Abstract
Background
MicroRNAs influence the growth and metastasis of breast cancer (BC) by regulating their target genes. Our study aims to screen and identify miRNAs that are closely related to the development of breast cancer, and explore the role of these miRNAs and their target genes in breast cancer.
Methods
Bioinformatics tools were applied to screen breast cancer-associated miRNAs and predict their potential target genes. Serum miRNAs were measured using RT-PCR. The correlation between miRNA expression and different clinicopathological features of BC patients was analyzed. Receiver operating characteristic (ROC) curve was used to evaluate the diagnostic value. GEPIA, Kaplan-Meier Plotter, TIMER, and TISIDB databases were used to validate the expression levels and their prognostic value, as well as their target gene associated with immune infiltrating cells and immune checkpoints.
Results
Breast cancer-associated serum miR-338-3p and miR-501-3p were screened and verified for the first time. Serum miR-501-3p was elevated in BC and was closely linked to the ki-67 index and histological grade. CDKN2C, as a potential target gene of miR-501-3p, was enriched in the cGMP-PKG signaling pathway. Serum miR-338-3p was reduced in BC and was strongly linked to lymph node metastasis and histological grading. ACTR2, CDH1, COL1A1, RBBP5, RRM1, and TPM3, as potential target genes of miR-338-3p, were enriched in MAPK, PI3K-Akt, and RAS signaling pathways. These target genes were found to be linked to breast cancer prognosis, immune infiltrating cells, and immune checkpoint inhibitors. Analysis of ROC curve showed that serum miR-501-3p combined with serum miR-338-3p had a high diagnostic value in breast cancer (AUC: 0.89, 95% CI: 0.821–0.958).
Conclusion
Serum miR-501-3p combined with serum miR-338-3p show obvious clinical significance in the diagnosis and prognosis of breast cancer, which suggests that they may act as novel diagnostic biomarkers for breast cancer.
Keywords: breast cancer, miR-501-3p, miR-338-3p, serum biomarker, bioinformatics analysis
Introduction
According to statistics, breast cancer (BC) is a malignant tumor that seriously endangers women’s health.1,2 Its incidence has surpassed lung cancer as the most prevalent cancer worldwide.3,4 The early symptoms of this disease are not obvious, and the best treatment period is often missed because the diagnosis is not timely. Thus, further study of the molecular mechanism of breast cancer and finding potential therapeutic targets and biomarkers is essential.
As we all know, microRNAs (miRNAs) influence the biological processes of disease by regulating multiple oncogenes or tumor suppressor genes,5 which may be an important cause of tumor pathogenesis. Recent studies have proved that miRNA expression is strongly associated with the biological behavior and clinicopathological features of breast cancer.6 For example, miR-92b-3p is increased in breast cancer, and its expression is also associated with tumor size, differentiation degree, TNM stage, lymphatic node metastasis, and survival status.7 miRNA-621 can make BC sensitive to paclitaxel plus carboplatin by inhibiting FBXO11 and enhancing p53 activity.8 The above results show that miRNA has a critical part in the diagnosis, prognosis, and treatment of BC. In addition, miRNAs can be stably present and more easily detected in the blood,9 therefore, serum miRNAs are considered promising biomarkers for BC.
In this study, miR-501-3p and miR-338-3p were preliminarily screened as target miRNAs by bioinformatics methods and analyzed their potential targets and possible mechanisms for the first time. In addition, we also detected the serum miR-501-3p and miR-338-3p in BC patients to explore their relationship with clinicopathological features of breast cancer patients and diagnostic value.
Materials and Methods
Serum Samples and Study Subjects
A vacuum centrifuge tube containing blood samples was centrifuged at 3500 r/min for 5 min. The serum was collected in 1.5 mL RNase-free tubes and immediately stored in a −80°C freezer until further analysis.
68 female patients with breast disease and 34 healthy patients with normal physical examination and no previous breast disease or other diseases were selected as subjects from October 2021 to May 2022 in the Department of Breast and Thyroid Surgery of Weifang People’s Hospital of Shandong Province. Inclusion criteria for patients with breast diseases: all patients were confirmed by pathological examination; no antibiotics were used within 3 months before admission; did not receive chemoradiotherapy or other immunotherapy before surgery; no history of other cancers, serious cardiovascular, lung, kidney, or other diseases; patients with complete clinical data.
The flow chart of bioinformatics analysis in this study is shown in Figure 1.
Figure 1.
Flow chart of this study.
Datasets and Screening Target miRNAs
The GSE41922, GSE83270, and GSE45666 datasets were downloaded and screened for differentially expressed miRNAs using the GEO2R tool. The GSE41922 dataset included serum samples from 32 BC patients and 22 normal healthy individuals; the GSE83270 dataset included whole blood samples from 6 BC patients and 6 normal healthy controls; the GSE4566 dataset included 101 breast cancer tissues and 15 paraneoplastic tissue samples. The cutoff criterion to filter miRNAs in GSE41922 and GSE45666 datasets as P<0.05, |log fold change (FC)|≥1; and the GSE83270 datasets as P<0.05, |log fold change (FC)|≥1.5. Among different miRNAs, we selected the two most valuable miRNAs that were simultaneously low or high expressed in two or more datasets as candidate miRNAs based on the relevant literature.
Bioinformatics Analysis
Potential target genes of candidate miRNAs were identified using the intersection of three databases (TargetScan, http://www.targetscan.org, mirwalk, http://mirwalk.uni-hd.de/, and mirDIP, http://ophid.utoronto.ca/). GO enrichment and KEGG signaling pathway was performed using the DAVID database (https://david.ncifcrf.gov/) with a screening criterion of P<0.05. In addition, The STRING database (https://string-db.org/) was used to predict target gene interactions and Cytoscape software was used to screen the top 50 hub target genes. GEPIA (http://gepia.cancer-pku.cn/) website was used to analyze differential expression of target genes of miRNA in breast cancer. The prognostic value of key target genes of miRNA in BC according to relapse-free survival (RFS) was assessed by The Kaplan-Meier Plotter (https://kmplot.com/analysis/) website. The expression levels of key target genes of miRNA in BC were assessed by the TIMER database (https://cistrome.shinyapps.io/timer/) with six tumor-infiltrating immune cells (CD8+ T cells, CD4+ T cells, macrophages, B cells, dendritic cells, neutrophils) were correlated. In order to further understand the relationship between candidate genes and tumor immunity, the TISIDB database (http://cis.hku.hk/TISIDB/) was used to investigate the relationship between the key target genes and immune checkpoint inhibitors.
