Abstract
Neutralizing antibodies (nAbs) are being increasingly used as passive antiviral reagents in prophylactic and therapeutic modalities and to guide viral vaccine design. In vivo, nAbs can mediate antiviral functions through several mechanisms, including neutralization, which is defined by in vitro assays in which nAbs block viral entry to target cells, and antibody effector functions, which are defined by in vitro assays that evaluate nAbs against viruses and infected cells in the presence of effector systems. Interpreting in vivo results in terms of these in vitro assays is challenging but important in choosing optimal passive antibody and vaccine strategies. Here, I review findings from many different viruses and conclude that, although some generalizations are possible, deciphering the relative contributions of different antiviral mechanisms to the in vivo efficacy of antibodies currently requires consideration of individual antibody–virus interactions.
Subject terms: Antibodies, Infection
Assays for, and mechanisms of action of, neutralizing antibodies against viruses are considered here as tools to interpret and predict the activity of such antibodies in vivo, administered passively or induced by vaccination or infection.
Introduction
The COVID-19 pandemic has introduced the concept of neutralizing antibodies (nAbs) to the lay public and raised the profile of such antibodies in the scientific community generally. As is the case for many viruses, there is a fairly good correlation between levels of serum nAbs, as identified by in vitro assays, and protection against infection and disease for SARS-CoV-2, the virus that causes COVID-19. However, there are many misconceptions about nAbs and how they function. The term ‘neutralizing’ might be taken to indicate that these antibodies function in vivo predominantly by neutralization (as defined below), but this is not necessarily the case. Furthermore, there is great heterogeneity in the mechanism(s) of action of individual nAbs, even those that are apparently closely related. Much information has accumulated recently on nAbs to a wide range of viruses, which can be used to gain a better understanding of their activities.
There are several definitions of neutralization, two of which are widely accepted. One is “the loss of infectivity which ensues when antibody molecule(s) bind to a virus particle, and usually occurs without the involvement of any other agency”1. The phrase “without the involvement of any other agency” (although, rarely, complement is included) indicates that neutralization is typically measured in vitro by incubating antibodies, virus particles and target cells together and demonstrating reduced infection. A second definition of neutralization is “the reduction in viral infectivity by the binding of antibodies to the surface of viral particles (virions), thereby blocking a step in the viral replication cycle that precedes virally encoded transcription or synthesis”2. For enveloped viruses, this block occurs before virus entry into a host cell but for non-enveloped viruses, it can occur after entry2,3. In both cases, neutralization according to these definitions can be measured in in vitro assays. However, the term ‘neutralization’ can also be used to describe antiviral activities of antibodies such as protection in vivo, which may or may not involve neutralization in terms of blocking viral entry. This has created considerable confusion in the literature when antiviral (‘neutralizing’) activity in vivo is mediated by antibodies that do not block viral entry in typical neutralization assays in vitro — in other words, non-neutralizing antibodies (nnAbs). For the most part, the field has chosen to avoid confusion by defining neutralization so that it can be assessed by typical in vitro assays, involving antibody, virus and target cells alone.
The ability of nAbs to block viral entry by enveloped viruses in vitro requires that the antibodies bind to functional entry molecules on the surface of infectious virions, typically envelope (Env) protein spikes. Binding to these functional structures also endows nAbs with many other potential antiviral activities that could be manifested in vivo but are not present in neutralization assays in vitro. Thus, for example, virions coated with nAbs could be taken up in vivo by phagocytic cells via receptors for the antibody Fc (crystallizable fragment) domain (FcRs) or trapped on FcR-bearing cells and prevented from accessing target cells. Alternatively, functional viral structures that are recognized by nAbs can be expressed on infected cells, rendering them potential targets for antibody-dependent cellular cytotoxicity (ADCC). Again, recognition of functional structures will enhance the ability of antibodies to inhibit cell–cell spread of a viral infection.
Of crucial importance to the topics discussed in this Review, I suggest that in some respects, therefore, the best way to think of a nAb may be as an antibody that binds to functional structures on the surface of virions and infected cells.
In vitro, under the right assay conditions, this can lead to neutralization. In vivo, there are several possible outcomes, including but not limited to neutralization. As I discuss here, neutralization may even make a non-decisive contribution to the in vivo efficacy of antibodies that are described as neutralizing in vitro.
This Review focuses on nAbs but it should be emphasized that some nnAbs also have Fc-dependent antiviral activities against virions and against infected cells that are manifested in vitro when effector cells are included, and in vivo in the prophylaxis or therapy of viral infections. The activities of nnAbs against molecules expressed on the surface of infected cells but not on virions, such as the NS1 protein of dengue virus, are readily understood. The activities of nnAbs against free virions could, in principle, involve two types of target. The first is functional entry molecules on the surface of virions, in which case the binding of nnAbs must, by definition, not interfere with viral entry. The second is non-functional molecules on the virion surface, for which binding of nnAbs also does not affect viral entry. Indeed, disassembly or conformational rearrangement of a proportion of functional molecules on infectious virions, by exposing epitopes that are not available on functional molecules, presents opportunities for nnAbs to trigger effector systems such as complement activation and phagocytosis. Similar mechanisms can operate to enable nnAbs to act against virus-infected cells. Generally, however, nAbs are much more effective than nnAbs in vivo, which explains the greater research focus on nAbs.
With these introductory considerations in mind, I review findings from many different viruses — with a particular focus on HIV and SARS-CoV-2 — and consider our current understanding of the mechanisms of antibody-mediated neutralization in vitro, the antiviral mechanisms of nAbs that become relevant if effector cells are included in typical in vitro assays and the mechanisms of nAb activity against viruses in vivo.
Mechanisms of neutralization in vitro
Neutralization activity is determined for antibodies in sera or for monoclonal antibodies (mAbs) using in vitro assays, which aim to predict the effect of the antibody in vivo in terms of protection or therapy. Understanding the mechanisms of neutralization may assist in these predictions and is of clear scientific interest in terms of advancing our general knowledge of the interactions between antibodies and viruses.
Ideally, to be predictive of an in vivo effect, in vitro neutralization assays should mimic the in vivo conditions under which nAbs encounter virus as closely as possible. However, neutralization assays often use conditions that are far removed from those in vivo. For example, pseudoviruses that express surface proteins of the virus of interest on the background of another virus such as vesicular stomatitis virus or murine leukaemia virus are frequently used. Target cell lines, containing genes for indicator molecules such as luciferase to report on infection and enable high-throughput screening in single-cycle assays, are often used rather than the primary cells that would be infected in vivo. Possible artefacts arising from the artificiality of such assays can be explored by comparing results from pseudovirus (clonal) infection of cell lines with those from authentic replicating (quasispecies) virus infection of primary cells. Differences in the efficacy of nAbs between these assays are often seen in terms of their neutralization titres — measured as the concentration of nAb giving 50%, 80% or 90% inhibition of virus replication (IC50, IC80 or IC90, respectively). Differences can also be reflected in other parameters such as the maximum neutralization achieved at high nAb concentrations; for example, a pseudovirus might be neutralized to 100% by a given nAb but an authentic virus to notably less than 100% owing to, in this example, a greater surface heterogeneity. Another factor to consider in an in vitro neutralization assay is the length of time for which the antibody is preincubated with the virus; IC50, IC80 or IC90 values will often depend on this parameter, which could vary considerably for different viruses, target cells and infection environments in vivo. Therefore, in seeking to understand nAb activity in vivo, it is important to consider the limitations of the neutralization assay used. Nevertheless, despite these cautionary notes, it is the case that many neutralization assays using pseudoviruses and target cell lines have proven effective at broadly predicting the behaviour of antiviral antibodies in vivo, including those against HIV and SARS-CoV-24–9. However, deviations from a directly proportional relationship between in vitro activity and in vivo activity are seen for some antibodies, for example for antibodies to HIV10–12 and influenza virus13 and for a combination of SARS-CoV-2 nAbs14.
Antibody-mediated neutralization in vitro is increasingly understood to operate by various mechanisms (Fig. 1). Any antibody that binds with sufficient affinity to a native structure on virions will have the potential to interfere with viral entry. Several mechanisms have been associated with preventing virion attachment to target cell receptors. These include binding to the viral receptor-binding site or in its vicinity and preventing attachment by steric obstruction, disassembling or changing the conformation of viral surface entry proteins, and aggregating virions. nAbs can also interfere with entry post-attachment and have effects post-entry, including in the host cell cytoplasm and during viral egress from host cells. Finally, nAbs can also enhance infection in vitro as discussed in Box 1.
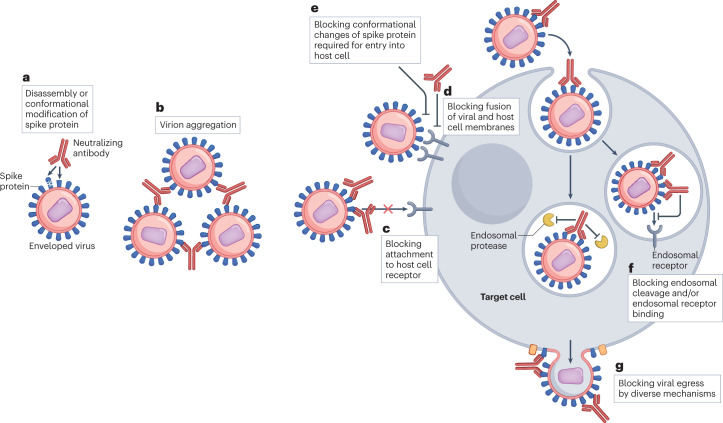
Fig. 1. Mechanisms of neutralization of an enveloped virus by neutralizing antibodies in vitro.
Antibody-mediated neutralization of enveloped viruses in vitro can operate by several mechanisms. a, Preventing the attachment of virions to host cell receptors by the disassembly or conformational modification of viral spike proteins. b, Aggregation of virions, impeding attachment to host cell receptors. c, Directly blocking binding of viral spike protein to host cell receptors through steric obstruction. d, Blocking the fusion of viral and host cell membranes by steric obstruction. e, Blocking conformational changes in spike protein required for virus entry into host cells. f, For viruses that enter endosomes, blocking entry into the cytoplasm by blocking endosomal cleavage and/or endosomal receptor binding. g, Blocking viral egress from the cell; although this is not strictly a mechanism of neutralization, it would be observed in multiple-round, but not single-round, neutralization assays and can occur through the aggregation of progeny virions at the surface of infected cells. The figure illustrates neutralization of an enveloped virus. For non-enveloped viruses, there is additionally intracytoplasmic neutralization involving the ubiquitin ligase TRIM21, which targets antibody-bound virus to the proteasome for degradation. Finally, the mechanisms that prevent viruses gaining entry to host cells can also prevent viruses spreading from one cell to another directly by inhibiting attachment of the infected cell to the uninfected cell and/or by inhibiting fusion of the membranes of the two infected cells.
Box 1 Antibody-dependent enhancement of infection.
Under some conditions, antibodies can enhance infection in classical neutralization assays through a phenomenon known as antibody-dependent enhancement (ADE)190–193. ADE is typically observed with neutralizing antibodies (nAbs) at sub-neutralizing concentrations and has most commonly been described for flaviviruses that can gain entry to crystallizable fragment (Fc) receptor (FcR)-bearing cells through interaction of virus-bound nAb with FcRs191. However, ADE has also been described for some antibody Fab regions and therefore can occur independently of FcRs193,194. The occupancy model of neutralization explains ADE as an effect that occurs at low occupancy of virion sites in the presence of permissive cells (for example, those bearing FcRs). As nAb concentrations increase, coating of the virus with nAb increases, eventually resulting in sufficient occupancy for neutralization. Interestingly, complement component C1q (which binds to the Fc portion of antibodies) has been shown to restrict ADE for flaviviruses in vitro and in vivo195, which might reflect more effective coating of virions because of the large size of the C1q molecule, more effective neutralization and a shift in ADE to lower nAb concentrations107. ADE is often viewed in the context of enhanced infection of permissive cells but nAb-complexed virus may also enter cells that are normally non-permissive, which may be followed by viral replication and the release of infectious progeny (productive infection) or may be a dead-end (non-productive infection or, more simply, viral ‘uptake’). Productive infection is much more likely to have in vivo consequences than non-productive infection. Non-productive ADE has been described for SARS-CoV and MERS-CoV in various cell lines in vitro196–201 and involved alternative receptor usage in non-permissive FcγRII-bearing cell lines202,203 and primary cells199. ADE, both productive and non-productive, has also been described for SARS-CoV-2204–206 involving FcR-mediated viral entry. In mice and in monkeys, antibodies to SAR-CoV-2 that mediated ADE in vitro protected effectively against virus in vivo204, which suggests that the in vitro phenomena are of limited importance. Studies showed that uptake of antibody-complexed SARS-CoV-2 occurred non-productively into human monocytes and macrophages via FcRs but that this could lead to inflammation that might contribute to COVID-19 pathogenesis205,207.
Although ADE can be readily demonstrated for several viruses in vitro, evidence in vivo is much more limited191. For dengue virus, clinical data suggest that the interaction of antibody-coated virus with FcRs on susceptible cells can enhance disease under certain conditions, in particular for sub-neutralizing concentrations of nAbs. Of note, disease severity resulting from secondary dengue virus infection seems to be related to antibody fucosylation and the strength of the interaction with FcRs; afucosylation, which increases binding to FcRs, leads to more severe disease208. For respiratory viruses such as respiratory syncytial virus and SARS-CoVs, there is also the possibility of vaccine-associated enhanced respiratory disease, in which immune complexes with viral proteins are deposited in the capillaries of the lungs, leading to tissue damage209.
