Abstract
Biosynthesis of many important polysaccharides (including peptidoglycan, lipopolysaccharide, and N-linked glycans) necessitates transport of lipid-linked oligosaccharides (LLO) across membranes from their cytosolic site of synthesis to their sites of utilization. Much of our current understanding of LLO transport comes from genetic, biochemical, and structural studies of the multidrug/oligosaccharidyl-lipid/polysaccharide (MOP) superfamily protein MurJ, which flips the peptidoglycan precursor lipid II. MurJ plays a pivotal role in bacterial cell wall synthesis and is an emerging antibiotic target. Here we review the mechanism of LLO flipping by MurJ including the structural basis for lipid II flipping and ion coupling. We then discuss inhibition of MurJ by antibacterials including Humimycins and the Phage M lysis protein, as well as how studies on MurJ could provide insight to other flippases both within and beyond the MOP superfamily.
Keywords: peptidoglycan, cell wall, lipid, transporter, antibiotic
INTRODUCTION
One of the hallmarks of life is compartmentalization of cellular contents by lipid membranes, allowing for segregation of metabolic pathways, protection from extracellular stressors, as well as generation of membrane potential and chemical gradients to power cellular processes. At the same time, evolution of the biological membrane also necessitated adaptations for transmembrane movement of otherwise impermeable molecules, often in the form of membrane-embedded transport proteins such as transporters or ion channels. Besides selective transport of ions and water-soluble small molecules across membranes, cells also require transmembrane transport of lipids—i.e., “flipping” from one side of the membrane to another. Lipid transport is a critical step in many biosynthetic processes including N-linked glycosylation in humans (1) and cell wall synthesis in bacteria (2). Lipid transporters also regulate membrane asymmetry, i.e., different composition of lipids on each side of the membrane, which is critical to processes ranging from apoptosis in humans (3) and antibiotic resistance conferred by the outer membrane of Gram-negative bacteria (4).
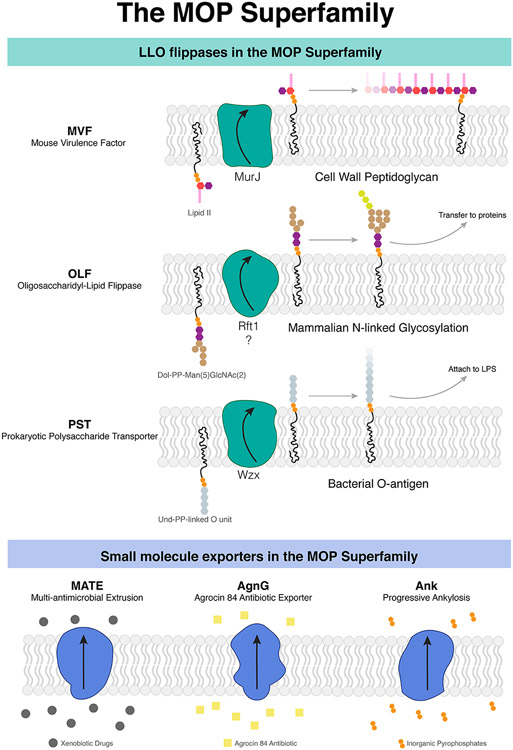
Lipid flippases are broadly classified as energy (ATP)-dependent and independent flippases (5). Many ATP-independent lipid flippases belong to the multidrug/oligosaccharidyl-lipid/polysaccharide (MOP) exporter superfamily (http://www.tcdb.org/search/result.php?tc=2.A.66). MOP proteins are characterized by a core transport domain consisting of 12-14 transmembrane helices (TMs) arranged in a fold distinct from other superfamilies. This superfamily has six functionally characterized subfamilies, of which three are known to mediate the transport of lipid-linked oligosaccharide (LLO) intermediates critical for many biosynthetic pathways (6) (Figure 1): the bacterial mouse virulence factor (MVF) family that transports lipid-linked cell wall precursors (7-9), the prokaryotic polysaccharide transporter (PST) family that transports lipid-linked cell surface polysaccharide precursors (10-14), and the eukaryotic oligosaccharidyl-lipid flippase (OLF) family that transports LLO precursors for N-linked glycosylation (15-18). The other three families—the multi-antimicrobial extrusion (MATE) family (19, 20), the Agrocin 84 exporter (AgnG) family (21), and the progressive ankylosis (Ank) family (22)—mediate the export of xenobiotic drugs, antibiotics, and inorganic pyrophosphate respectively (Figure 1).
Figure 1.
MOP superfamily proteins mediate the transport of lipids important for biosynthetic pathways as well as the expulsion of small molecules. (a) Three subfamilies (MVF, OLF, PST) comprise flippases that transport lipid substrates, while another three subfamilies (MATE, AgnG, and Ank) transport small molecules. (b) MurJ is the prototypical MOP flippase belonging to the MVF family. Abbreviations: AgnG, Agrocin 84; Ank, ankylosis; LLO, lipid-linked oligosaccharide; LPS, lipopolysaccharide; MATE, multidrug and toxic compound extrusion; MOP, multidrug/oligosaccharidyl-lipid/polysaccharide; MVF, mouse virulence factor; OLF, oligosaccharidyl-lipid flippase; PST, polysaccharide transporter.
In this review we focus on MurJ, the sole member of the MVF family and the first flippase member of the MOP superfamily to be structurally characterized. Pioneering genetic, biochemical, and structural studies on MurJ have provided much insight into the MOP flippase mechanism. Because MurJ is an emerging target for antibiotics development, we then review the remarkably diverse strategies of MurJ inhibition employed by small molecule, lipopeptide, and protein inhibitors. Finally, we discuss how knowledge of the MurJ mechanism and inhibition can be extended to other lipid flippases both within the MOP superfamily and beyond.
MURJ IS THE FLIPPASE FOR PEPTIDOGLYCAN PRECURSORS
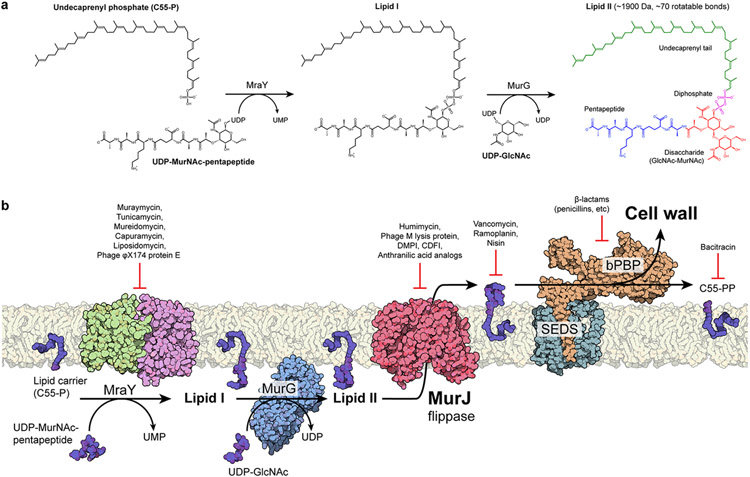
The importance of lipid transport in a biosynthetic pathway is exemplified by peptidoglycan biosynthesis (Figure 2). Peptidoglycan (PG) is a unique protective matrix enveloping the vast majority of bacterial cells, conferring rigidity and resistance to osmotic pressure as the main component of the bacterial cell wall. Peptidoglycan biosynthesis begins in the cytoplasm, where the water-soluble precursor UDP-MurNAc-pentapeptide is synthesized by the muramyl ligases MurA-F (23). The integral membrane enzyme MraY transfers the phospho-MurNAc-pentapeptide moiety from UDP-MurNAc-pentapeptide to the lipid carrier undecaprenyl-phosphate, forming lipid I (24, 25). This step is notable as it translocates the soluble precursors to the inner leaflet of the cytoplasmic membrane. Subsequently, MurG adds a GlcNAc moiety from UDP-GlcNAc, forming undecaprenyl-disphosphate-MurNAc-pentapeptide-GlcNAc, also known as lipid II (26). Lipid II must be subsequently flipped from the cytoplasmic side to the periplasmic side of the membrane. Once flipped, the GlcNAc-MurNAc-pentapeptide moiety of lipid II is incorporated into peptidoglycan by the glycosyltranferase activities of either shape, elongation, division and sporulation (SEDs) proteins (27-29) or bifunctional (class A) penicillin-binding proteins (aPBPs) (30). Glycan chains are then cross-linked between their stem peptides by transpeptidase activities of either bifunctional or monofunctional (class B) penicillin-binding proteins (bPBPs) (31). Finally, an undecaprenyl-diphosphate phosphatase regenerates the lipid carrier (32), which is then flipped back into the cytoplasm (33-35). These membrane-associated steps of peptidoglycan biosynthesis are known hot spots for antibiotic targeting (Figure 2).
Figure 2.
Membrane-associated steps of peptidoglycan biosynthesis are hot spots for antibiotic targeting. (a) Soluble cell wall precursors are synthesized in the cytosol and attached to the lipid carrier C55-P by MraY and MurG to form the lipid-linked intermediate lipid II. Lipid II is a large (~1,900 Da), flexible (~70 rotatable bonds), and anionic molecule that requires a dedicated transport protein to flip across the membrane. (b) On the periplasmic side of the cell membrane, a series of glycosyltransferase (SEDS proteins) and transpeptidases (class B PBPs) activities form the cell wall. The flippase MurJ forms the critical link between the cytosolic and periplasmic steps by flipping lipid II from the cytoplasmic side to the periplasmic side. Many of these steps are established targets of known antibiotics.
Abbreviations: C55-P, undecaprenyl phosphate; C55-PP, undecaprenyl diphosphate; CDFI, 2-(2-Chlorophenyl)-3- [1-(2,3-dimethylbenzyl)piperidin-4-yl]-5-fluoro-1H-indole; DMPI, 3-{1-[(2,3-Dimethylphenyl)methyl]piperidin-4-yl}-1-methyl-2- pyridin-4-yl-1H-indole; GlcNAc, N-acetylglucosamine; MurNAc, N-acetylmuramic acid; bPBP, class B penicillin-binding protein; SEDS, shape, elongation, division, sporulation; UDP, uridine diphosphate; UMP, uridine monophosphate.
