ABSTRACT
Animal embryos are provided by their mothers with a diverse nutrient supply that is crucial for development. In Drosophila, the three most abundant nutrients (triglycerides, proteins and glycogen) are sequestered in distinct storage structures: lipid droplets (LDs), yolk vesicles (YVs) and glycogen granules (GGs). Using transmission electron microscopy as well as live and fixed sample fluorescence imaging, we find that all three storage structures are dispersed throughout the egg but are then spatially allocated to distinct tissues by gastrulation: LDs largely to the peripheral epithelium, YVs and GGs to the central yolk cell. To confound the embryo's ability to sort its nutrients, we employ Jabba and mauve mutants to generate LD-GG and LD-YV compound structures. In these mutants, LDs are mis-sorted to the yolk cell and their turnover is delayed. Our observations demonstrate dramatic spatial nutrient sorting in early embryos and provide the first evidence for its functional importance.
Keywords: Drosophila, Developmental metabolism, Embryogenesis, Glycogen, Lipid droplets
Summary: In Drosophila embryos, Jabba is essential for correct sorting of glycogen granules and lipid droplets into different tissues by preventing inappropriate interactions between these organelles.
INTRODUCTION
After fertilization, animal embryos develop for extended periods without access to external nutrients. In oviparous species, for example, the young animal gains access to additional nutrients only after hatching, when it can feed independently. Therefore, mothers provide their embryos with nutrient reserves to enable embryonic development. In Drosophila, these reserves include neutral lipids (triglycerides and sterol esters), proteins (yolk proteins) and carbohydrates (predominantly glycogen) (Gandara and Wappner, 2018; Miyazawa and Aulehla, 2018; Song et al., 2019; Tennessen et al., 2014). These nutrients provide both energy supplies and carbon backbones for anabolic metabolism. Different nutrients are not present freely in the cytoplasm, but are segregated from each other and packaged into distinct structures (King, 1970). Neutral lipids are present in the center of lipid droplets (LDs), ubiquitous cellular organelles in which a single phospholipid layer surrounds a central hydrophobic core. Yolk proteins reside inside membranous yolk vesicles (YVs), oocyte- and embryo-specific LROs (lysosome-related organelles) delimited by a phospholipid bilayer. Glycogen forms so-called β particles (carbohydrate chains attached to the priming protein Glycogenin) that assemble into larger glycogen granules (GGs or α particles). These nutrients are utilized at different rates and at different embryonic stages (An et al., 2014; Gutzeit et al., 1994; Song et al., 2019; Tennessen et al., 2014). Most previous studies relied on biochemical analysis of whole embryos and thus could not address the spatial organization of these nutrients in the embryo. In this study, we address this issue and find that GGs, LDs and YVs undergo dramatic sorting in the first few hours of embryogenesis and that this sorting is a prerequisite for proper nutrient utilization.
Drosophila embryos are a syncytium for the first ∼2.5 h. During this period, the nuclei divide 13 times near synchronously before undergoing bulk cytokinesis. The first eight divisions (Stages 1 and 2) occur deep within the embryo, followed by the migration of the majority of nuclei to the surface, leaving a minority of nuclei in the interior. Arrival of nuclei at the surface and formation of the pole cells/germline (Stage 3) mark the beginning of the syncytial blastoderm. Over the next hour (Stage 4), the peripheral nuclei undergo their 9th-13th divisions (José-Antonio and Volker, 1985). Stage 5 encompasses a 1 h-long interphase where simultaneous cytokinesis (cellularization) generates ∼6000 diploid cells organized as an epithelium at the embryo's periphery and one central, syncytial yolk cell. The epithelium gives rise to all the tissues of the future larva and adult, whereas the yolk cell is a transient tissue that functions as a yolk protein depot as well as a positional cue during the maturation of ecto- and endoderm (José-Antonio and Volker, 1985; Reed et al., 2004). Gastrulation movements (Stage 6) and germband extension (Stages 7-10) then transform this 2D epithelial layer into a complex 3D body plan.
The embryo initially shows little or no spatial segregation of nutrients, with LDs and YVs homogeneously distributed (Welte et al., 1998). In larvae and adults, by contrast, nutrients are unevenly distributed, with specialized storage tissues dedicated to receiving and disseminating specific nutrients (e.g. fat body tissue for fat storage). Nutritional specialization is already evident at gastrulation when LDs and YVs are allocated to distinct cells: most LDs to the peripheral epithelium; YVs exclusively to the yolk cell (Welte, 2015a). This distribution persists for the rest of embryogenesis (Fig. 1A-C). Thus, one of the first organizational events in Drosophila embryogenesis differentially sorts nutrients, foreshadowing nutrient handling by specialized tissues in later life stages.
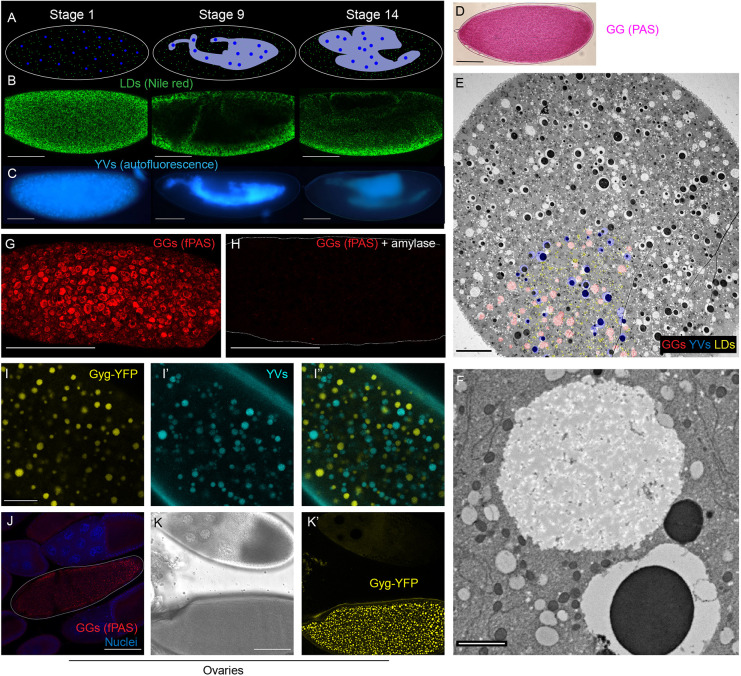
Fig. 1.
Visualization of embryonic nutrient stores. (A-C) Distribution of LDs and YVs at three embryonic stages (Stage 1, newly laid; Stage 9, ∼4.5 h old; Stage 14,∼11 h old). (A) Diagrammatic summary. LDs are in green, YVs in blue, yolk cell in gray. (B) Fixed embryos stained with Nile Red to label LDs and imaged with confocal microscopy. Scale bars: 100 µm. (C) Living embryos imaged by epifluorescence microscopy to reveal the distribution of the autofluorescent YVs. Scale bars: 100 µm. (D) fPAS staining of a newly laid embryo visualized by brightfield microscopy. Scale bar: 100 µm. (E) Cross-section of a Stage 1 embryo imaged by TEM; LDs, GGs and YVs are evenly distributed. A portion of the storage organelles is pseudo-colored, to orient the viewer, in the lower left: YVs blue, LDs yellow, GGs red. Scale bar: 20 µm. (F) TEM image of a GG, the large white circle top/middle of the image. Scale bar: 2 µm. (G,H) fPAS staining of newly laid embryos visualized by confocal microscopy; the embryo in H was pretreated with α-amylase for 2 h to digest glycogen. Scale bars: 100 µm. (I-I″) Confocal micrographs of a live Stage 2 embryo expressing Glycogenin-YFP. Scale bar: 20 µm. I shows Glycogenin-YFP, I′ yolk autofluorescence, and I″ the merged image. (J-K′) Glycogen accumulation in ovary follicles. Scale bars: 100 µm. (J) PAS- and DAPI-stained follicles. PAS signal is visible only in the Stage 13 oocyte (outlined), with no signal in the Stage 10 follicle above it. (K,K′) Follicles expressing Glycogenin-YFP. K shows brightfield image; K′, the YFP channel. GGs are not detectable at Stage 10 (top) but are prominent in Stage 14 (bottom).
How embryos spatially control the third major nutrient, glycogen, remains unknown. We therefore visualized GGs using Periodic Acid–Schiff (PAS) staining and fluorescence microscopy as well as a Glycogenin-YFP fusion. These approaches revealed GGs as highly dynamic, undergoing both morphological changes and redistribution before cellularization. By gastrulation, GGs are almost exclusively allocated to the yolk cell, like YVs. We also found that in embryos lacking the LD protein Jabba, LDs are tightly associated with GGs and are transported together to the yolk cell. These misallocated LDs are consumed more slowly than in the wild type and persist through hatching. We surmise that perturbed LD turnover is likely the result of LD misallocation, because delayed consumption is also observed in embryos in which LD misdistribution results from inappropriate interactions with YVs. We conclude that early nutrient sorting during Drosophila embryogenesis leads to optimal nutrient allocation, which may ensure that nutrients are utilized efficiently.