Conformational change
Many Env spikes are metastable — on the virion surface they are in a pre-fusion conformation, and interaction with host cell receptors triggers extensive structural changes that allow for the viral entry process. Several cases have been described in which nAbs bind to and trigger conformational changes in recombinant Env proteins that render them non-functional and potentially irreversibly prevent viral entry15 (Fig. 1a). Examples of the use of such a mechanism include antibodies to HIV16–18, SARS-CoV19 and SARS-CoV-220,21. For HIV, the mAb PGT121 binds to the V3-glycan site on the Env spike, which is distant from the site that binds the host cell receptor CD4, but this can allosterically inhibit binding of soluble CD4 and therefore may provide a neutralization mechanism22. PGT121 inhibits binding of soluble CD4 not only to recombinant Env trimers but also to cell surface Env. Nevertheless, this does not unambiguously prove that induction of conformational change is required for neutralization by this antibody. Indeed, it has been shown that nAbs to the V3-glycan site of Env bind to residues that associate with the host cell co-receptor CCR5 and neutralization may thus result from inhibition of CCR5 binding rather than inhibition of CD4 binding23. Although there is a tendency to refer definitively to ‘mechanisms of neutralization’ in the literature from structural studies on recombinant proteins, it is worth emphasizing that these may be hypotheses that would require detailed and difficult studies to be truly definitive. Furthermore, even structural studies on whole virus particles can be misleading. Thus, nAbs have been shown to induce conformational changes in picornaviruses but, for rhinovirus, the ability to induce such changes was not correlated with neutralization activity and it was concluded that the changes are epiphenomena that are associated with some, but not all, nAbs24.
Perhaps the best evidence of neutralization being dependent upon nAbs disassembling or altering the conformation of viral surface proteins is provided by antibodies that have been shown to irreversibly reduce viral infectivity before incubation with target cells. For example, studies have shown that the infectivity of several viruses, including dengue virus, West Nile virus (WNV) and HIV, decays with time and that nAbs can accelerate the rate of this decay16,17,25. For HIV, antibodies to the V3-glycan site of Env (mAb PGT128) and to the gp120–gp41 interface site (mAb 3BC315) have been shown to accelerate the rate of decay of infectious virus16,17. The observed dependence of neutralization efficacy on length of time for which nAb and virus are pre-incubated before adding to target cells in neutralization assays is a further indication that the nAb causes irreversible conformational change that leads to inactivation of the virus. By contrast, the broadly neutralizing antibody (bnAb) 2G12 (Box 2) was shown to reversibly neutralize HIV post-attachment, probably by interfering with virus–CCR5 interaction26.
In general, the instability of Env spikes is reflected in the strong antibody responses often observed to post-fusion conformations of Env in many viral infections. The disassembly of multisubunit viral surface proteins with time inevitably results in the exposure of large protein surfaces, previously cryptic in the native structure, that are likely to be immunogenic. In terms of vaccine design, maintaining the pre-fusion conformation of Env is an oft-encountered problem in the design of recombinant Env proteins as potential immunogens27–29. Furthermore, the ability of antibodies generated by priming immunization to trigger the disassembly of recombinant Env protein could generate non-native forms of Env in subsequent immunization steps that are then likely to elicit nnAbs rather than nAbs30.
Box 2 Broadly neutralizing antibodies.
A category of neutralizing antibodies that has received much attention in recent years is broadly neutralizing antibodies (bnAbs) to highly antigenically variable viruses such as HIV, hepatitis C virus and influenza virus210,211. bnAbs recognize the more conserved regions of the surfaces of highly variable viruses and are prized as guides for the design of vaccines to such viruses15,212–217. For HIV, the discovery of bnAbs provided evidence that an antibody-based vaccine to prevent AIDS might be possible and defined the sites to be targeted by rational design principles212–217. For influenza virus, the discovery of bnAbs to both the stem and the head region of haemagglutinin have underpinned one set of approaches to develop ‘universal’ flu vaccines210,211,218,219. Recently, the emergence of variants of SARS-CoV-2 and the need for antibodies and vaccines to prevent future coronavirus pandemics have prompted searches for bnAbs to coronaviruses, particularly to sarbecoviruses and, more generally, to beta-coronaviruses220–224. Increasingly, it is understood that antibodies can be found that recognize almost every part of the surface of complex proteins. For example, a large proportion of the underlying protein surface of the HIV Env spike can be recognized by bnAbs despite the dense coating of the spike with glycans. In comparison to the HIV Env spike, the spike protein of SARS-CoV-2 is less comprehensively covered by glycans. It remains to be determined to what extent antibodies recognize regions of the native spike protein but do not neutralize virus.
Aggregation to prevent virus attachment
The aggregation of virions by antibody binding may contribute to neutralization by reducing attachment of viruses to target cells2 (Fig. 1b). However, aggregated virions may still attach to some extent to target cells depending upon the nAb and assay conditions, and aggregation may affect a step downstream from viral attachment such as fusion or viral egress from infected cells (see below). The extent of aggregation is determined by the molar ratio of viral particles to nAbs. Virion aggregation has been proposed as one potential contributor to the incomplete neutralization of virus, by preventing full access of nAbs to virion particles in the aggregate, leaving a non-neutralizable fraction (also known as the persistent fraction)31. Aggregation of virions has recently been proposed to contribute to the neutralization of Chikungunya virus32.
Steric obstruction to prevent virus attachment
The size of an antibody (IgG) molecule relative to the size of a typical viral surface protein is often underestimated (Fig. 2); the relatively large antibody size means that binding directly to the binding site for a host cell receptor on a virion surface protein is not necessarily required to perturb the interaction of a membrane-bound receptor on the target cell with the virion-associated protein15,33–39 (Fig. 1c). A clear demonstration of the perturbatory effect associated with the relatively large size of the antibody molecule binding to a viral surface is shown by antibodies to the haemagglutinin (HA) protein of influenza virus that interfere with the catalytic activity of viral neuraminidase (NA)40. Another example is provided by antibodies to host molecules that have been incorporated into the membranes of HIV particles but are not required for viral entry. The phenomenon is noted particularly in the presence of complement and presumably arises, at least in part, from the obstruction of HIV Env trimers by antibodies bound to neighbouring host molecules41–43. Neutralization of HIV and simian immunodeficiency virus with short peptide tags introduced into the Env molecules at varying positions has also been described for the corresponding anti-tag antibodies35,44,45, which is consistent with neutralization through general steric obstruction rather than specific targeting of functional sites.
Fig. 2. Model to scale of antibody-mediated neutralization of HIV.
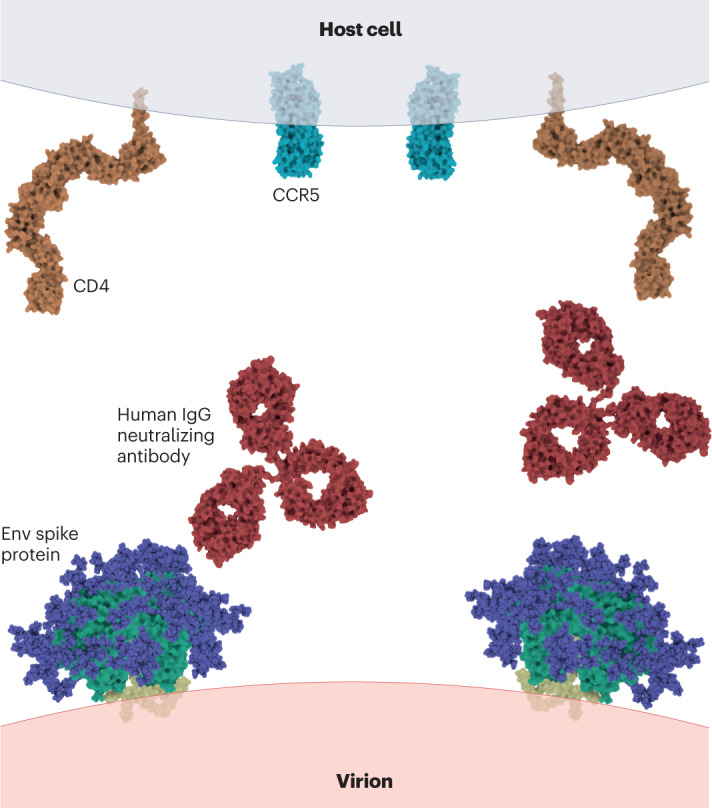
The large size of a human IgG neutralizing antibody (Protein Data Bank (PDB) ID: 1HZH) relative to the molecules that are involved in the entry of HIV into host cells — the viral envelope (Env) spike protein (PDB ID: 3J5M), and CD4 (PDB ID: 1WIO) and CCR5 (PDB ID: 1OPT) receptors on host cells — suggests that antibody can readily interfere with viral attachment to and/or fusion with the host cell through steric obstruction.
Post-attachment neutralization
Post-attachment neutralization is a major mechanism of neutralization for many viruses and antibodies. The fusion of viral and host cell membranes is a complex multistep process that can be sterically inhibited by the large bulk of the antibody molecule binding to, or in the vicinity of, the fusion machinery of the virus2,15 (Fig. 1d). For example, the transition of a spike protein from the pre-fusion to post-fusion conformation might be blocked by nAb (Fig. 1e). If the fusion process involves the interaction of multiple virion surface molecules with multiple host cell receptors, one might liken antibody-mediated inhibition of fusion to an object being inserted into a zipper2,46. Some viruses, such as WNV, can still gain entry to host cell endosomes with antibodies bound but are then inhibited by the antibodies from fusing with endosomal membranes and are therefore unable to gain access to the host cell cytoplasm47 and are destroyed in lysosomes (Fig. 1f). Other more specialized mechanisms have been suggested to operate for some viruses. In the case of Chikungunya virus, cryo-electron microscopy studies have shown that an antibody Fab (antigen-binding fragment) can bridge between two domains of the E2 glycoprotein of the viral E1–E2 complex, thereby preventing exposure of the fusion loop on the E1 protein and blocking fusion48. Another cryo-electron microscopy study of Chikungunya virus proposed that two nAbs bound between two protomers of E2 on one Env spike, thereby blocking viral fusion and also viral egress49. It should also be noted that many viruses have separate attachment and fusion proteins and that the efficacy of nAbs to the fusion proteins is consistent with post-attachment neutralization.
Inhibition of viral egress
Viral egress is another point in the virus life cycle at which nAbs can intercede (Fig. 1g). As this occurs after infection of a cell, it does not qualify as ‘neutralization’ according to the accepted definitions but it is nevertheless often considered together with the mechanisms for inhibition of viral entry. Inhibition of viral egress would be observed in a neutralization assay involving multiple rounds of viral replication but not in an assay involving a single round. Viral egress has been studied in influenza virus infection in particular. Inhibition of viral egress was shown to be mediated not only by nAbs to HA, but also by nnAbs to M2 and NA proteins on the surface of influenza virus-infected cells50. More recently, an HA head-binding antibody was shown to operate primarily by blocking viral egress51,52. The effects of this antibody were similar to those of a NA inhibitor by densely aggregating progeny virions on the surface of infected cells. Another antibody binding to a unique epitope on the HA head did not block receptor binding but inhibited viral egress by a similar aggregating mechanism53. Antibodies to influenza virus NA have been shown to inhibit viral egress and to offer protection in mice54,55. Of note, although antibodies to NA do not generally inhibit viral replication in single-cycle neutralization assays, they can do so in the case of modified viruses that use NA for entry into host cells56. Inhibition of viral egress has also been described for nAbs to Eastern equine encephalitis virus57, Sindbis virus57, Chikungunya virus32,49 and HIV58.
Viral surface protein breathing
The above discussion tends to assume that viral surface molecules are static but increasingly it is understood that such molecules can ‘breathe’ — in other words, undergo conformational relaxation over time that exposes novel epitopes or increases the exposure of existing epitopes. The ability of antibodies to bind to these exposed epitopes can then lead to neutralization. Neutralization by antibodies binding to internal or cryptic epitopes was first described for poliovirus59 and later for flaviviruses60, including WNV, dengue virus and hepatitis C virus61–63, and norovirus64. Potential consequences of ‘breathing’ of viral epitopes are a marked dependence of neutralization activity on the temperature and length of time for which virus and antibody are incubated together65.
Inhibition of cell–cell spread
nAbs are generally considered in the context of inhibiting infection of target cells by free virus particles in vitro but they can also inhibit the cell–cell spread of a virus. This can be shown in specifically designed assays, for which higher concentrations of nAbs are typically required than for inhibition of free virus66–73, although this is not always the case72. The requirement for increased nAb concentration to inhibit cell–cell spread may be associated with avidity effects of extended cell surfaces interacting with one another (in contrast to a viral surface with potentially less available surface for interaction), although other factors such as the restricted access of nAbs to interacting cell surfaces could also contribute74,75. The ability of nAbs to interfere with cell–cell spread could be important in many established viral infections, for example the spread of HIV between activated CD4+ T cells in tissues. It could also be important for protection against the initial stages of an infection that involves cell–cell spread, such as for herpes simplex virus infection.
