput cloning method may have high applicable value in developing therapeutic antibodies with diverse epitope. Indeed, we were able to obtain neutralizing mAbs against SARS-CoV-2 rapidly.
Finally, this method can also be used to discover antibodies for non-infectious diseases. Aducanumab, a human monoclonal antibody that selectively reduces Aβ plaques, was discovered by human B cells from healthy elderly [45]. Our method could also be used to discover new therapeutic antibodies against Aβ or other AD targets, with much high efficiency and diversity. In brief, this simple and high-throughput single-B-cell cloning method is potentially useful for discovering therapeutic antibodies for many diseases and next pandemic.
METHODS
ELISA analysis of plasma
The recombinant SARS-CoV-2 spike (Vazyme, CG203) was diluted to final concentrations of 2 μg/ml. Diluted spike coated onto 96-well plates and incubated at 4°C overnight. Samples were washed with PBST (PBS containing 0.05% Tween-20) and blocked with blocking buffer (PBS containing 5% skim milk) at 37°C for 2 h. Either serially diluted plasma samples or diluted purified mAbs were added 96-well plates and incubated at 37°C for 1 h, then washed with PBST for three times and incubated with secondary anti-human IgG labeled with horseradish peroxidase (HRP). Optical density (OD) was measured by a spectrophotometer at 450 nm.
Sorting of spike-specific memory B cells
B cells were purified magnetically (STEMCELL Technologies, 17954) and stained with anti-CD27-APC (Biolegend, 356410), anti-human IgM (Biolegend, 314512), anti-human IgG (Biolegend, 410708) and biotinylated spike protein. Spike-specific memory B cells were isolated by flow-cytometric sorting using a BD FACSAria ΙΙΙ flow cytometer (BD Biosciences) from pooled PBMCs. Flow-cytometric data were analyzed with FlowJo.
Single-B-cell RT-PCR and high-throughput cloning
Single B cells’ cDNA was prepared using Sc Reverse Transcriptase (Vazyme, N721) primed with oligo (dT) via RT-PCR. Expression cassettes of antibodies were amplified via two rounds of PCR. Variable region genes were amplified via Pre-PCR from single B cells’ cDNA using gene-specific primers at both the 5′ and 3′ ends. The PCR program for Pre-PCR: 95°C for 3 min, 30 cycles of 95°C for 15 s, 61°C for 15 s and 72°C for 30 s. Variable region genes, a human cytomegalovirus (HCMV) promoter fragment and an antibody constant region were combined and amplified to produce linear products using overlapping PCR: one linear PCR product encoding heavy chain and the other linear PCR product encoding light chain. The PCR program for overlapping PCR: 95°C for 3 min, 30 cycles of 95°C for 15 s, 58°C for 15 s and 72°C for 2 min. All of the PCR primers are listed in Supplementary Table S6. Purified overlapping PCR products (two separate linear PCR products) were co-transfected into ExpiCHO (Life Technologies) cells grown in 96-deep well plates (2 ml per well). Monoclonal antibodies were produced by transient transfection of ExpiCHO cells. Monoclonal antibodies’ concentrations were determined by Add&Read Human Fc kit (Vazyme DD2102–01). Antigen-specific ELISA and HTRF ACE2-RBD assay were used to detect the binding capacity and blocking activity of nAbs in transfected culture supernatants to SARS-CoV-2 RBD.
ELISA analysis of binding of antibodies to SARS-CoV-2 RBD
SARS-CoV-2 RBD (Vazyme, CG201) was diluted to final concentrations of 2 μg/ml, then coated onto 96-well plates and incubated at 4°C for overnight. Samples were washed with PBS-T one time and blocked with blocking buffer (PBS containing 5% skim milk and 2% BSA) at 37°C for 2 h. MAbs were diluted in serial dilutions starting at 5 μg/ml, added the plates and incubated at 37°C for 1 h. Wells were then incubated with secondary anti-human IgG labeled with HRP and TMB substrates. OD was measured by a spectrophotometer at 450 nm.