RESULTS
Lipid droplets and yolk vesicles are sorted to different tissues by cellularization
Previous studies had suggested that LDs and YVs are likely present throughout early Drosophila embryos (Welte, 2015a). However, they had relied on fluorescence microscopy of whole-mount samples. To determine unambiguously the distribution of LDs and YVs, we analyzed the two organelles in cross-sections using TEM. Both LDs and YVs were homogenously distributed throughout the depth of the embryo (Fig. 1E; in the lower left portion of the image, LDs are false colored in yellow, YVs in blue). Thus, early embryos start out with LDs and YVs intermixed.
In Stage 2 embryos, myosin-II mediated contractions of the cortex lead to large-scale circular flows (Deneke et al., 2019; Foe et al., 1993). At the periphery, cytoplasm flows from the pole along the cortex to the middle of the embryo where it descends towards the interior and then flows along the long axis of the embryo to re-emerge at the poles (Deneke et al., 2019) (schematized in Fig. S3F). YVs are known to be carried by this flow (Deneke et al., 2019). We injected embryos with an LD-specific dye and monitored them live by confocal microscopy (Movie 1). In Stage 2 embryos, LDs flowed in the expected pulsative manner along the periphery. In subcortical planes (Movie 2), individual particles could be followed for long distances, allowing us to quantify the flow of several organelles by particle image velocimetry (Kilwein and Welte, 2021) (see below). Because our imaging conditions do not allow us to image the middle of the embryo, we could not directly observe their flow in the embryo center. However, movies taken at 40 µm depth (Movie 1) are consistent with massive rearrangement of these interior LDs and their flow towards the poles.
Previous fixed-embryo analysis had found that by Stage 3 LDs are highly enriched in the peripheral ∼40 µm of the embryo (Arora et al., 2016; Welte et al., 1998), whereas YVs remain distributed throughout. We observed the same pattern in our movies: LDs became enriched at the periphery during Stage 2 and remained enriched there through Stage 5 (Movie 1). During Stage 4, YVs become depleted from this region (Foe et al., 1993), and by Stage 9 LDs and YVs have segregated into the epithelial cells versus the yolk cell (Welte, 2015a), respectively, a distribution that remains through the rest of embryogenesis (Fig. 1B,C) (Arora et al., 2016; Welte et al., 1998).
In summary, YVs and LDs are intermixed in the early embryo and segregate from each other in two steps. During Stage 2, LDs become enriched in the periphery, and during Stage 4 YVs become depleted from the same region. As a result, the two nutrient stores are allocated to distinct cells by cellularization, creating an early nutrient differentiation between the cells of the embryo (Fig. 1A).
Two novel, subcellular approaches to visualize glycogen granules by light microscopy
Does the third nutrient store (GGs) also undergo sorting? By electron microscopy, GGs appeared as large, weakly stained, membrane-less structures (Fig. 1F). TEM cross-sections of young embryos showed that GGs are evenly distributed early on, like LDs and YVs (Fig. 1E, GGs in red in the lower left). Thus, all three structures are present throughout the embryo at the start of development.
To visualize GGs by light microscopy, we employed PAS staining, a histological stain for visualizing carbohydrates. This approach works well at the organismal and tissue levels, but it does not reveal the subcellular organization into GGs. PAS-stained early embryos exhibit signal throughout the cytoplasm, but conventional imaging approaches do not reveal fine structure (Fig. 1D). However, when adapted for fluorescence microscopy (T. Kornberg, personal communication), fluorescent PAS (fPAS) signal revealed discrete granular structures of <1-5 µm diameter throughout the cytoplasm (Fig. 1G). When embryos were pretreated with α-amylase to degrade glycogen, fPAS signal was largely abolished (Fig. 1H), confirming that most fPAS signal in the early embryo represents glycogen. As an independent test, we analyzed fPAS signal during oogenesis, because glycogen specifically accumulates in late-stage oocytes (Stage 13 and 14) (King, 1970; Sieber et al., 2016). The fPAS signal recapitulated this pattern: only the oldest oocytes were fPAS positive (Fig. 1J). We conclude that fluorescent imaging with PAS staining labels glycogen, with sufficient contrast to resolve individual granules.
Live observation of YVs and LDs has revealed detailed information about their mechanism of motion (Welte, 2015a). Because β particles, the subunits of GGs, contain the priming protein Glycogenin at their center (Prats et al., 2018), we employed a protein trap line (Lowe et al., 2014) in which an additional coding YFP exon is inserted into the endogenous Glycogenin (Gyg) gene. By confocal microscopy, we observed structures ∼1-5 µm in diameter in Stage 2 embryos of this strain (Fig. 1I). Because YFP fluorescence is destroyed by PAS staining, we could not directly compare PAS-stained GGs and Glycogenin-YFP, but several lines of evidence strongly support that Glycogenin-YFP does indeed reveal GGs. First, the size, distribution and abundance of the YFP-positive structures were similar to the GGs visualized by PAS staining (Fig. 1G). Second, the only known abundant embryonic structures in this size range are YVs and GGs, and live imaging of embryos revealed that the autofluorescent YVs are distinct from the YFP structures (Fig. 1I-I″). Third, during oogenesis, YFP structures became distinct only in Stage 13 and Stage 14 oocytes (Fig. 1K,K′), just like GGs (Fig. 1J). Fourth, in certain mutants (see below), LDs are arranged around GGs (as observed by TEM or fPAS), and we observed the same association around Glycogenin-YFP granules (Fig. 3G′). Thus, Glycogenin-YFP reveals GGs.
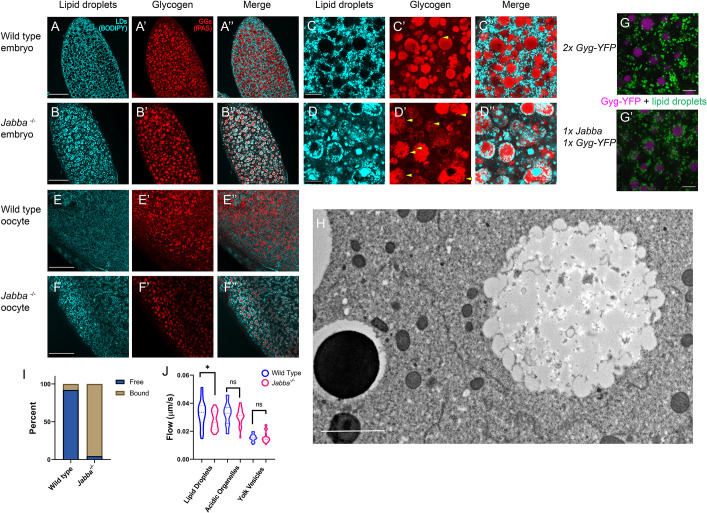
Fig. 3.
LDs and GGs aberrantly interact in Jabba mutant embryos. (A-F″) LDs (cyan, BODIPY) and GGs (fPAS, red). (A-B″) Wild-type (A-A″) and Jabba (B-B″) stage 2 embryos. Scale bars: 60 µm. (C-D″) The same embryos at a higher magnification. Scale bars: 10 µm. Arrowheads in C′,D′ indicate indentations in GGs occupied by LDs. (E-F″) Late-stage oocytes stained for LDs (cyan, BODIPY) and GGs (fPAS, red). Scale bars: 40 µm. GG-LD association is already visible in F″. (G,G′) LDs (green, LipidSpot 610) and GGs (magenta, Glycogenin-YFP) in Glycogenin-YFP (G) and 1×Glycogenin-YFP,1x Jabba (G′) embryos. 1x Jabba embryos display LD-GG association not found in the wild type. Scale bars: 5 µm. (H) TEM image of a Jabba mutant embryo showing a GG with its surface decorated by bound LDs. LDs are largely absent from the cytoplasm. Scale bar: 2 µm. (I) Quantification of the number of LDs bound to GGs versus free in wild-type and Jabba mutant embryos. Quantification of three TEM images per genotype (>400 LDs) revealed that the majority of LDs are not associated with GGs in the wild type, whereas ∼95% of LDs are bound to GGs in Jabba mutant embryos. (J) PIV analysis of the motion of LDs, lysosomes (acidic organelles) and YVs in wild-type and Jabba mutant embryos. The variation in speeds for a given organelle results from the pulses at the embryo cortex. Mean LD velocity in wild type versus Jabba: 0.034 versus 0.028 µm/s; P=0.0144. Mean velocity of acidic organelles: 0.032 versus 0.031 µm/s, P=0.406. Mean velocity of YVs: 0.015 versus 0.016 µm/s, P=0.815. Analysis based on at least three embryos per genotype, with nine measurements per embryo, i.e. n=at least 27. P-values generated by unpaired, two-tailed t-tests. *P<0.05. ns, not significant.