Intracytoplasmic neutralization
Antibody-mediated interception of virus after entry into the target cell is a more recently described mechanism of viral inactivation that involves the degradation of non-enveloped viruses, including adenoviruses, picornaviruses and rotavirus, facilitated by the ubiquitin ligase TRIM2176–78. TRIM21 interacts with a site at the CH2–CH3 domain interface of the Fc region of IgG bound to virus, which results in ubiquitylation of the antibody-bound virus complex and recruitment to the proteasome for degradation. Another type of neutralization, often described as intracellular — even though antibody and virus gain access from the extracellular space without traversing a membrane — can occur if antibody-containing endosomes fuse with virus-containing endosomes. For example, polymeric IgA can intercept Sendai virus or rotavirus during the process of transcytosis79–85.
Neutralization stoichiometry
Closely related to the mechanisms of antibody-mediated neutralization of viruses in vitro are the stoichiometry and kinetics of neutralization.
Antibody occupancy and neutralization
Early studies proposed that virions are neutralized by single antibody molecules based on single-hit kinetics. However, later studies noted flaws in this analysis and multiple-hit models, in which a single virion is bound by many antibody molecules, are now more generally accepted2,86,87. In fact, a body of evidence favours occupancy theories in which neutralization is related to the degree of occupancy of virus surface molecules by antibody. One version of the occupancy theory suggests that neutralization occurs once a virion is coated with a given density of antibody molecules. Support for this theory comes from an observed approximately linear relationship for several viruses, including poliovirus, influenza virus and WNV, between the surface area of a virus and the number of antibody molecules bound at neutralization87,88. The theory points to the large size of the antibody molecule relative to the typical size of viral surface entry molecules and argues that steric interference is likely to be important in inhibiting viral entry. It is derived from several observations, including the inability of antibodies to minor populations of surface proteins — for example, antibodies to NA on influenza viruses or to an altered conformation of the rabies G protein — to neutralize the corresponding viruses owing to insufficient coating of virions. In the case of rabies G protein, conversion of the minority form to a majority form leads to gain of neutralization activity by the relevant antibody89,90. In contrast to the simplest coating theories, nAbs to the flaviviruses WNV and dengue virus can be placed in groups of ‘high’ or ‘low’ occupancy for effective neutralization91. The group of nAbs requiring higher levels of occupancy for neutralization has been associated with cryptic epitopes and Env breathing.
Neutralizing antibody affinity versus avidity
Several other features are linked to the stoichiometry of neutralization, including antibody binding avidity, incomplete neutralization and neutralization synergy. As regards avidity, for some combinations of antibody and viral target, there are small differences in the neutralization potency of Fab compared with whole IgG, but in other cases whole IgG is much more potent than the corresponding Fab in neutralization assays in vitro92–95. These differences are likely to reflect monovalent binding of Fab compared with divalent binding of whole IgG, depending upon the spacing of targeted epitopes on the virion surface94–96. The Env spikes on HIV are sparsely distributed, which is consistent with neutralization by monovalent antibody binding in many cases, although it has also been proposed that divalent binding may occur in the context of clustered spikes associated with viral entry16. For SARS-CoV-2, divalent IgG binding has been shown to lead to more potent neutralization and more effective shedding of the S1 subunit, which will render Env spikes non-infectious, than does monovalent Fab binding21. Different antibody isotypes have different flexibilities that might affect how the two Fab arms of an antibody interact with virion surface proteins, both in terms of intrinsic affinity and avidity through bivalent binding. Furthermore, the length and character of the hinge region between Fab and Fc domains of an antibody can vary considerably, and for IgM and divalent or secretory IgA the molecular weight of the antibody is notably different from that of IgG. All of these factors might be expected to influence neutralization activity and, indeed, where they have been studied, this seems to be the case. For example, in the case of HIV, IgG3 has been shown to be generally more effective at neutralization than IgG1 of the same specificity, possibly related to the greater flexibility of IgG3 associated with a longer hinge region, which allows for more restricted epitopes to be accessed, although there are some exceptions97–100. The authors of one of these papers suggested that isotype switching could allow for HIV-infected individuals to respond to the escape of HIV from neutralization in a phenomenon they termed ‘switch redemption’98. Similarly, an IgA2 bnAb to HIV was shown to be more potent than the corresponding IgG1, possibly related to CH1 domain flexibility101. For SARS-CoV-2, an IgM antibody is much more potent than the corresponding IgG and confers intranasal protection against variant virus challenge in a mouse model, whereas the IgG antibody does not102.
Incomplete and synergistic neutralization
Incomplete (less than 100%) neutralization by mAbs at saturating concentrations is sometimes observed and may arise, for example, from glycan or conformational heterogeneity of Env spikes103–105. The effect can vary between virus strains, viruses grown in different cell types and different target cells. The consequences of incomplete neutralization could in principle be very serious, as a fraction of virions may still be infectious in the in vivo environment.
Neutralization synergy and, more often, additive neutralization have been observed when combinations of mAbs are studied2. An interesting recent example is provided by SARS-CoV-2, for which synergy between nAbs is clearly shown in vitro but is lost in vivo at the limited range of concentrations of antibody that have been assessed106. The discovery of nAbs that have synergistic neutralizing effects is a goal of attempts to generate cocktails of nAbs for prophylaxis or therapy106.
Antibody effector assays
By common definition, neutralization assays include only virus, antibody and target cell but, in vivo, several other factors could contribute to the antiviral activity of nAbs by recognizing clusters of nAbs bound to antigen on virions or infected cells (Fig. 3). These include complement and cells bearing FcRs, which can directly mediate antibody effector activities or indirectly, via antigen presentation, enhance cellular immunity. The creation of assays that accurately reflect in vivo conditions for antibody effector activity is perhaps even more difficult than for neutralization activity.
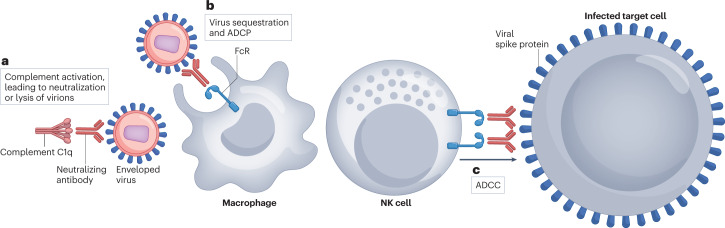
Fig. 3. Antiviral activities of neutralizing antibodies mediated by Fc-dependent effector functions.
a, In the presence of complement, neutralizing antibody (nAb)-coated virions are susceptible to enhanced neutralization (by steric hindrance) and to complement-mediated lysis. b, In the presence of crystallizable fragment receptor (FcR)-bearing effector cells, nAb-coated virions can be sequestered and prevented from binding to host cells or can be taken up by effector cells such as macrophages by antibody-dependent cellular phagocytosis (ADCP). c, Virus-infected cells coated with nAbs are susceptible to killing by effector cells such as natural killer (NK) cells through FcR-dependent, antibody-dependent cellular cytotoxicity (ADCC).
Complement
Most straightforward is the inclusion of complement in neutralization assays. In principle, one would expect that the binding of C1q (a protein of molecular mass ~400–450 kDa) to nAb bound to virions would enhance neutralization by increasing steric hindrance of viral attachment to or fusion with host cell membrane (Fig. 3a). Indeed, many examples have been described in which the inclusion of complement increases nAb potency in vitro107–110, and for WNV this was shown to be mechanistically associated with reducing the stoichiometry of antibody-mediated neutralization107. By contrast, for HIV, the addition of complement to in vitro assays has been associated with increased antibody-dependent enhancement (ADE)111 (Box 1) and enhanced HIV transmission112. In addition to steric hindrance, complement deposition can also mediate the lysis of virions and of infected cells (Fig. 3a). Indeed, as long ago as 1980, it was shown that complement could lyse murine leukaemia virus in vitro in conjunction with antibodies to the p15(E), but not the gp70, envelope protein113. The lytic activity was associated with the embedding of p15(E) in the virus membrane whereas gp70 projects further from the membrane. Generally though, interest in complement as a potential mechanism to enhance the antiviral activity of nAbs has been limited, in part because of early in vivo observations that nAbs against several viruses had similar levels of protection in complement-deficient mice and wild-type mice87. Furthermore, complement was suggested not to contribute to protection mediated by a nAb against simian–human immunodeficiency virus (SHIV) in non-human primates114, although other nAbs than the one used in that study may be more effective complement activators115.
Antibody-dependent cellular phagocytosis
In contrast to the role of complement, there is evidence from many viruses for the importance of FcR-mediated effector functions of nAbs in antiviral activity in vivo, particularly from studies comparing nAbs with variants in which FcR binding is eliminated or enhanced116–118. The main Fc-dependent effector functions of antibodies that have been studied are antibody-dependent cellular phagocytosis (ADCP) and ADCC. ADCP could, in principle, apply to both virions and infected cells complexed with nAb. Most interest has focused on ADCP of virions118,119, which can be assessed in various in vitro assays (Fig. 3b). One type of assay makes use of cell lines to phagocytose antibody-complexed, viral antigen-coated beads120. Such an assay has advantages for high-throughput screening but may not mimic in vivo conditions well. For example, the viral antigen is typically a recombinant protein rather than native protein from the viral membrane, the density of antigen molecules on the beads (which may be crucial for activation of antibody effector functions) may differ greatly from that on a virion, the size of beads is likely to be different from the size of virions and the phagocytosing cell line may have important differences from primary cells. Assays that are closer to in vivo conditions involve the phagocytosis of infectious virions by primary phagocytes119,121. The relative worth of different ADCP assays in predicting the in vivo efficacy of nAbs has not been well established. Indeed, as discussed below, this is very difficult to establish. Interestingly, HIV has been suggested to avoid ADCP owing to the irregular and relatively large spacing between Env spikes on the virion surface122.
Antibody-dependent cellular cytotoxicity
Infected cells complexed with antibody can be killed by ADCC (Fig. 3c). In vitro assays for ADCC assess the ability of antibodies to mediate target cell killing through interaction between the Fc domain of antigen-complexed nAb and FcR on an effector cell. Crosslinking of FcRs on effector cells leads to intracellular signalling that results in the release of cytotoxic factors to kill target cells and cytokines to inhibit viral spread. Effector activity is most often associated with natural killer cells but mononuclear phagocytes and neutrophils may also be important123. Ideally, as for the other antibody functions, an in vitro assay for ADCC would mimic in vivo conditions as closely as possible. Historically, assays have ranged from those using target cells with surface receptors that capture recombinant viral proteins and effector cell lines to those using virus-infected primary target cells and isolated effector cells from the same donor. There is a large potential for variation in results according to the assay used, and support for the authenticity of a given assay is best generated by showing a correlation with an in vivo outcome. Examples of the potential anomalies that can arise with ADCC assays have been described for HIV124,125. For example, target cells infected with primary HIV encoding functional Nef and Vpu accessory proteins, which downregulate CD4 expression on the surface of infected cells to evade host defences, are largely resistant to ADCC mediated by a class of antibodies specific for the CD4-induced epitope (CD4i) of gp120. However, in some assays that use target cells defective for Nef and/or Vpu expression, CD4 expressed by these cells is not downregulated and can capture shed gp120; these cells are much more sensitive to antibodies to CD4i, leading to an erroneous conclusion that these antibodies are likely to be effective in ADCC in vivo. Overall, any conclusion that a given antibody or serum is effective in mediating ADCC needs to be carefully examined in the context of the assay used.
Another assay to measure ADCC is the antibody-dependent cell-mediated virus inhibition assay. This assay infects a cell line with virus (typically HIV or SIV), removes free virus, adds antibody and effector cells, incubates for 7 days and then measures virus replication. The assay measures ADCC but also the effects of non-cytolytic antiviral mechanisms such as β-chemokine release from effector cells126. It also probably includes a contribution from antibody-mediated neutralization. Trogocytosis, whereby membrane from a virus-infected cell is transferred to an effector cell, may also involve Fc-dependent antibody interactions and have antiviral implications119,127,128.
Virus capture
In principle at least, virions bound by antibody could be captured on FcR-bearing cells in vivo and their contact with target cells reduced (Fig. 3b). For example, HIV entering the body through a mucosal surface will encounter few target cells in the form of activated CD4+ T cells initially and during a limited time period before its infectivity will be lost. One can hypothesize that if antibody-bound virus attaches to an FcR-bearing cell, this would reduce the likelihood of productive infection of a target cell irrespective of direct antiviral effects of the antibody. In vitro capture assays can provide a measure of the ability of immobilized antibody to capture virions129,130. Of interest for HIV, nnAbs can also capture infectious virions in such an assay, presumably by binding to non-functional structures on virions, such as gp41 ‘stumps’ left after gp120 has been shed129,131. However, most attempts to directly demonstrate notable protection by such antibodies by passive transfer in vivo have failed132,133, which suggests that, for this virus at least, any effect of sequestration is limited.
Antiviral antibody activities in vivo
The antiviral activities of nAbs as assessed by in vitro assays may operate in vivo but it is difficult to determine the relative contributions of the different mechanisms — direct neutralization, nAb-mediated effector functions against free virions or infected cells, or virus sequestration. Furthermore, the contributions of these different mechanisms may vary depending upon viral challenge dose, exposure conditions, nAb characteristics and a large number of other factors. The choice of animal models to explore the in vivo activity of nAbs and their mechanisms of action is an important one and differences between models and conditions can be anticipated. Nevertheless, certain common themes emerge (Fig. 4).
Fig. 4. Common themes associated with neutralizing antibody functions in vivo.