HTRF assay for ACE-RBD blocking antibodies
The final concentrations of Eu anti mouse Fc antibody (Vazyme, DD2303) and d2 anti his tag antibody (Vazyme, DD2302) were 0.2 and 2 μg/ml, respectively. Equal volumes of RBD-his, ACE2-mFc and diluted nAbs were mixed. The final concentrations of RBD-his and ACE2-mFc were at 5 and 2 nM, respectively. The nAbs were added in 3-fold serial dilutions starting at 10 μg/ml to 384 Microporous plates. Mixed donor and receptor were added into the 384 Microporous plate. After 2-h final incubation at room temperature, HTRF signals were measured using a Multimode Reader (Spark, Tecan) equipped and fluorescence detected at 620 and 665 nm. IC50 (HTRF) was defined as the nAbs concentration at which the combination of ACE2 and RBD decreased by 50%.
Antibody affinity characterization
Binding kinetics of SARS-CoV-2 RBD (Acro) and anti-SARS-CoV-2 RBD antibodies was determined by biolayer interferometry using the Octet RED96 system (FortéBio). The AHC biosensors were pre-equilibrated in a buffer containing 20 mM HEPES, pH 7.4 and 120 mM NaCl and 0.02% (v/v) Tween-20 for 10 min. Antibodies were loaded onto AHC biosensors for 120 s. Biosensors were coupled with gradient concentrations of SARS-CoV-2 RBD for 60S. The Biosensors dissociate in Sample Dilution Buffer for 180 s. Binding kinetics was evaluated using a 1:1 Langmuir binding model by ForteBio Data Analysis 11.0 software.
Pseudovirus neutralization assay
The pseudovirus neutralization assays were performed using lentivirus pseudoparticle and HEK-293 cell line, which highly express ACE2. Serially diluted mAbs (3-fold serial dilutions using Dulbecco's Modified Eagle's medium, DMEM) were mixed with SARS-CoV-2 pseudovirus in a 96-well plate and incubated for 1 h at 37 °C. Then, 50 μl (approximately 2 × 104 cells per well) of HEK-293-ACE2 were added into the 96-well plate and incubated for 48 h at 37°C. Titers were measured as luciferase activity in relative light units (Vazyme DD1204). The luciferase signal was measured using a SPARK microplate reader (Tecan). IC50 (half-maximal inhibitory concentrations) of the evaluated monoclonal antibodies were determined by luciferase activity using GraphPad Prism 7 (GraphPad Software).
Authentic SARS-CoV-2 PRNT
PRNT was performed using a clinical isolate of SARS-CoV-2 (BetaCoV/Beijing/IME-BJ01/2020). Briefly, 5-fold serial dilutions of mAbs were added into the same volume of approximate 100 PFU of SARS-CoV-2 and incubated for 1 h at 37°C. The mixture was then added into a monolayer of Vero cells in a 24-well plate and incubated for 1 h at 37°C. After that, the mixture was removed, and 0.5 ml of 1.0% (w/v) low melting point (LMP) agarose (Promega) in 2 × DMEM supplied with 4% (v/v) foetal bovine serum (FBS) was added onto the infected cells. After further incubation at 37°C supplied with 5% CO2 for 2 days, the wells were stained with 1% (w/v) crystal violet dissolved in 4% (v/v) formaldehyde to visualize the plaques. All the above experiments were performed in a Biosafety Level 3 facility. The values of PRNT50 were calculated using GraphPad Prism 7 (GraphPad Software).