The Expression Levels of miRNA in Serum
MiRNA of Serum was extracted according to the EasyPure® miRNA Kit reagent instructions (Beijing TransGen Biotech Co., Ltd). Total miRNA was reverse-transcribed into cDNA at 37°C for 60 min and 85°C for 5s using TransScript® miRNA first-strand cDNA Synthesis SuperMix. Finally, real-time fluorescence quantitative polymerase chain reaction was performed using PerfectStart® Green qPCR SuperMix kit (SYBR Green). Reaction conditions: 94°C for 30s, 94°C for 5s, and 60°C for 30s (45 cycles). All reactions were set up with two replicate wells to reduce errors caused by sample addition, etc. to affect the experimental results. Melt curve analysis was performed using ABI QuantStudio5 software to detect reaction specificity. The relative quantification method (with cel-miR-39-3p as a reference) was used to determine the expression level of miRNA in the serum. Relative expression was determined using 2-∆∆Ct. The primer was obtained from Sangon Biotech (Shanghai) Co., Ltd. The primer sequences were cel-miR-39-3p: 5’-UCA CCG GGU GUA AAU CAG CUU G-3’; miR-338-3p: 5’-UCC AGC AUC AGU GAU UUU GUU G-3’; miR-501-3p:5’-AAU GCA CCC GGG CAA GGA UUC U-3’.
Survival Analysis
An online analytical tool (Kaplan-Meier Plotter) was used to assess miRNA for breast cancer survival at 10 years. According to the median, they were divided into a high-expression group and a low-expression group, and P<0.05 was statistically significant.
Serum CA153 Quantification
The common tumor marker (CA153) of breast cancer was detected by Cobas E601 full-automatic electrochemistry luminescence immunity analyzer.
Statistical Methods
All data were carried out using SPSS 26.0 and GraphPad Prism 9.0. Shapiro Wilk was used to test the normality of the data. Univariate analysis of variance (ANOVA) was used for the three groups of data conforming to a normal distribution and with equal variances; otherwise, the non-parametric Mann–Whitney H-test was used. A t-test was used for the two groups of data conforming to a normal distribution, and a non-parametric Mann–Whitney U-test was used the other way around. ROC curves were used to assess the diagnostic efficacy of serum miRNAs and CA153 in breast cancer. In all data results, P < 0.05 was statistically significant.
Results
Clinicopathological Data
All patients were diagnosed and pathologically confirmed by clinicians. Including 24 cases in the benign tumor group (LC), all patients with breast fibroadenoma, aged 37–71 years, mean (49.17±10.09) years, and 44 cases in the breast cancer group (BC), aged 34–70 years, mean (51.67±9.98) years, all with invasive ductal carcinoma. Thirty-four healthy individuals, aged 34–71 years, with a mean of (51.07±9.64) were randomly selected for the healthy control group (HC) from the health screening center check-ups during the same period. All three groups were female. Age was not statistically significant in the three groups (P>0.05).
Screening Target miRNAs
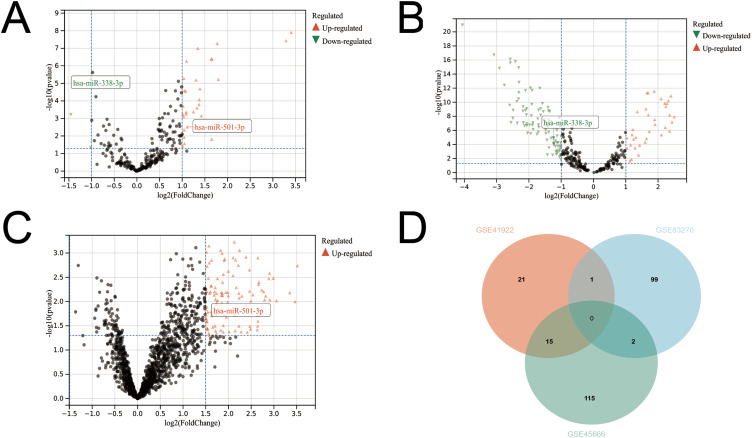
We performed a comprehensive analysis of three GEO datasets (GSE41922, GSE45666, and GSE83270). We found 37 differentially expressed miRNAs in the GSE41922 dataset, including 34 up-regulated and 3 down-regulated miRNAs (Figure 2A), 132 differentially expressed miRNAs in the GSE45666 dataset, including 36 up-regulated and 96 down-regulated miRNAs (Figure 2B), and 102 upregulated differentially expressed miRNAs in the GSE38270 dataset (Figure 2C). Taking the intersection of the three datasets two by two, we obtained a total of 18 miRNAs that were differentially expressed in both datasets (Figure 2D), including miR-10a/b, miR-143, miR-154, miR-671-5p, miR-451, miR-486-5p, miR-214, and miR-133b, highly expressed in the GSE41922 dataset but lowly expressed in the GSE45666 dataset, were excluded; miR-338-3p was simultaneously lowly expressed in the GSE41922 and GSE45666 datasets, and miR-501-3p was simultaneously highly expressed in the GSE41922 and GSE83270 datasets miR-140-3p/5p was highly expressed in GSE45666 and GSE83270 datasets, and miR-182, miR-185, miR-16-2, miR-96, and miR-7 were highly expressed in GSE41922 and GSE45666 datasets (Supplementary Tables 1–3). After combining the miRNAs with the literature, we found miR-338-3p and miR-501-3p play an important role in cancer, and no one has studied their clinical value in breast cancer sera. Therefore, we determined up-regulated miR-501-3p and down-regulated miR-338-3p as candidate miRNAs for subsequent analysis. In addition, the bar graphs of miR-501-3p in the GSE41922 and GSE83270 are shown in Supplementary Figure 1A and B. The bar figures of miR-338-3p in the GSE41922 and GSE45666 are shown in Supplementary Figure 1A and C.