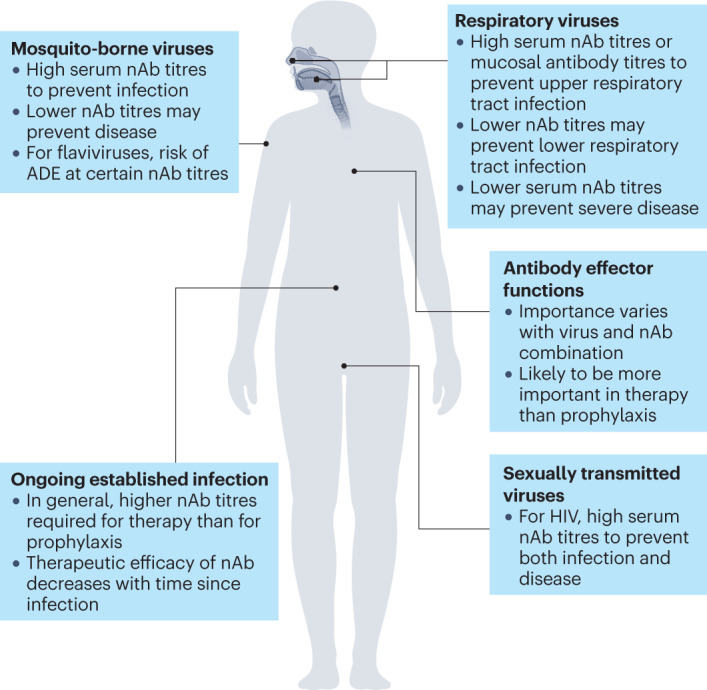
The activities of neutralizing antibodies (nAbs) against select virus types and routes of exposure are illustrated schematically. ADE, antibody-dependent enhancement.
Sterilizing immunity at high nAb titres
One common theme is that high serum titres of nAb (two to three orders of magnitude greater than in vitro nAb titres) are typically required to provide sterilizing immunity in vivo following viral challenge — in other words, to completely protect against establishment of infection through complete block of significant viral replication. From studies of HIV in small-animal models and SHIV in non-human primates, we refer to the ‘1:100 rule’, whereby the serum nAb concentration required to achieve 50% protection in vivo is 100 times the nAb concentration required to achieve 50% neutralization in vitro9. This would correspond to complete protection in vivo at nAb levels at least 500 times higher than the IC50 as measured in vitro or 200 times higher than the IC80. This pattern has largely been reproduced in many, although not all, studies in non-human primates using different SHIVs, different nAbs and different challenge routes12. More importantly, a similar pattern has been described for protection against HIV in humans by the bnAb VRC017,134. Similarly, a nAb to respiratory syncytial virus (RSV) was shown to offer essentially complete protection in the cotton rat model following intranasal challenge at a serum neutralizing titre of ~1:380135, and this helped to guide the antibody dose used to protect at-risk infants by prophylactic administration136,137. Another study of RSV infection in cotton rats showed 90% protection mediated by nAb at a serum neutralizing titre of 1:1,300138. High doses of SARS-CoV-2 mAbs were used in several COVID-19 prophylactic studies and in one study were shown to provide significant protection against detectable infection139.
However, it should be noted that the requirement for high nAb titres for sterilizing immunity is qualitative rather than strictly quantitative. For example, several bnAbs, including some directed to the base of the HIV Env spike, may be more effective in vivo than would be anticipated from their in vitro neutralization titres10–12. A similar deviation from a uniform relationship between neutralization and protection has been described for nAbs to influenza virus13 and for a combination of SARS-CoV-2 nAbs14. Effector functions that are triggered unequally by different nAbs but that are important for protection in vivo, or poor mimicry of in vivo neutralization by the in vitro assay, may be contributory factors.
Several explanations can be advanced for the apparent over-capacity in neutralization activity that is required for sterilizing immunity in many animal models and some human studies. If it is assumed that neutralization is the dominant protective mechanism, then for some viruses, animal models or challenge routes, it may be necessary for antibody to neutralize essentially every virus particle to achieve protection, which would require an excess of antibody. An alternative explanation is that the over-capacity is artefactual because nAb concentration is measured in serum but protection is mediated at tissue sites, for example mucosal surfaces, where the nAb concentration is only a fraction of that measured in serum46,140. Finally,protection may require an additional activity of nAbs distinct from neutralization. For example, it may be that activity against infected cells, as well as (or in some cases even instead of) activity against free virions, is needed to provide protection. This would be consistent with observations that nAb concentrations that are required, for example, to block cell–cell transmission are typically higher than those required for neutralization of free virus particles. One caveat that should also be noted is that many animal models involve relatively severe challenge conditions — for example, direct cerebral injection of virus — and so the extrapolation of any conclusions to natural exposure of humans should be treated with caution.
Protective advantage at lower nAb titres
A second common theme in the activity of nAbs in vivo is that prophylactic nAbs at concentrations less than those required to produce sterilizing immunity can nevertheless provide protective advantage. This may not be the case for viruses such as HIV, for which any breach of nAb-mediated defences may result in a latent infection, but for other viruses an advantage is gained even at low titres of nAbs. For example, nAbs to lymphocytic choriomeningitis virus in a mouse model141 and to Ebola virus in a guinea pig model142 do not provide sterilizing immunity but do prevent disease. For SARS-CoV-2, several studies suggest that breakthrough infections in nAb-treated individuals result in less severe symptoms than in individuals treated with placebo139,143,144. For RSV, nAbs are effective at preventing hospitalization but many treated infants are still infected and may still be symptomatic136,145.
Prophylaxis versus therapy
Other common themes are that nAbs are more effective in a prophylactic setting than as a therapeutic modality and that any therapeutic efficacy of a nAb decreases the later it is administered in an ongoing viral infection. These themes were first indicated by early animal studies87 and have since gained further support from human studies of RSV and SARS-CoV-2 infection. The RSV nAbs palivizumab and motavizumab are effective in prophylaxis but were not effective therapeutically in established RSV infection137,146. Several SARS-CoV-2 nAbs and cocktails of nAbs have shown efficacy early in infection but not in hospitalized patients with COVID-19 later in infection144,147–156. Therefore, the notable efficacy of nAbs in established Ebola virus infection in monkeys and humans, even when symptoms are present, is perhaps a surprise, albeit an encouraging one for nAb therapies, given the severity and rapid spread of Ebola virus infection in vivo157–159. Also of note, nAbs to HIV were shown to be effective in monkeys in controlling infection in some circumstances160,161, as well as subsequently in some humans, suggesting a potential role for nAbs in therapy and even cure of HIV162–164.
It should also be mentioned that the latest generation of nAbs (‘super-antibodies’), which have been isolated from donors with very high serum neutralization titres and/or by deeply mining antibody responses, are extremely potent and effective165. Conclusions drawn from earlier generations of nAbs may be challenged as super-antibodies are more widely used. In some respects, we are entering the fourth generation of nAbs: the first was serum antibodies, the second purified polyclonal antibodies, the third monoclonal antibodies, and now, super-antibodies.
Fc-dependent effector functions
One of the most well investigated aspects of nAb activities in vivo is the relative importance of Fc-dependent effector functions in protection and in therapy. Early animal models suggested that such functions were important in some cases but not others; for example, nAbs lacking the Fc region mediated effective protection only in some cases87. The importance of Fc-dependent antibody effector functions for protection against yellow fever virus in mice was indicated by the greater efficacy of an IgG2a antibody (which is a potent activator of effector functions) than an IgG1 antibody (a poor activator of effector functions) with the same Fab region166. As noted earlier, many of these early model studies that suggested the importance of Fc-dependent effector functions also found that protection was independent of complement, leading to a greater focus on FcR-mediated interactions87. More recently, the contributions of Fc-dependent effector functions to the antiviral activity of nAbs have been explored using antibodies engineered to have point substitutions in the Fc region that reduce, eliminate or enhance such functions116–118. An overall picture is emerging in which the contribution of effector functions to nAb-mediated protection depends on the individual antibody–virus combination, even for antibodies directed to the same viral protein. The contribution of nAb effector functions tends to be greater during established infection than during prophylaxis. For SHIV, FcR binding has been shown to contribute to protection from infection in monkeys for one bnAb but not for another more potent bnAb directed to a different site on the HIV Env protein114,167–169. In established SHIV infection in monkeys, antibody effector functions contribute to clearance of virus by nAbs but the effects are modest and enhanced FcR binding decreases rather than increases virus clearance170,171. In mouse models of HIV infection, increased FcR binding enhances virus control but only over certain ranges of nAb titre172. For influenza virus, nAbs to the HA stem region require Fc-dependent effector functions to mediate protection whereas nAbs to the HA head region do not173,174. For Ebola virus, protective antibodies show a range of dependencies on FcR interactions175. For SARS-CoV-2 infection in small-animal models, nAbs have been described that do or do not require Fc-dependent effector functions14,176. Therapy of SARS-CoV-2 infection is more dependent on antibody effector functions than is prophylaxis176.
The factors responsible for the differing dependence on Fc-dependent effector functions of different nAb–virus combinations are under-explored. For influenza virus, the ability of HA stem-targeting nAbs to induce ADCC but their rather weak neutralizing activity compared with nAbs to the HA head domain, which have weak interaction as immune complexes with FcRs but potent neutralizing activity, probably explains the dependence of anti-stem nAbs (but not anti-head nAbs) on Fc-dependent effector functions for protective efficacy173. Increased ADCC for antibodies recognizing epitopes that are closer to the target cell membrane has been described previously177. For HIV, the differing behaviour of nAbs to Env is not readily explained by differences in ADCC or FcR interactions as assessed in vitro, nor by differences in neutralization potency as experimental conditions were adjusted to control for such differences168. In general, though, some differences in effector function can be anticipated for nAbs that have different angles of approach to their epitopes and cluster differently on the virion or infected cell surface.
A dependence of antibody-mediated protection on intact Fc sites that mediate effector functions may be taken to indicate the importance of these functions but does not mean that neutralization activity does not also contribute. Clearly, one cannot simply knock out neutralizing activity from nAbs to carry out the appropriate control experiment. Therefore, one interpretation of nAb dependence on Fc effector functions might be that neutralization is important but not decisive, and that, in the absence of Fc-dependent effector functions, enough virus avoids neutralization to establish infection. The converse, the independence of nAb-mediated protection from Fc-dependent effector functions, is likely to be interpreted as support for a dominant role of neutralization in mediating protection but the models and conditions used should be carefully considered with respect to their relevance to humans. One elegant study directly investigated protection against human papilloma virus (HPV) in vivo in a mouse model178. Importantly, in this case, the details of the crucial nAb activities in vivo were complex and not fully captured by in vitro neutralization assays, which typically involve inhibition of HPV pseudovirus infection in an epithelial cell line by nAb179. In vivo, HPV infection requires initial association of virus with an acellular basement membrane, and it was shown that antibodies to L1, the major capsid protein of HPV, can interfere with infection at two stages. High concentrations of nAb to L1 prevent basement membrane binding, whereas low concentrations of nAb to L1 allow such association but prevent epithelial cell (keratinocyte) surface binding. Interestingly, viral capsids that were inhibited from binding to basement membrane were subsequently found as antibody–virus complexes in large cellular aggregates, suggesting a possible role for FcRs in their clearance. Antibodies to HPV L2 protein do not interfere with basement membrane association but inhibit epithelial cell binding.
An Fc-dependent phenomenon that has received recent attention in the context of viral infection, originally described in oncology, is the ‘vaccinal effect’ of antibodies in vivo180–184. It is suggested that immune complexes of viral proteins with nAbs or nnAbs engage FcRs on dendritic cells, leading to the presentation of viral antigens and enhanced T cell-mediated immunity. Vaccinal effects have been suggested following the passive administration of nAbs in non-human primates and humans160,163. For SARS-CoV-2, a therapeutic nAb with enhanced FcR affinity to generate a vaccinal effect has been proposed in preprint data (not peer reviewed)185.
Monoclonal versus polyclonal antibodies
Most of the above discussion refers to monoclonal nAbs, for which mechanistic effects and rules can be most readily determined. However, in vivo, following infection or vaccination, the nAbs generated are polyclonal. In fact, nAbs are almost always present as a small fraction of the total concentration of antibodies to a given viral surface protein expressed in isolation. Thus, serum neutralization assays from infection or vaccination are run in the presence of an excess of nnAbs, which is in contrast to monoclonal nAb assays. Where it has been studied, the presence of excess nnAbs does not seem to perturb the activity of nAbs in vitro129,186. This is consistent with only nAbs binding to crucial sites on functional virion structures whereas both nAbs and nnAbs bind to isolated, for example recombinant, viral surface proteins. As an example, only nAbs bind to the CD4-binding site of HIV Env trimer, whereas both nAbs and nnAbs bind to the same site on isolated gp120186,187.
The role of FcRs in protection against SARS-CoV-2 strains mediated by passively transferred immune sera and by vaccine-induced polyclonal antibodies was recently investigated in mice (preprint data; not peer reviewed)188. It was noted that protection mediated by passive transfer of immune sera was lost in mice lacking expression of activating FcRs, in particular FcγRIII, or depleted of alveolar macrophages. Furthermore, immunization with an mRNA vaccine led to loss of protection in the respiratory tract in mice lacking FcγRIII. Thus, FcR function seems to have a crucial role in protection mediated by the polyclonal antibodies provided by passive or active immunization, although as noted above, individual mAbs may not require FcR function for protection.