Authentic SARS-CoV-2 CPE assay
To evaluate the neutralization activity against the live SARS-CoV-2 virus, the live virus neutralization assay was performed in a BSL-3 laboratory of National Institute for Viral Disease Control and Prevention, China CDC. The prototype (F13 Strain), Beta (GD84 Strain), Delta (NCoV210097 strain) and Omicron (NPRC2.192100003) viruses were used in the live virus neutralization assays. The antibody or molecule was serially diluted and mixed with an equal volume of 100 TCID50 live SARS-CoV-2 virus. After incubation at 37°C for 2 h, the neutralized mixed solution was added into the well of the plate containing Vero cells with density of 2 × 105 per ml, which was then cultured at 37°C for 3 to 4 days. Both cell and virus controls were also set up as comparison. Subsequently, the inhibition of virus infection to the cells was observed, and the neutralizing antibody titer against the live SARS-CoV-2 virus was measured as the reciprocal of the serum dilution for 50% neutralization of viral infection.
Crystallization and structure solution
The purified SARS-CoV-2 RBD was mixed and incubated with the cleaved Fab of A8–1 (with a molar ratio of 1:1.2). RBD-A8–1 further purified by gelfiltration chromatography. Crystals were successfully obtained in 2% v/v Tacsimate pH 5.0, 0.1 M Sodium citrate tribasic dihydrate pH 5.6, 16% w/v polyethylene glycol 3,350. Diffraction data were collected at 100 K and at a wavelength of 1.071 Å on the BL17U1 beam line of the Shanghai Synchrotron Research Facility. Diffraction data were autoprocessed with aquarium pipeline and the data processing statistics are listed in Supplementary Table S3. The structure was solved by the molecular replacement method. Subsequent model building and refinement were performed using COOT and PHENIX, respectively. The structure file for RBD-A8–1 has been deposited in the Protein Data Bank (PDB) under accession number 7F7H.
Expression and purification of S-trimer
The gene containing the coding fragments of SARS-CoV-2 S ectodomain (residues 1–1208) or RBD domain (residues 319–541) (GenBank: MN908947) was synthesized and inserted into the mammalian expression vector pCAGGS. For the S gene, two proline substitutions at residues 986–987, a ‘GSAS’ substitution at the furin cleavage site and a C-terminal T4 fibritin folden site were introduced to facilitate the protein expression. A C-terminal twin-strep-tag II was inserted into both two constructs for protein purification. Then, either of the two constructed plasmids was transiently transfected into HEK293F cells using polyethylenimine. After incubation at 37°C for 3 days, the supernatant was harvested, filtered and purified by affinity chromatography using column packed with streptavidin-attached resin and further dialyzed into the buffer containing 20 mM Tris pH 8.0 and 200 mM NaCl.
Cryo-EM sample preparation and data collection
The purified S protein was mixed and incubated with the cleaved Fab of A5–10 or A34–2 (with a molar ratio of 1.0: 1.5) to obtain the S-Fab complexes; and purified RBD was mixed and incubated with both A5–10 Fab and A34–2 Fab to obtain the complex of RBD-A5–10-A34–2 (with a molar ratio of 1.0:2.0:2.0). Afterwards, 3-μl aliquot of either complex was deposited onto the glow-discharged holey carbon-coated gold grid, blotted for 7 s with a force value of 0, or 5 s with a force value of 2, but both in100% relative humidity and then plunged into the liquid ethane using Vitrobot (FEI). Cry-EM data sets were collected at 300 kV with a Titan Krios microscope (FEI). Movies for S-Fab-complexes or RBD-Fabs-complex were recorded using a K2 Summit direct detector with a defocus range between 1.5 and 2.7 μm or 1.8–3.0 μm, respectively. Automated single particle data acquisition was performed by SerialEM, with a pixel size of 1.04 Å or 0.83 Å, respectively.
Cryo-EM data processing
A total of 2913, 2689 and 4421 micrographs of the complexes of S-A5–10, S-A34–2 or RBD-Fabs were recorded for data processing. After motion correction, all the images were subjected to defocus value estimation by Gctf. Then, particles were autopicked and extracted for two-dimensional alignment and three-dimensional classification. The resulting models were performed postprocessing to obtain the final maps, which were determined by the gold-standard Fourier shell correlation and evaluated by ResMap. All procedures involved were carried out in Relion 3.0.