Figure 2.
Screening of differentially expressed miRNAs (A) GSE41922 dataset (B) GSE45666 dataset (C) GSE83270 dataset (D) Venn diagram of three datasets.
Bioinformatics Analysis of miRNA
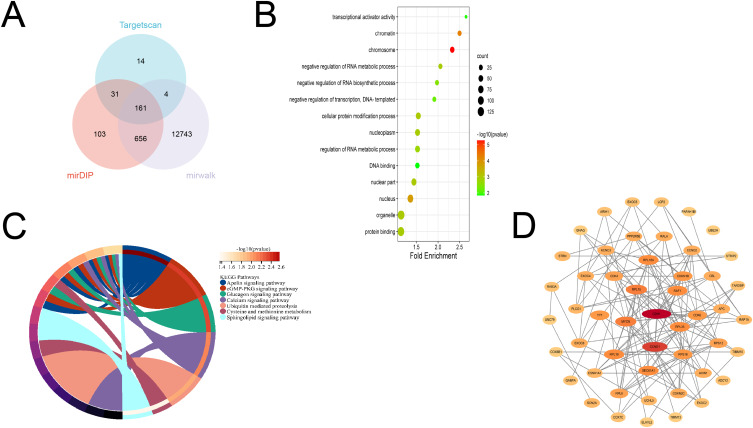
To explore the mechanism of miR-501-3p and miR-338-3p, firstly, their target genes were predicted using mirDIP, mirwalk, and targetscan databases. Next, we performed the functional annotation, KEGG enrichment analysis, and transcription factor prediction for common target genes. In addition, many studies have shown that genes with higher node connectivity (degree) may have a vital part in the network. Therefore, we used Cytoscape software to screen the top 50 hub genes in the PPI network.
miR-501-3p was predicted by the database to obtain 161 common target genes (Figure 3A). GO functional analysis showed (Figure 3B) that biological processes (BP) were involved in the negative regulation of RNA metabolism, negative regulation of transcription, and DNA template; cellular components (CC) were mainly involved in organelles, nuclear plasma, chromosomes; molecular functional aspects (MF) were mainly involved in protein binding, DNA binding, transcriptional activation activity. KEGG signaling pathway analysis (Figure 3C) showed that the target genes were involved in the cGMP-PKG signaling pathway, glucagon signaling pathway, and calcium signaling pathway. In addition, The PPI network uses Cytoscape for visualization. The top 50 hub genes of miR-501-3p were screened using node connectivity (degree) of Cytoscape software (Figure 3D and Supplementary Table 4), meaning that these target genes may play a more important role.
Figure 3.
Bioinformatics analysis of miR-501-3p (A) Venn diagram (B) GO (C) KEGG (D) Transcription factor prediction.
miR-338-3p was predicted by the database to obtain 266 common target genes. (Figure 4A) GO functional analysis showed (Figure 4B) that BP is enriched in the regulation of signal transduction, regulation of RNA metabolic process, and regulation of RNA polymerase II promoter transcription; the CC is enriched in the cytoplasm, nuclear fraction, and intracellular organelles; MF is mainly involved in protein serine/threonine kinase activity, phosphoprotein phosphatase KEGG signaling pathway analysis (Figure 4C) showed that the target genes were involved in MAPK, Ras, ErbB, PI3K-Akt, and other signaling pathways. In addition, the top 50 hub genes of miR-338-3p were screened using node connectivity (degree) of Cytoscape software (Figure 4D), indicating that these target genes may play a more important role.
Figure 4.
Bioinformatics analysis of miR-338-3p (A) Venn diagram (B) GO (C) KEGG (D) transcription factor prediction.
Expression Levels and Prognostic Value of Target Genes
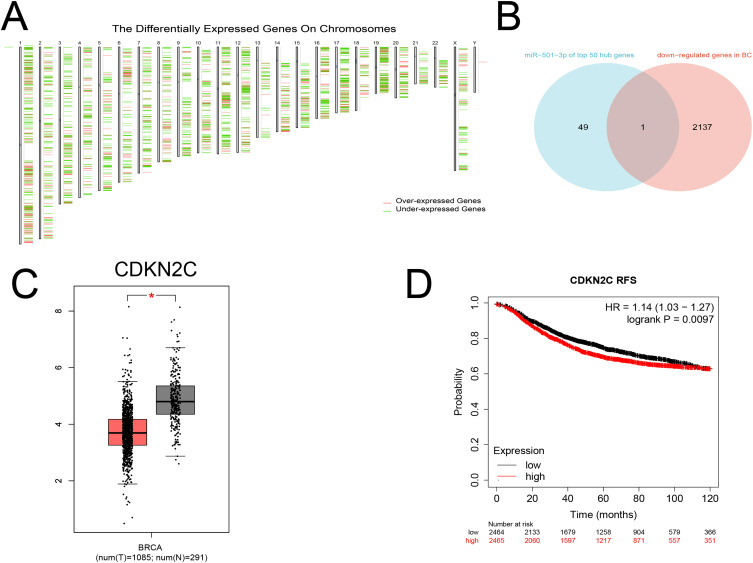
There is abundant evidence that miRNAs negatively regulate target genes, so the GEIPA database was used to identify differentially expressed genes between breast cancer samples and normal samples (screening criteria: |Log FC|>1, P<0.05), and obtained a total of 2138 down-regulated and 1418 up-regulated genes (Figure 5A).
Figure 5.