As a final test, we performed in vivo embryo centrifugation, a technique in which living syncytial embryos are centrifuged to separate their components by density (Cermelli et al., 2006; Tran and Welte, 2010). GGs as revealed by PAS staining represent the densest fraction, opposite the low-density neutral lipid (Fig. S1B); this fraction appeared clear in bright light (Fig. S1A), as expected for a lipid-free fraction. Similarly, in centrifuged Glycogenin-YFP embryos, YFP signal accumulated at the very bottom of the embryos, even below the autofluorescent YVs (Fig. S1B). In centrifuged oocytes, coalesced GGs also form a cap below tightly packed YVs (King, 1970).
Glycogen distribution changes during development leading to yolk cell allocation
By TEM, GGs were observed throughout the embryo at Stages 1 and 2, just like YVs and LDs (Fig. 1E). To determine whether GGs are also spatially sorted during embryogenesis, we performed fPAS staining on embryos of different ages. In Stage 1 embryos, glycogen was evenly distributed (Fig. 1G). By Stage 3, imaging in a subcortical plane showed a reduction in the number of GGs at the surface of the embryo (compare Fig. 2A with Fig. 1G). At Stage 5, fPAS signal was absent from the periphery; the embryo shown in Fig. 2B was imaged 40 µm below the surface (embryo is outlined in white). This developmental time course suggests that GGs become depleted from the peripheral regions.
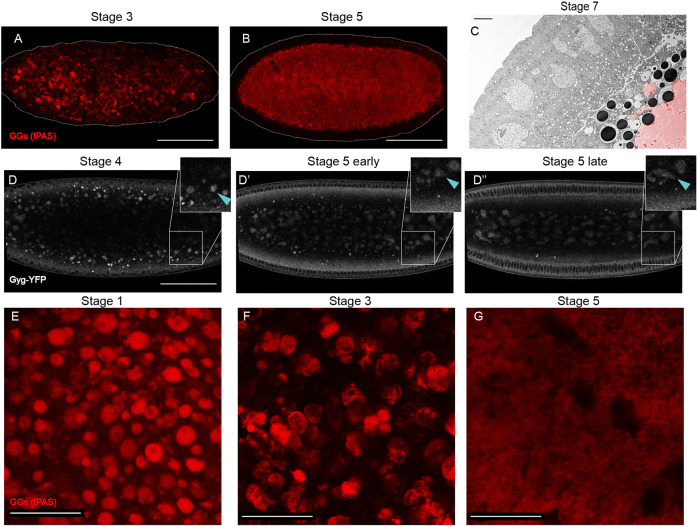
Fig. 2.
GGs change their morphology and distribution during the first 3 h of embryogenesis. (A,B) PAS staining of whole embryos, imaged by confocal microscopy. Scale bars: 100 µm. (A) Stage 3 embryo with GGs beginning to coalesce. (B) Stage 5 embryo with coalesced GGs localized to the embryo interior. (C) TEM image of a Stage 6 embryo. Coalesced glycogen (false-colored red) evident in the yolk cell, which occupies the bottom-right corner. Scale bar: 5 µm. (D-D″) Frames from a confocal time-lapse movie of a Glycogenin-YFP-expressing embryo (Movie 4). Note the inward movement of the GGs and their coalescence. Scale bar: 100 µm. Insets show magnified regions of the embryo periphery to highlight the morphological changes to GGs. Blue arrowhead indicates a single GG tracked through time as it coalesces with other GGs. (E-G) fPAS staining of embryos of different stages. Scale bars: 20 µm. The reduced signal in G is likely a result of the deeper focal plane because of inward motion of the GGs.
Live imaging of Glycogenin-YFP-expressing embryos identified discrete, mobile GGs in young embryos (Movie 2). Fig. 2D-D″ shows still images from a movie (Movie 3) for an embryo imaged ∼40 µm below the surface through Stages 4 to 5. Initially (Fig. 2D), GGs were enriched in a broad area just below the peripheral nuclei (black circles). Over time, GGs progressively moved into the interior. By the onset of Stage 5 (Fig. 2D′), GGs were largely absent from this area and most were in the center. Towards the end of Stage 5, the GGs were deep within the interior of the embryo (Fig. 2D″), well below the forming cells. This inward movement was confirmed by epifluorescence microscopy (Fig. S2A).
Thus, both fPAS and live imaging show redistribution of GGs into the interior of the embryo during Stages 3-5. This pattern suggests that after cellularization the GGs are allocated to the yolk cell and are absent from the surrounding epithelium. This distribution was confirmed by TEM imaging: the Stage 7 embryo in Fig. 2C has glycogen (false-colored in red) entirely in the yolk cell (see also Fig. 4B). fPAS staining of embryos in Stages 9 and 10 revealed that the majority of glycogen is indeed in the yolk cell (Fig. S2B). The overall signal was much reduced at those stages, and by Stage 11 we could no longer detect appreciable fPAS signal (Fig. S2C), suggesting that glycogen is being turned over. Glycogenin-YFP was also restricted to the yolk cell in later stages (Fig. S2C).
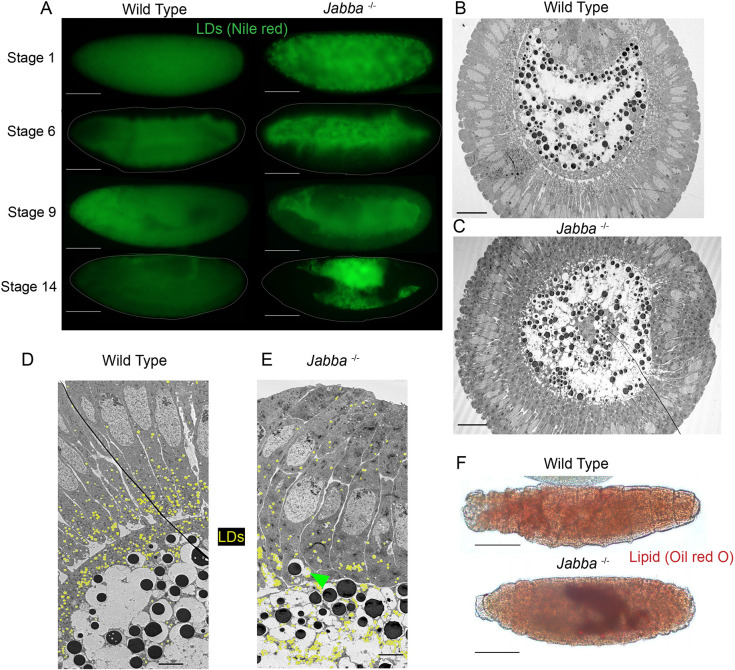
Fig. 4.
In Jabba mutant embryos, lipid droplets are mislocalized and their consumption is disrupted. (A) Wild-type and Jabba mutant embryos of various stages stained with Nile Red to detect LDs and imaged with epifluorescence microscopy. Scale bars: 100 µm. (B,C) TEM cross-sections of Stage 6 embryos. Scale bars: 100 µm. (B) Ventral furrow is at the top of the image. LDs are false-colored in yellow. (C) The ventral furrow is at the right of the image. (D,E) TEMs of Stage 6 embryos at higher magnification showing the border between the cellularized epithelium (right) and yolk cell (left). Scale bars: 5 µm. The green arrowhead in E shows a YV attached to an LD-GG complex, which has inappropriately localized to the epithelium. (F,G) L1 larva stained with Oil Red O to detect LDs. Residual LDs in the Jabba larva are visible in its gut. Scale bars: 80 µm.
GG morphology changes during development
Our fPAS and Glycogenin-YFP time courses suggest that GG morphology changes as the embryo develops. We therefore examined fPAS-stained embryos at higher magnification. Newly laid embryos were characterized by discrete GGs evenly distributed within the focal plane (Fig. 2E). In Stage 3 embryos, GGs were arranged much less homogeneously, forming clusters: frequently, multiple GGs were in close contact with each other, and there was glycogen-free space between clusters (Fig. 2F). This pattern implies that GGs not only move inwards (i.e. perpendicular to the focal plane shown), but also within the plane towards each other. Consistent with that notion, some of the GGs were no longer round, but appeared as oblong or more complicated aggregates, implying that GGs are coalescing. By Stage 5, fPAS signal formed a single, largely homogeneous mass (Fig. 2G) that only retained remnants of the granular structure at its outer edges (Fig. 2C). This single GG mass occupied the center of the embryo (Fig. 2B). Consistent with this, TEM imaging of Stage 7 embryos revealed a large, fused glycogen structure (Fig. 2C, red area).
We observed this same morphology shift in movies of Glycogenin-YFP embryos (Fig. 2D-D″; Movie 3; Fig. S2A). In Stage 4 embryos, GGs near the periphery appeared as largely discrete structures, whereas GGs deeper in the embryo tended to be larger and were misshapen, consistent with fusion. Over time, as the GGs shifted towards the center of the embryos, they formed larger, less spherical entities (Stage 5 early, Fig. 2D′; Fig. S2A), so that by the end of Stage 5, GG signal showed many amorphous structures in the interior of the embryo (Fig. 2D″; Fig. S2A). The insets in Fig. 2D-D″ track a single GG, indicated with a blue arrowhead, as it moved inward and closer to other GGs and then fused. In summary, GGs undergo a dramatic morphological shift as they migrate into the center of the embryo.