Cryo-EM data availability
The cryo-EM density maps of S-A5–10, S-A34–2 and RBD-A5–10-A34–2 have been deposited in the Electron Microscopy Data Bank under accession codes: EMD-31439, EMD-31435, EMD-31436, EMD-31437, EMD-31438 and EMD-31434 respectively. Other data will be available from the corresponding authors upon reasonable request.
AUTHORS’ CONTRIBUTIONS
Y.D. and B.L. conceived and designed the study. Y.L., G.Z. and H.L. are in charge of sample collection. Y.D. with assistance from X.X. isolated neutralizing mAbs. Y.D. with assistance from L.X. and P.D. conducted and analyzed pseudovirus neutralization experiment. Y.-Q.D. and K.X. with assistance from T.C., C.-F.Q. and G.W. conducted and analyzed the live virus neutralization experiment. Y.D. and J.L. with assistance from X.W. solved and analyzed crystal structure of antibody and RBD complex. Z.J. and P.L. assistance from K.W. and X.W. solved and analyzed Cryo-EM structure. Y.D. and B.L. wrote and edited the manuscript.
FUNDING
This work was supported by Vazyme Biotech and Tsinghua University.
CONFLICT OF INTEREST STATEMENT
Patent has been filed for some of the antibodies presented here. X.X., L.X. and P.D. are employees of Nanjing Vazyme Biotech, and declare that they have no other conflicts of interest that might be relevant to the contents of this article.
The remaining authors have nothing to disclose.
ANIMAL RESEARCH STATEMENT
No animal was used in this work.
ETHICS AND CONSENT STATEMENT
This study received approval from the Research Ethics Committee of the Fourth People’s Hospital of Taiyuan City (approval number: 2020–016). The study subjects agreed and signed the written informed consents for research use of their blood samples.
DATA AVAILABILITY STATEMENT
The authors confirm that the data supporting this study are available within the article and supplementary.
Supplementary Material
Contributor Information
Yang Dou, School of Pharmaceutical Sciences, IDG/McGovern Institute for Brain Research, Tsinghua University, Beijing 100084, China.
Ke Xu, NHC Key Laboratory of Biosafety, National Institute for Viral Disease Control and Prevention, Chinese Center for Disease Control and Prevention, Beijing 102206, China.
Yong-Qiang Deng, State Key Laboratory of Pathogen and Biosecurity, Beijing Institute of Microbiology and Epidemiology, AMMS, Beijing 100071, China.
Zijing Jia, National Laboratory of Macromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China.
Jun Lan, Beijing Advanced Innovation Center for Structural Biology, School of Life Sciences, Tsinghua University, Beijing 100084, China.
Xiaoyu Xu, Vazyme Biotech Co., Ltd., Nanjing, China.
Guorui Zhang, Department of Respiratory and Critical Care Medicine, The First Affiliated Hospital of Zhengzhou University, Zhengzhou 450052, China.
Tianshu Cao, National Laboratory of Macromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China.
Pan Liu, National Laboratory of Macromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China.
Xiangxi Wang, National Laboratory of Macromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China.
Xinquan Wang, Beijing Advanced Innovation Center for Structural Biology, School of Life Sciences, Tsinghua University, Beijing 100084, China.
Lingjie Xu, Vazyme Biotech Co., Ltd., Nanjing, China.
Pan Du, Vazyme Biotech Co., Ltd., Nanjing, China.
Cheng-Feng Qin, State Key Laboratory of Pathogen and Biosecurity, Beijing Institute of Microbiology and Epidemiology, AMMS, Beijing 100071, China.
Hong Liu, Department of Respiratory and Critical Care Medicine, The First Affiliated Hospital of Zhengzhou University, Zhengzhou 450052, China.
Yafeng Li, Shanxi Provincial Key Laboratory of Kidney Disease, The Fifth Hospital of Shanxi Medical University, Taiyuan 030012, China.
Guizhen Wu, NHC Key Laboratory of Biosafety, National Institute for Viral Disease Control and Prevention, Chinese Center for Disease Control and Prevention, Beijing 102206, China.