Candidate target genes of miR-501-3p and their prognostic value. (A) Differentially expressed genes. (B) Venn diagram for CDKN2C. (C) Expression levels of CDKN2C. (D) Correlation between CDKN2C with RFS. * P<0.05.
Bioinformatics analysis showed that miR-501-3p was increased in BC, so we intersected the top 50 hub genes in the PPI network with 2138 downregulated target genes to obtain a total of one downregulated key target gene (CDKN2C). (Figure 5B) In the GEPIA database, CDKN2C was under-expressed in breast cancer patient samples compared with normal samples (P<0.05) (Figure 5C). Kaplan-Meier Plotter showed that the RFS of the high CDKN2C expression group was lower than that of the low CDKN2C expression group (P<0.05) (Figure 5D).
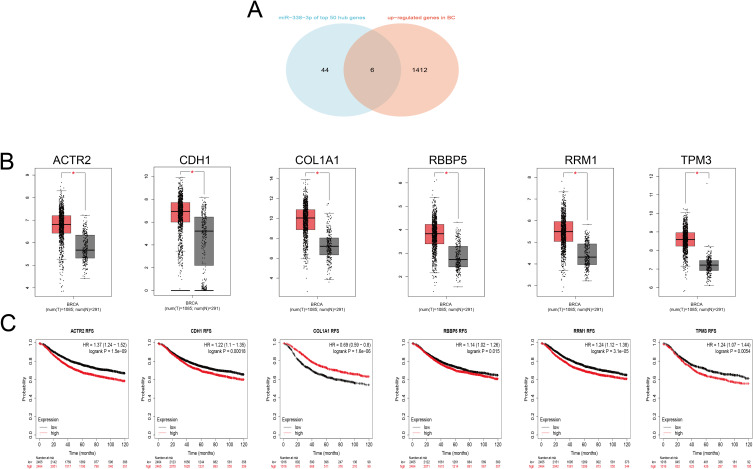
Bioinformatics analysis showed that miR-338-3p was decreased in BC, so we intersected the top 50 hub genes in the PPI network with 1418 upregulated target genes to obtain a total of six upregulated key target genes (ACTR2, CDH1, COL1A1, RBBP5, RRM1, TPM3). (Figure 6A) In the GEPIA database, compared with normal samples. These six genes were highly expressed in BC patient samples (p<0.05) (Figure 6B). Kaplan-Meier Plotter showed that the RFS of breast cancer patients in high expression groups of ACTR2, CDH1, RBBP5, RRM1, and TPM3 were lower than those in low expression group (P<0.05) (Figure 6C).
Figure 6.
Candidate target genes of miR-338-3p and their prognostic value. (A) Venn diagram for ACTR2, CDH1, COL1A1, RBBP5, RRM1, and TPM3. (B) Expression levels of ACTR2, CDH1, COL1A1, RBBP5, RRM1, and TPM3. (C) Correlation of ACTR2, CDH1, COL1A1, RBBP5, RRM1, and TPM3 with RFS. * P<0.05.
Correlation of Seven Key Target Genes with Tumor-Infiltrating Immune Cells
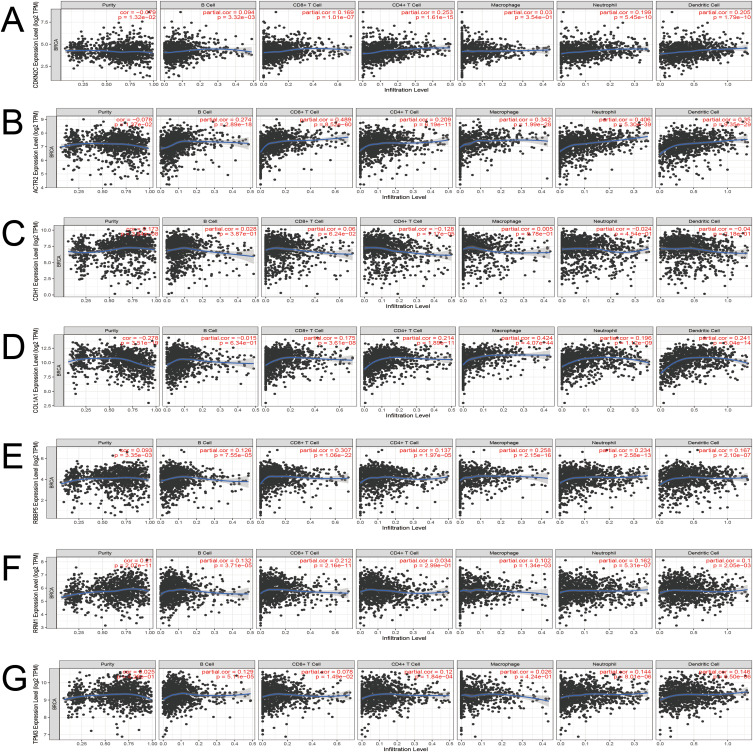
TIMER database analysis showed that ACTR2 expression was positively correlated with CD8+T, neutrophils, dendritic cells, and macrophages; RBBP5 expression was positively correlated with CD8+T; Other key target genes such as CDKN2C, CDH1, COL1A1, RRM1, and TPM3 were also associated with immune infiltrating cells at a low level (Figure 7). This suggests that ACTR2 and RBBP5 have a more important value in the treatment of BC, other target genes may play a smaller role. In conclusion, these target genes all play a regulatory role in BC tumor immunity.
Figure 7.
Association of (A) CDKN2C, (B) ACTR2, (C) CDH1, (D) COL1A1, (E) RBBP5, (F) RRM1, and (G) TPM3 with immunoinfiltrating cells.