The lipid II flippase bridges the cytosolic precursor synthesis steps with the periplasmic polymerization and crosslinking steps. The MOP superfamily protein MurJ (formerly known as MviN) was initially identified to be the flippase for peptidoglycan biosynthesis in 2008 by Ruiz (7) and Inoue et al (8). Ruiz (7) used a reductionist bioinformatics approach to identify genes present in endosymbiotic bacteria producing peptidoglycan but absent in non-producers, which was subsequently verified by radiolabel bioincorporation experiments in a conditional MurJ depletion strain. Meanwhile, Inoue et al (8) isolated a temperature-sensitive mutant of MurJ that swelled and burst at nonpermissive temperature and also accumulated peptidoglycan precursors. Follow up complementation and homology modeling studies showed that charged residues in a solvent-accessible central cavity are essential for MurJ activity in Escherichia coli, consistent with the idea that MurJ is a transporter protein (36, 37). Besides Escherichia coli, MurJ was also demonstrated to be essential for cell wall synthesis and viability in Burkholderia cenocepacia (38). A breakthrough came in 2014, when Ruiz, Bernhardt, and colleagues showed MurJ-dependent flipping of radiolabeled lipid II in E. coli cells and spheroplasts (9). They developed an in vivo lipid II flippase assay based on the fact that the Colicin M toxin cleaves the flipped lipid II and releases the soluble product (diphosphate-MurNAc-GlcNAc-pentapeptide) into the periplasm. Combined with the in vivo assay, they performed substituted cysteine accessibility method (SCAM) experiments and discovered that cells expressing the A29C mutant exhibit reduced lipid II flipping activity leading to cell lysis when treated with sodium 2-sulfonatoethyl methanethiosulfonate (MTSES) (9). Subsequent studies showed inactivation of the MurJ variant A29C with MTSES resulted in lipid II accumulation (39-41). Direct binding of lipid II by MurJ was detected by native mass spectrometry (42, 43). Because cell wall synthesis and cell division are coordinated processes in bacteria, MurJ activity appears to play a critical role in regulation of bacterial cell division—fluorescent microscopy studies showed that MurJ is recruited to the midcell septum together with other late localizing divisome proteins (including upstream enzymes MraY and MurG) and this recruitment is dependent on complete divisome assembly and both upstream lipid II synthesis and downstream FtsW/PBP3 activity in Escherichia coli (44). Recruitment of MurJ to the septum (and presumably MurJ activity there) drives local peptidoglycan incorporation coinciding with switch from a slow FtsZ-dependent stage to a fast FtsZ-independent stage of cytokinesis in Staphylococcus aureus (45).
While an increasing body of biochemical and genetic evidence has supported MurJ as the lipid II flippase, our understanding of this critical step was limited by a dearth of structural information. MATE drug exporters in the MOP superfamily have been proposed to utilize an alternating-access mechanism (46-50) driven by either a Na+ or H+ gradient (12, 46, 51, 52), but the mechanism and energy factor(s) that drive MurJ transport remain unknown. It has been shown that MurJ activity is dependent on membrane potential but not on the H+ gradient (40, 53), but involvement of either Na+ or anions has not been ruled out. Taken together, the transport mechanism of MurJ is an important question that would not only advance our understanding of peptidoglycan biosynthesis, but also of other membrane transport pathways mediated by MOP superfamily proteins including those in humans.
STRUCTURE AND MECHANISM OF MURJ
Architecture of MurJ
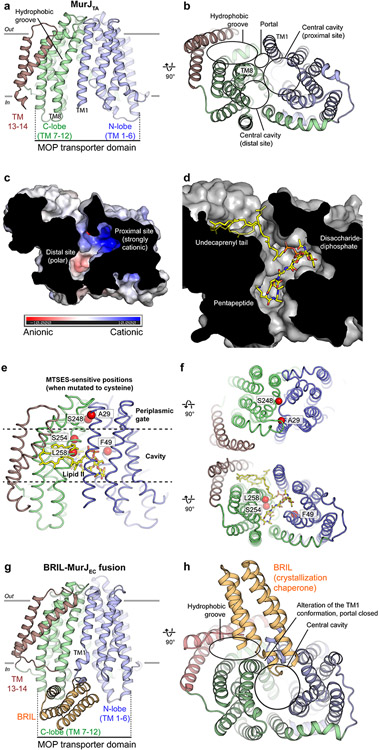
The first crystal structure of MurJ we published in 2017 provided an initial glimpse into its transport mechanism (54). We identified the MurJ protein from the hyperthermophile Thermosipho africanus (MurJTA) as a suitable candidate for structural studies. Expression of MurJTA was able to rescue E. coli growth upon depletion of endogenous MurJ (MurJEC), indicating MurJTA can complement MurJEC in function (54). Crystals of MurJTA were subsequently grown in lipidic cubic phase and a 2.0-Å resolution structure was determined using experimental phases from selenomethionine-substituted crystals (54). The crystal structure of MurJTA revealed an inward-facing conformation (Figure 3a), the first such structure in the MOP superfamily because all previously available crystal structures are of MATE drug exporters in the outward-facing conformation (47-50, 55). MurJ consists of 14 transmembrane helices (TMs)—the first 12 TMs forming the core transport domain common to all MOP superfamily proteins and 2 additional C-terminal helices (TMs 13-14) unique to MurJ. The core transport domain is made up of two helical bundles of 6 TMs each, the N-lobe (TMs 1-6) and C-lobe (TMs 7-12), which are arranged in an inward-facing N-shaped conformation instead of the outward-facing V-shaped conformation seen in MATE transporters. While the N-lobe and C-lobe are symmetric in MATE structures, this two-fold symmetry between two lobes is distorted in MurJ, especially in TMs 1, 2, 7, 8 that enclose the central cavity. TMs 13 and 14, not found in MATE drug exporters, form a hydrophobic groove that leads into the central cavity through a lateral membrane portal between TMs 1 and 8 (Figures 3a and 3b). The central cavity can be divided into a strongly cationic site adjacent to the portal (proximal site) and a polar site away from the portal (distal site) (Figure 3c). Directed mutagenesis and complementation experiments by us and others revealed the positive charges at the portal (Arg18) and proximal site (Arg24 and Arg255, that is Arg270 in E. coli MurJ) to be essential (36, 37, 54, 56). Furthermore, cysteine mutants placed at three positions in the cavity of E. coli MurJ result in cell lysis or cell shape defects upon treatment with the cysteine-reactive reagent MTSES (9). Based on these mutagenesis, chemical genetics, and in-silico docking data we proposed a model of lipid II binding (Figure 3d)—placing the undecaprenyl tail in the hydrophobic groove, the anionic GlcNac-MurNAc-diphosphate moiety in the cationic proximal site, and the pentapeptide in the polar distal site (54). To gain functional insight into the MurJ mechanism, we mapped the previously determined MTSES-sensitive cysteine substitutions that resulted in cell lysis or cell shape defects in E. coli (9) (Figure 3e). Two of the positions (A29 and S248) were located at the periplasmic gate, suggesting that adducts at these positions could have blocked closure of this gate. The other three positions (F49, S254, L258) were located in the central cavity and adducts could have blocked lipid II binding (Figure 3f).
Figure 3.
Structure of MurJ and putative mode of lipid II binding. (a) MurJTA was captured in an inward-facing conformation not previously seen in other MOP superfamily proteins. MurJ contains the 12-TM MOP transporter domain arranged in two 6-TM bundles (N-lobe and C-lobe) plus two additional C-terminal helices (TMs 13 and 14). (b) TMs 13 and 14 form a hydrophobic groove that enters into the central cavity through a lateral membrane portal between TM1 and TM8. The central cavity can be divided into a proximal site adjacent to the portal and a distal site away from the portal. (c) The proximal site is highly cationic, while the distal site is only polar. (d) Model of lipid II (yellow sticks) docked into the MurJTA structure, placing the undecaprenyl tail in the hydrophobic groove, the disaccharide-diphosphate in the proximal site, and the pentapeptide in the distal site. (e) Previously determined MTSES-sensitive positions in MurJEC that result in cell lysis or cell shape defects are mapped to the MurJTA structure as red spheres. ( f ) Ala29 and Ser248 are located at the periplasmic gate, and adducts likely obstructed the closure of this gate. In contrast, Phe49, Ser254, and Leu258 face toward the central cavity and adducts at these positions could have blocked lipid II binding. (g) The structure of MurJEC was determined by fusion with a crystallization chaperone in the absence of lipid II, whereas lipid II was present in the MurJTA crystallization conditions. The MOP transporter domain of this MurJEC structure is less wide than that of MurJTA. (h) MurJEC also contains a hydrophobic groove and central cavity, but the portal appears to be closed in this structure, possibly due to the BRIL-induced distortion of TM1 and/or the absence of lipid II.
Abbreviations: BRIL, thermostabilized cytochrome b562 RIL; MOP, multidrug/oligosaccharidyl-lipid/polysaccharide; MTSES, sodium 2-sulfonatoethyl methanethiosulfonate; MurJEC, Escherichia coli MurJ; MurJTA, Thermosipho africanus MurJ; TM, transmembrane helix.
A crystal structure of E. coli MurJ (MurJEC) was subsequently determined to 3.5-Å resolution by fusion to the crystallization chaperone BRIL (56) (Figures 3g and 3h). While this structure of MurJEC is similar to MurJTA, it was crystallized in the absence of lipid II in contrast to the presence of lipid II in MurJTA crystallization conditions. The MOP transporter domain of this MurJEC structure is ~10% narrower than that in MurJTA and the portal appears to be closed. Taken together, the authors concluded that this structure likely represents an apo state of MurJ (56). They also presented high-throughput mutagenesis data confirming the importance of the hydrophobic groove, central cavity, and cytoplasmic/periplasmic gates to MurJ function, noting that substitutions that altered the charge of the central cavity were particularly poorly tolerated consistent with importance of electrostatic interactions to lipid II binding and/or transport (56).