Lack of the LD protein Jabba results in tight association between LDs and GGs
Our analysis shows that LDs, GGs and YVs are initially intermixed, but are segregated to distinct tissues by cellularization: whereas most LDs end up in the peripheral epithelium, both YVs and GGs are allocated to the interior yolk cell. The enrichment of both protein and glycogen stores in the yolk cell is consistent with the idea of the yolk cell as a metabolically supportive tissue (José-Antonio and Volker, 1985; Reed et al., 2004; Rickoll, 1976). But why LDs are allocated differently and why the embryonic nutrient supply is spatially organized at all is unknown.
As a potential inroad into this problem, we re-examined embryos lacking the LD protein Jabba, one of the most abundant proteins on embryonic LDs (Li et al., 2012). Such embryos have normal triglyceride content but display abnormal LD distribution at Stage 4 (Li et al., 2012). When we imaged fixed Stage 1-2 embryos stained for LDs, wild-type embryos displayed the pattern expected from our TEM analysis (Fig. 1E); LDs were absent from many circular areas (presumably GGs and YVs), but distributed evenly throughout the remaining spaces (Fig. 3A). The pattern in Jabba mutants (two different alleles, JabbaDL and Jabbazl01) was dramatically different. Here, most LDs were accumulated in rings, with few LDs occupying the space between the rings (Fig. 3B). This aberrant distribution was not an artefact of fixation, as we saw the same pattern in embryos injected with LD dyes and imaged live (Fig. S3D, 0 s panel).
The pattern in Jabba mutants suggests that LDs accumulate around YVs or GGs. Co-detection of LDs and YVs revealed no association between them (Fig. S3E). In contrast, co-labeling of GGs and LDs showed that LDs surrounded GGs (Fig. 3A′-B″). This finding was further confirmed in TEM cross-sections (Fig. S4A). A similar, but less pronounced, association was observed when Jabba dosage was reduced (Fig. S3C, 1x Jabba). We employed this observation to confirm LD-GG association in living embryos. We injected LipidSpot 610 into embryos from mothers expressing Glycogenin-YFP and either one or two copies of the wild-type Jabba gene. LDs and GGs displayed minimal association in the otherwise wild-type background (Fig. 3G), whereas many LDs were present at or near the surface of GGs when Jabba dosage was reduced (Fig. 3G′). We conclude that when Jabba levels are reduced, LDs associate with GGs. This association could already be observed in oocytes, as soon as GGs are detected (Fig. 3E-F″). Finally, we detected no association between LDs and GGs when other LD proteins were missing, namely LSD-2 (also known as PLIN-2), Sturkopf (CG9186) or Klar (Fig. S3C). We conclude that Jabba uniquely suppresses inappropriate interactions between these storage structures.
Although we occasionally observed LDs deep within GGs of Jabba mutants, for the most part the LDs were arranged in a ring pattern with glycogen in the center (Fig. 3D″). In fact, LDs appeared embedded within the outer regions of the GGs; see, for example, Fig. 3D′,D″, where GGs display indentations/areas of exclusion in the fPAS signal (Fig. 3D′, arrowheads). These regions were filled with LDs (Fig. 3D″). Rarely were such associations or indentations observed in the wild type (Fig. 3C′, arrowheads). TEM analysis confirmed a tight LD-GG association: in Jabba mutant embryos, LDs appeared to contact GGs directly, with glycogen bulging out between LDs (3H; Fig. S3B). In the wild type, such association was observed rarely (Fig. 1E; Fig. S3A; Fig. 3I). This association was strong enough that when Jabba mutant embryos were centrifuged, the dramatic separation of LDs and GGs into distinct layers at opposite ends of the embryo observed in the wild type (Fig. S1B) was disrupted. In the Jabba mutant, glycogen signal was found to be intermixed with the LD layer in the region of lowest density, and pockets of LD signal were present within the glycogen layer in the highest-density region (Fig. S1C).
Consequences of LD-GG interactions on LD motility
To determine whether the LD-GG association in Jabba mutant embryos affects LD motility, we labeled LDs by injecting dyes into embryos, monitored their behavior live, and quantified their flow speeds by particle image velocimetry (Kilwein and Welte, 2021). In the wild type, LDs and acidic organelles flowed faster than YVs (Fig. 3J), presumably owing to their smaller size. Jabba mutant embryos displayed the expected ‘ring’ arrangement of LDs (Fig. S3D); these rings moved as a unit (Movie 5), reinforcing the idea that LD-GG complexes are stable structures. Compared with wild type, they displayed stuttered motion, minimal displacement (Fig. S3D) and lower average velocity (Fig. 3J). In contrast, YVs and acidic organelles showed similar mobility between the two genotypes (Fig. 3J). Thus, altered LD flow in Jabba mutant embryos does not represent a general defect in cytoplasmic streaming, but rather a specific disruption of LD motility, likely because of the much larger size of LD-GG complexes relative to individual LDs.
Consequences of LD-GG interactions on LD allocation
During wild-type development, GGs and LDs were initially intermixed and homogeneously distributed throughout the embryo (Fig. 1E, Fig. S4A). LDs became enriched at the periphery by Stage 3 (Movie 1), and by the end of Stage 5 GGs and LDs were segregated from each other and allocated to different tissues. In the absence of Jabba, GGs and LDs formed large composite structures that were also distributed throughout the early embryo (Fig. S4B). But because these composite structures travel together, segregation to distinct locations seemed no longer possible. Indeed, in our movies with labeled LDs (Movies 5, 6), the LDs trapped in GG:LDs complexes were far less mobile, engaging in delayed, stuttered motion and did not become enriched in the periphery as in the wild type (Movie 1). For post-cellularization embryos, TEM analysis revealed a massive redistribution of LDs in Jabba mutant embryos relative to wild type (Fig. 4B-E), with fewer LDs in the peripheral epithelium and more in the yolk cell. In the wild type, 72% of the 510 LDs scored were present in the epithelial cells, whereas in Jabba mutant embryos only 23% of the 696 LDs scored were present in epithelial cells.
In contrast, we only found minor differences in glycogen distribution between the two genotypes. By fPAS staining, wild-type and Jabba glycogen distribution was very similar (Fig. S2E). By TEM, the bulk of GGs in Jabba mutant embryos were appropriately localized to the yolk cell, just like in the wild type (Fig. 4B-E). There were a few instances of small LD-GG complexes segregated into blastoderm cells (Fig. 4E, green arrowhead). We conclude that the composite LD-GG structures in Jabba mutant embryos are allocated like GGs, leading to LD mislocalization.
As an independent approach, we detected LDs by Nile Red staining and fluorescence microscopy in whole-mount embryos (Fig. 4A). At early stages, the LD distribution already looked different: the signal was diffuse throughout the embryo in wild type and granular in Jabba mutants, presumably reflecting LD enrichment around GGs. At the beginning of gastrulation, Nile Red signal in the mutant was more prominent in the yolk cell, a pattern that became even more pronounced at later stages. By Stage 14, LD signal was absent from the yolk cell and present everywhere else, whereas in Jabba mutant embryos this pattern was reversed (Fig. 4A). We conclude that in Jabba mutant embryos the bulk of LDs are indeed mislocalized to the yolk cell.
In late-stage embryos, LD signal in Jabba mutant embryos was not only restricted to the yolk cell, but also displayed increased intensity. This difference even persisted post-hatching. LD staining using Oil Red O revealed strong signal in the gut lumen (the location of the yolk cell remnant) of newly hatched Jabba larvae (Fig. 4F), unlike wild type (Fig. 4F). Thus, Jabba mutants not only mislocalize their LDs, but fail to consume them properly.
LD interactions with YVs also lead to their mislocalization and persistence
LD consumption in the mutant might be impaired because Jabba is important for LD breakdown or because the yolk cell is not equipped to metabolize this high concentration of LDs. To distinguish between these possibilities, we took advantage of a recent report that embryos lacking the Mauve protein display an interaction between LDs and YVs (Lattao et al., 2021). Mauve is a resident protein on lysosome-related organelles (LROs), which is important for their maturation. YVs are a type of LRO and in the absence of Mauve display several phenotypes, including an association with LDs (Lattao et al., 2021). Might these YV-LD interactions affect nutrient sorting?
As strong disruption of Mauve severely impairs early embryonic development (Lattao et al., 2021), we generated mothers transheterozygous for two weak alleles, mauveRosario and mauve3. As reported for stronger allele combinations (Lattao et al., 2021), these embryos displayed YV size heterogeneity and autofluorescence variability (Fig. 5A) (Lattao et al., 2021), as well as LDs in close proximity to YV autofluorescence (Fig. 5A, arrows). As autofluorescence marks the lumen of YVs, this arrangement indicates direct association between LDs and YVs. In contrast, unlike for Jabba mutants (Fig. 3D″), GGs did not display the indentations typical for embedded LDs (Fig. 5B, arrows), and did not appear tightly associated with LDs.