Correlation of Seven Key Target Genes with Immune Checkpoint Inhibitors
To further understand the relationship between these seven key target genes and tumor immunity, we used the TISIDB database to analyze them for immune checkpoint inhibitors. The results show that CDKN2C was positively correlated with PD-1, LAG3, IDO1, and CTLA4. ACTR2 was positively correlated with CD724, PDCD1LG2, and TGFBR1. CDH1 was negatively correlated with CD244, CSF1R, PD-1, and VTCN1. COL1A1 has a significant positive correlation with TGFB1 and TGFBR1, but a low correlation with CSF1R and HAVCR2. RBBP5 is positively correlated with TGFB1 and TGFR1 but negatively correlated with LAG3 and PDCD1. RRM1 was negatively correlated with CD244, CSF1R, PD-1, and TGFB1. TPM3 has a low negative correlation with CSF1R, TGFB1, and KDR, and a low positive correlation with IL10RB. Our study confirms that these target genes are involved in the immune regulation of BC (Figure 8).
Figure 8.
Association of (A) CDKN2C, (B) ACTR2, (C) CDH1, (D) COL1A1, (E) RBBP5, (F) RRM1, and (G) TPM3 with Immune checkpoint inhibitors.
Notes: Heat map - target genes with all immune checkpoints, scatter map - target genes with the four most significant immune checkpoints.
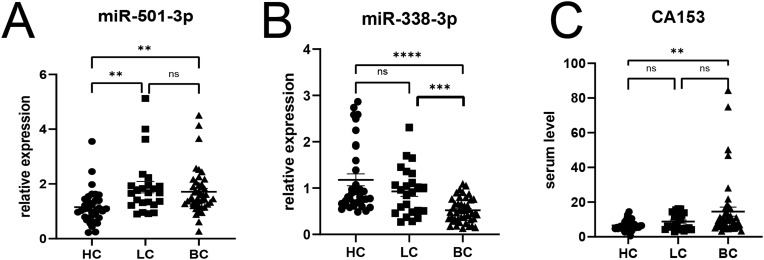
The Expression of miR-501-3p and miR-338-3p in Serum and Their Relationship with Clinicopathological Features
Serum miR-501-3p was higher in the BC group and benign tumor group than in the healthy control group (P<0.05) (Figure 9A). Serum miR-338-3p in the BC group was lower than that in the healthy control group and the benign tumor group, and the difference was statistically significant (P<0.05) (Figure 9B); while there was no statistical significance between the benign tumor group and the healthy control group (P>0.05). In addition, low expression of miR-338-3p was closely linked to lymph node metastasis and histological grading (P<0.05), and not with tumor size, stage, or age (P>0.05). High expression of miR-501-3p was closely correlated with histological grading and ki-67 index (P<0.05), and not with lymph node metastasis, stage, or age (P>0.05) (Table 1).
Figure 9.
Expression levels (A) miR-501-3p (B) miR-338-3p (C) CA153. ns p>0.05, ** P<0.01, ***P<0.001, ****P<0.0001.
Table 1.
miRNA and Clinicopathological Characteristics of Breast Cancer Patients
| Clinical Features | N | miR-338-3p (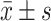 ) ) |
P | miR-501-3p [M (P25, P75)] | P |
|---|---|---|---|---|---|
| Age/Years | 0.972 | 0.459 | |||
| ≤50 | 21 | 0.52±0.25 | 1.62 (1.28, 2.06) | ||
| >50 | 23 | 0.52±0.27 | 1.45 (1.18, 1.98) | ||
| Diameter/cm | 0.889 | 0.496 | |||
| ≥3 | 15 | 0.51±0.32 | 1.44 (1.26, 1.83) | ||
| <3 | 29 | 0.52±0.23 | 1.60 (1.23, 2.02) | ||
| Lymph node metastasis | <0.05 | 0.945 | |||
| No | 28 | 0.61±0.27 | 1.45 (1.30, 1.98) | ||
| Yes | 16 | 0.36±0.12 | 1.59 (1.16, 2.10) | ||
| Histological grade | <0.05 | <0.05 | |||
| Medium/high differentiation | 25 | 0.59±0.26 | 1.26 (1.09, 1.72) | ||
| Under differentiation | 19 | 0.44±0.23 | 1.62 (1.39, 2.17) | ||
| TNM stage | 0.566 | 0.691 | |||
| T2+T3 | 17 | 0.49±0.22 | 1.60 (1.16, 2.25) | ||
| T1 | 27 | 0.54±0.28 | 1.45 (1.29, 1.90) | ||
| Ki-67/% | 0.693 | <0.05 | |||
| >30 | 24 | 0.53±0.24 | 1.82 (1.39, 2.38) | ||
| ≤30 | 20 | 0.50±0.28 | 1.35 (1.10, 1.61) |
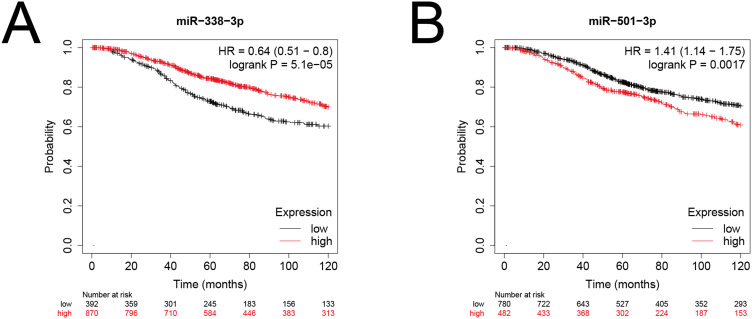
Survival Curve of miRNA
Both miR-338-3p and miR-501-3p are significantly associated with the prognosis of breast cancer patients (P<0.05). High expression of miR-338-3p was linked to a better prognosis, while high expression of miR-501-3p was linked to a poor prognosis (Figure 10).
Figure 10.
Prognostic value (A) miR-338-3p (B) miR-501-3p.
Expression Levels of Serum CA153
The expression level of serum CA153 in the BC group was higher than that in the healthy control group, and the difference was statistically significant (P<0.05). There was no significant difference between the benign tumor group and the breast cancer group and the healthy control group (P>0.05) (Figure 9C).