Co-evolution data showed strong couplings between the N-lobe and C-lobe, not only at the periplasmic gate but also the cytoplasmic gate, supporting the existence of an outward-facing state (56). Mapping of five positions previously determined to be sensitive to the membrane-impermeant cysteine-reactive reagent MTSES (9) indicated that the central cavity is accessible to the periplasm, again consistent with the presence of an outward-facing conformation. The two positions at the periplasmic gate (Ala29 and Ser248) were located at the interface between the N-lobe and C-lobe, suggesting that MTSES labeling at these positions would sterically block periplasmic gate closure and leave MurJ trapped in an outward-facing conformation (54). Our inward-facing MurJTA structure provides the molecular basis for the reduced flippase activity of the A29C mutant upon MTSES introduction (9) and also suggest that an outward-facing conformation is important for lipid II flipping (54). Further evidence for an outward-facing conformation was subsequently presented in cysteine crosslinking and thrombin proteolysis data (53). Taken together, we proposed that MurJ utilizes an alternating access mechanism between the inward- and outward-facing states for flipping the lipid II headgroup, while the lipid II tail stays outside the central cavity possibly in the hydrophobic groove. This mechanism is unlike what was proposed for the lipid-linked oligosaccharide flippase PglK, in which the outward-facing conformation alone is sufficient for flipping (57).
Conformational transitions of MurJ
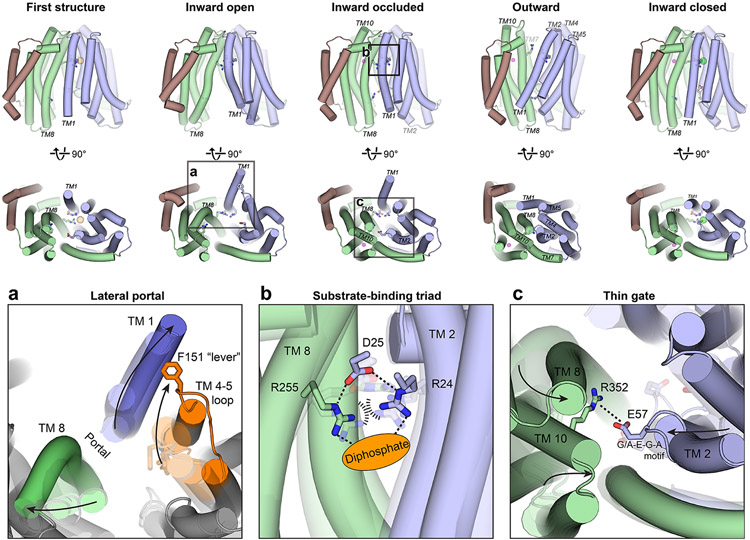
While MurJ utilizes alternating access for transport, neither the outward-facing conformation nor the conformation transitions that drive lipid II flipping were known. Crystal structures of MurJTA in 4 additional conformations were subsequently determined in the presence of lipid II—an outward-facing conformation and 3 inward-facing conformations distinct from the previously published structure (Figure 4 and Supplemental movie) (41). Superposition of the inward-facing structures revealed different sizes of the lateral portal ranging from closed to wide open, regulating accessibility of the central cavity from the membrane (Figure 4a). The Cα-Cα distance between Ser11 (TM 1) and Ser267 (TM 8) increased from 8.0 Å in the closed structure to 17.4 Å in the open structure. The initial inward-facing structure appeared to be an intermediate between these two conformations (Cα-Cα distance of 10.7 Å). Portal opening is controlled by the bending of TM 1 out into the membrane together with concomitant rearrangements in TM 8 and the TM 4-5 loop (Figure 4a). Notably, the conserved Phe151 (Phe157 in MurJEC) in the TM 4-5 loop acts as a lever to push TM 1 out into the membrane (41). Because lipid II is believed to access the central cavity of MurJ through the portal between TM 1 and TM 8 (54), the different sizes of the portal could regulate entry of the large lipid II headgroup, thereby providing a lateral-gating mechanism. Many residues in TM 1 and TM 4-5 loop are conserved in MurJ sequences but not MATE drug efflux pumps in the same superfamily, suggesting the lateral gate to be a specific adaptation for flippase function.
Figure 4.
Crystal structures of MurJTA in inward-open, inward-occluded, outward, and inward-closed states elucidate the conformational changes that mediate lipid II flipping. (a) The lateral membrane portal is opened in the inward-open conformation by the bending of TM1 out into the membrane and concomitant rearrangements in TM8 and the TM4–5 loop (orange), specifically those involving the Phe151 lever, allowing entry of the large lipid II headgroup into the cavity. Translucent outlines denote the other inward-facing conformations. (b) In the inward-occluded structure, Arg24 and Arg255 are brought into unusual proximity at the apex of the cavity, their electrostatic repulsion possibly stabilized by Asp25 and the anionic diphosphate moiety of lipid II (orange oval). TM1 is hidden in this panel for clarity. Translucent sticks denote these residues in the other inward-facing conformations. Arg255 is disengaged from this Arg24-Asp25-Arg255 triad in the outward-facing conformation. (c) Bending of TM2 and closing of the C-lobe forms a thin gate between Glu57 of the N-lobe and Arg352 of the C-lobe, possibly to occlude the lipid II headgroup from the cytosol. Translucent sticks denote these residues in the other inward-facing conformations.
Abbreviation: MurJTA, Thermosipho africanus MurJ; TM, transmembrane helix.
The inward-occluded conformation of MurJ exhibited a partially dilated membrane portal but was otherwise divergent from the rest of the inward-facing structures. The first defining feature of the inward-occluded conformation is located at the apex of the cavity, where Arg24 and Arg255 are brought into unusual proximity (just 2.9 Å apart at the closest) (Figure 4b). Since such close contact between two arginine side chains would experience unfavorable repulsion, we reasoned that they could be stabilized by electrostatic interactions with Asp25 and the diphosphate moiety of lipid II. Because of their conserved and essential nature (36, 54), as well as their coordinated rearrangement at the cavity apex in proximity to unmodeled density, we designated Arg24-Asp25-Arg255 to be the putative substrate-binding triad (41). The second defining feature is the thin gate formed between Glu57 of the conserved G/A-E-G-A motif in the N-lobe with Arg352 (Lys368 in MurJEC) on TM 10 of the C-lobe (Figure 4c). Formation of the thin gate is mediated by bending of the C-lobe towards the N-lobe, as well as by bending of TM 2 facilitated by the flexible G/A-E-G-A motif. Mutation of Glu57 or Arg352 to alanine led to loss of MurJTA function, supporting the importance of this thin gate for transport (41). Taken together, we proposed that the Arg24-Asp25-Arg255 triad at the apex of the cavity could serve to bind the diphosphate moiety of lipid II, while the Glu57-Arg352 thin gate concurrently occludes the bound lipid II headgroup from the cytosolic milieu.
Transition to the outward-facing conformation is associated with a rotation of the N- and C-lobes resulting in cytoplasmic gate closure and periplasmic gate opening (Figure 4 and Supplemental movie). The N-lobe rotated by ~15° from the inward-occluded structure while the C-lobe rotated by only ~7.5°, possibly because the latter has already bent inwards in the occluded structure. The fulcrum of this rotation is located at TM 7 bridging the N- and C-lobes, which was bent by 90° in the inward-occluded structure but straightened in the outward-facing structure (Figures 4 and 5). Aside from TM 7, TM 1 also straightens from its bent conformation in the inward-occluded structure, closing the lateral membrane portal. The outward-facing cavity was substantially shallower and narrower than the inward-facing cavity, supporting the idea that cavity shrinkage could be a mechanism to displace substrate into the periplasm, as was proposed for MATE transporters (46, 48). We observed unmodeled electron density in the hydrophobic groove leading into the outward-facing cavity, which might indicate the binding site of the undecaprenyl lipid tail of lipid II while the headgroup is on the periplasmic side.
Figure 5.
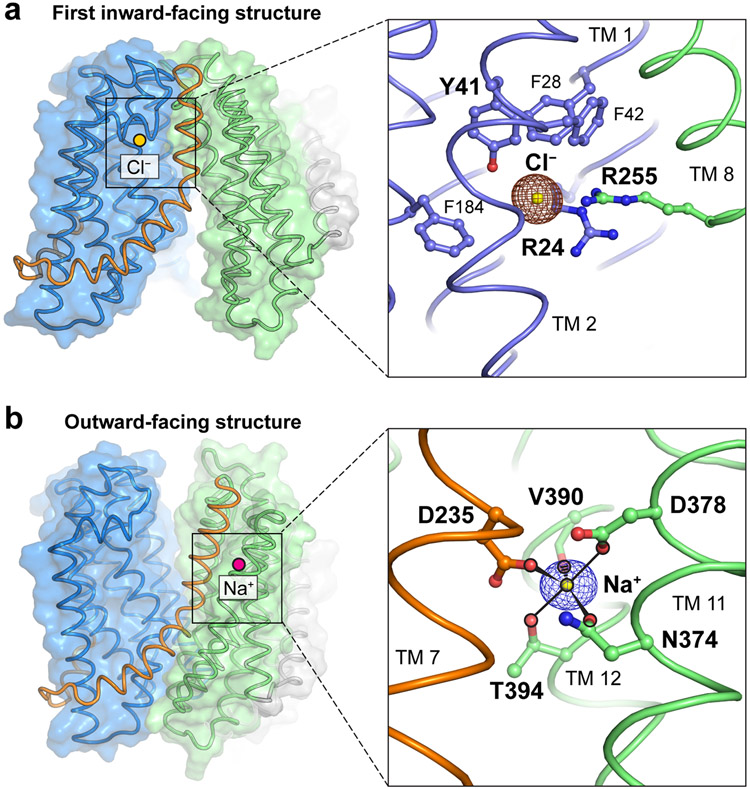
Ions captured in the MurJTA structure that might be involved in lipid II transport. (a) A chloride ion was observed in the first inward-facing (2.0-Å resolution, shown) and the inward-closed structures. Chloride was coordinated by Arg24, Tyr41, and Arg255, with the last being the only residue from the C-lobe and the rest from the N-lobe. Phe28, Phe42, and Phe184 are also in orientations suitable to form anion-quadrupole interactions with chloride. Anomalous difference electron density from a bromide soak is shown in brown mesh contoured to 4.5 σ. (b) A sodium ion was observed in the outward-facing (1.8-Å resolution, shown), inward-occluded, and inward-closed structures. Sodium was coordinated in trigonal bipyramidal geometry by Asn374, Asp378, Val390 (backbone carbonyl), Thr394, and Asp235, with the last being the only residue from the hinge helix TM7 (orange) and the rest from TM11/12. Omit electron density for sodium is shown in purple mesh contoured to 4.5 σ.
Abbreviations: MurJTA, Thermosipho africanus MurJ; TM, transmembrane helix.