Concluding remarks
It is in the nature of science to seek to discern patterns and establish rules but nAbs and viruses are very diverse and generalizations about the interplay between the two are to be treated with caution. Nevertheless, several general points can be made. The ability of an antibody to neutralize in vitro is probably the best-known predictor of its antiviral efficacy in vivo. This may be because of the importance of inhibiting viral entry to target cells (direct neutralization) but equally it may reflect the ability of the antibody to bind to common structures on virions and/or infected cells and therefore trigger antiviral functions such as Fc-dependent effector mechanisms, viral sequestration and inhibition of cell–cell spread of virus. In terms of Fc-dependent effector mechanisms, knowledge regarding the contribution of these mechanisms to nAb-mediated protection could be valuable, for example, in the design of antibodies for prophylaxis. Although trends in the relative importance of different antibody functions can be associated with certain specificities — for example, antibodies to the HA head compared with the HA stem of influenza virus — each antibody–virus combination should probably be considered individually. Results from animal models will inform the design of antibodies for use in humans, but only with evaluation under real-world conditions can the impact of, for example, effector functions on antibody prophylaxis or therapy be fully evaluated. In any case, the contribution of blocking viral entry to target cells to the overall antiviral activity of a nAb may, in many cases, remain undetermined.
Although neutralization in vitro is probably the best predictor of in vivo efficacy, there are numerous examples of deviation from a uniform relationship between neutralization and protection and/or therapy. This may be due to differential contributions of effector functions to antiviral activities, which might be anticipated for antibodies binding even to the same epitope with different geometries of interaction. Alternatively, it may be that the in vitro neutralization assay is not fully representative of in vivo conditions, as has been described in the context of HPV infection of mice178.
The efficacy of vaccination against viruses in clinical trials is often estimated by measuring serum neutralization. For SARS-CoV-2, it has recently been argued that T cell responses should also be assessed189. One can also argue that other parameters of nAbs such as effector function should be evaluated. However, the difficulty will be to generate in vitro assays that accurately reflect or predict in vivo activities and vaccine efficacy. With current knowledge, measuring neutralization by in vitro assays remains the most effective way to evaluate likely vaccine efficacy in most cases.
Acknowledgements
I thank the National Institute of Allergy and Infectious Diseases, the Bill and Melinda Gates Foundation, the International AIDS Vaccine Initiative, the Ragon Institute of Massachusetts General Hospital, Massachusetts Institute of Technology and Harvard University and the James B. Pendleton Charitable Trust for financial support. I also thank P. J. Klasse and L. Walker for a thorough reading of the manuscript, and R. Andrabi, B. Briney, L. Hangartner, E. Scherer and J. Voss for helpful comments.
Peer review
Peer review information
Nature Reviews Immunology thanks S. Kent and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Competing interests
The author declares no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Dimmock NJ. Update on the neutralization of animal viruses. Rev. Med. Virol. 1995;5:165–179. doi: 10.1002/rmv.1980050306. [DOI] [Google Scholar]
- 2.Klasse PJ. Neutralization of virus infectivity by antibodies: old problems in new perspectives. Adv. Biol. 2014;2014:157895. doi: 10.1155/2014/157895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McEwan WA, James LC. TRIM21-dependent intracellular antibody neutralization of virus infection. Prog. Mol. Biol. Transl. Sci. 2015;129:167–187. doi: 10.1016/bs.pmbts.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 4.Khoury DS, et al. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021;27:1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- 5.Gilbert PB, et al. Immune correlates analysis of the mRNA-1273 COVID-19 vaccine efficacy clinical trial. Science. 2022;375:43–50. doi: 10.1126/science.abm3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Earle KA, et al. Evidence for antibody as a protective correlate for COVID-19 vaccines. Vaccine. 2021;39:4423–4428. doi: 10.1016/j.vaccine.2021.05.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corey L, et al. Two randomized trials of neutralizing antibodies to prevent HIV-1 acquisition. N. Engl. J. Med. 2021;384:1003–1014. doi: 10.1056/NEJMoa2031738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pegu A, et al. Neutralizing antibodies to HIV-1 envelope protect more effectively in vivo than those to the CD4 receptor. Sci. Transl. Med. 2014;6:243ra288. doi: 10.1126/scitranslmed.3008992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parren PW, et al. Antibody protects macaques against vaginal challenge with a pathogenic R5 simian/human immunodeficiency virus at serum levels giving complete neutralization in vitro. J. Virol. 2001;75:8340–8347. doi: 10.1128/JVI.75.17.8340-8347.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hessell AJ, et al. Broadly neutralizing human anti-HIV antibody 2G12 is effective in protection against mucosal SHIV challenge even at low serum neutralizing titers. PLoS Pathog. 2009;5:e1000433. doi: 10.1371/journal.ppat.1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hessell AJ, et al. Broadly neutralizing monoclonal antibodies 2F5 and 4E10 directed against the human immunodeficiency virus type 1 gp41 membrane-proximal external region protect against mucosal challenge by simian-human immunodeficiency virus SHIVBa-L. J. Virol. 2010;84:1302–1313. doi: 10.1128/JVI.01272-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pegu A, et al. A meta-analysis of passive immunization studies shows that serum-neutralizing antibody titer associates with protection against SHIV challenge. Cell Host Microbe. 2019;26:336–346.e3. doi: 10.1016/j.chom.2019.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sutton TC, et al. In vitro neutralization is not predictive of prophylactic efficacy of broadly neutralizing monoclonal antibodies CR6261 and CR9114 against lethal H2 influenza virus challenge in mice. J. Virol. 2017;91:e01603–e01617. doi: 10.1128/JVI.01603-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schafer A, et al. Antibody potency, effector function, and combinations in protection and therapy for SARS-CoV-2 infection in vivo. J. Exp. Med. 2021;218:e20201993. doi: 10.1084/jem.20201993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murin CD, Wilson IA, Ward AB. Antibody responses to viral infections: a structural perspective across three different enveloped viruses. Nat. Microbiol. 2019;4:734–747. doi: 10.1038/s41564-019-0392-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pejchal R, et al. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science. 2011;334:1097–1103. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee JH, et al. Antibodies to a conformational epitope on gp41 neutralize HIV-1 by destabilizing the Env spike. Nat. Commun. 2015;6:8167. doi: 10.1038/ncomms9167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruprecht CR, et al. MPER-specific antibodies induce gp120 shedding and irreversibly neutralize HIV-1. J. Exp. Med. 2011;208:439–454. doi: 10.1084/jem.20101907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walls AC, et al. Unexpected receptor functional mimicry elucidates activation of coronavirus fusion. Cell. 2019;176:1026–1039.e15. doi: 10.1016/j.cell.2018.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wec AZ, et al. Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science. 2020;369:731–736. doi: 10.1126/science.abc7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan R, et al. Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies. Cell Res. 2021;31:517–525. doi: 10.1038/s41422-021-00487-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Julien JP, et al. Broadly neutralizing antibody PGT121 allosterically modulates CD4 binding via recognition of the HIV-1 gp120 V3 base and multiple surrounding glycans. PLoS Pathog. 2013;9:e1003342. doi: 10.1371/journal.ppat.1003342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sok D, et al. A prominent site of antibody vulnerability on HIV envelope incorporates a motif associated with CCR5 binding and its camouflaging glycans. Immunity. 2016;45:31–45. doi: 10.1016/j.immuni.2016.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith TJ. Antibody interactions with rhinovirus: lessons for mechanisms of neutralization and the role of immunity in viral evolution. Curr. Top. Microbiol. Immunol. 2001;260:1–28. doi: 10.1007/978-3-662-05783-4_1. [DOI] [PubMed] [Google Scholar]
- 25.Dowd KA, Pierson TC. Antibody-mediated neutralization of flaviviruses: a reductionist view. Virology. 2011;411:306–315. doi: 10.1016/j.virol.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Platt EJ, Gomes MM, Kabat D. Kinetic mechanism for HIV-1 neutralization by antibody 2G12 entails reversible glycan binding that slows cell entry. Proc. Natl Acad. Sci. USA. 2012;109:7829–7834. doi: 10.1073/pnas.1109728109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanders RW, et al. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J. Virol. 2002;76:8875–8889. doi: 10.1128/JVI.76.17.8875-8889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McLellan JS, et al. Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science. 2013;342:592–598. doi: 10.1126/science.1243283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wrapp D, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turner HL, et al. Disassembly of HIV envelope glycoprotein trimer immunogens is driven by antibodies elicited via immunization. Sci. Adv. 2021;7:eabh2791. doi: 10.1126/sciadv.abh2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mandel B. Neutralization of animal viruses. Adv. Virus Res. 1978;23:205–268. doi: 10.1016/S0065-3527(08)60101-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou QF, et al. Structural basis of Chikungunya virus inhibition by monoclonal antibodies. Proc. Natl Acad. Sci. USA. 2020;117:27637–27645. doi: 10.1073/pnas.2008051117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He RT, et al. Antibodies that block virus attachment to Vero cells are a major component of the human neutralizing antibody response against dengue virus type 2. J. Med. Virol. 1995;45:451–461. doi: 10.1002/jmv.1890450417. [DOI] [PubMed] [Google Scholar]
- 34.Bizebard T, et al. Structural studies on viral escape from antibody neutralization. Curr. Top. Microbiol. Immunol. 2001;260:55–64. doi: 10.1007/978-3-662-05783-4_4. [DOI] [PubMed] [Google Scholar]
- 35.Ren X, Sodroski J, Yang X. An unrelated monoclonal antibody neutralizes human immunodeficiency virus type 1 by binding to an artificial epitope engineered in a functionally neutral region of the viral envelope glycoproteins. J. Virol. 2005;79:5616–5624. doi: 10.1128/JVI.79.9.5616-5624.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yuan M, et al. Structural and functional ramifications of antigenic drift in recent SARS-CoV-2 variants. Science. 2021;373:818–823. doi: 10.1126/science.abh1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abernathy ME, Dam KA, Esswein SR, Jette CA, Bjorkman PJ. How antibodies recognize pathogenic viruses: structural correlates of antibody neutralization of HIV-1, SARS-CoV-2, and Zika. Viruses. 2021;13:2106. doi: 10.3390/v13102106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dingens AS, Arenz D, Weight H, Overbaugh J, Bloom JD. An antigenic atlas of HIV-1 escape from broadly neutralizing antibodies distinguishes functional and structural epitopes. Immunity. 2019;50:520–532.e3. doi: 10.1016/j.immuni.2018.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ugolini S, et al. Inhibition of virus attachment to CD4+ target cells is a major mechanism of T cell line-adapted HIV-1 neutralization. J. Exp. Med. 1997;186:1287–1298. doi: 10.1084/jem.186.8.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wohlbold TJ, et al. Hemagglutinin stalk- and neuraminidase-specific monoclonal antibodies protect against lethal H10N8 influenza virus infection in mice. J. Virol. 2016;90:851–861. doi: 10.1128/JVI.02275-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rizzuto CD, Sodroski JG. Contribution of virion ICAM-1 to human immunodeficiency virus infectivity and sensitivity to neutralization. J. Virol. 1997;71:4847–4851. doi: 10.1128/jvi.71.6.4847-4851.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spear GT, Olinger GG, Saifuddin M, Gebel HM. Human antibodies to major histocompatibility complex alloantigens mediate lysis and neutralization of HIV-1 primary isolate virions in the presence of complement. J. Acquir. Immune Defic. Syndr. 2001;26:103–110. doi: 10.1097/00126334-200102010-00001. [DOI] [PubMed] [Google Scholar]