Fig. 5.
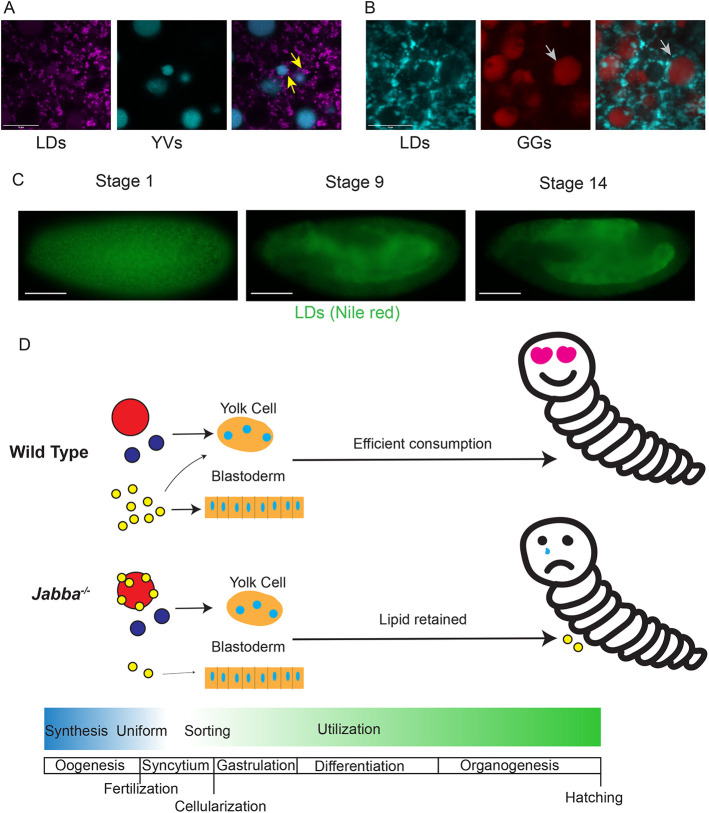
Disruption of LD localization affects LD consumption. (A) Live imaging of LDs (magenta, LipidSpot 610) and YVs (blue, autofluorescence) in newly laid mauve mutant embryos. LDs are associated with adjacent YVs (arrows). Note that the autofluorescence signal may not extend to the furthest edges of the YV owing to the layered internal structure of YVs. (B) Fixed imaging of LDs (blue, BODIPY) and GGs (red, fPAS) in mauve embryos. Arrows indicate a GG without LDs embedded in its outer surface. (A,B) Confocal microscopy. Scale bars: 10 µm. (C) LD (green, Nile Red) distribution in fixed mauve embryos. Stage 1 shows LD clustering, presumably around YVs, reminiscent of GG:LD cluster seen in Jabba mutants. Stages 9 and 14 show mislocalization of LDs to the yolk cell. Images by epifluorescence microscopy. Scale bars: 100 µm. (D) Model of nutrient sorting from oogenesis through embryogenesis, comparing wild type with the disruptions seen in Jabba mutants. Note that the timeline is not scaled to absolute developmental time, but is meant to show the brief life of nutrients through synthesis, distribution and consumption compared with other developmental events. Mislocalized LDs in Jabba mutants are speculated to negatively affect completion of embryogenesis or the larvae itself. Red circles, glycogen granules; blue circles, yolk vesicles; yellow circles, lipid droplets.
The mauveRosario/3 YV-LD association was an exciting opportunity to test our model that LDs are mis-sorted during cellularization if they are associated with a structure destined for yolk cell deposition, allowing us to utilize YVs instead of GGs. When the mutant embryos were stained for LDs (Fig. 5C), signal in Stage 1 was clustered instead of diffuse like in wild type (Fig. 4A). At Stage 9, the LD signal was predominately in the yolk cell, but also present in the other tissues (Fig. 5C). At Stage 14, LD staining was clearly enriched in the yolk cell (Fig. 5A) relative to wild type (Fig. 4A). Thus, mutations in two unrelated genes, Jabba and mauve, show that when LDs inappropriately interact with other nutrient structures, they are mislocalized to the yolk cell and are turned over more slowly.
DISCUSSION
Glycogen is a major energy store in animals, uniquely capable of providing energy rapidly, both aerobically and anaerobically. Although the functions of glycogen have been extensively investigated in adult tissues, its role during embryogenesis is less understood. In this study, we developed new tools to determine the spatial distribution of glycogen in Drosophila oocytes and embryos. Consistent with previous biochemical and electron microscopic analysis, we find that glycogen stores accumulate late in oogenesis and are organized into large, membrane-less structures. These GGs are evenly distributed throughout the early embryo and undergo two types of transitions during syncytial stages. First, they are displaced from the subcortical region during Stage 4, leading to their accumulation in the center and allocation to the yolk cell. Second, simultaneously, GGs fuse into larger and larger structures, so that by the end of Stage 5, most glycogen is present in a large superstructure in the yolk cell. LDs and YVs also start out with an even distribution and are then specifically allocated. After Stage 5, YVs are restricted to the yolk cell, like GGs, whereas LDs are predominantly sorted to the peripheral epithelial cells. In two different mutant conditions, we find physical interactions between LDs and either GGs or YVs. In both cases, LDs are misallocated to the yolk cell. In Jabba mutants, a portion of these LDs fail to be consumed during embryogenesis and persist into larval stages; in mauve mutants persistence through embryogenesis is milder, consistent with less severe mislocalization of LDs. These observations strongly suggest that mislocalization of LDs early in embryogenesis affects subsequent LD consumption.
GGs in Drosophila oocytes and embryos are akin to the α particles that mediate long-term glycogen storage in many tissues, including muscle, liver and the fat body (Brewer and Gentry, 2019; Prats et al., 2018). Their large size [which must correspond to thousands of β particles (King, 1970) as opposed to ∼30 in liver (Brewer and Gentry, 2019)] likely protects against glycogen breakdown and facilitates their physical movement for differential allocation. How α particles assemble and disassemble remains unclear. Proposed binding agents holding neighboring β particles together include covalent links between glycogen chains (Froese et al., 2015; Mitchell et al., 2010) or Glycogenin molecules at the surface of the β particles (Tan et al., 2018), in addition to the Glycogenin dimer in their cores. We speculate that the amorphous mass of glycogen in Stage 5 embryos represents partial dissolution of GGs into β particles, as a prelude to enzymatic breakdown of glycogen. Consistent with this notion, fPAS staining at Stages 9-10 becomes very weak (Fig. S2B,C) and biochemical measures of glycogen levels show a similar reduction (Tennessen et al., 2014). This timing of glycogen depletion overlaps with a proposed switch in embryonic metabolism from carbohydrate-based to triglyceride-based energy production (Tennessen et al., 2014). Disassembly into β particles might provide enhanced access for the cytosolic glycogen phosphorylase, responsible for most glycogen turnover in embryos (Yamada et al., 2019).
Going forward, Drosophila oocytes and embryos should be a powerful model for unraveling the mechanism of the conversion between α and β particles. GGs are large enough to be followed by light microscopy, their assembly and disassembly occurs quickly (each within ∼2 h or less), and Glycogenin-YFP allows live imaging of these processes. In fact, to our knowledge this is the first example of live imaging of glycogen in any system.
Although during oogenesis the three major nutrient stores are made at different times and through different mechanisms (King, 1970; Welte, 2015a), they all start out intermixed and homogeneously distributed in the early embryo. Nutrient sorting starts in Stage 2 when myosin-II driven cortex contractions establish large-scale cytoplasmic flows throughout the embryo. Flow speeds, as estimated from the behavior of YVs, are sufficient to spread out the interior nuclei along the entire anterior-posterior axis (Deneke et al., 2019). As LDs flow faster than YVs (Fig. 3J), these flows should be able to transport most LDs from the center to the poles. If these LDs are somehow captured at the periphery, reducing their return to the embryo center, this would explain their enrichment in the periphery by Stage 3. Consistent with an important contribution from cytoplasmic flow, the LD-GG aggregates in Jabba mutants flow with reduced speeds and LDs fail to enrich at the embryo surface (Movie 4). Thus, we propose that this cytoplasmic flow promotes the first step of nutrient sorting, when LDs accumulate peripherally.
By Stages 4 and 5, the nuclei at the embryo surface set up an array of radially oriented microtubules that traverse a ∼40 µm peripheral zone (Welte et al., 1998). These microtubules are proposed to push YVs into the interior (Foe et al., 1993), and GGs may be displaced by the same mechanism, as they deplete from this zone at the same time. LDs, in contrast, move bidirectionally along these microtubules, employing cytoplasmic dynein and kinesin-1 (Gross et al., 2000; Shubeita et al., 2008); this motion confines them to the microtubule zone (Arora et al., 2016). We propose that microtubules keep LDs in the peripheral zone, whereas they push GGs and YVs inward, resulting in the second sorting step. Our analysis of Jabba and mauve mutants reveals that successful sorting also requires that nutrient stores stay separated. When LDs are tightly associated with either GGs or YVs, sorting fails, and LDs are misallocated to the yolk cell.