Ion coupling of MurJ
While the conformation transitions that mediate lipid II flipping have been characterized, the energy-coupling mechanism of MurJ is still an open question. We observed a chloride ion bound in the N-lobe of the original inward-facing structure and in the inward-closed structure (Figure 5a). This ion is coordinated by Arg24, Tyr41, and Arg255, with only the latter residue coming from the C-lobe. Phe28, Phe42, and Phe184 are also in orientations suitable to form anion-quadrupole interactions with chloride. Spontaneous and specific binding of chloride to this position was observed during molecular dynamics simulations of the recent inward-facing PfMATE crystal structure in the same superfamily (PfMATE has Arg284 at the same location as Arg255 in MurJ), from which the authors suggested that chloride could be part of the transport mechanism (58). Because Arg255 is a member of the putative substrate-binding triad which is dissociated in the outward-facing conformation, we speculate that chloride binding could serve to re-engage Arg255 with the N-lobe and thus reset MurJ from the outward-facing state back to the inward-facing state. However, we caution against excessive interpretation of the chloride ion as current biochemical data only reveals a role of membrane potential in resetting MurJ to the inward-facing state (40). As such, the importance of chloride is yet to be tested.
In contrast to the unclear significance of the chloride ion, many MATE transporters in the MOP superfamily have been characterized to be driven by sodium ions (51, 52, 59-62). A sodium ion was bound in the C-lobe of the outward-facing (1.8 Å resolution, shown), inward-occluded, and inward-closed structures (Figure 5b). Sodium is coordinated in trigonal bipyramidal geometry, with an equatorial plane formed by Asp235, Asn374, Val390 (backbone carbonyl) and axial positions occupied by Asp378 and Thr394. The equatorial positions were intolerant of substitutions underlining their importance, while certain substitutions were tolerated at the axial positions. Asp235 is the only Na+-coordinating residue from TM 7, with the rest from either TM 11 or TM 12. The Na+ bound structures exhibited different degrees of TM 7 straightening, resulting in concomitant lowering of the N-terminal half of TM 7 and the TM 6-7 loop relative to the Na+ site, suggesting that Na+-induced conformational change at Asp235 could be propagated down TM 7. Interestingly, recent structural re-analysis of PfMATE crystal structures showed that they contain an identical Na+ site albeit in the N-lobe (48, 62), consistent with spectroscopic experiments on another MATE protein, NorM (52). Taken together, Na+ coordination appears to be associated with rearrangement of TM 7 in MurJ or TM 1 in PfMATE, suggesting a role of Na+ in inward-to-outward transition.
Transport mechanism of MurJ
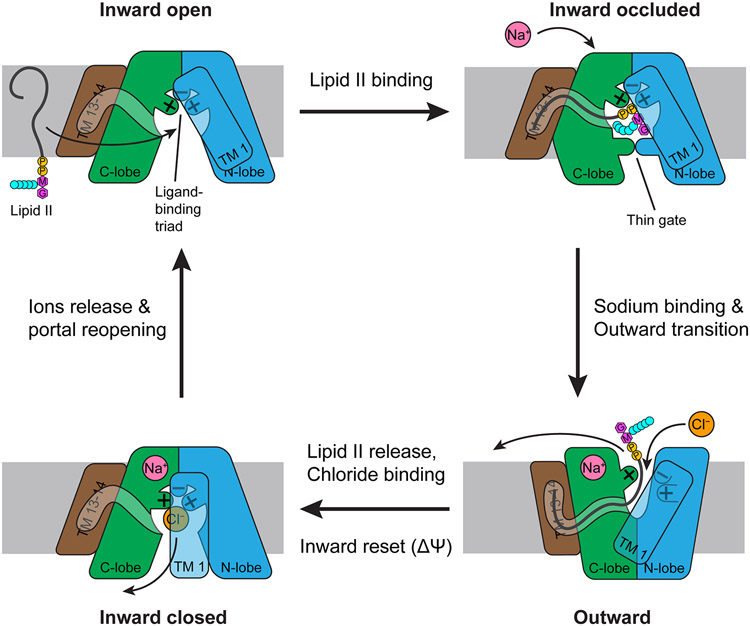
Taken together, the current understanding of MurJ function points to an intricate alternating-access mechanism exchanging lipid II with sodium and chloride ions, with involvement of membrane potential as previously proposed (40, 53) (Figure 6). In the inward-open state, lipid II enters the central cavity through the lateral membrane portal. The diphosphate moiety of lipid II is captured by the Arg24-Asp25-Arg255 triad positioned at the apex of the cavity, while the lipid tail is associated to the hydrophobic groove formed by TMs 13 and 14. The bound lipid II headgroup is occluded from the cytosol by the thin gate formed by Glu57 and Arg352, which poises the MurJ-lipid-II complex for outward transition. Binding of a sodium ion to the conserved site in the C-lobe tips the balance towards the outward-facing state. The shallow outward-facing cleft is too small to fit the lipid II headgroup, which is then released. We have observed unmodeled electron density in the hydrophobic groove and a short tunnel leading up to the outward-facing cleft but not inside the cleft itself (41), suggesting the undecaprenyl tail could remain associated with MurJTA even after the headgroup has been displaced from the outward-facing cleft. The binding and dissociation of such a large, flexible substrate might not follow simple single-step kinetics, and it is conceivable that the headgroup might be displaced first before the lipid tail follows. While this is a high (1.8 Å)-resolution structure, we cannot exclude the possibility that these densities could be a conglomeration of adventitious lipid molecules.
Figure 6.
Mechanism of lipid II flipping by MurJ. Lipid II enters the central cavity through a lateral portal. The Arg24/Asp25/Arg255 triad in the cavity binds the diphosphate moiety of lipid II, while the undecaprenyl tail fits into the hydrophobic groove formed by TMs 13 and 14. Lipid II is occluded from the cytosol by the Glu57-Arg352 thin gate, poised for outward transition. Upon sodium binding to the C-lobe, MurJ transits to the outward-facing state, disengaging Arg255 from Arg24 and Asp25. Because the outward-facing cleft is too narrow to accommodate the lipid II headgroup, lipid II is released. Chloride binding reassociates the Arg24/Asp25/Arg255 triad, resetting MurJ to an inward-facing apo state where the portal is closed. Membrane potential might also facilitate this inward reset. Release of sodium and chloride ions into the cytosol mediates reopening of the portal, completing the transport cycle. The net reaction is the export of lipid II, uptake of one sodium ion, and uptake of one chloride ion.
Abbreviation: TM, transmembrane helix.
After release of lipid II, MurJ is reset to the inward-closed state by membrane potential and/or chloride binding, which re-engages Arg255 with Arg24 and Asp25 and thus completes the substrate-binding triad once again. Usage of chloride could be evolutionarily advantageous for two reasons. First, it allows bacteria to tap into not just the inward sodium gradient but also the inward chloride gradient when living in high-salinity environments, providing additional driving force for lipid II transport. Second, uptake of chloride could help offset the positive charge buildup generated from export of the anionic lipid II molecule and uptake of sodium. Each lipid II molecule carries 3 to 4 negative charges (depending on whether lipid II is meso-diaminopimelate type or lysine type) from the cytoplasmic to the periplasmic side of the membrane. Factoring in sodium uptake, this would be buildup of 4 to 5 positive charges inside the cell relative to outside per MurJ transport cycle—uptake of chloride would reduce this back to 3 to 4 positive charges accumulated per cycle. Release of chloride and sodium ions into the cytosol mediates reopening of the portal, allowing entry of another lipid II molecule thus completing the transport cycle.
INHIBITION OF MURJ FOR ANTIBIOTICS DEVELOPMENT
MurJ is a promising but underexplored target for novel antibiotics development for the following reasons. First, MurJ is an essential protein in the PG synthesis pathway. Second, antibiotic development campaigns by pharmaceutical companies led to the identification of compounds that potentiate β-lactams and restore the antibacterial β-lactam efficacy. These compounds target either MurJ or a MurJ homolog. Third, natural products from human microbiome or a phage protein were discovered to inhibit MurJ. Taken together, MurJ is a good candidate as an antibiotic target and potentially useful for restoring the clinical utility of β-lactams. Here we discuss the inhibitors that have been identified to date.
Lipopeptides: Humimycins
Humimycins (human microbiome mycins) are lipopeptides that were predicted from gene clusters in the human microbiome and produced by chemical synthesis (63, 64). Humimycins are broadly active against Firmicutes (including Staphylococcus and Streptococcus) and potentiate and restore the activity of β-lactam against methicillin-resistant Staphylococcus aureus (MRSA) (63). SAV1754, a homolog of MurJ in Staphylococcus aureus and other Gram-positive bacteria, is the target of humimycin as mutations and overexpression of SAV1754 confers resistance. Humimycins are linear heptapeptides N-terminally capped with β-hydroxy myristic acid (Figure 7a). The position and stereochemistry of the β-hydroxylation is critical for antibiosis suggesting its involvement in binding to SAV1754. The peptide sequences also likely play important role of the inhibition because the entire heptapeptide showed little tolerance for changes with the central three residues (Tyr, Phe, Thr) being the most critical for antibacterial activity (64). Chu et al. subsequently optimized Humimycin A to the more potent inhibitor Humimycin 17S, against which no resistant mutations have arisen (64).
Figure 7.