- 43.Page M, et al. Complement-mediated virus infectivity neutralisation by HLA antibodies is associated with sterilising immunity to SIV challenge in the macaque model for HIV/AIDS. PLoS ONE. 2014;9:e88735. doi: 10.1371/journal.pone.0088735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laird ME, Desrosiers RC. Infectivity and neutralization of simian immunodeficiency virus with FLAG epitope insertion in gp120 variable loops. J. Virol. 2007;81:10838–10848. doi: 10.1128/JVI.00831-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pantophlet R, Wang M, Aguilar-Sino RO, Burton DR. The human immunodeficiency virus type 1 envelope spike of primary viruses can suppress antibody access to variable regions. J. Virol. 2009;83:1649–1659. doi: 10.1128/JVI.02046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brandenberg OF, et al. Predicting HIV-1 transmission and antibody neutralization efficacy in vivo from stoichiometric parameters. PLoS Pathog. 2017;13:e1006313. doi: 10.1371/journal.ppat.1006313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thompson BS, et al. A therapeutic antibody against West Nile virus neutralizes infection by blocking fusion within endosomes. PLoS Pathog. 2009;5:e1000453. doi: 10.1371/journal.ppat.1000453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Long F, et al. Cryo-EM structures elucidate neutralizing mechanisms of anti-Chikungunya human monoclonal antibodies with therapeutic activity. Proc. Natl Acad. Sci. USA. 2015;112:13898–13903. doi: 10.1073/pnas.1515558112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jin J, et al. Neutralizing monoclonal antibodies block Chikungunya virus entry and release by targeting an epitope critical to viral pathogenesis. Cell Rep. 2015;13:2553–2564. doi: 10.1016/j.celrep.2015.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gerhard W. The role of the antibody response in influenza virus infection. Curr. Top. Microbiol. Immunol. 2001;260:171–190. doi: 10.1007/978-3-662-05783-4_9. [DOI] [PubMed] [Google Scholar]
- 51.Dreyfus C, et al. Highly conserved protective epitopes on influenza B viruses. Science. 2012;337:1343–1348. doi: 10.1126/science.1222908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brandenburg B, et al. Mechanisms of hemagglutinin targeted influenza virus neutralization. PLoS ONE. 2013;8:e80034. doi: 10.1371/journal.pone.0080034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bangaru S, et al. A multifunctional human monoclonal neutralizing antibody that targets a unique conserved epitope on influenza HA. Nat. Commun. 2018;9:2669. doi: 10.1038/s41467-018-04704-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gilchuk IM, et al. Influenza H7N9 virus neuraminidase-specific human monoclonal antibodies inhibit viral egress and protect from lethal influenza infection in mice. Cell Host Microbe. 2019;26:715–728.e8. doi: 10.1016/j.chom.2019.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stadlbauer D, et al. Broadly protective human antibodies that target the active site of influenza virus neuraminidase. Science. 2019;366:499–504. doi: 10.1126/science.aay0678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gentles LE, Wan H, Eichelberger MC, Bloom JD. Antibody neutralization of an influenza virus that uses neuraminidase for receptor binding. Viruses. 2020;12:597. doi: 10.3390/v12060597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Williamson LE, et al. Therapeutic alphavirus cross-reactive E1 human antibodies inhibit viral egress. Cell. 2021;184:4430–4446.e22. doi: 10.1016/j.cell.2021.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dufloo J, et al. Broadly neutralizing anti-HIV-1 antibodies tether viral particles at the surface of infected cells. Nat. Commun. 2022;13:630. doi: 10.1038/s41467-022-28307-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li Q, Yafal AG, Lee YM, Hogle J, Chow M. Poliovirus neutralization by antibodies to internal epitopes of VP4 and VP1 results from reversible exposure of these sequences at physiological temperature. J. Virol. 1994;68:3965–3970. doi: 10.1128/jvi.68.6.3965-3970.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dowd KA, Pierson TC. The many faces of a dynamic virion: implications of viral breathing on flavivirus biology and immunogenicity. Annu. Rev. Virol. 2018;5:185–207. doi: 10.1146/annurev-virology-092917-043300. [DOI] [PubMed] [Google Scholar]
- 61.Dowd KA, Jost CA, Durbin AP, Whitehead SS, Pierson TC. A dynamic landscape for antibody binding modulates antibody-mediated neutralization of West Nile virus. PLoS Pathog. 2011;7:e1002111. doi: 10.1371/journal.ppat.1002111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sabo MC, et al. Hepatitis C virus epitope exposure and neutralization by antibodies is affected by time and temperature. Virology. 2012;422:174–184. doi: 10.1016/j.virol.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lok SM, et al. Binding of a neutralizing antibody to dengue virus alters the arrangement of surface glycoproteins. Nat. Struct. Mol. Biol. 2008;15:312–317. doi: 10.1038/nsmb.1382. [DOI] [PubMed] [Google Scholar]
- 64.Lindesmith LC, et al. Particle conformation regulates antibody access to a conserved GII.4 norovirus blockade epitope. J. Virol. 2014;88:8826–8842. doi: 10.1128/JVI.01192-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.VanBlargan LA, Goo L, Pierson TC. Deconstructing the antiviral neutralizing-antibody response: implications for vaccine development and immunity. Microbiol. Mol. Biol. Rev. 2016;80:989–1010. doi: 10.1128/MMBR.00024-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hooks JJ, Burns W, Hayashi K, Geis S, Notkins AL. Viral spread in the presence of neutralizing antibody: mechanisms of persistence in foamy virus infection. Infect. Immun. 1976;14:1172–1178. doi: 10.1128/iai.14.5.1172-1178.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pantaleo G, et al. Effect of anti-V3 antibodies on cell-free and cell-to-cell human immunodeficiency virus transmission. Eur. J. Immunol. 1995;25:226–231. doi: 10.1002/eji.1830250137. [DOI] [PubMed] [Google Scholar]
- 68.Schiffner T, Sattentau QJ, Duncan CJ. Cell-to-cell spread of HIV-1 and evasion of neutralizing antibodies. Vaccine. 2013;31:5789–5797. doi: 10.1016/j.vaccine.2013.10.020. [DOI] [PubMed] [Google Scholar]
- 69.Abela IA, et al. Cell-cell transmission enables HIV-1 to evade inhibition by potent CD4bs directed antibodies. PLoS Pathog. 2012;8:e1002634. doi: 10.1371/journal.ppat.1002634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Reh L, et al. Capacity of broadly neutralizing antibodies to inhibit HIV-1 cell-cell transmission is strain- and epitope-dependent. PLoS Pathog. 2015;11:e1004966. doi: 10.1371/journal.ppat.1004966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li H, Zony C, Chen P, Chen BK. Reduced potency and incomplete neutralization of broadly neutralizing antibodies against cell-to-cell transmission of HIV-1 with transmitted founder Envs. J. Virol. 2017;91:e02425-16. doi: 10.1128/JVI.02425-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Suphaphiphat K, et al. Broadly neutralizing antibodies potently inhibit cell-to-cell transmission of semen leukocyte-derived SHIV162P3. EBioMedicine. 2020;57:102842. doi: 10.1016/j.ebiom.2020.102842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dufloo J, Bruel T, Schwartz O. HIV-1 cell-to-cell transmission and broadly neutralizing antibodies. Retrovirology. 2018;15:51. doi: 10.1186/s12977-018-0434-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Duncan CJ, et al. High-multiplicity HIV-1 infection and neutralizing antibody evasion mediated by the macrophage-T cell virological synapse. J. Virol. 2014;88:2025–2034. doi: 10.1128/JVI.03245-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Igakura T, et al. Spread of HTLV-I between lymphocytes by virus-induced polarization of the cytoskeleton. Science. 2003;299:1713–1716. doi: 10.1126/science.1080115. [DOI] [PubMed] [Google Scholar]
- 76.Watkinson RE, McEwan WA, Tam JC, Vaysburd M, James LC. TRIM21 promotes cGAS and RIG-I sensing of viral genomes during infection by antibody-opsonized virus. PLoS Pathog. 2015;11:e1005253. doi: 10.1371/journal.ppat.1005253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bottermann M, Caddy SL. Virus neutralisation by intracellular antibodies. Semin. Cell Dev. Biol. 2022;126:108–116. doi: 10.1016/j.semcdb.2021.10.010. [DOI] [PubMed] [Google Scholar]
- 78.Fan W, et al. Swine TRIM21 restricts FMDV infection via an intracellular neutralization mechanism. Antivir. Res. 2016;127:32–40. doi: 10.1016/j.antiviral.2016.01.004. [DOI] [PubMed] [Google Scholar]
- 79.Bomsel M, et al. Intracellular neutralization of HIV transcytosis across tight epithelial barriers by anti-HIV envelope protein dIgA or IgM. Immunity. 1998;9:277–287. doi: 10.1016/S1074-7613(00)80610-X. [DOI] [PubMed] [Google Scholar]
- 80.Fujioka H, et al. Immunocytochemical colocalization of specific immunoglobulin A with Sendai virus protein in infected polarized epithelium. J. Exp. Med. 1998;188:1223–1229. doi: 10.1084/jem.188.7.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kato H, Kato R, Fujihashi K, McGhee JR. Role of mucosal antibodies in viral infections. Curr. Top. Microbiol. Immunol. 2001;260:201–228. doi: 10.1007/978-3-662-05783-4_11. [DOI] [PubMed] [Google Scholar]
- 82.Mazanec MB, Coudret CL, Fletcher DR. Intracellular neutralization of influenza virus by immunoglobulin A anti-hemagglutinin monoclonal antibodies. J. Virol. 1995;69:1339–1343. doi: 10.1128/jvi.69.2.1339-1343.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mazanec MB, Lamm ME, Lyn D, Portner A, Nedrud JG. Comparison of IgA versus IgG monoclonal antibodies for passive immunization of the murine respiratory tract. Virus Res. 1992;23:1–12. doi: 10.1016/0168-1702(92)90063-F. [DOI] [PubMed] [Google Scholar]
- 84.Caddy SL, et al. Intracellular neutralisation of rotavirus by VP6-specific IgG. PLoS Pathog. 2020;16:e1008732. doi: 10.1371/journal.ppat.1008732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gunaydin G, Yu S, Graslund T, Hammarstrom L, Marcotte H. Fusion of the mouse IgG1 Fc domain to the VHH fragment (ARP1) enhances protection in a mouse model of rotavirus. Sci. Rep. 2016;6:30171. doi: 10.1038/srep30171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Della-Porta AJ, Westaway EG. A multi-hit model for the neutralization of animal viruses. J. Gen. Virol. 1978;38:1–19. doi: 10.1099/0022-1317-38-1-1. [DOI] [PubMed] [Google Scholar]
- 87.Parren PW, Burton DR. The antiviral activity of antibodies in vitro and in vivo. Adv. Immunol. 2001;77:195–262. doi: 10.1016/S0065-2776(01)77018-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Klasse PJ, Burton DR. Antibodies to West Nile virus: a double-edged sword. Cell Host Microbe. 2007;1:87–89. doi: 10.1016/j.chom.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 89.Flamand A, Raux H, Gaudin Y, Ruigrok RW. Mechanisms of rabies virus neutralization. Virology. 1993;194:302–313. doi: 10.1006/viro.1993.1261. [DOI] [PubMed] [Google Scholar]
- 90.Raux H, Coulon P, Lafay F, Flamand A. Monoclonal antibodies which recognize the acidic configuration of the rabies glycoprotein at the surface of the virion can be neutralizing. Virology. 1995;210:400–408. doi: 10.1006/viro.1995.1356. [DOI] [PubMed] [Google Scholar]
- 91.Pierson TC, Diamond MS. A game of numbers: the stoichiometry of antibody-mediated neutralization of flavivirus infection. Prog. Mol. Biol. Transl. Sci. 2015;129:141–166. doi: 10.1016/bs.pmbts.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Parren PW, Burton DR. Antibodies against HIV-1 from phage display libraries: mapping of an immune response and progress towards antiviral immunotherapy. Chem. Immunol. 1997;65:18–56. doi: 10.1159/000319346. [DOI] [PubMed] [Google Scholar]
- 93.Klein JS, et al. Examination of the contributions of size and avidity to the neutralization mechanisms of the anti-HIV antibodies b12 and 4E10. Proc. Natl Acad. Sci. USA. 2009;106:7385–7390. doi: 10.1073/pnas.0811427106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Klein JS, Bjorkman PJ. Few and far between: how HIV may be evading antibody avidity. PLoS Pathog. 2010;6:e1000908. doi: 10.1371/journal.ppat.1000908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ekiert DC, et al. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science. 2011;333:843–850. doi: 10.1126/science.1204839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sharma A, et al. The epitope arrangement on flavivirus particles contributes to Mab C10’s extraordinary neutralization breadth across Zika and dengue viruses. Cell. 2021;184:6052–6066.e18. doi: 10.1016/j.cell.2021.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Richardson SI, et al. IgG3 enhances neutralization potency and Fc effector function of an HIV V2-specific broadly neutralizing antibody. PLoS Pathog. 2019;15:e1008064. doi: 10.1371/journal.ppat.1008064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Scheepers C, et al. Antibody isotype switching as a mechanism to counter HIV neutralization escape. Cell Rep. 2020;33:108430. doi: 10.1016/j.celrep.2020.108430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Richardson SI, et al. HIV broadly neutralizing antibodies expressed as IgG3 preserve neutralization potency and show improved Fc effector function. Front. Immunol. 2021;12:733958. doi: 10.3389/fimmu.2021.733958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Moyo-Gwete T, et al. Enhanced neutralization potency of an identical HIV neutralizing antibody expressed as different isotypes is achieved through genetically distinct mechanisms. Sci. Rep. 2022;12:16473. doi: 10.1038/s41598-022-20141-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tudor D, et al. Isotype modulates epitope specificity, affinity, and antiviral activities of anti-HIV-1 human broadly neutralizing 2F5 antibody. Proc. Natl Acad. Sci. USA. 2012;109:12680–12685. doi: 10.1073/pnas.1200024109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ku Z, et al. Nasal delivery of an IgM offers broad protection from SARS-CoV-2 variants. Nature. 2021;595:718–723. doi: 10.1038/s41586-021-03673-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.McCoy LE, et al. Incomplete neutralization and deviation from sigmoidal neutralization curves for HIV broadly neutralizing monoclonal antibodies. PLoS Pathog. 2015;11:e1005110. doi: 10.1371/journal.ppat.1005110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rogers TF, et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kim AS, Leaman DP, Zwick MB. Antibody to gp41 MPER alters functional properties of HIV-1 Env without complete neutralization. PLoS Pathog. 2014;10:e1004271. doi: 10.1371/journal.ppat.1004271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zost SJ, et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 2020;584:443–449. doi: 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mehlhop E, et al. Complement protein C1q reduces the stoichiometric threshold for antibody-mediated neutralization of West Nile virus. Cell Host Microbe. 2009;6:381–391. doi: 10.1016/j.chom.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Byrne AB, Talarico LB. Role of the complement system in antibody-dependent enhancement of flavivirus infections. Int. J. Infect. Dis. 2021;103:404–411. doi: 10.1016/j.ijid.2020.12.039. [DOI] [PubMed] [Google Scholar]