Presumably the nutrients stored in GGs, YVs and LDs all support the metabolic needs of the developing embryo. Why then are they allocated differently? One reason might be the different properties of their breakdown products. Glycogen and yolk proteins are broken down into glucose and amino acids, respectively; for example, levels of free alanine and glutamine rise as embryogenesis proceeds and YVs in the yolk cell are turned over (Von Der Crone-Gloor, 1959). Amino acids and glucose are water soluble, making diffusion through gap junctions or membrane transporters viable options for dissemination. Thus, the yolk cell can serve as a hub for glucose and amino acid distribution, as it remains connected via cytoplasmic bridges to the blastoderm through Stage 9 (Kuhn et al., 2015; Rickoll, 1976) and expresses numerous nutrient transporters. In contrast, free fatty acids generated from LD breakdown are poorly water soluble and potentially toxic (Chitraju et al., 2017; Nguyen et al., 2017); they are typically immediately channeled into specific intracellular pathways (Mashek et al., 2007). For example, efficient fatty acid transfer from LDs to mitochondria requires proximity and direct contact (Herms et al., 2015; Kilwein and Welte, 2019; Rambold et al., 2015). Thus, allocation of LDs predominantly to the periphery would allow efficient local energy production. Intriguingly, during zebrafish embryogenesis, LDs are initially highly enriched in the future yolk sac, but are imported into the embryo proper via cytoplasmic bridges and actin-myosin based motility (Dutta and Kumar Sinha, 2015; Gupta et al., 2017). Thus, the zebrafish embryo may also depend on a local LD supply to support its dividing cells.
What are the consequences of mislocalizing LDs to the yolk cell? Our data indicate that a fraction of these LDs persist through the end of embryogenesis. Recent work on embryonic glycogen metabolism suggests that even minor disruption of LD metabolism has the potential for widespread effects on embryogenesis. Embryos that either lack glycogen reserves or are unable to access them display widespread changes in their metabolome as well as hatching delays (Yamada et al., 2019). Given that fat contributes roughly ten times the energy of glucose (derived from glycogen) during Drosophila embryogenesis (Song et al., 2019), even the modest retention of LDs in Jabba mutants might have prominent effects on development. A full exploration of this possibility goes beyond the scope of this paper and is followed up in a separate manuscript (M.D.K., T. K. Dao and M.A.W., unpublished).
LDs and GGs co-exist not only in embryos, but also in mature tissues (e.g. muscle, intestinal epithelia, liver, fat body) (Prats et al., 2018; Walther and Farese, 2012; Welte, 2015b). It is conceivable that in embryos inappropriate interactions between LDs and GGs are particularly harmful because of the large size of GGs and the extensive cytoplasmic streaming, which presumably leads to many encounters between these organelles. It will therefore be interesting to determine whether mechanisms to keep LDs and GGs apart are specific to embryos or also important in other cells.
By devising novel imaging methods for glycogen storage structures, we have shown that Drosophila embryos dramatically reorganize their nutrients by cellularization, with distinct nutrients sorted into separate nascent tissues. The embryo employs multiple mechanisms to get its nutrient stores to the correct location, including cytoplasmic streaming, preventing inappropriate interactions, and microtubule-dependent transport. We also provide the first suggestion that correct spatial allocation of LDs is necessary for their efficient consumption. Together, these observations suggest that embryos need to achieve an optimal nutrient allocation to support subsequent steps in development (Fig. 5D) and that the spatial allocation of nutrients is essential to fully understand embryonic metabolism. The importance of this allocation is particularly remarkable as these nutrient stores exist only transiently and are consumed by the end of embryogenesis.
MATERIALS AND METHODS
Origin of fly strains
Oregon R was used as the wild-type strain. JabbaDL and Jabbazl01 were generated previously in the lab and are strong loss-of-function alleles with no Jabba protein detected in early embryos (Li et al., 2012). Df(2R)Exel7158/CyO carries a large deletion that encompasses Jabba and is used to reduce Jabba dosage; for simplicity, embryos from mothers carrying this deletion are referred to as 1x Jabba. This stock was obtained from the Bloomington Drosophila Stock Center (#7895; FlyBase: FBab0038053). The hypomorphic alleles mauve3 and mauveRosario were a gift from Ramona Lattao (Lattao et al., 2021). The YFP insertion in the Glycogenin locus was generated in a large genetic screen (Lowe et al., 2014) and was obtained from the Kyoto Drosophila Stock Center (#115562).
Microscopy
Laser-scanning confocal microscopy was performed on a Leica SP5 equipped with HyD detectors, using either a 40× objective to show most of the embryo, or a 63× objective for subregions. Epifluorescence imaging was performed on a Nikon Eclipse E600 using a 20× objective. All images were assembled in Adobe Illustrator.
Movies, except for Movie 4, were captured at 1 frame per 30 s and are displayed at 20 frames per second. Movie 4 was captured at 1 frame per 15 min and displayed at 1 frame per 0.8 s. The orientation of the embryo was chosen to maximize the amount of the embryo captured in frame. Video processing was performed in Fiji (NIH).
Sample sizes were as follows: for live imaging, at least three embryos were imaged per genotype per experiment; for fixed samples, the staining was performed at least twice; for TEM analysis, the core facility was given ten appropriately staged embryos per genotype per experiment, and then chose which were imaged based on staining success.
Exclusion criteria for imaging embryos were predetermined. Embryos not of the stage of interest, determined to have expired during preparation or image acquisition, or which were imaged in the incorrect orientation/focal depth were excluded.
PAS and LD staining
Embryos were collected on apple juice plates for the desired time range and dechorionated with 50% bleach and fixed for 20 min using a 1:1 mixture of heptane and 4% formaldehyde in PBS. To detect GGs, embryos were devitellinized using heptane/methanol and subsequently washed three times in 1×PBS/0.1% Triton X-100. Embryos were incubated first in 0.1 M phosphatidic acid (pH 6) for 1 h and then in 0.15% periodic acid in dH2O for 15 min. After one wash with dH2O, embryos were incubated in Schiff's reagent (Sigma-Aldrich) until the embryos went from uncolored, to pink, to red (∼2 min). The reaction was quenched with 5.6% sodium borate/0.25 normal HCl stop solution for at least 2 min with agitation. After replacing half the volume of the stop solution with an equal volume of 1×PBS/0.1% Triton X-100 to reintroduce detergent, the sample was shaken vigorously to free embryos stuck to the container or each other. For subsequent imaging, embryos were mounted in either Aqua-Poly/Mount, Polysciences, or glycerol (90% glycerol, 10% PBS). Mounted samples were imaged by scanning confocal microscopy using excitation setting for Alexa 633 (excitation at 633 nm) and with emission captured from 650nm-750 nm. Mounting media with ‘antifade’ or O2 scavenging additives were avoided. To determine the specificity of the PAS signal, fixed and devitellinized embryos were first incubated with α-amylase (porcine, Sigma-Aldrich, 0.2 mg/ml in PBS, incubated for 2 h) to specifically digest the α-(1,4) glycosidic linkages in glycogen.
To detect LDs by staining, the methanol step was omitted as it extracts neutral lipids. Instead, embryos were washed extensively with 1×PBS/0.1% Triton X-100 to remove residual heptane in a wire-mesh basket then transferred to a 1.7 ml microcentrifuge tube. They were then washed twice with 1×PBS/0.1% Triton X-100. If co-staining with PAS, the phosphatidic acid incubation step was carried out next, adding 1 µl of 1 mg/ml BODIPY 493/503 (Invitrogen), then proceeding with subsequent steps as normal. Red lipid dyes overlap Schiff's reagent's spectra and were therefore avoided for co-staining.
For staining of lipid droplets without PAS co-staining, the 1×PBS/0.1% Triton X-100 wash was replaced with 1×PBS/0.5% Triton X-100/10% bovine serum albumin (BSA)/0.02% sodium azide for a 1 h incubation. The solution was then replaced with fresh 1×PBS/0.5% Triton X-100/10% BSA/0.02% sodium azide and either 1 µl of 1 mg/ml BODIPY 493/503 in DMSO, 1 µl LipidSpot 610 (1000×) (Biotium) in DMSO, or 10 µl of 200 mg/ml (Sigma-Aldrich) in acetone was added.
To determine the distribution of LDs and GGs in centrifuged embryos, in vivo centrifugation was performed as described (Tran and Welte, 2010), followed by fixation and simultaneous LD-GG detection as above. For analyzing follicles, ovaries were dissected from females maintained on yeast at 25°C overnight. Samples were then fixed with 4% formaldehyde in PBS for 15 min, washed in 1×PBS/0.1% Triton X-100, and simultaneously stained for LDs and GGs as described above.