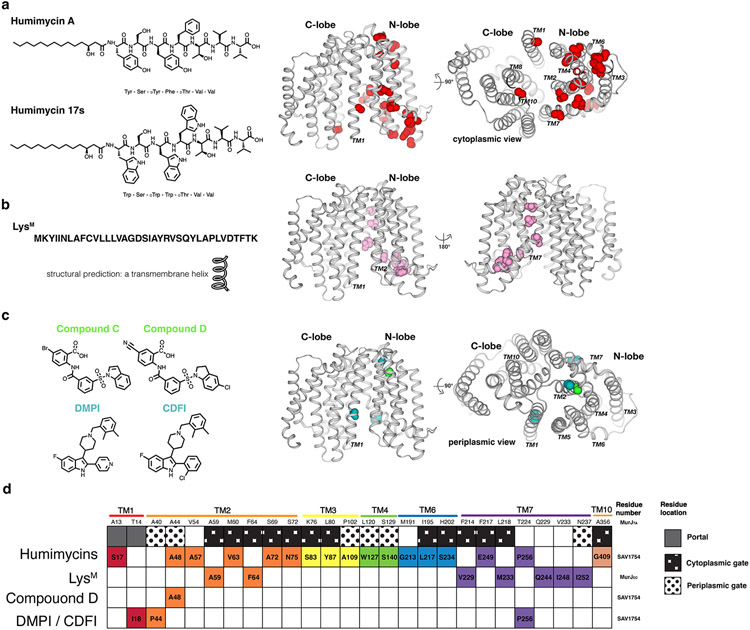
Inhibitors of MurJ. (a) Chemical structures of humimycins. The locations of resistance mutations mapped to the MurJTA structure are indicated as spheres. (b) Peptide sequence of LysM and resistance mutations to LysM mapped to the MurJTA structure. (c) Chemical structures of the putative small-molecule inhibitors DMPI, CDFI, Compound C, and Compound D. Resistance mutations to each were mapped to the MurJTA structure. (d) A barcode tabular summary of resistance mutations for each inhibitor and their locations on MurJ. Humimycins appeared to target the portal and cytoplasmic gate of MurJ, while LysM putatively targets the TM2–TM7 region of MurJ facing outside, toward the membrane. The mechanism of small-molecule inhibitors is less clear due to the small number of resistant mutants that have been isolated.
Abbreviations: CDFI, 2-(2-Chlorophenyl)-3-[1-(2,3-dimethylbenzyl)piperidin-4-yl]-5-fluoro-1H-indole; DMPI, 3-{1-[(2,3-Dimethylphenyl)methyl]piperidin-4-yl}-1-methyl-2-pyridin-4-yl-1H-indole; LysM, phage M lysis protein; MurJTA, Thermosipho africanus MurJ; TM, transmembrane helix.
Currently there is no direct biochemical study delineating the mechanism of action of humimycin. Mutations leading to resistance against humimycin A cluster near the cytoplasmic side of the N-lobe and partially overlap with the putative lipid II binding site (Figure 7a), so it is possible that humimycin acts by blocking closure of the cytoplasmic gate and/or is a competitive inhibitor for SAV1754.
Phage M lysis protein (LysM)
The bacteriophage lysis protein (LysM) of the E. coli levivirus Phage M, a small lytic phage, was discovered as a putative MurJ inhibitor by Chamakura et al (65). LysM is a 37-residue polypeptide that is predicted to contain a single transmembrane helix and predicted to be N-out and C-in membrane topology (Figure 7b) (66, 67). Expression of LysM induced morphological defects prior to lysis similarly cause by β-lactams suggesting LysM might be an inhibitor of cell wall synthesis. Mutations and overexpression of MurJ give resistance to LysM, pinpointing the target of LysM to MurJ. Colicin M assay showed that lipid II flipping is inhibited in the presence of LysM in vivo while Lipid II flipping in E.coli with the MurJ variant containing resistant mutations is not inhibited (65).
Resistance mutations cluster in TM2 and TM7 regions exposed to the surrounding membrane (Figure 7b). SCAM experiments on MurJ demonstrated that LysM induces conformation changes in MurJ and likely traps MurJ in an outward-facing conformation (65). Direct binding of LysM to MurJ remains to be demonstrated.
Small molecule inhibitors (I): Compound C & D
Compound C and D belong to a class of anthranilic acid derivative (Figure 7c). By analysis of incorporation of radiolabeled precursors of fatty acid, DNA, RNA protein or cell wall, treatment with compound C and compound D resulted in inhibition of cell wall biosynthesis (68) . One point mutation was identified on SA1575 (which is the same gene as SAV1754. SA1575 is based on S. aureus strain N315 genome annotation) that confer resistance to compound D suggesting these compounds inhibits SAV1754 (68).
Currently the mode of action is unknown. Compound C and D were reported before the structure of MurJ was published; mapping the resistance mutation A48T to the MurJTA structure shows that A48 is at the periplasmic side, away from the substrate binding site (Figure 7c). Based on this observation, it is unlikely that compounds C and D compete with lipid II for binding to MurJ. Rather, it is possible that Compound C and D bind to a site away from lipid II and allosterically inhibit MurJ by trapping it in a certain conformation. Whether compounds C and D bind to the site containing A48 remains to be determined. We caution against drawing conclusions about the inhibitory mechanism from a single resistant mutant because it could have secondary effects such as affecting protein folding.
Small molecule inhibitors (II): DMPI and CDFI
Huber et al. identified compounds DMPI and CDFI using high-throughput screening for compounds that can restore activity of β-lactams against MRSA(69). DMPI and CDFI are indole compounds (Figure 7c). When treated with these compounds, S. aureus cells displayed a significance reduction in peptidoglycan labeling by fluorescent-vancomycin, especially at the septum. DMPI and CDFI were found to target protein SAV1754, as overexpression of SAV1754 or specific point mutations in SAV1754 confer resistance to these compounds. Genetic inactivation of SAV1754 results in pronounced hypersensitivity to a wide range of beta-lactams, and even more than PBP2 inactivation, highlighting the druggability of MurJ and MurJ homologs as antibiotic targets (69). DMPI was subsequently used by Monteiro et al. to investigate the critical step of peptidoglycan synthesis for cytokinesis during cell division (45). They found that addition of DMPI blocked septal ring constriction and thus cytokinesis by inhibition of peptidoglycan synthesis (45). This indicates that DMPI could be a peptidoglycan synthesis inhibitor in vivo, making it a useful research tool for biochemical and cell biology studies.
Three point-mutations were identified to confer resistance to DMPI and CDFI. The three mutants do not confer resistance to compound D suggesting that these indole compounds likely have different inhibition mechanism from compound D (69). The three resistant mutation sites are far from each other on MurJ suggesting that not all three sites are involved in binding to DMPI and CDFI (Figure 7c). Similar to Compound C & D, DMPI and CDFI might also inhibit MurJ allosterically.
Summary of inhibition mechanisms
MurJ undergoes dynamic conformation changes, especially at the cytoplasmic gate, TM7 which rearranges during inward to outward transition and TM1 (closing the lateral membrane portal). Resistance mutations of all the putative inhibitors mostly cluster around cytoplasmic gate, TM7 and TM1. Because these regions (TM7, TM1, and the cytoplasmic gate) in MurJ are critical for lipid II flipping (Figure 7d), it is tempting to speculate these compounds inhibit MurJ by conformational locking. Unfortunately, there is no biochemical data available to further support the genetic studies on MurJ inhibition by these inhibitors. Given their diverse chemical structure, the inhibitory mechanism for MurJ is most likely to be different for each type of inhibitor. Whether these molecules act on MurJ as competitive or allosteric inhibitors is yet to be determined.
Intriguingly, these putative MurJ inhibitors are diverse in size and chemical structure, which include lipopeptide-like molecules, transmembrane helical-like peptides, and small molecules. This suggests that MurJ has various hot spots for inhibition and reinforces further the idea that MurJ is a promising target for novel antibiotic development. Despite low sequence similarity between Gram-negative MurJ and SAV1754 (~18%), Ruiz reported that the SAV1754 homolog from Streptococcus pyogenes (YtgP) complements E.coli strains depleted of MurJ (70), suggesting that there is structural similarity between Gram-negative and Gram-positive MurJ. This observation raises a possibility that inhibitors for SAV1754 could also be used for E.coli MurJ upon chemical structure optimization..
MURJ AND OTHER LIPID-LINKED OLIGOSACCHARIDE FLIPPASES
Amongst different types of lipid flippases with different lipid specificity, MurJ is a lipid-linked oligosaccharide (LLO) flippase. LLO are precursors for many critical cellular polysaccharides such as N-link glycan, peptidoglycan, lipopolysaccharide (LPS), and O-antigens. LLOs are formed by translocases that attach oligosaccharide precursors to a lipid carrier: undecaprenyl phosphate in bacteria and dolichyl phosphate in eukaryotes (71). The translocation of LLO across membrane is a common process in all domains of life, which is carried out by LLO flippases. There have been recent advances on structural and mechanistic studies of other types of LLO flippases, expanding our understanding on the principles of innerworkings of LLO flippases. Here we summarize current understanding on mechanisms of various LLO flippases. We will discuss the other LLO flippases in the same superfamily as of MurJ as well as LLO flippases from other superfamilies.
LLO flippases in the MOP superfamily
We first discuss LLO flippases from the same superfamily as MurJ before we expand our discussion to LLO flippases in other superfamilies.
Wzx proteins:
Wzx proteins (belonging to the PST family within the MOP superfamily) transport various undecaprenyl pyrophosphate (Und-PP)-linked oligosaccharides that are displayed on the cell surface (Figure 1) (14). For example, the O-antigen components of nearly all E.coli lipopolysaccharide are transported through Wzx (72). The Lam group performed an elegant experiment to monitor I− flux from liposome-reconstituted Pseudomonas aeruginsa Wzx (WzxPa) and found that WzxPa exhibited pH-dependent I− flux. They also found that mutation of the three acidic amino acids (E61, D269, and D359) reduced pH-dependent I− flux by WzxPa substantially, suggesting that these residues are involved in H+-coupled transport (12). Based on the observation, the Lam group proposed a H+-dependent antiporter mechanism for WzxPa. However, the underlying assumption for this antiporter proposal is that I− can be a surrogate for O-antigen, which has not been substantiated. Regardless, it is noteworthy that this study presented the involvement of both cation (H+) and halide anion (I−) in the O-antigen transport by WzxPa, which is analogous to the potential involvement of Na+ and Cl− in the lipid II flipping by MurJ (Figure 6). Earlier mutagenesis studies reported both positively and negatively charged residues essential for Wzx function (73, 74). Because most O-antigens are not charged, these charge residues may not participate in direct O-antigen binding but play a role in either ion coupling (e.g. D269) or conformational changes associated with transport.