- 109.Zinkernagel RM, et al. Neutralizing antiviral antibody responses. Adv. Immunol. 2001;79:1–53. doi: 10.1016/S0065-2776(01)79001-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Carroll MC. The complement system in regulation of adaptive immunity. Nat. Immunol. 2004;5:981–986. doi: 10.1038/ni1113. [DOI] [PubMed] [Google Scholar]
- 111.Robinson WE. Mechanism for complement-mediated, antibody-dependent enhancement of human immunodeficiency virus type 1 infection in MT2 cells is enhanced entry through CD4, CD21, and CXCR4 chemokine receptors. Viral Immunol. 2006;19:434–447. doi: 10.1089/vim.2006.19.434. [DOI] [PubMed] [Google Scholar]
- 112.Marasini B, et al. Mucosal AIDS virus transmission is enhanced by antiviral IgG isolated early in infection. AIDS. 2021;35:2423–2432. doi: 10.1097/QAD.0000000000003050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Oroszlan S, Nowinski RC. Lysis of retroviruses with monoclonal antibodies against viral envelope proteins. Virology. 1980;101:296–299. doi: 10.1016/0042-6822(80)90507-3. [DOI] [PubMed] [Google Scholar]
- 114.Hessell AJ, et al. Fc receptor but not complement binding is important in antibody protection against HIV. Nature. 2007;449:101–104. doi: 10.1038/nature06106. [DOI] [PubMed] [Google Scholar]
- 115.Goldberg BS, et al. Revisiting an IgG Fc loss-of-function experiment: the role of complement in HIV broadly neutralizing antibody b12 activity. mBio. 2021;12:e0174321. doi: 10.1128/mBio.01743-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Keeler SP, Fox JM. Requirement of Fc-Fc gamma receptor interaction for antibody-based protection against emerging virus infections. Viruses. 2021;13:1037. doi: 10.3390/v13061037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bournazos S, DiLillo DJ, Ravetch JV. The role of Fc-FcγR interactions in IgG-mediated microbial neutralization. J. Exp. Med. 2015;212:1361–1369. doi: 10.1084/jem.20151267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lu LL, Suscovich TJ, Fortune SM, Alter G. Beyond binding: antibody effector functions in infectious diseases. Nat. Rev. Immunol. 2018;18:46–61. doi: 10.1038/nri.2017.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tay MZ, Wiehe K, Pollara J. Antibody-dependent cellular phagocytosis in antiviral immune responses. Front. Immunol. 2019;10:332. doi: 10.3389/fimmu.2019.00332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ackerman ME, et al. A robust, high-throughput assay to determine the phagocytic activity of clinical antibody samples. J. Immunol. Methods. 2011;366:8–19. doi: 10.1016/j.jim.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tay MZ, et al. Antibody-mediated internalization of infectious HIV-1 virions differs among antibody isotypes and subclasses. PLoS Pathog. 2016;12:e1005817. doi: 10.1371/journal.ppat.1005817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gach JS, et al. Human immunodeficiency virus type-1 (HIV-1) evades antibody-dependent phagocytosis. PLoS Pathog. 2017;13:e1006793. doi: 10.1371/journal.ppat.1006793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Biburger M, Lux A, Nimmerjahn F. How immunoglobulin G antibodies kill target cells: revisiting an old paradigm. Adv. Immunol. 2014;124:67–94. doi: 10.1016/B978-0-12-800147-9.00003-0. [DOI] [PubMed] [Google Scholar]
- 124.Forthal DN, Finzi A. Antibody-dependent cellular cytotoxicity in HIV infection. AIDS. 2018;32:2439–2451. doi: 10.1097/QAD.0000000000002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Anand SP, Finzi A. Understudied factors influencing Fc-mediated immune responses against viral infections. Vaccines. 2019;7:103. doi: 10.3390/vaccines7030103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Forthal DN, Landucci G, Daar ES. Antibody from patients with acute human immunodeficiency virus (HIV) infection inhibits primary strains of HIV type 1 in the presence of natural-killer effector cells. J. Virol. 2001;75:6953–6961. doi: 10.1128/JVI.75.15.6953-6961.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kramski M, Parsons MS, Stratov I, Kent SJ. HIV-specific antibody immunity mediated through NK cells and monocytes. Curr. Hiv. Res. 2013;11:388–406. doi: 10.2174/1570162X113116660061. [DOI] [PubMed] [Google Scholar]
- 128.Pollara J, et al. Application of area scaling analysis to identify natural killer cell and monocyte involvement in the GranToxiLux antibody dependent cell-mediated cytotoxicity assay. Cytom. A. 2018;93:436–447. doi: 10.1002/cyto.a.23348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Poignard P, et al. Heterogeneity of envelope molecules expressed on primary human immunodeficiency virus type 1 particles as probed by the binding of neutralizing and nonneutralizing antibodies. J. Virol. 2003;77:353–365. doi: 10.1128/JVI.77.1.353-365.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cantin R, Martin G, Tremblay MJ. A novel virus capture assay reveals a differential acquisition of host HLA-DR by clinical isolates of human immunodeficiency virus type 1 expanded in primary human cells depending on the nature of producing cells and the donor source. J. Gen. Virol. 2001;82:2979–2987. doi: 10.1099/0022-1317-82-12-2979. [DOI] [PubMed] [Google Scholar]
- 131.Moore PL, et al. Nature of nonfunctional envelope proteins on the surface of human immunodeficiency virus type 1. J. Virol. 2006;80:2515–2528. doi: 10.1128/JVI.80.5.2515-2528.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Burton DR, et al. Limited or no protection by weakly or nonneutralizing antibodies against vaginal SHIV challenge of macaques compared with a strongly neutralizing antibody. Proc. Natl Acad. Sci. USA. 2011;108:11181–11186. doi: 10.1073/pnas.1103012108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Dugast AS, et al. Lack of protection following passive transfer of polyclonal highly functional low-dose non-neutralizing antibodies. PLoS ONE. 2014;9:e97229. doi: 10.1371/journal.pone.0097229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Burton DR. Amping up HIV antibodies. Science. 2021;372:1397–1398. doi: 10.1126/science.abf5376. [DOI] [Google Scholar]
- 135.Johnson S, et al. Development of a humanized monoclonal antibody (MEDI-493) with potent in vitro and in vivo activity against respiratory syncytial virus. J. Infect. Dis. 1997;176:1215–1224. doi: 10.1086/514115. [DOI] [PubMed] [Google Scholar]
- 136.The IMpact-RSV Study Group. Palivizumab, a humanized respiratory syncytial virus monoclonal antibody, reduces hospitalization from respiratory syncytial virus infection in high-risk infants. Pediatrics. 1998;102:531–537. doi: 10.1542/peds.102.3.531. [DOI] [PubMed] [Google Scholar]
- 137.Hu J, Robinson JL. Treatment of respiratory syncytial virus with palivizumab: a systematic review. World J. Pediatr. 2010;6:296–300. doi: 10.1007/s12519-010-0230-z. [DOI] [PubMed] [Google Scholar]
- 138.Zhu Q, et al. A highly potent extended half-life antibody as a potential RSV vaccine surrogate for all infants. Sci. Transl. Med. 2017;9:eaaj1928. doi: 10.1126/scitranslmed.aaj1928. [DOI] [PubMed] [Google Scholar]
- 139.Cohen MS, et al. Effect of bamlanivimab vs placebo on incidence of COVID-19 among residents and staff of skilled nursing and assisted living facilities: a randomized clinical trial. JAMA. 2021;326:46–55. doi: 10.1001/jama.2021.8828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.McKinley SA, et al. Modeling neutralization kinetics of HIV by broadly neutralizing monoclonal antibodies in genital secretions coating the cervicovaginal mucosa. PLoS ONE. 2014;9:e100598. doi: 10.1371/journal.pone.0100598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wright KE, Buchmeier MJ. Antiviral antibodies attenuate T-cell-mediated immunopathology following acute lymphocytic choriomeningitis virus infection. J. Virol. 1991;65:3001–3006. doi: 10.1128/jvi.65.6.3001-3006.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Parren PW, Geisbert TW, Maruyama T, Jahrling PB, Burton DR. Pre- and postexposure prophylaxis of Ebola virus infection in an animal model by passive transfer of a neutralizing human antibody. J. Virol. 2002;76:6408–6412. doi: 10.1128/JVI.76.12.6408-6412.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.O’Brien MP, et al. Subcutaneous REGEN-COV antibody combination to prevent Covid-19. N. Engl. J. Med. 2021;385:1184–1195. doi: 10.1056/NEJMoa2109682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.O’Brien MP, et al. Effect of subcutaneous casirivimab and imdevimab antibody combination vs placebo on development of symptomatic COVID-19 in early asymptomatic SARS-CoV-2 infection: a randomized clinical trial. JAMA. 2022;327:432–441. doi: 10.1001/jama.2021.24939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hammitt LL, et al. Nirsevimab for prevention of RSV in healthy late-preterm and term infants. N. Engl. J. Med. 2022;386:837–846. doi: 10.1056/NEJMoa2110275. [DOI] [PubMed] [Google Scholar]
- 146.Ramilo O, et al. Motavizumab treatment of infants hospitalized with respiratory syncytial virus infection does not decrease viral load or severity of illness. Pediatr. Infect. Dis. J. 2014;33:703–709. doi: 10.1097/INF.0000000000000240. [DOI] [PubMed] [Google Scholar]
- 147.RECOVERY Collaborative Group. Casirivimab and imdevimab in patients admitted to hospital with COVID-19 (RECOVERY): a randomised, controlled, open-label, platform trial. Lancet. 2022;399:665–676. doi: 10.1016/S0140-6736(22)00163-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Razonable RR, et al. Casirivimab-imdevimab treatment is associated with reduced rates of hospitalization among high-risk patients with mild to moderate coronavirus disease-19. eClinicalMedicine. 2021;40:101102. doi: 10.1016/j.eclinm.2021.101102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Gupta A, et al. Effect of sotrovimab on hospitalization or death among high-risk patients with mild to moderate COVID-19: a randomized clinical trial. JAMA. 2022;327:1236–1246. doi: 10.1001/jama.2022.2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.ACTIV-3/Therapeutics for Inpatients with COVID-19 (TICO) Study Group. Efficacy and safety of two neutralising monoclonal antibody therapies, sotrovimab and BRII-196 plus BRII-198, for adults hospitalised with COVID-19 (TICO): a randomised controlled trial. Lancet Infect. Dis. 2022;22:622–635. doi: 10.1016/S1473-3099(21)00751-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Weinreich DM, et al. REGN-COV2, a neutralizing antibody cocktail, in outpatients with Covid-19. N. Engl. J. Med. 2021;384:238–251. doi: 10.1056/NEJMoa2035002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Gupta A, et al. Early treatment for Covid-19 with SARS-CoV-2 neutralizing antibody sotrovimab. N. Engl. J. Med. 2021;385:1941–1950. doi: 10.1056/NEJMoa2107934. [DOI] [PubMed] [Google Scholar]
- 153.ACTIV-3/TICO LY-CoV555 Study Group. et al. A neutralizing monoclonal antibody for hospitalized patients with Covid-19. N. Engl. J. Med. 2021;384:905–914. doi: 10.1056/NEJMoa2033130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chen P, et al. SARS-CoV-2 neutralizing antibody LY-CoV555 in outpatients with Covid-19. N. Engl. J. Med. 2021;384:229–237. doi: 10.1056/NEJMoa2029849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Dougan M, et al. Bamlanivimab plus etesevimab in mild or moderate Covid-19. N. Engl. J. Med. 2021;385:1382–1392. doi: 10.1056/NEJMoa2102685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Gottlieb RL, et al. Effect of bamlanivimab as monotherapy or in combination with etesevimab on viral load in patients with mild to moderate COVID-19: a randomized clinical trial. JAMA. 2021;325:632–644. doi: 10.1001/jama.2021.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Corti D, et al. Protective monotherapy against lethal Ebola virus infection by a potently neutralizing antibody. Science. 2016;351:1339–1342. doi: 10.1126/science.aad5224. [DOI] [PubMed] [Google Scholar]
- 158.Pascal KE, et al. Development of clinical-stage human monoclonal antibodies that treat advanced Ebola virus disease in nonhuman primates. J. Infect. Dis. 2018;218:S612–S626. doi: 10.1093/infdis/jiy285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Iversen PL, et al. Recent successes in therapeutics for Ebola virus disease: no time for complacency. Lancet Infect. Dis. 2020;20:e231–e237. doi: 10.1016/S1473-3099(20)30282-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Barouch DH, et al. Therapeutic efficacy of potent neutralizing HIV-1-specific monoclonal antibodies in SHIV-infected rhesus monkeys. Nature. 2013;503:224–228. doi: 10.1038/nature12744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Shingai M, et al. Antibody-mediated immunotherapy of macaques chronically infected with SHIV suppresses viraemia. Nature. 2013;503:277–280. doi: 10.1038/nature12746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Nishimura Y, Martin MA. Of mice, macaques, and men: broadly neutralizing antibody immunotherapy for HIV-1. Cell Host Microbe. 2017;22:207–216. doi: 10.1016/j.chom.2017.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Caskey M, Klein F, Nussenzweig MC. Broadly neutralizing anti-HIV-1 monoclonal antibodies in the clinic. Nat. Med. 2019;25:547–553. doi: 10.1038/s41591-019-0412-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Mahomed S, et al. Safety and pharmacokinetics of monoclonal antibodies VRC07-523LS and PGT121 administered subcutaneously for HIV prevention. J. Infect. Dis. 2022;226:510–520. doi: 10.1093/infdis/jiac041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Walker LM, Burton DR. Passive immunotherapy of viral infections: ‘super-antibodies’ enter the fray. Nat. Rev. Immunol. 2018;18:297–308. doi: 10.1038/nri.2017.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Schlesinger JJ, Chapman S. Neutralizing F(ab′)2 fragments of protective monoclonal antibodies to yellow fever virus (YF) envelope protein fail to protect mice against lethal YF encephalitis. J. Gen. Virol. 1995;76:217–220. doi: 10.1099/0022-1317-76-1-217. [DOI] [PubMed] [Google Scholar]