Kang Wang, National Laboratory of Macromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China.
Bai Lu, School of Pharmaceutical Sciences, IDG/McGovern Institute for Brain Research, Tsinghua University, Beijing 100084, China.
References
- 1. Callaway, E, Ledford, H. How bad is Omicron? What scientists know so far. Nature 2021; 600: 197–9. 10.1038/d41586-021-03614-z. [DOI] [PubMed] [Google Scholar]
- 2. Hoffmann, M, Kleine-Weber, H, Schroeder, Set al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020; 181: 271, e278–80. 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Walls, AC, Park, YJ, Tortorici, MAet al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 2020; 181: 281, e286–92. 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang, Q, Zhang, Y, Wu, Let al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell 2020; 181: 894, e899–904. 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wrapp, D, Wang, N, Corbett, KSet al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020; 367: 1260–3. 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chi, X, Yan, R, Zhang, Jet al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science 2020; 369: 650–5. 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hansen, J, Baum, A, Pascal, KEet al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science 2020; 369: 1010–4. 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ju, B, Zhang, Q, Ge, Jet al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature 2020; 584: 115–9. 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- 9. Rogers, TF, Zhao, F, Huang, Det al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science 2020; 369: 956–63. 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wu, Y, Wang, F, Shen, Cet al. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science 2020; 368: 1274–8. 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shi, R, Shan, C, Duan, Xet al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature 2020; 584: 120–4. 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- 12. Cao, Y, Su, B, Guo, Xet al. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients' B cells. Cell 2020; 182: e16: 73–84. 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wec, AZ, Wrapp, D, Herbert, ASet al. Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science 2020; 369: 731–6. 10.1126/science.abc7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kreer, C, Zehner, M, Weber, Tet al. Longitudinal isolation of potent near-germline SARS-CoV-2-neutralizing antibodies from COVID-19 patients. Cell 2020; 182: 1663–73. 10.1016/j.cell.2020.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu, L, Wang, P, Nair, MSet al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature 2020; 584: 450–6. 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- 16. Walker, LM, Burton, DR. Passive immunotherapy of viral infections: 'super-antibodies' enter the fray. Nat Rev Immunol 2018; 18: 297–308. 10.1038/nri.2017.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Clargo, AM, Hudson, AR, Ndlovu, Wet al. The rapid generation of recombinant functional monoclonal antibodies from individual, antigen-specific bone marrow-derived plasma cells isolated using a novel fluorescence-based method. MAbs 2014; 6: 143–59. 10.4161/mabs.27044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yuan, M, Liu, H, Wu, NCet al. Structural basis of a shared antibody response to SARS-CoV-2. Science 2020; 369: 1119–23. 10.1126/science.abd2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ying, T, Prabakaran, P, du, Let al. Junctional and allele-specific residues are critical for MERS-CoV neutralization by an exceptionally potent germline-like antibody. Nat Commun 2015; 6: 8223. 10.1038/ncomms9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lan, J, Ge, J, Yu, Jet al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020; 581: 215–20. 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 21. Lv, Z, Deng, YQ, Ye, Qet al. Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody. Science 2020; 369: 1505–9. 10.1126/science.abc5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yao, HP, Sun, Y, Deng, YQet al. Rational development of a human antibody cocktail that deploys multiple functions to confer Pan-SARS-CoVs protection. Cell Res 2021; 31: 25–36. 