Interestingly, some WzxC variants (capsule colanic acid synthesis) lost their substrate specificity for colonic acid and instead can flip lipid II and thus complement E. coli MurJ depletion (75, 76), suggesting that WzxC is structurally similar to MurJ with low intrinsic substrate specificity. Although the structure of WzxC is not available, most of those variants were predicted located at the periplasmic gate based on homolog model, suggesting these variants might destabilize the inward-facing conformation and promote conformation transitions (76). Similar mutation leading to the loss of substrate specificity was identified in another member of the PST family, TacF. TacF is thought to be the teichoic acid flippase in Streptococcus pneumoniae (77, 78). A TacF variant was identified that can transport precursors with slight difference and this variant was predicted to located at the periplasmic side of TacF. The variant was proposed to destabilize the inward-facing state and bypass the need for substrate-induced conformational transition (76, 77). The apparent importance of the inward-facing state for substrate specificity is interesting, but because the observation was based on genetic studies, future biochemical and biophysical studies on an isolated system are necessary to test this interesting idea. Consistent with this hypothesis, some LLO transporters themselves appear to be interchangeable between different organisms despite their presumably different O-antigen compositions—the ATP-binding cassette (ABC) transporter superfamily LLO flippase PglK from Campylobacter jejuni (discussed in the section titled PglK) could complement the depletion of Wzx and restore LPS synthesis (79), showing the interchangeable role between LLO flippases. However, we caution that many of these substrate-switching experiments were performed under non-physiological conditions (overexpression of the complementing flippase and/or depletion of the endogenous flippase, the latter of which could result in elevation of substrate levels). The Reeves group reported exquisite specificity of Wzx proteins for their ideal substrates when the experiment was performed under near-physiological conditions using gene replacement (80).
Rft1:
The eukaryotic protein Rft1 belongs to the OLF family. Based on the yeast genetic analysis, Rft1 was proposed to be the LLO flippase that flip Man5GlcNAc2-PP-Dol from the cytoplasmic side to the luminal side of the endoplasmic reticulum (Figure 1) (15). However, this result was challenged by the observation that a Rft1-null low-eukaryotic organism (Trypanosoma brucei) grows normally (18). Despite the controversy (15, 16, 18), it remains conceivable that Rft1 is the LLO flippase involved in mammalian N-linked glycosylation, given the similarity between the membrane-associated steps of bacterial cell wall synthesis and the early steps of N-linked glycosylation, together with the homology between Rft1 and MurJ.
LLO flippases in other superfamilies
Here, we highlight a few LLO flippases belonging to other superfamilies of transporters that have been biochemically and structurally characterized.
LtaA, a member of the major facilitator superfamily (MFS), is the flippase for the translocation of the lipid-linked precursor of lipoteichoic acids in Gram-positive bacteria. LtaA transports the anchor lipid-linked-disaccharide gentiobiosyl-diacylglycerol (anchor-LLD) across the cytoplasmic membrane (81). Deletion of LtaA results in attenuated virulence in S. aureus. Recently, the structure of LtaA (PDB 6S7V) was reported (82). Similar to MurJ, the LtaA structure reveals a large amphiphilic central cavity that is expected to bind the lipid tail at C-terminal hydrophobic pocket and the disaccharide group at the N-terminal hydrophilic pocket (Figure 8). LtaA displays two lateral hydrophobic entrances, similar to the hydrophobic groove in MurJ. Also, it likely adopts an alternating-access mechanism. Using fluorescence quenching liposome assay, LtaA was shown to be a proton-coupled antiporter (82).
Figure 8.
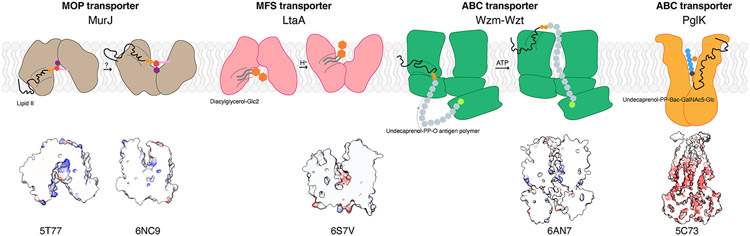
Diversity of LLO flipping mechanisms beyond the MOP superfamily. Cartoon representations of LLO flippases MurJ, LtaA, Wzm-Wzt and PglK with their flipping mechanisms. Dashed lines indicate a hydrophobic groove or opening. MurJ and LtaA utilize alternating access. Wzm-Wzt forms a continuous channel inside the transmembrane region to transport substrate. PglK was proposed to use an unconventional mechanism in which only the outer-facing conformation is involved in flipping. The lipid tail does not enter into the protein but interacts only with the helix on the periplasmic side while the headgroup enters the positive charged cavity. Shown below are electrostatic surface representations highlighting the cavity where substrates bind, with cationic surfaces in blue and anionic surfaces in red.
Abbreviations: ABC, ATP-binding cassette; LLO, lipid-linked oligosaccharide; MFS, major facilitator superfamily; MOP, multidrug/oligosaccharidyl-lipid/polysaccharide; PDB ID, Protein Data Bank identifier.
Wzm-Wzt, the ABC transporter responsible for flipping the lipid-linked polysaccharides for O-antigen synthesis, has been proposed to use a channel-like mechanism (Figure 8) (83-85). Unlike MurJ flipping monomer of Lipid II, Wzm-Wzt flips the long O-antigen polymers that have been synthesized on the cytoplasmic side prior to flipping (86, 87). The structural studies by the Zimmer group revealed that Wzm-Wzt forms a continuous transmembrane channel in the transmembrane domain that is spacious enough to accommodate an O-antigen glycan polymer. The presence of a small gate helix creates a positive pocket near the inner leaflet of the membrane that could interact with the pyrophosphate group of Und-PP and the first sugar unit, suggesting that this gate helix provides the substrate specificity (84). Lipid-linked O-antigen likely first interact with the small gate after which undecaprenol lipid tail flipping and the polymer insertion into the channel was speculated to take place spontaneously. The channel is lined with hydrophilic residues as well as aromatic residues, which likely to provide interaction with the sugar rings of the polymer. ATP hydrolysis results in the gate helix and TM-forming helixes moving toward the periplasmic side and this rigid body movement propagates the processive polymer translocation through the transmembrane channel (83, 88). Periplasmic channel exit forms later gates facing the outer leaflet, proposed to function as releasing the LLO (84, 85).
PglK, the flippase for substrate of N-link glycosylation in the bacteria Campylobacter jejuni, was proposed to adopt a novel flipping mechanism where only the outward-facing conformation plays a role in transport (57). The Locher group proposed the polyisoprenoid end of the LLO interacts with the hydrophobic grooves formed by the external helix at the periplasmic side (Figure 8). The headgroup of LLO enters the outward-facing translocation cavity and stabilized by the positively charged belt in the cavity. ATP hydrolysis-driven motion on the transmembrane domain squeeze the LLO headgroup to be released into the external side of the membrane (89). Mutagenesis studies showed the importance of the external helix on flippase function, but there is no direct evidence of the interaction of the lipid tail with the helix. Although structural and mutagenesis analysis supports the importance of the outward-facing state in LLO transport in PglK, an alternating-access mechanism has not been definitively ruled out for PglK.
Summary of LLO flippases
Both prokaryotes and eukaryotes use flippases to transport LLOs across cellular membranes. Our mechanistic understanding of LLO flipping comes from extensive biochemical and structural studies on MurJ, LtaA, Wzm-Wzt, and PglK. MurJ and LtaA utilize the classic alternating-access mechanism, while Wzm-Wzt and PglK adopt very distinct mechanisms (i.e., channel-like transportation and an outward-facing squeezing mechanism). These mechanisms for LLO flipping are likely conserved in other flippases that have yet to be identified or characterized. LLO flippases share similar features such as a charged cavity, pocket, or channel to facilitate transport of LLO polar head groups, hydrophobic grooves to capture the lipid tail, and a gating mechanism to pro- mote conformational changes. Interestingly, though, LLO molecules have evolved over time and exhibit great chemical diversity. This implies that differences in LLO transportation mechanisms and/or substrate specificity must also exist to transport different species of LLO molecules. Despite recent progress on structural and mechanistic studies of LLO flippases, no LLO flippase structures with their substrate have been reported. This is likely due to the highly flexible nature of LLOs. Therefore, the diverse proposed mechanisms for LLO flipping by different LLO flippases remains speculative. Capturing the structure of an LLO flippase bound to its substrate will help answer many open questions in the field regarding the mechanisms for LLO flipping.
CONCLUSIONS
The structural and biochemical characterization of MurJ have brought us considerable understanding of the design principles and transport mechanisms of lipid-linked oligosaccharide flippases. Recent advances to further this understanding have been very forthcoming, ranging from capability to detect specific versus annular lipid binding by mass spectrometry techniques (43), detection of transport intermediates by photo-crosslinking and biotin-tagging (90), and fluorescence anisotropy assays for high-throughput screening of binders (91). Nevertheless, many questions remain open, especially pertaining to how lipid II is actually bound by MurJ and the molecular basis of selectivity over other lipids which should not be transported (such as lipid I that is only missing a GlcNAc residue when compared to lipid II). We envision that more exciting developments are to come in this nascent field given its increasing importance as antibiotic targets. On a more fundamental level, these specialized flippase mechanisms could be the reason such elaborate polyisoprenoid-linked oligosaccharide biosynthesis pathways have evolved in the first place. The undecaprenyl tail of lipid II, if fully extended, is longer than the thickness of the membrane bilayer (25)—it is conceivable that the polyisoprenoid tail could itself be integral to the flipping mechanism such as by looping inside the flippase or in the membrane.
Supplementary Material
Supplemental movie. Conformational cycle of MurJ. The 5 crystal structures of MurJTA were interpolated in the following order: inward open, inward occluded, outward, inward closed, first inward structure, (loop back to inward open). The N-lobe (TMs 1-6) is shown in blue, the C-lobe (TMs 7-12) in green, and TMs 13-14 in brown. Front view of MurJ is shown on top while the corresponding view of the cavity from the cytosol is shown at the bottom. For simplicity, ions captured in the structures were not shown in this animation.
ACKNOWLEDGMENTS
The authors thank Ellene Mashalidis, Ziqiang Guan, and Satoshi Ichikawa for their contribution to the MurJ studies, as well as Natividad Ruiz for sharing E.coli strains. We gratefully acknowledge funding from the National Institutes of Health (R01GM120594 to S.-Y.L.). A.C.Y.K. is currently a Human Frontier Science Program (HFSP) fellow (LT000733/2020-L).