Live imaging
For live imaging involving dye injections, a previously published procedure was followed (Kilwein and Welte, 2021). In short, embryos were collected on apple juice plates for the desired time, hand-dechorionated, transferred to a coverslip with heptane glue, desiccated, and placed in Halocarbon oil 700. Embryos were then injected with BODIPY 493/503 (1 mg/ml), LysoTracker Red (1 mM) or LipidSpot 610 (1000×) and imaged on a Leica SP5 confocal microscope.
For live imaging of Glycogenin-YFP and YV autofluorescence, embryos were collected on apple juice plates for the desired time, hand-dechorionated, transferred to a coverslip with heptane glue, covered with Halocarbon oil 27, and imaged. To improve signal, flies were kept in the dark, and light exposure during embryo preparation was kept to a minimum.
TEM
Embryos were collected from 7- to 14-day-old flies, dechorionated in 3% sodium hypochlorite, and washed extensively with distilled water. Embryos were fixed in 4% paraformaldehyde/2% gluteraldehyde/PBS with an equal volume of heptane added. The vials were shaken then left on an agitator for 20 min. After fixation, embryos were washed extensively with 1×PBS/0.1% Triton X-100, then transferred onto a piece of double-sided tape, adhered, then submerged in 1×PBS/0.1% Triton X-100. The embryos were then gently hand-rolled using fine forceps until the vitelline membrane was removed. Embryos were transferred to a small glass vial. The embryos were then fixed a second time with 4% paraformaldehyde/2% gluteraldehyde/PBS, excluding the heptane, for 30 min. Embryos were then washed three times with 0.2 M sucrose in 0.1 M cacodylate buffer. They were washed an additional three times in 0.1 M sodium cacodylate before post-fixation in 1% osmium tetroxide for 2 h followed by uranyl acetate enhancement in 0.5% uranyl acetate overnight at 4°C. Specimens were washed and then dehydrated in a graded ethanol series, transitioned to propylene oxide and embedded in an Epon/Araldite resin. Thin sections were stained with 0.3% lead citrate and imaged on a Hitachi 7650 transmission electron microscope using an 11 MP Gatan Erlanshen CCD camera. TEM work was conducted at the Electron and Cryo Microscopy Resource in the Center for Advanced Research Technologies at the University of Rochester.
Quantification of TEM
To quantify the association of LDs and GGs, LDs were manually identified based on their appearance and size (diameter of 0.3-0.75 µm), and GGs were manually identified based on the staining pattern and diameter (2-7 µm). The two structures were considered to be associated if the distance between them was less than 30 nm (∼2 pixels).
Particle image velocimetry (PIV)
We performed PIV as described previously (Kilwein and Welte, 2021); this citation includes the Python script used to generate the data. The motion of acidic organelles and LDs was captured simultaneously in the same embryos. Embryos were collected, staged, mounted on a coverslip, and co-injected with BODIPY 493/503 and LysoTracker Red as described above. Three embryos per genotype were imaged at 25°C. Time series were captured by confocal microscopy at a rate of 1 frame per 30 s, within a superficial plane of the embryo. The raw time series were then analyzed, finding the eighth nuclear division by first finding the division where nuclei arrive at the periphery (the ninth division), then going back one contraction cycle. Ten sequential frames were taken from this division starting at the period of the highest lipid droplet motion determined empirically. The signal from the embryos was then isolated from these ten frames using a mask. These processed frames were then fed into a PIV algorithm based on OpenPIV, a python-based PIV implementation, generating nine frame transitions per time series. The output vectors for each transition were then processed to remove any vectors that failed to meet or exceeded the empirically determined vector boundaries. Then, directionality was removed, the vectors were averaged across the transition, pixels were converted to microns, and these averages were plotted on violin plots to show that pulses were being captured.
To perform PIV analysis for YVs, the same procedure was followed, with the exception that embryos were not injected, and embryos were illuminated with a 405 nm laser and yolk autofluorescence was captured by collecting emission in a 410-500 nm window.
Statistics
All statistics were performed using GraphPad Prism. P-values were calculated using two-tailed, unpaired Student's t-tests. At least three embryos were used per genotype. For the PIV statistical comparisons, three embryos were used per genotype contributing nine transitions per embryo, giving at least n=27 per genotype.
The first question we sought to answer with statistical tests was ‘Are the Jabba LD-GG complexes flowing slower then wild-type singular LDs?’. Having received a positive result from this question, we next asked ‘Is the flow generally slower in Jabba than wild type or is the diminished LD-GG speed due to complex formation?’, for which we used acidic organelles and YVs as unclustered controls. These are binary questions seeking to determine whether two populations of related numbers (flow speeds of organelles in each genotype) were different; thus, we chose to use Student's t-tests.
Supplementary Material
Acknowledgements
We thank FlyBase (supported by NHGRI Awards U41HG000739 and U24HG010859) for annotations and curations. We thank the Bloomington Drosophila Stock Center (supported by NIH Award P40OD018537), the Kyoto Drosophila Stock Center at Kyoto Institute of Technology and Ramona Lattao for fly stocks. We thank Zehra Ali-Murthy and Thomas Kornberg (University of California San Francisco) who provided us with the protocol for fPAS staining. TEM analysis was performed by Chad Galloway and Karen Bentley in the Electron and Cryo Microscopy Resource in the Center for Advanced Research Technologies at the University of Rochester. The Electron and Cryo Microscopy Resource is supported in part by institutional funding. We are grateful to Fredrick Gootkind for initial analysis of the LD clustering phenotype in Jabba mutants and Ethan Fisher for his work on fPAS staining.
Footnotes
Author contributions
Conceptualization: M.D.K., M.R.J., J.M.T., M.A.W.; Methodology: A.P.M.; Software: M.D.K.; Formal analysis: M.D.K., M.R.J., J.M.T.; Investigation: M.D.K., M.R.J., J.M.T., A.P.M.; Writing - original draft: M.D.K., M.A.W.; Writing - review & editing: M.D.K., M.R.J., J.M.T., A.P.M., M.A.W.; Supervision: M.A.W.; Project administration: M.A.W.; Funding acquisition: M.D.K., M.A.W.
Funding
This work was supported by National Institutes of Health grants (F31 HD100127 to M.D.K.; R01 GM102155 to M.A.W.). Deposited in PMC for release after 12 months.
Data availability
All relevant data can be found within the article and its supplementary information.