10.1038/s41422-020-00444-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhu, L, Deng, YQ, Zhang, RRet al. Double lock of a potent human therapeutic monoclonal antibody against SARS-CoV-2. Natl Sci Rev 2021; 8: nwaa297. https://doi.org/ARTN nwaa29710.1093/nsr/nwaa297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Korber, B, Fischer, WM, Gnanakaran, Set al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell 2020; 182: 812, e819–27. 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. van Dorp, L, Acman, M, Richard, Det al. Emergence of genomic diversity and recurrent mutations in SARS-CoV-2. Infect Genet Evol 2020; 83: 104351. 10.1016/j.meegid.2020.104351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hoffmann, M, Arora, P, Groß, Ret al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell 2021; 184: 2384, e2312–93. 10.1016/j.cell.2021.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang, P, Nair, MS, Liu, Let al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature 2021; 593: 130–5. 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 28. Wibmer, CK, Ayres, F, Hermanus, Tet al. SARS-CoV-2 501Y.V2 escapes neutralization by south African COVID-19 donor plasma. Nat Med 2021; 27: 622–5. 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- 29. Planas, D, Veyer, D, Baidaliuk, Aet al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature 2021; 596: 276–80. 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- 30. Cameroni, E, Bowen, JE, Rosen, LEet al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature 2022; 602: 664–70. 10.1038/s41586-021-04386-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cao, Y, Wang, J, Jian, Fet al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature 2022; 602: 657–63. 10.1038/s41586-021-04385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cele, S, Jackson, L, Khoury, DSet al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature 2022; 602: 654–6. 10.1038/s41586-021-04387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu, L, Iketani, S, Guo, Yet al. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature 2022; 602: 676–81. 10.1038/s41586-021-04388-0. [DOI] [PubMed] [Google Scholar]
- 34. Planas, D, Saunders, N, Maes, Pet al. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature 2022; 602: 671–5. 10.1038/s41586-021-04389-z. [DOI] [PubMed] [Google Scholar]
- 35. Callaway, E. Heavily mutated Omicron variant puts scientists on alert. Nature 2021; 600: 21. 10.1038/d41586-021-03552-w. [DOI] [PubMed] [Google Scholar]
- 36. Brekke, OH, Sandlie, I. Therapeutic antibodies for human diseases at the dawn of the twenty-first century. Nat Rev Drug Discov 2003; 2: 52–62. 10.1038/nrd984. [DOI] [PubMed] [Google Scholar]
- 37. Chan, AC, Carter, PJ. Therapeutic antibodies for autoimmunity and inflammation. Nat Rev Immunol 2010; 10: 301–16. 10.1038/nri2761. [DOI] [PubMed] [Google Scholar]
- 38. Zost, SJ, Gilchuk, P, Chen, REet al. Rapid isolation and profiling of a diverse panel of human monoclonal antibodies targeting the SARS-CoV-2 spike protein. Nat Med 2020; 26: 1422–7. 10.1038/s41591-020-0998-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gerard, A, et al. High-throughput single-cell activity-based screening and sequencing of antibodies using droplet microfluidics. Nat Biotechnol 2020; 38: 715–21. 10.1038/s41587-020-0466-7. [DOI] [PubMed] [Google Scholar]
- 40. Wang, K, Jia, Z, Bao, Let al. Memory B cell repertoire from triple vaccinees against diverse SARS-CoV-2 variants. Nature 2022; 603: 919–25. 10.1038/s41586-022-04466-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pinto, D, Park, YJ, Beltramello, Met al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature 2020; 583: 290–5. 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 42. Torres, JL, Ozorowski, G, Andreano, Eet al. Structural insights of a highly potent pan-neutralizing SARS-CoV-2 human monoclonal antibody. Proc Natl Acad Sci U S A 2022; 119: e2120976119. 10.1073/pnas.2120976119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liu, L, Iketani, S, Guo, Yet al. An antibody class with a common CDRH3 motif broadly neutralizes sarbecoviruses. Sci Transl Med 2022; 14: eabn6859. 10.1126/scitranslmed.abn6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Westendorf, K, Žentelis, S, Wang, Let al. LY-CoV1404 (bebtelovimab) potently neutralizes SARS-CoV-2 variants. Cell Rep 2022; 39: 110812. 10.1016/j.celrep.2022.110812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sevigny, J, Chiao, P, Bussière, Tet al. The antibody aducanumab reduces Abeta plaques in Alzheimer's disease. Nature 2016; 537: 50–6. 10.1038/nature19323. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting this study are available within the article and supplementary.