- 167.Hessell AJ, et al. Effective, low-titer antibody protection against low-dose repeated mucosal SHIV challenge in macaques. Nat. Med. 2009;15:951–954. doi: 10.1038/nm.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Hangartner L, et al. Effector function does not contribute to protection from virus challenge by a highly potent HIV broadly neutralizing antibody in nonhuman primates. Sci. Transl. Med. 2021;13:eabe3349. doi: 10.1126/scitranslmed.abe3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Parsons MS, et al. Fc-dependent functions are redundant to efficacy of anti-HIV antibody PGT121 in macaques. J. Clin. Invest. 2019;129:182–191. doi: 10.1172/JCI122466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Asokan M, et al. Fc-mediated effector function contributes to the in vivo antiviral effect of an HIV neutralizing antibody. Proc. Natl Acad. Sci. USA. 2020;117:18754–18763. doi: 10.1073/pnas.2008236117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Wang P, et al. Quantifying the contribution of Fc-mediated effector functions to the antiviral activity of anti-HIV-1 IgG1 antibodies in vivo. Proc. Natl Acad. Sci. USA. 2020;117:18002–18009. doi: 10.1073/pnas.2008190117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Bournazos S, et al. Broadly neutralizing anti-HIV-1 antibodies require Fc effector functions for in vivo activity. Cell. 2014;158:1243–1253. doi: 10.1016/j.cell.2014.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.DiLillo DJ, Tan GS, Palese P, Ravetch JV. Broadly neutralizing hemagglutinin stalk-specific antibodies require FcγR interactions for protection against influenza virus in vivo. Nat. Med. 2014;20:143–151. doi: 10.1038/nm.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Impagliazzo A, et al. A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science. 2015;349:1301–1306. doi: 10.1126/science.aac7263. [DOI] [PubMed] [Google Scholar]
- 175.Bournazos S, DiLillo DJ, Goff AJ, Glass PJ, Ravetch JV. Differential requirements for FcγR engagement by protective antibodies against Ebola virus. Proc. Natl Acad. Sci. USA. 2019;116:20054–20062. doi: 10.1073/pnas.1911842116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Winkler ES, et al. Human neutralizing antibodies against SARS-CoV-2 require intact Fc effector functions for optimal therapeutic protection. Cell. 2021;184:1804–1820.e16. doi: 10.1016/j.cell.2021.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Cleary KLS, Chan HTC, James S, Glennie MJ, Cragg MS. Antibody distance from the cell membrane regulates antibody effector mechanisms. J. Immunol. 2017;198:3999–4011. doi: 10.4049/jimmunol.1601473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Day PM, et al. In vivo mechanisms of vaccine-induced protection against HPV infection. Cell Host Microbe. 2010;8:260–270. doi: 10.1016/j.chom.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Buck CB, Pastrana DV, Lowy DR, Schiller JT. Generation of HPV pseudovirions using transfection and their use in neutralization assays. Methods Mol. Med. 2005;119:445–462. doi: 10.1385/1-59259-982-6:445. [DOI] [PubMed] [Google Scholar]
- 180.Hilchey SP, et al. Rituximab immunotherapy results in the induction of a lymphoma idiotype-specific T-cell response in patients with follicular lymphoma: support for a “vaccinal effect” of rituximab. Blood. 2009;113:3809–3812. doi: 10.1182/blood-2008-10-185280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.DiLillo DJ, Ravetch JV. Differential Fc-receptor engagement drives an anti-tumor vaccinal effect. Cell. 2015;161:1035–1045. doi: 10.1016/j.cell.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Bournazos S, Ravetch JV. Fcγ receptor function and the design of vaccination strategies. Immunity. 2017;47:224–233. doi: 10.1016/j.immuni.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Bournazos S, Corti D, Virgin HW, Ravetch JV. Fc-optimized antibodies elicit CD8 immunity to viral respiratory infection. Nature. 2020;588:485–490. doi: 10.1038/s41586-020-2838-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Junker F, Gordon J, Qureshi O. Fc gamma receptors and their role in antigen uptake, presentation, and T cell activation. Front. Immunol. 2020;11:1393. doi: 10.3389/fimmu.2020.01393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Cathcart AL, et al. The dual function monoclonal antibodies VIR-7831 and VIR-7832 demonstrate potent in vitro and in vivo activity against SARS-CoV-2. bioRxiv. 2021 doi: 10.1101/2021.03.09.434607. [DOI] [Google Scholar]
- 186.Herrera C, et al. Nonneutralizing antibodies to the CD4-binding site on the gp120 subunit of human immunodeficiency virus type 1 do not interfere with the activity of a neutralizing antibody against the same site. J. Virol. 2003;77:1084–1091. doi: 10.1128/JVI.77.2.1084-1091.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Pantophlet R, et al. Fine mapping of the interaction of neutralizing and nonneutralizing monoclonal antibodies with the CD4 binding site of human immunodeficiency virus type 1 gp120. J. Virol. 2003;77:642–658. doi: 10.1128/JVI.77.1.642-658.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Mackin SR, et al. Fcγ receptor-dependent antibody effector functions are required for vaccine protection against infection by antigenic variants of SARS-CoV-2. bioRxiv. 2022 doi: 10.1101/2022.11.27.518117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Wherry, E. J., Vardhana, S., Morice, W. & Baldo, L. Letter to FDA — T-cell assessment in vaccine. Twitterhttps://twitter.com/ejohnwherry/status/1517322366124187650?lang=en (2022).
- 190.Arvin AM, et al. A perspective on potential antibody-dependent enhancement of SARS-CoV-2. Nature. 2020;584:353–363. doi: 10.1038/s41586-020-2538-8. [DOI] [PubMed] [Google Scholar]
- 191.Bournazos S, Gupta A, Ravetch JV. The role of IgG Fc receptors in antibody-dependent enhancement. Nat. Rev. Immunol. 2020;20:633–643. doi: 10.1038/s41577-020-00410-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Halstead SB. Immune enhancement of viral infection. Prog. Allergy. 1982;31:301–364. [PubMed] [Google Scholar]
- 193.Sullivan NJ. Antibody-mediated enhancement of viral disease. Curr. Top. Microbiol. Immunol. 2001;260:145–169. doi: 10.1007/978-3-662-05783-4_8. [DOI] [PubMed] [Google Scholar]
- 194.Sullivan N, Sun Y, Li J, Hofmann W, Sodroski J. Replicative function and neutralization sensitivity of envelope glycoproteins from primary and T-cell line-passaged human immunodeficiency virus type 1 isolates. J. Virol. 1995;69:4413–4422. doi: 10.1128/jvi.69.7.4413-4422.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Mehlhop E, et al. Complement protein C1q inhibits antibody-dependent enhancement of flavivirus infection in an IgG subclass-specific manner. Cell Host Microbe. 2007;2:417–426. doi: 10.1016/j.chom.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Takada A, Kawaoka Y. Antibody-dependent enhancement of viral infection: molecular mechanisms and in vivo implications. Rev. Med. Virol. 2003;13:387–398. doi: 10.1002/rmv.405. [DOI] [PubMed] [Google Scholar]
- 197.Tirado SM, Yoon KJ. Antibody-dependent enhancement of virus infection and disease. Viral Immunol. 2003;16:69–86. doi: 10.1089/088282403763635465. [DOI] [PubMed] [Google Scholar]
- 198.Wan Y, et al. Molecular mechanism for antibody-dependent enhancement of coronavirus entry. J. Virol. 2020;94:e02015–e02019. doi: 10.1128/JVI.02015-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Yip MS, et al. Antibody-dependent infection of human macrophages by severe acute respiratory syndrome coronavirus. Virol. J. 2014;11:82. doi: 10.1186/1743-422X-11-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Maxmen A. How blood from coronavirus survivors might save lives. Nature. 2020;580:16–17. doi: 10.1038/d41586-020-00895-8. [DOI] [PubMed] [Google Scholar]
- 201.Yip MS, et al. Antibody-dependent enhancement of SARS coronavirus infection and its role in the pathogenesis of SARS. Hong. Kong Med. J. 2016;22:25–31. [PubMed] [Google Scholar]
- 202.Jaume M, et al. Anti-severe acute respiratory syndrome coronavirus spike antibodies trigger infection of human immune cells via a pH- and cysteine protease-independent FcγR pathway. J. Virol. 2011;85:10582–10597. doi: 10.1128/JVI.00671-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Kam YW, et al. Antibodies against trimeric S glycoprotein protect hamsters against SARS-CoV challenge despite their capacity to mediate FcγRII-dependent entry into B cells in vitro. Vaccine. 2007;25:729–740. doi: 10.1016/j.vaccine.2006.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Li D, et al. In vitro and in vivo functions of SARS-CoV-2 infection-enhancing and neutralizing antibodies. Cell. 2021;184:4203–4219.e32. doi: 10.1016/j.cell.2021.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Junqueira C, et al. FcγR-mediated SARS-CoV-2 infection of monocytes activates inflammation. Nature. 2022;606:576–584. doi: 10.1038/s41586-022-04702-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Zhou Y, et al. Enhancement versus neutralization by SARS-CoV-2 antibodies from a convalescent donor associates with distinct epitopes on the RBD. Cell Rep. 2021;34:108699. doi: 10.1016/j.celrep.2021.108699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Sefik E, et al. Inflammasome activation in infected macrophages drives COVID-19 pathology. Nature. 2022;606:585–593. doi: 10.1038/s41586-022-04802-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Bournazos S, et al. Antibody fucosylation predicts disease severity in secondary dengue infection. Science. 2021;372:1102–1105. doi: 10.1126/science.abc7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Graham BS. Rapid COVID-19 vaccine development. Science. 2020;368:945–946. doi: 10.1126/science.abb8923. [DOI] [PubMed] [Google Scholar]
- 210.Burton DR, Poignard P, Stanfield RL, Wilson IA. Broadly neutralizing antibodies present new prospects to counter highly antigenically diverse viruses. Science. 2012;337:183–186. doi: 10.1126/science.1225416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Corti D, Lanzavecchia A. Broadly neutralizing antiviral antibodies. Annu. Rev. Immunol. 2013;31:705–742. doi: 10.1146/annurev-immunol-032712-095916. [DOI] [PubMed] [Google Scholar]
- 212.Burton DR. Antibodies, viruses and vaccines. Nat. Rev. Immunol. 2002;2:706–713. doi: 10.1038/nri891. [DOI] [PubMed] [Google Scholar]
- 213.Burton DR. What are the most powerful immunogen design vaccine strategies? Reverse vaccinology 2.0 shows great promise. Cold Spring Harb. Perspect. Biol. 2017;9:a030262. doi: 10.1101/cshperspect.a030262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Kwong PD, Mascola JR. HIV-1 Vaccines based on antibody identification, B cell ontogeny, and epitope structure. Immunity. 2018;48:855–871. doi: 10.1016/j.immuni.2018.04.029. [DOI] [PubMed] [Google Scholar]
- 215.Haynes BF, Mascola JR. The quest for an antibody-based HIV vaccine. Immunol. Rev. 2017;275:5–10. doi: 10.1111/imr.12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Bjorkman PJ. Can we use structural knowledge to design a protective vaccine against HIV-1? HLA. 2020;95:95–103. doi: 10.1111/tan.13759. [DOI] [PubMed] [Google Scholar]
- 217.Burton DR. A vaccine for HIV type 1: the antibody perspective. Proc. Natl Acad. Sci. USA. 1997;94:10018–10023. doi: 10.1073/pnas.94.19.10018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Nachbagauer R, Palese P. Is a universal influenza virus vaccine possible? Annu. Rev. Med. 2020;71:315–327. doi: 10.1146/annurev-med-120617-041310. [DOI] [PubMed] [Google Scholar]
- 219.Nachbagauer R, et al. A chimeric hemagglutinin-based universal influenza virus vaccine approach induces broad and long-lasting immunity in a randomized, placebo-controlled phase I trial. Nat. Med. 2021;27:106–114. doi: 10.1038/s41591-020-1118-7. [DOI] [PubMed] [Google Scholar]
- 220.Burton DR, Topol EJ. Variant-proof vaccines — invest now for the next pandemic. Nature. 2021;590:386–388. doi: 10.1038/d41586-021-00340-4. [DOI] [PubMed] [Google Scholar]
- 221.Burton DR, Walker LM. Rational vaccine design in the time of COVID-19. Cell Host Microbe. 2020;27:695–698. doi: 10.1016/j.chom.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Dolgin E. Pan-coronavirus vaccine pipeline takes form. Nat. Rev. Drug Discov. 2022;21:324–326. doi: 10.1038/d41573-022-00074-6. [DOI] [PubMed] [Google Scholar]
- 223.Chen Y, et al. Broadly neutralizing antibodies to SARS-CoV-2 and other human coronaviruses. Nat. Rev. Immunol. 2022 doi: 10.1038/s41577-022-00784-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Zhou P, et al. Broadly neutralizing anti-S2 antibodies protect against all three human betacoronaviruses that cause deadly disease. Immunity. 2023;56:669–683. doi: 10.1016/j.immuni.2023.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]