Peer review history
The peer review history is available online at https://journals.biologists.com/dev/lookup/doi/10.1242/dev.201423.reviewer-comments.pdf
References
- An, P. N., Yamaguchi, M., Bamba, T. and Fukusaki, E. (2014). Metabolome analysis of Drosophila melanogaster during embryogenesis. PLoS ONE 9, e99519. 10.1371/journal.pone.0099519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora, G. K., Tran, S. L., Rizzo, N., Jain, A. and Welte, M. A. (2016). Temporal control of bidirectional lipid-droplet motion in Drosophila depends on the ratio of kinesin-1 and its co-factor Halo. J. Cell Sci. 129, 1416-1428. 10.1242/jcs.183426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer, M. K. and Gentry, M. S. (2019). Brain glycogen structure and its associated proteins: past, present and future. Adv. Neurobiol. 23, 17-81. 10.1007/978-3-030-27480-1_2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermelli, S., Guo, Y., Gross, S. P. and Welte, M. A. (2006). The lipid-droplet proteome reveals that droplets are a protein-storage depot. Curr. Biol. 16, 1783-1795. 10.1016/j.cub.2006.07.062 [DOI] [PubMed] [Google Scholar]
- Chitraju, C., Mejhert, N., Haas, J. T., Diaz-Ramirez, L. G., Grueter, C. A., Imbriglio, J. E., Pinto, S., Koliwad, S. K., Walther, T. C. and Farese, R. V., Jr. (2017). Triglyceride synthesis by DGAT1 protects adipocytes from lipid-induced ER stress during lipolysis. Cell Metab. 26, 407-418.e403. 10.1016/j.cmet.2017.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deneke, V. E., Puliafito, A., Krueger, D., Narla, A. V., De Simone, A., Primo, L., Vergassola, M., De Renzis, S. and Di Talia, S. (2019). Self-organized nuclear positioning synchronizes the cell cycle in Drosophila embryos. Cell 177, 925-941.e917. 10.1016/j.cell.2019.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta, A. and Kumar Sinha, D. (2015). Turnover of the actomyosin complex in zebrafish embryos directs geometric remodelling and the recruitment of lipid droplets. Sci. Rep. 5, 13915. 10.1038/srep13915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foe, V. E., Odell, G. M. and Edgar, B. A. (1993). Mitosis and Morphogenesis in the Drosophila embryo: Point and Counterpoint. In The Development of Drosophila Melanogaster (ed. Michael Bate A. M. A.), pp. 149-300. Cold Spring Harbor Laboratory Press. [Google Scholar]
- Froese, D. S., Michaeli, A., McCorvie, T. J., Krojer, T., Sasi, M., Melaev, E., Goldblum, A., Zatsepin, M., Lossos, A., Alvarez, R.et al. (2015). Structural basis of glycogen branching enzyme deficiency and pharmacologic rescue by rational peptide design. Hum. Mol. Genet. 24, 5667-5676. 10.1093/hmg/ddv280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandara, L. and Wappner, P. (2018). Metabo-Devo: A metabolic perspective of development. Mech. Dev. 154, 12-23. 10.1016/j.mod.2018.02.004 [DOI] [PubMed] [Google Scholar]
- Gross, S. P., Welte, M. A., Block, S. M. and Wieschaus, E. F. (2000). Dynein-mediated cargo transport in vivo. A switch controls travel distance. J. Cell Biol. 148, 945-956. 10.1083/jcb.148.5.945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, P., Martin, R., Knolker, H. J., Nihalani, D. and Kumar Sinha, D. (2017). Myosin-1 inhibition by PClP affects membrane shape, cortical actin distribution and lipid droplet dynamics in early Zebrafish embryos. PLoS ONE 12, e0180301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutzeit, H. O., Zissler, D., Grau, V., Liphardt, M. and Heinrich, U. R. (1994). Glycogen stores in mature ovarian follicles and young embryos of Drosophila: ultrastructural changes and some biochemical correlates. Eur. J. Cell Biol. 63, 52-60. [PubMed] [Google Scholar]
- Herms, A., Bosch, M., Reddy, B. J., Schieber, N. L., Fajardo, A., Ruperez, C., Fernandez-Vidal, A., Ferguson, C., Rentero, C., Tebar, F.et al. (2015). AMPK activation promotes lipid droplet dispersion on detyrosinated microtubules to increase mitochondrial fatty acid oxidation. Nat. Commun. 6, 7176. 10.1038/ncomms8176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- José-Antonio, C.-O. and Volker, H. (1985). The Embryonic Development of Drosophila Melanogaster: Springer-Verlag. [Google Scholar]
- Kilwein, M. D. and Welte, M. A. (2019). Lipid droplet motility and organelle contacts. Contact (Thousand Oaks) 145, dev131110. 10.1177/2515256419895688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilwein, M. D. and Welte, M. A. (2021). Visualizing cytoskeleton-dependent trafficking of lipid-containing organelles in Drosophila embryos. J. Vis. Exp. 178, e63291. 10.3791/63291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, R. C. (1970). Ovarian Development in Drosophila Melanogaster: Academic Press. [Google Scholar]
- Kuhn, H., Sopko, R., Coughlin, M., Perrimon, N. and Mitchison, T. (2015). The Atg1-Tor pathway regulates yolk catabolism in Drosophila embryos. Development 142, 3869-3878. 10.1242/dev.12541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lattao, R., Rangone, H., Llamazares, S. and Glover, D. M. (2021). Mauve/LYST limits fusion of lysosome-related organelles and promotes centrosomal recruitment of microtubule nucleating proteins. Dev. Cell 56, 1000-1013.e1006. 10.1016/j.devcel.2021.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z., Thiel, K., Thul, P. J., Beller, M., Kuhnlein, R. P. and Welte, M. A. (2012). Lipid droplets control the maternal histone supply of Drosophila embryos. Curr. Biol. 22, 2104-2113. 10.1016/j.cub.2012.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe, N., Rees, J. S., Roote, J., Ryder, E., Armean, I. M., Johnson, G., Drummond, E., Spriggs, H., Drummond, J., Magbanua, J. P.et al. (2014). Analysis of the expression patterns, subcellular localisations and interaction partners of Drosophila proteins using a pigP protein trap library. Development 141, 3994-4005. 10.1242/dev.111054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashek, D. G., Li, L. O. and Coleman, R. A. (2007). Long-chain acyl-CoA synthetases and fatty acid channeling. Future Lipidol 2, 465-476. 10.2217/17460875.2.4.465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, A., Sullivan, F. V., Richard, A. C., David, S., Angus, A. G.-W. and Robert, G. G. (2010). Nature of α and β particles in glycogen using molecular size distributions. Biomacromolecules 11, 1094-1100. 10.1021/bm100074p [DOI] [PubMed] [Google Scholar]
- Miyazawa, H. and Aulehla, A. (2018). Revisiting the role of metabolism during development. Development 145, dev131110. 10.1242/dev.131110 [DOI] [PubMed] [Google Scholar]
- Nguyen, T. B., Louie, S. M., Daniele, J. R., Tran, Q., Dillin, A., Zoncu, R., Nomura, D. K. and Olzmann, J. A. (2017). DGAT1-dependent lipid droplet biogenesis protects mitochondrial function during starvation-induced autophagy. Dev. Cell 42, 9-21.e25. 10.1016/j.devcel.2017.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prats, C., Graham, T. E. and Shearer, J. (2018). The dynamic life of the glycogen granule. J. Biol. Chem. 293, 7089-7098. 10.1074/jbc.R117.802843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambold, A. S., Cohen, S. and Lippincott-Schwartz, J. (2015). Fatty acid trafficking in starved cells: regulation by lipid droplet lipolysis, autophagy, and mitochondrial fusion dynamics. Dev. Cell 32, 678-692. 10.1016/j.devcel.2015.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, B. H., Wilk, R., Schock, F. and Lipshitz, H. D. (2004). Integrin-dependent apposition of Drosophila extraembryonic membranes promotes morphogenesis and prevents anoikis. Curr. Biol. 14, 372-380. 10.1016/j.cub.2004.02.029 [DOI] [PubMed] [Google Scholar]
- Rickoll, W. L. (1976). Cytoplasmic continuity between embryonic cells and the primitive yolk sac during early gastrulation in Drosophila melanogaster. Dev. Biol. 49, 304-310. 10.1016/0012-1606(76)90278-5 [DOI] [PubMed] [Google Scholar]
- Shubeita, G. T., Tran, S. L., Xu, J., Vershinin, M., Cermelli, S., Cotton, S. L., Welte, M. A. and Gross, S. P. (2008). Consequences of motor copy number on the intracellular transport of kinesin-1-driven lipid droplets. Cell 135, 1098-1107. 10.1016/j.cell.2008.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber, M. H., Thomsen, M. B. and Spradling, A. C. (2016). Electron Transport Chain Remodeling by GSK3 during Oogenesis Connects Nutrient State to Reproduction. Cell 164, 420-432. 10.1016/j.cell.2015.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, Y., Park, J. O., Tanner, L., Nagano, Y., Rabinowitz, J. D. and Shvartsman, S. Y. (2019). Energy budget of Drosophila embryogenesis. Curr. Biol. 29, R566-R567. 10.1016/j.cub.2019.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, X., Sullivan, M. A., Nada, S. S., Deng, B., Schulz, B. L. and Gilbert, R. G. (2018). Proteomic Investigation of the Binding Agent between Liver Glycogen beta Particles. ACS Omega 3, 3640-3645. 10.1021/acsomega.8b00119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen, J. M., Bertagnolli, N. M., Evans, J., Sieber, M. H., Cox, J. and Thummel, C. S. (2014). Coordinated metabolic transitions during Drosophila embryogenesis and the onset of aerobic glycolysis. G3 (Bethesda) 4, 839-850. 10.1534/g3.114.010652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran, S. L. and Welte, M. A. (2010). In-vivo centrifugation of Drosophila embryos. J. Vis. Exp. 40, e2005. 10.3791/2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Der Crone-Gloor, U. (1959). Quantitative untersuchung der Freien Aminosäuren und Polypeptide während der Embryonalentwicklung von Drosophila melanogaster. J. Insect Physiol. 3, 50-56. 10.1016/0022-1910(59)90058-7 [DOI] [Google Scholar]
- Walther, T. C. and Farese, R. V., Jr. (2012). Lipid droplets and cellular lipid metabolism. Annu. Rev. Biochem. 81, 687-714. 10.1146/annurev-biochem-061009-102430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welte, M. A. (2015a). As the fat flies: The dynamic lipid droplets of Drosophila embryos. Biochim. Biophys. Acta 1851, 1156-1185. 10.1016/j.bbalip.2015.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welte, M. A. (2015b). Expanding roles for lipid droplets. Curr. Biol. 25, R470-R481. 10.1016/j.cub.2015.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welte, M. A., Gross, S. P., Postner, M., Block, S. M. and Wieschaus, E. F. (1998). Developmental regulation of vesicle transport in Drosophila embryos: forces and kinetics. Cell 92, 547-557. 10.1016/S0092-8674(00)80947-2 [DOI] [PubMed] [Google Scholar]
- Yamada, T., Habara, O., Yoshii, Y., Matsushita, R., Kubo, H., Nojima, Y. and Nishimura, T. (2019). The role of glycogen in development and adult fitness in Drosophila. Development 146, dev176149. 10.1242/dev.176149 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.