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Rush JS. 2015. Role of Flippases in Protein Glycosylation in the Endoplasmic Reticulum. Lipid Insights 8: 45–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ruiz N. 2015. Lipid Flippases for Bacterial Peptidoglycan Biosynthesis. Lipid Insights 8: 21–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Segawa K, Nagata S. 2015. An Apoptotic 'Eat Me' Signal: Phosphatidylserine Exposure. Trends Cell Biol 25: 639–50 [DOI] [PubMed] [Google Scholar]
- 4.Henderson JC, Zimmerman SM, Crofts AA, Boll JM, Kuhns LG, et al. 2016. The Power of Asymmetry: Architecture and Assembly of the Gram-Negative Outer Membrane Lipid Bilayer. Annu Rev Microbiol 70: 255–78 [DOI] [PubMed] [Google Scholar]
- 5.Pomorski T, Menon AK. 2006. Lipid flippases and their biological functions. Cell Mol Life Sci 63: 2908–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hvorup RN, Winnen B, Chang AB, Jiang Y, Zhou XF, Saier MH Jr. 2003. The multidrug/oligosaccharidyl-lipid/polysaccharide (MOP) exporter superfamily. Eur J Biochem 270: 799–813 [DOI] [PubMed] [Google Scholar]
- 7.Ruiz N. 2008. Bioinformatics identification of MurJ (MviN) as the peptidoglycan lipid II flippase in Escherichia coli. Proc Natl Acad Sci U S A 105: 15553–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Inoue A, Murata Y, Takahashi H, Tsuji N, Fujisaki S, Kato J. 2008. Involvement of an essential gene, mviN, in murein synthesis in Escherichia coli. J Bacteriol 190: 7298–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sham L-T, Butler EK, Lebar MD, Kahne D, Bernhardt TG, Ruiz N. 2014. MurJ is the flippase of lipid-linked precursors for peptidoglycan biogenesis. Science 345: 220–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu D, Cole RA, Reeves PR. 1996. An O-antigen processing function for Wzx (RfbX): a promising candidate for O-unit flippase. J Bacteriol 178: 2102–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Islam ST, Fieldhouse RJ, Anderson EM, Taylor VL, Keates RA, et al. 2012. A cationic lumen in the Wzx flippase mediates anionic O-antigen subunit translocation in Pseudomonas aeruginosa PAO1. Mol Microbiol 84: 1165–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Islam ST, Eckford PD, Jones ML, Nugent T, Bear CE, et al. 2013. Proton-dependent gating and proton uptake by Wzx support O-antigen-subunit antiport across the bacterial inner membrane. MBio 4: e00678–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu MA, Morris P, Reeves PR. 2018. Wzx flippases exhibiting complex O-unit preferences require a new model for Wzx-substrate interactions. Microbiologyopen: e00655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hong Y, Liu MA, Reeves PR. 2018. Progress in Our Understanding of Wzx Flippase for Translocation of Bacterial Membrane Lipid-Linked Oligosaccharide. J Bacteriol 200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Helenius J, Ng DT, Marolda CL, Walter P, Valvano MA, Aebi M. 2002. Translocation of lipid-linked oligosaccharides across the ER membrane requires Rft1 protein. Nature 415: 447–50 [DOI] [PubMed] [Google Scholar]
- 16.Frank CG, Sanyal S, Rush JS, Waechter CJ, Menon AK. 2008. Does Rft1 flip an N-glycan lipid precursor? Nature 454: E3–4; discussion E4-5 [DOI] [PubMed] [Google Scholar]
- 17.Sanyal S, Frank CG, Menon AK. 2008. Distinct flippases translocate glycerophospholipids and oligosaccharide diphosphate dolichols across the endoplasmic reticulum. Biochemistry 47: 7937–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jelk J, Gao N, Serricchio M, Signorell A, Schmidt RS, et al. 2013. Glycoprotein biosynthesis in a eukaryote lacking the membrane protein Rft1. J Biol Chem 288: 20616–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kusakizako T, Miyauchi H, Ishitani R, Nureki O. 2020. Structural biology of the multidrug and toxic compound extrusion superfamily transporters. Biochim Biophys Acta Biomembr 1862: 183154. [DOI] [PubMed] [Google Scholar]
- 20.Morita Y, Kodama K, Shiota S, Mine T, Kataoka A, et al. 1998. NorM, a putative multidrug efflux protein, of Vibrio parahaemolyticus and its homolog in Escherichia coli. Antimicrob Agents Chemother 42: 1778–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim JG, Park BK, Kim SU, Choi D, Nahm BH, et al. 2006. Bases of biocontrol: sequence predicts synthesis and mode of action of agrocin 84, the Trojan horse antibiotic that controls crown gall. Proc Natl Acad Sci U S A 103: 8846–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nurnberg P, Thiele H, Chandler D, Hohne W, Cunningham ML, et al. 2001. Heterozygous mutations in ANKH, the human ortholog of the mouse progressive ankylosis gene, result in craniometaphyseal dysplasia. Nat Genet 28: 37–41 [DOI] [PubMed] [Google Scholar]
- 23.El Zoeiby A, Sanschagrin F, Levesque RC. 2003. Structure and function of the Mur enzymes: development of novel inhibitors. Mol Microbiol 47: 1–12 [DOI] [PubMed] [Google Scholar]
- 24.Ikeda M, Wachi M, Jung HK, Ishino F, Matsuhashi M. 1991. The Escherichia coli mraY gene encoding UDP-N-acetylmuramoyl-pentapeptide: undecaprenyl-phosphate phospho-N-acetylmuramoyl-pentapeptide transferase. J Bacteriol 173: 1021–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chung BC, Zhao J, Gillespie RA, Kwon DY, Guan Z, et al. 2013. Crystal structure of MraY, an essential membrane enzyme for bacterial cell wall synthesis. Science 341: 1012–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mengin-Lecreulx D, Texier L, Rousseau M, van Heijenoort J. 1991. The murG gene of Escherichia coli codes for the UDP-N-acetylglucosamine: N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase involved in the membrane steps of peptidoglycan synthesis. J Bacteriol 173: 4625–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meeske AJ, Riley EP, Robins WP, Uehara T, Mekalanos JJ, et al. 2016. SEDS proteins are a widespread family of bacterial cell wall polymerases. Nature [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cho H, Wivagg CN, Kapoor M, Barry Z, Rohs PD, et al. 2016. Bacterial cell wall biogenesis is mediated by SEDS and PBP polymerase families functioning semi-autonomously. Nat Microbiol: 16172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Emami K, Guyet A, Kawai Y, Devi J, Wu LJ, et al. 2017. RodA as the missing glycosyltransferase in Bacillus subtilis and antibiotic discovery for the peptidoglycan polymerase pathway. Nat Microbiol 2: 16253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jackson GE, Strominger JL. 1984. Synthesis of peptidoglycan by high molecular weight penicillin-binding proteins of Bacillus subtilis and Bacillus stearothermophilus. J Biol Chem 259: 1483–90 [PubMed] [Google Scholar]
- 31.Pollock JJ, Ghuysen JM, Linder R, Salton MR, Perkins HR, et al. 1972. Transpeptidase activity of Streptomyces D-alanyl-D carboxypeptidases. Proc Natl Acad Sci U S A 69: 662–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.El Ghachi M, Bouhss A, Blanot D, Mengin-Lecreulx D. 2004. The bacA gene of Escherichia coli encodes an undecaprenyl pyrophosphate phosphatase activity. J Biol Chem 279: 30106–13 [DOI] [PubMed] [Google Scholar]
- 33.Manat G, Roure S, Auger R, Bouhss A, Barreteau H, et al. 2014. Deciphering the metabolism of undecaprenyl-phosphate: the bacterial cell-wall unit carrier at the membrane frontier. Microb Drug Resist 20: 199–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.El Ghachi M, Howe N, Huang CY, Olieric V, Warshamanage R, et al. 2018. Crystal structure of undecaprenyl-pyrophosphate phosphatase and its role in peptidoglycan biosynthesis. Nat Commun 9: 1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Workman SD, Worrall LJ, Strynadka NCJ. 2018. Crystal structure of an intramembranal phosphatase central to bacterial cell-wall peptidoglycan biosynthesis and lipid recycling. Nat Commun 9: 1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Butler EK, Davis RM, Bari V, Nicholson PA, Ruiz N. 2013. Structure-function analysis of MurJ reveals a solvent-exposed cavity containing residues essential for peptidoglycan biogenesis in Escherichia coli. J Bacteriol 195: 4639–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Butler EK, Tan WB, Joseph H, Ruiz N. 2014. Charge requirements of lipid II flippase activity in Escherichia coli. J Bacteriol 196: 4111–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mohamed YF, Valvano MA. 2014. A Burkholderia cenocepacia MurJ (MviN) homolog is essential for cell wall peptidoglycan synthesis and bacterial viability. Glycobiology 24: 564–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qiao Y, Srisuknimit V, Rubino F, Schaefer K, Ruiz N, et al. 2017. Lipid II overproduction allows direct assay of transpeptidase inhibition by beta-lactams. Nat Chem Biol 13: 793–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rubino FA, Kumar S, Ruiz N, Walker S, Kahne DE. 2018. Membrane Potential Is Required for MurJ Function. J Am Chem Soc 140: 4481–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuk ACY, Hao A, Guan Z, Lee SY. 2019. Visualizing conformation transitions of the Lipid II flippase MurJ. Nat Commun 10: 1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bolla JR, Sauer JB, Wu D, Mehmood S, Allison TM, Robinson CV. 2018. Direct observation of the influence of cardiolipin and antibiotics on lipid II binding to MurJ. Nat Chem 10: 363–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bolla JR, Corey RA, Sahin C, Gault J, Hummer A, et al. 2020. A Mass-Spectrometry-Based Approach to Distinguish Annular and Specific Lipid Binding to Membrane Proteins. Angew Chem Int Ed Engl 59: 3523–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu X, Meiresonne NY, Bouhss A, den Blaauwen T. 2018. FtsW activity and lipid II synthesis are required for recruitment of MurJ to midcell during cell division in Escherichia coli. Mol Microbiol 109: 855–84 [DOI] [PubMed] [Google Scholar]
- 45.Monteiro JM, Pereira AR, Reichmann NT, Saraiva BM, Fernandes PB, et al. 2018. Peptidoglycan synthesis drives an FtsZ-treadmilling-independent step of cytokinesis. Nature 554: 528–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miyauchi H, Moriyama S, Kusakizako T, Kumazaki K, Nakane T, et al. 2017. Structural basis for xenobiotic extrusion by eukaryotic MATE transporter. Nat Commun 8: 1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He X, Szewczyk P, Karyakin A, Evin M, Hong WX, et al. 2010. Structure of a cation-bound multidrug and toxic compound extrusion transporter. Nature 467: 991–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tanaka Y, Hipolito CJ, Maturana AD, Ito K, Kuroda T, et al. 2013. Structural basis for the drug extrusion mechanism by a MATE multidrug transporter. Nature 496: 247–51 [DOI] [PubMed] [Google Scholar]