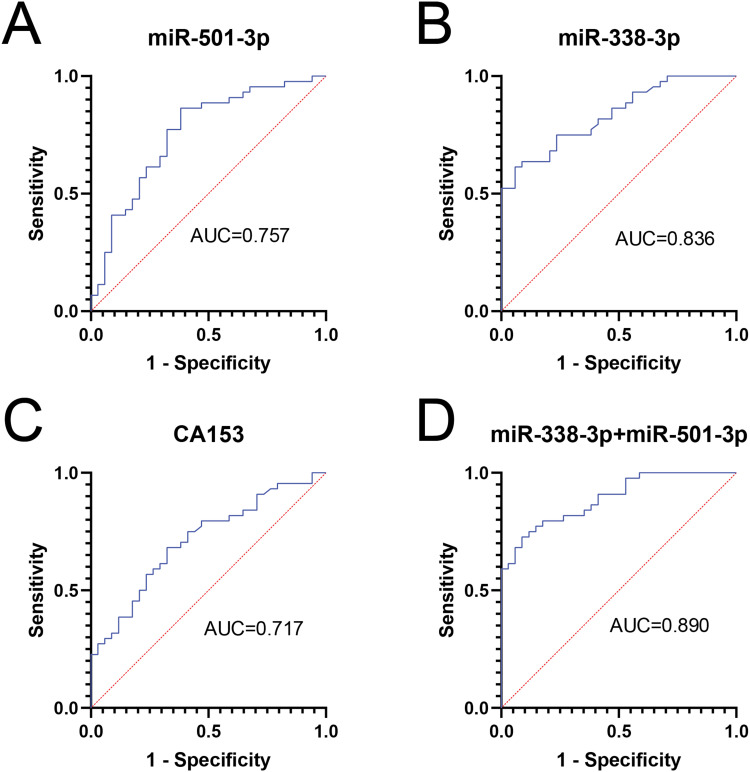
The Diagnostic Value of miR-501-3p, miR-338-3p, and CA153
ROC curves showed (Figure 11) that the AUC of serum miR-501-3p and miR-338-3p for the diagnosis of breast cancer were 0.757 (95% CI:0.65–0.87) and 0.836 (95% CI:0.75–0.92), respectively, with sensitivities of 86.4% and 61.4% and specificities were 61.8% and 94.1%, respectively (P<0.01). The AUC for the combined diagnosis of breast cancer was 0.890 (95% CI: 0.958), with sensitivities and specificities of 72.7% and 91.2%, respectively (P<0.01); the AUC for CA153 was 0.717 (95% CI: 0.60–0.83), with sensitivities and specificities of 70.5% and 67.6%, respectively (P <0.01). The combined AUC of the two miRNA was higher than that of CA153, with higher sensitivity and specificity, and had higher diagnostic values.
Figure 11.
The ROC curve (A) miR-501-3p (B) miR-338-3p (C) CA153 (D) miR-501-3p+miR-338-3p.
Discussion
Breast cancer is the number one cancer in the world,4 and nearly 2.26 million women are diagnosed with breast cancer every year, posing a serious threat to women’s health and causing great inconvenience to patients’ lives. Because the pathogenesis of breast cancer is exceptionally complex and the early symptoms are not very obvious, many patients often ignore it and do not go to the hospital in time, thus losing the best treatment period. Therefore, it is especially important to find highly sensitive and specific markers for the diagnosis of breast cancer.
With the rapid development of medical technology, liquid biopsy is in a crucial position and has an important clinical value for early diagnosis, personalized treatment, and prognosis of breast cancer. Increasing miRNAs are found to be differentially expressed in breast cancer patients and have good clinical applications. When combined with other molecules, they will be more useful for clinicians to identify the disease. In addition, miRNAs are stably present in serum and are more readily detectable. Therefore, serum miRNA is the most promising biomarker for breast cancer diagnosis.
In this study, we screened two miRNAs (miR-338-3p and miR-501-3p) and our results show that miRNAs can participate in the occurrence and development of various tumors by regulating malignant biological behaviors.10 Previous studies confirmed that miR-501-3p expression was upregulated in a variety of tumors. For example, Dai et al showed that high miR-501-3p expression was strongly linked to TNM stage and distant metastasis. In addition, its expression in patients with osteosarcoma may influence prognosis, and the mechanism may be that high expression of miR-501-3p inhibited lBCL7A expression to promote proliferation and invasion of osteosarcoma cells.11 In contrast, miR-338-3p expression is downregulated in a lot of cancers, Previous study showed that miR-338-3p can inhibit colorectal cancer proliferation and invasion by suppressing the expression of MACC1.12
Our data showed that serum miR-501-3p was upregulated in BC patients, while miR-338-3p was downregulated. Meanwhile, serum of both miR-501-3p and miR-338-3p is linked to the histological grade of breast cancer (P<0.05), indicating that they have a synergistic effect and are jointly involved in the malignant transformation and progression of BC. In addition, miR-501-3p was also closely associated with the ki-67 index (P<0.05), suggesting that it may act as an oncogene to boost cell proliferation and thus participate in the development of BC. miR-338-3p was also closely linked to lymph node metastasis of BC (P<0.05), suggesting that miR-338-3p has an essential part in the metastasis process of breast cancer. Nevertheless, the specific molecular mechanism is not well understood. It has been suggested that the HIF1α/miR-338-3p/EYA2 axis can control EGFR-mediated BC growth and metastasis.13 Another study showed that miR-338-3p could inhibit hypoxia-induced growth, migration, invasion, and epithelial-mesenchymal transition of breast cancer cells through ZEB2 and NF-ĸB/PI3K signaling pathways,14 suggested that miR-338-3p can be used as an oncogene to regulate the occurrence of breast cancer. We believe that detecting the levels of miR-501-3p and miR-338-3p can predict the degree of malignancy or progression and the occurrence of lymph node metastasis of BC.
MiRNAs influence tumor development by suppressing target genes. Thus, we performed target gene prediction for miR-501-3p and miR-338-3p, respectively, and then further analyzed their functions and signaling pathways. Further pathway analysis suggested that they might be closely related to the development of BC. We found that miR-501-3p was enriched in the cGMP-PKG and calcium signaling pathways; miR-338-3p is involved in Ras, PI3K-Akt, MAPK, ERBB, and other cancer signaling pathways. The most classical signaling pathway, Ras, is typically activated first and is a major target for pharmacological therapy in many illnesses.15 There is increasing evidence that the PI3K/AKT signaling pathway can be activated by miRNA, thus playing a part in the molecular mechanism of BC. Fan et al found that miR-204 inhibited the proliferation and metastasis of BC cells through the PI3K/AKT pathway.16 Similarly, the MAPK signaling pathway is also associated with breast carcinogenesis, for example, miR-188-5p can promote apoptosis and inhibit cell proliferation by regulating this pathway, and miR-122-5p can boost the invasion of triple-negative BC by regulating this pathway.17,18 In addition, all these pathways are strongly associated with the angiogenesis process, so we speculate that miR-338-3p may influence the development of breast cancer through MAPK/PI3K-Akt/Ras signaling pathway and thus regulate angiogenesis.
In our study, we also explored the expression and prognostic value of key target genes of miR-501-3p and miR-338-3p. We found that miR-501-3p-regulated CDKN2C was lowly expressed in BC, and its low expression was linked to better RFS. Meanwhile, we predicted that six genes regulated by miR-338-3p, ACTE2, CDH1, COL1A1, RBBP5, RRM1, and TPM3 were highly expressed in breast cancer. The high expression of ACTR2, CDH1, PBBP5, RRM1, and TPM3 was linked to poorer RFS. COL1A1 is located in chromosome 17q21.33 and belongs to the collagen family. It is highly expressed in various cancers and regulates biological processes such as cell proliferation, metastasis, and apoptosis.19 Liu et al found that increased COL1A1 expression level was linked to poor survival rate, especially in ER+ breast cancer patients. Patients with a high level of COL1A1 showed a better cisplatin-based chemotherapy response;20 furthermore, in vitro experiments confirmed that expression of COL1A1 promoted BC metastasis; this suggests that COL1A1 is a novel prognostic biomarker and a potential therapeutic target for BC. This is consistent with our results. RRM1, located at chromosome 11p15.5, is a key substance in the DNA damage repair system and can be involved in processes such as cell proliferation and cancer metastasis.21 Qu et al showed that RRM1 was highly expressed in triple-negative breast cancer and linked to TNM stage and histological grade.22 CDH1 is a calcium-dependent intercellular adhesion molecule and tumor suppressor gene, and studies have shown that hypermethylation of CDH1 is a potential new drug target for the personalized treatment of breast cancer.23 Yao et al found that TPM3 is highly expressed in platelets of breast cancer patients and linked to metastasis, and may be a biomarker for early diagnosis of metastatic breast cancer.24 However, the mechanism of CDKN2C, ACTR2, and RBBP5 in BC needs to be further investigated. CDKN2C may be a new target gene of miR-501-3p, while ACTR2 and RBBP5 may be new target genes of miR-338-3p.
Immunotherapy is currently considered to be an important cause of cancer progression and treatment, so we performed an immune analysis of target genes. Our study found that the expression of these key target genes is associated with different levels of immune infiltration and Immune checkpoint inhibitors, which confirms that they may influence the pathogenesis of BC and may have potential implications for BC immunotherapy.
Finally, ROC and survival curves were performed to further explore the accuracy of miR-501-3p and miR-338-3p as biomarkers for breast cancer. The AUCs of serum miR-501-3p and miR-338-3p for diagnosing breast cancer were 0.757 and 0.836, respectively, which were higher than that of CA153 (AUC of 0.717) and had some clinical diagnostic value. Many researchers believe that the sensitivity and specificity of individual marker assays are low and insufficient to achieve the clinical need for the diagnosis of diseases. Therefore, in addition to testing individual miRNAs and plotting ROC curves, we also performed a combined test of two miRNAs and found that their combined AUC for breast cancer diagnosis was 0.890, and its sensitivity and specificity were higher than that of CA153, which a higher diagnostic ability and can serve as a diagnostic biomarker for BC. Both miR-338-3p and miR-501-3p are significantly associated with the prognosis of breast cancer patients and can be used as prognostic biomarkers.
Limitations
This study had some limitations. In the future, we will expand the volume of specimens (specimens are still being collected) and compare the molecular typing (Luminal A, Luminal B, her-2-positive, triple-negative breast cancer) of breast cancer patients to analyze whether they can distinguish different molecular subtypes of breast cancer. The key target genes analyzed through bioinformatics have only been partially verified in the database and lack of experimental verification in vivo and in vitro. Therefore, in the future, we will further carry out experimental verification at the cellular level and in clinical samples to analyze their functions and provide effective means for the diagnosis and treatment of breast cancer.
Conclusion
MiR-338-3p may inhibit breast cancer development by regulating COL1A1, ACTR2, CDH1, RBBP5, RRM1, and TPM3; miR-501-3p may promote breast cancer development by inhibiting CDKN2C. Their key target genes are related to BC development, prognosis, and immunotherapy. In addition, serum miR-338-3p and miR-501-3p have very good diagnostic values. Thus, miR-338-3p and miR-501-3p have the potential as new therapeutic targets and biomarkers for BC.
Funding Statement
The present work was supported by the Youth Natural Science Foundation of Shandong Province (ZR2021QH102), the Science and Technology Development Plan Project of Weifang City (2021YX040), the Students’ Research Fund of Weifang Medical University (X2021615, X2021641, X2021186).
Abbreviations
BC, Breast cancer; DEMs, Differentially Expressed MicroRNAs; ROC, Receiver operating characteristic curve; AUC, The area under the Curve; GO, Gene Ontology; KEGG, Kyoto Encyclopaedia of Genes and Genomes; PPI, Protein-Protein Interaction network.
Data Sharing Statement
All data during the course of this study are included in this article.