- 49.Lu M, Radchenko M, Symersky J, Nie R, Guo Y. 2013. Structural insights into H+-coupled multidrug extrusion by a MATE transporter. Nat Struct Mol Biol 20: 1310–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu M, Symersky J, Radchenko M, Koide A, Guo Y, et al. 2013. Structures of a Na+-coupled, substrate-bound MATE multidrug transporter. Proc Natl Acad Sci U S A 110: 2099–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Steed PR, Stein RA, Mishra S, Goodman MC, McHaourab HS. 2013. Na(+)-substrate coupling in the multidrug antiporter norm probed with a spin-labeled substrate. Biochemistry 52: 5790–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Claxton DP, Jagessar KL, Steed PR, Stein RA, McHaourab HS. 2018. Sodium and proton coupling in the conformational cycle of a MATE antiporter from Vibrio cholerae. Proc Natl Acad Sci U S A 115: E6182–e90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kumar S, Rubino FA, Mendoza AG, Ruiz N. 2019. The bacterial lipid II flippase MurJ functions by an alternating-access mechanism. J Biol Chem 294: 981–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuk ACY, Mashalidis EH, Lee SY. 2017. Crystal structure of the MOP flippase MurJ in an inward-facing conformation. Nat Struct Mol Biol 24: 171–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mousa JJ, Yang Y, Tomkovich S, Shima A, Newsome RC, et al. 2016. MATE transport of the E. coli-derived genotoxin colibactin. Nature Microbiology 1: 15009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zheng S, Sham LT, Rubino FA, Brock KP, Robins WP, et al. 2018. Structure and mutagenic analysis of the lipid II flippase MurJ from Escherichia coli. Proc Natl Acad Sci U S A 115: 6709–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Perez C, Gerber S, Boilevin J, Bucher M, Darbre T, et al. 2015. Structure and mechanism of an active lipid-linked oligosaccharide flippase. Nature 524: 433–8 [DOI] [PubMed] [Google Scholar]
- 58.Zakrzewska S, Mehdipour AR, Malviya VN, Nonaka T, Koepke J, et al. 2019. Inward-facing conformation of a multidrug resistance MATE family transporter. Proc Natl Acad Sci U S A 116: 12275–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morita Y, Kataoka A, Shiota S, Mizushima T, Tsuchiya T. 2000. NorM of vibrio parahaemolyticus is an Na(+)-driven multidrug efflux pump. J Bacteriol 182: 6694–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Otsuka M, Yasuda M, Morita Y, Otsuka C, Tsuchiya T, et al. 2005. Identification of essential amino acid residues of the NorM Na+/multidrug antiporter in Vibrio parahaemolyticus. J Bacteriol 187: 1552–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jin Y, Nair A, van Veen HW. 2014. Multidrug transport protein norM from vibrio cholerae simultaneously couples to sodium- and proton-motive force. J Biol Chem 289: 14624–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ficici E, Zhou W, Castellano S, Faraldo-Gomez JD. 2018. Broadly conserved Na(+)-binding site in the N-lobe of prokaryotic multidrug MATE transporters. Proc Natl Acad Sci U S A 115: E6172–e81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chu J, Vila-Farres X, Inoyama D, Ternei M, Cohen LJ, et al. 2016. Discovery of MRSA active antibiotics using primary sequence from the human microbiome. Nat Chem Biol 12: 1004–06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chu J, Vila-Farres X, Inoyama D, Gallardo-Macias R, Jaskowski M, et al. 2018. Human Microbiome Inspired Antibiotics with Improved beta-Lactam Synergy against MDR Staphylococcus aureus. ACS Infect Dis 4: 33–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chamakura KR, Sham LT, Davis RM, Min L, Cho H, et al. 2017. A viral protein antibiotic inhibits lipid II flippase activity. Nat Microbiol 2: 1480–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rumnieks J, Tars K. 2012. Diversity of pili-specific bacteriophages: genome sequence of IncM plasmid-dependent RNA phage M. BMC Microbiol 12: 277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chamakura K, Young R. 2019. Phage single-gene lysis: Finding the weak spot in the bacterial cell wall. J Biol Chem 294: 3350–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mott JE, Shaw BA, Smith JF, Bonin PD, Romero DL, et al. 2008. Resistance mapping and mode of action of a novel class of antibacterial anthranilic acids: evidence for disruption of cell wall biosynthesis. J Antimicrob Chemother 62: 720–9 [DOI] [PubMed] [Google Scholar]
- 69.Huber J, Donald RG, Lee SH, Jarantow LW, Salvatore MJ, et al. 2009. Chemical genetic identification of peptidoglycan inhibitors potentiating carbapenem activity against methicillin-resistant Staphylococcus aureus. Chem Biol 16: 837–48 [DOI] [PubMed] [Google Scholar]
- 70.Ruiz N 2009. Streptococcus pyogenes YtgP (Spy_0390) complements Escherichia coli strains depleted of the putative peptidoglycan flippase MurJ. Antimicrob Agents Chemother 53: 3604–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mashalidis EH, Lee SY. 2020. Structures of Bacterial MraY and Human GPT Provide Insights into Rational Antibiotic Design. J Mol Biol 432: 4946–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Iguchi A, Iyoda S, Kikuchi T, Ogura Y, Katsura K, et al. 2015. A complete view of the genetic diversity of the Escherichia coli O-antigen biosynthesis gene cluster. DNA Res 22: 101–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Islam ST, Lam JS. 2013. Wzx flippase-mediated membrane translocation of sugar polymer precursors in bacteria. Environ Microbiol 15: 1001–15 [DOI] [PubMed] [Google Scholar]
- 74.Marolda CL, Li B, Lung M, Yang M, Hanuszkiewicz A, et al. 2010. Membrane topology and identification of critical amino acid residues in the Wzx O-antigen translocase from Escherichia coli O157:H4. J Bacteriol 192: 6160–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Elhenawy W, Davis RM, Fero J, Salama NR, Felman MF, Ruiz N. 2016. The O-Antigen Flippase Wzk Can Substitute for MurJ in Peptidoglycan Synthesis in Helicobacter pylori and Escherichia coli. PLoS One 11: e0161587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sham LT, Zheng S, Yakhnina AA, Kruse AC, Bernhardt TG. 2018. Loss of specificity variants of WzxC suggest that substrate recognition is coupled with transporter opening in MOP-family flippases. Mol Microbiol 109: 633–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Damjanovic M, Kharat AS, Eberhardt A, Tomasz A, Vollmer W. 2007. The essential tacF gene is responsible for the choline-dependent growth phenotype of Streptococcus pneumoniae. J Bacteriol 189: 7105–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Percy MG, Grundling A. 2014. Lipoteichoic acid synthesis and function in gram-positive bacteria. Annu Rev Microbiol 68: 81–100 [DOI] [PubMed] [Google Scholar]
- 79.Alaimo C, Catrein I, Morf L, Marolda CL, Callewaert N, et al. 2006. Two distinct but interchangeable mechanisms for flipping of lipid-linked oligosaccharides. EMBO J 25: 967–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hong Y, Reeves PR. 2014. Diversity of o-antigen repeat unit structures can account for the substantial sequence variation of wzx translocases. J Bacteriol 196: 1713–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Grundling A, Schneewind O. 2007. Genes required for glycolipid synthesis and lipoteichoic acid anchoring in Staphylococcus aureus. J Bacteriol 189: 2521–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang B, Liu X, Lambert E, Mas G, Hiller S, et al. 2020. Structure of a proton-dependent lipid transporter involved in lipoteichoic acids biosynthesis. Nat Struct Mol Biol 27: 561–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Caffalette CA, Corey RA, Sansom MSP, Stansfeld PJ, Zimmer J. 2019. A lipid gating mechanism for the channel-forming O antigen ABC transporter. Nat Commun 10: 824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bi Y, Mann E, Whitfield C, Zimmer J. 2018. Architecture of a channel-forming O-antigen polysaccharide ABC transporter. Nature 553: 361–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Caffalette CA, Zimmer J. 2021. Cryo-EM structure of the full-length WzmWzt ABC transporter required for lipid-linked O antigen transport. Proc Natl Acad Sci U S A 118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Raetz CR, Whitfield C. 2002. Lipopolysaccharide endotoxins. Annu Rev Biochem 71: 635–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hug I, Feldman MF. 2011. Analogies and homologies in lipopolysaccharide and glycoprotein biosynthesis in bacteria. Glycobiology 21: 138–51 [DOI] [PubMed] [Google Scholar]
- 88.Caffalette CA, Kuklewicz J, Spellmon N, Zimmer J. 2020. Biosynthesis and Export of Bacterial Glycolipids. Annu Rev Biochem 89: 741–68 [DOI] [PubMed] [Google Scholar]
- 89.Perez C, Mehdipour AR, Hummer G, Locher KP. 2019. Structure of Outward-Facing PglK and Molecular Dynamics of Lipid-Linked Oligosaccharide Recognition and Translocation. Structure 27: 669–78 e5 [DOI] [PubMed] [Google Scholar]
- 90.Rubino FA, Mollo A, Kumar S, Butler EK, Ruiz N, et al. 2020. Detection of Transport Intermediates in the Peptidoglycan Flippase MurJ Identifies Residues Essential for Conformational Cycling. J Am Chem Soc 142: 5482–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Boes A, Olatunji S, Mohammadi T, Breukink E, Terrak M. 2020. Fluorescence anisotropy assays for high throughput screening of compounds binding to lipid II, PBP1b, FtsW and MurJ. Sci Rep 10: 6280. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental movie. Conformational cycle of MurJ. The 5 crystal structures of MurJTA were interpolated in the following order: inward open, inward occluded, outward, inward closed, first inward structure, (loop back to inward open). The N-lobe (TMs 1-6) is shown in blue, the C-lobe (TMs 7-12) in green, and TMs 13-14 in brown. Front view of MurJ is shown on top while the corresponding view of the cavity from the cytosol is shown at the bottom. For simplicity, ions captured in the structures were not shown in this animation.