Ethics Approval
This study design was approved by the Medical Ethics Committee of Weifang People’s Hospital of Shandong Province (KYLL20230120-2) and all patients provided informed consent, in accordance with the Declaration of Helsinki.
Consent to Participate
Obtain informed consent from all participants.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare no competing interests.
References
- 1.Januškevičienė I, Petrikaitė V. Heterogeneity of breast cancer: the importance of interaction between different tumor cell populations. Life Sci. 2019;239:117009. doi: 10.1016/j.lfs.2019.117009 [DOI] [PubMed] [Google Scholar]
- 2.Akram M, Iqbal M, Daniyal M, Khan AU. Awareness and current knowledge of breast cancer. Biol Res. 2017;50(1):33. doi: 10.1186/s40659-017-0140-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hong R, Xu B. Breast cancer: an up-to-date review and future perspectives. Cancer Commun. 2022;42(10):913–936. doi: 10.1002/cac2.12358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 6.Cardinali B, Tasso R, Piccioli P, Ciferri MC, Quarto R, Del Mastro L. Circulating miRNAs in breast cancer diagnosis and prognosis. Cancers. 2022;14(9):2317. doi: 10.3390/cancers14092317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Du Y, Miao Z, Wang K, Lv Y, Qiu L, Guo L. Expression levels and clinical values of miR-92b-3p in breast cancer. World J Surg Oncol. 2021;19(1):239. doi: 10.1186/s12957-021-02347-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xue J, Chi Y, Chen Y, et al. MiRNA-621 sensitizes breast cancer to chemotherapy by suppressing FBXO11 and enhancing p53 activity. Oncogene. 2016;35(4):448–458. doi: 10.1038/onc.2015.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin H, Li C, Ge H, Jiang Y, Li Y. Circulating microRNA: a novel potential biomarker for early diagnosis of intracranial aneurysm ruptures a case control study. J Transl Med. 2013;11:296. doi: 10.1186/1479-5876-11-296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18(1):5–18. doi: 10.1038/nrc.2017.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dai J, Lu L, Kang L, Zhang J. MiR-501-3p promotes osteosarcoma cell proliferation, migration and invasion by targeting BCL7A. Hum Cell. 2021;34(2):624–633. doi: 10.1007/s13577-020-00468-x [DOI] [PubMed] [Google Scholar]
- 12.Zou T, Duan J, Liang J, et al. miR-338-3p suppresses colorectal cancer proliferation and progression by inhibiting MACC1. Int J Clin Exp Pathol. 2018;11(4):2256–2267. [PMC free article] [PubMed] [Google Scholar]
- 13.Liang Y, Xu X, Wang T, et al. The EGFR/miR-338-3p/EYA2 axis controls breast tumor growth and lung metastasis. Cell Death Dis. 2017;8(7):e2928. doi: 10.1038/cddis.2017.325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He J, Wang J, Li S, Li T, Chen K, Zhang S. Hypoxia-inhibited miR-338-3p suppresses breast cancer progression by directly targeting ZEB2. Cancer Sci. 2020;111(10):3550–3563. doi: 10.1111/cas.14589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Erickson KE, Rukhlenko OS, Posner RG, Hlavacek WS, Kholodenko BN. New insights into RAS biology reinvigorate interest in mathematical modeling of RAS signaling. Semin Cancer Biol. 2019;54:162–173. doi: 10.1016/j.semcancer.2018.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fan X, Fang X, Liu G, Xiong Q, Li Z, Zhou W. MicroRNA-204 inhibits the proliferation and metastasis of breast cancer cells by targeting PI3K/AKT pathway. J BUON. 2019;24(3):1054–1059. [PubMed] [Google Scholar]
- 17.Zhu X, Qiu J, Zhang T, et al. MicroRNA-188-5p promotes apoptosis and inhibits cell proliferation of breast cancer cells via the MAPK signaling pathway by targeting Rap2c. J Cell Physiol. 2020;235(3):2389–2402. doi: 10.1002/jcp.29144 [DOI] [PubMed] [Google Scholar]
- 18.Wang Z, Wang X. miR-122-5p promotes aggression and epithelial-mesenchymal transition in triple-negative breast cancer by suppressing charge multivesicular body protein 3 through mitogen-activated protein kinase signaling. J Cell Physiol. 2020;235(3):2825–2835. doi: 10.1002/jcp.29188 [DOI] [PubMed] [Google Scholar]
- 19.Li X, Sun X, Kan C, et al. COL1A1: a novel oncogenic gene and therapeutic target in malignancies. Pathol Res Pract. 2022;236:154013. doi: 10.1016/j.prp.2022.154013 [DOI] [PubMed] [Google Scholar]
- 20.Liu J, Shen JX, Wu HT, et al. Collagen 1A1 (COL1A1) promotes metastasis of breast cancer and is a potential therapeutic target. Discov Med. 2018;25(139):211–223. [PubMed] [Google Scholar]
- 21.Specks J, Lecona E, Lopez-Contreras AJ, Fernandez-Capetillo O, Single Conserved A. Residue mediates binding of the ribonucleotide reductase catalytic subunit RRM1 to RRM2 and is essential for mouse development. Mol Cell Biol. 2015;35(17):2910–2917. doi: 10.1128/MCB.00475-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gengbao Q, Qiang Z, Feng W, Pilin W, Tie Z. Expression and significance of ERCC1, TUBB3 and RRM1 in triple negative breast cancer tissue. J Xinjiang Med Univ. 2021;44(9):994–998+1005. [Google Scholar]
- 23.Huang R, Ding P, Yang F. Clinicopathological significance and potential drug target of CDH1 in breast cancer: a meta-analysis and literature review. Drug Des Devel Ther. 2015;9:5277–5285. doi: 10.2147/DDDT.S86929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yao B, Qu S, Hu R, et al. Delivery of platelet TPM3 mRNA into breast cancer cells via microvesicles enhances metastasis. FEBS Open Bio. 2019;9(12):2159–2169. doi: 10.1002/2211-5463.12759 [DOI] [PMC free article] [PubMed] [Google Scholar]